Abstract
Mosaicism refers to the occurrence of two or more genomes in an individual derived from a single zygote. Germline mosaicism is a mutation that is limited to the gonads and can be transmitted to offspring. Somatic mosaicism is a postzygotic mutation that occurs in the soma, and it may occur at any developmental stage or in adult tissues. Mosaic variation may be classified in six ways: (a) germline or somatic origin, (b) class of DNA mutation (ranging in scale from single base pairs to multiple chromosomes), (c) developmental context, (d) body location(s), (e) functional consequence (including deleterious, neutral, or advantageous), and (f) additional sources of mosaicism, including mitochondrial heteroplasmy, exogenous DNA sources such as vectors, and epigenetic changes such as imprinting and X-chromosome inactivation. Technological advances, including single-cell and other next-generation sequencing, have facilitated improved sensitivity and specificity to detect mosaicism in a variety of biological contexts.
Keywords: mosaicism, somatic mutation, germline mutation, mosaic aneuploidy
1. INTRODUCTION TO MOSAICISM
At the molecular level, the mutation of DNA is the main source of variation on which natural selection can operate. DNA variants may be inherited (transmitted through the germline) or occur by postzygotic mosaicism, mutations localized to a subset of cells. Mosaicism is defined as the presence of two or more different genomes within an individual derived from a single zygote. In the case of somatic mosaicism, mutations are acquired in somatic cells as postzygotic events. In germline mosaicism (also termed gonadal mosaicism), mutations selectively occur postzygotically in germ cells within the gonads. An individual with gonadal mosaicism may eventually transmit variants to offspring, including sporadic disease-causing mutations. Natural selection operates on heritable germline variation, adjusting population frequency as a consequence of fitness. Nonheritable somatic variants are under intraindividual selective pressure and can also have functional consequences on fitness.
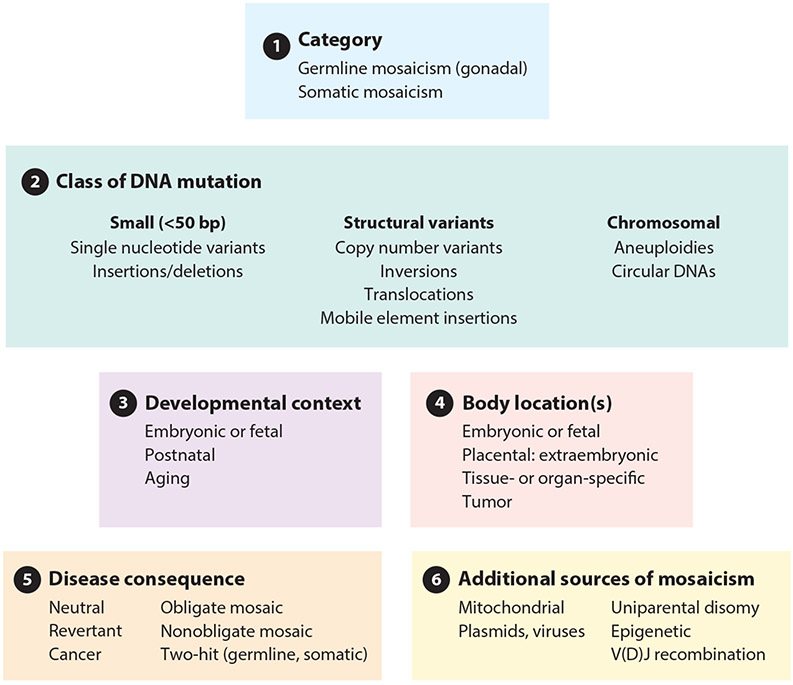
Mosaic variation is a fundamental biological process that has broad implications with regard to mutation, selection, development, and disease. Mosaicism shakes and shapes the human genome: Inevitably, we are all mosaics, and this impacts our identity. In this review, we classify mosaicism by six criteria (Figure 1). (a) We distinguish germline mosaicism from somatic mosaicism. (b) We describe classes of DNA change that are involved in mosaicism, from single nucleotide variants (SNVs) to structural variants (SVs), including affected whole chromosomes. We discuss the context of mosaic variation with regard to (c) the timing and (d) the body localization of mosaic events. (e) We next describe the consequences of mosaic variation, which include neutral, advantageous, and deleterious changes, (f) Finally, we discuss additional noncanonical mechanisms of mosaicism, including nonnuclear endogenous, exogenous, and functional epigenetic sources.
Figure 1.
Classification of mosaic variation. We classify mosaic variation as: (a) germline or somatic origin, (b) class of DNA mutation (ranging in scale from single base pairs to multiple chromosomes), (c) developmental context, (d) body location(s), (e) disease consequence, and (f) additional sources of mosaicism.
In addition to their classification, we consider the fundamental properties of mosaic variants. These properties include rates and nucleotide signatures (discussed in Section 3) as well as mechanisms by which meiotic errors, mitotic errors, and environmental insults contribute to the formation of mosaic variants. We also discuss experimental designs and technologies that have facilitated the discovery of mosaic variation.
Chimerism refers to the presence of two or more genetically distinct populations within an individual. In chimerism, these distinct populations derive from two (or more) separate zygotes. This distinguishes chimerism from mosaicism.
2. GERMLINE VERSUS SOMATIC MOSAICISM
Germline mosaicism occurs when an individual has genetically distinct populations of cells in the gonads (i.e., ovaries or testes) (26, 133). Wild-type and other populations of cells, having neutral, deleterious, or advantageous mosaic mutations, coexist in tissues. The term germline mosaicism is specific to the presence of distinct genomes in the germ cells of an individual. In contrast, germline variation as a genetics term ordinarily refers to a constitutional variant, that is, a DNA change (and in particular a mutation) that is transmitted through a parent’s germline to all somatic and germ cells of a child.
Germline mosaicism may arise during meiosis, as germ cells are formed postzygotically, or throughout a human life span by spontaneous mutagenesis. A schematic of embryonic through fetal development is shown in Figure 2a, with an example of germline mosaicism in Figure 2b.
Figure 2.
Occurrence of somatic mosaicism across embryonic and fetal development, including development from the morula (16-cell stage) and blastocyst. The embryo forms from the inner cell mass of the blastocyst, while the placenta develops from the outer cell mass (from the trophoblasts at the exterior of the blastocyst). Mutations are indicated with a lightning bolt symbol, (a) Euploid (wild type) development. A normal karyotype (46,XX for a female in this example) is indicated, (b Germline mosaicism. Mutation can occur in the germline (e.g., germ cell precursors) of an embryo. Mosaic mutation is limited to germ cells that may later be transmitted to offspring as inherited germline variations. The individual depicted here, harboring the germ cell mosaic, may eventually become a parent who is phenotypieally normal, (c) Postzygotic mosaic mutations. Whether involving aneuploidy (whole chromosome gains or losses) or single nucleotide variants, postzygotic mosaic mutations can manifest through the entire body of an individual, particularly if the mutation occurs early in development, or be constrained to particular regions or organs, as indicated here. Trisomies that are ordinarily lethal (trisomies 1–12, 14–17, 19–20, and 22) may persist in the mosaic state with variable clinical phenotypes, (d) Confined placental mosaicism. Mosaic mutations can occur solely in the placenta while the fetus has a normal karyotype. A postzygotic, somatic mutation may lead to a cell and its daughter cells having mosaicism (such as mos 46,XX/47,XX+21 for mosaic trisomy 21) in a subset of the placental cells. Chorionic villus sampling can indicate trisomy 21. Determining whether the fetus is euploid or trisomic requires a separate test, such as amniocentesis, (e) Uniparental disomy. A trisomy (such as 47,XX+15) can arise at an early embryonic stage, persisting as a mosaic in the placenta (mos 46,XX/47,XX+15). In the embryo, trisomic rescue may occur in which a third copy of the chromosome is deleted from the cell. Rescue produces disomy and a euploid state (46,XX) or disomy in which both copies of the chromosome derive from one parent [e.g., maternal uniparental disomy (UPD mat)]. Uniparental heterodisomy refers to a meiosis I error, and both copies of the chromosome are from one parent but from different homologs. Uniparental isodisomy results from a meiosis II error or occurs through postzygotic duplication (as shown here). In this example, the child is susceptible to developing Prader-Willi syndrome [Online Mendelian Inheritance in Man (OMIM) 176270], an imprinting disorder due to loss of the paternal copy of chromosome 15q11.2–q13. This figure was derived in part from a public domain image from the Human Placenta Project (https://www.nichd.nih.gov/sites/default/files/inline-images/HPP-placental-development.png) at the Eunice Kennedy Shriver National Institute of Child Health and Human Development (National Institutes of Health).
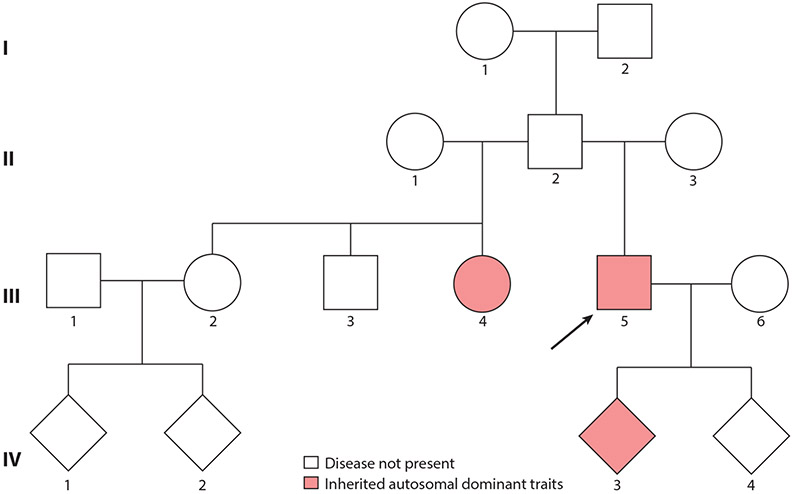
Germline mosaicism can be identified by pedigree analysis. In some families, a parent (such as the father, individual II.2, in Figure 3) appears phenotypically normal but has multiple children affected with a penetrant, autosomal dominant or X-linked disorder. Such a pedigree can be explained by germline mosaicism in which a proportion of germ cells harbor the damaging allele, which may be transmitted to progeny. Some disorders are particularly prone to germline mosaicism. One is Duchenne muscular dystrophy [Online Mendelian Inheritance in Man (OMIM) 310200], caused by mutations in the DMD gene encoding dystrophin (8). Another is osteogenesis imperfecta type 1 (OMIM 166200), caused by heterozygous mutations in the COL1A1 or COL1A2 gene (99). In Down syndrome, caused by trisomy 21, ~5% or more of cases are caused by germline mosaicism (26, 27).
Figure 3.
Inference of germline mosaicism by pedigree analysis. In this pedigree, individuals III.4 and III. 5 present with an inherited autosomal dominant trait. Nevertheless, the father, II.2, is healthy. Individuals III.4 and III.5 are born from two different mothers, suggesting that the father, who does not show a clinical phenotype, is a gonadal mosaic for the mutation carried by offspring III.4 and III. 5. The genetic risk of transmitting the autosomal dominant condition to generation III subjects differs depending on the proportion of mutated sperm and the disease mechanism (autosomal dominant in this example). Siblings III.2 and III. 3 are healthy, having inherited the normal paternal allele, and are at no risk of transmitting the disease to their children. Subjects III.4 and III. 5 carry the germline mutation inherited by their father and have the mutation in both their germ and somatic cells. As for any autosomal dominant trait, there is a 50% probability of transmitting the mutant allele to their children (e.g., individual IV.3 is affected but IV.4 is not). In males, it is possible to confirm the mutation as germline mosaic by searching for it in DNA from sperm and somatic cells, for example, in peripheral blood lymphocytes. The mutation is germline mosaic if it is detected in sperm but not in lymphocyte DNA. The arrow indicates individual III. 5 who is the proband in this pedigree.
Somatic mosaicism refers to postzygotic mutations that occur in the soma, not originating in germ cells. Somatic mosaicism is more abundant and pervasive in human disease than germline mosaicism and, therefore, the emphasis of this review. Mosaicism may occur in an individual’s germline, somatic cells, or both, depending on mutation developmental timing (112). An adult with a mosaic NF1 mutation in both the germline and soma has a risk of transmitting the mutation to offspring and the risk of acquiring the disease (neurofibromatosis; OMIM 162200) if the mutation is in susceptible cells.
3. CLASSES OF MOSAIC VARIATION
3.1. Single Nucleotide Variants and Insertions and Deletions
SNVs are alterations to a single base, while insertions and deletions (indels) are typically less than 50 bp. SNVs and indels result from errors in DNA damage, replication, and repair. DNA polymerase fidelity and repair have been estimated at ~0.27 to 0.99 errors per 109 nucleotides replicated per cell division (84). Errors in base excision repair, nucleotide excision repair, and transcription-coupled repair result in mosaic SNVs and indels (16). Ultraviolet radiation, environmental carcinogens, and X-ray radiation induce DNA lesions, which lead to somatic SNVs or indels (24). The Pan-Cancer Analysis of Whole Genomes Consortium has reported mutational signatures corresponding to systematic errors in DNA replication and DNA repair or caused by certain environmental exposures (2). They reported mutational signatures consisting of single base substitutions (n = 49 signatures), doublet base substitutions (n = 11 signatures), clustered base substitutions (n = 4 signatures), and indels (n = 17 signatures).
Rates of mosaic SNVs vary between cell types and tissues and across development. Early embryonic development and neurogenesis have increased rates of somatic SNVs compared to those of adulthood (7). High rates of somatic SNVs in development are thought to be attributable to the rapid cycling and reduction of G1 and G2 checkpoints, giving DNA repair machinery a shorter interval to repair single nucleotide lesions introduced during replication (79, 85, 98).
3.2. Copy Number and Neutral Variation
There are four main classes of structural variation (defined as DNA variants ≥50 bp): deletions, amplifications, inversions, and translocations (116,127). A typical human genome includes ~2,100 to 2,500 SVs affecting ~18 million bp of sequence (6, 116). These include large deletions (n ≈ 1,000), copy number variants (CNVs; n ≈ 160), insertions (such as Alu and L1; n ≈ 1,100), and inversions (n ≈ 10). Each of these SVs can occur in mosaic form (122).
Copy number neutral (CNN) variants alter genomic organization with unchanged nucleotide frequency. Inversions, translocations, and complex rearrangements are CNN SVs. CNN breakpoints are difficult to detect as they preferentially occur within segmental duplications and fragile sites (94). Mosaic structural abnormalities have been implicated in developmental disorders (22, 62). Like other somatic variations, somatic CNN mutations increase with age (44).
Monozygotic twins provide a useful model system to study mosaic variation. Mosaic CNVs have been consistently observed in monozygotic twin pairs (10, 32). Mosaic copy number loss and loss of heterozygosity (CNL-LOH) is the deletion of an allele from the heterozygous state resulting in a homozygous state (107). CNL-LOH can lead to the gain or loss of imprinting by duplication or loss of the unmethylated or methylated allele (65). Prevalence and size distribution of mosaic CNVs are dictated by detection sensitivity (experimental technology and design), mutation and repair mechanisms (localization and occurrence), and fitness consequence (for instance, large CNVs are more likely lethal).
3.3. Chromosomal Mosaicism
Chromosomal aneuploidy is a change in the chromosome copy number, such as monosomy (e.g., 45,X) or trisomy (e.g., 47,XY,+21). Three autosomal trisomies are compatible with postnatal survival: Down syndrome (chromosome 21), Patau syndrome (chromosome 13), and Edward syndrome (chromosome 18) (12). Most other autosomal aneuploidies are embryonic lethal, and studies of spontaneous abortions have revealed trisomies of all chromosomes (51, 54, 56). However, almost all trisomies and some monosomies can be viable in the mosaic form (11). Clinical phenotypes are often severe, typically including intrauterine growth retardation, congenital heart disease, and dysmorphism such as craniofacial abnormalities.
Cells can respond to the condition of trisomy by reverting to the diploid state. The process, called trisomic rescue, is caused by mitotic segregation errors (85). When one of three chromosome copies is lost, the resulting cell is disomic and may have one copy derived from each parent (a euploid state). However, in two-thirds of cases, the result is uniparental disomy (UPD) (36), shown schematically in Figure 2e. Both chromosomes can be inherited as the same allele from one parent (uniparental isodisomy), or both chromosomes are inherited from the same parent but with different alleles (uniparental heterodisomy) (85). UPD is responsible for several genetic imprinting disorders, including Prader-Willi syndrome and Angelman syndrome (59).
Chromosomal aneuploidies are a normal feature of human development. Aneuploidy occurs in 30–35% of neurons in neurotypical human brain development (12). Hepatocytes in the adult human liver are frequently polyploid (43). Mosaic aneuploidy affects cell survival, proliferation, and signaling (85). Mosaicism in the brain creates heterogeneous cell populations, increasing functional diversity but also brain disease risk (84). Autosomal trisomies can be mosaic, with ~1.3% to 5% of Down syndrome cases having mixed cell populations of disomic and trisomic chromosome 21 (3, 12,96).
3.4. Extrachromosomal Circular DNA
Damaged or deleted DNA can circularize and coexist in somatic cells as semistable extrachromosomal DNA. In humans, three classes of circular DNA exist: small extrachromosomal DNA, large, copy number–amplified extrachromosomal circular DNA, and ring chromosomes or neochromosomes (64). Extrachromosomal circular DNAs are found in many cancers and have been shown to promote oncogenesis (90). Circular DNAs are often monoallelic; enriched for genic or pseudogenic regions; and generated by nonhomologous end joining, replication-associated mechanisms, or microhomology-mediated DNA repair (64, 90). With regard to extrachromosomal DNA, 99% are <25 kb, with the largest being megabase-sized ring chromosomes or neochromosomes (90). Extrachromosomal DNA contributes to somatic rearrangements, and has been implicated in somatic reorganization in neuroblastoma (64).
3.5. Mobile Element Insertions
Mobile element insertions (MEIs) are the transposition or integration of transposable elements in the genome (61). As much as two-thirds of the human genome is composed of mobile elements, including Alu, L1 retrotransposons, short interspersed nuclear elements (SINEs), and SINE/VNTR/Alus (SVAs) (25, 113). Stochastic insertion of mobile elements, and often imprecise excision, generates structural variation in somatic cell genomes (113). L1 retrotransposons are active during embryonic development and neurogenesis, inducing mosaic copy number changes (40, 46). Although they are often transcriptionally silenced, a subset is active, driven by developmental queues (38). Cases of Rett syndrome, hemophilia, breast cancer, and other Mendelian disorders have been documented as a result of somatic MEIs (28, 113). About one somatic L1 retrotransposon insertion occurs per neuron (28), although L1 elements exhibit no identified genomic integration hotspots (38). The observation of mosaic MEIs in postmitotic neurons suggests active retrotransposition in neurogenesis and aging, increasing the likelihood of long-term consequences in the geriatric brain (38).
4. DEVELOPMENTAL CONTEXT OF SOMATIC MOSAICISM
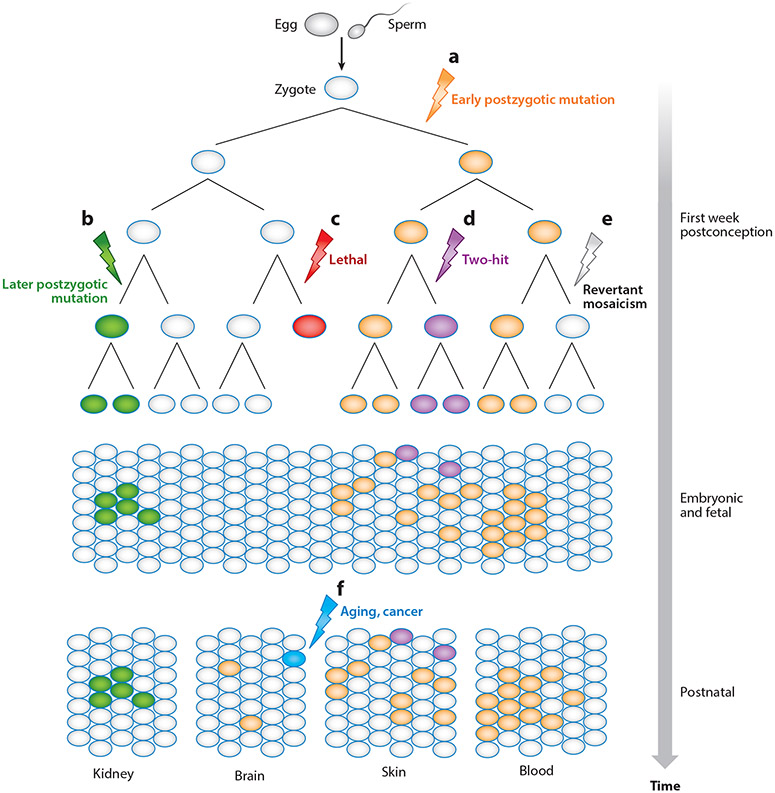
Somatic mosaicism occurs throughout the human life span. The molecular and phenotypic consequences of mosaicism depend on the timing of the mutation (112). Generally, somatic mutations occurring in early development are propagated through more cellular divisions, thereby affecting a greater cell proportion and more diverse tissues and body segments (Figure 4a). Mutations occurring later in development may affect more restricted regions of the body (Figure 4b). A somatic mutation that occurs earlier in development may lead to a nonviable embryo, while such a mutation in an adult may have no consequence on fitness (Figure 4c,f).
Figure 4.
Somatic mosaicism across development and body regions and schematic representation of somatic mosaicism through cellular development from early life (top) to adulthood (bottom). Lightning bolts represent the occurrence of a somatic mosaic mutation, (a) An early postzygotic mutation affects one of two parental alleles. If the affected cell can divide and proliferate, it is likely to affect a relatively large part of the body (see orange cells, bottom), (b) A later-occurring somatic mutation may result in fewer affected cells, for example, restricted, organ-specific mosaicism, (c) Lethal somatic mosaic mutation can occur, (d) Additional, independent mosaic mutations can occur, (e) Revertant mosaicism occurs when a cell retrogresses the mosaic mutation to wild type, (f) Somatic mosaic mutations may occur at any time in development, from postconception to old age. An acquired somatic mutation is indicated, such as may occur as part of the aging process or in response to a mutagen.
Maintenance of cellular homeostasis requires precise molecular regulation of the genome. Somatic mutations have cell-specific differential effects due to cell-type-specific genomic function. In concert with germline variation, somatic mutations at the same loci can have diverse functional consequences. In exons, somatic synonymous mutations are far more likely to be tolerated than nonsense or missense mutations (108). The genomic location of somatic mutation confers differential consequences. Noncoding regions are less highly conserved and predicted to be less functionally constrained than coding regions; therefore, somatic mutations at these loci are less likely to be damaging (101). Minor somatic contractions or expansions of repetitive elements are typically inconsequential to cellular fitness, although repetitive element somatic copy number changes result in human disease. For instance, facioscapulohumeral muscular dystrophy has been associated with a germline or postzygotic somatic contraction of 3.3-kb D4Z4 repeats below 10 copies (71,72).
Each individual is a unique, complex somatic mosaic. Somatic mutations result from genetic and extrinsic environmental exposures. Healthy tissue accumulates age-related somatic mutations, which increase exponentially in frequency with age (78, 80, 81). Age-related mosaic loss of the Y chromosome affects nearly 20% of elderly men, particularly after age 60 (132). Mosaic clones dynamically expand and contract over time, which suggests changing selective forces (43, 80). Age-related erosion of the genome has been attributed to reduced efficacy of genomic repair mechanisms (80). Aneuploidy has been demonstrated to increase with age (98). Gonadal mosaicism also increases with age, with mothers accruing ~0.37 mutations per year and fathers ~1.51 per year (57, 58). Somatic mutation may contribute to sporadic neurodegenerative disorders, such as Alzheimer and Parkinson’s diseases (77), whether in a causal role or by contributing to disease heterogeneity.
5. MOSAICISM ACROSS BODY ORGANS AND TISSUES
5.1. Confined Placental Mosaicism
Following zygote formation, the placenta and embryo differentiate from the outer cell mass and inner cell mass, respectively (59). Surprisingly, about 2% of viable pregnancies are confined placental mosaic (CPM), whereby mosaicism exists solely within the placenta (Figure 2d) (60). Detected by chorionic villus sampling during pregnancy, CPM aneuploidy is typically the result of mitotic chromosomal nondisjunction (14,43). Complications include increased incidence of preterm births, low birth weights, intrauterine growth restriction, and intrauterine death of a chromosomally normal fetus (59, 121).
5.2. Tissue-Specific Mosaicism
The vast majority of genetics studies focus on readily available tissue and cell sources, such as blood, transformed lymphoblastoid cell lines, and fibroblasts. Mosaic variation may occur in a tissue-specific manner in any body region. For example, trisomy 21 has been observed in myocardium and lung but not skin or lymphocytes tissue (131). It may be necessary to sample multiple tissues for low-level mosaicism. This is practical in postmortem specimens or in animal models.
5.3. V(D)J Recombination
During lymphocyte differentiation, genomic regions of early T and B cell progenitors undergo somatic V(D)J recombination. V(D)J recombination is a programmed mechanism of somatic mutation in the human immune system that creates a diverse repertoire of immunoglobulin and T cell receptors (9). To recognize the vast array of potential foreign or malignant molecules requiring immune system intervention, B and T cells undergo a programmed somatic recombination of hypervariable genomic segments to produce diverse signal recognition motifs. Recombination events that recognize healthy self-peptides or fail to recombine correctly undergo apoptosis (106). Although V(D)J recombination represents a canonical, programmed mechanism of somatic mosaicism, it has been implicated in several autoimmune disorders and cancer. Autoimmune disorders, such as rheumatoid arthritis and systemic lupus erythematosus, result from recombination events that produce autoantibodies that recognize nonpathogenic molecules, such as allergens or healthy self-peptides, attacking host cells and resulting in chronic inflammation and other disease phenotypes (9, 29). Aberrant recombination-activating gene (RAG)-initiated double-stranded breaks at loci that share high sequence identity with canonical recombination signal sites have been identified in lymphoid neoplasms (106).
6. FUNCTIONAL CONSEQUENCES OF SOMATIC MOSAICISM
6.1. Mosaicism in Human Disease
Somatic mutations result from a variety of mutational processes, spanning many classes of genomic variation. Likewise, human disease resulting from or contributed to by somatic mosaicism can be categorized into several classes of disease: obligatory somatic, nonobligatory somatic, second-hit somatic, and revertant somatic. Each class demonstrates a unique mechanism through which somatic genomic mutations manifest in human disease.
6.2. Obligatory Somatic Mosaicism
Dominant heterozygous mutations are often embryonic lethal. Depending on the developmental timing and affected cell, such mutations can be tolerated in the somatic mosaic state (50). The somatically acquired mutation occurs at a developmental time point and in a specific cell or cell type such that it may perturb biological pathways and systems but is not sufficiently disruptive to be nonviable. Therefore, human diseases that are constitutionally lethal but somatic mosaic viable are classified as obligatory somatic disorders.
Sturge-Weber syndrome (SWS; OMIM 185300), a neurocutaneous disorder accompanied by nonsyndromic port-wine stain birthmarks (OMIM 163000), is caused by a somatic activating missense mutation R183Q in the GNAQ gene encoding guanine nucleotide-binding protein q (109). SWS is never recurrent in a family (even involving monozygotic twins) and has no documented heritability, supporting the obligatory somatic pathogenic nature of the disease. Further, a constitutional mutation in a mouse model is embryonic lethal. Other obligate mosaic disorders include tuberous sclerosis 1 (caused by mutations in TSC1; OMIM 191100), focal cortical dysplasia type II (caused by mutations in MTOR; OMIM 607341), and Proteus syndrome (caused by mutations in AKT1; OMIM 176920).
6.3. Nonobligatory Somatic Mosaicism
Human genetic diseases that can result from either constitutional or somatic mutation are classified as nonobligatory somatic. Heterozygous dominant constitutional mutations alter protein function or dosage, resulting in disease. In some cases, such disease-causing mutations occur somatically. For example, neurofibromatosis 1 (NF1; OMIM 162200) is a commonly occurring autosomal dominant disorder. In some cases, segmental NF1 occurs, in which the clinical phenotype manifests in only a limited region of the body (48). The timing, cell type, and functional context of the somatically acquired mutation dictate the phenotypic consequences. In general, the spectrum of aberrant phenotypes for nonobligatory somatic diseases is diverse, and they often have an attenuated phenotype relative to constitutional disease as the mutation occurs at lower frequency throughout the body. Some proportion of de novo or isolated cases of dominant, heritable Mendelian disease can likely be attributed to somatic mosaicism.
6.4. Second-Hit Somatic Mosaicism
In 1971, Knudson (63) introduced a two-hit model of diseases such as retinoblastoma. He postulated the inheritance of a recessive, heterozygous, damaging mutation and the acquisition of an inactivating somatic mutation in the other allele. Together, these mutations lead to disease.
Second-hit somatic disease contributes to several neurocutaneous syndromes, including NF1 and tuberous sclerosis 1 (39,47). In each disorder, a constitutional, damaging mutation is inherited and accompanied by the somatic acquisition of a second damaging allele in a single gene. As evidenced by second-hit somatic neurocutaneous disorders, multiple lesions in multiple tissues may occur, suggesting that the acquisition of a secondary, somatic, damaging mutation in the other allele can occur multiple times independently and either somatic mutations occur more often than expected or these loci are at increased susceptibility to mutation (84).
6.5. Revertant Mosaicism
Revertant mosaicism is the spontaneous correction of a pathogenic mutation in a somatic cell (53). Spontaneous reversions produce wild-type and mutant cell populations that can modify or even alleviate disease, so-called natural gene therapy (68). The earlier a reversion occurs, the more likely a larger portion of cells will be phenotypically normal (Figure 4e), influencing disease severity. Partial revertant mosaicism can result in negligible or no clinical improvement. Reversion events can occur within a single cell lineage, multiply independently within a patient, or affect multiple cell types. Corrective mechanisms implicated in restoring wild-type phenotypes include back mutation, gene conversion, intragenic recombination, and second-site mutation (68). Each mechanism serves to restore protein function or abundance. Corrections of dominant Mendelian disorders such as Wiskott-Aldrich syndrome (OMIM 301000) and Fanconi anemia are not uncommon, occurring in up to 11% and 18% of cases, respectively (68).
6.6. Common Complex Diseases
The advent of inexpensive massively parallel sequencing enabled the sequencing of hundreds of thousands, if not millions, of individuals. Genome-wide association studies have investigated many common complex diseases, identifying loci that contribute to disease pathology. Interrogation of constitutional variation and its contribution to common complex diseases has been the major focus of these studies. However, SNP and next-generation sequencing (NGS) data have recently been interrogated to identify mosaic variation in common diseases. For example, mosaic variation is significantly increased in probands diagnosed with autism spectrum disorder relative to unaffected siblings (31,45, 66, 75).
6.7. Cancer
Cancer is the most extensively studied disease whose etiology stems from mosaicism. Studies into the mechanisms, pathways, and downstream effectors of cancer pathology have yielded significant insight into somatic mosaicism. As discussed above (Section 3.1), somatic mutation signatures have been identified and correlated with intracellular and extracellular factors (2). Rates, signatures, and mechanisms of mosaic variation have been identified for extrinsic environmental factors, such as ultraviolet radiation, smoking, and other toxins (103). Intrinsic cellular features contributing to somatic variation and cancer pathology, such as DNA replication, polymerase fidelity, and recombination hotspots in a variety of cellular states and contexts, have been extensively studied in the oncogenic context. Although much insight has been gained through investigations of cancer pathology, this information is only partially translatable to other classes of somatic mosaicism, given the complex context of investigation.
7. ADDITIONAL SOURCES OF MOSAICISM
While mosaicism is usually considered in terms of nuclear DNA, variation in nonnuclear DNA also represents a type of mosaicism.
7.1. Mitochondrial Heteroplasmy
Mitochondria experience tremendous oxidative stress and many replication cycles throughout the human life span. Short-lived, highly reactive oxygen species produced in mitochondria induce DNA damage by the oxidation of nucleoside bases (67). Somatic mutations accumulate due to the failure of the DNA damage response to repair oxidative damage, creating DNA lesions and strand breaks. The lower fidelity of the mitochondrial DNA polymerase gamma introduces somatic variation into mitochondrial DNA(mtDNA) (23). Pathogenic somatic point mutations have been discovered in the mtDNA of both healthy and diseased individuals (35).
Heteroplasmy is the occurrence of somatic mutations that affect only a portion of the mtDNA copies within a cell. Low-level heteroplasmic pathogenic mutations are often well tolerated due to the many nonpathogenic, wild-type mitochondrial copies. However, mitochondrial replication and turnover can increase the cellular proportion of somatic pathogenic mutations (114). Respiratory chain defects may occur after exceeding a threshold level of mutated mtDNA, typically >80%, resulting in human disease (33). Somatic heteroplasmic mutations have been implicated in late-onset degenerative disease including Parkinson’s and Alzheimer disease (114).
7.2. Pathogens and Gene Therapy Integration
Retroviruses insert a reverse-transcribed, double-stranded DNA copy of their genomic RNA into a host cell genome. Eight percent of the human genome is estimated to be of retroviral origin (69). Prominent human retroviral infections include human immunodeficiency virus and human T-lymphotropic virus (128). Similar to MEIs, retroviral integration can disrupt cellular pathways and homeostasis. Insertion in the vicinity of proto-oncogenes can drive overexpression, promoting tumorigenesis (82). Integration at or adjacent to regulatory elements, exonic coding regions, and other noncoding loci as well as expression of virus-encoded accessory genes can perturb cellular networks (74). Retroviral infection contributes to hematopoietic oncogenic malignancies (82). Retrovirus insertion or expression may distort cell signaling, transcriptional cascades, or functional genomic elements, resulting in genetically and phenotypically mosaic host cells and promoting human disease.
In vivo and in vitro gene therapies manipulate the human host cell genome to combat cancer and inherited Mendelian disorders. In vitro techniques, such as chimeric antigen receptor T cell therapy, extract cells from a patient and introduce a targeted somatic recombination (115). Modified cells are screened and reintroduced into the patient to ameliorate disease. In vivo approaches use modified adeno-associated viruses (AAVs) to deliver target therapeutic DNA to host cells. AAV gene therapy was first approved by the European Commission for the treatment of lipoprotein lipase deficiency (41). AAV serotypes have distinctive infectivity rates and tissue specificity (83). Therapeutic DNA integrates into the host genome by recombinant vector transduction. AAV target tissue, cell type, and DNA modification has low specificity, routinely producing off-target or unexpected somatic transductions (83). Random vector integration can lead to loss-or gain-of-function mutations altering cell functionality and homeostasis (21). The generation of AAV particles is imperfect, with random mutations introduced in synthesis yielding a mosaic of particles and integrating a mosaic of targets into the host. Gene-therapy-induced somatic mosaicism is persistent with >7 years of AAV transgene expression documented in liver and muscle (92). Off-target effects are of great concern, as AAVs could produce gonadal mosaicism through the modification of germ cell genomes. Gonadal mosaics can be passed to progeny as inherited constitutional variation, introducing new alleles into the population.
7.3. Epigenetic Mosaicism
An epigenetic mark is a heritable change that does not involve alterations in DNA sequence. It may involve physical modification of genomic DNA (such as methylation) or acetylation of histones. Epigenetic mosaicism is the presence of different epigenotypes in cells derived from a single zygote. Instability in the epigenome makes it difficult to discern normal epigenetic polymorphism, i.e., differential cell-type-specific epigenetic regulation, from epigenetic mosaicism. Histone modifications change rapidly in response to the cellular environment (97). Highly unstable histone modifications make it particularly difficult to discern epigenetic states and, therefore, mosaic epigenotypes (52). In humans, DNA methylation is the covalent methylation of cytosine nucleotides. It has unique patterns associated with the physiologic state and is stable and self-perpetuating at CpG islands (55). In vitro, spontaneous errors in methylation maintenance occur at a rate of 10−4 to 10−5 nucleotides in promoter CpG islands (55). Therefore, replication-dependent errors in the maintenance of the methylation state underlie DNA methylation drift, generating epigenetic mosaicism. Older monozygotic twin pairs have significantly greater DNA methylation discordance, consistent with the stochastic methylation drift that is at least partially independent of genotype (119). Environmental chemical exposures can also have effects on the epigenome. For instance, red blood cell folate level is positively correlated with CpG island methylation drift (124). Spatial and temporal exposure to environmental chemicals generates diverse, unique epigenotypes. Methylation drift contributes to autoimmune disease by unmasking hidden tissue antigens or hypomethylation-induced altered functionality (102).
7.4. Genomic Imprinting and X-Chromosome Inactivation
First initiated in the germline, genomic imprinting is the allele-specific epigenetic marking of parental alleles in a set of developmentally important genes (117). Approximately 100 imprinted genes have been identified in humans (91). Most imprinted genes are marked by DNA methylation and preferentially expressed from a single allele. Monoallelic expression suggests that mutation in a single allele is sufficient for pathogenesis (110). Genomic imprinting disorders result from the disturbance of imprinted methylation polymorphism, genetic variants, and UPD. Environmental factors affect DNA methylation; therefore, exposures may perturb imprinted gene regulation. Assisted reproductive technologies do not alter genomic nucleotide content; however, conceptions with assisted reproductive technologies are at increased risk of developing several imprinting disorders, which is suggestive of environment-induced imprinting defects (93). UPD of chromosomes containing imprinted loci changes the maternal and paternal gene dosages, possibly causing them to lack active copies of essential imprinted genes and leading to disease (37). Well-known imprinting disorders include Angelman syndrome, Prader-Willi syndrome, and Beckwith-Wiedemann syndrome (13).
X-chromosome inactivation (XCI) is the random silencing of an X chromosome in females, equalizing gene dosage between XX females and XY males (95). Inactivation occurs in early embryogenesis with an equal probability of silencing either X chromosome (129). A mosaic of paternal and maternal inactive X chromosomes is established as the silenced allele is propagated through cellular divisions. XCI mosaicism can be advantageous, alleviating deleterious X-linked mutations and contributing to physiological diversity (89). The ratio of expressed alleles differs within and between individuals due to stochasticity or mutations affecting selection (88). Skewed XCI, the unbalanced expression of damaging mutant alleles, can cause Wiskott-Aldrich syndrome and Lesch-Nyhan syndrome (88). Clinical severity is often influenced by the mosaic composition of X-linked heterozygous alleles.
8. MOSAIC DISCOVERY METHODS
8.1. Mosaic Variant Profiling
To comprehensively profile all classes of somatic genomic mosaic mutations would require a multitude of technologies and experimental designs. The robustness of somatic mutation discovery is highly technology dependent. Sequencing technologies vary widely in their sensitivity and specificity to detect different classes of somatic mutations. Here we discuss four technologies used to detect somatic mutations: cytogenetics, SNP arrays, NGS, and single-cell genomics.
8.2. Cytogenetics
Conventional and molecular cytogenetics are microscopy-based techniques to detect large chromosomal abnormalities in single cells. Conventional karyotype banding is the clinical gold standard for single-cell assessment of chromosomal mosaicism and structural aberrations including inversions, translocations, and copy number changes (126). Microscopic evaluation of staining resolves genome-wide 5–7-Mb structural abnormalities (73, 123). Detection resolution and fresh tissue requirement limitations of karyotyping drove the advancements of higher-resolution, diversified molecular cytogenetic techniques. Fluorescence in situ hybridization (FISH) is a molecular cytogenetic method in which fluorescently labeled DNA probes are hybridized to genomic complements. FISH probes can target repetitive sequences (centromeres and telomeres), specific sites, and whole chromosomes with multiprobe techniques (125). FISH resolves similar structural aberrations as conventional karyotyping does but at a higher resolution, accurately identifying genomic changes as small as ~2 kb but more typically those that are several hundred kilobases (123). Off-target probe hybridization limits the clinical applications and base resolutions of molecular cytogenetic techniques, especially for multiplex and site-specific methods, but can be mitigated with adequate controls (5).
Conventional cytogenetics has the ability to detect the mosaicism of large chromosomal variations by screening an appropriate number of metaphase spreads (e.g., n = 30). FISH techniques have the advantage that they can be used on interphase cells. The use of interphase FISH has been particularly useful for cancer cytogenetics, where mosaicism is common. FISH analyses of uncultured interphase cells are useful for prenatal testing with amniotic fluid or chorionic villus samples.
8.3. Single-Nucleotide Polymorphism Microarrays
SNP arrays are DNA microarrays containing oligonucleotide probes that target population SNPs. Reference and SNP alleles are spotted on a slide, and a fluorescently labeled test sample is hybridized. A sample genotype is determined by the differential signal intensity of alleles, and copy number changes are detected by the integration of signal intensities across probed loci. Medium-to large-sized mosaic copy number changes at >5% aneuploidy can be detected by the balance of allele intensities across SNPs (22, 76). SNP arrays can also detect mosaic trisomy and UPD (73). Arrays have difficulty detecting somatic mutations occurring at low variant allele frequency (e.g., < 5 %) and small events due to limited probe coverage. Arrays typically exclude highly repetitive regions, pseudogenes, and low-complexity regions due to off-target cohybridization, limiting mosaic variation detection in these regions.
8.4. Next-Generation Sequencing
DNA sequencing was revolutionized by the advent of massively parallel NGS. NGS technologies produce millions or billions of short, high-confidence genomic reads. Sequencing reads are typically 100–150 bp and may be either single-read or paired-end sequenced. Single-read sequencing is a consecutive string of nucleotides sequenced from a DNA molecule. Paired-end sequencing, in which both ends of a DNA fragment are sequenced, provides more robust mapping and variant phasing by leveraging the mapping and contained genomic variation between both reads (30). With direct evidence of physical linkage, paired-end reads are expected to map to nearby genomic locations, reducing the search space and improving mapping specificity. Paired-end read mapping can also be informative of structural variation as split or discordant read mapping provides evidence for inversions, duplications, and translocations. Phasing somatic mutations to a nearby heterozygous germline mutation on the same or paired-end read can allow its haplotype to be discerned. Somatic mutations are expected to occur on a single haplotype, and the phasing of candidates to neighboring SNPs can increase the sensitivity of the sequencing to detect true mosaic variation. NGS can discover all classes of mosaic variation across almost the entire genome. NGS can survey bulk cell populations, single cells, or clonally expanded colonies. Whole-exome sequencing (WES) and whole-genome sequencing (WGS) are two of the most commonly employed NGS methods. WES captures the protein-coding exons of the genome (~1–2% of the entire genome). Mosaic CNV discovery is hindered by exon capture bias in WES (87). WGS comprehensively assesses the entire genome with mild nonuniformity (1).
8.5. Single-Cell Sequencing
Bulk genome sequencing has limited resolution to detect low variant allele frequency (VAF) somatic mutations due to wild-type cell admixture (76). Sequencing depth and the utilization of aggregate constituent DNA hinder somatic mutation discovery and elucidation of the underlying subclonal phylogenetic architecture. Within single cells, 13–41 % have at least one megabase-scale postzygotic CNV (43). Assessments of the temporal acquisition and distribution of somatic variation are impractical in bulk genetic analyses. Single-cell sequencing (SCS) isolates, amplifies, and sequences DNA from single cells to allow the interrogation of somatic mosaicism at the finest biological scale. Approximately 90% of the genome is accessible by SCS (118). SCS enables the discovery of somatic mutations within individual cells, cell types, and tissues. High-resolution somatic mutation discovery by SCS indicates that there are >1,000 SNVs per neuron, much more than previously appreciated (43). Cosegregation single-cell somatic analysis facilitates the reconstruction of cell lineage, developmental occurrence, affected cell types, and body localization of mutations (86).
Many SCS techniques and methods of analysis have been developed to assess somatic mosaicism. SCS requires DNA amplification to obtain sufficient DNA to sequence each cell. Some single-cell DNA amplification methods, for example, isothermal multiple displacement amplification (MDA) and PCR-based techniques, suffer from major biases, including allelic dropout (ADO), amplification bias, and coverage nonuniformity. PCR-based methods have better amplification uniformity but approximately tenfold more single nucleotide errors, while MDA produces more broad, uniform genomic coverage but still can result in >30% ADO (118). Targeted bulk DNA sequencing is performed to validate somatic genomic mutations identified in SCS in order to exclude false-positive heterozygous constitutional variants and technical sequencing artifacts. Techniques that obviate bulk comparison have emerged in the form of rapid and cost-effective profiling of many single cells in parallel. Leveraging many single cells for validation, as detecting somatic mutations in multiple cells, minimizes sequencing artifacts and identifies constitutional variation (118).
8.6. Single-Molecule Sequencing
Often referred to as third-generation sequencing, single-molecule sequencing (SMS) directly sequences individual DNA molecules. SMS generates exceptionally long sequencing reads (>20 kb) from unamplified high-molecular-weight DNA (20, 34). Single-molecule long reads permit sequencing through repetitive elements, improved variant phasing, and detection of epigenetic modifications (4, 17). Variant phasing improves somatic mosaic variant discovery specificity by identifying and removing haplotype discordant mosaic candidates. Resolving allele phasing with SMS enabled the detection of revertant mosaicism in an exceptional case of keratitis-ichthyosis-deafness syndrome (49). Relative to short-read sequencing, long SMS has more precise genomic mapping, particularly to low-complexity or highly homologous regions such as repetitive elements and pseudogenes, enabling interrogation of somatic variation in a larger portion of the genome (4). By providing more accurate mapping, alignment across a greater portion of the genome, and the ability to span multiple breakpoints within a single read, SMS has improved researchers’ power to detect and resolve simple and complex somatic structural rearrangements (17). Long reads also enable the highly contiguous de novo assembly of a sample, reconstructing a personalized genome that incorporates individual de novo or inherited mutations that are absent from the human reference (111). Alignment to the de novo assembly eliminates many alignment biases induced by mapping to the human reference. Native DNA sequencing in SMS eliminates the amplification bias found in PCR-amplified short-read technologies (105). PCR amplification contributes to false-positive somatic mutations due to systematic biases including the generation of chimeric reads, slippage-induced repeat size variation, and skewed guanine-cytosine content nucleotide bias (4). Although considered to be the latest rendition of sequencing technologies, SMS has several weaknesses, including the generation of more indels, higher per-base nucleotide error rates, large DNA input requirements (>5 μg), variable sequence read lengths, errors in low-sequence complexity regions, and prohibitive cost (15). SMS is therefore presently ill-suited for calling mosaic indels and SNVs, low VAF somatic mutation, and sparse samples (100, 104).
8.7. Experimental Design
Experimental design for the discovery of mosaicism is highly dependent on the hypothesis. Common experimental designs include pedigrees, paired (tumor-normal), and single-sample (tumor-only) approaches. Here we discuss the trade-offs between each of these paradigms.
Pedigrees sequence the affected individual (proband) and related family members, including parents, siblings, cousins, and other extended familial relationships. The sequencing of additional related persons is utilized to improve the specificity of somatic variation discovery Trio pedigrees, including the mother, father, and affected child, are sequenced to take advantage of the high degree of genomic relatedness and allelic completeness. Often, siblings, e.g., quaternary pedigrees, are also included to further exclude sequencing artifacts and for phenotypic comparison. Somatic false-positive mutations, which are often germline population polymorphisms, and rare, shared familial genomic mutations, are removed through consideration of inherited alleles (84). This improves somatic variant calling specificity and enables the discovery of de novo variation in the proband.
Paired sample, also referred to as tumor-normal, analysis sequences two cell types or tissues from the patient: one apparently normal and the other diseased. Paired somatic variant discovery leverages the technical and patient-specific genomic architecture shared by the normal and affected samples to exclude shared germline variants and sequencing artifacts. Artifacts are introduced during sample preparation, sequencing, and alignment (130). Utilization of the unaffected sample to account for technical and other sample-related noise improves somatic genomic mutation discovery specificity. Of major concern, the paired sample design assumes the normal sample has the wild-type genotype, not harboring the suspected damaging somatic mutation. Although the amount of contamination in the unaffected sample can be accounted for in somatic variant discovery, the true percentage of contamination of the causal somatic mutation remains unknown. Sample contamination of 1.5% from another human source is common, adding further complexity to the estimation of contamination (30). Underestimation of contamination reduces sensitivity while overestimation may unmask many artifacts, dramatically increasing false-positive somatic variants.
Matched normal or familial samples are not always available; therefore, variant calling using a single sample, referred to as tumor-only design, can be necessary for the discovery of somatic mutations. In single-sample analyses, only an affected, diseased sample is sequenced. Single-sample analyses have no genotypically related samples to utilize for artifact discrimination, making it more difficult to identify somatic variation. The single-sample strategy is often used due to sample availability and cost reduction. Some advantages of the single-sample approach are increased sequencing depth for the detection of low VAF somatic mutations and increased power to detect recurrent somatic mutations through the sequencing of more affected cases as the design is less costly.
9. DISCUSSION
While the phenomenon of mosaicism has been known for about a century, only now are we beginning to appreciate its nature and the extent of its effects. Initially, mosaicism was recognized because of visible consequences, such as birthmarks or skin patterns. The emergence of cytogenetics led to the discovery of mosaicism in chromosomal copies. After the 1956 discovery that humans have 23 pairs of chromosomes (120), researchers learned by 1959 that Down syndrome is caused by trisomy of chromosome 21 (70). Just two years later, the first reports of mosaic trisomy 21 were published (19, 42). Since then, cytogenetics has continued to have a fundamental role in the detection of mosaicism.
By the mid-2000s, SNP arrays had emerged as a second essential method to detect mosaicism. The combination of copy number and genotype estimation allowed CNN phenomena such as UPD to be readily detected, even in segmental form in small genomic regions (e.g., <10,000 bp). Furthermore, SNP array data allowed the sensitive and specific detection of mosaic variants, even below 5% frequency.
In the past decade, the emergence of NGS technologies has facilitated the detection of mosaic variation in single cells and at base-pair resolution. We now recognize that almost all cells are mosaic. This tremendous diversity of genomic DNA content challenges the notion that each person has a stable genome over time. For obligate mosaic diseases, NGS technology has facilitated the discovery of dozens of genes that, when mutated somatically, lead to disease. For nonobligate mosaicism, such as Mendelian diseases that may sometimes manifest in the mosaic state, we can begin to estimate the prevalence. For common diseases, we are only beginning to assess the role of mosaic variation. For example, while neuropsychiatric diseases such as autism spectrum disorder and schizophrenia are highly heritable, a role for nonheritable, somatic mosaicism has recently been demonstrated.
Key aspects of mosaic variation include mechanism, timing, location, and functional consequence. We anticipate that deeper sampling across development, across body regions, and across clinical cohorts will continue to expand our appreciation of this diverse, fundamental, biological process. Somatic mosaicism may play an underestimated role in hereditary diseases that show reduced penetrance and/or variability of clinical expression. This is because mosaic variants with functional effects on the disease pathogenic pathway may or may not influence the phenotype, depending on the cells and tissue in which they are expressed. The same reasoning can be applied to the variegation of epigenetic silencing. For instance, the removal of XCI or imprinting with aging can unmask variants that may have a detrimental effect in certain tissues.
SUMMARY POINTS.
Mosaicism may occur in germ or somatic cells. Germline mosaicism is heritable and poses cryptic risk for spontaneous disease in progeny. Somatic mosaicism is nonheritable and sporadically contributes to cellular diversity and disease.
In parallel with inherited germline variation, mosaic DNA variants include those that are small (single nucleotide variants and indels) and large (structural variants and aneuploidies). These vary in their frequency of occurrence, methods of discovery, mechanism of formation, and functional consequence.
Mosaicism occurs at all stages of development. It appears to be a normal phenomenon during embryogenesis, postnatally, and with aging. Mosaic disease can be evident at birth, such as birthmarks associated with neurocutaneous disorders, or later in life, such as in cancer.
The location of mosaic mutations depends on the time of the initial mutation (e.g., embryologically earlier events tend to result in a greater extent of mosaicism across different cell types and organs) and has direct consequences on disease presentation.
The functional consequences of mosaicism are diverse. Although predominantly functionally neutral, mosaicism can be deleterious or advantageous. Mosaic variants cause (or contribute to) obligate mosaic disorders (disease-causing postzygotic mutations that would be embryonic lethal as constitutional mutations), nonobligate mosaic disorders (germline disorders than can also manifest as mosaic), and second-hit somatic disease (the combined effect of a spontaneous somatic mutation and an inherited germline variation).
Sources of mosaicism exist that are not limited to the mutation of the nuclear genome, such as mitochondrial heteroplasmy, viral vectors, or epigenetic mosaicism.
Although discussed independently, these six categories are exceedingly intertwined. For instance, rates of mosaic variation vary according to the class of mutation, developmental timing, environmental insults, type of mosaicism, and underlying genotype. The nature and extent of mosaicism in human health and disease are only beginning to be uncovered, with implications across a broad spectrum of disorders and increased appreciation of its effects in complex human disease.
FUTURE ISSUES.
There is an obligate role for mosaic variation in dozens of diseases, including single-gene disorders and mosaic aneuploidies. How can such conditions be better diagnosed, prevented, or treated? For example, there is often a very long lag between the identification of the symptoms of Sturge-Weber syndrome and its clinical diagnosis (18).
What is the role of mosaic variation in common, complex disease? With an increasing proportion of the population being sequenced, the interrogation of the contribution of somatic mosaicism in neurodevelopmental and neuropsychiatric disorders becomes possible. The interplay between mosaicism and inherited germline variation is difficult to discern given that even small populations of cells with damaging mutations can cause or contribute to disease.
One of greatest challenges in biology today is to understand the relationship between genotype and phenotype. Even for single-gene disorders there is great phenotypic variability observed for the same mutation. From a clinical perspective, this makes prognosis difficult. It is important to understand the role of mosaic variation in this context.
ACKNOWLEDGMENTS
J.P. was supported by the Intellectual and Developmental Disabilities Research Center (grant U54 HD079123) from the Eunice Kennedy Shriver National Institute of Child Health and Human Development and by a grant from the National Institutes of Mental Health (U01 MH106884).
Glossary
- Mutation
any change in a specific DNA sequence
- Postzygotic
referring to a pathogenic variant or abnormality in chromosome replication, segregation, and/or methylation that occurs after the fertilization of the ovum by the sperm, often leading to mosaicism
- Zygote
the resulting diploid cell from the fusion of two haploid gametes, the result of sexual reproduction
- Somatic mosaicism
two or more cells with different genetic compositions within an individual, which may or may not include the germline cells
- Germline mosaicism
two or more cells with different genetic compositions confined to the precursor (germline) cells of the egg or sperm
- Imprinting
the process by which maternally and paternally derived chromosomes are uniquely chemically modified (usually by methylation), leading to the different expression of a certain gene or genes on those chromosomes, depending on their parental origin
- Aneuploidy
one or more extra or missing chromosomes leading to an unbalanced chromosome complement, or any chromosome number that is not an exact multiple of the haploid number (i.e., 23)
- Uniparental disomy (UPD)
the situation in which both members of a chromosome pair or segments of a chromosome pair are inherited from one parent and neither is inherited from the other parent, resulting in an abnormal phenotype in some instances
- De novo
referring to a gene variant that does not occur in either parent; it is present for the first time in the proband
- Heteroplasmy
the presence within a single cell of both normal and mutated mitochondrial DNA
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Ajay SS, Parker SC, Abaan HO, Fajardo KV, Margulies EH. 2011. Accurate and comprehensive sequencing of personal genomes. Genome Res. 21:1498–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexandrov LB, Kim J, Haradhvala NJ, Huang MN, Tian Ng AW, et al. 2020. The repertoire of mutational signatures in human cancer. Nature 578:94–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antonarakis SE. 2017. Down syndrome and the complexity of genome dosage imbalance. Nat. Rev. Genet 18:147–63 [DOI] [PubMed] [Google Scholar]
- 4.Ardui S, Ameur A, Vermeesch JR, Hestand MS. 2018. Single molecule real-time (SMRT) sequencing comes of age: applications and utilities for medical diagnostics. Nucleic Acids Res. 46:2159–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arvey A, Hermann A, Hsia CC, Ie E, Freund Y, McGinnis W. 2010. Minimizing off-target signals in RNA fluorescent in situ hybridization. Nucleic Acids Res. 38:e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. 2015. A global reference for human genetic variation. Nature 526:68–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bae T, Tomasini L, Mariani J, Zhou B, Roychowdhury T, et al. 2018. Different mutational rates and mechanisms in human cells at pregastrulation and neurogenesis. Science 359:550–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bakker E, Van Broeckhoven C, Bonten EJ, van de Vooren MJ, Veenema H, et al. 1987. Germline mosaicism and Duchenne muscular dystrophy mutations. Nature 329:554–56 [DOI] [PubMed] [Google Scholar]
- 9.Bassing CH, Swat W, Alt FW. 2002. The mechanism and regulation of chromosomal V(D)J recombination. Cell 109(Suppl.):S45–55 [DOI] [PubMed] [Google Scholar]
- 10.Bruder CE, Piotrowski A, Gijsbers AA, Andersson R, Erickson S, et al. 2008. Phenotypically concordant and discordant monozygotic twins display different DNA copy-number-variation profiles. Am. J. Hum. Genet 82:763–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bunnell ME, Wilkins-Haug L, Reiss R. 2017. Should embryos with autosomal monosomy by preimplantation genetic testing for aneuploidy be transferred?: Implications for embryo selection from a systematic literature review of autosomal monosomy survivors. Prenat. Diagn 37:1273–80 [DOI] [PubMed] [Google Scholar]
- 12.Bushman DM, Chun J. 2013. The genomically mosaic brain: aneuploidy and more in neural diversity and disease. Semin. Cell Dev. Biol 24:357–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Butler MG. 2009. Genomic imprinting disorders in humans: a mini-review. J. Assist. Reprod. Genet 26:477–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Campbell IM, Shaw CA, Stankiewicz P, Lupski JR. 2015. Somatic mosaicism: implications for disease and transmission genetics. Trends Genet. 31:382–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carneiro MO, Russ C, Ross MG, Gabriel SB, Nusbaum C, DePristo MA. 2012. Pacific biosciences sequencing technology for genotyping and variation discovery in human data. BMC Genom. 13:375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carvalho CM, Lupski JR. 2016. Mechanisms underlying structural variant formation in genomic disorders. Nat. Rev. Genet 17:224–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chaisson MJ, Huddleston J, Dennis MY, Sudmant PH, Malig M, et al. 2015. Resolving the complexity of the human genome using single-molecule sequencing. Nature 517:608–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cho S, Maharathi B, Ball KL, Loeb JA, Pevsner J. 2019. Sturge-Weber syndrome patient registry: delayed diagnosis and poor seizure control. J. Pediatr 215:158–63.e6 [DOI] [PubMed] [Google Scholar]
- 19.Clarke CM, Edwards JH. 1961. 21-Trisomy/normal mosaicism in an intelligent child with some Mongoloid characters. Lancet 1:1028–30 [DOI] [PubMed] [Google Scholar]
- 20.Clarke J, Wu HC, Jayasinghe L, Patel A, Reid S, Bayley H. 2009. Continuous base identification for single-molecule nanopore DNA sequencing. Nat. Nanotechnol 4:265–70 [DOI] [PubMed] [Google Scholar]
- 21.Colella P, Ronzitti G, Mingozzi F. 2018. Emerging issues in AAV-mediated in vivo gene therapy. Mol. Ther. Methods Clin. Dev 8:87–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conlin LK, Kaur M, Izumi K, Campbell L, Wilkens A, et al. 2012. Utility of SNP arrays in detecting, quantifying, and determining meiotic origin of tetrasomy 12p in blood from individuals with Pallister-Killian syndrome. Am. J. Med. Genet. A 158A:3046–53 [DOI] [PubMed] [Google Scholar]
- 23.Copeland WC. 2010. The mitochondrial DNA polymerase in health and disease. Subcell. Biochem 50:211–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De S. 2011. Somatic mosaicism in healthy human tissues. Trends Genet. 27:217–23 [DOI] [PubMed] [Google Scholar]
- 25.de Koning AP, Gu W, Castoe TA, Batzer MA, Pollock DD. 2011. Repetitive elements may comprise over two-thirds of the human genome. PLOS Genet. 7:el002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Delhanty JD. 2011. Inherited aneuploidy: germline mosaicism. Cytogenet. Genome Res 133:136–40 [DOI] [PubMed] [Google Scholar]
- 27.Delhanty JD, SenGupta SB, Ghevaria H. 2019. How common is germinal mosaicism that leads to premeiotic aneuploidy in the female? J. Assisted Reprod. Genet 36:2403–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.D’Gama AM, Walsh CA. 2018. Somatic mosaicism and neurodevelopmental disease. Nat. Neurosci 21:1504–14 [DOI] [PubMed] [Google Scholar]
- 29.Dorner T, Lipsky PE. 2001. Immunoglobulin variable-region gene usage in systemic autoimmune diseases. Anhritis Rheum. 44:2715–27 [DOI] [PubMed] [Google Scholar]
- 30.Dou Y, Gold HD, Luquette LJ, Park PJ. 2018. Detecting somatic mutations in normal cells. Trends Genet. 34:545–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dou Y, Yang X, Li Z, Wang S, Zhang Z, et al. 2017. Postzygotic single-nucleotide mosaicisms contribute to the etiology of autism spectrum disorder and autistic traits and the origin of mutations. Hum. Mutat 38:1002–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dumanski JP, Piotrowski A. 2012. Structural genetic variation in the context of somatic mosaicism. Methods Mol. Biol 838:249–72 [DOI] [PubMed] [Google Scholar]
- 33.Durham SE, Samuels DC, Cree LM, Chinnery PF. 2007. Normal levels of wild-type mitochondrial DNA maintain cytochrome c oxidase activity for two pathogenic mitochondrial DNA mutations but not for m.3243→G. Am. J. Hum. Genet 81:189–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eid J, Fehr A, Gray J, Luong K, Lyle J, et al. 2009. Real-time DNA sequencing from single polymerase molecules. Science 323:133–38 [DOI] [PubMed] [Google Scholar]
- 35.Elliott HR, Samuels DC, Eden JA, Relton CL, Chinnery PF. 2008. Pathogenic mitochondrial DNA mutations are common in the general population. Am. J. Hum. Genet 83:254–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Engel E. 1980. A new genetic concept: uniparental disomy and its potential effect, isodisomy. Am. J. Med. Genet 6:137–43 [DOI] [PubMed] [Google Scholar]
- 37.Engel E. 1997. Uniparental disomy (UPD). Genomic imprinting and a case for new genetics (prenatal and clinical implications: the “Likon” concept). Ann. Genet 40:24–34 [PubMed] [Google Scholar]
- 38.Erwin JA, Marchetto MC, Gage FH. 2014. Mobile DNA elements in the generation of diversity and complexity in the brain. Nat. Rev. Neurosci 15:497–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eur. Chromosom. 16 Tuberous Scler. Consort. 1993. Identification and characterization of the tuberous sclerosis gene on chromosome 16. Cell 75:1305–15 [DOI] [PubMed] [Google Scholar]
- 40.Evrony GD, Cai X, Lee E, Hills LB, Elhosary PC, et al. 2012. Single-neuron sequencing analysis of L1 retrotransposition and somatic mutation in the human brain. Cell 151:483–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ferreira V, Petty H, Salmon F. 2014. Immune responses to AAV-vectors, the Glybera example from bench to bedside. Front. Immunol. 5:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fitzgerald PH, Lycette RR. 1961. Mosaicism in man, involving the autosome associated with mongolism. Heredity 16:509–12 [Google Scholar]
- 43.Forsberg LA, Gisselsson D, Dumanski JP. 2017. Mosaicism in health and disease—clones picking up speed. Nat. Rev. Genet 18:128–42 [DOI] [PubMed] [Google Scholar]
- 44.Forsberg LA, Rasi C, Razzaghian HR, Pakalapati G, Waite L, et al. 2012. Age-related somatic structural changes in the nuclear genome of human blood cells. Am. J. Hum. Genet 90:217–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Freed D, Pevsner J. 2016. The contribution of mosaic variants to autism spectrum disorder. PLOS Genet. 12:el006245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gajecka M. 2016. Unrevealed mosaicism in the next-generation sequencing era. Mol. Genet. Genom 291:513–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garcia-Linares C, Fernandez-Rodriguez J, Terribas E, Mercade J, Pros E, et al. 2011. Dissecting loss of heterozygosity (LOH) in neurofibromatosis type 1-associated neurofibromas: importance of copy neutral LOH. Hum. Mutat 32:78–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Garcia-Romero MT, Parkin P, Lara-Corrales I. 2016. Mosaic neurofibromatosis type 1: a systematic review. Pediatr. Dermatol 33:9–17 [DOI] [PubMed] [Google Scholar]
- 49.Gudmundsson S, Wilbe M, Ekvall S, Ameur A, Cahill N, et al. 2017. Revertant mosaicism repairs skin lesions in a patient with keratitis-ichthyosis-deafness syndrome by second-site mutations in connexin 26. Hum. Mol. Genet 26:1070–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Happle R 1987. Lethal genes surviving by mosaicism: a possible explanation for sporadic birth defects involving the skin. J. Am. Acad. Dermatol 16:899–906 [DOI] [PubMed] [Google Scholar]
- 51.Hassold T, Chen N, Funkhouser J, Jooss T, Manuel B, et al. 1980. A cytogenetic study of 1000 spontaneous abortions. Ann. Hum. Genet 44:151–78 [DOI] [PubMed] [Google Scholar]
- 52.Henikoff S, Shilatifard A. 2011. Histone modification: cause or cog? Trends Genet. 27:389–96 [DOI] [PubMed] [Google Scholar]
- 53.Hirschhorn R. 2003. In vivo reversion to normal of inherited mutations in humans. J. Med. Genet 40:721–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Iourov IY, Vorsanova SG, Yurov YB, Kutsev SI. 2019. Ontogenetic and pathogenetic views on somatic chromosomal mosaicism. Genes 10:379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Issa JP. 2014. Aging and epigenetic drift: a vicious cycle. J. Clin. Invest 124:24–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jackson-Cook C. 2011. Constitutional and acquired autosomal aneuploidy. Clin. Lab. Med 31:481–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jonsson H, Sulem P, Arnadottir GA, Palsson G, Eggertsson HP, et al. 2018. Multiple transmissions of de novo mutations in families. Nat. Genet 50:1674–80 [DOI] [PubMed] [Google Scholar]
- 58.Jonsson H, Sulem P, Kehr B, Kristmundsdottir S, Zink F, et al. 2017. Parental influence on human germline de novo mutations in 1,548 trios from Iceland. Nature 549:519–22 [DOI] [PubMed] [Google Scholar]
- 59.Kalousek DK. 1994. Confined placental mosaicism and uniparental disomy. Funct. Dev. Morphol 4:93–98 [PubMed] [Google Scholar]
- 60.Kalousek DK, Vekemans M. 1996. Confined placental mosaicism. J. Med. Genet 33:529–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kazazian HH Jr., Moran JV. 2017. Mobile DNA in health and disease. N Engl. J. Med 377:361–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.King DA, Jones WD, Crow YJ, Dominiczak AF, Foster NA, et al. 2015. Mosaic structural variation in children with developmental disorders. Hum. Mol. Genet. 24:2733–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Knudson AG Jr. 1971. Mutation and cancer: statistical study of retinoblastoma. PNAS 68:820–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Koche RP, Rodriguez-Fos E, Helmsauer K, Burkert M, MacArthur IC, et al. 2020. Extrachromosomal circular DNA drives oncogenic genome remodeling in neuroblastoma. Nat. Genet 52:29–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kotzot D. 1999. Abnormal phenotypes in uniparental disomy (UPD): fundamental aspects and a critical review with bibliography of UPD other than 15. Am. J. Med. Genet 82:265–74 [PubMed] [Google Scholar]
- 66.Krupp DR, Barnard RA, Duffourd Y, Evans SA, Mulqueen RM, et al. 2017. Exonic mosaic mutations contribute risk for autism spectrum disorder. Am. J. Hum. Genet 101:369–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kujoth GC, Hiona A, Pugh TD, Someya S, Panzer K, et al. 2005. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science 309:481–84 [DOI] [PubMed] [Google Scholar]
- 68.Lai-Cheong JE, McGrath JA, Uitto J. 2011. Revertant mosaicism in skin: natural gene therapy. Trends Mol. Med 17:140–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409:860–921 [DOI] [PubMed] [Google Scholar]
- 70.Lejeune J, Gautier M, Turpin R. 1959. Étude des chromosomes somatiques de neuf enfants mongoliens. C. R. Acad. Sci. Paris 248:1721–22 [PubMed] [Google Scholar]
- 71.Lemmers RJ, van der Vliet PJ, Klooster R, Sacconi S, Camano P, et al. 2010. A unifying genetic model for facioscapulohumeral muscular dystrophy. Science 329:1650–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lemmers RJ, van der Wielen MJ, Bakker E, Padberg GW, Frants RR, van der Maarel SM. 2004. Somatic mosaicism in FSHD often goes undetected. Ann. Neurol. 55:845–50 [DOI] [PubMed] [Google Scholar]
- 73.Levy B, Burnside RD. 2019. Are all chromosome microarrays the same? What clinicians need to know. Prenat. Diagn 39:157–64 [DOI] [PubMed] [Google Scholar]
- 74.Li W, Yang L, Harris RS, Lin L, Olson TL, et al. 2019. Retrovirus insertion site analysis of LGL leukemia patient genomes. BMC Med. Genom 12:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lim ET, Uddin M, De Rubeis S, Chan Y, Kamumbu AS, et al. 2017. Rates, distribution and implications of postzygotic mosaic mutations in autism spectrum disorder. Nat. Neurosci. 20:1217–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lim YH, Moscato Z, Choate KA. 2017. Mosaicism in cutaneous disorders. Annu. Rev. Genet 51:123–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lodato MA, Walsh CA. 2019. Genome aging: Somatic mutation in the brain links age-related decline with disease and nominates pathogenic mechanisms. Hum. Mol. Genet 28:R197–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Loftfield E, Zhou W, Graubard BI, Yeager M, Chanock SJ, et al. 2018. Predictors of mosaic chromosome Y loss and associations with mortality in the UK Biobank. Sci. Rep 8:12316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lynch M. 2010. Rate, molecular spectrum, and consequences of human mutation. PNAS 107:961–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Machiela MJ. 2019. Mosaicism, aging and cancer. Cum Opin. Oncol 31:108–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Machiela MJ, Zhou W, Sampson JN, Dean MC, Jacobs KB, et al. 2015. Characterization of large structural genetic mosaicism in human autosomes. Am. J. Hum. Genet 96:487–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Maeda N, Fan H, Yoshikai Y. 2008. Oncogenesis by retroviruses: old and new paradigms. Rev. Med. Virol 18:387–405 [DOI] [PubMed] [Google Scholar]
- 83.Mak KY, Rajapaksha IG, Angus PW, Herath CB. 2017. The adeno-associated virus—a safe and promising vehicle for liver-specific gene therapy of inherited and non-inherited disorders. Curr. Gene Ther 17:4–16 [DOI] [PubMed] [Google Scholar]
- 84.McConnell MJ, Moran JV, Abyzov A, Akbarian S, Bae T, et al. 2017. Intersection of diverse neuronal genomes and neuropsychiatric disease: The Brain Somatic Mosaicism Network. Science 356:eaal1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.McCoy RC. 2017. Mosaicism in preimplantation human embryos: when chromosomal abnormalities are the norm. Trends Genet. 33:448–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.McKenna A, Gagnon JA. 2019. Recording development with single cell dynamic lineage tracing. Development 146:dev169730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Meynert AM, Ansari M, FitzPatrick DR, Taylor MS. 2014. Variant detection sensitivity and biases in whole genome and exome sequencing. BMC Bioinform. 15:247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Migeon BR. 1998. Non-random X chromosome inactivation in mammalian cells. Cytogenet. Cell Genet 80:142–48 [DOI] [PubMed] [Google Scholar]
- 89.Migeon BR. 2006. The role of X inactivation and cellular mosaicism in women’s health and sex-specific diseases. JAMA 295:1428–33 [DOI] [PubMed] [Google Scholar]
- 90.Moller HD, Mohiyuddin M, Prada-Luengo I, Sailani MR, Hailing JF, et al. 2018. Circular DNA elements of chromosomal origin are common in healthy human somatic tissue. Nat. Commun 9:1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Monk D, Mackay DJG, Eggermann T, Maher ER, Riccio A. 2019. Genomic imprinting disorders: lessons on how genome, epigenome and environment interact. Nat. Rev. Genet 20:235–48 [DOI] [PubMed] [Google Scholar]
- 92.Nathwani AC, Tuddenham EG, Rangarajan S, Rosales C, McIntosh J, et al. 2011. Adenovirus-associated virus vector-mediated gene transfer in hemophilia B. N. Engl. J. Med 365:2357–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Niemitz EL, Feinberg AP. 2004. Epigenetics and assisted reproductive technology: a call for investigation. Am. J. Hum. Genet 74:599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.O’Keefe C, McDevitt MA, Maciejewski JP. 2010. Copy neutral loss of heterozygosity: a novel chromosomal lesion in myeloid malignancies. Blood 115:2731–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Panning B. 2008. X-chromosome inactivation: the molecular basis of silencing. J. Biol 7:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Papavassiliou P, Charalsawadi C, Rafferty K, Jackson-Cook C. 2015. Mosaicism for trisomy 21: a review. Am. J. Med. Genet. A 167A:26–39 [DOI] [PubMed] [Google Scholar]
- 97.Peters AH, Kubicek S, Mechtler K, O’Sullivan RJ, Derijck AA, et al. 2003. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol. Cell 12:1577–89 [DOI] [PubMed] [Google Scholar]
- 98.Potter H, Chial HJ, Caneus J, Elos M, Elder N, et al. 2019. Chromosome instability and mosaic aneuploidy in neurodegenerative and neurodevelopmental disorders. Front. Genet 10:1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pyott SM, Pepin MG, Schwarze U, Yang K, Smith G, Byers PH. 2011. Recurrence of perinatal lethal osteogenesis imperfecta in sibships: parsing the risk between parental mosaicism for dominant mutations and autosomal recessive inheritance. Genet. Med 13:125–30 [DOI] [PubMed] [Google Scholar]
- 100.Quail MA, Smith M, Coupland P, Otto TD, Harris SR, et al. 2012. A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genom. 13:341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rech GE, Sanz-Martin JM, Anisimova M, Sukno SA, Thon MR. 2014. Natural selection on coding and noncoding DNA sequences is associated with virulence genes in a plant pathogenic fungus. Genome Biol. Evol 6:2368–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Richardson BC. 2008. Epigenetics and autoimmunity. Overview. Autoimmunity 41:243–44 [DOI] [PubMed] [Google Scholar]
- 103.Risques RA, Kennedy SR. 2018. Aging and the rise of somatic cancer-associated mutations in normal tissues. PLOS Genet. 14:e1007108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Roberts RJ, Carneiro MO, Schatz MC. 2013. The advantages of SMRT sequencing. Genome Biol. 14:405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ross MG, Russ C, Costello M, Hollinger A, Lennon NJ, et al. 2013. Characterizing and measuring bias in sequence data. Genome Biol. 14:R51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Roth DB. 2014. V(D)J recombination: mechanism, errors, and fidelity. Microbiol. Spectr 2:MDNA3-0041-2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ryland GL, Doyle MA, Goode D, Boyle SE, Choong DY, et al. 2015. Loss of heterozygosity: What is it good for? BMC Med. Genom 8:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sharp PM, Averof M, Lloyd AT, Matassi G, Peden JF. 1995. DNA sequence evolution: the sounds of silence. Philos. Trans. R. Soc. B 349:241–47 [DOI] [PubMed] [Google Scholar]
- 109.Shirley MD, Tang H, Gallione CJ, Baugher JD, Frelin LP, et al. 2013. Sturge-Weber syndrome and port-wine stains caused by somatic mutation in GNAQ. N. Engl. J. Med 368:1971–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Soellner L, Begemann M, Mackay DJ, Gronskov K, Tumer Z, et al. 2017. Recent advances in imprinting disorders. Clin. Genet 91:3–13 [DOI] [PubMed] [Google Scholar]
- 111.Sohn JI, Nam JW. 2018. The present and future of de novo whole-genome assembly. Brief. Bioinform 19:23–40 [DOI] [PubMed] [Google Scholar]
- 112.Spinner NB, Conlin LK. 2014. Mosaicism and clinical genetics. Am. J. Med. Genet. C 166C:397–405 [DOI] [PubMed] [Google Scholar]
- 113.Stewart C, Kural D, Stromberg MP, Walker JA, Konkel MK, et al. 2011. A comprehensive map of mobile element insertion polymorphisms in humans. PLOS Genet. 7:e1002236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Stewart JB, Chinnery PF. 2015. The dynamics of mitochondrial DNA heteroplasmy: implications for human health and disease. Nat. Rev. Genet 16:530–42 [DOI] [PubMed] [Google Scholar]
- 115.Stock S, Schmitt M, Sellner L. 2019. Optimizing manufacturing protocols of chimeric antigen receptor T cells for improved anticancer immunotherapy. Int. J. Mol. Sci 20:6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sudmant PH, Rausch T, Gardner EJ, Handsaker RE, Abyzov A, et al. 2015. An integrated map of structural variation in 2,504 human genomes. Nature 526:75–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Surani MA. 1998. Imprinting and the initiation of gene silencing in the germ line. Cell 93:309–12 [DOI] [PubMed] [Google Scholar]
- 118.Szulwach KE, Chen P, Wang X, Wang J, Weaver LS, et al. 2015. Single-cell genetic analysis using automated microfluidics to resolve somatic mosaicism. PLOS ONE 10:e0135007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Talens RP, Christensen K, Putter H, Willemsen G, Christiansen L, et al. 2012. Epigenetic variation during the adult lifespan: cross-sectional and longitudinal data on monozygotic twin pairs. Aging Cell 11:694–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tjio JH, Levan A. 1956. The chromosome number of man. Hereditas 42:1–6 [Google Scholar]
- 121.Toutain J, Goutte-Gattat D, Horovitz J, Saura R. 2018. Confined placental mosaicism revisited: impact on pregnancy characteristics and outcome. PLOS ONE 13:e0195905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tubio JM. 2015. Somatic structural variation and cancer. Briefings Fund. Genom 14:339–51 [DOI] [PubMed] [Google Scholar]
- 123.Vorsanova SG, Yurov YB, Iourov IY. 2010. Human interphase chromosomes: a review of available molecular cytogenetic technologies. Mol. Cytogenet 3:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wallace K, Grau MV, Levine AJ, Shen L, Hamdan R, et al. 2010. Association between folate levels and CpG Island hypermethylation in normal colorectal mucosa. Cancer Prev. Res. 3:1552–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wan TS. 2014. Cancer cytogenetics: methodology revisited. Ann. Lab. Med 34:413–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wan TS, Ma ES. 2012. Molecular cytogenetics: an indispensable tool for cancer diagnosis. Chang Gung Med.J 35:96–110 [DOI] [PubMed] [Google Scholar]
- 127.Weischenfeldt J, Symmons O, Spitz F, Korbel JO. 2013. Phenotypic impact of genomic structural variation: insights from and for human disease. Nat. Rev. Genet 14:125–38 [DOI] [PubMed] [Google Scholar]
- 128.Weiss RA. 1984. Human retroviruses in health and disease. Princess Takamatsu Symp. 15:3–12 [PubMed] [Google Scholar]
- 129.Wutz A, Gribnau J. 2007. X inactivation Xplained. Curr. Opin. Genet. Dev 17:387–93 [DOI] [PubMed] [Google Scholar]
- 130.Xu C 2018. A review of somatic single nucleotide variant calling algorithms for next-generation sequencing data. Comput. Struct. Biotechnol. J 16:15–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yokoyama Y, Narahara K, Kamada M, Tsuji K, Seino Y. 1992. Tissue-specific mosaicism for trisomy 21 and congenital heart disease. J. Pediatr 121:80–82 [DOI] [PubMed] [Google Scholar]
- 132.Zhou W, Machiela MJ, Freedman ND, Rothman N, Malats N, et al. 2016. Mosaic loss of chromosome Y is associated with common variation near TCL1A. Nat. Genet 48:563–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zlotogora J. 1998. Germ line mosaicism. Hum. Genet 102:381–86 [DOI] [PubMed] [Google Scholar]