Abstract
SARS-CoV-2 variants raise concern regarding the mortality caused by COVID-19 epidemics. We analyse 88,375 cycle amplification (Ct) values from variant-specific RT-PCR tests performed between January 26 and March 13, 2021. We estimate that on March 12, nearly 85% of the infections were caused by the Alpha variant and that its transmission advantage over wild type strains was between 38 and 44%. We also find that tests positive for Alpha and Beta/Gamma variants exhibit significantly lower cycle threshold (Ct) values.
Context
At least three SARS-CoV-2 variants are currently major sources of concern: Alpha from lineage B.1.1.7 [2], [10], Beta from lineage B.1.351 [9], and Gamma from lineage P.1 [3]. Alpha and Gamma variants have been shown to be more contagious [2], [3], [10], while Beta and Gamma seem to evade immune responses [3], [9]. Although the mechanistic bases are still being investigated, the increased transmissibility could be driven by the N501Y mutation and the 69-70 deletion in the Spike protein [2], where also lies the E484K mutation related to immune escape [3], [9]. Current evidence regarding potential differences in cycle threshold (Ct) values are still unclear [3], [11] as how well they reflect viral loads [7], [8].
In France, using data from 11,916 tests performed on Jan 6 and 7, 2021, a study estimated that 3.3% of the infections were caused by the Alpha variant at that time [4]. Another study used 40,777 tests performed between Jan 26 and Feb 16, 2021, and estimated that on Feb 16, 55% of the infections were caused by Alpha, Beta, or Gamma variants [5].
Here, we estimate the spread of SARS-CoV-2 variants of concern using 123,867 variant-specific RT-PCRs performed in nasopharyngeal (NP) swabs between Jan 26 and Mar 19, 2021 in France and compare Ct values between variants (the dataset and technical specifications are detailed in the Appendix).
Alpha variant is dominant
Using the same methodology described in Haim-Boukobza et al. [5] and in the Appendix, we calculated the transmission advantage of each variant compared to the wild type strain after correction for several biases (region, sampling date, assay, and patient age). This inference was performed for individuals from 5 to 80 years old) and without the data from hospitals (to avoid sampling delay bias). For Alpha, the transmission advantage was 40% (95% confidence interval, CI: [38,42]%). For Beta-Gamma, the estimate was 28% (95%CI: [27,30]%).
We then estimated the proportion of new infections caused by each type of strain on Mar 19, 2021. At a national level, the estimate was 87% for Alpha and 5,4% for Beta/Gamma, but with strong regional heterogeneity (Figure S1).
Variants have lower Ct values
We analysed tests and for which the strain has been determined from 1 to 89 year old patients with Ct values lower than 30, i.e. 76,745 tests (91% of all the tests with Ct values).
We used a multiple linear regression to study variations in Ct values between strains. The covariates were the age, the sampling facility (hospital or city), the sampling date, and the region (including the interaction between the last two). We then performed a F-test of a model including the variant identity as a covariate, in interaction with age, against a model without it. The variant effect was found to be statistically significant while the model explained a small fraction of the Ct variability (adj. R=3.8%), which is consistent with these values being highly variable [1].
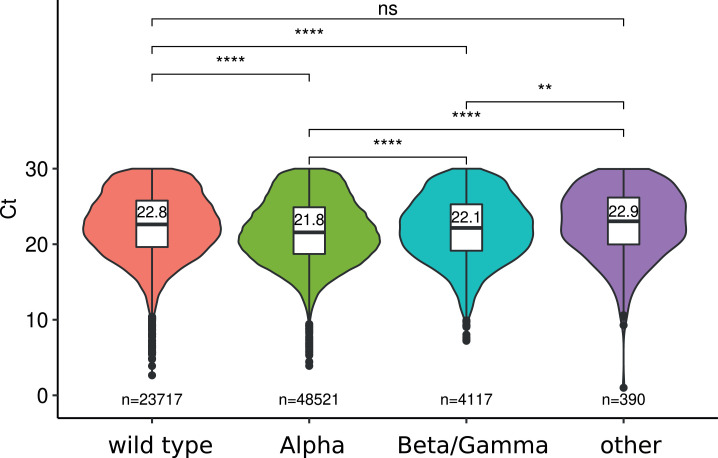
The virus strain effect was highly significant in the ANOVA (Figure 1 ). Samples from Alpha variants had a significantly smaller Ct than that from Beta/Gamma (21.8 vs. 22.1). Both had significantly smaller Ct values than wild type strains (22.8) and other variants.
Fig. 1.
Cycles threshold (Ct) value for SARS-CoV-2 strains. Median estimates based on the linear model are shown in the box plot, and number of tests in each class are shown in the bottom of the graph. ‘other’ indicate tests with the 69-70 deletion and without the N501Y mutation. Start indicate the significance level ( for a p-values strictly lower than and for a p-value of , ns: not significant).
The model also indicated a significant decrease of Ct with age, in line with existing data [1]. Interestingly, this decrease was significantly (more than half) stronger for Alpha, while the inference cannot exclude equivalent slopes for the other variants (note that this trend holds even on the non-Alpha variant subset of Ct values). The sampling date, the sampling region and their interaction were also found to be significant. Samples from hospitals has a slightly higher Ct, likely due to the fact that testing in the general population occurs approximately one week after infection and one week before potenital hospitalization. Therefore, hospitalized patients data is likely to reflect an older state of the epidemic.
Discussion
We show that variant of concern Alpha is now vastly dominant in France compared to wild type strains (87% vs. less than 10%). Beta or Gamma remain limited (approximately 5.4% of the new infections). These results are consistent with earlier reports of a marked transmission-advantage of the Alpha variant [2], [4], [5], [10].
By investigating the RT-PCR Ct values, which can inform us on clinical features of the infection [1], [8], we show that infections caused by variants significantly differ from that caused by wild type strains. That variants are associated with lower Ct values could be an indication of higher viral load, although care must be taken because of the biology of SARS-CoV-2 [7] and of the variability inherent to such values [1].
This result contrasts with earlier findings. One study did not find a significant result when comparing Ct values for tests with or without the S-gene target failure [11]. However, our results are based on a variant-specific PCR. Another study on the Gamma variant [3] did not find a significant difference after accounting for the symptom onset to sampling delay. However, their study was performed on a limited number of samples (). Finally, our estimates, based on a large number of interpretable samples with Ct lower than 30, suggest that differences in within-host replication and transcription between Alpha and non-Alpha variants might be lower than previous quantified [6].
Data availability
The data and scripts used for the analysis will be shared upon publication.
Funding Source
This work received no specific funding.
Ethical Approval
This study was approved by the Internal Review Board of the CHU of Montpellier (ClinicalTrial.gov identier NCT04738331).
Acknowledgements
We thank the ETE modelling team for discussion, as well as the CNRS, the IRD, the ANR, and the Région Occitanie for funding (PHYEPI grant).
Footnotes
Supplementary material associated with this article can be found, in the online version, at 10.1016/j.ijid.2021.09.076
Appendix A. Supplementary materials
Supplementary Raw Research Data. This is open data under the CC BY license http://creativecommons.org/licenses/by/4.0/
References
- 1.Alizon S., Selinger C., Sofonea M.T., Boukobza S., Giannoli J.M., Ninove L., Pillet S., Vincent T., Rougemont A.d., Tumiotto C., Solis M., Stephan R., Bressollette-Bodin C., Salmona M., L’Honneur A.S., Behillil S., Lefeuvre C., Dina J., Hantz S., Hartard C., Veyer D., Delagreverie H.M., Fourati S., Visseaux B., Henquell C., Lina B., Foulougne V., Burrel S. Epidemiological and clinical insights from SARS-CoV-2 RT-PCR cycle amplification values. medRxiv. 2021 doi: 10.1101/2021.03.15.21253653. [DOI] [Google Scholar]; 2021.03.15.21253653 Publisher: Cold Spring Harbor Laboratory Press
- 2.Davies N.G., Abbott S., Barnard R.C., Jarvis C.I., Kucharski A.J., Munday J.D., Pearson C.A.B., Russell T.W., Tully D.C., Washburne A.D., Wenseleers T., Gimma A., Waites W., Wong K.L.M., Zandvoort K.v., Silverman J.D., Group1 C.C.W., C.U. Consortium C.G.U.K., Diaz-Ordaz K., Keogh R., Eggo R.M., Funk S., Jit M., Atkins K.E., Edmunds W.J. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021 doi: 10.1126/science.abg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]; Publisher: American Association for the Advancement of Science Section: Research Article
- 3.Faria N.R., Mellan T.A., Whittaker C., Claro I.M., Candido D.d.S., Mishra S., Crispim M.A.E., Sales F.C.S., Hawryluk I., McCrone J.T., Hulswit R.J.G., Franco L.A.M., Ramundo M.S., Jesus J.G.d., Andrade P.S., Coletti T.M., Ferreira G.M., Silva C.A.M., Manuli E.R., Pereira R.H.M., Peixoto P.S., Kraemer M.U.G., Gaburo N., Camilo C.d.C., Hoeltgebaum H., Souza W.M., Rocha E.C., Souza L.M.d., Pinho M.C.d., Araujo L.J.T., Malta F.S.V., Lima A.B.d., Silva J.d.P., Zauli D.A.G., de F.A.C., Schnekenberg S.R.P., Laydon D.J., Walker P.G.T., Schlüter H.M., Santos A.L.P.d., Vidal M.S., Caro V.S.D., Filho R.M.F., Santos H.M.d., Aguiar R.S., Proença Modena J.L., Nelson B., Hay J.A., Monod M., Miscouridou X., Coupland H., Sonabend R., Vollmer M., Gandy A., Prete C.A., Nascimento V.H., Suchard M.A., Bowden T.A., Pond S.L.K., Wu C.H., Ratmann O., Ferguson N.M., Dye C., Loman N.J., Lemey P., Rambaut A., Fraiji N.A., S. Carvalho M.d.P.S., Pybus O.G., Flaxman S., Bhatt S., Sabino E.C., et al. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science. 2021 doi: 10.1126/science.abh2644. [DOI] [PMC free article] [PubMed] [Google Scholar]; Publisher: American Association for the Advancement of Science Section: Research Article
- 4.Gaymard A., Bosetti P., Feri A., Destras G., Enouf V., Andronico A., Burrel S., Behillil S., Sauvage C., Bal A., Morfin F., Werf S.V.D., Josset L., Covid-19 A.M.A., Group F.v.C., Blanquart F., Coignard B., Cauchemez S., Lina B. Early assessment of diffusion and possible expansion of SARS-CoV-2 Lineage 20I/501Y.V1 (B.1.1.7, variant of concern 202012/01) in France, January to March 2021. Eurosurveillance. 2021;26(9):2100133. doi: 10.2807/1560-7917.ES.2021.26.9.2100133. [DOI] [PMC free article] [PubMed] [Google Scholar]; Publisher: European Centre for Disease Prevention and Control
- 5.Haim-Boukobza S., Roquebert B., Trombert-Paolantoni S., Lecorche E., Verdurme L., Foulongne V., Selinger C., Michalakis Y., Sofonea M.T., Alizon S. Detecting rapid spread of SARS-cov-2 variants, france, january 26–february 16, 2021. Emerging infectious diseases. 2021;27(5):1496. doi: 10.3201/eid2705.210397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones T.C., Biele G., Mühlemann B., Veith T., Schneider J., Beheim-Schwarzbach J., Bleicker T., Tesch J., Schmidt M.L., Sander L.E., et al. Estimating infectiousness throughout SARS-cov-2 infection course. Science. 2021 doi: 10.1126/science.abi5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Michalakis Y., Sofonea M.T., Alizon S., Bravo I.G. Sars-cov-2 viral rna levels are not ǣviral loadǥ. Trends in Microbiology. 2021 doi: 10.1016/j.tim.2021.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Néant N., Lingas G., Hingrat Q.L., Ghosn J., Engelmann I., Lepiller Q., Gaymard A., Ferré V., Hartard C., Plantier J.C., Thibault V., Marlet J., Montes B., Bouiller K., Lescure F.X., Timsit J.F., Faure E., Poissy J., Chidiac C., Raffi F., Kimmoun A., Etienne M., Richard J.C., Tattevin P., Garot D., Moing V.L., Bachelet D., Tardivon C., Duval X., Yazdanpanah Y., Mentré F., Laouénan C., Visseaux B., Guedj J., C.I. Groups f.t.F.C., Study F.C. Modeling SARS-CoV-2 viral kinetics and association with mortality in hospitalized patients from the French COVID cohort. PNAS. 2021;118(8) doi: 10.1073/pnas.2017962118. [DOI] [PMC free article] [PubMed] [Google Scholar]; Publisher: National Academy of Sciences Section: Biological Sciences
- 9.Tegally H., Wilkinson E., Lessells R.J., Giandhari J., Pillay S., Msomi N., Mlisana K., Bhiman J.N., von Gottberg A., Walaza S., et al. Sixteen novel lineages of SARS-cov-2 in south africa. Nature Medicine. 2021;27(3):440–446. doi: 10.1038/s41591-021-01255-3. [DOI] [PubMed] [Google Scholar]
- 10.Volz E., Mishra S., Chand M., Barrett J.C., Johnson R., Geidelberg L., Hinsley W.R., Laydon D.J., Dabrera G., O’Toole A., et al. Assessing transmissibility of SARS-cov-2 lineage b. 1.1. 7 in england. Nature. 2021;593(7858):266–269. doi: 10.1038/s41586-021-03470-x. [DOI] [PubMed] [Google Scholar]
- 11.Walker P.G.T., Whittaker C., Watson O.J., Baguelin M., Winskill P., Hamlet A., Djafaara B.A., Cucunubá Z., Mesa D.O., Green W., Thompson H., Nayagam S., Ainslie K.E.C., Bhatia S., Bhatt S., Boonyasiri A., Boyd O., Brazeau N.F., Cattarino L., Cuomo-Dannenburg G., Dighe A., Donnelly C.A., Dorigatti I., Elsland S.L.v., FitzJohn R., Fu H., Gaythorpe K.A.M., Geidelberg L., Grassly N., Haw D., Hayes S., Hinsley W., Imai N., Jorgensen D., Knock E., Laydon D., Mishra S., Nedjati-Gilani G., Okell L.C., Unwin H.J., Verity R., Vollmer M., Walters C.E., Wang H., Wang Y., Xi X., Lalloo D.G., Ferguson N.M., Ghani A.C., et al. The impact of COVID-19 and strategies for mitigation and suppression in low- and middle-income countries. Science. 2020;369(6502):413–422. doi: 10.1126/science.abc0035. [DOI] [PMC free article] [PubMed] [Google Scholar]; Publisher: American Association for the Advancement of Science Section: Research Article
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Raw Research Data. This is open data under the CC BY license http://creativecommons.org/licenses/by/4.0/
Data Availability Statement
The data and scripts used for the analysis will be shared upon publication.