Abstract
Mammalian genomes have distinct levels of spatial organization and structure that have been hypothesized to play important roles in transcription regulation. Although much has been learned about these architectural features with ensemble techniques, single-cell studies are showing a new universal trend: Genomes are stochastic and dynamic at every level of organization. Stochastic gene expression, on the other hand, has been studied for years. In this review, we probe whether there is a causative link between the two phenomena. We specifically discuss the functionality of chromatin state, topologically associating domains (TADs), and enhancer biology in light of their stochastic nature and their specific roles in stochastic gene expression. We highlight persistent fundamental questions in this area of research.
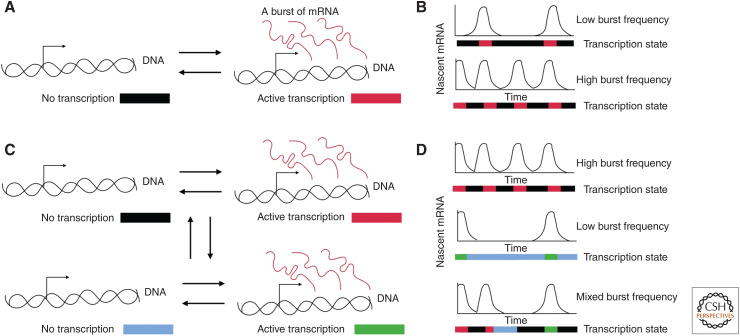
Gene expression varies from cell to cell through programmed and stochastic processes (Elowitz et al. 2002; Swain et al. 2002; Raj and van Oudenaarden 2008). This phenomenon has been shown extensively based on observations of RNA and protein copy number per cell. Live-cell imaging of RNA production from single genes reveals the dynamic nature of this process in human cells (Rodriguez et al. 2019), and single-cell RNA sequencing shows the pervasiveness of copy number variations (Chen et al. 2018b; Larsson et al. 2019). At the molecular level, many genes toggle between active and inactive states, and the resulting “transcriptional bursting” is indeed often described by a two-state model, although more complicated models are sometimes needed to quantitatively describe gene expression at the single-cell level (Fig. 1; Shahrezaei and Swain 2008; Tantale et al. 2016; Rodriguez and Larson 2020; Tunnacliffe and Chubb 2020). Heterogeneous gene expression has been linked to changes in cell phenotype, cancer, and aging (Raj and van Oudenaarden 2008). Additionally, transcription variation can serve as a readout of the underlying biochemistry: The characterization of specific perturbations on transcription with mathematical modeling can lead to a deeper understanding of the mechanism. Therefore, understanding the mechanisms that control the properties of a transcriptional burst is vital. For example, laboratories have used the bursting paradigm to dissect the role of trans-acting factors, noncoding RNA (ncRNA), and cis-acting motifs in modulating transcription (Suter et al. 2011; Hornung et al. 2012; Donovan et al. 2019; Stavreva et al. 2019). Recent studies have also begun to probe the connection between enhancers and transcriptional bursting in cell lines and even whole organisms (Fukaya et al. 2016; Chen et al. 2018a; Lim et al. 2018; Alexander et al. 2019). More broadly, a study in single cells observed a distinct multimodal distribution of burst frequencies that was directly correlated with chromatin “state” (heterochromatin or euchromatin) (Su et al. 2020). Thus, an emerging theme in the study of stochastic gene regulation is the potential role of nuclear architecture for modulating transcription dynamics. This link between genomic structure and transcriptional regulation as revealed through single-cell studies is the subject of this review.
Figure 1.
Transcription varies in time. (A) The two-state model for transcription in which a gene can switch between an inactive state and active state leading to bursts of RNA. (B) Illustration of a gene with a low burst frequency and a high burst frequency. The colors underneath each time trace indicate the underlying state of the gene through time. (C) A multistate model of transcription in which a gene can switch between states with different burst frequencies. The gene can occupy one of four different states illustrated with the different colors. (D) The diverse time traces produced by a multistate model of transcription dynamics. The colors underneath the time traces show the actual state of the gene.
Although much has been learned about genomic organization and functionality at the ensemble level using methods such as Hi-C, ChIA-PET, ATAC-seq, etc., single-cell Hi-C and imaging studies are revolutionizing our understanding of these structures and how they vary between cells. Specific properties of the genome have been shown to play a pivotal role in directing transcription. From 10 kb bacterial DNA loops causing the temporal buildup and release of supercoiling leading to transcriptional bursts (Chong et al. 2014) to the various ways chromatin is able to regulate the binding of transcription factors (TFs) to direct transcription (Bulger and Groudine 2010)—the importance of understanding chromatin structure and its regulatory mechanisms has been clearly shown with our enhanced comprehension of disease (Krijger and De Laat 2016). A new trend is that genomic structures are probabilistic at almost every level of organization, and this stochasticity is suggestively linked to gene expression (Finn and Misteli 2019). Here we discuss some of the methodologies for deciphering single-cell genomic structure while highlighting possible links to stochastic gene expression. Specifically, we review both the evidence for genomic structure in single cells at the level of A/B “compartments” and “topologically associating domains (TADs)” and also the potential for such structures to regulate transcription. Similarly, we highlight what our knowledge of stochastic gene expression might reveal about the functionality of genomic structures and highlight critical questions for the field.
ENSEMBLE STUDIES OF GENOME STRUCTURE
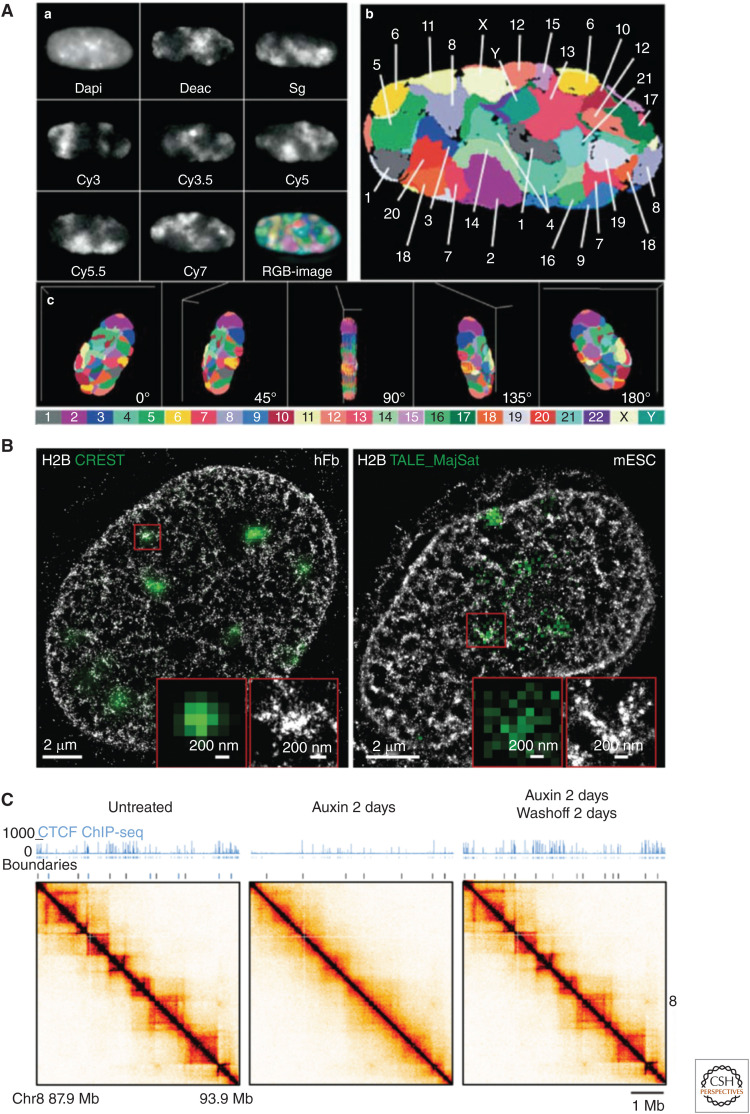
Nonrandom nuclear organization is visible at multiple length scales. At the highest level of organization, individual chromosomes occupy chromosomal territories within the nucleus (Fig. 2A; Bolzer et al. 2005; Stevens et al. 2017; Tan et al. 2018), and chromatin is separated into two major conformations or states: euchromatin and heterochromatin. Euchromatin is predominantly made up of “open” gene rich fibers (Stevens et al. 2017) and is often enriched in the center of the nucleus (Gilbert et al. 2004). Heterochromatin fibers have a condensed structure (Ricci et al. 2015), a low density of genes with little expression, and often make direct contact with the nuclear lamina (Guelen et al. 2008; Kind and van Steensel 2010). These chromatin states are enriched in particular histone modifications, DNA modifications, and proteins (Jenuwein and Allis 2001; Janssen et al. 2018), all of which play a role in the formation and propagation of the compartments; the driving forces behind these compartments are still an active area of investigation (Ganai et al. 2014; Strom et al. 2017; van Steensel and Belmont 2017; Abramo et al. 2019; Falk et al. 2019; Wang et al. 2019).
Figure 2.
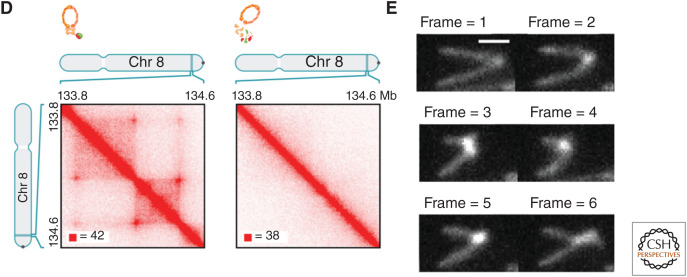
Hierarchical nuclear organization. (A) Individual chromosomes occupy distinct territories. (Panel A reprinted from Bolzer et al. 2005 under the terms of a Creative Commons Attribution License.) (B) Superresolution imaging of chromatin (labeled H2B) showing a wide distribution of densities, with Crest and TALE_ MajSat (heterochromatin markers), showing that dense clusters of chromatin correspond to heterochromatin. (Panel B from Ricci et al. 2015; reprinted, with permission, from Elsevier © 2015.) (C) CTCF is needed for ensemble topologically associated domain (eTAD) formation in ensemble Hi-C maps. Here, CTCF is degraded in the presence of auxin illustrated with the ChIP-seq data. (Panel C from Nora et al. 2017; reprinted, with permission, from Elsevier © 2017.) (D) Cohesin is needed for eTAD formation in ensemble Hi-C maps. Here, auxin eliminates functional cohesion. (Panel D from Rao et al. 2017; reprinted, with permission, from Elsevier © 2017.) (E) Direct visualization of loop extrusion with cohesion. (Panel E from Davidson et al. 2019; reprinted, with permission, from the American Association for the Advancement of Science © 2019.)
With ensemble Hi-C contact maps, chromosomal regions can also be classified into two compartments (A and B), defined by their propensity to have higher contact frequencies between DNA segments within the same compartment than with loci in the other compartment (Lieberman-Aiden et al. 2009). These A and B compartments were shown to directly correspond to the euchromatin (A) and heterochromatin (B) states (Bickmore and Van Steensel 2013), allowing one to correlate a particular ensemble chromatin state to segments of DNA. However, as we discuss below, these compartments have a different interpretation in single cells.
On a smaller scale, another prominent feature of ensemble Hi-C contact maps are ∼1 Mb regions that show an enrichment in pairwise contacts termed topologically associating domains (TADs) (Fig. 2C,D). For clarity, here we refer to these structures as eTADs (ensemble TADs) when derived from ensemble techniques. eTADs have an average size of 1 Mb with a range of 100 kb to 5 Mb (Dixon et al. 2012; Nora et al. 2012; Sexton et al. 2012; Rao et al. 2014), with a twofold enrichment in pairwise contacts within an eTAD compared to interactions with regions outside of eTADs (Dixon et al. 2012; Hou et al. 2012; Nora et al. 2012; Sexton et al. 2012; Nagano et al. 2013). The difference in contact frequency is also reflected in the distances between loci, with smaller distances between loci that share an ensemble domain (Bintu et al. 2018; Szabo et al. 2018; Finn et al. 2019; Mateo et al. 2019). Whereas some eTADs are invariant throughout various cells and tissues, others have been shown to form in a tissue-specific manner (Rao et al. 2014; Bintu et al. 2018; Mateo et al. 2019), and can also be correlated with cell fate (Bonev et al. 2017).
The central mechanism behind the formation of eTADs is “loop extrusion,” an active process in the cell that occurs through the interplay between the architecture proteins CTCF and cohesin (Mizuguchi et al. 2014; Nora et al. 2017; Rao et al. 2017). Originally believed to be specific to vertebrates, this mechanism has now been shown to also be at work in Drosophila (Mateo et al. 2019). Note fly embryos lacking CTCF are still able to develop (Gambetta and Furlong 2018), suggesting that CTCF-mediated eTADs may not be necessary for function, at least in these organisms (Rowley et al. 2017). The simplest form of the loop extrusion model posits the following: The Nipbl-Mau4 complex loads cohesin rings onto specific Nipbl sites and extrudes the chromatin (using energy from ATP) until coming into contact with a pair of CTCF-binding sites in a convergent orientation, forming a “stable” complex (Rao et al. 2014; Sanborn et al. 2015; Fudenberg et al. 2016; Gassler et al. 2017; Vian et al. 2018). The cohesin release factor, WAPL, can aid in the dissociation of cohesin at all stages (Haarhuis et al. 2017), and certain TFs likely influence this process, especially for cell-type-specific eTADs (Phanstiel et al. 2017). Support for this model comes from many different studies, which describe the fusion of neighboring eTADs with the deletion of a CTCF-binding site (Lupiáñez et al. 2015; Sanborn et al. 2015; Mateo et al. 2019) and the elimination of eTADs upon the depletion of cohesin or CTCF (Fig. 2C,D; Nora et al. 2017; Rao et al. 2017; Bintu et al. 2018). Inversions of CTCF sites likewise “flip” the topological structure in regard to the favorable convergent CTCF orientation (Guo et al. 2015). There are also interdependencies between CTCF sites, evidenced by the observation that deletion of one site will influence the binding occupancy of neighboring CTCF sites (Narendra et al. 2015). Last, the extrusion process has now been directly visualized in vitro (Fig. 2E; Davidson et al. 2019; Kim et al. 2019), solidifying a central concept of the model.
Variations on these general architectural motifs have also been reported. Most studies have focused on pairwise interactions between segments of DNA, but even at the ensemble level a more complicated picture is emerging. Cooperative three-way interactions between three CTCF-binding sites have been shown to take place, leading to the formation of complex topological structures (Narendra et al. 2015). At an even finer scale (as small as that of a single locus), small compartments (sub-eTADs) have been shown to form within the eTADs (Rowley et al. 2017; Hsieh et al. 2020).
Taken together, nuclear organization is evident at almost any length scale that can be experimentally interrogated, but the function of these architectures remains an enduring question in cell biology.
SINGLE-CELL STUDIES OF GENOME STRUCTURE
Possible clues to the function of nuclear architecture might come from observing these same structures in single cells. Two main methodologies allow one to quantify heterogeneous chromosomal structure in individual cells: single-cell Hi-C and DNA fluorescence-based in situ hybridization (DNA-FISH). The number of labeled DNA segments for traditional DNA-FISH is limited by the number of colors one can image, greatly limiting the technique. This limitation can be partially overcome with multiple rounds of hybridization, leading to chromatin-tracing techniques that rely on determining the positions of many different chromosomal positions inside each cell after each iterative hybridization. Additionally, DNA-FISH-based methodologies can be augmented with RNA-FISH, allowing one to simultaneously quantify how transcriptional state relates to chromosomal structure.
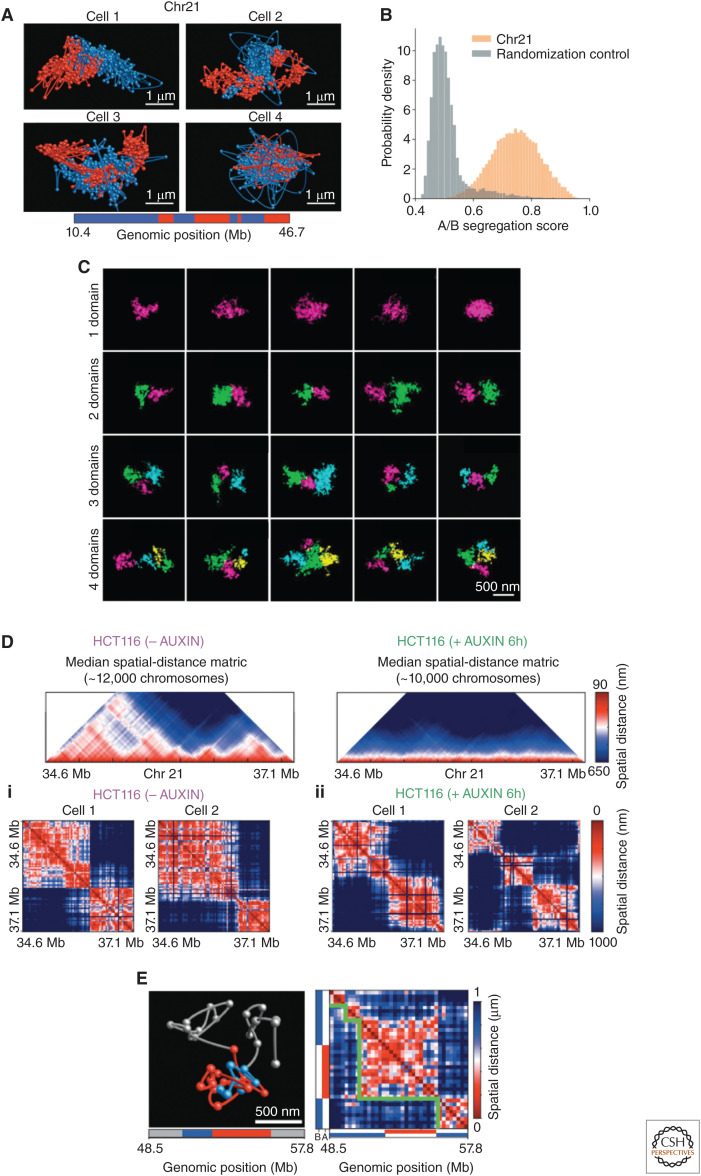
Both single-cell Hi-C and DNA-FISH have revealed that the higher-order organization of the chromatin varies from cell to cell in the overall folding of the chromosome and the specific interfaces between chromosomes (Nagano et al. 2013; Stevens et al. 2017; Tan et al. 2018). A seminal study in this area came from Boettiger et al., which was the first to show that compartment-folding patterns are fundamentally different from each other and that the volumes of specific chromosomal regions can vary among cells (Boettiger et al. 2016). More recently, Su et al. showed that global chromosomal structure takes on a variety of different conformations (Fig. 3A; Su et al. 2020). Moreover, they extended the technique by looking at nascent RNA in parallel, imaging more than 1000 different genomic loci and more than 1000 genes in individual cells (Su et al. 2020). Similar approaches (but on a smaller scale) have also been performed in Drosophila (Cardozo Gizzi et al. 2019; Larson 2019; Mateo et al. 2019). In general, sequencing-based approaches and microscopy-based approaches are in rough agreement. However, it is now clear that the static picture of chromosomal structures as distinct, well-separated, stable features is an oversimplification. In reality, these structures are dynamic and these structures relate to transcriptional activity (Cardozo Gizzi et al. 2019).
Figure 3.
Chromosome organization varies in single cells. (A) Diverse structures of whole individual chromosomes with the loci that occupy the ensemble-A and -B compartments shown in red and blue. (Panel A from Su et al. 2020; reprinted, with permission, from Elsevier © 2020.) (B) The degree of separation of chromatin for individual chromosomes in the two ensemble-A and -B states compared with a random control. (Panel B from Su et al. 2020; reprinted, with permission, from Elsevier © 2020.) (C) Direct visualization of chromatin with electron microscopy showing the large amount of stochasticity of individual topologically associating domains (TADs)—each assigned domain is color-coded. (Panel C is reprinted from Trzaskoma et al. 2020 under a Creative Commons Attribution 4.0 International License.) (D) Individual TADs still form in the absence of cohesin. The first row shows the ensemble median distances with (no auxin) and without cohesin (auxin). (Panel D from Bintu et al. 2018; reprinted, with permission, from the American Association for the Advancement of Science © 2018.) (i) TADs in individual cells with cohesion, and (ii) without cohesin. (E) TADs in individual cells can contain loci assigned to the ensemble-A and -B compartments. (Panel E from Su et al. 2020; reprinted, with permission, from Elsevier © 2020.)
First, truly quantifying whether a locus is in a heterochromatin or euchromatin state in individual cells is still a central goal of the field. Specifically, in terms of chromatin state, assigning a locus to an A or B compartment in single cells can only be performed with additional assumptions. For example, the DNA sequences of euchromatin are CpG-rich (Xie et al. 2017), and, consequently, the local density of CpG has been used as a proxy of chromatin state in single-cell Hi-C studies (Tan et al. 2018). Nonetheless, in individual cells, Wang et al. (2016) showed that the loci assigned to the ensemble-A compartments and ensemble-B compartments, on average per cell, do generally separate from each other. This separation was also apparent in Su et al. where they were able to trace an entire chromosome. However, large variations were observed in the arrangement of ensemble-A and -B loci in individual cells (Fig. 3A; Su et al. 2020). Furthermore, even when averaged over entire chromosomes, the separation between the two types of loci (assigned at the ensemble level) showed overlap with the randomized control, suggesting that the level of intermixing at the single locus level is substantial (Fig. 3B). With additional assumptions, the single-cell Hi-C work of Tan et al. was able to show that chromosomal regions could occupy either A and B compartments (Tan et al. 2018). This result is also supported with the superresolution work of Szabo et al. (2018) where heterogeneity was seen in the higher-order structure: “ranging from a compact conformation to rarer unfolded chromosomes.” Interestingly, autosomal alleles varied in their compartments to the same extent as that between cells, suggesting the noise is intrinsic, that is, not attributable to the varying amounts of specific proteins between cells (Tan et al. 2018).
Second, a major question that the first single-cell Hi-C experiments sought to investigate was whether TADs in individual cells were consistently formed in the locations of the eTADs (Nagano et al. 2013). Specifically, the enrichment of contacts at the eTAD locations in individual cells was quantified and compared with that of the ensemble. Interestingly, they found no difference, suggesting that the TADs that form eTADs were consistently formed in individual cells. Here we should note that the limited number of contacts per cell in single-cell Hi-C data often lead to the use of additional assumptions to reach conclusions. The logic of this particular analysis was dependent on the formation (or absence) of complete TADs (at the locations of the eTADS) in individual cells. Interestingly, with further single-cell Hi-C experiments (Flyamer et al. 2017; Stevens et al. 2017; Tan et al. 2018), various superresolution DNA-FISH methodologies (Bintu et al. 2018; Szabo et al. 2018; Cardozo Gizzi et al. 2019; Finn et al. 2019; Mateo et al. 2019; Su et al. 2020), and now with high-resolution electron microscopy technologies (Peddie and Collinson 2014; Trzaskoma et al. 2020), the formation of TADs and their barriers were found to be extremely stochastic in individual cells. For example, a 1.7 Mb genomic region observed at the single-cell level clearly showed one domain in 25% of cells, two domains in 39% of cells, three domains in 31% of cells, and four or five domains in 5% of cells. Furthermore, even for the individual chromosome regions that had the same number of domains there were clearly different folding patterns (Fig. 3C; Trzaskoma et al. 2020), suggesting again a very dynamic and stochastic structure. Additionally, TAD formation at different alleles in individual cells was shown to be independent (Finn et al. 2019), indicating that the noise in TAD formation is also intrinsic.
These probabilistic structures do still have structural properties that emerge with different analyses. Of particular note, the eTADs were found to result from cohesin introducing biases in which boundaries could form (Bintu et al. 2018). Surprisingly, even in the absence of cohesin, TADs were still formed, but with no bias in boundary formation (Fig. 3D). This result could be interpreted as suggesting that the forces directing the chromatin to A and B compartments do not play a significant role in the formation of TADs. Furthermore, TADs can have both ensemble-A loci and ensemble-B loci, again suggesting the ensemble compartmentalization may not play a dominating role in TAD formation (Fig. 3E; Su et al. 2020). Individual TADs appear to show globular structures (Bintu et al. 2018; Szabo et al. 2018) and Drosophila Polycomb TADs have been shown to be extremely compact and organized like that of a random coil (Mateo et al. 2019). Notably, Polycomb proteins are important for propagating and maintaining chromatin modifications, and these modifications were shown to play a role in “strong long-range” chromosomal contacts (Bonev et al. 2017) and form “loops” with “patches” of the Polycomb chromatin forming the contact (Hsieh et al. 2020; see Aranda et al. 2015 for information on Polycomb proteins and their influence on TAD formation). Last, the fact that TADs are probabilistic structures causes their DNA to frequently contact their neighboring eTADs; stochastic boundary formation can cause DNA segments within neighboring eTADs to be located within the same TAD (Bintu et al. 2018; Finn et al. 2019).
In summary, it appears that there is a great deal of heterogeneity in the overall conformation of chromosomes, chromatin state, and TAD structure. Understanding how this heterogeneous structural genomic landscape influences transcription and cellular decisions thus becomes a central question of future studies (Misteli 2020).
TRANSCRIPTION IN THE LIGHT OF STOCHASTIC GENOMIC STRUCTURES
Does the stochastic nature of the genome have any influence on transcription or vice versa? This is still a very much open question, and the answer ultimately lies in the functionality of genomic structure. Here we focus primarily on compartment- and TAD-level organization and what new methodologies reveal about this question.
The role of the local surroundings of a gene on transcriptional regulation has a long history, starting first with the suggestion that location of a gene in a physically compact region of chromatin might be indicative of low activity (Schultz 1947; Raser and O'Shea 2004; Raj et al. 2006). Studies hoping to investigate how the local chromatin state influences transcription sought to investigate perturbations to nuclear position. Multiple experiments have redirected/tethered specific chromosomal regions to the different regions of the nucleus and have quantified its impact on transcription. Interestingly, whether or not transcription was influenced was context specific; repositioning an interior gene to the periphery of the nucleus did not always lead to its inactivation (Williams et al. 2006; Finlan et al. 2008; Kumaran and Spector 2008; Reddy et al. 2008; Therizols et al. 2014; Wijchers et al. 2016). In terms of chromosomal territories, regions of active transcription are enriched in trans-chromosomal contacts (Lieberman-Aiden et al. 2009; Yaffe and Tanay 2011), owing to active transcription at the interfaces of chromosomes (Nagano et al. 2013; Stevens et al. 2017). To date, to the best of our knowledge, it is unclear whether general rules can be inferred about the role of chromosome interfaces or the nuclear periphery in influencing transcriptional state or bursting behavior.
Moreover, even within the context dependence and nondeterministic role of nuclear position on transcription, the higher-order structure could have a long timescale impact on the variation of transcription owing to the constraints on chromatin mobility. The diffusion of DNA-bound fluorescently tagged TFs (Robinett et al. 1996) has shown that the majority of chromatin exhibit confined diffusion (Chubb et al. 2002). Memory of past positions and specific dynamics are evident well beyond 20 min with the dynamic behavior specific to individual cells (Alexander et al. 2019), suggesting it would take a long time for specifically bound TFs to reach a new location through chromatin diffusion. Here we should also note that with transcriptional activation, chromatin was able to move directionally from the periphery to the center of the nucleus (Chuang et al. 2006). Last, the repositioning of chromatin to different nuclear positions was shown to largely depend on mitosis (Kumaran and Spector 2008). It is therefore tempting to state that if changing nuclear position does change the chromatin state, the characteristic timescales of switching between different transcriptional states would be on the order of the cell cycle.
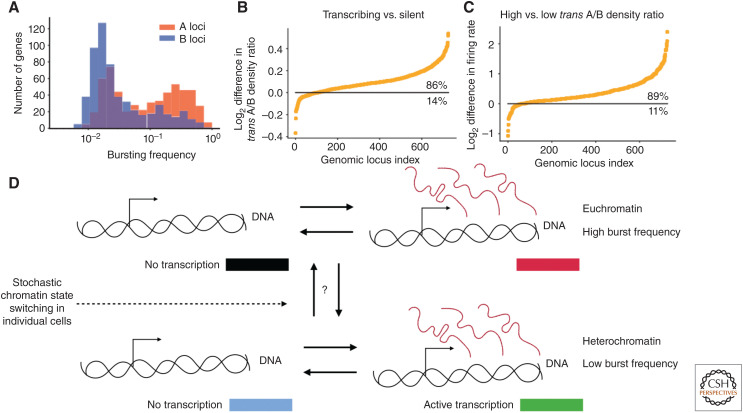
At the compartment/euchromatin/heterochromatin level, global methodologies found that the influence of ensemble heterochromatin on bursting was noisy expression resulting from a low burst frequency (Dey et al. 2015). This result was reemphasized by the finding that genes that occupied the two ensemble chromatin states had a bimodal distribution of burst frequencies (Fig. 4A). The power of this recent work is the ability to directly quantify how the stochastic nature of the chromatin state at the single-cell level influences transcription. Specifically, instead of assigning loci to A and B compartments in individual cells, they quantified the density of trans-ensemble-A loci relative to trans-ensemble-B loci for each gene, excluding the ensemble chromatin state of the gene in question, and investigated whether there was a link to its transcriptional state. This “trans A/B density” can be thought of as an approximation for whether or not a gene is in an A or B state in individual cells. Importantly, there was a strong correlation between the trans A/B density and transcription, where 86% of genes showed an enrichment in trans A/B density when they were transcribing and 89% showed a higher burst frequency with a higher trans A/B ratio (Fig. 4B,C; Su et al. 2020).
Figure 4.
Relationship between transcriptional bursting and compartmentalization. (A) The bimodal bursting frequency distribution of genes naturally found in the ensemble-A and -B chromatin states. (B) The difference in the trans A/B ratio (the local density of trans-ensemble-A loci to trans-ensemble-B loci in individual cells around each gene) when a gene was transcribing versus when it was not, showing there is a strong correlation between chromatin state and transcription state in individual cells. (C) Similar to B but with the difference in bursting rate on the y-axis between high and low trans A/B density, showing a strong correlation of chromatin state in individual cells and burst frequency. (Figures and data in A–C from Su et al. 2020; reprinted, with permission, from Elsevier © 2020.) (D) Hypothetical model of transcription for a gene with two different burst frequencies owing to variations in chromatin state within individual cells (the same as Fig. 1A).
These results suggest the modulation of burst frequency through chromatin state could be the mechanism leading to correlations in the steady-state RNA levels between genomically co-located genes (Raj et al. 2006; Sun and Zhang 2019; Ibragimov et al. 2020), and should be further quantified in terms of specific mechanism. Nonetheless, these results suggest a transcriptional model with multiple burst frequencies for genes that stochastically switch between chromatin states (Fig. 4D). A potential mechanism where the stochasticity of the chromatin state could be propagated through to transcription could be enhancer activation and deactivation (see below), possibly through specific histone modifications/chromatin conformations that are concomitant with activation of enhancers (Krijger and De Laat 2016). Last, if the chromatin state is what drives burst frequency, could one use transcriptional bursts as a monitor of single-cell chromatin state?
FUNCTIONALITY OF TOPOLOGICALLY ASSOCIATING DOMAINS (TADs) AND ENSEMBLE TADs (eTADs)
Topological domains arise through the interplay between cohesin and cis- and trans-acting boundary elements. CTCF is perhaps the best characterized of these trans-acting factors (Ghirlando and Felsenfeld 2016), consistent with its role as an insulator (Bell and Felsenfeld 2000). Thus, in relation to the previously discussed higher-order structure of the genome, the transcriptional states of individual eTADs are often believed to be partitioned into either heterochromatin or euchromatin (Le Dily et al. 2014; Wang et al. 2016; Wijchers et al. 2016) and correlated with chromatin state (Sexton et al. 2012; Rao et al. 2014; Ulianov et al. 2016). The functionality of eTADs is believed to be in (1) promoting specific intracontacts such as specific enhancer–promoter interactions, and (2) limiting the contacts between adjacent eTADs, avoiding enhancer cross talk. Indeed, the expression of tissue-specific expression is correlated with eTAD structure (Symmons et al. 2014) and perturbations of individual eTADs lead to the dysregulation of expression (Northcott et al. 2014; Lupiáñez et al. 2015; Flavahan et al. 2016; Franke et al. 2016; Hnisz et al. 2016; Krijger and De Laat 2016; Weischenfeldt et al. 2017; Mateo et al. 2019) providing strong support for this view.
Yet, experiments that have globally eliminated eTAD structure through cohesin (Rao et al. 2017; Vian et al. 2018) or CTCF (Nora et al. 2017) depletion have shown minimal ensemble transcription changes. Similarly, although eTADs might be expected to constrain the spread of chromatin state, the compartmentalization of chromatin into the ensemble-A and -B states was maintained and even strengthened with the depletion of eTADs (Nora et al. 2017; Rao et al. 2017; Vian et al. 2018). Also, the concept of eTADs directing specific DNA contacts and preventing others is a difficult model to grasp, considering TADs have been shown to be stochastic structures with ill-defined boundaries in individual cells. Furthermore, as pointed out in the work of Ghavi-Helm et al., most studies that found a strong correlation of perturbations to eTAD structure and transcription generally started with a phenotype (like cancer) and “worked backward to explain the misexpression” (Northcott et al. 2014; Lupiáñez et al. 2015; Flavahan et al. 2016; Franke et al. 2016; Hnisz et al. 2016; Weischenfeldt et al. 2017). Therefore, the general influence of eTAD structure on transcription appears minimal. Indeed, in their work, they were able to quantify the effects of a large number of unbiased chromosomal perturbations using balancer chromosomes in Drosophila and found that only a small fraction of genes was sensitive to disruptions in chromosome topology (Ghavi-Helm et al. 2019).
Indeed, recent data suggest a more nuanced view. Within TADs, one observes smaller “sub-TADs.” The boundaries of these sub-eTADs are enriched in TFs, coactivators, and RNA polymerase II (RNAP II), and the boundary strengths of sub-TADs are directly proportional to transcription activity (Hsieh et al. 2020). Importantly, RNAP II inhibition (elongation and initiation) did not perturb these structures, suggesting it is the other factors involved in transcription directing the formation of these sub-eTAD structures (Hsieh et al. 2020). Here, we should note that there is a similar relationship between eTADs and active transcription (Du et al. 2017; Hug et al. 2017; Ke et al. 2017; Hsieh et al. 2020) and some TFs have been shown to form eTAD boundaries (Hug et al. 2017; Weintraub et al. 2017). Additionally, recent investigations showed that active transcription within an eTAD seemed to unfold the region (evident at the ensemble level) (Cardozo Gizzi et al. 2019), directly linking eTAD structure to transcription, likely attributable to the unfolding of chromosomal domains with supercoiling (Naughton et al. 2013).
Distinguishing the role of compartments versus TADs as the primary determinant of transcriptional regulation is emerging as a central concept in the field. The interesting recent work of Luppino et al. (2020) was able to show that cohesin is responsible for the intermixing of neighboring eTADs and through this intermixing, cohesin modulates the burst frequency of genes near the borders of eTADs, suggesting an interesting direction of research. Also, the strengthening of A and B compartmentalization seen with cohesin depletion (Rao et al. 2017; Vian et al. 2018) could indicate that eTADs play a role in the stochasticity of A and B compartmentalization and the higher organization of the genome. However, modeling studies of Nuebler et al. (2018) suggest that it is the activity of loop extrusion that leads to the active mixing of the chromatin and the weakening of A and B compartmentalization, potentially leading to this stochasticity and bringing into question the specific role of eTADs. Still, this stochasticity in A and B compartmentalization would not necessarily lead to changes in mean expression levels but could in turn influence the noise and (potentially) the correlations between certain genes, but this remains to be seen. Overall, much needs to be probed in terms of TAD and eTAD structure as a general process controlling gene expression and whether their stochastic nature influences enhancer biology.
ENHANCER-DEPENDENT TRANSCRIPTION ACTIVATION
An appealing connection between genomic structure and transcription centers on the concept of the enhancer. Enhancers were initially discovered in 1981 on plasmids with the observation that DNA elements “far” from the promoter greatly influenced transcription (Banerji et al. 1981; Benoist and Chambon 1981; Schaffner 2015). Enhancers were then shown in metazoans (mouse) in 1983 (Banerji et al. 1983) and are now believed to be a central mechanism of transcription regulation. Mammalian genomes are predicted to have hundreds of thousands of enhancers (The 1000 Genomes Project Consortium et al. 2012; Shen et al. 2012), and the majority of genes have more than one enhancer (Fishilevich et al. 2017) presenting an enormous challenge for dissecting their mechanism of action. Enhancers can act synergistically, thus raising expression to a higher extent than their sum, repress the action of neighboring enhancers, and can show a degree of “hierarchical logic” (Long et al. 2016); for example, the activation of one enhancer is necessary for the activation of others. Additionally, some enhancers seem to act on any gene, while some show specificity (Furlong and Levine 2018). Still, given their dominant role in regulation, the variations within the steps of enhancer-mediated transcriptional activation are a clear knob for the cell to control the timescales of transcriptional bursting and the extent of noise within expression (Larsson et al. 2019).
At the molecular level, the “parts list” of enhancers has been well characterized. DNA sequences of enhancers are enriched in accessible DNA (Buenrostro et al. 2013) and the H3K4me1 and H3K27ac histone modifications (Rada-Iglesias et al. 2011). A majority of enhancers are cell/tissue specific, in which the expression of specific TFs and chromatin modifications are believed to lead to the specific activation of the enhancer. Coactivators (Mediator, BRG1, and p300), TFs (Spitz and Furlong 2012), enhancer RNA, and RNAP II binding are all characteristics of active enhancers (Long et al. 2016). The confluence of these factors has engendered a view that for proper enhancer activation, multiple different TFs bind cooperatively and displace the nucleosomes. An alternative first step is that pioneer factors can bind and modify specific chromatin sites paving the way for the future factors to bind and work (Zaret and Carroll 2011), although it is unclear whether pioneering activity is a unique activity reserved for certain proteins (Voss et al. 2011). Nevertheless, the majority of experiments have indeed shown that coactivators trigger transcription through guided recruitment (Hilton et al. 2015; Stampfel et al. 2015).
ENHANCER–PROMOTER PROXIMITY: THE QUESTION OF “RANGE OF ACTION”
If there is a general mechanism of enhancer transcription activation, it has yet to be clearly shown. A variety of different models have been proposed, but given a lack of direct experimental evidence, we do not exhaustively discuss them here (Bulger and Groudine 2010; Furlong and Levine 2018). However, enhancer-activated transcription does seem to be dependent on the physical proximity of a promoter and its enhancer; whether or not direct contact between a promoter and an enhancer is needed for transcription activation is still a matter of debate. Proximity is believed to lead to an increase in coactivators and TFs around the promoter, increasing the likelihood of a transcriptional burst (Rodriguez and Larson 2020). Indeed, the dominating mechanistic role of enhancers in stochastic gene expression has been shown in living cells, where the major effect of enhancer function is modulating burst frequency (Bartman et al. 2016; Fukaya et al. 2016; Larsson et al. 2019; Rodriguez et al. 2019). Different mechanisms have different degrees of proximity needed for activation; regardless of the exact mechanism, the degree of proximity (or “range of action” for the enhancer) needed for transcription must be relatively small (hundreds of nm) to ensure precision as argued in Furlong and Levine (2018).
This simple model has been quite useful, as enhancer–promoter proximity has been extremely important for our understanding of oncogene dysregulation. Specific examples include, but are not limited to (Fang et al. 2020) (1) A single chromosomal rearrangement that moves the GATA2 enhancer proximal to EVI1 (a stem-cell regulator) directly results in leukemia (Gröschel et al. 2014); (2) The up-regulation of MYC in some forms of Burkitt lymphoma is attributable to the repositioning of immunoglobin heavy chain enhancer (Dalla-Favera et al. 1982); (3) Mutations within the superenhancer of the LMO1 oncogene was shown to modulate its TF-binding sites leading to neuroblastoma (Oldridge et al. 2015); and (4) Copy number variations of superenhancers around various oncogenes (KLF5, USP12, PARD6B, and MYC) lead to their dysregulation in 12 different tumor types, providing strong evidence that perturbations to enhancer biology is a common driving force behind cancer.
The genomic distances between enhancers and their target genes can be very large, such as the Shh limb enhancer (ZRS), which is >1 Mb away from the target gene (Lettice et al. 2003). Therefore, understanding how chromosomal rearrangements direct/limit enhancers to a particular promoter and which conformations actually trigger transcription have become central questions. eTADs are believed to play a role in directing enhancers as perturbations in individual eTAD structures can lead to large changes in gene expression and disease (Northcott et al. 2014; Lupiáñez et al. 2015; Flavahan et al. 2016; Franke et al. 2016; Hnisz et al. 2016; Krijger and De Laat 2016; Weischenfeldt et al. 2017; Mateo et al. 2019). Additional support for the previous statement is clearly shown in the pivotal work of Symmons et al. where they monitored the expression level of a minimal promoter with the lacZ gene randomly inserted into hundreds of different positions within the mouse genome (Symmons et al. 2014; developed in Ruf et al. 2011). Notably, a large majority of these insertions showed tissue-specific expression similar to that of the neighboring genes’ tissue-specific expression. Further investigation found the regions over which the reporter gene showed similar expression was directly correlated with the eTADs, suggesting that the “region of action” for the enhancers directing the tissue-specific expression was confined to eTADs (Symmons et al. 2014).
Directed looping is an attractive model for modulating proximity through chromosomal conformation. For example, in the work of Guo et al. (2015), ∼50% of their identified enhancers had a CTCF-binding site nearby, suggesting that upon TAD formation, the CTCF-adjacent enhancer could be repositioned for transcription activation. Looping between a promoter and its enhancer has been shown to directly increase mean expression levels (Deng et al. 2012, 2014; Williamson et al. 2016; Morgan et al. 2017), attributable to an increase in burst frequency (Bartman et al. 2016). Also, the oscillatory expression of circadian genes was shown to correlate with promoter–enhancer contacts (Mermet et al. 2018). Similar to a looping mechanism, cohesin could promote a direct enhancer–promoter contact through loop extrusion, which has been proposed as a mechanism to constrain enhancer–promoter interactions to the same eTAD (Dixon et al. 2012). Last, in ensemble Hi-C experiments, enriched contacts of enhancer–promoters have been observed, suggesting looping could promote direct contact (Rao et al. 2014). However, the majority of active enhancer–promoters did not show an enrichment in contact frequency (Rao et al. 2014; Long et al. 2016), suggesting that for the most general form of enhancer regulation, direct contact is not needed. The work of Benabdallah et al. 2019. clearly demonstrated this phenomenon by showing the exact opposite. Expression of Shh was correlated with its promoter–enhancer distances increasing, bringing into question this general model. Thus, obstacles to a unified view of eTAD functionality may be the gaps in our understanding of enhancer biology.
CONNECTIONS BETWEEN ENHANCER–PROMOTER PROXIMITY AND BURSTING REVEALED IN SINGLE-CELL STUDIES
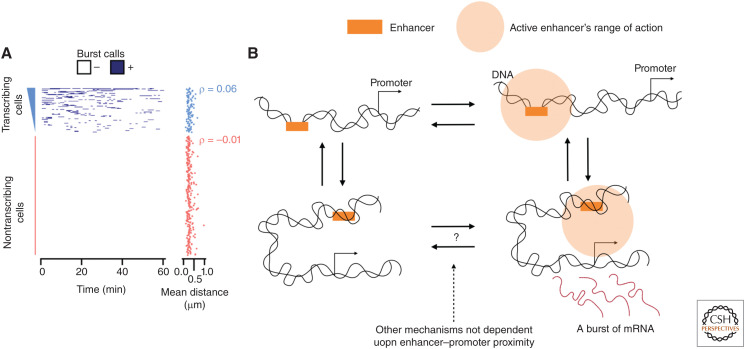
Ideally, an experimentalist would want to monitor DNA conformation and nascent RNA production in living cells with millisecond time resolution and nanometer spatial precision. Current microscopy technology does not yet allow these experiments. However, the progress in recent years in simultaneously imaging both DNA and RNA has been substantial. Pivotal live-cell experiments have provided invaluable information about the dynamics of chromosomal enhancer transcription regulation (Larson et al. 2011). Two studies have now shown that a single enhancer is able to activate two adjacent genes at the same time, arguing against direct enhancer–promoter contact for activation (Fukaya et al. 2016; Lim et al. 2018). In the work of Chen et al. (2018a) (Drosophila), it was clearly shown that loop formation was needed to allow for enhancer–promoter proximity activation. Upon loop formation, the enhancer–promoter distances showed a Gaussian distribution with a mean of ∼375 nm, whereas the unlooped conformation showed a Gaussian distribution with a mean of ∼720 nm. Interestingly, for transcription activation, the promoter–enhancer distances showed a Gaussian distribution with a slightly smaller mean of ∼330 nm, suggesting once a promoter is within this distance the enhancer could trigger transcription. Note, the investigators attributed this observation to DNA compaction with transcriptional activation, which is opposite to the finding of Cardozo Gizzi et al. (2019). Similarly, high-resolution work within embryonic stem cells partially supports this result, where they investigated the placement of an enhancer close to a promoter. This experiment resulted in an enhancer–promoter distance distribution with a mean of 300 nm and an extended tail up to 750 nm. Importantly, they found that there was no dependence between transcription and enhancer–promoter distance (Fig. 5A), suggesting that for this system the promoter is always within the range of action of the enhancer (Alexander et al. 2019). Yet, the bursting behavior of the gene was still extremely variable, in which the majority (>65%) of the cells were nontranscribing for a period of >60 min (Fig. 5A). If the bursts within individual cells were independent events, it is clear that the transcribing cells and nontranscribing cells did not show the same bursting frequency. Taken together, these results suggest that a large proportion of the variation observed within transcription is not dependent on the modulation of enhancer–promoter proximity and is likely governed by other mechanisms, such as enhancer activation, local variations in TF concentrations, etc.
Figure 5.
Enhancer-driven transcription. (A) The transcription state over time for individual cells and the distance between the enhancer and promoter to the right showing little correlation with transcription. (Panel A reprinted from Alexander et al. 2019 under the terms of a Creative Commons Attribution License.) (B) Model of transcription with the range of action of the enhancer taken into consideration, showing that if enhancer–promoter proximity is within the range of action of the enhancer, other mechanisms must be responsible for the stochastic nature of transcription.
In addition to the live-cell studies, the single-cell imaging work of Mateo et al. (2019) was able to directly visualize the 3D structure of a local region of the chromosome as well as the nascent RNA from the different genes within the region, providing the experimental means of investigating enhancer–promoter proximity and chromosomal structure. The enhancer–promoter distances between the two were predictive of nascent transcription, but only weakly. Importantly, the DNA segments that were predictive of transcription were not specific to the enhancer location but seemed to be confined to certain domains of the chromosome. Additionally, genes were active when separated from their enhancers and inactive when in proximity to their enhancers (Mateo et al. 2019). Again, we offer the interpretation that a large amount of the stochasticity is not directly dictated by enhancer–promoter proximity as long as the promoter is within the range of action of the enhancer.
OUTLOOK
In summary, the genome is stochastic at every level of organization. Stochastic A and B compartmentalization in individual cells is propagated through to transcription, but the specifics of this process are unclear. Enhancers seem to have a range of action around 300 nm potentially related to local ensemble chromosomal domains. Even so, enhancer-regulated transcription has a large amount of variation that is not dependent on fluctuations in enhancer–promoter proximity. In Figure 5B, we provide a hypothetical model of transcription to show these findings.
In terms of the higher-order structure of the chromosome, the lack of studies investigating A and B compartmentalization in individual cells and transcription clearly indicates the need for further work. A primary experimental impediment is that there is still no clear way to define the chromatin state in individual cells. Additionally, the conflicting results surrounding the importance of TADs and eTADs indicate that understanding what they do and how they work is still a central question within the field. For instance, how do these probabilistic structures convey the range of action of enhancers to the ensemble domains? There have been proposals that enhancers are able to nucleate “phase-separated” droplets, suggesting that maybe some of the probabilistic structures could create or limit this phenomenon. Although, even this recent idea has to be questioned as the expression of Shh was shown to be extremely resistant to perturbations of the eTAD structure (Williamson et al. 2019).
Overall, it should be noted that our understanding of stochastic genomic organization is dependent on very new technologies, and only time will tell whether our current model of the stochastic genome is attributable to real biological variability or experimental noise. Still, we have shown here that there are plausible links between the stochastic chromosome and stochastic transcription. Presently, single-cell studies are much better at visualizing proteins such as TFs and RNA than the high-resolution structure of the chromatin fiber. Future efforts should be aimed at dissecting the contributions of the myriad factors involved in transcriptional regulation as they work in the dynamic milieu of the nucleus.
Footnotes
Editors: Ana Pombo, Martin W. Hetzer, and Tom Misteli
Additional Perspectives on The Nucleus available at www.cshperspectives.org
REFERENCES
- Abramo K, Valton AL, Venev SV, Ozadam H, Fox AN, Dekker J. 2019. A chromosome folding intermediate at the condensin-to-cohesin transition during telophase. Nat Cell Biol 21: 1393–1402. 10.1038/s41556-019-0406-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander JM, Guan J, Li B, Maliskova L, Song M, Shen Y, Huang B, Lomvardas S, Weiner OD. 2019. Live-cell imaging reveals enhancer-dependent Sox2 transcription in the absence of enhancer proximity. eLife 8: e41769. 10.7554/eLife.41769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranda S, Mas G, Di Croce L. 2015. Regulation of gene transcription by Polycomb proteins. Sci Adv 1: e1500737. 10.1126/sciadv.1500737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerji J, Rusconi S, Schaffner W. 1981. Expression of a β-globin gene is enhanced by remote SV40 DNA sequences. Cell 27: 299–308. 10.1016/0092-8674(81)90413-X [DOI] [PubMed] [Google Scholar]
- Banerji J, Olson L, Schaffner W. 1983. A lymphocyte-specific cellular enhancer is located downstream of the joining region in immunoglobulin heavy chain genes. Cell 33: 729–740. 10.1016/0092-8674(83)90015-6 [DOI] [PubMed] [Google Scholar]
- Bartman CR, Hsu SC, Hsiung CCS, Raj A, Blobel GA. 2016. Enhancer regulation of transcriptional bursting parameters revealed by forced chromatin looping. Mol Cell 62: 237–247. 10.1016/j.molcel.2016.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell AC, Felsenfeld G. 2000. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405: 482–485. 10.1038/35013100 [DOI] [PubMed] [Google Scholar]
- Benabdallah NS, Williamson I, Illingworth RS, Kane L, Boyle S, Sengupta D, Grimes GR, Therizols P, Bickmore WA. 2019. Decreased enhancer-promoter proximity accompanying enhancer activation. Mol Cell 76: 473–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoist C, Chambon P. 1981. In vivo sequence requirements of the SV40 early promoter region. Nature 290: 304–310. 10.1038/290304a0 [DOI] [PubMed] [Google Scholar]
- Bickmore WA, Van Steensel B. 2013. Genome architecture: domain organization of interphase chromosomes. Cell 152: 1270–1284. 10.1016/j.cell.2013.02.001 [DOI] [PubMed] [Google Scholar]
- Bintu B, Mateo LJ, Su JH, Sinnott-Armstrong NA, Parker M, Kinrot S, Yamaya K, Boettiger AN, Zhuang X. 2018. Super-resolution chromatin tracing reveals domains and cooperative interactions in single cells. Science 362: eaau1783. 10.1126/science.aau1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettiger AN, Bintu B, Moffitt JR, Wang S, Beliveau BJ, Fudenberg G, Imakaev M, Mirny LA, Wu CT, Zhuang X. 2016. Super-resolution imaging reveals distinct chromatin folding for different epigenetic states. Nature 529: 418–422. 10.1038/nature16496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolzer A, Kreth G, Solovei I, Koehler D, Saracoglu K, Fauth C, Müller S, Eils R, Cremer C, Speicher MR, et al. 2005. Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. PLoS Biol 3: e157. 10.1371/journal.pbio.0030157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonev B, Mendelson Cohen N, Szabo Q, Fritsch L, Papadopoulos GL, Lubling Y, Xu X, Lv X, Hugnot JP, Tanay A, et al. 2017. Multiscale 3D genome rewiring during mouse neural development. Cell 171: 557–572.e24. 10.1016/j.cell.2017.09.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. 2013. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10: 1213–1218. 10.1038/nmeth.2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulger M, Groudine M. 2010. Enhancers: the abundance and function of regulatory sequences beyond promoters. Dev Biol 339: 250–257. 10.1016/j.ydbio.2009.11.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardozo Gizzi AM, Cattoni DI, Fiche JB, Espinola SM, Gurgo J, Messina O, Houbron C, Ogiyama Y, Papadopoulos GL, Cavalli G, et al. 2019. Microscopy-based chromosome conformation capture enables simultaneous visualization of genome organization and transcription in intact organisms. Mol Cell 74: 212–222.e5. 10.1016/j.molcel.2019.01.011 [DOI] [PubMed] [Google Scholar]
- Chen H, Levo M, Barinov L, Fujioka M, Jaynes JB, Gregor T. 2018a. Dynamic interplay between enhancer–promoter topology and gene activity. Nat Genet 50: 1296–1303. 10.1038/s41588-018-0175-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Teichmann SA, Meyer KB. 2018b. From tissues to cell types and back: single-cell gene expression analysis of tissue architecture. Ann Rev Biomed Data Sci 1: 29–51. 10.1146/annurev-biodatasci-080917-013452 [DOI] [Google Scholar]
- Chong S, Chen C, Ge H, Xie XS. 2014. Mechanism of transcriptional bursting in bacteria. Cell 158: 314–326. 10.1016/j.cell.2014.05.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang CH, Carpenter AE, Fuchsova B, Johnson T, de Lanerolle P, Belmont AS. 2006. Long-range directional movement of an interphase chromosome site. Curr Biol 16: 825–831. 10.1016/j.cub.2006.03.059 [DOI] [PubMed] [Google Scholar]
- Chubb JR, Boyle S, Perry P, Bickmore WA. 2002. Chromatin motion is constrained by association with nuclear compartments in human cells. Curr Biol 12: 439–445. 10.1016/S0960-9822(02)00695-4 [DOI] [PubMed] [Google Scholar]
- Dalla-Favera R, Bregni M, Erikson J, Patterson D, Gallo RC, Croce CM. 1982. Human c-myc onc gene is located on the region of chromosome 8 that is translocated in Burkitt lymphoma cells. Proc Natl Acad Sci 79: 7824–7827. 10.1073/pnas.79.24.7824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson IF, Bauer B, Goetz D, Tang W, Wutz G, Peters JM. 2019. DNA loop extrusion by human cohesin. Science 366: 1338–1345. 10.1126/science.aaz3418 [DOI] [PubMed] [Google Scholar]
- Deng W, Lee J, Wang H, Miller J, Reik A, Gregory PD, Dean A, Blobel GA. 2012. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell 149: 1233–1244. 10.1016/j.cell.2012.03.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Rupon JW, Krivega I, Breda L, Motta I, Jahn KS, Reik A, Gregory PD, Rivella S, Dean A, et al. 2014. Reactivation of developmentally silenced globin genes by forced chromatin looping. Cell 158: 849–860. 10.1016/j.cell.2014.05.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey SS, Foley JE, Limsirichai P, Schaffer DV, Arkin AP. 2015. Orthogonal control of expression mean and variance by epigenetic features at different genomic loci. Mol Syst Biol 11: 806. 10.15252/msb.20145704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485: 376–380. 10.1038/nature11082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan BT, Huynh A, Ball DA, Patel HP, Poirier MG, Larson DR, Ferguson ML, Lenstra TL. 2019. Live-cell imaging reveals the interplay between transcription factors, nucleosomes, and bursting. EMBO J 38: e100809. 10.15252/embj.2018100809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Zheng H, Huang B, Ma R, Wu J, Zhang X, He J, Xiang Y, Wang Q, Li Y, et al. 2017. Allelic reprogramming of 3D chromatin architecture during early mammalian development. Nature 547: 232–235. 10.1038/nature23263 [DOI] [PubMed] [Google Scholar]
- Elowitz MB, Levine AJ, Siggia ED, Swain PS. 2002. Stochastic gene expression in a single cell. Science 297: 1183–1186. 10.1126/science.1070919 [DOI] [PubMed] [Google Scholar]
- Falk M, Feodorova Y, Naumova N, Imakaev M, Lajoie BR, Leonhardt H, Joffe B, Dekker J, Fudenberg G, Solovei I, et al. 2019. Heterochromatin drives compartmentalization of inverted and conventional nuclei. Nature 570: 395–399. 10.1038/s41586-019-1275-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang C, Rao S, Crispino JD, Ntziachristos P. 2020. Determinants and role of chromatin organization in acute leukemia. Leukemia 34: 2561–2575. 10.1038/s41375-020-0981-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlan LE, Sproul D, Thomson I, Boyle S, Kerr E, Perry P, Ylstra B, Chubb JR, Bickmore WA. 2008. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet 4: e1000039. 10.1371/journal.pgen.1000039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn EH, Misteli T. 2019. Molecular basis and biological function of variability in spatial genome organization. Science 365: eaaw9498. 10.1126/science.aaw9498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn EH, Pegoraro G, Brandão HB, Valton AL, Oomen ME, Dekker J, Mirny L, Misteli T. 2019. Extensive heterogeneity and intrinsic variation in spatial genome organization. Cell 176: 1502–1515.e10. 10.1016/j.cell.2019.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishilevich S, Nudel R, Rappaport N, Hadar R, Plaschkes I, Stein TI, Rosen N, Kohn A, Twik M, Safran M, et al. 2017. GeneHancer: genome-wide integration of enhancers and target genes in GeneCards. Database (Oxford) 2017: bax028. 10.1093/database/bax028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavahan WA, Drier Y, Liau BB, Gillespie SM, Venteicher AS, Stemmer-Rachamimov AO, Suvà ML, Bernstein BE. 2016. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature 529: 110–114. 10.1038/nature16490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flyamer IM, Gassler J, Imakaev M, Brandão HB, Ulianov SV, Abdennur N, Razin SV, Mirny LA, Tachibana-Konwalski K. 2017. Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte-to-zygote transition. Nature 544: 110–114. 10.1038/nature21711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke M, Ibrahim DM, Andrey G, Schwarzer W, Heinrich V, Schöpflin R, Kraft K, Kempfer R, Jerković I, Chan WL, et al. 2016. Formation of new chromatin domains determines pathogenicity of genomic duplications. Nature 538: 265–269. 10.1038/nature19800 [DOI] [PubMed] [Google Scholar]
- Fudenberg G, Imakaev M, Lu C, Goloborodko A, Abdennur N, Mirny LA. 2016. Formation of chromosomal domains by loop extrusion. Cell Rep 15: 2038–2049. 10.1016/j.celrep.2016.04.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaya T, Lim B, Levine M. 2016. Enhancer control of transcriptional bursting. Cell 166: 358–368. 10.1016/j.cell.2016.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlong EEM, Levine M. 2018. Developmental enhancers and chromosome topology. Science 361: 1341–1345. 10.1126/science.aau0320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambetta MC, Furlong EEM. 2018. The insulator protein CTCF is required for correct Hox gene expression, but not for embryonic development in Drosophila. Genetics 210: 129–136. 10.1534/genetics.118.301350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganai N, Sengupta S, Menon GI. 2014. Chromosome positioning from activity-based segregation. Nucleic Acids Res 42: 4145–4159. 10.1093/nar/gkt1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassler J, Brandão HB, Imakaev M, Flyamer IM, Ladstätter S, Bickmore WA, Peters JM, Mirny LA, Tachibana K. 2017. A mechanism of cohesin-dependent loop extrusion organizes zygotic genome architecture. EMBO J 36: 3600–3618. 10.15252/embj.201798083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghavi-Helm Y, Jankowski A, Meiers S, Viales RR, Korbel JO, Furlong EEM. 2019. Highly rearranged chromosomes reveal uncoupling between genome topology and gene expression. Nat Genet 51: 1272–1282. 10.1038/s41588-019-0462-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghirlando R, Felsenfeld G. 2016. CTCF: making the right connections. Genes Dev 30: 881–891. 10.1101/gad.277863.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert N, Boyle S, Fiegler H, Woodfine K, Carter NP, Bickmore WA. 2004. Chromatin architecture of the human genome: gene-rich domains are enriched in open chromatin fibers. Cell 118: 555–566. 10.1016/j.cell.2004.08.011 [DOI] [PubMed] [Google Scholar]
- Gröschel S, Sanders MA, Hoogenboezem R, de Wit E, Bouwman BAM, Erpelinck C, van der Velden VHJ, Havermans M, Avellino R, van Lom K, et al. 2014. A single oncogenic enhancer rearrangement causes concomitant EVI1 and GATA2 deregulation in leukemia. Cell 157: 369–381. 10.1016/j.cell.2014.02.019 [DOI] [PubMed] [Google Scholar]
- Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, et al. 2008. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453: 948–951. 10.1038/nature06947 [DOI] [PubMed] [Google Scholar]
- Guo Y, Xu Q, Canzio D, Shou J, Li J, Gorkin DU, Jung I, Wu H, Zhai Y, Tang Y, et al. 2015. CRISPR inversion of CTCF sites alters genome topology and enhancer/promoter function. Cell 162: 900–910. 10.1016/j.cell.2015.07.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haarhuis JHI, van der Weide RH, Blomen VA, Yáñez-Cuna JO, Amendola M, van Ruiten MS, Krijger PHL, Teunissen H, Medema RH, van Steensel B, et al. 2017. The cohesin release factor WAPL restricts chromatin loop extension. Cell 169: 693–707.e14. 10.1016/j.cell.2017.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton IB, D'Ippolito AM, Vockley CM, Thakore PI, Crawford GE, Reddy TE, Gersbach CA. 2015. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol 33: 510–517. 10.1038/nbt.3199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Weintraub AS, Day DS, Valton AL, Bak RO, Li CH, Goldmann J, Lajoie BR, Fan ZP, Sigova AA, et al. 2016. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science 351: 1454–1458. 10.1126/science.aad9024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung G, Bar-Ziv R, Rosin D, Tokuriki N, Tawfik DS, Oren M, Barkai N. 2012. Noise–mean relationship in mutated promoters. Genome Res 22: 2409–2417. 10.1101/gr.139378.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou C, Li L, Qin ZS, Corces VG. 2012. Gene density, transcription, and insulators contribute to the partition of the Drosophila genome into physical domains. Mol Cell 48: 471–484. 10.1016/j.molcel.2012.08.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh THS, Cattoglio C, Slobodyanyuk E, Hansen AS, Rando OJ, Tjian R, Darzacq X. 2020. Resolving the 3D landscape of transcription-linked mammalian chromatin folding. Mol Cell 78: 539–553.e8. 10.1016/j.molcel.2020.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug CB, Grimaldi AG, Kruse K, Vaquerizas JM. 2017. Chromatin architecture emerges during zygotic genome activation independent of transcription. Cell 169: 216–228.e19. 10.1016/j.cell.2017.03.024 [DOI] [PubMed] [Google Scholar]
- Ibragimov AN, Bylino OV, Shidlovskii YV. 2020. Molecular basis of the function of transcriptional enhancers. Cells 9: 1620. 10.3390/cells9071620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen A, Colmenares SU, Karpen GH. 2018. Heterochromatin: guardian of the genome. Ann Rev Cell Dev Biol 34: 265–288. 10.1146/annurev-cellbio-100617-062653 [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. 2001. Translating the histone code. Science 293: 1074–1080. 10.1126/science.1063127 [DOI] [PubMed] [Google Scholar]
- Ke Y, Xu Y, Chen X, Feng S, Liu Z, Sun Y, Yao X, Li F, Zhu W, Gao L, et al. 2017. 3D chromatin structures of mature gametes and structural reprogramming during mammalian embryogenesis. Cell 170: 367–381.e20. 10.1016/j.cell.2017.06.029 [DOI] [PubMed] [Google Scholar]
- Kim Y, Shi Z, Zhang H, Finkelstein IJ, Yu H. 2019. Human cohesin compacts DNA by loop extrusion. Science 366: 1345–1349. 10.1126/science.aaz4475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind J, van Steensel B. 2010. Genome–nuclear lamina interactions and gene regulation. Curr Opin Cell Biol 22: 320–325. 10.1016/j.ceb.2010.04.002 [DOI] [PubMed] [Google Scholar]
- Krijger PHL, De Laat W. 2016. Regulation of disease-associated gene expression in the 3D genome. Nat Rev Mol Cell Biol 17: 771–782. 10.1038/nrm.2016.138 [DOI] [PubMed] [Google Scholar]
- Kumaran RI, Spector DL. 2008. A genetic locus targeted to the nuclear periphery in living cells maintains its transcriptional competence. J Cell Biol 180: 51–65. 10.1083/jcb.200706060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson DR. 2019. Structure and function in Drosophila chromosomes: visualizing topological domains. Mol Cell 74: 3–4. 10.1016/j.molcel.2019.03.017 [DOI] [PubMed] [Google Scholar]
- Larson DR, Zenklusen D, Wu B, Chao JA, Singer RH. 2011. Real-time observation of transcription initiation and elongation on an endogenous yeast gene. Science 332: 475–478. 10.1126/science.1202142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson AJM, Johnsson P, Hagemann-Jensen M, Hartmanis L, Faridani OR, Reinius B, Segerstolpe Å, Rivera CM, Ren B, Sandberg R. 2019. Genomic encoding of transcriptional burst kinetics. Nature 565: 251–254. 10.1038/s41586-018-0836-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Dily F, Baù D, Pohl A, Vicent GP, Serra F, Soronellas D, Castellano G, Wright RHG, Ballare C, Filion G, et al. 2014. Distinct structural transitions of chromatin topological domains correlate with coordinated hormone-induced gene regulation. Genes Dev 28: 2151–2162. 10.1101/gad.241422.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettice LA, Heaney SJH, Purdie LA, Li L, de Beer P, Oostra BA, Goode D, Elgar G, Hill RE, de Graaff E. 2003. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum Mol Genet 12: 1725–1735. 10.1093/hmg/ddg180 [DOI] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326: 289–293. 10.1126/science.1181369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim B, Heist T, Levine M, Fukaya T. 2018. Visualization of transvection in living Drosophila embryos. Mol Cell 70: 287–296.e6. 10.1016/j.molcel.2018.02.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long HK, Prescott SL, Wysocka J. 2016. Ever-changing landscapes: transcriptional enhancers in development and evolution. Cell 167: 1170–1187. 10.1016/j.cell.2016.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupiáñez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, Horn D, Kayserili H, Opitz JM, Laxova R, et al. 2015. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 161: 1012–1025. 10.1016/j.cell.2015.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luppino JM, Park DS, Nguyen SC, Lan Y, Xu Z, Yunker R, Joyce EF. 2020. Cohesin promotes stochastic domain intermingling to ensure proper regulation of boundary-proximal genes. Nat Genet 52: 840–848. 10.1038/s41588-020-0647-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateo LJ, Murphy SE, Hafner A, Cinquini IS, Walker CA, Boettiger AN. 2019. Visualizing DNA folding and RNA in embryos at single-cell resolution. Nature 568: 49–54. 10.1038/s41586-019-1035-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermet J, Yeung J, Hurni C, Mauvoisin D, Gustafson K, Jouffe C, Nicolas D, Emmenegger Y, Gobet C, Franken P, et al. 2018. Clock-dependent chromatin topology modulates circadian transcription and behavior. Genes Dev 32: 347–358. 10.1101/gad.312397.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. 2020. The self-organizing genome: principles of genome architecture and function. Cell 183: 28–45. 10.1016/j.cell.2020.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi T, Fudenberg G, Mehta S, Belton JM, Taneja N, Folco HD, FitzGerald P, Dekker J, Mirny L, Barrowman J, et al. 2014. Cohesin-dependent globules and heterochromatin shape 3D genome architecture in S. pombe. Nature 516: 432–435. 10.1038/nature13833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan SL, Mariano NC, Bermudez A, Arruda NL, Wu F, Luo Y, Shankar G, Jia L, Chen H, Hu JF, et al. 2017. Manipulation of nuclear architecture through CRISPR-mediated chromosomal looping. Nat Commun 8: 15993. 10.1038/ncomms15993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano T, Lubling Y, Stevens TJ, Schoenfelder S, Yaffe E, Dean W, Laue ED, Tanay A, Fraser P. 2013. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature 502: 59–64. 10.1038/nature12593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra V, Rocha PP, An D, Raviram R, Skok JA, Mazzoni EO, Reinberg D. 2015. CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science 347: 1017–1021. 10.1126/science.1262088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naughton C, Avlonitis N, Corless S, Prendergast JG, Mati IK, Eijk PP, Cockroft SL, Bradley M, Ylstra B, Gilbert N. 2013. Transcription forms and remodels supercoiling domains unfolding large-scale chromatin structures. Nat Struct Mol Biol 20: 387–395. 10.1038/nsmb.2509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. 2012. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485: 381–385. 10.1038/nature11049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Goloborodko A, Valton AL, Gibcus JH, Uebersohn A, Abdennur N, Dekker J, Mirny LA, Bruneau BG. 2017. Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell 169: 930–944.e22. 10.1016/j.cell.2017.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcott PA, Lee C, Zichner T, Stütz AM, Erkek S, Kawauchi D, Shih DJH, Hovestadt V, Zapatka M, Sturm D, et al. 2014. Enhancer hijacking activates GFI1 family oncogenes in medulloblastoma. Nature 511: 428–434. 10.1038/nature13379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuebler J, Fudenberg G, Imakaev M, Abdennur N, Mirny LA. 2018. Chromatin organization by an interplay of loop extrusion and compartmental segregation. Proc Natl Acad Sci 115: E6697–E6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldridge DA, Wood AC, Weichert-Leahey N, Crimmins I, Sussman R, Winter C, McDaniel LD, Diamond M, Hart LS, Zhu S, et al. 2015. Genetic predisposition to neuroblastoma mediated by a LMO1 super-enhancer polymorphism. Nature 528: 418–421. 10.1038/nature15540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peddie CJ, Collinson LM. 2014. Exploring the third dimension: volume electron microscopy comes of age. Micron 61: 9–19. 10.1016/j.micron.2014.01.009 [DOI] [PubMed] [Google Scholar]
- Phanstiel DH, Van Bortle K, Spacek D, Hess GT, Saad Shamim M, Machol I, Love MI, Lieberman Aiden E, Bassik MC, Snyder MP. 2017. Static and dynamic DNA loops form AP-1-bound activation hubs during macrophage development. Mol Cell 67: 1037–1048.e6. 10.1016/j.molcel.2017.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. 2011. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470: 279–283. 10.1038/nature09692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, van Oudenaarden A. 2008. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell 135: 216–226. 10.1016/j.cell.2008.09.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S. 2006. Stochastic mRNA synthesis in mammalian cells. PLoS Biol 4: e309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SSP, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. 2014. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159: 1665–1680. 10.1016/j.cell.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SSP, Huang SC, St Hilaire BG, Engreitz JM, Perez EM, Kieffer-Kwon KR, Sanborn AL, Johnstone SE, Bascom GD, Bochkov ID, et al. 2017. Cohesin loss eliminates all loop domains. Cell 171: 305–320.e24. 10.1016/j.cell.2017.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raser JM, O'Shea EK. 2004. Control of stochasticity in eukaryotic gene expression. Science 304: 1811–1814. 10.1126/science.1098641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy KL, Zullo JM, Bertolino E, Singh H. 2008. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature 452: 243–247. [DOI] [PubMed] [Google Scholar]
- Ricci MA, Manzo C, García-Parajo MF, Lakadamyali M, Cosma MP. 2015. Chromatin fibers are formed by heterogeneous groups of nucleosomes in vivo. Cell 160: 1145–1158. 10.1016/j.cell.2015.01.054 [DOI] [PubMed] [Google Scholar]
- Robinett CC, Straight A, Li G, Willhelm C, Sudlow G, Murray A, Belmont AS. 1996. In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J Cell Biol 135: 1685–1700. 10.1083/jcb.135.6.1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez J, Larson DR. 2020. Transcription in living cells: molecular mechanisms of bursting. Annu Rev Biochem 89: 189–212. 10.1146/annurev-biochem-011520-105250 [DOI] [PubMed] [Google Scholar]
- Rodriguez J, Ren G, Day CR, Zhao K, Chow CC, Larson DR. 2019. Intrinsic dynamics of a human gene reveal the basis of expression heterogeneity. Cell 176: 213–226.e18. 10.1016/j.cell.2018.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley MJ, Nichols MH, Lyu X, Ando-Kuri M, Rivera ISM, Hermetz K, Wang P, Ruan Y, Corces VG. 2017. Evolutionarily conserved principles predict 3D chromatin organization. Mol Cell 67: 837–852.e7. 10.1016/j.molcel.2017.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruf S, Symmons O, Uslu VV, Dolle D, Hot C, Ettwiller L, Spitz F. 2011. Large-scale analysis of the regulatory architecture of the mouse genome with a transposon-associated sensor. Nat Genet 43: 379–386. 10.1038/ng.790 [DOI] [PubMed] [Google Scholar]
- Sanborn AL, Rao SSP, Huang SC, Durand NC, Huntley MH, Jewett AI, Bochkov ID, Chinnappan D, Cutkosky A, Li J, et al. 2015. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci 112: E6456–E6465. 10.1073/pnas.1518552112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffner W. 2015. Enhancers, enhancers—from their discovery to today's universe of transcription enhancers. Biol Chem 396: 311–327. 10.1515/hsz-2014-0303 [DOI] [PubMed] [Google Scholar]
- Schultz J. 1947. The nature of heterochromatin. Cold Spring Harb Symp Quant Biol 12: 179–191. 10.1101/SQB.1947.012.01.021 [DOI] [Google Scholar]
- Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, Parrinello H, Tanay A, Cavalli G. 2012. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 148: 458–472. 10.1016/j.cell.2012.01.010 [DOI] [PubMed] [Google Scholar]
- Shahrezaei V, Swain PS. 2008. Analytical distributions for stochastic gene expression. Proc Natl Acad Sci 105: 17256–17261. 10.1073/pnas.0803850105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, et al. 2012. A map of the cis-regulatory sequences in the mouse genome. Nature 488: 116–120. 10.1038/nature11243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F, Furlong EEM. 2012. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet 13: 613–626. 10.1038/nrg3207 [DOI] [PubMed] [Google Scholar]
- Stampfel G, Kazmar T, Frank O, Wienerroither S, Reiter F, Stark A. 2015. Transcriptional regulators form diverse groups with context-dependent regulatory functions. Nature 528: 147–151. 10.1038/nature15545 [DOI] [PubMed] [Google Scholar]
- Stavreva DA, Garcia DA, Fettweis G, Gudla PR, Zaki GF, Soni V, McGowan A, Williams G, Huynh A, Palangat M, et al. 2019. Transcriptional bursting and co-bursting regulation by steroid hormone release pattern and transcription factor mobility. Mol Cell 75: 1161–1177.e11. 10.1016/j.molcel.2019.06.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens TJ, Lando D, Basu S, Atkinson LP, Cao Y, Lee SF, Leeb M, Wohlfahrt KJ, Boucher W, O'Shaughnessy-Kirwan A, et al. 2017. 3D structures of individual mammalian genomes studied by single-cell Hi-C. Nature 544: 59–64. 10.1038/nature21429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom AR, Emelyanov AV, Mir M, Fyodorov DV, Darzacq X, Karpen GH. 2017. Phase separation drives heterochromatin domain formation. Nature 547: 241–245. 10.1038/nature22989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su JH, Zheng P, Kinrot SS, Bintu B, Zhuang X. 2020. Genome-scale imaging of the 3D organization and transcriptional activity of chromatin. Cell 182: 1641–1659.e26. 10.1016/j.cell.2020.07.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M, Zhang J. 2019. Chromosome-wide co-fluctuation of stochastic gene expression in mammalian cells. PLoS Genet 15: e1008389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter DM, Molina N, Gatfield D, Schneider K, Schibler U, Naef F. 2011. Mammalian genes are transcribed with widely different bursting kinetics. Science 332: 472–474. 10.1126/science.1198817 [DOI] [PubMed] [Google Scholar]
- Swain PS, Elowitz MB, Siggia ED. 2002. Intrinsic and extrinsic contributions to stochasticity in gene expression. Proc Natl Acad Sci 99: 12795–12800. 10.1073/pnas.162041399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symmons O, Uslu VV, Tsujimura T, Ruf S, Nassari S, Schwarzer W, Ettwiller L, Spitz F. 2014. Functional and topological characteristics of mammalian regulatory domains. Genome Res 24: 390–400. 10.1101/gr.163519.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo Q, Jost D, Chang JM, Cattoni DI, Papadopoulos GL, Bonev B, Sexton T, Gurgo J, Jacquier C, Nollmann M, et al. 2018. TADs are 3D structural units of higher-order chromosome organization in Drosophila. Science Adv 4: eaar8082. 10.1126/sciadv.aar8082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L, Xing D, Chang CH, Li H, Xie XS. 2018. Three-dimensional genome structures of single diploid human cells. Science 361: 924–928. 10.1126/science.aat5641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tantale K, Mueller F, Kozulic-Pirher A, Lesne A, Victor JM, Robert MC, Capozi S, Chouaib R, Bäcker V, Mateos-Langerak J, et al. 2016. A single-molecule view of transcription reveals convoys of RNA polymerases and multi-scale bursting. Nat Commun 7: 12248. 10.1038/ncomms12248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The 1000 Genomes Project Consortium; Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. 2012. An integrated map of genetic variation from 1,092 human genomes. Nature 491: 56–65. 10.1038/nature11632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therizols P, Illingworth RS, Courilleau C, Boyle S, Wood AJ, Bickmore WA. 2014. Chromatin decondensation is sufficient to alter nuclear organization in embryonic stem cells. Science 346: 1238–1242. 10.1126/science.1259587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trzaskoma P, Ruszczycki B, Lee B, Pels KK, Krawczyk K, Bokota G, Szczepankiewicz AA, Aaron J, Walczak A, Śliwińska MA, et al. 2020. Ultrastructural visualization of 3D chromatin folding using volume electron microscopy and DNA in situ hybridization. Nat Commun 11: 2120. 10.1038/s41467-020-15987-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunnacliffe E, Chubb JR. 2020. What is a transcriptional burst? Trends Genet 36: 288–297. 10.1016/j.tig.2020.01.003 [DOI] [PubMed] [Google Scholar]
- Ulianov SV, Khrameeva EE, Gavrilov AA, Flyamer IM, Kos P, Mikhaleva EA, Penin AA, Logacheva MD, Imakaev MV, Chertovich A, et al. 2016. Active chromatin and transcription play a key role in chromosome partitioning into topologically associating domains. Genome Res 26: 70–84. 10.1101/gr.196006.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B, Belmont AS. 2017. Lamina-associated domains: links with chromosome architecture, heterochromatin, and gene repression. Cell 169: 780–791. 10.1016/j.cell.2017.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vian L, Pękowska A, Rao SSP, Kieffer-Kwon KR, Jung S, Baranello L, Huang SC, El Khattabi L, Dose M, Pruett N, et al. 2018. The energetics and physiological impact of cohesin extrusion. Cell 173: 1165–1178.e20. 10.1016/j.cell.2018.03.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss TC, Schiltz RL, Sung MH, Yen PM, Stamatoyannopoulos JA, Biddie SC, Johnson TA, Miranda TB, John S, Hager GL. 2011. Dynamic exchange at regulatory elements during chromatin remodeling underlies assisted loading mechanism. Cell 146: 544–554. 10.1016/j.cell.2011.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Su JH, Beliveau BJ, Bintu B, Moffitt JR, Wu CT, Zhuang X. 2016. Spatial organization of chromatin domains and compartments in single chromosomes. Science 353: 598–602. 10.1126/science.aaf8084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Gao Y, Zheng X, Liu C, Dong S, Li R, Zhang G, Wei Y, Qu H, Li Y, et al. 2019. Histone modifications regulate chromatin compartmentalization by contributing to a phase separation mechanism. Mol Cell 76: 646–659.e6. 10.1016/j.molcel.2019.08.019 [DOI] [PubMed] [Google Scholar]
- Weintraub AS, Li CH, Zamudio AV, Sigova AA, Hannett NM, Day DS, Abraham BJ, Cohen MA, Nabet B, Buckley DL, et al. 2017. YY1 is a structural regulator of enhancer-promoter loops. Cell 171: 1573–1588.e28. 10.1016/j.cell.2017.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weischenfeldt J, Dubash T, Drainas AP, Mardin BR, Chen Y, Stütz AM, Waszak SM, Bosco G, Halvorsen AR, Raeder B, et al. 2017. Pan-cancer analysis of somatic copy-number alterations implicates IRS4 and IGF2 in enhancer hijacking. Nat Genet 49: 65–74. 10.1038/ng.3722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijchers PJ, Krijger PHL, Geeven G, Zhu Y, Denker A, Verstegen MJAM, Valdes-Quezada C, Vermeulen C, Janssen M, Teunissen H, et al. 2016. Cause and consequence of tethering a subTAD to different nuclear compartments. Mol Cell 61: 461–473. 10.1016/j.molcel.2016.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RRE, Azuara V, Perry P, Sauer S, Dvorkina M, Jørgensen H, Roix J, McQueen P, Misteli T, Merkenschlager M, et al. 2006. Neural induction promotes large-scale chromatin reorganisation of the Mash1 locus. J Cell Sci 119: 132–140. 10.1242/jcs.02727 [DOI] [PubMed] [Google Scholar]
- Williamson I, Lettice LA, Hill RE, Bickmore WA. 2016. Shh and ZRS enhancer colocalisation is specific to the zone of polarising activity. Development 143: 2994–3001. 10.1242/dev.139188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson I, Kane L, Devenney PS, Flyamer IM, Anderson E, Kilanowski F, Hill RE, Bickmore WA, Lettice LA. 2019. Developmentally regulated Shh expression is robust to TAD perturbations. Development 146: dev179523. 10.1242/dev.179523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie WJ, Meng L, Liu S, Zhang L, Cai X, Gao YQ. 2017. Structural modeling of chromatin integrates genome features and reveals chromosome folding principle. Sci Rep 7: 2818. 10.1038/s41598-017-02923-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe E, Tanay A. 2011. Probabilistic modeling of Hi-C contact maps eliminates systematic biases to characterize global chromosomal architecture. Nat Genet 43: 1059–1065. 10.1038/ng.947 [DOI] [PubMed] [Google Scholar]
- Zaret KS, Carroll JS. 2011. Pioneer transcription factors: establishing competence for gene expression. Genes Dev 25: 2227–2241. 10.1101/gad.176826.111 [DOI] [PMC free article] [PubMed] [Google Scholar]