Summary
Current approaches for oral health care rely on procedures that are unaffordable to impoverished populations, whereas aerosolized droplets in the dental clinic and poor oral hygiene may contribute to spread of several infectious diseases including COVID‐19, requiring new solutions for dental biofilm/plaque treatment at home. Plant cells have been used to produce monoclonal antibodies or antimicrobial peptides for topical applications to decrease colonization of pathogenic microbes on dental surface. Therefore, we investigated an affordable method for dental biofilm disruption by expressing lipase, dextranase or mutanase in plant cells via the chloroplast genome. Antibiotic resistance gene used to engineer foreign genes into the chloroplast genome were subsequently removed using direct repeats flanking the aadA gene and enzymes were successfully expressed in marker‐free lettuce transplastomic lines. Equivalent enzyme units of plant‐derived lipase performed better than purified commercial enzymes against biofilms, specifically targeting fungal hyphae formation. Combination of lipase with dextranase and mutanase suppressed biofilm development by degrading the biofilm matrix, with concomitant reduction of bacterial and fungal accumulation. In chewing gum tablets formulated with freeze‐dried plant cells, expressed protein was stable up to 3 years at ambient temperature and was efficiently released in a time‐dependent manner using a mechanical chewing simulator device. Development of edible plant cells expressing enzymes eliminates the need for purification and cold‐chain transportation, providing a potential translatable therapeutic approach. Biofilm disruption through plant enzymes and chewing gum‐based delivery offers an effective and affordable dental biofilm control at home particularly for populations with minimal oral care access.
Keywords: therapeutic enzymes, plant‐derived biopharmaceuticals, dental biofilm control, topical drug delivery
Introduction
Aerosolized microbes generated during dental procedures and mechanical plaque/biofilm removal that has been recognized as potential threat for the spread of several infectious diseases (Bennett et al., 2000) has now received greater attention during the current COVID‐19 global pandemic (Xu et al., 2020). The saliva and dorsum of the tongue are major sources of SARS‐CoV‐2 and stability of the virus in the aerosol and its spread in the aerosolized form has been widely reported (Van Doremalen et al., 2020; World Health Organization [WHO], 2020; Xu et al., 2020). Therefore, WHO and dental associations including American Dental Association recommended suspension of aerosol‐generating procedures in the clinic except for emergencies (Bennett et al., 2000; Van Doremalen et al., 2020; WHO, 2020; Xu et al., 2020). Furthermore, COVID‐19 patients have shown high accumulation of pathogenic oral bacteria, whereas poor oral hygiene which disproportionately afflicts impoverished populations, may be a risk factor for COVID‐19 (Patel and Sampson, 2020). Accumulation of microbes on teeth leads to the formation of intractable dental biofilms (plaque) that cause oral diseases such as dental caries (tooth decay) requiring costly clinical interventions at the dental office. Hence, development of alternative methods for plaque control at home is of paramount importance and urgency.
Topical application of therapeutic proteins/antibodies has been recognized as a potential alternative in this perspective. Due to several advantages of using plants as a recombinant protein production platform, this has been explored for prevention or control of several infectious diseases by topical delivery. For example, anti‐herpes simplex virus 2 (HSV‐2) monoclonal antibody produced in soybean was as effective as mammalian cell product in prevention of vaginal HSV‐2 infection in the mouse model, through vaginal delivery (Zeitlin et al., 1998). Antibody expressed in maize (anti‐2G12) and tobacco (anti‐ α CCR5) to neutralize human immunodeficiency virus (HIV) was developed as a microbicide to prevent transmission of HIV by topical vaginal delivery (Pogue et al., 2010; Ramessar et al., 2008; Yusibov et al., 2011). Other plant‐derived proteins developed as topical microbicides for prevention of HIV transmission are Griffithsin and Cyanovirin‐N. Both the proteins (lectins) produced recombinantly in transgenic rice endosperm and are non‐toxic to human cells as crude extract or in minimally processed form (Vamvaka et al., 2016a,b). Limited studies are available in this perspective for oral health care. Plantibody against Streptococcal antigen I/II that is secretory monoclonal antibody SIgA/G (Guy’s 13) has been produced in carrot cells and well demonstrated for passive immunization in human without any side effects (Ma et al., 1998). In another recent study, plant‐made antimicrobial peptide (protegrin or retrocyclin) has been demonstrated to prevent S. mutans biofilm formation on the hydroxyapatite surface (a tooth surrogate) (Liu et al., 2016). The antimicrobial peptides were also able to deliver into the gingival and periodontal cells around the teeth. Interestingly, cell‐penetrating efficiency of chloroplast made antimicrobial peptides were 13%–48% higher compared to other tested cell‐penetrating peptides (Liu et al., 2016).
Dental caries is one of the most neglected infectious diseases that afflict 2.3 billion people worldwide, costing >$140 billion in the US alone (Bowen et al., 2018; Hajishengallis et al., 2017; Peres et al., 2019; Vos et al., 2017). It is caused by pathogenic plaque accumulation under sugar‐rich conditions that erodes the tooth mineral leading to cavitation and further complications to the oral and systemic health (Bowen et al., 2018; Hajishengallis et al., 2017). In particular, severe childhood caries is highly prevalent (60%–90%) among underprivileged children with low socio‐economic status, and constitutes a major public health problem (Hajishengallis et al., 2017). Microbiological studies revealed a highly virulent mixed‐kingdom biofilm involving a synergistic interaction between S. mutans (a bacterium) and the fungal species Candida albicans (Falsetta et al., 2014; Hajishengallis et al., 2017). This interaction enhances biofilm formation comprised of a diverse mixture of bacterial clusters, yeast cells and hyphal forms of the fungi that are enmeshed in an extensive matrix of extracellular polymeric substances (EPS), providing a protected microenvironment while potentiating virulence in vivo (Falsetta et al., 2014). The mixed biofilm is structurally and functionally complex and challenging to eradicate using conventional modalities due to enhanced tolerance to antimicrobials (Koo et al., 2017). Despite high prevalence of dental caries, treatment alternatives are unavailable and current anti‐plaque agents have limited efficacy (Autio‐Gold, 2008). To address these challenges, we designed a novel low‐cost approach using edible plant‐derived heterologous proteins to target pathogenic microbes and the EPS scaffold that can be incorporated into an affordable chewing gum for easy‐to‐use topical delivery at home.
Recent advances in biofilm research have highlighted emerging combinatorial antibiofilm strategies targeting the EPS matrix and microbes together or independent EPS components (Koo et al., 2017). Glucanohydrolases such as dextranase and mutanase can break down the EPS glucans in cariogenic biofilms (Ren et al., 2019) but have limited effects on microbial function/growth and other components of the matrix. Lipase displayed antifouling properties possibly due to the enzymatic degradation of lipids which play key roles for cellular integrity, biofilm formation and extracellular matrix in both bacteria and fungi (Prabhawathi et al., 2014). Given that glucans and lipids are major EPS components (Bowen et al., 2018; Koo et al., 2017; Ren et al., 2019), we hypothesized that a synergistic enzymatic combination targeting the multicomponent mixed‐kingdom structure could lead to effective biofilm disruption.
Based on the aforementioned rationale, lipase, dextranase and mutanase were expressed independently in chloroplasts to facilitate high‐level expression and to improve the stability after prolonged storage at ambient temperature. It also eliminated the prohibitively expensive technologies currently used for protein drug production including fermentation, purification, cold chain for storage/transportation and the formulation to stabilize purified proteins (Daniell et al., 2019a,b; Daniell et al., 2020; Kumari et al., 2019; Park et al., 2020). Antibiofilm activities of the enzymes (either alone or in combination) and their performance in disrupting the EPS matrix and the biofilm structure were investigated. To assess feasibility of topical drug delivery, chewing gum tablets were made with green fluorescent protein (GFP) expressing plant cells and protein stability upon prolonged storage (up to 3 years) and release kinetics after mechanical chewing were quantified using GFP fluorescence. Altogether, this study provides a singular conceptual approach to address a major technical and societal challenge—an affordable and effective way to treat dental biofilms and improve oral hygiene at home.
Results
Generation and characterization of marker‐free lettuce transplastomic lines expressing dextranase, mutanase, lipase
The Smdex gene (2574 bp, gene designation from Kim et al., 2011) encoding dextranase was isolated from S. mutans strain ATCC 25175 genomic DNA using PCR (Figure S2A,B) and fused to the PG1 (encoding antimicrobial peptide Protegrin‐1). The mut gene (3780 bp, gene designation from Otsuka et al., 2015) of Paenibacillus sp. encoding mutanase was codon optimized in order to improve its translation efficiency in plant chloroplasts based on psbA genes from 133 plant species as described previously (Kwon et al., 2016). Within total 1260 codons of mut gene, 576 codons including 327 rare codons were replaced by more highly preferred codons, resulting in an increased AT content from 44% to 57% (Figure S3, A and B). The Smdex, mut and lipY (encoding lipase, gene designation from Deb et al., 2006) genes (native or codon optimized) were cloned into the newly designed marker‐free chloroplast vector pLsLF‐MF as described previously (Daniell et al., 2019a,b; Daniell et al., 2020; Kumari et al., 2019; Park et al., 2020) and the constructed plasmids were then delivered by gene gun into lettuce (Lactuca sativa) cv. Simpson Elite leaves (Ruhlman et al., 2010). The successful Marker‐free events were identified by screening shoots for presence of the transgene cassette but absence of the antibiotic resistance gene aadA by PCR using primers described in the Experimental procedures section (Figure 1a).
Figure 1.
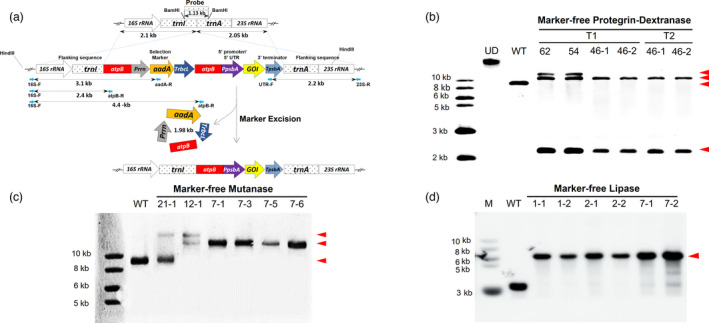
Generation of Marker‐free (MF) lettuce plants expressing dextranase, mutanase and lipase and evaluation of transgene integration, marker removal and homoplasmy. Schematic representation of the integration of two expression cassettes (gene of interest—GOI and selectable marker) into lettuce chloroplast genome via homologous recombination of flanking sequences: 16S rRNA‐trnI and tranA‐23S rRNA and subsequent removal of the antibiotic resistance gene via homologous recombination between two identical atpB regions. GOI represents PG1‐Smdex or mut or lipY. Probe indicates the DNA fragment region which was digested by Bam HI and used to detect hybridizing fragments in Southern blots. (a) Southern blots confirm PG1‐Smdex gene integration, marker removal and homoplasmy in transplastomic plants with 10.5 kb with 2.2 kb fragments, while 12.5 kb with 10.5 kb and 2.2 kb demonstrated heteroplasmy (with or without the aadA gene) after gDNA was digested with HindIII. Untransformed plant (WT) and undigested DNA (UD) were used as controls (b). In MF mutanase T0 generation, expected bands of 14.1 or 16.1 kb as a result of HindIII digested gDNA confirm mut gene integration in T0 generation plants, and the 14.1 kb band alone represents the homoplasmy (c). Expected band size of 5.6 kb obtained from SmaI digested gDNA confirms lipY gene integration, antibiotic marker gene removal and homoplasmy in lipase expressing T1 generation plants (d). Gene of interest band size is represented with arrowheads.
To characterize homoplasmic status of transplastomic lines, total plant gDNA was extracted from marker‐free Protegrin‐dextranase transplastomic plants, digested with HindIII, and probed with the DIG‐labeled trnI and trnA flanking sequence (Figure 1a). The 9.1 kb hybridizing fragment was only present in the untransformed wild‐type (WT) chloroplast genome, but not in the transplastomic lines, confirming their homoplasmic status (Figure 1b). Therefore, all copies (up to 10 000) of chloroplast genomes had the PG1‐Smdex gene cassette stably integrated, within the limits of detection. Two transplastomic lines (54 and 62) showed 10.5 and 12.5 kb bands, indicating a partial marker‐free removal process. All other transplastomic lines showed only the 10.5 kb hybridizing fragment, confirming a complete marker removal status. Most importantly, homoplasmic marker‐free PG1‐Smdex cassette was stably maintained in T1 and T2 generations of 46‐1 and 46‐2 lines in the absence of the antibiotic resistance gene (Figure 1b). The status of T0 transplastomic lettuce plants integrated with marker‐free mut construct was confirmed by southern blot using HindIII as well. Some lines (21‐1) showed both 16.1 kb and a 9.1 kb fragments suggesting a heteroplasmic status of chloroplast genomes (Figure 1c) with the marker gene. Additionally, line 12‐1 with the 14.1 kb and 16.1 kb bands, but without the 9.1 kb fragment suggested an incomplete removal of the antibiotic resistance gene. The presence of only the 14.1 kb fragment in all other lines confirmed their homoplasmic and marker‐free status of all chloroplast genomes (Figure 1c).
The stability and inheritance of lipY gene in T1 generation and its homoplasmic status was confirmed in the present study. The single hybridizing fragment of 5.6 kb in the transplastomic and 3.13 kb in untransformed WT plants after SmaI restriction digestion of gDNA was detected in Southern blot when probed with trnI/trnA genes, flanking the expression cassette (Figure 1d). The presence of a single larger 5.6 kb hybridizing fragment in all six tested transplastomic lines (compared to the 3.13 kb fragment in WT) confirmed the inheritance and stability of integrated lipY gene and the absence of marker gene in the T1 generation. Moreover, the absence of the 3.13 kb fragment (detected in WT) in each transplastomic plant confirmed their homoplasmic nature.
Characterization of plant‐derived glucanohydrolase and lipase activity
On the plate assay, protein crude extracts from all four tested leaf harvests (30 and 45 days of P‐46 and P‐47) and the purified commercial dextranase from Penicillium sp. (positive control) produced hallo rings on blue dextran, while no hallow formation was observed from untransformed WT plant extracts (Figure 2a). Expression level correlated with the maturity of leaves. In the quantitative assay, dextranase activity evaluated in transplastomic lines P‐46 and P‐47 at different stages of their growth (30 and 45 days) varied from 38.80 ± 2.04 to 59.24 ± 3.13 and 43.05 ± 2.32 to 60.68 ± 1.91 µmol/h/g dry weight, respectively, confirming again increased expression as leaves matured (Figure 2b). Enzyme release from freeze‐dried plant cells with or without sonication, showed similar enzyme activity from both preparations (Figure 2c), thereby eliminating the requirement for sonication for the release of protein from the plant powder, an important criterion for easy release of proteins from chewing gums described below. Similar levels of enzyme activity were observed in the plant extracts with or without protease inhibitor cocktail (PIC) used at the time of protein extraction (Figure 2d), suggesting that dextranase was resistant to proteases released in the plant crude extract during protein extraction. Statistical significance analysed by t‐test for dextranase enzyme activity was P < 0.001 (***). The calculated mutanase activity in mature leaves of transplastomic and untransformed WT lettuce were 33.68 ± 1.09 and 15.22 ± 0.43 µmol/h/g dry weight, respectively and statistical significance of mutanase was P < 0.05 (*, Figure 2e). Lower level of mutanase activity than dextranase is probably due to low level of expression in T0 generation but this typically increases 10–20‐folds in subsequent generations of transplastomic lines (Park et al., 2020).
Figure 2.
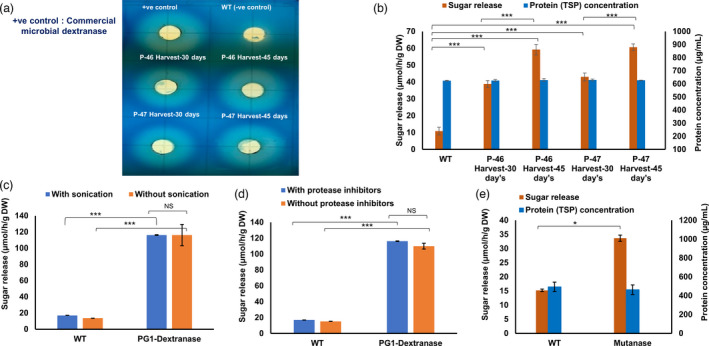
Chloroplast derived Marker‐free PG1‐dextranse and mutanase enzyme activity. Enzyme extracted from Marker‐free PG1‐dextranse lyophilized leaf powder from harvested leaves and evaluated for the qualitative assay against 0.5% blue dextran on plate (a), quantitative enzyme assay against 1% dextran (b), release of enzyme in the plant crude extract with or without sonication (c), and enzyme stability evaluation of protein extracted in presence/absence of protease inhibitors (d). Marker‐free mutanase enzyme activity (e). Enzyme activity calculated by measuring released reducing sugars and compared with the maltose standard. Assays were performed in triplicates and data represents mean and standard deviation. Statistical significance analysed by t‐test. Statistical significance was set at P < 0.05 (*), and P < 0.001 (***). WT represents untransformed wild‐type plant (–ve control); NS represents not significant.
Lipase activities in matured leaves of the transplastomic line and untransformed WT were 12 542.52 ± 257.03 and 522.76 ± 12.85 µmol/h/g dry weight, respectively with sonication and PIC in the extraction buffer. Lipase extracted without sonication but with PIC showed almost the same level of activity (Figure S1A), suggesting that the ultrasonic disruption was not required for enzyme release, making this a suitable candidate for the chewing gum approach. Interestingly, proteins extracted in the absence of PIC showed a 21% increase in the enzyme activity (Figure S1B), presumably due to the phenyl methyl sulfonyl fluoride (PMSF) or other components in PIC might have an inhibitory effect on the recombinant lipase.
Antibiofilm activity of plant derived and purified commercial lipase
Efficacy of the plant‐derived lipase was evaluated by employing a mixed‐kingdom biofilm model and a treatment regimen based on topical exposure (Figure S5). Purified commercial lipase of equivalent enzyme activity unit was tested as positive control. The data revealed that treatment with plant‐lipase extract or purified lipase significantly inhibited Candida (in cyan) hyphal formation, a key factor for cross‐kingdom interaction and biofilm development, and reduced bacterial (in green) and EPS glucans (in red) accumulation (Figure 3a). Fungal cells were mostly in yeast form (Figure 3a, yellow arrow heads) with less bacterial clusters and more dispersed cells (white arrow heads), whereas the EPS matrix formation was also disrupted.
Figure 3.
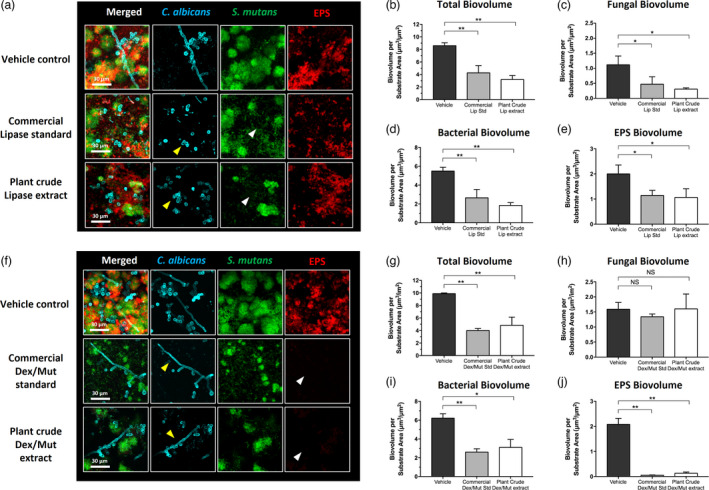
Antibiofilm effects of commercial and plant‐derived enzymes against bacterial‐fungal mixed biofilms. Commercial purified enzymes of the same activity unit as measured in the plant crude extracts (333.3 U/mL for lipase and 7.08/0.84 U/mL for dextranase/mutanase, respectively) were used as standards to evaluate the antibiofilm efficacy of the plant crude extracts. (a) Confocal images showing the antibiofilm efficacy of commercial and plant‐derived lipase. (b–e) Quantitative computational analysis of the confocal biofilm images treated with commercial and plant‐derived lipase. (f) Confocal images showing the antibiofilm efficacy of commercial and plant‐derived Dextranase/Mutanase combination. (g–j) Quantitative computational analysis of the confocal biofilm images treated with commercial and plant‐derived Dextranase/Mutanase combination. For multi‐channel confocal images, C. albicans cells (yeasts or hyphae) are depicted in cyan; S. mutans cells are depicted in green; The EPS glucan matrix is depicted in red. For the computational data, the title of each graph indicates the channel(s) used for individual analysis. *, P < 0.05; **, P < 0.01 (one‐way analysis of variance with Tukey’s multiple comparisons test). Enzyme units of lipase and dextranase/mutanase represent µmol of pNP and reducing sugar produced in 1 h, respectively.
We also performed quantitative computational analysis using the images acquired via confocal microscopy. The plant‐lipase crude extract significantly reduced the total biovolume of the mixed biofilm (Figure 3b, >50% reduction compared to vehicle‐control, P < 0.01). Further analysis of each fluorescence channel revealed reduction of both the fungal (Figure 3c, P < 0.05) and bacterial biovolume (Figure 3d, P < 0.05), consistent with the confocal images (Figure 3a). Notably, the plant‐lipase extract was as effective as the purified commercial lipase (if not slightly better) in disrupting biofilms. The total amount of EPS glucan was significantly reduced (Figure 3e, P < 0.05) in the treated biofilms. Altogether, the data suggest that the plant‐lipase extract potently inhibits fungal filamentation (a novel finding) that eventually reduced total biovolume of the mixed biofilm, revealing its potential to replace the commercial purified enzyme based on antibiofilm efficacy. Lipase ability to inhibit C. albicans hyphal formation was evaluated in C. albicans monoculture as well. As expected, similar finding was obtained (Figure S6).
Antibiofilm activity of plant‐derived and commercial purified dextranase and mutanase
To investigate whether the plant‐derived glucanohydrolases could be used as antibiofilm therapeutics, we performed the bioassays using a mixture of dextranase and mutanase (5:1 activity ratio) that was optimized previously using purified enzymes to provide maximum matrix‐degrading activity (Ren et al., 2019). Commercial purified dextranase and mutanase (at the same ratio) was used as positive‐control. The data showed that both plant‐dextranase/mutanase extract and the equivalent purified enzymes were highly effective in disrupting EPS glucans, resulting in near abrogation of glucan‐matrix in the mixed‐kingdom biofilm (Figure 3f, white arrow heads). Treatment with the dual‐enzyme formulation also resulted in less bacterial clusters accompanied by cellular dispersion (Figure 3f, green channel), thereby reducing the density of bacterial accumulation. However, topical exposure of plant‐derived or commercial dextranase/mutanase showed no effects on fungal hyphal production (Figure 3f, yellow arrow heads).
Further computational analysis confirmed the inhibitory effects exerted by glucanohydrolases. As expected, the EPS was effectively degraded compared to vehicle control (>90% reduction, P < 0.01; Figure 3j). Bacterial accumulation and overall biofilm volume were also significantly reduced in biofilms treated with commercial purified or plant‐derived glucanohydrolases (P < 0.01; Figure 3i and P < 0.01; Figure 3g). In contrast, Candida was minimally affected after the treatment by both enzyme preparations (P > 0.05; Figure 3h), consistent with limited effects on fungal hyphal formation. The data suggest that plant‐derived dextranase/mutanase can effectively disrupt EPS glucan matrix with equivalent potency to that of commercial enzymes. However, these glucanohydrolases display limited effects on fungal accumulation, indicating that combination with lipase may result in a more effective multitargeted approach against mixed‐kingdom biofilms.
Cumulative effect of dextranase/mutanase and lipase on biofilm accumulation
To develop a therapeutic solution for fungal‐bacterial mixed biofilms, we employed a combinatorial approach using dextranase/mutanase and lipase to enhance the antibiofilm efficacy. The data show that the topical treatment with dextranase/mutanase and lipase is remarkably effective, resulting in near‐complete suppression of mixed‐biofilm formation (Figure 4a). The multi‐enzymatic activity eliminated bacterial clustering with only few dispersed S. mutans (predominantly single cells) with minimal EPS glucan matrix (Figure 4a, magnified view in left panel). Notably, only few C. albicans yeast cells were attached on the surface whereas hyphal formation was abrogated (Figure 4a). Quantitative computational analysis confirmed the potent inhibition of fungal/bacterial and EPS biovolumes by the multi‐enzyme treatment (Figure 4b–e), which could not be achieved with dextranase/mutanase or lipase alone.
Figure 4.
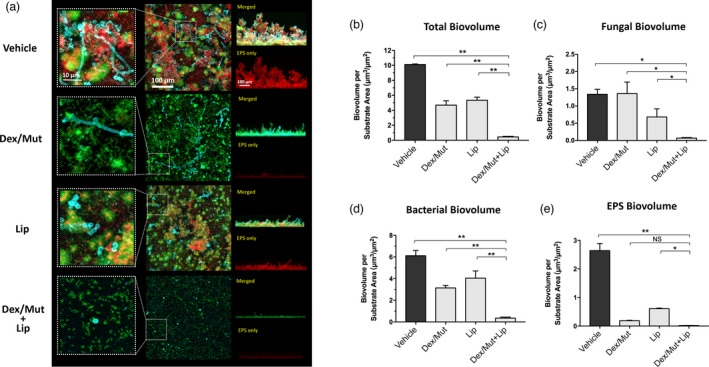
Prevention of fungal‐bacterial mixed biofilm by topical sequential treatment of commercial Lipase and Dextranse/Mutanse combination. Commercial enzymes of the optimum activity units for bioactivty (1000 U/mL for lipase and 525/105 U/mL for dextranase/mutanase, respectively) were used. (a) Three‐dimensional confocal images of the fungal‐bacterial mixed biofilm formed after the topical sequential treatments. C. albicans cells (yeasts or hyphae) are depicted in cyan; S. mutans cells are depicted in green; The EPS glucan matrix is depicted in red. Representative merged biofilm images are displayed in the middle panel, while a magnified (close‐up) view of each small box is positioned in the left panel. Lateral (side) views of each biofilm are displayed at the right panel (the merged image at the top and the EPS channel at the bottom). (b–e) Quantitative computational analysis of the confocal images. The title of each graph indicates the channel(s) used for individual analysis. *P < 0.05; **P < 0.01 (one‐way analysis of variance with Tukey’s multiple comparisons test). Enzyme unit of lipase and dextranase/mutanase represent µmol of pNP and reducing sugar produced in 1 h, respectively.
To further investigate the impact of the treatment on the biofilm accumulation, we determined the Total Biofilm Inhibition (TBI) index (Figure 5a), which evaluates the combined net effects on the reduction of fungal colony forming unit (CFU), bacterial CFU and biomass (dry‐weight), as detailed in the Experimental procedures section. The combination treatment (dextranase/mutanase and lipase) resulted in significantly lower TBI (0.007 ± 0.003) than either dextranase/mutanase (0.665 ± 0.070) or lipase alone (0.158 ± 0.050) (Figure 5a), suggesting synergistic inhibitory effects. We also assessed cell viability via in situ confocal imaging and fluorescent labeling of live and dead bacterial/fungal cells within intact mixed‐kingdom biofilms. Interestingly, in addition to dispersion of bacterial clusters and inhibition of fungal hyphae (Figure 5b, upper panel), the biofilms treated with dextranase/mutanase and lipase harbored mostly dead C. albicans in yeast form (Figure 5b, lower panel, white arrow heads). In contrast, fungal cells (yeast and/or hyphae) in biofilms treated with dextranase/mutanase or lipase alone were mostly viable. Altogether, we demonstrate a feasible approach using EPS‐degrading enzymes to potently disrupt mixed‐kingdom biofilms by reducing both the microbes and matrix components.
Figure 5.
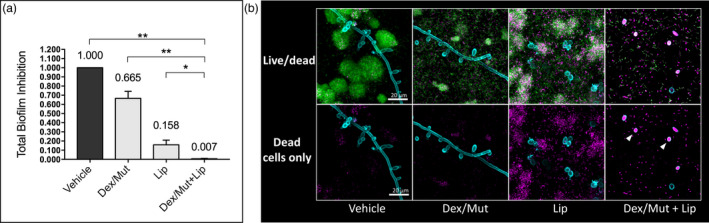
Viability of the fungal‐bacterial mixed biofilm after sequential treatments with commercial Lipase and Dextranse/Mutanse combination. (a) Total Biofilm Inhibition (TBI) index of the treatments. TBI = I fungal CFU × I bacterial CFU × IDW, where Inhibition of bacterial/fungal CFU or ICFU = (CFUtreatment/CFUcontrol) × 100%, and Inhibition of Dry Weight or IDW = (DWtreatment/DWcontrol) × 100%. *, P <0.05; **, P <0.01 (one‐way analysis of variance with Tukey's multiple comparisons test). (b) Live/dead staining of the fungalbacteria mixed biofilms. Live cells are depicted in green; Dead cells are depicted in magenta. C.albicans cell wall is depicted in cyan to indicate the location of fungal cells. The optimum activity units (U) were used for commercial purified lipase (1000U/mL) and Dex/Mut (525/105U/mL) in the experiments. Enzyme unit of lipase and dextranase/mutanase represent μmol of pNP and reducing sugar produced in 1h, respectively.
Chewing gums, protein stability and functionality
We tested the feasibility of incorporating the plant‐derived proteins in chewing gum as an alternative, easy‐to‐use and more affordable delivery approach (Figure 6a,b). In order to study release of proteins impregnated in chewing gums, we utilized plant cells expressing GFP in ground powder form after lyophilization (Gupta et al., 2015; Lee et al., 2011). Transplastomic lettuce expressing GFP‐protegrin was grown in a Fraunhofer cGMP hydroponic facility, lyophilized and powdered as described previously (Daniell et al., 2019a,b; Daniell et al., 2020; Kumari et al., 2019; Park et al., 2020; Su et al., 2015). Chewing gum tablets using ground plant powder were made by MastiX LLC, (Hunt Valley, MD) through compression process that is advantageous over traditional gum manufacturing process which requires higher temperature (93°C) and extrusion/rolling that introduces variability in the concentration of the active ingredient. Gum tablets chews and performs exactly like conventional chewing gum based on taste, softness and compression.
Figure 6.
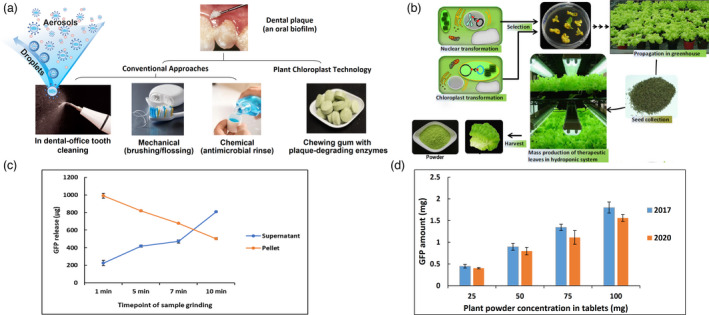
A proposed anti‐plaque chewing gum concept containing enzymes expressed in chloroplasts. (a) Current plaque control modalities and the chewing gum prototype using the chloroplast technology. Conventional mechanical brushing/flossing requires appropriate cumbersome techniques and suffers from low compliance. Chemical approaches e.g. antimicrobial mouthwash has limited efficacy against cariogenic dental plaque and are costly. Dental clinic tooth cleaning generates significant amounts of droplets and aerosols, posing potential risks of infection transmission, including COVID‐19. (b) Steps in creation and mass production of lettuce plants expressing enzymes. (c) Estimation of the GFP release from the 25 mg chewing gum tablet. The GFP released in saliva and the remaining pellet after grinding at 1, 5, 7, and 10 min time points. (d) GFP activity remained stable in chewing gum tablets containing 25, 50, 75 and 100 mg lyophilized plant powder after storage at room temperature for 3 years
There was no detectable loss of protein in gum tablets preparation process based on GFP quantification (Figure S7). Release of GFP from gum tablets was studied using a Universal Mechanical Testing Machine by placing chewing gum pellets in 10 mL of artificial saliva in a polycarbonate chamber and loaded cyclically in compression using a piston attached to a load cell (Figure S8) under conditions described in Supplementary Figures and table (Figure S9A–C, and Table S1). GFP‐protegrin concentration in saliva increased from 225 µg/mL in 1 min to 809 µg/mL in 10 min in the supernatant and decreased from 988 µg/mL at 1 min in the pellet to 502 µg/mL at 10 min, confirming steady release during the chewing process (Figure 6c). Quantification of GFP in gum tablets showed that GFP was stable in gum tablets when stored at ambient temperature for 3 years (Figure 6d), indicating a potential long shelf life of the final product.
Discussion
In order to address the high cost of production and delivery of recombinant therapeutic proteins, the Daniell laboratory has pioneered a novel concept to express protein drugs (PDs) in plant chloroplasts and orally deliver them through bioencapsulation within plant cells (Daniell et al., 2016; Daniell et al., 2021; Daniell et al., 2019a,b). Thin lettuce leaves facilitate rapid removal of water through lyophilization and offer an ideal system for expression and delivery of PDs. This platform is currently advanced to deliver therapeutic proteins in the clinic, yet it remains unexplored in dental medicine. Mechanical disruption using manual or electric toothbrushes can remove dental plaque, but they are cumbersome to use with low compliance while being costly, whereas current antimicrobial agents to treat cariogenic plaque are inefficient due to the presence of EPS matrix (Autio‐Gold, 2008; Koo et al., 2017; Ren et al., 2019). Matrix‐degrading enzymes can effectively target the biofilm structure and enhance antimicrobial killing, while simultaneously weakening its mechanical stability, promoting bacterial removal (Autio‐Gold, 2008; Koo et al., 2017; Ren et al., 2019). However, the development of clinically feasible approaches is hindered by high costs for mass production as enzymes require complex microbial fermentation and expensive purification procedures. Here, we report for the first time the expression of mutanase and dextranase in plant cells and synergistic efficacy when combined with plant‐derived lipase for oral biofilm prevention. Although microbial dextranase has been extensively used in food industry with demonstrated safety (Purushe et al., 2012), dextranase has never been expressed in plant cells. Importantly, creation of marker‐free edible plant cells expressing enzymes with high yield and stability eliminates the need for purification, providing a translatable therapeutic approach.
Current chemical modalities to treat biofilm‐associated infections are primarily targeting individual bacterial or fungal components despite increasing evidence highlighting the importance of the extracellular EPS matrix in biofilm antimicrobial tolerance (Koo et al., 2017). In addition, targeting EPS can also disrupt the viscoelastic properties of biofilms and further weaken its mechanical stability, promoting bacterial dispersal or removal (Ren et al., 2019). Our data show that the lipase, dextranase and mutanase crude plant extracts have potent antibiofilm efficacy that can replace the costly purified enzyme standards of equivalent enzyme units. Although lipase has been approved by FDA to treat several metabolic disorders it remains underexplored to treat dental plaque. In the present study, lipase showed remarkable reduction in fungal load within treated biofilm by abrogating hyphal formation, a crucial step in cross‐kingdom biofilm development as hyphae provide a scaffold for co‐species adhesion and EPS accumulation (Prabhawathi et al., 2014). The precise role of lipid in biofilm formation remains unclear, but recent data suggest that lipids appear to be important membrane components modulating fungal morphogenesis and hyphal elongation (Rella et al., 2016). Hence, it is possible that lipase treatment could inhibit the filamentous Candida growth impacting cross‐kingdom biofilm scaffolding and accumulation. Conversely, plant‐derived dextranase and mutanase degraded EPS glucan effectively inhibiting the matrix development and bacterial accumulation. However, fungal cells were unaffected by dextranase and mutanase whereas detectable EPS was still present post‐treatment, indicating that inclusion of lipase was crucial to achieve the near‐complete suppression of the mixed‐kingdom biofilm.
The lack of intrinsic antimicrobial activity of therapeutic enzymes such as dextranase and mutanase has been an important limitation for their application alone as antibiofilm therapeutics (Koo et al., 2017). Interestingly, lipase has been shown to have antimicrobial properties possibly through degradation of the membrane lipids (Prabhawathi et al., 2014; Seghal Kiran et al., 2014). Using high‐resolution confocal microscopy, we found that the combination of glucanohydrolases and lipase displayed enhanced antimicrobial activity against both species in the biofilms, whereas no significant killing was observed with either glucanohydrolases or lipase alone. The data also revealed that degradation of EPS glucans by dextranase and mutanase can locally expose the embedded S. mutans and C. albicans cells within the treated biofilm, which could provide access for lipase to the fungal/bacterial cell surface causing microbial death. Thus, the three‐enzyme combination treatment suppressed the pathogenic fungal‐bacterial biofilm formation via a complementary mechanism whereby the effective EPS degradation by glucanohydrolases with antimicrobial effects of lipase in situ potentiated antibiofilm efficacy. Further studies are needed to elucidate the detailed mechanisms by which the multi‐enzyme approach targets the mixture of different bacterial and fungal cells and degrades the complex biofilm structure spatiotemporally. Because the combination of chloroplast dextranase, mutanase and lipase is able to degrade both the EPS matrices and display antimicrobial properties, it would be interesting to evaluate this combined effect on the pre‐formed biofilm in future studies.
The use of chewing gum supplemented with antimicrobial and antibiofilm therapeutics to improve treatments of dental diseases has been proposed for decades as a promising alternative in oral health care applications (Wessel et al., 2016). Yet, there are no such products currently available owing to high costs for inclusion of therapeutic additives and practical formulation development. Due to their prominent biofilm‐degrading activity, dextranase and mutanase have been previously added to chewing gum to specifically target formed biofilm (Murai et al., 1975; Kolahi and Abrishami, 2013). However, suboptimal level of enzymes for antibiofilm efficacy, enzymatic stability issues and lack of antimicrobial activity impacted clinical translation (Koo et al., 2017; Liu et al., 2016). Furthermore, recent data indicate that the combination of dextranase and mutanase at specific amounts and ratios (as employed here) is required to maximize EPS degradation, whereas inclusion of antibacterial agents is needed to display antimicrobial action against pre‐formed biofilms (Ren et al., 2019). Here, we demonstrated the feasibility of using chewing gum to release plant‐derived proteins in saliva solution following mechanical bite forces. Furthermore, the proteins were stable in chewing gum stored at ambient temperature (up to 3 years), indicating a practical and easy‐to‐use topical delivery platform. Altogether, we provide a conceptual framework for plant made biofilm‐degrading enzymes and chewing gum‐based protein delivery that may lead to an innovative and affordable approach of controlling dental biofilm. This strategy could help address the societal issue of inequity to access dental care services by enabling a low‐cost technology for improvement of oral health, which may be also relevant during the current global crisis caused by COVID‐19.
Experimental procedures
Codon optimization of the mut gene
Daniell lab previously developed a codon usage reference table for codon optimization based on codon usage frequency of psbA gene in 133 plant species (Kwon et al., 2016). Native mutanase coding mut gene nucleotide sequence from Paenibacillus sp. was codon optimized by replacing less preferred codons with more preferred ones, which eventually generated codon usage frequency close to that in reference psbA gene. Rare codons in the native mut gene sequence with a frequency <5% in the reference were replaced by more preferred codons (Figure S4).
PG1‐Smdex and mut gene (co) cloning in pLsLF‐marker‐free chloroplast vector
The native Smdex gene sequence from S. mutans (ATCC 25175) fused with PG1 (Protegrin‐1 encoding) at downstream and codon optimized mut gene were synthesized (GenScript Biotech, Piscataway Township, NJ) (Liu et al., 2016). The PG1‐Smdex and synthesized mut (co) genes were cloned into pLsLF‐marker‐free vector using NdeI and PshAI restriction sites and transformed into TOP10 E. coli cells. Daniell lab lettuce chloroplast transformation vector pLsLF was used as a backbone, which contains spectinomycin‐resistant gene (aadA, aminoglycoside 3´‐adenylytransferase gene) as selectable marker. To design a marker‐removable vector, the 649‐bp long direct repeat DNA sequence, derived from atpB promoter and 5′ UTR (Daniell et al., 2019a,b; Kumari et al., 2019), was PCR amplified using lettuce total genomic DNA as a template and then the sequence‐confirmed direct repeats were cloned to flank aadA expression cassette. For the insertion of single‐digested atpB fragments into the vector backbone, NEBuilder HiFi DNA (NEB, Ipswich, MA) assembly kit was used to avoid the possible ligation of the fragments in a reverse direction. The successful insertions were confirmed by restriction digestion and expression of PG1‐dextranase and mutanase in E. coli was confirmed by Western blot. The pLsLF‐MF‐PG1‐dextranase and pLsLF‐MF‐mutanase (co) plasmid were extracted using PureYieldTM plasmid Midiprep System (Promega, Madison, WI) and used for subsequent particle bombardment.
Generation and molecular characterization of marker‐free transplastomic lettuce lines
The pLsLF‐MF‐PG1‐dextranase and pLsLF‐MF‐mutanase (co) plasmids were transformed into 1‐month‐old lettuce (L. sativa) leaves by bombardment as previously described (Lee et al., 2011). After the bombardment, lettuce leaves were cut into small pieces (<1 cm2) and grown on regeneration media containing spectinomycin (50 mg/mL) as described previously (Daniell et al., 2019a,b; Kumari et al., 2019; Lee et al., 2011; Ruhlman et al., 2010). The integration of pLsLF‐MF‐PG1‐dextranase and pLsLF‐MF‐mutanase (co) vectors in regenerated shoots were validated by Southern blot and PCR using specific primers sets:
16S‐F, 5′‐CAGCAGCCGCGGTAATACAGAGGATGCAAGC
aadA‐R, 5′‐CCGCGTTGTTTCATCAAGCCTTACGGTCACC;
atpB‐R, 5′‐GAATTAACCGATCGACGTGCTAGCGGACATT;
UTR‐F, 5′‐ AGGAGCAATAACGCCCTCTTGATAAAAC
23S‐R, 5′‐TGCACCCCTACCTCCTTTATCACTGAGC
The PCR positive leaves were subjected to the second round of selection on regeneration media containing spectinomycin (50 mg/L). Any regenerated shoots showing bleached leaves were immediately evaluated by PCR analysis to confirm the excision of aadA gene and were then transferred to spectinomycin‐free rooting media to induce roots. Once the roots were formed, homoplasy was confirmed by Southern blots as described below. Expression of enzymes in homoplasmic lines were confirmed using Southern blots and enzyme assays as described previously (Daniell et al., 2019a,b; Kumari et al., 2019; Ruhlman et al., 2010; Verma et al., 2008).
Southern blotting of marker‐free lettuce plants
For Southern blotting, 2 μg total genomic DNA from untransformed WT, T1 and T2 generation marker‐free plants integrated with PG1‐Smdex, lipY and T0 generation integrated with mut (co) were digested by suitable restriction enzymes and separated in 0.8% agarose gel, transferred onto the nylon membranes (Nytran, GE Healthcare), and probed as described previously (Kumari et al., 2019; Kwon et al., 2018). Seeds from untransformed WT and previously developed three independent T0 transplastomic lipase (Kumari et al., 2019) expressing plants were germinated on ½ MS medium without any antibiotics. The germinated seedlings were transferred and grown in magenta box. The genomic DNA from leaves of two different T1 plants in each of the three independent events (in total six plants) and WT was isolated, and digested with SmaI restriction enzyme (New England Biolabs, Hertfordshire, UK).
Plant‐derived enzymes activity assay
Leaves from marker‐free transplastomic plants expressing lipase, PG1‐dextranase, mutanase and untransformed WT plants were stored at −80°C and freeze‐dried in a lyophilizer as described previously (Daniell et al., 2019a,b). Lyophilized leaves were ground into fine powder in a coffee blender and used as the source material for the proteins/enzyme extraction. Total soluble proteins (TSP) was extracted by suspending 50 mg of plant powder in 1 ml plant extraction buffer (respective buffer of each enzymes and EDTA‐free protease inhibitor cocktail (Thermo Scientific, Waltham, MA, USA) and kept on a mixer (Eppendorf) at 4°C for 1 h. Samples were sonicated, centrifuged at 9391 g for 30 min and the supernatant (TSP) was collected. TSP was quantified by Bio‐Rad protein assay dye (Bio‐Rad, Hercules, CA, USA) by following the manufacturer’s protocol. Bovine serum albumin (BSA) protein was used as standard. The impact of sonication during extraction of TSP and the role of PIC in the extraction buffer was evaluated by independent experiments. The stability of recombinant enzyme in each crude extract was evaluated by activity assay and compared.
Dextranase assay
Leaves harvested at two different time points (30 and 45 days) from two independent marker‐free PG1‐dextranase transplastomic events (Plant‐46 and Plant‐47) were lyophilized, ground into powder and evaluated for the enzyme activity. Blue dextran plate assay was performed for qualitative enzyme activity analysis. Blue dextran substrate (from Leuconostoc mesenteroides, Sigma, Louise, MO, USA) and agar were added into 100 mm sodium acetate buffer (pH 5.5) at final concentration of 0.5% and 1.25% respectively. The suspension was boiled, mixed properly and poured into the plate. After solidification, small wells were created and 50 µg of plant crude extract (TSP) from PG1‐dextranase transplastomic plants and WT plant (as negative control) was loaded into the wells. The plate was incubated at 37°C overnight. Enzymatic activity of PG1‐dextranase was visualized in the form of a hallo caused by the breakdown of blue dextran around the well. Purified dextranase enzyme from Penicillium sp. (Sigma) was used as positive control.
Enzyme assay was performed to quantify dextranase activity in the crude extract of transplastomic PG1‐dextranase plants. TSP of PG1‐dextranase transplastomic and WT plants (50 µL) was incubated with 50 µL of dextranase (1% in 100 mm sodium acetate pH 5.5), incubated at 37°C for 1h and the released sugar was estimated by dinitrosilycilic acid method (Kumari et al., 2019) using maltose as standards. The enzymatic assay was performed in triplicates and the data are presented as mean and standard deviation. The enzyme activity was represented as sugar released µmol/h/g dry weight of plant powder. The importance of sonication for the release of PG1‐dextranase enzyme from the plant powder and PIC for the stability of the released enzyme in the crude plant extract was also evaluated. Plant powder of 45 days old leaves from plant‐46 was used for these experiments.
Lipase assay
Lipase enzyme assay was performed using the method described previously (Kumari et al., 2019). Briefly, TSP was extracted from the lyophilized plant powder in 100 mm sodium phosphate buffer (pH 8.0) and was quantified. In the assay, 50 µL of TSP and 100 mm p‐nitrophenyl butyrate (5 µL) was mixed in 450 µL reaction buffer (100 mm sodium phosphate buffer pH 8 and 0.9% NaCl) and incubated at 37°C for 10 min. Released p‐nitrophenol (hydrolysed product of p‐nitrophenyl butyrate) was estimated by using different known concentrations of p‐nitrophenol as standards. The enzyme assay was performed in triplicate and data are presented as mean and standard deviation. The enzyme activity was represented as p‐nitrophenol released µmol/h/g dry weight. The effect of sonication during the protein extraction and stability of enzyme in the absence of PIC was also examined for plant‐derived lipase as described earlier.
Mutanase assay
Lyophilized plant powder was extracted with a ratio of 50 mg powder/1 mL extraction buffer [0.1 m sodium acetate buffer (pH 5.5)], followed by vortex homogenization at 4°C for 1 h (Eppendorf 5432) and sonication for 3 cycles (5 s on, 10 s off, 80% amplitude). The homogenate was centrifuged at 9391 g for 30 min and the supernatant was transfer into a new tube. Then the supernatant was dialysed as follows: 10 mL of plant protein extraction was sealed in 15 cm of semi‐permeable membrane (MWCO: 20 KD) and placed in 2 L of extraction buffer for 4 h. This buffer was replaced with fresh buffer and incubated at 4°C overnight (16 h). The total protein concentration was determined by Bradford assay. The activity of mutanase in the dialysed plant crude was determined by enzymatic assays as described with some modifications (Kopec et al., 1997; Verma et al., 2008). Crude extracts of transplastomic plants were incubated with the substrate (mutan or α‐1, 3‐linked glucans) in 0.1 M sodium acetate buffer (pH 5.5) at 37°C for 60 min. The amount of reducing sugar released was determined by the Somogyi‐Nelson method (Somogyi, 1945). Crude extracts of untransformed plant (WT) with equal protein concentrations were used to check the endogenous mutanase activity from plant cells. One unit (U) of mutanase activity was defined as the amount of enzyme that releases 1.0 μmol of reducing sugar from mutan per hour at pH 5.5 at 37°C. Mutanase (EC3.2.1.59) purified from Bacillus sp. fermentation (Amano Enzyme, Japan) was used as the commercial enzyme standard.
Preparation of chewing gum and GFP stability and release assay
Chewing gum tablets containing ground plant powder were prepared by MastiX LLC, (Hunt Valley, MD) by compression process but not the traditional gum manufacturing process which requires higher temperature and extrusion/rolling that introduces variability in the concentration of the active ingredient. Gum tablets contained the gum base (27.71%), sorbitol (17.18%), maltitol (14.78%), xylitol (13.86%), isomalt (10.07%), natural and artificial flavors (7.21%), magnesium stearate (2.95%), silicon dioxide (0.82%), stevia (0.42%) and plant cell powder (5%) in order to offer the best flavor, taste, softness and compression. The gum tablet chews and performs exactly like the conventional chewing gum based on physical characteristics.
For stability assay, 125 mg of gum tablet was ground with 500 µL protein extraction buffer [(0.2 M Tris HCl pH 8.0, 0.1 M NaCl, 0.01 M EDTA, 0.4 M sucrose, 0.2% Triton X supplemented with 2% PMSF and a PIC (Pierce, Waltham, MA, USA)], followed by sonication at 80% amplitude for 5 s for two times. After sonication, the samples were centrifuged at 13 523 g at 4 °C for 10 min. Then 100 µL of supernatant was loaded to a fluorescent microplate reader where the GFP was detected at 485 nm (excitation) and 538 nm (emission) using the commercial GFP (Vector Laboratories, San Diego, CA, USA) as a standard. Release of GFP from gum tablets was studied using a Universal Mechanical Testing Machine equipped with Merlin software (Instron Model 5564, Norwood, MA) in cyclic loading mode. Chewing gum tablets (25 mg) were placed in 10 mL of artificial saliva (Pickering Laboratories, Mountain view, CA) in a polycarbonate chamber and loaded cyclically in compression using a piston attached to a load cell (Figure S8A,B). A load range of −1.5 to −500N for intervals of 1, 5, 7 and 10 min and cycles of 55, 287, 364 and 591 (Table S1) to simulate human chewing. Figures S9 (A, B and C) show loading and unloading cycles for different time durations. Parameters for simulated chewing are summarized in Table S1. A wide range of bite forces have been reported for adult humans. Values ranging from 1300N to 285N have been reported (Takaki et al., 2014; Yong, 2010). Varga et al. (2011) reported mean bite force values of 522N for males and 465N for females with normal occlusion. A representative compressive load of 500N was selected for this study. The GFP concentrations in both the supernatant and the pellet after 1, 5, 7, and 10 min were determined by the same above‐mentioned method.
Microorganisms used in biofilm studies and growth conditions
Candida albicans SC5314, a well‐characterized fungal strain and Streptococcus mutans UA159 serotype c (ATCC 700610), a cariogenic dental pathogen and well‐characterized EPS producer were used in cross‐kingdom biofilm experiments. To prepare the inoculum used in this study, C. albicans (yeast form) and S. mutans cells were grown to mid‐exponential phase in ultrafiltered (10‐kDa molecular‐mass cutoff membrane; Millipore, MA) tryptone‐yeast extract broth (UFTYE; 2.5% tryptone and 1.5% yeast extract) with 1% (wt/vol) glucose at 37°C and 5% CO2 as described previously (Falsetta et al., 2014).
In vitro biofilm model and topical treatment regimen
Biofilms were formed using saliva‐coated hydroxyapatite (sHA) disc model as detailed previously (Falsetta et al., 2014; Hwang et al., 2017). Briefly, the hydroxyapatite discs (surface area, 2.7 ± 0.2 cm2; Clarkson Chromatography Products, Inc., South Williamsport, PA) coated with filter‐sterilized, clarified whole saliva were vertically suspended in a 24‐well plate using a custom‐made disc holder (Figure S5A), mimicking the dental enamel surface. The fungal‐bacterial inoculum containing approximately 2 × 106 CFU/mL of S. mutans and 2 × 104 CFU/mL of C. albicans (in yeast form) at mid‐exponential growth phase in UFTYE (pH 7.0) with 1% (wt/vol) sucrose; this proportion of the microorganisms is similar to that found in saliva samples collected from children with early childhood caries (ECC) (de Carvalho et al., 2006). Biofilms were maintained at 37°C under 5% CO2. To access the antibiofilm efficacy of the plant crude extracts, we developed a topical treatment regimen for feasible clinical applications (Figure S5B). The sHA surface (a tooth surrogate) was topically treated with plant leaf crude extract for 60 min at 37°C to mimic the first application after toothbrushing. The sHA disks were inoculated with the culture medium containing the bacterial‐fungal inoculum and the mixed biofilms were allowed to initiate under cariogenic (sucrose‐rich) conditions for 6 h. The second topical treatment was then performed using the same plant crude extract with 60 min exposure to mimic a repeated application. After that, the treated sHA disks were transferred back to the culture medium for continued biofilm development.
Microbiological analysis of the biofilms
Biofilms were grown until 19 h and were subject to microbiological analyses, including the total number of viable cells (CFU) of bacteria/fungi and the total biomass on each sHA disk (dry weight) as detailed elsewhere (Hwang et al., 2017). Briefly, at 19 h, the biofilms were harvested from the sHA disks and homogenized via optimized sonication procedure, which does not kill fungal or bacterial cells while providing maximum recoverable counts (Koo et al., 2013). Aliquots of biofilm suspensions were serially diluted and plated on blood agar plates and the plates were grown at 37°C under 5% CO2 for 2 days. Bacterial and fungal viability in the biofilm was accessed by determining their respective CFU recovered on the blood agar plates. The amount of biofilm dry weight (biomass) was also determined.
Three‐dimensional confocal biofilm imaging and quantitative analysis
The impact of the topical treatments was assessed by examining the 3D architecture and the spatial distribution of Gtf‐derived EPS glucans and fungal/bacterial cells within live biofilms using our well‐established protocols optimized for biofilm imaging and quantification (Falsetta et al., 2014; Hwang et al., 2017). Briefly, the EPS glucan matrix was labelled via incorporation of AlexaFluor 647 dextran conjugate (Molecular Probes Inc., Eugene, OR) throughout the biofilm formation. This labelling method is highly specific for Gtf‐derived α‐glucan as the fluorescently labelled dextran serves as a primer for glucan synthesis which is directly incorporated into the EPS matrix over the course of biofilm development. S. mutans was stained with Syto 9 (Molecular Probes), while C. albicans cell wall was labeled with Concanavalin A‐tetramethyl rhodamine conjugate (Molecular Probes). High‐resolution confocal imaging was performed using confocal laser scanning fluorescence microscope (LSM800 with Airyscan, Zeiss, Germany) equipped with a 20× (1.0 numerical aperture) water immersion objective. Each biofilm was scanned at five randomly selected areas, and confocal image series were generated by optical sectioning at each of these positions. Computational analysis of confocal images using the advanced biofilm 3‐dimensional analysis tool BiofilmQ (https://drescherlab.org/data/biofilmQ) was conducted to determine the biovolumes of bacteria, fungi and EPS in order to complement our microbiological analysis (Hartmann et al., 2021). ImageJ (FIJI) was used for post‐acquisition image processing and creating 3‐dimensional renderings of biofilm architecture (Schindelin et al., 2012).
In situ cell viability staining and imaging
To investigate the impact of the treatments on bacterial/fungal cell viability within the mixed biofilm, we performed in situ cell viability staining followed by detailed imaging at single‐cell resolution using cell membrane integrity as a biomarker for viable cells. We used TOTO‐3 (Molecular Probes), a cell impermeable dimeric cyanine acid dye as a dead cell indicator for both bacterial and fungal cells because of its high affinity for nucleic acids (Chiaraviglio and Kirby, 2014). Thus, when the microbial cells are killed and the plasma membrane integrity are compromised, these probes will enter cells, bind to nucleic acids, and exhibit a strong fluorescence. Biofilms were stained using the optimized 1 µm of TOTO‐3 in 0.9% sodium chloride at 37°C for 10 min and were counterstained with 0.65 µm SYTO9 (a cell‐permeable dye). Concanavalin A‐tetramethylrhodamine conjugate was used to label fungal cell wall as described previously Falsetta et al., 2014. The biofilms were sequentially scanned (488/640 nm lasers for SYTO9/TOTO‐3, then 561 nm laser for Concanavalin A‐tetramethylrhodamine) and the fluorescence emitted was collected using optimum emission wavelength filters (Zeiss LSM800 confocal microscope with Airyscan). Each biofilm was scanned at least three randomly selected areas. ImageJ FIJI was used for image processing and to create representative multi‐channel images (Schindelin et al., 2012).
Conflict of interest
Both corresponding authors have filed patents on biofilm disruption using plant‐derived enzymes but have no specific financial conflict to disclose. All other authors no conflict of interest.
Author contributions
HD conceived this project, designed experiments, interpreted data and wrote/edited several versions of this manuscript. HK designed all biofilm investigations, interpreted data and wrote/edited several versions. RS characterized lipase/dextranase plant enzyme, interpreted data, wrote part of introduction, results and methods section for lipase/dextranase enzyme assays. ZR designed and performed biofilm investigations, interpreted data and wrote/edited the manuscript. YS characterized dextranase/mutanase plant enzymes and wrote the corresponding methods and results. SL created dextranase/mutanase plants and KCK/BS created dextranase/mutanase chloroplast vectors with codon optimized genes. FM designed GFP release from chewing gum and VR performed these experiments.
Supporting information
Figure S1 Chloroplast derived lipase activity against p‐Nitrophenyl butyrate.
Figure S2 Complete coding DNA sequence (A) and translated amino acid sequence (B) of Smdex gene from Streptococcus mutans strain ATCC 25175.
Figure S3 Codon optimized Paenibacillus sp. mut gene (A) and translated amino acid sequence (B).
Figure S4 Paenibacillus sp. native mut gene codon frequency vs. codon optimized gene frequency.
Figure S5 In vitro cariogenic dental biofilm model and the topical treatment regimen used.
Figure S6 Inhibition of hyphal formation in the C. albicans monoculture as a result of lipase topical treatment.
Figure S7 Evaluation of stability of GFP during preparation of gum tablets.
Figure S8 Experimental set up showing recessed polycarbonate chamber (A) and loading piston (B).
Figure S9 Loading and unloading curves for chewing gum at different time intervals (1min (A), 5 min (B), and 7 min(C)).
Table S1 Parameters for cyclic loading of chewing gum set in Merlin Software.
Acknowledgements
This research was supported by funding from NIH grant R01 HL 107904, R01 HL 109442, R01 HL 133191 to Henry Daniell, and biofilm evaluation was supported by NIH grant R01DE018023, DE025220 to Hyun Koo.
Singh, R. , Ren, Z. , Shi, Y. , Lin, S. , Kwon, K.‐C. , Balamurugan, S. , Rai, V. , Mante, F. , Koo, H. and Daniell, H. (2021) Affordable oral health care: dental biofilm disruption using chloroplast made enzymes with chewing gum delivery. Plant Biotechnol. J., 10.1111/pbi.13643
Contributor Information
Hyun Koo, Email: koohy@upenn.edu.
Henry Daniell, Email: hdaniell@upenn.edu.
References
- Autio‐Gold, J. (2008) The role of chlorhexidine in caries prevention. Oper. Dent. 33, 710–716. [DOI] [PubMed] [Google Scholar]
- Bennett, A.M. , Fulford, M.R. , Walker, J.T. , Bradshaw, D.J. , Martin, M.V. and Marsh, P.D. (2000) Microbial aerosols in general dental practice. Br. Dent. J. 189, 664–667. [DOI] [PubMed] [Google Scholar]
- Bowen, W.H. , Burne, R.A. , Wu, H. and Koo, H. (2018) Oral biofilms: pathogens, matrix, and polymicrobial interactions in microenvironments. Trends Microbiol. 26, 229–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Carvalho, F.G. , Silva, D.S. , Hebling, J. , Spolidorio, L.C. and Spolidorio, D.M. (2006) Presence of mutans streptococci and Candida spp. in dental plaque/dentine of carious teeth and early childhood caries. Arch. Oral Biol. 51, 1024–1028. [DOI] [PubMed] [Google Scholar]
- Chiaraviglio, L. and Kirby, J.E. (2014) Evaluation of impermeant, DNA‐binding dye fluorescence as a real‐time readout of eukaryotic cell toxicity in a high throughput screening format. Assay Drug Dev. Technol. 12, 219–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell, H. , Chan, H.T. and Pasoreck, E.K. (2016) Vaccination via chloroplast genetics: affordable protein drugs for the prevention and treatment of inherited or infectious human diseases. Annu. Rev. Genet. 50, 595–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell, H. , Jin, S.X. , Zhu, X.G. , Gitzendanner, M.A. , Soltis, D.E. and Soltis, P.S. (2021) Green giant – a tiny chloroplast genome with mighty power to produce high‐value proteins: history and phylogeny. Plant Biotechnol. J. 19, 430–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell, H. , Kulis, M. and Herzog, R.W. (2019a) Plant cell‐made protein antigens for induction of oral tolerance. Biotechnol. Adv. 37, 107413. 10.1016/j.biotechadv.2019.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell, H. , Ribeiro, T. , Lin, S. , Saha, P. , McMichael, C. , Chowdhary, R. and Agarwal, A. (2019b) Validation of leaf and microbial pectinases: commercial launching of a new platform technology. Plant Biotechnol. J. 17, 1154–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell, H. , Mangu, V. , Yakubov, B. , Park, J. , Habibi, P. , Shi, Y. , Gonnella, P.A. et al. (2020) Investigational new drug enabling angiotensin oral‐delivery studies to attenuate pulmonary hypertension. Biomaterials, 233, 119750. 10.1016/j.biomaterials.2019.119750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deb, C. , Daniel, J. , Sirakova, T.D. , Abomoelak, B. , Dubey, V.S. and Kolattukudy, P.E. (2006) A novel lipase belonging to the hormone‐sensitive lipase family induced under starvation to utilize stored triacylglycerol in Mycobacterium tuberculosis . J. Biol. Chem. 281, 3866–3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falsetta, M.L. , Klein, M.I. , Colonne, P.M. , Scott‐Anne, K. , Gregoire, S. , Pai, C.H. , Gonzalez‐Begne, M. et al. (2014) Symbiotic relationship between Streptococcus mutans and Candida albicans synergizes virulence of plaque biofilms in vivo . Infect. Immun. 82, 1968–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, K. , Kotian, A. , Subramanian, H. , Daniell, H. and Ali, H. (2015) Activation of human mast cells by retrocyclin and protegrin highlight their immunomodulatory and antimicrobial properties. Oncotarget, 6, 28573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis, E. , Parsaei, Y. , Klein, M.I. and Koo, H. (2017) Advances in the microbial etiology and pathogenesis of early childhood caries. Mol. Oral Microbiol. 32, 24–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann, R. , Jeckel, H. , Jelli, E. , Singh, P.K. , Vaidya, S. , Bayer, M. , Rode, D.K. et al. (2021) Quantitative image analysis of microbial communities with BiofilmQ. Nat. Microbiol. 6, 151–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, G. , Liu, Y. , Kim, D. , Li, Y. , Krysan, D.J. and Koo, H. (2017) Candida albicans mannans mediate Streptococcus mutans exoenzyme GtfB binding to modulate cross‐kingdom biofilm development in vivo . PLoS Pathog. 13, e1006407. 10.1371/journal.ppat.1006407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y.M. , Shimizu, R. , Nakai, H. , Mori, H. , Okuyama, M. , Kang, M.S. , Fujimoto, Z. et al. (2011) Truncation of N‐and C‐terminal regions of Streptococcus mutans dextranase enhances catalytic activity. Appl. Microbiol. Biotechnol. 91, 329–339. [DOI] [PubMed] [Google Scholar]
- Kolahi, J. and Abrishami, M. (2013) Mutanase‐containing chewing gum: a new potential approach for prevention of dental caries. Dental Hypotheses, 4, 53–54. [Google Scholar]
- Koo, H. , Allan, R.N. , Howlin, R.P. , Stoodley, P. and Hall‐Stoodley, L. (2017) Targeting microbial biofilms: current and prospective therapeutic strategies. Nat. Rev. Microbiol. 15, 740–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo, H. , Hayacibara, M.F. , Schobel, B.D. , Cury, J.A. , Rosalen, P.L. , Park, Y.K. , Vacca‐Smith, A.M. et al. (2013) Inhibition of Streptococcus mutans biofilm accumulation and polysaccharide production by apigenin and tt‐farnesol. J. Antimicrob. Chemother. 52, 782–789. [DOI] [PubMed] [Google Scholar]
- Kopec, L.K. , Vacca‐Smith, A.M. and Bowen, W.H. (1997) Structural aspects of glucans formed in solution and on the surface of hydroxyapatite. Glycobiology, 7, 929–934. [DOI] [PubMed] [Google Scholar]
- Kumari, U. , Singh, R. , Ray, T. , Rana, S. , Saha, P. , Malhotra, K. and Daniell, H. (2019) Validation of leaf enzymes in the detergent and textile industries: launching of a new platform technology. Plant Biotechnol. J. 17, 1167–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon, K.C. , Chan, H.T. , León, I.R. , Williams‐Carrier, R. , Barkan, A. and Daniell, H. (2016) Codon optimization to enhance expression yields insights into chloroplast translation. Plant Physiol. 172, 62–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon, K.C. , Sherman, A. , Chang, W.J. , Kamesh, A. , Biswas, M. , Herzog, R.W. and Daniell, H. (2018) Expression and assembly of largest foreign protein in chloroplasts: oral delivery of human FVIII made in lettuce chloroplasts robustly suppresses inhibitor formation in haemophilia A mice. Plant Biotechnol. J. 16, 1148–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S.B. , Li, B. , Jin, S. and Daniell, H. (2011) Expression and characterization of antimicrobial peptides Retrocyclin‐101 and Protegrin‐1 in chloroplasts to control viral and bacterial infections. Plant Biotechnol. J. 9, 100–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , Kamesh, A.C. , Xiao, Y. , Sun, V. , Hayes, M. , Daniell, H. and Koo, H. (2016) Topical delivery of low‐cost protein drug candidates made in chloroplasts for biofilm disruption and uptake by oral epithelial cells. Biomaterials, 105, 156–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J.K.C. , Hikmat, B.Y. , Wycoff, K. , Vine, N.D. , Chargelegue, D. , Yu, L. , Hein, M.B. et al. (1998) Characterization of a recombinant plant monoclonal secretory antibody and preventive immunotherapy in humans. Nat. Med. 4, 601–606. [DOI] [PubMed] [Google Scholar]
- Murai, S. , Igarashi, A. , Ito, K. , Kanamori, S. and Otani, K. (1975) On the inhibitory effect of dental plaque in man by use of dextranase‐containing chewing‐gum (author's transl). Nihon Shishubyo Gakkai kaishi, 17, 74–79. [DOI] [PubMed] [Google Scholar]
- Otsuka, R. , Imai, S. , Murata, T. , Nomura, Y. , Okamoto, M. , Tsumori, H. , Kakuta, E. et al. (2015) Application of chimeric glucanase comprising mutanase and dextranase for prevention of dental biofilm formation. Microbiol. Immunol. 59, 28–36. [DOI] [PubMed] [Google Scholar]
- Park, J. , Yan, G. , Kwon, K.C. , Liu, M. , Gonnella, P.A. , Yang, S. and Daniell, H. (2020) Oral delivery of novel human IGF‐1 bioencapsulated in lettuce cells promotes musculoskeletal cell proliferation, differentiation and diabetic fracture healing. Biomaterials, 233, 119591. 10.1016/j.biomaterials.2019.119591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel, J. and Sampson, V. (2020) The role of oral bacteria in COVID‐19. Lancet Microbe, 1, e105. 10.1016/S2666-5247(20)30057-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peres, M.A. , Macpherson, L.M.D. , Weyant, R.J. , Daly, B. , Venturelli, R. , Mathur, M.R. , Listl, S. et al. (2019) Oral diseases: a global public health challenge. Lancet, 394, 249–260. [DOI] [PubMed] [Google Scholar]
- Pogue, G.P. , Vojdani, F. , Palmer, K.E. , Hiatt, E. , Hume, S. , Phelps, J. , Long, L. et al. (2010) Production of pharmaceutical‐grade recombinant aprotinin and a monoclonal antibody product using plant‐based transient expression systems. Plant Biotechnol. J. 8, 638–654. [DOI] [PubMed] [Google Scholar]
- Prabhawathi, V. , Boobalan, T. , Sivakumar, P.M. and Doble, M. (2014) Antibiofilm properties of interfacially active lipase immobilized porous polycaprolactam prepared by LB technique. PLoS One, 9, e96152. 10.1371/journal.pone.0096152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purushe, S. , Prakash, D. , Nawani, N.N. , Dhakephalkar, P. and Kapadnis, B. (2012) Biocatalytic potential of an alkalophilic and thermophilic dextranase as a remedial measure for dextran removal during sugar manufacture. Bioresour. Technol. 115, 2–7. [DOI] [PubMed] [Google Scholar]
- Ramessar, K. , Rademacher, T. , Sack, M. , Stadlmann, J. , Platis, D. , Stiegler, G. , Labrou, N. et al. (2008) Cost‐effective production of a vaginal protein microbicide to prevent HIV transmission. Proc. Natl Acad. Sci. USA, 105, 3727–3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rella, A. , Farnoud, A.M. and Del Poeta, M. (2016) Plasma membrane lipids and their role in fungal virulence. Prog. Lipid Res., 61, 63–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, Z. , Kim, D. , Paula, A.J. , Hwang, G. , Liu, Y. , Li, J. , Daniell, H. et al. (2019) Dual‐targeting approach degrades biofilm matrix and enhances bacterial killing. J. Dent. Res. 98, 322–330. [DOI] [PubMed] [Google Scholar]
- Ruhlman, T. , Verma, D. , Samson, N. and Daniell, H. (2010) The role of heterologous chloroplast sequence elements in transgene integration and expression. Plant Physiol. 152, 2088–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin, J. , Arganda‐Carreras, I. , Frise, E. , Kaynig, V. , Longair, M. , Pietzsch, T. , Preibisch, S. et al. (2012) Fiji: an open‐source platform for biological‐image analysis. Nat. Methods, 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seghal Kiran, G. , Nishanth Lipton, A. , Kennedy, J. , Dobson, A.D. and Selvin, J. (2014) A halotolerant thermostable lipase from the marine bacterium Oceanobacillus sp. PUMB02 with an ability to disrupt bacterial biofilms. Bioengineered, 5, 305–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somogyi, M. (1945) A new reagent for the determination of sugars. J. Biol. Chem. 160, 61–68. [Google Scholar]
- Su, J. , Zhu, L. , Sherman, A. , Wang, X. , Lin, S. , Kamesh, A. , Norikane, J.H. et al. (2015) Low cost industrial production of coagulation factor IX bioencapsulated in lettuce cells for oral tolerance induction in hemophilia B. Biomaterials, 70, 84–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaki, P. , Vieira, M. and Bommarito, S. (2014) Maximum bite force analysis in different age groups. Int Arch Otorhinolaryngol, 18, 272–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vamvaka, E. , Arcalis, E. , Ramessar, K. , Evans, A. , O'Keefe, B.R. , Shattock, R.J. , Medina, V. et al. (2016b) Rice endosperm is cost‐effective for the production of recombinant griffithsin with potent activity against HIV. Plant Biotechnol. J. 14, 1427–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vamvaka, E. , Evans, A. , Ramessar, K. , Krumpe, L.R.H. , Shattock, R.J. , O’Keefe, B.R. , Christou, P. and et al. (2016a) Cyanovirin‐N produced in rice endosperm offers effective pre‐exposure prophylaxis against HIV‐1 BaL infection in vitro. Plant Cell Rep. 35, 1309–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doremalen, N. , Bushmaker, T. , Morris, D.H. , Holbrook, M.G. , Gamble, A. , Williamson, B.N. , Tamin, A. et al. (2020) Aerosol and surface stability of SARS‐CoV‐2 as compared with SARS‐CoV‐1. N. Engl. J. Med. 382, 1564–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga, S. , Spalj, S. , Lapter Varga, M. , Anic Milosevic, S. , Mestrovic, S. and Slaj, M. (2011) Maximum voluntary molar bite force in subjects with normal occlusion. Eur. J. Orthodont. 33, 427–433. [DOI] [PubMed] [Google Scholar]
- Verma, D. , Samson, N.P. , Koya, V. and Daniell, H. (2008) A protocol for expression of foreign genes in chloroplasts. Nat. Protoc. 3, 739–758. [DOI] [PubMed] [Google Scholar]
- Vos, T. , Abajobir, A.A. , Abate, K.H. , Abbafati, C. , Abbas, K.M. , Abd‐Allah, F. , Abdulkader, R.S. et al. (2017) Global, regional, and national incidence, prevalence, and years lived with disability for 328 diseases and injuries for 195 countries, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet, 390, 1211–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel, S.W. , van der Mei, H.C. , Maitra, A. , Dodds, M.W. and Busscher, H.J. (2016) Potential benefits of chewing gum for the delivery of oral therapeutics and its possible role in oral healthcare. Expert Opin. Drug Deliv. 13, 1421–1431. [DOI] [PubMed] [Google Scholar]
- WHO . (2020) Transmission of SARS‐CoV‐2: implications for infection prevention precautions: scientific brief, 09 July 2020 (No. WHO/2019‐nCoV/Sci_Brief/Transmission_modes/2020.3). World Health Organization. [Google Scholar]
- Xu, R. , Cui, B. , Duan, X. , Zhang, P. , Zhou, X. and Yuan, Q. (2020) Saliva: potential diagnostic value and transmission of 2019‐nCoV. Int. J. Oral Sci. 12, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong, E.D. (2010) Human jaws are surprisingly strong and efficient. https://www.nationalgeographic.com/science/phenomena/2010/06/22/who‐are‐you‐calling‐weak‐human‐jaws‐are‐surprisingly‐strong‐and‐efficient
- Yusibov, V. , Streatfield, S.J. and Kushnir, N. (2011) Clinical development of plant‐produced recombinant pharmaceuticals: vaccines, antibodies and beyond. Hum Vaccin. 7, 313–321. [DOI] [PubMed] [Google Scholar]
- Zeitlin, L. , Olmsted, S.S. , Moench, T.R. , Co, M.S. , Martinell, B.J. , Paradkar, V.M. , Russell, D.R. et al. (1998) A humanized monoclonal antibody produced in transgenic plants for immunoprotection of the vagina against genital herpes. Nat. Biotechnol. 16, 1361–1364. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Chloroplast derived lipase activity against p‐Nitrophenyl butyrate.
Figure S2 Complete coding DNA sequence (A) and translated amino acid sequence (B) of Smdex gene from Streptococcus mutans strain ATCC 25175.
Figure S3 Codon optimized Paenibacillus sp. mut gene (A) and translated amino acid sequence (B).
Figure S4 Paenibacillus sp. native mut gene codon frequency vs. codon optimized gene frequency.
Figure S5 In vitro cariogenic dental biofilm model and the topical treatment regimen used.
Figure S6 Inhibition of hyphal formation in the C. albicans monoculture as a result of lipase topical treatment.
Figure S7 Evaluation of stability of GFP during preparation of gum tablets.
Figure S8 Experimental set up showing recessed polycarbonate chamber (A) and loading piston (B).
Figure S9 Loading and unloading curves for chewing gum at different time intervals (1min (A), 5 min (B), and 7 min(C)).
Table S1 Parameters for cyclic loading of chewing gum set in Merlin Software.


