Abstract
Sulfides and persulfides/polysulfides (R-Sn-R’, n > 2; R-Sn-H, n > 1) are endogenously produced metabolites that are abundant in mammalian and human cells and tissues. The most typical persulfides that are widely distributed among different organisms include various reactive persulfides—low-molecular-weight thiol compounds such as cysteine hydropersulfide, glutathione hydropersulfide, and glutathione trisulfide as well as protein-bound thiols. These species are generally more redox-active than are other simple thiols and disulfides. Although hydrogen sulfide (H2S) has been suggested for years to be a small signaling molecule, it is intimately linked biochemically to persulfides and may actually be more relevant as a marker of functionally active persulfides. Reactive persulfides can act as powerful antioxidants and redox signaling species and are involved in energy metabolism. Recent evidence revealed that cysteinyl-tRNA synthetases (CARSs) act as the principal cysteine persulfide synthases in mammals and contribute significantly to endogenous persulfide/polysulfide production, in addition to being associated with a battery of enzymes including cystathionine β-synthase, cystathionine γ-lyase, and 3-mercaptopyruvate sulfurtransferase, which have been described as H2S-producing enzymes. The reactive sulfur metabolites including persulfides/polysulfides derived from CARS2, a mitochondrial isoform of CARS, also mediate not only mitochondrial biogenesis and bioenergetics but also anti-inflammatory and immunomodulatory functions. The physiological roles of persulfides, their biosynthetic pathways, and their pathophysiology in various diseases are not fully understood, however. Developing basic and high precision techniques and methods for the detection, characterization, and quantitation of sulfides and persulfides is therefore of great importance so as to thoroughly understand and clarify the exact functions and roles of these species in cells and in vivo.
Keywords: Sulfides, Persulfides/polysulfides, Methodology, Hydrogen sulfide, Reactive sulfur metabolites
1. Introduction
1.1. Basic aspects of sulfur species in biology
Numerous diatomic and triatomic species have been identified as important endogenously synthesized biological signaling agents that are involved in a multitude of physiological functions. Nitric oxide (NO), carbon monoxide, and hydrogen sulfide (H2S) are the primary members of this group [1-4]; other small-molecule species may be included as well [5, 6]. At room temperature and atmospheric pressure, these molecules exist as gases and have therefore been referred to as gasotransmitters, although they are solutes and not gases when they function physiologically (thus, this term is misleading and ill-conceived). As a member of this group, H2S is a molecule of significant research interest. Although first characterized as a toxic sewer gas more than 300 years ago [7], during the past 25 years H2S has been touted as a biological mediator involved in, for example, brain NMDA neurochemistry [8], regulation of vascular tone [9, 10], regulation of KATP channels [11], and mediation of inflammation [12]. In many cases, reports of the biological actions of H2S are primarily descriptive because the biochemical mechanisms by which H2S acts remain for the most part undetermined.
One challenging aspect of investigating the mechanisms associated with H2S biochemistry and physiology is that H2S reacts in a biological milieu, and the specific species responsible for the observed biological effects are not always clear. That is, H2S can react with numerous other biologically relevant molecules, and the products of these reactions may be the actual biological mediators. Moreover, the purity of pharmacologically administered H2S (or sulfide salts) is also a problem [13], especially because impurities themselves have been hypothesized to be biological mediators [14]. In fact, many biological effects attributed to H2S may be due to other sulfide-related reactive species (also called reactive sulfur species [15], which were hypothesized to be physiologically important even before the idea of H2S signaling functions emerged [16]. Thus, to begin to clarify the mechanisms of H2S (or reactive sulfur species) activity, determining what chemical species exist in an H2S-containing solution or system becomes important, and, at the very least, attempts should be made to correlate the levels of these compounds with observed activities. Therefore, before a discussion of the methodologies for H2S and reactive sulfur species detection and measurement, reviewing the relevant chemistry of these molecules and the potentially confounding problems associated with their detection becomes worthwhile.
1. 2. Chemistry of persulfide/polysulfide species
H2S is the simplest of all thiols (RSH) and as such reacts/behaves analogously to other RSHs in biological solutions (vide infra). However, some distinctions between H2S and most RSH species are worth mentioning. Unlike other biologically relevant RSHs [e.g. cysteine (CysSH), glutathione (GSH)], H2S is extremely volatile, with a boiling point of −60 °C. Thus, H2S will volatilize from open vessels, which results in a change in not only H2S levels but, as will be discussed later, also levels of species that are in dynamic equilibrium with it [17]. H2S is also more acidic than most RSHs. The pKa value of H2S is 6.8 (at 37 °C), which means that the hydrosulfide anion (HS−) is the predominant species (approximately 80%) at the physiological pH of 7.4. In comparison, the pKa values of CysSH, GSH, and other biologically relevant RSHs are in the range of 8–9, which indicates that their -SH group is predominantly protonated at physiological pH. Finally, inasmuch as H2S has two dissociable protons, it can form two bonds via reactions that replace both protons (e.g. possibly resulting in a bridging sulfur). Thus, we may expect electrophilic trapping of H2S to result in the consumption of two equivalents of the trapping agent and give products with a bridging sulfur between the two electrophilic traps.
Although the biological chemistry of H2S is extremely diverse [18–20], our emphasis here is on only the chemistry that potentially affects analytical methodology. H2S can participate in redox and substitution reactions similar to those of other RSH species. Studies have established that RSH will react with disulfides (RSSRs) and sulfenic acids (RSOHs) via substitution reactions and result in redistribution products (Reactions 1 and 2) [21].
| (1) |
| (2) |
H2S will demonstrate chemical properties analogous to those of RSSR and RSOH, in both cases generating the corresponding hydropersulfide (RSSH) and RSH (Reactions 3 and 4, which are analogous to Reactions 1 and 2 with R’ = H; note that the protonation states of H2S and RSSH reflect the predominant anionic species at physiological pH) [22–25].
| (3) |
| (4) |
The dynamic equilibrium of Reaction 3 intimately links H2S and RSSH on opposite sides of the equilibrium expression, which means that in a biological system, where RSSR and RSH are prevalent, both H2S and RSSH will likely coexist. This relationship between H2S and RSSH also means that, as alluded to above, in an experimental system in which H2S is allowed to escape, the equilibrium of Reaction 3 will result in depletion of RSSH. Moreover, addition of RSH to an experimental system will also lead to RSSH depletion, according to Reaction 3. The kinetics of Reaction 3 in a purely chemical system reportedly varied, with rate constants in the range 1–103 M−1 s−1, depending on the nature of the RSSR [25, 26]. Reaction 3, as written, is approximately thermoneutral and under expected physiological conditions (in which RSH should be at a much higher concentration compared with that of RSSR) will be uphill [27]. However, subsequent reactions of the perthiolate anion (RSS−) can drive this reaction forward (vide infra).
RSSH can be either nucleophilic or electrophilic, depending on the protonation state. When deprotonated, anionic RSS− is very nucleophilic, akin to the thiolate anion (RS−), and when protonated, RSSH is electrophilic, like RSSR. Thus, pH and pKa values of RSSH are factors that are critical to its biochemical fate. The pKa values of RSSH were reportedly 1–4 pKa units less than those of the corresponding RSH. For example, the pKa value of glutathione hydropersulfide (GSSH) was 5.45, significantly lower than the pKa value of GSH, at 8.94 [28]. The dual reactivity of RSS− and RSSH means that solutions of RSSH are unstable (at neutral pH) because both RSS− and RSSH will be in equilibrium with each other and can react. Both sulfur atoms of RSSH were reportedly potential electrophilic centers [29] and therefore several products can be expected from reactions with nucleophiles (e.g. Reactions 5 and 6).
| (5) |
| (6) |
Additional reactions of the products of Reactions 5 and 6 are possible; this chemistry is extremely complex and will result in multiple possible products. Indeed, the speciation of both organic and inorganic polysulfide molecules is a complex and potentially confounding issue with regard to detection and experimental interpretation [30].
Despite the fact that RSSH is oxidized with respect to RSH, RSS− and RSSH are now well established as much more reducing and nucleophilic compared with the corresponding RS− and RSH [24, 25, 28, 31, 32]. Therefore, generation of RSSH in Reaction 3 should lead to additional chemical properties related to its nucleophilicity. That is, nucleophilic RSS− should be able to react readily with other electrophilic species in solution such as RSSR (Reaction 7) (note that Reaction 7 is analogous to Reaction 6 above because RSSH and RSSR are similarly electrophilic).
| (7) |
Combining Reactions 3 and 7 gives Reaction 8
| (8) |
The reaction of H2S with RSSR in a purely chemical system led to detectable levels of RSH, RSSR, and RSSSR [33], a result that is consistent with the relevance of Reaction 8. RSSH was not detected, which may lead some to believe that it was not involved in this chemistry. However, RSSH can be electrophilically trapped in these reactions, which indicates its intermediate position (and low steady-state level) in this chemistry.
Hydropolysulfide species (RSnH, n > 1), some of which were alluded to above, are good reductants, superior to the corresponding monosulfide RSH species. If we use RSS− as an example, RSS− readily reduced ferric hemes (Fe(III) heme) to ferrous hemes (Fe(II) heme) under conditions in which the corresponding RSH did not [24, 32, 34] (Reaction 9).
| (9) |
The perthiyl radical (RSS·) formed in such reactions appeared to dimerize only to the tetrasulfide (RSSSSR) under the conditions used in these studies (Reaction 10) [32, 35].
| (10) |
RSSHs were also shown to be good hydrogen atom donors. For example, Chauvin and coworkers [35] reported that RSSH readily transferred a hydrogen atom to reactive one-electron oxidants (e.g. ROO·, RS·, RO·), thus quenching their oxidizing potential. Bianco et al. [32] also showed that RSSH can reduce relatively stable nitroxide radicals and thereby quench their radical characteristic [32]. The ability of RSS− and RSSH to act as one-electron reductant or hydrogen atom donors is due to the stability of RSS·. RSS· is significantly more stable than RS· (the one-electron oxidized RSH species). The relative stability of the two radicals is easily seen by comparing the S-H bond dissociation energies (BDEs) of RS-H and RSS-H. The S-H BDE for RS-H is approximately 92 kcal/mol, whereas the S-H BDE for RSS-H is approximately 70 kcal/mol [36]. This significant difference can be attributed to the greater inherent stability of RSS· versus that of RS·. The stability of RSS· means that it will be a poor oxidant. Indeed, RSS· is so stable that it does not even have the tendency to react with NO and O2 [32], which is a paramagnetic molecule known for its ability to react with and quench radical species [37]. Dimerization of RSS· to give RSSSSR (Reaction 10) represents a mechanism for the generation of high-order dialkylpolysulfides (RSSnR, n > 1). This discussion of the polysulfide RSSH should also pertain to higher order hydropolysulfides (e.g. RSSSH, RSSSSH), which will also be good reductants and hydrogen atom donors and precursors of even higher order dialkylpolysulfides.
The novel chemical and biochemical properties of persulfides/polysulfides compared with the corresponding RSHs/RSSRs have led to hypotheses regarding their potential biological utility. For example, given that RSSH is readily oxidized compared with RSH and yet is a superior reductant may indicate that it may be important for a biological response to oxidative stress [15]. That is, during cellular oxidative stress RSH is oxidatively converted to RSSH, which as a superior reductant can combat the initial stress. Although this interpretation is speculative at this time, certain reports support it [33, 38–43], Of course, as an oxidized species, RSSH can also be toxic at high levels and will likely be highly regulated. For example, Lin and coworkers [44] reported that cysteine hydropersulfide (CysSSH) was exported by some cells when RSSH levels were significantly increased, possibly to avoid toxicity. As an interesting result, GSSH was not appreciably exported, although it existed at a much higher intracellular concentration compared with CysSSH. Thus, GSSH may be compartmentalized intracellularly, akin to glutathione disulfide [45], to avoid potentially deleterious inadvertent oxidation of crucial proteins. From an analytical perspective, the export and retention (possibly compartmentalized) of persulfide/polysulfide species become important factors in experimental design and interpretation.
In prokaryotic systems, effects similar to those reported for eukaryotes have been described. For example, Escherichia coli bacteria are similarly protected from electrophilic toxicity by increased intracellular CysSSH levels, as previously seen with eukaryotic cells, and can also export CysSSH [46]. The novel polysulfide species (e.g. CysSSH and GSSH) have also been implicated as precursors for the generation of structurally important protein polysulfides that regulate bacterial transcription [47] and confer antibiotic resistance [48, 49], among other effects. Clearly, as recognition of the occurrence of some of the above-mentioned polysulfide species (as well as others) increases, other biological effects are bound to be discovered in both eukaryotic and prokaryotic systems.
Persulfide and polysulfide species are apparently ubiquitous and are important in biology. Moreover, their presumed signaling, regulatory, and protective functions indicate that they are likely to be highly regulated. As alluded to above, the potential complexity of the myriad reactions that link H2S chemically with persulfides/polysulfides makes the specific detection and assignment of actual biological effectors and mediators difficult. As an example of past problems associated with H2S and polysulfide detection, some earliest reports of such attempts indicated that plasma and/or tissue H2S levels are in the 30–100 μM range, but subsequent rapid analysis that avoided H2S extraction resulted in more realistic measurements that were 3 orders of magnitude lower [50]. The 30–100 μM range for H2S levels is certainly incorrect because the human nose is quite sensitive to H2S, and solutions with these levels would have a strong and foul odor, which is not the case [50]. A likely reason for the high levels of H2S detected in the early studies is that H2S is in dynamic equilibrium (vide supra) with numerous, non-volatile persulfides/polysulfides, and detection, which will consume H2S, draws the equilibrium toward H2S, thus depleting these other species. Another technical pitfall leading to inaccurate detection of free H2S in biological samples is the use of electrophilic alkylating agents that can trap various persulfides/polysulfides (as discussed later). In fact, polysulfide stability was recently described as being greatly affected by numerous factors, which can be attenuated by appropriately designed trapping reagents [51–53]; this issue is discussed below in “Section 2. Reactive sulfur metabolome.” Thus, hypothesizing that the 30–100 μM range represents H2S equivalents that are incorporated in persulfides/polysulfides and that actual free H2S levels are normally very low may be reasonable. Understanding the complexities and issues associated with the detection and quantitation of sulfide-associated persulfides/polysulfides is thus important.
2. Reactive sulfur metabolome
2. 1. Overview
Various analytical approaches have been developed to measure reactive sulfur species. The fluorescent reagent monobromobimane (MBB) has been widely used to determine levels of different RSHs by alkylation [54]. H2S also reacts with MBB by means of double S-alkylation, which forms the specific reaction product sulfide dibimane (SDB). On the basis of this reaction, a refined high-performance liquid chromatography (HPLC) method was devised to detect bioavailable total sulfide, acid-labile sulfide, and bound sulfane sulfur [55, 56]. With a 2.0 nM limit of detection, this MBB method is sensitive and reliable for use with biological samples. The method has been utilized by many researchers and applied in cell, animal model, and clinical studies. The MBB method has enabled measurement of, for example, sulfide bioavailability, with sulfide used as a biomarker that was confirmed by receiver-operator characteristic analysis for cardiovascular disease and Alzheimer’s disease [57, 58]; determination of sulfide concentration differences between oral squamous cell carcinoma compared with adjacent benign oral mucosae [59]; and measurement of sulfide concentrations as a clinical indicator in patients with COVID-19 compared with healthy controls [60]. Thus, the MBB/HPLC method is both useful and powerful for measuring bioavailable sulfide in a wide range of biological conditions.
To ensure accurate quantitation of endogenously formed sulfides and persulfides/polysulfides, innovative methods have also been designed to measure the sulfur metabolome by using liquid chromatography-tandem mass spectrometry (LC-MS/MS) analyses that make use of new iodoacetamide (IAM) sulfur-derivatizing agents, such as β-(4-hydroxyphenyl)ethyl iodoacetamide (HPE-IAM) and TME-IAM (N-iodoacetyl L-tyrosine methyl ester) [51, 52]. Polysulfides are inherently reactive and thus likely unstable in biological systems. However, HPE-IAM and TME-IAM can stabilize them. Protection against hydrolysis-dependent degradation of polysulfides can be mediated by the inhibitory activity of the hydroxyphenyl residue of HPE and to a much greater extent by TME in each IAM derivative. A plausible mechanism for the inhibition of hydrolysis by these reagents was described in detail in recent articles from our laboratory [52, 53, 61]. In addition, various stable isotope-labeled internal standards for all sulfide and polysulfide derivatives have been prepared so that quantitative mass spectrometry of these diverse and unstable sulfur metabolites produced in vivo and in crude biological extracts can be achieved.
2. 2. Sulfide MBB HPLC method
2. 2. 1. Materials
MBB (Sigma-Aldrich, Cat. No. B4380)
Sodium sulfide (Alfa Aesar, Cat. No. 65122)
2-Mercaptoethanol (2-ME) (Sigma-Aldrich, Cat. No. M7154)
Ethyl acetate (Sigma-Aldrich, Cat. No. 650528)
Methanol (Sigma-Aldrich, Cat. No. 34860)
Alltech Prevail SPE C18 cartridge (Grace, Cat. No. 605430)
Sulfosalicylic acid (Sigma-Aldrich, Cat. No. S2130)
Acetonitrile (Sigma-Aldrich, Cat. No. 34851)
Trifluoroacetic acid (TFA, Thermo Scientific, Cat. No. 28903)
BD Vacutainer (Becton–Dickinson, Cat. No. 366703)
1-mL plastic syringe
PCR tube (Molecular BioProducts, Cat. No. 34129)
25-gauge spinal needle (BD, Cat. No. 405180)
30-gauge needle (BD, Cat. No. 305106)
2. 2. 2. Equipment
HPLC system: Shimadzu Prominence HPLC (LC-20) with a fluorescence detector (model RF-10AXL or RF-20AXL). The HPLC column used is an Agilent Eclipse XDB-C18 column (4.6 × 250 mm, 100Å) with a guard column (4.6 × 12.5 mm, 80 Å). The data processing software is LC Solution (Version 1.23)
2. 2. 3. Procedures
- Bioavailable Sample Preparation
- To measure free sulfide, 30 μL of sample is directly used for MBB derivatization.
- To measure acid-labile sulfide, 50 μL of sample is transferred to a Vacutainer tube, after which 450 μL of 100 mM phosphate buffer [pH 2.6, 0.1 mM diethylenetriaminepentaacetic acid (DTPA)] is added.
- To measure total sulfide, 50 μL of sample is transferred to a Vacutainer tube, after which 450 μL of 100 mM phosphate buffer [pH 2.6, 0.1 mM DTPA and 1 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP)] is added.
- All Vacutainer tubes are put onto a rocker and rocked for 30 min.
- All solutions in the Vacutainer tubes are removed by using a 1-mL syringe with a spinal needle so that the solution is removed without inverting the tube.
- Volatilized H2S is then trapped by adding 500 μL of 100 mM Tris-HCl buffer (pH 9.5, 0.1 mM DTPA).
- All Vacutainer tubes are placed on a rocker and rocked for 30 min.
- After 30 min, samples are ready for derivatization reactions with MBB.
- MBB Derivatization
- A 30-μ1 sample is added to a PCR tube containing 70 μL of 100 mM Tris-HCl buffer (pH 9.5, 0.1 mM DTPA), followed by addition of 50 μL of 10 mM MBB (dissolved in deoxygenated acetonitrile) and is then incubated for 30 min at room temperature.
- The reaction is stopped by adding 50 μL of 200 mM 5-sulfosalicylic acid for 10 min on ice. Samples are spun at 3300 g for 5 min to pellet insoluble material.
- All derivatized samples should be kept in 4 °C until being analyzed by using RP-HPLC with a fluorescence detector.
- SDB Standards Preparation
- A 4-mL sample of 6 mM sodium sulfide (or non-radioactive Na234S for MS analysis) is added to a 50-mL tube with 10 mL of 100 mM deoxygenated Tris-HCl buffer (pH 9.5, 0.1 mM DTPA) followed by addition of 5 mL of 10 mM MBB solution.
- After 30 min of incubation in a 1% O2 hypoxic chamber at room temperature, 1 mL of 2-ME is added to quench excess MBB.
- SDB is extracted by using 10 mL of ethyl acetate, is evaporated by using a nitrogen stream, and is then purified by means of an Alltech Prevail SPE C18 cartridge.
- After evaporation of solvent in the fractions containing pure SDB, dry purified SDB powder can be weighed and then prepared as standard solutions with different concentrations.
- The concentration of purified SDB is verified by using spectrophotometry, with the extinction coefficient values of 4883.257 and 4694.125 dm3 mol−1 cm−1 at 370 nm (methanol solution) and 380 nm (HCl solution), respectively, to calculate an average SDB concentration. The presence of SDB can also be confirmed by LC-MS [62].
- HPLC Analysis
- A 10-μL sample is injected into the RP-HPLC system with an XDB-C18 column (4.6 × 250 mm, 80 Å) and a guard column (4.6 × 12.5 mm, 80 Å), with the fluorescence detector setting (excitation wavelength 390 nm, emission wavelength 475 nm).
- The mobile phase for elution of SDB consists of water (A, 0.1% TFA, v/v) and 99.9% pure acetonitrile (B, 0.1% TFA, v/v), which is applied in the following gradient elution at 0.6 mL/min flow rate:
| Time (min) | % A | % B |
|---|---|---|
| 0 | 85 | 15 |
| 5 | 65 | 35 |
| 16 | 45 | 55 |
| 23 | 30 | 70 |
| 24 | 10 | 90 |
| 26 | 10 | 90 |
| 28 | 85 | 15 |
The retention times of SDB and MBB are approximately 16.4 and 17.8 min, respectively.
The amount of SDB is determined by using linear plots of the HPLC peak area of known SDB concentration.
2. 2. 4. Typical results observed
Bioavailable sulfides include sulfide, acid-labile sulfide, and bound sulfane sulfur pools, and they are measured as discrete values by using RP-HPLC equipped with a fluorescence detector after generation of SDB via selective liberation, trapping, and sulfide derivatization. The amount of H2S can be measured from linear plots of the HPLC peak areas of SDB vs. known concentrations of SDB standards. The bound sulfane sulfur level is calculated by subtracting the acid-labile sulfide value from the total sulfide value (Fig. 1).
Fig. 1.
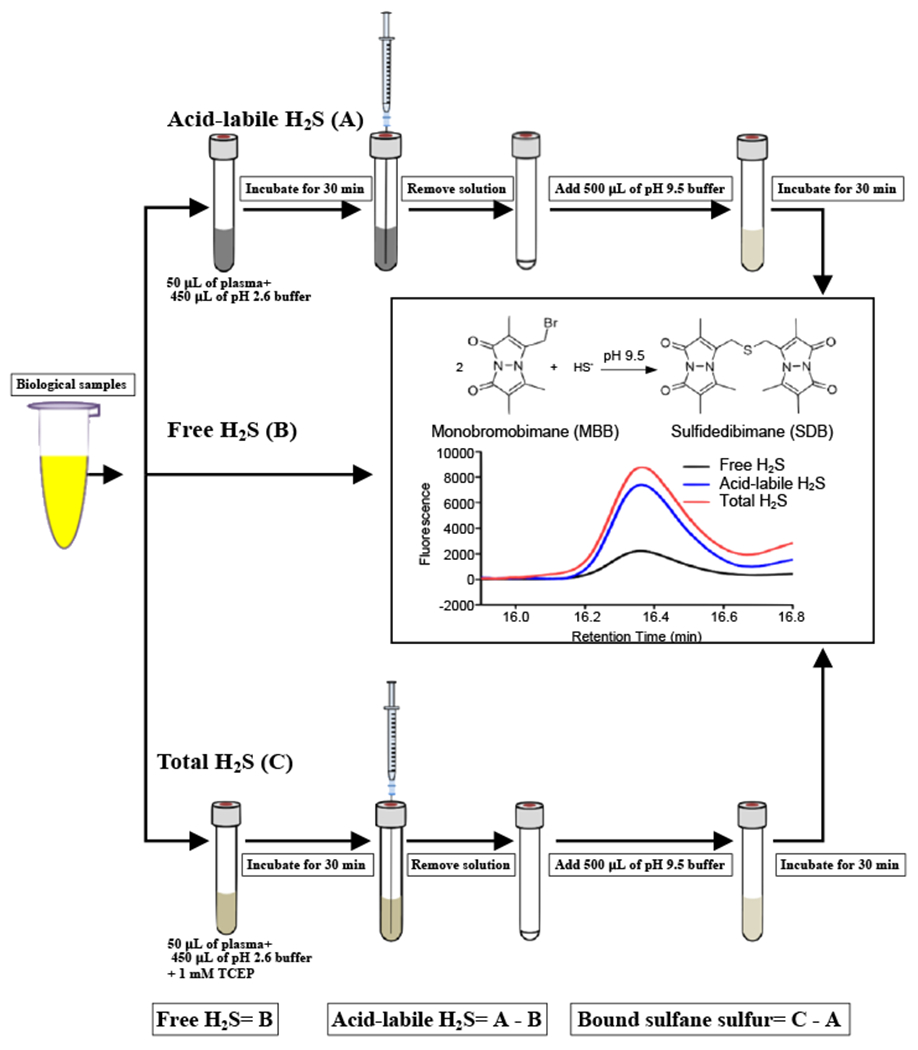
Overview of the MBB method. After derivatization of sulfide, SDB is formed and separated by using RP-HPLC with a gradient elution and is then analyzed by fluorescence detection (excitation wavelength 390 nm, emission wavelength 475 nm).
2. 2. 5. Caveats and considerations
Bioavailable sulfide may exist in three biological forms: free H2S, acid-labile sulfide, and bound sulfane sulfur [55, 63–65]. At pH 7.4 and 37 °C, 18.5% of free H2S exists as H2S gas and the remainder is almost all HS−, with a negligible contribution of sulfide anion (S2−) [17]. Acid-labile sulfide consists of sulfur in the iron-sulfur clusters present in iron-sulfur proteins [64]. Bound sulfane sulfur includes thiosulfate, persulfides, polysulfides, and their related metabolites [65]. These bioavailable sulfides can be directly or indirectly measured by means of RP-HPLC with a fluorescence detector, and the procedure involves selective liberation, trapping, and derivatization of H2S.
H2S is easily lost from biological samples through volatilization and oxidation, especially at micromolar or lower levels of sulfide. Hence, sample preparation must be carefully and consistently performed for accurate measurement. Several important facts must be remembered, including: 1) Photolysis of MBB will result in formation of fluorescent bimane [66], so using dark amber vials and dim room lighting is essential. 2) Sulfide derivatization is affected by the pH of the reaction buffer, oxygen concentration, and trace metals. 3) H2S readily binds to glass, so high-quality polypropylene plastic tubes should be used for sample preparation. 4) As prolonged derivatization with MBB may result in artificial formation of SDB possibly by shifting the equilibrium (Reaction 3 shown in Section 1. 2.; [53]), the incubation time should be optimized appropriately for each assay.
2. 3. HPE-IAM-based LC-MS/MS analysis
2. 3. 1. Materials
CysSH (Nacalai Tesque, Cat. No. 10313-84)
GSH (Nacalai Tesque, Cat. No. 17050-01)
NOC7 (1-hydroxy-2-oxo-3-(N-methyl-3-aminopropyl)-3-methyl-1-triazene; Dojindo Laboratories, Cat. No. N377)
HPE-IAM (CHEM-IMPEX INT’L INC., Cat. No. 23038)
DMSO (dimethyl sulfoxide; Nacalai Tesque, Cat. No. 09659-14)
Methanol (for LC/MS; FUJIFILM Wako Pure Chemical Corporation, Cat. No. 134-14523)
Sodium acetate (NaOAc; FUJIFILM Wako Pure Chemical Corporation, Cat. No. 192-01075)
Formic acid (FUJIFILM Wako Pure Chemical Corporation, Cat. No. 067-04531)
Bicinchoninic acid (BCA) protein assay kit (Nacalai Tesque, Cat. No. 06385)
HCl (Nacalai Tesque, Cat. No. 18321-05)
SDS (sodium dodecyl sulfate; FUJIFILM Wako Pure Chemical Corporation, Cat. No. 194-13985)
Tris (Nacalai Tesque, Cat. No. 35434-05)
PBS (phosphate-buffered saline, pH 7.4; Nacalai Tesque, Cat. No. 07906-82)
Dulbecco’s Modified Eagle’s Medium (DMEM) (FUJIFILM Wako Pure Chemical Corporation, Cat. No. 044-29765)
Isoflurane (FUJIFILM Wako Pure Chemical Corporation, Cat. No. 26675-46-7)
BCA protein assay kit (Nacalai Tesque, Cat. No. 06385-00)
2. 3. 2. Equipment
Shimadzu Nexera ultra-high-performance liquid chromatography (UHPLC) system (Shimadzu)
Speed Vac Concentrator (EYELA, Cat. No. CVE-3100)
YMC-Triart C18 column (50 × 2.0 mm inner diameter)
Triple quadrupole mass spectrometer LCMS-8060 (Shimadzu)
2. 3. 3. Procedures
- Synthesis of HPE-IAM adduct standards: CysSH/CysSSnH and GSH/GSSnH adduct standards (n > 1) with HPE-IAM are synthesized according to our previous report [49]. Stable isotope-labeled HPE-IAM adducts are synthesized by reacting each persulfide/polysulfide with NaH34S instead of sodium hydrosulfide (NaHS) or with d4-labeled HPE-IAM instead of HPE-IAM.
- CysSH (0.5 mM) or GSH (0.5 mM) is incubated with an equal concentration of NOC7 (0.5 mM) in 10 mM Tris-HCl buffer (pH 7.4) at room temperature for 5 min.
- An equal concentration of NaHS (0.5 mM) is used for additional incubation at room temperature for 5 min.
- Incubation is continued with 25 mM HPE-IAM at 37 °C for 20 min.
- The alkylation reaction is stopped with 1% formic acid, followed by appropriate dilution.
- Each HPE-IAM adduct is separated by using the HPLC system under the following conditions: YMC-Triart C18 column (50 × 2.0 mm inner diameter, 3 μm); flow rate: 0.2 mL/min; mobile phase A: 0.1% formic acid; mobile phase B: 100% methanol; gradient: linear gradient (from 5% to 90% B, for 15 min); monitor (absorbance): 275 nm (for HPE-IAM adduct); temperature: 40 °C.
- Each HPE-IAM adduct is concentrated by using the Speed Vac Concentrator.
- The quality and molecular weight of each HPE-IAM adduct are confirmed by using HPLC and LC-MS (scan).
- The concentration of each newly synthesized HPE-IAM adduct standard is measured on the basis of the signal intensity of the known amounts of stable isotope-labeled internal standards spiked before injection into the HPLC system (Fig. 2).
- Cell culture: This section describes how to prepare cell samples (the representative experiment is performed with HEK293T cells) for persulfide metabolome analysis.
- HEK293T cells, seeded in a 24-well plate at 0.5 mL at a density of 2.5 χ 105 cells per well in the usual culture medium, are cultured to 90% confluence in a 37 °C, 5% CO2 humidified incubator.
- Medium is replaced with serum-free DMEM, and cells are incubated for 1–3 h.
- Serum-free DMEM is removed, and cells are quickly rinsed with cold PBS.
- Cell lysate preparation
- The rinsed cells are treated with 0.2 mL of 5 mM HPE-IAM in 70% methanol/30 mM acetate buffer (pH 6.5) and are collected by scraping into a 1.5-mL microcentrifuge tube.
- The sample is sonicated on ice with a tip sonicator at 30 W and 20 kHz, three times with 10-s pulses and 5-s intervals.
- The sonication fluid is then incubated at 37 °C for 20 min, followed by centrifugation (10,000 g for 10 min at 4 °C) to collect supernatants. The supernatants are immediately processed by using LC-MS/MS.
- The precipitates remaining after centrifugation are resuspended with 0.1% SDS in PBS and are sonicated on ice with a tip sonicator at 30 W and 20 kHz, three times with 10-s pulses and 5-s intervals.
- The protein concentration of the precipitates is determined by using the BCA protein assay kit.
- Tissue preparation
- Mice are anesthetized with isoflurane and are euthanized by cervical dislocation.
- Mice are dissected to harvest tissues (e.g. liver, lung, kidney, spleen, and heart). The tissues are washed with cold PBS and then cut into tiny pieces with surgical scissors.
- Sliced pieces of tissue are weighed to give about 10 mg for each sample, after which they are transferred to a cryotube and saved in liquid nitrogen until use.
- Cold 80% methanol/30 mM acetate buffer (pH 6.5) solution containing 5 mM HPE-IAM is added to the cryotube containing a tissue sample. The sample is homogenized with a Dounce tissue grinder on ice.
- The homogenates are transferred to a 1.5-mL microcentrifuge tube and incubated at 37 °C for 20 min. After centrifugation at 18,000 g for 10 min at 4 °C, supernatants are collected and immediately processed by using LC-MS/MS.
- The precipitates remaining after centrifugation are resuspended with 0.1% SDS in PBS and are sonicated on ice with a tip sonicator at 30 W and 20 kHz, three times with 10-s pulses with 5-s intervals.
- The protein concentration of the precipitates is determined by using the BCA protein assay kit.
- LC-MS/MS analysis
- Supernatants of the lysates derived from cells or tissues are diluted with 0.1% formic acid containing known amounts of isotope-labeled internal standards, after which they are analyzed via liquid chromatography-electrospray ionization-tandem mass spectrometry (LC-ESI-MS/MS) to determine persulfide content.
- The triple quadrupole mass spectrometer LCMS-8060 coupled to the Nexera UHPLC system is used to perform LC-ESI-MS/MS.
- Persulfide derivatives are separated by means of the Nexera UHPLC system with a YMC-Triart C18 column (50 × 2.0 mm inner diameter) under the following elution conditions: mobile phase A (0.1% formic acid) with a linear gradient of mobile phase B (0.1% formic acid in methanol) from 5% to 90% for 15 min at a flow rate of 0.2 mL/min at 40 °C.
- MS spectra are obtained at each temperature of the ESI probe, desolvation line, and heat block at 300, 250, and 400 °C, respectively; and the nebulizer, heating, and drying nitrogen gas flows are set to 3, 10, and 10 L/min, respectively.
- Various persulfide derivatives are identified and quantified by means of the multiple reaction monitoring (MRM) parameter.
Fig. 2.
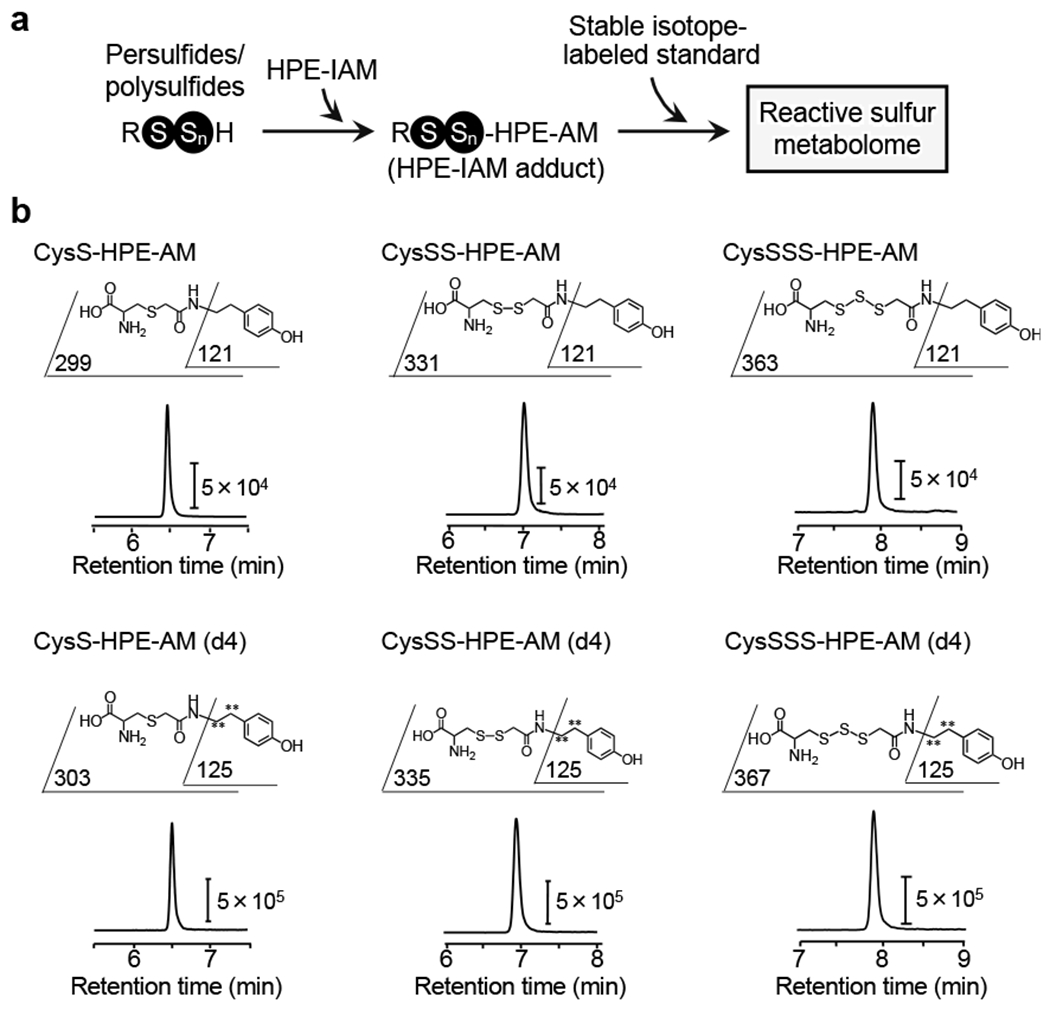
Schematic representation of HPE-IAM-based LC-MS/MS analysis. The LC-ESI-MS/MS approach with HPE-IAM trapping (a) and LC-ESI-MS/MS profiles including each fragmentation pattern (upper panels) of the CysSSnH standard for each HPE-IAM adduct (b). CysSH, CysSSnH [i.e. CysSSH and cysteine hydrotrisulfide (CysSSSH)], and other related sulfide compounds are derivatized with HPE-IAM, whose MRM parameters appear in Table 1, and are used for our LC-ESI-MS/MS analysis.
2. 3. 4. Typical results observed
LC-ESI-MS/MS with HPE-IAM as a trapping agent was used to investigate the reactive sulfur metabolome (Fig. 2a). Figure 2b shows typical chromatograms of the CysSSnH standard (namely, CysSn-HPE-AM, n = 1–3). Table 1 summarizes the MRM parameters for the HPE-IAM derivatives.
Table 1.
MRM parameters of various persulfide derivatives with HPE-IAM used in LC-ESI-MS/MS analyses.
| Analyte | Polarity | Precursor ion (m/z) | Product ion (m/z) | Collision energy (V) |
|---|---|---|---|---|
| CysS-HPE-AM | + | 298.9 | 121.0 | −29 |
| CysS-HPE-AM (d4) | + | 302.9 | 125.0 | −29 |
|
| ||||
| CysSS-HPE-AM | + | 330.8 | 121.0 | −32 |
| CysSS-HPE-AM (d4) | + | 334.8 | 125.0 | −32 |
|
| ||||
| CysSSS-HPE-AM | + | 362.8 | 121.0 | −29 |
| CysSSS-HPE-AM (d4) | + | 366.8 | 125.0 | −29 |
|
| ||||
| GS-HPE-AM | + | 484.9 | 356.3 | −18 |
| GS-HPE-AM (d4) | + | 488.9 | 360.3 | −18 |
|
| ||||
| GSS-HPE-AM | + | 516.9 | 388.2 | −18 |
| GSS-HPE-AM (d4) | + | 520.9 | 392.2 | −18 |
|
| ||||
| GSSS-HPE-AM | + | 549.1 | 420.1 | −29 |
| GSSS-HPE-AM (d4) | + | 553.1 | 424.1 | −29 |
|
| ||||
| Bis-S-HPE-AM | + | 388.9 | 121.0 | −30 |
| Bis-S-HPE-AM (d8) | + | 396.9 | 125.0 | −30 |
|
| ||||
| Bis-SS-HPE-AM | + | 420.9 | 121.0 | −23 |
| Bis-SS-HPE-AM (d8) | + | 428.9 | 125.0 | −23 |
|
| ||||
| Bis-SSS-HPE-AM | + | 452.9 | 121.0 | −37 |
| Bis-SSS-HPE-AM (d8) | + | 460.9 | 125.0 | −37 |
|
| ||||
| HSO3-HPE-AM | − | 258.1 | 121.0 | 20 |
| HSO3-HPE-AM (d4) | − | 262.1 | 121.0 | 20 |
|
| ||||
| HS2O3-HPE-AM | − | 290.0 | 208.2 | 14 |
| HS2O3-HPE-AM (d4) | − | 294.0 | 212.2 | 14 |
|
| ||||
| GSSG | − | 611.2 | 306.2 | 23 |
| GSSG[13C4, 15N2] | − | 617.2 | 309.2 | 23 |
|
| ||||
| GSSSG | − | 643.2 | 272.4 | 26 |
| GS34SSG | − | 645.2 | 272.4 | 26 |
|
| ||||
| GSSSSG | − | 675.2 | 305.8 | 25 |
| GS34S34SSG | − | 679.2 | 305.8 | 25 |
2. 4. TME-IAM-based LC-MS/MS analysis
2. 4. 1. Materials: Materials other than HPE-IAM described in “Section 2. 3.
HPE-IAM-based LC-MS/MS analysis” are used for analysis with TME-IAM described here.
TME-IAM (N-iodoacetyl l-tyrosine methyl ester iodoacetamide; kindly provided by Prof. Hideshi Ihara, Osaka Prefectural University)
2. 4. 2. Procedure
Synthesis of TME-IAM and stable TME-IAM adduct standards: TME-IAM adducts standards and stable isotope-labeled TME-IAM adducts are synthesized by reacting each persulfide with TME-IAM and TME-IAM(d2) instead of HPE-IAM, as described in “ Section 2. 3 . HPE-IAM-based LC-MS/MS analysis. ”
Cell culture: Cell culture is performed as described in “ Section 2. 3 . HPE-IAM-based LC-MS/MS analysis” without modifications.
Cell lysate preparation and tissue preparation: Cell lysates are prepared with TME-IAM instead of HPE-IAM as described in “ Section 2. 3 . HPE-IAM-based LC-MS/MS analysis.”
LC-MS/MS detection: LC-MS/MS detection is performed with TME-IAM adducts instead of HPE-IAM adducts as described in “ Section 2. 3 . HPE-IAM-based LC-MS/MS analysis.” Various persulfide derivatives are identified and quantified by means of MRM. Table 2 summarizes the MRM parameters for the TME-IAM derivatives.
Table 2.
MRM parameters of various persulfide derivatives with TME-IAM used in LC-ESI-MS/MS analyses.
| Analyte | Polarity | Precursor ion (m/z) | Product ion (m/z) | Collision energy (V) |
|---|---|---|---|---|
| CysS-TME-AM | + | 357.2 | 136.1 | −27 |
| CysS-TME-AM (d2) | + | 359.2 | 137.1 | −27 |
|
| ||||
| CysSS-TME-AM | + | 389.2 | 174.0 | −25 |
| CysSS-TME-AM (d2) | + | 391.2 | 175.0 | −25 |
|
| ||||
| CysSSS-TME-AM | + | 421.1 | 136.0 | −38 |
| CysSSS-TME-AM (d2) | + | 423.1 | 137.0 | −38 |
|
| ||||
| GS-TME-AM | + | 543.0 | 414.0 | −19 |
| GS-TME-AM (d2) | + | 545.0 | 416.0 | −19 |
|
| ||||
| GSS-TME-AM | + | 575.0 | 446.0 | −20 |
| GSS-TME-AM (d2) | + | 577.0 | 448.0 | −20 |
|
| ||||
| GSSS-TME-AM | + | 607.0 | 478.0 | −21 |
| GSSS-TME-AM (d2) | + | 609.0 | 480.0 | −21 |
|
| ||||
| Bis-S-TME-AM | + | 505.0 | 445.0 | −19 |
| Bis-S-TME-AM (d4) | + | 509.0 | 449.0 | −19 |
|
| ||||
| Bis-SS-TME-AM | + | 537.0 | 477.0 | −19 |
| Bis-SS-TME-AM (d4) | + | 541.0 | 481.0 | −19 |
|
| ||||
| Bis-SSS-TME-AM | + | 569.0 | 509.0 | −19 |
| Bis-SSS-TME-AM (d4) | + | 573.0 | 513.0 | −19 |
|
| ||||
| HSO3-TME-AM | − | 316.0 | 80.0 | 42 |
| HSO3-TME-AM (d2) | − | 318.0 | 80.0 | 42 |
|
| ||||
| HS2O3-TME-AM | − | 348.0 | 232.0 | 15 |
| HS2O3-TME-AM (d2) | − | 350.0 | 234.0 | 15 |
2. 4. 3. Typical results observed
LC-ESI-MS/MS with TME-IAM instead of HPE-IAM as a trapping agent that suppresses hydrolysis-dependent degradation of persulfides was used to investigate the reactive sulfur metabolome. Figure 3 shows the endogenous formation of persulfides/polysulfides such as CysSSH, CysSSSH, GSSH, and glutathione hydrotrisulfide (GSSSH) in the mouse tissues, found by using TME-IAM-based LC-MS/MS analysis. TME-IAM labelling resulted in higher amounts of various persulfides/polysulfides formed in the tissues than HPE-IAM labelling.
Fig. 3.
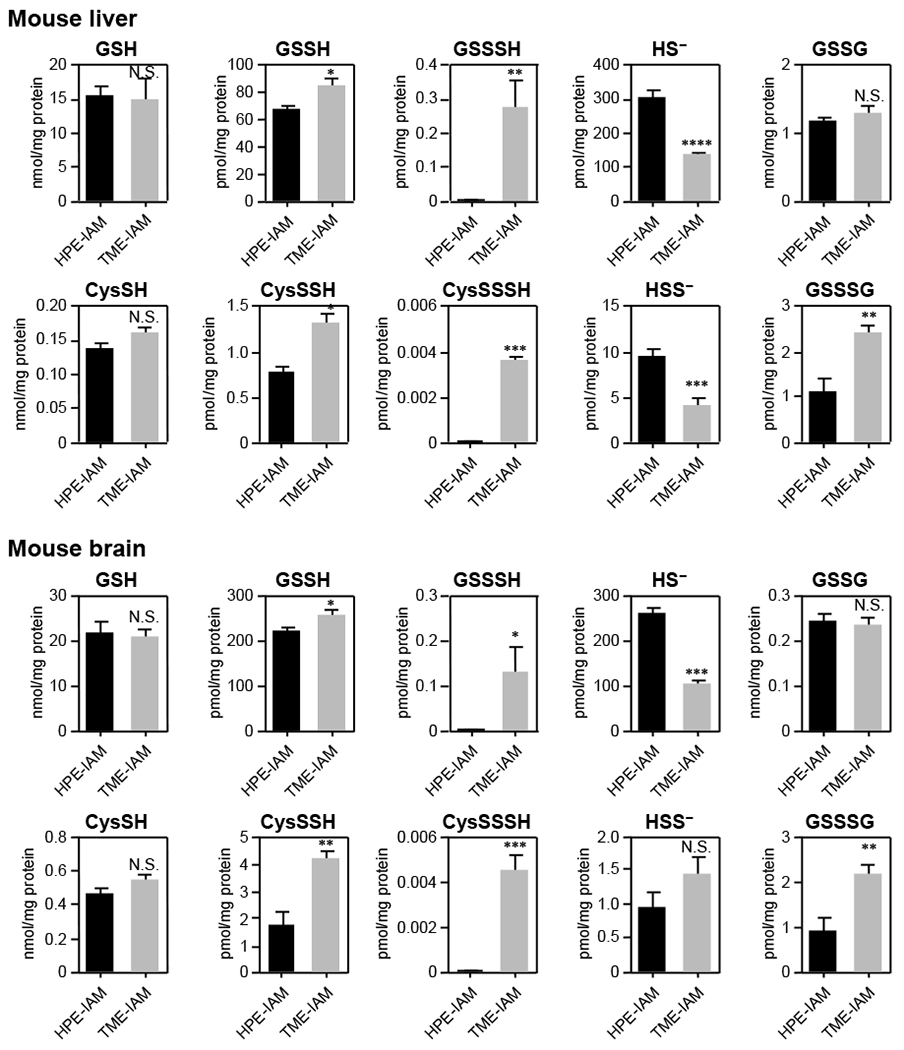
Quantitation of the amount of endogenous persulfides/polysulfides in mouse tissues. Data are mean values ± S.D. *p < 0.05, **p < 0.01, ***p < 0.001, N.S., not significant. Student’s t-test.
2. 5. Identification of protein persulfidation
2. 5. 1. Materials
Pronase (Merck, Cat No. 53702)
PD SpinTrap G-25 column (Cytiva, Cat. No. 11783329)
40 mM NaOAc buffer (pH 5.5)
RIPA buffer (10 mM Tris-HCl, 1% NP-40, 0.1% sodium deoxycholate, 0.1% SDS, 150 mM NaCl, pH 7.4)
HPE-IAM
BCA protein assay kit (Nacalai Tesque, Cat. No. 06385)
2. 5. 2. Procedure
Samples of recombinant protein, mouse tissues, and cultured cells are homogenized in RIPA buffer containing 5 mM HPE-IAM by using a homogenizer and centrifuge (15,000 g, 5 min, 4 °C).
Supernatants are collected and applied to the PD SpinTrap G-25 column equilibrated with a 40 mM NaOAc buffer (pH 5.5), after which samples are centrifuged (800 g, 2 min, 4 °C) to remove low-molecular-weight compounds.
The high-molecular-weight fraction is incubated with 5 mM HPE-IAM for alkylation of protein RSH groups, and the protein concentration is quantified by using the BCA protein assay kit.
Protein samples at the concentration 1.0 mg/mL are reacted with the Pronase digestion solution (7.2 mg/mL Pronase in 40 mM NaOAc buffer, pH 5.5) at 37 °C for 3 h.
Samples are diluted with 0.1% formic acid and then centrifuged at 15,000 g for 10 min at 4 °C, after which supernatants are collected.
Supernatants are processed by using LC-MS/MS (refer to “Section 2. 3. HPE-IAM-based LC-MS/MS analysis”).
2. 5. 3. Typical results observed
The levels of persulfidation in proteins are identified by means of LC-ESI-MS/MS and the Pronase digestion method. As Fig. 4 shows, CysSSnH formed in recombinant proteins [e.g. alcohol dehydrogenase 5 (ADH5) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH)] was analyzed and quantified by using the Pronase digestion method, which revealed that more than 60% of CysSH residues were persulfidated.
Fig. 4.
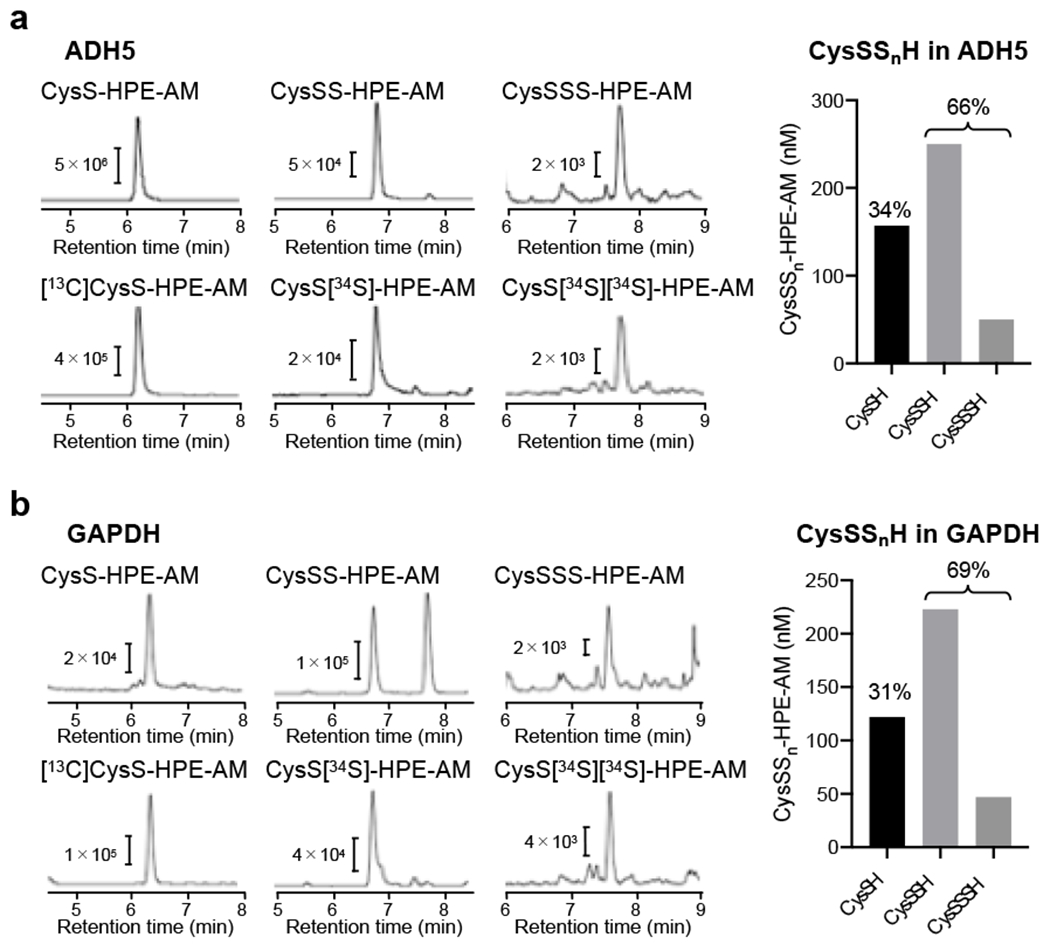
CysSSnH formation (persulfidation) in various proteins. (a) LC-ESI-MS/MS chromatograms obtained from analysis of CysSSnH formation in recombinant ADH5 (left panel). The amounts of CysSSnH detected in ADH5 (right panel). (b) LC-ESI-MS/MS chromatograms obtained from analysis of CysSSnH in recombinant GAPDH (left panel) and their quantitation (right panel).
3. Reactive sulfur proteome
3. 1. Overview
Protein persulfidation occurs not only via post-translational modifications but also via the incorporation of CysSSH produced by cysteinyl-tRNA synthetase (CARS) into tRNA during translation as a part of the translation process [51]. Co-translational protein persulfidation, therefore, has been implicated in the regulation of protein function and in structural maintenance. Thus, we developed a method for precise detection of persulfidated proteins. For example, by using a biotin-linked cyanoacetate (CN-biotin) assay and proteome technology, we can identify specific polysulfides in proteins recovered from cells and tissues. Moreover, the capture method of the biotin-polyethylene glycol (PEG)-conjugated maleimide-labeling gel shift assay (PMSA) can identify the specific persulfidated proteins. PMSA with several different electrophilic compounds can show persulfidation levels by observing the mobility of target proteins. Analysis with immunoprecipitation and mass spectrometry can assess persulfidation levels and modifications of persulfidated CysSH residues in greater detail. Several other methods for detecting protein polysulfidation have been proposed. For example, the protein persulfide detection protocol (ProPerDP) identifies protein polysulfidation by alkylation with a biotin-tagged IAM, whose principles and characteristics are discussed elsewhere and thus not explained here in detail [67–70].
3. 2. Non-targeted method
3. 2. 1. CN-biotin assay
3. 2. 1. 1. Materials
MSBT (2-methylsulfonyl benzothiazole, Santa Cruz Biotechnology, Cat. No. sc-274013)
CN-biotin (kindly provided by Prof. Ming Xian, Brown University)
NeutrAvidin horseradish peroxidase (HRP)
RIPA buffer
3. 2. 1. 2. Procedure
Cultured cells are homogenized with RIPA buffer and are centrifuged at 20,000 χ g for 10 min at 4 °C).
Supernatants are reacted with 10 mM MSBT in 20 mM Tris-HCl (pH 7.4) at 37 °C for 30 min.
The samples are then reacted with 20 mM CN-biotin at 37 °C for 30 min.
The samples are denatured with SDS sample buffer including 5% 2-ME at 95 °C for 5 min.
The samples are subjected to 2D silver staining and 2D Western blotting with NeutrAvidin HRP.
Silver stain-positive spots corresponding to HRP-positive protein spots (persulfidated proteins) are cut from the silver staining gel.
Proteins are identified by using common proteome analysis techniques.
3. 2. 1. 3. Typical results observed
To screen for persulfidated proteins in cells, the tag-switch assay was developed, in which nucleophilic protein sulfhydryls (simple RSHs and hydropersulfides/polysulfides) are tagged with the electrophile MSBT. Among all tagged RSH species, only those with a reactive sulfur residue present in polysulfides are selectively modified by a CN-biotin–labeled probe. Treatment with an avidin-conjugated enzyme-linked chemiluminescence probe then allows detection. This proteomic analysis indeed allowed identification of several persulfidated proteins (Fig. 5). Proteins involved in sulfur metabolism and known to possess protein-bound cysteine persulfides were found along with other persulfidated proteins that are unrelated to sulfur metabolism, including protein disulfide isomerase, heat shock proteins, aldo-keto reductase, GAPDH, enolase, and phosphoglycerate kinase [38]. These proteins and enzymes may thus function as sensors and effectors regulated by protein persulfidation as a post-translational modification.
Fig. 5.
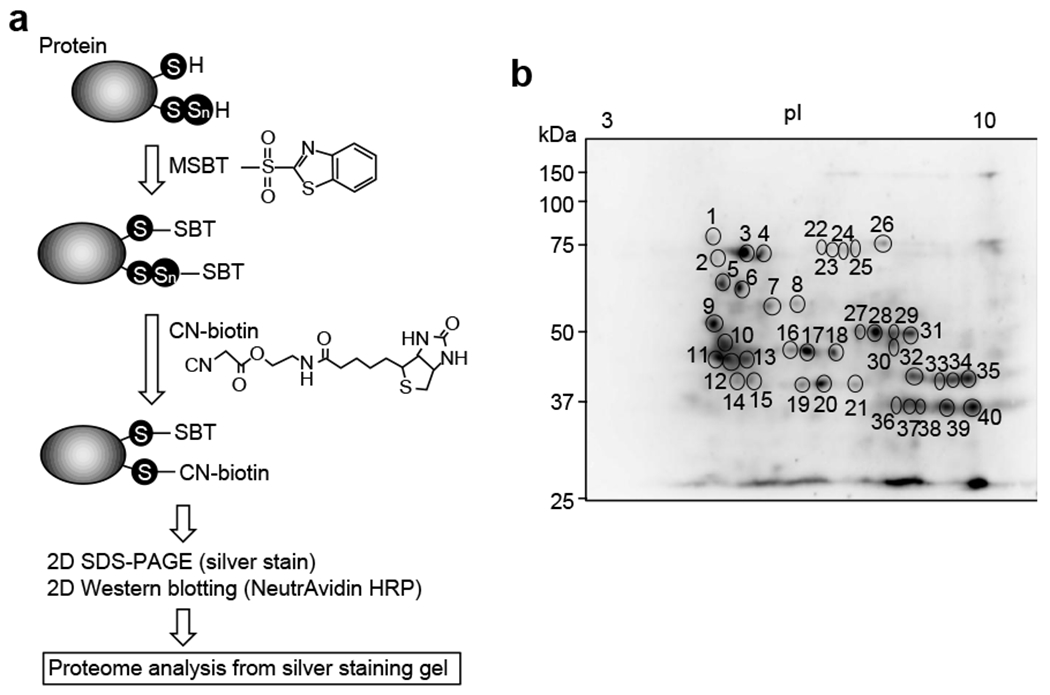
Reactive sulfur proteome using CN-biotin assay. (a) Schematic illustration of the CN-biotin assay. (b) Protein-cysteine persulfidation identified as spots on membranes transferred from 2D gels. Some labeled spots in the image were subjected to persulfide proteomics, which resulted in identification of several persulfidated proteins.
3. 3. Targeted methods
3. 3. 1. Biotin-PEG-MAL capture method
3. 3. 1. 1. Materials
Biotin-PEG36-MAL (MW: 2023.42; Dojindo Laboratories)
Streptavidin Mag Sepharose
RIPA buffer (10 mM Tris-HCl, 1% NP-40, 0.1% sodium deoxycholate, 0.1% SDS, 150 mM NaCl, pH 7.4)
TBS buffer (50 mM Tris-HCl, 0.2 M NaCl, pH 7.4)
26-gauge needle
3. 3. 1. 2. Procedure
Cultured cells are seeded at a density of 6.0 × 105 cells/mL, 2 mL/well in a 6-well plate, and are incubated at 37 °C (to 95% confluence).
Cells are washed three times with cold PBS.
Cells are harvested with 500 μL of cold-RIPA buffer including 1 mM biotin-PEG36-MAL.
Samples are homogenized by using 20 passages through a 26-gauge needle with a 1-mL syringe and are centrifuged at 20,000 × g for 30 min at 4 °C, after which supernatants are collected.
The protein concentration is quantitated by using the BCA protein assay kit.
The supernatants at a concentration of 5 mg/mL are incubated with 1 mM biotin-PEG36-MAL in RIPA buffer at 37 °C for 1 h.
The reaction solution is applied to Streptavidin Mag Sepharose resin equilibrated with RIPA buffer and incubated at room temperature for 1 h with rotation.
Beads are washed three times with TBS buffer using a magnetic rack.
Samples are eluated with 10 mM 2-ME in RIPA buffer at 37 °C for 1 h.
Samples are then subjected to SDS-PAGE and Western blotting is performed.
3. 3. 1. 3. Typical results observed
The biotin-PEG-MAL capture method described is utilized to detect protein persulfidation in cultured cells (Fig. 6a). CARSs, which are associated with other enzymes including cystathionine β-synthase (CBS), cystathionine γ-lyase (CSE), and 3-mercaptopyruvate sulfurtransferase, which were previously described as H2S-producing enzymes [16], were observed to act as the principal cysteine persulfide synthases in mammals and to contribute significantly to endogenous persulfide/polysulfide production [51]. Two isoforms of CARS exist, which also contribute to production of persulfide synthase, i.e. cytosolic CARS1 and mitochondrial CARS2. As seen in Fig. 6b, this biotin capture method detected protein persulfidation of dynamin-related protein 1 (Drp1) and ADH5 in HEK293T cells and revealed marked suppression of this persulfidation by both CARS2 knockout (KO) and additional CARS1/2 double knockdown respectively. Because CARS2 is a major source of endogenous persulfides [51], these results uneqivocally confirm specific detection of persulfidated proteins such as Drp1 by this biotin capture method.
Fig. 6.
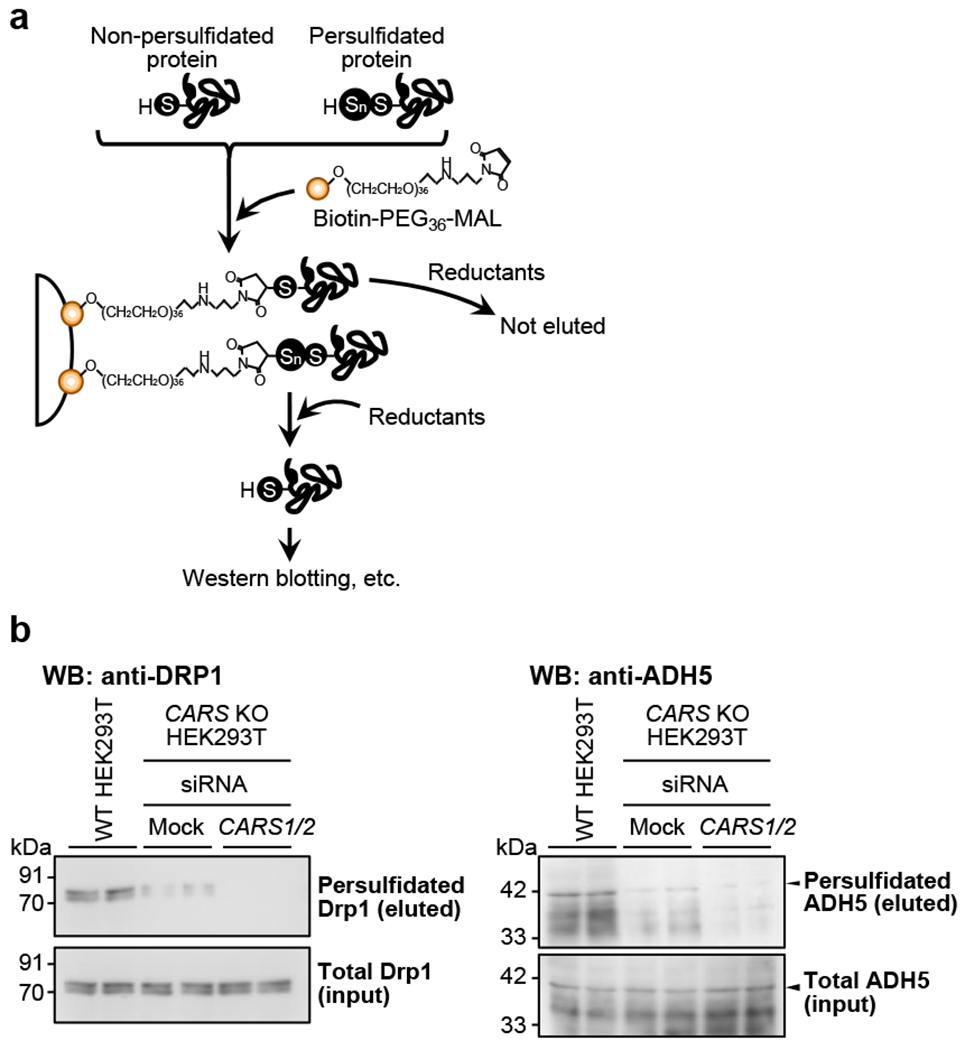
The biotin-PEG-MAL capture method. (a) Schematic illustration of the biotin-PEG-MAL capture method for quantitative identification of endogenous persulfidated proteins, which are isolated by reductive treatment of biotin-PEG36-MAL-bound avidin beads that capture persulfidated proteins. (b) Specific detection with Western blotting (WB) follows use of the capture method. These typical immunoblots show endogenous protein persulfidation of Drp1 (b, left panel) and ADH5 (b, right panel) in HEK293T cells. WT: wild type.
3. 3. 2. PMSA (PEG-maleimide labeling gel shift assay)
3. 3. 2. 1. Materials
8NcG (8-nitro-cGMP; synthesized in our laboratory)
ASBT (aminosulfonyl benzothiazole, MW: 213.98)
MSBT (MW: 213.3)
IAM (MW: 184.96; Nacalai Tesque, Cat. No. 19302-54)
MBB (MW: 271.11; Sigma-Aldrich, Cat. No. M1813)
MMTS (S-methyl methanethiosulfonate, MW: 126.2; Sigma-Aldrich, Cat. No. 208795)
NEM (N-ethylmaleimide, MW: 125.13; Nacalai Tesque, Cat. No. 15512-24)
DTP (4,4′-dithiopyridine, MW: 220.31; Nacalai Tesque, Cat. No. 14108-51)
DTNB (5,5′-dithiobis(2-nitrobenzoic acid), MW: 396.35; Sigma-Aldrich, Cat. No. D8130)
Biotin-PEG36-MAL (MW: 2023.42)
TCEP (Nacalai Tesque, Cat. No. 06342-21)
RIPA buffer
PD SpinTrap G-25 column (GE Healthcare, Cat. No. 28918004)
3. 3. 2. 2. Procedure
- For recombinant proteins
- Recombinant proteins are applied to the PD SpinTrap G-25 column equilibrated with RIPA buffer to remove reductants.
- The desalted proteins (0.3 mg/mL) are incubated with 1 mM biotin-PEG36-MAL in RIPA buffer at 37 °C for 1 h.
- The reaction solution is subsequently incubated with the following alkylating reagents, each at 3 mM: 8NcG, ASBT, MSBT, IAM, MBB, MMTS, NEM, DTP, and DTNB at 37 °C for 1 h.
- The sample is subjected to SDS-PAGE and Coomassie Brilliant Blue (CBB) staining.
- For cell lysates
- Cultured cells are washed with cold PBS and placed in ice-cold RIPA buffer that contains 1 mM TCEP and proteinase inhibitor cocktail, if necessary.
- The cells are homogenized by 20 passages through a 26-gauge needle with a 1-mL syringe on ice and are then centrifuged at 20,000 g at 4 °C for 25 min.
- The supernatant is incubated at 37 °C for 1 h in the presence of 1 mM TCEP, followed by application to the PD SpinTrap G-25 column for desalting.
- The eluate is incubated with biotin-PEG36-MAL and alkylating reagents as described above in the procedure “For recombinant proteins,” after which Western blotting is performed.
3. 3. 2. 3. Typical results observed and caveats for considerations
The unique properties and reactivities of polysulfides allowed us to develop several analytical techniques aimed at determining endogenous production of low-molecular-weight and protein-bound polysulfides (Fig. 7). A convenient method for specific detection of persulfidated proteins that we developed is the PMSA [51]. This PMSA demonstrated extensive protein-bound CysSH persulfidation (Fig. 7), not only for recombinant proteins, prepared in an E. coli cell expression system, but also for endogenous proteins expressed in mammalian cells. This method can detect both reduced and oxidized forms of polysulfides, but not RSOH and other RSH modifications such as glutathionylation.
Fig. 7.
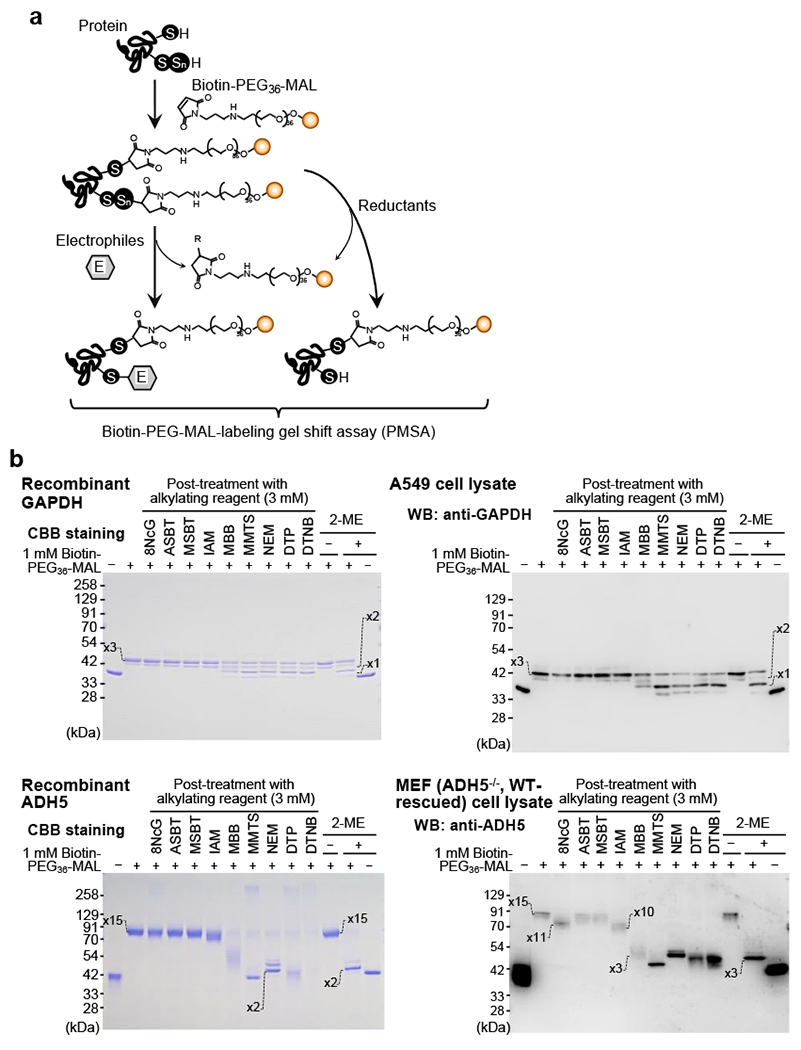
Method for detection of specific persulfidated proteins. (a) Schematic illustration of the PMSA for identification of persulfidated proteins by using biotin-PEG36-MAL and different electrophilic compounds. These typical immunoblots show endogenous protein persulfidation of GAPDH (b, upper panels) and ADH5 (b, lower panels) in recombinant protein (b, left panels) and cultured cells (b, right panels). Numbers shown in the gels indicate the numbers of biotin-PEG36-MAL labels in the protein, which indicate the numbers of polysulfidated CysSH residues in the protein.
Another critical pitfall that has been overlooked concerns sulfonyl fluoride derivatives such as phenylmethylsulfonyl fluoride (PMSF) and 4-(2-aminoethyl)benzenesulfonyl fluoride (AEBSF), which have been widely used as serine protease inhibitors and are often included in commercially available proteinase inhibitor cocktails. These sulfonyl fluoride derivatives unexpectedly react with and readily decompose various polysulfides of low-molecular-weight compounds and proteins during sample preparation and storage of cells and tissues as well as processing for metabolomic and proteomic analyses (our unpublished observations). Therefore, use of these sulfonyl fluoride reagents is not appropriate or applicable to the detection of low-molecular-weight and protein-bound polysulfides.
4. Reactive sulfur fluorescence imaging
4. 1. Overview
In 2013, Xian et al. described the SSP series of fluorescent probes for detection of persulfides: SSP2, SSP3, and SSP4 (Fig. 8a) [71]. These SSP probes utilize sulfur transfer chemistry from a persulfide to the probe, and when an RSH group in the probe is converted to an RSSH, the reactive group is eliminated by ester decomposition and releases a fluorescent species (Fig. 8b). SSP4 is a highly reactive green fluorescent probe in the SSP series and is widely used for imaging of various persulfides (Fig. 9). Xian et al. also developed another fluorescent probe, PSP-3, that has high reactivity with dihydrogen disulfide (Fig. 8a) [72]. This probe reacts not only with dihydrogen disulfide but also other hydropolysulfides, HSSnH. In SSP and PSP-3 staining, a cationic surfactant, cetyltrimethylammonium bromide (CTAB), is used to introduce the probe into cells (Fig. 9). Therefore, SSP and PSP-3 staining cannot be applied to live-cell imaging. In contrast, Umezawa et al. developed a hydropolysulfide probe, QS10 (QuantSSH-10; the number indicates the Kd value), for live-cell and real-time imaging of persulfides based on Förster resonance energy transfer (Figs. 10 and 11) [73].
Fig. 8.
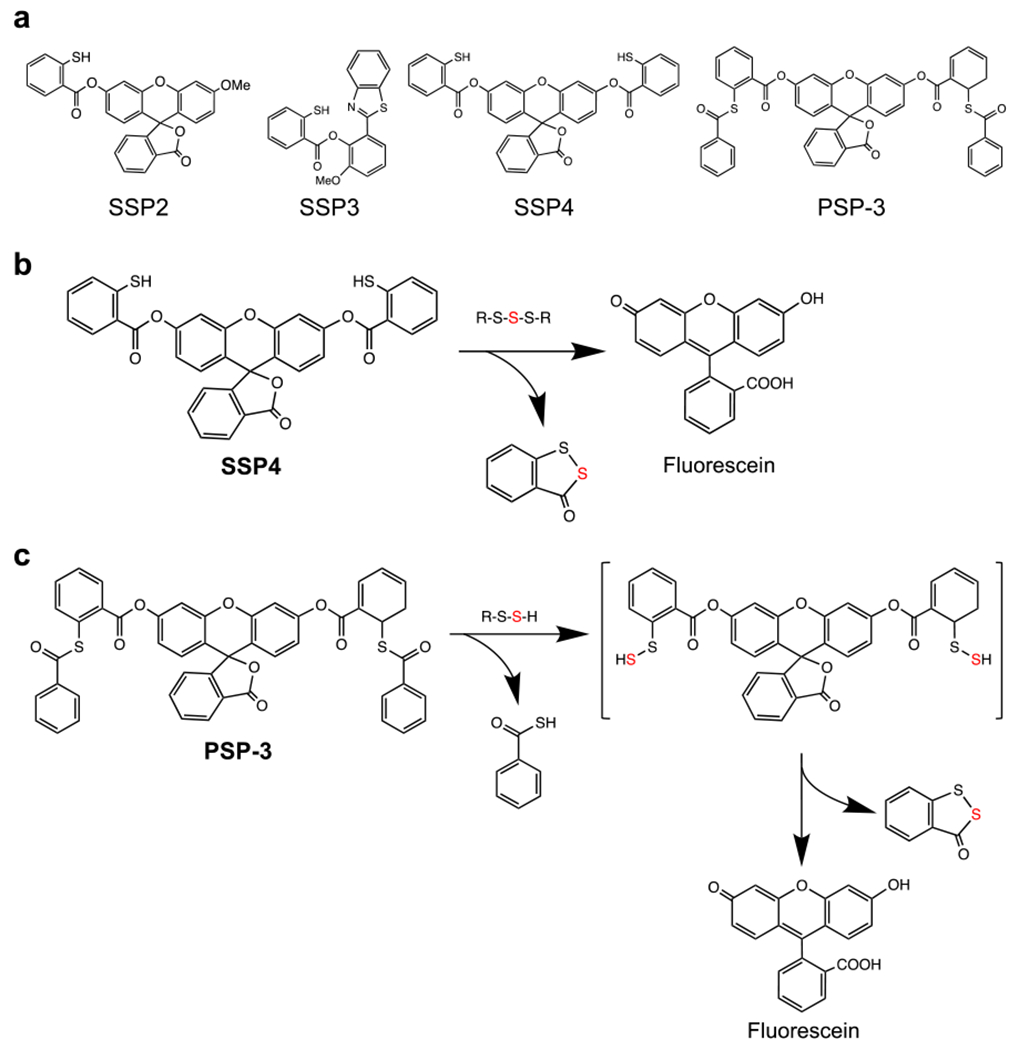
Chemical structures and reactions of the SSP series of fluorescent probes. (a) Structures of SSP2, SSP3, SSP4, and PSP-3. (b, c) Illustration of the reactions of SSP4 (b) and PSP-3 (c) with persulfides.
Fig. 9.
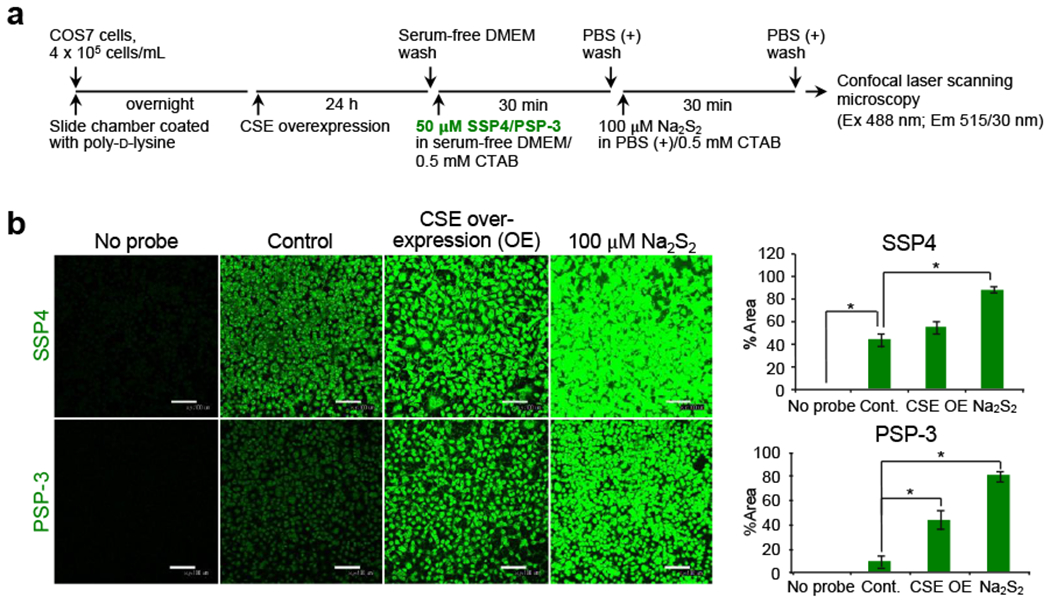
Representative persulfide fluorescence imaging with SSP4 and PSP-3 probes. (a) Illustration of the SSP4 and PSP-3 staining protocol. COS7 cells overexpressing CSE are subjected to fluorescence staining. Na2S2 (100 μM) is used as a positive control. (b) Fluorescence images (left panel) and fluorescence intensities (right panel) of SSP4 and PSP-3 staining. Data are mean values ± S.D. *p < 0.05, one-way ANOVA with Tukey’s test.
Fig. 10.
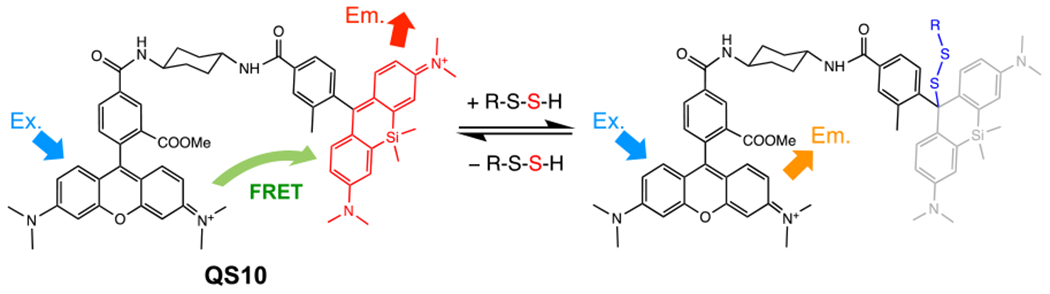
Chemical structure of the QS10 probe and the reaction of QS10 with persulfides for live-cell and real-time imaging of persulfides. Ex., excitation; Em., emission.
Fig. 11.
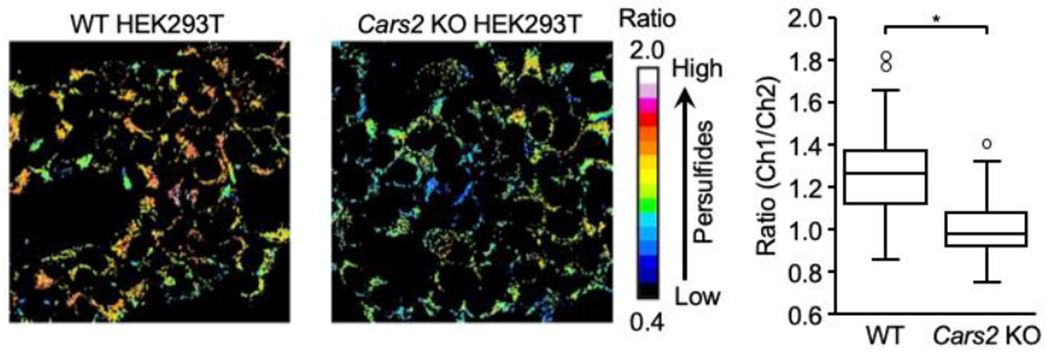
Representative persulfide fluorescence imaging with the QS10 probe. WT and Cars2 KO HEK293T cells are subjected to fluorescence staining. Fluorescence images (left panel) and a box plot of ratio values for each (right panel). Data are mean values ± S.D. *p < 0.05, Student’s t-test.
Other probes such as HSip-1 and SSip-1 can be used to measure H2S [74, 75]. However, some nonspecific fluorescence responses may occur because of the possible reaction of these probes with persulfides other than H2S, which may preclude precise detection of each sulfide species with these probes.
4. 2. SSP4 and PSP-3 staining
4. 2. 1. Materials
SSP4 (Dojindo, Cat. No. SB10) is dissolved in DMSO at a concentration of 2.5 mM; aliquots can be stored as a stock solution at −20 °C for 1 year. The SSP4 stock solution is diluted in DMEM containing 0.5 mM CTAB to a final concentration of 50 μM. SSP2 and SSP3 are not commercially available.
PSP-3 is dissolved in DMSO at a concentration of 2.5 mM; aliquots can be stored as a stock solution at −20 °C for 1 year. The SSP4 stock solution is diluted in DMEM containing 0.5 mM CTAB to a final concentration of 50 μM.
CTAB (FUJIFILM Wako Pure Chemical Corporation, Cat. No. 036-02102) is dissolved in ethanol at a concentration of 100 mM; aliquots can be stored as a stock solution at −20 °C for 1 year.
Na2S2 (sodium disulfide; Dojindo, Cat. No. SB02) is freshly dissolved in Milli-Q water at a concentration of 100 mM. The Milli-Q water should be degassed with argon gas before use.
Na2S3 (sodium trisulfide; Dojindo, Cat. No. SB03) is freshly dissolved in Milli-Q water at a concentration of 100 mM. The Milli-Q water should be degassed with argon gas before use.
Na2S4 (sodium tetrasulfide; Dojindo, Cat. No. SB04) is freshly dissolved in Milli-Q water at a concentration of 100 mM. The Milli-Q water should be degassed with argon gas before use.
PBS (+) (8.1 mM Na2HPO4, 1.47 mM KH2PO4, 2.7 mM KCl, 137 mM NaCl, 0.9 mM CaCl2, 0.33 mM MgCl2, pH 7.4) is stored in 4 °C for 1 year.
Poly-D-lysine (Sigma-Aldrich, Cat. No. P6407)
8-well slide chamber (Matsunami, Cat. No. SCS-N08)
4. 2. 2. Equipment
Confocal laser scanning microscope (Nikon C2 Plus, with NIS-Elements Version 5.01 software)
4. 2. 3. Procedure
Before cell seeding, an 8-well slide chamber is coated with poly-D-lysine (0.25 mL per well) for more than 1 h and then washed twice with PBS.
Cultured cells are seeded at a density of 4 × 105 cells per well and are cultured in 5% CO2 at 37 °C.
Cells are washed once gently with serum-free DMEM, followed by incubation with 50 μM SSP4 or PSP-3 in serum-free DMEM containing 0.5 mM CTAB at room temperature for 20 min. Cells that are not treated with SSP4 are used as a negative control.
After excess probes are removed from the cells and washing the cells with PBS, the cells are incubated in PBS for 30 min at 37 °C.
The cells are then washed twice with PBS, followed by measurement of fluorescence in images at an excitation wavelength of 488 nm and emission wavelength of 515/30 nm with a confocal laser scanning microscope.
Fluorescence intensities (a.u.) of images are calculated by using ImageJ software (National Institutes of Health).
4. 2. 4. Typical results observed
Generation of CysSSH (and CysSSSH) in COS7 and other cells depended greatly on levels of CSE and/or CBS proteins [38], because overexpression of CSE or CBS led to dramatic increases in SSP and PSP-3 fluorescence intensity of CysSSH and CysSSSH (Fig. 9).
4. 3. QS10 staining protocol
4. 3. 1. Materials
Pluronic F-127 (20% solution in DMSO; Invitrogen, Cat. No. P3000MP) is freshly diluted in DMEM before use.
QS10 is diluted in DMEM containing 0.01–0.04% of Pluronic F-127 to a final concentration of 1–5 μM.
DMEM (FUJIFILM Wako Pure Chemical Corporation, Cat. No. 044-29765)
4. 3. 2. Procedure
Cultured cells are incubated and loaded with 1–5 μM QS10 probe for 10 min after exchange of medium to serum-free DMEM.
Cell fluorescence is measured in real time by time-lapse ratio imaging at an excitation wavelength of 550 nm and detection wavelength of 560–640 nm [Ch1: TMR (tetramethylrhodamine) and Cy3 specifications]/650–750 nm (Ch2: Cy5 specifications) with a confocal laser scanning microscope.
4. 3. 3. Typical results observed
By using the QS10 staining protocol, intracellular RSSH formation can be identified by real-time live imaging with A549 cells and other common cell lines widely used in the laboratory (Fig. 11).
5. Summary
Investigation of the prevalence, (patho)physiology, biological mechanisms of action, and potential therapeutic applications of the polysulfide species described herein requires methods for their detection and quantitation in biological systems. The ephemeral nature and reactivity of many polysulfide species also necessitate a fundamental understanding of the detailed chemical biology to fully evaluate and appreciate their biological utility and function. The complex and intertwined chemistry that connects all of these species, as well as RSHs, requires special analytical techniques. Fortunately, the recent development of the various analytical techniques described herein provides methodology amenable to use in biological systems to begin to evaluate the physiology and biochemistry of these persulfide and polysulfide species. With these analytical tools (and others) in hand, the field is now primed to advance. Discovering what (patho)physiological roles and functions the polysulfide species have and how this knowledge can be used for the development of potential therapies and strategies for promoting life science and human health promises to be exciting.
Highlights.
Reactive sulfur species and per/polysulfides are abundant endogenous products.
Precise analytical technique is essential for understanding biology of polysulfides.
With analytical tools in hand, the reactive sulfur field is now primed to advance.
Polysulfide analyses, therapies and strategies promote science and human health.
Acknowledgements
This work was supported by an Institutional Development Award (IDeA) from the National Institutes of General Medical Sciences of the NIH under grant GM121307, and by National Heart, Lung, and Blood Institute grant HL149264 to C.G.K; by Grants-in-Aid for Scientific Research [(S), (B), (C), Early-Career Scientists, Challenging Exploratory Research] from the Ministry of Education, Science, Sports, and Technology (MEXT), Japan, to T.M. (19K07554), T.T. (20K15983), T.I. (20K07306), H.M. (20H04832), M.M. (19K07341) and T.A. (18H05277 and 20K21496); by the Japan Science and Technology Agency (JST), CREST, Japan to T.A. (JPMJCR2024); and by the Japan Agency for Medical Research and Development (AMED), Japan, to H.M. (JP21gm5010002).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Pryor WA, Houk KN, Foote CS, Fukuto JM, Ignarro LJ, Squadrito GL, Davies KJA, Free radical biology and medicine, it’s a gas, man, Am. J. Physiol. Regul. Integr. Comp. Physiol 291 (2006) R491–R511. [DOI] [PubMed] [Google Scholar]
- [2].Mustafa AK, Gadalla MM, Snyder SH, Signaling by gasotransmitters, Sci. Signal 2 (2009) 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Fukuto JM, Carrington SJ, Tantillo DJ, Harrison JG, Ignarro LJ, Freeman BA, Chen A, Wink DA, Small molecule signaling agents: the integrated chemistry and biochemistry of nitrogen oxides, oxides of carbon, dioxygen, hydrogen sulfide, and their derived species, Chem. Res. Toxicol 25 (2012) 769–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Basudhar D, Ridnour LA, Cheng R, Kesarwala AH, Heinecke J, Wink DA, Biological signaling by small molecule inorganics, Coord. Chem. Rev 306 (2016) 708–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].DeMartino AW, Zigler DF, Fukuto JM, Ford PC, Carbon disulfide. Just toxic or also bioregulatory and/or therapeutic? Chem. Soc. Rev 46 (2017) 21–39. [DOI] [PubMed] [Google Scholar]
- [6].Steiger AK, Zhao Y, Pluth MD, Emerging roles of carbonyl sulfide in chemical biology: sulfide transporter or gasotransmitter? Antiox. Redox Signal 28 (2018) 1516–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Czabo C, A timeline of hydrogen sulfide (H2S) research: from environmental toxin to biological mediator, Biochem. Pharmacol 149 (2018) 5–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Abe K, Kimura H, The possible role of hydrogen sulfide as an endogenous neuromodulator, J. Neurosci 16 (1996) 1066–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Benavides GA, Sqaudrito GL, Mills RW, Patel HD, Isbell TS, Patel RP, Darley-Usmar VM, Doeller JE, Kraus DW, Hydrogen sulfide mediates the vasoactivity of garlic, Proc. Natl. Acad. Sci. USA 104 (2007) 17977–17982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Stubbert D, Prysyazhna O, Rudyk O, Scotcher J, Burgoyne JR, Eaton P, Protein kinase G Iα oxidation paradoxically underlies blood pressure lowering by the reductant hydrogen sulfide, Hypertension 64 (2014) 1344–1351. [DOI] [PubMed] [Google Scholar]
- [11].Zhao W, Zhang J, Lu Y, Wang L, The vasorelaxation effect of H2S as a novel endogenous gaseous KATP channel opener, EMBO J. 20 (2001) 6008–6016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Wallace JL, Ferraz JGP, Muscara MN, Hydrogen sulfide: an endogenous mediator of resolution of inflammation and injury, Antiox. Redox Signal 17 (2012) 58–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Nagy P, Palinkas Z, Nagy A, Budai B, Toth I, Vasas A, Chemical aspects of hydrogen sulfide measurements in physiological samples, Biochim. Biophys. Acta 1840 (2014) 876–891. [DOI] [PubMed] [Google Scholar]
- [14].Toohey JI, Sulphane sulphur in biological systems: a possible regulatory role, Biochem. J 264 (1989) 625–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Fukuto JM, Ignarro LJ, Nagy P, Wink DA, Kevil CG, Feelisch M, Cortese-Krott MM, Bianco CL, Kumagai Y, Hobbs AJ, Lin J, Akaike T, Biological hydropersulfides and related polysulfides: a new concept and perspective in redox biology, FEES Lett. 592 (2018) 2140–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Toohey JI, Sulfur signaling: is the agent sulfide or sulfane? Anal. Biochem 413 (2011) 1–7. [DOI] [PubMed] [Google Scholar]
- [17].Hughes MN, Centelles MN, Moore KP, Making and working with hydrogen sulfide: the chemistry and generation of hydrogen sulfide in vitro and its measurement in vivo: a review, Free Radic. Biol. Med 47 (2009) 1346–1353. [DOI] [PubMed] [Google Scholar]
- [18].Li Q, Lancaster JR Jr., Chemical foundations of hydrogen sulfide biology, Nitric Oxide 35 (2013) 21–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ono K, Akaike T, Sawa T, Kumagai Y, Wink DA, Tantillo DJ, Hobbs AJ, Nagy P, Xian M, Lin J, Fukuto JM, The redox chemistry and chemical biology of H2S, hydropersulfides and derived species: implications of their possible biological activity and utility, Free Radic. Biol. Med 77 (2014) 82–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Filipovic MR, Zivanovic J, Alvarez B, Baneijee R, Chemical biology of H2S signaling through persulfidation, Chem. Rev 118 (2018) 1253–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Nagy P, Kinetics and mechanisms of thiol-disulfide exchange covering direct substitution and thiol oxidation-mediated pathways, Antiox. Redox Signal 18 (2013) 1623–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Rao GS, Gorin G, Reaction of cystine with sodium sulfide in sodium hydroxide solution, J. Org. Chem 24 (1959) 749–753. [Google Scholar]
- [23].Cavallini D, Federici G, Barboni E, Interactions of proteins with sulfide, Eur. J. Biochem 14 (1970) 169–174. [DOI] [PubMed] [Google Scholar]
- [24].Francoleon NE, Carrington SJ, Fukuto JM, The reaction of H2S with oxidized thiols: generation of persulfides and implications to H2S biology, Arch. Biochem. Biophys 516 (2011) 146–154. [DOI] [PubMed] [Google Scholar]
- [25].Cuevasanta E, Lange M, Bonanata J, Coitino EL, Ferrer-Sueta G, Filipovic MR, Alvarez B, Reaction of hydrogen sulfide with disulfide and sulfenic acid to form the strongly nucleophilic persulfide, J. Biol. Chem 290 (2015) 26866–26880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Vasas A, Doka E, Fabian I, Nagy P, Kinetic and thermodynamic studies on the disulfide-bond reducing potential of hydrogen sulfide, Nitric Oxide 46 (2015) 93–101. [DOI] [PubMed] [Google Scholar]
- [27].Koppenol WH, Bonds PL, Signaling by sulfur-containing molecules. Quantitative aspects, Arch. Biochem. Biophys 617 (2017) 3–8. [DOI] [PubMed] [Google Scholar]
- [28].Benchoam D, Semelak JA, Cuevasanta E, Mastrogiovanni M, Grassano JS, Ferrer-Sueta G, Zeida A, Trujillo M, Moller MN, Estrin DA, Alvarez B, Acidity and nucleophilic reactivity of glutathione persulfide, J. Biol. Chem 295 (2020) 15466– 15481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Saund SS, Sosa V, Henriquez S, Nguyen QNN, Bianco CL, Soeda S, Millikin R, White C, Le H, Ono K, Tantillo DJ, Kumagai Y, Akaike T, Lin J, Fukuto JM, The chemical biology of hydropersulfides (RSSH): chemical stability, reactivity and redox roles, Arch. Biochem. Biophys 588 (2015) 15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Bogdandi V, Ida T, Sutton TR, Bianco C, Ditroi T, Koster G, Henthorn HA, Minnion M, R Toscano J, van der Vliet A, Pluth MD, Feelisch M, Fukuto JM, Akaike T, Nagy R, Speciation of reactive sulfur species and their reactions with alkylating agents: do we have any clue about what is present inside the cell? Br. J. Pharmacol 176 (2018) 646–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Everett SA, Wardman P, Perthiols as antioxidants: radical-scavenging and prooxidative mechanisms, Methods Enzymol. 251 (1995) 55–69. [DOI] [PubMed] [Google Scholar]
- [32].Bianco CL, Chavez TA, Sosa V, Saund SS, Nguyen QNN, Tantillo DJ, Ichimura AS, Toscano JP, Fukuto JM, The chemical biology of the persulfide (RSSH)/perthiyl (RSS•) redox couple and possible role in biological redox signaling, Free Radio. Biol. Med 101 (2016) 20–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Bianco CL, Akaike T, Ida T, Nagy P, Bogdandi V, Toscano JP, Kumagai Y, Henderson CF, Goddu RN, Lin J, Fukuto JM, The reaction of hydrogen sulfide with disulfides: formation of a stable trisulfide and implications for biological systems. Br J. Pharmacol 176 (2019) 671–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Galardon E, Huguet F, Herrero C, Ricoux R, Artaud I, Padovani D, Reactions of persulfides with the heme cofactor of oxidized myoglobin and microperoxidase 11: reduction or coordination, Dalton Trans. 46 (2017) 7939–7946. [DOI] [PubMed] [Google Scholar]
- [35].Chauvin J-PR, Griesser M, Pratt DA, Hydropersulfides: H-atom transfer agents par excellence, J. Am. Chem. Soc 139 (2017) 6484–6493. [DOI] [PubMed] [Google Scholar]
- [36].Benson SW, Thermochemistry and kinetics of sulfur-containing molecules and radicals, Chem. Rev 78 (1978) 23–35. [Google Scholar]
- [37].Padmaja S, Huie RE, The reaction of nitric oxide with organic peroxyl radicals, Biochem. Biophys. Res. Commun 195 (1993) 539–544. [DOI] [PubMed] [Google Scholar]
- [38].Ida T, Sawa T, Ihara H, Kasamatsu S, Kunieda K, Tsuchiya Y, Watanabe Y, Kumagai Y, Nishida M, Suematsu M, Motohashi H, Fujii S, Matsunaga T, Yamamoto M, Ono K, Devarie-Baez NO, Xian M, Fukuto JM, Akaike T, Reactive cysteine persulfides and S-polythiolation regulate oxidative stress and redox signaling, Proc. Natl. Acad. Sci. USA 111 (2014) 7606–7611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Ezerinia D, Takano Y, Hanaoka K, Urano Y, P Dick T, N-Acetyl cysteine functions as a fast-acting antioxidant by triggering intracellular H2S and sulfane sulfur production, Cell Chem. Biol 25 (2018) 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Hankins RA, Suarez SI, Kalk MA, Green NM, Harty MN, Lukesh II JC, An innovative hydrogen peroxide-sensing scaffold and insight towards its potential as an ROS-activated persulfide donor, Angew. Chem 59 (2020) 22238–22245. [DOI] [PubMed] [Google Scholar]
- [41].Wang Y, Dillon KM, Li Z, Winckler EW, Matson JB, Alleviating cellular oxidative stress through treatment with superoxide-triggered persulfide prodrugs, Angew. Chem 59 (2020) 16698–16704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Shen X, Kolluru GK, Yuan S, Kevil CG, Measurement of H2S in vivo and in vitro by the monobromobimane method, Methods Enzymol. 554 (2015) 31–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Kolluru GK, Shen X, Kevil CG, Reactive sulfur species: a new redox player in cardiovascular pathophysiology, Arterioscler. Thromb. Vasc. Biol 40 (2020) 874–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Lin J, Akiyama M, Bica I, Long FT, Henderson CF, Goddu RN, Saurez Y, Baker B, Ida T, Shinkai Y, Nagy R, Akaike T, Fukuto JM, Kumagai Y, The uptake and release of polysuflur cysteine species by cells: physiological and toxicological implications, Chem. Res. Toxicol 32 (2019) 447–455. [DOI] [PubMed] [Google Scholar]
- [45].Morgan B, Ezerina D, Amoako TNE, Riemer J, Seedorf M, Dick TP, Multiple glutathione disulfide removal pathways mediate cytosolic redox homeostasis, Nat. Chem. Biol 9 (2013) 119–125. [DOI] [PubMed] [Google Scholar]
- [46].Henderson CF, Bica I, Long FT, Irwin DD, Stull CH, Baker BW, Suarez-Vega V, Taugher ZM, Fletes ED, Bartleson JM, Humphrey ML, Alvarez L, Akiyama M, Kumagai Y, Fukuto JM, Lin J, Cysteine trisulfide protects E. coli from electrophile-induced death through the generation of cysteine hydropersulfide, Chem. Res. Toxicol 33 (2020) 678–686. [DOI] [PubMed] [Google Scholar]
- [47].Capdevila DA, Walsh BJC, Zhang Y, Dietrich C, Gonzalez-Gutierrez G, R Giedroc D, Structural basis for persulfide-sensing specificity in a transcriptional regulator, Nat. Chem. Biol 17 (2021) 65–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Xuan G, Lu C, Xu H, Chen Z, Li K, Liu H, Liu H, Xia Y, Xun L, Sulfane sulfur is an intrinsic signal activating MexR-regulated antibiotic resistance in Pseudomonas aeruginosa, Mol. Microbiol 114 (2020) 1038–1048. [DOI] [PubMed] [Google Scholar]
- [49].Walsh BJC, Giedroc DP, H2S and reactive sulfur signaling at the host-bacterial pathogen interface, J. Biol. Chem 295 (2020) 13150–13168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Furne J, Saeed A, Levitt MD, Whole tissue hydrogen sulfide concentrations are orders of magnitude lower than presently accepted values, Am. J. Physiol. Regul. Integr. Comp. Physiol 295 (2008) R1479–R1485. [DOI] [PubMed] [Google Scholar]
- [51].Akaike T, Ida T, Wei F-Y, Nishida M, Kumagai Y, Alam M, Ihara H, Sawa T, Matsunaga T, Kasamatsu S, Nishimura A, Morita M, Tomizawa K, Nishimura A, Watanabe S, Inaba K, Shima H, Tanuma N, Jung M, Fujii S, Watanabe Y, Ohmuraya M, Nagy R, Feelisch M, Fukuto JM, Motohashi H, Cysteinyl-tRNA synthetase governs cysteine polysulfidation and mitochondrial bioenergetics, Nat. Commun 8 (2017) 1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Kasamatsu S, Ida T, Koga T, Asada K, Motohashi H, Ihara H, Akaike T, High-precision sulfur metabolomics innovated by a new specific probe for trapping reactive sulfur species, Antioxid. Redox Signal 34 (2021) 1407–1419. [DOI] [PubMed] [Google Scholar]
- [53].Sawa T, Takata T, Matsunaga T, Jung M, Ihara H, Motohashi H, Akaike T, Chemical biology of reactive sulfur species: hydrolysis equilibrium determinant of the physiological functions, Antioxid. Redox Signal (2021) in press, 10.1089/ars.2021.0170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Newton GL, Dorian R, Fahey RC, Analysis of biological thiols: derivatization with monobromobimane and separation by reverse-phase high-performance liquid chromatography, Anal. Biochem 114 (1981) 383–387. [DOI] [PubMed] [Google Scholar]
- [55].Shen X, Peter EA, Bir S, Wang R, Kevil CG, Analytical measurement of discrete hydrogen sulfide pools in biological specimens, Free Radic. Biol. Med 52 (2012) 2276–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Shen X, Pattillo CB, Pardue S, Bir SC, Wang R, Kevil CG, Measurement of plasma hydrogen sulfide in vivo and in vitro, Free Radic. Biol. Med 50 (2011) 1021–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Rajpal S, Katikaneni P, Deshotels M, Pardue S, Glawe J, Shen X, Akkus N, Modi K, Bhandari R, Dominic P, Reddy P, Kolluru GK, Kevil CG, Total sulfane sulfur bioavailability reflects ethnic and gender disparities in cardiovascular disease, Redox Biol. 15 (2018) 480–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Disbrow E, Stokes KY, Ledbetter C, Patterson J, Kelley R, Pardue S, Reekes T, Larmeu L, Batra V, Yuan S, Cvek U, Trutschl M, Kilgore P, Alexander JS, Kevil CG, Plasma hydrogen sulfide: a biomarker of Alzheimer’s disease and related dementias, Alzheimers Dement. March12 (2021) doi: 10.1002/alz.12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Meram AT, Chen J, Patel S, Kim DD, Shirley B, Covello P, Coppola D, Wei EX, Ghali G, Kevil CG, Shackelford RE, Hydrogen sulfide is increased in oral squamous cell carcinoma compared to adjacent benign oral mucosae, Anticancer Res. 38 (2018) 3843–3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Dominic P, Ahmad J, Bhandari R, Pardue S, Solorzano J, Jaisingh K, Watts M, Bailey SR, Orr AW, Kevil CG, Kolluru GK, Decreased availability of nitric oxide and hydrogen sulfide is a hallmark of COVID-19, Redox Biol. 43 (2021) 101982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Hamid HA, Tanaka A, Ida T, Nishimura A, Matsunaga T, Fujii S, Morita M, Sawa T, Fukuto JM, Nagy P, Tsutsumi R, Motohashi H, Ihara H, Akaike T, Polysulfide stabilization by tyrosine and hydroxyphenyl-containing derivatives that is important for a reactive sulfur metabolomics analysis, Redox Biol. 21 (2019) 101096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Shen X, Chakraborty S, Dugas TR, Kevil CG, Hydrogen sulfide measurement using sulfide dibimane: critical evaluation with electrospray ion trap mass spectrometry, Nitric Oxide 41 (2014) 97–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Ishigami M, Hiraki K, Umemura K, Ogasawara Y, Ishii K, Kimura H, A source of hydrogen sulfide and a mechanism of its release in the brain, Antioxid. Redox Signal 11 (2009) 205–214. [DOI] [PubMed] [Google Scholar]
- [64].Johnson DC, Dean DR, Smith AD, Johnson MK, Structure, function, and formation of biological iron-sulfur clusters. Annu. Rev. Biochem 74 (2005) 247–281. [DOI] [PubMed] [Google Scholar]
- [65].Ubuka T, Assay methods and biological roles of labile sulfur in animal tissues, J. Chromatogr. B 781 (2002) 227–249. [DOI] [PubMed] [Google Scholar]
- [66].Kosower NS, Kosower EM, Thiol labeling with bromobimanes, Methods Enzymol. 143 (1987) 76–84. [DOI] [PubMed] [Google Scholar]
- [67].Dóka É, Pader I, Bíró A, Johansson K, Cheng Q, Ballagó K, Prigge JR, Pastor-Flores D, Dick TP, Schmidt EE, Arnér ESJ, Nagy P, A novel persulfide detection method reveals protein persulfide- and polysulfide-reducing functions of thioredoxin and glutathione systems, Sci. Adv 2 (2016) e1500968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Dóka É, Ida T, Dagnell M, Abiko Y, Luong NC, Balog N, Takata T, Espinosa B, Nishimura A, Cheng Q, Funato Y, Miki H, Fukuto JM, Prigge JR, Schmidt EE, Arnér ESJ, Kumagai Y, Akaike T, Nagy P, Control of protein function through oxidation and reduction of persulfidated states, Sci. Adv 6 (2020) eaax8358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Fan K, Chen Z, Liu H, Evidence that the ProPerDP method is inadequate for protein persulfidation detection due to lack of specificity, Sci. Adv 6 (2020) eabb6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Dóka É, Arnér ESJ, Schmidt EE, Dick TP, van der Vliet A, Yang J, Szatmári R, Ditrói T, Wallace JL, Cirino G, Olson K, Motohashi H, Fukuto JM, Pluth MD, Feelisch M, Akaike T, Wink DA, Ignarro LJ, Nagy P, Comment on “Evidence that the ProPerDP method is inadequate for protein persulfidation detection due to lack of specificity”, Sci. Adv 7 (2021) eabe7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Chen W, Liu C, Peng B, Zhao Y, Pacheco A, Xian M, New fluorescent probes for sulfane sulfurs and the application in bioimaging, Chem. Sci 4 (2013) 2892–2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Chen W, Rosser EW, Matsunaga T, Pacheco A, Akaike T, Xian M, The development of fluorescent probes for visualizing intracellular hydrogen polysulfides, Angew. Chem 127 (2015) 14167–14171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Umezawa K, Kamiya M, Urano Y, A reversible fluorescent probe for real-time live-cell imaging and quantification of endogenous hydropolysulfides, Angew. Chem 127 (2018) 14167–14171. [DOI] [PubMed] [Google Scholar]
- [74].Sasakura K, Hanaoka K, Shibuya N, Mikami Y, Kimura Y, Komatsu T, Ueno T, Terai T, Kimura H, Nagano T, Development of a highly selective fluorescence probe for hydrogen sulfide, J. Am. Chem. Soc 133 (2011) 18003–18005. [DOI] [PubMed] [Google Scholar]
- [75].Takano Y, Hanaoka K, Shimamoto K, Miyamoto R, Komatsu T, Ueno T, Terai T, Kimura H, Nagano T, Urano Y, Development of a reversible fluorescent probe for reactive sulfur species, sulfane sulfur, and its biological application, Chem. Commun 53 (2017) 1064–1067. [DOI] [PubMed] [Google Scholar]


