Abstract
In 2020, the emergence of SARS-CoV-2 caused a global public health crisis with significant mortality rates and a large socioeconomic burden. The rapid spread of this new virus has led to the appearance of new variants, making the characterization and monitoring of genetic diversity necessary to understand the population dynamics and evolution of the virus. Here, a population-genetics-based study was performed starting with South American genome sequences available in the GISAID database to investigate the genetic diversity of SARS-CoV-2 on this continent and the evolutionary mechanisms that modulate it.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00705-021-05258-w.
Introduction
Many human infectious diseases have emerged because of the pathogen’s ability to jump hosts [1, 2]; this is the case with betacoronaviruses [3]. Like SARS-CoV and MERS-CoV [4, 5], a new member of the genus Betacoronavirus termed SARS-CoV-2 [6] emerged as a zoonotic virus [7, 8]. It was recorded for the first time at the end of 2019 in Wuhan, China [9], and it is responsible for the COVID-19 pandemic. The genome of SARS-CoV-2 is a 29.9-kb, positive-sense, single-stranded RNA with 10 ORFs and several overlapping genes [8, 10, 11]. Despite its high structural and genetic similarity to SARS-CoV, the rapid spread and different clinical manifestations have raised new questions about the evolution of this new virus.
In South America (SA), the virus arrived at approximately the end of February 2020 from the European continent, followed by multiple additional introductions [12–16]. Despite control measures by different governments to reduce infections, the virus spread across the continent and, as was expected, new variants emerged due to mutation, increasing viral genetic diversity.
Genetic diversity is a relevant factor for viruses to adapt to changing environments [17]. In the case of infectious pathogens, DNA/RNA sequence analysis can be useful for understanding what factors (mutation, natural selection, genetic drift, recombination, migration [18, 19]) shape the genetic diversity patterns in natural populations, and this a key tool for understanding species evolution, monitoring the emergence of new variants, associating viral genetic variations with disease, and assessing the spread of an outbreak [19, 20]. Considering that population-genetics-based studies integrating sequence data from SA are limited, here, we analyze the diversity of SARS-CoV-2 from seven SA countries using methods based on genealogy, population genetics, and coalescent theory.
Materials and methods
Data collection
A total of 2098 SARS-CoV-2 genome sequences from SA (Argentina, 38; Brazil, 1070; Chile, 183; Colombia, 175; Ecuador, 90; Peru, 384; Surinam, 71; Uruguay, 75; Venezuela, 12) submitted by December 2020 were downloaded from the database of the Global Initiative on Sharing Avian Influenza Data (GISAID, https://www.gisaid.org/, Online Resource 1). These sequences were screened, and only those with less than 1% of the data missing were used. Additionally, virus genome reference sequences of the Wuhan SARS-CoV-2 (GenBank accession number NC_045512.2), bat BtCoV RaTG13 (China National Center for Bioinformation accession number GWHABKP00000001), bat BtCoV RmYN02 (GISAID accession number EPI_ISL_412977) and pangolin MP789 (GenBank accession number MT121216.1) were obtained.
Sequence alignments and analysis of genetic diversity
Only entries in which missing data or ambiguous nucleotides accounted for less than 1% of the genome sequence were included in the dataset. Ultimately, 1141 genome sequences (Argentina, 33; Brazil, 426; Chile, 162; Colombia, 107; Ecuador, 67; Peru, 303; Suriname, 43) were aligned with the Wuhan SARS-CoV-2, bat BtCoV RaTG13, BtCoV RmYN02, and pangolin MP789 coronavirus genome reference sequences using MUSCLE [21], followed by manual editing. The 5′ and 3′ ends of the aligned sequences were excised, resulting in an alignment consisting of 29,404 sites from ORF1a to ORF10. Genome sequences from Uruguay and Venezuela were omitted because they were submitted as partial genome sequences or had more than 1% of the data missing.
Several measures of DNA sequence polymorphism within or between populations were estimated, including the number of segregating sites (Ss), the number of singleton (S) and parsimony-informative sites (Ps), the number of haplotypes (H, the set of single-nucleotide polymorphisms found in the genome), the nucleotide polymorphism per site (θW), the nucleotide diversity per site (π) applying the Jukes-Cantor correction [22], and the average number of nucleotide differences between any two sequences (κ). A sliding window for nucleotide diversity with a window length of 100 sites and a step size of 25 sites was used. All of these measurements were made with DnaSP v.6 software [23]; gaps or sites with missing data were excluded. In addition, the Phylogenetic Assignment of Named Global Outbreak Lineages (PANGOLIN) tool v.3.1.5 [24] was used to identify the SARS-CoV-2 lineages in the data set.
Assessing molecular evolution
Natural selection, genetic drift, mutation, recombination, and gene flow are the essential explanatory factors used for understanding the population genetic basis of evolutionary change [18, 19]. Several tests were performed to assess these evolutionary forces. Tajima’s D [25] and Fu and Li’s D*/F* [26] frequency spectrum tests were performed to assess the null hypothesis that all mutations are selectively neutral [27]. In addition, haplotype-based tests, such as K, Hd [28], and Fu’s Fs [29], were also implemented. Statistical significance for the frequency spectrum and the haplotype-based test was evaluated using coalescence simulations, obtaining confidence intervals. All of these procedures were carried out using DnaSP v.6 software. Tajima’s D and Fu’s Fs were also computed on Arlequin v.3.1 software [30].
Natural selection at the inter- and intraspecies level was assessed using the coding alignment. First, the molecular evolution rate was inferred by partitioning the alignment at codon positions 1, 2, and 3 to obtain the relative substitution rates at all three codon positions using a Bayesian approach in BEAST v.1.7 software [31]. Then, the selection signatures (positive and purifying) at individual sites (codons) were detected using the single-likelihood ancestor counting (SLAC) [32], mixed-effects model of evolution (MEME) [33], and fast, unconstrained Bayesian approximation (FUBAR) methods [34]. A sliding window for omega rates (ω = dN-dS) was also used with the SLAC data. Because recombination can bias the inference of positive selection sites [35, 36], the genetic algorithm for recombination detection (GARD) method [37] was used before running the aforementioned tests. Later, the effective number of codons (ENC) [38] and the codon bias index (CBI) [39] were estimated to measure the selective pressure at the translational level [40, 41].
On the other hand, linkage disequilibrium (LD) was assessed by using the ZNS statistic [42]; this is the average of R2 over all pairwise comparisons. Linear regression between LD and nucleotide distance was then performed to ascertain whether recombination could have occurred. Recombination was also evaluated using the GARD algorithm [37] and RDP v.5 software [43].
Demographic history in South America
Statistically significant values in the neutrality test could be due to demographic factors. Therefore, the demographic history of SARS-CoV-2 in SA was assessed by computing the distribution of the observed number of differences between pairs of haplotypes (the mismatch distribution) using Arlequin software. The expected values for a demographic expansion were calculated and plotted against the observed values. Both the pure demographic expansion and the spatial expansion models were used to compute the sum of square deviations (SSD) between the observed and expected mismatch as a test statistic, as well as the raggedness index of the observed distribution defined by Harpending [44]. In addition, a Bayesian skyline plot (BSP) was used to examine changes in the effective population size (Ne) through time using the Bayesian Markov chain Monte Carlo (MCMC) approach available in BEAST v.1.7 software. BEAUti v.1.7 was used to generate a BEAST XML file for BSP using the sample collection dates, the generalized time-reversible (GTR) substitution model with gamma site heterogeneity (+G), base frequency estimates (+F), and a strict clock. The MCMC length chain was run for 50 million generations. Tracer v1.5 [45] was used check the effective sample size (ESS) as well as to construct the BSP plot for each population.
Genetic structure indices based on the allelic content of haplotypes and their frequencies [46] were estimated by molecular variance analysis (AMOVA). The pairwise FST index was computed for all population pairs; this can be used as short-term genetic distances between populations [47, 48]. We also tested the hypothesis of individuals’ random distribution between population pairs by performing an exact test of population differentiation [49]. These analyses were performed using Arlequin software. NETWORK v.10 software was then used for constructing a median-joining network [50] to evaluate possible mutational pathways giving rise to SARS-CoV-2 haplotypes (the set of single-nucleotide polymorphisms found on the genome), their distribution, and frequencies. The network was inferred with the star contraction [51] and post-processing MP options [52] to delete all superfluous median vectors and links not contained in the shortest trees in the network. In addition, the GTR+G+F model was used to infer a maximum-likelihood (ML) phylogenetic tree using the IQ-TREE web server [53]. Topology reliability was evaluated by bootstrap analysis, using 1,000 replicates.
Diversity of the spike, E, and M proteins
The viral spike, E, and M proteins are exposed to the immune system and are therefore the main proteins of interest for vaccine development. Thus, the diversity of these proteins at the amino acid level was assessed independently, since different substitutions within them may alter their antigenicity, which is a potential immune escape mechanism.
Results
SARS-CoV-2 genetic diversity in SA
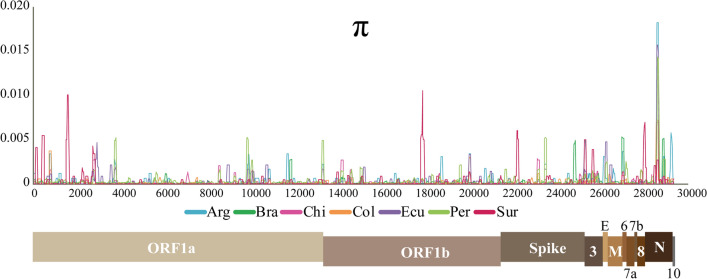
A total of 1141 genome sequences from seven countries in SA were analyzed to evaluate the genetic diversity of SARS-CoV-2. In SA, there were 1260 segregating sites (Ss), 679 haplotypes (H), and 57 lineages (L). Of the Ss, 65.6% were singleton sites and 34.4% were parsimony-informative sites. The average number of nucleotide differences (κ = 8) was like previous estimates around the world [54, 55]. The diversity estimators displayed low values (θW = 5.7 × 10-3 and π = 2.8 × 10-4), with Peru, Argentina, and Suriname having the viral populations with the highest nucleotide sequence diversity; Colombia had the lowest diversity (Table 1). Because the sample collection dates differed from one country to another, these estimators were calculated again using only the sequences obtained between March and May 2020, and the results were similar to those reported above. The nucleotide diversity sliding window showed that the most diverse regions of the genome were located within the ORF1a, ORF3, E, ORF6, and N regions (Fig. 1).
Table 1.
Estimators of SARS-CoV-2 genetic diversity in South America
| n | Sites | Ss | S | Ps | M | H | L | θw | π | κ | Tajima | Fu and L | Fu | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D | D* | F* | Fs | ||||||||||||
| South America | |||||||||||||||
| 1142 | 28,945 | 1260 | 827 | 433 | 1278 | 679 | 57 | 0.0057 | 0.00028 | 8.10 | − 2.78 | − 19.2 | − 10.22 | < − 30.0 | |
| Estimators per country | |||||||||||||||
| Argentina | 33 | 29,109 | 68 | 41 | 27 | 68 | 28 | 7 | 0.0006 | 0.00032 | 9.31 | − 1.65 | − 2.61* | − 2.51* | − 15.17 |
| Brazil | 426 | 29,369 | 547 | 420 | 127 | 550 | 300 | 14 | 0.0028 | 0.00022 | 6.46 | − 2.82+ | − 15.31+ | − 9.85+ | − 32.28 |
| Chile | 162 | 29,397 | 153 | 104 | 49 | 153 | 79 | 16 | 0.0009 | 0.00020 | 5.90 | − 2.50+ | − 7.82* | − 6.40* | − 80.93+ |
| Colombia | 107 | 29,314 | 121 | 85 | 36 | 121 | 58 | 15 | 0.0008 | 0.00015 | 4.40 | − 2.66+ | − 6.62* | − 5.90* | − 33.15+ |
| Ecuador | 67 | 29,409 | 71 | 40 | 31 | 71 | 26 | 11 | 0.0005 | 0.00024 | 7.06 | − 1.78+ | − 3.57+ | − 3.44+ | − 5.202 |
| Peru | 303 | 29,393 | 522 | 318 | 204 | 524 | 186 | 23 | 0.0028 | 0.00039 | 11.46 | − 2.69+ | − 9.87* | − 7.16* | − 24.63+ |
| Suriname | 43 | 29,409 | 70 | 39 | 31 | 70 | 35 | 2 | 0.0006 | 0.00030 | 8.82 | − 1.60 | − 2.71* | − 2.75* | − 21.97++ |
n number of sequences analyzed, Sites number of sites analyzed, Ss number of polymorphic segregating sites, S singleton sites, Ps parsimony-informative sites, M number of mutations, H number of haplotypes, L number of lineages, θw nucleotide polymorphism, π nucleotide diversity, κ average number of nucleotide differences
*p < 0.05, **p < 0.02, +p < 0.01, ++p < 0.001
Fig. 1.
Sliding window of nucleotide diversity. SARS-CoV-2 diversity in seven South American countries was assessed by computing the nucleotide diversity per site. Window length, 100 bp; step size, 25 bp. A SARS-CoV-2 genome model is given below the sliding window displaying the ORFs.
Regarding lineages, the earliest A and B lineages were present in some countries. A was found in Chile, while B was present in Brazil, Chile, Colombia, and Peru. The lineages A.1, A.2, and A.5 were found in Chile, Colombia, and Peru, whereas the B.1 and B.1.1 lineages were found in all of the countries (Online Resource 2, Supplementary Table S1). The country with the most lineages was Peru, while Suriname was the country with the fewest lineages (Table 1 and Online Resource 2, Supplementary Table S1).
Assessing molecular evolution
Several tests were performed to determine the impact of evolutionary forces on SARS-CoV-2 diversity in SA. Tajima’s D and Fu and Li’s D*/F* neutrality tests showed statistically significant negative values (Table 1, Online Resource 2, Supplementary Table S2). The haplotype number and haplotype diversity did not show significant differences from those expected under neutrality. On the other hand, Fu’s Fs estimator had statistically significant negative values. All of these estimators were then calculated locus by locus. Tajima’s D, Fu and Li’s D*/F*, and Fu’s Fs showed statistically significant negative values in almost all loci. Moreover, the haplotype number was higher than expected under neutrality at some of them (Online Resource 2, Supplementary Table S3).
Comparison of the relative substitution rates at the three codon positions showed that the third position had a much higher evolution rate (about 12-fold faster) than the other two codon positions (Online Resource 3). Furthermore, codon-based methods showed that more than 2660 codons were affected by purifying selection (Fig. 2 and Online Resource 4). However, positive selection signals were also found throughout the genome (Fig. 2 and Online Resource 4). Most of the positively selected codons were found in the regions encoding the non-structural proteins ORF1a (22 sites), ORF1b (9 sites), and ORF3 (9 sites), as well as in the region encoding the spike protein (9 sites). Most of these changes were non-conservative (i.e., the amino acid was replaced by one with different physicochemical properties), with D614G in the spike protein, G204R in N, and V/L3606F in ORF1 being the most frequent polymorphisms. Moreover, S84L was detected in ORF8, which is a reversion to the ancestral state. Finally, seven codons under positive selection showed substitutions corresponding to interspecies changes, all of which were non-conservative substitutions. These were in the regions that encode the spike (3 sites), ORF1a (1 site), M (1 site), ORF8 (1 sites), and N (1 sites) proteins.
Fig. 2.
Sliding window for the ω (dN - dS) rate. The nonsynonymous (dN) and synonymous (dS) substitution rates throughout the SARS-CoV-2 genome were determined based on SLAC data. A SARS-CoV-2 genome model is given below the sliding window displaying the 10 ORFs. Values greater than zero indicate a positive selection, while values below zero indicate purifying selection.
The ENC and CBI were estimated to assess whether the low genetic diversity in SARS-CoV-2 could be (in part) due to selective pressure at the translational level. The average ENC value was 45.4, while the CBI value was 0.38 in all countries. Finally, some estimators were calculated to determine if recombination had taken place in the SARS-CoV-2 genomes in SA. The ZNS values were relatively low (Online Resource 2, Supplementary Table S2). Additionally, no correlation was observed between the LD and nucleotide distance (Online Resource 5A). Using the GARD method as well as the programs in RDP, no intraspecies recombination signals were found. However, RDP software did find recombination signals at the interspecies level (Online Resource 5B).
Demographic history in South America
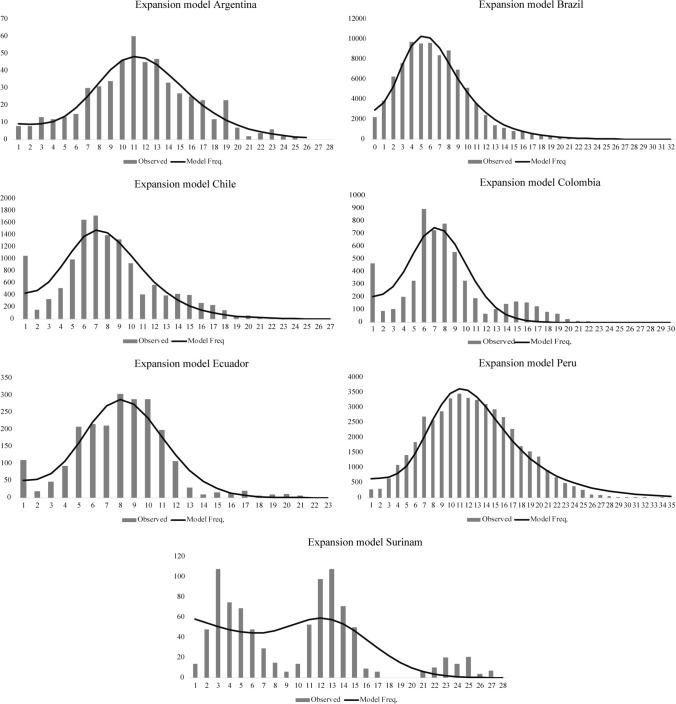
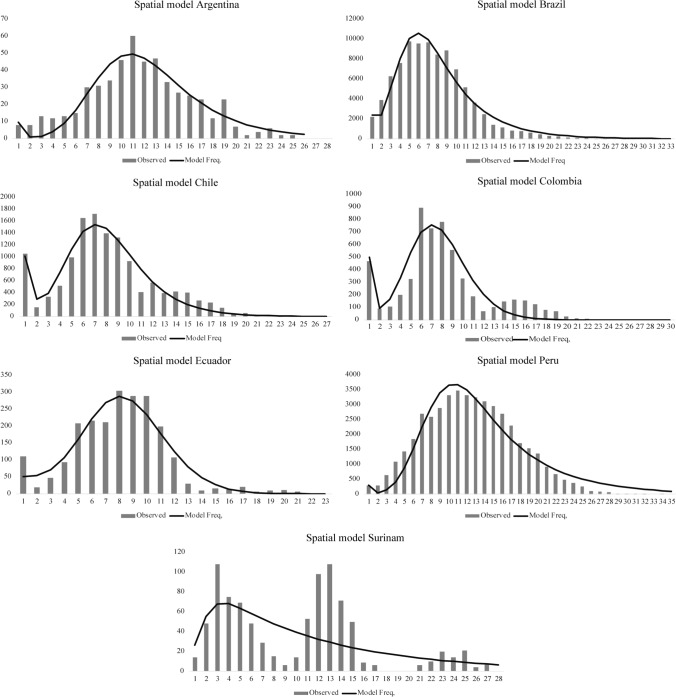
The demographic models based on the distribution of the observed number of differences between pairs of haplotypes in the sample displayed a unimodal distribution for each country (Figs. 3 and 4). The population data from Argentina, Brazil, and Peru showed that the observed distribution fit the expectations under the pure demographic expansion model (Fig. 3). Meanwhile, the data from Chile and Colombia fit the spatial expansion model better (Fig. 4). In Suriname, the observed data showed a multimodal distribution (Figs. 3 and 4).
Fig. 3.
Mismatch distribution for the pure demographic expansion model. Frequency distributions of the observed number of pairwise nucleotide differences between haplotypes for seven South American countries are shown. The solid line is the theoretical distribution under the assumption of pure demographic expansion.
Fig. 4.
Mismatch distribution for the spatial expansion model. Frequency distributions of the observed number of pairwise nucleotide differences between haplotypes for seven South American countries are shown. The solid line is the theoretical distribution under the assumption of spatial population expansion.
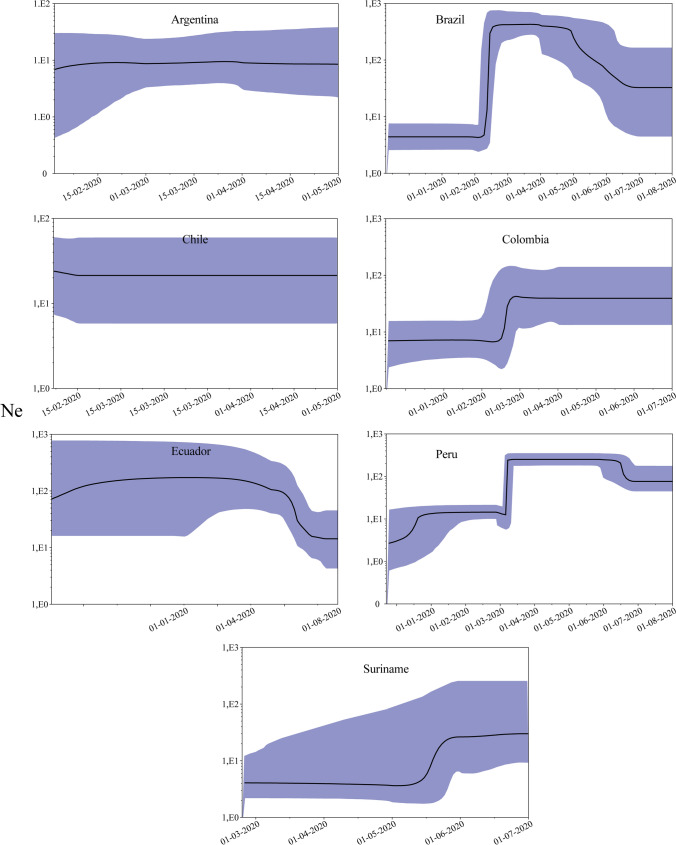
The effective virus population size (Ne) changes were then inferred within each country (Fig. 5). An increase in Ne was observed throughout January and February 2020; Brazil and Peru showed a drastic increase. The lowest Ne was observed in Suriname. In that country, Ne showed an increase around May 15. This behavior seems to agree with World Health Organization (WHO) statistics on the average number of cases reported daily for each country.
Fig. 5.
Bayesian skyline plot (BSP) of the SARS‐CoV‐2 outbreak. BSP assessed changes in effective population size (Ne) in seven South American countries. The y‐axis indicates Ne, and the x‐axis shows the time in days/months. The solid black line represents the estimated median value, and the blue shades indicate the 95% highest posterior density.
In order to assess the population structure, an AMOVA analysis was performed evaluating the significance of the covariance components associated with the different possible levels of genetic structure. Given that the sequences recovered from the GISAID database were collected at different times, those collected during the same period for all countries (March to May) were chosen to perform this analysis. Both within and among viral populations, genetic structure indices showed statically significant values (Table 2). Eighty percent of the estimated variation in SARS-CoV-2 in South America was due to variation within populations, while differences between them could explain about 20% of the variation. The degree of genetic differentiation between populations was inferred using the FST index. Comparisons between populations showed statistically significant values higher than 0.05, indicating a slight genetic differentiation (Table 2). The exact test of population differentiation also displayed statistical support for genetic differentiation in SA. This also was evident when the lineage data were considered. There were several non-shared lineages circulating within each country (Online Resource 2, Supplementary Table S1).
Table 2.
Analysis of molecular variance (AMOVA) analysis and population genetic differentiation estimated from March to May 2020 using the fixation index (FST)
| Source of variation | Sum of squares | Variance components | Percentage variation | |
|---|---|---|---|---|
| Among populations | 503.17 | 0.73 | 19.73++ | |
| Within populations | 2717.86 | 2.97 | 80.27++ | |
| Total | 3221.03 | 3.70 |
| Arg | Bra | Chi | Col | Ecu | Per | |
|---|---|---|---|---|---|---|
| Arg | ++ | ++ | ++ | ++ | ||
| Bra | 0.14 | ++ | ++ | ++ | ++ | |
| Chi | 0.08 | 0.25 | ++ | ++ | ++ | |
| Col | 0.13 | 0.32 | 0.02 | + | ++ | |
| Ecu | 0.03 | 0.13 | 0.08 | 0.16 | ** | |
| Per | 0.06 | 0.19 | 0.11 | 0.15 | 0.03 |
Arg Argentina, Bra Brazil, Chi Chile, Col Colombia, Ecu Ecuador, Per Peru
**p < 0.02; +p < 0.01; ++p < 0.00001.
Likewise, the network results showed three high-frequency haplotypes (Online Resource 6A). The first one included the Wuhan-Hu-1 haplotype and sequences belonging to the B, B.1, B.1.8, B.1.9, B.1.110, B.1.111, B.1.67, B.1.195, B.1.223, B.1.225, B.1.547, B.3, and B.6 lineages. The second high-frequency haplotype included sequences from the B.1, B11, B.1.1.28, B.1.1.33, and N.4 lineages. The last high-frequency haplotype included sequences from the B.1, B.1.1, B.1.1.10, B.1.1.28, B.1.1.33, B.1.1.231, B.1.1.388, and B.59 lineages. Others derived from these three major haplotypes formed a star-like shape in the network. The low representation of haplotypes from Suriname is due to the fact that only 12 cases were reported from March to May, and only three sequences were available in the GISAID database. By including all of the sequences recovered by December 2020, a new haplotype network was inferred (Online Resource 6B) that preserved the topology shown in Online Resource 6A. It was also evidenced that all of the Suriname haplotypes (which cluster into the B.1.219 and N.2 lineages) seem to be exclusive to that country, showing a clear structuring. The ML phylogenetic tree agreed with these results (Online Resource 6C).
Diversity of the spike, E, and M proteins
SARS-CoV-2 has four structural proteins that are essential for viral assembly, attachment, and entry into host cells. The spike protein is a 1273-amino-acid (aa) glycoprotein that is involved in the interaction with human receptors. One hundred fourteen haplotypes (“alleles”) of this protein were observed in South America, 109 of which had the D614G mutation (Online Resource 7). Haplotype 1 (Wuhan-Hu-1) had a low frequency, while haplotype 2 (with the 614G mutation) within the B.1 lineage was the most frequent in all populations except in Suriname, where haplotype 109 (E281V and D614G) within the B.1.219 lineage was the major haplotype.
Unlike the spike protein, M (222 aa) and E (75 aa) showed fewer haplotypes, with six and three, respectively (Online Resource 8). The Wuhan-Hu-1 haplotype was the most frequent in all countries for both the M and E proteins. However, haplotype 4 of M (D3G) reached 20.9% in Ecuador and 12.9% in Peru, and haplotype 2 of E (V58F) reached 18.2% in Argentina.
Discussion
Natural selection, genetic drift, and demographic processes play an important role in shaping the genetic background of natural populations, and the genetic diversity within them is a determining factor for survival of the species. Hence, characterization and monitoring of polymorphisms are key to understanding the dynamics and evolution of pathogens [19, 20]. The emergence of SARS-CoV-2 in humans is a relatively recent event that has triggered the current pandemic. The virus was reported in SA on February 26, when the first case of COVID-19 was registered in a 61-year-old traveler arriving in Brazil from Italy. A few days later, more cases of COVID-19 were reported in other countries in the region. Since then, it has spread to all SA countries.
A high diversity of lineages was found in SA, with B.1 (20.6%), B.1.1.33 (19.3%), B.1.1 (13.5%), and B.1.1.28 (11.4%) being the most frequent lineages. However, the genetic diversity could be higher than the lineage diversity, since some mutations might be present in some but not all sequences within lineage. For instance, in the P.2 lineage, the spike protein substitutions are E484K, D614G, V1176F, and F565L, but the last of these substitutions has been detected in some sequences but not all. Thus, SARS-CoV-2 genetic diversity was investigated. The first reports show low diversity and a low frequency of recurrent mutations [56–58]. According to our analysis, singleton sites (S, rare variants carried by a unique sequence) were 1.9-fold higher than parsimony-informative sites (Ps, recurring variants in two or more sequences), contrasting with the first reports, where S was three times higher than Ps [56–58]. This suggests an increase in the frequency of some variants that emerged after the appearance of this new betacoronavirus.
Various estimators can be calculated from the number of segregating sites (Ss) and the number of sequences to assess the genetic diversity of populations. Although nucleotide polymorphism (or Watterson estimator θW) was considered the best estimator of SARS-CoV-2 diversity [57], the nucleotide diversity (π) is a better estimator, since it considers the Ss frequency, and it is not influenced by the sample size. The nucleotide diversity in SA (π = 0.00028) was lower than in other continents/countries (Africa, π = 0.00047; Oceania, π = 0.00046; United States of America, π = 0.00044; Europe, π = 0.00043; Asia, π = 0.00042 [59]). All of these values were lower than those observed for other human viruses (H1N1 = 0.004-0.012 [60, 61]; H3N2 = 0.004 [60]; hepatitis B virus = 0.015 [62]; parainfluenza virus = 0.002 [63]), which could be due to the fact that SARS-CoV-2 is a new human virus.
The nucleotide diversity sliding window showed that the most diverse regions were located at the ORF1a, ORF3, and N loci (Fig. 1), agreeing with previous reports [59]. The viral populations from Peru, Argentina, and Suriname had the highest nucleotide diversity in SA. The covid performance (https://interactives.lowyinstitute.org/features/covid-performance/) gauges the relative performance of countries at different points in the pandemic. No correlation between nucleotide diversity (or lineage diversity) and covid performance was observed. Argentina (π = 0.0003), Brazil (π = 0.0002), and Colombia (π = 0.0001) got low scores (<10), while Peru (π = 0.0004), Ecuador, and Chile (π = 0.0002) had a score >30. The diversity observed could have resulted from different numbers of introductions [12–16] and the subsequent mutations emerging after arrival of the virus in each country, some of which have led to new lineages and variants of epidemiological interest [64].
The genetic diversity observed in natural populations results from five evolutionary forces: mutation, genetic drift, natural selection, recombination, and migration [18, 19]. Several tests were carried out to determine the impact of these forces on SARS-CoV-2 diversity in SA by the first months of the pandemic. Neutrally tests showed statistically significant negative values (Table 1). This could be the outcome of directional selection, selective sweep, or a population expansion. Selection is expected to affect diversity at some regions within loci. In contrast, a demographic process is expected to affect diversity throughout the genome.
This latter scenario is supported by the distribution of polymorphisms as well as by locus-by-locus analysis. The polymorphisms distribution in each population (the mismatch distribution) displayed a unimodal pattern (Figs. 3 and 4); populations that have experienced demographic growth in the recent past show this behavior [65]. Furthermore, statistically significant negative values for Fu’s Fs by country and by locus (Table 1 and Online Resource 2, Supplementary Table S3) indicate an excess in the number of recent mutations, supporting a demographic expansion. This is expected, since COVID-19 is a pandemic. However, Suriname showed a multimodal distribution, which could be due to either a population in equilibrium, co-circulation of multiple lineage, migration, or a population admixture.
Population expansion is expected to occur rapidly after the arrival of the virus. To address this scenario, changes in the effective population size (Ne) was inferred (Fig. 5). Ne increased before the first reported cases (February 26–March 13), suggesting that the virus may have arrived at SA in early January/mid-February. Phylogenetic analysis conducted in Colombia supports this [14, 16]. Once again, Suriname showed a different behavior. There, an increase in Ne seems to have occurred in mid-May although the first case was reported on March 13. Thus, Ne increase does not correspond to the arrival of the virus but to the control measures taken by the local government. They adopted social distancing and isolation only one day after the first case. This response appears to have contained virus dispersion. Both the Ne and case number increase coincide with the parliamentary elections, when citizens went out to vote on May 25 [66].
In contrast to Suriname, a drastic Ne increase was observed in Ecuador, Peru, or Brazil. From the time of the arrival of the virus in Brazil, the central government was discouraging the measures of physical distancing and lockdown brought in by state governors, and this, together with the complicated social situation, led to an increase in the number of cases [67], triggering an increase in the viral Ne. The aforesaid may suggest the effectiveness of early isolation to contain viral spread, and this should be considered in future outbreaks or new potential pandemics.
On the other hand, a population structuring is probable, considering the spatial expansion model observed in SA and since founder effects and genetic drift appear to be the main forces in SARS-CoV-2 evolution. Both within and among viral populations, genetic structure indices showed statically significant values (Table 2). The degree of genetic differentiation between populations using the FST index and population differentiation exact test showed low but statistically significant values, suggesting a slight genetic differentiation. Network and ML tree results and the presence of several non-shared lineages support this (Online Resource 2, Supplementary Table S1 and Online Resource 6).
Purifying selection also modulates SARS-CoV-2 evolution. The high evolutionary rate observed in the third codon position and the large number of sites with a dN - dS value < 0 confirm this (Fig. 2 and Online Resources 3 and 4). However, positive selection (+Sel) cannot be completely ruled out. Codon-based methods identified sites under +Sel at the intraspecies level. Most of them were found in regions encoding nonstructural proteins (Online Resource 4). Furthermore, the amino acid replacements change physicochemical properties. It is not clear why these proteins are the most diverse and had +Sel signatures. Thus, these variants should be monitored. Likewise, it needs to be evaluated through functional assays whether these replacements influence viral processes. Additionally, seven non-conservative amino acid replacements among SARS-CoV-2, bat CoV, and pangolin CoV displayed +Sel signatures (Online Resource 4). These could be ancient adaptive changes of a progenitor of SARS-CoV-2, as has been suggested previously [68].
Analysis of the structural spike protein displayed 114 unique haplotypes (“alleles”) in SA (Online Resource 7), and some sites were under +Sel, which could be due to diversifying selection, as the immune system exerts selective pressure or directional selection as an adaptation to the host. Within this last scenario, position D614G stands out, which was identified as being under positive selection in all of the populations analyzed here. Spike haplotypes containing both variants were found in SA, with the G mutation being the most frequent. The 614G polymorphism appears to affect viral infectivity and transmissibility [69–71]. Considering that a) under drift, polymorphism is expected to be fixed or get lost randomly, b) this variant had positive selection signals, and c) 614G has almost completely replaced the 614D variant worldwide, it is feasible to think that this mutation has been fixed by directional selection.
Recently, several SARS-CoV-2 variants and/or lineages have emerged and are circulating worldwide. Some of them are considered Variants of Interest (VOI: Epsilon [B.1.427 and B.1.429], Eta [B.1.525], Iota [B.1.526], Kappa [B.1.617.1], Zeta [P.2]), Variants of Concern (VOC: Alpha [B.1.1.7], Beta [B.1.351], Delta [B.1.617.2], Gamma [P.1]) or Variants of Note (A.23.1 and B.1.525). Fifty-seven different lineages from the dataset analyzed were found in SA, but none of these variants were detected. However, several spike mutations defining the Gamma and Zeta variants (i.e., E484K, F565L, D614G, and V1176F of Zeta and P26S, E484K, and D614G of Gamma) were already circulating in SA before the first report of these variants (Online Resource 7). Thus, virus isolates (or lineages) containing some of these mutations were their ancestors. For instance, the Gamma variant is a sublineage of B.1.1.28, which has been observed to be circulating in Brazil since March 2020. Additionally, other mutations (i.e., L5F, T95I, Q677H, P681R, and K1191N) observed in other VOIs or VOCs were present in the dataset analyzed here (Online Resource 7), suggesting a potential in SA for the emergence of new VOI or VOC, since they have been defined by multiple substitutions that usually arise independently within different lineages (convergent mutation) [72].
On the other hand, selection at the transcriptional level can also modulate diversity [40, 41]. To assess this scenario, ENC and CBI values were computed. ENC values ranges from 61 (all of the synonymous codons for each amino acid are used equally) to 20 (only one codon per amino acid). Likewise, CBI values range from 0 (uniform use of synonymous codons) to 1 (maximum codon bias). Since average values were higher than 45 and lower than 0.4, respectively, selective pressure at the transcriptional level was ruled out. Finally, recombination is an evolutionary force that is important for many viruses [73, 74]. Recombination signals have been observed previously in the SARS-CoV-2 genome [57, 75]. In addition, phylogenetic evidence of recombination has been reported when novel bat coronaviruses closely related to SARS-CoV-2 were considered [76]. Here, recombination events were observed at the interspecies level (Online Resource 5B), but not within the SARS-CoV-2 genome. No correlation was observed between the LD and nucleotide distance (Online Resource 5A). Moreover, GARD and RDP did not show intraspecies recombination signals. Thus, although recombination has taken place in coronaviruses, this mechanism is not yet a relevant factor in the evolution of the SARS-CoV-2 genome in SA.
SARS-CoV-2 has four structural proteins that are necessary for virus assembly, receptor binding, and entry into host cells, making them potential vaccine candidates. However, mutations within these proteins could alter their antigenicity. The sequences of the M and E proteins were found to be highly conserved in SA, and therefore, they might be considered good vaccine candidates. However, they appear to be poorly immunogenic [77], and their use as vaccine candidates is therefore limited. On the other hand, the spike glycoprotein, which plays an important role in the interaction with the ACE2 receptor, is the main target of the immune system and has therefore been used in vaccine design [77]. The vaccines developed so far use the full-length spike sequence or just the RBD domain. Mutations in RDB (D354, Y364, and F367) have been reported in several variants from Asia, Europe, and the USA, suggesting that these could impact antigenicity [62]. Mutational scanning analysis has detected several escape mutations within the N-terminal spike domain and RBD at positions 148, 150-151, 406, 409, 439, 443-452, 455-456, 472-473, 477, 483-486, 490, 493, 499, and 503 [72]. In SA, some of these sites (or sites close to them) as well as other sites throughout the protein showed polymorphism (Online Resource 7). It is not yet clear whether these variations in SA strains impact antigenicity. If so, the Sputnik and Janssen vaccines could be highly effective in SA, as RBD had a low degree of polymorphism among the sequences analyzed here. Conversely, the long-term effectiveness of the Pfizer, Moderna, and AstraZeneca vaccines, which induce antibodies directed against different regions of the spike [77], could be affected by the diversity found in the spike protein. Further studies are necessary to evaluate whether spike polymorphism will undergo a balancing selection process, maintaining an excess of variants at intermediate frequencies in the population as an immune evasion mechanism or due to selective pressure exerted by vaccination.
Supplementary Information
Below is the link to the electronic supplementary material.
Online Resource 1 Acknowledgement data source (XLSX 28 KB)
Online Resource 2 Supplementary Tables S1–S3 (PDF 165 KB)
Online Resource 3 The relative substitution rates at all three codon positions (TIF 911 KB)
Online Resource 4 Positions in the SARS-CoV-2 genome under natural selection in South America. The codons under positive or purifying (negative) selection were identified by codon-based methods for each population independently. The physicochemical properties of the amino acid replacements were assessed using the PAM250 matrix (Dayhoff and Schwartz, 1978). *, interspecies; +, reversion to ancestral character; italics, conservative change; bold, non-conservative change (XLSX 139 KB)
Online Resource 5 Recombination analysis. A. Relationship between linkage disequilibrium (LD) and nucleotide distance estimated for each South America country. The red line represents the regression line between LD and nucleotide distance. B. Recombination events found at the interspecies level (TIF 1563 KB)
Online Resource 6 Median-joining network for South America haplotypes and ML tree. A. Median-joining network for South America haplotypes. The figure shows the SARS-CoV-2 haplotypes identified in seven South American countries between March and May 2020. B. Median-joining network for South America haplotypes. The figure shows the SARS-CoV-2 haplotypes identified in seven South America countries using all sequences available by December 2020 in the GISAID database. For both networks, the star contraction option was chosen for simplifying interpretation of the network. Each node is a haplotype, and its size indicate its frequency. The lines connecting the haplotypes represent the different mutational paths explaining the relationship and evolutionary origin. The early A and B lineages are shown. C. SARS-CoV-2 phylogenetic tree inferred by maximum likelihood using genome sequences from South America. The topology was obtained using the GTR + G + F model. Lineage data are provided (TIF 2850 KB)
Online Resource 7 Spike haplotype alignment. The alignment shows the haplotypes (“alleles”) found in the spike protein. The numbers at the top indicate the amino acid positions where segregating sites were observed. The numbering is based on the SARS-CoV-2 Wuhan reference genome sequence (GenBank accession number NC_045512.2). The dots indicate identical amino acids. Red highlighted sites indicate the mutations observed in Variants of Interest or Variants of Concern (TIF 2931 KB)
Online Resource 8 M and E haplotype alignment. The alignment shows the haplotypes (“alleles”) found in M (A) and E (B) proteins. The numbers at the top indicate the amino acid positions where segregating sites were observed. The numbering is based on the SARS-CoV-2 Wuhan reference genome sequence (GenBank accession number NC_045512.2). The dots indicate identical amino acids (TIF 1133 KB)
Acknowledgements
We would like to thank the authors, laboratories, and institutions responsible for generating and sharing via GISAID the SARS-CoV-2 genome data. Special thanks to Gypsy Español, who reviewed this document. This research is dedicated to the memory of Professor Leopoldo Arrieta Violet, who died recently. We will always be grateful to Professor Leopoldo.
Author contributions
SPB: data collection, analysis and writing of the original draft of the manuscript. DG-O: conceptualization, design, analysis, and writing-review of the manuscript. All authors read and approved the final manuscript.
Funding
This work was carried out within the framework of project number BPIN 202010075 supported by the Sistema General de Regalias from Colombia.
Availability of data and material
The genome sequence data on which this research is based is available in the GISAID database.
Declarations
Conflict of interest
The authors have no competing interests to declare.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Sindy P. Buitrago, Email: biopao.k18@gmail.com
Diego Garzón-Ospina, Email: degarzon@gmail.com.
References
- 1.McArthur DB. Emerging infectious diseases. Nurs Clin N Am. 2019;54:297–311. doi: 10.1016/j.cnur.2019.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang LF, Crameri G. Emerging zoonotic viral diseases. Rev Sci Tech. 2014;33:569–581. doi: 10.20506/rst.33.2.2311. [DOI] [PubMed] [Google Scholar]
- 3.Lu G, Wang Q, Gao GF. Bat-to-human: spike features determining 'host jump' of coronaviruses SARS-CoV, MERS-CoV, and beyond. Trends Microbiol. 2015;23:468–478. doi: 10.1016/j.tim.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coleman CM, Frieman MB. Emergence of the Middle East respiratory syndrome coronavirus. PLoS Pathog. 2013;9:e1003595. doi: 10.1371/journal.ppat.1003595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thomas PA. Severe acute respiratory syndrome. Indian J Med Microbiol. 2003;21:152–160. doi: 10.1016/S0255-0857(21)03064-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coronaviridae Study Group of the International Committee on Taxonomy of V The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lau SKP, Luk HKH, Wong ACP, Li KSM, Zhu L, He Z, Fung J, Chan TTY, Fung KSC, Woo PCY. Possible bat origin of severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis. 2020;26:1542–1547. doi: 10.3201/eid2607.200092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, Bi Y, Ma X, Zhan F, Wang L, Hu T, Zhou H, Hu Z, Zhou W, Zhao L, Chen J, Meng Y, Wang J, Lin Y, Yuan J, Xie Z, Ma J, Liu WJ, Wang D, Xu W, Holmes EC, Gao GF, Wu G, Chen W, Shi W, Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bogoch II, Watts A, Thomas-Bachli A, Huber C, Kraemer MUG, Khan K. Pneumonia of unknown aetiology in Wuhan, China: potential for international spread via commercial air travel. J Travel Med. 2020 doi: 10.1093/jtm/taaa008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, Yuan ML, Zhang YL, Dai FH, Liu Y, Wang QM, Zheng JJ, Xu L, Holmes EC, Zhang YZ. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pavesi A. New insights into the evolutionary features of viral overlapping genes by discriminant analysis. Virology. 2020;546:51–66. doi: 10.1016/j.virol.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Candido DDS, Watts A, Abade L, Kraemer MUG, Pybus OG, Croda J, de Oliveira W, Khan K, Sabino EC, Faria NR. Routes for COVID-19 importation in Brazil. J Travel Med. 2020 doi: 10.1093/jtm/taaa042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Castillo AE, Parra B, Tapia P, Lagos J, Arata L, Acevedo A, Andrade W, Leal G, Tambley C, Bustos P, Fasce R, Fernandez J. Geographical distribution of genetic variants and lineages of SARS-CoV-2 in Chile. Front Public Health. 2020;8:562615. doi: 10.3389/fpubh.2020.562615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramirez JD, Florez C, Munoz M, Hernandez C, Castillo A, Gomez S, Rico A, Pardo L, Barros EC, Castaneda S, Ballesteros N, Martinez D, Vega L, Jaimes JE, Cruz-Saavedra L, Herrera G, Patino LH, Teheran AA, Gonzalez-Reiche AS, Hernandez MM, Sordillo EM, Simon V, van Bakel H, Paniz-Mondolfi A. The arrival and spread of SARS-CoV-2 in Colombia. J Med Virol. 2021;93:1158–1163. doi: 10.1002/jmv.26393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Candido DS, Claro IM, de Jesus JG, Souza WM, Moreira FRR, Dellicour S, Mellan TA, du Plessis L, Pereira RHM, Sales FCS, Manuli ER, Theze J, Almeida L, Menezes MT, Voloch CM, Fumagalli MJ, Coletti TM, da Silva CAM, Ramundo MS, Amorim MR, Hoeltgebaum HH, Mishra S, Gill MS, Carvalho LM, Buss LF, Prete CA, Jr, Ashworth J, Nakaya HI, Peixoto PS, Brady OJ, Nicholls SM, Tanuri A, Rossi AD, Braga CKV, Gerber AL, de CGAP. Gaburo N, Jr, Alencar CS, Ferreira ACS, Lima CX, Levi JE, Granato C, Ferreira GM, Francisco RS, Jr, Granja F, Garcia MT, Moretti ML, Perroud MW, Jr, Castineiras T, Lazari CS, Hill SC, de Souza Santos AA, Simeoni CL, Forato J, Sposito AC, Schreiber AZ, Santos MNN, de Sa CZ, Souza RP, Resende-Moreira LC, Teixeira MM, Hubner J, Leme PAF, Moreira RG, Nogueira ML, Brazil-Uk Centre for Arbovirus Discovery DG, Epidemiology Genomic N. Ferguson NM, Costa SF, Proenca-Modena JL, Vasconcelos ATR, Bhatt S, Lemey P, Wu CH, Rambaut A, Loman NJ, Aguiar RS, Pybus OG, Sabino EC, Faria NR. Evolution and epidemic spread of SARS-CoV-2 in Brazil. Science. 2020;369:1255–1260. doi: 10.1126/science.abd2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laiton-Donato K, Villabona-Arenas CJ, Usme-Ciro JA, Franco-Munoz C, Alvarez-Diaz DA, Villabona-Arenas LS, Echeverria-Londono S, Cucunuba ZM, Franco-Sierra ND, Florez AC, Ferro C, Ajami NJ, Walteros DM, Prieto F, Duran CA, Ospina-Martinez ML, Mercado-Reyes M. Genomic epidemiology of severe acute respiratory syndrome coronavirus 2, Colombia. Emerg Infect Dis. 2020;26:2854–2862. doi: 10.3201/eid2612.202969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arber W. Genetic variation: molecular mechanisms and impact on microbial evolution. FEMS Microbiol Rev. 2000;24:1–7. doi: 10.1111/j.1574-6976.2000.tb00529.x. [DOI] [PubMed] [Google Scholar]
- 18.Casillas S, Barbadilla A. Molecular population genetics. Genetics. 2017;205:1003–1035. doi: 10.1534/genetics.116.196493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moya A, Holmes EC, Gonzalez-Candelas F. The population genetics and evolutionary epidemiology of RNA viruses. Nat Rev Microbiol. 2004;2:279–288. doi: 10.1038/nrmicro863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grubaugh ND, Ladner JT, Lemey P, Pybus OG, Rambaut A, Holmes EC, Andersen KG. Tracking virus outbreaks in the twenty-first century. Nat Microbiol. 2019;4:10–19. doi: 10.1038/s41564-018-0296-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jukes TH, Cantor C. Evolution of protein molecules. In: Munro HN, editor. Mammalian protein metabolism. New York: Academic Press; 1969. [Google Scholar]
- 23.Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, Sánchez-Gracia A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol Biol Evol. 2017;34:3299–3302. doi: 10.1093/molbev/msx248. [DOI] [PubMed] [Google Scholar]
- 24.Rambaut A, Holmes EC, O'Toole A, Hill V, McCrone JT, Ruis C, du Plessis L, Pybus OG. Addendum: a dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol. 2021;6:415. doi: 10.1038/s41564-021-00872-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fu YX, Li WH. Statistical tests of neutrality of mutations. Genetics. 1993;133:693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kimura M. The neutral theory of molecular evolution. Cambridge New York: Cambridge University Press; 1983. [Google Scholar]
- 28.Depaulis F, Veuille M. Neutrality tests based on the distribution of haplotypes under an infinite-site model. Mol Biol Evol. 1998;15:1788–1790. doi: 10.1093/oxfordjournals.molbev.a025905. [DOI] [PubMed] [Google Scholar]
- 29.Fu YX. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 1997;147:915–925. doi: 10.1093/genetics/147.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform Online. 2007;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- 31.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kosakovsky Pond SL, Frost SDW. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol Biol Evol. 2005;22:1208–1222. doi: 10.1093/molbev/msi105. [DOI] [PubMed] [Google Scholar]
- 33.Murrell B, Wertheim JO, Moola S, Weighill T, Scheffler K, Kosakovsky Pond SL. Detecting individual sites subject to episodic diversifying selection. PLoS Genet. 2012;8:e1002764. doi: 10.1371/journal.pgen.1002764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murrell B, Moola S, Mabona A, Weighill T, Sheward D, Kosakovsky Pond SL, Scheffler K. FUBAR: a fast, unconstrained bayesian approximation for inferring selection. Mol Biol Evol. 2013;30:1196–1205. doi: 10.1093/molbev/mst030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anisimova M, Nielsen R, Yang Z. Effect of recombination on the accuracy of the likelihood method for detecting positive selection at amino acid sites. Genetics. 2003;164:1229–1236. doi: 10.1093/genetics/164.3.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arenas MPD. The influence of recombination on the estimation of selection from coding sequence alignments. In: Fares MA, editor. Natural selection: methods and applications. Boca Raton: CRC Press/Taylor Francis; 2014. pp. 112–125. [Google Scholar]
- 37.Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SD. Automated phylogenetic detection of recombination using a genetic algorithm. Mol Biol Evol. 2006;23:1891–1901. doi: 10.1093/molbev/msl051. [DOI] [PubMed] [Google Scholar]
- 38.Wright F. The 'effective number of codons' used in a gene. Gene. 1990;87:23–29. doi: 10.1016/0378-1119(90)90491-9. [DOI] [PubMed] [Google Scholar]
- 39.Morton BR. Chloroplast DNA codon use: evidence for selection at the psb A locus based on tRNA availability. J Mol Evol. 1993;37:273–280. doi: 10.1007/bf00175504. [DOI] [PubMed] [Google Scholar]
- 40.Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes. J Mol Biol. 1981;146:1–21. doi: 10.1016/0022-2836(81)90363-6. [DOI] [PubMed] [Google Scholar]
- 41.Shields DC, Sharp PM, Higgins DG, Wright F. "Silent" sites in Drosophila genes are not neutral: evidence of selection among synonymous codons. Mol Biol Evol. 1988;5:704–716. doi: 10.1093/oxfordjournals.molbev.a040525. [DOI] [PubMed] [Google Scholar]
- 42.Kelly JK. A test of neutrality based on interlocus associations. Genetics. 1997;146:1197–1206. doi: 10.1093/genetics/146.3.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martin DP, Murrell B, Golden M, Khoosal A, Muhire B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015;1:vev003. doi: 10.1093/ve/vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harpending HC. Signature of ancient population growth in a low-resolution mitochondrial DNA mismatch distribution. Hum Biol. 1994;66:591–600. [PubMed] [Google Scholar]
- 45.Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. Posterior summarization in Bayesian phylogenetics using tracer 1.7. Syst Biol. 2018;67:901–904. doi: 10.1093/sysbio/syy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reynolds J, Weir BS, Cockerham CC. Estimation of the coancestry coefficient: basis for a short-term genetic distance. Genetics. 1983;105:767–779. doi: 10.1093/genetics/105.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Slatkin M. A measure of population subdivision based on microsatellite allele frequencies. Genetics. 1995;139:457–462. doi: 10.1093/genetics/139.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raymond M, Rousset F. An exact test for population differentiation. Evolution. 1995;49:1280–1283. doi: 10.1111/j.1558-5646.1995.tb04456.x. [DOI] [PubMed] [Google Scholar]
- 50.Bandelt HJ, Forster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- 51.Forster P, Torroni A, Renfrew C, Rohl A. Phylogenetic star contraction applied to Asian and Papuan mtDNA evolution. Mol Biol Evol. 2001;18:1864–1881. doi: 10.1093/oxfordjournals.molbev.a003728. [DOI] [PubMed] [Google Scholar]
- 52.Polzin T, Daneshmand SV. On Steiner trees and minimum spanning trees in hypergraphs. Oper Res Lett. 2003;31:12–20. doi: 10.1016/S0167-6377(02)00185-2. [DOI] [Google Scholar]
- 53.Trifinopoulos J, Nguyen LT, von Haeseler A, Minh BQ. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016;44:W232–235. doi: 10.1093/nar/gkw256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Power JP, Lawlor E, Davidson F, Holmes EC, Yap PL, Simmonds P. Molecular epidemiology of an outbreak of infection with hepatitis C virus in recipients of anti-D immunoglobulin. Lancet. 1995;345:1211–1213. doi: 10.1016/s0140-6736(95)91993-7. [DOI] [PubMed] [Google Scholar]
- 55.van Dorp L, Acman M, Richard D, Shaw LP, Ford CE, Ormond L, Owen CJ, Pang J, Tan CCS, Boshier FAT, Ortiz AT, Balloux F. Emergence of genomic diversity and recurrent mutations in SARS-CoV-2. Infect Genet Evol. 2020;83:104351. doi: 10.1016/j.meegid.2020.104351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dearlove B, Lewitus E, Bai H, Li Y, Reeves DB, Joyce MG, Scott PT, Amare MF, Vasan S, Michael NL, Modjarrad K, Rolland M. A SARS-CoV-2 vaccine candidate would likely match all currently circulating variants. Proc Natl Acad Sci USA. 2020;117:23652–23662. doi: 10.1073/pnas.2008281117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chaw SM, Tai JH, Chen SL, Hsieh CH, Chang SY, Yeh SH, Yang WS, Chen PJ, Wang HY. The origin and underlying driving forces of the SARS-CoV-2 outbreak. J Biomed Sci. 2020;27:73. doi: 10.1186/s12929-020-00665-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu Q, Zhao S, Shi CM, Song S, Zhu S, Su Y, Zhao W, Li M, Bao Y, Xue Y, Chen H. Population genetics of SARS-CoV-2: disentangling effects of sampling bias and infection clusters. Genom Proteom Bioinform. 2020 doi: 10.1016/j.gpb.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Flores-Alanis A, Cruz-Rangel A, Rodriguez-Gomez F, Gonzalez J, Torres-Guerrero CA, Delgado G, Cravioto A, Morales-Espinosa R. Molecular epidemiology surveillance of SARS-CoV-2: mutations and genetic diversity one year after emerging. Pathogens. 2021 doi: 10.3390/pathogens10020184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.López-Labrador FX, Natividad-Sancho A, Pisareva M, Komissarov A, Salvatierra K, Fadeev A, Moya A, Grudinin M, Díez-Domingo J, Afanasieva O, Konovalova N, Sominina A, Puig-Barberà J. Genetic characterization of influenza viruses from influenza-related hospital admissions in the St. Petersburg and Valencia sites of the Global Influenza Hospital Surveillance Network during the 2013/14 influenza season. J Clin Virol. 2016;84:32–38. doi: 10.1016/j.jcv.2016.09.006. [DOI] [PubMed] [Google Scholar]
- 61.Martinez-Hernandez F, Jimenez-Gonzalez DE, Martinez-Flores A, Villalobos-Castillejos G, Vaughan G, Kawa-Karasik S, Flisser A, Maravilla P, Romero-Valdovinos M. What happened after the initial global spread of pandemic human influenza virus A (H1N1)? A population genetics approach. Virol J. 2010;7:196. doi: 10.1186/1743-422x-7-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Phan T. Genetic diversity and evolution of SARS-CoV-2. Infect Genet Evol. 2020;81:104260. doi: 10.1016/j.meegid.2020.104260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Beck ET, He J, Nelson MI, Bose ME, Fan J, Kumar S, Henrickson KJ. Genome sequencing and phylogenetic analysis of 39 human parainfluenza virus type 1 strains isolated from 1997–2010. PLoS ONE. 2012;7:e46048. doi: 10.1371/journal.pone.0046048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Munoz M, Patino LH, Ballesteros N, Paniz-Mondolfi A, Ramirez JD. Characterizing SARS-CoV-2 genome diversity circulating in South American countries: signatures of potentially emergent lineages? Int J Infect Dis. 2021;105:329–332. doi: 10.1016/j.ijid.2021.02.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rogers AR, Harpending H. Population growth makes waves in the distribution of pairwise genetic differences. Mol Biol Evol. 1992;9:552–569. doi: 10.1093/oxfordjournals.molbev.a040727. [DOI] [PubMed] [Google Scholar]
- 66.Araya H (2020) Elections Suriname May 2020. First elections in Latin America at the time of COVID-19. Minsait, Buenos Aires. https://www.minsait.com/ideasfordemocracy/en/elections-suriname-may-2020-covid19
- 67.Lancet, COVID-19 in Brazil: "so what?". Lancet. 2020;395:1461. doi: 10.1016/S0140-6736(20)31095-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.MacLean OA, Lytras S, Weaver S, Singer JB, Boni MF, Lemey P, Kosakovsky Pond SL, Robertson DL. Natural selection in the evolution of SARS-CoV-2 in bats created a generalist virus and highly capable human pathogen. PLoS Biol. 2021;19:e3001115. doi: 10.1371/journal.pbio.3001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, Hengartner N, Giorgi EE, Bhattacharya T, Foley B, Hastie KM, Parker MD, Partridge DG, Evans CM, Freeman TM, de Silva TI, McDanal C, Perez LG, Tang H, Moon-Walker A, Whelan SP, LaBranche CC, Saphire EO, Montefiori DC. Tracking changes in SARS-CoV-2 Spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827.e819. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nakashima A, Takeya M, Kuba K, Takano M, Nakashima N. Virus database annotations assist in tracing information on patients infected with emerging pathogens. Inform Med Unlocked. 2020;21:100442. doi: 10.1016/j.imu.2020.100442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Plante JA, Liu Y, Liu J, Xia H, Johnson BA, Lokugamage KG, Zhang X, Muruato AE, Zou J, Fontes-Garfias CR, Mirchandani D, Scharton D, Bilello JP, Ku Z, An Z, Kalveram B, Freiberg AN, Menachery VD, Xie X, Plante KS, Weaver SC, Shi PY. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2020 doi: 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Harvey WT, Carabelli AM, Jackson B, Gupta RK, Thomson EC, Harrison EM, Ludden C, Reeve R, Rambaut A, Consortium C-GU. Peacock SJ, Robertson DL. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jackwood MW, Boynton TO, Hilt DA, McKinley ET, Kissinger JC, Paterson AH, Robertson J, Lemke C, McCall AW, Williams SM, Jackwood JW, Byrd LA. Emergence of a group 3 coronavirus through recombination. Virology. 2010;398:98–108. doi: 10.1016/j.virol.2009.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Perez-Losada M, Arenas M, Galan JC, Palero F, Gonzalez-Candelas F. Recombination in viruses: mechanisms, methods of study, and evolutionary consequences. Infect Genet Evol. 2015;30:296–307. doi: 10.1016/j.meegid.2014.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Motayo BO, Oluwasemowo OO, Olusola BA, Akinduti PA, Arege OT, Obafemi YD, Faneye AO, Isibor PO, Aworunse OS, Oranusi SU. Evolution and genetic diversity of SARS-CoV-2 in Africa using whole genome sequences. Int J Infect Dis. 2020;103:282–287. doi: 10.1016/j.ijid.2020.11.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhou H, Chen X, Hu T, Li J, Song H, Liu Y, Wang P, Liu D, Yang J, Holmes EC, Hughes AC, Bi Y, Shi W. A novel bat coronavirus closely related to SARS-CoV-2 contains natural insertions at the S1/S2 cleavage site of the spike protein. Curr Biol. 2020;30(2196–2203):e2193. doi: 10.1016/j.cub.2020.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dai L, Gao GF. Viral targets for vaccines against COVID-19. Nat Rev Immunol. 2021;21:73–82. doi: 10.1038/s41577-020-00480-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Online Resource 1 Acknowledgement data source (XLSX 28 KB)
Online Resource 2 Supplementary Tables S1–S3 (PDF 165 KB)
Online Resource 3 The relative substitution rates at all three codon positions (TIF 911 KB)
Online Resource 4 Positions in the SARS-CoV-2 genome under natural selection in South America. The codons under positive or purifying (negative) selection were identified by codon-based methods for each population independently. The physicochemical properties of the amino acid replacements were assessed using the PAM250 matrix (Dayhoff and Schwartz, 1978). *, interspecies; +, reversion to ancestral character; italics, conservative change; bold, non-conservative change (XLSX 139 KB)
Online Resource 5 Recombination analysis. A. Relationship between linkage disequilibrium (LD) and nucleotide distance estimated for each South America country. The red line represents the regression line between LD and nucleotide distance. B. Recombination events found at the interspecies level (TIF 1563 KB)
Online Resource 6 Median-joining network for South America haplotypes and ML tree. A. Median-joining network for South America haplotypes. The figure shows the SARS-CoV-2 haplotypes identified in seven South American countries between March and May 2020. B. Median-joining network for South America haplotypes. The figure shows the SARS-CoV-2 haplotypes identified in seven South America countries using all sequences available by December 2020 in the GISAID database. For both networks, the star contraction option was chosen for simplifying interpretation of the network. Each node is a haplotype, and its size indicate its frequency. The lines connecting the haplotypes represent the different mutational paths explaining the relationship and evolutionary origin. The early A and B lineages are shown. C. SARS-CoV-2 phylogenetic tree inferred by maximum likelihood using genome sequences from South America. The topology was obtained using the GTR + G + F model. Lineage data are provided (TIF 2850 KB)
Online Resource 7 Spike haplotype alignment. The alignment shows the haplotypes (“alleles”) found in the spike protein. The numbers at the top indicate the amino acid positions where segregating sites were observed. The numbering is based on the SARS-CoV-2 Wuhan reference genome sequence (GenBank accession number NC_045512.2). The dots indicate identical amino acids. Red highlighted sites indicate the mutations observed in Variants of Interest or Variants of Concern (TIF 2931 KB)
Online Resource 8 M and E haplotype alignment. The alignment shows the haplotypes (“alleles”) found in M (A) and E (B) proteins. The numbers at the top indicate the amino acid positions where segregating sites were observed. The numbering is based on the SARS-CoV-2 Wuhan reference genome sequence (GenBank accession number NC_045512.2). The dots indicate identical amino acids (TIF 1133 KB)
Data Availability Statement
The genome sequence data on which this research is based is available in the GISAID database.