Abstract
Summary
Hydrogen–Deuterium eXchange coupled to mass spectrometry is a powerful tool for the analysis of protein dynamics and interactions. Bottom-up experiments looking at deuterium uptake differences between various conditions are the most common. These produce multi-dimensional data that can be challenging to depict in a single visual format. Each user must also set significance thresholds to define meaningful differences and make these apparent in data presentation. To assist in this process, we have created HD-eXplosion, an open-source, web-based application for the generation of chiclet and volcano plots with statistical filters. HD-eXplosion fills a void in available software packages and produces customizable plots that are publication quality.
Availability and implementation
The HD-eXplosion application is available at http://hd-explosion.utdallas.edu. The source code can be found at https://github.com/HD-Explosion.
1 Introduction
Hydrogen–Deuterium eXchange (HDX) with mass spectrometry is an analytical approach for monitoring protein conformation and interfaces in solution (Pirrone et al., 2015; Wales and Engen, 2006). Most HDX experiments are bottom-up, measuring deuterium incorporation into backbone amides at the peptide level. Deuterium uptake is measured over time to uncover kinetic insights that relate to amide H-bonding and solvent accessibility (Masson et al., 2019; Wales and Engen, 2006). Data interpretation however is complicated, as uneven loss of deuterium and difficulty in creating a fully deuterated control, limit the ability to measure underlying rates of exchange. HDX is more often used comparatively, looking at a protein in multiple states, such as with and without a ligand (Chalmers et al., 2006, 2011; Iacob et al., 2015). In these experiments, the data must be interrogated to determine if deuterium uptake of a peptide varies between states (Hageman and Weis, 2019). Our web-based application, HD-eXplosion, assists this process by generating and applying statistical thresholds to certain plots of comparative HDX data.
One challenge inherent in a comparative HDX experiment is the multi-dimensionality of the final data. Pertinent variables include peptide boundaries, exposure time, protein state, deuterium uptake and some measure of reproducibility (Brown and Wilson, 2017; Masson et al., 2017). Given the large number of variables, it is near impossible to encapsulate all the information in a single aggregate graphic. Various plots are available and user choice may be biased by the availability and ease-of-use of computational tools. Example computation tools include MEMHDX (Hourdel et al., 2016) and Deuteros (Lau et al., 2019). HD-eXplosion provides such computational tools for two plots, the chiclet plot and the volcano plot, to the HDX community. It has a user-friendly interface that bypasses the need for coding skills or even program installation.
Another challenge in a comparative HDX experiment is setting thresholds for what constitutes a meaningful difference in deuterium uptake (Masson et al., 2019). While the field agrees that replicates are required, there lacks a consensus on statistical handling. The most basic metric is the magnitude of the difference in deuterium uptake. This has frequently been used in isolation without regard to peptide length or error. Several laboratories have also touted the use of P-values that incorporate the standard deviation of replicate measurements. Recently it has been shown that both metrics should be applied if false positives and false negatives are to be avoided (Hageman and Weis, 2019). In line with this, HD-eXplosion can apply two statistical thresholds when creating chiclet and volcano plots. It allows users complete control in setting these thresholds, although a P-value less than 0.01 and a difference in deuterium uptake greater than the pooled standard deviation are recommended values (Hageman and Weis, 2019). HD-eXplosion calculates P-values, but the deuteration cutoff needs to be determined offline.
The chiclet plot, named after the candy-coated chewing gum, was pioneered by the Engen laboratory (Kerres et al., 2017). It is a grid colored according to the difference in deuterium uptake. One axis lists the peptides ordered from the N- to the C-terminus of the protein, while the other is exposure time. Chiclet plots show where differences in deuterium uptake occur and display data from multiple time points in a single figure. The main drawbacks of chiclet plots are the size of the images, especially for large proteins, and an inability to see measurement error. To date, chiclet plots have mostly used the difference in deuterium uptake as a single threshold and have been created manually using spreadsheets. HD-eXplosion can be used to create customizable chiclet plots with two thresholds of significance in an automated fashion.
Volcano plots are widely used to visualize sequencing and -omics data and were championed in the HDX arena by the Weis laboratory (Hageman and Weis, 2019; Liu et al., 2020). They are 2-D scatter plots with each point representing a peptide at a specific time. The x-axis is the difference in deuterium uptake and the y-axis is the calculated P-value. A dashed box within the plot shows significance cutoffs. Volcano plots display thresholds in the context of the data distribution and show all peptides at all time points in a compact graph. The primary disadvantage of volcano plots is that the relative location of the peptide in the protein sequence and its change over time are not immediately apparent. Few of the currently available HDX programs have volcano plots as an option for output. HD-eXplosion can quickly create volcano plots with user-selected cutoffs and generate a list of significant peptides.
An additional feature in HD-eXplosion is the extraction of a list of peptides shared between datasets. This has utility for direct comparison of biological replicates or when a protein is independently studied in multiple states. The extracted list can be used to create chiclet and volcano plots. HD-eXplosion is thus an accessible web-based application for the generation of publication-quality chiclet and volcano plots with statistical thresholds.
An additional feature in HD-eXplosion is the extraction of a list of peptides shared between datasets. This has utility for direct comparison of biological replicates or when a protein is independently studied in multiple states. The extracted list can be used to create chiclet and volcano plots. HD-eXplosion is thus an accessible web-based application for the generation of publication-quality chiclet and volcano plots with statistical thresholds.
2 Materials and methods
HD-eXplosion is an open-source web-based application written in HTML, Javascript and Python. It uses Flask (1.1.1) for the application backend, Numpy (1.18.2) for statistics (Oliphant, 2006), Pandas (1.0.3) for data formatting and Matplotlib (3.2.1) for data plotting.
3 Results
HD-eXplosion is an online tool for the generation of chiclet and volcano plots with statistical thresholds. The webpage is organized into panels for uploading data, setting plot parameters and previewing/exporting generated plots. Data can be uploaded as a CSV file that was directly exported from DynamX or manually prepared. Shared peptides can be extracted from up to five different CSV files and the resulting peptide list downloaded. Data are submitted to the server with the ‘Submit’ button. To generate customized plots, several parameters can be defined (Fig. 1). For chiclet plots, the user selects the protein, comparison, color range and step, layout features and statistical filter. For volcano plots, the user selects the protein, time point(s), comparison, range of both axes, layout features and significance cutoffs. The P-value used is based on a Welch’s t-test (Hageman and Weis, 2019), which tests whether two populations with different variances have the same means. The number of replicates is also specified. Insignificant differences will be shown as white in the chiclet plot and will be separated by cutoff lines in the volcano plot. Plots are previewed in the webpage or full screen after pressing the ‘Show’ button, and can be downloaded as PNG, EPS or CSV files. The list of significant peptides in the volcano plot can also be downloaded. The webpage easily resets with the ‘Start Over’ button. More extensive guidelines are included in a pdf attached to the ‘Instructions’ button and a demo file is included. HD-eXplosion provides a user-friendly interface for the rapid generation of chiclet and volcano plots with statistical filtering. It is easily implemented, and we hope will be of value to the HDX community.
Fig. 1.
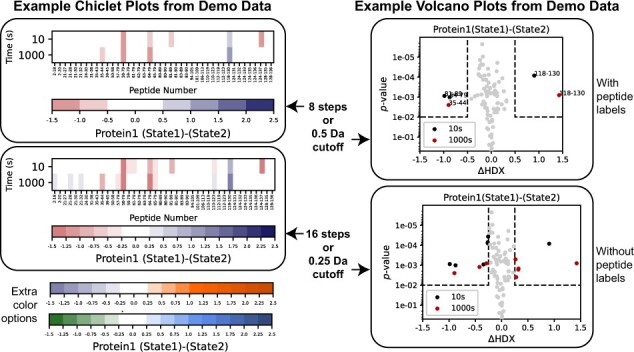
Example chiclet and volcano plots generated with demo data on HD-eXplosion. The top plots have equivalent statistical filters; P-value < 0.01 and a difference in deuterium uptake greater than |0.5|Da. The bottom plots also have equivalent statistical filters; P-value < 0.01 and a difference in deuterium uptake great than |0.25|Da. Some customization options are demonstrated. For example, alternative color schemes for the chiclet plot and peptide labels for the volcano plot. The demo data can be downloaded from the website
Funding
This work was supported by the NIH National Institute of General Medical Sciences (GM133751) to S. D’Arcy.
Conflict of Interest: none declared.
Contributor Information
Naifu Zhang, Department of Chemistry and Biochemistry, The University of Texas at Dallas, Richardson, TX 75080, USA.
Sheena D’Arcy, Department of Chemistry and Biochemistry, The University of Texas at Dallas, Richardson, TX 75080, USA.
References
- Brown K.A., Wilson D.J. (2017) Bottom-up hydrogen deuterium exchange mass spectrometry: data analysis and interpretation. Analyst, 142, 2874–2886. [DOI] [PubMed] [Google Scholar]
- Chalmers M.J. et al. (2006) Probing protein ligand interactions by automated hydrogen/deuterium exchange mass spectrometry. Anal. Chem., 78, 1005–1014. [DOI] [PubMed] [Google Scholar]
- Chalmers M.J. et al. (2011) Differential hydrogen/deuterium exchange mass spectrometry analysis of protein–ligand interactions. Expert Rev. Proteomics, 8, 43–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hageman T.S., Weis D.D. (2019) Reliable identification of significant differences in differential hydrogen exchange-mass spectrometry measurements using a hybrid significance testing approach. Anal. Chem., 91, 8008–8016. [DOI] [PubMed] [Google Scholar]
- Hourdel V. et al. (2016) MEMHDX: an interactive tool to expedite the statistical validation and visualization of large HDX-MS datasets. Bioinformatics, 32, 3413–3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacob R.E. et al. (2015) Hydrogen/deuterium exchange mass spectrometry applied to IL-23 interaction characteristics: potential impact for therapeutics. Expert Rev. Proteomics, 12, 159–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerres N. et al. (2017) Chemically induced degradation of the oncogenic transcription factor BCL6. Cell Rep., 20, 2860–2875. [DOI] [PubMed] [Google Scholar]
- Lau A.M.C. et al. (2019) Deuteros: software for rapid analysis and visualization of data from differential hydrogen deuterium exchange-mass spectrometry. Bioinformatics, 35, 3171–3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. et al. (2020) FACT caught in the act of manipulating the nucleosome. Nature, 577, 426–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson G.R. et al. (2017) An overview of hydrogen deuterium exchange mass spectrometry (HDX-MS) in drug discovery. Expert Opin. Drug Discov., 12, 981–994. [DOI] [PubMed] [Google Scholar]
- Masson G.R. et al. (2019) Recommendations for performing, interpreting and reporting hydrogen deuterium exchange mass spectrometry (HDX-MS) experiments. Nat. Methods, 16, 595–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliphant T.E. (2006) Guide to NumPy. Trelgol Publishing, USA. [Google Scholar]
- Pirrone G.F. et al. (2015) Applications of hydrogen/deuterium exchange MS from 2012 to 2014. Anal. Chem., 87, 99–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wales T.E., Engen J.R. (2006) Hydrogen exchange mass spectrometry for the analysis of protein dynamics. Mass Spectrom. Rev., 25, 158–170. [DOI] [PubMed] [Google Scholar]


