Abstract
Oxytocin has garnered much interest due to its role in affective states, social behaviors, and diverse physiological functions. However, approaches for measuring endogenous oxytocin concentrations have generated considerable controversy and debate. Common procedures for measuring oxytocin often produce uncorrelated results, and the detected concentrations frequently vary across two orders of magnitude. These findings have led some researchers to argue that immunoassays of plasma oxytocin may be unreliable and nonspecific, particularly when samples are not first processed using an extraction procedure. Here, we assess the specificity of oxytocin immunoassays using plasma samples from wildtype (WT) and oxytocin knockout (KO) mice. Plasma samples from both genotypes were measured using immunoassay and were measured with or without a solid phase extraction. Using a commercially available kit from Arbor Assays, we demonstrate that both techniques generate a clear contrast between genotypes, with wildtype samples containing high concentrations of oxytocin (unextracted mean = 468 pg/ml; extracted mean = 381 pg/ml), while knockout samples measured below the lower limit of detection. Analytical validations demonstrated good parallelism and spike recovery for both methods. Furthermore, the same wildtype samples measured with both procedures were highly correlated (r = 0.950), although unextracted samples measured at significantly higher concentrations (p = 2.0 × 10−7, Cohen’s d = 2.65). To test the generalizability of these results across immunoassay kits, we performed additional assays with kits from Cayman Chemical and Enzo Life Sciences. The Cayman Chemical kit produced results similar to Arbor Assays with a clean signal differentiating WT and KO plasma, both with and without an extraction step. The Enzo kit also differentiated the genotypes, with correlation between extracted and unextracted samples, but was considerably more susceptible to interference without the extraction, as evidenced by false positive signal in KO plasma samples. The extent to which these results generalize to other species remains unknown and challenging to assess.
Keywords: oxytocin, immunoassay, ELISA, extraction, sample matrix, knockout
Graphical Abstract
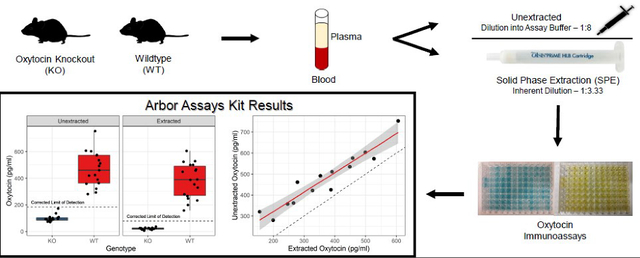
1. Introduction
Although the neurohormone oxytocin was discovered over a century ago, it has attracted increasing scientific and popular attention in recent decades due to growing recognition of its roles in emotional states (Carter, 2017; Uvnäs-Moberg, 1998), social behavior and cognition (Caldwell et al., 2017; Heinrichs et al., 2009) and their development (Hammock, 2015), as well as a wide range of other physiological processes (Carter et al., 2020). Despite a proliferation of studies on oxytocin—ranging from behavioral genetics to pharmacology—there is surprisingly little consensus on how to measure endogenous oxytocin and how to interpret the often discordant results of various methods (MacLean et al., 2019).
Oxytocin is primarily synthesized in the brain and is released both peripherally as a hormone and within the central nervous system as a neurotransmitter (Buijs and Heerikhuize, 1982) and neuromodulator (Ludwig, 1998; Stoop, 2012), sometimes in a coordinated fashion and sometimes independently (reviewed in Jurek and Neumann, 2018; Landgraf and Neumann, 2004). Oxytocin can therefore be measured in a variety of biological matrices, including cerebrospinal fluid, blood plasma, saliva, milk, and urine; however, some have argued that cerebrospinal fluid is the most relevant matrix when assessing associations with behavior, in part because of assumptions about the blood-brain barrier (e.g. Neumann and Landgraf, 2012). Nevertheless, oxytocin also plays important roles in the periphery, many of which feed back to the central nervous system (e.g. Iwasaki et al., 2015; Tabbaa and Hammock, 2020). Additionally, intranasal oxytocin studies suggest that inhaled oxytocin can reach the central nervous system (reviewed in Quintana et al., 2018), and a route for active transport into the brain has recently been discovered in mice (Yamamoto et al., 2019). Thus, reliable methods for assessing oxytocin in peripheral matrices present potentially powerful approaches for studying the oxytocin system.
Unfortunately, the methods used to measure peripheral oxytocin continue to be mired in controversy (Leng and Sabatier, 2016; MacLean et al., 2019; McCullough et al., 2013; Szeto et al., 2012). This is in part due to the diversity of methods utilized and the inconsistency of results across them (e.g. Christensen et al., 2014; Robinson et al., 2014; Szeto et al., 2012). To date, the most common approaches have involved immunoassay, and methods of sample preparation range from direct measurement of unextracted samples to liquid-liquid and solid-phase extractions (SPE), or more recently reduction-alkylation procedures that liberate protein-bound oxytocin (Brandtzaeg et al., 2016). However, plasma samples assayed with and without extraction often produce uncorrelated results (Leng and Sabatier, 2016; Robinson et al., 2014), and the concentrations detected in unextracted samples are frequently two orders of magnitude greater than those with extracted samples (Szeto et al., 2012). The use of unextracted samples has been heavily criticized on a number of grounds, including arguments that 1) these concentrations are physiologically unrealistic and 2) these high concentrations simply reflect interference from molecules other than oxytocin. Regarding the first point, we now know that oxytocin does occur at high concentrations in plasma but that it frequently binds to other molecules (Brandtzaeg et al., 2016) that may be lost in most extractions. Regarding the second point, interference in immunoassay is always an important consideration given that the identity of the analyte is never determined with certainty.
Immunoassays are susceptible to multiple types of interference, a broad term used to describe processes that impede the accurate measurement of the analyte, resulting in artificially high or low measurements. One important factor is the specificity of measurement, which depends not only on properties of the antibody itself but also on properties of the intended analyte (here, oxytocin), the composition of the sample matrix (here, plasma), and the various reagents used in the immunoassay procedure (Tate and Ward, 2004). Immunoassays rely on competitive binding between the sample analyte and a labeled antigen conjugate; the analyte itself is not measured directly, but rather the concentration is calculated based on the binding of the conjugate, which produces a measurable signal through the release of the label, whether color (absorption format, EIA or ELISA), visible light (chemiluminescent format, CLIA), or gamma radiation (radioassay format, RIA) (Engvall and Perlmann, 1971; Schroeder et al., 1976; Yalow and Berson, 1960). Thus, anything that disrupts the binding of the antibodies can result in erroneously high measurement. One such disruption, termed heterophilic interference, involves the binding of other antibodies in the sample matrix disrupting the intended antibody-antigen binding (Bolstad et al., 2013; Boscato and Stuart, 1988). Another disruption, termed cross-reactivity, can be caused by the binding of other chemicals that resemble the analyte of interest (Tate and Ward, 2004), potentially including metabolites of the analyte (MacLean et al., 2019). Other reagents used in the immunoassay procedure can also affect measurement not only during the incubation period (Tate and Ward, 2004), but also during development, either producing (Porstmann et al., 1981) or minimizing (Saini et al., 1995) interference by affecting the readout mechanism (i.e. color production for EIA). Diagnosing and identifying these and other potential sources of interference presents a challenge, particularly when working with complex biological matrices.
Here, we address whether immunoassays for plasma oxytocin suffer from interference by comparing performance with plasma samples from wildtype and oxytocin knockout mice using both extracted and unextracted samples. First, given that oxytocin knockout mice should have no circulating oxytocin, we expected to find much lower oxytocin concentrations in knockout than wildtype mice; we did not expect measurements of zero given the sensitivity limitations of these assays as well as the potential for interference. Second, because plasma from oxytocin knockout mice should still contain other common plasma proteins—putative sources of interference—we expected that any signal in knockout mouse samples substantially above the assay’s lower limit of detection would constitute evidence for immunoassay interference. Third, given that commercially available kits use different antibodies and chemistries, we expected that the results might differ based on the specific immunoassay kit; we therefore tested three commonly used oxytocin kits from the companies Arbor Assays, Enzo Life Sciences, and Cayman Chemical.
2. Methods
2.1. Subjects and Sample Collection
Oxttm1Zuk mice (Nishimori et al., 1996) were maintained on a C57BL/6J background and bred at Florida State University. All breeding and sample collection procedures were approved by the Institutional Animal Care and Use Committee following the Guide for the Care and Use of Laboratory Animals. Oxt+/− breeder pairs were continuously housed. The first morning of the appearance of a litter was noted as postnatal day 0 (P0). Mice were weaned, tagged, and tailed for genotyping on P21 and group housed by sex. After genotyping, same-sex mice were re-housed by genotype, so that only mice of the same genotype were housed together. Mice were housed on a 12:12 L:D cycle in open wire-top caging with wood chip bedding and provided ad libitum food (LabDiet Rodent 5001) and water. In total, samples from 83 mice of this strain (WT: 38, KO: 45) were used in this study.
Adult C57BL/6J wildtype mice were purchased from Jackson Labs (total n = 18).
Adult mice were euthanized with CO2. Approximately 400 ml of trunk blood was collected into a chilled microfuge tube containing 20 μl of Anticoagulant Citrate Dextrose Solution A (Becton Dickinson). Tubes were centrifuged for 10 minutes at 2000g in a refrigerated centrifuge at 4°C. Plasma was transferred to a microcentrifuge tube pre-chilled in dry ice.
Samples were shipped frozen to the University of Arizona on dry ice and stored at −80°C until time of assay. No genotype information was included with the shipments.
2.1.1. Genotyping
DNA from tail samples was genotyped for Oxt alleles using a common forward primer (5’-TCAGAGATTGAACAAGACGCC) and specific reverse primers for WT (5’-TCAGAGCCAGTAAGCCAAGC) and KO (5’-ACTTGTGTAGCGCCAAGTGC). Using a hot start and 40 cycles of 30 seconds each of 94°C, 57°C, and 72°C, the primers generated a wildtype allele of approximately 500 bp and a knockout allele of approximately 180 bp.
2.1.2. Samples in Each Analysis
A pilot study was first conducted with 11 samples to determine the optimal dilution factor to use with the Arbor Assays kit (4 female, 7 male; 5 WT, 6 KO) (see appendix, Table A1, Figure A1). Assays reported in the main Arbor Assays analyses included samples from 38 subjects (unextracted (UE): 23 female, 11 male; solid phase extraction (SPE): 21 female, 13 male); due to limited sample volume in some instances, only 30 of these subjects were assayed using both methods (21 female, 9 male). See Table A3 for subject information. Plasma samples from an additional eight C57BL/6J, six Oxt+/+ individuals, and nine Oxt−/− were used for the analytical validations and as pools to calculate inter- and intra-assay CVs as well as extraction efficiencies. Sample pools for the three-way kit comparison used extra volume from these and other samples, totaling approximately 50 individuals per genotype. Samples from an additional 19 individuals were assayed on the Enzo kit (Oxt+/+: 1 female, 9 male; Oxt−/−: 1 female, 8 male) using both unextracted and extracted methods (Table A4). Ten of these individuals (Oxt+/+: 1 female, 4 male; Oxt−/−: 1 female, 4 male) were also assayed on the Cayman kit, unextracted only (Table A5). Samples and individuals used in each analysis are summarized in Table 1.
Table 1:
Summary of sample pools and individuals used for each analysis. Some individuals are used in multiple analyses and are counted here separately.
| Indiv. Used | ||||||
|---|---|---|---|---|---|---|
| Kit | Analysis | Strain | Reported in | # in Pool | Female | Male |
|
| ||||||
| Arbor | ||||||
| Pilot Study | Oxt +/+ | Appendix | 2 | 3 | ||
| Oxt −/− | 2 | 4 | ||||
| Pools for Inter- and Intra-assay CVs | Oxt +/+ | 2.32 | 6 | |||
|
| ||||||
| Individual Samples - Unextracted | Oxt +/+ | 3.1–3.3 | 11 | 7 | ||
| Oxt −/− | 3.1 | 12 | 4 | |||
| Individual Samples - Extracted | Oxt +/+ | 3.1–3.3 | 9 | 9 | ||
| Oxt −/− | 3.1 | 12 | 4 | |||
|
| ||||||
| UE Parallelism (1) & Spike Recovery | C57BL/6J | 8 | ||||
| UE Parallelism (2) | C57BL/6J | 10 | ||||
| SPE Parallelism & Spike Recovery | Oxt +/+ | 3.4 | 6 | |||
| KO Parallelism, Spike Recovery & Extraction Efficiency | Oxt −/− | 9 | ||||
|
| ||||||
| All | Kit Comparison Pools | Oxt +/+ | 3.5.1 | ~50 | ||
| Oxt −/− | ~50 | |||||
|
| ||||||
| Enzo | Individual Samples | Oxt +/+ | 3.5.2 | 1 | 9 | |
| Oxt −/− | 1 | 8 | ||||
|
| ||||||
| Cayman | Individual Samples (Subset of Enzo Individuals) | Oxt +/+ | 3.5.3 | 1 | 4 | |
| Oxt −/− | 1 | 4 | ||||
2.2. Statistical Software
Statistical analyses were conducted in R version 4.0.2 (R Core Team, 2020). Data manipulation and visualization were conducted primarily using the tidyverse (Wickham et al., 2019). All statistical tests were two-sided, and effect sizes were calculated using the package rstatix (Kassambara, 2020). Statistical details are reported for each experiment below.
2.3. Experiment 1 – Arbor Assays Kit
2.3.1. Oxytocin Assays
Oxytocin was first measured using the Arbor Assays Oxytocin EIA kit (Catalog #K048-H5). We did not use the manufacturer-provided extraction solution, but otherwise followed the recommended assay protocol. The reported sensitivity for this kit is 17.0 pg/ml and the lower limit of detection is 22.9 pg/ml. Due to sample dilution inherent in each method (UE = 1:8; SPE = 1:3.33), the lower limits of detection (LLOD) using the methods reported here are multiplied by the dilution factor of each method (LLODUE = 183.2 pg/ml; LLODSPE = 76.3 pg/ml). These corrected concentrations are reported in the text for those samples which measured above the lower limit of detection; all measured concentrations are reported in the appendix. Arbor Assays also reports that cross-reactivity is 94.3% for isotocin, 88.4% for mesotocin, and less than 0.15% for vasotocin and arginine vasopressin.
2.3.2. Coefficients of Variation
The genotype comparison was assessed across two plates. Intra-assay coefficients of variation (CVs) were calculated by running two sets of duplicates (four wells) of a pooled unextracted wildtype (C57BL/6J) sample at two dilutions (factor of 2); the inter-assay CVs were calculated by running these same samples on each plate. Mean intra-assay CVs were 6.46% and 1.02% for the high and low value samples, respectively; inter-assay CVs were 7.97% and 5.16% for the high and low value samples, respectively. For individual samples (table A3) that measured above the limit of detection (n = 18), the average CV across duplicates was 6.24% for unextracted samples and 5.93% for extracted samples.
2.3.3. Analytical Validation
Using a pool of plasma samples from known wildtype individuals (C57BL/6J), we performed parallelism and spike recovery for unextracted samples. Spike recovery was assessed using samples at a 1:8 dilution, the same dilution factor used for unextracted samples in all other assays (see appendix, Table A1, Figure A1). Spiked samples consisted of 90% sample matrix (plasma diluted into assay buffer at 1:8) and 10% synthetic oxytocin in assay buffer (kit standards 1–5); for the unextracted samples, one spiked sample was excluded due to a poor CV (>20%). Percent recovery was calculated as (observed / expected) × 100, where the expected values were measured independently by adding each spike to assay buffer. Parallelism was assessed via serial dilution of a plasma sample and calculation of the coefficient of variation on corrected concentrations at each dilution (Andreasson et al., 2015). A second parallelism with a different pool of wildtype individuals (C57BL/6J) was used to replicate the unextracted parallelism. The same procedure was used for a validation of the extraction method, using a separate pool of plasma samples from known wildtype individuals (Oxt+/+); one sample in this serial dilution was excluded from the results due to a poor CV (>20%).
A similar procedure was also performed on pools of unextracted knockout samples to assess the measurement of knockout samples at less dilution and to ensure that spiked oxytocin measured as expected in knockout samples. Since we were only interested in dilutions more concentrated than those used in the main results, we examined four dilutions: 100%, 50%, 25%, and 12.5%. One of five spike recovery samples was excluded from the results due to a poor CV (>20%).
2.3.4. Solid Phase Extraction
Solid phase extraction (SPE) was performed using OASIS PRiME HLB 1 cc cartridges (Waters Corporation, Milford, MA, USA, Part Number: 186008055), which utilize a reversed-phase Hydrophilic-Lipophilic Balance chemistry, and a positive pressure manifold (Biotage PRESSURE+48). Samples were diluted into an equal volume of 0.1% Trifluoroacetic acid (TFA) in water (75 μl each), vortexed for 30 s, and centrifuged at 10,000 rpm (RCF = 9632 × g) for 5 minutes. SPE cartridges were conditioned first with 1 ml acetonitrile (ACN) with 0.1% trifluoroacetic acid (TFA) and then 1 ml 0.1% TFA in water. The entire sample supernatant was applied to the cartridge. Each cartridge was then washed with 1 ml 10% ACN, 0.1% TFA. Finally, samples were eluted with 1 ml 30% ACN, 0.1% TFA. This elution is at a lower percent organic content than in previously published studies; for more information on the development of this method, see the appendix. The eluted samples were placed at −80°C overnight and lyophilized the next day using a centrivap (Labconco model #7810016) with the following settings: centrivap unheated, cold trap −80–85°C, vacuum 0.3–0.4 mbar. Samples were reconstituted in 250 μl assay buffer at the time of assay, resulting in a dilution factor of 1:3.33.
2.3.5. Extraction Efficiency
To assess the extraction efficiency of this method, we spiked knockout samples with oxytocin standard from the Arbor Assays kit. These samples were aliquoted into two portions, one of which was extracted as described above, while the other was frozen at −20°C overnight and assayed unextracted the next day. This was done at two different concentrations, and the spike was never more than 90% of the sample volume. Reported percent recoveries represent the proportion of the total oxytocin measured in the unextracted sample that was recovered in the matching extracted sample.
2.3.6. Statistical Analyses
For an assessment of the study’s power based on pilot data, see the appendix.
One outlier was identified in the unextracted assay as being outside 1.5x the interquartile range of the wildtype samples. The results below are presented excluding this outlier; however, inclusion did not affect the overall pattern of results (see appendix).
Shapiro-Wilk tests revealed that the extracted wildtype and knockout samples did not deviate significantly from a normal distribution (pWT = 0.98; pKO = 0.62). Shapiro-Wilk tests of the unextracted samples revealed that the knockout sample distribution was not normally distributed (p = 0.005), while the wildtype samples did not deviate significantly from a normal distribution after outlier exclusion (p = 0.717). Given these results and the unequal variances between genotypes (see Figure 1), Welch’s t-tests were used to compare the extracted samples, while a non-parametric Wilcoxon-Mann-Whitney test was used to compare unextracted samples. As the difference in concentrations between the two methods did not significantly differ from a normal distribution (Shapiro-Wilk p = 0.96), a paired-sample t-test was used to compare the means of each method.
Figure 1:
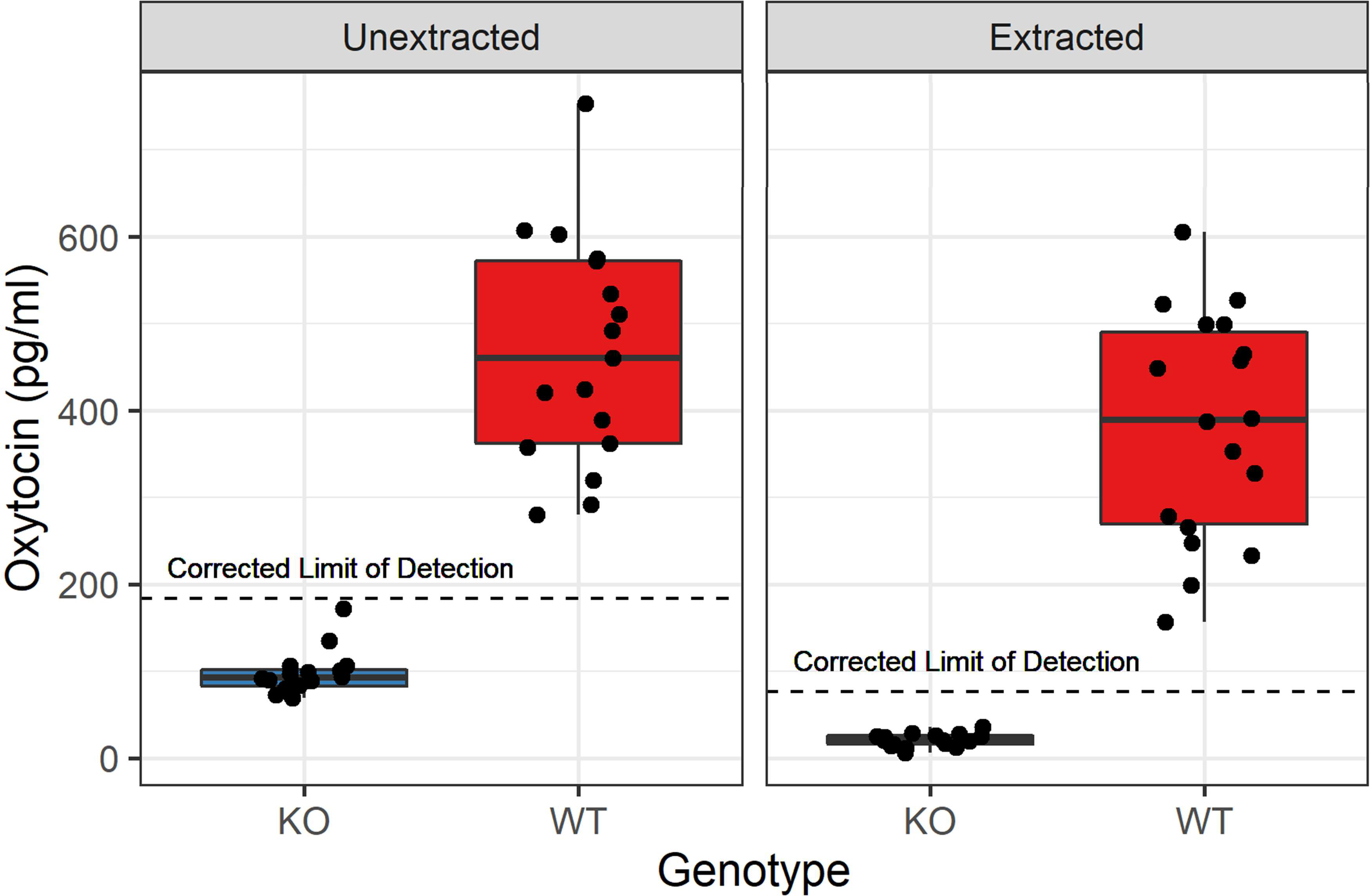
Comparison of oxytocin concentrations assayed with solid phase extraction (right) and without extraction (left), using the Arbor Assays kit. Knockout (KO) samples all measured below the lower limit of detection, whereas wildtype (WT) samples measured much higher, with considerable individual variation; both effects are large and significant (Wilcoxon-Mann-Whitney WUE = 0, pUE = 1.7 × 10−9, r = 0.853; t(17.1)SPE = −11.7, pSPE = 1.3 × 10−9, Cohen’s d = 3.91). Note that the corrected lower limit of detection indicated is the measurable limit of detection for the assay multiplied by the dilution factor for each method; values below this should not be interpreted.
To explore potential sex differences among the wildtype individuals, we ran four linear models using the lm function in R, predicting measured plasma oxytocin in extracted and unextracted samples, both with and without controlling for body mass (see appendix for model details).
2.4. Experiment 2 – Enzo Life Sciences and Cayman Chemical Kits
2.4.1. Assay Information
To assess the generalizability of our results to other oxytocin immunoassay kits with different antibodies and chemistries, we attempted to replicate some of our findings with two commonly used assay kits from Enzo Life Sciences (Catalog # ADI-900-153A-0001) and Cayman Chemical (Catalog #500440). Company-reported sensitivities for these kits are 15 pg/ml for Enzo and 20 pg/ml for Cayman. The lower limit of detection is not reported by Enzo, so we used the sensitivity as the lower threshold for interpreting values from each method (UE: 120 pg/ml, SPE: 50 pg/ml); on the Cayman kit, the lower limit of detection is 16.4 pg/ml (personal communication) (accounting for dilution: LLODUE = 131.2, LLODSPE = 54.7 pg/ml). Corrected concentrations are reported in the text for those samples which measured above each kit’s respective threshold (LLOD or sensitivity); all measured concentrations are reported in the appendix. The reported cross-reactivities for the Enzo kit are: mesotocin 7.0%, Arg8-vasotocin 7.5%, and <0.02% for all other reported compounds including Arg8-vasopressin. Reported cross-reactivities for the Cayman kit are 100% for mesotocin and isotocin and <0.01% for all other reported compounds including Arg8-vasopressin.
The unextracted samples were diluted identically to the previous experiments for both kits (1:8 in kit-specific assay buffer). It should be noted that these dilutions were not optimized for these kits, and further dilution might produce different results. Extracted samples on the Enzo kit were processed identically to the procedure reported above (but were not run on the Cayman kit due to limited space).
2.4.2. Three-way Pooled Sample Comparison: HLB and MCX Extractions
For the three-way kit comparison, the HLB-extracted samples were processed at higher volumes so that a single sample pool could be split three ways across the three assays. 250 μl of each plasma pool was diluted into an equal volume of 0.1% TFA in water. The procedure proceeded identically as above, except 2 ml of 30% ACN, 0.1% TFA was used to elute the samples to accommodate the higher sample volume. The eluate was then vortexed and split into three 600 μl aliquots (remaining volume discarded), frozen at −80°C, lyophilized, and each aliquot was resuspended in 250 μl of the appropriate assay buffer (as provided in each kit). This process resulted in the same dilution factor as above (1:3.33).
For the three-way kit comparison only, we also used an additional solid phase extraction method, using OASIS PRiME MCX 1 cc cartridges (Waters Corporation, Milford, MA, USA, Part Number: 186008917). These cartridges use a mixed-mode cation exchange chemistry involving both reverse-phase and ion retention, which can be leveraged to produce a more selective extraction. Since we did not know whether HLB-extracted samples would exhibit interference on the additional kits, the MCX extraction method was added to this comparison with the expectation that it would produce a cleaner sample that might minimize interference.
For these samples, 250 μl of plasma were diluted into 250 μl of loading buffer (200 mM ammonium formate, with phosphoric acid to achieve pH 5), vortexed for 30 s, and centrifuged at 10,000 rpm (RCF = 9632 × g) for 5 minutes. The cartridges were first conditioned with 1 ml methanol (MeOH) and 1 ml of deionized water (Thermo Scientific #751628). The entire supernatant of the sample was then loaded onto each cartridge. Each cartridge was washed with successive 1 ml washes: 1 ml deionized water, 4 ml wash buffer (60% loading buffer, 40% MeOH), 1 ml deionized water. Finally, each sample was eluted in 2 ml of elution buffer (50% MeOH, 50% ammonium hydroxide solution, pH 12). Identically to the HLB samples for this experiment, each eluate was then vortexed and split into three 600 μl aliquots, frozen, evaporated (due to the melting point of MeOH, this was not cold enough to lyophilize, although the same procedure was used), and each sample was resuspended in 250 μl of the appropriate assay buffer. This process resulted in the same dilution factor as with the HLB cartridges (1:3.33).
2.4.3. Statistical Analyses
Shapiro-Wilk tests revealed that the extracted wildtype and knockout samples measured on the Enzo kit did not deviate significantly from a normal distribution (pWT = 0.59; pKO = 0.39). Shapiro-Wilk tests of the unextracted samples measured on the Enzo kit revealed that the knockout sample distribution was not normally distributed (p = 4.7 × 10−5), while the wildtype samples did not deviate significantly from a normal distribution after outlier exclusion (p = 0.09). Given these results and the unequal variances between genotypes (see Figure 5A), Welch’s t-tests were used to compare the extracted samples, while a non-parametric Wilcoxon-Mann-Whitney test was used to compare unextracted samples. Shapiro-Wilk tests also revealed that the unextracted samples assayed on the Cayman kit did not deviate significantly from a normal distribution (pWT = 0.89; pKO = 0.32), so a Welch’s t-test was used to compare genotypes.
Figure 5:
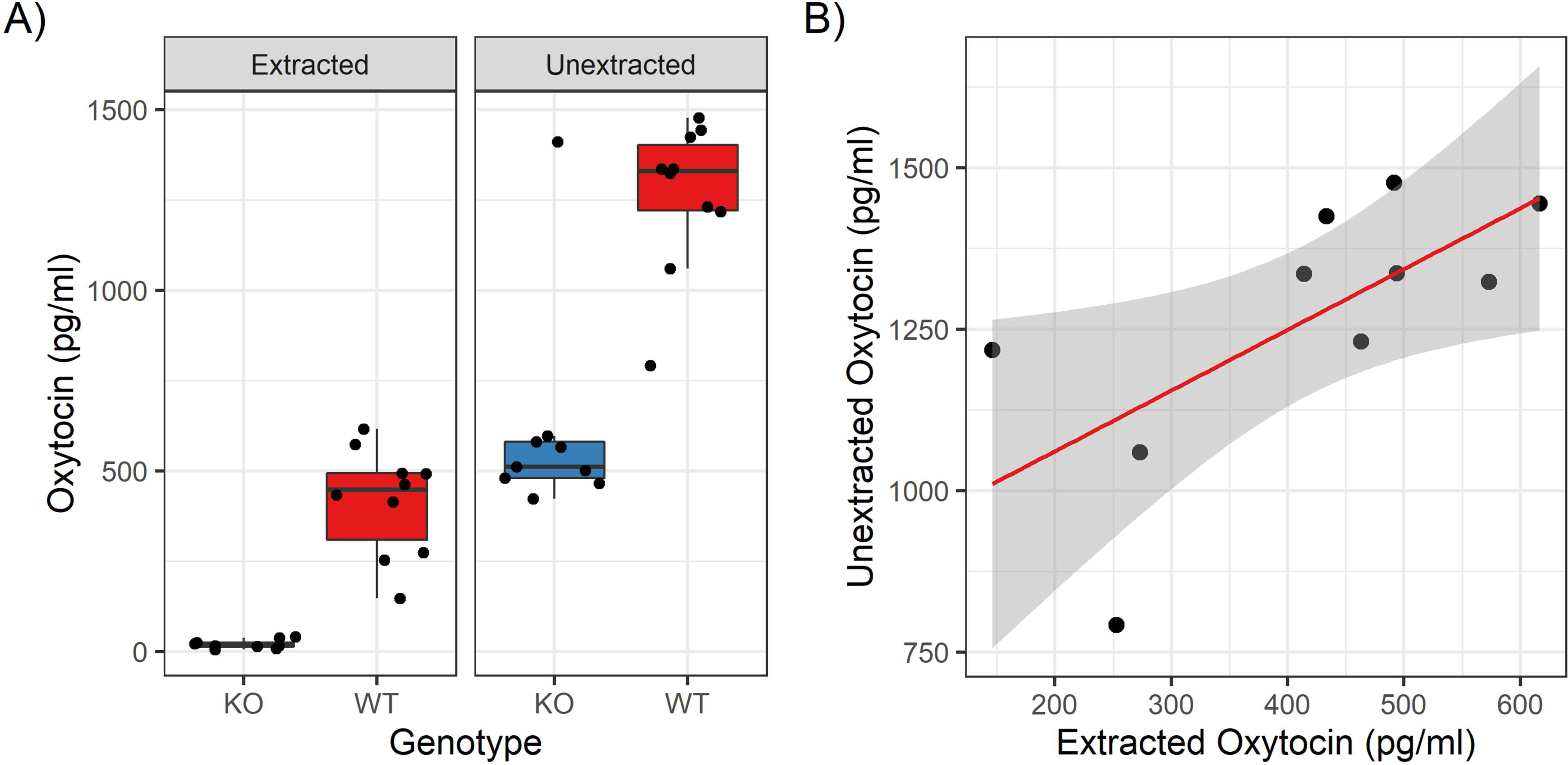
Results on the Enzo kit. A) The genotype contrast is shown for both methods. There is a clean distinction for extracted samples (t(9.1) = 8.38, p = 1.38 × 10−5, Cohen’s d = 3.75), but while unextracted samples are statistically different (Wilcoxon-Mann-Whitney W = 83, p = 9.7 × 10−4, r = 0.712), there is considerable interference evidenced by high values for knockout samples. B) Correlation for the same samples measured both with and without extraction (r = 0.676).
3. Results
3.1. Genotype Comparison
Using the Arbor Assays kit, oxytocin concentrations in plasma samples from wildtype mice averaged 468 pg/mL without extraction (n = 34) and 381 pg/mL following solid phase extraction (n = 34). Oxytocin concentrations from knockout mice were all below the lower limit of detection (LLOD) using both approaches. Although specific concentrations below the LLOD should not be interpreted, the knockout measurements were significantly lower than those of wildtype mice using both unextracted (UE) and solid phase extraction (SPE) methods (Wilcoxon-Mann-Whitney WUE = 0, pUE = 1.7 × 10−9, r = 0.853; t(17.1)SPE = −11.7, pSPE = 1.3 × 10−9, Cohen’s d = 3.91) (Figure 1, Table A2).
3.2. Method Comparison
For samples with sufficient sample volume (n = 30), we assayed the same samples both with and without SPE. Since knockout samples measured below the limit of detection on both assays, those values were deemed meaningless for the purpose of method correlation. Among the wildtype samples (n = 14), the results were very strongly correlated (r = 0.95; Figure 2), although unsurprisingly, the unextracted concentrations were systematically higher than the extracted concentrations (t(13) = 9.92, p = 2.0 × 10−7, Cohen’s d = 2.65).
Figure 2:
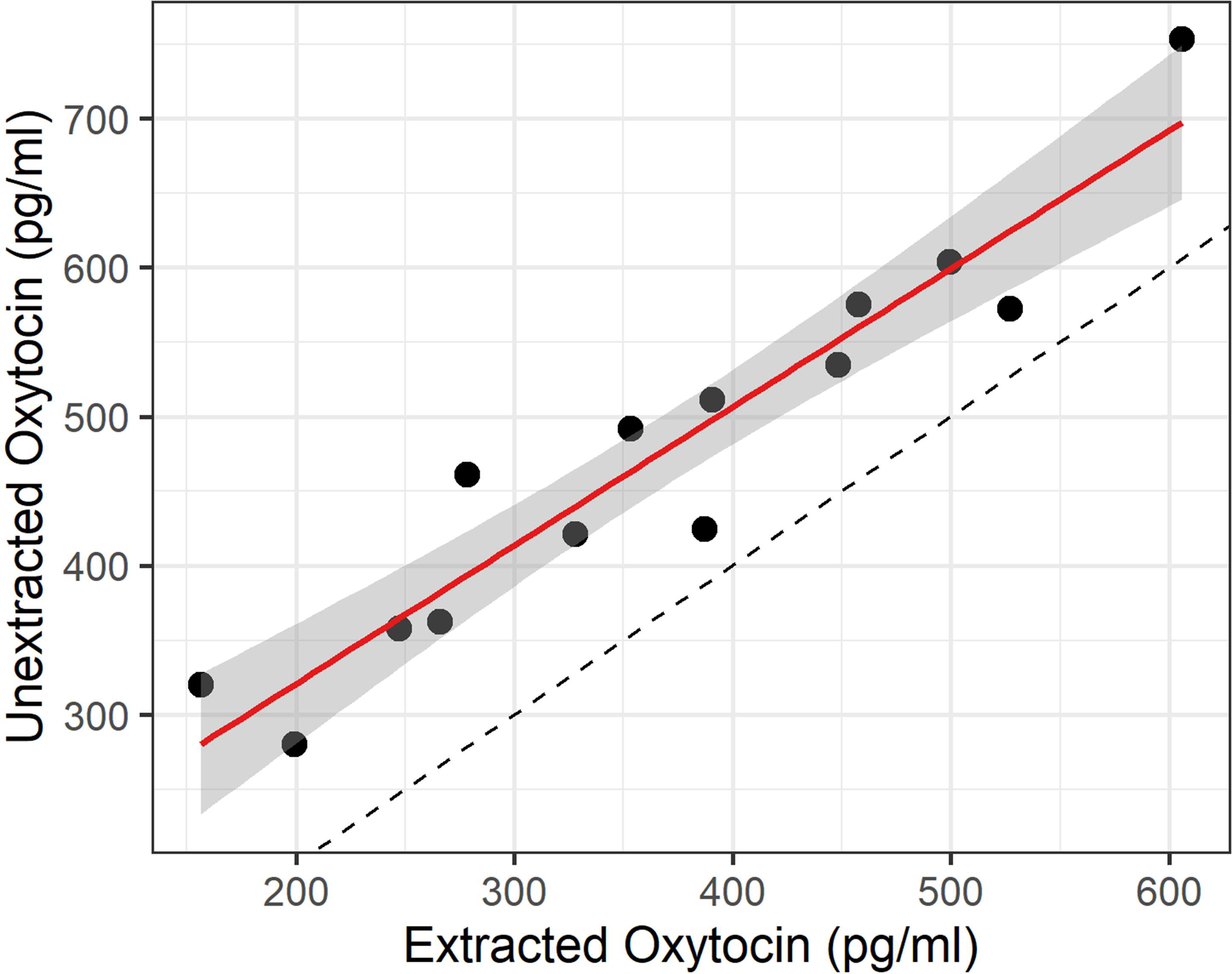
Relationship between unextracted and extracted measures for wildtype samples measured with both methods using the Arbor Assays kit (n = 14). The solid red line shows the linear regression, while the dotted black line indicates an ideal 1:1 correspondence. The results of the two methods are highly correlated (r = 0.95), although unextracted values are systematically higher than the extracted values (t(13) = 9.92, p = 2.0 × 10−7, Cohen’s d = 2.65).
3.3. Individual Differences
Although males trended towards lower plasma oxytocin levels on average, sex was not a statistically significant predictor of unextracted plasma oxytocin levels as measured on the Arbor Assays kit (βsex = −78.17, SE = 63.01, t(15) = −1.241, p = 0.234). Among extracted samples on the Arbor Assays kit, sex was a significant predictor—with males having lower plasma oxytocin levels (βsex = −125.54, SE = 54.94, t(16) = −2.285, p = 0.036)—but this effect was no longer statistically significant after controlling for body mass (βsex = −13.296, SE = 28.59, t(15) = −0.465, p = 0.649; see table A2 for full model summaries).
3.4. Analytical Validations
We also performed analytical validation of both methods, assessing parallelism and spike recovery. Our unextracted mouse plasma pool produced a dilution series that measured in parallel with the standard curve (Figure 3A). The CV of the corrected concentrations for the dilutions within the assay range was 20.9%; excluding the undiluted sample, this value dropped to 16.4% and excluding the 1:2 dilution, this value dropped further, to 7.4%. This suggested better parallelism at a dilution of more than 1:2, which we confirmed with a second parallelism on an additional sample pool. The CV of corrected concentrations for four dilutions ranging from 13–44% was 6.9%, although it was much higher (28.4%) when including the undiluted and 66.7% samples (see table A6). The average spike recovery for the unextracted pool (diluted 1:8) across four different spike concentrations was 106.12% (range: 103.45 – 111.05%).
Figure 3:
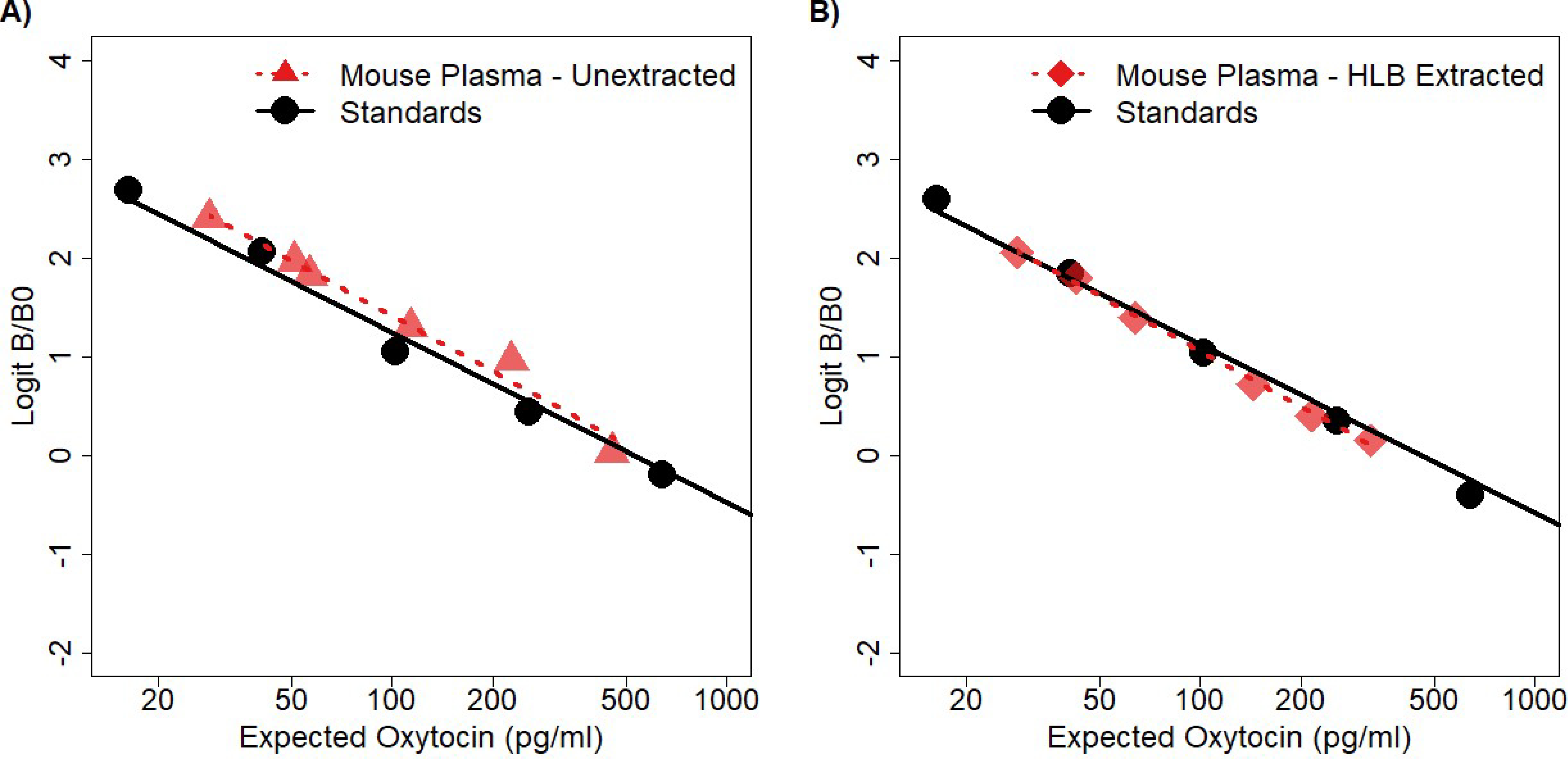
Parallelisms for both the unextracted method and solid phase extraction using the Arbor Assays kit. A) Unextracted mouse plasma measured at 5 serial dilutions (factor of two), as well as the spike recovery baseline (90% of the 1:8 dilution). The 1:32 dilution was excluded because it measured below the lowest standard. Visual inspection indicates that the samples diluted in parallel with the standard curve. The CV of the corrected concentration over the 5 serial dilutions within the assay range was 20.9% and dropped to 7.4% when excluding the first two (rightmost) dilutions. B) Extracted mouse plasma measured at 6 serial dilutions (factor of 2:3, one additional dilution excluded for poor duplicate CV). Visual inspection indicates that the samples diluted in parallel with the standard curve. The CV of the corrected concentration over all the dilutions was 3.87%.
Our extracted wildtype mouse plasma dilution series was also parallel with the standard curve (Figure 3B), and the CV of the corrected concentrations was 3.87%. The average spike recovery across five different spike concentrations was 98.0% (range: 90.1 – 102.3%). Extraction efficiencies for this method were assessed by measuring spiked knockout samples. The extraction efficiencies were 97.2% and 92.5% for the high- and low-concentration samples, respectively.
To address the potential concern that we had simply chosen a sufficient dilution to prevent detection of interference in the knockout samples—and especially given the wildtype parallelism results at dilutions of less than 1:2—we measured an unextracted knockout pool both undiluted and at three dilutions. All three dilutions measured below the lower limit of detection of the assay. Only the undiluted sample produced a meaningful measurement: 53.8 pg/ml. The average spike recovery in a knockout sample pool (diluted 1:8) across four different spike concentrations was 105.8% (range: 92.0 – 113.2%).
3.5. Extension to Other Immunoassay Kits
3.5.1. Three-way Pooled Sample Comparison
To extend the generalizability of these results to kits with other chemistries—and perhaps most importantly, different antibodies—we performed a similar test of the Enzo Life Sciences and Cayman Chemical kits on a smaller scale. The mixed cation exchange (MCX) extraction method was also used with the expectation that it might minimize any interference observed with other methods. Using identical pooled samples across kits, the HLB- and MCX-extracted samples all showed relative consistency across kits for both wildtype and knockout samples, with all of the latter samples measuring below the kits’ sensitivities or lower limits of detection (Figure 4). For unextracted samples, the results were more divergent. Although the Arbor and Cayman kits showed no signs of interference in the knockout samples (both well below the kits’ reported sensitivities), the Enzo kit exhibited considerable interference (812.9 pg/ml). The wildtype pool also measured correspondingly higher on the Enzo kit (1245.6 pg/ml) than on the other two kits (Arbor: 429.7 pg/ml, Cayman: 474.7 pg/ml).
Figure 4:
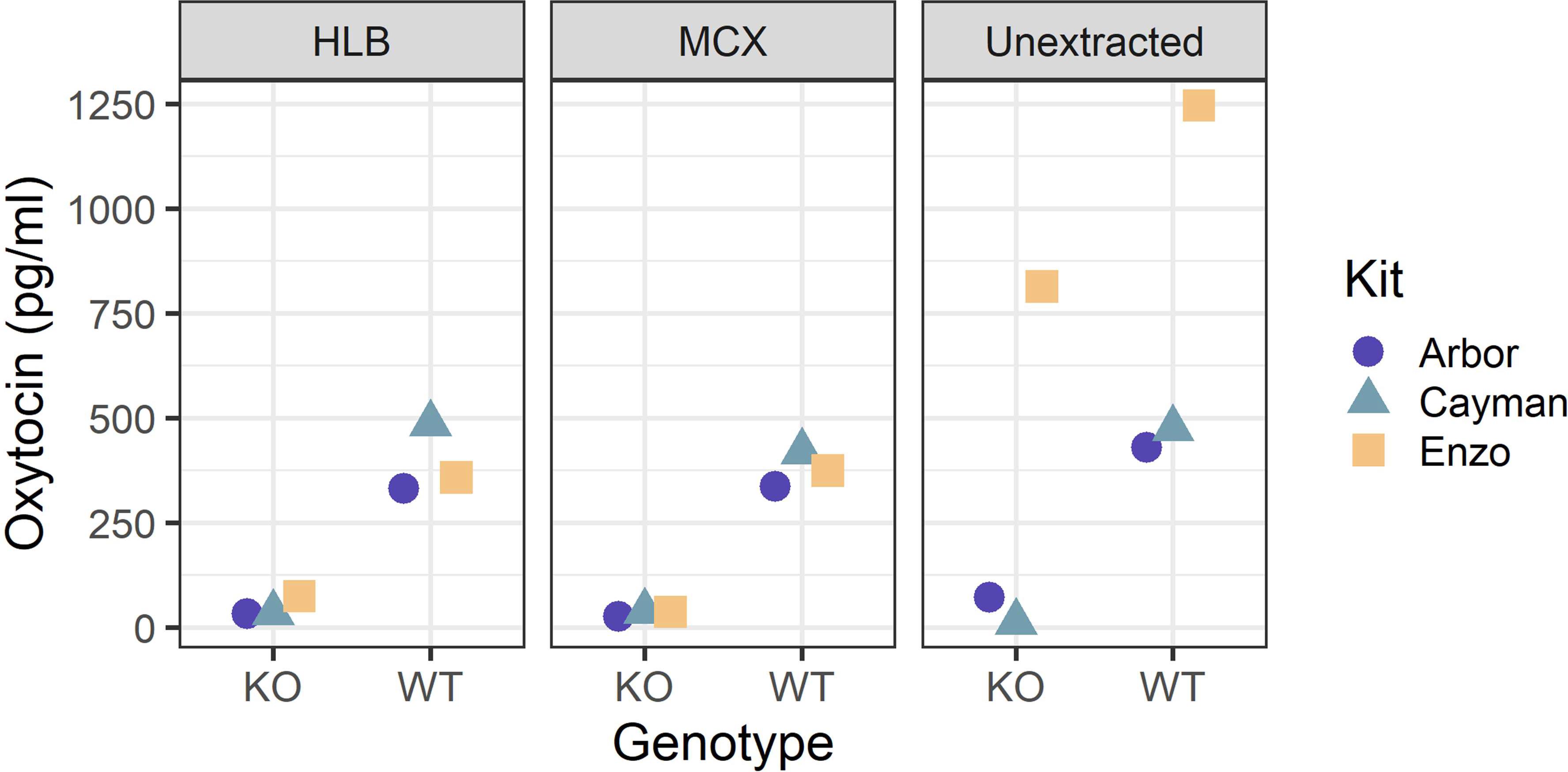
The same sample pools were processed three different ways (HLB extraction, MCX extraction, unextracted), aliquoted, and measured on each of the three immunoassay kits (Arbor, Cayman, Enzo). Reported concentrations are corrected for the dilution of the method. The Enzo kit exhibited considerable interference for the unextracted knockout sample, working at a 1:8 dilution. In all other cases, knockout samples measured below the lower limit of detection or sensitivity of the kit.
3.5.2. Enzo Life Sciences Kit
To assess whether the interference observed on the Enzo kit was consistently additive or whether it interfered with the correlation between methods, we measured 19 additional individual samples on the Enzo kit using both the HLB extraction and unextracted methods. For extracted samples, the wildtype samples had an average concentration of 415.8 pg/ml, while the knockout samples were all under the reported sensitivity of the assay (Figure 5A, Table A3), a significant difference (t(9.1) = 8.38, p = 1.38 × 10−5, Cohen’s d = 3.75). However, for unextracted samples, the knockout samples measured at 615.0 pg/ml on average, with the wildtype samples measuring correspondingly higher as well, averaging 1264.2 pg/ml. As a group, the wildtype samples still measured significantly higher than the knockout samples (Wilcoxon-Mann-Whitney W = 83, p = 9.7 × 10−4, r = 0.712), although it is worth noting that one knockout sample measured at a level indistinguishable from wildtype samples (Figure 5A, Table A3). Further, among wildtype samples the correlation between the two methods (r = 0.676) was not as strong as for the samples measured on the Arbor Assays kit (Figure 5B).
3.5.3. Cayman Chemical Kit
Finally, 10 of the individual samples assayed on the Enzo kit were also assayed—unextracted only—on the Cayman Chemical kit. As with the Arbor kit, but in contrast to the Enzo kit, the knockout samples all measured below the sensitivity of the assay (Figure A5a). Wildtype samples averaged 563.2 pg/ml (Table A4), and the genotype contrast was significant (t(4.1) = 4.072, p = 0.01, Cohen’s d = 2.58). Additionally, the results for the same unextracted wildtype samples on the Enzo and Cayman kits (n = 5) were moderately correlated (r = 0.658), however visual inspection of the data (Figure A3b) suggests more data is necessary to properly assess the correspondence between kits.
4. Discussion
We measured endogenous oxytocin concentrations in oxytocin knockout and wildtype mouse plasma samples, finding a strong contrast between genotypes using both unextracted and extracted samples measured with the Arbor Assays immunoassay kit. Specifically, while wildtype individuals exhibited high levels of endogenous oxytocin, samples from oxytocin knockout individuals were all below the limit of detection for both methods. This finding suggests that oxytocin can be measured in unextracted, diluted (1:8) mouse plasma samples without interference using this kit. To further confirm the validity of immunoassay, we performed parallelism and spike recovery using both methods with satisfactory results, indicating that both extracted and unextracted samples can be assayed reliably and accurately. It should be noted, however, that while a knockout plasma pool displayed acceptable spike recovery when assessed at a 1:8 dilution, it exhibited signs of interference when undiluted. Furthermore, parallelisms for unextracted sample pools were best when excluding samples measured at less than a 1:2 dilution. This suggests that a dilution of more than 1:2 is needed to reliably assay unextracted plasma oxytocin in mice; it is currently unclear whether this required dilution would be similar in other species.
Additionally, in wildtype samples, extracted and unextracted oxytocin measurements were highly correlated on the Arbor Assays kit, despite concentrations in unextracted samples being systematically higher. The high correlation we observed is surprising given previous reports that extracted and unextracted measures have generally not been correlated (Christensen et al., 2014; Leng and Sabatier, 2016; Robinson et al., 2014; Szeto et al., 2012). Further, the concentrations we detected in extracted samples were much higher than is typically reported (McCullough et al., 2013). This finding may result from the improved extraction protocol employed here (supplemental materials), which uses a relatively low percentage of organic solvent for elution of oxytocin. However, it is also possible that the high concentrations detected were influenced by the method of euthanasia (CO2), which is reported to cause a large release of pituitary hormones into circulation (Reed et al., 2009). Although these results are encouraging, it is possible that they are limited to specific details of these methods, including the SPE cartridges, the extraction protocol, and the exact dilution of unextracted samples.
Each method described here has distinct advantages. Unextracted (diluted) samples can be assayed with a very small volume of sample and involve reduced materials, equipment, and labor costs. Furthermore, an extraction step introduces additional opportunities for human and technical errors in processing. However, extracted samples are less likely to exhibit interference regardless of the matrix or kit used. They should therefore be used as a benchmark to compare methods, and unextracted samples should not be assumed to be free of interference without rigorous testing.
Although average plasma oxytocin levels were lower in males than in females, this difference was only statistically significant for extracted samples. For both extracted and unextracted samples, the effect of sex shrank—and statistical significance for extracted samples disappeared—after controlling for body mass. This supports the idea that larger individuals and species may have lower plasma oxytocin levels, potentially due to allometric scaling between pituitary and circulating blood volume (Bienboire-Frosini et al., 2017; Kjeld and Ólafsson, 2008), and calls into question studies that have not controlled for body mass (e.g. Marazziti et al., 2019). However, it should be noted that the effect of body mass was not itself statistically significant in these models, and thus the links between body mass, sex, and plasma oxytocin levels remain an important question for further study. Additionally, while these experiments explored the validity of oxytocin measurements on the level of genotype and individual differences, research measuring endogenous oxytocin levels often explores differences in response to experimental stimuli; further work should thus investigate the sensitivity of these methods to various stimuli associated with endogenous oxytocin release.
We attempted to extend our findings with the Arbor Assays kit to other commercially available oxytocin immunoassay kits with mixed results. The Cayman Chemical kit showed comparable contrasts between wildtype and knockout samples, although we did not test it as thoroughly, while the Enzo Life Sciences kit was susceptible to interference, as evidenced by the high measurements for knockout samples. Furthermore, the correlation between extracted and unextracted measurements was considerably weaker with the Enzo kit compared to the Arbor kit, suggesting that this interference may not be strictly additive. Nonetheless, the oxytocin concentrations in extracted and unextracted samples were positively correlated using the Enzo kit, suggesting relatively strong signal from the oxytocin molecule relative to the background noise. Dilution is an important factor in immunoassays, however, and it is possible that further dilution of samples (beyond the 1:8 working dilution used here) would minimize the interference observed on the Enzo kit.
Our findings indicate that differences between kits are extremely important, as has been found in head-to-head comparisons of immunoassays for other analytes (e.g. Kinn Rød et al., 2017). There are multiple ways in which the chemistries of each kit differ, but we suggest the most likely reason for the observed difference is due to the distinct antibodies used by each company, the effects of which can also be seen in their different reported cross-reactivities. It is also important to note that in some cases the antibodies used by a given company may change over time (e.g. Enzo kit ADI-901-153A-0001 replacing ADI-901-153). In the case of the Enzo kit, it is not uncommon to see this assay referred to as a product of Assay Designs (a company that was acquired by Enzo Life Sciences in 2009), although the original kit and antibody used by Assay Designs is no longer manufactured. Our divergent results across kits underscore the importance of validating the specific method used on a given kit, with updated validations required any time a key component of the kit is changed (e.g. the antibody).
Finally, while our results support the validity of unextracted assays for mouse plasma diluted 1:8 using the Arbor Assays kit, and provide preliminary support for this approach with the Cayman Chemical kit, the extent to which these findings generalize to other matrices (e.g. urine, saliva, CSF) remains to be determined. Given the presence of different proteins, metabolites, etc. in each biological fluid, we anticipate that results may differ in other sample matrices. Similarly, it is important to note that extrapolation from these results to species other than mice requires caution. We anticipate that oxytocin concentrations—and thus suitable dilutions—will vary across species (Bienboire-Frosini et al., 2017), potentially related to their size (Kjeld and Ólafsson, 2008), physiology (Ding et al., 2019; Wang et al., 2015), and socioecology (Finkenwirth et al., 2016; Kramer et al., 2004; Nagasawa et al., 2015; Snowdon et al., 2010).
Furthermore, our findings reveal the potential for interference from some components of plasma, but the specific source and mechanism of the interference we observed remains unknown. Although common mammalian plasma proteins evolved early in vertebrate evolution (Doolittle, 1987), plasma composition and protein binding characteristics vary considerably among mammals (Martinez, 2011). Thus, until the specific source of interference can be identified, our ability to extrapolate these results to other species remains limited. Nonetheless, lacking knockout models in other species, this may be the best practicable test of assay interference, and our findings provide prima facie evidence regarding the specificity and analytical validity of several of the methods presented herein.
5. Conclusions
Despite concerns about the validity of measuring oxytocin in unextracted plasma samples, our results indicate both analytical and biological validity of this approach using the Arbor Assays kit. This supports the notion that oxytocin can be meaningfully measured in unextracted plasma samples using these methods, although dilution is required for optimal performance. Our results represent a critical advance for oxytocin research, as the ability to use unextracted samples affords faster, easier, and less expensive assays, and removes an additional step that can introduce technical error. Although further work is needed to assess the generalizability of these results—for example, to other matrices and species—the use of oxytocin knockout mouse samples presents a fruitful avenue of research for the further development and validation of oxytocin assays. In contrast, our results using the Enzo kit indicate caution is warranted when interpreting results from unextracted plasma samples using the current version of this kit. This finding also demonstrates the importance of differences between kits and highlights the importance of rigorously validating immunoassay methods in oxytocin research.
Supplementary Material
Highlights.
Mouse plasma oxytocin can be reliably measured without extraction by immunoassay.
Unextracted plasma samples must be diluted for accurate measurement by immunoassay.
With the reported methods, extracted and unextracted samples are highly correlated.
Results are dependent on the immunoassay, potentially due to antibody differences.
Some commercially available assay kits are susceptible to matrix interference.
Acknowledgements:
This material is based upon work supported by the Eunice Kennedy Shriver National Institute of Child Health & Human Development of the National Institutes of Health under award number R21HD095217 and by the National Science Foundation Graduate Research Fellowship Program under grant number DGE-1746060. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Institutes of Health or the National Science Foundation.
Funding:
This study was funded in part by the National Institutes of Health (R21HD095217). G.E.G. was additionally funded by the NSF Graduate Research Fellowship Program (DGE-1746060). The Laboratory for the Evolutionary Endocrinology of Primates (LEEP) was funded by the University of Arizona College of Social and Behavioral Sciences, School of Anthropology, Institute for the Environment, Provost’s Office, and Bio-5 Institute.
Footnotes
Declarations of interest: none.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
6. References
- Andreasson U, Perret-Liaudet A, van Waalwijk van Doorn LJC,Blennow K, Chiasserini D, Engelborghs S, Fladby T, Genc S, Kruse N, Kuiperij HB, Kulic L, Lewczuk P, Mollenhauer B, Mroczko B, Parnetti L, Vanmechelen E, Verbeek MM, Winblad B, Zetterberg H, Koel-Simmelink M, Teunissen CE, 2015. A practical guide to immunoassay method validation. Front. Neurol 6, 1–8. 10.3389/fneur.2015.00179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienboire-Frosini C, Chabaud C, Cozzi A, Codecasa E, Pageat P, 2017. Validation of a commercially available enzyme immunoassay for the determination of oxytocin in plasma samples from seven domestic animal species. Front. Neurosci 11. 10.3389/fnins.2017.00524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad N, Warren DJ, Nustad K, 2013. Heterophilic antibody interference in immunometric assays. Best Pract. Res. Clin. Endocrinol. Metab 27, 647–661. 10.1016/j.beem.2013.05.011 [DOI] [PubMed] [Google Scholar]
- Boscato LM, Stuart MC, 1988. Heterophilic antibodies: A problem for all immunoassays. Clin. Chem 34, 27–33. 10.1093/clinchem/34.1.27 [DOI] [PubMed] [Google Scholar]
- Brandtzaeg OK, Johnsen E, Roberg-Larsen H, Seip KF, MacLean EL, Gesquiere LR, Leknes S, Lundanes E, Wilson SR, 2016. Proteomics tools reveal startlingly high amounts of oxytocin in plasma and serum. Sci. Rep 6, 1–7. 10.1038/srep31693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buijs RM, Heerikhuize JJV, 1982. Vasopressin and oxytocin release in the brain - a synaptic event. Brain Res 252, 71–76. 10.1016/0006-8993(82)90979-9 [DOI] [PubMed] [Google Scholar]
- Caldwell HK, Aulino EA, Freeman AR, Miller TV, Witchey SK, 2017. Oxytocin and behavior: Lessons from knockout mice. Dev. Neurobiol 77, 190–201. 10.1002/dneu.22431 [DOI] [PubMed] [Google Scholar]
- Carter CS, 2017. The oxytocin-vasopressin pathway in the context of love and fear. Front. Endocrinol. (Lausanne). 8, 1–12. 10.3389/fendo.2017.00356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter CS, Kenkel WM, Maclean EL, Wilson SR, Perkeybile AM, Yee JR, Ferris CF, Nazarloo HP, Porges SW, Davis JM, Connelly JJ, Kingsbury MA, 2020. Is oxytocin “nature’s medicine”? Pharmacol. Rev 72, 829–861. 10.1124/pr.120.019398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen JC, Shiyanov PA, Estepp JR, Schlager JJ, 2014. Lack of association between human plasma oxytocin and interpersonal trust in a prisoner’s dilemma paradigm. PLoS One 9, 1–22. 10.1371/journal.pone.0116172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding C, Leow MKS, Magkos F, 2019. Oxytocin in metabolic homeostasis: implications for obesity and diabetes management. Obes. Rev 20, 22–40. 10.1111/obr.12757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle RF, 1987. The Evolution of the Vertebrate Plasma Proteins. Biol. Bull. 172, 269–283. [Google Scholar]
- Engvall E, Perlmann P, 1971. Enzyme-linked immunosorbent assay (ELISA) quantitative assay of immunoglobulin G. Immunochemistry 8, 871–874. 10.1016/0161-5890(71)90063-0 [DOI] [PubMed] [Google Scholar]
- Finkenwirth C, Martins E, Deschner T, Burkart JM, 2016. Oxytocin is associated with infant-care behavior and motivation in cooperatively breeding marmoset monkeys. Horm. Behav 80, 10–18. 10.1016/j.yhbeh.2016.01.008 [DOI] [PubMed] [Google Scholar]
- Hammock EAD, 2015. Developmental perspectives on oxytocin and vasopressin. Neuropsychopharmacology 40, 24–42. 10.1038/npp.2014.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrichs M, von Dawans B, Domes G, 2009. Oxytocin, vasopressin, and human social behavior. Front. Neuroendocrinol 30, 548–557. 10.1016/j.yfrne.2009.05.005 [DOI] [PubMed] [Google Scholar]
- Iwasaki Y, Maejima Y, Suyama S, Yoshida M, Arai T, Katsurada K, Kumari P, Nakabayashi H, Kakei M, Yada T, 2015. Peripheral oxytocin activates vagal afferent neurons to suppress feeding in normal and leptin-resistant mice: a route for ameliorating hyperphagia and obesity. Am. J. Physiol. - Regul. Integr. Comp. Physiol 308, R360–R369. 10.1152/ajpregu.00344.2014 [DOI] [PubMed] [Google Scholar]
- Jurek B, Neumann ID, 2018. The oxytocin receptor: From intracellular signaling to behavior. Physiol. Rev 98, 1805–1908. 10.1152/physrev.00031.2017 [DOI] [PubMed] [Google Scholar]
- Kassambara A, 2020. rstatix: Pipe-Friendly Framework for Basic Statistical Tests.
- Kinn Rød AM, Harkestad N, Jellestad FK, Murison R, 2017. Comparison of commercial ELISA assays for quantification of corticosterone in serum. Sci. Rep 7, 1–5. 10.1038/s41598-017-06006-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjeld M, Ólafsson Ö, 2008. Allometry (scaling) of blood components in mammals: Connection with economy of energy? Can. J. Zool 86, 890–899. 10.1139/Z08-061 [DOI] [Google Scholar]
- Kramer KM, Cushing BS, Carter CS, Wu J, Ottinger MA, 2004. Sex and species differences in plasma oxytocin using an enzyme immunoassay. Can. J. Zool 82, 1194–1200. 10.1139/Z04-098 [DOI] [Google Scholar]
- Landgraf R, Neumann ID, 2004. Vasopressin and oxytocin release within the brain: A dynamic concept of multiple and variable modes of neuropeptide communication. Front. Neuroendocrinol 25, 150–176. 10.1016/j.yfrne.2004.05.001 [DOI] [PubMed] [Google Scholar]
- Leng G, Sabatier N, 2016. Measuring Oxytocin and Vasopressin: Bioassays, Immunoassays and Random Numbers. J. Neuroendocrinol 28, 1–13. 10.1111/jne.12413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig M, 1998. Dendritic release of vasopressin and oxytocin. J. Neuroendocrinol 10, 881–895. 10.1046/j.1365-2826.1998.00279.x [DOI] [PubMed] [Google Scholar]
- MacLean EL, Wilson SR, Martin WL, Davis JM, Nazarloo HP, Carter CS, 2019. Challenges for measuring oxytocin: The blind men and the elephant? Psychoneuroendocrinology 107, 225–231. 10.1016/j.psyneuen.2019.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marazziti D, Baroni S, Mucci F, Piccinni A, Moroni I, Giannaccini G, Carmassi C, Massimetti E, Osso LD, 2019. Sex-Related Differences in Plasma Oxytocin Levels in Humans. Clin. Pract. Epidemiol. Ment. Heal 15, 58–63. 10.2174/1745017901915010058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez MN, 2011. Factors influencing the use and interpretation of animal models in the development of parenteral drug delivery systems. AAPS J 13, 632–649. 10.1208/s12248-011-9303-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough ME, Churchland PS, Mendez AJ, 2013. Problems with measuring peripheral oxytocin: Can the data on oxytocin and human behavior be trusted? Neurosci. Biobehav. Rev. 37, 1485–1492. 10.1016/j.neubiorev.2013.04.018 [DOI] [PubMed] [Google Scholar]
- Nagasawa M, Mitsui S, En S, Ohtani N, Ohta M, Sakuma Y, Onaka T, Mogi K, Kikusui T, 2015. Oxytocin-gaze positive loop and the coevolution of human-dog bonds. Science (80-.). 348, 333–336. 10.1126/science.1261022 [DOI] [PubMed] [Google Scholar]
- Neumann ID, Landgraf R, 2012. Balance of brain oxytocin and vasopressin: Implications for anxiety, depression, and social behaviors. Trends Neurosci 35, 649–659. 10.1016/j.tins.2012.08.004 [DOI] [PubMed] [Google Scholar]
- Nishimori K, Young LJ, Guo Q, Wang Z, Insel TR, Matzuk MM, 1996. Oxytocin is required for nursing but is not essential for parturition or reproductive behavior. Proc. Natl. Acad. Sci. U. S. A 93, 11699–11704. 10.1073/pnas.93.21.11699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porstmann B, Porstmann T, Nugel E, 1981. Comparison of Chromogens for the Determination of Horseradish Peroxidase as a Marker in Enzyme Immunoassay. Clin. Chem. Lab. Med 19, 435–440. 10.1515/cclm.1981.19.7.435 [DOI] [PubMed] [Google Scholar]
- Quintana DS, Smerud KT, Andreassen OA, Djupesland PG, 2018. Evidence for intranasal oxytocin delivery to the brain: Recent advances and future perspectives. Ther. Deliv 9, 515–525. 10.4155/tde-2018-0002 [DOI] [PubMed] [Google Scholar]
- R Core Team, 2020. R: A Language and Environment for Statistical Computing.
- Reed B, Varon J, Chait BT, Kreek MJ, 2009. Carbon dioxide-induced anesthesia results in a rapid increase in plasma levels of vasopressin. Endocrinology 150, 2934–2939. 10.1210/en.2008-1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson KJ, Hazon N, Lonergan M, Pomeroy PP, 2014. Validation of an enzyme-linked immunoassay (ELISA) for plasma oxytocin in a novel mammal species reveals potential errors induced by sampling procedure. J. Neurosci. Methods 226, 73–79. 10.1016/j.jneumeth.2014.01.019 [DOI] [PubMed] [Google Scholar]
- Saini PK, Webert DW, Judkins JC, 1995. Role of Sodium Azide in Reducing Nonspecific Color Development in Enzyme Immunoassays. J. Vet. Diagnostic Investig 7, 509–514. 10.1177/104063879500700415 [DOI] [PubMed] [Google Scholar]
- Schroeder HR, Vogelhut PO, Carrico RJ, Boguslaski RC, Buckler RT, 1976. Competitive protein binding assay for biotin monitored by chemiluminescence. Anal. Chem 48, 1933–1937. 10.1021/ac50007a032 [DOI] [PubMed] [Google Scholar]
- Snowdon CT, Pieper BA, Boe CY, Cronin KA, Kurian AV, Ziegler TE, 2010. Variation in oxytocin is related to variation in affiliative behavior in monogamous, pairbonded tamarins. Horm. Behav 58, 614–618. 10.1016/j.yhbeh.2010.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoop R, 2012. Neuromodulation by Oxytocin and Vasopressin. Neuron 76, 142–159. 10.1016/j.neuron.2012.09.025 [DOI] [PubMed] [Google Scholar]
- Szeto A, Ph D, Mccabe PM, Ph D, Nation DA, Ph D, Benjamin A, Rossetti MA, Mccullough ME, Ph D, Schneiderman N, Ph D, Mendez AJ, Ph D, 2012. Evaluation of enzyme immunoassay and radioimmunoassay methods for the measurement of plasma oxytocin. Psychosom. Med 73, 393–400. 10.1097/PSY.0b013e31821df0c2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabbaa M, Hammock EAD, 2020. Orally administered oxytocin alters brain activation and behaviors of pre-weaning mice. Horm. Behav 118, 104613. 10.1016/j.yhbeh.2019.104613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate J, Ward G, 2004. Interferences in immunoassay. Clin. Biochem. Rev 25, 477–480. [PMC free article] [PubMed] [Google Scholar]
- Uvnäs-Moberg K, 1998. Oxytocin may mediate the benefits of positive social interaction and emotions. Psychoneuroendocrinology 23, 819–835. 10.1016/S0306-4530(98)00056-0 [DOI] [PubMed] [Google Scholar]
- Wang P, Yang HP, Tian S, Wang L, Wang SC, Zhang F, Wang YF, 2015. Oxytocin-secreting system: A major part of the neuroendocrine center regulating immunologic activity. J. Neuroimmunol 289, 152–161. 10.1016/j.jneuroim.2015.11.001 [DOI] [PubMed] [Google Scholar]
- Wickham H, Averick M, Bryan J, Chang W, McGowan LD, François R, Grolemund G, Hayes A, Henry L, Hester J, Kuhn M, Pedersen TL, Miller E, Bache SM, Müller K, Ooms J, Robinson D, Seidel DP, Spinu V, Takahashi K, Vaughan D, Wilke C, Woo K, Yutani H, 2019. Welcome to the {tidyverse}. J. Open Source Softw 4, 1686. 10.21105/joss.01686 [DOI] [Google Scholar]
- Yalow RS, Berson SA, 1960. Immunoassay of Endogenous Plasma Insulin in Man. J. Clin. Invest 39, 1157–1175. 10.1172/JCI104130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Liang M, Munesue S, Deguchi K, Harashima A, Furuhara K, Yuhi T, Zhong J, Akther S, Goto H, Eguchi Y, Kitao Y, Hori O, Shiraishi Y, Ozaki N, Shimizu Y, Kamide T, Yoshikawa A, Hayashi Y, Nakada M, Lopatina O, Gerasimenko M, Komleva Y, Malinovskaya N, Salmina AB, Asano M, Nishimori K, Shoelson SE, Yamamoto H, Higashida H, 2019. Vascular RAGE transports oxytocin into the brain to elicit its maternal bonding behaviour in mice. Commun. Biol 2. 10.1038/s42003-019-0325-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


