Abstract
Nonalcoholic fatty liver disease (NAFLD) is a common metabolic dysfunction leading to hepatic steatosis. However, NAFLD's global impact on the liver lipidome is poorly understood. Using high-resolution shotgun mass spectrometry, we quantified the molar abundance of 316 species from 22 major lipid classes in liver biopsies of 365 patients, including nonsteatotic patients with normal or excessive weight, patients diagnosed with NAFL (nonalcoholic fatty liver) or NASH (nonalcoholic steatohepatitis), and patients bearing common mutations of NAFLD-related protein factors. We confirmed the progressive accumulation of di- and triacylglycerols and cholesteryl esters in the liver of NAFL and NASH patients, while the bulk composition of glycerophospho- and sphingolipids remained unchanged. Further stratification by biclustering analysis identified sphingomyelin species comprising n24:2 fatty acid moieties as membrane lipid markers of NAFLD. Normalized relative abundance of sphingomyelins SM 43:3;2 and SM 43:1;2 containing n24:2 and n24:0 fatty acid moieties, respectively, showed opposite trends during NAFLD progression and distinguished NAFL and NASH lipidomes from the lipidome of nonsteatotic livers. Together with several glycerophospholipids containing a C22:6 fatty acid moiety, these lipids serve as markers of early and advanced stages of NAFL.
Supplementary key words: NAFLD, shotgun lipidomics, lipid biomarkers, sphingomyelins, liver biopsies, NAFL;, NASH, biclustering analysis, steatosis, liver lipidome
Abbreviations: ALT, alanine aminotransferase; AP, alkaline phosphatase; AST, aspartate transaminase; AUC, area under the curve; CE, cholesteryl ester; Cer, ceramide; Chol, cholesterol; DG, diacylglycerol; FFA, free fatty acid; GGT, gamma-glutamyl transferase; GPL, glycerophospholipid; HO, healthy obese; LPA, lyso-phosphatidic acid; LPC, lyso-phosphatidylcholine; LPE, lysophosphatidylethanolamine; LPI, lyso-phosphatidylinositol; NAFLD, nonalcoholic fatty liver disease; NASH, nonalcoholic steatohepatitis; NC, normal control; PA, phosphatidic acid; PC, phosphatidylcholine; PC O-, ether phosphatidylcholine; PE, phosphatidylethanolamine; PE O-, ether phosphatidylethanolamine; PG, phosphatidylglycerol; PI, phosphatidylinositol; PR, precision recall; PS, phosphatidylserine; ROC, receiver operator characteristic; SM, sphingomyelin; TG, triacylglycerol
Nonalcoholic fatty liver disease (NAFLD) is a metabolic dysfunction histologically characterized by hepatic fat accumulation (hepatic steatosis) in the absence of heavy alcohol consumption in past medical history (1). NAFLD affects up to 30% of adults and up to 80% of obese and diabetic individuals worldwide (2). The prevalence and severity of NAFLD are higher in men although in postmenopausal women, the NAFLD rate increases (3). NAFLD is subdivided into nonalcoholic fatty liver (NAFL), nonalcoholic steatohepatitis (NASH), cirrhosis, and hepatocellular carcinoma (4, 5). In contrast to NASH, hepatic steatosis in NAFL may occur with no significant inflammation.
The molecular background and pathophysiology of NAFL and why and how it progresses to NASH are poorly understood (6, 7, 8, 9). While the intracellular accumulation of triacylglycerols (TG), diacylglycerols (DG) and free fatty acids (FFA) is a metabolic hallmark of NAFLD, it is unclear whether the disease also alters a broader scope of lipids (10, 11, 12, 13, 14, 15, 16). Mutations in NAFLD risk factors, e.g., PNPLA3 or MBOAT7, affect the liver lipidome (17, 18, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31). It is also conceivable that lipidome remodeling may contribute to or be associated with the onset and propagation of NAFLD (15). However, lipidomic evidence is mostly semiquantitative and based on limited lipid class coverage (32). Previous studies were often focused on the composition of energy storage lipids (12, 13, 33), while membrane and signaling lipids received less attention (28, 34, 35). Chiappini et al. (36) used TOF-SIMS imaging to analyze 104 lipid species in biopsies of 61 NAFLD patients and suggested a NASH lipidomic signature comprising 32 lipid species, although the analysis was biased to most abundant and best ionized classes, e.g., phosphatidylcholines (PC) and TG. The study by Gorden et al. (13) covered 186 lipids in liver biopsies and plasma of 91 patients and revealed a combination of plasma biomarkers (including polyunsaturated glycero- and glycerophospholipids together with long-chain ceramides (Cer)) that distinguished NASH from NAFL. However, lipid markers and their fold changes between disease states reported by independent studies were not concordant. Also, if NAFLD globally alters the liver lipidome and if changes in the abundance of glycerophospho- and sphingolipids (for convenience, here we termed them as membrane lipids) corroborate major clinical indices and correlate with disease severity is an open question, particularly because no reference values for individual liver lipids and their physiological variation are available.
To better understand how NAFLD is transforming the human liver lipidome, we assembled a cohort of 365 histologically characterized biopsies reflecting its progression from nonsteatotic obesity to overt NASH, together with appropriate nonsteatotic and nonobese controls. We then used shotgun mass spectrometry (37, 38) to systematically quantify the molar abundance of 316 species from 22 major lipid classes that encompassed membrane and energy storage lipids including cholesterol. Biclustering analysis of the curated lipidomics dataset recognized several signatures comprising specific membrane lipids that enabled patient's stratification at different stages of NAFLD independently of progressive accumulation of DG and TG.
Materials and Methods
Cohort recruitment, study design, and ethic approval
The study protocol accords the ethical guidelines of the 1975 Declaration of Helsinki and was approved by the authority of Universität Kiel (D425/07, A111/99) before the study commenced. All patients had given their written informed consent. In total, 365 individuals (124 males; 241 females) from 17 to 85 years of age and whose BMI was in the range of 14.8–83.6 (kg/m2) were recruited within the time period of 2007–2016. NASH and NAFL were defined by the NAFLD activity score (NAS) as described (39). Phenotyping of the entire cohort was performed using standardized histology protocol (17) in a blinded fashion by a board-certified surgical pathologist (C. R.) having the specialization in hepatopathology (details are in supplemental data, Histology).
Alcohol consumption was assessed by self-reporting; subjects with average alcohol consumption of more than 30 g/day in men or 20 g/day in women (an equivalent of three and two standard alcoholic drinks per day, respectively) were not enrolled (40). The collected metadata included age, sex, BMI, blood test, including gamma-glutamyl transferase (GGT), medication taken by each patient, and mutation status of the following genes: PNPLA3, TM6SF2, MBOAT7, HSD17B13, SERPINA1 (SERPINA S and SERPINA Z). Evidence for insulin resistance was not available.
Common chemicals and lipid standards
Synthetic lipid standards (see supplemental data, Common chemicals and lipid standards for complete list) were purchased from Avanti Polar Lipids (Alabaster, AL, USA). Individual standards were mixed and diluted with methyl-tert-butyl ether (MTBE)/methanol (MeOH) 10:3 (v/v) (see supplemental data, Common chemicals and lipid standards for details).
Annotation of lipid classes and species
Glycerolipids are referred to TG and DG; glycerophospholipids and lyso-glycerophospholipids to phosphatidic acids (PA), phosphatidylinositols (PI), phosphatidylserines (PS), phosphatidylglycerols (PG), phosphatidylethanolamines (PE), phosphatidylcholines (PC), ether phosphatidylethanolamines (PE O-), ether phosphatidylcholines (PC O-), lyso-phosphatidic acids (LPA), lyso-phosphatidylinositols (LPI), lyso-phosphatidylcholines (LPC), and lysophosphatidylethanolamines (LPE); sphingolipids to ceramides (Cer) and sphingomyelins (SM); sterols to cholesterol (Chol) and cholesteryl esters (CE). Species of glycero- and glycerophospholipids and cholesteryl esters are annotated as <lipid class> <total number of carbon atoms> : <total number of double bonds> in both (or, for lyso-lipids and cholesteryl esters, in one) fatty acid or fatty alcohol moieties (moiety). Sphingolipids are annotated as <lipid class> <total number of carbon atoms> : <total number of double bonds>; <total number of hydroxyl groups> at the ceramide backbone.
Sample preparation for shotgun lipidomics
Biopsies (wet weight of 4.2–21.9 mg) were shock-frozen in liquid nitrogen ensuring an ex vivo time of less than 40 s and stored at –80°C freezer. Prior to lipid extraction, the tissues were homogenized in 300 μl of isopropanol using zirconium beads; the total protein content was determined by Pierce 660 assay (Thermo Fisher Scientific, USA). Lipids were extracted from aliquots containing an equivalent of 50 μg of total protein by adding 700 μl of MTBE/MeOH 10:3 (v/v) containing the internal standard mix (41, 42) (see details in supplemental data, Sample preparation). After evaporation of the organic phase, lipid extracts were reconstituted in 600 μl of 2:1 (v/v) MeOH / CHCl3 and stored at –20°C. Ten microliters of a lipid extract were diluted with 90 μl of the spray solution (4:2:1 isopropanol/MeOH/CHCl3 (v/v/v) containing 7.5 mM ammonium formate) for mass spectrometric analysis. Samples were analyzed in technical duplicates.
Lipid identification and quantification by shotgun mass spectrometry
The mass spectrometric analysis was performed on a Q Exactive instrument (Thermo Fisher Scientific, Bremen, Germany) equipped with a robotic nanoflow ion source TriVersa NanoMate (Advion BioSciences, Ithaca, NY) using nanoelectrospray chips with spraying nozzle diameter of 4.1 μm. The ion source was controlled by the Chipsoft 8.3.1 software (Advion BioSciences). Ionization voltage was +0.96 kV in positive and −0.96 kV in negative mode; backpressure was 1.25 psi in both modes (43). Temperature of the ion transfer capillary was 200 °C; S-lens RF level was 50%. FT MS spectra were acquired within the range of m/z 400–1,000 in positive and m/z 350–1,000 in negative ion mode at the target mass resolution of R m/z 200=140,000; automated gain control (AGC) of 3 × 106 and maximal injection time of 3 s. In both modes t-SIM spectra were acquired at the same mass resolution and m/z range as above; AGC of 5 × 104; maximum injection time of 650 ms; width of isolation window of 20 Th. The inclusion list of masses targeted by t-SIM started at m/z 355 in negative and m/z 405 in positive ion mode and other masses were computed by adding 10 Da increment (i.e., m/z 355, 365, 375) up to m/z 1,005. Free cholesterol was quantified by parallel reaction monitoring (PRM) FT MS/MS (41, 44, 45) during the same analysis. The number of micro-scans was set to 1; width of precursor isolation window of 0.8 Th; normalized collision energy (nCE): 12.5%; AGC: 5 × 104 and maximum injection time of 3 s.
Raw t-SIM spectra were subjected to repetition rate filtering by PeakStrainer software (46) and then stitched together by SIMStitcher software (47). Lipids were identified by LipidXplorer software (48) by accurately determined m/z (mass accuracy better than 5 ppm) and quantified by comparing the isotopically corrected abundances of their molecular ions with the abundances of internal standards of the same lipid class. All internal standards were detected in all samples. MS2 validation of selected species by HCD FT MS/MS was applied as described in supplemental data, MS2 validation.
Raw data processing
The LipidXplorer output was processed using several steps. Technical replicates were averaged and spectra of QC samples (details in supplemental data, Pilot sub-cohort and Quality Control) set aside. In each sample lipid abundances for which the standard deviation (SD) exceeded 40% were set to Not-a-Number (NaN). Lipids whose abundance exceed the minimal value determined for this species by less than 2-fold were exempted from SD filtering (supplemental data, Raw data processing and supplemental Fig. S1). Also, cholesterol values were not SD-filtered because they were determined by PRM. Next, we grouped the patient samples according to their disease status (normal control, healthy obese, NAFL, NASH, and none). The abundance of lipid species that were detected in less than 15% of all patient samples of the same group was set to zero. Finally, we applied plate bias correction using the ComBat approach (49) as detailed in supplemental data, Raw data processing. The final dataset comprised the molar abundance of 316 lipid species from 22 lipid classes in 365 patients (supplemental dataset S1).
Bioinformatics and data mining
Biclustering analysis was performed using the isa (50) and qubic (51) algorithms. To eliminate redundancy of biclusters, we screened them using a consensus approach (see supplemental data, Bioinformatics and data mining).
Classifications were performed using random forest classifiers from the “randomForest” R package. Hundred classifiers were trained per scenario on bootstrapped lipidomics data. The models were evaluated using precision recall (PR) and receiver operator characteristics (ROC) area under the curve (AUC) values on the test set. Feature importance was computed using gini index averaged over all bootstrapped models (For further details, see supplemental data, Bioinformatics and data mining).
Lipidomics impact of mutations
The impact of mutations was probed within each group (supplemental data, Genotyping), in which the lipidomes of homozygotes, heterozygotes, and normal genotypes were compared pairwise (details are in supplemental data, Statistical analysis of mutation-correlated lipidome changes).
Results
Study cohort
Percutaneous or surgical liver biopsies were taken from in total of 365 Caucasian patients (124 males and 241 females) admitted to the University Hospital Schleswig-Holstein, Kiel, Germany during 2007–2016. Control biopsies were collected during gastrointestinal surgery for pathologies having no direct association with NAFLD. Each biopsy was examined by the same surgical pathologist (C. R.) (17) and assessed according to the NAFLD activity score (NAS) (39).
Patients were further sorted into four basal and one additional group (52) as outlined below. Biopsies were classified according to the presence of steatosis and inflammation as the main criteria to differentiate simple steatosis (NAFL) from steatohepatitis (NASH) (52). Therefore, individuals whose biopsies were having histological fat content ≤5%; BMI <30 kg/m2; no histologically proven liver pathology; no lobular inflammation and absence of significant (stage 2 or higher) fibrosis formed the normal control (NC) group. Healthy obese (HO) group comprised patients having no NAFLD per clinical and histological evidence, yet their BMI exceeded 30 kg/m2. Patients were assigned to NASH and NAFL groups if the histological examination revealed steatosis with or without inflammation, respectively. In total, 337 patients were sorted into NC (n = 49), HO (n = 51), NAFL (n = 143), and NASH (n = 94) groups (Table 1; supplemental Table S1, age and BMI distribution is in supplemental Fig. S4).
Table 1.
Clinicopathological characteristics of the study cohort
| Group | Number of patients |
NAS Parametersa |
Fibrosisa | Mean age, years | Mean BMI kg/m2 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Total | Female | Male | NAS Fat | NAS Ballooning | NAS Inflammation | NAS Average | ||||
| NC | 49 | 25 | 24 | 0 | 0 | 0 | 0 | 0 | 67.7 | 24.1 |
| HO | 51 | 47 | 4 | 0 | 0 | 0 | 0 | 0 | 42.3 | 46.7 |
| NAFL | 143 | 94 | 49 | 1 | 0 | 0 | 0 | 0.35 | 45.6 | 47.1 |
| NASH | 94 | 62 | 32 | 2 | 0 | 1 | 4 | 1 | 45.1 | 49.3 |
| None | 28 | 13 | 15 | n.a. | n.a. | n.a. | n.a. | n.a. | 57.1 | 33.7 |
Median values, in arbitrary units assigned according to (39).
Within the cohort, 28 patients who did not receive NAS because of serious liver conditions, such as histologically confirmed cancer with abnormal and higher than control levels of GGT, alkaline phosphatase (AP), and bilirubin, were assigned to the additional group “None.” Their lipidomes helped to delineate the impact of a broader spectrum of liver diseases different from NAFLD. The impact of obesity (53) was evaluated by comparing lipidomes of NC and HO groups. Since mean BMI of HO, NAFL, and NASH groups was similar, their lipidomes could be compared with no further adjustment. The mean age of NC group was 12 years higher than of NAFL/NASH/None groups because of fewer younger people undergoing gastrointestinal surgery.
Shotgun lipidomics of liver biopsies
Although surgical biopsies were similar in size, they varied in weight by as much as 5-fold and also differed in fat and fibrotic content (Table 1). Since they were unique and histologically inhomogeneous, each specimen was analyzed as a single sample with no independently processed replicates. In an aliquot of each biopsy lysate, we first determined the total protein content and used it for subsequent normalization of molar lipid abundances.
To quantify the lipidomes, we employed high-resolution shotgun Fourier transform mass spectrometry (FT MS) that relies on direct nanoflow infusion of total lipid extracts (37, 38). During shotgun analysis, internal standards spiked into biopsy lysates prior lipid extraction are ionized together with endogenous lipids and enable their absolute (molar) quantification (17, 42, 54, 55, 56). Because of high and variable content of TG together with abundant chemical background, we analyzed liver extracts by the method of t-SIM (for targeted single ion monitoring) (47). In each analysis FT MS spectra were successively acquired from partially overlapping m/z windows of 20 Th and then these spectra were stitched together into a master spectrum by SIMStitcher software. Although typical t-SIM acquisition (supplemental Fig. S5) required ca. 11 min per sample (compared with less than 1 min required for conventional FT MS analysis), it increased the number of quantifiable lipid species by ca. 45% (57).
To ensure spectra acquisition consistency and enable batch correction of lipid abundances, we created a QC standard by pooling equal aliquots of 36 extracts that, according to a preliminary experiment, reflected extreme values of the total lipid content and represented both genders, stages of inflammation and fibrosis. In each aliquot of the QC sample, we quantified 255 lipid species from 19 lipid classes. The molar abundance of more than 98% of lipid species differed by less than 5% and only cholesterol (quantified by the method of PRM (57, 58, 59)) and a few TG species were detected with higher SD (supplemental Fig. S2).
The final dataset covering 365 biopsies comprised normalized molar abundances (in pmol per μg of total protein) of 316 lipid species from 22 lipid classes: Out of the total of 316 lipid species, only 114 were detected in more than 98% of biopsies (supplemental dataset S2).
Liver lipidome of the normal control group
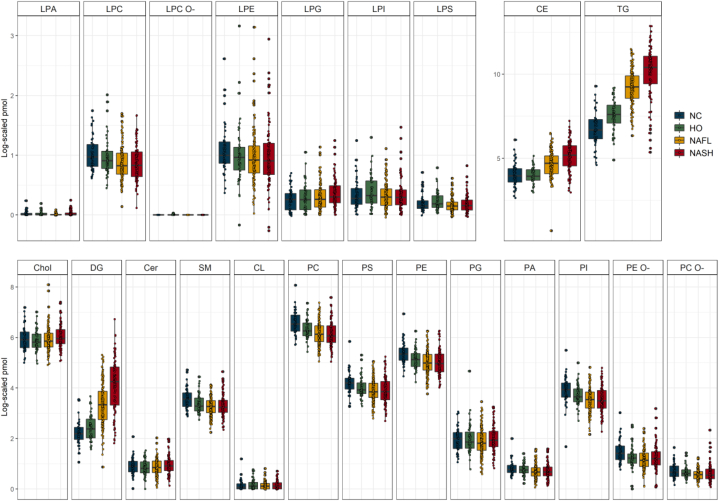
The NC group (n = 49) combined patients with no apparent liver pathology and obesity, whose lipidome we assumed as basal (Fig. 1). The mol% of lipid classes generally corroborates previous reports (13) (supplemental Table S2), although we detected considerably more Cer, PS, SM, and lyso-lipids.
Figure 1.
Lipid class composition (in pmol per μg of total protein; log2 scaled ) of liver biopsies in the four main groups of patients. The total abundance of each lipid class was calculated by adding up molar abundances of lipid species. Boxing highlights values between 25% and 75% quartiles; vertical lines connect minimum and maximum values excluding outliers. Filled circles stand for the lipid class abundance in individual biopsies (average of three technical replicates); black lines within boxes indicate median values. Color coding is shown in the inset at the right-hand side.
Since the liver is a hub of lipoprotein biosynthesis, we further examined if the NC lipidome differs from plasma lipidome (42, 56, 60). While both lipidomes have similar mol% proportions of PC, SM, and Chol, plasma contained 10-fold more CE (supplemental Fig. S6). Liver lipidome contains a larger variety of lyso-species (liver-specific are LPG, LPI, LPS, LPA), while LPC is more abundant in mol%. TG and DG are also more abundant in the liver, as well as PE, PG, PI, PS, and Cer. Lipid class profiles in the liver showed no pronounced gender bias (supplemental Fig. S6) that was apparent in healthy plasma (42).
Mutation-associated changes of the liver lipidome
We further evaluated how common mutations in NAFLD risk genes: TM6SF2 variant rs58542926 (hcv89463510), PNPLA3 variant rs738409 (hcv7241), MBOAT7 variant rs641738 (hcv8716820), SERPINA1 variant PiZ (Glu342Lys) rs28929474 (hcv34508510), SERPINA1 variant PiS affected the liver lipidome. Within NC and NASH groups liver lipidomes of homozygotes, heterozygotes and normal genotype carriers were compared pairwise (Table 2). Note that, for consistency, we compared absolute (in moles per μg of total protein) rather than relative (in mol% within each class) lipid abundances, although the latter could reveal more affected species (17) (supplemental Dataset S1).
Table 2.
PNPLA3 and MBOAT7 mutation-dependent changes of lipidome in NASH and NC groups
| Lipida | Mutation | Group | Mutation Status | Abundance Ratio |
|---|---|---|---|---|
| CE 22:2 | PNPLA3 rs738409 | NASH | Heterozygote/No mutation | 4.0b |
| TG 56:7 | PNPLA3 rs738409 | NASH | Heterozygote/No mutation | 6.8c |
| TG 56:7 | PNPLA3 rs738409 | NASH | Homozygote/No mutation | 15.1c |
| TG 56:7 | PNPLA3 rs738409 | NASH | Homozygote/Heterozygote | 2.2b |
| PI 36:1 | MBOAT7 rs641738 | NC | Heterozygote/No mutation | 2.2b |
| PI 36:1 | MBOAT7 rs641738 | NC | Homozygote/No mutation | 4.5d |
Significantly changed lipid in the two compared groups.
Significance P < 0.05.
Significance P < 0.005.
Significance P < 0.01.
MBOAT7 mutation-dependent changes were only detected in the NC group and confined to PI 36:1 (Table 2), consistent with its phosphatidylinositol O-acyltransferase activity and the role in NAFLD pathogenesis (17). In the NASH group PNPLA3 rs738409 mutation elevated the levels of CE 22:2 and TG 56:7, while the total abundance of CE and TG classes did not change (supplemental Figs. S7, S8A,B) (19, 20, 29, 31, 61, 62, 63).
Condition-associated changes in lipid classes
There was no marked difference between the total abundance of lipid classes in disease groups compared with NC (Fig. 1), except significantly higher levels of TG and, to lesser extent, of DG and CE in NAFL and NASH patients (supplemental Figs. S9–S11). Although the mean BMI of HO patients was almost 2-fold higher compared with NC and was as high as in NAFL and NASH patients, the abundance of TG in HO and NC was similar.
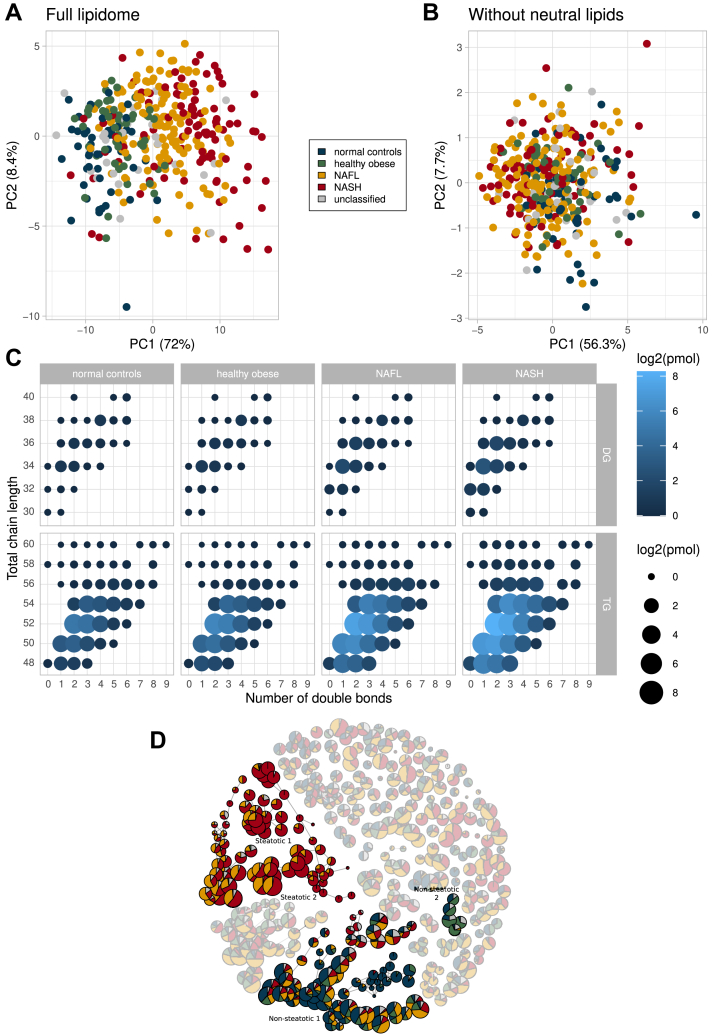
In principal component analysis (PCA) plots of patients' lipidomes, the gradient across PC1 (principal component 1) from NC (at the left) to NASH (at the right) (Fig. 2A) reflected the increased abundance of glycerolipids and CE (Fig. 2B). Interestingly, the profiles of hydrocarbon chain length and unsaturation of fatty acid moieties in TG and DG species were the same in all patient groups (Fig. 2C).
Figure 2.
Segregation of patient groups by the composition of lipidomes. A: PCA plot for full lipidomes of liver biopsies of 365 patients, whose disease group is indicated by color (coding scheme is in the inset); (B) PCA plot of lipidomes from which glycerolipids and CE were omitted. C: Length and unsaturation of fatty acid moieties in TG and DG species. Circle size and color reflect lipid abundances. D: Similarity network of biclusters. Node size is proportional to the number of patients in the bicluster. Highlighted are four network components (annotated) comprising the largest number of connected biclusters attributed to steatotic (in red and amber for NASH and NAFL) or nonsteatotic patients (in blue and green for NC and HO).
We further looked if TG and DG accumulated in the liver of NAFL and NASH patients were compositionally different from the adipose tissue. The ten most abundant TG species in liver (current dataset) and white adipose tissue (WAT) (64, 65) (supplemental Table S3) were almost the same suggesting that TG metabolism in tissues is not organ-specific. However, they differed from TG in plasma of obese patients (66), presumably because in tissues TG accumulation sequesters lipotoxic FFA, while plasma TG are packed into lipoproteins for transporting via bloodstream (65).
Molecular stratification of patient groups by biclustering
Lipid class composition of liver biopsies (Figs. 1, 2) appears to be conserved and, apart from major increase in glycerolipids and CE, offers limited molecular insight or diagnostic perspective. We reasoned that better molecular stratification of patients could rely on clusters of individual species spanning multiple lipid classes whose molar abundance coherently changes with the disease progression. Specifically, we employed a biclustering ensemble approach (Bioinformatics and data mining), in which lipidome compositions are clustered and the results consolidated into networks of connected components comprising lipid compositions specific for a selection of patients (Fig. 2D; supplemental Figs. S13, S14). Lipid compositions and meta-data could be compared across biclusters and reveal lipid signatures specific for a disease condition or patient group. Within all patients' lipidomes, we recognized two components comprising higher proportion of steatotic (NASH and NAFL) and two components with mostly nonsteatotic (NC and HO) patients (Fig. 2D, supplemental Table S4). Their lipidomes were clearly separated by PCA (supplemental Fig. S12A).
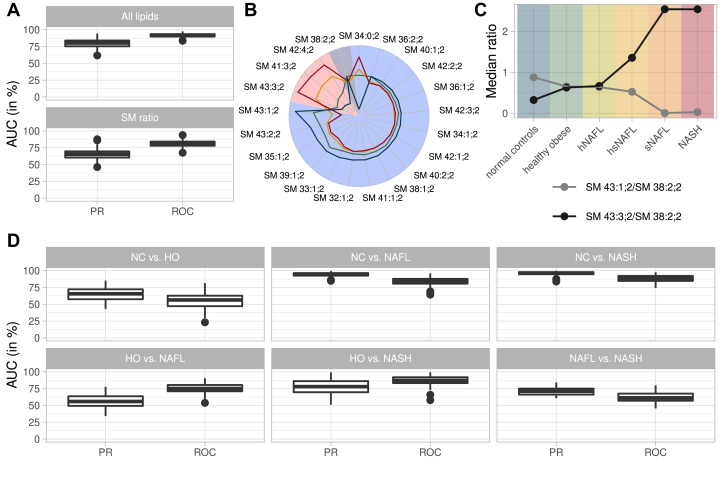
We first tested if lipids identified by biclustering could detail the trend toward steatosis and identify markers distinguishing nonsteatotic from steatotic groups by training random forest-based classifiers. These groups were not separated by common clinical indices, e.g., total bilirubin, GGT, AP, alanine aminotransferase (ALT), and aspartate transaminase (AST) (supplemental Fig. S15B), but were readily distinguished by lipidome compositions (Fig. 3A). Classification performance was evaluated using the precision recall and ROCs (supplemental Fig. S16).
Figure 3.
Classification of disease groups by lipid markers. A: Classification of steatotic and nonsteatotic patients reported as an area under the curve (AUC) for precision recall (PR) and receiver operator characteristics (ROC). Classification based on all lipids (upper panel) and only by the ratio of SM 43:1;2 and SM 43:3;2 normalized to SM 38:2;2 (lower panel). B: Changes in the abundance of SM species in steatotic (red background) and nonsteatotic (blue background) components. SM 38:2;2 belongs to none of the two components (gray background). Lines indicate disease progression: NC (blue), HO (green), NAFL (yellow), and NASH (red) patient groups. Axes indicate relative abundances of SM species; data points per species sum up to 100%. C: Medians of the ratios of the abundance of SM 43:1;2 to SM 38:2;2 and SM41:3;2 to SM 38:2;2 for the patient groups, including subgroups of NAFL (D) Classifications of all patient groups against each other using the two SM ratios.
Lipids most significant for the classification were mono- and diunsaturated TG and DG (e.g., TG 50:1, TG 52:2, or DG 34:1) (supplemental Fig. S15A). To further test if unsaturation of fatty acid moieties in TG plays a role, we trained two classifiers with TG having in total 1–4 and 5–8 double bonds, respectively. Both models performed significantly better than random. However, the median Precision Recall Area Under the Curve (PR AUC) of the model with more saturated TG showed better performance (supplemental Fig. S15C, D).
Taken together, our analysis revealed several mono- and diunsaturated DG and TG species as lipid markers of steatosis while common membrane lipids (including glycerophospholipids (GPL), Chol, or Cer) were not in the markers list with one notable exception of two SM species (supplemental Fig. S15A) that also showed clear NAFLD-dependent profile (Fig. 3B).
Sphingomyelin species sharing n24:2 fatty acid moiety are same-class membrane lipid markers of NAFLD
We plotted the relative abundances of all SM species found in steatotic and nonsteatotic groups (Fig. 3B). The abundance of species from the nonsteatotic group (such as SM 42:2;2, SM 40:1;2, SM 42:1;2) either changed marginally or, as for SM 43:1;2, markedly decreased with NAFLD progression with the exception of SM 34:0;2 (Fig. 3B & supplemental Fig. S17).
In contrast, the abundance of SM 41:3;2, SM 43:3;2, and SM 42:4;2 from the steatotic group followed the opposite trend: it consistently increased with NAFLD progression. To elaborate on this finding, we first validated their identification by high-resolution HCD FT MS/MS (supplemental Materials and Methods, MS2 validation). We note that these lipids were previously identified in human plasma by the method of LC-MSn. Major species of SM 41:3;2 and SM 43:3;2 were recognized as SM d17:1/n24:2 and SM d19:1/n24:2, respectively (67); and SM 42:4;2 was SM d18:2/n24:2 (60), suggesting that these three SM may be sharing n24:2 fatty acid moiety. In contrast to SM 41:3;2, another compositionally related yet more saturated sphingomyelin SM 43:1;2 was attributed to nonsteatotic group and its abundance dropped with NAFLD progression.
To validate the molecular composition of liver sphingomyelins, we subjected a few combined extracts of steatotic and nonsteatotic biopsies to targeted LC-MSn (67) (method details are in supplemental data, Identification of SM molecular species by LC-MSn). The analysis (supplemental Table S5) confirmed that SM 34:0;2 belongs to dihydrosphingomyelins, the lipid class associated with fat deposition in organs, including the liver and pancreas (68). SM 42:4;2 and SM 43:1;2 comprised unsaturated (n24:2) and saturated (n24:0) fatty acid moieties, respectively (supplemental Fig. S19; supplemental Table S5). Because of their low abundance, LC-MSn analysis of SM 43:3;2 and SM 41:3;2 was inconclusive.
We noticed that the abundance of SM 41:3;2 and SM 43:3;2 (both comprising n24:2) as compared with SM 41:1;2 (comprising n24:0) followed opposite trends (Fig. 3B), and we examined if their ratio to some unchanged SM species could distinguish different stages of NAFLD. We computed the ratios of SM 43:1;2 (decreasing in NAFLD) and of SM 43:3;2 (increasing in NAFLD) to SM 38:2;2 (SM d18:2/n20:0; see supplemental Table S5) that did not associate with steatotic and nonsteatotic groups and whose abundance was not affected by NAFLD (Fig. 3B). These ratios could be determined directly from a shotgun spectrum of a total lipid extract without prior adjustment to the abundance of internal standards or biopsy size (Fig. 3C). Strikingly, they distinguished nonsteatotic and steatotic subcohorts with only slightly lower specificity than all lipids (Fig. 3A). SM-based classification was unaffected by obesity: it marginally distinguished NC and HO, but readily delineated NC from NAFL and NASH (Fig. 3D). To corroborate these findings, we further examined mol% profiles of species of SM and Cer classes in the four patient groups (supplemental Figs. S17, S18). They confirmed the notable decrease in the marker SM 41:1:2 as well as of the most abundant SM 34:1;2 together with the concomitant increase of SM 41:3;2 and SM 43:3;2 in NAFL and NASH subgroups, as compared with NC and HO. This, however, did not affect the abundance of matching Cer species: the mean value of Cer 34:1;2 was unchanged and none of Cer with odd number of carbon atoms was detectable.
We noted that SM ratios were less specific when we compared HO and NAFL (Fig. 3D). At the same time, HO and NASH were distinguished notably better albeit there was no global difference between their lipidomes except higher abundance of neutral lipids. Therefore, we hypothesized that the cohort of NAFL patients may be compositionally heterogeneous and consist of smaller, yet compositionally distinct subcohorts reflecting some intermediate stages of NAFLD pathogenesis. In this way, SM ratios and likely other lipid markers could reflect the transition from initial and, likely, reversible stage(s) of NAFL towards NASH.
Heterogeneity of NAFL lipidome and its molecular markers
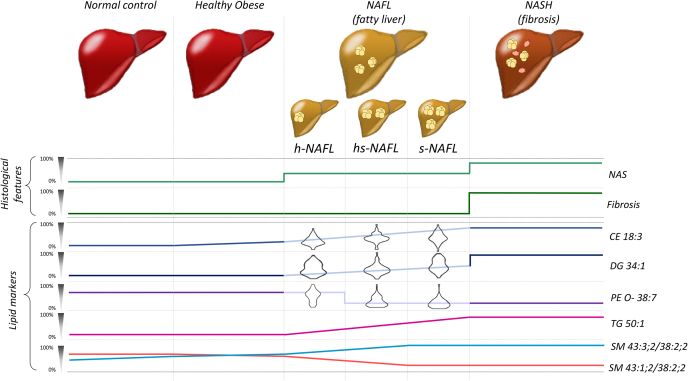
We noticed that a component “Non-steatotic 1” (Fig. 2C) covers the largest number of nonsteatotic (NC+HO) patients, but also some patients with NAFL and NASH. Similarly, “Steatotic 2” mainly covers steatotic (NAFL and NASH) patients and almost no patients of NC and HO groups. We hypothesized that lipidomes of some NAFL patients may have higher similarity to the lipidomes of HO or of NASH. Based on the similarity of lipidome compositions, we used biclustering to divide NAFL patients into four subgroups that clustered together with nonsteatotic (h- for healthy) individuals, with NASH (s- for sick) individuals or neither h- nor s- (hs-) (Fig. 4). To test if this lipidome-based clustering reflects the disease progression also within the NAFL group, we compared the median values of individual histopathological indices (e.g., liver fat mass, ballooning, fibrosis, and inflammation) that were used for calculating NAS. We observed clear disease-related trends for each index and also for the total NAS (supplemental Table S6). Within NAFL subgroups, fat and ballooning expectantly increased from h-NAFL to s-NAFL, whereas fibrosis and inflammation did not, indicating that developing steatosis has not yet led to NASH. Expectantly, BMI did not change progressively between the subgroups.
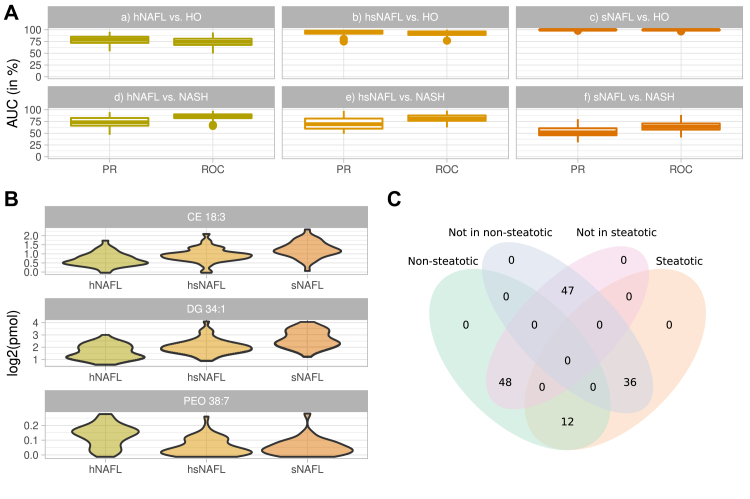
Figure 4.
Classification of NAFL subgroups. A: Classification of NAFL subgroups against HO and NASH groups by ratios of SM 43:1;2 to SM 38:2;2 and SM41:3;2 to SM 38:2;2 whose trend lines are shown in Fig. 3C. B: Violin plots of log2-scaled abundances of CE 18:3, DG 34:1, and PE O- 38:7 in the three NAFL subgroups. Color coding of in panels A and B is the same as in Fig. 3C. C: Distribution of NAFL patients in bicluster components indicating the representative number of patients in each subgroup.
Next, we subjected the patients subgroups to random-forest classification according to the following scenarios: (a) h-NAFL versus HO, (b) hs-NAFL versus HO, (c) s-NAFL versus. HO, (d) s-NAFL versus NASH and (e) hs-NAFL versus NASH, (f) h-NAFL versus NASH on the full lipidome (supplemental Fig. S20). As anticipated, we observed that the classifications performance increased from (a) to (c) and from (d) to (f) indicating that the lipidome of NAFL subgroups consistently changed with the disease progression. The change of SM ratios in subgroups of NAFL and also in four major groups of patients visualized this trend (Fig. 3C).
In these classifications DG 34:1, but also CE 18:3 and CE 18:2 were the most discriminating markers (supplemental Fig. S21). We next tested, if h-NAFL, hs-NAFL, and s-NAFL patients, selected solely by comparing their lipidomes, are also distinguishable histologically? If so, what marker lipids could reflect the transition from h-NAFL to s-NAFL and further toward NASH? The examination of histological indices indicated gradual increase in both inflammation and steatosis (supplemental Table S6). However, none of them reached typical NASH values, apart from % of accumulated fat. In their lipidomes, the abundance of CE 18:3 and DG 34:1 gradually increased from h-NAFL to s-NAFL (Fig. 4B).
We also classified lipidomes of NAFL patients from the steatotic bicluster component against all remaining NAFL patients and identified PE O- 38:7 as the most discriminating marker (supplemental Fig. S22). Examination by HCD FT MS/MS identified it as plasmalogen PE O- 16:1/22:6 (supplemental Fig. S23 and supplemental Materials and Methods, MS2 validation) whose abundance dropped already in hs-NAFL subgroup down to s-NAFL level. Another three polyunsaturated glycerophospholipids: LPE 22:6, PC 38:6, and PC 40:7 showed similar classification power (supplemental Fig. S20). Their sum formula suggests that, similar to PE O- 38:7 and LPE 22:6, they also comprise C22:6 fatty acid moiety. We argue that stepwise decrease in the abundance of C22:6 - containing glycerophospholipids might indicate the onset of transition of h-NAFL toward s-NAFL, despite that there was only a marginal difference between histological indices of h-NAFL and hs-NAFL subgroups (supplemental Table S6). Interestingly, the classifications based on the SM ratios showed similar specificity (supplemental Fig. S17).
Taken together, our analysis distinguished nonsteatotic (NC and HO) and steatotic (NAFL and NASH) cohorts by the accumulation of storage lipid markers TG 50:1, DG 34:1, CE 18:3 and, independently, by the ratios of sphingomyelins SM 43:3;2 and SM 41:3;2 SM to the reference SM 38:2;2. Furthermore, the SM ratios together with coherently changing C22:6 - containing glycerophospholipids (most specifically PE O- 38:7, but also LPE 22:6, PC 38:6 and PC 40:7) revealed the compositional heterogeneity of the NAFL lipidome and how it changes, once the progressing disease gradually remodels HO-like lipidome toward NASH (Fig. 5).
Figure 5.
Schematic trends of abundances of lipidomics and histological markers during NAFLD. The magnitude of change (in %) of lipid (Figs. 3 and 4) and histological (supplemental Table S4) markers scaled to the difference between NC and NASH. Cartoon images of the liver indicate progressive accumulation of fat eventually leading to inflammation and fibrosis.
Discussion
Excessive accumulation of TG is the histological hallmark of NAFLD. While this alone links NAFLD to altered lipid metabolism, little was known if and how NAFLD globally alters the liver lipidome, particularly its membrane and signaling complements.
Our study confirmed the accumulation of neutral lipids along with NAFLD progression. Interestingly, the molecular composition of TG and DG species did not change and, altogether, was similar to white adipose tissue (WAT). Accordingly, biclustering analysis identified monounsaturated TAG 50:1 and DAG 34:1, but also unsaturated CE 18:3 as most specific markers of NAFLD progression whose abundance steadily increases from HO to NASH (Figure 3, Figure 4, Figure 5). The impact of five risk genes implicated in NAFLD development corroborated the molecular specificity of PNPLA3 and MBOAT7, but did not alter the liver lipidome globally.
Interestingly, the levels of glycerophospholipids and of sphingolipids, e.g., Cer and (with a few notable exceptions) SM were not perturbed significantly (Fig. 1). Also, no apparent association with altered SM/Cer metabolism emerged from the transcriptomics analysis (69). Although liver inflammation and apoptosis in NASH are supposed to increase the levels of Cer also by cleaving SM (9, 13, 32, 70, 71), we did not observe it in the biopsies. However, if SM directly contributed to or are associated with steatosis and its transformation to NASH was an open question (72). Specific bidirectional changes of the four SM species supported the robust same lipid class disease state classification. Conveniently, ratios of SM markers could be computed from raw intensities of corresponding molecular peaks: they did not depend on the biopsy size and did not require the full lipidome quantification. To the best of our knowledge, the prospective marker lipids SM 43:3;2, SM 41:3;2, and SM 42:4;2 were not previously spotted in the pathophysiological context of metabolic syndrome. Therefore, it seems promising to use targeted quantification to follow their levels in plasma as it might yield a convenient marker for independent and noninvasive stratification of NAFLD.
Sphingolipids with n24:2 moities are produced from linoleic (C18:2) fatty acid (73). CE 18:2 is the most abundant cholesteryl ester in both the liver and plasma, and it is also enriched in NAFLD patients. It is therefore conceivable that enhanced biosynthesis of SM comprising n24:2 reflects increasing availability of free linoleic acid, while the abundance of SM with saturated or monounsaturated fatty acid moieties tends to decrease (Fig. 3B). However, relative changes of the abundance of SM species between steatotic and nonsteatotic conditions are not proportional to their absolute (molar) content and are also sphingosine-backbone dependent. We noticed that two out of three marker SM comprise sphingosine backbone having odd number of carbon atoms. Therefore, it would be interesting to assess the contribution of microbiome lipids (reviewed in (74)) or de novo synthesis from branched amino acids (75).
One of the most intriguing findings of this work was the lipidomics evidence of the NAFL cohort heterogeneity (Figure 3, Figure 4, Figure 5) that was not apparent from the histological examination (Table 1). In the NAFL subgroups that we conveniently termed as hs- and s-NAFL the levels of plasmalogen PE O- 38:7, but also PC 38:6, PC 40:7 and LPI 22:6 (all comprising C22:6 moiety) dropped down to the level typical for NASH patients. In the same NAFL subgroups the dynamics of histopathological indices (supplemental Table S6) was concordant with progressive steatosis with no hallmarks of fibrosis and inflammation. We therefore speculate that altered SM ratios together with decreased levels of C22:6-containing glycerophospholipids might indicate a turning point in NAFLD pathogenesis, where the transition from NAFL to NASH becomes irreversible.
While at this stage we cannot offer a suitable mechanistic explanation, we hypothesize that depletion of C22:6 containing GPL might affect the levels of PPARa (76) and SREBP1c (77) and, eventually, promote steatosis and inflammation. Also, we cannot rule out that the decreased abundance of these lipids hints at enhanced oxidative stress, despite that shotgun profiling revealed no notable accumulation of oxidized TG and glycerophospholipids.
Irrespective of NAFLD stage, the organism strived to maintain the compositional identity of the hepatocyte membrane lipidome and changes were limited in both molecular scope and magnitude. The unexpected link between species-specific SM metabolism and NAFLD progression, together with NAFL-specific change in the levels of C22:6 - containing glycerophospholipids could contribute to both patients stratification and mechanistic understanding of the lipidome regulation during the disease.
Last but not least, this work created an open and transparent lipidomic resource that could be expanded with or cross-validated by further independent studies reporting molar abundances of liver lipids, irrespective of their research objectives and employed analytical methods. Studies design and reported evidence should enable direct comparison of lipidomics data, rather than their context-dependent interpretations and trends. Eventually, this may help to establish reference values of molar lipid abundances and use them for molecular diagnostics of a broad spectrum of liver pathologies. It will also complement ongoing efforts to create harmonized lipidomic resources for liquid and solid biopsies sharing similar principles of collecting and organizing the data (41, 44, 55, 60, 78).
Data availability
All data concerned with this study are presented within the manuscript, supplemental data, and supplemental datasets.
Supplemental data
This article contains supplemental data (13, 17, 39, 42, 49, 50, 51, 64, 66, 67, 79, 80, 81, 82, 83, 84, 85, 86).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
The data that support the findings of this study are available from the corresponding author upon reasonable request. We thank Prof Juergen Graessler for his advice on the statistical analysis of the data. We are grateful to Prof Kai Simons, Dr Christian Klose, Dr Sider Penkov, and members of Shevchenko lab for fruitful discussions and experimental support.
Author contributions
O. V., T. D. R., J. Hampe, J. K. P., and A. S. conceptualization; O. V., O. K., A. P., J. A. H. W., J. M. A., C. H., M. B., V. R. T., S. B., and J. K. P. data curation; T. D. R., A. P., J. A. H. W., J. M. A., and C. H. formal analysis; H. C. K., Ü. C., E. K., J. Hampe, J. K. P., and A. S. funding acquisition; O. V., T, D. R., O. K., Y. W., T. Z., and J. Hartler investigation; T. D. R., O. K., J. A. H. W., K. S., Y. W., J. Hartler, H. C. K., J. K. P., and A. S. methodology; O. V., M. B., J. Hampe, and A. S. project administration; M. B., S. B., C. R., W. v. S., J. G., and C. S. resources; T. D. R., A. P., J. M. A., C. H., and J. K. P. software; H. C. K., C. R., Ü. C., E. K., W. v. S., J. G., C. S., J. Hampe, J. K. P., and A. S. supervision; O. V., O. K., K. S., and Y. W. validation; O. V., T. D. R., and A. P. visualization; O. V., T. D. R., J. K. P., and A. S. writing–original draft; M. B., V. R. T., S. B., J. Hartler, H. C. K., Ü. C., E. K., and C. S. writing–review and editing.
Funding and additional information
This study was supported by the German Ministry of Research and Education (BMBF) through the Liver Systems Medicine (LiSyM) network grant (to J. H., E. K., and A. S.); Lipidomics and Informatics for Life Sciences (LIFS) unit of de.nBi consortium (to A. S.); TRR83 grant from Deutsche Forschungsgemeinschaft (DFG) to A. S. and U. C.; German Federal Ministry of Education and Research (BMBF) grant to the German Center for Diabetes Research (DZD e.V.) for U. C. T. D. R. and J. K. P. were supported by the Bavarian State Ministry of Science and the Arts coordinated by the Bavarian Research Institute for Digital Transformation (bidt). H. C. K. is supported by the Austrian Federal Ministry of Education, Science and Research (grant BMWFW-10.420/0005-WF/V/3c/2017) and by HRSM project “Explorative Lipidomics seltener und chronischer Krankheiten.” J. H. gratefully acknowledges financial support by the University of Graz.
Contributor Information
Josch Konstantin Pauling, Email: josch.pauling@wzw.tum.de.
Andrej Shevchenko, Email: shevchenko@mpi-cbg.de.
Supplemental data
References
- 1.Marjot T., Moolla A., Cobbold J.F., Hodson L., Tomlinson J.W. Nonalcoholic fatty liver disease in adults: current concepts in etiology, outcomes, and management. Endocr. Rev. 2020;41:66–117. doi: 10.1210/endrev/bnz009. [DOI] [PubMed] [Google Scholar]
- 2.Musso G., Gambino R., De Michieli F., Cassader M., Rizzetto M., Durazzo M., Fagà E., Silli B., Pagano G. Dietary habits and their relations to insulin resistance and postprandial lipemia in nonalcoholic steatohepatitis. Hepatology. 2003;37:909–916. doi: 10.1053/jhep.2003.50132. [DOI] [PubMed] [Google Scholar]
- 3.Lonardo A., Nascimbeni F., Ballestri S., Fairweather D., Win S., Than T.A., Abdelmalek M.F., Suzuki A. Sex differences in nonalcoholic fatty liver disease: state of the art and identification of research gaps. Hepatology. 2019;70:1457–1469. doi: 10.1002/hep.30626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Friedman S.L., Neuschwander-Tetri B.A., Rinella M., Sanyal A.J. Mechanisms of NAFLD development and therapeutic strategies. Nat. Med. 2018;24:908–922. doi: 10.1038/s41591-018-0104-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hardy T., Oakley F., Anstee Q.M., Day C.P. Nonalcoholic fatty liver disease: pathogenesis and disease spectrum. Annu. Rev. Pathol. Mech. Dis. 2016;11:451–496. doi: 10.1146/annurev-pathol-012615-044224. [DOI] [PubMed] [Google Scholar]
- 6.Paschos P., Paletas K. Non alcoholic fatty liver disease two-hit process: multifactorial character of the second hit. Hippokratia. 2009;13:128. [PMC free article] [PubMed] [Google Scholar]
- 7.Kleiner D.E., Brunt E.M., Wilson L.A., Behling C., Guy C., Contos M., Cummings O., Yeh M., Gill R., Chalasani N., Neuschwander-Tetri B.A., Diehl A.M., Dasarathy S., Terrault N., Kowdley K. Association of histologic disease activity with progression of nonalcoholic fatty liver disease. JAMA Netw. Open. 2019;2:e1912565. doi: 10.1001/jamanetworkopen.2019.12565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lamotte A., Leclercq I., Lanthier N. The mechanisms of steatosis pathogenesis during NASH development. Acta Gastroenterol. Belg. 2020;83 [Google Scholar]
- 9.Simon J., Ouro A., Ala-Ibanibo L., Presa N., Delgado T.C., Martínez-Chantar M.L. Sphingolipids in non-alcoholic fatty liver disease and hepatocellular carcinoma: ceramide turnover. Int. J. Mol. Sci. 2020;21:40. doi: 10.3390/ijms21010040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tamura S., Shimomura I. Contribution of adipose tissue and de novo lipogenesis to nonalcoholic fatty liver disease. J. Clin. Invest. 2005;115:1139–1142. doi: 10.1172/JCI24930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Listenberger L.L., Han X., Lewis S.E., Cases S., Farese R.V., Ory D.S., Schaffer J.E. Triglyceride accumulation protects against fatty acid-induced lipotoxicity. Proc. Natl. Acad. Sci. U. S. A. 2003;100:3077–3082. doi: 10.1073/pnas.0630588100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alkhouri N., Dixon L.J., Feldstein A.E. Lipotoxicity in nonalcoholic fatty liver disease: not all lipids are created equal. Expert Rev. Gastroenterol. Hepatol. 2009;3:445–451. doi: 10.1586/egh.09.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorden D.L., Myers D.S., Ivanova P.T., Fahy E., Maurya M.R., Gupta S., Min J., Spann N.J., McDonald J.G., Kelly S.L., Duan J., Sullards M.C., Leiker T.J., Barkley R.M., Quehenberger O. Biomarkers of NAFLD progression: a lipidomics approach to an epidemic. J. Lipid Res. 2015;56:722–736. doi: 10.1194/jlr.P056002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanders F.W.B., Griffin J.L. De novo lipogenesis in the liver in health and disease: more than just a shunting yard for glucose. Biol. Rev. 2016;91:452–468. doi: 10.1111/brv.12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knebel B., Fahlbusch P., Dille M., Wahlers N., Hartwig S., Jacob S., Kettel U., Schiller M., Herebian D., Koellmer C., Lehr S., Müller-Wieland D., Kotzka J. Fatty liver due to increased de novo lipogenesis: alterations in the hepatic peroxisomal proteome. Front. Cell Dev. Biol. 2019;7:248. doi: 10.3389/fcell.2019.00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cohen J.C., Horton J.D., Hobbs H.H. Human fatty liver disease: old questions and new insights. Science. 2011;332:1519–1523. doi: 10.1126/science.1204265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thangapandi V.R., Knittelfelder O., Brosch M., Patsenker E., Vvedenskaya O., Buch S., Hinz S., Hendricks A., Nati M., Herrmann A., Rekhade D.R., Berg T., Matz-Soja M., Huse K., Klipp E. Loss of hepatic Mboat7 leads to liver fibrosis. Gut. 2021;70:940–950. doi: 10.1136/gutjnl-2020-320853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krawczyk M., Rau M., Schattenberg J.M., Bantel H., Pathil A., Demir M., Kluwe J., Boettler T., Lammert F., Geier A. Combined effects of the PNPLA3 rs738409, TM6SF2 rs58542926, and MBOAT7 rs641738 variants on NAFLD severity: a multicenter biopsy-based study. J. Lipid Res. 2017;58:247–255. doi: 10.1194/jlr.P067454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stickel F., Hampe J. Genetic determinants of alcoholic liver disease. Gut. 2012;61:150–159. doi: 10.1136/gutjnl-2011-301239. [DOI] [PubMed] [Google Scholar]
- 20.Stickel F., Moreno C., Hampe J., Morgan M.Y. The genetics of alcohol dependence and alcohol-related liver disease. J. Hepatol. 2017;66:195–211. doi: 10.1016/j.jhep.2016.08.011. [DOI] [PubMed] [Google Scholar]
- 21.Jonas W., Schürmann A. Genetic and epigenetic factors determining NAFLD risk. Mol. Metab. 2020;50:101111. doi: 10.1016/j.molmet.2020.101111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trépo E., Valenti L. Update on NAFLD genetics: from new variants to the clinic. J. Hepatol. 2020;72:1196–1209. doi: 10.1016/j.jhep.2020.02.020. [DOI] [PubMed] [Google Scholar]
- 23.Strnad P., Buch S., Hamesch K., Fischer J., Rosendahl J., Schmelz R., Brueckner S., Brosch M., Heimes C.V., Woditsch V. Heterozygous carriage of the alpha1-antitrypsin Pi∗ Z variant increases the risk to develop liver cirrhosis. Gut. 2019;68:1099–1107. doi: 10.1136/gutjnl-2018-316228. [DOI] [PubMed] [Google Scholar]
- 24.Mancina R.M., Dongiovanni P., Petta S., Pingitore P., Meroni M., Rametta R., Borén J., Montalcini T., Pujia A., Wiklund O. The MBOAT7-TMC4 variant rs641738 increases risk of nonalcoholic fatty liver disease in individuals of European descent. Gastroenterology. 2016;150:1219–1230. doi: 10.1053/j.gastro.2016.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Di Sessa A., Umano G.R., Cirillo G., Del Prete A., Iacomino R., Marzuillo P., Del Giudice E.M. The membrane-bound O-Acyltransferase7 rs641738 variant in pediatric nonalcoholic fatty liver disease. J. Pediatr. Gastroenterol. Nutr. 2018;67:69–74. doi: 10.1097/MPG.0000000000001979. [DOI] [PubMed] [Google Scholar]
- 26.Thabet K., Asimakopoulos A., Shojaei M., Romero-Gomez M., Mangia A., Irving W.L., Berg T., Dore G.J., Grønbæk H., Sheridan D. MBOAT7 rs641738 increases risk of liver inflammation and transition to fibrosis in chronic hepatitis C. Nat. Commun. 2016;7:1–8. doi: 10.1038/ncomms12757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thabet K., Chan H.L.Y., Petta S., Mangia A., Berg T., Boonstra A., Brouwer W.P., Abate M.L., Wong V.W., Nazmy M. The membrane-bound O-acyltransferase domain-containing 7 variant rs641738 increases inflammation and fibrosis in chronic hepatitis B. Hepatology. 2017;65:1840–1850. doi: 10.1002/hep.29064. [DOI] [PubMed] [Google Scholar]
- 28.Luukkonen P.K., Zhou Y., Hyötyläinen T., Leivonen M., Arola J., Orho-Melander M., Orešič M., Yki-Järvinen H. The MBOAT7 variant rs641738 alters hepatic phosphatidylinositols and increases severity of non-alcoholic fatty liver disease in humans. J. Hepatol. 2016;65:1263–1265. doi: 10.1016/j.jhep.2016.07.045. [DOI] [PubMed] [Google Scholar]
- 29.Kawano Y., Cohen D.E. Mechanisms of hepatic triglyceride accumulation in non-alcoholic fatty liver disease. J. Gastroenterol. 2013;48:434–441. doi: 10.1007/s00535-013-0758-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buch S., Stickel F., Trépo E., Way M., Herrmann A., Nischalke H.D., Brosch M., Rosendahl J., Berg T., Ridinger M., Rietschel M., McQuillin A., Frank J., Kiefer F., Schreiber S. A genome-wide association study confirms PNPLA3 and identifies TM6SF2 and MBOAT7 as risk loci for alcohol-related cirrhosis. Nat. Genet. 2015;47:1443–1448. doi: 10.1038/ng.3417. [DOI] [PubMed] [Google Scholar]
- 31.Sliz E., Sebert S., Würtz P., Kangas A.J., Soininen P., Lehtimäki T., Kähönen M., Viikari J., Männikkö M., Ala-Korpela M., Raitakari O.T., Kettunen J. NAFLD risk alleles in PNPLA3, TM6SF2, GCKR and LYPLAL1 show divergent metabolic effects. Hum. Mol. Genet. 2018;27:2214–2223. doi: 10.1093/hmg/ddy124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Montefusco D.J., Allegood J.C., Spiegel S., Cowart L.A. Non-alcoholic fatty liver disease: insights from sphingolipidomics. Biochem. Biophys. Res. Commun. 2018;504:608–616. doi: 10.1016/j.bbrc.2018.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang H., Quiroga A.D., Lehner R. Analysis of lipid droplets in hepatocytes. Methods Cell Biol. 2013;116:107–127. doi: 10.1016/B978-0-12-408051-5.00007-3. [DOI] [PubMed] [Google Scholar]
- 34.Lovric A., Granér M., Bjornson E., Arif M., Benfeitas R., Nyman K., Ståhlman M., Pentikäinen M.O., Lundbom J., Hakkarainen A., Sirén R., Nieminen M.S., Lundbom N., Lauerma K., Taskinen M.R. Characterization of different fat depots in NAFLD using inflammation-associated proteome, lipidome and metabolome. Sci. Rep. 2018;8:1–14. doi: 10.1038/s41598-018-31865-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alonso C., Noureddin M., Lu S.C., Mato J.M. Biomarkers and subtypes of deranged lipid metabolism in nonalcoholic fatty liver disease. World J. Gastroenterol. 2019;25:3009–3020. doi: 10.3748/wjg.v25.i24.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chiappini F., Coilly A., Kadar H., Gual P., Tran A., Desterke C., Samuel D., Duclos-Vallée J.C., Touboul D., Bertrand-Michel J., Brunelle A., Guettier C., Le Naour F. Metabolism dysregulation induces a specific lipid signature of nonalcoholic steatohepatitis in patients. Sci. Rep. 2017;7:1–17. doi: 10.1038/srep46658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ryan E., Reid G.E. Chemical derivatization and ultrahigh resolution and accurate mass spectrometry strategies for “shotgun” lipidome analysis. Acc. Chem. Res. 2016;49:1596–1604. doi: 10.1021/acs.accounts.6b00030. [DOI] [PubMed] [Google Scholar]
- 38.Schwudke D., Schuhmann K., Herzog R., Bornstein S.R., Shevchenko A. Shotgun lipidomics on high resolution mass spectrometers. Cold Spring Harb. Perspect. Biol. 2011;3:a004614. doi: 10.1101/cshperspect.a004614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kleiner D.E., Brunt E.M., Van Natta M., Behling C., Contos M.J., Cummings O.W., Ferrell L.D., Liu Y., Torbenson M.S., Unalp-Arida A., Yeh M., McCullough A.J., Sanyal A.J., Nonalcoholic Steatohepatitis Clinical Research Network Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41:1313–1321. doi: 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
- 40.Danielsson A.K., Lundin A., Andréasson S. Using mobile phone technology to treat alcohol use disorder: study protocol for a randomized controlled trial. Trials. 2018;19:1–8. doi: 10.1186/s13063-018-3137-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sales S., Knittelfelder O., Shevchenko A. Lipidomics of human blood plasma by high-resolution shotgun mass spectrometry. Methods Mol. Biol. 2017;1619:203–212. doi: 10.1007/978-1-4939-7057-5_16. [DOI] [PubMed] [Google Scholar]
- 42.Sales S., Graessler J., Ciucci S., Al-Atrib R., Vihervaara T., Schuhmann K., Kauhanen D., Sysi-Aho M., Bornstein S.R., Bickle M., Cannistraci C.V., Ekroos K., Shevchenko A. Gender, contraceptives and individual metabolic predisposition shape a healthy plasma lipidome. Sci. Rep. 2016;6:1–14. doi: 10.1038/srep27710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schuhmann K., Almeida R., Baumert M., Herzog R., Bornstein S.R., Shevchenko A. Shotgun lipidomics on a LTQ Orbitrap mass spectrometer by successive switching between acquisition polarity modes. J. Mass Spectrom. 2012;47:96–104. doi: 10.1002/jms.2031. [DOI] [PubMed] [Google Scholar]
- 44.Vvedenskaya O., Wang Y., Ackerman J.M., Knittelfelder O., Shevchenko A. Analytical challenges in human plasma lipidomics: a winding path towards the truth. Trends Anal. Chem. 2019;120:115277. [Google Scholar]
- 45.Knittelfelder O., Prince E., Sales S., Fritzsche E., Wöhner T., Brankatschk M., Shevchenko A. Sterols as dietary markers for Drosophila melanogaster. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2020;1865:158683. doi: 10.1016/j.bbalip.2020.158683. [DOI] [PubMed] [Google Scholar]
- 46.Schuhmann K., Thomas H., Ackerman J.M., Nagornov K.O., Tsybin Y.O., Shevchenko A. Intensity-independent noise filtering in FT MS and FT MS/MS spectra for shotgun lipidomics. Anal. Chem. 2017;89:7046–7052. doi: 10.1021/acs.analchem.7b00794. [DOI] [PubMed] [Google Scholar]
- 47.Schuhmann K., Srzentić K., Nagornov K.O., Thomas H., Gutmann T., Coskun Ü., Tsybin Y.O., Shevchenko A. Monitoring membrane lipidome turnover by metabolic 15N labeling and shotgun ultra-high-resolution orbitrap Fourier transform mass spectrometry. Anal. Chem. 2017;89:12857–12865. doi: 10.1021/acs.analchem.7b03437. [DOI] [PubMed] [Google Scholar]
- 48.Herzog R., Schwudke D., Schuhmann K., Sampaio J.L., Bornstein S.R., Schroeder M., Shevchenko A. A novel informatics concept for high-throughput shotgun lipidomics based on the molecular fragmentation query language. Genome Biol. 2011;12:1–25. doi: 10.1186/gb-2011-12-1-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson W.E., Li C., Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 50.Bergmann S., Ihmels J., Barkai N. Iterative signature algorithm for the analysis of large-scale gene expression data. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2003;67:031902. doi: 10.1103/PhysRevE.67.031902. [DOI] [PubMed] [Google Scholar]
- 51.Li G., Ma Q., Tang H., Paterson A.H., Xu Y. QUBIC: a qualitative biclustering algorithm for analyses of gene expression data. Nucleic Acids Res. 2009;37:e101. doi: 10.1093/nar/gkp491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brosch M., Kattler K., Herrmann A., von Schönfels W., Nordström K., Seehofer D., Damm G., Becker T., Zeissig S., Nehring S., Reichel F., Moser V., Thangapandi R.V., Stickel F., Baretton G. Epigenomic map of human liver reveals principles of zonated morphogenic and metabolic control. Nat. Commun. 2018;9:1–11. doi: 10.1038/s41467-018-06611-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sarwar R., Pierce N., Koppe S. Obesity and nonalcoholic fatty liver disease: current perspectives. Diabetes Metab. Syndr. Obes. Targets Ther. 2018;11:533–542. doi: 10.2147/DMSO.S146339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Surma M.A., Herzog R., Vasilj A., Klose C., Christinat N., Morin-Rivron D., Simons K., Masoodi M., Sampaio J.L. An automated shotgun lipidomics platform for high throughput, comprehensive, and quantitative analysis of blood plasma intact lipids. Eur. J. Lipid Sci. Technol. 2015;117:1540–1549. doi: 10.1002/ejlt.201500145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang Y., Hinz S., Uckermann O., Hönscheid P., von Schönfels W., Burmeister G., Hendricks A., Ackerman J.M., Baretton G.B., Hampe J., Brosch M., Schafmayer C., Shevchenko A., Zeissig S. Shotgun lipidomics-based characterization of the landscape of lipid metabolism in colorectal cancer. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2020;1865:158579. doi: 10.1016/j.bbalip.2019.158579. [DOI] [PubMed] [Google Scholar]
- 56.Bowden J.A., Heckert A., Ulmer C.Z., Jones C.M., Koelmel J.P., Abdullah L., Ahonen L., Alnouti Y., Armando A.M., Asara J.M., Bamba T., Barr J.R., Bergquist J., Borchers C.H., Brandsma J. Harmonizing lipidomics: NIST interlaboratory comparison exercise for lipidomics using SRM 1950-Metabolites in frozen human plasma. J. Lipid Res. 2017;58:2275–2288. doi: 10.1194/jlr.M079012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Knittelfelder O., Traikov S., Vvedenskaya O., Schuhmann A., Segeletz S., Shevchenko A., Shevchenko A. Shotgun lipidomics combined with laser capture microdissection: a tool to analyze histological zones in cryosections of tissues. Anal. Chem. 2018;90:9868–9878. doi: 10.1021/acs.analchem.8b02004. [DOI] [PubMed] [Google Scholar]
- 58.Higashi T., Shimada K. Derivatization of neutral steroids to enhance their detection characteristics in liquid chromatography-mass spectrometry. Anal. Bioanal. Chem. 2004;378:875–882. doi: 10.1007/s00216-003-2252-z. [DOI] [PubMed] [Google Scholar]
- 59.Casanovas A., Hannibal-Bach H.K., Jensen O.N., Ejsing C.S. Shotgun lipidomic analysis of chemically sulfated sterols compromises analytical sensitivity: recommendation for large-scale global lipidome analysis. Eur. J. Lipid Sci. Technol. 2014;116:1618–1620. doi: 10.1002/ejlt.201400451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Quehenberger O., Dennis E.A. The human plasma lipidome. N. Engl. J. Med. 2011;365:1812–1823. doi: 10.1056/NEJMra1104901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bruschi F.V., Claudel T., Tardelli M., Caligiuri A., Stulnig T.M., Marra F., Trauner M. The PNPLA3 I148M variant modulates the fibrogenic phenotype of human hepatic stellate cells. Hepatology. 2017;65:1875–1890. doi: 10.1002/hep.29041. [DOI] [PubMed] [Google Scholar]
- 62.BasuRay S., Wang Y., Smagris E., Cohen J.C., Hobbs H.H. Accumulation of PNPLA3 on lipid droplets is the basis of associated hepatic steatosis. Proc. Natl. Acad. Sci. U. S. A. 2019;116:9521–9526. doi: 10.1073/pnas.1901974116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Luukkonen P.K., Nick A., Hölttä-Vuori M., Thiele C., Isokuortti E., Lallukka-Brück S., Zhou Y., Hakkarainen A., Lundbom N., Peltonen M., Orho-Melander M., Orešič M., Hyötyläinen T., Hodson L., Ikonen E. Human PNPLA3-I148M variant increases hepatic retention of polyunsaturated fatty acids. JCI Insight. 2019;4:e127902. doi: 10.1172/jci.insight.127902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Al-Sari N., Suvitaival T., Mattila I., Ali A., Ahonen L., Trost K., Henriksen T.F., Pociot F., Dragsted L.O., Legido-Quigley C. Lipidomics of human adipose tissue reveals diversity between body areas. PLoS One. 2020;15:e0228521. doi: 10.1371/journal.pone.0228521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Alves-Bezerra M., Cohen D.E. Triglyceride metabolism in the liver. Compr. Physiol. 2017;8:1–8. doi: 10.1002/cphy.c170012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Graessler J., Schwudke D., Schwarz P.E.H., Herzog R., Schevchenko A., Bornstein S.R. Top-down lipidomics reveals ether lipid deficiency in blood plasma of hypertensive patients. PLoS One. 2009;4:e6261. doi: 10.1371/journal.pone.0006261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hartler J., Armando A.M., Trötzmüller M., Dennis E.A., Köfeler H.C., Quehenberger O. Automated annotation of sphingolipids including accurate identification of hydroxylation sites using MSn data. Anal. Chem. 2020;92:14054–14062. doi: 10.1021/acs.analchem.0c03016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu Z.E., Fraser K., Kruger M.C., Sequeira I.R., Yip W., Lu L.W., Plank L.D., Murphy R., Cooper G.J.S., Martin J.-C., Hollingsworth K.G., Poppitt S.D. Untargeted metabolomics reveals plasma metabolites predictive of ectopic fat in pancreas and liver as assessed by magnetic resonance imaging: the TOFI_Asia study. Int. J. Obes. 2021;45:1844–1854. doi: 10.1038/s41366-021-00854-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sen P., Govaere O., Sinioja T., McGlinchey A., Geng D., Ratziu V., Bugianesi E., Schattenberg J.M., Vidal-Puig A., Allison M. Quantitative genome-scale analysis of human liver reveals dysregulation of glycosphingolipid pathways in progressive nonalcoholic fatty liver disease. medRxiv. 2021 doi: 10.1101/2021.02.09.21251354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Iqbal J., Walsh M.T., Hammad S.M., Hussain M.M. Sphingolipids and lipoproteins in health and metabolic disorders. Trends Endocrinol. Metab. 2017;28:506–518. doi: 10.1016/j.tem.2017.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Aguilera-Romero A., Gehin C., Riezman H. Sphingolipid homeostasis in the web of metabolic routes. Biochim. Biophys. Acta. 2014;1841:647–656. doi: 10.1016/j.bbalip.2013.10.014. [DOI] [PubMed] [Google Scholar]
- 72.Bikman B.T., Summers S.A. Sphingolipids and hepatic steatosis. Adv. Exp. Med. Biol. 2011;721:87–97. doi: 10.1007/978-1-4614-0650-1_6. [DOI] [PubMed] [Google Scholar]
- 73.Edagawa M., Sawai M., Ohno Y., Kihara A. Widespread tissue distribution and synthetic pathway of polyunsaturated C24:2 sphingolipids in mammals. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2018;1863:1441–1448. doi: 10.1016/j.bbalip.2018.09.002. [DOI] [PubMed] [Google Scholar]
- 74.Adolph T.E., Grander C., Moschen A.R., Tilg H. Liver–microbiome axis in health and disease. Trends Immunol. 2018;39:712–723. doi: 10.1016/j.it.2018.05.002. [DOI] [PubMed] [Google Scholar]
- 75.Wallace M., Green C.R., Roberts L.S., Lee Y.M., McCarville J.L., Sanchez-Gurmaches J., Meurs N., Gengatharan J.M., Hover J.D., Phillips S.A. Enzyme promiscuity drives branched-chain fatty acid synthesis in adipose tissues. Nat. Chem. Biol. 2018;14:1021–1031. doi: 10.1038/s41589-018-0132-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jump D.B. N-3 polyunsaturated fatty acid regulation of hepatic gene transcription. Curr. Opin. Lipidol. 2008;19:242–247. doi: 10.1097/MOL.0b013e3282ffaf6a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Botolin D., Wang Y., Christian B., Jump D.B. Docosahexaneoic acid (22:6,n-3) regulates rat hepatocyte SREBP-1 nuclear abundance by Erk- and 26S proteasome-dependent pathways. J. Lipid Res. 2006;47:181–192. doi: 10.1194/jlr.M500365-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Burla B., Arita M., Arita M., Bendt A.K., Cazenave-Gassiot A., Dennis E.A., Ekroos K., Han X., Ikeda K., Liebisch G. MS-based lipidomics of human blood plasma: a community-initiated position paper to develop accepted guidelines. J. Lipid Res. 2018;59:2001–2017. doi: 10.1194/jlr.S087163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li P., Wu Q., Burges C.J. McRank: learning to rank using multiple classification and gradient boosting. Adv. Neural Inf. Process. Syst. 2008:897–904. [Google Scholar]
- 80.Triebl A., Trötzmüller M., Hartler J., Stojakovic T., Köfeler H.C. Lipidomics by ultrahigh performance liquid chromatography - high resolution mass spectrometry and its application to complex biological samples. J. Chromatogr. B. 2017;1053:72–80. doi: 10.1016/j.jchromb.2017.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fauland A., Köfeler H.C., Trötzmüller M., Knopf A., Hartler J.J., Eberl A., Chitraju C., Lankmayr E., Spener F. A comprehensive method for lipid profiling by liquid chromatography-ion cyclotron resonance mass spectrometry. J. Lipid Res. 2011;52:2314–2322. doi: 10.1194/jlr.D016550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hartler J., Triebl A., Ziegl A., Trötzmüller M., Rechberger G.N., Zeleznik O.A., Zierler K.A., Torta F., Cazenave-Gassiot A., Wenk M.R. Deciphering lipid structures based on platform-independent decision rules. Nat. Methods. 2017;14:1171–1174. doi: 10.1038/nmeth.4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hartler J., Trötzmüller M., Chitraju C., Spener F., Köfeler H.C., Thallinger G.G. Lipid Data Analyzer: unattended identification and quantitation of lipids in LC-MS data. Bioinformatics. 2011;27:572–577. doi: 10.1093/bioinformatics/btq699. [DOI] [PubMed] [Google Scholar]
- 84.Hanczar B., Nadif M. Using the bagging approach for biclustering of gene expression data. Neurocomputing. 2011;74:1595–1605. [Google Scholar]
- 85.Strnad P., Buch S., Hamesch K., Fischer J., Rosendahl J., Schmelz R., Brueckner S., Brosch M., Heimes C.V., Woditsch V., Scholten D., Nischalke H.D., Janciauskiene S., Mandorfer M., Trauner M. Heterozygous carriage of the alpha1-antitrypsin Pi∗ Z variant increases the risk to develop liver cirrhosis. Gut. 2019;68:1099–1107. doi: 10.1136/gutjnl-2018-316228. [DOI] [PubMed] [Google Scholar]
- 86.Stickel F., Buch S., Nischalke H.D., Weiss K.H., Gotthardt D., Fischer J., Rosendahl J., Marot A., Elamly M., Casper M. Genetic variants in PNPLA3 and TM6SF2 predispose to the development of hepatocellular carcinoma in individuals with alcohol-related cirrhosis. Am. J. Gastroenterol. 2018;113:1475–1483. doi: 10.1038/s41395-018-0041-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data concerned with this study are presented within the manuscript, supplemental data, and supplemental datasets.