Abstract
Mcm proteins play an essential role in eukaryotic DNA replication, but their biochemical functions are poorly understood. Recently, we reported that a DNA helicase activity is associated with an Mcm4-Mcm6-Mcm7 (Mcm4,6,7) complex, suggesting that this complex is involved in the initiation of DNA replication as a DNA-unwinding enzyme. In this study, we have expressed and isolated the mouse Mcm2,4,6,7 proteins from insect cells and characterized various mutant Mcm4,6,7 complexes in which the conserved ATPase motifs of the Mcm4 and Mcm6 proteins were mutated. The activities associated with such preparations demonstrated that the DNA helicase activity is intrinsically associated with the Mcm4,6,7 complex. Biochemical analyses of these mutant Mcm4,6,7 complexes indicated that the ATP binding activity of the Mcm6 protein in the complex is critical for DNA helicase activity and that the Mcm4 protein may play a role in the single-stranded DNA binding activity of the complex. The results also indicated that the two activities of DNA helicase and single-stranded DNA binding can be separated.
The minichromosome maintenance (Mcm) protein family, consisting of Mcm2, Mcm3, Mcm4, Mcm5, Mcm6, and Mcm7 proteins, is required for the DNA replication in eukaryotes (7, 25–27, 51). All of the six Mcm proteins play an essential role in yeast cell growth (8, 9, 13, 15, 18, 35, 38, 40, 47, 48, 56). They are also one of the components of the replication licensing system in Xenopus that limits the occurrence of DNA replication to only once in a single cell cycle (6, 33, 37). The assembly of Mcm proteins onto the replication origin in Saccharomyces cerevisiae (2, 49) and their genetic interactions with ORC, Cdc6p, Cdc45p, and Cdc7p/Dbf4p kinase have been reported (10, 11, 16, 17, 34, 57). These findings suggest that the Mcm proteins play a role in the initiation of DNA replication.
Several lines of evidence indicate that these six Mcm proteins interact with one another to form a heterohexameric complex (1, 28, 43). In addition, the subcomplexes, including Mcm2-Mcm4-Mcm6-Mcm7 (Mcm2,4,6,7), Mcm3,5, and Mcm4,6,7, have been detected in extracts from various organisms (5, 21, 29, 41, 46, 47, 50). These findings suggest that Mcm4,6,7 proteins form a relatively stable core complex and that the other Mcm proteins are loosely associated with this complex. All six Mcm proteins contain DNA-dependent ATPase motifs in their central domain, including motifs A and B which are probably involved in the nucleotide binding (30). These motifs in Mcm proteins are well conserved from yeast to mammals, suggesting that they play an indispensable role in DNA replication. Recently, we detected DNA helicase activity associated with a 600-kDa human Mcm4,6,7 complex purified from HeLa cells (20). The addition of Mcm2 protein to this complex inhibited the helicase activity by blocking the formation of the complex (22). These findings suggest that the Mcm4,6,7 complex is involved in the initiation of DNA replication as a DNA-unwinding enzyme. However, the issue of whether the DNA helicase activity is intrinsically associated with the Mcm4,6,7 complex remained to be unequivocally established.
In this study, biochemical analyses with the mouse Mcm4, Mcm6, and Mcm7 protein complexes prepared from baculovirus-infected cells were undertaken to address whether the DNA helicase activity was an inherent activity of this complex and to determine the physiological significance of this activity. An Mcm4,6,7 complex was purified from extracts of insect cells coinfected with recombinant baculoviruses containing the wild-type mouse Mcm2, Mcm4, Mcm6, and Mcm7 genes. The purified Mcm4,6,7 complex contained both single-stranded DNA-dependent ATPase and the DNA helicase activities. Studies with the mutant Mcm complexes showed that mutations in the ATP binding motifs of Mcm6 protein preferentially affected the ATP binding of the Mcm4,6,7 complex and resulted in loss of the DNA helicase activity. A mutation in the ATP binding motifs of the Mcm4 protein affected the single-stranded DNA-binding activity of the complex, which moderately inhibited the DNA helicase activity. These results indicate that the Mcm4,6,7 complex contains an intrinsic DNA helicase activity and that the Mcm4 and Mcm6 proteins play distinct roles in the function of the helicase activity.
MATERIALS AND METHODS
Cloning of wild-type and mutant forms of Mcm genes into baculovirus transfer vectors.
DNAs containing full-length mouse Mcm2, Mcm4, Mcm6, and Mcm7 genes cloned into the plasmid pBluescript II SK(−) (Stratagene) (kindly provided by H. Kimura, University of Oxford, Oxford, United Kingdom) were used in this study, and their GenBank/EMBL accession numbers are D86725, D26089, D86726, and D26091, respectively. To facilitate the purification of Mcm2, Mcm4, Mcm6, and Mcm7 proteins in High 5 insect cells, sequences encoding a six-histidine tag were added to the DNAs containing the Mcm4 and Mcm7 genes by PCR. First, to amplify the N-terminal fragment of the Mcm4 gene product (amino acids 1 to 148 as an EcoRI-HindIII fragment) and the N-terminal fragment of the Mcm7 gene product (amino acids 1 to 63 as an EcoRI-SalI fragment), oligonucleotides 5′-GAGAGAGAATTCATGGGACATCATCATCATCATCACGGATCGTCCCCGGCATCCACCCCG-3′ (the engineered EcoRI restriction site is underlined) and 5′-TTACATGTTGCCACATTCAC-3′ were used as primers for the amplification of the Mcm4 gene and oligonucleotides 5′ - GAGAGAGAAT TC ATGGGACATCATCATCATCATCACGGAGCGC TTAAGGACTACGCGATC-3′ and 5′-TGAGTAGCGCTTGGCATTCTCGC-3′ were used as primers for the amplification of the Mcm7 gene; the primers were designed with DNASISmac software (Hitachi). The PCR products comprising the Mcm4 gene product N-terminal fragment were ligated to the Mcm4 C-terminal fragment (amino acids 149 to 862 as a HindIII-EcoRI fragment). The resultant full-size Mcm4 gene containing the His6 tag and full-size Mcm6 gene were subcloned into the EcoRI and BamHI sites, respectively, of the pAcUW31 vector (Pharmingen), which contains both the baculovirus p10 and polyhedrin promoters. The PCR products comprising the Mcm7 gene product N-terminal fragment were ligated to the C-terminal fragment (amino acids 64 to 821 as a SalI-EcoRI fragment), and the resultant full-size Mcm7 gene containing the His6 tag and full-size Mcm2 gene were sequentially subcloned into the EcoRI and BamHI sites, respectively, of pAcUW31.
Site-directed mutagenesis of the Mcm4 and Mcm6 genes was conducted with the QuikChange site-directed mutagenesis kit (Stratagene). The oligonucleotide 5′ - CAATGGGATATGC TGCATCGC TGCG T T TGACAAAATGAATGAAAG-3′ was used as the primer to prepare the DE572AA Mcm4 mutant in plasmid pBluescript II SK(−) where the C-terminal fragment of the Mcm4 gene had been cloned; the oligonucleotides 5′-GGTGTCTGTTGTATTGCTGCATTTGATAAGATGGAC-3′ and 5′-GGTGATCCAAGTACAGCTGCGGCCCAATTTCTCAAGCACGTGG-3′ were used as the primers to prepare the DE459AA and KS401AA Mcm6 mutants, respectively, in plasmid pBluescript II SK(−) where the full-size Mcm6 gene had been cloned.
The mutagenized Mcm4 C-terminal fragment (HindIII-EcoRI fragment) and the His-tagged N-terminal fragment of the Mcm4 gene were ligated into the baculovirus vector pAcUW31 where the full-size Mcm6 gene had been cloned. Similarly, the mutagenized Mcm6 gene and the full-size Mcm4 gene containing the histidine tag were subcloned into the baculovirus vector pAcUW31. The Mcm4DE6DE double mutant was prepared by ligating both the His-tagged N-terminal fragment and the mutagenized C-terminal fragment of the Mcm4 gene into pAcUW31 where the mutagenized Mcm6 gene had been cloned. The nucleotide sequences of wild-type and all mutated DNAs were confirmed by DNA sequencing in an Applied Biosystems automated sequencer (PRISM 377; Perkin-Elmer).
Expression of wild-type and mutant forms of Mcm2,4,6, and Mcm7 proteins in insect cells.
Sf9 insect cells were cultured in Grace’s insect medium (Gibco Life Technologies) supplemented with 10% fetal calf serum (Gibco), 20 μg of gentamicin (Sigma) per ml, and Fungizone (amphotericin B) (Gibco). To generate recombinant baculovirus for expressing the Mcm2,7 and Mcm4,6 proteins, BaculoGold Autographa californica nuclear polyhedrosis virus DNA (BaculoGold AcNPV; Pharmingen) and each of the cloned Mcm2,7 or Mcm4,6 genes were cotransfected into Sf9 cells and recombinant baculoviruses were isolated by plaque purification as recommended by the manufacturer. Recombinant viruses expressing the Mcm proteins were identified by infecting 2 × 106 cells with the plaque supernatant and then performing an immunoblot analysis of the cell lysate. For the expression of Mcm2, Mcm4, Mcm6, and Mcm7 proteins, 1.2 × 108 High 5 insect cells (Invitrogen), which were plated in eight dishes (diameter, 150 mm), were coinfected with the recombinant baculoviruses carrying the Mcm2,7 and Mcm4,6 genes at a multiplicity of infection of approximately 10 (1.0 ml of each virus stock per dish) and then collected at 42 to 46 h postinfection.
Purification of wild-type and mutant Mcm proteins.
The recombinant Mcm proteins in infected cell lysate were purified by Ni-nitrilotriacetic acid (NTA) affinity column chromatography as follows. The infected cells were washed once in ice-cold phosphate-buffered saline and then suspended in 8 ml of lysis buffer consisting of 10 mM Tris-HCl (pH 7.5), 130 mM NaCl, 1% Triton X-100, 10 mM NaF, 10 mM Na phosphate buffer, 10 mM Na4P2O7, 16 μg of benzamidine HCl per ml, 10 μg of phenanthroline per ml, 10 μg of aprotinin per ml, 10 μg of leupeptin per ml, 10 μg of pepstatin A per ml, and 1 mM phenylmethylsulfonyl fluoride (PMSF). After incubation for 40 min on ice, insoluble material was removed by centrifugation at 40,000 rpm (50.2Ti rotor; Beckman) for 40 min at 4°C. To 1 volume of the clarified lysate, 1/10 volume of Ni-NTA-agarose that had been washed twice with 10 bead volumes of buffer A (50 mM Na phosphate buffer [pH 6.0], 300 mM NaCl, 10% glycerol) was added, and the mixture was incubated for 1 h at 4°C on a rocking platform. The beads were then collected by centrifugation and stringently washed with buffer A containing 30 mM imidazole until the absorbance at 280 nm A280 of the supernatant was less than 0.01. Next the beads were washed once with buffer B (50 mM Na phosphate buffer [pH 8.0], 300 mM NaCl, 10% glycerol) containing 30 mM imidazole, and the proteins bound to the beads were eluted by adding 1 bead volume of buffer B containing 100 mM imidazole. This was followed by incubation for 2 min at room temperature on a rocking platform and removal of the beads by centrifugation. The proteins were subsequently eluted from the beads with 200 and 400 mM imidazole.
Further purification of Mcm proteins was carried out by histone-Sepharose column chromatography as described previously (20). The Mcm-containing fractions eluted from Ni-NTA-agarose were combined and then loaded onto a histone H3/H4-Sepharose column equilibrated with 0.3 M NaCl, and proteins bound to the column were eluted with a linear gradient from 0.3 to 2 M NaCl. The fractions mainly containing Mcm4,6,7 proteins were pooled and then concentrated about 10-fold with Centricon 30 (Amicon). The concentrated sample was diluted to 0.15 M NaCl with buffer containing 20 mM Tris-HCl (pH 7.5), 0.5 mM EDTA, 10% glycerol, 1 mM dithiothreitol (DTT), 0.1 mM PMSF, and 0.01% Triton X-100 and then concentrated to approximately 1 mg/ml with a Centricon 30 apparatus. The concentrated Mcm proteins were further fractionated by glycerol gradient centrifugation at 36,000 rpm for 14 h (TLS55 rotor; Beckman) in a 15 to 30% linear glycerol gradient containing 0.15 M NaCl, 20 mM Tris-HCl (pH 7.5), 0.5 mM EDTA, 1 mM DTT, 0.1 mM PMSF, and 0.01% Triton X-100. Five drop fractions were collected from the bottom of the centrifugation tube. Proteins in each fraction were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (10% polyacrylamide) as well as by polyacrylamide gel electrophoresis (5% polyacrylamide) under nondenaturing conditions, and they were stained with silver or Coomassie brilliant blue G-250 (GelCode blue-staining reagent; Pierce). Thyroglobulin (669 kDa) and ferritin (440 kDa) (Pharmacia) were used as protein molecular mass markers in the 5% native gel. The human Mcm4,6,7 complex of 600 kDa was purified as reported previously (20). The Mouse Mcm2 gene was cloned into the pAcHLT-A vector (Pharmingen), and the His-tagged Mcm2 protein, which was produced in High 5 cells, was purified as reported previously (22).
DNA helicase and ATPase activities.
A 17-mer oligonucleotide (5′-GTTTTCCCAGTCACGAC-3′; −40 primer for M13 dideoxynucleotide sequencing [U. S. Biochemical Corp.]) was labeled at the 5′ end with polynucleotide kinase in the presence of [γ-32P]ATP, and the resultant 17-mer oligonucleotide was annealed to single-stranded M13mp18 DNA as described previously (20). Aliquots of the purified DNA were used for the DNA helicase assay as described below. Approximately 5 fmol of the annealed oligomer was incubated at 37°C for 1 h with Mcm proteins in 50 mM Tris-HCl (pH 7.9), 20 mM β-mercaptoethanol, 10 mM ATP, 0.5 mg of bovine serum albumin per ml, and 10 mM Mg(CH3COO)2 instead of MgCl2. The reaction was terminated by adding SDS to a final concentration of 0.2%, and an aliquot was electrophoresed on a 12% acrylamide gel in Tris-borate-EDTA (TBE). The labeled oligomer in the gel was detected by autoradiography or by using a Bio-Image analyzer (BAS2000; Fuji).
To assay ATPase activity, Mcm proteins were incubated at 37°C for 1 h with 2 μCi of [γ-32P]ATP (3,000 Ci/mmol) in the same solution used for measuring the DNA helicase activity but containing 10 mM ATP in the presence of 5 μg of single-stranded DNA (heat denatured). Then, 0.5 μl of the reaction mixture was spotted on a polyethyleneimine-cellulose thin-layer chromatography plate (Cellulose F; Merck). Chromatography was carried out at 4°C in 0.8 M LiCl for 2 h. The radioactivity on the plate was detected by using a Bio-Image analyzer.
UV-mediated cross-linking of ATP.
The cross-linking mixture, in a final volume of 20 μl, contained 20 mM HEPES buffer (pH 7.5), 10% glycerol, 0.1 mM DTT, 10 μCi of [α-32P]ATP (3,000 μCi/mmol), and the given amounts of Mcm proteins purified by glycerol gradient centrifugation. After the mixture had been incubated for 10 min on ice, UV (254 nm) irradiation was carried out in a microcentrifugation tube at 4°C for 30 min at a distance of 5 cm (Mineralight; UVP) (20). Upon termination of UV exposure, 0.8 μl of 100 mM ATP and 20 μg of bovine serum albumin were added to the mixture. Trichloroacetic acid was added to a final concentration of 10%, and the mixture was incubated for 10 min on ice. The precipitate was recovered by centrifugation and was washed once with acetone containing 0.5% hydrochloric acid and then twice with acetone. Proteins were separated by SDS-PAGE (10% polyacrylamide), and labeled proteins were visualized by using a Bio-Image analyzer.
Gel shift analysis.
A 37-mer oligonucleotide (5′-AATTCGAGCTCGGTACCCGGGGATCCTCTAGAGTCGA-3′) was labeled at its 5′ end in the presence of [γ-32P]ATP with T4 polynucleotide kinase. Mcm proteins were incubated with the labeled 37-mer oligonucleotide (0.15 pmol) at 37°C in a buffer consisting of 10 mM creatine phosphate (sodium salt), 5 mM ATP, 5 mM MgCl2, 0.3 mM DTT, 0.01% Triton X-100, and 15 mM potassium phosphate (pH 7.7). After 30 min, 10% glutaraldehyde was added to the reaction mixture at a final concentration of 0.1% and the incubation was continued for 10 min. The reaction mixture was analyzed with a 5% polyacrylamide gel under nondenaturing conditions. The gel was dried on DE81 paper (Whatman), and the radioactivity was analyzed by using a Bio-Image analyzer.
Immunodetection of Mcm proteins.
After electrophoresis in an SDS-polyacrylamide gel, proteins were transferred to a nitrocellulose membrane by electrophoresis in 49 mM Tris–38 mM glycine–0.037% SDS–20% methanol at 15 V for 1 h. The membrane was immersed in 5% skim milk plus TBS (50 mM Tris-HCl [pH 7.5], 0.15 M NaCl) and then incubated with anti-Mcm4 rabbit antibodies (20) or anti-Mcm6 antibodies. The anti-Mcm6 antibodies were prepared by immunizing rabbits with full-size mouse Mcm6 protein that was produced by the baculovirus expression system. After incubation with anti-rabbit antibodies conjugated with horseradish peroxidase (Bio-Rad), the membrane was treated with chemiluminescence detection reagent (SuperSignal West PicoChemiluminescent Substrate; Pierce) and exposed to X-ray film. The proteins that had been electrophoresed under nondenaturing conditions were transferred to a membrane after incubation with 49 mM Tris–38 mM glycine–0.25% SDS at 80°C for 1 h. The membrane was then processed as described above.
RESULTS
Expression and purification of the Mcm2,4,6,7 complex.
During the course of overexpression of the Mcm proteins in Sf9 cells, the Mcm4 and Mcm7 proteins were produced as insoluble forms while Mcm2 and Mcm6 were recovered as soluble forms. To facilitate the isolation and purification of soluble Mcm protein complexes, the His6 tag was added to the N terminus of the Mcm4 and Mcm7 proteins and the four Mcm proteins (Mcm2, Mcm4, Mcm6, and Mcm7) were expressed simultaneously. Recombinant baculoviruses expressing the His6-Mcm4 and Mcm6 proteins, under the control of the p10 promoter and the polyhedrin promoter, respectively, were constructed. Viruses expressing the His6-Mcm7 and Mcm2 proteins were also constructed.
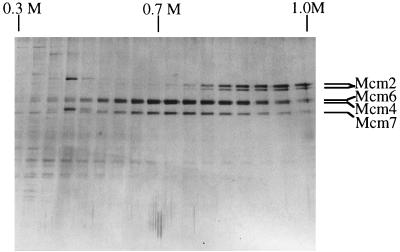
To express Mcm2,4,6,7 proteins, High 5 insect cells were coinfected with two recombinant baculoviruses, each carrying the Mcm4,6 and Mcm2,7 genes. The recombinant Mcm2,4,6,7 proteins were recovered from the lysed cells and purified by Ni-NTA affinity column chromatography. Nearly equal amounts of the four Mcm proteins were detected in the eluate of the Ni column, which was analyzed by SDS-PAGE, and the proteins were stained with Coomassie briliant blue (data not shown). The partially purified Mcm2,4,6,7 protein complex was further purified by a histone H3/H4-Sepharose column chromatography, as described previously (20). SDS-PAGE of proteins in the fractions eluted from the histone-Sepharose column is shown in Fig. 1. The peaks of the Mcm4, Mcm6, and Mcm7 proteins were detected in the histone column fractions eluted with 0.75 M NaCl, while the peak of Mcm2 protein was detected in fractions eluted with 1 M NaCl. We previously suggested that Mcm2 is the only Mcm protein that binds to histone (22). For this reason, the Mcm proteins probably bind to the histone column as an Mcm2,4,6,7 heterotetramer, particularly through the interaction between Mcm2 and histone H3, and the Mcm4,6,7 proteins may elute from the column earlier than the Mcm2 protein. However, the possibility that the Mcm4,6,7 complex itself has a weak affinity for histone H3/H4 remains to be tested.
FIG. 1.
Purification of the recombinant Mcm2, Mcm4, Mcm6, and Mcm7 proteins by histone-Sepharose column chromatography. The recombinant proteins were produced in High 5 insect cells coinfected with recombinant baculoviruses carrying the Mcm2-his-Mcm7 and his-Mcm4-Mcm6 genes. Mcm proteins in the lysed cell extracts were purified by Ni-NTA affinity column chromatography followed by histone-Sepharose column chromatography. Proteins eluted from the histone column were subjected to SDS-PAGE (10% polyacrylamide) and stained with silver. Bands of the Mcm2, Mcm4, Mcm6, and Mcm7 proteins are indicated.
Characterization of DNA helicase and ATPase activities of wild-type Mcm4,6,7 complex.
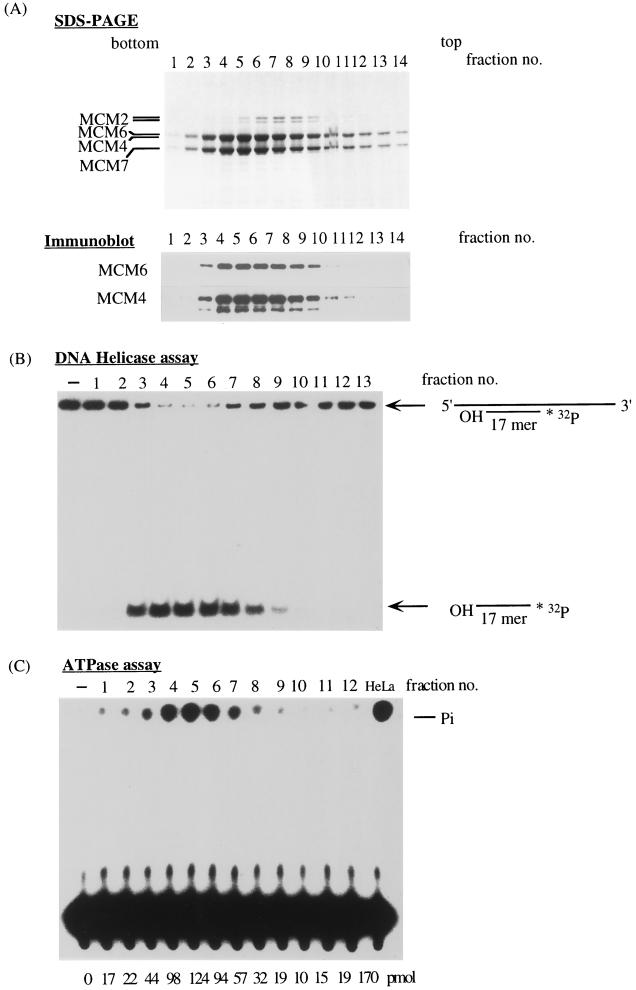
The histone-Sepharose fractions that contained predominantly the Mcm4,6,7 complex were pooled and further fractionated by glycerol gradient centrifugation. The proteins in these fractions were then analyzed by SDS-PAGE and detected by staining with silver (Fig. 2A, top). Proteins present in these fractions were also analyzed by immunoblotting with antibodies against the Mcm6 protein (Fig. 2A, bottom), and the membrane was then reprobed with the antibodies against the Mcm4 protein (Fig. 2A, bottom). The data obtained indicated that the Mcm4, Mcm6, and Mcm7 proteins were cosedimented and peaked at approximately 350 kDa.
FIG. 2.
Purified Mcm4,6,7 complex has both DNA helicase and ATPase activities. (A) The histone-Sepharose fractions that mainly contain Mcm4, Mcm6, and Mcm7 proteins were pooled and further fractionated by glycerol gradient centrifugation. Proteins in the fractions were analyzed by SDS-PAGE and stained with silver or immunoblotted with antibodies to Mcm6 or Mcm4, as indicated. (B) DNA helicase activity that displaces 32P-labeled 17-mer oligonucleotides annealed with M13 DNA was examined in the gradient fractions. The positions of the annealed oligomer and the released oligomer are indicated. (C) The ATPase activity of the Mcm4,6,7 protein complex was measured in the presence of single-stranded DNA, and the released 32P was detected by thin-layer chromatography. The ATPase activity of the Mcm4,6,7 protein complex purified from HeLa cells was measured (HeLa). The released phosphate (Pi) (picomoles) was calculated and is indicated at the bottom.
Although the amount of Mcm7 protein in the peak fractions appeared to exceed those of Mcm4 and Mcm6 protein in this experiment, staining of proteins in the peak fractions with Coomassie brilliant blue suggested that the amount of Mcm7 protein did not differ greatly from that of other Mcm proteins (see Fig. 4A). These results suggest that these proteins form a stoichiometric complex. The DNA helicase and ATPase activities present in the separated glycerol gradient fractions were examined (Fig. 2B and C). The DNA helicase activity was measured with a labeled 17-mer oligonucleotide annealed to a single-stranded circular M13 DNA. The results show that the DNA helicase activity cosedimented with Mcm4,6,7 (Fig. 2B). The peak of the helicase activity was detected at 350 kDa (fractions 4 to 6), and the activity in these fractions was almost proportional to the amount of the Mcm proteins present. The ATPase activity in the glycerol gradient fractions was measured in the presence of single-stranded DNA (Fig. 2C). Similar to the DNA helicase activity, the ATPase activity cosedimented with the Mcm proteins. These results suggest that the purified recombinant mouse Mcm4,6,7 protein complex possesses both DNA helicase activity and ATPase activity, which is consistent with the results obtained with the purified human Mcm4,6,7 complex (20).
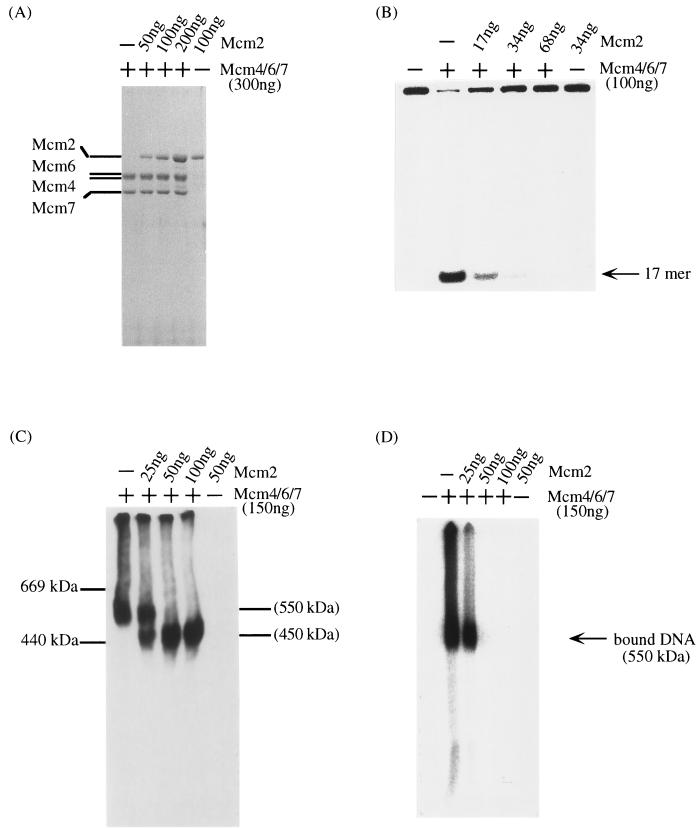
FIG. 4.
Inhibition of Mcm4,6,7 DNA helicase activity by Mcm2. Increasing amounts of His6-tagged mouse Mcm2 proteins purified from baculovirus-infected cells were incubated with the recombinant Mcm4,6,7 complex in 50 mM Tris-HCl (pH 7.9)–20 mM 2-mercaptoethanol–5 mM MgCl2–5 mM ATP–0.01% Triton X-100 for 30 min at 37°C. Aliquots of these reaction mixtures were analyzed by SDS-PAGE (A), for the activity of DNA helicase (B), by native gel electrophoresis (C), and for the activity of single-stranded DNA binding (D). The amount of Mcm2 and the presence of the Mcm4,6,7 complex are indicated at the top of each panel. In panel A, proteins were stained with Coomassie brilliant blue. In panel C, Mcm4 protein in Mcm complexes was detected with anti-Mcm4 antibodies as described in Materials and Methods.
Mcm4,6,7 complex binds single-stranded DNA.
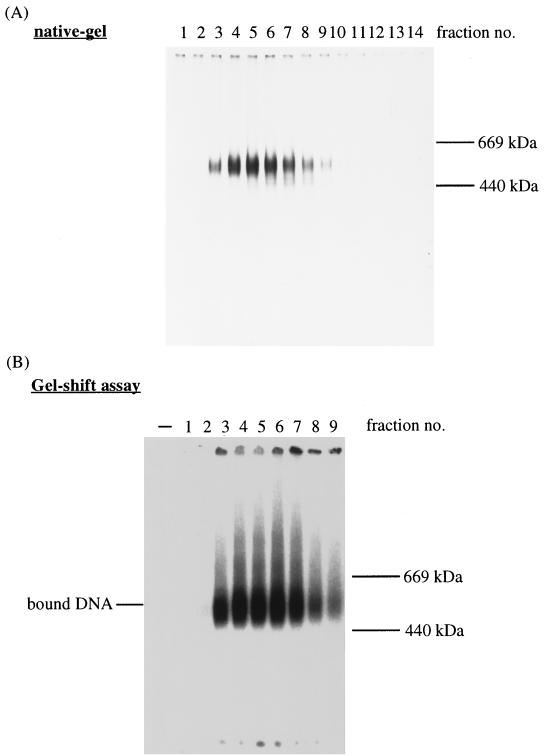
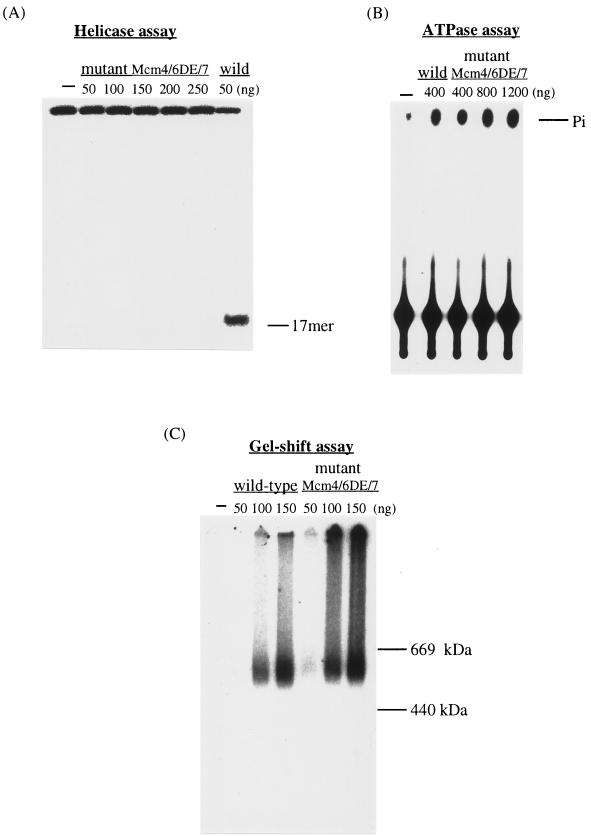
We next examined the formation of the purified Mcm4,6,7 protein complex and the single-stranded DNA binding activity of the complex (Fig. 3). The Mcm proteins present in glycerol gradient fractions were electrophoresed under nondenaturing conditions to analyze the complex (Fig. 3A). In this native gel, a major protein band of approximately 550 kDa was detected (fractions 4 to 6), which is slightly smaller than the 600-kDa human Mcm4,6,7 complex (20). The structure of the Mcm4,6,7 complex is estimated in Discussion.
FIG. 3.
The Mcm4,6,7 protein complex can bind single-stranded DNA. (A) Proteins in the gradient fractions were electrophoresed on a 5% native polyacrylamide gel and stained with silver. As markers, thyroglobulin (669 kDa) and ferritin (440 kDa) were electrophoresed. (B) A gel shift assay was carried out with the Mcm4,6,7 protein complexes present in the gradient fractions. 32P-labeled 37-mer oligonucleotides were incubated with the fractions. After cross-linking, DNA-protein complexes were separated by native PAGE (5% polyacrylamide). Autoradiography of the dried gel was performed.
The single-stranded DNA binding activity of the purified Mcm protein complex was investigated as described in Materials and Methods (Fig. 3B). When each glycerol gradient fraction was incubated with the labeled 37-mer single-stranded DNA, a band that migrated at the expected position of ∼550 kDa was detected. The binding of the Mcm complex with the 37-mer oligonucleotide does not require ATP (data not shown). The intensity of the band appeared to be proportional to the amount of the 550-kDa Mcm4,6,7 complex present in the glycerol gradient fraction added (Fig. 3A). These results indicate that the recombinant Mcm4,6,7 complex can bind the 37-mer single-stranded DNA, consistent with the finding that both DNA helicase and the single-stranded DNA-dependent ATPase activities are cofractionated with the 550-kDa Mcm4,6,7 complex.
Comparison of native and recombinant Mcm4,6,7 DNA helicases.
The results described above suggest that the recombinant Mcm4,6,7 complex has DNA helicase activity similar to that of the native human Mcm4,6,7 complex. To further address this point, the DNA helicase activity of these two complexes was examined in more detail. The specific activities of the DNA helicase of the recombinant mouse and the native human Mcm4,6,7 complex were comparable; approximately 100 ng of Mcm4,6,7 complex was required to displace 5 fmol of the annealed 17-mer oligonucleotides for 30 min under standard conditions (data not shown). This means that about a 40-fold molar excess of protein, if it forms a hexamer, compared to the 17-mer oligonucleotide, was necessary to displace the 17-mer. Thus, since we added substantial amounts of Mcm4,6,7 complex compared to the 17-mer, it is difficult to conclude that this reaction is catalytic. However, this activity appears to satisfy the criteria of a DNA helicase; the DNA helicase activity of Mcm4,6,7 complex is dependent on the presence of hydrolyzable ATP, and the data suggest that the complex migrates along single-stranded DNA in the 3′-to-5′ direction (20).
We reported that the incubation of Mcm4,6,7 complex with Mcm2 leads to the inhibition of DNA helicase activity, which is associated with the change from a 600-kDa Mcm4,6,7 complex to a 450-kDa Mcm2,4,6,7 complex (22). Similary, incubation of the recombinant mouse Mcm4,6,7 complex with the Mcm2 protein resulted in inhibition of the DNA helicase activity (Fig. 4A and B) and in the conversion of the 550-kDa Mcm4,6,7 complex to the 450-kDa complex, which was confirmed by using anti-Mcm4 antibodies (Fig. 4C). The 450-kDa complex did not possess the single-stranded DNA binding activity (Fig. 4D). An identical effect of the Mcm2 protein on the human Mcm4,6,7 complex was observed (22, 23). These results indicate that the recombinant mouse Mcm4,6,7 complex has DNA helicase activity which is essentially the same as that of the native human Mcm4,6,7 complex.
Biochemical characterization of mutant Mcm4,6,7 complexes.
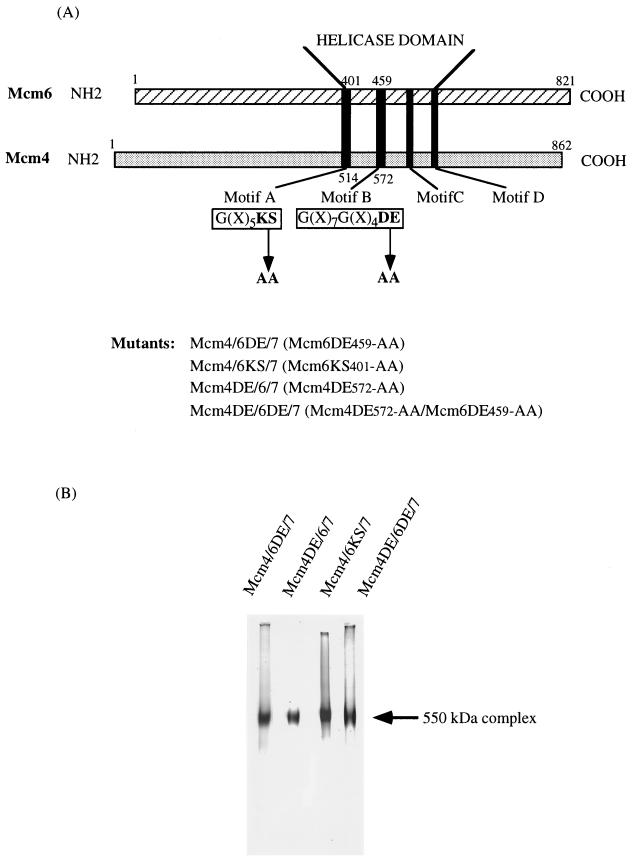
A series of mutations that changed specific amino acids located in the conserved DNA-dependent ATPase motifs of the Mcm proteins were carried out (Fig. 5A) (30). Aspartic and glutamic acid (DE) residues in motif B, which were highly conserved among various ATPases, were changed to alanine residues in the Mcm6 (DE459-AA) or Mcm4 (DE572-AA) protein. In addition, the highly conserved lysine and serine (KS) residues in motif A were converted to alanine residues in the Mcm6 protein (KS401-AA). These amino acids have been implicated in DNA binding, ATP binding, and ATP hydrolysis in other ATPases (31, 32, 52, 54).
FIG. 5.
Mutations introduced into Mcm4 and Mcm6 proteins for the formation of various mutant Mcm4,6,7 complexes. (A) A schematic presentation of the Mcm4 and Mcm6 proteins depicts the DNA-dependent ATPase motifs A, B, C, and D in the conserved regions, and the mutagenized amino acids in motifs A and B are indicated. A set of mutants of Mcm complexes constructed are shown. (B) Purified mutant Mcm4,6,7 complexes were electrophoresed on 5% native gels, and the proteins were stained with silver.
The Mcm4 and/or Mcm6 protein, mutagenized at these particular sites, were coexpressed in insect cells with other wild-type Mcm proteins. The mutant Mcm complexes of Mcm2,4,6DE-AA,7, Mcm2,4DE-AA,6,7, Mcm2,4DE-AA,6DE-AA,7, and Mcm2,4,6KS-AA,7 were purified by the same procedure used for the isolation of the wild-type complex. After glycerol gradient centrifugation, a 550-kDa Mcm4,6,7 complex containing the mutated Mcm4 and/or Mcm6 protein was isolated (Fig. 5B), although the recovery of the Mcm4,6,7 complex varied somewhat among the mutant Mcm complexes (data not shown). These results suggest that the mutations in these conserved amino acids in the ATPase motifs of the Mcm4 and Mcm6 proteins did not significantly affect the assembly of Mcm proteins into the 550-kDa Mcm4,6,7 complex.
The mutant Mcm4,6,7 complex (Mcm4,6DE-AA,7) in which DE were converted to AA in motif B of Mcm6 protein was characterized first. The DNA helicase and ATPase activities of the mutant Mcm4,6,7 complex were compared with those of the wild type. The mutant Mcm complex did not show any DNA helicase activity even when substantially higher levels of the complex compared to the wild-type complex were added to the reaction mixtures (Fig. 6A). On the other hand, the levels of ATPase and single-stranded DNA binding activities detected with the Mcm4,6DE-AA,7 mutant complex were nearly comparable to those detected with the wild-type complex (Fig. 6B and C). Thus, the DE in motif B of the Mcm6 protein is essential for the DNA helicase activity of the Mcm4,6,7 complex, but this mutation hardly affected the ATPase and the single-stranded DNA binding activities.
FIG. 6.
A defect in the DNA helicase activity of the Mcm4,6,7 complex containing mutated Mcm6. The DNA helicase (A), ATPase (B), and single-stranded DNA binding (C) activities of the mutant Mcm4,6,7 complex where DE in motif B of the Mcm6 protein was converted to AA (Mcm4,6DE-AA,7) were measured and compared with those of the wild-type Mcm4,6,7 complex (wild). Increasing amounts of the mutant complex were added to the reaction mixtures as indicated. Pi, phosphate.
ATP binding activity of the mutant Mcm complex.
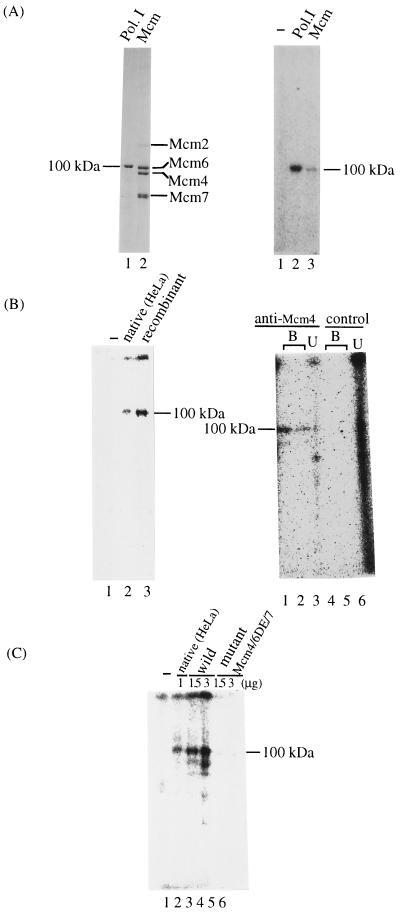
Binding and hydrolysis of nucleotide triphosphates are necessary for helicase activity. It has been shown that either the Mcm4 or the Mcm6 protein, in the native human Mcm4,6,7 complex, can be affinity labeled with ATP (20). To clarify which Mcm protein binds ATP with high affinity, the human Mcm4,6,7 complex was incubated with [α-32P]ATP, subjected to UV irradiation, and then separated by SDS-PAGE (Fig. 7A). As a marker, Escherichia coli DNA polymerase I (molecular mass, 103 kDa) was affinity labeled to provide a 100-kDa band marker on SDS-PAGE. A major band of 100 kDa was detected after incubating the human Mcm4,6,7 complex with [α-32P]ATP. Comparison of the electrophoretic mobility of Mcm proteins and DNA polymerase I suggested that the labeled 100-kDa protein is the Mcm6 protein. Furthermore, the 100-kDa band was also detected by affinity labeling the recombinant mouse Mcm4,6,7 complex of the wild type with [α-32P]ATP (Fig. 7B). The 100-kDa ATP-labeled protein band formed with the recombinant Mcm4,6,7 complex was immunodepleted with respect to the anti-Mcm4 antibodies but not to the control antibodies. This finding suggests that the Mcm6 protein in the Mcm4,6,7 complex has high affinity for ATP. However, this finding does not rule out the possibility that Mcm4 and Mcm7 proteins have a lower affinity for ATP. It is also possible that the lack of cross-linking of radiolabeled ATP to the Mcm4 and Mcm7 proteins is due to technical problems.
FIG. 7.
Mutation in motif B of the Mcm6 protein results in the reduction of ATP binding activity of the Mcm4,6,7 complex. (A) Native Mcm4,6,7 complex of HeLa cells (lane 2) and E. coli DNA polymerase I (Pol. I) (lane 1) were electrophoresed in an SDS–8% polyacrylamide gel and then stained with silver (left). [α-32P]ATP was incubated in the absence (lane 1) or the presence of Mcm proteins (1.5 μg, lane 3) or polymerase I (lane 2) under UV irradiation, and the proteins were electrophoresed through an SDS–8% polyacrylamide gel (right). The cross-linked proteins were detected by using a Bio-Image Analyzer. (B) [α-32P]ATP was incubated in the absence (lane 1) or the presence of native HeLa (lane 2) or wild-type recombinant Mcm4,6,7 complex (lane 3) as in panel A. Half of the reaction mixture was analyzed directly by SDS-PAGE (10% polyacrylamide) (left); the other half was immunodepleted with anti-Mcm4 antibody beads (lanes 1 to 3) or control beads (lanes 4 to 6). Proteins bound (lanes B) and unbound (lanes U) to the beads were electrophoresed (right). (C) Similar experiments were conducted on the mutant Mcm4,6DE-AA,7 complex. [α-32P]ATP was incubated in the absence (lane 1) or presence (lane 2) of native HeLa cells, increasing amounts of wild-type Mcm4,6,7 complex (lanes 3 and 4), or the mutant Mcm4,6DE-AA,7 complex (lanes 5 and 6) as indicated under UV irradiation. Proteins were analyzed by SDS-PAGE (10% polyacrylamide).
Since the mutant Mcm complex (Mcm6DE-AA) showed no DNA helicase activity, we investigated the ATP binding activity of this complex. The ATP binding activity of the mutant complex was markedly reduced compared to that of the wild-type complex (Fig. 7C). These results suggest that the DE mutation in motif B of the Mcm6 protein affects the DNA helicase activity of the Mcm4,6,7 complex by lowering the affinity of the complex for ATP. However, it is also possible that the Mcm6DE-AA mutation affects DNA-unwinding activity by altering the Mcm6 protein structure and hence the interaction of Mcm6 with Mcm4 and Mcm7.
Characteristics of the other mutants of the Mcm4,6,7 complex.
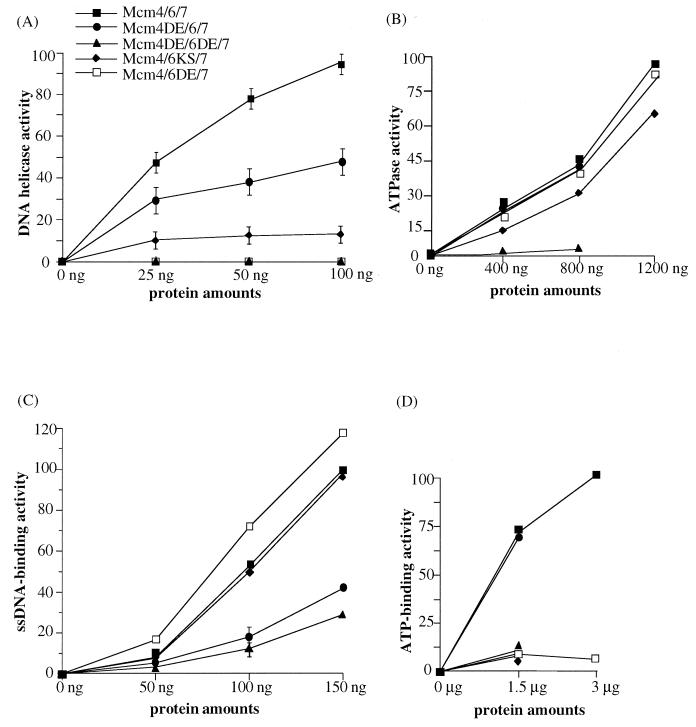
The results shown in Fig. 6 and 7 indicated that the DE-to-AA changes in motif B of the Mcm6 protein of the Mcm4,6,7 complex affected both the DNA helicase and ATP binding activities but did not affect the ATPase or the single-stranded DNA binding activities of the complex. We next asked whether other amino acid changes in the conserved ATPase domain of the other proteins would affect the activities of the Mcm4,6,7 complex. These include the changes DE to AA in motif B of the Mcm4 protein, KS to AA in motif A of the Mcm6 protein, and the double mutant in which the changes DE to AA were made in motif B of both the Mcm4 and Mcm6 proteins. These Mcm4,6,7 mutant complexes were purified, and the influence of these changes on the DNA helicase, DNA-dependent ATPase, single-stranded DNA binding, and ATP binding activities were examined. The DNA helicase activities of these mutants are shown in Fig. 8A and Table 1. The Mcm4,6,7 complexes containing the mutated Mcm4DE or Mcm6KS retained DNA helicase activity, but the specific activity was lower than that of the wild-type Mcm4,6,7 complex. No DNA helicase activity was detected in the complex containing mutations in both Mcm6DE and Mcm4DE.
FIG. 8.
Characterization of various mutant forms of Mcm4,6,7 complex. The activities of DNA helicase (A), ATPase (B), single-stranded DNA binding (C), and ATP binding (D) were investigated. The designations of the mutant Mcm4,6,7 complexes are described in the legend to Fig. 5 and in the text. The reactions were performed under standard conditions with various amounts of the wild-type and mutant Mcm4,6,7 complexes. Each activity was quantitated, and the activities of the wild type and mutant complexes are expressed in relation to the activity of the wild-type complex at the highest dose, where this activity was regarded as 100. In panel A, 3.7 fmol of 17-mer was displaced in the presence of the highest dose of the wild-type complex. In panel B, 184 pmol of phosphate was released in the presence of the highest dose of the wild-type complex. Values from two independent experiments are shown as vertical bars, and their average in several Mcm complexes is plotted in panels A and C.
TABLE 1.
Summary of the biochemical properties of mutants of MCM4,6,7 complexesa
| Mutant | DNA helicase activity | ATPase activity | Single-stranded DNA binding activity | ATP binding activity |
|---|---|---|---|---|
| MCM4,6,7 (wild type) | ++++ | ++++ | ++++ | ++++ |
| MCM4,6DE,7 | − | ++++ | ++++ | − |
| MCM4DE,6,7 | ++ | ++++ | ++ | ++++ |
| MCM4DE,6DE,7 | − | − | + | − |
| MCM4,6KS,7 | + | +++ | ++++ | − |
The assays were performed as described in Materials and Methods. The activity of mutant MCM complexes is shown by the number of plus signs; the activity of the wild type complex is indicated as ++++ in each assay. No activity is shown as a minus sign.
The ATPase activity of the three Mcm4,6,7 mutants was also examined (Fig. 8B and Table 1). The Mcm4,6,7 complexes containing the mutated Mcm4DE or the mutated Mcm6DE exhibited wild-type levels of ATPase activity. A slightly reduced ATPase activity was detected in the Mcm4,6,7 complex in which Mcm6KS was mutated. The ATPase activity of the double mutant, Mcm4DE and Mcm6DE, was significantly reduced.
We next compared the single-stranded DNA binding activities of the various mutants (Fig. 8C and Table 1). The two mutants which showed either a decrease in helicase activity (Mcm4DE) or no helicase activity (Mcm4DE6DE) also showed a reduced DNA binding activity compared to the wild-type Mcm4,6,7 complex. The mutant complex containing Mcm6KS possessed single-stranded DNA binding activity comparable to that of the wild-type complex, and the other mutant containing Mcm6DE showed a level of activity slightly higher than that of the wild-type complex. Thus, the DE residues of motif B of Mcm4 but not of Mcm6 may play a significant role in the single-stranded DNA binding activity of the complex. However, it is also possible that the DNA binding defect in Mcm4DE is due to a coincidental alteration of Mcm4 protein structure and/or interaction of Mcm4 with Mcm6 and 7 proteins.
The ATP binding activity of the mutated Mcm4,6,7 complexes was investigated (Fig. 8D and Table 1). The activity of the Mcm4DE mutant was similar to that of the wild-type Mcm4,6,7 complex, whereas the other Mcm6 mutants with mutation in either motif A or motif B exhibited almost no ATP binding activity. The Mcm4,6,7 complex mutated in both the DE of Mcm4 and the DE of Mcm6 also exhibited no ATP binding activity. These results suggest that Mcm6 plays an important role in ATP binding, consistent with the observation that the Mcm6 protein has a high affinity for ATP (Fig. 7).
Based on the biochemical activities observed with the mutated Mcm complexes, the following conclusion can be drawn. The loss of ATP binding activity observed with the Mcm6DE or KS mutant complex leads to the inactivation of the DNA helicase activity. In addition, the results with the two Mcm6 mutants indicated that the mutations can affect the ATP binding, ATPase, and single-stranded DNA binding activities differently. Similarly, the mutations in the Mcm4 protein uncoupled the single-stranded DNA binding activity from the ATPase and ATP binding activities. Finally, the results suggest that defects in the ATP binding or the single-stranded DNA binding activities lead to loss of the DNA helicase activity of the Mcm4,6,7 complex.
DISCUSSION
DNA helicase activity consists of a set of subactivities including nucleotide binding, DNA binding, and ATP hydrolysis; coordination of these activities is required to unwind duplex DNA (36). The present results indicate that the recombinant mouse Mcm4, Mcm6, and Mcm7 proteins, which form a complex, exhibit both DNA helicase and ATPase activities as well as the ability to bind ATP and single-stranded DNA. Moreover, analyses of the Mcm complexes mutated in conserved ATPase motifs demonstrated that the Mcm4,6,7 complex contains intrinsic DNA helicase activity. These studies also suggested that the Mcm4 and Mcm6 proteins play different roles in the functions of the Mcm4,6,7 helicase.
Structure and function of the Mcm4,6,7 helicase.
The human Mcm complex consisting of apparently equal amounts of the Mcm4, Mcm6, and Mcm7 proteins, which sediment at approximately 350 kDa by glycerol gradient centrifugation, form a 600-kDa complex after protein cross-linking in SDS-polyacrylamide gels (20). Based on the molecular mass of each Mcm protein, the 600-kDa complex is thought to consist of two molecules each of Mcm4, Mcm6, and Mcm7 proteins. The human Mcm4,6,7 complex was also detected at 600 kDa by gel electrophoresis under nondenaturing conditions (22), and the recombinant mouse Mcm4,6,7 complex was detected at 550 kDa (Fig. 3A). On incubation of the human 600-kDa complex at 37°C under certain conditions, one smaller complex of approximately 400 kDa, in addition to the 600-kDa complex, was detected by native gel electrophoresis. These two complexes consisted of Mcm4, Mcm6, and Mcm7 proteins, as shown by two-dimensional SDS-PAGE (data not shown). These results suggest that the smaller complex is a trimer of Mcm4,6,7 proteins and the 600-kDa complex is a dimer of the trimer (hexamer). However, more definitive experiments are necessary to conclude whether the Mcm4,6,7 complex, which has DNA helicase activity, functions as a hexamer. Most DNA helicases usually form dimers or hexamers (4, 19, 36). The RecBCD helicase may be a dimer of the trimer composed of the RecB, RecC, and RecD proteins (12). This structure appears to be similar to that of the Mcm4,6,7 helicase.
Whether the Mcm4,6,7 complex functions as a replicative helicase responsible for the movement of the replication fork remains an open question. In S. cerevisiae, Mcm proteins are assembled onto an ORC origin complex with the assistance of a loading factor, Cdc6p, in an orderly fashion prior to the initiation of DNA replication in vivo (2, 49). Furthermore, the association of the Mcm proteins during the replication fork movement in S. cerevisiae suggests that they not only are required for the initiation of DNA replication but also function as a replicative helicase to unwind the duplex DNA at replication forks (2, 42). However, our results show that the human as well as the recombinant mouse Mcm4,6,7 complex can displace only oligonucleotides shorter than 30-mer (reference 20 and data not shown). This result would argue against its role as the fork helicase. In vivo, however, the DNA helicase activity of the Mcm4,6,7 complex could be enhanced by its association with the other replication proteins such as Cdc45p, a single-stranded DNA binding protein (replication protein A) and protein kinases. Another possibility is that the Mcm helicase is required only at the initial step of DNA unwinding at the origin region and that other DNA helicases such as the Werner-syndrome helicase (55) or DNA helicase B (39, 45) subsequently unwind the duplex DNA. Consistent with this notion, the prokaryotic enhancer binding protein NTRC, whose ATPase motifs are homologous to Mcm proteins (30), activates transcription by catalyzing an open-complex formation by RNA polymerase (3, 53).
Mcm2 can disassemble the Mcm4,6,7 complex.
Our mutational analyses of the recombinant Mcm proteins demonstrate that the Mcm4,6,7 complex has intrinsic DNA helicase activity. The Mcm2 protein inhibits the DNA helicase activity of the recombinant Mcm4,6,7 protein complex by converting the 550-kDa Mcm4,6,7 complex to the 450-kDa complex, which probably consisted of the Mcm2, Mcm4, Mcm6, and Mcm7 proteins. Since Mcm proteins bind to chromatin as a heterohexamer (1, 28, 43), these findings raise the possibility that the removal of Mcm2 protein from the Mcm2–7 heterohexamer is required for the activation of the Mcm4,6,7 helicase at the onset of DNA replication. Adachi et al. (1) reported that DNA helicase activity was not detected in the Mcm2–7 heterohexameric complex purified from Schizosaccharomyces pombe. Consistent with this result, we have obtained preliminary data showing that the mouse Mcm2–7 heterohexameric complex hardly binds single-stranded DNA (data not shown). Both Cdk2/cyclin and Cdc7/Dbf4 protein kinases, which are required for the initiation of DNA replication (24), may phosphorylate Mcm2 in the heterohexamer to facilitate the assembly of the Mcm4,6,7 proteins. However, more experiments are required to establish whether the dissociation of Mcm proteins leads to the formation of the Mcm4,6,7 core complex in vivo. Several groups have reported results suggesting that the Mcm proteins bound to chromatin are present mainly as a heterohexameric complex (14, 46). In addition to its potential regulatory role in the Mcm4,6,7 DNA helicase function, the mouse Mcm2 protein can bind histone H3 in vitro (21, 22) and has a nuclear localization activity (22, 28). Therefore, it is possible that the Mcm2 plays a role in transporting newly synthesized Mcm4, Mcm6, and Mcm7 proteins to the nucleus and tethering the Mcm proteins to chromatin in vivo, although the observation that only Mcm2 protein among the members can be dissociated from chromatin under low ionic conditions argues against the role of Mcm2 in the chromatin tethering (44). All six of the Mcm2–7 proteins are essential for cell growth in yeast (8, 9, 13, 15, 18, 35, 38, 40, 47, 48, 56), and they all contain conserved DNA-dependent ATPase motifs in the central domain. The biochemical function of Mcm3 and Mcm5 proteins remains to be determined.
ATPase motifs I and II are both important for Mcm functions.
The presence of the conserved DNA-dependent ATPase motifs in each of the Mcm proteins suggests that these motifs are critical for the protein function and also suggest that Mcm proteins function as a DNA-unwinding protein (30). Although the role of these motifs in the DNA helicase function has been analyzed for several helicases by site-directed mutagenesis, their effects on the complex of Mcm proteins have not been previously examined. The results of our experiments are summarized in Table 1. The mutations in motifs A and B of Mcm6 that change the lysine and serine residues to alanine residues (KS401-AA) and the aspartic and glutamic acid residues to alanine residues (DE459-AA) resulted in the reduction or loss, respectively, of the DNA helicase activity of the Mcm4,6,7 complex. These mutant Mcm complexes lacked ATP binding activity but were capable of hydrolyzing ATP (Fig. 6 to 8), suggesting that these two activities can be differentiated in the action of DNA helicase. It is conceivable that Mcm4 and Mcm7 proteins have a lower affinity for ATP and that ATP bound to these Mcm proteins is hydrolyzed in these mutant complexes. This result is similar to that observed with a mutant of the RecBCD complex in which mutation of the conserved ATPase motif of the RecD protein resulted in a decrease in the level of DNA helicase activity and loss of the ATP binding activity (31, 32). A mutant complex of Mcm4DE,6,7, in which the aspartic and glutamic acid residues of Mcm4 protein were changed to alanine residues, showed reduced DNA helicase and single-stranded DNA binding activities. However, neither the ATP binding nor the ATPase activity was affected by these changes (Fig. 8). Similar results were observed with mutants of T7 primase/helicase (52). These results indicated that the ability to hydrolyze ATP is not a sufficient property for exhibiting DNA helicase activity. In addition, both the ATP binding and the single-stranded DNA binding are regarded as necessary functions for DNA helicase activity, since mutants lacking helicase activity exhibited decreased levels of the ATP binding and single-stranded DNA binding activities. Based on these results, as well as those observed with the double mutant of Mcm4 and Mcm6 proteins, we conclude that the ATPase motifs in these proteins are crucial for the DNA helicase function of the Mcm4,6,7 complex.
Mutants defective in the Mcm4,6,7 complex formation have not been identified. Protein-protein interactions that stabilize the complex may occur over a large surface area; hence, multiple mutations over these regions would be required to affect complex formation to a significant extent.
Mcm4 and Mcm6 proteins may play different roles in DNA helicase function.
Detailed biochemical analyses of the mutants of the Mcm4,6,7 helicase have defined the role of the Mcm4 and Mcm6 proteins in the helicase action. Based on the results obtained from site-directed mutagenesis of the conserved ATPase motifs of the Mcm6 protein, we suggest that Mcm6 plays an important role in the ATP binding activity, which is likely to be required for the DNA helicase activity of the Mcm4,6,7 complex (Table 1 and Fig. 8). The mutation of Mcm4 in the Mcm4,6,7 complex reduced the single-stranded DNA binding activity of the complex but did not affect the ATPase or the ATP binding activity. The decrease of the DNA helicase activity in the Mcm4 mutant could be due to the reduced single-stranded DNA binding activity. These results suggest that Mcm4 and Mcm6 play distinct roles in the Mcm4,6,7 helicase function. Since the Mcm4,6,7 complex in which both Mcm4 and Mcm6 proteins were mutated is defective in all activities, there may be a synergistic effect of these two mutations. The ATPase activity was impaired only in the double mutant of Mcm4DE6DE.
The model of a rolling mechanism of DNA helicase action for DNA unwinding suggests that a functional helicase performs the DNA unwinding by the successive reactions of single-stranded DNA binding, interaction with the double-stranded DNA region through ATP binding, and ATP hydrolysis (36). According to this model, Mcm4 and Mcm6 proteins may be required for the translocation of the Mcm4,6,7 complex at the beginning of the helicase-catalyzed DNA unwinding. This model can also explain why the ATP hydrolysis activity alone is insufficient to unwind DNA.
The DNA-dependent ATPase motifs of Mcm proteins are conserved from yeast to mammalian cells, suggesting that they play important roles in Mcm functions in cellular DNA replication. Our biochemical analyses of the recombinant Mcm4,6,7 complexes suggest that the DNA helicase activity of this complex is involved in DNA replication in vivo. To address this point directly, we plan to express these mutant Mcm proteins in mammalian cells to examine their effects on cellular DNA replication.
ACKNOWLEDGMENTS
We thank Jerard Hurwitz for critical revision of the manuscript and Hiroshi Kimura for providing the anti-Mcm4 antibody and cDNA for Mcm proteins.
This work was supported in part by a grant-in-aid for scientific research on priority areas from the Ministry of Education, Science and Culture of Japan.
REFERENCES
- 1.Adachi Y, Usukura J, Yanagida M. A globular complex formation by Nda1 and the other five members of the MCM protein family in fission yeast. Genes Cells. 1997;2:467–479. doi: 10.1046/j.1365-2443.1997.1350333.x. [DOI] [PubMed] [Google Scholar]
- 2.Aparicio O M, Weinstein D M, Bell S P. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;91:59–69. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- 3.Austin S, Dixon R. The prokaryotic enhancer binding protein NTRC has an ATPase activity which is phosphorylation and DNA dependent. EMBO J. 1992;11:2219–2228. doi: 10.1002/j.1460-2075.1992.tb05281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borowiec J A. DNA helicases. In: DePamphilis M L, editor. DNA replication in eukaryotic cells. Vol. 1. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1996. pp. 545–574. [Google Scholar]
- 5.Burkhart R, Schulte D, Hu D, Musahl C, Göhring F, Knippers R. Interactions of human nuclear proteins P1Mcm3 and P1Cdc46. Eur J Biochem. 1995;228:431–438. [PubMed] [Google Scholar]
- 6.Chong J P, Mahbubani H M, Khoo C Y, Blow J J. Purification of an MCM-containing complex as a component of the DNA replication licensing system. Nature. 1995;375:418–421. doi: 10.1038/375418a0. [DOI] [PubMed] [Google Scholar]
- 7.Chong J P, Thömmes P, Blow J J. The role of MCM/P1 proteins in the licensing of DNA replication. Trends Biochem Sci. 1996;21:102–106. [PubMed] [Google Scholar]
- 8.Coxon A, Maundrell K, Kearsey S E. Fission yeast cdc21+ belongs to a family of proteins involved in an early step of chromosome replication. Nucleic Acids Res. 1992;20:5571–5577. doi: 10.1093/nar/20.21.5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dalton S, Whitbread L. Cell cycle-regulated nuclear import and export of Cdc47, a protein essential for initiation of DNA replication in budding yeast. Proc Natl Acad Sci USA. 1995;92:2514–2518. doi: 10.1073/pnas.92.7.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donovan S, Harwood J, Drury L S, Diffley J F. Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc Natl Acad Sci USA. 1997;94:5611–5616. doi: 10.1073/pnas.94.11.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dutta A, Bell S P. Initiation of DNA replication in eukaryotic cells. Annu Rev Cell Dev Biol. 1997;13:293–332. doi: 10.1146/annurev.cellbio.13.1.293. [DOI] [PubMed] [Google Scholar]
- 12.Dykstra C C, Palas K M, Kushner S R. Purification and characterization of exonuclease V from Escherichia coli K-12. Cold Spring Harbor Symp Quant Biol. 1984;49:463–467. doi: 10.1101/sqb.1984.049.01.052. [DOI] [PubMed] [Google Scholar]
- 13.Forsburg S L, Sherman D A, Ottilie S, Yasuda J R, Hodson J A. Mutational analysis of Cdc19p, a Schizosaccharomyces pombe MCM protein. Genetics. 1997;147:1025–1041. doi: 10.1093/genetics/147.3.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujita M, Kiyono T, Hayashi Y, Ishibashi M. In vivo interaction of human MCM heterohexameric complexes with chromatin. Possible involvement of ATP. J Biol Chem. 1997;272:10928–10935. doi: 10.1074/jbc.272.16.10928. [DOI] [PubMed] [Google Scholar]
- 15.Gibson S I, Surosky R T, Tye B K. The phenotype of the minichromosome maintenance mutant mcm3 is characteristic of mutants defective in DNA replication. Mol Cell Biol. 1990;10:5707–5720. doi: 10.1128/mcb.10.11.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grallert B, Nurse P. The ORC1 homolog orp1 in fission yeast plays a key role in regulating onset of S phase. Genes Dev. 1996;10:2644–2654. doi: 10.1101/gad.10.20.2644. [DOI] [PubMed] [Google Scholar]
- 17.Hardy C F, Dryga O, Seematter S, Pahl P M, Sclafani R A. mcm5/cdc46-bob1 bypasses the requirement for the S phase activator Cdc7p. Proc Natl Acad Sci USA. 1997;94:3151–3155. doi: 10.1073/pnas.94.7.3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hennessy K M, Lee A, Chen E, Botstein D. A group of interacting yeast DNA replication genes. Genes Dev. 1991;5:958–969. doi: 10.1101/gad.5.6.958. [DOI] [PubMed] [Google Scholar]
- 19.Hingorani M M, O’Donnell M. Toroidal proteins: running rings around DNA. Curr Biol. 1998;8:R83–R86. doi: 10.1016/s0960-9822(98)70052-1. [DOI] [PubMed] [Google Scholar]
- 20.Ishimi Y. A DNA helicase activity is associated with an MCM4, -6, and -7 protein complex. J Biol Chem. 1997;272:24508–24513. doi: 10.1074/jbc.272.39.24508. [DOI] [PubMed] [Google Scholar]
- 21.Ishimi Y, Ichinose S, Omori A, Sato K, Kimura H. Binding of human minichromosome maintenance proteins with histone H3. J Biol Chem. 1996;271:24115–24122. doi: 10.1074/jbc.271.39.24115. [DOI] [PubMed] [Google Scholar]
- 22.Ishimi Y, Komamura Y, You Z, Kimura H. Biochemical function of mouse minichromosome maintenance 2 protein. J Biol Chem. 1998;273:8369–8375. doi: 10.1074/jbc.273.14.8369. [DOI] [PubMed] [Google Scholar]
- 23.Ishimi, Y. Unpublished data.
- 24.Jallepalli P V, Kelly T J. Cyclin-dependent kinase and initiation at eukaryotic origins: a replication switch? Curr Opin Cell Biol. 1997;9:358–363. doi: 10.1016/s0955-0674(97)80008-7. [DOI] [PubMed] [Google Scholar]
- 25.Kearsey S E, Labib K. MCM proteins: evolution, properties, and role in DNA replication. Biochim Biophys Acta. 1998;1398:113–136. doi: 10.1016/s0167-4781(98)00033-5. [DOI] [PubMed] [Google Scholar]
- 26.Kearsey S E, Labib K, Maiorano D. Cell cycle control of eukaryotic DNA replication. Curr Opin Genet Dev. 1996;6:208–214. doi: 10.1016/s0959-437x(96)80052-9. [DOI] [PubMed] [Google Scholar]
- 27.Kearsey S E, Maiorano D, Holmes E C, Todorov I T. The role of MCM proteins in the cell cycle control of genome duplication. Bioessays. 1996;18:183–190. doi: 10.1002/bies.950180305. [DOI] [PubMed] [Google Scholar]
- 28.Kimura H, Ohtomo T, Yamaguchi M, Ishii A, Sugimoto K. Mouse MCM proteins: complex formation and transportation to the nucleus. Genes Cells. 1996;1:977–993. doi: 10.1046/j.1365-2443.1996.840284.x. [DOI] [PubMed] [Google Scholar]
- 29.Kimura H, Takizawa N, Nozaki N, Sugimoto K. Molecular cloning of cDNA encoding mouse Cdc21 and CDC46 homologs and characterization of the products: physical interaction between P1(MCM3) and CDC46 proteins. Nucleic Acids Res. 1995;23:2097–2104. doi: 10.1093/nar/23.12.2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koonin E V. A common set of conserved motifs in a vast variety of putative nucleic acid-dependent ATPases including MCM proteins involved in the initiation of eukaryotic DNA replication. Nucleic Acids Res. 1993;21:2541–2547. doi: 10.1093/nar/21.11.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Korangy F, Julin D A. Alteration by site-directed mutagenesis of the conserved lysine residue in the ATP-binding consensus sequence of the RecD subunit of the Escherichia coli RecBCD enzyme. J Biol Chem. 1992;267:1727–1732. [PubMed] [Google Scholar]
- 32.Korangy F, Julin D A. A mutation in the consensus ATP-binding sequence of the RecD subunit reduces the processivity of the RecBCD enzyme from Escherichia coli. J Biol Chem. 1992;267:3088–3095. [PubMed] [Google Scholar]
- 33.Kubota Y, Mimura S, Nishimoto S, Takisawa H, Nojima H. Identification of the yeast MCM3-related protein as a component of Xenopus DNA replication licensing factor. Cell. 1995;81:601–609. doi: 10.1016/0092-8674(95)90081-0. [DOI] [PubMed] [Google Scholar]
- 34.Lei M, Kawasaki Y, Young M R, Kihara M, Sugino A, Tye B K. Mcm2 is a target of regulation by Cdc7-Dbf4 during the initiation of DNA synthesis. Genes Dev. 1997;11:3365–3374. doi: 10.1101/gad.11.24.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang D T, Hodson J A, Forsburg S L. Reduced dosage of a single fission yeast MCM protein causes genetic instability and S phase delay. J Cell Sci. 1999;112:559–567. doi: 10.1242/jcs.112.4.559. [DOI] [PubMed] [Google Scholar]
- 36.Lohman T M, Bjornson K P. Mechanisms of helicase-catalyzed DNA unwinding. Annu Rev Biochem. 1996;65:169–214. doi: 10.1146/annurev.bi.65.070196.001125. [DOI] [PubMed] [Google Scholar]
- 37.Madine M A, Khoo C Y, Mills A D, Laskey R A. MCM3 complex required for cell cycle regulation of DNA replication in vertebrate cells. Nature. 1995;375:421–424. doi: 10.1038/375421a0. [DOI] [PubMed] [Google Scholar]
- 38.Maiorano D, Van Assendelft G B, Kearsey S E. Fission yeast cdc21, a member of the MCM protein family, is required for onset of S phase and is located in the nucleus throughout the cell cycle. EMBO J. 1996;15:861–872. [PMC free article] [PubMed] [Google Scholar]
- 39.Matsumoto K, Seki M, Masutani C, Tada S, Enomoto T, Ishimi Y. Stimulation of DNA synthesis by mouse DNA helicase B in a DNA replication system containing eukaryotic replication origins. Biochemistry. 1995;34:7913–7922. doi: 10.1021/bi00024a016. [DOI] [PubMed] [Google Scholar]
- 40.Miyake S, Okishio N, Samejima I, Hiraoka Y, Toda T, Saitoh I, Yanagida M. Fission yeast genes nda1+ and nda4+, mutations of which lead to S-phase block, chromatin alteration and Ca2+ suppression, are members of the CDC46/MCM2 family. Mol Biol Cell. 1993;4:1003–1015. doi: 10.1091/mbc.4.10.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Musahl C, Schulte D, Burkhart R, Knippers R. A human homologue of the yeast replication protein Cdc21. Interactions with other Mcm proteins. Eur J Biochem. 1995;230:1096–1101. doi: 10.1111/j.1432-1033.1995.tb20660.x. [DOI] [PubMed] [Google Scholar]
- 42.Newlon C S. Putting it all together: building a prereplicative complex. Cell. 1997;91:717–720. doi: 10.1016/s0092-8674(00)80459-6. [DOI] [PubMed] [Google Scholar]
- 43.Richter A, Knippers R. High-molecular-mass complexes of human minichromosome-maintenance proteins in mitotic cells. Eur J Biochem. 1997;247:136–141. doi: 10.1111/j.1432-1033.1997.00136.x. [DOI] [PubMed] [Google Scholar]
- 44.Richter A, Baack M, Holthoff H P, Ritzi M, Knippers R. Mobilization of chromatin-bound Mcm proteins by micrococcal nuclease. Biol Chem. 1998;379:1181–1187. doi: 10.1515/bchm.1998.379.8-9.1181. [DOI] [PubMed] [Google Scholar]
- 45.Seki M, Kohda T, Yano T, Tada S, Yanagisawa J, Eki T, Ui M, Enomoto T. Characterization of DNA synthesis and DNA-dependent ATPase activity at a restrictive temperature in temperature-sensitive tsFT848 cells with thermolabile DNA helicase B. Mol Cell Biol. 1995;15:165–172. doi: 10.1128/mcb.15.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sherman D A, Pasion S G, Forsburg S L. Multiple domains of fission yeast cdc19p (MCM2) are required for its association with the core MCM complex. Mol Biol Cell. 1998;9:1833–1845. doi: 10.1091/mbc.9.7.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sherman D A, Forsburg S L. Schizosaccharomyces pombe Mcm3p, an essential nuclear protein, associates tightly with Nda4p (Mcm5p) Nucleic Acids Res. 1998;26:3955–3960. doi: 10.1093/nar/26.17.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takahashi K, Yamada H, Yanagida M. Fission yeast minichromosome loss mutants mis cause lethal aneuploidy and replication abnormality. Mol Biol Cell. 1994;5:1145–1158. doi: 10.1091/mbc.5.10.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tanaka T, Knapp D, Nasmyth K. Loading of an Mcm protein onto DNA replication origins is regulated by Cdc6p and CDKs. Cell. 1997;90:649–660. doi: 10.1016/s0092-8674(00)80526-7. [DOI] [PubMed] [Google Scholar]
- 50.Thömmes P, Kubota Y, Takisawa H, Blow J J. The RLF-M component of the replication licensing system forms complexes containing all six MCM/P1 polypeptides. EMBO J. 1997;16:3312–3319. doi: 10.1093/emboj/16.11.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tye B K. The Mcm2-3-5 proteins: are they replication licensing factor? Trends Cell Biol. 1994;4:160–166. doi: 10.1016/0962-8924(94)90200-3. [DOI] [PubMed] [Google Scholar]
- 52.Washington M T, Rosenberg A H, Griffin K, Studier F W, Patel S S. Biochemical analysis of mutant T7 primase/helicase proteins defective in DNA binding, nucleotide hydrolysis, and the coupling of hydrolysis with DNA unwinding. J Biol Chem. 1996;271:26825–26834. doi: 10.1074/jbc.271.43.26825. [DOI] [PubMed] [Google Scholar]
- 53.Weiss D S, Batut J, Klose K E, Keener J, Kustu S. The phosphorylated form of the enhancer-binding protein NTRC has an ATPase activity that is essential for activation of transcription. Cell. 1991;67:155–167. doi: 10.1016/0092-8674(91)90579-n. [DOI] [PubMed] [Google Scholar]
- 54.Weng Y, Czaplinski K, Peltz S W. Genetic and biochemical characterization of mutations in the ATPase and helicase regions of the Upf1 protein. Mol Cell Biol. 1996;16:5477–5490. doi: 10.1128/mcb.16.10.5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yan H, Chen C Y, Kobayashi R, Newport J. Replication focus-forming activity 1 and the Werner syndrome gene product. Nat Genet. 1998;19:375–378. doi: 10.1038/1263. [DOI] [PubMed] [Google Scholar]
- 56.Yan H, Gibson S, Tye B K. Mcm2 and Mcm3, two proteins important for ARS activity, are related in structure and function. Genes Dev. 1991;5:944–957. doi: 10.1101/gad.5.6.944. [DOI] [PubMed] [Google Scholar]
- 57.Zou L, Mitchell J, Stillman B. CDC45, a novel yeast gene that functions with the origin recognition complex and Mcm proteins in initiation of DNA replication. Mol Cell Biol. 1997;17:553–563. doi: 10.1128/mcb.17.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]