Significance
d-amino acids are increasingly recognized as important signaling molecules in the mammalian central nervous system. Cysteine is the amino acid with the fastest in vitro spontaneous racemization rate, but its d-stereoisomer has not been examined. Here, we establish the presence of endogenous d-cysteine in the mammalian brain. Using sensitive and specific assays, we delineate its actions as a negative regulator of growth factor signaling during cortical development and identify a putative binding partner mediating these effects. By describing the newest member of the d-amino acid family, we open an avenue of research into the functions of these multifaceted signaling molecules.
Keywords: D-cysteine, serine racemase, FoxO, neural progenitor cell, cortical development
Abstract
d-amino acids are increasingly recognized as important signaling molecules in the mammalian central nervous system. However, the d-stereoisomer of the amino acid with the fastest spontaneous racemization ratein vitro in vitro, cysteine, has not been examined in mammals. Using chiral high-performance liquid chromatography and a stereospecific luciferase assay, we identify endogenous d-cysteine in the mammalian brain. We identify serine racemase (SR), which generates the N-methyl-d-aspartate (NMDA) glutamate receptor coagonist d-serine, as a candidate biosynthetic enzyme for d-cysteine. d-cysteine is enriched more than 20-fold in the embryonic mouse brain compared with the adult brain. d-cysteine reduces the proliferation of cultured mouse embryonic neural progenitor cells (NPCs) by ∼50%, effects not shared with d-serine or l-cysteine. The antiproliferative effect of d-cysteine is mediated by the transcription factors FoxO1 and FoxO3a. The selective influence of d-cysteine on NPC proliferation is reflected in overgrowth and aberrant lamination of the cerebral cortex in neonatal SR knockout mice. Finally, we perform an unbiased screen for d-cysteine–binding proteins in NPCs by immunoprecipitation with a d-cysteine–specific antibody followed by mass spectrometry. This approach identifies myristoylated alanine-rich C-kinase substrate (MARCKS) as a putative d-cysteine–binding protein. Together, these results establish endogenous mammalian d-cysteine and implicate it as a physiologic regulator of NPC homeostasis in the developing brain.
Biochemical stereospecificity permits the extraordinary precision of enzymatic reactions and receptor–ligand interactions. On account of the selective incorporation of l-amino acids into proteins, d-amino acids were long considered “unnatural” isomers, occurring endogenously only in microorganisms and invertebrates (1). However, since the first report of free endogenous d-aspartate in human tissues in 1986 (2), d-amino acids have come to be appreciated as physiologic signaling molecules in mammals, particularly in the central nervous system (3).
The physiological roles of d-aspartate and d-serine, the first two d-amino acids established in mammals (2, 4), remain the most well characterized. Both molecules act at N-methyl-d-aspartate-type glutamate receptors (NMDARs), with d-serine serving as the major endogenous ligand for the coagonist (“glycine”) site (5), while d-aspartate binds the glutamate site in addition to neuroendocrine functions (6–8). Metabolism of diverse d-amino acids is mediated by analogous pathways: both d-serine and d-aspartate are generated by pyridoxal 5′-phosphate-dependent enzymes, serine racemase (SR) and aspartate racemase, respectively (9, 10), and degraded by the flavin adenine dinucleotide-dependent oxidoreductases d-amino acid oxidase (DAAO) and d-aspartate oxidase (11–13).
The d-stereoisomers of a majority of proteogenic amino acids have been identified in the mammalian brain (14). Despite this widespread research into the presence and function of mammalian d-amino acids, the endogenous d-stereoisomer of the amino acid with the fastest in vitro spontaneous racemization rate, cysteine (15), has not been examined. In the current study we identify endogenous d-cysteine in the mammalian brain. d-cysteine is greatly enriched in embryonic mouse brain, with levels exceeding 4 mM, and is physiologically regulated by SR. d-cysteine, but not d-serine, diminishes proliferation of cultured embryonic neural progenitor cells through Akt-mediated disinhibition of the transcription factors FoxO1 and FoxO3a. These effects have functional consequences in vivo, as neonatal SR−/− mice exhibit drastic cortical overgrowth and aberrant layering. Lastly, we use unbiased proteomics to uncover putative d-cysteine–interacting proteins, an approach that identifies myristoylated alanine-rich C-kinase substrate (MARCKS) as a candidate binding partner.
Results
Distribution of d-Cysteine in the Mammalian Brain.
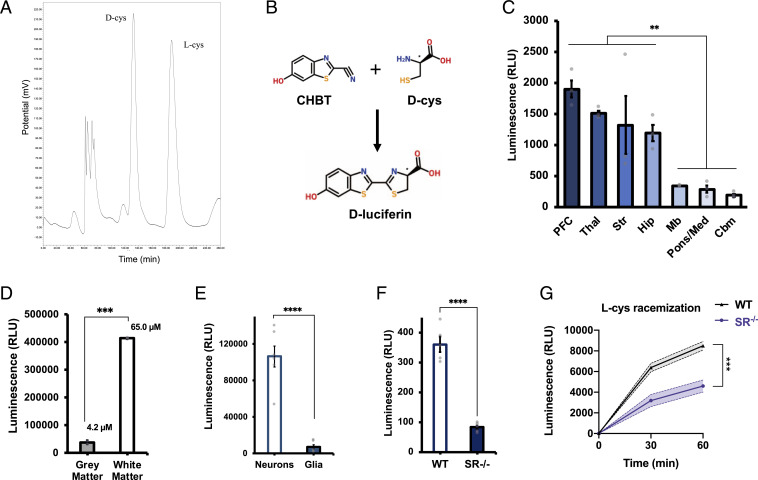
To search for endogenous d-cysteine in mammals, we developed assays to separate and measure l- and d-cysteine in tissue samples. High-performance liquid chromatography (HPLC) is the gold standard for resolution of stereoisomeric mixtures (16). We accordingly developed a method for labeling of cysteine enantiomers followed by chiral HPLC. Fluorescent labeling of free thiols with 4-(aminosulfonyl)-7-fluoro-2,1,3-benzoxadiazole (ABD-F) (17) followed by enantiomeric separation using the teicoplanin-based CHIROBIOTIC T chiral column (SI Appendix, Fig. S1A) allows for robust differentiation of l- and d-cysteine in purified solution and tissue lysates (Fig. 1A and SI Appendix, Fig. S1 B and C). This method establishes the presence of endogenous d-cysteine in the mammalian central nervous system. d-cysteine is present at levels of 50 µM in the adult mouse brain (Fig. 1A), a concentration approximately one-fifth that of brain d-serine (4). We also detect free d-cysteine in human cerebrospinal fluid at a concentration of 79 µM (SI Appendix, Fig. S1C).
Fig. 1.
Identification of endogenous d-cysteine in the mammalian brain. (A) Representative chromatogram from HPLC analysis of adult mouse brain extract. (B) Reaction scheme for d-cysteine (d-cys) luciferase assay. Condensation of endogenous d-cys with exogenously added CHBT yields d-luciferin, which is oxidized by d-luciferin–specific luciferase from the firefly P. pyralis to produce light. (C) Luciferase assay measurements of relative d-cys levels in lysates of various regions of adult mouse brain (n = 3). PFC, prefrontal cortex; Thal, thalamus; Str, striatum; Hip, hippocampus; Mb, midbrain; Pons/Med, pons/medulla; and Cbm, cerebellum. (d) Luciferase assay of d-cys levels in postmortem samples of human gray and white matter (n = 2). (E) Luciferase assay of d-cys in primary cortical neuronal and mixed glial cultures (n = 6). (F) Luciferase assay of d-cys in brain lysates from adult wild-type (WT) and serine racemase knockout (SR−/−) mice (n = 5). (G) l-cysteine racemization assay in brain lysates from adult wild-type and SR−/− mice (n = 4). Data are graphed as mean ± SEM **P < 0.01, ***P < 0.001, ****P < 0.0001, ANOVA with post hoc Tukey’s test (C), two-tailed unpaired Student’s t test (D–F), or ANOVA with post hoc Sidak’s test (G).
While HPLC allows for sensitive determination of absolute d-cysteine concentration in biological samples, the long elution times of this assay limit its throughput. To circumvent this issue, we took advantage of the use of d-cysteine in the industrial synthesis of d-luciferin (18) to develop a rapid and specific bioluminescence assay for measuring free d-cysteine. Condensation of d-cysteine with 2-cyano-6-hydroxybenzothiazole (CHBT) yields d-luciferin, which is then oxidized to oxyluciferin upon addition of adenosine triphosphate (ATP) and luciferase from the firefly Photinus pyralis to generate light (Fig. 1B). The stereospecificity of the P. pyralis luciferase for d-luciferin prevents the generation of luminescence by l-cysteine. This assay readily detects low micromolar concentrations of d-cysteine (SI Appendix, Fig. S1D) and is highly specific for d-cysteine, with less than 1% cross-reactivity against l-cysteine, d-serine, and other biological thiols (SI Appendix, Fig. S1E).
We leveraged the simplicity of the luciferase assay to assess the distribution of d-cysteine in the mammalian brain. d-cysteine is enriched in regions of mouse forebrain (prefrontal cortex, thalamus, striatum, and hippocampus) compared with midbrain and hindbrain regions (pons/medulla and cerebellum), though differences between forebrain regions were not significant (Fig. 1C). In postmortem human brain, d-cysteine levels are over 10-fold higher in white matter than in gray matter (Fig. 1D). Extrapolation from standard curve values yields concentrations of 65.0 µM and 4.2 µM in white and gray matter, respectively. d-cysteine appears to be primarily localized to neurons, as its levels in cultured murine primary cortical neurons (PCNs) were 15 times greater than those found in primary mixed glial cultures from mouse cortex (Fig. 1E).
Cysteine and serine are structurally similar: cysteine contains a sulfhydryl group bound to its β-carbon, while serine contains a chemically related hydroxyl group at the same position. Accordingly, we hypothesized that SR, which generates d-serine from l-serine, might also serve as a cysteine racemase. In support of this hypothesis, we found that levels of d-cysteine are reduced by 80% in the brains of adult SR null (SR−/−) mice (Fig. 1F). SR appears to possess cysteine racemase activity, as the ability of brain lysates to racemize exogenously added l-cysteine to d-cysteine is significantly blunted upon deletion of SR (Fig. 1G). These data suggest that SR may serve as a biosynthetic enzyme for d-cysteine.
Brain d-Cysteine Is Developmentally Regulated.
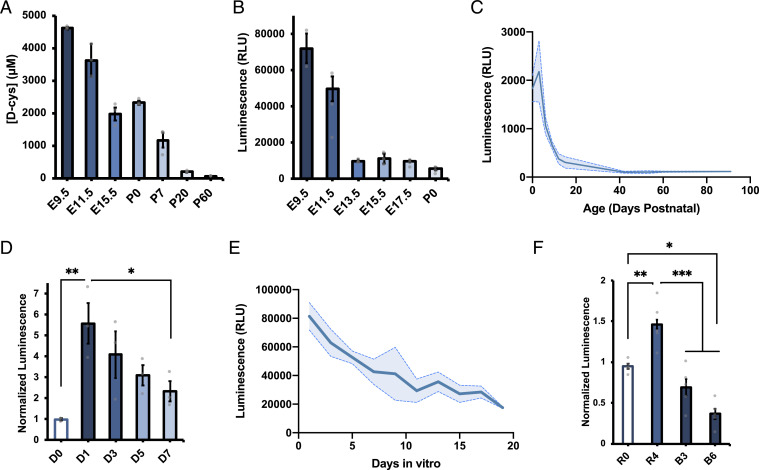
We next interrogated potential developmental regulation of brain d-cysteine. Separation of cysteine enantiomers by HPLC revealed that d-cysteine, but not l-cysteine, is dramatically enriched in the embryonic mouse brain compared with the adult (Fig. 2A and SI Appendix, Fig. S1F). Brain d-cysteine concentrations are as high as 4.5 mM at embryonic day 9.5 (E9.5) and decline steadily to adult levels of 50 µM. Measurement of relative d-cysteine levels at higher temporal resolution by luciferase assay highlighted two time periods during which brain d-cysteine levels drop dramatically: during midgestation (E9.5 to E13.5) and over the first 2 wk of postnatal life (Fig. 2 B and C). By contrast, d-cysteine levels are relatively stable from E13.5 to birth and during early adult life.
Fig. 2.
Brain d-cysteine levels are developmentally regulated. (A–C) d-cys levels in brain lysates from mice of indicated ages determined by (A) HPLC (main group effect ****P < 0.0001, ANOVA) and (B and C) luciferase assay (***P < 0.001, ANOVA) (n = 3). (D) Luciferase assay measurements of relative d-cys levels in primary cortical NPCs that were either left undifferentiated (D0) or differentiated for the indicated number of days (n = 3). (E) Luciferase assay of d-cys levels in PCNs cultured for the indicated number of days after isolation (n = 3), ****P < 0.0001, ANOVA. (F) Luciferase assay of d-cys levels in SK-N-SH cells left undifferentiated (R0) or differentiated with retinoic acid for 4 d (R4) followed by BDNF (B3, B6) (n = 5). Data are graphed as mean ± SEM *P < 0.05, **P < 0.01, ***P < 0.001, ANOVA with post hoc Tukey’s test.
The two developmental periods during which d-cysteine levels decrease markedly coincide with major milestones in forebrain development. Beginning at E9, neuroepithelial cells, which constitute the major neural stem cell (NSC) population in the early embryonic central nervous system, begin to differentiate into radial glial cells, the primary neural progenitor cell (NPC) population of the developing brain (19). Similarly, radial glia and other NPCs execute their final neurogenic divisions over the course of the first week of postnatal life (20). These correlations suggest that d-cysteine may be involved in cell fate decisions in the developing forebrain. In order to explore this hypothesis, we isolated and cultured NPCs from E13.5 mouse cortex and monitored cellular d-cysteine concentrations during induced differentiation into postmitotic neurons. Differentiation elicited a robust increase in d-cysteine levels 24 h after initiation of differentiation, which gradually declined over the following week (Fig. 2d). This decrease was mimicked in PCNs allowed to mature in vitro for 19 d, during which time d-cysteine levels decreased by fourfold (Fig. 2E).
To confirm our findings in another context and assess their potential conservation in human cells, we utilized the SK-N-SH human neuroblastoma cell line, whose subclone SH-SY5Y has been used as a model for neuronal differentiation (21). In this two-stage system, cells are treated with retinoic acid, which induces cell-cycle exit and initiates neurite outgrowth (i.e., differentiation phase), before treatment with brain-derived neurotrophic factor (BDNF), which further stimulates neurite outgrowth and arborization (maturation phase) (22). Retinoic acid–mediated differentiation increased cellular d-cysteine content by 50% after 4 d; induction of maturation with BDNF decreased d-cysteine levels to below untreated values after 6 d (Fig. 2F). Collectively, these observations point to a role for d-cysteine in neuronal cell fate decisions.
d-Cysteine Inhibits NPC Proliferation through Modulation of the Akt-FoxO Signaling Axis.
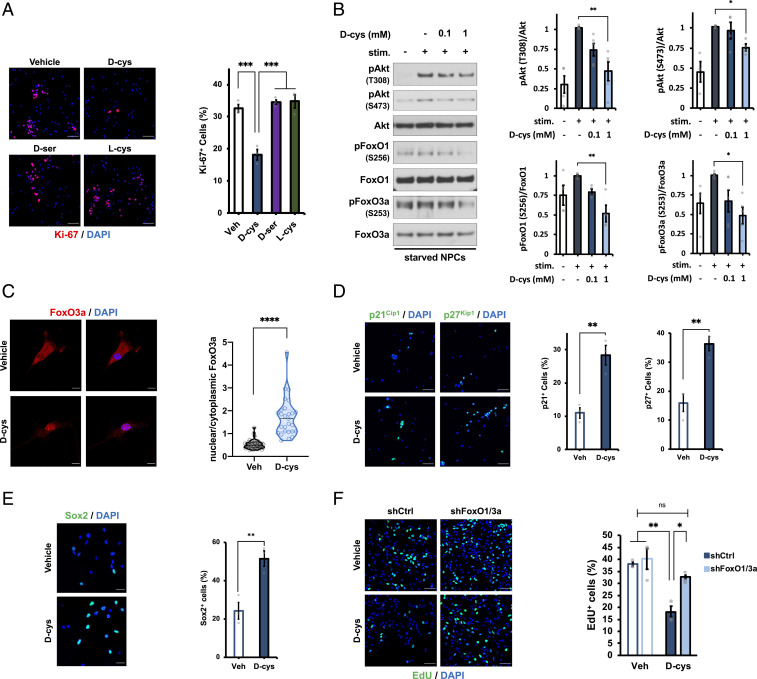
Prenatal brain development reflects a balance between NPC proliferation (self-renewal) and differentiation (neurogenesis). Because NPCs are the predominant brain cell type during embryonic life, during which d-cysteine is present at high levels, we explored the influence of d-cysteine on these two fundamental properties of NPC homeostasis. Treatment with 1 mM d-cysteine, the concentration found in perinatal brain, for 48 h reduced proliferation of cultured NPCs by ∼50% as determined by staining for the mitotic marker Ki-67 and incorporation of the nucleotide analog 5-ethynyl-2′-deoxyuridine (EdU), which labels dividing cells (SI Appendix, Fig. S2 A and B). Importantly, these effects were not elicited by equivalent concentrations of d-serine or l-cysteine (Fig. 3A). The lack of influence of l-cysteine on NPC proliferation appears to be due to inefficient racemization to d-cysteine in cultured NPCs (SI Appendix, Fig. S2C). Promotion of cell-cycle exit by d-cysteine was not associated with an induction of neuronal differentiation, as d-cysteine treatment did not significantly alter the numbers of newborn or mature neurons present in the cultures as monitored by doublecortin and MAP2 immunofluorescence, respectively (SI Appendix, Fig. S2 E and F).
Fig. 3.
D-cysteine inhibits neural progenitor cell proliferation through activation of FoxOs. (A) Representative images and quantification of immunostaining for Ki-67 and DAPI in NPCs treated with saline (vehicle) or 1 mM of the indicated amino acid for 48 h (n = 3). Data were quantified as the number of Ki-67–positive (Ki-67+) cells as a percent of the total number of (DAPI+) cells. (Scale bar, 50 µm.) (B) Representative immunoblots and quantification of phospho- and total Akt, FoxO1, and FoxO3a in NPCs. Cells were starved overnight followed by treatment with vehicle or the indicated concentration of d-cys for 1 h, followed by stimulation with complete medium for 5 min. Data are expressed as the normalized ratio of phosphorylated to total protein in each condition (n = 4 to 5). (C) Immunostaining for FoxO3a and DAPI in NPCs treated with vehicle or 1 mM d-cys for 48 h. Data are expressed as the ratio of the mean FoxO3a fluorescence intensity in the nucleus (DAPI-positive area) compared with the cytoplasm (n = 25 to 35 cells per group). (Scale bar, 10 µm.) (D and E) Immunostaining for (D) p21Cip1, p27Kip1, (E) Sox2, and DAPI in NPCs treated with vehicle or 1 mM d-cys for 48 h. Data were quantified as in A (n = 3). (Scale bar, 50 µm in D and 20 µm in E.) (F) EdU and DAPI staining in NPCs. Cells were infected with lentivirus producing shRNA against a nontargeting control (shCtrl) or shRNAs targeting FoxO1 and FoxO3a for 72 h before treatment with vehicle or 1 mM d-cys for 48 h, followed by addition of 10 µM EdU for 2 h. Data were quantified as in A (n = 3). (Scale bar, 50 µm.) Bar graphs depict mean ± SEM; violin plots show median (dark line) ± interquartile range (dashed lines). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, ns = P > 0.05, ANOVA with post hoc Tukey’s test (A, B, F) or two-tailed unpaired Student’s t test (C–E).
Kimura and coworkers (23) recently reported that exogenous d-cysteine can serve as a metabolic precursor for hydrogen sulfide (H2S) upon oxidation by DAAO. To determine whether d-cysteine inhibits NPC proliferation via H2S production, we treated NPCs with d-cysteine and the DAAO inhibitor (24) 6-chloro-1,2-benzisoxazol-3(2H)-one (CBIO). Consistent with DAAO functioning as a physiologic degradative enzyme for d-cysteine, CBIO treatment alone phenocopied the antiproliferative effect of d-cysteine and potentiated its influence on NPC proliferation (SI Appendix, Fig. S2D). This result suggests that the antiproliferative actions of d-cysteine in NPCs are independent of H2S production.
Cell proliferation is regulated by myriad signaling pathways, many of which converge on protein kinase B/Akt (25). We therefore hypothesized that d-cysteine inhibits Akt signaling. Accordingly, 1-h d-cysteine pretreatment dose-dependently inhibited the induction of Akt phosphorylation at Thr308 and Ser473 elicited by growth factor supplement stimulation (26) of growth factor–starved NPCs (Fig. 3B). Akt can phosphorylate and thereby regulate the activity of diverse proteins. We assessed the influence of d-cysteine on several of the most prominent Akt targets. The most dramatic influence of d-cysteine was on phosphorylation of the forkhead box O transcription factors FoxO1 and FoxO3a, which are negatively regulated by Akt (27). In accordance with the 50% reduction of NPC proliferation, 1 mM d-cysteine reduced phosphorylation of FoxO1 (at Ser256) and FoxO3a (Ser253) by ∼50% (Fig. 3B). d-cysteine also reduced phosphorylation of glycogen synthase kinase 3β (GSK-3β) (28), albeit to a much lesser extent (SI Appendix, Fig. S3A). By contrast, d-cysteine did not affect the activity of the mammalian target of rapamycin complex 1 (mTORC1) (29–31) as determined by phosphorylation of its targets p70 S6 kinase (S6K) and eukaryotic translation initiation factor 4E-binding protein 1 (4E-BP1) (SI Appendix, Fig. S3A). A 48-h treatment with 1 mM d-cysteine was sufficient to reduce Akt, FoxO1, and FoxO3a phosphorylation in the absence of starvation or growth factor manipulations (SI Appendix, Fig. S2C).
We next focused on the actions of d-cysteine upon FoxO-mediated signaling in NPCs. FoxOs are also inactivated by serum and glucocorticoid-regulated kinase 1 (SGK1) (32), another target of which is N-myc downstream regulated gene 1 (NDRG1) (33). d-cysteine reduced NDRG1 phosphorylation at Thr346 in NPCs (SI Appendix, Fig. S2B), suggesting that SGK1 activity is inhibited and may thus contribute to the activation of FoxOs by d-cysteine. SGK1 and Akt are both downstream of phosphoinositide 3-kinase (PI3K); thus, d-cysteine may act upstream of Akt and influence PI3K activity to modulate NPC proliferation (34, 35).
Akt-mediated phosphorylation of FoxOs promotes their binding to 14-3-3 chaperones, which sequester FoxOs in the cytoplasm and thereby inhibit their transcriptional activity (27). Accordingly, d-cysteine, which inhibits Akt-mediated FoxO phosphorylation, stimulates the nuclear translocation of FoxO3a (Fig. 3C) and induces expression of FoxO target genes, including the cyclin-dependent kinase inhibitors p21Cip1 and p27Kip1, which mediate cell cycle exit (36), and the pluripotency factor Sox2 (37), which is essential for the maintenance of NPC identity (38) (Fig. 3 D and E). FoxOs are also central regulators of programmed cell death (27, 32), a process essential for proper brain development (39–41). Thus, d-cysteine stimulated caspase 3 cleavage, an essential step in apoptosis (40), after 48 h (SI Appendix, Fig. S3D). Crucially, lentiviral shRNA-mediated double knockdown of FoxO1 and FoxO3a to 28% and 40% of control levels, respectively (SI Appendix, Fig. S3E), prevented the inhibition of NPC proliferation by d-cysteine (Fig. 3F), demonstrating that FoxO-dependent transcription is the crucial effector of d-cysteine signaling in NPCs.
Expansion and Aberrant Lamination of the Cerebral Cortex in Perinatal SR−/− Mice.
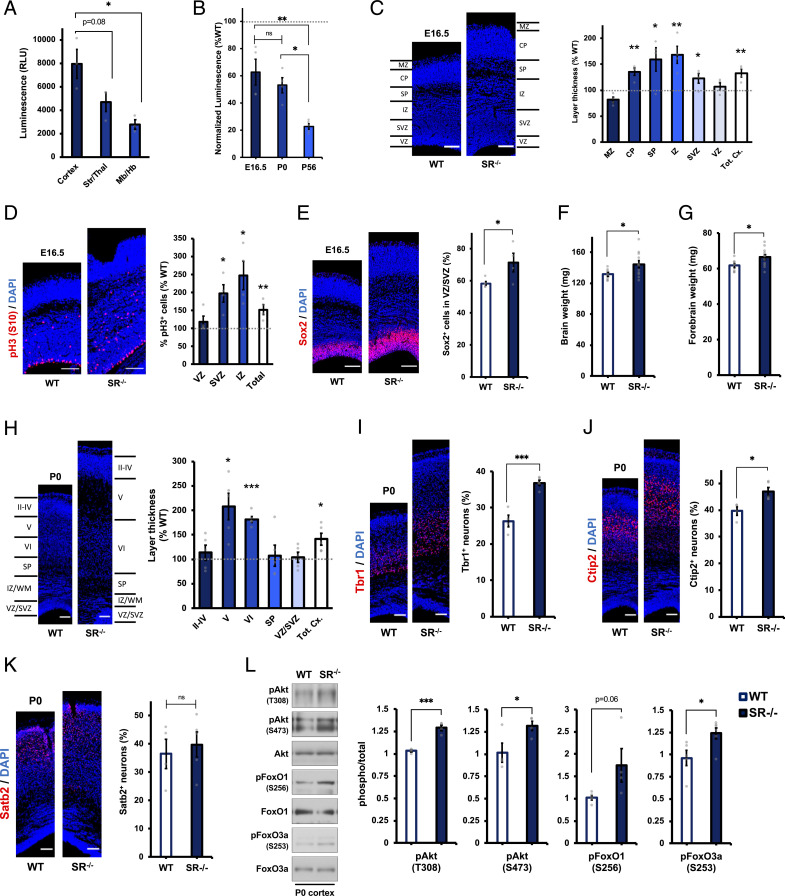
In light of the finding that d-cysteine, but not d-serine, regulates cortical NPC proliferation, we made use of SR−/− mice as a model to explore the influence of d-cysteine upon cortical development in vivo. We first established that d-cysteine is enriched in the neonatal cortex compared with other brain regions (Fig. 4A). We next confirmed loss of d-cysteine in the SR−/− brain at developmentally relevant time points. In contrast to adult SR−/− mice, whose brains contain nearly 80% less d-cysteine than wild-type controls, E16.5 and postnatal day 0 (P0) SR−/− brains had only 40 to 50% less d-cysteine than wild-type brains (Fig. 4B), suggesting that SR regulates brain d-cysteine levels in an age-dependent manner. However, we suspected that reducing d-cysteine levels by half would still be sufficient to induce neurodevelopmental alterations. Indeed, cortices of E16.5 SR−/− mice were 32% thicker than wild-type counterparts, with expansion being particularly pronounced in neurogenic zones (cortical plate, subplate, and intermediate zone [IZ]) but also evident in the subventricular zone (SVZ) (Fig. 4C). Proliferative NPCs reside in the intermediate, subventricular, and ventricular zones (VZ) of the developing cortex (19). We found increased fractions of proliferative cells, as determined by phosphohistone H3 (S10) staining, which marks mitotic cells, in SR−/− cortex (Fig. 4D), consistent with a function for d-cysteine as an endogenous inhibitor of NPC proliferation. Hyperproliferation was especially evident in the IZ but was also observed in the SVZ. Loss of d-cysteine, by disinhibiting NPC proliferation, would be expected to result in an expansion of the NPC population. Thus, a greater fraction of VZ/SVZ cells stained positive for the NPC marker Sox2 in E16.5 SR−/− cortices (Fig. 4E).
Fig. 4.
Aberrant cortical architecture in embryonic and neonatal SR−/− mice. (A) Luciferase measurements of d-cys levels in lysates from cortex, striatum/thalamus (Str/Thal), or midbrain/hindbrain (Mb/Hb) of P0 wild-type (WT) mice (n = 3). (B) Luciferase measurements of d-cys levels in SR−/− mouse brain lysates at the indicated ages. Data are normalized to luciferase measurements in age-matched wild-type controls (n = 4 to 5). (C) Representative DAPI staining and quantification of cortical zone thickness in coronal cortical sections from E16.5 WT and SR−/− mice. Layer thicknesses in SR−/− mice are normalized to the mean thickness of the corresponding layer in WT mice (n = 4 to 5). MZ, marginal zone; CP, cortical plate; SP, subplate; IZ, intermediate zone, SVZ, subventricular zone; VZ, ventricular zone; and Tot. Cx., total cortical thickness. (D) Immunostaining for phosphohistone H3 (pH3, S10) and DAPI in E16.5 WT and SR−/− cortical sections. Data are expressed as the ratio of pH3+ cells to total (DAPI+) cells in the indicated layer of SR−/− mice, normalized to WT controls (n = 4). (E) Immunostaining for Sox2 and DAPI in E16.5 WT and SR−/− cortical sections. Data are expressed as the ratio of Sox2+ cells to total (DAPI+) cells in the VZ/SVZ (n = 4 to 5). (F and G) Weight of (F) whole brains and (G) forebrains of P0 WT and SR−/− mice (n = 8 to 11). (H) Representative DAPI staining and quantification of cortical layer thickness in P0 WT and SR−/− mice, quantified as in C (n = 4 to 5). (I–K) Immunostaining for (I) Tbr1, (J) Ctip2, (K) Satb2, and DAPI in P0 WT and SR−/− cortical sections. Data are quantified as the ratio of marker-positive cells to the total number of (DAPI+) cells (n = 4 to 5). (L) Representative immunoblots and quantification of phospho- and total Akt, FoxO1, and FoxO3a in cortical lysates from P0 WT and SR−/− mice. Data are expressed as the normalized ratio of phosphorylated to total protein (n = 5). (Scale bars, 100 µm.) Data are graphed as mean ± SEM *P < 0.05, **P < 0.01, ***P < 0.001, ns = P > 0.05, ANOVA with post hoc Tukey’s test (A and B) or two-tailed unpaired Student’s t test (C–L).
We continued our examination of developmental aberrations in neonatal (P0) SR−/− mice, which exhibited increased weight of both whole brain and forebrain (Fig. 4 F and G). Cortical thickening was even more pronounced in P0 brain, with a 41% increase in overall cortical thickness in SR−/− mice. This expansion was accounted for by a near doubling of the thickness of deep cortical layers (layers V and VI), while the thickness of layers II to IV was unaffected (Fig. 4H). This expansion would most likely be due to an overproduction of early-born deep-layer neurons, which could result from an increase in the size of the progenitor pool as observed above. Accordingly, a greater percentage of SR−/− cortical cells stained positive for Tbr1 and Ctip2, markers of subplate/layer VI and layer V neurons (42, 43), respectively (Fig. 4 I and J). By contrast, numbers of Satb2+ later-born upper-layer neurons (44) were unchanged (Fig. 4K). Exaggerated proliferation and expansion of the cerebral cortex in SR−/− mice appear to be attributable to the same molecular mechanisms impacted by d-cysteine in NPCs in vitro, as phosphorylation of Akt, FoxO1, and FoxO3a was increased in neonatal SR−/− cortex (Fig. 4L). Together, these data support the hypothesis that loss of d-cysteine promotes Akt-mediated expansion of the cortical NPC population and thereby increases cortical size through overproduction of deep-layer neurons.
Characterization of Myristoylated Alanine-Rich C-Kinase Substrate as a Putative d-Cysteine–Binding Protein.
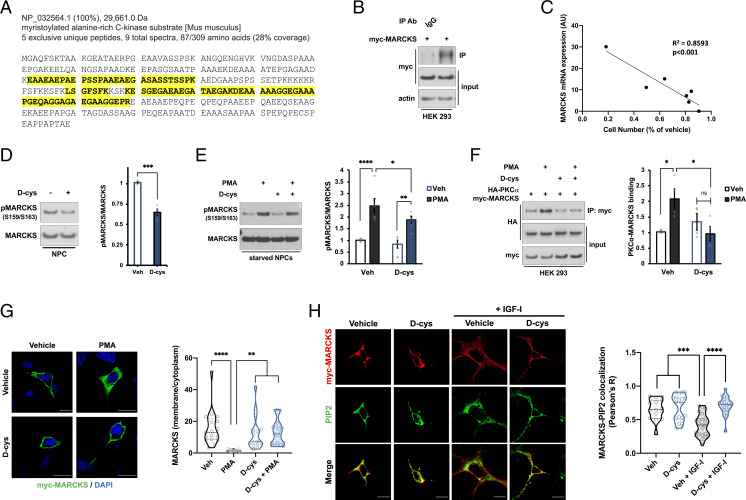
In order to further clarify the molecular functions of d-cysteine, we employed an unbiased proteomic approach to screen for potential d-cysteine–binding partners in NPCs. We immunoprecipitated proteins from d-cysteine–treated NPCs with either an anti–d-cysteine antibody or a control preimmune immunoglobulin G (IgG), after which proteins eluted by d-cysteine were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS). We filtered identified proteins by their frequency of occurrence in the contaminant repository for affinity purification (CRAPome) (45). We noted an increase in immunoprecipitated proteins with CRAPome frequency ≥0.4; accordingly, we prioritized proteins below this threshold as putative true-positive hits (SI Appendix, Fig. S4A). A total of 44 proteins met this criterion, among which were the myristoylated alanine-rich C-kinase substrate (MARCKS) and its homolog MARCKSL1 (Fig. 5A and SI Appendix, Fig. S4 B–E). We were particularly intrigued by the presence of MARCKS, as it is known to interface with PI3K signaling (46). To validate our mass spectrometric results, we performed anti–d-cysteine immunoprecipitation followed by Western blot in human embryonic kidney (HEK 293) cells overexpressing myc-tagged MARCKS. MARCKS was immunoprecipitated by the anti–d-cysteine antibody, but not the control IgG (Fig. 5B), thus confirming our LC-MS/MS data.
Fig. 5.
MARCKS is a putative d-cysteine–interacting protein. (A) Amino acid sequence of mouse MARCKS protein. Peptides identified by mass spectrometric analysis of anti–d-cys immunoprecipitates are highlighted in yellow. (B) Immunoblots of lysates (input) and immunoprecipitates (IP) from HEK 293 cells overexpressing myc-MARCKS. Lysates were immunoprecipitated with either preimmune rabbit immunoglobulin (IgG) or rabbit anti–d-cys antibody (d-cys). (C) Correlation between MARCKS mRNA expression [as determined by the Human Protein Atlas (84), available at http://www.proteinatlas.org] and antiproliferative effect of d-cys, as determined by the ratio of cell number after treatment with 1 mM d-cys for 48 h compared with vehicle-treated cells, in a panel of human cancer cell lines (n = 3). (D and E) Immunoblots and quantification of MARCKS phosphorylation in NPCs that were either (D) treated with vehicle or 1 mM d-cys for 48 h (n = 4) or (E) starved overnight followed by 20-min treatment with vehicle, 1 mM d-cys, and/or 250 nM PMA as indicated (n = 5). (F) Immunoblots and quantification of lysates (input) and anti-myc immunoprecipitates (IP) from HEK 293 cells overexpressing HA-PKCα and myc-MARCKS constructs. Cells were starved overnight followed by 20-min treatment with vehicle, 1 mM d-cys, and/or 500 nM PMA as indicated. Data are expressed as ratio of HA-PKCα in the IP fraction to HA-PKCα in the input fraction (n = 4). (G) Representative images and quantification of immunostaining for myc and DAPI in HEK 293 cells overexpressing myc-MARCKS and treated as in F. Data are expressed as the ratio of the mean myc-MARCKS fluorescence intensity in the membrane compared to the cytoplasm (n = 15 to 20 cells per group). (Scale bar, 20 µm.) (H) Immunostaining for myc and PIP2 in HEK 293 cells overexpressing myc-MARCKS. Cells were starved overnight before treatment with vehicle or 1 mM d-cys for 1 h followed by treatment with 100 ng/mL IGF-I for 10 min. Data are expressed as the Pearson correlation coefficient between myc and PIP2 stains at the plasma membrane (n = 15 to 20 cells per group). (Scale bar, 15 µm.) Bar graphs depict mean ± SEM; violin plots show median (dark line) ± interquartile range (dashed lines). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, ns = P > 0.05, sum of squares F test (C), two-tailed unpaired Student’s t test (D), or ANOVA with post hoc Tukey’s test (E–H).
Basic bioinformatic analyses of published datasets (47, 48) revealed that Marcks mRNA is enriched in progenitor compared with neuronal cell populations in E14.5 mouse brain (SI Appendix, Fig. S4G). Like d-cysteine, MARCKS mRNA expression decreases in the brain over the course of embryonic and postnatal development (SI Appendix, Fig. S4H). By contrast, transcript expression of the NMDAR GluN1 subunit, which acts as the receptor for d-serine, is enriched in neuronal populations and increases over developmental time (SI Appendix, Fig. S4 G and H). Srr mRNA is also enriched in neuronal populations, consistent with the greater loss of d-cysteine in adult versus perinatal SR−/− mice (SI Appendix, Fig. S4G). Importantly, MARCKS mRNA expression was strongly correlated (R2 = 0.8593, P < 0.001) with the magnitude of proliferation inhibition by d-cysteine in a panel of human cancer cell lines (Fig. 5C and SI Appendix, Fig. S4F). These data indicate that MARCKS is present in the cell types upon which d-cysteine exerts its actions and displays a developmental expression profile consistent with that of d-cysteine, supporting the notion that it may be a physiologically relevant d-cysteine–interacting protein.
We next investigated the impact of d-cysteine on MARCKS-mediated signaling. A 48-h treatment with 1 mM d-cysteine reduced MARCKS phosphorylation at Ser159/163 in NPCs (Fig. 5D). As its name suggests, MARCKS is the major substrate of protein kinase C (PKC) (49). Upon activation by calcium, diacylglycerol, or other stimuli, PKC binds and phosphorylates MARCKS, an action which translocates MARCKS from the plasma membrane, where it is basally localized, into the cytoplasm (50). d-cysteine reduced phosphorylation of MARCKS elicited by the PKC activator phorbol 12-myristate 13-acetate (PMA) in starved NPCs (Fig. 5E) and prevented the binding of overexpressed myc-tagged MARCKS and HA-tagged PKCα induced by PMA in starved HEK 293 cells (Fig. 5F). d-cysteine further prevented the ability of PMA to stimulate myc-MARCKS internalization in HEK 293 cells (Fig. 5G).
MARCKS is known to associate with the lipid phosphatidylinositol 4,5-bisphosphate (PIP2) at the membrane during inactive (starved) cellular conditions (46). Upon growth factor stimulation, activation of MARCKS by PKC and other kinases dissociates it from PIP2, freeing PIP2 to be phosphorylated by PI3K to form phosphatidylinositol 3,4,5-trisphosphate (PIP3), the essential molecule for Akt activation (51, 52). Stimulation of starved HEK 293 cells with insulin-like growth factor I (IGF-I) decreases the colocalization of PIP2 and myc-MARCKS at the membrane, and d-cysteine completely blocks this decrease (Fig. 5H). Thus, d-cysteine causes MARCKS to sequester PIP2 away from PI3K in response to growth factor signaling, providing a mechanistic basis for inhibition of Akt signaling by d-cysteine. While preliminary, these findings suggest that MARCKS may be a physiologic interactor of d-cysteine.
Discussion
d-amino acids, once thought to be unnatural isomers, are an important class of endogenous signaling molecules whose roles in mammalian neurobiology continue to be unearthed. Our present work describes the newest member of this family, d-cysteine. By developing sensitive and specific assays for the detection of d-cysteine, we report the identification of endogenous d-cysteine in mammals. We elucidate its actions in cortical development and delineate a signaling axis and putative binding partner mediating these effects. Our findings not only have implications for the understanding of the functions of d-amino acids in the nervous system, but also provide insight into the regulation of NPC homeostasis in the developing brain by small molecules.
The regional distribution of d-cysteine in the adult brain closely parallels that of SR and d-serine (9, 53), with enrichment in forebrain regions, including the prefrontal cortex and hippocampus. Conversely, d-cysteine localizations are reciprocal to those of DAAO (53). These data are consistent with conceptualizations of SR and DAAO as biosynthetic and degradative enzymes for d-cysteine, respectively (23). It will be of interest to explore the extent to which modulation of d-cysteine levels contributes to previously observed phenotypes of SR−/− and DAAO−/− mice.
In contrast to their similar regional localizations, the temporal distributions of d-cysteine and d-serine are inversely related. Brain d-cysteine content drops drastically during early postnatal life, while d-serine levels increase over the same period (54). Interestingly, d-aspartate shows a similar developmental expression profile as d-cysteine (2, 54, 55). SR expression parallels that of d-serine in mouse brain (54), though its transcript expression, along with d-serine levels (55), is mostly stable during prenatal development in human brain (SI Appendix, Fig. S4H). SR is further enriched in neuronal compared with progenitor populations in the embryonic mouse brain (SI Appendix, Fig. S4G), and while adult SR−/− mice lack 80% of brain d-cysteine, d-cysteine levels are decreased by only 40% and 50% in embryonic and neonatal SR−/− brains, respectively. Further, our HPLC analysis shows that the developmental trajectory of l-cysteine expression is neither inverse nor parallel to that of d-cysteine. Collectively, these observations imply the existence of SR/cysteine racemization-independent pathways for d-cysteine production in the developing brain.
Our analyses of d-cysteine actions in NPCs suggest its major function is to inhibit their proliferation. This finding appears somewhat contradictory, as d-cysteine is present at high levels during developmental periods in which NSC/NPC proliferation is widespread. However, as proliferative growth factors are present at high levels at these time points (19, 20, 56), the existence of endogenous inhibitors or “brakes” on growth factor signaling serves a homeostatic function, as evidenced by macrocephaly and neurodevelopmental disorders in patients and animal models containing loss-of-function mutations in negative regulators of growth factor signaling cascades (57–59). By inactivating Akt, d-cysteine may serve as such a brake, in a manner similar to the phosphatase and tensin homolog (PTEN) and other inhibitors of growth factor/Akt signaling (60, 61). Our findings support a model wherein, as the fraction of proliferative cells decreases over development due to an increase in terminally differentiated cells (neurons), the proliferation-limiting effects of d-cysteine become progressively less necessary and its production is decreased. It will be interesting in future studies to determine whether high d-cysteine levels persist in neurogenic niches of the adult brain and whether d-cysteine and SR may accordingly regulate the balance between quiescence and proliferation of adult NSCs.
We identified the transcription factors FoxO1 and FoxO3a as the crucial effectors of d-cysteine in NPCs. FoxOs are established negative regulators of NPC proliferation and neurogenesis (62, 63). While we did observe increased numbers of deep-layer cortical neurons in SR−/− mice, apparently due to an expanded NPC pool, d-cysteine did not have a direct effect on neurogenic differentiation of NPCs in vitro. However, by activating FoxOs, d-cysteine may inhibit differentiation in response to extrinsic signals, particularly by inducing expression of Sox2, which opposes neurogenesis (38). FoxOs and Sox2 specify the stem cell identity of populations including NPCs (37, 38, 62, 63). Accordingly, d-cysteine may itself function to maintain NSC/NPC identity. This hypothesis aligns well with our finding that d-cysteine levels decrease rapidly from E9.5 to E13.5 and P0 to P7, two periods of widespread NSC/NPC differentiation (19, 20). Importantly, further work will be required to determine whether d-cysteine acts on NPCs in a paracrine or autocrine/cell-autonomous manner in vivo.
We found that d-cysteine, but not d-serine, reduces proliferation of NPCs. Thus, the neurodevelopmental aberrations observed in SR−/− mice, which stem from expansion of the NPC population and involve enhanced Akt activation and consequent FoxO inhibition, appear to be attributable to d-cysteine. However, our data do not exclude a role for d-serine in related cellular events during corticogenesis. Interestingly, p21Cip1 gene expression was found to be decreased in the cortex of adult SR−/− mice (64), consistent with stimulation of FoxO-mediated p21Cip1 transcription by d-cysteine.
Our proteomic analysis of anti–d-cysteine immunocomplexes identified MARCKS as a putative d-cysteine interactor. MARCKS has established roles in forebrain development, and its germline knockout is embryonic lethal in mice (65). MARCKS−/− mice display aberrant cortical lamination and incomplete fusion of the cerebral hemispheres as well as area-specific defects in NPC proliferation and migration (66). Sequestration of PIP2 by MARCKS regulates PI3K-dependent signaling (51) and thus provides a mechanistic link between d-cysteine–MARCKS interactions and d-cysteine–mediated Akt inhibition. We previously demonstrated that SR is physiologically inhibited by PIP2 (67), suggesting the potential existence of an autoregulatory feedback loop for d-cysteine production and signaling through MARCKS. Future studies will be required to delineate the consequences of d-cysteine–MARCKS binding on cortical development in vivo.
We considered the possibility that the NMDAR might serve as a d-cysteine receptor, given the structural similarity between d-cysteine and the NMDAR coagonist d-serine (5). However, we did not identify any NMDAR subunits in our anti–d-cysteine immunoprecipitates, and the developmental expression profile of the d-serine–binding GluN1 subunit (68) is reciprocal to that of d-cysteine (SI Appendix, Fig. S4 G and H). Our data do not exclude the possibility of NMDAR-mediated functions of d-cysteine in the adult brain. However, in addition to being present at much lower levels than d-serine (50 µM compared with 250 µM), d-cysteine exhibits 20-fold lower affinity for forebrain synaptic NMDAR sites (69). H2S is known to facilitate NMDAR neurotransmission (70). Accordingly, d-cysteine may also influence NMDAR function indirectly through DAAO-mediated production of H2S (23), as exogenous d-cysteine has been shown to exert H2S-dependent dendritogenic effects in cultured cerebellar neurons (71).
The strongest link between d-amino acids and human neuropsychiatric phenotypes comes from schizophrenia. Polymorphisms in the SRR gene, as well as decreased d-serine levels, have been identified in multiple cohorts of patients with schizophrenia (72–75), and SR−/− mice have been found to recapitulate many of the molecular and behavioral correlates of schizophrenia (76). While these phenotypes have primarily been attributed to impaired d-serine–mediated NMDAR neurotransmission during adulthood (77), schizophrenia is increasingly appreciated as a neurodevelopmental disorder (78–81). Alterations in MARCKS expression in schizophrenia have also been described (82, 83). Accordingly, it will be of great interest to determine d-cysteine levels in patients with schizophrenia and to explore the extent to which d-cysteine–dependent neurodevelopmental aberrations underlie schizophrenia-related phenotypes of SR−/− mice.
Materials and Methods
Separation of l- and d-Cysteine by High-Performance Liquid Chromatography.
l- and d-cysteine from tissue lysates were separated using the CHIROBIOTIC T chiral HPLC column (5-µm particle size, L × D 25 cm × 4.6 mm) (Supelco) attached to a Waters 2690 alliance separations module connected to a fluorescence detector. Solvent was 20 mM ammonium acetate (80% vol/vol)-methanol (20% vol/vol) followed by isocratic elution. The flow rate was 0.05 mL/min. Cysteine stereoisomers were detected by fluorescence at excitation λ 380 nm and emission λ 510 nm. The column was maintained at room temperature. d-cysteine from brain lysates was extracted by homogenization in 100 mM HEPES buffer pH 7.5 using a handheld homogenizer on ice with 10 to 15 strokes. The homogenates were then lysed using a Branson sonicator on ice with six pulses of 10 to 15 s each. The samples were then centrifuged at 16,000 × g for 30 min at 4 °C. After centrifugation the supernatant was harvested in a prechilled tube. Proteins in the supernatant were precipitated by adding 12 N HCl to the lysate to a final concentration of 2 N. The sample was cooled on ice for 30 min and centrifuged at 16,000 × g for 30 min at 4 °C. The supernatant containing cysteine was harvested. The supernatant was neutralized by adding one volume 10 N NaOH followed by addition of 1 volume of ABD-F in 200 mM sodium borate buffer pH 8.0 containing 1 mM ethylenediaminetetraacetic acid (EDTA) and 0.1 volume of 10% tri-n-butylphosphine in acetonitrile to label the free thiol of cysteine. The mixture was incubated at 50 °C for 5 min and vortexed thoroughly after incubation. The mixture was placed on ice and 0.01 volume of 2.5 N HCl added to neutralize the sample. The sample was then diluted 1:1 with HPLC-grade water and filtered using a Millex LG 0.2-µm syringe filter (Millipore) into an amber HPLC vial. The fluorescently labeled sample was used for injection (10 µL volume) on the preequilibrated chiral HPLC column and the labeled thiols were detected by fluorescence. The areas under the respective peaks were quantified based on a standard curve of purified l- and d-cysteine standards.
d-Cysteine Luciferase Assay.
Tissue samples were homogenized on ice in five volumes of lysis buffer (500 mM tricine, 100 mM MgSO4, 2 mM EDTA, 1% Triton X-100, pH 7.8) supplemented immediately before homogenization/lysis with 10 mM tris(2-carboxyethyl)phosphine (TCEP), 200 mM K2CO3, and 0.1 mg/mL CHBT (Santa Cruz). In the case of cultured cells, culture media were aspirated, cells were washed twice with ice-cold phosphate-buffered saline (PBS), and cells were harvested on ice in the above solution at a volume of 250 µL per 35-mm dish. Following sonication, samples were incubated at 30 °C for 10 min with shaking to allow for d-luciferin formation. Samples were then cooled on ice and cleared by centrifugation at 16,000 × g at 4 °C for 30 min. Supernatants were transferred to fresh tubes and neutralized with 12 N HCl to a pH of 7.8. ATP was added to a final concentration of 5 mM, after which d-luciferin–specific Quantilum Recombinant Luciferase (Promega) was added to a final concentration of 25 µg/mL. Samples were transferred to opaque 96-well plates and incubated at room temperature in the dark for 5 min, at which point luminescence was measured on a SpectraMax M3 microplate reader (Molecular Devices) with a 500-ms integration time. Luminescence values were normalized to total protein concentration, obtained from supernatants using a colorimetric protein assay (Bio-Rad).
l-Cysteine Racemization Assay.
A total of 20 µg of protein from precleared brain lysates was incubated in luciferase assay buffer (see above) with 50 µM pyridoxal 5′-phosphate (Sigma) for 10 min at 37 °C prior to addition of 1 mM l-cysteine and incubation at 37 °C for times ranging from 0 to 60 min. Samples were then placed on ice and reactions were quenched by addition of 10 mM TCEP, 200 mM K2CO3, and 0.1 mg/mL CHBT. Samples were then incubated at 30 °C for 10 min and cooled on ice. Samples were then neutralized with HCl and processed for luminescence measurements as described above.
Supplementary Material
Acknowledgments
We are grateful to Roxanne Barrow, Lauren Albacarys, Adele Snowman, Lynda Hester, and Susan McTeer (S.H.S. laboratory) for their assistance; Tatiana Boronina and Bob Cole (Johns Hopkins School of Medicine Mass Spectrometry and Proteomics Core) for assistance with mass spectrometry experiments; and members of the S.H.S. laboratory for helpful discussion. We thank the Rocky Mountain Multiple Sclerosis Center for supplying postmortem human brain tissue samples. This work was supported by US Public Health Service/National Institute of Mental Health Grant R01MH180501.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2110610118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Corrigan J. J., D-amino acids in animals. Science 164, 142–149 (1969). [DOI] [PubMed] [Google Scholar]
- 2.Dunlop D. S., Neidle A., McHale D., Dunlop D. M., Lajtha A., The presence of free D-aspartic acid in rodents and man. Biochem. Biophys. Res. Commun. 141, 27–32 (1986). [DOI] [PubMed] [Google Scholar]
- 3.Snyder S. H., Kim P. M., D-amino acids as putative neurotransmitters: Focus on D-serine. Neurochem. Res. 25, 553–560 (2000). [DOI] [PubMed] [Google Scholar]
- 4.Hashimoto A., et al., The presence of free D-serine in rat brain. FEBS Lett. 296, 33–36 (1992). [DOI] [PubMed] [Google Scholar]
- 5.Mothet J.-P., et al., D-serine is an endogenous ligand for the glycine site of the N-methyl-D-aspartate receptor. Proc. Natl. Acad. Sci. U.S.A. 97, 4926–4931 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Erreger K., et al., Subunit-specific agonist activity at NR2A-, NR2B-, NR2C-, and NR2D-containing N-methyl-D-aspartate glutamate receptors. Mol. Pharmacol. 72, 907–920 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Schell M. J., Cooper O. B., Snyder S. H., D-aspartate localizations imply neuronal and neuroendocrine roles. Proc. Natl. Acad. Sci. U.S.A. 94, 2013–2018 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang A. S., et al., D-aspartate regulates melanocortin formation and function: Behavioral alterations in D-aspartate oxidase-deficient mice. J. Neurosci. 26, 2814–2819 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wolosker H., Blackshaw S., Snyder S. H., Serine racemase: A glial enzyme synthesizing D-serine to regulate glutamate-N-methyl-D-aspartate neurotransmission. Proc. Natl. Acad. Sci. U.S.A. 96, 13409–13414 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim P. M., et al., Aspartate racemase, generating neuronal D-aspartate, regulates adult neurogenesis. Proc. Natl. Acad. Sci. U.S.A. 107, 3175–3179 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krebs H. A., Metabolism of amino-acids: Deamination of amino-acids. Biochem. J. 29, 1620–1644 (1935). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pollegioni L., Sacchi S., Murtas G., Human D-amino acid oxidase: Structure, function, and regulation. Front. Mol. Biosci. 5, 107 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simonic T., et al., cDNA cloning and expression of the flavoprotein D-aspartate oxidase from bovine kidney cortex. Biochem. J. 322, 729–735 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weatherly C. A., et al., D-amino acid levels in perfused mouse brain tissue and blood: A comparative study. ACS Chem. Neurosci. 8, 1251–1261 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Man E. H., Bada J. L., Dietary D-amino acids. Annu. Rev. Nutr. 7, 209–225 (1987). [DOI] [PubMed] [Google Scholar]
- 16.Ilisz I., Aranyi A., Pataj Z., Péter A., Enantiomeric separation of nonproteinogenic amino acids by high-performance liquid chromatography. J. Chromatogr. A 1269, 94–121 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Toyo’oka T., Imai K., New fluorogenic reagent having halogenobenzofurazan structure for thiols: 4-(aminosulfonyl)-7-fluoro-2,1,3-benzoxadiazole. Anal. Chem. 56, 2461–2464 (1984). [Google Scholar]
- 18.Seliger H. H., McELROY W. D., White E. H., Field G. F., Stereo-specificity and firefly bioluminescence, a comparison of natural and synthetic luciferins. Proc. Natl. Acad. Sci. U.S.A. 47, 1129–1134 (1961). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kriegstein A., Alvarez-Buylla A., The glial nature of embryonic and adult neural stem cells. Annu. Rev. Neurosci. 32, 149–184 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dwyer N. D., et al., Neural stem cells to cerebral cortex: Emerging mechanisms regulating progenitor behavior and productivity. J. Neurosci. 36, 11394–11401 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaplan D. R., Matsumoto K., Lucarelli E., Thiele C. J.; Eukaryotic Signal Transduction Group , Induction of TrkB by retinoic acid mediates biologic responsiveness to BDNF and differentiation of human neuroblastoma cells. Neuron 11, 321–331 (1993). [DOI] [PubMed] [Google Scholar]
- 22.Laurent B., et al., A specific LSD1/KDM1A isoform regulates neuronal differentiation through H3K9 demethylation. Mol. Cell 57, 957–970 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shibuya N., et al., A novel pathway for the production of hydrogen sulfide from D-cysteine in mammalian cells. Nat. Commun. 4, 1366 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Ferraris D., et al., Synthesis and biological evaluation of D-amino acid oxidase inhibitors. J. Med. Chem. 51, 3357–3359 (2008). [DOI] [PubMed] [Google Scholar]
- 25.Manning B. D., Cantley L. C., AKT/PKB signaling: Navigating downstream. Cell 129, 1261–1274 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alessi D. R., et al., Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J. 15, 6541–6551 (1996). [PMC free article] [PubMed] [Google Scholar]
- 27.Brunet A., et al., Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 96, 857–868 (1999). [DOI] [PubMed] [Google Scholar]
- 28.Cross D. A. E., Alessi D. R., Cohen P., Andjelkovich M., Hemmings B. A., Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature 378, 785–789 (1995). [DOI] [PubMed] [Google Scholar]
- 29.Sancak Y., et al., PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol. Cell 25, 903–915 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Inoki K., Li Y., Zhu T., Wu J., Guan K. L., TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat. Cell Biol. 4, 648–657 (2002). [DOI] [PubMed] [Google Scholar]
- 31.Burnett P. E., Barrow R. K., Cohen N. A., Snyder S. H., Sabatini D. M., RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc. Natl. Acad. Sci. U.S.A. 95, 1432–1437 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brunet A., et al., Protein kinase SGK mediates survival signals by phosphorylating the forkhead transcription factor FKHRL1 (FOXO3a). Mol. Cell. Biol. 21, 952–965 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murray J. T., et al., Exploitation of KESTREL to identify NDRG family members as physiological substrates for SGK1 and GSK3. Biochem. J. 384, 477–488 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burgering B. M. T., Coffer P. J., Protein kinase B (c-Akt) in phosphatidylinositol-3-OH kinase signal transduction. Nature 376, 599–602 (1995). [DOI] [PubMed] [Google Scholar]
- 35.Park J., et al., Serum and glucocorticoid-inducible kinase (SGK) is a target of the PI 3-kinase-stimulated signaling pathway. EMBO J. 18, 3024–3033 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Medema R. H., Kops G. J. P. L., Bos J. L., Burgering B. M. T., AFX-like Forkhead transcription factors mediate cell-cycle regulation by Ras and PKB through p27kip1. Nature 404, 782–787 (2000). [DOI] [PubMed] [Google Scholar]
- 37.Zhang X., et al., FOXO1 is an essential regulator of pluripotency in human embryonic stem cells. Nat. Cell Biol. 13, 1092–1099 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Graham V., Khudyakov J., Ellis P., Pevny L., SOX2 functions to maintain neural progenitor identity. Neuron 39, 749–765 (2003). [DOI] [PubMed] [Google Scholar]
- 39.Blaschke A. J., Staley K., Chun J., Widespread programmed cell death in proliferative and postmitotic regions of the fetal cerebral cortex. Development 122, 1165–1174 (1996). [DOI] [PubMed] [Google Scholar]
- 40.Kuida K., et al., Decreased apoptosis in the brain and premature lethality in CPP32-deficient mice. Nature 384, 368–372 (1996). [DOI] [PubMed] [Google Scholar]
- 41.Kuida K., et al., Reduced apoptosis and cytochrome c-mediated caspase activation in mice lacking caspase 9. Cell 94, 325–337 (1998). [DOI] [PubMed] [Google Scholar]
- 42.Hevner R. F., et al., Tbr1 regulates differentiation of the preplate and layer 6. Neuron 29, 353–366 (2001). [DOI] [PubMed] [Google Scholar]
- 43.Arlotta P., et al., Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron 45, 207–221 (2005). [DOI] [PubMed] [Google Scholar]
- 44.Britanova O., Akopov S., Lukyanov S., Gruss P., Tarabykin V., Novel transcription factor Satb2 interacts with matrix attachment region DNA elements in a tissue-specific manner and demonstrates cell-type-dependent expression in the developing mouse CNS. Eur. J. Neurosci. 21, 658–668 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Mellacheruvu D., et al., The CRAPome: A contaminant repository for affinity purification-mass spectrometry data. Nat. Methods 10, 730–736 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Glaser M., et al., Myristoylated alanine-rich C kinase substrate (MARCKS) produces reversible inhibition of phospholipase C by sequestering phosphatidylinositol 4,5-bisphosphate in lateral domains. J. Biol. Chem. 271, 26187–26193 (1996). [DOI] [PubMed] [Google Scholar]
- 47.Loo L., et al., Single-cell transcriptomic analysis of mouse neocortical development. Nat. Commun. 10, 134 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miller J. A., et al., Transcriptional landscape of the prenatal human brain. Nature 508, 199–206 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stumpo D. J., Graff J. M., Albert K. A., Greengard P., Blackshear P. J., Molecular cloning, characterization, and expression of a cDNA encoding the “80- to 87-kDa” myristoylated alanine-rich C kinase substrate: A major cellular substrate for protein kinase C. Proc. Natl. Acad. Sci. U.S.A. 86, 4012–4016 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thelen M., Rosen A., Nairn A. C., Aderem A., Regulation by phosphorylation of reversible association of a myristoylated protein kinase C substrate with the plasma membrane. Nature 351, 320–322 (1991). [DOI] [PubMed] [Google Scholar]
- 51.Ziemba B. P., Burke J. E., Masson G., Williams R. L., Falke J. J., Regulation of PI3K by PKC and MARCKS: Single-molecule analysis of a reconstituted signaling pathway. Biophys. J. 110, 1811–1825 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stokoe D., et al., Dual role of phosphatidylinositol-3,4,5-trisphosphate in the activation of protein kinase B. Science 277, 567–570 (1997). [DOI] [PubMed] [Google Scholar]
- 53.Schell M. J., Molliver M. E., Snyder S. H., D-serine, an endogenous synaptic modulator: Localization to astrocytes and glutamate-stimulated release. Proc. Natl. Acad. Sci. U.S.A. 92, 3948–3952 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang L.-Z., Zhu X.-Z., Spatiotemporal relationships among D-serine, serine racemase, and D-amino acid oxidase during mouse postnatal development. Acta Pharmacol. Sin. 24, 965–974 (2003). [PubMed] [Google Scholar]
- 55.Hashimoto A., et al., Embryonic development and postnatal changes in free D-aspartate and D-serine in the human prefrontal cortex. J. Neurochem. 61, 348–351 (1993). [DOI] [PubMed] [Google Scholar]
- 56.Raballo R., et al., Basic fibroblast growth factor (Fgf2) is necessary for cell proliferation and neurogenesis in the developing cerebral cortex. J. Neurosci. 20, 5012–5023 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Groszer M., et al., Negative regulation of neural stem/progenitor cell proliferation by the Pten tumor suppressor gene in vivo. Science 294, 2186–2189 (2001). [DOI] [PubMed] [Google Scholar]
- 58.Buxbaum J. D., et al., Mutation screening of the PTEN gene in patients with autism spectrum disorders and macrocephaly. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 144B, 484–491 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.D’Gama A. M., et al., Somatic mutations activating the mTOR pathway in dorsal telencephalic progenitors cause a continuum of cortical dysplasias. Cell Rep. 21, 3754–3766 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stambolic V., et al., Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell 95, 29–39 (1998). [DOI] [PubMed] [Google Scholar]
- 61.Jeong D., et al., LRIG1-mediated inhibition of EGF receptor signaling regulates neural precursor cell proliferation in the neocortex. Cell Rep. 33, 108257 (2020). [DOI] [PubMed] [Google Scholar]
- 62.Paik J. H., et al., FoxOs cooperatively regulate diverse pathways governing neural stem cell homeostasis. Cell Stem Cell 5, 540–553 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Renault V. M., et al., FoxO3 regulates neural stem cell homeostasis. Cell Stem Cell 5, 527–539 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Balu D. T., Basu A. C., Corradi J. P., Cacace A. M., Coyle J. T., The NMDA receptor co-agonists, D-serine and glycine, regulate neuronal dendritic architecture in the somatosensory cortex. Neurobiol. Dis. 45, 671–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stumpo D. J., Bock C. B., Tuttle J. S., Blackshear P. J., MARCKS deficiency in mice leads to abnormal brain development and perinatal death. Proc. Natl. Acad. Sci. U.S.A. 92, 944–948 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weimer J. M., et al., MARCKS modulates radial progenitor placement, proliferation and organization in the developing cerebral cortex. Development 136, 2965–2975 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mustafa A. K., et al., Glutamatergic regulation of serine racemase via reversal of PIP2 inhibition. Proc. Natl. Acad. Sci. U.S.A. 106, 2921–2926 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Laurie D. J., Seeburg P. H., Regional and developmental heterogeneity in splicing of the rat brain NMDAR1 mRNA. J. Neurosci. 14, 3180–3194 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Monahan J. B., Corpus V. M., Hood W. F., Thomas J. W., Compton R. P., Characterization of a [3H]glycine recognition site as a modulatory site of the N-methyl-D-aspartate receptor complex. J. Neurochem. 53, 370–375 (1989). [DOI] [PubMed] [Google Scholar]
- 70.Abe K., Kimura H., The possible role of hydrogen sulfide as an endogenous neuromodulator. J. Neurosci. 16, 1066–1071 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Seki T., et al., d-Cysteine promotes dendritic development in primary cultured cerebellar Purkinje cells via hydrogen sulfide production. Mol. Cell. Neurosci. 93, 36–47 (2018). [DOI] [PubMed] [Google Scholar]
- 72.Goltsov A. Y., et al., Polymorphism in the 5′-promoter region of serine racemase gene in schizophrenia. Mol. Psychiatry 11, 325–326 (2006). [DOI] [PubMed] [Google Scholar]
- 73.Morita Y., et al., A genetic variant of the serine racemase gene is associated with schizophrenia. Biol. Psychiatry 61, 1200–1203 (2007). [DOI] [PubMed] [Google Scholar]
- 74.Labrie V., et al., Serine racemase is associated with schizophrenia susceptibility in humans and in a mouse model. Hum. Mol. Genet. 18, 3227–3243 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bendikov I., et al., A CSF and postmortem brain study of D-serine metabolic parameters in schizophrenia. Schizophr. Res. 90, 41–51 (2007). [DOI] [PubMed] [Google Scholar]
- 76.Balu D. T., et al., Multiple risk pathways for schizophrenia converge in serine racemase knockout mice, a mouse model of NMDA receptor hypofunction. Proc. Natl. Acad. Sci. U.S.A. 110, E2400–E2409 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Balu D. T., Coyle J. T., The NMDA receptor ‘glycine modulatory site’ in schizophrenia: D-serine, glycine, and beyond. Curr. Opin. Pharmacol. 20, 109–115 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gulsuner S.et al.; Consortium on the Genetics of Schizophrenia (COGS); PAARTNERS Study Group , Spatial and temporal mapping of de novo mutations in schizophrenia to a fetal prefrontal cortical network. Cell 154, 518–529 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jaffe A. E.et al.; BrainSeq Consortium , Developmental and genetic regulation of the human cortex transcriptome illuminate schizophrenia pathogenesis. Nat. Neurosci. 21, 1117–1125 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Selemon L. D., Zecevic N., Schizophrenia: A tale of two critical periods for prefrontal cortical development. Transl. Psychiatry 5, e623 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Birnbaum R., Weinberger D. R., Genetic insights into the neurodevelopmental origins of schizophrenia. Nat. Rev. Neurosci. 18, 727–740 (2017). [DOI] [PubMed] [Google Scholar]
- 82.Pinner A. L., Haroutunian V., Meador-Woodruff J. H., Alterations of the myristoylated, alanine-rich C kinase substrate (MARCKS) in prefrontal cortex in schizophrenia. Schizophr. Res. 154, 36–41 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Huckins L. M.et al.; CommonMind Consortium; Schizophrenia Working Group of the Psychiatric Genomics Consortium; iPSYCH-GEMS Schizophrenia Working Group , Gene expression imputation across multiple brain regions provides insights into schizophrenia risk. Nat. Genet. 51, 659–674 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Thul P. J., et al., A subcellular map of the human proteome. Science 356, eaal3321 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.