Significance
Annual and perennial species differ in their timing and intensity of flowering, but the underlying mechanisms are poorly understood. We hybridized closely related annual and perennial plants and used genetics, transgenesis, and genomics to characterize differences in the activity and function of their flowering-time genes. We identify a gene encoding a transcription factor that moved between chromosomes and is retained in the annual but absent from the perennial. This gene strongly delays flowering, and we propose that it has been retained in the annual to compensate for reduced activity of closely related genes. This study highlights the value of using direct hybridization between closely related plant species to characterize functional differences in fast-evolving reproductive traits.
Keywords: Arabis alpina, flowering, MADS AFFECTING FLOWERING, introgression lines
Abstract
The timing of reproduction is an adaptive trait in many organisms. In plants, the timing, duration, and intensity of flowering differ between annual and perennial species. To identify interspecies variation in these traits, we studied introgression lines derived from hybridization of annual and perennial species, Arabis montbretiana and Arabis alpina, respectively. Recombination mapping identified two tandem A. montbretiana genes encoding MADS-domain transcription factors that confer extreme late flowering on A. alpina. These genes are related to the MADS AFFECTING FLOWERING (MAF) cluster of floral repressors of other Brassicaceae species and were named A. montbretiana (Am) MAF-RELATED (MAR) genes. AmMAR1 but not AmMAR2 prevented floral induction at the shoot apex of A. alpina, strongly enhancing the effect of the MAF cluster, and MAR1 is absent from the genomes of all A. alpina accessions analyzed. Exposure of plants to cold (vernalization) represses AmMAR1 transcription and overcomes its inhibition of flowering. Assembly of the tandem arrays of MAR and MAF genes of six A. alpina accessions and three related species using PacBio long-sequence reads demonstrated that the MARs arose within the Arabis genus by interchromosomal transposition of a MAF1-like gene followed by tandem duplication. Time-resolved comparative RNA-sequencing (RNA-seq) suggested that AmMAR1 may be retained in A. montbretiana to enhance the effect of the AmMAF cluster and extend the duration of vernalization required for flowering. Our results demonstrate that MAF genes transposed independently in different Brassicaceae lineages and suggest that they were retained to modulate adaptive flowering responses that differ even among closely related species.
Annual and perennial species occur in many plant families. Annual plants and some perennials are monocarpic (flowering once in their life cycle), characterized by a massive flowering and typically produce many seeds before the whole plant senesces. By contrast, most perennials live for many years, show delayed reproduction, and are polycarpic (flowering multiple times in their life cycle) (1, 2). Therefore, annuals and perennials differ in the timing, duration, and intensity of reproduction. In both annuals and perennials, environmental cues regulate the timing of floral induction, the initial reproductive stage of higher plants, or the maturation of floral buds to ensure that flowers mature at the optimal time during the seasonal cycle to produce progeny and maximize fitness (1, 3, 4). Here, we use interspecies crosses between annual and perennial Brassicaceae species to characterize genetic differences that contribute to their distinct patterns of flowering-time control.
Annuals and perennials diverge in response to environmental pressures in relatively short evolutionary time scales, giving rise to sister annual and perennial species (4–6). Nevertheless, the genetic mechanisms underlying the divergence of these life histories are poorly understood. In Sorghum and Mimulus, a small number of genetic loci were described to have major effects in differentiating the life history of sister species (4, 7). By contrast, the separation of annual and perennial Oryza species was genetically complex (8). In Mimulus, a large inversion contributed to the divergence of annual and perennial species, which involved the differentiation of a locus with a large effect on flowering time (4, 9, 10). In the Brassicaceae, a key floral repressor gene is differentially transcribed between annuals and perennials, and this difference evolved several times to confer differences in the duration of flowering (5, 11, 12). Alterations in transcriptional patterns of key regulators have been shown to underlie rapid evolution of developmental traits in other systems and might play a broader role in divergence of annuals and perennials. In addition, gene regulatory networks can diverge rapidly through duplication or deletion of genes that encode central regulators of phenotypic traits (13, 14). Although this has not been described in the context of annual and perennial species, the reduction in genome size and genomic alterations that occurred during the evolution of annual Arabidopsis thaliana L. from its perennial progenitor, suggests that differences in gene content might also contribute to the evolution of annualism (15).
We have used the Arabis genus of the Brassicaceae as a model system to study divergence of annual and perennial species. Arabis alpina L. was established as a model perennial species because it is amenable to forward genetics (12) and subsequently, its sequenced genome was assembled (16, 17). Phylogenetic reconstruction showed that this species is sister to annual Arabis montbretiana Boiss, which enables comparisons between closely related annuals and perennials (18). Furthermore, because A. alpina belongs to the same family as A. thaliana, regulatory pathways that have been described in detail in A. thaliana can be relatively easily tested for their conservation or divergence in A. alpina. Flowering of the reference accession A. alpina Pajares only occurs after exposure to an extended cold period that mimics winter conditions, called vernalization. Characteristic perennial flowering patterns have been described in A. alpina. For example, the plant flowers after vernalization but then reverts to vegetative growth, which limits the duration of a flowering episode (12). The PERPETUAL FLOWERING 1 (PEP1) gene (12), which encodes a MADS-domain transcription factor orthologous to A. thaliana FLOWERING LOCUS C (FLC) (19, 20), plays a central role in conferring these traits. In the reference accession A. alpina Pajares, PEP1 represses flowering prior to vernalization, is transcriptionally repressed during cold treatment when flowering occurs, and is reactivated after exposure to cold to restrict the duration of flowering. This reactivation does not occur to the same extent in annuals such as A. montbretiana and A. thaliana, allowing them to flower indefinitely (5, 11, 19–22).
The construction of introgression lines (ILs) is a powerful genetic approach to identify genes that confer phenotypic differences between related species. In these lines, chromosomal segments from a donor parent are introduced by hybridization and backcrossing into a recipient parent. The effect of donor-parent chromosomal segments on the phenotypes of the recipient parent can then be determined. ILs can subsequently be used to rapidly develop secondary F2 populations for positional cloning of causal genes and quantitative trait loci (QTL) that underlie phenotypes of interest, including flowering time (23, 24). To facilitate the genetic study of traits modified during the divergence of annuals and perennials, an introgression line population was developed after hybridization of A. alpina and A. montbretiana (5, 11). The annual A. montbretiana was used as donor parent. Flowering of this species is accelerated by vernalization, but it does flower without vernalization (5, 11). Chromosomal segments from the annual donor parent were introduced into the perennial background, using the obligate vernalization requiring A. alpina Pajares genotype or the pep1-1 mutant (12), and the plants screened for altered phenotypes related to flowering. Here we describe the characterization of one flowering locus identified by this approach.
We used transgenesis, long-read genomic sequencing, and RNA-sequencing (RNA-seq) to study a locus of A. montbretiana that strongly delays flowering of A. alpina. We identify a gene related to the MADS AFFECTING FLOWERING (MAF) cluster of floral repressors that has transposed to a new location in the Arabis genus. This transposed MAF-RELATED gene is present in the annuals A. montbretiana and Arabis nova subsp. Iberica Mart. ex Talavera, but absent from perennial A. alpina. We analyze the function and evolution of this gene and discuss the broader diversification of MAF genes in the Brassicaceae and the relevance of the transposed copy to the divergence of life history.
Results
Identification of A. montbretiana MAR Genes that Delay Flowering of A. alpina.
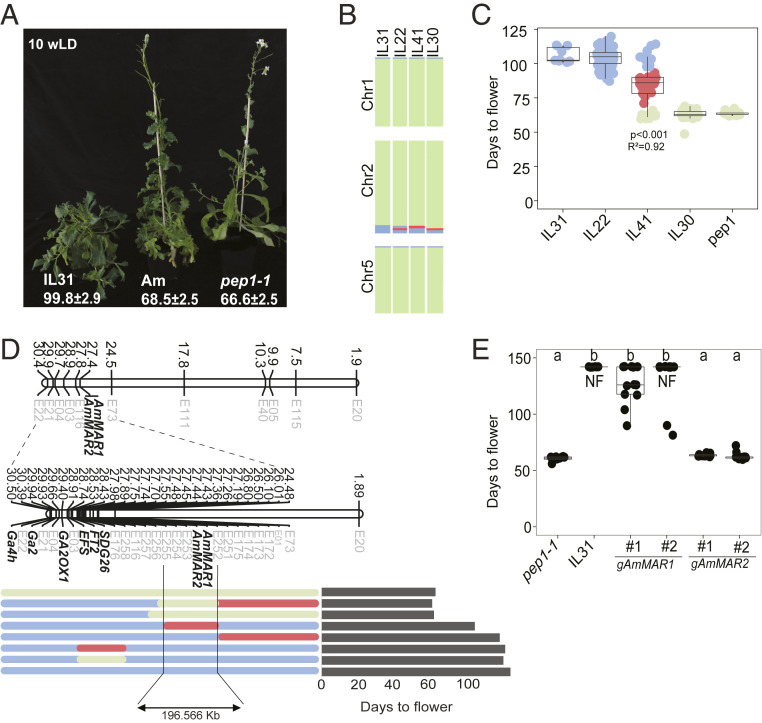
The interspecific introgression library obtained by crossing annual A. montbretiana and perennial A. alpina was screened for plants showing altered reproductive traits. Several near-isogenic lines (NILs) containing segments of chromosome 2 of A. montbretiana in the A. alpina pep1-1 background flowered much later after germination than the A. alpina pep1-1 parent (Fig. 1A and Materials and Methods). Three lines carrying partially heterozygous introgressed segments of A. montbretiana chromosome 2 were selected (IL22, IL30, and IL41) for association studies (Fig. 1B). In addition, a line with a homozygous introgression (IL31) was used as a late-flowering control. Each line was self-fertilized and the progeny were scored for flowering time and genotyped using molecular markers designed on the basis of polymorphisms between A. alpina and A. montbretiana (Fig. 1C and Dataset S1). These data indicated that IL31 and IL22 were homozygous for the locus causing late flowering, whereas IL41 was heterozygous and IL30 did not contain the locus (Fig. 1C). Thus, the A. montbretiana locus that conferred late flowering was present within the heterozygous segment of introgressed DNA in IL41 that was absent in IL30 (Fig. 1B). Furthermore, the late-flowering phenotype segregated among the progeny of IL41 in a 1:2:1 ratio, demonstrating that it is caused by a single codominant locus (P < 0.01, df = 2, χ2 = 0.142, n.s. [non-significant]).
Fig. 1.
Phenotypic characterization of late-flowering plants identified from introgressing A. montbretiana genomic segments into A. alpina pep1. (A) Plants grown for 10 wk after germination. Days to flowering (DTF) after gemination are indicated on the Bottom. IL31 was later flowering than A. montbretiana (Am) and A. alpina pep-1-1. (B) Schematic representation of the genotypes of the IL used for association analysis. Different lines segregating for different fragments of chromosome 2 were chosen. Only chromosomes with introgressions from A. montbretiana are represented. (C) DTF for the ILs represented in B. n = 10 in parental lines, 70 in ILs. Only IL41 segregated for the flowering-time phenotype. The total phenotypic variation and P value are indicated for IL41. Individual plants are represented and color coded by the genotype of the most-associated molecular marker. (D) Physical map of the candidate region, showing markers (faint font) and flowering-time genes (bold font), as well as their physical positions on A. montbretiana chromosome 2 in megabases. The genotype of informative recombinants on chromosome 2 and the flowering time of each line is indicated on the Right. The candidate region that confers late flowering is located between markers E252 and E255, comprising ∼196 kb. (E) DTF of transgenic plants containing the genomic locus of each candidate gene in pep1-1. Only plants containing AmMAR1 showed a late-flowering phenotype. n = 10 to 12 plants. NF, non flowering plants at the end of the experiment. Flowering phenotype is measured in days from germination to the first open flower. Letters indicate statistically significant differences determined by multiple pairwise comparisons using Tukey’s least significant difference (LSD) test (P ≤ 0.05). In all panels, alleles from the recurrent parent (A. alpina pep1-1) are colored green, alleles from the donor (A. montbretiana) are in blue, and heterozygous regions are marked in red.
The position and size of the introgressions were defined at higher resolution by whole-genome sequencing of IL31 and IL41 (Dataset S2). Comparison of the introgressed A. montbretiana sequences in these lines allowed the identification of a genomic segment of about 1,617,567 bp that was associated with late flowering. This introgressed segment replaced a region of 2,339,533 bp in the A. alpina genome (Dataset S2). Analysis of recombinants identified in the progeny of IL41 allowed the region carrying the locus to be positioned between markers E252 and E255 on chromosome 2 of A. montbretiana (Fig. 1D and Dataset S1), a region of 196,566 bp. Comparison of the genomic sequences of this region with the orthologous region of A. alpina chromosome 2 revealed that 50 A. alpina genes were replaced by 41 genes from A. montbretiana. The A. montbretiana genes included a tandem duplication of two genes encoding MADS-domain transcription factors that were related to MADS AFFECTING FLOWERING proteins of A. thaliana. MAFs were previously shown to be repressors of floral transition (25, 26), but their locations on chromosome 1 (FLOWERING LOCUS M/MAF1) and chromosome 5 (MAF2–MAF5) of A. thaliana are not syntenic with the two genes on chromosome 2 of A. montbretiana. Therefore, we named the A. montbretiana genes AmMAF-RELATED (MAR) 1 and AmMAR2.
To test whether the AmMAR genes caused late flowering of pep1-1, transgenic plants were obtained that carried the genomic locus of each gene. Two independent T3 homozygous lines containing a single-locus insertion were selected for each gene construct. The transgenic pep1-1 plants carrying the genomic locus of AmMAR1 showed strongly delayed flowering, whereas those carrying AmMAR2 did not (Fig. 1E and SI Appendix, Fig. S1A). In addition, AmMAR1 transgenic plants flowered first on secondary shoots, and growth of the main shoot arrested in most plants. Therefore, the A. montbretiana AmMAR1 gene confers late flowering in the pep1-1 background, and is likely responsible for the late-flowering phenotype of the introgression lines.
MAR Genes Are Inactive in A. alpina Pajares and Arose in the Arabis Genus.
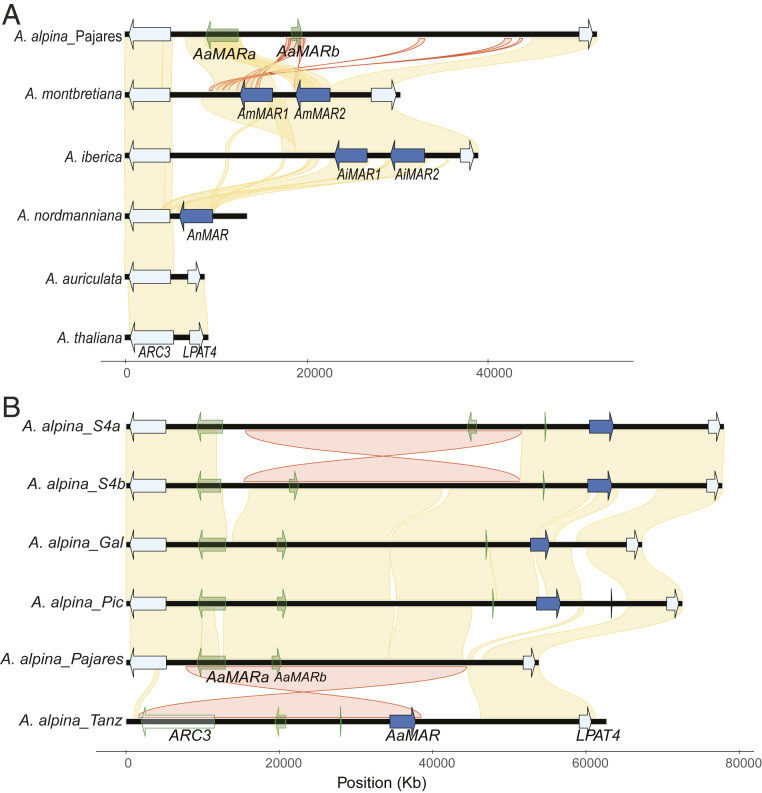
To understand why introgression of the AmMAR genes caused late flowering of A. alpina pep1-1, and to determine the evolutionary divergence of the MAR locus, the genome sequences of A. montbretiana and A. alpina Pajares were compared with those of the annuals A. nova subsp. Iberica, Arabis auriculata Lam., Arabis nordmanniana Rupr., and A. thaliana. The synteny analysis considered the common flanking genes between all species: AtARC3 (ACCUMULATION AND REPLICATION OF CHLOROPLAST 3, AT1G75010) and AtLPAT4 (LYSOPHOSPHATIDYL ACYLTRANSFERASE 4, AT1G75020). This analysis revealed that neither A. thaliana nor A. auriculata, which is a member of a sister clade to A. alpina (18), contains sequences related to MAR1 and MAR2 between the orthologs of AtARC3 and AtLPAT4 (Fig. 2A). In A. montbretiana, AmMAR1 and AmMAR2 are present as a tandem duplication between AtARC3 and AtLPAT4 (Fig. 2A). The same genome structure is observed in A. nova subsp. Iberica, which is closely related to A. montbretiana. In A. alpina Pajares, the reference accession for the species (16, 17), the orthologs of AtARC3 and AtLPAT4 are ∼40 kb apart, which includes two sequences related to MAR genes. However, within this interval no MAR genes predicted to encode full-length proteins were detected. The absence of active MAR genes in A. alpina Pajares might explain why the introgression of the active AmMAR genes causes late flowering of A. alpina pep1-1, while the presence of MAR pseudogenes that do not encode full-length proteins suggests that active MAR genes were lost in A. alpina Pajares following divergence from the lineage leading to A. montbretiana. The tetraploid species A. nordmanniana diverged from A. montbretiana after A. auriculata (18). Although no contig containing ARC3 and LPAT4 was found in the A. nordmanniana genome, analysis of short-read sequences showed that this species contains at least one gene closely related to AmMAR1 and AmMAR2 that is physically linked to AnARC3 (Fig. 2A). Finally, no MAR genes were detected in any genome available from more distantly related Brassicaceae species. Therefore, these analyses indicate that the MAR genes arose after divergence of the A. alpina/A. montbretiana/A. nordmanniana lineage from A. auriculata and were subsequently lost in A. alpina Pajares.
Fig. 2.
Comparison of MAR gene clusters among Arabis species and A. alpina accessions. (A) Synteny analysis between A. thaliana and different Arabis species. MAR genes arose after A. auriculata diverged in the Arabis clade. (B) Synteny analysis for A. alpina accessions collected across the geographical range of the species. A copy of a MAR gene that is predicted to be functional was identified. This copy is absent in the A. alpina Pajares accession. Common genes with Arabidopsis (ARC3 and LPAT4) were taken as flanking genes for the analysis. Syntenic regions are colored in yellow and inversions in red. The accessions were collected in their natural habitats: A. alpina_S4a and S4b are from Scandinavia; A. alpina_Gal and Pic are from the French Alps; A. alpina_Pajares is the Spanish A. alpina reference accession, and A. alpina_Tanz is from Tanzania. In both panels, genes shown by arrows outlined or filled in with green represent predicted truncated nonfunctional proteins.
The presence of two MAR pseudogenes in A. alpina Pajares (AaMARa and AaMARb) at the syntenic position to AmMAR1/2 (Fig. 2A) raised the possibility that other accessions of A. alpina might retain active MAR genes. To test this, the region between AaARC3 and AaLPAT4 was assembled from five other accessions of A. alpina using PacBio reads (Fig. 2B). These accessions were collected across the wide geographical range of the species and included one accession from Tanzania, two from Scandinavia, and two from France, whereas Pajares was collected in northern Spain. Analysis of the genome segments of these accessions showed that they all contained one AaMAR gene that encoded a full-length protein (AaMAR). In addition, all European accessions contained both pseudogenes present in Pajares, whereas the Tanzanian accession contained a single pseudogene (Fig. 2B). This analysis suggests that most A. alpina accessions contain an active AaMAR gene, and that the Pajares lineage probably lost it recently.
MAR Proteins Are Most Closely Related to the MAF1-LIKE Clade.
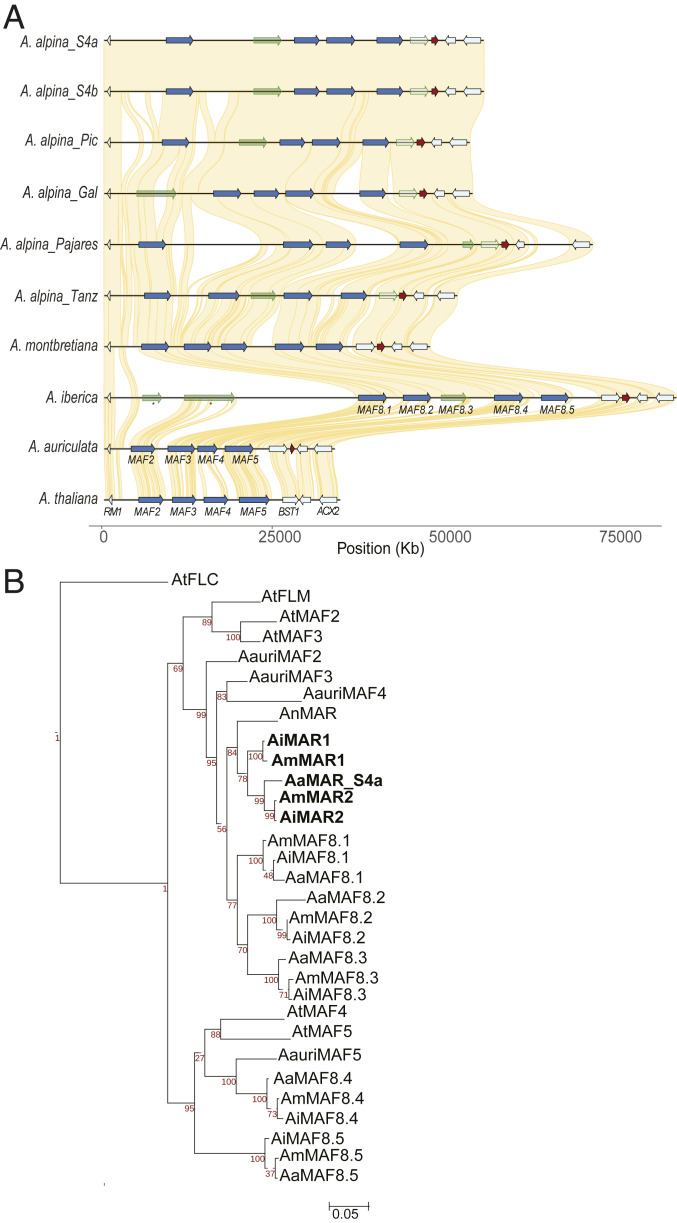
The MAF genes of A. thaliana consist of a tandem array of four genes on chromosome 5 (MAF2–MAF5) and MAF1 (also called FLM) on chromosome 1. To determine whether these are conserved within the Arabis genus, the MAF clusters at the syntenic position on chromosome 8 of A. montbretiana, A. nova subsp. Iberica, A. auriculata, and six accessions of A. alpina were analyzed (Fig. 3A). A. auriculata contained four MAF genes in the cluster in a similar arrangement to that in A. thaliana, whereas A. montbretiana contained five full-length genes. A. nova subsp. Iberica and the A. alpina accessions all contained the five genes orthologous to those of A. montbretiana, but in each genome, at least one gene did not encode the full-length protein (Fig. 3A). Thus, the MAF cluster is conserved in Arabis, with some variation in copy number among species. No MAF1 ortholog was detected in any Arabis species.
Fig. 3.
Dynamic variation in the MAF cluster among A. alpina accessions and related species. (A) Synteny analysis of the MAF cluster located on chromosome 8 from different accessions collected across the geographical range of A. alpina. Genes outlined or filled in green represent genes predicted to code nonfunctional proteins. Arabis MAF genes were numbered following their order on the chromosome. Common genes were taken as flanking genes for the analysis. A. alpina accessions were described in Fig. 2. (B) Maximum likelihood tree based on nucleotide alignment of the coding sequences of FLC clade members. Abbreviations: At (A. thaliana), Aauri (A. auriculata), An (A. nordmanniana), Ai (A. nova subsp. Iberica), Am (A. montbretiana), and Aa (A. alpina).
To assess the relatedness of the MAR genes to the MAF genes present in the chromosome 8 cluster, a phylogenetic tree was constructed using the coding sequences of the genes from the MAF cluster of A. thaliana and the Arabis species, as well as the MAR genes of A. montbretiana, A. nova subsp. Iberica, A. nordmanniana, and A. alpina (Fig. 3B). The MAF genes clustered into two major clades, one represented by MAF1/2/3 and the other by MAF4/5 of A. thaliana, as previously described (27). All of the Arabis species contained MAF genes in each clade as observed in other Brassicaceae species, suggesting that the biological functions represented by both clades are widely conserved within the family. A. montbretiana contained three genes in the MAF1/2/3 clade and two in the MAF4/5 clade (Fig. 3 A and B). In five of the A. alpina accessions, one of the genes in the MAF1/2/3 clade, either MAF8.1, MAF8.2, or MAF8.3, seems to be mutated and is predicted to encode a truncated protein, whereas in A. alpina Pajares, MAF8.5 from the MAF4/5 clade was mutated (Fig. 3 A and B). A. auriculata contained only one gene in the MAF4/5 clade, suggesting this may have duplicated in A. montbretiana after divergence from A. auriculata, and that duplication occurred independently in the A. thaliana lineage (Fig. 3B).
The MAR genes are located within the clade containing A. thaliana MAF1/2/3. Therefore, the MAR genes probably arose in the Arabis lineage after divergence of A. auriculata and A. alpina/A. montbretiana by transposition of a gene from the MAF1/2/3 group (referred to below as MAF1-LIKE) from the chromosome 8 cluster to chromosome 2. The A. nordmanniana MAR gene is present in a separate subclade to AmMAR1/2, suggesting that they arose by tandem duplication after the divergence of A. montbretiana from the lineage leading to A. nordmanniana. By contrast, the active MAR gene present in most A. alpina accessions clearly associates with AmMAR2.
Divergence of the MAR Genes from the MAF Cluster.
To understand the evolution of MAF/MAR gene sequences, the dN/dS ratio (ω) was compared among different lineages (28). Only the branch leading from AnMAR to the rest of the MAR cluster was statistically supported as showing a variable evolutionary rate compared to all other branches and showed a ω-value greater than 1 (SI Appendix, Fig. S2 and Dataset S3), supporting the notion that the clade containing most of the MAR genes is under positive selection. Furthermore, when the amino acid sites in the proteins were analyzed, the branch leading from AnMAR to the other MAR genes was again significantly different, indicating that some amino acids might be under positive selection. Among these, V43I and D61S have the highest values (Dataset S4 and SI Appendix, Fig. S3). These sites are within the MADS domain that is required for DNA binding. Therefore, the MAR1 and MAR2 genes of A. montbretiana and Arabis nova subsp. Iberica may have diversified from AnMAR by selection at specific residues.
The residues 43I and 61S are identical in AmMAR1 and AmMAR2, therefore variation at these residues does not explain the distinct functions of AmMAR1 and AmMAR2. AmMAR1 and AmMAR2 contain 20 nonsynonymous mutations, including 5 and 12 in the MADS-box and the K-box domains, respectively (SI Appendix, Fig. S4). The divergent region in the K domain is within the leucine-zipper motif of the second helix of the domain, and the positions of two leucine residues (position 119 in AmMAR2, and 124 in AmMAR1), previously proposed to be involved in dimerization of SEP3 (29), are altered. Thus, these changes might affect the ability of AmMAR1 and AmMAR2 to interact with partner proteins and explain the apparent specificity of AmMAR1 in delaying flowering.
MAR1 Represses Flowering of the Primary Inflorescence, Reduces Shoot Elongation, and Confers a Vernalization Response.
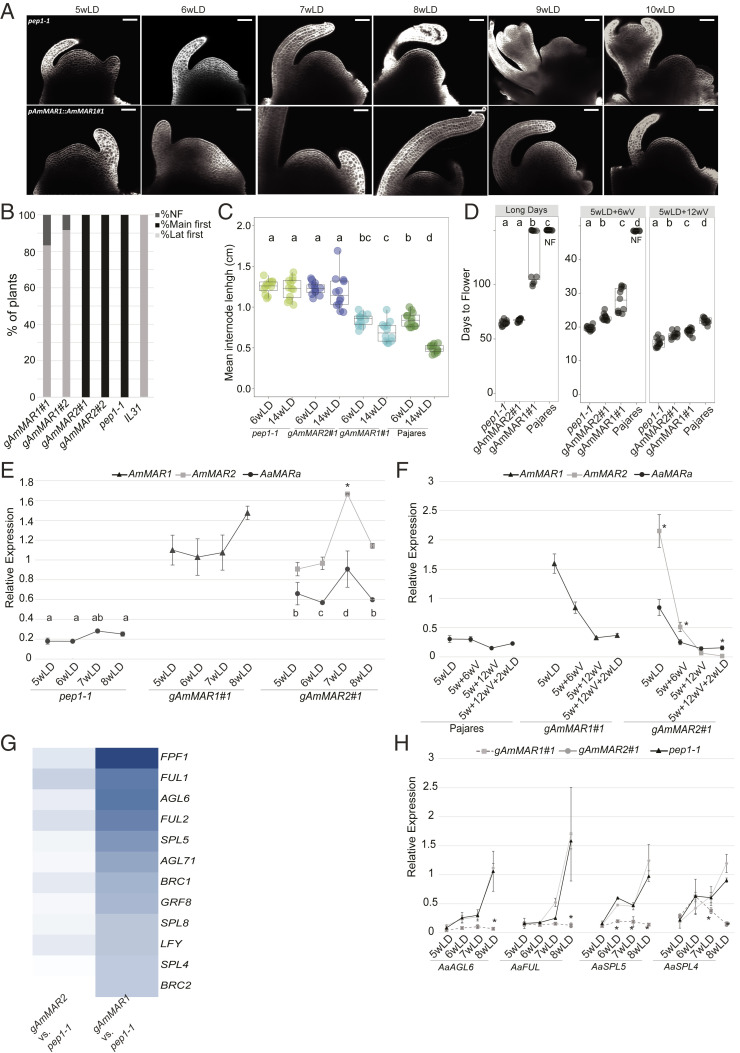
The role of AmMAR1 in the repression of floral transition was characterized in more detail. Microscopic analysis of the shoot apical meristem demonstrated that pep1-1 formed well-developed floral primordia by 9 wk after germination, whereas the morphology of the shoot apex of pAmMAR1::gAmMAR1 plants remained vegetative 10 wk after germination (Fig. 4A). In pep1-1 and pAmMAR2::gAmMAR2 transgenic plants, flowers always appeared first at the shoot apex, whereas most pAmMAR1::gAmMAR1 plants produced flowers and seeds from secondary inflorescences, and no flowers were visible at the shoot apex (Fig. 4B and SI Appendix, Fig. S1B). Therefore, the repression of flowering caused by AmMAR1 appears to be stronger at the shoot apical meristem than on lateral branches. In addition to delaying flowering, AmMAR1 reduced plant height and the length of internodes in the primary shoot (Fig. 4C and SI Appendix, Fig. S1C). Internode length of wild-type A. alpina Pajares plants was also shorter than that of pep1-1 mutants (Fig. 4C) (30), indicating that AmMAR1 and PEP1 have similar effects on plant height as well as flowering time.
Fig. 4.
AmMAR1 represses floral transition of pep1-1 mutants under LD. (A) Meristem morphology in pep1-1 and gAmMAR1::gAmMAR1 at different time points in long-day conditions. Transgenic plants carrying AmMAR1 are not induced to flower after 10 wk in long days, when pep1-1 mutants have well-developed floral buds. (Scale bar: 50 µm.) (B) Percentage of plants flowering first on the main and lateral shoots. Plants carrying AmMAR1 flowered mainly on secondary lateral shoots, whereas pep1-1 and AmMAR2 plants flowered first on the main shoot. Lat. first, first flower formed on lateral shoot; main first, first flower formed on main shoot; NF, not flowering. (C) Mean internode length for Pajares and late-flowering transgenic plants carrying AmMAR1, compared with that of pep1-1 and pAmMAR2::gAmMAR2 after 6 and 14 wk in long days. Late-flowering plants had shorter internodes. The error bars represent the SD; n = 12. (D) Comparison of the flowering time of transgenic MAR lines, perennial A. alpina Pajares, and pep1-1 without vernalization in long days and after two vernalization periods. pAmMAR1::gAmMAR1 flowers late without vernalization, the reference Pajares never flowers without vernalization, and pep1-1 flowers perpetually. The differences in flowering time are strongly reduced by vernalization and are smaller after vernalization for 12 wk. (E) Levels of AaMARa, AmMAR1, and AmMAR2 mRNA in the main inflorescence apex of AmMAR1 and AmMAR2 transgenic lines and pep1-1 growing for 8 wk in long days. (F) The level of mRNA of PEP1, AaMARa, AmMAR1, and AmMAR2 in apices without vernalization or on exposure to different vernalization periods. Expression of AmMAR genes is strongly repressed after 6 wk of vernalization (n = 12). (G) Heat map of DEGs for flowering time according to the log2-fold change (log2FC) values for pAmMAR1::gAmMAR1 and pAmMAR2::gAmMAR2. All DEGs are listed in Dataset S3. (H) Expression level of selected DEGs in shoot apices after growth for 5 to 8 wk in long days. The data represent the means of two biological replicates, and error bars represent the SD. Asterisks above or below the datapoints indicate significant differences determined by multiple pairwise comparisons using Tukey’s honestly significant difference (HSD) test (P ≤ 0.05). qPCR data are the mean of two biological replicates, and error bars represent the SD. Asterisks above the datapoints indicate significant differences determined by multiple pairwise comparisons using Tukey’s HSD test (P ≤ 0.05).
The effect of MAF genes on flowering of A. thaliana can be overcome by vernalization (26); therefore, the flowering time of the MAR transgenic plants was tested after vernalization. Five-week-old pAmMAR1::gAmMAR1 plants vernalized for only 6 wk flowered within 32 d after vernalization, whereas the Pajares reference accession did not flower (Fig. 4D). After this short vernalization treatment, pAmMAR1::gAmMAR1 all flowered at the shoot apex and on lateral branches, although pAmMAR1::gAmMAR1 flowered later than pAmMAR2::gAmMAR2 and pep1-1 plants and pAmMAR2::gAmMAR2 flowered slightly later than pep1-1 (Fig. 4D). After 12 wk of vernalization, Pajares also flowered and the differences in flowering among genotypes were smaller (Fig. 4D). Thus, the strong repression of flowering caused by pAmMAR1::gAmMAR1 can be largely overcome by short vernalization treatments of 6 wk.
To understand further the function of AmMAR1 and AmMAR2, their mRNA levels were analyzed in different tissues and at various times after germination. Each mRNA was highly expressed in the corresponding transgenic plant in all tissues and time points tested, with the lowest expression in cotyledons (SI Appendix, Fig. S5A). In pep1-1, the AmMAR1 primers amplified the transcript formed from one of the A. alpina Pajares pseudogenes, and the abundance of this transcript was greater in pAmMAR2::gAmMAR2 transgenic plants, suggesting that AmMAR2 can directly or indirectly activate this gene (SI Appendix, Fig. S5A). Apices of the transgenic plants and pep1-1 were then analyzed between 5 and 8 wk after germination, when the plants underwent floral induction. The mRNA level of AmMAR1 and AmMAR2 remained high throughout the time course (Fig. 4E), demonstrating that AmMAR2 expression is not reduced during floral induction of pAmMAR2::gAmMAR2 plants, and that AmMAR1 mRNA is present in apices of pAmMAR1::gAmMAR1 plants when their phenotypes start to diverge from those of pep1-1. In the first few lateral shoots of the primary stems of the transgenic plants, high levels of AmMAR1 and AmMAR2 mRNAs were also detected, similar to those found in the primary shoot apex at these time points (SI Appendix, Fig. S5B).
The delay in flowering of pAmMAR1::gAmMAR1 compared with pep1-1 was overcome by vernalization (Fig. 4D); therefore, the mRNA levels of AmMAR1 and AmMAR2 were quantified in apices before and after exposure to 6 and 12 wk of vernalization (Fig. 4F). The expression of both genes was strongly repressed by the end of the 12-wk vernalization treatment (Fig. 4F) and remained low 2 wk after vernalization. After 6 wk of vernalization, AmMAR1 and AmMAR2 mRNA abundance was strongly reduced (Fig. 4F), indicating that the early flowering of pAmMAR1::gAmMAR1 after vernalization is probably due to repression of AmMAR1 transcription.
Effects of AmMAR1 on Gene Expression.
To determine the effect of AmMAR1 on gene expression at the shoot apex, the transcriptomes of both transgenic lines and pep1-1 were determined by RNA-sEq 6 wk after germination. At this time point, the apex of the pep1-1 mutant was at an early stage of floral transition, whereas pAmMAR1::gAmMAR1 transgenic plants remained vegetative (Fig. 4A). As expected, more differentially expressed genes (DEGs) were found between pAmMAR1::gAmMAR1 and pep1-1 than between pAmMAR2::gAmMAR2 and pep1-1, and only 17 were common between both comparisons (SI Appendix, Fig. S6A and Dataset S5). We performed Gene Ontology (GO) enrichment analysis for the 167 DEGs in pAmMAR1::gAmMAR1. GO terms related to meristem maintenance, positive regulation of flower development, and regulation of the vegetative phase, were significantly enriched in this analysis (SI Appendix, Fig. S6B). Notably, expression of most DEGs was reduced in pAmMAR1::gAmMAR1 compared with pep1-1 (Fig. 4G). One of the most down-regulated genes in pAmMAR1::gAmMAR1 was FLOWERING PROMOTING FACTOR 1 (AaFPF1), which in A. thaliana is up-regulated during floral transition and causes early flowering when overexpressed from a heterologous promoter (31). Furthermore, the expression of several genes encoding orthologs of transcription factors with established roles in floral induction and floral meristem identity of A. thaliana was also reduced in pAmMAR1::gAmMAR1 (Dataset S5). These data indicate that MAR1 represses the expression of many genes involved in the early stages of floral transition.
To test differences in the dynamics of flowering-time gene expression, RNA was harvested from apices of pAmMAR1::gAmMAR1, pAmMAR2::gAmMAR2, and pep1-1 plants grown for 5 to 8 wk under long days. Four DEGs identified by RNA-seq were analyzed by qRT-PCR in all three genotypes across the time course in main and lateral shoots (Fig. 4H and SI Appendix, Fig. S6C). The mRNA abundance of AaFUL, AaAGL6, AaSPL4, and AaSPL5 all increased during the time course in pAmMAR2::gAmMAR2 and pep1-1, but remained at low levels in pAmMAR1::gAmMAR1 (Fig. 4H). In lateral shoots, a slight increase in AaAGL6, AaSPL4, and AaSPL5 was observed in the final time point (SI Appendix, Fig. S6C). These data support the conclusion that AmMAR1 strongly blocks the early stages of the floral transition at the primary shoot apex.
Comparative Analysis of the Responses of MAF and MAR Gene Expression to Vernalization in A. thaliana, A. montbretiana, and A. alpina.
In A. thaliana, MAF genes extend the duration of vernalization required for flowering and differ in their rate of repression by vernalization (25, 26, 32). However, the expression of all MAF genes during vernalization has not been quantitatively tested and the extent to which their responses to vernalization are conserved in different Brassicaceae species is unknown. To address these issues, RNA-seq was used to determine the mRNA levels of FLC, MAF, and MAR genes during vernalization in A. thaliana, A. montbretiana, and A. alpina Pajares, as well as in pep1-1 mutants not exposed to vernalization (Fig. 5 and Dataset S6).
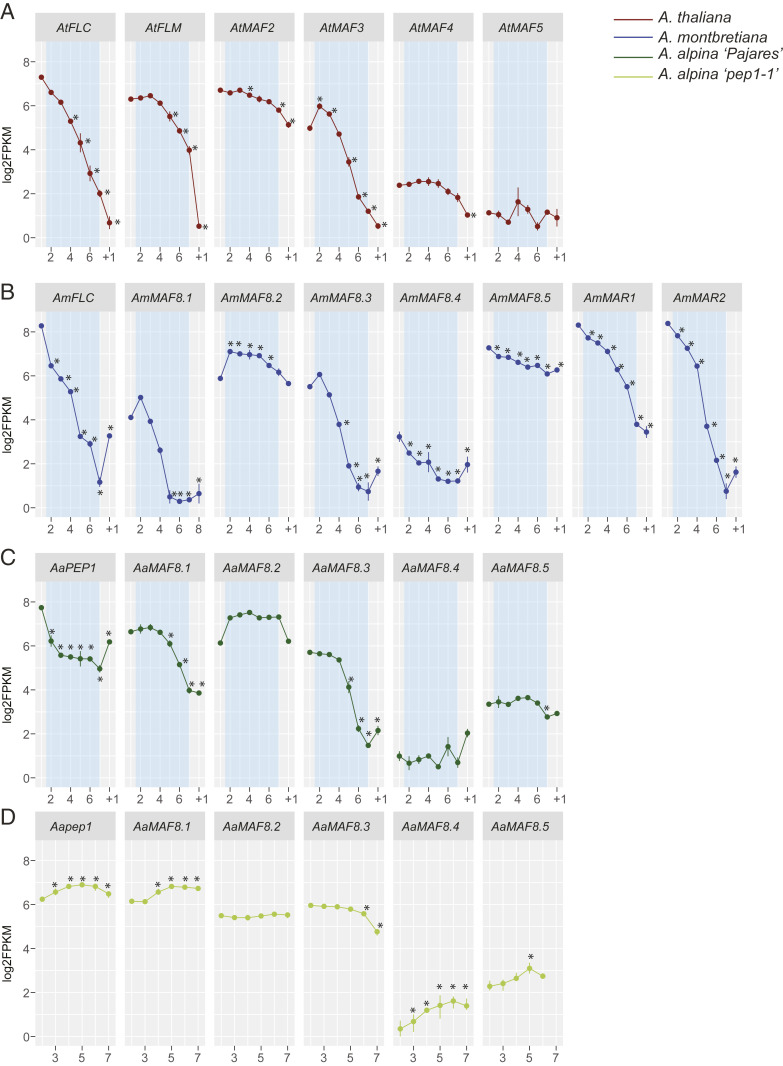
Fig. 5.
The transcriptional dynamics in different genotypes of MAF and MAR gene expression in response to vernalization. Log2 FPKM (fragments per kilobase of transcript per million mapped reads) for orthologs of FLC, MAF, and MAR genes before, during, and after vernalization for A. thaliana (A), A. montbretiana (B), and A. alpina Pajares (C), and A. alpina pep1-1 growth in long days with no vernalization harvested weekly from 2 to 7 wk after germination (D). For A–C, shoot apices were harvested before vernalization (2 wk for Arabidopsis and A. montbretiana or 6 wk for A. alpina Pajares), during vernalization treatment (2, 4, 6, 8, 10, and 12 wk), and 1 wk after plants were returned to warm temperatures. The blue box indicates the duration of vernalization. The genes are ordered according to their chromosomal position and not their evolutionary relationships. Asterisks indicate significant differences calculated using DESeq2 of the fold changes versus the first time point. AaMARa, which does not encode an active protein, is not represented.
In A. thaliana, the mRNAs of AtFLC and AtMAF1/2/3 were present at high levels prior to vernalization, and the mRNA abundance of AtFLC, AtMAF1 (FLM), and AtMAF3 decreased rapidly in cold, whereas that of AtMAF2 decreased more slowly. By contrast, AtMAF4/5 mRNAs were expressed at lower levels, and the AtMAF4 mRNA level was reduced slowly by cold (Fig. 5A). These results are in broad agreement with previous reports (25, 26). Remarkably similar patterns were detected in A. alpina and A. montbretiana, although with significant differences (Fig. 5 B and C). In A. montbretiana, expression of AmFLC, two of the AmMAF1-LIKE genes (AmMAF8.1 and AmMAF8.3), and AmMAR2 was rapidly reduced during vernalization, whereas expression of AmMAR1 and AmMAF8.2 fell more slowly (Fig. 5B). The mRNAs of the MAF4-LIKE genes also reduced only slowly during vernalization, similar to that of the A. thaliana genes in this group. In A. alpina, the PEP1 and AaMAF1-LIKE genes were generally less responsive to vernalization (Fig. 5C). During vernalization, the abundance of PEP1 and AaMAF8.1 mRNA was reduced slowly and was still relatively high after the 12-wk vernalization treatment (Fig. 5C). In the pep1-1 mutant, which was not exposed to vernalization, the expression of AaMAF8.1, -8.2, and -8.3 was almost unchanged through the time course, showing that the repression of these genes in A. alpina Pajares during vernalization is due to exposure to cold (Fig. 5D). Collectively, these results indicate that in different lineages of the Brassicaceae, FLC and most of the MAF1-related genes are highly expressed prior to vernalization and repressed during vernalization, although the rate of reduction may differ between species, as observed in the relatively slow rate of repression of these genes in A. alpina. In addition, the transcriptional patterns of the AmMAR genes are most similar to those of the MAF1-LIKE genes, consistent with their position in the phylogeny, but AmMAR1 is expressed more highly than the AmMAF genes prior to vernalization and is repressed by vernalization at a slower rate than AmFLC or AmMAR2.
Discussion
We used interspecies hybridization, long-read genomic sequencing, and RNA-seq to identify a tandem array of genes encoding MAR MADS-domain transcription factors that arose in the Arabis genus, strongly repress flowering at the shoot apex, and differ in their arrangement among closely related species as well as among accessions of A. alpina. Our data emphasize the importance of transposed MAF gene copies in flowering-time regulation and show that these have arisen independently in different lineages of the Brassicaceae. We discuss our data in the context of duplication and retention of plant genes and their significance in life-history divergence.
Tandem Duplication and Transposition of MAF Genes in the Brassicaceae.
Gene duplication contributes to the appearance of novel traits during plant evolution (33–35). Duplicates arise by whole-genome duplication or different mechanisms of single-gene duplications such as transposition or unequal crossover at meiosis to create a tandem duplication. Tandem duplicates have often been misannotated as single-copy genes because of the difficulty of assembling them from short-read sequencing data (33). The PacBio long-read sequences used here enabled the assembly of the tandem arrays of MAF and MAR genes from several Arabis species. The MAF gene cluster occurs at a syntenic position to the one in A. thaliana (26) and Brassica rapa (27), whereas the MAR cluster arose by transposition within Arabis.
Ancestral genome and phylogenetic reconstructions indicated that the MAF gene cluster arose from tandem ancestral MAF1-LIKE and MAF4-LIKE genes that were present in the stem group of the core Brassicaceae (27). In A. thaliana, four MAF genes are present in the cluster (26, 36): two derived from the ancestral MAF1-LIKE gene and two from the MAF4-LIKE gene, and these duplications are proposed to have occurred in lineage I leading to A. thaliana (27). Among accessions of A. thaliana, naturally occurring mutations in this cluster, particularly in MAF2 and MAF3, cause earlier flowering (36, 37). In Arabis, which is in lineage IV of the Brassicaceae and diverged from the lineage leading to A. thaliana around 23 million years ago (38, 39), the MAF cluster has amplified independently. A. auriculata and A. alpina Pajares, the reference accession, each contain three genes derived from the ancestral MAF1-LIKE gene, and one derived from the ancestral MAF4-LIKE gene, whereas A. montbretiana contains an extra MAF4-LIKE gene that is also present in other accessions of A. alpina. In Brassica species in lineage II of the Brassicaceae, descendants of MAF1-LIKE and MAF4-LIKE are also present in the MAF gene cluster (27, 40). Therefore, amplification and loss of MAF genes within syntenic tandem arrays have occurred frequently and independently in different lineages of the Brassicaceae.
In addition to the tandem array of MAF genes, transposed copies occur on other chromosomes. In A. thaliana, AtMAF1 (also called AtFLM) is located on a different chromosome from the AtMAF cluster and confers late flowering (41, 42). AtMAF1 is proposed to have arisen by transposition of an ancestral MAF1-LIKE gene out of the AtMAF cluster. No ortholog of AtMAF1 was detected in the genomes of Brassica species, suggesting that transposition occurred in lineage I (27, 40). Similarly, in Arabis species, an ortholog of AtMAF1 is absent, but independent transposition of a MAF1-LIKE gene occurred, generating the MAR genes at a position that is not syntenic with AtMAF1. The transposed MAR gene then duplicated, creating a tandem array of AmMAR1 and AmMAR2 in A. montbretiana and A. nova spp. Iberica, although only AaMAR2 is present in A. alpina. The presence of transposed copies in Brassica is more difficult to determine because of the genome triplication and reassortment that occurred in that lineage. Nevertheless, our analysis of the Arabis genus clearly demonstrates that in addition to amplification of the syntenic MAF cluster, recent, independent transposition events have generated additional copies of MAF1-LIKE genes at different locations in separate Brassicaceae lineages.
Functions of MAF Genes and the Retention of Duplicate Copies.
Single-gene duplication occurs frequently through unequal crossover or transposition (33, 43), but most duplicates are subsequently lost and are either deleted from the genome or become pseudogenes (44, 45). The syntenic MAF clusters of all species tested in the core group of the Brassicaceae contain descendants of both the ancestral MAF1-LIKE and MAF4-LIKE genes, suggesting that these two gene lineages have distinct functions and are retained by selection (26, 40). The MAF1-LIKE genes are present in multiple copies in all genomes tested. Several explanations have been proposed for the retention of gene copies, including gene dosage, subfunctionalization, and neofunctionalization (33, 46). In A. thaliana, genetic analysis demonstrated that all of the members of the MAF1-LIKE group, AtMAF1, AtMAF2, and AtMAF3, as well as AtMAF4 in the MAF4-LIKE group, delay flowering, because mutation of each gene causes earlier flowering (25, 26). Notably, the single mutation with the strongest effect is maf1, in which the transposed copy is inactivated and the genes in the MAF cluster are still active (25, 41). Moreover, each maf mutation has a stronger early-flowering phenotype at 16 °C than at 23 °C, suggesting that the genes are particularly important in delaying flowering at low temperatures (25, 41). Similarly, the MAF genes were proposed to enhance the duration of vernalization required to promote flowering, because maf mutants flowered after exposure to shorter vernalization treatments (25, 26). Combining mutations in AtMAF1, AtMAF2, and the related floral repressor AtFLC caused an extreme early-flowering phenotype even at 16 °C (25). Thus, the AtMAF genes are partially redundant with each other and with AtFLC, suggesting that they have an additive effect on floral repression. However, they probably also have qualitatively distinct effects. For example, in contrast to transcription of AtFLC, that of AtMAF2 is repressed slowly or not at all by vernalization (26), and this may confer its capacity to extend the duration of vernalization required for flowering. Moreover, AtMAF1 and AtMAF2 transcripts are differentially spliced at higher temperatures, reducing the activity of the gene and allowing earlier flowering (32, 47, 48). Overall, the AtMAF genes play important roles in delaying and modulating flowering time in response to changes in temperature, in the context of vernalization or ambient temperature changes. Therefore, the retention of MAF genes after duplication might be a consequence of selection for altered flowering time in response to changes in environmental temperatures (27).
In Arabis, AmMAR1 confers extreme late flowering in pep1-1 mutants that also contain the full AaMAF cluster. Therefore, the transposed copy delays flowering more strongly than the ancestral MAF cluster, as described for AtMAF1 in an A. thaliana flc mutant (25). The AaMAF cluster alone has a relatively weak effect on flowering time in the absence of PEP1 and MAR1, as shown by the early-flowering phenotype of the pep1-1 mutant and our observation in RNA-seq analysis that the whole AaMAF cluster is expressed in apices of pep1-1 mutants (Fig. 5D).
Nonsynonymous amino acid changes and altered transcriptional patterns may contribute to the stronger delay of flowering caused by AmMAR1 in A. alpina than the ancestral AaMAF cluster. Genome-wide analyses in rice and A. thaliana previously demonstrated that dispersed gene duplicates, such as those generated by transposition, tend to be more diverged in gene expression pattern than tandem duplicates (49, 50). Consistent with these observations, in our RNA-seq analysis AmMAR1 transcripts were more abundant prior to vernalization than those of any of the other AmMAF genes and this increased expression may contribute to the stronger phenotypic effect of the transposed copy. However, we also found that the rate of nonsynonymous changes in the MAR clade after divergence from AnMAR was higher than in the ancestral MAF clade, consistent with accelerated protein evolution after transposition. Particularly, the described changes in the K domain between MAR1 and MAR2 may alter protein interactions and enhance the effect of MAR1 on flowering time.
Transgenic A. alpina pep1-1 mutants carrying AmMAR2 did not flower later than the parental plants without vernalization; however, these plants did flower slightly later than pep1-1 mutants after short vernalization periods. Therefore, MAR2 may have a weaker but significant role in modulating flowering time, comparable to genes in the AtMAF cluster of A. thaliana. MAR2 is also present in all A. alpina accessions (except Pajares), as well as in A. montbretiana and A. nova subsp. Iberica, suggesting that it has been retained by selection during the divergence and diversification of these species.
MAF Genes and the Divergence of Annual and Perennial Life History.
Flowering is generally more strongly repressed in perennial Brassicaceae species than in their annual counterparts; therefore, it was unexpected that annual A. montbretiana and A. nova subsp. Iberica have retained MAR1, a strong repressor of flowering, whereas the gene is absent in all accessions of perennial A. alpina tested. Whereas AmMAR1 effectively prevents flowering of pep1-1 on the main shoot, A. montbretiana flowers in the absence of vernalization when the AmMAR, AmMAF, and AmFLC genes would be expected to be expressed. How the effect of these repressors on floral transition is overcome in A. montbretiana is unknown, but probably other flowering pathways act independently of these repressors to bypass their effect on gene expression and floral transition. Such interactions among pathways have been extensively analyzed in A. thaliana (51). Extensive genetic analysis in A. montbretiana will be required to determine the effect of AmMAR1 loss of function, and this will require the development of transformation protocols and reverse genetics for this species.
The RNA-seq analysis suggests the selection pressure to retain MAR1 in the annuals may be explained by compensation among other members of the MAF family. In A. montbretiana, the MAF1-LIKE genes in the MAF cluster that are repressed by vernalization, AmMAF8.1 and AmMAF8.3, are expressed at relatively low levels prior to vernalization and are rapidly repressed during vernalization. By contrast, AmMAR1 is expressed four- to fivefold higher than these genes prior to vernalization and its expression is reduced more slowly during vernalization. Therefore, AmMAR1 may be retained in A. montbretiana to compensate for the lower level of expression of the related genes in the MAF cluster. Similarly, AtMAF2 was proposed to extend the duration of vernalization required for flowering of A. thaliana (25, 26). The requirement for MAF1 in A. alpina may be weaker because AaMAF8.1 is expressed at a higher level than its ortholog in A. montbretiana prior to vernalization, and its repression during vernalization is much more gradual. Applying CRISPR-Cas9 for reverse genetic analysis to test the contribution of MAF genes to flowering time and vernalization response in the Arabis species would help resolve the contributions of individual genes.
The FLC ortholog PEP1 is related to the MAF genes and plays a central role in the perennial life cycle of A. alpina by restricting floral induction to a short time period at the end of vernalization (12, 52). Our comparative RNA-seq analysis showed that the FLC orthologs are much more rapidly repressed by cold in the annuals A. thaliana and A. montbretiana than in perennial A. alpina, and that in the latter, PEP1 is still significantly expressed after 12 wk of vernalization. The duration of vernalization response in A. alpina may therefore be strongly determined by PEP1 expression, and this may ensure that the plant requires longer vernalization treatments for flowering to occur than in the annuals. This suggestion is consistent with previous observations that FLC alleles of A. thaliana that differ in the rate of repression by vernalization determine the duration of vernalization required for flowering (53), and that short vernalization treatments do not allow full inflorescence development in A. alpina (22). Our analysis of the FLC and MAF orthologs of these annual and perennial species, therefore, suggests that the selection pressure to retain MAR1 may be stronger in the annuals than in the perennial, because of differences in the patterns of expression of their respective FLC orthologs and MAF gene clusters. In this case, because PEP1 and vernalization response play a central role in the perennial life history of A. alpina, the absence of MAR1 in this species would be an indirect consequence of its perennial life history.
Materials and Methods
Plant Material and Growth Conditions.
The perennial A. alpina reference accession Pajares, the pep1-1 mutant, and annual A. montbretiana (accession BM7968, provided by Birol Mutlu, Turkey) were used as parent lines to generate the populations used in this study (12, 18). Parental A. montbretiana flowers without vernalization around 68 d after germination, but contains an active FLC ortholog, and its flowering time is accelerated by vernalization treatment (5, 11). The A. alpina Pajares parent shows an obligate vernalization requirement, while the pep1-1 mutant flowers without vernalization (12). An F1 population was obtained for the cross A. montbretiana × A. alpina Pajares and was then backcrossed to both Pajares and pep1-1. Up to 35 plants for each family were self-fertilized to generate BC1S1 seeds and genotyped by genotyping by sequencing (GBS). Using this information, 44 lines with introgressed fragments of A. montbretiana fixed in the pep1-1 background were obtained.
For flowering-time experiments, seeds were stratified in darkness for 3 to 5 d at 4 °C. Plants were then grown in the glasshouse under long days (LDs) (16 h light:8 h dark) at a light intensity of 200 to 500 µmol m−2 s−1 at 22 °C. Vernalization was performed in a short days (SDs) growth chamber at 4 °C and a light intensity of 14 µmol m−2 s−1. Days to flower (DTF) was measured for each genotype as days to the first open flower from germination. All experiments were performed with at least 12 plants.
Marker Development, Genotyping, and Whole-Genome Sequencing.
To estimate and characterize the annual introgressions, lines of interest were subjected to whole-genome sequencing using Illumina HiSeq3000 with150 bp (paired-end reads, Project no. PRJNA532504, biosamples SAMN18581157 [IL31] and SAMN18581158 [IL41]). The cleaned reads were mapped to the pooled genomes of A. alpina (Pajares) V5.1 (16) and A. montbretiana V3.1 using BWA (54) and the number of read pairs that mapped to each annotated gene was determined. The counts were normalized based on the total number of reads mapped and the length of the gene, to obtain fragments per kilobase of transcript per million mapped reads (FPKM) values. To determine which regions of the A. montbretiana genome were introgressed into A. alpina, we first identified syntenic blocks between the two genomes by performing whole-proteome Blast (55) searches (e value) using the output for the DAGCHAINER program (56). Finally, the FPKM ratio between the FPKM of the matching genes (Aa/Am) in the syntenic blocks was used to determine whether the gene was homozygous A. alpina (2), homozygous A. montbretiana (0.5), or heterozygous.
Once introgressions were defined, plants were genotyped using primer pairs specifically designed within the introgressed intervals (Dataset S1).
RNA Extraction and qRT-PCR Analysis.
RNA was extracted using the RNeasy Plant Mini Kit (Qiagen) and treated with DNA-free DNase (Ambion). Total RNA (1.5 μg) was used to synthesize cDNA with SuperScript IV Reverse Transcriptase (Invitrogen) with oligo(dT)18 as a primer. Transcript levels were quantified by quantitative PCR in a LightCycler 480 (Roche) and iQ SYBR Green Supermix detection system (Bio-Rad). Each data point was derived from two biological and three technical replicates and represents the mean ± SD. LightCycler melting curves were obtained for the reactions, revealing single peak melting curves for most amplification products. AaRAN3 and AaUBI were used for normalization (57). The sequences of the primers used in this work are listed in Dataset S1.
RNA-Seq Analysis.
RNA-seq studies were used to study 1) differential gene expression in apices of 6-wk-old pep1-1 plants carrying AmMAR1 or AmMAR2 transgenes (PRJNA730091); 2) the expression of MAF genes in shoot apices before vernalization (2 wk for Arabidopsis and A. montbretiana or 6 wk for A. alpina Pajares), during vernalization treatment (2, 4, 6, 8, 10, and 12 wk), and 1 wk after plants were returned to warm temperatures (PRJNA730701); and 3) expression of MAF genes in the absence of PEP1 in shoot apices of pep1-1 plants at 2, 3, 4, 5, 6, and 7 wk after germination in long days (PRJNA728651). In all RNA-seq experiments, apices of 12 to 20 plants were harvested at each time point for each biological replicate (three for time frames 1 and 3 [above]; two for time frame 2 [above]).
RNA was isolated as described above and RNA integrity was confirmed on an Agilent BioAnalyzer. Library preparation and sequencing were performed at the Max Planck Genome Center Cologne, Germany (https://mpgc.mpipz.mpg.de/home/). Poly(A) RNA was isolated from 1 µg of total RNA using NEBNext Poly(A) mRNA Magnetic Isolation Module (New England Biolabs) and used for library construction with NEBNext Ultra Directional RNA Library Prep Kit for Illumina (New England Biolabs). RNA-seq was performed on an Illumina HiSEq. 3000 system with 150-bp single-read lengths.
RNA-seq–based expression levels were quantified using Salmon (58). Batch effects were identified in the vernalization dataset and corrected using the sva R package (59). FPKM normalization for visualization was obtained using the fpkm function of the DESeq2 package (60), which uses robust scaling factors rather than the total fragment depth. Differential expression analysis was performed with DESeq2.
Synteny and Phylogenetic Analysis.
For synteny analysis, nucleotide alignments between each sequence pair compared were generated using lastal (61). The initial alignment blocks were further processed using SyRI (62), resulting in a minimum set of alignments reflecting colinearity or rearrangements between the sequences.
To generate maximum likelihood (ML) trees, initial amino acid alignments generated with MUSCLE were processed by removing poorly conserved regions using trimAl (-automated1) (63, 64). The trimmed amino acid alignments were converted to codon alignments by replacing each amino acid with its corresponding codon from the original nucleotide sequence and multiplying gaps by 3. jModelTest (65) was used to determine the most appropriate substitution model for subsequent maximum likelihood tree reconstruction based on the Bayesian information criterion (BIC). The final ML tree was generated using PhyML (66) with parameters suggested by jModelTest, including 100 bootstrapping replicates. Phylogenetic trees were rendered after midpoint rooting using the ete3 python package (67).
For dN/dS analysis, the original MUSCLE alignment was trimmed by removing positions with more than 20% gaps and then converted into a codon alignment suitable for PAML (28). Parameter settings for the branch and branch site tests were set according to the PAML manual.
Supplementary Material
Acknowledgments
This work was supported by funding to G.C. from the Deutsche Forschugsgemeinschaft through Cluster of Excellence CEPLAS (EXC 2048/1 Project ID: 390686111) and from the European Research Council with the Advanced Grant HyLife (ERC-2013-339113). The laboratory of G.C. receives core funding from the Max Planck Society. We thank Dr. Koch MA (Heidelberg University), and Dr. Martin-Bravo S (University of Seville), for generously providing materials. We thank J. Chandler, M. Albani, and A. Fulgione for their critical reading of the manuscript.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2109204118/-/DCSupplemental.
Data Availability
Sequencing raw data have been deposited in NCBI (PRJNA728651: Whole-genome sequencing of IL PRJNA728651: RNA-seq time series of A. alpina pep1-1 apices grown under LD conditions; PRJNA730091: Effects of annual AmMAR1/AmMAR2 on gene expression; PRJNA730701: Gene regulation underlying the vernalization response in annual and perennial plants; PRJNA731145: Whole-genome sequencing of A. montbretiana Boiss; MZ736051-MZ736067: chr8 MAF region PacBio assembly). All study data are included in the article and/or SI Appendix. There are no additional data underlying this work.
References
- 1.Albani M. C., Coupland G., Comparative Analysis of Flowering in Annual and Perennial Plants. Curr. Top. Dev. Biol., Plant Development, Timmermans M. C. P., Ed. (Academic Press, 2010), vol. 91, pp. 323–348. [DOI] [PubMed] [Google Scholar]
- 2.Friedman J., The evolution of annual and perennial plant life histories: Ecological correlates and genetic mechanisms. Annu. Rev. Ecol. Evol. Syst. 51, 461–481 (2020). [Google Scholar]
- 3.Barrett S. C., The evolution of plant sexual diversity. Nat. Rev. Genet. 3, 274–284 (2002). [DOI] [PubMed] [Google Scholar]
- 4.Hu F. Y., et al., Convergent evolution of perenniality in rice and sorghum. Proc. Natl. Acad. Sci. U.S.A. 100, 4050–4054 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kiefer C., et al., Divergence of annual and perennial species in the Brassicaceae and the contribution of cis-acting variation at FLC orthologues. Mol. Ecol. 26, 3437–3457 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stebbins G., Flowering Plants: Evolution Above the Species Level (Belknap Press of Harvard Univeristy Press, Cambridge, 1974). [Google Scholar]
- 7.Lowry D. B., Willis J. H., A widespread chromosomal inversion polymorphism contributes to a major life-history transition, local adaptation, and reproductive isolation. PLoS Biol. 8, e1000500 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grillo M. A., et al., Genetic architecture for the adaptive origin of annual wild rice, oryza nivara. Evolution 63, 870–883 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Lowry D. B., et al., The case for the continued use of the genus name Mimulus for all monkeyflowers. Taxon 68, 617–623 (2019). [Google Scholar]
- 10.Lowry D. B., Rockwood R. C., Willis J. H., Ecological reproductive isolation of coast and inland races of Mimulus guttatus. Evolution 62, 2196–2214 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hyun Y., et al., A regulatory circuit conferring varied flowering response to cold in annual and perennial plants. Science 363, 409–412 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Wang R., et al., PEP1 regulates perennial flowering in Arabis alpina. Nature 459, 423–427 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Carroll S. B., Evolution at two levels: On genes and form. PLoS Biol. 3, e245 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castelán-Muñoz N., et al., MADS-box genes are key components of genetic regulatory networks involved in abiotic stress and plastic developmental responses in plants. Front. Plant Sci. 10, 853 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu T. T., et al., The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat. Genet. 43, 476–481 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiao W. B., et al., Improving and correcting the contiguity of long-read genome assemblies of three plant species using optical mapping and chromosome conformation capture data. Genome Res. 27, 778–786 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Willing E. M., et al., Genome expansion of Arabis alpina linked with retrotransposition and reduced symmetric DNA methylation. Nat. Plants 1, 1–7 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Karl R., Kiefer C., Ansell S. W., Koch M. A., Systematics and evolution of Arctic-Alpine Arabis alpina (Brassicaceae) and its closest relatives in the eastern Mediterranean. Am. J. Bot. 99, 778–794 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Michaels S. D., Amasino R. M., FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11, 949–956 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sheldon C. C., et al., The FLF MADS box gene: A repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11, 445–458 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bergonzi S., et al., Mechanisms of age-dependent response to winter temperature in perennial flowering of Arabis alpina. Science 340, 1094–1097 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Lazaro A., Obeng-Hinneh E., Albani M. C., Extended vernalization regulates inflorescence fate in Arabis alpina by stably silencing PERPETUAL FLOWERING1. Plant Physiol. 176, 2819–2833 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Finkers R., et al., The construction of a Solanum habrochaites LYC4 introgression line population and the identification of QTLs for resistance to Botrytis cinerea. Theor. Appl. Genet. 114, 1071–1080 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yano M., et al., Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12, 2473–2484 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gu X., et al., Arabidopsis FLC clade members form flowering-repressor complexes coordinating responses to endogenous and environmental cues. Nat. Commun. 4, 1–10 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ratcliffe O. J., Kumimoto R. W., Wong B. J., Riechmann J. L., Analysis of the Arabidopsis MADS AFFECTING FLOWERING gene family: MAF2 prevents vernalization by short periods of cold. Plant Cell 15, 1159–1169 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Theißen G., Rümpler F., Gramzow L., Array of MADS-box genes: Facilitator for rapid adaptation? Trends Plant Sci. 23, 563–576 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Yang Z., PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Lai X., Daher H., Galien A., Hugouvieux V., Zubieta C., Structural basis for plant MADS transcription factor oligomerization. Comput. Struct. Biotechnol. J. 17, 946–953 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tilmes V., et al., Gibberellins act downstream of Arabis PERPETUAL FLOWERING1 to accelerate floral induction during vernalization. Plant Physiol. 180, 1549–1563 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kania T., Russenberger D., Peng S., Apel K., Melzer S., FPF1 promotes flowering in Arabidopsis. Plant Cell 9, 1327–1338 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Airoldi C. A., McKay M., Davies B., MAF2 is regulated by temperature-dependent splicing and represses flowering at low temperatures in parallel with FLM. PLoS One 10, e0126516 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Panchy N., Lehti-Shiu M., Shiu S. H., Evolution of gene duplication in plants. Plant Physiol. 171, 2294–2316 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van de Peer Y., Fawcett J. A., Proost S., Sterck L., Vandepoele K., The flowering world: A tale of duplications. Trends Plant Sci. 14, 680–688 (2009). [DOI] [PubMed] [Google Scholar]
- 35.Wang Y., Wang X., Paterson A. H., Genome and gene duplications and gene expression divergence: A view from plants. Ann. N. Y. Acad. Sci. 1256, 1–14 (2012). [DOI] [PubMed] [Google Scholar]
- 36.Caicedo A. L., Richards C., Ehrenreich I. M., Purugganan M. D., Complex rearrangements lead to novel chimeric gene fusion polymorphisms at the Arabidopsis thaliana MAF2-5 flowering time gene cluster. Mol. Biol. Evol. 26, 699–711 (2009). [DOI] [PubMed] [Google Scholar]
- 37.Rosloski S. M., Jali S. S., Balasubramanian S., Weigel D., Grbic V., Natural diversity in flowering responses of Arabidopsis thaliana caused by variation in a tandem gene array. Genetics 186, 263–276 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hohmann N., Wolf E. M., Lysak M. A., Koch M. A., A time-calibrated road map of Brassicaceae species radiation and evolutionary history. Plant Cell 27, 2770–2784 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nikolov L. A., et al., Resolving the backbone of the Brassicaceae phylogeny for investigating trait diversity. New Phytol. 222, 1638–1651 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Li H., et al., Genome-wide identification of flowering-time genes in Brassica species and reveals a correlation between selective pressure and expression patterns of vernalization-pathway genes in Brassica napus. Int. J. Mol. Sci. 19, 3632 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scortecci K. C., Michaels S. D., Amasino R. M., Identification of a MADS-box gene, FLOWERING LOCUS M, that represses flowering. Plant J. 26, 229–236 (2001). [DOI] [PubMed] [Google Scholar]
- 42.Werner J. D., et al., Quantitative trait locus mapping and DNA array hybridization identify an FLM deletion as a cause for natural flowering-time variation. Proc. Natl. Acad. Sci. U.S.A. 102, 2460–2465 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.DeBolt S., Copy number variation shapes genome diversity in Arabidopsis over immediate family generational scales. Genome Biol. Evol. 2, 441–453 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hanada K., Zou C., Lehti-Shiu M. D., Shinozaki K., Shiu S. H., Importance of lineage-specific expansion of plant tandem duplicates in the adaptive response to environmental stimuli. Plant Physiol. 148, 993–1003 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maere S., et al., Modeling gene and genome duplications in eukaryotes. Proc. Natl. Acad. Sci. U.S.A. 102, 5454–5459 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maere S., Van de Peer Y., “Duplicate retention after small- and large-scale duplications” in Evolution after Gene Duplication, Dittmar K., Liberles D., Eds. (John Wiley & Sons, 2010), pp. 31–56. [Google Scholar]
- 47.Capovilla G., Symeonidi E., Wu R., Schmid M., Contribution of major FLM isoforms to temperature-dependent flowering in Arabidopsis thaliana. J. Exp. Bot. 68, 5117–5127 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sureshkumar S., Dent C., Seleznev A., Tasset C., Balasubramanian S., Nonsense-mediated mRNA decay modulates FLM-dependent thermosensory flowering response in Arabidopsis. Nat. Plants 2, 1–7 (2016). [DOI] [PubMed] [Google Scholar]
- 49.Casneuf T., De Bodt S., Raes J., Maere S., Van de Peer Y., Nonrandom divergence of gene expression following gene and genome duplications in the flowering plant Arabidopsis thaliana. Genome Biol. 7, 1–13 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li Z, et al., Expression pattern divergence of duplicated genes in rice. BMC Bioinformatics 10 (suppl 6), S8 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kinoshita A., Richter R., Genetic and molecular basis of floral induction in Arabidopsis thaliana. J. Exp. Bot. 71, 2490–2504 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Madrid E., Chandler J. W., Coupland G., Gene regulatory networks controlled by FLOWERING LOCUS C that confer variation in seasonal flowering and life history. J. Exp. Bot. 72, 4–14 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Coustham V., et al., Quantitative modulation of polycomb silencing underlies natural variation in vernalization. Science 337, 584–587 (2012). [DOI] [PubMed] [Google Scholar]
- 54.Li H., Durbin R., Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- 56.Haas B. J., Delcher A. L., Wortman J. R., Salzberg S. L., DAGchainer: A tool for mining segmental genome duplications and synteny. Bioinformatics 20, 3643–3646 (2004). [DOI] [PubMed] [Google Scholar]
- 57.Stephan L., Tilmes V., Hülskamp M., Selection and validation of reference genes for quantitative Real-Time PCR in Arabis alpina. PLoS One 14, e0211172 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Patro R., Duggal G., Love M. I., Irizarry R. A., Kingsford C., Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leek J. T., Johnson W. E., Parker H. S., Jaffe A. E., Storey J. D., The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28, 882–883 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 1–21 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kiełbasa S. M., Wan R., Sato K., Horton P., Frith M. C., Adaptive seeds tame genomic sequence comparison. Genome Res. 21, 487–493 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Goel M., Sun H., Jiao W.-B., Schneeberger K., SyRI: Finding genomic rearrangements and local sequence differences from whole-genome assemblies. Genome Biol. 20, 1–13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Capella-Gutiérrez S., Silla-Martínez J. M., Gabaldón T., trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Edgar R. C., MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Posada D., jModelTest: Phylogenetic model averaging. Mol. Biol. Evol. 25, 1253–1256 (2008). [DOI] [PubMed] [Google Scholar]
- 66.Guindon S., Delsuc F., Dufayard J. F., Gascuel O., Estimating maximum likelihood phylogenies with PhyML. Methods Mol. Biol. 537, 113–137 (2009). [DOI] [PubMed] [Google Scholar]
- 67.Huerta-Cepas J., Serra F., Bork P., ETE 3: Reconstruction, analysis, and visualization of phylogenomic data. Mol. Biol. Evol. 33, 1635–1638 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing raw data have been deposited in NCBI (PRJNA728651: Whole-genome sequencing of IL PRJNA728651: RNA-seq time series of A. alpina pep1-1 apices grown under LD conditions; PRJNA730091: Effects of annual AmMAR1/AmMAR2 on gene expression; PRJNA730701: Gene regulation underlying the vernalization response in annual and perennial plants; PRJNA731145: Whole-genome sequencing of A. montbretiana Boiss; MZ736051-MZ736067: chr8 MAF region PacBio assembly). All study data are included in the article and/or SI Appendix. There are no additional data underlying this work.