The crystal structure of DynU16, a protein identified in the dynemicin-biosynthetic gene cluster of Micromonospora chersina, was determined using iodide phasing and reveals a di-domain helix-grip fold.
Keywords: START/Bet v1 domain, dynemicin, helix-grip fold, DynU16
Abstract
The 1.5 Å resolution crystal structure of DynU16, a protein identified in the dynemicin-biosynthetic gene cluster, is reported. The structure adopts a di-domain helix-grip fold with a uniquely positioned open cavity connecting the domains. The elongated dimensions of the cavity appear to be compatible with the geometry of a linear polyene, suggesting the involvement of DynU16 in the upstream steps of dynemicin biosynthesis.
1. Introduction
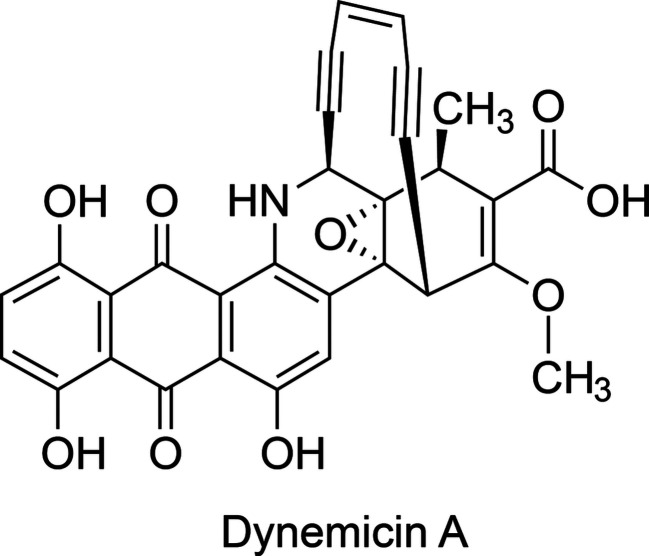
Dynemicin A (DYN; Fig. 1 ▸) is a bacterial-derived metabolite possessing a ten-membered 1,5-diyn-3-ene core that achieves double-stranded DNA scission via radical-mediated hydrogen abstraction (Konishi et al., 1990 ▸; Sluka et al., 1987 ▸; Elbaum et al., 1995 ▸). An anthraquinone (ANQ) moiety is appended to the core, with the quinone carbonyl O atoms serving as the activation sites for the reaction cascade, resulting in the formation of a phenyl biradical intermediate following core Bergman cyclization (Elbaum et al., 1995 ▸; Nicolaou et al., 1988 ▸; Sugiura et al., 1990 ▸). The DYN-biosynthetic gene cluster was first reported in 2008, illuminating the conserved core-assembling elements encoded in other enediyne pathways (Gao & Thorson, 2008 ▸); however, many of the enzymes do not possess substantial homology to proteins in the Protein Data Bank (Berman et al., 2003 ▸), raising more questions about DYN bioconstruction. The enediyne core is synthesized by a type I highly reducing iterative polyketide synthase (iPKS), yielding a linear conjugated polyene upon release from the iPKS by a thioesterase (Gao & Thorson, 2008 ▸; Ahlert et al., 2002 ▸; Liew et al., 2010 ▸). It was recently discovered that the iPKS also participates in the synthesis of the ANQ carbon skeleton, revealing the dual functionality encoded in the iPKS (Cohen & Townsend, 2018 ▸). The iPKS duality provides additional evidence supporting the notion that the tailoring enzymes govern the carbon-skeleton chemodiversity, as opposed to the iPKS and associated thioesterase (Horsman et al., 2010 ▸). The downstream cyclases mediating enediyne-core cyclization have not been identified, and several potential candidates appear to be membrane-bound based on bioinformatic analysis, which impeded their functional and structural characterization. Fortunately, a subset of DYN enzymes share homology with enzymes encoded by aromatic polyketide pathways, which use cytoplasmic mono-domain and di-domain cyclase/aromatases to produce diverse decaketides.
Figure 1.
The chemical structure of dynemicin A, showing the enediyne ‘warhead’ above the anthraquinone-fused core. The figure was generated with ChemDraw (version 16; PerkinElmer Informatics).
Here, we report, at a resolution of 1.5 Å, the crystal structure of DynU16, a putative di-domain protein homologous to StAR-related lipid-transfer (START/Bet v1-like) domain-containing proteins. Comparative bioinformatics to enediyne-biosynthetic gene clusters reveals homologues in the biosynthetic pathways of anthraquinone-fused enediynes. Moreover, DynU16 shares 30% sequence identity with CalU16, a calicheamicin mono-domain resistance protein; however, DynU16 possesses an additional domain, suggesting divergence in function from the self-sacrificing functionalities (Elshahawi et al., 2014 ▸; Singh et al., 2006 ▸; Biggins et al., 2003 ▸). In search of a di-domain homologue, an aromatase BexL was identified from the BE-7585A aromatic polyketide pathway (Caldara-Festin et al., 2015 ▸). In contrast to the type II PKS-associated di-domain cyclase/aromatases (CYC/AROs) BexL and StfQ, the DynU16 structure reveals an extended cavity bridging the domains. With only two reported di-domain structures from type II PKS systems, the DynU16 structure expands our limited knowledge.
2. Materials and methods
2.1. Macromolecule cloning, expression and purification
The DynU16 gene was codon-optimized for Escherichia coli expression and synthesized by Integrated DNA Technology (Coralville, Iowa, USA). The DynU16 gene was cloned into a pET-28a-derived expression plasmid via Polymerase Incomplete Primer Extension (PIPE) methods (Klock & Lesley, 2009 ▸). The PCR products were digested using the DpnI enzyme, mixed and annealed at 315 K. The annealed DynU16 and expression plasmid was transformed into NEB High-Efficiency E. coli cells (New England BioLabs, Ipswich, Massachusetts, USA) and plated onto kanamycin LB agar plates. Individual colonies were prepared and sequence-verified using Sanger sequencing (Genewiz, South Plainfield, New Jersey, USA). Positive clones were transformed into NEB E. coli BL21(DE3) expression cells.
A single colony was grown in LB medium supplemented with 50 µg ml−1 kanamycin overnight at 310 K. The cultured cells were inoculated into LB medium containing 50 µg ml−1 kanamycin and grown at 310 K until the OD600 reached ∼0.6. Protein synthesis was induced with 1.0 mM isopropyl β-d-1-thiogalactopyranoside (IPTG; GoldBio, St Louis, Missouri, USA) at 291 K for 18 h. The cells were harvested by centrifugation at 5000 rev min−1 at 277 K and stored at 193 K. For purification, the cells were thawed and resuspended in binding buffer consisting of 50 mM HEPES pH 7.5, 200 mM NaCl, 10 mM imidazole. Cell disruption was performed by sonication using a Qsonica sonicator programmed to 40% amplitude with iterative sonications. The lysed cells were separated by centrifugation at 18 000 rev min−1 for 45 min and the supernatant was incubated with 3 ml Nickel–NTA Superflow resin equilibrated with binding buffer (Qiagen, Hilden, Germany) on a rotating platform at 277 K. The protein-bound resin was washed with binding buffer supplemented with 30 mM imidazole and subsequently eluted with 200 mM imidazole. DynU16 was incubated with His-tagged TEV protease for 6 h at room temperature and further purified by reverse-phase nickel IMAC chromatography followed by size-exclusion chromatography on a HiLoad 16/600 column (GE Healthcare, Piscataway, New Jersey, USA). The protein concentration was determined from the absorbance at 280 nm using a calculated molar extinction coefficient of 75 970 M −1 cm−1). Macromolecule-production information is summarized in Table 1 ▸.
Table 1. Macromolecule-production information.
| Source organism | Micromonospora chersina |
| DNA source | E. coli codon-optimized gene synthesis, Integrated DNA Technology (Coralville, Iowa, USA) |
| I-PIPE forward primer | catgaaaacctgtacttccaatccATGCCAGATGGGTATTTGAAGCTT |
| I-PIPE reverse primer | ggatccgttatccacttccaatattttaCGGCAATTCTTGGCGGGTACG |
| V-PIPE forward primer | CGTACCCGCCAAGAATTGCCGTAAAATATTGGAAGTGGATAACGGATCC |
| V-PIPE reverse primer | AAGCTTCAAATACCCATCTGGCATGGATTGGAAGTACAGGTTTTCATG |
| Expression vector† | pET His6 TEV LIC cloning vector (1B) |
| Expression host | E. coli BL21(DE3) |
| Complete amino-acid sequence of the construct produced‡ | mgsshhhhhhenlyfq/sMPDGYLKLPDDWVRVMVSVPAPVDEVWEAVTDPRRVAQWFGHLSAPMTTGASTRVDFGDGDFFDIEVDHVEPRDRLLFRWSFLGVGPECQVGWTLTGGAEATTLTVDDSCPGRPGSEVAQLKAGWLDFVGRLARYLETGKPSRYDWRQEIDGSVVLPNGSWHPLREETVVDWLPIATNGAGPGWFFVVDEEGPRRFTLRDWQLDRERALTFAVEIPGARTVTACQVRTEPGERGRTLSVSHQGWHRLGLSDLQERTLRHRFAATWTAALSLAEECARTRQELP |
The pET His6 TEV LIC cloning vector (1B) was a gift from Scott Gradia (Addgene plasmid #29653).
The expression and purification tag is shown in lower case, with the TEV recognition site underlined and the cleavage site indicated with a slash.
2.2. Crystallization and derivatization
DynU16 was concentrated to 15 mg ml−1 using an Amicon Ultra-15 centrifugal filter (Merck KGaA, Darmstadt, Germany) and subjected to high-throughput crystallization screening, which was performed using a Mosquito LCP robot (SPT Labtech) with the commercially available screens NeXtal JCSG Core Suites I–IV. Crystals were observed in well B13 of Core Suite III (Table 2 ▸, Supplementary Fig. S1). The crystals were cryoprotected with 20% glycerol and flash-cooled in liquid nitrogen. A heavy-atom derivative was produced by soaking crystals in 1 M potassium iodide for 1.5 min prior to cryoprotection with 10% glycerol and flash-cooling.
Table 2. Crystallization.
| Method | Vapor diffusion, sitting drop |
| Plate type | MRC 2 well UVP (SwissSci) |
| Temperature (K) | 293 |
| Protein concentration (mg ml−1) | 15 |
| Buffer composition of protein solution | 20 mM HEPES pH 7.5, 200 mM NaCl |
| Composition of reservoir solution | 100 mM Tris–HCl pH 8.5, 200 mM MgCl2, 20%(w/v) PEG 8000 |
| Volume and ratio of drop | 1 µl:1 µl |
| Volume of reservoir (µl) | 200 |
2.3. Data collection and processing
The crystals were sent to the LS-CAT 21-ID-D beamline at the Advanced Photon Source (APS) for screening and data collection. A native data set was collected from a crystal that diffracted to a resolution of 1.5 Å and was indexed in space group P3121. The data were integrated and scaled with XDS (Kabsch, 2010 ▸) within the xia2 (Winter, 2010 ▸) data-processing pipeline (Table 3 ▸). Data were collected from the heavy-atom-soaked DynU16 crystals at a wavelength of 2.07 Å to enhance the anomalous signal from iodide; the crystals diffracted to 2.3 Å resolution.
Table 3. Data collection and processing.
Values in parentheses are for the outer shell.
| Native | KI soak | |
|---|---|---|
| Diffraction source | APS beamline 21-ID-D | APS beamline 21-ID-D |
| Wavelength (Å) | 1.00 | 2.07 |
| Temperature (K) | 100 | 100 |
| Detector | Dectris EIGER X 9M pixel | Dectris EIGER X 9M pixel |
| Crystal-to-detector distance (mm) | 180.1 | 100.0 |
| Rotation range per image (°) | 0.2 | 0.2 |
| Total rotation range (°) | 348 | 321 |
| Space group | P3121 | P3121 |
| a, b, c (Å) | 73.17, 73.17, 123.94 | 72.46, 72.46, 125.11 |
| Resolution range (Å) | 44.3–1.50 (1.54–1.50) | 56.1–2.28 (2.42–2.28) |
| Total No. of reflections | 1009253 (20881) | 269410 (27463) |
| No. of unique reflections | 58838 (2572) | 17898 (2772) |
| Completeness (%) | 95.4 (57.0) | 99.4 (96.9) |
| Multiplicity | 17.2 (8.1) | 15.0 (9.9) |
| Mosaicity (°) | 0.07 | 0.10 |
| 〈I/σ(I)〉 | 31.2 (1.6)† | 12.1 (1.6) |
| R meas | 0.044 (1.069) | 0.149 (0.903) |
| CC1/2 | 1.00 (0.78) | 1.00 (0.87) |
| Overall B factor from Wilson plot (Å2) | 34 | 49 |
〈I/σ(I)〉 falls below 2.0 at 1.55 Å resolution.
2.4. Structure solution and refinement
DynU16 phases were determined by single isomorphous replacement with anomalous scattering (SIRAS) using SHELXC/D/E (Sheldrick, 2010 ▸) for anomalous signal assessment and phase calculations within the HKL2MAP graphical interface (Pape & Schneider, 2004 ▸). ARP/wARP was used for initial modeling building (Langer et al., 2008 ▸). Coot (Emsley et al., 2010 ▸) and phenix.refine (Liebschner et al., 2019 ▸) were subsequently used for rebuilding and structure refinement (Table 4 ▸). The structure was visualized using a collaborative 3D graphics system (Yennamalli et al., 2014 ▸). The super algorithm in PyMOL (version 2.4.1; Schrödinger) was used for r.m.s.d. calculations. The structural biology software applications used in this project were compiled and configured by SBGrid (Morin et al., 2013 ▸). Coordinates and structure factors were deposited in the worldwide Protein Data Bank (Berman et al., 2003 ▸) as PDB entry 6v04.
Table 4. Structure refinement.
Values in parentheses are for the outer shell.
| PDB entry | 6v04 |
| Resolution range (Å) | 34.6–1.50 (1.54–1.50) |
| Completeness (%) | 95.3 |
| σ Cutoff | F > 0σ(F) |
| No. of reflections, working set | 56687 (2134) |
| No. of reflections, test set | 2087 (79) |
| Final R cryst | 0.162 (0.277) |
| Final R free | 0.188 (0.328) |
| No. of non-H atoms | |
| Protein | 2129 |
| Ions (Na/Mg/Cl) | 1/1/3 |
| Water | 282 |
| Total | 2416 |
| R.m.s. deviations | |
| Bonds (Å) | 0.005 |
| Angles (°) | 0.8 |
| Average B factors (Å2) | |
| Protein | 35 |
| Ions (Na/Mg/Cl) | 26/23/33 |
| Water | 44 |
| All-atom clashscore | 1.6 |
| Ramachandran plot | |
| Most favored (%) | 96.3 |
| Allowed (%) | 100 |
3. Results and discussion
3.1. Overall structure
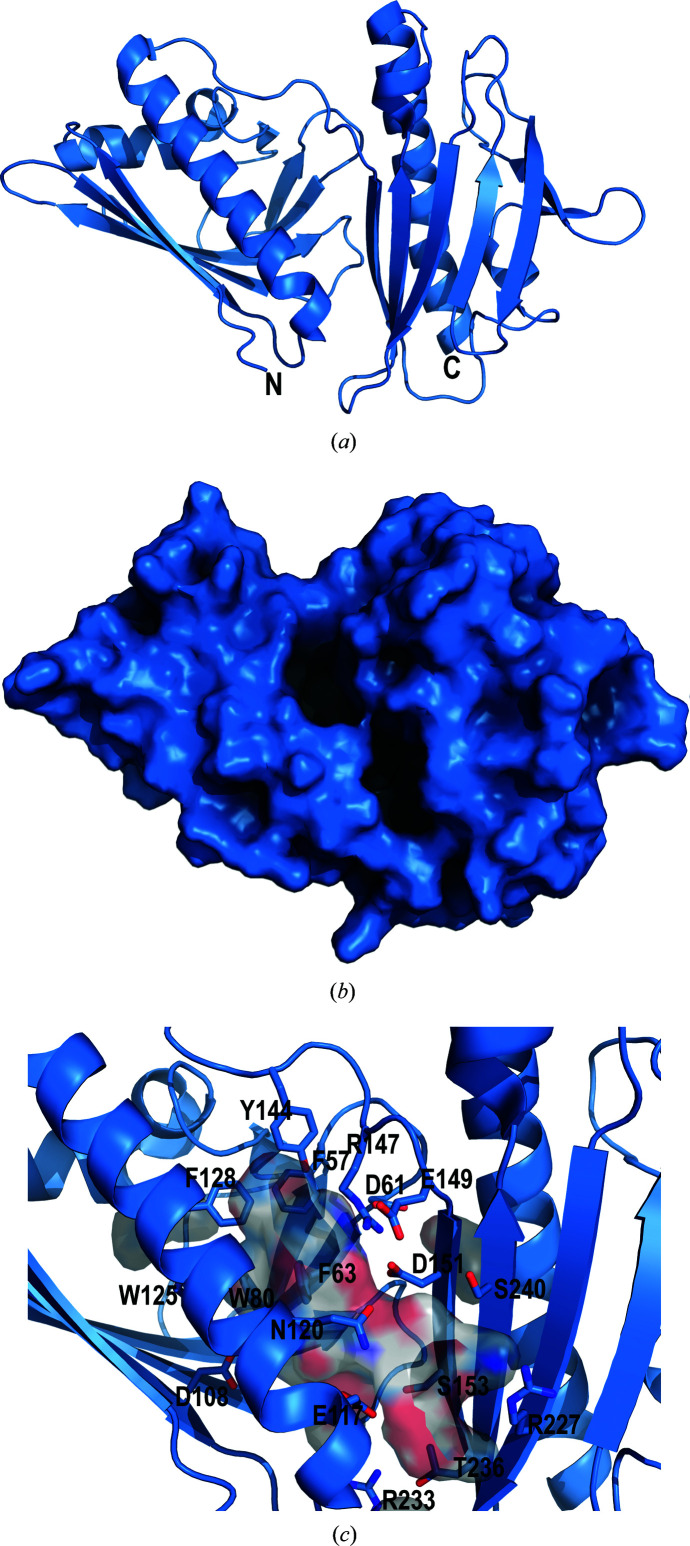
The DynU16 structure was determined at a nominal resolution of 1.5 Å using SIRAS, revealing a di-domain helix-grip fold (Fig. 2 ▸ a, Table 4 ▸). The final model included 269 protein residues, 282 water molecules and five ions. Residues 0 (left from the tag after cleavage) to 8 at the N-terminus and 278–283 at the C-terminus were disordered and were not modeled. The N-terminal domain comprises an antiparallel β-sheet, two short α-helices and one extended α-helix. The N-terminal (residues 1–139) and C-terminal (residues 150–283) domains are connected by a flexible ten-residue linker (140–149). The sequence and structure similarity between the N- and C-terminal domains suggests that the second domain was acquired through a gene-duplication event. A structure-based backbone-atom alignment of 313 atoms in the N- and C-terminal domains was calculated, giving a root-mean-square atomic position deviation of 2.0 Å with 15% sequence identity over 107 aligned residues (Supplementary Fig. S2). Assessment of the C-terminal domain suggests divergence from the canonical START/Bet v1 fold observed in the N-terminal domain. The C-terminal domain possesses eight instead of six antiparallel β-strands and has adopted additional loops.
Figure 2.
(a) DynU16 (PDB entry 6v04) adopts a helix-grip fold with a di-domain architecture. (b) The surface view shows the cavity formed between the N-terminal domain (left) and the C-terminal domain (right). (c) The elongated cavity is composed of aromatic residues localized towards the N-terminal domain and polar residues towards the C-terminal domain. The aromatic residues are positioned to stabilize the hydrophobic polyene substrate. The side chains lining the cavity are shown as sticks in cartoon representation, with C atoms shown in sky blue, N atoms in dark blue and O atoms in red. The surface of the cavity is shown to illustrate its size between the domains and the two openings. The figure was generated with PyMOL (version 2.4.1; Schrödinger).
3.2. Putative cavity
The cavity bridges the N- and C-terminal domains, with approximate dimensions of 22 × 12 × 8 Å (Figs. 2 ▸ b and 2 ▸ c). One end of the pocket comprises the polar residues Ser153, Ser240, Arg147, Arg227, Arg233, Glu117, Glu149, Asp61, Asp108, Asp151 and Gln120, along with Leu121. The back of the pocket has two clustered cysteines, Cys89 and Cys110, that are not engaged in a disulfide bond and could be involved in substrate stabilization. The opposing region of the cavity is composed of aromatic residues, including Phe57, Phe63, Phe82, Phe128, Trp80, Trp125 and Tyr144. The abundance of aromatic residues localized in one region of the cavity suggests a possible role in carbocation intermediate stabilization if cyclization is catalyzed in the cavity (Dougherty, 1996 ▸). The cavity-forming residues frequently reside in loops or β-strands, potentially increasing cavity flexibility and enabling contraction and expansion to accommodate substantial substrate rearrangements. The shape and size of the cavity appears to support the binding of a linear or perhaps a cyclic polyene (Fig. 2 ▸ c). The cavity openings (Fig. 2 ▸ b) could accommodate either a free polyene or one tethered to an acyl carrier protein (ACP) domain of the iPKS. However, it has not yet been established whether the enediyne core is synthesized attached to the ACP of the iPKS (Cohen & Townsend, 2018 ▸) or whether the intermediates remain free after they have been cleaved from the iPKS by the thioesterase (Annaval et al., 2017 ▸).
3.3. Sequence and structure comparison
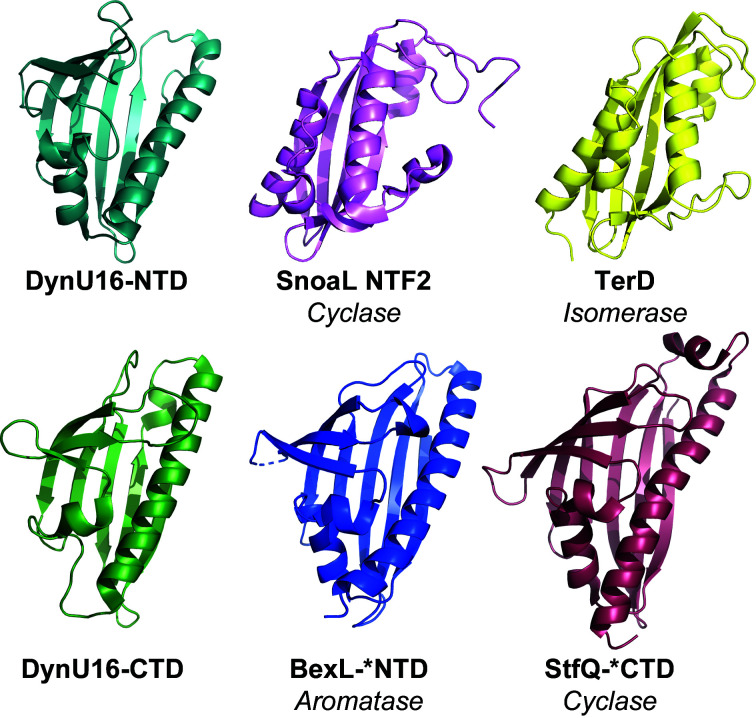
Prior to the structural determination of DynU16, a limited number of structures of di-domain START/Bet v1 enzymes had been reported. BexL and StfQ were identified in type II PKS systems and operate on nonreduced and partially reduced decaketide substrates, respectively. BexL possesses aromatase activity localized to the N-terminal domain and a pseudocatalytic C-terminal domain. StfQ possesses a pseudo-catalytic N-terminal domain and cyclase activity localized to the C-terminal domain. In the cases of BexL and StfQ, the pseudo-catalytic domains are hypothesized to provide both solubility and structural support (Caldara-Festin et al., 2015 ▸). Due to the limited information on the START-domain family, folds reminiscent of the START helix-grip fold were explored. The nuclear transport factor 2 (NTF2)-like cyclase α+β structure adopted by SnoaL and the isomerase TsrD provide additional evidence for the speculation that DynU16 harbors cyclase or dehydratase functionality (Fig. 3 ▸; Caldara-Festin et al., 2015 ▸; Sultana et al., 2004 ▸). The biosynthesis of the nine-membered enediyne antibiotic C-1027 requires SgcJ, which is a monodomain NTF2-like superfamily member (Huang et al., 2016 ▸).
Figure 3.
Structural comparison of the N-terminal domain (NTD) and C-terminal domain (CTD) of DynU16 with similar folds encompassing diverse functionalities including cyclization, aromatization and isomerization. The DynU16 helix-grip fold shares distant structural homology to the START di-domain polyketide-biosynthesis cyclase StfQ (PDB entry 4xrt; Caldara-Festin et al., 2015 ▸) and aromatase BexL (PDB entry 4xrw; Caldara-Festin et al., 2015 ▸). Additionally, the helix-grip fold resembles enzymes in the NTF2-domain family, including SnoaL, a mono-domain cyclase (PDB entry 1sjw; Sultana et al., 2004 ▸), and TsrD, a mono-domain isomerase (PDB entry 5x7l; Y. Song, Z. Lin, W. Deng & W. Liu, unpublished work). The catalytic domains selected for illustration are marked with an asterisk. The figure was generated with PyMOL (version 2.4.1; Schrödinger).
The NCBI BLAST platform (Altschul et al., 1997 ▸) was used to explore the sequence space of related enzymes (Supplementary Fig. S3). The search identified 20 uncharacterized di-domain homologues with sequence identity equal to or exceeding 30% and 90% coverage, with E values of less than 2 × 10−40. Using ESPript (Robert & Gouet, 2014 ▸), an extended motif in the N-terminal domain, RxxFxDGDFF, was identified amongst related family members. Structural mapping of the conserved motif locates it to the connection between β-strands 3 and 4 in proximity to the cavity. This region of the cavity aligns with the polyketide-binding location in the bifunctional mono-domain ARO/CYC enzymes TcmN and WhiE (Lee et al., 2012 ▸). Assessment of the interactions of the motif with nearby residues identifies a number of salt links between Arg147 and Asp151 and between Arg54 and Asp56 and van der Waals interactions between Phe63 and Trp80, suggesting that this motif may be involved in stabilizing the enzyme during catalysis. Other cyclase systems have been examined for their associated energetics, revealing highly exothermic reactions that could destabilize the enzyme without the proper network of stabilizing forces (Thoma et al., 2004 ▸). Additionally, conserved residues were identified in the C-terminal domain, identifying a region that potentially mediates protein–protein interactions within the biosynthetic network. While an actual enzymatic assay is needed to confirm the speculated function of DynU16, we conclude that it is likely that this protein participates in the biosynthesis of dynemicin as a cyclase or perhaps at least as a dehydratase of an extended or cyclic ANQ precursor.
Supplementary Material
PDB reference: DynU16, 6v04
Supplementary Figures. DOI: 10.1107/S2053230X21008943/rf5034sup1.pdf
Acknowledgments
This work used research resources provided by the University of Kentucky Center of Biomedical Research Excellence (COBRE) in Pharmaceutical Research and Innovation (CPRI; NIH P20 GM130456), College of Pharmacy and the National Center for Advancing Translational Sciences (UL1TR000117 and UL1TR001998). This research used resources of the Advanced Photon Source, a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (Grant 085P1000817). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the National Science Foundation.
Funding Statement
This work was funded by National Institutes of Health, National Institute of General Medical Sciences grants GM115261 and T32GM0082280 to George N. Phillips Jr, Jon S. Thorson, and Sarah K. Alvarado; National Institutes of Health, National Cancer Institute grant CA217255 to George N. Phillips Jr, Jon S. Thorson, and Steven G. Van Lanen; National Science Foundation, BioXFEL Science and Technology Center grant 1231306 to George N. Phillips Jr.
References
- Ahlert, J., Shepard, E., Lomovskaya, N., Zazopoulos, E., Staffa, A., Bachmann, B. O., Huang, K., Fonstein, L., Czisny, A., Whitwam, R. E., Farnet, C. M. & Thorson, J. S. (2002). Science, 297, 1173–1176. [DOI] [PubMed]
- Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997). Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed]
- Annaval, T., Rudolf, J. D., Chang, C. Y., Lohman, J. R., Kim, Y., Bigelow, L., Jedrzejczak, R., Babnigg, G., Joachimiak, A., Phillips, G. N. Jr & Shen, B. (2017). ACS Omega, 2, 5159–5169. [DOI] [PMC free article] [PubMed]
- Berman, H. M., Henrick, K. & Nakamura, H. (2003). Nat. Struct. Mol. Biol. 10, 980. [DOI] [PubMed]
- Biggins, J. B., Onwueme, K. C. & Thorson, J. S. (2003). Science, 301, 1537–1541. [DOI] [PubMed]
- Caldara-Festin, G., Jackson, D. R., Barajas, J. F., Valentic, T. R., Patel, A. B., Aguilar, S., Nguyen, M., Vo, M., Khanna, A., Sasaki, E., Liu, H.-W. & Tsai, S.-C. (2015). Proc. Natl Acad. Sci. USA, 112, E6844–E6851. [DOI] [PMC free article] [PubMed]
- Cohen, D. R. & Townsend, C. A. (2018). Nat. Chem. 10, 231–236. [DOI] [PMC free article] [PubMed]
- Dougherty, D. A. (1996). Science, 271, 163–168. [DOI] [PubMed]
- Elbaum, D., Porco, J. A. Jr, Stout, T. J., Clardy, J. & Schreiber, S. L. (1995). J. Am. Chem. Soc. 117, 211–225.
- Elshahawi, S. I., Ramelot, T. A., Seetharaman, J., Chen, J., Singh, S., Yang, Y., Pederson, K., Kharel, M. K., Xiao, R., Lew, S., Yennamalli, R. M., Miller, M. D., Wang, F., Tong, L., Montelione, G. T., Kennedy, M. A., Bingman, C. A., Zhu, H., Phillips, G. N. Jr & Thorson, J. S. (2014). ACS Chem. Biol. 9, 2347–2358. [DOI] [PMC free article] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Gao, Q. & Thorson, J. S. (2008). FEMS Microbiol. Lett. 282, 105–114. [DOI] [PMC free article] [PubMed]
- Horsman, G. P., Chen, Y., Thorson, J. S. & Shen, B. (2010). Proc. Natl Acad. Sci. USA, 107, 11331–11335. [DOI] [PMC free article] [PubMed]
- Huang, T., Chang, C. Y., Lohman, J. R., Rudolf, J. D., Kim, Y., Chang, C., Yang, D., Ma, M., Yan, X., Crnovcic, I., Bigelow, L., Clancy, S., Bingman, C. A., Yennamalli, R. M., Babnigg, G., Joachimiak, A., Phillips, G. N. Jr & Shen, B. (2016). J. Antibiot. 69, 731–740. [DOI] [PMC free article] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Klock, H. E. & Lesley, S. A. (2009). Methods Mol. Biol. 498, 91–103. [DOI] [PubMed]
- Konishi, M., Ohkuma, H., Tsuno, T., Oki, T., VanDuyne, G. D. & Clardy, J. (1990). J. Am. Chem. Soc. 112, 3715–3716.
- Langer, G., Cohen, S. X., Lamzin, V. S. & Perrakis, A. (2008). Nat. Protoc. 3, 1171–1179. [DOI] [PMC free article] [PubMed]
- Lee, M. Y., Ames, B. D. & Tsai, S. C. (2012). Biochemistry, 51, 3079–3091. [DOI] [PMC free article] [PubMed]
- Liebschner, D., Afonine, P. V., Baker, M. L., Bunkóczi, G., Chen, V. B., Croll, T. I., Hintze, B., Hung, L.-W., Jain, S., McCoy, A. J., Moriarty, N. W., Oeffner, R. D., Poon, B. K., Prisant, M. G., Read, R. J., Richardson, J. S., Richardson, D. C., Sammito, M. D., Sobolev, O. V., Stockwell, D. H., Terwilliger, T. C., Urzhumtsev, A. G., Videau, L. L., Williams, C. J. & Adams, P. D. (2019). Acta Cryst. D75, 861–877.
- Liew, C. W., Sharff, A., Kotaka, M., Kong, R., Sun, H., Qureshi, I., Bricogne, G., Liang, Z.-X. & Lescar, J. (2010). J. Mol. Biol. 404, 291–306. [DOI] [PubMed]
- Morin, A., Eisenbraun, B., Key, J., Sanschagrin, P. C., Timony, M. A., Ottaviano, M. & Sliz, P. (2013). eLife, 2, e01456. [DOI] [PMC free article] [PubMed]
- Nicolaou, K. C., Ogawa, Y., Zuccarello, G., Schweiger, E. J. & Kumazawa, T. (1988). J. Am. Chem. Soc. 110, 4866–4868.
- Pape, T. & Schneider, T. R. (2004). J. Appl. Cryst. 37, 843–844.
- Robert, X. & Gouet, P. (2014). Nucleic Acids Res. 42, W320–W324. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2010). Acta Cryst. D66, 479–485. [DOI] [PMC free article] [PubMed]
- Singh, S., Hager, M. H., Zhang, C., Griffith, B. R., Lee, M. S., Hallenga, K., Markley, J. L. & Thorson, J. S. (2006). ACS Chem. Biol. 1, 451–460. [DOI] [PubMed]
- Sluka, J. P., Horvath, S. J., Bruist, M. F., Simon, M. I. & Dervan, P. B. (1987). Science, 238, 1129–1132. [DOI] [PubMed]
- Sugiura, Y., Shiraki, T., Konishi, M. & Oki, T. (1990). Proc. Natl Acad. Sci. USA, 87, 3831–3835. [DOI] [PMC free article] [PubMed]
- Sultana, A., Kallio, P., Jansson, A., Wang, J.-S., Niemi, J., Mäntsälä, P. & Schneider, G. (2004). EMBO J. 23, 1911–1921. [DOI] [PMC free article] [PubMed]
- Thoma, R., Schulz-Gasch, T., D’Arcy, B., Benz, J., Aebi, J., Dehmlow, H., Hennig, M., Stihle, M. & Ruf, A. (2004). Nature, 432, 118–122. [DOI] [PubMed]
- Winter, G. (2010). J. Appl. Cryst. 43, 186–190.
- Yennamalli, R., Arangarasan, R., Bryden, A., Gleicher, M. & Phillips, G. N. Jr (2014). J. Appl. Cryst. 47, 1153–1157. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: DynU16, 6v04
Supplementary Figures. DOI: 10.1107/S2053230X21008943/rf5034sup1.pdf