Abstract
The outbreak of severe pneumonia at the end of 2019 was proved to be caused by the SARS-CoV-2 virus spreading out the world. And COVID-19 spread rapidly through a terrible transmission way by human-to-human, which led to many suspected cases waiting to be diagnosed and huge daily samples needed to be tested by an effective and rapid detection method. With an increasing number of COVID-19 infections, medical pressure is severe. Therefore, more efficient and accurate diagnosis methods were keen urgently established. In this review, we summarized several methods that can rapidly and sensitively identify COVID-19; some of them are widely used as the diagnostic techniques for SARS-CoV-2 in various countries, some diagnostic technologies refer to SARS (Severe Acute Respiratory Syndrome) or/and MERS (Middle East Respiratory Syndrome) detection, which may provide potential diagnosis ideas.
Keywords: COVID-19, Severe respiratory disease, SARS-CoV-2 virus, Rapid and accurate methods, Diagnostic approach
Introduction
Since the end of 2019, an ongoing pandemic referred to as Corona Virus Disease 2019 (COVID-19) was reported via a human-to-human transmission manner [1, 2]. According to the complete genome sequences revealed by scientists of the National Institute of Viral Disease Control and Prevention (IVDC) [3], the pneumonia disease causative agent is SARS-CoV-2, which has been identified as a novel coronavirus classified as a β lineage betacoronaviruses, including MERS-CoV, SARS-CoV, SARS-related coronavirus (SARSr-CoV), etc. [4]. Compared with the genome of SARS-CoV-2, the genome of the novel virus is 79.5% identical to SARS-CoV, while the homology of the Bat-CoV-RaTG13 gene, from the bat, reaches 96%, indicating that COVID-19 may cross-species barrier and originate in bats [5]. Considering the rare direct contact between bats and humans, the intermediate hosts are still unknown. Just as SARS-CoV and MERS-CoV, this novel coronavirus can spread across species and between human beings, with a transmission rate (R0) of 2.2% [6], similar to SARS (3%) [7], indicating that just as easy to spread as SARS-CoV. Up to April 19, 2021, it has been reported that 1421 million total cases were confirmed positive to COVID-19, and 3 million people were died around the world, including 103,317 confirmed cases and 4856 death tolls in China. Travel-related cases were reported in many countries, and the more serious of which are the United States, India, Brazil, and France as reported. Given the rapid transmission, high contagion, and high pathogenicity of SARS-CoV-2, it is in dire need of accurate and rapid detection of COVID-19. Therefore, it is critical for a sensitive and specificity platform to detect, track, and limit pathogenicity and infectious novel coronavirus. Here, based on the biological and epidemiological characteristics as well as clinical data of COVID-19, we introduce several kinds of valuable methods for accurate and rapid identification of COVID-19, including next-generation sequencing (NGS), nucleic acid assay, serological diagnostics, capillary electrophoresis (CE), biosensors for detecting SARS-CoV-2 and lung computed tomography scan.
Diagnostic Approaches of COVID-19
The bronchoalveolar lavage fluid of COVID-19 infected patients was collected in Beijing hospitals and was cultured to isolate the virus. Then, the genome sequences of SARS-CoV-2 were obtained via Illumina second-generation sequencing combine with nanopore third-generation sequencing technology [3, 4], which belong to two kinds of method of next-generation sequencing (NGS). The sequence has been submitted to GISAID (accession no. EPI_ISL_402124). Although NGS can identify the mutation within the viral genome and has played critical roles in identification, typing, traceability, and diagnosing SARS-CoV-2, it is not suitable for routine clinical detection of COVID-19 due to its disadvantages of long sequencing time and high requirements on laboratory equipment, as well as its substantial financial burden. After acquiring the genomic sequence of SARS-CoV-2, several laboratory diagnostic technologies were developed to characterize and identify COVID-19. Here, we will introduce several commonly used diagnostic techniques in our daily life.
Structure of SARS-CoV-2 is shown in Fig. 1 [8].
Fig. 1.
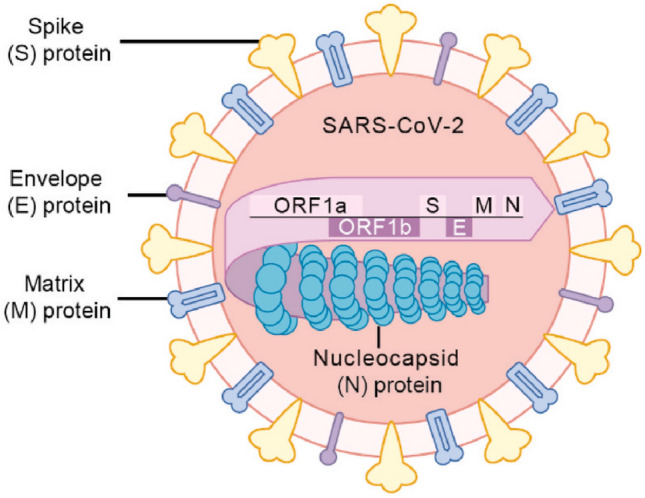
Structure of SARS-CoV-2.
Source cited with permission from Ref. [8]. 2020, Cell
Nucleic Acid Assay of SARS-CoV-2
Reverse Transcription Polymerase Chain Reaction (RT-PCR)
PCR was first introduced by Kary Mullis in 1985 [9]. The basic principle of PCR is similar to the DNA replication process, and its specificity depends on the oligonucleotide primers that complement each end of the target sequence. Typically, PCR includes three basic reaction steps: Denaturation of template DNA; Annealing (renaturation) of template DNA and primer; The extension of primers, DNA template, according to a principle of complementary base pairing and half reserved copy, synthesis of a new and complementary to the template DNA half reserved copy chain, repeat the cycle of degeneration–annealing–extension can get more “half reserved copy chain”, and the new chain can be the template of next replication cycle again. After 40–50 cycles, the target gene can be amplified several million times.
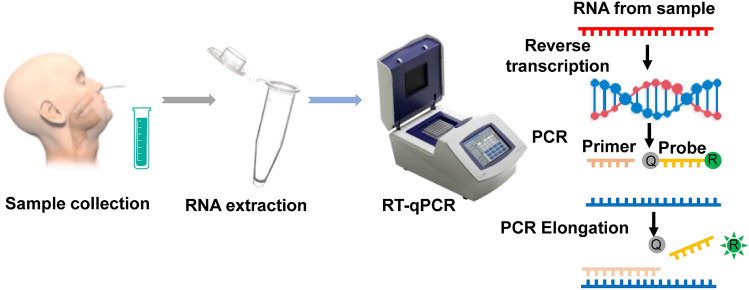
RT-PCR is a kind of basic gene detection method, which refers to adding fluorescent probes or fluorescent dyes to the conventional PCR reaction system, and tracking the product to achieve a quantitative analysis of the starting template (Fig. 2). RT-PCR with simple operation, intuitive results, strong specificity, high sensitivity, rapid [10–14], and quantitative detection with accuracy and repeatability, lower testing cost and the whole detection process were closed, which has solved the PCR contamination problem and has been widely used into scientific research and clinical diagnosis [10, 12–16]. The high sensitivity of PCR allows it being applied into terrible conditions, including the test samples partially degraded, formalin-fixed, or paraffin embedding materials [17–19]. The PCR has been widely used in clinical as a gold standard for disease detection, diagnosis, and quantification in the past decades. Recently, RT-PCR is widely used in detecting various respiratory virus infections: influenza viruses A, B and C, parainfluenza viruses 1–4 (PIV-1, -2, -3 and -4), human respiratory syncytial virus (hRSV), HCoV OC43 and 229E, human rhinoviruses (hRV), adenoviruses as well as some human enteroviruses (hEV. [15], SARS-CoV, MERS-CoV, H7N9, etc. In the past two decades, PCR methods play vital roles in virus diagnosing, prevention, and treatment [10, 16, 20–25].
Fig. 2.
The progress of the nucleic acid test, RT-qPCR.
Obtaining permission from Ref. [29]. 2021, Talanta
The quantitative RT-PCR (RT-qPCR) is the primary diagnostic technique for COVID-19 [14]. Based on the full-length genome sequences, the conserved regions of open reading frames 1ab (ORF1ab) and nucleoprotein (N) genes and the conserved regions of human housekeeping gene RNase P were selected, and the specific primers/TaqMan probes were designed for detecting COVID-19 by RT-PCR (Table 1) (http://nmdc.cn/#/nCoV).
Table 1.
The primers and Taqman probes for the detection of COVID-19
| Gene | Sequence (5´–3´) |
|---|---|
| ORF1ab | |
| Forward primer | CCCTGTGGGTTTTACACTTAA |
| Reverse primer | ACGATTGTGCATCAGCTGA |
| TaqMan probe | 5'-FAM-CCGTCTGCGGTATGTGGAAAGGTTATGG-BHQ1-3' |
| N gene | |
| Forward primer | GGGGAACTTCTCCTGCTAGAAT |
| Reverse primer | CAGACATTTTGCTCTCAAGCTG |
| TaqMan probe | 5'-FAM-TTGCTGCTGCTTGACAGATT-TAMRA-3' |
European Centre for Disease Prevention and Control also rapidly established a fluorescent RT-PCR system used for detecting COVID-19. Real-time fluorescent RT-PCR was included in the diagnosis and prognosis guidelines for SARS-CoV-2 according to the Technical Guidelines for COVID-19 Laboratory Testing (Fifth Edition) by the Chinese Center for Disease Control and Prevention. The National Medical Products Administration (NMPA) in China has approved RT-qPCR technology as detection method for COVID-19. ORF1ab and N genes were selected as targets to detect their conserved regions, and the results of the new coronavirus were more reliable by dual control targets. The selection of the conserved regions of the human housekeeping gene RNase P as the internal reference gene, which can monitor the collection of samples and nucleic acid extraction process in the test and effectively avoid false-negative results caused by sample quality problems. The sensitivity of SARS-CoV-2 detecting reagents was up to 1000 copies/mL [26]. Multiple real-time fluorescent RT-PCR systems have significant advantage: the higher sensitivity. The higher the probability of correctly detecting the samples at low concentration, the lower false-negative rate occurred. Real-time fluorescent RT-PCR diagnosis of COVID-19 has made a foundation for the epidemiological study of the novel coronavirus [11, 20, 23, 25, 27, 28].
However, there are still some problems with the detection sensitivity of RT-PCR. The possible reasons leading to false-negative are: (1) The virus load during sampling is too low. The theoretical detection limit was 1copy/reaction, that is, 200 copies/mL. When the viral load is lower than this concentration, false-negative results might occur; (2) Poor sampling quality [11, 16, 27]. There are few specimens of alveolar lavage fluid and deep cough sputum, and the common specimen type is nasopharyngeal swab; (3) RNA virus nucleic acid is not stable [11, 27], vulnerable to transport conditions, inactivation conditions, and experimental supplies; (4) The quality of detection kits is unstable. The quality of the vital components of the nucleic acid detection kit developed and produced in an emergency is fluctuate, such as the quality of the probe, primer, and enzyme, or the quality control standard is not in place during the urgent mass production process, which leads to the instability of the quality of the kit. The lower detection sensitivity may lead to many suspected cases not being confirmed by etiology and not being treated timely, and finally family cluster infection. The collection of samples is dangerous to the sampler, and the virus isolation and reverse transcription should be limited in the corresponding operating platform and operated by professional technicians; improper operation or insufficient laboratory conditions may lead to false-positive/-negative results. The whole process needs several hours [27].
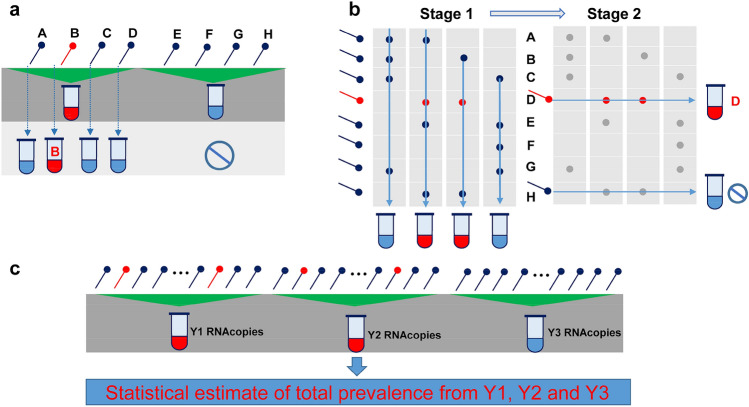
Group testing provides a rapid pathway in increasing high-throughput screening by combining the test pool samples (illustrated in Fig. 3). Brian Cleary et al. confirmed that their theoretical results of group testing providing that the combined nasopharyngeal specimens from the number of samples could accurately acquire the test results and solve the critical concerns about the loss of sensitivity and the feasibility of implementation by system illustration [30]. Increasing the detection efficiency within the shortage of the supplement of diagnosing reagents and consumables and the limitations of financial support in the short term can help limit the transmission of COVID-19.
Fig. 3.
Group testing designs. a, b The test results used to identify positive samples; c estimate the prevalence in the community.
The figure reproduced with permission from Ref. [30]. 2021, Sci Transl Med
CRISPR-Based Fluorescent Diagnosis System for COVID-19 (COVID-19 CRISPR-FDS)
CRISPR-Cas/gRNA is a widespread technique in molecular biology, and is applied into sensitively detecting nucleic acids and human pathogens [31–36]. CRISPR-FDS method contains three related procedures: (1) RNA isolation from samples; (2) Amplification of sample DNA; (3) Detecting the fluorescent signal by microplate reader [31]. The target regions of coronavirus RNA isolated from nasopharyngeal specimens were amplified by one-step RT-PCR/RT-RPA, and the amplicons samples were transferred into the gRNA/Cas12a-based CRISPR system, and then the fluorescence signal was detected by microplate reader; the target amplicon was recognized by the gRNA/Cas12a complex and regulated by a target-specific synthetic gRNA, inducing the target amplicon was explicitly cleaved by the gRNA/Cas12a complex, and the reporter oligo non-specifically cleaved, fluorescein and a quencher molecule were modified at each terminus and presented with a fluorescent signal. CRISPR-FDS technique provides a relatively short period to acquire the testing reports: about 50 min from the RNA was extracted from virus samples to the testing reports, which saving a lot of detection time. The reagents and equipment of the CRISPR-FDS method are available in most clinical laboratories, especially the entire automatic operation of sample viral RNA extracted to test report can satisfy the needs of high-throughput testing. Many studies recently reported that RNA-targeting CRISPR-associated enzyme Cas13 in detecting nucleic acids presents rapid and convenient advantages [36–38] and can be applied for the diagnosis of SRAS-CoV-2. However, these novel methods also face the viral RNA needed to be extracted, the quality of the sample, and the requirement of professional technical expertise, proper operation, and sufficient laboratory conditions, which limit their use in professional places.
Digital PCR
Digital PCR (dPCR) is first introduced in the 1990s [39] and as a robust PCR technique, provide precise and accurate absolute quantitation of target samples by diluting and partitioning the samples into numerous compartments [40]. dPCR is increasingly adapted for the quantitation of nucleic acid and is particularly suitable for low-level detection [41–43]. Various dPCR platforms used for commercialization with different technical characteristics have been developed, such as microfluidic chamber-based, droplet-based, micro-well chip-based, microchannels, and droplet-based crystal dPCR [43, 44]. The principle of dPCR is to partition the traditional PCR reaction mixture to the level of single compartments, and then a single DNA template, which was maximally diluted, was amplified, then the fraction of the fluorescing droplets signal was generated, which is precisely derived from a single template and calculated using the Poisson distribution [29, 45]. Since the outbreak of COVID-19, dPCR was used for diagnosing SARS-CoV-2, and many reports have proved that dPCR has a lower limit of detection (LOD) for ORF1ab and N genes of COVID-19, even more than 500 times lower than the traditional quantitative real-time PCR assay reports [46, 47]. Micro-wells-based dPCR assay of COVID-19 has acquired EUA from the US FDA [29]. However, there are still some limitations of dPCR applied into diagnosing SARS-CoV-2, such as requiring specialized micro-wells and cyclic heating with professional instruments, which proved that dPCR is not applicable for high-throughput screening in the diagnose of SARS-CoV-2 [29, 48].
Nucleic Acid Mass Spectrometry (NC-MS) Assay
Matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF) technology is the basis of NC-MS, which as a new technology platform, can quickly and accurately determine molecular weight and structure. It is used to determine the molecular mass and purity of biological macromolecules such as polypeptide proteins, nucleic acids, and polysaccharides. In 2005, Alicij et al. used MALDI-TOF mass spectrometry to obtain the M095L and M151R proteins in Myxoma virus particles [49]. And Youn et al. quantitatively detected single-base mutations by hybridizing PNA probes with MALDI-TOF [50]. Four years later, Xiu et al. used mCoV-MS (based on MALDI-TOF MS system) to detect potential pathogen CoV in coronaviruses and beta coronaviruses, providing a basis for detecting unknown coronaviruses [51]. Recently, Liu et al. used MALDI-TOF MS for simultaneous detecting and genotyping of ten viruses in ducks with detection limits ranging from 1.3 to 7.8 copies/μL [52]. MALDI-TOF MS has a significant increase in virus detection due to its fast speed and simple analysis. And it is an effective tool for diagnosing respiratory infections.
Isothermal Amplification (IA) Technology
Unlike the conventional PCR amplification technology, IA technology does not need precise temperature control instruments, and can achieve amplification reactions at room temperature [53]. IA technology mainly includes seven detection technologies: Loop-Mediated Isothermal Amplification (LAMP), Helicase-Dependent Amplification (HDA), Rolling Circle Amplification (RCA), Nuclear Acid Sequence-Based Amplification (NASBA), Recombinase Polymerase Amplification (RPA), Strand Displacement Amplification (SDA), and Crossing Priming Amplification (CPA). LAPM technology is widely applied into clinical molecular diagnosis as well as other fields due to the advantages of short analysis time (it does not require thermal denaturation of DNA double strands in advance) and high amplification rate (it is amplifying the specific sequences of nucleic acid templates with a series of 4/6 unique primers, which can be amplified 1 × 109–1 × 1010 times in 1 h) [54, 55]. Pyrolysate precipitation detection, fluorescence detection, and gel electrophoresis detection are often used to detect LAMP amplification products [56]. There are many examples of LAMP applications. By combining LAMP with reverse transcription (RT-LAMP) methods for detecting RNA viruses, such as SARS [57], respiratory syncytial virus [58, 59], influenza virus [60, 61], Ebola virus [62], West Nile virus [63] and yellow fever virus [64], have been established. It is worth mentioning that the nucleic acid visualization technology recently established by Pei et al., combining reverse transcription LAMP technology with vertical flow visualization bar, can be successfully applied into detecting the N gene of MERS-CoV. Compared with the previous RT-PCR method, it has more accessible detection technology and lower detection afford. This method required for 6 designed primers and labeled them with biotin to visualize the detection results. It only takes 35 min from the beginning to the test reports, faster than the RT-PCR analysis. While this method has certain disadvantages: the detection sensitivity is lower compared with RT-PCR [65]. There is a challenge and a new direction for us. The Chinese Center for Disease Control and Prevention of Viral Diseases has successfully developed novel COVID-19 nucleic acid IA detection kits, producing results in 8–15 min. Chengdu Boao Jingxin Biotechnology Co., Ltd., West China Hospital of Sichuan University and Tsinghua University, have jointly designed and developed a nucleic acid detection kit (constant temperature amplification chip method) that can be used to detect the new coronavirus (COVID-19), which has been approved by the State Drug Administration. Attention needs to be paid to the contamination of nucleic acid amplified fragments when researching detection methods of COVID-19.
Nucleic Acid Point of Care Testing (POCT) Technology
POCT is a testing system that integrates sample preparation and testing. Nucleic acid POCT technology integrates functional modules for nucleic acid isolation, amplification, detection, as well as pre-packages of each reagent required to react in a detection cassette. This detection method can decrease the risk of transmission of infectious diseases during the detection process and avoid cross-contamination. Except the unprocessed reagents are added by operators, it can automatically perform the procedure of nucleic acid extraction, amplification, signal acquisition, subsequent result analysis quickly. The requirements for the research environment and testing personnel's capabilities are lower than the methods mentioned above, and it can realize self-testing in a community hospital or family [66]. The detection methods of POCT can be applied to the following three types: rapid immunological detection reagents; nucleic acid-based molecular diagnostic products; and miniaturized portable dry biochemical, immunofluorescence, dry blood gas, and chemiluminescence diagnostic products. The application of nucleic acid POCT technology is of great significance to alleviate the testing pressure of large medical institutions and improve the nucleic acid detection capability of primary medical institutions [67]. Many nucleic acid POCT detection instruments with the supporting detection reagents have been developed, such as Meikang Biological, Da'an Gene, Shanghai Tellgen Cooperation, Wanfu Biological, BGI, Xilong Science, etc. The state has accelerated the examination and approval process based on ensuring quality. Rapid, sensitive, and specific POCT methods are still urgently needed due to the timely diagnosis of infected patients and effective contact tracing the potential cases of COVID-19 infection [68].
Serological Diagnostics
Immunoassays based on serological/antibody are providing rapid and effective approaches for the screening of various pathogenic infections [24], and present with high-throughput capability and not too harsh requirements of samples than viral RNA-based assays [14].
Colloidal Gold Immunochromatographic Assay
Due to RT-qPCR's time-consuming properties, a new method, colloidal gold immunochromatography, was developed to address these issues. Similar to SARS-CoV, IgM antibodies of SARS-CoV-2 are produced rapidly in the early stages of infection and have a short maintenance time, which can be used as an early indicator of COVID-19 infection; whereas IgG antibodies can produce later and maintain for long periods, which can be used as the previous infection in blood tests [5, 69].
A newly developed SARS-CoV-2 IgG–IgM-coated antibody detection kit with significant specificity and high sensitivity was recently reported [70]. Three kinds of secondary antibodies, anti-human-IgM, anti-human-IgG, and anti-rabbit-IgG, were fixed on NC membrane as assay M, G and control line (C line), respectively. The recombinant antigen (MK201027) of SARS-CoV-2 conjugated with colloidal gold nanoparticles and AuNP-rabbit IgG conjugates were previously prepared as a conjugate pad. When IgM and IgG are present in test samples, red and pink lines form in the M and G regions due to colloidal gold, respectively. The C line turns red, which means a negative sample. The M or G line or both lines presented the existence of IgM or IgG of COVID-19 or both antibodies in the test samples. If the C line does not display in red, it means the test failed and needs to be assayed again.
Those kits tested blood samples from a total of 525 patients, of which 352 were positive (397 clinically confirmed positive), and 116 were negative (128 clinically confirmed negative), revealing with a sensitivity is 88.66% and the specificity is 90.63% [71]. The IgM and IgG antibodies against the SARS-CoV-2 in patients’ blood can be detected simultaneously so that patients at different stages of infection can be diagnosed. More importantly, the results can be obtained within 15 min, which obviously shortens the time and significantly facilitates case diagnosis. Meanwhile, this antibody detection kit detects antibodies in the blood more accurately than RT-qPCR for detecting the upper respiratory tract because the lesion area locats in the lungs [72]. And, no particular instrument and environmental requirements are another significant advantages of this technology [70].
On March 1, 2020, the NMPA of China approved several detection kits for SARS-CoV-2 IgG–IgM, addressing the urgent need to test a large number of clinical samples.
However, the validity of the COVID-19 IgG–IgM-coated antibody detection kits still require more clinical data. Besides, there remains another problem that the combined antibody test kit cannot detect SARS-CoV-2 during the window period due to insufficient antibodies produced in the body [73]. The sensitivity of this method is inadequate. After adding samples, the principle relies on chromatography, that is, dispersing outward. If the materials were not up to standard baseline or with other qualities, the detection results would be affected. False negatives can also occur if the patient was infected for a short time or has low virus levels at the sampling site. In addition, due to its dependence on antigens and antibodies, this method is crucial for selecting coated antigens/antibodies in early development, and the specificity and sensitivity of products developed in a short duration need to be confirmed. Therefore, more researches are still need to be done, and this assay can only provide a method of fast screening test combined with RT-qPCR to identify COVID-19.
Enzyme-Linked Immunosorbent Assay (ELISA)
Unique binding to the antibody of antigen is the basis of ELISA, a simple, sensitive, and rapid technique for detecting the antigen or antibody of interest samples attached to a solid surface [74] (illustrated in Fig. 4). ELISA reveals tremendous commercial value in many aspects, such as laboratory research, the biomarkers diagnostic of the disease, and quality control in industries [75]. ELISA can detect biomarkers in two different ways: the direct way, an enzyme-linked antibody can recognize the antigen in the test samples; the indirect way, the antigen is combined on specific microplates and then bound by a primary antibody, subsequently the primary antibody is detected by a secondary enzyme-labeled antibody [76]. To date, several commercial ELISA detection kits have been developed and applied to diagnosing SARS-CoV-2. Jie Xiang et al. reported an ELISA kit that can direct measure IgM. The mouse anti-human IgM monoclonal antibody was coated in the test microplate, and IgM in serum samples was detected by an enzyme-labeled antibody; IgG was caught by the indirect method, the recombinant antigen of SARS-CoV-2 was coated on the surface of a microplate, and then the HRP-conjugated monoclonal mouse antibody for SARS-CoV-2 competitively was bound to IgG in serum samples [77]. Sheikhzadeh et al. reported that N protein-based ELISA assayed 208 cases of plasma samples, including confirmed cases (n = 82) and suspected cases (n = 58). ELISA detecting results indicated that the median time for detecting IgM and IgA is 5 days, IgG is 14 days after symptom onset [78]. And many reports proved that the specificity and sensitivity of ELISA are 100% and 87.3%, respectively, with a LOD of 100 ng/mL [70, 79]. Mohammad M. Sajadi group reported 3 ELISA methods: nucleocapsid IgG, trimer spike IgA and IgG, which were applied into detecting anti-SARS-CoV-2 antibodies, evaluating the potency with 4 kinds of commercial ELISA kits, and the sensitivity and specificity. They found that in-house tests exhibited the highest combined sensitivity and specificity [80]. Unite RT-qPCR with ELISA can increase the precision of COVID-19 cases, and effectively inhibit the dissemination and spread of COVID-19 [78].
Fig. 4.
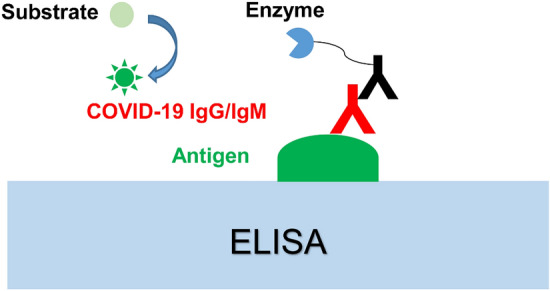
Conventional antibody tests of COVID-19 by ELISA.
Obtaining permission from Ref. [29]. 2021, Talanta
While the guidelines of FDA, other international professional groups and many reports illustrated that serology tests might not be suitable as a diagnosis method for SRAS-CoV-2, due to the positive serology assay results indicate, to some extent, that individuals have been infected with COVID-19 [73, 81–83]. The immunoassays for detecting IgM and IgG were proved to be a faster and more economical way than the nucleic acid assay like RT-PCR; however, there are also limitation such as cross-reactivity leading to false positivity [73]. In fact, in the clinical diagnosis of SRAS-CoV-2, serologic testing may as a complement of PCR-based diagnostic testing and used for confirming the diagnosis combined with the clinical characterizations or/and nucleic acid test results. On the other hand, in epidemiologic studies, serology tests of SRAS-CoV-2 may serve as a monitor in detecting the immune response to COVID-19 when injected with the vaccine, diagnosing convalescent plasma donors, or learning the prevalence of COVID-19 in public [73, 84, 85]
Biosensors for Detecting COVID-19
Sensors include chemical/biological receptors and transducers. Biosensors are based on bioanalytical devices which convert the selectivity features of biological reactions into identify signal results [86]. The devices of biosensors include a bio-reactor (such as enzyme, antibodies, microorganisms, cell receptors, nucleic acids, etc. [86]), transducer (like nanomaterials), and a computer display system [87]. The biosensors have been adapted for many parts, such as viruses, bacteria, proteins, etc., and proved to be with high sensitivity, high throughput abilities, and portability, which attracting highly attention [88]. In the past few decades, there are several types of based biosensors: Optical based, Electrochemical based, Piezoelectric based, and Thermal based biosensors [89], which are applied to detecting respiratory viral infections, such as Influenza A virus, SARS-CoV, MERS [90]. Novel biosensors adapted for detecting RNA viruses include CRISPR–Cas9-based paper strip, nucleic acid-based biosensor, optical biosensor, aptamer-based biosensor, surface plasmon resonance (SPR), etc. [90], performing practical, sensitive, authentic, portable, and specific in the diagnosis of COVID-19.
Surface Plasmon Resonance(SPR) Sensor
Collective oscillation of electrons on a metal surface is the basis of SPR sensors. It is widely adapted for monitoring the molecular interactions, such as protein–protein, protein–RNA, protein–DNA, etc., rapidly and more sensitively [91, 92]. Conventional SPR-based biosensor detecting platforms present with many advantages in diagnosing the interactions between two molecules [92]. Many studies have reported that SPR sensors have been applied into the detection of Dengue, Ebola, and Zika viruses. It is reported that the LOD of Dengue protein is 0.08 pmol/L [92, 93]. Abdelhadi Djaileb and coworkers [94] reported an SPR sensor successfully applied to the detection of nucleocapsid antibodies in undiluted patients serum, which is specific against the COVID-19. The SPR sensor conjugated with a peptide monolayer and functionalized with SARS-CoV-2 nucleocapsid recombinant protein presents against anti-SARS-CoV-2 antibodies in nanomolar level, and the real progress from the undiluted human serum samples to analysis results by this portable SPR instrument is within 15 min. Qiu and coworkers [95] reported a SARS-CoV-2 biosensor, based on the principle of RT-PCR, in diagnoses of SARS-CoV and SARS-CoV-2, and the specific sequences oligonucleotides were selected as the thiol-complementary DNA receptor. A dual-functional plasmonic biosensor combining the plasmonic photothermal (PPT) effect and localized surface plasmon resonance (LSPR) sensing transduction was reported, providing an alternative and promising manner in the diagnosis of COVID-19 by nucleic acid hybridization. The dual-functional LSPR biosensor was proved to be of high sensitivity in viral sequences, including RdRp, ORF1ab, and E genes toward the selected SARS-CoV-2 sequences, with the LOD of 0.22 pmol/L. Especially, this LSPR biosensor can precisely detect the specific target in a multigene mixture. Later, a label-free field-effect transistor-based genosensor via a specific spike antibody to detect SARS-CoV-2 antigen protein was reported and evaluated in different samples, such as the universal transport medium, nasopharyngeal swabs, and clinical samples, with satisfactory results [86]. Therefore, under certain circumstances, SPR is an alternative detecting platform for rapidly diagnosing clinical samples, and releasing the RT-PCR-based test burdens. However, both the SPR and LSPR systems have some drawbacks, such as complexity or noise signals, which should be noticed.
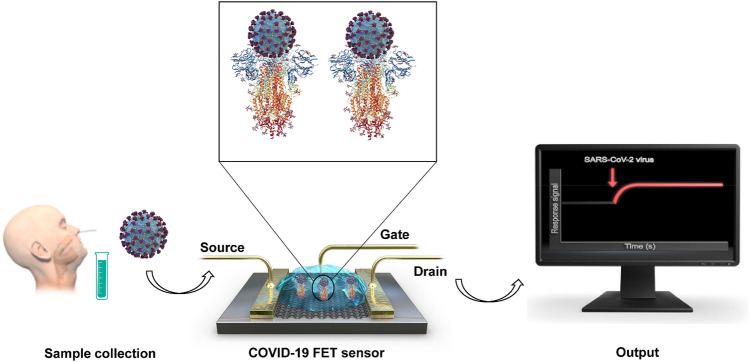
Field-Effect Transistor (FET)-Based Biosensor
FET-based biosensors as a kind of electric biosensors have been applied to monitoring the surface potential changes before/after the target molecule binding to the biorecognition element coated on the highly conductive chip surface [96]. It is reported that the first FET-based biosensor device includes a three-electrode structure containing the drain, source, and gate [96, 97] (illustrated in Fig. 5). The devices of FET-based biosensor need a spot of analytes and are proved to be with highly sensitive and instantaneous measurements advantages [98–100]. With the development of FET-based biosensors, they have been applied to diagnosing the viral disease, point-of-care testing, on-site detection, and detecting small molecules [98]. FET-based biosensors are based on semiconductor materials such as graphene, titanium oxide, etc., which can increase the efficacy and sensitivity in different device [101]. Recently, Changkyun Park and Seung Il Kim groups reported a FET-based biosensor device applied to detecting clinical samples infected with SARS-CoV-2. Their sensor immobilized graphene sheets of the FET with a specific antibody against S protein of SARS-CoV-2, which successfully performed into detecting antigen protein, cultured virus, and nasopharyngeal swab samples from clinical cases. The graphene-based FET device was proved to be sensitive in detecting the S protein of SARS-CoV-2 at concentration of 1 fg/mL in phosphate buffer solution and in clinical transport medium of 100 fg/mL. And the device was successfully applied to detecting COVID-19 in clinical samples (including 19 cases of patients and other healthy population), with the LODs of 16 pfu/mL and 242 copies/mL, respectively. Zhang et al. report a graphene FET (Gr-FET) immunosensor proved to be with high selective [102]; the immunosensor can rapidly identify the spike protein S1 of COVID-19 (about 2 min) and accurately capture the virus in a real-time manner with the LOD lower to 0.2 pmol/L.
Fig. 5.
The schematic diagram of FET based biosensor.
Reproduced with permission from Ref. [98]. 2020, ACS Nano
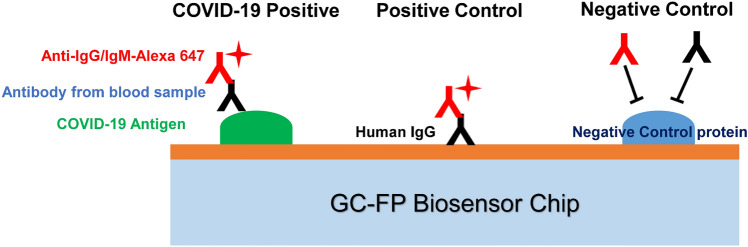
Grating-Coupled Fluorescent Plasmonics (GC-FP) Biosensor
There are also many other biosensors applied to the diagnosis of COVID-19. Nathaniel C. Cady and coworkers developed a multiplexed GC-FP biosensor [103] (illustrated in Fig. 6). They reported that their GC-FP biosensor platform can detect the specific antibody–antigen interactions for multiple targets in a single specimen, with 100% selectivity and sensitivity (n = 23) for the serum IgG against the COVID-19 antigens. The GC-FP biosensor proved to be with a linear response for serum specimens with a lower dilution ratio of 1:1600, and performed better than ELISA. The efficacy of the GC-FP platform with other specimen matrices, such as 63 cases of dried blood spot specimens was evaluated. The results were proved to be with high sensitivity (86.7%) and selectivity (100%) for the diagnostic of COVID-19 infection.
Fig. 6.
The schematic diagram of GC-FP biosensor chips, and the principle of assessing the blood samples with the antibody of COVID-19.
Reproduced with permission from Ref. [103]. 2021, Biosens Bioelectron
Capillary Electrophoresis(CE)
CE is a type of liquid-phase separation technology that uses a high-voltage electric field as a driving force, and a capillary as a separate channel to achieve separation based on differences in the mobility and distribution behavior of each component of a sample [104]. Under the action of a high-voltage electric field, the positive and negative ions in the solution move at different rates so that the outflow rates are different. Then the separation effects are achieved. CE requires only a small amount of specimens to achieve efficient and rapid analysis, and it is easy to implement automation. And with its colloidal nature, considerable molecular weight, and amphoteric dissociation characteristics, the virus is very suitable for CE detection [105]. In 2004, Zhou et al. designed a microfluidic chip system, which combined with CE technology and successfully improved the positive accuracy rate of SARS-CoV detection (94.44%). It is worth noting that the detection time is only 50–60 min [106]. CE technology can detect multiple viruses simultaneously, which will play an essential role in achieving rapid patient shunting, optimizing resource allocation, and shortening the diagnosis time before the diagnosis of COVID-19.
Computed Tomography (CT) Scan
CT scan is a medical platform that captures cross-sectional images via combined multiple X-ray measurements in different angles. CT, especially the high-resolution CT, has been performed with protocols of 1–5 mm slice thickness, presents the symptoms correlates with lung lesions, at the same time reveals the radiological changes in the lungs [107]. It is reported that 76.4% of COVID-19 cases were diagnosed with pneumonia upon admission [108]. The typical clinical symptoms of lungs include patchy bilateral shadows or/and ground-glass opacity in COVID-19 patients by CT scan. Ai et al. [109] reported that the diagnostic value and consistency of CT scans in compared with RT-PCR in the diagnostic of COVID-19: from 1014 cases of patients test by CT scan and RT-PCR assay, the positive results of RT-PCR and chest CT scans are 59% and 88%, respectively, proving lung CT with high sensitivity for the diagnostic of COVID-19. Recently, a 3D deep scan framework applied to diagnosing the pathological change of the diagnosed SARS-CoV-2 patients was reported, with 96% specificity and 90% sensitivity when evaluation of SARS-CoV-2 infected samples [110, 111]. However, a CT scan cannot diagnose the causes of viral pneumonia, only reveal the radiological change of lungs, and shows lower sensitivity than RT-PCR [112]. Therefore, RT-PCR-based techniques are remaining as the recommended techniques in the diagnostic of COVID-19, while the combination of RT-PCR and CT scan or other techniques are proved to be more effective tools in enhancing the sensitivity of COVID-19 diagnosis [113].
Summary and Outlook
Accurate and rapid identification of COVID-19 is critically important for timely treating patients and preventing COVID-19 spread-out. These laboratory diagnostic technologies should meet clinical testing requirements: strongly specific, highly sensitive, incredibly convenient, rapid, and economical. In addition to the above methods described, there are still needs for other ways to diagnose COVID-19. Some conventional techniques like CT and diagnosis of clinical symptoms are usually not accurate to identify SARS-CoV-2, due to there can be various causative antigens that result in respiratory tract viral infection like the influenza virus, RSV, coronavirus, human adenovirus (hAdV), and hRV [14]. So far, RT-qPCR is the most widely used diagnostic technique in various countries. Other diagnostic techniques are still urgently needed for the diagnosis of COVID-19, including immunofluorescence assay (IFA), and ELISA-based immunoassay, which is still not available alone for the diagnosis of SRAS-CoV-2, and as the complementary methods to PCR-based diagnostic testing. Other faster and more accurate clinical diagnostic tests are highly expected. Hopefully, the pandemics will be overcome as soon as possible with the advent of vaccines of SARS-CoV-2.
Acknowledgements
The project is supported by the Shandong Natural Science Foundation (ZR2018ZC0233) and the Taishan Scholar Program at Shandong Province.
Author Contributions
Gao-Pan Dong, Ying-Ai Sun and Zheng Zhang draft the manuscript. Gao-Pan Dong and Xiu-Juan Guo reproduce the figures. Min-Yong Li and Lu-Pei Du revised the manuscript. The final manuscript was approved by all authors.
Funding
The Shandong Natural Science Foundation (ZR2018ZC0233) and the Taishan Scholar Program at Shandong Province.
Availability of Data and Material
Not applicable.
Code Availability
Microsoft Office 2016, Adobe Illustrator CS6 software.
Declarations
Conflict of interest
The authors declare no conflicts of interest.
References
- 1.Chan JF, Yuan S, Kok KH, To KK, Chu H, Yang J, Xing F, Liu J, Yip CC, Poon RW, Tsoi HW, Lo SK, Chan KH, Poon VK, Chan WM, Ip JD, Cai JP, Cheng VC, Chen H, Hui CK, Yuen KY. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395(10223):514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, Ren R, Leung KSM, Lau EHY, Wong JY, Xing X, Xiang N, Wu Y, Li C, Chen Q, Li D, Liu T, Zhao J, Liu M, Tu W, Chen C, Jin L, Yang R, Wang Q, Zhou S, Wang R, Liu H, Luo Y, Liu Y, Shao G, Li H, Tao Z, Yang Y, Deng Z, Liu B, Ma Z, Zhang Y, Shi G, Lam TTY, Wu JT, Gao GF, Cowling BJ, Yang B, Leung GM, Feng Z. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med. 2020;382(13):1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tan W, Zhao X, Ma X, Wang W, Nie P, Xu W, Gao GF, Wu G. A novel coronavirus genome identified in a cluster of pneumonia cases—Wuhan, China 2019–2020. China CDC Weekly. 2020;2(4):61–62. doi: 10.46234/ccdcw2020.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li X, Wang W, Zhao X, Zai J, Zhao Q, Li Y, Chaillon A. Transmission dynamics and evolutionary history of 2019-nCoV. J Med Virol. 2020;92(5):501–511. doi: 10.1002/jmv.25701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang S, Xia S, Ying T, Lu L. A novel coronavirus (2019-nCoV) causing pneumonia-associated respiratory syndrome. Cell Mol Immunol. 2020;17(5):554. doi: 10.1038/s41423-020-0372-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oberfeld B, Achanta A, Carpenter K, Chen P, Gilette NM, Langat P, Said JT, Schiff AE, Zhou AS, Barczak AK, Pillai S. SnapShot: COVID-19. Cell. 2020;181(4):954–954.e1. doi: 10.1016/j.cell.2020.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saiki RK, Scharf S, Faloona F, Mullis KB, Horn GT, Erlich HA, Arnheim N. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985;230(4732):1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- 10.Lin C, Ye R, Xia YL. A meta-analysis to evaluate the effectiveness of real-time PCR for diagnosing novel coronavirus infections. Genet Mol Res. 2015;14(4):15634–15641. doi: 10.4238/2015.December.1.15. [DOI] [PubMed] [Google Scholar]
- 11.Poon LL, Chan KH, Wong OK, Yam WC, Yuen KY, Guan Y, Lo YM, Peiris JS. Early diagnosis of SARS coronavirus infection by real time RT-PCR. J Clin Virol. 2003;28(3):233–238. doi: 10.1016/j.jcv.2003.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poon LL, Wong BW, Chan KH, Ng SS, Yuen KY, Guan Y, Peiris JS. Evaluation of real-time reverse transcriptase PCR and real-time loop-mediated amplification assays for severe acute respiratory syndrome coronavirus detection. J Clin Microbiol. 2005;43(7):3457–3459. doi: 10.1128/JCM.43.7.3457-3459.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mahony JB, Richardson S. Molecular diagnosis of severe acute respiratory syndrome: the state of the art. J Mol Diagn. 2005;7(5):551–559. doi: 10.1016/S1525-1578(10)60587-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang N, Wang L, Deng X, Liang R, Su M, He C, Hu L, Su Y, Ren J, Yu F, Du L, Jiang S. Recent advances in the detection of respiratory virus infection in humans. J Med Virol. 2020;92(4):408–417. doi: 10.1002/jmv.25674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bellau-Pujol S, Vabret A, Legrand L, Dina J, Gouarin S, Petitjean-Lecherbonnier J, Pozzetto B, Ginevra C, Freymuth F. Development of three multiplex RT-PCR assays for the detection of 12 respiratory RNA viruses. J Virol Methods. 2005;126(1–2):53–63. doi: 10.1016/j.jviromet.2005.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Corman V, Eckerle I, Bleicker T, Zaki A, Landt O, Eschbachbludau M, Van Boheemen S, Gopal R, Ballhause M, Bestebroer TM, Muth D, Müller MA, Drexler JF, Zambon M, Osterhaus AD, Fouchier RM, Drosten C. Detection of a novel human coronavirus by real-time reverse-transcription polymerase chain reaction. Euro Surveill. 2012;17(39):20285. doi: 10.2807/ese.17.39.20285-en. [DOI] [PubMed] [Google Scholar]
- 17.Bizouarn F. Clinical applications using digital PCR. Methods Mol Biol. 2014;1160:189–214. doi: 10.1007/978-1-4939-0733-5_16. [DOI] [PubMed] [Google Scholar]
- 18.Lo YM, Chan KC. Setting Up a Polymerase Chain Reaction Laboratory. Methods Mol Biol. 2006;336:11–18. doi: 10.1385/1-59745-074-X:11. [DOI] [PubMed] [Google Scholar]
- 19.Clinical Applications of PCR. Second Edition, (Edited by Y. M. Dennis Lo, Rossa W. K. Chiu, K. C. Allen Chan) Humana Press Inc. 2006.
- 20.Essaidi-Laziosi M, Lyon M, Mamin A, Fernandes Rocha M, Kaiser L, Tapparel C. A new real-time RT-qPCR assay for the detection, subtyping and quantification of human respiratory syncytial viruses positive- and negative-sense RNAs. J Virol Methods. 2016;235:9–14. doi: 10.1016/j.jviromet.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 21.Sakthivel SK, Whitaker B, Lu X, Oliveira DB, Stockman LJ, Kamili S, Oberste MS, Erdman DD. Comparison of fast-track diagnostics respiratory pathogens multiplex real-time RT-PCR assay with in-house singleplex assays for comprehensive detection of human respiratory viruses. J Virol Methods. 2012;185(2):259–266. doi: 10.1016/j.jviromet.2012.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, Chen J, Jie Z, Qiu H, Xu K, Xu X, Lu H, Zhu W, Gao Z, Xiang N, Shen Y, He Z, Gu Y, Zhang Z, Yang Y, Zhao X, Zhou L, Li X, Zou S, Zhang Y, Li X, Yang L, Guo J, Dong J, Li Q, Dong L, Zhu Y, Bai T, Wang S, Hao P, Yang W, Zhang Y, Han J, Yu H, Li D, Gao GF, Wu G, Wang Y, Yuan Z, Shu Y. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med. 2013;368(20):1888–1897. doi: 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]
- 23.Lu X, Whitaker B, Sakthivel SK, Kamili S, Rose LE, Lowe L, Mohareb E, Elassal EM, Al-sanouri T, Haddadin A, Erdman DD. Real-time reverse transcription-PCR assay panel for middle east respiratory syndrome coronavirus. J Clin Microbiol. 2014;52(1):67–75. doi: 10.1128/JCM.02533-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huh HJ, Kim JY, Kwon HJ, Yun SA, Lee MK, Ki CS, Lee NY, Kim JW. Performance evaluation of the PowerChek MERS (upE & ORF1a) real-time PCR kit for the detection of middle east respiratory syndrome coronavirus RNA. Ann Lab Med. 2017;37(6):494–498. doi: 10.3343/alm.2017.37.6.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Niu P, Lu R, Lan J, Liu G, Wang W, Tan W. Development of novel multiplex real-time RT-PCR assays for detection of MERS-CoV infection. Chin J Virol. 2016;32(3):349–354. [PubMed] [Google Scholar]
- 26.Elpaeva EA, Komissarov AB, Pisareva MM, Grudinin MP, Kiselev OI. New method for determining hepatitis b virus resistance mutations m204i/v to nucleos(t)ide analogues in patients with chronic hepatitis b. Infekci I Immunitet. 2015;5(3):265–272. [Google Scholar]
- 27.Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, Bleicker T, Brünink S, Schneider J, Schmidt ML, Mulders DG, Haagmans BL, van der Veer B, van den Brink S, Wijsman L, Goderski G, Romette JL, Ellis J, Zambon M, Peiris M, Goossens H, Reusken C, Koopmans MP, Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3):2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poon LL, Wong OK, Chan KH, Luk W, Yuen KY, Peiris JS, Guan Y. Rapid diagnosis of a coronavirus associated with severe acute respiratory syndrome (SARS) Clin Chem. 2003;49(6 Pt 1):953–955. doi: 10.1373/49.6.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen L, Zhang G, Liu L, Li Z. Emerging biosensing technologies for improved diagnostics of COVID-19 and future pandemics. Talanta. 2021;225:121986. doi: 10.1016/j.talanta.2020.121986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cleary B, Hay JA, Blumenstiel B, Harden M, Cipicchio M, Bezney J, Simonton B, Hong D, Senghore M, Sesay AK, Gabriel S, Regev A, Mina MJ. Using viral load and epidemic dynamics to optimize pooled testing in resource-constrained settings. Sci Transl Med. 2021;13(589):eabf1568. doi: 10.1126/scitranslmed.abf1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang Z, Tian D, Liu Y, Lin Z, Lyon CJ, Lai W, Fusco D, Drouin A, Yin X, Hu T, Ning B. Ultra-sensitive and high-throughput CRISPR-p owered COVID-19 diagnosis. Biosens Bioelectron. 2020;164:112316. doi: 10.1016/j.bios.2020.112316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bruch R, Baaske J, Chatelle C, Meirich M, Madlener S, Weber W, Dincer C, Urban GA. CRISPR/Cas13a-powered electrochemical microfluidic biosensor for nucleic acid amplification-free miRNA diagnostics. Adv Mater. 2019;31(51):e1905311. doi: 10.1002/adma.201905311. [DOI] [PubMed] [Google Scholar]
- 33.Bruch R, Urban GA, Dincer C. CRISPR/Cas Powered Multiplexed Biosensing. Trends Biotechnol. 2019;37(8):791–792. doi: 10.1016/j.tibtech.2019.04.005. [DOI] [PubMed] [Google Scholar]
- 34.Hajian R, Balderston S, Tran T, deBoer T, Etienne J, Sandhu M, Wauford NA, Chung JY, Nokes J, Athaiya M, Paredes J, Peytavi R, Goldsmith B, Murthy N, Conboy IM, Aran K. Detection of unamplified target genes via CRISPR-Cas9 immobilized on a graphene field-effect transistor. Nat Biomed Eng. 2019;3(6):427–437. doi: 10.1038/s41551-019-0371-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen JS, Ma E, Harrington LB, Da Costa M, Tian X, Palefsky JM, Doudna JA. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360(6387):436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Freije CA, Myhrvold C, Boehm CK, Lin AE, Welch NL, Carter A, Metsky HC, Luo CY, Abudayyeh OO, Gootenberg JS, Yozwiak NL, Zhang F, Sabeti PC. Programmable inhibition and detection of RNA viruses using Cas13. Mol Cell. 2019;76(5):826–837. doi: 10.1016/j.molcel.2019.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wright AV, Nuñez JK, Doudna JA. Biology and Applications of CRISPR Systems: harnessing Nature's Toolbox for Genome Engineering. Cell. 2016;164(1–2):29–44. doi: 10.1016/j.cell.2015.12.035. [DOI] [PubMed] [Google Scholar]
- 38.Gootenberg JS, Abudayyeh OO, Lee JW, Essletzbichler P, Dy AJ, Joung J, Verdine V, Donghia N, Daringer NM, Freije CA, Myhrvold C, Bhattacharyya RP, Livny J, Regev A, Koonin EV, Hung DT, Sabeti PC, Collins JJ, Zhang F. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356(6336):438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sykes PJ, Neoh SH, Brisco MJ, Hughes E, Condon J, Morley AA. Quantitation of targets for PCR by use of limiting dilution. Biotechniques. 1992;13(3):444–449. [PubMed] [Google Scholar]
- 40.Kanagal-Shamanna R. Digital PCR: principles and applications. Methods Mol Biol. 2016;1392:43–50. doi: 10.1007/978-1-4939-3360-0_5. [DOI] [PubMed] [Google Scholar]
- 41.Lo YM, Lun FM, Chan KC, Tsui NB, Chong KC, Lau TK, Leung TY, Zee BC, Cantor CR, Chiu RW. Digital PCR for the molecular detection of fetal chromosomal aneuploidy. Proc Natl Acad Sci U S A. 2007;104(32):13116–13121. doi: 10.1073/pnas.0705765104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanders R, Huggett JF, Bushell CA, Cowen S, Scott DJ, Foy CA. Evaluation of digital PCR for absolute DNA quantification. Anal Chem. 2011;83(17):6474–6484. doi: 10.1021/ac103230c. [DOI] [PubMed] [Google Scholar]
- 43.Pomari E, Piubelli C, Perandin F, Bisoffi Z. Digital PCR: a new technology for diagnosis of parasitic infections. Clin Microbiol Infect. 2019;25(12):1510–1516. doi: 10.1016/j.cmi.2019.06.009. [DOI] [PubMed] [Google Scholar]
- 44.Dong L, Meng Y, Sui Z, Wang J, Wu L, Fu B. Comparison of four digital PCR platforms for accurate quantification of DNA copy number of a certified plasmid DNA reference material. Sci Rep. 2015;5:13174. doi: 10.1038/srep13174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morley AA. Digital PCR: a brief history. Biomol Detect Quantif. 2014;1(1):1–2. doi: 10.1016/j.bdq.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suo T, Liu X, Feng J, Guo M, Hu W, Guo D, Ullah H, Yang Y, Zhang Q, Wang X, Sajid M, Huang Z, Deng L, Chen T, Liu F, Xu K, Liu Y, Zhang Q, Liu Y, Xiong Y, Chen G, Lan K, Chen Y. ddPCR: a more accurate tool for SARS-CoV-2 detection in low viral load specimens. Emerg Microbes Infect. 2020;9(1):1259–1268. doi: 10.1080/22221751.2020.1772678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dong L, Zhou J, Niu C, Wang Q, Pan Y, Sheng S, Wang X, Zhang Y, Yang J, Liu M, Zhao Y, Zhang X, Zhu T, Peng T, Xie J, Gao Y, Wang D, Dai X, Fang X. Highly accurate and sensitive diagnostic detection of SARS-CoV-2 by digital PCR. Talanta. 2021;224:121726. doi: 10.1016/j.talanta.2020.121726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Salipante SJ, Jerome KR. Digital PCR-An Emerging Technology with Broad Applications in Microbiology. Clin Chem. 2020;66(1):117–123. doi: 10.1373/clinchem.2019.304048. [DOI] [PubMed] [Google Scholar]
- 49.Zachertowska A, Brewer D, Evans DH. Characterization of the major capsid proteins of myxoma virus particles using MALDI-TOF mass spectrometry. J Virol Methods. 2005;132(1–2):1–12. doi: 10.1016/j.jviromet.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 50.Yeo YJ, Roh K, Bang JY, Lee EH, Park HS, Kim DE. Quantitative detection of single base mutation by combining PNA hybridization and MALDI-TOF mass analysis. Chem Commun (Camb) 2013;49(36):3754–3756. doi: 10.1039/c3cc00070b. [DOI] [PubMed] [Google Scholar]
- 51.Xiu L, Zhang C, Wu Z, Peng J. Establishment and application of a universal coronavirus screening method using MALDI-TOF mass spectrometry. Front Microbiol. 2017;8:1510. doi: 10.3389/fmicb.2017.01510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu N, Wang L, Cai G, Zhang D, Lin J. Establishment of a simultaneous detection method for ten duck viruses using MALDI-TOF mass spectrometry. J Virol Methods. 2019;273:113723. doi: 10.1016/j.jviromet.2019.113723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wong YP, Othman S, Lau YL, Radu S, Chee HY. Loop-mediated isothermal amplification (LAMP): a versatile technique for detection of micro-organisms. J Appl Microbiol. 2018;124(3):626–643. doi: 10.1111/jam.13647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28(12):e63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nagamine K, Hase T, Notomi T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol Cell Probes. 2002;16(3):223–229. doi: 10.1006/mcpr.2002.0415. [DOI] [PubMed] [Google Scholar]
- 56.Mori Y, Nagamine K, Tomita N, Notomi T. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem Biophys Res Commun. 2001;289(1):150–154. doi: 10.1006/bbrc.2001.5921. [DOI] [PubMed] [Google Scholar]
- 57.Hong TC, Mai QL, Cuong DV, Parida M, Minekawa H, Notomi T, Hasebe F, Morita K. Development and evaluation of a novel loop-mediated isothermal amplification method for rapid detection of severe acute respiratory syndrome coronavirus. J Clin Microbiol. 2004;42(5):1956–1961. doi: 10.1128/JCM.42.5.1956-1961.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shirato K, Nishimura H, Saijo M, Okamoto M, Noda M, Tashiro M, Taguchi F. Diagnosis of human respiratory syncytial virus infection using reverse transcription loop-mediated isothermal amplification. J Virol Methods. 2007;139(1):78–84. doi: 10.1016/j.jviromet.2006.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ushio M, Yui I, Yoshida N, Fujino M, Yonekawa T, Ota Y, Notomi T, Nakayama T. Detection of respiratory syncytial virus genome by subgroups-A, B specific reverse transcription loop-mediated isothermal amplification (RT-LAMP) J Med Virol. 2005;77(1):121–127. doi: 10.1002/jmv.20424. [DOI] [PubMed] [Google Scholar]
- 60.Imai M, Ninomiya A, Minekawa H, Notomi T, Ishizaki T, Tashiro M, Odagiri T. Development of H5-RT-LAMP (loop-mediated isothermal amplification) system for rapid diagnosis of H5 avian influenza virus infection. Vaccine. 2006;24(44–46):6679–6682. doi: 10.1016/j.vaccine.2006.05.046. [DOI] [PubMed] [Google Scholar]
- 61.Mahony J, Chong S, Bulir D, Ruyter A, Mwawasi K, Waltho D. Multiplex loop-mediated isothermal amplification (M-LAMP) assay for the detection of influenza A/H1, A/H3 and influenza B can provide a specimen-to-result diagnosis in 40 min with single genome copy sensitivity. J Clin Virol. 2013;58(1):127–131. doi: 10.1016/j.jcv.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 62.Xu C, Wang H, Jin H, Feng N, Zheng X, Cao Z, Li L, Wang J, Yan F, Wang L, Chi H, Gai W, Wang C, Zhao Y, Feng Y, Wang T, Gao Y, Lu Y, Yang S, Xia X. Visual detection of Ebola virus using reverse transcription loop-mediated isothermal amplification combined with nucleic acid strip detection. Arch Virol. 2016;161(5):1125–1133. doi: 10.1007/s00705-016-2763-5. [DOI] [PubMed] [Google Scholar]
- 63.Cao Z, Wang H, Wang L, Li L, Jin H, Xu C, Feng N, Wang J, Li Q, Zhao Y, Wang T, Gao Y, Lu Y, Yang S, Xia X. Visual detection of west nile virus using reverse transcription loop-mediated isothermal amplification combined with a vertical flow visualization strip. Front Microbiol. 2016;7:554. doi: 10.3389/fmicb.2016.00554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Parida M, Posadas G, Inoue S, Hasebe F, Morita K. Real-time reverse transcription loop-mediated isothermal amplification for rapid detection of West Nile virus. J Clin Microbiol. 2004;42(1):257–263. doi: 10.1128/JCM.42.1.257-263.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huang P, Wang H, Cao Z, Jin H, Chi H, Zhao J, Yu B, Yan F, Hu X, Wu F, Jiao C, Hou P, Xu S, Zhao Y, Feng N, Wang J, Sun W, Wang T, Gao Y, Yang S, Xia X. A rapid and specific assay for the detection of MERS-CoV. Front Microbiol. 2018;9:1101. doi: 10.3389/fmicb.2018.01101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Park S, Zhang Y, Lin S, Wang TH, Yang S. Advances in microfluidic PCR for point-of-care infectious disease diagnostics. Biotechnol Adv. 2011;29(6):830–839. doi: 10.1016/j.biotechadv.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kost GJ, Katip P, Curtis CM. Strategic point-of-care requirements of hospitals and public health for preparedness in regions at risk. Point Care. 2012;11(2):114–118. doi: 10.1097/POC.0b013e31825a2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hellewell J, Abbott S, Gimma A, Bosse NI, Jarvis CI, Russell TW, Munday JD, Kucharski AJ, Edmunds WJ, Centre for the Mathematical Modelling of Infectious Diseases COVID-19 Working Group. Funk S, Eggo RM. Feasibility of controlling COVID-19 outbreaks by isolation of cases and contacts. Lancet Glob Health. 2020;8(4):e488–e496. doi: 10.1016/S2214-109X(20)30074-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee HK, Lee BH, Seok SH, Baek MW, Lee HY, Kim DJ, Na YR, Noh KJ, Park SH, Kumar DN, Kariwa H, Nakauchi M, Heo SJ, Park JH. Production of specific antibodies against SARS-coronavirus nucleocapsid protein without cross reactivity with human coronaviruses 229E and OC43. J Vet Sci. 2010;11(2):165–167. doi: 10.4142/jvs.2010.11.2.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li Z, Yi Y, Luo X, Xiong N, Liu Y, Li S, Sun R, Wang Y, Hu B, Chen W, Zhang Y, Wang J, Huang B, Lin Y, Yang J, Cai W, Wang X, Cheng J, Chen Z, Sun K, Pan W, Zhan Z, Chen L, Ye F. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J Med Virol. 2020;92(9):1518–1524. doi: 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen Z, Zhang Z, Zhai X, Li Y, Lin L, Zhao H, Bian L, Li P, Yu L, Wu Y, Lin G. Rapid and sensitive detection of anti-SARS-CoV-2 IgG, using lanthanide-doped nanoparticles-based lateral flow immunoassay. Anal Chem. 2020;92(10):7226–7231. doi: 10.1021/acs.analchem.0c00784. [DOI] [PubMed] [Google Scholar]
- 72.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan. China Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fang B, Meng QH. The laboratory's role in combating COVID-19. Crit Rev Clin Lab Sci. 2020;57(6):400–414. doi: 10.1080/10408363.2020.1776675. [DOI] [PubMed] [Google Scholar]
- 74.Lin AV. Indirect ELISA. Methods Mol Biol. 2015;1318:51–59. doi: 10.1007/978-1-4939-2742-5_5. [DOI] [PubMed] [Google Scholar]
- 75.Konstantinou GN. Enzyme-Linked Immunosorbent Assay (ELISA) Methods Mol Biol. 2017;1592:79–94. doi: 10.1007/978-1-4939-6925-8_7. [DOI] [PubMed] [Google Scholar]
- 76.Sheikhzadeh E, Eissa S, Ismail A, Zourob M. Diagnostic techniques for COVID-19 and new developments. Talanta. 2020;220:121392. doi: 10.1016/j.talanta.2020.121392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xiang J, Yan M, Li H, Liu T, Shen C. Evaluation of enzyme-linked immunoassay and colloidal gold- immunochromatographic assay kit for detection of novel coronavirus (SARS-Cov-2) causing an outbreak of pneumonia (COVID-19). 2020. 10.1101/2020.02.27.20028787.
- 78.Guo L, Ren L, Yang S, Xiao M, Chang D, Yang F, Dela Cruz CS, Wang Y, Wu C, Xiao Y, Zhang L, Han L, Dang S, Xu Y, Yang QW, Xu SY, Zhu HD, Xu YC, Jin Q, Sharma L, Wang L, Wang J. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19) Clin Infect Dis. 2020;71(15):778–785. doi: 10.1093/cid/ciaa310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li R, Pei S, Chen B, Song Y, Zhang T, Yang W, Shaman J. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV-2) Science. 2020;368(6490):489–493. doi: 10.1126/science.abb3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rikhtegaran Tehrani Z, Saadat S, Saleh E, Ouyang X, Constantine N, DeVico AL, Harris AD, Lewis GK, Kottilil S, Sajadi MM. Performance of nucleocapsid and spike-based SARS-CoV-2 serologic assays. PLoS ONE. 2020;15(11):e0237828. doi: 10.1371/journal.pone.0237828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.U.S. Food and Drug Administration. FAQs on Testing for SARS-CoV-2. https://www.fda.gov/medical-devices/emergency-situations-medical-devices/faqs-testing-sars-cov-2#serology. Accessed 25 Mar 2021.
- 82.World Heath Organization. Laboratory testing for 2019 novel coronavirus (2019-nCoV) in suspected human cases. https://www.who.int/publications-detail/laboratory-testing-for-2019-novel-coronavirus-in-suspected-human-cases-20200117. Accessed 19 Mar 2020.
- 83.Centers for Disease Control and Prevention. Serology Testing for COVID-19. https://www.cdc.gov/coronavirus/2019-ncov/lab/serologytesting.html. Accessed 3 Nov 2020.
- 84.Duan K, Liu B, Li C, Zhang H, Yu T, Qu J, Zhou M, Chen L, Meng S, Hu Y, Peng C, Yuan M, Huang J, Wang Z, Yu J, Gao X, Wang D, Yu X, Li L, Zhang J, Wu X, Li B, Xu Y, Chen W, Peng Y, Hu Y, Lin L, Liu X, Huang S, Zhou Z, Zhang L, Wang Y, Zhang Z, Deng K, Xia Z, Gong Q, Zhang W, Zheng X, Liu Y, Yang H, Zhou D, Yu D, Hou J, Shi Z, Chen S, Chen Z, Zhang X, Yang X. Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc Natl Acad Sci U S A. 2020;117(17):9490–9496. doi: 10.1073/pnas.2004168117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Okba NMA, Müller MA, Li W, Wang C, GeurtsvanKessel CH, Corman VM, Lamers MM, Sikkema RS, de Bruin E, Chandler FD, Yazdanpanah Y, Le Hingrat Q, Descamps D, Houhou-Fidouh N, Reusken CBEM, Bosch BJ, Drosten C, Koopmans MPG, Haagmans BL. Severe acute respiratory syndrome Coronavirus 2-specific antibody responses in coronavirus disease patients. Emerg Infect Dis. 2020;26(7):1478–1488. doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ribeiro BV, Cordeiro TAR, Oliveira e Freitas GR, Ferreira LF, Franco DL. Biosensors for the detection of respiratory viruses: a review. Talanta open. 2020;2:100007. doi: 10.1016/j.talo.2020.100007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Behera S, Rana G, Satapathy S, Mohanty M, Pradhan S, Panda MK, Ningthoujam R, Hazarika BN, Singh YD. Biosensors in diagnosing COVID-19 and recent development. Sens Int. 2020;1:100054. doi: 10.1016/j.sintl.2020.100054. [DOI] [Google Scholar]
- 88.Bhunia AK, Taitt CR, Kim, MS. High throughput screening strategies and technology platforms for detection of pathogens: an introduction. In: High throughput screening for food safety assessment; 2015. p. 1–9.
- 89.Sin ML, Mach KE, Wong PK, Liao JC. Advances and challenges in biosensor-based diagnosis of infectious diseases. Expert Rev Mol Diagn. 2014;14(2):225–244. doi: 10.1586/14737159.2014.888313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Samson R, Navale GR, Dharne MS. Biosensors: frontiers in rapid detection of COVID-19. 3 Biotech. 2020;10(9):385. doi: 10.1007/s13205-020-02369-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kurt H, Eyüpoğlu AE, Sütlü T, Budak H, Yüce M. Plasmonic selection of ssDNA aptamers against fibroblast growth factor receptor. ACS Comb Sci. 2019;21(8):578–587. doi: 10.1021/acscombsci.9b00059. [DOI] [PubMed] [Google Scholar]
- 92.Yüce M, Filiztekin E, Özkaya KG. COVID-19 diagnosis—a review of current methods. Biosens Bioelectron. 2021;172:112752. doi: 10.1016/j.bios.2020.112752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Saylan Y, Erdem Ö, Ünal S, Denizli A. An alternative medical diagnosis method: biosensors for virus detection. Biosensors (Basel) 2019;9(2):65. doi: 10.3390/bios9020065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Djaileb A, Charro B, Jodaylami MH, Thibault V, Coutu J, Stevenson K, Forest S, S.Live L, Boudreau D, N.Pelletier J, Masson JF. A rapid and quantitative serum test for SARS-CoV-2 antibodies with portable surface plasmon resonance sensing. 2020. 10.26434/chemrxiv.12118914.
- 95.Qiu G, Gai Z, Tao Y, Schmitt J, Kullak-Ublick GA, Wang J. Dual-functional plasmonic photothermal biosensors for highly accurate severe acute respiratory syndrome Coronavirus 2 DETECTION. ACS Nano. 2020;14(5):5268–5277. doi: 10.1021/acsnano.0c02439. [DOI] [PubMed] [Google Scholar]
- 96.Nehra A, Pal Singh K. Current trends in nanomaterial embedded field effect transistor-based biosensor. Biosens Bioelectron. 2015;74:731–743. doi: 10.1016/j.bios.2015.07.030. [DOI] [PubMed] [Google Scholar]
- 97.Bergveld P. Development of an ion-sensitive solid-state device for neurophysiological measurements. IEEE Trans Biomed Eng. 1970;17(1):70–71. doi: 10.1109/TBME.1970.4502688. [DOI] [PubMed] [Google Scholar]
- 98.Seo G, Lee G, Kim MJ, Baek SH, Choi M, Ku KB, Lee CS, Jun S, Park D, Kim HG, Kim SJ, Lee JO, Kim BT, Park EC, Kim SI. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14(4):5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- 99.Janissen R, Sahoo PK, Santos CA, da Silva AM, von Zuben AAG, Souto DEP, Costa ADT, Celedon P, Zanchin NIT, Almeida DB, Oliveira DS, Kubota LT, Cesar CL, Souza AP, Cotta MA. InP nanowire biosensor with tailored biofunctionalization: ultrasensitive and highly selective disease biomarker detection. Nano Lett. 2017;17(10):5938–5949. doi: 10.1021/acs.nanolett.7b01803. [DOI] [PubMed] [Google Scholar]
- 100.Liu J, Chen X, Wang Q, Xiao M, Zhong D, Sun W, Zhang G, Zhang Z. Ultrasensitive monolayer MoS(2) field-effect transistor based DNA sensors for screening of down syndrome. Nano Lett. 2019;19(3):1437–1444. doi: 10.1021/acs.nanolett.8b03818. [DOI] [PubMed] [Google Scholar]
- 101.Chen X, Liu Y, Fang X, Li Z, Pu H, Chang J, Chen J, Mao S. Ultratrace antibiotic sensing using aptamer/graphene-based field-effect transistors. Biosens Bioelectron. 2019;126:664–671. doi: 10.1016/j.bios.2018.11.034. [DOI] [PubMed] [Google Scholar]
- 102.Zhang X, Qi Q, Jing Q, Ao S, Zhang Z, Ding M, Wu M, Liu K, Wang W, Ling Y. Electrical probing of COVID-19 spike protein receptor binding domain via a graphene field-effect transistor. 2020.
- 103.Cady NC, Tokranova N, Minor A, Nikvand N, Strle K, Lee WT, Page W, Guignon E, Pilar A, Gibson GN. Multiplexed detection and quantification of human antibody response to COVID-19 infection using a plasmon enhanced biosensor platform. Biosens Bioelectron. 2021;171:112679. doi: 10.1016/j.bios.2020.112679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kostal V, Arriaga EA. Recent advances in the analysis of biological particles by capillary electrophoresis. Electrophoresis. 2008;29(12):2578–2586. doi: 10.1002/elps.200700917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kremser L, Blaas D, Kenndler E. Virus analysis using electromigration techniques. Electrophoresis. 2009;30(1):133–140. doi: 10.1002/elps.200800590. [DOI] [PubMed] [Google Scholar]
- 106.Zhou Z, Liu D, Zhong R, Dai Z, Wu D, Wang H, Du Y, Xia Z, Zhang L, Mei X. Determination of SARS-coronavirus by a microfluidic chip system. Electrophoresis. 2004;25(17):3032–3039. doi: 10.1002/elps.200305966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Waller JV, Kaur P, Tucker A, Lin KK, Diaz MJ, Henry TS, Hope M. Diagnostic tools for Coronavirus disease (COVID-19): comparing CT and RT-PCR viral nucleic acid testing. AJR Am J Roentgenol. 2020;215(4):834–838. doi: 10.2214/AJR.20.23418. [DOI] [PubMed] [Google Scholar]
- 108.Jiang F, Deng L, Zhang L, Cai Y, Cheung CW, Xia Z. Review of the clinical characteristics of Coronavirus disease 2019 (COVID-19) J Gen Intern Med. 2020;35(5):1545–1549. doi: 10.1007/s11606-020-05762-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ai T, Yang Z, Hou H, Zhan C, Chen C, Lv W, Tao Q, Sun Z, Xia L. Correlation of chest CT and RT-PCR testing for Coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020;296(2):E32–E40. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Li Y, Xia L. Coronavirus disease 2019 (COVID-19): role of chest CT in diagnosis and management. AJR Am J Roentgenol. 2020;214(6):1280–1286. doi: 10.2214/AJR.20.22954. [DOI] [PubMed] [Google Scholar]
- 111.Fang Y, Zhang H, Xie J, Lin M, Ying L, Pang P, Ji W. Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology. 2020;296(2):E115–E117. doi: 10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yu Z, Li X, Sun H, Wang J, Zhao T, Chen H, Ma Y, Zhu S, Xie Z. Rapid identification of COVID-19 severity in CT scans through classification of deep features. Biomed Eng Online. 2020;19(1):63–000. doi: 10.1186/s12938-020-00807-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tenda ED, Yulianti M, Asaf MM, Yunus RE, Septiyanti W, Wulani V, Pitoyo CW, Rumende CM, Setiati S. The importance of chest CT scan in COVID-19. Acta Med Indones. 2020;52(1):68–73. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.
Microsoft Office 2016, Adobe Illustrator CS6 software.