Abstract
Glutaredoxins are members of a superfamily of thiol disulfide oxidoreductases involved in maintaining the redox state of target proteins. In Saccharomyces cerevisiae, two glutaredoxins (Grx1 and Grx2) containing a cysteine pair at the active site had been characterized as protecting yeast cells against oxidative damage. In this work, another subfamily of yeast glutaredoxins (Grx3, Grx4, and Grx5) that differs from the first in containing a single cysteine residue at the putative active site is described. This trait is also characteristic for a number of glutaredoxins from bacteria to humans, with which the Grx3/4/5 group has extensive homology over two regions. Mutants lacking Grx5 are partially deficient in growth in rich and minimal media and also highly sensitive to oxidative damage caused by menadione and hydrogen peroxide. A significant increase in total protein carbonyl content is constitutively observed in grx5 cells, and a number of specific proteins, including transketolase, appear to be highly oxidized in this mutant. The synthetic lethality of the grx5 and grx2 mutations on one hand and of grx5 with the grx3 grx4 combination on the other points to a complex functional relationship among yeast glutaredoxins, with Grx5 playing a specially important role in protection against oxidative stress both during ordinary growth conditions and after externally induced damage. Grx5-deficient mutants are also sensitive to osmotic stress, which indicates a relationship between the two types of stress in yeast cells.
Reactive oxygen compounds, such as hydrogen peroxide, the superoxide anion, and the hydroxyl radical derived from the latter, exert toxic effects on diverse cellular molecules, including the oxidation of protein thiol groups (10). Cells have developed a number of protective mechanisms against this oxidant effect on proteins, the thiol-disulfide oxidoreductase activities of thioredoxins and glutaredoxins being among the more significant of these (9, 17, 18, 34). While thioredoxin directly reduces protein disulfide groups with NADPH as the hydrogen donor, the tripeptide thiol glutathione (l-γ-glutamyl-l-cysteinyl-glycine) in its reduced form (GSH) acts as the hydrogen donor for the reduction of protein disulfides by glutaredoxin (17). It has been proposed elsewhere that thioredoxin and glutaredoxin systems are essential for maintaining the adequate redox state of proteins in the intracellular environment and thus for regulating various cellular activities (1, 9, 17). However, only ribonucleotide reductase and 3′-phosphoadenylylsulfate reductase have been firmly recognized as in vivo targets for both systems (2, 28, 37). Even in this case, not all Escherichia coli glutaredoxins seem to participate in protection against oxidation of these substrates (28, 51). Thus, many of the in vivo targets of thioredoxins and glutaredoxins are still to be elucidated (1). Nevertheless, the presence of both thioredoxins and glutaredoxins in different organisms, together with the conservation of their active sites through evolution (17, 18), points to their important role as intracellular protein antioxidants (1).
Two genes encoding glutaredoxins (GRX1 and GRX2) in Saccharomyces cerevisiae have been characterized elsewhere (12, 29). Grx2 accounts for most of the glutaredoxin activity during exponential growth (29). The GRX1 and GRX2 gene products are highly homologous to rice, pig, and human glutaredoxins, as well as to two E. coli glutaredoxins (18, 29). Cell growth is not affected in individual and double grx1 grx2 mutants in either rich or minimal medium. On the other hand, while grx1 mutant cells are particularly sensitive to oxidative stress caused by menadione (a generator of superoxide anions), the grx2 mutant is hypersensitive to hydrogen peroxide (29), suggesting separate roles for Grx1 and Grx2 proteins in protection against several types of oxidative stress. Yeast gluthatione reductase (encoded by GLR1) regulates levels of GSH in the cells, providing the substrate for glutaredoxin. Thus, it is also necessary for protection against oxidative stress, as shown by the sensitivity phenotype of glr1 mutants to reactive oxygen species (15, 42). Yeast mutants with mutations in GSH1 (which codes for glutathione synthetase) do not grow unless glutathione is added to the medium (54), and diethylmaleate-induced gluthatione depletion causes growth arrest (53). These observations indicate that GSH is necessary for cell proliferation, being required for glutaredoxin-mediated reduction of protein disulfide bonds and/or performing additional essential roles in cell metabolism. With respect to the thioredoxin system in S. cerevisiae, neither of the two individual mutants with mutations in the thioredoxin genes (TRX1 and TRX2) presents any defects in cell growth. This contrasts with the case of the double trx1 trx2 mutant, which grows poorly even though deoxyribonucleotide levels in the cell remain unaltered (40, 41), thus pointing to additional functions of the thioredoxin system besides the role it plays in ribonucleotide reductase activity. The fact that the thioredoxin and glutaredoxin systems display at least partially overlapping functions in maintaining the physiological redox state of yeast proteins is supported by the observation that glutathione reductase function is absolutely necessary for cell growth in aerobic conditions in a trx1 trx2 mutant background. This is probably due to accumulation of toxic levels of oxidized glutathione in glr1 trx1 trx2 mutant cells, while the single glr1 mutant displays normal vegetative growth (42).
Besides thioredoxin and glutaredoxin, other cellular activities protect cells from oxidative damage (19, 38). Some of the responsible genes are under the control of the Yap1 transcription factor (15, 24, 25, 49). However, protection against oxidation is also related to other types of stresses. Thus, the expression of CTT1 (coding for cytosolic catalase) is induced not only by oxidative damage but also by heat and osmotic stresses, through the action of the Msn2 and Msn4 zinc-finger transcription factors on the STRE elements present in the CTT1 promoter (32, 33, 47). A number of different mutants which are hypersensitive to oxidative damage also displayed increased sensitivity to osmotic stress (23). One of the mutations in these mutants corresponds to the SKN7 gene, which codes for a response regulator in a two-component regulatory system that can be activated alternatively by osmotic stress (via the Sln1 phosphorelay) or by oxidative stress and which regulates the expression of a number of genes including the TRX2 gene coding for a thioredoxin (6, 21, 27, 39). Nevertheless, the molecular basis that explains the relationship between osmotic and oxidative stress still remains to be characterized.
In the course of the S. cerevisiae genome sequencing project, a family of three previously unknown open reading frames (ORFs) with homology to glutaredoxin genes has emerged. In this work, we present data confirming that this family of GRX3, GRX4, and GRX5 genes code for proteins with glutaredoxin activity and show evidence for a role of these genes in the defense against certain types of stress and for a functional interaction among them and with GRX2. Finally, we emphasize the importance of Grx5 in such defense functions.
MATERIALS AND METHODS
Strains and growth conditions.
Yeast strains used in this work are described in Table 1. CML235 (MATa ura3-52 leu2Δ1 his3Δ200) and CML236 (like CML236 but MATα) were employed as wild-type strains. E. coli DH5α was used as a host for DNA cloning. Yeast cells were grown at 30°C in yeast extract-peptone-dextrose (YPD) medium or, when indicated, in SD minimal medium with adequate auxotrophic supplements (3) and glucose (at a 2% concentration) or glycerol (at 3%) as a carbon source.
TABLE 1.
Strains used in this worka
| Strain | Relevant genotype | Comments |
|---|---|---|
| CML235 | Wild type | |
| CML236 | Wild type | |
| MML15 | MATa grx3::kanMX4 | Deletion in CML235 |
| MML16 | MATα grx3::kanMX4 | Deletion in CML236 |
| MML17 | MATa grx4::kanMX4 | Deletion in CML235 |
| MML18 | MATα grx4::kanMX4 | Deletion in CML236 |
| MML19 | MATa grx5::kanMX4 | Deletion in CML235 |
| MML20 | MATα grx5::kanMX4 | Deletion in CML236 |
| MML37 | MATa grx3::kanMX4 grx5::kanMX4 | From a cross, MML15 × MML20 |
| MML39 | MATa grx4::kanMX4 grx5::kanMX4 | From a cross, MML17 × MML20 |
| MML41 | MATa grx3::kanMX4 grx4::kanMX4 | From a cross, MML15 × MML18 |
| MML42 | MATα grx3::kanMX4 grx4::kanMX4 | From a cross, MML15 × MML18 |
| MML44 | MATa grx2::LEU2 | Deletion in CML235 |
| MML45 | MATa grx2::LEU2 grx3::kanMX4 | From a cross, MML44 × MML16 |
| MML47 | MATa grx2::LEU2 grx4::kanMX4 | From a cross, MML44 × MML18 |
| MML57 | MATα grx5::kanMX4 tetO2(GRX5)::kanMX4 | From MML20, by transformation with the pCM224 cassette (4) |
| MML58 | MATa grx3::kanMX4 grx4::kanMX4 grx5::kanMX4 tetO2(GRX5)::kanMX4 | From a cross, MML41 × MML57 |
| MML59 | MATa grx2::LEU2 grx3::kanMX4 grx4::kanMX4 | From a cross, MML42 × MML44 |
Wild-type strains CML235 (MATa ura3-52 leu2Δ1 his3Δ200) and CML236 (MATα ura3-52 leu2Δ1 his3Δ200) are spores from FY1679 (diploid, MATa/α ura3-52/ura3-52 leu2Δ1/+ his3Δ200/+ trp1Δ63/+; from B. Dujon, Pasteur Institute, Paris, France). The other strains have been obtained during this work.
Gene disruptions and other genetic methods.
Standard methods (3) were used for plasmid DNA preparation and manipulation and also for bacterial transformations. Crosses between yeast strains, sporulation, and tetrad analyses were carried out as described in reference 20.
To delete GRX3, GRX4, or GRX5 in the wild-type CML235 and CML236 strains, we made use of the kanMX4 cassette from pFA6a-kanMX4, according to the short flanking homology strategy (52). A similar approach was used for disrupting GRX2 with LEU2 as a marker, except that a pFA6a-kanMX4 derivative (plasmid pCM376, containing the yeast LEU2 gene and flanking regions instead of kanMX4) was used for amplification of the disruption cassette. In all cases, the DNA cassette was amplified by PCR with Expand High-Fidelity enzyme (Boehringer), followed by DNA transformation of yeast cells (4). Oligonucleotides for cassette amplification were designed in such a way that most of the targeted gene was disrupted upon transformation with the amplified DNA. Thus, for the disrupted GRX2 gene, only 9 bp of the original ORF remains at the 5′ end and 11 bp remains at the 3′ end; for GRX3, 2 and 19 bp remain, respectively; and for GRX4, 10 and 9 bp remain, respectively. For GRX5, the deletion covers from base +25 (origin at +1) up to the stop codon. Deletions were confirmed by PCR.
Sensitivity to stress conditions.
Exponentially growing cells at about 107 cells per ml were treated with the respective compound, which was directly added to the growth medium at the concentrations and during the intervals indicated for each experiment. Untreated cultures were incubated in parallel over the same periods. Viability was determined by colony counts on YPD plates (each dilution three times) after 3 days of incubation at 30°C. Total cell number was determined from formaldehyde-fixed samples, by using an Epics XL flow cytometer (Coulter).
Analysis of cell wall-altered phenotype.
Agents used as indicators for cell wall alterations were tested by spotting 4-μl samples of 1/8 serial dilutions of cultures exponentially grown in YPD at 30°C (initial concentration of 107 cells per ml) on YPD plates containing the respective agent and monitoring growth after 3 days at 30°C. The following compounds and concentration ranges were employed: calcofluor white, 25 to 75 μg/ml; sodium dodecyl sulfate (SDS), 0.05 to 0.2%; and caffeine, 5 to 20 mM.
Northern blot analyses.
RNA purification, electrophoresis, probe labelling with digoxigenin, hybridization, and signal detection were carried out as previously described (11). Signals were quantified with the Lumi-Imager equipment (Boehringer) software. Probes for the GRX3, GRX4, and GRX5 genes were generated by PCR from genomic DNA, by using oligonucleotides designed to amplify fragments covering the entire ORF without adjacent sequences.
Preparation of cell extracts and determination of enzyme activities.
Extracts were prepared from yeast cells exponentially growing in YPD medium at 30°C by collecting, washing, and finally resuspending them (at 1:100 of the original volume) in 20 mM imidazole buffer (pH 7.0) plus 2 mM EDTA and protease inhibitors (2 mM phenylmethylsulfonyl fluoride, 0.2 mM tolylsulfonyl phenylalanyl chloromethyl ketone [TPCK], and 2 μM pepstatin, final concentrations). Cells were broken by repeated vortexing in cold conditions with an equivalent volume of glass beads (0.6-mm diameter; Sigma), followed by low-speed centrifugation (4,000 × g for 5 min at 4°C). This supernatant was again centrifuged at 30,000 × g for 40 min at 4°C, and the final supernatant was kept for further analyses.
Glutaredoxin (GSH disulfide oxidoreductase) activity was measured by the reduction of the mixed disulfide formed between β-hydroxyethyl disulfide and glutathione, according to reference 18. Cell extracts were heated at 85°C for 5 min to inactivate glutathione reductase, thioredoxin reductase, and other interfering activities. Glutathione reductase activity was determined as previously described (14), following the decrease in absorbance (340 nm) due to the oxidation of NADPH. Transketolase activity was determined as described in reference 22. The protein concentration was measured by the Bradford method.
Quantification of protein carbonyl groups.
The protein carbonyl content in crude extracts was determined according to the dinitrophenylhydrazine derivatization method (26). Quantification was carried out with a Zorbax GF-250 high-pressure liquid chromatography gel filtration column at flow rate of 1 ml/min at 30°C. Absorbance at 276 and 370 nm was monitored with a Waters 996 diode array detector.
Other analytical and preparative protein methods.
Analytical SDS-polyacrylamide gel electrophoresis and immunodetection of peptides bound to 2,4-dinitrophenylhydrazones (DNPs) were carried out as previously described (48). Anti-DNP antibodies (supplied by DAKO) were employed at a 1:4,000 dilution. Preparative electrophoresis, peptide mapping (after limited proteolysis with endoproteinase V8 from Staphylococcus aureus), and sequencing were conducted as described in reference 50.
Sequence analyses.
FASTA analysis (as provided by the Munich Information Centre for Protein Sequences [35]) was initially carried out to compare each pair of protein sequences. Multiple protein alignments were calculated with the ClustalW package (16). The Sequence Space algorithm (8) was applied to regions where significant alignments could be established, in order to classify sequences into groups according to their similarity. The original algorithm was implemented in a Mathematica package, and all computations were performed with this program.
RESULTS
A new glutaredoxin family in S. cerevisiae.
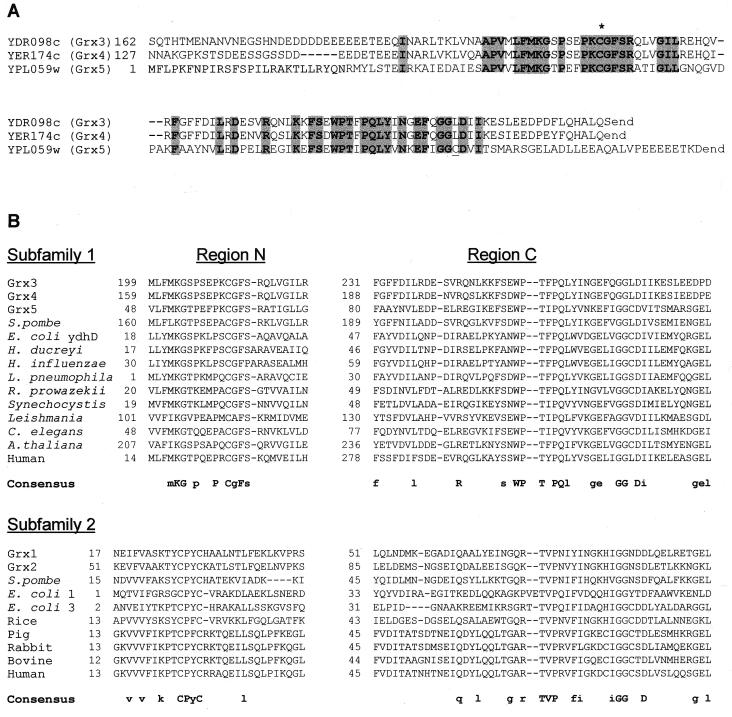
Analysis of the yeast genome revealed the existence of a family of three ORFs (YDR098c, YER174c, and YPL059w) whose products display significant homology to known glutaredoxins. ORFs YDR098c and YER174c have N-terminal extensions that are absent in YPL059w (Fig. 1A). In the homologous region, the predicted protein sequences of all three ORFs display 29% identity, which increases to 71% when only YDR098c and YER174c are considered. The highest homology concentrates in two separate regions (Fig. 1A). The most N-terminal of these regions includes a common cysteine residue. In contrast with other glutaredoxins from yeasts or other prokaryotic or eukaryotic organisms (18, 29), a motif of two cysteine residues separated by two additional amino acids does not occur in the above three ORF products, although YPL059c contains a second cysteine in the most C-terminal homology region.
FIG. 1.
Comparative analysis of glutaredoxin sequences. (A) Alignment of the S. cerevisiae Grx3, Grx4, and Grx5 amino acid sequences deduced from the nucleotide sequences of their respective ORFs. Common residues in the three sequences are shaded. The N-terminal extensions of Grx3 and Grx4 are not represented. The asterisk marks the common cysteine residue present in all three sequences. A second cysteine present in Grx5 is underlined. (B) Sequence analysis of relevant regions of 23 different glutaredoxin proteins. Regions N and C are respectively the most N- and C-terminal regions of the molecules for which significant alignments can be established. Sequences outside these two regions are not represented. Subfamilies 1 and 2 are initially defined according to the consensus sequences indicated in the figure. For the consensus sequences, residues identical in all members of each subfamily are represented in uppercase letters, while those common to at least 75% of them are in lowercase letters. More details about these sequences can be obtained from reference 36. H. ducreyi, Haemophilus ducreyi; H. influenzae, Haemophilus influenzae; L. pneumophila, Legionella pneumophila; R. prowazekii, Rickettsia prowazekii; C. elegans, Caenorhabditis elegans.
Comparisons were extended to a total of 23 putative glutaredoxin protein sequences present in the databases (Fig. 1B). This allowed us to define two subfamilies of glutaredoxins based on the sequence patterns of the two regions of highest homology, here referred to as regions N and C. Homology in region C (the most C-terminal) extends to all members of the two subfamilies, with a total of four residues conserved in all 23 proteins. In contrast, alignment in region N (the most N-terminal) was significant only when applied within each subfamily. Members of subfamily 1 all contain the motif PXCG/AFS/P (X being nondefined), with no other cysteine residue being present in this region, while subfamily 2 is defined by the above-mentioned motif CPY/FC. This motif partially defines the characterized active site of some glutaredoxins (43, 55), all of which are included in subfamily 2. No equivalent studies have been reported for subfamily 1 members. Interestingly, glutaredoxins of both subfamilies coexist in organisms ranging from bacteria (i.e., E. coli) to higher eukaryotes (such as humans) (Fig. 1B). In the case of S. cerevisiae, the previously characterized GRX1- and GRX2-encoded glutaredoxins (29) are ascribed to subfamily 2, while the products of YDR098c, YER174c, and YPL059w are subfamily 1 members. From the homology patterns and also from the determination of enzyme activities (see below), we propose to rename these last three ORFs GRX3, GRX4, and GRX5, respectively.
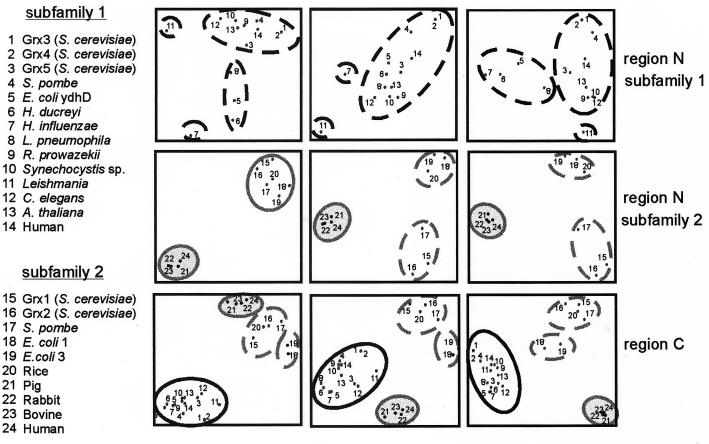
The Sequence Space approach (8) was used for a more detailed comparative analysis of the 23 glutaredoxin sequences, separately for regions N and C (Fig. 2). When this method was used to analyze region C in the whole set of sequences, the previously defined subfamily 1 clustered separately from the remaining sequences. Inside this cluster, Grx5 appears closer to glutaredoxins from multicellular eukaryotes than to yeast Grx3 and Grx4, which are positioned almost together. The 10 sequences of subfamily 2 were divided into three clusters corresponding respectively to bacterial molecules, mammalian molecules, and a cluster of yeast (S. cerevisiae Grx1 and Grx2 and Schizosaccharomyces pombe) and rice glutaredoxins. This same division in subfamily 2 glutaredoxins was confirmed from analysis of region N, with the difference that rice glutaredoxin mapped closer to E. coli glutaredoxins. Sequence analysis of region N in subfamily 1 confirmed the relative distance between Grx5 and Grx3/4. Although these three proteins are all positioned in the same cluster, the Grx5 sequence comes closer to Arabidopsis thaliana or human glutaredoxins than to Grx3 and Grx4.
FIG. 2.
Sequence Space analysis of the glutaredoxin family. Principal component analyses of the protein sequences are shown (from left to right) on the resulting 1-2, 1-3, and 2-3 discriminant axes (8). Analyses were carried out separately for region N in subfamily 1, region N in subfamily 2, and region C in the whole glutaredoxin family. Each point in the plots represents an individual sequence identified by a number. Distances between points are proportional to sequence divergence. Sequence clusters are defined according to proximity in the resulting plots (continuous lines). These clusters were tentatively divided into subsets of sequences (dashed lines) when the results on the three dimensions suggested the existence of relevant subgroups. See the Fig. 1 legend for genus abbreviations.
A triple grx3 grx4 grx5 mutant is not viable.
In order to genetically characterize the new glutaredoxins, null individual mutants were obtained for each of the GRX3, GRX4, and GRX5 loci and also double mutants derived from their respective crosses. Cultures of the grx3 or grx4 mutants displayed the same growth phenotype as the wild type both in rich and in SD-glucose minimal medium at the temperature intervals ranging from 15 to 37°C. In contrast, in grx5 mutant cells growth rate was decreased by a factor of 1.6 compared to wild-type cells in YPD medium at 30°C (Table 2). Moreover, this mutant grew poorly in SD-glucose medium at 30°C and was unable to grow when the temperature was increased to 37°C. The grx5 mutation was also linked to the inability to grow in YP-glycerol medium. Growth characteristics were even more affected in the grx3 grx5 double mutant (although not in the grx4 grx5 and grx3 grx4 mutants) with respect to grx5 mutant cells (Table 2).
TABLE 2.
Enzyme activities and protein oxidation levels in grx mutantsa
| Strain | Relevant genotype | Doubling time (min)b | Glutaredoxin activityc | Glutathione reductase activityc | Carbonyl contentd
|
||
|---|---|---|---|---|---|---|---|
| Control | Menadione | Hydrogen peroxide | |||||
| CML235 | Wild type | 90 ± 4 | 51 ± 4 | 102 ± 10 | 0.69 ± 0.05 | 1.28 ± 0.05 | 0.96 ± 0.05 |
| MML15 | grx3 | 94 ± 1 | 30 ± 3 | 101 ± 6 | 0.78 ± 0.06 | 1.53 ± 0.05 | 1.10 ± 0.05 |
| MML17 | grx4 | 92 ± 5 | 28 ± 3 | 97 ± 6 | 0.75 ± 0.03 | 1.38 ± 0.05 | 1.08 ± 0.05 |
| MML19 | grx5 | 145 ± 11 | 22 ± 1 | 96 ± 6 | 0.99 ± 0.07 | 2.10 ± 0.05 | 1.63 ± 0.05 |
| MML37 | grx3 grx5 | 208 ± 9 | 19 ± 4 | 101 ± 10 | 1.16 ± 0.10 | ND | ND |
| MML39 | grx4 grx5 | 147 ± 3 | 21 ± 4 | 99 ± 9 | 1.15 ± 0.12 | ND | ND |
| MML41 | grx3 grx4 | 150 ± 16 | 44 ± 7 | 98 ± 12 | 0.89 ± 0.09 | ND | ND |
| MML44 | grx2 | 88 ± 2 | 14 ± 3 | ND | 0.81 ± 0.03 | 1.50 ± 0.10 | 0.95 ± 0.05 |
| MML45 | grx2 grx3 | 92 ± 1 | 19 ± 4 | ND | 0.89 ± 0.11 | ND | ND |
| MML47 | grx2 grx4 | 90 ± 2 | 16 ± 4 | ND | 1.09 ± 0.14 | ND | ND |
| MML59 | grx2 grx3 grx4 | 159 ± 7 | 8 ± 2 | ND | 1.25 ± 0.15 | ND | ND |
Values are shown as means ± standard deviations. Three independent determinations were done in all cases. ND, not determined.
In exponential cultures in YPD liquid medium at 30°C.
Nanomoles of NADPH oxidized per minute per milligram of protein.
Nanomoles of carbonyl groups per milligram of protein, in untreated cells (control) or cells treated for 1 h with 20 mM menadione or 2.5 mM hydrogen peroxide.
In order to obtain the multiple mutant disrupted in all three glutaredoxin genes of subfamily 1, a grx3 grx4 mutant strain was crossed with a grx5 mutant. Fifty tetrads derived from the resulting diploid were analyzed, and yet no grx3 grx4 grx5 multiple mutant could be isolated, in contrast to the other possible genotype combinations. To confirm that the above combination was not viable, we employed the tetO promoter substitution cassette (4) to substitute the chromosomal GRX5 promoter for the doxycycline-regulatable tetO promoter in a grx3 grx4 mutant background. The resulting strain was able to grow in the absence of doxycycline but arrested growth in the presence of the antibiotic (data not shown), confirming that inactivation of the three glutaredoxins was lethal for yeast cells.
Glutaredoxin-reduced activity causes protein oxidative damage in grx3, grx4, and grx5 mutants under ordinary growth conditions.
In order to prove that GRX3, GRX4, and GRX5 code for proteins with glutaredoxin activity, we measured enzyme levels in the respective single mutants (Table 2). In grx2 and grx3 mutants, glutaredoxin activity decreased by 40% with respect to wild-type cells. It is remarkable that although grx5 mutant cells showed growth defects not observed in the grx3 or grx4 mutants, the decrease in glutaredoxin levels in grx5 mutant cells was only slightly higher than that in the other two mutants. While in the grx3 grx4 double mutant there seemed to be a compensatory effect in activity levels relative to the respective single mutants, glutaredoxin activity in mutants affected in GRX5 plus one of the other two genes was similar to that in the single grx5 mutant. Glutathione reductase activity, measured as a control, maintained equivalent levels in all the strains tested (Table 2).
It has been proposed that glutaredoxins participate in the maintenance of an adequate intracellular concentration of thiols, which play an antioxidant role in the cell (9, 34). Therefore, we tested whether the previously mentioned deletion mutants have higher basal levels of protein oxidative damage than wild-type cells. For this purpose, we measured the protein carbonyl content in crude extracts from cells grown in YPD medium. This parameter has been widely used to assess minimal values of protein damage under oxidative stress conditions (26, 48, 50). As shown in Table 2, single grx3 and grx4 mutants displayed a moderate increase in carbonyl content with respect to wild-type cells. This increase was more severe in the grx5 mutant. In the case of the double mutants, the carbonyl content was slightly increased in grx3 grx4 mutant cells and markedly increased in grx5 mutant cells that also contained inactivating mutations in GRX3 or GRX4. The effect of inactivation of the already-known glutaredoxin GRX2 gene on protein oxidative damage was then checked in the same way (Table 2). In this case, the decreased levels of glutaredoxin enzymatic activity in the grx2 mutant cells were also reflected in an increase in protein carbonyl content of about 15% with respect to wild-type cells. This was of the same magnitude as that in the grx3 and grx4 mutant cells but clearly less than the values obtained for the grx5 mutant. In the case of the double mutants, a 50% increase was observed for grx2 grx3 mutant cells, and one of 30% was observed in the case of the grx2 grx4 mutant.
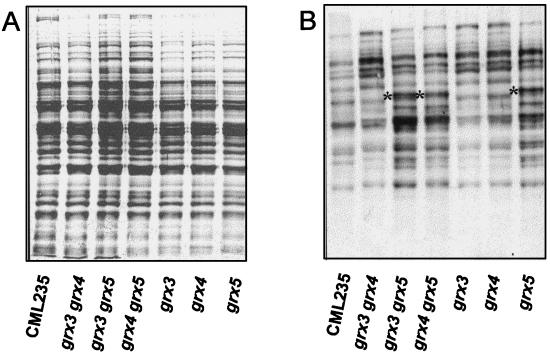
To test whether the observed increases in protein carbonyl content in mutant cells affected the whole protein pool or only some proteins, we used Western blot analysis to compare the patterns of oxidized proteins exhibited by wild-type and mutant strains (Fig. 3). All the bands observed in wild-type cells that have to be considered background levels of protein oxidation increase in all mutants. Furthermore, in the mutants lacking GRX5 at least one band appeared to be specifically oxidized (indicated by an asterisk in Fig. 3). This was not observed in the other mutant strains. By using crude extracts from grx5 mutant cells, this protein band was purified to homogeneity by preparative electrophoresis, and its N terminus was sequenced. The protein was identified as transketolase. This was further confirmed by the N-terminal sequence of one oxidized peptide obtained after limited proteolysis of the whole protein with endoproteinase V8. Further extending these results, transketolase activity was measured in extracts from wild-type and grx5 mutant cells. The latter exhibited only about 25% of the activity present in wild-type cells (20 versus 83 mU/mg of protein), confirming that oxidation leads to enzyme inactivation.
FIG. 3.
Protein oxidative damage under normal growth conditions of wild type and grx single and double mutants. MATa strains were employed. Cultures of wild type (CML235) and single and double mutants were grown in YPD liquid medium at 30°C until an optical density at 600 nm of 1 was reached. The crude extracts obtained were analyzed by Western blotting with anti-DNP antibodies (B). A parallel run stained with Coomassie brilliant blue is shown in panel A. Each lane contained 20 μg of total protein. Asterisks mark the identified transketolase band (see text for details).
Grx5 glutaredoxin plays a central role in protection against induced oxidative and hyperosmotic stresses.
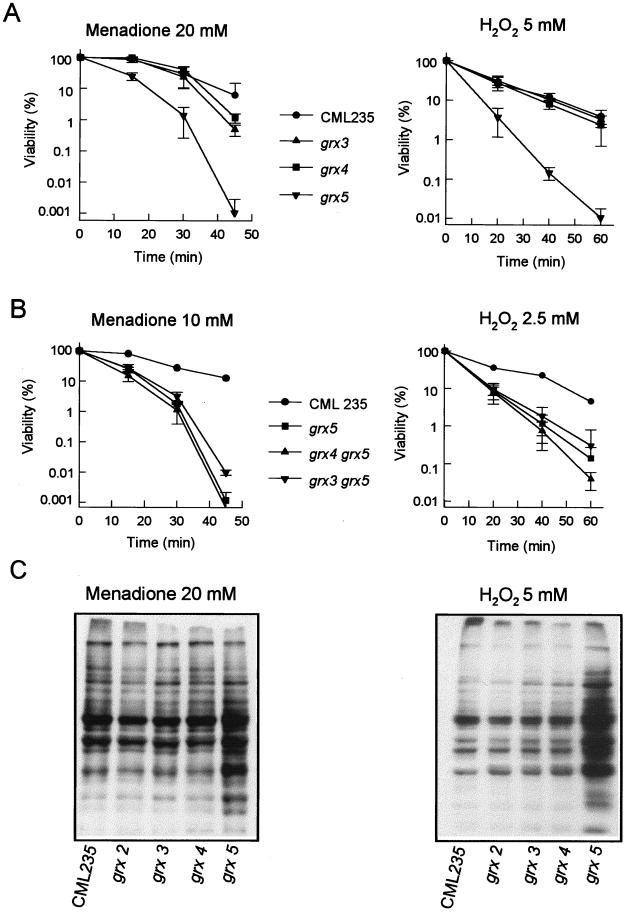
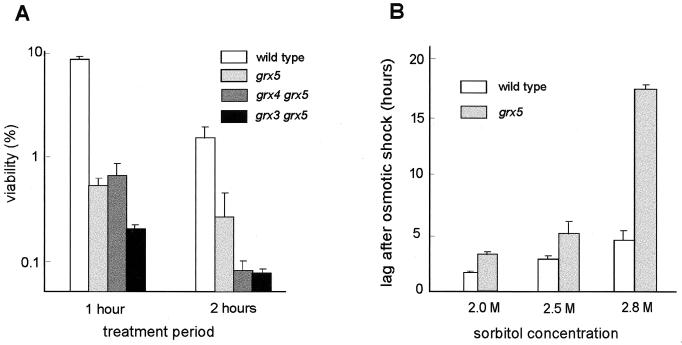
Once it was demonstrated that the products of GRX3, GRX4, and GRX5 are required for maintaining normal glutaredoxin levels in the cell, we next studied their role in protection against an externally induced oxidative stress. The effect of hydrogen peroxide and menadione (a generator of superoxide radicals) on viability was tested when they were applied to exponentially growing cells. Disruption of GRX3 and GRX4 had only a moderate effect on sensitivity to menadione and no effect on sensitivity to hydrogen peroxide, while disruption of GRX5 caused a dramatic increase in sensitivity to both oxidants (Fig. 4A). The grx3 grx5 and grx4 grx5 double mutants were not markedly more sensitive to menadione and hydrogen peroxide than were grx5 single mutants (Fig. 4B). In fact, survival after long-term treatment was slightly higher in grx3 grx5 mutant cells than in grx5 mutants, although this may be an effect of the lower growth rate of the double mutant.
FIG. 4.
Sensitivity of S. cerevisiae grx mutants to oxidative agents. MATa strains were employed. (A) Cultures of wild-type (CML235) and single mutant strains growing exponentially in YPD liquid medium at 30°C were exposed to the indicated agents and concentrations, and viable numbers (relative to time zero values) were determined at different times. (B) As in panel A, except that lower agent concentrations were used to determine sensitivity of double mutants compared to wild-type and single mutant strains. (C) Protein oxidative damage in wild type and glutaredoxin mutants under stress conditions. Cultures of wild type and single glutaredoxin mutants were grown in YPD liquid medium at 30°C, and at an optical density at 600 nm of 1, menadione or hydrogen peroxide was added to the cultures at the final concentration of 20 or 5 mM, respectively. After 60 min of treatment, the cultures were harvested by centrifugation and crude extracts were obtained. Analyses by Western blotting with anti-DNP antibodies were conducted as described in Materials and Methods. Each lane contained 10 μg of total protein.
Protein damage promoted by adding 20 mM menadione or 5 mM hydrogen peroxide to growing cells was analyzed by Western blotting (Fig. 4C). In these conditions, the grx3 and grx4 mutants revealed only a moderate increase in the level of protein oxidation with respect to wild-type cells, while a heavily oxidized protein band pattern was exhibited by the grx5 mutant. For comparison, we included the already-described grx2 mutant, which displayed a protein oxidation pattern similar to those observed in wild-type cells and grx3 and grx4 mutants. In agreement with the data presented in Table 2, the overall increase in carbonyl content in mutant cells was not due to a qualitative difference of oxidation in particular protein bands but to an increase in oxidative damage in most protein bands present in all the stressed strains.
Other authors have shown that some yeast mutants hypersensitive to oxidants are also more sensitive to osmotic stress (23). We therefore tested the sensitivity of grx5 mutant cells to hypertonic conditions. This mutation increased sensitivity to high concentrations of KCl more than 10-fold, and the sensitivity was even higher in the double grx3 grx5 mutant (Fig. 5A). To show that this effect was not caused by ion toxicity, we tested the effect of sorbitol at a concentration of 2 M or higher on transitory cell division arrest after the osmotic shock. In these conditions, growth was also more affected in grx5 mutant cells than in the wild-type strain (Fig. 5B), confirming that the Grx5 product protects not only against oxidative stress but also against different types of hyperosmotic stress. On the other hand, none of the grx3, grx4, or grx5 single mutants was more sensitive than wild-type cells to heat shock (shift from 25 to 37°C [data not shown]).
FIG. 5.
Sensitivity of S. cerevisiae grx mutants to hyperosmotic treatments. (A) Exponentially growing wild-type (CML235) and mutant (MATa type) cells in YPD medium at 30°C were supplemented with 2 M KCl, and cell viability (made relative to parallel untreated cultures) was determined at the indicated times. (B) Exponentially growing cells in YPD medium were treated with sorbitol at the final concentrations indicated, and incubation was continued under these conditions. Total cell numbers were measured at subsequent periods. Bars represent the lag periods after sorbitol addition during which cell division remained arrested before cultures resumed growth.
The hypersensitivity of grx5 mutant cells to osmotic stress could have been caused by the effect of reactive oxygen species on cell wall architecture. To analyze this possibility, we tested the sensitivity of wild-type and grx5 mutant cells to a number of especially toxic agents for cells altered in cell wall structure (31). grx5 mutant cells did not show increased sensitivity (relative to wild-type cells) to calcofluor white, SDS, or caffeine (data not shown), thus eliminating the possibility of explaining increased osmotic sensitivity as being a direct consequence of hyperoxidation of cell wall molecules.
Grx2 and Grx5 functions can substitute for each other.
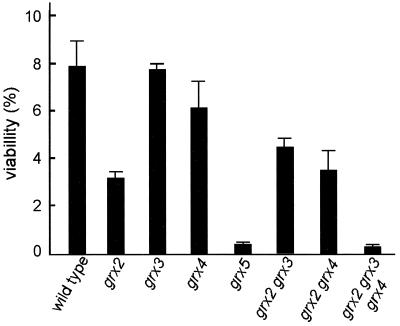
Grx2 has been reported to account for most of the glutathione-dependent oxidoreductase activity of glutaredoxins in yeast cells and to play an important role in protection against hydrogen peroxide, but not against menadione (29). It was possible to obtain grx2 grx3 and grx2 grx4 mutant strains by standard genetic crosses from their respective single mutants, and they had significantly reduced oxidoreductase activity compared with single grx3 or grx4 mutants (see above and Table 2). However, no double grx2 grx5 mutant could be obtained from a total of 40 tetrads analyzed. We conclude that this mutant combination is lethal and therefore that Grx2 activity can functionally substitute, at least in these particular conditions, for the loss of activity in grx5 mutant cells. Loss of GRX2 caused a less-than-threefold increase in sensitivity to hydrogen peroxide stress in cells grown in SD-glucose medium, and this was of the same order as that in the double grx2 grx3 and grx2 grx4 mutants (Fig. 6). Differences in sensitivity between wild-type and grx2 mutant cells were even smaller in cultures grown in YPD medium (data not shown).
FIG. 6.
Effect of oxidative stress (5 mM hydrogen peroxide for 1 h) on cell viability of grx mutants (MATa strains) compared to that of wild-type cells (strain CML235). Cells were grown exponentially at 30°C in SD medium plus glucose, and after treatments, they were plated on YPD solid medium in order to determine viability. Bars indicate the percentages of viable cells relative to those in parallel untreated cultures.
A triple grx2 grx3 grx4 mutant was subsequently obtained by standard genetic crosses. Loss of the three genes caused about the same effect on cell growth rate in rich medium as the loss of the single GRX5 gene (Table 2), although the multiple mutant grew more efficiently in minimal medium than did the grx5 mutant (data not shown). Simultaneous disruption of GRX2, GRX3, and GRX4 caused a 50% reduction in total cellular glutaredoxin activity compared to the single grx2 mutant or the double grx2 grx3 and grx2 grx4 mutants. Correspondingly, total protein carbonylation was higher in the triple grx2 grx3 grx4 mutant than in the other single and double mutants (Table 2), and sensitivity to hydrogen peroxide was higher in grx2 grx3 grx4 mutant cells than in the single grx2 mutant and of the same order as that in the grx5 mutant (Fig. 6). We can conclude that although Grx2 and Grx5 can functionally substitute for each other, loss of Grx5 has more severe effects on cell physiology than loss of Grx2 alone and that in order to observe effects comparable to those of the loss of Grx5, it is necessary to simultaneously eliminate Grx2, Grx3, and Grx4.
Expression of the GRX3-GRX4-GRX5 gene family in response to stresses.
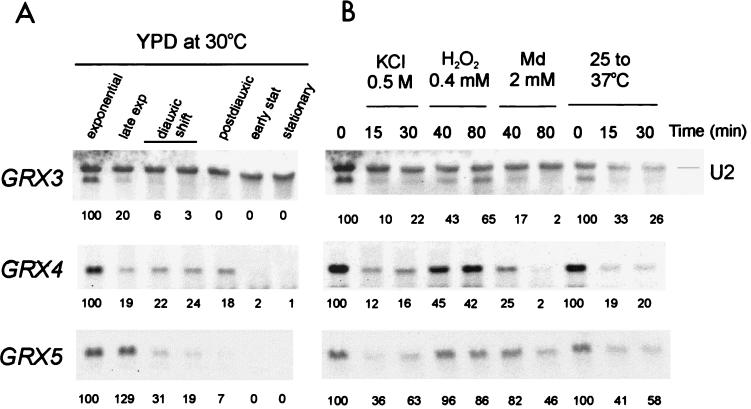
The transcriptional pattern of GRX3, GRX4, and GRX5 was measured under several conditions (Fig. 7). Maximum expression for the three genes occurred during the exponential growth phase. As cells traversed the diauxic shift, transcript levels progressively decreased to under detectable levels in stationary phase. However, the rate of mRNA disappearance was different for each of the three genes. GRX3 mRNA rapidly became undetectable, while GRX4 expression was still detectable until the postdiauxic stage (Fig. 7). The expression of the three genes was not inducible under any of the three stresses applied (osmotic, oxidative with hydrogen peroxide or menadione, and heat). In fact, all three types of stress caused a reduction in the respective mRNA levels. This was moderate for GRX5 and more intense for the other two transcripts. We can therefore conclude that the role of the Grx5 glutaredoxin in protection against oxidative and osmotic stresses does not depend on transcriptional changes induced by the respective type of stress.
FIG. 7.
Northern blot analyses of GRX3, GRX4, and GRX5 expression. Samples were taken at different stages of the population growth curve in YPD liquid medium at 30°C (A) or after treatment of mid-exponential-phase cells (at 30°C except for heat shock) with KCl (0.5 M), hydrogen peroxide (0.4 mM), menadione (2 mM), or heat shock for the indicated times (B). Small nuclear U2 mRNA is shown as the loading control. Numbers under the lanes indicate the mRNA levels for each time point, relative to the mid-exponential-phase sample. For heat shock analysis, the time zero sample corresponds to exponential cultures at 25°C.
DISCUSSION
Glutaredoxins are important for maintaining the reducing status of thiol groups in proteins (1, 9, 45). Together with thioredoxins, they are members of a superfamily of proteins that exert their activity through a disulfide exchange reaction involving one or two cysteine residues at the active site. The initially characterized members of the glutaredoxin family contained a CXXC active site, with XX being PY in most cases. Studies of phage T4 (43), pig (55), and E. coli (44) glutaredoxins have shown that of the two cysteine residues, only the most N-terminal is absolutely essential for enzyme activity, while mutants with mutations in the more C-terminal cysteine retain part of their GSH oxidoreductase activity (7, 43, 44, 55). Glutaredoxin-mediated protein glutathionylation has been explained in terms of the participation of a single cysteine (43). Studies involving an E. coli glutaredoxin mutated at the second cysteine residue indicate that both cysteine residues are required for reduction of protein disulfides (such as that in ribonucleotide reductase) through a dithiol mechanism, while the deglutathionylation of protein substrates would employ a monothiol mechanism (7) that could play an important physiological role at the endoplasmic reticulum for the maintenance of native protein conformation (30). Here we define a group of three new glutaredoxins (Grx3 to Grx5) in S. cerevisiae that structurally differ from the subfamily containing the CXXC motif at the active site and that constitute a separate subfamily including members ranging from bacterial to human glutaredoxins. Members of the latter subfamily contain the motif CG/AFS/P. The fact that this is the only conserved cysteine residue present in the members of this subfamily suggests that it may be part of the active site of the enzyme. Most members of this new subfamily also contain one or two basic amino acid residues separated by a few positions from the cysteine residue toward the C end. The presence of one or two basic residues close to the active site is also characteristic of the first subfamily, and it has been suggested that it might be needed for the thioltransferase reaction due to the enhancement of the S nucleophilicity of the reactive cysteine (55). This analogy reinforces the role of the CG/AFS/P motif in the reactivity of the glutaredoxins of the new subfamily.
The Grx3, Grx4, and Grx5 yeast glutaredoxins display sequence differences, though all three are members of the single-cysteine subfamily. Grx5 lacks part of an N-terminal domain present in Grx3 and Grx4. Application of the Sequence Space method (which allows sequence clustering based on amino acid conservation) has shown the Grx5 sequence to be closer to plant or mammalian glutaredoxin sequences than to Grx3 or Grx4. This method also permits us to observe that Grx1 and Grx2 yeast glutaredoxins are structurally separated from the Grx3/4/5 group. In the C-terminal region of homology, Grx5 contains the IGGC motif, which is absent in the other four yeast glutaredoxins but is present in the mammalian members of the cysteine-pair subfamily. The glycine pair in the above motif is common to all glutaredoxins of both subfamilies and might contribute to bringing an aspartic acid residue close to the active site cleft. The role of this conserved aspartic acid has been shown to be essential in the case of pig glutaredoxin (43).
Cell growth rate is not affected by single mutations in GRX1 to GRX4 (reference 29 and this work). In contrast, grx5 mutant cells are constitutively affected in growth pattern (lower growth rate in rich medium, poor growth in minimal medium, and no growth in glycerol medium). Simultaneously, the grx5 mutant has a higher basal protein carbonyl content than the other single glutaredoxin mutants. Since carbonyl content is employed as a measure of oxidative protein damage, the above observations could be interpreted as indicating that the Grx5 glutaredoxin has an important role in protection against oxidative damage of proteins during exponential growth. This could be correlated with the role of glutaredoxins in the homeostatic maintenance of intracellular thiols, which are necessary for several antioxidant activities in the cell (9, 34). In E. coli, protein oxidative damage is higher during respiratory growth conditions (50), and this also appears to be the case in S. cerevisiae (46), which would explain the inability of grx5 cells to grow on glycerol when it is the only carbon source. The correlation cannot be extended, however, to all situations involving respiratory metabolism. Thus, during the postdiauxic growth stage, GRX5 expression decreases and the viability of the grx5 mutant is not affected. In this situation, yeast cells perhaps employ alternative protection strategies against oxidative damage.
When cells are oxidatively stressed with menadione or hydrogen peroxide, the accumulation of protein damage is much higher in grx5 mutant cells than in wild type or in the other grx mutant strains. This again correlates with the extreme effect of these situations on grx5 mutant viability. Therefore, in those conditions in which an external oxidative stress is applied, there is a close relationship between the extent of protein carbonylation and the effect on cell growth, and these data confirm that Grx5 may be the most important glutaredoxin in protecting exponentially growing yeast cells against oxidative protein damage not only under normal growth conditions but also during induced stress. In carrying out this antioxidant function, Grx5 does not discriminate between the effects caused by menadione and those caused by hydrogen peroxide, in contrast with the protective role that Grx2 performs exclusively against hydrogen peroxide (29).
This relationship between protein carbonylation levels and growth defects has one exception. Simultaneous lack of Grx2, Grx3, and Grx4 has a more profound effect on constitutive protein oxidation than on cell growth. Also, the relationship cannot be strictly extrapolated to explain the relative contribution of each glutaredoxin species to overall cellular glutaredoxin activity. Total GSH oxidoreductase activity due to Grx5 alone seems to be similar to that of Grx3 or Grx4 but less than that of Grx2. However, the rate of growth is affected much more in grx5 mutant cells. These differential effects of the grx mutations on growth could be explained by the fact that specific yeast glutaredoxins could identify individual protein substrates instead of acting as general GSH oxidoreductases. While inactivation of each GRX3, GRX4, or GRX5 gene causes a general increase in oxidation levels of cell proteins, in the case of grx5 mutant cells some individual protein bands (detected by Western blot immunoassay) are more prominently oxidized. Among these, transketolase, which is not detectable as an oxidized species in wild-type cells, appears to be particularly oxidized only in strains carrying the grx5 mutation, even in a nonstressed situation. The finding that transketolase is especially susceptible to oxidative stress in yeast cells is relevant considering that it has been shown recently that E. coli transketolase activity is negatively affected in superoxide dismutase-deficient mutants, as well as in hyperoxia conditions (5). The presence of carbonyl groups in transketolase and the inactivation of the enzyme could both be a consequence of the highly oxidized environment created inside grx5 mutant cells. Subsequently, since transketolase is involved in the pentose phosphate pathway, inactivation of this enzyme might lead to a depletion of NADPH levels, which would account for the lowered antioxidant capacity. Furthermore, this situation would block the possibility of redirecting carbohydrate metabolism to the regeneration of NADPH at the expense of glycolysis, which is what happens in wild-type cells a few minutes after hydrogen peroxide exposure (13). Under such circumstances, cell viability would obviously be compromised. Through depletion of erythrose-4-phosphate (which requires transketolase for its synthesis), superoxide dismutase deficiency causes auxotrophy for aromatic amino acids in E. coli (5). We tested whether the growth deficiency in grx5 mutant cells in minimal medium was relieved by the addition of aromatic amino acids, but this was not the case (data not shown). Thus, although transketolase inactivation may contribute to growth deficiency in this particular situation, inactivation of other as-yet-uncharacterized proteins must be essential for the phenotype of Grx5-deficient cells.
Mutations in GRX5 add to the list of oxidation-sensitive mutants which are also hypersensitive to osmotic stress (23). From our studies, the idea of a direct oxidative effect on cell wall architecture in grx5 mutant cells should be discarded. Signal transduction pathways responding to hyperoxidative and hyperosmotic signals are interconnected in yeast. The pathway interrelationship is exemplified by Skn7, which is a signal transducer whose activity can be regulated by osmotic and oxidative stresses (6, 21, 27, 39). For the moment, no evidence to suggest that GRX5 is a target for the pathways regulated by oxidative or osmotic signals exists, as the expression of GRX5 is not induced by these stresses. Alternatively, the susceptibility of shared components of both types of pathways to the protein-hyperoxidation situation created in Grx5-deficient cells would result in sensitivity to oxidative and osmotic stresses.
The growth and stress sensitivity phenotypes of grx double mutants, together with the lethality of the grx2 grx5 and grx3 grx4 grx5 mutations, point to a central role of Grx5 in the regulation of the basal redox state of a number of functionally important proteins during exponential growth. Although we have not considered Grx1 glutaredoxin, as it has been shown to play only a minor role in exponential conditions (29), we have observed that a multiple grx1 grx2 grx3 grx4 mutant is viable (our unpublished observations). These results could be explained by the existence of two different protein populations whose redox status could be separately regulated by the Grx1/2 and the Grx3/4 groups, respectively, while Grx5 would be able to act on both groups of protein substrates. Alternatively, the dithiol Grx1 and Grx2 enzymes and the monothiol Grx3, Grx4, and Grx5 enzymes could perform different thiol oxidoreductase activities, the first group reducing protein disulfides through a dithiol mechanism and the second group deglutathionylating glutathione-modified proteins through a monothiol mechanism (7, 30). Yeast cells would be unable to survive in the absence of the monothiol mechanism, but they would still be viable in the absence of the GSH-related dithiol one. In any case, Grx5 alone would be sufficient for maintaining the protein redox state, as it is able to replace the function of other glutaredoxins, at least when these are absent. This would also apply for an externally induced oxidative stress. In summary, Grx5 would act as a housekeeper for the adequate protein redox state during normal growth and as the agent responsible for the elimination of externally induced oxidative damage. Understanding the role of Grx5 and the other glutaredoxins will give us a better knowledge of how yeast cells protect themselves against constitutive and induced protein oxidative damage.
ACKNOWLEDGMENTS
We thank Lidia Piedrafita for excellent technical assistance, María Angeles de la Torre and Eloi Garí for comments on the manuscript, and Silvia Atrian for suggestions on the Sequence Space method.
This work was supported by the European Union (contract BIO4-CT97-2294), the Spanish Ministry of Education and Culture (project PB97-1456), and the Generalitat de Catalunya (projects SGR97/00087 to E.H. and SGR98-00012 to J.R.).
REFERENCES
- 1.Aslund F, Beckwith J. Bridge over troubled waters: sensing stress by disulfide bond formation. Cell. 1999;96:751–753. doi: 10.1016/s0092-8674(00)80584-x. [DOI] [PubMed] [Google Scholar]
- 2.Aslund F, Ehn B, Miranda-Vizuete A, Pueyo C, Holmgren A. Two additional glutaredoxins exist in Escherichia coli: glutaredoxin 3 as a hydrogen donor for ribonucleotide reductase in a thioredoxin/glutaredoxin 1 double mutant. Proc Natl Acad Sci USA. 1994;91:9813–9817. doi: 10.1073/pnas.91.21.9813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, et al. Current protocols in molecular biology. New York, N.Y: Wiley-Interscience; 1989. [Google Scholar]
- 4.Bellí G, Garí E, Aldea M, Herrero E. Functional analysis of yeast essential genes using a promoter-substitution cassette and the tetracycline-regulatable dual expression system. Yeast. 1998;14:1127–1138. doi: 10.1002/(SICI)1097-0061(19980915)14:12<1127::AID-YEA300>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 5.Benov L, Fridovich I. Why superoxide imposes an aromatic amino acid auxotrophy on Escherichia coli. J Biol Chem. 1999;274:4202–4206. doi: 10.1074/jbc.274.7.4202. [DOI] [PubMed] [Google Scholar]
- 6.Brown J L, Bussey H, Stewart R C. Yeast Skn7p functions in a eukaryotic two-component regulatory pathway. EMBO J. 1994;13:5186–5194. doi: 10.1002/j.1460-2075.1994.tb06849.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bushweller J H, Aslund F, Wuthrich K, Holmgren A. Structural and functional characterization of the mutant Escherichia coli glutaredoxin (C14-S) and its mixed disulfide with glutathione. Biochemistry. 1992;31:9288–9293. doi: 10.1021/bi00153a023. [DOI] [PubMed] [Google Scholar]
- 8.Casari G, Sander C, Valencia A. A method to predict functional residues in proteins. Struct Biol. 1995;2:171–178. doi: 10.1038/nsb0295-171. [DOI] [PubMed] [Google Scholar]
- 9.Cotgreave I A, Gerdes R G. Recent trends in glutathione biochemistry-glutathione-protein interactions: a molecular link between oxidative stress and cell proliferation? Biochem Biophys Res Commun. 1998;242:1–9. doi: 10.1006/bbrc.1997.7812. [DOI] [PubMed] [Google Scholar]
- 10.Davies K J A. Protein damage and degradation by oxygen radicals. J Biol Chem. 1987;262:9895–9901. [PubMed] [Google Scholar]
- 11.Gallego C, Garí E, Colomina N, Herrero E, Aldea M. The Cln3 cyclin is down-regulated by translational repression and degradation during G1 arrest caused by nitrogen deprivation in budding yeast. EMBO J. 1997;16:7196–7206. doi: 10.1093/emboj/16.23.7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gan Z R. Cloning and sequencing of a gene encoding yeast thioltransferase. Biochem Biophys Res Commun. 1992;187:949–955. doi: 10.1016/0006-291x(92)91289-3. [DOI] [PubMed] [Google Scholar]
- 13.Godon C, Lagniel G, Lee J, Buhler J-M, Kieffer S, Perrot M, Boucherie H, Toledano M B, Labarre J. The H2O2 stimulon in Saccharomyces cerevisiae. J Biol Chem. 1998;273:22480–22489. doi: 10.1074/jbc.273.35.22480. [DOI] [PubMed] [Google Scholar]
- 14.Goldberg D M, Spooner R J. Glutathione reductase. In: Bergmeyen H V, editor. Methods of enzymatic analysis. 3rd ed. Vol. 3. Deerfield Beach, Fla: Verlag Chemie; 1983. pp. 258–265. [Google Scholar]
- 15.Grant C M, Collinson L P, Roe J H, Dawes I W. Yeast glutathione reductase is required for protection against oxidative stress and is a target gene for yAP-1 transcriptional regulation. Mol Microbiol. 1996;21:171–179. doi: 10.1046/j.1365-2958.1996.6351340.x. [DOI] [PubMed] [Google Scholar]
- 16.Higgins D J, Thompson J D, Gibson T J. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 1996;266:383–389. doi: 10.1016/s0076-6879(96)66024-8. [DOI] [PubMed] [Google Scholar]
- 17.Holmgren A. Thioredoxin and glutaredoxin systems. J Biol Chem. 1989;254:13963–13966. [PubMed] [Google Scholar]
- 18.Holmgren A, Aslund F. Glutaredoxin. Methods Enzymol. 1995;252:283–292. doi: 10.1016/0076-6879(95)52031-7. [DOI] [PubMed] [Google Scholar]
- 19.Jamieson D J. Oxidative stress responses of the yeast Saccharomyces cerevisiae. Yeast. 1998;14:1511–1527. doi: 10.1002/(SICI)1097-0061(199812)14:16<1511::AID-YEA356>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 20.Kaiser C, Michaelis S, Mitchell A. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 21.Ketela T, Brown J L, Stewart R C, Bussey H. Yeast Skn7p activity is modulated by the Sln1p-Ypd1p osmosensor and contributes to regulation of the HOG pathway. Mol Gen Genet. 1998;259:372–378. doi: 10.1007/s004380050824. [DOI] [PubMed] [Google Scholar]
- 22.Kochetov G A. Transketolase from yeast, rat liver, and pig liver. Methods Enzymol. 1982;90:209–217. doi: 10.1016/s0076-6879(82)90128-8. [DOI] [PubMed] [Google Scholar]
- 23.Krems B, Charizanis C, Entian K-D. Mutants of Saccharomyces cerevisiae sensitive to oxidative and osmotic stress. Curr Genet. 1995;27:427–434. doi: 10.1007/BF00311211. [DOI] [PubMed] [Google Scholar]
- 24.Kuge S, Jones N. YAP1 dependent activation of TRX2 is essential for the response of activation of Saccharomyces cerevisiae to oxidative stress by hydroperoxides. EMBO J. 1994;13:655–664. doi: 10.1002/j.1460-2075.1994.tb06304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuge S, Jones N, Nomoto A. Regulation of yAP-1 nuclear localization in response to oxidative stress. EMBO J. 1997;16:1710–1720. doi: 10.1093/emboj/16.7.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levine R L, Williams J A, Stadtman E R, Schacter E. Carbonyl assays for determination of oxidatively-modified proteins. Methods Enzymol. 1994;233:346–357. doi: 10.1016/s0076-6879(94)33040-9. [DOI] [PubMed] [Google Scholar]
- 27.Li S, Ault A, Malone C L, Raitt D, Dean S, Johnston L H, Deschenes R J, Fassler J S. The yeast histidine protein kinase, Sln1p, mediates phosphotransfer to two response regulators, Ssk1p and Skn7p. EMBO J. 1998;17:6952–6962. doi: 10.1093/emboj/17.23.6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lillig C H, Prior A, Schwenn J D, Aslund F, Ritz D, Vlamis-Gardikas A, Holmgren A. New thioredoxins and glutaredoxins as electron donors of 3′-phosphoadenylylsulfate reductase. J Biol Chem. 1999;274:7695–7698. doi: 10.1074/jbc.274.12.7695. [DOI] [PubMed] [Google Scholar]
- 29.Luikenhuis S, Perrone G, Dawes I W, Grant C M. The yeast Saccharomyces cerevisiae contains two glutaredoxin genes that are required for protection against reactive oxygen species. Mol Biol Cell. 1998;9:1081–1091. doi: 10.1091/mbc.9.5.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lunström-Ljung J, Vlamis-Gardikas A, Aslund F, Holmgren A. Reactivity of glutaredoxins 1, 2 and 3 from Escherichia coli and protein disulfide isomerase glutathionyl-mixed disulfides in ribonuclease A. FEBS Lett. 1999;443:85–88. doi: 10.1016/s0014-5793(98)01698-6. [DOI] [PubMed] [Google Scholar]
- 31.Lussier M, White A M, Sheraton J, Dipaolo T, Treadwell J, Southard S B, Horestein C I, Chen-Weiner J, Ram A F J, Kapteyn J C, Roemer T, Vo D H, Bondoc A C, All J, Zhong W, Sdicu A M, Davies J, Klis F M, Robbins P W, Bussey H. Large scale identification of genes involved in cell surface biosynthesis and architecture in Saccharomyces cerevisiae. Genetics. 1997;147:435–450. doi: 10.1093/genetics/147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marchler C, Schüller C, Adam G, Ruis H. A Saccharomyces cerevisiae UAS element controlled by protein kinase A activates transcription in response to a variety of stress conditions. EMBO J. 1993;12:1997–2003. doi: 10.1002/j.1460-2075.1993.tb05849.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martínez-Pastor M T, Marchler G, Schüller C, Marchler-Bauer A, Ruis H, Estruch F. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress-response element (STRE) EMBO J. 1996;15:2227–2235. [PMC free article] [PubMed] [Google Scholar]
- 34.Meister A. Glutathione-ascorbic acid antioxidant system in animals. J Biol Chem. 1994;269:9397–9400. [PubMed] [Google Scholar]
- 35.Mewes H W, Heumann K, Kaps A, Pfeiffer F, Stocker S, Frishman D. MIPS: a database for genomes and protein sequences. Nucleic Acids Res. 1999;27:44–48. doi: 10.1093/nar/27.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.MIPS Yeast Genome Database. 30 August 1999, posting date. [Online.] Munich Information Center for Protein Sequences. http://www.mips.biochem.mpg.de/proj/yeast. [14 September 1999, last date accessed.]
- 37.Miranda Vizuete A, Martínez Galisteo E, Aslund F, López Barea J, Pueyo C, Holmgren A. Null thioredoxin and glutaredoxin Escherichia coli K-12 mutants have no enhanced sensitivity to mutagens due to a new GSH-dependent hydrogen donor and high increases in ribonucleotide reductase activity. J Biol Chem. 1994;269:16631–16637. [PubMed] [Google Scholar]
- 38.Moradas-Ferreira P, Costa V, Piper P, Mager W. The molecular defences against reactive oxygen species. Mol Microbiol. 1996;19:651–658. doi: 10.1046/j.1365-2958.1996.403940.x. [DOI] [PubMed] [Google Scholar]
- 39.Morgan B A, Banks G R, Toone W M, Raitt D, Kuge S, Johnston L H. The Skn7 response regulator controls gene expression in the oxidative stress response of the budding yeast Saccharomyces cerevisiae. EMBO J. 1997;16:1035–1044. doi: 10.1093/emboj/16.5.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muller E G D. Deoxyribonucleotides are maintained at normal levels in a yeast thioredoxin mutant defective in DNA synthesis. J Biol Chem. 1994;269:24466–24471. [PubMed] [Google Scholar]
- 41.Muller E G D. A redox-dependent function of thioredoxin is necessary to sustain a rapid rate of DNA synthesis in yeast. Arch Biochem Biophys. 1995;318:356–361. doi: 10.1006/abbi.1995.1240. [DOI] [PubMed] [Google Scholar]
- 42.Muller E G D. A glutathione reductase mutant of yeast accumulates high levels of oxidized glutathione and requires thioredoxin for growth. Mol Biol Cell. 1996;7:1805–1813. doi: 10.1091/mbc.7.11.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nikkola M, Gleason F L, Saarinen M, Joelson T, Björnberg O, Eklund H. A putative glutathione-binding site in T4 glutaredoxin investigated by site-directed mutagenesis. J Biol Chem. 1991;266:16105–16112. [PubMed] [Google Scholar]
- 44.Nordstrand K, Aslund F, Meunier S, Holmgrem A, Otting G, Berndt K D. Direct NMR observation of the Cys-14 thiol proton of reduced Escherichia coli glutaredoxin-3 supports the presence of an active site thiol-thiolate hydrogen bond. FEBS Lett. 1999;449:196–200. doi: 10.1016/s0014-5793(99)00401-9. [DOI] [PubMed] [Google Scholar]
- 45.Prinz W A, Aslund F, Holmgren A, Beckwith J. The role of the thioredoxin and glutaredoxin pathways in reducing protein disulfide bonds in the Escherichia coli cytoplasm. J Biol Chem. 1997;272:15661–15667. doi: 10.1074/jbc.272.25.15661. [DOI] [PubMed] [Google Scholar]
- 46.Ros, J., and E. Cabiscol. Unpublished observations.
- 47.Schüller C, Brewster J L, Alexander M R, Gustin M C, Ruis H. The HOG pathway controls osmotic regulation of transcription via the stress response element (STRE) of the Saccharomyces cerevisiae CTT1 gene. EMBO J. 1994;13:4382–4389. doi: 10.1002/j.1460-2075.1994.tb06758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shacter E, Williams J A, Lim M, Levine R L. Differential susceptibility of plasma proteins to oxidative modification. Examination by Western blot immunoassay. Free Radic Biol Med. 1994;17:429–437. doi: 10.1016/0891-5849(94)90169-4. [DOI] [PubMed] [Google Scholar]
- 49.Stephen D W S, Rivers S L, Jamieson D J. The role of the YAP1 and YAP2 genes in the regulation of the adaptative oxidative stress responses of Saccharomyces cerevisiae. Mol Microbiol. 1995;16:415–423. doi: 10.1111/j.1365-2958.1995.tb02407.x. [DOI] [PubMed] [Google Scholar]
- 50.Tamarit J, Cabiscol E, Ros J. Identification of the major oxidatively damaged proteins in Escherichia coli cells exposed to oxidative stress. J Biol Chem. 1998;273:3027–3032. doi: 10.1074/jbc.273.5.3027. [DOI] [PubMed] [Google Scholar]
- 51.Vlamis-Gardikas A, Aslund F, Spyrou G, Bergman T, Holmgren A. Cloning, overexpression, and characterization of glutaredoxin 2, an atypical glutaredoxin from Escherichia coli. J Biol Chem. 1997;272:11236–11243. doi: 10.1074/jbc.272.17.11236. [DOI] [PubMed] [Google Scholar]
- 52.Wach A, Brachat A, Pöhlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;13:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 53.Wanke V, Accorsi K, Porro D, Esposito F, Russo T, Venoni M. In budding yeast, reactive oxygen species induce both RAS-dependent and RAS-independent cell cycle-specific arrest. Mol Microbiol. 1999;32:753–764. doi: 10.1046/j.1365-2958.1999.01391.x. [DOI] [PubMed] [Google Scholar]
- 54.Wu A, Moye-Rowley W S. GSH1, which encodes γ-glutamylcysteine synthetase, is a target gene for yAP-1 transcriptional regulation. Mol Cell Biol. 1994;14:5832–5839. doi: 10.1128/mcb.14.9.5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang Y, Wells W W. Identification and characterization of the functional amino acids at the active center of pig liver thioltransferase by site-directed mutagenesis. J Biol Chem. 1991;266:12759–12765. [PubMed] [Google Scholar]