Abstract
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic has become a global health concern. Various SARS-CoV-2 vaccines have been developed and are being used for vaccination worldwide. However, no therapeutic agents against coronavirus disease 2019 (COVID-19) have been developed so far; therefore, new therapeutic agents are urgently needed. In the present study, we evaluated several hepatitis C virus direct-acting antivirals as potential candidates for drug repurposing against COVID-19. Theses include asunaprevir (a protease inhibitor), daclatasvir (an NS5A inhibitor), and sofosbuvir (an RNA polymerase inhibitor). We found that asunaprevir, but not sofosbuvir and daclatasvir, markedly inhibited SARS-CoV-2-induced cytopathic effects in Vero E6 cells. Both RNA and protein levels of SARS-CoV-2 were significantly decreased by treatment with asunaprevir. Moreover, asunaprevir profoundly decreased virion release from SARS-CoV-2-infected cells. A pseudoparticle entry assay revealed that asunaprevir blocked SARS-CoV-2 infection at the binding step of the viral life cycle. Furthermore, asunaprevir inhibited SARS-CoV-2 propagation in human lung Calu-3 cells. Collectively, we found that asunaprevir displays broad-spectrum antiviral activity and therefore might be worth developing as a new drug repurposing candidate for COVID-19.
Keywords: asunaprevir, COVID-19, drug repurposing, hepatitis C virus, SARS-CoV-2
INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the causative agent of coronavirus disease 2019 (COVID-19) (Ciotti et al., 2019). SARS-CoV-2 belongs to the Coronaviridae family which includes alphacoronavirus (alpha-CoV), betacoronavirus (beta-CoV), gammacoronavirus (gamma-CoV), and deltacoronavirus (delta-CoV) (Chu et al. 2020; Coleman and Frieman, 2014; Cui et al., 2019; Lefkowitz et al., 2018; Machhi et al., 2020; Woo et al., 2010). SARS-CoV-2 is an enveloped, single-stranded, positive-sense RNA virus with a genome of ~30 kb (Corbett et al., 2020; Hassan et al., 2020; Mulligan et al., 2020). Its genomic RNA has a 5′ cap and 3′ polyA tail. The nonstructural proteins, ORF1a and ORF1b, are translated from the genomic RNA. Discontinuous subgenomic mRNAs encode surface spike (S), envelope (E), integral membrane (M), and nucleocapsid (N) proteins and six accessory proteins (3a, 6, 7a, 7b, 8, and 10) (Manfredonia and Incarnato, 2021). The S protein is a homotrimeric glycoprotein that binds to the angiotensin-converting enzyme 2 (ACE2) receptor and mediates cell-virus membrane fusion (Duan et al., 2020); therefore, it is the main target for neutralizing antibodies against viral infection and represents the core immunogen constituent of vaccine designs. SARS-CoV-2 has evolved a four-amino acid insertion, RRAR, at positions 682-685, which represents a potential furin cleavage site. This characteristic is distinct from all other coronaviruses; thus, it may contribute to the higher transmissibility of this novel coronavirus.
The COVID-19 pandemic has posed an unprecedented challenge to public health and society, and has led to millions of deaths worldwide in a year (World Health Organization, 2020). Although various vaccines are limitedly available worldwide, no therapeutic (antiviral) agents are currently available for the treatment of COVID-19. Therefore, it is necessary to study the antiviral efficacy of existing highly safe drugs for emergency use in treating COVID-19 (Guy et al., 2020). Drug repurposing, also known as drug reprofiling or repositioning, is an emerging strategy in which U.S. Food and Drug Administration (FDA)-approved existing drugs are redeployed to control difficult-to-treat diseases (Guy et al., 2020; Law et al., 2013; Trivedi et al., 2020). By employing drug repurposing, we explored the benefits of hepatitis C virus (HCV) direct-acting antivirals (DAAs) for emergency use in controlling the COVID-19 pandemic.
The recent development of new oral DAAs has revolutionized HCV therapy. Numerous promising HCV DAAs have been developed, and many of these are being used for clinical treatment. More than 95% of HCV patients have a sustained viral response (SVR) by using some of these DAAs. Sofosbuvir, the most representative hepatitis C treatment agent, targets HCV nonstructural 5B (NS5B) RNA-dependent RNA polymerase and blocks RNA replication (Gane et al., 2013; Murakami et al., 2010). Daclatasvir targets HCV NS5A protein and inhibits HCV propagation (Gao et al., 2010; Nettles et al., 2011). Asunaprevir binds to the active site of the HCV NS3 serine protease and inhibits NS3 protease-mediated polypotein maturation (Lok et al., 2012; Suzuki et al., 2013). These drugs have been selected through clinical trials to make them suitable for the treatment of HCV patients.
In the present study, we assessed the anti-SARS-CoV-2 effects of sofosbuvir, asunaprevir, and daclatasvir. Of these three agents, asunaprevir exerted a strong antiviral effect against SARS-CoV-2 in both Vero E6 and Calu-3 cell lines. Based on our in vitro data, asunaprevir may be a promising drug candidate for the clinical treatment of COVID-19.
MATERIALS AND METHODS
Cell culture
All cell lines, including Vero E6, Calu-3, Huh7.5, and HEK293T cells, were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin in 5% CO2 at 37°C as reported previously (Lim and Hwang, 2011).
Preparation of infectious SARS-CoV-2
SARS-CoV-2 (NCCP-43331) was provided by the National Culture Collection for Pathogens, South Korea. It was cultured in Vero E6 cells in a biosafety level 3 (BSL-3) facility, the Korea Zoonosis Research Institute, Jeonbuk National University. Viral stocks were prepared by propagation in Vero E6 cells in DMEM supplemented with 2% FBS, 1% penicillin-streptomycin, and HEPES (Invitrogen, USA). Viral titers were determined by the 50% tissue culture infectious dose (TCID50) assay.
Preparation of infectious HCV
A monolayer of Huh7.5 cells was washed twice in PBS, trypsinized, and centrifuged at 850 rpm for 3 min (Lim et al., 2021). The cells were resuspended in 1 ml of Opti-MEM (Invitrogen), centrifuged at 1,000 rpm, and resuspended in 360 µl of Cytomix (120 mM KCl2, 0.15 mM CaCl2, 10 mM K2HPO4, 25 mM HEPES, 2 mM EDTA, and 5 mM MgCl2; pH 7.6) containing 2 mM ATP and 5 mM glutathione. The cells were mixed with 10 µg of Jc1 RNA and electroporated using the Gene Pulser Xcell system (Bio-Rad Laboratories, USA) in a 4-mm-gap cuvette. Cell culture supernatants containing de-novo HCV particles were harvested at 4 days after electroporation using syringe filter unit with a 0.45 µm pore size (Millipore, Germany). Huh7.5 cells were infected with the Jc1 virus at a multiplicity of infection (MOI) of 0.5 (Lim et al., 2011).
Pseudoparticle production and entry assay
HEK293T cells were transfected with 1 µg of SARS-CoV-2 spike expression plasmid (pGBW-m4137382; Addgene, USA), 3.1 µg of Gag/Pol packaging plasmid, and 3.1 µg of transfer vector encoding the firefly luciferase reporter protein using polyethyleneimine (Park et al., 2015). At 48 h after transfection, supernatants containing SARS-CoV-2 pseudoparticle (SARS-CoV-2pps) were collected. A pseudoparticle entry assay was performed by inoculating HEK293T cells with SARS-CoV-2pps for 36 h. Following this, a luciferase assay was performed using the Bio-GloTM Luciferase Assay System (Promega, USA).
Quantification of SARS-CoV-2 RNA and HCV RNA
cDNA was synthesized from total cellular RNAs isolated from Vero E6 cells using a cDNA synthesis kit (Toyobo, Japan) according to the manufacturer’s instructions. Quantitative real-time polymerase chain reaction (qRT-PCR) was performed using the CFX Connect real-time system (Bio-Rad Laboratories) with the following primers: sense, 5′-GTG AAA TGG TCA TGT GTG GCG G-3′ and antisense, 5′-CAA ATG TTA AAA ACA CTA TTA GCA TA-3′ for SARS-CoV-2 polymerase; sense, 5′-TGA CAG CAG TCG GTT GGA GCG-3′ and antisense, 5′-GAC TTC CTG TAA CAA CGC ATC TCA TA-3′ for actin; and sense, 5′-TTA GTA TGA GTG TCG TAC AGC CTC CAG-3′ and antisense, 5′-GGC ATA GAG TGG GTT TAT CCA AGA AAG G-3′ for HCV.
Immunoblot assay
Cells were lysed in a cell culture lysis reagent (Promega). Immunoblot assays were performed as described previously (Lim et al., 2021). The SARS-CoV-2 nucleoprotein was detected using an anti-nucleoprotein antibody (Sino Biological, China). An anti-actin antibody was purchased from Sigma-Aldrich (USA).
TCID50 assay
The TCID50-assay was performed to determine the infectious titer of cultured SARS-CoV-2. Vero E6 cells were seeded on 96-well plates overnight and infected with a 10-fold serial dilution of the virus-containing supernatants. At 5 days postinfection, the number of SARS-CoV-2-infected cells was counted under a microscope and the TCID50 value/ml was determined, as previously reported (Reed and Muench, 1938).
Water-soluble tetrazolium salt (WST) assay
Vero E6 cells seeded on a 96-well plate were treated with sofosbuvir (MedChemExpress, USA), daclatasvir (MedChemExpress), or asunaprevir (MedChemExpress). At the indicated time points, cell viability was measured using 30 µl of WST (Dail Lab, Korea), as reported previously (Choi et al., 2020).
Statistical analysis
Data are presented as the mean ± SD. Student’s t-test was used for statistical analysis (Microsoft Excel 2016; Microsoft, USA). The asterisks in the figures indicate significant differences (*P < 0.05; **P < 0.01; and ***P < 0.001), and ns indicates not significant.
RESULTS
Asunaprevir inhibits SARS-CoV-2 propagation
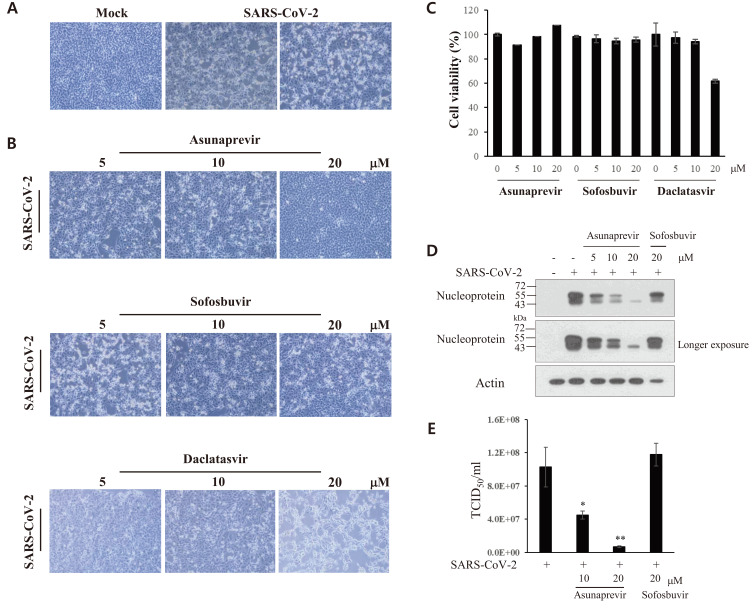
To assess the viral cytopathogenicity of SARS-CoV-2, Vero E6 cells were either mock-infected or infected with SARS-CoV-2 and the cytopathic effect (CPE) was monitored by light microscopy. As shown in Fig. 1A, a distinct CPE was noted at 2 days postinfection. To assess whether HCV DAAs displayed any anti-SARS-CoV-2 activity, Vero E6 cells were inoculated with a mixture of SARS-CoV-2 (MOI = 0.01) and asunaprevir, sofosbuvir, or daclatasvir for 1 h. Cell morphology was examined at 48 h postinfection. As shown in Fig. 1B, the SARS-CoV-2-induced CPE was not altered by treatment with either sofosbuvir or dacalatasvir. Surprisingly, it was markedly inhibited by 10 µM asunaprevir, and was completely blocked in the presence of 20 μM asunaprevir. Treatment with up to 20 μM asunaprevir had no effect on the viability of Vero E6 cells (Fig. 1C). Furthermore SARS-CoV-2 protein levels decreased on treatment with asunaprevir in a dose-dependent manner (Fig. 1D). The TCID50 value was then determined by infecting naive Vero E6 cells with serially-diluted virus harvested from supernatants (Fig. 1A). As shown in Fig. 1E, asunaprevir significantly decreased virion release from SARS-CoV-2-infected Vero E6 cells.
Fig. 1. Asunaprevir inhibits SARS-CoV-2 propagation.
(A) Vero E6 cells were either mock-infected or infected with SARS-CoV-2 (MOI = 0.01). At day 2 postinfection, the CPE in Vero E6 cells was visualized under a light microscope. (B) Vero E6 cells were infected with SARS-CoV-2 (MOI = 0.01) for 1 h with the indicated concentrations of asunaprevir, sofosbuvir, or daclatasvir. The SARS-CoV-2-infected cells were further cultured in media containing each DAA. At day 2 postinfection, the CPE induced by SARS-CoV-2 in Vero E6 cells was visualized under a light microscope. (C) Vero E6 cells were treated with the indicated concentrations of asunaprevir, sofosbuvir, or daclatasvir. At 48 h posttreatment, cytotoxicity was measured by a WST assay. (D) Asunaprevir inhibited SARS-CoV-2 protein levels in Vero E6 cells. Vero E6 cells were either mock-infected or infected with SARS-CoV-2 (MOI = 0.01) for 1 h in the absence or presence of asunaprevir and further cultured for 2 days in media containing various concentrations of asunaprevir. SARS-CoV-2 protein levels were determined by an immunoblot assay using the indicated antibodies. Sofosbuvir was used as a negative control. (E) Vero E6 cells were infected with serially diluted SARS-CoV-2-containing culture supernatants harvested from panel B. At day 5 postinfection, the TCID50 value was determined, as reported previously (Lim et al., 2011). *P < 0.05; **P < 0.01.
Verification of anti-viral activities of selected DAAs in HCV-infected cells
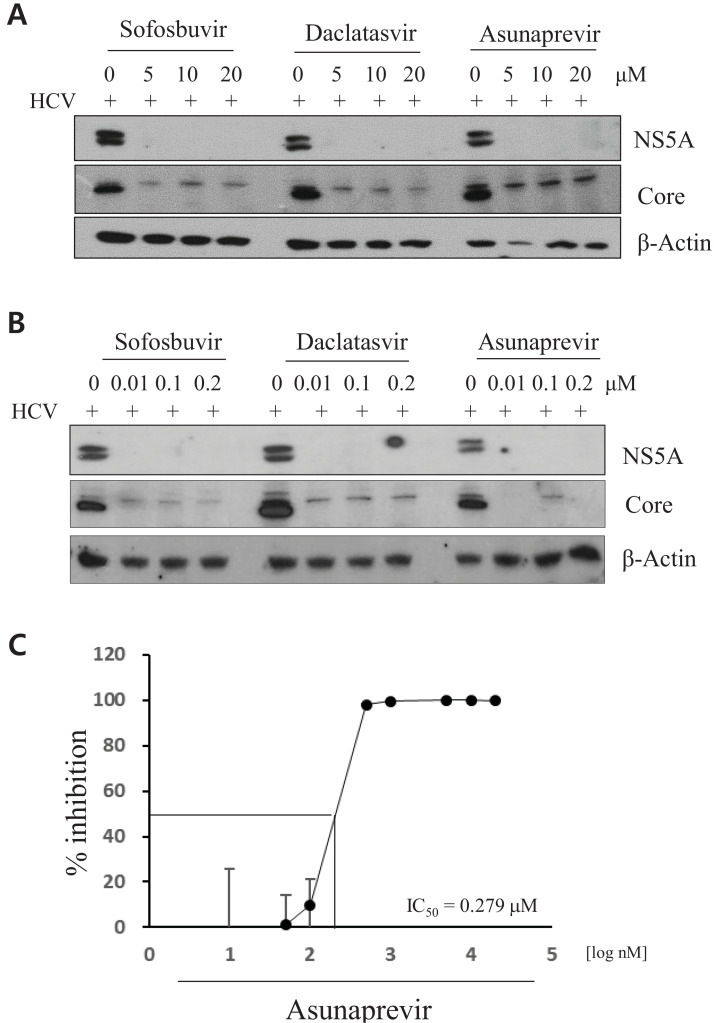
To verify the activities of selected DAAs in HCV-infected cells, Huh7.5 cells were infected with cell culture-derived HCV (HCVcc) for 4 h in the presence of various concentrations of sofosbuvir, daclatasvir, or asunaprevir. At 48 h postinfection, HCV protein levels were measured by immunoblotting using the indicated antibodies. The antiviral activities of DAAs were assessed using the same concentrations of DAAs, as described in Fig. 1. Indeed, HCV protein expression levels markedly decreased on treatment with DAAs. As shown in Fig. 2A, HCV protein levels equally decrteased on treatment with three different DAAs. Notably, compared with core protein expression, NS5A protein expression was completely inhibited by DAAs. As these DAAs are specific inhibitors of HCV, the antiviral activities of DAAs were also assessed by treating HCV-infected cells with lower concentrations of DAAs. HCV protein levels profoundly decreased on treatment with as low as 0.01 μM of each DAA (Fig. 2B). To further determine the half-maximal inhibitory concentration (IC50) for the anti-HCV activity of asunaprevir, Huh7.5 cells infected with Jc1 were treated with various concentrations of asunaprevir. Following this, HCV RNA levels were measured by RT-PCR. Asunaprevir inhibited HCV propagation with an IC50 value of 0.279 μM in Huh7.5 cells (Fig. 2C).
Fig. 2. Antiviral activities of DAAs in HCV-infected cells.
(A and B) Huh7.5 cells were infected with HCV Jc1 for 4 h. Following this, the culture medium was replaced with fresh medium in the presence of the indicated drugs. At 48 h posttreatment, viral proteins were detected by an immunoblot assay using the indicated antibodies. (C) Huh7.5 cells infected with HCV Jc1 were treated with various concentrations of asunaprevir. At 48 h postinfection, total RNA was extracted and the IC50 value was determined on the basis of HCV RNA levels measured by qRT-PCR.
Confirmation of anti-SARS-CoV-2 activity of asunaprevir
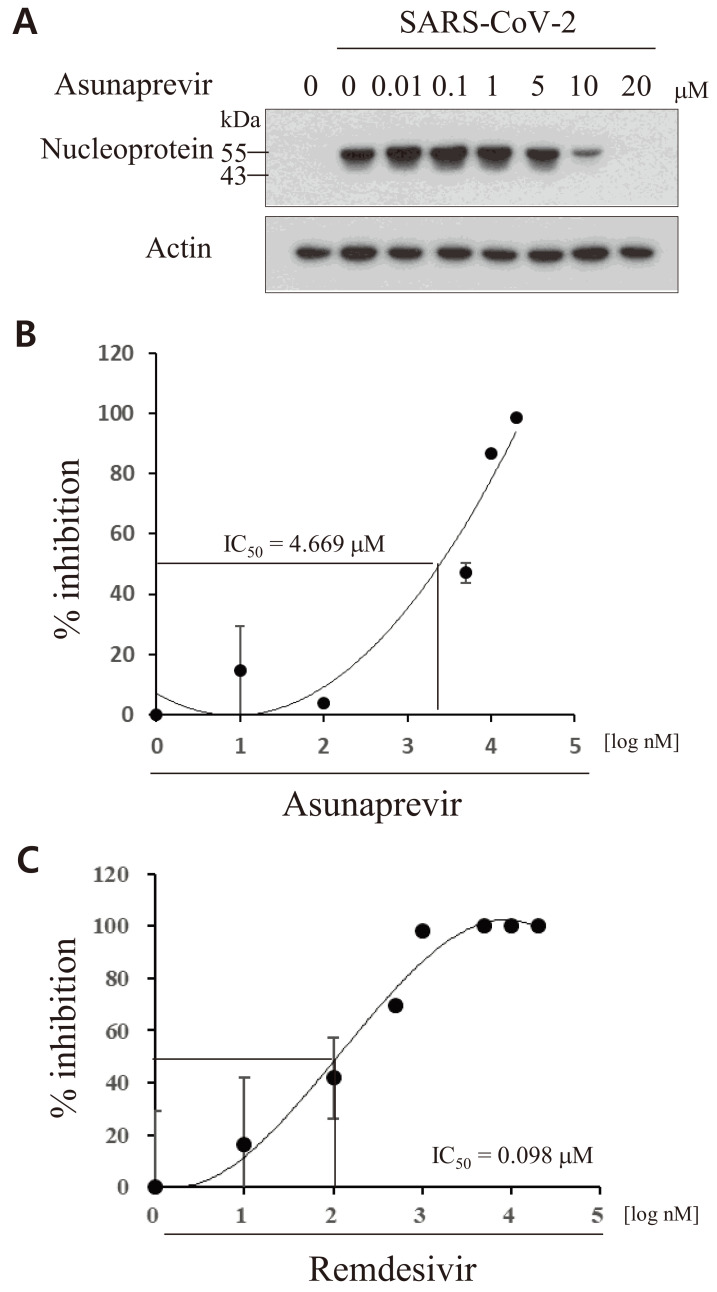
To further characterize the antiviral effect of asunaprevir against SARS-CoV-2, the IC50 for the on-target activity of asunaprevir was determined. Vero E6 cells were inoculated with a mixture of SARS-CoV-2 (MOI = 0.01) and asunaprevir for 1 h and further cultured for 24 h in the presence of asunaprevir. SARS-CoV-2 protein levels decreased on treatment with asunaprevir in a dose-dependent manner (Fig. 3A). As shown in Fig. 3B, asunaprevir blocked SARS-CoV-2 infection in Vero E6 cells with an IC50 value of 4.669 μM. The IC50 value of remdesivir was 0.098 μM in Vero E6 cells (Fig. 3C). Remdesivir was used as a positive control as it is an FDA-approved anti-SARS-CoV-2 drug.
Fig. 3. Verification of anti-SARS-CoV-2 activity of asunaprevir.
(A) Vero E6 cells were either mock-infected or infected with SARS-CoV-2 (MOI = 0.1) in the absence or presence of asunaprevir for 1 h. At 24 h postinfection, SARS-CoV-2 nucleoprotein levels were determined by an immunoblot assay. (B) Vero E6 cells were infected with SARS-CoV-2 in the presence of various concentrations of asunaprevir. At 24 h postinfection, total RNA was extracted and the IC50 value was determined on the basis of SARS-CoV-2 RNA levels measured by qRT-PCR. (C) Vero E6 cells were infected with SARS-CoV-2 in the presence of various concentrations of remdesivir. At 24 h postinfection, the IC50 value was determined as described in panel B.
Asunaprevir inhibits SARS-CoV-2 propagation at the entry step of the viral life cycle
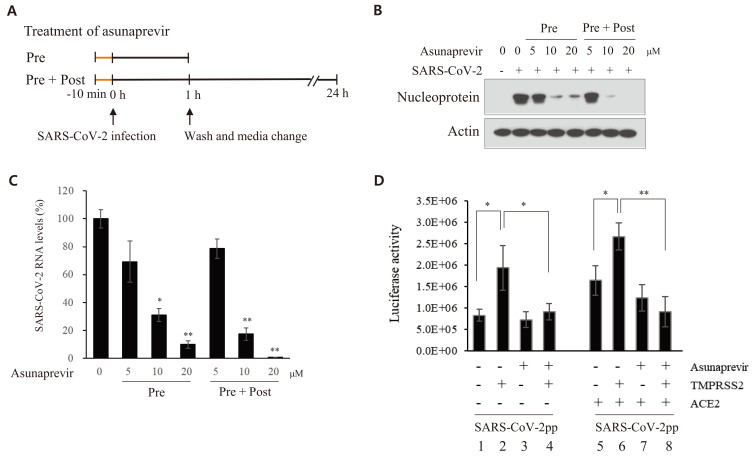
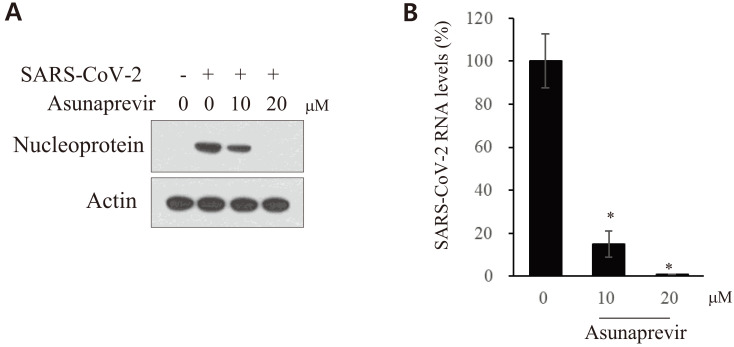
To determine the effect of asunaprevir on the life cycle of SARS-CoV-2, Vero E6 cells were treated with asunaprevir under two different experimental conditions (Fig. 4A). The cells were preincubated with asunaprevir for 10 min and then inoculated with SARS-CoV-2 for 1 h in the presence of asunaprevir. The culture medium was replaced with fresh medium in the absence (pretreatment) or presence (pretreatment + posttreatment) of asunaprevir for 24 h. As shown in Fig. 4B, SARS-CoV-2 nucleoprotein levels markedly decreased on treatment with 10 μM asunaprevir under the both conditions. Notably, higher antivial activity was noted on pretreatment + posttreatment with asunaprevir than on only pretreatment. The RNA levels of SARS-CoV-2 were also assessed. Total RNA was isolated from SARS-CoV-2-infected Vero E6 cells treated with asunaprevir as described in Fig. 4A. Following this, viral RNA levels were determined by qRT-PCR. SARS-CoV-2 RNA levels under two different conditions significantly decreased on treatment with asunaprevir in the same manner (Fig. 4C). The spike protein plays a pivotal role in SARS-CoV-2 infection. To determine whether asunaprevir blocks SARS-CoV-2 at the infection step of the life cycle, a viral entry assay was performed using SARS-CoV-2pps carrying a luciferase gene. HEK293T cells were transfected with the indicated combinations of ACE2 and TMPRSS2 expression plasmids. The cells were either left untreated or pretreated with asunaprevir and then infected with pseudoparticles in the absence or presence of asunaprevir. As shown in Fig. 4D, overexpression of either TMPRSS2 or ACE2 increased luciferase activity. Furthermore, luciferase activity significantly decreased on treatment with asunaprevir in cells transfected with TMPRSS2 but not in cells transfected with ACE2 alone (Fig. 4D, lane 4 vs lane 7). These data suggest that TMPRSS2 plays a key role in SARS-CoV-2 infection at the entry step of the viral life cycle. Furthermore, ACE2-induced luciferase activity significantly increased in cells transfected with TMPRSS2 and that this activity significantly decreased on treatment with by asunaprevir (Fig. 4D, lane 6 vs lane 8). Most importantly, ACE2- and TMPRSS2-mediated luciferase activity significantly decreased to the basal level on treatment with asunaprevir, indicating that asunaprevir inhibits SARS-CoV-2 propagation at the entry step of the viral life cycle.
Fig. 4. Asunaprevir inhibits SARS-CoV-2 propagation at the entry step of the viral life cycle.
(A) A schematic illustration of the experimental design. (B) Vero E6 cells were pretreated with various concentrations of asunaprevir for 10 min and then inoculated with SARS-CoV-2 for 1 h in the presence of asunaprevir. The culture medium was replaced with fresh medium in the absence of asunaprevir (pretreatment [Pre]). Alternatively, Vero E6 cells were pretreated with various concentrations of asunaprevir for 10 min and then inoculated with SARS-CoV-2 for 1 h in the presence of asunaprevir. The culture medium was replaced with fresh medium in the presence (pretrteatment + posttreatment [Pre + Post]) of various concentrations of asunaprevir. At 24 h postinfection, SARS-CoV-2 protein levels were determined by an immunoblot assay. (C) Total RNA was extracted from Vero E6 cells treated as described in panel B and SARS-CoV-2 RNA levels were determined by qRT-PCR. (D) HEK293T cells were transfected with vector, TMPRSS2, or ACE2 plasmid, or were cotransfected with TMPRSS2 and ACE2 for 12 h in the absence or presence of 20 µM asunaprevir. SARS-CoV-2pps were mixed with 20 µM asunaprevir for 1 h, following which cells were infected with the pseudoparticle mixture for 1 h. At 36 h postinfection, the cells were harvested and viral entry was determined by luciferase activity. *P < 0.05; **P < 0.01.
Asunaprevir decreases SARS-CoV-2 propagation in human lung cells
Calu-3 is a human lung cancer cell line that has been used to verify in vitro data for drug development. To further confirm the antiviral effect of asunaprevir on SARS-CoV-2 infection in lung cells, Calu-3 cells were inoculated with SARS-CoV-2 at an MOI of 0.1 in the presence of various concentrations of asunaprevir. As shown in Fig. 5A, SARS-CoV-2 nucleoprotein levels decreased on treatment with 10 μM asunaprevir and were undetectable on treatment with 20 μM asunaprevir. Consistently, SARS-CoV-2 RNA levels significantly decreased on treatment with asunaprevir (Fig. 5B), verifying that asunaprevir truly suppresses SARS-CoV-2 propagation in human lung cells.
Fig. 5. Asunaprevir decreases SARS-CoV-2 propagation in Calu-3 cells.
(A) Calu-3 cells were either mock-infected or infected with SARS-CoV-2 (MOI = 0.1) for 1 h in the presence of various concentrations of asunaprevir. At 48 h postinfection, SARS-CoV-2 protein levels were determined by an immunoblot assay. (B) Total RNA was extracted from Calu-3 cells treated as described in panel A. Following this, SARS-CoV-2 RNA levels were determined by qRT-PCR. *P < 0.05.
DISCUSSION
SARS-CoV-2 has caused a coronavirus pandemic and COVID-19 caused by SARS-CoV-2 has become a global public health concern. Although vaccines are now available for immunization against SARS-CoV-2, no anti-SARS-CoV-2 drugs have been developed so far. COVID-19 pandemic has already caused millions of deaths; therefore the development of antiviral drugs to treat SARS-CoV-2 infection is urgently needed. Recently, drug repurposing has been tried to stop the rapid spread of SARS-CoV-2 (Guy et al., 2020; Yousefi et al., 2021; Zhou et al., 2020). Drug repurposing is a technique used to treat emerging diseases by finding new uses for existing drugs. Therefore, it has the advantage of substantially reducing the drug development timeline and costs for the treatment of COVID-19. As SARS-CoV-2 is an RNA virus, we explored the possibility of drug repurposing of HCV DAAs to treat COVID-19. Numerous HCV DAAs are already approved by the FDA for safety in human trials; therefore, we used these drugs to assess their anti-SARS-CoV-2 effects. There are a number of HCV DAAs that have been demonstrated to be potent anti-HCV drugs. Of these HCV DAAs, we selected sofosbuvir, asunaprevir, and daclatasvir and assessed their anti-SARS-CoV-2 effects. Sofosbuvir is a nucleotide analog inhibitor of HCV NS5B, an RNA-dependent RNA polymerase, which is the key enzyme mediating HCV RNA replication. Asunaprevir is a potent serine protease inhibitor of HCV NS3, and daclatasvir is an NS5A inhibitor.
Remdesivir, an adenosine analog, has shown broad-spectrum efficacy against a panel of viruses, including SARS-CoV (Agostini et al., 2018), MERS-CoV (Sheahan et al., 2017), Ebola (Warren et al., 2016), and SARS-CoV-2 (Grein et al., 2020). As remdesivir is an RNA polymerase inhibitor, we expected sofosbuvir, an HCV RNA polymerase inhibitor, to be a good therapeutic candidate against SARS-CoV-2 infection. Unexpectedly, sofosbuvir exhibited no anti-SARS-CoV-2 activity. Similarly, daclatasvir showed no inhibitory activity against SARS-CoV-2. Surprisingly, asunaprevir exhibited strong anti-SARS-CoV-2 activity at both RNA and protein levels. Light microscopy data further confirmed that SARS-CoV-2-induced cytopathogenicity was remarkably inhibited by asunaprevir in Vero E6 cells. However, why an HCV protease inhibitor and not an HCV RNA polymerase inhibitor exhibited antiviral activity against SARS-CoV-2 remains unclear. We speculated that either the target site or the enzymatic activity of asunaprevir shares some protease similarity between HCV and SARS-CoV-2 RNA viruses, leading to broad spectrum antiviral activity.
ACE2 is an important cellular receptor that binds to the spike protein of SARS-CoV-2 (Wang, 2020). In the present study, we found that SARS-CoV-2pp entry did not decrease significantly on treatment with asunaprevir in cells transfected with ACE2; however, it decreased significantly on treatment with asunaprevir in cells expressing TMPRSS2. SARS-CoV-2 propagation requires three major proteases. TMPRSS2 is a serine protease involved in the early infection stage of the SARS-CoV-2 life cycle. It induces the cleavage of the spike protein to facilitate the entry of coronavirus (Matsuyama et al., 2010). SARS-CoV-2 encodes M3CLpro (also known as Mpro, the main cysteine protease of coronavirus) and PLpro (papain-like protease) enzymes. These two cysteine proteases catalyze the proteolysis of polyproteins translated from the viral genome into nonstructural proteins; therefore, inhibiting the activity of these proteases impedes viral replication (Ghahremanpour et al., 2020). Hence, it is intriguing how asunaprevir, a potent HCV NS3 protease inhibitor, inhibits the entry of SARS-CoV-2. As asunaprevir blocked the entry step of the SARS-CoV-2 life cycle and this inhibition was found to be significantly increased in cells overexpressing TMPRSS2, asunaprevir may target TMPRSS2 activity. Moreover, as asunaprevir targets the HCV NS3 serine protease (Soumana et al., 2014), it is possible that asunaprevir could bind to the TMPRSS2 serine protease. It remains unclear whether TMPRSS2 and HCV NS3 have structural similarities in their serine proteases. Collectively, further studies are required to demonstrate the mode of action by which asunaprevir affects the entry of SARS-CoV-2.
The present findings suggest that the drug repurposing of asunaprevir, a primary HCV DAA, can help treat SARS-CoV-2 infection. Although the IC50 value of asunaprevir for SARS-CoV-2 is much higher than that of asunaprevir for HCV, we found that asunaprevir exhibits broad-spectrum antiviral activity against RNA viruses. This finding indicates that asunaprevir may be a promising candidate for drug repurposing against COVID-19. However, further studies are needed to confirm the safety and efficacy of asunaprevir in patients with COVID-19.
ACKNOWLEDGMENTS
The pathogen resources (NCC43331) for this study were provided by the National Culture Collection for Pathogens.
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (2021R1A2C2003275). This work, including the efforts of Yun-Sook Lim, was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (2019R1A2C1086914).
Footnotes
AUTHOR CONTRIBUTIONS
All authors have given approval to the final version of the manuscript. Y.S.L. performed experiments, analyzed data, and wrote the manuscript. L.P.N. performed experiments. G.H.L., S.G.L., K.S.L., and B.K. provided reagents and expertise. Y.S.L. and S.B.H. designed experiments and secured funding. S.B.H. supervised the study and wrote the manuscript.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Agostini M.L., Andres E.L., Sims A.C., Graham R.L., Sheahan T.P., Lu X., Smith E.C., Case J.B., Feng J.Y., Jordan R., et al. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. Bio. 2018;9:e00221–18. doi: 10.1128/mBio.00221-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J.W., Kim J.W., Nguyen L.P., Nguyen H.C., Park E.M., Choi D.H., Han K.M., Kang S.M., Tark D., Lim Y.S., et al. Nonstructural NS5A protein regulates LIM and SH3 domain protein 1 to promote hepatitis C virus propagation. Mol. Cells. 2020;43:469–478. doi: 10.14348/molcells.2020.0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu D.K.W., Pan Y., Cheng S.M.S., Hui K.P.Y., Krishnan P., Liu Y., Ng D.Y.M., Wan C.K.C., Yang P., Wang Q., et al. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin. Chem. 2020;66:549–555. doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciotti M., Angeletti S., Minieri M., Giovannetti M., Benvenuto D., Pascarella S., Sagnelli C., Bianchi M., Bernardini S., Ciccozzi M. COVID-19 outbreak: an overview. Chemotherapy. 2019;64:215–223. doi: 10.1159/000507423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman C.M., Frieman M.B. Coronaviruses: important emerging human pathogens. J. Virol. 2014;88:5209–5212. doi: 10.1128/JVI.03488-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett K.S., Flynn B., Foulds K.E., Francica J.R., Boyoglu-Barnum S., Werner A.P., Flach B., O'Connell S., Bock K.W., Minai M., et al. Evaluation of the mRNA-1273 vaccine against SARS-CoV-2 in nonhuman primates. N. Engl. J. Med. 2020;383:1544–1555. doi: 10.1056/NEJMoa2024671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan L., Zheng Q., Zhang H., Niu Y., Lou Y., Wang H. The SARS-CoV-2 spike glycoprotein biosynthesis, structure, function, and antigenicity: implications for the design of spike-based vaccine immunogens. Front. Immunol. 2020;11:576622. doi: 10.3389/fimmu.2020.576622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gane E.J., Stedman C.A., Hyland R.H., Ding X., Svarovskaia E., Symonds W.T., Hindes R.G., Berrey M.M. Nucleotide polymerase inhibitor sofosbuvir plus ribavirin for hepatitis C. N. Engl. J. Med. 2013;368:34–44. doi: 10.1056/NEJMoa1208953. [DOI] [PubMed] [Google Scholar]
- Gao M., Nettles R.E., Belema M., Snyder L.B., Nguyen V.N., Fridell R.A., Serrano-Wu M.H., Langley D.R., Sun J.H., O'Boyle D.R., 2nd, et al. Chemical genetics strategy identifies an HCV NS5A inhibitor with a potent clinical effect. Nature. 2010;465:96–100. doi: 10.1038/nature08960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghahremanpour M.M., Tirado-Rives J., Deshmukh M., Ippolito J.A., Zhang C.H., de Vaca I.C., Liosi M.E., Anderson K.S., Jorgensen W.L. Identification of 14 known drugs as inhibitors of the main protease of SARS-CoV-2. ACS Med. Chem. Lett. 2020;11:2526–2533. doi: 10.1021/acsmedchemlett.0c00521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grein J., Ohmagari N., Shin D., Diaz G., Asperges E., Castagna A., Feldt T., Green G., Green M.L., Lescure F.X., et al. Compassionate use of remdesivir for patients with severe Covid-19. N. Engl. J. Med. 2020;382:2327–2336. doi: 10.1056/NEJMoa2007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy R.K., Dipaola R.S., Romanelli F., Dutch R.E. Rapid repurposing of drugs for COVID-19. Science. 2020;368:829–830. doi: 10.1126/science.abb9332. [DOI] [PubMed] [Google Scholar]
- Hassan A.O., Kafai N.M., Dmitriev I.P., Fox J.M., Smith B.K., Harvey I.B., Chen R.E., Winkler E.S., Wessel A.W., Case J.B., et al. A single-dose intranasal ChAd vaccine protects upper and lower respiratory tracts against SARS-CoV-2. Cell. 2020;183:169–184.e13. doi: 10.1016/j.cell.2020.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law G.L., Tisoncik-Go J., Korth M.J., Katze M.G. Drug repurposing: a better approach for infectious disease drug discovery? Curr. Opin. Immunol. 2013;25:588–592. doi: 10.1016/j.coi.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefkowitz E.J., Dempsey D.M., Hendrickson R.C., Orton R.J., Siddell S.G., Smith D.B. Virus taxonomy: the database of the International Committee on Taxonomy of Viruses (ICTV) Nucleic Acids Res. 2018;46(D1):D708–D717. doi: 10.1093/nar/gkx932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim Y.S., Hwang S.B. Hepatitis C virus NS5A protein interacts with phosphatidylinositol 4-kinase type IIIalpha and regulates viral propagation. J. Biol. Chem. 2011;286:11290–11298. doi: 10.1074/jbc.M110.194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim Y.S., Mai H.N., Nguyen L.P., Kang S.M., Tark D., Hwang S.B. Adenosylhomocysteinase like 1 interacts with nonstructural 5A and regulates hepatitis C virus propagation. J. Microbiol. 2021;59:101–109. doi: 10.1007/s12275-021-0470-8. [DOI] [PubMed] [Google Scholar]
- Lim Y.S., Tran H.T., Park S.J., Yim S.A., Hwang S.B. Peptidyl-prolyl isomerase Pin1 is a cellular factor required for hepatitis C virus propagation. J. Virol. 2011;85:8777–8788. doi: 10.1128/JVI.02533-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lok A.S., Gardiner D.F., Lawitz E., Martorell C., Everson G.T., Ghalib R., Reindollar R., Rustgi V., McPhee F., Wind-Rotolo M., et al. Preliminary study of two antiviral agents for hepatitis C genotype 1. N. Engl. J. Med. 2012;366:216–224. doi: 10.1056/NEJMoa1104430. [DOI] [PubMed] [Google Scholar]
- Machhi J., Herskovitz J., Senan A.M., Dutta D., Nath B., Oleynikov M.D., Blomberg W.R., Meigs D.D., Hasan M., Patel M., et al. The natural history, pathobiology, and clinical manifestations of SARS-CoV-2 infections. J. Neuroimmune Pharmacol. 2020;15:359–386. doi: 10.1007/s11481-020-09944-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfredonia I., Incarnato D. Structure and regulation of coronavirus genomes: state-of-the-art and novel insights from SARS-CoV-2 studies. Biochem. Soc. Trans. 2021;49:341–352. doi: 10.1042/BST20200670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S., Nagata N., Shirato K., Kawase M., Takeda M., Taguchi F. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J. Virol. 2010;84:12658–12664. doi: 10.1128/JVI.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan M.J., Lyke K.E., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Raabe V., Bailey R., Swanson K.A., et al. Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature. 2020;586:589–593. doi: 10.1038/s41586-020-2639-4. [DOI] [PubMed] [Google Scholar]
- Murakami E., Tolstykh T., Bao H., Niu C., Steuer H.M., Bao D., Chang W., Espiritu C., Bansal S., Lam A.M., et al. Mechanism of activation of PSI-7851 and its diastereoisomer PSI-7977. J. Biol. Chem. 2010;285:34337–34347. doi: 10.1074/jbc.M110.161802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nettles R.E., Gao M., Bifano M., Chung E., Persson A., Marbury T.C., Goldwater R., DeMicco M.P., Rodriguez-Torres M., Vutikullird A., et al. Multiple ascending dose study of BMS-790052, a nonstructural protein 5A replication complex inhibitor, in patients infected with hepatitis C virus genotype 1. Hepatology. 2011;54:1956–1965. doi: 10.1002/hep.24609. [DOI] [PubMed] [Google Scholar]
- Park C., Min S., Park E.M., Lim Y.S., Kang S., Suzuki T., Shin E.C., Hwang S.B. Pim kinase interacts with nonstructural 5A protein and regulates hepatitis C virus entry. J. Virol. 2015;89:10073–10086. doi: 10.1128/JVI.01707-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed L.J., Muench H. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 1938;27:493–497. doi: 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
- Sheahan T.P., Sims A.C., Graham R.L., Menachery V.D., Gralinski L.E., Case J.B., Leist S.R., Pyrc K., Feng J.Y., Trantcheva I., et al. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci. Transl. Med. 2017;9:eaal3653. doi: 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soumana D.I., Ali A., Schiffer C.A. Structural analysis of asunaprevir resistance in HCV NS3/4A protease. ACS Chem. Biol. 2014;9:2485–2490. doi: 10.1021/cb5006118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y., Ikeda K., Suzuki F., Toyota J., Karino Y., Chayama K., Kawakami Y., Ishikawa H., Watanabe H., Hu W., et al. Dual oral therapy with daclatasvir and asunaprevir for patients with HCV genotype 1b infection and limited treatment options. J. Hepatol. 2013;58:655–662. doi: 10.1016/j.jhep.2012.09.037. [DOI] [PubMed] [Google Scholar]
- Trivedi J., Mohan M., Byrareddy S.N. Drug repurposing approaches to combating viral infections. J. Clin. Med. 2020;9:3777. doi: 10.3390/jcm9113777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.Y., et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894–904.e9. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren T.K., Jordan R., Lo M.K., Ray A.S., Mackman R.L., Soloveva V., Siegel D., Perron M., Bannister R., Hui H.C., et al. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature. 2016;531:381–385. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Huang Y., Lau S.K., Yuen K.Y. Coronavirus genomics and bioinformatics analysis. Viruses. 2010;2:1804–1820. doi: 10.3390/v2081803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization, author. [Retrieved December 8, 2020];Coronavirus disease (COVID-19) situation reports. 2020 https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports/
- Yousefi H., Mashouri L., Okpechi S.C., Alahari N., Alahari S.K. Repurposing existing drugs for the treatment of COVID-19/SARS-CoV-2 infection: a review describing drug mechanisms of action. Biochem. Pharmacol. 2021;183:114296. doi: 10.1016/j.bcp.2020.114296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Wang F., Tang J., Nussinov R., Cheng F. Artificial intelligence in COVID-19 drug repurposing. Lancet Digit. Health. 2020;2:e667–e676. doi: 10.1016/S2589-7500(20)30192-8. [DOI] [PMC free article] [PubMed] [Google Scholar]