Abstract
In eukaryotic cells, proteins are targeted to their final subcellular locations with precise timing. A key underlying mechanism is the active transport of cognate mRNAs, which in many systems can be linked intimately to membrane trafficking. A prominent example is the long‐distance endosomal transport of mRNAs and their local translation. Here, we describe current highlights of fundamental mechanisms of the underlying transport process as well as of biological functions ranging from endosperm development in plants to fungal pathogenicity and neuronal processes. Translation of endosome‐associated mRNAs often occurs at the cytoplasmic surface of endosomes, a process that is needed for membrane‐assisted formation of heteromeric protein complexes and for accurate subcellular targeting of proteins. Importantly, endosome‐coupled translation of mRNAs encoding mitochondrial proteins, for example, seems to be particularly important for efficient organelle import and for regulating subcellular mitochondrial activity. In essence, these findings reveal a new mechanism of loading newly synthesised proteins onto endocytic membranes enabling intimate crosstalk between organelles. The novel link between endosomes and mitochondria adds an inspiring new level of complexity to trafficking and organelle biology.
Keywords: endosomes, local translation, microtubules, mitochondria, organelle, RNA transport
Subject Categories: Membranes & Trafficking, RNA Biology, Translation & Protein Quality
This review highlights mRNA transport and translation on endosomes in different physiological contexts, and the role this process has in protein complex formation and subcellular protein targeting, including of mitochondrial proteins.
Glossary
- ALS
amyotrophic lateral sclerosis
- BicD
bicaudal D
- Cdc
cell division control
- CMT2B
charcot‐marie‐tooth type 2B
- CSRP1
cysteine and glycine‐rich protein 1
- EEA1
early endosomal antigen 1
- Egl
egalitarian
- EJC
exon junction complex
- ELAV
embryonic lethal abnormal vision
- ER
endoplasmic reticulum
- FERRY
five‐subunit endosomal Rab5 and RNA/ribosome intermediary
- FMRP
fragile X mental retardation protein
- FYVE
Fab 1, YOTB, Vac 1, and EEA1
- IDRs
intrinsically disordered regions
- KH
K homology domain
- LAMP1
Lysosomal‐associated membrane protein 1
- LB2
Lamin B2
- MAMs
mitochondria‐associated ER membranes
- MLLE
MademoiseLLE domain
- mRNPs
messenger ribonucleoproteins
- MTS
mitochondrial targeting sequence
- NAC
nascent chain‐associated complex
- nRCCs
Mitochondrial respiratory chain complexes
- NSF
N‐ethylmaleimide‐sensitive factor
- NTC
NineTeen complex
- PAM2
PAB1C‐associated motif
- PARKIN
RBR E3 ubiquitin protein ligase
- PI3P
phosphatidylinositol 3‐phosphate
- PINK1
PTEN‐induced serine/threonine kinase 1
- RBP
RNA‐binding protein
- RRMs
RNA recognition motifs
- SFPQ
splicing factor proline‐ and glutamine‐rich
- STRIPAK
Striatin‐interacting phosphatase and kinase complex
- SYN2
synaptojanin 2
- SYNJ2BP
SYN2 binding protein
- TOM
translocase of the mitochondrial outer membrane complex
- UTR
untranslated region
- vRNA
viral genomic RNA
Introduction
A major evolutionary invention is the subcellular organisation of the eukaryotic cell, relying on defined compartments for division of labour. The nucleus, for example, harbours the genetic information, the endoplasmic reticulum (ER) is essential for protein secretion, and mitochondria act as cellular powerhouses. However, despite the apparent separation by different functionalities, there is intensive transport and communication between organelles. For example, most mRNAs encoding mitochondrial proteins are synthesised in the nucleus and translated in the cytosol, and subsequently, the translation products are targeted to distinct mitochondrial subcompartments (Pfanner et al, 2019). Furthermore, interorganelle contacts contribute to the synthesis of numerous lipids that undergo bidirectional transfers between the ER and mitochondria (Tamura et al, 2020).
To orchestrate communication between organelles, the cells heavily depend on intracellular trafficking of membranes, proteins and mRNAs. Outbound membrane and protein trafficking is mediated by secretion via the ER, Golgi apparatus and secretory vesicles. For inbound trafficking or endocytosis, protein‐containing plasma membrane portions are internalised via early and late endosomes for degradation in the vacuole/lysosome compartment or recycling to the plasma membrane (Huotari & Helenius, 2011). Importantly, the intracellular trafficking is tightly intertwined with the transport of mRNAs. For instance, mRNAs are targeted to the ER and their membrane‐coupled translation results in the entry of proteins into the ER to follow the secretory pathway (Akopian et al, 2013; Kramer et al, 2019). mRNA delivery and local translation on the surface of mitochondria can occur by distinct pathways ensuring import of a specific subset of mitochondrial proteins (Hansen & Herrmann, 2019; Bykov et al, 2020). However, the currently known examples appear to be only the tip of the iceberg and new cases are being uncovered steadily.
In this review, we describe novel mechanistic insights and implications for the co‐transport of mRNAs with endosomes and discuss how local translation at the cytoplasmic surface of early and late endosomes is linked to mitochondrial protein import. Originally, endosomal mRNA trafficking was found as an alternative hitchhiking pathway for long‐distance transport in infectious hyphae of the plant pathogen Ustilago maydis (Haag et al, 2015; Box 1). However, endosome‐coupled RNA transport seems to be more widespread than previously anticipated, as the process is important for plant endosperm development and neuronal functions (Tian et al, 2020a; Fernandopulle et al, 2021). Malfunction of membrane‐coupled mRNA transport has been implicated in neuronal diseases such as Charcot‐Marie‐Tooth type 2B neuropathy (Cioni et al, 2019; Liao et al, 2019). These biological processes exemplify how membrane, protein and mRNA trafficking communicate intensively. Thus, these seemingly separate fields of research studying endocytosis, post‐transcriptional regulation and mitochondrial biology need to converge in the future. For more information on specific topics in the individual fields, we refer to excellent recent reviews (Holt & Bullock, 2009; Das et al, 2019; Hansen & Herrmann, 2019; Pfanner et al, 2019; Bykov et al, 2020; Das et al, 2021; Fernandopulle et al, 2021).
Box 1: The eukaryotic model organism Ustilago maydis .
Ustilago maydis is the causative agent of corn smut disease and serves as a fungal model system for biotrophic phytopathogens, DNA repair, and cell biology (Holliday, 2004; Vollmeister et al, 2012; Lanver et al, 2017). The fact that highly polarised growth of infectious hyphae is strongly dependent on endosomal transport along microtubules makes it an ideal system to study membrane‐coupled mRNA transport and translation (Haag et al, 2015). The dimeric key transcription factor regulating formation of infectious hyphae is called bEast (bE) and bWest (bW; in analogy with the reunion of Berlin). This central regulator is only active as heterodimer with the respective subunits derived from different mating partners (Vollmeister et al, 2012). Thereby, only after cell fusion an active bE/bW heterodimer can be formed. The regulation by heterodimerisation elegantly links transcription factor activation with mating. The resulting transcriptional activator is necessary and sufficient to arrest the cell cycle and elicit the hyphal growth programme with a defined axis of polarity: i.e. cells elongate at the apical growth pole and insert septa at the basal pole (Fig 3). This knowledge was used to design laboratory strains expressing an active bE/bW heterodimer under control of promoters regulatable by the nitrogen or carbon source in the medium (Brachmann et al, 2001). Thus, hyphal growth can be efficiently and highly reproducibly elicited in culture, independent of a mating partner. The resulting monokaryotic hyphae resemble their dikaryotic counterpart in all aspects of polarised hyphal growth (Brachmann et al, 2001; Baumann et al, 2012; Pohlmann et al, 2015).
Membrane‐free versus membrane‐coupled mRNA transport
As noted above, the transport of mRNAs is a pronounced subcellular mechanism to orchestrate spatiotemporal protein expression. Currently, the main mechanisms described for mRNA transport function independent of membranes. Key RNA‐binding proteins (RBPs) recognize specific RNA localisation elements within cargo mRNAs. Such transporter RBPs join with accessory RBPs to form large ribonucleoprotein complexes called mRNPs, which are the actual transport units (Martin & Ephrussi, 2009; Das et al, 2019; Abouward & Schiavo, 2020). The transport mRNPs are linked to molecular motors by tethering proteins for their active transport along the actin or microtubule cytoskeleton (Tekotte & Davis, 2002; Mofatteh & Bullock, 2017; Das et al, 2019).
Elegant in vitro studies deciphering minimal transport systems in fungi, animal and mammalian systems revealed important fundamental principles. In Saccharomyces cerevisiae, actin‐dependent mRNA transport is important to regulate mating type‐switching during cell division (Niessing et al, 2018). The minimal system for processive mRNA transport consists of the RNA‐binding proteins She2p and She3p that cooperate to bind RNA localisation elements within the ASH1 cargo mRNA. Binding of a She2p tetramer induces a pronounced conformational change in the mRNA and the presence of a She3p dimer is essential for specific RNA recognition (Edelmann et al, 2017). The dimeric She3p configuration allows recruitment of two Myo4p myosin motors forming a dimeric myosin complex that is essential for processive movement along actin filaments towards the daughter cell (Heym et al, 2013; Sladewski et al, 2013).
In Drosophila melanogaster, microtubule‐dependent mRNA transport is crucial during embryogenesis to express determinants for patterning of embryonic segments locally (Bullock & Ish‐Horowicz, 2001; Wilkie & Davis, 2001). The RNA‐binding protein Egalitarian (Egl), for example, binds defined RNA localisation elements in pair‐rule transcripts. The binding of two Egl per cargo mRNA promotes the engagement of BicD and releases the autoinhibition of this adaptor protein. Activated BicD recruits the dynein motor and its accessory complex dynactin more efficiently resulting in enhanced processive movement along microtubules with velocities observed in vivo (McClintock et al, 2018; Sladewski et al, 2018).
In neurons, mRNA transport is vital for a variety of functions ranging from directed axonal outgrowth to synaptic plasticity (Das et al, 2019; Holt et al, 2019; Dalla Costa et al, 2021; Fernandopulle et al, 2021). The nucleus in the soma of these highly polarised cells is separated substantially from distal dendrites and axons. This holds particularly true for axons that can extend up to a metre in length. Thus, long‐distance mRNA transport needs to provide all parts of the neuronal cells with transcripts, and localised translation is a crucial determinant for the local proteome (Zappulo et al, 2017; Cagnetta et al, 2018; Das et al, 2019; Poulopoulos et al, 2019; Wang et al, 2019). Among the best‐studied examples is the transport of β‐actin mRNA taking place in both dendrites and axons (Zhang et al, 2001; Tiruchinapalli et al, 2003). Newly translated actin is most likely important for morphological changes of expanding synapses and dendritic spines (Yoon et al, 2016). The first in vitro transport‐competent minimal system from humans revealed that the neuronal RNA‐binding protein APC (adenomatous polyposis coli; Preitner et al, 2014) binds specific guanine‐rich RNA sequences. The resulting RNA/protein complex is tethered to kinesin‐2 via the adaptor protein KAP3 for microtubule‐dependent movement of, e.g., β‐actin mRNAs towards the plus ends of microtubules. Both, APC and cargo mRNAs, promote transport efficiency (Baumann et al, 2020). Thus, various in vitro studies with minimal systems recapitulate the natural transport processes independent of membranes with physiological run length and velocities comparable to those observed in living cells (Edelmann et al, 2017; McClintock et al, 2018; Sladewski et al, 2018; Baumann et al, 2020).
However, in vivo the process of mRNA transport appears to be more complex, since (i) higher‐order mRNP transport granules are formed and (ii) alternative membrane‐dependent transport pathways exist (Béthune et al, 2019; preprint: Cohen et al, 2021; preprint: Harbauer et al, 2021, see below). The formation of higher‐order RNP granules in the form of biomolecular condensates is known for neuronal mRNA transport (Brangwynne et al, 2009; Kato & McKnight, 2018). Such RNP transport granules are comparable to other cellular biomolecular condensates like the nucleolus or RNP stress granules. All are formed by liquid–liquid phase separation involving RNA/RNA interactions (Tauber et al, 2020). Assembly is supported by RBPs acting as RNA chaperones. They often contain intrinsically disordered regions (IDRs) that promote formation of additional protein/RNA and protein/protein interactions (Protter & Parker, 2016). These larger mRNP granules might constitute key units of transport in vivo (Fernandopulle et al, 2021) and could explain how numerous mRNAs and RBPs are co‐transported in the cell with a limited set of motor and adaptor proteins (Pushpalatha & Besse, 2019). Interestingly, these membrane‐less RNP granules can be linked to membrane‐enclosed organelles (Liao et al, 2019; Pushpalatha & Besse, 2019 see below), providing even higher‐order transport structures consisting of both membrane‐less and membrane‐enclosed subcellular units.
Hitchhiking on membrane‐enclosed organelles is an alternative transport mode and has been described for various organelle membranes like ER, mitochondria as well as endosomes (Béthune et al, 2019; preprint: Cohen et al, 2021; preprint: Harbauer et al, 2021; see below). These alternative modes of mRNA transport will be extensively discussed in the remaining part of the review with a special focus on endosomal transport. In summary, in a living cell, there are various transport mechanisms for mRNA delivery that are dependent and independent of membranes. Outstanding questions are what the individual contributions of each transport pathway to cellular RNA trafficking are, and what the function of the close association with membranes is. To address these critical questions, we need to uncover the specific routes cargo mRNAs take in vivo and to disclose the underlying mechanisms in the most detailed fashion possible.
Mechanistic insight into endosomal mRNA transport and membrane‐coupled translation in U. maydis
The birth of an important new concept in mRNA transport has been the discovery of co‐transport of mRNAs with membranes. Among the best‐studied systems is endosomal mRNA transport in fungi (Haag et al, 2015; Béthune et al, 2019; Fig 1) that was disclosed during polar growth of infectious hyphae from U. maydis (Vollmeister et al, 2012, Box 1). In this fungus, the switch from yeast to hypha is an essential prerequisite for infection. The resulting highly polarised cells depend heavily on active motor‐mediated transport. Loss of microtubule‐dependent transport causes defects in the unipolar hyphal growth programme. A characteristic feature of aberrant growth is the formation of a second growth pole, resulting in bipolar hyphae (Vollmeister & Feldbrügge, 2010; Haag et al, 2015).
Figure 1. Model depicting endosomal mRNA transport in Ustilago maydis .
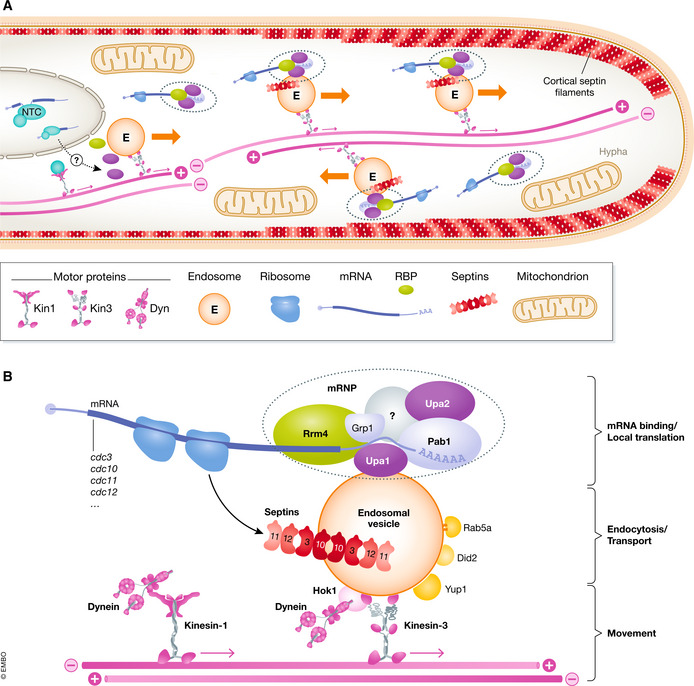
(A) Overview of an infectious hypha. Endosomes (E) shuttle along antiparallel microtubule bundles (pink). Plus‐end directed transport is mediated by Kin3, whereas dynein transports endosomes to the minus ends. Cargo mRNPs (dashed ovals) are assembled most likely close to the nucleus involving components of the NineTeen complex (NTC, light blue) deposited during splicing. Local translation (ribosomes in dark blue) of cargo mRNAs like septins results in septin complex formation (red octamer). These complexes are transported towards the growth pole to generate higher‐order septin filaments with gradients emanating from the growing tip (red line). Endosomal mRNP transport appears to be also important for import of proteins into mitochondria (see Fig 3). (B) Close‐up of endosomes shown above. Components important for movement, endocytosis, transport and localised translation are shown (further details are given in the text).
Major carriers for long‐distance transport are endosomes that shuttle bidirectionally along microtubules throughout the hyphae, between the basal septum and the hyphal tip, passing the nucleus in the centre (Fig 1A). Microtubules are bundled in antiparallel arrays stretched throughout the whole hyphae, with unipolar regions at the plus ends of hyphal poles (Steinberg et al, 2001, Fig 1A and B). Processive movement of endosomes is mainly achieved by the plus‐end directed Kinesin‐3 type motor protein Kin3 (Schuster et al, 2011b). Minus‐end‐directed transport is mediated by cytoplasmic split dynein Dyn1/2 (Straube et al, 2001). The adaptor protein Hok1 coordinates motor attachment to endosomes (Bielska et al, 2014b), and the conventional kinesin Kin2 is needed to transport dynein back to the plus ends of microtubules for motor recycling (Lenz et al, 2006; Schuster et al, 2011a; Fig 1B). As expected, loss of any of these motors results in aberrant bipolar growth due to stalling of endosomal movement (Steinberg, 2012).
The shuttling endosomes display characteristics of early endosomes, because they are positive for the small GTPase Rab5a and the SNARE Yup1 (soluble N‐ethylmaleimide‐sensitive factor attachment protein receptors; Wedlich‐Söldner et al, 2000). Early endosomes are classically known to function during endocytosis (Steinberg, 2014; Haag et al, 2017). However, according to a more modern view, early endosomes function as multipurpose platforms also involved in signalling and transport of protein complexes, such as peroxisomes and mRNPs (Bielska et al, 2014a; Guimaraes et al, 2015; Salogiannis & Reck‐Peterson, 2016; Béthune et al, 2019). These functions need to be precisely regulated. The ESCRT regulator Did2 coordinates accurate maturation of endosomes in U. maydis (Haag et al, 2017). Loss of Did2 renders Rab5a‐positive early endosomes Rab7‐positive, so that they adopt the membrane identity of late endosomes. This changes the motor composition and results in a switch from motile early endosomes to static late endosomes. Thus, without Did2, endocytosis and mRNP transport are disturbed, indicating that the ESCRT regulator orchestrates these two vital functions of the endosomal multipurpose platform (Haag et al, 2017; Fig 1B).
The core components of endosomal mRNA transport in U. maydis
The link between endosome and mRNA transport was discovered by studying the key RNA‐binding protein Rrm4. This post‐transcriptional regulator shuttles intensively along microtubules and binds thousands of mRNAs (see below; Baumann et al, 2012; Baumann et al, 2014; Olgeiser et al, 2019). Importantly, Rrm4 hitchhikes on the cytoplasmic surface of Rab5a‐positive endosomes and loss of Rrm4 function results in the formation of aberrant bipolar hyphae. Comparable defects in hyphal growth are also observed during malfunction of microtubules or endosomal movement suggesting that the transport of mRNPs constitutes an essential function of endosomes during regulation of highly polarised growth (Becht et al, 2006; König et al, 2009). Endosomal mRNA transport is mediated by a defined core machinery consisting of factors like Rrm4, whose loss of function results in disturbed mRNA transport causing distinct defects in hyphal growth. The key factor Rrm4 contains three N‐terminal RRMs (RNA recognition motifs) with a spacing typical for embryonic lethal abnormal vision (ELAV)‐type RBPs. These RRMs are functionally important for RNA binding (Becht et al, 2006; Olgeiser et al, 2019), and RRM3 recognises specifically the core sequence UAUG in cargo mRNAs (Olgeiser et al, 2019). At its C‐terminus, Rrm4 contains two MademoiseLLE domains (MLLE, also known as PABC, poly(A)‐binding protein C‐terminal domain, Becht et al, 2006; Müller et al, 2019), which serve as a protein‐protein interaction pocket for specific interaction with two PAM2‐like motifs (PAB1C‐associated motif, Xie et al, 2014) present in the adaptor protein Upa1. The second core component Upa1 tethers mRNPs containing Rrm4 to the cytoplasmic surface of endosomes (Pohlmann et al, 2015, Fig 1). Attachment is mediated via its FYVE (Fab 1, YOTB, Vac 1, and EEA1) zing finger domain for specific interaction with PI(3)P lipids (phosphatidylinositol 3‐phosphate, Stenmark et al, 2002). Loss of Upa1 causes defects in Rrm4 shuttling, which in turn results in disturbed mRNA transport and aberrant bipolar hyphal growth (Pohlmann et al, 2015). Besides the two PAM2‐like motifs, Upa1 contains a single PAM2 motif for the interaction with the poly(A)‐binding protein Pab1, an RBP with four RRMs that also carries an MLLE domain at its C‐terminus. Notably, the MLLE domains of Rrm4 and Pab1 have different specificities for the PAM2 or the PAM2‐like motifs in Upa1 (Pohlmann et al, 2015).
An additional core component of endosomal mRNP transport is the scaffold protein Upa2 containing four PAM2 motifs for interaction with Pab1 and a conserved C‐terminal GWW sequence for interaction with a currently unknown component on the endosomal surface. Loss of Upa2 also interferes with mRNP transport resulting in the formation of bipolar hyphae (Jankowski et al, 2019). Similarly, the small glycine‐rich protein Grp1 shuttles intensively on transport endosomes. However, Grp1 is not an essential core component of endosomal mRNPs and rather serves as RNA chaperone accompanying cargo mRNAs (Olgeiser et al, 2019). Loss of Grp1 results in the formation of unipolar hyphae with defects in response to cell wall stress (Olgeiser et al, 2019). Both Upa2 and Grp1 contain intrinsically disordered regions, opening up the possibility that biomolecular condensation is important for the formation of endosomal transport mRNPs in U. maydis. This assumption is supported by recent findings demonstrating that the fungal RNA‐binding protein Whi3 from Ashbya gossypii containing glutamine‐rich disordered regions forms biomolecular condensates associated with ER membranes. Importantly, membrane association of Whi3 controls the size of RNP condensates suggesting that interaction with lipid bilayers is crucial for regulating the precise formation and composition of these condensates in vivo (preprint: Snead et al, 2021). Similarly, Upa2 and Grp1 containing intrinsically disordered regions might influence formation of such RNP condensates at endosomal surfaces. In summary, the core endosomal mRNP consists of the key factors Rrm4, Upa1 and Upa2 and accessory RBPs such as Grp1 and Pab1 (Figs 1B and 2A).
Figure 2. Schematic comparison of endosomal RNA transport in fungi, plants and animals.
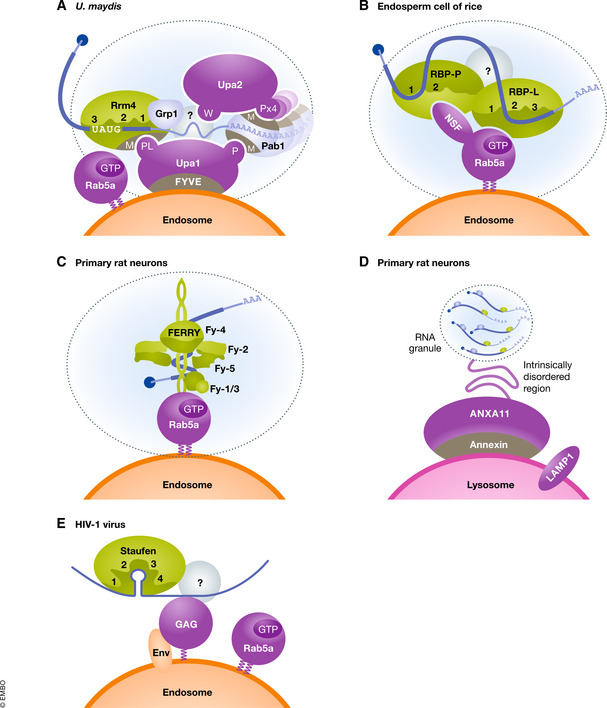
(A–E) On the cytoplasmic surface of transport endosomes or lysosomes, mRNPs are attached by different factors (purple) to endosomes. Key RNA‐binding proteins (green) interact with cargo RNA (blue). mRNAs are symbolised by CAP (blue circle) and a poly(A) tail (B and D are adapted from Tian et al, 2020b and Liao et al, 2019, respectively. Panel (C) is adapted from Schuhmacher et al, 2021 and Quentin et al, 2021 (both preprints). Panels (D, E) are proposed based on current literature (further details are given in the text).
Potential loading mechanism of mRNPs for endosomal transport in U. maydis
Currently, it is unclear how mRNPs are loaded on endosomes in U. maydis. Interestingly, a potential link between splicing and mRNP transport was discovered by studying Num1 (Kellner et al, 2014; Zhou et al, 2018). This regulatory protein is a homologue of splice factor SPF27 in humans that functions as core component of the evolutionarily conserved Prp19/CDC5 complex during intron removal (also called NTC, NineTeen complex; Grote et al, 2010, Fig 1A). Besides its nuclear function, Num1 also localises in the cytoplasm in the vicinity of microtubules and interacts with conventional kinesin Kin1 (Kellner et al, 2014; Zhou et al, 2018; Fig 1A). Loss of Num1 causes defects in processive movement of endosomes and associated mRNPs (Kellner et al, 2014). Although its precise function in endosomal mRNP transport is not solved, an attractive hypothesis is that Num1 is loaded onto mRNPs during splicing in the nucleus. Upon export to the cytoplasm, Num1 interacts with the kinesin motor close to microtubules and participates in loading of mRNPs onto endosomes (Fig 1A).
The link to the nuclear history of mRNPs and their cytoplasmic remodelling is well‐known from transport processes in other organisms (Mofatteh & Bullock, 2017). For example, in S. cerevisiae, mRNP transport is initiated in the nucleus when the nuclear RBP Loc1p interacts with localisation elements of ASH1 mRNA (Long et al, 2001; Niedner et al, 2013). During entry into the cytoplasm, the mRNP is remodelled and the SHE machinery containing She2p/She3p takes over the interaction with the RNA localisation elements during transport (Long et al, 2001; Niedner et al, 2013). In D. melanogaster, the exon junction complex (EJC) is deposited on mRNAs during splicing (Obrdlik et al, 2019). EJC deposition is particularly important for the transport of oskar mRNPs, since splicing creates the oskar mRNA localisation element by exon/exon junction and deposition of the EJC is needed for correct kinesin‐mediated transport (Ghosh et al, 2012; Simon et al, 2015). Hence, these examples illustrate how post‐transcriptional processes like splicing and mRNA transport are tightly intertwined to orchestrate spatiotemporal expression.
Endosomal transport of translationally active mRNAs in U. maydis
Studying cargo mRNAs of Rrm4 by combining in vivo UV crosslinking (iCLIP) and fluorescent protein (FP)‐based RNA live imaging in U. maydis (Box 2) revealed that key cargo mRNAs encode the four septins Cdc3, Cdc10, Cdc11 and Cdc12 (König et al, 2009; Zander et al, 2016; Olgeiser et al, 2019). Besides these septin mRNAs, their translation products are also shuttling on transport endosomes, (Baumann et al, 2012; Baumann et al, 2014; Zander et al, 2016; Fig 1). Interestingly, endosomal septin transport is needed for assembly of the four septin subunits into hetero‐octameric complexes on the surface of endosomes. The resulting complexes are transported towards the hyphal tip and form higher‐order septin filaments with a gradient emanating from the growth pole (Baumann et al, 2014; Zander et al, 2016; Fig 1). In the absence of endosomal septin mRNA transport, hetero‐octameric septin complexes no longer shuttle on endosomes and the formation of septin filaments as well as the tip gradient is disturbed. Instead, aberrant accumulations of small septin rings are visible in the cell (Zander et al, 2016).
Box 2: Main techniques used to study RNA transport and local translation.
Monitoring RNA localisation and transport
Apex‐Seq: Proximity labelling of mRNAs combined with RNA sequencing or mass spectrometry analysis to examine their localisation and interaction with proteins (Fazal et al, 2019; Padron et al, 2019). In this technique, the ascorbate peroxidase APEX2 is targeted to subcellular compartments or fused to known RBPs for specific and direct biotin labelling of nearby mRNAs using biotin‐phenol.
FISH: RNA fluorescence in situ hybridisation (RNA‐FISH) or single‐molecule FISH (smFISH) enable the specific and quantitative detection of endogenous mRNAs by using fluorescently labelled antisense oligonucleotides. Most of the knowledge about the subcellular localisation of mRNAs has been obtained using FISH methodology. However, these methods only provide a static view of mRNA localisation because cells need to be fixed prior to analysis (Bassell et al, 1994; Maekiniemi et al, 2020).
Fluorescent protein (FP)‐based RNA live imaging: Enables in vivo labelling of specific single mRNAs and provides dynamic spatiotemporal information. mRNAs are visualised with bacteriophage‐derived RBPs fused to fluorescent proteins that specifically bind to unique RNA hairpins integrated into the 3′ UTR (Buxbaum et al, 2015). Three systems are mainly used that exploit the RNA‐binding characteristics of these RBPs to distinctive RNA hairpins: the MS2 (Bertrand et al, 1998), PP7 (Larson et al, 2011) and λN system (Daigle & Ellenberg, 2007). For the application of this method, it must be ensured that the integrated hairpins and the tagging do not affect the stability or translation of the mRNAs.
Molecular Beacon technology for live imaging: Molecular beacons (MBs) offer another method to analyse the localisation of specific endogenous mRNAs in vivo without any modification of the mRNA itself. MBs are single‐stranded antisense oligonucleotide probes, which can form a stem‐loop structure with a fluorophore and a quencher (Tyagi & Kramer, 1996). Upon hybridisation to the mRNA of interest, a conformational change releases the quencher and fluorescent signal appears (Bratu et al, 2003; Donlin‐Asp et al, 2021). Although MBs are not yet widely exploited, they give great insight into the localisation of mRNAs especially for low abundant RNA molecules (Bratu, 2003).
Cy3‐UTP labelling for live imaging: Fluorescently labelled uridine‐5′‐triphosphate (UTP) analogs, e.g. Cy3‐UTP or Cy5‐UTP, can be injected into cells to label endogenous nascent RNAs in vivo. The UTP analogs are incorporated into RNA (including mRNA and rRNA) during its synthesis and can subsequently be monitored by fluorescence microscopy (Piper et al, 2015; Cioni et al, 2019). This method also allows the imaging of RNAs without major modification and reduces, e.g. the possibility of influencing the stability of the RNA. However, the feasibility to inject UTP analogs is a prerequisite for the application of this method.
Analysing local translation
Proximity‐based ribosome profiling: Enables ribosome profiles for specific organelles such as ER or mitochondria. For this purpose, the deep‐sequencing‐based technique is combined, for example, with the expression of a biotin ligase (BirA), which can be fused to a localisation element specific for organelles such as ER or mitochondria. As a result, ribosomes located in close proximity are labelled and can be specifically purified. mRNAs that have been protected from nuclease digestion by these ribosomes can subsequently be identified using RNA sequencing. This method allows to quantify the level of new protein synthesis in close proximity to, e.g., organelles and gives information on the ribosome position on bound mRNAs (Ingolia et al, 2009).
SunTag approach: Enables the visualisation of translation of mRNA molecules of interest in vivo. FP‐based RNA live imaging is combined with fluorescent tagging of the nascent peptide chain. To this end, a repeating peptide array is fused to the ORF of interest recruiting a fluorescently labelled single‐chain variable antibody fragment (Tanenbaum et al, 2014). Thereby, imaging of single mRNA molecules simultaneously with their nascent peptides is possible and the technique is successfully used in different cell types (Wang et al, 2016; Cioni et al, 2019).
Image‐based transcriptomic analysis: Combination of smFISH, puromycin treatment and indirect immunofluorescence to study the localisation of translationally active mRNAs in proximity of membranous organelles. Puromycin dislocates mRNAs from translating ribosomes by initiating premature termination. Therefore, translation‐dependent mRNA localisation can be studied (preprint: Popovic et al, 2020).
The fact that septin mRNAs, translation products and ribosomes associate with endosomes in an Rrm4‐dependent manner strongly suggests that endosome‐coupled translation mediates septin complex formation and their long‐distance transport (Baumann et al, 2014; Zander et al, 2016; Fig 1B). Aberrant cytoplasmic translation of septin subunits with promiscuous membrane‐binding properties causes mistargeting and problems in complex formation (Zander et al, 2016). In essence, one critical function of endosomal mRNP transport is the loading of newly synthesised proteins onto the cytoplasmic surface of endosomes. Hence, membrane‐coupled co‐translation and co‐movement of complex subunits promote formation and transport of heteromeric septin complexes.
Spatiotemporal regulation of complex assembly by co‐translational mechanisms is an ancient process already operational in prokaryotes. The bacterial organisation of polycistronic mRNAs promotes the local translation of subunits and likely favours the co‐translational assembly of these subunits (Shieh et al, 2015; Kramer et al, 2019). Eukaryotes follow a different strategy to reach the same goal. Monocistronic mRNAs of eukaryotes are spatially and temporally organised in RNA operons and regulons by the action of RBPs binding preferentially in the 3′UTR to orchestrate co‐localisation of mRNAs and cognate translation products like subunits of protein complexes (Keene, 2007; Blackinton & Keene, 2014). Thereby, co‐translational assembly of complexes is promoted. Consistently, in S. cerevisiae, ribosome profiling revealed that nine out of 12 multi‐protein complexes, like fatty acid synthase (FAS) and translation initiation factor eIF2, are co‐translationally assembled (Shiber et al, 2018; Schwarz & Beck, 2019). In the case of the proteasomal subunits Rpt1 and Rpt2, mRNAs and translation products co‐localise in defined assembly particles for co‐translational complex formation (Panasenko et al, 2019). Thus, co‐translational assembly of protein complexes promoted by mRNA co‐localisation appears to be a widespread mechanism.
Distributive function of endosomal mRNA transport in U. maydis
Generating a transcriptome‐wide view revealed that Rrm4 binds thousands of mRNAs mainly in the 3′UTR (Baumann et al, 2014; Olgeiser et al, 2019). All four septin mRNAs, for example, are bound in the 3′ UTR, which is consistent with co‐translational assembly and transport. The fact that thousands of mRNAs can be bound by Rrm4 suggests intensive bulk endosomal transport of mRNAs. Consistently, the polyA‐binding protein Pab1 also shows intensive endosomal co‐shuttling (König et al, 2009). Bulk mRNA transport reflects an important distributive function of endosomal mRNP transport, avoiding gradients of mRNAs around the nucleus and supplying all areas of the hyphae with sufficient transcripts. The function in mRNA distribution also explains why most cargo mRNAs shuttle bidirectionally along microtubules, a very common feature of long‐distance transport also observed during oocyte development and neuronal transport.
In oocytes from D. melanogaster, mRNAs are moving back and forth along microtubules with a small bias towards the posterior cortex (Zimyanin et al, 2008). Furthermore, at later stages of oocyte development, gradient formation is avoided by intensive kinesin‐dependent ooplasmic streaming actively mixing the complete ooplasm (Lu et al, 2018). In neurons, bulk mRNA transport is also very prominent. Most mRNPs move bidirectionally in dendrites and axons (Cajigas et al, 2012; Shigeoka et al, 2016; Zappulo et al, 2017; Tushev et al, 2018; Das et al, 2019). This transport phenomenon was explained in dendrites by the sushi belt model: mRNPs are transported bidirectionally at all times to supply synapses with mRNAs for local translation. Hence “hungry” synapses are served on demand by deposition of specific mRNPs (Doyle & Kiebler, 2011).
It was also demonstrated in U. maydis for the first time that translationally active polyribosomes are co‐transported intensively on endosomes (Baumann et al, 2014; Higuchi et al, 2014). Thereby, not only mRNAs but also ribosomes are transported over long distances to distal areas in highly polarised cells. This supports the hypothesis of a distributive function of endosomal transport to enable efficient translation throughout the entire cell (Baumann et al, 2014; Higuchi et al, 2014). Noteworthy, distal ribosomes can also be remodelled locally after deposition in neurons, as described recently for distinct ribosomal proteins, which were locally translated at growth poles of axons (Shigeoka et al, 2016).
In summary, main functions of endosomal mRNA transport are the distribution of mRNAs throughout cells, the transport of translation products and associated ribosomes, as well as the loading of newly synthesised proteins onto the surface of transport endosomes. Thereby, the entire cell is supplied with vital components for optimal spatiotemporal protein expression at all times.
Conservation and biological functions of endosomal mRNA transport
How widespread is endosomal transport and translation of mRNAs? Besides U. maydis, endosome‐mediated mRNA trafficking is also operational in other fungi (Müller et al, 2019; Stein et al, 2020), plants (Tian et al, 2020b) and humans (Cioni et al, 2019; Liao et al, 2019). The phylogenetic analysis of the core endosomal mRNP transport machinery from U. maydis revealed that homologues of the components are present in other Basidiomycota. In addition, the orthologues are also found in distantly related Chytridiomycota and Mucoromycota (Müller et al, 2019). Interestingly, Rrm4 and Upa1 orthologues of Rhizophagus irregularis—a distantly related symbiont forming arbuscular mycorrhiza within plant cells (Mucoromycota)—shuttle on transport endosomes when expressed in hyphae of U. maydis. This suggests that core endosomal mRNP transport is widespread and functionally conserved in fungi (Müller et al, 2019).
By contrast, no orthologues of the core endosomal transport machinery are found in Ascomycota, while RNA transport is well‐known in S. cerevisiae and C. albicans (Elson et al, 2009; Niessing et al, 2018). Thus, endosomal transport might be carried out by different factors in Ascomycetes. One such transport component could be the RNA‐binding protein Gul1/Ssd1 that shuttles on endosomes in Sordaria macrospora and its close relative Neurospora crassa (Herold et al, 2019; Stein et al, 2020). Since the poly(A)‐binding protein Pab1 also shuttles on endosomes, mRNP transport via endosomes is highly likely in these fungi (Stein et al, 2020). Remarkably, we also identified a Gul1 orthologue as a potential interaction partner of Rrm4 that shuttles on mRNP transport endosomes in U. maydis (M Tulinski, K Müntjes, M Feldbrügge; unpublished observation). This points towards a conserved, ancient and widespread endosomal transport machinery in fungi. Noteworthy, Gul1 was identified as a target of the STRIPAK “striatin‐interacting phosphatase and kinase” complex, an evolutionarily conserved signalling complex implicated in several human diseases, like different types of cancer and cardiovascular disorders (Kück et al, 2019; Stein et al, 2020). In humans, the STRIPAK complex has been hypothesised to modulate dynein activity in axons and might thereby influence vesicle and autophagosome transport (Neisch et al, 2017). These findings provide new hints on how post‐translational regulation might influence endosomal mRNP transport.
In endosperm cells of developing rice seeds, mRNAs encoding the storage proteins glutelin and prolamine are segregated to distinct, specific subdomains of the cortical ER, thereby enabling the localisation of the encoded storage proteins in separate endomembrane compartments in the mature seed (Tian et al, 2020a). Specific localisation elements present in the glutelin and prolamine mRNAs are recognised by the RNA‐binding proteins RBP‐P and RBP‐L, which might mediate mRNP transport (Hamada et al, 2003; Washida et al, 2009; Washida et al, 2012). These proteins contain two and three RRMs, respectively, and carry a glycine‐rich intrinsically disordered region at their C‐terminus (Tian et al, 2018). Both RBPs interact with each other and in addition, RBP‐P binds to N‐ethylmaleimide‐sensitive factor (NSF), an ATPase involved in the regulation of membrane fusion. RBP‐L interacts with the GTP‐bound form of the small GTPase Rab5a, a marker for early endosomes resulting in a quaternary complex containing mRNA (Fig 2B). The mRNP complex is formed at the cytoplasmic surface of early endosomes containing the glutelin cargo mRNA. Loss of function mutations in rab5a results in mistargeting of the mRNP complex verifying that endosomal transport of translationally inactive mRNAs is crucial for the efficient targeting of the mRNA to the cortical ER (Tian et al, 2020b). This uncovers an alternative mode of mRNP attachment to endosomes via direct Rab5a interaction (Fig 2B). In essence, endosomal RNA transport in plants also include RRM proteins and is needed for organelle communication between endosomes and ER.
Endosomal mRNA transport and endosome‐coupled translation are also quite common in animal cells. Initially, this process was found in axons of retinal ganglion cells from Xenopus laevis, but it is also operational in rat and zebrafish neurons, as well as in various types of cultured mammalian cells (Cioni et al, 2019; Liao et al, 2019; preprint: Popovic et al, 2020 preprint: Schuhmacher et al, 2021). Studies of mRNP transport in axons of retinal ganglion cells showed that Cy3‐UTP injections resulted in labelling of endogenous mRNAs (Box 2) that were co‐transported with Rab5a‐positive, early and Rab7a‐positive, late endosomes (Cioni et al, 2019). Movement was mainly observed on motile late endosomes. Interfering with Rab5a and Rab7a function did not strongly influence the bulk mRNP transport (Cioni et al, 2019), suggesting the presence of alternative transport pathways. Importantly, the authors found evidence for membrane‐coupled translation at the surface of late endosomes. Prominent examples are mRNAs encoding mitochondrial proteins, suggesting a function as endosomal platform to orchestrate mitochondrial protein import (Cioni et al, 2019). Consistently, studying human Rab5 effector proteins revealed the presence of the novel FERRY protein complex potentially linking endosomal mRNA transport, the translation machinery and mitochondrial protein import in neurons (preprint: Schuhmacher et al, 2021). This stable five‐membered complex, consisting of Fy1 to Fy5, interacts in vitro specifically with the activated GTP‐bound form of Rab5a. Structural studies revealed a clamp‐like elongated structure with arm‐like appendages protruding from a central Fy4 dimer. One of the extensions composed of the Fy2 coiled‐coil region interacts with Rab5 most likely on the endosomal surface (preprint: Quentin et al, 2021; Fig 2C).
Another example for membrane‐coupled mRNA transport was uncovered in neurons from rats and zebrafish, studying LAMP1‐positive endosomes of the lysosomal and endolysosomal system, referred to as lysosomes in short. Highly comparable to mRNA transport on the cytoplasmic surface of endosomes, RNP granules hitchhike on LAMP1‐positive lysosomal vesicles (Fig 2D). Transport mRNPs containing cargo like β‐actin mRNA are linked to lysosomes by annexin ANXA11 (Liao et al, 2019). This membrane‐associated factor contains an N‐terminal intrinsically disordered region for interaction with RNP granules and a C‐terminal annexin domain for calcium‐dependent lipid binding. Remarkably, a new concept was disclosed by demonstrating that the IDR of ANAX11 enables co‐transport of mRNPs in form of biomolecular condensates. Hence, ANAX11 serves as a novel tethering factor linking membrane‐less RNP granules with membrane‐bound organelles (Liao et al, 2019; Fig 2D). Mutations associated with amyotrophic lateral sclerosis (ALS) in ANXA11 interfere with mRNP transport suggesting a link to neuronal disease (Liao et al, 2019).
Importantly, the link between mRNAs and endosomes is not restricted to neurons, since image‐based transcriptomic analysis (Box 2) of candidate mRNAs demonstrated endosomal association also in cultured mammalian cells (preprint: Popovic et al, 2020). Studying a selection of mRNAs encoding various endosomal and endomembrane regulators revealed the presence of twelve mRNAs specifically associated with endosomes. Biochemical purification of endosomes combined with RNAseq experiments verified that all twelve mRNAs were present in the endosomal fraction (preprint: Popovic et al, 2020). Endosomal localisation of mRNAs encoding FYVE proteins like EEA1 was dependent on translation, and the cysteine and glycine‐rich protein 1 (CSRP1) appears to be involved in translational regulation (preprint: Popovic et al, 2020). Hence, local translation might function in efficient subcellular endosomal targeting of EEA1, a process that is comparable with local translation of the membrane‐associated septins in U. maydis (see above).
Investigating membrane association of mRNPs in animal and human systems revealed the presence of mRNAs on the cytoplasmic surface of Rab5‐positive early endosomes, Rab7‐positive late endosomes and LAMP1‐positive lysosomal vesicles (Cioni et al, 2019; Liao et al, 2019; preprint: Popovic et al, 2020; preprint: Schuhmacher et al, 2021; see below). For LAMP1‐positive lysosomal vesicles, the association of cargo mRNAs via the adaptor protein ANAX11 is important for membrane‐coupled long‐distance transport in highly polarised axonal cells (Liao et al, 2019). These examples indicate that mRNAs are associated with numerous types of membranes of the endocytic transport pathway. However, the precise biological function appears to be specific in the different systems. In case of early and late endosomes, local translation might be more important to load newly synthesised proteins on membranes of the endocytic system, whereas LAMP1‐positive vesicles are used for long‐distance mRNA trafficking. Furthermore, since it has been observed that some neuronal LAMP1‐positive lysosomes lack major lysosomal hydrolases, the presence of different subpopulations of lysosomes with variable functions is conceivable (Cheng et al, 2018). This is clearly an emerging research field and additional examples and more mechanistic insights are needed to draw final conclusions.
Previously, it was found that viral RNAs hijack the endosomal machinery for maturation and transport towards the plasma membrane (Basyuk et al, 2020). This indicates that the endosomal transport is not restricted to mRNAs. The genomic RNA of Murine Leukaemia Virus (MLV) is co‐transported with early endosomes, where the membrane‐associated viral Gag protein recognises the genomic vRNA and functions as a tethering factor (Basyuk et al, 2003; Fig 2E). Similarly, the HIV‐1 genomic vRNA travels intracellularly on a dynein‐dependent LAMP1‐positive endosomes. Membrane association is mediated by myristoylated Gag protein (Lehmann et al, 2009) and cellular factors like the double‐strand RNA‐binding protein Staufen 1 might form vRNPs resembling natural occurring mRNPs (Mouland et al, 2001; Chatel‐Chaix et al, 2004; Fig 2E). It is noteworthy that the transport of vRNA on endosomal membrane directly influences viral production (Lehmann et al, 2009).
More recently, it was shown that also precursors of miRNAs (pre‐miRNAs) are transported on late endosomes/lysosomes in axons of retinal ganglion cells from X. laevis (Corradi & Baudet, 2020; Corradi et al, 2020). The transported pre‐miRNAs are delivered to the growth cone, where they are spatially processed to active miRNAs after exposure with repellent cues. Thereby, the translation of specific transcripts encoding, for example, tubulin beta 3 class III (TUBB3) is silenced. This process is needed for growth cone steering and correct formation of neuronal circuits (Corradi & Baudet, 2020; Corradi et al, 2020). Interestingly, small RNAs also shuttle bidirectionally suggesting a distributive function via these trafficking mechanism.
In summary, endosomal RNA transport is a widespread mechanism present in fungi, plants and animals. Cargos are various types of RNAs, like mRNAs, vRNAs and pre‐miRNAs. Evidence is provided that mRNAs are translated at the cytoplasmic surface of endosome to determine precise subcellular targeting of the cognate translation products. The new link of mitochondrial proteins in X. laevis and U. maydis to endosome‐coupled translation (see below) opens up new possibilities to interlink different organelles and regulate subcellular mitochondrial functions.
Targeting of proteins to mitochondria in a pre‐, co‐, or post‐translational manner
More than 99% of all mitochondrial proteins are encoded in the nucleus and many of those are synthesised in the cytosol and transported post‐translationally to distinct locations within mitochondria using N‐terminal, C‐terminal or internal targeting signals (Pfanner et al, 2019). There is clear evidence that a substantial subset of mitochondrial proteins is targeted to this organelle of endosymbiotic origin via pre‐translational and co‐translational pathways. Efficient targeting of mitochondrial proteins optimises most likely mitochondrial function locally and dynamically (Williams et al, 2014; Fazal et al, 2019). The corresponding pathways are briefly introduced as follows: (i) the classical co‐translational pathway depends on the interaction of an N‐terminal mitochondrial targeting sequence (MTS) in the nascent mitochondrial protein with mitochondrial binding partners during translation, (ii) direct mRNA anchoring to the mitochondrial surface by mitochondria‐associated RNA‐binding proteins followed by in situ translation, (iii) the ER surface‐mediated protein‐targeting pathway (ER‐SURF), (iv) and endosome‐coupled translation in the vicinity of mitochondria.
The classical co‐translational targeting pathway was suggested already in the 1970s by Ron Butow et al based on the observation of cytosolic 80S ribosomes that were located on the mitochondrial outer membrane where mRNAs encoding mitochondrial proteins are translated (Kellems & Butow, 1972; Kellems et al, 1974). In S. cerevisiae, the mitochondrial outer membrane protein OM14 is one receptor for ribosomes that interacts with the nascent chain‐associated complex (NAC) and thereby supports co‐translational import (Lesnik et al, 2014; Béthune et al, 2019). In this setting, the MTS emerging from the ribosome exit tunnel directly interacts with the translocase of the outer membrane (TOM) complex, so that the protein import occurs co‐translationally. Consistently, electron cryotomography showed that ribosomes at mitochondrial membranes face with their exit tunnels towards the mitochondrial membrane (Gold et al, 2017). Active translation of mRNAs by such ribosomes was demonstrated by proximity‐based ribosome profiling studies (Vardi‐Oknin & Arava, 2019; Williams et al, 2014; Fazal et al, 2019; Box2).
Besides this pathway, there is increasing evidence that mRNAs can be targeted to mitochondria by mitochondria‐associated RNA‐binding proteins. Consistently, it has been demonstrated that localisation elements in the 3′ UTR are crucial for targeting ATP2 and Oxa1 to mitochondria (Gadir et al, 2011; Béthune et al, 2019). The mitochondrial‐associated RBP Puf3p from S. cerevisiae, for example, binds numerous mRNAs encoding mitochondrial proteins in their 3′ UTR and thereby promotes mitochondrial protein import (Gerber et al, 2004; Bykov et al, 2020). This was further supported by a systematic approach using proximity labelling of mRNAs with APEX‐Seq (Box 2) demonstrating the presence of mRNAs encoding distinct sets of mitochondrial proteins in the vicinity of mitochondria in a manner that is independent of translation (Fazal et al, 2019).
Targeting mRNAs to mitochondria also enables precise control of translation. In D. melanogaster, mRNAs of nuclear‐encoded subunits of mitochondrial respiratory chain complexes (nRCCs) are translationally repressed in the cytosol. These mRNAs are recruited to mitochondria by PINK1, which interacts with Tom20, a major mitochondrial import receptor at the outer membrane of mitochondria. Upon mRNA binding, PINK1 together with PARKIN stimulate the degradation of translational repressors and thereby activate translation of the nRCC mRNAs. Thereby, PINK1 might not only regulate translation of specific mitochondrial proteins but also ensures that translation occurs in the vicinity of the TOM complex (Gehrke et al, 2015). Since PINK1 stabilisation and PARKIN recruitment is taking place at the outer membrane of dysfunctional mitochondria, it appears that this PINK1‐dependent mechanism might be of particular importance for localised mitochondrial quality control. It could also contribute to the pathogenesis of Parkinson's disease beyond the known roles of PINK1 and PARKIN in mitophagy (Kumar & Reichert, 2021).
An important question is how mitochondrial functions are regulated in highly polarised cells like neurons and hyphae. Recent findings indicate that organelle co‐transport is important. One example is mRNA hitchhiking on mitochondria as shown for PINK1 mRNA (preprint: Harbauer et al, 2021). As elaborated above, the short‐lived PINK1 protein needs to accumulate on the surface of mitochondria to orchestrate mitophagy. How is a constant supply of newly synthesised protein guaranteed for mitochondria that are far away from the nucleus? A neuron‐specific variant of Synaptojanin 2 (SYN2) containing an RRM type RNA‐binding domain binds PINK1 mRNA in complex with the SYN2 binding protein (SYNJ2BP). Mitochondrial association of the PINK1 mRNA depends on translation as well as the synergistic action of the MTS and sequence elements at the beginning of the ORF. Therefore, mRNA transport in dendrites and axons is achieved (preprint: Harbauer et al, 2021). Interestingly, mitochondrial hitchhiking is not restricted to PINK1 mRNA, as Cox7c mRNA (encoding cytochrome c oxidase subunit 7c; respiration chain complex IV) is also actively transported on the surface of mitochondria in an MTS‐dependent manner in axons (preprint: Cohen et al, 2021). One critical RBP‐binding mRNAs encoding mitochondrial proteins is CLUH that was shown to couple mitochondrial biogenesis with mitophagy and mTORC1 signalling (Pla‐Martin et al, 2020).
Besides mRNA co‐transport on the surface of mitochondria, new insights disclosed an intimate link of mitochondrial protein import with communicating organelles like ER and endosomes. In case of the ER‐SURF pathway, certain proteins with the final destination mitochondria are found initially at the ER surface (Hansen et al, 2018; Bykov et al, 2020). Targeting of mitochondrial proteins might be supported by ER‐coupled translation, since mRNAs encoding mitochondrial proteins are found at the ER (Jan et al, 2014). Here, they are translated and the resulting proteins are transferred to mitochondria. The ER‐localised Hsp40/DnaJ‐like protein Djp1 is required for protein translocation into the mitochondria (Hansen et al, 2018). Protein transfer from ER to mitochondria presumably occurs at ER‐mitochondrial contact sites, also termed as mitochondria‐associated ER membranes (MAMs). MAMs are well‐known interorganelle sites that are needed, e.g. for lipid transport and Ca2+ exchange (Lackner, 2019). In addition to mitochondrial and ER surfing, another post‐translational pathway promoting mitochondrial protein import depends on local translation of mitochondrial proteins from nuclear‐encoded mRNAs at the cytoplasmic surface of endosomes (see next section).
Endosome‐coupled translation and mitochondrial protein import
Endosome‐coupled translation constitutes a novel concept mediating efficient mitochondrial protein import. This process appears to be particularly important in highly polarised cells like fungal hyphae and neurons (Cioni et al, 2019; Olgeiser et al, 2019; preprint: Schuhmacher et al, 2021), where subcellular mitochondrial function needs to be maintained at a large distance from the nucleus (Fig 3). Local mitochondrial activity might be particularly important in neurons. This notion is based on the observation that local translation of mitochondrial proteins at activated synapses mediates efficient mitochondrial protein import to fuel high local energy demands in dendrites (Rangaraju et al, 2019; Kuzniewska et al, 2020).
Figure 3. Schematic comparison of endosome‐coupled translation and mitochondrial protein import.
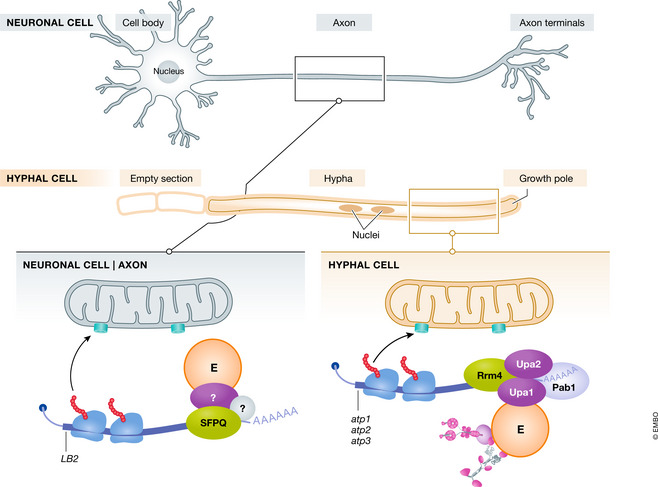
At the top, a highly polarised neuron and a fungal hypha are depicted. In both cases, a defined axis of polarity is established with the growth pole indicated on the right. Local translation of mRNAs encoding mitochondrial proteins at the surface of late endosomes (E) in neurons (left) is compared to membrane‐coupled translation at the surface of early endosomes (E) in hyphae (right). The TOM complex is depicted as a pore in the mitochondrial membrane. Symbols are used as in the other figures, and details are given in the text.
In axons of retinal ganglion cells from X. laevis local mitochondrial activity is needed to coordinate branching. Focal hotspots of translation have been identified in the vicinity of mitochondria throughout axon length (Spillane et al, 2013), and mRNAs are co‐transported with early and late endosomes. The latter serve as platforms of local translation in axons (Cioni et al, 2019; Fig 3). Consistently, ribosomal proteins as well as the RBP SFPQ (splicing factor proline‐ and glutamine‐rich) co‐localise with Rab7a‐positive endosomes (Cioni et al, 2019). SFPQ controls an RNA regulon involved in axon functionality (Cosker et al, 2016).
Notably, late endosomes carrying mRNAs encoding mitochondrial proteins often pause at mitochondria (Cioni et al, 2019; Fig 3). For example, an important target mRNA for endosomal transport encodes the intermediate filament protein lamin B2 (LB2), an essential factor for axon maintenance. LB2 conventionally localises at the nuclear membrane. However, in axons, LB2 is targeted to mitochondria where it determines mitochondrial activity (Yoon et al, 2012). According to results obtained with the in vivo SunTag fluorescent tagging system (Wang et al, 2016; Wu et al, 2016; Yan et al, 2016; Box 2), LB2 mRNA is associated with late endosomes and translationally active on these endosomes in the vicinity of mitochondria (Cioni et al, 2019). This observation suggests that endosomal‐coupled translation of LB2 mRNA targets the translation product to mitochondria. A second example is the mRNA encoding voltage‐dependent anion‐selective channel protein 2 (VDAC2), which functions in exchanging solutes across the outer mitochondrial membrane (Naghdi & Hajnoczky, 2016). VDAC2 is translated on endosomes near mitochondria. Hence, it appears to be a common process that mRNAs encoding mitochondrial proteins are translated on the surface of endosomes often in proximity to mitochondria (Cioni et al, 2019; Fig 3). Remarkably, Rab7a mutations characteristic for CMT2B Charcot‐Marie‐Tooth type 2B disease interfere with endosome‐coupled translation at late endosomes (Cioni et al, 2019). Expressing these mutant alleles resulted in highly elongated mitochondria and altered mitochondrial membrane potential indicating a functional influence on mitochondrial morphology and bioenergetics.
Intriguingly, a novel link between endosomes, RNA and mitochondrial biology has been recently discovered studying the novel Rab5a effector complex FERRY (preprint: Quentin et al, 2021; preprint: Schuhmacher et al, 2021; Fig 2C). Co‐purification experiments using the FERRY complex as molecular handle revealed the presence of mRNAs and ribosomal proteins on endosomes suggesting a role in endosome‐coupled transport and translation of mRNAs (preprint: Schuhmacher et al, 2021). No specific RNA‐binding domain has been identified yet, but several RNA contact sites are present along the elongated structure of the FERRY complex (preprint: Quentin et al, 2021). mRNAs that encode mitochondrial proteins, like mrpl41 mRNA encoding ribosomal protein 41 of the large subunit of 70S mitochondrial ribosomes, are enriched on endosomes. Based on results obtained with smFISH experiments, mRNAs, endosomes and mitochondria are found in close proximity in primary rat hippocampal neurons. Although endosomal mRNA movement has not been demonstrated, the FERRY complex has the potential to serve as a novel anchor for endosomal mRNA transport in neurons. In three of five cases, namely Fy1 (Tbck), Fy2 (Ppp1r21) and Fy3 (C12orf4), mutations in these human proteins cause brain malfunctions (preprint: Quentin et al, 2021; preprint: Schuhmacher et al, 2021; Fig 2C). In summary, early and late endosome‐coupled translation might determine mitochondrial function and be linked to neuronal diseases.
Studying the proteome of membrane‐associated fractions in hyphae from U. maydis showed that loss of endosomal mRNA transport altered the amount of mitochondrial proteins. Accumulation of the complex I component Nuo2 and the F1FO ATP synthase subunit Atp4 was reduced in comparison with wildtype, while ATPase family gene 3 (Afg3) was increased in rrm4Δ strains lacking endosomal mRNA transport (Koepke et al, 2011). Afg3 is a subunit of the m‐AAA protease located in the inner membrane and functions in the quality control of mitochondrial complex assembly by mediating degradation of non‐assembled mitochondrial inner membrane proteins (Arlt et al, 1998). This exemplifies a molecular link between endosomal mRNA transport and the biogenesis of mitochondrial respiratory chain complexes. Consistently, uncovering transcriptome‐wide Rrm4 target mRNAs revealed that mRNAs encoding mitochondrial proteins such as most subunits of the F1FO ATP synthase are recognised by Rrm4. Intriguingly, the mRNAs are predominantly bound by Rrm4 precisely at the stop codon linking transport with translational termination (Olgeiser et al, 2019).
Thus, mRNAs encoding mitochondrial proteins might be co‐translationally transported exposing the N‐terminal MTS for a potential direct handover to the mitochondrial import apparatus (Fig 3), yet might be delayed for translational termination. Based on this hypothesis, Rrm4 would pause translation just before termination by binding to the stop codon, until the translation product reaches its destination. Hence, spatial regulation of translation would be another function for endosome‐coupled transport to mitochondria (Fig 3). Consistently, it was found in S. cerevisiae that translation speed was important for mRNA localisation to mitochondria. The coding sequence of mitochondrial protein TIM50 contains seven consecutive prolines slowing down translation, which increases the possibility for the MTS to interact with its receptor on the surface. Therefore, the localisation to mitochondria is enhanced (Tsuboi et al, 2020). In essence, we observe a link of endosomal mRNA transport to mitochondria both in axons and fungal hyphae suggesting evolutionary conversation (Fig 3). The linkage could either be due to convergent or divergent evolution. At present, this is unknown, but we favour the idea that endosomal mRNA transport is an ancient process, since it is present in all fungal phyla, plants and mammalian cells. Therefore, also the link to mitochondrial biology might be ancient and more widespread than currently anticipated.
Consistently, potential homologues of components of the FERRY complex were found in fungi (preprint: Schuhmacher et al, 2021). Along this line, we speculate that the process of membrane‐coupled transport of mRNAs to mitochondria is not restricted to polarised cells. During the oogenesis of X. laevis, nuclear mRNAs destined to the “vegetal pole” are enriched at a peculiar mitochondrial cloud (Kloc et al, 1996; Chang et al, 2004). This region is rich in active mitochondria and located between germinal vesicles and the plasma membrane (Wilding et al, 2001). A target mRNA encodes XNOA36 (Vaccaro et al, 2012), the homolog of the mammalian NOA36, which associates dynamically with outer membranes of mitochondria (de Melo et al, 2009). Since the subcellular region is also rich in ER, another potential link between RNA, membrane trafficking and mitochondrial function is conceivable.
Concluding remarks and outlook
mRNP trafficking constitutes a highly complex intracellular process with various transport solutions for distinct sets of cargo mRNAs. Besides the classical system consisting of membrane‐free mRNP transport, it now becomes apparent that alternative pathways exist like organelle hitchhiking on mitochondria, the ER and endosomes (Béthune et al, 2019; preprint: Cohen et al, 2021; preprint: Harbauer et al, 2021; preprint: Schuhmacher et al, 2021). Why are several sophisticated modes of transport systems operational? In this context, it is important to reiterate that each translation product has a well‐defined final subcellular location: β‐actin is needed at the cellular growth poles, septins need to assemble in heteromeric complexes delivered to specific subcellular sites and mitochondrial proteins end up in mitochondria. Hence, the most parsimonious explanation for the existence of various transport pathways is the presence of tailor‐made solutions that are best adapted to the individual need of the cargo mRNA. In analogy, the cell can choose between car, truck, plane or ship type vehicles for transportation. Another advantage of using additional membrane‐coupled pathways is to avoid traffic jams for protein import into organelles by fully exploiting other membrane surfaces. In this way, mRNAs or co‐translational intermediates can be stored or parked nearby.
One of the emerging alternative transport mechanisms is endosomal mRNA transport, an ancient, conserved mechanism, present in fungi, plants and animals. Various types of RNAs are transported, and the main function is spatiotemporal expression of proteins. More precisely, endosomal transport and translation of mRNAs are needed for (i) equal distribution of mRNAs and ribosomes, (ii) heteromeric complex assembly, (iii) ER targeting and (iv) mitochondrial protein import. The use of organellar membranes as local platforms for membrane‐coupled translation (Béthune et al, 2019) mediates targeting to other organelles, and this appears to be an efficient mechanism regulating local activities in the eukaryotic cell. Currently, it is restricted to ER and endosomal membranes; however, links to secretory vesicles and mitochondrial membranes have been described (Zabezhinsky et al, 2016; Hansen et al, 2018). We believe that the link between mRNA transport, localised translation and organelle communication is a widespread phenomenon. Outstanding questions that need to be addressed are listed in Box 3.
Box 3: In need of answers.
How widespread is endosome‐coupled mRNA transport and translation? Additional examples of membrane‐coupled trafficking in other cell types and organisms are needed to answer this question.
What is the diversity of subcellular organelles endosomes communicate with? Besides peroxisomes and mitochondria, the link to other subcellular organelles should be analysed.
What is the composition of the endosome‐associated transcriptome in different cell types and at different subcellular locations? The respective inventory of endosomal mRNAs using tailored biochemical purification and RNA sequencing approaches need to be determined.
How are mRNPs molecularly linked to endosomes? Structure function analyses of adaptor proteins or RBPs are required.
How is endosomal RNA biology regulated and dynamically adapted to different growth and stress conditions? We need to identify key post‐translational modifications in RBPs and study the action of the modifying enzymes.
How are mRNPs loaded and unloaded from endosomes? What are the interaction partners of key RBPs and how are these factors post‐translationally regulated?
How does membrane‐coupled translation orchestrate subcellular mitochondrial function? Biosensors and optogenetics can be used to study the influence of translation on local mitochondrial activity.
Finally, we would like to point out that the intimate link of subcellular organelles like ER and endosomes with mitochondria of endosymbiont origin is also inspiring to reflect on the origin of endosymbiosis. It is now well accepted that genetic information of the initial endosymbiont was transferred to the nucleus and that protein‐targeting signals enable the necessary import of proteins into the endosymbiotic organelle. Additionally, RNA‐centric targeting signals are also required to direct RNAs in the vicinity of mitochondria for efficient protein import to orchestrate organelle function. Surprisingly, import of distinct mitochondrial proteins is promoted by membrane‐coupled translation of cognate mRNAs at the surface of neighbouring organelles such as endosomes, demonstrating intensive networking of intracellular organelles. It is conceivable that for a successful incorporation of an endosymbiont in the local environment of its host, it is beneficial to establish intricate modes of communication with the host intracellular organelles like nucleus, ER and endosomes. In essence, studying the complex life of RNAs does not only increase our understanding of fundamental processes of the eukaryotic cell but also bridges numerous research areas ranging from RNA, membrane and mitochondrial biology to evolutionary biology.
Author contributions
KM was responsible for the figures. All authors contributed to writing and revising of the manuscript and approved the final version.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgements
We thank laboratory members for critical reading of the manuscript. The work was funded by grants from the Deutsche Forschungsgemeinschaft under Germany's Excellence Strategy EXC‐2048/1—Project ID 39068111 to MF; Project ID 267205415 – SFB 1208 to MF and ASR, DFG‐FOR2333 FE448/10‐2, DFG‐FOR5116 FE448/15‐1 and DFG‐FE 448/11‐1 to MF as well as DFG‐GRK2576, DFG‐GRK 2578 and DFG‐RE1575/2‐1 to ASR. Open Access funding enabled and organized by ProjektDEAL.
EMBO reports (2021) 22: e52445.
See the Glossary for abbreviations used in this article.
References
- Abouward R, Schiavo G (2020) Walking the line: mechanisms underlying directional mRNA transport and localisation in neurons and beyond. Cell Mol Life Sci 78: 2665–2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akopian D, Shen K, Zhang X, Shan SO (2013) Signal recognition particle: an essential protein‐targeting machine. Annu Rev Biochem 82: 693–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlt H, Steglich G, Perryman R, Guiard B, Neupert W, Langer T (1998) The formation of respiratory chain complexes in mitochondria is under the proteolytic control of the m‐AAA protease. EMBO J 17: 4837–4847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassell GJ, Powers CM, Taneja KL, Singer RH (1994) Single mRNAs visualized by ultrastructural in situ hybridization are principally localized at actin filament intersections in fibroblasts. J Cell Biol 126: 863–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basyuk E, Galli T, Mougel M, Blanchard JM, Sitbon M, Bertrand E (2003) Retroviral genomic RNAs are transported to the plasma membrane by endosomal vesicles. Dev Cell 5: 161–174 [DOI] [PubMed] [Google Scholar]
- Basyuk E, Rage F, Bertrand E (2020) RNA transport from transcription to localized translation: a single molecule perspective. RNA Biol 18: 1221–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann S, Pohlmann T, Jungbluth M, Brachmann A, Feldbrügge M (2012) Kinesin‐3 and dynein mediate microtubule‐dependent co‐transport of mRNPs and endosomes. J Cell Sci 125: 2740–2752 [DOI] [PubMed] [Google Scholar]
- Baumann S, König J, Koepke J, Feldbrügge M (2014) Endosomal transport of septin mRNA and protein indicates local translation on endosomes and is required for correct septin filamentation. EMBO Rep 15: 94–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann S, Komissarov A, Gili M, Ruprecht V, Wieser S, Maurer SP (2020) A reconstituted mammalian APC‐kinesin complex selectively transports defined packages of axonal mRNAs. Sci Adv 6: eaaz1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becht P, König J, Feldbrügge M (2006) The RNA‐binding protein Rrm4 is essential for polarity in Ustilago maydis and shuttles along microtubules. J Cell Sci 119: 4964–4973 [DOI] [PubMed] [Google Scholar]
- Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM (1998) Localization of ASH1 mRNA particles in living yeast. Mol Cell 2: 437–445 [DOI] [PubMed] [Google Scholar]
- Béthune J, Jansen RP, Feldbrügge M, Zarnack K (2019) Membrane‐associated RNA‐binding proteins orchestrate organelle‐coupled translation. Trends Cell Biol 29: 178–188 [DOI] [PubMed] [Google Scholar]
- Bielska E, Higuchi Y, Schuster M, Steinberg N, Kilaru S, Talbot NJ, Steinberg G (2014a) Long‐distance endosome trafficking drives fungal effector production during plant infection. Nat Commun 5: 5097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielska E, Schuster M, Roger Y, Berepiki A, Soanes DM, Talbot NJ, Steinberg G (2014b) Hook is an adapter that coordinates kinesin‐3 and dynein cargo attachment on early endosomes. J Cell Biol 204: 989–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackinton JG, Keene JD (2014) Post‐transcriptional RNA regulons affecting cell cycle and proliferation. Sem Cell Dev Biol 34: 44–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann A, Weinzierl G, Kämper J, Kahmann R (2001) Identification of genes in the bW/bE regulatory cascade in Ustilago maydis . Mol Microbiol 42: 1047–1063 [DOI] [PubMed] [Google Scholar]
- Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, Julicher F, Hyman AA (2009) Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324: 1729–1732 [DOI] [PubMed] [Google Scholar]
- Bratu DP (2003) Molecular beacons light the way: imaging native mRNAs in living cells. Discov Med 3: 44–47 [PubMed] [Google Scholar]
- Bratu DP, Cha BJ, Mhlanga MM, Kramer FR, Tyagi S (2003) Visualizing the distribution and transport of mRNAs in living cells. Proc Natl Acad Sci USA 100: 13308–13313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullock SL, Ish‐Horowicz D (2001) Conserved signals and machinery for RNA transport in Drosophila oogenesis and embryogenesis. Nature 414: 611–616 [DOI] [PubMed] [Google Scholar]
- Buxbaum AR, Haimovich G, Singer RH (2015) In the right place at the right time: visualizing and understanding mRNA localization. Nat Rev Mol Cell Biol 16: 95–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bykov YS, Rapaport D, Herrmann JM, Schuldiner M (2020) Cytosolic events in the biogenesis of mitochondrial proteins. Trends Biochem Sci 45: 650–667 [DOI] [PubMed] [Google Scholar]
- Cagnetta R, Frese CK, Shigeoka T, Krijgsveld J, Holt CE (2018) Rapid cue‐specific remodeling of the nascent axonal proteome. Neuron 99: 29–46.e24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajigas IJ, Tushev G, Will T, tom Dieck S, Fuerst N, Schuman E (2012) The local transcriptome in the synaptic neuropil revealed by deep sequencing and high‐resolution imaging. Neuron 74: 453–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang P, Torres J, Lewis RA, Mowry KL, Houliston E, King ML (2004) Localization of RNAs to the mitochondrial cloud in Xenopus oocytes through entrapment and association with endoplasmic reticulum. Mol Biol Cell 15: 4669–4681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatel‐Chaix L, Clement JF, Martel C, Beriault V, Gatignol A, DesGroseillers L, Mouland AJ (2004) Identification of Staufen in the human immunodeficiency virus type 1 Gag ribonucleoprotein complex and a role in generating infectious viral particles. Mol Cell Biol 24: 2637–2648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng XT, Xie YX, Zhou B, Huang N, Farfel‐Becker T, Sheng ZH (2018) Revisiting LAMP1 as a marker for degradative autophagy‐lysosomal organelles in the nervous system. Autophagy 14: 1472–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cioni J‐M, Lin JQ, Holtermann AV, Koppers M, Jakobs MAH, Azizi A, Turner‐Bridger B, Shigeoka T, Franze K, Harris WAet al (2019) Late endosomes act as mRNA translation platforms and sustain mitochondria in axons. Cell 176: 56–72.e15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen B, Golani‐Armon A, Altman T, Savulescu AF, Mhlanga MM, Perlson E, Arava Y (2021) Mitochondria serve as axonal shuttle for Cox7c mRNA through mechanism that involves its mitochondrial targeting signal. bioRiv 10.1101/2021.05.19.444640 [PREPRINT] [DOI] [Google Scholar]
- Corradi E, Baudet ML (2020) In the right place at the right time: miRNAs as key regulators in developing axons. Int J Mol Sci 21: 8726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corradi E, Dalla Costa I, Gavoci A, Iyer A, Roccuzzo M, Otto TA, Oliani E, Bridi S, Strohbuecker S, Santos‐Rodriguez Get al (2020) Axonal precursor miRNAs hitchhike on endosomes and locally regulate the development of neural circuits. EMBO J 39: e102513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosker KE, Fenstermacher SJ, Pazyra‐Murphy MF, Elliott HL, Segal RA (2016) The RNA‐binding protein SFPQ orchestrates an RNA regulon to promote axon viability. Nat Neurosci 19: 690–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle N, Ellenberg J (2007) LambdaN‐GFP: an RNA reporter system for live‐cell imaging. Nat Methods 4: 633–636 [DOI] [PubMed] [Google Scholar]
- Dalla Costa I, Buchanan CN, Zdradzinski MD, Sahoo PK, Smith TP, Thames E, Kar AN, Twiss JL (2021) The functional organization of axonal mRNA transport and translation. Nat Rev Neurosci 22: 77–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Singer RH, Yoon YJ (2019) The travels of mRNAs in neurons: do they know where they are going? Curr Opin Neurobiol 57: 110–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Vera M, Gandin V, Singer RH, Tutucci E (2021) Intracellular mRNA transport and localized translation. Nat Rev Mol Cell Biol 22: 483–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlin‐Asp PG, Polisseni C, Klimek R, Heckel A, Schuman EM (2021) Differential regulation of local mRNA dynamics and translation following long‐term potentiation and depression. Proc Natl Acad Sci USA 118: E2017578118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle M, Kiebler MA (2011) Mechanisms of dendritic mRNA transport and its role in synaptic tagging. EMBO J 30: 3540–3552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelmann FT, Schlundt A, Heym RG, Jenner A, Niedner‐Boblenz A, Syed MI, Paillart J‐C, Stehle R, Janowski R, Sattler Met al (2017) Molecular architecture and dynamics of ASH1 mRNA recognition by its mRNA‐transport complex. Nat Struct Mol Biol 24: 152–161 [DOI] [PubMed] [Google Scholar]
- Elson SL, Noble SM, Solis NV, Filler SG, Johnson AD (2009) An RNA transport system in Candida albicans regulates hyphal morphology and invasive growth. PLoS Genet 5: e1000664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazal FM, Han S, Parker KR, Kaewsapsak P, Xu J, Boettiger AN, Chang HY, Ting AY (2019) Atlas of subcellular RNA localization revealed by APEX‐Seq. Cell 178: 473–490.e426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandopulle MS, Lippincott‐Schwartz J, Ward ME (2021) RNA transport and local translation in neurodevelopmental and neurodegenerative disease. Nat Neurosci 24: 622–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadir N, Haim‐Vilmovsky L, Kraut‐Cohen J, Gerst JE (2011) Localization of mRNAs coding for mitochondrial proteins in the yeast Saccharomyces cerevisiae . RNA 17: 1551–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrke S, Wu Z, Klinkenberg M, Sun Y, Auburger G, Guo S, Lu B (2015) PINK1 and Parkin control localized translation of respiratory chain component mRNAs on mitochondria outer membrane. Cell Metab 21: 95–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber AP, Herschlag D, Brown PO (2004) Extensive association of functionally and cytotopically related mRNAs with Puf family RNA‐binding proteins in yeast. PLoS Biol 2: e79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Marchand V, Gaspar I, Ephrussi A (2012) Control of RNP motility and localization by a splicing‐dependent structure in oskar mRNA. Nat Struct Mol Biol 19: 441–449 [DOI] [PubMed] [Google Scholar]
- Gold VA, Chroscicki P, Bragoszewski P, Chacinska A (2017) Visualization of cytosolic ribosomes on the surface of mitochondria by electron cryo‐tomography. EMBO Rep 18: 1786–1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grote M, Wolf E, Will CL, Lemm I, Agafonov DE, Schomburg A, Fischle W, Urlaub H, Luhrmann R (2010) Molecular architecture of the human Prp19/CDC5L complex. Mol Cell Biol 30: 2105–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimaraes SC, Schuster M, Bielska E, Dagdas G, Kilaru S, Meadows BR, Schrader M, Steinberg G (2015) Peroxisomes, lipid droplets, and endoplasmic reticulum "hitchhike" on motile early endosomes. J Cell Biol 211: 945–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag C, Steuten B, Feldbrügge M (2015) Membrane‐coupled mRNA trafficking in fungi. Annu Rev Microbiol 69: 265–281 [DOI] [PubMed] [Google Scholar]
- Haag C, Pohlmann T, Feldbrügge M (2017) The ESCRT regulator Did2 maintains the balance between long‐distance endosomal transport and endocytic trafficking. PLoS Genet 13: e1006734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada S, Ishiyama K, Sakulsingharoj C, Choi SB, Wu Y, Wang C, Singh S, Kawai N, Messing J, Okita TW (2003) Dual regulated RNA transport pathways to the cortical region in developing rice endosperm. Plant Cell 15: 2265–2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KG, Aviram N, Laborenz J, Bibi C, Meyer M, Spang A, Schuldiner M, Herrmann JM (2018) An ER surface retrieval pathway safeguards the import of mitochondrial membrane proteins in yeast. Science 361: 1118–1122 [DOI] [PubMed] [Google Scholar]
- Hansen KG, Herrmann JM (2019) Transport of proteins into mitochondria. Protein J 38: 330–342 [DOI] [PubMed] [Google Scholar]
- Harbauer AB, Wanderoy S, Hees JT, Gibbs W, Ordonez M, Cai Z, Cartoni R, Ashrafi G, Wang C, He Zet al (2021) Neuronal mitochondria transport Pink1 mRNA via Synaptojanin 2 to support local mitophagy. bioRiv 10.1101/2021.05.19.444778 [PREPRINT] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold I, Kowbel D, Delgado‐Alvarez DL, Garduno‐Rosales M, Mourino‐Perez RR, Yarden O (2019) Transcriptional profiling and localization of GUL‐1, a COT‐1 pathway component, in Neurospora crassa . Fungal Genet Biol 126: 1–11 [DOI] [PubMed] [Google Scholar]
- Heym RG, Zimmermann D, Edelmann FT, Israel L, Okten Z, Kovar DR, Niessing D (2013) In vitro reconstitution of an mRNA‐transport complex reveals mechanisms of assembly and motor activation. J Cell Biol 203: 971–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi Y, Ashwin P, Roger Y, Steinberg G (2014) Early endosome motility spatially organizes polysome distribution. J Cell Biol 204: 343–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday R (2004) Early studies on recombination and DNA repair in Ustilago maydis . DNA Repair 3: 671–682 [DOI] [PubMed] [Google Scholar]
- Holt CE, Bullock SL (2009) Subcellular mRNA localization in animal cells and why it matters. Science 326: 1212–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt CE, Martin KC, Schuman EM (2019) Local translation in neurons: visualization and function. Nat Struct Mol Biol 26: 557–566 [DOI] [PubMed] [Google Scholar]
- Huotari J, Helenius A (2011) Endosome maturation. EMBO J 30: 3481–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS (2009) Genome‐wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324: 218–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan CH, Williams CC, Weissman JS (2014) Principles of ER cotranslational translocation revealed by proximity‐specific ribosome profiling. Science 346: 1257521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowski S, Pohlmann T, Baumann S, Müntjes KM, Devan SK, Zander S, Feldbrügge M (2019) The multi PAM2 protein Upa2 functions as novel core component of endosomal mRNA transport. EMBO Rep 24: e47381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, McKnight SL (2018) A solid‐state conceptualization of information transfer from gene to message to protein. Annu Rev Biochem 87: 351–390 [DOI] [PubMed] [Google Scholar]
- Keene JD (2007) RNA regulons: coordination of post‐transcriptional events. Nat Rev Genet 8: 533–543 [DOI] [PubMed] [Google Scholar]
- Kellems RE, Butow RA (1972) Cytoplasmic‐type 80 S ribosomes associated with yeast mitochondria. I. Evidence for ribosome binding sites on yeast mitochondria. J Biol Chem 247: 8043–8050 [PubMed] [Google Scholar]
- Kellems RE, Allison VF, Butow RA (1974) Cytoplasmic type 80 S ribosomes associated with yeast mitochondria. II. Evidence for the association of cytoplasmic ribosomes with the outer mitochondrial membrane in situ . J Biol Chem 249: 3297–3303 [PubMed] [Google Scholar]
- Kellner N, Heimel K, Obhof T, Finkernagel F, Kämper J (2014) The SPF27 homologue Num1 connects splicing and kinesin 1‐dependent cytoplasmic trafficking in Ustilago maydis . PLoS Genet 10: e1004046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloc M, Larabell C, Etkin LD (1996) Elaboration of the messenger transport organizer pathway for localization of RNA to the vegetal cortex of Xenopus oocytes. Dev Biol 180: 119–130 [DOI] [PubMed] [Google Scholar]
- Koepke J, Kaffarnik F, Haag C, Zarnack K, Luscombe NM, König J, Ule J, Kellner R, Begerow D, Feldbrügge M (2011) The RNA‐binding protein Rrm4 is essential for efficient secretion of endochitinase Cts1. Mol Cell Proteom 10: 011211–011215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- König J, Baumann S, Koepke J, Pohlmann T, Zarnack K, Feldbrügge M (2009) The fungal RNA‐binding protein Rrm4 mediates long‐distance transport of ubi1 and rho3 mRNAs. EMBO J 28: 1855–1866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer G, Shiber A, Bukau B (2019) Mechanisms of cotranslational maturation of newly synthesized proteins. Annu Rev Biochem 88: 337–364 [DOI] [PubMed] [Google Scholar]
- Kück U, Radchenko D, Teichert I (2019) STRIPAK, a highly conserved signaling complex, controls multiple eukaryotic cellular and developmental processes and is linked with human diseases. Biol Chem 400: 1005–1022 [DOI] [PubMed] [Google Scholar]
- Kumar R, Reichert AS (2021) Common principles and specific mechanisms of mitophagy from yeast to humans. Int J Mol Sci 22: 4363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzniewska B, Cysewski D, Wasilewski M, Sakowska P, Milek J, Kulinski TM, Winiarski M, Kozielewicz P, Knapska E, Dadlez Met al (2020) Mitochondrial protein biogenesis in the synapse is supported by local translation. EMBO Rep 21: e48882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner LL (2019) The expanding and unexpected functions of mitochondria contact sites. Trends Cell Biol 29: 580–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanver D, Tollot M, Schweizer G, Lo Presti L, Reissmann S, Ma L‐S, Schuster M, Tanaka S, Liang L, Ludwig Net al (2017) Ustilago maydis effectors and their impact on virulence. Nat Rev Microbiol 15: 409–421 [DOI] [PubMed] [Google Scholar]
- Larson DR, Zenklusen D, Wu B, Chao JA, Singer RH (2011) Real‐time observation of transcription initiation and elongation on an endogenous yeast gene. Science 332: 475–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann M, Milev MP, Abrahamyan L, Yao XJ, Pante N, Mouland AJ (2009) Intracellular transport of human immunodeficiency virus type 1 genomic RNA and viral production are dependent on dynein motor function and late endosome positioning. J Biol Chem 284: 14572–14585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz JH, Schuchardt I, Straube A, Steinberg G (2006) A dynein loading zone for retrograde endosome motility at microtubule plus‐ends. EMBO J 25: 2275–2286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesnik C, Cohen Y, Atir‐Lande A, Schuldiner M, Arava Y (2014) OM14 is a mitochondrial receptor for cytosolic ribosomes that supports co‐translational import into mitochondria. Nat Commun 5: 5711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao YC, Fernandopulle MS, Wang G, Choi H, Hao L, Drerup CM, Patel R, Qamar S, Nixon‐Abell J, Shen Yet al (2019) RNA granules hitchhike on lysosomes for long‐distance transport, using annexin A11 as a molecular tether. Cell 179: 147–164.e120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long RM, Gu W, Meng X, Gonsalvez G, Singer RH, Chartrand P (2001) An exclusively nuclear RNA‐binding protein affects asymmetric localization of ASH1 mRNA and Ash1p in yeast. J Cell Biol 153: 307–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Lakonishok M, Serpinskaya AS, Kirchenbuechler D, Ling SC, Gelfand VI (2018) Ooplasmic flow cooperates with transport and anchorage in Drosophila oocyte posterior determination. J Cell Biol 217: 3497–3511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekiniemi A, Singer RH, Tutucci E (2020) Single molecule mRNA fluorescent in situ hybridization combined with immunofluorescence in S. cerevisiae: dataset and quantification. Data Brief 30: 105511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin KC, Ephrussi A (2009) mRNA localization: gene expression in the spatial dimension. Cell 136: 719–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock MA, Dix CI, Johnson CM, McLaughlin SH, Maizels RJ, Hoang HT, Bullock SL (2018) RNA‐directed activation of cytoplasmic dynein‐1 in reconstituted transport RNPs. eLife 7: e36312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Melo IS, Iglesias C, Benitez‐Rondan A, Medina F, Martinez‐Barbera JP, Bolivar J (2009) NOA36/ZNF330 is a conserved cystein‐rich protein with proapoptotic activity in human cells. Biochim Biophys Acta 1793: 1876–1885 [DOI] [PubMed] [Google Scholar]
- Mofatteh M, Bullock SL (2017) SnapShot: subcellular mRNA localization. Cell 169: 178.e171 [DOI] [PubMed] [Google Scholar]
- Mouland AJ, Xu H, Cui H, Krueger W, Munro TP, Prasol M, Mercier J, Rekosh D, Smith R, Barbarese Eet al (2001) RNA trafficking signals in human immunodeficiency virus type 1. Mol Cell Biol 21: 2133–2143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J, Pohlmann T, Feldbrügge M (2019) Core components of endosomal mRNA transport are evolutionarily conserved in fungi. Fungal Genet Biol 126: 12–16 [DOI] [PubMed] [Google Scholar]
- Naghdi S, Hajnoczky G (2016) VDAC2‐specific cellular functions and the underlying structure. Biochim Biophys Acta 1863: 2503–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neisch AL, Neufeld TP, Hays TS (2017) A STRIPAK complex mediates axonal transport of autophagosomes and dense core vesicles through PP2A regulation. J Cell Biol 216: 441–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedner A, Muller M, Moorthy BT, Jansen RP, Niessing D (2013) Role of Loc1p in assembly and reorganization of nuclear ASH1 messenger ribonucleoprotein particles in yeast. Proc Natl Acad Sci USA 110: E5049–E5058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niessing D, Jansen RP, Pohlmann T, Feldbrügge M (2018) mRNA transport in fungal top models. Wiley Interdiscip Rev RNA 9: e1453 [DOI] [PubMed] [Google Scholar]
- Obrdlik A, Lin G, Haberman N, Ule J, Ephrussi A (2019) The transcriptome‐wide landscape and modalities of EJC binding in adult Drosophila . Cell Rep 28: 1219–1236.e1211 [DOI] [PubMed] [Google Scholar]
- Olgeiser L, Haag C, Boerner S, Ule J, Busch A, Koepke J, König J, Feldbrügge M, Zarnack K (2019) The key protein of endosomal mRNP transport Rrm4 binds translational landmark sites of cargo mRNAs. EMBO Rep 20: e46588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padron A, Iwasaki S, Ingolia NT (2019) Proximity RNA labeling by APEX‐seq reveals the organization of translation initiation complexes and repressive RNA granules. Mol Cell 75: 875–887.e875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panasenko OO, Somasekharan SP, Villanyi Z, Zagatti M, Bezrukov F, Rashpa R, Cornut J, Iqbal J, Longis M, Carl SHet al (2019) Co‐translational assembly of proteasome subunits in NOT1‐containing assemblysomes. Nat Struct Mol Biol 26: 110–120 [DOI] [PubMed] [Google Scholar]
- Pfanner N, Warscheid B, Wiedemann N (2019) Mitochondrial proteins: from biogenesis to functional networks. Nat Rev Mol Cell Biol 20: 267–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper M, Lee AC, van Horck FP, McNeilly H, Lu TB, Harris WA, Holt CE (2015) Differential requirement of F‐actin and microtubule cytoskeleton in cue‐induced local protein synthesis in axonal growth cones. Neural Dev 10: 3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pla‐Martin D, Schatton D, Wiederstein JL, Marx MC, Khiati S, Kruger M, Rugarli EI (2020) CLUH granules coordinate translation of mitochondrial proteins with mTORC1 signaling and mitophagy. EMBO J 39: e102731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlmann T, Baumann S, Haag C, Albrecht M, Feldbrügge M (2015) A FYVE zinc finger domain protein specifically links mRNA transport to endosome trafficking. eLife 4: e06041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic D, Nijenhuis W, Kapitein LC, Pelkmans L (2020) Co‐translational targeting of transcripts to endosomes. bioRxiv 10.1101/2020.07.17.208652 [PREPRIT] [DOI] [Google Scholar]
- Poulopoulos A, Murphy AJ, Ozkan A, Davis P, Hatch J, Kirchner R, Macklis JD (2019) Subcellular transcriptomes and proteomes of developing axon projections in the cerebral cortex. Nature 565: 356–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preitner N, Quan J, Nowakowski DW, Hancock ML, Shi J, Tcherkezian J, Young‐Pearse TL, Flanagan JG (2014) APC is an RNA‐binding protein, and its interactome provides a link to neural development and microtubule assembly. Cell 158: 368–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protter DSW, Parker R (2016) Principles and properties of stress granules. Trends Cell Biol 26: 668–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pushpalatha KV, Besse F (2019) Local translation in axons: when membraneless RNP granules meet membrane‐bound organelles. Front Mol Biosci 6: 129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quentin D, Schuhmacher JS, Klink BU, Lauer J, Shaikh TR, Huis in t't Veld PJ, Welp LM, Urlaub H, Zerial M, Raunser S (2021) Structure of the human FERRY Rab5 effector complex. bioRiv 10.1101/2021.06.21.449265 [PREPRINT] [DOI] [PubMed] [Google Scholar]
- Rangaraju V, Lauterbach M, Schuman EM (2019) Spatially stable mitochondrial compartments fuel local translation during plasticity. Cell 176: 73–84.e15 [DOI] [PubMed] [Google Scholar]
- Salogiannis J, Reck‐Peterson SL (2016) Hitchhiking: a non‐canonical mode of microtubule‐based transport. Trends Cell Biol 27: 141–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuhmacher JS, Dieck ST, Christoforidis S, Landerer C, Hersemann L, Seifert S, Giner A, Toth‐Petroczy A, Kalaidzidis Y, Schumann EMet al (2021) The novel Rab5 effector FERRY links early endosomes with the translation machinery. BioRiv. 10.1101/2021.06.20.449167 [DOI] [PubMed] [Google Scholar]
- Schuster M, Kilaru S, Ashwin P, Lin C, Severs NJ, Steinberg G (2011a) Controlled and stochastic retention concentrates dynein at microtubule ends to keep endosomes on track. EMBO J 30: 652–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster M, Kilaru S, Fink G, Collemare J, Roger Y, Steinberg G (2011b) Kinesin‐3 and dynein cooperate in long‐range retrograde endosome motility along a non‐uniform microtubule array. Mol Biol Cell 22: 3645–3657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz A, Beck M (2019) The benefits of cotranslational assembly: a structural perspective. Trends Cell Biol 29: 791–803 [DOI] [PubMed] [Google Scholar]
- Shiber A, Doring K, Friedrich U, Klann K, Merker D, Zedan M, Tippmann F, Kramer G, Bukau B (2018) Cotranslational assembly of protein complexes in eukaryotes revealed by ribosome profiling. Nature 561: 268–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh YW, Minguez P, Bork P, Auburger JJ, Guilbride DL, Kramer G, Bukau B (2015) Operon structure and cotranslational subunit association direct protein assembly in bacteria. Science 350: 678–680 [DOI] [PubMed] [Google Scholar]
- Shigeoka T, Jung H, Jung J, Turner‐Bridger B, Ohk J, Lin JQ, Amieux PS, Holt CE (2016) Dynamic axonal translation in developing and mature visual circuits. Cell 166: 181–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon B, Masiewicz P, Ephrussi A, Carlomagno T (2015) The structure of the SOLE element of oskar mRNA. RNA 21: 1444–1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sladewski TE, Bookwalter CS, Hong MS, Trybus KM (2013) Single‐molecule reconstitution of mRNA transport by a class V myosin. Nat Struct Mol Biol 20: 952–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sladewski TE, Billington N, Ali MY, Bookwalter CS, Lu H, Krementsova EB, Schroer TA, Trybus KM (2018) Recruitment of two dyneins to an mRNA‐dependent Bicaudal D transport complex. eLife 7: e36306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snead WT, Gerbich TM, Seim I, Hu Z, Gladfelter AS (2021) Membrane surface regulate assembly of a ribonucleoprotein condensate. bioRxiv 10.1101/2021.1104.441251 [PREPRINT] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spillane M, Ketschek A, Merianda TT, Twiss JL, Gallo G (2013) Mitochondria coordinate sites of axon branching through localized intra‐axonal protein synthesis. Cell Rep 5: 1564–1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein V, Blank‐Landeshammer B, Müntjes K, Marker R, Teichert I, Feldbrügge M, Sickmann A, Kück U (2020) The STRIPAK signaling complex regulates dephosphorylation of GUL1, an RNA‐binding protein that shuttles on endosomes. PLoS Genet 16: e1008819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg G, Wedlich‐Söldner R, Brill M, Schulz I (2001) Microtubules in the fungal pathogen Ustilago maydis are highly dynamic and determine cell polarity. J Cell Sci 114: 609–622 [DOI] [PubMed] [Google Scholar]
- Steinberg G (2012) The transport machinery for motility of fungal endosomes. Fungal Genet Biol 49: 675–676 [DOI] [PubMed] [Google Scholar]
- Steinberg G (2014) Endocytosis and early endosome motility in filamentous fungi. Curr Opin Microbiol 20: 10–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark H, Aasland R, Driscoll PC (2002) The phosphatidylinositol 3‐phosphate‐binding FYVE finger. FEBS Lett 513: 77–84 [DOI] [PubMed] [Google Scholar]
- Straube A, Enard W, Berner A, Wedlich‐Söldner R, Kahmann R, Steinberg G (2001) A split motor domain in a cytoplasmic dynein. EMBO J 20: 5091–5100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura Y, Kawano S, Endo T (2020) Lipid homeostasis in mitochondria. Biol Chem 401: 821–833 [DOI] [PubMed] [Google Scholar]
- Tanenbaum ME, Gilbert LA, Qi LS, Weissman JS, Vale RD (2014) A protein‐tagging system for signal amplification in gene expression and fluorescence imaging. Cell 159: 635–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauber D, Tauber G, Parker R (2020) Mechanisms and regulation of RNA condensation in RNP granule formation. Trends Biochem Sci 45: 764–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekotte H, Davis I (2002) Intracellular mRNA localization: motors move messages. Trends Genet 18: 636–642 [DOI] [PubMed] [Google Scholar]
- Tian L, Chou HL, Zhang L, Hwang SK, Starkenburg SR, Doroshenk KA, Kumamaru T, Okita TW (2018) RNA‐binding protein RBP‐P is required for glutelin and prolamine mRNA localization in rice endosperm cells. Plant Cell 30: 2529–2552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Chou HL, Fukuda M, Kumamaru T, Okita TW (2020a) mRNA localization in plant cells. Plant Physiol 182: 97–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Doroshenk KA, Zhang L, Fukuda M, Washida H, Kumamaru T, Okita T (2020b) Zipcode RNA‐binding proteins and membrane trafficking proteins cooperate to transport glutelin mRNAs in rice endosperm. Plant Cell 32: 2566–2581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiruchinapalli DM, Oleynikov Y, Kelic S, Shenoy SM, Hartley A, Stanton PK, Singer RH, Bassell GJ (2003) Activity‐dependent trafficking and dynamic localization of zipcode binding protein 1 and beta‐actin mRNA in dendrites and spines of hippocampal neurons. J Neurosci 23: 3251–3261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboi T, Viana MP, Xu F, Yu J, Chanchani R, Arceo XG, Tutucci E, Choi J, Chen YS, Singer RHet al (2020) Mitochondrial volume fraction and translation duration impact mitochondrial mRNA localization and protein synthesis. eLife 9: e57814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tushev G, Glock C, Heumuller M, Biever A, Jovanovic M, Schuman EM (2018) Alternative 3' UTRs modify the localization, regulatory potential, stability, and plasticity of mRNAs in neuronal compartments. Neuron 98: 495–511.e496 [DOI] [PubMed] [Google Scholar]
- Tyagi S, Kramer FR (1996) Molecular beacons: probes that fluoresce upon hybridization. Nat Biotechnol 14: 303–308 [DOI] [PubMed] [Google Scholar]
- Vaccaro MC, Wilding M, Dale B, Campanella C, Carotenuto R (2012) Expression of XNOA 36 in the mitochondrial cloud of Xenopus laevis oocytes. Zygote 20: 237–242 [DOI] [PubMed] [Google Scholar]
- Vardi‐Oknin D, Arava Y (2019) Characterization of factors involved in localized translation near mitochondria by ribosome‐proximity labeling. Front Cell Dev Biol 7: 305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmeister E, Feldbrügge M (2010) Posttranscriptional control of growth and development in Ustilago maydis . Curr Opin Microbiol 13: 693–699 [DOI] [PubMed] [Google Scholar]
- Vollmeister E, Schipper K, Baumann S, Haag C, Pohlmann T, Stock J, Feldbrügge M (2012) Fungal development of the plant pathogen Ustilago maydis . FEMS Microbiol Rev 36: 59–77 [DOI] [PubMed] [Google Scholar]
- Wang C, Han B, Zhou R, Zhuang X (2016) Real‐time imaging of translation on single mRNA transcripts in live cells. Cell 165: 990–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XI, You X, Langer JD, Hou J, Rupprecht F, Vlatkovic I, Quedenau C, Tushev G, Epstein I, Schaefke Bet al (2019) Full‐length transcriptome reconstruction reveals a large diversity of RNA and protein isoforms in rat hippocampus. Nat Commun 10: 5009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washida H, Sugino A, Kaneko S, Crofts N, Sakulsingharoj C, Kim D, Choi SB, Hamada S, Ogawa M, Wang Cet al (2009) Identification of cis‐localization elements of the maize 10‐kDa delta‐zein and their use in targeting RNAs to specific cortical endoplasmic reticulum subdomains. Plant J 60: 146–155 [DOI] [PubMed] [Google Scholar]
- Washida H, Sugino A, Doroshenk KA, Satoh‐Cruz M, Nagamine A, Katsube‐Tanaka T, Ogawa M, Kumamaru T, Satoh H, Okita TW (2012) RNA targeting to a specific ER sub‐domain is required for efficient transport and packaging of alpha‐globulins to the protein storage vacuole in developing rice endosperm. Plant J 70: 471–479 [DOI] [PubMed] [Google Scholar]
- Wedlich‐Söldner R, Bölker M, Kahmann R, Steinberg G (2000) A putative endosomal t‐SNARE links exo‐ and endocytosis in the phytopathogenic fungus Ustilago maydis . EMBO J 19: 1974–1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilding M, Carotenuto R, Infante V, Dale B, Marino M, Di Matteo L, Campanella C (2001) Confocal microscopy analysis of the activity of mitochondria contained within the 'mitochondrial cloud' during oogenesis in Xenopus laevis . Zygote 9: 347–352 [DOI] [PubMed] [Google Scholar]
- Wilkie GS, Davis I (2001) Drosophila wingless and pair‐rule transcripts localize apically by dynein‐mediated transport of RNA particles. Cell 105: 209–219 [DOI] [PubMed] [Google Scholar]
- Williams CC, Jan CH, Weissman JS (2014) Targeting and plasticity of mitochondrial proteins revealed by proximity‐specific ribosome profiling. Science 346: 748–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Eliscovich C, Yoon YJ, Singer RH (2016) Translation dynamics of single mRNAs in live cells and neurons. Science 352: 1430–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Kozlov G, Gehring K (2014) The "tale" of poly(A) binding protein: the MLLE domain and PAM2‐containing proteins. Biochim Biophys Acta 1839: 1062–1068 [DOI] [PubMed] [Google Scholar]
- Yan X, Hoek TA, Vale RD, Tanenbaum ME (2016) Dynamics of translation of single mRNA molecules in vivo . Cell 165: 976–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon BC, Jung H, Dwivedy A, O'Hare CM, Zivraj KH, Holt CE (2012) Local translation of extranuclear lamin B promotes axon maintenance. Cell 148: 752–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon YJ, Wu B, Buxbaum AR, Das S, Tsai A, English BP, Grimm JB, Lavis LD, Singer RH (2016) Glutamate‐induced RNA localization and translation in neurons. Proc Natl Acad Sci USA 113: E6877–E6886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabezhinsky D, Slobodin B, Rapaport D, Gerst JE (2016) An essential role for COPI in mRNA localization to mitochondria and mitochondrial function. Cell Rep 15: 540–549 [DOI] [PubMed] [Google Scholar]
- Zander S, Baumann S, Weidtkamp‐Peters S, Feldbrügge M (2016) Endosomal assembly and transport of heteromeric septin complexes promote septin cytoskeleton formation. J Cell Sci 129: 2778–2792 [DOI] [PubMed] [Google Scholar]
- Zappulo A, van den Bruck D, Ciolli Mattioli C, Franke V, Imami K, McShane E, Moreno‐Estelles M, Calviello L, Filipchyk A, Peguero‐Sanchez Eet al (2017) RNA localization is a key determinant of neurite‐enriched proteome. Nat Commun 8: 583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HL, Eom T, Oleynikov Y, Shenoy SM, Liebelt DA, Dictenberg JB, Singer RH, Bassell GJ (2001) Neurotrophin‐induced transport of a beta‐actin mRNP complex increases beta‐actin levels and stimulates growth cone motility. Neuron 31: 261–275 [DOI] [PubMed] [Google Scholar]
- Zhou L, Obhof T, Schneider K, Feldbrügge M, Nienhaus GU, Kämper J (2018) Cytoplasmic transport machinery of the SPF27 homologue Num1 in Ustilago maydis . Sci Rep 8: 3611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimyanin VL, Belaya K, Pecreaux J, Gilchrist MJ, Clark A, Davis I, St Johnston D (2008) In vivo imaging of oskar mRNA transport reveals the mechanism of posterior localization. Cell 134: 843–853 [DOI] [PMC free article] [PubMed] [Google Scholar]


