Abstract
The CRISPR/Cas9 system is a widely used tool for genome editing in plants. In Arabidopsis (Arabidopsis thaliana), egg cell-specific promoters driving Cas9 expression have been applied to reduce the proportion of T1 transformants that are chimeras; however, this approach generally leads to relatively low mutagenesis rates. In this study, a GLABRA2 mutation-based visible selection (GBVS) system was established to enrich nonchimeric mutants among T1 plants generated by an egg cell-specific CRISPR/Cas9 system. GBVS generally enhanced mutation screening, increasing the frequency by 2.58- to 7.50-fold, and 25%–48.15% of T1 plants selected through the GBVS system were homozygous or biallelic mutants, which was 1.71- to 7.86-fold higher than the percentage selected using the original system. The mutant phenotypes of T2 plants were not obviously affected by the glabrous background for all four target genes used in this study. Additionally, the nonchimeric pyrabactin resistance 1 (PYR1)/PYR1-like 1 (PYL1) and PYL2 triple mutant pyr1/pyl1/pyl2 could be obtained in the T1 generation with a ratio of 26.67% when GBVS was applied. Collectively, our results show that compared with the known CRISPR/Cas9 systems, the GBVS system described here saves more time and labor when used for the obtainment of homozygous or biallelic monogenic mutants and nonchimeric polygenic mutants in Arabidopsis.
Homozygous or biallelic Arabidopsis mutants enriched through GLABRA2-based visible selection in CRISPR/Cas9-edited T1 plants produced seeds suitable for phenotyping.
Introduction
Streptococcus-derived clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated 9 (Cas9) systems have been widely applied to modify the genomes of various organisms, including diverse plant species (Gaillochet et al., 2021). Arabidopsis (Arabidopsis thaliana) is a valuable model plant for investigating the mechanisms of plant development and stress responses (Koornneef and Meinke, 2010). Most of the knowledge obtained from Arabidopsis could potentially be translated to other plants. Thus, facilitating the screening of mutants generated by the CRISPR/Cas9 system in Arabidopsis can efficiently accelerate plant research. However, there are still persisting challenges related to the relatively low efficiency of heritable mutation when using the CRISPR/Cas9 system in Arabidopsis (Khumsupan et al., 2019).
When using the cauliflower mosaic virus (CaMV) 35S promoter to drive Cas9 expression in Arabidopsis, most mutants identified in the first generation are chimeras with nonheritable mutations (Feng et al., 2014). This may be due to the weak activity of the CaMV 35S promoter in germ line cells and zygotes, and high activity in somatic cells (Feng et al., 2014). The high somatic mutation rate results in a heavy workload for scientists detecting heritable mutations in each generation. To solve this problem, several highly efficient promoters, such as the ubiquitin, ribosomal protein S5A, and Yao promoters, have been employed in Arabidopsis (Yan et al., 2015; Tsutsui and Higashiyama, 2017; Ramona Grützner, 2020; Wolabu et al., 2020). Compared with the 35S promoter, these promoters dramatically elevate the proportion of T1 plants with mutations, especially heritable mutations. Meanwhile, to evaluate gene-targeting efficiency, Cas9 was fused with fluorescence protein to indicate the expression level of Cas9 by fluorescence intensity (Wang and Chen, 2019). However, there is still a high rate of somatic mutations.
As egg cells are the targets of Agrobacterium tumefaciens-mediated floral dip transformation in Arabidopsis (Clough and Bent, 1998; Ye et al., 1999), germ line- and zygote-specific genome editing might be apt to create nonchimeric mutants. Thus, egg cell-specific promoters of the egg cell secretion protein 1 (EC1) genes and the male gametocyte-specific promoter of sporocyteless (SPL) were used to drive Cas9 expression (Wang et al., 2015; Mao et al., 2016). Mutations were rarely detected in pSPL:Cas9 T1 plants but were abundant in T2 plants (Mao et al., 2016). pEC1:Cas9 usually created nonchimeric mutants in the T1 generation, but its efficiency was relatively low (Wang et al., 2015).
In multiplex genome edited plants, multiple mutations at different sites were more frequently detected than single site mutations, indicating that co-editing is a common phenomenon (Zhang et al., 2016; Wang et al., 2019). These results hint that some target site mutations can be used as proxies for high expression of Cas9. Thus, adding co-editing markers to the CRISPR/Cas system should be an easier strategy to enrich and select cells or plants edited at target sites. This strategy has recently been applied to enrich mutants generated by cytosine base editors and adenine base editors (Zhang et al., 2019; Xu et al., 2020).
Several genes, such as GLABRA1 (GL1) and GL2, are responsible for trichome establishment; mutations in these genes result in plants with glabrous leaves and stems (Szymanski et al., 2000). Many genes have been studied in no-trichome backgrounds, and mutation of these trichome-determining genes has a minor effect on the exploration of other gene functions (Liu and Zhu, 1997; Zhu et al., 2002; Yamasaki et al., 2007). The glabrous phenotype is clearly visible and has been used to evaluate gene-targeting efficiency (Miki et al., 2018). Thus, genes determining trichome development might serve as co-editing markers to enrich for Cas9-generated indel (insertion and deletion) mutants.
In this study, we established a GL2 mutation-based visible selection (GBVS) system to improve the screening efficiency of egg cell-specific CRISPR/Cas9 system in Arabidopsis. Homozygous or biallelic mutants of target sites of interest can be easily selected in the T1 generation, and then these plants or their progenies can be used for phenotyping. Nonchimeric polygenic mutant could also be efficiently identified. Our results indicate that GBVS is a labor- and time-conserving strategy when using the CRISPR/Cas9 system to generate nonchimeric mutations at target sites in Arabidopsis.
Results
Design of the GBVS system
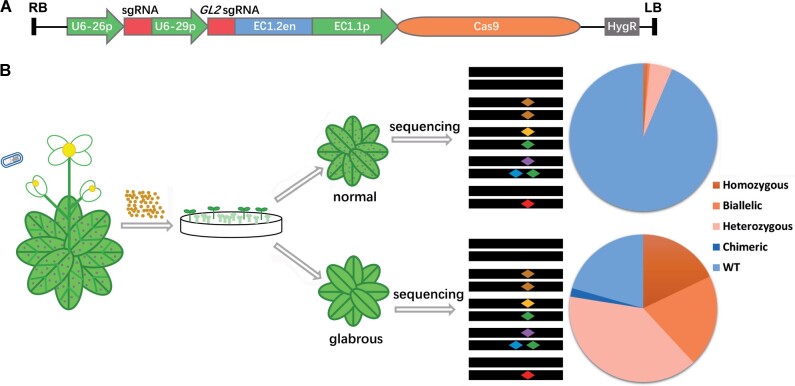
To facilitate the screening for Cas9-created Arabidopsis T1 nonchimeric mutants using a visible marker, we employed GL2, mutations of which produce an obviously glabrous phenotype on true leaves (Szymanski et al., 2000). GL2 acts downstream of the previously reported co-editing marker GL1 (Hahn et al., 2017; Li et al., 2020). Compared with GL1, GL2 may regulate fewer cellular processes. Somatic mutation is the key factor that leads to the generation of chimeric Arabidopsis mutants in the T1 generation. Thus, we used pHEE401E, in which the enhancer of the egg cell-specific promoter EC1.2 fused to the EC1 promoter drives the expression of Cas9 (Wang et al., 2015). The single guide RNA (sgRNA) for GL2, driven by the AtU6 promoter, was introduced to pHEE401E to create a new vector designated pEC1-GL2 (Figure 1A).
Figure 1.
Schematic showing the strategy for enrichment of homozygous and biallelic mutants in T1 plants using GBVS. A, The structure of the pEC1-GL2 plasmid. The expression of sgRNAs for GL2 and the target locus was driven by the U6-29 and U6-26 promoters, respectively. The expression of Cas9 was driven by the EC1 promoter (EC1.2 enhancer plus EC1.1 promoter). HygR, hygromycin resistance gene; RB and LB, T-DNA right and left borders, respectively. B, Outline of the GBVS strategy. pEC1-GL2 containing an sgRNA for the target locus is transformed into Col-0 plants. T1 transgenic lines are isolated on selection media and are shown as green, large seedlings. The glabrous (leaves with no trichomes, which are represented by dots) T1 plants are sequenced for indels in target gene. We assume mutations in the target gene will be enriched or reduced in glabrous and normal plants, respectively. Different colored diamonds indicate different mutations in the target site. Percentages of normal and glabrous T1 plants with WT sequences and different types of target site mutations are indicated. Portions of the images were modified from the Microsoft PowerPoint clip art database.
The GBVS strategy is shown in Figure 1B. The sgRNA for target gene of interest is cloned into pEC1-GL2 and the final construct is transferred into Arabidopsis. Stable T1 transgenic lines are isolated on selection media and transplanted to soil and grown for 2 weeks. Then the glabrous plants are sequenced to identify mutation events at target sites. We predict that indels in the target gene will be enriched in glabrous plants and reduced in normal plants.
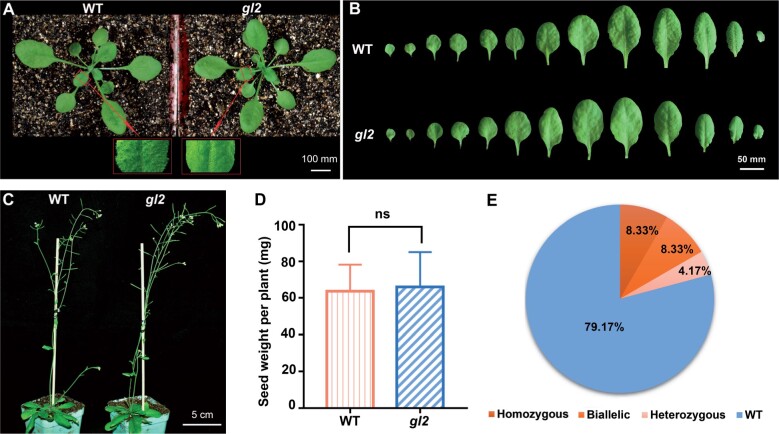
To build an efficient GBVS system, we chose a previously reported sgRNA targeting GL2 to construct pEC1-GL2 (Supplemental Figure S1; Mao et al., 2016), and transformed Arabidopsis with this vector containing only the GL2 sgRNA. 24 T1 plants were obtained, and four of them showed a glabrous phenotype (Figure 2A) but no other obvious growth defects (Figure 2, B–D). GL2 in all 24 lines was sequenced. As summarized in Figure 2E, 8.33% of the T1 plants were homozygotes, 8.33% were biallelic mutants, 4.17% were heterozygotes and 79.17% were wild-type (WT) plants (Figure 2E). No chimera was detected. The mutation efficiency of GL2 in the T1 generation was 20.83%. The four glabrous mutants were all homozygous or biallelic mutants (Supplemental Figure S2). All these results suggest that GL2 is an appropriate co-editing marker and the mutation efficiency of this GL2 target site is reasonable for a proxy.
Figure 2.
GL2 is an effective co-editing marker. A, The glabrous phenotype of the gl2 mutant. 3-week-old plants were photographed. Part of the seventh leaf of both WT and gl2 were enlarged to show the trichomes clearly. B, The 1st to 13th rosette leaves of representative 2-month-old WT and gl2 mutant plants. C, Representative 2-month-old WT and gl2 mutant plants. D, Seed weight per plant for WT and the gl2 mutant (n = 20); E, Percentages of WT, homozygous, heterozygous, and biallelic mutations at the GL2 site identified in 24 T1 plants. Data are shown as mean ± sd (n = 22). ns, no significant difference, P > 0.05 determined by Student’s t test.
GBVS enriches homozygous and biallelic mutations at target sites
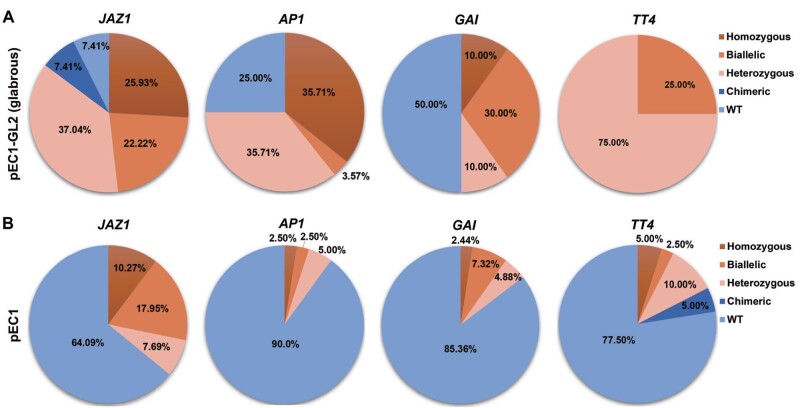
To demonstrate the enrichment for indels at target sites and test if homozygotes or biallelic mutants could be easily selected in the T1 generation using GBVS, sgRNAs targeting four genes, jasmonate-zim-domain protein 1 (JAZ1), apetala 1 (AP1), gibberellic acid insensitive (GAI), and transparent testa 4 (TT4), were individually cloned into pEC1-GL2 and transformed into Arabidopsis (Supplemental Figure S1). More than 100 T1 plants (total) were obtained for each target site, and 3.48%–24.56% of T1 plants showed a glabrous phenotype (glabrous; Supplemental Table S1). All T1 plants were sequenced to identify indels in these target sites. GBVS significantly improved the selection of mutations in target genes. For example, GBVS enhanced the screening efficiency from 32.77% in the total group to 92.59% for JAZ1, and from 20.18% to 75.00% for AP1 (Figure 3A; Supplemental Figure S3A). The GAI and TT4 loci in the total group had low indel frequencies, 7.26% and 7.83%, respectively, which increased by 6.89- and 12.78-fold with GBVS (Figure 3A; Supplemental Figure S3A). The ratios of homozygous and biallelic mutants in the target sites obtained using the GBVS system also dramatically increased, by 3.82-fold for JAZ1, 4.07-fold for AP1, 9.92-fold for GAI, and 7.18-fold for TT4 (Figure 3A; Supplemental Figure S3A). We further analyzed the indel frequencies at target sites in the T1 plants with normal trichomes (normal), and found that only 2.33%–15.22% of T1 plants were mutated, mostly heterozygotes (Supplemental Figure S3B). For example, the mutagenesis rates for the GAI locus were 7.26% in the total group, 50% in the glabrous group, and 3.51% in the normal group. Half of the GAI mutations in the glabrous group were homozygous or biallelic, while only 0.88% of those in the normal group were homozygous and biallelic (Figure 3A; Supplemental Figure S3). To further verify the mutation enrichment of GBVS, we transformed pEC1-GL2 vector harboring the sgRNA for JAZ1 and GAI target sites again and got similar results shown that GBVS facilitated the mutation screening of T1 plants (Supplemental Figure S4).
Figure 3.
Mutation types and frequencies at target sites with or without GBVS. A, Percentages of WT and different types of mutation at four target sites selected using GBVS (pEC1-GL2 glabrous). B, Percentages of WT and different types of mutation at four target sites using the original system (pEC1).
The indel frequency might be affected when co-expressing two sgRNAs. To test whether the co-expression of sgRNAs targeting GL2 and the other target locus affects the generation of indels, we compared the frequency of indels created using the pEC1-GL2 and pEC1 (pHEE401E, which we renamed as pEC1 for comparing with pEC1-GL2). The indel frequency for TT4 was lower when the two sgRNAs were expressed using pEC1-GL2 than when only the target locus sgRNA was expressed using pEC1, decreasing from 22.50% to 7.83% (Supplemental Table S2). Unexpectedly, the indel frequency for AP1 was dramatically higher when using the pEC1-GL2 vector (Supplemental Table S2). These results indicate that targeting two loci simultaneously does affect the mutation efficiency.
Compared with the pEC1 group, the frequency of indel generation after applying GBVS was significantly higher for all four sites (Supplemental Table S2). GBVS enhanced the screening efficiency from 35.91% to 92.59% for JAZ1, from 10.00% to 75.00% for AP1, from 14.64% to 50.00% for GAI, and from 22.50% to 100% for TT4 (Figure 3; Supplemental Table S2). We further analyzed the ratio of homozygous and biallelic mutants in T1 plants before and after applying GBVS. A great enrichment of homozygous or biallelic mutants was observed for all four loci using GBVS (Figure 3). The ratio of homozygous and biallelic mutants of target sites was increased 1.71-fold for JAZ1, 7.86-fold for AP1, 4.10-fold for GAI, and 3.33-fold for TT4 after applying GBVS (Figure 3). This result indicates that using GBVS for the screening of homozygous and biallelic mutants could save lots of time and effort. For example, we had to sequence 20 T1 plants to obtain one homozygous or biallelic ap1 mutant with original system, while with GBVS, sequencing three T1 glabrous plants was sufficient (Figure 3).
As meristem-specific promoters could cause a high indel frequency (Yan et al., 2015), we compared the mutagenesis rate between GBVS and a meristem-expressed CRISPR/Cas9 system (pYao), in which the expression of Cas9 is driven by the YAO promoter. The pYao group had the highest indel frequency for all loci among the three vectors we used in this study, while most T1 plants were chimeras or heterozygotes (Supplemental Table S2 and Supplemental Figure S5). After the application of GBVS, the glabrous group had higher ratios of homozygous or biallelic mutants, and even higher mutagenesis rates than the pYao group at most sites (Figure 3A; Supplemental Figure S5). These results implied that GBVS effectively enriched mutations, especially homozygous or biallelic mutations, at target sites in T1 plants.
The homozygous or biallelic mutations obtained through the GBVS system are heritable
To ensure that the mutations at target sites in the T1 homozygous or biallelic mutants generated by the GBVS system were heritable, we checked the mutation status of 220 progenies of 11 different T1 lines. These lines included six homozygotes (#19, #33, and #41 for JAZ1, #10 and #59 for AP1, and #3 for GAI), and five biallelic mutants (#4 for JAZ1, #33 for AP1, #27 and #56 for GAI, and #8 for TT4). All T2 plants showed the glabrous phenotype, which means that the mutations in GL2 for all 11 T1 lines were heritable. We then sequenced the corresponding target loci in the T2 plants, and found that the same mutation was faithfully passed down in all of them (Table 1). No heterozygote or chimera was detected. These results demonstrated that all homozygous and biallelic mutations we generated with the GBVS system were heritable.
Table 1.
Segregation patterns of homozygous and biallelic T1 mutations
| Line | T1 |
T2 |
||||||
|---|---|---|---|---|---|---|---|---|
| Mutation Type | Indels | Number of Lines Tested | M1 (Homozygote) | M2 (Homozygote) | M1 and M2 (Biallele) | Number of New Mutations | Number of Reversions | |
| jaz1 #4 | Biallele | M1: D (TTGACA) | 20 | 5 | 6 | 9 | 0 | 0 |
| M2: I (A) | ||||||||
| jaz1 #19 | Homozygote | M1: D (ACAGAACTTCCTATTG) | 20 | 20 | 0 | 0 | 0 | 0 |
| jaz1 #33 | Homozygote | M1: D (CATTGAC) | 20 | 20 | 0 | 0 | 0 | 0 |
| jaz1 #41 | Homozygote | M1: D (ACAGAACTTCCTATTG) | 20 | 20 | 0 | 0 | 0 | 0 |
| ap1 #10 | Homozygote | M1: D (T) | 20 | 20 | 0 | 0 | 0 | 0 |
| ap1 #33 | Biallele | M1: I (T) | 20 | 6 | 5 | 9 | 0 | 0 |
| M2: I (C) | ||||||||
| ap1 #59 | Homozygote | M1: D (T) | 20 | 20 | 0 | 0 | 0 | 0 |
| gai #3 | Homozygote | M1: D (GTT) | 20 | 20 | 0 | 0 | 0 | 0 |
| gai #27 | Biallele | M1: D (T) | 20 | 5 | 7 | 8 | 0 | 0 |
| M2: D (AGCTGTTCT) | ||||||||
| gai #56 | Biallele | M1: D (G) | 20 | 4 | 9 | 7 | 0 | 0 |
| M2: D (TT) | ||||||||
| tt4 #8 | Biallele | M1: I (A) | 20 | 6 | 6 | 8 | 0 | 0 |
| M1: D (CAA) | ||||||||
Twenty T2 seedlings from each T1 homozygous or biallelic line were genotyped by sequencing. M1 and M2 indicate the type of mutation in T1 plants. D indicates deletion and I indicates insertion at target sites.
GL2 mutations have no obvious effect on mutant phenotyping of target sites
In addition to the target sites, the GL2 gene is also mutated when using the GBVS system. Previous reports indicate that the glabrous background has minor effect on the exploration of gene function, and many genes have been studied in glabrous backgrounds in which GL1 is mutated (Liu and Zhu, 1997; Zhu et al., 2002; Yamasaki et al., 2007). Thus, we predicted that the GL2 mutation would not impede the functional studies of target genes.
To test this prediction, we evaluated the effects of GL2 mutation on several different processes involved in plant development and responses to hormone treatment: flower development, root growth, seed color, response to jasmonic acid (JA), and response to gibberellic acid (GA). As shown in Supplemental Figure S6, there was no obvious difference in flower development or seed color between WT and the gl2 mutant. We also grew WT and gl2 mutant plants on 1/2 Murashige and Skoog (MS) plates supplemented with or without 50 µM JA. Both WT and the gl2 mutant had shorter primary roots on the JA plate than on the control plate as expected (Supplemental Figure S7, A and B). The primary roots of the two genotypes looked similar on both plates, and no significant differences were detected after analyzing root length (Supplemental Figure S7, A and C). We observed no obvious difference between WT and the gl2 mutant when seedlings were treated with GA (Supplemental Figure S8). These data indicate that the GL2 mutation does not affect plant root growth or response to plant hormones. Taken together, these results demonstrate that mutation of GL2 has no obvious effect on these plant developmental and stress-responsive processes.
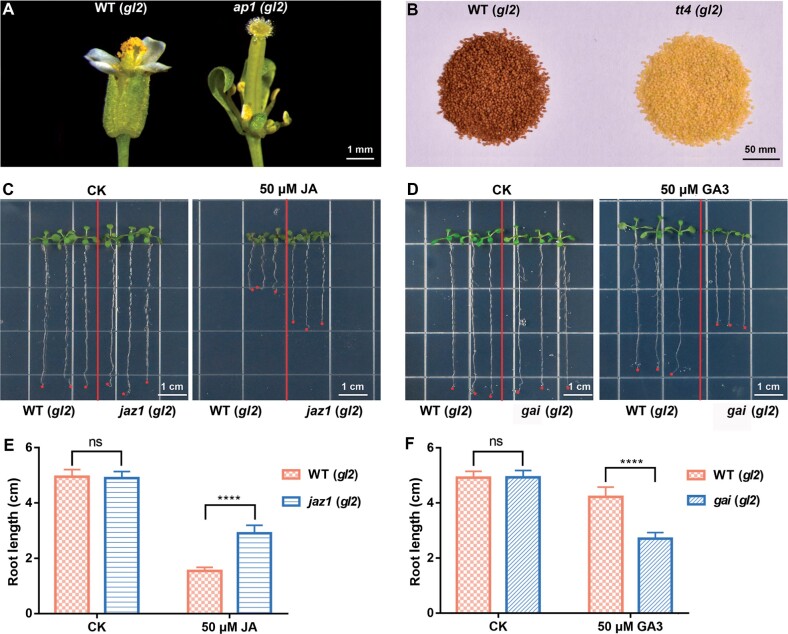
We next checked the phenotypes of the tt4 and ap1 mutants in the gl2 background. AP1 is a floral homeotic gene, and the ap1 mutant lacks petals (Irish and Sussex, 1990). Consistent with previous reports, all flowers of ap1 #10 showed an abnormal floral developmental phenotype (Figure 4A). The tt4 mutation disrupts the synthesis of brown pigment in the seed coat, thus the mutant seeds are yellow instead of dark brown (Shirley et al., 1995). As shown in Figure 4B, all the T1 seeds of tt4 #8 were yellow as expected, consistent with the finding that all these seeds are homozygous or biallelic for mutations in the TT4 locus (Table 1).
Figure 4.
The homozygous and biallelic mutants obtained using GBVS are suitable for phenotyping. All genotypes used in this figure are in the gl2 background. A, Representative flowers of WT and ap1 T2 mutant. B, Seeds of WT and the tt4 T1 mutant. C, Representative seedlings of WT and the jaz1 T2 mutant grown on 1/2 MS plates with or without 50 µM JA. D, Representative seedlings of WT and the gai T2 mutant grown on 1/2 MS plates with or without 50-µM GA. E, The analyses of primary root length of WT and jaz1 mutant seedlings grown on 1/2 MS plates with or without 50 µM JA. F, The analyses of primary root length of WT and gai mutant seedlings grown on 1/2 MS plates with or without 50 µM GA. Data are shown as mean ± sd (n = 15). ns, no significant difference; ****P < 0.0001 determined by Student’s t test.
The phenotypes of the jaz1 and gai mutants were also examined under the gl2 background. JAZ1 belongs to a transcription repressor family involved in JA signaling. The JAZ1 sgRNA, which was designed to target the coding region of the JA-associated domain, generates mutants failing to respond to exogenous JA (Thines et al., 2007). As expected, the T2 seedlings of jaz1 #33 were less sensitive to JA when grown on 1/2 MS plates supplemented with 50 µM JA (Figure 4, C and E). GAI is a transcriptional repressor of gibberellin signaling; thus, the gai mutants might be more sensitive to exogenous GA (Peng et al., 1997). As shown in Figure 4D, a high concentration GA repressed primary root growth, and the progenies of gai #27 had shorter primary roots than WT seedlings on GA plates (Figure 4, D and F). Collectively, these results indicate that the mutation of GL2 has no obvious effects on traits that might affect the exploration of gene function. Thus, the GL2 is a suitable co-editing marker for identifying mutations in other genes.
GBVS enhances the screening of nonchimeric polygenic mutant
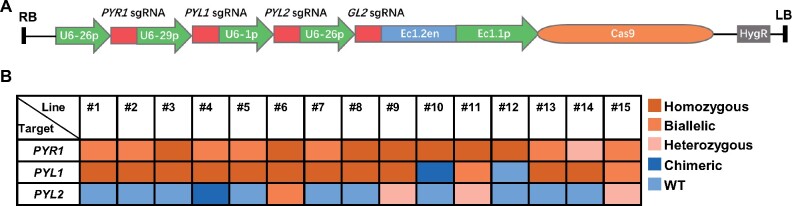
One of the biggest advantages of the CRISPR/Cas9 system is the ability to simultaneously target multiple sites, so we further explored the effectiveness of GBVS for multiplexing by targeting three genes from the abscisic acid (ABA) receptor family, the pyrabactin resistance 1 (PYR1), PYR1-like 1 (PYL1), and PYL2 (Gonzalez-Guzman et al., 2012). The sgRNAs for these genes were expressed from different sgRNA transcriptional units in one construct (Figure 5A; Supplemental Figure S9).
Figure 5.
Mutation types at each target site of multiplex edited T1 plants with GBVS. A, The structure of the pEC1-GL2 plasmid with multiple sgRNAs. The expression of sgRNAs for GL2 and the target loci (PYR1, PYL1, and PYL2) was driven by the U6-26, U6-29, and U6-1 promoters, respectively. HygR, hygromycin resistance gene; RB and LB, T-DNA right and left borders, respectively. B, Mutation types of PYR1, PYL1, and PYL2 loci were analyzed in individual T1 plants showing glabrous phenotype.
Fifteen T1 plants showed a glabrous phenotype among the 128 transformants of this multiple sgRNA vector, and were further sequenced to identify indels in the corresponding target sites. The sequencing results showed that PYR1 mutation was presented in all 15 plants (seven homozygotes, seven biallelic mutants, and one heterozygote), PYL1 mutation showed in 14 plants (11 homozygotes, 2 biallelic mutants, and 1 chimera), and PYL2 mutation happened in five plants (one biallelic mutant, three heterozygotes, and one chimera; Figure 5B). Among them, five plants were mutated in all target sites, four were nonchimeric mutants (26.67%), and one was homozygous or biallelic mutant (6.67%) for all three sites. We further randomly analyzed the indel frequency at target sites in 20 T1 plants with normal trichomes and found that only two plants were nonchimeric (10%) and none of them was homozygous or biallelic for all three target sites (Supplemental Figure S10). Compared with T1 plants with normal trichomes, GBVS also increased the ratios of nonchimeric mutants in these target sites by 4.00-fold for PYR1, 2.17-fold for PYL1, and 1.78-fold for PYL2 (Figure 5B; Supplemental Figure S10). Collectively, nonchimeric polygenic mutants could be enriched with GBVS in T1 plants.
Discussion
The egg cell-specific CRISPR/Cas9 system generally has a relatively low mutagenesis rate in Arabidopsis, especially for some target sites, such as AP1 in this study. Here, we introduced GBVS to solve this problem. Different from a previously published paper, in which GL1 was used as co-editing marker, we used the EC1 promoter instead of the ubiquitin promoter to drive the expression of Cas9 to reduce the frequency of somatic mutations (Li et al., 2020). The majority of T1 mutants we obtained in this study were homozygous, biallelic, or heterozygous.
The GBVS strategy efficiently increased, by 2.58- to 7.50-fold, the screening efficiency at the tested target sites. We also found a 1.71- to 7.86-fold increase of the frequency of homozygous and biallelic mutants identified among edited T1 plants using the GBVS strategy. These T1 homozygous and biallelic mutants or their T2 progenies could be used for phenotyping, which would save scientists at least 3 months. No obvious difference between WT and the gl2 mutant was observed for the investigated developmental and hormone-responsive processes, further demonstrating that GL2 is an excellent co-editing selection marker for CRISPR/Cas9 system. Based on these results, we assume GL2 could also serve as a co-editing marker for base editors, especially in Arabidopsis. We propose that the GBVS strategy is suitable for various purposes, such as the exploration of gene functions, as 12% of Arabidopsis genes have no mutation in T-DNA collections (O'Malley and Ecker, 2010). In addition, the GBVS system is especially important for functional studies of noncoding regions, including cis-elements in promoters, upstream open reading frames in the 5′-untranslated region (UTR), and small RNA genes, as current mutant libraries can hardly meet the strict requirements of these regions. The GBVS system established here could also save time and effort for scientists who desire to generate mutant pools to explore the functions of gene families or search for key cis-elements in promoter regions regulating genes of interest. We also observed that the frequencies of GL2 mutation-caused glabrous phenomenon were highly variable (from 3.48% to 24.56%) when co-edited with different target sites. This might due to the variation of sgRNA expression among independent transformation events. Thus, to increase GBVS efficiency, strong RNA polymerase II promoter could be applied to drive the expression of sgRNA precursors, in which sgRNA was flanked with tRNAs, self-cleaving ribozyme sequences or Csy4-recognized hairpin (Gao and Zhao, 2014; Xie et al., 2015; Cermak et al., 2017).
Compared with the previous pEC system and other CRISPR/Cas systems using highly efficient promoters, GBVS efficiently increases the screening efficiency of nonchimeric mutants, which makes the identification of homozygous or biallelic mutants easier in T1 plants. The GBVS system we established here is also very convenient, as it enriches mutants with a visible phenotype as a marker; no additional treatment or equipment is required, unlike other co-editing marker systems (Zhang et al., 2019; Xu et al., 2020). However, GBVS may not be suitable for all target loci. For example, GBVS may not be applicable to the mutation enrichment of many genes involved in plant defense, as trichome plays an important role in this process. And the high editing efficiency in gl2 mutant plants may be accompanied by multiple-copy insertions, which would make it more difficult to get rid of the transgenes in progenies.
In addition to GL2, we assume that genes for which mutants have visible phenotypes could be applied as co-editing markers for the CRISPR/Cas9 systems. These phenotypes include, but are not limited to, alterations in leaf shape, leaf number, leaf color, flowering time, hypocotyl length, plant height, and root hairs. For example, peapod2 (PPD2) and elongated hypocotyl 5 (HY5) can serve as co-editing markers in Arabidopsis, as the ppd2 mutant has propeller-like rosettes with increased lamina size and dome-shaped leaves, and hy5 has a longer hypocotyl than WT under light conditions (Oyama et al., 1997; Baekelandt et al., 2018). Genes that are involved in plant responses to the environment are suitable candidates too, such as abscisic acid insensitive 4 (ABI4). The abi4 mutant has reduced sensitivity to ABA inhibition of seed germination (Leon et al., 2012) and can germinate in the presence of 5-µM ABA; thus, the seeds harvested from T0 plants could first be selected on ABA plates to identify edited lines. One criterion for choosing the co-editing marker is that it does not affect subsequent studies of gene function. An alternative way is removing the mutation in the co-edited marker by crossing before functional study of the target gene.
There should be mutations that cause visible phenotypes in all plants, such as the glabrous phenotype of the soybean (Glycine max) Gmnap1 and the high anthocyanin level in the tomato high pigment 2 mutant (Mustilli et al., 1999; Tang et al., 2020). Thus, visible phenotype-based selection could be applied to the whole plant kingdom, especially to species or inbred lines that are sensitive to selection using typical marker genes, such as antibiotic resistance genes and herbicide resistance genes. For some vegetative propagated plants, such as potato (Solanum tuberosum) and sweet potato (Ipomoea batatas), or some trees that have a long juvenile period, such as pear (Pyrus communis) and apple (Malus domestica), using a co-editing marker could enrich mutations in target sites and avoid the use of antibiotic genes, which are generally opposed by the public.
Conclusions
Mutations at target sites generated using the CRISPR/Cas9 system can be enriched using GBVS in Arabidopsis. GBVS enhanced the screening efficiency by 2.58- to 7.50-fold, and more than one quarter of T1 plants had homozygous or biallelic mutations, which could be passed down to the next generation. We further confirmed that mutations in GL2 have no obvious effect on exploring the functions of co-edited genes. Nonchimeric triple mutants could also be identified in T1 plant with a ratio of 26.67% when GBVS applied. We assume that visible phenotype-based selection, taking GBVS as an example, could be adapted to all plant species to increase the screening efficiency of CRISPR/Cas9 systems.
Materials and methods
Vector construction
To construct the pEC1-GL2 vector, which targeted the GL2 locus as a co-editing marker, GL2 sgRNA was introduced into GL2-2Tar-R primer. Then primer pair GL2-2Tar-F/R was used to amplify a sequence containing the U6 terminator-U6 promoter-GL2 sgRNA element using pCBC-DT1T2 plasmid as a template (Xing et al., 2014; Supplemental Table S3). By Gibson Assembly, this element was ligated into the pHEE401E binary vector (Wang et al., 2015) digested by restriction endonuclease BsaI. We added the BsaI restriction site to the GL2-2Tar-F/R primers, so the final vector still retained the BsaI cutting site, allowing for the insertion of sgRNAs for target loci of interest.
To construct the pEC1 vectors for targeting JAZ1, AP1, GAI, or TT4, different pairs of DNA oligos coding for the designed sgRNAs were annealed to form double-stranded DNA (dsDNA). Then dsDNA was ligated into the pHEE401E plasmid, which was digested with BsaI, to generate the corresponding sgRNA construct. The sequences of the synthesized DNA oligos and all the primers used in this study are listed in Supplemental Table S3. For the pEC1-GL2 constructs for targeting JAZ1, AP1, GAI, or TT4, oligo pairs were annealed to generate dsDNAs, which were cloned into the BsaI sites of separate pEC1-GL2 vectors.
To construct the pYao vector, the primer pair Yaop-F/R was used to amplify the pYao promoter from the pYao-Cas9 plasmid (Yan et al., 2015). The amplicon was cloned into pHEE401E digested by SpeI and XbaI. For the pYao constructs targeting JAZ1, AP1, GAI, or TT4, each oligo pair was annealed to generate dsDNA, which was cloned into separate pYao plasmids digested by BsaI to generate the corresponding sgRNA construct.
To construct the pEC1-GL2 vector for targeting PYR1, PYL1 and PYL2, their sgRNAs were introduced into 4DT-1F, 4DT-1R and 4DT-2F, or 4DT-2R primers, respectively. The 4DT–1F/R primer pair was used to amplify PYR1 sgRNA-U6 terminator-U6 promoter-PYL1 sgRNA fragment from pCBC-DT1T2 plasmid (Xing et al., 2014). The PYL1 sgRNA-U6 terminator-U6 promoter-PYL2 sgRNA fragment was amplified from pCBC-DT2T3 plasmid using 4DT-2F/R (Xing et al., 2014). Then these two PCR fragments were ligated into pEC1 plasmid digested with BsaI by Gibson Assembly to create a middle construct. A KpnI restriction site was contained in 4DT-2R primer, thus the U6 terminator-U6 promoter-GL2 sgRNA element amplified from pCBC-DT3T4 by 4DT-3F/4DT-3R could be cloned into KpnI site of middle construct to produce the final construct targeting all four genes (Xing et al., 2014).
Plant growth conditions and transformation
Arabidopsis (Arabidopsis thaliana) ecotype Columbia (Col-0) was used in this study, and all plants were grown under 16-h light/8-h dark at 22°C. Agrobacterium tumefaciens-mediated transformation of Col-0 was performed as per the floral dip method described previously (Clough and Bent, 1998). Seeds harvested from the A. tumefaciens-infected plants were sterilized with 2.5% (v/v) plant preservative mixture for 2 d at 4°C and selected on 1/2 strength MS medium containing 25 mg·L–1 hygromycin, plus 100 mg·L–1 carbenicillin to inhibit A. tumefaciens growth. The T1 transformants were transplanted to soil 2 weeks later.
DNA extraction, PCR, and sequencing analysis
Genomic DNA was extracted by grinding leaves using DNA extraction buffer (50 mM Tris–HCl [pH 7.5], 300 mM NaCl, 300 mM sucrose) in 300 μL centrifuge tubes. Then the tubes were heated at 95°C for 10 min, followed by centrifugation for 5 min at 2,500g. The supernatants were transferred to new tubes and used as PCR templates. PCR was performed to identify mutations in T1 and T2 plants using the primer pairs listed in Supplemental Table S3. The PCR products were sequenced, and then the sequencing results were decoded online (http://skl.scau.edu.cn/dsdecode/; Liu et al., 2015).
Phenotype analysis of gl2, jaz1, and gai mutants
The seeds of WT, gl2 #21, jaz1 #33, and gai #27 were sterilized and sown on vertically oriented 1/2 MS plates supplemented with or without 50 μM MeJA or 50 μM GA3 as indicated. Photographs were taken 10 d later. Root length of seedlings was measured using ImageJ. Student’s t tests and two-tail t tests were performed using GraphPad Prism 7.0 software.
Accession numbers
GL2: At1g79840; AP1: At1g69120; TT4: At5g13930; JAZ1: At1g19180; GAI: At1g14920; PYR1: At4g17870; PYL1: At5g46790; PYL2: At2g26040; sequences for plasmids used in this paper can be found in Supplemental Data Set S1.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. The target sites of genes for monogenic mutation used in this study.
Supplemental Figure S2. DNA sequences of the gl2 mutants.
Supplemental Figure S3. Analyses of the frequencies of mutations for all transgenic T1 plants (total) and those with normal trichomes (normal) group obtained using the pEC1-GL2 vector.
Supplemental Figure S4. Analyses of the frequencies of JAZ1 and GAI mutations for all transgenic T1 plants (total) and glabrous (glabrous) group obtained using the pEC1-GL2 vector.
Supplemental Figure S5. Analyses of the frequencies of mutation in T1 plants obtained using the pYao vector.
Supplemental Figure S6. Mutation of GL2 does not affect flower development or seed color.
Supplemental Figure S7. Mutation of GL2 does not affect plant response to JA.
Supplemental Figure S8. Mutation of GL2 does not affect plant response to GA.
Supplemental Figure S9. The target sites of genes for polygenic mutation used in this study.
Supplemental Figure S10. Mutation types at each target sites of multiplex edited T1 plants with normal trichomes.
Supplemental Table S1. Numbers of all transgenic T1 plants and glabrous T1 plants for each target gene obtained using the pEC-GL2 vector.
Supplemental Table S2. Mutation frequencies of four target genes using different CRISPR-Cas9 systems.
Supplemental Table S3. Primers used in this study.
Supplemental Data Set S1. Sequences for plasmids used in this paper.
Supplementary Material
Acknowledgments
We thank Qijun Chen from China Agricultural University for sharing the pHEE401E plasmid, and Qi Xie from the Institute of Genetics and Developmental Biology, CAS for sharing the pYao-Cas9 plasmid.
Funding
This work was supported by grants from Taishan Scholar Foundation of Shandong Province (tsqn202103160) and Shandong Science and Technology Innovation Funds for Huawei Zhang, and Natural Science Foundation of Shandong province (ZR2020MC026) and the Qilu Scholarship from Shandong University (11200087963080) for Lijing Liu.
Conflict of interest statement. None declared.
H.Z. conceived this study. H.Z. and L.L. supervised the research. X.K. and W.P. performed all experiments and analyzed the data with help from N.X. and T.Z. L.L. and H.Z. wrote the manuscript with input from all authors. H.Z. agrees to serve as the author responsible for contact and ensures communication.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/General-Instructions) is: Huawei Zhang (huawei.zhang@pku-iaas.edu.cn).
References
- Baekelandt A, Pauwels L, Wang Z, Li N, De Milde L, Natran A, Vermeersch M, Li Y, Goossens A, Inze D, et al. (2018) Arabidopsis leaf flatness is regulated by PPD2 and NINJA through repression of CYCLIN D3 genes. Plant Physiol 178: 217–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermak T, Curtin SJ, Gil-Humanes J, Cegan R, Kono TJY, Konecna E, Belanto JJ, Starker CG, Mathre JW, Greenstein RL, et al. (2017) A multipurpose toolkit to enable advanced genome engineering in plants. Plant Cell 29: 1196–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Feng Z, Mao Y, Xu N, Zhang B, Wei P, Yang DL, Wang Z, Zhang Z, Zheng R, Yang L, et al. (2014) Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc Natl Acad Sci U S A 111: 4632–4637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillochet C, Develtere W, Jacobs TB (2021) CRISPR screens in plants: approaches, guidelines, and future prospects. Plant Cell 33: 794–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao YB, Zhao YD (2014) Self- processing of ribozyme- flanked RNAs into guide RNAs in vitro and in vivo for CRISPR- mediated genome editing. J Integr Plant Biol 56: 343–349 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Guzman M, Pizzio GA, Antoni R, Vera-Sirera F, Merilo E, Bassel GW, Fernandez MA, Holdsworth MJ, Perez-Amador MA, Kollist H, et al. (2012) Arabidopsis PYR/PYL/RCAR receptors play a major role in quantitative regulation of stomatal aperture and transcriptional response to abscisic acid. Plant Cell 24: 2483–2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn F, Mantegazza O, Greiner A, Hegemann P, Eisenhut M, Weber AP (2017) An efficient visual screen for CRISPR/Cas9 activity in Arabidopsis thaliana. Front Plant Sci 8: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irish VF, Sussex IM (1990) Function of the apetala-1 gene during Arabidopsis floral development. Plant Cell 2: 741–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khumsupan P, Donovan S, McCormick AJ (2019) CRISPR/Cas in Arabidopsis: overcoming challenges to accelerate improvements in crop photosynthetic efficiencies. Physiol Plant 166: 428–437 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Meinke D (2010) The development of Arabidopsis as a model plant. Plant J 61: 909–921 [DOI] [PubMed] [Google Scholar]
- Leon P, Gregorio J, Cordoba E (2012) ABI4 and its role in chloroplast retrograde communication. Front Plant Sci 3: 304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Vavrik C, Danna CH (2020) Proxies of CRISPR/Cas9 activity to aid in the identification of mutagenized Arabidopsis plants. G3 (Bethesda) 10: 2033–2042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zhu JK (1997) An Arabidopsis mutant that requires increased calcium for potassium nutrition and salt tolerance. Proc Natl Acad Sci USA 94: 14960–14964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Xie X, Ma X, Li J, Chen J, Liu YG (2015) DSDecode: a web-based tool for decoding of sequencing chromatograms for genotyping of targeted mutations. Mol Plant 8: 1431–1433 [DOI] [PubMed] [Google Scholar]
- Mao Y, Zhang Z, Feng Z, Wei P, Zhang H, Botella JR, Zhu JK (2016) Development of germ-line-specific CRISPR-Cas9 systems to improve the production of heritable gene modifications in Arabidopsis. Plant Biotechnol J 14: 519–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki D, Zhang WX, Zeng WJ, Feng ZY, Zhu JK (2018) CRISPR/Cas9-mediated gene targeting in Arabidopsis using sequential transformation. Nat Commun 9: 1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustilli AC, Fenzi F, Ciliento R, Alfano F, Bowler C (1999) Phenotype of the tomato high pigment-2 mutant is caused by a mutation in the tomato homolog of DEETIOLATED1. Plant Cell 11: 145–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Malley RC, Ecker JR (2010) Linking genotype to phenotype using the Arabidopsis unimutant collection. Plant J 61: 928–940 [DOI] [PubMed] [Google Scholar]
- Oyama T, Shimura Y, Okada K (1997) The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev 11: 2983–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Carol P, Richards DE, King KE, Cowling RJ, Murphy GP, Harberd NP (1997) The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev 11: 3194–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramona Grützner PM, Horn C, Mortensen S, Cram EJ, Lee-Parsons CWT, Stuttmann J, Marillonnet S (2020) High-efficiency genome editing in plants mediated by A Cas9 gene containing multiple introns. Plant Commun 2: 100135 [DOI] [PMC free article] [PubMed]
- Shirley BW, Kubasek WL, Storz G, Bruggemann E, Koornneef M, Ausubel FM, Goodman HM (1995) Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J 8: 659–671 [DOI] [PubMed] [Google Scholar]
- Szymanski DB, Lloyd AM, Marks MD (2000) Progress in the molecular genetic analysis of trichome initiation and morphogenesis in Arabidopsis. Trends Plant Sci 5: 214–219 [DOI] [PubMed] [Google Scholar]
- Tang K, Yang S, Feng X, Wu T, Leng J, Zhou H, Zhang Y, Yu H, Gao J, Ma J, et al. (2020) GmNAP1 is essential for trichome and leaf epidermal cell development in soybean. Plant Mol Biol 103: 609–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J (2007) JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448: 661–665 [DOI] [PubMed] [Google Scholar]
- Tsutsui H, Higashiyama T (2017) pKAMA-ITACHI vectors for highly efficient CRISPR/Cas9-mediated gene knockout in Arabidopsis thaliana. Plant Cell Physiol 58: 46–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Liu Q, Shen Y, Hua Y, Wang J, Lin J, Wu M, Sun T, Cheng Z, Mercier R, et al. (2019) Clonal seeds from hybrid rice by simultaneous genome engineering of meiosis and fertilization genes. Nat Biotechnol 37: 283–286 [DOI] [PubMed] [Google Scholar]
- Wang J, Chen H (2019) A novel CRISPR/Cas9 system for efficiently generating Cas9-free multiplex mutants in Arabidopsis. aBIOTECH 1: 6–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZP, Xing HL, Dong L, Zhang HY, Han CY, Wang XC, Chen QJ (2015) Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol 16: 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolabu TW, Park JJ, Chen M, Cong L, Ge Y, Jiang Q, Debnath S, Li G, Wen J, Wang Z (2020) Improving the genome editing efficiency of CRISPR/Cas9 in Arabidopsis and Medicago truncatula. Planta 252: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K, Minkenberg B, Yang Y (2015) Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc Natl Acad Sci USA 112: 3570–3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing HL, Dong L, Wang ZP, Zhang HY, Han CY, Liu B, Wang XC, Chen QJ (2014) A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol 14: 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Yang Y, Liu Y, Kang G, Wang F, Li L, Lv X, Zhao S, Yuan S, Song J, et al. (2020) Discriminated sgRNAs-based SurroGate system greatly enhances the screening efficiency of plant base-edited cells. Mol Plant 13: 169–180 [DOI] [PubMed] [Google Scholar]
- Yamasaki H, Abdel-Ghany SE, Cohu CM, Kobayashi Y, Shikanai T, Pilon M (2007) Regulation of copper homeostasis by micro-RNA in Arabidopsis. J Biol Chem 282: 16369–16378 [DOI] [PubMed] [Google Scholar]
- Yan L, Wei S, Wu Y, Hu R, Li H, Yang W, Xie Q (2015) High-efficiency genome editing in Arabidopsis using YAO promoter-driven CRISPR/Cas9 system. Mol Plant 8: 1820–1823 [DOI] [PubMed] [Google Scholar]
- Ye GN, Stone D, Pang SZ, Creely W, Gonzalez K, Hinchee M (1999) Arabidopsis ovule is the target for Agrobacterium in planta vacuum infiltration transformation. Plant J 19: 249–257 [DOI] [PubMed] [Google Scholar]
- Zhang R, Liu J, Chai Z, Chen S, Bai Y, Zong Y, Chen K, Li J, Jiang L, Gao C (2019) Generation of herbicide tolerance traits and a new selectable marker in wheat using base editing. Nat Plants 5: 480–485 [DOI] [PubMed] [Google Scholar]
- Zhang ZJ, Mao YF, Ha S, Liu WS, Botella JR, Zhu JK (2016) A multiplex CRISPR/Cas9 platform for fast and efficient editing of multiple genes in Arabidopsis. Plant Cell Rep 35: 1519–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Gong Z, Zhang C, Song CP, Damsz B, Inan G, Koiwa H, Zhu JK, Hasegawa PM, Bressan RA (2002) OSM1/SYP61: a syntaxin protein in Arabidopsis controls abscisic acid-mediated and nonabscisic acid-mediated responses to abiotic stress. Plant Cell 14: 3009–3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.