Abstract
Cyanobacteriochromes (CBCRs) are spectrally diverse photosensors from cyanobacteria distantly related to phytochromes that exploit photoisomerization of linear tetrapyrrole (bilin) chromophores to regulate associated signaling output domains. Unlike phytochromes, a single CBCR domain is sufficient for photoperception. CBCR domains that regulate the production or degradation of cyclic nucleotide second messengers are becoming increasingly well characterized. Cyclic di-guanosine monophosphate (c-di-GMP) is a widespread small-molecule regulator of bacterial motility, developmental transitions, and biofilm formation whose biosynthesis is regulated by CBCRs coupled to GGDEF (diguanylate cyclase) output domains. In this study, we compare the properties of diverse CBCR-GGDEF proteins with those of synthetic CBCR-GGDEF chimeras. Our investigation shows that natural diversity generates promising candidates for robust, broad spectrum optogenetic applications in live cells. Since light quality is constantly changing during plant development as upper leaves begin to shade lower leaves—affecting elongation growth, initiation of flowering, and responses to pathogens, these studies presage application of CBCR-GGDEF sensors to regulate orthogonal, c-di-GMP-regulated circuits in agronomically important plants for robust mitigation of such deleterious responses under natural growing conditions in the field.
Natural diversity of light-regulated diguanylate cyclases outperforms rational design for generating promising candidates for robust, broad spectrum optogenetic applications in live cells.
Introduction
Photosensory proteins respond to color, intensity, direction, and/or duration of light by triggering biochemical pathways for real-time adaptation to fluctuating environmental conditions. Found in abundance in photosynthetic organisms, photosensors play diverse roles in regulating photosynthesis-associated gene expression, cell growth, movement, reproduction, and other processes (Möglich et al., 2010; Gomelsky and Hoff, 2011). Cyanobacteria possess a particularly rich variety of photosensory proteins that exploit the photochemical properties of associated retinals, flavins, and linear tetrapyrroles (bilins; Fiedler et al., 2005; Mandalari et al., 2013; Schuergers et al., 2017). The bilin-binding phytochrome superfamily is among the most spectrally diverse, absorbing light throughout the photosynthetically active spectral range (Ikeuchi and Ishizuka, 2008; Anders and Essen, 2015; Rockwell and Lagarias, 2017; Villafani et al., 2020; Fushimi and Narikawa, 2021). All members of this superfamily share a conserved cGMP-specific phosphodiesterase (PDE)/Adenylyl cyclase/FhlA (GAF) domain that harbors a photoactive linear tetrapyrrole (bilin) chromophore that is typically covalently attached to a conserved Cys residue via a thioether linkage to the bilin A-ring (Rockwell et al., 2006; Rockwell and Lagarias, 2010, 2017). Reversible conversion between two spectrally distinct photostates via photoisomerization of the 15,16-double bond of the bilin chromophore is a hallmark of these proteins. Canonical phytochrome photoconversion is achieved via absorption of red or far-red light, which triggers protein structural rearrangements to affect downstream signal transduction (Nagano, 2016; Rensing et al., 2016; Viczian et al., 2017; Beattie et al., 2018; Pham et al., 2018).
Cyanobacteriochromes (CBCRs) are cyanobacterial proteins with bilin-binding GAF domains distantly related to those of canonical phytochromes. CBCRs exhibit broader spectral range than phytochromes, absorbing from the near ultraviolet to the near infrared, and also require only the GAF domain for assembly with bilin and for full, reversible photoconversion (Hirose et al., 2008; Ikeuchi and Ishizuka, 2008; Rockwell et al., 2011, 2012, 2016; Ma et al., 2012; Hirose et al., 2013; Narikawa et al., 2014). Like phytochromes, CBCRs regulate diverse C-terminal output domains with a range of biochemical activities. These include two-component transmitter modules, methyl-accepting chemotaxis domains, and enzymes that mediate the synthesis or degradation of second messengers such as cAMP, cGMP, and c-di-GMP (Ikeuchi and Ishizuka, 2008; Rockwell and Lagarias, 2010; Blain-Hartung et al., 2018; Wiltbank and Kehoe, 2019). Owing to their small size (<200 amino acids), the photosensory “GAF-only” modules of CBCRs are ideal targets for development of optogenetic tools (Tabor et al., 2011; Ramakrishnan and Tabor, 2016; Blain-Hartung et al., 2017, 2018; Fernandez-Rodriguez et al., 2017; Fushimi et al., 2017a). By contrast, canonical phytochromes require two additional domains that flank the GAF domain for full photosensory function (Wu and Lagarias, 2000). Phytochrome-based regulators of cyclic nucleotide levels have been extensively studied as optogenetic reagents (Tarutina et al., 2006; Gasser et al., 2014; Ryu and Gomelsky, 2014; Ryu et al., 2014; Gourinchas et al., 2017; Lindner et al., 2017; Etzl et al., 2018; Gourinchas et al., 2018; Stuven et al., 2019), but optogenetic applications of CBCR-regulated nucleotidyl cyclases have been examined only recently (Blain-Hartung et al., 2017, 2018; Fushimi et al., 2017a). The present work addresses regulatory control of GGDEF domains (exhibiting diguanylate cyclase or DGC activity) by CBCRs to inform rational design of light-regulated nucleotidyl cyclases that respond to a non-overlapping range of light colors.
Cyclic di-GMP (c-di-GMP) is a widespread bacterial second messenger known to regulate prokaryotic biofilm production, cell cycle, virulence, and motility, among other processes (Ryjenkov et al., 2005; Romling et al., 2013). c-di-GMP also functions as an agonist of well-described transcription factors, riboswitches, and human immune receptors (Karaolis et al., 2005; Sudarsan et al., 2008; Smith et al., 2009; Christen et al., 2010; Shu et al., 2012; Ryu and Gomelsky, 2014; Chen et al., 2017). C-di-GMP regulated pathways appear to be absent in most eukaryotes, so co-expression of light-regulated DGCs with c-di-GMP-dependent PDEs, such as the blue-light-activated diguanylate PDEs, BldP (Ryu et al., 2017) or PA2133 (Pu et al., 2018), should prove an effective means for greater control of c-di-GMP-mediated responses therein.
Such approaches would benefit from a panel of light-regulated DGCs activated under different light conditions. The optical properties of different organisms are not well suited to a single light-regulated DGC: for example, red- or far-red activation is preferable in mammalian cells but will trigger a broad range of endogenous photobiology in plants (Fernandez-Milmanda and Ballare, 2021). To develop a suitable body of reagents, we here report two approaches. In one, we started with the previously described C-terminal CBCR-GGDEF bidomain (Blain-Hartung et al., 2017) of Tlr0924 (SesA) from the cyanobacterium Thermosynechococcus elongatus (Tlr0924Δ: Figure 1, A), whose DGC activity is stimulated by blue light and repressed by green light (Enomoto et al., 2014; Enomoto et al., 2015; Blain-Hartung et al., 2017). We then replaced the blue/green CBCR domain of Tlr0924Δ with CBCR domains with different spectral sensitivities. In the other approach, we identified, cloned, and expressed spectrally diverse CBCR-GGDEF candidates from sequenced cyanobacterial genomes. Our studies establish that natural diversity is a powerful alternative to rational design of broad spectrum CBCR-based DGCs, provide insight into the structural basis of light-regulation of DGC activity, and describe additional CBCR-GGDEF proteins with greater spectral diversity.
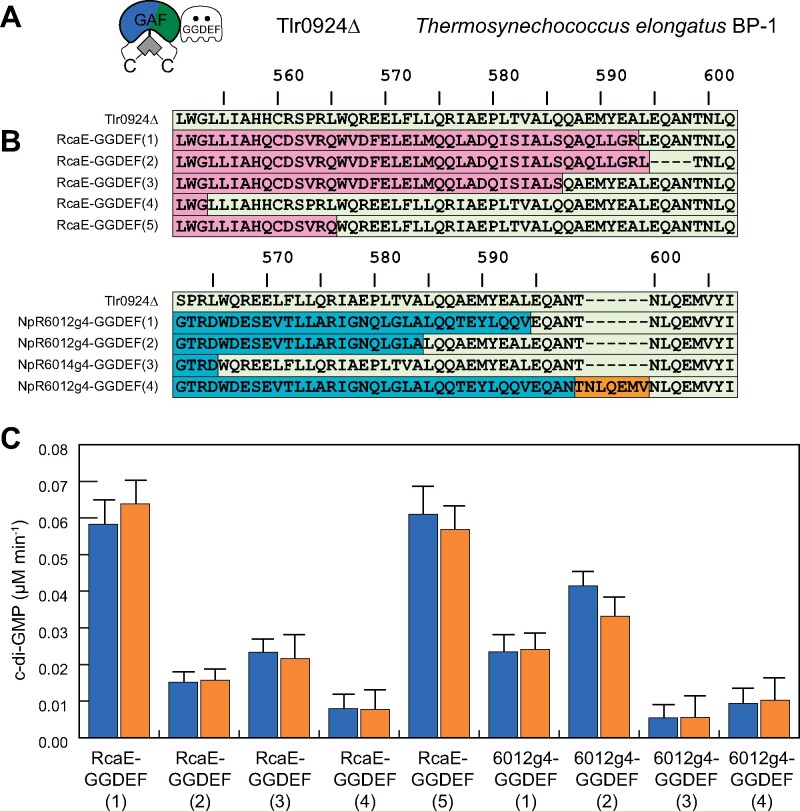
Figure 1.
Fusion of CBCR “GAF” domains with DGC “GGDEF” domain of Tlr0924 (SesA). A, Domain architecture of Tlr0924Δ. B, Alignment of fusion protein constructs, color-coded by coding sequence (Tlr0924, pale green; RcaE, pink; NpR6012g4, cyan; dark orange box, short repeated motif). The alignment starts with the last β strand of the GAF domain (LWGLLIAH in Tlr0924Δ), with the GAF domain ending in the vicinity of VALQQ in the same sequence and with the GGDEF domain in the vicinity of NLQ in the same sequence. Numbering is based on full-length Tlr0924 (GenBank accession BAC08476). C, DGC activity of fusion protein constructs (indicated as in panel B) in 15Z (blue) and 15E (orange) photostates. Error bars are drawn at one standard deviation (n = 3). No construct exhibited significant light-regulated DGC activity (P < 0.05).
Results
Rational design of chimeric CBCR-GGDEF proteins fails to yield robust light-regulated DGCs
We initially engineered GAF-GGDEF fusions of the red/green CBCR NpR6012g4 (Rockwell et al., 2012c) and the green/red CBCR RcaE (Hirose et al., 2013) with the GGDEF domain of Tlr0924 (Blain-Hartung et al., 2017). These constructs were designed to replace the linker region between the two domains with variable lengths of linker regions C-terminal to the light-sensing GAF domain and N-terminal to the GGDEF output domain (Figure 1, B). While all fusion proteins retained spectral properties of the parent CBCR (Supplemental Figure S1), none displayed light-regulated synthesis of c-di-GMP despite striking differences in basal DGC activity (Figure 1, C). These results underscore the difficulty of generating robust light-regulated chimeras by domain fusion of light-sensing input domains with diverse signal output domains, even when linker length and chemical properties are taken in consideration (Möglich and Moffat, 2010). Indeed, our previously unsuccessful attempts to alter the amino acid composition and length of the linker region between the CBCR and GGDEF domains of Tlr0924Δ demonstrate that light-regulated signal output is exquisitely controlled by critical structural features of the linker region (Blain-Hartung et al., 2017). The present studies show that the regulatory function of the linker is context dependent, no doubt reflecting its co-evolution with associated CBCR and GGDEF domains (Gourinchas et al., 2018).
Bioinformatic analysis reveals that CBCR-DGC sensors are widespread in cyanobacteria
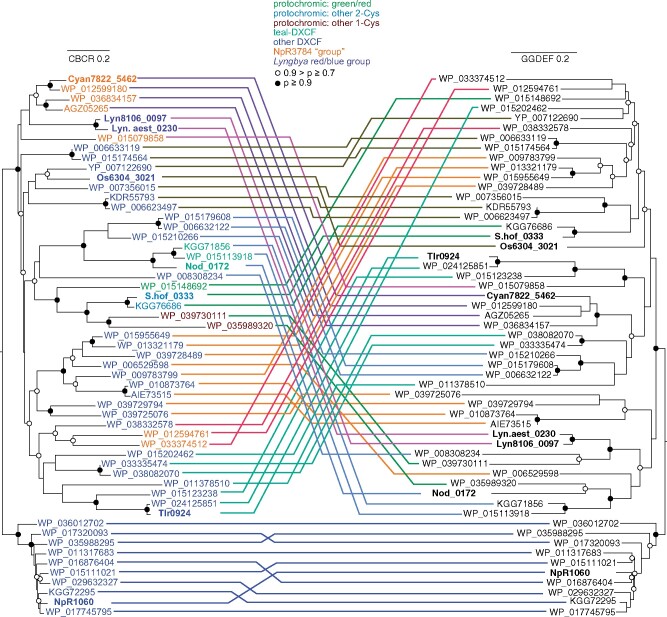
As an alternative to rational design, we worked from previous analyses of CBCR diversity (Rockwell et al., 2008, 2015) to generate a list of 55 proteins containing recognizable CBCR GAF domains immediately N-terminal to GGDEF domains containing fully conserved catalytic GG(D/E)EF motifs (Ryjenkov et al., 2005), mimicking the domain arrangement of Tlr0924. CBCR, linker, and DGC GGDEF regions were aligned separately for these proteins (Supplemental Figures S2–S4). The linker region was quite small, but the CBCR and GGDEF alignments were used to infer maximum-likelihood phylogenies. These phylogenies are presented in a tanglegram representation (Figure 2). In Figure 2, CBCR names are colored based on the type of CBCR domain (Rockwell et al., 2015b). The isolated CBCR and GGDEF domains in the respective trees are connected by arbitrarily colored lines; in this fashion, co-evolving clusters can be identified. CBCR sequence clusters containing NpR1060 and Cyan7822_5462 each correspond to a monophyletic group of GGDEF domains, providing support for co-evolution of the CBCR and GGDEF domains in these lineages. Other cases do not show such co-evolution of the GAF-GGDEF pair, suggesting that gene fusion events have also played a significant role in the evolution of these sensors.
Figure 2.
Tanglegram view of CBCR (left) and GGDEF (right) maximum-likelihood phylogenies. Bold lettering indicates proteins further described in paper. Color of CBCR lettering matches subfamily seen at top of tanglegram. Connecting lines are colored to facilitate comparison of the two trees; instances of co-evolution should thus yield coherent groups of connecting lines. The cluster of proteins which includes NpR1060 was chosen as an arbitrary outgroup and is placed at the bottom of both phylogenies. Scale bars for each tree indicate 0.2 substitutions per position.
The majority of the CBCR-GGDEF proteins (35 of 55) contain the DXCF signature sequence found in Tlr0924Δ (Supplemental Figure S2). The cysteine residue within this motif forms a second thioether linkage to the C10 position of the bilin chromophore in one or both photostates, shortening the conjugated systems and blue shifting the CBCR spectrum (Ishizuka et al., 2011; Rockwell et al., 2012a, 2012b; Burgie et al., 2013). Thus, the presence of this DXCF motif indicates that these proteins most likely will detect violet or blue light in one or both photostates. An even larger fraction of these proteins (43 of 55) also possess a c-di-GMP-binding RxxD inhibition site located directly upstream of the GG(D/E)F motif (Supplemental Figure S3) and hence would be expected to exhibit product inhibition, potentially providing another layer of feedback and regulation in native systems (Ryjenkov et al., 2005; Romling et al., 2013). Inhibition sites can present another layer of feedback and regulation in native systems. We expected that the linker region between the CBCR and GGDEF domain would contain conserved motifs important to light regulation of these enzymes. Consistent with its importance for DGC activity (Blain-Hartung, 2017, p. 31), the linker position corresponding to Leu177 of Tlr0924Δ is 96% conserved (Supplemental Figure S4). Positioned on the opposite side of a possible amphipathic helix, Glu179 in Tlr0924Δ is not conserved; however, the polar and hydrophilic nature of this residue is preserved in all of the proteins (Supplemental Figure S4). Such observations lend credence to the hypothesis that this region forms an amphipathic helix (Blain-Hartung et al., 2017). The consensus sequence from this linker alignment is also computationally predicted to adopt an α-helix secondary structure, consistent with the presence of regularly spaced hydrophobic and hydrophilic residues and the absence of Pro residues. Insertions in the linker occur in roughly 15% of the proteins (Supplemental Figure S4).
CBCR-regulated DGC candidates encompass a predicted broad spectral range
Seven proteins from this phylogenetic analysis were selected for more detailed analysis because of their predicted spectral diversity and phylogenetic divergence (Figure 3): Oscillatoria acuminata PCC 6304_3021 (hereafter, Os6304_3021), WP_033336739 from Scytonema hofmannii (hereafter, S.hof_0333), Npun_R1060 from Nostoc punctiforme PCC 73102 (hereafter, NpR1060; Rockwell et al., 2012a), WP_009782402 from Lyngbya sp. strain PCC 8106 (hereafter, Lyn8106_0097), WP_023065890 from Lyngbya aestuarii strain BL J (hereafter, Lyn.aest_0230), Cyan7822_5462 from Gloeothece verrucosa PCC 7822 (formerly Cyanothece sp. strain PCC 7822; Mares et al., 2019), and WP_017296945 from Nodosilinea nodulosa PCC 7104 (hereafter, Nod_0172).
Figure 3.
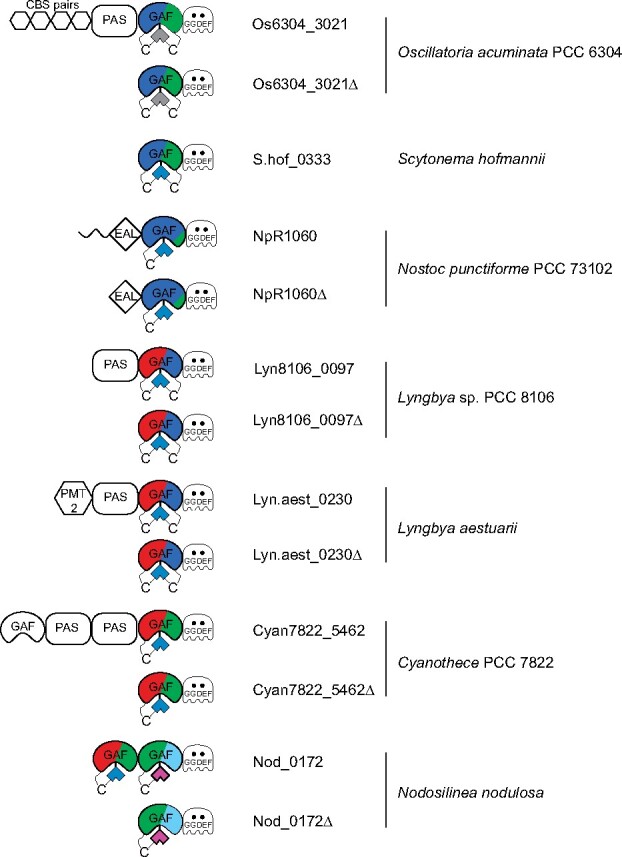
Jellybean-style domain diagram displaying selected CBCR-regulated DGC proteins and CBCR-GGDEF truncations. Domain abbreviations: CBS pairs (cystathionine beta synthase domain repeats); EAL (c-di-GMP-dependent PDE); GAF (cGMP-specific PDE/adenylyl cyclase/FhlA); GGDEF (diguanylate cyclase); PAS (Per-Arnt-Sim); PMT2 (protein-O-mannosyltransferase-2). CBCR photocycles indicated by color.
CBCR-GGDEF truncations (Os6304_3021Δ, Lyn8106_0097Δ, Lyn.aest_0230Δ, Cyan7822_5462Δ) were co-expressed in the bacterium Escherichia coli engineered to produce the PCB chromophore, as was full-length S.hof_0333. Truncated versions of NpR1060 (NpR1060Δ) with EAL-GAF-GGDEF domains and of Nod_0172 (GAF1-GAF2-GGDEF) with both of the tandem CBCR sensors were also examined. NpR1060, Os6304_3021, and S.hof_0333 possessed CBCRs with a DXCF motif similar to Tlr0924 (Supplemental Figure S2), whereas the others did not contain known second Cys residues and were initially predicted to exhibit red/green photocycles (Figure 3).
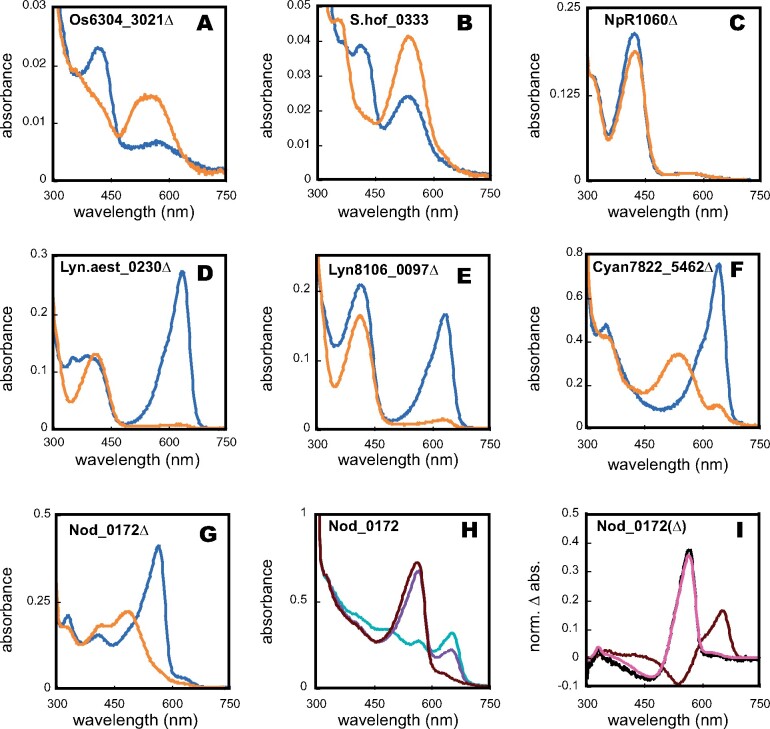
The spectral and photochemical properties of recombinant versions of these proteins were determined (Figure 4 and Table 1), and denaturation assays were performed to assign the d-ring configuration of both photostates. As expected, the DXCF CBCRs Os6304_3021Δ, S.hof_0333, and NpR1060Δ exhibited 15Z dark states absorbing violet or blue light, similar to that of the DXCF CBCR Tlr0924 (Figure 4, A–C, blue traces; Blain-Hartung et al., 2017). Os6304_3021 and S.hof_0333 also exhibited green-absorbing 15E photoproducts (Figure 4, A and B, orange traces). By contrast, the 15E photoproduct of NpR1060Δ retained a blue-absorbing 15E state (Figure 4, C, orange trace), consistent with the behavior of the GAF-only construct reported previously (Rockwell et al., 2012a). The 15E photoproduct of NpR1060Δ possessed an absorbance “shoulder” around 550 nm, allowing green light to trigger its 15E to 15Z conversion back to the dark state (Rockwell et al., 2012a).
Figure 4.
Spectral analysis of selected CBCR-GGDEF proteins. Absorbance spectra of (A) Os6304_3021Δ; (B) S.hof_0333; (C) NpR1060Δ; (D) Lyn.aest_0230Δ; (E) Lyn8106_0097Δ; (F) Cyan7822_5462Δ; (G) Nod_0172Δ; (H) Nod_0172; and (I) Nod_0172 (GAF1) and Nod_0172Δ. For panels A–G, blue traces represent 15Z states and orange traces represent 15E states. For panel H, the teal trace represents the sample irradiated with saturating teal light (500 ± 10 nm). Subsequent irradiation with violet light (400 ± 35 nm; violet trace) or red light (650 ± 20 nm; black red trace) is shown. Panel I depicts the photochemical difference spectra of GAF1 (black red trace), GAF2 (coral trace; obtained by subtraction of the violet trace from the red trace in panel H), and that of Nod_0172Δ (black trace).
Table 1.
Spectral properties of purified CBCR-GGDEF proteins
| Construct | 15Z dark-adapted state | 15E photoproduct state |
|---|---|---|
| Abs max (nm) | Abs max (nm) | |
| Os6304_3021Δ | 423 | 540 |
| S.hof_0333 | 413 | 538 |
| NpR1060Δ | 422 | 424;562 (sh) |
| Lyn.aest_0230Δ | 638 | 412 |
| Lyn8106_0097Δ | 636 | 412 |
| Cyan7822_5462Δ | 646 | 542 |
| Nod_0172Δ | 566 | 486 |
| Nod_0172 (GAF1) | 656 | 538 |
| Nod_0172 (native spec.) | 656 and 566 | 538 and 486 |
| Construct | Difference spectra Abs max (nm) | Difference spectra Abs min (nm) |
| Nod_0172 – 580 nm | 654 | 540 |
| Nod_0172 – 650 nm | 568 | 470 |
| Nod_0172 (GAF1) | 656 | 538 |
Lyn.aest_0230Δ and Lyn8106_0097Δ both exhibited red-absorbing 15Z dark states with peaks at 638 and 636 nm, respectively (Figure 4, D and E). In both cases, red light triggered a profound spectral shift into the blue region (λmax = 412 nm), similar to the red/blue photocycle of AM1_1186g2 (Narikawa et al., 2014). This result implicated the presence of second Cys residues in both Lyngbya sensors responsible for forming a covalent linkage to the C10 methine bridge in the 15E photoproduct. Lyn.aest_0230Δ and Lyn8106_0097Δ possess a conserved Cys residue located N-terminal to the DXCF motif (Supplemental Figure S2, highlighted in red) and C-terminal to the insertion loop containing the second Cys residue of insert-Cys CBCRs (Rockwell et al., 2011). This Cys residue is thus potentially positioned to form a second linkage and is apparently unique to these proteins. To assess the role of this cysteine residue in spectral tuning of the two Lyngbya sensors, we expressed Lyn.aest_0230Δ(C73A) and Lyn8106_0097Δ(C53A) variants that lack this Cys residue. Both variants retained red-absorbing 15Z dark states, but failed to produce blue-absorbing photoproduct states (Supplemental Figure S5, A and B), instead giving photoproducts absorbing in the green to orange region of the spectrum (520–586 nm). Based on this result, we conclude that this candidate second Cys residue is essential for formation of the blue-absorbing photoproduct, presumably via second linkage formation. These proteins thus provide another example of independent evolution of a dual-Cys CBCR photocycle (Rockwell et al., 2011, 2017; Narikawa et al., 2014).
We next examined Cyan7822_5462 and the GAF-GAF-GGDEF tridomain Nod_0172, also predicted to have red/green CBCR domains (Figure 3). Cyan7822_5462Δ exhibited the expected red/green photocycle, with respective absorbance maxima of its 15Z- and 15E-states of 656 and 542 nm (Figure 4, F). The spectrum of the 15Z-dark state of full-length Nod_0172 was more complex due to the presence of two photoactive CBCR domains in tandem (Figure 4, G). Since one of these exhibited a peak in the red (λmax = 656 nm) and the other in the green (λmax = 564 nm), Nod_0172 contained a mixture of 15Z and 15E states for both GAF domains due to exposure to ambient light during purification. To confirm the photocycle of GAF1 and GAF2 of Nod_0172, we separately expressed the N-terminal CBCR GAF domain, Nod_0172(GAF1), and the C-terminal GAF2-DGC bidomain Nod_0172Δ. Nod_0172(GAF1) exhibited a canonical red/green CBCR photocycle (Supplemental Figure S5, A) accounting for the red-absorbing peak in the full-length protein. Nod_0172Δ exhibited a green/teal photocycle (Figure 4, G) resembling that of NpR5113g1 (Rockwell et al., 2012a) and FdDpxA (Wiltbank and Kehoe, 2016).
To determine whether the two CBCR GAF domains affect each other’s spectral properties, we more closely examined the photocycle of full-length Nod_0172, which contains a predicted red/green CBCR, a predicted DXCF CBCR, and a GGDEF domain (Figure 3). To do so, we first irradiated Nod_0172 with saturating 500 nm light to fully convert GAF1 to its red-absorbing 15Z state and GAF2 to its green-absorbing 15Z state (Figure 4, H, green trace). Irradiation with violet light (Figure 4, H, violet trace) triggered conversion of both domains due to overlap with the Soret region, whereas illumination with red light (Figure 4, H, red trace) triggered apparently specific conversion of the red/green domain. Subtracting the green trace from the red trace in Figure 4, H afforded a difference spectrum that was identical to the normalized difference spectrum of GAF1 alone (Figure 4, I, red trace). Removal of the residual difference spectrum of GAF1 from the (violet product—red product) difference spectrum of Nod_0172 by subtraction of the violet trace from the red trace in Figure 4, H yielded a GAF2 difference spectrum (Figure 4, I, green trace) that was also superimposable on that of the GAF2-GGDEF construct Nod_0172Δ (Figure 4, I, black trace). These results indicate that the steady-state photochemical properties of both CBCR GAF domains are unaltered in the context of the full-length Nod_0172 sensor.
Light dependent regulation of c-di-GMP
To analyze the enzymatic properties of the newly identified CBCR-associated DGCs, in vitro DGC assays were performed in both 15E and 15Z photostates. C-di-GMP levels were measured using a quantitative RP-HPLC assay described previously (Blain-Hartung et al., 2017). Most of the CBCR-DGC proteins exhibited statistically significant light-regulated DGC activity. In all such cases, the average DGC activity of the 15E lit states was greater than those of the 15Z dark state (Table 2). Of the three DGCs that lack the RxxD inhibition (I) site (Supplemental Figure S4 and see above), Os6304_3021Δ and NpR1060Δ exhibited the greatest overall activity in both photostates as expected (Figure 5, A and Table 2). By contrast, S.hof_0333 exhibited only low, poorly light-regulated DGC activity. We also examined whether c-di-GMP breakdown products were produced by NpR1060Δ, because this construct retains an N-terminal “EAL” c-di-GMP PDE domain with apparently functional consensus motifs. Neither linearized di-GMP nor GMP was detected, indicating that NpR1060Δ was not an active c-di-GMP PDE under these conditions. Rates of c-di-GMP production by Lyn8106_0097Δ and Lyn.aest_0230Δ were significantly lower (Figure 5, B and Table 2), consistent with the presence of functional autoinhibition regulated by the I site; both were more active under red light, indicating that the 15E state of the CBCR results in higher DGC activity. Similar to the blue/green DGC S.hof_0333, the red/green Cyan7822_5462Δ did not exhibit statistically significant differences in DGC activity for the two photostates when accounting for sample deviation (Figure 5, B and Table 2).
Table 2.
DGC activity of CBCR-GGDEF proteins
| Protein | Length (a.a.) | Z-state c-di-GMP (µmol min−1)/illumination wavelength (nm)a | E-state c-di-GMP (µmol min−1)/illumination wavelength (nm)a | Fold difference |
|---|---|---|---|---|
| Os6304_3021Δ | 346 | 0.6 (±0.1)/520 | 1.42 (±0.15)/445 | 2.4 |
| S.hof_0333 | 396 | 0.07 (±0.03)/520 | 0.09 (±0.08)/445 | 1.3 |
| NpR1060Δ | 647 | 1.10 (±0.17)/520 | 3.05 (±0.37)/445 | 2.8 |
| Lyn8106_0097Δ | 342 | 0.06 (±0.03)/445 | 0.30 (±0.07)/630 | 5 |
| Lyn_aest_0230Δ | 362 | 0.25 (±0.07)/445 | 0.38 (±0.05)/630 | 1.5 |
| Cyan7822_5462Δ | 346 | 0.09 (±0.04)/520 | 0.14 (±0.04)/630 | 1.5 |
| Nod_0172Δ | 370 | 0.04 (±0.03)/420 | 0.31 (±0.08)/520 | 7.8 |
| Tlr0924Δb | 347 | 0.06 (0.02)/420 | 0.54 (±0.02)/520 | 9 |
| BphG1c | 931 | 0.5d | 6.25d | 11 |
Values (±SD, n = 3) are shown.
Estimated values.
Figure 5.
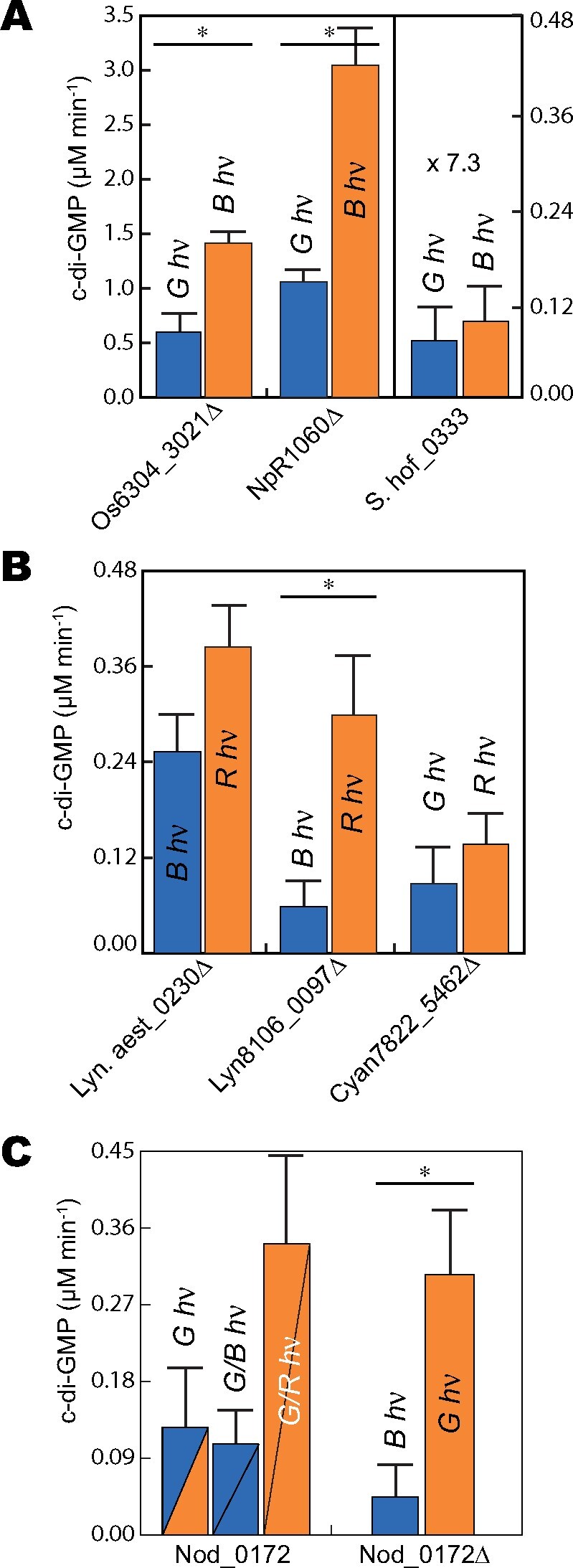
DGC activities of selected CBCR-GGDEF proteins. A, NpR1060Δ, Os6304_3021Δ, and S.hof_0333; B, Lyn.aest_0230Δ, Lyn8106_0097Δ, and Cyan7822_5462Δ; C, Nod_0172 and Nod_0172Δ. Total protein concentration of each protein is 5 µM. For panels A and B, the color of light used for sample irradiation is indicated above each bar column, and those columns are colored blue or orange for samples maintained in 15Z or 15E states, respectively. For panel C, the color of light used for sample irradiation is indicated above/within each bar column. Nod_0172 samples labeled G/B and G/R were first irradiated with saturating green light and then maintained under blue or red, respectively. Nod_0172 bar columns are divided by a line and are colored blue or orange to represent the photostate of GAF1 (upper left) and GAF2 (lower right). Error bars were calculated as standard deviation of all replicates (n = 3); Statistically significant differences between photostates are indicated with asterisks (P ≤ 0.025; Student’s t test). G hν, green light; B hν, blue light; R hν, red light.
Unexpectedly, the tandem CBCR-DGC construct Nod_0172 was most active under red light, whereas green or blue irradiation reduced its activity (Figure 5, C and Table 3). Since red light maintains the N-terminal GAF1 domain in its 15E “lit” state but should not activate GAF2, this result suggests that light regulation of the C-terminal DGC activity is mainly dependent on the photostate of the N-terminal GAF1 domain. However, the red source used in these experiments partially overlaps with the green-absorbing dark state of GAF2. Thus, this broader red light would also generate the lit state of GAF2, instead raising the possibility that maximal activity of the tandem construct requires activation of both domains. The DGC activity of the truncated Nod_0172Δ protein is increased by green light to a level comparable to that seen in Nod_0172 under red light (Figure 5, C). Taken together, this result suggests that both the GAF1 photostate and the GAF2 photostate influence the DGC activity of the full-length protein, as is seen in the complex phytochrome/CBCR chimera All2699 (Ma et al., 2012).
Table 3.
DGC activity of Nod_0172 (GAF-GAF-GGDEF)
| Protein | Length (a.a.) | c-di-GMP (µmol min−1)/illumination wavelength (nm)a | c-di-GMP (µmol min−1)/illumination wavelength (nm)a | c-di-GMP (µmol min−1)/illumination wavelength (nm)a | Fold difference |
|---|---|---|---|---|---|
| Nod_0172 | 559 | 0.13 (±0.07)/520 | 0.11 (±0.03)/420 | 0.34 (±0.10)/630 | 1.0:0.85:2.6 |
Values (±SD, n = 3) are shown.
Light-dependent regulation of cell motility in E. coli
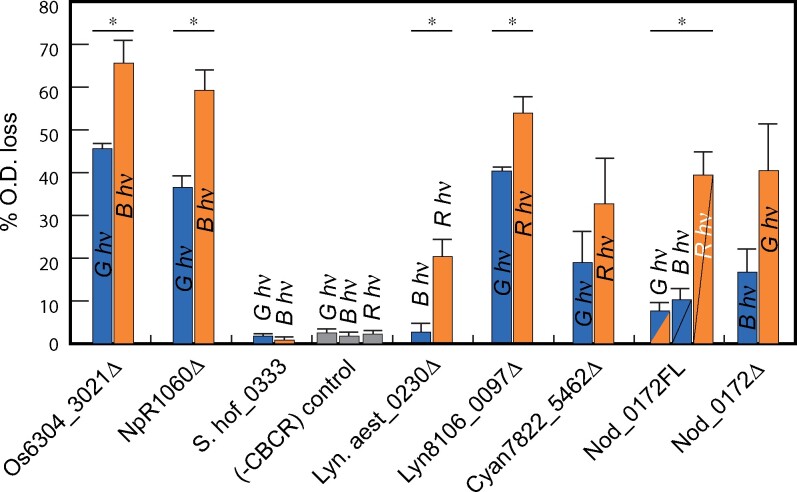
Harnessing light to regulate cellular events is a key tenet of optogenetic and synthetic biology applications. For this purpose, we analyzed the ability of selected CBCR-DGC constructs to control motility of E. coli cells by specific wavelengths of light. C-di-GMP is a well-known negative regulator of bacterial cell motility that triggers the transition to stress-resistant states known as biofilms (Romling et al., 2013). In T. elongatus BP-1, three blue/green CBCR-DGC sensors regulate c-di-GMP levels to trigger cell aggregation and settling (Enomoto et al., 2015; Enomoto and Ikeuchi, 2020): SesA/Tlr0924, SesB/Tlr1999, and SesC/Tlr0911. We previously developed an E. coli cell settling assay to test the ability of Tlr0924Δ and variants to regulate heterologous cell aggregation in response to the ambient light conditions (Blain-Hartung et al., 2017), presumably relying on endogenous E. coli c-di-GMP signaling pathways to trigger cell–cell aggregation. Small cultures are grown under defined light conditions and then allowed to rest in darkness; settling is then assayed by measuring optical density (OD) at the top of the culture. We used the same settling assay to examine the light-regulated activities of the newly identified CBCR-GGDEF proteins in vivo.
The activity of each construct was estimated by the percentage loss of OD of the culture. Os6304_3021Δ and NpR1060Δ expression yielded the most dramatic cell settling phenotypes (Figure 6), consistent with their more robust in vitro activities (Figure 5, A). The relative activity of both constructs was always greater under blue light relative to green light, again consistent with the activity of these constructs in vitro. Expression of S.hof_0333 failed to stimulate cell settling, consistent with its feeble DGC activity in vitro (Figure 6). For cultures expressing Lyn8106_0097Δ and Lyn.aest_0230Δ, greater cell settling was seen under red light relative to blue light (Figure 6). The fold difference between red- versus blue-light activities was greater for Lyn.aest_0230Δ than for Lyn8106_0097Δ, whereas in vitro DGC measurements showed the opposite trend (Figure 5, B). Finally, cells containing the red/green sensor Cyan7822_5462Δ settled at slightly greater rates under red light than under green light. This difference was not statistically significant, consistent with in vitro assay data (Figure 6).
Figure 6.
Selected CBCR-GGDEF proteins exhibit DGC activity in vivo. Cultures were grown in a shaking incubator under filtered light (see the “Materials and methods” section for filters), and expression of selected CBCR-GGDEF proteins was induced using 0.2% arabinose + 1 mM IPTG (Os6304_3021Δ and ΔNpR1060: 0.05% arabinose + 1 mM IPTG). Settling measured as percent OD loss of cultures in darkness after growth. Color of bars indicates the photostate (blue, 15Z; orange, 15E), with Nod_0172FL colored as in Figure 5. Error bars are drawn at one standard deviation (n = 3). Each CBCR-GGDEF construct was assessed for light-controlled cell settling using Student’s t test for unpaired data, with no assumption of equal variances. Cases exhibiting significant light effects (P < 0.025) are indicated with asterisks.
Expression of Nod_0172Δ produced settling phenotypes that largely corresponded with its in vitro DGC activity: green light enhanced cell settling significantly more than blue light (Figure 6). In addition, the tandem CBCR-DGC Nod_0172 construct resulted in the greatest degree of cell settling after exposure to red light, while significantly less settling was seen upon exposure to green or blue light (Figure 6). Overall, we have demonstrated that the majority of these chosen proteins not only exhibit light-regulated DGC activity in vitro, but also function in vivo for regulation of c-di-GMP signaling by red, blue, or green light.
Discussion
The second messenger c-di-GMP regulates well-characterized genetic pathways via transcription factors, riboswitches, and other factors (Jenal et al., 2017). Hence, some of the CBCR-GGDEF bidomain sensors described here may hold considerable promise for broad spectrum optogenetic control of engineered genetic pathways responsive to c-di-GMP in live cells (Figure 6). However, the lack of detailed structural information for CBCR-GGDEF bidomains retards progress toward successful rational design of robust light-regulated DGCs. Our failure to generate viable fusions with the GGDEF domain of Tlr0924/SesA using well-characterized model CBCRs RcaE and NpR6012g4 highlights these limitations. By mining the natural diversity of native cyanobacterial CBCR-GGDEF proteins, we have instead demonstrated that extant diversity may also provide a wide spectral palette of light-regulated DGCs.
CBCR-GGDEF diversity
Based on bioinformatic data mining, we created multiple sequence alignments of 55 candidate CBCR-GGDEF bidomains, a list that is by no means complete and that is still continuously expanding. These alignments help to identify residues that may be essential for light regulated signal transduction. The helical linker was of especial interest because previous studies on Tlr0924Δ (Blain-Hartung et al., 2017), on the engineered phytochrome-regulated DGC BphS (Ryu and Gomelsky, 2014; Ryu et al., 2014; Ryu et al., 2017), and on the natural phytochrome-regulated DGC IsPadC from Idiomarina species strain A28L (Gourinchas et al., 2017, 2018) established the importance of the interdomain linker for transmission of the light signal. Similar considerations also apply to phytochrome-regulated PDEs such as LAPD (Gasser et al., 2014) and DdPAC (Stuven et al., 2019). In this regard, conserved blocks of residues in the linker region are located immediately downstream and upstream of the CBCR and GGDEF domains, respectively (Supplemental Figure S4). A nearly fully conserved leucine residue in the interdomain linker of Tlr0924Δ, Leu177, is known to be essential for DGC activity (Blain-Hartung et al., 2017). LOV and BLUF domain-containing proteins also leverage helical linkers for light signal transmission to C-terminal output domains (Moglich and Moffat, 2007; Moglich et al., 2009; Masuda, 2012; Ziegler and Moglich, 2015). Our unsuccessful attempts to exchange CBCR domains to create functional CBCR-GGDEF chimeras suggest that linkers function in the context of specific CBCRs which have co-evolved to function together with their cognate GGDEF domains.
DXCF CBCR-GGDEFs are the most widespread
To an extent, photosensory properties can be predicted via conserved motifs in multiple sequence alignments of CBCRs. The majority of the CBCR-GGDEFs possess the DXCF motif found in the canonical blue/green family of CBCRs (Rockwell et al., 2011). Interestingly, the tanglegram analysis suggests that a minority of CBCR-GGDEF proteins possess co-evolved GAF and GGDEF domains (Figure 2), while the majority arose from some combination of gene-transfer and gene-fusion events. The preponderance of DXCF CBCR representatives suggests a strong adaptive value of DGCs that sense higher energy blue light, which is more damaging than longer wavelength light due to an abundance of blue-absorbing photosensitizers in cells (Schuergers et al., 2017). However, blue/green CBCRs could also indirectly sense depletion of blue light by neighboring cells, consistent with a proposed role for such blue/green CBCRs as shade sensors in microbial mats (Enomoto and Ikeuchi, 2020). C-di-GMP levels mediated by the blue/green CBCR-DGCs Tlr0924, Os6304_3021, or S.hof_0333 would be expected to increase under low cell densities, triggering a settling response in unfiltered light environments.
Our tanglegram phylogenetic analysis shows that NpR1060 is a representative member of a clade of GGDEF domains that clearly co-evolved with a specific clade of DXCF CBCRs (Figure 2). NpR1060Δ exhibits an unusual photocycle with very weak green light absorption in the 15E lit state (Figure 4). This observation indicates that the photochemical quantum yield for the reverse conversion of blue-absorbing 15E photoproduct of NpR1060Δ by blue light is considerably smaller than that of the forward photoconversion. The weak absorption of green light can be rationalized by increased stability of the 15E lit state’s second thioether linkage, yielding a photoproduct equilibrium that disfavors the cleaved green-absorbing photoproduct. This unusual spectral behavior potentially implicates a distinct photobiological role for this clade of DGCs, because they are less sensitive to inhibition by green light than prototypical blue/green CBCRs.
A new class of dual cysteine red/blue CBCRs evolved within the CBCR-regulated DGC family
Lyn8106_0097Δ and Lyn.aest_0230Δ were found to be atypical red/blue DGCs. The previously described red/blue CBCR AM1_1186g2 is descended from canonical red/green CBCRs (Narikawa et al., 2014; Rockwell et al., 2017) as part of the XRG lineage (Fushimi et al., 2017b). By contrast, Lyn8106_0097Δ and Lyn.aest_0230Δ possess a different second cysteine residue and are not descended from canonical red/green CBCRs; instead, they may derive from the alternative NpR3784 lineage (Rockwell et al., 2015a). Hence, these CBCR-GGDEF proteins represent a distinct two-cysteine CBCR subfamily which can sense blue light. We propose designating such proteins as the RB2 subfamily, reflecting prior discovery of red/blue CBCRs that utilize a different second Cys residue (Narikawa et al., 2014). Replacement of the second Cys with Ala via site-directed mutagenesis did not prevent photoisomerization but did prevent formation of the blue-absorbing 15E state. The large spectral difference between the maximum 15Z and 15E absorbance in members of the RB2 subfamily provides little overlap between the “activating” and “repressing” light conditions, making these proteins interesting candidates for orthogonal multi-channel applications in optogenetics and synthetic biology.
Signal transduction via multiple CBCR domains
The tandem CBCR-GGDEF Nod_0172 provides an opportunity to examine the regulatory roles of tandem CBCR domains using readily assayed DGC output activity. Red light elicits the most active state of the enzyme by simultaneously triggering efficient conversion of the N-terminal GAF1 to its 15E state and incomplete photoconversion of the C-terminal GAF2 due to partial spectral overlap with the green-absorbing 15Z dark state. Shorter wavelengths of light are less effective in activating Nod_0172 because of the complex interplay between forward and reverse photoconversion. By contrast, green light elicits the highest activity of Nod_0172Δ, which lacks the GAF1 domain that functions as an inhibitor of DGC activity under green light. Many cyanobacterial photoreceptors contain several CBCR domains in tandem followed by a C-terminal output, so Nod_0172 may prove a suitable model system for understanding how these proteins integrate varied light conditions into a unified output response. Detailed structural analysis of the multi-domain phytochrome-GGDEF (IsPadC) suggested a “violin” model in which light regulated DGC activity is propagated through multiple structural domains by creating a dynamic set of conformations which favor activation/deactivation (Gourinchas et al., 2017). If this model holds in multi-domain CBCR proteins, each separate domain may play a key role in creating the correct structural orientation for dimerization and activation in response to varying light stimuli (Lim et al., 2014).
CBCR-regulated DGC encompass a broad spectral range and provide good lead candidates for second messenger-based optogenetic reagent development
We have demonstrated the benefit of examining the natural diversity of light-regulated enzymes that generate a second messenger not used by eukaryotes. While many of the proteins are not ready-made optogenetic tools, these proteins that both regulate c-di-GMP levels in a light-regulated manner and trigger cell settling when heterologously expressed in E. coli have not been examined before now. As such, this work lays the groundwork for further investigations into understanding the structural underpinnings of CBCR-GGDEF systems, which in turn will inform future efforts for optimizing light-regulated activities and/or for informed design of robust light-regulated fusion proteins. Optogenetic applications favor small constructs such as CBCR-GGDEFs, and the spectrally and enzymatically diversity of these reversible modules may prove well suited to controlling genetic circuits by utilizing a wide color palette of light from the near UV to the near IR and beyond.
Materials and methods
Bioinformatics
CBCR-GGDEF protein sequences were identified by BLASTP searches of the GenBANK (https://www.ncbi.nlm.nih.gov/) and DOE-IMG (https://genome.jgi.doe.gov/portal/) databases using the GAF-GGDEF bidomain sequence of Tlr0924 as query. CBCR and GGDEF alignments (Supplemental Figures S2–S4) were constructed using MAFFT (Katoh and Standley, 2013) as previously described (Rockwell and Lagarias, 2017). For structurally informed maximum-likelihood phylogenetic analysis, PDB accessions 4FOF and 4GLQ (CBCR-GAF domains) and 3I5A, 3IGN, 4ZMM, and 4ZVF (DGC-GGDEF domains) were used as references to generate input files for PhyML-structure (Guindon et al., 2005), and phylogenetic analyses were carried out as described (Rockwell and Lagarias, 2017); notably, this procedure removes the reference sequences prior to calculating the phylogeny so that both calculations use identical sets of full-length sequences. Phylogenetic trees were visualized using FigTree v1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/) and then used for construction of a single “tanglegram” representation using Adobe Illustrator. Additional sequence alignments of linker regions were constructed independently using MUSCLE (Edgar, 2004). Secondary structure predictions were performed using Phyre2 (http://www.sbg.bio.ic.ac.uk/).
Protein expression and spectral analysis
Fusion proteins were created using a protocol adapted from the sequence and ligation independent cloning (SLIC) method (Li and Elledge, 2012). Os6304_3021Δ and NpR1060Δ were cloned from genomic DNA as described previously (Rockwell et al., 2012a). Synthetic DNA constructs of S.hof_0333, Nod_0172, Cyan7822_5462Δ, Lyn8106_0097Δ, and Lyn.Aest_0230Δ were codon optimized by and purchased from GenScript. Proteins were expressed as intein-CBD fusion proteins in E. coli engineered to produce PCB using a dual-plasmid system as described previously (Gambetta and Lagarias, 2001; Rockwell et al., 2012a). After cell lysis, CBCR-GGDEF fusion proteins were affinity purified on a chitin column (NEB) in accordance with the manufacturer’s directions and dialyzed into TKKG buffer (25 mM TES-KOH, pH 7.5; 25 mM KCl; 10% v/v glycerol). Absorption spectra were recorded in TKKG buffer at 25°C on a Cary 50 spectrophotometer equipped with a thermostatted cell holder (Quantum Northwest) that was illuminated to photosaturation from above with a 3-mW green (532 nm) laser pointer or with a water-filtered 75-W xenon source passed through bandpass filters. Bandpass filters used for saturating photoconversion were 650 nm center/40 nm width, 600 nm/40 nm, 550 nm/70 nm, 500 nm/25 nm, 450 nm/25 nm, 400 nm/70 nm, 334 nm/10 nm, and 280 nm/10 nm. The BCA protein assay (Pierce) was used to determine protein concentration with bovine serum albumin as a protein standard (Smith et al., 1985). Denaturation assays to determine the configuration of each photostate were performed as described previously (Rockwell et al., 2012a).
In vitro DGC assays
DGC assays were performed in triplicate as described previously (Blain-Hartung et al., 2017). Purified protein in DGC assay buffer (0.5 M Tris–HCl buffer pH 7.6 containing 0.05 M NaCl, 0.01 M MgCl2, and 0.5 mM EDTA) was incubated at a final concentration of 5 µM for 30 min at 37°C under continuous exposure to saturating blue, green, or red light. Light conditions were achieved using cool white fluorescent tubes filtered through LEE plastic (#071 Tokyo Blue, #090 Dark Yellow Green, and #164 Flame Red) with a light fluence rate of 4–5 µmol m−2 s−1. Photoconversion was assessed by absorption spectroscopy prior to assay initiation. To initiate catalysis, GTP (Thermo Fisher Scientific) was added to a final concentration of 1 mM. Assays were terminated by placing in a 100°C thermal block for 5 min followed by centrifugation at 15,000 × g for 5 min. Control measurements were performed to ensure that c-di-GMP recovery was not affected by boiling. A 15 µL aliquot of reaction mixture was analyzed using reverse phase high-performance liquid chromatography (RP-HPLC) as described previously (Blain-Hartung et al., 2017). A standard curve of c-di-GMP (Biolog) was used and integrated peak area calculations were performed using Agilent ChemStation software.
Cell aggregation assays
Escherichia coli cultures co-expressing CBCR-GGDEF and PCB biosynthesis constructs were exposed to continuous red, blue, or green light using LEE filters after induction with arabinose and isopropyl β-d-1-thiogalactopyranoside, as described previously (Blain-Hartung et al., 2017). The light fluence rate in the culture shaker was 4–5 µmol m−2 s−1. After incubation for 16 h, cell cultures were immediately placed in darkness without shaking. To analyze the degree of cell aggregation, initial OD600 and initial OD600/final OD600 measurements were performed as described previously (Blain-Hartung et al., 2017).
Accession numbers
Accession numbers from which the constructs used for these studies can be found in Supplemental Table S1.
Supplemental data
Supplemental Figure S1. Spectral analysis of selected fusion CBCR-DGC proteins.
Supplemental Figure S2. CBCR GAF domain alignment.
Supplemental Figure S3. DGC GGDEF domain alignment.
Supplemental Figure S4. CBCR-DGC linker alignment.
Supplemental Figure S5 . Spectroscopic characterization of second cysteine mutant constructs of novel R/B CBCR proteins and the GAF1-only construct, Nod_0172(GAF1).
Supplemental Table S1. Accession numbers and amino acids of GAF-GGDEF constructs.
Supplementary Material
Acknowledgments
The authors thank Ms. Shelley Martin for technical assistance and helpful discussions.
Funding
This work was supported by grants from the Chemical Sciences, Geosciences, and Biosciences Division, Office of Basic Energy Sciences, Office of Science, U.S. Department of Energy (DOE DE-FG02-09ER16117) to J.C.L., from the National Institute of General Medical Sciences (RO1-M068552 and R35-GM139598) to J.C.L. and (T32-GM07377) to M.B.-H. The content is solely the responsibility of the authors and does not necessarily represent the official views of the Department of Energy or the National Institutes of Health.
Conflict of interest statement. None declared.
M.B.-H. designed, performed, and analyzed the experiments, and wrote the manuscript. N.C.R. designed, performed, and analyzed the experiments, and edited the manuscript. J.C.L. interpreted the data and edited the manuscript. All authors approved the final version of the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: J. Clark Lagarias (jclagarias@ucdavis.edu).
References
- Anders K, Essen LO (2015) The family of phytochrome-like photoreceptors: diverse, complex and multi-colored, but very useful. Curr Opin Struct Biol 35: 7–16 [DOI] [PubMed] [Google Scholar]
- Beattie GA, Hatfield BM, Dong HL, McGrane RS (2018) Seeing the light: The roles of red- and blue-light sensing in plant microbes. Annu Rev Phytopathol 56: 41–66 [DOI] [PubMed] [Google Scholar]
- Blain-Hartung M, Rockwell NC, Moreno MV, Martin SS, Gan F, Bryant DA, Lagarias JC (2018) Cyanobacteriochrome-based photoswitchable adenylyl cyclases (cPACs) for broad spectrum light regulation of cAMP levels in cells. J Biol Chem 293: 8473–8483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blain-Hartung MD, Rockwell NC, Lagarias JC (2017) Light-regulated synthesis of cyclic-di-GMP by a bidomain construct of the cyanobacteriochrome Tlr0924 (SesA) without stable dimerization. Biochemistry 56: 6145–6154 [DOI] [PubMed] [Google Scholar]
- Burgie ES, Walker JM, Phillips GN Jr, Vierstra RD (2013) A photo-labile thioether linkage to phycoviolobilin provides the foundation for the blue/green photocycles in DXCF-cyanobacteriochromes. Structure 21: 88–97 [DOI] [PubMed] [Google Scholar]
- Chen Y, Xia J, Su Z, Xu G, Gomelsky M, Qian G, Liu F (2017) Lysobacter PilR, the regulator of type IV pilus synthesis, controls antifungal antibiotic production via a cyclic di-GMP pathway. Appl Environ Microbiol 83: e03397–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christen M, Kulasekara HD, Christen B, Kulasekara BR, Hoffman LR, Miller SI (2010) Asymmetrical distribution of the second messenger c-di-GMP upon bacterial cell division. Science 328: 1295–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto G, Ikeuchi M (2020) Blue-/green-light-responsive cyanobacteriochromes are cell shade sensors in red-light replete niches. iScience 23: 100936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto G, Ni Ni W, Narikawa R, Ikeuchi M (2015) Three cyanobacteriochromes work together to form a light color-sensitive input system for c-di-GMP signaling of cell aggregation. Proc Natl Acad Sci USA 112: 8082–8087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto G, Nomura R, Shimada TNi-Ni-WinNarikawa R, Ikeuchi M (2014) Cyanobacteriochrome SesA is a diguanylate cyclase that induces cell aggregation in Thermosynechococcus. J Biol Chem 289: 24801–24809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etzl S, Lindner R, Nelson MD, Winkler A (2018) Structure-guided design and functional characterization of an artificial red light-regulated guanylate/adenylate cyclase for optogenetic applications. J Biol Chem 293: 9078–9089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Milmanda GL, Ballare CL (2021) Shade avoidance: Expanding the color and hormone palette. Trends Plant Sci 26: 509–523. [DOI] [PubMed] [Google Scholar]
- Fernandez-Rodriguez J, Moser F, Song M, Voigt CA (2017) Engineering RGB color vision into Escherichia coli. Nat Chem Biol 5: 706–708 [DOI] [PubMed] [Google Scholar]
- Fiedler B, Borner T, Wilde A (2005) Phototaxis in the cyanobacterium Synechocystis sp. PCC 6803: role of different photoreceptors. Photochem Photobiol 81: 1481–1488 [DOI] [PubMed] [Google Scholar]
- Fushimi K, Enomoto G, Ikeuchi M, Narikawa R (2017a) Distinctive properties of dark reversion kinetics between two red/green-type cyanobacteriochromes and their application in the photoregulation of cAMP synthesis. Photochem Photobiol 93: 681–691 [DOI] [PubMed] [Google Scholar]
- Fushimi K, Ikeuchi M, Narikawa R (2017b) The expanded red/green cyanobacteriochrome lineage: An evolutionary hot spot. Photochem Photobiol 93: 903–906 [DOI] [PubMed] [Google Scholar]
- Fushimi K, Narikawa R (2021) Phytochromes and cyanobacteriochromes: Photoreceptor molecules incorporating a linear tetrapyrrole chromophore. In Yawo H, Kandori H, Koizumi A, Kageyama R, eds, Optogenetics. Advances in Experimental Medicine and Biology, Ed 2021/01/06 Vol 1293. Springer, Singapore, pp 167–187 [DOI] [PubMed] [Google Scholar]
- Gambetta GA, Lagarias JC (2001) Genetic engineering of phytochrome biosynthesis in bacteria. Proc Natl Acad Sci USA 98: 10566–10571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser C, Taiber S, Yeh CM, Wittig CH, Hegemann P, Ryu S, Wunder F, Moglich A (2014) Engineering of a red-light-activated human cAMP/cGMP-specific phosphodiesterase. Proc Natl Acad Sci USA 111: 8803–8808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomelsky M, Hoff WD (2011) Light helps bacteria make important lifestyle decisions. Trends Microbiol 19: 441–448 [DOI] [PubMed] [Google Scholar]
- Gourinchas G, Etzl S, Gobl C, Vide U, Madl T, Winkler A (2017) Long-range allosteric signaling in red light-regulated diguanylyl cyclases. Sci Adv 3: e1602498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourinchas G, Heintz U, Winkler A (2018) Asymmetric activation mechanism of a homodimeric red light regulated photoreceptor. eLife 7: e34815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Lethiec F, Duroux P, Gascuel O (2005) PHYML online—a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res 33: W557–W559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose Y, Rockwell NC, Nishiyama K, Narikawa R, Ukaji Y, Inomata K, Lagarias JC, Ikeuchi M (2013) Green/red cyanobacteriochromes regulate complementary chromatic acclimation via a protochromic photocycle. Proc Natl Acad Sci USA 110: 4974–4979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose Y, Shimada T, Narikawa R, Katayama M, Ikeuchi M (2008) Cyanobacteriochrome CcaS is the green light receptor that induces the expression of phycobilisome linker protein. Proc Natl Acad Sci USA 105: 9528–9533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi M, Ishizuka T (2008) Cyanobacteriochromes: a new superfamily of tetrapyrrole-binding photoreceptors in cyanobacteria. Photochem Photobiol Sci 7: 1159–1167 [DOI] [PubMed] [Google Scholar]
- Ishizuka T, Kamiya A, Suzuki H, Narikawa R, Noguchi T, Kohchi T, Inomata K, Ikeuchi M (2011) The cyanobacteriochrome, TePixJ, isomerizes its own chromophore by converting phycocyanobilin to phycoviolobilin. Biochemistry 50: 953–961 [DOI] [PubMed] [Google Scholar]
- Jenal U, Reinders A, Lori C (2017) Cyclic di-GMP: second messenger extraordinaire. Nat Rev Microbiol 15: 271–284 [DOI] [PubMed] [Google Scholar]
- Karaolis DK, Rashid MH, Chythanya R, Luo W, Hyodo M, Hayakawa Y (2005) c-di-GMP (3′–5′-cyclic diguanylic acid) inhibits Staphylococcus aureus cell–cell interactions and biofilm formation. Antimicrob Agents Chemother 49: 1029–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30: 772–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MZ, Elledge SJ (2012) SLIC: a method for sequence- and ligation-independent cloning. In Peccoud J, ed., Gene Synthesis. Methods in Molecular Biology (Methods and Protocols), Vol 852. Humana Press, Totowa, NJ, pp 51–59 [DOI] [PubMed] [Google Scholar]
- Lim S, Rockwell NC, Martin SS, Dallas JL, Lagarias JC, Ames JB (2014) Photoconversion changes bilin chromophore conjugation and protein secondary structure in the violet/orange cyanobacteriochrome NpF2164g3. Photochem Photobiol Sci 13: 951–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner R, Hartmann E, Tarnawski M, Winkler A, Frey D, Reinstein J, Meinhart A, Schlichting I (2017) Photoactivation mechanism of a bacterial light-regulated adenylyl cyclase. J Mol Biol 429: 1336–1351 [DOI] [PubMed] [Google Scholar]
- Ma Q, Hua HH, Chen Y, Liu BB, Kramer AL, Scheer H, Zhao KH, Zhou M (2012) A rising tide of blue-absorbing biliprotein photoreceptors: characterization of seven such bilin-binding GAF domains in Nostoc sp. PCC7120. FEBS J 279: 4095–4108 [DOI] [PubMed] [Google Scholar]
- Mandalari C, Losi A, Gärtner W (2013) Distance-tree analysis, distribution and co-presence of bilin- and flavin-binding prokaryotic photoreceptors for visible light. Photochem Photobiol Sci 12: 1144–1157 [DOI] [PubMed] [Google Scholar]
- Mares J, Johansen JR, Hauer T, Zima J, Ventura S, Cuzman O, Tiribilli B, Kastovsky J (2019) Taxonomic resolution of the genus Cyanothece (Chroococcales, Cyanobacteria), with a treatment on Gloeothece and three new genera, Crocosphaera, Rippkaea, and Zehria. J Phycol 55: 578–610 [DOI] [PubMed] [Google Scholar]
- Masuda S (2012) Light detection and signal transduction in the BLUF photoreceptors. Plant Cell Physiol 54: 171–179 [DOI] [PubMed] [Google Scholar]
- Moglich A, Ayers RA, Moffat K (2009) Design and signaling mechanism of light-regulated histidine kinases. J Mol Biol 385: 1433–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moglich A, Moffat K (2007) Structural basis for light-dependent signaling in the dimeric LOV domain of the photosensor YtvA. J Mol Biol 373: 112–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möglich A, Moffat K (2010) Engineered photoreceptors as novel optogenetic tools. Photochem Photobiol Sci 9: 1286–1300 [DOI] [PubMed] [Google Scholar]
- Möglich A, Yang X, Ayers RA, Moffat K (2010) Structure and function of plant photoreceptors. Annu Rev Plant Biol 61: 21–47 [DOI] [PubMed] [Google Scholar]
- Nagano S (2016) From photon to signal in phytochromes: similarities and differences between prokaryotic and plant phytochromes. J Plant Res 129: 123–135 [DOI] [PubMed] [Google Scholar]
- Narikawa R, Enomoto GNi-Ni-WinFushimi K, Ikeuchi M (2014) A new type of dual-Cys cyanobacteriochrome GAF domain found in cyanobacterium Acaryochloris marina, which has an unusual red/blue reversible photoconversion cycle. Biochemistry 53: 5051–5059 [DOI] [PubMed] [Google Scholar]
- Pham VN, Kathare PK, Huq E (2018) Phytochromes and phytochrome interacting factors. Plant Physiol 176: 1025–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu L, Yang S, Xia A, Jin F (2018) Optogenetics manipulation enables prevention of biofilm formation of engineered Pseudomonas aeruginosa on surfaces. ACS Synth Biol 7: 200–208 [DOI] [PubMed] [Google Scholar]
- Ramakrishnan P, Tabor JJ (2016) Repurposing Synechocystis PCC6803 UirS-UirR as a UV-violet/green photoreversible transcriptional regulatory tool in E. coli. ACS Synth Biol 5: 733–740 [DOI] [PubMed] [Google Scholar]
- Rensing SA, Sheerin DJ, Hiltbrunner A (2016) Phytochromes: more than meets the eye. Trends Plant Sci 21: 543–546 [DOI] [PubMed] [Google Scholar]
- Rockwell NC, Lagarias JC (2010) A brief history of phytochromes. ChemPhysChem 11: 1172–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell NC, Lagarias JC (2017) Ferredoxin-dependent bilin reductases in eukaryotic algae: Ubiquity and diversity. J Plant Physiol 217: 57–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell NC, Lagarias JC (2017) Phytochrome diversification in cyanobacteria and eukaryotic algae. Curr Opin Plant Biol 37: 87–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell NC, Martin SS, Feoktistova K, Lagarias JC (2011) Diverse two-cysteine photocycles in phytochromes and cyanobacteriochromes. Proc Natl Acad Sci USA 108: 11854–11859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell NC, Martin SS, Gan F, Bryant DA, Lagarias JC (2015a) NpR3784 is the prototype for a distinctive group of red/green cyanobacteriochromes using alternative Phe residues for photoproduct tuning. Photochem Photobiol Sci 14: 258–269 [DOI] [PubMed] [Google Scholar]
- Rockwell NC, Martin SS, Gulevich AG, Lagarias JC (2012a) Phycoviolobilin formation and spectral tuning in the DXCF cyanobacteriochrome subfamily. Biochemistry 51: 1449–1463 [DOI] [PubMed] [Google Scholar]
- Rockwell NC, Martin SS, Lagarias JC (2012b) Mechanistic insight into the photosensory versatility of DXCF cyanobacteriochromes. Biochemistry 51: 3576–3585 [DOI] [PubMed] [Google Scholar]
- Rockwell NC, Martin SS, Lagarias JC (2012c) Red/green cyanobacteriochromes: sensors of color and power. Biochemistry 51: 9667–9677 [DOI] [PubMed] [Google Scholar]
- Rockwell NC, Martin SS, Lagarias JC (2015b) Identification of DXCF cyanobacteriochrome lineages with predictable photocycles. Photochem Photobiol Sci 14: 929–941 [DOI] [PubMed] [Google Scholar]
- Rockwell NC, Martin SS, Lagarias JC (2016) Identification of cyanobacteriochromes detecting far-red light. Biochemistry 55: 3907–3919 [DOI] [PubMed] [Google Scholar]
- Rockwell NC, Martin SS, Lagarias JC (2017) There and back again: Loss and reacquisition of two-cys photocycles in cyanobacteriochromes. Photochem Photobiol 93: 741–754 [DOI] [PubMed] [Google Scholar]
- Rockwell NC, Njuguna SL, Roberts L, Castillo E, Parson VL, Dwojak S, Lagarias JC, Spiller SC (2008) A second conserved GAF domain cysteine is required for the blue/green photoreversibility of cyanobacteriochrome Tlr0924 from Thermosynechococcus elongatus. Biochemistry 47: 7304–7316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell NC, Su YS, Lagarias JC (2006) Phytochome structure and signaling mechanisms. Annu Rev Plant Biol 57: 837–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romling U, Galperin MY, Gomelsky M (2013) Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev 77: 1–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryjenkov DA, Tarutina M, Moskvin OV, Gomelsky M (2005) Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: Insights into biochemistry of the GGDEF protein domain. J Bacteriol 187: 1792–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu MH, Fomicheva A, Moskvin OV, Gomelsky M (2017) Optogenetic module for dichromatic control of c-di-GMP signaling. J Bacteriol 199: e00014–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu MH, Gomelsky M (2014) Near-infrared light responsive synthetic c-di-GMP module for optogenetic applications. ACS Synth Biol 3: 802–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu MH, Kang IH, Nelson MD, Jensen TM, Lyuksyutova AI, Siltberg-Liberles J, Raizen DM, Gomelsky M (2014) Engineering adenylate cyclases regulated by near-infrared window light. Proc Natl Acad Sci USA 111: 10167–10172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuergers N, Mullineaux CW, Wilde A (2017) Cyanobacteria in motion. Curr Opin Plant Biol 37: 109–115 [DOI] [PubMed] [Google Scholar]
- Shu C, Yi G, Watts T, Kao CC, Li P (2012) Structure of STING bound to cyclic di-GMP reveals the mechanism of cyclic dinucleotide recognition by the immune system. Nat Struct Mol Biol 19: 722–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KD, Lipchock SV, Ames TD, Wang J, Breaker RR, Strobel SA (2009) Structural basis of ligand binding by a c-di-GMP riboswitch. Nat Struct Mol Biol 16: 1218–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PK, Krohn RI, Hemanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olsen BJ, Klenk DC (1985) Measurement of protein using bicinchoninic acid. Anal Biochem 150: 76–85 [DOI] [PubMed] [Google Scholar]
- Stuven B, Stabel R, Ohlendorf R, Beck J, Schubert R, Moglich A (2019) Characterization and engineering of photoactivated adenylyl cyclases. Biol Chem 400: 429–441 [DOI] [PubMed] [Google Scholar]
- Sudarsan N, Lee ER, Weinberg Z, Moy RH, Kim JN, Link KH, Breaker RR (2008) Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science 321: 411–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor JJ, Levskaya A, Voigt CA (2011) Multichromatic control of gene expression in Escherichia coli. J Mol Biol 405: 315–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarutina M, Ryjenkov DA, Gomelsky M (2006) An unorthodox bacteriophytochrome from Rhodobacter sphaeroides involved in turnover of the second messenger c-di-GMP. J Biol Chem 281: 34751–34758 [DOI] [PubMed] [Google Scholar]
- Viczian A, Klose C, Adam E, Nagy F (2017) New insights of red light-induced development. Plant Cell Environ 40: 2457–2468 [DOI] [PubMed] [Google Scholar]
- Villafani Y, Yang HW, Park YI (2020) Color sensing and signal transmission diversity of cyanobacterial phytochromes and cyanobacteriochromes. Mol Cells 43: 509–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiltbank LB, Kehoe DM (2016) Two cyanobacterial photoreceptors regulate photosynthetic light harvesting by sensing teal, green, yellow, and red light. mBio 7: e02130–02115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiltbank LB, Kehoe DM (2019) Diverse light responses of cyanobacteria mediated by phytochrome superfamily photoreceptors. Nat Rev Microbiol 17: 37–50 [DOI] [PubMed] [Google Scholar]
- Wu SH, Lagarias JC (2000) Defining the bilin lyase domain: Lessons from the extended phytochrome superfamily. Biochemistry 39: 13487–13495 [DOI] [PubMed] [Google Scholar]
- Ziegler T, Moglich A (2015) Photoreceptor engineering. Front Mol Biosci 2: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.