Abstract
Myrosinases are β-thioglucoside glucosidases that are unique to the Brassicales order. These enzymes hydrolyze glucosinolates to produce compounds that have direct antibiotic effects or that function as signaling molecules in the plant immune system, protecting plants from pathogens and insect pests. However, the effects of jasmonic acid (JA), a plant hormone that is crucial for plant disease resistance, on myrosinase activity remain unclear. Here, we systematically studied the effects of JA on myrosinase activity and explored the associated internal transcriptional regulation mechanisms. Exogenous application of JA significantly increased myrosinase activity, while the inhibition of endogenous JA biosynthesis and signaling reduced myrosinase activity. In addition, some myrosinase genes in Arabidopsis (Arabidopsis thaliana) were upregulated by JA. Further genetic and biochemical evidence showed that transcription factor FAMA interacted with a series of JASMONATE ZIM-DOMAIN proteins and affected JA-mediated myrosinase activity. However, among the JA-upregulated myrosinase genes, only THIOGLUCOSIDE GLUCOHYDROLASE 1 (TGG1) was positively regulated by FAMA. Further biochemical analysis showed that FAMA bound to the TGG1 promoter to directly mediate TGG1 expression in conjunction with Mediator complex subunit 8 (MED8). Together, our results provide evidence that JA acts as an important signal upstream of the FAMA/MED8–TGG1 pathway to positively regulate myrosinase activity in Arabidopsis.
The interaction of FAMA transcription factor and jasmonic acid response proteins regulates myrosinase activity by affecting the expression of THIOGLUCOSIDE GLUCOHYDROLASE 1.
Introduction
Glucosinolates are secondary metabolites that are found only in plants in the Brassicales order (Canistro et al., 2004; Wang et al., 2011; Fernandez-Calvo et al., 2020). Although these compounds themselves do not have biological activity, they can be decomposed via myrosinase catalysis into many types of metabolites that are deterrent or toxic to insects and pathogens (Andersson et al., 2009). Glucosinolates and myrosinases are normally physically separated at the tissue level or at the single-cell level in plants. However, these components can be brought together as a result of tissue damage caused by insects or pathogens. Glucosinolate–myrosinase interactions release large amounts of biologically active hydrolysis products (Sugiyama and Hirai, 2019; Bhat et al., 2020; Kayum et al., 2020; Mocniak et al., 2020).
In Arabidopsis (Arabidopsis thaliana), myrosinases are divided into two categories based on conserved amino acid residues: the QE- and the EE-type myrosinases. The QE-type myrosinases contain Gln and Glu residues in the catalytic site. In Arabidopsis, there are six QE-type myrosinases, which are encoded by the thioglucoside glucohydrolase (TGG) genes (TGG1–6). TGG1 and TGG2 are expressed in the aboveground parts of the plant, and their encoded enzymes decompose aliphatic glucosinolates and indole glucosinolates (Barth and Jander, 2006); TGG3 and TGG6 are pseudogenes in Arabidopsis Col-0 (Xue et al., 1995); and TGG4 and TGG5 are only expressed in plant roots and are associated with auxin synthesis (Xue et al., 1995; Fu et al., 2016). A previous study showed that, despite the replacement of the Gln residue in the PENETRATION2 (PEN2) protein by Glu, PEN2 still exerted myrosinase activity against indol-3-ylmethyl glucosinolate (I3G) and its 4-methoxy analog (Bednarek et al., 2009). Similarly, PYK10 (BGLU23) myrosinase activity was maintained despite the substitution of the Gln residue by Glu (Nakano et al., 2017). Proteins containing the two glutamic acid residues conserved in PEN2 and PYK10 are designated EE-type myrosinases (Sugiyama and Hirai, 2019). There are 16 EE-type myrosinase genes in Arabidopsis: BGLU18–BGLU33. Some of these genes are involved in specific stresses. BGLU18, one of the primary plant proteins inducing ER bodies, can be upregulated by jasmonic acid (JA) or wounding (Lee et al., 2006; Ogasawara et al., 2009; Yamada et al., 2011; Nakazaki et al., 2019). BGLU19 is induced by high salt stress, and BGLU19 mutants are tolerant to salt stress (Cao et al., 2017). PEN2 plays an important role in plant resistance to pathogens (Bednarek et al., 2009). Both BGLU28 and BGLU30 are induced by sulfur depletion (Hirai et al., 2003; Matsushima et al., 2003; Hirai and Saito, 2004); BGLU30 is also induced by extended darkness or senescence (Fujiki et al., 2001; Lee et al., 2007). BGLU33 mutants are more sensitive to salt stress than wild-type (Xu et al., 2012). In addition, these proteins have myrosinase activities with substrate selectivity. In vitro hydrolysis of ABA glucose ester (ABA-GE) showed that BGLB18 and BGLB33 exhibit ABA-GE hydrolyzing activity in vitro (Lee et al., 2006; Xu et al., 2012). The hydrolysis of 4-methoxyindol-3-ylmethyl glucosinolate (4MI3G) was remarkably retarded in the bglu18 pyk10 mutant compared with that in wild-type, indicating that 4MI3G is hydrolyzed by BGLU18 and/or PYK10 (Nakazaki et al., 2019). In vitro glucosinolate hydrolysis experiments showed that PYK10 (also known as BGLU23) and its closest homologlucosinolates (BGLU21 and BGLU22) specifically hydrolyzed scopolin and some substrates whose aglycone moiety is similar to scopolin (e.g. esculin and 4-MU-glucoside; Ahn et al., 2010). However, they did not hydrolyze sinigrin (Ahn et al., 2010). In addition to coumarin glucosides, in vitro activity testing of recombinant PYK10 protein toward I3G showed that PYK10 exhibited a high hydrolysis activity toward I3G (Nakano et al., 2017). Recent research on glucosinolate hydrolysis experiments with pyk10 bglu21 double mutant showed PYK10 and/or BGLU21 constitute the main source of myrosinase activity against aliphatic and indolic glucosinolates in young Arabidopsis seedlings (Yamada et al., 2020). Heterologous expression of a form of PEN2 lacking 64 residues from the C-terminal region could hydrolysis I3G and 4MI3G in vitro (Bednarek et al., 2009).
JA is an important signal molecule that regulates many plant physiological processes. In Arabidopsis, JA directly regulates root growth, plant fertility, anthocyanin accumulation, senescence, and stress resistance (Wasternack and Hause, 2013; Wasternack and Feussner, 2018). Previous studies have thoroughly characterized the JA signal transduction pathway. First, JA-Ile, the active form of JA, is recognized by the F-box protein COI1 in the nucleus (Fonseca et al., 2009; Sheard et al., 2010). COI1 interacts with the Jas domain of JASMONATE ZIM-DOMAIN (JAZ) proteins and degrades JAZ proteins through the 26S proteasome (Chini et al., 2007; Thines et al., 2007). JAZ proteins also directly interact with a series of specific transcription factors to inhibit their transcriptional activities, thus inhibiting specific physiological functions of JA (Fernandez-Calvo et al., 2011; Hu et al., 2013; Zhai et al., 2015; Chini et al., 2016; Howe et al., 2018). When JAZ proteins are degraded, transcription factors are released, reactivating JAZ transcription and triggering the JA signal. For example, the basic helix-loop-helix (bHLH)-class transcription factors MYC2, MYC3, and MYC4, as well as transcription factors TARGET OF EAT1 (TOE1) and TOE2, regulate plant root growth, resistance responses, and flowering by directly interacting with JAZ proteins (Fernandez-Calvo et al., 2011; Zhai et al., 2015). This suggests that JA regulates plant-specific responses via direct physical interaction with JAZ proteins and specific transcription factors that influence the expression of downstream genes.
FAMA is a bHLH transcription factor that was first shown to play an important role in stomatal development (Ohashi-Ito and Bergmann, 2006; Serna, 2007). FAMA and two other bHLH transcription factors, SPCH and MUTE, regulate three different stages of stomatal development (Lampard and Bergmann, 2007). In fama mutants, stomatal development is abnormal, and the plant is sterile (Ohashi-Ito and Bergmann, 2006). FAMA has more recently been shown to participate in the development of myrosin cells. The expression levels of TGG1 and TGG2 in fama mutants were substantially lower than expression levels in controls (Li and Sack, 2014; Shirakawa et al., 2014). Recently, we showed that, in conjunction with MED8, FAMA regulated plant resistance to Botrytis cinerea (Li et al., 2018). Therefore, FAMA may have multiple key functions in plants.
Although studies have shown that JA induces the expression of myrosinase genes (e.g. BGLU18, PYK10, and TGG1; Capella et al., 2001; Matsushima et al., 2002; Ogasawara et al., 2009), there is no evidence that JA-signaling specifically regulates myrosinase activity. Here, we systematically investigate the effect of JA on myrosinase activity using molecular and genetic methods. We found that treatment with exogenous methyl jasmonate (MeJA) significantly increased myrosinase activity, but that myrosinase activity was reduced in JA-synthesis and JA-signaling mutants. Our mechanistic study showed that JAZ proteins interacted with FAMA and inhibited the transcription function of FAMA to regulate TGG1 expression. MED8 was also involved in this process. Our results provide evidence that JA affects myrosinase activity by regulating the FAMA/MED8–TGG1 pathway.
Results
The JA signal positively regulates myrosinase activity
To verify the role of the plant hormone JA in glucosinolate hydrolysis, 21-d-old Arabidopsis Col-0 seedlings were treated with MeJA, and myrosinase activity was detected after treatment. We found that, as treatment time increased, plants treated with MeJA exhibited higher levels of myrosinase activity than the untreated controls, and that, as MeJA concentration increased from 0 to 100 µM, myrosinase activity increased concomitantly (Figure 1A). This suggested that MeJA increased myrosinase activity, in a time- and concentration-dependent manner.
Figure 1.
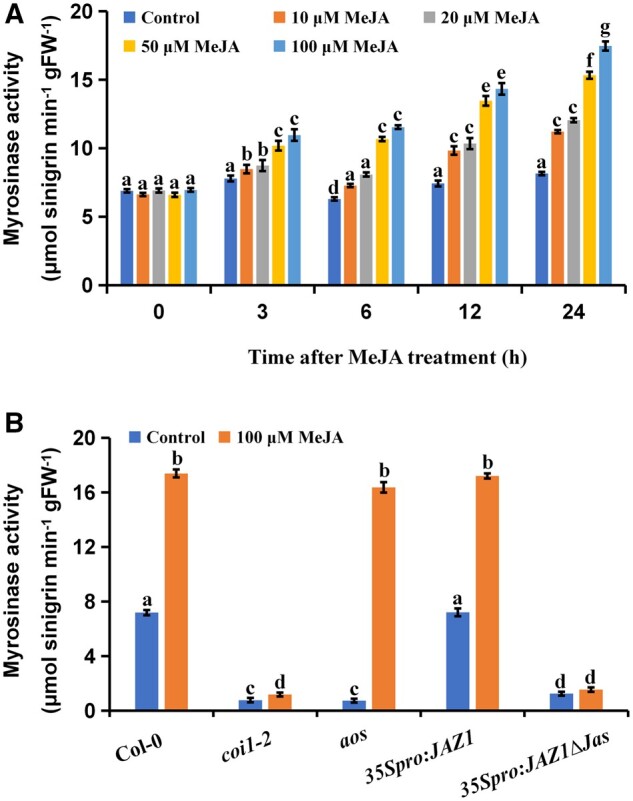
The JA signal positively regulates myrosinase activity. A and B, Myrosinase activity of 21-d-old seedlings at different times after treatment with different concentrations of MeJA (A) and of 21-d-old seedlings of different genetic materials (for JA synthesis and signal molecules) after treating with 100 μM of MeJA for 24 h (B). Values are means ± sem of 8–16 plants. The experiments were repeated at least 3 times with similar results. Different letters represent significant differences (P < 0.05, Student’s t test).
To verify the role of JA in glucosinolate hydrolysis, we performed glucosinolate hydrolysis experiments using Arabidopsis treated with various concentrations of MeJA for 72 h. We found that the glucosinolate contents of plants treated with MeJA decreased faster, and the rate of glucosinolate decrease was proportional to the concentration of MeJA (Supplemental Figure S1, A and B). That is, plants treated with higher concentrations of MeJA exhibited higher rates of glucosinolate hydrolysis (Supplemental Figure S1, A and B). This further indicated that JA promoted glucosinolate hydrolysis.
We investigated the role played by JA biosynthesis and signaling in the regulation of myrosinase activity. As allene oxide synthase (AOS) is an important enzyme in JA synthesis (Park et al., 2002), we tested whether AOS levels affected myrosinase activity. We found that myrosinase activity in aos mutants was low, but that aos mutants treated with MeJA exhibited similar levels of myrosinase activity to MeJA-treated wild-type Arabidopsis seedlings (Figure 1B). This suggested that AOS affected myrosinase activity, and that JA played an important role in the regulation of myrosinase activity.
The F-box protein COI1 is the JA receptor and a key positive regulator in the JA signaling pathway (Xie et al., 1998). Previous studies have shown that myrosinase activity in coi1 mutants is decreased (Capella et al., 2001). Consistent with this, we found that myrosinase activity decreased by 88.9% in the coi1-2 mutant compared with the wild-type (Figure 1B). To determine whether other molecular elements in the JA pathway are also involved in the regulation of myrosinase activity, we examined the effects of JAZ1, which acts as a transcriptional repressor of JA-responsive genes (Chini et al., 2007; Thines et al., 2007), on myrosinase activity. For this purpose, we detected myrosinase activity in 35Spro:JAZ1 and 35Spro:JAZ1Δ JAS plants, which ectopically express the JAZ1 cDNA and a Jas-domain deletion version of JAZ1 cDNA, respectively (Zhai et al., 2015). Myrosinase activity was significantly reduced in 35Spro:JAZ1Δ JAS plants, but was similar to that of the wild-type in 35Spro:JAZ1 plants (Figure 1B). These results suggested that the inhibition of the JA pathway led to the inhibition of myrosinase activity. Along with our glucosinolate hydrolysis experiment, this experiment provides further evidence that JA biosynthesis and signaling are important for glucosinolate hydrolysis (Supplemental Figure S1, C and D).
Effects of JA on myrosinase gene expression
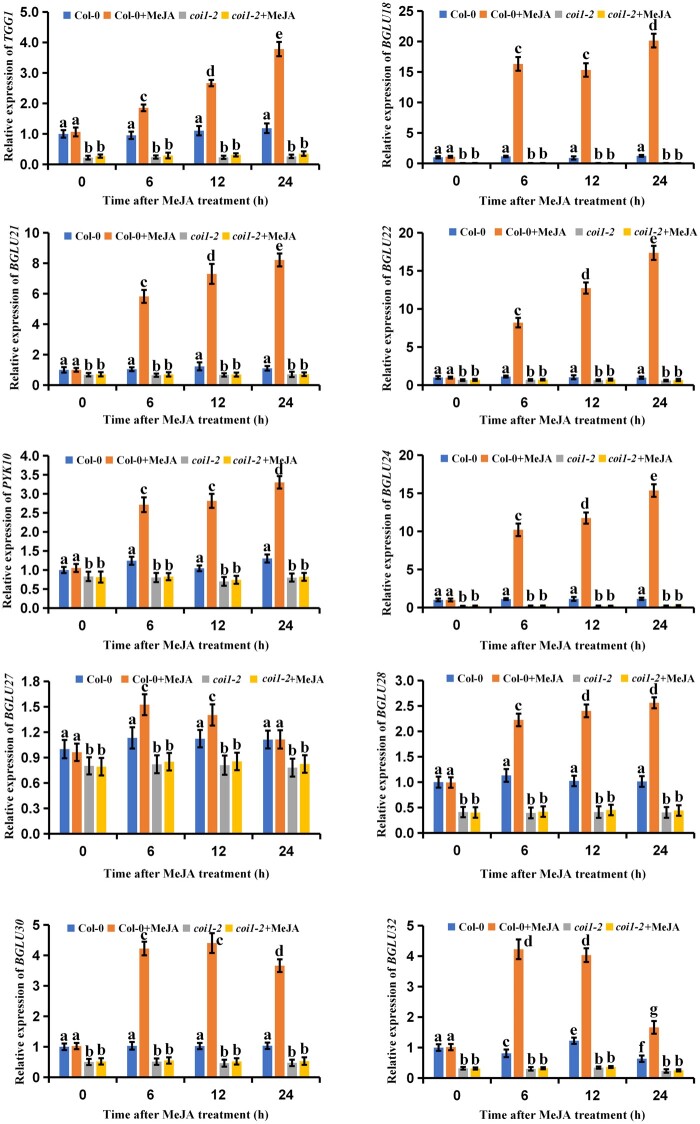
We then tested whether JA affected myrosinase gene expression. A total of 22 myrosinase genes have been reported in Arabidopsis (Sugiyama and Hirai, 2019), but two of these (TGG3 and TGG6) have been identified as pseudogenes in Arabidopsis Col-0 (Xue et al., 1995). Thus, we quantified changes in the relative expression levels of the remaining 20 myrosinase genes after MeJA treatment. We found that TGG1, BGLU18, BGLU21, BGLU22, PYK10, BGLU24, BGLU27, BGLU28, BGLU30, and BGLU32 were significantly upregulated after JA treatment in the wild-type Arabidopsis (Figure 2). Surprisingly, TGG2 and PEN2 were significantly downregulated by JA (Supplemental Figure S2), suggesting that JA might negatively regulate TGG2 and PEN2 expression levels. To confirm the regulatory effects of JA on myrosinase genes, we also quantified myrosinase gene expression in the coi1-2 mutants. We found that the JA-driven upregulation of the myrosinase genes was reduced in the JA-treated coi1-2 mutants as compared to the JA-treated wild-type plants (Figure 2). Conversely, TGG2 and PEN2 were upregulated in the JA-treated coi1-2 mutants as compared to the JA-treated wild-type plants (Supplemental Figure S2). Thus, these results suggest that JA positively regulates the expression of selected myrosinase genes in Arabidopsis.
Figure 2.
Effects of JA on myrosinase gene expression. Twenty-one-day-old seedlings of wild-type (Col-0) and coi1-2 were treated with 100 μM of MeJA for the indicated time periods. The treated plants were harvested for total RNA extraction and RT-qPCR assays. Means ± sem are relative values obtained from three technical replicates. Different letters represent significant differences (P < 0.05, Student’s t test).
A series of JAZ proteins interact with the FAMA transcription factor
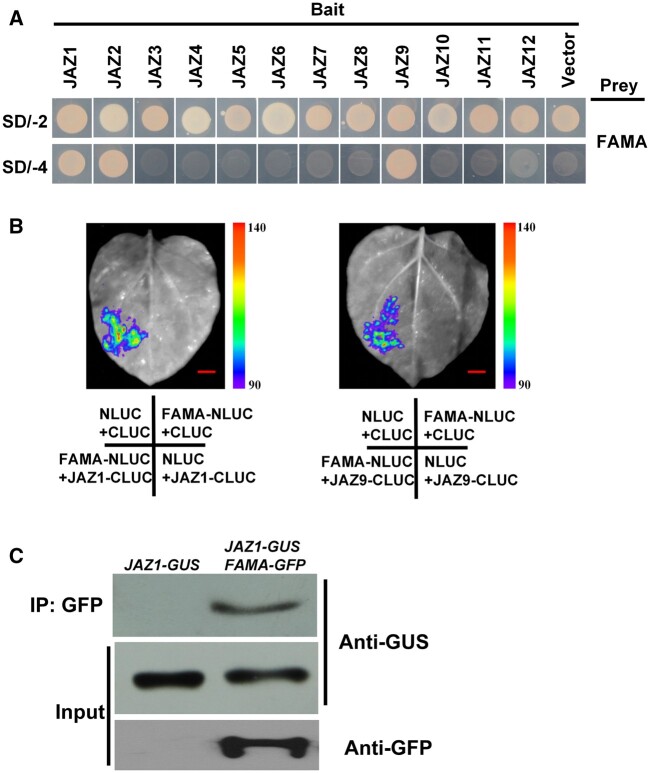
It has previously been shown that certain plant-specific responses are regulated via the direct physical interaction of JAZ proteins with specific transcription factors (Fernandez-Calvo et al., 2011; Zhai et al., 2015; Howe et al., 2018). Thus, we screened the proteins interacting with JAZ1 using a yeast two-hybrid (Y2H) system to find these potential transcription factors involved in the process of JA-regulated myrosinase activity. Among the identified JAZ1-interacting proteins, we focused on a bHLH transcription factor, FAMA, which was previously reported to regulate the expression of TGG1 (Li and Sack, 2014; Shirakawa et al., 2014). FAMA and two other transcription factors (MUTE and SPCH) are critically involved in stomatal development (Lampard and Bergmann, 2007; Ortega et al., 2019). Therefore, we used the Y2H system to test whether JAZ1 specifically interacted with FAMA, MUTE, and SPCH. We verified that JAZ1 specifically interacted with FAMA (Figure 3A), but that JAZ1 did not interact with either MUTE or SPCH (Supplemental Figure S3). We then examined the interactions between other JAZ family proteins and FAMA. In the Y2H system, JAZ1, JAZ2, and JAZ9 interacted with the FAMA protein, while other members of the JAZ family did not (Figure 3A).
Figure 3.
A subset of JAZs interact with FAMA. Y2H assay to detect interactions of FAMA with the 12 JAZ proteins. Yeast cells cotransformed with pGADT7-FAMA, or pGADT7-AP2 (preys) and pGBKT7-JAZ1-12 (bait) were grown on yeast synthetic dropout lacking Leu and Trp (SD/-2) as transformation control or on selective media lacking Ade, His, Leu, and Trp (SD/-4) to test protein interactions. pGADT7-FAMA, and pGADT7-AP2 cotransformed with pGBDT7 vector were included as a control. Split-LUC assays showing that JAZ1 and JAZ9 can interact with FAMA in N. benthamiana leaves. Representative images of N. benthamiana leaves 72 h after infiltration are shown. The images were digitally extracted for comparison. The right part of pseudocolor bar shows the range of luminescence intensity in each image. The bottom part indicates the infiltrated constructs. Bars = 10 mm. Three biological replicates were performed, and similar results were obtained. C, Co-IP assays to verify the interaction of JAZ1 with FAMA in vivo. Protein extracts from transgenic plants carrying both 35Spro:JAZ1-GUS and ProFAMA:FAMA-GFP (FAMA-GFP JAZ1-GUS) or from transgenic plants harboring 35Spro:JAZ1-GUS (JAZ-GUS) were immunoprecipitated with GFP antibody, and immunoprecipitated proteins were analyzed by immunoblotting using anti-GUS and anti-GFP antibodies. The experiments were repeated 3 times with similar results.
We verified the interaction between JAZ proteins and FAMA using a split-luciferase (Split-LUC) complementation assay. We inserted the FAMA gene into an nLUC vector, and inserted the JAZ1 and JAZ9 genes into a cLUC vector. Then, JAZ1 and JAZ9 were co-injected with FAMA into Nicotiana benthamiana for transient co-expression. LUC fluorescence was observed in N. benthamiana leaves co-injected with FAMA and JAZ1 or JAZ9, but not leaves injected with FAMA, JAZ1, or JAZ9 alone (Figure 3B). This indicated that JAZ1 and JAZ9 also interacted with FAMA in vivo.
The interaction between JAZ1 and FAMA was further verified using co-immunoprecipitation (Co-IP) assays. Transgenic Arabidopsis carrying the FAMA-GFP plasmid were hybridized with transgenic Arabidopsis carrying the 35Spro:JAZ1-GUS plasmid, and total protein from the F1 offspring was extracted and co-precipitated with the GFP antibody. Subsequent immunoblotting with the GUS antibody showed that the JAZ1-GUS protein was expressed in the immunoprecipitated cells (Figure 3C). This indicated that FAMA interacted with JAZ1 in plant cells (Figure 3C).
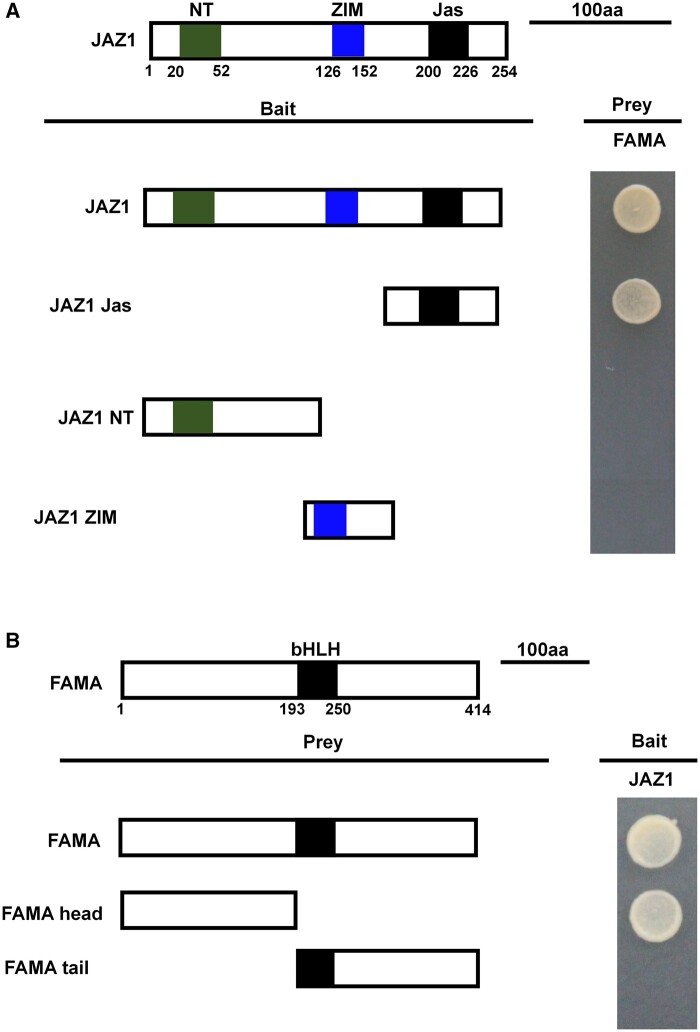
The domains implicated in the JAZ1–FAMA interaction
The JAZ protein harbors three primary domains (Pauwels and Goossens, 2011). In general, the JAS domain interacts with COI1 and various transcription factors, the NT domain interacts with various other transcription factors, and the ZIM domain interacts with other JAZ proteins or with NINJA (Pauwels and Goossens, 2011). To verify the specific JAZ1 domain(s) that interacted with FAMA, we truncated the JAZ1 protein such that each truncated protein included only one primary domain: JAZ1 Jas, JAZ1 NT, or JAZ1 ZIM. We then used these truncated proteins as bait in FAMA interaction analyses. Only the truncated protein containing the JAZ1 Jas domain interacted with FAMA (Figure 4A), indicating that the JAS domain, but neither the NT domain nor the ZIM domain, interacted with FAMA.
Figure 4.
Mapping of the protein domains involved in the interaction of FAMA with JAZ1, using Y2H assays. A, Based on the schematic protein structure of JAZ1, full-length JAZ1, or its derivatives (pGBKT7-JAZ1 or pGBKT7-JAZ1 derivatives) were tested for interactions with FAMA (pGADT7-FAMA). Yeast cells cotransformed with pGBKT7-JAZ1 or pGBKT7-JAZ1 derivatives (baits) and pGADT7-FAMA (prey) were grown on yeast synthetic dropout lacking Leu and Trp (SD/-2) as transformation control, or on selective media lacking Ade, His, Leu, and Trp (SD/-4) to test protein interactions. The different truncations of JAZ1 are represented. B, Based on the schematic protein structure of FAMA, full-length FAMA, or its derivatives (pGADT7-FAMA or pGADT7-FAMA derivatives) were tested for interactions with JAZ1 (pGBKT7-JAZ1). Yeast cells cotransformed with pGADT7-FAMA or pGADT7-FAMA derivatives (prey) and pGBKT7-JAZ1 (bait) were grown on yeast synthetic dropout lacking Leu and Trp (SD/-2) as transformation control, or on selective media lacking Ade, His, Leu, and Trp (SD/-4) to test protein interactions. The different domains of FAMA are represented.
To similarly identify which part of the FAMA protein interacted with JAZ1, we split the FAMA protein into an N-terminal fragment lacking the bHLH domain (FAMA-head) and a C-terminal fragment with the bHLH domain (FAMA-tail). We then used Y2H assays to test which of these fragments interacted with JAZ1-BD. We found that the FAMA-head fragment interacted with JAZ1, but the FAMA-tail fragment did not (Figure 4B), indicating that the domain of JAZ1–FAMA interaction was located at the N-terminal, before the bHLH domain.
FAMA positively regulated JA-mediated myrosinase activity
To understand the physiological significance of the interaction between FAMA and JAZ1 with respect to JA-mediated myrosinase activity, we first quantified FAMA expression in Arabidopsis after treatment with 10–100 µM MeJA. FAMA was upregulated with respect to the untreated control in all plants treated with MeJA (Figure 5A). We also quantified FAMA expression in the coi1-2 mutants. We found that FAMA expression was reduced in the coi1-2 mutants as compared to the wild-type plants, irrespective of MeJA treatment (Figure 5B). These results suggest that JA positively regulates FAMA expression. We then tested the effects of FAMA on myrosinase activity. We used four transgenic Arabidopsis lines previously designed in our laboratory: fama-1 and fama-2, in which FAMA is inhibited, and OE-3 and OE-7, in which FAMA is overexpressed (Li et al., 2018). As previously reported, FAMA was upregulated in 5-d-old OE-3 and OE-7 plants as compared to the wild-type controls, but downregulated in 21-d-old OE-3 and OE-7 plants compared to the wild-type controls (Supplemental Figure S4, A and B; Li et al., 2018). In addition, in 21-d-old OE-3 and OE-7 plants, myrosinase activity and TGG1 expression were also reduced compared with controls, irrespective of MeJA treatment (Supplemental Figure S4, C and D). Conversely, in 5-d-old OE-3 and OE-7 plants, myrosinase activity and TGG1 expression were increased compared with controls, irrespective of MeJA treatment (Figures 5, B and 6). The 5-dy-old fama-1 and fama-2 mutants exhibited inverse responses to MeJA treatment as compared to the 5-d-old OE-3 and OE-7 mutants (Figures 5, B and 6; Supplemental Figure S5). Similar trends were observed in hydrolysis experiments using aliphatic glucosinolates and indole glucosinolates (Supplemental Figure S5). Thus, the regulation of myrosinase activity by FAMA is a complex and precise process in plants.
Figure 5.
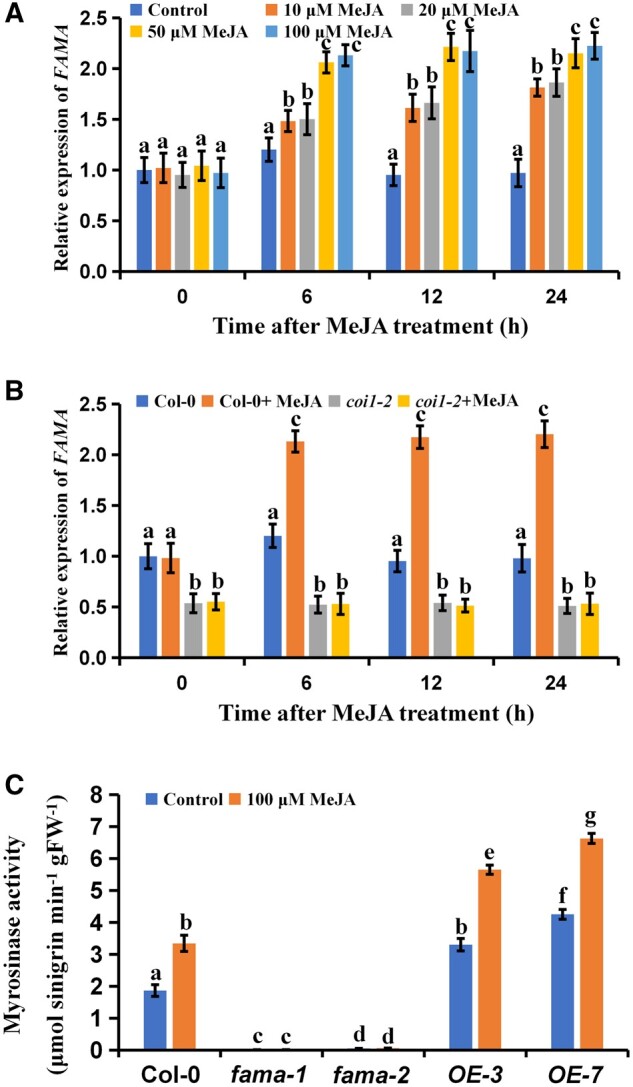
FAMA positively regulates JA-mediated myrosinase activity. Expression levels of FAMA of 5-d-old seedlings at different times after treatment with different concentrations of MeJA. Means ± sem are relative values obtained from three technical replicates; different letters represent significant differences (P < 0.05, Student’s t test). Five-day-old seedlings of wild-type (Col-0) and coi1-2 were treated with 100 μM of MeJA for the indicated time periods. The treated plants were harvested for total RNA extraction and RT-qPCR assays. Means ± sem are relative values obtained from three technical replicates. Different letters represent significant differences (P < 0.05, Student’s t test). C, Myrosinase activity of five-day-old seedlings of the indicated genotypes after treating with 100 μM of MeJA for 24 h. Values are means ± sem of 8–16 plants. The experiments were repeated at least 3 times with similar results. Different letters represent significant differences (P < 0.05, Student’s t test).
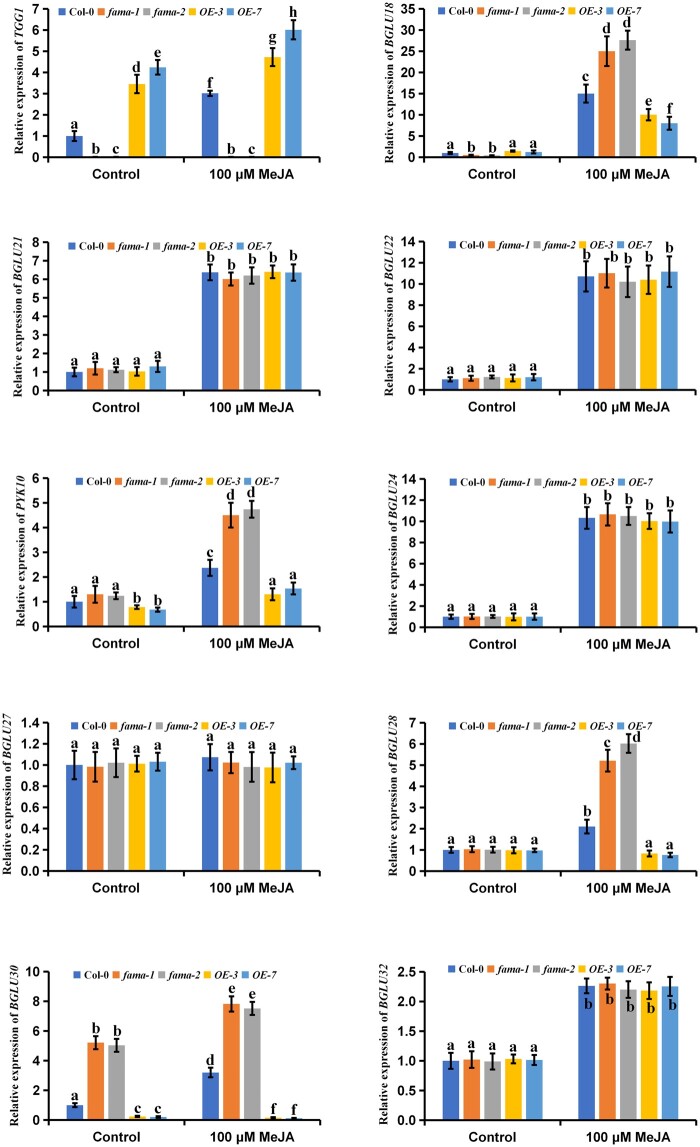
Figure 6.
Effects of FAMA on myrosinase gene expression. Five-day-old seedlings of the indicated genotypes after treating with 100 μM of MeJA for 24 h. The treated plants were harvested for total RNA extraction and RT-qPCR assays. Means ± sem are relative values obtained from three technical replicates. Different letters represent significant differences (P < 0.05, Student’s t test).
FAMA alters the expression patterns of JA-upregulated myrosinase genes
We next investigated the effects of FAMA on the expression profiles of JA-upregulated myrosinase genes, including TGG1, BGLU18, BGLU21, BGLU22, PYK10, BGLU24, BGLU27, BGLU28, BGLU30, and BGLU32. The results showed that TGG1 was significantly upregulated in the FAMA-overexpressing plants, while BGLU18, PYK10, BGLU28, and BGLU30 were significantly downregulated; and in the fama-1 and fama-2 mutants, the expression patterns showed opposite trend compared with that in the FAMA-overexpressing plants (Figure 6). The expression levels of the genes BGLU21, BGLU22, BGLU24, BGLU27, and BGLU32 were not affected by FAMA expression or MeJA treatment (Figure 6). Together, these results indicated that FAMA had gene-specific effects on the expression patterns of JA-upregulated myrosinase genes.
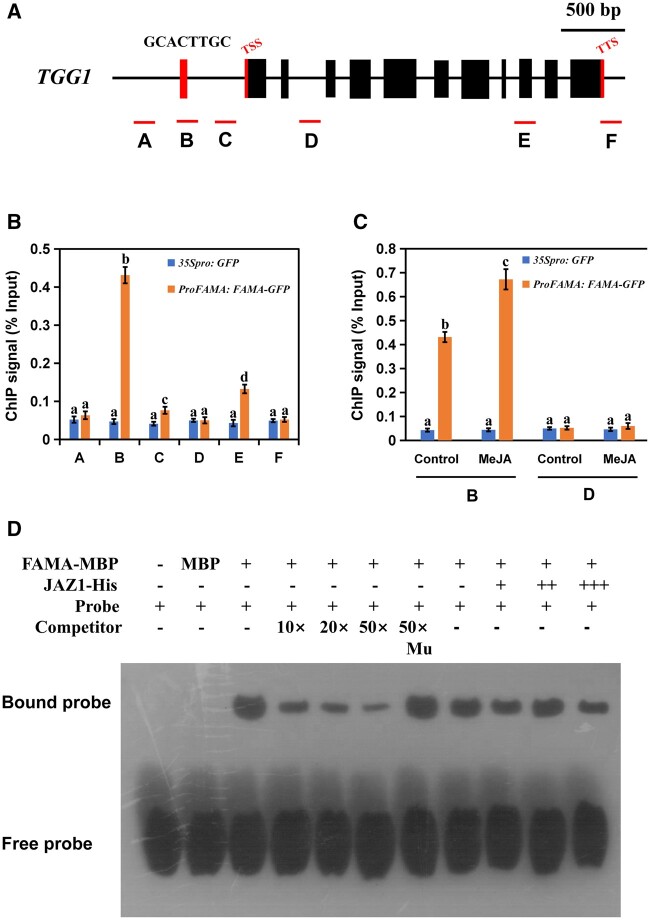
FAMA binds to the TGG1 promoter
To verify that FAMA induced TGG1 expression by binding to its promoter, we performed chromatin immunoprecipitation (ChIP) using previously developed transgenic Arabidopsis lines carrying the ProFAMA: FAMA-GFP plasmid (Li et al., 2018). We identified a G-box-like sequence (GCACTTGC) in the TGG1 promoter region (Figure 7A). We therefore selected this site, two other promoter regions, the coding region, and the 3′-UTR of TGG1 for ChIP-PCR assays. The results of these assays showed that FAMA was significantly enriched in the G-box-like region of the promoter, as well as in exon 9 (Figure 7B). Treatment with MeJA significantly increased FAMA accumulation on the TGG1 promoter (Figure 7C). These results indicated that FAMA was able to bind to the TGG1 promoter, which was also regulated by JA.
Figure 7.
FAMA could occupy the G-box-like region in the promoter of TGG1. Schematic diagram of TGG1 indicating the amplicons and probe used for the ChIP-qPCR assay and EMSA. A–C are located in the promoter region of TGG1; D is located in the second intron region of TGG1; E contains the ninth exon and part of the intron region of TGG1; F is located in the 3′-UTR region of TGG1. Positions of the transcription start site and transcription termination site are indicated with thin red bars. B, ChIP-qPCR assays showing that FAMA associates with the TGG1 locus. The chromatin of transgenic plants expressing ProFAMA: FAMA-GFP or 35Spro: GFP was immunoprecipitated with an anti-GFP antibody, and 35Spro: GFP plants served as control. Immunoprecipitated chromatin was analyzed by RT-qPCR using primers corresponding to the amplicons represented by the schematic diagram of TGG1 (A). ChIP signal is displayed as the percentage of total input DNA. Means ± sem are relative values obtained from three technical replicates; different letters represent significant differences (P < 0.05, Student’s t test). C, Dynamic recruitment of FAMA to the TGG1 locus. ChIP assays were performed as in (B), except that ProFAMA: FAMA-GFP and 35Spro: GFP plants were treated with 100 μM of MeJA for 30 min before cross-linking. Means ± sem are relative values obtained from three technical replicates; different letters represent significant differences (P < 0.05, Student’s t test). D, EMSA showing that the FAMA–maltose binding protein (MBP) fusion protein binds to the DNA probes of TGG1 in vitro. Biotin-labeled probes were incubated with FAMA–MBP protein or FAMA–MBP and JAZ1-His proteins, and the free and bound DNAs were separated on an acrylamide gel. As indicated, the unlabeled probe and unlabeled mutant probe were used as competitors. Mu, mutated probe in which the 5′-GCACTTGC-3′ motif was replaced with 5′-TTTTTTTT-3′. A gradient concentration of JAZ1-His was applied (0.5 mg for +; 1.0 mg for ++; 1.5 mg for +++).
To verify the binding of FAMA to the TGG1 promoter, we performed electrophoretic mobility shift assays (EMSAs) using the G-box-like sequence of TGG1 as a probe. We found that FAMA bound the TGG1 G-box-like sequence in vitro, and that this binding was not affected by the presence of the JAZ1 protein (Figure 7D). This may be because other factors have synergistic effects on JAZ1 as it interacts with FAMA.
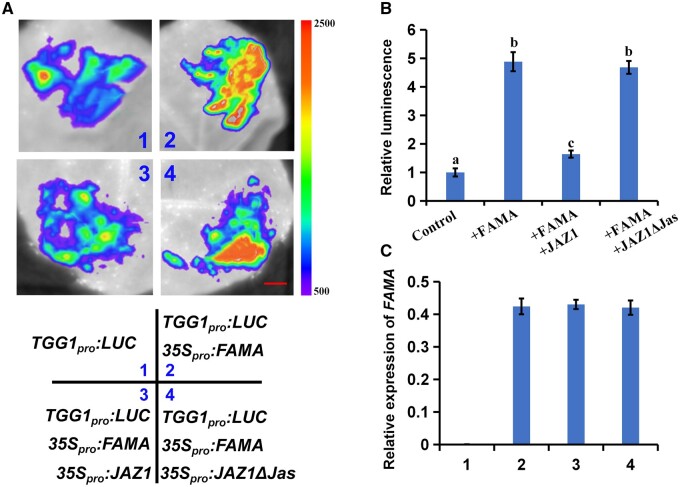
JAZ1 inhibits the transcriptional regulation of FAMA
As transcriptional inhibitors, JAZ proteins downregulate transcription factors in the JA pathway (Chini et al., 2007; Thines et al., 2007). Therefore, we speculated that JAZ1 might also inhibit the transcriptional regulation of FAMA. To test this hypothesis, we examined the effects of JAZ1 on the transcription factor function of FAMA in N. benthamiana. We constructed the LUC reporter vector ProTGG1: LUC, and injected ProTGG1: LUC into N. benthamiana leaves for transient expression. We then measured the fluorescence intensity in N. benthamiana leaves. When ProTGG1: LUC and 35Spro: FAMA were co-expressed in N. benthamiana, the fluorescence intensity of the co-expressed plants was significantly stronger than that of plants injected with ProTGG1: LUC alone (Figure 8, A and B). However, when ProTGG1: LUC was co-expressed with 35Spro: FAMA and 35Spro: JAZ1, the fluorescence intensity was the same as that of plants expressing ProTGG1: LUC alone, while the fluorescence intensity of plants expressing 35Spro: JAZ1 ΔJas was the same as those of plants co-expressing ProTGG1: LUC and 35Spro: FAMA (Figure 8, A and B). These results indicated that FAMA regulates TGG1 expression, and that JAZ1 inhibits the transcriptional regulation of FAMA.
Figure 8.
JAZ1 represses the transcriptional function of FAMA in N. benthamiana. A, Transient expression assays showing that JAZ1 counteracts the function of FAMA in activating TGG1 expression. Representative images of N. benthamiana leaves 72 h after infiltration are shown. The right part of pseudocolor bar shows the range of luminescence intensity in each image. The bottom part indicates the infiltrated constructs. Bars = 10 mm. B, Quantitative analysis of luminescence intensity in (A). Means ± sem are relative values obtained from three independent determinations; different letters represent significant differences (P < 0.05, Student’s t test). C, RT-qPCR analysis of FAMA expression in the infiltrated leaf areas shown in (A). Total RNA was extracted from leaves of N. benthamiana coinfiltrated with the indicated constructs. Means ± sem are relative values obtained from three independent determinations.
Transgenic FAMA expression reversed the reduced myrosinase activity of the coi1-2 mutant
In conjunction with corroborating what is known to date about the molecular mechanisms underlying JA-mediated gene transcription, our results were also consistent with a system in which JAZ proteins accumulate in the coi1-2 mutant and repress FAMA function, inhibiting TGG1 expression. This downregulation of TGG1 in the coi1-2 mutant explains the reduced myrosinase activity observed in this mutant. We, therefore, speculated that FAMA overexpression might induce TGG1 expression and rescue myrosinase activity in coi1-2. To test this, we crossed the FAMA overexpression line OE-7 with the coi1-2 mutant to obtain OE-7/coi1-2 plants. We then detected myrosinase activity and TGG1 gene expression levels in the wild-type, coi1-2, OE-7, and OE-7/coi1-2 plants. Consistent with expectations, myrosinase activity, and TGG1 transcription levels were significantly lower in the coi1-2 plants and significantly higher in the OE-7 plants, as compared to the wild-type (Figure 9, A and B). However, examination of myrosinase activity and TGG1 transcription levels showed that, in contrast with coi1-2 plants, myrosinase activity and TGG1 transcription levels in the OE-7/coi1-2 plants were much higher than in wild-type plants, irrespective of MeJA treatment (Figure 9, A and B). This indicated that the ectopic expression of FAMA rescued the reduced enzyme activity associated with the coi1-2 mutant. This possibility was supported by our tests of aliphatic- and indole-glucosinolates hydrolysis (Supplemental Figure S6, A and B).
Figure 9.
Transgenic expression of FAMA rescues the reduced enzyme activity of coi1-2 mutant. A and B, Myrosinase activity (A) and expression levels of TGG1 (B) of 5-d-old seedlings of the indicated genotypes after treating with 100 μM of MeJA for 24 h. Values are means ± sem of 8–16 plants. The experiments were repeated at least 3 times with similar results. Different letters represent significant differences (P < 0.05, Student’s t test).
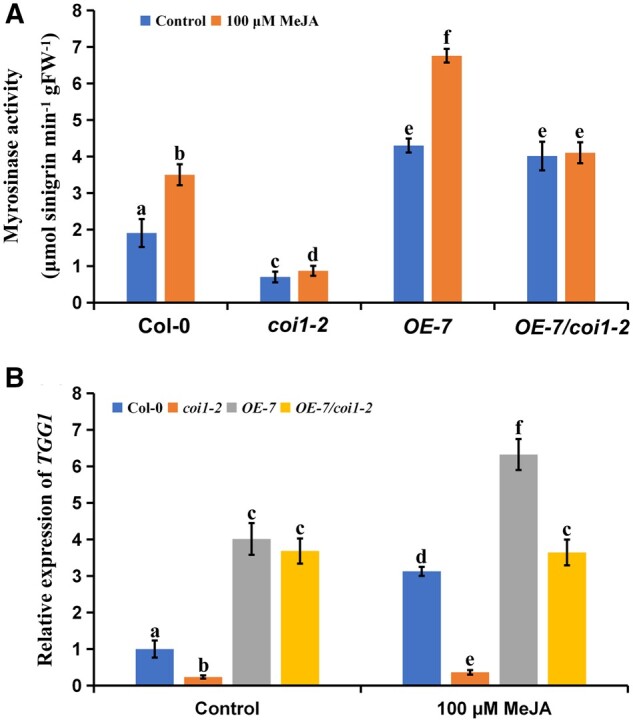
MED8 is involved in the JA-mediated regulation of myrosinase activity
Previously, we showed that FAMA interacts with MED8 to regulate plant resistance to B. cinerea (Li et al., 2018). We therefore also investigated whether MED8 affected myrosinase activity using the med8 mutant. We found that myrosinase activity and TGG1 gene expression levels in the med8 mutant were significantly lower than those in the controls, irrespective of MeJA treatment (Figure 10, A and B). Further glucosinolate hydrolysis experiments also indicated that MED8 played a positive role in the JA-mediated regulation of myrosinase activity (Supplemental Figure S7, A and B). We further tested whether MED8 could bind the TGG1 promoter and found that, consistent with our FAMA results, MED8 bound to the G-box-like region of the TGG1 promoter, and that MeJA treatment promoted this binding (Figure 10C). In addition, the binding of FAMA or MED8 to the TGG1 promoter was significantly reduced in the med8 or fama-2 mutants (Figure 10, D and E). Therefore, our results suggested that both MED8 and FAMA participate in the JA-mediated regulation of myrosinase activity.
Figure 10.
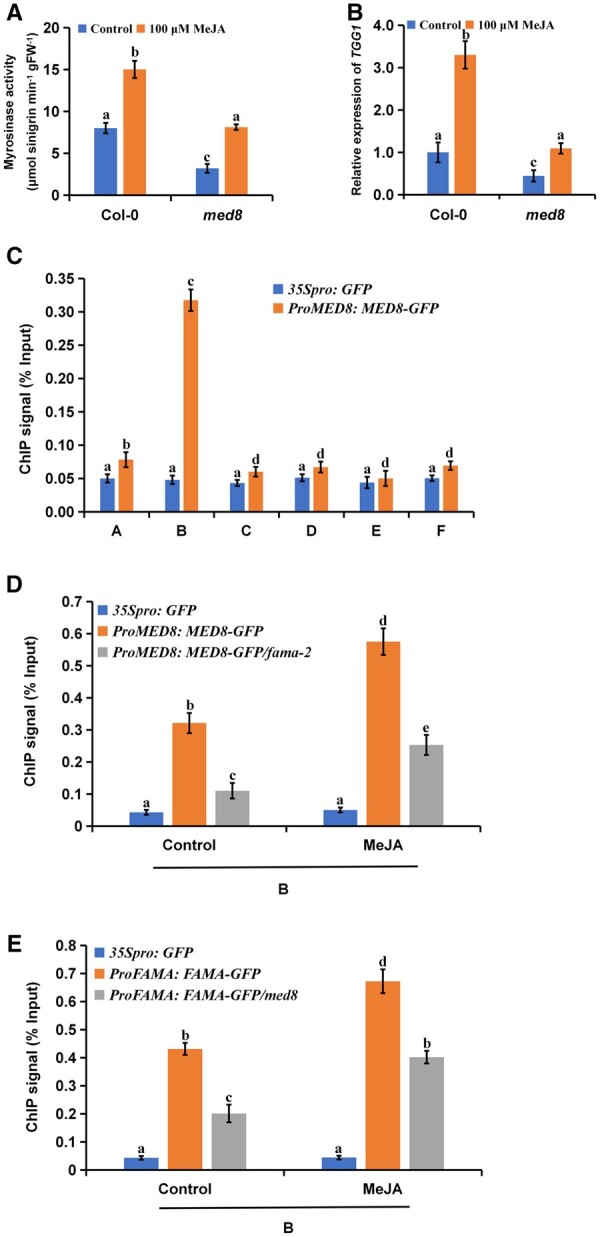
MED8 positively regulates JA-mediated myrosinase activity. A and B, Myrosinase activity (A) and expression levels of TGG1 (B) of 21-d-old seedlings of the indicated genotypes after treating with 100 μM of MeJA for 24 h. Values are means ± sem of 8–16 plants. The experiments were repeated at least 3 times with similar results. Different letters represent significant differences (P < 0.05, Student’s t test). C, ChIP-qPCR assays showing that MED8 associates with the TGG1 locus. The chromatin of transgenic plants expressing ProMED8: MED8-GFP or 35Spro: GFP was immunoprecipitated with an anti-GFP antibody, and 35Spro: GFP plants served as control. Immunoprecipitated chromatin was analyzed by RT-qPCR using primers corresponding to the amplicons represented by the schematic diagram of TGG1 (Figure 7A). ChIP signal was displayed as the percentage of total input DNA. Means ± sem are relative values obtained from three technical replicates; different letters represent significant differences (P < 0.05, Student’s t test). D, FAMA affecting the recruitment of MED8 to the TGG1 locus. ChIP assays were performed as in (C), except that ProMED8: MED8-GFP, ProMED8: MED8-GFP/fama-2, and 35Spro: GFP plants were treated with Control or 100 μM of MeJA for 30 min before cross-linking. E, MED8 affecting the recruitment of FAMA to the TGG1 locus. ChIP assays were performed as in (C), except that ProFAMA: FAMA-GFP, ProFAMA: FAMA-GFP/med8, and 35Spro: GFP plants were treated with Control or 100 μM of MeJA for 30 min before cross-linking. D and E, Means ± sem are relative values obtained from three technical replicates; different letters represent significant differences (P < 0.05, Student’s t test).
Discussion
Glucosinolates play important roles in plant resistance to insect pests and pathogens (Frerigmann and Gigolashvili, 2014; Vela-Corcia et al., 2019; Ting et al., 2020), and the plant hormone JA regulates glucosinolate metabolism (Mewis et al., 2005; Dombrecht et al., 2007; Liu et al., 2010). Although several previous studies have explored the regulatory effects of JA on glucosinolate synthesis (Liu et al., 2010; Mitreiter and Gigolashvili, 2021), the roles played by JA in the regulation of glucosinolate hydrolysis remain unclear. Here, we elucidated the mechanisms underlying the JA-mediated regulation of glucosinolate hydrolysis with several different lines of evidence. First, exogenous MeJA treatment significantly increased myrosinase activity (Figure 1A). Second, the blockage of endogenous JA biosynthesis and signaling decreased myrosinase activity (Figure 1B). Third, exogenous MeJA treatment induced the expression of myrosinase genes (Figure 2). We, therefore, concluded that JA positively regulates myrosinase activity in Arabidopsis. Our FAMA binding results were consistent with this conclusion.
In Arabidopsis, TGG1 and TGG2 have redundant functions: these proteins contribute jointly to myrosinase activity in aerial plant parts (Barth and Jander, 2006). However, only TGG1 was induced by JA (Figure 2); JA inhibited the expression of TGG2 (Supplemental Figure S2). This indicated that TGG1 and TGG2 play different roles during the JA-mediated regulation of myrosinase activity. We also found that FAMA bound the promoters of both TGG1 and TGG2 (Figure 7; Supplemental Figure S8). MeJA treatment improved the ability of FAMA to bind to the TGG1 promoter, but decreased the ability of FAMA to bind to TGG2 promoter (Figure 7; Supplemental Figure S8). This suggested that JA might induce another regulator to compete with FAMA for the binding site on the TGG2 promoter, thus downregulating TGG2. To further clarify the mechanisms underlying the regulatory effects of JA on myrosinase activity, the factors that regulate TGG2 expression should be thoroughly characterized in subsequent studies.
There are two types of myrosinases in Arabidopsis: the QE- and the EE-type myrosinases (Sugiyama and Hirai, 2019). In conjunction with previous studies, our results indicated that JA regulated the expression levels of both types of myrosinase genes (Figure 2). However, the relationship between the QE- and EE-type myrosinases has yet to be explored. Our results suggested that there might be antagonism between these two types of myrosinases whose activities are regulated by JA. For example, the transcription factor FAMA positively regulated JA-mediated TGG1 gene expression, but negatively regulated the JA-mediated expression of the BGLU18, PYK10, BGLU28, and BGLU30 genes (Figure 6). In addition, FAMA did not directly bind to the promoters of BGLU18, PYK10, or BGLU28 (Supplemental Figure S9), indicating that FAMA may inhibit other factors that induce the expression of these three genes. Interestingly, we found that FAMA bound the promoter of BGLU30 (Supplemental Figure S10), indicating that FAMA might compete with other factors to bind to the promoter of BGLU30, thus inhibiting the expression of BGLU30. In short, FAMA inhibited the expression of EE-type myrosinases genes at a variety of levels. Thus, our results suggested that JA does not regulate the activity levels of the QE- and the EE-type myrosinases independently. Future studies should attempt to identify additional factors involved in the JA-mediated expression of BGLU18, PYK10, BGLU28, and BGLU30; the relationships between these as-yet unknown factors and FAMA are critical for the characterization of the JA-associated network through which myrosinase activity is regulated. Interestingly, we found that FAMA did not affect JA-mediated BGLU21 or BGLU22 gene expression (Figure 6), both of which are homologs of the PRK10 gene (Nakano et al., 2017). This may be because BGLU21 and BGLU22 are expressed only in plant roots (Ahn et al., 2010), while the FAMA gene is expressed in aboveground parts (Ohashi-Ito and Bergmann, 2006).
The molecular mechanisms underlying the regulation by JA of specific physiological processes can be clarified by identifying pairs of JAZ proteins and transcription factors. For example, previous studies have shown that JAZ proteins directly target several bHLH transcription factors (i.e. MYC2, MYC3, MYC4, ICE1, and ICE2), the WD-repeat/bHLH/MYB transcriptional complexes (e.g. GL3, EGL3, GL1, MYB21, and MYB24), ETHYLENE INSENSITIVE3, two APETALA2 transcription factors (TOE1 and TOE2), ROOT HAIR DEFECTIVE 6 (RHD6), and RHD6 LIKE1 to regulate various JA-mediated processes (Qi et al., 2011; Song et al., 2011; Hu et al., 2013; Zhai et al., 2015; Chini et al., 2016; Howe et al., 2018; Han et al., 2020). Here, we also found that FAMA interacted with JAZ proteins, and that JAZ proteins repressed the transcriptional effects of FAMA on TGG1 (Figure 3; Figure 8). Moreover, the JAZ–FAMA pairing had important genetic and phenotypic effects on the mediation of myrosinase activity by JA. Thus, this study clarified the role played by JA in the regulation of myrosinase activity and elucidated the underlying molecular mechanisms.
FAMA, along with MUTE and SPCH, was initially described in relation to the regulation of various stages of stomatal development (Lampard and Bergmann, 2007). Recent studies have shown that JA is also involved in the regulation of stomatal development: JA inhibits stomatal development by regulating the expression of FAMA, MUTE, and SPCH, suggesting that FAMA negatively regulates JA-mediated stomatal development (Han et al., 2018). This was inconsistent with our results, which indicated that FAMA positively regulates JA-mediated myrosinase activity. Thus, FAMA may play a variety of different roles in various physiological processes involving JA. Interestingly, we found that JAZ proteins did not interact with MUTE or SPCH (Supplemental Figure S3), the other two important transcription factors in stomatal development (Torii et al., 2007), and that TGG1 gene expression levels were similar among the wild-type, mute, and spch plants (Shirakawa et al., 2014). These results suggested that the interaction between JAZ proteins and FAMA might not be important for stomatal development, but instead might specifically help to regulate myrosinase activity.
Recent studies have shown that FAMA is also involved in the development of myrosin cells, which are idioblasts that store myrosinases (Li and Sack, 2014; Shirakawa et al., 2014). In Arabidopsis, myrosin cells play an important role in pest and disease resistance (Chhajed et al., 2020). However, there is as of yet no evidence that JA is associated with myrosin cells. Our results showed that JA induced the expression of FAMA (Figure 5A), and that, in the coi1-2 mutant, FAMA expression was significantly reduced compared with the wild-type (Figure 5B). Previous studies have shown that FAMA overexpression increases the number of myrosin cells in hypocotyls and roots, and that myrosin cell numbers are substantially reduced in fama mutants (Li and Sack, 2014; Shirakawa et al., 2014). Our work, in conjunction with these previous results, indirectly indicates that JA may positively regulate the development of myrosin cells. Further genetic and cellular data are needed to more precisely define the role of JA in the development of myrosin cells.
The transcription mediator complex plays an important role in transcriptional regulation (Backstrom et al., 2007; Chen et al., 2012; Zhai and Li, 2019). For example, mediator complex subunit 25 (MED25), the most important transcription mediator complex subunit in the JA pathway, regulates JA-mediated root length inhibition and disease resistance (An et al., 2017; Wang et al., 2019; You et al., 2019; Wu et al., 2020). Previously, we demonstrated that MED8 regulates plant resistance to B. cinerea by interacting with FAMA (Li et al., 2018). Here, we found that MED8 was also involved in JA-mediated regulation of myrosinase activity (Figure 10). This suggested that MED8 and FAMA may work in concert to affect various plant physiological processes.
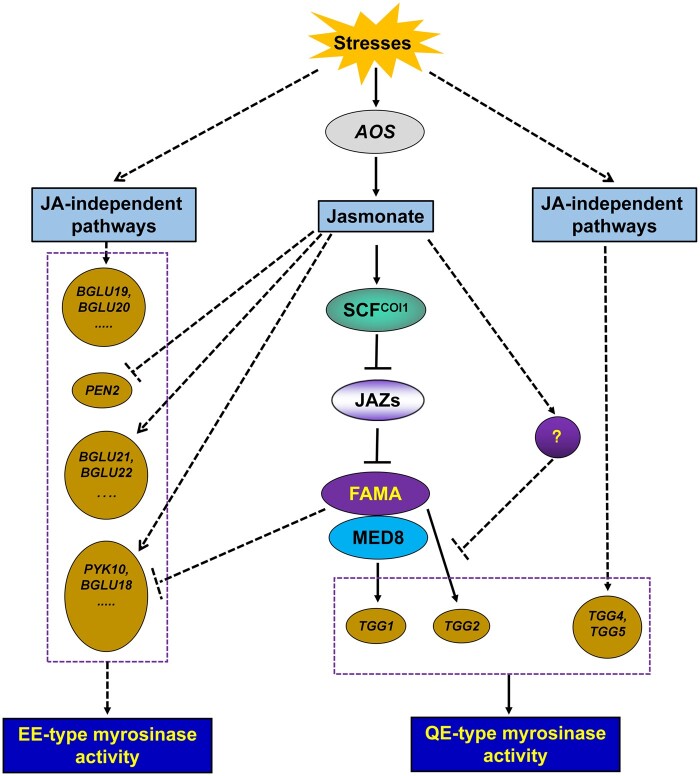
Based on our results, we propose a simple working model of the molecular mechanism underlying the JA-mediated FAMA/MED8–TGG1 pathway in the regulation of myrosinase activity (Figure 11). Under normal growth conditions, JAZ repressors physically interact with the FAMA transcription factor, attenuating its transcriptional function, and thereby repressing the expression of the downstream TGG1 gene. However, when plants are attacked by diseases or insect pests, endogenous JA is produced, which promotes glucosinolate synthesis. JA-Ile is recognized by the receptor COI1, leading to COI1 activation. Activated COI1 then degrades JAZ proteins, triggering FAMA transcriptional activity and activating downstream TGG1 gene. This in turn promotes glucosinolate hydrolysis. However, many uncertainties remain, such as the role of TGG2 in the JA-mediated regulation of myrosinase activity, the relationship between the two JA-mediated myrosinase activities (i.e. QE or EE), and the mechanisms underlying the coordination of JA-mediated stomatal development and myrosinase activity by FAMA. The answers to these questions will help us to understand the physiological processes mediated by JA in plants.
Figure 11.
A model of JA-dependent and JA-independent signaling pathways in regulating the enzyme activity of myrosinase. When plants are attacked by diseases and insect pests, stressed plants could induce the expression of myrosinase genes to resist the stresses. There are two signaling pathways to mediate the expression of myrosinase genes: One is the JA-independent signaling pathway which regulates the expression of BGLU19, BGLU20, TGG4, and so on; the other one is the JA-dependent signaling pathway. For the JA-dependent signaling pathway in regulating the expression of myrosinase genes, when plants are attacked by diseases and insect pests, endogenous JA will be produced through inducing AOS expression. JA-Ile can be recognized by the receptor COI1 (SCFCOI1) and activate COI1. The activated COI1 can degrade the JAZ proteins, release the transcriptional activity of FAMA, and activate the downstream TGG1 gene expression to promote the synthesis of myrosinase, to promote the hydrolysis of glucosinolate. JA also mediates myrosinase activity by inducing or inhibiting the expression of other myrosinase genes. The final outcome of jasmonate regulation is the enhancement of myrosinase activity. Purple boxes represent the two types of myrosinases; the solid lines represent the pathways that have been clarified in this study; dashed lines represent the pathways that need further study; the question mark bubble represents the important unknown element in the regulation of TGG2 expression by JA, which needs further exploration.
Conclusions
We demonstrated that JA mediates myrosinase activity by regulating TGG1 expression in conjunction with the JAZ–FAMA/MED8–TGG1 pathway: FAMA and MED8 bind the TGG1 promoter and regulate TGG1 expression, while JAZ proteins repress the transcriptional activity of FAMA. The results of this study help to extend and deepen our understanding of the roles played by JA in plants.
Materials and methods
Plant materials and growth conditions
Arabidopsis ecotype Col-0 was used as the wild-type. Some of the plant materials used in this study were previously described: coi1-2 (Xu et al., 2002); 35Spro: JAZ1 and 35Spro: JAZ1ΔJas (Zhai et al., 2015); aos (Park et al., 2002); 35Spro: GFP (Li et al., 2011); fama-1, fama-2, OE-3 (ProFAMA: FAMA-GFP 3#), OE-7 (ProFAMA: FAMA-GFP 7#), med8, ProMED8: MED8-GFP, ProMED8: MED8-GFP/fama-2, and ProFAMA: FAMA-GFP/med8 (Li et al., 2018). OE-7/coi1-2 was generated by crossing the parental single homozygous lines.
Plants were grown initially on half-strength Murashige and Skoog (MS) agar plates for 10 d and then transferred to sterile soil in plastic trays at 22°C with a 16-h light/8-h dark photoperiod (light intensity 120 μM photons m−2 s−1). Nicotiana benthamiana was grown in sterile soil under a 16-h light (28°C)/8-h dark (22°C) photoperiod.
Measurement of total myrosinase activity
Total myrosinase activity was measured as described by Barth and Jander, with slight modifications (Barth and Jander, 2006). Plant tissues were collected and ground into a powder in dry ice. The powder was then added to 0.3 mL of pre-chilled sodium phosphate buffer (33 mM; pH 7.0) and vortexed well. The mixture was centrifuged at 10,000 g at 4°C for 10 min. Then, 200 μL of the supernatant was added to a Sephadex G-50 (Sigma-Aldrich, St. Louis, MO, USA) column; Sephadex columns were filled with resin that was packed in 33 mM sodium phosphate buffer (pH 7.0) overnight at 4°C. After supernatant addition, columns were centrifuged at 720 g at 4°C for 1 min. Then, 100 μL of filtrate was collected and diluted with water to a final volume of 200 μL. Next, 0.34 mM sinigrin and 0.3 mM ascorbic acid were separately diluted to 200 μL with the diluted filtrate mixture. The new mixtures were plated in 96-well plates, which were analyzed at 25°C for 15 min with a microplate reader at 227 nm; data were collected every 10 s. Myrosinase activity was calculated following Barth and Jander (2006).
Measurement of glucosinolate breakdown
Glucosinolate breakdown experiments were performed as described by Barth and Jander, with minor modifications (Barth and Jander, 2006). Leaves were collected from the indicated plants (80 mg fresh weight). Samples collected 0 min after treatment were immediately snap-frozen in liquid nitrogen. Samples collected 1 min after treatment were ground with a plastic pestle in 400 μL of water for 0.25 min, then held an additional 0.75 min at room temperature before snap-freezing in liquid nitrogen. Samples collected at all other time points were ground with a plastic pestle in 400 μL of water for 0.25 min. Next, 60 μL of 100% (v/v) methanol was added to each sample and vortexed several times. Each mixture was immediately heated to 75°C for 15 min to inactivate myrosinase and to extract glucosinolates. Finally, 10 μL of 1.25 mM sinigrin was mixed with extracted glucosinolates as an internal standard. The desulfoglucosinolates in the mixtures were quantified using high-performance liquid chromatography following Barth and Jander (2006). Glucosinolate hydrolysis rate was calculated as (glucosinolate content before reaction − glucosinolate content after reaction)/reaction time.
RNA analysis
Total RNA extraction was performed as previously described (Li et al., 2018). Reverse transcription quantitative PCR (RT-qPCR) was performed as previously described (Yu et al., 2020). All primers used for RT-qPCR are listed in Supplemental Table S1.
Y2H assays
The full-length coding sequence of JAZ1 was amplified with the primers listed in Supplemental Table S1. The PCR products were enzyme-digested and cloned into the pGBKT7 vector. Y2H screening was performed using the pGADT7-based Arabidopsis cDNA library described previously (Zhai et al., 2015). FAMA was identified by screening.
The interaction between JAZs and FAMA was verified following a previous study (Li et al., 2018). Briefly, 12 JAZs, including JAZ1, and the associated domain were cloned into separate pGBKT7 vectors. FAMA and its derivatives were cloned into the pGADT7 vector. Primers used for vector construction are listed in Supplemental Table S1. Interactions were observed after incubation with the yeast strain Saccharomyces cerevisiae AH109 for 3 d at 30°C.
Transient expression assay in N. benthamiana leaves
The transient expression assays were performed in N. benthamiana leaves as previously described (Li et al., 2018). Briefly, for Split-LUC complementation assay, FAMA was cloned into vector pCAMBIA1300-nLUC, and JAZ1 or JAZ9 was cloned into vector pCAMBIA1300-cLUC. Primers are summarized in Supplemental Table S1. For transcriptional activation assays, the TGG1 promoter was amplified using Gateway-compatible primers. The PCR products were cloned by pENTR Directional TOPO cloning kits (Invitrogen, Waltham, MA, USA) and recombined with the binary vector pGWB35 to generate the reporter construct TGG1pro: LUC. The FAMA effector construct was 35Spro: FAMA-GFP (35Spro: FAMA). We used a low-light cooled CCD imaging apparatus (NightOWL II LB983 with indigo software) to capture the LUC image and to assess luminescence intensity. The leaves were sprayed with 0.5 mM luciferin and placed in darkness for 3 min prior to luminescence detection. The camera was cooled to −110°C and the images were taken with an exposure time of 2 min. Relative LUC activity was equivalent to luminescence intensity/0.2 mm2 leaf area. Relative luminescence was the ratio of relative LUC activity between different experimental groups and control groups.
Co-IP assays
To further investigate the interaction between JAZ1 and FAMA, Arabidopsis carrying the 35Spro: JAZ1-GUS plasmid was crossed with plants carrying the ProFAMA: FAMA-GFP plasmid (Zhai et al., 2015; Li et al., 2018). Five-day-old F1 ProFAMA: FAMA-GFP (JAZ1-GUS FAMA-GFP) seedlings were used in Co-IP assays; 5-d-old seedlings carrying only 35Spro: JAZ1-GUS were used as controls. All plants were harvested and ground in liquid nitrogen. The ground samples were added to extraction buffer containing 50 mM Tris–HCl (pH 7.5) 150 mM NaCl, 0.1% (v/v) Triton X-100, 0.2% (v/v) Nonidet P-40, 0.6 mM phenylmethylsulfonyl fluoride (PMSF), and 20 μΜ MG132, as well as 1×Roche protease inhibitor cocktail. After protein extraction, 20 μL protein G plus agarose (Santa Cruz Biotechnology, Dallas, TX, USA) was added to 3-mg protein extracts to reduce nonspecific immunoglobulin binding. After 1 h of incubation, the supernatant was transferred to a new tube. GFP antibody-bound agarose beads (Medical & Biological Laboratories Co., Ltd., Nagoya, Japan) were then added to each reaction and incubated for 1 h at 4°C. The precipitated samples were washed at least 3 times with lysis buffer. Samples were then eluted by adding 1×SDS protein loading buffer and boiling for 5 min. Total and immunoprecipitated proteins were analyzed by immunoblotting using anti-GUS and anti-GFP antibodies.
EMSAs
The full-length coding sequences of FAMA and JAZ1 were amplified and cloned into pMAL-c2 and pET32a vectors, respectively. Primers are summarized in Supplemental Table S1. The recombinant FAMA and JAZ1 proteins were expressed in Escherichia coli. Oligonucleotide probes were synthesized and labeled with biotin at the 3′-end (Invitrogen). EMSAs were performed using a LightShift Chemiluminescent EMSA kit (Thermo Scientific, Waltham, MA, USA) following a previous study (Zhai et al., 2015). Briefly, the synthesized probes were incubated with or without indicated proteins in incubation buffer (1×binding buffer, 2.5% (v/v) glycerol, 50 mM KCl, 5 mM MgCl2, and 10 mM EDTA) at room temperature for 20–30 min. Unlabeled probes were added to the reactions as competition assays. The probe sequence was 5′-CCCGTTTCCTCACATCTGCACTTGCTAAGCCAACCTTGGCTTACTTATGACCACC-3′, and the mutated probe sequence was 5′-CCCGTTTCCTCACATCTTTTTTTTTTAAGCCAACCTTGGCTTACTTATGACCACC-3′.
ChIP-PCR assays
Seedlings of several previously constructed transgenic lines (ProFAMA: FAMA-GFP, ProMED8: MED8-GFP, ProMED8: MED8-GFP/fama-2, ProFAMA: FAMA-GFP/med8, and 35Spro: GFP; Li et al., 2018) were grown in MS medium. Five-day-old seedlings of each line were either treated with 50 μM of MeJA for 24 h or left untreated as controls. After 24 h, 1.5 g samples of treated and control plants were collected and cross-linked in 1% (v/v) formaldehyde. Following cross-linking, a GFP antibody (Abcam, Cambridge, UK) was used to immunoprecipitate the chromatin protein–DNA complex, and the precipitated DNA was purified using a PCR purification kit (Qiagen, Hilden, Germany) in preparation for real-time RT-qPCR analysis. All ChIP-PCR assays were performed 3 times. Chromatin precipitated from 35Spro: GFP seedlings were considered the negative control, while chromatin isolated before precipitation was used as the input control. Primers used for this analysis are listed in Supplemental Table S1.
Accession numbers
The accession numbers of the genes mentioned in this article are given below and their sequence data can be found in Arabidopsis genome initiative (www.arabidopsis.org): COI1 (AT2G39940), JAZ1 (AT1G19180), JAZ2 (AT1G74950), JAZ3 (AT3G17860), JAZ4 (AT1G48500), JAZ5 (AT1G17380), JAZ6 (AT1G72450), JAZ7 (AT2G34600), JAZ8 (AT1G30135), JAZ9 (AT1G70700), JAZ10 (AT5G13220), JAZ11 (AT3G43440), JAZ12 (AT5G20900), AOS (AT5G42650), FAMA (AT3G24140), SPCH (AT5G53210), MUTE (AT3G06120), TGG1 (AT5G26000), TGG2 (AT5G25980), TGG3 (AT5G48375), TGG4 (AT1G47600), TGG5 (AT1G51470), TGG6 (AT1G51490), BGLU18 (AT1G52400), BGLU19 (AT3G21370), BGLU20 (AT1G75940), PYK10 (AT3G09260), BGLU21 (AT1G66270), BGLU22 (AT1G66280), BGLU24 (AT5G28510), BGLU25 (AT3G03640), PEN2 (AT2G44490), BGLU27 (AT3G60120), BGLU28 (AT2G44460), BGLU29 (AT2G44470), BGLU30 (AT3G60140), BGLU31 (AT5G24540), BGLU32 (AT5G24550), BGLU33 (AT2G32860), and MED8 (AT2G03070).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1 The JA signal positively regulates the hydrolysis rate of glucosinolates.
Supplemental Figure S2 Expression levels of JA-unaffected and JA-downregulated myrosinase genes.
Supplemental Figure S3 JAZ1 did not interact with MUTE or SPCH.
Supplemental Figure S4 The phenotypes of 21-d-old FAMA mutants and overexpression plants in JA-regulated myrosinase activity.
Supplemental Figure S5 FAMA positively regulates the hydrolysis rate of glucosinolates.
Supplemental Figure S6 Transgenic expression of FAMA rescues the reduced hydrolysis rate of glucosinolates of coi1-2 mutant.
Supplemental Figure S7 MED8 positively regulates the hydrolysis rate of glucosinolates.
Supplemental Figure S8 JA repressed the occupation of FAMA on the G-box-like region in the promoter of TGG2.
Supplemental Figure S9 FAMA did not bind the promoters of BGLU18, PYK10, or BGLU28.
Supplemental Figure S10 FAMA bound the promoters of BGLU30.
Supplemental Table S1 List of primers used in this article.
Supplementary Material
Acknowledgments
We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript. We also thank funding from the National Natural Science Foundation of China (31600986); Li Dak Sum Yip Yio Chin Kenneth Li Marine Biopharmaceutical Development Fund; National 111 Project of China; and partly sponsored by K.C. Wong Magna Fund in Ningbo University.
Funding
This work was supported by the National Natural Science Foundation of China (31600986); Li Dak Sum Yip Yio Chin Kenneth Li Marine Biopharmaceutical Development Fund; National 111 Project of China; and partly sponsored by K.C. Wong Magna Fund in Ningbo University.
Conflict of interest statement: None declared.
Q.F. performed all the biochemical experiments, such as yeast-two hybrid assays, electrophoretic mobility shift assay, and part of physiological experiments, such as myrosinase activity and glucosinolate breakdown. L.L. performed most of the physiological experiments, such as myrosinase activity and glucosinolate breakdown, analyzed the data, made the figures and wrote the first draft of the MS. Y.L. performed part of reverse transcription quantitative PCR. X.S. revised the MS. X.L. contributed in designing experiments, guided the first two authors, and finalized the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Xiaohui Li (lixiaohui@nbu.edu.cn).
References
- Ahn YO, Shimizu B, Sakata K, Gantulga D, Zhou C, Bevan DR, Esen A (2010) Scopolin-hydrolyzing beta-glucosidases in roots of Arabidopsis. Plant Cell Physiol 51: 132–143 [DOI] [PubMed] [Google Scholar]
- An C, Li L, Zhai Q, You Y, Deng L, Wu F, Chen R, Jiang H, Wang H, Chen Q, Li C (2017) Mediator subunit MED25 links the jasmonate receptor to transcriptionally active chromatin. Proc Natl Acad Sci US A 114: E8930–E8939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson D, Chakrabarty R, Bejai S, Zhang J, Rask L, Meijer J (2009) Myrosinases from root and leaves of Arabidopsis thaliana have different catalytic properties. Phytochemistry 70: 1345–1354 [DOI] [PubMed] [Google Scholar]
- Backstrom S, Elfving N, Nilsson R, Wingsle G, Bjorklund S (2007) Purification of a plant mediator from Arabidopsis thaliana identifies PFT1 as the Med25 subunit. Mol Cell 26: 717–729 [DOI] [PubMed] [Google Scholar]
- Barth C, Jander G (2006) Arabidopsis myrosinases TGG1 and TGG2 have redundant function in glucosinolate breakdown and insect defense. Plant J 46: 549–562 [DOI] [PubMed] [Google Scholar]
- Bednarek P, Pislewska-Bednarek M, Svatos A, Schneider B, Doubsky J, Mansurova M, Humphry M, Consonni C, Panstruga R, Sanchez-Vallet A, et al. (2009) A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 323: 101–106 [DOI] [PubMed] [Google Scholar]
- Bhat R, Faiz S, Ali V, Khajuria M, Mukherjee D, Vyas D (2020) Effect of temperature and insect herbivory on the regulation of glucosinolate-myrosinase system in Lepidium latifolium. Physiol Plant 172: 53–63 [DOI] [PubMed] [Google Scholar]
- Canistro D, Croce CD, Iori R, Barillari J, Bronzetti G, Poi G, Cini M, Caltavuturo L, Perocco P, Paolini M (2004) Genetic and metabolic effects of gluconasturtiin, a glucosinolate derived from cruciferae. Mutat Res 545: 23–35 [DOI] [PubMed] [Google Scholar]
- Cao YY, Yang JF, Liu TY, Su ZF, Zhu FY, Chen MX, Fan T, Ye NH, Feng Z, Wang LJ, et al. (2017) A phylogenetically informed comparison of GH1 hydrolases between Arabidopsis and rice response to stressors. Front Plant Sci 8: 350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella AN, Menossi M, Arruda P, Benedetti CE (2001) COI1 affects myrosinase activity and controls the expression of two flower-specific myrosinase-binding protein homologues in Arabidopsis. Planta 213: 691–699 [DOI] [PubMed] [Google Scholar]
- Chen R, Jiang H, Li L, Zhai Q, Qi L, Zhou W, Liu X, Li H, Zheng W, Sun J, Li C (2012) The Arabidopsis mediator subunit MED25 differentially regulates jasmonate and abscisic acid signaling through interacting with the MYC2 and ABI5 transcription factors. Plant Cell 24: 2898–2916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhajed S, Mostafa I, He Y, Abou-Hashem M, El-Domiaty M, Chen S (2020) Glucosinolate biosynthesis and the glucosinolate–myrosinase system in plant defense. Agronomy 10: 1786 [Google Scholar]
- Chini A, Fonseca S, Fernandez G, Adie B, Chico JM, Lorenzo O, Garcia-Casado G, Lopez-Vidriero I, Lozano FM, Ponce MR, et al. (2007) The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671 [DOI] [PubMed] [Google Scholar]
- Chini A, Gimenez-Ibanez S, Goossens A, Solano R (2016) Redundancy and specificity in jasmonate signalling. Curr Opin Plant Biol 33: 147–156 [DOI] [PubMed] [Google Scholar]
- Dombrecht B, Xue GP, Sprague SJ, Kirkegaard JA, Ross JJ, Reid JB, Fitt GP, Sewelam N, Schenk PM, Manners JM, et al. (2007) MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19: 2225–2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Calvo P, Chini A, Fernandez-Barbero G, Chico JM, Gimenez-Ibanez S, Geerinck J, Eeckhout D, Schweizer F, Godoy M, Franco-Zorrilla JM, et al. (2011) The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23: 701–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Calvo P, Inigo S, Glauser G, Vanden Bossche R, Tang M, Li B, De Clercq R, Nagels Durand A, Eeckhout D, et al. (2020) FRS7 and FRS12 recruit NINJA to regulate expression of glucosinolate biosynthesis genes. New Phytol 227: 1124–1137 [DOI] [PubMed] [Google Scholar]
- Fonseca S, Chini A, Hamberg M, Adie B, Porzel A, Kramell R, Miersch O, Wasternack C, Solano R (2009) (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat Chem Biol 5: 344–350 [DOI] [PubMed] [Google Scholar]
- Frerigmann H, Gigolashvili T (2014) MYB34, MYB51, and MYB122 distinctly regulate indolic glucosinolate biosynthesis in Arabidopsis thaliana. Mol Plant 7: 814–828 [DOI] [PubMed] [Google Scholar]
- Fu L, Wang M, Han B, Tan D, Sun X, Zhang J (2016) Arabidopsis myrosinase genes AtTGG4 and AtTGG5 are root-tip specific and contribute to auxin biosynthesis and root-growth regulation. Int J Mol Sci 17: 892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki Y, Yoshikawa Y, Sato T, Inada N, Ito M, Nishida I, Watanabe A (2001) Dark-inducible genes from Arabidopsis thaliana are associated with leaf senescence and repressed by sugars. Physiol Plant 111: 345–352 [DOI] [PubMed] [Google Scholar]
- Han X, Hu Y, Zhang G, Jiang Y, Chen X, Yu D (2018) Jasmonate negatively regulates stomatal development in Arabidopsis cotyledons. Plant Physiol 176: 2871–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Zhang M, Yang M, Hu Y (2020) Arabidopsis JAZ proteins interact with and suppress RHD6 transcription factor to regulate jasmonate-stimulated root hair development. Plant Cell 32: 1049–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai MY, Fujiwara T, Awazuhara M, Kimura T, Noji M, Saito K (2003) Global expression profiling of sulfur-starved Arabidopsis by DNA macroarray reveals the role of O-acetyl-l-serine as a general regulator of gene expression in response to sulfur nutrition. Plant J 33: 651–663 [DOI] [PubMed] [Google Scholar]
- Hirai MY, Saito K (2004) Post-genomics approaches for the elucidation of plant adaptive mechanisms to sulphur deficiency. J Exp Bot 55: 1871–1879 [DOI] [PubMed] [Google Scholar]
- Howe GA, Major IT, Koo AJ (2018) Modularity in Jasmonate Signaling for Multistress Resilience. Annu Rev Plant Biol 69: 387–415 [DOI] [PubMed] [Google Scholar]
- Hu Y, Jiang L, Wang F, Yu D (2013) Jasmonate regulates the inducer of cbf expression-C-repeat binding factor/DRE binding factor1 cascade and freezing tolerance in Arabidopsis. Plant Cell 25: 2907–2924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayum MA, Nath UK, Park JI, Hossain MR, Kim HT, Kim HR, Nou IS (2020) Glucosinolate profile and Myrosinase gene expression are modulated upon Plasmodiophora brassicae infection in cabbage. Funct Plant Biol 48: 103–118 [DOI] [PubMed] [Google Scholar]
- Lampard GR, Bergmann DC (2007) A shout-out to stomatal development: how the bHLH proteins SPEECHLESS, MUTE and FAMA regulate cell division and cell fate. Plant Signal Behav 2: 290–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, He K, Stolc V, Lee H, Figueroa P, Gao Y, Tongprasit W, Zhao H, Lee I, Deng XW (2007) Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19: 731–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KH, Piao HL, Kim HY, Choi SM, Jiang F, Hartung W, Hwang I, Kwak JM, Lee IJ, Hwang I (2006) Activation of glucosidase via stress-induced polymerization rapidly increases active pools of abscisic acid. Cell 126: 1109–1120 [DOI] [PubMed] [Google Scholar]
- Li H, Jiang H, Bu Q, Zhao Q, Sun J, Xie Q, Li C (2011) The Arabidopsis RING finger E3 ligase RHA2b acts additively with RHA2a in regulating abscisic acid signaling and drought response. Plant Physiol 156: 550–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Sack FD (2014) Myrosin idioblast cell fate and development are regulated by the Arabidopsis transcription factor FAMA, the auxin pathway, and vesicular trafficking. Plant Cell 26: 4053–4066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Yang R, Chen H (2018) The Arabidopsis thaliana Mediator subunit MED8 regulates plant immunity to Botrytis cinerea through interacting with the basic helix-loop-helix (bHLH) transcription factor FAMA. PLoS One 13: e0193458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Jiang H, Ye S, Chen WP, Liang W, Xu Y, Sun B, Sun J, Wang Q, Cohen JD, Li C (2010) The Arabidopsis P450 protein CYP82C2 modulates jasmonate-induced root growth inhibition, defense gene expression and indole glucosinolate biosynthesis. Cell Res 20: 539–552 [DOI] [PubMed] [Google Scholar]
- Matsushima R, Hayashi Y, Kondo M, Shimada T, Nishimura M, Hara-Nishimura I (2002) An endoplasmic reticulum-derived structure that is induced under stress conditions in Arabidopsis. Plant Physiol 130: 1807–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima R, Kondo M, Nishimura M, Hara-Nishimura I (2003) A novel ER-derived compartment, the ER body, selectively accumulates a beta-glucosidase with an ER-retention signal in Arabidopsis. Plant J 33: 493–502 [DOI] [PubMed] [Google Scholar]
- Mewis I, Appel HM, Hom A, Raina R, Schultz JC (2005) Major signaling pathways modulate Arabidopsis glucosinolate accumulation and response to both phloem-feeding and chewing insects. Plant Physiol 138: 1149–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitreiter S, Gigolashvili T (2021) Regulation of glucosinolate biosynthesis. J Exp Bot 72: 70–91 [DOI] [PubMed] [Google Scholar]
- Mocniak LE, Elkin K, Bollinger JM Jr (2020) Lifetimes of the aglycone substrates of specifier proteins, the autonomous iron enzymes that dictate the products of the glucosinolate-myrosinase defense system in brassica plants. Biochemistry 59: 2432–2441 [DOI] [PubMed] [Google Scholar]
- Nakano RT, Pislewska-Bednarek M, Yamada K, Edger PP, Miyahara M, Kondo M, Bottcher C, Mori M, Nishimura M, Schulze-Lefert P, et al. (2017) PYK10 myrosinase reveals a functional coordination between endoplasmic reticulum bodies and glucosinolates in Arabidopsis thaliana. Plant J 89: 204–220 [DOI] [PubMed] [Google Scholar]
- Nakazaki A, Yamada K, Kunieda T, Sugiyama R, Hirai MY, Tamura K, Hara-Nishimura I, Shimada T (2019) Leaf endoplasmic reticulum bodies identified in Arabidopsis rosette leaves are involved in defense against herbivory. Plant Physiol 179: 1515–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogasawara K, Yamada K, Christeller JT, Kondo M, Hatsugai N, Hara-Nishimura I, Nishimura M (2009) Constitutive and inducible ER bodies of Arabidopsis thaliana accumulate distinct beta-glucosidases. Plant Cell Physiol 50: 480–488 [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito K, Bergmann DC (2006) Arabidopsis FAMA controls the final proliferation/differentiation switch during stomatal development. Plant Cell 18: 2493–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega A, de Marcos A, Illescas-Miranda J, Mena M, Fenoll C (2019) The tomato genome encodes SPCH, MUTE, and FAMA candidates that can replace the endogenous functions of their Arabidopsis orthologs. Front Plant Sci 10: 1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Halitschke R, Kim HB, Baldwin IT, Feldmann KA, Feyereisen R (2002) A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J 31: 1–12 [DOI] [PubMed] [Google Scholar]
- Pauwels L, Goossens A (2011) The JAZ proteins: a crucial interface in the jasmonate signaling cascade. Plant Cell 23: 3089–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi T, Song S, Ren Q, Wu D, Huang H, Chen Y, Fan M, Peng W, Ren C, Xie D (2011) The jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 23: 1795–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serna L (2007) bHLH proteins know when to make a stoma. Trends Plant Sci 12: 483–485 [DOI] [PubMed] [Google Scholar]
- Sheard LB, Tan X, Mao H, Withers J, Ben-Nissan G, Hinds TR, Kobayashi Y, Hsu FF, Sharon M, Browse J, et al. (2010) Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468: 400–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirakawa M, Ueda H, Nagano AJ, Shimada T, Kohchi T, Hara-Nishimura I (2014) FAMA is an essential component for the differentiation of two distinct cell types, myrosin cells and guard cells, in Arabidopsis. Plant Cell 26: 4039–4052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Qi T, Huang H, Ren Q, Wu D, Chang C, Peng W, Liu Y, Peng J, Xie D (2011) The Jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect Jasmonate-regulated stamen development in Arabidopsis. Plant Cell 23: 1000–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama R, Hirai MY (2019) Atypical myrosinase as a mediator of glucosinolate functions in plants. Front Plant Sci 10: 1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J (2007) JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448: 661–665 [DOI] [PubMed] [Google Scholar]
- Ting HM, Cheah BH, Chen YC, Yeh PM, Cheng CP, Yeo FKS, Vie AK, Rohloff J, Winge P, Bones AM, et al. (2020) The role of a glucosinolate-derived nitrile in plant immune responses. Front Plant Sci 11: 257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii KU, Kanaoka MM, Pillitteri LJ, Bogenschutz NL (2007) Stomatal development: three steps for cell-type differentiation. Plant Signal Behav 2: 311–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vela-Corcia D, Aditya Srivastava D, Dafa-Berger A, Rotem N, Barda O, Levy M (2019) MFS transporter from Botrytis cinerea provides tolerance to glucosinolate-breakdown products and is required for pathogenicity. Nat Commun 10: 2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Li S, Li Y, Xu Y, Wang Y, Zhang R, Sun W, Chen Q, Wang XJ, Li C, et al. (2019) MED25 connects enhancer-promoter looping and MYC2-dependent activation of jasmonate signalling. Nat Plants 5: 616–625 [DOI] [PubMed] [Google Scholar]
- Wang H, Wu J, Sun S, Liu B, Cheng F, Sun R, Wang X (2011) Glucosinolate biosynthetic genes in Brassica rapa. Gene 487: 135–142 [DOI] [PubMed] [Google Scholar]
- Wasternack C, Feussner I (2018) The oxylipin pathways: biochemistry and function. Annu Rev Plant Biol 69: 363–386 [DOI] [PubMed] [Google Scholar]
- Wasternack C, Hause B (2013) Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann Bot 111: 1021–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Deng L, Zhai Q, Zhao J, Chen Q, Li C (2020) Mediator subunit MED25 couples alternative splicing of JAZ genes with fine-tuning of jasmonate signaling. Plant Cell 32: 429–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie DX, Feys BF, James S, Nieto-Rostro M, Turner JG (1998) COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094 [DOI] [PubMed] [Google Scholar]
- Xu L, Liu F, Lechner E, Genschik P, Crosby WL, Ma H, Peng W, Huang D, Xie D (2002) The SCF(COI1) ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell 14: 1919–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu ZY, Lee KH, Dong T, Jeong JC, Jin JB, Kanno Y, Kim DH, Kim SY, Seo M, Bressan RA, et al. (2012) A vacuolar beta-glucosidase homolog that possesses glucose-conjugated abscisic acid hydrolyzing activity plays an important role in osmotic stress responses in Arabidopsis. Plant Cell 24: 2184–2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue J, Jorgensen M, Pihlgren U, Rask L (1995) The myrosinase gene family in Arabidopsis thaliana: gene organization, expression and evolution. Plant Mol Biol 27: 911–922 [DOI] [PubMed] [Google Scholar]
- Yamada K, Goto-Yamada S, Nakazaki A, Kunieda T, Kuwata K, Nagano AJ, Nishimura M, Hara-Nishimura I (2020) Endoplasmic reticulum-derived bodies enable a single-cell chemical defense in Brassicaceae plants. Commun Biol 3: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Hara-Nishimura I, Nishimura M (2011) Unique defense strategy by the endoplasmic reticulum body in plants. Plant Cell Physiol 52: 2039–2049 [DOI] [PubMed] [Google Scholar]
- You Y, Zhai Q, An C, Li C (2019) LEUNIG_HOMOLOG mediates MYC2-dependent transcriptional activation in cooperation with the coactivators HAC1 and MED25. Plant Cell 31: 2187–2205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Chen X, Jiang M, Li X (2020) Two marine natural products, penicillide and verrucarin J, are identified from a chemical genetic screen for neutral lipid accumulation effectors in Phaeodactylum tricornutum. Appl Microbiol Biotechnol 104: 2731–2743 [DOI] [PubMed] [Google Scholar]
- Zhai Q, Li C (2019) The plant Mediator complex and its role in jasmonate signaling. J Exp Bot 70: 3415–3424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Q, Zhang X, Wu F, Feng H, Deng L, Xu L, Zhang M, Wang Q, Li C (2015) Transcriptional mechanism of jasmonate receptor COI1-mediated delay of flowering time in Arabidopsis. Plant Cell 27: 2814–2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.