Abstract
The DNA mismatch repair pathway is well known for its role in correcting biosynthetic errors of DNA replication. We report here a novel role for mismatch repair in signaling programmed cell death in response to DNA damage induced by chemical carcinogens. Cells proficient in mismatch repair were highly sensitive to the cytotoxic effects of chemical carcinogens, while cells defective in either human MutS or MutL homologs were relatively insensitive. Since wild-type cells but not mutant cells underwent apoptosis upon treatment with chemical carcinogens, the apoptotic response is dependent on a functional mismatch repair system. By analyzing p53 expression in several pairs of cell lines, we found that the mismatch repair-dependent apoptotic response was mediated through both p53-dependent and p53-independent pathways. In vitro biochemical studies demonstrated that the human mismatch recognition proteins hMutSα and hMutSβ efficiently recognized DNA damage induced by chemical carcinogens, suggesting a direct participation of mismatch repair proteins in mediating the apoptotic response. Taken together, these studies further elucidate the mechanism by which mismatch repair deficiency predisposes to cancer, i.e., the deficiency not only causes a failure to repair mismatches generated during DNA metabolism but also fails to direct damaged and mutation-prone cells to commit suicide.
DNA mismatch repair (MMR) is a critical pathway for the maintenance of genomic integrity. In Escherichia coli, methyl-directed MutHLS-dependent MMR ensures chromosome fidelity by correcting mispairs (both base-base and insertion-deletion mispairs) that result from biosynthetic errors and homologous recombination (51). In humans, a MutHLS-homologous MMR pathway has been characterized. Both the E. coli and the human pathway involve mismatch recognition (by MutS and MutL or their homologs), repair excision (by exonucleases), and resynthesis (by replicative DNA polymerases). A key feature in both systems is that the repair is strand specific. In E. coli, the repair is always targeted to the newly synthesized strand. This strand specificity is ensured by the function of MutH, an endonuclease that recognizes hemimethylated GATC sequences and makes a strand break in the newly synthesized, as-yet-unmethylated strand. Although the signal for strand discrimination in human cells is unknown, human MMR can be directed to a specific strand in vitro by a single-strand break in the DNA substrate (30, 67).
Defects in the human MMR proteins (encoded by the mutS homolog MSH2 and mutL homologs MLH1, PMS1, and PMS2) are associated with hereditary nonpolyposis colorectal cancer (HNPCC) (for reviews, see references 38, 40, and 53). Mutations in these HNPCC-associated genes and other MMR genes (e.g., MSH3 and MSH6) have also been identified in some fraction of sporadic cancers (8, 31, 35, 45, 74). Biochemical studies have demonstrated that functional eukaryotic MutS and MutL homologs are heterodimers. MSH2 interacts with MSH6 and MSH3 to form MutSα and MutSβ, respectively. Like the E. coli MutS protein, MutSα and MutSβ are mismatch recognition proteins (1, 17, 26, 32, 33, 46, 57). MLH1 interacts with PMS2 (PMS1 in Saccharomyces cerevisiae) to form MutLα (43, 61). A second eukaryotic MutL heterodimer containing MLH1 and PMS1 (MLH3 in yeast) has recently been proposed (19).
In addition to mismatch correction, MMR proteins participate in other cellular functions, such as transcription-coupled nucleotide excision repair (NER) (49, 50), meiotic chromosome synapsis (6, 7), the extension of heteroduplex intermediates during mitotic and meiotic recombination (11, 13, 14), and the recognition of Holliday junctions and other branched structures (2, 3, 69). Recently, human MutSα (hMutSα) has been shown to recognize a variety of DNA lesions that were previously believed to be processed only by the NER pathway (62). These lesions include O6-methylguanine (18), cisplatin (18), UV-induced photoproducts (54, 68), 2-aminofluorene (AF), and N-acetyl-2-aminofluorene (AAF) (42). These findings suggest that MMR proteins may be involved in the response to DNA damage induced by exposure to physical and chemical carcinogens. However, the molecular mechanism by which MMR proteins mediate this process is unclear.
Environmental carcinogens play an active role in tumorigenesis by covalently modifying DNA and inducing mutations in genes critical for the control of cellular growth. To explore the role of MMR in chemical carcinogenesis, we tested the ability of human MMR proteins to recognize and repair DNA adducts induced by several environmental carcinogens, such as aromatic amines and polycyclic aromatic hydrocarbons (PAHs). These two classes of carcinogens are widely distributed in our environment as byproducts of tobacco smoke, fuel combustion, dyes, and cooked meat, and they can be metabolically activated to highly reactive, mutagenic, and carcinogenic species (reviewed in references 27 and 29). The model compound for aromatic amines is N-acetoxy-AAF (AAAF), which reacts with DNA with a high degree of specificity at the C-8 position of deoxyguanine to form the major adduct N-(2′-deoxyguanosine-8-yl)-N-acetyl-2-aminofluorene (dG-AAF). The well-studied metabolites of PAHs are benzo[a]pyrene-7,8-dihydrodiol-9,10-epoxides (B[a]PDE). These dihydrodiol epoxides can exist as a pair of diastereoisomers, designated as syn or anti derivatives. Each diastereoisomer in turn can be further resolved into two enantiomers, specified as (+) and (−). B[a]PDE stereoisomers react preferentially with the exocyclic amino group of guanine (48, 56, 63).
In this study, we demonstrate that MMR-proficient cells are highly sensitive to the lethal cytotoxic effects of some chemical carcinogens, while cells defective in either some hMutS homologs or hMutL homologs are resistant to carcinogen-induced lethality. The cell death induced in wild-type cells occurs as an active programmed cell death, suggesting the involvement of MMR in triggering carcinogen-induced apoptosis. In vitro biochemical experiments show that hMutS homologs efficiently bind to individual B[a]PDE adducts. Thus, the interaction between DNA adducts and MMR proteins (hMutS and hMutL) may act as a signal for programmed cell death.
MATERIALS AND METHODS
Cell lines and nuclear extracts.
Lymphoblastoid lines TK6, WI-L2-NS, and MT1 were grown in RPMI 1640 medium (GIBCO) supplemented with 10% fetal bovine serum (HyClone) as described previously (37). Colorectal tumor cell lines HCT116 and HCT116-3-6 were grown in McCoy’s 5A medium with 10% fetal bovine serum. HeLa S3 cells were cultured as described previously (30). Nuclear extracts from all cell lines were prepared as described previously (30, 37, 59).
Construction of oligonucleotides containing B[a]PDE adducts.
Four oligodeoxynucleotide 11-mers containing (+)-trans-, (−)-trans-, (+)-cis-, or (−)-cis-anti-B[a]PDE stereoisomers were synthesized, purified, and characterized as described previously (60). These B[a]PDE-containing oligonucleotides were used to construct oligonucleotide 50-mer duplexes as depicted in Fig. 1A. Each of the B[a]PDE-modified oligomers (oligomer A, with a modification on the guanine residue of the oligonucleotide 5′-CCATCG*CTACC-3′; the asterisk represents the B[a]PDE adduct) was annealed to the centrally located complementary sequence in a 50-mer (oligomer C [5′-GCAGATCTGGCCTGATTGCGGTAGCGATGGAGCCGTAACAGTACGTAGTC-3′]) and ligated with another oligonucleotide (oligomer B [5′-GACTACGTACTGTTACGGCT-3′]) 5′ to the modified oligonucleotide. The ligated product was elongated by using the 50-mer as a template in the presence of deoxynucleoside triphosphates and the Klenow fragment of DNA polymerase I as described previously (42). The resulting 50-mer DNA duplexes containing carcinogen adducts were purified by polyacrylamide gel electrophoresis as described previously (76). A 50-mer homoduplex without B[a]PDE modification and a heteroduplex containing an A-C mismatch were also similarly constructed by using oligonucleotides 5′-CCATCGCTACC-3′ and 5′-CCATCACTACC-3′, respectively.
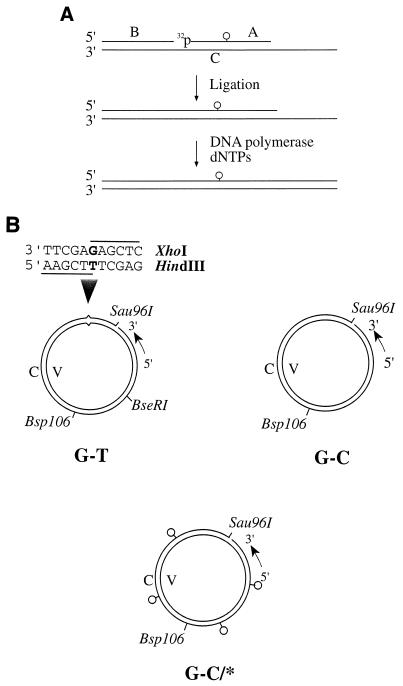
FIG. 1.
DNA substrates. (A) Construction of a 50-mer oligonucleotide containing carcinogen adducts. A 32P-labeled B[a]PDE-containing 11-mer (oligonucleotide A [5′-CCATCG*CTACC-3′]) was ligated with oligonucleotide B (5′-GACTACGTACTGTTACGGCT-3′) after they were annealed to a complementary 50-mer (oligomer C [5′-GCAGATCTGGCCTGATTGCGGTAGCGATGGAGCCGTAACAGTACGTAGTC-3′). The ligated oligomer was then elongated to 50 nucleotides by using oligonucleotide C as the template. The 50-mer homoduplex (G-C) and heteroduplex (A-C) were also constructed by using oligonucleotides 5′-CCATCGCTACC-3′ and 5′-CCATCACTACC-3′, respectively, as oligonucleotide A. dNTPs, deoxynucleoside triphosphates. (B) Circular DNA substrates. Each 6,440-bp circular substrate contained a strand break at the Sau96I site in the complementary (C) strand. Substrate G-T was a heteroduplex containing a single G-T mismatch, substrate G-C was a homoduplex, and substrate G-C/* was a homoduplex modified by chemical carcinogens in the complementary strand. The small circles represent carcinogen-DNA adducts. Several restriction sites relevant to this study are also shown.
Purification of hMutSα and hMutSβ.
hMutSα and hMutSβ were purified from nuclear extracts of HeLa S3 cells as described previously (17, 22, 42). The purified hMutSα and hMutSβ were nearly homogeneous as judged by Coomassie brilliant blue staining (data not shown). Western blot analysis using antibodies against hMSH2, hMSH3, and hMSH6 confirmed that hMutSα consists of hMSH2 (105 kDa) and hMSH6 (160 kDa), while hMutSβ consists of hMSH2 and hMSH3 (125 kDa) (data not shown).
Band shift analysis.
Band shift assays were performed as described previously (42) in 25-μl reaction mixtures containing 0.5 pmol of a 32P-labeled oligonucleotide duplex, 66 ng (about 0.25 pmol) of hMutSα or hMutSβ, 0.4 μg of double-stranded f1MR3 DNA (competitor DNA), 10 mM HEPES-KOH (pH 7.5), 110 mM KCl, 1 mM EDTA, 1 mM dithiothreitol, and 4% glycerol. Reaction mixtures were incubated on ice for 20 min, followed by the addition of 5 μl of 50% sucrose (wt/vol). The samples were then fractionated at room temperature through a 6% nondenaturing polyacrylamide gel in 6.7 mM Tris-acetate (pH 7.5)–1 mM EDTA with buffer recirculation. Bands were detected by autoradiography.
Treatment of cells with chemical carcinogens.
The desired amount of AAAF or B[a]PDE dissolved in dimethyl sulfoxide was added to cells suspended in culture medium. Cells were then incubated at 37°C in the presence of 5% CO2 for 1 h. The exposure was terminated by pelleting the cells and resuspending them in fresh medium. The final concentration of dimethyl sulfoxide was always less than 0.1% (vol/vol) and did not contribute to toxicity (data not shown). For cells growing in suspension, such as TK6, WI-L2-NS, and MT1 cells, both carcinogen-treated and untreated cells were plated in 96-well plates at a density of 1 to 3 cells/well in 0.2 ml of medium in the presence of TK6 feeder cells 105 cells/well), which were gamma irradiated (137Cs) with a total of 4.2 kilorads. In preliminary experiments, we determined that immediately after receiving 2.1 kilorads of gamma irradiation, cells were incapable of proliferation and could no longer be cloned. Cells were maintained at 37°C in a humidified atmosphere containing 5% CO2 until clone formation was visible. For cells growing in a monolayer (e.g., HCT116 and HCT116-3-6 cells), ∼50 carcinogen-treated or untreated cells were plated in six-well plates and cultured at 37°C in a 5% CO2 atmosphere. After incubation for 2 weeks, clones were counted under a microscope.
Construction of mismatch- or carcinogen adduct-containing substrates.
Heteroduplexes containing a single G-T mismatch (substrate G-T in Fig. 1B) were prepared as described previously (64) by hybridizing f1MR1 single-stranded DNA with Sau96I-linearized f1MR3 double-stranded DNA. A similar protocol was used to construct heteroduplexes containing multiple mismatches by using M13 and fd phage DNAs, which differ by 3% at the nucleotide level. To construct duplexes containing carcinogen adducts, double-stranded f1MR3 DNA was linearized with the restriction endonuclease Sau96I and treated with the desired amount of carcinogen at 37°C for 1 h. Unreacted carcinogen was removed by ether extraction and ethanol precipitation as described previously (12). DNA samples were resuspended in Tris-EDTA buffer and hybridized with unmodified f1MR3 single-stranded DNA to form homoduplexes with carcinogen adducts in the complementary strand. The average number of DNA adducts per molecule was determined by quantitative PCR based on the fact that these bulky DNA adducts either inhibit DNA synthesis or induce apurinic or apyrimidinic (AP) sites (in the case of B[a]PDE), which undergo β-elimination at a high temperature. In either case, shortened PCR products are expected, and these can be detected by quantitative analysis as described previously (47, 73). Homoduplex DNAs (substrate G-C in Fig. 1B) without carcinogen modification were prepared from f1MR3 double-stranded and single-stranded DNAs by a similar method.
Competitive repair.
Unless otherwise indicated, in vitro competitive repair was performed by using HeLa cell nuclear extracts. Competitive repair was assayed at 37°C for 15 min in a 15-μl reaction mixture containing 50 μg of nuclear extract and 100 ng of the G-T heteroduplex in the presence of various amount of competitor substrates (an M13-fd heteroduplex and a carcinogen-modified or unmodified homoduplex). Specific interaction of individual substrates with MMR proteins was monitored by their ability to inhibit the repair of a G-T mismatched heteroduplex in HeLa cell nuclear extracts. The repair of the G-T heteroduplex was evaluated by the assay for the production of two restriction fragments derived from cleavage of the repaired molecules with HindIII and Bsp106 or HindIII and BseRI, depending on the competitor substrates (see Fig. 1B).
Apoptosis analysis.
Chemical carcinogen-induced apoptosis was determined by the analysis of DNA fragmentation by using agarose gel electrophoresis and the terminal deoxynucleotidyltransferase (TdT)-mediated dUTP nick end labeling (TUNEL) method (21, 55). Briefly, cells were incubated with medium alone (control) or with medium containing 10 μM AAAF or 0.3 μM B[a]PDE at 37°C for 1 h. The supernatants were removed and replaced with fresh medium. After 24 or 48 h of culture, an aliquot of the cells was harvested, and DNA was isolated from these cells by protease K digestion and phenol-chloroform extraction, electrophoresed through 1.6% agarose gels, and visualized under UV light in the presence of ethidium bromide. The remaining cells were fixed with 1% (wt/vol) paraformaldehyde in phosphate-buffered saline and labeled with propidium iodide (PI) and fluorescein isothiocyanate (FITC)-dUTP in the presence of TdT; all these materials were provided in the APO-DIRECT kit (PharMingen, San Diego, Calif.). The cells were then analyzed by flow cytometry.
Chemicals and reagents.
AAAF and B[a]PDE were obtained from Chemsyn Science Laboratories, Lenexa, Kans. [γ-32P]ATP was obtained from Dupont. Oligonucleotides were purchased from Gibco-BRL Laboratories. Restriction enzymes were obtained from New England Biolabs. Antibodies against hMSH2 and hMSH6 were purchased from Oncogene Sciences (Boston, Mass.) and Serotec (Raleigh, N.C.), respectively. The hMSH3 antibody was the generous gift of Paul Modrich (Duke University, Durham, N.C.). The p53 antibody was from Santa Cruz Biotechnology (Santa Cruz, Calif.). Western blot analysis was performed with the ECL kit from Amersham Life Science.
RESULTS
Recognition of B[a]PDE adducts by hMutSα and hMutSβ.
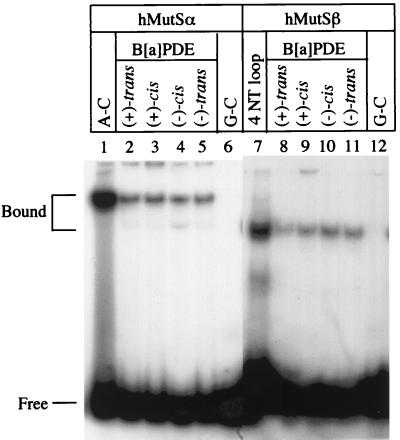
In previous work it was demonstrated that hMutSα recognizes DNA adducts induced by aromatic amines (42). To determine whether DNA MMR plays any role in PAH-induced mutagenesis and carcinogenesis, we tested the ability of hMutSα to recognize a 50-bp oligodeoxynucleotide duplex containing a single adduct of B[a]PDE at a guanine residue (see Fig. 1). Substrates that include four different antistereoisomers of B[a]PDE, i.e., (+)-trans-anti-B[a]PDE, (−)-trans-anti-B[a]PDE, (+)-cis-anti-B[a]PDE, and (−)-cis-anti-B[a]PDE, were constructed. As shown in Fig. 2, hMutSα efficiently binds to oligonucleotide duplexes containing each of the B[a]PDE adducts (lanes 2 to 5). Although the affinity of hMutSα for the B[a]PDE substrates was lower than that for an A-C mismatched substrate (Fig. 2, lane 1), it was significantly higher than the affinity to a homoduplex, whose interaction with hMutSα was almost undetectable (lane 6).
FIG. 2.
Binding of hMutSα and hMutSβ to DNA containing B[a]PDE. Unless otherwise indicated, gel shift assays were performed in 25-μl reaction mixtures containing 0.5 pmol of 32P-labeled oligonucleotide duplexes, 0.25 pmol of purified hMutSα or hMutSβ, 0.4 mg of double-stranded-f1MR3 DNA (competitor DNA), 10 mM HEPES-KOH (pH 7.5), 110 mM KCl, 1 mM EDTA, 1 mM dithiothreitol, and 4% glycerol. After 20 min of incubation on ice, 5 μl of 50% sucrose was added. The samples were fractionated at room temperature through a 6% nondenaturing polyacrylamide gel in 6.7 mM Tris-acetate (pH 7.5)–1 mM EDTA with buffer recirculation. NT, nucleotide.
Unlike hMutSα, which recognizes both base-base mismatches and insertion-deletion mispairs, hMutSβ recognizes only insertion-deletion mispairs (22, 57). To determine whether hMutSβ plays a role in chemical carcinogenesis, a similar approach was used to analyze the interaction between hMutSβ and DNA duplexes containing adducts of B[a]PDE. Like hMutSα, hMutSβ demonstrated a high affinity for the heteroduplex containing a loop of 4 nucleotides (Fig. 2, lane 7) and for each of the B[a]PDE adducts (lanes 8 to 11) but interacted poorly with an identical unmodified homoduplex (lane 12). These results indicate that hMutSβ is capable of recognizing DNA damage induced by bulky chemical carcinogens.
ATP has been shown to inhibit the interaction of hMutSα with mismatch-containing heteroduplexes in gel shift analyses (10, 17, 25, 42), since it drives molecular translocations (4, 10) or molecular switches (25). Thus, gel shift experiments were carried out in the presence of ATP to determine its effect on DNA-adduct binding by hMutSα and hMutSβ. The binding of hMutSα and hMutSβ to each of the B[a]PDE adducts was totally inhibited by the presence of 1 mM ATP (data not shown), suggesting that hMutS proteins interact with the carcinogen adducts and DNA mismatches in a similar manner.
Carcinogen adducts block strand-specific MMR.
The experiments described above demonstrate that hMutSα and hMutSβ recognize and bind to carcinogen-DNA adducts. To further test the specific interaction between carcinogen-DNA adducts and MMR proteins, a competition MMR assay was performed. Strand-specific MMR is assayed in vitro by using a circular heteroduplex containing a single mismatch located within two overlapping restriction endonuclease recognition sites (see Fig. 1B) (30). The presence of heterology within the recognition sites renders the heteroduplex resistant to cleavage by both enzymes. Thus, repair of the mismatch can be monitored by the sensitivity of the heteroduplex to one endonuclease or the other. This assay was carried out by using a G-T mismatched DNA duplex in the presence of chemically adducted DNA or other DNA competitor substrates. If carcinogen-induced DNA damage specifically interacts with MMR proteins, then it is predicted that chemically adducted substrates will effectively compete with the G-T heteroduplex for MMR proteins and inhibit repair.
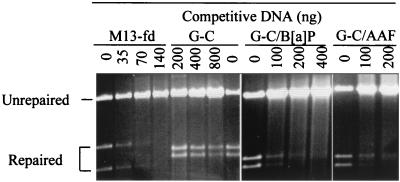
Figure 3 shows the repair of a G-T heteroduplex by HeLa nuclear extracts in the presence of an M13-fd heteroduplex, a G-C homoduplex, and B[a]PDE- or AAAF-adducted DNA competitors. Repair was completely inhibited by 70 ng of the M13-fd heteroduplex, which contains about 192 mismatches per molecule (3% heterology [69]). Repair was only slightly reduced even in the presence of 800 ng of the homoduplex DNA. This result is consistent with the prediction that DNA molecules that are MMR substrates will be efficient competitors in this assay. Figure 3 also shows that repair of the G-T heteroduplex was totally blocked by 200 ng of either AAAF- or B[a]PDE-modified homoduplexes, which contained about 10 adducts per molecule in each case as determined by quantitative PCR analysis (see Materials and Methods for details). This result indicates that DNA adducts of AAAF and B[a]PDE actively compete with the G-T heteroduplex for MMR proteins.
FIG. 3.
Inhibition of MMR reaction by DNA-containing carcinogen adducts. G-T mismatch repair was performed as described previously (30) by using 50 μg of HeLa nuclear extracts and 100 ng of a heteroduplex containing a single G-T mismatch. Repair assays were performed in reaction mixtures containing 100 ng of the G-T mismatched heteroduplex, the indicated competitor DNA, and 50 μg of HeLa nuclear extracts. The reaction mixtures were incubated at 37°C for 15 min. After phenol and ether extraction, DNA substrates were recovered by ethanol precipitation and digested with restriction endonucleases to score the repair of the G-T heteroduplex. For the homoduplex (G-C) or carcinogen-modified homoduplexes (G-C/B[a]PDE, and G-C/AAAF), restriction endonucleases HindIII and Bsp106 were used to detect the repair of the G-T substrate, producing 3.1- and 3.3-kb fragments (see Fig. 1). For the M13-fd heteroduplex, HindIII and BseRI (producing 2.8- and 3.6-kb fragments) were used since Bsp106 cleaves the M13-fd substrate into fragments of the same size as those obtained by HindIII and Bsp106 cleavage of the repaired G-T substrate.
MMR-proficient cells are sensitive to chemical carcinogen-induced cytotoxicity.
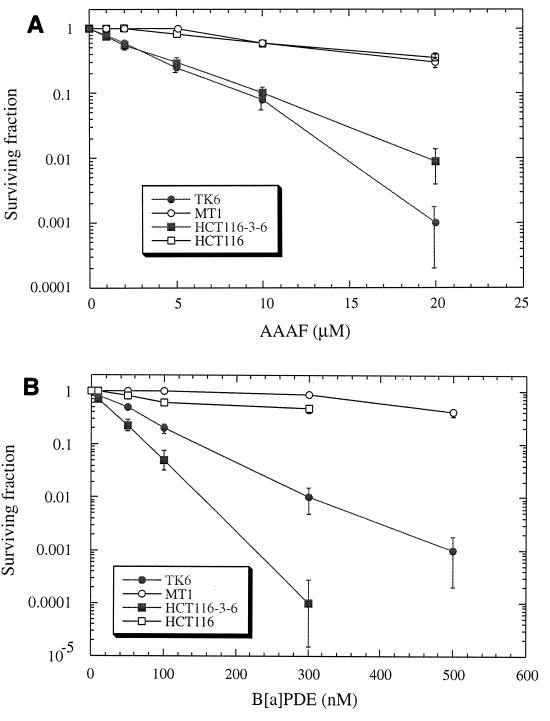
Because hMutSα and hMutSβ recognize DNA damage induced by chemical carcinogens, it was reasoned that the MMR pathway could influence the biological impact of exposure to these agents. Thus, the cytotoxic effects of exposure to B[a]PDE and AAAF were determined in MMR-proficient and MMR-deficient cells. The MMR-deficient cell lines used were MT1 (MSH6 defective), and HCT116 (MLH1 defective); the corresponding wild-type cell lines are TK6 and HCT116-3-6. MT1 was derived from the lymphoblastoid cell line TK6 by frameshift mutagenesis and is tolerant to N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) (23); HCT116-3-6 was derived from colorectal tumor cell line HCT116 by transfer of chromosome 3, which carries the wild-type MLH1 gene (39).
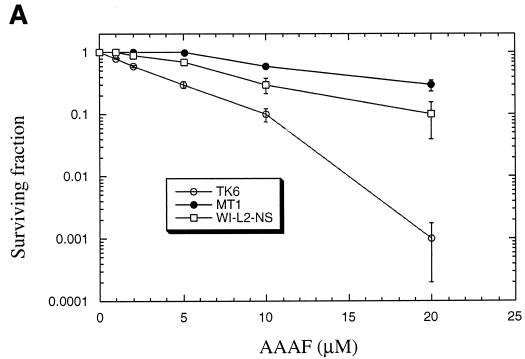
Cells were treated with AAAF, followed by clonogenic survival assays as described in Materials and Methods. Figure 4A shows that the MMR-deficient MT1 cells exhibited severalfold-greater resistance to killing by AAAF than the wild-type TK6 cells. In the presence of 20 μM AAAF, almost all TK6 cells were killed, but more than 30% of the MT1 cells survived. With the MLH1-deficient HCT116 cell line and its wild-type counterpart HCT116-3-6, similar results were observed. MMR-deficient HCT116 cells showed higher levels of clonal survival than repair-proficient HCT116-3-6 cells following exposure to each concentration of AAAF. MT1 cells are defective in hMutSα, and HCT116 cells are deficient in hMutLα (43). This result indicates that both hMutS and hMutL complexes are involved in AAAF-induced cell death. As observed with AAAF treatment, MT1 and HCT116 cells exhibited severalfold-greater resistance to killing after treatment with B[a]PDE than the corresponding wild-type cells (Fig. 4B).
FIG. 4.
Sensitivity of MMR-proficient cells to the cytotoxicity of chemical carcinogens. Graphs show the clonogenic survival of MMR-proficient (TK6 and HCT116-3-6) and MMR-deficient (MT1 and HCT116) cells following exposure to AAAF (A) or B[a]PDE (B). Standard deviations were calculated from three independent experiments.
Carcinogen-induced cell death is mediated by apoptosis.
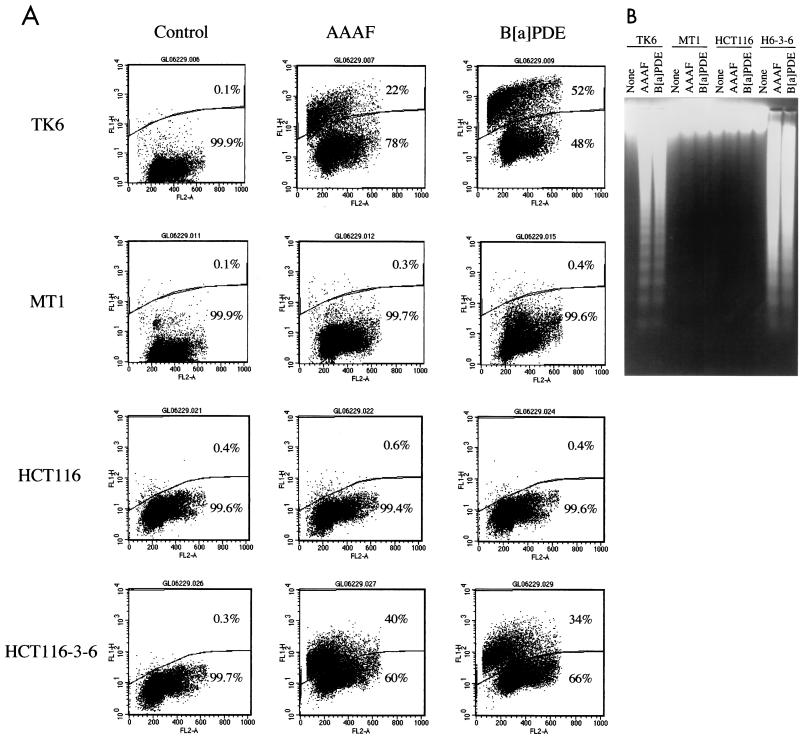
The mechanism of carcinogen-induced cell death was determined by measuring DNA degradation after treatment with AAAF or B[a]PDE. Cells were incubated for 1 h with 10 μM AAAF or 0.3 μM B[a]PDE, concentrations that are sufficient to kill more than 90% of wild-type cells in clonogenic analysis, but that kill less than 25% of MMR-deficient cells (see Fig. 4). After removal of the carcinogens, cells were cultured in fresh media for 24 or 48 h and harvested for DNA fragmentation analysis. Figure 5A shows the level of cell death 24 h after carcinogen treatment as determined by the TUNEL assay described in Materials and Methods. Cells undergoing DNA fragmentation (positive in the TUNEL assay) were labeled with FITC-dUTP by TdT and were believed to be apoptotic cells (positive in the TUNEL assay). Figure 5A shows histograms of these analyses, with FITC-dUTP staining and PI staining displayed in the vertical and horizontal axes in each of the histograms, respectively. Upon treatment with AAAF or B[a]PDE, 20 to 50% of MMR-proficient cells (TK6 and HCT116-3-6) were TUNEL positive within 24 h, while no significant increase in TUNEL staining occurred in repair-deficient cells (MT1 and HCT116). Similar results were also obtained when cells were grown for 48 h after carcinogen treatments (data not shown).
FIG. 5.
Chemical carcinogens induce apoptosis. (A) TUNEL analysis. Cells were incubated with medium alone (control) or with medium containing 10 μM AAAF or 0.3 μM B[a]PDE at 37°C for 1 h and were then cultured in fresh medium for 24 h before being harvested for flow cytometry analysis as described in Materials and Methods. The cells above the gate are apoptotic (TUNEL positive), while those below the gate are nonapoptotic (TUNEL negative). The percentages of cells in each category are shown. FL2-A (horizontal axis) indicates PI staining, and FL1H (vertical axis) indicates FITC-dUTP staining. (B) DNA fragmentation in TK6 and HCT116-3-6 (H6-3-6) cells induced by AAAF and B[a]PDE. Cells were treated as described above. Genomic DNA was isolated by protease K digestion and phenol-chloroform extraction, fractionated through 1.6% agarose gels, and visualized under UV light in the presence of ethidium bromide. None, control.
To further confirm that cell death in MMR-proficient cells occurs through an apoptotic pathway, direct visualization of DNA fragmentation, a hallmark of apoptosis, was carried out. Genomic DNA was isolated from individual cell lines after treatment with chemical carcinogens and was fractionated in 1.6% agarose gels. As shown in Fig. 5B, DNA fragmentation was observed in MMR-proficient TK6 and HCT116-3-6 cells, but not in mutant MT1 and HCT116 cells, after treatment with AAAF or B[a]PDE. These results are consistent with those of the TUNEL assay described above, suggesting that carcinogen-induced cell death occurs by apoptosis and is an MMR-dependent process.
p53 involvement.
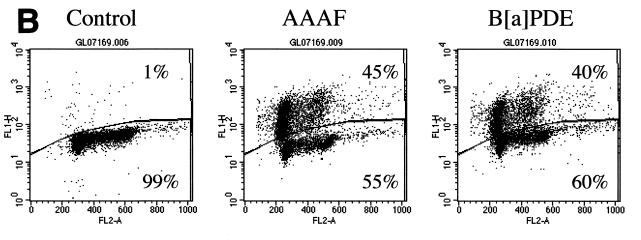
p53 plays a critical role in apoptosis. To determine if p53 is involved in carcinogen-induced apoptosis, the p53-defective lymphoblastoid cell line WI-L2-NS was subjected to clonogenic analysis after AAAF or B[a]PDE treatment. The WI-L2-NS line was derived from the same origin as the TK6 line but harbors a point mutation in the p53 gene at codon 237, resulting in a methionine-to-isoleucine change (9, 72). This mutation prevents WI-L2-NS cells from undergoing apoptotic death by radiation (44, 75). Nevertheless, WI-L2-NS cells possess strand-specific MMR activity comparable to that of the wild-type TK6 cells (25a). Clonogenic survival analysis following treatment with AAAF or B[a]PDE indicated that WI-L2-NS cells were more resistant to killing by either carcinogen than TK6 cells but more sensitive than MT1 cells (Fig. 6A). TUNEL and DNA fragmentation analyses were also performed. As demonstrated in Fig. 6B, TUNEL-positive cells were observed among WI-L2-NS cells after treatment with either AAAF or B[a]PDE. These results are similar to those observed with TK6 cells. To directly visualize DNA fragmentation, DNA from carcinogen-treated WI-L2-NS cells was fractionated through an agarose gel. Unlike the findings for TK6 cells, where distinct DNA fragments were displayed (Fig. 5B), smeared DNA was observed in AAAF- or B[a]PDE-treated WI-L2-NS cells (Fig. 6C). It is not clear whether this reflects p53-independent apoptosis or random DNA degradation due to necrotic cell death.
FIG. 6.
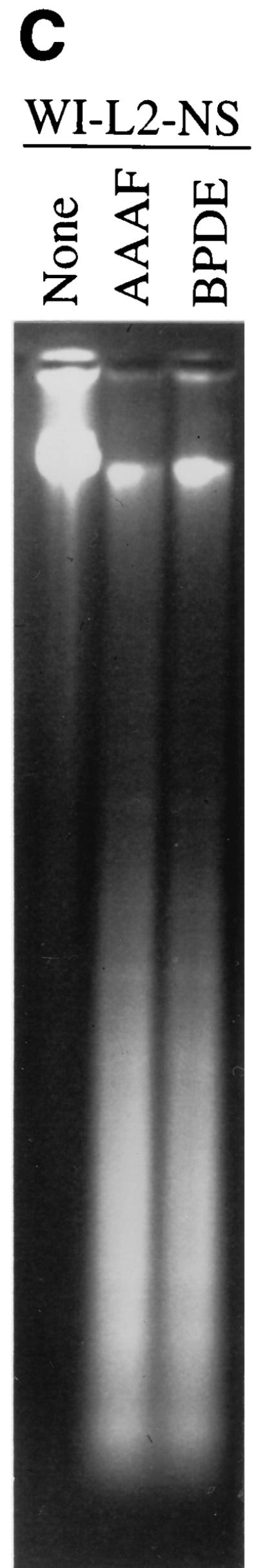
Response of p53-defective WI-L2-NS cells to treatments with chemical carcinogens. (A) clonogenic survival; (B) TUNEL analysis; (C) DNA fragmentation analysis. None, control.
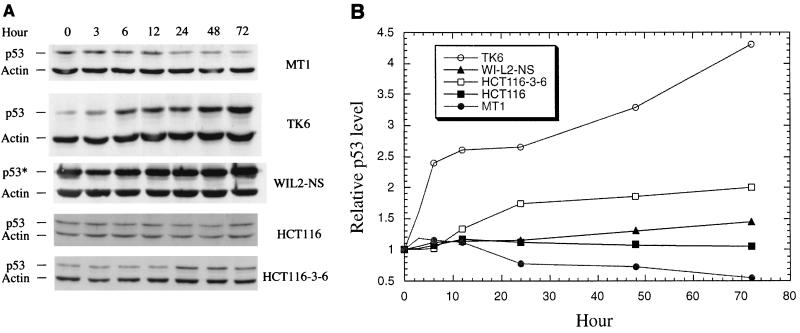
The induction of p53 is always associated with apoptosis induced by DNA damage (reviewed in reference 41). We therefore investigated the expression of p53 in cells treated with AAAF or B[a]PDE. After 1 h of incubation with the carcinogen, cells were cultured in fresh media for various periods of time, and cell extracts were prepared for Western blot analysis. Both TK6 and MT1 cells expressed significant amounts of p53. However, the p53 level in TK6 cells apparently increased steadily post-AAAF treatment, while this increase was not observed in MT1 cells (Fig. 7). Although the p53 in WI-L2-NS cells is mutated, there seemed to be a slight increase in the level of the mutated protein in the cells after AAAF treatment. The difference in p53 expression between HCT116-3-6 and HCT116 cells was not as obvious as that observed between TK6 and MT1 cells (Fig. 7). The relative p53 levels for each pair of cell lines after B[a]PDE treatment were similar to those observed in AAAF experiments (data not shown).
FIG. 7.
Expression of p53 in MMR-proficient and -deficient cells after treatment with AAAF. (A) Western blot analysis. After AAAF treatment, cells were cultured in fresh medium for the indicated times. Actin was used as an internal control, and equal amounts of protein were loaded for each sample. Both p53 protein and actin were detected by chemiluminescence on a Western blot by using antibodies against p53 and actin (Sigma), respectively. p53*, mutant p53 protein. (B) Relative amounts of p53. The p53 level at each point was normalized by its corresponding actin level as well as by the amount of p53 at time zero.
DISCUSSION
Mutations in DNA are the primary cause of human cancer and mainly result from two sources: errors in DNA metabolism and damage induced by endogenous or exogenous mutagens or carcinogens. It is generally accepted that MMR is responsible for correcting errors of DNA metabolism (51), while base excision repair and NER are mainly responsible for the repair of carcinogen-induced DNA damage (62). However, recent studies from several groups suggest that MMR is also involved in the repair of DNA damage that was previously thought to be processed only by excision repair pathways. Indirect evidence has shown that MutS and MutL mutants of E. coli, or their homologs in humans, are also defective in transcription-coupled NER of UV-induced pyrimidine dimers (49, 50). More direct evidence comes from studies demonstrating specific interactions between purified hMutSα and DNA containing damage induced by many agents. These agents include MNNG, 6-thioguanine, cisplatin, UV, AF, and AAF (18, 42, 54, 65, 68). In this study, we have extended this list to B[a]PDE, one of the most carcinogenic compounds identified to date. In addition, we also show that, like hMutSα, hMutSβ specifically recognizes bulky DNA adducts induced by B[a]PDE, indicating that hMutSβ shares the same specificity with hMutSα with respect to its interactions with carcinogen-DNA adducts. The binding of hMutS heterodimers to carcinogen-DNA adducts can be blocked by the presence of ATP, which has also been demonstrated for the binding of hMutSα to DNA mismatches (10, 17, 25). These findings suggest that hMutS homologs interact with carcinogen-DNA adducts and DNA mismatches through a similar mechanism. The specific interaction between hMutS homologs and carcinogen-DNA adducts is further confirmed by the demonstration that carcinogen-DNA adducts effectively inhibit strand-specific MMR in HeLa nuclear extracts (Fig. 3).
This work clearly demonstrates for the first time that MMR-proficient cells (TK6 and HCT116-3-6 cells) are much more sensitive to killing by AAAF and B[a]PDE than the corresponding MMR-defective mutant cells (MT1 and HCT116 cells). It is known that these carcinogens exert their carcinogenic effects by covalently modifying genomic DNA to form DNA adducts. Removal of these adducts by a DNA repair system is very important for genomic integrity. Therefore, one would expect cells defective in DNA repair to be sensitive to the action of environmental carcinogens. Indeed, this is the case for NER (reviewed in reference 20). In addition to tolerance to chemical carcinogens, MMR mutants have previously been shown to be resistant to the cytotoxic effects of many common chemotherapeutic drugs such as MNNG, 6-thioguanine, cisplatin, adriamycin, doxorubicin, and temozolomide (5, 15, 16, 28, 37, 39, 65).
We demonstrate here that upon treatment with chemical carcinogens, MMR-proficient cells undergo programmed cell death as judged by TUNEL analysis as well as by gel-based DNA fragmentation analysis. These two analyses distinguish between two cell death pathways: apoptosis and necrosis (70, 71). In contrast to the formation of DNA ladders by apoptotic cells, necrotic cells are characterized by a smear of low-molecular-weight DNA molecules due to random cleavage of DNA by nonspecific DNases (70, 71). TUNEL analysis is very sensitive in detecting in situ DNA degradation but cannot distinguish DNA fragmented during apoptosis and necrosis. The gel-based DNA fragmentation assay allows conclusive identification of the specific cell death mechanism. Based on these analyses, the involvement of p53 in carcinogen-induced apoptosis was investigated by comparing cell lines with different p53 backgrounds for their DNA fragmentation patterns and p53 expression levels. The lymphoblastoid cell lines TK6, MT1, and WI-L2-NS are derived from the same donor (9, 72), but they differ in their p53 and MMR phenotypes: TK6 cells are wild type for both MMR and p53; MT1 cells are defective in MMR but wild type for p53 (15); WI-L2-NS cells possess normal MMR activity (25a) but carry a point mutation in exon 7 of the p53 gene, which abolishes the function of p53 (9, 72). MT1 cells are negative in both TUNEL and gel-based DNA fragmentation analyses. Although both TK6 and WI-L2-NS cells are positive in the TUNEL assay, the gel-based DNA degradation assay revealed that cell death in these two lines may occur through different mechanisms. DNA fragmentation was clearly observed in TK6 cells but not in WI-L2-NS cells. Instead, a smeared DNA was recovered from WI-L2-NS cells. Although we cannot rule out the possibility that WI-L2-NS cells die through apoptosis, the nature of DNA degradation in the cell line is very similar to that of cells undergoing necrosis. If the latter is the case, chemical-carcinogen-induced apoptosis in these cells requires both functional MMR and p53. If WI-L2-NS cells do die by apoptosis, the cell death is mediated by factors independent of p53. However, p53 may contribute to the promotion of DNA laddering in TK6 cells.
We further addressed the involvement of p53 in this process by measuring p53 levels in different cell lines after treatments with chemical carcinogens. Upon exposure to AAAF or B[a]PDE, TK6 cells, but not MT1 cells, showed increased levels of p53 protein. Even though the p53 gene is mutated in WI-L2-NS cells, there was a slight increase in the level of the mutant protein. Therefore, it seems that there is a close correlation between the expression of p53 (both functional and mutated forms) and a functional MMR system. Thus, it appears that chemical-carcinogen-induced apoptosis is dependent on both functional MMR and p53 in these cells. The correlation in these cell lines was also reported when they were treated with temozolomide (15), an alkylating chemotherapeutic agent. By analyzing the cellular response to MNNG and cisplatin in mice with defects in the MSH2 or p53 gene, Toft et al. (66) have recently shown that the MSH2-dependent apoptotic response is mediated through a p53-dependent pathway. Taken together, these findings indicate that MMR-mediated apoptosis in response to chemical agents can occur through a p53-dependent pathway.
However, this conclusion cannot explain the apoptotic response in HCT116-3-6 cells, which possess both a functional MMR system and wild-type p53. Unlike the findings for TK6 cells, where p53 levels increased severalfold (Fig. 7) within 3 days after carcinogen treatments in comparison with both untreated cells and treated MT1 cells, p53 levels in HCT116-3-6 increased only slightly compared with those in HCT116 cells (Fig. 7). This suggests that apoptosis in this cell line, although dependent on MMR, may not be mediated by a p53-dependent pathway. In fact, while this article was being revised, Gong et al. (24) demonstrated that cisplatin-induced apoptosis occurs only in HCT116-3-6 cells, not in HCT116 cells, indicating the involvement of MMR in cisplatin-induced apoptosis. They also found that apoptosis in HCT116-3-6 cells is mediated not by p53 but by p73 (24), a p53-related protein that can also induce apoptosis (34). It is possible that the chemical-carcinogen-induced cell death described here may also be regulated by a p73-dependent manner. Although we do not know the difference in apoptotic response between the MT1-TK6 pair and the HCT116–HCT116-3-6 pair, these two pairs of cells were derived from different origins. First, MT1 and TK6 lines are lymphoblastoid cells, while HCT116 and HCT116-3-6 lines are colorectal tumor cells. Second, in the HCT116–HCT116-3-6 pair, the MMR-proficient HCT116-3-6 line was derived from the mutant HCT116 line by chromosome transfer (39), but the mutant MT1 line of the MT1-TK6 pair was derived from the wild-type TK6 line by treatment of the latter with a mutagen and selection for MNNG-resistant cells (23). Therefore, MT1 cells may harbor an enormous number of mutations that are not present in the parental cells and that affect apoptotic response. Nevertheless, this study, together with previous investigations, reveals that MMR deficiency confers cancer predisposition not only through failed repair of DNA mismatches generated from DNA metabolism but also through the failure to signal apoptosis in cells with damaged DNA.
The molecular mechanism by which MMR triggers carcinogen-induced apoptosis is not known. Since cells defective in either hMutS homologs or hMutL homologs fail to engage chemical-induced apoptosis, both hMutS and hMutL homologs must be involved in the process. Thus, several models have been proposed (for reviews see references 36 and 52). A favored candidate is the futile-repair model. Chemical carcinogens modify DNA to form carcinogen-DNA adducts, which could pair with appropriate bases or introduce mispairs during DNA replication. Under either circumstance, hMutS homologs can recognize these unusual pairs as “mismatches” to provoke a strand-specific MMR reaction. However, since MMR is always targeted to the newly synthesized strand, the offending adducts, which are located in the template strand, cannot be removed by strand-specific MMR and thus continue to produce unusual base pairs upon DNA resynthesis. As a result, the repair cycle is reinitiated. Such futile repair may be translated into an apoptotic signal. Alternatively, cell death may be due to the binding of MMR proteins (at least hMutS and hMutL homologs) to DNA adducts, which either leads to a checkpoint response or blocks other DNA transactions such as replication, transcription, and proper damage repair.
Our gel shift experiments suggest that both hMutSα and hMutSβ may be involved in signaling p53- or p73-dependent apoptosis, as judged by their ability to recognize DNA containing carcinogen adducts. However, it is worth noting that the MT1 cell line, which supposedly has wild-type hMutSβ but is defective in hMutSα (17, 58), still fails to commit suicide in response to chemical-carcinogen cytotoxicity. This result suggests that either hMutSβ is not involved in the apoptotic response or the hMSH3 gene in MT1 cells contains an unidentified mutation that affects apoptotic response. Further investigations are required to distinguish between these hypotheses.
ACKNOWLEDGMENTS
We thank David Scicchitano for stimulating discussions at the beginning of this project, Dao-Hong Zhou for help in analyzing the flow cytometry data, and Scott McCulloch for comments on the manuscript.
This work is supported by grants CA72856 (to G.-M.L.) and CA20851 (to N.E.G.) from the National Cancer Institute, by grant 4755 (to G.-M.L.) from the Council for Tobacco Research Inc., and by funds (to G.-M.L.) from the Lucille P. Markey Trust.
REFERENCES
- 1.Acharaya S, Wilson T, Gradia S, Kane M F, Guerrette S, Marsischky G T, Kolodner R, Fishel R. hMSH2 forms specific mispair-binding complexes with hMSH3 and hMSH6. Proc Natl Acad Sci USA. 1996;93:13629–13634. doi: 10.1073/pnas.93.24.13629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alani E, Lee S, Kane M F, Griffith J, Kolodner R D. Saccharomyces cerevisiae MSH2, a mispaired base recognition protein, also recognizes Holliday junctions in DNA. J Mol Biol. 1997;265:289–301. doi: 10.1006/jmbi.1996.0743. [DOI] [PubMed] [Google Scholar]
- 3.Alani E, Reenan R A, Kolodner R D. Interaction between mismatch repair and genetic recombination in Saccharomyces cerevisiae. Genetics. 1994;137:19–39. doi: 10.1093/genetics/137.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allen D J, Makhov A, Grilley M, Taylor J, Thresher R, Modrich P, Griffith J D. MutS mediates heteroduplex loop formation by a translocation mechanism. EMBO J. 1997;16:4467–4476. doi: 10.1093/emboj/16.14.4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anthoney D A, McIlwrath A J, Gallagher W M, Edlin A R M, Brown R. Microsatellite instability, apoptosis, and loss of p53 function in drug-resistant tumor cells. Cancer Res. 1996;56:1374–1381. [PubMed] [Google Scholar]
- 6.Baker S M, Bronner C E, Zhang L, Plug A W, Robatzek M, Warren G, Elliott E A, Yu J, Ashley T, Arnheim N, et al. Male mice defective in the DNA mismatch repair gene PMS2 exhibit abnormal chromosome synapsis in meiosis. Cell. 1995;82:309–319. doi: 10.1016/0092-8674(95)90318-6. [DOI] [PubMed] [Google Scholar]
- 7.Baker S M, Plug A W, Prolla T A, Bronner C E, Harris A C, Yao X, Christie D M, Monell C, Arnheim N, Bradley A, Ashley T, Liskay R M. Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nat Genet. 1996;13:336–342. doi: 10.1038/ng0796-336. [DOI] [PubMed] [Google Scholar]
- 8.Benachenhou N, Guiral S, Gorska-Flipot I, Labuda D, Sinnett D. High resolution deletion mapping reveals frequent allelic losses at the DNA mismatch repair loci hMLH1 and hMSH3 in non-small cell lung cancer. Int J Cancer. 1998;77:173–180. doi: 10.1002/(sici)1097-0215(19980717)77:2<173::aid-ijc1>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 9.Bill C A, Yu Y, Miselis N R, Little J B, Nickoloff J A. A role for p53 in DNA end rejoining by human cell extracts. Mutat Res. 1997;385:21–29. doi: 10.1016/s0921-8777(97)00040-2. [DOI] [PubMed] [Google Scholar]
- 10.Blackwell L J, Martik D, Bjornson K P, Bjornson E S, Modrich P. Nucleotide-promoted release of hMutSalpha from heteroduplex DNA is consistent with an ATP-dependent translocation mechanism. J Biol Chem. 1998;273:32055–32062. doi: 10.1074/jbc.273.48.32055. [DOI] [PubMed] [Google Scholar]
- 11.Borts R H, Leung W Y, Kramer W, Kramer B, Williamson M, Fogel S, Haber J E. Mismatch repair-induced meiotic recombination requires the PMS1 gene product. Genetics. 1990;124:573–584. doi: 10.1093/genetics/124.3.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown W C, Romano L J. Benzo[a]pyrene adducts inhibit translocation by the gene 4 protein of bacteriophage T7. J Biol Chem. 1989;263:6748–6754. [PubMed] [Google Scholar]
- 13.Chambers S R, Hunter N, Louis E J, Borts R H. The mismatch repair system reduces meiotic homeologous recombination and stimulates recombination-dependent chromosome loss. Mol Cell Biol. 1996;16:6110–6120. doi: 10.1128/mcb.16.11.6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen W, Jinks-Robertson S. The role of the mismatch repair machinery in regulating mitotic and meiotic recombination between diverged sequences in yeast. Genetics. 1999;151:1299–1313. doi: 10.1093/genetics/151.4.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D’Atri S, Tentori L, Lacal P M, Graziani G, Pagani E, Benincasa E, Zambruno G, Bonmassar E, Jiricny J. Involvement of the mismatch repair system in temozolomide-induced apoptosis. Mol Pharmacol. 1998;54:334–341. doi: 10.1124/mol.54.2.334. [DOI] [PubMed] [Google Scholar]
- 16.Drummond J T, Anthoney A, Brown R, Modrich P. Cisplatin and adriamycin resistance are associated with MutLα and mismatch repair deficiency in an ovarian tumor cell line. J Biol Chem. 1996;271:19645–19648. doi: 10.1074/jbc.271.33.19645. [DOI] [PubMed] [Google Scholar]
- 17.Drummond J T, Li G M, Longley M J, Modrich P. Isolation of an hMSH2–p160 heterodimer that restores DNA mismatch repair to tumor cells. Science. 1995;268:1909–1912. doi: 10.1126/science.7604264. [DOI] [PubMed] [Google Scholar]
- 18.Duckett D R, Drummond J T, Murchie A I H, Reardon J T, Sancar A, Lilley D M, Modrich P. Human MutSα recognizes damaged DNA base pairs containing O6-methylguanine, O4-methylthymine, or the cisplatin-d(GpG) adduct. Proc Natl Acad Sci USA. 1996;93:6443–6447. doi: 10.1073/pnas.93.13.6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flores-Rozas H, Kolodner R D. The Saccharomyces cerevisiae MLH3 gene functions in MSH3-dependent suppression of frameshift mutations. Proc Natl Acad Sci USA. 1998;95:12404–12409. doi: 10.1073/pnas.95.21.12404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friedberg E C, Walker G C, Siede W. DNA repair and mutagenesis. Washington, D.C.: ASM Press; 1995. [Google Scholar]
- 21.Gavrieli Y, Sherman Y, Ben-Sasson S A. Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. J Cell Biol. 1992;119:493–501. doi: 10.1083/jcb.119.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Genschel J, Littman S J, Drummond J T, Modrich P. Isolation of MutSβ from human cells and comparison of the mismatch repair specificities of MutSβ and MutSα. J Biol Chem. 1998;273:19895–19901. doi: 10.1074/jbc.273.31.19895. [DOI] [PubMed] [Google Scholar]
- 23.Goldmacher V S, Cuzick R A, Thilly W G. Isolation and partial characterization of human cell mutants differing in sensitivity to killing and mutation by methylnitrosourea and N-methy-N′-nitro-nitrosoguanidine. J Biol Chem. 1986;261:12462–12471. [PubMed] [Google Scholar]
- 24.Gong J G, Costanzo A, Yang H Q, Melino G, Kaelin W G, Jr, Levrero M, Wang J Y. The tyrosine kinase c-Abl regulates p73 in apoptotic response to cisplatin-induced DNA damage. Nature. 1999;399:806–809. doi: 10.1038/21690. [DOI] [PubMed] [Google Scholar]
- 25.Gradia S, Acharya S, Fishel R. The human mismatch recognition complex hMSH2-hMSH6 functions as a novel molecular switch. Cell. 1997;91:995–1005. doi: 10.1016/s0092-8674(00)80490-0. [DOI] [PubMed] [Google Scholar]
- 25a.Gu, L., and G.-M. Li. Unpublished data.
- 26.Habraken Y, Sung P, Prakash L, Prakash S. Enhancement of MSH2–MSH3-mediated mismatch recognition by the yeast MLH1–PMS1 complex. Curr Biol. 1997;7:790–793. doi: 10.1016/s0960-9822(06)00337-x. [DOI] [PubMed] [Google Scholar]
- 27.Hall M, Grover P L. Polycyclic aromatic hydrocarbons: metabolism, activation and tumor initiation. San Diego, Calif: Academic Press; 1990. [Google Scholar]
- 28.Hawn M T, Umar A, Carethers J M, Marra G, Kunkel T A, Boland C R, Koi M. Evidence for a connection between the mismatch repair system and the G2 cell cycle checkpoint. Cancer Res. 1995;55:3721–3725. [PubMed] [Google Scholar]
- 29.Heflich R H, Neft R E. Genetic toxicity of 2-acetylaminofluorene, 2-aminofluorene and some of their metabolites and model metabolites. Mutat Res. 1994;318:73–114. doi: 10.1016/0165-1110(94)90025-6. [DOI] [PubMed] [Google Scholar]
- 30.Holmes J, Clark S, Modrich P. Strand-specific mismatch correction in nuclear extracts of human and Drosophila melanogaster cell lines. Proc Natl Acad Sci USA. 1990;87:5837–5841. doi: 10.1073/pnas.87.15.5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Homfray T F, Cottrell S E, Ilyas M, Rowan A, Talbot I C, Bodmer W F, Tomlinson I P. Defects in mismatch repair occur after APC mutations in the pathogenesis of sporadic colorectal tumours. Hum Mutat. 1998;11:114–120. doi: 10.1002/(SICI)1098-1004(1998)11:2<114::AID-HUMU3>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 32.Iaccarino I, Palombo F, Drummond J, Totty N F, Hsuan J J, Modrich P, Jiricny J. MSH6, a Saccharomyces cerevisiae protein that binds to mismatches as a heterodimer with MSH2. Curr Biol. 1996;6:484–486. doi: 10.1016/s0960-9822(02)00516-x. [DOI] [PubMed] [Google Scholar]
- 33.Johnson R E, Kovvali G K, Prakash L, Prakash S. Requirement of the yeast MSH3 and MSH6 genes for MSH2-dependent genomic stability. J Biol Chem. 1996;271:7285–7288. doi: 10.1074/jbc.271.13.7285. [DOI] [PubMed] [Google Scholar]
- 34.Jost C A, Marin M C, Kaelin W G., Jr p73 is a human p53-related protein that can induce apoptosis. Nature. 1997;389:191–194. doi: 10.1038/38298. [DOI] [PubMed] [Google Scholar]
- 35.Kane M F, Loda M, Gaida G M, Lipman J, Mishra R, Goldman H, Jessup J M, Kolodner R. Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res. 1997;57:808–811. [PubMed] [Google Scholar]
- 36.Karran P, Bignami M. DNA damage tolerance, mismatch repair and genome instability. Bioessays. 1994;16:833–839. doi: 10.1002/bies.950161110. [DOI] [PubMed] [Google Scholar]
- 37.Kat A, Thilly W G, Fang W H, Longley M J, Li G M, Modrich P. An alkylation-tolerant, mutator human cell line is deficient in strand-specific mismatch repair. Proc Natl Acad Sci USA. 1993;90:6424–6428. doi: 10.1073/pnas.90.14.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kinzler K W, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 39.Koi M, Umar A, Chauhan D P, Cherian S P, Carethers J M, Kunkel T A, Boland C R. Human chromosome 3 corrects mismatch repair deficiency and microsatellite instability and reduces N-methyl-N′-nitro-N-nitrosoguanidine tolerance in colon tumor cells with homozygous hMLH1 mutation. Cancer Res. 1994;54:4308–4312. [PubMed] [Google Scholar]
- 40.Kolodner R. Biochemistry and genetics of eukaryotic mismatch repair. Genes Dev. 1996;10:1433–1442. doi: 10.1101/gad.10.12.1433. [DOI] [PubMed] [Google Scholar]
- 41.Leonard C J, Canman C E, Kastan M B. The role of p53 in cell-cycle control and apoptosis: implications for cancer. J. B. Philadelphia, Pa: Lippincott; 1995. [PubMed] [Google Scholar]
- 42.Li G-M, Wang H, Romano L J. Human MutSα specifically binds to DNA containing aminofluorene and acetylaminofluorene adducts. J Biol Chem. 1996;271:24084–24088. [PubMed] [Google Scholar]
- 43.Li G M, Modrich P. Restoration of mismatch repair to nuclear extracts of H6 colorectal tumor cells by a heterodimer of human MutL homologs. Proc Natl Acad Sci USA. 1995;92:1950–1954. doi: 10.1073/pnas.92.6.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Little J B, Nagasawa H, Keng P C, Yu Y, Li C Y. Absence of radiation-induced G1 arrest in two closely related human lymphoblast cell lines that differ in p53 status. J Biol Chem. 1995;270:11033–11036. doi: 10.1074/jbc.270.19.11033. [DOI] [PubMed] [Google Scholar]
- 45.Liu B, Nicolaides N C, Markowitz S, Willson J K V, Parsons R E, Jen J, de la Chapelle A, Hamilton S R, Kinzler K, Vogelstein B. Mismatch repair gene defects in sporadic colorectal cancers with microsatellite instability. Nat Genet. 1995;9:48–55. doi: 10.1038/ng0195-48. [DOI] [PubMed] [Google Scholar]
- 46.Marsischky G T, Filosi N, Kane M F, Kolodner R. Redundancy of Saccharomyces cerevisiae MSH3 and MSH6 in MSH2-dependent mismatch repair. Genes Dev. 1996;10:407–420. doi: 10.1101/gad.10.4.407. [DOI] [PubMed] [Google Scholar]
- 47.McCarthy M J, Rosenblatt J I, Lloyd R S. A modified quantitative polymerase chain reaction assay for measuring gene-specific repair of UV photoproducts in human cells. Mutat Res. 1996;363:57–66. doi: 10.1016/0921-8777(95)00061-5. [DOI] [PubMed] [Google Scholar]
- 48.Meehan T, Straub K, Calvin M. Benzo[alpha]pyrene diol epoxide covalently binds to deoxyguanosine and deoxyadenosine in DNA. Nature. 1977;269:725–727. doi: 10.1038/269725a0. [DOI] [PubMed] [Google Scholar]
- 49.Mellon I, Champe G N. Products of DNA mismatch repair genes mutS and mutL are required for transcription-coupled nucleotide-excision repair of the lactose operon in Escherichia coli. Proc Natl Acad Sci USA. 1996;93:1292–1297. doi: 10.1073/pnas.93.3.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mellon I, Rajpal D K, Koi M, Boland C R, Champe G N. Transcription-coupled repair deficiency and mutations in human mismatch repair genes. Science. 1996;272:557–560. doi: 10.1126/science.272.5261.557. [DOI] [PubMed] [Google Scholar]
- 51.Modrich P. Mechanisms and biological effects of mismatch repair. Annu Rev Genet. 1991;25:229–253. doi: 10.1146/annurev.ge.25.120191.001305. [DOI] [PubMed] [Google Scholar]
- 52.Modrich P. Strand-specific mismatch repair in mammalian cells. J Biol Chem. 1997;272:24727–24730. doi: 10.1074/jbc.272.40.24727. [DOI] [PubMed] [Google Scholar]
- 53.Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu Rev Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 54.Mu D, Tursun M, Duckett D R, Drummond J T, Modrich P, Sancar A. Recognition and repair of compound DNA lesions (base damage and mismatch) by human mismatch repair and excision repair systems. Mol Cell Biol. 1997;17:760–769. doi: 10.1128/mcb.17.2.760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Negoescu A, Lorimier P, Labat-Moleur F, Frouet C, Robert C, Guillermet C, Brambilla C, Brambilla E. In situ apoptotic cell labeling by the TUNEL method: improvement and evaluation on cell preparations. J Histochem Cytochem. 1996;44:959–968. doi: 10.1177/44.9.8773561. [DOI] [PubMed] [Google Scholar]
- 56.Osborne M R, Beland F A, Harvey R G, Brookes P. The reaction of (+/−)-7alpha, 8beta-dihydroxy-9beta, 10beta-epoxy-7,8,9,10-tetrahydrobenzo(a)pyrene with DNA. Int J Cancer. 1976;18:364–368. doi: 10.1002/ijc.2910180315. [DOI] [PubMed] [Google Scholar]
- 57.Palombo F, Iaccarino I, Nakajima E, Ikejima M, Shimada T, Jiricny J. hMutSbeta, a heterodimer of hMSH2 and hMSH3, binds to insertion/deletion loops in DNA. Curr Biol. 1996;6:1181–1184. doi: 10.1016/s0960-9822(02)70685-4. [DOI] [PubMed] [Google Scholar]
- 58.Papadopoulos N, Nicolaides N C, Liu B, Parsons R E, Palombo F, D’Arrigo A, Markowitz S, Willson J K V, Kinzler K, Jiricny J, Vogelstein B. Mutations in GTBP in genetically unstable cells. Science. 1995;268:1914–1917. doi: 10.1126/science.7604266. [DOI] [PubMed] [Google Scholar]
- 59.Parsons R, Li G M, Longley M J, Fang W H, Papadopoulos N, Jen J, de la Chapelle A, Kinzler K W, Vogelstein B, Modrich P. Hypermutability and mismatch repair deficiency in RER+ tumor cells. Cell. 1993;75:1227–1236. doi: 10.1016/0092-8674(93)90331-j. [DOI] [PubMed] [Google Scholar]
- 60.Pirogov N, Shafirovich V, Kolbanovskiy A, Solntsev K, Courtney S A, Amin S, Geacintov N E. Role of hydrophobic effects in the reaction of a polynuclear aromatic diol epoxide with oligodeoxynucleotides in aqueous solutions. Chem Res Toxicol. 1998;11:381–388. doi: 10.1021/tx980006q. [DOI] [PubMed] [Google Scholar]
- 61.Prolla T A, Christie D M, Liskay R M. Dual requirement in yeast DNA mismatch repair for MLH1 and PMS1, two homologs of the bacterial mutL gene. Mol Cell Biol. 1994;14:407–415. doi: 10.1128/mcb.14.1.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sancar A. Mechanisms of DNA excision repair. Science. 1994;266:1954–1956. doi: 10.1126/science.7801120. [DOI] [PubMed] [Google Scholar]
- 63.Straub K M, Meehan T, Burlingame A L, Calvin M. Identification of the major adducts formed by reaction of benzo(a)pyrene diol epoxide with DNA in vitro. Proc Natl Acad Sci USA. 1977;74:5285–5289. doi: 10.1073/pnas.74.12.5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Su S-S, Lahue R S, Au K G, Modrich P. Mispair specificity of methyl-directed DNA mismatch correction in vitro. J Biol Chem. 1988;263:6829–6835. [PubMed] [Google Scholar]
- 65.Swann P F, Waters T R, Moulton D C, Xu Y Z, Zheng Q, Edwards M, Mace R. Role of postreplication DNA mismatch repair in the cytotoxic action of thioguanine. Science. 1996;273:1109–1111. doi: 10.1126/science.273.5278.1109. [DOI] [PubMed] [Google Scholar]
- 66.Toft N J, Winton D J, Kelly J, Howard L A, Dekker M, te Riele H, Arends M J, Wyllie A H, Margison G P, Clarke A R. Msh2 status modulates both apoptosis and mutation frequency in the murine small intestine. Proc Natl Acad Sci USA. 1999;96:3911–3915. doi: 10.1073/pnas.96.7.3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Umar A, Boyer J C, Thomas D C, Nguyen D C, Risinger J I, Boyd J, Ionov Y, Perucho M, Kunkel T A. Defective mismatch repair in extracts of colorectal and endometrial cancer cell lines exhibiting microsatellite instability. J Biol Chem. 1994;269:14367–14370. [PubMed] [Google Scholar]
- 68.Wang H, Lawrence C W, Li G M, Hays J B. Specific binding of human MSH2. MSH6 mismatch-repair protein heterodimers to DNA incorporating thymine- or uracil-containing UV light photoproducts opposite mismatched bases. J Biol Chem. 1999;274:16894–16900. doi: 10.1074/jbc.274.24.16894. [DOI] [PubMed] [Google Scholar]
- 69.Worth L, Jr, Clark S, Radman M, Modrich P. Mismatch repair proteins MutS and MutL inhibit RecA-catalyzed strand transfer between diverged DNAs. Proc Natl Acad Sci USA. 1994;91:3238–3241. doi: 10.1073/pnas.91.8.3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wyllie A H. Cell death: a new classification separating apoptosis from necrosis. New York, N.Y: Chapman and Hall; 1981. [Google Scholar]
- 71.Wyllie A H, Kerr J F, Currie A R. Cell death: the significance of apoptosis. Int Rev Cytol. 1980;68:251–306. doi: 10.1016/s0074-7696(08)62312-8. [DOI] [PubMed] [Google Scholar]
- 72.Xia F, Wang X, Wang Y, Tsang N, Yandell D W, Kelsey K, Liber L. Altered p53 status correlates with differences in sensitivity to radiation-induced mutation and apoptosis in two closely related human lymphoblast lines. Cancer Res. 1995;55:12–15. [PubMed] [Google Scholar]
- 73.Yakes F M, Van Houten B. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc Natl Acad Sci USA. 1997;94:514–519. doi: 10.1073/pnas.94.2.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yamamoto H, Sawai H, Weber T K, Rodriguez B M, Perucho M. Somatic frameshift mutations in DNA mismatch repair and proapoptosis genes in hereditary nonpolyposis colorectal cancer. Cancer Res. 1998;58:997–1003. [PubMed] [Google Scholar]
- 75.Yu Y, Little J B. p53 is involved in but not required for ionizing radiation-induced caspase-3 activation and apoptosis in human lymphoblast cell lines. Cancer Res. 1998;58:4277–4281. [PubMed] [Google Scholar]
- 76.Zou Y, Liu T M, Geacintov N E, Van Houten B. Interaction of UvrABC nuclease system with a DNA duplex containing a single stereoisomer of dG-(+)- or dG-(−)-anti-BPDE. Biochemistry. 1995;34:13582–13593. doi: 10.1021/bi00041a038. [DOI] [PubMed] [Google Scholar]