Abstract
Background
Acne inversa/hidradenitis suppurativa (HS) is a chronic, recurrent inflammatory disease of the skin that can significantly affect patients' quality of life. The etiology and pathogenesis of HS are unclear and gene mutations might play a role.
Summary
The primary focus of the review is on aggregating the gene mutations reported, summarizing the structure of γ-secretase and analyzing and speculating about the mechanism and the underlying relations between gene mutation and functional changes of protein. The systematic literature review was done by searching the PubMed, Embase, and Web of Science databases. γ-Secretase is an intramembrane protease complex responsible for the intramembranous cleavage of more than 30 type-1 transmembrane proteins including amyloid precursor protein and Notch receptors. The protein complex consists of four hydrophobic proteins: presenilin, presenilin enhancer-2 (PSENEN), nicastrin, and anterior pharynx defective 1 (APH1). To date, 57 mutations of γ-secretase genes have been reported in 70 patients or families worldwide, including 39 in NCSTN, 14 in PSENEN, and 4 in PSEN1, of which 17 are frameshifts, 15 result in nonsense mutations, 13 in missense mutations, and 12 are splice site mutations. Given the structure of γ-secretase and analysis of related mutation loci of NCSTN, PSENEN, and PSEN1, mutations in γ-secretase genes could affect activation of presenilin, prevent substrate binding, and hinder intramembrane cleavage of select proteins.
Keywords: Hidradenitis suppurativa/acne inversa, Genetics, γ-Secretase
Introduction
Acne inversa (OMIM 142690) or triad occlusion, hidradenitis suppurativa (HS) is a chronic, recurrent inflammatory disease of the skin, characterized by painful, inflamed lesions predominantly on flexural areas. The disease usually presents after puberty and mostly in the third and fourth decades. The prevalence of HS ranges among 0.05∼8% in different studies, and recently a prevalence of 0.7–1.2% was claimed in the US and European population [1, 2]. Diagnostic criteria include typical lesions (painful nodules, abscesses, sinus tracts, bridged scars, or open comedones), typical sites (axillae, groin, perineal region, perianal region, infra- and intermammary folds, or buttocks), and chronic and recurrent courses [3].
More than one-third of HS patients have family histories, and two large HS case series revealed the familial ratio to be 35% [4, 5]. These HS kindreds may follow autosomal dominant inheritance with incomplete penetrance [6]. Familial HS cases are supposed to present a more severe phenotype; however, a large case series of 302 patients revealed a negative association between family history and severity of HS [5]. Buttock involvements tend to be more common in familial HS [7].
The physiological and psychological consequences of HS can significantly affect patients' quality of life. However, the specific etiology and pathogenesis of HS are still unclear and therapies including antibiotics, antiandrogens, and surgery applied in clinical courses could only make partial improvements. Mutations of γ-secretase have been found in familial HS of 6 Chinese kindreds 10 years ago [8], and thereafter a great many novel gene mutations have been reported in recent years. The prevalence of γ-secretase was uncommon, as estimated to be 6.4% in a large HS cohort [9]. HS patients with γ-secretase gene mutation might suffer from severe and widespread skin lesions [10]. This review, therefore, focused on studies concentrating on the genetics of γ-secretase in HS in the period 2010–2020. The primary focus of the review is on aggregating the gene mutations reported, summarizing the structure of γ-secretase and analyzing and speculating about the mechanism and the underlying relations between gene mutation and functional changes of protein.
Methods
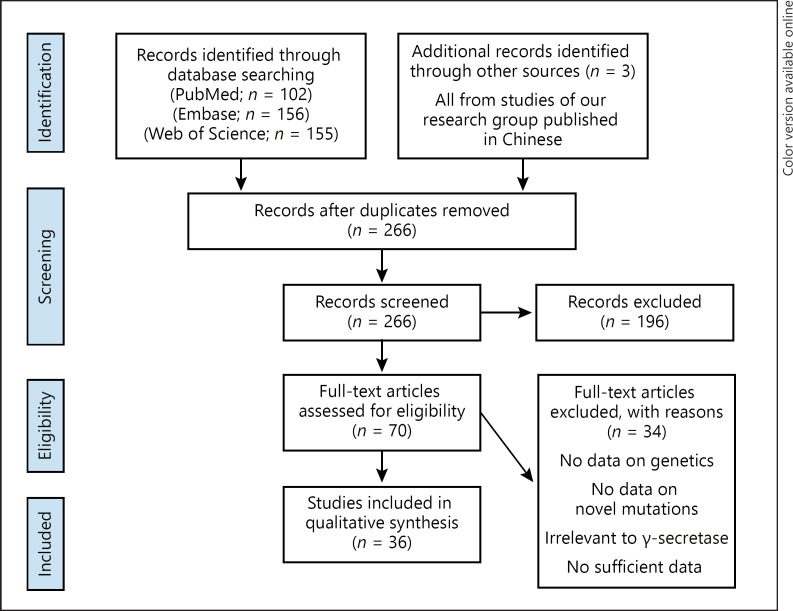
We conducted a systematic literature review according to the Preferred Reporting Items for Systematic Review and Meta-Analysis (PRISMA) [11]. The search was conducted in PubMed, Embase, and Web of Science, last updated September 2020, using the following search terms: “hidradenitis suppurativa” AND “mutation.” Studies were considered for inclusion provided that they reported mutations of γ-secretase in HS patients or family. Additionally, three studies of our research group published in Chinese were included. Papers irrelevant to γ-secretase or reporting insufficient data were excluded. We extracted data on the genetic features from each study independently (Fig. 1 and online suppl. Table at www.karger.com/doi/10.1159/000512455).
Fig. 1.
Flowchart of Methods.
Main Text
Gene Mutations
Heterozygous mutations in γ-secretase genes PSENEN, PSEN1, and NCSTN were first described in 6 Chinese multiplex kindreds, and mutations have since been reported in 64 patients or families worldwide, including 19 Chinese [12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25], 7 Caucasian [9, 26], 5 Japanese [27, 28, 29, 30, 31], 4 Jewish Ashkenazi [32], 3 French [33], 2 British [34], 2 Indian [35, 36], 1 African American [37], 1 German [36],1 Dutch [38], 1 South Welsh [39] kindreds, and 2 kindreds with unknown origin [40, 41], 7 sporadic cases (2 British, 2 Caucasian 1 Afro-Caribbean, 1 Chinese, and 1 unknown origin) [9, 26, 42, 43], and 9 patients (1 German, 1 French, 1 Singaporean, 3 South Welsh, and 3 unknown) [36, 39, 44, 45] with unknown familial history. To date, 57 mutations of γ-secretase genes have been reported including 39 in NCSTN, 14 in PSENEN, and 4 in PSEN1, of which 17 are frameshifts, 15 result in nonsense mutations, 13 in missense mutations, and 12 are splice site mutations (shown in Table 1).
Table 1.
All the γ-secretase mutations in HS that have been reported
| Gene | Mutation | Protein |
|---|---|---|
| NCSTN | c.210_211delAG | p.T70fsX18 |
| c.218delC | p.P73LfsX15 | |
| c.450_459del | p.S151QfsX48 | |
| c.487delC | p.Q163SfsX39 | |
| c.687_688insCC | p.C230PfsX31 | |
| c.978delG | p.M326IfsX30 | |
| c.1325_1326insGTTGTTCTGTAGTGGC | p.D443LfsX6 | |
| c.1752delG | p.E584DfsX44 | |
| c.1912_1915delCAGT | p.S638fsX32 | |
| c.97G>A | p.G33R | |
| c.182G>T | p.G61V | |
| c.223G>A | p.V75I | |
| c.553G>A | p.D185N | |
| c.632C>G | p.P211R | |
| c.647A>C | p.Q216P | |
| c.887A>G | p.E296G | |
| c.944C>T | p.A315V | |
| c.1229C>T | p.A410V | |
| c.1727G>T | p.G576V | |
| c.1768A>G | p.S590AfsX3 | |
| c.349C>T | p.R117X | |
| c.477C>A | p.C159X | |
| c.497C>A | p.S166X | |
| c.617C>A | p.S206X | |
| c.1258C>T | p.Q420X | |
| c.1300C>T | p.R434X | |
| c.1285C>T | p.R429X | |
| c.1381delC | p.L461X | |
| c.1695T>G | p.Y565X | |
| c.1702C>T | p.Q568X | |
| c.1747C>T | p.R583X | |
| c.1876C>T | p.R626X | |
| c.582+1delG | ||
| c.996+7G>A | ||
| c.1101+1G>A | ||
| c.1101+10A>G | ||
| c.1180–5C>G | ||
| c.1352+1G>A | ||
| c.1551+1G>A | p.A486_T517del | |
|
| ||
| PSENEN | c.43_56del | p.C15fs |
| c.66delG | p.F23LfsX46 | |
| c.66_67insG | p.F23VfsX98 | |
| c.229_230insCACC | p.I77HfsX45 | |
| c.279delC | p.F94SfsX51 | |
| c.304T>A | p.X102RextX50 | |
| c.194T>G | p.L65R | |
| c.35T>A | p.L12X | |
| c.115C>T | p.R39X | |
| c.168T>G | p.Y56X | |
| c.62–1G>C | ||
| c.166+2T>C | ||
| c.167–2A>G | ||
| g.1412T>C | ||
|
| ||
| PSEN1 | c.725delC | p.P242LfsX11 |
| c.1167_1168insGA | p.S390EfsX20 | |
| c.953A>G | p.E318G | |
γ-Secretase
γ-Secretase is an intramembrane protease complex with 20 transmembrane domains (TMs), responsible for the intramembranous cleavage of more than 30 type-1 transmembrane proteins including amyloid precursor protein and Notch receptors [10, 46]. The protein complex is assembled from four hydrophobic proteins, presenilin, presenilin enhancer-2 (PSENEN), nicastrin, and anterior pharynx defective 1 (APH1), which are encoded by PSEN1/PSEN2, PSENEN, NCSTN, and APH1A/ APH1B, respectively. Importantly, presenilin can be encoded by PSEN1 or PSEN2 and APH1 can be encoded by APH-1a or APH-1b, but PSEN1 and PSEN2 (or APH-1a and APH-1b) never coexist in any one γ-secretase complex. In addition, APH-1a and APH-1b are alternatively spliced; thus, at least six different combinations might exist in human beings [47, 48].
Presenilin
Among the four protein subunits, presenilin provides the catalytic core to γ-secretase complexes. Presenilin participates in multiple fundamental cellular events, including cell differentiation, intracellular signaling, and membrane trafficking and various biological functions especially the generation of amyloid-β peptides, given that Psen1 knockout mice displayed a Notch phenotype and a tough block in amyloid-β peptide production [47, 49]. Presenilin is best known for its role in early-onset genetic Alzheimer's disease and non-Alzheimer's dementias; notably over 170 mutations have been identified in PSEN1 for Alzheimer's disease [48]. Differently, four mutations of PSEN1 have been reported in HS patients so far, including one mutation considered to be a novel SNP. Presenilin comprises 9 TMs with N-terminus and C-terminus facing the cytosol and extracellular sides, including a highly flexible and mobile TM2. The mutations of presenilin reported in HS located at the ectodomain (ECD) between TM5 and TM6 (loop5), cytoplasmic loop domain (loop6), and TM7, respectively. Within presenilin, the site of autocatalytic cleavage locates in the sequences between TM6 and TM7, and on TM9, the two catalytic residues, Asp257 and Asp385, might play a role in substrate recognition [46]. Thus, the missense mutation c.953A>G locating between TM6 and TM7 might probably affect the catalytic function of presenilin.
PSENEN
PSENEN is a small membrane protein required for the endoproteolysis and activation of proteolytic subunit presenilin [10]. It has three TMs with N-terminus and C-terminus facing the cytosol and extracellular sides, respectively. PSENEN interacts with both nicastrin and presenilin through van der Waals contacts. All three TMs bind to TM4 of presenilin, including the hydrophobic residues (Leu71, Ile75, and Phe78) from TM3. In addition, Phe94, in the extracellular side of presenilin enhancer-2, also interacts with TM4 of presenilin [46]. Of all the 14 mutations (including 4 splice site mutations) reported in PSENEN, 4 frameshift, 3 nonsense mutations, and 1 missense mutation are likely located within the TMs and 2 frameshift mutations in ECD. Most of the frameshift mutations (p.C15fs, p.F23LfsX46, p.F23VfsX98, p.I77HfsX45, and notably p.F94SfsX51), nonsense mutations (p.Leu12X, p.Arg39X, and p.Y56X), and missense mutation (p.L65R) are involved in the interactions with presenilin, which may affect activation of presenilin.
Interestingly, 85.7% (12/14) of PSENEN gene mutations mentioned below were reported to present with a “clinical subphenotype,” Dowling-Degos disease (DDD), a rare autosomal dominant disorder characterized by reticulate hyperpigmentation at the flexural areas [50, 51]. The co-manifestation of HS and DDD was first reported in 1990 [52], with more HS-DDD cases reported; however, the co-occurrence phenomena appear to be more than coincidental. In vivo evidence revealed PSENEN played a role in melanocyte migration and differentiation, given pigmentation abnormalities, especially altered distribution of pigmentation found in zebrafish with PSENEN gene knock-down [36]. Therefore, PSENEN mutations give rise to HS and increase the susceptibility to comorbid DDD simultaneously [51].
APH1
In the protease complex, APH1 is the most stable component, serving as the initial scaffold, and it might play a structural role in the assembly and maturation of γ-secretase [53]. The APH1 proteins contain 7 TMs, with the N-terminus facing the lumen and C-terminus the cytosol [47]. No mutation of APH1 in HS has been reported.
Nicastrin
Nicastrin is the largest and essential component of γ-secretase, and 39 of 57 mutations reported in HS are located in NCSTN [54]. It is a type I transmembrane glycoprotein consisting of a large lobe, a small lobe, and a lone TM, which closely stacks against TM1/TM5/TM7 of Aph-1 [46]. Nicastrin ECD contains at least 11 glycans; the presenilin-dependent nicastrin hyperglycosylation is essential for nicastrin maturation/localization to cell surface and recognizing the N-terminus of substrate protein but is not required for γ-secretase activity [46, 55]. The large ECD of nicastrin forms the bulk of the extracellular portion of γ-secretase, and the ECD undergoes a remarkable conformational change during assembly into γ-secretase complex [46, 47].
Nicastrin TM is involved in γ-secretase assembly, while ECD is crucial for substrate binding. Nicastrin ECD could specifically and directly recognize and bind substrates. Within the DAP domain of nicastrin ECD, Glu333 and Tyr337 are two essential residues required for substrate recruitment, and γ-secretase activity was “abolished” when 28 amino acids (residues 312–340) of DAP domain were deleted by researchers [46, 53].
The structure of nicastrin reported by Bai et al. in 2015 [46] revealed a hydrophilic pocket covered by a Lid. This pocket contributes to substrate recruitment and recognition precedes opening of the Lid for substrates binding. The Lid contains five aromatic residues (Phe145, Tyr148, Tyr152, Phe156, Trp164), and the two important residues Glu333 and Tyr337 and many charged and polar amino acids including four arginine residues (Arg281, Arg285, Arg429, and Arg432) nestle in the pocket. Outside, two glycans on Asn55 and Asn435 “sandwiched” the pocket. Conformation of Lid changes from closed to open during substrate binding, requiring disruptions of interactions between related residues (for instance, van der Waals contacts between Trp164 and Pro424, Phe448, and Gln420) and rotation of the large lobe with Phe287 as the central pivot in a greasy pocket (formed by Phe103, Leu171, Phe176, and Ile180) from the small lobe.
It is noteworthy that all the missense, nonsense, and frameshift mutations and one splice site mutation reported in the γ-secretase gene NCSTN are located within the ECD of nicastrin, with 9 mutations in the DAP domain and 9 affecting the DAP domain. Given the conclusion that nicastrin ECD is involved in substrate recruitment, recognition, and binding, we speculate that mutations of NCSTN in HS change the structure of nicastrin ECD, causing damage of the “pocket” or hindering the opening of the Lid, consequently preventing substrate binding and intramembrane cleavage of select proteins by γ-secretase, thus affecting regulation of intracellular signaling pathways [49]. Among the frameshift and nonsense mutations, 13 mutations (p.T70fsX18, p.R73LfsX15, p.R117X, p.S151QfsX48, p.C159X, p.Q163SfsX39, p.S166X, p.S206X, p.C230PfsX31, p.M326IfsX30, p.Q420X, p.R429X, and R434X) are supposed to abolish or have an impact on the structure of the Lid and/or pocket and obstruct the conformation change of nicastrin.
Conclusion
Mutations in γ-secretase might play a key role in the etiology of HS, especially familial HS. Till now, 57 mutations of γ-secretase genes, including 39 mutations in NCSTN, have been reported in 70 patients or families worldwide. Given the structure of γ-secretase, mutations in γ-secretase genes could affect activation of presenilin, prevent substrate binding, and hinder intramembrane cleavage of select proteins.
Key Message
To date, 57 mutations of γ-secretase genes have been reported in hidradenitis suppurativa.
Conflict of Interest Statement
The authors have no conflicts of interest to declare.
Funding Sources
This work was supported by the CAMS Innovation Fund for Medical Sciences (CIFMS; grant No. 2016-I2M-1-003).
Author Contributions
Zhongshuai Wang was responsible for drafting the manuscript as well as the acquisition and analysis of the literature; Yan Yan and Baoxi Wang revised the manuscript and approved the final version.
Supplementary Material
Supplementary data
References
- 1.Jemec GB, Kimball AB. Hidradenitis suppurativa: epidemiology and scope of the problem. J Am Acad Dermatol. 2015 Nov;73((5 Suppl 1)):S4–7. doi: 10.1016/j.jaad.2015.07.052. [DOI] [PubMed] [Google Scholar]
- 2.Nguyen TV, Damiani G, Orenstein LAV, Hamzavi I, Jemec G. Hidradenitis Suppurativa: An Update on Epidemiology, Phenotypes, Diagnosis, Pathogenesis, Comorbidities and Quality of Life. Journal of the European Academy of Dermatology and Venereology: JEADV. 2020 doi: 10.1111/jdv.16677. [DOI] [PubMed] [Google Scholar]
- 3.Ingram JR, Collier F, Brown D, Burton T, Burton J, Chin MF, et al. British Association of Dermatologists guidelines for the management of hidradenitis suppurativa (acne inversa) 2018. Br J Dermatol. 2019 May;180((5)):1009–17. doi: 10.1111/bjd.17537. [DOI] [PubMed] [Google Scholar]
- 4.Canoui-Poitrine F, Le Thuaut A, Revuz JE, Viallette C, Gabison G, Poli F, et al. Identification of three hidradenitis suppurativa phenotypes: latent class analysis of a cross-sectional study. J Invest Dermatol. 2013 Jun;133((6)):1506–11. doi: 10.1038/jid.2012.472. [DOI] [PubMed] [Google Scholar]
- 5.Canoui-Poitrine F, Revuz JE, Wolkenstein P, Viallette C, Gabison G, Pouget F, et al. Clinical characteristics of a series of 302 French patients with hidradenitis suppurativa, with an analysis of factors associated with disease severity. J Am Acad Dermatol. 2009 Jul;61((1)):51–7. doi: 10.1016/j.jaad.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 6.Ingram JR. The Genetics of Hidradenitis Suppurativa. Dermatol Clin. 2016 Jan;34((1)):23–8. doi: 10.1016/j.det.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 7.Ingram JR, Wood M, John B, Butler R, Anstey AV. Absence of pathogenic γ-secretase mutations in a South Wales cohort of familial and sporadic hidradenitis suppurativa (acne inversa) Br J Dermatol. 2013 Apr;168((4)):874–6. doi: 10.1111/bjd.12048. [DOI] [PubMed] [Google Scholar]
- 8.Wang B, Yang W, Wen W, Sun J, Su B, Liu B, et al. Gamma-secretase gene mutations in familial acne inversa. Science. 2010 Nov;330((6007)):1065–1065. doi: 10.1126/science.1196284. [DOI] [PubMed] [Google Scholar]
- 9.Duchatelet S, Miskinyte S, Delage M, Ungeheuer MN, Lam T, Benhadou F, et al. Low Prevalence of GSC Gene Mutations in a Large Cohort of Predominantly Caucasian Patients with Hidradenitis Suppurativa. J Invest Dermatol. 2020 Oct;140((10)):2085–2088.e14. doi: 10.1016/j.jid.2019.10.025. [DOI] [PubMed] [Google Scholar]
- 10.Pink AE, Simpson MA, Desai N, Trembath RC, Barker JN. γ-Secretase mutations in hidradenitis suppurativa: new insights into disease pathogenesis. J Invest Dermatol. 2013 Mar;133((3)):601–7. doi: 10.1038/jid.2012.372. [DOI] [PubMed] [Google Scholar]
- 11.Moher D, Liberati A, Tetzlaff J, Altman DG, PRISMA Group Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ. 2009 Jul;339(jul21 1):b2535. doi: 10.1136/bmj.b2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Y, Gao M, Lv YM, Yang X, Ren YQ, Jiang T, et al. Confirmation by exome sequencing of the pathogenic role of NCSTN mutations in acne inversa (hidradenitis suppurativa) J Invest Dermatol. 2011 Jul;131((7)):1570–2. doi: 10.1038/jid.2011.62. [DOI] [PubMed] [Google Scholar]
- 13.Li CR, Jiang MJ, Shen DB, Xu HX, Wang HS, Yao X, et al. Two novel mutations of the nicastrin gene in Chinese patients with acne inversa. Br J Dermatol. 2011 Aug;165((2)):415–8. doi: 10.1111/j.1365-2133.2011.10372.x. [DOI] [PubMed] [Google Scholar]
- 14.Zhang C, Wang L, Chen L, Ren W, Mei A, Chen X, et al. Two novel mutations of the NCSTN gene in Chinese familial acne inverse. J Eur Acad Dermatol Venereol. 2013 Dec;27((12)):1571–4. doi: 10.1111/j.1468-3083.2012.04627.x. [DOI] [PubMed] [Google Scholar]
- 15.Ma SS, Yu YX, Yu GQ, Zhang FR. Identification of one novel mutation of the NCSTN gene in one Chinese acne inversa family. Zhonghua Pifuke Yixue Zazhi. 2014;32((2)):126–8. [Google Scholar]
- 16.Jiao T, Dong H, Jin L, Wang S, Wang J. A novel nicastrin mutation in a large Chinese family with hidradenitis suppurativa. Br J Dermatol. 2013 May;168((5)):1141–3. doi: 10.1111/bjd.12135. [DOI] [PubMed] [Google Scholar]
- 17.Zhou C, Wen GD, Soe LM, Xu HJ, Du J, Zhang JZ. Novel Mutations in PSENEN Gene in Two Chinese Acne Inversa Families Manifested as Familial Multiple Comedones and Dowling-Degos Disease. Chin Med J (Engl) 2016 Dec;129((23)):2834–9. doi: 10.4103/0366-6999.194648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang S, Meng J, Jiang M, Zhao J. Characterization of a Novel Mutation in the NCSTN Gene in a Large Chinese Family with Acne Inversa. Acta Derm Venereol. 2016 Mar;96((3)):408–9. doi: 10.2340/00015555-2259. [DOI] [PubMed] [Google Scholar]
- 19.Wu C, Yang J, Zhang S, Li J, Jin H, Zhang X. A novel NCSTN gene mutation in a Chinese family with acne inversa. Mol Genet Genomics. 2018 Dec;293((6)):1469–75. doi: 10.1007/s00438-018-1475-9. [DOI] [PubMed] [Google Scholar]
- 20.Zhang X, Su H, Qin Y, Li C, Xiao X, Xu H, et al. Acne inversa in a family: a clinical survey and genetic mutation analysis. Zhonghua Pifuke Zazhi. 2016;49:508–10. [Google Scholar]
- 21.Xiao X, Xu H, Hui Y, Zhang X. Y. H, Li C, Wang B: mutation analysis of the nicastrin gene in a Chinese pedigree with acne inversa. Int J Dermatol Venereol. 2016;42:201–4. [Google Scholar]
- 22.Liu Y, Xu H, Hui Y, Zhang X, Xiao X, Li C, et al. Mutations in subunits of γ-secretase gene in one family with hidradenitis suppurativa. Chin J Lepr Skin Dis. 2015;31:323–5. [Google Scholar]
- 23.Shi TW, Bai N, Zhang JA, Lu F, Chen XB, Kong XD, et al. Mutations in the γ-secretase genes PSEN1, PSENEN, and NCSTN in a family with acne inversa. Eur J Dermatol. 2018 Jun;28((3)):374–6. doi: 10.1684/ejd.2018.3244. [DOI] [PubMed] [Google Scholar]
- 24.Xu H, Xiao X, Hui Y, Zhang X, He Y, Li C, et al. Phenotype of 53 Chinese individuals with nicastrin gene mutations in association with familial hidradenitis suppurativa (acne inversa) Br J Dermatol. 2016 Apr;174((4)):927–9. doi: 10.1111/bjd.14268. [DOI] [PubMed] [Google Scholar]
- 25.Wu J, Ge H, Fan Y, Zhen Q, Tang L, Sun L. Novel mutation of the NCSTN gene identified in a Chinese acne inversa family. Ann Dermatol. 2020;32((3)):237–42. doi: 10.5021/ad.2020.32.3.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu M, Davis JW, Idler KB, Mostafa NM, Okun MM, Waring JF. Genetic analysis of NCSTN for potential association with hidradenitis suppurativa in familial and nonfamilial patients. Br J Dermatol. 2016 Aug;175((2)):414–6. doi: 10.1111/bjd.14482. [DOI] [PubMed] [Google Scholar]
- 27.Takeichi T, Matsumoto T, Nomura T, Takeda M, Niwa H, Kono M, et al. A novel NCSTN missense mutation in the signal peptide domain causes hidradenitis suppurativa, which has features characteristic of an autoinflammatory keratinization disease. Br J Dermatol. 2019;•••:3. doi: 10.1111/bjd.18445. [DOI] [PubMed] [Google Scholar]
- 28.Nomura Y, Nomura T, Suzuki S, Takeda M, Mizuno O, Ohguchi Y, et al. A novel NCSTN mutation alone may be insufficient for the development of familial hidradenitis suppurativa. J Dermatol Sci. 2014 May;74((2)):180–2. doi: 10.1016/j.jdermsci.2014.01.013. [DOI] [PubMed] [Google Scholar]
- 29.Nomura Y, Nomura T, Sakai K, Sasaki K, Ohguchi Y, Mizuno O, et al. A novel splice site mutation in NCSTN underlies a Japanese family with hidradenitis suppurativa. Br J Dermatol. 2013 Jan;168((1)):206–9. doi: 10.1111/j.1365-2133.2012.11174.x. [DOI] [PubMed] [Google Scholar]
- 30.Kan T, Takahagi S, Shindo H, Tanaka A, Kawai M, Hide M. A unique clinical phenotype of a patient bearing a newly identified deletion mutation in the PSENEN gene along with the pathogenic serum desmoglein-1 antibody. Clin Exp Dermatol. 2018 Apr;43((3)):329–32. doi: 10.1111/ced.13326. [DOI] [PubMed] [Google Scholar]
- 31.Nishimori N, Hayama K, Kimura K, Fujita H, Tadashi T. A novel g-secretase gene mutation in a Japanese family with hidradenitis. J Invest Dermatol. 2017;137((10)):S201. [Google Scholar]
- 32.Pavlovsky M, Sarig O, Eskin-Schwartz M, Malchin N, Bochner R, Mohamad J, et al. A phenotype combining hidradenitis suppurativa with Dowling-Degos disease caused by a founder mutation in PSENEN. Br J Dermatol. 2018 Feb;178((2)):502–8. doi: 10.1111/bjd.16000. [DOI] [PubMed] [Google Scholar]
- 33.Miskinyte S, Nassif A, Merabtene F, Ungeheuer MN, Join-Lambert O, Jais JP, et al. Nicastrin mutations in French families with hidradenitis suppurativa. J Invest Dermatol. 2012 Jun;132((6)):1728–30. doi: 10.1038/jid.2012.23. [DOI] [PubMed] [Google Scholar]
- 34.Pink AE, Simpson MA, Brice GW, Smith CH, Desai N, Mortimer PS, et al. PSENEN and NCSTN mutations in familial hidradenitis suppurativa (Acne Inversa) J Invest Dermatol. 2011 Jul;131((7)):1568–70. doi: 10.1038/jid.2011.42. [DOI] [PubMed] [Google Scholar]
- 35.Ratnamala U, Jhala D, Jain NK, Saiyed NM, Raveendrababu M, Rao MV, et al. Expanding the spectrum of γ-secretase gene mutation-associated phenotypes: two novel mutations segregating with familial hidradenitis suppurativa (acne inversa) and acne conglobata. Exp Dermatol. 2016 Apr;25((4)):314–6. doi: 10.1111/exd.12911. [DOI] [PubMed] [Google Scholar]
- 36.Ralser DJ, Basmanav FB, Tafazzoli A, Wititsuwannakul J, Delker S, Danda S, et al. Mutations in γ-secretase subunit-encoding PSENEN underlie Dowling-Degos disease associated with acne inversa. J Clin Invest. 2017 Apr;127((4)):1485–90. doi: 10.1172/JCI90667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen S, Mattei P, You J, Sobreira NL, Hinds GA. γ-Secretase Mutation in an African American Family With Hidradenitis Suppurativa. JAMA Dermatol. 2015 Jun;151((6)):668–70. doi: 10.1001/jamadermatol.2014.5306. [DOI] [PubMed] [Google Scholar]
- 38.Vossen A, van Straalen KR, Swagemakers SMA, de Klein J, Stubbs AP, Venter DJ, van der Zee HH, van der Spek PJ, Prens EP. A novel nicastrin mutation in a three-generation Dutch family with hidradenitis suppurativa: a search for functional significance. Journal of the European Academy of Dermatology and Venereology: JEADV. 2020 doi: 10.1111/jdv.16310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ingram JR, Wood M, John B, Butler R, Anstey AV. Absence of pathogenic γ-secretase mutations in a South Wales cohort of familial and sporadic hidradenitis suppurativa (acne inversa) Br J Dermatol. 2013 Apr;168((4)):874–6. doi: 10.1111/bjd.12048. [DOI] [PubMed] [Google Scholar]
- 40.LeGall C, Puca L, Madrange M, Bal E, Choukair Z, Hamel Y, et al. Impairment of notch 1 signaling is a common defect in lesions from patients with hidradenitis suppurativa. J Invest Dermatol. 2017;137((10)):S226. [Google Scholar]
- 41.Garcovich S, Tricarico PM, Nait-Meddour C, Giovanardi G, Peris K, Crovella S, et al. Novel nicastrin mutation in hidradenitis suppurativa-Dowling-Degos disease clinical phenotype: more than just clinical overlap? Br J Dermatol. 2020 Oct;183((4)):758–9. doi: 10.1111/bjd.19121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pink AE, Simpson MA, Desai N, Dafou D, Hills A, Mortimer P, et al. Mutations in the γ-secretase genes NCSTN, PSENEN, and PSEN1 underlie rare forms of hidradenitis suppurativa (acne inversa) J Invest Dermatol. 2012 Oct;132((10)):2459–61. doi: 10.1038/jid.2012.162. [DOI] [PubMed] [Google Scholar]
- 43.Liu Y, Miao T, Ma J, Shao L, Luo S, Li Y, et al. PSENEN c.66delG in sporadic acne inversa. Eur J Dermatol. 2016 Jun;26((3)):298–9. doi: 10.1684/ejd.2016.2742. [DOI] [PubMed] [Google Scholar]
- 44.Haines RL, Common JE, Teo D, Tang MB, Lane EB. Sequencing of the γ-secretase complex in Singaporean patients with acne inversa reveals a novel mutation in nicastrin, but suggests other mechanisms must be present. Br J Dermatol. 2012;166:e33. [Google Scholar]
- 45.Gonzalez-Villanueva I, Montoyo IP, Alvarez-Chinchilla P, Gutierrez M, Ramirez JCP. Hidradenitis suppurativa/Dowling-Degos disease (DDD) phenotype associated with mutations in NCSTN gene. Experimental dermatology. 2019;28((063)):32–32. [Google Scholar]
- 46.Bai XC, Yan C, Yang G, Lu P, Ma D, Sun L, et al. An atomic structure of human γ-secretase. Nature. 2015 Sep;525((7568)):212–7. doi: 10.1038/nature14892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tolia A, De Strooper B. Structure and function of gamma-secretase. Semin Cell Dev Biol. 2009 Apr;20((2)):211–8. doi: 10.1016/j.semcdb.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 48.Bergmans BA, De Strooper B. gamma-secretases: from cell biology to therapeutic strategies. Lancet Neurol. 2010 Feb;9((2)):215–26. doi: 10.1016/S1474-4422(09)70332-1. [DOI] [PubMed] [Google Scholar]
- 49.Oikawa N, Walter J. Presenilins and γ-Secretase in Membrane Proteostasis. Cells. 2019 Mar;8((3)):8. doi: 10.3390/cells8030209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li W, Xu H, He Y, Lin L, Li C. Comorbidities or different entities? Phenotype variability associated with PSENEN mutations. Br J Dermatol. 2019 Jan;180((1)):221–2. doi: 10.1111/bjd.17111. [DOI] [PubMed] [Google Scholar]
- 51.Li C, Li W, Xu H, Zhang X, Su B, Zhang W, et al. PSENEN Mutation Carriers with Co-manifestation of Acne Inversa (AI) and Dowling-Degos Disease (DDD): is AI or DDD the Subphenotype? J Invest Dermatol. 2017 Oct;137((10)):2234–6. doi: 10.1016/j.jid.2017.05.021. [DOI] [PubMed] [Google Scholar]
- 52.Weber LA, Kantor GR, Bergfeld WF. Reticulate pigmented anomaly of the flexures (Dowling-Degos disease): a case report associated with hidradenitis suppurativa and squamous cell carcinoma. Cutis. 1990 Jun;45((6)):446–50. [PubMed] [Google Scholar]
- 53.Shah S, Lee SF, Tabuchi K, Hao YH, Yu C, LaPlant Q, et al. Nicastrin functions as a gamma-secretase-substrate receptor. Cell. 2005 Aug;122((3)):435–47. doi: 10.1016/j.cell.2005.05.022. [DOI] [PubMed] [Google Scholar]
- 54.Li Y, Liew LS, Li Q, Kang C. Structure of the transmembrane domain of human nicastrin-a component of γ-secretase. Sci Rep. 2016 Jan;6((1)):19522. doi: 10.1038/srep19522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Herreman A, Van Gassen G, Bentahir M, Nyabi O, Craessaerts K, Mueller U, et al. gamma-Secretase activity requires the presenilin-dependent trafficking of nicastrin through the Golgi apparatus but not its complex glycosylation. J Cell Sci. 2003 Mar;116((Pt 6)):1127–36. doi: 10.1242/jcs.00292. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data