Abstract
Clinical studies of circulating tumor cells (CTC) have stringent demands for high capture purity and high capture efficiency. Nanostructured surfaces have been shown to significantly increase the capture efficiency yet suffer from low capture purity. Here we introduce a dual-functional lipid coating on nanostructured surfaces. The lipid coating serves both as an effective passivation layer that helps prevent nonspecific cell adhesion and as a functionalized layer for antibody-based specific cell capture. In addition, the fluidity of lipid bilayers enables antibody clustering that enhances the cell–surface interaction for efficient cell capture. As a result, the lipid-coating method helps promote both the capture efficiency and capture purity of nanostructure-based CTC capture.
Graphical Abstract
INTRODUCTION
Circulating tumor cells (CTCs) hold great potential for a better understanding of cancer metastasis and easier monitoring of cancer progression and treatment response.1–3 However, a big challenge for CTC-based clinical studies is to capture CTCs in both high purity and high efficiency due to the extremely low concentration of CTCs in the bloodstream (~1–100 CTCs per billions of blood cells).4,5 To meet this challenge, two broadly defined strategies have emerged in recent years to improve CTC capture: one is enriching CTCs according to their physical properties that can distinguish them from blood cells (e.g., size, deformability, and electric charges);6–9 the other is affinity-based CTC selection using protein markers expressed by CTCs but not by blood cells (e.g., epithelial cell adhesion molecule, EpCAM).4,10,11 Methods based on the physical properties can offer fast processing speed/high throughput but suffer from low purity and concerns of missing CTC populations as CTCs are highly heterogeneous in size, shape, and possibly other physical properties.5 In comparison, an affinity-based strategy is more specific for capturing CTCs. However, its capture efficiency and capture purity strongly depend on surface properties for effective antibody immobilization and cell interaction.
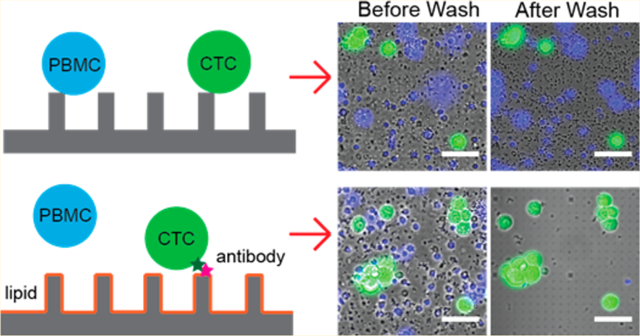
Surfaces with nanostructures have recently shown great promise in improving affinity-based CTC capture in two aspects: providing a larger surface area for antibody immobilization and presenting nanotopography that promotes cell–surface interaction.12–14 For example, for CTC numeration, a silicon nanowire-based NanoVelcro platform has been shown to outperform the sensitivity of the FDA approved CellSearch assay.15 It has also been combined with aptamer- or polymer-based surface functionalization to recover CTCs for downstream analysis.16–21 Similarly, surfaces with vertical quartz nanowire arrays had been shown to achieve superior capture efficiency compared to flat surfaces for CTCs as well as on T lymphocytes.22–24 Besides nanowires, graphene oxide nanosheets,25 polymer nanofibers, and nanodots were also reported to enhance CTC capture by providing high surface area and nanotopography.26–28 However, despite their improved capture efficiency, surfaces with nanostructures encounter a critical challenge of low capture purity. Recent studies show that nanoroughened surfaces without antibody modification promoted the nonspecific adhesion of cells.29,30 The capture efficiency by nanoroughened surfaces was similar around ~93% either with or without the presence of background peripheral blood mononuclear cells (PBMC), but the capture purity dropped from 84 to 14% when background PBMC was present.29 Therefore, a surface-modification method to improve the capture purity is vital for nanostructure-based CTC capture.
Surface modification methods critically control both antibody functionalization and surface passivation for cell interaction. For antibody immobilization on surfaces with nanostructures, most approaches were based on covalent chemistry (e.g., NHS/maleimide reaction or aldehyde cross-linking).26,27,31 However, they were known to have limited yield, modest uniformity, and poor repeatability.32 For surface passivation, nonspecific bound cells were usually removed by mechanical washing with limited removal efficiency.31 A few works used PEG for surface passivation, which required extra chemical modification and still inherited the limitation of covalent chemistry as discussed above.25
In this work, we introduce supported lipid bilayers (SLBs) as a surface coating in quartz nanopillar-based CTC capture, which serves dual functions: (1) enabling easy and uniform surface functionalization with antibodies and (2) providing effective surface passivation to prevent the nonspecific binding of background cells.33,34 The lipid bilayer can be easily functionalized by doping lipids with functional groups, such as biotinylated lipid molecules for streptavidin–biotin-based antibody functionalization. Besides, the fluidic nature of the lipid bilayer enables the antibodies to redistribute upon antigen binding to enhance the capture strength spontenously.35–37 Furthermore, lipid bilayers are a natural passivation layer. Supported lipid bilayers have been shown to effectively prevent cells from binding to underlying surfaces.38 They can be easily formed in vitro by applying small unilamellar vesicles (SUVs) to the hydrophilic surface under physiological conditions.36,39 For nanostructure-based CTC capture, we demonstrate that lipid bilayers can simultaneously increase the capture purity and the capture efficiency by providing sufficient surface passivation and enhanced antibody–cell interaction through antibody clustering.
EXPERIMENTAL SECTION
Fabrication of Quartz Nanopillar Arrays.
Quartz nanopillar arrays were fabricated on square quartz coverslips (Technical Glass Products, Inc.). The coverslips were first spin-coated with 300 nm of ZEP-520 (ZEON Chemicals), followed by E-Spacer 300Z (Showa Denko). The desired patterns were exposed by electron-beam lithography (Raith150) and developed in xylene. The mask was then formed by the sputter deposition of 100 nm Cr, followed by lift-off in acetone. The nanostructures was generated by reactive ion etching with CHF3 and O2 chemistry (AMT 8100 etcher, Applied Materials). The diameters and pitches of nanopillar arrays are shown in Table S1.
Preparation of Lipid Vesicles.
The composition of lipid vesicles is egg-PC (Avanti) mixed with 0.5 mol % of Texas Red-1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine (Texas Red-DHPE) (Invitrogen) and 1 mol % of 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(biotinyl) (Biotin-DOPE) (Avanti). Lipids were dissolved in chloroform and mixed in a fixed molar ratio as described above. The lipid solution was then blown dried in clean glass tubes with pure nitrogen followed by vacuum drying overnight to remove the remaining chloroform. The dried lipid was stored in −20 C° to prevent oxidization. When preparing lipid vesicles, 1 mg of the lipid mixture was suspended in 400 μL of PBS buffer and extruded through a 100 nm Nucelpore membrane filter with the use of an Avanti polar lipid extruder and Hamilton syringes as the extruder device. The lipid vesicle solution is stored in 4 °C for up to 1 month.
Forming Antibody -Functionalized Supported Lipid Bilayers on Glass and Quartz Nanopillar Substrates.
The glass and quartz nanopillar chips were cleaned with oxygen plasma in a plasma cleaner (Harrick Plasma) for 1 h to remove any remaining impurities on the surfaces. The cleaned chips were assembled with PDMS substrates. The lipid vesicle solution was diluted 5-fold in PBS buffer, loaded into channels, incubated for 15 min to form supported lipid bilayer, and washed with 0.5 mL of PBS buffer three times to remove free vesicles. Streptavidin (100 nM, Invitrogen) solution was then added and incubated for 30 min, followed by another half hour of incubation of 20 nM biotinylated antibodies to make an antibody-functionalized supported lipid bilayer. PBS buffer was used to remove unbound molecules after every step. Two biotinylated antibodies were used in this research article: anti-EpCAM (R&D Systems) for capturing EpCAM-expressed cells and anti-EGFR (Gift from S. S. Jeffery laboratory, Stanford) for capturing EGFR-expressed cells.
Fluorescence Recovery after Photobleaching (FRAP) Experiment.
The FRAP experiments were performed using a Leica DMI6000 B microscope. Photobleaching of a 21-μm-diameter spot was performed using an LED light source (Lumencor SOLAR-SE-I, 42 mW/cm2). Raw images were taken every 12 s (5 images per minute) and exported as tagged image file format (TIFF) files using MAG Biosystem Capture-Scope software. For intensity analysis, images were analyzed using ImageJ software. The intensity of the bleached area was calibrated using double normalization involving an acquisition–bleaching correction:40
For every image, the intensity of the bleached area was subtracted from the background intensity. To remove the effect of acquisition bleaching, the intensity of the bleached area was corrected by the intensity of the unbleached area. The diffusion coefficient was calculated according to the established equation:41
w is the radius of the bleached area, and τ1/2 is the half-life time of fluorescence recovery. τ1/2 was calculated by fitting the calibrated intensity to a single-exponential recovery:36
Ieq and I0 are the fluorescence intensity at equilibrium time and initial time, respectively. To take the sidewall area of nanopillars into consideration, the calculated diffusion coefficients are normalized to a flat surface and divided by the surface area ratio (Table S2).
Cell Culture.
HeLa, MCF7, and MDA-MB-231 cell lines were used to evaluate the capture and separation efficiencies of the supported lipid bilayers. Cells were cultured under ordinary cell culture conditions (37 C°, 5% CO2) in a polystyrene tissue culture plate (Falcon). Dulbecco’s Modified Eagle’s Medium (DMEM) (Hyclone, Thermo Scientific) supplemented with 10% fetal bovine serum (FBS) (Gibco, Life Technologies), 100 units/mL penicillin, and 100 μg/mL streptomycin (Gibco, Life Technologies) was used as the culture medium. Before the experiment, cells were prestained with 1 μM Calcein-AM (Life Technologies) or 1 μg/mL Hoechst 33342 (Life Technologies) for 10 min at 37 C° and washed with PBS buffer twice. Then cells were treated with 0.25% trypsin (Gibco, Life Technologies) for 3 min at 37 C° to detach from the tissue culture plate. The detached cells were resuspended in PBS buffer, and the densities of the cells were measured with a hemocytometer.
Isolated PBMCs Sample Preparation and Experiment.
The human whole blood was provided by the Stanford Blood Center. Blood was drawn from healthy donors and collected in 6 mL blood collection tubes containing K2-EDTA as an anticoagulant (BD Biosciences). Peripheral blood mononuclear cells (PBMCs) were isolated by density gradient separation with Ficoll-Paque (GE Healthcare Bio-Sciences). The isolated PBMCs were stained with 1 μg/mL Hoechst 33342 for 15 min at RT and then washed twice with PBS buffer. Every 1 mL of human whole blood results in 100 μL of isolated PBMCs after the isolation process. Prepared MCF7 cells were then used to spike the PBMCs with an MCF7/PBMC ratio of between 1:10 and 1:100. MCF7-spiked PBMC samples were then infused into the prepared quartz nanopillar chips with anti-EpCAM-functionalized lipid bilayers and incubated for 15 min in RT. The unbound cells were removed with a PBS buffer. Images of nanopillar arrays were taken before and after the washing process to calculate the capture efficiency of MCF7 cells and the capture purity.
RESULTS AND DISCUSSION
The lipid-based functionalization of nanopillars consists of three steps (illustrated in Figure 1a): (1) the formation of a supported lipid bilayer on freshly cleaned quartz by the spontaneous rupture of small unilamellar vesicles that are composed of phosphatidylcholine doped with 1 mol % biotinylated phosphoethanolamine (biotin-DOPE, Avanti); (2) the binding of streptavidin (SA) on biotinylated lipids; and (3) the binding of biotinylated antibody on an SA-bound supported lipid bilayer. We performed this procedure on a quartz nanopillar with a 500 nm diameter (SEM, see Figure 1b). The whole incubation process takes <75 min, which is much faster than covalent methods (~4 h).27 Visualizing the supported lipid bilayer by adding 0.5 mol % of fluorescence-labeled lipids (Texas Red-DHPE, Invitrogen) to the lipid mixture (Figure 1c), we clearly see that the supported lipid bilayer was uniformly formed on the flat areas between nanopillars with no visible bright puncta or dark defects. In comparison, the fluorescence at nanopillar locations is much brighter and outlines a ring shape due to the projected fluorescence signal from the nanopillar sidewall as expected.42 Quantification of the fluorescence of several nanopillars shows consistent intensities among all of them (Figure 1d), indicating a conformal lipid bilayer coating on all of the nanopillar surfaces. Furthermore, we used Alexa488-labeled streptavidin (SA-488) to visualize the uniformity of the proteins attached to the supported lipid bilayer. As expected, SA-488 gives similar uniformity as the lipid marker (Texas Red-DHPE) (Figure 1e), suggesting uniform protein binding to the lipid bilayer. Compared to traditional approaches that rely on hours of covalent chemical reaction, the lipid bilayer provides facile surface functionalization in about an hour.
Figure 1.
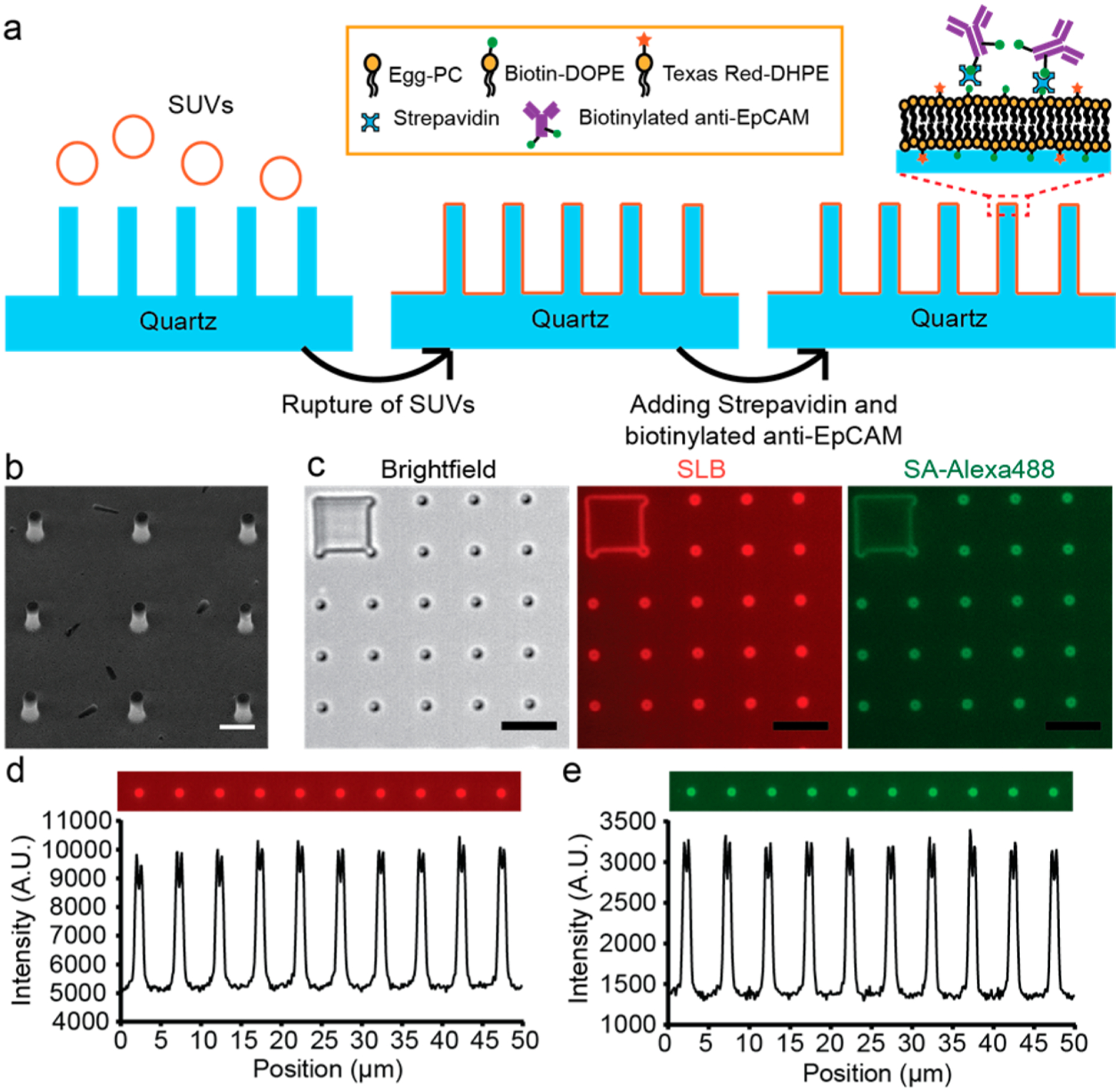
Lipid-based functionalization of antibodies on quartz nanopillar arrays. (a) Schematic illustration of the lipid-based antibody functionalization procedure on a quartz nanopillar array. (b) Scanning electron microscopy (SEM) image of a quartz nanopillar array with 500 nm diameter, 3 μm spacing, and 1.3 μm height, tilting 30°. (Scale bar 1 μm.) (c) Bright field and fluorescence images of the supported lipid bilayer (SLB) and SA-Alexa488 on a quartz nanopillar array. The bright spots in the fluorescence images indicate that lipid bilayers also formed on the side wall of quartz nanopillars. (Scale bars 5 μm.) (d, e) Fluorescence intensity profile of SLB (d) and SA-Alexa488 (e) on quartz nanopillar arrays. The intensity shows that both SLB and SA-Alexa488 are distributed uniformly on each nanopillar.
To examine the effect of supported lipid bilayers on surface passivation and functionalization, we performed cell capture using cancer cell lines (MCF7, Hela, and MDA-MB-231). As shown in Figure 2a, surface passivation with a lipid bilayer completely prevented the nonspecific binding of MCF7, an EpCAM-positive breast cancer cell line, to the surface (0.7 ± 0.6% binding on a lipid-bilayer-coated quartz glass vs 91.1 ± 3.5% binding on bare quartz glass). Next, we functionalized the lipid bilayer with anti-EpCAM antibody and measured the capture efficiencies for MCF7 cells vs HeLa cells. MCF7 cells express high levels of EpCAM on their surface, and HeLa cells have very low EpCAM expression and are often used as an EpCAM-negative cell model.12,26,27 After being washed, the anti-EpCAM-functionalized lipid bilayer captures MCF7 cells with 86.8 ± 8.9% efficiency but captured almost no (1.1 ± 0.5%) HeLa cells (Figure 2b). This proves that the anti-EpCAM-functionalized lipid bilayer successfully prevents nonspecific cell adhesion (HeLa) while retaining the specific capture of cells (MCF7) with high efficiency.
Figure 2.
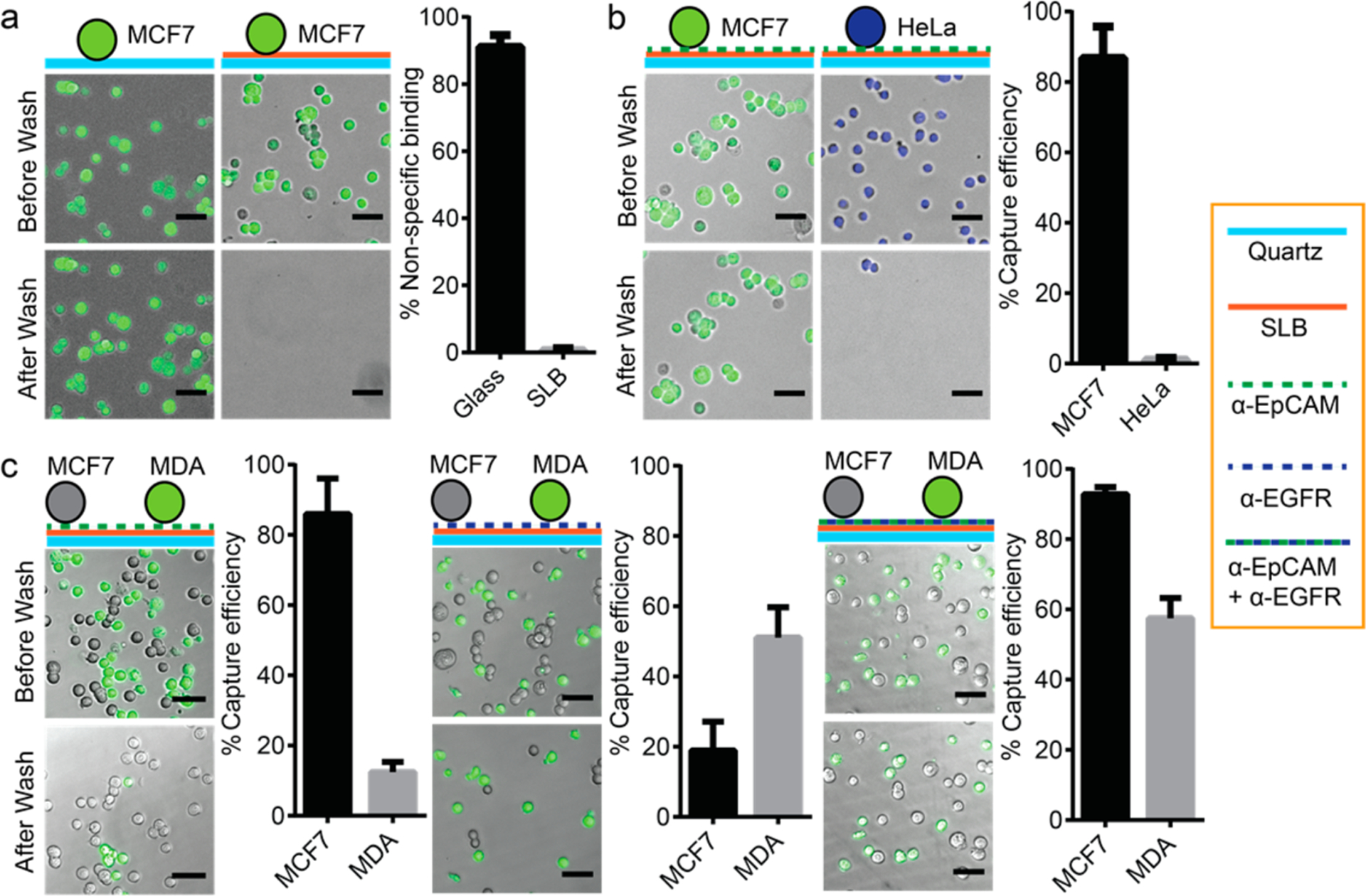
Nonspecific cell adhesion and antigen-specific cell capture on an antibody-functionzlied supported lipid bilayer. (a) A supported lipid bilayer significantly reduces the nonspecific cell adhesion of MC7 cells stained with Celcein-AM (green). 91.1 ± 3.5% of MCF7 cells were captured on a bare glass surface. However, only 0.7 ± 0.6% of MCF7 cells were captured by supported lipid bilayers. (b) Anti-EpCAM-functionalized supported lipid bilayers specifically captured EpCAM-expressed cells. 86.8 ± 8.9% of MCF7 cells (EpCAM-positive) stained with Celcein-AM (green) were captured by anti-EpCAM-functionalized supported lipid bilayers. However, only 1.1 ± 0.5% of HeLa cells (EpCAM-negative) stained with Hoechst33342 (blue) were captured. (c) Heterogeneous cell capture was demonstrated using a multiantibody-functionalized supported lipid bilayer. An anti-EpCAM-functionalized supported lipid bilayer captured 86.0 ± 10.1% of MCF7 cells (high-level EpCAM expression) but only 12.4 ± 3.0% of MDA-MB-231 cells (low-level EpCAM expression) stained with Celcein-AM (green). Meanwhile, an anti-EGFR-functionalized supported lipid bilayer captured 51.1 ± 8.7% of MDA-MB-231 cells (high-level EGFR expression) but only 18.9 ± 8.2% of MCF7 cells (low-level EGFR expression). With a supported lipid bilayer functionalized with both anti-EpCAM and anti-EGFR, both MCF7 and MDA-MB-231 cells could be captured simultaneously with capture efficiencies of 92.8 ± 2.0 and 57.5 ± 5.8%, respectively (All scale bars are 50 μm.) Every experiment was repeated five times (n = 5), and error bars represent the standard deviation (SD).
The ease of lipid bilayers in antibody immobilization can further facilitate the capture of cells with distinct properties in light of addressing the challenge of heterogeneous CTC populations.43 As a demonstration, we tested two breast cancer cell lines with different surface marker profiles: MCF7, a cell line expressing high levels of EpCAM, and MDA-MB-231, a cell line expressing lower levels of EpCAM but higher levels of EGFR (epidermal growth factor receptor) compared to MCF7.44 The two cell line cells were premixed, and MDA-MB-231 cells were stained with calcein-AM dye to distinguish them from MCF7 cells. As shown in Figure 2c, lipid bilayers functionalized with anti-EpCAM captured MCF7 cells with 86.0 ± 10.1% efficiency but only 12.4 ± 3.0% efficiency for MDA-MB-231 cells. In comparison, lipid bilayers functionalized with anti-EGFR captured MDA-MB-231 cells with 51.1 ± 8.7% efficiency but only 18.9 ± 8.2% efficiency for MCF7 cells. By using lipid bilayers functionalized with two antibodies (i.e., anti-EpCAM and anti-EGFR), both MCF7 and MDA-MB-231 cells were captured with 92.8 ± 2.0 and 57.5 ± 5.8% capture efficiency, respectively. The capture efficiency of MCF7 on a dual-antibody-functionalized lipid bilayer was comparable to those functionalized only by single anti-EpCAM. A similar situation was also found for MDA-MB-231 captured on dual-antibody-functionalized lipid bilayers compared to single anti-EGFR-based capture. Thus, the presence of different antibodies on the same lipid bilayer did not interfere with the other’s capture ability (Figure 2c). This observation demonstrates that supported lipid bilayer-based multiantibody functionalization supports heterogeneous CTC capture.
Besides presenting antibody uniformly for capture, the lipid bilayer also enhances cell capture by its natural fluidity. Fluorescence recovery after photobleaching (FRAP) experiments (Figure S1a, Movie S1 and S2) showed that lipid molecules moved freely along the lipid bilayer, ensuring an even distribution of lipids by diffusion. FRAP experiments were tested on nanopillar arrays with different diameters and pitches (Table S1). The measured diffusion coefficients of lipid bilayers are independent of quartz nanopillar dimensions after being normalized to the surface area (Figure S1c, Table S2), indicating that the lipid fluidity on quartz nanopillars is similar to that on a flat surface, which is consistent with previous reports.42 FRAP experiments on SA-488 also showed that proteins functionalized on lipid diffuse freely as well, although with a lower diffusion coefficient than for the small lipid molecules (Figure S1b,c).
The interaction between the fluid antibody layer and cells was then examined by capturing MCF7 on quartz nanopillar arrays functionalized with SA-488 and anti-EpCAM. As shown in the first row of Figure 3a, puncta of SA-488 appeared after 20 min of incubation, suggesting that SA-488 formed clusters beneath the captured MCF7 cell. The clustering of SA-488 is also seen on a flat lipid-coated anti-EpCAM-functionalized surface (Figure S2). In comparison, no puncta were observed on control samples where MCF7 cells were incubated on the lipid–bilayer surface without anti-EpCAM (Figure 3a, bottom row) and neither were the puncta formed on the glass surface without lipid bilayers (Figure 3a, middle row), which indicates that the fluidity of lipid bilayers is necessary for puncta formation (Figure 3a, middle and bottom rows; Figure 3b, green and red lines). The continuous growth of the antibody cluster was clearly observed during cell capture (Movie S3), with the fluorescence intensity profile shown in Figure 3b. The formation of SA-488 puncta is due to the clustering of the anti-EpCAM bound to EpCAM antigens on the cell membrane, which is similar to the clustering effect of membrane-bound receptors as previously reported.45–48 Compared to covalent-chemistry-based methods, the fluidity of antibodies on supported lipid bilayers not only ensures the even distribution of antibodies but also enables the clustering of antibodies under captured cells, which further strengthens the cell–surface interaction for efficient cell capture.
Figure 3.
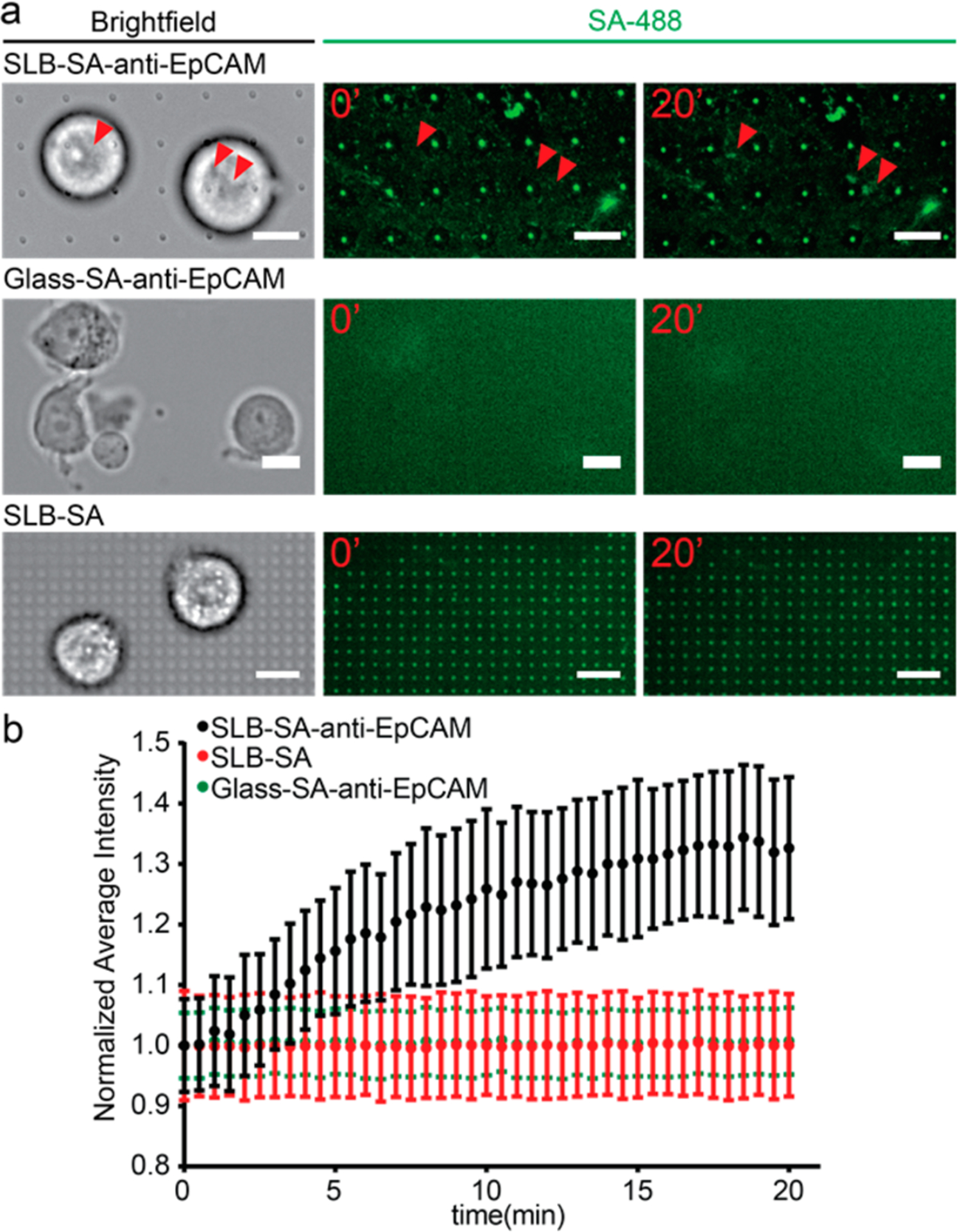
Clustering of anti-EpCAM around captured MCF7 cells enabled by the antibody–antigen interaction and the fluidity of supported lipid bilayers. (a) MCF7 cells on different surfaces: supported lipid bilayer-coated quartz nanopillar arrays functionalized with SA-488 and anti-EpCAM (upper), bare glass coated with SA-488 and anti-EpCAM (middle), and supported lipid bilayer-coated quartz nanopillar arrays functionalized with SA-488 only (bottom). Only MCF7 on lipid bilayers with SA-488 and anti-EpCAM shows the clustering of SA-488 (red arrows). (Scale bars 20 μm.) (b) The average intensity change of SA-488 beneath the MCF7 cells on different surfaces. The average intensity of the cluster area increased about 30% in 20 min on SLB with SA-488/anti-EpCAM (black). However, no clustering of SA-488 was observed under MCF7 cells on surface-absorbed SA-488/anti-EpCAM (green) or the supported lipid bilayer with SA-488 only (red).
The capture purity and efficiency based on lipid passivation and antibody clustering were then further evaluated using a human blood sample spiked with fluorescently labeled MCF7 cells. For the capture of spiked MCF7 cells in whole blood (Figure S3a), the efficiency was 72.8 ± 3.8% when diluting the blood sample 100-fold. Without blood dilution, however, spiked MCF7 cells were barely captured, which is mainly due to the overwhelming numbers of blood cells blocking the interaction between MCF7 and anti-EpCAM, and thus no clustering could happen (Figure S3b).
To avoid surface blocking caused by dense blood cells and to improve the capture throughput, we performed blood fractionation. Among different components in the whole blood sample, red blood cells (RBC) are the major cause of blocking due to its abundancy of 109 cell/mL, but it is easy to remove thanks to their significant difference in density and size compared to CTCs.7,49 On the contrary, peripheral blood mononuclear cells (PBMCs) often challenge the CTC capture purity as impurities since they have similar physical properties to CTCs and are prone to surface attachment.5,31 We performed PBMC isolation not only to avoid the blocking effect but also to significantly reduce the sample size. The simple size after PBMC isolation decreased to 1/10 of its original volume (100 μL of PBMC solution per 1 mL of whole blood). The capture efficiency and purity of MCF7 used to spike isolated PBMC samples were tested using quartz nanopillar arrays (a snapshot of the pillar chip is shown in Figure 4a) with and without lipid functionalization (Figure 4b). Both surfaces gave high capture efficiencies (86.6 ± 10.6 and 92.5 ± 0.8%, respectively) (Figure 4c). Therefore, PBMCs did not exhibit a noticeable blocking effect on MCF7 capture (Figure S4). However, the lipid bilayer coating drastically increased the capture purity from 3.2 ± 0.4 to 71.3 ± 17.2% (Figure 4c). Lipid bilayer-coated nanopillar arrays with different diameters and pitch size produced similar results with respect to both the capture efficiency and capture purity of the lipid-coated flat surface (Figure S5). These results prove that supported lipid bilayer-based antibody functionalization enables the capture of specific cancer cells on nanopillars while significantly preventing the nonspecific binding of PBMCs from blood samples.
Figure 4.
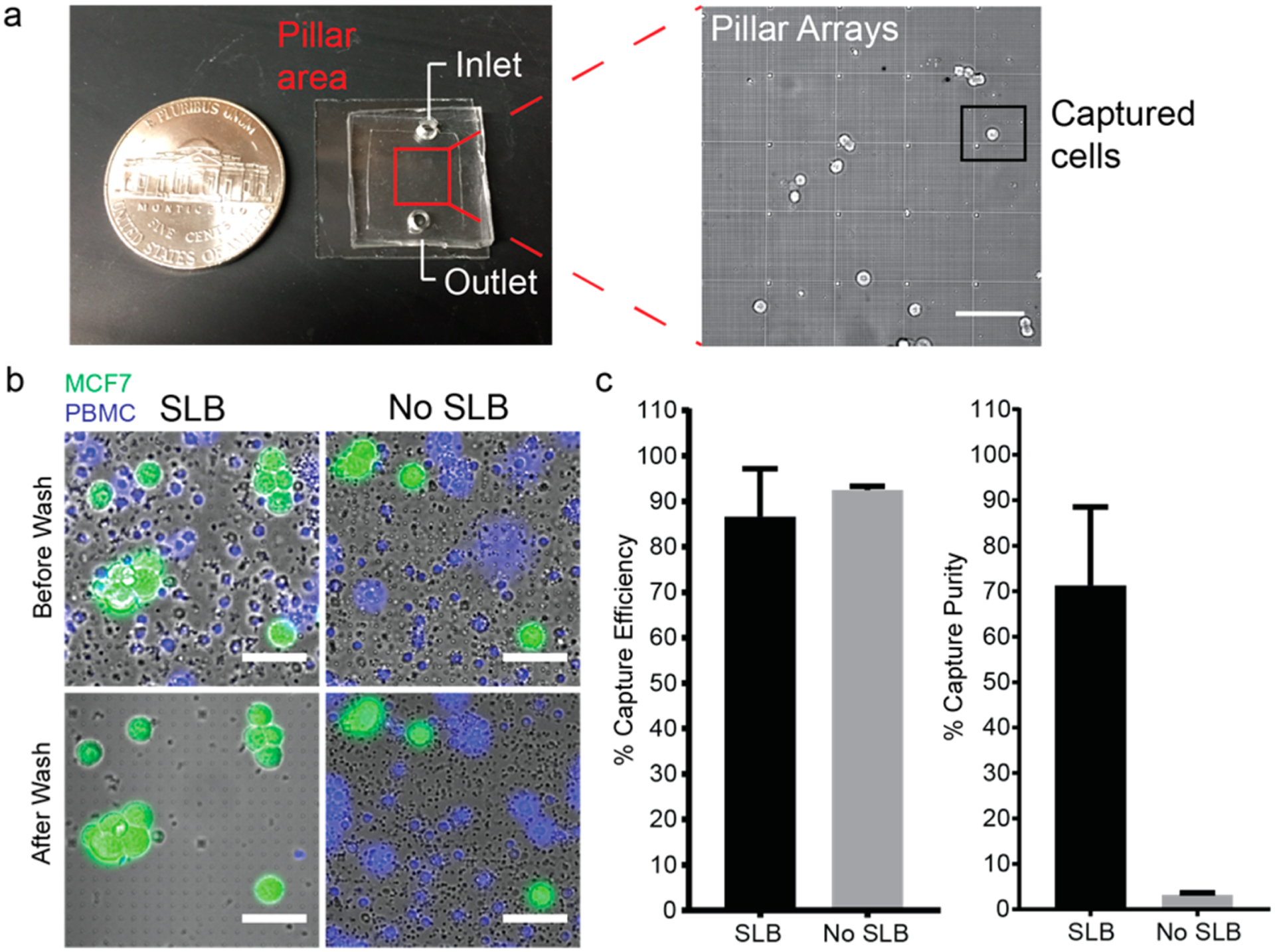
Antibody-functionalized supported lipid bilayer selectively captured spiked MCF7 cells in PBMCs on quartz nanopillar arrays. (a, Left) Snapshot of the nanopillar chip. The nanopillar chip is covered with PDMS to create a chamber for both surface functionalization and cell capture. The pillar arrays are located in the center of the chip (marked by a red square). (Right) Zone-in picture of nanopillar arrays with captured MCF7 cells. (Scale bar 100 μm.) (b) Anti-EpCAM-lipid-functionalized nanopillar array-captured MCF7 stained with Celcein-AM (green), preventing the nonspecific adhesion of PBMCs stained with Hoechst 33342 (blue). (Scale bars 50 μm.) (c) Capture efficiency (left) and capture purity (right) of spiked MCF7 cells in isolated PBMCs on supported lipid bilayer-functionalized quartz nanopillar arrays and bare quartz nanopillar arrays. The capture efficiencies of supported lipid bilayer-functionalized nanopillar arrays and bare quartz nanopillar arrays are 86.6 ± 10.6 and 92.5 ± 0.8%, respectively. The purities of both surfaces are 71.3 ± 17.2 and 3.2 ± 0.4%, respectively. (Every experiment was repeated three times (n = 3), and error bars represent the standard deviation (SD).
CONCLUSIONS
We demonstrated in this work that supported lipid bilayers constitute a dual-functional coating on nanostructures to facilitate CTC capture in two aspects: (1) ensure high capture purity by preventing nonspecific cell adhesion and 2) boost the capture efficiency by presenting antibody uniformly and allowing antibody clustering upon cell capture to strengthen the cell–surface interaction. With this coating, the capture purity on nanopillars was significantly increased, more than 20-fold, with uncompromised capture efficiency even when PBMC was present in the background. In conclusion, the combination of supported lipid bilayer-based surface functionalization and nanostructure-based cell capture provides an attractive solution for fulfilling the needs of a CTC study on high capture efficiency and high capture purity.
Supplementary Material
ACKNOWLEDGMENTS
The fabrication and characterization of nanopillar substrates were conducted at the Stanford Nanofabrication Facility and Stanford Nano Shared Facility. The MCF-7 cell line and MDA-MB-231 cell line were gifts from the Snyder laboratory and Jeffery laboratory at Stanford University, respectively. The human blood sample was from the Stanford Blood Center. This work was supported by the NSF (CAREER award no. 1055112), the NIH (grant no. NS057906), a Searle Scholar award, a Packard Science and Engineering Fellowship (to B.C.), a CCNE-T pilot grant (to B.C. and Y.C.), and a Studying Abroad Scholarship (to H.-Y.L.).
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.langmuir.6b03903.
Quartz nanopillar arrays’ geometry and their relative surface area. Additional supporting results of whole blood experiments, blocking effect of PBMCs, and the effect of pillar geometry. (PDF)
FRAP of a supported lipid bilayer. (AVI)
FRAP of SA-488 on a supported lipid bilayer. (AVI)
Clustering of SA-488 beneath the captured MCF7 cells. (AVI)
The authors declare no competing financial interest.
REFERENCES
- (1).de Bono JS; Scher HI; Montgomery RB; Parker C; Miller MC; Tissing H; Doyle GV; Terstappen LWWM; Pienta KJ; Raghavan D Circulating Tumor Cells Predict Survival Benefit from Treatment in Metastatic Castration-Resistant Prostate Cancer. Clin. Cancer Res 2008, 14, 6302–6309. [DOI] [PubMed] [Google Scholar]
- (2).Cohen SJ; Punt CJA; Iannotti N; Saidman BH; Sabbath KD; Gabrail NY; Picus J; Morse M; Mitchell E; Miller MC; Doyle GV; Tissing H; Terstappen LWMM; Meropol NJ Relationship of Circulating Tumor Cells to Tumor Response, Progression-Free Survival, and Overall Survival in Patients With Metastatic Colorectal Cancer. J. Clin. Oncol 2008, 26, 3213–3221. [DOI] [PubMed] [Google Scholar]
- (3).Maheswaran S; Haber DA Circulating Tumor Cells: A Window into Cancer Biology and Metastasis. Curr. Opin. Genet. Dev 2010, 20, 96–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Alix-Panabieres C; Pantel K Circulating Tumor Cells: Liquid Biopsy of Cancer. Clin. Chem 2013, 59, 110–118. [DOI] [PubMed] [Google Scholar]
- (5).Harouaka R; Kang Z; Zheng S-Y; Cao L Circulating Tumor Cells: Advances in Isolation and Analysis, and Challenges for Clinical Applications. Pharmacol. Ther 2014, 141, 209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Gupta V; Jafferji I; Garza M; Melnikova VO; Hasegawa DK; Pethig R; Davis DW ApoStream, a New Dielectrophoretic Device for Antibody Independent Isolation and Recovery of Viable Cancer Cells from Blood. Biomicrofluidics 2012, 6, 24133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Sarioglu AF; Aceto N; Kojic N; Donaldson MC; Zeinali M; Hamza B; Engstrom A; Zhu H; Sundaresan TK; Miyamoto DT; Luo X; Bardia A; Wittner BS; Ramaswamy S; Shioda T; Ting DT; Stott SL; Kapur R; Maheswaran S; Haber DA; Toner M A Microfluidic Device for Label-Free, Physical Capture of Circulating Tumor Cell Clusters. Nat. Methods 2015, 12, 685–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Lee HJ; Oh JH; Oh JM; Park J-M; Lee J-G; Kim MS; Kim YJ; Kang HJ; Jeong J; Kim SI; Lee SS; Choi J-W; Huh N Efficient Isolation and Accurate In Situ Analysis of Circulating Tumor Cells Using Detachable Beads and a High-Pore-Density Filter. Angew. Chem., Int. Ed 2013, 52, 8337–8340. [DOI] [PubMed] [Google Scholar]
- (9).Hou HW; Warkiani ME; Khoo BL; Li ZR; Soo RA; Tan DS-W; Lim W-T; Han J; Bhagat AAS; Lim CT Isolation and Retrieval of Circulating Tumor Cells Using Centrifugal Forces. Sci. Rep 2013, 3, 1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Becker TM; Caixeiro NJ; Lim SH; Tognela A; Kienzle N; Scott KF; Spring KJ; De Souza P New Frontiers in Circulating Tumor Cell Analysis: A Reference Guide for Biomolecular Profiling toward Translational Clinical Use. Int. J. Cancer 2014, 134, 2523–2533. [DOI] [PubMed] [Google Scholar]
- (11).Balic M; Lin H; Williams A; Datar RH; Cote RJ Progress in Circulating Tumor Cell Capture and Analysis: Implications for Cancer Management. Expert Rev. Mol. Diagn 2012, 12, 303–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Wang S; Wan Y; Liu Y Effects of Nanopillar Array Diameter and Spacing on Cancer Cell Capture and Cell Behaviors. Nanoscale 2014, 6, 12482–12489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Yoon HJ; Kozminsky M; Nagrath S Emerging Role of Nanomaterials in Circulating Tumor Cell Isolation and Analysis. ACS Nano 2014, 8, 1995–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Kwak M; Kim D-J; Lee M-R; Wu Y; Han L; Lee S-K; Fan R Nanowire Array Chips for Molecular Typing of Rare Trafficking Leukocytes with Application to Neurodegenerative Pathology. Nanoscale 2014, 6, 6537–6550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Lu YT; Zhao L; Shen Q; Garcia MA; Wu D; Hou S; Song M; Xu X; OuYang WH; OuYang WW-L; Lichterman J; Luo Z; Xuan X; Huang; Chung LWK; Rettig M; Tseng H-R; Shao C; Posadas EM NanoVelcro Chip for CTC Enumeration in Prostate Cancer Patients. Methods 2013, 64, 144–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Wang S; Wang H; Jiao J; Chen K-JJ; Owens GE; Kamei KI; Sun J; Sherman DJ; Behrenbruch CP; Wu H; Tseng H-R Three-Dimensional Nanostructured Substrates toward Efficient Capture of Circulating Tumor Cells. Angew. Chem., Int. Ed 2009, 48, 8970–8973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Wang S; Liu K; Liu J; Yu ZT-F; Xu X; Zhao L; Lee T; Lee EK; Reiss J; Lee Y-K; Chung LWK; Huang J; Rettig M; Seligson D; Duraiswamy KN; Shen CK-F; Tseng H-R Highly Efficient Capture of Circulating Tumor Cells by Using Nanostructured Silicon Substrates with Integrated Chaotic Micromixers. Angew. Chem., Int. Ed 2011, 50, 3084–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Hou S; Zhao H; Zhao L; Shen Q; Wei KS; Suh DY; Nakao A; Garcia MA; Song M; Lee T; Xiong B; Luo S-C; Tseng H-R; Yu H-H Capture and Stimulated Release of Circulating Tumor Cells on Polymer-Grafted Silicon Nanostructures. Adv. Mater 2013, 25, 1547–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Chen L; Liu X; Su B; Li J; Jiang L; Han D; Wang S Aptamer-Mediated Efficient Capture and Release of T Lymphocytes on Nanostructured Surfaces. Adv. Mater 2011, 23, 4376–4380. [DOI] [PubMed] [Google Scholar]
- (20).Shen Q; Xu L; Zhao L; Wu D; Fan Y; Zhou Y; Ouyang W-H; Xu X; Zhang Z; Song M; Lee T; Garcia MA; Xiong B; Hou S; Tseng H-R; Fang X Specific Capture and Release of Circulating Tumor Cells Using Aptamer-Modified Nanosubstrates. Adv. Mater 2013, 25, 2368–2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Hou S; Zhao L; Shen Q; Yu J; Ng C; Kong X; Wu D; Song M; Shi X; Xu X; OuYang W-H; He R; Zhao X-Z; Lee T; Brunicardi FC; Garcia MA; Ribas A; Lo RS; Tseng H-R Polymer Nanofiber-Embedded Microchips for Detection, Isolation, and Molecular Analysis of Single Circulating Melanoma Cells. Angew. Chem., Int. Ed 2013, 52, 3379–3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Kim ST; Kim D-J; Kim T-J; Seo D-W; Kim T-H; Lee S-Y; Kim K; Lee K-M; Lee S-K Novel Streptavidin-Functionalized Silicon Nanowire Arrays for CD4+ T Lymphocyte Separation. Nano Lett. 2010, 10, 2877–2883. [DOI] [PubMed] [Google Scholar]
- (23).Park G-S; Kwon H; Kwak DW; Park SY; Kim M; Lee J-H; Han H; Heo S; Li XS; Lee JH; Kim YH; Lee J-G; Yang W; Cho HY; Kim SK; Kim K Full Surface Embedding of Gold Clusters on Silicon Nanowires for Efficient Capture and Photothermal Therapy of Circulating Tumor Cells. Nano Lett. 2012, 12, 1638–1642. [DOI] [PubMed] [Google Scholar]
- (24).Lee S-K; Kim D-J; Lee G; Kim G-S; Kwak M; Fan R Specific Rare Cell Capture Using Micro-Patterned Silicon Nanowire Platform. Biosens. Bioelectron 2014, 54, 181–188. [DOI] [PubMed] [Google Scholar]
- (25).Yoon HJ; Kim TH; Zhang Z; Azizi E; Pham TM; Paoletti C; Lin J; Ramnath N; Wicha MS; Hayes DF; Simeone DM; Nagrath S Sensitive Capture of Circulating Tumour Cells by Functionalized Graphene Oxide Nanosheets. Nat. Nanotechnol 2013, 8, 735–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Sekine J; Luo S-C; Wang S; Zhu B; Tseng H-R; Yu H-H Functionalized Conducting Polymer Nanodots for Enhanced Cell Capturing: The Synergistic Effect of Capture Agents and Nanostructures. Adv. Mater 2011, 23, 4788–4792. [DOI] [PubMed] [Google Scholar]
- (27).Zhang N; Deng Y; Tai Q; Cheng B; Zhao L; Shen Q; He R; Hong L; Liu W; Guo S; Liu K; Tseng H-R; Xiong B; Zhao X-Z Electrospun TiO 2 Nanofiber-Based Cell Capture Assay for Detecting Circulating Tumor Cells from Colorectal and Gastric Cancer Patients. Adv. Mater 2012, 24, 2756–2760. [DOI] [PubMed] [Google Scholar]
- (28).Han W; Allio BA; Foster DG; King MR Nanoparticle Coatings for Enhanced Capture of Flowing Cells in Microtubes. ACS Nano 2010, 4, 174–180. [DOI] [PubMed] [Google Scholar]
- (29).Chen W; Weng S; Zhang F; Allen S; Li X; Bao L; Lam HWR; Macoska JA; Merajver SD; Fu J Nanoroughened Surfaces for Efficient Capture of Circulating Tumor Cells without Using Capture Antibodies. ACS Nano 2013, 7, 566–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Chen W; Allen SG; Reka AK; Qian W; Han S; Zhao J; Bao L; Keshamouni VG; Merajver SD; Fu J Nanoroughened Adhesion-Based Capture of Circulating Tumor Cells with Heterogeneous Expression and Metastatic Characteristics. BMC Cancer 2016, 16, 614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Lee S-K; Kim G-S; Wu Y; Kim D-J; Lu Y; Kwak M; Han L; Hyung J-H; Seol J-K; Sander C; Gonzalez A; Li J; Fan R Nanowire Substrate-Based Laser Scanning Cytometry for Quantitation of Circulating Tumor Cells. Nano Lett. 2012, 12, 2697–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Wong LS; Khan F; Micklefield J Selective Covalent Protein Immobilization: Strategies and Applications. Chem. Rev 2009, 109, 4025–4053. [DOI] [PubMed] [Google Scholar]
- (33).Kam L; Boxer S Cell Adhesion to Protein-Micropatterned-Supported Lipid Bilayer Membranes. J. Biomed. Mater. Res 2001, 55, 487–495. [DOI] [PubMed] [Google Scholar]
- (34).Andersson A-S; Glasmästar K; Sutherland D; Lidberg U; Kasemo B Cell Adhesion on Supported Lipid Bilayers. J. Biomed. Mater. Res., Part A 2003, 64, 622–629. [DOI] [PubMed] [Google Scholar]
- (35).Wu J-C; Tseng P-Y; Tsai W-S; Liao M-Y; Lu S-H; Frank CW; Chen J-S; Wu H-C; Chang Y-C Antibody Conjugated Supported Lipid Bilayer for Capturing and Purification of Viable Tumor Cells in Blood for Subsequent Cell Culture. Biomaterials 2013, 34, 5191–5199. [DOI] [PubMed] [Google Scholar]
- (36).Lin W-C; Yu C-H; Triffo S; Groves JT Supported Membrane Formation, Characterization, Functionalization, and Patterning for Application in Biological Science and Technology. Curr. Protoc. Chem. Biol 2010, 2, 235–269. [DOI] [PubMed] [Google Scholar]
- (37).Chen J-Y; Tsai W-S; Shao H-J; Wu J-C; Lai J-M; Lu S-H; Hung T-F; Yang C-T; Wu LC; Chen J-S; Lee W-H; Chang Y-C Sensitive and Specific Biomimetic Lipid Coated Microfluidics to Isolate Viable Circulating Tumor Cells and Micro-emboli for Cancer Detection. PLoS One 2016, 11, e0149633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Groves J; Mahal L; Bertozzi C Control of Cell Adhesion and Growth with Micropatterned Supported Lipid Membranes. Langmuir 2001, 17, 5129–5133. [Google Scholar]
- (39).Königsberg R; Obermayr E; Bises G; Pfeiler G; Gneist M; Wrba F; de Santis M; Zeillinger R; Hudec M; Dittrich C Detection of EpCAM Positive and Negative Circulating Tumor Cells in Metastatic Breast Cancer Patients. Acta Oncol. 2011, 50, 700–710. [DOI] [PubMed] [Google Scholar]
- (40).Phair RD; Gorski SA; Misteli T Measurement of Dynamic Protein Binding to Chromatin In Vivo, Using Photobleaching Microscopy. Methods Enzymol. 2003, 375, 393–414. [DOI] [PubMed] [Google Scholar]
- (41).Axelrod D; Koppel DE; Schlessinger J; Elson E; Webb WW Mobility Measurement by Analysis of Fluorescence Photobleaching Recovery Kinetics. Biophys. J 1976, 16, 1055–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Dabkowska AP; Niman CS; Piret G; Persson H; Wacklin HP; Linke H; Prinz CN; Nylander T Fluid and Highly Curved Model Membranes on Vertical Nanowire Arrays. Nano Lett. 2014, 14, 4286–4292. [DOI] [PubMed] [Google Scholar]
- (43).Scatena R; Bottoni P; Giardina B Circulating Tumour Cells and Cancer Stem Cells: A Role for Proteomics in Defining the Interrelationships between Function, Phenotype and Differentiation with Potential Clinical Applications. Biochim. Biophys. Acta, Rev. Cancer 2013, 1835, 129–143. [DOI] [PubMed] [Google Scholar]
- (44).Powell AA; Talasaz AH; Zhang H; Coram MA; Reddy A; Deng G; Telli ML; Advani RH; Carlson RW; Mollick JA; Sheth S; Kurian AW; Ford JM; Stockdale FE; Quake SR; Pease RF; Mindrinos MN; Bhanot G; Dairkee SH; Davis RW; Jeffrey SS Single Cell Profiling of Circulating Tumor Cells: Transcriptional Heterogeneity and Diversity from Breast Cancer Cell Lines. PLoS One 2012, 7, e33788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Triffo SB; Huang HH; Smith AW; Chou ET; Groves JT Monitoring Lipid Anchor Organization in Cell Membranes by PIE-FCCS. J. Am. Chem. Soc 2012, 134, 10833–10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Huang C-J; Cho N-J; Hsu C-J; Tseng P-Y; Frank CW; Chang Y-C Type I Collagen-Functionalized Supported Lipid Bilayer as a Cell Culture Platform. Biomacromolecules 2010, 11, 1231–1240. [DOI] [PubMed] [Google Scholar]
- (47).Huang C-J; Tseng P-Y; Chang Y-C Effects of Extracellular Matrix Protein Functionalized Fluid Membrane on Cell Adhesion and Matrix Remodeling. Biomaterials 2010, 31, 7183–7195. [DOI] [PubMed] [Google Scholar]
- (48).Hartman NC; Nye JA; Groves JT Cluster Size Regulates Protein Sorting in the Immunological Synapse. Proc. Natl. Acad. Sci. U. S. A 2009, 106, 12729–12734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Rosenberg R; Gertler R; Friederichs J; Fuehrer K; Dahm M; Phelps R; Thorban S; Nekarda H; Siewert JR Comparison of Two Density Gradient Centrifugation Systems for the Enrichment of Disseminated Tumor Cells in Blood. Cytometry 2002, 49, 150–158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


