Abstract
Retinoic acid receptor-related orphan receptor-α (RORα) is a member of the orphan nuclear receptor family and functions as a transcriptional activator in response to circadian changes. Circadian rhythms are complex cellular mechanisms regulating diverse metabolic, inflammatory, and tumorigenic gene expression pathways that govern cyclic cellular physiology. Disruption of circadian regulators, including RORα, plays a critical role in tumorigenesis and facilitates the development of inflammatory hallmarks. Although RORα contributes to overall fitness among anticancer, anti-inflammatory, lipid homeostasis, and circadian clock mechanisms, the molecular mechanisms underlying the mode of transcriptional regulation by RORα remain unclear. Nonetheless, RORα has important implications for pharmacological prevention of cancer, inflammation, and metabolic diseases, and understanding context-dependent RORα regulation will provide an innovative approach for unraveling the functional link between cancer metabolism and rhythm changes.
Subject terms: Transcriptional regulatory elements, Cell signalling
Receptor proteins: adopting an orphan to understand and treat disease
The protein known as retinoic acid receptor-related orphan receptor α (RORα) activates specific genes in response to the daily cellular changes associated with circadian rhythms, and disruption of its activity is associated with cancer, inflammation and metabolic diseases. Proteins like RORα are called orphan receptors because the molecules that bind to them to initiate their activities are unknown or unconfirmed. Ji Min Lee at Kangwon National University, Chuncheon, South Korea and colleagues review current understanding of the activities of RORα, including the molecular mechanisms underlying its likely involvement in various diseases. They highlight the possible significance of manipulating RORα activity as a route towards new treatments for several diseases including cancer. Pharmaceutical companies are exploring the therapeutic possibilities of molecules that can bind to and activate RORα, especially natural products.
Introduction
Retinoic acid receptor-related orphan receptor-α (RORα) belongs to the orphan nuclear receptor (NR) superfamily. The RORα gene, which is located on human chromosome 15q22.2, generates by alternative splicing four human isoforms comprising a conserved DNA-binding domain (DBD), ligand-binding domain (LBD), hinge domain, and distinct N-terminal domain. RORα is expressed in the liver, skin, lung, adipose tissue, brain, and muscle. All four isoforms, RORα1–4, are expressed in humans, whereas only RORα1 and RORα4 are expressed in mice. Expression of these isoforms varies and the isoforms regulate specific functions in several physiological processes1,2. The RORα recognizes a specific DNA sequence, the ROR response element (RORE), which comprises the AGGTCA consensus motif followed by an A/T-rich region, to regulate target gene transcription via interaction with coactivators and corepressors3–5. For most NRs, ligand binding is a critical step that triggers a conformational change in the receptor and the possibility of being replaced by a coactivator. Although cognate endogenous ligands of RORα have not been clearly identified, cholesterol and cholesterol intermediates have been suggested. Crystal structure analysis has shown that these ligands reversibly bind to RORα, but whether cholesterol and cholesterol intermediates are relevant physiological ligands remains unclear. In addition to natural ligands, SR3335 and SR1078 constitute developed inverse agonists of RORα. Selective synthetic SR3335 inhibits expression of RORα target genes, such as the gluconeogenic enzymes G6pc and Pepck6, and effects of SR1078 on p53 regulation, adipose tissue inflammation, renal ischemia, and allergic asthma have been studied7–10.
The naturally occurring mutant mouse strain “staggerer” (also known as Rorαsg/sg)11 was discovered by positional cloning12; it lacks up to 90% of the Purkinje cells (PCs) found in wild-type (WT) mice and has a short lifespan. The defective phenotype of staggerer mice is due to deletion of the LBD region of the RORα gene, which causes a shift in the reading frame and prevents adequate translation. These mice exhibit ataxia, motor deficiencies, irregular circadian rhythm, hyperinflammation, osteopenia, atherosclerosis, and muscle atrophy. Moreover, the similar phenotypes of RORα null-mutant (RORα−/−)13 and Rorαsg/sg indicate that RORα is an essential regulator of PC differentiation and cerebellar function. Furthermore, RORα is involved in various physiological processes, such as the circadian clock, inflammation, tumorigenesis, and metabolic diseases, and it is a tumor suppressor in many types of cancer. Its expression is downregulated in cancers of the skin, colon, prostate, liver, and breast, as well as in melanoma14–19. Decreased RORα expression correlates with cancer aggressiveness, short overall survival, and poor prognosis. In addition, RORα modulates cyclic expression of BMAL1, a key transcription factor (TF) that regulates circadian rhythms. RORα contributes to the inflammatory response, M1/M2 polarization, and the nuclear factor-κB (NF-κB) pathway20–22 and regulates lipid and glucose metabolic gene expression, implying its involvement in energy homeostasis.
The regulatory mechanisms of RORα in many physiological processes are bidirectional. Its cis-acting regulation is based on direct recruitment of RORα to the RORE of target gene promoters. However, RORα often affects transcriptional programs in transaction mode through the antagonistic mechanisms of other TFs along with altered coregulator recruitment15,23. This review summarizes and discusses the current understanding of RORα function and molecular mechanisms in various pathophysiological processes, including cancer, inflammation, cerebellar development, circadian rhythm, and lipid homeostasis. This review also highlights potential targets for treating various diseases.
Anticancer mechanisms involving RORα
Oncogenes are abnormally activated during tumor progression, which might involve signal-dependent determination of target gene expression profiles24–27. Cancer is associated with gene mutations, abnormal gene expression due to gene duplication, unbalanced numbers of chromosomes, or chromatin remodeling. These aberrant genetic changes can increase expression of oncogenes encoding tumor-promoting proteins or decrease that of genes encoding tumor-suppressor proteins28–30. Gene mutations and altered gene expression are generally connected. For instance, a gene mutation that activates an oncogene functions as a negative regulator of tumor-suppressor gene expression, which leads to a decrease in tumor suppressors. Therefore, understanding gene mutations and changes in expression is important to fully comprehend cancer.
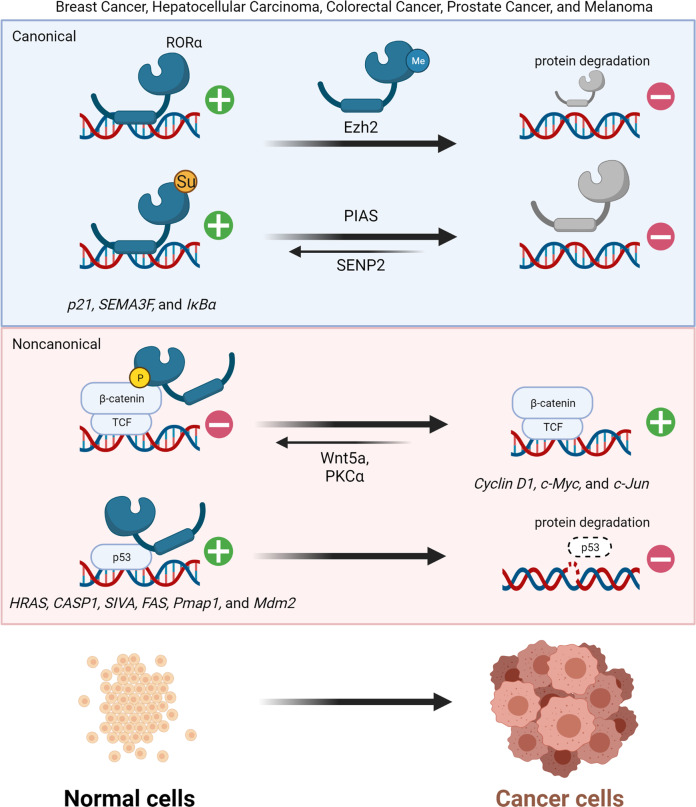
TFs interact with large complexes of coregulators in a gene-specific manner. Specific sets of TFs and coregulators are required to control subsets of genes in a given cell type. Impaired physiological pathways are important in the progression of cancer and a link between TFs and coregulators in the regulation of specific physiological pathways has been investigated31,32. However, under specific signaling, TFs indirectly regulate the expression of other target genes through protein–protein interactions as coregulators and this type of regulation selectively determines which genes in tumorigenesis or tumor-suppressive pathways are expressed. Increases or decreases in posttranslational modifications (PTMs), PTM crosstalk, changes in binding partners within complexes, nongenomic actions, and regulation by nuclear export signals can lead to RORα functioning as a TF that directly regulates target genes via its DBD and as a coregulator via protein–protein interactions in noncanonical pathways (Fig. 1). This type of transregulation by RORα as a coregulator allows for more sophisticated anticancer regulation than that achieved solely by increased RORα expression.
Fig. 1. Regulatory mechanism of RORα through canonical and noncanonical pathways in cancer.
Posttranslational modification by SUMOylation stimulates RORα to function as a direct TF on target gene promoters. Phosphorylation alters the direction of RORα functioning as a coregulator. Different signals can also change the roles of RORα-transregulating activities, leading to recruitment of p53-dependent target gene promoters as coactivators.
Target genes associated with several diseases, including cancer, are transcriptionally regulated by RORα. Transcriptional regulation via PTM mediated by RORα is affected by various cellular signaling pathways33. SUMOylation is a PTM in which a small ubiquitin-like modifier (SUMO) is added to target proteins, to change their binding affinity34. SUMO E3 ligase protein inhibitor of activated STAT SUMOylates RORα, whereas SUMO-specific peptidase 2 deSUMOylates it. SUMOylation upregulates RORα transcriptional activity and activates tumor-suppressive target genes35.
The classical ability of RORα to regulate target gene expression is key for activation of tumor-suppressive target genes (Fig. 1)36, although a transcription-independent function of RORα via a noncanonical pathway can potentiate tumor-suppressive roles. These functions involve direct interaction of RORα with molecules in the Wnt/β-catenin pathway, allowing RORα to transrepress Wnt pathways in colorectal cancer22. The Wnt signaling pathway can be classified as canonical Wnt/β-catenin or noncanonical Wnt/Ca2+ signaling37,38. As a corepressor, RORα is responsible for downregulating expression of Wnt/β-catenin target genes. In the noncanonical Wnt pathway, Wnt5a activates protein kinase Cα (PKCα) via phosphorylation and phosphorylation of RORα by activated PKCα can serve as an intersection between the noncanonical and canonical Wnt pathways. Phosphorylated RORα transrepresses β-catenin in the canonical Wnt/β-catenin pathway and downregulates expression of cyclin D1, Axin, c-Jun, and c-Myc15. In addition, RORα contributes to the intracellular DNA damage response, which is a checkpoint that determines cell fate, progression to a tumor or cell cycle blockade, and apoptosis during the early stage of tumorigenesis39. Under normal conditions, low levels of p53 are maintained by E3 ubiquitin ligases, such as Mdm2, but p53 expression is stabilized during DNA damage responses40,41. Moreover, expression of RORα is increased by the DNA damage signal and it is then recruited to specific genomic sites by interacting with p53 as a coregulator. The target genes of p53 comprise those with HAUSP (herpes virus-associated ubiquitin-specific protease)-dependent (Hras, Casp1, and Siva) and -independent (Fas, Pmap1, and Mdm2) promoters, with RORα functioning as a coactivator for both types42.
The mechanisms through which RORα acts as a canonical TF via its DBD and coregulator differ considerably. The action of RORα can be determined by the types of PTMs involved, such as SUMOylation or phosphorylation. The fact that RORα regulates expression of the master tumor-suppressor p53 and increases p53 protein stability suggests that agonists enhance the effects of RORα and create synergistic effects in two layers of RORα downstream signaling.
Anti-inflammatory mechanisms involving RORα
Acute and chronic inflammation are major causes of cancer and understanding of their relationships is increasing43. The fundamental mediators between inflammation and cancer include cytokines, noncoding RNAs, and NF-κB TFs44, and the effects of these mediators principally cause anti- or protumorigenic inflammatory responses45. NF-κB is a key mediator of inflammation and tumorigenesis, and its functional link is supported by experimental evidence explaining the mutual negative regulation of NF-κB and the tumor-suppressor RORα22,42,46,47.
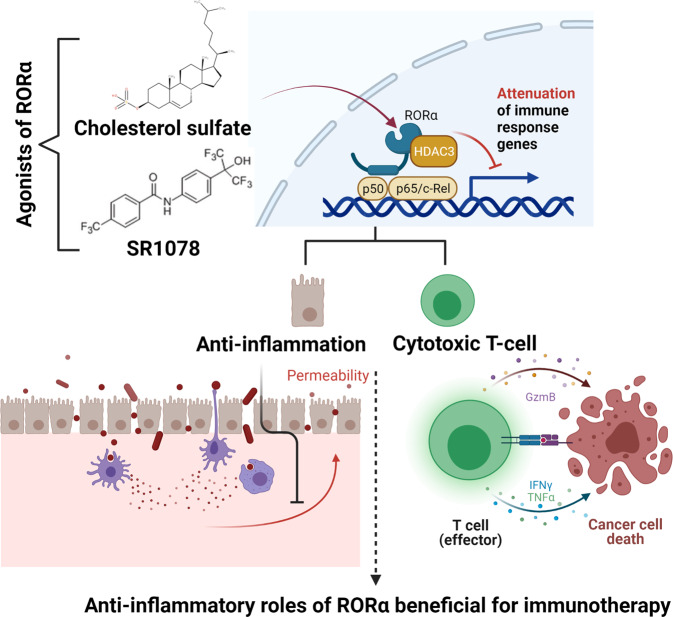
Anti-inflammatory repression through NR crosstalk (transrepression) occurs in a transacting manner without direct binding of NR to DNA response elements. Studies of RORα along with its anti-inflammatory roles in inflammatory bowel disease and ulcerative colitis have specifically advanced our understanding of the underlying molecular anti-inflammatory transrepression mechanism. The mode of action involves concomitant direct interactions of RORα with proinflammatory NF-κB and corepressor HDAC3 (Fig. 2). Recruitment of HDAC3 via RORα is responsible for the transrepressive function of NF-κB in intestinal inflammation by attenuating recruitment of histone acetyltransferase BRD4/CBP complexes and the potential application of RORα transrepression activity for therapeutics indicates new avenues for treating intestinal inflammatory diseases. The development of innovative RORα ligands would help promote anti-inflammatory effects on cancer-mediated inflammation and reduce undesirable side effects.
Fig. 2. Attenuation of inflammation by RORα.
Attenuated activation of NF-κB target genes by RORα contributes to survival and tumorigenesis prevention. Infectious agents lead to activation of NF-κB signaling, resulting in changes in the microenvironment to invoke immune responses and increase permeability in tissues, such as the intestine. Under ROR-mediated NF-κB target gene suppression, the functions of damaged cells are restored to reduce hyperinflammation. The anti-inflammatory roles of RORα further induce effective cytotoxic function in CD8+ T cells, resulting in cancer cell death. Activation of RORα with selective agonists, such as SR1078 and cholesterol sulfate, result in stimulatory effector responses of CD8+ T cells.
Metabolic changes have been observed in cancer cells and the tumor microenvironment (TME)48, and the mechanisms underlying these changes related to RORα have recently been identified. For example, RORα responds to metabolic changes and influences cholesterol synthesis pathways in cytotoxic T cells by inhibiting NF-κB target genes. The contributions of these activities to cytotoxic CD8+ T cells have been verified using RORα agonists49. Pharmacological stimulation of RORα using cholesterol sulfate and the synthetic agonist SR1078 activates downstream target genes of RORα, whereas the synthetic specific antagonist SR3335 functions as a selective inhibitor of RORα. Compared to SR1078 acting as a dual agonist of RORα and RORγ, cholesterol sulfate is a natural selective ligand of RORα. Given that directly targeting RORα via its LBD is a preferred approach for minimizing off-target effects, the selective agonist cholesterol sulfate has therapeutic anticancer effects. Cholesterol sulfate functions to regulate the effector responses of CD8+ T cells by reducing cholesterol esterification in an NF-κB suppression-dependent manner under RORα activation (Fig. 2). Immunosurveillance is an effective antitumor immune response achieved by innate and adaptive components of the immune system; however, cancer can evade immune cytotoxicity. Cytotoxic CD8+ T cells are considered the major effectors of antitumor immunity. The repressive abilities of RORα on NF-κB target genes, such as Acat1/2 and Abca1, induce CD8+ T cells to secrete cytotoxic cytokines, including tumor necrosis factor-α and interferon-γ. Anti-inflammatory drugs have considerable importance for potential use in cancer chemoprevention. Indeed, anti-inflammatory drugs modulate the cytotoxic effector functions of antitumor immunity, and the attenuated inflammation and increased cytotoxic effector function promoted by activated RORα are promising for the development of a versatile therapeutic approach. Furthermore, as a safer and more effective therapy than currently available classical anti-inflammatory agents, this approach may support antitumor immune-potentiating activity because of an increased selective agonistic effect on RORα signaling in the TME.
Control of cerebellar development and circadian rhythms by RORα
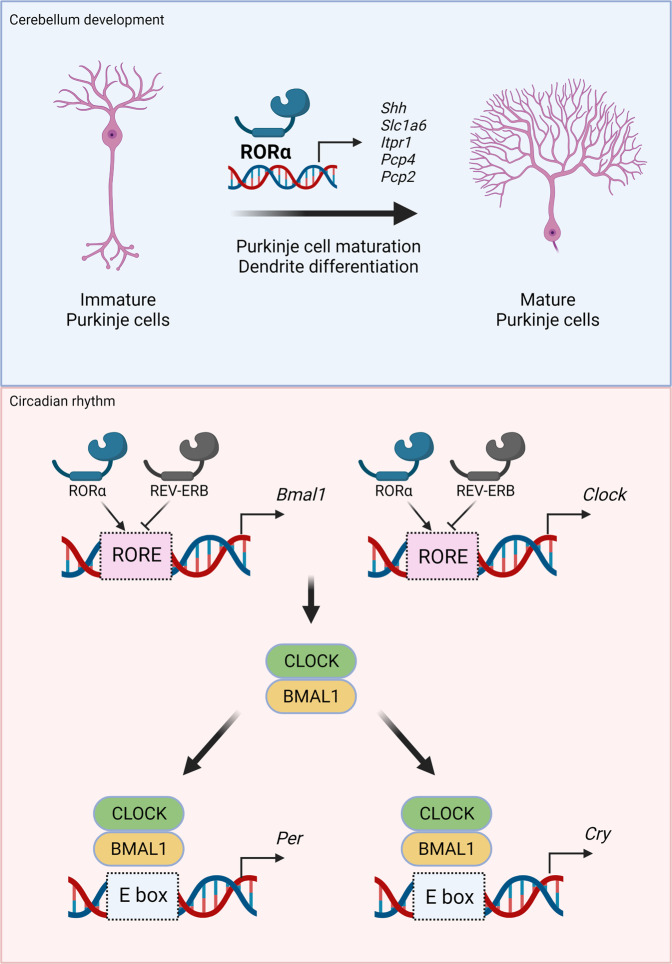
Abundant RORα is expressed in cerebellar PCs, retinal ganglion cells, and thalamic nuclei. The size of the agranular cerebellum is reduced and PCs are not arranged the same way in RORα−/− mice as in WT mice. The atypical cerebellar phenotypes in RORα−/− and Rorαsg/sg mice are identical and these defects cause abnormal cerebellar development and cerebellar ataxia because of cell-autonomous problems with PC development. Phenotypic analyses of genetically modified mice have revealed that RORα is a prerequisite for the maturation and innervation characteristics of adult PCs, suggesting that RORα is a vital determinant of survival and appropriate differentiation of PCs50. PCs are representative models for investigating dendrite development, as well as the function of circulating sex steroids and neurosteroids in neuronal death. Neuronal loss in Rorα+/sg leads to a decline in circulating sex steroids and cerebellar neurosteroids, thus inducing PC death during normal aging51. Embryonic knockdown using a miRNA against RORα results in PC mislocation and primitive dendrites. These results indicate that RORα is important for PC migration and primitive dendrite regression. Therefore, appropriate expression of RORα is required for the formation and maintenance of dendrites and spines in young and adult PCs, respectively52. Moreover, genome-wide analysis has revealed that RORα controls the transcriptional program of cerebellar development via direct recruitment of target genes with specific coactivators, including β-catenin, p300, and tip60. After directly binding to the promoters of Shh, Slc1a6, Itpr1, Pcp4, and Pcp2, RORα recruits various combinations of coactivators (Fig. 3). In additon, in RORα−/− mice, granular precursor proliferation and survival are rescued when recombinant proteins restore reduced Shh expression to normal levels53. Taken together, RORα maintains granular and PC functions in the cerebellum from embryonic development to adulthood.
Fig. 3. Transcriptional regulation of cerebellum development and the circadian clock via RORα and REV-ERBs.
RORα controls Purkinje cell maturation through transcriptional activation of several target genes, such as Shh, Slc1a6, Itpr1, Pcp4, and Pcp1. Direct recruitment of RORα to target gene promoters positively regulates adequate expression for dendritic differentiation of Purkinje cells. Both RORα and REV-ERBs competitively control BMAL1 and CLOCK gene expression via RORE; RORα and REV-ERBs positively and negatively regulate BMAL1 and CLOCK, respectively. The BMAL1 and CLOCK generated cooperate at the E BOX to induce downstream target gene expression of PER and CRY, maintaining the master axis of the circadian rhythm.
The circadian clock is a periodic biological system that interconnects the diurnal sleep–wake cycle, metabolism, body temperature control, and immune response. Therefore, circadian rhythm dysfunction correlates with the onset of metabolic diseases, cancer, and neuropsychiatric disorders. The master axis of the circadian rhythm is generated by a specific feedback loop involving BMAL1 expression and CLOCK activation of cryptochrome (CRY1 and CRY2) and period (PER1, PER2, and PER3) genes. BMAL1 and CLOCK TF heterodimer formation is critical for controlled expression as a negative regulatory mechanism. RORα participates in regulating the core loop by transcriptional regulation of the BMAL1 gene with REV-ERBs54–56 and the dynamic interplay between RORα and REV-ERBs modulates BMAL1 expression and provides a feedback loop for timely expression. RORα works competitively with REV-ERBs by sharing the same DNA response element in the BMAL1 promoter (Fig. 3); expression of BMAL1 is silenced and increased by REV-ERBs and RORα, respectively. Oscillation by these two NRs plays a crucial role in enabling elaborate regulation of BMAL1 expression. In particular, PGC-1α is a coactivator of RORα in BMAL1 regulation; it interacts with RORα via the LXXLL motif to recruit RORE in the proximal BMAL1 promoter along with p300 and GCN5. The function of PGC-1α depends on RORE and the ability of RORα to induce BMAL1 expression is abolished in PGC-1α-null hepatocytes57. The CLOCK target gene depends on both RORα and REV-ERBs. Chromatin immunoprecipitation assays have revealed that RORα and REV-ERBs bind to two functional ROREs in the promoter of CLOCK and control BMAL1 expression58. Receptor-interacting protein 140 (RIP140) supports RORα transcriptional activity as a positive activator of the BMAL1 gene, whereas REV-ERBs repress RIP140 expression and directly interact with RORα. As RIP140 exerts no effects in the absence of RORα, RIP140 might ROR-dependently participate in the feedback mechanism of the circadian clock and BMAL1 expression59.
Overall, circadian clock genes might be associated with the onset and risk of breast, prostate, and colorectal cancers and lung carcinoma60–62. Cancer predisposition statistically correlates with genetic variations in the circadian pathway via adaptive rank truncated product (ARTP)-based gene expression and pathways60. RORα ranks top among genes associated with the risk of breast and lung cancer. Moreover, RORα plays dual roles and has a functional link between circadian rhythms and cancer pathogenesis.
Role of RORα in control of lipid homeostasis
A coordinated transcriptional network regulates expression of lipid and glucose metabolic target genes for energy homeostasis. In particular, changes in clock genes result in abnormal metabolic phenotypes in models in vivo. The roles of RORα in metabolism have been characterized in RORα−/− mice. Triglyceride (TG) and total plasma cholesterol levels, as well as ApoCIII, APOA1, and APOA2 expression, are lower in Rorαsg/sg mice than in WT mice. In addition, expression of the cholesterol transporters Abca1 and Abca8/G1 is attenuated in Rorαsg/sg mice, suggesting that the phenotype of impaired high-density lipoprotein biosynthesis is caused by decreased expression of ABCA163. Reduced TGs in Rorαsg/sg mice are due to downregulation of transcriptional regulators responsible for fatty acid biosynthesis and lipogenesis, including SREBP1c, FAS, and ABCG1. Indeed, Rorαsg/sg mice are resistant to standard high-fat diet (HFD)-induced obesity and weight gain. Although heart and liver tissue weights do not significantly differ between WT and Rorαsg/sg mice, epididymal and inguinal fat depots are distinct. Moreover, PCK1 expression is reduced in the livers of Rorαsg/sg mice after a 6 h fast. The gluconeogenic enzyme PCK1 is a rate-determining factor in the adaptive response of the fasting period. Hepatic gluconeogenesis and adipose tissue glyceroneogenesis are positively controlled by RORα and its regulation is mediated by PCK164. Expression of thermogenic genes, such as UCP1, is increased in brown and subcutaneous inguinal adipose compartments of Rorαsg/sg mice, suggesting increased thermal control and cold tolerance in these mice65. This process is associated with increased oxygen consumption and energy expenditure.
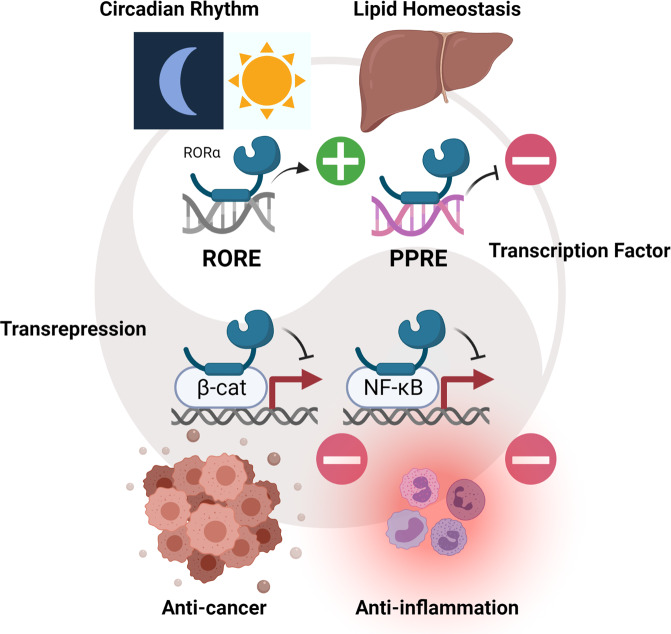
Nevertheless, a HFD significantly increases body weight and the obesity phenotype in liver-specific Rorα conditional knockout mice (RORαLKO) compared with RORα-floxed mice (RORαf/f). White and brown fat depots and the area of adipocytes in visceral fat tissue are significantly increased in RORαLKO mice, and the livers of HFD-fed RORαLKO mice are enlarged and paler than those in HFD-fed RORαf/f mice. Oil red O staining and hepatic TG analysis have revealed significantly increased lipid levels in HFD-fed RORαLKO mice. Genes associated with lipogenesis, gluconeogenesis, and lipid sequestration are abundantly expressed in RORαLKO mice fed an HFD, suggesting that RORα negatively regulates HFD-induced steatosis. Furthermore, genome-wide analysis showed that RORα is involved in PPARα-dependent target gene transcription. The target genes of PPARγ, such as Cd36, Plin2, and Scd1, are elevated in HFD-fed RORαLKO mice compared with HFD-fed RORαf/f mice. The absence of RORα leads to enhanced PPARγ recruitment to target genes and subsequent elevated expression of downstream target genes, indicating that RORα suppresses peroxisome proliferator-activated receptor-γ (PPARγ)-dependent target gene expression. In addition, RORα is directly recruited to the peroxisome proliferator-activated receptor element (PPRE) of a target gene promoter with HDAC3 for repression and competes with PPARγ (Fig. 4). Indeed, metabolic defects, such as body weight gain, hepatic steatosis, and lipid metabolism in HFD-fed RORαLKO mice, are mitigated by PPARγ antagonism with GW9662. Thus, RORα is a crucial factor in maintaining diet-induced hepatic lipid homeostasis through suppression of PPARγ signaling. The phenotypes of RORα and RORγ double-knockout mice in the lipogenic response have been determined66, and the ROR family subtype RORγ functions in hepatic gluconeogenesis. Generating double-floxed liver-specific, double-knockout (LDKO) mice by injecting targeted AAV-TBG-Cre has revealed that although expression of circadian clock genes, such as Bmal1, Cry1, and Npas2, is downregulated in LDKO mice, that of Fasn, Elovl6, and Acaca, which are involved in hepatic lipid and fatty acid metabolism, is enhanced. Both RORα and RORγ show functional redundancy in target gene expression in the mouse liver. Moreover, RORα and RORγ control Insig2a expression to repress SREBP1c, which is required for cholesterol synthesis and hepatic lipid metabolism at the end of the feeding phase in the dark. Consistent with this finding, the significantly increased hepatic TG levels and intensely Oil Red O-stained liver sections in ROR LDKO mice indicate that ROR regulates the homeostasis of lipogenic metabolism. The activity of RORα toward liver X receptor α (LXRα) in hepatic steatosis is antagonistic42. Activation of AMPK is induced by RORα, which leads to simultaneous reductions in LXRα protein levels and inhibition of transcriptional activity (Fig. 4). As LXRα is a positive regulator of hyperglycemia and hepatic fatty acid synthesis67, expression of lipogenic genes, such as Srepb-1c, Fas, and Accα, is diminished and the antilipogenic effects of RORα have been validated in HFD-induced fatty liver. Orally administered ROR-activating compounds drive AMPK activation as well as LXRα inhibition in the liver of HFD models, decreasing TG and body weight. Moreover, RORα expression correlates with the onset of metabolic diseases such as HFD-induced nonalcoholic steatohepatitis, coronary heart disease with concomitant abdominal obesity, and metabolic syndrome21,68–70. Therefore, RORα may be considered a key target in the development of agents to treat various metabolic diseases.
Fig. 4. Regulation of RORα in lipid homeostasis.
Transcriptional regulation of lipid homeostasis mediated by PPAR is inhibited by RORα. Transcriptional activation of PPARγ with PGC-1α and transcriptional repression of RORα with HDAC3 balance each other at the promoters of PPAR-dependent target genes. Simultaneously, AMPK activation and antagonized LXRα expression by RORα results in reduced expression of lipogenic genes, such as SREBP-1 and FAS. Downregulated SREBP-1 results in decreased fatty acid oxidation and lipogenic effects.
Conclusions
Circadian and metabolic disruptions are associated with an increased incidence of specific types of cancer and inflammatory diseases; however, the relationships between causes and mediators remain unclear71,72. As RORα regulates cancer, inflammation, circadian rhythm, and metabolism, it not only functions as a transcriptional effector but also as a sensitive sensor that captures and reconstructs distinct changes in the cellular environment into transcriptional responses73. Elucidating the signaling pathways of second messengers, hormones, and chromatin configurations, which modulate interactions between RORα and its binding partners, is important. Such knowledge is relevant to the identification and development of novel compounds that increase or decrease the binding affinity of RORα and its complexes, and direct the activity of cofactors toward disease crosstalk pathways.
The role of RORα as a TF or a coregulator is important for both fine-tuning transcriptional networks and further regulating many aspects of disease progression or suppression74,75. A controlled balance between direct TFs and coregulators in transcriptional programs can promote or inhibit target gene expression, and defining and harmonizing the signaling pathways that modulate gene expression between TFs and coregulators is crucial. The fact that different target genes of the same regulator RORα require distinctive combinations of TFs and coregulators is due to distinct chromatin environments and signaling cascades. The specific DNA sequence to which a TF binds can affect the selection of complex combinations and their actions. Variations in upstream signaling may influence recruitment of RORα as a coregulator. In addition, the coregulator activity of RORα may enhance other TF activities and bias expression of pathway-specific genes with unwanted side effects because of the gene- and pathway-specific actions of TF-coregulator combinations (Fig. 5).
Fig. 5. Target gene activation or repression by RORα and functions of RORα.
Circadian rhythm, lipid homeostasis, anticancer, and anti-inflammatory effects are integrated by processes in which RORα plays key roles. Direct activation of RORα via RORE may result in balanced circadian clock gene expression and different responses might be repressed by the same TF through binding of different PPREs. Evidence indicates that RORα functions in transrepression through other TFs such as β-catenin and NF-κB, which are master regulators of tumorigenesis and inflammation, respectively. The roles of RORα in health and disease are likely to depend on which cellular response is switched on and the context in which target selection occurs via RORα.
Overall, targeting individual pathways using a direct inhibitor of RORα as a TF by suppressing the DBD or LBD only inhibits one or two pathways. Disease progression can be decelerated, but other pathways in which RORα functions as a coregulator and transregulator of target genes may be upregulated as compensation, thus generating resistance to therapy. If a defined role of RORα as a coregulator is targeted and its function prevented, combined pathways will be concurrently inhibited. Such a strategy blocks the alternate pathway of compensatory upregulation, decreasing the onset of acquired drug resistance. In this scheme, a small-molecule drug designed to block a specific action of RORα, such as a PKCα agonist acting as a coregulator, can lead to simultaneous inhibition of an array of Wnt/β-catenin downstream target genes. As PTMs designating the action of RORα as a TF or a coregulator are a result of prior specific enzymatic reactions, drugs can inhibit or activate modifying enzymes. These approaches are important for understanding the regulation of specific noncanonical pathways in which drugs do not modulate RORα via LBDs, suggesting that a mechanism-based drug administration strategy may be useful for specifically inhibiting cancer activity.
Future investigation in these areas will be critical for defining the sensory and physiological codes of the dual RORα roles as a TF and coregulator, and recognizing the potential of dual concepts as therapeutic targets. Only a few signaling pathways or PTMs that regulate the activities of specific recruitment of RORα as a direct TF or coregulator have been elucidated thus far. The specific signaling pathways and enzymes responsible for conducting the signaling cascade in terms of TFs or coregulators and the inhibitors that would be applicable for clinical applications await further investigation in the future.
Many efforts to find ligands of RORα, such as the discovery of endogenous ligands and the chemical synthesis for synthetic ligand design, have been applied to verify its importance α in various biological systems and disease models. Although T0901317, first known as an inverse agonist of RORα, was revealed to be a nonspecific ligand for several NRs, it contributed to the development of a new synthetic ligand. In addition, SR1078 showed efficacy in p53 stabilization and subsequent apoptosis, and may be utilized in cancer treatment in the future. In addition, the inverse agonist SR1001, which interferes with the interaction between receptor and coactivator, has been proven to be effective for treating autoimmune disease. Many researchers have attempted to discover a potent and selective synthetic ligand of RORα and verify its efficacy in disease models. Although RORα is a very promising target that can be applied to circadian rhythm, cancer, and metabolic and immune diseases, there are no drugs precisely targeting RORα on the market. Several pharmaceutical companies are dedicated to the discovery and development of using natural products as selective agonists of RORα. For example, GENFIT and BICOLL identified the first potent bioavailable natural product neoruscogenin as a specific agonist of RORα, and they are developing neoruscogenin as a therapeutic drug for autoimmune disorders in the preclinical stage76. Therefore, the development of drugs targeting RORα in the future will be a significant and valuable achievement for treating related diseases.
Acknowledgements
This study was supported by the Creative Research Initiatives Program (Research Center for Epigenetics Code and Diseases, NRF-2017R1A3B1023387) and Samsung Research Funding & Incubation Center of Samsung Electronics (SRFC-MA1502-51) to S.H.B., the Basic Science Research Program (NRF-2021R1C1C1008780) to J.M.L., and the Basic Science Research Program (NRF-2020R1C1C1010489) to H.K., from the National Research Foundation (NRF) grant funded by the Korean government. The figures were generated using BioRender, with a paid license for publication.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Hyunkyung Kim, Email: hyunkkim@korea.ac.kr.
Sung Hee Baek, Email: sbaek@snu.ac.kr.
References
- 1.Park SC, Park IG, Kim H, Lee JM. N-terminal domain mediated regulation of RORalpha1 inhibits invasive growth in prostate cancer. Int. J. Mol. Sci. 2019;20:1684. doi: 10.3390/ijms20071684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Song H, et al. Isoform-specific lysine methylation of RORalpha2 by SETD7 is required for association of the TIP60 coactivator complex in prostate cancer progression. Int. J. Mol. Sci. 2020;21:1622. doi: 10.3390/ijms21051622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giguère V, McBroom LD, Flock G. Determinants of target gene specificity for ROR alpha 1: monomeric DNA binding by an orphan nuclear receptor. Mol. Cell. Biol. 1995;15:2517–2526. doi: 10.1128/MCB.15.5.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harding HP, Atkins GB, Jaffe AB, Seo WJ, Lazar MA. Transcriptional activation and repression by RORalpha, an orphan nuclear receptor required for cerebellar development. Mol. Endocrinol. 1997;11:1737–1746. doi: 10.1210/mend.11.11.0002. [DOI] [PubMed] [Google Scholar]
- 5.Atkins GB, et al. Coactivators for the orphan nuclear receptor RORalpha. Mol. Endocrinol. 1999;13:1550–1557. doi: 10.1210/mend.13.9.0343. [DOI] [PubMed] [Google Scholar]
- 6.Kumar N, et al. Identification of SR3335 (ML-176): a synthetic RORalpha selective inverse agonist. ACS Chem. Biol. 2011;6:218–222. doi: 10.1021/cb1002762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y, Solt LA, Kojetin DJ, Burris TP. Regulation of p53 stability and apoptosis by a ROR agonist. PLoS ONE. 2012;7:e34921. doi: 10.1371/journal.pone.0034921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Y, et al. Retinoic acid receptor-related orphan receptor alpha stimulates adipose tissue inflammation by modulating endoplasmic reticulum stress. J. Biol. Chem. 2017;292:13959–13969. doi: 10.1074/jbc.M117.782391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cai J, Jiao X, Fang Y, Yu X, Ding X. The orphan nuclear receptor RORalpha is a potential endogenous protector in renal ischemia/reperfusion injury. FASEB J. 2019;33:5704–5715. doi: 10.1096/fj.201802248R. [DOI] [PubMed] [Google Scholar]
- 10.Lee JE, et al. A critical regulation of Th2 cell responses by RORalpha in allergic asthma. Sci. China Life Sci. 2021;64:1326–1335. doi: 10.1007/s11427-020-1825-3. [DOI] [PubMed] [Google Scholar]
- 11.Hamilton BA, et al. Disruption of the nuclear hormone receptor RORalpha in staggerer mice. Nature. 1996;379:736–739. doi: 10.1038/379736a0. [DOI] [PubMed] [Google Scholar]
- 12.Gold DA, Gent PM, Hamilton BA. ROR alpha in genetic control of cerebellum development: 50 staggering years. Brain Res. 2007;1140:19–25. doi: 10.1016/j.brainres.2005.11.080. [DOI] [PubMed] [Google Scholar]
- 13.Steinmayr M, et al. Staggerer phenotype in retinoid-related orphan receptor alpha-deficient mice. Proc. Natl Acad. Sci. USA. 1998;95:3960–3965. doi: 10.1073/pnas.95.7.3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dai J, Brooks Y, Lefort K, Getsios S, Dotto GP. The retinoid-related orphan receptor RORalpha promotes keratinocyte differentiation via FOXN1. PLoS ONE. 2013;8:e70392. doi: 10.1371/journal.pone.0070392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee JM, et al. RORalpha attenuates Wnt/beta-catenin signaling by PKCalpha-dependent phosphorylation in colon cancer. Mol. Cell. 2010;37:183–195. doi: 10.1016/j.molcel.2009.12.022. [DOI] [PubMed] [Google Scholar]
- 16.Moretti RM, Montagnani Marelli M, Sala A, Motta M, Limonta P. Activation of the orphan nuclear receptor RORalpha counteracts the proliferative effect of fatty acids on prostate cancer cells: crucial role of 5-lipoxygenase. Int. J. Cancer. 2004;112:87–93. doi: 10.1002/ijc.20387. [DOI] [PubMed] [Google Scholar]
- 17.Byun JK, et al. Retinoic acid-related orphan receptor alpha reprograms glucose metabolism in glutamine-deficient hepatoma cells. Hepatology. 2015;61:953–964. doi: 10.1002/hep.27577. [DOI] [PubMed] [Google Scholar]
- 18.Xiong G, Wang C, Evers BM, Zhou BP, Xu R. RORalpha suppresses breast tumor invasion by inducing SEMA3F expression. Cancer Res. 2012;72:1728–1739. doi: 10.1158/0008-5472.CAN-11-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Slominski AT, et al. On the role of classical and novel forms of vitamin D in melanoma progression and management. J. Steroid Biochem. Mol. Biol. 2018;177:159–170. doi: 10.1016/j.jsbmb.2017.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kopmels B, et al. Evidence for a hyperexcitability state of staggerer mutant mice macrophages. J. Neurochem. 1992;58:192–199. doi: 10.1111/j.1471-4159.1992.tb09295.x. [DOI] [PubMed] [Google Scholar]
- 21.Han YH, et al. RORalpha induces KLF4-mediated M2 polarization in the liver macrophages that protect against nonalcoholic steatohepatitis. Cell Rep. 2017;20:124–135. doi: 10.1016/j.celrep.2017.06.017. [DOI] [PubMed] [Google Scholar]
- 22.Oh SK, et al. RORalpha is crucial for attenuated inflammatory response to maintain intestinal homeostasis. Proc. Natl Acad. Sci. USA. 2019;116:21140–21149. doi: 10.1073/pnas.1907595116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim K, et al. RORalpha controls hepatic lipid homeostasis via negative regulation of PPARgamma transcriptional network. Nat. Commun. 2017;8:162. doi: 10.1038/s41467-017-00215-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Power TE, Robinson J. Cancers related to genetic mutations: important psychosocial issues for Canadian family physicians. Can. Fam. Physician. 2006;52:1425–1431. [PMC free article] [PubMed] [Google Scholar]
- 25.Tollis M, Schneider-Utaka AK, Maley CC. The evolution of human cancer gene duplications across mammals. Mol. Biol. Evol. 2020;37:2875–2886. doi: 10.1093/molbev/msaa125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thompson SL, Compton DA. Chromosomes and cancer cells. Chromosome Res. 2011;19:433–444. doi: 10.1007/s10577-010-9179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nair SS, Kumar R. Chromatin remodeling in cancer: a gateway to regulate gene transcription. Mol. Oncol. 2012;6:611–619. doi: 10.1016/j.molonc.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anderson MW, Reynolds SH, You M, Maronpot RM. Role of proto-oncogene activation in carcinogenesis. Environ. Health Perspect. 1992;98:13–24. doi: 10.1289/ehp.929813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rak J, Yu JL, Klement G, Kerbel RS. Oncogenes and angiogenesis: signaling three-dimensional tumor growth. J. Investig. Dermatol. Symp. Proc. 2000;5:24–33. doi: 10.1046/j.1087-0024.2000.00012.x. [DOI] [PubMed] [Google Scholar]
- 30.Pedraza-Farina LG. Mechanisms of oncogenic cooperation in cancer initiation and metastasis. Yale J. Biol. Med. 2006;79:95–103. [PMC free article] [PubMed] [Google Scholar]
- 31.Gupta S, Takebe N, Lorusso P. Targeting the Hedgehog pathway in cancer. Ther. Adv. Med. Oncol. 2010;2:237–250. doi: 10.1177/1758834010366430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sever R, Brugge JS. Signal transduction in cancer. Cold Spring Harb. Perspect. Med. 2015;5:a006098. doi: 10.1101/cshperspect.a006098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berrabah W, Aumercier P, Lefebvre P, Staels B. Control of nuclear receptor activities in metabolism by post-translational modifications. FEBS Lett. 2011;585:1640–1650. doi: 10.1016/j.febslet.2011.03.066. [DOI] [PubMed] [Google Scholar]
- 34.Flotho A, Melchior F. Sumoylation: a regulatory protein modification in health and disease. Annu. Rev. Biochem. 2013;82:357–385. doi: 10.1146/annurev-biochem-061909-093311. [DOI] [PubMed] [Google Scholar]
- 35.Hwang EJ, et al. SUMOylation of RORalpha potentiates transcriptional activation function. Biochem. Biophys. Res. Commun. 2009;378:513–517. doi: 10.1016/j.bbrc.2008.11.072. [DOI] [PubMed] [Google Scholar]
- 36.Giguère V, et al. Isoform-specific amino-terminal domains dictate DNA-binding properties of ROR alpha, a novel family of orphan hormone nuclear receptors. Genes Dev. 1994;8:538–553. doi: 10.1101/gad.8.5.538. [DOI] [PubMed] [Google Scholar]
- 37.Katoh M. Canonical and non-canonical WNT signaling in cancer stem cells and their niches: cellular heterogeneity, omics reprogramming, targeted therapy and tumor plasticity (Review) Int. J. Oncol. 2017;51:1357–1369. doi: 10.3892/ijo.2017.4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang L, Wang H, Duan X, Dai P, Li J. Comprehensive analysis of the canonical and non-canonical Wnt signaling pathways in gastric cancer. Dig. Dis. Sci. 2019;64:2830–2842. doi: 10.1007/s10620-019-05606-6. [DOI] [PubMed] [Google Scholar]
- 39.Bartkova J, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 40.Yin Y, Stephen CW, Luciani MG, Fahraeus R. p53 Stability and activity is regulated by Mdm2-mediated induction of alternative p53 translation products. Nat. Cell Biol. 2002;4:462–467. doi: 10.1038/ncb801. [DOI] [PubMed] [Google Scholar]
- 41.Brooks CL, Gu W. p53 ubiquitination: Mdm2 and beyond. Mol. Cell. 2006;21:307–315. doi: 10.1016/j.molcel.2006.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim EJ, et al. Retinoic acid receptor-related orphan receptor alpha-induced activation of adenosine monophosphate-activated protein kinase results in attenuation of hepatic steatosis. Hepatology. 2012;55:1379–1388. doi: 10.1002/hep.25529. [DOI] [PubMed] [Google Scholar]
- 43.Buckley CD, et al. Fibroblasts regulate the switch from acute resolving to chronic persistent inflammation. Trends Immunol. 2001;22:199–204. doi: 10.1016/S1471-4906(01)01863-4. [DOI] [PubMed] [Google Scholar]
- 44.Chen F, Castranova V, Shi X, Demers LM. New insights into the role of nuclear factor-kappaB, a ubiquitous transcription factor in the initiation of diseases. Clin. Chem. 1999;45:7–17. doi: 10.1093/clinchem/45.1.7. [DOI] [PubMed] [Google Scholar]
- 45.Baldwin AS., Jr The NF-kappa B and I kappa B proteins: new discoveries and insights. Annu. Rev. Immunol. 1996;14:649–683. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- 46.Marx J. Cancer research. Inflammation and cancer: the link grows stronger. Science. 2004;306:966–968. doi: 10.1126/science.306.5698.966. [DOI] [PubMed] [Google Scholar]
- 47.Han S, et al. ROR alpha protects against LPS-induced inflammation by down-regulating SIRT1/NF-kappa B pathway. Arch. Biochem. Biophys. 2019;668:1–8. doi: 10.1016/j.abb.2019.05.003. [DOI] [PubMed] [Google Scholar]
- 48.Barry AE, et al. Hepatic stellate cells and hepatocarcinogenesis. Front. Cell. Dev. Biol. 2020;8:709. doi: 10.3389/fcell.2020.00709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee IK, et al. RORalpha regulates cholesterol metabolism of CD8(+) T cells for anticancer immunity. Cancers (Basel) 2020;12:1733. doi: 10.3390/cancers12071733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen XR, et al. Mature Purkinje cells require the retinoic acid-related orphan receptor-alpha (RORalpha) to maintain climbing fiber mono-innervation and other adult characteristics. J. Neurosci. 2013;33:9546–9562. doi: 10.1523/JNEUROSCI.2977-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Janmaat S, et al. Age-related Purkinje cell death is steroid dependent: RORalpha haplo-insufficiency impairs plasma and cerebellar steroids and Purkinje cell survival. Age (Dordr.). 2011;33:565–578. doi: 10.1007/s11357-010-9203-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takeo YH, Kakegawa W, Miura E, Yuzaki M. RORalpha regulates multiple aspects of dendrite development in cerebellar Purkinje cells in vivo. J. Neurosci. 2015;35:12518–12534. doi: 10.1523/JNEUROSCI.0075-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gold DA, et al. RORalpha coordinates reciprocal signaling in cerebellar development through sonic hedgehog and calcium-dependent pathways. Neuron. 2003;40:1119–1131. doi: 10.1016/S0896-6273(03)00769-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Akashi M, Takumi T. The orphan nuclear receptor RORalpha regulates circadian transcription of the mammalian core-clock Bmal1. Nat. Struct. Mol. Biol. 2005;12:441–448. doi: 10.1038/nsmb925. [DOI] [PubMed] [Google Scholar]
- 55.Guillaumond F, Dardente H, Giguère V, Cermakian N. Differential control of Bmal1 circadian transcription by REV-ERB and ROR nuclear receptors. J. Biol. Rhythms. 2005;20:391–403. doi: 10.1177/0748730405277232. [DOI] [PubMed] [Google Scholar]
- 56.Forman BM, et al. Cross-talk among ROR alpha 1 and the Rev-erb family of orphan nuclear receptors. Mol. Endocrinol. 1994;8:1253–1261. doi: 10.1210/mend.8.9.7838158. [DOI] [PubMed] [Google Scholar]
- 57.Liu C, Li S, Liu T, Borjigin J, Lin JD. Transcriptional coactivator PGC-1alpha integrates the mammalian clock and energy metabolism. Nature. 2007;447:477–481. doi: 10.1038/nature05767. [DOI] [PubMed] [Google Scholar]
- 58.Crumbley C, Wang Y, Kojetin DJ, Burris TP. Characterization of the core mammalian clock component, NPAS2, as a REV-ERBalpha/RORalpha target gene. J. Biol. Chem. 2010;285:35386–35392. doi: 10.1074/jbc.M110.129288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Poliandri AH, et al. Modulation of clock gene expression by the transcriptional coregulator receptor interacting protein 140 (RIP140) J. Biol. Rhythms. 2011;26:187–199. doi: 10.1177/0748730411401579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mocellin S, Tropea S, Benna C, Rossi CR. Circadian pathway genetic variation and cancer risk: evidence from genome-wide association studies. BMC Med. 2018;16:20. doi: 10.1186/s12916-018-1010-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gu D, et al. Circadian clock pathway genes associated with colorectal cancer risk and prognosis. Arch. Toxicol. 2018;92:2681–2689. doi: 10.1007/s00204-018-2251-7. [DOI] [PubMed] [Google Scholar]
- 62.Qiu MJ, et al. Research on circadian clock genes in common abdominal malignant tumors. Chronobiol. Int. 2019;36:906–918. doi: 10.1080/07420528.2018.1477792. [DOI] [PubMed] [Google Scholar]
- 63.Lau P, et al. The orphan nuclear receptor, RORalpha, regulates gene expression that controls lipid metabolism: staggerer (SG/SG) mice are resistant to diet-induced obesity. J. Biol. Chem. 2008;283:18411–18421. doi: 10.1074/jbc.M710526200. [DOI] [PubMed] [Google Scholar]
- 64.Kadiri S, et al. The nuclear retinoid-related orphan receptor-alpha regulates adipose tissue glyceroneogenesis in addition to hepatic gluconeogenesis. Am. J. Physiol. Endocrinol. Metab. 2015;309:E105–E114. doi: 10.1152/ajpendo.00518.2014. [DOI] [PubMed] [Google Scholar]
- 65.Lau P, et al. Roralpha deficiency and decreased adiposity are associated with induction of thermogenic gene expression in subcutaneous white adipose and brown adipose tissue. Am. J. Physiol. Endocrinol. Metab. 2015;308:E159–E171. doi: 10.1152/ajpendo.00056.2014. [DOI] [PubMed] [Google Scholar]
- 66.Zhang Y, et al. The hepatic circadian clock fine-tunes the lipogenic response to feeding through RORalpha/gamma. Genes Dev. 2017;31:1202–1211. doi: 10.1101/gad.302323.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schultz JR, et al. Role of LXRs in control of lipogenesis. Genes Dev. 2000;14:2831–2838. doi: 10.1101/gad.850400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chai C, et al. Agonist of RORA attenuates nonalcoholic fatty liver progression in mice via up-regulation of microRNA 122. Gastroenterology. 2020;159:999–1014.e9. doi: 10.1053/j.gastro.2020.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Panteleeva AA, et al. Expression of genes encoding nuclear factors PPARgamma, LXRbeta, and RORalpha in epicardial and subcutaneous adipose tissues in patients with coronary heart disease. Bull. Exp. Biol. Med. 2021;170:654–657. doi: 10.1007/s10517-021-05126-2. [DOI] [PubMed] [Google Scholar]
- 70.Panteleeva AA, et al. [The expression of genes encoding ABCA1 and ABCG1 transporters and PPARgamma, LXRbeta, and RORalpha transcriptional factors in subcutaneous and visceral adipose tissue in women with metabolic syndrome] Mol. Biol. (Mosk.) 2021;55:64–74. doi: 10.1134/S0026893321010131. [DOI] [PubMed] [Google Scholar]
- 71.Verlande A, Masri S. Circadian clocks and cancer: timekeeping governs cellular metabolism. Trends Endocrinol. Metab. 2019;30:445–458. doi: 10.1016/j.tem.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Masri S, Sassone-Corsi P. The emerging link between cancer, metabolism, and circadian rhythms. Nat. Med. 2018;24:1795–1803. doi: 10.1038/s41591-018-0271-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang Y, Luo XY, Wu DH, Xu Y. ROR nuclear receptors: structures, related diseases, and drug discovery. Acta Pharmacol. Sin. 2015;36:71–87. doi: 10.1038/aps.2014.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Han SJ, Lonard DM, O’Malley BW. Multi-modulation of nuclear receptor coactivators through posttranslational modifications. Trends Endocrinol. Metab. 2009;20:8–15. doi: 10.1016/j.tem.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Filtz TM, Vogel WK, Leid M. Regulation of transcription factor activity by interconnected post-translational modifications. Trends Pharmacol. Sci. 2014;35:76–85. doi: 10.1016/j.tips.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Helleboid S, et al. The identification of naturally occurring neoruscogenin as a bioavailable, potent, and high-affinity agonist of the nuclear receptor RORalpha (NR1F1) J. Biomol. Screen. 2014;19:399–406. doi: 10.1177/1087057113497095. [DOI] [PubMed] [Google Scholar]