Abstract
Cadmium (Cd) accumulates with aging and is elevated in long-lived species. Metallothioneins (MTs), small cysteine-rich proteins involved in metal homeostasis and Cd detoxification, are known to be related to longevity. However, the relationship between Cd accumulation, the role of MTs, and aging is currently unclear. Specifically, we do not know if long-lived species evolved an efficient metal stress response by upregulating their MT levels to reduce the toxic effects of environmental pollutants, such as Cd, that accumulate over their longer life span. It is also unknown if the number of MT genes, their expression, or both protect the organisms from potentially damaging effects during aging. To address these questions, we reanalyzed several cross-species studies and obtained data on MT expression and Cd accumulation in long-lived mouse models. We confirmed a relationship between species maximum life span in captive mammals and their Cd content in liver and kidney. We found that although the number of MT genes does not affect longevity, gene expression and protein amount of specific MT paralogs are strongly related to life span in mammals. MT expression rather than gene number may influence the high Cd levels and longevity of some species. In support of this, we found that overexpression of MT-1 accelerated Cd accumulation in mice and that tissue Cd was higher in long-lived mouse strains with high MT expression. We conclude that long-lived species have evolved a more efficient stress response by upregulating the expression of MT genes in presence of Cd, which contributes to elevated tissue Cd levels.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11357-021-00393-3.
Keywords: Aging, Comparative biogerontology, Cadmium, Mammals, Metallothionein, Longevity
Introduction
Cadmium (Cd) is a toxic transition metal with ubiquitous distribution in the earth’s crust and is enriched in the food supply due to bioaccumulation and significant anthropogenic pollution [64]. Cd has no known biological function and it is classified as a group I carcinogen by the International Agency for Research on Cancer (IARC). Recent studies have linked modestly elevated urinary Cd excretion — a reliable measure of overall exposure — with cancer, cardiovascular disease, and all-cause mortality [47]. Cadmium accumulation, especially in liver and kidney, has also been associated with age-related pathologies such as hypertension, cardiovascular dysfunction, osteoporosis, and telomere-shortening [1, 62, 88]. Consistent with a relationship between Cd and aging, Cd accumulation has been found elevated in the liver and kidney of long-lived mammals [51] and accumulated with age in the kidney of horses [39]. Long-lived species may accumulate more Cd than short-lived species due to greater consumption of food (and thus Cd) and the extremely long half-life of Cd in the organism (estimated from 10 to 30 years in humans) [38]. Although there is no unifying theory to explain the toxicity of Cd and its role in aging, it is believed that even low-level exposure to environmental Cd contributes to age-related renal and vascular dysfunction through disruption of zinc (Zn) and redox homeostasis [20, 62]. Increased Cd levels in long-lived species suggest that lower uptake of this metal is not used as a protection mechanism against Cd toxicity by organisms that live longer, although fibroblasts of long-lived species were found more resistant to Cd [31]. Therefore, considering that Cd has no known biological function, it is toxic and its tissue levels were found to be elevated in long-lived species [51]; it would be expected that species that live longer would have some protective mechanism to sequester this metal and reduce its negative effects on these organisms, allowing them to live longer.
One of the best characterized intracellular responses to Cd is the induction of metallothionein (MT) gene expression, coding for cysteine-rich metal-binding proteins. These multifunctional proteins not only protect from Cd and heavy metal toxicity but also regulate copper (Cu) and zinc (Zn) homeostasis, act as antioxidants and as redox sensors [14, 55], and are functionally related to the insulin signaling pathway [29, 34, 72, 75]. The MT gene superfamily has undergone numerous gene duplication and loss events across vertebrates. Number of paralogs, isoforms, pseudogenes, and polymorphism at MT genes is therefore highly variable across species, with the highest number of MT genes occurring in humans and the lowest number in fish and amphibians [34, 72]. This variation in terms of number of functional genes across species has resulted in subfunctionalization, with different MT proteins having distinct binding affinities for metals and some of them being ubiquitous and some being tissue specific [34, 75]. The presence of several paralogs with similar or redundant functions is a challenge for genomic and transcriptomic comparative research, especially when a reference genome is not available [10, 56, 82]. This challenge also affects studies aimed at establishing the genetic determinants of longevity, and a targeted or focused approach on a single gene belonging to a gene family is therefore usually preferred in functional studies.
Multiple lines of evidence suggest that MTs are related to longevity. MT expression is elevated in long-lived worms, flies [75], many long-lived mouse models, and during aging [76]. Several studies have highlighted that overexpression or induction of MTs extends mouse life span [52, 84], whereas double knockout of MT-1 and MT-2 shortens life span [41] and promotes cancer [53]. In humans, single nucleotide polymorphisms in MT genes are linked to age-related diseases [46] and exceptional longevity [13]. Moreover, some MT isoforms are elevated in peripheral blood mononuclear cells from centenarian offspring compared to controls and this is associated with altered Zn homeostasis [25].
However, the role of MTs in aging, especially their relationship with Cd accumulation during aging and in long-lived species, is not fully understood. Cd accumulation varies among tissues, sexes, age, and species [44, 58, 59]. For example, in non-human primates and horses, Cd concentration was highly variable but generally higher in older individuals, and in kidney than liver, while Zn was found higher in liver than kidney [39, 58]. MTs have higher affinity for Cd than Zn — thus displacing Zn when Cd is available [62] — so that lower Zn concentrations correlate with higher expression of MTs accumulating Cd [39, 58]. Furthermore, in horses while the level of MTs followed Cd concentration in the kidney, no clear relationship between MTs and Cd levels was observed in the liver [39]. Indeed, liver is the primary target of Cd in acute toxicity, whereas kidneys are the respective major target of Cd in chronic toxicity [67]. These data suggest that Cd accumulation during aging may be mediated by higher expression of MTs in the kidney but not in the liver. Longer-lived species may therefore have increased MT gene expression in comparison to shorter-lived species to allow them to deal with increased Cd accumulation experienced during aging. However, to our knowledge, there are currently no data testing this hypothesis across species with distinct life spans. Furthermore, considering that distinct species have different numbers of MT genes (paralogs), we currently do not know if gene expansion in this gene family may be related to increased longevity — i.e., species with a higher number of MT paralogs can live longer because of improved metal homeodynamic processes — and if Cd accumulation during aging similarly affects the expression of different MT paralogs or if some MT paralogs are more responsive than others.
To address these hypotheses, here we combine meta-analyses of already published cross-species datasets on Cd accumulation and MT expression with our own data on Cd accumulation in long-lived mice and with comparative genomic approaches on the number of MT paralogs across vertebrates. Our results confirm that independently of age at sampling, Cd accumulates during aging in mammalian species living in controlled environments. We found that MT gene expression and protein levels but not MT paralog numbers are related to increased life span in mammals and that Cd accumulates with aging as a consequence of the increased MT expression.
Progressive loss of metallostasis (metal homeostasis) appears to be a conserved phenomenon across aging of multiple species [3, 6, 26], and there is evidence that alterations in metal abundance modulate life span in most common animal models used in aging studies, such as rats [68], mice [52], worms [42], and flies [85]. Our work reveals that MT gene expression and protein level rather than MT gene number play a crucial role in the maintenance of metallostasis suggesting that these proteins could be a useful target for interventions to improve life span. To our knowledge, our work represents the first case in which we specifically tease apart the contribution of distinct paralogs vs. their gene expression and protein levels for genes known to be involved in aging-related processes.
Methods
Data collection from repositories and databases
Species maximum life span and body mass data
Species maximum life span (MLS) and body mass were determined from the AnAge database [78] except in a few cases when MLS and body mass data were estimated from other sources — i.e., bird banding and published literature (Supplementary data 1).
Cd accumulation
We searched the literature for datasets which measured species-specific Cd accumulation in liver and kidney in mammals and birds kept under standardized conditions (captivity) or living in the wild. We identified four datasets with at least 10 species each to allow statistical analysis (Table 1). Information about studied vertebrate group (mammals or birds or both), sex, and life stage of the animals in each dataset are listed in Table 1. Age at sampling — when available — was used to take into account the influence of different sampled ages on Cd accumulation.
Table 1.
Partial correlation between Cd accumulation and maximum life span taking body mass into account in captive (zoo) and wild animals
| Dataset | NKN1 | CEETV dataset2 | Frank 19863 | Eisler’s compendium |
|---|---|---|---|---|
| Vertebrate | Mammal and bird species | Mammal and bird species | Mammal and bird species | Mammal and bird species |
| Method | AAS | Various | AAS | Various |
| Geography | Japan | N. America | Sweden | Marine |
| Age | Adult | Adult | Any | Adult |
| Status | Captive | Wild | Wild | Wild |
| Sex | Combined/split | Combined | Combined | Combined |
| Kidney | ||||
| Mammals (n)* | 53 | 11 | 11 | 25 |
| Pearson r | 0.51 (< 0.001) | 0.78 (< 0.01) | 0.12 (0.72) | 0.08 (0.69) |
| Birds (n)* | Not used | 31 | 29 | 0 |
| Pearson r | NA | 0.23 (0.20) | − 0.30 (0.11) | NA |
| Males (n)* | 32 | NA | NA | NA |
| Pearson r | 0.43 (< 0.05) | NA | NA | NA |
| Females (n)* | 44 | NA | NA | NA |
| Pearson r | 0.62 (< 0.0001) | NA | NA | NA |
| Liver | ||||
| Mammals (n)* | 53 | 11 | 11 | 20 |
| Pearson r | 0.50 (< 0.001) | 0.36 (0.27) | 0.25 (0.45) | − 0.06 (0.79) |
| Birds (n)* | Not used | 39 | 29 | 11 |
| Pearson r | NA | 0.42 (0.42) | − 0.24 (0.21) | 0.39 (0.23) |
| Males (n)* | 32 | NA | NA | NA |
| Pearson r | 0.55 (< 0.01) | NA | NA | NA |
| Females (n)* | 44 | NA | NA | NA |
| Pearson r | 0.58 (< 0.0001) | NA | NA | NA |
1Animals died of predominantly natural causes. In this study, we pooled and reanalyzed data from Ninomiya et al. [58], Koizumi et al. [44], and Ninomiya et al. [59]. For our analyses, we did not use the birds included in one of these studies as only five species were included
2CEETV is a “contaminant database,” hence results may be more heterogeneous than for other datasets
3Animals were found dead and the post-mortem delay is not known, making interpretation difficult
*Most, but not all, species are represented by more than one individual; thus for some dataset, the sum of the numbers reported for females and males may be higher than the total number (see Supplementary Information 1)
All p-values are indicated in parentheses. Significant p-values (< 0.05) are in bold. NA, not enough data available; AAS, atomic absorption spectrometry
We reanalyzed cadmium data published by others [44, 58, 59], who reported Cd levels in animals from zoos. The data from wild animals were obtained by using the CEETV database (http://www.pwrc.usgs.gov/contaminants-online/), the Compendium of Trace Metals and Marine Biota [21], and from the study of Frank [23]. We selected the above-mentioned studies and dataset to ensure the consistency of data, including data points expressed as wet-weight, from adult animals and unpolluted areas. Data used for the following analyses can be found as Supplementary data 11.
MT paralog number
Numbers of MT paralogs per species were obtained for mammals with a genome available from ENSEMBL release 96 [35] and with a coverage equal or higher than 6X. We determined the number of MT paralogs by running a search on ENSEMBL for each MT for each species. When MT annotation or gene number in ENSEMBL was doubtful, a BLASTX search was performed on NCBI using the non-annotated coding sequence of the gene. For the analysis of MT paralog number and MLS, we only considered species for which MT paralog number was consistent between ENSEMBL and NCBI search. Data were obtained for 31 species of mammals (Supplementary data 1).
MT gene expression and MT protein level
We searched the literature for cross-species studies of MT with more than 10 species to allow statistical analyses. We obtained MT expression from RNA-Seq studies of non-human primates [65] and from a study including mostly mammals [24]. Peng et al. [65] provided FPKM values (fragments per kilobase of exon model per million reads mapped), a normalized estimation of gene expression, for RNA-Seq data from up to 12 species that were mapped to human genes, allowing us to correlate gene expression with non-human primate life span. For the analyses, we summed the FPKM values for the eight functional MT-1 isoforms, MT-2, and MT-3 in each species and excluded MT-4, which has specialized functions in epithelial tissues [20]. We pooled the expression data for the different MT paralogs as all of them are expressed ubiquitously in the body, respond to Cd accumulation and are stress inducible and cytoprotective, and involved in the so-called multistress resistance theory of aging [12, 20, 57, 63, 69]. Since MT expression was skewed towards very high values in some species and some organs, we further normalized the data by log10-transformation. We also considered several other biases including misclassification of MT transcripts due to poor genome annotation. However, most correlations were unchanged whether we included MT pseudogenes or MT-3 in our analysis and were independent of the gene annotation framework chosen (Supporting Information Table S1). Although Peng et al. [65] had expression data across different tissues, we report here only results for kidney and brain. Correlation between MT gene expression and MLS in other tissues is reported in Supporting Information Table S1.
Raw data from Fushan et al. [24] were downloaded from the GEO database [4]: GSE30352 (H. sapiens, M. musculus), GSE29629 (M. fascicularis), and GSE43013 (M. musculus and all other species). We only considered species available in ENSEMBL release 96 [35]. Assembly genomes, gtf files, and gene length information were obtained from ENSEMBL. Sequencing reads were filtered and trimmed with trimmomatic v0.39 and were then aligned to the reference genomes using STAR 2.7.0d [17]. Read counts were obtained from STAR, while species and genes that had fewer than 25 read counts were excluded from further analysis. BiomaRt [19] or UNIPROT [80]were used to query ENSEMBL and identify genes annotated as MTs. DESeq2 [50] was used to normalize the read counts, taking the gene lengths into account, before correlating individual MT genes with MLS. Read counts were used to calculate FPKM values and pooled analysis of MT transcripts was performed as described above for the Peng et al. [65] dataset. To assess the influence of each single MT gene (paralog) expression on MLS, we also used gene expression data from each paralog separately.
Data on MT protein levels were obtained from the literature from papers using the Cd-saturation and silver-saturation method [70]. For this purpose, we obtained data from mammalian liver from Henry et al. [33] and also built a pooled dataset using data obtained only by Cd-saturation from Teranishi et al. [79], Koizumi et al. [43], and Ninomiya et al. [58] (data from liver and kidney of primates and horses).
Analysis of Cd and MT in tissue samples from long-lived mice
Long-lived mice strains
All samples used in this study consisted of leftover samples stored in mouse tissues biobanks. These samples originated from different studies which were originally approved by the respective local ethical committees.
Ames dwarf
Long-lived Ames mice lack plasma growth hormone, prolactin, and thyrotropin as a consequence of a loss of function mutation at Prop-1 locus [8]. Liver and kidney tissues from long-lived Ames dwarf (Prop1 df/df) and littermate control mice were obtained from cryopreserved samples at the Research Institute of Wildlife Ecology [81]. Mice of the same sex were pair-housed and the absence of infections was continuously monitored through sentinel mice. All animals were females, with the exception of one dwarf, had been fed rodent chow (v118, Ssniff) ad libitum, and were sacrificed between 12 and 17 months of age.
Calorie-restricted and rapamycin-fed mice
Calorie restriction [66] and rapamycin feeding [32] are widely accepted models of interventions to extend mouse life span. Cryopreserved liver tissues from female calorie-restricted (CR) and rapamycin-fed UM-HET3 mice as well as from their respective controls (also only females) were kindly provided by Richard Miller (University of Michigan). These mice were housed under specific pathogen free conditions and received Purina 5LG6 chow [32]. Treatments started from the age of 4 months with one group receiving 60% of ad libitum caloric intake and another 14 ppm of microencapsulated rapamycin mixed in chow. Animals were sacrificed at 12 months of age.
GHRKO mice
GH receptor gene-disrupted knockout mice are characterized by a significant reduction of body size and life span extension [5]. Liver and kidney tissues from male GHR − / − , MDR2 + / − mice (shortened for simplicity to GHRKO from herein) and littermate controls on a 129 Sv/C57BL/6 genetic background were obtained from cryopreserved samples at the University of Vienna. MDR2 + / − mice were reported to be disease-free and used previously as littermate controls of GHR − / − , MDR2 − / − mice [74]. Moreover, the mouse p-glycoprotein MDR2 is involved in phospholipid and bile acid transport with no known role in Cd transport. These mice were housed at the Medical University of Vienna and received rodent chow ad libitum (V1126-000, Ssniff) [74]. Animals were sacrificed at 2 months of age.
MT-Tg mice
MT-1-overexpressing mice (MT-Tg) were originally developed to study physiological mechanisms of protection from Cd [37] and were later discovered to display a higher median and maximum life span (approx. 18% and 13%, respectively) compared to controls [52]. MT-Tg mice have 10- to 20-fold higher basal levels of MT protein in pancreas, liver, and stomach, as well as 2- to sixfold higher MT protein levels in other organs (kidney, intestine, uterus, testes, spleen, heart, and lung) than control mice [37]. Liver and kidney samples from male MT-Tg on a C57BL/6 J background were obtained from cryopreserved samples at the INRCA animal facility (Ancona, Italy). These mice were housed under standard conditions and received rodent chow (Mucedola 4RF24) as described previously [52]. Cryopreserved samples from 24-month-old male C57BL/6 from the Mouse Clinic for Cancer and Aging (Groningen) were used as controls. These mice were housed under standard conditions and received HopeFarms AM-II chow which was later switched to SDS RM1 diet.
Data on Cd content
Cd content data from long-lived mice (and their respective controls; every study/mouse listed below also has a strain-specific control that is not long-lived) are all newly obtained for this study. Liver and kidney samples were digested with 1.5 ml of HNO3 (Merck, Suprapur) in a microwave oven and analyzed by graphite furnace atomic absorption spectrometry (Hitachi Z 8200 Polarized Zeeman AAS) as described previously [28]. The reference material we used was Seronorm Trace Elements Urine L-2 (Lot: 1,403,081). The mean recovery of the reference material was 115 ± 36% across several measurements (n = 8) and all samples were above the limit of detection of 0.25 ng Cd/ml.
MT expression data
For the long-lived Ames mice, MT expression data were newly obtained by qPCR [CFX Connect RT-PCR (Bio-Rad), sybr green] following the protocol of Li et al. [48]. Data on MT gene expression obtained by qPCR for rapamycin fed as well as from CR mice were kindly provided by XL (University of Michigan,PCR conditions and primer sequences can be found in the Supplementary methodological Information). We referred to published data for all other strains and their respective controls including GHRKO [77] and MT-Tg [37] mice.
Statistical analyses
Relationship between Cd tissue levels and maximum life span
Since Cd tissue levels increase with both higher maximum life span and body mass, it is not possible to disentangle these effects without further corrections [73]. We therefore first ran a partial correlation — Pearson correlation — on Cd concentration versus MLS by controlling for body mass (all variables were log10-transformed). We also repeated these analyses comparing females and males across species when data were available (Table 1, Fig. 1B). Finally, because an earlier study attributed elevated Cd in long-lived species to sampling bias, i.e., longer-lived species were also chronologically older [51], we performed an adjustment for age at sampling as information about the age at which sampled animals died or were euthanized were available to us, either as known age at sampling (majority of the cases) or as a numeric estimate provided by the authors. We ran a partial correlation — Pearson correlation — on Cd concentration versus MLS by controlling for age at sampling (all traits log10-transformed). If — as previously reported — Cd accumulates with age independently on body mass and age at sampling, we expect that species with higher MLS than predicted by body mass would also have higher tissue Cd levels.
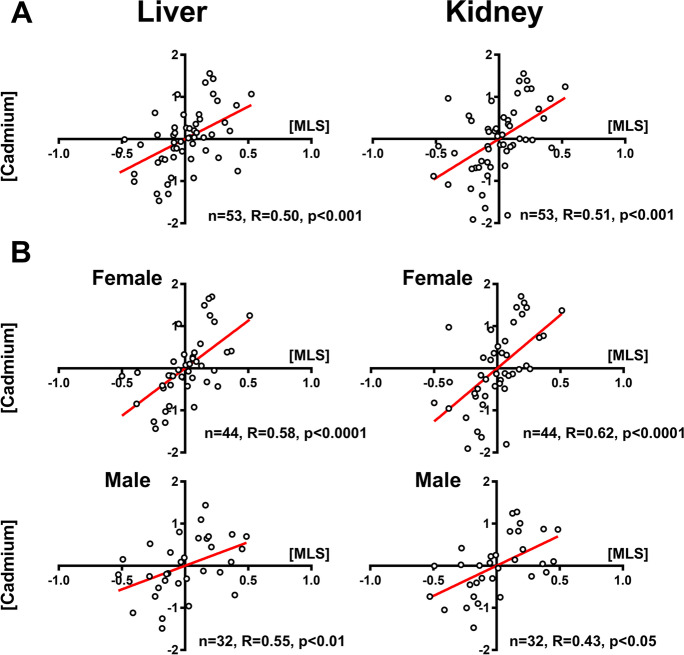
Fig. 1.
Partial correlation of cadmium levels on maximum life span (MLS) taking into account body mass for data obtained on zoo animals. Cadmium (Cd) content in liver and kidney correlates with maximum life span (MLS) in mammals (A). This partial correlation is confirmed when data for females and males are analyzed separately (B). The dataset is pooled from Ninomiya et al. [58], Koizumi et al. [44], and Ninomiya et al. [59] (see Supplementary Information 1). Tissue Cd content and species MLS were log10-transformed. Sample size, Pearson correlation coefficients, and p-values are indicated for each analysis. Regression lines for plots with p < 0.05 are indicated in red
Furthermore, we took into account the influence of phylogenetic relationships on the non-independence of the data — closely related species are expected to show more similarity in traits than phylogenetically distant ones [36] — for datasets with 20 species or more (Table 1), when studying the influence of MLS on Cd accumulation. This approach was applied to data of Cd accumulation in liver and kidney in mammals and birds (see Table 1 for dataset used for this). As body mass and MLS are often linked traits — increased body mass is often associated with longer life spans — we performed an allometric correction using the mammalian equation for all the mammal datasets and a bird equation for the bird datasets [15] and used the log10-transformed corrected MLS data for the analyses. Comparative analyses were carried out using the phylogenetic generalized least squares method (PGLS) in R computing environment using the CAPER package [60]. For the analyses on mammals, we used the phylogenetic tree of Bininda-Emonds et al. [7], while for the analyses on the bird datasets, we used two separate bird trees used by Jetz et al. [40]. The analyses were done using both trees independently,results were consistent independently of which tree was used.
Relationship between metallothionein paralogs number, MT expression, MT protein amount, and maximum life span
The influence of MT paralog number for each mammal species considered in this study (Supplementary data 1) on MLS was assessed by taking into account the phylogenetic relationships among the species. Briefly, we used the MLS values after applying the specific allometric correction for mammals to take into account the influence of variation in body mass on the data. Adjusted MLS data were log-transformed before the analysis. We then ran a comparative analysis using the phylogenetic generalized least squares method (PGLS) as described above.
To study the influence of MT gene expression, we used RNA-Seq data [24, 65] and MT protein levels obtained by Cd-saturation and silver-saturation method [33, 43, 58, 79]; data were analyzed separately for each dataset running a correlation analysis with MLS. Furthermore, the influence of the number of expressed MT paralogs on MLS taking into account phylogenetic relationships was estimated using PGLS for the only dataset available with at least 15 species [24]. Phylogenetic relationships were not taken into account for datasets with fewer than 15 species as these were not sufficient to ensure enough statistical power [11]. Moreover, for data from Fushan et al. [24], we built a heatmap of gene expression data for each MT paralog taking into account the influence of MLS across three organs (n = 4 to 15, pheatmap package [45]). The results are shown as a heatmap according to the strength of the relationship (log2-fold change).
Relationship between metallothionein expression and Cd tissue levels in long-lived mice
Most data were normally distributed justifying the use of Pearson for correlation studies. However, we adopted a more conservative approach in all correlation studies by running both Pearson and Spearman rank correlation (only Pearson correlations as shown as results in this manuscript since the results from both methods are concordant). Cd levels in mouse liver and kidney were compared by Welchs’s t-test or ANOVA with Dunnett post-hoc test using Prism software (v7.02). Two outliers were identified by Grubbs’ test (α = 0.05) and excluded from analyses.
Results
Metadata analysis of cadmium accumulation and species maximum life span
We reanalyzed the data on cadmium accumulation (Supplementary data 1) from three published studies [44, 58, 59] on captive animals and confirmed that long-lived mammals accumulated significantly more Cd in the liver and in the kidney than short-lived ones (Table 1 and Fig. 1A). This correlation was consistent and of similar magnitude in both sexes (Table 1 and Fig. 1B), even though females had higher mean Cd levels in the liver than males as also previously observed [44]. An intersex dichotomy of Cd accumulation with aging has been observed also in human retina [83]. These findings may suggest that males and females tune MT in response to the increased inflammatory state of aging in a different way. We considered the influence of age at sampling on estimates of correlation between MLS and sampling age on Cd accumulation, as species with longer life span may have been sampled at older age. We found that independently of age at sampling, the Cd accumulation in the kidney and liver remain significant (Fig. S1, Supplementary data 2).
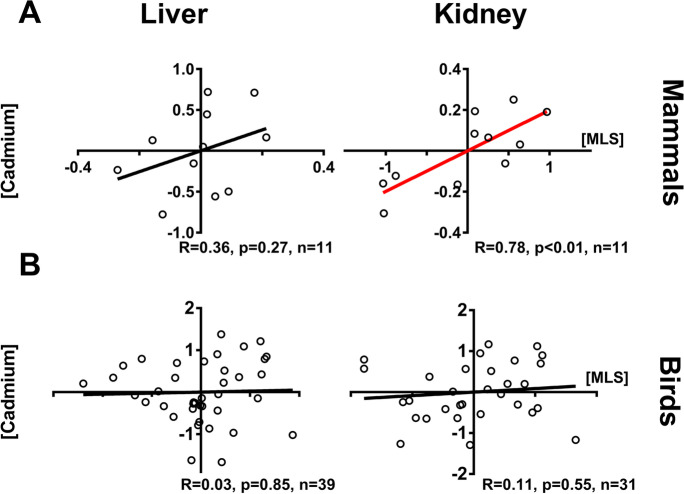
To understand if the results obtained were supported also when wild animals (mammals and birds) were sampled, we used the Contaminant Exposure and Effects Terrestrial Vertebrates (CEETV) database from the U.S. Geological Survey and data from other sources [21, 23] (Table 1 and Supplementary data 1). In mammals, there was a significant correlation between kidney Cd accumulation and MLS, but the same result was not obtained for liver (Fig. 2A, CEETV data). In birds, we saw no correlation between either liver or kidney Cd and MLS (Fig. 2B, CEETV data). The analysis of additional dataset on wild birds and mammals [21, 23] (Table 1 and Supplementary data 1) supported the lack of associations between MLS and tissue Cd (Table 1; Supplementary data 2, Fig. S2), in contrast to what observed in captive animals.
Fig. 2.
Partial correlation of cadmium levels on maximum life span (MLS) taking into account body mass for data obtained on wild animals based on CEETV database (see “Methods” and Supplement 1). Renal cadmium (Cd) content correlates with maximum life span (MLS) in wild mammals whereas the correlation is not significant in liver (A). No significant correlations are seen in birds (B). Tissue Cd content and species MLS were log10-transformed. Sample size, Pearson correlation coefficients, and p-values are indicated for each analysis. Regression lines for plots with p < 0.05 are indicated in red
Analyses conducted taking into account the influence of phylogenetic relationships on the observed variation in Cd accumulation in liver and kidney in captive mammals indicate that this variation is only slightly influenced by phylogeny and partially influenced by life span (λML ~ 0.4, significantly different from λML = 1 in both cases with a p-value < < 0.001; N = 52, t-value = 2.6 and p-value = 0.01, adjusted R2 = 10% in kidney; t-value = 2.42 and p-value = 0.02, adjusted R2 = 9% in liver; dataset NKN Supplementary data 1). The results suggest that other factors, together with phylogeny and variation in longevity in mammals, can contribute to variation in cadmium accumulation. In contrast, we found that in wild birds, phylogenetic relationships are an important factor in explaining variation in Cd accumulation in liver (λML = 0.86, significantly different from λML = 0 and from λML = 1 in both cases with a p-value < < 0.001) and kidney (λML = 0.74, significantly different from λML = 0 with a p-value = 0.002 and from λML = 1 with a p-value < < 0.001), while variation in life span is not (N = 39 for liver, N = 31 for kidney; t-value = 0.3 for liver and 0.06 for kidney, and p-value > 0.75 in both cases; dataset CEETV Supplementary data 1). Unfortunately, due to the relatively low number of species available for mammals from wild animals (< 15, see Supplementary data 1), the relationship of Cd accumulation and longevity taking phylogeny into account in wild mammals cannot be carried out.
Relationship between metallothionein paralogs number, MT expression, MT protein amount, and maximum life span
In order to understand the relationship between higher MT expression and longevity, we assessed the relationship between MT paralog number and MLS across vertebrates and between higher MT expression and MLS in mammals.
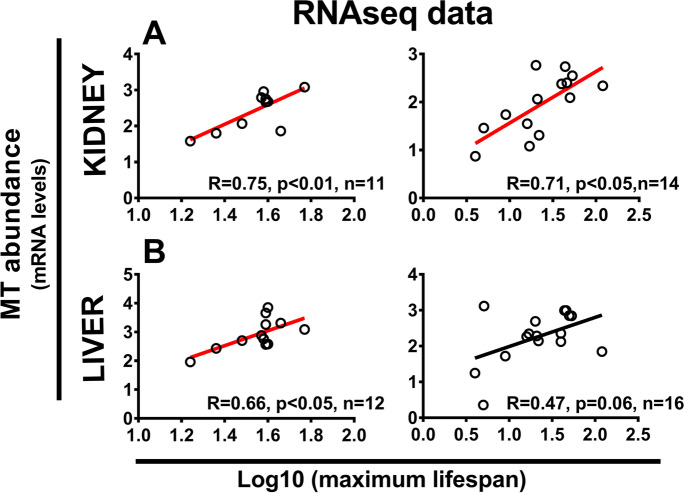
We found that the variation in number of MT paralogs in mammals (Supplementary data 1) strongly depends on phylogenetic relationships (λML = 1, significantly different from λML = 0 with a p-value < < 0.001) and that variation in longevity is not significantly influenced by the number of MT paralogs (N = 31, t-value = 0.70 and p-value = 0.49). We then looked at the relationship between MT gene expression levels and MLS. The analysis of previously collected RNA-Seq data from which we obtained MT gene expression data for MT 1–3 paralogs indicated a significant correlation between kidney MT gene expression and mammalian MLS [Fig. 3; data from Peng et al. [65] and Fushan et al. [24]]. Data from Peng et al. [65], but not Fushan et al. [24], also indicate a significant relationship between MT expression in liver and MLS in mammals (Fig. 3). Although data from Fushan et al. [24] were not found to be significant, the correlation between hepatic MT and MLS in mammals became significant when the analysis was limited to placental mammals (short-tailed opossum excluded,Supplementary data 2, Fig. S3).
Fig. 3.
Pooled metallothionein (MT) expression in kidney (A) and liver (B) is positively correlated with species maximum life span (MLS). Data based on RNA-seq pooled expression of all MT-1, MT-2, and MT-3 genes plotted against primate MLS (left side; data from [65] or mammalian MLS (right side,data from [24]. The species MLS and MT expression levels were log10-transformed and Pearson correlation coefficients are shown. Regression lines for plots with p < 0.05 are indicated in red
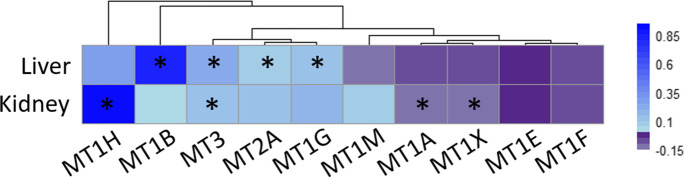
We then tested whether individual expressed MT paralogs or total number of expressed MT paralogs are responsible for the observed relationship between MT expression and MLS. We found that, once phylogenetic relationships are taken into account, while the number of paralog itself is not important, the number and type of MT paralog genes that are expressed influences MLS [N = 15, t-value = 4.9 and p-value < < 0.001, R2 = 64%, data for liver from Fushan et al. [24]]. We found that across liver and kidney MT-3, MT-2A, and MT-1G expression showed the most consistent association with MLS [Fig. 4; data from Fushan et al. [24]]. Furthermore, MT-2A and MT-1G expression was also associated with MLS across both RNA-Seq datasets combined [[24, 65],Table 3, Table S2, Supplementary data 2].
Fig. 4.
Based on the study by Fushan et al. [24], we determined the association (the log2 fold-change of MT counts per unit of MLS in days, multiplied by 1000 for better readability) between individual MT paralogs and species MLS across two organs (n = 4 to 17). The results are shown as a heatmap according to the strength of the relationship (log2-fold change). Full data provided in Table S2. *Indicates significant associations (p < 0.05)
Table 3.
Pearson correlation between gene expression for each MT paralog and MLS in liver and kidney of non-human primates
| Peng et al. [65] | ||
|---|---|---|
| Liver (n = 12 species) | Kidney (n = 11 species) | |
| MT3 | 0.29 | 0.10 |
| MT2A | 0.51 | 0.55 |
| MT1A | 0.51 | − 0.41 |
| MT1B | 0.26 | 0.44 |
| MT1E | 0.59 | 0.64 |
| MT1F | 0.45 | 0.59 |
| MT1G | 0.35 | 0.34 |
| MT1H | 0.61 | 0.62 |
| MT1M | 0.28 | 0.26 |
| MT1X | 0.79 | 0.81 |
p < 0.05 is underlined and bolded
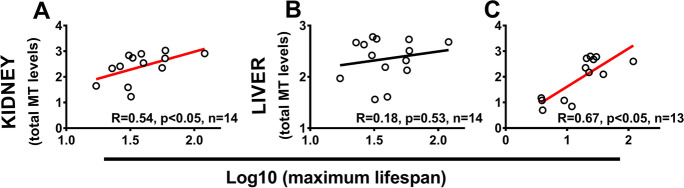
Finally, we also tested the relationship between MT proteins and MLS by using the available literature data on MT protein levels estimated by the Cd-saturation method [data from Henry et al. [33], and combined data from Koizumi et al. [43],Teranishi et al. [79],Ninomiya et al. [58]]. In primates and horses (one species represented by equus caballus), a relationship between kidney MT protein levels — but not liver — and MLS was observed (Fig. 5A and B). When different species of mammals are considered [33], liver MT protein levels were found to be correlated with MLS (Fig. 5C).
Fig. 5.
Relationship between MT protein levels, as estimated by cadmium-saturation method, and MLS in mammals (A–C). Combined data (MT measured only by Cd-saturation method) for horse and primates from Teranishi et al. [79], Koizumi et al. [43], and Ninomiya et al. [58] for kidney (A) and liver (B). C Data for liver across different mammalian species from a dataset where MT have been measured with both silver and Cd-saturation method [33]. Regression lines for plots with p < 0.05 are indicated in red. Data are not corrected for phylogeny due to the relatively small sample size
Elevated tissue cadmium in long-lived mouse models overexpressing MTs
In order to further corroborate the results reported above — which are all based on the analysis of already available data — on the relationship between MT gene expression, longevity, and Cd accumulation, we combined new data on Cd accumulation with published and new data on MT expression in long-lived mouse models and their controls (Table 2).
Table 2.
Metallothionein (MT) in long-lived mouse models (fold-change vs. controls)
| Model | MT | Sex | Strain | Age | Method | Reference |
|---|---|---|---|---|---|---|
| Kidney | ||||||
| GHRKO | 1.7 | Male | C57BL/6 J | Adult | qPCR | Swindell et al. [77] |
| Ames dwarf | 1.9 | Male | Mixed | 12 mo | qPCR | This manuscript |
| MT-Tg | 4.7 | Male | C57BL/6 J | 7–10 wks | Cd/Hb | Iszard et al. [37] |
| Liver | ||||||
| Rapamycin | 0.1 | Female | UM-HET3 | 12 mo | qPCR | Miller et al. (unpublished) |
| CR | 1.8 | Female | UM-HET3 | 12 mo | qPCR | Miller et al. (unpublished) |
| GHRKO | 6.2 | Male | C57BL/6 J | 12 mo | qPCR | Swindell et al. [77] |
| Ames dwarf | 7.7 | Female | Mixed | 3 mo | qPCR | This manuscript |
| MT-Tg | 8 | Male | C57BL/6 J | 7–10 wks | Protein | Iszard et al. [37] |
This table provides estimates from the literature (unless they are from this work as indicate in the column Reference) of MT expression in long-lived mice and their control. We present data for animals of the same sex as those we used for Cd analyses. Although we do not have data for kidney MT in female Ames dwarf, we assume it is higher based on the liver data. MT represents the mean of changes in MT-1 and MT-2. Cd/Hb, Cd/hemoglobin assay; mo, months; d, days; wks, weeks
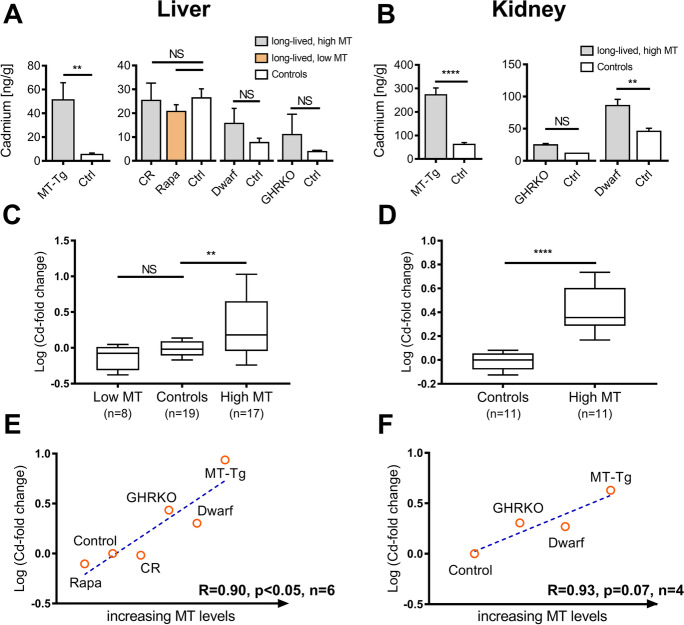
Hepatic cadmium levels in 24-month-old MT-Tg mice (mice overexpressing MTs) were elevated ninefold compared to these age-matched controls (Fig. 6A). Renal Cd was also higher in MT-Tg mice (fourfold compared to controls, Fig. 6B) and in Ames dwarf mice (twofold compared to controls, Fig. 6B). In addition, renal and hepatic Cd levels in our male MT-Tg mice were exceptionally high when compared to somewhat younger control mice from our colony (data not shown) and mice in the study of Koizumi et al. [44]. To further insight on this association between Cd and MT, we grouped all published and newly collected data based on MT expression and compared tissue Cd levels among high or low MT expressing strains (including CR and rapamycin-fed mice). This result suggests that mice with higher reported MT expression (Table 2) may also have higher Cd levels (Fig. 6C, D). We also noted strong correlations when we plotted the ranks of MT expression (from the lowest to the highest MT expression) data against the fold-change in Cd as compared to matched controls. The correlation was significant in liver (p < 0.05, Fig. 6E), but not kidney (p = 0.07, Fig. 6F), possibly due to the limited sample size. Finally, we found that female and male Ames dwarf mice have elevated hepatic and renal MT expression across their entire life span, when compared to their littermate controls (Fig. S4-S6, Supplementary data 2). This result suggests that Cd accumulation during aging is not responsible for the higher MT expression observed in long-lived mice compared to controls, but that higher Cd accumulation in longer-lived species instead occurs because these species have higher levels of MT expression. Since results from earlier studies were inconsistent in females, we also verified that 3-month-old female Ames dwarf mice had higher liver MT expression than female controls (Fig. S6, Supplementary data 2).
Fig. 6.
Cadmium (Cd) content in liver (A, C, E) and kidney (B, D, F) of long-lived mice is elevated compared to age-matched controls. Significance was determined by Welchs’s T-test or one-way ANOVA where appropriate: **p < 0.01; ****p < 0.0001; NS, not significant; error bars show standard error of the mean. A Hepatic cadmium in 24-month-old MT transgenic mice (MT; n = 4) compared to controls (n = 5). In separate experiments, we compared hepatic cadmium in 8-week-old growth-hormone receptor knockout mice (GHRKO, n = 3) vs. controls (ctrl, n = 2), hepatic cadmium in 12-month-old rapamycin (Rapa) treated or calorie-restricted (CR) animals vs. controls (n = 7–8), and hepatic cadmium in middle aged Ames dwarf mice (n = 3) vs. controls (n = 5). B Renal cadmium in 24-month-old MT transgenic mice (MT-Tg; n = 5) compared to controls (n = 5). In separate experiments, we compared hepatic cadmium in 8-week-old growth-hormone receptor knockout mice (GHRKO, n = 2) vs. controls (ctrl, n = 1) and hepatic cadmium in middle aged Ames dwarf mice (n = 4) vs. controls (n = 5). C, D Pooled and normalized cadmium content in long-lived mice as compared to controls (log fold-change). Data was stratified by MT expression. Box and whisker plots show median, interquartile range, and 10–90% confidence interval. E, F MT expression (rank) was plotted against tissue cadmium content (log fold-change compared to controls). Animals were rank-ordered by their metallothionein expression as shown in Table 2. The blue dashed lines represent linear regression and Pearson correlation coefficients are shown
Discussion
Long-lived species can accumulate more Cd as a result of increased MT expression
In many species, including humans, aging leads to a remarkable accumulation of cadmium in the liver and kidney [1]. Although this accumulation is linked to several age-related diseases, the mechanisms are not well understood. If Cd toxicity could limit species life span, long-lived species may have evolved ways to accumulate less Cd or ways to cope with Cd-related stress. Evidence that cells from long-lived species are more resistant to Cd toxicity has been previously provided [30], but a study on the elemental composition of mammalian organs argued against a reduction of Cd in tissues of long-lived species [51]. Through a combination of metadata analysis and the collection and analysis of new data, to our knowledge, our study is the first to provide a link between the observed accumulation of Cd and the role of metallothioneins in long-lived species. Specifically, we found evidence that long-lived mammalian species have an enhanced capacity to accumulate more Cd in their tissues possibly due to higher expressed MT genes and MT proteins, but not as a consequence of an increased number of metallothionein genes (paralogs).
Although before this study, it was suggested that metallothioneins are associated with aging [13, 25, 52, 75, 76, 84], the mechanisms behind the role of these genes, if any, in healthy aging are still unknown. Aging is characterized by a progressive shrinkage of the homeodynamic space [16] and the stress response is a key component of this space by initiating a series of events for maintenance, repair, adaptation, remodeling, and survival.
It has been hypothesized that MTs may contribute to longevity by protecting the cells from oxidative stress. Indeed, oxidants and electrophiles readily react with the sulfhydryl groups of MTs releasing zinc ions [54] which, in turn, bind to the metal-responsive transcription factor MTF-1 to activate the metal and oxidative cellular response [2]. Here, we provide evidence for a potential role of gene expression response to heavy metals in healthy aging. Our work perfectly fits with the observation that short-lived mammals have a less active MTF-1 (display defects in heavy metal response) than the longer lived humans and bats [71] and supports the hypothesis that an efficient metal homeodynamic process is a key aspect of longevity.
Liver and kidney cadmium are elevated in long-lived mammals
A previous study found a positive relationship between hepatic and renal Cd and MLS in young adult animals [51]. An important limitation of this study, however, was the exclusive use of males, which may have lower Cd levels than females at any given age [44]. We found that Cd accumulation during aging occurs in both sexes and that the association persists even after adjusting for multiple sources of bias. Cd accumulation is therefore much higher in species that have longer life spans, independently of their age at sampling. However, although a relationship between Cd accumulation and longevity could be observed across different mammalian taxonomic groups kept in captivity, a similar relationship was not found in wild birds or wild mammals. Conceivably, in the wild, MLS-related Cd accumulation may be influenced by environmental exposure [9, 61]. At our knowledge, we are the only study that even considered wild animals in this context and further investigation is needed to clarify this issue.
Metallothionein gene expression and protein level are elevated in long-lived animals
MTs are neglected players in organismal multi-stress resistance [75]. Despite their small size, MTs effectively protect from genotoxic and cytotoxic damage through sequestration of reactive metals and by acting as direct antioxidants [12]. Although MTs are known to be related to longevity and are elevated, at the mRNA level, in some of the mouse models that have an extended life span, their mechanistic role in aging is still poorly understood.
Here, we show that high MT gene expression and elevated protein levels are indeed linked to longevity. Our results indicate that basal expression of MT genes in the kidney is consistently associated with MLS. The relationship between MLS and MT expression in the liver based on our study seems to be less clear and dependent on the dataset used; however, a comparison of the mouse and naked mole rat transcriptomes indicated elevated hepatic MT-2 associated with longevity [86]. Previous work indicates that in mice, MT expression displays higher interindividual variability in the liver than the kidney [87], suggesting that the different results obtained for the different dataset used may be due to a decreased synthesis or faster degradation of mRNA in the liver than in the kidney and thus to higher fluctuation of hepatic MT expression.
One of the novelties of our work is showing that MT expression of specific paralogs and not number of paralogs itself is related to MLS. Although we initially hypothesized that a higher number of MT genes would allow higher MT expression through a gene dosage effect and therefore correlate with species MLS, the analysis of available RNA-Seq data uncovered a relationship between the expression of specific MT paralogs and MLS (Table 3, Table S2). Future studies should further test this hypothesis that higher expression of specific paralogs and not gene dosage is related to MLS.
Given the elevated Cd and MT levels in tissues of long-lived species, how can we reconcile our data with reduced Cd uptake in fibroblasts from long-lived species [18]. Interestingly, both altered Cd transport and elevated MTs may explain the well-described correlation between species longevity and fibroblast resistance to Cd [31]. We propose that these two protective mechanisms operate to varying degrees in different tissue and cell types. In liver and kidney, Cd import and sequestration predominate, while fibroblasts avoid Cd accumulation and induce MTs when needed. Consistent with this model, MTF-1 from longer-lived species is more efficient at inducing MTs in vitro [71].
Disentangling the effects of age on metallothioneins and cadmium accumulation
Since Cd accumulates with age and induces MTs, this may create a spurious correlation between MLS and MTs when longer-lived animals are sampled at older ages [51]. Here, we provide several counterarguments to this, in favor of a direct relationship between MTs and MLS. Even in tissues that do show age-related Cd accumulation, like liver and kidney, this is not fully explained by chronologic age. One of the datasets we analyzed [44, 58, 59] provided a unique opportunity to adjust for age at sampling, because the authors noted the age at which they acquired tissue samples. Our analysis indicates that Cd accumulation is linked to longevity per se and does not simply reflect chronologic age.
Studies in which Cd was added to the diet of MT knockout mice suggested that MT would indeed trap Cd, but similar studies in MT overexpressing mice — MT-Tg mice — did not [49]. To further investigate the relationship between Cd accumulation, MT expression, and longevity, we measured hepatic and renal Cd in long-lived mice and control strains fed with normal diet (i.e., not enriched in Cd) and correlated to levels of MT expression. Isolated overexpression of MT-1 extended life span [52] and greatly increased Cd accumulation in the liver and kidney in support of our hypothesis. GHRKO, Ames dwarf, and calorie-restricted (CR) mice have elevated MT expression, whereas rapamycin-fed animals have reduced MT expression. Cd accumulation in these models correlated with MT expression. Our work also suggests that rapamycin-fed mice have the lowest expression of MT compared to the other long-lived mouse models herein studied. However, this is not completely surprising as rapamycin can suppress hepatic Nrf2 signaling [22], which in turn contribute to enhance MT expression [27]. This suggests that overexpression of MTs, albeit important, is not a universal trait of longevity.
In summary in this study, we (1) confirm a relationship between species maximum life span and their tissue Cd content in captivity, (2) demonstrate that gene and protein expression of MT relate to life span in mammals, and (3) show that elevated tissue Cd levels in long-lived mice are related to high MT expression. Based on this, we propose that long-lived species evolved an efficient metal stress response upregulating their MT levels and accumulating Cd as a consequence (Fig. 7). We have preliminary evidence that this process may be observed both at macroevolutionary (across mammals) and microevolutionary (within mice) scale, but currently, we do not have data to extrapolate if this relationship would persist across different vertebrate species and not just in mammals, neither we have data on MT expression and variation in longevity within the same species beyond mice. Hence, our findings support a relationship between Cd, MT, and MLS in mammals and provide the necessary background to further investigate if the observed pattern is maintained across vertebrates with different life span. Furthermore, our results provide the foundation to carry out future work on how altered gene expression and protein levels — including the influence of microRNAs and silencing RNAs on them — of different MTs may impact longevity.
Fig. 7.
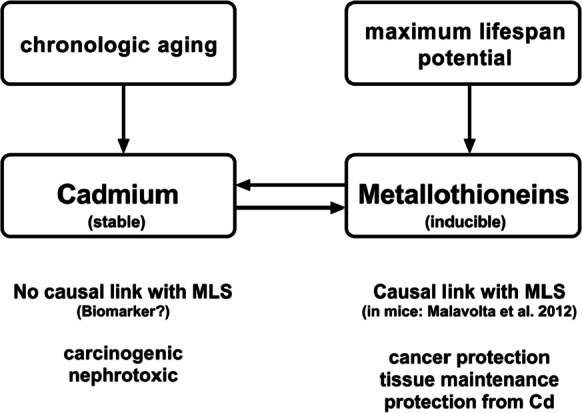
The main interactions between cadmium (Cd), metallothioneins (MTs), chronologic age, and maximum life span (MLS) are shown in the context of our working model. Since Cd is retained in tissues and induces MTs, it can serve as a measure of cumulative MT expression
Limitation of the study
A limitation of the study is that some of the data used in this study and obtained from public databases of wild animals consisted of one individual sampled as representative for the entire species. Another limitation regards the reduced sample size of some groups used to study MT and Cd in long-lived mice.
Supplementary information
(XLSX 72 kb)
(DOCX 620 kb)
(DOCX 16 kb)
Acknowledgements
We wish to thank Emilio Casanova, Richard Miller, and Thomas Weichhart for providing mouse tissues. Old mice were provided by Gerald de Haan and Ronald van Os through the Mouse Clinic for Cancer and Aging (MCCA), funded by a Large Infrastructure grant from the Netherlands Organization for Scientific Research (NWO). Finally, we are thankful to Isabella Capellini for discussion on the results of the phylogenetic comparative analyses and to Vincent Lynch for general discussion about metallothioneins and aging. We are thankful to Richard Miller for useful comments on this article. YC is grateful to the National Science Foundation (NSF) for supporting work related to this project.
Author contribution
Conception of the work: M.M., K.N., and M.P.
Data collection: K.P., E.S., R.G., H.B., Y.C.
Data analysis and interpretation: Y.C., K.P., C.G., E.S., M.M., C.S., P.G.
Drafting the article: Y.C., K.P., M.M.
Critical revision: Y.C., K.P., M.M., C.G., T.V., H.B.
Funding
This study was supported by Ricerca Corrente funding from Italian Ministry of Health to MP, MM, and RG as well as from Austrian Science Fund FWF (Grant: P22323-B17 and V 197-B17) to TV.
Dclarations
Ethics approval
All samples used in this study consisted of leftover samples stored in mouse tissues biobanks. These samples originated from different studies which were originally approved by the respective local ethical committees.
Data availability statement
The data that support the findings of this study are available in the Supplementary data 1 of this article.
Conflict of interest
The authors declare no competing interests.
Footnotes
Kamil Pabis and Ylenia Chiari shared first authorship.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Kamil Pabis, Email: kamil.pabis@gmail.com.
Ylenia Chiari, Email: ychiari@gmu.edu.
Claudia Sala, Email: claudia.sala3@unibo.it.
Elisabeth Straka, Email: elisabeth_straka@gmx.at.
Robertina Giacconi, Email: r.giacconi@inrca.it.
Mauro Provinciali, Email: m.provinciali@inrca.it.
Xinna Li, Email: xinna@umich.edu.
Holly Brown-Borg, Email: holly.brown.borg@und.edu.
Karin Nowikovsky, Email: karin.nowikovsky@meduniwien.ac.at.
Teresa G. Valencak, Email: teresa.valencak@vetmeduni.ac.at
Claudia Gundacker, Email: claudia.gundacker@meduniwien.ac.at.
Paolo Garagnani, Email: paolo.garagnani2@unibo.it.
Marco Malavolta, Email: m.malavolta@inrca.it.
References
- 1.Åkesson A, Barregard L, Bergdahl IA, et al Non-renal effects and the risk assessment of environmental cadmium exposure 2014. [DOI] [PMC free article] [PubMed]
- 2.Andrews GK. Regulation of metallothionein gene expression by oxidative stress and metal ions. Biochem Pharmacol. 2000;59:95–104. doi: 10.1016/S0006-2952(99)00301-9. [DOI] [PubMed] [Google Scholar]
- 3.Arredondo M, González M, Latorre M Copper. In: Malavolta M, Mocchegiani E (eds) Trace elements and minerals in health and longevity. 2018. pp 35–62.
- 4.Barrett T, Wilhite SE, Ledoux P, et al. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res. 2013 doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartke A Somatic growth, aging, and longevity. npj Aging Mech. Dis. 2017. [DOI] [PMC free article] [PubMed]
- 6.Beattie JH, Malavolta M, Korichneva I Zinc. 2018. pp 99–131.
- 7.Bininda-Emonds ORP, Cardillo M, Jones KE, et al. The delayed rise of present-day mammals. Nature. 2007 doi: 10.1038/nature05634. [DOI] [PubMed] [Google Scholar]
- 8.Brown-Borg HM, Borg KE, Meliska CJ, Bartke A. Dwarf mice and the ageing process. Nature. 1996;384:33–33. doi: 10.1038/384033a0. [DOI] [PubMed] [Google Scholar]
- 9.Buchwalter DB, Cain DJ, Martin CA, et al. Aquatic insect ecophysiological traits reveal phylogenetically based differences in dissolved cadmium susceptibility. Proc Natl Acad Sci U S A. 2008;105:8321–8326. doi: 10.1073/pnas.0801686105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carruthers M, Yurchenko AA, Augley JJ, et al. De novo transcriptome assembly, annotation and comparison of four ecological and evolutionary model salmonid fish species. BMC Genomics. 2018 doi: 10.1186/s12864-017-4379-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiari Y, Glaberman S, Serén N, et al. Phylogenetic signal in amphibian sensitivity to copper sulfate relative to experimental temperature. Ecol Appl. 2015 doi: 10.1890/14-0439.1. [DOI] [PubMed] [Google Scholar]
- 12.Chiaverini N, De Ley M Protective effect of metallothionein on oxidative stress-induced DNA damage. Free Radic. Res. 2010. [DOI] [PubMed]
- 13.Cipriano C, Malavolta M, Costarelli L, et al. Polymorphisms in MT1a gene coding region are associated with longevity in Italian Central female population. Biogerontology. 2006;7:357–365. doi: 10.1007/s10522-006-9050-x. [DOI] [PubMed] [Google Scholar]
- 14.Cousins RJ. Absorption, transport, and hepatic metabolism of copper and zinc: special reference to metallothionein and ceruloplasmin. 1985. [DOI] [PubMed]
- 15.de Magalhães JP, Costa J, Church GM, et al. An analysis of the relationship between metabolism, developmental schedules, and longevity using phylogenetic independent contrasts. J Gerontol A Biol Sci Med Sci. 2007;62:149–160. doi: 10.1093/gerona/62.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Demirovic D, Rattan SIS. Establishing cellular stress response profiles as biomarkers of homeodynamics, health and hormesis. Exp Gerontol. 2013 doi: 10.1016/j.exger.2012.02.005. [DOI] [PubMed] [Google Scholar]
- 17.Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013 doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dostál L, Kohler WM, Penner-Hahn JE, et al. Fibroblasts from long-lived rodent species exclude cadmium. J Gerontol A Biol Sci Med Sci. 2015 doi: 10.1093/gerona/glu001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Durinck S, Spellman PT, Birney E, Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protoc. 2009 doi: 10.1038/nprot.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dziegiel P. Metallothioneins in normal and cancer cells, advances in anatomy, embryology and cell biology. J Neurol Neurosurg Psychiatry. 2016. [DOI] [PubMed]
- 21.Eisler R. Compendium of trace metals and marine biota. 2010
- 22.Fok WC, Chen Y, Bokov A, et al. Mice fed rapamycin have an increase in lifespan associated with major changes in the liver transcriptome. PLoS ONE. 2014 doi: 10.1371/journal.pone.0083988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frank A. In search of biomonitors for cadmium: cadmium content of wild Swedish fauna during 1973–1976. Sci Total Environ. 1986 doi: 10.1016/0048-9697(86)90009-4. [DOI] [PubMed] [Google Scholar]
- 24.Fushan AA, Turanov AA, Lee SG, et al. Gene expression defines natural changes in mammalian lifespan. Aging Cell. 2015 doi: 10.1111/acel.12283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giacconi R, Costarelli L, Piacenza F, et al. Zinc-induced metallothionein in centenarian offspring from a large European population: the MARK-AGE project. J Gerontol A Biol Sci Med Sci. 2018 73. 10.1093/gerona/glx192 [DOI] [PubMed]
- 26.Grubić Kezele T Iron. 2018. pp 1–34.
- 27.Gu J, Cheng Y, Wu H, et al. Metallothionein is downstream of Nrf2 and partially mediates sulforaphane prevention of diabetic cardiomyopathy. Diabetes. 2017 doi: 10.2337/db15-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gundacker C, Pietschnig B, Wittmann KJ, et al. Smoking, cereal consumption, and supplementation affect cadmium content in breast milk. J Expo Sci Environ Epidemiol. 2007;17:39–46. doi: 10.1038/sj.jes.7500518. [DOI] [PubMed] [Google Scholar]
- 29.Hall JA, McElwee MK, Freedman JH. Identification of ATF-7 and the insulin signaling pathway in the regulation of metallothionein in C. elegans suggests roles in aging and reactive oxygen species. PLoS One. 2017. 10.1371/journal.pone.0177432 [DOI] [PMC free article] [PubMed]
- 30.Harper JM, Salmon AB, Leiser SF, et al. Skin-derived fibroblasts from long-lived species are resistant to some, but not all, lethal stresses and to the mitochondrial inhibitor rotenone. Aging Cell. 2007 doi: 10.1111/j.1474-9726.2006.00255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harper JM, Wang M, Galecki AT, et al. Fibroblasts from long-lived bird species are resistant to multiple forms of stress. J Exp Biol. 2011 doi: 10.1242/jeb.054643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harrison DE, Strong R, Sharp ZD, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Henry RB, Liu J, Choudhuri S, Klaassen CD. Species variation in hepatic metallothionein. Toxicol Lett. 1994;74:23–33. doi: 10.1016/0378-4274(94)90071-X. [DOI] [PubMed] [Google Scholar]
- 34.Hidalgo J, Chung R, Penkowa M, Vašák M. 10. Structure and function of vertebrate metallothioneins. 2009:pp 279–317.
- 35.Hunt SE, McLaren W, Gil L, et al. Ensembl variation resources. Database (Oxford) 2018 doi: 10.1093/database/bay119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hylton A, Chiari Y, Capellini I, et al. Mixed phylogenetic signal in fish toxicity data across chemical classes. Ecol Appl. 2018 doi: 10.1002/eap.1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iszard MB, Liu J, Liu Y, et al. Characterization of metallothionein-I-transgenic mice. Toxicol Appl Pharmacol. 1995;133:305–312. doi: 10.1006/taap.1995.1155. [DOI] [PubMed] [Google Scholar]
- 38.Järup L, Åkesson A Current status of cadmium as an environmental health problem. Toxicol Appl Pharmacol. 2009. [DOI] [PubMed]
- 39.Jeffery EH, Noseworthy R, Cherian MG. Age dependent changes in metallothionein and accumulation of cadmium in horses. Comp Biochem Physiol Part C, Comp. 1989 doi: 10.1016/0742-8413(89)90242-9. [DOI] [PubMed] [Google Scholar]
- 40.Jetz W, Thomas GH, Joy JB, et al. The global diversity of birds in space and time. Nature. 2012 doi: 10.1038/nature11631. [DOI] [PubMed] [Google Scholar]
- 41.Kadota Y, Aki Y, Toriuchi Y, et al. Deficiency of metallothionein-1 and -2 genes shortens the lifespan of the 129/Sv mouse strain. Exp Gerontol. 2015;66:21–24. doi: 10.1016/j.exger.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 42.Klang IM, Schilling B, Sorensen DJ, et al. Iron promotes protein insolubility and aging in C. elegans. Aging (Albany NY) 2014: 10.18632/aging.100689 [DOI] [PMC free article] [PubMed]
- 43.Koizumi N, Inoue Y, Ninomiya R, et al. Relationship of cadmium accumulation to zinc or copper concentration in horse liver and kidney. Environ Res. 1989 doi: 10.1016/S0013-9351(89)80025-8. [DOI] [PubMed] [Google Scholar]
- 44.Koizumi N, Murata K, Hayashi C, et al. High cadmium accumulation among humans and primates: comparison across various mammalian species—a study from Japan. Biol Trace Elem Res. 2008 doi: 10.1007/s12011-007-8048-9. [DOI] [PubMed] [Google Scholar]
- 45.Kolde R. pheatmap : pretty heatmaps. R Packag version. 2015;1:8. [Google Scholar]
- 46.Krizkova S, Kepinska M, Emri G, et al. Microarray analysis of metallothioneins in human diseases—a review. J. Pharm Biomed Anal. 2016. [DOI] [PubMed]
- 47.Larsson SC, Wolk A Urinary cadmium and mortality from all causes, cancer and cardiovascular disease in the general population: systematic review and meta-analysis of cohort studies. Int. J. Epidemiol. 2016. [DOI] [PubMed]
- 48.Li X, Bartke A, Berryman DE, et al. Direct and indirect effects of growth hormone receptor ablation on liver expression of xenobiotic metabolizing genes. Am J Physiol - Endocrinol Metab. 2013 doi: 10.1152/ajpendo.00304.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu J, Klaassen CD. Absorption and distribution of cadmium in metallothionein-I transgenic mice. Fundam Appl Toxicol. 1996;29:294–300. doi: 10.1006/faat.1996.0034. [DOI] [PubMed] [Google Scholar]
- 50.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014 doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ma S, Lee S-G, Kim EB, et al. Organization of the mammalian ionome according to organ origin, lineage specialization, and longevity. Cell Rep. 2015;13:1319–1326. doi: 10.1016/j.celrep.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Malavolta M, Basso A, Piacenza F, et al. Survival study of metallothionein-1 transgenic mice and respective controls (C57BL/6J): influence of a zinc-enriched environment. Rejuvenation Res. 2012;15:140–143. doi: 10.1089/rej.2011.1261. [DOI] [PubMed] [Google Scholar]
- 53.Malavolta M, Orlando F, Piacenza F, et al. Metallothioneins, longevity and cancer: comment on “deficiency of metallothionein-1 and -2 genes shortens the lifespan of the 129/Sv mouse strain”. Exp Gerontol. 2016;73:28–30. doi: 10.1016/j.exger.2015.11.014. [DOI] [PubMed] [Google Scholar]
- 54.Maret W. The redox biology of redox-inert zinc ions. Free Radic Biol Med. 2019. [DOI] [PubMed]
- 55.Maret W, Krȩzel A, Krezel A. Cellular zinc and redox buffering capacity of metallothionein/thionein in health and disease. In: Molecular medicine (Cambridge, Mass.): 2007. pp 371–5 [DOI] [PMC free article] [PubMed]
- 56.McKinney GJ, Waples RK, Seeb LW, Seeb JE. Paralogs are revealed by proportion of heterozygotes and deviations in read ratios in genotyping-by-sequencing data from natural populations. Mol Ecol Resour. 2017 doi: 10.1111/1755-0998.12613. [DOI] [PubMed] [Google Scholar]
- 57.Miura N, Koizumi S. Heavy metal responses of the human metallothionein isoform genes. Yakugaku Zasshi. 2007. [DOI] [PubMed]
- 58.Ninomiya R, Koizumi N, Murata K. Concentrations of cadmium, zinc, copper, iron, and metallothionein in liver and kidney of nonhuman primates. Biol Trace Elem Res. 2002 doi: 10.1385/BTER:87:1-3:095. [DOI] [PubMed] [Google Scholar]
- 59.Ninomiya R, Koizumi N, Murata K. Metal concentrations in the liver and kidney of aquatic mammals and penguins. Biol Trace Elem Res. 2004 doi: 10.1385/BTER:97:2:135. [DOI] [PubMed] [Google Scholar]
- 60.Orme D, Freckleton R, Thomas G, et al (2014) Caper: comparative analyses of phylogenetics and evolution in R. R Packag. version 0.5.2/ r121
- 61.Øverjordet IB, Gabrielsen GW, Berg T, et al. Effect of diet, location and sampling year on bioaccumulation of mercury, selenium and cadmium in pelagic feeding seabirds in Svalbard. Chemosphere. 2015 doi: 10.1016/j.chemosphere.2014.10.060. [DOI] [PubMed] [Google Scholar]
- 62.Pabis K, Gundacker C, Giacconi R, et al. Zinc supplementation can reduce accumulation of cadmium in aged metallothionein transgenic mice. Chemosphere. 2018 doi: 10.1016/j.chemosphere.2018.08.017. [DOI] [PubMed] [Google Scholar]
- 63.Palumaa P, Tammiste I, Kruusel K, et al. Metal binding of metallothionein-3 versus metallothionein-2: lower affinity and higher plasticity. Biochim Biophys Acta - Proteins Proteomics. 2005 doi: 10.1016/j.bbapap.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 64.Pastorelli AA, Campanella L, Coppa A, Stacchini P. Exposure to cadmium and lead in an agropastoral iron age population. Int J Osteoarchaeol. 2016 doi: 10.1002/oa.2403. [DOI] [Google Scholar]
- 65.Peng X, Thierry-Mieg J, Thierry-Mieg D, et al. Tissue-specific transcriptome sequencing analysis expands the non-human primate reference transcriptome resource (NHPRTR) Nucleic Acids Res. 2015 doi: 10.1093/nar/gku1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pugh TD, Oberley TD, Weindruch R. Dietary intervention at middle age: caloric restriction but not dehydroepiandrosterone sulfate increases lifespan and lifetime cancer incidence in mice. Cancer Res. 1999. [PubMed]
- 67.Sabolić I, Breljak D, Škarica M, Herak-Kramberger CM. Role of metallothionein in cadmium traffic and toxicity in kidneys and other mammalian organs. BioMetals. 2010. [DOI] [PubMed]
- 68.Sahm A, Bens M, Szafranski K, et al. Long-lived rodents reveal signatures of positive selection in genes associated with lifespan. PLoS Genet. 2018 doi: 10.1371/journal.pgen.1007272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sato M, Bremner I. Oxygen free radicals and metallothionein. Free Radic Biol Med. 1993 doi: 10.1016/0891-5849(93)90029-T. [DOI] [PubMed] [Google Scholar]
- 70.Scheuhammer AM, Cherian MG. Quantification of metallothioneins by a silver-saturation method. Toxicol Appl Pharmacol. 1986 doi: 10.1016/0041-008X(86)90277-2. [DOI] [PubMed] [Google Scholar]
- 71.Schmidt K, Steiner K, Petrov B, et al. Short-lived mammals (shrew, mouse) have a less robust metal-responsive transcription factor than humans and bats. Biometals. 2016 doi: 10.1007/s10534-016-9926-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Serén N, Glaberman S, Carretero MA, Chiari Y. Molecular evolution and functional divergence of the metallothionein gene family in vertebrates. J Mol Evol. 2014 doi: 10.1007/s00239-014-9612-5. [DOI] [PubMed] [Google Scholar]
- 73.Speakman JR. Correlations between physiology and lifespan—two widely ignored problems with comparative studies. Aging Cell. 2005. [DOI] [PubMed]
- 74.Stiedl P, Mcmahon R, Blaas L, et al. Growth hormone resistance exacerbates cholestasis-induced murine liver fibrosis. Hepatology. 2015 doi: 10.1002/hep.27408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Swindell WR Metallothionein and the biology of aging. Ageing Res Rev. 2011. [DOI] [PMC free article] [PubMed]
- 76.Swindell WR, Johnston A, Sun L, et al. Meta-profiles of gene expression during aging: limited similarities between mouse and human and an unexpectedly decreased inflammatory signature. PLoS ONE. 2012 doi: 10.1371/journal.pone.0033204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Swindell WR, Masternak MM, Bartke A. In vivo analysis of gene expression in long-lived mice lacking the pregnancy-associated plasma protein A (PappA) gene. Exp Gerontol. 2010;45:366–374. doi: 10.1016/j.exger.2010.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tacutu R, Craig T, Budovsky A, et al. Human ageing genomic resources: integrated databases and tools for the biology and genetics of ageing. Nucleic Acids Res. 2012;41:D1027–D1033. doi: 10.1093/nar/gks1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Teranishi A, Ninomiya R, Koizumi N. Relationship of metallothionein to cadmium and to zinc in human liver and kidney. In: Metallothionein IV. Birkhäuser Basel, Basel: 1999 pp 485–488
- 80.The UniProt Consortium. UniProt: a worldwide hub of protein knowledge The UniProt Consortium. Nucleic Acids Res. 2019. [DOI] [PMC free article] [PubMed]
- 81.Valencak TG, Ruf T. Phospholipid composition and longevity: lessons from Ames dwarf mice. Age (Omaha) 2013 doi: 10.1007/s11357-013-9533-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vijay N, Poelstra JW, Künstner A, Wolf, JBW. Challenges and strategies in transcriptome assembly and differential gene expression quantification. A comprehensive in silico assessment of RNA-seq experiments. In: Molecular Ecology. 2013. [DOI] [PubMed]
- 83.Wills NK, Ramanujam VMS, Chang J, et al. Cadmium accumulation in the human retina: effects of age, gender, and cellular toxicity. Exp Eye Res. 2008;86:41–51. doi: 10.1016/j.exer.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 84.Yang X, Doser TA, Fang CX, et al. Metallothionein prolongs survival and antagonizes senescence-associated cardiomyocyte diastolic dysfunction: role of oxidative stress. FASEB J. 2006;20:1024–1026. doi: 10.1096/fj.05-5288fje. [DOI] [PubMed] [Google Scholar]
- 85.Yang X, Han Y, Mu Y, et al. Multigenerational effects of cadmium on the lifespan and fertility of Drosophila melanogaster. Chemosphere. 2020 doi: 10.1016/j.chemosphere.2019.125533. [DOI] [PubMed] [Google Scholar]
- 86.Yu C, Li Y, Holmes A, et al. RNA sequencing reveals differential expression of mitochondrial and oxidation reduction genes in the long-lived naked mole-rat when compared to mice. PLoS ONE. 2011 doi: 10.1371/journal.pone.0026729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang D, Jin T, Xu Y, qiao, , et al. Diurnal-and sex-related difference of metallothionein expression in mice. J Circadian Rhythms. 2012 doi: 10.1186/1740-3391-10-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zota AR, Needham BL, Blackburn EH, et al. Associations of cadmium and lead exposure with leukocyte telomere length: findings from National Health And Nutrition Examination Survey, 1999–2002. Am J Epidemiol. 2015 doi: 10.1093/aje/kwu293. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX 72 kb)
(DOCX 620 kb)
(DOCX 16 kb)
Data Availability Statement
The data that support the findings of this study are available in the Supplementary data 1 of this article.