Abstract
The dramatically expanding coronavirus disease 2019 (COVID-19) needs multiple effective countermeasures. Neutralizing nanobodies (Nbs) are a potential therapeutic strategy for treating COVID-19. Here, we characterize several receptor binding domain (RBD)-specific Nbs isolated from an Nb library derived from an alpaca immunized with the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike glycoprotein (S); among them, three Nbs exhibit picomolar potency against SARS-CoV-2 live virus, pseudotyped viruses, and circulating SARS-CoV-2 variants. To improve their efficacy, various configurations of Nbs are engineered. Nb15-NbH-Nb15, a trimer constituted of three Nbs, is constructed to be bispecific for human serum albumin (HSA) and RBD of SARS-CoV-2. Nb15-NbH-Nb15 exhibits single-digit ng/ml neutralization potency against the wild-type and Delta variants of SARS-CoV-2 with a long half-life in vivo. In addition, we show that intranasal administration of Nb15-NbH-Nb15 provides effective protection for both prophylactic and therapeutic purposes against SARS-CoV-2 infection in transgenic hACE2 mice. Nb15-NbH-Nb15 is a potential candidate for both the prevention and treatment of SARS-CoV-2 through respiratory administration.
Keywords: SARS-CoV-2, nanobody, intranasal, Nb15, bispecific, hACE2 mice, Delta, potent neutralizing, Nb15-NbH-Nb15, VHH
Graphical abstract
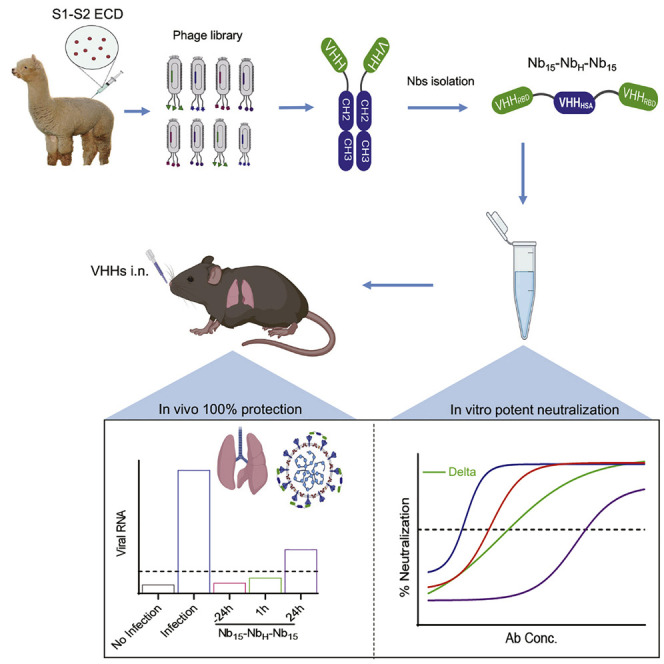
Wu et al. report Nb15-NbH-Nb15 with a heterotrimeric bispecific configuration, which exhibits potent neutralization potency against SARS-CoV-2 and its variants, including the prevalent Delta variant, in vitro and provides in vivo protection against SARS-CoV-2 infection in hACE2 transgenic mice by intranasal delivery.
Introduction
As of February 1st, 2021, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused more than 100 million confirmed cases and over 2.2 million deaths globally. The containment of the expanding coronavirus disease 2019 (COVID-19) pandemic needs multiple countermeasures. Prophylactic vaccines have been recently approved (Dai and Gao, 2020), and a number of SARS-CoV-2-neutralizing monoclonal antibodies (mAbs) that target the receptor binding domain (RBD) of the spike protein (Baum et al., 2020; Ju et al., 2020; Liu et al., 2020; Rogers et al., 2020; Shi et al., 2020) were identified, which could be developed as either therapeutic or prophylactic agents.
In addition to conventional antibodies, camelids also generate heavy-chain-only antibodies (HCAbs), constituting a single variable domain (nanobody [Nb]) specific for binding antigens (Hamers-Casterman et al., 1993). This single variable domain, referred as a single-domain antibody, variable heavy-chain domains of heavy-chain antibody (VHH), or Nb, has higher affinity, thermal stability, and chemostability than most antibodies (Jovčevska and Muyldermans, 2019; Steeland et al., 2016) and can be constructed easily into multivalent formats devoid of Fc, which will overcome potential deleterious antibody-dependent enhancement (ADE) of infection observed in some viral infections, including dengue virus, HIV, and SARS-CoV (Luo et al., 2018; Taylor et al., 2015; Tirado and Yoon, 2003). Their favorable biophysical properties have led to the development of several Nbs as therapeutics against viral infection, such as severe fever with thrombocytopenia syndrome virus (SFTSV) (Wu et al., 2020a) and respiratory syncytial virus (RSV) (Cunningham et al., 2020; Detalle et al., 2015; Van Heeke et al., 2017).
SARS-CoV-2 is transmitted by the upper respiratory tract (Zhou et al., 2020), and an analysis of clinical specimens showed that SARS-CoV-2 was detected with the highest viral copies in multiple sites of the respiratory tract, whereas few viral copies are in the blood (Wang et al., 2020). These data indicate that biotherapeutic agents directly delivered by the respiratory route to the sites of infection would be an attractive alternative to systemic routes of administration. Parenteral inoculation of a biotherapeutic antibody is a particularly ineffective way to deliver drugs to the respiratory tract. Indeed, a study on mepolizumab (anti-interleukin-5 mAb) demonstrated that only 0.2% of the dose administered reached the lung by systemic administration (Hart et al., 2001). In addition, the therapeutic effect of pulmonary delivery of human immunoglobulins for controlling RSV in cotton rats was shown to be 160 times more effective than that by the parenteral administration (Prince et al., 1987). Thus, pulmonary delivery may be superior to parenteral administration for the treatment of respiratory tract infections.
A key requirement for pulmonary delivery is the stability of the biologic product so that it endures the degrading environment, and thus, the drug will have to be formulated to maintain its structural integrity and bioactivity through the upper respiratory tract and the lungs. Nbs are delivered directly to the lungs by an inhaler given their small size, simple and robust structure, high thermal stability, and solubility. For instance, ALX-0171, a homotrimeric Nbs, is highly effective at reducing nasal and lung RSV titers by the pulmonary administration of inhalation (Cunningham et al., 2020; Detalle et al., 2015).
To date, several Nbs against SARS-CoV-2 were reported for their in vitro activities; however, only few Nbs have recently been evaluated in vivo (Dong et al., 2020; Hanke et al., 2020; Huo et al., 2020; Nambulli et al., 2021; Pymm et al., 2021; Schoof et al., 2020; Wu et al., 2020b; Xiang et al., 2020). In the current report, anti-sera specific for RBD were elicited by immunizing an alpaca with the SARS-CoV-2 spike glycoprotein (S). Nbs specific for RBD were isolated from a phage library displaying Nbs. We identified three Nbs exhibiting potent neutralization activity against live virus and a panel of SARS-CoV-2 pseudotyped viruses. To improve the efficacy and stability, various configurations of Nbs were engineered. Nb15-NbH-Nb15, a bispecific format constituted of three Nbs, was constructed to be trivalent and bispecific for the RBD of SARS-CoV-2 and human serum albumin (HSA). This bispecific antibody exhibited potent inhibitory activity against the wild type and variants of SARS-CoV-2, including the currently circulating variants, such as the predominant mutant viruses in the United Kingdom and South Africa with an N501Y mutation. In addition, we showed that intranasal (i.n.) administration of Nb15-NbH-Nb15 provided 100% protection in both the prevention and treatment of SARS-CoV-2 -infected transgenic hACE2 mice. Nb15-NbH-Nb15 is a potential candidate for both the prevention and treatment of SARS-CoV-2.
Results
Anti-sera response elicited by the S protein
One alpaca was immunized with the extracellular domain of the SARS-CoV-2 spike protein (S1+S2 ECD, S) (Figure S1A). Compared to the pre-immunized serum (blank serum), the anti-serum after the third immunization exhibited specific serologic activities against SARS-CoV-2 S and RBD proteins with binding titers of 2.19 × 106 and 7.29 × 104, respectively (Figures S1B and S1C). The immunized serum showed potent neutralization activity against the pseudotyped SARS-CoV-2 with a half-maximal neutralization dilution (ND50) of ∼9,600 (Figure S1D). These data indicate that potent anti-serum specific for RBD with robust neutralization against SARS-CoV-2 was induced in the immunized alpaca.
Isolation of Nbs with potent neutralization activity against SARS-CoV-2
To isolate monoclonal Nbs, a C9-Nb library, a phage library displaying Nbs from the immunized alpaca, was constructed with a size of 2.0 × 109, 100% sequence diversity, and 96% in-frame rate as validated by PCR and sequencing (Figure S2A). Nbs specific for the SARS-CoV-2 S protein were isolated through three rounds of biopanning on the C9-Nb phage library by the S protein. The panned library was analyzed by phage ELISA for binding with the S protein, and we found an incremental increase of the optical density 450 (OD450) readout from 0.79 before enrichment to 1.6, 2.4, and 2.8 after the first, second, and third rounds of enrichment, respectively (Figure S2B), indicating successful enrichment. To verify whether the enriched library contained specific S-reactive phages, 40 and 46 clones were selected from the libraries after the second and third rounds, respectively, of enrichment for single-phage ELISA. The percentage of positive clones was 57.5% and 69.6% for the second and third rounds, respectively (Figure S2C). Among these positive binders, 21 distinct Nb sequences were identified according to the sequencing results (Table S1). For a further characterization, these 21 Nbs were expressed in mammalian cells by fusing the Nb gene with a human Fc1, which was cloned into the pCDNA3.4 vector to express the Nb-Fc antibody (named as Nb-Fc) (Figure S3A). ELISA results showed that all 21 Nb-Fcs reacted with the S protein; among them, 14 Nb-Fcs displayed specific binding with the SARS-CoV-2 RBD protein (Figure S3B). These results were validated by bio-layer interferometry (BLI), wherein 14 Nb-Fcs exhibited specific binding to RBD with dissociation constant (KD) values ranging from 4.25 to 37.6 nM (Figures S3C and S3D). A neutralization analysis showed potent inhibition of pseudotyped SARS-CoV-2 by culture supernatants of RBD-specific Nb15-Fc, Nb22-Fc, and Nb31-Fc (Figure S3E).
Epitope analysis of Nb-Fcs
The purified Nb15-Fc, Nb22-Fc, and Nb31-Fc exhibited dose-dependent binding with the RBD protein by ELISA (Figure S4A). In addition, Nb15-Fc, Nb22-Fc, and Nb31-Fc likely reacted with the conformational structure, as their bindings with reduced RBD protein were almost completely abolished (Figure S4B). The kinetic binding of Nb15-Fc, Nb22-Fc, and Nb31-Fc with the RBD protein ranged from a KD of 1.13 to 1.76 nM, indicating tightly clustered binding characteristics (Figures S4C–S4E), which was substantiated by the superimposed ELISA binding curves (Figure S4A). These three Nb-Fcs were next evaluated for epitope specificity in a competition assay by BLI by using the RBD protein as a capture antigen. The results revealed that the pre-bound Nb-Fcs efficiently blocked the further binding of the other two Nb-Fcs to the RBD protein, suggesting that all three Nb-Fcs likely recognize an overlapping epitope (Figures S5A–S5C). Together, Nb15-Fc, Nb22-Fc, and Nb31-Fc recognize a quaternary and overlapping epitope on RBD with nanomolar affinities.
Nb-Fcs exhibit potent neutralization against SARS-CoV-2 and variants
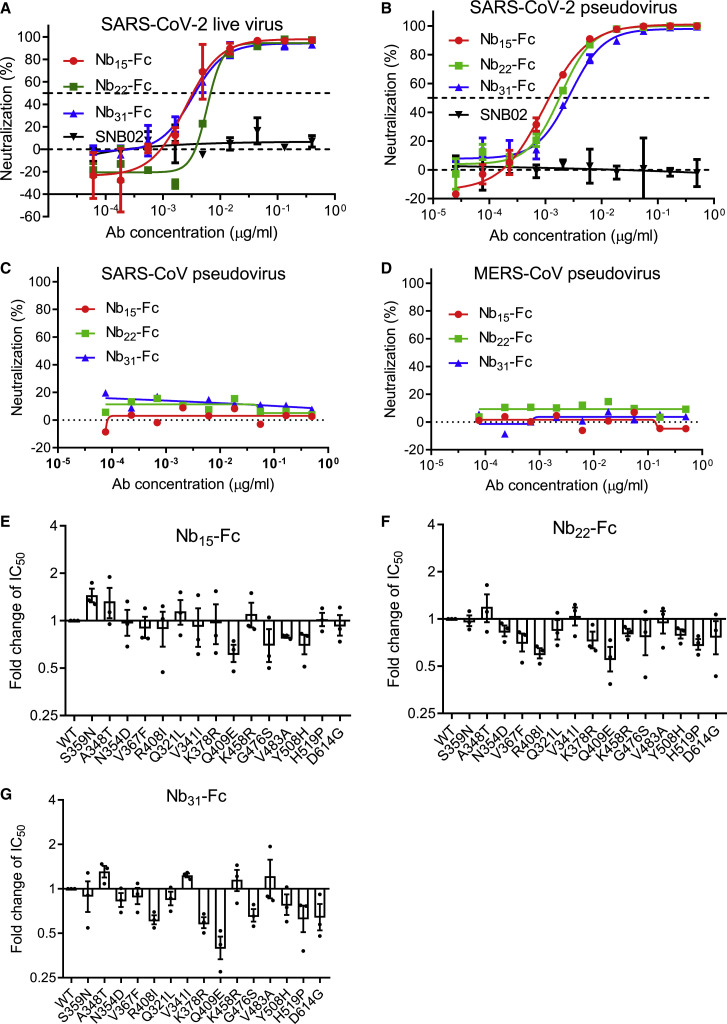
The neutralizing activity of Nb15-Fc, Nb22-Fc, and Nb31-Fc against SARS-CoV-2 live virus was investigated in Vero E6 cells. All three Nb-Fcs exhibited potent neutralization activity with the half maximal inhibitory concentration (IC50) values in the range of 0.0033–0.0068 μg/ml (41.3-75 pM) and IC90 in the range of 0.0156–0.0235 μg/ml (195–293.8 pM) (Figure 1 A; Figures S6A–S6E). The neutralizing potency was validated by the SARS-CoV-2 pseudovirus neutralization assay with consistent results, with IC50 values of 0.0008, 0.0018, and 0.0023 μg/ml (10, 22.5, and 28.8 pM), respectively (Figure 1B; Table S2). The IC50 values are comparable to those of the ultrapotent neutralizing antibodies or Nbs reported (Robbiani et al., 2020; Rogers et al., 2020; Schoof et al., 2020; Xiang et al., 2020; Zost et al., 2020). The cross neutralization of these Nbs against other coronaviruses was also evaluated in a pseudovirus assay, and the results showed that these three Nb-Fcs did not inhibit either Middle East Respiratory Syndrome (MERS)-CoV or SARS-CoV pseudovirus (Figures 1C and 1D) but inhibited 15 representative variants of SARS-CoV-2 that are identified to represent over 7,000 distinct viral genomes (Baum et al., 2020). As expected, Nb15-Fc, Nb22-Fc, and Nb31-Fc also inhibited the replication of recently arising SARS-CoV-2 variants with a D614G mutation with similar potency (Figures 1E–1G; Table S2). This evidence demonstrates the neutralizing activity of the Nb-Fcs against multiple SARS-CoV-2 variants and suggests that the Nb-Fcs target at a highly conserved epitope on the RBD protein. Taken together, these three Nb-Fcs exhibited excellent neutralization potency against the original and the representative variants of SARS-CoV-2, whereas they did not inhibit MERS-CoV and SARS-CoV infection. Given the overlapped epitope recognized by the three Nb-Fcs, Nb15-Fc with the highest neutralization potency was selected for further investigation.
Figure 1.
Characterizing the potency and breadth of neutralization conferred by Nb-Fcs
(A and B) The neutralization potency of Nb-Fcs was detected based on an authentic SARS-CoV-2 plaque reduction neutralization test (A) and the pseudotyped SARS-CoV-2 neutralization assay (B). SNB02 was taken as a negative isotype control antibody (Nb fused with human Fc1).
(C–G) Nb-Fcs were tested for the neutralization against the pseudovirus infection of SARS-CoV (C) and MERS-CoV (D). The pseudovirus of 15 SARS-CoV-2 variants identified from circulating viral sequences were tested to evaluate the neutralization potency conferred by Nb15-Fc (E), Nb22-Fc (F), and Nb31-Fc (G). The y axis shows the ratio of IC50 of indicated SARS-CoV-2 variant/IC50 of SARS-CoV-2 wild-type (WT) conferred by Nb-Fcs. The names of SARS-CoV-2 variants with an amino acid point mutation based on the wild type of SARS-CoV-2 are indicated. Data are represented as mean ± SEM. All experiments were repeated at least twice.
Construction and characterization of multiple-valent Nb15s
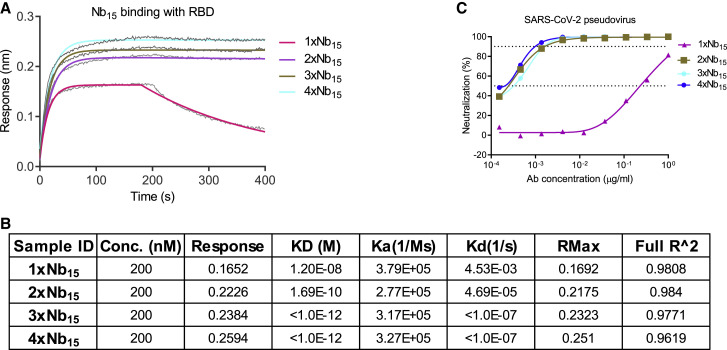
To improve potency, prolong in vivo half-life, and avoid potential Fc-mediated ADE, a number of dimeric and trimeric configurations of Nbs were engineered. Monomer (1 × Nb15), homodimer (2 × Nb15), homotrimer (3 × Nb15), and homotetramer (4 × Nb15) formats were constructed and analyzed by BLI. The binding of these constructs to the RBD protein showed an increasing KD ranging from 12 to <0.001 nM as the valence increased (Figures 2 A and 2B). Multivalent formats of Nb15 were evaluated for neutralization against pseudotyped SARS-CoV-2 infection in vitro. Monomeric 1 × Nb15 exhibited low inhibitory activity with an IC50 of 307 ng/ml (2.3 nM), whereas the bi-, tri-, and tetravalent configurations exhibited higher neutralization potency than the monomer but comparable potency among the multimers with IC50 values of 2.8, 3.5, and 2.3 ng/ml (11, 9.0, and 4.3 pM), respectively (Figure 2C; Table 1 ), suggesting that increasing valence does not confer improved anti-viral activity. As such, 3 × Nb15 was selected for further functional exploration.
Figure 2.
Characterization of Nb15s with multivalent or various formats
(A) The binding curve of multivalent Nb15s with RBD protein detected by BLI.
(B) The table summary of the binding of Nb15s with RBD protein tested by BLI.
(C) Multivalent Nb15s and various formats were evaluated for neutralization potency against pseudotyped SARS-CoV-2 infection.
Table 1.
Summary of various Nbs inhibiting pseudotyped SARS-CoV-2
| Nbs | IC50 (mean ± SD μg/ml) | IC80 (mean ± SD μg/ml) | IC90 (mean ± SD μg/ml) |
|---|---|---|---|
| 1 × Nb15 | 0.3074 ± 0.0237 | 0.5059 ± 0.0699 | 0.622 ± 0.0969 |
| 2 × Nb15 | 0.0003 ± 0 | 0.0008 ± 0.0001 | 0.0011 ± 0.0001 |
| 3 × Nb15 | 0.0004 ± 0 | 0.001 ± 0 | 0.0014 ± 0.0001 |
| 4 × Nb15 | 0.0002 ± 0.0001 | 0.0008 ± 0.0001 | 0.0011 ± 0.0002 |
| Nb15-NbH | 0.5529 ± 0.0889 | 1.071 ± 0.1754 | 1.374 ± 0.2263 |
| NbH-Nb15 | 0.1974 ± 0.004 | 0.3469 ± 0.0533 | 0.4344 ± 0.0822 |
| Nb15-Nb15-NbH | 0.0251 ± 0.0058 | 0.0419 ± 0.0051 | 0.0517 ± 0.0047 |
| NbH-Nb15-Nb15 | 0.0008 ± 0.0001 | 0.0017 ± 0.0003 | 0.0022 ± 0.0004 |
| Nb15-NbH-Nb15 | 0.0004 ± 0 | 0.0009 ± 0 | 0.0013 ± 0 |
| Nb15-Fc | 0.0009 ± 0.0001 | 0.0019 ± 0.0001 | 0.0025 ± 0.0002 |
Nb15-NbH-Nb15, heterotrimeric and bispecific for RBD and HSA, exhibits potent neutralization against SARS-CoV-2
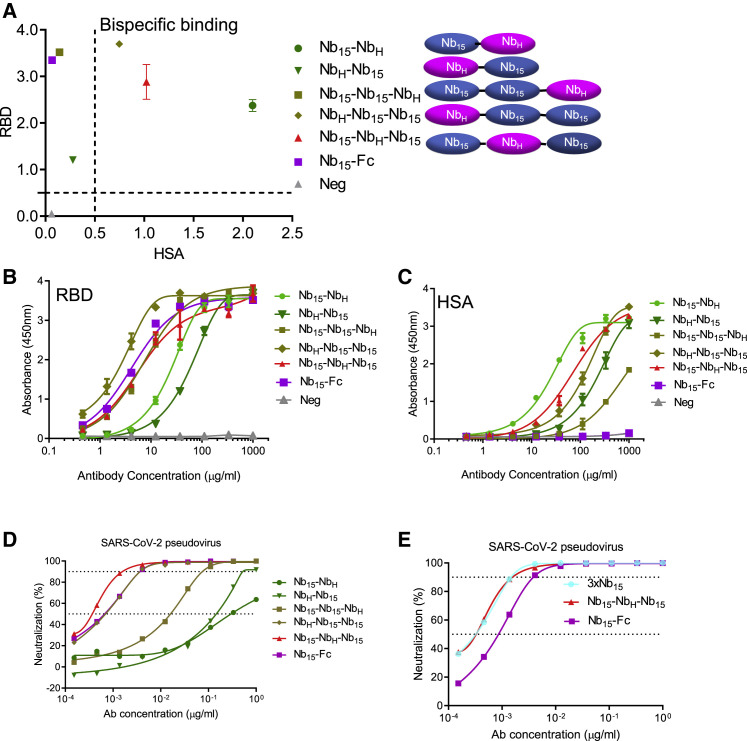
In order to improve Nb15 efficacy and stability in vivo, we constructed bispecific Nbs consisting of one Nb specific for HSA (NbH) developed by our lab and one or two Nb15s specific for RBD with (G4S)3 as the linker between each Nb (Figure 3 A) and analyzed their binding and viral inhibitory activities. In addition to the heterodimeric configuration of Nb-NbH that was previously reported (Van Roy et al., 2015), various configurations of Nbs were engineered as depicted in Figure 3A. An ELISA analysis showed that all combinations containing Nb15 reacted with the RBD protein; among them, heterotrimeric Nb15-Nb15-NbH, NbH-Nb15-Nb15, and Nb15-NbH-Nb15 exhibited better binding with the RBD protein than heterodimeric Nb15-NbH and NbH-Nb15 configurations (Figures 3A and 3B). Furthermore, Nb15-NbH, NbH-Nb15-Nb15, and Nb15-NbH-Nb15 were the top HSA binders as compared to other configurations (Figures 3A and 3C). Bispecific Nbs in various configurations were tested for the inhibition of SARS-CoV-2 infection; among them, Nb15-NbH-Nb15 exhibited the most potent neutralization of the pseudotyped virus with an IC50 of 0.4 ng/ml (9.0 pM) (Figure 3D; Table 1). We next compared Nb15-NbH-Nb15 with homotrimer Nbs (3 × Nb15) or Nb-Fc for their binding and anti-viral activities and found that 3 × Nb15, Nb15-NbH-Nb15, and Nb15-Fc exhibited comparable potency with IC50 values of 0.4, 0.4, and 0.9 ng/ml (9.0, 9.0, and 11.3 pM), respectively (Figure 3E; Table 1).
Figure 3.
Design and characterization of bispecific Nbs
(A) Various Nbs at 37 μg/ml binding to RBD, and HSA protein identified by ELISA.
(B and C) The binding curve of Nbs interacting with RBD protein (B) and HSA protein (C) identified by ELISA.
(D) The neutralizing potency of bispecific Nbs against SARS-CoV-2 pseudovirus measured by neutralization assay.
(E) The neutralizing potency of Nb15-Fc, 3 × Nb15, and Nb15-NbH-Nb15 against SARS-CoV-2 pseudovirus infection measured by neutralization assay.
Data are represented as mean ± SEM. All experiments were repeated at least twice.
Pharmacokinetics and delivery of Nb constructs
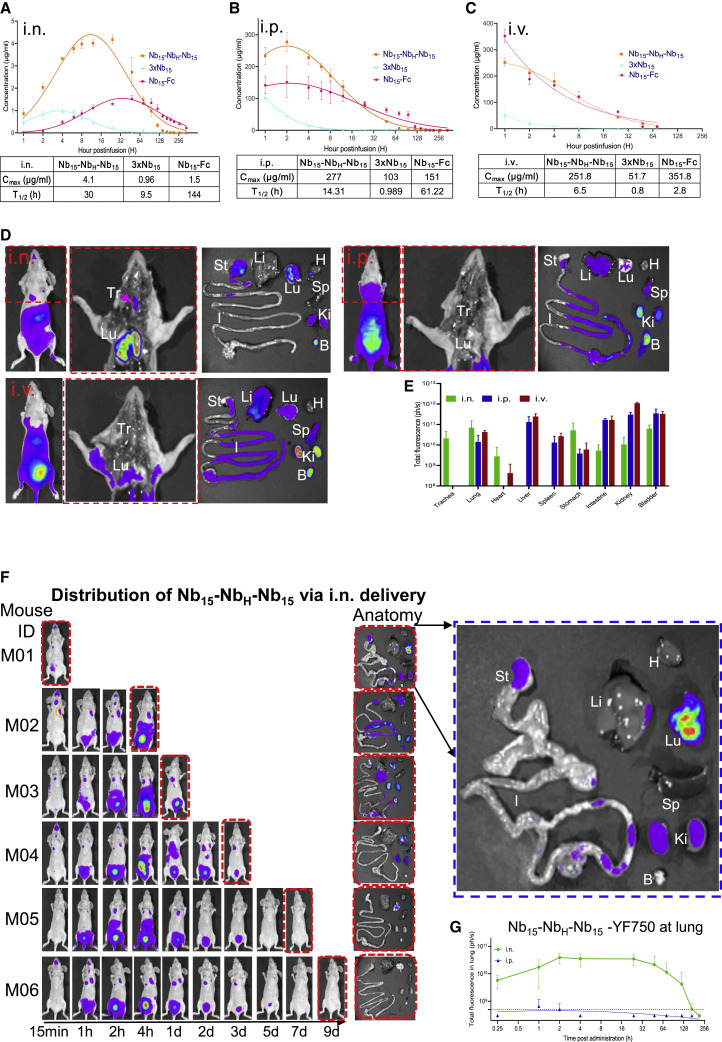
Given that 3 × Nb15, Nb15-NbH-Nb15, and Nb15-Fc exhibited comparable neutralization activity (Figure 3E), these three constructs were evaluated for their in vivo pharmacokinetic activity. The results showed that, when administrated by i.n., intraperitoneal (i.p.), or intravenous (i.v.) routes, 3 × Nb15 was rapidly metabolized as compared to Nb15-Fc and Nb15-NbH-Nb15 (Figures 4 A–4C); therefore, 3 × Nb15 was ruled out for further analysis. To determine the tissue distribution of Nbs, YF®750 SE-labeled Nb15-NbH-Nb15 (Nb15-NbH-Nb15-YF750) were administered by i.n., i.p., or i.v. routes in mouse model. The results revealed that the fluorescence in trachea could be detected only when Nb15-NbH-Nb15-YF750 was administered i.n. Furthermore, the fluorescence intensity was higher in lungs when Nb15-NbH-Nb15-YF750 was administered i.n. (6.9 × 1010 ph/s) than that when administered i.p. or i.v. (1.4 × 1010 and 4.3 × 1010 ph/s, respectively) (Figures 4D and 4E). In addition, the results also showed that Nb15-NbH-Nb15 could reach lungs and was sustained for more than 168 h (7 days) when administrated i.n.; in contrast, the fluorescence could only be detected between 1 and 2 h after i.p. infusion (Figures 4F and 4G). These results suggest that i.n. administration of Nb15-NbH-Nb15 will be a favorable route for the antibody to reach the nasopharynx and lungs where SARS-CoV-2 replicates. Therefore, to avoid the potential ADE associated by Fc in the Nb-Fc, we selected Nb15-NbH-Nb15 for further efficacy evaluation in vivo.
Figure 4.
Pharmacokinetics of Nb15s in vivo
(A–C) Bioavailability and T1/2 (half-life) of Nb15s in BALB/c mice. Nb15 variants were intranasally (i.n.) administered into mice (n = 3, female) at 200 μg (average of 10 mg/kg mice) (A), intraperitoneally (i.p.) administered into mice (n = 3, female) at 400 μg (average of 20 mg/kg mice) (B), and intravascularly (i.v.) administered into mice (n = 3, female) at 400 μg (average of 20 mg/kg mice) (C). Serum concentrations of the Nbs were determined at indicated time points by ELISA. Nb15 variants are colored as follows: Nb15-Fc (red), Nb15-NbH-Nb15 (orange), and 3 × Nb15 (cyan). Cmax, maximum observed plasma concentration. Data are represented as mean ± SEM.
(D) Spatial distribution of Nb15-NbH-Nb15YF750 1 h after infusion into mice (n = 3 in each group) by i.n., i.p., and i.v. routes was detected by a NightOwl LB 983 system. The middle figure in the red dashed line shows the dissected image of the left mouse in the red dashed line. The right figure shows the organs from dissected mice that were imaged immediately after sacrifice. Tr, trachea; Lu, lung; H, heart; Li, liver; Sp, spleen; St, stomach; I, large and small intestine; Ki, kidneys; B, bladder.
(E) The fluorescence intensity (ph/s) summary of each organ in (D) was quantified and presented as the mean ± SEM.
(F) Pharmacokinetics of Nb15-NbH-Nb15-YF150 by i.n. administration at the indicated time point. Mice were sacrificed at the indicated time point for the analysis of fluorescence intensity in various organs labeled as in (D). The blue dashed line figure shows the enlarged image of the individual figure indicated by corresponding arrows.
(G) Nude mice (n = 3–6) were administered with Nb15-NbH-Nb15-YF750 by the i.n. or i.p. route. The fluorescence intensity at the lung location as shown in the yellow dashed line circle of M02 in (F) was measured at the indicated time point. Data are represented as mean ± SEM.
In vitro characterization of Nb15-NbH-Nb15
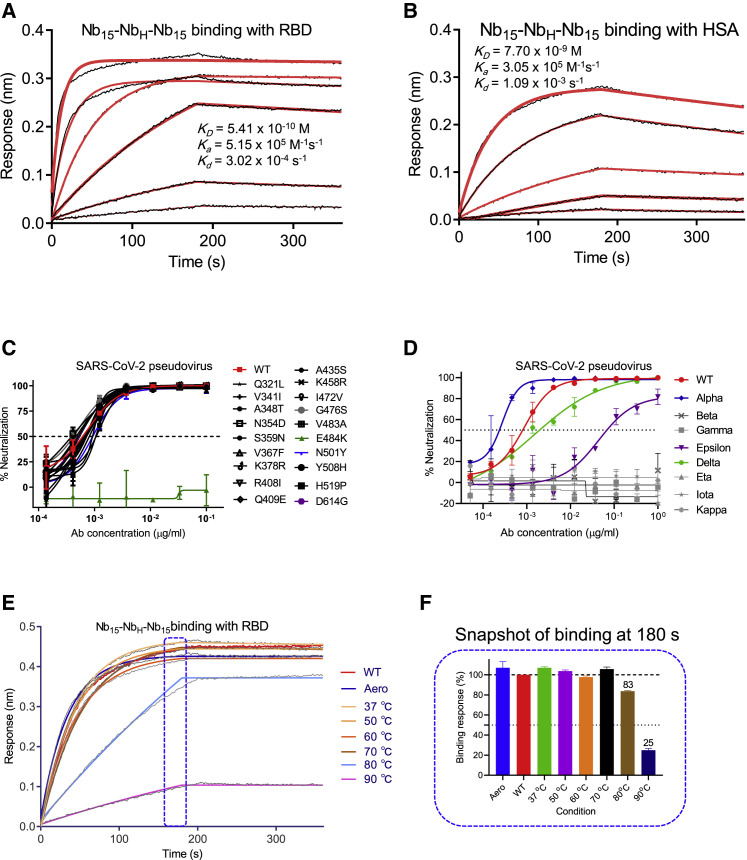
Nb15-NbH-Nb15 was further characterized in vitro. Nb15-NbH-Nb15 exhibited specific binding to RBD and HSA with KD values of 0.54 and 7.7 nM, respectively (Figures 5 A and 5B). In addition, Nb15-NbH-Nb15 also showed specific binding with murine serum albumin (MSA) with KD values of 14.5 nM (Figure S7), indicating that mice can be used as an animal model to investigate the half-life of Nb15-NbH-Nb15. Furthermore, Nb15-NbH-Nb15 exhibited sub-ng/ml (pM) potency against both the wild type (WHU01) and 18 out of 19 currently circulating SARS-CoV-2 mutant variants tested (Figure 5C; Table S2). Importantly, Nb15-NbH-Nb15 showed comparable potency against the pseudotyped SARS-CoV-2 variants with D614G and N501Y mutations that circulate predominantly in the United Kingdom and South Africa. Moreover, Nb15-NbH-Nb15 exhibited neutralization against pseudotyped variants of the wild type, Alpha (N501Y), Epsilon (L452R), and Delta (L452R and T478K) with IC50 values of 0.4 ng/ml, 0.26 ng/ml, 88.95 ng/ml, and 5.16 ng/ml (9.0, 5.9, 2001, and 116 pM), respectively. However, it failed to neutralize Gamma (E484K and K417T), Beta (E484K and K417N), and other circulating variants (Figure 5D). Nb15-NbH-Nb15 also showed excellent thermal stability by retaining 100% and 83% activities even at 70°C and 80°C for 1 h, respectively (Figures 5E and 5F; Table S3). Furthermore, Nb15-NbH-Nb15 retained 100% activity after aerosolization, indicating the potential application as a nebulized drug (Figures 5E and 5F; Table S3).
Figure 5.
Functional characterization of Nb15-NbH-Nb15
(A and B) Kinetic binding curve of Nb15-NbH-Nb15 at the concentrations 300 nM, 100 nM, 33.3 nM, 11.1 nM, 3.7 nM, and 1.2 nM with RBD (A) and HSA (B) by BLI. Binding curves are colored black, and the fit of the data to a 1:1 binding model is colored red.
(C) The neutralization curve of Nb15-NbH-Nb15 inhibiting SARS-CoV-2 pseudovirus and its variants with amino acid point mutation as indicated. WT, wild-type SARS-CoV-2 pseudovirus of 2019-nCoV WHU01 (accession ID: MN988668.1).
(D) The neutralization curve of Nb15-NbH-Nb15 inhibiting SARS-CoV-2 pseudovirus of circulating variants. WT was indicated as in (C). Alpha, B.1.1.7 variant reported in United Kingdom; Beta, B.1.351 variant reported in S. Africa; Gamma, P.1 variant reported in Brazil; Epsilon, B.1.429 variant reported in California; Delta, B.1.617.2 variant reported in India; Eta, B.1.525 variant reported in UK; Iota, B.1.526 variant reported in New York; Kappa, B.1.617.1 variant reported in India.
(E) Binding curve of RBD with Nb15-NbH-Nb15 at the concentration of 133 nM (5 μg/ml) before (no treatment, WT) or after aerosolization (Aero) or after treatment at the indicated temperature, including 37°C, 50°C, 60°C, 70°C, 80°C, and 90°C for 1 h. Binding curves are colored black, and the fit of the data to a 1:1 binding model is colored as indicated.
(F) Snapshot of the relative binding response of the highest binding response at the indicated condition/the highest response of RBD with Nb15-NbH-Nb15 in the no-treatment condition (WT). The relative binding of WT was normalized as 100%. Data are represented as mean ± SEM. All experiments were repeated at least twice.
In vivo anti-SARS-CoV-2 activity of Nb15-NbH-Nb15
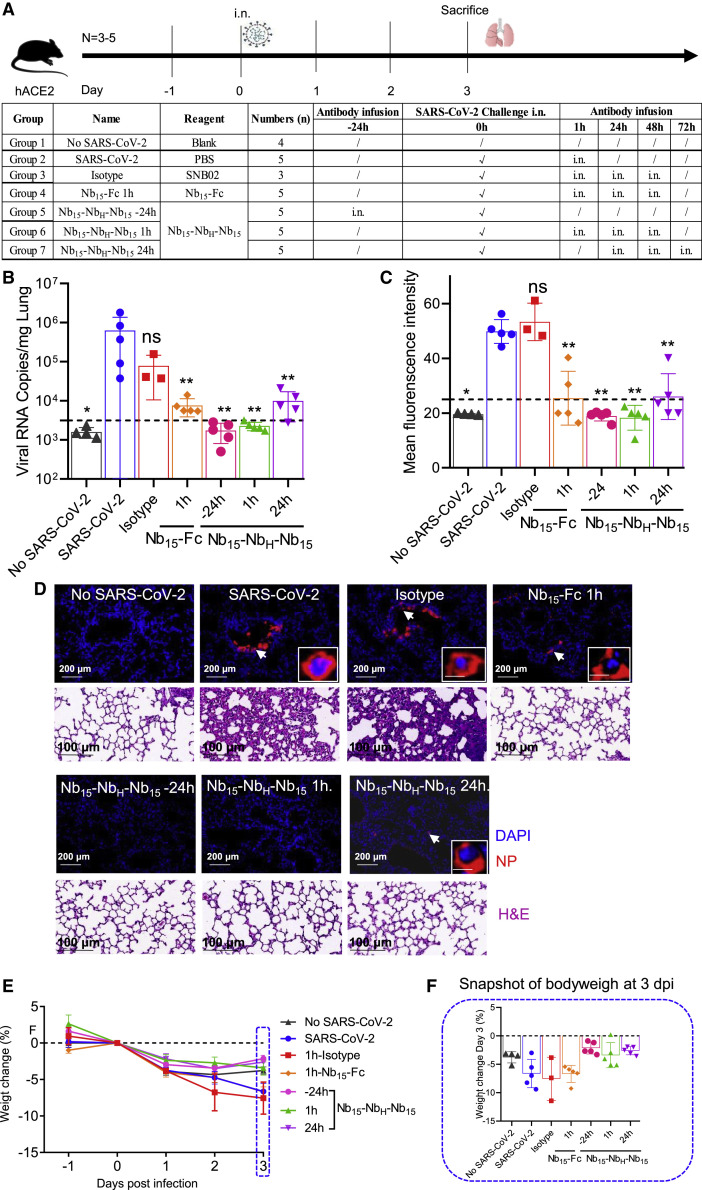
To evaluate the efficacy of Nb15-NbH-Nb15 in vivo, hACE2 transgenic mice were challenged with 1 × 105 PFU SARS-CoV-2 (Strain IVCAS 6.7512; Zhou et al., 2020), and Nb15-NbH-Nb15 was administrated i.n. either before or after viral challenge for prophylactic or therapeutic efficacy (Figure 6 A). Viral RNA was detected in the lungs of control mice (6.28 × 105 copies/mg on average in SARS-CoV-2 group, n = 5) and the isotype-treated control mice (7.8 × 104 copies/mg on average in isotype group, n = 3). For the prophylactic group, no viral RNA or infected cells were detected in 100% (5/5) of the mice when 250-μg (average of 10 mg/kg) Nb15-NbH-Nb15 was administrated i.n. 24 h before SARS-CoV-2 infection (Nb15-NbH-Nb15 −24h group, n = 5), as evidenced by real-time PCR and immunofluorescence staining (Figures 6B–6D). A total of 100% of mice were also completely protected when 250-μg Nb15-NbH-Nb15 as administrated i.n. 1 h postinfection, as no viral RNA and infected cells were detected in all infected mice (Nb15-NbH-Nb15 1-h group, n = 5) (Figures 6B–6D). Significantly lower SARS-CoV-2 RNA copies (9.98 × 103 copies/mg on average) were detected in the lungs of the mice treated with Nb15-NbH-Nb15 i.n. 24 h postinfection (Nb15-NbH-Nb15 24 h group, n = 5) than those in the control mice (6.28 × 105 copies/mg on average in SARS-CoV-2 group and 7.8 × 104 copies/mg in isotype control) (Figures 6B–6D). Nb15-Fc inhibited viral replication and reduced the viral copy number (average of 7.59 × 103 copies/mg in Nb15-Fc 1-h group, n = 5) but failed to provide complete protection under the same conditions as Nb15-NbH-Nb15 (Figures 6B–6D). Furthermore, a histopathological analysis of lung tissues showed that SARS-CoV-2 challenge induced severe lung lesions, as shown by the infiltration of inflammatory cells and thickened alveolar septa (Figure 6D). In contrast, the lungs of the mice receiving Nb15-NbH-Nb15 or Nb15-Fc treatment showed no apparent pathological changes (Figure 6D). Together, Nb15-NbH-Nb15 at an average of 10 mg/kg administrated i.n. 24 h before or 1 h after challenge provided complete protection against SARS-CoV-2 infection and significantly inhibited SARS-CoV-2 replication when the antibody was administrated 24 h postinfection. Nb15-Fc used at an average of 10 mg/kg administrated i.n. 1 h after challenge significantly reduced viral load but failed to provide complete protection. We noted that those mice receiving Nb15s treatment showed less weight loss than the control mice but did not achieve a statistical difference (Figures 6E and 6F). These results indicate that Nb15-NbH-Nb15, when used early during infection, conferred a higher protection efficacy than that used at a later time point. In summary, the Nb15-NbH-Nb15 configuration administered i.n. was superior to Nb15-Fc and exhibited both prophylactic and therapeutic efficacy against SARS-CoV-2 challenge.
Figure 6.
The efficacy of Nb15s evaluated in hACE2 transgenic mice challenged by SARS-CoV-2
(A) Experimental schedule of Nb15s in the prevention and treatment of SARS-CoV-2 infection. Bottom, table summary of groups (n = 3–5 mice) with different treatments.
(B) Viral loads in lungs among 7 groups were measured by qRT-PCR. The name of each group in the x axis was indicated as in the table in (A). Each dot represents one mouse. The limit of detection was 3,160 copies/mg referenced to blank control (no-SARS-CoV-2 group).
(C) Sections of lungs were analyzed by immunofluorescence staining by using antibodies specific to SARS-CoV-2 nucleocapsid protein (NP) in red and DAPI (4′,6-diamidino-2-phenylindole) for nuclei in blue, respectively. The fluorescence signal intensity of red was taken as a quantitative indicator for viral infection, which was calculated by ImageJ software.
(D) Representative sections of lung in (C) were visualized under the ×20 objective at the indicated scale bar (200 μm). The insets are enlarged images of individual cells indicated by corresponding arrows at the indicated scale bar as 10 μm. H&E staining was conducted to analyze the lung inflammation and observed at the indicated scale bar as 100 μm.
(E) Body weights of mice among the above 7 groups were recorded. Each line represents data from one group.
(F) Snapshot of body weight on 3 days post infection in (E) was plotted. Data are represented as mean ± SEM; Mann-Whitney test was performed to compare treatment group with the SARS-CoV-2 control group. ns, no significance; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. Data of (B), (C), (E), and (F) are represented as mean ± SEM. All experiments of (B) and (C) were repeated twice.
Discussion
In this study, three potent neutralizing Nb-Fcs were isolated from a phage display platform derived from an alpaca immunized with the SARS-CoV-2 S protein. These three RBD-specific Nb-Fcs exhibited potent inhibitory activities against 15 mutant variants of pseudotyped SARS-CoV-2 with IC50 values in the range of 0.5-2.8 ng/ml (Figures 1E–1G; Table S2). The IC50 values are comparable to those of the most potent neutralizing antibodies or Nbs reported (Robbiani et al., 2020; Rogers et al., 2020; Schoof et al., 2020; Xiang et al., 2020; Zost et al., 2020). These 15 representative variants of SARS-CoV-2 are identified to represent over 7,000 distinct viral genomes (Baum et al., 2020). As expected, recently arising variants with a D614G mutation were also sensitive to the neutralization by Nb15-Fc, Nb22-Fc, and Nb31-Fc with similar sensitivity (Figures 1E–1G; Table S2). This evidence demonstrates the neutralizing activity of these Nbs against multiple SARS-CoV-2 variants and suggests that the Nb-Fcs target at a highly conserved epitope on the RBD protein.
To improve potency, prolong in vivo half-life, and avoid potential Fc-mediated ADE, NbH specific for HSA and MSA was used to construct a trimeric Nb15 (Nb15-NbH-Nb15) for SARS-CoV-2, and the resulting Nb15-NbH-Nb15 exhibited the highest neutralization potency with the IC50 value of 0.4 ng/ml among other configurations, including Nb15-NbH, a configuration reported earlier (Van Roy et al., 2015). Interestingly, we found that Nb15-NbH and NbH-Nb15 with the same components exhibited distinct neutralization potencies with IC50 values of 552.3 ng/ml and 197.4 ng/ml, respectively. In addition, NbH-Nb15-Nb15 and Nb15-Nb15-NbH also displayed distinct neutralization potencies with IC50 values of 25.1 ng/ml and 8 ng/ml, respectively (Figure 3D; Table 1), indicating that the Nb configuration has an effect on the neutralizing activity. In addition, the heterotrimeric bispecific configuration is superior to the bispecific heterodimer. We also noted that the bi-, tri-, and tetravalent configurations exhibited comparable potency with IC50 values of 2.8, 3.5, and 2.3 ng/ml (11, 9.0, and 4.3 pM), respectively. The neutralizing potency did not correspond to the valence increase when there are two or more than two Nb15s, although monomeric 1 × Nb15 had a much lower inhibitory activity (Figure 2C; Table 1). We noted that Nb15-NbH-Nb15 shows higher potency than NbH-Nb15-Nb15, Nb15-Nb15-NbH, and all the homomultimers, suggesting that the position of NbH plays important roles in neutralization activity. We speculate that in Nb15-NbH-Nb15, NbH may space out the two Nb15s to either avoid cross intereference with each other or allow better binding of the trimeric Nb to S proteins on the viral particle. Furthermore, Nb15-NbH-Nb15 displayed comparable neutralizing potency as those of Nb15-Fc and 3 × Nb15 and higher neutralization potency and longer half-life than 3 × Nb15 in vivo when delivered via i.n., i.p. or i.v. routes. Altogether, Nb15-NbH-Nb15 was established as a potentially promising construct, suggesting a format of nanobodies with improved potency and half-life in vivo.
Importantly, Nb15-NbH-Nb15 exhibited neutralizing activities against 18 out of 19 pseudotyped SARS-CoV-2 variants except the E484K mutant variant that we tested. Moreover, Nb15-NbH-Nb15 exhibited neutralization against pseudotyped variants of wild type (Wuhan-Hu-01), Alpha (N501Y, B1.1.7 variant, United Kingdom), Epsilon (L452R, B1.429 variant, California), and Delta (L452R and T478K, B.1.617.2 variant, India), which has become predominant and contributed to the current wave of infection in many countries. Delta has been designated as a variant of concern (VOC) and is believed to be 60% more transmissible than variant Alpha (Planas et al., 2021). Some mAbs, including bamlavinimab, lost binding to the Spike and no longer neutralized variant Delta (Planas et al., 2021). Nb15-NbH-Nb15 exhibited potent neutralization against variant Delta (IC50 values of 5.16 ng/ml) with about 20-fold less potent neutralization against Alpha (IC50 values of 0.26 ng/ml) (Figure 5D). However, it failed to neutralize P.1 (E484K and K417T, Gamma, Brazil), B1.351 (E484K and K417N, Beta, South Africa), B.1.617.1 (L452R and E484Q, India), and other circulating variants (Figure 5D), suggesting a E484K/Q mutation variant could be resistant to the neutralization of Nb15-NbH-Nb15.
Although several neutralizing antibodies against SARS-CoV-2 are in various stages of clinical trials or have been approved as an emergency therapy, most of these antibodies are of limited efficacy. SARS-CoV-2 is present mainly in the nasopharynx and lungs (Gallo et al., 2020; Higgins et al., 2020). Differing from many previously reported therapies with a systemic delivery route, Nb15-NbH-Nb15 was delivered to the site of infection. Direct administration to the airways is likely to provide faster and more robust antiviral activity in the respiratory tract, where the virus gains entry and replicates (Cunningham et al., 2020; Higgins et al., 2020), as i.n. delivery has been shown to result in fast and efficient drug delivery to the main site of SARS-CoV-2 infection, i.e., the upper and lower respiratory tract (Higgins et al., 2020). Indeed, the therapeutic effect of topical administration of ALX-0171 displayed promising results in reducing the RSV viral load (Cunningham et al., 2020; Detalle et al., 2015). The current study demonstrated the in vivo efficacy of Nbs against SARS-CoV-2 infection by i.n. delivery.
In summary, compared to the configurations of Nb-NbH, Nb-Fc, and Nb homotrimer, a potential promising construct of Nb15-NbH-Nb15 exhibited a higher neutralization activity and longer half-life in vivo. Our results provide a strategy for the construction of multivalent Nbs with improved potency and half-life in vivo. Nb15-NbH-Nb15 exhibited highly potent antiviral activity against a large panel of SARS-CoV-2 clinical variants except E484K/Q mutants. Furthermore, direct delivery of Nb15-NbH-Nb15 to the airways/lungs by the i.n. route proved an effective mode of drug delivery, and the outstanding thermal stability of Nb15-NbH-Nb15 is an additional advantage. We suggest that respiratory delivery of Nb15-NbH-Nb15 is a promising route for the prevention and treatment of SARS-CoV-2 infection and thus warrants further clinical evaluation by i.n. delivery.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-human IgG conjugated with an IRDye 800CW | Rockland | Cat#926-32232 |
| Anti-rabbit IgG conjugated with an IRDye 800CW | Rockland | Cat#925-32211 |
| Goat anti-llama IgG (H+L) secondary antibody with HRP | Novus | Cat# NB7242 |
| Anti-M13 bacteriophage antibody with HRP | Sino Biological | Cat#11973-MM05T-H |
| Rabbit anti-SARS-CoV-N IgG | Sino Biological | Cat#40143-R019 |
| HRP-conjugated goat anti-rabbit IgG (H+L) antibody | Jackson ImmunoResearch | Cat#111-035-003 |
| SARS-CoV/SARS-CoV-2 Nucleocapsid Antibody, Mouse mAb | Sino Biological | Cat#40143-MM05 |
| SNB02 | Y-Clone | Cat#YL2018-003 |
| Nanobody against HSA (NbH) | Abrev | Cat#AR2020-010 |
| Bacterial and virus strains | ||
| TG1 bacteria | Y-Clone | N/A |
| M13KO7 helper phage | Invitrogen | Cat#18311019 |
| SARS-CoV-2 live virus of Beta/Shenzhen/SZTH-003/2020 for cell assay | Shenzhen Third People’s Hospital | EPI_ISL_406594 at GISAID |
| SARS-CoV-2 pseudovirus and its variants | This Paper | N/A |
| SARS-CoV pseudovirus | This Paper | N/A |
| Mers-CoV Pseudovirus | This Paper | N/A |
| SARS-CoV-2 live virus for animal study | Wuhan Institute of Virology | IVCAS 6.7512 |
| Biological samples | ||
| Nb Phage library | This Paper | C9-Nb-lib |
| Chemicals, peptides, and recombinant proteins | ||
| SARS-CoV-2 Spike protein (S1+S2 ECD, S) | Sino Biological | Cat#40589-V08B1 |
| SARS-CoV-2 RBD protein | Sino Biological | Cat#40592-V08H |
| Freund’s complete adjuvant | Sigma | Cat#F5881-10ML |
| Freund’s incomplete adjuvant | Sigma | Cat#F5506-10M |
| 3,3′,5,5′-Tetramethylbenzidine | Sigma | Cat#54827-17-7 |
| Ficoll-Paque Plus | GE | Cat#17-1140-02 |
| TRIzol reagent | Life Technologies | Cat#15-596-018 |
| KPL TrueBlue Peroxidase Substrates | SeraCare Life Sciences Inc. | Cat#5510-0030 |
| 10% neutral buffered formalin | Sigma | Cat# Z2902 |
| Far infrared dye YF®750 SE | US EVERBRIGHT INC | YS0056 |
| Critical commercial assays | ||
| Anti-human Fc (AHC) biosensors | Fortebio | Cat#18-5060 |
| AR2G biosensor | Fortebio | Cat#18-5092 |
| Amino coupling kit | Fortebio | Cat#18-5095 |
| RT-PCR Prime Script Kit | Takara | Cat#PR005B |
| Experimental models: Cell lines | ||
| HEK293T-ACE2 cells | Yeasen Biotech | Cat# 41107ES0 |
| Huh7 cells | ATCC | CCL-185 |
| 293T cells | ATCC | CRL-3216 |
| 293-F cells | Thermo Fisher Scientific | R79007 |
| Experimental models: Organisms/strains | ||
| Transgenic hACE2 mice (C57BL/6J) | GemPharmatech | Cat T037630 |
| Alpaca | Y-Clone | AR-0019 |
| BALB/c | Qing Long Shan Animal Center | Qls02-0202 |
| Nude mice | Qing Long Shan Animal Center | Qls03-0102 |
| Oligonucleotides | ||
| TaqMan probe (5′-FAM− CAGGT GGAACCTCATCAGGAGATGC −MGB-3′) |
This paper | N/A |
| Primer of orf1ab gene of SARS-CoV-2 Forward: (5′- GTGARATGGTCATGTGTGGCGG −3′′) | This paper | N/A |
| Primer of orf1ab gene of SARS-CoV-2 Reverse: (5′- CARATGTTAAASACACTATTAGCATA −3′) | This paper | N/A |
| Recombinant DNA | ||
| pCDNA3.1-S encoding spike protein of variant coronavirus | This paper | N/A |
| pCDNA3.4 eukaryotic expression vector | Invitrogen | Cat#A14697 |
| HIV-1 NL4-3 ΔEnv Vpr Luciferase Reporter Vector (pNL4-3.Luc.R-E-) | HIV AIDS Reagent Program | Cat#3418 |
| phV1 phagemid plasmid | Y-Clone | N/A |
| pcDNA3.4-Nbs | This paper | N/A |
| Software and algorithms | ||
| GraphPad Prism 8.01 | GraphPad Software Inc. | https://www.graphpad.com/scientific-software/prism/; RRID:SCR_002798 |
| ImageJ | https://imagej.nih.gov/ij/ | https://imagej.nih.gov/ij/; RRID: SCR_001935 |
| Indigo imaging software | Berthold | https://www.berthold.com/en/bioanalytic/products/in-vivo-imaging-systems/nightowl-lb983/ |
| OriginPro 8.5 software | OriginLab | https://www.originlab.com/; RRID:SCR_014212 |
| Others | ||
| 96-well plates | Corning | Cat# 9018 |
| Protein G | Thermo Fisher Scientific | Cat# 20399 |
| Ni-NTA | Thermo Fisher Scientific | Cat#R901100 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Zhiwei Wu (wzhw@nju.edu.cn).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Alpaca used in the immunization
An alpaca (Female, 2-3 years old, cat.# AR-0019) was purchased from Xuzhou Animal Center (Xuzhou, China). Animal experiments were done by veterinarian with the authorization of animal operation and in accordance with the China law for animal protection.
hACE2 mice in the evaluating efficacy of Nbs in vivo.
A total of 31 8-week-old male transgenic hACE2 mice (C57BL/6J) (cat.# T037630) were purchased from GemPharmatech Co., Ltd. (Nanjing, China). All of the hACE2 mice were handled in Biosafety Level 3 animal facilities in accordance with the recommendations for care and use of the Institutional Review Board of Wuhan Institute of Virology of the Chinese Academy of Sciences (Ethics Number: WIVA11202003).
BALB/c mice
BALB/c (6-8 weeks of age, Female, cat.# Qls02-0202) was purchased from Qing Long Shan Animal Center (Nanjing, China). The BALB/c mice were kept in ventilated cages and given access to standard pellet feed and water ad libitum following Nanjing University Laboratory Animal center’s standard operational procedures.
Nude mice in spatial distribution of Nbs in vivo
Nude mice (18-22 g, female, Qls03-0102) were purchased from Qing Long Shan Animal Center (Nanjing, China). The nude mice were kept in ventilated cages and given access to standard pellet feed and water ad libitum following Nanjing University Laboratory Animal center’s standard operational procedures.
Cell lines
293T cells (ATCC), HEK293T-ACE2 cells (Cat# 41107ES0, Yeasen Biotech) and Huh7 cells (ATCC) were cultured in Dulbecco’s Modified Eagle Medium (DMEM) (Thermo Fisher Scientific) containing 10% Fetal bovine serum, 100 U/mL penicillin and 2 mM L-glutamine and were incubated at 37°C in 5% CO2 setting. 293-F cells (Thermo Fisher Scientific) were cultured in Expi293TM Expression Medium and were incubated in a 37°C incubator and 8% CO2 setting on an orbital shaker platform at 130 rpm according to the manufacturer’s instructions.
Study approval
The study and the protocol for this research were approved by the Center for Public Health Research, Medical School, Nanjing University. All animal experimental procedures without infection were approved by the Committee on the Use of Live Animals by the Ethics Committee of Nanjing University. All of the animals infected by SARS-CoV-2 were handled in Biosafety Level 3 animal facilities in accordance with the recommendations for care and use of the Institutional Review Board of Wuhan Institute of Virology of the Chinese Academy of Sciences (Ethics Number: WIVA11202003). All the authors declare their compliance with publishing ethics.
Method details
SDS-PAGE and western blotting (WB)
The purified protein or antibody was separated by electrophoresis in a 7.5%–12% polyacrylamide gel. The separated protein or antibody was revealed either using Coomassie blue or transferred to PVDF membrane for WB analysis under reducing or non-reducing conditions with β-mercaptoethanol. The membrane was first blocked and then incubated overnight at 4°C or 37°C for one hour with diluted plasma or antibody, followed by incubation with the secondary antibody of either anti-human IgG or anti-rabbit IgG conjugated with an IRDye 800CW (cat.# 926-32232, Rockland). Protein bands were visualized using the Odyssey Image System (Li-COR).
ELISA analysis
Anti-sera titer and antibody characterization or antibody quantification in vivo were examined by ELISA as reported in our previously published method (Wu et al., 2019) with modifications. In brief, the protein was coated to high protein-binding ELISA plates (Corning) at a concentration of 0.5 μg/ml, 100 μL per well at 37°C for 2 hours (h) or 4°C overnight. After washing, blocking buffer with 5% non-fat milk in PBS was added and incubated at 37°C for 1 h. After washing 2-4 times, 100 μL serially diluted anti-serum or purified antibody was added and incubated at 37°C for 1.5 h. Following washing, goat anti-llama IgG (H+L) secondary antibody with HRP (Novus, cat.# NB7242, 1:10000 dilution) was added and incubated at 37°C for 1 h. Accordingly, 3,3′,5,5′-Tetramethylbenzidine (TMB, Sigma) substrate was added at 37°C for 10 minutes (min); and the reaction was stopped by adding 10 μL 0.2 M H2SO4. The optical densities at 450 nm were measured using the Infinite 200 (Tecan, Ramsey, MN, USA). Antibody titers were defined as the highest dilution when the diluted serum produced at least 2.1-fold optical density readout as compared to the control serum sample at the same dilution.
Alpaca immunization
250 μg the extracellular domain of SARS-CoV-2 spike protein fused with His tag (S1+S2 ECD, S, cat.# 40589-V08B1, Sino Biological) was emulsified with 250 μL Freund’s complete adjuvant (F5881-10ML, Sigma) to immunize an alpaca (Female, 2-3 years old, Y-Clone, China). On day 14 and 28, the alpaca was boosted twice with 250 μg S protein in 250 μL Freund’s incomplete adjuvant (F5506-10ML, Sigma). One week following the 2nd immunization, we collected the blood samples to measure anti-serum titer. One week after the 3rd immunization, 100 mL of blood was collected to measure anti-serum titer and construct a phage library displaying Nb.
Construction of a phage library displaying Nbs
Nb phage library was constructed following our previously published method with some modifications (Wu et al., 2020a). In brief, PBMCs were isolated from 100 mL blood of immunized alpaca using a lymphocyte separation solution (cat.# 17-1140-02, Ficoll-Paque Plus, GE). RNA was extracted and reverse transcribed into cDNA by oligo (dT) and random hexamers as primers using the TRIzol kit (cat.# 15596018, Life Technologies), following manufacturer’s instruction. The alpaca Nb gene was amplified with the combination of primers and cloned into phV1 phagemid plasmid (Y-Clone, Ltd., China) to transform TG1 bacteria.
Panning Nb phage library and phage ELISA
Affinity selection for S-binding recombinant phages was performed as previously reported with the following modifications (Jähnichen et al., 2010). The Nb-phagemid-transformed bacteria were rescued with M13KO7 helper phage (cat.# 18311019, Invitrogen), and precipitated with PEG/NaCl. The phage Nb antibody library was enriched three times with 50 μg/ml of S protein. The enriched phage was eluted, transformed, and selected for the monoclonal phage to be evaluated by phage ELISA.
Phage ELISA
200 ng S or RBD protein in coating buffer (pH 9.6) was used to coat 96-well plates (cat.# 9018, Corning) at 4°C overnight. After washing, the plates were blocked with blocking buffer (3% BSA in PBST) for 1 h at 37°C, and then incubated with library phages or single clone phage in bacterial supernatant at 4°C for 1.5 h. After washing, an anti-M13 bacteriophage antibody with HRP (1:10000 dilution, cat.# 11973-MM05T-H, Sino Biological) was added and incubated at 37°C for 1 h. Accordingly, TMB substrate(Sigma) was added at 37°C for 10 min; 10 μL 0.2 M H2SO4 was added to stop the reaction. Optical densities were measured at 450 nm using the Infinite 200 (Tecan, Ramsey, MN, USA). Clones with readout at 450 nm > 0.5 were sequenced.
Expression and purification of Nbs with different formats
To facilitate the purification and prolong the half-life of the Nb antibody, the Fc1 gene (CH2-CH3) of the human monoclonal antibody was fused with the Nb gene (Nb-Fc), as our previously published method (Wu et al., 2020a). In addition, to improve the activity of Nb, we constructed Nbs with various configurations wherein (GGGGS)3 linkers were introduced between Nbs in dimeric and trimeric forms. Nanobody specific to HSA, NbH (Table S1, Abrev biotechnology, China), was developed from an alpaca receiving the immunization of HSA. To facilitate protein purification, a 6xHis-tag was fused to the N terminus of the Nbs of monomeric, dimeric or trimeric configuration. The Nbs with different configurations were finally cloned into the pcDNA3.4 eukaryotic expression vector (Invitrogen), which were transfected into 293F cells to produce Nbs with different configurations. Nb fused with Fc, or His tag was purified using Protein G (cat.# 20399, Thermo Scientific) and Ni-NTA (cat.# R901100, Thermo Fisher Scientific), respectively.
Neutralization activity of Nbs against pseudovirus
Pseudovirus neutralization assay was performed as previously described with the following modifications (Ju et al., 2020). SARS-CoV-2, SARS-CoV, and MERS-CoV pseudoviruses were produced by co-transfection of pNL4-3.Luc.R-E-, an HIV-1 NL4-3 luciferase reporter vector that contains defective Nef, Env and Vpr (HIV AIDS Reagent Program), and pCDNA3.1 (Invitrogen) expression vectors encoding the respective spike proteins (GenBank: MN988668.1 for SARS-CoV-2, AAP13567.1 for SARS-CoV, AFS88936.1 for MERS-CoV) into 293T cells (ATCC). Pseudovirus containing supernatants were collected after 48 h, and viral titers were measured by luciferase assay in relative light units (Bright-Glo Luciferase Assay Vector System, Promega Biosciences). S genes of SARS-CoV-2 variants with indicated mutations based on the human codon optimized S gene (GenBank: MN988668.1) were synthesized, and the corresponding pseudoviruses were produced following above protocol. For neutralization assay, SNB02 (Nb-Fc) against SFTSV (Wu et al., 2020a) served as a control. Neutralization assays were performed by incubating pseudoviruses with serial dilutions of purified Nbs or serum at 37°C for 1 h. HEK293T-ACE2 cells (cat.# 41107ES03, Yeasen Biotech Co., Ltd. China) for SARS-CoV-2 and SARS-CoV, Huh7 cells (ATCC) for MERS-CoV (approximately 1.5 × 104 per well) were then added in duplicate to the virus-antibody mixture. Half-maximal inhibitory dilution (ND50) of the evaluated sera or half-maximal inhibitory concentrations (IC50) of the evaluated Nbs were determined by luciferase activity 48 h after exposure to virus-antibody mixture, and analyzed by GraphPad Prism 8.01 (GraphPad Software Inc.).
Neutralization activity of Nbs against live SARS-CoV-2
SARS-CoV-2 focus reduction neutralization test was performed in a certified Biosafety Level 3 laboratory, as previously described with the following modifications (Ju et al., 2020). Briefly, a clinical isolate (Beta/Shenzhen/SZTH-003/2020, EPI_ISL_406594 at GISAID) previously obtained from a nasopharyngeal swab of an infected patient was used for the analysis. Serial concentrations of Nbs were mixed with 75 μL of SARS-CoV-2 (8 × 103 focus forming unit/ml, FFU/ml) in 96-well microwell plates and incubated at 37°C for 1 h. The mixtures were then transferred to 96-well plates seeded with Vero E6 cells and incubated at 37°C for 1 h. Next, the inoculums were removed prior to the addition of the overlay media (100 μL MEM containing 1.6% carboxymethylcellulose, CMC) and the plates were then incubated at 37°C for 24 h. Cells were fixed with 4% paraformaldehyde solution for 30 min, and then the overlays were removed. Cells were permeabilized with 0.2% Triton X-100 and incubated with cross-reactive rabbit anti-SARS-CoV-N IgG (Sino Biological, Inc) for 1 h at room temperature before the addition of HRP-conjugated goat anti-rabbit IgG (H+L) antibody (Jackson ImmunoResearch) and further incubated at room temperature. The foci were stained with KPL TrueBlue Peroxidase substrates (SeraCare Life Sciences Inc.) and were counted with an EliSpot reader (Cellular Technology Ltd.).
Affinity determination by Bio-Layer Interferometry (BLI)
Affinity assays were performed on a ForteBio OctetRED 96 biolayer interferometry instrument (Molecular Devices ForteBio LLC, Fremont, CA) at 25°C with shaking at 1,000 rpm. To measure the affinity of Nbs with human Fc tag, anti-human Fc (AHC) biosensors (cat.# 18-5060, Fortebio) were hydrated in water for 30 min prior to 60 s (sec) incubation in a kinetic buffer (PBS, 0.02% (v/v) Tween-20, pH 7.0). Either Nb-Fc in cell supernatant or purified Nb-Fcs were loaded in a kinetic buffer for 200 s prior to baseline equilibration for 200 s in a kinetic buffer. Association of SARS-CoV-2 RBD in a two-fold dilution series from 20 nM to 2.5 nM was performed prior to dissociation for 180 s. To measure the affinity of Nbs without Fc tag, RBD protein was coupled to AR2G biosensor (cat.# 18-5092, Fortebio) via BLI instrument according to the instructions of the amino coupling kit. Association of Nbs in a serial dilution was performed prior to dissociation for 180 s. After each cycle, the biosensors were regenerated via 3 short pulses of 5 s each of 100 mM pH 2.7 glycine-HCL followed by running buffer. The data were baseline subtracted before fitting performed using a 1:1 binding model and the ForteBio data analysis software. K D, Ka and Kd values were evaluated with a global fit applied to all data.
Epitope binning by BLI
The epitope binning assay was performed with AR2G biosensor (cat.# 18-5092, Fortebio) following the manufacturer’s protocol ‘in-tandem assay’ as previously reported (Rogers et al., 2020). After loading the RBD protein, a saturating concentration of antibody or Nbs (50 μg/ml) as the first antibody was added for 300 s following with the baseline step with 30 s immersion in 0.02% PBST. The second competing concentration of antibody or Nb (50 μg/ml) was then added for 300 s to measure binding in the presence of the first saturating antibody or Nb. GraphPad was used to illustrate the time-response course of two antibodies binding to RBD protein.
Pharmacokinetics of Nbs in vivo
Purified Nbs were injected intranasally (i.n.), intraperitoneally (i.p.) or intravascularly into BALB/c (Qing Long Shan Animal Center, Nanjing, China) at a dose of 10-20 mg/kg. ELISA was used to measure the serum concentration of Nbs. The T1/2 of Nbs was computed as ln (2)/k, where k is a rate constant expressed reciprocally of the x axis time units by the one phase decay equation or plateau followed one phase decay in the GraphPad software.
Spatial distribution of Nbs in vivo
Nbs were labeled with far infrared dye YF®750 SE (US EVERBRIGHT INC, YS0056) (named as Nbs-YF750). Purified Nbs-YF750 were injected i.n., i.p. or i.v. into nude mice (18-22 g, Qing Long Shan Animal Center, Nanjing, China) at a dose of 10-20 mg/kg. Images were observed at Ex:740 nm/Em:780 nm by NightOWL LB 983 (Berthold, Germany) at the indicated time point. Images were analyzed using Indigo imaging software Ver. A 01.19.01.
Evaluating the efficacy of Nbs in SARS-CoV-2 infected hACE2 mice
A total of 31 8-week-old male transgenic hACE2 mice (C57BL/6J) (cat.# T037630, GemPharmatech Co., Ltd., Nanjing, China) were challenged with 1x 105 PFU SARS-CoV-2 (IVCAS 6.7512; Zhou et al., 2020) per mouse as previously reported (Ma et al., 2020) with following modifications. The mice were split into seven groups (n = 3-5) for either prophylactic or therapeutic evaluation, as described in Figure 5A. Mice without any challenge and treatment served as blank control (No SARS-CoV-2, n = 4). Mice challenged with SARS-CoV-2 were taken as infection control (SARS-CoV-2, n = 5). 250 μg SNB02 (Y-Clone, China), an anti-SFTSV antibody constructed by Nb fused with human Fc1 (Nb-Fc) (Wu et al., 2020a), was intranasally injected 1 h after infection and served as an isotype treated control (Isotype). For the prophylactic group, mice were intranasally injected with Nb15-NbH-Nb15 at a dose of 250 μg/mouse (average of 10 mg/kg) 24 h before infection (Nb15-NbH-Nb15 −24h, n = 5). For the therapeutic group, mice were intranasally injected with N Nb15-NbH-Nb15 at a dose of 250 μg/mouse (average of 10 mg/kg) 1 h or 24 h after infection (named as Nb15-NbH-Nb15 1h and Nb15-NbH-Nb15 24 h, n = 5, respectively). As a comparison, Nb15-Fc at a dose of 250 μg/mouse (average of 10 mg/kg) was intranasally injected 1 h after infection (Nb15-Fc 1 h). Body weight of every mouse was measured daily. Transgenic hACE2 mice typically clear virus within five days after SARS-CoV-2 and viral RNA copies reached a peak at 3 dpi (Bao et al., 2020). Accordingly, the mice were sacrificed at 3 days post infection (dpi), and the lungs were collected for viral load determination and tissue sections for hematoxylin and eosin (H&E) and immunofluorescence staining.
Viral load measurement by quantitative RT-PCR
Viral load was detected by quantitative real-time PCR (qRT-PCR) on RNA extracted from the supernatant of lung homogenates as described previously (Cao et al., 2020). Briefly, lung homogenates were prepared by homogenizing perfused whole lung using an electric homogenizer. The supernatant was collected, and total RNA was extracted. Each RNA sample was reverse transcribed to 50 μL cDNA with RT-PCR Prime Script Kit (Takara). The cDNA (5 μl) was used in a 25 μL qRT-PCR reaction with the TaqMan Universal PCR Master Mix (Life Technologies), a TaqMan probe (5′-FAM− CAGGTGGAACCTCATCAGGAGATGC −MGB-3′), and primers designed to target the orf1ab gene of SARS-CoV-2 (5′- GTGARATGGTCATGTGTGGCGG −3′ and 5′- CARATGTTAAASACACTATTAGCATA −3′). The samples were run in triplicate on an ABI 7900 Real-Time System (Applied Biosystems, Thermo Fisher Scientific). The following cycling conditions were used: 1 cycle of 50°C for 2 min, 1 cycle of 95°C for 10 min, and 40 cycles of 95°C for 15 s and 58°C for 1 min. The virus titer was determined by comparison with a standard curve generated using RNA extracted from a serially diluted reference viral stock. All experiments were performed in a Biosafety Level 3 facility.
Immunofluorescence staining of SARS-CoV-2-infected cells in tissues
Lung tissues were immersed in 10% neutral buffered formalin (cat.# Z2902, Sigma) for 24 h. After the formalin fixation, the tissues were placed in 70% ethanol (Merck) and subsequently embedded with paraffin. Tissue sections (4-μm thick) were used for immunofluorescence staining for SARS-CoV-2 detection using the Coronavirus nucleocapsid antibody (cat. 40143-MM05, Sino Biological). Images were obtained by OLYMPUS IX73 using HCImage Live ( × 64) software and analyzed by ImageJ (NIH).
Quantification and statistical analysis
All statistical analyses were performed using GraphPad Prism 8.01 software (GraphPad) or OriginPro 8.5 software (OriginLab). ANOVA or Mann-Whitney test was performed for group comparisons. p < 0.05 was considered as statistically significant with mean ± SEM or mean ± SD. All of the statistical details of experiments can be found in the figure legends.
Acknowledgments
This work was supported by National Science Foundation of China (NSFC) (no. 81803414 and 31970149), the Major Research and Development Project (2018ZX10301406), Nanjing University-Ningxia University Collaborative Project (grant no. 2017BN04), Jiangsu Province Natural Science Foundation for Young Scholar (grant no. BK20170653), Key Natural Science Foundation of Jiangsu Province (grant no. ZDA2020014), Jiangsu Province “Innovative and Entrepreneurial talent” and Six Talent Peaks Project of Jiangsu Province, the Emergency Prevention and Control Capacity Program for New Severe Infectious Diseases of National Institute for Viral Disease Control and Prevention, and the 135 Strategic Program of Chinese Academy of Sciences, the Science and Technology Innovation Committee of Shenzhen Municipality (JCYJ20180228162229889).
Author contributions
X.W. conducted most experiments, analyzed the data, and wrote the manuscript draft. L.C. conducted all the neutralization experiments. B.H., L.Z., S.X., H.S., D.Z., H.Y., and W.N. provided technical assistance and did animal experiments. M.F., Y.L., P.Y., and Q.H. evaluated the efficacy of Nbs in SARS-CoV-2-infected transgenic hACE2 mice. Z.W. designed the study, directed and financially supported the study, and revised the manuscript. All authors critically reviewed the draft manuscript and approved the final version.
Declaration of interests
The authors declared no competing interests. X.W. and Z.W. are listed as inventors on a China patent (CN202110120326.8) related with the Nbs for the treatment of SARS-CoV-2 infection.
Published: October 6, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109869.
Supplemental information
Data and code availability
-
•
The data that support the findings of this study are available upon request from the lead contact.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Bao L., Deng W., Huang B., Gao H., Liu J., Ren L., Wei Q., Yu P., Xu Y., Qi F., et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020;583:830–833. doi: 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- Baum A., Fulton B.O., Wloga E., Copin R., Pascal K.E., Russo V., Giordano S., Lanza K., Negron N., Ni M., et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020;369:1014–1018. doi: 10.1126/science.abd0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X., Zheng Y., Geng C., et al. Potent Neutralizing Antibodies against SARS-CoV-2 Identified by High-Throughput Single-Cell Sequencing of Convalescent Patients’ B Cells. Cell. 2020;182:73–84.e16. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham S., Piedra P.A., Martinon-Torres F., Szymanski H., Brackeva B., Dombrecht E., Detalle L., Fleurinck C., Cunningham S., Piedra P.A., et al. Nebulised ALX-0171 for respiratory syncytial virus lower respiratory tract infection in hospitalised children: a double-blind, randomised, placebo-controlled, phase 2b trial. Lancet Respir. Med. 2020;9:21–32. doi: 10.1016/S2213-2600(20)30320-9. [DOI] [PubMed] [Google Scholar]
- Dai L., Gao G.F. Viral targets for vaccines against COVID-19. Nat. Rev. Immunol. 2020;21:73–82. doi: 10.1038/s41577-020-00480-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detalle L., Stohr T., Palomo C., Piedra P.A., Gilbert B.E., Mas V., Millar A., Power U.F., Stortelers C., Allosery K., et al. Generation and Characterization of ALX-0171, a Potent Novel Therapeutic Nanobody for the Treatment of Respiratory Syncytial Virus Infection. Antimicrob. Agents Chemother. 2015;60:6–13. doi: 10.1128/AAC.01802-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J., Huang B., Jia Z., Wang B., Gallolu Kankanamalage S., Titong A., Liu Y. Development of multi-specific humanized llama antibodies blocking SARS-CoV-2/ACE2 interaction with high affinity and avidity. Emerg. Microbes Infect. 2020;9:1034–1036. doi: 10.1080/22221751.2020.1768806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo O., Locatello L.G., Mazzoni A., Novelli L., Annunziato F. The central role of the nasal microenvironment in the transmission, modulation, and clinical progression of SARS-CoV-2 infection. Mucosal Immunol. 2020;14:305–316. doi: 10.1038/s41385-020-00359-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamers-Casterman C., Atarhouch T., Muyldermans S., Robinson G., Hamers C., Songa E.B., Bendahman N., Hamers R. Naturally occurring antibodies devoid of light chains. Nature. 1993;363:446–448. doi: 10.1038/363446a0. [DOI] [PubMed] [Google Scholar]
- Hanke L., Vidakovics Perez L., Sheward D.J., Das H., Schulte T., Moliner-Morro A., Corcoran M., Achour A., Karlsson Hedestam G.B., Hällberg B.M., et al. An alpaca nanobody neutralizes SARS-CoV-2 by blocking receptor interaction. Nat. Commun. 2020;11:4420. doi: 10.1038/s41467-020-18174-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart T.K., Cook R.M., Zia-Amirhosseini P., Minthorn E., Sellers T.S., Maleeff B.E., Eustis S., Schwartz L.W., Tsui P., Appelbaum E.R., et al. Preclinical efficacy and safety of mepolizumab (SB-240563), a humanized monoclonal antibody to IL-5, in cynomolgus monkeys. J. Allergy Clin. Immunol. 2001;108:250–257. doi: 10.1067/mai.2001.116576. [DOI] [PubMed] [Google Scholar]
- Higgins T.S., Wu A.W., Illing E.A., Sokoloski K.J., Weaver B.A., Anthony B.P., Hughes N., Ting J.Y. Intranasal Antiviral Drug Delivery and Coronavirus Disease 2019 (COVID-19): A State of the Art Review. Otolaryngol. Head Neck Surg. 2020;163:682–694. doi: 10.1177/0194599820933170. [DOI] [PubMed] [Google Scholar]
- Huo J., Le Bas A., Ruza R.R., Duyvesteyn H.M.E., Mikolajek H., Malinauskas T., Tan T.K., Rijal P., Dumoux M., Ward P.N., et al. Neutralizing nanobodies bind SARS-CoV-2 spike RBD and block interaction with ACE2. Nat. Struct. Mol. Biol. 2020;27:846–854. doi: 10.1038/s41594-020-0469-6. [DOI] [PubMed] [Google Scholar]
- Jähnichen S., Blanchetot C., Maussang D., Gonzalez-Pajuelo M., Chow K.Y., Bosch L., De Vrieze S., Serruys B., Ulrichts H., Vandevelde W., et al. CXCR4 nanobodies (VHH-based single variable domains) potently inhibit chemotaxis and HIV-1 replication and mobilize stem cells. Proc. Natl. Acad. Sci. USA. 2010;107:20565–20570. doi: 10.1073/pnas.1012865107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovčevska I., Muyldermans S. The Therapeutic Potential of Nanobodies. BioDrugs. 2019;34:11–26. doi: 10.1007/s40259-019-00392-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju B., Zhang Q., Ge J., Wang R., Sun J., Ge X., Yu J., Shan S., Zhou B., Song S., et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- Liu L., Wang P., Nair M.S., Yu J., Rapp M., Wang Q., Luo Y., Chan J.F., Sahi V., Figueroa A., et al. Potent neutralizing antibodies directed to multiple epitopes on SARS-CoV-2 spike. bioRxiv. 2020 doi: 10.1101/2020.06.17.153486. [DOI] [PubMed] [Google Scholar]
- Luo F., Liao F.L., Wang H., Tang H.B., Yang Z.Q., Hou W. Evaluation of Antibody-Dependent Enhancement of SARS-CoV Infection in Rhesus Macaques Immunized with an Inactivated SARS-CoV Vaccine. Virol. Sin. 2018;33:201–204. doi: 10.1007/s12250-018-0009-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Zou F., Yu F., Li R., Yuan Y., Zhang Y., Zhang X., Deng J., Chen T., Song Z., et al. Nanoparticle Vaccines Based on the Receptor Binding Domain (RBD) and Heptad Repeat (HR) of SARS-CoV-2 Elicit Robust Protective Immune Responses. Immunity. 2020;53:1315–1330.e9. doi: 10.1016/j.immuni.2020.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambulli S., Xiang Y., Tilston-Lunel N.L., Rennick L.J., Sang Z., Klimstra W.B., Reed D.S., Crossland N.A., Shi Y., Duprex W.P. Inhalable Nanobody (PiN-21) prevents and treats SARS-CoV-2 infections in Syrian hamsters at ultra-low doses. Sci. Adv. 2021;7:eabh0319. doi: 10.1126/sciadv.abh0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planas D., Veyer D., Baidaliuk A., Staropoli I., Guivel-Benhassine F., Rajah M.M., Planchais C., Porrot F., Robillard N., Puech J., et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021;596:276–280. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- Prince G.A., Hemming V.G., Horswood R.L., Baron P.A., Chanock R.M. Effectiveness of topically administered neutralizing antibodies in experimental immunotherapy of respiratory syncytial virus infection in cotton rats. J. Virol. 1987;61:1851–1854. doi: 10.1128/jvi.61.6.1851-1854.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pymm P., Adair A., Chan L.J., Cooney J.P., Mordant F.L., Allison C.C., Lopez E., Haycroft E.R., O’Neill M.T., Tan L.L., et al. Nanobody cocktails potently neutralize SARS-CoV-2 D614G N501Y variant and protect mice. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2101918118. e2101918118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S., et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers T.F., Zhao F., Huang D., Beutler N., Burns A., He W.T., Limbo O., Smith C., Song G., Woehl J., et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369:956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoof M., Faust B., Saunders R.A., Sangwan S., Rezelj V., Hoppe N., Boone M., Billesbølle C.B., Puchades C., Azumaya C.M., et al. QCRG Structural Biology Consortium An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike. Science. 2020;370:1473–1479. doi: 10.1126/science.abe3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- Steeland S., Vandenbroucke R.E., Libert C. Nanobodies as therapeutics: big opportunities for small antibodies. Drug Discov. Today. 2016;21:1076–1113. doi: 10.1016/j.drudis.2016.04.003. [DOI] [PubMed] [Google Scholar]
- Taylor A., Foo S.S., Bruzzone R., Dinh L.V., King N.J., Mahalingam S. Fc receptors in antibody-dependent enhancement of viral infections. Immunol. Rev. 2015;268:340–364. doi: 10.1111/imr.12367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirado S.M., Yoon K.J. Antibody-dependent enhancement of virus infection and disease. Viral Immunol. 2003;16:69–86. doi: 10.1089/088282403763635465. [DOI] [PubMed] [Google Scholar]
- Van Heeke G., Allosery K., De Brabandere V., De Smedt T., Detalle L., de Fougerolles A. Nanobodies® as inhaled biotherapeutics for lung diseases. Pharmacol. Ther. 2017;169:47–56. doi: 10.1016/j.pharmthera.2016.06.012. [DOI] [PubMed] [Google Scholar]
- Van Roy M., Ververken C., Beirnaert E., Hoefman S., Kolkman J., Vierboom M., Breedveld E., ’t Hart B., Poelmans S., Bontinck L., et al. The preclinical pharmacology of the high affinity anti-IL-6R Nanobody® ALX-0061 supports its clinical development in rheumatoid arthritis. Arthritis Res. Ther. 2015;17:135. doi: 10.1186/s13075-015-0651-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., Tan W. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA. 2020;323:1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Ma X., Li Y., Xu Y., Zheng N., Xu S., Nawaz W., Wu Z. Induction of neutralizing antibodies by human papillomavirus vaccine generated in mammalian cells. Antib. Ther. 2019;2:45–53. doi: 10.1093/abt/tbz004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Li Y., Huang B., Ma X., Zhu L., Zheng N., Xu S., Nawaz W., Xu C., Wu Z. A single-domain antibody inhibits SFTSV and mitigates virus-induced pathogenesis in vivo. JCI Insight. 2020;5:e136855. doi: 10.1172/jci.insight.136855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Li C., Xia S., Tian X., Kong Y., Wang Z., Gu C., Zhang R., Tu C., Xie Y., et al. Identification of Human Single-Domain Antibodies against SARS-CoV-2. Cell Host Microbe. 2020;27:891–898.e5. doi: 10.1016/j.chom.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y., Nambulli S., Xiao Z., Liu H., Sang Z., Duprex W.P., Schneidman-Duhovny D., Zhang C., Shi Y. Versatile and multivalent nanobodies efficiently neutralize SARS-CoV-2. Science. 2020;370:1479–1484. doi: 10.1126/science.abe4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost S.J., Gilchuk P., Case J.B., Binshtein E., Chen R.E., Nkolola J.P., Schäfer A., Reidy J.X., Trivette A., Nargi R.S., et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature. 2020;584:443–449. doi: 10.1038/s41586-020-2548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The data that support the findings of this study are available upon request from the lead contact.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.