Abstract
Breast cancer is an individually unique, multi-faceted and chameleonic disease, an eternal challenge for the new era of high-integrated precision diagnostic and personalized oncomedicine. Besides traditional single-omics fields (such as genomics, epigenomics, transcriptomics and metabolomics) and multi-omics contributions (proteogenomics, proteotranscriptomics or reproductomics), several new “-omics” approaches and exciting proteomics subfields are contributing to basic and advanced understanding of these “multiple diseases termed breast cancer”: phenomics/cellomics, connectomics and interactomics, secretomics, matrisomics, exosomics, angiomics, chaperomics and epichaperomics, phosphoproteomics, ubiquitinomics, metalloproteomics, terminomics, degradomics and metadegradomics, adhesomics, stressomics, microbiomics, immunomics, salivaomics, materiomics and other biomics. Throughout the extremely complex neoplastic process, a Breast Cancer Cell Continuum Concept (BCCCC) has been modeled in this review as a spatio-temporal and holistic approach, as long as the breast cancer represents a complex cascade comprising successively integrated populations of heterogeneous tumor and cancer-associated cells, that reflect the carcinoma’s progression from a “driving mutation” and formation of the breast primary tumor, toward the distant secondary tumors in different tissues and organs, via circulating tumor cell populations. This BCCCC is widely sustained by a Breast Cancer Proteomic Continuum Concept (BCPCC), where each phenotype of neoplastic and tumor-associated cells is characterized by a changing and adaptive proteomic profile detected in solid and liquid minimal invasive biopsies by complex proteomics approaches. Such a profile is created, beginning with the proteomic landscape of different neoplastic cell populations and cancer-associated cells, followed by subsequent analysis of protein biomarkers involved in epithelial-mesenchymal transition and intravasation, circulating tumor cell proteomics, and, finally, by protein biomarkers that highlight the extravasation and distant metastatic invasion. Proteomics technologies are producing important data in breast cancer diagnostic, prognostic, and predictive biomarkers discovery and validation, are detecting genetic aberrations at the proteome level, describing functional and regulatory pathways and emphasizing specific protein and peptide profiles in human tissues, biological fluids, cell lines and animal models. Also, proteomics can identify different breast cancer subtypes and specific protein and proteoform expression, can assess the efficacy of cancer therapies at cellular and tissular level and can even identify new therapeutic target proteins in clinical studies.
Keywords: Proteomics, biomarkers, breast cancer cell continuum concept, breast cancer proteomic continuum concept
Breast cancer proteomics in the multi-omics era
Breast cancer is a highly heterogeneous malignant disease with various functional phenotypes [1] that create an extremely challenging puzzle of histo-morphologic, proteomic, and genomic features. Taken together, these features will determine prognosis, predict tumor response to different targeted or immune therapies and contribute towards deciding on the optimal method for follow-up [2]. The biology of breast cancer also depends on the complex tumor microenvironment (TME), where a plethora of cellular and tissular factors, such as epithelial and non-epithelial neoplastic cells, tumor-associated stromal cells, infiltrating immune cells and other adjacent normal or abnormal non-neoplastic cells connected by the vascular network and extracellular matrix (ECM) interact and determine clinical behaviour [3]. As such, the resulting tumor will always have some individually unique characteristics [4], a feature of breast cancer that partly explains resistance to systemic treatment.
Nowadays, as we are going through “emerging era of high-integrated precision diagnostics” [5], proteomics, the study of the large set of proteins expressed by an organelle, a cell type, a tissue or an organism at a given time [6], has become more and more important in both diagnosis and treatment. A protein is the basic unit of cell function and biological pathway [7]. Proteomics information is essential to classify the functional subtypes and stages of the breast cancer, to decipher its tumorigenesis mechanisms, cancer behaviour and aggressiveness, to predict recurrence, to assess and reduce the cancer cell resistance, to choose and monitor the most appropriate breast cancer treatment. Breast cancer proteomics complements genomics [8], transcriptomics [9], metabolomics [10-13], and epigenomics [14-16]. In recent years, several other research areas have emerged, many of which have been applied in breast cancer research: lipidomics [17-19], gut and breast metagenomics in order to understand the microbiome’s role in breast carcinogenesis [20-22], estrobolomics that studies the aggregate of estrogen-metabolizing enteric bacterial genes [23], toxicogenomics that studies the structure and the genome output as well as its responses to adverse xenobiotic exposure [24-26], pharmacogenomics that focused on the study of the role of the genome in drug response [27-29], phylogenomics that studies evolutionary history of cancer [30], interactomics, which deciphers the complex interactions between tumor molecules, especially protein-protein interactions (PPIs) [31-33] and protein interaction networks (PINs), and it is involved in breast cancer prognostic modeling [33], and connectomics that studies the network interactions between the various components of the TME [34]. Integrated proteotranscriptomics of breast cancer deciphers new disease characteristics [35], revealing the biological basis of intraoperative radiotherapy-treated tumors [36], and subtyping of triple-negative breast cancers (TNBC) [37]. Integrated proteomics, transcriptomics and glycomics helps to elucidate the biological pathways involved in breast cancer metastasis [38]. Histopathology, proteogenomics and transcriptomics data integrate multiple biological information from breast cancer samples in generating novel potential predictive biomarkers [39]. Proteomics can be situated downstream of genomics into an analytic flow that describes the translation from the genome characteristics to the phenotypes and functions of breast cancer cells [40].
Several promising proteomics subfields also contribute to advanced understanding of these “multiple diseases that are termed breast cancer” [3] at a molecular level: secretomics that identifies the secreted proteins in the TME [41-43]; matrisomics that studies the protein profiling in the tumor ECM; exosomics that focuses on the nanostructures released by cells such as exosomes from human breast milk [44]; angiomics defined as vascular proteomics of angiogenesis [45]; phosphoproteomics that studies the phosphorylation-based post-translational modifications of proteins (PTMs) [46-48]; metalloproteomics focused on the expression of metalloproteins from different metalloproteomes, including the study of the therapeutic role of matrix metalloproteinases (MMPs) in breast cancer [49]; ubiquitinomics that includes all ubiquitinated proteins [50]; chaperomics that studies the chaperones, co-chaperones, adaptors, and folding enzymes regulating the cellular homeostasis together with the protein degradation systems [51]; epichaperomics based on epichaperomes involved in proteomic alterations associated with malignancy [52]; degradomics and terminomics dedicated to the protease degradome and terminome [53]; metadegradomics defined as the N-terminome analysis of proteases in tissues and organs [54]; adhesomics that focuses on the study of cell-to-cell and cell-to-ECM adhesion proteins [55]; stressomics that studies the protein expression in response to stress; microbiomics based on the composition and role of the gut, breast, milk [56], urogenital and skin microbiome [57] as a risk factor in breast cancer. The next-generation of breast cancer “-omics” includes immunomics [58], nutrigenomics [59], and other biomics approaches to explore the organism level [3]. For example, salivaomics [60], tearomics, and milkomics could be several promising fields that include proteomics technologies as a source of non-invasive biomarkers in early detection, disease monitoring and prognosis assessment for breast cancer. While the single-level omics approaches have contributed to the identification of cancer-specific molecular aberrations or to the classification of tumors, the onco-multi-omics approaches assess cancer cells and tissues in multiple ways which allow for the deciphering of molecular mechanisms involved in carcinogenesis and increases the likelihood of validating new biomarkers that shape the field of personalized oncomedicine [61].
Protein separation technologies and mass spectrometry (MS) assessments, especially matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS), have become essential tools in this field [62], enabling the identification and quantification of large sets of proteins that can be monitored simultaneously in a single sample. MS-based proteomics analysis contributes to demonstrate the intertumor heterogeneity across different breast cancers subtypes and compared to healthy controls, which can impact cancer treatment [63], enhancing the precision and accuracy in discovery and validation of candidate protein biomarkers. MALDI-mass spectrometry imaging (MSI) allows the analysis of heterogeneous tumor samples to differentiate cancer from healthy regions, in order to remove the need for laser capture microdissection [64]. Thus, histology-directed MALDI-MSI may help to elucidate the molecular origins of cancer and the tissue transformation under breast oncogenic stress [65]. MALDI-TOF MS is also a good analytical tool to distinguish between specific plasma peptidome of breast cancer patients and healthy controls [66], but it is also suitable for molecular profiling of solid tumors [67]. MALDI-TOF and surface-enhanced laser desorption/ionization (SELDI)-TOF MS offer proteomic-based profiling analyses of tumor for discovering and validating novel biomarkers of breast cancer [9]. In order to detect and quantify cancer-related expression of proteins and their isoforms, large-scale integration of genomic, bottom-up and top-down proteomic data for the comparative analysis of human-in-mouse xenograft models of basal-like and luminal human breast cancer have been published [68]. Together, MS-based proteomics, the enzyme-linked immunosorbent assay (ELISA) and immunohistochemistry (IHC) techniques are widely involved in plasma-, tissue- or cell-based investigation for enzymatic and non-enzymatic PTMs as biomarkers in breast cancer [69]. Thus, MS is the most flexible and general tool available today for the PTMs study, which can “open a window on a normally hidden cross-section of the proteome” [70]. A general proteomics experiment is presented in Figure 1, while the principles of bottom-up and top-down proteomics are presented in Figure 2. A workflow strategy for identification of two types of PTMs (glycosylation and phosphorylation) is shown in Figure 3. Finally, strategies for proteins quantification in a proteomics experiment are shown in Figure 4.
Figure 1.
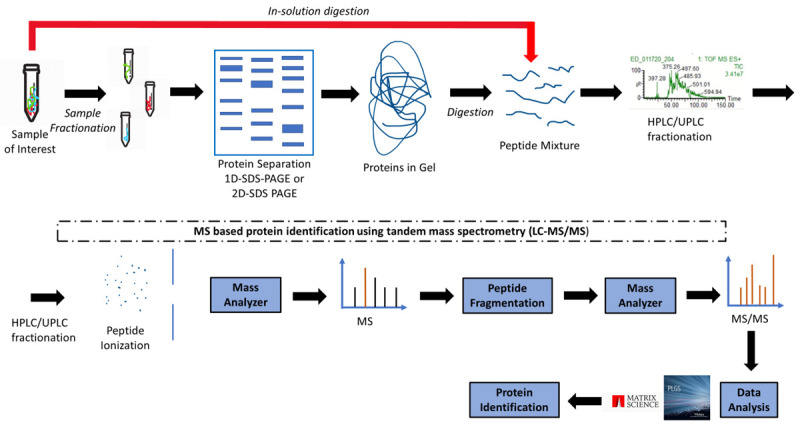
Schematic of a general proteomic workflow. A sample can be fractionated (i.e., by electrophoresis) and then digested by trypsin (in-gel digestion), or digested in-solution by trypsin. The peptides mixture is then ionized (with or without separation by reversed phase chromatography). Peptides are then ionized and their corresponding m/z is measured in the MS mode under low collision energy, or fragmented and then measured in MS/MS mode under high collision energy. Data analysis using protemics software leads to identification of a peptide that is part of a protein, thus also identifying the protein.
Figure 2.
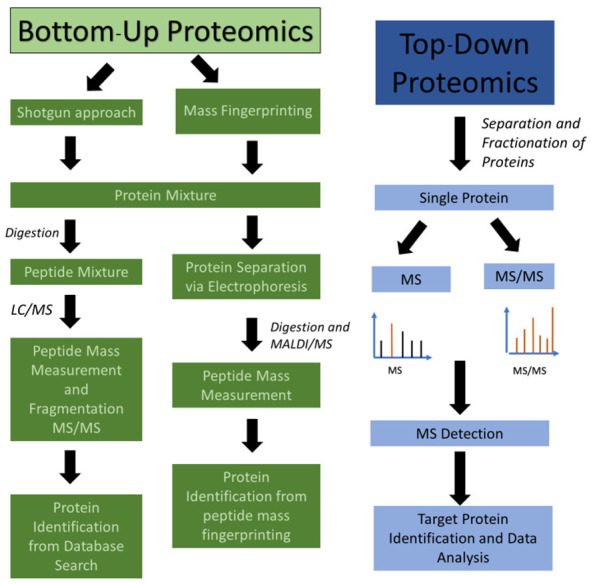
Bottom-up and top-down proteomics. In bottom-up proteomics, the protein mixtures are digested and the peptide mixtures are analyzed by LC-MS and LC-MS/MS (shotgun approach) or separated by electrophoresis and then individual proteins are digested and analyzed by MALDI-MS in a method called peptide mass fingerprinting. In top-down proteomics, the individual proteins (or a mixture of proteins) are analyzed for molecular mass in MS mode or fragmented to provide partial fragments in MS/MS mode. Using this approach, the target protein’s mass is identified and its amino acid sequence confirmed by MS/MS fragmentation.
Figure 3.
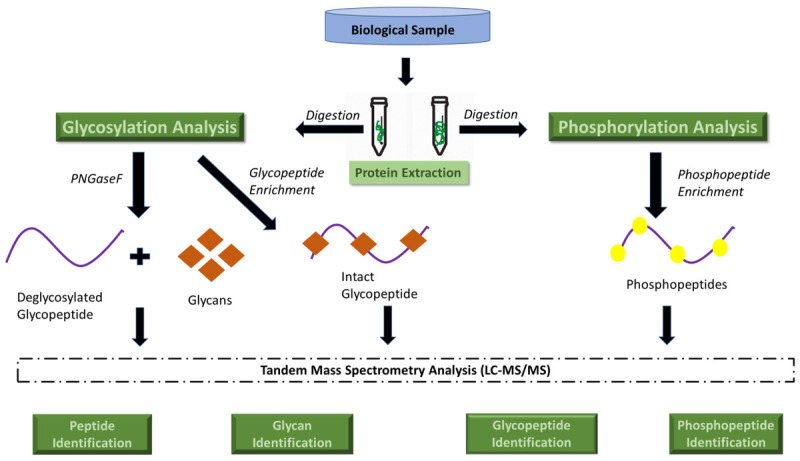
General strategy for identification of two major PTMs: phosphorylation and glycosylation.
Figure 4.
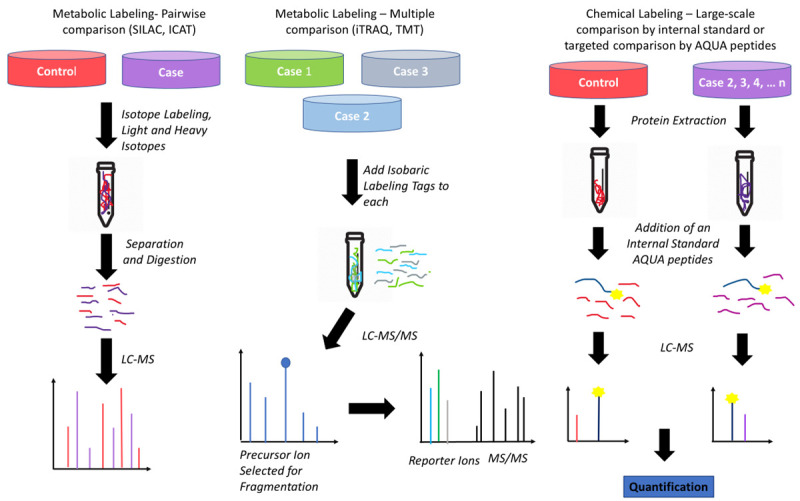
Examples of methods for quantitative proteomics using labeled tags. The samples labeled case and control are mixed at the protein level prior fractionation and digestion (e.g., using classical SILAC or ICAT methods) or first labeled at the protein level with a tag (i.e. with iTRAQ or TMT) for and then mixed and further fractionated and digested. In this case, more than one labeled condition can be used (e.g., case 1, 2, 3). In the last case, chemical labeling happens at the peptide level, i.e., after the samples were fractionated and digested. Note that this method can target all peptides (global acetylation and deuterated acetylation), or specific peptides, i.e. using absolute quantitation (AQUA) peptides. Note that AQUA is an internal standard peptide custom-built to quantify a particular peptide. Note that internal standard peptides (other than AQUA peptides) can be used and applied to all proteomics methods discussed in this figure.
Breast cancer subtypes
From an anatomical stand point, breasts are dynamic modified tubuloalveolar apocrine sweat glands [71], composed of skin, subcutaneous tissue, parenchyma, and stroma, which are considered vestigial organs in male [72]. Parenchymal architecture of the mammary gland involves a network of branching ducts and terminal secretory lobules. There are 15 to 20 lobes radiating out from the breast nipple, and each lobe is made by 20 to 40 lobules, which consists of about 20 to 30 clustered alveoli or acini containing mammary secretory epithelial cells [71]. All acini that open into the same terminal duct form the terminal ductal lobulo-alveolar/lobular unit (TDLU) where most breast tumors arise [73]. Mammary lobules are the functional units of the breast, ensuring the synthesis of milk. A lactiferous duct drains each lobe. The preponderance of glandular tissue in the upper outer quadrant (UOQ) of the breast makes this anatomical region the most common site for the development of the breast carcinoma [74], while the lower inner quadrant (LIQ) was cited with a lowest frequency of tumorigenic process [75].
Histologically, both breast ducts and TDLUs are bilayered with two main breast cell populations, displaying specific patterns in their normal protein profiles [76]: an apically oriented luminal epithelial layer lining the lumen and ensuring the secretion of milk during lactation, with polarized cuboidal cells expressing cytokeratin 8 (CK8), CK18, as well as the estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor-2 (HER2) [77], and an outermost contractile semi-continuous and spindle-shaped myoepithelial/basal layer/myoepithelium resting on the basement membrane that separates the breast epithelium and the surrounding stroma [78], expressing p63 in the nuclei [79], and cytoplasmic CK5, CK14, and smooth muscle actin (SMA) that sustains the contractile function allowing the milk ejection [77]. Compared with its more common female counterpart, in situ male breast carcinoma (MBC) is a rare malignancy, emphasizing different histopathologic differences that reflect the gender-specific anatomy of the breast [80]. MBC express many of the same biomarkers as female breast cancer (FBC) [81], but the androgen receptor (AR) is expressed in the majority of MBC [82].
Breast cancer classification has been the focus of numerous worldwide efforts [63]. A recent integrative scheme for assessing breast cancer subtypes has been published [2]. The main types of breast cancer are classified as in situ/non-invasive (IS) and invasive or infiltrative carcinoma, both of which can have a ductal or a lobular origin: ductal carcinoma in situ (DCIS), lobular carcinoma in situ (LCIS), invasive lobular carcinoma (ILC), and invasive ductal carcinoma (IDC). Based on discriminatory protein profiles obtained by MALDI MS analysis, breast cancers have been additionally divided into five main subtypes: luminal A (LA) and luminal B (LB), characterized by the expression of luminal/epithelial markers [83], showing positive estrogen receptor (ER+) and/or positive progesterone receptor (PR/PgR+) expression [2], the HER2-enriched tumors, with a human epidermal growth factor receptor 2 (HER2/neu tyrosine kinase receptor) overexpression, the normal breast-like group closest to the molecular profile of a normally mammary gland, and triple negative breast cancers (TNBC), that does not express ER, PR or HER2, characterized by poor prognosis, high recurrence, poor overall survival and no well-defined molecular targeted treatment [84]. Recent classification based on histopathology and gene expression additionally defines four TNBC subtypes: basal-like 1, basal-like 2, mesenchymal-like, and luminal androgen receptor-like (LAR), without significant differences in prognosis [64]. TNBC with basal-like features (TNBC-BL) is a subtype defined by a proteomic landscape similar to the basal/myoepithelial cells of the breast. This subtype is often associated with overexpression of tumor protein p53 as a biomarker of poor prognosis, due to several mutations in p53 gene that inhibit its function as a transcriptional repressor of several oncogenes [85], and Ki67, an important proliferation biomarker [86]. Also, TNBC-BL is more aggressive and has higher rates of lymph node metastasis than the non-BL type [87]. A study based on sequential windowed acquisition of all theoretical fragment ion spectra (SWATH) MS proteotype patterns showed that the TNBC are the most heterogeneous [88]. Results obtained from the transcriptoproteomics approaches have suggested another classification of TNBC: molecular apocrine (C1-characterized by luminal and androgen-regulated proteins), basal-like immune-suppressed (C2-associated with invasion and ECM) and basal-like immune response (C3-associated with interferon pathway and immunoglobulins) [37]. Similarly, the “-omics” approach has also identified a novel LA breast cancer subtype characterized by increased phosphoinositide 3-kinase (PI3K) signalling, a critical transduction system linking oncogenes and different receptor classes to many important cellular functions and considered one of the most activated signalling pathway in human cancer [89], highlighting the additional value of clinical proteomics in breast cancer to discover specific features not available by genomic approaches [63]. The majority of MBC are classified as luminal A subtype [82]; based on the differences in molecular profiles and outcome, luminal M1 and luminal M2 stable subgroups have been characterized as different from the subgroups described in female breast cancer (FBC) [90].
Therefore, the tumour tissue proteome can be considered to be a “complex orchestra of proteins” [91] involved in many essential pathological pathways: tumour initiation and progression, neo-angiogenesis, metabolic reprogramming, stem cell maintenance, response resistance to chemo- and radiation therapy, apoptosis [92], and metastasis steps, such as the epithelial-mesenchymal transition (EMT), local tissue invasion, intravasation, homing, mesenchymal-epithelial transition (MET), extravasation, priming, licensing, initiation and progression at the pre-metastatic niche level [93], and during metastatic niche formation [94].
Breast cancer cell continuum concept (BCCCC) & breast cancer proteomic continuum concept (BCPCC)
During the whole extremely complex neoplastic process, a Breast Cancer Cell Continuum Concept (BCCCC) could be modeled based on the idea that successively integrated populations of heterogeneous tumor and cancer associated-cells progress from a “driving mutation” that leads to the primary tumor, toward the distant secondary tumors in different tissues and organs, via circulating tumor cell populations (CTCs) as an intermediate stage. The complexity of cancer is a consequence of the spatial and temporal genetic, epigenetic and phenotypic heterogeneity of tumor cells that compose the tumor tissue, and of the bidirectional relationship with their surrounding tissue microenvironment [95]. There are a multitude of data suggesting that a plethora of extrinsic and intrinsic breast cancer risk factors [96] may influence the histo-molecular profile of the normal breast tissue even before cancer develops [97] or during the neoplastic progression, resulting in plasticity of the tumor under stress: age [98], gender [99], ethnicity [100], genetics [101], thoracic irradiation [102], lifestyle and obesity [103], alcohol consumption [104] and cigarette smoking [105], diet and environmental contaminants [106], exposure to oral contraceptives [107], hormonal replacement therapy and hormonal treatment in transgender people, especially in trans-women [108], breastfeeding [100], nulliparity [109] and time since last birth [110], height and early-life body size, menopausal status/later onset of menopause, menstrual phase, and personal history of breast atypical hyperplasia [111].
The BCCCC is useful for understanding the step by step differentiation pattern and the versatile behaviour of neoplastic cells and their adjacent cell-related and microenvironmental adaptations. Most tumors originate in a unique cell, due to an aberrant genetic event that differentially affects cellular functions by altering the expression and activity of normal proteins and their isoforms [112], according to their host cell characteristics [113]. During tumorigenesis, cancer cells acquire additional aberrations, each tumor becoming a puzzle of multiple sub-clones as consequence of “passenger mutations” [112]. Intratumoral heterogeneity is a consequence of these cell subpopulations with different features, such as modified phenotypes, adapted proteomic profiles, behaviour, tumorigenicity, treatment resistance and metastatic potential [112]. Proteomics datasets based on MS may highlight the complex relationship between genomic alterations and cancer cell phenotypes, being extensively involved in “bridging the gap between genotype and phenotype” [40]. Thus, the BCCCC can be widely sustained by a Breast Cancer Proteomic Continuum Concept (BCPCC), whereas each phenotype of neoplastic and tumor-associated cells is characterized by a changing and adaptive proteomic profile detected in solid or liquid biopsies, by complex proteomics approaches, beginning with the proteomic landscape of different neoplastic and cancer-associated cells, followed by subsequent analysis of protein biomarkers involved in epithelial-mesenchymal transition (EMT) and intravasation, circulating tumor cells (CTCs) proteomics, and finally by protein biomarkers that highlight the extravasation and distant metastatic invasion. Both BCCCC and BCPCC rely on the analysis of cancer cells and their microenvironment, which in turn is shaped by various cellular and non-cellular factors. This complex interaction is unique to each individual tumor and despite constant on-going research none of the current theories is universally accepted in the scientific community. Even most breast cancers arise form a mammary ductal cell, available data suggest that any type of cell within the breast tissue is susceptible to neoplastic switch and that each individual type of non-cancer cell plays an important role in shaping the future TME and, subsequently, cancer progression and metastasis. In the following paragraphs, the key cellular and non-cellular players involved in BCCCC and BCPCC will be discussed.
Cells of origin in breast cancer
Stem cells
Stem cells are able to self-renew and to differentiate into any cell type of the organism [114]. The adult breast stem (BSCs) and progenitor cells disseminated among normal mature cells support the capacity of the mammary gland to structurally modify and functionally adapt during menstrual cycle, pregnancy and lactation [115]. There is a multidirectional relationship between stem, progenitor and differentiated cells and any abnormality in these interactions could lead to differentiation of normal stem cells into cancer stem cells (CSCs) or to de-differentiation of progenitor or mature epithelial cells into CSCs [116]. BSCs possess infinite proliferation, self-renewal, multi-directional differentiation, long-term survival and expansion [73], while progenitor cells are characterized by a high proliferative potential; both stem and progenitor cells have been proposed as cells of origin in breast cancers [73]. CSCs are a putative source of metastasis-initiating cell (MIC) subpopulations [117].
There are two theories that explain the origin of breast cancer stem cells (BCSCs). The first one is based on the fact that the BCSCs arise from either mammary stem cells (MaSCs) or progenitor cells that retain their immortal proprieties through morphogenesis; the second hypothesis suggests that the BCSCs are created through the reprogramming of mature cells that after being exposed to damaging environmental factors acquire genetic alterations, inducing their de-differentiation and regaining the stem-like proprieties that lead to new generations of BCSCs [118]. Two additional hybrid models, wherein these two concepts merge, have been proposed to explain the tumor heterogeneity and plasticity: the BCSCs hypothesis suggests that only BCSCs are drivers of tumor growth with unlimited capacity for proliferation and metastasis, and the clonal evolution model is based on the concept that cancer arises from any differentiated cell of mammary tissue, which acquires BCSCs characteristics through various mutations, as an adaptive response to environmental stress mediated by the TME [116]. The cancer stem cell model sustains that the cellular diversity and tumor hierarchy are generated by the BCSCs [119], which are characterized by a high heterogeneity, different breast cancers emphasizing distinct subtypes and frequencies of BCSCs, involving numerous cellular biomarkers and regulatory signal pathways, and contributing to tumorigenesis and treatment resistance [119]. BCSCs can self-renew and asymmetrically divide to more differentiated cancer cells [120] and almost all epithelial tumors contain cancer stem-like cells [121]. A limited subpopulation of tumor-initiating cells (TICs), the CSCs also called tumor-propagating cells, is recognized to drive tumorigenesis, whereas they maintain a high self-renewal and differentiation potential during several generations in xenotransplants and ma-nifest the capability to remake the primary tumor intraheterogeneity by asymmetric division [122], and to induce the initiation, progression, metastasis and recurrence of tumor [123], also increasing colony formation and sphere forming ability [121]. Additionally, a recent work suggests that CSCs can also be the cells of origin for non-tumorigenic-differentiated stromal cells, such as cancer-associated fibroblasts (CAFs), tumor endothelial cells (TECs), tumor-associated adipocytes (TAAs) and tumor-associated macrophages (TAMs), into an experiment that used induced pluripotent stem cells (iPSCs), which were reprogrammed from normal cells [124].
Nevertheless, the eradication of BCSCs represents a promising insight in breast cancer molecular-targeted therapy [125]. A comprehensive review on the proteome of BCSCs emphasizes the main classes of specific protein biomarkers [126]: cell surface proteins, such as cluster of differentiation CD24/CD44, CD90/Thy-1 that was overexpressed in human malignant breast cancer lines, while CD14 was highly expressed as a stem cell biomarker in a normal breast cell line [127], human cripto-1 (Cr-1), also known as teratoma/teratocarcinoma-derived growth factor 1 (TDGF-1) involved in stem cell maintenance and malignant progression [128], and the epithelial cell adhesion molecule (EpCAM), which also contributes to the promotion of bone metastases [129]; signalling pathways proteins, such as those involved in the Notch pathway that has an important participation in BCSCs survival and self-renewal [130] as well as in breast cancer cells growth, migration, metastasis and angiogenesis [131], sonic hedgehog (Shh) and Wnt/β-catenin or Wnt/CTNNB1 [132] pathways involved in maintaining the stem characteristics of BCSCs [133], nuclear factor-κB (NF-κB) and phosphoinositide 3-kinase/Akt/mammalian target of rapamycin (PI3K/Akt/mTOR) signalling pathways [134], Hippo tumor suppressor pathway [135], the Janus kinase and signal transducer and activator of transcription (JAK-STAT) pathway that may represent a characteristic of BCSCs [136], and transforming growth factor (TGF)/SMAD pathway [137]; detoxifying proteins, such as ATP-binding cassette (ABC) transporters and aldehyde dehydrogenase (ALDH) that increase the cell resistance to anti-cancer drugs; communication proteins, such as connexines (Cx) involved in gap junctional communication [138], chemokine receptors like CXC chemokine receptor type 4 (CXCR4/CD184), a specific receptor for stromal-derived-factor-1 (SDF-1/CXCL12) involved in cell survival, proliferation, migration, and metastasis [139], different growth factor receptors, such as epidermal growth factor receptor (EGFR), fibroblast growth factor receptor (FGFR) [140], insulin-like growth factor receptor (IGFR) [141], insulin receptor (IR) [142], platelet-derived growth factor receptor (PDGF), transforming growth factor beta (TGF-β) receptor [137] or tyrosine kinase (c-Kit) receptors, and adhesion molecules, such as cadherins and integrins (ITGs); transcription factors, such as octamer-binding transcription factor 4 (OCT4), a stemness biomarker [143], SRY-box transcription factor 2 (SOX2) that promotes cell proliferation and metastasis in TNBC [144], Nanog protein that acts in maintaining the undifferentiated characteristics of pluripotent stem cells and can be used as a biomarker for prognostic prediction in breast ductal carcinoma [145], Krűppel-like factor 4 (KLF4) that maintains stem-like characteristics and promotes cell migration and invasion [146], and c-MYC protein that mediates cancer stem-like cells and EMT changes in TNBC [147].
In order to identify a stem cell niche residing in ducts and progenitor cells zones in lobules, two biomarkers have been used: cytokeratin CK14, that marks the myoepithelium, and CK19, a cytoskeletal protein that is overexpressed in breast cancer cells [148], which allows for the changeover of one type of cytoskeleton in the other [149]; the mammary stem cell activity could be also emphasized on the basis of surface markers, such as CD49f/α6-integrin and EpCAM that is expressed in basal membrane of normal epithelial cells but is upregulated [150] and identified as a biomarker for CSCs [129]. The keratin profiles suggest that CK19+/CK14+ stem cells reside in ducts [149]. The cell surface expression of the adhesion molecules CD44+ and CD24-/low Lineage- tumorigenic cells as well as phenotypically diverse populations of non-tumorigenic cells have been emphasized using a human breast cancer model, where the cells were grown in immunocompromised mice [151]. A high CD44+/CD24-/low ratio associated with cell proliferation and tumorigenesis, and ALDH1+, a strong indicator for cell migration and metastasis [117], are widely used as BCSCs biomarkers, from primary tumors to CTCs and disseminated tumor cells (DTCs). Normal and neoplastic human breast epithelial cells with increased ALDH activity have stem/progenitor proprieties [152]. The BCSCs are also involved in resistance to chemotherapy and radiotherapy [153]. The phenotypic plasticity enables BCSCs to switch between mesenchymal and epithelial-like states; the BCSCs in EMT process express CD44+/CD24-/low, while during the MET state express ALDH1 as biomarker, this transition between these two aformentioned states being essential for their ability to invade and grow at the metastatic primary and distant sites [154]. The overexpression of CD24, as a highly glycosylated mucin-like antigen, was associated with a poor prognosis in LA and TNBC [155], and also in HER2-enriched and BL breast cancer [156]. Serum CD44 can be an independent prognostic biomarker in primary breast cancer [157]. Tumors with high expression of CD44+/CD24-/low were shown to quickly relapse through bone metastases [158]. Many other protein biomarkers have been discovered both on cell surface and in the cytoplasm of BCSCs [120]. Protein C receptor (Procr) is a surface biomarker on MaSCs from the basal layer of the normal mammary gland and is expressed in some TNBC subgroups, where it is implicated in tumor progression [159] via stimulation of multiple signalling pathways such as Procr-dependent ERK and PI3k/Akt/mTOR through Src kinase and transactivation of IGF-1R [160]. A dysregulation of pathways of stemness and self-renewal involved in cancer invasiveness, blood spreading and metastases, such as Wnt, PI3k/Akt/FOXO, TGF-β and Notch was also described [161]. Cells that express CD133+ or prominin 1 (PROM1), a transmembrane glycoprotein found in epithelial and non-epithelial cells, have a greater colony-forming efficiency, higher proliferative rate and greater ability to form non-invasive and invasive breast tumors, supporting CD133 as a suitable biomarker for the identification of BCSCs in the most aggressive subtypes of cancer [162]. In invasive breast cancer, CD133 expression was positively associated with proliferation biomarkers including p16, cyclin E and Ki67, correlated with higher expression of other BCSCs biomarkers, such as CD24/CD44, SOX10, ALDH1A3 and α6-integrin [163]. Stem-like and EMT-like breast cancer cells show the highest expression of SOX10, a member of family transcription factors that induce preferential differentiation [164]. In metastatic breast cancer models and cancer cell lines, hypoxia-inducible factors (HIFs) are well known to be mediators of tumor growth, EMT and metastasis, as well as promoters in maintaining of BCSCs activity; α6-integrin is directly regulated by HIFs and mediates interactions with ECM [165].
Luminal epithelial cells
The clonal evolution model is based on the concept that cancer arises from any differentiated cell type of mammary tissue which acquires BCSCs characteristics via mutations and signals received from the TME [116]. Breast cancer can progress from non-neoplastic luminal cells through the epithelial hyperplasia, atypical ductal hyperplasia (ADH) and atypical lobular hyperplasia (ALH), ductal and lobular carcinoma in situ (CIS) and ductal and lobular invasive carcinoma (IC) [166]. Most human solid tumors originate in epithelial cells that are interconnected by tight and adherens junctions and desmosomes [167], which minimize the epithelial cell mobility [168]. The desmosomal proteins play an essential role in neoplastic progression or suppression [169], the loss of cell-cell adhesion being an essential step in progression of metastatic cascade [170]. During the EMT process, the epithelial cells dissolve cell-cell adhesions, reorganize their cytoskeleton, and transform into spindle-shape mesenchymal cells with migratory and invasive behaviour, emphasizing an overexpression of mesenchymal biomarkers, such as neural cadherin (N-cadherin), vimentin, fibroblast-specific protein (FSP1), and smooth muscle α-actin (SMA) [168]. A poor relapse-free survival was associated with a high preoperative expression of N-cadherin in peripheral blood or breast tumor tissues [171]. In addition, two novel metastasis inducers in breast cancer, DnaJ homolog subfamily B member 4 (DNAJB4) and CD81 have been identified, concluding that their suppression results in decreased cell migration and reduces primary tumor growth, extravasation and lung metastasis [172].
The epithelial cadherin (E-cadherin) is normally expressed in adherens junctions of breast epithelial tissue and it is downregulated in breast cancer, the absence of its expression being frequently observed [173]; additionally, a low or the loss of E-cadherin expression is associated with lymph node metastasis in IDC and a poor prognosis [174]. The E-cadherin-to-N-cadherin switch is a hallmark of EMT [168]. E-cadherin expression in lymph node metastases is lower than in primary sites [175]. β-catenin expression and location might serve as well as a biomarker in human [176] and mice [177] breast cancer. Perturbations in β-catenin proteasomal destruction result in its cytoplasmic accumulation and translocation in the nucleus, where it promotes the transcription of many oncogenes [178], contributing to tumor growth, cell migration and metastasis. In TNBC, β-catenin expression was associated with poor survival [179]. Mucins (MUCs) are large transmembrane glycoproteins that are expressed on the apical surface of almost all glandular epithelial cells and overexpressed in many adenocarcinomas, metastatic tissues, CTCs, and serum samples (as free or antibody-complexed mucin forms) from patients with breast cancer [180], where their abnormally high expression indicates a poor prognosis [181]. In cells that undergo tumorigenic changes, mucins suffer glycosylation as the main PTM and are translocated in cytoplasm where, following a proteolytic cleavage and nuclear translocation of MUC1 cytoplasmic domain (MUC1-CD), they can drive transcription of pro-invasive genes [182]. Together, MUC1, β-catenin and proto-oncogen tyrosine-protein kinase Src initiate changes in cytoskeleton and adhesive capability of transformed cells and, furthermore, by its interactions with intercellular adhesion molecule 1 (ICAM-1/CD54) and EGFR, activate their pro-metastatic capacity [182].
Another relevant protein family for luminal cells are CKs. In healthy breast tissues, both myoepithelial and luminal epithelial cells express the high-molecular-weight keratin CK5/6 in different quantities [183]. In normal breast tissue, CK7, CK8, CK18 and CK19 are expressed in the ductal epithelium, CKs IHC playing a key role in diagnosis, classification and prognostication of breast carcinoma; CK7 and CK8 showed the highest expression in all different subtypes of breast carcinoma, in particular for high-grade tumors [184]. Primary breast carcinomas showed changes in CK expression during metastatic progression to lymph nodes [185]. CK5/6 and clone 34βE12 in conjunction with ER may be helpful for a correct diagnosis of various stages of breast cancer [186]. Syndecan-1 (SDC1) is involved in the differentiation and prognosis of breast cancer [187] and breast cancer metastasis in the brain, supporting breast cancer cells migration across the blood-brain-barrier (BBB) [188]. The aberrant expression of stromal fibroblast-derived SDC1 stimulates breast carcinoma cells and induces stromal ECM fibers alignment, creating an invasive-permissive microenvironment [189]. Therefore, SDC1 is a novel molecular biomarker for inflammatory TNBC, modulating the cancer stem cell phenotype via IL6/STAT3, Notch and EGFR signalling pathways [190]. The majority of claudin (CLDN)-low breast tumors are aggressive TNBC characterized by low expression of genes involved in tight junctions and epithelial cell-to-cell adhesion proteins, such as claudins (CLDN) 3, 4 and 7, occludin (OCLN) and E-cadherin [191], associated with mesenchymal and stemness cells features [192]. CLDN-low breast cancers are characterized by low to absent expression of luminal biomarkers and closely resembles MaSCs, that leads to the idea that the MaSCs could be the cell of origin for this breast cancer subtype [193]. CLDN 5 is expressed in tight junctions of both epithelial and endothelial cells, and its expression was associated with cell motility and metastasis of human breast cancer [194]. The subfamilies of WASP/WAVE family, WASP and WASP verprolin-homologous protein (WAVE), are involved in control of actin polymerization through activation of the actin-related protein (Arp) 2/3 complex, which is essential for the formation of the small specialized actin-based protrusions called invadopodia [195], necessary for breast cancer cell motility to provide migration and invasion [196]. The introduction of CLDN 20 in breast cancer cell lines MDA-MB-231 and MCF7 resulted in aggressive tumor cells and reduced the trans-epithelial resistance [197]. The expression of OCLN as well as CLDN is lost in human breast cancer and presents a relationship with bone metastasis, OCLN leading to complex changes in human cancer cells [198]. The abnormal localization of OCLN and CLDN in mammary epithelial monolayers leads to apoptosis [199]. On the other hand, the desmoglein 2 (Dsg2) may act as a tumor suppressor [170], and human desmocollin 3 (Dsc3) shows a significant downregulation in breast cancer cell lines and primary breast tumor [200]. p53 protein is a multifunctional transcription factor that controls a great number of genes involved in cell cycle control, apoptosis, DNA repair and angiogenesis [201]; p53 promotes tumor cell death, its down-regulation and mutations contributing to tumorigenesis and cancer progression [201]. The serological high level of the mutant p53 is a promising biomarker for the assessment of the prognosis of breast cancer patients from early stages of malignancy [201]. Additionally, matrix metalloproteinases (MMPs) that degrade and modify cell-ECM and cell-cell adhesion junction proteins were found to be up-regulated in breast cancer and are currently associated with carcinogenesis, cancer cell invasion and metastasis. The tumor cells produce MMP enzymes and growth factors that may facilitate invasion of neoplastic cells [202].
Within normal breast epithelium of premenopausal women there are only 3-7% ER+ cells, 12-29% PR+ cells, and 3% Ki67+ cells [97]. ER and PR are expressed in approximatelly 80% and 60-70% of breast carcinoma respectivelly, while HER2 oncoprotein is overexpressed in 15 to 20% of primary breast carcinoma [112]. LA and LB subtypes are defined by the ER positive expression that responds to the antiestrogen therapy, and TNBC (ussually basal-like) lacks expression of ER, PR, and HER2 [119]. Postmenopausal women have higher expression of ER and lower expression of Ki67 when compared with premenopausal women [203]. Signal transducer and activator of transcription 3 (STAT3), an important member of the transcription factors family, is aberrantly active in many breast cancers [204]. Mammary epithelial cells secrete and absorb nipple aspirate fluid (NAF) that could contain exfoliated epithelial cells from the ductal/lobular system [111]. The use of NAF analysis as a breast-specific liquid biopsy is necessary for the early detection of breast cancer by biomarker profiling [84]. The presence of epithelial cells within NAF as a consequence of excessive growth and the exfoliation of epithelium is associated with breast cancer risk and could be a useful biomarker [205].
Myoepithelial cells
Basal/spindle/myoepithelial cells (MECs) are contractile hybrid of both smooth muscle cells, characterized by smooth muscle actin (SMA) and smooth muscle myosin (SMM) as biomarkers, and epithelial cells, with E-cadherin-mediated cell-cell junctions, as well as epithelial CKs as intermediate filaments; the loss of MEC layer marks the invasive cancer [206]. MECs and basement membrane reduce the invasion of tumor cells [202]. MECs are currently identified by IHC stains with numerous antibodies against SMA, high molecular weight cytokeratin (HMWCK), smooth muscle myosin heavy chain (SMMHC), the nuclear protein p63, CK5/6 [78], CK14, CK17, S100 protein [207], muscle-specific actin (MSA), the contractile protein calponin, p75 neutrophin receptor (nerve growth factor receptor), P-cadherin, mammary serine protease inhibitor (maspin), and CD10 [76]. p63 is normally expressed in the nuclei of breast MECs of normal ducts and lobules, strongly expressed in metaplastic carcinomas, and negative expressed in Phyllodes tumors and sarcomas [79]. CK5/6 appears to be more sensitive than CK14 for the basal subtype of breast carcinoma [184]. The normal MECs supress stromal invasion of tumor cells by the secretion of different anti-invasive and anti-angiogenic molecules, while the disruption of the MEC layer is followed by a release of growth and angiogenetic factors such as the VEGF, which plays a key role in lymphangiogenesis [208]. Breast adenomyoepithelioma is a rare type of breast cancer characterized by both epithelial and myoepithelial proliferation. In spindle-cell lesions, HMWCK, CK5/6 and CK14 are overexpressed, while S100 is downregulated; on the contrary, in clear cell lesions, HMWCK showed significant lower IHC staining, with diffusely positive epithelial cells and completely negative MECs image, that could be useful in diagnosing of adenomyoepithelioma [207].
Breast tumor microenvironment (TME)
Mammary parenchyma is surrounded by a stroma that contains fibrous and fat stromal pad, consisting of nutritional and supporting connective tissue. Fibrous stroma is organized in suspensory Cooper ligaments, consisting of dense fibrous connective tissue, whose shortening combined with an inflammatory oedematous skin gives rise to the histopathological “peau d’orange” appearance in breast carcinoma [209]. Fatty stroma consists of the adipose tissue of the breast and it is situated interlobularly rather than intralobularly.
Tumor tissue consists of tumor cells and stromal cells that build the TME also infiltrated by immune and vascular cells. Thus, the TME becomes an heterogeneous “ecosystem”, a veritable “fertilizing malignant tumor soil” allowing “the fast-growing tumor seeds” [210]. It is composed of mesenchymal cell populations, infiltrating immune cells, and matrix components involved in neoplastic progression [211], and promoting tumor angiogenesis [212]. The normal tissue microenvironment acts a barrier to tumorigenesis, but a plethora of factors, such as a low pH, reactive oxygen species (ROS), chemokines, cytokines, and hypoxia can disrupt the local homeostasis, promoting proinflammatory signals [213]. In some conditions, the TME can switch the immune cells from an antitumor behaviour into a novel permissive phenotype, which supports tumor growth and progression [214]. ECM is a 3D scaffold made up of collagen I and III, proteoglycans, hyaluronic acid, fibronectin, and tenascins [71] as well as cellular components, such as immune and inflammatory cells, endothelial cells, fibroblasts, adipocytes and bone marrow-derived cells [215]. The ECM supports epithelial development and survival by supressing apoptosis, and maintains the normal epithelial cells polarity, associated with low cell proliferation and tumorigenesis [215]. The vascular and lymphatic vessels develop in association with the breast epithelial tree, providing essential pathways for dissemination of neoplastic cells [202]. Blood vessels in breast cancer may emphasize dilatation, tortuosity, and abnormal perivascular coverage, partly due to the phenotypical and molecular alterations of pericytes [216].
Breast cancer is an inflammatory microenvironment that is supported by infiltrating immune cells, such as natural killer cells (NK) and neutrophils or cytotoxic T cells as well as naïve and memory T cells [217]. Various cells identified in tumor tissue can increase tumorigenicity by inhibition of antitumor immune responses, can contribute to angiogenesis and can initiate the tumor cell extravasation via EMT [218]. Eosinophils, monocytes, and B lymphocytes are associated with a good prognosis. Macrophages, mast cells, and eosinophils regulate invasion in the breast fat pad and are necessary for ductal differentiation. Interactions between tumor cells and surrounding stroma contribute to carcinogenesis [112]. Thus, the TME plays a determinant role on tumor survival, proliferation and metastasis [218]. During the tumor progression, the BCSCs and cancer cells originating in abnormal luminal and/or contractile myoepithelial cells break through the basement membrane; the BCSCs settle in a niche of tumor/cancer-associated macrophages (TAMs/CAMs), tumor/cancer-associated fibroblasts (TAFs/CAFs) [138], tumor/cancer-associated mast cells (TAMCs/CAMCs) and cancer-associated adipocytes (CAAs) possibly originating, based on a recent hypothetical view, in BCSCs [124]. The cancer cells communicate with each other or with tumor stromal- and infiltrating-associated cells, via gap junctions. Currently, two metastatic dispersion pathways are most often described: the hematogenic path, where the cancer cells and BCSCs enter the blood, process initiated by gap junction communication with endothelial cells that are themselves sealed by tight junctions and communicate by gap junctions, and the lymphogenic path where BCSCs or cancer cells enter lymphatic vessels directly at their open beginnings [138]. Endothelial progenitor cells (EPCs), mature endothelial cells, pericytes, fibroblasts, and immune mediators express a number of proinflammatory cytokines, such as tumor necrosis factor (TNF), and growth factors that interact with each other or with ECM components to control the major events that occur during angiogenesis [219]. The tumor-stroma communication occurs directly between stromal cells or via the autocrine and paracrine secretome and consists of several proteins that are secreted into ECM by tumor cells and cancer-associated stromal cells [43]. In the following subchapters, the key cellular players involved in breast cancer TME will be discussed.
Tumor-associated mesenchymal stem cells (TAMSCs)
Mesenchymal stem cells, also named mesenchymal stromal cells (MSCs), are self-renewing multipotent progenitor cells, found in bone marrow, adipose and others connective tissues, that are capable to differentiating into mesodermal cells, such as adipocytes [220] and adipose tissue-derived stem cells (ASCs) [221]. MSCs migrate towards the site of developing tumors and become integral components of the TME where they modulate the behaviour of other cells, actively supporting the tumor initiation, growth, metastasis and angiogenesis, especially under the stimulation of platelet-derived growth factor (PDGF) isoforms [221] and regulating responses to chemotherapy and radiotherapy by producing growth factors, chemokines and cytokines [222]. MSCs have been considered as cells with “double-bladed effects”, emphasizing protumor and antitumor activity [223]. The interaction between breast cancer cells and MSCs results in an increased proliferation and metabolic activity of breast cancer cells [224]. TAMSCs are a putative source for cancer/tumor-associated fibroblasts (CAFs/TAFs) [225]. The paracrine signalling from breast cancer cells converts MSCs into TAMSCs; the aggressive breast cancer cells engulf MSCs, in contrast to non-cancerous cells, and this cannibalistic behavior generates tumor cells with increased stemness, invasive and metastatic potential [226]. ECM remodelling by senescent MSCs has been associated with increased proliferation and motility of BCSCs, suggesting the role of this more invasive phenotype in age-related cancer progression [227].
Breast cancer-associated fibroblasts (CAFs)
Fibroblasts constitute the major cellular population of normal breast stroma and are responsible for elaboration of most of the components of healthy connective tissue, such as fibronectin and tenascin, which affect cell adhesion and proliferation, ECM deposition, MMPs production, and growth factor synthesis. CAFs/TAFs, the predominant cell type in the TME of the most aggressive cancers [212], differentiate from fibroblasts, MSCs, epithelial cells, and other cell types; the blockade of the interaction between PDGFRβ and TGFβR is a putative strategy to prevent TGFβ-mediated differentiation of MSCs into CAFs [228]. A fibroblast heterogeneity is recognized in primary tumors [229]: reactive fibroblasts, peritumoral fibroblasts, myofibroblasts and CAFs. CAFs are spindle-shape, α-smooth muscle actin (SMA)-positive fibroblasts, involved in neoplastic initiation, proliferation, invasion and metastasis; CAFs have demonstrated an overexpression of α-SMA, p53, podoplanin (PDPN) that promotes inflammation, cancer-associated thrombosis and regulates proliferation, contractility, migration, EMT of tumor cells, and remodeling of the ECM [230], CD10 that supports breast cancer dissemination and progression by its relationship with CSCs [231], fibroblast activation protein (FAP) often used as a pro-tumorigenic stromal biomarker in cancer [232], MMPs and tenascin-C (TNC) that play an essential role in cell proliferation, migration and tumour cells invasion [233]. PDGFα/β and caveolin 1 (Cav-1) downregulation [234] induces breast cancer cells proliferation and inhibition of apoptosis [235]. The cooperation between neoplastic cells and their stroma, and the important role of active CAFs that support their cancer-promoting function through paracrine effects are well-known [215]. Several CAFs subpopulations have been identified in metastatic lymph nodes: CAF-S1 stimulates neoplastic migration and initiates EMT through the chemokine CXCL12 and TGFβ pathways, and CAF-S4 induces neoplastic cell invasion by Notch signalling [229]. Myofibroblasts synthetize some factors which may stimulate proliferation and infiltration of neoplastic cells, such as IGF-2 and hepatocyte growth factor (HGF); the overexpression of Ki67 and HER2 within myofibroblasts from ductal breast cancer stroma is predictive for a worse prognosis [202]. The stromal expression of α-SMA in stromal myofibroblasts has been correlated with worse clinical outcome in invasive breast cancer [236]. The stromal loss of CD34 expression and acquisition of SMA myofibroblastic profile may represent a signal for tumor invasiveness in IDC; TGF-β is involved in inhibition of epithelial cells growth, but it stimulates the mesenchymal cell proliferation and activate the transformation of fibroblasts and CAFs into myofibroblasts, facilitating the breast neoplastic invasion [237]. CAFs are involved in recruitment of pro-angiogenic myeloid immune cells, such as macrophages and MDSCs, promoting angiogenesis and metastasis [212].
Breast cancer-associated adipocytes (CAAs)
Adipocytes are considered to play a key role in the inflammatory TME; normal adipocytes acquire an altered phenotype by upregulation of their beige-brown characteristics [238], an increased catabolism and change functionality when they become cancer-associated adypocytes (CAAs) under the influence of invasive cancer cells that intercommunicate with CAAs via exosomes [211]. Thus, the CAAs secrete inflammatory factors that modify the behaviour of breast cancer cells, promoting proliferation, invasion, metastasis and angiogenesis [211,239]. Adipocytes induce a transport-associated major vault protein (MVP)-related multi-drug resistant phenotype in breast cancer cells, which could contribute to obesity-related resistance to chemotherapy [240]; additionally, they secrete VEGF and regulate angiogenesis in mammary gland [241]. Secretions of dysfunctional adipocytes can alter the gene expression profile, induce hypoxia, and inhibit apoptosis [91]. Adipocytes release adipokines that are strongly involved in obesity-related tumorigenesis [242]. A high serum level of adiponectin, the only one adipokine that has demonstrated antitumor proprieties, might decrease the risk of postmenopausal breast cancer [243], and may serve as a risk biomarker in breast cancer [244]. Adiponectin has antiproliferative and proapoptotic effects in ERα-negative cells, while in obese patients it acts as a growth factor in ERα-positive breast cancer cells [245]. The adipocyte-mediated proliferation and migration of breast cancer cells was observed to be supressed when an inhibition of the PI3K/AKT/mTOR signalling pathway occurs, supporting the importance of interactions between breast cancer cells and adypocytes in the TME [246,247].
Breast tumor-associated macrophages (TAMs)
Macrophages are differentiated from monocytes recruited by tumor-derived cytokines [43]. Macrophages collaborate with B and T lymphocytes based on release of cytokines, chemokines, reactive radicals and other proteins [217]. TAMs/CAMs play a key role in cancer-induced lymphangiogenesis via upregulation of the VEGF-C expression that acts by the vascular endothelial growth factor receptor 3 (VEGFR-3) on the surface of lymphatic endothelial cells, contributing to their proliferation, lymphatic invasion and metastatic progression in breast cancer [248]. At the interface between pericytes, endothelial cells and macrophages, the deficiency of neural/glial NG2 proteoglycan, a pericyte biomarker in myeloid-specific NG2 null mice, is associated with the lack of ability to recruit macrophages to tumor site and other sites of inflammation and this absence of macrophages deprives pericytes of a signal that is essential for their propriety to interact with endothelial cells [249]. A new subpopulation of circulating cells that are not tumor cells but rather TAMs has been identified by use of CD45 and CD68 hematopoietic biomarkers [250].
Breast tumor-associated mast cells (TAMCs)
Mast cells (MCs) are bone marrow-derived granulated immune cells that localize at the margins or infiltrate the stroma surrounding solid tumors, including breast cancer [251], becoming tumor/cancer-associated mast cells (TAMCs/CAMCs). To promote the inflammation, inhibit tumor growth, and induce apoptosis of cancer cells, TAMCs secrete a wide variety of cytokines such as IL-1, IL-4, IL-8, IL-6, MCP-3, MCP-4, IFN-γ, LTB4, TGF-β, and chymase, while the IL-10, histamine, TNF-α, and adenosine manifests immunosuppressive capabilities [252]. Also, MCs release proangiogenetic factors, such as VEGF, FGF-2, PDGF, NGF, TGF-β, IL-8, heparin, tryptase and chymase, directly associated with neovascularization [252]. TAMCs stimulate other inflammatory cells to release angiogenetic factors in TME from diverse breast cancer subtypes [217], and support tumor invasiveness and vascularization by releasing MMPs [252]. Hormone receptors (HR: ER and PR) and HER2 status strongly influence the MCs dynamics, the intratumoral MCs increasing as density and distribution in aggressive tumors, as a worse prognostic factor [253]. In LA and LB breast tumors, significant high number of intratumoral chymase- and tryptase-positive MCs were observed when compared to TNBC and HER2+ cancers; consequently, a higher infiltration of TAMCs was associated with lower tumor grade, higher ER and PR expression, lower proliferation rate as well as the failure of HER2 overexpression [254]. Their pro- or antitumorigenic functions are still controversial [255].
Breast tumor-associated neutrophils (TANs)
Neutrophils, the most abundant circulating white blood cells, are immune cells that reflect a state of host inflammation as a hallmark of tumor; they participate, especially by attenuating immune system, in tumor initiation, growth, proliferation and metastasis [256]. TANs, as significant cells of the TME, are present in most TNBC and are associated with aggressiveness and a poor prognosis [257]. Neutrophils are attracted at tumor site by CXCR2 ligands and release MMPs that mediate angiogenesis, nitric oxide synthase (iNOS), arginase 1 (ARG1), ROS, reactive nitrogen species (RNS), and proteases that promote tumoral cell proliferation, EMT or modulate tumor cell lysis by T-lymphocytes [43]. There are evidences that the neutrophil to lymphocyte ratio (NLR) can be associated with adverse breast cancer prognosis and survival [258]. In cancer tissue, neutrophils are often associated with granulocytic myeloid-derived suppressor cells (gMDSCs) that share similar phenotype and express the same cell surface biomarkers with mature neutrophils [259].
Breast tumor-associated myeloid-derived suppressor cells (MDSCs)
MDSCs are myeloid cells, such as immature macrophages, granulocytes, dendritic cells (DCs), responsible for cancer-associated immunosuppressive action on T cells, DCs, and NK cells [260], tumor growth, recurrence, and breast cancer bone metastases [261]. MDSCs accumulate under influence of VEGF, TGFβ1, granulocyte-macrophage colony-stimulating factor (GM-CSF) [262], ILs and prostaglandin, and release iNOS, ARG1 and ROS, as well as TGFβ1 and MMPs when infiltrate primary cancer lesions, promoting angiogenesis and invasion. The overexpression of VEGF, TGFβ1, and IL-10 secreted by the numerous MDSCs infiltrated in TME induces EMT of tumor cells and promotes metastasis after the stress of surgical removal of the primary breast cancer [263]. MDSCs commonly express CD33 and CD11b as cell surface biomarkers and can be stimulatory to tumor-resident T regulatory (Treg) cells and TAMs [260].
Other stromal infiltrating immune cells
Stromal tumor-infiltrating lymphocytes (sTILs) are associated with a positive long-term prognosis [264], being important prognostic and predictive biomarkers in TNBC and HER2+ breast cancers [265]. CD8+ and CD4+ T cells as well as Treg cells expressing the transcription factor Foxp3 (FOXP3 +) [266] have been better correlated with a favourable outcome [264]. Biomarkers of B cells are also associated with longer survival in all subtypes of breast cancer [267]. It seems that the metastatic breast tumors in lung showed more sTILs than other metastatic sites and matched primary tumors [268]. DCs can be exploited by immune-based therapies in breast cancer due to their ability for antigen-cross-presentation and activation of lymphocytes against cancer cells [269], supporting the induction and maintenance of antitumor responses [214]. The accumulation of eosinophils in the TME, called tumor-associated tissue eosinophilia (TATE), was noted in 32.5% luminal, 5% HER2+, and 15% TNBC subtypes [270], TATE being reported as a putative prognostic biomarker for a better outcome of cancer patients due to the regulatory functions of eosinophils towards other immune cells in the TME and their direct cytotoxic functions against tumor cells [271].
Breast tumor endothelial cells (TECs)
Endothelium plays a key role in metastasis formation [272]. TECs stimulate expansion and activation of the pericyte precursor cell population via the pericyte activating factors, such as PDGFβ, VEGF, TGF-β, and angiopoietins (Angpts), as well as by signalling pathways involving Notch and ephrins [273]. The exocytosis mediated by Weibel-Palade bodies (WPBs), EC-specific organelles that contain the proangiogenic factor Angpt-2, and angiopoietin secretion are regulated during angiogenesis to limit pericyte coverage and blood vessels remodeling [274]. The early metastasis in mouse was correlated with glycocalyx disruption and endothelial inflammation; during the late metastasis, alterations in hemostasis were emphasized by increased plasminogen activator inhibitor (PAI-1) and von Willebrand factor (vWF), as well as a rise in adrenomedullin (ADM) and significant fall in adiponectin concentration [272]. The circulating endothelial progenitor cells (CEPCs) enter the blood stream from the bone marrow and subsequently migrate to sites of tumor vascularization [219]. The alterations in CEPCs counts could be a putative biomarker for monitoring response to chemotherapy [219].
Breast tumor vessels associated-pericytes
Pericytes are mural vascular smooth muscle cells surrounding the capillaries and post-capillary venules, adjacent to endothelial cells [275]. Pericytes are capable of tumor homing and are key components of the TME [276]. They play an essential role in angiogenesis, due to their close connection and interaction with the endothelial cells, that stabilize the newly formed endothelial tubes, modulate blood flow and vascular permeability, and regulate endothelial proliferation, differentiation, migration and survival [277]. Aberrations in pericyte-endothelial cell signalling networks may be involved in tumor angiogenesis and metastasis [278]. In vivo, the mammary epithelial cells undergo a spontaneous type of EMT called epithelial-to-pericyte transition (EPT), and consequently, the majority of EPT-resulting cancer-derived pericytes express pericyte biomarkers, such as neural/glial antigen 2 (NG2) and SMA, exhibit close vascular association, and seem to constitute a great population of tumor vessels associated-pericytes, necessary for vascular stabilization and cancer growth [168]. NG2 controls pericyte proliferation and mobility, acting as an auxiliary receptor that enhances signalling through integrins (ITGs) and tyrosine kinase growth factor receptors; NG2 also activates integrin signalling in endothelial cells that leads to formation and maturation of endothelial cell junctions [249].
Disseminated breast cancer cells
Disseminated breast cancer cells are detectable in the peripheral blood of patients with different malignancies as circulating tumor cells (CTCs) and in the bone marrow or lymph nodes as disseminated tumor cells (DTCs) [279]. Ubiquitous in epithelial malignancy, apoptosis resistant circulating tumor-associated cells (C-TACs) and their circulating ensembles of tumor-associated cells (C-ETACs) that comprise fibroblasts, tumor emboli and immune cells are well-known for the thrombosis risk and aggressive metastasis and could be considered as a systemic hallmark of cancer [280]. Single-CTC and CTC-clusters are rare, heterogeneous cells or bulk of neoplastic cells that detach from a primary solid tumor and metastatic sites and circulate in peripheral blood during cancer progression as an intermediate stage of metastatic process [112]. CTCs are considered as a new generation of “liquid biomarkers” [281], serving as a “liquid biopsy” that reflects the activity of the primary tumor [282]. Even if the spread of CTCs does not always lead to metastasis [282], the metastatic travel begins within the primary tumor with an EMT process that leads to mobilisation and intravasation of neoplastic epithelial cells and ends with a MET process, that leads to extravasation and invasion in distant tissues and organs, where, after a putative dormancy time, the tumor cells regain their epithelial phenotype, grow and become detectable within secondary foci of disease [283]. CTCs can be detected in peripheral blood even during the early stages of the tumoral disease [161]. The detection, isolation, capture, and characterization of CTCs as “minimally invasive multifunctional biomarkers” [284] have been correlated with an inferior prognosis, but they are also useful for assessing the effects of anti-cancer therapies [285]. The CTC molecular features highly interchange during invasion time [250]. In bloodstream, CTCs are exposed to immune system attacks, high oxygen level, high pressure and eventually anti-cancer therapeutic molecules [95], such that most CTCs die in circulation [286]. CTCs interact and activate platelets, which contribute to cancer cell survival and proliferation, being also involved in the early metastatic niche formation by release of different growth factors and chemokines, such as VEGF, CXCL5 and CXCL7 [218]. A major proportion of CTCs of metastatic breast cancer patients shows EMT and BCSCs characteristics [287]. EMT has been related to the differentiation of BCSCs that confers intratumoral heterogeneity and chemoresistance [288].
Among CTCs, it is possible to discriminate an epithelial CTC phenotype, mesenchymal CTCs, and a hybrid epithelial/mesenchymal bi-phenotype [161]. The CTC population can also comprise a subset of more dangerous cells, named circulating cancer stem cells (CSCs), endowed with self-renewal, multipotency and tumorigenic features [250]. In breast cancer patients, CTCs and DTCs can be simultaneous detected in blood and bone marrow, respectively [289]. DTCs in the bone marrow microenvironment display a quiescent mesenchymal-, osteoblast- or osteoclast-like phenotype, their long-time dormant state being a source of cancer relapse after cancer therapy [286]. CTC-clusters arising from oligoclonal groupings of primary tumor cells held together through plakoglobin-dependent cell-to-cell adhesion and seem to possess 23 to 50 fold increased metastatic potential and a shorter half-life in circulation [290]. Targeted single-CTC proteomics offers a unique, complementary understanding of CTCs biology and assesses their clinical impact [291]. The EMT may be induced by the TME, following the release of several growth factors or in response to hypoxia, leading to a local spread of cancer cells [289]. Several biomarkers are expressed by mesenchymal-like CTCs: oncogenic serine-threonine kinase 2 (AKT2), phosphoinositide 3-kinase (PI3Kα), and twist-related protein 1 (TWIST1) [161]. AKT2 promotes cell migration and invasion via vimentin induction [292], the overexpression of AKT2 promoting lung metastasis [293]. N-cadherin is also expressed in mesenchymal-like CTCs [250]. In breast cancer patients, the main protein biomarkers for the epithelial phenotype of CTCs are EpCAM, E-cadherin, CK8, CK9, and CK19 [294], zonula occludens (ZO), and the epithelial splicing regulator 1 (ESPR1) proteins [250]. Normally expressed in the basal membrane of epithelial cells, EpCAM is involved in cell adhesion, migration, proliferation and differentiation. It can also be found in DTCs and CTCs [150], CTCs being usually isolated from the blood by targeting the EpCAM as a biomarker [295]. The label-dependent techniques used for isolation of CTCs also exploit other surface antigens, such as HER2 and MUC1 [282], normally secreted by the luminal surface of the glandular epithelia [296]. HER2+ CTCs were identified in all stages, even in early breast cancer, with a higher detection rate in metastatic breast cancer [297], where the HER2-targeted therapy could reduce the overall CTCs count [298]. The main biomarkers for the stem-like CTCs (circulating-CSC population) are: ALDH1, CD44 and ABC transporters involved in chemoresistance [250,299]. A novel CTCs subpopulation that has both epithelial and hematopoietic cells characteristics has been described by use of EpCAM and CKs as epithelial biomarkers, and CD45 and CD68 as hematopoietic biomarkers [295].
Pre-metastatic and metastatic breast cancer niches
Metastasis is the spread of neoplastic cells from primary tumors toward distant organs [283], based on the ability and adaptability of cancer cells to create new niches and survive within different secondary sites [300]. Breast cancer predominantly spreads to the lymph nodes, lung, brain, bone and liver [93]. Metastasis is a multistage process, during which neoplastic cells express chameleonic phenotypes, continuously acquiring and adapting their specific proteomic profile to a precise stage during the cancer progression. The pre-metastatic niche, a permissive microenvironment prepared for colonization of tumor cells in specific organs [301], is defined based on molecular interaction between primary tumor that secrete primary tumor-derived factors, which partially directs the choice of organ for metastasis formation, and the distant tissues, by myeloid cells and local stromal players; the metastatic niche results from the interaction of cancer cells with the local microenvironment when arriving in a distant organ [302]. Tumor and endothelial cells secrete chemokines, which are responsible for leukocytes attraction, contributing to tumor cell extravasation from the blood [218]. The local inflammatory microenvironment stimulate tumor cells to produce tumor-derived secreted factors (TDSFs) (such as VEGF, TNF-α, TGF-β and IL) that attract the bone marrow-derived cells and stimulate their migration to pre-metastatic niche [301]. Thus, monocytes, macrophages and neutrophils are recruited to the early metastatic foci, increasing cancer cell extravasation, preventing neoplastic cell destruction by NK cells or transmitting survival signals to the tumor cells [218]. Primary tumors release exosomes responsible for cell-cell communication and that were found in bloodstream, urine, saliva and milk [303], expressing programmed death ligand 1 (PD-L1) and causing the immune escape of neoplastic cells [301]. Exosomes also transfer oncoproteins, attract macrophages and induce their reprogramming into the pre-metastatic niche [43]. ECM is a key component of metastatic niches. Homing of metastatic breast cancer cells within areas rich in osteoblasts and blood microvessels from trabecular regions of long bones is independent of their ER status [304]. The molecular profile of the ECM in distant organs supports cancer cell attachment, reactivates their survival signalling pathways, assures the nutrients availability and tumor metabolism, and coordinates the interaction with pro-tumor immune cells [302].
Tumor-tissue proteomics and cancer-associated biomarkers in breast cancer biopsies
Proteome profiling of fresh frozen (FF) or formalin-fixed paraffin-embedded (FFPE) breast tumor samples obtained by invasive surgical removal (lumpectomy or mastectomy) is useful for detecting and validating novel biomarkers in breast cancer diagnosis, staging and prognosis [305]. Among tumor-associated biomarkers currently in clinical use, the expression of hormone receptors (ER, PR, HER2), and urokinase-type plasminogen activator (uPA) that is an ECM-degrading protease involved in cancer invasion and metastasis interacting with its inhibitor plasminogen-activator type-1 (PAI-1) is essential and can be determined especially by ELISA and IHC detection [84]. Both uPA and PAI-1 are validated as prognostic biomarkers available for lymph node-negative breast cancer [306]. Cysteine-rich intestinal protein 1 (CRIP1) expression was proposed as a biomarker in HER2+ breast cancer patients [307]. PA28α/β proteins that are involved in breast cancer cell migration, invasion and metastasis [308,309], and a cleaved form of thymosin β-4 (TMSB4) specific to tumor-associated stroma have been identified using a MALDI MSI approach. Additionally, TMSB10 was overexpressed in breast cancer cells and tissues, promoting proliferation and migration of breast cancer cells; consequently, TMSB10 could be a diagnostic biomarker and a potential therapeutic target for breast cancer therapy [310]. For TNBC, MALDI-TOF MSI has identified many protein expression as potential prognostic biomarkers: pleckstrin homology domain-containing family G member 2 (PLEKHG2), ubiquitin protein ligase E3 component N-recognin 4 (UBR4)/p600, aminoimidazole carboxamide ribonucleotide transformylase/inosine monophosphate cyclohydrolase (ATIC), MUC4 [311], collagen type I alpha 1 chain (COL1A1) [312] and COL1A2, both upregulated in invasive breast cancer, and cytoplasmic and nuclear SRY-box transcription factor 11 (SOX11) involved in cell growth and invasion, used as a biomarker of poor prognosis in BL breast cancers [64]; the nuclear SOX11 could be a potential target for breast cancer therapy [313]. The tumor cells cytokeratin profile is quite versatile, the tumors often ceasing to express CKs which are normally present in their cells of origin [314]; however, the basal CK5/6, CK14, and CK17 are often used as positive biomarkers in classifying TNBC-BL [315]. In FFPE biopsies from TNBC patients, the expression of CK5/6 and HER1 was shown to predict poor survival and unfavourable response to chemotherapy [316]. CKs expression has been also assessed in human hair follicles [317]. Hair proteome has been investigated using a 2D LC-MS/MS approach, highlighting the PTMs of keratins [318], which can be manifested in response to cancer [319]. Specific hair CKs, such as cytokeratin 81 (KRT81) that was expressed in breast cancer cells, were shown to contribute to their migration and invasion [320]. Inositol polyphosphate 4-phosphatase type II (INPP4B) [321], an emerging tumor suppressor in breast cancer [322,323], and cyclin-dependent kinase 1 (CDK1) [324] have been found as relevant for breast cancer classification; the downregulation of CDK15 expression also promotes breast cancer cell invasion and metastasis [308]. Surprisingly, COL1A1 changes its orientation from parallel to perpendicular aligned straight fibers at the tumor-stromal boundary as a predictor of poor patient outcome, while tenascin C (TNC) and thrombospondin 2 (TSP-2) significantly co-localized with aligned collagen fibers in IDC tissues [325]. TSP-1 has been identified as an inhibitor of angiogenesis and metastasis, but it also manifests an opposite prometastatic role, i.e., in lung, depending on the molecular characteristics of the TME [326,327] or mediates in vitro cell mobility within mouse mammary tumor cells [328]. Apolipoprotein A1 (APOA1) and ApoB are risk factors for intraocular metastasis in patients with breast cancer [329]. Gelsolin (GELS) was downregulated in breast cancer tissues and was linked with metastasis and death [330]. Heat shock protein 90-beta (HSP90B), elongation factor 1-alpha 1 (EF1A1) that is significantly downregulated in invasive breast carcinoma [331], Na+/H+ exchange regulatory cofactor (NHRF1) and peroxiredoxin-1 (PRDX1), which downregulation significantly impaired the growth of breast cancer cell lines, have been also investigated as biomarkers [332]. Other most relevant proteomic signatures were: lysosomal protease cathepsin D (CTSD), a biomarker of poor prognosis in breast cancer [333], calreticulin (CALR) that was found to be upregulated mainly in ERα-breast cancer [334], ATP synthase subunit beta (ATPB) and HSP60/CH60 positive expression that was found to be significantly correlated with advanced stage of the tumor, lymph node metastasis and older patient’s age [335]. Coronin 1c and F-actin proteins are highly expressed in breast cancer tissues; F-actin protein expression was upregulated in patients with lymph node metastasis [336]. The ubiquitin binding protein SHARPIN belonging to the linear ubiquitin assembly complex (LUBAC), which consists of three potential diagnostic biomarkers for breast cancer [337], is highly express in human breast cancer [338], facilitating p53 protein degradation; p53 acts as a tumor suppressor and could be a prognostic biomarker for breast malignancy [339]. S100 calcium-binding protein P (S100P) promotes the aggressive features of breast cancer cells and can be used to predict the therapeutic effect of chemotherapy in HER2+ patients [340]; a novel truncated form of S100P was positively associated with larger tumor size and higher histological grade [341]. The epithelial calcium-binding protein S100A8 [342] may be associated with lymph node metastasis of breast cancer and could be considered as a biomarker for breast cancer progression [343,344]. The overall survival of breast cancer patients with positive expression of nucleobindin-2 (NUCB-2) was shorter [345]. The alphaB-crystallin (CRYAB/HSPB5) expression was upregulated in solid epithelial cancers and was also identified in DTCs and CTCs [150], suggesting that this protein could be used as a predictor of breast cancer metastasis to the lymph nodes [346] and the brain [347]. The prevalence of ALDH1A2+/CD44+ tumor cells is significantly associated with worse prognosis and poor clinical outcome in breast cancer patients [348]. The phosphoglycerate kinase-1 (PGK1) overexpression is a predictor of poor survival and a new prognostic biomarker of chemoresistance to paclitaxel [349], and stromal analysis of galectin expression has showed this contribution to cell survival after chemotherapy [350] and heterogeneity of breast cancer [351]. The hypoxic breast cancer TME induces an increasing activity of the hypoxia-inducible factors (HIFs), HIF-1 and HIF-2 [83], that are involved in EMT, angiogenesis and cell proliferation [94]. Several exosomal proteins were also identified as potential breast cancer biomarkers: glucose transporter 1 (GLUT1) [352,353], glypican 1 (GPC-1) and GPC-4 that act as tumor suppressors [354], a disintegrin and metalloproteinase domain-containing protein 10 (ADAM10) that when co-expressed with the amyloid precursor protein (APP) predicts a worse survival in patients with non-luminal breast cancers [355], and ADAM29, whose expression and mutations significantly influenced proliferation, migration and invasion of in vitro breast cancer cell lines [356]. Caveolin-1 (Cav1), a constituent protein of caveolae, plays a critical role in carcinogenesis, tumor progression through oxidative stress modulation [357], and chemotherapy or radiotherapy resistance [235], even though some authors reported contradictory results concerning the downregulation of stromal Cav-1 that promotes tumor survival and predicts a poor tumor prognosis, according to the TME characteristics [358]. Using an IHC approach applied to FF samples of male breast cancer (MBC), the N-acetyltransferase-1 (NAT1) was identified as possible prognostic biomarker for MBC, its positivity being correlated with a better outcome [90].
Liquid biopsies proteomics and humoral biomarkers in breast cancer
Serological protein biomarkers
Plasma testing is extensively used for proteomic analysis. It is relatively non-invasive, has low costs and does not require special equipment, even though the levels of tumour-derived proteins detected in plasma are often much lower than that those of tissues [359]. There are more than 8000 estimated proteins in the plasma proteome [360], and several tumor-derived proteins have been identified even during the first stages of tumorigenesis. The most commonly used current proteomic technologies for plasma proteome profiling are: sample fractionation and separation, such as dye, metal and antibody affinity chromatography, MALDI-TOF MS, liquid chromatography tandem mass spectrometry (LC-MS/MS), and multiplex protein quantitation [361]. A plasma peptidome-degradome profile from breast cancer patients and healthy controls was obtained via tandem Fourier transform mass spectrometry (FT-MS/MS) [362]. Additionally, a SER-ological Proteome Analysis (SERPA) was performed by 2D electrophoresis separation, immunoblotting, image analysis, and MS in order to multiply affinity protein profiling (MAPPing) into a breast cancer immunomics approach to identify autoantibody signatures produced in response to the tumoral process [58]. Recently, a plasma label-free Nano-LC-MS/MS approach identified several systemic molecular features and possible plasma biomarkers for LA, LB, HER2+ and TNBC patients [363]. The integration of proteomic profiles from mouse models of breast cancer and from human breast cancer cell lines may help isolate proteins released by tumor cells into the blood [364]. Using quantitative seroproteomics, four panels of serum proteins involved in the immune response, lipid metabolism, platelet degranulation, fibrinolysis, blood coagulation, glycolysis and cancer signalling pathways, with specificity for aforementioned subtypes of breast cancer have been identified [365].
MS-based serum proteomics, antibody detection and IHC techniques are essential for identifying enzymatic and non-enzymatic PTMs as potentially useful biomarkers for breast cancer [69]. Glycosylation, one of the most common PTMs in breast cancer [366], changes the expression of tumor-associated carbohydrate antigens (TACAs) that are involved not only in cell-cell and cell-ECM adhesion, but also in migration, proliferation, neoplastic growth and cancer aggressiveness [367]. Glycosylated-proteins and their specific glycoforms represent more than half of the cancer biomarkers and MS-based N-glycoproteomics techniques have developed an important contribution in this field [366]. MUC1 is the most intensively researched tumor-associated antigen [181]. Normally, its expression is detectable on the apical surface of the glandular epithelial cells and ductal epithelium but, when cells undergo tu-morigenic changes, they can produce glycosylated mucins that lose their localization emphasized by IHC staining [180], and become localized throughout the cell [182]. The transmembrane glycoprotein MUC1 is aberrantly glycosylated and overexpressed in different epithelial tumors; tumor-associated MUC1 participates in intracellular signal transduction pathways and controls the expression of target genes at transcriptional and post-transcriptional levels [368], and it has been shown to be involved in metastatic progression [181]. In breast cancer patients, the MUC1 overexpression was detected both in breast cancer tissue samples by IHC staining as well as free and antibody-complexed forms in serum samples by ELISA assay, the expression of free IgG and IgM antibodies against MUC1 being also concomitantly identified [369]. Currently, there are two soluble forms of MUC1 that are used in breast cancer clinical practice: the carbohydrate/cancer antigens CA 27.29 and CA 15-3 [296]. Additionally, circulating truncated extracellular HER2 is used for the detection of breast cancer [84]. The preoperative serum levels of carcinoembryonic antigen (CEA), the cancer associated carbohydrate antigen CA 125, and CA 15-3 were significantly higher in patients with breast cancer than controls subjects, the high level of serum CA 125 being a predictive marker for breast cancer outcomes and correlates with aggressive molecular subtypes [370]. CEA and CA 125 showed benefit over CA 15-3 alone in metastatic breast cancer (MBC) and all three biomarkers should be considered in MBC [371]. Two serum/soluble CK fragments, the tissue polypeptide antigen (TPA) and the tissue polypeptide-specific antigen (TPS), combined with CEA or not, have been suggested as biomarkers in the detection of distant metastasis [372]; TPA and TPS were found to be better as markers for monitoring chemotherapy response in metastatic breast cancer than CA 15-3 [373]. The serum specific antigen for CK19 fragments, CYFRA 21-1, was shown to be indicator of recurrence and treatment efficacy in breast cancer patients [374]. Serum CA 15-3, HSP90A and PAI-1 could be considered as early prognostic biomarkers [375]. THBS1 and bromodomain and WD repeat-containing protein 3 (BRWD3) were shown to be increased in plasma from breast cancer patients [376]. Apolipoproteins (APOs) are involved in autophagy, oxidative stress, and drug resistance [377]. In early and advanced stages of breast cancer, APOA2 overexpression were detected in plasma; APOC3 was detected in patients with early and intermediate stages of breast cancer, while clusterin (CLU/APOJ) was present at higher levels in intermediate stages of breast cancer [378]. The expression of TMSB10 is also significantly elevated in serum of breast cancer patients as a consequence of its upregulated expression in breast cancer cells and tissues and can be used as a serum biomarker for the diagnosis of breast cancer [310]. Apoptosis disruption is an important feature of cancer [296]; cellular byproducts from tumor cells that undergo apoptosis arrive in the systemic circulation at low concentrations, where they can be assessed in serum or plasma as soluble biomarkers of apoptosis [379]. Fas/APO1/CD95/TNFRSF6 and Fas Ligand (FasL) [380] act as major regulators of apoptosis by activation of caspase cascade involved in tumor cell death [381,382]. The soluble form of FAS (sFas) shows significantly higher serum concentrations in metastasized breast cancer patients [383], and the FasL could be used as a circulating apoptosis prognostic biomarker in breast carcinoma [379]. Circulating granzyme B and cytochrome c that increase following chemotherapy were also cited as apoptosis biomarkers in breast cancer [384]. Survivin (Sur) is an important member of the inhibitors of apoptosis protein (IAP) family, present in nucleus, cytoplasm, mitochondria and TME as an exosome-bound surviving protein [385]. Sur overexpression in neoplastic tissue is linked to lower survival rates in breast cancer patients [386], suggesting it as a potential cancer biomarker [387].
Proteomic profiles of cancer-derived exosomes (EVs) secreted by various breast cancer cell lines may be present in body biofluids such as blood, urine, saliva and breast milk, being indicative of cancer molecular subtype; their use as non-invasive biomarkers for diagnosis and management of breast cancer could be feasible thanks to the MS-based proteomics that enable an accurate EVs proteome profiling [388]. 10000 phosphopeptides and more than 100 phosphoproteins as candidate biomarkers from human plasma EVs were detected in higher quantities in breast cancer patients, using label-free quantitative proteomics [389]. A complex proteomic approach of two representative breast cancer lines led to the identification of 270 proteins, of which EGF-like repeats and discoidin I-like domain-containing 3 (EDIL-3) protein involved in cancer progression and metastatic breast cancer are of particular interest; fibronectin (FN) was also selected as a candidate biomarker due to its increased presence on the surface of EVs secreted by breast cancer cells, and it decreases after tumor surgical removal [390]. Developmental endothelial locus-1 (Del-1) on circulating EVs is a candidate biomarker for identification of breast cancer patients with early-stage breast cancer; also, it has been suggested that it can distinguish breast carcinoma from noncancerous and benign breast lesions [391]. Macrophage migration inhibitory factor (MIF), a small protein secreted by tumor [392], T cells, macrophages, endothelial and other epithelial cells [393] inhibits the migration of macrophages. MIF and its main extracellular receptor CD74 or its co-receptor CXCR4 are overexpressed in breast cancer cells and TME, promoting breast cancer cells survival, inhibition of autophagy or protection of tumor cells from apoptosis; neo-angiogenesis, breast cancer endothelial transmigration and prevalence of immune suppressive cells in the TME are also promoted by MIF [393]. The aza-derivatives of resveratrol, a natural polyphenolic compound found in grape skin and wine, were found to be inhibitors of MIF tautomerase, MIF biological activity and cell growth into MCF-7 breast carcinoma line, using a MALDI-TOF MS approach [392]. A quantitative proteomic MS immunoassay (MSIA) was developed and validated for discovery and quantification of different proteoformes of MIF in serum samples [394].
Angiogenesis is a significant process involved in tumor development and metastasis. Several signalling pathways, such as NF-κB, STAT3, PI3K/Akt, and p38 MAPK, have been associated with the interrelated inflammatory TME and angiogenesis process [395]. Neo-vascularization requires EpCAMs, such as cadherins, integrins (ITGs), selectins, immunoglobulin and CD44 molecules [396]. The endothelial selectin (E-selectin) induces MET and stem cells traits in DTCs in the bone and lung metastasis of breast cancer [397]. Intracellular adhesion molecule-1 (ICAM-1) is a transmembrane protein belonging to the immunoglobulin superfamily, which also has also a soluble isoform (sICAM-1) derived by proteolytic cleavage of membrane-bound ICAM-1 [398]. Normally present in endothelial cells, leukocytes, macrophages, monocytes and other immune cells, but also on the surface of cancer cells, CAMs mediate the adhesion on the surface of the vascular endothelial cells and transendothelial migration of activated leukocytes. The levels of soluble vascular cell adhesion molecule (sVCAM-1) and sICAM-1 in the serum of breast cancer patients were significantly higher than in healthy controls, suggesting that the soluble forms of these adhesion proteins may be useful serum biomarkers in clinical setting [396]. Platelet endothelial cell adhesion molecule-1 (PCAM-1), a CAM expressed on platelets, leukocytes and endothelial cells, has been implicated in the late progression of metastatic tumors, its activity appearing to be modulated by the TME and tumor cell proliferation [399]. The vimentin expression in peripheral blood was also higher in metastatic breast cancer [171]. A recently published study demonstrated that the abnormal levels of circulating thrombotic biomarkers, such as prothrombin fragment 1+2 (F 1+2), D-dimer and fibrinogen, may represent novel non-invasive factors for improving prediction of breast cancer recurrence risk in resected breast cancer patients [400]. The plasma fibrinogen level could be a possible biomarker for assessing the clinical response to chemotherapy and metastasis after surgery in advanced breast cancer patients [401].
Urinary protein biomarkers
Human urine is one of the most interesting biofluids for clinical proteomic studies, advances in MS leading to the identification of thousands of proteins and peptides in urine [402]. Urine peptidomics and uroproteome alterations study represent alternative approaches for discovering candidate biomarkers, especially in early cancer diagnosis [403], discriminating between breast cancer patients from healthy controls [404]. Urinary zinc-alpha2-glycoprotein (ZA2G), leucine-rich alpha2-glycoprotein (LRG), retinol-binding protein A4 (RBP4), also emphasizing significant higher serum levels in patients with breast cancer [405], annexin A1 (ANXA1), ganglioside GM2 activator (SAP3/GM2A), Src substrate cortactin (SRC8/CTTN), gelsolin (GELS/GSN), kininogen-1 (KNG1), CO9, clusterin (CLUS) that when overexpressed might be a predictive factor for recurrence [406], whereas ceruloplasmin (CERU), and α1-antitrypsin (A1AT) have been proposed as candidate biomarkers that could discriminate HER2-enriched subtype breast cancer from the healthy controls [404]. Circulating plasma gelsolin (pGSN), which is carried by exosomes [407], is a secreted iosoform of GSN that was identified in urine and other biofluids and it was recognized as a putative biomarker of the inflammatory environment [408]. A label-free LC-MS/MS approach was used to detect some urine protein biomarkers, such as extracellular matrix protein 1 (EMP1), microtubule associated serine/threonine kinase member 4 (MAST4), and filaggrin (FLG), correlating the urinary proteomic profiles with different stages of breast cancer or early detection and monitoring progression in invasive breast cancer; leucine-rich repeat-contain protein 36 (LRC36), MAST4 and uncharacterized protein CI131 were associated with preinvasive DCIS, dynein heavy chain 8 (DYH8), hemoglobin subunit alpha (HBA), pepsin A (PEPA), FLG and multimerin-2 (MMRN2), which is required for blood vessels homeostasis and stabilization, its upregulation being correlated with an increased endothelial cell permeability [409], were associated with early invasive breast cancer, while AGRIN that promotes cell proliferation, migration and oncogenic signalling [410], neuronal growth regulator 1 (NEGR1) that is downregulated in various cancers and its overexpression significantly reduces the tumorigenicity in some cancer lines [411], fibrinogen alpha chain (FIBA) and keratin KIC10 have been associated with metastatic breast cancer [402].
Salivary protein biomarkers
Saliva is an easily accessible biofluid that assesses the presence and characteristics of cancer in real-time health monitoring and high-impact preventive medicine [412]. In saliva, more than 2300 proteins and peptides have been identified, which represents only 30% of the blood proteome; furthermore, salivaomics, including human salivary proteomics, is widely involved in biomarker discovery [413]. In IDC, a catalogue of altered salivary proteins includes: genomic integrity related proteins, molecular chaperones/HSPs, cell growth and apoptosis related proteins, immunity related proteins, cytoskeleton, membrane and calcium binding related proteins, metabolism related and oral anti-microbial related proteins [414]. VEGF, EGF and CEA are useful in breast cancer salivary detection [415]. Usually, saliva contains all biomarkers present in blood, but in lower quantities [416]. CA 125 level in saliva was higher than in serum, but serum and salivary levels were significantly higher in women with untreated breast cancer [417]. Salivary expression of the HER2 protein receptor may not be yet a viable alternative to breast cancer tissue assessment because of the lack of a statistically significant difference between the HER2-positive and HER2-negative groups, and between case and control groups [418]. The presence of CA 15-3 in human saliva was performed using an Au/ZnO thin film surface plasmon resonance (SPR)-based biosensor immunoassay [419]. Autoantibodies against HER2 and MUC1 detected both in serum and saliva could be useful in breast cancer screening [420]. Additionally, proteomic profiling by 2D-DIGE and protein immunoblotting revealed that the carbonic anhydrase VI (CA6) and psoriasin (S100A7) expression was significantly different between breast cancer patients and control samples [421]; S100A7 is highly and more frequent expressed in DCIS than in invasive breast carcinomas (IBC), is associated with poor prognostic in both DCIS and IBC [422], promoting aggressive features in breast cancer [423].
Tear fluid protein biomarkers
Almost 2000 tear proteins have been reported in human by tearomics approach; approximately 500-600 plasma proteins were also identified in tear fluid, this overlap allowing detection of systemic diseases through tear proteomics [360]. A SELDI-TOF MS approach identified several non-invasive protein biomarkers within tear fluid, allowing the discrimination between breast cancer patients and heathy controls [424], and a MALDI-TOF/TOF-driven semi-quantitative proteomic study compared tear protein levels in primary IBC with healthy controls [425]. In breast cancer patients, several proteins involved in immune response, such as inflammation-related protein S100A8 and complement factor C1Q1, and in different metabolic processes, such as ALDH3A and triosephosphate isomerase (TPI) [426], have modified expression.
Peritoneal, pleural and pericardial fluid biomarkers
Cytopathological investigation of malignant pleural, peritoneal and pericardial effusions occurs in different clinical settings; in females, the most common metastatic tumors detected in pleural effusions were breast primary tumors, most commonly IDC, followed by ILC; among peritoneal effusions, most commonly was ILC, followed by IDC, and for pericardial effusions, the most commonly was IDC [427]. Malignant pleural effusion (MPE) occurs in 2-11% of patients with breast cancer, the tumor cells disseminating into the pleural cavity via lymphatic vessels; the presence of nuclear antigen Ki-67 as a cellular proliferation biomarker in MPE predicts a worse outcome [428]. MMP-2 and MMP-9 expression was investigated and might be used as novel and additive diagnostic biomarker in peritoneal and pleural fluid of patients with metastatic breast cancer, by zymography and ELISA assays [429]. The peritoneal fluid can be also used for determining primary tumor sites; large clusters of cells correlated with only CK7 positivity could indicate a malignancy originating from the breast [430].
Breast-specific liquid biopsies
Nipple aspirate fluid (NAF), a secretome originating in mammary ductal cells that contains proteins associated with the TME [42], and ductal lavage (DL) fluid, in contrast to a breast biopsy, are non-invasive, inexpensive and feasible sources of biomarkers that in combination with proteomic profiling technology are highly specific to breast cancer [430]. Proteins are important constituents of NAF and are often expressed in higher concentrations than in plasma [84]. The NAF proteome was described using 2D-PAGE followed by a MALDI-TOF MS approach [432]. ELISA was used to emphasize some proteins in NAF, such as SOD-1, CRP, chitinase-3-like protein 1 (YKL40), cathepsin D (CTSD), basic fibroblast growth factor (bFGF) [433], gross cystic disease fluid protein (GCDFP)-15, ApoD and α1-acid glycoprotein (AAG) [432]. Differential NAF proteomic expression was detected by SELDI-TOF and MALDI-TOF MS between healthy and breast cancer patients, with identification of a β-casein-like peptide associated with breast cancer [434]. A proteolytic fragment of alpha1-antitrypsin (A1A/AAT) has been shown to be elevated in NAF of breast cancer patients [435]. NAF containing antibodies against tumor-specific MUC1 peptide derived from the MUC1-ECD can be a potential biomarker to predict tumor aggressiveness in breast cancer [436]. CA2, catalase, and PRDX2 have been identified in serum samples and in nipple discharge (ND) and proposed as new candidate breast cancer biomarkers, following a 2D nano-LC/ESI-MS/MS coupled with Western blot analysis and sandwich ELISA approach [437]. Significant overexpression of PAI-1, also detected in plasma, have been reported in NAF of women with breast cancer [84].
There are several studies based on SELDI-MS that have performed a comparative proteomic analysis of NAF from healthy and breast cancer patients [438,439], and detected several breast cancer biomarkers in NAF [440,441]. Thus, the NAF proteome composition is widely related to breast cancer behaviour, based on a higher glucose consumption, oncogene activation, silencing of tumor suppression genes, innate immune system activation or ECM remodeling and degradation, and can be correlated with invasiveness, aggressiveness and metastatic potential [42]. By 2D-LC and gel-based proteomic methods, 151 proteins were identified in the aqueous phase of human colostrum [442]. HPLC and LC-MS techniques have contributed to purify the lactoferrin (Lf), an iron-binding protein from different sources of bovine colostrum, in order to treat with Lf the triple negative MDA-MB-231 and the ER-positive MCF-7 breast cancer cell lines; results showed that Lf has the capacity to inhibit the in vitro growth of aforementioned cell lines, inducing cytotoxicity, reduction in cell proliferation, apoptosis and changes at transcriptional level [443]. The soluble TNF-related apoptosis-inducing ligand (sTRAIL), a member of the TNF protein superfamily of cytokines from human colostrum and milk, is present at really high levels, mediating the antitumor activity of human milk by apoptosis and cell proliferation control [444]. Human breast milk contains a lot of biologically active components, including milk proteome made by proteins, enzymes/proteases, glycoproteins, and endogenous peptides [115], a diverse microbiome, and a heterogeneous population of epithelial and immune cells [445]. Breast milk is an appropriate cancer microenvironment for identifying breast cancer biomarkers; MS-based proteomics on human breast milk may offer a novel perspective to identify an increased risk of developing breast cancer [446]. A comparative proteomic analysis of human milk samples from breastfeeding mothers with cancer, who were diagnosed either before or after milk donation, and from healthy women, has been published based on combinatorial electrophoresis and MS-based proteomics [447]. Alternatively, the fine-needle aspiration biopsy guided by ultrasonography (US-FNAB) can be used to detect axillary lymph node metastasis; CYFRA 21-1 levels in FNAB represent an important marker in detecting metastatic deposits of breast cancer that complements cytology and CK19 IHC as diagnostic tool [448].
Conclusions and perspectives
In this review, we have proposed a synthetic Breast Cancer Cell Continuum Concept (BCCCC) based on a Breast Cancer Proteomic Concept (BCPCC) to reach a better understanding of the breast cancer tumorigenic cascade at a histo-molecular level, from a driving mutation to metastatic foci development by the help of classic methods (i.e., ELISA and IHC) and advanced proteomics technologies, such as MS-based approaches. The BCCCC comprises many successively integrated populations of heterogeneous neoplastic and non-tumorigenic-associated cells, such as breast cancer stem cells (BCSCs) and/or de-differentiated mammary tumor cells, tumor-associated mesenchymal stem cells (TAMSCs), tumor-associated fibroblasts (TAFs), tumor-associated macrophages (TAMs), tumor-associated mast cells (TAMCs), tumor-associated neutrophils (TANs), cancer-associated adipocytes (CAAs), other stromal infiltrating immune cells, circulating tumor cells (CTCs), disseminated breast cancer cells (DTCs), circulating ensembles of tumor-associated cells (C-ETACs), and distant cell populations driving the metastatic breast niches. The Breast Cancer Proteomic Continuum Concept (BCPCC), whereas each phenotype of neoplastic and tumor-associated cells is characterized by a changing and adaptive proteomic profile detected in solid or liquid, non-invasive biopsies by complex proteomics approaches, begins with the proteomic landscape of different neoplastic and cancer-associated cells, followed by subsequent analysis of protein biomarkers involved in epithelial-mesenchymal transition (EMT) and intravasation, CTCs proteomics, and finally by protein biomarkers that highlight the extravasation and distant metastatic invasion. Both BCCCC and BCPCC rely on the analysis of cancer cells and their TME, which is shaped by various cellular and non-cellular factors. Targeted proteomics and its related subfields validate and monitor proteins and their proteoforms as diagnostic, prognostic or predictive biomarkers that lead to a high-integrated precision diagnostic and personalized oncomedicine for this challenging cancer characterized by a remarkable intraindividual and interindividual.
Acknowledgements
We would like to thank the past and current members of the Biochemistry & Proteomics Lab for their contribution to research, colegiality and pleasant environment in the lab. This manuscript is dedicated to the memory of Prof. Dr. Vlad Artenie from the Al. I. Cuza University of Iași, Romania.
Disclosure of conflict of interest
None.
References
- 1.Elzamly S, Badri N, Padilla O, Dwivedi AK, Alvarado LA, Hamilton M, Diab N, Rock C, Elfar A, Teleb M, Sanchez L, Nahleh Z. Epithelial-mesenchymal transition markers in breast cancer and pathological responseafter neoadjuvant chemotherapy. Breast Cancer (Auckl) 2018;12:1178223418788074. doi: 10.1177/1178223418788074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mueller C, Haymond A, Davis JB, Williams A, Espina V. Protein biomarkers for subtyping breast cancer and implications for future research. Expert Rev Proteomics. 2018;15:131–152. doi: 10.1080/14789450.2018.1421071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coleman WB. Next generation breast cancer omics. Am J Pathol. 2017;187:2130–2132. doi: 10.1016/j.ajpath.2017.07.011. [DOI] [PubMed] [Google Scholar]
- 4.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481:306–313. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davatzikos C, Rathore S, Bakas S, Pati S, Bergman M, Kalarot R, Sridharan P, Gastounioti A, Jahani N, Cohen E, Akbari H, Tunc B, Doshi J, Parker D, Hsieh M, Sotiras A, Li H, Ou Y, Doot RK, Bilello M, Fan Y, Shinohara RT, Yushkevich P, Verma R, Kontos D. Cancer imaging phenomics toolkit: quantitative imaging analytics for precision diagnostics and predictive modeling of clinical outcome. J Med Imaging (Bellingham) 2018;5:011018. doi: 10.1117/1.JMI.5.1.011018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chandramouli K, Qian PY. Proteomics: challenges, techniques and possibilities to overcome biological sample complexity. Hum Genomics Proteomics. 2009;2009:239204. doi: 10.4061/2009/239204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ren J, Wang B, Li J. Integrating proteomic and phosphoproteomic data for pathway analysis in breast cancer. BMC Syst Biol. 2018;12:130. doi: 10.1186/s12918-018-0646-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guerin M, Goncalves A, Toiron Y, Baudelet E, Audebert S, Boyer JB, Borg JP, Camoin L. How may targeted proteomics complement genomic data in breast cancer? Expert Rev Proteomics. 2017;14:43–54. doi: 10.1080/14789450.2017.1256776. [DOI] [PubMed] [Google Scholar]
- 9.Qin XJ, Ling BX. Proteomic studies in breast cancer (review) Oncol Lett. 2012;3:735–743. doi: 10.3892/ol.2012.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Silva C, Perestrelo R, Silva P, Tomás H, Câmara JS. Breast cancer metabolomics: from analytical platforms to multivariate data analysis. A review. Metabolites. 2019;9:102. doi: 10.3390/metabo9050102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Z, Li Z, Li H, Jiang Y. Metabolomics: a promising diagnostic and therapeutic implement for breast cancer. Onco Targets Ther. 2019;12:6797–6811. doi: 10.2147/OTT.S215628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dougan MM, Li Y, Chu LW, Haile RW, Whittemore AS, Han SS, Moore SC, Sampson JN, Andrulis IL, John EM, Hsing AW. Metabolomic profiles in breast cancer: a pilot case-control study in the breast cancer family registry. BMC Cancer. 2018;18:532. doi: 10.1186/s12885-018-4437-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alakwaa FM, Savelieff MG. Bioinformatics analysis of metabolomics data unveils association of metabolic signatures with methylation in breast cancer. J Proteome Res. 2020;19:2879–2889. doi: 10.1021/acs.jproteome.9b00755. [DOI] [PubMed] [Google Scholar]
- 14.Jovanovic J, Rønneberg JA, Tost J, Kristensen V. The epigenetics of breast cancer. Mol Oncol. 2010;4:242–254. doi: 10.1016/j.molonc.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Romagnolo DF, Daniels KD, Grunwald JT, Ramos SA, Propper CR, Selmin OI. Epigenetics of breast cancer: modifying role of environmental and bioactive food compounds. Mol Nutr Food Res. 2016;60:1310–1329. doi: 10.1002/mnfr.201501063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lo PK, Sukumar S. Epigenomics and breast cancer. Pharmacogenomics. 2008;9:1879–1902. doi: 10.2217/14622416.9.12.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan F, Zhao H, Zeng Y. Lipidomics: a promising cancer biomarker. Clin Transl Med. 2018;7:21. doi: 10.1186/s40169-018-0199-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perrotti F, Rosa C, Cicalini I, Sacchetta P, Del Boccio P, Genovesi D, Pieragostino D. Advances in lipidomics for cancer biomarkers discovery. Int J Mol Sci. 2016;17:1992. doi: 10.3390/ijms17121992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fichtali K, Bititi A, Elghanmi A, Ghazi B. Serum lipidomic profiling in breast cancer to identify screening, diagnostic, and prognostic biomarkers. Biores Open Access. 2020;9:1–6. doi: 10.1089/biores.2018.0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu J, Liao M, Yao Z, Liang W, Li Q, Liu J, Yang H, Ji Y, Wei W, Tan A, Liang S, Chen Y, Lin H, Zhu X, Huang S, Tian J, Tang R, Wang Q, Mo Z. Breast cancer in postmenopausal women is associated with an altered gut metagenome. Microbiome. 2018;6:136. doi: 10.1186/s40168-018-0515-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Costantini L, Magno S, Albanese D, Donati C, Molinari R, Filippone A, Masetti R, Merendino N. Characterization of human breast tissue microbiota from core needle biopsies through the analysis of multi hypervariable 16S-rRNA gene regions. Sci Rep. 2018;8:16893. doi: 10.1038/s41598-018-35329-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Banerjee J, Mishra N, Dhas Y. Metagenomics: a new horizon in cancer research. Meta Gene. 2015;5:84–9. doi: 10.1016/j.mgene.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Komorowski AS, Pezo RC. Untapped “-omics”: the microbial metagenome, estrobolome, and their influence on the development of breast cancer and response to treatment. Breast Cancer Res Treat. 2020;179:287–300. doi: 10.1007/s10549-019-05472-w. [DOI] [PubMed] [Google Scholar]
- 24.Horcajadas JA, Gosálvez J. Preface. Reproductomics. In: Horcajadas JA, Gosálvez J, editors. Academic Press; 2018. pp. xi–xvi. [Google Scholar]
- 25.Zarbl H. Toxicogenomic analyses of genetic susceptibility to mammary gland carcinogenesis in rodents: implications for human breast cancer. Breast Dis. 2007;28:87–105. doi: 10.3233/bd-2007-28109. [DOI] [PubMed] [Google Scholar]
- 26.Ning B, Su Z, Mei N, Hong H, Deng H, Shi L, Fuscoe JC, Tolleson WH. Toxicogenomics and cancer susceptibility: advances with next-generation sequencing. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev. 2014;32:121–158. doi: 10.1080/10590501.2014.907460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jeibouei S, Akbari ME, Kalbasi A, Aref AR, Ajoudanian M, Rezvani A, Zali H. Personalized medicine in breast cancer: pharmacogenomics approaches. Pharmgenomics Pers Med. 2019;12:59–73. doi: 10.2147/PGPM.S167886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ingle J. Pharmacogenetics and pharmacogenomics of endocrine agents for breast cancer. Breast Cancer Res. 2008;10(Suppl 4):S17. doi: 10.1186/bcr2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodríguez-Vicente AE, Lumbreras E, Hernández JM, Martín M, Calles A, Otín CL, Algarra SM, Páez D, Taron M. Pharmacogenetics and pharmacogenomics as tools in cancer therapy. Drug Metab Pers Ther. 2016;31:25–34. doi: 10.1515/dmpt-2015-0042. [DOI] [PubMed] [Google Scholar]
- 30.Albuquerque TAF, Drummond do Val L, Doherty A, de Magalhães JP. From humans to hydra: patterns of cancer across the tree of life. Biol Rev Camb Philos Soc. 2018;93:1715–1734. doi: 10.1111/brv.12415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Korwar AM, Bhonsle HS, Ghole VS, Gawai KR, Koppikar CB, Kulkarni MJ. Proteomic profiling and interactome analysis of ER-positive/HER2/neu negative invasive ductal carcinoma of the breast: towards proteomics biomarkers. OMICS. 2013;17:27–40. doi: 10.1089/omi.2012.0054. [DOI] [PubMed] [Google Scholar]
- 32.Gulfidan G, Turanli B, Beklen H, Sinha R, Arga KY. Pan-cancer mapping of differential protein-protein interactions. Sci Rep. 2020;10:3272. doi: 10.1038/s41598-020-60127-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lim S, Park Y, Hur B, Kim M, Han W, Kim S. Protein Interaction Network (PIN)-based breast cancer subsystem identification and activation measurement for prognostic modeling. Methods. 2016;110:81–89. doi: 10.1016/j.ymeth.2016.06.015. [DOI] [PubMed] [Google Scholar]
- 34.Parekh V, Jacobs M. Tumor connectomics: mapping the intra-tumoral complex interaction network. 2019 doi: 10.3390/cancers14061481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang W, Zhou M, Dorsey TH, Prieto DA, Wang XW, Ruppin E, Veenstra TD, Ambs S. Integrated proteotranscriptomics of breast cancer reveals globally increased protein-mRNA concordance associated with subtypes and survival. Genome Med. 2018;10:94. doi: 10.1186/s13073-018-0602-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shahani M, Shakeri J, Akbari ME, Arefnezhad B, Tafti A, Zali H, Nafisi N, Hashemi M, Rezaei-Tavirani M, Mohammadpour S, Salami SAR, Mirzai HR, Samsami M, Ezabady SHJ, Akbari A. Transcriptomic and proteomic approaches reveal biological basis of intraoperative radiotherapy-treated tumor bed modification in breast cancer patients: a pilot study. J Proteomics. 2020;212:103596. doi: 10.1016/j.jprot.2019.103596. [DOI] [PubMed] [Google Scholar]
- 37.Jézéquel P, Guette C, Lasla H, Gouraud W, Boissard A, Guérin-Charbonnel C, Campone M. iTRAQ-based quantitative proteomic analysis strengthens transcriptomic subtyping of triple-negative breast cancer tumors. Proteomics. 2019;19:e1800484. doi: 10.1002/pmic.201800484. [DOI] [PubMed] [Google Scholar]
- 38.Peng W, Zhu R, Zhou S, Mirzaei P, Mechref Y. Integrated transcriptomics, proteomics, and glycomics reveals the association between up-regulation of sialylated N-glycans/integrin and breast cancer brain metastasis. Sci Rep. 2019;9:17361. doi: 10.1038/s41598-019-53984-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhan X, Cheng J, Huang Z, Han Z, Helm B, Liu X, Zhang J, Wang TF, Ni D, Huang K. Correlation analysis of histopathology and proteogenomics data for breast cancer. Mol Cell Proteomics. 2019;18:S37–S51. doi: 10.1074/mcp.RA118.001232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ellis MJ, Gillette M, Carr SA, Paulovich AG, Smith RD, Rodland KK, Townsend RR, Kinsinger C, Mesri M, Rodriguez H, Liebler DC Clinical Proteomic Tumor Analysis Consortium (CPTAC) Connecting genomic alterations to cancer biology with proteomics: the NCI Clinical Proteomic Tumor Analysis Consortium. Cancer Discov. 2013;3:1108–1112. doi: 10.1158/2159-8290.CD-13-0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Song P, Kwon Y, Joo JY, Kim DG, Yoon JH. Secretomics to discover regulators in diseases. Int J Mol Sci. 2019;20:3893. doi: 10.3390/ijms20163893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brunoro GVF, Carvalho PC, Barbosa VC, Pagnoncelli D, De Moura Gallo CV, Perales J, Zahedi RP, Valente RH, Neves-Ferreira AGDC. Differential proteomic comparison of breast cancer secretome using a quantitative paired analysis workflow. BMC Cancer. 2019;19:365. doi: 10.1186/s12885-019-5547-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.da Cunha BR, Domingos C, Stefanini ACB, Henrique T, Polachini GM, Castelo-Branco P, Tajara EH. Cellular interactions in the tumor microenvironment: the role of secretome. J Cancer. 2019;10:4574–4587. doi: 10.7150/jca.21780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de la Torre Gomez C, Goreham RV, Bech Serra JJ, Nann T, Kussmann M. “Exosomics”-a review of biophysics, biology and biochemistry of exosomes with a focus on human breast milk. Front Genet. 2018;9:92. doi: 10.3389/fgene.2018.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kassack M. Pharmacogenomics, the search for individualized therapies. In: Licinio J, Wong ML, editors. Weinheim, Germany: Wiley-VCH Verlag GmbH; 2002. p. 559. [Google Scholar]
- 46.Zagorac I, Fernandez-Gaitero S, Penning R, Post H, Bueno MJ, Mouron S, Manso L, Morente MM, Alonso S, Serra V, Muñoz J, Gómez-López G, Lopez-Acosta JF, Jimenez-Renard V, Gris-Oliver A, Al-Shahrour F, Piñeiro-Yañez E, Montoya-Suarez JL, Apala JV, Moreno-Torres A, Colomer R, Dopazo A, Heck AJR, Altelaar M, Quintela-Fandino M. In vivo phosphoproteomics reveals kinase activity profiles that predict treatment outcome in triple-negative breast cancer. Nat Commun. 2018;9:3501. doi: 10.1038/s41467-018-05742-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Metodiev M, Alldridge L. Phosphoproteomics: a possible route to novel biomarkers of breast cancer. Proteomics Clin Appl. 2008;2:181–194. doi: 10.1002/prca.200780011. [DOI] [PubMed] [Google Scholar]
- 48.Gökmen-Polar Y, True JD, Vieth E, Gu Y, Gu X, Qi GD, Mosley AL, Badve SS. Quantitative phosphoproteomic analysis identifies novel functional pathways of tumor suppressor DLC1 in estrogen receptor positive breast cancer. PLoS One. 2018;13:e0204658. doi: 10.1371/journal.pone.0204658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Queiroz Jd, Santos Fd, Braga C, Leite A, Buzalaf M, Zara L, Okoshi M, Padilha CD, Padilha PD. Metalloproteomic approach of mercury in breast milk samples of lactating women in the Amazon region, Brazil. (LB254) FASEB J. 2014;28:LB254. [Google Scholar]
- 50.Kessler B. Ubiquitin - omics reveals novel networks and associations with human disease. Curr Opin Chem Biol. 2013;17:59–65. doi: 10.1016/j.cbpa.2012.12.024. [DOI] [PubMed] [Google Scholar]
- 51.Ellis RJ. Chaperomics: in vivo GroEL function defined. Curr Biol. 2005;15:R661–663. doi: 10.1016/j.cub.2005.08.025. [DOI] [PubMed] [Google Scholar]
- 52.Rodina A, Wang T, Yan P, Gomes ED, Dunphy MPS, Pillarsetty N, Koren J, Gerecitano JF, Taldone T, Zong H, Caldas-Lopes E, Alpaugh M, Corben A, Riolo M, Beattie B, Pressl C, Peter RI, Xu C, Trondl R, Patel HJ, Shimizu F, Bolaender A, Yang C, Panchal P, Farooq MF, Kishinevsky S, Modi S, Lin O, Chu F, Patil S, Erdjument-Bromage H, Zanzonico P, Hudis C, Studer L, Roboz GJ, Cesarman E, Cerchietti L, Levine R, Melnick A, Larson SM, Lewis JS, Guzman ML, Chiosis G. The epichaperome is an integrated chaperome network that facilitates tumour survival. Nature. 2016;538:397–401. doi: 10.1038/nature19807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rogers LD, Overall CM. Proteolytic post-translational modification of proteins: proteomic tools and methodology. Mol Cell Proteomics. 2013;12:3532–3542. doi: 10.1074/mcp.M113.031310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Doucet A, Butler GS, Rodríguez D, Prudova A, Overall CM. Metadegradomics: toward in vivo quantitative degradomics of proteolytic post-translational modifications of the cancer proteome. Mol Cell Proteomics. 2008;7:1925–51. doi: 10.1074/mcp.R800012-MCP200. [DOI] [PubMed] [Google Scholar]
- 55.Rizwan A, Cheng M, Bhujwalla ZM, Krishnamachary B, Jiang L, Glunde K. Breast cancer cell adhesome and degradome interact to drive metastasis. NPJ Breast Cancer. 2015;1:15017. doi: 10.1038/npjbcancer.2015.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eslami-S Z, Majidzadeh-A K, Halvaei S, Babapirali F, Esmaeili R. Microbiome and breast cancer: new role for an ancient population. Front Oncol. 2020;10:120. doi: 10.3389/fonc.2020.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen J, Douglass J, Prasath V, Neace M, Atrchian S, Manjili M, Shokouhi S, Habibi M. The microbiome and breast cancer: a review. Breast Cancer Res Treat. 2019;178:493–496. doi: 10.1007/s10549-019-05407-5. [DOI] [PubMed] [Google Scholar]
- 58.Hardouin J, Lasserre JP, Sylvius L, Joubert-Caron R, Caron M. Cancer immunomics. Ann N Y Acad Sci. 2007;1107:223–30. doi: 10.1196/annals.1381.024. [DOI] [PubMed] [Google Scholar]
- 59.Dwivedi S, Shukla S, Goel A, Sharma P, Khattri S, Pant K. Nutrigenomics in breast cancer. 2014 [Google Scholar]
- 60.Kaczor-Urbanowicz KE, Martín Carreras-Presas C, Kaczor T, Tu M, Wei F, Garcia-Godoy F, Wong DT. Emerging technologies for salivaomics in cancer detection. J Cell Mol Med. 2017;21:640–647. doi: 10.1111/jcmm.13007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chakraborty S, Hosen MI, Ahmed M, Shekhar HU. Onco-Multi-OMICS approach: a new frontier in cancer research. Biomed Res Int. 2018;2018:9836256. doi: 10.1155/2018/9836256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Aslam B, Basit M, Nisar MA, Khurshid M, Rasool MH. Proteomics: technologies and their applications. J Chromatogr Sci. 2017;55:182–196. doi: 10.1093/chromsci/bmw167. [DOI] [PubMed] [Google Scholar]
- 63.Yanovich G, Agmon H, Harel M, Sonnenblick A, Peretz T, Geiger T. Clinical proteomics of breast cancer reveals a novel layer of breast cancer classification. Cancer Res. 2018;78:6001–6010. doi: 10.1158/0008-5472.CAN-18-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Phillips L, Gill AJ, Baxter RC. Novel prognostic markers in triple-negative breast cancer discovered by MALDI-mass spectrometry imaging. Front Oncol. 2019;9:379. doi: 10.3389/fonc.2019.00379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Neagu AN. Proteome imaging: from classic to modern mass spectrometry-based molecular histology. 2019. pp. 55–98. [DOI] [PubMed] [Google Scholar]
- 66.Yang J, Xiong X, Liu S, Zhu J, Luo M, Liu L, Zhao L, Qin Y, Song T, Huang C. Identification of novel serum peptides biomarkers for female breast cancer patients in Western China. Proteomics. 2016;16:925–934. doi: 10.1002/pmic.201500321. [DOI] [PubMed] [Google Scholar]
- 67.Kang HS, Lee SC, Park YS, Jeon YE, Lee JH, Jung SY, Park IH, Jang SH, Park HM, Yoo CW, Park SH, Han SY, Kim KP, Kim YH, Ro J, Kim HK. Protein and lipid MALDI profiles classify breast cancers according to the intrinsic subtype. BMC Cancer. 2011;11:465. doi: 10.1186/1471-2407-11-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ntai I, LeDuc RD, Fellers RT, Erdmann-Gilmore P, Davies SR, Rumsey J, Early BP, Thomas PM, Li S, Compton PD, Ellis MJC, Ruggles KV, Fenyö D, Boja ES, Rodriguez H, Townsend RR, Kelleher NL. Integrated bottom-up and top-down proteomics of patient-derived breast tumor xenografts. Mol Cell Proteomics. 2016;15:45–56. doi: 10.1074/mcp.M114.047480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jin H, Zangar RC. Protein modifications as potential biomarkers in breast cancer. Biomark Insights. 2009;4:191–200. doi: 10.4137/bmi.s2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weidenauer L, Wang T, Joshi S, Chiosis G, Quadroni MR. Proteomic interrogation of HSP90 and insights for medical research. Expert Rev Proteomics. 2017;14:1105–1117. doi: 10.1080/14789450.2017.1389649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Aranda-Gutierrez A, Diaz-Perez H. Histology, mammary glands. 2019 [PubMed] [Google Scholar]
- 72.Werth AJ. Vestiges of the natural history of development: historical holdovers reveal the dynamic interaction between ontogeny and phylogeny. Evolution: Education and Outreach. 2014;7:12. [Google Scholar]
- 73.Chen X, Liu Q, Song E. Mammary stem cells: angels or demons in mammary gland? Signal Transduct Target Ther. 2017;2:16038. doi: 10.1038/sigtrans.2016.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee A. Why is carcinoma of the breast more frequent in the upper outer quadrant? A case series based on needle core biopsy diagnoses. Breast. 2005;14:151–2. doi: 10.1016/j.breast.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 75.Rummel S, Hueman MT, Costantino N, Shriver CD, Ellsworth RE. Tumour location within the breast: does tumour site have prognostic ability? Ecancermedicalscience. 2015;9:552. doi: 10.3332/ecancer.2015.552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dewar R, Fadare O, Gilmore H, Gown A. Best practices in diagnostic lmmunohistochemistry myoepithelial markers in breast pathology. Arch Pathol Lab Med. 2011;135:422–429. doi: 10.5858/2010-0336-CP.1. [DOI] [PubMed] [Google Scholar]
- 77.Tharmapalan P, Mahendralingam M, Berman HK, Khokha R. Mammary stem cells and progenitors: targeting the roots of breast cancer for prevention. EMBO J. 2019;38:e100852. doi: 10.15252/embj.2018100852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jean-Marc G, Menet E, Tardivon A, Cherel P, Vanel D. Normal and pathological breast, the histological basis. Eur J Radiol. 2005;54:6–14. doi: 10.1016/j.ejrad.2004.11.020. [DOI] [PubMed] [Google Scholar]
- 79.Koker M, Kleer C. p63 expression in breast cancer: a highly sensitive and specific marker of metaplastic carcinoma. Am J Surg Pathol. 2004;28:1506–1512. doi: 10.1097/01.pas.0000138183.97366.fd. [DOI] [PubMed] [Google Scholar]
- 80.Anderson WF, Devesa SS. In situ male breast carcinoma in the surveillance, epidemiology, and end results database of the national cancer institute. Cancer. 2005;104:1733–1741. doi: 10.1002/cncr.21353. [DOI] [PubMed] [Google Scholar]
- 81.Humphries MP, Sundara Rajan S, Honarpisheh H, Cserni G, Dent J, Fulford L, Jordan LB, Jones JL, Kanthan R, Litwiniuk M, Di Benedetto A, Mottolese M, Provenzano E, Shousha S, Stephens M, Kulka J, Ellis IO, Titloye AN, Hanby AM, Shaaban AM, Speirs V. Characterisation of male breast cancer: a descriptive biomarker study from a large patient series. Sci Rep. 2017;7:45293. doi: 10.1038/srep45293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zografos E, Gazouli M, Tsangaris G, Marinos E. The significance of proteomic biomarkers in male breast cancer. Cancer Genomics Proteomics. 2016;13:183–190. [PubMed] [Google Scholar]
- 83.Gilkes DM, Semenza GL. Role of hypoxia-inducible factors in breast cancer metastasis. Future Oncol. 2013;9:1623–1636. doi: 10.2217/fon.13.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shaheed SU, Tait C, Kyriacou K, Linforth R, Salhab M, Sutton C. Evaluation of nipple aspirate fluid as a diagnostic tool for early detection of breast cancer. Clin Proteomics. 2018;15:3. doi: 10.1186/s12014-017-9179-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dimas-González J, Lagunas V, Díaz-Chávez J, López-Arellano M, Muñoz-Camacho J, Terán Porcayo M, Lagunas-Martínez A. Overexpression of p53 protein is a marker of poor prognosis in Mexican women with breast cancer. Oncol Rep. 2017;37:3026–3036. doi: 10.3892/or.2017.5553. [DOI] [PubMed] [Google Scholar]
- 86.Elkablawy MA, Albasri AM, Mohammed RA, Hussainy AS, Nouh MM, Alhujaily AS. Ki67 expression in breast cancer. Correlation with prognostic markers and clinicopathological parameters in Saudi patients. Saudi Med J. 2016;37:137–141. doi: 10.15537/smj.2016.2.12285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kutomi G, Ohmura T, Suzuki Y, Kameshima H, Shima H, Takamaru T, Satomi F, Otokozawa S, Mori M, Hirata K. Clinicopathological characteristics of basal type breast cancer in triple-negative breast cancer. Asian Pac J Cancer Prev. 2012;3:836–840. [Google Scholar]
- 88.Bouchal P, Schubert OT, Faktor J, Capkova L, Imrichova H, Zoufalova K, Paralova V, Hrstka R, Liu Y, Ebhardt HA, Budinska E, Nenutil R, Aebersold R. Breast cancer classification based on proteotypes obtained by SWATH mass spectrometry. Cell Rep. 2019;28:832–843. e837. doi: 10.1016/j.celrep.2019.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liu P, Cheng H, Roberts TM, Zhao JJ. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat Rev Drug Discov. 2009;8:627–644. doi: 10.1038/nrd2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Johansson I, Nilsson C, Berglund P, Lauss M, Ringnér M, Olsson H, Luts L, Sim E, Thorstensson S, Fjällskog ML, Hedenfalk I. Gene expression profiling of primary male breast cancers reveals two unique subgroups and identifies N-acetyltransferase-1 (NAT1) as a novel prognostic biomarker. Breast Cancer Res. 2012;14:R31. doi: 10.1186/bcr3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chu DT, Nguyen T, Nguyen Le Bao T, Tran DK, Nguyen TT, Thanh V, Quang T, Bui Le M, Pham H, Ngoc V, Kushekhar K. The effects of adipocytes on the regulation of breast cancer in the tumor microenvironment: an update. Cells. 2019;8:857. doi: 10.3390/cells8080857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Leong S, McKay MJ, Christopherson RI, Baxter RC. Biomarkers of breast cancer apoptosis induced by chemotherapy and TRAIL. J Proteome Res. 2012;11:1240–50. doi: 10.1021/pr200935y. [DOI] [PubMed] [Google Scholar]
- 93.Medeiros B, Allan AL. Molecular mechanisms of breast cancer metastasis to the lung: clinical and experimental perspectives. Int J Mol Sci. 2019;20:2272. doi: 10.3390/ijms20092272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liu ZJ, Semenza GL, Zhang HF. Hypoxia-inducible factor 1 and breast cancer metastasis. J Zhejiang Univ Sci B. 2015;16:32–43. doi: 10.1631/jzus.B1400221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rossi E, Zamarchi R. Single-cell analysis of circulating tumor cells: how far have we come in the-omics era? Front Genet. 2019;10:958. doi: 10.3389/fgene.2019.00958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kamińska M, Ciszewski T, Łopacka-Szatan K, Miotła P, Starosławska E. Breast cancer risk factors. Prz Menopauzalny. 2015;14:196–202. doi: 10.5114/pm.2015.54346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Oh H, Eliassen AH, Beck AH, Rosner B, Schnitt SJ, Collins LC, Connolly JL, Montaser-Kouhsari L, Willett WC, Tamimi RM. Breast cancer risk factors in relation to estrogen receptor, progesterone receptor, insulin-like growth factor-1 receptor, and Ki67 expression in normal breast tissue. NPJ Breast Cancer. 2017;3:39. doi: 10.1038/s41523-017-0041-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.McGuire A, Brown JA, Malone C, McLaughlin R, Kerin MJ. Effects of age on the detection and management of breast cancer. Cancers. 2015;7:908–929. doi: 10.3390/cancers7020815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Brinton LA, Key TJ, Kolonel LN, Michels KB, Sesso HD, Ursin G, Van Den Eeden SK, Wood SN, Falk RT, Parisi D, Guillemette C, Caron P, Turcotte V, Habel LA, Isaacs CJ, Riboli E, Weiderpass E, Cook MB. Prediagnostic sex steroid hormones in relation to male breast cancer risk. J. Clin. Oncol. 2015;33:2041–50. doi: 10.1200/JCO.2014.59.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Anstey EH, Shoemaker ML, Barrera CM, O’Neil ME, Verma AB, Holman DM. Breastfeeding and breast cancer risk reduction: implications for black mothers. Am J Prev Med. 2017;53:S40–S46. doi: 10.1016/j.amepre.2017.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Khushalani JS, Qin J, Ekwueme DU, White A. Awareness of breast cancer risk related to a positive family history and alcohol consumption among women aged 15-44 years in United States. Prev Med Rep. 2019;17:101029. doi: 10.1016/j.pmedr.2019.101029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Benveniste MF, Gomez D, Carter BW, Betancourt Cuellar SL, Shroff GS, Benveniste APA, Odisio EG, Marom EM. Recognizing radiation therapy-related complications in the chest. Radiographics. 2019;39:344–366. doi: 10.1148/rg.2019180061. [DOI] [PubMed] [Google Scholar]
- 103.Picon-Ruiz M, Morata-Tarifa C, Valle-Goffin JJ, Friedman ER, Slingerland JM. Obesity and adverse breast cancer risk and outcome: mechanistic insights and strategies for intervention. CA Cancer J Clin. 2017;67:378–397. doi: 10.3322/caac.21405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sinclair J, McCann M, Sheldon E, Gordon I, Brierley-Jones L, Copson E. The acceptability of addressing alcohol consumption as a modifiable risk factor for breast cancer: a mixed method study within breast screening services and symptomatic breast clinics. BMJ Open. 2019;9:e027371. doi: 10.1136/bmjopen-2018-027371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kispert S, McHowat J. Recent insights into cigarette smoking as a lifestyle risk factor for breast cancer. Breast Cancer (Dove Medical Press) 2017;9:127–132. doi: 10.2147/BCTT.S129746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fucic A, Mantovani A. Gender and age related modulation of xenoestrogen-induced tumorigenesis. Open Biotechnol J. 2015:9. [Google Scholar]
- 107.White ND. Hormonal contraception and breast cancer risk. Am J Lifestyle Med. 2018;12:224–226. doi: 10.1177/1559827618754833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.de Blok CJM, Wiepjes CM, Nota NM, van Engelen K, Adank MA, Dreijerink KMA, Barbé E, Konings IRHM, den Heijer M. Breast cancer risk in transgender people receiving hormone treatment: nationwide cohort study in the Netherlands. BMJ. 2019;365:l1652. doi: 10.1136/bmj.l1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fortner RT, Sisti J, Chai B, Collins LC, Rosner B, Hankinson SE, Tamimi RM, Eliassen AH. Parity, breastfeeding, and breast cancer risk by hormone receptor status and molecular phenotype: results from the Nurses’ Health Studies. Breast Cancer Res. 2019;21:40. doi: 10.1186/s13058-019-1119-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sun X, Nichols HB, Tse CK, Bell MB, Robinson WR, Sherman ME, Olshan AF, Troester MA. Association of parity and time since last birth with breast cancer prognosis by intrinsic subtype. Cancer Epidemiol Biomarkers Prev. 2016;25:60–7. doi: 10.1158/1055-9965.EPI-15-0864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hornberger J, Chen SC, Li Q, Kakad P, Quay SC. Proliferative epithelial disease identified in nipple aspirate fluid and risk of developing breast cancer: a systematic review. Curr Med Res Opin. 2015;31:253–262. doi: 10.1185/03007995.2014.988209. [DOI] [PubMed] [Google Scholar]
- 112.Turashvili G, Brogi E. Tumor heterogeneity in breast cancer. Front Med (Lausanne) 2017;4:227. doi: 10.3389/fmed.2017.00227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lawrence RT, Perez EM, Hernández D, Miller CP, Haas KM, Irie HY, Lee SI, Blau CA, Villén J. The proteomic landscape of triple-negative breast cancer. Cell Rep. 2015;11:630–644. doi: 10.1016/j.celrep.2015.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zakrzewski W, Dobrzyński M, Szymonowicz M, Rybak Z. Stem cells: past, present, and future. Stem Cell Res Ther. 2019;10:68. doi: 10.1186/s13287-019-1165-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhu J, Dingess KA. The functional power of the human milk proteome. Nutrients. 2019;11:1834. doi: 10.3390/nu11081834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gökmen-Polar Y, Nakshatri H, Badve S. Biomarkers for breast cancer stem cells: the challenges ahead. Biomark Med. 2011;5:661–671. doi: 10.2217/bmm.11.57. [DOI] [PubMed] [Google Scholar]
- 117.Rodriguez-Torres M, Allan AL. Aldehyde dehydrogenase as a marker and functional mediator of metastasis in solid tumors. Clin Exp Metastasis. 2016;33:97–113. doi: 10.1007/s10585-015-9755-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sin WC, Lim CL. Breast cancer stem cells-from origins to targeted therapy. Stem Cell Investig. 2017;4:96. doi: 10.21037/sci.2017.11.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhou J, Chen Q, Zou Y, Chen H, Qi L, Chen Y. Stem cells and cellular origins of breast cancer: updates in the rationale, controversies, and therapeutic implications. Front Oncol. 2019;9:820. doi: 10.3389/fonc.2019.00820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hwang-Verslues WW, Lee WH, Lee EY. Biomarkers to target heterogeneous breast cancer stem cells. J Mol Biomark Diagn. 2012;(Suppl 8):6. doi: 10.4172/2155-9929.S8-006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ghuwalewala S, Ghatak D, Das P, Dey S, Sarkar S, Alam N, Panda CK, Roychoudhury S. CD44(high)CD24(low) molecular signature determines the cancer stem cell and EMT phenotype in oral squamous cell carcinoma. Stem Cell Res. 2016;16:405–417. doi: 10.1016/j.scr.2016.02.028. [DOI] [PubMed] [Google Scholar]
- 122.Zhang M, Lee AV, Rosen JM. The cellular origin and evolution of breast cancer. Cold Spring Harb Perspect Med. 2017;7:a027128. doi: 10.1101/cshperspect.a027128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Li W, Ma H, Zhang J, Zhu L, Wang C, Yang Y. Unraveling the roles of CD44/CD24 and ALDH1 as cancer stem cell markers in tumorigenesis and metastasis. Sci Rep. 2017;7:13856. doi: 10.1038/s41598-017-14364-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Osman A, Afify SM, Hassan G, Fu X, Seno A, Seno M. Revisiting cancer stem cells as the origin of cancer-associated cells in the tumor microenvironment: a hypothetical view from the potential of iPSCs. Cancers (Basel) 2020;12:879. doi: 10.3390/cancers12040879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Jaggupilli A, Elkord E. Significance of CD44 and CD24 as cancer stem cell markers: an enduring ambiguity. Clin Dev Immunol. 2012;2012:708036. doi: 10.1155/2012/708036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Scatena R, Bottoni P, Pontoglio A, Giardina B. The proteomics of cancer stem cells. Potential clinical applications for innovative research in oncology. Proteomics Clin Appl. 2011;5:590–602. doi: 10.1002/prca.201000142. [DOI] [PubMed] [Google Scholar]
- 127.Lobba ARM, Forni MF, Carreira AC, Sogayar MC. Differential expression of CD90 and CD14 stem cell markers in malignant breast cancer cell lines. Cytometry A. 2012;81:1084–1091. doi: 10.1002/cyto.a.22220. [DOI] [PubMed] [Google Scholar]
- 128.Castro NP, Fedorova-Abrams ND, Merchant AS, Rangel MC, Nagaoka T, Karasawa H, Klauzinska M, Hewitt SM, Biswas K, Sharan SK, Salomon DS. Cripto-1 as a novel therapeutic target for triple negative breast cancer. Oncotarget. 2015;6:11910–11929. doi: 10.18632/oncotarget.4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hiraga T, Ito S, Nakamura H. EpCAM expression in breast cancer cells is associated with enhanced bone metastasis formation. Int J Cancer. 2016;138:1698–1708. doi: 10.1002/ijc.29921. [DOI] [PubMed] [Google Scholar]
- 130.Kontomanolis EN, Kalagasidou S, Pouliliou S, Anthoulaki X, Georgiou N, Papamanolis V, Fasoulakis ZN. The notch pathway in breast cancer progression. ScientificWorldJournal. 2018;2018:2415489. doi: 10.1155/2018/2415489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhou W, Wang G, Guo S. Regulation of angiogenesis via notch signaling in breast cancer and cancer stem cells. Biochim Biophys Acta. 2013;1836:304–20. doi: 10.1016/j.bbcan.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.van Schie EH, van Amerongen R. Aberrant WNT/CTNNB1 signaling as a therapeutic target in human breast cancer: weighing the evidence. Front Cell Dev Biol. 2020;8:25. doi: 10.3389/fcell.2020.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Li X, Wang X, Xie C, Zhu J, Meng Y, Chen Y, Li Y, Jiang Y, Yang X, Wang S, Chen J, Qi Z, Geng S, Wu J, Zhong C, Zhao Y. Sonic hedgehog and Wnt/β-catenin pathways mediate curcumin inhibition of breast cancer stem cells. Anticancer Drugs. 2018;29:208–215. doi: 10.1097/CAD.0000000000000584. [DOI] [PubMed] [Google Scholar]
- 134.Brilliant A, Brilliant Y, Sazonov S. WNT, hedgehog and NOTCH signaling pathways in triple negative breast cancer with high and low content of cancer stem cells. Ann Oncol. 2019;30:iii13–iii14. [Google Scholar]
- 135.Maugeri-Saccà M, De Maria R. Hippo pathway and breast cancer stem cells. Crit Rev Oncol Hematol. 2016;99:115–122. doi: 10.1016/j.critrevonc.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 136.Dandawate PR, Subramaniam D, Jensen RA, Anant S. Targeting cancer stem cells and signaling pathways by phytochemicals: novel approach for breast cancer therapy. Semin Cancer Biol. 2016;40-41:192–208. doi: 10.1016/j.semcancer.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bellomo C, Caja L, Moustakas A. Transforming growth factor β as regulator of cancer stemness and metastasis. Br J Cancer. 2016;115:761–769. doi: 10.1038/bjc.2016.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Beckmann A, Hainz N, Tschernig T, Meier C. Facets of communication: gap junction ultrastructure and function in cancer stem cells and tumor cells. Cancers (Basel) 2019;11:288. doi: 10.3390/cancers11030288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Xu C, Zhao H, Chen H, Yao Q. CXCR4 in breast cancer: oncogenic role and therapeutic targeting. Drug Des Devel Ther. 2015;9:4953–64. doi: 10.2147/DDDT.S84932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Piasecka D, Braun M, Kitowska K, Mieczkowski K, Kordek R, Sadej R, Romanska H. FGFs/FGFRs-dependent signalling in regulation of steroid hormone receptors - implications for therapy of luminal breast cancer. J Exp Clin Cancer Res. 2019;38:230. doi: 10.1186/s13046-019-1236-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Farabaugh SM, Boone DN, Lee AV. Role of IGF1R in breast cancer subtypes, stemness, and lineage differentiation. Front Endocrinol (Lausanne) 2015;6:59. doi: 10.3389/fendo.2015.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Rostoker R, Abelson S, Bitton-Worms K, Genkin I, Ben-Shmuel S, Dakwar M, Orr ZS, Caspi A, Tzukerman M, LeRoith D. Highly specific role of the insulin receptor in breast cancer progression. Endocr Relat Cancer. 2015;22:145–157. doi: 10.1530/ERC-14-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Cho Y, Kang HG, Kim SJ, Lee S, Jee S, Ahn SG, Kang MJ, Song JS, Chung JY, Yi EC, Chun KH. Post-translational modification of OCT4 in breast cancer tumorigenesis. Cell Death Differ. 2018;25:1781–1795. doi: 10.1038/s41418-018-0079-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Liu P, Tang H, Song C, Wang J, Chen B, Huang X, Pei X, Liu L. SOX2 promotes cell proliferation and metastasis in triple negative breast cancer. Front Pharmacol. 2018;9:942. doi: 10.3389/fphar.2018.00942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Emadian Saravi O, Naghshvar F, Torabizadeh Z, Sheidaei S. Immunohistochemical expression of Nanog and its relation with clinicopathologic characteristics in breast ductal carcinoma. Iran Biomed J. 2019;23:184–189. doi: 10.29252/.23.3.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Yu F, Li J, Chen H, Fu J, Ray S, Huang S, Zheng H, Ai W. Kruppel-like factor 4 (KLF4) is required for maintenance of breast cancer stem cells and for cell migration and invasion. Oncogene. 2011;30:2161–2172. doi: 10.1038/onc.2010.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Yin S, Cheryan VT, Xu L, Rishi AK, Reddy KB. Myc mediates cancer stem-like cells and EMT changes in triple negative breast cancers cells. PLoS One. 2017;12:e0183578. doi: 10.1371/journal.pone.0183578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Fujisue M, Nishimura R, Okumura Y, Tashima R, Nishiyama Y, Osako T, Toyozumi Y, Arima N. Clinical significance of CK19 negative breast cancer. Cancers (Basel) 2012;5:1–11. doi: 10.3390/cancers5010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Villadsen R, Fridriksdottir AJ, Rønnov-Jessen L, Gudjonsson T, Rank F, LaBarge MA, Bissell MJ, Petersen OW. Evidence for a stem cell hierarchy in the adult human breast. J Cell Biol. 2007;177:87–101. doi: 10.1083/jcb.200611114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Mohtar MA, Syafruddin SE, Nasir SN, Low TY. Revisiting the roles of pro-metastatic EpCAM in cancer. Biomolecules. 2020;10:255. doi: 10.3390/biom10020255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–8. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, Jacquemier J, Viens P, Kleer CG, Liu S, Schott A, Hayes D, Birnbaum D, Wicha MS, Dontu G. ALDH1 Is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Rabinovich I, Sebastião APM, Lima RS, Urban CDA, Junior ES, Anselmi KF, Elifio-Esposito S, De Noronha L, Moreno-Amaral AN. Cancer stem cell markers ALDH1 and CD44+/CD24- phenotype and their prognosis impact in invasive ductal carcinoma. Eur J Histochem. 2018;62:2943. doi: 10.4081/ejh.2018.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Luo M, Clouthier S, Deol Y, Liu S, Nagrath S, Azizi E, Wicha M. Breast cancer stem cells: current advances and clinical implications. Methods Mol Biol. 2015;1293:1–49. doi: 10.1007/978-1-4939-2519-3_1. [DOI] [PubMed] [Google Scholar]
- 155.Kwon MJ, Han J, Seo JH, Song K, Jeong HM, Choi JS, Kim YJ, Lee SH, Choi YL, Shin YK. CD24 overexpression is associated with poor prognosis in luminal A and triple-negative breast cancer. PLoS One. 2015;10:e0139112. doi: 10.1371/journal.pone.0139112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Jing X, Cui X, Liang H, Hao C, Yang Z, Li X, Yang X, Han C. CD24 is a potential biomarker for prognosis in human breast carcinoma. Cell Physiol Biochem. 2018;48:111–119. doi: 10.1159/000491667. [DOI] [PubMed] [Google Scholar]
- 157.Kong Y, Lyu N, Wu J, Tang H, Xie X, Yang L, Li X, Wei W, Xie X. Breast cancer stem cell markers CD44 and ALDH1A1 in serum: distribution and prognostic value in patients with primary breast cancer. J Cancer. 2018;9:3728–3735. doi: 10.7150/jca.28032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Abraham BK, Fritz P, McClellan M, Hauptvogel P, Athelogou M, Brauch H. Prevalence of CD44+/CD24-/low cells in breast cancer may not be associated with clinical outcome but may favor distant metastasis. Clin Cancer Res. 2005;11:1154–1159. [PubMed] [Google Scholar]
- 159.Wang D, Hu X, Liu C, Jia Y, Bai Y, Cai C, Wang J, Bai L, Yang R, Lin C, Liu YR, Li S, Qiao F, Yao L, Chen L, Ge G, Jiang H, Li D, Li L, Chen J, Shao ZM, Zeng YA. Protein C receptor is a therapeutic stem cell target in a distinct group of breast cancers. Cell Res. 2019;29:832–845. doi: 10.1038/s41422-019-0225-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Wang D, Liu C, Wang J, Jia Y, Hu X, Jiang H, Shao ZM, Zeng YA. Protein C receptor stimulates multiple signaling pathways in breast cancer cells. J Biol Chem. 2018;293:1413–1424. doi: 10.1074/jbc.M117.814046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Scioli MG, Storti G, D’Amico F, Gentile P, Fabbri G, Cervelli V, Orlandi A. The role of breast cancer stem cells as a prognostic marker and a target to improve the efficacy of breast cancer therapy. Cancers (Basel) 2019;11:1021. doi: 10.3390/cancers11071021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Brugnoli F, Grassilli S, Alqassab Y, Capitani S, Bertagnolo V. CD133 in breast cancer cells: more than a stem cell marker. J Oncol. 2019;2019:1–8. doi: 10.1155/2019/7512632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Joseph C, Arshad M, Kurozomi S, Althobiti M, Miligy IM, Al-izzi S, Toss MS, Goh FQ, Johnston SJ, Martin SG, Ellis IO, Mongan NP, Green AR, Rakha EA. Overexpression of the cancer stem cell marker CD133 confers a poor prognosis in invasive breast cancer. Breast Cancer Res Treat. 2019;174:387–399. doi: 10.1007/s10549-018-05085-9. [DOI] [PubMed] [Google Scholar]
- 164.Dravis C, Spike Benjamin T, Harrell JC, Johns C, Trejo Christy L, Southard-Smith EM, Perou Charles M, Wahl Geoffrey M. Sox10 regulates stem/progenitor and mesenchymal cell states in mammary epithelial cells. Cell Rep. 2015;12:2035–2048. doi: 10.1016/j.celrep.2015.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Brooks DL, Schwab LP, Krutilina R, Parke DN, Sethuraman A, Hoogewijs D, Schörg A, Gotwald L, Fan M, Wenger RH, Seagroves TN. ITGA6 is directly regulated by hypoxia-inducible factors and enriches for cancer stem cell activity and invasion in metastatic breast cancer models. Mol Cancer. 2016;15:26. doi: 10.1186/s12943-016-0510-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Mukhopadhyay P, Chakraborty S, Ponnusamy MP, Lakshmanan I, Jain M, Batra SK. Mucins in the pathogenesis of breast cancer: implications in diagnosis, prognosis and therapy. Biochim Biophys Acta. 2011;1815:224–240. doi: 10.1016/j.bbcan.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Capaldo CT, Farkas AE, Nusrat A. Epithelial adhesive junctions. F1000Prime Rep. 2014;6:1. doi: 10.12703/P6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Lu J, Shenoy AK. Epithelial-to-pericyte transition in cancer. Cancers. 2017;9:77. doi: 10.3390/cancers9070077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Zhou G, Yang L, Gray A, Srivastava AK, Li C, Zhang G, Cui T. The role of desmosomes in carcinogenesis. Onco Targets Ther. 2017;10:4059–4063. doi: 10.2147/OTT.S136367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Davies E, Cochrane RA, Hiscox S, Jiang W, Sweetland HM, Mansel R. The role of desmoglein 2 and E-cadherin in the invasion and motility of human breast cancer cells. Int J Oncol. 1997;11:415–419. doi: 10.3892/ijo.11.2.415. [DOI] [PubMed] [Google Scholar]
- 171.Masuda T, Ueo H, Kai Y, Noda M, Hu Q, Sato K, Fujii A, Hayashi N, Tsuruda Y, Otsu H, Kuroda Y, Eguchi H, Ohno S, Mimori K, Ueo H. N-Cadherin mRNA levels in peripheral blood could be a potential indicator of new metastases in breast cancer: a pilot study. Int J Mol Sci. 2020;21:511. doi: 10.3390/ijms21020511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Uretmen Kagiali ZC, Sanal E, Karayel Ö, Polat AN, Saatci Ö, Ersan PG, Trappe K, Renard BY, Önder TT, Tuncbag N, Şahin Ö, Ozlu N. Systems-level analysis reveals multiple modulators of epithelial-mesenchymal transition and identifies DNAJB4 and CD81 as novel metastasis inducers in breast cancer. Mol Cell Proteomics. 2019;18:1756–1771. doi: 10.1074/mcp.RA119.001446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Singhai R, Patil VW, Jaiswal SR, Patil SD, Tayade MB, Patil AV. E-Cadherin as a diagnostic biomarker in breast cancer. N Am J Med Sci. 2011;3:227–233. doi: 10.4297/najms.2011.3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Yang L, Wang XW, Zhu LP, Wang HL, Wang B, Zhao Q, Wang XY. Significance and prognosis of epithelial-cadherin expression in invasive breast carcinoma. Oncol Lett. 2018;16:1659–1665. doi: 10.3892/ol.2018.8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Fulga V, Rudico L, Balica AR, Cimpean AM, Saptefrati L, Margan MM, Raica M. Differential expression of E-Cadherin in primary breast cancer and corresponding lymph node metastases. Anticancer Res. 2015;35:759–765. [PubMed] [Google Scholar]
- 176.Wang Z, Zhang H, Hou J, Niu J, Ma Z, Zhao H, Liu C. Clinical implications of β-catenin protein expression in breast cancer. Int J Clin Exp Pathol. 2015;8:14989–14994. [PMC free article] [PubMed] [Google Scholar]
- 177.Zhang D, Fei F, Li S, Zhao Y, Yang Z, Qu J, Zhang X, Yin Y, Zhang S. The role of β-catenin in the initiation and metastasis of TA2 mice spontaneous breast cancer. J Cancer. 2017;8:2114–2123. doi: 10.7150/jca.19723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Shang S, Hua F, Hu ZW. The regulation of β-catenin activity and function in cancer: therapeutic opportunities. Oncotarget. 2017;8:33972–33989. doi: 10.18632/oncotarget.15687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Pai SG, Carneiro BA, Mota JM, Costa R, Leite CA, Barroso-Sousa R, Kaplan JB, Chae YK, Giles FJ. Wnt/beta-catenin pathway: modulating anticancer immune response. J Hematol Oncol. 2017;10:101. doi: 10.1186/s13045-017-0471-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Siroy A, Abdul-Karim FW, Miedler J, Fong N, Fu P, Gilmore H, Baar J. MUC1 is expressed at high frequency in early-stage basal-like triple-negative breast cancer. Hum Pathol. 2013;44:2159–2166. doi: 10.1016/j.humpath.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Jing X, Liang H, Hao C, Yang X, Cui X. Overexpression of MUC1 predicts poor prognosis in patients with breast cancer. Oncol Rep. 2019;41:801–810. doi: 10.3892/or.2018.6887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Horm TM, Schroeder JA. MUC1 and metastatic cancer: expression, function and therapeutic targeting. Cell Adh Mig. 2013;7:187–198. doi: 10.4161/cam.23131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Otterbach F, Bànkfalvi À, Bergner S, Decker T, Krech R, Boecker W. Cytokeratin 5/6 immunohistochemistry assists the differential diagnosis of atypical proliferations of the breast. Histopathology. 2000;37:232–240. doi: 10.1046/j.1365-2559.2000.00882.x. [DOI] [PubMed] [Google Scholar]
- 184.Shao MM, Chan SK, Yu AM, Lam CC, Tsang JY, Lui PC, Law BK, Tan PH, Tse GM. Keratin expression in breast cancers. Virchows Arch. 2012;461:313–322. doi: 10.1007/s00428-012-1289-9. [DOI] [PubMed] [Google Scholar]
- 185.Joosse SA, Hannemann J, Spötter J, Bauche A, Andreas A, Müller V, Pantel K. Changes in keratin expression during metastatic progression of breast cancer: impact on the detection of circulating tumor cells. Clin Cancer Res. 2012;18:993–1003. doi: 10.1158/1078-0432.CCR-11-2100. [DOI] [PubMed] [Google Scholar]
- 186.Khazai L, Rosa M. Use of immunohistochemical stains in epithelial lesions of the breast. Cancer Control. 2015;22:220–225. doi: 10.1177/107327481502200214. [DOI] [PubMed] [Google Scholar]
- 187.Szatmári T, Ötvös R, Hjerpe A, Dobra K. Syndecan-1 in cancer: implications for cell signaling, differentiation, and prognostication. Dis Markers. 2015;2015:796052. doi: 10.1155/2015/796052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Sayyad MR, Puchalapalli M, Vergara NG, Wangensteen SM, Moore M, Mu L, Edwards C, Anderson A, Kall S, Sullivan M, Dozmorov M, Singh J, Idowu MO, Koblinski JE. Syndecan-1 facilitates breast cancer metastasis to the brain. Breast Cancer Res Treat. 2019;178:35–49. doi: 10.1007/s10549-019-05347-0. [DOI] [PubMed] [Google Scholar]
- 189.Chute C, Yang X, Meyer K, Yang N, O’Neil K, Kasza I, Eliceiri K, Alexander C, Friedl A. Syndecan-1 induction in lung microenvironment supports the establishment of breast tumor metastases. Breast Cancer Res. 2018;20:66. doi: 10.1186/s13058-018-0995-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Ibrahim SA, Gadalla R, El-Ghonaimy EA, Samir O, Mohamed HT, Hassan H, Greve B, El-Shinawi M, Mohamed MM, Götte M. Syndecan-1 is a novel molecular marker for triple negative inflammatory breast cancer and modulates the cancer stem cell phenotype via the IL-6/STAT3, Notch and EGFR signaling pathways. Mol Cancer. 2017;16:57. doi: 10.1186/s12943-017-0621-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Dias K, Dvorkin-Gheva A, Hallett RM, Wu Y, Hassell J, Pond GR, Levine M, Whelan T, Bane AL. Claudin-low breast cancer; Clinical & pathological characteristics. PLoS One. 2017;12:e0168669. doi: 10.1371/journal.pone.0168669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Pommier RM, Sanlaville A, Tonon L, Kielbassa J, Thomas E, Ferrari A, Sertier AS, Hollande F, Martinez P, Tissier A, Morel AP, Ouzounova M, Puisieux A. Comprehensive characterization of claudin-low breast tumors reflects the impact of the cell-of-origin on cancer evolution. Nat Commun. 2020;11:3431. doi: 10.1038/s41467-020-17249-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Zhang M, Lee AV, Rosen JM. The cellular origin and evolution of breast cancer. Cold Spring Harb Perspect Med. 2017;7:a027128. doi: 10.1101/cshperspect.a027128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Escudero-Esparza A, Jiang WG, Martin TA. Claudin-5 is involved in breast cancer cell motility through the N-WASP and ROCK signalling pathways. J Exp Clin Cancer Res. 2012;31:43. doi: 10.1186/1756-9966-31-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Hebbrecht T, Van Audenhove I, Zwaenepoel O, Verhelle A, Gettemans J. VCA nanobodies target N-WASp to reduce invadopodium formation and functioning. PLoS One. 2017;12:e0185076. doi: 10.1371/journal.pone.0185076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Frugtniet B, Jiang WG, Martin TA. Role of the WASP and WAVE family proteins in breast cancer invasion and metastasis. Breast Cancer (Dove Medical Press) 2015;7:99–109. doi: 10.2147/BCTT.S59006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Martin TA, Lane J, Ozupek H, Jiang WG. Claudin-20 promotes an aggressive phenotype in human breast cancer cells. Tissue Barriers. 2013;1:e26518. doi: 10.4161/tisb.26518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Martin T, Jordan N, Davies E, Jiang W. Metastasis to bone in human cancer is associated with loss of occludin expression. Anticancer Res. 2016;36:1287–1293. [PubMed] [Google Scholar]
- 199.Beeman N, Webb PG, Baumgartner HK. Occludin is required for apoptosis when claudin-claudin interactions are disrupted. Cell Death Dis. 2012;3:e273. doi: 10.1038/cddis.2012.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Klus G, Rokaeus N, Bittner M, Chen Y, Korz D, Sukumar S, Schick A, Szallasi Z. Down-regulation of the desmosomal cadherin desmocollin 3 in human breast cancer. Int J Oncol. 2001;19:169–174. doi: 10.3892/ijo.19.1.169. [DOI] [PubMed] [Google Scholar]
- 201.Balogh GA, Mailo D, Nardi H, Corte MM, Vincent E, Barutta E, Lizarraga G, Lizarraga P, Montero H, Gentili R. Serological levels of mutated p53 protein are highly detected at early stages in breast cancer patients. Exp Ther Med. 2010;1:357–361. doi: 10.3892/etm_00000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Surowiak P, Suchocki S, Györffy B, Gansukh T, Wojnar A, Maciejczyk A, Pudełko M, Zabel M. Stromal myofibroblasts in breast cancer: relations between their occurrence, tumor grade and expression of some tumour markers. Folia Histochem Cytobiol. 2006;44:111–6. [PubMed] [Google Scholar]
- 203.Kang KS, Morita I, Cruz A, Jeon YJ, Trosko J, Chang CC. Expression of estrogen receptors in a normal human breast epithelial cell type with luminal and stem cell characteristics and its neoplastically transformed cell lines. Carcinogenesis. 1997;18:251–257. doi: 10.1093/carcin/18.2.251. [DOI] [PubMed] [Google Scholar]
- 204.Segatto I, Baldassarre G, Belletti B. STAT3 in breast cancer onset and progression: a matter of time and context. Int J Mol Sci. 2018;19:2818. doi: 10.3390/ijms19092818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Buehring GC, Letscher A, McGirr KM, Khandhar S, Che LH, Nguyen CT, Hackett AJ. Presence of epithelial cells in nipple aspirate fluid is associated with subsequent breast cancer: a 25-year prospective study. Breast Cancer Res Treat. 2006;98:63–70. doi: 10.1007/s10549-005-9132-5. [DOI] [PubMed] [Google Scholar]
- 206.Liu H. Application of immunohistochemistry in breast pathology: a review and update. Arch Pathol Lab Med. 2014;138:1629–1642. doi: 10.5858/arpa.2014-0094-RA. [DOI] [PubMed] [Google Scholar]
- 207.Moritani S, Ichihara S, Yatabe Y, Hasegawa M, Iwakoshi A, Hosoda W, Narita M, Nagai Y, Asai M, Ujihira N, Yuba Y, Jijiwa M. Immunohistochemical expression of myoepithelial markers in adenomyoepithelioma of the breast: a unique paradoxical staining pattern of high-molecular weight cytokeratins. Virchows Arch. 2015;466:191–198. doi: 10.1007/s00428-014-1687-2. [DOI] [PubMed] [Google Scholar]
- 208.Kong LL, Yang NZ, Shi LH, Zhao GH, Zhou W, Ding Q, Wang MH, Zhang YS. The optimum marker for the detection of lymphatic vessels. Mol Clin Oncol. 2017;7:515–520. doi: 10.3892/mco.2017.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Wiggett WS, Louw M, Karusseit VO. The histology of peau d’orange in breast cancer - what are the implications for surgery? S Afr J Surg. 2012;50:75–78. doi: 10.7196/sajs.1103. [DOI] [PubMed] [Google Scholar]
- 210.Liu T, Zhou L, Li D, Andl T, Zhang Y. Cancer-associated fibroblasts build and secure the tumor microenvironment. Front Cell Dev Biol. 2019;7:60. doi: 10.3389/fcell.2019.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Wu Q, Li B, Li Z, Li J, Sun S, Sun S. Cancer-associated adipocytes: key players in breast cancer progression. J Hematol Oncol. 2019;12:95. doi: 10.1186/s13045-019-0778-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Zonneville J, Safina A, Truskinovsky AM, Arteaga CL, Bakin AV. TGF-β signaling promotes tumor vasculature by enhancing the pericyte-endothelium association. BMC Cancer. 2018;18:670. doi: 10.1186/s12885-018-4587-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Sammarco G, Varricchi G, Ferraro V, Ammendola M, De Fazio M, Altomare DF, Luposella M, Maltese L, Currò G, Marone G, Ranieri G, Memeo R. Mast cells, angiogenesis and lymphangiogenesis in human gastric cancer. Int J Mol Sci. 2019;20:2106. doi: 10.3390/ijms20092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.da Cunha A, Michelin MA, Murta EF. Pattern response of dendritic cells in the tumor microenvironment and breast cancer. World J Clin Oncol. 2014;5:495–502. doi: 10.5306/wjco.v5.i3.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Aboussekhra A. Role of cancer-associated fibroblasts in breast cancer development and prognosis. Int J Dev Biol. 2011;55:841–849. doi: 10.1387/ijdb.113362aa. [DOI] [PubMed] [Google Scholar]
- 216.Kim J. Pericytes in breast cancer. Adv Exp Med Biol. 2019;1147:93–107. doi: 10.1007/978-3-030-16908-4_3. [DOI] [PubMed] [Google Scholar]
- 217.Segovia-Mendoza M, Morales-Montor J. Immune tumor microenvironment in breast cancer and the participation of estrogen and its receptors in cancer physiopathology. Front Immunol. 2019;10:348. doi: 10.3389/fimmu.2019.00348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Ponert JM, Schwarz S, Haschemi R, Müller J, Pötzsch B, Bendas G, Schlesinger M. The mechanisms how heparin affects the tumor cell induced VEGF and chemokine release from platelets to attenuate the early metastatic niche formation. PLoS One. 2018;13:e0191303. doi: 10.1371/journal.pone.0191303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Tanaka S, Ueno T, Ishiguro H, Morita S, Toi M. The lack of increases in circulating endothelial progenitor cell as a negative predictor for pathological response to neoadjuvant chemotherapy in breast cancer patients. NPJ Precis Oncol. 2017;1:6. doi: 10.1038/s41698-017-0006-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Ullah M, Akbar A, Ng NN, Concepcion W, Thakor AS. Mesenchymal stem cells confer chemoresistance in breast cancer via a CD9 dependent mechanism. Oncotarget. 2019;10:3435–3450. doi: 10.18632/oncotarget.26952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Salha S, Gehmert S, Brébant V, Anker A, Loibl M, Prantl L, Gehmert S. PDGF regulated migration of mesenchymal stem cells towards malignancy acts via the PI3K signaling pathway. Clin Hemorheol Microcirc. 2018;70:543–551. doi: 10.3233/CH-189319. [DOI] [PubMed] [Google Scholar]
- 222.Shi Y, Du L, Lin L, Wang Y. Tumour-associated mesenchymal stem/stromal cells: emerging therapeutic targets. Nat Rev Drug Discov. 2017;16:35–52. doi: 10.1038/nrd.2016.193. [DOI] [PubMed] [Google Scholar]
- 223.Javan MR, Khosrojerdi A, Moazzeni SM. New insights into implementation of mesenchymal stem cells in cancer therapy: prospects for anti-angiogenesis treatment. Front Oncol. 2019;9:840. doi: 10.3389/fonc.2019.00840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Maffey A, Storini C, Diceglie C, Martelli C, Sironi L, Calzarossa C, Tonna N, Lovchik R, Delamarche E, Ottobrini L, Bianco F. Mesenchymal stem cells from tumor microenvironment favour breast cancer stem cell proliferation, cancerogenic and metastatic potential, via ionotropic purinergic signalling. Sci Rep. 2017;7:13162. doi: 10.1038/s41598-017-13460-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Blache U, Horton ER, Xia T, Schoof EM, Blicher LH, Schönenberger A, Snedeker JG, Martin I, Erler JT, Ehrbar M. Mesenchymal stromal cell activation by breast cancer secretomes in bioengineered 3D microenvironments. Life Sci Alliance. 2019;2:e201900304. doi: 10.26508/lsa.201900304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Chen YC, Gonzalez ME, Burman B, Zhao X, Anwar T, Tran M, Medhora N, Hiziroglu AB, Lee W, Cheng YH, Choi Y, Yoon E, Kleer CG. Mesenchymal stem/stromal cell engulfment reveals metastatic advantage in breast cancer. Cell Rep. 2019;27:3916–3926. e3915. doi: 10.1016/j.celrep.2019.05.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Ghosh D, Mejia Pena C, Quach N, Xuan B, Lee AH, Dawson MR. Senescent mesenchymal stem cells remodel extracellular matrix driving breast cancer cells to a more-invasive phenotype. J Cell Sci. 2020;133:jcs232470. doi: 10.1242/jcs.232470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Aoto K, Ito K, Aoki S. Complex formation between platelet-derived growth factor receptor β and transforming growth factor β receptor regulates the differentiation of mesenchymal stem cells into cancer-associated fibroblasts. Oncotarget. 2018;9:34090–34102. doi: 10.18632/oncotarget.26124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Pelon F, Bourachot B, Kieffer Y, Magagna I, Mermet-Meillon F, Bonnet I, Costa A, Givel AM, Attieh Y, Barbazan J, Bonneau C, Fuhrmann L, Descroix S, Vignjevic D, Silberzan P, Parrini MC, Vincent-Salomon A, Mechta-Grigoriou F. Cancer-associated fibroblast heterogeneity in axillary lymph nodes drives metastases in breast cancer through complementary mechanisms. Nat Commun. 2020;11:404. doi: 10.1038/s41467-019-14134-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Quintanilla M, Montero-Montero L, Renart J, Martín-Villar E. Podoplanin in inflammation and cancer. Int J Mol Sci. 2019;20:707. doi: 10.3390/ijms20030707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Louhichi T, Saad H, Dhiab MB, Ziadi S, Trimeche M. Stromal CD10 expression in breast cancer correlates with tumor invasion and cancer stem cell phenotype. BMC Cancer. 2018;18:49. doi: 10.1186/s12885-017-3951-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Puré E, Blomberg R. Pro-tumorigenic roles of fibroblast activation protein in cancer: back to the basics. Oncogene. 2018;37:4343–4357. doi: 10.1038/s41388-018-0275-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Wawrzyniak D, Grabowska M, Głodowicz P, Kuczyński K, Kuczyńska B, Fedoruk-Wyszomirska A, Rolle K. Down-regulation of tenascin-C inhibits breast cancer cells development by cell growth, migration, and adhesion impairment. PLoS One. 2020;15:e0237889. doi: 10.1371/journal.pone.0237889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Buchsbaum RJ, Oh SY. Breast cancer-associated fibroblasts: where we are and where we need to go. Cancers. 2016;8:19. doi: 10.3390/cancers8020019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Qian XL, Pan YH, Huang QY, Shi YB, Huang QY, Hu ZZ, Xiong LX. Caveolin-1: a multifaceted driver of breast cancer progression and its application in clinical treatment. Onco Targets Ther. 2019;12:1539–1552. doi: 10.2147/OTT.S191317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Yamashita M, Ogawa T, Zhang X, Hanamura N, Kashikura Y, Takamura M, Yoneda M, Shiraishi T. Role of stromal myofibroblasts in invasive breast cancer: stromal expression of alpha-smooth muscle actin correlates with worse clinical outcome. Breast Cancer. 2012;19:170–176. doi: 10.1007/s12282-010-0234-5. [DOI] [PubMed] [Google Scholar]
- 237.Catteau X, Simon P, Noël JC. Myofibroblastic stromal reaction and lymph node status in invasive breast carcinoma: possible role of the TGF-β1/TGF-βR1 pathway. BMC Cancer. 2014;14:499. doi: 10.1186/1471-2407-14-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Wu Q, Li J, Li Z, Sun S, Zhu S, Wang L, Wu J, Yuan J, Zhang Y, Sun S, Wang C. Exosomes from the tumour-adipocyte interplay stimulate beige/brown differentiation and reprogram metabolism in stromal adipocytes to promote tumour progression. J Exp Clin Cancer Res. 2019;38:223. doi: 10.1186/s13046-019-1210-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 239.Rybinska I, Agresti R, Trapani A, Tagliabue E, Triulzi T. Adipocytes in breast cancer, the thick and the thin. Cells. 2020;9:560. doi: 10.3390/cells9030560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Lehuédé C, Li X, Dauvillier S, Vaysse C, Franchet C, Clement E, Esteve D, Longué M, Chaltiel L, Le Gonidec S, Lazar I, Geneste A, Dumontet C, Valet P, Nieto L, Fallone F, Muller C. Adipocytes promote breast cancer resistance to chemotherapy, a process amplified by obesity: role of the major vault protein (MVP) Breast Cancer Research. 2019;21:7. doi: 10.1186/s13058-018-1088-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Inman JL, Robertson C, Mott JD, Bissell MJ. Mammary gland development: cell fate specification, stem cells and the microenvironment. Development. 2015;142:1028–1042. doi: 10.1242/dev.087643. [DOI] [PubMed] [Google Scholar]
- 242.Gelsomino L, Naimo GD, Catalano S, Mauro L, Andò S. The emerging role of adiponectin in female malignancies. Int J Mol Sci. 2019;20:2127. doi: 10.3390/ijms20092127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Liu LY, Wang M, Ma ZB, Yu LX, Zhang Q, Gao DZ, Wang F, Yu ZG. The role of adiponectin in breast cancer: a meta-analysis. PLoS One. 2013;8:e73183. doi: 10.1371/journal.pone.0073183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Gu L, Cao C, Fu J, Li Q, Li DH, Chen MY. Serum adiponectin in breast cancer: a meta-analysis. Medicine. 2018;97:e11433. doi: 10.1097/MD.0000000000011433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Naimo GD, Gelsomino L, Catalano S, Mauro L, Andò S. Interfering role of ERα on adiponectin action in breast cancer. Front Endocrinol (Lausanne) 2020;11:66. doi: 10.3389/fendo.2020.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Porta C, Paglino C, Mosca A. Targeting PI3K/Akt/mTOR signaling in cancer. Front Oncol. 2014;4:64. doi: 10.3389/fonc.2014.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Park JY, Kang SE, Ahn KS, Um JY, Yang WM, Yun M, Lee SG. Inhibition of the PI3K-AKT-mTOR pathway suppresses the adipocyte-mediated proliferation and migration of breast cancer cells. J Cancer. 2020;11:2552–2559. doi: 10.7150/jca.37975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Ding M, Fu X, Tan H, Wang R, Chen Z, Ding S. The effect of vascular endothelial growth factor C expression in tumor-associated macrophages on lymphangiogenesis and lymphatic metastasis in breast cancer. Mol Med Rep. 2012;6:1023–1029. doi: 10.3892/mmr.2012.1043. [DOI] [PubMed] [Google Scholar]
- 249.Stallcup W. The NG2 proteoglycan in pericyte biology. Adv Exp Med Biol. 2018;1109:5–19. doi: 10.1007/978-3-030-02601-1_2. [DOI] [PubMed] [Google Scholar]
- 250.Barriere G, Fici P, Gallerani G, Fabbri F, Zoli W, Rigaud M. Circulating tumor cells and epithelial, mesenchymal and stemness markers: characterization of cell subpopulations. Ann Transl Med. 2014;2:109. doi: 10.3978/j.issn.2305-5839.2014.10.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Anca Maria C, Tamma R, Ruggieri S, Nico B, Toma A, Ribatti D. Mast cells in breast cancer angiogenesis. Crit Rev Oncol Hematol. 2017;115:23–26. doi: 10.1016/j.critrevonc.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 252.Komi DEA, Redegeld FA. Role of mast cells in shaping the tumor microenvironment. Clin Rev Allergy Immunol. 2020;58:313–325. doi: 10.1007/s12016-019-08753-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Carpenco E, Ceauşu Ra, Cimpean Am, Gaje PN, Saptefraţi L, Fulga V, David V, Raica M. Mast cells as an indicator and prognostic marker in molecular subtypes of breast cancer. In Vivo. 2019;33:743–748. doi: 10.21873/invivo.11534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Glajcar A, Szpor J, Pacek A, Tyrak KE, Chan F, Streb J, Hodorowicz-Zaniewska D, Okoń K. The relationship between breast cancer molecular subtypes and mast cell populations in tumor microenvironment. Virchows Arch. 2017;470:505–515. doi: 10.1007/s00428-017-2103-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Aponte López A, Fuentes-Pananá E, Cortes-Muñoz D, Muñoz CS. Mast Cell, the neglected member of the tumor microenvironment: role in breast cancer. J Immunol Res. 2018;2018:2584243. doi: 10.1155/2018/2584243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Ocana A, Nieto-Jiménez C, Pandiella A, Templeton AJ. Neutrophils in cancer: prognostic role and therapeutic strategies. Mol Cancer. 2017;16:137. doi: 10.1186/s12943-017-0707-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Soto-Perez-de-Celis E, Chavarri-Guerra Y, Leon-Rodriguez E, Gamboa-Dominguez A. Tumor-associated neutrophils in breast cancer subtypes. Asian Pac J Cancer Prev. 2017;18:2689–2693. doi: 10.22034/APJCP.2017.18.10.2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Gago-Dominguez M, Matabuena M, Redondo CM, Patel SP, Carracedo A, Ponte SM, Martínez ME, Castelao JE. Neutrophil to lymphocyte ratio and breast cancer risk: analysis by subtype and potential interactions. Sci Rep. 2020;10:13203. doi: 10.1038/s41598-020-70077-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Wu L, Saxena S, Awaji M, Singh RK. Tumor-associated neutrophils in cancer: going pro. Cancers. 2019;11:564. doi: 10.3390/cancers11040564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Markowitz J, Wesolowski R, Papenfuss T, Brooks TR, Carson WE 3rd. Myeloid-derived suppressor cells in breast cancer. Breast Cancer Res Treat. 2013;140:13–21. doi: 10.1007/s10549-013-2618-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Danilin S, Merkel AR, Johnson JR, Johnson RW, Edwards JR, Sterling JA. Myeloid-derived suppressor cells expand during breast cancer progression and promote tumor-induced bone destruction. Oncoimmunology. 2012;1:1484–1494. doi: 10.4161/onci.21990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Bunt SK, Hanson EM, Sinha P, Srivastava MK, Clements VK, Ostrand-Rosenberg S. Chapter 17 - tumor-associated myeloid-derived suppressor cells. In: Prendergast GC, Jaffee EM, editors. Cancer Immunotherapy. Burlington: Academic Press; 2007. pp. 309–331. [Google Scholar]
- 263.Ma X, Wang M, Yin T, Zhao Y, Wei X. Myeloid-derived suppressor cells promote metastasis in breast cancer after the stress of operative removal of the primary cancer. Front Oncol. 2019;9:855. doi: 10.3389/fonc.2019.00855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Gao G, Wang Z, Qu X, Zhang Z. Prognostic value of tumor-infiltrating lymphocytes in patients with triple-negative breast cancer: a systematic review and meta-analysis. BMC Cancer. 2020;20:179. doi: 10.1186/s12885-020-6668-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Kos Z, Roblin E, Kim RS, Michiels S, Gallas BD, Chen W, van de Vijver KK, Goel S, Adams S, Demaria S, Viale G, Nielsen TO, Badve SS, Symmans WF, Sotiriou C, Rimm DL, Hewitt S, Denkert C, Loibl S, Luen SJ, Bartlett JMS, Savas P, Pruneri G, Dillon DA, Cheang MCU, Tutt A, Hall JA, Kok M, Horlings HM, Madabhushi A, van der Laak J, Ciompi F, Laenkholm AV, Bellolio E, Gruosso T, Fox SB, Araya JC, Floris G, Hudeček J, Voorwerk L, Beck AH, Kerner J, Larsimont D, Declercq S, Van den Eynden G, Pusztai L, Ehinger A, Yang W, AbdulJabbar K, Yuan Y, Singh R, Hiley C, Bakir MA, Lazar AJ, Naber S, Wienert S, Castillo M, Curigliano G, Dieci MV, André F, Swanton C, Reis-Filho J, Sparano J, Balslev E, Chen IC, Stovgaard EIS, Pogue-Geile K, Blenman KRM, Penault-Llorca F, Schnitt S, Lakhani SR, Vincent-Salomon A, Rojo F, Braybrooke JP, Hanna MG, Soler-Monsó MT, Bethmann D, Castaneda CA, Willard-Gallo K, Sharma A, Lien HC, Fineberg S, Thagaard J, Comerma L, Gonzalez-Ericsson P, Brogi E, Loi S, Saltz J, Klaushen F, Cooper L, Amgad M, Moore DA, Salgado R International Immuno-Oncology Biomarker Working Group. Pitfalls in assessing stromal tumor infiltrating lymphocytes (sTILs) in breast cancer. NPJ Breast Cancer. 2020;6:17. doi: 10.1038/s41523-020-0156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Plitas G, Konopacki C, Wu K, Bos PD, Morrow M, Putintseva EV, Chudakov DM, Rudensky AY. Regulatory T cells exhibit distinct features in human breast cancer. Immunity. 2016;45:1122–1134. doi: 10.1016/j.immuni.2016.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Hellwig B. Highlight report: tumor infiltrating lymphocytes in breast cancer. EXCLI J. 2019;18:129–131. [PMC free article] [PubMed] [Google Scholar]
- 268.Lee M, Heo SH, Song I, Rajayi H, Park H, Park IA, Kim YA, Lee H, Gong G, Lee HJ. Presence of tertiary lymphoid structures determines the level of tumor-infiltrating lymphocytes in primary breast cancer and metastasis. Mod Pathol. 2018;32:70–80. doi: 10.1038/s41379-018-0113-8. [DOI] [PubMed] [Google Scholar]
- 269.Palucka K, Coussens LM, O’Shaughnessy J. Dendritic cells, inflammation, and breast cancer. Cancer J. 2013;19:511–516. doi: 10.1097/PPO.0000000000000007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Chouliaras K, Tokumaru Y, Asaoka M, Oshi M, Attwood KM, Yoshida K, Ishikawa T, Takabe K. Prevalence and clinical relevance of tumor-associated tissue eosinophilia (TATE) in breast cancer. Surgery. 2021;169:1234–1239. doi: 10.1016/j.surg.2020.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Simon S, Utikal J, Umansky V. Opposing roles of eosinophils in cancer. Cancer Immunol Immunother. 2019;68:823–833. doi: 10.1007/s00262-018-2255-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Suraj J, Kurpińska A, Zakrzewska A, Sternak M, Stojak M, Jasztal A, Walczak M, Chlopicki S. Early and late endothelial response in breast cancer metastasis in mice: simultaneous quantification of endothelial biomarkers using a mass spectrometry-based method. Dis Model Mech. 2019;12:dmm036269. doi: 10.1242/dmm.036269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Ribatti D, Nico B, Crivellato E. The role of pericytes in angiogenesis. Int J Dev Biol. 2011;55:261–268. doi: 10.1387/ijdb.103167dr. [DOI] [PubMed] [Google Scholar]
- 274.Cossutta M, Darche M, Carpentier G, Houppe C, Ponzo M, Raineri F, Vallée B, Gilles M-E, Villain D, Picard E, Casari C, Denis C, Paques M, Courty J, Cascone I. Weibel-palade bodies orchestrate pericytes during angiogenesis. Arterioscler Thromb Vasc Biol. 2019;39:1843–1858. doi: 10.1161/ATVBAHA.119.313021. [DOI] [PubMed] [Google Scholar]
- 275.Attwell D, Mishra A, Hall CN, O’Farrell FM, Dalkara T. What is a pericyte? J Cereb Blood Flow Metab. 2016;36:451–5. doi: 10.1177/0271678X15610340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Ribeiro A, Okamoto O. Combined effects of pericytes in the tumor microenvironment. Stem Cells Int. 2015;2015:868475. doi: 10.1155/2015/868475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Lu C, Sood A. Role of pericytes in angiogenesis. 2008:117–132. [Google Scholar]
- 278.Raza A, Franklin M, Dudek A. Pericytes and vessel maturation during tumor angiogenesis and metastasis. Am J Hematol. 2010;85:593–598. doi: 10.1002/ajh.21745. [DOI] [PubMed] [Google Scholar]
- 279.Mohme M, Riethdorf S, Pantel K. Circulating and disseminated tumour cells - mechanisms of immune surveillance and escape. Nat Rev Clin Oncol. 2017;14:155–167. doi: 10.1038/nrclinonc.2016.144. [DOI] [PubMed] [Google Scholar]
- 280.Akolkar D, Patil D, Crook T, Limaye S, Page R, Datta V, Patil R, Sims C, Ranade A, Fulmali P, Fulmali P, Srivastava N, Devhare P, Apurwa S, Patel S, Patil S, Adhav A, Pawar S, Ainwale A, Datar R. Circulating ensembles of tumor associated cells: a redoubtable new systemic hallmark of cancer. Int J Cancer. 2020;146:3485–3494. doi: 10.1002/ijc.32815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Kamal M, Razaq W, Leslie M, Adhikari S, Tanaka T. Circulating tumor cells in breast cancer: a potential liquid biopsy. 2017 [Google Scholar]
- 282.Jin KT, Chen XY, Lan HR, Wang SB, Ying XJ, Abdi SM, Wang W, Hu ZM, Mou XZ. Current progress in the clinical use of circulating tumor cells as prognostic biomarkers. Cancer Cytopathol. 2019;127:739–749. doi: 10.1002/cncy.22189. [DOI] [PubMed] [Google Scholar]
- 283.Cominetti MR, Altei WF, Selistre-de-Araujo HS. Metastasis inhibition in breast cancer by targeting cancer cell extravasation. Breast Cancer (Dove Med Press) 2019;11:165–178. doi: 10.2147/BCTT.S166725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Yap TA, Lorente D, Omlin A, Olmos D, de Bono JS. Circulating tumor cells: a multifunctional biomarker. Clin Cancer Res. 2014;20:2553–2568. doi: 10.1158/1078-0432.CCR-13-2664. [DOI] [PubMed] [Google Scholar]
- 285.Danila DC, Pantel K, Fleisher M, Scher HI. Circulating tumors cells as biomarkers: progress toward biomarker qualification. Cancer J. 2011;17:438–450. doi: 10.1097/PPO.0b013e31823e69ac. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Sai B, Xiang J. Disseminated tumour cells in bone marrow are the source of cancer relapse after therapy. J Cell Mol Med. 2018;22:5776–5786. doi: 10.1111/jcmm.13867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Aktas B, Tewes M, Fehm T, Hauch S, Kimmig R, Kasimir-Bauer S. Stem cell and epithelial-mesenchymal transition markers are frequently overexpressed in circulating tumor cells of metastatic breast cancer patients. Breast Cancer Res. 2009;11:R46. doi: 10.1186/bcr2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Woosley AN, Dalton AC, Hussey GS, Howley BV, Mohanty BK, Grelet S, Dincman T, Bloos S, Olsen SK, Howe PH. TGFβ promotes breast cancer stem cell self-renewal through an ILEI/LIFR signaling axis. Oncogene. 2019;38:3794–3811. doi: 10.1038/s41388-019-0703-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Bidard FC, Proudhon C, Pierga JY. Circulating tumor cells in breast cancer. Mol Oncol. 2016;10:418–430. doi: 10.1016/j.molonc.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Aceto N, Bardia A, Miyamoto DT, Donaldson MC, Wittner BS, Spencer JA, Yu M, Pely A, Engstrom A, Zhu H, Brannigan BW, Kapur R, Stott SL, Shioda T, Ramaswamy S, Ting DT, Lin CP, Toner M, Haber DA, Maheswaran S. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell. 2014;158:1110–1122. doi: 10.1016/j.cell.2014.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Sinkala E, Sollier-Christen E, Renier C, Rosàs-Canyelles E, Che J, Heirich K, Duncombe TA, Vlassakis J, Yamauchi KA, Huang H, Jeffrey SS, Herr AE. Profiling protein expression in circulating tumour cells using microfluidic western blotting. Nat Commun. 2017;8:14622. doi: 10.1038/ncomms14622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Hinz N, Jücker M. Distinct functions of AKT isoforms in breast cancer: a comprehensive review. Cell Commun Signal. 2019;17:154. doi: 10.1186/s12964-019-0450-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Riggio M, Perrone MC, Polo ML, Rodriguez MJ, May M, Abba M, Lanari C, Novaro V. AKT1 and AKT2 isoforms play distinct roles during breast cancer progression through the regulation of specific downstream proteins. Sci Rep. 2017;7:44244. doi: 10.1038/srep44244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Keyvani S, Karimi N, Orafa Z, Bouzari S, Oloomi M. Assessment of cytokeratin-19 gene expression in peripheral blood of breast cancer patients and breast cancer cell lines. Biomark Cancer. 2016;8:57–63. doi: 10.4137/BIC.S38229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Lustberg MB, Balasubramanian P, Miller B, Garcia-Villa A, Deighan C, Wu Y, Carothers S, Berger M, Ramaswamy B, Macrae ER, Wesolowski R, Layman RM, Mrozek E, Pan X, Summers TA, Shapiro CL, Chalmers JJ. Heterogeneous atypical cell populations are present in blood of metastatic breast cancer patients. Breast Cancer Res. 2014;16:R23. doi: 10.1186/bcr3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Gam LH. Breast cancer and protein biomarkers. World J Exp Med. 2012;2:86–91. doi: 10.5493/wjem.v2.i5.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Chen W, Zhang J, Huang L, Chen L, Zhou Y, Tang D, Xie Y, Wang H, Huang C. Detection of her2-positive circulating tumor cells using the liquidbiopsy system in breast cancer. Clin Breast Cancer. 2019;19:e239–e246. doi: 10.1016/j.clbc.2018.10.009. [DOI] [PubMed] [Google Scholar]
- 298.Deutsch TM, Riethdorf S, Fremd C, Feisst M, Nees J, Fischer C, Hartkopf AD, Pantel K, Trumpp A, Schütz F, Schneeweiss A, Wallwiener M. HER2-targeted therapy influences CTC status in metastatic breast cancer. Breast Cancer Res Treat. 2020;182:127–136. doi: 10.1007/s10549-020-05687-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Begicevic RR, Falasca M. ABC transporters in cancer stem cells: beyond chemoresistance. Int J Mol Sci. 2017;18:2362. doi: 10.3390/ijms18112362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Hebert JD, Myers SA, Naba A, Abbruzzese G, Lamar J, Carr SA, Hynes RO. Proteomic profiling of the ECM of xenograft breast cancer metastases in different organs reveals distinct metastatic niches. Cancer Res. 2020;80:1475–1485. doi: 10.1158/0008-5472.CAN-19-2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Guo Y, Ji X, Liu J, Fan D, Zhou Q, Chen C, Wang W, Wang G, Wang H, Yuan W, Ji Z, Sun Z. Effects of exosomes on pre-metastatic niche formation in tumors. Mol Cancer. 2019;18:39. doi: 10.1186/s12943-019-0995-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Doglioni G, Parik S, Fendt SM. Interactions in the (Pre)metastatic niche support metastasis formation. Front Oncol. 2019;9:219. doi: 10.3389/fonc.2019.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Sauter E, Reidy D. How exosomes in human breast milk may influence breast cancer risk. Translational Cancer Research. 2017;6:S1384–S1388. [Google Scholar]
- 304.Allocca G, Hughes R, Wang N, Brown HK, Ottewell PD, Brown NJ, Holen I. The bone metastasis niche in breast cancer-potential overlap with the haematopoietic stem cell niche in vivo. J Bone Oncol. 2019;17:100244. doi: 10.1016/j.jbo.2019.100244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Chung L, Shibli S, Moore K, Elder EE, Boyle FM, Marsh DJ, Baxter RC. Tissue biomarkers of breast cancer and their association with conventional pathologic features. Br J Cancer. 2013;108:351–360. doi: 10.1038/bjc.2012.552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Duffy MJ, McGowan PM, Harbeck N, Thomssen C, Schmitt M. uPA and PAI-1 as biomarkers in breast cancer: validated for clinical use in level-of-evidence-1 studies. Breast Cancer Res. 2014;16:428. doi: 10.1186/s13058-014-0428-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Ludyga N, Englert S, Pflieger K, Rauser S, Braselmann H, Walch A, Auer G, Höfler H, Aubele M. The impact of Cysteine-Rich Intestinal Protein 1 (CRIP1) in human breast cancer. Mol Cancer. 2013;12:28. doi: 10.1186/1476-4598-12-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Li S, Dai X, Gong K, Song K, Tai F, Shi J. PA28α/β promote breast cancer cell invasion and metastasis via down-regulation of CDK15. Front Oncol. 2019;9:1283. doi: 10.3389/fonc.2019.01283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Dekker TJA, Balluff BD, Jones EA, Schöne CD, Schmitt M, Aubele M, Kroep JR, Smit VTHBM, Tollenaar RAEM, Mesker WE, Walch A, McDonnell LA. Multicenter matrix-assisted laser desorption/ionization mass spectrometry imaging (maldi msi) identifies proteomic differences in breast-cancer-associated stroma. J Proteome Res. 2014;13:4730–4738. doi: 10.1021/pr500253j. [DOI] [PubMed] [Google Scholar]
- 310.Zhang X, Ren D, Guo L, Wang L, Wu S, Lin C, Ye L, Zhu J, Li J, Song L, Lin H, He Z. Thymosin beta 10 is a key regulator of tumorigenesis and metastasis and a novel serum marker in breast cancer. Breast Cancer Res. 2017;19:15. doi: 10.1186/s13058-016-0785-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Mukhopadhyay P, Lakshmanan I, Ponnusamy M, Chakraborty S, Jain M, Pai P, Smith L, Lele S, Batra S. MUC4 overexpression augments cell migration and metastasis through EGFR family proteins in triple negative breast cancer cells. PLoS One. 2013;8:e54455. doi: 10.1371/journal.pone.0054455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Liu J, Shen JX, Wu HT, Li XL, Wen XF, Du CW, Zhang GJ. Collagen 1A1 (COL1A1) promotes metastasis of breast cancer and is a potential therapeutic target. Discov Med. 2018;25:211–223. [PubMed] [Google Scholar]
- 313.Liu DT, Peng-Zhao , Han JY, Lin FZ, Bu XM, Xu QX. Clinical and prognostic significance of SOX11 in breast cancer. Asian Pac J Cancer Prev. 2014;15:5483–5486. doi: 10.7314/apjcp.2014.15.13.5483. [DOI] [PubMed] [Google Scholar]
- 314.Heatley M, Maxwell P, Whiteside C, Toner P. Cytokeratin intermediate filament expression in benign and malignant breast disease. J Clin Pathol. 1995;48:26–32. doi: 10.1136/jcp.48.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Thike AA, Iqbal J, Cheok PY, Chong AP, Tse GM, Tan B, Tan P, Wong NS, Tan PH. Triple negative breast cancer: outcome correlation with immunohistochemical detection of basal markers. Am J Surg Pathol. 2010;34:956–964. doi: 10.1097/PAS.0b013e3181e02f45. [DOI] [PubMed] [Google Scholar]
- 316.Abdelrahman AE, Rashed HE, Abdelgawad M, Abdelhamid MI. Prognostic impact of EGFR and cytokeratin 5/6 immunohistochemical expression in triple-negative breast cancer. Ann Diagn Pathol. 2017;28:43–53. doi: 10.1016/j.anndiagpath.2017.01.009. [DOI] [PubMed] [Google Scholar]
- 317.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 318.Lee YJ, Rice RH, Lee YM. Proteome analysis of human hair shaft. From protein identification to posttranslational modification. Mol Cell Proteomics. 2006;5:789–800. doi: 10.1074/mcp.M500278-MCP200. [DOI] [PubMed] [Google Scholar]
- 319.Adeola H, van Wyk J, Arowolo A, Khumalo N. Human hair as a testing substrate in the era of precision medicine: potential role of ‘omics-based approaches. 2018 [Google Scholar]
- 320.Nanashima N, Horie K, Yamada T, Shimizu T, Tsuchida S. Hair keratin KRT81 is expressed in normal and breast cancer cells and contributes to their invasiveness. Oncol Rep. 2017;37:2964–2970. doi: 10.3892/or.2017.5564. [DOI] [PubMed] [Google Scholar]
- 321.Reed DE, Shokat KM. INPP4B and PTEN loss leads to PI-3,4-P2 accumulation and inhibition of PI3K in TNBC. Mol Cancer Res. 2017;15:765–775. doi: 10.1158/1541-7786.MCR-16-0183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Bertucci MC, Mitchell CA. Phosphoinositide 3-kinase and INPP4B in human breast cancer. Ann N Y Acad Sci. 2013;1280:1–5. doi: 10.1111/nyas.12036. [DOI] [PubMed] [Google Scholar]
- 323.Vo TT, Fruman DA. INPP4B is a tumor suppressor in the context of PTEN deficiency. Cancer Discov. 2015;5:697–700. doi: 10.1158/2159-8290.CD-15-0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Sepideh I, Afshin N, Mohammad HF, Afshin N, Gholamreza A, Hamed M, Mehdi Y, Farhad JN. CDK1 in breast cancer: implications for theranostic potential. Anticancer Agents Med Chem. 2020;20:758–767. doi: 10.2174/1871520620666200203125712. [DOI] [PubMed] [Google Scholar]
- 325.Tomko LA, Hill RC, Barrett A, Szulczewski JM, Conklin MW, Eliceiri KW, Keely PJ, Hansen KC, Ponik SM. Targeted matrisome analysis identifies thrombospondin-2 and tenascin-C in aligned collagen stroma from invasive breast carcinoma. Sci Rep. 2018;8:12941. doi: 10.1038/s41598-018-31126-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Huang T, Sun L, Yuan X, Qiu H. Thrombospondin-1 is a multifaceted player in tumor progression. Oncotarget. 2017;8:84546–84558. doi: 10.18632/oncotarget.19165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Yee KO, Connolly CM, Duquette M, Kazerounian S, Washington R, Lawler J. The effect of thrombospondin-1 on breast cancer metastasis. Breast Cancer Res. 2009;114:85–96. doi: 10.1007/s10549-008-9992-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Ndishabandi D, Duquette C, Billah GE, Reyes M, Duquette M, Lawler J, Kazerounian S. Thrombospondin-1 modulates actin filament remodeling and cell motility in mouse mammary tumor cells in vitro. Discoveries (Craiova) 2014;2:e31. doi: 10.15190/d.2014.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Liu JX, Yuan Q, Min YL, He Y, Xu QH, Li B, Shi WQ, Lin Q, Li QH, Zhu PW, Shao Y. Apolipoprotein A1 and B as risk factors for development of intraocular metastasis in patients with breast cancer. Cancer Manag Res. 2019;11:2881–2888. doi: 10.2147/CMAR.S191352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Baig R, Mahjabeen I, Sabir M, Masood N, Ali K, Malik F, Kayani M. Mutational spectrum of gelsolin and its down regulation is associated with breast cancer. Dis Markers. 2013;34:71–80. doi: 10.3233/DMA-120952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Hassan MK, Kumar D, Naik M, Dixit M. The expression profile and prognostic significance of eukaryotic translation elongation factors in different cancers. PLoS One. 2018;13:e0191377. doi: 10.1371/journal.pone.0191377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Bajor M, Zych AO, Graczyk-Jarzynka A, Muchowicz A, Firczuk M, Trzeciak L, Gaj P, Domagala A, Siernicka M, Zagozdzon A, Siedlecki P, Kniotek M, O’Leary PC, Golab J, Zagozdzon R. Targeting peroxiredoxin 1 impairs growth of breast cancer cells and potently sensitises these cells to prooxidant agents. Br J Cancer. 2018;119:873–884. doi: 10.1038/s41416-018-0263-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Ketterer S, Mitschke J, Ketscher A, Schlimpert M, Reichardt W, Baeuerle N, Hess ME, Metzger P, Boerries M, Peters C, Kammerer B, Brummer T, Steinberg F, Reinheckel T. Cathepsin D deficiency in mammary epithelium transiently stalls breast cancer by interference with mTORC1 signaling. Nat Commun. 2020;11:5133. doi: 10.1038/s41467-020-18935-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Cruz-Ramos E, Sandoval-Hernández A, Tecalco-Cruz AC. Differential expression and molecular interactions of chromosome region maintenance 1 and calreticulin exportins in breast cancer cells. J Steroid Biochem Mol Biol. 2019;185:7–16. doi: 10.1016/j.jsbmb.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 335.Bodoor K, Abu-Sheikha A, Matalka I, Alzoubi H, Batiha O, Abu-Awad A, Abu Jalboush S, Fayyad L, Qadiri E, Jarun Y, Albatayneh K, Haddad Y. Immunohistochemical analysis of heat shock proteins in triple negative breast cancer: HSP60 expression is a marker of poor prognosis. Eur J Gynaecol Oncol. 2018:36. [Google Scholar]
- 336.Shao J, Zhang H, Wang Z. Coronin 1c and F-actin promote metastasis of breast cancer. Med Sci Monit. 2018;24:5980–5987. doi: 10.12659/MSM.908929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Kharman-Biz A, Gao H, Ghiasvand R, Haldosen LA, Zendehdel K. Expression of the three components of linear ubiquitin assembly complex in breast cancer. PLoS One. 2018;13:e0197183. doi: 10.1371/journal.pone.0197183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Yang H, Yu S, Wang W, Li X, Hou Y, Liu Z, Shi Y, Mu K, Niu G, Xu J, Wang H, Zhu J, Zhuang T. SHARPIN facilitates p53 degradation in breast cancer cells. Neoplasia. 2017;19:84–92. doi: 10.1016/j.neo.2016.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Sadighi S, Zokaasadi M, Kasaeian A, Maghsudi S, Jahanzad I, Kamranzadeh Fumani H. The effect of immunohistochemically detected p53 accumulation in prognosis of breast cancer; a retrospective survey of outcome. PLoS One. 2017;12:e0182444. doi: 10.1371/journal.pone.0182444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Cong Y, Cui Y, Wang S, Jiang L, Cao J, Zhu S, Birkin E, Lane J, Ruge F, Jiang WG, Qiao G. Calcium-binding protein S100P promotes tumor progression but enhances chemosensitivity in breast cancer. Front Oncol. 2020;10:566302. doi: 10.3389/fonc.2020.566302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Al-Wajeeh AS, Salhimi SM, Al-Mansoub MA, Khalid IA, Harvey TM, Latiff A, Ismail MN. Comparative proteomic analysis of different stages of breast cancer tissues using ultra high performance liquid chromatography tandem mass spectrometer. PLoS One. 2020;15:e0227404. doi: 10.1371/journal.pone.0227404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Bao YI, Wang A, Mo J. S100A8/A9 is associated with estrogen receptor loss in breast cancer. Oncol Lett. 2016;11:1936–1942. doi: 10.3892/ol.2016.4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Zhong JM, Li J, Kang AD, Huang SQ, Liu WB, Zhang Y, Liu ZH, Zeng L. Protein S100-A8: a potential metastasis-associated protein for breast cancer determined via iTRAQ quantitative proteomic and clinicopathological analysis. Oncol Lett. 2018;15:5285–5293. doi: 10.3892/ol.2018.7958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Wang D, Liu G, Wu B, Chen L, Zeng L, Pan Y. Clinical significance of elevated S100A8 expression in breast cancer patients. Front Oncol. 2018;8:496. doi: 10.3389/fonc.2018.00496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Zeng L, Zhong J, He G, Li F, Li J, Zhou W, Liu W, Zhang Y, Huang S, Liu Z, Deng X. Identification of nucleobindin-2 as a potential biomarker for breast cancer metastasis using iTRAQ-based quantitative proteomic analysis. J Cancer. 2017;8:3062–3069. doi: 10.7150/jca.19619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Zeng L, Deng X, Zhong J, Yuan L, Tao X, Zhang S, Zeng Y, He G, Tan P, Tao Y. Prognostic value of biomarkers EpCAM and αB-crystallin associated with lymphatic metastasis in breast cancer by iTRAQ analysis. BMC Cancer. 2019;19:831. doi: 10.1186/s12885-019-6016-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Caporossi D, Parisi A, Fantini C, Grazioli E, Cerulli C, Dimauro I. AlphaB-crystallin and breast cancer: role and possible therapeutic strategies. Cell Stress Chaperones. 2021;26:19–28. doi: 10.1007/s12192-020-01175-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Qiu Y, Pu T, Guo P, Wei B, Zhang Z, Zhang H, Zhong X, Zheng H, Chen L, Bu H, Ye F. ALDH+/CD44+ cells in breast cancer are associated with worse prognosis and poor clinical outcome. Exp Mol Pathol. 2016;100:145–150. doi: 10.1016/j.yexmp.2015.11.032. [DOI] [PubMed] [Google Scholar]
- 349.Sun S, Liang X, Zhang X, Liu T, Shi Q, Song Y, Jiang Y, Wu H, Lu X, Pang D. Phosphoglycerate kinase-1 is a predictor of poor survival and a novel prognostic biomarker of chemoresistance to paclitaxel treatment in breast cancer. Br J Cancer. 2015;112:1332–1339. doi: 10.1038/bjc.2015.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Boutas I, Potiris A, Brenner W, Lebrecht A, Hasenburg A, Kalantaridou S, Schmidt M. The expression of galectin-3 in breast cancer and its association with chemoresistance: a systematic review of the literature. Arch Gynecol Obstet. 2019;300:1113–1120. doi: 10.1007/s00404-019-05292-9. [DOI] [PubMed] [Google Scholar]
- 351.Grosset AA, Labrie M, Vladoiu MC, Yousef EM, Gaboury L, St-Pierre Y. Galectin signatures contribute to the heterogeneity of breast cancer and provide new prognostic information and therapeutic targets. Oncotarget. 2016;7:18183–18203. doi: 10.18632/oncotarget.7784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Hussein YR, Bandyopadhyay S, Semaan A, Ahmed Q, Albashiti B, Jazaerly T, Nahleh Z, Ali-Fehmi R. Glut-1 expression correlates with basal-like breast cancer. Transl Oncol. 2011;4:321–327. doi: 10.1593/tlo.11256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Barbosa AM, Martel F. Targeting glucose transporters for breast cancer therapy: the effect of natural and synthetic compounds. Cancers (Basel) 2020;12:154. doi: 10.3390/cancers12010154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Munir J, Van Ngu T, Na Ayudthaya PD, Ryu S. Downregulation of glypican-4 facilitates breast cancer progression by inducing cell migration and proliferation. Biochem Biophys Res Commun. 2020;526:91–97. doi: 10.1016/j.bbrc.2020.03.064. [DOI] [PubMed] [Google Scholar]
- 355.Tsang JYS, Lee MA, Chan TH, Li J, Ni YB, Shao Y, Chan SK, Cheungc SY, Lau KF, Tse GMK. Proteolytic cleavage of amyloid precursor protein by ADAM10 mediates proliferation and migration in breast cancer. EBioMedicine. 2018;38:89–99. doi: 10.1016/j.ebiom.2018.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Zhao M, Jia W, Jiang WG, Wang P, Du G, Cheng S, Song M. ADAM29 expression in human breast cancer and its effects on breast cancer cells in vitro. Anticancer Res. 2016;36:1251–1258. [PubMed] [Google Scholar]
- 357.Wang S, Wang N, Zheng Y, Zhang J, Zhang F, Wang Z. Caveolin-1: an oxidative stress-related target for cancer prevention. Oxid Med Cell Longev. 2017;2017:7454031. doi: 10.1155/2017/7454031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Chen D, Che G. Value of caveolin-1 in cancer progression and prognosis: emphasis on cancer-associated fibroblasts, human cancer cells and mechanism of caveolin-1 expression (review) Oncol Lett. 2014;8:1409–1421. doi: 10.3892/ol.2014.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Peng L, Cantor DI, Huang C, Wang K, Baker MS, Nice EC. Tissue and plasma proteomics for early stage cancer detection. Mol Omics. 2018;14:405–423. doi: 10.1039/c8mo00126j. [DOI] [PubMed] [Google Scholar]
- 360.Zhou L, Beuerman RW. The power of tears: how tear proteomics research could revolutionize the clinic. Expert Rev Proteomics. 2017;14:189–191. doi: 10.1080/14789450.2017.1285703. [DOI] [PubMed] [Google Scholar]
- 361.Huang Z, Ma L, Huang C, Li Q, Nice EC. Proteomic profiling of human plasma for cancer biomarker discovery. Proteomics. 2017;17 doi: 10.1002/pmic.201600240. [DOI] [PubMed] [Google Scholar]
- 362.Shen Y, Tolić N, Liu T, Zhao R, Petritis BO, Gritsenko MA, Camp DG, Moore RJ, Purvine SO, Esteva FJ, Smith RD. Blood peptidome-degradome profile of breast cancer. PLoS One. 2010;5:e13133. doi: 10.1371/journal.pone.0013133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Corrêa S, Panis C, Binato R, Herrera AC, Pizzatti L, Abdelhay E. Identifying potential markers in breast cancer subtypes using plasma label-free proteomics. J Proteomics. 2017;151:33–42. doi: 10.1016/j.jprot.2016.07.030. [DOI] [PubMed] [Google Scholar]
- 364.Pitteri SJ, Faca VM, Kelly-Spratt KS, Kasarda AE, Wang H, Zhang Q, Newcomb L, Krasnoselsky A, Paczesny S, Choi G, Fitzgibbon M, McIntosh MW, Kemp CJ, Hanash SM. Plasma proteome profiling of a mouse model of breast cancer identifies a set of up-regulated proteins in common with human breast cancer cells. J Proteome Res. 2008;7:1481–1489. doi: 10.1021/pr7007994. [DOI] [PubMed] [Google Scholar]
- 365.Gajbhiye A, Dabhi R, Taunk K, Jagadeeshaprasad MG, RoyChoudhury S, Mane A, Bayatigeri S, Chaudhury K, Santra MK, Rapole S. Multipronged quantitative proteomics reveals serum proteome alterations in breast cancer intrinsic subtypes. J Proteomics. 2017;163:1–13. doi: 10.1016/j.jprot.2017.05.007. [DOI] [PubMed] [Google Scholar]
- 366.Zhang Y, Jiao J, Yang P, Lu H. Mass spectrometry-based N-glycoproteomics for cancer biomarker discovery. Clin Proteomics. 2014;11:18. doi: 10.1186/1559-0275-11-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Cazet A, Julien S, Bobowski M, Burchell J, Delannoy P. Tumour-associated carbohydrate antigens in breast cancer. Breast Cancer Res. 2010;12:204. doi: 10.1186/bcr2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Nath S, Mukherjee P. MUC1: a multifaceted oncoprotein with a key role in cancer progression. Trends Mol Med. 2014;20:332–342. doi: 10.1016/j.molmed.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Croce M, Larrain M, Demichelis S, Gori J, Price M, Segal-Eiras A. Tissue and serum MUC1 mucin detection in breast cancer patients. Breast Cancer Res Treat. 2003;81:195–207. doi: 10.1023/A:1026110417294. [DOI] [PubMed] [Google Scholar]
- 370.Fang C, Cao Y, Liu X, Zeng XT, Li Y. Serum CA125 is a predictive marker for breast cancer outcomes and correlates with molecular subtypes. Oncotarget. 2017;8:63963–63970. doi: 10.18632/oncotarget.19246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Gaughran G, Aggarwal N, Shadbolt B, Stuart-Harris R. The utility of the tumor markers CA15.3, CEA, CA-125 and CA19.9 in metastatic breast cancer. Breast Cancer Management. 2020;9:BMT50. [Google Scholar]
- 372.Kucera R, Topolcan O, Fiala O, Kinkorova J, Treska V, Zedníková I, Slouka D, Simanek V, Safanda M, Babuska V. The role of TPS and TPA in the diagnostics of distant metastases. Anticancer Res. 2016;36:773–777. [PubMed] [Google Scholar]
- 373.Sjöström J, Alfthan H, Joensuu H, Stenman UH, Lundin J, Blomqvist C. Serum tumour markers CA 15-3, TPA, TPS, hCG β and TATI in the monitoring of chemotherapy response in metastatic breast cancer. Scand J Clin Lab Invest. 2001;61:431–441. doi: 10.1080/00365510152567068. [DOI] [PubMed] [Google Scholar]
- 374.Nakata B, Ogawa Y, Ishikawa T, Ikeda K, Kato Y, Nishino H, Hirakawa K. Serum CYFRA 21-1 is one of the most reliable tumor markers for breast carcinoma. Cancer. 2000;89:1285–1290. doi: 10.1002/1097-0142(20000915)89:6<1285::aid-cncr13>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 375.Kazarian A, Blyuss O, Metodieva G, Gentry-Maharaj A, Ryan A, Kiseleva EM, Prytomanova OM, Jacobs IJ, Widschwendter M, Menon U, Timms JF. Testing breast cancer serum biomarkers for early detection and prognosis in pre-diagnosis samples. Br J Cancer. 2017;116:501–508. doi: 10.1038/bjc.2016.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Suh EJ, Kabir MH, Kang UB, Lee JW, Yu J, Noh DY, Lee C. Comparative profiling of plasma proteome from breast cancer patients reveals thrombospondin-1 and BRWD3 as serological biomarkers. Exp Mol Med. 2012;44:36–44. doi: 10.3858/emm.2012.44.1.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Ren L, Yi J, Li W, Zheng X, Liu J, Wang J, Du G. Apolipoproteins and cancer. Cancer Med. 2019;8:7032–7043. doi: 10.1002/cam4.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Lobo MD, Moreno FB, Souza GH, Verde SM, Moreira RA, Monteiro-Moreira AC. Label-free proteome analysis of plasma from patients with breast cancer: stage-specific protein expression. Front Oncol. 2017;7:14. doi: 10.3389/fonc.2017.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Singh R, Meena R, Tiwary S, Khanna S, Khanna R. Fas ligand as a circulating apoptosis marker in carcinoma breast. ResearchGate. 2018;7:7–14. [Google Scholar]
- 380.Volpe E, Sambucci M, Battistini L, Borsellino G. Fas-fas ligand: checkpoint of t cell functions in multiple sclerosis. Front Immunol. 2016;7:382–382. doi: 10.3389/fimmu.2016.00382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Ueno T, Toi M, Tominaga T. Circulating soluble fas concentration in breast cancer patients. Clin Cancer Res. 1999;5:3529–3533. [PubMed] [Google Scholar]
- 382.Matsumoto H, Murakami Y, Kataoka K, Notomi S, Mantopoulos D, Trichonas G, Miller J, Gregory M, Ksander B, Rothstein A, Vavvas D. Membrane-bound and soluble fas ligands have opposite functions in photoreceptor cell death following separation from the retinal pigment epithelium. Death Dis. 2015;6:e1986. doi: 10.1038/cddis.2015.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Fersching Dmi, Nagel D, Siegele B, Salat C, Heinemann V, Holdenrieder S, Stoetzer OJ. Apoptosis-related biomarkers sFAS, MIF, ICAM-1 and PAI-1 in serum of breast cancer patients undergoing neoadjuvant chemotherapy. Anticancer Res. 2012;32:2047–2058. [PubMed] [Google Scholar]
- 384.Kadam CY, Abhang SA. Chapter five - apoptosis markers in breast cancer therapy. In: Makowski GS, editor. Advances in Clinical Chemistry. Elsevier; 2016. pp. 143–193. [DOI] [PubMed] [Google Scholar]
- 385.Khan S, Ferguson Bennit H, Asuncion Valenzuela MM, Turay D, Diaz Osterman CJ, Moyron RB, Esebanmen GE, Ashok A, Wall NR. Localization and upregulation of survivin in cancer health disparities: a clinical perspective. Biologics. 2015;9:57–67. doi: 10.2147/BTT.S83864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Jaiswal PK, Goel A, Mittal RD. Survivin: a molecular biomarker in cancer. Indian J Med Res. 2015;141:389–397. doi: 10.4103/0971-5916.159250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Veiga GLD, Silva RDMD, Pereira EC, Azzalis LA, Alves BDCA, Gehrke FS, Gascón TM, Fonseca FLA. The role of Survivin as a biomarker and potential prognostic factor for breast cancer. Rev Assoc Med Bras (1992) 2019;65:893–901. doi: 10.1590/1806-9282.65.6.893. [DOI] [PubMed] [Google Scholar]
- 388.Rontogianni S, Synadaki E, Li B, Liefaard MC, Lips EH, Wesseling J, Wu W, Altelaar M. Proteomic profiling of extracellular vesicles allows for human breast cancer subtyping. Commun Biol. 2019;2:325. doi: 10.1038/s42003-019-0570-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Chen IH, Xue L, Hsu CC, Paez JSP, Pan L, Andaluz H, Wendt MK, Iliuk AB, Zhu JK, Tao WA. Phosphoproteins in extracellular vesicles as candidate markers for breast cancer. Proc Natl Acad Sci U S A. 2017;114:3175–3180. doi: 10.1073/pnas.1618088114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Moon PG, Lee JE, Cho YE, Lee SJ, Chae YS, Jung JH, Kim IS, Park HY, Baek MC. Fibronectin on circulating extracellular vesicles as a liquid biopsy to detect breast cancer. Oncotarget. 2016;7:40189–40199. doi: 10.18632/oncotarget.9561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Moon PG, Lee JE, Cho YE, Lee SJ, Jung JH, Chae YS, Bae HI, Kim YB, Kim IS, Park HY, Baek MC. Identification of developmental endothelial locus-1 on circulating extracellular vesicles as a novel biomarker for early breast cancer detection. Clin Cancer Res. 2016;22:1757–1766. doi: 10.1158/1078-0432.CCR-15-0654. [DOI] [PubMed] [Google Scholar]
- 392.Fujita Y, Islam R, Sakai K, Kaneda H, Kudo K, Tamura D, Aomatsu K, Nagai T, Kimura H, Matsumoto K, de Velasco MA, Arao T, Okawara T, Nishio K. Aza-derivatives of resveratrol are potent macrophage migration inhibitory factor inhibitors. Invest New Drugs. 2012;30:1878–1886. doi: 10.1007/s10637-011-9749-7. [DOI] [PubMed] [Google Scholar]
- 393.Richard V, Kindt N, Saussez S. Macrophage migration inhibitory factor involvement in breast cancer (review) Int J Oncol. 2015;47:1627–1633. doi: 10.3892/ijo.2015.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Sherma ND, Borges CR, Trenchevska O, Jarvis JW, Rehder DS, Oran PE, Nelson RW, Nedelkov D. Mass spectrometric immunoassay for the qualitative and quantitative analysis of the cytokine macrophage migration inhibitory factor (MIF) Proteome Sci. 2014;12:52. doi: 10.1186/s12953-014-0052-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Zhou W, Yang L, Nie L, Lin H. Unraveling the molecular mechanisms between inflammation and tumor angiogenesis. Am J Cancer Res. 2021;11:301–317. [PMC free article] [PubMed] [Google Scholar]
- 396.Thielemann A, Baszczuk A, Kopczyński Z, Nowak A, Grodecka-Gazdecka S. The clinical usefulness of assessing the concentration of cell adhesion molecules sVCAM-1 and sICAM-1 in the serum of women with primary breast cancer. Contemp Oncol (Pozn) 2014;18:252–259. doi: 10.5114/wo.2014.43492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Harjes U. E-selectin fills two needs for metastasis. Nature Reviews Cancer. 2019;19:301. doi: 10.1038/s41568-019-0151-7. [DOI] [PubMed] [Google Scholar]
- 398.Otto VI, Damoc E, Cueni LN, Schürpf T, Frei R, Ali S, Callewaert N, Moise A, Leary JA, Folkers G, Przybylski M. N-Glycan structures and N-glycosylation sites of mouse soluble intercellular adhesion molecule-1 revealed by MALDI-TOF and FTICR mass spectrometry. Glycobiology. 2006;16:1033–1044. doi: 10.1093/glycob/cwl032. [DOI] [PubMed] [Google Scholar]
- 399.Abraham V, Cao G, Parambath A, Lawal F, Handumrongkul C, Debs R, DeLisser HM. Involvement of TIMP-1 in PECAM-1-mediated tumor dissemination. Int J Oncol. 2018;53:488–502. doi: 10.3892/ijo.2018.4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Giaccherini C, Marchetti M, Masci G, Verzeroli C, Russo L, Celio L, Sarmiento R, Gamba S, Tartari CJ, Diani E, Vignoli A, Malighetti P, Spinelli D, Tondini C, Barni S, Giuliani F, Petrelli F, D’Alessio A, Gasparini G, De Braud F, Santoro A, Labianca R, Falanga A HYPERCAN Investigators. Thrombotic biomarkers for risk prediction of malignant disease recurrence in patients with early stage breast cancer. Haematologica. 2020;105:1704–1711. doi: 10.3324/haematol.2019.228981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Mei Y, Liu H, Sun X, Li X, Zhao S, Ma R. Plasma fibrinogen level may be a possible marker for the clinical response and prognosis of patients with breast cancer receiving neoadjuvant chemotherapy. Tumor Biol. 2017;39:1010428317700002. doi: 10.1177/1010428317700002. [DOI] [PubMed] [Google Scholar]
- 402.Beretov J, Wasinger VC, Millar EKA, Schwartz P, Graham PH, Li Y. Proteomic analysis of urine to identify breast cancer biomarker candidates using a label-free LC-MS/MS approach. PLoS One. 2015;10:e0141876. doi: 10.1371/journal.pone.0141876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Bauça JM, Martínez-Morillo E, Diamandis E. Peptidomics of urine and other biofluids for cancer diagnostics. Clin Chem. 2013;60:1052–61. doi: 10.1373/clinchem.2013.211714. [DOI] [PubMed] [Google Scholar]
- 404.Gajbhiye A, Dabhi R, Taunk K, Vannuruswamy G, RoyChoudhury S, Adhav R, Seal S, Mane A, Bayatigeri S, Santra MK, Chaudhury K, Rapole S. Urinary proteome alterations in HER2 enriched breast cancer revealed by multipronged quantitative proteomics. Proteomics. 2016;16:2403–2418. doi: 10.1002/pmic.201600015. [DOI] [PubMed] [Google Scholar]
- 405.Jiao C, Cui L, Ma A, Li N, Si H. Elevated Serum levels of retinol-binding protein 4 are associated with breast cancer risk: a case-control study. PLoS One. 2016;11:e0167498. doi: 10.1371/journal.pone.0167498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Yom CK, Woo HY, Min SY, KanG SY, Kim HS. Clusterin overexpression and relapse-free survival in breast cancer. Anticancer Res. 2009;29:3909–3912. [PubMed] [Google Scholar]
- 407.Bai Y, Guo J, Liu Z, Li Y, Jin S, Wang T. The role of exosomes in the female reproductive system and breast cancers. Onco Targets Ther. 2020;13:12567–12586. doi: 10.2147/OTT.S281909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Piktel E, Levental I, Durnaś B, Janmey PA, Bucki R. Plasma gelsolin: indicator of inflammation and its potential as a diagnostic tool and therapeutic target. Int J Mol Sci. 2018;19:2516. doi: 10.3390/ijms19092516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Pellicani R, Poletto E, Andreuzzi E, Paulitti A, Doliana R, Bizzotto D, Braghetta P, Colladel R, Tarticchio G, Sabatelli P, Bucciotti F, Bressan G, Iozzo RV, Colombatti A, Bonaldo P, Mongiat M. Multimerin-2 maintains vascular stability and permeability. Matrix Biol. 2020;87:11–25. doi: 10.1016/j.matbio.2019.08.002. [DOI] [PubMed] [Google Scholar]
- 410.Chakraborty S, Lakshmanan M, Swa HL, Chen J, Zhang X, Ong YS, Loo LS, Akıncılar SC, Gunaratne J, Tergaonkar V, Hui KM, Hong W. An oncogenic role of agrin in regulating focal adhesion integrity in hepatocellular carcinoma. Nat Commun. 2015;6:6184. doi: 10.1038/ncomms7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Kim H, Hwang JS, Lee B, Hong J, Lee S. Newly identified cancer-associated role of human neuronal growth regulator 1 (NEGR1) J Cancer. 2014;5:598–608. doi: 10.7150/jca.8052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Aro K, Wei F, Wong DT, Tu M. Saliva liquid biopsy for point-of-care applications. Front Public Health. 2017;5:77. doi: 10.3389/fpubh.2017.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Shah S. Salivaomics: the current scenario. J Oral Maxillofac Pathol. 2018;22:375–381. doi: 10.4103/jomfp.JOMFP_171_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Streckfus CF, Bigler L. A catalogue of altered salivary proteins secondary to invasive ductal carcinoma: a novel in vivo paradigm to assess breast cancer progression. Sci Rep. 2016;6:30800. doi: 10.1038/srep30800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Brooks MN, Wang J, Li Y, Zhang R, Elashoff D, Wong DT. Salivary protein factors are elevated in breast cancer patients. Mol Med Rep. 2008;1:375–378. [PMC free article] [PubMed] [Google Scholar]
- 416.Abrao Nemeir I, Saab J, Hleihel W, Errachid A, Jafferzic-Renault N, Zine N. The advent of salivary breast cancer biomarker detection using affinity sensors. Sensors (Basel) 2019;19:2373. doi: 10.3390/s19102373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417.Agha-Hosseini F, Mirzaii-Dizgah I, Rahimi A, Seilanian-Toosi M. Correlation of serum and salivary CA125 levels in patients with breast cancer. J Contemp Dent Pract. 2009;10:E001–008. [PubMed] [Google Scholar]
- 418.Laidi F, Bouziane A, Lakhdar A, Khabouze S, Rhrab B, Zaoui F. Salivary expression of soluble HER2 in breast cancer patients with positive and negative HER2 status. Onco Targets Ther. 2014;7:1285–1289. doi: 10.2147/OTT.S64230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Liang YH, Chang CC, Chen CC, Chu-Su Y, Lin CW. Development of an Au/ZnO thin film surface plasmon resonance-based biosensor immunoassay for the detection of carbohydrate antigen 15-3 in human saliva. Clin Biochem. 2012;45:1689–93. doi: 10.1016/j.clinbiochem.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 420.Laidi F, Bouziane A, Errachid A, Zaoui F. Usefulness of salivary and serum auto-antibodies against tumor biomarkers HER2 and MUC1 in breast cancer screening. Asian Pac J Cancer Prev. 2016;17:335–339. doi: 10.7314/apjcp.2016.17.1.335. [DOI] [PubMed] [Google Scholar]
- 421.Zhang L, Xiao H, Karlan S, Zhou H, Gross J, Elashoff D, Akin D, Yan X, Chia D, Karlan B, Wong DT. Discovery and preclinical validation of salivary transcriptomic and proteomic biomarkers for the non-invasive detection of breast cancer. PLoS One. 2010;5:e15573. doi: 10.1371/journal.pone.0015573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Krop I, März A, Carlsson H, Li X, Bloushtain-Qimron N, Hu M, Gelman R, Sabel M, Schnitt S, Ramaswamy S, Kleer C, Enerbäck C, Polyak K. A putative role for psoriasin in breast tumor progression. Cancer Res. 2006;65:11326–11334. doi: 10.1158/0008-5472.CAN-05-1523. [DOI] [PubMed] [Google Scholar]
- 423.West NR, Watson PH. S100A7 (psoriasin) is induced by the proinflammatory cytokines oncostatin-M and interleukin-6 in human breast cancer. Oncogene. 2010;29:2083–2092. doi: 10.1038/onc.2009.488. [DOI] [PubMed] [Google Scholar]
- 424.Lebrecht A, Boehm D, Schmidt M, Koelbl H, Schwirz RL, Grus FH. Diagnosis of breast cancer by tear proteomic pattern. Cancer Genomics Proteomics. 2009;6:177–182. [PubMed] [Google Scholar]
- 425.Böhm D, Keller K, Pieter J, Boehm N, Wolters D, Siggelkow W, Lebrecht A, Schmidt M, Kölbl H, Pfeiffer N, Grus FH. Comparison of tear protein levels in breast cancer patients and healthy controls using a de novo proteomic approach. Oncol Rep. 2012;28:429–438. doi: 10.3892/or.2012.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.Keller K, Pieter J, Böhm D, Boehm N, Wolters D, Kölbl H, Pfeiffer N, Grus F. Proteomic analysis of tear fluid of breast cancer patients and healthy subjects shows differences in protein expression levels. 2011 [Google Scholar]
- 427.Dermawan JKT, Policarpio-Nicolas ML. Malignancies in pleural, peritoneal, and pericardial effusions: a 17-year single-institution review from 30 085 specimens. Arch Pathol Lab Med. 2020;144:1086–1091. doi: 10.5858/arpa.2019-0429-OA. [DOI] [PubMed] [Google Scholar]
- 428.Skok K, Hladnik G, Grm A, Crnjac A. Malignant pleural effusion and its current management: a review. Medicina (Kaunas) 2019;55:490. doi: 10.3390/medicina55080490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429.Noh S, Jung JJ, Jung M, Kim KH, Lee HY, Wang B, Cho J, Kim TS, Jeung HC, Rha SY. Body fluid MMP-2 as a putative biomarker in metastatic breast cancer. Oncol Lett. 2012;3:699–703. doi: 10.3892/ol.2012.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.Park CK, Malinowski DP, Cho NH. Diagnostic algorithm for determining primary tumor sites using peritoneal fluid. PLoS One. 2018;13:e0199715. doi: 10.1371/journal.pone.0199715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Li J, Zhao J, Yu X, Lange J, Kuerer H, Krishnamurthy S, Schilling E, Khan SA, Sukumar S, Chan DW. Identification of biomarkers for breast cancer in nipple aspiration and ductal lavage fluid. Clin Cancer Res. 2005;11:8312–8320. doi: 10.1158/1078-0432.CCR-05-1538. [DOI] [PubMed] [Google Scholar]
- 432.Alexander H, Stegner AL, Wagner-Mann C, Du Bois GC, Alexander S, Sauter ER. Proteomic analysis to identify breast cancer biomarkers in nipple aspirate fluid. Clin Cancer Res. 2004;10:7500–7510. doi: 10.1158/1078-0432.CCR-04-1002. [DOI] [PubMed] [Google Scholar]
- 433.Shidfar A, Fatokun T, Ivancic D, Chatterton RT, Khan SA, Wang J. Protein biomarkers for breast cancer risk are specifically correlated with local steroid hormones in nipple aspirate fluid. Horm Cancer. 2016;7:252–259. doi: 10.1007/s12672-016-0264-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Sauter ER, Davis W, Qin W, Scanlon S, Mooney B, Bromert K, Folk WR. Identification of a β-casein-like peptide in breast nipple aspirate fluid that is associated with breast cancer. Biomark Med. 2009;3:577–588. doi: 10.2217/bmm.09.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435.Zhou J, Trock B, Tsangaris TN, Friedman NB, Shapiro D, Brotzman M, Chan-Li Y, Chan DW, Li J. A unique proteolytic fragment of alpha1-antitrypsin is elevated in ductal fluid of breast cancer patient. Breast Cancer Res Treat. 2010;123:73–86. doi: 10.1007/s10549-009-0625-5. [DOI] [PubMed] [Google Scholar]
- 436.Menekse E, McKolanis J, Finn O, McAuliffe P, Johnson R, Soran A. Anti-MUC1 antibody in nipple aspirate fluids correlates with tumor aggressiveness in breast cancer: a feasibility study. Dis Markers. 2015;2015:179689. doi: 10.1155/2015/179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.Kurono S, Kaneko Y, Matsuura N, Oishi H, Noguchi S, Kim SJ, Tamaki Y, Aikawa T, Kotsuma Y, Inaji H, Matsuura S. Identification of potential breast cancer markers in nipple discharge by protein profile analysis using two-dimensional nano-liquid chromatography/nanoelectrospray ionization-mass spectrometry. Proteomics Clin Appl. 2016;10:605–613. doi: 10.1002/prca.201500016. [DOI] [PubMed] [Google Scholar]
- 438.Noble JL, Dua RS, Coulton GR, Isacke CM, Gui GP. A comparative proteinomic analysis of nipple aspiration fluid from healthy women and women with breast cancer. Eur J Cancer. 2007;43:2315–2320. doi: 10.1016/j.ejca.2007.08.009. [DOI] [PubMed] [Google Scholar]
- 439.Pawlik T, Fritsche H, Coombes K, Xiao L, Krishnamurthy S, Hunt K, Pusztai L, Chen JN, Clarke C, Arun B, Hung MC, Kuerer H. Significant differences in nipple aspirate fluid protein expression between healthy women and those with breast cancer demonstrated by time-of-flight mass spectrometry. Breast Cancer Res Treat. 2005;89:149–157. doi: 10.1007/s10549-004-1710-4. [DOI] [PubMed] [Google Scholar]
- 440.Paweletz CP, Trock B, Pennanen M, Tsangaris T, Magnant C, Liotta LA, Petricoin EF 3rd. Proteomic patterns of nipple aspirate fluids obtained by SELDI-TOF: potential for new biomarkers to aid in the diagnosis of breast cancer. Dis Markers. 2001;17:301–307. doi: 10.1155/2001/674959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441.He J, Gornbein J, Shen D, Lu M, Rovai L, Shau H, Katz J, Whitelegge J, Faull K, Chang H. Detection of breast cancer biomarkers in nipple aspirate fluid by SELDI-TOF and their identification by combined liquid chromatography-tandem mass spectrometry. Int J Oncol. 2007;30:145–154. [PubMed] [Google Scholar]
- 442.Palmer DJ, Kelly VC, Smit AM, Kuy S, Knight CG, Cooper GJ. Human colostrum: identification of minor proteins in the aqueous phase by proteomics. Proteomics. 2006;6:2208–2216. doi: 10.1002/pmic.200500558. [DOI] [PubMed] [Google Scholar]
- 443.Sharma A, Shandilya UK, Sodhi M, Mohanty AK, Jain P, Mukesh M. Evaluation of milk colostrum derived lactoferrin of sahiwal (Bos indicus) and karan fries (Cross-Bred) cows for its anti-cancerous potential. Int J Mol Sci. 2019;20:6318. doi: 10.3390/ijms20246318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444.Davanzo R, Zauli G, Monasta L, Vecchi Brumatti L, Abate M, Ventura G, Rimondi E, Secchiero P, Demarini S. Human colostrum and breast milk contain high levels of TNF-related apoptosis-inducing ligand (TRAIL) J Hum Lact. 2012;29:23–5. doi: 10.1177/0890334412441071. [DOI] [PubMed] [Google Scholar]
- 445.Witkowska-Zimny M, Kaminska-El-Hassan E. Cells of human breast milk. Cell Mol Biol Lett. 2017;22:11. doi: 10.1186/s11658-017-0042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 446.Aslebagh R, Ngounou A, Channaveerappa D, Arcaro K, Darie C. Proteomics study of human breast milk for breast cancer biomarkers discovery. FASEB J. 2015;29:567.26. [Google Scholar]
- 447.Aslebagh R, Channaveerappa D, Pentecost B, Arcaro K, Darie C. Combinatorial electrophoresis and mass spectrometry-based proteomics in breast milk for breast cancer biomarker discovery. Adv Exp Med Biol. 2019;1140:451–467. doi: 10.1007/978-3-030-15950-4_26. [DOI] [PubMed] [Google Scholar]
- 448.Liscia D, Detoma P, Zanchetta M, Anro P, Molinar D, Favettini E, Paduos A. The use of CYFRA 21-1 for the detection of breast cancer axillary lymph node metastases in needle washouts of fine-needle aspiration biopsies. Appl Immunohistochem Mol Morphol. 2015;25:190–195. doi: 10.1097/PAI.0000000000000287. [DOI] [PubMed] [Google Scholar]


