Abstract
TNBG-5602, a new synthesized derivative of tetrazanbigen, is a potential chemotherapeutic agent against cancer. However, its underlying mechanism is complex and still unknown. In this investigation, the anticancer effects of TNBG-5602 were determined in vitro and in vivo. Small RNA retroviral library plasmids that overexpress 19-bp fragments were used to generate TNBG-5602-resistant cells. After validation, the overexpressed 19-bp fragments were sequenced using next-generation sequencing (NGS) in the drug-resistant cells. Furthermore, the relationship of TNBG-5602, phosphatase and tensin homolog deleted on Chromosome 10 (PTEN), and the phosphatidylinositol 3 kinase (PI3K)/protein kinase B (Akt) pathway was explored. The results showed that TNBG-5602 can effectively inhibit cancer cell proliferation and induce apoptosis in vitro and in vivo. Drug-resistant cells were screened using the small RNA library. Compared with naïve cells, drug-resistant cells were more resistant to TNBG-5602 in vitro and in vivo. NGS results revealed that the second highest overexpressed 19-bp fragment perfectly matched the PTEN gene, so the expression of PTEN in various cells and tissues was verified. Further research showed that exogenous overexpression of PTEN strengthened the anticancer effects of TNBG-5602 on p-Akt expression, whereas silencing of PTEN weakened these effects in naïve cells. Taken together, by using this library, we confirmed that PTEN is the target gene to the anticancer effects of TNBG-5602 via the PI3K/Akt pathway.
Keywords: TNBG-5602, small RNA retroviral library, anticancer, PTEN, PI3K/Akt pathway
Introduction
Cancer is a major public health concern and the leading cause of death worldwide. In 2017, 1,688,780 new cancer cases and 600,920 cancer deaths occurred in the United States [1]. Over the past few decades, cancer treatments, especially chemotherapy, have been developed quickly. The drugs have evolved from broad spectrum to target and from chemical to biological. However, all these drugs have some common limits, such as drug resistance. Therefore, there is a great clinical need to explore new antineoplastic agents with improved curative effects and fewer side effects. For each potential chemotherapeutic drug, it is particularly important to confirm its anticancer effects and determine its specific underlying mechanism.
Tetrazanbigen (TNBG) is a novel synthesized derivative of isoquinoline that was synthesized by our group (Chinese Patent 2001; CN1124251A). We previously reported that TNBG can strongly inhibit the proliferation of multiple human cancer cells and that the anticancer effects of TNBG may be associated with disturbing the lipid metabolism balance [2]. However, its water solubility is poor. Therefore, a series of TNBG analogues with improved water solubility were designed and synthesized [3,4]. Among these derivatives, TNBG-5602 showed good water solubility (as much as 152.95 μg/ml at pH 7.4) and a strong antiproliferative effect in many cancer cells, especially liver cancer cells [3,5]. Further study showed that the toxicity of TNBG-5602 was low (LD50=172 mg/kg in mice). In a xenograft liver cancer model, TNBG-5602 could remarkably inhibit the growth of tumors [5]. These results suggest that TNBG-5602 is a potential anticancer drug.
Generally, researchers connect the effects of a drug with signaling pathways, core genes, long noncoding RNAs, microRNAs, and other cellular components and processes to explore the possible mechanism of action of a compound. We used a novel approach in which a small RNA retroviral library of plasmids overexpresses 19-bp fragments to silence some specific genes and generate drug-resistant cells. After sequencing, the overexpressed 19-bp fragments were defined, and the targeted genes were located after a National Center of Biotechnology Information (NCBI) blast search.
In this study, we evaluated the anticancer activity of TNBG-5602 in naïve liver cancer cells; screened out drug-resistant cells using the retrovirus library; located the target gene, phosphatase and tensin homolog deleted on chromosome 10 (PTEN), by sequencing and a blast search; and explored the relationship between PTEN and the effects of TNBG-5602, as well as the pathway potentially involved. We found that PTEN is vitally important to the anticancer function of TNBG-5602 and might be mediated by the phosphatidylinositol 3 kinase (PI3K)/protein kinase B (Akt) signaling pathway.
Materials and methods
Chemicals and preparations
TNBG-5602 was synthesized by the Department of Medicinal Chemistry at Chongqing Medical University, China, and an aqueous solution with different concentrations of TNBG-5602 was used for this study. All primary antibodies in this study were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). LY294002 (Cat No. T2008) was purchased from Targetmol Co., Ltd. (Shanghai, China). Cell counting kit 8 (CCK-8) was purchased from 7Sea PharmTech (Shanghai, China). Small RNA retroviral library plasmids and the pCL-Ampho plasmid were donated by Tongchuan He (Molecular Oncology Laboratory, Department of Orthopedic Surgery and Rehabilitation Medicine, The University of Chicago Medical Center, Chicago, IL, USA).
Cell lines and culture
The normal human hepatic cell line LO2 and human liver cancer cell lines HepG2, QGY-7701, and SMMC-7721 were obtained from the Cell Bank of the Shanghai Institute of Biochemistry and Cell Biology (Chinese Academy of Sciences, China). 293PA (293 Phoenix Ampho) cells were purchased from the American Type Culture Collection (Manassas, VA, USA). The cells were cultured with Dulbecco’s modified Eagle medium containing 10% fetal bovine serum and maintained at 37°C and 5% CO2.
Crystal violet staining
Briefly, exponentially growing cells were seeded in 24-well plates and received different treatments for 48 h. After treatment, the cells were washed with phosphate-buffered saline (PBS) three times and stained with crystal violet solution for 20 min at 25°C. Finally, the plates were washed with water and scanned.
Cell counting kit 8 assays
The cell counting kit 8 (CCK-8) assay was used to assess cell viability. Briefly, cells were seeded in 96-well plates at a density of 4×103 cells/well and treated with different concentrations of TNBG-5602 (0 μM, 5 μM, 10 μM, 15 μM and 20 μM) for a scheduled length of time (24 h, 48 h or 72 h). To each well, 10 μl CCK-8 reagent was added, and then the plates were incubated at 37°C for 2 h. The results were measured at the optical absorbance at 450 nm using a microplate reader.
Cell cycle and apoptosis analysis by flow cytometry
The cells were seeded in 6-well plates and treated with different concentrations of TNBG-5602 (0 μM, 5 μM, 10 μM, and 15 μM) for 48 h. For cell cycle analysis, cells were collected and washed twice with ice-cold (4°C) PBS and fixed with ice-cold 70% ethanol overnight. After centrifugation, the precipitates were resuspended in PBS and incubated with propidium iodide (20 mg/ml) and RNase (1 mg/ml) for 30 min, followed by flow cytometry analysis. For apoptosis analysis, cells were collected (including the supernatant) and washed twice with ice-cold (4°C) PBS, followed by incubation with Annexin V-EGFP and propidium iodide following the manufacturer’s instructions (KeyGen Biotech, Nanjing, China). Then, the cells were analyzed using flow cytometry.
Western blot assay
Cells were seeded in 6-well plates and treated based on the experimental design. Cells were lysed with lysis buffer, mixed with loading buffer, and boiled for 10 min. Then, the samples were separated using sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to polyvinylidene fluoride membranes. After incubation with 5% bovine serum albumin for 1 h, the membranes were incubated with the corresponding primary antibodies (1:1000 dilutions) and incubated for 2 h, washed four times with TBS-T solution, and incubated with secondary antibody for 30 min. After incubation, the membranes were washed four more times with TBS-T, and the expressions of proteins were detected by the SuperSignal West Pico Substrate (Pierce, Rockford, IL, USA).
Construction of drug-resistant cells by small RNA retroviral library plasmids
Small RNA retroviral library plasmids were used to overexpress 19-bp fragments in cells. The fragments were synthesized by 4 deoxynucleotide triphosphates (dNTPs) randomly in advance and inserted in the library plasmids. In theory, the variety of fragments could reach the number of 419. Briefly, small RNA retroviral library plasmids and pCL-Ampho were transfected into 293PA (293 Phoenix Ampho) cells with lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) to produce retrovirus. After culture for 36 h, 48 h, 60 h, or 72 h, the medium was harvested, filtered, and mixed. Once the naïve QGY-7701 cells were seeded in 10-cm dishes, the medium containing retrovirus was added to infect the cells for 4 h. After infection, the cells were treated with blasticidin (BSD, a screening gene located in the small RNA retroviral library plasmids) to eliminate cells that had not been infected with library retrovirus. To generate drug-resistant cells, a lethal dose (25 μM) of TNBG-5602 was added, and cells were screened 5 to 8 times. The drug-resistant cells were named QGY-7701R.
Genomic DNA extraction and next-generation sequencing
The genomic DNA of cells was obtained using gDNA extracting solution (250 μl solution containing 5 μl NaOH [10 M], 0.5 μl EDTA-Na [0.5 M; pH=8.0] and 244.5 μl ddH2O). Next-generation sequencing (NGS) was performed on genomic DNAs by the Beijing Genomics Institute. Control cell groups included both HT29 and QGY-7701 cells. Control cells were infected with library retrovirus (obtained as mentioned above) but only screened using BSD, not TNBG-5602, 3 to 5 times.
Xenograft tumor model of human liver cancer and histological evaluation
The animal experiments in this study were approved by the Institutional Animal Care and Use Committee of Chongqing Medical University. Athymic nude mice (male, 4-6 weeks old) were ordered from the Animal Centre of Chongqing Medical University (Chongqing, China). Xenograft human liver cancer models were successfully established by subcutaneously injecting naïve QGY-7701 cells or drug-resistant QGY-7701R cells into the flanks of athymic nude mice. Briefly, cells were harvested and resuspended in cold PBS (4°C) at 5×107 cells/ml. The cell suspension was injected into the left flanks of athymic mice (0.1 ml/injection). Tumor growth was monitored every day. When a tumor with a size of approximately 15 mm3 was detected [2], the mice were randomly divided into control and treatment groups (n=5). The mice of the treatment group were treated with different doses of TNBG-5602 (4.3, 8.6, and 17.2 mg/kg, equivalent to the 1/40, 1/20, and 1/10 of the LD50) by peritoneal injection, once a day for 3 weeks; the mice in the control group were treated with the same volume of normal saline. At the end of the third week, all nude mice were sacrificed. All tumor samples were retrieved, weighed, and then fixed with 10% formalin and embedded in paraffin. Serial sections of the embedded samples were stained with hematoxylin and eosin [6].
Human tissue specimens
This protocol was approved by the Ethics Committee of Chongqing Medical University, and informed consent forms were signed by all patients before surgery. Liver tumor and peritumor tissues of 12 patients who underwent surgery were obtained from the Department of Hepatobiliary Surgery at The First Affiliated Hospital of Chongqing Medical University. None of these patients had received chemotherapy or radiotherapy before surgery. All tissues were stored in liquid nitrogen.
Total RNA extraction and real-time quantitative polymerase chain reaction analysis
The cells were seeded in dishes and treated based on the experimental design. Total RNA from the cells and clinical liver tumor and peritumor tissues was extracted with TRIzol reagent (Invitrogen) and reverse transcribed to generate cDNAs. These cDNAs were then used as templates to detect the expression level of the target genes with real-time quantitative polymerase chain reaction analysis (RT-qPCR). GAPDH, with the sequences (5’→3’) CAACGAATTTGGCTACAGCA and AGGGGAGATTCAGTGTGGTG, was used as a control to normalize the results. The primer sequences (5’→3’) of PTEN were TAAAGGCACAAGAGGCCCTA and CGCCACTGAACATTGGAATA.
Cell immunofluorescent staining
Cells were seeded in 48-well plates and treated according based on the experimental design for 48 h. Next, they were washed with PBS and fixed with 4% methanol (4°C) for 20 min, then washed again with PBS, and permeabilized with 0.3% Triton X-100 for 10 min. After being blocked with 5% bovine serum albumin at room temperature for 30 min, the cells were incubated with the primary antibody anti-PTEN (Santa Cruz Biotechnology) overnight (4°C), with the corresponding immunoglobulin G used as a negative control. Then, the cells were incubated with fluorescein isothiocyanate conjugated secondary antibodies for 1 h. After being washed with PBS, the cells were stained with 4’,6-diamidino-2-phenylindole (0.1 μg/ml). Fluorescent images were recorded under an inverted microscope.
Construction of recombinant adenovirus
A recombinant adenoviruses that overexpresses PTEN (also called AdPTEN) and small interference RNA (siRNA) fragments for the silencing of PTEN (also called AdsiPTEN) were constructed using the AdEasy system [7,8]. Briefly, the coding sequence of human PTEN and red fluorescence protein (RFP) were amplified, and the siRNA fragments for PTEN knockdown were synthesized commercially. These fragments were then cloned into shuttle vectors. Next, the shuttle vectors were recombined with BJ/Adeasy-1 cells, and the correct recombinant vectors were linearized and transfected into 293PA (293 Phoenix Ampho) cells for the packaging of recombinant adenoviruses. The recombinant adenovirus expressing RFP only (AdRFP) was used as the vector control.
Statistical analysis
All quantitative data in this study are presented as the means ± standard deviation unless otherwise indicated. Statistical analyses were performed with SPSS 19.0. Data were examined for normality by skewness/kurtosis. A nonnormal distribution was transformed into a normal distribution by Blom’s formula. Significant differences in the results were analyzed using one-way analysis of variance with Tukey test for equal variances and Games-Howell test for unequal variances. A P value less than 0.05 was considered as statistically significant.
Results
Antiproliferative and apoptosis-promoting effects of TNBG-5602 on cancer cells
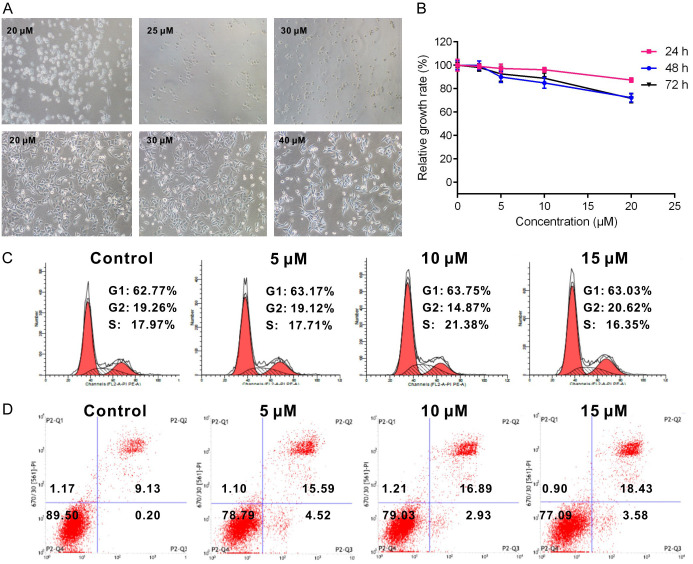
To validate whether TNBG-5602 can be used as a chemotherapeutic agent for cancer, the antiproliferation and apoptosis-promoting effects of TNBG-5602 were explored. Treatment with TNBG-5602 effectively inhibited the proliferation of three liver cancer cell lines in a time- and concentration-dependent manner, as revealed by results of crystal violet staining and the CCK-8 assay. The effect of TNBG-5602 on QGY-7701 cells was more pronounced than that on the other cells (Figure 1A and 1B). Therefore, QGY-7701 cells were selected for the following tests.
Figure 1.
The anticancer effects of TNBG-5602 on liver cancer cells. TNBG-5602 showed antiproliferation and apoptosis-promoting effects on liver cancer cells, especially QGY-7701 cells. A. Crystal violet staining of different cancer cells after treatment with TNBG-5602. B. The proliferation assays of cancer cells after treatment with TNBG-5602 by CCK-8 (*P<0.05, vs. control; **P<0.01, vs. control). C. The cell cycle arrest effect of TNBG-5602 in QGY-7701 cells analyzed by flow cytometer. D. The effect of TNBG-5602 on the expression of PCNA in QGY-7701 cells tested by western blot. E. The apoptosis-promoting effect induced by TNBG-5602 in QGY-7701 cells. F. The effect of TNBG-5602 on the expression of Bcl-2 and Bad in QGY-7701 cells. GAPDH was used as the loading control.
Treatment with TNBG-5602 induced the arrest of cell cycle in QGY-7701 cells at the G1 phase (Figure 1C). Additionally, TNBG-5602 significantly suppressed the expression of proliferating cell nuclear antigen (PCNA), a marker of cell proliferation [9] (Figure 1D). According to results of flow cytometric analyses, TNBG-5602 increased the apoptotic rate of QGY-7701 cells in a concentration-dependent manner (Figure 1E). TNBG-5602 decreased the expression of B-cell lymphoma gene 2 (Bcl-2) but increased expression of Bcl-xl/Bcl-2-associated death promoter (Bad) (Figure 1F). Furthermore, the apoptosis-promoting effect of TNBG-5602 was concentration dependent.
Verification of drug-resistant cells in vitro
With the use of the small RNA library, TNBG-5602-resistant cells called QGY-7701R were obtained. Compared with primary QGY-7701 cells, QGY-7701-R were more resistant to TNBG-5602 (Figures 2A, 1B and 2B). Unlike naïve cells, no significant differences in cell cycle and apoptosis rate of drug-resistant QGY-7701R cells were found between the control and experimental groups (Figures 2C, 2D, 1C and 1E).
Figure 2.
Verification of resistance to novel isoquinoline derivative TNBG-5602 in vitro. Compared with QGY-7701 cells, drug-resistant QGY-7701R cells were more resistant to TNBG-5602 in vitro. A. Cell viability under different concentrations of TNBG-5602 in naïve and drug-resistant cells (upper: naïve QGY-7701 cells; lower: drug-resistant QGY-7701R cells). B. The proliferation assays of drug-resistant cells after treatment with TNBG-5602 by CCK-8. C, D. The cell cycle and apoptosis rate assays of drug-resistant QGY-7701R cells after treatment with TNBG-5602.
Verification of drug-resistant cells in vivo
Xenograft human liver cancer models were successfully established using naïve QGY-7701 cells or drug-resistant QGY-7701R cells. After treatment with TNBG-5602, the sizes and weights of tumor masses formed using naïve QGY-7701 cells were significantly reduced (Figure 3A and 3B). However, no obvious reduction was found in masses formed using drug-resistant cells (Figure 3C and 3D). Furthermore, the experimental groups of naïve cells exhibited more necrotic cells than the control group, whereas drug-resistant cells had less (Figure 3E).
Figure 3.

Verification of resistance to novel isoquinoline derivative TNBG-5602 in naïve and drug-resistant cells in vivo. A, B. Ectopic tumor masses of naïve QGY-7701 cells retrieved from nude mice treated with TNBG-5602 (low, 4.3 mg/kg; middle, 8.6 mg/kg; high, 17.2 mg/kg; P<0.05, compared with control; P<0.01, compared with control; x̅±s; n=5). C, D. Ectopic tumor masses of drug-resistant QGY-7701R cells retrieved from nude mice treated with TNBG-5602 (N.S., no significance, compared with control; x̅±s; n=5). E. Hematoxylin and eosin staining results of ectopic tumor established by naïve and drug-resistant cells (upper, naïve QGY-7701 cells, ectopic tumor; lower, drug-resistant QGY-7701R cells, ectopic tumor). Representative results are shown (×200).
NGS results of drug-resistant cells’ genomic DNA
Random 19-bp fragments, whose sequences theoretically reached 419, were inserted into small RNA retroviral library plasmids. After being infected by the library retrovirus and screened with BSD and TNBG-5602, drug-resistant cells called QGY-7701R were obtained. The characteristics of the drug-resistant cells were identified in vitro and in vivo, and NGS was used to determine the specific sequence of overexpressed 19-bp fragment that was inserted into the genomic DNA by retrovirus. The results of NGS are shown in Table 1.
Table 1.
Most frequently expressed 19-bp fragments in drug-resistant cells
| No.a | Tag | iMEFn19 cells | UACC-62n19 cells | QGY-7701R cells |
|---|---|---|---|---|
| n (%) | n (%) | n (%) | ||
| 1 | TTCTGCTCTGTCGTATCTG | 0 (0.00) | 0 (0.00) | 1,004,306 (13.87) |
| 2b | TATATCTTCACCTTTAGCT | 0 (0.00) | 0 (0.00) | 537,615 (7.43) |
| 3 | ATAAGTATGCAAGAATATG | 0 (0.00) | 0 (0.00) | 445,149 (6.15) |
| … | … | … | … | … |
| 5266c | Total | 211,364 (5.30) | 209,003 (5.46) | 6,647,889 (91.83) |
Data expressed as n (%).
The three most frequently expressed 19-bp fragments are listed.
Through a blast search with NCBI, the fragment was determined to perfectly match the PTEN gene (query cover, 100%; identify, 100%).
The fragments read by NGS were listed from the highest to the lowest.
From the 1st to the 5266th one, all the fragments sequenced were calculated totally and presented as n (%).
Effects of TNBG-5602 on PTEN in hepatocellular carcinoma tissues and cells
After NGS, the second most enriched 19-bp fragment (with the sequence TATATCTTCACCTTTAGCT) was found to match the PTEN gene 100% after a blast search with NCBI. The overexpressed fragment could potentially disturb the translation of PTEN mRNA in QGY-7701R cells. To verify this, the mRNA levels of PTEN in naïve and drug-resistant cells were detected using RT-qPCR, and the results showed reduced PTEN mRNA levels in the QGY-7701R drug-resistant cells (Figure 4A). Due to suppression by the fragment (listed in Table 1), PTEN in QGY-7701R cells had lower expression, which may result in drug resistance. To verify this, mRNA and protein levels of PTEN in clinical hepatocellular carcinoma (HCC; 12 cases) and peritumor (10 cases) tissues, as well as those in the human normal hepatic cell line LO2 and three cancer cells lines, were determined. As expected, compared with the control group, the mRNA level of PTEN in HCC tissues and cancer cell lines was significantly reduced (Figure 4B and 4C), as well as the endogenous protein level of PTEN in cancer cell lines (Figure 4D). Meanwhile, after TNBG-5602 treatment, the mRNA and protein levels of PTEN were remarkably increased in a concentration-dependent manner in naïve cells (Figure 4E and 4F), and similar results were found with immunofluorescence staining (Figure 4G) and in ectopic human liver tumor tissues formed by naïve cells (Figure 4H).
Figure 4.

The transformation of PTEN in various cells, tissues, and treatment states. A. RT-qPCR verification of PTEN expression in naïve and drug-resistant cells (**P<0.01, vs. naïve cells). B. The mRNA expression of PTEN in liver tumor tissues and peritumor tissues (**P<0.01, vs. peritumor). C, D. Endogenous mRNA and protein expression of PTEN in normal and cancer cells (**P<0.01, vs. LO2). E, F. The mRNA and protein expression of PTEN change under treatment of TNBG-5602 in naïve cells (**P<0.01, vs. control). G. Immunofluorescence staining of PTEN in naïve cells treated with TNBG-5602. H. The protein expression of PTEN in ectopic tumors of naïve cells (low, 4.3 mg/kg; middle, 8.6 mg/kg; high, 17.2 mg/kg).
Effects of PTEN on the antiproliferation effect of TNBG-5602
The role of PTEN in the anticancer effect of TNBG-5602 was investigated. Adenovirus was used to silence and overexpress of PTEN gene in the following experiments. The function of adenovirus (AdsiPTEN or AdPTEN) was verified on the expression of both mRNA and protein in naïve cells (Figure 5A and 5B). Unsurprisingly, the exogenous overexpression of PTEN substantially enhanced the antiproliferation effect of TNBG-5602, while silencing of PTEN weakens this effect in QGY-7701 cells (Figure 5C and 5D).
Figure 5.

The effects of PTEN on anticancer activity of TNBG-5602. A, B. The mRNA and protein expression verification of PTEN treated with AdsiPTEN or AdPTEN (*P<0.05, **P<0.01, compared with the AdRFP group). C, D. The anticancer activity of TNBG-5602 influenced by silencing or overexpressing the PTEN gene (*P<0.05, **P<0.01).
Effects of TNBG-5602 on the PI3K/Akt pathway
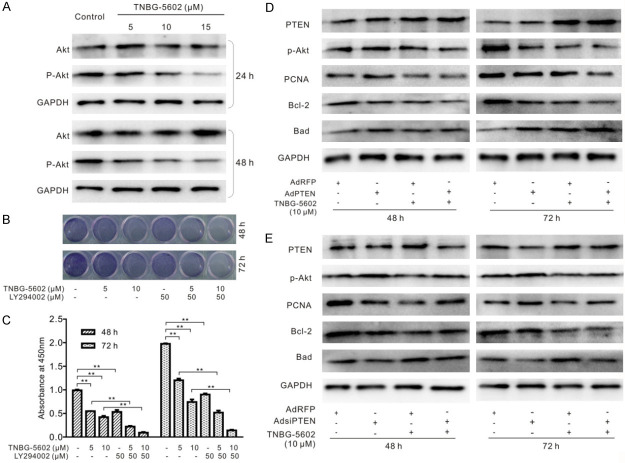
PTEN is an important negative regulator for PI3K/Akt signaling. Considering the relationship between PTEN and TNBG-5602 shown in Figure 5, we speculated that the anticancer effect of TNBG-5602 might be mediated by inactivating the PI3K/Akt pathway via upregulating PTEN. Compared with the control cells, the expression level of p-Akt declined after treatment with TNBG-5602 in a concentration-dependent manner (Figure 6A). Meanwhile, the antiproliferation effect of TNBG-5602 on naïve cells was significantly enhanced by the PI3K inhibitor LY294002 (Figure 6B and 6C). Furthermore, the effect of TNBG-5602 on the level of p-Akt, PCNA, Bcl-2, and Bad was strengthened by PTEN overexpression (Figure 6D) but weakened by PTEN silencing (Figure 6E).
Figure 6.
The role of the PI3K/Akt pathway in the anticancer activity of TNBG-5602 combined with PTEN silencing or overexpression. A. The transformation of Akt and p-Akt induced by TNBG-5602. B, C. The PI3K inhibitor (LY294002) enhanced the antiproliferative effect of TNBG-5602 (**P<0.01). D, E. The effect of silencing or overexpression of PTEN on the protein levels of p-Akt, PCNA, Bcl-2, and Bad affected by TNBG-5602. GAPDH was used as the loading control.
Discussion
Many chemical compounds containing heterocyclic rings, such as quinoline, pyrane, pyridine, and thiazole, have been shown to be potential antitumor agents. TNBG is a de novo-synthesized compound derivative of isoquinoline with a sterol structure. In our previous study, we demonstrated the effects of TNBG against cell proliferation on human cancer in vitro and in vivo [2]. However, the solubility of TNBG in water is poor, limiting the clinical use of TNBG. Thus, we designed and synthesized a series of TNBG derivatives to improve its water solubility. In this study, we used TNBG-5602, one of these derivatives with desirable solubility in water (as much as 152.95 μg/ml at pH 7.4).
First, we explored the anticancer effects of TNBG-5602 on liver cancer cell lines. Our results showed that TNBG-5602 can inhibit the proliferation of liver cancer cell lines, especially QGY-7701 cells, in a concentration-dependent manner. Because of this finding, QGY-7701 cells were used as naïve cancer cells in subsequent experiments. Apoptosis is one of the fundamental processes in carcinogenesis and responses to anticancer treatment. An imbalance between the expression of pro- and anti-apoptotic proteins of the BCL-2 family induced by the majority of genetic modifications in HCC has been previously observed [10,11]. Our study showed that TNBG-5602 downregulated the expression of Bcl-2 and upregulated the expression of Bad in naïve cells. These results support TNBG-5602 as a promising compound for tumor therapy. However, the molecular mechanism underlying this anticancer activity still must be elucidated.
Instead of the traditional approach to characterizing potential anticancer agents, we used small RNA retroviral library plasmids to generate drug-resistant cells. The plasmids were designed to overexpress random 19-bp fragments to silence some specific genes in cells after infection with retrovirus. The fragments were randomly synthesized before insertion into the library plasmids, and the variety could theoretically reach to the number of 419. After treatment with the library retrovirus, BSD can be used first to screen out cells that have not been infected by retrovirus. These cells can be used as controls. To generate drug-resistant cells, a lethal dose (25 μM) of TNBG-5602 was added to screen 5 to 8 times. The drug-resistant cells were called QGY-7701R.
After the drug-resistant cells were screened, we verified the effect with naïve cells in vitro and in vivo. To our surprise, compared with naïve cells, drug-resistant QGY-7701R cells showed better tolerance to TNBG-5602 in terms of proliferation and apoptosis. Moreover, no significance difference was found in xenograft tumor models formed by QGY-7701R under TNBG-5602 treatment. Meanwhile, the anticancer effect of TNBG-5602 was verified again in naïve cells in vivo. These findings indicate that drug-resistant QGY-7701R cells were accurately screened by the retrovirus library.
As designed, the overexpressed 19-bp fragments, responsible for the drug resistance, were inserted into the genomic DNA of QGY-7701R cells. NGS was used to determine the specific sequences of fragments that induced drug resistance. After NGS, we found that the second most frequent fragment listed in Table 1 perfectly matched the PTEN gene (query cover, 100%; identify, 100%). Therefore, we hypothesized that PTEN expression is suppressed in drug-resistant cells, lower expression of PTEN is crucial for cell survival, and higher expression of PTEN would enhance the effect of TNBG-5602.
To verify these hypotheses, we detected mRNA expression in naïve and drug-resistant cells, as well as tumor and peritumor tissues. As expected, the mRNA or protein level of PTEN was downregulated in QGY-7701R cells, as well as HCC tissues and liver cancer cell lines. After treatment with TNBG-5602, the mRNA and protein levels of PTEN in naïve cells were increased in a concentration-dependent manner. Furthermore, the relationship between them was validated by immunofluorescence.
Considering the importance of the PTEN gene, we successfully constructed recombinant adenoviruses named AdPTEN and AdsiPTEN, which overexpress and silence PTEN expression, respectively. Our results show that the antiproliferation effect of TNBG-5602 was substantially strengthened by the exogenous overexpression of PTEN but weakened by the silencing of PTEN in naïve cells.
PTEN is one of the most frequently inactivated tumor suppressor genes and was originally identified on chromosome 10q23 in 1997 [12,13]. A primary function of PTEN is to negatively regulate PI3K/Akt signaling. The PI3K/Akt pathway plays important roles in regulating cell proliferation, apoptosis, survival, metabolism, and migration and is involved in many cancers [14-18]. Meanwhile, the activation of Akt plays a pivotal role in the PI3K/Akt pathway by phosphorylating a variety of substrates. Thus, we speculated that the anticancer effect of TNBG-5602 may be mediated by inactivating the PI3K/Akt pathway via upregulating PTEN. As expected, the expression level of p-Akt was decreased after treatment with TNBG-5602 in a concentration-dependent manner; however, the antiproliferation effect of TNBG-5602 on naïve cells was significantly enhanced by the PI3K inhibitor LY294002. Moreover, the effect of TNBG-5602 on the levels of p-AKt, PCNA, Bcl-2, and Bad was strengthened by PTEN overexpression but weakened by PTEN silencing.
As reported, PTEN exhibits phosphatase activity toward phosphoinositide, resulting in PIP3 dephosphorylation to form PIP2 and inactivation of the PI3K signaling pathway [19-22]. The loss or mutation of PTEN can drive a malignant phenotype in a variety of human cancers, such as endometrial carcinoma, breast cancer, glioblastoma multiforme, and liver cancers. Even a small reduction in PTEN expression can elicit cancer phenotypes [23,24]. By contrast, the overexpression of PTEN results in a constitutively augmented tumor suppressive state.
The NGS results showed that silenced PTEN by overexpressed 19-bp fragments, which were inserted into genomic DNA, was responsible for drug resistance. PTEN was suppressed in QGY-7701R cells, HCC tissues, and liver cancer cell lines. However, upregulated PTEN strengthened the effects of TNBG-5602 on the levels of p-Akt. Therefore, the antiproliferation and apoptosis-inducing effects of TNBG-5602 may be a result of upregulated PTEN blocking the phosphorylation of Akt. However, further studies should be conducted to determine (1) the exact mechanisms of TNBG-5602, PTEN, and the PI3K/Akt pathway; (2) the function of other fragments enriched in drug-resistant cells; and (3) long noncoding RNAs or microRNAs involved in the effects of TNBG-5602.
Acknowledgements
This work was supported by a research grant from the Natural Science Foundation of China (grant no. NSFC81802162), Chongqing Science and Technology Commission (grant no. cstc2017jcyjAX0262 and cstc2017jcyjAX0228), and Science and Technology Foundation of Chongqing Education Commission (grant no. KJ1600201).
Disclosure of conflict of interest
None.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Zheng X, Li W, Lan Z, Yang X, Li L, Yuan Y, Xia Z, Chen X, Zhang X, Yu Y. Antitumour effects of tetrazanbigen against human hepatocellular carcinoma QGY-7701 through inducing lipid accumulation in vitro and in vivo. J Pharm Pharmacol. 2015;67:1593–1602. doi: 10.1111/jphp.12467. [DOI] [PubMed] [Google Scholar]
- 3.Chen X, Jun GY, Yu Y, Yuan Z. Synthesis and evaluation of new sterol derivatives as potential antitumor agents. Rsc Advances. 2018;8:26528–26537. doi: 10.1039/c8ra04152k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gan L, Gan Z, Dan Y, Li Y, Zhang P, Chen S, Ye Z, Pan T, Wan C, Hu X, Yu Y. Tetrazanbigen derivatives as Peroxisome Proliferator-Activated Receptor Gamma (PPARγ) partial agonists: design, synthesis, structure-activity relationship, and anticancer activities. J Med Chem. 2021;64:1018–1036. doi: 10.1021/acs.jmedchem.0c01512. [DOI] [PubMed] [Google Scholar]
- 5.Hu X, Wan C, Gan Z, Liu R, Chen Y, Wang J, Gan L, Li Y, He B, Yu Y. TNBG-5602, a novel derivative of quinoxaline, inhibits liver cancer growth via upregulating peroxisome proliferator-activated receptor gamma in vitro and in vivo. J Pharm Pharmacol. 2019;71:1684–1694. doi: 10.1111/jphp.13159. [DOI] [PubMed] [Google Scholar]
- 6.Zhang RX, Li Y, Tian DD, Liu Y, Nian W, Zou X, Chen QZ, Zhou LY, Deng ZL, He BC. Ursolic acid inhibits proliferation and induces apoptosis by inactivating Wnt/beta-catenin signaling in human osteosarcoma cells. Int J Oncol. 2016;49:1973–1982. doi: 10.3892/ijo.2016.3701. [DOI] [PubMed] [Google Scholar]
- 7.He TC, Zhou S, da Costa LT, Yu J, Kinzler KW, Vogelstein B. A simplified system for generating recombinant adenoviruses. Proc Natl Acad Sci U S A. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo J, Deng ZL, Luo X, Tang N, Song WX, Chen J, Sharff KA, Luu HH, Haydon RC, Kinzler KW, Vogelstein B, He TC. A protocol for rapid generation of recombinant adenoviruses using the AdEasy system. Nat Protoc. 2007;2:1236–1247. doi: 10.1038/nprot.2007.135. [DOI] [PubMed] [Google Scholar]
- 9.Wang SC. PCNA: a silent housekeeper or a potential therapeutic target? Trends Pharmacol Sci. 2014;35:178–186. doi: 10.1016/j.tips.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 10.Fabregat I, Roncero C, Fernández M. Survival and apoptosis: a dysregulated balance in liver cancer. Liver Int. 2007;27:155–162. doi: 10.1111/j.1478-3231.2006.01409.x. [DOI] [PubMed] [Google Scholar]
- 11.Ikeda M, Mitsunaga S, Ohno I, Hashimoto Y, Takahashi H, Watanabe K, Umemoto K, Okusaka T. Systemic chemotherapy for advanced hepatocellular carcinoma: past, present, and future. Diseases. 2015;3:360–381. doi: 10.3390/diseases3040360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, Puc J, Miliaresis C, Rodgers L, McCombie R, Bigner SH, Giovanella BC, Ittmann M, Tycko B, Hibshoosh H, Wigler MH, Parsons R. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 13.Steck PA, Pershouse MA, Jasser SA, Yung WK, Lin H, Ligon AH, Langford LA, Baumgard ML, Hattier T, Davis T, Frye C, Hu R, Swedlund B, Teng DH, Tavtigian SV. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat Genet. 1997;15:356–362. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- 14.Stambolic V, Suzuki A, de la Pompa JL, Brothers GM, Mirtsos C, Sasaki T, Ruland J, Penninger JM, Siderovski DP, Mak TW. Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell. 1998;95:29–39. doi: 10.1016/s0092-8674(00)81780-8. [DOI] [PubMed] [Google Scholar]
- 15.Sun H, Lesche R, Li DM, Liliental J, Zhang H, Gao J, Gavrilova N, Mueller B, Liu X, Wu H. PTEN modulates cell cycle progression and cell survival by regulating phosphatidylinositol 3,4,5,-trisphosphate and Akt/protein kinase B signaling pathway. Proc Natl Acad Sci U S A. 1999;96:6199–6204. doi: 10.1073/pnas.96.11.6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Furnari FB, Lin H, Huang HS, Cavenee WK. Growth suppression of glioma cells by PTEN requires a functional phosphatase catalytic domain. Proc Natl Acad Sci U S A. 1997;94:12479–12484. doi: 10.1073/pnas.94.23.12479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li J, Simpson L, Takahashi M, Miliaresis C, Myers MP, Tonks N, Parsons R. The PTEN/MMAC1 tumor suppressor induces cell death that is rescued by the AKT/protein kinase B oncogene. Cancer Res. 1998;58:5667–5672. [PubMed] [Google Scholar]
- 18.Raftopoulou M, Etienne-Manneville S, Self A, Nicholls S, Hall A. Regulation of cell migration by the C2 domain of the tumor suppressor PTEN. Science. 2004;303:1179–1181. doi: 10.1126/science.1092089. [DOI] [PubMed] [Google Scholar]
- 19.Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 20.Cully M, You H, Levine AJ, Mak TW. Beyond PTEN mutations: the PI3K pathway as an integrator of multiple inputs during tumorigenesis. Nat Rev Cancer. 2006;6:184–192. doi: 10.1038/nrc1819. [DOI] [PubMed] [Google Scholar]
- 21.Baker SJ. PTEN enters the nuclear age. Cell. 2007;128:25–28. doi: 10.1016/j.cell.2006.12.023. [DOI] [PubMed] [Google Scholar]
- 22.Worby CA, Dixon JE. PTEN. Annu Rev Biochem. 2014;83:641–669. doi: 10.1146/annurev-biochem-082411-113907. [DOI] [PubMed] [Google Scholar]
- 23.Trotman LC, Niki M, Dotan ZA, Koutcher JA, Di Cristofano A, Xiao A, Khoo AS, Roy-Burman P, Greenberg NM, Van Dyke T, Cordon-Cardo C, Pandolfi PP. Pten dose dictates cancer progression in the prostate. PLoS Biol. 2003;1:E59. doi: 10.1371/journal.pbio.0000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alimonti A, Carracedo A, Clohessy JG, Trotman LC, Nardella C, Egia A, Salmena L, Sampieri K, Haveman WJ, Brogi E, Richardson AL, Zhang J, Pandolfi PP. Subtle variations in Pten dose determine cancer susceptibility. Nat Genet. 2010;42:454–458. doi: 10.1038/ng.556. [DOI] [PMC free article] [PubMed] [Google Scholar]