SUMMARY
Psychological stress (PS) is associated with systemic inflammation and accelerates inflammatory disease progression (e.g., atherosclerosis). The mechanisms underlying stress-mediated inflammation and future health risk are poorly understood. Monocytes are key in sustaining systemic inflammation, and recent studies demonstrate that they maintain the memory of inflammatory insults, leading to a heightened inflammatory response upon rechallenge. We show that PS induces remodeling of the chromatin landscape and transcriptomic reprogramming of monocytes, skewing them to a primed hyperinflammatory phenotype. Monocytes from stressed mice and humans exhibit a characteristic inflammatory transcriptomic signature and are hyperresponsive upon stimulation with Toll-like receptor ligands. RNA and ATAC sequencing reveal that monocytes from stressed mice and humans exhibit activation of metabolic pathways (mTOR and PI3K) and reduced chromatin accessibility at mitochondrial respiration-associated loci. Collectively, our findings suggest that PS primes the reprogramming of myeloid cells to a hyperresponsive inflammatory state, which may explain how PS confers inflammatory disease risk.
In brief
Barrett et al. investigate the impact of psychological stress on monocytes. They report that psychological stress remodels the chromatin landscape of monocytes and the myeloid transcriptome, with stress-mediated changes skewing monocytes to a primed-hyperinflammatory phenotype. This study provides mechanistic insight into how psychological stress confers risk to inflammation-based disorders.
Graphical abstract
INTRODUCTION
Clinical and experimental studies have linked psychological stress (PS) and alterations in the immune and inflammatory systems (Fleshner and Crane, 2017; Powell et al., 2013). Epidemiological studies have demonstrated that biomarkers of inflammation, such as proinflammatory cytokines (e.g., interleukin [IL]-6 and IL-1β) and acute phase proteins (e.g., C-reactive protein), are elevated in patients with post-traumatic stress and anxiety disorders compared to healthy controls (Dimsdale, 2008). PS is also associated with increased rates of disease comorbidities in which immune dysregulation is involved, including atherosclerotic cardiovascular disease (CVD), metabolic syndrome, and autoimmune diseases. For example, work or life stress can increase the risk of coronary heart disease and stroke on both acute and chronic timescales (Esler, 2017; Golbidi et al., 2015; Iso et al., 2002; Kivimäki and Steptoe, 2018; Rosengren et al., 2004). Post-traumatic stress disorder (i.e., in combat veterans) confers an increased long-term risk of atherosclerotic events (Ahmadi et al., 2011), while events such as earthquakes (Itoh et al., 2014) and the World Cup soccer championship (Wilbert-Lampen et al., 2008) increase the incidence of acute myocardial infarction (MI). Permanent stress at work or home is associated with a ≈2-fold increased risk of MI (Rosengren et al., 2004), and high levels of perceived stress at the time of MI increase the risk of death within the following 2 years by 40% (Arnold et al., 2012). The mechanisms by which stress contributes to increased CVD risk are largely unknown; however, chronic inflammation has emerged as an underappreciated CV risk factor, with a risk profile comparable to classical factors such as diabetes and hypertension (Furman et al., 2019).
In mouse models of chronic PS, enhanced sympathetic nervous system activity increases autonomic tone in the bone marrow, resulting in a transient expansion of innate immune effector cells in the circulation and hematopoietic reservoirs (Heidt et al., 2014). Mice exposed to variable stressors such as crowding, isolation, cage tilt, and changes in bedding have increased norepinephrine levels in the bone marrow, which acts via β3-adrenergic receptors on bone marrow niche cells to reduce chemokine (C-X-C motif) ligand (CXCL)12 levels (Heidt et al., 2014). Consequently, the percentage of cycling hematopoietic stem and progenitor cells increases, thereby increasing bone marrow production and the release of monocytes and neutrophils. Mice that genetically lack the β3-adrenergic receptor exhibit a blunted hematopoietic response to stress, as their CXCL12 levels and bone marrow leukocyte output remain unchanged (Heidt et al., 2014). These data correlate with reports of increased myeloid cell production after stress exposure (Powell et al., 2013; Xu et al., 2020) and after injection of epinephrine into mice (Spiegel et al., 2007). Monocyte-derived macrophages play key roles in atherosclerosis initiation and progression and contribute to the destabilization of plaques that precipitate acute CV events (Moore et al., 2013). Exposure to variable stressors aggravates atherosclerosis progression in hyperlipidemic Apoe−/− mice (Heidt et al., 2014), and this was associated with a more inflamed plaque phenotype, with higher macrophage numbers and increased protease activity reminiscent of vulnerable plaques in humans (Heidt et al., 2014). These data suggest that stress modulates the myeloid lineage both within and beyond the bone marrow niche.
Recent studies have shown that cells of the innate immune system, including monocytes and macrophages, harbor features of immunological memory that promote a heightened response upon sequential pathogen challenge. During inflammatory priming, a primary stimulus alters the functional state of the cell, facilitating a hyperinflammatory response following a secondary challenge (Divangahi et al., 2021). Distinct from trained immunity, in which functional reprogramming of the myeloid system can be sustained up to several months, immune priming refers to exposure to the primary and secondary stimuli in relatively quick succession (e.g., ex vivo stimulation of cells within the first month of receiving a vaccination). During immune training, the stable accumulation of epigenetic marks (e.g., DNA methylation, histone modifications) at immune genes and enhancer elements contributes to cellular reprogramming, allowing trained genes to be more robustly transcribed in response to future challenges. Epigenetic changes occur in tandem with the rewiring of cellular metabolic pathways such as glycolysis, oxidative phosphorylation, fatty acid metabolism, and amino acid metabolism (Cheng et al., 2014; Mitroulis et al., 2018; Netea et al., 2016). In contrast to trained immunity, relatively little is known about epigenetic or metabolic changes that occur in cells during priming. Given the capacity of primed innate immune cells to mount a robust response to secondary stimuli (Divangahi et al., 2021), it is conceivable that metabolic reprogramming and changes to chromatin accessibility at inflammatory loci may occur during priming, which ultimately increases the capacity of innate immune cells.
A growing body of evidence suggests that the ability of the innate immune system to mount a faster and enhanced response to secondary infection is not always beneficial. A heightened inflammatory potential can have detrimental effects in immune-mediated and chronic inflammatory diseases, such as CVD. Recent studies in mice and humans showed that a western-style diet or oxidized forms of low-density lipoprotein (LDL) can induce innate immune training that leads to augmented myeloid inflammatory responses and exacerbation of atherosclerosis (Bekkering et al., 2014; Christ et al., 2018). We reasoned that PS may reprogram the hematopoietic compartment and monocytes toward a primed, proinflammatory phenotype. In a murine model of chronic variable stress, we find that PS induces the transcriptomic and epigenomic reprogramming of monocytes, enhancing their innate immune responses to secondary stimuli. We then translated and extended these findings to myeloid cells collected from a cohort of women with reported low and high PS. Notably, we find a common transcriptomic signature of inflammatory and metabolic gene expression in high-stress mice and humans, which translates to an exacerbated inflammatory immune response in humans with high PS.
RESULTS
Chronic variable stress activates hematopoiesis
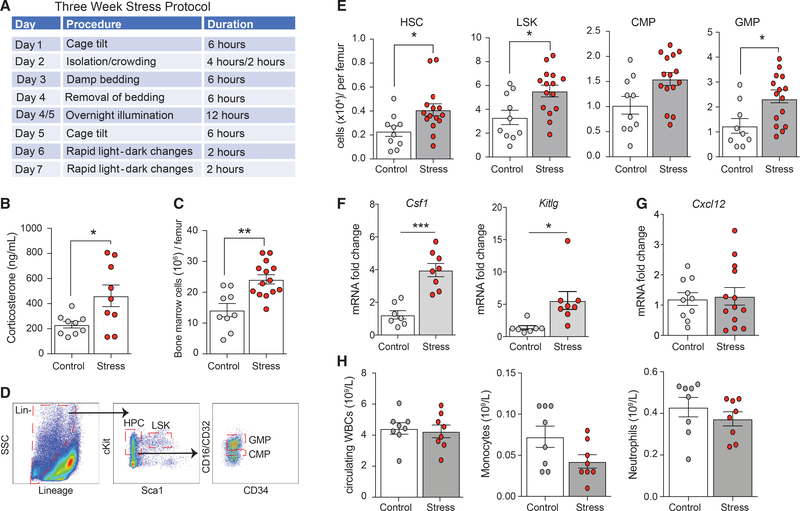
To examine the impact of stress on hematopoiesis and subsequent immune function, we subjected female mice to variable stressors validated by behavioral neuroscience studies (Heidt et al., 2014), including crowding, isolation, cage tilt, and changes in bedding, differing in duration and type over a 3-week period (Figure 1A). Compared to controls, stressed mice had increased circulating levels of the stress hormone corticosterone at the end of the 3-week protocol (p < 0.05; Figure 1B). As previously described (Heidt et al., 2014), we observed the activation of hematopoiesis in stressed mice, as manifested by an increased number of bone marrow cells, compared to nonstressed control mice (p < 0.01; Figure 1C). Characterization of stem and progenitor cells in the bone marrow revealed higher numbers of hematopoietic stem cells (HSCs), Lin−Sca-1+c-Kit+ progenitors (LSKs), and granulocyte macrophage progenitors (GMPs) in stressed versus nonstressed mice (Figures 1D and 1E). This was accompanied by increased expression genes encoding the myeloid expansion factors, macrophage colony-stimulating factor (Csf1), and stem cell factor (Kitlg) (Figure 1F). No significant change in Cxcl12 (Figure 1G), a negative regulator of hematopoietic stem and progenitor cell (HSPC) proliferation (Tzeng et al., 2011), was found. Despite this activation of the bone marrow compartment in response to chronic stress, we observed similar numbers of leukocytes, monocytes, and neutrophils in the blood of stressed and control mice (Figure 1H). No changes were found in weight, glucose, insulin, or lipid levels between control and stressed mice (Figure S1).
Figure 1. Chronic variable stress activates hematopoiesis in mice.
(A) Experimental outline of 3-week chronic variable stress protocol.
(B) Circulating corticosterone levels in control (white circles) and stressed (red circles) mice at the end of the 3-week chronic variable stress protocol.
(C and D) Total bone marrow cell count (C) and characterization of hematopoietic and stem cell populations by flow cytometry (D) in the bone marrow of stressed and control mice.
(E) Levels of hematopoietic progenitor cells (HPCs), Lin−Sca1+cKit+ (LSK), common myeloid progenitors (CMPs), and granulocyte macrophage progenitors (GMPs) in the bone marrow of stressed and control mice.
(F and G) Relative mRNA levels of (F) the cytokine macrophage colony-stimulating factor (Mcsf) and stem cell factor (Scf), and (G) the retention factor Cxcl12 in the bone marrow of stressed and control mice.
(H) Circulating white blood cell (WBC), monocyte, and neutrophil counts in stressed and control mice. Data are the means ± SEMs. p values were calculated using a Mann-Whitney t test; *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001. n = 3–15 mice/group.
Chronic variable stress primes monocytes to a proinflammatory, metabolically altered phenotype
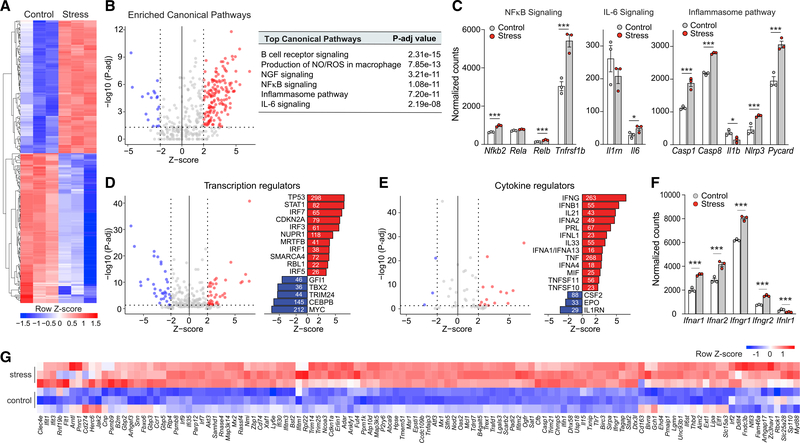
To investigate whether chronic stress reprograms monocytic reservoirs, we isolated monocytes from the bone marrow of stressed and nonstressed control mice and performed transcriptomic profiling by RNA sequencing (RNA-seq) and subsequent computational analysis. Unsupervised hierarchical clustering of genes differentially expressed in stressed and control mice showed that bone marrow monocytes markedly reprogram transcriptional responses after stress (Figure 2A; Table S1). Among the top differentially expressed genes in response to stress, we observed an enrichment in canonical pathways associated with proinflammatory signaling and production of reactive oxygen and nitrogen species (Figure 2B; Table S2), including differential expression of genes involved in nuclear factor κB (NF-κB) signaling (Nfkb2, Relb, and Tnfrsf1b), IL-6 signaling, and the inflammasome pathway (Casp1, Casp8, Il1b, Nlrp3, Pycard) (Figure 2C). To identify factors driving the differential expression of genes in monocytes from stressed and control mice, we performed upstream regulator analysis using Ingenuity Pathway Analysis (IPA), which predicts potential upstream regulators, including transcription factors and cytokines demonstrated experimentally to alter the affected gene pathways (Figures 2D and 2E). This analysis identified the enrichment of binding sites for transcription factors associated with interferon (IFN) signaling (signal transducer and activator of transcription 1 [STAT1], IFN regulatory factor 7 [IRF7], IRF3, IRF1), cell-cycle regulation and DNA repair (CDKN2A, TP53, MRTFB, RBL1), and the stress response (NUPR1) in genes upregulated by stress. Furthermore, cytokines predicted to positively regulate stress-mediated monocyte transcriptional perturbations included IFNs (IFNG, IFNB1, IFNA2, IFNL1, IFNA1, IFNK), tumor necrosis factor (TNF), and IL-12, while the IL-1 receptor antagonist was predicted to be downregulated (Figure 2E). In monocytes from stressed compared to control mice, the role of IFN signaling in mediating stress-induced transcriptional changes was further supported by increased mRNA expression of IFN receptors (Figure 2F) and IFN target genes (Figure 2G). These data indicate that the bone marrow compartment responds strongly to chronic stress, increasing myelopoiesis and transcriptional reprogramming of bone marrow monocytes to a proinflammatory phenotype.
Figure 2. Chronic variable stress leads to transcriptome reprogramming in mouse monocytes.
(A) RNA-seq heatmap displaying differentially expressed transcripts (adjusted p < 0.05 and |log2FC| > 1.5) between monocytes isolated from mice exposed to chronic variable stress and nonstressed control mice (n = 3 mice/group).
(B) Top canonical pathways of genes upregulated in monocytes from stressed versus control mice (p < 0.01).
(C) Normalized expression counts of genes in the NF-κB, IL-6, and inflammasome signaling pathways in monocytes from stressed and control mice.
(D and E) Ingenuity Pathway Analysis of predicted upstream (D) transcriptional and (E) cytokine regulators of genes differentially expressed in monocytes from stressed versus control mice. The number next to each bar represents genes in our dataset that have a measurement direction consistent with the activation of that transcriptional regulator.
(F) Normalized expression counts of interferon receptor-related genes in monocytes from stressed and control mice.
(G) Heatmap showing RNA-seq normalized gene expression values of interferon-stimulated genes (row Z score) in monocytes isolated from the bone marrow of stressed or control mice (n = 3). Data in (C) and (F) are means ± SEMs. p values were calculated using a Mann-Whitney test; *p ≤ 0.05, ***p ≤ 0.001.
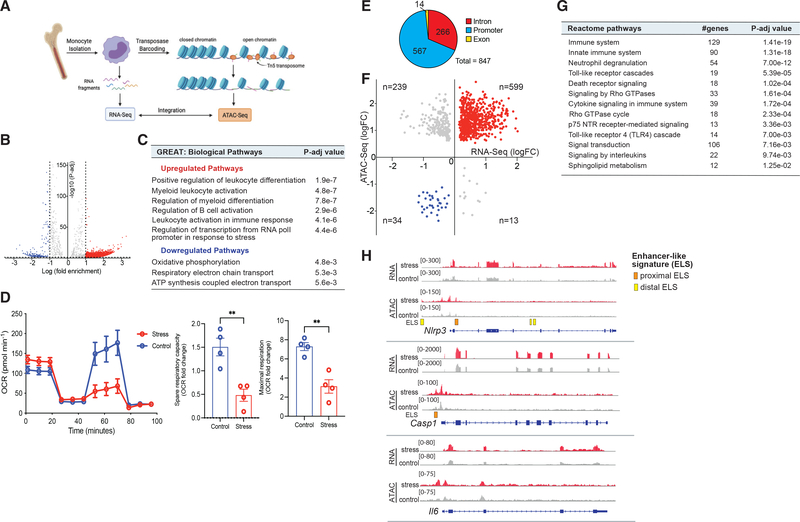
To investigate whether chronic stress altered the chromatin landscape of monocytes, we performed genome-wide profiling of open chromatin by assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-seq) in bone marrow monocytes from stressed and control mice (Figure 3A). ATAC-seq measures DNA accessibility using hyperactive Tn5 transposase, which preferentially inserts sequencing adaptors in areas of open chromatin (Figure 3A). Chronic stress induced marked changes in the chromatin landscape of bone marrow monocytes, with 3,922 regions of more accessible chromatin and 508 regions of reduced chromatin accessibility compared to controls (adjusted p < 0.05; Table S3). Genomic regions enrichment of annotations tool (GREAT) analysis of loci with increased accessibility following chronic stress identified gene pathways associated with Gene Ontology (GO) terms such as myeloid leukocyte activation, regulation of myeloid differentiation, leukocyte activation involved in the immune response, and regulation of transcription from RNA polymerase II (RNAPol II) promoter in response to stress (Figures 3B and 3C; Table S4). By contrast, genes in pathways associated with oxidative phosphorylation and mitochondrial respiration were less accessible in monocytes from stressed mice compared to control mice (Figures 3B and 3C; Table S4). Consistent with stress-induced metabolic reprogramming, Seahorse extracellular flux analysis confirmed that monocytes from stressed mice exhibited a decreased oxygen consumption rate (OCR), spare respiratory capacity, and maximal respiration (Figure 3D), reflective of reduced oxidative phosphorylation.
Figure 3. Epigenetic reprogramming of inflammatory gene loci by stress.
(A) Experimental design of assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-seq) of bone marrow monocytes from control and stressed mice and integration with RNA-seq.
(B) Volcano plot showing chromatin loci identified by ATAC-seq to be more (red) or less (blue) accessible in bone marrow monocytes from mice exposed to chronic variable stress versus control (n = 6, 3 mice/phenotype).
(C) Biological pathways were identified using the genomic regions enrichment of annotations tool (GREAT) analysis of loci with increased and decreased accessibility following chronic stress.
(D) Oxygen consumption rate (OCR) of monocytes isolated from the bone marrow of stressed or control mice (n = 4) and sequentially treated with oligomycin, carbonyl cyanide p-trifluoro-methoxyphenylhydrazone (FCCP), and rotenone plus antimycin (Rtn/AA). Quantification of spare and maximal respiration.
(E) Pie chart showing distribution of open chromatin loci in genes overlapping in RNA-seq and ATAC-seq datasets.
(F) Integration scatterplot showing overlap between chromatin accessibility (ATAC-seq) and gene expression (RNA-seq) in monocytes from stressed versus nonstressed mice. Differentially expressed (DE) genes adjusted p value < 0.05, FC > 1.5.
(G) Reactome pathways of overlapping ATAC-seq peaks and RNA-seq reads that were activated or inhibited in stressed mice relative to control mice.
(H) RNA-seq (top) and ATAC-seq (bottom) reads in bone marrow monocytes at the Nlrp3, Casp1, and Il6 loci. Enhancer-like sequences (ELSs) are indicated by boxed regions at bottom. Data in (D) are means ± SEMs. p values were calculated using a Mann-Whitney t test; **p ≤ 0.01 n = 4 mice/group.
To determine whether the changes in open chromatin regions from the ATAC-seq analysis correlated with gene expression changes found in stressed monocytes by RNA-seq, we analyzed the overlap of these datasets. The integration of peak annotation of open chromatin regions and transcriptomic RNA-seq analyses revealed a considerable overlap of open regions and transcripts differentially expressed in stressed mice compared to control mice: 429 genes that were expressed at significantly higher levels in bone marrow monocytes from stressed mice (adjusted p < 0.05, fold change [FC] > 1.5) also showed ≥1 regions of greater chromatin accessibility compared to control monocytes (Figures 3E and 3F), and 17 genes that were expressed at significantly lower levels in monocytes from stressed mice (adjusted p < 0.05, FC < −1.5) also had ≥1 associated closed chromatin regions (Figure 3E). The majority of ATAC-seq peaks in these loci occurred in genomic regions designated as promoters (60%) and introns (35%), consistent with stress-activated chromatin alterations in gene regulatory regions (Figure 3F). Pathway analysis of these loci revealed enrichment of genes linked to inflammation, cell proliferation, and morphology-associated pathways, including Toll-like receptor (TLR) cascades, cytokine signaling, G alpha signaling, and Rho GTPase cycle pathways (Figure 3G). Of interest, promoter regions of genes linked to inflammasome-IL-1 production (Nlrp3 and Casp1) and Il6 showed greater open chromatin domains in monocytes from stressed mice, consistent with their higher level of RNA expression, compared to monocytes from control mice (Figure 3H). These data indicate that stress priming of monocytes mediates changes to chromatin accessibility at inflammatory loci. Reminiscent of changes that occur during innate immune training, these findings indicate that PS can skew myeloid cells toward a primed hyperinflammatory state.
Stress-mediated priming reprograms myeloid cells to a hyperinflammatory phenotype
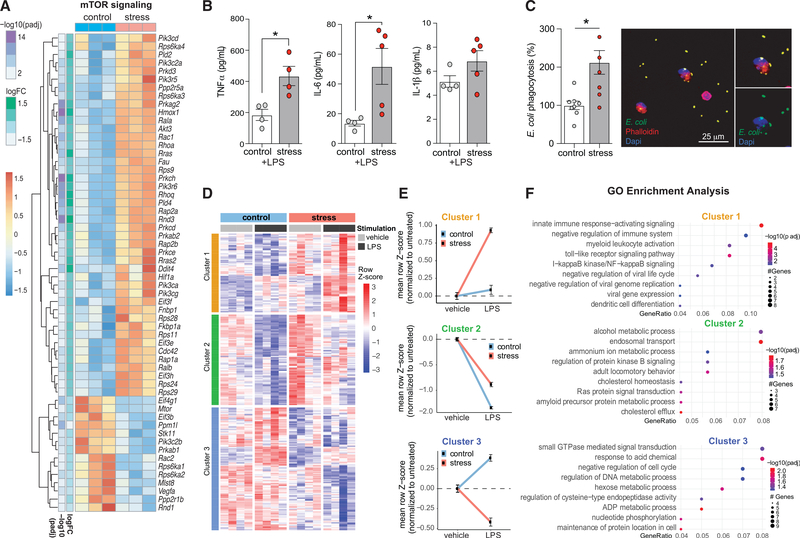
Metabolic rewiring of myeloid cells and their progenitors has been shown to be a driving force in mediating a hyperinflammatory immune response (Christ et al., 2018; Netea et al., 2016). In particular, activation of the Akt-phosphatidylinositol 3-kinase (PI3K) and mammalian target of rapamycin (mTOR) signaling pathway has been shown to mediate a shift from oxidative phosphorylation to aerobic glycolysis and subsequent histone modifications that are integral to innate immune training by microbial and sterile ligands (Bekkering et al., 2018; Cheng et al., 2014; Mitroulis et al., 2018). However, whether these alterations occur in stress-primed monocytes is unknown. Consistent with metabolic reprogramming, transcriptomic profiling of bone marrow monocytes from stressed versus control mice showed an increased expression of genes associated with Akt-PI3K (Figure S2) and mTOR signaling (Figure 4A) and the suppression of pathways linked to fatty acid oxidation (Figure S3. Hypoxia-inducible factor 1α (HIF-1α) is one of the transcription factors activated by mTOR that plays an important role in stimulating glycolysis and innate immune training, and we observed the upregulation of HIF-1α target genes, including Hif1a itself, as well as Hmox1, Fau, Pik3c2a, Pik3r5, Prkce, Eif3e, and Eif3f (Figure 4A). PI3K-mTOR-HIF-1α signaling has been shown to be an important regulator of trained cytokine responses (Cheng et al., 2014). Thus, we next investigated whether chronic variable stress induced functional reprogramming of the myeloid compartment in the bone marrow. Bone marrow monocytes isolated from stressed and control mice were stimulated ex vivo with lipopolysaccharide (LPS) for 6 h as a model of “secondary challenge,” and secretion of cytokines assessed. Monocytes from stressed mice produced higher levels of TNF-α and IL-6 in response to LPS challenge compared to monocytes from control mice (Figure 4B), consistent with a primed hyperinflammatory monocyte phenotype. To understand whether stressed monocytes exhibit a functional enhancement of phagocytosis, monocytes from stressed and control mice were exposed to fluorescently labeled Escherichia coli for 2 h. We observed a 2-fold increase in bacterial uptake in monocytes from stressed mice compared to controls, indicating functional enhancement of phagocytic capacity (Figure 4C).
Figure 4. Stress primes monocytes toward a hyperinflammatory phenotype characterized by alterations to metabolic processes, cytokine production, and efferocytotic capacity.
Heatmap showing the RNA-seq gene expression profile of the mammalian target of rapamycin (mTOR) signaling pathway in monocytes from mice exposed to chronic variable stress or nonstressed mice (n = 3/phenotype).
(B) Lipopolysaccharide (LPS) stimulated (10 ng/mL; 6 h) cytokine response of bone marrow monocytes isolated from mice exposed to chronic variable stress or nonstressed mice (n = 4–5 mice/phenotype).
(C) Phagocytosis of E. coli bioparticles by bone marrow monocytes isolated from stressed and nonstressed mice (n = 6 per phenotype).
(D) Gene expression heatmap of monocytes isolated from stressed and nonstressed mice, and subsequently stimulated with LPS (n = 4/group); DE genes defined from effect of stress treatment factor alone (297 genes, p < 0.01) segregate into 3 clusters using hierarchical clustering (cluster 1 = 80 genes, cluster 2 = 92 genes, cluster 3 = 125 genes).
(E) Mean expression of genes in each cluster for stress and nonstressed (control) mice in both basal and LPS stimulation.
(F) GO enrichment analysis was performed separately for each cluster using the ClusterProfiler package from BioConductor, using an adjusted p value cutoff of 0.05.
Data in (B) and (C) are means ± SEMs. p values were calculated using a Mann-Whitney test; *p ≤ 0.05.
We next performed RNA-seq profiling of untreated and LPS-stimulated monocytes from stressed and control mice (n = 4/group). To understand differential gene expression driven by stress, we used a multifactorial linear model. This analysis resulted in 297 genes that were differentially expressed between control and stressed mice (Figure 4D; p < 0.01; Table S5). Using hierarchical clustering of the expression patterns, we found that the expression profile of these 297 genes clustered into 3 distinct expression patterns (Figure 4E). We then performed GO enrichment pathway analysis of each gene block to understand the characteristics of each cluster (Figure 4F). Following normalization to basal expression, we found that monocytes from stressed mice had a hyperresponsive expression of genes linked to innate immune pathways following LPS stimulation (cluster 1; Figures 4D–4F). Consistent with metabolic reprogramming, the genes in cluster 2 showed changes in pathways associated with metabolism that were blunted in stressed mice relative to control mice following LPS challenge (cluster 2; Figures 4D–4F). Finally, the genes in cluster 3 showed discordant changes in expression following inflammatory challenge in monocytes from control and stressed mice that were related to cell-cycle regulation, metabolic processes, and small guanosine triphosphatase (GTPase) signal transduction (cluster 3; Figures 4D–4F). These data indicate that chronic variable stress induces monocyte reprogramming leading to altered responsiveness upon secondary challenge, consistent with innate immune priming.
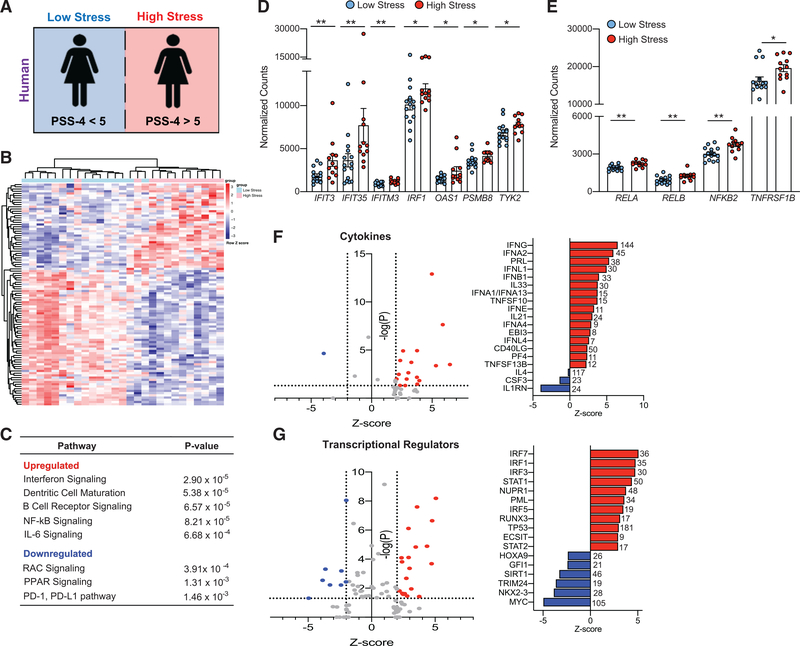
Stress alters the human blood transcriptional profile
In our murine model, we found that stress-induced priming skewed the myeloid transcriptome and chromatin accessibility landscape toward a hyperinflammatory phenotype. To assess the clinical relevance of these findings, we analyzed whole-blood RNA-seq from 27 women with and without MI enrolled into the Heart Attack Research Program (HARP), who were queried regarding their perceived stress over the preceding 4 weeks using the validated 4-item Perceived Stress Scale (PSS-4) questionnaire (Cohen et al., 1983) (demographics, Figure 5A; Table S6). In this age-matched cohort, the blood transcriptome profile of women with high (PSS-4 ≥5) stress (Redmond et al., 2013) (n = 12, PSS-4 median [interquartile range, IQR]: 7.5 [5.5–9]) was distinct from that of women who reported low (PSS-4 <5) stress (n = 15, PSS-4 median [IQR]: 3 [2–4]), with 1,959 genes (p < 0.05) differentially expressed in subjects reporting high versus low stress, after regressing out gene expression effects from MI status (Figure 5B; Table S7). IPA of differentially expressed genes found upregulation of inflammatory pathways also enriched in our mouse study, including IFN, IL-6, and NF-κB signaling, supporting a stress-induced hyperinflammatory state (Figure 5C; Table S8). Accordingly, we confirmed the increased expression of numerous IFN-stimulated genes (Figure 5D), as well as components of the NF-κB complex and the TNF receptor (Figure 5E) in the blood transcriptome of the high-stress cohort relative to those with low stress. After excluding women without MI, a similar enrichment pattern of pathways associated with IFN, IL-6, and NF-κB signaling was observed between high and low PS (Figure S4). Analysis of upstream regulators of genes differentially expressed in women with high and low stress identified IFNs (IFNG, INFA1, IFNA2, IFNA4, IFNB1, IFNL1, IFNE, IFNL4) and TNF family (TNFSF10, TNFSF13B) cytokines as predicted to be activated, and the IL-1 receptor antagonist predicted to be inhibited (Figure 5F). Consistent with this, transcriptional regulators predicted to drive the upregulation of inflammatory genes in stressed women were transcription factors associated with IFN signaling (STAT1, IRF7, IRF1, IRF3, IRF5), cell-cycle regulation and DNA repair (TP53, PML, PTTG1), macrophage activation (SPI1), and chromatin remodeling (SMARCA4) (Figure 5G). Among transcriptional regulators that were predicted to be decreased were GFI, a transcriptional repressor that antagonizes NF-κB activation and cell proliferation (Sharif-Askari et al., 2010); TRIM24, a negative regulator of P53 (Allton et al., 2009); and SIRT1, a negative regulator of mTOR signaling (Figure 5G) (Ghosh et al., 2010). These data are consistent with the hyperinflammatory primed myeloid phenotype found in stressed mice.
Figure 5. Stress alters the human blood transcriptional profile.
(A) Study design: whole blood was collected from women in PAXgene Blood RNA tubes, and RNA isolated and analyzed by RNA-seq. At the time of enrollment, the stress of the participants was determined by the validated Perceived Stress Scale 4 (PSS-4) questionnaire. Subjects were stratified into those with low stress (PSS-4 < 5, n = 15) and those with high stress (PSS-4 > 5, n = 12). See demographics in Table S6.
(B) Heatmap of the top 100 DE transcripts between low- and high-stress subjects, ranked by p value. Subjects and genes were hierarchically clustered with each row representing a single gene and each column representing an individual subject.
(C) Canonical pathways upregulated and downregulated as determined by Ingenuity Pathway Analysis (IPA) of significantly DE transcripts in low-stress versus high-stress subjects (p < 0.05).
(D and E) Expression of (D) interferon-related and (E) NF-κB-related transcripts DE in subjects with low and high stress.
(F and G) IPA upstream analysis of predicted (F) cytokines and (G) transcriptional regulators of genes differentially expressed between subjects with high and low stress. The number next to each bar represents genes in our dataset that have a measurement direction consistent with the activation of that cytokine or transcriptional regulator. Data in (D) and (E) are means ± SEMs. p values were calculated using a Mann-Whitney t test; *p ≤ 0.05, **p ≤ 0.01.
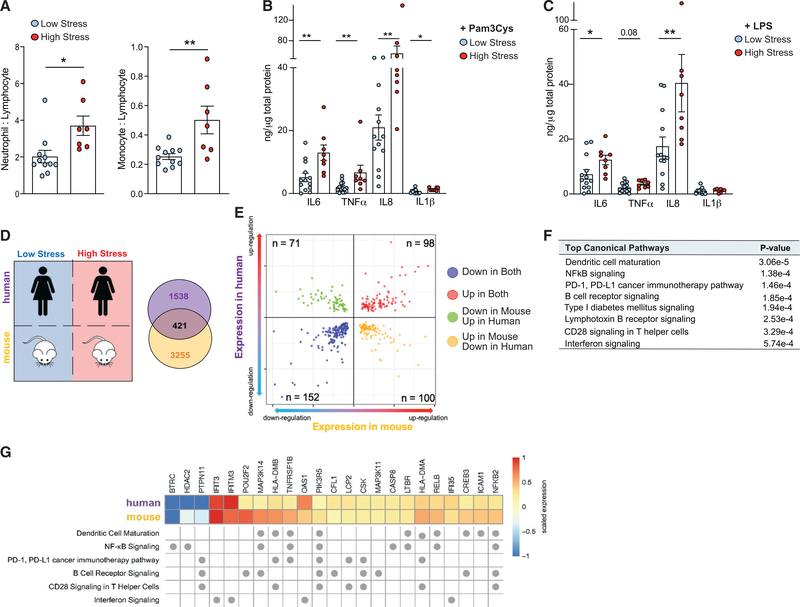
Monocytes from stressed individuals are hyperinflammatory, consistent with innate immune priming
To determine whether stress leads to altered functional immune responses, we collected adhesion-purified monocytes isolated from peripheral blood mononuclear cells (PBMCs) from women with high (n = 8, PSS-4 median [IQR]: 9.5 [9–11]) and low (n = 13, PSS-4 median [IQR]: 1 [1–3]) perceived stress. Subjects with high stress had higher white blood cell (WBC) counts, with absolute higher counts for neutrophils and monocytes (demographics, Table 1). In addition, the neutrophil:lymphocyte ratio and the monocyte:lymphocyte ratio, markers of subclinical inflammation, were higher in subjects with high stress compared to subjects with low stress (p = 0.01, p = 0.006, respectively; Figure 6A). Upon challenge with the TLR ligands Pam3Cys and LPS, monocytes from subjects with high stress produced higher levels of proinflammatory cytokines (TNF-α, IL-6, IL-8, and IL-1β) compared to low stress (Figures 6B and 6C; Figure S5).
Table 1.
Demographics of PBMC subjects
| Demographics | Low stress | High stress |
|---|---|---|
|
| ||
| N | 13 | 8 |
| PSS-4 score (median, IQR) | 1 (1–3) | 9.5 (9–11)*** |
| Age, y (median, IQR) | 69 (58–72) | 55 (48–61) |
| BMI (median, IQR) | 28.9 (28.2–34.9) | 24.3 (23.3–25.2) |
| Race (White, %) | 54.5 | 75 |
| Ethnicity (Hispanic, %) | 18 | 0 |
| Diabetes (%) | 50 | 12.5 |
| Hypertension (%) | 72.7 | 50 |
| Dyslipidemia (%) | 63.6 | 25 |
| Aspirin (%) | 90.9 | 100 |
| White blood cells (109/L, median, IQR) | 6.2 (5.2–7.5) | 9.4 (7.5–11.5)* |
| Neutrophils (109/L, median, IQR) | 3.5 (2.9–4.2) | 6.1 (4.8–8.5)* |
| Monocytes (109/L, median, IQR) | 0.5 (0.4–0.5) | 0.98 (0.52–1.05)** |
| Lymphocytes (109/L, median, IQR) | 1.8 (1.5–2.3) | 1.76 (1.4–2.3) |
| NLR (median, IQR) | 1.6 (1.6–2.1)* | 3.5 (2.6–5.1)** |
| MLR (median, IQR) | 0.25 (0.20–0.29)* | 0.46 (0.31–0.72)** |
NLR, Neutrophil:lymphocyte ratio; MLR, monocyte:lymphocyte ratio.
p < 0.05
p < 0.01
p < 0.001, as determined by an unpaired t test.
Figure 6. Stress priming induces hyperinflammatory immune phenotype characterized by increased cytokine production.
(A) Ratios of neutrophils to lymphocytes and monocytes to lymphocytes in peripheral blood of women enrolled in the HARP study (demographics, Table 1) stratified by low stress (PSS-4 < 5, n = 11) and high stress (PSS-4 > 5, n = 7).
(B and C) Cytokine production in monocyte purified PBMCs from low stress and high stress subjects following stimulation with (B) Pam3Cys (10 μg/mL) or (C) LPS (10 ng/mL). Data are means ± SEMs. p values were calculated using a Mann-Whitney test; *p ≤ 0.05 relative to the low-stress group.
(D) Comparison of human RNA-seq to mouse RNA-seq between high and low stress. Venn diagram depicting the 421-gene overlap in stress-induced differential gene expression in humans and mice. DE genes are defined by p < 0.05 in the human RNA-seq and adjusted p < 0.05 in the mouse RNA-seq.
(E) Two-way volcano plot depicting overlap between human DE genes (y axis) and mouse DE genes (x axis); red dots indicate genes upregulated (n = 98) and blue dots indicate genes downregulated (n = 152) in both datasets.
(F) Top common pathways of genes with concordant expression directionality in myeloid cells from chronically stressed humans and mice as determined by IPA (n = 250, 98 genes upregulated, 152 genes downregulated).
(G) Heatmap of human and mouse mRNA expression values for genes found in the top enriched pathways common to high-stress mice and humans. Fold change of mRNA expression is scaled to represent relative maximum (1) and minimum (−1) expression of genes within each organism. Pathway assignment of genes is indicated by dots below the heatmap.
Effects of stress on transcription common to humans and mice
To determine the overlap in our findings between humans and mice subjected to chronic stress, we integrated the RNA-seq datasets from myeloid cells isolated from humans and mice to identify genes commonly dysregulated by stress (Figure 6D). This analysis revealed 421 genes differentially expressed in response to PS in both humans and mice, with 98 upregulated and 152 downregulated transcripts with common directionality of expression across the cohorts (Figure 6E). Pathway analysis of genes with concordant directionality in expression in both species revealed enrichment in proinflammatory pathways, including IFN signaling, dendritic cell maturation, and NF-κB signaling (Figure 6F). IFN signaling was among the top canonical pathways enriched in both mouse and human, with the upregulation of IFIT3, IFITM3, IFI35, and OAS1 driving this association (Figure 6G). Other genes upregulated in stressed mice and humans included those coding for components of the TNF and IL-1 signaling cascades, such as the TNF receptors 1 and 2 (TNFRSF1, TNFRSF2), the NF-κB components p100 and RELB (NFKB2, RELB), the NF-κB-inducing kinase (MAP3K14) that participates in pathways involved in NF-κB activation, production of reactive oxygen and nitrogen species, and dendritic cell maturation (Figure 6G). These data identify common pathways upregulated in mice and humans and is consistent with stress-conferred innate immune priming.
DISCUSSION
PS is a risk factor for various human diseases, particularly CVD and other chronic inflammatory conditions. Many studies have reported associations of chronic stress with markers of inflammation (Fleshner and Crane, 2017; Powell et al., 2013), but the mechanisms by which stress influences the immune system remain poorly understood. Our study shows that chronic stress has enduring consequences on the amplitude of myeloid cell inflammatory responses in mice and humans due to inflammatory priming (Divangahi et al., 2021). Specifically, we found that stress induces a hyperinflammatory state in the myeloid cells of mice and humans, which is characterized by broad changes in chromatin architecture at inflammatory and metabolic gene loci, increased inflammatory gene expression, and augmented cytokine production upon secondary challenge. Notably, chronic stress induces the Akt-mTOR-HIF-1 and IFN signaling pathways known to be critical mediators of the metabolic and epigenetic rewiring of myeloid cells. IFN, a known mediator of epigenetic changes at inflammatory loci, was the top canonical pathway enriched in both humans and mice. Overall, these results indicate that like microbial danger signals, chronic stress imprints priming that confers a hyperresponsive state. While this may be beneficial to the organism in mounting an enhanced response to microbial infections, stress-induced priming has the potential to be maladaptive in the development of chronic inflammatory and autoimmune conditions.
Our study adds to the evidence that systemic perturbations associated with an unhealthy lifestyle can induce durable reprogramming of innate immune cells that contribute to a state of chronic inflammation. Like chronic stress, western diet feeding of mice was found to trigger hematopoietic expansion and functionally reprogram myeloid cell responses to microbial stimuli even after cessation of the high-fat, high-cholesterol diet (Christ et al., 2018). The lasting effects of the western diet were mediated by the epigenomic reprogramming of myeloid progenitor cells in the bone marrow, which led to increased responsiveness upon LPS stimulation. Notably, many of the monocytic inflammatory pathways we found to be upregulated in stress-primed myeloid cells were also observed in western diet-trained myeloid cells, including type I IFN response genes and the IL-6 pathway (Christ et al., 2018). Metabolic signaling has also been shown to be an important regulator of inflammatory memory responses in primed myeloid cells, resulting in a sensitized state with a lower threshold for activation, leading to an exaggerated inflammatory response (Crişan et al., 2017; Keane et al., 2021; Ullevig et al., 2012).
In addition to promoting glycolysis, activation of the Akt-mTOR signaling pathway is important for histone modifications, whose persistence facilitates the heightened inflammatory response upon subsequent stimulation. In addition, mTOR-dependent translation has previously been reported to be an essential aspect of macrophage priming in aging and neurodegeneration (Keane et al., 2021). Unbiased transcriptomic profiling of bone marrow monocytes isolated from stressed and control mice by RNA-seq showed an increased expression of genes associated with Akt-PI3K, mTOR-, and HIF-1α-signaling pathways and the suppression of signaling pathways linked to fatty acid oxidation after chronic stress. This shift in cellular metabolic signaling was reinforced by decreased chromatin accessibility at loci involved in oxidative phosphorylation and respiratory electron chain transport, which were the pathways most significantly inhibited at the epigenomic level. The increased expression of genes in PI3K- and mTOR-signaling pathways was also observed in monocytes from stressed mice stimulated ex vivo with LPS, which was paralleled by the increased expression of genes involved in myeloid immune responses such as NF-κB signaling, phagocytosis, and production of reactive oxygen and nitrogen species. Bone marrow monocytes from stressed mice showed heightened functional responses upon ex vivo challenge, including more robust secretion of proinflammatory cytokines and phagocytosis of bacteria.
Functional annotation analysis of differentially expressed genes identified a strong signature of IFN-stimulated gene expression in both stressed mice and humans, with both type I (IFN-α, IFN-β) and type II (IFN-γ) IFNs predicted to be upstream regulators. IFNs can induce pervasive genome-wide changes in histone acetylation at promoter and enhancer sites of genes encoding IFN-stimulated genes, as well as canonical NF-κB target genes such as proinflammatory cytokines, allowing sustained transcription (Barrat et al., 2019; Qiao et al., 2013). This chromatin remodeling is mediated by the sustained occupancy of the STAT1 and IRF transcription factors (Barrat et al., 2019; Qiao et al., 2013), which were among the top predicted transcriptional regulators of stress-induced genes in both mice and humans. IFNs can also drive HSC proliferation to expand myeloid cell progenitors in the bone marrow (Chavakis et al., 2019), as we observed in mice exposed to chronic stress, and can mediate the mTOR-HIF-1α-dependent metabolic switch from oxidative phosphorylation to glycolysis in dendritic cells (Cheng et al., 2016; Pantel et al., 2014).
Limitations of the study
Our study details how PS alters the functional, metabolic, and transcriptomic landscape of myeloid cells in mice and humans. Both murine and human studies included only females; thus, it is unknown whether observed phenotypes are directly translatable to males who experience high levels of PS. In both the murine and human cohorts, cells were profiled soon after the induction of PS (mice), or stress questionnaire (humans). In each case, the transcriptome of each cohort did not return to basal levels before the introduction of a secondary stimulus; thus, we cannot confidently ascertain whether the proinflammatory state of the myeloid cells is sustained over time. Future studies will aim to assess whether following the initial inflammatory priming by PS the cells return to baseline and whether the hyperinflammatory phenotype is sustained as assessed by RNA- and ATAC-seq, and metabolic profiling following a secondary challenge. In addition, studies should be extended to further validate functional changes in monocyte responses beyond the phagocytosis, metabolism, and limited panel of cytokines that we examined. Furthermore, it will be important to determine how these functional changes may contribute to the progression of inflammatory-based diseases. The cohort of women enrolled in this study were administered a validated stress questionnaire (PSS-4) that assessed their chronic perceived stress levels within the past month. Within this cohort, several women had experienced an MI, an acute event likely to alter the blood transcriptome on its own. To account for this limitation, we used a bioinformatics approach (multifactor linear modeling) to investigate the differential expression driven by the stress factor alone, after accounting for any differences mediated by MI status.
Our work shows that PS primes myeloid cells toward a hyperinflammatory phenotype characterized by broad changes in the transcriptome and chromatin accessibility. Our data indicate that metabolic reprogramming may underlie the changes induced by stress-conferred priming due to the suppression of oxidative phosphorylation, activation of cellular metabolic pathways, including mTOR and PI3Kinase pathways, and reduced chromatin accessibility at loci associated with mitochondrial respiration. However, it also raises questions about the mechanisms involved in sensing this complex endogenous trigger and how this is translated by the innate immune system. Stress-mediated crosstalk between the brain and immune system is mediated by both the hypothalamic-pituitary-adrenal axis, which activates glucocorticoid production, and the sympathetic-adrenal-medullary axis, which drives catecholamine release to prepare the body for fight-or-flight responses. Noradrenaline released during chronic stress by the sympathetic nervous system signals via the β3-adrenergic receptor to upregulate HSC proliferation (Heidt et al., 2014). Whereas acute exposure of human monocytes to catecholamines is immunosuppressive, a recent study showed that noradrenaline can induce a hyperresponsive immune phenotype in vitro (van der Heijden et al., 2020). Noradrenaline reduces monocyte secretion of TNF-α and IL-6 after 24 h, yet restimulation of noradrenaline primed monocytes 6 days later with LPS augmented cytokine secretion compared to controls (van der Heijden et al., 2020). The potential contribution of noradrenaline in innate immune priming by chronic variable stress requires further investigation. Therapeutic and behavioral intervention strategies may be beneficial in counteracting the excessive and hyperresponsive inflammation associated with chronic stress. Lipoprotein-based nanobiologics containing the mTOR inhibitor rapamycin have been shown to reverse b-glucan-stimulated trained immunity in vitro, including decreasing H3K4me3 at the promoters of inflammatory and glycolysis-related genes and normalizing cytokine induction upon secondary challenge (Braza et al., 2018). However, whether such approaches would be beneficial in the context of stress-induced priming remains to be established. While there is considerable interest in the clinical translation of therapies targeting hyperinflammation due to innate immune memory, such strategies will need to be approached with caution so as not to compromise the host defense against pathogens.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Prof. Kathryn Moore (Kathryn.moore@nyulangone.org).
Materials availability
This study did not generate new unique reagents
Data and code availability
RNA-seq and ATAC-seq data have been deposited at GEO under accession # GSE167536 and are publicly available as of the date of publication. Accession numbers are listed in the Key resources table.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| ckit antibody | Biolegend | Cat # 105839, RRID:AB_2629798 |
| Sca1 antibody | Biolegend | Cat # 108120, RRID:AB_493273 |
| CD16/CD32 antibody | Biolegend | Cat # 101320, RRID:AB_1574975 |
| CD34 antibody | BD Biosciences | Cat # 551387, RRID:AB_394176 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| LPS | Invivogen | Cat # tlrl-pb5lps |
|
| ||
| Critical commercial assays | ||
|
| ||
| Vybrant Phagocytosis Assay kit | Molecular Probes | Cat # V6694 |
| MACS monocyte isolation kit | Miltenyi | Cat # 130-100-629 |
| Human Inflammatory Cytokine Cytometric Bead Array (CBA) | BD Biosciences | Cat # 551811 |
| XF MitoStress kit | Seahorse Biosciences | Cat # 101706-100 |
| PAXgene plastic 2.5 mL 16x100mm | QIAGEN | Cat # 762165 |
| QIAsymphony PAXgene Blood RNA Kit | QIAGEN | Cat # 762635 |
| Corticosterone ELISA kit | Abcam | Cat # ab108821 |
| TruSeq RNA Library Prep Kit v2 | Illumina | RS-122-2001 |
| Tagment DNA Enzyme and Buffer kit | Illumina | 20034197 |
| Mouse IL-1β Enhanced Sensitivity Flex Set | BD Biosciences | Cat # 562278 |
| Mouse IL-6 Enhanced Sensitivity Flex Set | BD Biosciences | Cat # 562236 |
| Mouse MCP-1 Flex Set | BD Biosciences | Cat # 558342 |
|
| ||
| Deposited data | ||
|
| ||
| Raw and analyzed data | This paper | GEO: GSE167536 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| C57BL/6 mice | JAX labs | Stock #00064 |
|
| ||
| Oligonucleotides | ||
|
| ||
| Primers, murine csf; for CCTTCTTCGACATGGCTGGG | This paper | ThermoFisher |
| Primers, murine csf; rev GTTCTGACACCTCCTTGGCA | This paper | ThermoFisher |
| Primers, murine kitlg; for GCGGGAATCCTGTGACTGA | This paper | ThermoFisher |
| Primers, murine kitlg; rev CATCCCGGCGACATAGTTGA | This paper | ThermoFisher |
| Primers, murine cxcl12; for GCCAACGTCAAGCATCTGAA | This paper | ThermoFisher |
| Primers, murine cxcl12; re TTGTTGTTCTTCAGCCGTGC | This paper | ThermoFisher |
|
| ||
| Software and algorithms | ||
|
| ||
| Basepair | Basepair | https://www.basepairtech.com |
| GENEWIZ, LLC | South Painfield, NJ, USA | https://www.genewiz.com |
| FastQC (v.0.11.7) | Andrews, 2010 | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ |
| fastx_trimmer (v.0.0.13) | FASTX-Toolkit | http://hannonlab.cshl.edu/fastx_toolkit/ |
| Trimmomatic | Bolger et al., 2014 | http://www.usadellab.org/cms/?page=trimmomatic |
| STAR (v.2.6.1d) | Dobin et al., 2013 | https://github.com/alexdobin/STAR |
| featureCounts (v.1.6.3) | Subread | http://subread.sourceforge.net/ |
| DESeq2 (v.1.24.0) | Love et al., 2014 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| pheatmap (v.1.0.12) | Cran | https://cran.r-project.org/web/packages/pheatmap/index.html |
| biomaRt (v.2.40.5) | Durinck et al., 2005, 2009 | https://bioconductor.org/packages/release/bioc/html/biomaRt.html |
| ggplot2 (v.3.3.3) | Wickham, 2016 | https://cran.r-project.org/web/packages/ggplot2/index.html |
| Ingenuity Pathway Analysis (IPA) | QIAGEN Redwood City, CA | https://digitalinsights.qiagen.com/products-overview/discovery-insights-portfolio/analysis-and-visualization/qiagen-ipa/ |
| ClusterProfiler | Wu et al., 2021; Yu et al., 2012 | https://bioconductor.org/packages/release/bioc/html/clusterProfiler.html |
| REVIGO | Supek et al., 2011 | http://revigo.irb.hr/ |
| bcl2fastq Conversion software (v.2.20) | Illumina | https://support.illumina.com/sequencing/sequencing_software/bcl2fastq-conversion-software.html |
| Bowtie2 (v.2.3.4.1) | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| Sambamba (v.0.6.7) | Tarasov et al., 2015 | https://lomereiter.github.io/sambamba/ |
| MACS (in Python v.2.7.3) | Zhang et al., 2008 | https://github.com/macs3-project/MACS |
| NucleoATAC | Schep et al., 2015 | https://github.com/GreenleafLab/NucleoATAC |
| deepTools (v.2.3.3) | Ramírez et al., 2016 | https://github.com/deeptools/deepTools |
| DiffBind (v.3.3.0) | Stark and Brown 2011; Ross-Innes et al., 2012 | https://bioconductor.org/packages/release/bioc/html/DiffBind.html |
| Genomic Regions Enrichment of Annotations Tool (GREAT) | McLean et al., 2010 | http://great.stanford.edu/public/html/ |
| Integrative Genomics Viewer (IGV) (v.2.5.2) | Robinson et al., 2011; Thorvaldsdottir et al., 2013 | https://software.broadinstitute.org/software/igv/ |
| GraphPad Prism8 (GraphPad Sofware) | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| Python (v.2.7.3) | Python Software Foundation | https://www.python.org |
| R (v.4.0.2) | The R Project for Statistical Computing | https://www.r-project.org/ |
| RStudio (v.1.3.1093) | RStudio | https://www.rstudio.com/ |
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
In this study female C57BL/6 mice at 7 weeks of age were exposed to a validated protocol of chronic variable stress for 3 weeks. Mice were purchased from Jackson Laboratory (stock #00064). Mice were randomly allocated to treatment groups. Experimental procedures were done in accordance with the US Department of Agriculture Animal Welfare Act and the US Public Health Service Policy on Humane Care and Use of Laboratory Animals and were approved by the New York University School of Medicine’s Institutional Animal Care and Use Committee.
For the human stress study, women were enrolled into the Heart Attack Research Program (HARP) study. Participants electively referred for invasive coronary angiography without acute coronary syndrome and did not have obstructive coronary artery disease were identified as disease controls. The study was performed in accordance with policies of the New York University School of Medicine Institutional Review Board, and informed consent was obtained from each subject. Demographics of the patient cohort are presented in Table 1 of this paper.
METHOD DETAILS
Study design
The overall objective of our study was to investigate the role of stress to myeloid cells, and to establish if stress could mediate innate immune training. To investigate this, we used female C57BL/6 mice exposed to a validated protocol of chronic variable stress for 3 weeks (Figure 1A) (Heidt et al., 2014), followed by a rest period of up to 48 h, and isolated monocytes from stressed and stress-free controls. Sources of mouse data included the following: analysis of harvested tissues and cells by RT-qPCR, RNA-seq, ATAC-seq, histology, flow cytometry, in vitro stimulation assays (E. coli, LPS). Mice for RNA-seq and ATAC-seq were randomly selected from the larger cohort of mice. Sample processing and statistical analysis were performed concurrently on experimental and control groups using identical conditions. For all the experiments, sample sizes were determined by our previous data, prior literature, and power calculation to ensure sufficient sample sizes to allow the detection of statistically significant differences. Analyses (including cytokine measures, RT-qPCR) were performed blinded to exposure group.
Mouse monocyte isolation for RNA sequencing
Monocytes were isolated from the tibia and femurs of mice by centrifugation of bones at 300 g to collect total bone marrow cell suspensions, lysis of red blood cells (ACK lysing buffer, Thermo Fisher Scientific), and collection of purified monocytes by negative selection using magnetic-activated cell sorting (MACS, Miltenyi Biotec). Purified monocytes were lysed in TRIzol (Invitrogen), and RNA was isolated from bone marrow-derived monocytes using Direct-zol RNA isolation MicroPrep columns (Zymo Research). RNA was used to generate barcoded cDNA libraries using the NEBNext Ultra RNA library prep (New England BioLabs) with rRNA depletion. Indexed libraries were sequenced in the 2×150bp configuration on the Illumina HiSeq2500 platform by GENEWIZ, LLC. (South Plainfield, NJ, USA).
Mouse RNA-seq data processing and bioinformatics analyses
RNA-seq data was processed with Basepair software (https://www.basepairtech.com) using their default parameters to generate raw counts for mm10 transcripts. RNA-seq data was analyzed with R package DESeq2 (v.1.24.0) using a pairwise comparison of stress versus control samples (n = 6, 3 in each group, mice were randomly selected from the larger cohort of mice). Only genes with a CPM (counts per million) > 5 in at least 2 samples were included. Statistical significance was calculated using Wald test. A heatmap of the top 266 DE genes after a cutoff of adjusted p-value < 0.05 and absolute log (fold change) > 1.5 was generated using R package pheatmap (v.1.0.12) on Z-scaled normalized counts (Figure 2A). Ingenuity Pathway Analysis (IPA®, QIAGEN Redwood City, CA) was used to obtain top canonical pathways (Figure 2B), transcription regulators (Figure 2D) and cytokine regulators (Figure 2E) (BH adjusted p-value < 0.05 and pathway Z-score > 2), using the full list of DESeq2 results ranked by significance and log fold change.
Mouse chromatin isolation for ATAC sequencing
Chromatin profiling was performed by ATAC-seq as described previously (Buenrostro et al., 2013). Briefly, 50,000 to 100,000 cells were washed in cold PBS and lysed in cold lysis buffer (10 mM Tris-HCl, pH = 7.4, 10 mM NaCl, 3 mM MgCl2, 0.1% IGEPAL CA-630). Next, transposition reaction (Illumina) was performed at 42°C for 45 min. After purification of the DNA with the MinElute PCR purification kit (QIAGEN), material was amplified for 5 cycles using NEB Next Ultra DNA Library Prep (New England BioLabs). Additional PCR cycles were evaluated by real time PCR. Final product was cleaned by Ampure Beads at a 1.5 × ratio, and checked with Kapa-Roche quantification kit and size was verified on Tapestation high sensitivity Screentape. Normalized libraries were sequenced on a HiSeq2500 1T in a 50bp/50bp Paired end run at the Genome Technology Center at NYU, using the TruSeq SBS Kit v3 (Illumina). An average of 47 × 106 paired reads were generated per sample.
Mouse ATAC-seq data processing and bioinformatics analyses
Per-read per-sample FASTQ files were generated using the bcl2fastq Conversion software (v.2.20) to convert per-cycle BCL base call files outputted by the sequencing instrument into the FASTQ format. The alignment program, Bowtie2 (v.2.3.4.1), was used for mapping sequenced reads from the mouse samples to the mouse reference genome mm10 and the application Sambamba (v.0.6.7) was utilized to remove duplicate reads. The algorithm, MACS (in Python v.2.7.3), was used to call peaks of signal for annotated genomic features and, similarly, the Python package NucleoATAC (Schep et al., 2015) was functioned to call nucleosome positions. The computeMatrix and plotProfile tools in the deepTools suite (v.2.3.3) (Ramírez et al., 2016) were utilized for generation of signal profile plots. For the differential peak statistical comparisons between control and stress groups of samples (n = 6, 3 in each group), the DiffBind package (Bioconductor v.3.3.0) in the R statistical programming environment was used. Pathway enrichment of ATAC peaks was generated using the genomic regions enrichment of annotations tool (GREAT) (Figure 3D), using the entire genome as a background set (McLean et al., 2010). Integrative Genomics Viewer (v.2.5.2) was used to visualize individual RNA-seq and ATAC-seq tracks, summed across replicates.
Mouse LPS RNA isolation and sequencing
Murine bone marrow-derived monocytes from stressed and control mice were isolated as described above using the MACS monocyte isolation kit (Miltenyi Biotec). Each monocyte sample represents a pool of monocytes from 4 individual mice. 100,000 cells were seeded in a 96 wells plates and incubated for 2h in RPMI 1640 (Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (FBS, Life Technologies) and 1% penicillin/streptomycin (PS, Life Technologies) prior to a 6h exposure to lipopolysaccharide (LPS, 10 ng/mL) or vehicle control in RPMI 1640 containing 2% FBS and 1% PS. Subsequently, cells were lysed using TRIzol (Invitrogen) and RNA was extracted using Direct-zol RNA isolation MicroPrep columns (Zymo Research). RNA was used to generate barcoded cDNA libraries using the NEBNext Ultra RNA library prep (New England BioLabs) with rRNA depletion. Indexed libraries were sequenced in the 2×150bp configuration on the Illumina HiSeq2500 platform by GENEWIZ, LLC. (South Plainfield, NJ, USA).
Mouse LPS RNA-seq data processing and bioinformatics analyses
Quality control of sequencing reads was assessed using FastQC (v.0.11.7). Low-quality base noise in the beginning of the reads was trimmed using FASTX-Toolkit fastx_trimmer (v.0.0.13) using flags -Q33 -f 11. Reads were mapped to mouse reference genome mm10 using STAR (v.2.6.1d). Genomic features were assigned using Subread featureCounts (v.1.6.3). RNA-seq data from untreated and LPS stimulated monocytes from non-stressed and stressed mice (n = 16, 4 in each group) was analyzed with R package DESeq2 (v.1.24.0) using a multi-factorial linear model (~day_batch + stress_treatment + LPS_stimulation) that accounts for both the stress treatment (stress versus control) and the LPS stimulation (LPS versus untreated), as well as any potential batch effect from the pool collection date. Only genes with a CPM (counts per million) > 5 in at least 2 samples were included. Statistical significance was calculated using Wald test on the stress_treatment factor alone, after regressing out effects from day batch and LPS stimulation, giving 297 DE genes (p-value < 0.01). Hierarchical clustering was done on Z-score scaled normalized counts using default Pearson correlation with R package heatmap (v.1.0.12) (Figure 4D). The mean of scaled normalized counts of each of the four groups (control untreated, control with LPS, stressed untreated, and stressed with LPS) in each of the three clusters was calculated and normalized to untreated group in control and stress separately (Figure 4E). GO enrichment analysis was performed separately for each cluster using the ClusterProfiler package from BioConductor, using an adjusted p value cutoff of 0.05. Enrichment results were then filtered to reduce redundancy using REVIGO under default parameters (Supek et al., 2011) (Xu et al., 2020) (Figure 4F).
Human studies
For the MI study, women presenting with MI were enrolled into the Heart Attack Research Program (HARP) study. Participants electively referred for invasive coronary angiography without acute coronary syndrome and did not have obstructive coronary artery disease were identified as disease controls. Patients and controls had blood collected at the time of coronary angiography. Briefly, subjects > 21 years old on aspirin were recruited from New York University Langone Medical Center, or Bellevue Hospital. Major exclusion criteria were use of NSAIDs (other than aspirin) in the past week, antithrombotic therapy, or any known hemorrhagic diathesis. At time of enrollment validated measures of stress (Perceived Stress Scale [PSS-4]) were assessed (Cohen et al., 1983).
Human blood transcriptome profiling
Whole blood was collected into PAXgene Blood RNA tubes (PreAnalytiX GmbH– BD Biosciences, Mississauga, ON, Canada), and RNA was isolated. The quality and yield of the isolated RNA was determined with an Agilent 2100 Bioanalyzer (Agilent, Palo Alto, CA) prior to RNA-sequencing. RNA library preps were made and sequences as single-end mode at the Genome Technology Center at NYU using Illumina HiSeq4000. White blood cell counts between low and high stress subjects were not significantly different (low stress, median [IQR]: 7.1 [6.1, 8.6], high stress, median [IQR]: 7.2 [5.5, 12.8]) thus no adjustments for cell count were made across samples.
Human RNA-seq data processing and bioinformatics analyses
RNA-seq data processing was performed at the NYU Langone’s Applied Bioinformatics Laboratories using their standard RNA-seq pipeline. Briefly, quality control was assessed using FastQC (v.0.11.7). Reads were trimmed using trimmomatic (v.0.36). Reads were mapped to human reference genome hg19 using STAR (v.2.6.1d) (Dobin et al., 2013). Genomic features were assigned using featureCounts (v.1.6.3). RNA-seq data was analyzed with R package DESeq2 (v.1.24.0) (Love et al., 2014) using a multi-factor linear model (~stress_level + MI_status) that accounts for both “stress level” (low versus high) and “MI status” (MI versus control). Statistical significance was calculated using Wald test on the stress_level factor alone, after regressing out effects from MI status (n = 27, 12 high stress, 15 low stress). Heatmap of top 100 DE genes ranked by significance was generated using R package pheatmap (v.1.0.12) on Z-scaled normalized counts (Figure 5B). IPA was used to obtain up and downregulated pathways (Figure 5C), cytokines (Figure 5F) and transcriptional regulators (Figure 5G), using the full list of DESeq2 results ranked by significance and log fold change.
Human and mouse RNA-seq comparison
Top mouse DE genes (5926 genes, adjusted p-value < 0.05) from the stress versus control comparison were mapped using biomaRt R package (v.2.40.5) (Durinck et al., 2005) to obtain 3676 human ortholog genes (MGI.symbol to HGNC.symbol). The intersection between these 3676 mouse to human ortholog genes and the top human DE genes (1959 genes, p-value < 0.05, baseMeans ~4 or larger) resulted in 421 genes found in both datasets. These genes were plotted in a “two-way volcano plot” (Figure 6E) using ggplot2 (v.3.3.3) where x axis and y axis show their log fold change directionality in the mouse and human dataset respectively. IPA was used to obtain top canonical pathways enriched in genes with concordant directionality (Figure 6F) (n = 250, 98 genes upregulated in both datasets and 152 genes downregulated in both datasets), using log fold changes and p-values from the mouse dataset. Comparison of mRNA expression of genes in the top canonical pathways (Figure 6G) was done by scaling the fold change expression of the genes to represent the strongest upregulation with a 1 (maximum relative expression between genes) and the strongest downregulation with a −1 (minimum relative expression between genes), with 0 representing the true 0 fold change. This scaling was done for each organism to allow direct comparison.
White blood cell counts
Circulating murine leukocytes were quantified using the Complete Blood Cell (CBC) Element HT5 machine (HESKA).
Cytokine analysis
Monocytes were isolated, and sorted using the MACS monocyte isolation kit (Miltenyi Biotec), from the bone marrow of mice and maintained in RPMI 1640 medium (ThermoFisher) with 10% FBS for 2 hours. Media was then removed and replaced with media containing either LPS (10 ng/μL) for 6 hours. TNF-α, IL-6, and IL-1β levels were measured in the cell supernatant using the Cytometric Bead Array (CBA) multiplex immunoassay (BD Bioscience) according to manufacturer’s instructions.
Murine bone marrow-derived monocytes from stressed and control mice were isolated as described above using the MACS monocyte isolation kit (Miltenyi Biotec). 100,000 cells were seeded in a 96 wells plates and incubated for 2h in RPMI 1640 (Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (FBS, Life Technologies) and 1% penicillin/streptomycin (PS, Life Technologies) prior to a 6h exposure to lipopolysaccharide (LPS, 10 ng/mL) or vehicle control in RPMI 1640 containing 2% FBS and 1% PS. TNF-α, IL-6, and IL-1β levels were measured in the cell supernatant using the Cytometric Bead Array (CBA) multiplex immunoassay (BD Bioscience).
RNA Isolation and qPCR
Total RNA was isolated using TRIzol reagent (Invitrogen) and Direct-zol RNA MiniPrep or MicroPrep columns (Zymo Research). RNA was reverse transcribed using iScript cDNA Synthesis Kit (Bio-Rad Laboratories), and quantitative PCR (qPCR) analysis was conducted using KAPA SYBR green Supermix (KAPA Biosystems) according to the manufacturer’s instructions and quantified on a Quantstudio 3 (Applied Biosystems). Fold change in mRNA expression was calculated using the comparative cycle method (2−ΔΔCt) normalized to the housekeeping gene Hprt. Primers used in this study are presented in the resource table.
Phagocytosis assay
Murine bone marrow-derived monocytes from stressed and control mice were isolated as described above using the MACS monocyte isolation kit (Miltenyi Biotec). 100,000 cells were seeded in a 96 wells plates 100,000 cells were seeded in a 96 wells plates and incubated for 12 h in RPMI 1640 (Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (FBS, Life Technologies) and 1% penicillin/streptomycin (PS, Life Technologies). Phagocytic capacity of the cells in response to E. Coli bioparticles was then assessed using the Vybrant Phagocytosis Assay kit (Molecular Probes).
Human monocyte stimulation and cytokine production
Blood was collected at time of study enrollment in sodium citrate (3.2%) tubes and diluted with HBSS before layering on top of 50 mL Ficoll-Paque (GE Healthcare) in SepMate tubes (Stemcell). After centrifugation, the layer of mononuclear cells was washed and resuspended in RPMI 1640 supplemented with 10% FBS, and frozen in liquid nitrogen. On the day of stimulation, PBMCs were adhesion purified for monocytes on tissue culture plates for 2 h, as previously described (Bekkering et al., 2019). 500,000 monocytes/well were stimulated ex vivo in duplicate for 6 h with either 10 ng/mL LPS, 10 μg/mL Pam3Cys, or buffer control to study ex vivo cytokine production capacity. Cytokine production (IL-6, IL-1B, TNFα, IL-8) was determined in supernatants using BD Cytometric Bead Array. Supernatant cytokine concentrations are expressed relative to total cell protein concentration in each well as determined by a bicinchoninic acid protein assay (Pierce). All ex vivo stimulation and subsequent cytokine analyses were performed on the same day.
QUANTIFICATION AND STATISTICAL ANALYSIS
The statistical significance of differences was evaluated with the Student’s t test or two-way analysis of variance (ANOVA) for multiple group comparisons followed by Sidak’s multiple comparison test. P values of less than 0.05 were considered significant. Analyses were performed using GraphPad Prism8 (GraphPad Software). Numbers of replicates and statistical tests are indicated in each Figure legend.
ADDITIONAL RESOURCES
Heart Attack Research Program (HARP) study, ClinicalTrials.gov Identifier: NCT03022552.
Supplementary Material
Highlights.
Psychological stress reprograms monocytes to a hyperinflammatory primed phenotype
Stress-mediated priming activates transcriptomic and metabolic pathways in monocytes
Stress-mediated priming alters inflammatory responses in mice and humans
ACKNOWLEDGMENTS
This work was supported by grants from the NIH (R35HL135799 to K.J.M., P01HL131478 to K.J.M., E.A.F., F.S., and M.N., R01HL084312 to K.J.M. and E.A.F., R35HL144993 to J.S.B., and T32HL098129 to C.v.S. and A.A.C.N.), the American Heart Association (16SFRN28730002 to J.S.B., J.S.H., H.R.R., and T.M.S.; 19CDA34630066 to C.v.S.; and 118CDA34110203 to T.J.B.), and the American Society of Hematology (18-A0–00-1001884 to T.J.B.).
DECLARATION OF INTERESTS
M.N. has received funds or material research support from Lilly, Alnylam, Biotronik, CSL Behring, GlycoMimetics, GSK, Medtronic, Novartis, and Pfizer, as well as consulting fees from Biogen, Gimv, IFM Therapeutics, Molecular Imaging, Sigilon, and Verseau Therapeutics. H.R.R. receives in-kind donations for research from Abbott Vascular, Siemens, and BioTelemetry. All of the other authors declare no competing interests.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109595.
REFERENCES
- Ahmadi N, Hajsadeghi F, Mirshkarlo HB, Budoff M, Yehuda R, and Ebrahimi R (2011). Post-traumatic stress disorder, coronary atherosclerosis, and mortality. Am. J. Cardiol 108, 29–33. [DOI] [PubMed] [Google Scholar]
- Allton K, Jain AK, Herz HM, Tsai WW, Jung SY, Qin J, Bergmann A, Johnson RL, and Barton MC (2009). Trim24 targets endogenous p53 for degradation. Proc. Natl. Acad. Sci. USA 106, 11612–11616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S (2010). FastQC: a quality control tool for high throughput sequence data, Available online at. http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- Arnold SV, Smolderen KG, Buchanan DM, Li Y, and Spertus JA (2012). Perceived stress in myocardial infarction: long-term mortality and health status outcomes. J. Am. Coll. Cardiol. 60, 1756–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrat FJ, Crow MK, and Ivashkiv LB (2019). Interferon target-gene expression and epigenomic signatures in health and disease. Nat. Immunol. 20, 1574–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekkering S, Quintin J, Joosten LA, van der Meer JW, Netea MG, and Riksen NP (2014). Oxidized low-density lipoprotein induces long-term proinflammatory cytokine production and foam cell formation via epigenetic reprogramming of monocytes. Arterioscler. Thromb. Vasc. Biol. 34, 1731–1738. [DOI] [PubMed] [Google Scholar]
- Bekkering S, Arts RJW, Novakovic B, Kourtzelis I, van der Heijden CDCC, Li Y, Popa CD, Ter Horst R, van Tuijl J, Netea-Maier RT, et al. (2018). Metabolic Induction of Trained Immunity through the Mevalonate Pathway. Cell 172, 135–146.e9. [DOI] [PubMed] [Google Scholar]
- Bekkering S, Stiekema LCA, Bernelot Moens S, Verweij SL, Novakovic B, Prange K, Versloot M, Roeters van Lennep JE, Stunnenberg H, de Winther M, et al. (2019). Treatment with Statins Does Not Revert Trained Immunity in Patients with Familial Hypercholesterolemia. Cell Metab. 30, 1–2. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, and Usadel B (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braza MS, van Leent MMT, Lameijer M, Sanchez-Gaytan BL, Arts RJW, Pérez-Medina C, Conde P, Garcia MR, Gonzalez-Perez M, Brahmachary M, et al. (2018). Inhibiting Inflammation with Myeloid Cell-Specific Nanobiologics Promotes Organ Transplant Acceptance. Immunity 49, 819–828.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, and Greenleaf WJ (2013). Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavakis T, Mitroulis I, and Hajishengallis G (2019). Hematopoietic progenitor cells as integrative hubs for adaptation to and fine-tuning of inflammation. Nat. Immunol. 20, 802–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng SC, Quintin J, Cramer RA, Shepardson KM, Saeed S, Kumar V, Giamarellos-Bourboulis EJ, Martens JH, Rao NA, Aghajanirefah A, et al. (2014). mTOR- and HIF-1α-mediated aerobic glycolysis as metabolic basis for trained immunity. Science 345, 1250684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng SC, Scicluna BP, Arts RJ, Gresnigt MS, Lachmandas E, Giamarellos-Bourboulis EJ, Kox M, Manjeri GR, Wagenaars JA, Cremer OL, et al. (2016). Broad defects in the energy metabolism of leukocytes underlie immunoparalysis in sepsis. Nat. Immunol. 17, 406–413. [DOI] [PubMed] [Google Scholar]
- Christ A, Günther P, Lauterbach MAR, Duewell P, Biswas D, Pelka K, Scholz CJ, Oosting M, Haendler K, Baßler K, et al. (2018). Western Diet Triggers NLRP3-Dependent Innate Immune Reprogramming. Cell 172, 162–175.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S, Kamarck T, and Mermelstein R (1983). A global measure of perceived stress. J. Health Soc. Behav. 24, 385–396. [PubMed] [Google Scholar]
- Crişan TO, Cleophas MCP, Novakovic B, Erler K, van de Veerdonk FL, Stunnenberg HG, Netea MG, Dinarello CA, and Joosten LAB (2017). Uric acid priming in human monocytes is driven by the AKT-PRAS40 autophagy pathway. Proc. Natl. Acad. Sci. USA 114, 5485–5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimsdale JE (2008). Psychological stress and cardiovascular disease. J. Am. Coll. Cardiol. 51, 1237–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divangahi M, Aaby P, Khader SA, Barreiro LB, Bekkering S, Chavakis T, van Crevel R, Curtis N, DiNardo AR, Dominguez-Andres J, et al. (2021). Trained immunity, tolerance, priming and differentiation: distinct immunological processes. Nat. Immunol. 22, 2–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durinck S, Moreau Y, Kasprzyk A, Davis S, De Moor B, Brazma A, and Huber W (2005). BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics 21, 3439–3440. [DOI] [PubMed] [Google Scholar]
- Durinck S, Spellman PT, Birney E, and Huber W (2009). Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 4, 1184–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esler M (2017). Mental stress and human cardiovascular disease. Neurosci. Biobehav. Rev 74 (Pt B), 269–276. [DOI] [PubMed] [Google Scholar]
- Fleshner M, and Crane CR (2017). Exosomes, DAMPs and miRNA: Features of Stress Physiology and Immune Homeostasis. Trends Immunol. 38, 768–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furman D, Campisi J, Verdin E, Carrera-Bastos P, Targ S, Franceschi C, Ferrucci L, Gilroy DW, Fasano A, Miller GW, et al. (2019). Chronic inflammation in the etiology of disease across the life span. Nat. Med. 25, 1822–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh HS, McBurney M, and Robbins PD (2010). SIRT1 negatively regulates the mammalian target of rapamycin. PLoS ONE 5, e9199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golbidi S, Frisbee JC, and Laher I (2015). Chronic stress impacts the cardiovascular system: animal models and clinical outcomes. Am. J. Physiol. Heart Circ. Physiol. 308, H1476–H1498. [DOI] [PubMed] [Google Scholar]
- Heidt T, Sager HB, Courties G, Dutta P, Iwamoto Y, Zaltsman A, von Zur Muhlen C, Bode C, Fricchione GL, Denninger J, et al. (2014). Chronic variable stress activates hematopoietic stem cells. Nat. Med. 20, 754–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iso H, Date C, Yamamoto A, Toyoshima H, Tanabe N, Kikuchi S, Kondo T, Watanabe Y, Wada Y, Ishibashi T, et al. (2002). Perceived mental stress and mortality from cardiovascular disease among Japanese men and women: the Japan Collaborative Cohort Study for Evaluation of Cancer Risk Sponsored by Monbusho (JACC Study). Circulation 106, 1229–1236. [DOI] [PubMed] [Google Scholar]
- Itoh T, Nakajima S, Tanaka F, Nishiyama O, Matsumoto T, Endo H, Sakai T, Nakamura M, and Morino Y (2014). Impact of the Japan earthquake disaster with massive Tsunami on emergency coronary intervention and inhospital mortality in patients with acute ST-elevation myocardial infarction. Eur. Heart J. Acute Cardiovasc. Care 3, 195–203. [DOI] [PubMed] [Google Scholar]
- Keane L, Antignano I, Riechers SP, Zollinger R, Dumas AA, Offermann N, Bernis ME, Russ J, Graelmann F, McCormick PN, et al. (2021). mTOR-dependent translation amplifies microglia priming in aging mice. J. Clin. Invest. 131, e132727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kivimäki M, and Steptoe A (2018). Effects of stress on the development and progression of cardiovascular disease. Nat. Rev. Cardiol. 15, 215–229. [DOI] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, and Bejerano G (2010). GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28, 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitroulis I, Ruppova K, Wang B, Chen LS, Grzybek M, Grinenko T, Eugster A, Troullinaki M, Palladini A, Kourtzelis I, et al. (2018). Modulation of Myelopoiesis Progenitors Is an Integral Component of Trained Immunity. Cell 172, 147–161.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore KJ, Sheedy FJ, and Fisher EA (2013). Macrophages in atherosclerosis: a dynamic balance. Nat. Rev. Immunol. 13, 709–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea MG, Joosten LA, Latz E, Mills KH, Natoli G, Stunnenberg HG, O’Neill LA, and Xavier RJ (2016). Trained immunity: a program of innate immune memory in health and disease. Science 352, aaf1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantel A, Teixeira A, Haddad E, Wood EG, Steinman RM, and Longhi MP (2014). Direct type I IFN but not MDA5/TLR3 activation of dendritic cells is required for maturation and metabolic shift to glycolysis after poly IC stimulation. PLoS Biol. 12, e1001759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell ND, Sloan EK, Bailey MT, Arevalo JM, Miller GE, Chen E, Kobor MS, Reader BF, Sheridan JF, and Cole SW (2013). Social stress up-regulates inflammatory gene expression in the leukocyte transcriptome via β-adrenergic induction of myelopoiesis. Proc. Natl. Acad. Sci. USA 110, 16574–16579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Y, Giannopoulou EG, Chan CH, Park SH, Gong S, Chen J, Hu X, Elemento O, and Ivashkiv LB (2013). Synergistic activation of inflammatory cytokine genes by interferon-γ-induced chromatin remodeling and toll-like receptor signaling. Immunity 39, 454–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, and Manke T (2016). deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44 (W1), W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redmond N, Richman J, Gamboa CM, Albert MA, Sims M, Durant RW, Glasser SP, and Safford MM (2013). Perceived stress is associated with incident coronary heart disease and all-cause mortality in low- but not high-income participants in the Reasons for Geographic And Racial Differences in Stroke study. J. Am. Heart Assoc. 2, e000447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, and Mesirov JP (2011). Integrative genomics viewer. Nat. Biotechnol. 29, 24–26. 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosengren A, Hawken S, Ounpuu S, Sliwa K, Zubaid M, Almahmeed WA, Blackett KN, Sitthi-amorn C, Sato H, and Yusuf S; INTERHEART Investigators (2004). Association of psychosocial risk factors with risk of acute myocardial infarction in 11119 cases and 13648 controls from 52 countries (the INTERHEART study): case-control study. Lancet 364, 953–962. [DOI] [PubMed] [Google Scholar]
- Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ, Brown GD, Gojis O, Ellis IO, Green AR, et al. (2012). Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 481, 389–393. 10.1038/nature10730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schep AN, Buenrostro JD, Denny SK, Schwartz K, Sherlock G, and Greenleaf WJ (2015). Structured nucleosome fingerprints enable high-resolution mapping of chromatin architecture within regulatory regions. Genome Res. 25, 1757–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif-Askari E, Vassen L, Kosan C, Khandanpour C, Gaudreau MC, Heyd F, Okayama T, Jin J, Rojas ME, Grimes HL, et al. (2010). Zinc finger protein Gfi1 controls the endotoxin-mediated Toll-like receptor inflammatory response by antagonizing NF-kappaB p65. Mol. Cell. Biol. 30, 3929–3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegel A, Shivtiel S, Kalinkovich A, Ludin A, Netzer N, Goichberg P, Azaria Y, Resnick I, Hardan I, Ben-Hur H, et al. (2007). Catecholaminergic neurotransmitters regulate migration and repopulation of immature human CD34+ cells through Wnt signaling. Nat. Immunol. 8, 1123–1131. [DOI] [PubMed] [Google Scholar]
- Stark R, and Brown G (2011). DiffBind: differential binding analysis of ChIP-Seq peak data. http://bioconductor.org/packages/release/bioc/vignettes/DiffBind/inst/doc/DiffBind.pdf.
- Supek F, Bošnjak M, Škunca N, and Šmuc T (2011). REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 6, e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarasov A, Vilella AJ, Cuppen E, Nijman IJ, and Prins P (2015). Sambamba: fast processing of NGS alignment formats. Bioinformatics 31, 2032–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdóttir H, Robinson JT, and Mesirov JP (2013). Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 14, 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng YS, Li H, Kang YL, Chen WC, Cheng WC, and Lai DM (2011). Loss of Cxcl12/Sdf-1 in adult mice decreases the quiescent state of hematopoietic stem/progenitor cells and alters the pattern of hematopoietic regeneration after myelosuppression. Blood 117, 429–439. [DOI] [PubMed] [Google Scholar]
- Ullevig S, Zhao Q, Lee CF, Seok Kim H, Zamora D, and Asmis R (2012). NADPH oxidase 4 mediates monocyte priming and accelerated chemotaxis induced by metabolic stress. Arterioscler. Thromb. Vasc. Biol. 32, 415–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Heijden CDCC, Groh L, Keating ST, Kaffa C, Noz MP, Kersten S, van Herwaarden AE, Hoischen A, Joosten LAB, Timmers HJLM, et al. (2020). Catecholamines Induce Trained Immunity in Monocytes In Vitro and In Vivo. Circ. Res. 127, 269–283. [DOI] [PubMed] [Google Scholar]
- Wickham H (2016). ggplot2: Elegant Graphics for Data Analysis (New York: Springer-Verlag; ), ISBN 978–3-319–24277-4. https://ggplot2.tidyverse.org. [Google Scholar]
- Wilbert-Lampen U, Leistner D, Greven S, Pohl T, Sper S, Völker C, Güthlin D, Plasse A, Knez A, Küchenhoff H, and Steinbeck G (2008). Cardiovascular events during World Cup soccer. N. Engl. J. Med. 358, 475–483. [DOI] [PubMed] [Google Scholar]
- Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, Feng T, Zhou L, Tang W, Zhan L, et al. (2021). clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. The Innovation 2, 100141. 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Lee SK, Zhang D, and Frenette PS (2020). The Gut Microbiome Regulates Psychological-Stress-Induced Inflammation. Immunity 53, 417–428.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Wang LG, Han Y, and He QY (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, and Liu XS (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq and ATAC-seq data have been deposited at GEO under accession # GSE167536 and are publicly available as of the date of publication. Accession numbers are listed in the Key resources table.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| ckit antibody | Biolegend | Cat # 105839, RRID:AB_2629798 |
| Sca1 antibody | Biolegend | Cat # 108120, RRID:AB_493273 |
| CD16/CD32 antibody | Biolegend | Cat # 101320, RRID:AB_1574975 |
| CD34 antibody | BD Biosciences | Cat # 551387, RRID:AB_394176 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| LPS | Invivogen | Cat # tlrl-pb5lps |
|
| ||
| Critical commercial assays | ||
|
| ||
| Vybrant Phagocytosis Assay kit | Molecular Probes | Cat # V6694 |
| MACS monocyte isolation kit | Miltenyi | Cat # 130-100-629 |
| Human Inflammatory Cytokine Cytometric Bead Array (CBA) | BD Biosciences | Cat # 551811 |
| XF MitoStress kit | Seahorse Biosciences | Cat # 101706-100 |
| PAXgene plastic 2.5 mL 16x100mm | QIAGEN | Cat # 762165 |
| QIAsymphony PAXgene Blood RNA Kit | QIAGEN | Cat # 762635 |
| Corticosterone ELISA kit | Abcam | Cat # ab108821 |
| TruSeq RNA Library Prep Kit v2 | Illumina | RS-122-2001 |
| Tagment DNA Enzyme and Buffer kit | Illumina | 20034197 |
| Mouse IL-1β Enhanced Sensitivity Flex Set | BD Biosciences | Cat # 562278 |
| Mouse IL-6 Enhanced Sensitivity Flex Set | BD Biosciences | Cat # 562236 |
| Mouse MCP-1 Flex Set | BD Biosciences | Cat # 558342 |
|
| ||
| Deposited data | ||
|
| ||
| Raw and analyzed data | This paper | GEO: GSE167536 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| C57BL/6 mice | JAX labs | Stock #00064 |
|
| ||
| Oligonucleotides | ||
|
| ||
| Primers, murine csf; for CCTTCTTCGACATGGCTGGG | This paper | ThermoFisher |
| Primers, murine csf; rev GTTCTGACACCTCCTTGGCA | This paper | ThermoFisher |
| Primers, murine kitlg; for GCGGGAATCCTGTGACTGA | This paper | ThermoFisher |
| Primers, murine kitlg; rev CATCCCGGCGACATAGTTGA | This paper | ThermoFisher |
| Primers, murine cxcl12; for GCCAACGTCAAGCATCTGAA | This paper | ThermoFisher |
| Primers, murine cxcl12; re TTGTTGTTCTTCAGCCGTGC | This paper | ThermoFisher |
|
| ||
| Software and algorithms | ||
|
| ||
| Basepair | Basepair | https://www.basepairtech.com |
| GENEWIZ, LLC | South Painfield, NJ, USA | https://www.genewiz.com |
| FastQC (v.0.11.7) | Andrews, 2010 | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ |
| fastx_trimmer (v.0.0.13) | FASTX-Toolkit | http://hannonlab.cshl.edu/fastx_toolkit/ |
| Trimmomatic | Bolger et al., 2014 | http://www.usadellab.org/cms/?page=trimmomatic |
| STAR (v.2.6.1d) | Dobin et al., 2013 | https://github.com/alexdobin/STAR |
| featureCounts (v.1.6.3) | Subread | http://subread.sourceforge.net/ |
| DESeq2 (v.1.24.0) | Love et al., 2014 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| pheatmap (v.1.0.12) | Cran | https://cran.r-project.org/web/packages/pheatmap/index.html |
| biomaRt (v.2.40.5) | Durinck et al., 2005, 2009 | https://bioconductor.org/packages/release/bioc/html/biomaRt.html |
| ggplot2 (v.3.3.3) | Wickham, 2016 | https://cran.r-project.org/web/packages/ggplot2/index.html |
| Ingenuity Pathway Analysis (IPA) | QIAGEN Redwood City, CA | https://digitalinsights.qiagen.com/products-overview/discovery-insights-portfolio/analysis-and-visualization/qiagen-ipa/ |
| ClusterProfiler | Wu et al., 2021; Yu et al., 2012 | https://bioconductor.org/packages/release/bioc/html/clusterProfiler.html |
| REVIGO | Supek et al., 2011 | http://revigo.irb.hr/ |
| bcl2fastq Conversion software (v.2.20) | Illumina | https://support.illumina.com/sequencing/sequencing_software/bcl2fastq-conversion-software.html |
| Bowtie2 (v.2.3.4.1) | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| Sambamba (v.0.6.7) | Tarasov et al., 2015 | https://lomereiter.github.io/sambamba/ |
| MACS (in Python v.2.7.3) | Zhang et al., 2008 | https://github.com/macs3-project/MACS |
| NucleoATAC | Schep et al., 2015 | https://github.com/GreenleafLab/NucleoATAC |
| deepTools (v.2.3.3) | Ramírez et al., 2016 | https://github.com/deeptools/deepTools |
| DiffBind (v.3.3.0) | Stark and Brown 2011; Ross-Innes et al., 2012 | https://bioconductor.org/packages/release/bioc/html/DiffBind.html |
| Genomic Regions Enrichment of Annotations Tool (GREAT) | McLean et al., 2010 | http://great.stanford.edu/public/html/ |
| Integrative Genomics Viewer (IGV) (v.2.5.2) | Robinson et al., 2011; Thorvaldsdottir et al., 2013 | https://software.broadinstitute.org/software/igv/ |
| GraphPad Prism8 (GraphPad Sofware) | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| Python (v.2.7.3) | Python Software Foundation | https://www.python.org |
| R (v.4.0.2) | The R Project for Statistical Computing | https://www.r-project.org/ |
| RStudio (v.1.3.1093) | RStudio | https://www.rstudio.com/ |
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.