We demonstrate universal questionnaire-free screening for autism with significant reduction in false positives.
Abstract
Here, we develop digital biomarkers for autism spectrum disorder (ASD), computed from patterns of past medical encounters, identifying children at high risk with an area under the receiver operating characteristic exceeding 80% from shortly after 2 years of age for either sex, and across two independent patient databases. We leverage uncharted ASD comorbidities, with no requirement of additional blood work, or procedures, to estimate the autism comorbid risk score (ACoR), during the earliest years when interventions are the most effective. ACoR has superior predictive performance to common questionnaire-based screenings and can reduce their current socioeconomic, ethnic, and demographic biases. In addition, we can condition on current screening scores to either halve the state-of-the-art false-positive rate or boost sensitivity to over 60%, while maintaining specificity above 95%. Thus, ACoR can significantly reduce the median diagnostic age, reducing diagnostic delays and accelerating access to evidence-based interventions.
INTRODUCTION
Autism spectrum disorder (ASD) is a developmental disability associated with significant social and behavioral challenges. Although ASD may be diagnosed as early as the age of 2 (1), children frequently remain undiagnosed until after the fourth birthday (2). With genetic and metabolomic tests (3–6) still in their infancy, a careful review of behavioral history and a direct observation of symptoms are currently necessary (7, 8) for a clinical diagnosis. Starting with a positive initial screen, a confirmed diagnosis of ASD is a multistep process that often takes 3 months to 1 year, delaying entry into time-critical intervention programs. While lengthy evaluations (9), cost of care (10), lack of providers (11), and lack of comfort in diagnosing ASD by primary care providers (11) are all responsible to varying degrees (12), one obvious source of this delay is the number of false positives produced in the initial ASD-specific screening tools in use today. For example, a large-scale study of the modified checklist for autism in toddlers with follow-up (M-CHAT/F) (13) (n = 20,375), which is used commonly as a screening tool (8, 14), demonstrated that it has an estimated sensitivity of 38.8%, specificity of 94.9%, and positive predictive value (PPV) of 14.6%. Thus, currently, out of every 100 children with ASD, M-CHAT/F flags about 39, and out of every 100 children it flags, about 85 are false positives, exacerbating wait times and queues (12). Automated screening that might be administered with no specialized training, requires no behavioral observations, and is functionally independent of the tools used in current practice, has the potential for immediate transformative impact on patient care.
While the neurobiological basis of autism remains poorly understood, a detailed assessment conducted by the U.S. Centers for Disease Control and Prevention (CDC) demonstrated that children with ASD experience higher-than-expected rates of many diseases (1). These include conditions related to dysregulation of immune pathways such as eczema, allergies, and asthma, as well as ear and respiratory infections, gastrointestinal problems, developmental issues, severe headaches, migraines, and seizures (15, 16). In the present study, we exploit these comorbidities to estimate the risk of childhood neuropsychiatric disorders on the autism spectrum. We refer to the risk estimated by our approach as the autism comorbid risk score (ACoR). Using only sequences of diagnostic codes from past doctor’s visits, our risk estimator reliably predicts an eventual clinical diagnosis, or the lack thereof, for individual patients. Thus, the key clinical contribution of this study is the formalization of subtle comorbidity patterns as a reliable screening tool to potentially improve wait times for diagnostic evaluations by significantly reducing the number of false positives encountered in initial screens in current practice.
A screening tool that tracks the risk of an eventual ASD diagnosis, based on the information already being gathered during regular doctor’s visits, and which may be implemented as a fully automated background process requiring no time commitment from providers has the potential to reduce avoidable diagnostic delays at no additional burden of time, money, and personnel resources. Use of patterns emergent in the diagnostic history to estimate risk might help reduce the subjective component in questionnaire-based screening tools, resulting in (i) reduced effect of potential language and cultural barriers in diverse populations and (ii) possibly better identification of children with milder symptoms (8). Furthermore, being functionally independent of the M-CHAT/F, we show that there is a clear advantage to combining the outcomes of the two tools: We can take advantage of any population stratification induced by the M-CHAT/F scores to significantly boost combined screening performance (see Materials and Methods and section S8).
Use of sophisticated analytics to identify children at high risk is a topic of substantial current interest, with independent progress being made by several groups (17–23). Many of these approaches focus on analyzing questionnaires, with recent efforts demonstrating the use of automated pattern recognition in video clips of toddler behavior. However, the inclusion of older kids above the age of 5 years and small cohort sizes might limit the immediate clinical adoption of these approaches for universal screening.
Laboratory tests for ASD have also begun to emerge, particularly leveraging detection of abnormal metabolites in plasma (3, 6) and salivary poly-omic RNA (5). However, as before, inclusion of older children limits applicability in screening, where we need a decision at 18 to 24 months. In addition, such approaches, while instrumental in deciphering ASD pathophysiology, might be too expensive for universal adoption at this time.
In contrast to ACoR, the above approaches require additional data or tests. Use of comorbidity patterns derived from past electronic health records (EHRs) either has been limited to establishing correlative associations (24, 25) or has substantially underperformed (26) [area under the curve (AUC) ≦65 % ] compared to our results.
MATERIALS AND METHODS
We view the task of predicting ASD diagnoses as a binary classification problem: Sequences of diagnostic codes are classified into positive and control categories, where “positive” refers to children eventually diagnosed with ASD, as indicated by the presence of a clinical diagnosis [International Classification of Diseases, Ninth Revision, with Clinical Modification (ICD-9-CM) code 299.X] in their medical records. Of the two independent sources of clinical incidence data used in this study, the primary source used to train our predictive pipeline is the Truven Health Analytics MarketScan Commercial Claims and Encounters Database for the years 2003 to 2012 (27), currently maintained by IBM Watson Health (referred to in this study as the Truven dataset). This U.S. national database merges data contributed by over 150 insurance carriers and large self-insurance companies, and comprises over 4.6 billion inpatient and outpatient service claims and almost 6 billion diagnosis codes. We extracted histories of patients within the age of 0 to 6 years and excluded patients for whom one or more of the following criteria fails: (i) at least one code pertaining to one of the 17 disease categories we use (see later for discussion of disease categories) is present in the diagnostic history and (ii) the first and last available record for a patient are at least 15 weeks apart. These exclusion criteria ensure that we are not considering patients who have too few observations. In addition, during validation runs, we restricted the control set to patients observable in the databases to those whose last record is not before the first 200 weeks of life. The characteristics of excluded patients are shown in Table 1. We trained with over 30 million diagnostic records (16,649,548 for males and 14,318,303 for females with 9835 unique codes). While the Truven database is used for both training and out-of-sample cross-validation with held-back data, our second independent dataset comprising de-identified diagnostic records for children treated at the University of Chicago Medical Center between the years of 2006 and 2018 (the UCM dataset) aids in further cross-validation. We considered children between the ages of 0 and 6 years and applied the same exclusion criteria as the Truven dataset. The number of patients used from the two databases is shown in Table 1. Our datasets are consistent with documented ASD prevalence and median diagnostic age [3 years in the claims database versus 3 years 10 months to 4 years in the United States (28)] with no significant geospatial prevalence variation (see fig. S1).
Table 1. Patient counts in de-identified data and the fraction of datasets excluded by our exclusion.
criteria. Dataset sizes are after the exclusion criteria are applied.
| Truven | UCM | |||
| Distinct patients | 115,805,687 | 69,484 | ||
| Male | Female | Male | Female | |
| ASD diagnosis count* |
12,146 | 3018 | 307 | 70 |
| Control count* | 2,301,952 | 2,186,468 | 20,249 | 17,386 |
| AUC at 125 weeks | 82.3% | 82.5% | 83.1% | 81.37% |
| AUC at 150 weeks | 84.79% | 85.26% | 82.15% | 83.39% |
| Excluded fraction of the datasets | ||||
| Positive category | 0.0002 | 0.0 | 0.0160 | 0.0 |
| Control category | 0.0045 | 0.0045 | 0.0413 | 0.0476 |
| Average number of diagnostic codes in excluded patients (corresponding number in included patients) | ||||
| Positive category | 4.33 (35.93) | 0.0 (36.07) | 2.6 (9.75) | 0.0 (10.18) |
| Control category | 1.57 (17.06) | 1.48 (15.96) | 2.32 (6.8) | 2.07 (6.79) |
*Cohort sizes are smaller than the total number of distinct patients due to the following exclusion criteria: (i) At least one code within our complete set of tracked diagnostic codes is present in the patient record, and (ii) time lag between first and last available record for a patient is at least 15 weeks.
Table 2. Engineered features (total count, 165).
| Feature type* | Description | No. of features |
| [Disease category]Δ | Likelihood defect (see Materials and Methods) |
17 |
| [Disease category]0 | Likelihood of control model (see Materials and Methods) |
17 |
| [Disease category]proportion | Occurrences in the encoded sequence/length of the sequence |
17 |
| [Disease category]streak | Maximum length of adjacent occurrences of [disease category] |
51 |
| [Disease category]prevalence |
Maximum, mean, and variance of occurrences in the encoded sequence/total number of diagnostic codes in the mapped sequence |
51 |
| Feature mean, feature variance, and feature maximum for difference of control and case models |
Mean, variance, and maximum of the [disease category]Δ values |
3 |
| Feature mean, feature variance, and feature maximum for control models |
Mean, variance, and maximum of the [disease category]0 values |
3 |
| Streak | Maximum, mean, and variance of the length of adjacent occurrences of [disease category] |
3 |
| Intermission | Maximum, mean, and variance of the length of adjacent empty weeks |
3 |
*Disease categories are described in table S1.
The significant diversity of diagnostic codes (6089 distinct ICD-9 codes and 11,522 distinct ICD-10-CM codes in total in the two datasets), along with the sparsity of codes per sequence and the need to make good predictions as early as possible, makes this a difficult learning problem. We proceed by partitioning the disease spectrum into 17 broad categories, e. g., infectious diseases, immunologic disorders, endocrinal disorders, etc. Each patient is then represented by 17 distinct time series, each tracking an individual disease category. At the population level, these disease-specific sparse stochastic time series are compressed into specialized Markov models (separately for the control and the treatment cohorts) to identify the distinctive patterns pertaining to elevated ASD risk. Each of these inferred models is a probabilistic finite state automaton (PFSA) (29). See section S10 for details on PFSA inference (30, 31). The number of states and connectivities in the PFSA models are explicitly inferred from data, with the algorithms generating higher-resolution models when needed and opting for a lower resolution otherwise automatically. This results in a framework with fewer nominal free parameters compared to standard neural network or deep learning architectures: The simplest deep learning model we investigated has more than 185,000 trainable parameters, while our final pipeline has 13,744. A smaller set of free parameters implies a smaller sample complexity, i. e., can be learned with less data, which, at least in the present application, gives us better performance relative to the standard architectures we investigated (see fig. S5).
After inferring models of diagnostic history, we need to quantify how such models diverge in the positive cohort versus those underlying the control cohort. We use a novel approach to evaluate subtle deviations in stochastic observations known as the sequence likelihood defect (SLD) (32), to quantify similarity of observed time series of diagnostic events to the control versus the positive cohorts for individual patients. This novel stochastic inference approach provides significant boost to the overall performance of our predictors; with only state-of-the-art machine learning, the predictive performance is significantly worse [see sections S2 and S3, as well as reported performance in the literature for predicting ASD risk from EHR data with standard algorithms (26)].
We briefly outline the SLD computation: To reliably infer the cohort type of a new patient, i. e., the likelihood of a diagnostic sequence being generated by the corresponding cohort model, we generalize the notion of Kullbeck-Leibler (KL) divergence (33, 34) between probability distributions to a divergence (G‖H) between ergodic stationary categorical stochastic processes (35) G and H as
| () | 1 |
where ∣x∣ is the sequence length, and pG(x) and pH(x) are the probabilities of sequence x being generated by the processes G and H, respectively, and the log-likelihood of x being generated by a process G is defined as follows:
| () | 2 |
The cohort type for an observed sequence x, which is actually generated by the hidden process G, can be formally inferred from observations based on the following provable relationships (section S10, theorems S6 and S7)
| () | 3a |
| () | 3b |
where ℋ(·) is the entropy rate of a process (33). Equation 3 shows that the computed likelihood has an additional nonnegative contribution from the divergence term when we choose the incorrect generative process. Thus, if a patient is eventually going to be diagnosed with ASD, then we expect that the disease-specific mapped series corresponding to her diagnostic history be modeled by the corresponding model in the positive cohort. Denoting the model corresponding to disease category j for positive and control cohorts as respectively, we can compute the SLD (Δj) as
| () | 4 |
With the inferred models and the individual diagnostic history, we estimate the SLD measure on the right-hand side of Eq. (4). The higher this likelihood defect, the higher the similarity of diagnosis history to that of children with autism.
In addition to the category-specific Markov models, we use a range of engineered features that reflect various aspects of the diagnostic histories, including the proportion of weeks in which a diagnostic code is generated, the maximum length of consecutive weeks with codes, and the maximum length of weeks with no codes (see table S2 for complete description), resulting in a total of 165 different features that are evaluated for each patient. With these inferred patterns included as features, we train a second-level predictor that learns to map individual patients to the control or the positive groups based on their similarity to the identified Markov models of category-specific diagnostic histories and the other engineered features (see section S1 for detailed mathematical details).
Since we need to infer the Markov models before the calculation of the likelihood defects, we need two training sets: one that is used to infer the models and one that subsequently trains the final classifier with features derived from the inferred models along with other engineered features. Thus, the analysis proceeds by first carrying out a random three-way split of the set of unique patients (in the Truven dataset) into Markov model inference (25%), classifier training (25%), and test (50%) sets. The approximate sample sizes of the three sets are as follows: ≈700,000 for each of the training sets, and ≈1.5 million for the test set. The features used in our pipeline may be ranked in order of their relative importance (see Fig. 1B for the top 15 features), by estimating the loss in performance when dropped out of the analysis. We verified that different random splits do not adversely affect performance. The UCM dataset in its entirety is used as a test set, with no retraining of the pipeline.
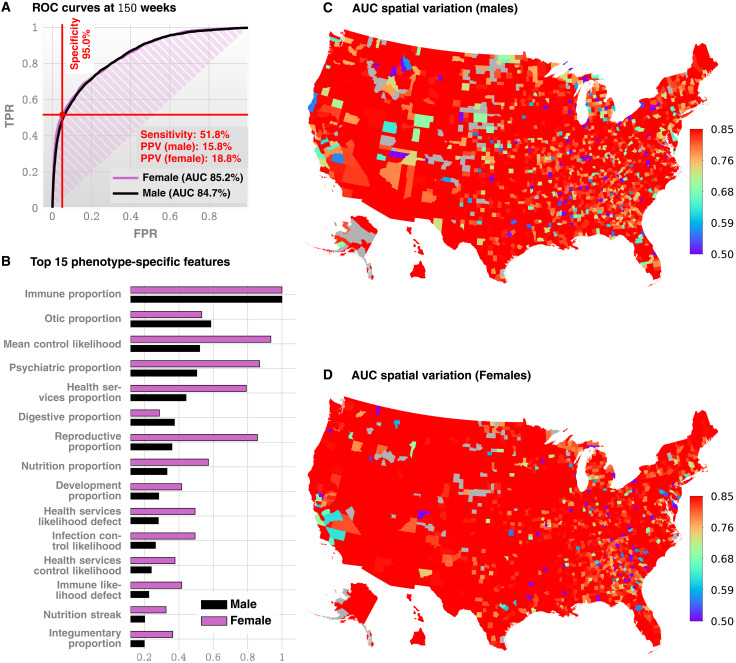
Fig. 1. Standalone predictive performance of ACoR.
(A) Receiver operating characteristic (ROC) curves for males and females (Truven data shown, UCM is similar, see Fig. 2A). (B) Feature importance inferred by our prediction pipeline. The detailed description of the features is given in Table 2. The most important feature is related to immunologic disorders, and we note that in addition to features related to individual disease categories, we also have the mean control likelihood (rank 3), which may be interpreted as the average likelihood of the diagnostic patterns corresponding to the control category as opposed to the positive category. (C and D) Spatial variation in the achieved predictive performance at 150 weeks, measured by AUC, for males and females, respectively. Gray areas lack data on either positive or negative cases. These county-specific AUC plots show that the performance of the algorithm has relatively weak geospatial dependence, which is important in the light of the current uneven distribution of diagnostic resources. Not all counties have nonzero number of ASD patients; high performance in those counties reflects a small number of false positives with zero false negatives.
Our pipeline maps medical histories to a raw indicator of risk. Ultimately, to make crisp predictions, we must choose a decision threshold for this raw score. In this study, we base our analysis on maximizing the F1 score, defined as the harmonic mean of sensitivity and specificity, to make a balanced trade-off between type 1 and type 2 errors (see section S4). The relative risk is then defined as the ratio of the raw risk to the decision threshold, and a value >1 predicts a future ASD diagnosis. Our two-step learning algorithm outperforms standard tools and achieves a stable performance across datasets strictly superior to documented M-CHAT/F.
The independence of our approach from questionnaire-based screening implies that we can further boost our performance by conditioning the sensitivity/specificity trade-offs on individual M-CHAT/F scores. In particular, we leverage the population stratification induced by M-CHAT/F to improve combined performance. Here, a combination of ACoR with M-CHAT/F refers to the conditional tuning of the sensitivity/specificity for ACoR in each subpopulation such that the overall performance is maximized. To describe this approach briefly, we assume that there are m subpopulations with the sensitivities and specificities achieved, and the prevalences in each subpopulation are given by si, ci, and ρi, respectively, with i ∈ {1, ⋯, m}. Let βi be the relative size of each subpopulation. Then, we have (see section S8A)
| (5a) |
| (5b) |
where we have denoted
| () | 5c |
and s, c, and ρ are the overall sensitivity, specificity, and prevalence, respectively. Knowing the values of γi, , we can carry out an m-dimensional search to identify the feasible choices of si, ci pairs for each i, such that some global constraint is satisfied, e. g., minimum values of specificity, sensitivity, and PPV. We consider four subpopulations defined by M-CHAT/F score brackets (13), and if the screen result is considered a positive (high risk, indicating the need for a full diagnostic evaluation) or a negative, i. e., low risk: (i) score ≤2 screening ASD negative, (ii) score [3 to 7] screening ASD negative on follow-up, (iii) score [3 to 7] screening ASD positive on follow-up, and (iv) score ≥8 screening ASD positive (see table S3). The “follow-up” in the context of M-CHAT/F refers to the re-evaluation of responses by qualified personnel. We use published data from the Children’s Hospital of Philadelphia (CHOP) study (13) on the relative sizes and the prevalence statistics in these subpopulations to compute the feasible conditional choices of our operating point to vastly supersede standalone M-CHAT/F performance. The CHOP study is the only large-scale study of M-CHAT/F we are aware of with sufficient follow-up after the age of 4 years to provide a reasonable degree of confidence in the sensitivity of M-CHAT/F.
Two limiting operating conditions are of particular interest in this optimization scheme, (i) where we maximize PPV under some minimum specificity and sensitivity (denoted as the high precision or the HP operating point) and (ii) where we maximize sensitivity under some minimum PPV and specificity (denoted as the high recall or the HR operating point). Taking these minimum values of specificity, sensitivity, and PPV to be those reported for M-CHAT/F, we identify the feasible set of conditional choices in a four-dimensional decision space that would significantly outperform M-CHAT/F in universal screening. The results are shown in Fig. 1B.
We carried out a battery of tests to ensure that our results are not significantly affected by class imbalance (since our control cohort is orders of magnitude larger) or systematic coding errors, e. g., we verified that restricting the positive cohort to children with at least two distinct ASD diagnostic codes in their medical histories instead of one has little impact on out-of-sample predictive performance (fig. S6B).
Use of de-identified patient data for the UCM dataset was approved by the Institutional Review Board (IRB) of the University of Chicago. An exemption was granted because of the de-identified nature of the data (IRB Committee: Biological Sciences Division, contact: A. Horst, ahorst@medicine.bsd.uchicago.edu. IRB#: Predictive Diagnoses IRB19-1040).
RESULTS
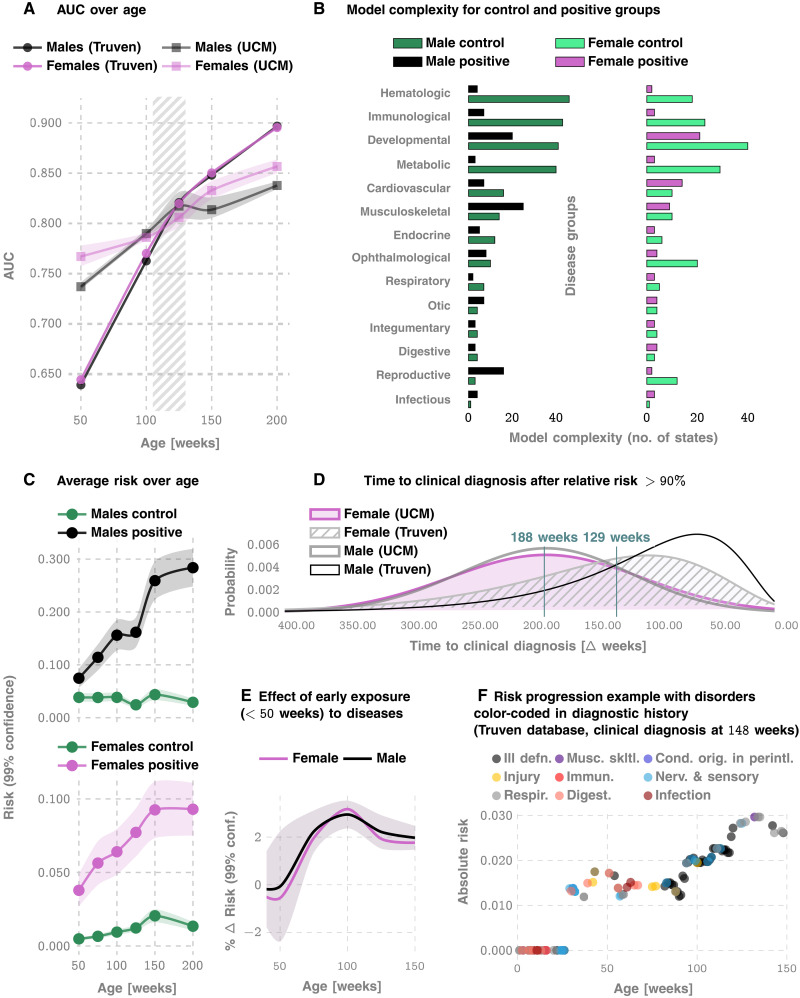
We measure our performance using several standard metrics including the AUC, sensitivity, specificity, and the PPV. For the prediction of the eventual ASD status, we achieve an out-of-sample AUC of 82.3 and 82.5% for males and females, respectively, at 125 weeks for the Truven dataset. In the UCM dataset, our performance is comparable: 83.1 and 81.3% for males and females, respectively (Figs. 1 and 2). Our AUC is shown to improve approximately linearly with patient age: Fig. 2A illustrates that the AUC reaches 90% in the Truven dataset at the age of 4. Note that Fig. 2C suggests that the risk progressions are somewhat monotonic. In addition, the computed confidence bounds suggest that the odds of a child with low risk up to 100 or 150 weeks of age abruptly shifting to a high risk trajectory are low. Thus, “borderline” cases need to be surveilled longer, while robust decisions may be obtained in other cases much earlier.
Fig. 2. More details on standalone predictive performance of ACoR and variation of inferred risk.
(A) AUC achieved as a function of patient age, for the Truven and UCM datasets. The shaded area outlines the 2 to 2.5 years of age and shows that we achieve >80% AUC for either sex from shortly after 2 years. (B) How inferred models differ between the control versus the positive cohorts. (C) How the average risk changes with time for the control and the positive cohorts. Note that the risk progressions are somewhat monotonic, and the computed confidence bounds suggest that the odds of a child with low risk up to 100 or 150 weeks of age abruptly shifting to a high-risk trajectory are low. (D) The distribution of the prediction horizon: The time to a clinical diagnosis after inferred relative risk crosses 90%. (E) shows that for each new disease code for a low-risk child, ASD risk increases by approximately 2% for either sex. (F) The risk progression of a specific, ultimately autistic male child in the Truven database. ill defn., symptoms, signs, and ill-defined conditions; musc. skltl., diseases of the musculoskeletal system and connective tissue; cond. orig. in perintl., certain conditions originating in the perinatal period; immun., endocrine, nutritional and metabolic diseases, and immunity disorders; nerv. & sensory, diseases of the nervous system and sense organs; respir., respiratory disorders; and digest., digestive disorders. On average, models become less complex, implying that the exposures become more statistically independent.
Recall that the UCM dataset is used purely for validation, and good performance on these independent datasets lends strong evidence for our claims. Furthermore, applicability in new datasets without local retraining makes it readily deployable in clinical settings.
Enumerating the top 15 predictive features (Fig. 1B), ranked according to their automatically inferred weights (the feature “importances”), we found that while infections and immunologic disorders are the most predictive, there is a significant effect from each of the 17 disease categories. Thus, the comorbid indicators are distributed across the disease spectrum, and no single disorder is uniquely implicated (see also Fig. 2F). Predictability is relatively agnostic to the number of local cases across U.S. counties (Fig. 1, C and D), which is important in light of the current uneven distribution of diagnostic resources (12, 36) across states and regions.
Unlike individual predictions that only become relevant over 2 years, the average risk over the populations is clearly different from around the first birthday (Fig. 2C), with the risk for the positive cohort rapidly rising. Also, we see a saturation of the risk after ≈3 years, which corresponds to the median diagnosis age in the database. Thus, if a child is not diagnosed up to that age, then the risk falls, since the probability of a diagnosis in the population starts to go down after this age. While average discrimination is not useful for individual patients, these reveal important clues as to how the risk evolves over time. In addition, while each new diagnostic code within the first year of life increases the risk burden by approximately 2% irrespective of sex (Fig. 2E), distinct categories modulate the risk differently; e. g., for a single random patient illustrated in Fig. 2F, infections and immunological disorders dominate early, while diseases of the nervous system and sensory organs, as well as ill-defined symptoms, dominate the latter period.
Given these results, it is important to ask how much earlier we can trigger an intervention. On average, the first time the relative risk (risk divided by the decision threshold set to maximize F1 score; see Materials and Methods) crosses the 90% threshold precedes diagnosis by ≈188 weeks in the Truven dataset and ≈129 weeks in the UCM dataset. This does not mean that we are leading a possible clinical diagnosis by over 2 years; a significant portion of this delay arises from families waiting in queue for diagnostic evaluations. Nevertheless, since delays are rarely greater than 1 year (12), we are still likely to produce valid red flags significantly earlier than the current practice.
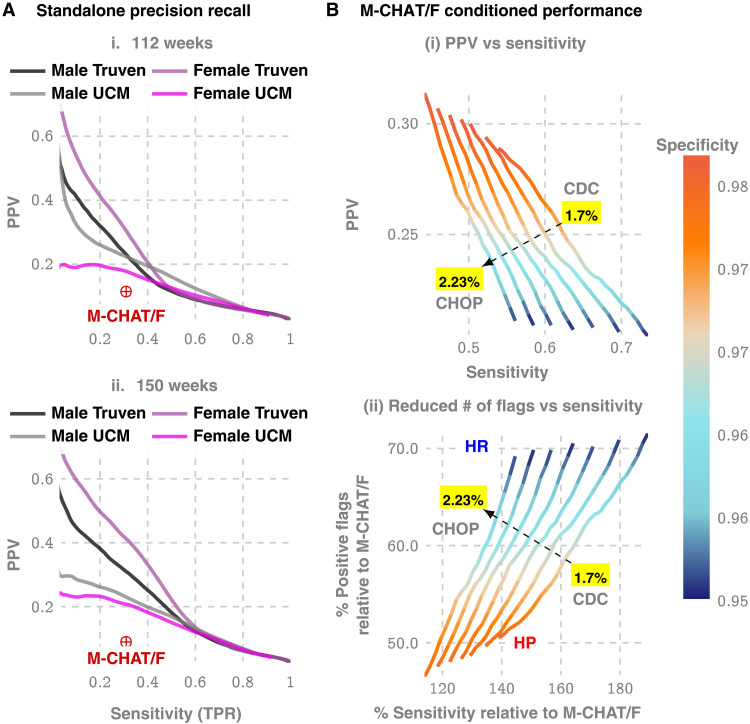
Our approach produces a strictly superior PPV, exceeding M-CHAT/F PPV by at least 14% (14.1 to 33.6%) when sensitivity and specificity are held at comparable values (approximately 38 and 95%) around the age of 26 months (≈112 weeks, note that 26 consecutive months is approximately 112 to 113 weeks). Figure 3A and Table 3 show the out-of-sample PPV versus sensitivity curves for the two databases, stratified by sex, computed at 100, 112, and 100 weeks. A single illustrative operating point is also shown on the receiver operating characteristic (ROC) curve in Fig. 1C, where, at 150 weeks, we have a sensitivity of 51.8% and a PPV of 15.8 and 18.8% for males and females, respectively, both at a specificity of 95%.
Fig. 3. Metrics relevant to clinical practice: PPV versus sensitivity trade-offs.
(A) Precision/recall curves, i. e., the trade-off between PPV and sensitivity for standalone operation with ACoR. (B) How we can boost ACoR performance using population stratification from the distribution of M-CHAT/F scores in the population, as reported by the CHOP study (13). This is possible because ACoR and M-CHAT/F use independent information (comorbidities versus questionnaire responses). Note that the population prevalence affects this optimization, and hence we have a distinct curve for each prevalence value (1.7% is the CDC estimate, while 2.23% is reported by the CHOP study). The two extreme operating zones marked as high precision (HP) and high recall (HR): if we choose to operate in HR, then we do not reduce the number of positive screens by much but maximize sensitivity, while by operating in HP, we increase sensitivity by 20 to 40% (depending on the prevalence) but double the PPV achieved in current practice. In contrast, when choosing to maximize sensitivity by operating in the HR zone, we only cut down positive flags to about 70% of what we get with M-CHAT/F, but boost sensitivity by 50 to 90% (reaching sensitivities over 70%). Note that, in all these zones, we maintain specificity above 95%, which is the current state of art, implying that by doubling the PPV, we can halve the number of positive screens currently reported, thus potentially sharply reducing the queues and wait times.
Table 3. Standalone PPV achieved at 100, 112, and 150 weeks for each dataset and gender [M-CHAT/F: sensitivity = 38:8%; specificity = 95%; and PPV = 14:6% between 16 and 26 months (≈112 weeks)].
| Weeks | Specificity | Sensitivity | PPV | Gender | Dataset |
| 100 | 0.92 | 0.39 | 0.14 | F | UCM |
| 100 | 0.95 | 0.39 | 0.19 | M | UCM |
| 100 | 0.93 | 0.39 | 0.13 | F | Truven |
| 100 | 0.91 | 0.39 | 0.10 | M | Truven |
| 112 | 0.93 | 0.39 | 0.16 | F | UCM |
| 112 | 0.95 | 0.39 | 0.20 | M | UCM |
| 112 | 0.96 | 0.39 | 0.22 | F | Truven |
| 112 | 0.95 | 0.39 | 0.17 | M | Truven |
| 150 | 0.94 | 0.39 | 0.19 | F | UCM |
| 150 | 0.98 | 0.39 | 0.34 | F | Truven |
| 150 | 0.97 | 0.39 | 0.26 | M | Truven |
| 150 | 0.97 | 0.39 | 0.26 | M | UCM |
Beyond standalone performance, independence from standardized questionnaires implies that we stand to gain substantially from a combined operation. With the recently reported population stratification induced by M-CHAT/F scores (13) (table S4), we can compute a conditional choice of sensitivity for our tool, in each subpopulation [M-CHAT/F score brackets, 0 to 2, 3 to 7 (negative assessment), 3 to 7 (positive assessment), and >8], leading to a significant performance boost. With such conditional operation, we get a PPV close to or exceeding 30% at the HP operating point across datasets (>33% for Truven and >28% for UCM) or a sensitivity close to or exceeding 50% for the HR operating point (>58% for Truven and >50% for UCM), when we restrict specificities to above 95% [see Table 4, Fig. 3B (i), and fig. S10]. Compared with standalone M-CHAT/F performance [Fig. 3B (ii)], we show that for any prevalence between 1.7 and 2.23%, we can double the PPV without losing sensitivity at >98% specificity, or increase the sensitivity by ∼50% without sacrificing PPV and keeping specificity ≧94%.
Table 4. Personalized operation conditioned on M-CHAT/F scores at 26 months.
NEG, negative; POS, positive.
| M-CHAT/F outcome | Global performance (Truven) | Global performance (UCM) | Prevalence* | |||||||
| 0–2 NEG | 3–7 NEG | 3–7 POS | ≥8 POS | Specificity | Sensitivity | PPV | Specificity | Sensitivity | PPV | |
| Specificity choices | ||||||||||
| 0.2 | 0.54 | 0.83 | 0.98 | 0.95 | 0.585 | 0.209 | 0.95 | 0.505 | 0.186 | 0.022 |
| 0.21 | 0.53 | 0.83 | 0.98 | 0.95 | 0.586 | 0.208 | 0.95 | 0.506 | 0.184 | 0.022 |
| 0.42 | 0.87 | 0.98 | 0.99 | 0.98 | 0.433 | 0.331 | 0.98 | 0.347 | 0.284 | 0.022 |
| 0.48 | 0.87 | 0.97 | 0.99 | 0.98 | 0.432 | 0.331 | 0.98 | 0.355 | 0.289 | 0.022 |
| 0.38 | 0.54 | 0.94 | 0.98 | 0.95 | 0.736 | 0.203 | 0.95 | 0.628 | 0.178 | 0.017 |
| 0.3 | 0.55 | 0.94 | 0.98 | 0.95 | 0.737 | 0.203 | 0.95 | 0.633 | 0.179 | 0.017 |
| 0.58 | 0.96 | 0.98 | 0.99 | 0.98 | 0.492 | 0.302 | 0.98 | 0.373 | 0.247 | 0.017 |
| 0.59 | 0.96 | 0.98 | 0.99 | 0.98 | 0.491 | 0.303 | 0.98 | 0.372 | 0.248 | 0.017 |
| 0.46 | 0.92 | 0.97 | 0.99 | 0.977 | 0.534 | 0.291 | 0.977 | 0.448 | 0.256 | 0.017 |
| 0.48 | 0.92 | 0.97 | 0.99 | 0.978 | 0.533 | 0.292 | 0.978 | 0.448 | 0.257 | 0.017 |
*Prevalence reported by CDC is 1.7%, while the CHOP study reports a value of 2.23%. The results of our optimization depend on the prevalence estimate.
DISCUSSION
In this study, we operationalize a documented aspect of ASD symptomology in that it has a wide range of comorbidities (15, 16, 37) occurring at above-average rates (8). Association of ASD with epilepsy (38), gastrointestinal disorders (39–44), mental health disorders (45), insomnia, decreased motor skills (46), allergies including eczema (39–44), and immunologic (37, 47–53) and metabolic (43, 54, 55) disorders is widely reported. These studies, along with support from large-scale exome sequencing (56, 57), have linked the disorder to putative mechanisms of chronic neuroinflammation, implicating immune dysregulation and microglial activation (49, 52, 58–61) during important brain developmental periods of myelination and synaptogenesis. However, these advances have not yet led to clinically relevant diagnostic biomarkers. Majority of the comorbid conditions are common in the control population, and rate differentials at the population level do not automatically yield individual risk (62).
ASD genes exhibit extensive phenotypic variability, with identical variants associated with diverse individual outcomes not limited to ASD, including schizophrenia, intellectual disability, language impairment, epilepsy, neuropsychiatric disorders, and also typical development (63). In addition, no single gene can be considered “causal” for more than 1% of cases of idiopathic autism (64).
Despite these hurdles, laboratory tests and potential biomarkers for ASD have begun to emerge (3, 5, 6). These tools are still in their infancy and have not demonstrated performance in the 18- to 24-month age group. In the absence of clinically useful biomarkers, current screening in pediatric primary care visits uses standardized questionnaires to categorize behavior. This is susceptible to potential interpretative biases arising from language barriers, as well as social and cultural differences, often leading to systematic underdiagnosis in diverse populations (8). In this study, we use time-stamped sequence of past disorders to elicit crucial information on the developing risk of an eventual diagnosis, and formulate the autism comorbid risk score. The ACoR is largely free from aforementioned biases (see performance comparison in out-of-sample populations stratified by race, gender, and ethnicity in fig.S2, where we find no significant differences in predictive performances across racial and ethnic boundaries) and yet significantly outperforms the tools in current practice. Our ability to maintain high performance in diverse populations is an important feature of ACoR; it is not obvious a priori that ACoR resists inheriting or compounding the systemic biases that might exist in diagnostic procedures. Nevertheless, our investigations with the UCM dataset show that we have no significant differences in predictive performance in African-American, white, or multiracial subpopulations. While the average performance among Hispanic children was somewhat lower, the differences were not significant.
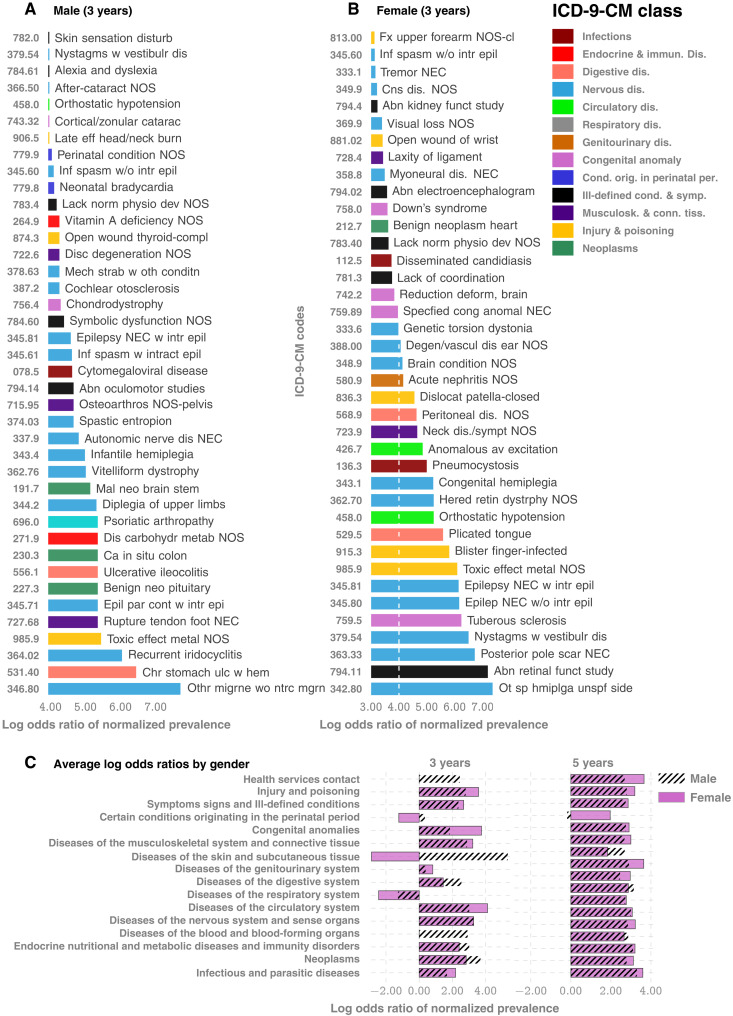
Going beyond screening performance, this approach provides a new tool to uncover clues to ASD pathobiology, potentially linking the observed vulnerability to diverse immunological, endocrinological, and neurological impairments to the possibility of allostatic stress load disrupting key regulators of CNS organization and synaptogenesis. Charting individual disorders in the comorbidity burden further reveals novel associations in normalized prevalence—the odds of experiencing a specific disorder, particularly in the early years (age <3 years), normalized over all unique disorders experienced in the specified time frame. We focus on the true positives in the positive cohort and the true negatives in the control cohort to investigate patterns that correctly disambiguate ASD status. On these lines, Fig. 4 and fig. S7 outline two key observations: (i) negative associations: some diseases that are negatively associated with ASD with respect to normalized prevalence, i. e., having those codes relatively overrepresented in one’s diagnostic history favors ending up in the control cohort; and (ii) impact of sex: there are sex-specific differences in the impact of specific disorders, and given a fixed level of impact, the number of codes that drive the outcomes is substantially higher in number in males (Fig. 4, A versus B).
Fig. 4. Comorbidity patterns.
(A and B) Difference in occurrence frequencies of diagnostic codes between true-positive and true-negative predictions. The dashed line in (B) shows the abscissa lower cutoff in (A), illustrating the lower prevalence of codes in females. (C) Log-odds ratios for ICD-9-CM disease categories at different ages. The negative associations disappear when we consider older children, consistent with the literature that lacks studies on very young cohorts.
Some of the disorders that show up in Fig. 4 (A and B) are unexpected, e. g., congenital hemiplegia or diplegia of the upper limbs indicative of either cerebral palsy (CP) or a spinal cord/brain injury, neither of which has a direct link to autism. However, this effect is easily explainable: Since only about 7% of the children with CP are estimated to have a co-occurring ASD (65, 66), and with the prevalence of CP significantly lower (1 in 352 versus 1 in 59 for autism), it follows that only a small number of children (approximately 1.17%) with autism have co-occurring CP. Thus, with significantly higher prevalence in children diagnosed with autism compared to the general population (1.7 versus 0.28%), CP codes show up with higher odds in the true-positive set. Other patterns are harder to explain. For example, fig. S7A shows that the immunological, metabolic, and endocrine disorders are almost completely risk-increasing, and respiratory diseases (fig. S7B) are largely risk-decreasing. On the other hand, infectious diseases have roughly equal representations in the risk-increasing and risk-decreasing classes (fig. S7C). The risk-decreasing infectious diseases tend to be due to viral or fungal organisms, which might point to the use of antibiotics in bacterial infections and the consequent dysbiosis of the gut microbiota (41, 55) as a risk factor.
Any predictive analysis of ASD must address if we can discriminate ASD from general developmental and behavioral disorders. The Diagnostic and Statistical Manual of Mental Disorders (DSM-5) established a single category of ASD to replace the subtypes of autistic disorder, Asperger syndrome, and pervasive developmental disorders (8). This aligns with our use of diagnostic codes from ICD-9-CM 299.X as specification of an ASD diagnosis and use standardized mapping to 299.X from ICD-10-CM codes when we encounter them. For other psychiatric disorders, we get high discrimination reaching AUCs over 90% at 100 to 125 weeks of age (fig. S6A), which establishes that our pipeline is indeed largely specific to ASD.
Can our performance be matched by simply asking how often a child is sick? While the density of ICD codes in a child’s medical history is higher for those eventually diagnosed with autism (see Table 1), we found this to be a crude measure. While somewhat predictive, achieving AUC ≈75% in the Truven database at 150 weeks (see fig. S6), code density by itself does not have a stable performance across the two databases, with a particularly poor performance in validation in the UCM database. Furthermore, adding code density as a feature shows no appreciable improvement in our pipeline.
We also investigated the impact of removing all psychiatric codes (ICD-9-CM 290-319, and corresponding ICD-10-CM) from the patient histories to eliminate the possibility that our performance is only reflective of prior psychiatric evaluation results. Training and validation on this modified data found no appreciable difference in performance (see fig. S9). In addition, we found that including information on prescribed medications and medical procedures in addition to diagnostic codes did not improve results.
A key limitation of our approach is that automated pattern recognition strategies are not guaranteed to reveal true causal precursors of future diseases. Thus, we must further scrutinize the patterns we find to be predictive. This is particularly true for the inferred negative associations of some disorders with the future odds of an ASD diagnosis; these might be indicative of collider bias (67), suggesting spurious associations arising from conditioning on the common effect of unrelated causes. Here, the putative collider in our framework is the presence of the child in a clinic or a medical facility for a medical issue. In the future, we will investigate the possibility of identifying more transparent and interpretable risk precursors via postprocessing of our inferred models.
Our approach is also affected by the uncurated nature of medical history, which invariably includes coding mistakes and other artifacts, e. g. , potential for overdiagnosis of children on the borderline of the diagnostic criteria due to clinicians’ desire to help families access service and biases arising from changes in diagnostic practices over time (68). Discontinuities in patient medical histories from change in provider networks can also introduce uncertainties in risk estimates, and socioeconomic status of patients and differential access to care might skew patterns in EHR databases. Despite these limitations, the design of a questionnaire-free component to ASD screening that systematically leverages comorbidities has far-reaching consequences, by potentially slashing the false positives and wait times and removing systemic underdiagnosis issues among females and minorities.
In the future, we will also explore the impact of maternal medical history and the use of calculated risk to trigger blood work to look for expected transcriptomic signatures of ASD. Last, the analysis developed here applies to phenotypes beyond ASD, thus opening the door to the possibility of general comorbidity-aware risk predictions from EHR databases.
One of our immediate future goals is to prospectively validate ACoR in a clinical setting. Ultimately, we hope to see widespread adoption of this new tool. A key hurdle to adoption is a general lack of trust in nontransparent decision algorithms, in particular in the light of the biases, misclassifications, misinterpretations, and subjectivity in the training datasets (69). We hope that results from prospective trials and the key advantages laid out in this study will help alleviate such concerns.
Acknowledgments
We acknowledge Professor Andrey Rzhetsky for inspiring us to investigate the notion of uncharted comorbidity patterns modulating autism risk. Funding: This work is funded, in part, by the Defense Advanced Research Projects Agency (DARPA) project number HR00111890043/P00004. The claims made in this study do not reflect the position or the policy of the U.S. Government. The UCM dataset is provided by the Clinical Research Data Warehouse (CRDW) maintained by the Center for Research Informatics (CRI) at the University of Chicago. The Center for Research Informatics is funded by the Biological Sciences Division, the Institute for Translational Medicine/CTSA (NIH UL1 TR000430) at the University of Chicago. Competing interests: The authors declare that they have no competing interests. Author contributions: D.O. implemented the algorithm and ran validation tests. D.O., Y.H., J.v.H., and I.C. carried out mathematical modeling and algorithm design. P.J.S., M.E.M., and I.C. interpreted results and guided research. I.C. procured funding and wrote the paper. Data and materials availability: All data and models needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The Truven and UCM datasets cannot be made available due to ethical and patient privacy considerations. Preliminary software implementation of the pipeline is available at https://github.com/zeroknowledgediscovery/ehrzero, and installation in standard Python environments may be done from https://pypi.org/project/ehrzero/. To enable fast execution, some more compute-intensive features are disabled in this version. Results from this software are for demonstration purposes only and must not be interpreted as medical advice or serve as replacement for such.
Supplementary Materials
This PDF file includes:
Tables S1 to S4
Figs. S1 to S14
Sections S1 to S13
References
REFERENCES AND NOTES
- 1.Centers for Disease Control and Prevention, Data & statistics on autism spectrum disorder (2019), https://cdc.gov/ncbddd/autism/data.html.
- 2.Schieve L. A., Tian L. H., Baio J., Rankin K., Rosenberg D., Wiggins L., Maenner M. J., Yeargin-Allsopp M., Durkin M., Rice C., King L., Kirby R. S., Wingate M. S., Devine O., Population attributable fractions for three perinatal risk factors for autism spectrum disorders, 2002 and 2008 autism and developmental disabilities monitoring network. Ann. Epidemiol. 24, 260–266 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Howsmon D. P., Kruger U., Melnyk S., James S. J., Hahn J., Classification and adaptive behavior prediction of children with autism spectrum disorder based upon multivariate data analysis of markers of oxidative stress and dna methylation. PLoS Comput. Biol. 13, e1005385 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li G., Lee O., Rabitz H., High efficiency classification of children with autism spectrum disorder. PLOS ONE 13, e0192867 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hicks S. D., Rajan A. T., Wagner K. E., Barns S., Carpenter R. L., Middleton F. A., Validation of a salivary rna test for childhood autism spectrum disorder. Front. Genet. 9, 534 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith A. M., Natowicz M. R., Braas D., Ludwig M. A., Ney D. M., Donley E. L. R., Burrier R. E., Amaral D. G., A metabolomics approach to screening for autism risk in the children’s autism metabolome project. Autism Res. 13, 1270–1285 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Volkmar F., Siegel M., Woodbury-Smith M., King B., McCracken J., State M.; American Academy of Child and Adolescent Psychiatry (AACAP) Committee on Quality Issues (CQI) , Practice parameter for the assessment and treatment of children and adolescents with autism spectrum disorder. J. Am. Acad. Child Adolesc. Psychiatry 53, 237–257 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Hyman S. L., Levy S. E., Myers S. M.; Council on children with disabilities, section on developmental and behavioral pediatrics , Identification, evaluation, and management of children with autism spectrum disorder. Pediatrics 145, e20193447 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Kalb L. G., Freedman B., Foster C., Menon D., Landa R., Kishfy L., Law P., Determinants of appointment absenteeism at an outpatient pediatric autism clinic. J. Dev. Behav. Pediatr. 33, 685–697 (2012). [DOI] [PubMed] [Google Scholar]
- 10.Bisgaier J., Levinson D., Cutts D. B., Rhodes K. V., Access to autism evaluation appointments with developmental-behavioral and neurodevelopmental subspecialists. Arch. Pediatr. Adolesc. Med. 165, 673–674 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Fenikilé T. S., Ellerbeck K., Filippi M. K., Daley C. M., Barriers to autism screening in family medicine practice: A qualitative study. Prim. Health Care Res. Dev. 16, 356–366 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Gordon-Lipkin E., Foster J., Peacock G., Whittling down the wait time: Exploring models to minimize the delay from initial concern to diagnosis and treatment of autism spectrum disorder. Pediatr. Clin. North Am. 63, 851–859 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guthrie W., Wallis K., Bennett A., Brooks E., Dudley J., Gerdes M., Pandey J., Levy S. E., Schultz R. T., Miller J. S., Accuracy of autism screening in a large pediatric network. Pediatrics 144, e20183963 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Robins D. L., Casagrande K., Barton M., Chen C. M. A., Dumont-Mathieu T., Fein D., Validation of the modified checklist for autism in toddlers, revised with follow-up (M-CHAT-R/F). Pediatrics 133, 37–45 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tye C., Runicles A. K., Whitehouse A. J. O., Alvares G. A., Characterizing the interplay between autism spectrum disorder and comorbid medical conditions: An integrative review. Front Psychiatry 9, 751 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kohane I. S., McMurry A., Weber G., MacFadden D., Rappaport L., Kunkel L., Bickel J., Wattanasin N., Spence S., Murphy S., Churchill S., The co-morbidity burden of children and young adults with autism spectrum disorders. PLOS ONE 7, e33224 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hyde K. K., Novack M. N., LaHaye N., Parlett-Pelleriti C., Anden R., Dixon D. R., Linstead E., Applications of supervised machine learning in autism spectrum disorder research: A review. Rev. J. Autism. Dev. Disord. 6, 128–146 (2019). [Google Scholar]
- 18.Abbas H., Garberson F., Liu-Mayo S., Glover E., Wall D. P., Multi-modular AI approach to streamline autism diagnosis in young children. Sci. Rep. 10, 5014 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duda M., Daniels J., Wall D. P., Clinical evaluation of a novel and mobile autism risk assessment. J. Autism Dev. Disord. 46, 1953–1961 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duda M., Kosmicki J., Wall D., Testing the accuracy of an observation-based classifier for rapid detection of autism risk. Transl. Psychiatry 4, e424 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fusaro V. A., Daniels J., Duda M., DeLuca T. F., D’Angelo O., Tamburello J., Maniscalco J., Wall D. P., The potential of accelerating early detection of autism through content analysis of youtube videos. PLOS ONE 9, e93533 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wall D. P., Dally R., Luyster R., Jung J.-Y., DeLuca T. F., Use of artificial intelligence to shorten the behavioral diagnosis of autism. PloS one 7, e43855 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wall D. P., Kosmicki J., Deluca T., Harstad E., Fusaro V. A., Use of machine learning to shorten observation-based screening and diagnosis of autism. Transl. Psychiatry 2, e100 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doshi-Velez F., Ge Y., Kohane I., Comorbidity clusters in autism spectrum disorders: An electronic health record time-series analysis. Pediatrics 133, e54–e63 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bishop-Fitzpatrick L., Movaghar A., Greenberg J. S., Page D., DaWalt L. S., Brilliant M. H., Mailick M. R., Using machine learning to identify patterns of lifetime health problems in decedents with autism spectrum disorder. Autism Res. 11, 1120–1128 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lingren T., Chen P., Bochenek J., Doshi-Velez F., Manning-Courtney P., Bickel J., Wildenger Welchons L., Reinhold J., Bing N., Ni Y., Barbaresi W., Mentch F., Basford M., Denny J., Vazquez L., Perry C., Namjou B., Qiu H., Connolly J., Abrams D., Holm I. A., Cobb B. A., Lingren N., Solti I., Hakonarson H., Kohane I. S., Harley J., Savova G., Electronic health record based algorithm to identify patients with autism spectrum disorder. PLOS ONE 11, e0159621 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.T. H. A. M. C. C. IBM MarketScan®. Formerly, E. Database, Marketscan research data, https://marketscan.truvenhealth.com/marketscanportal/Portal.aspx. [accessed 10 August 2021].
- 28.Baio J., Wiggins L., Christensen D. L., Maenner M. J., Daniels J., Warren Z., Kurzius-Spencer M., Zahorodny W., Rosenberg C. R., White T., Durkin M. S., Imm P., Nikolaou L., Yeargin-Allsopp M., Lee L.-C., Harrington R., Lopez M., Fitzgerald R. T., Hewitt A., Pettygrove S., Constantino J. N., Vehorn A., Shenouda J., Hall-Lande J., Van Naarden Braun K., Dowling N. F., Prevalence of autism spectrum disorder among children aged 8 years—Autism and Developmental Disabilities Monitoring Network, 11 sites, United States, 2014. MMWR Surveill Summ. 67, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chattopadhyay I., Ray A., Structural transformations of probabilistic finite state machines. Int. J. Control 81, 820–835 (2008). [Google Scholar]
- 30.Chattopadhyay I., Lipson H., Abductive learning of quantized stochastic processes with probabilistic finite automata. Philos Trans Royal Soc. A 371, 20110543 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Chattopadhyay I., Lipson H., Data smashing: Uncovering lurking order in data. J. Royal Soc. Interf. 11, 20140826 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Y. Huang, I. Chattopadhyay, Data smashing 2.0: Sequence likelihood (sl) divergence for fast time series comparison. arXiv:1909.12243 (2019).
- 33.T. M. Cover, J. A. Thomas, Elements of Information Theory (Wiley Series in Telecommunications and Signal Processing) (Wiley-Interscience, 2006). [Google Scholar]
- 34.Kullback S., Leibler R. A., On information and sufficiency. Ann. Math. Statist. 22, 79–86 (1951). [Google Scholar]
- 35.J. Doob, Stochastic Processes (John Wiley & Sons, 1953), Wiley Publications in Statistics. [Google Scholar]
- 36.Althouse L. A., Stockman III J. A., Pediatric workforce: A look at pediatric nephrology data from the American Board of Pediatrics. J. Pediatr. 148, 575–576 (2006). [DOI] [PubMed] [Google Scholar]
- 37.Zerbo O., Leong A., Barcellos L., Bernal P., Fireman B., Croen L. A., Immune mediated conditions in autism spectrum disorders. Brain Behav. Immun. 46, 232–236 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Won H., Mah W., Kim E., Autism spectrum disorder causes, mechanisms, and treatments: Focus on neuronal synapses. Front. Mol. Neurosci. 6, 19 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu G., Snetselaar L. G., Jing J., Liu B., Strathearn L., Bao W., Association of food allergy and other allergic conditions with autism spectrum disorder in children. JAMA Netw. Open 1, e180279 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Young A., Campbell E., Lynch S., Suckling J., Powis S. J., Aberrant Nf-κB expression in autism spectrum condition: A mechanism for neuroinflammation. Front. Psychiatry 2, 27 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fattorusso A., Di Genova L., Dell’Isola G. B., Mencaroni E., Esposito S., Autism spectrum disorders and the gut microbiota. Nutrients 11, 521 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Diaz Heijtz R., Wang S., Anuar F., Qian Y., Björkholm B., Samuelsson A., Hibberd M. L., Forssberg H., Pettersson S., Normal gut microbiota modulates brain development and behavior. Proc. Natl. Acad. Sci. U.S.A. 108, 3047–3052 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rose S., Bennuri S. C., Murray K. F., Buie T., Winter H., Frye R. E., Mitochondrial dysfunction in the gastrointestinal mucosa of children with autism: A blinded case-control study. PLOS ONE 12, e0186377 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sajdel-Sulkowska E. M., Makowska-Zubrycka M., Czarzasta K., Kasarello K., Aggarwal V., Bialy M., Szczepanska-Sadowska E., Cudnoch-Jedrzejewska A., Common genetic variants link the abnormalities in the gut-brain axis in prematurity and autism. Cerebellum 18, 255–265 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kayser M. S., Dalmau J., Anti-NMDA receptor encephalitis in psychiatry. Curr. Psychiatry Rev. 7, 189–193 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dadalko O. I., Travers B. G., Evidence for brainstem contributions to autism spectrum disorders. Front. Integr. Neurosci. 12, 47 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamashita Y., Makinodan M., Toritsuka M., Yamauchi T., Ikawa D., Kimoto S., Komori T., Takada R., Kayashima Y., Hamano-Iwasa K., Tsujii M., Matsuzaki H., Kishimoto T., Anti-inflammatory effect of ghrelin in lymphoblastoid cell lines from children with autism spectrum disorder. Front. Psych. 10, 152 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shen L., Feng C., Zhang K., Chen Y., Gao Y., Ke J., Chen X., Lin J., Li C., Iqbal J., Zhao Y., Wang W., Proteomics study of peripheral blood mononuclear cells (PBMCs) in autistic children. Front. Cell. Neurosci. 13, 105 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ohja K., Gozal E., Fahnestock M., Cai L., Cai J., Freedman J. H., Switala A., el-Baz A., Barnes G. N., Neuroimmunologic and neurotrophic interactions in autism spectrum disorders: Relationship to neuroinflammation. Neuromolecular Med. 20, 161–173 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gładysz D., Krzywdzińska A., Hozyasz K. K., Immune abnormalities in autism spectrum disorder—Could they hold promise for causative treatment? Mol. Neurobiol. 55, 6387–6435 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Theoharides T., Tsilioni I., Patel A., Doyle R., Atopic diseases and inflammation of the brain in the pathogenesis of autism spectrum disorders. Transl. Psychiatry 6, e844 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Young A. M., Chakrabarti B., Roberts D., Lai M.-C., Suckling J., Baron-Cohen S., From molecules to neural morphology: Understanding neuroinflammation in autism spectrum condition. Mol. Autism. 7, 9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Croen L. A., Qian Y., Ashwood P., Daniels J. L., Fallin D., Schendel D., Schieve L. A., Singer A. B., Zerbo O., Family history of immune conditions and autism spectrum and developmental disorders: Findings from the study to explore early development. Autism Res. 12, 123–135 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vargason T., McGuinness D. L., Hahn J., Gastrointestinal symptoms and oral antibiotic use in children with autism spectrum disorder: Retrospective analysis of a privately insured U.S. population. J. Autism Dev. Disord. 49, 647–659 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fiorentino M., Sapone A., Senger S., Camhi S. S., Kadzielski S. M., Buie T. M., Kelly D. L., Cascella N., Fasano A., Blood–brain barrier and intestinal epithelial barrier alterations in autism spectrum disorders. Mol. Autism. 7, 49 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Satterstrom F. K., Kosmicki J. A., Wang J., Breen M. S., de Rubeis S., An J. Y., Peng M., Collins R., Grove J., Klei L., Stevens C., Reichert J., Mulhern M. S., Artomov M., Gerges S., Sheppard B., Xu X., Bhaduri A., Norman U., Brand H., Schwartz G., Nguyen R., Guerrero E. E., Dias C., Betancur C., Cook E. H., Gallagher L., Gill M., Sutcliffe J. S., Thurm A., Zwick M. E., Børglum A. D., State M. W., Cicek A. E., Talkowski M. E., Cutler D. J., Devlin B., Sanders S. J., Roeder K., Daly M. J., Buxbaum J. D., Aleksic B., Anney R., Barbosa M., Bishop S., Brusco A., Bybjerg-Grauholm J., Carracedo A., Chan M. C. Y., Chiocchetti A. G., Chung B. H. Y., Coon H., Cuccaro M. L., Curró A., Dalla Bernardina B., Doan R., Domenici E., Dong S., Fallerini C., Fernández-Prieto M., Ferrero G. B., Freitag C. M., Fromer M., Gargus J. J., Geschwind D., Giorgio E., González-Peñas J., Guter S., Halpern D., Hansen-Kiss E., He X., Herman G. E., Hertz-Picciotto I., Hougaard D. M., Hultman C. M., Ionita-Laza I., Jacob S., Jamison J., Jugessur A., Kaartinen M., Knudsen G. P., Kolevzon A., Kushima I., Lee S. L., Lehtimäki T., Lim E. T., Lintas C., Lipkin W. I., Lopergolo D., Lopes F., Ludena Y., Maciel P., Magnus P., Mahjani B., Maltman N., Manoach D. S., Meiri G., Menashe I., Miller J., Minshew N., Montenegro E. M. S., Moreira D., Morrow E. M., Mors O., Mortensen P. B., Mosconi M., Muglia P., Neale B. M., Nordentoft M., Ozaki N., Palotie A., Parellada M., Passos-Bueno M. R., Pericak-Vance M., Persico A. M., Pessah I., Puura K., Reichenberg A., Renieri A., Riberi E., Robinson E. B., Samocha K. E., Sandin S., Santangelo S. L., Schellenberg G., Scherer S. W., Schlitt S., Schmidt R., Schmitt L., Silva I. M. W., Singh T., Siper P. M., Smith M., Soares G., Stoltenberg C., Suren P., Susser E., Sweeney J., Szatmari P., Tang L., Tassone F., Teufel K., Trabetti E., Trelles M. P., Walsh C. A., Weiss L. A., Werge T., Werling D. M., Wigdor E. M., Wilkinson E., Willsey A. J., Yu T. W., Yu M. H. C., Yuen R., Zachi E., Agerbo E., Als T. D., Appadurai V., Bækvad-Hansen M., Belliveau R., Buil A., Carey C. E., Cerrato F., Chambert K., Churchhouse C., Dalsgaard S., Demontis D., Dumont A., Goldstein J., Hansen C. S., Hauberg M. E., Hollegaard M. V., Howrigan D. P., Huang H., Maller J., Martin A. R., Martin J., Mattheisen M., Moran J., Pallesen J., Palmer D. S., Pedersen C. B., Pedersen M. G., Poterba T., Poulsen J. B., Ripke S., Schork A. J., Thompson W. K., Turley P., Walters R. K., Large-scale exome sequencing study implicates both developmental and functional changes in the neurobiology of autism. Cell 180, 568–584.e23 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gaugler T., Klei L., Sanders S. J., Bodea C. A., Goldberg A. P., Lee A. B., Mahajan M., Manaa D., Pawitan Y., Reichert J., Ripke S., Sandin S., Sklar P., Svantesson O., Reichenberg A., Hultman C. M., Devlin B., Roeder K., Buxbaum J. D., Most genetic risk for autism resides with common variation. Nat. Genet. 46, 881–885 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vargas D. L., Nascimbene C., Krishnan C., Zimmerman A. W., Pardo C. A., Neuroglial activation and neuroinflammation in the brain of patients with autism. Ann. Neurol. 57, 67–81 (2005). [DOI] [PubMed] [Google Scholar]
- 59.Wei H., Zou H., Sheikh A. M., Malik M., Dobkin C., Brown W. T., Li X., IL-6 is increased in the cerebellum of autistic brain and alters neural cell adhesion, migration and synaptic formation. J. Neuroinflammation 8, 52 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Young A. M., Campbell E., Lynch S., Suckling J., Powis S. J., Aberrant NF-kappaB expression in autism spectrum condition: A mechanism for neuroinflammation. Front. Psych. 2, 27 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hughes H. K., Mills Ko E., Rose D., Ashwood P., Immune dysfunction and autoimmunity as pathological mechanisms in autism spectrum disorders. Front. Cell. Neurosci. 12, 405 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pearce N., The ecological fallacy strikes back. J. Epidemiol. Community Health 54, 326–327 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Murdoch J. D., State M. W., Recent developments in the genetics of autism spectrum disorders. Curr. Opin. Genet. Dev. 23, 310–315 (2013). [DOI] [PubMed] [Google Scholar]
- 64.Hu V. W., The expanding genomic landscape of autism: Discovering the ‘forest’ beyond the ‘trees’. Future Neurol 8, 29–42 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Centers for Disease Control and Prevention, Prevalence of cerebral palsy, co-occurring autism spectrum disorders, and motor functioning (2020), https://cdc.gov/ncbddd/cp/features/prevalence.html.
- 66.Christensen D., van Naarden Braun K., Doernberg N. S., Maenner M. J., Arneson C. L., Durkin M. S., Benedict R. E., Kirby R. S., Wingate M. S., Fitzgerald R., Yeargin-Allsopp M., Prevalence of cerebral palsy, co-occurring autism spectrum disorders, and motor functioning—Autism and Developmental Disabilities Monitoring Network, USA, 2008. Dev. Med. Child Neurol. 56, 59–65 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Berkson J., Limitations of the application of fourfold table analysis to hospital data. Biometrics 2, 47–53 (1946). [PubMed] [Google Scholar]
- 68.Rødgaard E.-M., Jensen K., Vergnes J.-N., Soulières I., Mottron L., Temporal changes in effect sizes of studies comparing individuals with and without autism: A meta-analysis. JAMA Psychiat. 76, 1124–1132 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cheng M., Nazarian S., Bogdan P., There is hope after all: Quantifying opinion and trustworthiness in neural networks. Front. Artif. Intell. 3, 54 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Developmental Disabilities Monitoring Network Surveillance Year 2010 Principal Investigators; Centers for Disease Control and Prevention (CDC) , Prevalence of autism spectrum disorder among children aged 8 years—Autism and Developmental Disabilities Monitoring Network, 11 sites, United States, 2010. MMWR Surveill. Summ. 63, 1–21 (2014). [PubMed] [Google Scholar]
- 71.Bolton P. F., Golding J., Emond A., Steer C. D., Autism spectrum disorder and autistic traits in the Avon Longitudinal Study of Parents And Children: Precursors and early signs. J. Am. Acad. Child Adolesc. Psychiatry 51, 249–260.e25 (2012). [DOI] [PubMed] [Google Scholar]
- 72.J. A. Bondy, U. S. R. Murty, Graph Theory with Applications (Macmillan London, 1976), vol. 290. [Google Scholar]
- 73.Breiman L., Random forests. Mach. Learn. 45, 5–32 (2001). [Google Scholar]
- 74.Chattopadhyay I., Ray A., Structural transformations of probabilistic finite state machines. Int. J. Control 81, 820–835 (2008). [Google Scholar]
- 75.Chlebowski C., Green J. A., Barton M. L., Fein D., Using the childhood autism rating scale to diagnose autism spectrum disorders. J. Autism Dev. Disord. 40, 787–799 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.J. Doob, Stochastic Processes (Wiley, 1990), Wiley Publications in Statistics. [Google Scholar]
- 77.Esler A. N., Bal V. H., Guthrie W., Wetherby A., Weismer S. E., Lord C., The autism diagnostic observation schedule, toddler module: Standardized severity scores. J. Autism Dev. Disord. 45, 2704–2720 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Falkmer T., Anderson K., Falkmer M., Horlin C., Diagnostic procedures in autism spectrum disorders: A systematic literature review. Eur. Child Adolesc. Psychiatry 22, 329–340 (2013). [DOI] [PubMed] [Google Scholar]
- 79.Friedman J. H., Stochastic gradient boosting. Comput. Stat. Data Anal. 38, 367–378 (2002). [Google Scholar]
- 80.G. H. Hardy, Divergent Series (AMS Chelsea Publishing, 2000), vol. 334. [Google Scholar]
- 81.Hochreiter S., Schmidhuber J., Long short-term memory. Neural Comput. 9, 1735–1780 (1997). [DOI] [PubMed] [Google Scholar]
- 82.J. E. Hopcroft, Introduction to Automata Theory, Languages, and Computation (Pearson Education India, 2008). [Google Scholar]
- 83.Jarquin V. G., Wiggins L. D., Schieve L. A., Van Naarden-Braun K., Racial disparities in community identification of autism spectrum disorders over time; Metropolitan Atlanta, Georgia, 2000–2006. J. Dev. Behav. Pediatr. 32, 179–187 (2011). [DOI] [PubMed] [Google Scholar]
- 84.Johnson C. P., Myers S. M.; Council on Children With Disabilities , Identification and evaluation of children with autism spectrum disorders. Pediatrics 120, 1183–1215 (2007). [DOI] [PubMed] [Google Scholar]
- 85.L. C. Kai, Markov Chains: With Stationary Transition Probabilities (Springer-Verlag, 1967). [Google Scholar]
- 86.Kleinman J. M., Ventola P. E., Pandey J., Verbalis A. D., Barton M., Hodgson S., Green J., Dumont-Mathieu T., Robins D. L., Fein D., Diagnostic stability in very young children with autism spectrum disorders. J. Autism Dev. Disord. 38, 606–615 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.A. Klenke, Probability Theory: A Comprehensive Course (Springer Science & Business Media, 2013). [Google Scholar]
- 88.Kozlowski A. M., Matson J. L., Horovitz M., Worley J. A., Neal D., Parents’ first concerns of their child’s development in toddlers with autism spectrum disorders. Dev. Neurorehabil. 14, 72–78 (2011). [DOI] [PubMed] [Google Scholar]
- 89.Lord C., Risi S., DiLavore P. S., Shulman C., Thurm A., Pickles A., Autism from 2 to 9 years of age. Arch. Gen. Psychiatry 63, 694–701 (2006). [DOI] [PubMed] [Google Scholar]
- 90.Granger C. W. J., Joyeux R., An introduction to long-memory time series models and fractional differencing. J. Time Ser. Anal. 1, 15–29 (1980). [Google Scholar]
- 91.Matthews A. G. d. G., Hensman J., Turner R., Ghahramani Z., On sparse variational methods and the Kullback-Leibler divergence between stochastic processes. J. Mach. Learn. Res. 51, 231 (2016). [Google Scholar]
- 92.L. Gilotty, Early screening for autism spectrum (NIMH, 2019); www.nimh.nih.gov/funding/grant-writing-and-application-process/concept-clearances/2018/early-screening-for-autism-spectrum.shtml.
- 93.A. N. Trahtman, The road coloring and Cerny conjecture, in Proceedings of Prague Stringology Conference (Citeseer, 2008), vol. 1, p. 12. [Google Scholar]
- 94.M. Vidyasagar, Hidden Markov Processes: Theory and Applications to Biology (Princeton University Press, 2014), vol. 44. [Google Scholar]
- 95.Zwaigenbaum L., Bauman M. L., Choueiri R., Kasari C., Carter A., Granpeesheh D., Mailloux Z., Smith Roley S., Wagner S., Fein D., Pierce K., Buie T., Davis P. A., Newschaffer C., Robins D., Wetherby A., Stone W. L., Yirmiya N., Estes A., Hansen R. L., McPartland J. C., Natowicz M. R., Early intervention for children with autism spectrum disorder under 3 years of age: Recommendations for practice and research. Pediatrics 136, S60–S81 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 to S4
Figs. S1 to S14
Sections S1 to S13
References