RGX-202, a small-molecule creatine transporter SLC6A8 inhibitor, suppresses colorectal cancer and modulates human creatine levels.
Abstract
Colorectal cancer (CRC) is a leading cause of cancer mortality. Creatine metabolism was previously shown to critically regulate colon cancer progression. We report that RGX-202, an oral small-molecule SLC6A8 transporter inhibitor, robustly inhibits creatine import in vitro and in vivo, reduces intracellular phosphocreatine and ATP levels, and induces tumor apoptosis. RGX-202 suppressed CRC growth across KRAS wild-type and KRAS mutant xenograft, syngeneic, and patient-derived xenograft (PDX) tumors. Antitumor efficacy correlated with tumoral expression of creatine kinase B. Combining RGX-202 with 5-fluorouracil or the DHODH inhibitor leflunomide caused regressions of multiple colorectal xenograft and PDX tumors of distinct mutational backgrounds. RGX-202 also perturbed creatine metabolism in patients with metastatic CRC in a phase 1 trial, mirroring pharmacodynamic effects on creatine metabolism observed in mice. This is, to our knowledge, the first demonstration of preclinical and human pharmacodynamic activity for creatine metabolism targeting in oncology, thus revealing a critical therapeutic target.
INTRODUCTION
Colorectal cancer (CRC) is the second leading cause of cancer death in men and women in the United States (1) and a major cause of mortality worldwide. In 2020, roughly 148,000 individuals were diagnosed with CRC and 53,200 died from the disease. Early-stage CRCs are primarily treated by surgical resection. For larger tumors and those that have spread to lymph nodes, 5-fluorouracil (5-FU)–based chemotherapy regimens are administered in the postsurgical “adjuvant” setting to reduce the risk of metastatic relapse. For cancers that have spread to distant organs such as the liver—the primary site of metastatic relapse—systemic 5-FU–containing chemotherapy regimens are given. Targeted therapies including antibodies that engage the epidermal growth factor receptor (EGFR) or vascular endothelial growth factor (VEGF) can, in combination with chemotherapeutics, provide modest survival benefits (2, 3). However, most metastatic patients succumb to their disease, with a meager 5-year survival rate of 14% (1). Recently, innovative small molecules that covalently bind and inhibit the KRAS G12C oncogenic driver variant, which is present in approximately 4% of colorectal tumors, have elicited clinical responses in patients harboring this mutant allele (4–8). Other approaches include the development of inhibitors targeting downstream KRAS signaling in RAS-mutant tumors (9, 10). Limitations of such therapies include the emergence of resistance (11), their lack of efficacy against other KRAS mutant alleles present in ~40% of CRC patients, and the expected lack of efficacy in KRAS wild-type tumors (8). Additional approaches are thus needed to tackle the breadth of CRC.
Cancers evolve multiple mechanisms to sustain proliferative growth and survival. One such adaptive mechanism is altered metabolism, commonly referred to as metabolic rewiring. By altering the flux of metabolites in various metabolic pathways, cancer cells enhance biosynthesis of anabolic building blocks required for growth such as nucleotides, amino acids, and lipids (12). Certain metabolites, however, can become limiting during cancer progression—requiring their extracellular import through metabolic transporters. The search for such metabolic dependencies is an active area of cancer research (13, 14).
We previously reported that during progression of CRC to liver metastasis, cancer cells up-regulate creatine kinase brain-type (CKB) and secrete CKB into the extracellular space (15). CKB phosphorylates the metabolite creatine using adenosine 5′-triphosphate (ATP), thereby generating phosphocreatine. The γ-phosphate of phosphocreatine has ~50% greater free energy than the γ-phosphate of ATP (16). Phosphocreatine thus serves as a rapidly mobilizable high-energy phosphate reserve that can yield ATP in a reaction that does not require oxygen. Because of its high energy content, phosphocreatine is stored at high levels in metabolically active tissues such as muscle, brain, and kidney—allowing tight maintenance of organismal ATP levels, which are critical for a vast number of metabolic and homeostatic processes (17). Although extracellular ATP levels are normally low, the death of cancer cells and stromal cells substantially increases extracellular ATP concentrations in the tumor microenvironment, which have been observed to exceed 700 μM (18). Excess of ATP over circulating creatine levels, which range from 9 to 90 μM, thus favors phosphocreatine generation in the tumor microenvironment—a reaction mediated by tumor-secreted CKB. Phosphocreatine import by SLC6A8 enhanced tumoral ATP levels and accordingly promoted cell survival under hypoxia (15). Extracellular phosphocreatine supplementation rescued the metastatic defect and in vitro survival phenotype of CKB-depleted cells (15). Gastrointestinal tumors, such as CRC and pancreatic cancers, are highly hypoxic (19–21), as are metastases formed by these cancers. This metabolic axis provides a mechanism to support tumor growth in hypoxic environments. Moreover, the expression levels of CKB and SLC6A8 are associated with increased CRC metastasis in patients (15).
Here, we identify the SLC6A8 transporter as a therapeutic target in CRC. We show that an orally bioavailable small-molecule creatine mimetic (RGX-202) significantly inhibits SLC6A8 and suppresses the growth of CRC tumors in syngeneic, xenograft, and patient-derived xenograft (PDX) models and caused tumor cell apoptosis in vivo. RGX-202 also strongly suppressed liver metastasis formation. We show that this therapy is effective in both KRAS wild-type tumors and tumors bearing various KRAS mutations, including the KRAS G12D allele, which is not currently druggable by clinical stage KRAS inhibitors. Antitumor efficacy by RGX-202 correlated with increased CKB expression in tumors. RGX-202 exhibited greater activity in combination with multiple standard-of-care regimens, including 5-FU and gemcitabine, relative to single-agent efficacy. We also show that RGX-202 synergizes with the dihydroorotate dehydrogenase (DHODH) enzyme inhibitor leflunomide, an oral compound previously shown to suppress CRC cell growth under hypoxia by repressing nucleotide biosynthesis (22). Metabolic profiling studies revealed that RGX-202 suppresses intracellular phosphocreatine, creatine, and ATP levels. Last, we observe a drug exposure–dependent increase of creatine in the blood and urine of mice and in patients treated with oral RGX-202 therapy, confirming creatine transporter inhibition in patients and supporting further clinical testing of RGX-202 in later-stage clinical trials.
RESULTS
Oral RGX-202 suppresses in vivo creatine import and depletes cellular phosphocreatine and ATP levels
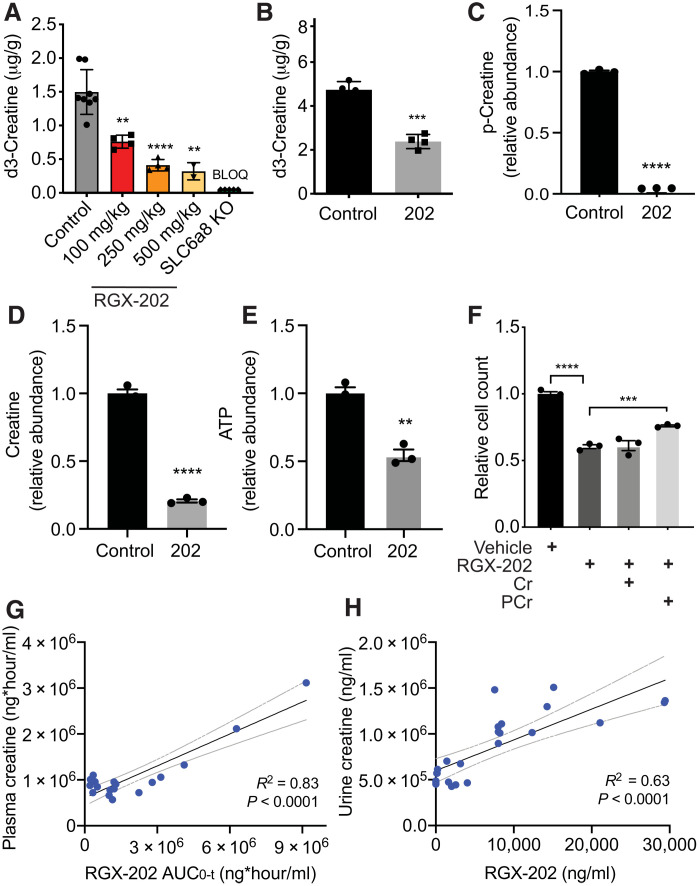
RGX-202 (β-guanidinoproprionic acid) is a creatine mimetic that competitively inhibits cellular creatine import by inhibiting the creatine transporter SLC6A8 (23, 24). RGX-202 and its pharmaceutically optimized form RGX-202-01, which is suitable for clinical administration, were used here to determine the impact of SLC6A8 inhibition on CRC progression. To assess the degree to which RGX-202 inhibits SLC6A8, we monitored creatine import in vivo (23, 25). We administered increasing concentrations of RGX-202 followed by injection of deuterium-labeled creatine (d3-creatine) into wild-type and SLC6A8 knockout mice. Mouse tissue was extracted and analyzed for levels of d3-creatine by liquid chromatography–tandem mass spectrometry (LC-MS/MS) (23). RGX-202 treatment inhibited tissue uptake of d3-creatine in a dose-dependent manner by up to 75% at 500 mg/kg (Fig. 1A). d3-creatine levels in SLC6A8 knockout animals were below the lower limit of quantification, confirming that creatine import is exclusively mediated by SLC6A8 (Fig. 1A). To determine whether RGX-202 could inhibit tumoral SLC6A8, we conducted similar studies in mice bearing syngeneic UN-KPC-961 pancreatic tumors. RGX-202 at 800 mg/kg supplemented in the diet for 35 days suppressed tumoral d3-creatine import by 50% (Fig. 1B). These findings reveal that administration of the creatine mimetic RGX-202 significantly suppresses creatine import into tissues in vivo.
Fig. 1. RGX-202 reduces cellular and tumoral creatine and phosphocreatine levels.
(A) RGX-202 or vehicle control was administered to C57BL/6 wild-type or SLC6A8 knockout (KO) mice before injection of d3-creatine (1 mg/kg). Hearts were extracted and d3-creatine levels were quantified by LC-MS/MS; n = 3 to 8 independent experiments; P = 0.0016 (100 mg/kg), P < 0.0001 (250 mg/kg), and P = 0.0014 (500 mg/kg). BLOQ, below limit of quantification. (B) UN-KPC-961 pancreatic tumor-bearing B6129SF1/J mice were fed a control or RGX-202–supplemented diet (800 mg/kg) for 35 days. D3-creatine (1 mg/kg) was injected, tumors were extracted, and d3-creatine levels were quantified by LC-MS/MS; n = 4 per group. (C to E) Lvm3b cells (3 × 105) were treated with vehicle control or 10 mM RGX-202 under hypoxia (0.5% O2) for 24 hours. Phosphocreatine (C), creatine (D), and ATP (E) levels were analyzed by LC-HRMS; n = 3; representative of three independent experiments. (F) Growth of Lvm3b cells incubated in hypoxia (0.5% oxygen) in the presence of RGX-202 (10 mM), creatine (Cr), or phosphocreatine (PCr) at 10 μM; n = 3 per group. P values are based on two-sided t test. Means ± SEM are shown (A to F). (G and H) Correlation analysis of plasma creatine and RGX-202 exposure (AUC0-t) (G) or average urine creatine and RGX-202 concentrations (H) measured over 24 hours in mice receiving either a control or RGX-202-01–supplemented diet at 100, 400, or 1200 mg/kg for 10 days; n = 6 mice per group. Gray lines denote 95% confidence interval.
We next assessed the impact of creatine transport inhibition on cellular levels of creatine, phosphocreatine, and ATP. LC-MS/MS analysis of the highly metastatic CRC cell line LS174T Lvm3b (Lvm3b) treated with RGX-202 under hypoxia revealed a nearly complete depletion of phosphocreatine (>99%), greater than 79% reduction in cellular creatine and a substantial (46%) reduction in intracellular ATP levels relative to control cells (Fig. 1, C to E). These observations are consistent with our previous findings, demonstrating that short hairpin RNA (shRNA)–mediated depletion of the SLC6A8 transporter reduced phosphocreatine import and reduced ATP levels in CRC cells under hypoxia (15).
We next evaluated the growth of Lvm3b cells upon RGX-202 treatment in the context of hypoxia. RGX-202 significantly reduced cell growth, and this effect was partially rescued by supplementation with high-dose phosphocreatine (Fig. 1F). The addition of creatine alone did not rescue the cell growth phenotype, consistent with a model whereby phosphocreatine serving as a high-energy phosphate source promotes growth when cells are exposed to hypoxic stress inherent to the CRC tumor and hepatic microenvironments (15). Overall, these findings support a model whereby RGX-202–induced depletion of intracellular creatine and phosphocreatine leads to a consequential depletion of ATP, a critical energetic metabolite. The impairment of both phosphocreatine and ATP energetic metabolites poses a barrier for growth and survival under hypoxia and energetic stress.
We hypothesized that we should observe changes in creatine concentration in the circulation of RGX-202–treated mice if RGX-202 inhibits cellular import of creatine. We administered increasing doses of RGX-202-01 (100, 400, and 1200 mg/kg) and measured RGX-202 and creatine in the plasma by LC-MS/MS. We observed an RGX-202 exposure–dependent increase in circulating plasma creatine levels, manifested by accumulation of plasma creatine upon blockade of SLC6A8 in tissues (Fig. 1G). We also observed a substantial exposure-dependent increase in urinary creatine levels in the same animals (Fig. 1H), consistent with inhibition of creatine import into tissues causing increased creatine in circulation and subsequent excretion in urine, thus reducing creatine levels available to tumors.
SLC6A8 inhibition exhibits broad antitumor activity against diverse primary and metastatic CRCs
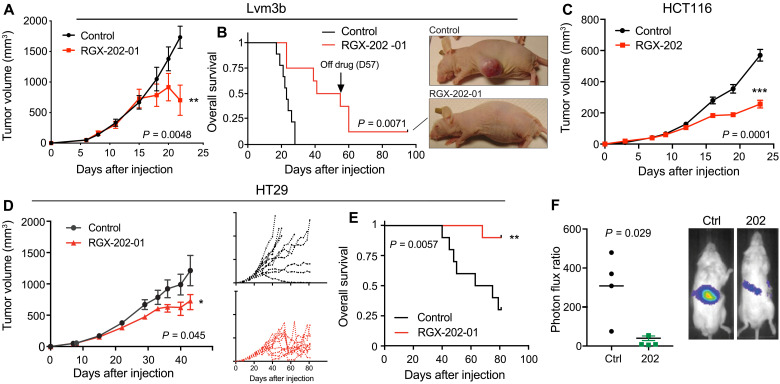
Primary tumors exhibit hypoxia, especially upon progression to larger sizes, and must gain metabolic adaptations to the hypoxic microenvironment to survive (19). Approximately 40% of CRCs harbor mutations in the KRAS oncogene. KRAS mutations have been shown to confer increased metabolic demand in cancer cells, including increased dependency on energy-producing metabolic pathways (26). We thus hypothesized that these tumors might exhibit sensitivity to the effects of SLC6A8 blockade by RGX-202. We implanted highly aggressive metastatic Lvm3b (KRAS G12D) cells subcutaneously into athymic nude mice and began RGX-202-01 treatment after the tumors became palpable (>30 mm3). Oral administration of RGX-202-01 caused a ~50% tumor growth inhibition (TGI) (Fig. 2A). The effects of RGX-202-01 emerged after tumors reached larger sizes (>500 mm3), consistent with the previously demonstrated role of creatine metabolism and SLC6A8 in hypoxic survival (15). RGX-202-01–treated mice exhibited a significant improvement in survival, experiencing a doubling of median survival time from 23 to 48 days. One of the nine immunodeficient mice experienced a complete tumor regression response, and the tumor remained undetectable >38 days after treatment termination (Fig. 2B). We also observed antitumor efficacy in HCT116 (KRAS G13D) as well as HT29 (KRAS wild-type) human CRC tumors upon oral RGX-202-01 administration (Fig. 2, C to E). Similar to Lvm3b, treatment of HT29 tumors induced regressions in 7 of 10 mice upon tumors reaching a large size (>900 mm3) and treatment substantially prolonged overall survival (Fig. 2, D and E). Depletion of the SLC6A8 transporter in CRC cells was previously shown to substantially reduce liver metastatic colonization (15). We thus sought to determine whether inhibiting SLC6A8 could suppress CRC metastasis. Highly metastatic Lvm3b cells, which harbored a luciferase reporter gene, were injected into the portal circulation of nonobese diabetic severe combined immunodeficient (NOD-SCID) mice via intrasplenic injections, and metastatic colonization to the liver was quantified by bioluminescent imaging. RGX-202 treatment reduced liver metastatic colonization by eightfold (Fig. 2F and fig. S1A). To determine whether the effect of SLC6A8 inhibition is cell autonomous, we pretreated KRAS G12D mutant PANC1 human pancreatic cancer cells in vitro with RGX-202 for 48 hours and subsequently injected these cells into the spleens of NSG mice. Pretreatment with RGX-202 reduced pancreatic cancer liver metastatic colonization by fivefold (fig. S1, B and C), demonstrating that depleting cellular creatine/phosphocreatine levels via SLC6A8 inhibition significantly inhibits liver metastasis in a tumor cell–autonomous manner.
Fig. 2. SLC6A8 inhibition exhibits antitumor activity against primary and metastatic CRCs.
(A and B) Subcutaneous tumor growth (A) and Kaplan-Meier survival curves (B) by 1 × 106 Lvm3b cells in athymic nude mice. Daily oral gavage of RGX-202-01 (200 mg/kg) started when tumors reached ~50 mm3; n = 8 to 9 per group. Pictures show a control and RGX-202-01–treated mouse. (C) Subcutaneous tumor growth by 0.5 × 106 HCT116 cells in athymic nude mice, receiving a control or RGX-202–supplemented diet (800 mg/kg) starting when tumors became palpable; n = 5 per group. (D and E) Subcutaneous tumor growth (D) and Kaplan-Meier survival curves (E) by 2 × 106 HT29 cells in athymic nude mice receiving a control or RGX-202-01–supplemented diet (800 mg/kg) when tumors reached ~55 mm3; insets represent individual tumor growth curves; n = 10 per group. (F) Bioluminescence plot of liver colonization by 5 × 105 Lvm3b cells after intrasplenic injections into NOD-SCID mice imaged on day 14; n = 4 per group. Means ± SEM are shown (A, C, and D). P values are based on two-sided t tests (A, C, and D), log-rank Mantel-Cox test (B and E), or Mann-Whitney test (F). Photo credit: Celia Andreu-Agullo, Inspirna.
To confirm that RGX-202 efficacy is dependent on the CKB/SLC6A8 pathway, we conducted liver metastasis assays with control or CKB CRISPR knockout Lvm3b CRC cells. Depletion of CKB in CRC cells resulted in 83% inhibition of liver metastasis, comparable to the effects mediated by RGX-202 (86%) (fig. S1, D and E). The similarity of the magnitude of these effects is consistent with CKB and SLC6A8 comprising a signaling pathway, per our previous findings, and demonstrates that the CKB/SLC6A8 pathway is critical for metastasis by Lvm3b cells. An additional 10% antitumor effect was observed when RGX-202 was administered to mice harboring CKB-depleted tumors. This additional minor effect could stem from RGX-202–mediated suppression of import of phosphocreatine derived from a host-derived creatine kinase or phosphocreatine released from dying colon cancer or host cells.
RGX-202 induces apoptosis and inhibits tumor cell proliferation
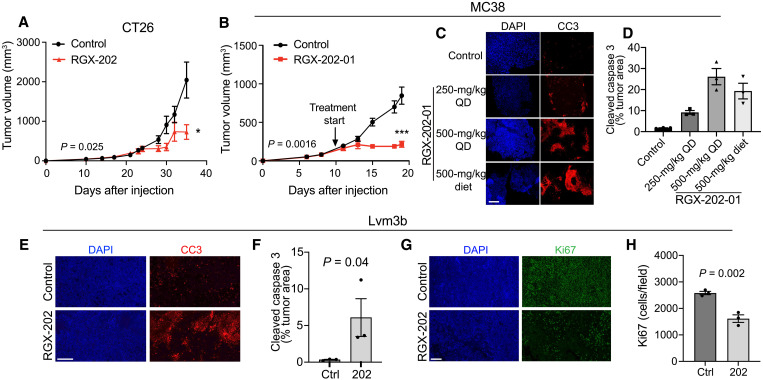
To determine whether SLC6A8 inhibition could suppress progression of murine CRC in an immunocompetent model, we implanted KRAS G12D mutant CT26 murine CRC cells into syngeneic mice. RGX-202 significantly inhibited CT26 tumor growth (Fig. 3A). This approach is also effective in treating larger tumors. We treated murine KRAS wild-type MC38 CRC tumors with RGX-202-01 after volumes reached ~150 mm3 and observed substantially inhibited tumor growth in this immunocompetent model (Fig. 3B). Drug treatment significantly enhanced in vivo tumor apoptosis in a dose-dependent manner as quantified by cleaved caspase 3 (CC3) immunofluorescence staining (Fig. 3, C and D). Increased apoptosis was also observed in RGX-202-01–treated Lvm3b and HCT15 tumors (Fig. 3, E and F, and fig. S2, A to C). We next asked whether RGX-202 modulates tumor cell proliferation by quantifying Ki67-positive cells. RGX-202-01 treatment inhibited tumor cell proliferation in both the Lvm3b and HCT15 tumor models (Fig. 3, G and H, fig. S2, B and C). In contrast, no effect on apoptosis or proliferation was observed in NCI-H508, a xenograft that did not respond to RGX-202-01 treatment (fig. S2, D to F). These results indicate that antitumor responses were associated with inhibition of tumor cell proliferation and induction of tumor apoptosis.
Fig. 3. RGX-202 induces tumor cell apoptosis and inhibits proliferation.
(A) Subcutaneous tumor growth by 0.5 × 106 CT26 cells in BALB/c mice receiving a control or RGX-202-01–supplemented diet (500 mg/kg) starting at a tumor size of ~100 mm3; n = 7 to 8 per group. (B) Subcutaneous tumor growth by 0.5 × 106 MC38 cells in C57BL/6 mice receiving a control or RGX-202-01–supplemented diet (500 mg/kg) starting at a tumor size of ~150 mm3; n = 7 per group. (C and D) MC38 tumors treated with RGX-202-01 by oral gavage [once daily (QD), 250 mg/kg, P = 0.0011; 500 mg/kg, P = 0.0032] or through diet (diet, P = 0.0092) for 9 days were immunostained for cleaved caspase 3 (CC3) (C). Quantification of the percentage of tumor area stained with CC3; n = 4 per group ± SEM (D). (E to H) Representative images and quantification of Lvm3b tumors extracted from mice treated with either RGX-202-01 or control diet and immunostained for CC3 (E and F) or Ki67 (G and H) and counterstained with 4′,6-diamidino-2-phenylindole (DAPI); n = 3 tumors per group. Means ± SEM are shown; P values are based on two-sided t tests (D, F, and H) and one-sided t tests (D and F). Scale bars, 200 μm (C, E, and G).
SLC6A8 inhibition exhibits broad activity against CRC PDX models
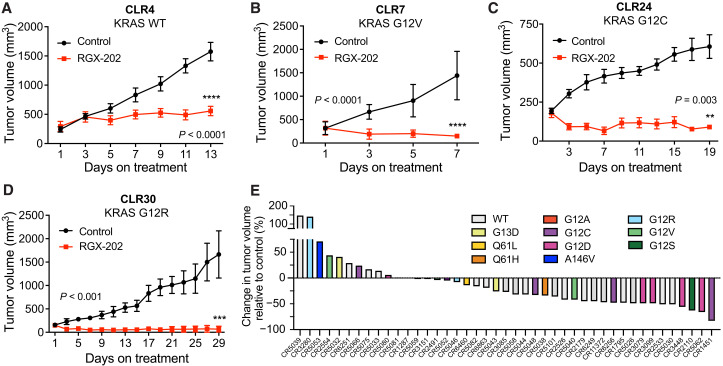
PDXs are thought to better recapitulate human tumor pathology and clinical drug responses (27–30). We therefore assessed the efficacy of SLC6A8 therapeutic inhibition on the growth of established (100 to 250 mm3) human KRAS wild-type and KRAS mutant CRC PDX tumors. Oral RGX-202 administration caused a 65% reduction in the growth of the CLR4 KRAS wild-type PDX model (Fig. 4A). Moreover, treatment also elicited tumor regressions in three PDX models harboring distinct KRAS mutations (CLR7 KRAS G12V, CLR24 KRAS G12C, and CLR30 KRAS G12R; Fig. 4, B to D). To further characterize the range of efficacy of RGX-202-01 across diverse subtypes of CRC tumors, we conducted a mouse PDX trial based on a 1 × 1 trial design where two mice become inoculated with the same PDX model and one mouse receives RGX-202-01 and the other receives control treatment (31). Such a trial design was developed to mimic a patient clinical trial and has shown reproducibility and translatability of therapeutic responses (31). We included 43 well-documented CRC PDXs of various mutational backgrounds including both KRAS wild-type and KRAS mutant subtypes (table S1). Of these models, 49% of tumors harboring diverse KRAS mutant subtypes exhibited single-agent antitumor efficacy greater than 30% relative to matched control animals (Fig. 4E). RGX-202-01 treatment also demonstrated antitumor efficacy in 30% of BRAF mutant tumors (two of seven) (table S1). These findings reveal that SLC6A8 inhibition by oral RGX-202 treatment exhibits single-agent antitumor efficacy against a broad range of CRC tumor subtypes and suggest potential for clinical benefit in both KRAS wild-type and KRAS mutant cancers.
Fig. 4. RGX-202 inhibits growth of KRAS mutant and wild-type CRC PDXs.
(A to D) Subcutaneous tumor growth by ~25 mm3 PDX fragments implanted in athymic nude mice receiving a control or RGX-202–supplemented diet (800 mg/kg) starting at tumor sizes of ~100 mm3 (CLR4), 250 mm3 (CLR7), 200 mm3 (CLR24), and 150 mm3 (CLR30); n = 10 per group. (E) Waterfall plot of responses to RGX-202-01 across 43 colorectal PDX models; each bar represents an individual PDX, and colors represent KRAS mutations. Mice were treated with control or RGX-202-01–supplemented diet at ~400 mg/kg for 21 days. Means ± SEM are shown; P values are based on two-sided t tests (A to D).
SLC6A8 inhibition synergizes with 5-FU and oral leflunomide therapy
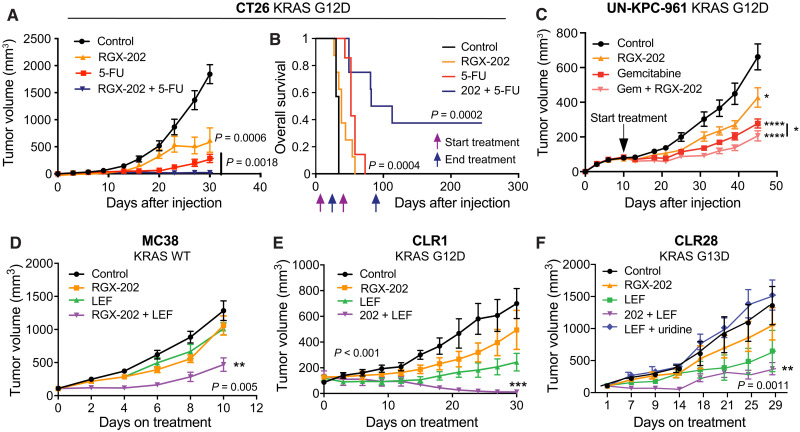
We next determined whether SLC6A8 inhibition could cooperate or synergize with other therapeutics. 5-FU is the backbone of many chemotherapeutic regimens used in CRC. We treated palpable CT26 subcutaneous tumors in immunocompetent syngeneic mice with vehicle control, oral RGX-202, 5-FU, or a combination regimen comprising RGX-202 and 5-FU. While single-agent RGX-202 or 5-FU significantly suppressed tumor growth (66% inhibition RGX-202, 85% inhibition 5-FU), combined RGX-202/5-FU caused a 99% tumor growth reduction (Fig. 5A). Survival studies revealed that combined RGX-202/5-FU therapy markedly enhanced survival of mice, causing 40% of mice to experience complete regression responses and long-term (>240 days) survival (Fig. 5B) despite cessation of treatment at day 85, 160 days before termination of the study. RGX-202-01 also elicited enhanced antitumor efficacy in combination with 5-FU and irinotecan, a standard-of-care regimen for metastatic CRC (fig. S3A). We also observed that combined treatment of established UN-KPC-961 (KrasG12D;Trp53R172H) pancreatic cancer tumors with RGX-202 and gemcitabine, a deoxycytidine analog and an approved pancreatic cancer therapeutic, elicited greater tumor suppression relative to either gemcitabine or RGX-202 alone, although the enhanced effects were modest compared to combination effects observed in CRC (Fig. 5C). We had previously shown that under hypoxia, the growth of metastatic CRC tumors is highly dependent on pyrimidine nucleotide biosynthetic pathways and that these cells become sensitized to inhibition of the DHODH enzyme, which catalyzes a critical step during pyrimidine nucleotide biosynthesis (22, 32). Treatment of mice with the oral DHODH inhibitor leflunomide, which is used as a rheumatoid arthritis drug, inhibited CRC primary tumor and metastatic progression (22). Combined treatment of established MC38 KRAS wild-type syngeneic CRC tumors with RGX-202/leflunomide elicited synergistic activity relative to single-agent treatment with either drug (Fig. 5D). We then tested CLR1 KRAS G12D mutant and CLR28 KRAS G13D mutant PDXs, which exhibited similar synergistic tumor regressions upon combined administration of RGX-202/leflunomide relative to tumor growth suppression responses by either agent alone (Fig. 5, E and F). To determine whether the effect of leflunomide on tumor growth repression was secondary to pyrimidine nucleotide depletion, we included a cohort in the CLR28 model that received leflunomide and uridine, a pyrimidine nucleotide. Uridine supplementation rescued tumor growth inhibition by leflunomide, consistent with pyrimidine nucleotide levels being limiting for tumor growth (Fig. 5F).
Fig. 5. RGX-202 synergizes with 5-FU and leflunomide.
(A and B) Subcutaneous tumor growth (A) and Kaplan-Meier survival curves (B) of 1.5 × 105 CT26 cells in BALB/c mice receiving a control, RGX-202–formulated diet (800 mg/kg), 5-FU (50 mg/kg per week), or a combination of RGX-202 and 5-FU; n = 7 to 8 per group. (C) Subcutaneous tumor growth of 2.5 × 106 UN-KPC-961 cells in B6129SF1/J mice receiving control, RGX-202–formulated diet (800 mg/kg, P = 0.021), gemcitabine (100 mg/kg, intraperitoneally, P < 0.0001), or a combination of RGX-202 and gemcitabine (P < 0.0001); n = 10 per group; *P (combination) = 0.04, one-tailed t test. (D) Subcutaneous tumor growth of 1 × 106 MC38 cells in C57BL/6 mice receiving a control, RGX-202–supplemented diet (200 mg/kg), leflunomide (LEF) (2.5 mg/kg), or combination of RGX-202 and leflunomide; n = 10 per group. (E and F) Subcutaneous tumor growth by CLR1 (E) or CLR28 (F) PDX fragments implanted into athymic nude mice. Treatment with a control diet, RGX-202–supplemented diet (800 mg/kg), a combination of RGX-202 and leflunomide (7.5 mg/kg), or leflunomide and uridine [1 g/kg, (F)] started when tumors reached ~100 mm3; n = 6 per group. Means ± SEM are shown; P values are based on two-sided t tests (A, C, and D to F).
RGX-202–treated animals did not show any adverse effects except for lack of body weight gain in animals that were fed a RGX-202–formulated diet but not when mice received treatment by oral gavage (fig. S3B). Overall, our findings reveal that RGX-202 is well tolerated as an oral therapy and can cooperate or synergize with the standard-of-care agent 5-FU as well as the U.S. Food and Drug and Administration–approved rheumatologic oral drug leflunomide.
Tumoral CKB expression as a predictive biomarker of SLC6A8 inhibition response
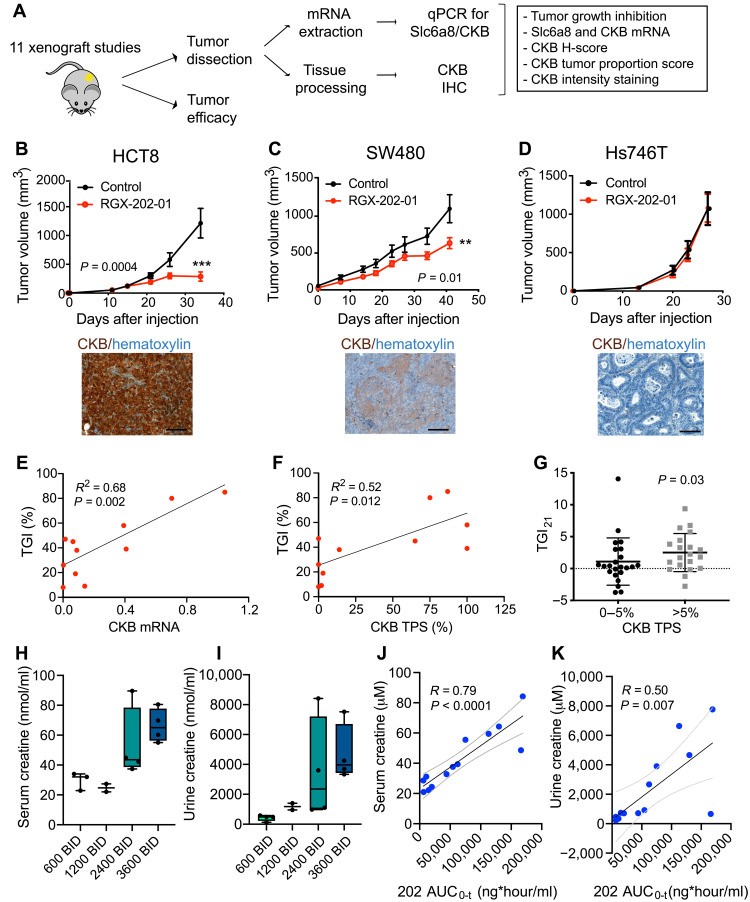
We next sought to identify a predictive biomarker for therapeutic response to SLC6A8 inhibition by RGX-202. Our observations of therapeutic efficacy upon SLC6A8 inhibition in both KRAS mutant, KRAS wild-type, and mismatch repair mutant CRC lines suggested that sensitivity to SLC6A8 inhibition was not solely attributable to oncogenic or genomic instability mutational backgrounds (table S2). We previously showed that the CKB enzyme is released from CRC cells and acts upstream of the SLC6A8 transporter by generating the energetic metabolite phosphocreatine, which is imported through the SLC6A8 transporter (15). Extracellular phosphocreatine supplementation rescued hypoxic survival impairment caused by CKB depletion in an SLC6A8-dependent manner (15). CKB expression was also shown to be repressed by microRNA-mediated silencing, raising the possibility that variation in its expression may predict sensitivity to therapeutic targeting of this axis (15). We therefore tested a collection of gastrointestinal cell lines as xenografts for responsiveness to RGX-202-01 (see table S2 for details on the cell lines). An independent cohort of animals was simultaneously inoculated with the same xenografts, and tumors were allowed to reach ~500 mm3, at which point tumoral SLC6A8 and CKB gene expression was assessed by quantitative polymerase chain reaction (qPCR) and CKB protein expression by immunohistochemistry (IHC) (Fig. 6A). SLC6A8 was not tested by IHC due to the lack of availability of an SLC6A8-specific antibody. Included in this assessment were tumors that responded to treatment and those that did not (Fig. 6, B to D; fig. S4; and table S2). We observed that therapeutic responses to RGX-202-01 positively correlated with increased CKB mRNA expression (Fig. 6E) but not SLC6A8 mRNA expression (fig. S5A). Consistent with this, the extent of CKB protein expression—as assessed by tumor proportion score (TPS)—also positively correlated with the magnitude of therapeutic response (Fig. 6F). We observed that tumors expressing elevated CKB protein levels (Fig. 6B) exhibited greater tumor growth repression responses relative to those expressing reduced CKB levels (Fig. 6D and fig. S5, B and C). To determine whether this observation could be recapitulated in an independent dataset, we assessed the association of CKB protein expression with therapeutic responses in the aforementioned mouse RGX-202-01 1 × 1 PDX trial. We observed a similar predictive association of elevated CKB protein expression by IHC with antitumor response (Fig. 6G). Creatine kinases are dimeric enzymes that consist of two subunits, CKB (brain type) or CKM (muscle type). Three different isoenzymes therefore exist: CK-MM, CK-BB, and CK-MB. We thus asked whether any of the other isoforms might be contributing to efficacy and measured CKM expression in tumors by qPCR. Analysis of the 11 aforementioned xenografts revealed significantly lower expression of CKM compared to CKB (fig. S5D), and no correlation of CKM expression with antitumor efficacy was observed (fig. S5E). We also assessed expression of the two mitochondrial creatine kinases CKMT1 and CKMT2. While both enzymes were detectable, expression of CKMT2 was significantly lower than CKB expression, with 4 of 11 xenografts having undetectable CKMT2 expression. We did not observe an association of CKMT expression with response to RGX-202 treatment (fig. S5, D, F, and G). These results demonstrate that elevated tumoral CKB mRNA and protein expression levels predicts enhanced responsiveness to SLC6A8 inhibition by RGX-202, consistent with CKB being an upstream component of the CKB/SLC6A8 phosphocreatine metabolic axis in CRC and a gene that exhibits posttranscriptional expression modulation and variation in cancer. Consistent with the proposed mechanism of action of RGX-202, we observed a positive correlation of response to RGX-202-01 with induction of CC3, a marker of apoptosis, and a negative correlation of response to RGX-202-01 with the proliferation marker Ki67 (fig. S5, H and I).
Fig. 6. Tumoral CKB expression and creatine levels as predictive and pharmacodynamic biomarkers of SLC6A8 inhibition.
(A) Experimental design. (B to D) Tumor growth by 5 × 106 HCT8 (B), 5 × 106 SW480 (C), and 2 × 106 Hs746T (D) cells subcutaneously injected into athymic nude mice receiving a control or RGX-202-01–supplemented diet (800 mg/kg). Pictures show IHC of CKB expression (brown) and hematoxylin (blue) in the control tumors; n = 8 to 10 per group. Means ± SEM are shown; P values based on two-sided t tests. (E and F) Linear regression analyses of TGI as a function of CKB mRNA (E) or CKB tumor proportion score (TPS) (F). Each dot represents the mean value of data points from one xenograft model; n = 5 to 7 tumors per model. (G) Nonparametric analysis of TGI on day 21 relative to control from the 43 PDX models, stratified into models with low (0 to 5%) or >5% CKB TPS. (H and I) Box-and-whisker plot representing the median of creatine concentrations in the serum (H) and urine (I) on day 15 of treatment; n = 2 to 4 patients per group. BID, twice daily. (J and K) Correlation analysis of serum (J) or urine (K) creatine concentration and RGX-202 blood exposure (AUC0-t). Dashed lines denote 95% confidence interval; n = 13 patients (H to K). Scale bars, 100 μm (B to D).
To assess what fraction of patients with metastatic CRC harbor tumors with elevated CKB protein expression, we immunohistochemically stained 23 human metastatic CRC specimens for CKB. We observed CKB positivity (TPS >5%) in the majority (56%; 13 of 23) of the tumor tissue samples (fig. S6, A and B). Overall, these findings reveal CKB as a potential patient stratification biomarker for clinical development of SLC6A8 inhibitors. We next evaluated CKB expression in a tissue microarray containing specimens from 10 human tissues (fig. S6C). We observed the highest CKB expression with a 100% TPS in the brain and heterogeneous expression patterns in several tissues including colon, appendix, bladder, kidney, myometrium, and pancreas. We did not detect CKB protein expression in the spleen, liver, and testis. These results are consistent with gene expression analysis using the Genotype-Tissue Expression (GTEx) database (fig. S6D) and previously published data (33).
RGX-202 increases serum and urinary creatine excretion in patients with cancer
Motivated by the robust preclinical efficacy of RGX-202-01, we initiated a multicenter phase 1a/b clinical trial in patients with advanced gastrointestinal cancers that had progressed on standard-of-care regimens (ClinicalTrials.gov, NCT03597581). In the phase 1a dose escalation stage of the study, patients received oral RGX-202-01 treatment at doses ranging from 600 to 3600 mg twice daily on a continuous regimen. Blood and urine samples were collected from 13 patients on day 15 during the first cycle (28 days), and bioanalytic analyses were conducted by an independent contract research laboratory using commercially available proprietary assays. Consistent with our observations in mice where RGX-202-01–mediated suppression of creatine uptake in tumors and tissues led to increased circulating and urinary excretion levels of creatine, we observed that patient serum and urine creatine levels increased upon RGX-202-01 administration to patients with gastrointestinal cancer (Fig. 6, H and I). Creatine concentrations in both serum and urine showed positive correlations with systemic exposure to RGX-202-01 (Fig. 6, J and K). These findings provide a proof of concept for therapeutic targeting of creatine metabolism in humans, mirroring our experimental observations in mice.
DISCUSSION
The creatine/phosphocreatine bioenergetic shuttle is a critical system that allows highly metabolic tissues to respond to energetic stress quickly through the generation of high-energy ATP in a reaction that does not require oxygen. CRC and pancreatic cancers are particularly hypoxic malignancies (19–21). The ability of such cancers to overexpress and release CKB as a means of generating extracellular phosphocreatine for import through the SLC6A8 transporter enables cancer cells to enhance availability of high-energy phosphate for ATP generation in the context of hypoxic and metabolic stresses. SLC6A8 inhibition by RGX-202 reduced tumor growth in numerous syngeneic, xenograft, and PDX mouse models and also suppressed liver metastasis. Antitumor efficacy was associated with enhanced tumor apoptosis and reduced tumor cell proliferation, consistent with previous findings supporting a critical role for creatine metabolism in CRC progression and hypoxic survival (15). While we have provided evidence revealing that RGX-202 targets the SLC6A8 creatine/phosphocreatine axis, we cannot exclude the possibility that RGX-202 may mediate additional antitumor effects via additional target(s).
Our work reveals that mutational background is not a significant predictor of response to SLC6A8 inhibition and that RGX-202 impairs the growth of a broad set of CRC tumors of distinct KRAS and mismatch repair mutant backgrounds, suggesting that rather diverse CRC tumors exploit creatine/phosphocreatine metabolism to drive progression. Consistent with a critical role for CKB in this pathway, we find that CKB expression levels are associated with response to RGX-202. Tumors with high CKB expression responded better than those with reduced levels, suggesting that CKB-overexpressing tumors are more dependent on phosphocreatine as an energy source and thus more sensitive to its depletion. These findings support the potential for assessing patient tumor CKB expression as a molecular biomarker for patient stratification in clinical trials.
Our data also reveal that inhibition of SLC6A8 and blockade of cellular creatine/phosphocreatine uptake lead to increased creatine excretion in the urine, both in mice and in patients. Urinary creatine levels positively associated with blood RGX-202 concentrations and represent a direct measure of SLC6A8 inhibition. The opportunity for noninvasive monitoring of the pharmacodynamics of creatine transport inhibition through urinalysis of creatine and potentially additional metabolites provides a simple and rapid method for assessing target engagement in patients.
Our findings reveal both single-agent activity and combination efficacy of RGX-202 in CRC. Because 5-FU is the mainstay chemotherapeutic used in CRC, we tested the activity of RGX-202 in combination with 5-FU and 5-FU/irinotecan. Our data show synergistic effects of SLC6A8 inhibition and complete tumor regressions with these standard-of-care regiments. Similarly, we observed robust antitumor efficacy when combining RGX-202 with the DHODH inhibitor leflunomide, an oral therapy that was previously shown to suppress CRC growth under hypoxia (22). The synergistic effects observed likely reflect the impairment of these two orthogonal metabolic pathways—creatine/phosphocreatine and pyrimidine biosynthesis—that appear critical for the growth and survival of CRC cells under hypoxia. Collectively, these results provide a rationale for incorporating RGX-202 into combination regimens including standard-of-care agents or those targeting nucleotide synthesis. Future studies could investigate the impact of this therapy in the adjuvant setting, as a means of suppressing CRC metastatic progression.
Overall, this work supports the potential for therapeutic targeting of creatine metabolism in CRC through the inhibition of the SLC6A8 transporter. While we have demonstrated pharmacodynamic effects on creatine metabolism in patients upon RGX-202 treatment, ongoing trials are testing the efficacy of this therapeutic approach in patients. To this end, this treatment approach is currently being tested in a multicenter national phase 1b/2 clinical trial in CRC (ClinicalTrials.gov, NCT03597581).
MATERIALS AND METHODS
Experimental design
The number of samples for each group was chosen based on knowledge on intragroup variation and expected effect size. Sample sizes for in vitro experiments were chosen based on previous knowledge on intragroup variation. Data were collected until reaching a predetermined end point (in vitro assays, PDX trial, and clinical sample analysis) or tumor burden exceeded >2000 mm3. No data were excluded. Replications were performed as noted in the text and figure legends. Replications for all presented experiments were successful. Samples were allocated randomly if possible. Animals were randomized before start of treatment, and mice were sex- and age-matched. No blinding was performed in the in vivo experiments due to cage labeling requirements. Quantification of CKB, CC3, and Ki67 signal by IHC was blinded.
Animal strains
All mouse experiments and procedures were approved by the Institutional Animal Care and Use Committee (IACUC) at the New York Blood Center, The Rockefeller University, and Crown Biosciences. C57BL/6 (JAX stock #000664, RRID: IMSR_JAX:000664), NOD-SCID (JAX stock #001303), athymic nude (J:NU, JAX stock #007850), NOD scid gamma (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ, JAX stock #005557), and B6129SF1/J (JAX stock #101043) mice were purchased from the Jackson Laboratory. BALB/c mice (stock #028) were purchased from Charles River. SLC6A8 knockout mice were originally obtained from the laboratory of M. R. Skelton (34) and bred in-house. BALB/c nude mice were purchased from Beijing AniKeeper Biotech Co. Ltd. (Beijing, China).
Primary tumor growth studies
For primary tumor growth experiments, cells [suspended in 50 μl of phosphate-buffered saline (PBS)] were mixed 1:1 with Matrigel (BD Biosciences, Bedford, MA, 356231) and subcutaneously injected unilaterally or bilaterally into the lower flank of 6- to 8-week-old sex-matched mice. Upon detection of tumor volumes reaching the size indicated in each figure, mice were randomly assigned to a drug treatment or a control cohort. RGX-202 was administered through formulated drug chow (Purina 5001, Research Diet, New Brunswick, NJ) at indicated doses or formulated in sterile drinking water for oral gavage. Control cohorts received either regular chow (Purina 5001) or vehicle control. Where xenograft models were tested for biomarker analysis, 10 to 15 animals were inoculated with the indicated cell lines. Out of these animals, 7 to 10 mice were assigned to the efficacy portion and were treated for the duration of the experiment. The remaining three to five animals were sacrificed when tumors reached ~500 mm3. Protein and RNA analysis of CKB expression was conducted on the control tumors. Tumor growth measurements were taken using a digital caliper on the days indicated throughout the course of the experiment. Tumor volume was calculated using the following formula: volume = (longest diameter)/2 × (shortest diameter)2. For survival analysis, mice were euthanized when the total tumor burden approached IACUC guidelines, with a tumor burden exceeding 2000 mm3 in volume. TGI was calculated using the following formula: TGI (%) = ((∆Ci − ∆C0) – (∆Ti − ∆T0))/(∆Ci − ∆C0)*100%.
PDX studies
Implantation of PDX’s was conducted as described before (22). Briefly, 20- to 30-mm3 tumor fragments were subcutaneously implanted bilaterally into the flank of 6- to 8-week-old age-matched athymic nude mice under general anesthesia with ketamine (100 mg/kg) (Henry Schein Animal Health, Melville, NY, 995-2949) and xylazine (10 mg/kg) (Sigma-Aldrich, St. Louis, MO, X1251).
Metastasis assays
Experiments were conducted by intrasplenic injection of 5 × 105 luciferase-labeled Lvm3b or CKB CRISPR knockout or CRISPR control Lvm3b cells suspended in 50 μl of PBS into 6- to 8-week-old NOD-SCID mice or NSG mice, respectively, that were anesthetized through intraperitoneal injection of ketamine (100 mg/kg)/xylazine (10 mg/kg) solution. The day after tumor cell inoculation, mice were randomly assigned to a control or RGX-202 treatment group. Control mice received intraperitoneal injections of 200 μl of PBS, and treatment mice received 200 μl of 0.5 M RGX-202 (~650 mg/kg). Treatment continued daily until the end of the experiment. In the PANC1 experiment, cells were grown in D10F complete medium and pretreated in vitro with or without RGX-202 at a dose of 10 mM (1.31 mg/ml) RGX-202 for 48 hours before injection into the mice. There was no treatment of the mice after the cells were injected. Bioluminescence measurements were conducted once per week for the duration of the experiment. One hundred microliters of d-luciferin [1 g in 60 ml of sterile Dulbecco’s PBS (DPBS); Thermo Fisher Scientific, Waltham, MA, 88292] was injected into the venous sinus, and the bioluminescence signal over the liver was measured using the IVIS Spectrum In Vivo Imaging System (PerkinElmer). Photon flux ratio is the ratio of bioluminescence signal at a given time point to the signal on day 0.
Drug treatments
5-FU (Millipore Sigma, St. Louis, MO, F6627) was administered in sterile 0.9% NaCl by intraperitoneal injections once per week as indicated in the figures. Gemcitabine (Millipore Sigma, St. Louis, MO, G6423) was administered at 100 mg/kg per week, intraperitoneally, in PBS. Irinotecan (50 mg; Millipore Sigma, St. Louis, MO, I1406) was administered at 15 mg/kg per week, intraperitoneally, in 2.5% dimethyl sulfoxide (DMSO) and 97.5% NaCl 0.9%. Leflunomide was formulated in DMSO and administered by daily intraperitoneal injections at 2.5 or 7.5 mg/kg at 0.5 ml/kg. Uridine was formulated in sterile water at 800 mg/kg and administered by daily intraperitoneal injections.
Cell culture
UN-KPC-961 cells were obtained from S. K. Batra at Eppley Institute for Research in Cancer (Omaha, Nebraska). COLO 205 (CCL-222), HCT15 (CCL-225), HT29 (HTB-38), SW480 (CCL-228), HCT116 (CCL-247), HCT8 (CCL-244), NCI-H508 (CCL-253), HepG2 (HB-8065), Hs746T (HTB-135), CT26 (CRL-2638), LS-174T (CL-88), PANC1 (CRL-1469), and NCI-N87 (CRL-5822) cell lines were purchased from the American Type Culture Collection (ATCC) (Baltimore, MD) and maintained in standard conditions following the providers’ instructions. MC38 cells were purchased from Kerafast (Boston, MA). Lvm3b was generated by in vivo selection from the parental cell line LS-174T (ATCC, Baltimore, MD) (15). UN-KPC-961 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, Langley, OK, 11960-044), 7.5% sodium bicarbonate (Gibco, Langley, OK, 25080-094), 1% penicillin-streptomycin (Lonza, 17-745E), 10% fetal bovine serum (FBS) (Sigma-Aldrich, St. Louis, MO, F4135), 200 mM l-glutamine (Gibco, Langley, OK, 25030081), 1 mM Hepes (Gibco, Langley, OK, 15630080), and gentamicin (50 mg/ml) (Gibco, Langley, OK, 15750078). MC38 cells were cultured in DMEM, 10% FBS, and 1 mM Hepes.
A 1 × 1 PDX trial (HuTrial)
Details on the models are found in table S1. PDX models were inoculated with a PDX cell suspension or a tumor fragment as follows. Cryogenic vials containing PDX tumor cells were thawed, and cells were washed in RPMI 1640, counted, and resuspended in cold RPMI 1640 at a concentration of 50,000 to 100,000 viable cells/50 ml. Cell suspensions were mixed with an equal volume of Cultrex extracellular matrix (ECM). One hundred microliters of the cell suspension in ECM medium was subcutaneously injected into the rear flank of five female NOD-SCID mice per model. Alternatively, tumor fragments from stock mice were harvested and used for inoculation into mice. Five 6- to 8-week-old female BALB/c nude mice per model were inoculated subcutaneously in the right flank with a primary human tumor fragment (2 to 3 mm in diameter) for tumor development. Two of these five mice were randomized into two groups (one mouse per group) when average tumor volume reached 100 to 150 mm3 and treatment with either control chow (Purina 5001) or RGX-202–formulated chow (in Purina 5001, Research Diet, New Brunswick, NJ) at ~400 mg/kg started within 24 hours after randomization. Randomization was performed based on the “Matched distribution” method (StudyDirector software, version 3.1.399.19). TGI on day 21 (TGI21) was calculated using the following formula: TGI21 = ((C21 − C0)/C21) – ((T21 − T0)/T0). The PDX trial was conducted at Crown Biosciences (San Diego, CA and Taicang Jiangsu Province, China).
Mouse urine and plasma creatine analysis
Six- to 8-week-old female CD1 mice were fed with control or RGX-202-01–formulated chow (Purina 5001, Research Diet, New Brunswick, NJ) at 100, 400, and 1200 mg/kg for 10 days. Bleeds were collected in EDTA-coated blood collection tubes (Thermo Fisher Scientific, 02-669-33), and plasma was separated upon centrifugation. Plasma and urine samples were collected from both the control and RGX-202-01–treated cohorts on day 10 (240, 246.5, and 254.5 hours) and day 11 (264 hours) time points. Samples were flash-frozen and stored at −80°C until bioanalysis for RGX-202 and creatine levels at Pharmaron (Beijing, China). Mice plasma and urine analysis were conducted as a non-GLP (good laboratory practice) study according to standard operating procedures at Pharmaron (Beijing, China). Briefly, the concentration of RGX-202-001 was determined using the surrogate analyte RGX-202-13C115N2 by LC-MS/MS. Lower limit of quantification was determined to be 50.0 ng/ml. The in-life portion of this study was conducted at L2P Research.
Creatine-(methyl-d3) uptake in the tumors and heart
Three to four B6129SF1/J male mice harboring UN-KPC-961 tumors that were treated with either control chow or RGX-202–formulated chow at ~800 mg/kg for 35 days received one dose of creatine-(methyl-d3) (1 mg/kg) (Cambridge Isotope Laboratories, Tewksbury, MA, DLM-1302-0.25) by intraperitoneal injection. One and a half hours later, the animals were euthanized and the tumors and hearts were extracted, flash-frozen in liquid nitrogen, and submitted to Seventh Wave Laboratories for bioanalysis.
Three to four 6- to 9-week-old C57BL/6J male wild-type mice received one dose of RGX-202 at 100, 250, or 500 mg/kg, respectively, in sterile 0.9% NaCl. Seven minutes following injection of RGX-202 or vehicle control, one dose of d3-creatine was administered at 1 mg/kg in sterile 0.9% NaCl by intraperitoneal injection. SLC6A8 knockout mice received one dose of d3-creatine. One hour later, the mice were anesthetized using 2.5% isoflurane and the animal was perfused with 5 to 10 ml of DPBS that was injected into the left heart chamber. The heart was then extracted and flash-frozen in liquid nitrogen. Heart samples were stored at −80°C until being submitted to Seventh Wave Laboratories for analysis. Hearts and tumors were homogenized with a 70/30 methanol/water solution at a 3:1 (v/w) ratio. Twenty microliters of the homogenate was diluted with 1 μl of internal standard solution (creatine-d5 in methanol). The samples were centrifuged, and the supernatant was analyzed. A fit-for-purpose, semiquantitative LC-MS/MS method was developed at Seventh Wave Laboratories for the determination of d3-creatine and creatinine in heart and tumor tissue homogenate. Creatinine concentrations were used to normalize the signal of d3-creatine across samples.
Real-time PCR analysis of tumor samples
RNA was extracted from 10 mg of flash-frozen vehicle or RGX-202-01–treated tumors using a Total RNA Purification kit (Norgen Biotek, Thorold, Canada, 37500) according to the manufacturer’s instructions. Ribonuclease-free deoxyribonuclease I (Norgen Biotek, Thorold, Canada, 25710) was used to remove genomic DNA. Complementary DNA (cDNA) synthesis was performed with 600 ng of total RNA using the Verso cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, MA, AB-1453/B) according to protocol. qPCR was performed using TaqMan Fast Universal PCR Master Mix (2×), No AmpErase UNG (Thermo Fisher Scientific, catalog no. 4366073), and TaqMan probes (20×) with 1 μl of 1:5 diluted cDNA.
Predesigned TaqMan gene expression assays for CKB, SLC6A8, CKM, CKMT1, CKMT2, and GUSB (Hs00176484_m1, Hs00940515_m1, Hs00176490_m1, Hs00179727_m1, Hs00176502_m1, and Hs00939627_m1; Thermo Fisher Scientific, Waltham, MA) were used to perform the reactions in a StepOnePlus Real-time PCR system (Applied Biosystems). GUSB was used as an internal control.
A standard curve for normalization was generated by preparing serial dilutions as follows. Undiluted cDNA from the HCT116 cell line at a stock concentration of 30 ng/μl was used to prepare dilutions of 12.5, 4.17, 1.25, 0.42, 0.125, and 0.013 ng. A 1:5 diluted cDNA sample of HCT116 cell line was used as a reference that was run in all qPCR plates to account for plate-to-plate variability. All standards and samples were tested in triplicate or quadruplicate.
Histology, IHC, and immunofluorescence
Tumors were fixed overnight with 4% paraformaldehyde (EMS, Hatfield, PA, 50-980-497) at 4°C. After two washes with PBS (Gibco, 10010023), tissue was embedded in paraffin following standard protocols. Tumors were sectioned using a microtome (Leica) and 5-μm-thick sections were mounted on Superfrost Plus Microscope Slides (Thermo Fisher Scientific, 22-037-246). The tissue sections were blocked first for 30 min in Background Blocking reagent (Innovex, NB306). A rabbit monoclonal anti-creatine kinase B–type antibody (Abcam, ab92452) was used in 1:400 dilution. The incubation with the primary antibody was done for 5 hours followed by 60 min with biotinylated goat anti-rabbit immunoglobulin G (IgG) (Vector Labs, PK6101) in 5.75 μg/ml, and Blocker D, streptavidin–horseradish peroxidase (HRP), and DAB detection kit (Ventana Medical Systems) were used according to the manufacturer’s instructions. Slides were counterstained with hematoxylin and coverslipped with Permount (Thermo Fisher Scientific). Antibody specificity was determined by stainings conducted on HCT116 cells transiently transfected with CKB small interfering RNAs (siRNAs). A tumor microarray containing 10 human healthy tissues in duplicate was purchased (OriGene, CT565861).
The detection of Ki67 Al 488+ and CC3 CF 594 by immunofluorescence was performed as follows: After 32 min of heat and CC1 (Cell Conditioning 1; Ventana Medical Systems, 950-500) retrieval, the tissue sections were blocked first for 30 min in Background Blocking reagent (Innovex, NB306). A mouse monoclonal anti-Ki67 antibody (DAKO, M7240) was used in concentrations of 0.5 μg/ml. The incubation with the primary antibody was done for 5 hours followed by biotinylated goat anti-mouse secondary antibody (5.75 μg/ml) (MOM Kit BMK-2202, Vector Labs). Blocker D and streptavidin-HRP D (part of DAB Map kit, Ventana Medical Systems), followed by incubation with tyramide Alexa Fluor 488 (Invitrogen, T20922), were prepared according to the manufacturer’s instruction in 1:150 for 16 min. A rabbit polyclonal anti-CC3 (Cell Signaling Technology, 9661) was used in a concentration of 0.1 mg/ml. The incubation with the primary antibody was done for 5 hours followed by 60-min incubation with biotinylated goat anti-rabbit IgG (Vector Labs, PK6101) in a concentration of 5.75 μg/ml. Blocker D, streptavidin-HRP, and tyramide-CF594 (92174, Biotium) were prepared according to the manufacturer’s instruction in 1:1500 dilution for 16 min. After staining, slides were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (5 mg/ml; Sigma-Aldrich, D9542) for 10 min and mounted with Mowiol. All stainings were performed at the Molecular Cytology Core Facility of Memorial Sloan Kettering Cancer Center using Discovery XT processor (Ventana Medical System, Roche, Indianapolis, IN).
Analysis of tumor samples
Six to eight tumors per xenograft model were assigned a score of 0 to 3 based on intensity of CKB staining. Score 0 is interpreted as negative for protein expression, while scores 1, 2, and 3 are interpreted as positive staining for each core, with 3 being the maximal intensity. For each positive sample, the area (% of tumor) corresponding to each intensity staining was recorded to allow for calculation of percentage tumor positivity (TPS). A weighted overall staining score (H-score) was calculated as (percentage area of 1 + staining × 1) + (percentage area of 2 + staining × 2) + (percentage area of 3 + staining × 3). Tumor sections were excluded from the analysis if that section was predominantly necrotic. In these cases, additional tumor sections were stained to reach a minimum of n = 3 to 4 sections per tumor, 3 tumors per cohort.
Image quantification
Quantification of the number of Ki67-positive cells and percentage of CC3 per area were performed using ImageJ (version 1.50i). Three tumors per group were selected, and three to five fields per tumor were chosen for quantification.
Hypoxia cell growth assay
Lvm3b cells were seeded at 300,000 cells per well in a six-well plate. After 24-hour incubation under normoxia, the cells were cultured under hypoxia (0.5% oxygen, 5% CO2, 37°C) for 96 hours in the presence of RGX-202, creatine (Sigma-Aldrich, #C3630), or phosphocreatine (Sigma-Aldrich, #237911) at 10 μM. The medium was replaced, and incubation continued up to 120 hours when cells were counted.
Metabolite extraction and liquid chromatography
Lvm3b cells were plated at 3 × 105 cells per well in triplicate in RPMI 1640 in the presence of dialyzed FBS, 2 mM glutamine, and 6 mM glucose and allowed to adhere to the plate for 24 hours. Cells are treated with either control or 10 mM RGX-202 for 24 hours in 0.5% O2. Cells were washed with ice-cold 0.9% NaCl and harvested in ice-cold 80:20 LC-MS methanol:water (v/v). Samples were vortexed vigorously and centrifuged at 20,000g at maximum speed at 4°C for 10 min. The supernatant was transferred to new tubes. Samples were then dried to completion using a nitrogen dryer. All samples were reconstituted in 30 μl of 2:1:1 LC-MS water:methanol:acetonitrile. The injection volume for polar metabolite analysis was 5 μl. Metabolite extraction and subsequent LC coupled to high-resolution MS (LC-HRMS) for polar metabolites of cells were carried out using Q Exactive Plus.
Liquid chromatography
A ZIC-pHILIC 150 × 2.1 mm (5-μm particle size) column (EMD Millipore) was used on a Vanquish Horizon UHPLC system for compound separation at 40°C. The autosampler tray was held at 4°C. Mobile phase A is water with 20 mM ammonium carbonate and 0.1% ammonium hydroxide (pH 9.3), and mobile phase B is 100% acetonitrile. The gradient is linear as follows: 0 min, 90% B; 22 min, 40% B; 24 min, 40% B; 24.1 min, 90% B; and 30 min, 90% B. The following rate was 0.15 ml/min. All solvents are LC-MS grade and purchased from Thermo Fisher Scientific.
Mass spectrometry
The Q Exactive Plus MS (Thermo Fisher Scientific) is equipped with a heated electrospray ionization (HESI) probe, and the relevant parameters include heated capillary, 250°C; HESI probe, 350°C; sheath gas, 40; auxiliary gas, 15; sweep gas, 0; and spray voltage, 3.0 kV. A full scan ranging from 55 to 825 m/z (mass/charge ratio) was used. The resolution was set at 70,000. The maximum injection time was 80 ms. Automated gain control (AGC) was targeted at 1 × 106 ions. Maximum injection time was 20 ms. Raw data collected from LC-Q Exactive Plus MS were processed on Skyline (https://skyline.ms/project/home/software/Skyline/begin.view) using a 5-ppm (part per million) mass tolerance and an input file of m/z and detected retention time of metabolites from an in-house library of chemical standards. The output file including detected m/z and relative intensities in different samples was obtained after data processing. Quantitation and statistics were calculated using Microsoft Excel, GraphPad Prism 8.1, and RStudio 1.0.143.
Clinical specimen analysis
Serum specimens were drawn from patients on cycle 1 day 15 at 0, 0.5, 1, 1.5, 2, 4, 6, 8, 10, and 12 hours after administration of RGX-202-01 and were analyzed for RGX-202 using an analytically validated LC-MS/MS method at Syneos Health. Urine specimens were collected on cycle 1 day 15 at 0, 4, and 12 hours after administration of RGX-202; average values are presented in the figure. Serum sample for creatine analysis was collected at 12 hours after administration. Creatine levels were analyzed by Q2 solutions at contract research laboratories.
Patient details
In the RGX-202-01 phase 1a/b human study, adult patients of both sexes over the age of 18 years were enrolled. Data from 13 patients were analyzed. The study is currently accruing and ongoing. The protocol was approved by all site Institutional Review Boards, and all patients signed informed consents before any screening procedures were obtained.
Clinical study design
This phase 1a/b study is open label, multicenter, and single arm, whose primary objective is to determine the maximum tolerated dose, or maximum tested dose at which multiple dose-limiting toxicities (DLTs) are not observed, of RGX-202-01. Inclusion and exclusion criteria are stipulated, requiring all patients to have a pathologic confirmation of a locally advanced or metastatic solid tumor or lymphoma that has been deemed refractory to standard therapies. Patients may not have any other active malignancy that could confound the study end points. Patients must not have a history of pancreatitis, active hepatitis B or C, or any illness/social situation in the opinion of the treating investigator that would limit compliance with the study requirements. Patients are not allowed to be treated with any other antineoplastic therapies while on study. Typical phase 1 study parameters are required for performance status, hematologic, and other organ function measurements. Patients are required to use acceptable contraceptive methods while on study and for a specified time period thereafter. Concomitant medications are restricted only if they pose a clinical risk of drug-drug interactions. Treatment for any condition with corticosteroids is not allowed unless at doses less than 10 mg daily prednisone equivalents. Patients were treated with RGX-202-01 at 600, 1200, 2400, or 3600 mg twice per day continuous dosing according to the patient cohort to which they were enrolled. Patients continued treatment with RGX-202-01 until treatment intolerance or progression of disease. The primary end point is incidence of DLTs, which is evaluated by the Inspirna sponsor medical monitor in collaboration with all treating clinical investigators. Secondary end points include pharmacokinetic measurements of RGX-202 and its metabolites in plasma and urine. Exploratory end points include measuring of serum and urine levels of creatine. Ultimately, efficacy end points will be obtained on a large sample size of patients in disease-specific expansion cohorts. DLTs are defined as any of the following toxicities occurring during the first 4 weeks of treatment that are not clearly related to another cause (i.e., disease progression): any grade ≥3 nonhematologic AE (adverse events) with the exceptions of grade 3 nausea, vomiting, diarrhea, constipation, fever, fatigue, or skin rash in which there has been suboptimal prophylaxis and management that resolves to grade 2 within 72 hours; grade 4 thrombocytopenia, or grade 3 thrombocytopenia with grade >1 bleeding or requirement for platelet transfusion; grade 4 neutropenia; grade ≥3 febrile neutropenia; grade ≥3 transaminase [AST (aspartate aminotransferase)/ALT (alanine aminotransferase)] elevation; any toxicity resulting in >25% held/skipped doses during the cycle; and any other substantial toxicity considered by the Investigator Sponsor’s medical representatives to be dose-limiting.
Statistical analysis
Significance of tumor growth curve comparisons was carried out using two-sided t tests. Metastasis assays were analyzed using the nonparametric Mann-Whitney U test. The Mantel-Cox log-rank test was used for statistical comparisons in survival analyses. Statistical analysis of creatine and RGX-202 area under the curve (AUC) correlations in human patients and mice was carried out using simple linear regression with GraphPad Prism 8. Statistical comparisons of CC3 and Ki67 signal, in vitro and in vivo d3-creatine analysis, qPCR, and IHC quantification were carried out using two-sided t tests. T test was used to compare metabolite abundance with Bonferroni’s multiple test correction where indicated. Throughout all figures: *P < 0.05, **P < 0.01, and ***P < 0.001, ****P < 0.0001. Significance was concluded at P < 0.05.
Acknowledgments
We are grateful to members of our laboratories for providing insightful comments on past versions of this manuscript. We thank E. Martinez for guidance on in vitro and in vivo creatine-d3 experiments and D. Darst for intellectual and operational guidance in RGX-202 development. We thank S. Wald for oversight of RGX-202-01 production. We thank C. Bailey, C. Sun, K. Hodges, and their teams at Crown Biosciences for performing the PDX trial. We thank S. Anaganti at L2P Research for conducting the in-life portion of the mouse creatine experiment. We thank Seventh Wave Laboratories, Q2 Solutions, and Syneos Health for bioanalytical analysis. Funding: This work was funded by SKI 05160 General Fund (to A.B. and K.M.-T.), NCI award CCSG P30 CA008748-53 (to A.B. and K.M.-T.), Robertson Foundation Pre-Clinical Award (to H.S.T. and N.Y.), Black Family Center for Research on Human Cancer Metastasis (to N.Y.), The National Center for Advancing Translational Sciences UL1TR001866 (to N.Y.), Meyer Clinical Scholar Support (to N.Y.), The Rockefeller University Clinical Scholar Endowment Fund (to N.Y.), Faculty Scholars grant from the Howard Hughes Medical Institute (to S.F.T.), and The Black Family Foundation (to S.F.T.). Author contributions: Conceptualization: I.K., M.F.T., and S.F.T. Experimentation and analysis: I.K., N.Y., C.A.-A., H.S.T., S.S., S.T., A.B., F.C.G., J.M.L., and R.B. Supervision: I.K., C.A.-A., N.Y., F.C.G., K.M.-T., M.F.T., and S.F.T. Design and supervision of the clinical trial: M.F.T., M.S., R.W., S.R., S.L.S., and S.F.T. Clinical investigators: J.C.B., J.S., M.F., A.J.M., A.E.H., L.S.R., and A.C. Writing—original draft: I.K., N.Y., C.A.-A., and S.F.T. Writing—review and editing: I.K. and S.T. Competing interests: M.F.T. and S.F.T. are cofounders and shareholders of Inspirna. M.F.T. is a board of director and S.F.T. is a member of Inspirna’s scientific advisory board. M.F.T., I.K., F.C.G., C.A.-A., S.T., S.S., F.C.G., R.B., R.W., and S.R. are stock/option holders of Inspirna. M.F.T., I.K., F.C.G., C.A.-A., S.T., and R.W. are current and S.S., F.C.G., and S.R. are past employees of Inspirna. S.L.S. and R.B. are consultants to Inspirna. J.M.L. and S.T. are inventors of a patent related to this work: WO/2014/071067 (PCT/US2013/067860)—2013; patent published, The Rockefeller University. N.Y. and S.T. are inventors of a patent related to this work: WO2021062157A1 (PCT/US2020/05272)—2020; patent published, The Rockefeller University. Inspirna, M.F.T., I.K., and F.C.G. are inventors of a patent related to this work: PCT/US2020/064317—2020; PCT application filed. Inspirna, I.K., and M.F.T. are inventors of a patent related to this work: U.S. patent #63/223926; 2021; provisional patent application filed. Inspirna filed published patent applications PCT/US2013/067860, PCT/US2017/020266, PCT/US2020/064317, and PCT/US2020/05272 related to this work. The other authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. A material transfer agreement was put in place between S.F.T.’s laboratory at the Rockefeller University and Inspirna Inc.
Supplementary Materials
This PDF file includes:
Figs. S1 to S6
Tables S1 and S2
REFERENCES AND NOTES
- 1.Siegel R. L., Miller K. D., Goding Sauer A., Fedewa S. A., Butterly L. F., Anderson J. C., Cercek A., Smith R. A., Jemal A., Colorectal cancer statistics, 2020. CA Cancer J. Clin. 70, 145–164 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Bridgewater J. A., Pugh S. A., Maishman T., Eminton Z., Mellor J., Whitehead A., Stanton L., Radford M., Corkhill A., Griffiths G. O., Falk S., Valle J. W., O’Reilly D., Siriwardena A. K., Hornbuckle J., Rees M., Iveson T. J., Hickish T., Garden O. J., Cunningham D., Maughan T. S., Primrose J. N.; New EPOC investigators , Systemic chemotherapy with or without cetuximab in patients with resectable colorectal liver metastasis (New EPOC): Long-term results of a multicentre, randomised, controlled, phase 3 trial. Lancet Oncol. 21, 398–411 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Welch S., Spithoff K., Rumble R. B., Maroun J.; Gastrointestinal Cancer Disease Site Group , Bevacizumab combined with chemotherapy for patients with advanced colorectal cancer: A systematic review. Ann. Oncol. 21, 1152–1162 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Canon J., Rex K., Saiki A. Y., Mohr C., Cooke K., Bagal D., Gaida K., Holt T., Knutson C. G., Koppada N., Lanman B. A., Werner J., Rapaport A. S., Miguel T. S., Ortiz R., Osgood T., Sun J. R., Zhu X., McCarter J. D., Volak L. P., Houk B. E., Fakih M. G., O’Neil B. H., Price T. J., Falchook G. S., Desai J., Kuo J., Govindan R., Hong D. S., Ouyang W., Henary H., Arvedson T., Cee V. J., Lipford J. R., The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature 575, 217–223 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Patricelli M. P., Janes M. R., Li L. S., Hansen R., Peters U., Kessler L. V., Chen Y., Kucharski J. M., Feng J., Ely T., Chen J. H., Firdaus S. J., Babbar A., Ren P., Liu Y., Selective inhibition of oncogenic KRAS output with small molecules targeting the inactive state. Cancer Discov. 6, 316–329 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Hallin J., Engstrom L. D., Hargis L., Calinisan A., Aranda R., Briere D. M., Sudhakar N., Bowcut V., Baer B. R., Ballard J. A., Burkard M. R., Fell J. B., Fischer J. P., Vigers G. P., Xue Y., Gatto S., Fernandez-Banet J., Pavlicek A., Velastagui K., Chao R. C., Barton J., Pierobon M., Baldelli E., Patricoin E. F. III, Cassidy D. P., Marx M. A., Rybkin I. I., Johnson M. L., Ou S. I., Lito P., Papadopoulos K. P., Janne P. A., Olson P., Christensen J. G., The KRASG12C inhibitor MRTX849 provides insight toward therapeutic susceptibility of KRAS-mutant cancers in mouse models and patients. Cancer Discov. 10, 54–71 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ostrem J. M., Shokat K. M., Direct small-molecule inhibitors of KRAS: From structural insights to mechanism-based design. Nat. Rev. Drug Discov. 15, 771–785 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Arrington A. K., Heinrich E. L., Lee W., Duldulao M., Patel S., Sanchez J., Garcia-Aguilar J., Kim J., Prognostic and predictive roles of KRAS mutation in colorectal cancer. Int. J. Mol. Sci. 13, 12153–12168 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kinsey C. G., Camolotto S. A., Boespflug A. M., Guillen K. P., Foth M., Truong A., Schuman S. S., Shea J. E., Seipp M. T., Yap J. T., Burrell L. D., Lum D. H., Whisenant J. R., Gilcrease G. W. 3rd, Cavalieri C. C., Rehbein K. M., Cutler S. L., Affolter K. E., Welm A. L., Welm B. E., Scaife C. L., Snyder E. L., McMahon M., Protective autophagy elicited by RAF→MEK→ERK inhibition suggests a treatment strategy for RAS-driven cancers. Nat. Med. 25, 620–627 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bryant K. L., Stalnecker C. A., Zeitouni D., Klomp J. E., Peng S., Tikunov A. P., Gunda V., Pierobon M., Waters A. M., George S. D., Tomar G., Papke B., Hobbs G. A., Yan L., Hayes T. K., Diehl J. N., Goode G. D., Chaika N. V., Wang Y., Zhang G. F., Witkiewicz A. K., Knudsen E. S., Petricoin E. F. III, Singh P. K., Macdonald J. M., Tran N. L., Lyssiotis C. A., Ying H., Kimmelman A. C., Cox A. D., Der C. J., Combination of ERK and autophagy inhibition as a treatment approach for pancreatic cancer. Nat. Med. 25, 628–640 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xue J. Y., Zhao Y., Aronowitz J., Mai T. T., Vides A., Qeriqi B., Kim D., Li C., de Stanchina E., Mazutis L., Risso D., Lito P., Rapid non-uniform adaptation to conformation-specific KRAS(G12C) inhibition. Nature 577, 421–425 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.N. S. Chandel, Navigating Metabolism (Cold Spring Harbor Laboratory Press, 2015), 248 pp. [Google Scholar]
- 13.Garcia-Bermudez J., Williams R. T., Guarecuco R., Birsoy K., Targeting extracellular nutrient dependencies of cancer cells. Mol Metab 33, 67–82 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeBerardinis R. J., Chandel N. S., Fundamentals of cancer metabolism. Sci. Adv. 2, e1600200 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loo J. M., Scherl A., Nguyen A., Man F. Y., Weinberg E., Zeng Z., Saltz L., Paty P. B., Tavazoie S. F., Extracellular metabolic energetics can promote cancer progression. Cell 160, 393–406 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.D. L. Nelson, M. M. Cox, Lehninger Principles of Biochemistry (W. H. Freeman, ed. 7, 2017). [Google Scholar]
- 17.Wyss M., Kaddurah-Daouk R., Creatine and creatinine metabolism. Physiol. Rev. 80, 1107–1213 (2000). [DOI] [PubMed] [Google Scholar]
- 18.Pellegatti P., Raffaghello L., Bianchi G., Piccardi F., Pistoia V., Di Virgilio F., Increased level of extracellular ATP at tumor sites: In vivo imaging with plasma membrane luciferase. PLOS ONE 3, e2599 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brahimi-Horn M. C., Chiche J., Pouyssegur J., Hypoxia and cancer. J. Mol. Med. 85, 1301–1307 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Harris A. L., Hypoxia—A key regulatory factor in tumour growth. Nat. Rev. Cancer 2, 38–47 (2002). [DOI] [PubMed] [Google Scholar]
- 21.Mathonnet M., Perraud A., Christou N., Akil H., Melin C., Battu S., Jauberteau M. O., Denizot Y., Hallmarks in colorectal cancer: Angiogenesis and cancer stem-like cells. World J. Gastroenterol. 20, 4189–4196 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamaguchi N., Weinberg E. M., Nguyen A., Liberti M. V., Goodarzi H., Janjigian Y. Y., Paty P. B., Saltz L. B., Kingham T. P., Loo J. M., de Stanchina E., Tavazoie S. F., PCK1 and DHODH drive colorectal cancer liver metastatic colonization and hypoxic growth by promoting nucleotide synthesis. eLife 8, e52135 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fitch C. D., Shields R. P., Payne W. F., Dacus J. M., Creatine metabolism in skeletal muscle. 3. Specificity of the creatine entry process. J. Biol. Chem. 243, 2024–2027 (1968). [PubMed] [Google Scholar]
- 24.Fitch C. D., Chevli R., Inhibition of creatine and phosphocreatine accumulation in skeletal muscle and heart. Metabolism 29, 686–690 (1980). [DOI] [PubMed] [Google Scholar]
- 25.Walker J. B., Creatine: Biosynthesis, regulation, and function. Adv. Enzymol. Relat. Areas Mol. Biol. 50, 177–242 (1979). [DOI] [PubMed] [Google Scholar]
- 26.Suzuki T., Kishikawa T., Sato T., Takeda N., Sugiura Y., Seimiya T., Sekiba K., Ohno M., Iwata T., Ishibashi R., Otsuka M., Koike K., Mutant KRAS drives metabolic reprogramming and autophagic flux in premalignant pancreatic cells. Cancer Gene Ther. (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hidalgo M., Amant F., Biankin A. V., Budinska E., Byrne A. T., Caldas C., Clarke R. B., de Jong S., Jonkers J., Maelandsmo G. M., Roman-Roman S., Seoane J., Trusolino L., Villanueva A., Patient-derived xenograft models: An emerging platform for translational cancer research. Cancer Discov. 4, 998–1013 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosfjord E., Lucas J., Li G., Gerber H. P., Advances in patient-derived tumor xenografts: From target identification to predicting clinical response rates in oncology. Biochem. Pharmacol. 91, 135–143 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Siolas D., Hannon G. J., Patient-derived tumor xenografts: Transforming clinical samples into mouse models. Cancer Res. 73, 5315–5319 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tentler J. J., Tan A. C., Weekes C. D., Jimeno A., Leong S., Pitts T. M., Arcaroli J. J., Messersmith W. A., Eckhardt S. G., Patient-derived tumour xenografts as models for oncology drug development. Nat. Rev. Clin. Oncol. 9, 338–350 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao H., Korn J. M., Ferretti S., Monahan J. E., Wang Y., Singh M., Zhang C., Schnell C., Yang G., Zhang Y., Balbin O. A., Barbe S., Cai H., Casey F., Chatterjee S., Chiang D. Y., Chuai S., Cogan S. M., Collins S. D., Dammassa E., Ebel N., Embry M., Green J., Kauffmann A., Kowal C., Leary R. J., Lehar J., Liang Y., Loo A., Lorenzana E., Robert McDonald E. III, McLaughlin M. E., Merkin J., Meyer R., Naylor T. L., Patawaran M., Reddy A., Roelli C., Ruddy D. A., Salangsang F., Santacroce F., Singh A. P., Tang Y., Tinetto W., Tobler S., Velazquez R., Venkatesan K., Von Arx F., Wang H. Q., Wang Z., Wiesmann M., Wyss D., Xu F., Bitter H., Atadja P., Lees E., Hofmann F., Li E., Keen N., Cozens R., Jensen M. R., Pryer N. K., Williams J. A., Sellers W. R., High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat. Med. 21, 1318–1325 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Martinez-Reyes I., Cardona L. R., Kong H., Vasan K., McElroy G. S., Werner M., Kihshen H., Reczek C. R., Weinberg S. E., Gao P., Steinert E. M., Piseaux R., Budinger G. R. S., Chandel N. S., Mitochondrial ubiquinol oxidation is necessary for tumour growth. Nature 585, 288–292 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uhlen M., Zhang C., Lee S., Sjostedt E., Fagerberg L., Bidkhori G., Benfeitas R., Arif M., Liu Z., Edfors F., Sanli K., von Feilitzen K., Oksvold P., Lundberg E., Hober S., Nilsson P., Mattsson J., Schwenk J. M., Brunnstrom H., Glimelius B., Sjoblom T., Edqvist P. H., Djureinovic D., Micke P., Lindskog C., Mardinoglu A., Ponten F., A pathology atlas of the human cancer transcriptome. Science 357, eaan2507 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Skelton M. R., Schaefer T. L., Graham D. L., Degrauw T. J., Clark J. F., Williams M. T., Vorhees C. V., Creatine transporter (CrT; Slc6a8) knockout mice as a model of human CrT deficiency. PLOS ONE 6, e16187 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S6
Tables S1 and S2