Introduction
Acute kidney injury (AKI) is a major complication associated with COVID-19 and occurs in up to 76% of patients in the intensive care unit.1,2 The mortality rate of patients with COVID-19 who developed AKI (COVID-AKI) is >10 times higher than those who did not. Although candidate AKI markers exist, the etiology of COVID-AKI is multifactorial requiring agnostic approaches for identification of analytes early in hospital course to provide insights into biomarkers and mechanisms associated with COVID-AKI and COVID-19 infection.
Research on COVID-19–associated effects on the urinary proteome is limited, and kidney dysfunction has not been reported.3 Approximately 70% of the proteins detected in the urine are produced in the kidney with a significant amount filtered from the blood. We hypothesize that the changes in protein abundances in the urine could lead to the discovery of protein markers associated with COVID-19 or COVID-AKI and provide mechanistic insights to improve understanding.
Results and Discussion
We analyzed urine samples from 14 participants (6 COVID-AKI, 3 COVID-no AKI, and 5 no COVID-no AKI) (Supplementary Materials and Methods, Supplementary Figure 1A and Supplementary Table S1). To account for the large variation in protein content across urine specimens, we used a bicinchoninic acid to measure peptide concentration after protein digestion (Supplementary Materials and Methods). All peptide samples are then normalized to the same concentration before the analysis to allow for relative quantitation of differences in the urinary proteome (Supplementary Materials and Methods).
The Urine Proteome of COVID-AKI
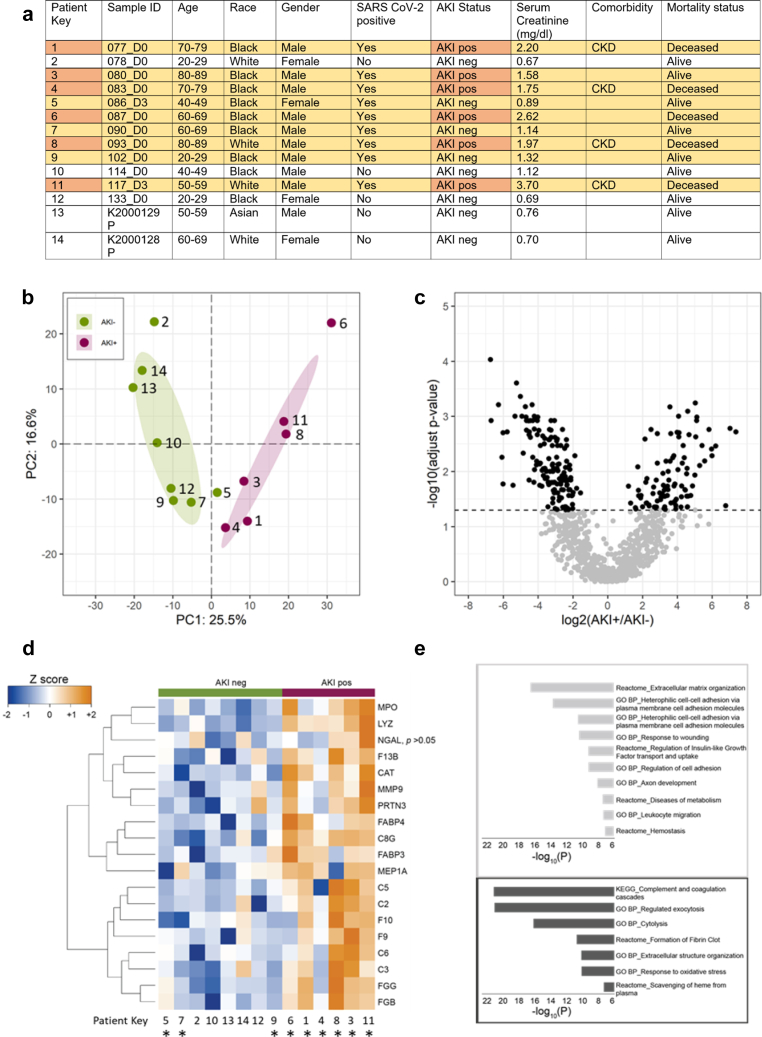
After confirming the quality of urine for analyte discovery (Supplementary Materials and Methods, Supplementary Table S2 and Supplementary Figure S1), we evaluated whether underlying variance could distinguish between AKI positive (AKI+) (6 COVID-AKI) from other samples without AKI (8 AKI negative); all AKI samples were from patients with COVID-19. The first 2 principal components accounted for 42.1% of the variance and clearly separated the 2 groups (Figure 1b), thereby suggesting a prominent impact by AKI on the urine proteome of patients with COVID-19. There was a statistically significant increase in 97 proteins and decrease in 140 proteins in the AKI+ group (adjust P < 0.05; Figure 1c and Supplementary Figure S2A). Hierarchical clustering and pathway enrichment analysis revealed that the top up-regulated pathways were complement activation, coagulation cascades, and regulated exocytosis (Figure 1d and e). Specifically, complement components C2, C3, C5, C6, and C8G were significantly increased in the AKI+ group affecting complement initiation (Figure 1d). Notably, decay-accelerating factor CD55 decreased 2.5-fold in the AKI+ group. This suggests reduced capacity to regulate complement activation, thereby exacerbating injury. This finding is supported by exacerbation of AKI in mice with deletion of CD55 after ischemia–reperfusion injury.4 Activation of the complement system is associated with severe COVID-19. The kidney cell protein sC5b-9, often associated with respiratory failure, is up-regulated in the plasma in the patients with COVID-19 with moderate to severe kidney injury. Comparison of COVID-19 samples with and without AKI also revealed enrichment of proteins associated with the complement pathway (Supplementary Materials and Methods, Supplementary Table S2 columns X–Z). The possibility that immunoglobulin complexes (found at higher levels in individuals with severe COVID-19) might conceivably be recruiting this complement to the nephron could explain the mechanism for AKI and severe failure requiring dialysis. Detection of different components of the complement pathways in the urine of patients with COVID-AKI is new. It could serve as a benchmark to compare how SARS-CoV-2 pathogenicity evolves during different waves of COVID-19 and paves the way for comparative evaluation with non–COVID-AKI in the future. Thus, urine is an easily accessible biospecimen to monitor the state of the complement system in patients with COVID-19.
Figure 1.
Characteristics and laboratory data of urine samples and donors, PCA separation, significant proteins, and affected pathways associated with COVID-AKI. (a) Characteristics and laboratory data of the study participants. The patient key is provided in the leftmost column. (b) The impact of COVID-AKI on the urinary proteome. PCA plot of urine proteomics data annotated with subject AKI status; note, all AKI+ patients had COVID-19. Patient key numbers provided for identification. (c) Volcano plot of the log2 fold changes of protein abundances and their statistical significance in urine owing to AKI status. The dashed line indicates adjusted P = 0.05. (d) Heatmap of select urine proteins in AKI+ and AKI− groups. The “∗” denotes patients with COVID-19 infected with SARS-CoV-2. The protein abundances (each row displayed as Z-score) were significantly changed owing to the AKI (limma test, adjusted P < 0.05). (e) The enriched reactome pathways, KEGG pathways, or GO BP were displayed for down-regulation (light gray) and up-regulation (dark gray) in the urine of patients with COVID-19 with AKI. AKI, acute kidney injury; AKI−, acute kidney injury negative; AKI+, acute kidney injury positive; COVID-AKI, AKI owing to COVID-19; GO BP, Gene Ontology biological processes; KEGG, Kyoto Encyclopedia of Genes and Genomes; PC, principal component; PCA, principal component analysis.
Furthermore, coagulation factors F9, F10, F13b, fibrinogen beta chain, and fibrinogen gamma chain were significantly increased in the AKI+ group (Figure 1d). Several markers of regulated exocytosis, including myeloperoxidase, catalase, lysozyme, leukotriene A4 hydrolase, proteinase 3, and matrix metallopeptidase 9, were also increased in this group (Figure 1d). The top down-regulated pathways included extracellular matrix organization and heterophilic cell-cell adhesion (Figure 1e), indicating tubular damage.
In addition, we detected that several known urinary biomarkers of AKI, including NGAL; FABP3, FABP4; MEP1A; and RBP4, increased in the urine of the patients with COVID-AKI (Figure 1d and Supplementary Table S2). Nevertheless, proteome analysis revealed a significant decrease in UMOD and EGF in this group. Urinary UMOD and EGF have been found to be inversely correlated with AKI, and their decrease is strongly associated with AKI severity. These results further validate our proteome approach and are indicative of tubular injury affecting secretion (NGAL, UMOD, EGF) or defective absorption by proximal tubules (FABP3, FABP4). A novel observation was a >30-fold increase in IGFBP6 in patients with COVID-AKI. Relatively little is known on IGFBP6 in AKI, but it has previously been found to correlate with reduced kidney function and renal failure.5 Additional studies using larger cohorts of urine samples across conditions are needed to confirm changes in these biomarkers and to compare with other causes of AKI, covariates, and outcomes.
The Urine Proteome Associated With COVID-19 in the Absence of AKI
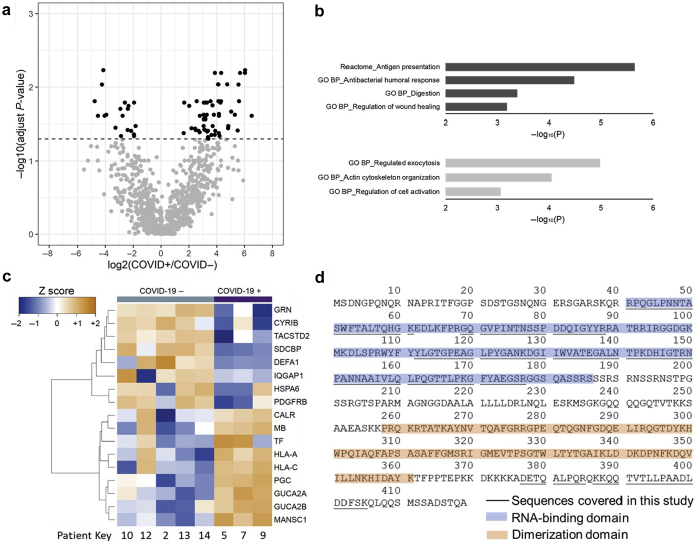
Because AKI leads to significant changes in the abundances of many urinary proteins, it is difficult to identify the changes that are only associated with COVID-19. We therefore compared the urine proteomes of COVID-19 positive (COVID+) (n = 3) and negative (COVID−) (n = 5) patients without AKI (Supplementary Figure S2B). The abundances of 57 proteins significantly increased, and 19 proteins were significantly decreased in the urine of COVID+ patients (adjust P < 0.05, Figure 2a and Supplementary Table S2). Furthermore, a large percentage of the proteins with significantly increased levels were immunoglobulins (49.1%). The remaining proteins with increased expression were involved in the antigen presentation pathway. Proteins with significantly decreased expression were enriched in the regulated exocytosis pathway (Figure 2b). Specifically, the average abundances of major MHC-I proteins HLA-A and HLA-C increased >3.6-fold in the urine owing to COVID-19. The MHC complex presents antigens at the cell surface to immune cells when cells are infected by viruses. MHC-peptide complexes have been detected in different human body fluids, including the serum and urine.6 The down-regulation of exocytosis was marked by the significantly decreased abundances of DEFA1, GRN, HSPA6, SDCBP, IQGAP1, and CYRIB (Figure 2c).
Figure 2.
The impact of COVID-19 infections on the urinary proteome. The urine donors were all negative for AKI. (a) Volcano plot of the log2 fold changes of protein abundances and their statistical significance owing to COVID-19 infections. The dashed line indicates adjusting P = 0.05. (b) Up-regulated (dark gray) and down-regulated (light gray) reactome pathways GO BP enriched owing to COVID-19 infections. (c) Heatmap of selected urinary proteins that displayed increased or decreased abundances in urine owing to COVID-19 (limma test, adjusting P < 0.05). (d) Sequence coverage of SARS-CoV-2 nucleoprotein. Peptides of SARS-CoV-2 proteins were detected in 2 urine samples. The 2 patients developed acute kidney injury along with severe COVID-19 symptoms. Details of peptide sequences were summarized in Supplementary Table S2. AKI, acute kidney injury; GO BP, Gene Ontology biological processes.
Furthermore, there were 4 proteins with significantly increased abundances in COVID+ group compared with COVID− group, including MB, CA1, MANSC1, and ABRACL (Figure 2c and Supplementary Table S2 columns AA–AC). In addition, 5 proteins were significantly decreased in COVID+, including GRN, CREB3L3, MUC1, CD320, and DLST (Figure 2c and Supplementary Table S2 columns AA–AC). These newly identified proteins could potentially be used as more sensitive biomarkers to evaluate the COVID-19 infection. Moreover, the data reveal that several markers are extrarenal in COVID+ patients (Figure 2), including 5.68-fold increase in myoglobin (heart specific) and 3.92-fold increase in zymogen pepsinogen C (lung, esophagus, and stomach injury), which are promising targets for noninvasive urine proteome profiling to detect and monitor kidney-independent organ damage.
The SARS-CoV-2 virion is not typically shed in the urine, but it has tropism to kidney cell types.7, 8, 9 We evaluated whether urine proteomics can detect SARS-CoV-2 proteins. Interestingly, peptides from SARS-CoV-2 nucleoprotein were detected in the urine samples collected from 2 severely ill patients with COVID-AKI who died (Supplementary Materials and Methods, Figure 1a and Supplementary Table S1). The peptide identification was confirmed by searching with both MaxQuant and MS-GF+ software. Near-complete sequence coverage of the RNA binding domain was achieved by single dimension liquid chromatography coupled to tandem mass spectrometry analysis. Sequences near the N-terminus of the SARS-CoV-2 nucleoprotein were also observed (Figure 2d). Owing to the relatively low-count peptide-spectrum matches of SARS-CoV-2 peptides observed, the concentration of SARS-CoV-2 nucleoprotein is predicted to be low in these urine samples (Supplementary Table S3).
Although our study is a proof-of-concept investigation with a relatively small number of patient samples, we were able to establish methods for high-quality urine proteomics of patients with COVID-19 in a clinical setting. We report potential biomarkers of COVID-19 infection and COVID-AKI which could be useful for detecting and monitoring the development of COVID-19 infection or AKI. In particular, the complement and coagulation cascade pathways and exocytosis pathway were highly up-regulated in the patients with COVID-AKI. Furthermore, proteins associated with antigen presentation were significantly correlated with COVID-19 infection. We propose a list of potential novel biomarker proteins (i.e., MB, CA1, MANSC1, ABRACL, GRN, CREB3L3, DEFA1, MUC1, DLST, and IGFBP6) and of previously reported AKI biomarker proteins (i.e., NGAL, FABP3, FABP4, MEP1A, and RBP4) for further validation of their specificity, sensitivity, and severity. A subset of our patients with COVID-AKI developed AKI on preexisting chronic kidney disease, which could also affect the urine proteins found enriched in this group. Parsing the effect of chronic kidney disease status in COVID-19 would require larger cohorts; however, we observe that the enrichment of complement and coagulation pathway proteins was found in the patients with COVID-AKI with and without chronic kidney disease (Figure 1d and Supplementary Table S1). Taken together, these exciting preliminary results clearly reveal the need for further investigation in an expanded clinical cohort that includes patients with non–COVID-19-associated AKI.
Disclosure
CRP reports being a member of the advisory board of and owning equity in Renalytix AI and serving as a consultant for Genfit. AV reports serving as a consultant for NxStage Medical Inc. and Astute Medical Inc. These entities did not support research in this study. All the other authors declared no competing interest.
Acknowledgments
This study used samples obtained from the COVID-19 biorepository of the Washington University School of Medicine, which is supported by the following: the Barnes-Jewish Hospital Foundation; the Siteman Cancer Center grant P30 CA091842 from the National Cancer Institute of the National Institutes of Health; and the Washington University Institute of Clinical and Translational Sciences grant UL1TR002345 from the National Center for Advancing Translational Sciences of the National Institutes of Health. We thank the George M. O’Brien Center of the Johns Hopkins University for performing the kidney injury biomarker assays (grant #P30DK079310). Parts of this research were performed using EMSL, a national scientific user facility sponsored by the Department of Energy's Office of Biological and Environmental Research and located at PNNL. The proteomics work is supported by NIH Common Fund, through the Office of Strategic Coordination/Office of the NIH Director under award UG3HL145593, and NIH grant U01DK124020. The content is solely the responsibility of the authors and does not necessarily represent the view of the National Institutes of Health. We thank the Kidney Translational Research Center (KTRC) of the Division of Nephrology at the Washington University in St. Louis for supporting the processing, preservation, and storage of the urine samples. We are grateful to Charles Goss, Adriana Rauseo Acevedo, and Rachel Presti for their efforts in data management and patient recruitments. We also thank Jana Chiang for her contributions in editing, formatting, and reviewing the content.
Footnotes
Supplementary Materials and Methods. Includes information on the methods used for sample acquisition and patient inclusion, urine processing and storage, proteomics sample preparation, LC-MS/MS-based proteomics analysis, proteomics data processing, correlation analysis of patients with COV-AKI compared with those without AKI, and where the data are available to download.
Figure S1. Pearson correlation of urinary proteomics across the studied samples.
Figure S2. Distribution of P values before adjustment. (A) Statistical results of limma test that compared the intensities of urine proteins based on the status of acute kidney injury (AKI). (B) Statistical results of limma test that compared the intensities of urine proteins based on the COVID-19 infection in the AKI-negative group.
Table S1. Master sheet of research participants.
Table S2. Statistical analysis results of urinary proteomics.
Table S3. SARS-CoV-2 peptides detected in 2 urine samples by LC-MS/MS. The proteomics data were searched with MSGF+ software.
STROBE Statement (PDF).
Contributor Information
Paul D. Piehowski, Email: paul.piehowski@pnnl.gov.
Sanjay Jain, Email: sanjayjain@wustl.edu.
Supplementary Material
Supplementary Materials and Methods. Includes information on the methods used for sample acquisition and patient inclusion, urine processing and storage, proteomics sample preparation, LC-MS/MS-based proteomics analysis, proteomics data processing, correlation analysis of patients with COV-AKI compared with those without AKI, and where the data are available to download.
Figure S1. Pearson correlation of urinary proteomics across the studied samples.
Figure S2. Distribution of P values before adjustment. (A) Statistical results of limma test that compared the intensities of urine proteins based on the status of acute kidney injury (AKI). (B) Statistical results of limma test that compared the intensities of urine proteins based on the COVID-19 infection in the AKI-negative group.
Table S1. Master sheet of research participants.
Table S2. Statistical analysis results of urinary proteomics.
Table S3. SARS-CoV-2 peptides detected in 2 urine samples by LC-MS/MS. The proteomics data were searched with MSGF+ software.
STROBE Statement (PDF)
References
- 1.Pei G., Zhang Z., Peng J., et al. Renal involvement and early prognosis in patients with COVID-19 pneumonia. J Am Soc Nephrol. 2020;31:1157–1165. doi: 10.1681/ASN.2020030276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vijayan A., Humphreys B.D. SARS-CoV-2 in the kidney: bystander or culprit? Nat Rev Nephrol. 2020;16:703–704. doi: 10.1038/s41581-020-00354-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Y., Wang Y., Liu H., et al. Urine proteome of COVID-19 patients. Urine (Amst) 2021;2:1–8. doi: 10.1016/j.urine.2021.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamada K., Miwa T., Liu J., et al. Critical protection from renal ischemia reperfusion injury by CD55 and CD59. J Immunol. 2004;172:3869–3875. doi: 10.4049/jimmunol.172.6.3869. [DOI] [PubMed] [Google Scholar]
- 5.Van Doorn J., Ringeling A.M., Shmueli S.S., et al. Circulating levels of human insulin-like growth factor binding protein-6 (IGFBP-6) in health and disease as determined by radioimmunoassay. Clin Endocrinol (Oxf) 1999;50:601–609. doi: 10.1046/j.1365-2265.1999.00694.x. [DOI] [PubMed] [Google Scholar]
- 6.Boehm T., Zufall F. MHC peptides and the sensory evaluation of genotype. Trends Neurosci. 2006;29:100–107. doi: 10.1016/j.tins.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 7.Xiao F., Sun J., Xu Y., et al. Infectious SARS-CoV-2 in feces of patient with severe COVID-19. Emerg Infect Dis. 2020;26:1920–1922. doi: 10.3201/eid2608.200681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun J., Zhu A., Li H., et al. Isolation of infectious SARS-CoV-2 from urine of a COVID-19 patient. Emerg Microbes Infect. 2020;9:991–993. doi: 10.1080/22221751.2020.1760144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Puelles V.G., Lutgehetmann M., Lindenmeyer M.T., et al. Multiorgan and renal tropism of SARS-CoV-2. N Engl J Med. 2020;383:590–592. doi: 10.1056/NEJMc2011400. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.