Abstract
Fungal mitochondrial genomes encode genes involved in crucial cellular processes, such as oxidative phosphorylation and mitochondrial translation, and the molecule has been used as a molecular marker for population genetics studies. Coccidioides immitis and C. posadasii are endemic fungal pathogens that cause coccidioidomycosis in arid regions across both American continents. To date, approximately 150 Coccidioides isolates have been sequenced to infer patterns of variation in nuclear genomes. However, less attention has been given to the mitochondrial genomes of Coccidioides. In this report, we describe the assembly and annotation of mitochondrial reference genomes for two representative strains of C. posadasii and C. immitis, as well as assess population variation among 77 selected genomes. The sizes of the circular-mapping molecules are 68.2 Kb in C. immitis and 75.1 Kb in C. posadasii. We identify 14 mitochondrial protein-coding genes common to most fungal mitochondria, which are largely syntenic across different populations and species of Coccidioides. Both Coccidioides species are characterized by a large number of group I and II introns, harboring twice the number of elements as compared to closely related Onygenales. The introns contain complete or truncated ORFs with high similarity to homing endonucleases of the LAGLIDADG and GIY-YIG families. Phylogenetic comparisons of mitochondrial and nuclear genomes show extensive phylogenetic discordance suggesting that the evolution of the two types of genetic material is not identical. This work represents the first assessment of mitochondrial genomes among isolates of both species of Coccidioides, and provides a foundation for future functional work.
Keywords: Valley fever, Coccidioides, Onygenales, coccidioidomycosis, mitochondrial genomes, phylogenetics, fungal pathogen, group I and II introns
Introduction
Fungal mitochondrial genomes exist as either linear or circular-mapping molecules and range in size from ∼17.6 kb (e.g., Schizosaccharomyces pombe Genbank ID MK618090.1) to well over 200 kb [e.g., 272,238 bp in Morchella importuna (Liu et al. 2020)]. Fungal mitochondrial genomes usually encode proteins involved in oxidative phosphorylation—the main source of ATP production of the cell—as well as two ribosomal RNA subunits, and a set of tRNAs involved in mitochondrial ribosome translation. More specifically, most fungal mitochondrial protein-coding genes fall into the following classes: seven subunits of ubiquinone oxidoreductase [nad; six for Saccharomycotina and fission yeasts (Bullerwell et al. 2003)], cytochrome b (cob), three subunits of cytochrome oxidase (cox) and up to three ATP synthase subunits (atp; the presence of atp8 and atp9 varies among fungal taxa) (Aguileta et al. 2014). A gene encoding a ribosomal protein subunit (rps3) is present in most fungal mitochondrial genomes (Korovesi et al. 2018). Fungal mitochondrial protein-coding genes may contain additional genes encoding structural RNAs such as ribosomal RNAs (small and large subunit rRNAs rns and rnl), the RNA subunit of RNase P (rnpB), and variable numbers of tRNAs, and are more variable across fungal lineages (Aguileta et al. 2014). Notably, the nad genes do not have this organizational structure, and tend to be arranged in operon-like structures, with some of the genes overlapping without discernable intergenic regions (e.g., nad4L situated upstream of nad5, overlapping by one to a dozen or more nucleotides) (Aguileta et al. 2014).
Mitochondrial genes in fungi contain highly variable numbers and arrangements of group I and II introns that are inserted in protein-coding as well as rRNA genes (Pogoda et al. 2019). For example, Endoconidiophora species contain more than 80 mitochondrial introns (Zubaer et al. 2018), which can create gene annotation challenges especially when transcriptome data are not available. Both intron groups may contain complete or truncated ORFs that encode either homing endonucleases of the LAGLIDADG and GIY-YIG families, or reverse transcriptases/maturases (Lang et al. 2007). If present, these proteins can direct intron transfers within mitochondrial genomes of genetically compatible fungal species, or less frequently across genera and kingdom boundaries (Stoddard 2011). Mitochondrial DNA (mtDNA)-encoded genes are particularly prone to crossing species boundaries via introgression even when nuclear gene flow is restricted (Sloan et al. 2017). Intron transfer via homing endonucleases may involve genetic co-conversion of flanking exon sequences. Therefore, phylogenetic inference using mtDNA for genes with many introns [e.g., cox1, cob and rnl (Bovers et al. 2009; Aguileta et al. 2014)] may reflect replacement of coding regions from ongoing intron invasion.
In this study, we focus on describing and annotating the mitogenomes of Coccidioides immitis and C. posadasii (Ascomycota, Onygenales), which are fungal species endemic to both American continents, and the causative agents of coccidioidomycosis (Barker et al. 2019). This disease is most frequently reported in the “Lower Sonoran Life Zone” (LSLZ) in California, Arizona, Texas, and northwestern Mexico (Fisher et al. 2007; Benedict et al. 2019). However, the organism is found throughout arid and semi-arid areas beyond the limits of the LSLZ (Kollath et al. 2019). Both species have complex biogeographic distribution patterns (Fisher et al. 2001; Teixeira et al. 2019a). Coccidioides immitis has been found in California and Baja Mexico as well in eastern Washington state, and each region harbors unique genotypes (Engelthaler et al. 2016; McCotter et al. 2019; Teixeira et al. 2019b). Coccidioides posadasii is present throughout the southwestern United States in Arizona, Utah, New Mexico, Texas; and in less well-described regions in Central and South America. The species is structured by an Arizona population, a Texas/Mexico/South America (TX/MX/SA) population, and a Caribbean population (Teixeira et al. 2019a).
Notably, nucDNA studies have found extensive differentiation between species of Coccidioides with some evidence for gene flow between species (Neafsey et al. 2010; Maxwell et al. 2019). However, no studies have addressed whether or not mtDNA reflects the divergence of nucDNA, or if mtDNA has moved between Coccidioides species or among populations. The only precedent suggests that the two species can be discriminated based on polymorphisms found at the first intron of the cox1 gene (Hamm et al. 2019). Therefore, to address these gaps in our knowledge we: (i) describe the full circular mitogenomes of C. posadasii and C. immitis, (ii) compare their core genes, structural RNAs, and introns of group I and II with other Onygenales fungal species, and (iii) compare the evolutionary histories of the mtDNA and nucDNA genomes of these medically important fungal pathogens.
Materials and methods
Mitochondrial genome assembly and annotation
Paired end Illumina sequence reads from 20 Coccidioides immitis and 57 C. posadasii were retrieved from the Sequence Read Archive (SRA) and accessions and details are listed in Table 1. Following cleaning and quality-clipping of reads with Trimmomatic v0.35, we assembled the genomes of C. posadasii Tucson-2 and C. immitis WA221 using the SPAdes Genome Assembler v3.14.0 (Bankevich et al. 2012) with a kmer sizes 61, 91, and 127. We identified mitochondrial contigs in this initial assembly using similarity searches with expected mtDNA-encoded fungal proteins (cob, cox1, nad1, nad2, nad3, nad4, nad4L, nad5, nad6, atp6, and atp9 of Allomyces macrogynus; NCBI Reference Sequence NC_001715.1). To minimize assembly error we (i) used Rcorrector (Song and Florea 2015) read correction, (ii) reduced the number of Illumina reads to a target kmer coverage of the mtDNA between 30 and 50x, (iii) reads mapping against the identified mitochondrial contigs were identified with Bowtie2 (Langmead and Salzberg 2012), which were then (iv) reassembled with Spades, resulting in preliminary (uncorrected) mitogenome assemblies. Finally, (v) all reads of the reduced 30–50x read set were aligned back to the preliminary assembly with Bowtie2 and analyzed for kmer coverage with Bedtools v2.29.2 (Quinlan 2014). We identified incorrectly assembled reads, defined by kmer frequency values of two or lower (likely the result of hybrid reads, originating from ligation of unrelated genomic DNA fragments during library construction), and removed them from the final assemblies. The circularity of the final mitogenomes assemblies was verified by identifying overlapping regions present on both ends of the final contigs. For both species, we obtained single circular-mapping closed contigs that carry the expected full set of fungal mitochondrial genes.
Table 1.
SRA accession number, isolate ID, species, and place of isolation for each strain used for nuclear and mitochondrial genetic analyses
| SRA accession number | Isolate ID | Species | Country of isolation |
|---|---|---|---|
| SRR6830881 | 4545-MICE | Coccidioides posadasii | Venezuela |
| SRR6830884 | 4542 | Coccidioides posadasii | Venezuela |
| SRR6830886 | 3796 | Coccidioides posadasii | Venezuela |
| SRR6830885 | 2566 | Coccidioides posadasii | Venezuela |
| SRR6830888 | JTORRES | Coccidioides posadasii | Venezuela |
| SRR6830887 | 3490 | Coccidioides posadasii | Mexico |
| SRR3468076 | 730332_Guatemala | Coccidioides posadasii | Guatemala |
| SRR3468077 | 730333_Guatelama | Coccidioides posadasii | Guatemala |
| SRR3468078 | 730334_Guatemala | Coccidioides posadasii | Guatemala |
| SRR3468067 | B0858_Guatemala | Coccidioides posadasii | Guatemala |
| SRR3468068 | B10757_Nevada | Coccidioides posadasii | United States |
| SRR3468069 | B10813_Texas | Coccidioides posadasii | United States |
| SRR3468070 | B1249_Guatemala | Coccidioides posadasii | Guatemala |
| SRR3468072 | B5773_Brazil | Coccidioides posadasii | Brazil |
| SRR3468053 | Coahuila_2 | Coccidioides posadasii | Mexico |
| SRR3468050 | Colorado_Springs_1 | Coccidioides posadasii | United States |
| SRR3468075 | GT002_Texas | Coccidioides posadasii | United States |
| SRR3468074 | GT017_Paraguay | Coccidioides posadasii | Paraguay |
| SRR3468066 | Michoacan_1 | Coccidioides posadasii | Mexico |
| SRR3468064 | Nuevo_Leon_1 | Coccidioides posadasii | Mexico |
| SRR3468065 | Nuevo_Leon_2 | Coccidioides posadasii | Mexico |
| SRR3468054 | Phoenix_1 | Coccidioides posadasii | United States |
| SRR3468055 | Phoenix_2 | Coccidioides posadasii | United States |
| SRR3468056 | Phoenix_3 | Coccidioides posadasii | United States |
| SRR3468057 | Phoenix_4 | Coccidioides posadasii | United States |
| SRR3468058 | Phoenix_5 | Coccidioides posadasii | United States |
| SRR3468059 | Phoenix_6 | Coccidioides posadasii | United States |
| SRR3468061 | Phoenix_7 | Coccidioides posadasii | United States |
| SRR3468062 | Phoenix_8 | Coccidioides posadasii | United States |
| SRR3468063 | Phoenix_9 | Coccidioides posadasii | United States |
| SRR3468048 | San_Antonio_1 | Coccidioides posadasii | United States |
| SRR3468051 | Sonora_1 | Coccidioides posadasii | Mexico |
| SRR3468052 | Sonora_2 | Coccidioides posadasii | Mexico |
| SRR3468033 | Tucson_10 | Coccidioides posadasii | United States |
| SRR3468034 | Tucson_11 | Coccidioides posadasii | United States |
| SRR3468035 | Tucson_12 | Coccidioides posadasii | United States |
| SRR3468036 | Tucson_13 | Coccidioides posadasii | United States |
| SRR3468037 | Tucson_14 | Coccidioides posadasii | United States |
| SRR3468039 | Tucson_15 | Coccidioides posadasii | United States |
| SRR3468040 | Tucson_16 | Coccidioides posadasii | United States |
| SRR3468041 | Tucson_17 | Coccidioides posadasii | United States |
| SRR3468042 | Tucson_18 | Coccidioides posadasii | United States |
| SRR3468043 | Tucson_19 | Coccidioides posadasii | United States |
| SRR3468044 | Tucson_20 | Coccidioides posadasii | United States |
| SRR3468045 | Tucson_21 | Coccidioides posadasii | United States |
| SRR3468046 | Tucson_22 | Coccidioides posadasii | United States |
| SRR3468047 | Tucson_23 | Coccidioides posadasii | United States |
| SRR3468073 | Tucson_24 | Coccidioides posadasii | United States |
| SRR3468032 | Tucson_9 | Coccidioides posadasii | United States |
| SRR3468031 | Tucson_8 | Coccidioides posadasii | United States |
| SRR3468030 | Tucson_7 | Coccidioides posadasii | United States |
| SRR3468029 | Tucson_6 | Coccidioides posadasii | United States |
| SRR3468028 | Tucson_5 | Coccidioides posadasii | United States |
| SRR3468026 | Tucson_4 | Coccidioides posadasii | United States |
| SRR3468025 | Tucson_3 | Coccidioides posadasii | United States |
| SRR3468024 | Tucson_2 | Coccidioides posadasii | United States |
| SRR3468023 | Tucson_1 | Coccidioides posadasii | United States |
| SRR1292228 | 212 | Coccidioides immitis | United States |
| SRR1292225 | 202 | Coccidioides immitis | United States |
| SRR1292226 | 205 | Coccidioides immitis | United States |
| SRR1292227 | 211 | Coccidioides immitis | United States |
| SRR1292224 | Washington_1 | Coccidioides immitis | United States |
| SRR3468022 | B0727_Argentina | Coccidioides immitis | Argentina |
| SRR3468018 | Guerrero_1 | Coccidioides immitis | Mexico |
| SRR3468019 | San_Diego_1 | Coccidioides immitis | United States |
| SRR3468021 | Coahuila_1 | Coccidioides immitis | Mexico |
| SRR3468015 | SJV_1 | Coccidioides immitis | United States |
| SRR3468016 | SJV_2 | Coccidioides immitis | United States |
| SRR3468027 | SJV_3 | Coccidioides immitis | United States |
| SRR3468038 | SJV_4 | Coccidioides immitis | United States |
| SRR3468049 | SJV_5 | Coccidioides immitis | United States |
| SRR3468060 | SJV_6 | Coccidioides immitis | United States |
| SRR3468071 | SJV_7 | Coccidioides immitis | United States |
| SRR3468079 | SJV_8 | Coccidioides immitis | United States |
| SRR3468080 | SJV_9 | Coccidioides immitis | United States |
| SRR3468081 | SJV_10 | Coccidioides immitis | United States |
| SRR3468017 | SJV_11 | Coccidioides immitis | United States |
To compare the Coccidioides mtDNA assembles with other fungi, we retrieved full mitochondrial assemblies from other Onygenales available in the NCBI GenBank database: Histoplasma capsulatum H143 (GG692467.1) Paracoccidioides brasiliensis Pb18 [AY955840.1, (Cardoso et al. 2007)], Blastomyces dermatitidis ATCC 18188 (GG753566.1), Epidermophyton flocossum ATCC 26072 [AY916130.1, (Tambor et al. 2006)], Trichophyton rubrum BMU 01672 [FJ385026.1 (Wu et al. 2009)], and Ascosphaera apis ARSEF 7405 [AZGZ01000045.1 (Shang et al. 2016)]. Other close relatives of Coccidioides (e.g., Uncinocarpus) had fragmented mitogenomes (Whiston et al. 2012) and were thus not included in our analyses.
Mitochondrial genes as well as introns of group I and group II, tRNAs, RNaseP RNA (rnpB), and the small and large subunit rRNAs (rns and rnl) for Coccidioides and other related Onygenalean fungi were annotated using the MFannot pipeline (https://github.com/BFL-lab/Mfannot). Coccidioides annotations were manually inspected and intron boundaries were checked and adjusted by aligning available RNAseq data (Whiston et al. 2012) with respective mitochondrial assemblies using Bowtie 2 (Langmead and Salzberg 2012). The assemblies and annotations were deposited in GenBank (MW722165-MW722166) and were visualized with the OGDRAW pipeline (Greiner et al. 2019).
Single nucleotide polymorphism identification
To identify Single Nucleotide Polymorphisms (SNPs) from the 77 Coccidioides isolates, we mapped Illumina paired-end reads to the assembled mitochondrial references of either C. posadasii strain Tucson2 or C. immitis strain WA221 in a species-dependent manner using Burrows-Wheeler Aligner (BWA) v 0.7.7 (Li and Durbin 2009). Indels were realigned to the species reference genomes using GATK RealignerTargetCreator and IndelRealigner tools [GATK toolkit v 3.3-0 (McKenna et al. 2010)]. Next, we used the UnifiedGenotyper package. We only included SNPs not located in duplicated loci [as identified by NUCmer (Kurtz et al. 2004)], with more than 10X coverage, and with a minor allele frequency of at least 10%. We used the same approach to call SNPs for the nuclear genomes (Teixeira et al. 2019a).
Phylogenetic analysis
We generated maximum likelihood concatenated trees for mtDNA and for nucDNA using methods implemented in IQ-TREE software (Nguyen et al. 2015) using -m MFP option [ModelFinder (Kalyaanamoorthy et al. 2017)] for model selection. To measure branch confidence, we used 1000 ultrafast bootstraps (Minh et al. 2013) and a Shimodaira–Hasegawa-like approximate likelihood ratio test (SH-aLRT) (Anisimova et al. 2011). We compared the topology of the two trees using FigTree v1.4.2—http://tree.bio.ed.ac.uk/software/figtree/, and scored the disagreements the two topologies using TOPD/FMTS v 4.6 (Puigbò et al. 2007) and the R function tree.dist [library phangorn (Schliep 2011)]. We calculated the Split Distance (SD) distance score, a metric that ranges between 0 (full concordance) and 1 (complete non-concordance). The SD is the ratio between the observed Robinson-Foulds (RF) symmetric difference and the maximum value of the RF distance (2n-6). Since we had 77 tips, the maximum number of the RF distance is 148. SD ranges from 0 (all partitions present in the two focal trees) to 1 (no shared partitions between the two trees).
Population genetics
We used fastSTRUCTURE v1.0 (Raj et al. 2014) to investigate potential genetic structure at the mitochondrial level within each of the two Coccidioides species. We used unlinked SNPs from the final .vcf SNP set using PLINK v1.07 using the –make-bed option. We repeated the same procedure using K (number of populations) values that range from two to eight. We used chooseK.py to determine the most likely scenario for the number of genetic clusters.
Data availability
SNP data for each strain analyzed for nuclear and mitochondrial trees, raw phylogenetic trees files (Figure 2) and unlinked SNPs and allele frequencies used as input for structure analysis (Figure 4) are available at Zenodo (doi:10.5281/zenodo.4592315).
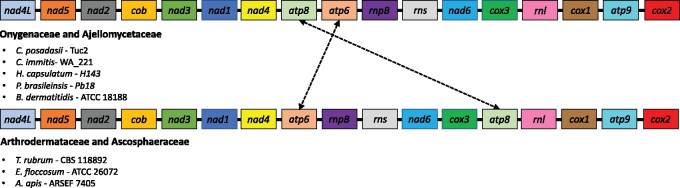
Figure 2.
Mitochondrial gene content and synteny among Onygenalean fungi. Genes are color-coded (see legend) and are displayed according to position on the genome. The mitogenomes are highly syntenic but atp8 gene positioning differs between Onygenaceae/Ajellomycetaceae and Arthrodermataceae/Ascosphaeraceae.
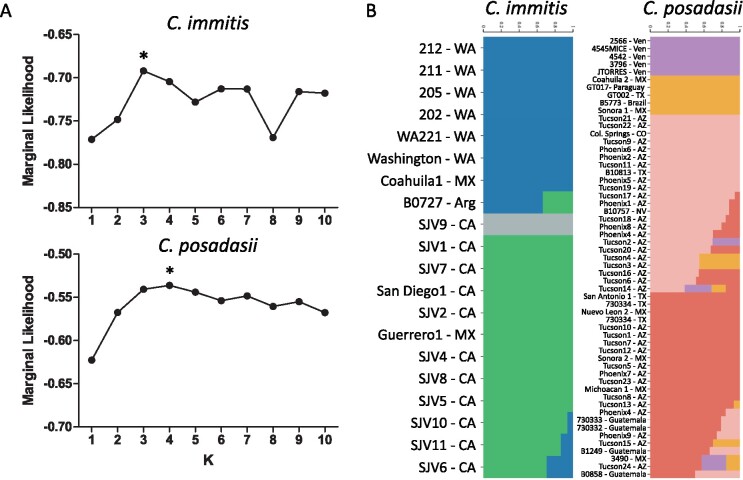
Figure 4.
Population structure within C. immitis and C. posadasii based on mitochondrial-derived alleles. (A) Marginal likelihood plots for both C. immitis and C. posadasii species and the best K (or number of populations) selected within each species is marked with asterisks and was K = 2 and K = 4, respectively. (B) Proportion of admixture of each C. immitis or C. posadasii. Heterozygous or non-admixed strains are represented by solid bar while admixed individuals are displayed as multicolored bars. The proportion of each homozygous population can be observed by the scale on the top of each plot.
Results
Coccidioides spp. mitogenome
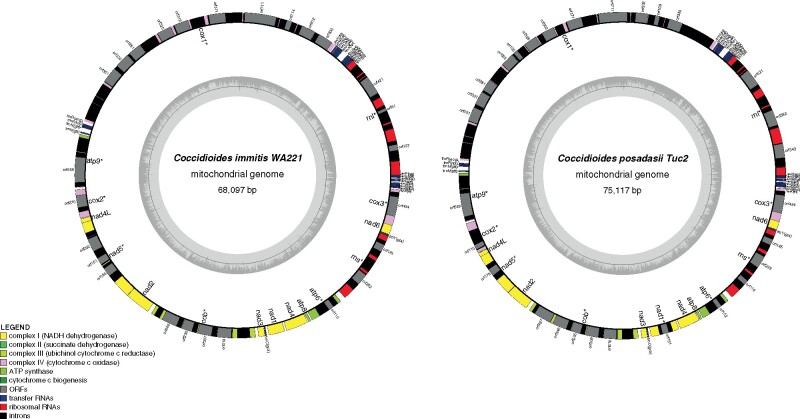
We assembled complete circular mtDNA molecules for each of the two species of Coccidioides. The two assemblies differ in size: the mitogenome is 68.6 Kb in C. immitis and 75.1 Kb in C. posadasii (Figure 1). This observation is consistent with documented variation in mitogenome size among species of the Onygenales (Table 2). The length of the mtDNA of both species of Coccidioides are on the larger end of the continuum of sizes of mitogenomes. The mitogenomes of Coccidioides harbor 14 protein-coding genes responsible for the formation of ubiquinone oxidoreductase, cytochrome b, cytochrome oxidase, and ATP synthase protein complexes (Figures 1 and 2). The two ribosomal small and large subunit rRNA genes (rns and rnl), RNase P RNA (rnpB) and 26 tRNAs were identified. The gene composition and synteny are conserved between Coccidioides (Onygenaceae), Blastomyces, Histoplasma, and Paracoccidioides (all Ajellomycetaceae) (Figure 1; Van Dyke et al. 2019). The position of the gene atp8 differs between the Onygenaceae/Ajellomycetaceae species and other species of the Onygenales, such as dermatophytes (Trichophyton rubrum and Epidermophyton floccosum—Arthrodermataceae), and the bee-pathogenic fungus Ascosphaera apis (Ascosphaeraceae, Figure 2). We observed no gene gain or loss of core mitochondrial genes within Onygenalean fungi (Figure 2).
Figure 1.
Circular maps of C. immitis and C. posadasii mitogenomes. The assembled and annotated genome features were converted into GenBank format and loaded into the OGDraw pipeline for physical visualization of the coding and non-coding elements of the mitochondrial genomes.
Table 2.
Numbers of introns and classes among Onygenalean fungi
| C. immitis | C. posadasii | H. capsulatum | P. brasiliensis | B. dermatitidis | E. flocossum | T. rubrum | A. apis | |
|---|---|---|---|---|---|---|---|---|
| Mitochondrial genome size (bp) | 68,597 | 75,194 | 39,129 | 71,335 | 51,071 | 30,910 | 26,985 | 118,650 |
| Intron IB (complete) | 15 | 15 | 3 | 5 | 6 | 3 | 0 | 8 |
| Intron IB (extra insertion) | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| Intron IB (5′, partial) | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| Intron IB (3′, partial) | 1 | 2 | 1 | 0 | 2 | 0 | 0 | 1 |
| Intron IA | 3 | 3 | 1 | 0 | 1 | 1 | 2 | 1 |
| Intron IA (5′, partial) | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| Intron I (derived, A) | 1 | 1 | 0 | 0 | 2 | 1 | 0 | 5 |
| Intron I (derived, B1) | 2 | 2 | 0 | 4 | 0 | 0 | 0 | 11 |
| Intron ID | 4 | 3 | 1 | 0 | 1 | 1 | 0 | 5 |
| Intron IC1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| Intron IC2 | 4 | 4 | 0 | 1 | 1 | 0 | 0 | 5 |
| Intron I (derived, B2) | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 1 |
| Intron II (domainV) | 4 | 5 | 1 | 3 | 2 | 0 | 0 | 3 |
| Intron II, derived | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Total | 36 | 39 | 9 | 13 | 16 | 6 | 2 | 44 |
The larger size of the mitogenome in Coccidioides is due to the presence of introns and intron-encoded open reading frames (ORFs) in both Coccidioides species, reflected in the increase in the frequency of intron type I and intron type II (Table 2, Figure 1) compared with other Ajellomycetaceae fungi. In fact, Coccidioides harbors twice the number of elements found in B. dermatitidis. The dermatophyte genera, Epidermophyton and Trichophyton, have only six and two intron elements respectively, whereas C. immitis and C. posadasii contain 39 elements, respectively (Table 2). The introns found in the Coccidioides mtDNA contain complete or truncated ORFs with high similarity to homing endonucleases of the LAGLIDADG and GIY-YIG families (Table 2). Both species contain 15 complete copies of Intron IB (Table 2). Ascopharaceae apis also has a large mitogenome (118.65 Kb) and a high number of intron-type I, specifically the intron I—derived, B1 element (Table 2). The frequency and distribution of intron-types I and II in the genes nad5, cob, and cox1 differ between C. immitis and C. posadasii (Figure 1). The difference in genome size between Coccidioides species stems from the increased number of copies of intron I and intron IB (Table 2) and repetitive DNA between ORF 330 and tRNA-H (Figure 1) in C. posadasii.
mtDNA and nucDNA phylogenetic trees
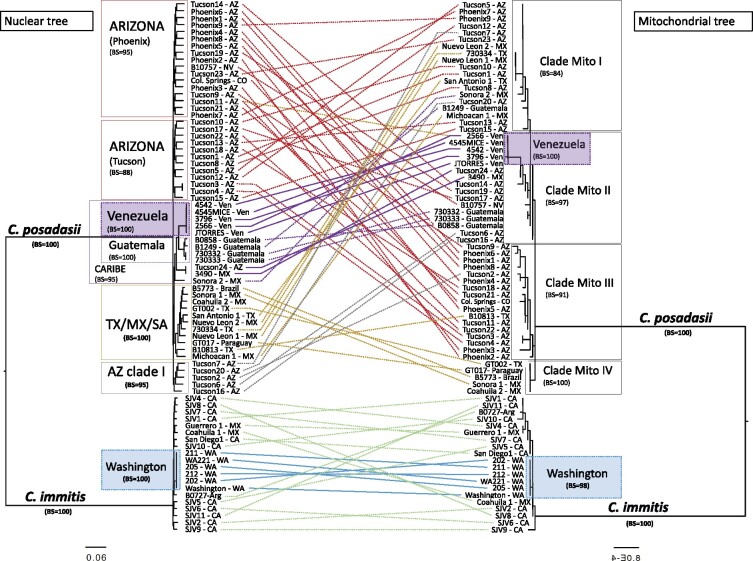
To determine whether the nuclear and the mitochondrial genomes show the same evolutionary trajectories, we compared the mitochondrial phylogeny with the whole genome species phylogeny. To score the differences in partitions produced between mtDNA and nucDNA trees, we used a normalized RF symmetric distance (Split distance, SD). If two trees are completely congruent the split distance score is 0, whereas with complete tree disagreement the score is 1. In the case of the nucDNA vs mtDNA tree comparison, the split distance equals 0.92 [138/150] thus indicating that the topologies are largely inconsistent with each other. Both topologies support species divergence between C. immitis and C. posadasii (i.e., no isolates being assigned to a different species; see Figure 3) but little to no concordance within species, especially within C. posadasii. Nonetheless, the lengths of the branches for each of the two species are much shorter than expected. This is unexpected because the mutation rate of mtDNA is larger than the mutation rate of nucDNA. In other fungi, mutation rates are orders of magnitude larger than in nuclear genes (Chou and Leu 2010). We assumed a nuclear mutation rate of 1 × 10−11 per site per generation (the order of magnitude in Aspergillus flavus, A. fumigatus, and A. nidulans) and a mitochondrial mutation rate of 0.76 to 1.6 × 10−7 per site per generation, which is a rate that seems to be consistent across most eukaryotes (Konrad et al. 2017). These rates suggest that the divergence of the nuclear genomes occurred approximately 1 × 109 generations, while the divergence of the mitochondrial genomes occurred in approximately 1 × 104 generations. This result suggests ancient exchange of mitochondrial genomes. The species-level concordance indicates that the mtDNA exchange event is old enough that each species has reaccumulated sufficient substitutions to differentiate the two species.
Figure 3.
Tree topology comparisons of Coccidioides nucDNA (left panel) and mtDNA (right panel) phylogenomic trees. Phylogenetic tree branches are proportional to the nucleotide divergence (see scale) and the main clades are highlighted. Bootstrap support was calculated, and branch support was added to the corresponding clade. The terminal taxa are color-coded according to their placement on the nucDNA tree and taxa are connected between mtDNA and nucDNA phylogenomic trees to visualize concordance (solid lines) vs discordance (dotted lines).
Next, we studied the arrangement of variation within each of the two species. Even though the species-level split was consistent between nucDNA and mtDNA, the topologies were largely inconsistent with each other for within-species comparisons. For C. posadasii, previous phylogenetic analyses of nuclear markers have shown that at least three distinct populations exist: isolates from Arizona (clinical and environmental), isolates from Texas, Brazil, Argentina, and Mexico, and a third set of isolates from the Caribbean region (Engelthaler et al. 2016; Teixeira et al. 2019a). Using mtDNA markers, we identified four clusters that do not follow this strong biogeographic pattern. For example, the isolates B10813, Tucson2 and Tucson20 have conflicting phylogenetic distributions, as previously these were placed within the Texas/Mexico/SouthAmerica or AZ clade I using nuclear markers (Figure 3). Mito1 is composed of 19 isolates from four populations as defined by nuclear markers (Arizona, Texas/Mexico/SouthAmerica, Caribbean and AZ clade I); Mito2 is composed of 16 isolates composed mostly of isolates from the Caribbean region—including a monophyletic group from Venezuela, and four isolates from Arizona and two from AZ clade I; Mito3 is composed of 17 isolates, mostly from Arizona (Arizona and AZ clade I) but with one isolate from Texas (Texas/Mexico/SouthAmerica population), and Mito4 is composed of fiveisolates, all from the Texas/Mexico/SouthAmerica population. These four clusters were also confirmed by FastStructure (Figure 4). For C. immitis both nucDNA and mtDNA phylogenies revealed a clade composed of isolates from Washington, which is genetically distinct from the rest of C. immitis (Figure 3). No other consistent pattern of clustering was observed for the remaining C. immitis individuals comparing the two phylogenies (Figure 3).
Using marginal likelihoods to assess the number of populations, three groups were observed within C. immitis, and four groups were supported in C. posadasii (Figure 4A). For C. immitis the three groups are (1) a group containing the Washington state, Argentina, and Coahuila1 isolates (Figure 4B, blue); (2) a group with a single strain, SJV9 (Figure 4B, grey); and (3) a third group composed of isolates from San Joaquin Valley, San Diego and Mexico (Figure 4B, green). Within C. posadasii, the four groups supported are: (1) a group containing all the Venezuela isolates (Figure 4B, purple); (2) a group formed with strains from Texas/Mexico/South America nucDNA clade, and corresponding to the clade Mito IV in the phylogenetic tree (Figure 4B, yellow); (3) a group formed by strains from Arizona and mostly belonging to the Tucson nucDNA clade (Figure 4B, pink); and (4) a group composed of the remaining isolates from Guatemala, Arizona, Mexico, and Texas (Figure 4B, salmon). This last group does not correspond to the population structure defined for nucDNA genes. We observed some admixed genotypes within C. immitis, and several within the C. posadasii species suggesting that gene flow in the mitogenome might occur within these groups (Figure 4B, isolates with mixed color bars).
Discussion
Mitochondrial genomes are responsible for key aspects of cellular metabolism. In this article, we report the assembled mitochondrial genomes of two species of the human pathogen Coccidioides and study the extent of genetic variation in the mtDNA of these two species. Our results have two implications. First, the assembly of the full circular genomes allows us to study conservation of gene order, content, and size among the Onygenales. Second, the study of polymorphism within and among species allows us to determine whether mtDNA markers are suitable for species and population identification. We discuss each of these implications as follows.
Gene order conservation
Gene content in mitochondria is remarkably conserved across the tree of life. However, fungal genomes show differences in the order of gene arrangement. Our results suggest that there is complete synteny in the mtDNA genome of the species from the Onygenaceae family in spite of over 100 million years of divergence. Howevr, this synteny is not conserved for the entire Onygenales order. Members of the family Arthrodermataceae and Ascosphaeraceae show similar gene order, but reveal a different order compared with Onygenaceae and Ajellomycetaceae (Figure 2). The atp6-atp8 pair is a highly conserved syntenic unit across the Eurotiomycetes (Aguileta et al. 2014), but the pair is disrupted by genomic rearrangements found within Arthrodermataceae and Ascosphaeraceae. This level of conservation over 150 million years [the crown age for the Onygenaceae family (Sharpton et al. 2009; Rouxel et al. 2011)] is comparable with the conservation of mtDNA order in a different group of ascomycetes, the species from the order Hypocreales [crown age: 179 MYA, (Sung et al. 2008; Rouxel et al. 2011)], but dissimilar to the level of turnover that occurs among other fungi (Aguileta et al. 2014). This apparent difference in the rate of evolution of gene order in mtDNA across taxa could be real, or be caused by unequal taxonomic sampling. The mechanisms for fungal mitochondrial gene order evolution remain largely unknown, but have been attributed primarily to mitochondrial non-homologous recombination (Beaudet et al. 2013). Gene order turnover and stasis could then be attributed to differences in the rate of mtDNA recombination. While this is a feasible explanation, only a systematic study with balanced sampling across fungi will resolve whether there are differences in the rate of evolution in gene order in fungal mitochondrial genomes.
Mitochondrial polymorphism
Molecular markers from mitochondrial genomes are extensively used as molecular markers in speciation studies (Dupuis et al. 2012). Before this study, the mitochondrial genome and the phylogenetic relationships of mitochondrial haplotypes in Coccidioides remained unstudied. Our results suggest that mtDNA can be used to distinguish between Coccidioides species, and that species-level diagnostic efforts using mtDNA variation are well-founded (Hamm et al. 2019). Nonetheless, mtDNA markers might lead to incorrect assignments at the intraspecies population level due to extensive genealogical incongruence within species, particularly within C. posadasii. Conflicting phylogenetic histories in mtDNA and nucDNA genealogies have been observed in other pathogenic fungi. For example, Paracoccidioides brasiliensis and P. restrepiensis appear to be polyphyletic using mtDNA markers, and the tree topologies differ from those obtained from nucDNA markers (Turissini et al. 2017). The incongruence between mtDNA and nucDNA in Paracoccidioides species seems to be the result of interspecific hybridization (Turissini et al. 2017). The remnants of such admixture events are only observed in the mtDNA as most of the nucDNA shows no evidence of gene exchange (Mavengere et al. 2020). Genealogical discordance between mitochondrial and nuclear gene genealogies also occurs in the Cryptococcus gattii species complex and seems to be caused by interspecies introgression (Bovers et al. 2009). Blocks of mtDNA with mixed ancestry have been observed in Candida (Anderson et al. 2001), Verticillum (Depotter et al. 2018), and Phellinus (Lee et al. 2019). The exchange of mtDNA across fungal species remains largely understudied, but represents an important source of genetic variation and evolutionary history. Additional admixture analyses within Coccidioides species are required to further evaluate the ancestral traces between different genotypes. Certainly, there are nuclear mitochondrial (NUMTs) genes found in the nuclear genome, and mapping analysis will be inconclusive unless the bona fide NUMT sequences have significantly diverged, or if long reads clearly identify the inserts in nucDNA to correct short read error with confidence. Although our SNP analysis method removed low coverage SNPs and rare alleles, and coverage of the mitogenome was very high, it remains a formal possibility that some of the SNPs are due to NUMTs. Thus, future mtDNA and nucDNA analysis in both species of Coccidioides should consider long read sequences to more fully explore this issue.
Pioneering studies of fungal mitochondrial polymorphisms suggest a lower rate of nucleotide substitutions in the mitochondrial genomes compared with mammals (Clark-Walker 1991). Population demographics and ecological factors can modulate the lower amount of mtDNA mutations in fungal species (Basse 2010). Despite the high levels of divergence between nuclear-mitochondrial trees within C. posadasii, we have observed low mitochondrial diversity within some genetic groups; there is a little to no mitochondrial genetic variation between the individuals Tucson5, Phoenix7, Phoenix9, Tucson12, Tucson7, and Tucson23 within the C. posadasii Mito1 clade compared with the genome tree. In contrast, the individuals from the Venezuela group also show a lower intraclade variation but are concordant nuclear-mitochondrial tree topologies. This pattern is also observed in other fungal species. Within the two dominant C. gatti lineages VGI and VGII, the mitochondrial sequence divergence within groups was about three times lower compared with nuclear genes (Xu et al. 2009). This observation suggests that selective sweeps in fungal mitogenomes are potentially due to uniparental inheritance of mitochondrial DNA due to the dominance of a few mitotypes in natural fungal populations.
Mitochondrial genomes may be associated with hybrid incompatibilities. Among hybrids between different species [e.g., Saccharomyces (Lee et al. 2008; Chou et al. 2010; Chou and Leu 2010); Drosophila (Montooth et al. 2010; Clancy et al. 2011)], and even within species [e.g., Caenorhabditis (Chang et al. 2016), Tigriopus copepods (Barreto et al. 2018; Lima et al. 2019)], epistatic interactions between the nucDNA and the mtDNA from progenitors can lead to hybrid breakdown, which in turn might be of importance for the persistence of species in the face of secondary contact and gene flow. The interaction between mtDNA and nucDNA plays a role on the fitness of fungal interspecific hybrids (Olson and Stenlid 2001; Giordano et al. 2018). Notably, alleles involved in reproductive isolation are more likely to show gene genealogies that deviate from the species tree, which makes mtDNA gene genealogies an unreliable proxy of evolutionary history in cases in which there are mito-nuclear incompatibilities. Understanding whether or not these interactions are pervasive across fungi will require the production of interspecific hybrids and the systematic study of their fitness, a task currently not feasible for Coccidioides (Burt et al. 1996; Chou and Leu 2010).
Future directions
The endozoan lifestyles of Coccidioides spp. (Taylor and Barker 2019) suggest that mitochondrial genes involved in thermal adaptation and oxidative stress could be under strong directional selection. Mitochondria are the primary site of ATP production, and are possibly associated with organismal thermal tolerance; however, general principles governing these patterns remain undefined (Chung and Schulte 2020). For example, the size of pathogenic nematode mitogenomes from different thermal habitats is significantly smaller in endothermic host species comparing with ectothermic hosts suggesting that host adaptation might be a significant impact on mitochondrial evolution (Lagisz et al. 2013). Coccidioides posadasii is more heat tolerant than C. immitis (Mead et al. 2020). In Saccharomyces, polymorphisms in the cox1 gene of mtDNA are associated with adaptation to variable temperatures. The yeast mitochondrial genome might be a hotspot in the evolution of thermal adaptation in Saccharomyces species (Baker et al. 2019; Li et al. 2019). In Cryptococcus, highly virulent strains show an increased ability to replicate within macrophages, which is in turn associated with an unusual mitochondrial morphology and upregulation of genes encoded by the mitochondrial genome and associated with mitochondrial activities (Ma et al. 2009; Ma and May 2010). Whether mtDNA variation is associated with thermal fitness or virulence in Coccidioides remains to be tested. Our genome assembly and measurements of variation also open the door to study the evolutionary drivers of mitochondrial variation in Coccidioides. Genealogical discordance is higher with C. posadasii than within C. immitis which might suggest differences in the rate of mtDNA recombination between species. These potential differences in recombination might indicate that the genomes of the two Coccidioides species might be under different selection regimes, which in turn might also differ from other fungi (Sandor et al. 2018).
Conclusions
The assembly and measurement of polymorphism in the mitogenome of Coccidioides will facilitate deeper investigations into the impact of mitochondrial evolution in Coccidioides’ niche adaptation, with particular emphasis on mammalian host co-evolution and oxidative stress responses. The results shown here will also aid in the study of the evolutionary drivers of mitogenome evolution in fungi.
Acknowledgments
The authors thank the Translational Genomics Institute and the Pathogen and Microbiome Institute and Northern Arizona University computing resources for data processing support as well as Dr. John Taylor for helpful discussion and edits.
M.T., J.E.S., B.F.L., D.R.M., and B.M.B wrote and edited the article; M.T., D.R.M. and B.F.L conducted data analysis.
Funding
This work was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC; RGPIN-2017-05411) and by the Fonds de Recherche Nature et Technologie, Quebec (FQRNT; 2018-PR-206806) (to BFL ); J.E.S. is a CIFAR Fellow in the program Fungal Kingdom: Threats and Opportunities and was supported by University of California MRPI grants (MRP-17-454959) “UC Valley Fever Research Initiative” and (VFR-19-633952) “Investigating fundamental gaps in Valley Fever knowledge” and United States Department of Agriculture—National Institute of Food and Agriculture Hatch Project (CA-R-PPA-5062-H); B.M.B. was supported by the National Institute of Allergy and Infectious Diseases (R21 AI128536), State of Arizona Technology and Research Initiative Fund , and the Flinn Foundation; D.R.M. was supported by the National Intitutes of Health (R01AI153523).
Conflicts of interest
None declared.
Contributor Information
Marcus de Melo Teixeira, Pathogen and Microbiome Institute, Northern Arizona University, Flagstaff, AZ 86011, USA; Faculty of Medicine, University of Brasília-DF, Brasília, Federal District 70910-3300, Brazil.
B Franz Lang, Robert Cedergren Centre for Bioinformatics and Génomiques, Département de Biochimie, Université de Montréal, Montréal, Quebec H3C 3J7, Canada.
Daniel R Matute, Biology Department, University of North Carolina, Chapel Hill, NC 27599, USA.
Jason E Stajich, Institute for Integrative Genome Biology, University of California, Riverside, CA 92521, USA; Department of Microbiology and Plant Pathology, University of California, Riverside, CA 92521, USA.
Bridget M Barker, Pathogen and Microbiome Institute, Northern Arizona University, Flagstaff, AZ 86011, USA.
Literature cited
- Aguileta G, de Vienne DM, Ross ON, Hood ME, Giraud T, et al. 2014. High variability of mitochondrial gene order among fungi. Genome Biol Evol. 6:451–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JB, Wickens C, Khan M, Cowen LE, Federspiel N, et al. 2001. Infrequent genetic exchange and recombination in the mitochondrial genome of Candida albicans. J Bacteriol. 183:865–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisimova M, Gil M, Dufayard JF, Dessimoz C, Gascuel O. 2011. Survey of branch support methods demonstrates accuracy, power, and robustness of fast likelihood-based approximation schemes. Syst Biol. 60:685–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker EP, Peris D, Moriarty RV, Li XC, Fay JC, et al. 2019. Mitochondrial DNA and temperature tolerance in lager yeasts. Sci Adv. 5:eaav1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker BM, Litvintseva AP, Riquelme M, Vargas-Gastelum L. 2019. Coccidioides ecology and genomics. Med Mycol. 57:S21–S29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreto FS, Watson ET, Lima TG, Willett CS, Edmands S, et al. 2018. Genomic signatures of mitonuclear coevolution across populations of Tigriopus californicus. Nat Ecol Evol. 2:1250–1257. [DOI] [PubMed] [Google Scholar]
- Basse CW. 2010. Mitochondrial inheritance in fungi. Curr Opin Microbiol. 13:712–719. [DOI] [PubMed] [Google Scholar]
- Beaudet D, Terrat Y, Halary S, de la Providencia IE, Hijri M. 2013. Mitochondrial genome rearrangements in Glomus species triggered by homologous recombination between distinct mtDNA haplotypes. Genome Biol Evol. 5:1628–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedict K, McCotter OZ, Brady S, Komatsu K, Sondermeyer Cooksey GL, et al. 2019. Surveillance for Coccidioidomycosis—United States, 2011-2017. MMWR Surveill Summ. 68:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovers M, Hagen F, Kuramae EE, Boekhout T. 2009. Promiscuous mitochondria in Cryptococcus gattii. FEMS Yeast Res. 9:489–503. [DOI] [PubMed] [Google Scholar]
- Bullerwell CE, Leigh J, Forget L, Lang BF. 2003. A comparison of three fission yeast mitochondrial genomes. Nucleic Acids Res. 31:759–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt A, Carter DA, Koenig GL, White TJ, Taylor JW. 1996. Molecular markers reveal cryptic sex in the human pathogen Coccidioides immitis. Proc Natl Acad Sci U S A. 93:770–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso MA, Tambor JH, Nobrega FG. 2007. The mitochondrial genome from the thermal dimorphic fungus Paracoccidioides brasiliensis. Yeast. 24:607–616. [DOI] [PubMed] [Google Scholar]
- Chang C-C, Rodriguez J, Ross J. 2016. Mitochondrial–nuclear epistasis impacts fitness and mitochondrial physiology of interpopulation Caenorhabditis briggsae hybrids. G3 (Bethesda). 6:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou J-Y, Hung Y-S, Lin K-H, Lee H-Y, Leu J-Y. 2010. Multiple molecular mechanisms cause reproductive isolation between three yeast species. PLOS Biol. 8:e1000432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou J-Y, Leu J-Y. 2010. Speciation through cytonuclear incompatibility: Insights from yeast and implications for higher eukaryotes. Bioessays. 32:401–411. [DOI] [PubMed] [Google Scholar]
- Chung DJ, Schulte PM. 2020. Mitochondria and the thermal limits of ectotherms. J Exp Biol. 223:jeb227801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy DJ, Hime GR, Shirras AD. 2011. Cytoplasmic male sterility in Drosophila melanogaster associated with a mitochondrial CYTB variant. Heredity. 107:374–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark-Walker GD. 1991. Contrasting mutation rates in mitochondrial and nuclear genes of yeasts versus mammals. Curr Genet. 20:195–198. [DOI] [PubMed] [Google Scholar]
- Depotter JRL, Beveren F. V, van den Berg GCM, Wood TA, Thomma BPHJ, et al. 2018. Nuclear and mitochondrial genomes of the hybrid fungal plant pathogen Verticillium longisporum display a mosaic structure. bioRxiv. 249565. [Google Scholar]
- Dupuis JR, Roe AD, Sperling FA. 2012. Multi-locus species delimitation in closely related animals and fungi: one marker is not enough. Mol Ecol. 21:4422–4436. [DOI] [PubMed] [Google Scholar]
- Engelthaler DM, Roe CC, Hepp CM, Teixeira M, Driebe EM, et al. 2016. Local population structure and patterns of western hemisphere dispersal for Coccidioides spp. the fungal cause of Valley Fever. MBio. 7:e00550–00516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher FS, Bultman MW, Johnson SM, Pappagianis D, Zaborsky E. 2007. Coccidioides niches and habitat parameters in the southwestern United States: a matter of scale. Ann N Y Acad Sci. 1111:47–72. [DOI] [PubMed] [Google Scholar]
- Fisher MC, Koenig GL, White TJ, San-Blas G, Negroni R, et al. 2001. Biogeographic range expansion into South America by Coccidioides immitis mirrors New World patterns of human migration. Proc Natl Acad Sci U S A. 98:4558–4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano L, Sillo F, Garbelotto M, Gonthier P. 2018. Mitonuclear interactions may contribute to fitness of fungal hybrids. Sci Rep. 8:1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47:W59–W64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamm PS Hutchison MI Leonard P Melman S Natvig DO. 2019. First Analysis of Human Coccidioides Isolates from New Mexico and the Southwest Four Corners Region: Implications for the Distributions of C. posadasii and C. immitis and Human Groups at Risk. JoF. 5:74. 10.3390/jof5030074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14:587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollath DR, Miller KJ, Barker BM. 2019. The mysterious desert dwellers: Coccidioides immitis and Coccidioides posadasii, causative fungal agents of coccidioidomycosis. Virulence. 10:222–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konrad A, Thompson O, Waterston RH, Moerman DG, Keightley PD, et al. 2017. Mitochondrial mutation rate, spectrum and heteroplasmy in Caenorhabditis elegans spontaneous mutation accumulation lines of differing population size. Mol Biol Evol. 34:1319–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korovesi AG, Ntertilis M, Kouvelis VN. 2018. Mt-rps3 is an ancient gene which provides insight into the evolution of fungal mitochondrial genomes. Mol Phylogenet Evol. 127:74–86. [DOI] [PubMed] [Google Scholar]
- Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, et al. 2004. Versatile and open software for comparing large genomes. Genome Biol. 5:R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagisz M, Poulin R, Nakagawa S. 2013. You are where you live: parasitic nematode mitochondrial genome size is associated with the thermal environment generated by hosts. J Evol Biol. 26:683–690. [DOI] [PubMed] [Google Scholar]
- Lang BF, Laforest MJ, Burger G. 2007. Mitochondrial introns: a critical view. Trends Genet. 23:119–125. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H-H, Ke H-M, Lin C-YI, Lee TJ, Chung C-L, et al. 2019. Evidence of extensive intraspecific noncoding reshuffling in a 169-kb mitochondrial genome of a basidiomycetous fungus. Genome Biol Evol. 11:2774–2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H-Y, Chou J-Y, Cheong L, Chang N-H, Yang S-Y, et al. 2008. Incompatibility of nuclear and mitochondrial genomes causes hybrid sterility between two yeast species. Cell. 135:1065–1073. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XC, Peris D, Hittinger CT, Sia EA, Fay JC. 2019. Mitochondria-encoded genes contribute to evolution of heat and cold tolerance in yeast. Sci Adv. 5:eaav1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima TG, Burton RS, Willett CS. 2019. Genomic scans reveal multiple mito-nuclear incompatibilities in population crosses of the copepod Tigriopus californicus. Evolution. 73:609–620. [DOI] [PubMed] [Google Scholar]
- Liu W, Cai Y, Zhang Q, Chen L, Shu F, et al. 2020. The mitochondrial genome of Morchella importuna (272.2 kb) is the largest among fungi and contains numerous introns, mitochondrial non-conserved open reading frames and repetitive sequences. Int J Biol Macromol. 143:373–381. [DOI] [PubMed] [Google Scholar]
- Ma H, Hagen F, Stekel DJ, Johnston SA, Sionov E, et al. 2009. The fatal fungal outbreak on Vancouver Island is characterized by enhanced intracellular parasitism driven by mitochondrial regulation. Proc Natl Acad Sci U S A. 106:12980–12985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, May RC. 2010. Mitochondria and the regulation of hypervirulence in the fatal fungal outbreak on Vancouver Island. Virulence. 1:197–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavengere H, Mattox K, Teixeira MM, Sepúlveda VE, Gomez OM, et al. 2020. Paracoccidioides genomes reflect high levels of species divergence and little interspecific gene flow. (Preprint posted 2020 July 17). bioRxiv. doi:10.1101/2020.07.16.204636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell CS, Mattox K, Turissini DA, Teixeira MM, Barker BM, et al. 2019. Gene exchange between two divergent species of the fungal human pathogen. Coccidioides . Evolution. 73:42–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCotter OZ, Benedict K, Engelthaler DM, Komatsu K, Lucas KD, et al. 2019. Update on the epidemiology of coccidioidomycosis in the United States. Med Mycol. 57:S30–S40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, et al. 2010. The genome analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead HL Hamm PS Shaffer IN Teixeira MdM Wendel CS, et al. 2020. Differential Thermotolerance Adaptation between Species of Coccidioides. JoF. 6:366. 10.3390/jof6040366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh BQ, Nguyen MA, von Haeseler A. 2013. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 30:1188–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montooth KL, Meiklejohn CD, Abt DN, Rand DM. 2010. Mitochondrial–nuclear fixed incompatibilities between species of Drosophila. Evolution. 64:3364–3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neafsey DE, Barker BM, Sharpton TJ, Stajich JE, Park DJ, et al. 2010. Population genomic sequencing of Coccidioides fungi reveals recent hybridization and transposon control. Genome Res. 20:938–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson Å, Stenlid J. 2001. Mitochondrial control of fungal hybrid virulence. Nature. 411:438–438. [DOI] [PubMed] [Google Scholar]
- Pogoda CS, Keepers KG, Nadiadi AY, Bailey DW, Lendemer JC, et al. 2019. Genome streamlining via complete loss of introns has occurred multiple times in lichenized fungal mitochondria. Ecol Evol. 9:4245–4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigbò P, Garcia-Vallvé S, McInerney JO. 2007. TOPD/FMTS: a new software to compare phylogenetic trees. Bioinformatics. 23:1556–1558. [DOI] [PubMed] [Google Scholar]
- Quinlan AR. 2014. BEDTools: the Swiss-army tool for genome feature analysis. Curr Prot Bioinform. 47:11.12.11–11.12.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, Stephens M, Pritchard JK. 2014. fastSTRUCTURE: variational inference of population structure in large SNP data sets. Genetics. 197:573–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouxel T Grandaubert J Hane JK Hoede C Van De Wouw AP, et al. 2011. Effector diversification within compartments of the Leptosphaeria maculans genome affected by Repeat-Induced Point mutations. Nat Commun. 2:202. 10.1038/ncomms1189PMC: 21326234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandor S, Zhang Y, Xu J. 2018. Fungal mitochondrial genomes and genetic polymorphisms. Appl Microbiol Biotechnol. 102:9433–9448. [DOI] [PubMed] [Google Scholar]
- Schliep KP. 2011. phangorn: phylogenetic analysis in R. Bioinformatics. 27:592–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Xiao G, Zheng P, Cen K, Zhan S, et al. 2016. Divergent and convergent evolution of fungal pathogenicity. Genome Biol Evol. 8:1374–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpton TJ, Stajich JE, Rounsley SD, Gardner MJ, Wortman JR, et al. 2009. Comparative genomic analyses of the human fungal pathogens Coccidioides and their relatives. Genome Res. 19:1722–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan DB, Havird JC, Sharbrough J. 2017. The on-again, off-again relationship between mitochondrial genomes and species boundaries. Mol Ecol. 26:2212–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Florea L. 2015. Rcorrector: efficient and accurate error correction for Illumina RNA-seq reads. GigaSci. 4:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoddard BL. 2011. Homing endonucleases: from microbial genetic invaders to reagents for targeted DNA modification. Structure. 19:7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung GH, Poinar GO Jr, Spatafora JW. 2008. The oldest fossil evidence of animal parasitism by fungi supports a Cretaceous diversification of fungal-arthropod symbioses. Mol Phylogenet Evol. 49:495–502. [DOI] [PubMed] [Google Scholar]
- Tambor JH, Guedes RF, Nobrega MP, Nobrega FG. 2006. The complete DNA sequence of the mitochondrial genome of the dermatophyte fungus Epidermophyton floccosum. Curr Genet. 49:302–308. [DOI] [PubMed] [Google Scholar]
- Taylor JW, Barker BM. 2019. The endozoan, small-mammal reservoir hypothesis and the life cycle of Coccidioides species. Med Mycol. 57:S16–S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira MM, Alvarado P, Roe CC, Thompson GR 3rd, Patane JSL, et al. 2019a. Population structure and genetic diversity among isolates of Coccidioides posadasii in Venezuela and surrounding regions. mBio. 10:e01976-19. 10.1128/mBio.01976-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira MM, Barker BM, Stajich JE. 2019b. Improved reference genome sequence of Coccidioides immitis strain WA_211, isolated in Washington state. Microbiol Resour Announc. 8:e00149-19. 10.1128/MRA.00149-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turissini DA, Gomez OM, Teixeira MM, McEwen JG, Matute DR. 2017. Species boundaries in the human pathogen Paracoccidioides. Fungal Genet Biol. 106:9–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyke MCC, Teixeira MM, Barker BM. 2019. Fantastic yeasts and where to find them: the hidden diversity of dimorphic fungal pathogens. Curr Opin Microbiol. 52:55–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiston E, Zhang Wise H, Sharpton TJ, Jui G, Cole GT, et al. 2012. Comparative transcriptomics of the saprobic and parasitic growth phases in Coccidioides spp. PloS One. 7:e41034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Yang J, Yang F, Liu T, Leng W, et al. 2009. Recent dermatophyte divergence revealed by comparative and phylogenetic analysis of mitochondrial genomes. BMC Genomics. 10:238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Yan Z, Guo H. 2009. Divergence, hybridization, and recombination in the mitochondrial genome of the human pathogenic yeast Cryptococcus gattii. Mol Ecol. 18:2628–2642. 10.1111/j.1365-294X.2009.04227.x 19457185 [DOI] [PubMed] [Google Scholar]
- Zubaer A, Wai A, Hausner G. 2018. The mitochondrial genome of Endoconidiophora resinifera is intron rich. Sci Rep. 8:17591. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
SNP data for each strain analyzed for nuclear and mitochondrial trees, raw phylogenetic trees files (Figure 2) and unlinked SNPs and allele frequencies used as input for structure analysis (Figure 4) are available at Zenodo (doi:10.5281/zenodo.4592315).
Figure 2.
Mitochondrial gene content and synteny among Onygenalean fungi. Genes are color-coded (see legend) and are displayed according to position on the genome. The mitogenomes are highly syntenic but atp8 gene positioning differs between Onygenaceae/Ajellomycetaceae and Arthrodermataceae/Ascosphaeraceae.
Figure 4.
Population structure within C. immitis and C. posadasii based on mitochondrial-derived alleles. (A) Marginal likelihood plots for both C. immitis and C. posadasii species and the best K (or number of populations) selected within each species is marked with asterisks and was K = 2 and K = 4, respectively. (B) Proportion of admixture of each C. immitis or C. posadasii. Heterozygous or non-admixed strains are represented by solid bar while admixed individuals are displayed as multicolored bars. The proportion of each homozygous population can be observed by the scale on the top of each plot.