Abstract
The MADS box organ identity gene AGAMOUS (AG) controls several steps during Arabidopsis thaliana flower development. AG cDNA contains an open reading frame that lacks an ATG triplet to function as the translation initiation codon, and the actual amino terminus of the AG protein remains uncharacterized. We have considered the possibility that AG translation can be initiated at a non-AUG codon. Two possible non-AUG initiation codons, CUG and ACG, are present in the 5′ region of AG mRNA preceding the highly conserved MADS box sequence. We prepared a series of AG genomic constructs in which these codons are mutated and assayed their activity in phenotypic rescue experiments by introducing them as transgenes into ag mutant plants. Alteration of the CTG codon to render it unsuitable for acting as a translation initiation site does not affect complementation of the ag-3 mutation in transgenic plants. However, a similar mutation of the downstream ACG codon prevents the rescue of the ag-3 mutant phenotype. Conversely, if an ATG is introduced immediately 5′ to the disrupted ACG codon, the resulting construct fully complements the ag-3 mutation. The AG protein synthesized in vitro by initiating translation at the ACG position is active in DNA binding and is of the same size as the AG protein detected from floral tissues, whereas AG polypeptides with additional amino-terminal residues do not appear to bind DNA. These results indicate that translation of AG is initiated exclusively at an ACG codon and prove that non-AUG triplets may be efficiently used as the sole translation initiation site in some plant cellular mRNAs.
The recognition by eukaryotic ribosomes of the translation initiation codon generally proceeds by a scanning mechanism in which the 40S ribosomal subunit migrates from the 5′ end of the mRNA molecule, stopping at the first AUG triplet that is found in a favorable sequence context for initiation of translation (25, 27, 31). An AUG codon indeed serves as the translation start site in the vast majority of eukaryotic genes, but evidence accumulated over the past years indicates that the translation of several animal and viral mRNAs can be or is initiated at codons which differ from AUG in one base, like CUG, ACG, and GUG (17, 28). These alternative initiation codons can form only two base pairs, instead of three, with the anticodon of the Met-tRNAiMet (the initiating amino acid of the polypeptides produced as a result of these noncanonical translation events has been shown to be methionine [2, 48, 61]). The weakened codon-anticodon interaction, however, might be compensated for by contacts with nearby nucleotides, since translation initiation at non-AUG codons requires that such codons be in an optimal or favorable sequence context (28, 33).
Most of the cases so far described in which a non-AUG codon acts as the start site for eukaryotic mRNA translation are analogous to translation by leaky scanning (27), in that two different protein products are produced: the synthesis of one polypeptide is initiated at the non-AUG codon, and another is synthesized from a downstream AUG, usually in frame with the non-AUG codon. The non-AUG-initiated product is often the less abundant of the two (11, 36, 47). The functional relevance of the non-AUG-initiated polypeptides has not often been demonstrated. In some cases, the alternative, non-AUG-initiated translation products, such as murine fibroblast growth factor 3 (FGF-3 or int-2), human FGF-2, mammalian Hck, or mammalian Bag-1, (1, 7, 37, 47), have been shown to have altered properties, for example, a different subcellular localization. Examples of alternative translation products that have been shown to have particular biological roles come mostly from viruses, like the ACG-initiated adeno-associated virus capsid protein B (45) or the CUG-initiated murine leukemia virus cell surface antigen gp85gag (50). Equine infectious anemia virus Tat protein is synthesized by initiation at a CUG codon, which is bypassed by some ribosomes (leaky scanning), allowing translation of the downstream rev cistron of the bicistronic tat-rev mRNA (8).
Eukaryotic cellular mRNAs whose translation is initiated exclusively at a non-AUG codon, however, are rare. These include the mRNAs for the human (and murine) translational regulator p97 (20), the human transcriptional enhancer factor 1 (62), and the rat (and human) bZIP transcription factor HLF326 (13).
AGAMOUS (AG) is an Arabidopsis thaliana MADS box homeotic gene that functions in determining the identity of the reproductive organs of the flower (stamens and carpels), in suppressing the indeterminate growth of the floral meristem (i.e., in limiting floral growth to the production of four whorls of organs), and in helping to maintain its floral identity (5, 6, 42, 43, 46, 63). Plants homozygous for a strong ag mutant allele have flowers in which the third-whorl stamens are converted to petals, while another flower, that will reiterate the same organ pattern, develops in place of the fourth-whorl carpels (5, 6, 63). MADS box genes represent a large multigene family in plants, and many of them are involved in different steps of flower development (reviewed in reference 53). MADS box genes code for MADS domain-containing transcription factors. The MADS domain is a conserved DNA-binding/dimerization region that is essential for protein function (35, 44, 52), and it is usually located at the amino terminus of the plant MADS proteins. However, AG, most of its orthologs from other species, and some other MADS domain proteins highly related to it, such as Arabidopsis AGL1 and AGL5, bear an amino-terminal extension of uncharacterized function (N region) that is variable in sequence and in length (53).
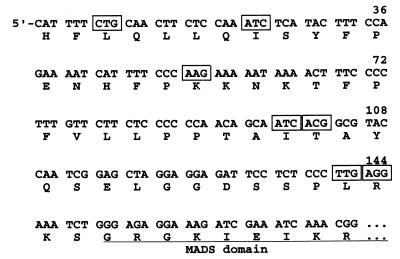
In spite of extensive studies on the functions of AG, its translation start site (and therefore the amino terminus of the AG protein) remains undetermined. As shown in Fig. 1, AG mRNA (deduced from the longest AG cDNA clones that were identified [63]) does not contain any AUG codon upstream of the highly conserved MADS box coding sequence that could serve as the translation initiation site; instead, the predicted AG open reading frame extends to the 5′ end of the mRNA. Those features of the AG cDNA clones suggested that they were not full length and that AG mRNA could have a secondary structure that prevented the synthesis of complete cDNA molecules (63). We have revisited that hypothesis, and we show here that AG translation is initiated exclusively at an ACG codon.
FIG. 1.
Sequence of the 5′ region of AG cDNA (63). The deduced amino acid sequence is shown below the nucleotide sequence, with residues that form part of the highly conserved MADS domain underlined. Triplets 5′ to the MADS box coding sequence that differ from ATG in only one nucleotide are boxed.
MATERIALS AND METHODS
Plasmid construction. (i) Plasmids for in vitro transcription and translation.
pSPUTK-AG is a pSPUTK (Stratagene)-derived plasmid to produce AG in in vitro transcription-translation reactions (51, 52). To construct pSPUTK-AG, the wild-type sequence 5′-CATTTT at the beginning of the AG cDNA was changed by means of a PCR amplification to 5′-ATGGGG, in order to provide an artificial initiating ATG codon. pSPUTK-AGATG1 contains an AG insert that starts at nucleotide (nt) 7 of AG cDNA in which the CTG codon at positions 7 to 9 of wild-type AG cDNA was changed to ATG. Similarly, the AG insert of pSPUTK-AGATG2 starts at nucleotide 100 of AG cDNA, and the ACG codon at positions 100 to 102 of wild-type AG cDNA was changed to ATG. In pSPUTK-AG and pSPUTK-AGATG2, the respective engineered ATG codon forms part of the NcoI site used for cloning the AG fragment into pSPUTK, and it is therefore embedded in an optimal sequence context for translation initiation (accATGg [30, 31]). For pSPUTK-AGATG1 construction, a BglII site was used, and the engineered ATG codon is in a suboptimal context (tttATGc). In all cases, the AG construct is immediately preceded by the Xenopus β-globin leader sequence present in that vector.
(ii) Plasmids for plant transformation.
A plasmid (pfAG6a) that contains an AG construct composed of approximately 6 kb of upstream sequences, genomic sequences through the large second intron, cDNA sequences through the rest of the coding region, and approximately 600 bp of 3′ sequences has been described previously (59). A PstI-BamHI genomic fragment from this AG construct, comprising from 1 kb upstream of exon 1 to most of the second intron, was subcloned into pSPORT1 (pSP1-AGG; GIBCO-BRL) and used as template in site-directed mutagenesis reactions performed according to the QuickChange (Stratagene) protocol. The changes introduced by site-directed mutagenesis in this AG genomic fragment were as follows: CTGm (the CTG codon that is found at positions 7 to 9 of wild-type AG cDNA was changed to CTT by using the sense oligodeoxynucleotide 5′-CTTACCTTCCATTTTCTTCAACTTCTCCAAATC); ACGm (the ACG codon that is found at positions 100 to 102 of wild-type AG cDNA was changed to ACC, using the sense oligodeoxynucleotide 5′-GGAGCAGCAATCACCGCGTACCAATCGG); ATGACGm (the sequence ATCACG [nt 97 to 102 of wild-type AG cDNA] was changed to ATGACC, using the sense oligodeoxynucleotide 5′-GCTTTGGAGCAGCAATGACCGCGTACCAATCGGAGC). (Underlining in all sequences indicates the mutation that is introduced with these oligonucleotides.) Fragments containing the pertinent mutations were introduced back into pSP1-AGG, replacing the wild-type sequences in the following single or combined changes: AGCTGm, AGACGm, AGATGACGm, AGCTGmACGm, and AGCTGmATGACGm. Plasmids were verified by sequencing for both the presence of the desired mutations and the absence of unwanted changes. The mutated PstI-BamHI fragments were cloned back into pfAG6a, and the final constructs were transferred to the plant transformation vector pCGN1547 (39).
In vitro translation and DNA binding assays.
AG, AGATG1, and AGATG2 proteins were synthesized from plasmids pSPUTK-AG, pSPUTK-AGATG1, and pSPUTK-AGATG2 by using the TnT coupled transcription-translation reticulocyte lysate system (Promega). [35S]methionine-labeled proteins were analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis on 12.5% gels. The DNA-binding activities of pSPUTK-AG-, pSPUTK-AGATG1-, and pSPUTK-AGATG2-derived proteins were tested in electrophoretic mobility shift assays, which were performed as described previously (51, 52).
Agrobacterium-mediated plant transformation and strain constructions.
The AG constructs in the pCGN1547 vector were transformed into Agrobacterium tumefaciens ASE. A. thaliana (Landsberg erecta [Ler]) plants that were either ag-3/+ or +/+ (F1 progeny from ag-3/+ parents) were transformed by vacuum infiltration (3), and transformants were selected by plating the seeds on kanamycin plates. For all of these AG constructs, we obtained an efficiency of transformation much lower than what we regularly obtain with other plasmids, perhaps owing to the large size of the AG insert. The genotype of the T1 plants at the AG locus (+/+, ag-3/+, or ag-3/ag-3) was determined by PCR primer-introduced restriction analysis of the products of reactions that amplified the relevant AG sequences from leaf tissue (procedure performed as described previously [54, 56]). Primers used for genotyping were 5′-GAAGTATTACCCGAATCCGCCCCAAGAAG and 5′-GTCGATTTCAGAAAATAAGAGCTC. The PCR product obtained with these oligodeoxynucleotides is 150 bp long, and it is cleaved by BslI to leave a 130-bp-long fragment if the amplified sequence is wild-type. T1 plants that were ag-3/+ or +/+ were allowed to self-fertilize or crossed to heterozygous ag-3 plants by manual cross-pollination to eventually obtain transgenic lines homozygous for the ag-3 mutation.
Plants were grown on a 1.5:1:1 soil:perlite:vermiculite mix under constant cool-white fluorescent light at 23°C.
Immunological detection of AG protein.
Floral buds (up to stage 10 [60]) of each plant were collected in 1.5-ml microcentrifuge tubes, homogenized in 2 volumes of 2× SDS sample buffer (0.125 M Tris-HCl [pH 6.8], 4% SDS, 10% β-mercaptoethanol, 20% sucrose, 0.02% bromophenol blue) and sonicated for 30 s with an ultrasonic cell disruptor (Microson, Westborough, Mass.). After sonication, samples were boiled for 2 min and centrifuged at 12,000 × g for 10 min, and the supernatant was collected for analysis. Proteins were separated on SDS–12.5% polyacrylamide gels and blotted onto nitrocellulose membranes (0.45-μm pore size; Millipore). For immunodetection of AG, blots were blocked in Tris-buffered saline (20 mM Tris-HCl [pH 7.5], 137 mM NaCl, 0.1% [vol/vol] Tween 20) containing 5% (wt/vol) nonfat dry milk powder and incubated with anti-AG antiserum (1:700 dilution) (21). Blots were washed, incubated with goat anti-rabbit horseradish peroxidase-conjugated secondary antibody, and developed by using an enhanced chemiluminescence immunodetection system (Amersham Life Science) according to procedures recommended by the manufacturer.
Nucleotide sequence accession number.
The genomic sequence of the AG locus has been made available through the Arabidopsis Genome Initiative (GenBank accession no. AL021711).
RESULTS
Possible AG initiation codons.
5′-RACE (rapid amplification of 5′ cDNA ends) experiments specifically designed to overcome problems caused by RNA secondary structure failed to extend the known AG sequence for more than a few nucleotides, which neither provided an ATG codon nor interrupted the open reading frame (ORF) (data not shown). The genomic sequence corresponding to the 5′ region of AG mRNA was determined for A. thaliana ecotypes Nossen and Wassilewskija to confirm that the absence of an initiating ATG codon was not a peculiarity of the previously characterized A. thaliana Ler (63) and Columbia (GenBank accession no. AL021711) AG alleles. In all cases, the sequence of that region lacked a suitable ATG initiation codon (data not shown). We therefore considered the possibility that AG translation can start at a non-AUG codon.
There are seven codons that differ from AUG at a single position in frame with, and 5′ to, the MADS box coding sequence in AG mRNA (Fig. 1). We considered two of them as potential translation initiation sites: a CUG codon (nt 7 to 9 of AG mRNA) and an ACG codon (nt 100 to 102 of AG mRNA) (Fig. 1). These two codons were chosen out of the seven because the majority of non-AUG translation initiation cases so far identified in eukaryotes involved CUG, ACG, or GUG triplets (4, 17). In addition, transient expression studies of translation initiation in plant protoplasts using fusions of non-AUG codons to a reporter gene ORF lacking an AUG showed that whereas CUG and ACG could act as initiation codons with 30 and 15% of the efficiency of an AUG in those assays, respectively, AUC, AAG, UUG, and AGG (also present in the AG sequence [Fig. 1]) exhibited only 0.1 to 3% efficiency (16).
Determination of the AG initiation codon in in vitro studies.
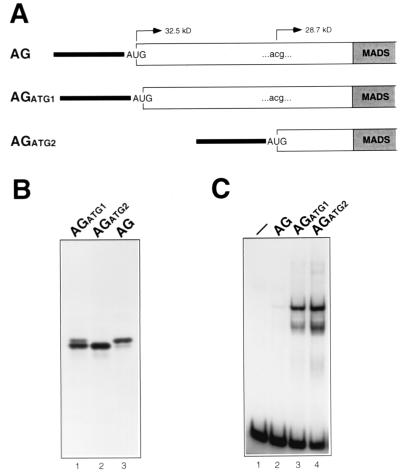
AG translation was first investigated in vitro with the purpose of assaying the DNA-binding activity of AG proteins initiated at the CUG and ACG positions (and therefore differing in the amino-terminal extension to the MADS domain, which is 31 amino acids longer in the AG form initiated at the CUG position [Fig. 1]), and of developing an AG molecular weight marker useful for studying the AG protein synthesized in vivo. In plasmids used for in vitro transcription and translation, an ATG codon had been engineered either at the position corresponding to the 5′ end of AG cDNA (AG construct), at the CTG position (AGATG1 construct), or at the ACG position (AGATG2 construct) (Fig. 2A). In vitro transcription-translations programmed with the AG construct generated two products, the higher-molecular-weight polypeptide being the more abundant of the two (Fig. 2B, lane 3). Two products were also produced from the AGATG1 construct, although in this case the amount of the smaller polypeptide exceeded that of the larger one (Fig. 2B, lane 1). Finally, only the lower-molecular-weight AG product was synthesized in translations programmed with the AGATG2 construct (Fig. 2B, lane 2). We interpret these in vitro results to mean that in RNA produced from the AG construct, translation started mostly at the engineered AUG codon (which is in an optimal context [accAUGg] [30, 31]), but polypeptides whose synthesis was initiated downstream at the ACG codon (which also is in a favorable sequence context [atcACGg]) were also produced. In contrast, in AGATG1 RNA, the engineered AUG codon is in a suboptimal context, uuuAUGc; thus, leaky scanning was enhanced and translation was initiated more often at the downstream ACG codon. Finally, the synthesis of AG polypeptides by initiating translation exclusively at the ACG position was facilitated by the AGATG2 construct, in which the ACG codon is changed to ATG and there are no upstream ATG triplets. The ACG codon present in AG mRNA therefore appears to function as an efficient initiation codon in the rabbit reticulocyte lysate in vitro system, since the amount of AG protein synthesized from the ACG codon after it is reached by leaky scanning ribosomes (AGATG1 construct) is, although lower, comparable to that produced from an ATG codon engineered at that position in an optimal context (AGATG2 construct [Fig. 2B, lanes 1 and 2).
FIG. 2.
Analysis of AG proteins synthesized in vitro by initiating translation at positions corresponding to the 5′ end of AG cDNA (AG), the CTG codon (AGATG1), and the ACG codon (AGATG2). (A) Schematic representation of the 5′ region of AG RNAs synthesized from pSPUTK-AG, pSPUTK-AGATG1, and pSPUTK-AGATG2. The β-globin leader sequence, derived from pSPUTK, is represented as a black line, and the AG sequence is represented as a bar (in gray for the MADS domain coding sequence). The ACG100–102 codon, present in pSPUTK-AG and pSPUTK-AGATG1, is indicated. (B) Products synthesized by in vitro translating AGATG1, AGATG2, and AG RNAs. (C) DNA-binding activity of the products synthesized by in vitro translating AG, AGATG1, and AGATG2 RNAs. A control with unprogrammed reticulocyte lysate is shown (lane 1).
The DNA-binding capabilities of these different AG forms were tested in electrophoretic mobility shift assays (Fig. 2C). DNA binding reactions performed with AG, AGATG1, and AGATG2 translation mixtures caused the same mobility shift of the free probe, and the intensity of the shifted band (Fig. 2C) correlated with the amount of the lower-molecular-weight AG polypeptide produced in the in vitro translation reactions (Fig. 2B). These results suggest that the AG product synthesized from the ACG position was active in DNA binding, whereas the AG polypeptides whose synthesis was initiated either at the CUG position or at the 5′ end of AG cDNA were not.
AG translation in vivo is initiated exclusively at an ACG codon.
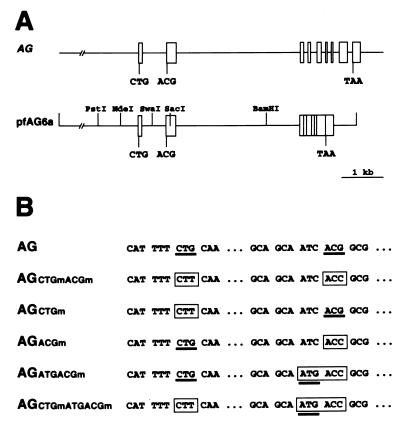
To determine which codon is used for initiation of AG translation in vivo, we engineered a series of mutations in an AG genomic construct and used the different mutant variants to generate transgenic lines to assay whether they could complement the ag-3 mutation. The AG construct used (pfAG6a) contains genomic sequences from approximately 6 kb upstream of the 5′ end of AG cDNA through the large second intron, cDNA sequences through the rest of the coding region, and genomic 3′ sequences (Fig. 3A) (59). Therefore, the AG transgenes are expressed from the AG promoter, no assumption is made about the transcription initiation site, and the 5′ region of the synthesized transcripts would be expected to be the same as that of wild-type AG mRNA. The engineered mutations consisted of changing the CTG and ACG codons (to CTT and ACC, respectively) to render them unsuitable for acting as a translation initiation site. These mutations were introduced together (AGCTGmACGm) or separately (AGCTGm and AGACGm) into the AG construct (Fig. 3B). An additional mutation was introduced to generate an ATG triplet immediately 5′ to the created ACC codon (AGATGACGm and AGCTGmATGACGm constructs [Fig. 3B]).
FIG. 3.
Mutations engineered in an AG genomic construct to determine the AG translation initiation codon in vivo. (A) Scheme of the AG genomic region and of the AG construct in pfAG6a. Exons are represented as boxes and introns as lines. Positions of the CTG and ACG triplets (in exons 1 and 2, respectively) and of the TAA stop codon are indicated. Restriction sites used for the introduction of the different mutations used in this study into pfAG6a are indicated. (B) Mutagenesis of the CTG and ACG codons. For each construct, mutated codons are boxed, and triplets that could conceivably act as a translation initiation codon are underlined.
Arabidopsis flowers consist of four sepals, four petals, six stamens, and two fused carpels, from the outermost to the innermost whorl (Fig. 4A). Plants homozygous for the strong ag-3 allele have flowers in which the third-whorl stamens are converted to petals while another flower, that will reiterate the same organ pattern, develops in place of the fourth-whorl carpels (Fig. 4B). Plants that were transgenic for the wild-type version of the AG construct described above and homozygous for the ag-3 mutation showed complete rescue of the mutant phenotype (AG ag-3 lines [Fig. 4C and reference 59]). However, when the AG transgene carried mutations at both the CTG and ACG codons, it was unable to complement the ag-3 mutation (AGCTGmACGm ag-3 lines [Fig. 4D]). The two engineered mutations, CTGm and ACGm, were also tested individually. Whereas disruption of the CTG codon did not affect the capability of the corresponding AG construct to complement the ag-3 mutation (AGCTGm ag-3 transgenic lines [Fig. 4E]), flowers of AGACGm ag-3 lines exhibited the characteristic ag-3 mutant phenotype. The effect of the ACGm mutation is not the result of an amino acid change in the sequence of the AG protein if translation were to be initiated upstream of the ACG codon, because the mutation was a change in the ACG codon to ACC (Fig. 3B), and both triplets code for the same amino acid, threonine. Another possible explanation for the failure of the AGACGm construct to rescue the ag-3 mutant phenotype is that the nucleotide change affected RNA stability and/or processing. To rule out this possibility, an ATG codon was engineered immediately upstream of the ACC triplet (AGATGACGm and AGCTGmATGACGm constructs [Fig. 3B]). If the ACG codon is indeed the translation initiation site in AG mRNA and the ACGm mutation does not affect RNA properties, this additional change could represent a compensatory mutation (it would allow translation to start immediately upstream of the ACG codon position) and restore the ability of the original AGACGm and AGCTGmACGm constructs to rescue the mutant phenotype when present as transgenes in ag-3 plants. In fact, AGATGACGm ag-3 and AGCTGmATGACGm ag-3 lines bear flowers with wild-type phenotypes (Fig. 4G and H).
FIG. 4.
AG RNA translation is initiated at an ACG codon in vivo. (A) Wild-type (wt) A. thaliana (Ler) flower. (B) ag-3 homozygous flower, showing petals and sepals in place of stamens and carpels. (C) AG ag-3 flower showing a wild-type phenotype. The ag-3 mutation is complemented by an AG transgene (pfAG6a construct). (D) AGCTGmACGm ag-3 flower consisting of sepals and petals only. The ag-3 mutant phenotype is not rescued by the AGCTGmACGm transgene. (E) AGCTGm ag-3 flower. The ag-3 mutation is complemented: one sepal and one petal have been removed to reveal the stamens and carpels. (F) AGACGm ag-3 flower showing the ag-3 mutant phenotype. (G) AGATGACGm ag-3 flower. The ag-3 mutation is complemented by the AGATGACGm transgene. One sepal and one petal have been removed. (H) AGCTGmATGACGm ag-3 flower showing complete rescue of the ag-3 mutant phenotype by the AGCTGmATGACGm transgene.
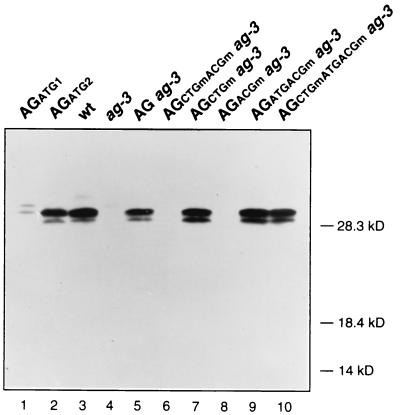
We characterized the AG protein(s) synthesized in the different lines by using an AG-specific antiserum (21). The AG polypeptides detected in extracts from wild-type flowers appear to be the same as those produced by in vitro translation of AGATG2 synthetic RNA: a major band that corresponds to initiation of protein synthesis at the ACG position and a minor smaller band that could be either a truncated product or the result of spurious initiation events (Fig. 5, lanes 2 and 3). No AG polypeptides were detected in extracts from floral buds of ag-3, AGCTGmACGm ag-3, and AGACGm ag-3 lines (Fig. 5, lanes 4, 6, and 8), in agreement with the mutant phenotype exhibited by those flowers. The AG products that accumulated in AGCTGm ag-3, AGATGACGm ag-3, and AGCTGmATGACGm ag-3 flowers were the same size as those from AG ag-3 and wild-type flowers (Fig. 5), in agreement with the rescue of the ag mutant phenotype that those lines showed and further supporting the conclusion that AG translation in vivo is initiated exclusively at the ACG100–102 codon.
FIG. 5.
Immunological detection of AG protein. Samples used in the Western blot are proteins synthesized by in vitro translation of AGATG1 and AGATG2 synthetic RNAs (lanes 1 and 2) and extracts from flowers (up to stage 10) of wild-type (Ler), ag-3, AG ag-3, AGCTGmACGm ag-3, AGCTGm ag-3, AGACGm ag-3, AGATGACGm ag-3, and AGCTGmATGACGm ag-3 plants.
DISCUSSION
mRNA translation in eukaryotes, and in particular translation initiation site recognition, is largely explained by the scanning model (25, 27, 31). In general, and in contrast to prokaryotic translation, eukaryotic translation is stringent in using only an AUG codon as the initiation site. Over the past several years, however, unconventional cases of mRNAs that are not translated according to all of the rules of the scanning process have been described, and the extent to which translation might deviate from the model appears to vary among different eukaryotic organisms, for example, between mammals and yeast. Translation initiation at non-AUG codons has been described for several cellular mRNAs from mammals and from Drosophila, as well as for several animal viruses. Mammal and insect non-AUG codon recognition is largely dependent on the alternative codon being in an optimal sequence context, which by itself is an important component of the scanning model. In contrast, yeast appears to be unable to efficiently use codons other than AUG as translation initiators (10, 12, 19), perhaps because sequence context in yeast has a very minor role in AUG recognition (9), which would leave the 3-bp codon-anticodon interaction as the only requirement for protein synthesis initiation (19).
Exceptions in plants to several rules of the scanning model have been described predominantly for RNAs derived from plant viruses (reviewed in references 14 and 55). In particular, the available information about initiation by non-AUG codons in plants is very limited: only two natural RNAs, derived from rice tungro bacilliform virus (RTBV) (15) and from soil-borne wheat mosaic virus (SBWMV) (58), have been shown to use noncanonical initiation codons. In the case of RTBV, an AUU codon that is reached by ribosome shunt, instead of by the normal process of scanning, is used as the initiation site for ORF1 (15). The efficiency of translation initiation at that codon at its natural position, downstream of the long RTBV leader sequence, reaches 5 to 10% of that provided by an AUG codon engineered at the same position (15). In SBWMV RNA2, inefficient alternative initiation at a CUG codon results in the synthesis of minor amounts of a 25-kDa protein that is an N-terminally extended version of the 19-kDa capsid protein, which is synthesized from a downstream in frame AUG codon (58). It has also been shown that an AUU codon present in the 5′ untranslated leader (Ω sequence) of tobacco mosaic virus can provide an alternative initiation site when this 5′ leader is used to enhance translation of a heterologous ORF in transgenic potato plants (57). Finally, transient expression studies in plant protoplasts using fusions of non-AUG codons embedded in an optimal sequence context to a CAT ORF lacking an AUG showed that some of those codons, most notably CUG and ACG, could act as initiation sites (16). Of these examples, only the ribosome shunt-mediated non-AUG initiation of RTBV RNA translation has been shown to be of physiological significance: it appears necessary to facilitate the translation by leaky scanning of additional downstream ORFs (15).
The results described here expand this limited repertoire by showing that the Arabidopsis AG mRNA, which is of importance in the life cycle of the plant, since in the absence of AG function reproductive organs do not develop, is translated exclusively from an ACG codon. The genetic approach that we have used to show the non-AUG initiation of AG translation rules out the possibility of an artifactual result, as has been found on several occasions in which non-AUG initiation events were deduced solely from cDNA sequences and in vitro data (32).
Two features of AG mRNA might account for its efficient use of an ACG codon as the translation initiation site. The first is that ACG100–102 is in an optimal sequence context (23, 26, 33). The second is the potential to adopt a secondary structure downstream of the ACG codon, where inverted repeats are present (Fig. 6A), that could facilitate its recognition. It has been shown that recognition by mammalian ribosomes of an AUG codon in a suboptimal context, as well as that of non-AUG codons in general, is enhanced by the presence of a downstream stem-loop (hairpin) structure (29). In fact, this appears to be a common feature for many of the mRNAs whose translation has been shown to initiate at non-AUG codons (30).
FIG. 6.
Comparison of AG and homologous genes. (A) Sequence of the 5′ region of AG (63) and BAG1 (38) cDNAs. The respective initiation codons, ACG and ATG, are boxed. Inverted repeated sequences that could potentially form a secondary structure in AG mRNA are indicated by arrows; nucleotide identities between AG and BAG1 sequences are indicated by asterisks. The beginning of the MADS box coding sequence is also indicated. (B) Amino acid sequence of the amino-terminal region of AG, BAG1, PLE (from Antirrhinum; GenBank accession no. S53900), NAG1 (from tobacco; L23925), TAG1 (from tomato; L26295), FBP6 and pMADS3 (from petunia; X68675 and X72916, respectively), SLM1 (from Silene latifolia, white campion; X80488), RAP1 (from Rumex acetosa, sorrel; X89107), CUM (from cucumber; AF035438), GAGA1 and GAGA2 (from Gerbera hybrida; AJ009722 and AJ009723), and ZAG1 and ZMM2 (from maize; L18924 and L81162). The amino acid sequence of these AG-related proteins is derived from the conceptual translation of the corresponding nucleotide sequences. All of these genes are homologous to AG and exhibit comparable expression patterns, but functional evidence showing that they are indeed AG orthologs is not available for all of them. For example, FBP6 may not be an AG cognate homolog (24). In some plant species, two homologous genes with overlapping but nonidentical activities are required for the functions that in Arabidopsis are carried out by AG (for example ZAG1 and ZMM2 in maize [40]).
The AG homolog from Brassica napus (a species from the same family as Arabidopsis, Brassicaceae), BAG1 (38), bears an ATG as initiation codon at exactly the same position (with respect to the downstream MADS box coding sequence) as the initiator ACG triplet is in AG (Fig. 6A). The 5′ untranslated regions of the two RNAs are nearly identical, and the similarity between AG and BAG1 extends throughout their entire lengths, including the 17-aa N-terminal extension to the MADS domain defined in this study (Fig. 6) (38). In fact, the position of the ATG initiation codon is also conserved among several other AG homologs from different species, such as NAG1, TAG1, FBP6, and pMADS3 (Fig. 6B). It therefore appears that the ACG codon that initiates translation in AG originated from a mutation in the ATG codon of the corresponding ancestral gene. That such a mutation was not severely deleterious could have been due to the above-mentioned features of AG mRNA that might facilitate its initiation of translation, since they are also present in BAG1 mRNA (Fig. 6A) and therefore likely in the common ancestor gene. In contrast, certain human genetic diseases are caused by the mutation of an ATG initiator codon to triplets like ACG, CTG, or GTG (49; see also reference 32). Since the ACG triplet as initiation codon for AG translation is found in several A. thaliana ecotypes, perhaps this mutation could be used to study the evolutionary relationships within the Arabidopsis genus.
The identification of the N terminus of the AG protein and the observation that an AG form initiated at the 5′ end of AG mRNA (by an artificial ATG codon introduced at that position) is inactive in binding to DNA, prompt a reevaluation of the published literature on AG function, including our own. In vitro, ACG100–102 is capable of acting as the AG initiation codon even in the presence of an upstream in frame AUG triplet (Fig. 2B). It therefore appears likely that AG constructs in which an ATG codon was engineered at the 5′ end of the cDNA were capable of retaining AG function because, in addition to the artifactual form of AG, the correct AG protein was synthesized from the downstream ACG triplet. These constructs include the pSPUTK-AG plasmid that we have used before (51, 52), constructs for AG expression in Escherichia coli and subsequent DNA binding assays (44), as well as constructs for ectopic expression of AG in transgenic plants under the control of the 35S promoter (transgenic lines of the 35S-ATG-1 series [41, 43]). In fact, all of those constructs were derived from the observation that 35S-ATG-1 lines showed ectopic AG activity (41), which was interpreted as evidence for the functionality of the AG protein synthesized from the artifactual ATG codon introduced at the 5′ end of AG cDNA. Based on the similarity between AG and BAG1, however, some of the other constructs that have been used to characterize AG and its function were engineered to have an ATG codon in place of ACG100–102, and the N-terminal extension of the corresponding proteins was therefore identical to that found in vivo (18, 22, 34, 35, 41, 51, 54).
Finally, the observation that a non-AUG triplet can act as the only functional initiation codon for the translation of an Arabidopsis mRNA adds yet another level of complexity to the accurate annotation of the sequence of the Arabidopsis genome. In fact, the AG locus is among the sequences already determined and annotated by the Arabidopsis Genome Initiative, but the translation initiation site has not been correctly predicted. The extent to which non-AUG translation initiation is used in plants is, although perhaps low, unknown. However, our results suggest that searching for non-AUG initiation sites for some plant proteins may be a necessary part of Arabidopsis post-genome sequence analysis.
ACKNOWLEDGMENTS
We are grateful to Vijaya Rao for helping with Arabidopsis transformations and with plant genotyping and maintenance; to Eva Ziegelhoffer for looking after our plants when needed; and to Catherine Baker, Chiou-Fen Chuang, Jennifer Fletcher, Carolyn Ohno, Doris Wagner, and Eva Ziegelhoffer for valuable comments on the manuscript.
This work was supported by National Institutes of Health grant GM45697 to E.M.M.; T.I. was supported by a postdoctoral fellowship from the Japan Society for the Promotion of Science.
REFERENCES
- 1.Acland P, Dixon M, Peters G, Dickson C. Subcellular fate of the Int-2 oncoprotein is determined by choice of initiation codon. Nature. 1990;343:662–665. doi: 10.1038/343662a0. [DOI] [PubMed] [Google Scholar]
- 2.Becerra S P, Rose J A, Hardy M, Baroudy B M, Anderson C W. Direct mapping of adeno-associated virus capsid proteins B and C: a possible ACG initiation codon. Proc Natl Acad Sci USA. 1985;82:7919–7923. doi: 10.1073/pnas.82.23.7919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bechtold N, Ellis J, Pelletier G. In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci. 1993;316:1194–1199. [Google Scholar]
- 4.Boeck R, Kolakofsky D. Positions +5 and +6 can be major determinants of the efficiency of non-AUG initiation codons for protein synthesis. EMBO J. 1994;13:3608–3617. doi: 10.1002/j.1460-2075.1994.tb06668.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowman J L, Smyth D R, Meyerowitz E M. Genes directing flower development in Arabidopsis. Plant Cell. 1989;1:37–52. doi: 10.1105/tpc.1.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bowman J L, Smyth D R, Meyerowitz E M. Genetic interactions among floral homeotic genes of Arabidopsis. Development. 1991;112:1–20. doi: 10.1242/dev.112.1.1. [DOI] [PubMed] [Google Scholar]
- 7.Bugler B F, Amalric F, Prats H. Alternative initiation of translation determines cytoplasmic or nuclear localization of basic fibroblast growth factor. Mol Cell Biol. 1991;11:573–577. doi: 10.1128/mcb.11.1.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carroll R, Derse D. Translation of equine infectious anemia virus bicistronic tat-rev mRNA requires leaky ribosome scanning of the tat CTG initiation codon. J Virol. 1993;67:1433–1440. doi: 10.1128/jvi.67.3.1433-1440.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cigan A M, Pabich E K, Donahue T F. Mutational analysis of the HIS4 translational initiator region in Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:2964–2975. doi: 10.1128/mcb.8.7.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clements J M, Laz T M, Sherman F. Efficiency of translation initiation by non-AUG codons in Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:4533–4536. doi: 10.1128/mcb.8.10.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Curran J, Kolakofsky D. Ribosomal initiation from an ACG codon in the Sendai virus P/C mRNA. EMBO J. 1988;7:245–251. doi: 10.1002/j.1460-2075.1988.tb02806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donahue T F, Cigan A M. Genetic selection for mutations that reduce or abolish ribosomal recognition of the HIS4 translational initiator region. Mol Cell Biol. 1988;8:2955–2963. doi: 10.1128/mcb.8.7.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Falvey E, Fleury-Olela F, Schibler U. The rat hepatic leukemia factor HLF gene encodes two transcriptional activators with distinct circadian rhythms, tissue distributions and target preferences. EMBO J. 1995;14:4307–4317. doi: 10.1002/j.1460-2075.1995.tb00105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fütterer J, Hohn T. Translation in plants—rules and exceptions. Plant Mol Biol. 1996;32:159–189. doi: 10.1007/BF00039382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fütterer J, Potrykus I, Bao Y, Li L, Burns T M, Hull R, Hohn T. Position-dependent ATT initiation during plant pararetrovirus rice tungro bacilliform virus translation. J Virol. 1996;70:2999–3010. doi: 10.1128/jvi.70.5.2999-3010.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gordon K, Fütterer J, Hohn T. Efficient initiation of translation at non-AUG triplets in plant cells. Plant J. 1992;2:809–813. [PubMed] [Google Scholar]
- 17.Hann S R. Regulation and function of non-AUG-initiated proto-oncogenes. Biochimie. 1994;76:880–886. doi: 10.1016/0300-9084(94)90190-2. [DOI] [PubMed] [Google Scholar]
- 18.Huang H, Mizukami Y, Hu Y, Ma H. Isolation and characterization of the binding sequences for the product of the Arabidopsis floral homeotic gene AGAMOUS. Nucleic Acids Res. 1993;21:4769–4776. doi: 10.1093/nar/21.20.4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang H-k, Yoon H, Hannig E M, Donahue T F. GTP hydrolysis controls stringent selection of the AUG start codon during translation initiation in Saccharomyces cerevisiae. Genes Dev. 1997;11:2396–2413. doi: 10.1101/gad.11.18.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Imataka H, Olsen H S, Sonenberg N. A new translational regulator with homology to eukaryotic translation initiation factor 4G. EMBO J. 1997;16:817–825. doi: 10.1093/emboj/16.4.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ito T, Takahashi N, Shimura Y, Okada K. A serine/threonine protein kinase gene isolated by an in vivo binding procedure using the Arabidopsis floral homeotic gene product, AGAMOUS. Plant Cell Physiol. 1997;38:248–258. doi: 10.1093/oxfordjournals.pcp.a029160. [DOI] [PubMed] [Google Scholar]
- 22.Jack T, Sieburth L, Meyerowitz E M. Targeted misexpression of AGAMOUS in whorl 2 of Arabidopsis flowers. Plant J. 1997;11:825–839. doi: 10.1046/j.1365-313x.1997.11040825.x. [DOI] [PubMed] [Google Scholar]
- 23.Joshi C P, Zhou H, Huang X, Chiang V L. Context sequences of translation initiation codon in plants. Plant Mol Biol. 1997;35:993–1001. doi: 10.1023/a:1005816823636. [DOI] [PubMed] [Google Scholar]
- 24.Kater M M, Colombo L, Franken J, Busscher M, Masiero S, van Lookeren Campagne M M, Angenent G C. Multiple AGAMOUS homologs from cucumber and petunia differ in their ability to induce reproductive organ fate. Plant Cell. 1998;10:171–182. doi: 10.1105/tpc.10.2.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kozak M. How do eucaryotic ribosomes select initiation regions in messenger RNA? Cell. 1978;15:1109–1123. doi: 10.1016/0092-8674(78)90039-9. [DOI] [PubMed] [Google Scholar]
- 26.Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kozak M. The scanning model for translation: an update. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kozak M. Context effects and inefficient initiation at non-AUG codons in eucaryotic cell-free translation systems. Mol Cell Biol. 1989;9:5073–5080. doi: 10.1128/mcb.9.11.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kozak M. Downstream secondary structure facilitates recognition of initiator codons by eukaryotic ribosomes. Proc Natl Acad Sci USA. 1990;87:8301–8305. doi: 10.1073/pnas.87.21.8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kozak M. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J Biol Chem. 1991;266:19867–19870. [PubMed] [Google Scholar]
- 31.Kozak M. Determinants of translational fidelity and efficiency in vertebrate mRNAs. Biochimie. 1994;76:815–821. doi: 10.1016/0300-9084(94)90182-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kozak M. Interpreting cDNA sequences: some insights from studies on translation. Mamm Genome. 1996;7:563–574. doi: 10.1007/s003359900171. [DOI] [PubMed] [Google Scholar]
- 33.Kozak M. Recognition of AUG and alternative initiator codons is augmented by G in position +4 but is not generally affected by the nucleotides in positions +5 and +6. EMBO J. 1997;16:2482–2492. doi: 10.1093/emboj/16.9.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krizek B A, Meyerowitz E M. Mapping the protein regions responsible for the functional specificities of the Arabidopsis MADS domain organ identity proteins. Proc Natl Acad Sci USA. 1996;93:4063–4070. doi: 10.1073/pnas.93.9.4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krizek B A, Riechmann J L, Meyerowitz E M. Use of the APETALA1 promoter to assay the in vivo function of chimeric MADS box genes. Sex Plant Reprod. 1999;12:14–26. [Google Scholar]
- 36.Lemaire P, Vesque C, Schmitt J, Stunnenberg H, Frank R, Charnay P. The serum-inducible mouse gene Krox-24 encodes a sequence-specific transcriptional activator. Mol Cell Biol. 1990;10:3456–3467. doi: 10.1128/mcb.10.7.3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lock P, Ralph S, Stanley E, Boulet I, Ramsay R, Dunn A. Two isoforms of murine hck, generated by utilization of alternative translation initiation codons, exhibit different patterns of subcellular localization. Mol Cell Biol. 1991;11:4363–4370. doi: 10.1128/mcb.11.9.4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mandel M A, Bowman J L, Kempin S A, Ma H, Meyerowitz E M, Yanofsky M F. Manipulation of flower structure in transgenic tobacco. Cell. 1992;71:133–143. doi: 10.1016/0092-8674(92)90272-e. [DOI] [PubMed] [Google Scholar]
- 39.McBride K E, Summerfelt K R. Improved binary vectors for Agrobacterium-mediated plant transformation. Plant Mol Biol. 1989;14:269–276. doi: 10.1007/BF00018567. [DOI] [PubMed] [Google Scholar]
- 40.Mena M, Ambrose B A, Meeley R B, Briggs S P, Yanofsky M F, Schmidt R J. Diversification of C-function activity in maize flower development. Science. 1996;274:1537–1540. doi: 10.1126/science.274.5292.1537. [DOI] [PubMed] [Google Scholar]
- 41.Mizukami Y, Ma H. Ectopic expression of the floral homeotic gene AGAMOUS in transgenic Arabidopsis plants alters floral organ identity. Cell. 1992;71:119–131. doi: 10.1016/0092-8674(92)90271-d. [DOI] [PubMed] [Google Scholar]
- 42.Mizukami Y, Ma H. Separation of AG function in floral meristem determinacy from that in reproductive organ identity by expressing antisense AG RNA. Plant Mol Biol. 1995;28:767–784. doi: 10.1007/BF00042064. [DOI] [PubMed] [Google Scholar]
- 43.Mizukami Y, Ma H. Determination of Arabidopsis floral meristem identity by AGAMOUS. Plant Cell. 1997;9:393–408. doi: 10.1105/tpc.9.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mizukami Y, Huang H, Tudor M, Hu Y, Ma H. Functional domains of the floral regulator AGAMOUS: characterization of the DNA binding domain and analysis of dominant negative mutations. Plant Cell. 1996;8:831–845. doi: 10.1105/tpc.8.5.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muralidhar S, Becerra S P, Rose J A. Site-directed mutagenesis of adeno-associated virus type 2 structural protein initiation codons: effects on regulation of synthesis and biological activity. J Virol. 1994;68:170–176. doi: 10.1128/jvi.68.1.170-176.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Okamuro J K, den Boer B G W, Lotys-Prass C, Szeto W, Jofuku K D. Flowers into shoots: photo and hormonal control of a meristem identity switch in Arabidopsis. Proc Natl Acad Sci USA. 1996;93:13831–13836. doi: 10.1073/pnas.93.24.13831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Packham G, Brimmel M, Cleveland J L. Mammalian cells express two differently localized Bag-1 isoforms generated by alternative translation initiation. Biochem J. 1997;328:807–813. doi: 10.1042/bj3280807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peabody D S. Translation initiation at non-AUG triplets in mammalian cells. J Biol Chem. 1989;264:5031–5035. [PubMed] [Google Scholar]
- 49.Pirastu M, Saglio G, Chang J C, Cao A, Yan Y W. Initiation codon mutation as a cause of α thalassemia. J Biol Chem. 1984;259:12315–12317. [PubMed] [Google Scholar]
- 50.Prats A-C, De Billy G, Wang P, Darlix J-L. CUG initiation codon used for the synthesis of a cell surface antigen coded by the murine leukemia virus. J Mol Biol. 1989;205:363–372. doi: 10.1016/0022-2836(89)90347-1. [DOI] [PubMed] [Google Scholar]
- 51.Riechmann J L, Krizek B A, Meyerowitz E M. Dimerization specificity of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA, and AGAMOUS. Proc Natl Acad Sci USA. 1996;93:4793–4798. doi: 10.1073/pnas.93.10.4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Riechmann J L, Wang M, Meyerowitz E M. DNA-binding properties of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA, and AGAMOUS. Nucleic Acids Res. 1996;24:3134–3141. doi: 10.1093/nar/24.16.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Riechmann J L, Meyerowitz E M. MADS domain proteins in plant development. Biol Chem. 1997;378:1079–1101. [PubMed] [Google Scholar]
- 54.Riechmann J L, Meyerowitz E M. Determination of floral organ identity by Arabidopsis MADS domain homeotic proteins AP1, AP3, PI, and AG is independent of their DNA-binding specificity. Mol Biol Cell. 1997;8:1243–1259. doi: 10.1091/mbc.8.7.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rohde W, Gramstat A, Schmitz J, Tacke E, Prüfer D. Plant viruses as model systems for the study of non-canonical translation mechanisms in higher plants. J Gen Virol. 1994;75:2141–2149. doi: 10.1099/0022-1317-75-9-2141. [DOI] [PubMed] [Google Scholar]
- 56.Sablowski R W M, Meyerowitz E M. Temperature-sensitive splicing in the floral homeotic mutant apetala3-1. Plant Cell. 1998;10:1453–1463. doi: 10.1105/tpc.10.9.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schmitz J, Prüfer D, Rohde W, Tacke E. Non-canonical translation mechanisms in plants: efficient in vitro and in planta initiation at AUU codons of the tobacco mosaic virus enhancer sequence. Nucleic Acids Res. 1996;24:257–263. doi: 10.1093/nar/24.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shirako Y. Non-AUG translation initiation in a plant RNA virus: a forty-amino-acid extension is added to the N terminus of the soil-borne wheat mosaic virus capsid protein. J Virol. 1998;72:1677–1682. doi: 10.1128/jvi.72.2.1677-1682.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sieburth L E, Running M P, Meyerowitz E M. Genetic separation of third and fourth whorl functions of AGAMOUS. Plant Cell. 1995;7:1249–1258. doi: 10.1105/tpc.7.8.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smyth D R, Bowman J L, Meyerowitz E M. Early flower development in Arabidopsis. Plant Cell. 1990;2:755–767. doi: 10.1105/tpc.2.8.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Taira M, Iizasa T, Shimada H, Kudoh J, Shimizu N, Tatibana M. A human testis-specific mRNA for phosphoribosylpyrophosphate synthetase that initiates from a non-AUG codon. J Biol Chem. 1990;265:16491–16497. [PubMed] [Google Scholar]
- 62.Xiao J H, Davidson I, Matthes H, Garnier J-M, Chambon P. Cloning, expression, and transcriptional properties of the human enhancer factor TEF-1. Cell. 1991;65:551–568. doi: 10.1016/0092-8674(91)90088-g. [DOI] [PubMed] [Google Scholar]
- 63.Yanofsky M F, Ma H, Bowman J L, Drews G N, Feldmann K A, Meyerowitz E M. The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature. 1990;346:35–39. doi: 10.1038/346035a0. [DOI] [PubMed] [Google Scholar]