Summary
Transcription start site (TSS) selection influences transcript stability and translation as well as protein sequence. Alternative TSS usage is pervasive in organismal development, is a major contributor to transcript isoform diversity in humans, and is frequently observed in human diseases including cancer. In this review, we discuss the breadth of techniques that have been used to globally profile TSSs and the resulting insights into gene regulation, as well as future prospects in this area of inquiry.
Keywords: TSS, transcription, transcription initiation, transcriptome, promoter, CAGE, nanoCAGE, nAnT-iCAGE, RAMPAGE, STRIPE-seq, CAGEr, TSRexploreR, CAGEfightR
Transcription start site (TSS) selection influences transcript stability and diversity in development and disease. In this Protocol Review, Policastro and Zentner comprehensively discuss molecular and computational techniques for global profiling of TSSs, providing a practical resource for researchers.
Introduction
The first base of a gene to be transcribed by an RNA polymerase, corresponding to the 5′-most base of the resulting transcript, is referred to as the transcription start site (TSS). Within a given gene promoter, there is generally not a single TSS, but rather a cluster of TSSs, referred to as a transcription start region (TSR). Furthermore, a gene might have multiple TSRs interspersed throughout the locus, indicating the presence of alternative promoters. The phenomenon of alternative transcription initiation is widespread in biology. For instance, several studies have described large-scale shifts in patterns of initiation during development (Batut et al., 2013; Danks et al., 2018; Zhang et al., 2017). This is perhaps most strikingly apparent in zebrafish, wherein the maternal and zygotic forms of more than 900 transcripts display differential TSS usage (Haberle et al., 2014), a phenomenon that appears to be conserved in mice (Cvetesic et al., 2020). Alternative promoter usage has also been reported to be widespread in human cancers, and use of alternative promoters is predictive of patient survival in some cases (Demircioğlu et al., 2019).
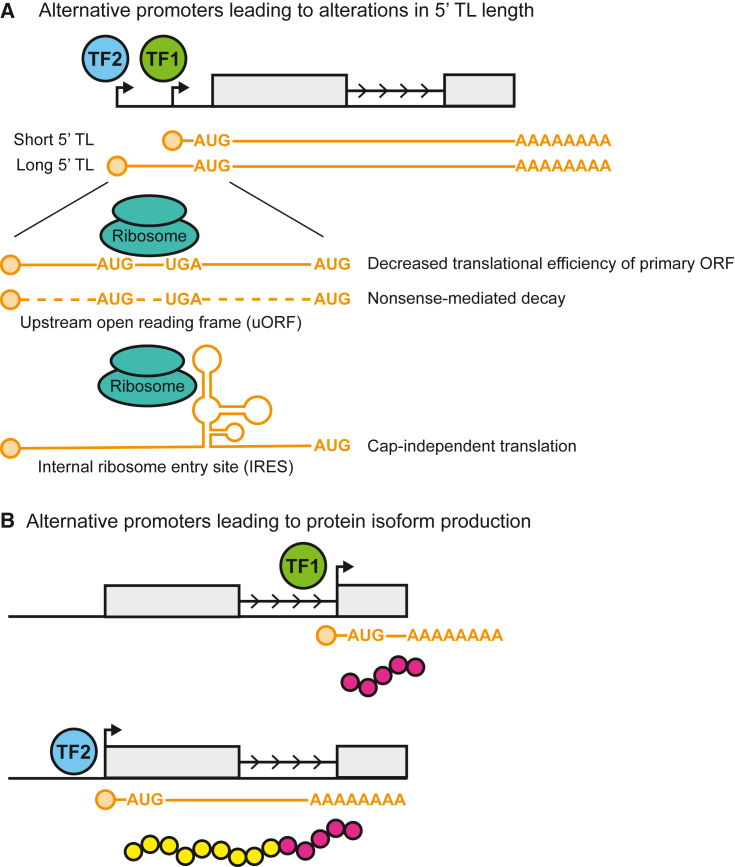
Broadly speaking, TSS shifting impacts gene regulation by altering the length of 5′ transcript leaders (5′ TLs). The 5′ TLs have been shown to play a large role in modulating both the stability and translation of mRNAs (Figure 1A) (Arribere and Gilbert, 2013; Dieudonné et al., 2015; Malabat et al., 2015; Rojas-Duran and Gilbert, 2012; Wang et al., 2016). In many cases, 5′ TLs encode upstream open reading frames (uORFs), which are short peptide-coding regions upstream of the primary ORF of a given transcript. uORFs might repress translation by preventing ribosomes from reaching the start codon of the primary ORF and might also lead to nonsense-mediated decay if the uORF stop codon is recognized as premature (Barbosa et al., 2013). A striking example of counteraction of uORF-mediated repression is observed in Arabidopsis thaliana, wherein exposure of seedlings to blue light causes downstream shifts in the TSS usage of 220 uORF-containing genes (Kurihara et al., 2018). Secondary structures within the 5′ TL, such as G-quadruplexes and pseudoknots, can also influence transcript stability and translation, and internal ribosome entry sites (IRESs) can promote cap-independent translation (Leppek et al., 2018).
Figure 1.
Effects of TSS selection on gene expression
(A) Possible effects of 5′ TL lengthening on transcript stability and translation. Transcription factor (TF) 1 specifies use of a proximal promoter, leading to a transcript with a short 5′ TL, while TF2 activates an upstream promoter that produces a transcript with a long 5′ TL. The extended 5′ TL may contain a uORF, which can act as a “sponge” for ribosomes by preventing them from reaching the transcript's primary ORF and may also lead to destruction of the transcript via NMD if the uORF stop codon is recognized as premature. The 5′ TL may also contain an IRES, enabling cap-independent translation. We note that these 5′ TL features are not mutually exclusive and direct interested readers to a recent comprehensive review on the roles of 5′ TLs in gene regulation (Leppek et al., 2018).
(B) Production of transcripts encoding distinct protein isoforms by TF-mediated activation of alternative promoters.
In addition to altering transcript regulation via 5′ TL length modulation, alternative TSS selection can give rise to transcript isoforms with distinct protein-coding potential. Indeed, it is estimated that the contributions of alternative promoters and transcription termination sites to transcript isoform diversity exceeds that of alternative splicing in multiple contexts, including normal human tissues (Reyes and Huber, 2018; Shabalina et al., 2014). Studies in plants have provided notable examples of alternative TSS selection giving rise to distinct transcript and protein isoforms. In maize, transcription of the mybr35 gene, encoding a MYB-family transcription factor, initiates from downstream TSSs in shoots, which gives rise to a protein lacking an N-terminal zinc finger domain. In contrast, upstream TSSs are used in roots, generating a full-length protein (Mejía-Guerra et al., 2015). In Arabidopsis, exposure of seedlings to red light globally alters TSS usage, leading to the production of N-terminally truncated protein isoforms with distinct subcellular localizations (Ushijima et al., 2017).
Although much work has been done on the impact of TSS selection on the regulation of the resulting transcript, it is becoming increasingly clear that transcription itself can inhibit TSS usage (Gowthaman et al., 2020). This phenomenon, termed transcriptional interference, refers to suppression in cis of transcription from one transcript unit by the act of RNAPII transcription of a second overlapping transcription unit. For instance, initiation from upstream TSSs might inhibit the use of downstream TSSs, a process termed tandem transcriptional interference (tTI). tTI might be induced by transcription of an mRNA isoform with an extended 5′ TL (Chen et al., 2017; Chia et al., 2017; Hollerer et al., 2019; Jorgensen et al., 2020; Nielsen et al., 2019) or an upstream noncoding RNA (Lin et al., 2018). In some cases, the long mRNA isoform might contain a uORF in its 5′ TL, downregulating translation (Chen et al., 2017; Chia et al., 2017; Hollerer et al., 2019), leading to a model of integrated transcriptional and translational repression.
Because of the importance of TSS selection in numerous biological contexts, a large number of methods for global TSS profiling have been developed. In this review, we describe the general enzymatic approaches used for this purpose, and the specific techniques that have used them. We also lay out computational strategies for TSS mapping data analysis. Last, we discuss current challenges and future prospects for the field, with a particular focus on single-cell mapping of heterogeneity in TSS usage.
Molecular approaches to global TSS mapping
Oligo-capping
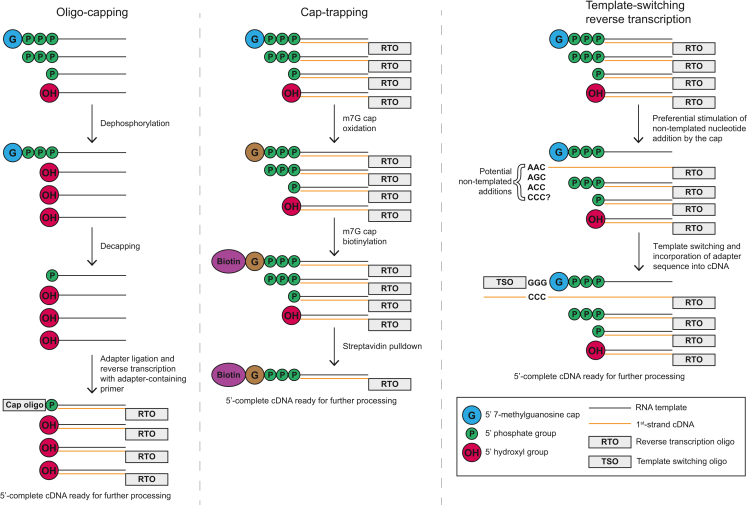
Oligo-capping was originally developed to facilitate recovery of 5′-complete cDNAs (Kazuo and Sumio, 1994; Suzuki and Sugano, 2003). Oligo-capping involves enzymatic removal of the 7-methylguanosine (m7G) cap of mRNAs (and other RNA species such as long noncoding RNAs, pre-micro RNAs, and enhancer RNAs) followed by ligation of a synthetic oligonucleotide (Figure 2A). In practice, the original oligo-capping protocol first uses Bacterial Alkaline Phosphatase (BAP) to hydrolyze the 5′ phosphates of uncapped RNA molecules, preventing subsequent adapter ligation. Tobacco Acid Pyrophosphatase (TAP) is then used to remove m7G caps, leaving a 5′ monophosphate suitable for adapter ligation. Prior to the widespread adoption of high-throughput sequencing, oligo-capping was used for an iteration of 5′ Serial Analysis of Gene Expression (5′ SAGE) (Hashimoto et al., 2004), wherein transcript 5′ sequences (5′ SAGE tags) are concatemerized, cloned, and sequenced. Oligo-capping was then adapted to high-throughput sequencing by ligation of 5′ tags to Solexa sequencing adapters and sequencing on the Illumina GA platform (Tsuchihara et al., 2009; Wakaguri et al., 2008), an approach later termed TSS-seq (Yamashita et al., 2011). Since the original implementation of TSS-seq, numerous TSS mapping methods have employed oligo-capping. Firstly, the Paired-End Analysis of TSSs (PEAT) approach (Ni et al., 2010) adapted oligo-capping for paired-end sequencing. Other oligo-capping approaches varied the enzymes used for RNA 5′ end processing. CapSeq (Gu et al., 2012), Transcript Leader sequencing (TL-seq) (Arribere and Gilbert, 2013), and Transcript IsoForm sequencing (TIF-seq) (Pelechano et al., 2013) used Calf Intestinal alkaline Phosphatase (CIP) instead of BAP for dephosphorylation of uncapped RNA, given that it can be heat inactivated, whereas Start-seq employed RNA 5′ Polyphosphatase (Nechaev et al., 2010). Start-seq and CapSeq also added a Terminator 5′ phosphate-dependent exonuclease (TEX) treatment to further reduce uncapped RNA (predominantly rRNA) levels, whereas TIF-seq provides both 5′and 3′ transcript end sequences thanks to a circular ligation step in the protocol. Simultaneous Mapping of RNA Ends by sequencing (SMORE-seq) enables 5′and 3′ transcript end mapping through sequential adapter ligation and also omits the phosphatase treatment prior to decapping, enabling capture of RNA degradation intermediates (Park et al., 2014).
Figure 2.
General approaches for TSS mapping
In oligo-capping, total RNA is first treated enzymatically to dephosphorylate uncapped RNAs. Caps are then removed, leaving 5′ monophosphates compatible with ligation. The cap oligo is ligated to the decapped RNAs and reverse transcription is performed, yielding 5′-complete cDNA ready for further processing. In cap-trapping, RNA:cDNA hybrids are chemically treated to oxidize RNA caps, which are then biotinylated. Streptavidin purification is then used to selectively enrich capped hybrids for further processing. In TSRT, total RNA is reverse transcribed, and the cap stimulates the addition of nontemplated nucleotides to the 3′ end of the first-strand cDNA. A TSO then interacts with the additional nucleotides and reverse transcriptase incorporates the complement of the TSO sequence into the first-strand cDNA, resulting in 5′-complete cDNA ready for further processing.
See Table 1 for advantages and disadvantages of each approach and Table 2 for RNA input requirements.
One note for oligo-capping methods is that TAP is no longer commercially available, so alternative enzymes are required. The E.coli RNA 5′ pyrophosphohydrolase RppH has been successfully used for decapping in Start-seq (Scheidegger et al., 2019), and a recombinant fusion of the Schizosaccharomyces pombe Dcp1-Dcp2 to its activator Edc1 has also shown promise in this regard (Paquette et al., 2018). Oligo-capping methods also tend to have high input requirements. For instance, it was reported that 30 μg of total RNA was necessary to construct an Arabidopsis PEAT library (Morton et al., 2014). Lastly, oligo-capping methods might suffer from sequence and/or structure biases in the RNA ligases used to add adapters (Baldrich et al., 2020; Fuchs et al., 2015; Hafner et al., 2011; Jayaprakash et al., 2011).
Cap-trapping
In addition to oligo-capping, early efforts to generate libraries of 5′-complete cDNAs led to the development of the cap-trapping approach (Carninci et al., 1996), in which the m7G cap is oxidized and biotinylated to allow streptavidin purification of 5′-complete cDNAs after reverse transcription (Figure 2B). In addition to its use in the generation of large cDNA libraries (Kawai et al., 2001; Okazaki et al., 2002), cap-trapping serves as the basis of Cap Analysis of Gene Expression (CAGE), perhaps the most widely known TSS mapping method. In the initial iteration of CAGE (Kodzius et al., 2006; Shiraki et al., 2003), reverse transcription and cap-trapping are performed, followed by ligation of a 5′ linker containing XmaJI and MmeI restriction sites. After second-strand synthesis, the double-stranded cDNA is digested with MmeI. MmeI is a Type IIs restriction enzyme that cuts 20 and 18 nucleotides downstream of the 3′ end of its recognition site, generating an asymmetric overhang to which a second adapter containing an XmaJI site can be ligated. XmaJI cleavage then releases the ligated cDNA fragments (referred to as CAGE tags), which are concatemerized, cloned, and sequenced by using the RIKEN Integrated Sequence Analysis (RISA) system (Shibata et al., 2000). A further iteration of CAGE, DeepCAGE, adapted the method to the 454 Life Sciences GS20 pyrosequencer (Balwierz et al., 2009; Valen et al., 2009). The first no-amplification version of CAGE, HeliScopeCAGE, greatly reduced input requirements and removed PCR biases by directly sequencing first-strand cDNA with the HeliScope Genetic Analysis System (Kanamori-Katayama et al., 2011). Switching to the EcoP15I restriction enzyme allowed for longer CAGE tags, facilitating more confident mapping to the genome (Takahashi et al., 2012), and eventually the need for restriction enzymes was removed in no-amplification non-tagging CAGE for Illumina sequencers (nAnT-iCAGE) (Murata et al., 2014). The major limitation of standard CAGE methods has been their high input requirements, though the nAnT-iCAGE protocol has been adapted to nanogram levels of RNA input via the use of capped, selectively degradable carrier RNA as Super-Low-Input Carrier CAGE (SLIC-CAGE) (Cvetesic et al., 2018).
Whereas CAGE relies on chemical modification of the cap structure to facilitate isolation of 5′-complete cDNAs, the recently published Multiplexed Affinity Purification of Capped RNA (MAPCap) method instead uses an antibody against m7G to isolate capped RNA prior to reverse transcription (Bhardwaj et al., 2019). Notably, MAPCap was reported to work well with as little as 100 ng RNA, suggesting that it is suitable for TSS mapping in precious samples. Natural proteins could also be used to isolate capped RNA for TSS mapping: a high-affinity mutant of the eukaryotic translation initiation factor 4E (eIF4E) (Choi and Hagedorn, 2003) has successfully been used to isolate mRNA (Blower et al., 2013) and nascent RNA (Matveeva et al., 2019) for sequencing.
Template-switching reverse transcription
Much like oligo-capping and cap-trapping, template-switching reverse transcription (TSRT) was initially leveraged as a means to generate 5′-complete cDNA molecules (Schmidt and Mueller, 1999; Zhu et al., 2001). TSRT leverages the propensity of Moloney murine leukemia virus (MMLV)-based reverse transcriptases to add 1–3 nontemplated bases to the 5′ end of a cDNA molecule. The prevailing view is that TSRT adds 1–3 predominantly cytosine residues, which can then serve as a handle for annealing of a template-switching oligo (TSO) with a three-riboguanosine overhang, allowing direct incorporation of a sequencing adapter at the 5′ end of the first-strand cDNA (Figure 2C). Interestingly, recent work indicates that the CCC overhang is a relatively rare result of TSRT (Wulf et al., 2019). Given that TSOs with riboguanosine overhangs have long been effective in TSRT despite the apparent paucity of CCC addition, it stands to reason that the mechanism of template switching does not necessarily rely on base pairing between templates.
Prior to the widespread adoption of high-throughput sequencing, TSRT was used to profile budding yeast TSSs in a modification of 5′ SAGE (Zhang and Dietrich, 2005). TSRT-based TSS mapping was brought into the high-throughput sequencing era via the original iteration of nanoCAGE (hereafter referred to as nanoCAGE [2010]) and CAGEscan (Plessy et al., 2010). After TSRT, nanoCAGE (2010) uses semi-suppressive PCR to reduce the prevalence of small artifactual fragments, followed by EcoP15I cleavage, adapter ligation, and library PCR, similar to first-generation CAGE approaches. CAGEscan simplified the nanoCAGE (2010) protocol by only requiring library PCR after semi-suppressive PCR, and also enabled paired-end sequencing of CAGE tags. CAGEscan, originally designed for the Illumina Genome AnalyzerIIX system, was subsequently adapted to the more modern Illumina HiSeq 2000 platform as NanoCAGE-XL (Cumbie et al., 2015). A further iteration of nanoCAGE (hereafter referred to as nanoCAGE [2017]), took advantage of Tn5 tagmentation to reduce input requirements and improve library size distribution, and added an optional TEX treatment step to reduce the prevalence of uncapped RNAs (such as rRNA) in the final libraries (Poulain et al., 2017). RNA Annotation and Mapping of Promoters for the Analysis of Gene Expression (RAMPAGE) combined TSRT with cap-trapping to provide additional specificity for capped transcripts (Batut et al., 2013). TSRT was later adapted to mapping of TSSs at single-cell resolution in methods such as C1-CAGE (Kouno et al., 2019), Tn5Prime (Cole et al., 2018), and low-input Parallel Analysis of RNA Ends (nanoPARE) (Schon et al., 2018). Finally, Survey of TRanscription Initiation and Promoter Elements with high-throughput sequencing (STRIPE-seq) (Policastro et al., 2020) removed the need for semi-suppressive PCR or tagmentation for modest RNA input amounts, and further reduced the prevalence of artifactual reads through improved oligo design and methodological optimizations.
TSRT-based methods are vulnerable to artifactual TSSs arising from a process termed strand invasion, wherein the TSO hybridizes to the first-strand cDNA before polymerization is complete, resulting in an artificially truncated cDNA molecule (Tang et al., 2013). Such artifacts can be reduced by introducing a spacer sequence into the TSO between the ribo-G overhang and the remaining sequence (e.g., TATAGGG in STRIPE-seq). This enables more confident filtering of such artifacts, as there are fewer matches to such a sequence within transcripts versus GGG. TSRT can also result in TSO chaining, wherein once RT reaches the 5′ end of the TSO, it performs another round of non-templated nucleotide addition, allowing another TSO to bind. The prevalence of the resulting TSO concatemers can be reduced by 3′ modification of the TSO with non-natural nucleotides (Kapteyn et al., 2010) or biotin (Turchinovich et al., 2014).
Mapping TSSs from nascent RNA
The methods discussed to this point profile the TSSs of stable, mature transcripts. However, in some cases, capture of TSSs from nascent transcripts might be desired. For instance, this is useful for unstable transcripts, which would be underrepresented in the steady-state RNA pool of the cell because of their rapid turnover after synthesis. The first iterations of nascent RNA 5′ end capture involved modifications of the Global Run-On sequencing (GRO-seq) and Precision Run-On sequencing (PRO-seq) protocols. The GRO-seq protocol involves isolation of nuclei, extension of nascent RNA, and simultaneous incorporation of the ribonucleotide analog 5-bromouridine 5′-triphosphate (BrUTP), and pulldown of nascent RNA with an anti-BrUTP antibody. PRO-seq, on the other hand, employs four parallel run-on reactions, each of which contains one of four biotinylated nucleotides, resulting in the incorporation of the biotinylated base and subsequent stalling of transcription, and streptavidin pulldown to capture nascent RNA (Kwak et al., 2013). GRO/PRO-cap (Core et al., 2014; Kwak et al., 2013) are modifications to the GRO/PRO-seq protocols designed to capture TSSs of nascent transcripts wherein, after the run-on reactions, nascent RNA is treated with TEX in the case of GRO-cap, and for both methods the subsequent addition of an oligo-capping strategy to the protocol. CAGE has also been extended to mapping TSSs from nascent RNA as Native Elongating Transcript sequencing and CAGE (NET-CAGE) (Hirabayashi et al., 2019). NET-CAGE combines CAGE with an approach to nascent RNA isolation used in 3′NT (Weber et al., 2014) and one iteration of mammalian NET-seq (Mayer et al., 2015). Both methods take advantage of the extraordinary resistance of the RNAPII:DNA:RNA ternary complex to harsh conditions such as high salt and urea (Cai and Luse, 1987; Wuarin and Schibler, 1994) to isolate chromatin-associated nascent RNAs.
Computational processing considerations
There are a few important points of consideration for preprocessing of TSS mapping data. TSS mapping data require precise alignment of reads to the genome, and for this task the STAR aligner (Dobin et al., 2013) is most often used. Another preprocessing consideration is the presence of PCR duplicates. Although more common in low-input methods such as nanoCAGE and STRIPE-seq, PCR duplicates can lead to inaccurate TSS and TSR quantification in all methods. In some methods (e.g., STRIPE-seq), a random sequence called a unique molecular identifier (UMI) is included in the R1 read, which for single-end sequencing data allows for the computational removal of PCR duplicates, as reads with the same genomic position and UMI are not expected to occur frequently by chance. For this task, UMI-tools (Smith et al., 2017) is recommended because of its ability to correct for PCR and sequencing errors that could lead to incorrect bases called in UMIs. For paired-end data, the read not anchored to the TSS tends to be somewhat randomly positioned because of random priming of RT or tagmentation. This can be used to remove PCR duplicates by using Samtools (Li et al., 2009), as unique reads would not be expected to have the same R1 and R2 positions by chance.
After PCR duplicate removal, there are two main steps shared by most methods. First, the 5′-most aligned bases should be processed for various artifacts. In TSRT-based methods, it is expected that 1–3 nontemplated bases will be added to the 3′ end of the cDNA. During alignment, these are generally marked as soft-clipped (that is, they are present in the read but are not part of the read’s alignment to the genome). After removal of soft-clipped bases, additional processing is necessary. Reverse transcription of capped RNA in cap-trapping and TSRT-based methods often leads to the addition of a C to the cDNA, likely templated by the m7G cap itself, which results in a G at this position in the R1 sequencing read. If this base does not match the genome during alignment, it will be soft-clipped; however, it is possible (particularly in mammalian genomes, where promoters tend to be GC-rich [Fenouil et al., 2012]) that this cap-templated base will match the genome and it is thus impossible to determine if it represents the true TSS. Both TSRexploreR (Policastro et al., 2021) and CAGEr (Haberle et al., 2015) implement probability-based stochastic removal of mapped 5′ Gs to mitigate this artifact. It is worthwhile to note that analysis of data generated by oligo-capping methods such as TSS-seq do not require G correction, as the native cap is removed prior to reverse transcription.
Data generated by various TSS mapping techniques can also be used to quantify transcript abundance. Thus, an RNA-seq-like analysis of such data can be performed, using software such as HTSeq (Anders et al., 2014) or featureCounts (Liao et al., 2019) to quantify counts at the gene level, or software such as RSEM (Li and Dewey, 2011), Salmon (Patro et al., 2017), or Kallisto (Bray et al., 2016) to estimate counts at the transcript level. For transcript quantification, it is recommended to extend the transcript sequences upstream roughly 100 to 200 base pairs (bp), given that the TSS-containing read will often be fully or partially upstream of the annotated 5′ TL, complicating analysis.
Software and analytical considerations
After preprocessing of TSS mapping data, there exists a wide breadth of tools available to explore and analyze the data (Figure 3). Because of the large number of possible analyses, this section is not intended to be comprehensive, but rather a showcase of important and interesting computational considerations. For more information, refer to the vignettes and papers for CAGEr (Haberle et al., 2015), TSRexploreR (Policastro et al., 2021), CAGEfighteR (Thodberg et al., 2019), and other tools mentioned to learn the full breadth of available analyses.
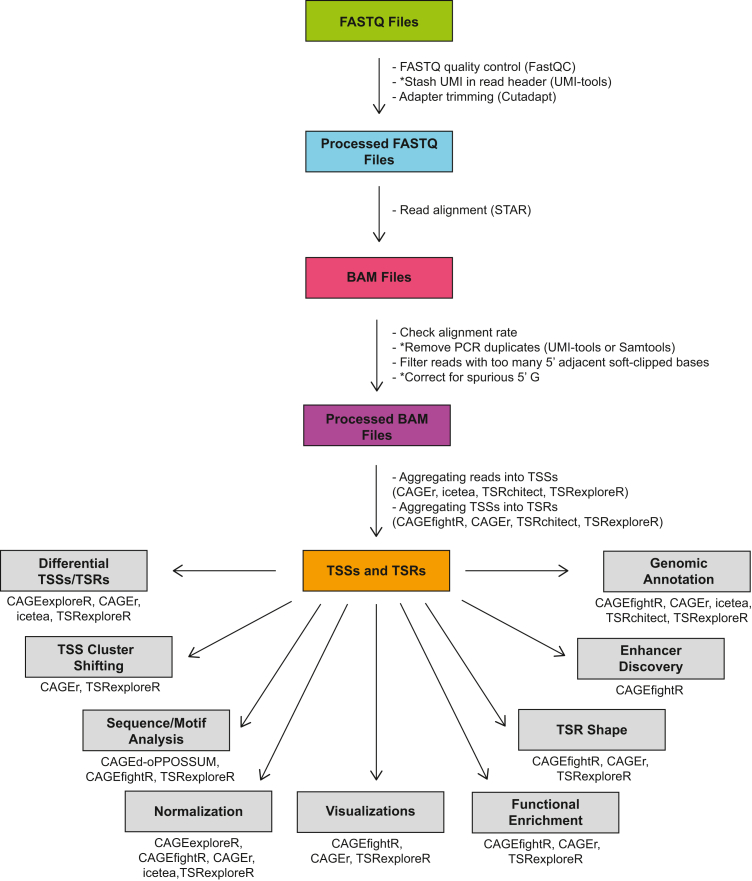
Figure 3.
Computational processing of TSS mapping data
A general workflow for processing and analysis of TSS mapping data is shown, with software that can be used for each step indicated. Asterisks indicate optional steps. More information on each piece of software listed here can be found at the following URLs: FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc); UMI-tools (https://github.com/CGATOxford/UMI-tools); Cutadapt (https://cutadapt.readthedocs.io/en/stable); STAR (https://github.com/alexdobin/STAR); Samtools (http://www.htslib.org); CAGEr (https://www.bioconductor.org/packages/release/bioc/html/CAGEr.html); icetea (https://www.bioconductor.org/packages/release/bioc/html/icetea.html); TSRchitect (https://www.bioconductor.org/packages/release/bioc/html/TSRchitect.html); TSRexploreR (https://zentnerlab.github.io/TSRexploreR/index.html); CAGEexploreR (https://github.com/edimont/CAGExploreR); CAGEd-oPPOSSUM (http://cagedop.cmmt.ubc.ca/CAGEd_oPOSSUM/); CAGEfightR (https://www.bioconductor.org/packages/release/bioc/html/CAGEfightR.html).
Calling TSSs from raw data
After processing and alignment of sequencing reads, the first step in analyzing TSS mapping data is aggregating read 5′ ends into TSS positions. Although conceptually simple, all global TSS mapping technologies suffer from spurious background reads over gene bodies, and so some form of thresholding is necessary to remove background TSS signal while retaining true TSSs. Thus, a number of thresholding approaches have been put forth. In CAGEr (Haberle et al., 2015) and CAGEfightR (Thodberg et al., 2019), read 5′ ends are first aggregated into CAGE-detected TSSs without thresholding and normalized (discussed below). In CAGEr, TSS filtering is performed during clustering, wherein the user specifies the number of samples that must have at least n normalized counts at a given TSS position for it to be considered for TSS clustering. Similarly, CAGEfightR allows users to discard TSSs not meeting the above-described sample number and count thresholds prior to clustering. In TSRexploreR, a genome annotation is used to determine the fraction of TSSs within a specified distance from an annotated TSS, as well as the number of features (genes or transcripts) with at least one unique TSS position (Policastro et al., 2021). We found that the promoter proximal TSS fraction increases markedly as the threshold is increased from a single read, likely indicating loss of weak TSSs within gene bodies. This gain in promoter proximal TSS fraction levels off as the threshold increases, and the number of features with a unique TSS decreases as weaker promoter proximal TSSs are progressively eliminated. We therefore suggest selecting a threshold within the inflection point of the promoter proximal fraction curve to balance removal of likely artifacts with retention of weak true TSSs. We found that, in STRIPE-seq data from yeast and human cells, a threshold of three raw counts per TSS is a suitable threshold for TSS retention (Policastro et al., 2020).
Normalization
An important step in the analysis of TSS mapping data is normalization. CAGEr and CAGEfighteR utilize transcripts per million normalization (Haberle et al., 2015; Thodberg et al., 2019), and CAGEr additionally allows for power-law normalization due to CAGE datasets being previously shown to follow a power-law distribution (Balwierz et al., 2009). TSRexploreR, on the other hand, utilizes either counts per million or DESeq2/edgeR normalization (Policastro et al., 2021). There are two important considerations for TSS normalization: the number of total sequenced reads, and compositional bias. The total number of sequenced reads (or library size) affects read quantification because of variations in counts per feature and the chance for lowly expressed features to drop out at lower sequencing depths. Compositional bias is a consequence of read counts being relative and not absolute values. At a fixed library size, as the number of reads captured for a set of genes increases, fewer reads are available to capture other sets of genes, which can give a false impression of altered expression between conditions. These considerations are similar to those experienced in bulk RNA-seq analysis, so software such as DESeq2 (Love et al., 2014) or edgeR (Robinson et al., 2010) can be used to correct for them. DESeq2 uses a geometric mean approach and edgeR uses the trimmed mean of M-values approach (Robinson and Oshlack, 2010), both of which will correct for library sequencing depth and compositional bias.
Clustering TSSs into TSRs
A number of different methods for clustering TSSs into TSRs have been developed. The simplest approach, here referred to as naive distance clustering, is available in tools such as CAGEr, TSRexploreR, TSRchitect (Raborn et al., 2017), and CAGEfightR (Thodberg et al., 2019). In this approach, TSSs meeting a specific score threshold and within a specified distance of one another (commonly 20–25 bp) are clustered into TSRs. CAGEr additionally offers a parametric clustering (PARACLU) algorithm that aims to find regions within chromosomes of maximal local TSS density (Frith et al., 2008). Reproducible clustering (RECLU) is a further iteration of PARACLU introduced by the functional annotation of the mammalian genome (FANTOM) consortium that incorporated a number of improvements, including irreproducible discovery rate analysis to enable assessment of TSR reproducibility (Ohmiya et al., 2014). Hypergeometric Optimization of Motif EnRichment (HOMER) employs a chromatin immunoprecipitation sequencing (ChIP-seq)-like clustering algorithm to cluster densities of TSSs, and can optionally perform additional quality control and filtering steps by using an available genome annotation (Duttke et al., 2019). Integrating Cap Enrichment with Transcript Expression Analysis (icetea), the software accompanying MAPCap, calls peaks with a ChIP-seq-like window-based method utilizing a negative binomial distribution (Bhardwaj et al., 2019). ADAPT-CAGE adopted a machine learning approach by using support vector machines and stochastic gradient boosting, whereby DNA shape features, motifs, and CAGE signal are used to discern strong putative TSSs from background (Georgakilas et al., 2020).
Differential TSSs and TSRs
Differential TSS and TSR state between conditions has received much interest because of the implications for transcript expression and isoform behavior. CAGEexploreR utilizes a user-supplied set of promoters to find differential promoter usage (Dimont et al., 2014), and a classification approach has been proposed to find differential TSS distributions (shape changes) between conditions (Liang et al., 2014). CAGEr and TSRexploreR use a more traditional approach, employing either edgeR or DESeq2 for differential TSS or TSR analysis (Haberle et al., 2015; Policastro et al., 2021).
TSS cluster shifting
It is increasingly clear that large-scale shifts in TSS usage are commonplace during development, disease, and in response to environmental perturbations. Shifting of TSS clusters is defined as TSS density, shifting a relatively short but meaningful distance (often ∼100 bases) upstream or downstream between conditions. For instance, a comprehensive CAGE study of zebrafish development uncovered over 900 transcripts with shifted TSRs corresponding to the transition between maternal and zygotic gene expression (Haberle et al., 2014). Shifting of TSSs at thousands of genes is also seen in budding yeast strains bearing various mutations in RNAPII and general transcription factors (Qiu et al., 2020), and also during certain meiotic and mitotic cell cycle stages (Chia et al., 2021).
Because of the emerging importance of TSS clustering shifting, various computational methods have been developed to detect this phenomenon. Generally, such approaches merge TSRs within a given distance to generate a set of consensus regions in which shifting will be assessed. As the distribution of TSSs within a TSR is essentially a discrete probability distribution, computational detection of TSS shifts might be approached as testing for differences between two such distributions. CAGEr introduced the first such method, which performs two distinct calculations. First, a shift score is calculated based on the maximal difference between the empirical cumulative distribution functions of the two TSS distributions. The magnitude of the shift score is reported to reflect the degree to which the TSS signal in the compared distributions is nonoverlapping (e.g., a CAGEr shift score of 0.4 indicates that 40% of the initiation between the two samples does not overlap). Second, a two-sample Kolmogorov-Smirnov (KS) test is performed to test for a significant difference between distributions. The KS test allows detection of shifts in TSS “mass” within a given TSR when the positions of initiation are largely or completely overlapping. Although this approach has been used to great effect in detecting TSS shifts in various contexts, there are limitations. The shift score does not capture shifts in TSS distribution that take place in largely overlapping positions, and its scale is somewhat unintuitive, ranging from negative infinity to 1 with a sign that does not reflect the directionality of the shift. Calculation of the KS test is also independent of the shift score, and the discrete nature of TSS distributions violates the KS test assumption of continuous distributions.
To address these limitations, we implemented an alternative approach based on the earth mover’s distance (EMD) that we termed earth mover’s score (EMS) Policastro et al., 2021. EMS has an intuitive scale that spans from −1 to 1: in accordance with conventions for denoting sequence positions in relation to a reference point, a negative EMS indicates an upstream shift and a positive EMS indicates a downstream shift. Furthermore, the p value is computed directly from the test statistic by using a permutation test, which facilitates agreement between EMS and p value. One limitation of the EMS is that a very small score, with attendant lack of significance, could mask “balanced” shifts, meaning expansion or contraction of a TSS cluster in which the movement of TSS density is symmetrical in both directions. To capture these cases, we also report standard unsigned EMD, which reports the overall difference between two distributions without regard to direction, and a corresponding permuted p value and false discovery rate threshold. EMD spans from 0 to 1, with 0 indicating identical TSS positions and 1 indicating no overlap of TSS positions. Balanced shifts are marked by an EMS score near 0, often without a significant p value, and a significant EMD with high magnitude. It is possible that these balanced shifts could be related to changes in peak shape (broad versus peaked), so more exploration of this phenomenon is required.
TSR shape
Early global studies found that TSRs can often be functionally classified by their overall shape. The first detailed global study of TSR shape generated human and mouse CAGE data for the FANTOM consortium (Carninci et al., 2006). In this paper, TSRs were classified into one of four shape categories (single dominant peak [SP], broad [BP], multimodal [MU], or broad with dominant peak [PB]) on the basis of the interquartile range (the distance between the TSS positions encompassing specified upper and lower quantiles of a TSR’s signal) and distance between TSSs. They found that TATA box-containing promoters tended to have more peaked TSS distributions, whereas TATA-less, CpG-rich promoters had broader TSS distributions on average. A study of Drosophila melanogaster TSRs derived from EST data used a simpler classification scheme wherein TSRs with a single TSS were considered peaked and those with more than one were annotated as broad (Rach et al., 2009). In addition to the TATA box, peaked promoters were also associated with the Initiator element (Inr), Downstream Promoter Element (DPE), and the Motif Ten Element (MTE). In contrast, broad promoters were enriched for the DNA Replication Element (DRE), and the Ohler 1, 6, and 7 elements. The original PEAT paper introduced another classification strategy wherein a smoothed density estimate was fit to each TSR followed by condensation of each TSR to the shortest width containing 95% of its constituent reads and classification into three patterns (narrow with peak, broad with peak [BP], or weak peak) on the basis of the width of the smoothed TSS density (Ni et al., 2010). This classification scheme yielded results similar to those of Rach et al. (2009) in terms of motif enrichment in peaked versus broad promoters.
The most popular contemporary TSR shape measurement is perhaps the shape index (SI). SI quantifies the number of TSSs at each position within a TSR. The scale of SI ranges from negative infinity to one, with scores > −1 classified as peaked and ≤ −1 as broad (Hoskins et al., 2011). SI is a more robust measure of TSR shape than TSR width, as it is not sensitive to low-scoring outlier TSSs. Furthermore, the continuous nature of SI allowed description of a continuum of TSR shapes. Application of SI to Drosophila CAGE data revealed many of the same motifs as Rach et al., 2009 and Ni et al. (2010), but additionally found the Pause Button and GAGA motifs enriched in peaked promoters and the NDM1 and DMv1 motifs in broad promoters. Furthermore, it was found that genes with broad promoters tended to be constitutively expressed through development, whereas peaked promoters tended to be activated at certain times during development Hoskins et al., 2011. TSRchitect (Raborn et al., 2021) introduced the Modified Shape Index, which ranges from −1 to 1 rather than negative infinity to 1. Another approach devised a two-step clustering method that utilized a dissimilarity metric called generalized minimum distance of distributions (GM-distance) which is a modified form of minimum distance of pair assignments, and a peakedness score (Zhao et al., 2011). Using this strategy, three TSR cluster shapes emerged: scattered, dense, and ultradense, with most dense and ultradense TSRs corresponding to the SP class in Carninci et al., 2006, and the scattered TSR corresponding to BP, MU, and PB. In addition to motif findings similar to Carninci et al., 2006, exploration of various ChIP-seq datasets showed interesting correlations such as H3K4 methylation, H2A.Z, and H3K79me3 being more associated with scattered promoters, and H3K27me3, H3K9me3, and DNA methylation being more associated with dense and ultradense promoters.
Future prospects
Single-cell TSS profiling
The past several years have seen a rapid proliferation of techniques for measuring transcript levels (Ramsköld et al., 2012; Tang et al., 2009), DNA methylation (Guo et al., 2013), chromosome conformation (Nagano et al., 2013), chromatin accessibility (Buenrostro et al., 2015; Cusanovich et al., 2015), and protein-DNA interactions (Bartosovic et al., 2021; Grosselin et al., 2019; Rotem et al., 2015; Wu et al., 2021) in single cells. However, little work has been done on cell-to-cell variation in TSS usage: to our knowledge, only two such studies have been performed. Tn5Prime (Cole et al., 2018) uses TSRT of RNA from a single lysed cell followed by tagmentation with Tn5 for library construction. In the second study, nanoCAGE (2017) was combined with the C1 microfluidic platform to yield C1 CAGE (Kouno et al., 2019). The resulting analysis of TSS usage in 136 single cells revealed heterogeneity in the transcriptional response of cells to transforming growth factor (TGF)-β as well as unidirectional enhancer transcription in each cell, suggesting that the bidirectional transcription often observed at enhancers is a result of sampling a population of cells.
Moving forward, how else might single-cell TSS profiling be performed? A number of scRNA-seq approaches use TSRT, already in wide use as a means for TSS mapping. Smart-seq3 (and its predecessors) (Hagemann-Jensen et al., 2020) and Single-cell Tagged Reverse Transcription (STRT) (Islam et al., 2011) use TSRT on single isolated cells, and STRT has been tested for TSS profiling from bulk RNA (Adiconis et al., 2018). The 10X Genomics Chromium microfluidic platform uses TSRT for 5′-centric gene expression profiling, suggesting that a combination of cell isolation and TSRT on the Chromium instrument with a TSS-focused library preparation protocol (e.g., STRIPE-seq) could yield single-cell TSS maps. Further development of such approaches will undoubtedly yield further insight into cell-to-cell variability in gene regulation.
Long-read sequencing
Although short-read sequencing (e.g., on Illumina platforms) is the standard readout for functional genomics methods, long-read sequencing technologies (Oxford NanoPore Technologies (ONT) and Pacific Biosciences Single-Molecule Real-Time (SMRT) sequencing) have gained popularity for applications such as improving existing genome assemblies and assessing structural variation in chromosomes (Logsdon et al., 2020). ONT and SMRT have also been used to determine full-length transcript sequences via cDNA sequencing (Byrne et al., 2017; Sharon et al., 2013), and ONT is capable of direct RNA sequencing for both transcriptome profiling (Garalde et al., 2018; Workman et al., 2019) and detection of modified bases (Leger et al., 2019; Liu et al., 2019). For TSS analysis, long-read sequencing would be advantageous in that a single read would contain both a transcript's TSS and complete coding sequencing, enabling one-to-one assignment of a TSS to its corresponding transcript. However, it has been observed that ONT direct RNA-seq reads, originating from a transcript's 3′ end, are often truncated before the transcript's true TSS (Workman et al., 2019). This might arise from electrical abnormalities because of enzyme stalling during RNA translocation or voltage spikes of unknown origin. Extremely rapid translocation of the 5′-most 10–15 nucleotides of a transcript though a pore also prevents reading of these terminal nucleotides. Thus, dedicated methods are still required to confidently detect TSSs when ONT direct RNA-seq is performed.
Concluding remarks
Despite the development of a wide variety of techniques for global TSS mapping over the past few decades, such methods are integrated into global studies of gene expression far less frequently than approaches for quantifying transcript levels (e.g., RNA-seq) or mapping protein-DNA interactions (e.g., ChIP-seq, CUT&RUN). Given that heterogeneity in TSS usage is a major driver of transcript isoform diversity (Reyes and Huber, 2018; Shabalina et al., 2014), likely plays important roles in development (Cvetesic et al., 2020; Haberle et al., 2014), is involved in the response to environmental stimuli (Kurihara et al., 2018; Lu and Lin, 2019; Ushijima et al., 2017), and is altered in cancer (Demircioğlu et al., 2019), we argue that TSS mapping techniques can provide insights into gene regulation complementary or inaccessible to those obtained with other more commonly used techniques. Indeed, CAGE has been extensively used alongside methods such as RNA-seq and ChIP-seq in the context of large consortia such as FANTOM (Forrest et al., 2014) and encyclopedia of DNA elements (ENCODE) (ENCODE Project Consortium, 2012) groups, where it has provided great insight into promoter-level gene regulation. Furthermore, many TSS mapping techniques can also provide information on transcript levels comparable to those obtained with various RNA-seq approaches, increasing the cost efficiency of each experiment. In some cases, widespread adoption of TSS mapping techniques might have been hampered by barriers of cost and/or technical difficulty. For instance, we calculated the per-sample cost of nAnT-iCAGE and SLIC-CAGE to be > $100 USD, with protocols spanning multiple days (Policastro et al., 2020), whereas a commercial nAnT-iCAGE kit (https://cage-seq.com/cage_kit/index.html) has a cost of 25,000 JPY (∼$230 USD) per sample. However, these methods are currently regarded as the gold standards for TSS mapping thanks to their sensitivity, resolution, and low bias (Cvetesic et al., 2018) and so are preferred for applications in which high sensitivity is essential. More routine profiling of TSS usage can easily be performed with TSRT-based methods, which trade a moderate degree of sensitivity for reduced cost and simpler, faster protocols (Policastro et al., 2020). Given the range of methods available, we surmise that any researchers interested in profiling transcription initiation will be able to find a method that suits their needs. To facilitate methodological comparisons, we provide Table 1, which outlines the general advantages and disadvantages associated with each of the three enzymatic approaches discussed here as well as salient features of specific techniques, and Table 2, which lists reported RNA input amounts for each protocol.
Table 1.
Advantages and disadvantages of TSS mapping approaches
| Enzymatic approach | Methods | General comments | Method-specific features |
|---|---|---|---|
| Oligo-capping | TSS-seq, PEAT, CapSeq, TL-seq, TIF-seq, Start-seq, SMORE-seq | Removal of m7G cap prior to reverse transcription reduces prevalence of the 5′ G artifact, thus providing high TSS specificity. However, oligo-capping methods generally have high total RNA input requirements (see Table 2), complex protocols, and may suffer from the sequence biases of ligases used to attach oligo caps. |
|
| Cap-trapping | nAnT-iCAGE, SLIC-CAGE, MAPCap | Oligo-capping methods generally have lower input requirements than cap-trapping methods (see Table 2) and provide high spatial resolution and sensitivity but suffer from the 5′ G artifact due to reverse transcription of capped RNA. Cap-trapping-based protocols are relatively complex and can be expensive. |
|
| Template-switching reverse transcription | nanoCAGE-XLa, nanoCAGE 2017a, RAMPAGEa Tn5Prime, nanoPAREa, STRIPE-seq |
TSRT-based approaches generally have the lowest input requirements of all TSS mapping methods (SLIC-CAGE excepted, see Table 2). Their protocols tend to be simpler than those of oligo-capping and cap-trapping methods. NanoCAGE 2017, Tn5Prime, and nanoPARE use Tn5 tagmentation for library preparation, while STRIPE-seq uses stringent bead purifications to optimize library size distribution. These methods may suffer from reduced sensitivity in complex transcriptomes and are susceptible to the 5′ G artifact. In addition, several TSRT-based methods use custom sequencing primers, complicating pooling with other types of libraries. |
|
Indicates that a custom sequencing primer is required for libraries of this type.
Table 2.
Reported input requirements of TSS mapping methods
| Method | Enzymatic approach | Reported inputs |
|---|---|---|
| TSS-seq | Oligo-capping | 200 μg total RNA (Yamashita et al., 2011); 500 ng poly(A)+ RNA (Malabat et al., 2015) |
| PEAT | Oligo-capping | 1–2 μg poly(A)+ RNA (Ni et al., 2010); 30 μg total RNA (Morton et al., 2014) |
| CapSeq | Oligo-capping | 500 ng–2 μg total RNA (Gu et al., 2012) |
| TL-seq | Oligo-capping | 1 μg poly(A)+ RNA (Arribere and Gilbert, 2013) |
| TIF-seq | Oligo-capping | 60 μg total RNA (Pelechano et al., 2013) |
| SMORE-seq | Oligo-capping | 500 ng poly(A)+ RNA (Park et al., 2014) |
| nAnT-iCAGE | Cap-trapping | 5 μg total RNA (Murata et al., 2014) |
| SLIC-CAGE | Cap-trapping | 1–100 ng total RNA brought up to 5 μg with carrier (Cvetesic et al., 2018) |
| MAPCap | Cap-trapping | 100 ng–5 μg total RNA (Bhardwaj et al., 2019) |
| nanoCAGE-XL | Template-switching reverse transcription | 200 ng rRNA-depleted RNA (Cumbie et al., 2015); 7.5 μg total RNA (Adiconis et al., 2018) |
| nanoCAGE 2017 | Template-switching reverse transcription | 50–500 ng total RNA (Poulain et al., 2017); Single cell (C1 CAGE [Kouno et al., 2019]) |
| RAMPAGE | Template-switching reverse transcription/cap-trapping | 5 μg total RNA (Batut et al., 2013) |
| Tn5Prime | Template-switching reverse transcription | Single cell – 5 ng total RNA (Cole et al., 2018) |
| nanoPARE | Template-switching reverse transcription | 10 pg (single-cell equivalent) – 5 ng total RNA (Schon et al., 2018) |
| STRIPE-seq | Template-switching reverse transcription | 50–250 ng total RNA (Policastro et al., 2020) |
Acknowledgments
Work in the Zentner Lab was supported by NIH grant R35GM128631 to G.E.Z.
Declaration of interests
R.A.P. and G.E.Z. are employees of eGenesis, Inc. The topics covered in this review are not related to any current work at the company.
References
- Adiconis X., Haber A.L., Simmons S.K., Levy Moonshine A., Ji Z., Busby M.A., Shi X., Jacques J., Lancaster M.A., Pan J.Q., et al. Comprehensive comparative analysis of 5′-end RNA-sequencing methods. Nat. Methods. 2018;15:505–511. doi: 10.1038/s41592-018-0014-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P.T., Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2014;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arribere J.A., Gilbert W.V. Roles for transcript leaders in translation and mRNA decay revealed by transcript leader sequencing. Genome Res. 2013;23:977–987. doi: 10.1101/gr.150342.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldrich P., Tamim S., Mathioni S., Meyers B. Ligation Bias Is a Major Contributor to Nonstoichiometric Abundances of Secondary siRNAs and Impacts Analyses of microRNAs. BioRxiv. 2020 doi: 10.1101/2020.09.14.296616. 2020.09.14.296616. [DOI] [Google Scholar]
- Balwierz P.J., Carninci P., Daub C.O., Kawai J., Hayashizaki Y., Van Belle W., Beisel C., van Nimwegen E. Methods for analyzing deep sequencing expression data: constructing the human and mouse promoterome with deepCAGE data. Genome Biol. 2009;10:R79. doi: 10.1186/gb-2009-10-7-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa C., Peixeiro I., Romão L. Gene expression regulation by upstream open reading frames and human disease. PLoS Genet. 2013;9:e1003529. doi: 10.1371/journal.pgen.1003529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartosovic M., Kabbe M., Castelo-Branco G. Single-cell CUT&Tag profiles histone modifications and transcription factors in complex tissues. Nat. Biotechnol. 2021;39:825–835. doi: 10.1038/s41587-021-00869-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batut P., Dobin A., Plessy C., Carninci P., Gingeras T.R. High-fidelity promoter profiling reveals widespread alternative promoter usage and transposon-driven developmental gene expression. Genome Res. 2013;23:169–180. doi: 10.1101/gr.139618.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj V., Semplicio G., Erdogdu N.U., Manke T., Akhtar A. MAPCap allows high-resolution detection and differential expression analysis of transcription start sites. Nat. Commun. 2019;10:3219. doi: 10.1038/s41467-019-11115-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower M.D., Jambhekar A., Schwarz D.S., Toombs J.A. Combining different mRNA capture methods to analyze the transcriptome: analysis of the Xenopus laevis transcriptome. PLoS One. 2013;8:e77700. doi: 10.1371/journal.pone.0077700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray N.L., Pimentel H., Melsted P., Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- Buenrostro J.D., Wu B., Litzenburger U.M., Ruff D., Gonzales M.L., Snyder M.P., Chang H.Y., Greenleaf W.J. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature. 2015;523:486–490. doi: 10.1038/nature14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne A., Beaudin A.E., Olsen H.E., Jain M., Cole C., Palmer T., DuBois R.M., Forsberg E.C., Akeson M., Vollmers C. Nanopore long-read RNAseq reveals widespread transcriptional variation among the surface receptors of individual B cells. Nat. Commun. 2017;8:16027. doi: 10.1038/ncomms16027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H., Luse D.S. Transcription initiation by RNA polymerase II in vitro. Properties of preinitiation, initiation, and elongation complexes. J. Biol. Chem. 1987;262:298–304. [PubMed] [Google Scholar]
- Carninci P., Kvam C., Kitamura A., Ohsumi T., Okazaki Y., Itoh M., Kamiya M., Shibata K., Sasaki N., Izawa M., et al. High-efficiency full-length cDNA cloning by biotinylated CAP trapper. Genomics. 1996;37:327–336. doi: 10.1006/geno.1996.0567. [DOI] [PubMed] [Google Scholar]
- Carninci P., Sandelin A., Lenhard B., Katayama S., Shimokawa K., Ponjavic J., Semple C.A.M., Taylor M.S., Engström P.G., Frith M.C., et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- Chen J., Tresenrider A., Chia M., McSwiggen D.T., Spedale G., Jorgensen V., Liao H., van Werven F.J., Ünal E. Kinetochore inactivation by expression of a repressive mRNA. Elife. 2017;6:e27417. doi: 10.7554/eLife.27417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia M., Tresenrider A., Chen J., Spedale G., Jorgensen V., Ünal E., van Werven F.J. Transcription of a 5’ extended mRNA isoform directs dynamic chromatin changes and interference of a downstream promoter. Elife. 2017;6:e27420. doi: 10.7554/eLife.27420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia M., Li C., Marques S., Pelechano V., Luscombe N.M., van Werven F.J. High-resolution analysis of cell-state transitions in yeast suggests widespread transcriptional tuning by alternative starts. Genome Biol. 2021;22:34. doi: 10.1186/s13059-020-02245-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y.H., Hagedorn C.H. Purifying mRNAs with a high-affinity eIF4E mutant identifies the short 3′ poly(A) end phenotype. Proc. Natl. Acad. Sci. 2003;100:7033–7038. doi: 10.1073/pnas.1232347100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole C., Byrne A., Beaudin A.E., Forsberg E.C., Vollmers C. Tn5Prime, a Tn5 based 5′ capture method for single cell RNA-seq. Nucleic Acids Res. 2018;46:e62. doi: 10.1093/nar/gky182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core L.J., Martins A.L., Danko C.G., Waters C.T., Siepel A., Lis J.T. Analysis of nascent RNA identifies a unified architecture of initiation regions at mammalian promoters and enhancers. Nat. Genet. 2014;46:1311–1320. doi: 10.1038/ng.3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumbie J.S., Ivanchenko M.G., Megraw M. NanoCAGE-XL and CapFilter: an approach to genome wide identification of high confidence transcription start sites. BMC Genomics. 2015;16:597. doi: 10.1186/s12864-015-1670-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusanovich D.A., Daza R., Adey A., Pliner H., Christiansen L., Gunderson K.L., Steemers F.J., Trapnell C., Shendure J. Multiplex single cell profiling of chromatin accessibility by combinatorial cellular indexing. Science. 2015;348:910–914. doi: 10.1126/science.aab1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvetesic N., Borkowska M., Hatanaka Y., Yu C., Vincent S.D., Müller F., Tora L., Leitch H.G., Hajkova P., Lenhard B. Global regulatory transitions at core promoters demarcate the mammalian germline cycle. BioRxiv. 2020 doi: 10.1101/2020.10.30.361865. [DOI] [Google Scholar]
- Cvetesic N., Leitch H.G., Borkowska M., Müller F., Carninci P., Hajkova P., Lenhard B. SLIC-CAGE: high-resolution transcription start site mapping using nanogram-levels of total RNA. Genome Res. 2018;28:1943–1956. doi: 10.1101/gr.235937.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danks G.B., Navratilova P., Lenhard B., Thompson E.M. Distinct core promoter codes drive transcription initiation at key developmental transitions in a marine chordate. BMC Genomics. 2018;19:164. doi: 10.1186/s12864-018-4504-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demircioğlu D., Cukuroglu E., Kindermans M., Nandi T., Calabrese C., Fonseca N.A., Kahles A., Lehmann K.-V., Stegle O., Brazma A., et al. A pan-cancer transcriptome analysis reveals pervasive regulation through alternative promoters. Cell. 2019;178:1465–1477.e17. doi: 10.1016/j.cell.2019.08.018. [DOI] [PubMed] [Google Scholar]
- Dieudonné F.-X., O’Connor P.B.F., Gubler-Jaquier P., Yasrebi H., Conne B., Nikolaev S., Antonarakis S., Baranov P.V., Curran J. The effect of heterogeneous Transcription Start Sites (TSS) on the translatome: implications for the mammalian cellular phenotype. BMC Genomics. 2015;16:986. doi: 10.1186/s12864-015-2179-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimont E., Hofmann O., Ho Sui S.J., Forrest A.R.R., Kawaji H., Hide W., the FANTOM Consortium CAGExploreR: an R package for the analysis and visualization of promoter dynamics across multiple experiments. Bioinformatics. 2014;30:1183–1184. doi: 10.1093/bioinformatics/btu125. [DOI] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duttke S.H., Chang M.W., Heinz S., Benner C. Identification and dynamic quantification of regulatory elements using total RNA. Genome Res. 2019;29:1836–1846. doi: 10.1101/gr.253492.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenouil R., Cauchy P., Koch F., Descostes N., Cabeza J.Z., Innocenti C., Ferrier P., Spicuglia S., Gut M., Gut I., et al. CpG islands and GC content dictate nucleosome depletion in a transcription-independent manner at mammalian promoters. Genome Res. 2012;22:2399–2408. doi: 10.1101/gr.138776.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrest A.R.R., Kawaji H., Rehli M., Kenneth Baillie J., de Hoon M.J.L., Haberle V., Lassmann T., Kulakovskiy I.V., Lizio M., Itoh M., et al. A promoter-level mammalian expression atlas. Nature. 2014;507:462–470. doi: 10.1038/nature13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frith M.C., Valen E., Krogh A., Hayashizaki Y., Carninci P., Sandelin A. A code for transcription initiation in mammalian genomes. Genome Res. 2008;18:1–12. doi: 10.1101/gr.6831208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs R.T., Sun Z., Zhuang F., Robb G.B. Bias in ligation-based small RNA sequencing library construction is determined by adaptor and RNA structure. PLoS One. 2015;10:e0126049. doi: 10.1371/journal.pone.0126049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garalde D.R., Snell E.A., Jachimowicz D., Sipos B., Lloyd J.H., Bruce M., Pantic N., Admassu T., James P., Warland A., et al. Highly parallel direct RNA sequencing on an array of nanopores. Nat. Methods. 2018;15:201–206. doi: 10.1038/nmeth.4577. [DOI] [PubMed] [Google Scholar]
- Georgakilas G.K., Perdikopanis N., Hatzigeorgiou A. Solving the transcription start site identification problem with ADAPT-CAGE: a Machine Learning algorithm for the analysis of CAGE data. Sci. Rep. 2020;10:877. doi: 10.1038/s41598-020-57811-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowthaman U., García-Pichardo D., Jin Y., Schwarz I., Marquardt S. DNA processing in the context of noncoding transcription. Trends Biochem. Sci. 2020;45:1009–1021. doi: 10.1016/j.tibs.2020.07.009. [DOI] [PubMed] [Google Scholar]
- Grosselin K., Durand A., Marsolier J., Poitou A., Marangoni E., Nemati F., Dahmani A., Lameiras S., Reyal F., Frenoy O., et al. High-throughput single-cell ChIP-seq identifies heterogeneity of chromatin states in breast cancer. Nat. Genet. 2019;51:1060–1066. doi: 10.1038/s41588-019-0424-9. [DOI] [PubMed] [Google Scholar]
- Gu W., Lee H.-C., Chaves D., Youngman E.M., Pazour G.J., Conte D., Mello C.C. CapSeq and CIP-TAP identify pol II start sites and reveal capped small RNAs as C. elegans piRNA precursors. Cell. 2012;151:1488–1500. doi: 10.1016/j.cell.2012.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Zhu P., Wu X., Li X., Wen L., Tang F. Single-cell methylome landscapes of mouse embryonic stem cells and early embryos analyzed using reduced representation bisulfite sequencing. Genome Res. 2013;23:2126–2135. doi: 10.1101/gr.161679.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberle V., Li N., Hadzhiev Y., Plessy C., Previti C., Nepal C., Gehrig J., Dong X., Akalin A., Suzuki A.M., et al. Two independent transcription initiation codes overlap on vertebrate core promoters. Nature. 2014;507:381–385. doi: 10.1038/nature12974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberle V., Forrest A.R.R., Hayashizaki Y., Carninci P., Lenhard B. CAGEr: precise TSS data retrieval and high-resolution promoterome mining for integrative analyses. Nucleic Acids Res. 2015;43:e51. doi: 10.1093/nar/gkv054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M., Renwick N., Brown M., Mihailović A., Holoch D., Lin C., Pena J.T.G., Nusbaum J.D., Morozov P., Ludwig J., et al. RNA-ligase-dependent biases in miRNA representation in deep-sequenced small RNA cDNA libraries. RNA. 2011;17:1697–1712. doi: 10.1261/rna.2799511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemann-Jensen M., Ziegenhain C., Chen P., Ramsköld D., Hendriks G.-J., Larsson A.J.M., Faridani O.R., Sandberg R. Single-cell RNA counting at allele and isoform resolution using Smart-seq3. Nat. Biotechnol. 2020;38:708–714. doi: 10.1038/s41587-020-0497-0. [DOI] [PubMed] [Google Scholar]
- Hashimoto S., Suzuki Y., Kasai Y., Morohoshi K., Yamada T., Sese J., Morishita S., Sugano S., Matsushima K. 5′-end SAGE for the analysis of transcriptional start sites. Nat. Biotechnol. 2004;22:1146–1149. doi: 10.1038/nbt998. [DOI] [PubMed] [Google Scholar]
- Hirabayashi S., Bhagat S., Matsuki Y., Takegami Y., Uehata T., Kanemaru A., Itoh M., Shirakawa K., Takaori-Kondo A., Takeuchi O., et al. NET-CAGE characterizes the dynamics and topology of human transcribed cis -regulatory elements. Nat. Genet. 2019;51:1369–1379. doi: 10.1038/s41588-019-0485-9. [DOI] [PubMed] [Google Scholar]
- Hollerer I., Barker J.C., Jorgensen V., Tresenrider A., Dugast-Darzacq C., Chan L.Y., Darzacq X., Tjian R., Ünal E., Brar G.A. Evidence for an integrated gene repression mechanism based on mRNA isoform toggling in human cells. G3 GenesGenomesGenetics. 2019;9:1045–1053. doi: 10.1534/g3.118.200802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins R.A., Landolin J.M., Brown J.B., Sandler J.E., Takahashi H., Lassmann T., Yu C., Booth B.W., Zhang D., Wan K.H., et al. Genome-wide analysis of promoter architecture in Drosophila melanogaster. Genome Res. 2011;21:182–192. doi: 10.1101/gr.112466.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam S., Kjällquist U., Moliner A., Zajac P., Fan J.-B., Lönnerberg P., Linnarsson S. Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome Res. 2011;21:1160–1167. doi: 10.1101/gr.110882.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaprakash A.D., Jabado O., Brown B.D., Sachidanandam R. Identification and remediation of biases in the activity of RNA ligases in small-RNA deep sequencing. Nucleic Acids Res. 2011;39:e141. doi: 10.1093/nar/gkr693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen V., Chen J., Vander Wende H., Harris D.E., McCarthy A., Breznak S., Wong-Deyrup S.W., Chen Y., Rangan P., Brar G.A., et al. Tunable transcriptional interference at the endogenous alcohol dehydrogenase gene locus in Drosophila melanogaster. G3 GenesGenomesGenetics. 2020;10:1575–1583. doi: 10.1534/g3.119.400937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamori-Katayama M., Itoh M., Kawaji H., Lassmann T., Katayama S., Kojima M., Bertin N., Kaiho A., Ninomiya N., Daub C.O., et al. Unamplified cap analysis of gene expression on a single-molecule sequencer. Genome Res. 2011;21:1150–1159. doi: 10.1101/gr.115469.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapteyn J., He R., McDowell E.T., Gang D.R. Incorporation of non-natural nucleotides into template-switching oligonucleotides reduces background and improves cDNA synthesis from very small RNA samples. BMC Genomics. 2010;11:413. doi: 10.1186/1471-2164-11-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai J., Shinagawa A., Shibata K., Yoshino M., Itoh M., Ishii Y., Arakawa T., Hara A., Fukunishi Y., Konno H., et al. Functional annotation of a full-length mouse cDNA collection. Nature. 2001;409:685–690. doi: 10.1038/35055500. [DOI] [PubMed] [Google Scholar]
- Kazuo M., Sumio S. Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides. Gene. 1994;138:171–174. doi: 10.1016/0378-1119(94)90802-8. [DOI] [PubMed] [Google Scholar]
- Kodzius R., Kojima M., Nishiyori H., Nakamura M., Fukuda S., Tagami M., Sasaki D., Imamura K., Kai C., Harbers M., et al. CAGE: cap analysis of gene expression. Nat. Methods. 2006;3:211–222. doi: 10.1038/nmeth0306-211. [DOI] [PubMed] [Google Scholar]
- Kouno T., Moody J., Kwon A.T.-J., Shibayama Y., Kato S., Huang Y., Böttcher M., Motakis E., Mendez M., Severin J., et al. C1 CAGE detects transcription start sites and enhancer activity at single-cell resolution. Nat. Commun. 2019;10:360. doi: 10.1038/s41467-018-08126-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara Y., Makita Y., Kawashima M., Fujita T., Iwasaki S., Matsui M. Transcripts from downstream alternative transcription start sites evade uORF-mediated inhibition of gene expression in Arabidopsis. Proc. Natl. Acad. Sci. 2018;115:7831–7836. doi: 10.1073/pnas.1804971115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak H., Fuda N.J., Core L.J., Lis J.T. Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science. 2013;339:950–953. doi: 10.1126/science.1229386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leger A., Amaral P.P., Pandolfini L., Capitanchik C., Capraro F., Barbieri I., Migliori V., Luscombe N.M., Enright A.J., Tzelepis K., et al. RNA Modifications Detection by Comparative Nanopore Direct RNA Sequencing. BioRxiv. 2019:843136. doi: 10.1038/s41467-021-27393-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leppek K., Das R., Barna M. Functional 5′ UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat. Rev. Mol. Cell Biol. 2018;19:158–174. doi: 10.1038/nrm.2017.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang K., Suzuki Y., Kumagai Y., Nakai K. Analysis of changes in transcription start site distribution by a classification approach. Gene. 2014;537:29–40. doi: 10.1016/j.gene.2013.12.038. [DOI] [PubMed] [Google Scholar]
- Liao Y., Smyth G.K., Shi W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 2019;47:e47. doi: 10.1093/nar/gkz114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D., Hiron T.K., O’Callaghan C.A. Intragenic transcriptional interference regulates the human immune ligand MICA. EMBO J. 2018;37:e97138. doi: 10.15252/embj.201797138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Begik O., Lucas M.C., Ramirez J.M., Mason C.E., Wiener D., Schwartz S., Mattick J.S., Smith M.A., Novoa E.M. Accurate detection of m6A RNA modifications in native RNA sequences. Nat. Commun. 2019;10:4079. doi: 10.1038/s41467-019-11713-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logsdon G.A., Vollger M.R., Eichler E.E. Long-read human genome sequencing and its applications. Nat. Rev. Genet. 2020;21:597–614. doi: 10.1038/s41576-020-0236-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z., Lin Z. Pervasive and dynamic transcription initiation in Saccharomyces cerevisiae. Genome Res. 2019;29:1198–1210. doi: 10.1101/gr.245456.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malabat C., Feuerbach F., Ma L., Saveanu C., Jacquier A. Quality control of transcription start site selection by nonsense-mediated-mRNA decay. Elife. 2015;4:e06722. doi: 10.7554/eLife.06722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matveeva E.A., Al-Tinawi Q.M.H., Rouchka E.C., Fondufe-Mittendorf Y.N. Coupling of PARP1-mediated chromatin structural changes to transcriptional RNA polymerase II elongation and cotranscriptional splicing. Epigenetics Chromatin. 2019;12:1–18. doi: 10.1186/s13072-019-0261-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer A., di Iulio J., Maleri S., Eser U., Vierstra J., Reynolds A., Sandstrom R., Stamatoyannopoulos J.A., Churchman L.S. Native elongating transcript sequencing reveals human transcriptional activity at nucleotide resolution. Cell. 2015;161:541–554. doi: 10.1016/j.cell.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejía-Guerra M.K., Li W., Galeano N.F., Vidal M., Gray J., Doseff A.I., Grotewold E. Core promoter plasticity between maize tissues and genotypes contrasts with predominance of sharp transcription initiation sites. Plant Cell. 2015;27:3309–3320. doi: 10.1105/tpc.15.00630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton T., Petricka J., Corcoran D.L., Li S., Winter C.M., Carda A., Benfey P.N., Ohler U., Megraw M. Paired-end analysis of transcription start sites in Arabidopsis reveals plant-specific promoter signatures. Plant Cell. 2014;26:2746–2760. doi: 10.1105/tpc.114.125617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata M., Nishiyori-Sueki H., Kojima-Ishiyama M., Carninci P., Hayashizaki Y., Itoh M. In: Transcription Factor Regulatory Networks: Methods and Protocols. Miyamoto-Sato E., Ohashi H., Sasaki H., Nishikawa J., Yanagawa H., editors. Springer; 2014. Detecting expressed genes using CAGE; pp. 67–85. [DOI] [PubMed] [Google Scholar]
- Nagano T., Lubling Y., Stevens T.J., Schoenfelder S., Yaffe E., Dean W., Laue E.D., Tanay A., Fraser P. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature. 2013;502:59–64. doi: 10.1038/nature12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechaev S., Fargo D.C., dos Santos G., Liu L., Gao Y., Adelman K. Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of pol II in Drosophila. Science. 2010;327:335–338. doi: 10.1126/science.1181421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni T., Corcoran D.L., Rach E.A., Song S., Spana E.P., Gao Y., Ohler U., Zhu J. A paired-end sequencing strategy to map the complex landscape of transcription initiation. Nat. Methods. 2010;7:521–527. doi: 10.1038/nmeth.1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen M., Ard R., Leng X., Ivanov M., Kindgren P., Pelechano V., Marquardt S. Transcription-driven chromatin repression of Intragenic transcription start sites. PLoS Genet. 2019;15:e1007969. doi: 10.1371/journal.pgen.1007969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmiya H., Vitezic M., Frith M.C., Itoh M., Carninci P., Forrest A.R., Hayashizaki Y., Lassmann T., The FANTOM Consortium RECLU: a pipeline to discover reproducible transcriptional start sites and their alternative regulation using capped analysis of gene expression (CAGE) BMC Genomics. 2014;15:269. doi: 10.1186/1471-2164-15-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki Y., Furuno M., Kasukawa T., Adachi J., Bono H., Kondo S., Nikaido I., Osato N., Saito R., Suzuki H., et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- Paquette D.R., Mugridge J.S., Weinberg D.E., Gross J.D. Application of a Schizosaccharomyces pombe Edc1-fused Dcp1–Dcp2 decapping enzyme for transcription start site mapping. RNA. 2018;24:251–257. doi: 10.1261/rna.062737.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D., Morris A.R., Battenhouse A., Iyer V.R. Simultaneous mapping of transcript ends at single-nucleotide resolution and identification of widespread promoter-associated non-coding RNA governed by TATA elements. Nucleic Acids Res. 2014;42:3736–3749. doi: 10.1093/nar/gkt1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R., Duggal G., Love M.I., Irizarry R.A., Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelechano V., Wei W., Steinmetz L.M. Extensive transcriptional heterogeneity revealed by isoform profiling. Nature. 2013;497:127–131. doi: 10.1038/nature12121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plessy C., Bertin N., Takahashi H., Simone R., Salimullah M., Lassmann T., Vitezic M., Severin J., Olivarius S., Lazarevic D., et al. Linking promoters to functional transcripts in small samples with nanoCAGE and CAGEscan. Nat. Methods. 2010;7:528–534. doi: 10.1038/nmeth.1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Policastro R.A., Raborn R.T., Brendel V.P., Zentner G.E. Simple and efficient profiling of transcription initiation and transcript levels with STRIPE-seq. Genome Res. 2020;30:910–923. doi: 10.1101/gr.261545.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Policastro R.A., McDonald D.J., Brendel V.P., Zentner G.E. Flexible analysis of TSS mapping data and detection of TSS shifts with TSRexploreR. NAR Genom. Bioinform. 2021;3:lqab051. doi: 10.1093/nargab/lqab051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulain S., Kato S., Arnaud O., Morlighem J.-É., Suzuki M., Plessy C., Harbers M. In: Promoter Associated RNA: Methods and Protocols. Napoli S., editor. Springer; 2017. NanoCAGE: a method for the analysis of coding and noncoding 5′-capped transcriptomes; pp. 57–109. [Google Scholar]
- Qiu C., Jin H., Vvedenskaya I., Llenas J.A., Zhao T., Malik I., Visbisky A.M., Schwartz S.L., Cui P., Čabart P., et al. Universal promoter scanning by Pol II during transcription initiation in Saccharomyces cerevisiae. Genome Biol. 2020;21:132. doi: 10.1186/s13059-020-02040-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raborn R.T., Sridharan K., Brendel V.P. TSRchitect: promoter identification from large-scale TSS profiling data. 2017. https://dx.org/doi:10.18129/b9.bioc.TSRchitect
- Raborn R.T., Brendel V.P., Sridharan K. Bioconductor version: Release; 2021. TSRchitect: Promoter Identification from Large-Scale TSS Profiling Data; p. 3.12. [Google Scholar]
- Rach E.A., Yuan H.-Y., Majoros W.H., Tomancak P., Ohler U. Motif composition, conservation and condition-specificity of single and alternative transcription start sites in the Drosophila genome. Genome Biol. 2009;10:R73. doi: 10.1186/gb-2009-10-7-r73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsköld D., Luo S., Wang Y.-C., Li R., Deng Q., Faridani O.R., Daniels G.A., Khrebtukova I., Loring J.F., Laurent L.C., et al. Full-Length mRNA-Seq from single cell levels of RNA and individual circulating tumor cells. Nat. Biotechnol. 2012;30:777–782. doi: 10.1038/nbt.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes A., Huber W. Alternative start and termination sites of transcription drive most transcript isoform differences across human tissues. Nucleic Acids Res. 2018;46:582–592. doi: 10.1093/nar/gkx1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas-Duran M.F., Gilbert W.V. Alternative transcription start site selection leads to large differences in translation activity in yeast. RNA. 2012;18:2299–2305. doi: 10.1261/rna.035865.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotem A., Ram O., Shoresh N., Sperling R.A., Goren A., Weitz D.A., Bernstein B.E. Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state. Nat. Biotechnol. 2015;33:1165–1172. doi: 10.1038/nbt.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheidegger A., Dunn C.J., Samarakkody A., Koney N.K.-K., Perley D., Saha R.N., Nechaev S. Genome-wide RNA pol II initiation and pausing in neural progenitors of the rat. BMC Genomics. 2019;20:477. doi: 10.1186/s12864-019-5829-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt W.M., Mueller M.W. CapSelect: a highly sensitive method for 5′ CAP-dependent enrichment of full-length cDNA in PCR-mediated analysis of mRNAs. Nucleic Acids Res. 1999;27 doi: 10.1093/nar/27.21.e31. e31–i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schon M.A., Kellner M.J., Plotnikova A., Hofmann F., Nodine M.D. NanoPARE: parallel analysis of RNA 5′ ends from low-input RNA. Genome Res. 2018;28:1931–1942. doi: 10.1101/gr.239202.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabalina S.A., Ogurtsov A.Y., Spiridonov N.A., Koonin E.V. Evolution at protein ends: major contribution of alternative transcription initiation and termination to the transcriptome and proteome diversity in mammals. Nucleic Acids Res. 2014;42:7132–7144. doi: 10.1093/nar/gku342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon D., Tilgner H., Grubert F., Snyder M. A single-molecule long-read survey of the human transcriptome. Nat. Biotechnol. 2013;31:1009–1014. doi: 10.1038/nbt.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata K., Itoh M., Aizawa K., Nagaoka S., Sasaki N., Carninci P., Konno H., Akiyama J., Nishi K., Kitsunai T., et al. RIKEN integrated sequence analysis (RISA) system—384-format sequencing pipeline with 384 multicapillary sequencer. Genome Res. 2000;10:1757–1771. doi: 10.1101/gr.152600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraki T., Kondo S., Katayama S., Waki K., Kasukawa T., Kawaji H., Kodzius R., Watahiki A., Nakamura M., Arakawa T., et al. Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc. Natl. Acad. Sci. 2003;100:15776–15781. doi: 10.1073/pnas.2136655100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T.S., Heger A., Sudbery I. UMI-tools: modelling sequencing errors in Unique Molecular Identifiers to improve quantification accuracy. Genome Res. 2017;27:491–499. doi: 10.1101/gr.209601.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y., Sugano S. In: Generation of CDNA Libraries: Methods and Protocols. Ying S.-Y., editor. Humana Press); 2003. Construction of a full-length enriched and a 5′-end enriched cDNA library using the oligo-capping method; pp. 73–91. [DOI] [PubMed] [Google Scholar]
- Takahashi H., Lassmann T., Murata M., Carninci P. 5′ end–centered expression profiling using cap-analysis gene expression and next-generation sequencing. Nat. Protoc. 2012;7:542–561. doi: 10.1038/nprot.2012.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D.T.P., Plessy C., Salimullah M., Suzuki A.M., Calligaris R., Gustincich S., Carninci P. Suppression of artifacts and barcode bias in high-throughput transcriptome analyses utilizing template switching. Nucleic Acids Res. 2013;41:e44. doi: 10.1093/nar/gks1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F., Barbacioru C., Wang Y., Nordman E., Lee C., Xu N., Wang X., Bodeau J., Tuch B.B., Siddiqui A., et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- Thodberg M., Thieffry A., Vitting-Seerup K., Andersson R., Sandelin A. CAGEfightR: analysis of 5′-end data using R/Bioconductor. BMC Bioinformatics. 2019;20:487. doi: 10.1186/s12859-019-3029-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchihara K., Suzuki Y., Wakaguri H., Irie T., Tanimoto K., Hashimoto S., Matsushima K., Mizushima-Sugano J., Yamashita R., Nakai K., et al. Massive transcriptional start site analysis of human genes in hypoxia cells. Nucleic Acids Res. 2009;37:2249–2263. doi: 10.1093/nar/gkp066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turchinovich A., Surowy H., Serva A., Zapatka M., Lichter P., Burwinkel B. Capture and amplification by tailing and switching (CATS) RNA Biol. 2014;11:817–828. doi: 10.4161/rna.29304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushijima T., Hanada K., Gotoh E., Yamori W., Kodama Y., Tanaka H., Kusano M., Fukushima A., Tokizawa M., Yamamoto Y.Y., et al. Light controls protein localization through phytochrome-mediated alternative promoter selection. Cell. 2017;171:1316–1325.e12. doi: 10.1016/j.cell.2017.10.018. [DOI] [PubMed] [Google Scholar]
- Valen E., Pascarella G., Chalk A., Maeda N., Kojima M., Kawazu C., Murata M., Nishiyori H., Lazarevic D., Motti D., et al. Genome-wide detection and analysis of hippocampus core promoters using DeepCAGE. Genome Res. 2009;19:255–265. doi: 10.1101/gr.084541.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakaguri H., Yamashita R., Suzuki Y., Sugano S., Nakai K. DBTSS: database of transcription start sites, progress report 2008. Nucleic Acids Res. 2008;36:D97–D101. doi: 10.1093/nar/gkm901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Hou J., Quedenau C., Chen W. Pervasive isoform-specific translational regulation via alternative transcription start sites in mammals. Mol. Syst. Biol. 2016;12:875. doi: 10.15252/msb.20166941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber C.M., Ramachandran S., Henikoff S. Nucleosomes are context-specific, H2A.Z-modulated barriers to RNA polymerase. Mol. Cell. 2014;53:819–830. doi: 10.1016/j.molcel.2014.02.014. [DOI] [PubMed] [Google Scholar]
- Workman R.E., Tang A.D., Tang P.S., Jain M., Tyson J.R., Razaghi R., Zuzarte P.C., Gilpatrick T., Payne A., Quick J., et al. Nanopore native RNA sequencing of a human poly(A) transcriptome. Nat. Methods. 2019;16:1297–1305. doi: 10.1038/s41592-019-0617-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S.J., Furlan S.N., Mihalas A.B., Kaya-Okur H.S., Feroze A.H., Emerson S.N., Zheng Y., Carson K., Cimino P.J., Keene C.D., et al. Single-cell CUT&Tag analysis of chromatin modifications in differentiation and tumor progression. Nat. Biotechnol. 2021;39:819–824. doi: 10.1038/s41587-021-00865-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuarin J., Schibler U. Physical isolation of nascent RNA chains transcribed by RNA polymerase II: evidence for cotranscriptional splicing. Mol. Cell. Biol. 1994;14:7219–7225. doi: 10.1128/mcb.14.11.7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulf M.G., Maguire S., Humbert P., Dai N., Bei Y., Nichols N.M., Corrêa I.R., Guan S. Non-templated addition and template switching by Moloney murine leukemia virus (MMLV)-based reverse transcriptases co-occur and compete with each other. J. Biol. Chem. 2019;294:18220–18231. doi: 10.1074/jbc.RA119.010676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita R., Sathira N.P., Kanai A., Tanimoto K., Arauchi T., Tanaka Y., Hashimoto S., Sugano S., Nakai K., Suzuki Y. Genome-wide characterization of transcriptional start sites in humans by integrative transcriptome analysis. Genome Res. 2011;21:775–789. doi: 10.1101/gr.110254.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Dietrich F.S. Mapping of transcription start sites in Saccharomyces cerevisiae using 5′ SAGE. Nucleic Acids Res. 2005;33:2838–2851. doi: 10.1093/nar/gki583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Dimont E., Ha T., Swanson D.J., Hide W., Goldowitz D., the FANTOM Consortium Relatively frequent switching of transcription start sites during cerebellar development. BMC Genomics. 2017;18:461. doi: 10.1186/s12864-017-3834-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Valen E., Parker B.J., Sandelin A. Systematic clustering of transcription start site landscapes. PLoS One. 2011;6:e23409. doi: 10.1371/journal.pone.0023409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y.y., Machleder E.m., Chenchik A., Li R., Siebert P.d. Reverse transcriptase template switching: a SMART™ approach for full-length cDNA library construction. BioTechniques. 2001;30:892–897. doi: 10.2144/01304pf02. [DOI] [PubMed] [Google Scholar]