Abstract
Objectives
In early January 2021 an outbreak of nosocomial cases of coronavirus disease 2019 (COVID-19) emerged in Western France; RT-PCR tests were repeatedly negative on nasopharyngeal samples but positive on lower respiratory tract samples. Whole-genome sequencing (WGS) revealed a new variant, currently defining a novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) lineage B.1.616. In March, the WHO classified this as a ‘variant under investigation’ (VUI). We analysed the characteristics and outcomes of COVID-19 cases related to this new variant.
Methods
Clinical, virological, and radiological data were retrospectively collected from medical charts in the two hospitals involved. We enrolled those inpatients with: (a) positive SARS-CoV-2 RT-PCR on a respiratory sample, (b) seroconversion with anti-SARS-CoV-2 IgG/IgM, or (c) suggestive symptoms and typical features of COVID-19 on a chest CT scan. Cases were categorized as B.1.616, a variant of concern (VOC), or unknown.
Results
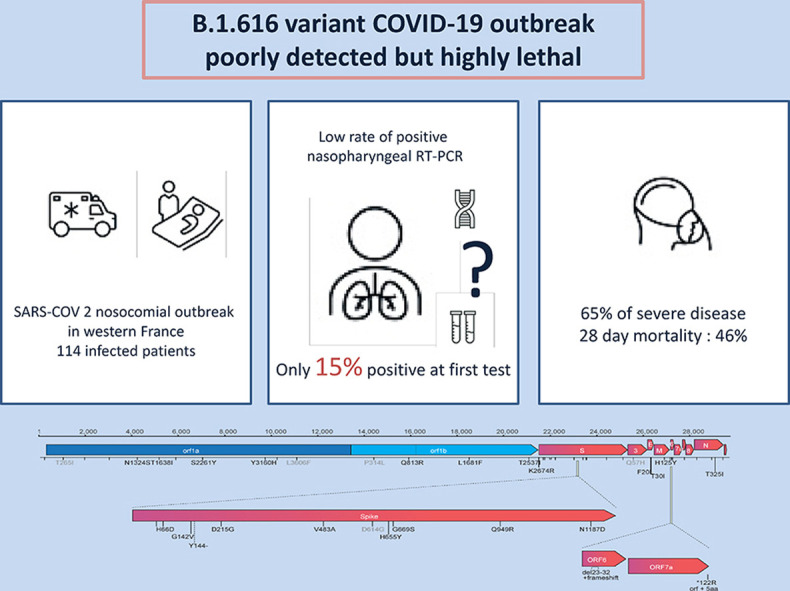
From 1st January to 24th March 2021, 114 patients fulfilled the inclusion criteria: B.1.616 (n = 39), VOC (n = 32), and unknown (n = 43). B.1.616-related cases were older than VOC-related cases (81 years, interquartile range (IQR) 73–88 versus 73 years, IQR 67–82, p < 0.05) and their first RT-PCR tests were rarely positive (6/39, 15% versus 31/32, 97%, p < 0.05). The B.1.616 variant was independently associated with severe disease (multivariable Cox model HR 4.0, 95%CI 1.5–10.9) and increased lethality (28-day mortality 18/39 (46%) for B.1.616 versus 5/32 (16%) for VOC, p = 0.006).
Conclusion
We report a nosocomial outbreak of COVID-19 cases related to a new variant, B.1.616, which is poorly detected by RT-PCR on nasopharyngeal samples and is associated with high lethality.
Keywords: Coronavirus infections/virology, Coronavirus infections/epidemiology, COVID-19, SARS-CoV-2 variants, Severity of illness index
Graphical abstract
Introduction
At the end of 2020, novel concerns were raised with the detection of rapidly spreading severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants of concern (VOCs) associated with increased transmissibility, increased severity and/or immune escape properties [[1], [2], [3], [4], [5]]. In January 2021, an outbreak of cases highly suggestive of coronavirus disease 2019 (COVID-19) despite negative RT-PCR tests on repeated nasopharyngeal samples was reported at the Lannion Hospital in Western France. Of note, when applied on lower respiratory tract samples, the performance of RT-PCR tests was preserved, suggesting that failure to detect this variant on nasopharyngeal samples was due to a viral load below the limit of detection in the upper respiratory tract, rather than to genomic mismatches between routine RT-PCR targets and this variant; this was confirmed by genomic data. Whole-genome sequencing (WGS) from lower respiratory tract samples of several cases identified a previously unknown variant of SARS-CoV-2 belonging to clade GH/20C (GISAID/Nextstrain nomenclatures) carrying several amino acid substitutions or deletions in the spike (S) protein, which received the B.1.616 Pango lineage designation. In March, the French public health agency, the National Reference Centre for respiratory viruses, and the WHO classified B.1.616 as a variant under investigation (VUI) [5,6]. We aimed to characterize this variant in terms of its virological features, clinical presentation, and outcomes.
Methods
Setting and patients
We conducted a retrospective cohort study of all patients who were diagnosed with SARS-CoV-2 infection at the Lannion and Saint-Brieuc hospitals from 1st January to 24th March 2021. SARS-CoV-2 infection was defined by at least one of the following: (a) positive SARS-CoV-2 RT-PCR on a respiratory sample, (b) seroconversion based on paired sera tested for anti-SARS-CoV-2 IgG/IgM, and (c) suggestive symptoms and typical features of COVID-19 on chest CT scan [7].
COVID-19 cases were categorized in one of the following groups: B.1.616, VOC, or unknown. The B.1.616 group included patients with B.1.616 infection documented by WGS (confirmed B.1.616 case), and patients for whom the SARS-CoV-2 isolate could not be characterized but who were close contacts of at least one patient with documented B.1.616 infection (probable B.1.616 case). Close contact was defined as household, occupational, or nosocomial (hospitalization in the same ward). Infection was considered as related to this contact when patients developed symptoms or first positive test at least 48 h after the first contact. Since all cases of B.1.616 infection confirmed by WGS lived in the Lannion district, this place was considered as the epidemic area.
The VOC group included all cases due to VOCs B.1.1.7, B.1.351 and P.1. Routine screening for these VOCs has been performed by RT-PCR targeting the N501Y mutation (common to B.1.1.7, B.1.351 and P.1) and 69–70 deletion (B.1.1.7-specific) in all patients with a positive RT-PCR after 10th February 2021. COVID-19 patients not fulfilling the criteria for the first two groups were categorized in the ‘unknown’ group. These patients were included in the study for clinical, biological, and radiological description. However, since it was not possible to assign them to either the B.1.616 group or the VOC group, they were not included in the primary analysis. Cases were categorized as nosocomial if symptoms appeared at least 2 days after hospital admission [8]. Patient management and virological methods are reported in the Supplementary Material.
Outcomes
Primary outcome was severity defined as a score >5 in the WHO clinical progression scale, which is achieved when patients require non-invasive ventilation or high-flow oxygen [9]. Secondary outcomes were ICU admission and 28 day-mortality.
Statistical analysis
Statistical analysis was performed with R 4.0.5. Categorical variables were expressed as numbers (per cent) and compared with the χ2 test and Fisher's exact test, as appropriate. Continuous variables were expressed as medians (interquartile range (IQR)) and compared with the Mann–Whitney U test or Kruskall–Wallis test, as appropriate. Kaplan–Meier survival curves were compared with the log-rank test. Multivariable analysis was made using the Cox proportional-hazard regression model. Variables associated with outcome with p < 0.2 on univariate analysis or those considered clinically relevant were included in the multivariate analysis. All tests were two-sided, and p < 0.05 was considered statistically significant.
Ethics
Patients or closest relatives were informed of the retrospective collection of data and could refuse to participate. The French infectious diseases society ethics committee (CER-MIT) approved the study (N° COVID 2021-06). Written informed consent was waived.
Results
Genomic characteristics of B.1.616
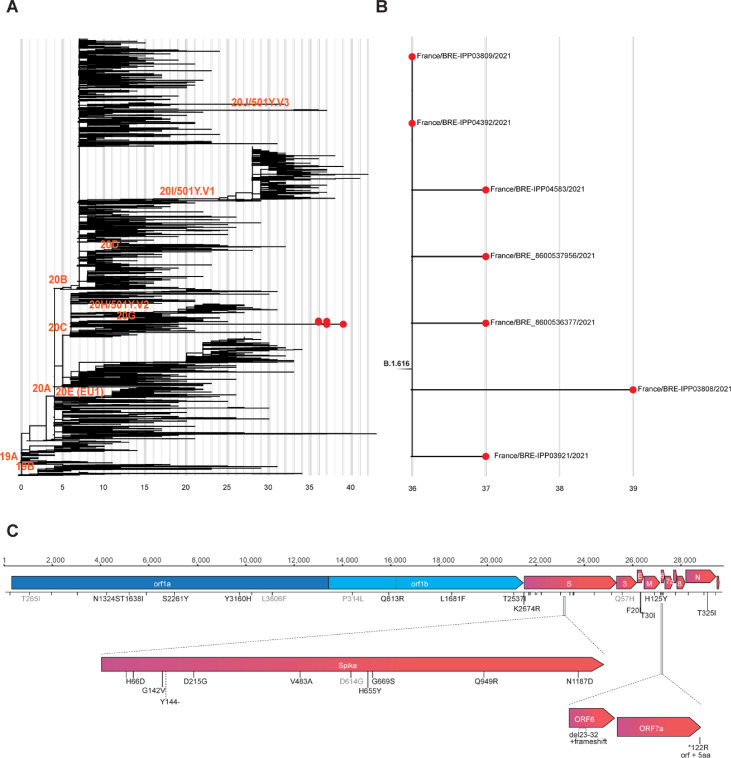
Phylogenetic analysis revealed an original variant carrying a unique constellation of mutations, which received the B.1.616 Pango lineage designation [1] (Fig. 1 ). It is characterized by nine amino acid changes and one deletion in the S protein in comparison with the original Wuhan strain (H66D, G142V, Y144del, D215G, V483A, D614G, H655Y, G669S, Q949R, N1187D), several unique amino-acid changes in the E, M, and N proteins, in ORF1ab and ORF3, as well as by a deletion and frameshift in ORF6 and replacement of the stop codon of ORF7a resulting in a five-amino-acid extension at its C-terminus (Supplementary Material Table S1). Interestingly, some mutations (Y144– and H655Y) in the S protein have been observed in VOCs B.1.1.7 and P.1, respectively. The V483A is located in the receptor binding motif next to residue 484, for which the E484K change found in several VOCs has been associated with reduced neutralization by post-infection and post-vaccination antibodies [[10], [11], [12]]. ORF6 and ORF7a are two proteins that antagonize various steps of type I interferon production and signalling [[13], [14], [15]].
Fig. 1.
Phylogenetic analysis of the B.1.616 lineage of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and characteristic non-synonymous substitutions. (A) Subsampled global phylogenetic maximum likelihood tree of SARS-CoV-2 with annotated Nextstrain clades next to the corresponding nodes and tips highlighted only for sequences from the Pangolin B.1.616 lineage. (B) Detailed view of the B1.616 lineage. In (A) and (B) branch lengths correspond to the number of nucleotide substitutions (shown below the tree) from the reference Wuhan-Hu-1 strain (NC_045512). (C) Nucleotide and amino-acid substitutions from the reference Wuhan-Hu-1 strain shared among the sequences from Lannion, France are represented as ticks along the SARS-CoV-2 genome and are annotated with text if non-synonymous. Light grey text annotated amino-acid substitutions are not unique to the Lannion (B1.616) lineage.
Population
From 1st January to 24th March 2021, 268 patients were hospitalized with SARS-CoV-2 infection in Lannion or Saint Brieuc hospitals, of whom 86 lived in the B.1.616 epidemic area (Supplementary Material Fig. S1). CT scan was typical of COVID-19 in 58/86 patients (67%) and serology was positive in 31/86 (36%). SARS-CoV-2 was detected by RT-PCR in 50/86 patients (58%), including 14 with all B.1.616 genomic characteristic features (confirmed B.1.616). In addition, 25 patients developed COVID-19 not related to a VOC at least 48 h after close contact with a confirmed B.1.616 case (probable B.1.616 cases). These 39 patients (14 confirmed and 25 probable) constituted the B.1.616 group (Supplementary Material Fig. S1).
During the study period, 108 patients were hospitalized in a ward where at least one case of B.1.616 COVID-19 was admitted, for a total of 780 patient-days at risk: 37/108 (34%) developed COVID-19 symptoms after they were admitted for another reason and were categorized as nosocomial B.1.616 cases (12 confirmed, and 25 probable). Therefore, the B.1.616 incidence rate in these wards was estimated at 47/1000 patient-days at risk. In addition, 47/86 patients living in the epidemic area were hospitalized because of COVID-19, in the absence of any known contact with a B.1.616 case, of whom four were infected by a VOC (B.1.1.7, n = 3; B.1.351, n = 1), and 43 were assigned to the unknown group. Epidemiological curve and infection control measures are reported in Supplementary Material Fig. S2. The two B 1.616 cases not healthcare-associated were the wife of a confirmed case who visited him just before he was diagnosed with COVID-19, and a physician who developed infection after taking care of a WGS-confirmed case.
Finally, among the 182 COVID-19 inpatients living outside of the B.1.616 epidemic area, 39 were screened for VOCs, of whom 28 (72%) were infected with the B.1.1.7 (n = 27) or the B.1.351 (n = 1) variants. These 28 patients, combined with the four patients from the B.1.616 epidemic area infected with a VOC, were assigned to the VOC group.
Baseline characteristics are displayed in Table 1 . Briefly, COVID-19 cases in the B.1.616 and unknown groups were older than those in the VOC group (respectively, 81 years (73–88) versus 80 years (68–87) versus 73 years (67–82), p = 0.022). B.1.616 cases were less likely to be documented at first RT-PCR (15% versus 23% for unknown variant versus 97% for VOC, p < 0.001), even though time from symptoms onset to first RT-PCR test was shorter in the B.1.616 group (0 (0–2) versus 0 (0–5) versus 3 (1–5); p = 0.004) (Table 1). Although B.1.616 cases had more RT-PCR tests (p < 0.001) and more lower respiratory tract samples (p < 0.001), only 26 (66%) had at least one positive RT-PCR. Among the 13 patients classified as B.1.616 in the absence of any positive RT-PCR, 13 had typical radiological findings on CT scan, and four had SARS-CoV-2 seroconversion, with a median time from symptoms onset to positive serology of 7 days (5–8) (Supplementary Material Table S2). In addition, the median cycle threshold (CT) in nasopharyngeal samples was higher in the B.1.616 and the unknown variant groups, at 29 (27–36) and 35 (30–40), versus 19 (14–22) in the VOC group, p < 0.001. Supplementary Material Fig. S3 illustrates the diagnosis criteria in VOC and B.1.616 groups.
Table 1.
Baseline characteristics of inpatients with severe acute respiratory syndrome coronavirus 2 (SARS-COV-2) infection according to the variant responsible for the infection
| Variables | B.1.616 n = 39 |
Undetermined variant n = 43 |
VOC n = 32 |
p |
|---|---|---|---|---|
| Age, year (IQR) | 81 (73–88) | 80 (68–87) | 73 (67–82) | 0.022 |
| Male, n (%) | 21 (54%) | 24 (56%) | 15 (47%) | 0.732 |
| Body mass index, kg/m2 (IQR) | 25 (22–29) | 26 (23–28) | 31 (27–33) | 0.012 |
| Comorbidities | ||||
| No comorbidity, n (%) | 6 (18%) | 9 (19%) | 4 (13%) | 0.812 |
| Cardiovascular disease, n (%) | 22 (56%) | 20 (47%) | 11 (34%) | 0.560 |
| Chronic respiratory disease, n (%) | 8 (21%) | 9 (21%) | 5 (16%) | 0.824 |
| Chronic kidney failure, n (%) | 12 (31%) | 7 (16%) | 5 (16%) | 0.185 |
| Cirrhosis, n (%) | 4 (10%) | 4 (9%) | 1 (3%) | 0.518 |
| Neurological disease, n (%) | 11 (28%) | 5 (12%) | 10 (31%) | 0.082 |
| Cancer, n (%) | 12 (31%) | 11 (26%) | 10 (31%) | 0.826 |
| Immunodepression, n (%) | 4 (10%) | 1 (2%) | 5 (16%) | 0.120 |
| Diabetes, n (%) | 9 (23%) | 11 (26%) | 6 (19%) | 0.783 |
| Hypertension, n (%) | 22 (56%) | 19 (44%) | 22 (69%) | 0.105 |
| Healthcare-associated COVID-19, n (%) | 37 (95%) | 17 (40%) | 10 (31%) | <0.001 |
| Clinical findings | ||||
| Fever, n (%) | 24 (61%) | 27 (63%) | 21 (66%) | 0.397 |
| Temperature, °C | 38.2 (37.0–38.8) | 38 (37.7–38.5) | 37.8 (37.0–38.8) | 0.732 |
| Dyspnoea, n (%) | 26 (67%) | 27 (63%) | 14 (44%) | 0.118 |
| Oxygen saturation, % | 91 (90–94) | 93 (92–96) | 94 (89 - 96) | 0.171 |
| Cough, n (%) | 14 (36%) | 15 (35%) | 14 (44%) | 0.705 |
| Headache, n (%) | 2 (5%) | 3 (7%) | 1 (3%) | 0.876 |
| Delirium, n (%) | 6 (15%) | 5 (12%) | 2 (6%) | 0.534 |
| Fatigue, n (%) | 9 (23%) | 14 (33%) | 12 (38%) | 0.400 |
| Anosmia, n (%) | 1 (3%) | 2 (5%) | 0 | 0.777 |
| Digestive symptoms, n (%) | 4 (10%) | 5 (12%) | 1 (3%) | 0.417 |
| Rhinorrhoea, n (%) | 0 | 1 (2%) | 2 (6%) | 0.279 |
| No symptoms – n (%) | 0 | 4 (9%) | 4 (12%) | 0.053 |
| Biological findings | ||||
| Neutrophils, x 109/L | 5140 (3245–7775) | 4050 (3150–8110) | 4130 (1440–6700) | 0.166 |
| Lymphocytes, x 109/L | 650 (425–1055) | 730 (580–1050) | 780 (610–1060) | 0.526 |
| Serum C-reactive protein, mg/mL | 79 (43–122) | 51 (16–142) | 49 (12–92) | 0.163 |
| D-dimers, μg/L | 1330 (589–1714) | 722 (521–1609) | 1528 (930–1942) | 0.476 |
| Thorax computed tomography (CT) | ||||
| No CT scan, n | 9 (23%) | 18 (42%) | 12 (37%) | |
| <25%, n (%)a | 9 (23%) | 9 (21%) | 5 (16%) | 0.953 |
| 25–50%, n (%)a | 12 (35%) | 11 (26%) | 9 (28%) | |
| 50-75%, n (%)a | 8 (21%) | 5 (12%) | 5 (16%) | |
| >75%, n (%)a | 1 (3%) | 0 | 1 (3%) | |
| Pulmonary embolism, n (%) | 3 (8%) | 1 (2%) | 2 (6%) | 0.583 |
| Time from symptoms onset to first RT-PCR test, days (IQR) | 0 (0–2) | 0 (0–5) | 3 (1–5) | 0.004 |
| Time from symptoms onset to first SARS-CoV-2 detection, days (IQR) | 3 (2–9) | 4 (1–7) | 1 (0–4) | 0.059 |
| First RT-PCR positive for SARS-CoV-2, n (%) | 6 (15%) | 10 (23%) | 31 (97%) | <0.001 |
| At least one RT-PCR positive for SARS-CoV-2, n (%) | 26 (67%) | 24 (56%) | 32 (100%) | <0.001 |
| Number of RT-PCR tests per patient | <0.001 | |||
| 1 | 6 (15%) | 13 (30%) | 31 (97%) | |
| 2 | 12 (31%) | 17 (40%) | 1 (3%) | |
| >3 | 21 (54%) | 13 (30%) | 0 | |
| Site of sample (first positive RT-PCR only), n | <0.001 | |||
| Endotracheal aspirate | 0 | 2 (5%) | 0 | |
| Expectoration | 2 (5%) | 0 | 0 | |
| Bronchoalveolar lavage | 5 (13%) | 0 | 0 | |
| Nasopharyngeal swab | 13 (33%) | 22 (451%) | 32 (100%) | |
| Stool | 0 | 1 (2%) | 0 | |
| CT value (positive RT-PCR only) | ||||
| Nasopharyngeal sample | 29 (27–36) | 35 (30–40) | 19 (14–22) | <0.001 |
| Lower respiratory tract sample | 33 (33–35) | 45 (45–45) | 26 | 0.035 |
VOC, variant of concern.
Proportion of lung involved.
Outcomes
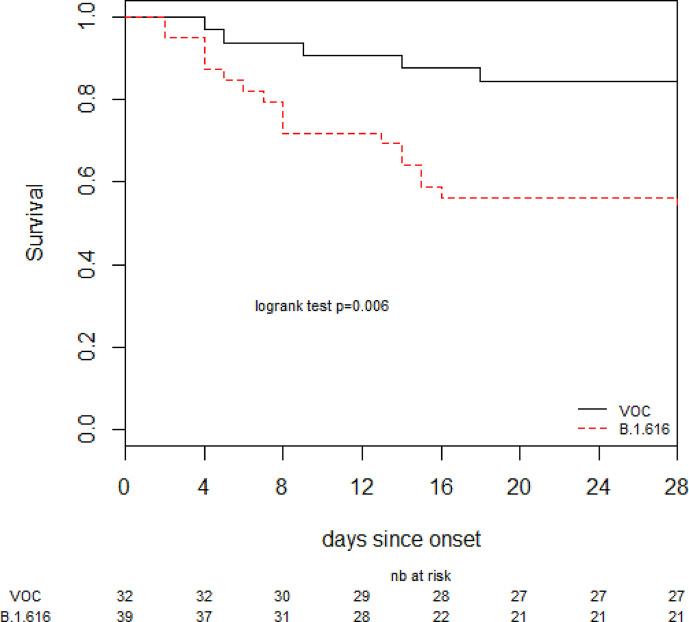
As planned, we compared the outcomes of B.1.616 and VOC cases. Patients with B.1.616 COVID-19 had worse clinical outcomes: 27 (65%) reached a WHO score >5 within 28 days from onset, 14 (36%) were admitted in the ICU, and 18 (46%) died within 28 days (Table 2 ). Variables independently associated with time to poor outcome in Cox proportional hazard regression (after adjusting for B.1.616, age, chronic respiratory disease, immunodepression, hypertension, and healthcare-associated COVID-19) were B.1.616 (HR 4.0 (1.5–10.9)), and chronic respiratory diseases (Table 3 ). Lethality was higher for B.1.616 at 18/39 (46%) versus 5/32 (16%) for VOC (p = 0.011), (Fig. 2 ), although risk of death within 28 days was not significant after adjustment for age and healthcare-associated infection (aHR 2.4, 95%CI 0.76–7.44).
Table 2.
Twenty-eight-day clinical outcomes of study patients according to the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variant responsible for the infection
| Variables | B.1.616 n = 39 |
Undetermined variant n = 43 |
VOC n = 32 |
p |
|---|---|---|---|---|
| Intensive care unit admission, n (%) | 14 (36%) | 5 (12%) | 10 (31%) | 0.028 |
| WHO score >5 within 28 days from onset, n (%) | 27 (65%) | 14 (40%) | 10 (31%) | <0.001 |
| 28 day-mortality, n (%) | 18 (46%) | 10 (23%) | 5 (16%) | 0.011 |
WHO, world health organization; VOC, variant of concern.
Table 3.
Risk factors for poor outcome (WHO >5) among the 39 B.1.616-related cases and the 32 VOC-related coronavirus disease 2019 (COVID-19) cases (Cox proportional hazard regression)
| Variable | Univariate analysis |
Multivariable analysis |
||
|---|---|---|---|---|
| HR | IC | HR | IC | |
| B.1.616 (versus VOC) | 3.0 | (1.4–6.1) | 4.0 | (1.5–10.9) |
| Age per supplementary 10 years | 1.0 | (0.8–1.2) | 0.9 | (0.6–1.2) |
| Male | 1.3 | (0.7–2.6) | ||
| Cardiovascular disease | 1.0 | (0.5– 1.9) | ||
| Chronic respiratory disease | 2.5 | (1.2–5.2) | 2.6 | (1.2–5.6) |
| Chronic kidney failure | 1.1 | (0.5–2.3) | ||
| Cirrhosis | 1.4 | (0.4–4.6) | ||
| Neurological issue | 0.7 | (0.3–1.6) | ||
| Cancer | 0.7 | (0.3–1.4) | ||
| Immunodepression | 0.3 | (0.1–1.4) | 0.6 | (0.1–2.9) |
| Diabetes | 1.0 | (0.4–2.1) | ||
| Hypertension | 0.5 | (0.3–1.1) | 0.7 | (0.3–1.6) |
| Healthcare-associated COVID | 1.4 | (0.7–2.7) | 0.9 | (0.3–2.1) |
VOC, variant of concern.
Fig. 2.
Survival curves. VOC, variant of concern.
We performed a sensitivity analysis in which the definition of nosocomial infection was restricted to patients who developed symptoms >8 days after admission (https://www.ecdc.europa.eu/en/covid-19/surveillance/surveillance-definitions); B.1.616 remained independently associated with severe disease (aHR 3.77, 95%CI 1.43–9.88) (Supplementary Material).
Discussion
The low rate of SARS-CoV-2 detection by RT-PCR tests on nasopharyngeal samples within a large cluster of COVID-19 cases led to the identification of a novel variant, B.1.616. Although COVID-19 cases due to this variant had clinical, biological and radiological findings in line with classical features of COVID-19, B.1.616 was associated with more severe disease. A clinical trial reported 39% of patients with severe disease who required high-flow oxygen or more invasive support at day 28, as compared to 65% for B.1.616 cases in our study, but that study was conducted prior to the emergence of VOCs, patients enrolled were younger (mean age 60 years), and had fewer comorbidities (81% with ≥1 comorbidity) [16]. In-hospital mortality ranged from 21% to 36% in previous studies, with higher mortality in nosocomial COVID-19 and in elderly patients [[17], [18], [19]]. The high mortality rate among B.1.616 cases in our study must be interpreted with caution, as patients enrolled combined both pejorative factors: age (median 81 years (73–88)) and healthcare acquisition (37/39, 95%). The B.1.616 group included both WGS-confirmed B.1.616 cases and COVID-19 cases following close contact with a confirmed case (probable B.1.616) in the absence of WGS confirmation. This conservative bias implies that comparisons with controls may lead to misjudgement of the actual differences, with the 25 probable cases being frails and therefore susceptible to increased morbidity. Other variants have been associated with an increase or a decrease in COVID-19 severity [4,20]. At the time of writing, four variants have been classified by the WHO as VOCs based on “evidence of an increase in transmissibility, more severe disease, significant reduction in neutralization by antibodies generated during previous infection or vaccination, reduced effectiveness of treatments or vaccines, or diagnostic detection failures” [5].
The second most salient feature of COVID-19 cases related to B.1.616 was the high number of negative or weakly positive RT-PCR tests on nasopharyngeal samples. Previous studies estimated that 10–16% of patients with COVID-19 have negative RT-PCR tests on nasopharyngeal samples [21,22]. A meta-analysis concluded that 1.8–33% of RT-PCR tests on first nasopharyngeal samples in COVID-19 patients are found negative [23]. In our study, 20/26 B.1.616 cases (77%) with at least one positive RT-PCR tested negative for RT-PCR on their first nasopharyngeal sample. Failure to detect B.1.616-related COVID-19 with the reference-standard diagnostic test most likely contributed to the emergence of several clusters, since implementation of specific infection control measures mostly relied on virological confirmation during the study period. Retrospective review of medical charts found that, in several cases, the diagnosis was suspected early in the COVID-19 course, but specific infection control measures were interrupted once RT-PCR tests returned negative. Failure to detect B.1.616 on nasopharyngeal samples is even more problematic for the screening of contacts. Indeed, the incidence of nosocomial COVID-19 was much higher during the early phase of the outbreak, before we realized the low yield of nasopharyngeal samples for this variant. Of note, although ‘diagnostic detection failures’ is one of the criteria for defining VOC, none of the four major VOCs nor any of the current VUIs meet this classification criterion. As RT-PCR assays used in France target at least two different viral genomic regions, it is unlikely that the B.1.616 strain would not be detected due to specific mismatches. Identification of this new lineage in lower respiratory tract samples confirmed the correct detection of the viral genome by commercially available assays used during the study period. Repeated failures to detect B.1.616 on nasopharyngeal samples were therefore most likely due to SARS-CoV-2 viral loads below the detection threshold in this site, rather than to suboptimal sensitivity of routine RT-PCR tests. Of note, RT-PCR was positive on nasopharyngeal samples for only 32% of patients with pneumonia due to SARS-CoV [24], and the Middle East respiratory syndrome-related coronavirus (MERS-CoV) is mostly detected in lower respiratory tract specimens, with high viral load, whereas nasopharyngeal specimens are poorly contributive [25].
In our study, among patients with a positive RT-PCR assay in the B.1.616 group, the sensitivity of one, two, three, and four tests on nasopharyngeal samples were, respectively, 6/39 (15%), 15/39 (38%), 16/39 (41%), and 21/39 (54%). RT-PCR tests on sputum, or bronchoalveolar lavage (BAL), were positive in 8/39 (20%) B.1.616 cases with previous negative nasopharyngeal RT-PCR tests. As samples from the lower respiratory tract are more difficult to obtain in frail patients, the real extent of the B.1.616-related COVID-19 outbreak in our institution has probably been underestimated. A large surveillance study, with sequencing of a representative sample of 15% of all RT-PCR-positive COVID-19 cases during the study period found no community-acquired B.1.616-related COVID-19 (Flash study#5, SpF, Paris, France, unpublished data), but the low detection in standard sampling may have contributed to this result.
Our study has limitations. First, the small sample size and the retrospective design both limit the statistical power. However, due to the fast pace of the pandemic, early communication on the characteristics of new variants is warranted, and this would be the first case series of B.1.616-related COVID-19. Second, B.1.616 confirmed cases were those for whom a deep respiratory sample was obtained, mostly motivated by disease severity, hence constituting a selection bias. Finally, the selection of controls may be an additional limitation: VOC-related cases were selected as controls mostly because these cases could be reliably classified as ‘non-B.1.616’. Since VOC screening was not performed up until 10th February, misclassification may have occurred, with inclusion of VOC cases among the 25 ‘probable B.1.616’ cases. However, the proportion of VOCs in this area was very low on 7th January, estimated at <1% [26]. Moreover, inclusion of VOC patients in the B.1.616 group would lead to underestimation of differences between groups. In addition, all consecutive cases with available VOC screening were enrolled in the control group, which limits selection bias.
Conclusion
We report a nosocomial outbreak of COVID-19 cases related to a new variant, B.1.616, characterized by poor detection with RT-PCR tests on nasopharyngeal samples despite having the typical clinical, radiological, and biological features of COVID-19. The novel variant reported here adds to the diversity of emerging SARS-CoV-2 variants with impact on early diagnosis and control. This work also highlights the difficulties of managing nosocomial cases when the reference-standard test fails to confirm the diagnosis. With new variants constantly emerging, one should remain attentive to any unusual clinical situation that could be linked to such emergence.
Author contributions
Conception and design: PF, SB, FXL, DC, SVDW, PT and NM. Acquisition of the data: PF, MJD, RV, PR, AG, NV, CM, SGDC, ED, VM, RB, MV, EG, VT, CG, CP, SVDW and NM. Analysis and interpretation: PF, SB, VE, BG, AB, AZ, MT, ESL, DC, SVDW, PT and NM. Drafting or revising the article: PF, SB, VE, BG, AB, AZ, MT, ESL, DC, SVDW, PT and NM. All authors approved the final version.
Transparency declaration
All authors have completed the Unified Competing Interest form (available on request from the corresponding author) and declare no support from any organization for the submitted work, and no financial relationships with any organizations that might have an interest in the submitted work in the previous 3 years. All authors have no other relationships or activities that could appear to have influenced the submitted work. SW has patents issued and pending for SARS-associated coronavirus diagnostics. The SW lab is funded by Institut Pasteur, CNRS, Université de Paris, Santé publique France, Labex IBEID (ANR-10-LABX-62-IBEID), REACTing (Research & Action Emerging Infectious Diseases), and by the H2020 project 101003589 (RECOVER). ESL acknowledges funding from the INCEPTION programme (Investissements d’Avenir grant ANR-16-CONV-0005). The authors declare that they have no conflicts of interest..
Acknowledgements
We thank Olivia Da Conceicao and Cylia Imekhlaf for their help in collecting data from patients' charts. We would like to thank all the healthcare workers, public health employees, and scientists involved in the COVID-19 response to this outbreak. We acknowledge the authors, originating and submitting laboratories of the sequences from GISAID (Supplementary Material Table S2). We avoided any direct analysis of genomic data not submitted as part of this paper and used this genomic data only as background. This work used the computational and storage services (Maestro cluster) provided by the IT department at Institut Pasteur, Paris.
Editor: L. Leibovici
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cmi.2021.09.035.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Rambaut A., Holmes E.C., O’Toole Á., Hill V., McCrone J.T., Ruis C., et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol. 2020;5:1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cele S., Gazy I., Jackson L., Hwa S.-H., Tegally H., Lustig G., et al. Escape of SARS-CoV-2 501Y.V2 from neutralization by convalescent plasma. Nature. 2021;593:142–146. doi: 10.1038/s41586-021-03471-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 4.Davies N.G., Jarvis C.I., CMMID COVID-19 Working Group. Edmunds W.J., Jewell N.P., Diaz-Ordaz K., et al. Increased mortality in community-tested cases of SARS-CoV-2 lineage B.1.1.7. Nature. 2021;593:270–274. doi: 10.1038/s41586-021-03426-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weekly epidemiological update on COVID-19—27 April 2021. https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---27-april-2021 n.d.
- 6.Nouveau variant détecté et sous surveillance en Bretagne. presse/2021/nouveau-variant-detecte-et-sous-surveillance-en-bretagne n.d.
- 7.Ye Z., Zhang Y., Wang Y., Huang Z., Song B. Chest CT manifestations of new coronavirus disease 2019 (COVID-19): a pictorial review. Eur Radiol. 2020;30:4381–4389. doi: 10.1007/s00330-020-06801-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rhee C., Baker M., Vaidya V., Tucker R., Resnick A., Morris C.A., et al. Incidence of nosocomial COVID-19 in patients hospitalized at a large US academic medical center. JAMA Netw Open. 2020;3 doi: 10.1001/jamanetworkopen.2020.20498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.WHO Working Group on the Clinical Characterisation and Management of COVID-19 infection A minimal common outcome measure set for COVID-19 clinical research. Lancet Infect Dis. 2020;20:e192–e197. doi: 10.1016/S1473-3099(20)30483-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greaney A.J., Loes A.N., Crawford K.H.D., Starr T.N., Malone K.D., Chu H.Y., et al. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe. 2021;29:463–476. doi: 10.1016/j.chom.2021.02.003. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Z., VanBlargan L.A., Bloyet L.-M., Rothlauf P.W., Chen R.E., Stumpf S., et al. Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe. 2021;29:477–488. doi: 10.1016/j.chom.2021.01.014. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Z., Schmidt F., Weisblum Y., Muecksch F., Barnes C.O., Finkin S., et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021;592:616–622. doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuen C.-K., Lam J.-Y., Wong W.-M., Mak L.-F., Wang X., Chu H., et al. SARS-CoV-2 nsp13, nsp14, nsp15 and orf6 function as potent interferon antagonists. Emerg Microbe Infect. 2020;9:1418–1428. doi: 10.1080/22221751.2020.1780953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li J.-Y., Liao C.-H., Wang Q., Tan Y.-J., Luo R., Qiu Y., et al. The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling pathway. Virus Res. 2020;286:198074. doi: 10.1016/j.virusres.2020.198074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xia H., Cao Z., Xie X., Zhang X., Chen J.Y.-C., Wang H., et al. Evasion of type I interferon by SARS-CoV-2. Cell Rep. 2020;33:108234. doi: 10.1016/j.celrep.2020.108234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosas I.O., Bräu N., Waters M., Go R.C., Hunter B.D., Bhagani S., et al. Tocilizumab in hospitalized patients with severe Covid-19 Pneumonia. N Engl J Med. 2021;384:1503–1516. doi: 10.1056/NEJMoa2028700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richardson S., Hirsch J.S., Narasimhan M., Crawford J.M., McGinn T., Davidson K.W., et al. Presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID-19 in the New York City area. JAMA. 2020;323:2052–2059. doi: 10.1001/jama.2020.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rickman H.M., Rampling T., Shaw K., Martinez-Garcia G., Hail L., Coen P., et al. Nosocomial transmission of coronavirus disease 2019: a retrospective study of 66 hospital-acquired cases in a London teaching hospital. Clin Infect Dis. 2021;72:690–693. doi: 10.1093/cid/ciaa816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mendes A., Serratrice C., Herrmann F.R., Genton L., Périvier S., Scheffler M., et al. Predictors of in-hospital mortality in older patients with COVID-19: the COVIDAge Study. J Am Med Dir Assoc. 2020;21:1546–1554.e3. doi: 10.1016/j.jamda.2020.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Young B.E., Fong S.-W., Chan Y.-H., Mak T.-M., Ang L.W., Anderson D.E., et al. Effects of a major deletion in the SARS-CoV-2 genome on the severity of infection and the inflammatory response: an observational cohort study. Lancet. 2020;396:603–611. doi: 10.1016/S0140-6736(20)31757-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.REMAP-CAP Investigators. Gordon A.C., Mouncey P.R., Al-Beidh F., Rowan K.M., Nichol A.D., et al. Interleukin-6 receptor antagonists in critically ill patients with Covid-19. N Engl J Med. 2021;384:1491–1502. doi: 10.1056/NEJMoa2100433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.RECOVERY Collaborative Group. Horby P., Lim W.S., Emberson J.R., Mafham M., Bell J.L., et al. Dexamethasone in hospitalized patients with Covid-19. N Engl J Med. 2021;384:693–704. doi: 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arevalo-Rodriguez I., Buitrago-Garcia D., Simancas-Racines D., Zambrano-Achig P., Del Campo R., Ciapponi A., et al. False-negative results of initial RT-PCR assays for COVID-19: a systematic review. PLoS One. 2020;15 doi: 10.1371/journal.pone.0242958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peiris J.S.M., Chu C.M., Cheng V.C.C., Chan K.S., Hung I.F.N., Poon L.L.M., et al. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet. 2003;361:1767–1772. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guery B., Poissy J., el Mansouf L., Séjourné C., Ettahar N., Lemaire X., et al. Clinical features and viral diagnosis of two cases of infection with Middle East Respiratory Syndrome coronavirus: a report of nosocomial transmission. Lancet. 2013;381:2265–2272. doi: 10.1016/S0140-6736(13)60982-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gaymard A., Bosetti P., Feri A., Destras G., Enouf V., Andronico A., et al. Early assessment of diffusion and possible expansion of SARS-CoV-2 Lineage 20I/501Y.V1 (B.1.1.7, variant of concern 202012/01) in France, January to March 2021. Euro Surveill. 2021;26 doi: 10.2807/1560-7917.ES.2021.26.9.2100133. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.