Abstract
δEF1, a representative of the zinc finger-homeodomain protein family, is a transcriptional repressor which binds E2-box (CACCTG) and related sequences and counteracts the activators through transrepression mechanisms. It has been shown that the N-proximal region of the protein is involved in the transrepression. Here we demonstrate that δEF1 has a second mechanism of transrepression recruiting CtBP1 or CtBP2 as its corepressor. A two-hybrid screen of mouse cDNAs with various portions of δEF1 identified these proteins, which bind to δEF1 in a manner dependent on the PLDLSL sequence located in the short medial (MS) portion of δEF1. CtBP1 is the mouse orthologue of human CtBP, known as the C-terminal binding protein of adenovirus E1A, while CtBP2 is the second homologue. Fusion of mouse CtBP1 or CtBP2 to Gal4DBD (Gal4 DNA binding domain) made them Gal4 binding site-dependent transcriptional repressors in transfected 10T1/2 cells, indicating their involvement in a transcriptional repression mechanism. When the MS portion of δEF1 was used to Gal4DBD and used to transfect cells, a strong transrepression activity was generated, but this activity was totally dependent on the PLDLSL sequence which served as the site for interaction with endogenous CtBP proteins, indicating that CtBP1 and -2 can act as corepressors. Exogenous CtBP1/2 significantly enhanced transcriptional repression by δEF1, and this enhancement was lost if the PLDLSL sequence was altered, demonstrating that CtBP1 and -2 act as corepressors of δEF1. In the mouse, CtBP1 is expressed from embryo to adult, but CtBP2 is mainly expressed during embryogenesis. In developing embryos, CtBP1 and CtBP2 are expressed broadly with different tissue preferences. Remarkably, their high expression occurs in subsets of δEF1-expressing tissues, e.g., cephalic and dorsal root ganglia, spinal cord, posterior-distal halves of the limb bud mesenchyme, and perichondrium of forming digits, supporting the conclusion that CtBP1 and -2 play crucial roles in the repressor action of δEF1 in these tissues.
Compelling evidence indicates that transcriptional repression is crucial for genetic regulation of a wide range of cellular processes (10). Among the variety of mechanisms which transcriptional repressors rely on (see reference 3 for a review), transrepression to counteract the effect of activators bound to nearby DNA sites is considered to be predominant.
There are two basic mechanisms of transrepression. In the first, the transcriptional regulator has an intrinsic repression domain, which is demonstrated by the fact that transplantation of the domain to heterologous DNA binding domain creates a new repressor protein. In the second mechanism, a portion of the regulatory protein serves as the binding site of a corepressor protein, so that the protein-corepressor complex functions to exert transrepression. As an established example, Groucho proteins which interact with DNA-binding transcriptional regulators carrying a WPRW(Y) amino acid sequence motif, e.g., Hairy and Runt of Drosophila, act as corepressors (1, 24). As demonstrated in this report, CtBP proteins comprise another class of corepressors, interacting with a subset of transcription factors through a short sequence motif, PLDLSL.
Human CtBP was first recognized as a cellular factor interacting with the C-terminal portion of adenovirus E1A protein (27). CtBP attenuates transcriptional activation and tumorigenicity which are attributed to the E1A protein (27, 32). dCtBP, the Drosophila homologue of CtBP, has been cloned and shown to bind to three transcriptional repressors, Hairy, Knirps, and Snail (20, 21). Recently, homologues of CtBP have been shown to bind to basic Krüppel-like factor (BKLF) (35) and the vertebrate homologue of Polycomb proteins XPc and HPC2 (31). Interaction of CtBP proteins with these negative transcriptional regulators raised the possibility that CtBPs act as corepressors, but direct proof of this possibility was not provided in these previous works.
δEF1 (7, 8), a representative member of zinc finger-homeodomain family transcription factors (4, 18, 36, 37), originally was identified as a binding protein of the lens-specific δ1-crystallin enhancer of the chicken (7) but later was found to be expressed in a variety of tissues of mesodermal and ectodermal origin in chicken and mouse embryos (8, 34). δEF1 carries two clusters of Krüppel-type C2H2 zinc fingers positioned close to N and C termini and a medially located homeodomain (8, 30). δEF1 and its homologues of various vertebrate species have been shown to repress transcription through binding to the consensus DNA sequence, CACCT (5, 9, 15, 23, 28, 29, 35). Both clusters of zinc fingers, but not the homeodomain, are involved in binding to CACCT (13, 28, 29). Binding of δEF1 to E2-box (CACCTG)-containing sequences would interfere with binding of various basic helix-loop-helix-type activators to the same sites (28). In addition, DNA-bound δEF1 exerts transrepression which is attributed, at least partly, to an intrinsic repression domain (N-proximal region [NR]) positioned close to the N terminus of δEF1 (29).
To gain further insight into the molecular basis of transcriptional repression by δEF1, we carried out two-hybrid cDNA screening using yeast cells for cellular proteins interacting with δEF1. Here we report identification of CtBP1 and CtBP2 of the mouse as corepressors of δEF1. These proteins act as repression domains when ligated to the Gal4 DNA binding domain (Gal4DBD), bind to the short medial (MS) portion of δEF1 containing the PLDLSL motif, and enhance transrepression by δEF1. In mouse embryos, high expression of CtBP1/2 genes occurs at sites corresponding to those of δEF1, supporting essential corepressor functions of CtBP1 and -2 for δEF1 action.
MATERIALS AND METHODS
Two-hybrid interactions in yeast cells.
The HybriZAP two-hybrid system (Stratagene) was used for cDNA screening. The bait plasmids were made by in-frame fusion of various portions of δEF1 cDNA to Gal4DBD sequence in pBD-gal4, after placing an EcoRI site just upstream of the ATG initiation codon of δEF1 cDNA. The N-terminal (N), long medial (ML), C-terminal (C), and MS portions of δEF1 (Fig. 1A) corresponded to the restriction fragments EcoRI(−5)-MunI(+1079), MunI(+1079)-NsiI(+2650), PvuII(+2277)-PvuII(+3325), and HindIII(+1501)-SalI(+2182), respectively, where the numbers indicate positions in the δEF1 open reading frame. The prey cDNA libraries used were those constructed from a mixture of random- and oligo(dT)-primed cDNAs of poly(A)+ RNAs isolated from 9.5- to 11.5-day mouse embryos, and MATCHMAKER Libraries of a later stage (17-day embryo), and adult brain purchased from Clontech. The HybriZAP phage cDNA library was amplified and converted to a pAD-gal4 plasmid library by helper phage-aided in vivo mass excision. Yeast transformants carrying each prey plasmid were generated, and the pAD-gal4 plasmid cDNA library was used for two-hybrid screening. Colonies were selected by histidine prototrophy in the presence of 3-aminotriazole (3-AT) and by expression of UAS-lacZ. Plasmid DNAs of the selected colonies were recovered and transformed into Escherichia coli to isolate the prey cDNA clones. The prey-bait interaction was confirmed by transforming the second yeast strain (SFY526) with the isolated bait and prey plasmids and examining for histidine prototrophy and β-galactosidase expression (26). CtBP cDNA fragments used for Fig. 2D were generated by PCR using appropriate primers.
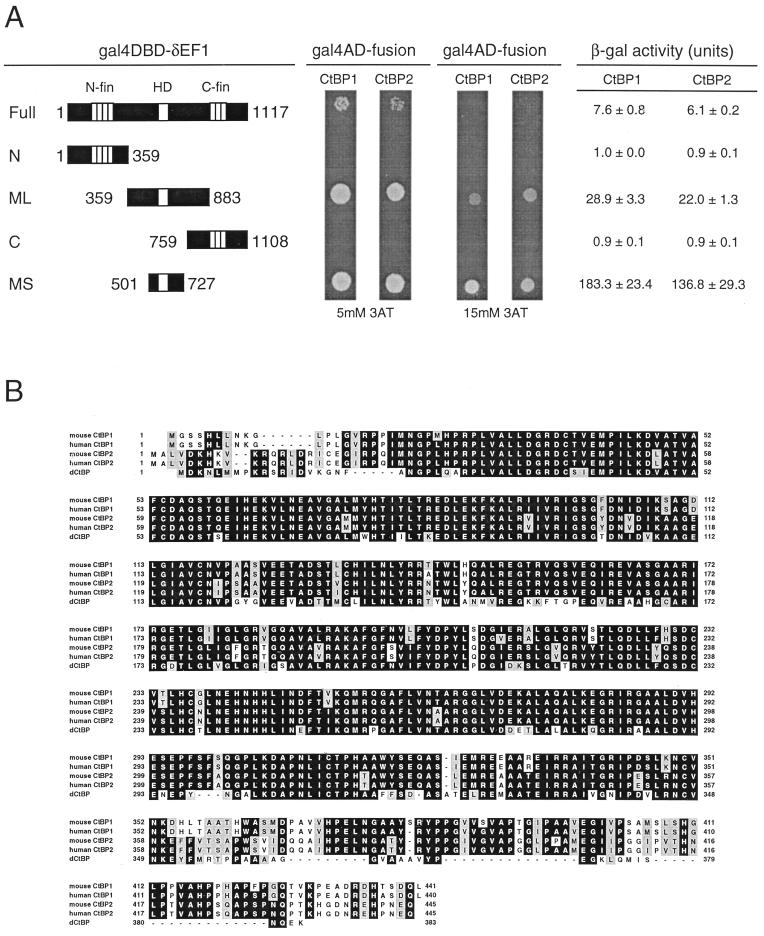
FIG. 1.
Identification of CtBP1 and CtBP2 as interacting factors with δEF1 and their primary structures. (A) Interaction of CtBP1 and CtBP2 with the middle portion of δEF1 in yeast cells. (Left) Portions of δEF1 protein fused to Gal4DBD. Amino acid residue numbers of the termini are indicated. N-fin, HD, and C-fin indicate N-proximal zinc finger cluster, homeodomain, and C-proximal zinc finger clusters, respectively. (Middle) Growth of yeast cells cotransformed with plasmids expressing Gal4AD-CtBP and Gal4DBD-δEF1 on plates containing 5 or 15 mM 3-AT. At 5 mM 3-AT, cells carrying Gal4DBD fused to MS-δEF1 or ML-δEF1 grew well, while those with full-length δEF1 (Full-δEF1) showed attenuated growth. At 15 mM 3-AT, growth of cells with Gal4DBD–Full-δEF1 was totally inhibited, and that of cells with Gal4DBD–ML-δEF1 was reduced. The results indicate that CtBP interacts with the MS portions of δEF1, but the transcriptional activation levels attained by bait-prey interaction are variable using MS, ML, and Full portions of δEF1 in the order MS > ML > Full. This was confirmed by measurement of β-galactosidase (β-gal) activity in a liquid culture of each yeast colony (right). (B) Alignment of amino acid sequences of mouse and human CtBP1, human CtBP2, and dCtBP. Identical amino acid residues are highlighted; similar residues are shaded. The first methionine codon of the mouse CtBP1 open reading frame which satisfies Kozak’s consensus was designated the initiation codon. In the mouse CtBP2 cDNA sequence, a stop codon immediately precedes the coding sequence. The human CtBP2 sequence is from the EST (expressed sequence tag) database. Nucleotide accession numbers for human CtBP1, human CtBP2, and dCtBP cDNAs are g1063638, g2909777, and g2950374, respectively.
FIG. 2.
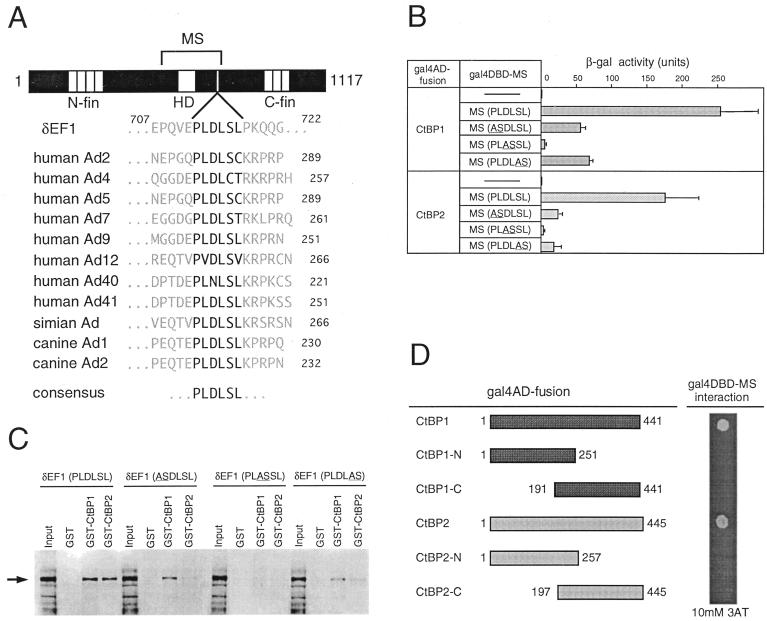
Binding of CtBP proteins is dependent on the PLDLSL sequence of δEF1. (A) Conservation of the CtBP binding motif PLDLSL among mouse δEF1 and adenovirus (Ad) E1A proteins. (B) β-Galactosidase (β-gal) activities generated by interaction between δEF1 (MS) and CtBP1/2 in the yeast two-hybrid assay. Yeast cells cotransformed with Gal4AD-CtBP1/CtBP2 and Gal4DBD-MS (normal or mutant ASDLSL, PLASSL, or PLDLAS were grown in liquid culture, and β-galactosidase activity was measured. (C) Binding of CtBP1 and CtBP2 to full-length δEF1 in vitro. GST, GST-CtBP1, or GST-CtBP2 bound to glutathione beads was mixed with in vitro-translated and N-terminally Xpress-tagged δEF1 (Input). The bound proteins were analyzed by Western blotting using an anti-Xpress antibody. The arrow indicates the position of Xpress-δEF1 on the blot. (D) Interaction of full-length or truncated forms of CtBP proteins with δEF1 in yeast cells. Full-length CtBP1/2 or fragments thereof (left) were fused with Gal4AD, and interaction with Gal4DBD-MS was assessed by growth of yeast cells on plates containing 10 mM 3-AT.
In vitro binding assay.
GST (glutathione S-transferase) fusion proteins of CtBP1 and CtBP2 were expressed in E. coli cells, using pGEX-4T-1 vector (Pharmacia), and purified as described (29). Mutant forms of δEF1 with amino acid substitutions (underlined), ASDLSL, PLASSL, and PLDLAS, were generated by using an ExSite PCR-based site-directed mutagenesis kit (Stratagene). Primers were designed to replace each pair of codons with NheI site (GCTAGC), coding for alanine and serine. Wild-type and mutant forms of δEF1 tagged with Xpress sequence at their N termini were synthesized in vitro, using pcDNA 3.1 vector (Invitrogen) and the TNT coupled rabbit reticulocyte lysate system (Promega). Thirty microliters of reticulocyte lysate containing the synthesized protein was added to 250 μl of the glutathione-Sepharose beads bound with GST or GST-CtBP1/2 suspended in TPBS (phosphate-buffered saline with 1% Tween 20) containing 0.01% bovine serum albumin and kept at 4°C for 1 h with gentle mixing. The beads were washed extensively with 0.01% bovine serum albumin–TPBS; the bound protein was released by boiling in sodium dodecyl sulfate-containing sample buffer for polyacrylamide gel electrophoresis and subjected to Western blotting using anti-Xpress antibody (Invitrogen).
Transcriptional repression by CtBP-Gal4DBD and MS-Gal4DBD fusion proteins.
The reporter plasmid p4xGAL-TK-Luc was constructed by inserting four copies of Gal4DBD sequence upstream of the herpes simplex virus thymidine kinase promoter (−197 to +56) of pTK-Luc (29). The effector plasmids for expression of Gal4DBD-CtBP fusion proteins were made by in-frame insertion of CtBP cDNAs downstream of the Gal4 sequence of pCMV/SV2-gal4DBD (14). 10T1/2 cells (25) grown in Dulbecco modified Eagle medium containing 10% fetal bovine serum were replated at 5 × 104 per 3.5-cm-diameter dish the day before transfection. The cells were transfected by a DNA-calcium phosphate coprecipitation method (2) with total 1.5 μg of DNA containing 0.5 μg of p4xGAL-TK-Luc, 0.2 μg of pSV-β-gal (Promega), 0, 0.02, 0.1, or 0.5 μg of pCMV/SV2-gal4DBD-CtBP1/2, insert-free pCMV/SV2-gal4DBD (to keep the molarity of pCMV/SV2 vector DNA constant), and pUC19. The cultures were fed with fresh medium after 8 h of transfection and harvested after 24 h. Luciferase and β-galactosidase activities in the cell extracts were measured as described by Sekido et al. (29). In the case of MS-Gal4DBD fusion proteins, the cDNA for the MS portion of δEF1 was fused to Gal4DBD in a similar fashion and used for transfection.
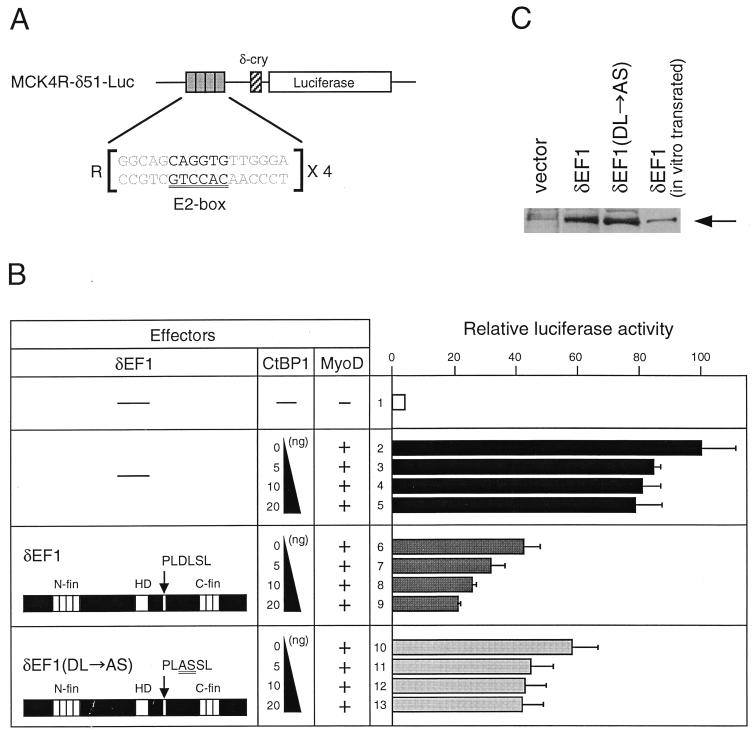
Corepression by CtBP in transcriptional repression by δEF1.
The reporter for the δEF1-dependent repression assay, MCK4R-δ51-Luc, was constructed by inserting the tetrameric E2 box sequence (4R) of the mouse muscle creatine kinase (MCK) enhancer into the BamHI site upstream of the promoter of the luciferase reporter δ51LucII (15). The 4R sequence was made by duplication of the 2R sequence described previously (28). The effector plasmids for this repression assay were made by cloning cDNA fragments of δEF1 or CtBP1/2 into the pcDNA3.1 vector. Transfection was done as described above; 1.5 μg of DNA contained 0.3 μg of MCK4R-δ51-Luc, 0.2 μg of pSV-β-gal, 0.25 μg of pcDNA3.1/δEF1, 0.75 μg of pβactmyoD (6, 28), 0, 5, 10, or 20 ng pcDNA3.1/CtBP1, insert-free pcDNA3.1 (to make the pcDNA3.1 molarity constant), and pUC19.
To ascertain that wild-type and mutant forms of δEF1 were synthesized in comparable amounts after transfection, nuclear extracts were prepared from transfected COS-7 cells (29) and subjected to Western blot analysis (11) using anti-δEF1 antibodies (8).
Northern blot analysis.
Five micrograms each of total RNAs derived from 9.5-, 10.5-, and 11.5-day embryos, adult tissues, and 10T1/2 mouse cells were analyzed by Northern blotting. Electrophoresis and hybridization were done as described by Higashi et al. (11) except that QuickHyb reagents (Stratagene) were used.
Whole-mount in situ hybridization.
Whole-mount in situ hybridization of mouse embryos was done according to a standard method (38) using 1.5% blocking reagent (Boehringer Mannheim). The PvuII-PstI fragment (1186 to 1916) of CtBP1 cDNA and the SacI fragment (356 to 1139) of CtBP2 cDNA were cloned in a Bluescript plasmid, and RNA probes were prepared by transcription of the linearized plasmids with T3 or T7 RNA polymerase (Stratagene), using digoxigenin-11-UTP (Boehringer Mannheim).
Nucleotide sequence accession number.
Accession numbers for the mouse CtBP1 and -2 sequences described in this report are AB033122 and AB033123, respectively.
RESULTS
δEF1 binds CtBP1 and CtBP2.
We searched for possible occurrence of δEF1-interacting proteins important for regulatory functions of this transcriptional regulator. cDNA fragments coding for the N, ML, MS, and C portions of δEF1 were fused to the Gal4 DBD sequence and used as bait plasmids for yeast two-hybrid screen (Fig. 1A). cDNAs of 9.5- to 11.5-day mouse embryo mRNAs were ligated to the Gal4 activation domain (Gal4AD) sequence and used as prey plasmids. For each bait plasmid, 3 × 106 transformants were screened for histidine prototrophy in a high (15 mM) concentration of 3-AT. Positive clones were obtained only when the MS portion was used as bait, at the frequency of 1 in 7 × 105 transformants. Screening of cDNAs of later-stage embryos and adult brain with the same MS bait identified additional MS-interacting clones, totaling eight independent cDNA clones.
All eight cDNAs belonged to one of two highly related sequences. One of them coded for a protein which is identical to human CtBP except for 6 amino acid positions in 440 amino acid residues (99% amino acid identity) and was designated the mouse orthologue, CtBP1. The other, CtBP2, also resembled CtBP but was more divergent (80% amino acid identity) than CtBP1. Search of the sequence database for CtBP-related sequences identified human CtBP2. Therefore, there are two CtBP proteins in humans and mice. Turner and Crossley have independently identified CtBP2 as an interacting protein of BKLF (35). The amino acid sequences of the mammalian CtBP1/2 and the homologues of D. melanogaster recently identified (20) are compared in Fig. 1B.
Interaction of CtBP proteins and δEF1.
In the two-hybrid screen, only the MS portion of δEF1 demonstrated interaction with CtBP. We tested whether longer portions of δEF1 including the full-length δEF1 interact with the cloned CtBP1/2. With 15 mM 3-AT, prototrophic growth was also observed in assays using the ML portion (Gal4DBD-ML) in combination with Gal4AD-CtBP1/2, but growth was slower than with MS portion (Fig. 1A), which may account for the failure of the ML fragment in selecting the prototroph clones in the initial two-hybrid screen. With a lower 3-AT concentration (5 mM), interaction of CtBP1/2 with the full-length δEF1 but never with subfragments N and C (Fig. 1A) or with Gal4DBD alone (data not shown), was also demonstrated. These differences in activation of genes by Gal4AD and Gal4DBD complexes formed by interaction between CtBP1/2 and different δEF1 subfragments were confirmed by β-galactosidase expression activated by the same complexes (Fig. 1A). The data clearly indicate that CtBP proteins bind to δEF1 at a site included in the MS portion. Further subdivisions of the MS portion indicated that only the part more proximal to the C terminus of the homeodomain is required for binding of CtBP proteins (data not shown). The mechanism for the lower activation levels in assays using the longer δEF1 portions in the bait plasmids is not well understood, but the observation raises the possibility that there is an intramolecular interaction to modulate binding of CtBP1/2 to the MS portion of δEF1.
In adenovirus E1A proteins, the CtBP binding site is mapped to a short region consisting of PLDLSL or related sequences (Fig. 2A), and mutant E1A proteins with alteration of this sequence cannot bind to CtBP1 (27). Examination of the amino acid sequence of the MS portion of δEF1 identified the sequence motif PLDLSL at its C-proximal side (Fig. 2A). To determine whether this motif is involved in CtBP binding, we measured two-hybrid interaction in yeast cells between Gal4AD-CtBP1/2 and the mutated MS portion fused to Gal4DBD, using the β-galactosidase gene as the reporter. As shown in Fig. 2B, mutations causing pairwise alterations of the PLDLSL amino acid sequence decreased the interaction with CtBP1/2; in particular, alteration of the sequence to PLASSL abolished the interaction as observed in the case of human CtBP1-E1A interaction (27). There were no substantial differences between CtBP1 and CtBP2 in the assay (Fig. 2B).
To demonstrate more directly that CtBP proteins bind to δEF1, Xpress-tagged full-length δEF1 or its mutant forms with the same sequence alterations as used for Fig. 2B were synthesized in vitro, and binding of GST-CtBP1/2 was examined by precipitation of the complex formed with glutathione beads (Fig. 2C). Wild-type Xpress-δEF1 was efficiently precipitated with GST-CtBP1 or GST-CtBP2, as indicated by Western blotting. This binding was significantly decreased by mutations ASDLSL and PLDLAS and was abolished by mutation PLASSL. Thus, full-length δEF1 binds CtBP proteins in a manner dependent on the integrity of the PLDLSL sequence.
We prepared cDNAs for N- and C-terminal halves of CtBP1 and CtBP2 and assessed their binding to the MS portion of δEF1 by the two-hybrid assay. These truncated forms of the CtBP proteins failed to interact with the MS portion (Fig. 2D), indicating that integrity of CtBP proteins is essential for binding to δEF1.
CtBP1 and CtBP2 have transrepression activity.
It has been demonstrated that binding of CtBP to adenovirus E1A protein attenuates the transactivation of genes by E1A (27), implying a negative regulatory function of CtBP1/2. We examined whether CtBP1/2 show transrepression activity when a DNA binding domain is supplied and they are bound to a specific DNA site. CtBP1 and CtBP2 were fused to Gal4DBD, and their effects on expression of the 4xGAL-TK-Luc construct were examined by transfection of 10T1/2 cells (Fig. 3A).
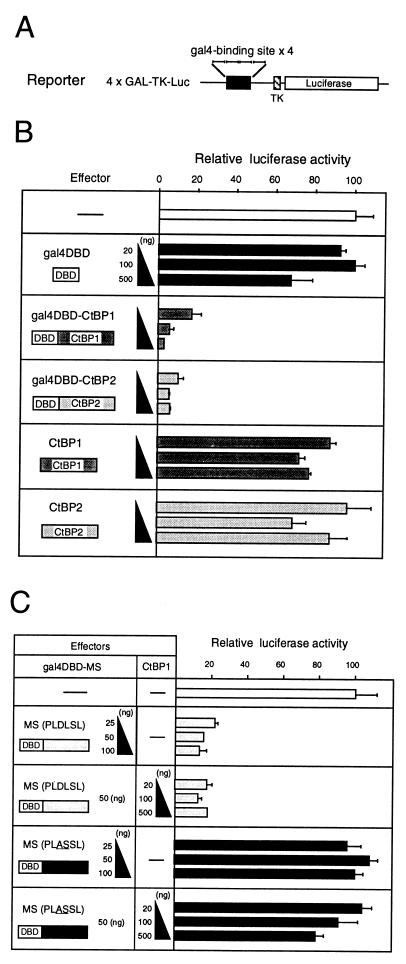
FIG. 3.
CtBP proteins exhibit transrepression when bound to DNA by fusion with Gal4DBD or by interaction with MS-Gal4DBD. (A) Structure of the luciferase reporter plasmid 4xGAL-TK-Luc. (B) Effects of gal4DBD-CtBP fusion proteins indicated at the left. The effect on transcription of the 4xGAL-TK-Luc reporter gene was assessed by expression of luciferase in transfected 10T1/2 cells (right panel; average of three transfections). CtBP1 or CtBP2 can actively repress the reporter activity only when fused to Gal4DBD; Gal4DBD or CtBP1/2 alone had no appreciable effect. (C) Effects of MS-gal4DBD fusion proteins and of exogenous CtBP1/2 on expression of 4xGAL-TK-Luc in 10T1/2 cells.
Luciferase expression was strongly repressed when Gal4DBD-CtBP1 or Gal4DBD-CtBP2 was cotransfected even at 20 ng of plasmid per transfection, whereas Gal4DBD alone or CtBP1/2 alone had no effect even at 500 ng per transfection (Fig. 3B). This result indicates that CtBP1 and CtBP2 have a potential for transcriptional transrepression that is exhibited only when DNA binding capacity is provided.
CtBP1 and CtBP2 act as corepressors.
The observations that the MS portion of δEF1 strongly binds CtBP1 and -2 and that CtBP1 and -2 show transrepression when a DNA binding domain is supplied argue for the model that CtBP proteins act as corepressors when bound to the MS portion and the portion is capable of DNA binding. This was tested by using fusion proteins made by ligating the MS portion to Gal4DBD. Transfection of 10T1/2 cells with wild-type MS-Gal4DBD very efficiently repressed expression of the reporter 4xGAL-TK-Luc, while CtBP binding-defective PLASDL mutant MS-Gal4DBD had no effect (Fig. 3C). The repression with MS-Gal4DBD occurred without exogenous CtBP1/2, and supplementation with exogenous CtBP1 did not alter the expression level of the reporter gene (Fig. 3C). Northern blot data (see Fig. 5) indicate that the 10T1/2 cells express both CtBP1 and CtBP2. It is likely that the CtBP proteins in 10T1/2 cells are abundant enough for high-affinity binding to the MS portion and act as corepressors of MS-Gal4DBD.
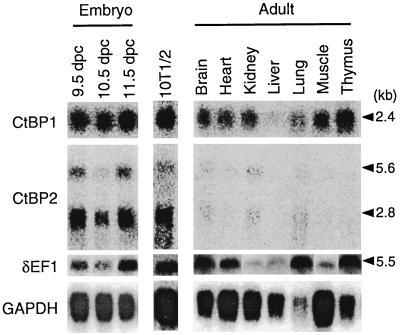
FIG. 5.
Northern blot analysis of the expression of CtBP1 and CtBP2 of the mouse in comparison with δEF1. GAPDH (glyceraldehyde-3-phosphate dehydrogenase) message is used to control the RNA loaded on the filter. CtBP1 has a transcript of 2.4 kb and is expressed from embryo to adult stages and widely among adult organs. CtBP2 transcript has two sizes, 2.8 kb (major) and 5.6 kb (minor), and expression in the embryo is much stronger than in the adult tissues. 10T1/2 cells express both CtBP1 and CtBP2 strongly, probably reflecting their origin of 13.5-day mouse embryo (25). δEF1 expression represented by the 5.6-kb transcript occurs among the various developmental stages, adult organs, and 10T1/2 cells.
CtBP1 and CtBP2 are corepressors of δEF1.
The above results strongly suggested that CtBP1 and -2 act as corepressors of intact δEF1. We thus set up an experiment in which the effect of CtBP1/2 on repression by δEF1 was examined. We previously demonstrated that δEF1 can repress E2-box-mediated gene activation by MyoD (28), and this system was used. An MCK minienhancer carrying an E2-box sequence which is bound by MyoD or δEF1 was tetramerized and placed upstream of a luciferase reporter gene (Fig. 4A). When this reporter was cotransfected with the MyoD effector vector, 25-fold activation of luciferase expression was observed (Fig. 4B, lanes 1 and 2). With a larger amount of MyoD vector, the activation level increased proportionately (data not shown), indicating that under this condition of transfection, not all E2-box sites are occupied by MyoD but some sites are still available for binding by δEF1. Expression of exogenous CtBP1 without exogenous δEF1 slightly lowered the MyoD-activated expression level (Fig. 4B, lanes 3 to 5), which is accounted for by the interaction of exogenous CtBP1 with endogenous δEF1 present in 10T1/2 cells (23, 24) (Fig. 5). The small repression by exogenous CtBP1/2 was observed only when the reporter gene contained the E2-box sequence (data not shown). Transfection of a moderate amount of δEF1 expression vector caused repression of the MyoD-activated luciferase expression to half of its level (Fig. 4B, lane 6), and this repression was further strengthened by exogenous CtBP1, resulting in decrease of the reporter expression to 20% of the MyoD-activated level (lanes 7 to 9). However, this effect of exogenous CtBP was diminished when the PLASSL mutant of δEF1 was used to repress MyoD-activated reporter (lanes 10 to 13), leaving only the same small repressing effect. This effect presumably originates from the interaction of exogenous CtBP1 with endogenous δEF1, which was also observed as weak repression in lanes 2 to 5. Essentially the same results were obtained with CtBP2 (data not shown). The data are fully rationalized on the assumption that wild-type and mutant forms of δEF1 are expressed with comparable efficiency after transfection. To verify this, COS-7 fibroblast cells were transfected with expression vectors for wild-type δEF1 and its PLASSL mutant, and nuclear extracts were analyzed by Western blotting. As shown in Fig. 4C, an equivalent amount of exogenous δEF1 proteins was detected.
FIG. 4.
CtBP proteins act as corepressors of δEF1. (A) Structure of the reporter plasmid (MCK4R-δ51-Luc) carrying four copies of the E2-box element (R) of the mouse MCK enhancer and basal promoter sequence (−51 to +57) of the chicken δ1-crystallin gene (15). (B) Effect of CtBP1 on transcriptional repression by δEF1. 10T1/2 cells were transfected with the reporter plasmid (0.3 μg) and with effector plasmids for expression of MyoD (0.75 μg), δEF1 or its mutant PLASSL (0.25 μg), and CtBP1. Relative luciferase expression levels averaged over three transfection experiments are indicated. (C) Western blot analysis of the wild-type and mutant (PLASSL) forms of δEF1 expressed in COS-7 cells. An equivalent amount of nuclear extract from the transfected cells was analyzed by Western blotting using anti-δEF1 antibodies. In vitro-translated δEF1 protein was included as the size marker. Exogenous δEF1 of mouse origin, both wild-type and PLASSL mutant forms, produced the same band intensities. The bands of exogenous δEF1 (mouse) were positioned slightly lower than that of endogenous simian δEF1 (open arrowhead), presumably reflecting the lack of exon 3 in rodent δEF1 (30).
An interesting difference in the action of CtBP1 and -2 as corepressors between MS-Gal4DBD fusion protein (Fig. 3C) and native δEF1 (Fig. 4B) is that endogenous CtBP1 and -2 were sufficient for full corepressor activity in the former case, while exogenous CtBP1 and -2 were effective corepressors in the latter. This difference presumably emanates from two major causes: first, δEF1 was overexpressed to counteract MyoD-mediated activation in the latter case, and second, perhaps full-length δEF1 has an intramolecular interaction which modulates CtBP binding, as suggested by the data for the two-hybrid assay (Fig. 1A).
Overall, the data indicate that exogenous CtBP proteins enhance the transrepression activity of δEF1, and this effect is dependent on the CtBP binding site of δEF1. This finding clearly demonstrates that CtBP1 and CtBP2 act as corepressors of δEF1.
Expression of CtBP1 and CtBP2 during mouse development.
Expression of the CtBP1 and CtBP2 genes was investigated by Northern blotting (Fig. 5) and by whole-mount in situ hybridization of the embryos (Fig. 6). The CtBP1 gene produced 2.4-kb transcript and was expressed throughout the developmental stages and in a wide range of adult tissues, while the CtBP2 gene produced transcripts of 2.8 kb (major) and 5.6 kb (minor), and strong expression was confined to the embryonic stages.
FIG. 6.
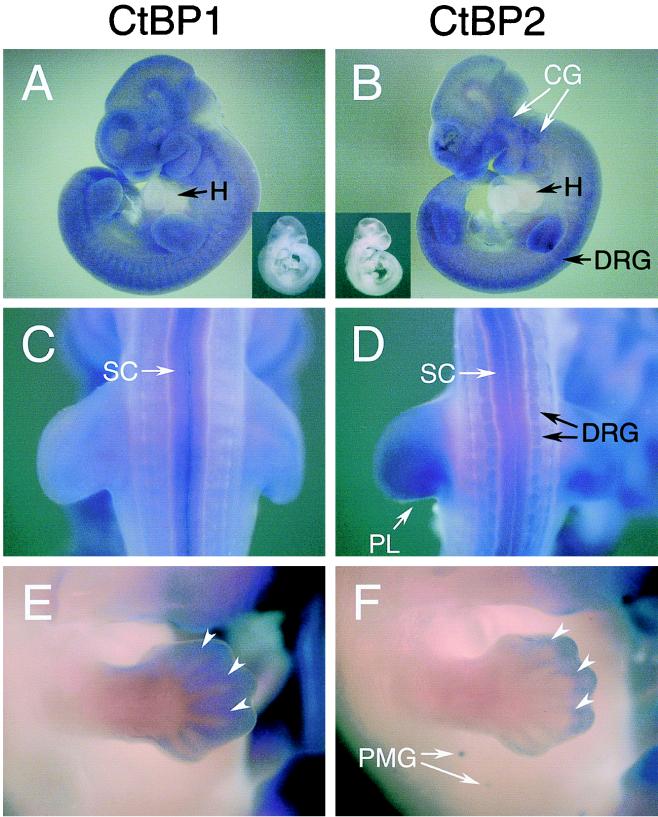
Whole-mount in situ hybridization analysis of CtBP1 and CtBP2 expression in mouse embryos. (A and B) Side views of 10.5-day embryos hybridized with CtBP1 antisense probe (A) and CtBP2 antisense probe (B). (C and D) Dorsal views of the same embryos. The insets show embryos hybridized with the corresponding sense probes. Major expression sites: CG, cephalic ganglia; DRG, dorsal root ganglia; PL, posterior-distal portion of the limb bud mesenchyme; SC, spinal cord; H, heart. The heart lacks expression of CtBP1/2. (E and F) Forelimbs and trunks of 12.5-day embryos hybridized with the CtBP1 (E) or CtBP2 (F) probe. Some of the perichondria are marked by arrowheads. PMG, primordia of mammary glands.
In 10.5-day embryos, CtBP1 was expressed broadly among various tissues (Fig. 6A). Only the spinal cord showed significant expression of CtBP1 (Fig. 6C). By contrast, CtBP2 was expressed predominantly in a few tissues (Fig. 6B and D) (cephalic ganglia, dorsal root ganglia, posterior-distal portion of the limb bud mesenchyme, and the spinal cord), all of which correspond to the major site of δEF1 expression around this stage (34). No expression of CtBP1/2 was detected in the heart (Fig. 6A and B).
In 12.5-day embryos, digits of the limbs were forming, and CtBP genes were highly expressed in the perichondria (Fig. 6E and F), the exact site of δEF1 expression (34). CtBP1 was expressed through the length of the digits (Fig. 6E), while CtBP2 was expressed only in the distal parts (Fig. 6F). Also, CtBP2 was uniquely expressed in the primordia of mammary gland (Fig. 6F).
Coincidence of major expression sites between δEF1 and CtBP genes strongly argues that corepressors CtBP1 and -2 play crucial roles in the regulatory functions of δEF1.
DISCUSSION
Significance of the interaction of CtBP in the regulatory activities of δEF1.
We identified CtBP1 and -2 as proteins interacting with the repressive transcriptional regulator δEF1 and demonstrated that they function as its authentic corepressor. We previously demonstrated that δEF1 has an intrinsic repression domain, NR, close to the N terminus (29) and thus is equipped with two different mechanisms of transrepression. In our previous work using an octamerized DC5 δ-crystallin minimal enhancer, repression of the enhancer activity in lens cells by δEF1 was dependent on the repression domain NR but not on the internal region bracketed by the zinc finger clusters (29), although CtBP activity is demonstrated in lens cells (data not shown). It is known that the DC5 enhancer is activated by a complex between Sox1/2/3 and δEF3 (14, 16, 17). However, in the case of repression of a tetramerized MCK minimal enhancer activated by MyoD, the NR and CtBP corepressor acted additively (Fig. 4), although the promoter and the transcribed region of the reporter gene were identical in the two experiments. It is therefore likely that multiple mechanisms of repression are differentially utilized depending on the context of the enhancer elements to be repressed. In support of this view, the region of AREB6 (human homologue of δEF1) effective in repression of the human T-cell leukemia virus type 1 promoter includes the corresponding CtBP binding site (12), while an activity of mouse δEF1 involved in repression of Ets-mediated gene activation was assigned to the third portion (23).
δEF1 knockout mice have two major defects, in thymocyte development and in skeleton development. Null mutants exhibit both defects (34), but mutants of the second allele lacking the C-proximal region showed only the thymocyte defect (11), indicating that the portion of δEF1 remaining in the latter mutants is responsible for the regulation of skeleton development. This portion includes the CtBP binding domain identified in this study. It is of interest to determine if CtBP interaction is crucial for the regulatory activities of δEF1 in skeletal development. Analysis of δEF1 knock-in mutants carrying mutations such as PLASSL (Fig. 2) which lack CtBP binding will provide an answer to this intriguing problem.
Binding interaction between CtBP and δEF1.
Adenovirus E1A proteins have PLDLSL or related amino acid sequences in the CtBP binding site (Fig. 2A), and amino acid alterations in this sequence either diminish or eliminate binding of CtBP proteins, arguing for involvement of this short sequence in binding of these proteins (27). In fact, the repressor proteins interacting with CtBP1/2 described so far have such an amino acid sequence motif as an essential element of the interacting site. A number of related motifs in the repressor proteins have been identified as candidate binding sites of CtBP1/2 (19, 22, 35). However, not all amino acid sequences with these motifs will necessarily be the authentic binding sites of CtBP. As shown in Fig. 2C, mutation of δEF1 (PLASSL) totally abolished binding of CtBP1/2, while there are other related motifs, e.g., 745 PLNLSC, present in the δEF1 protein, indicating that the 712 PLDLSL is the sole major binding site of CtBP1/2 in this protein.
Integrity of the major portion of the CtBP proteins seems to be required for establishing binding to the δEF1 protein. This argument stems from the observation that all positive cDNA clones of the initial two-hybrid screen for interaction with the MS region carried most of the coding sequence. In support of this view, division of the coding sequence into halves resulted in total loss of the binding to the MS portion of δEF1 (Fig. 2D).
Function of CtBP proteins in negative regulation.
CtBP is known to be the protein which binds adenovirus E1A and attenuates its transactivation potential. Recently, CtBP1 and -2 have also been identified as binding proteins of a few transcriptional repressors. dCtBP was identified as a binding protein of transcriptional repressors Knirps and Snail (20). Basic helix-loop-helix repressor protein Hairy also binds dCtBP at a site close to the C terminus (21). Examples other than δEF1 among vertebrate nuclear proteins which bind CtBP are BKLF (35) and vertebrate Polycomb homologues XPc and HPC2 (31). As CtBP1 and CtBP2 exhibit transrepression potential when a DNA binding domain is supplied (Fig. 3 and reference 35), it has been speculated that CtBP1 and -2 may act as corepressors of these transcriptional regulators. In this report, we have provided the first clear evidence that CtBP1 and -2 act as corepressors of δEF1 and augment transrepression activity of δEF1.
As discussed above, the requirement of corepressors CtBP1 and -2 in the overall repressor activity of δEF1 seems to depend on the context of the enhancer and on the activator protein which δEF1 antagonizes. It has been reported that CtBP1 binds histone deacetylase (33). It remains to be clarified whether this interaction is involved in the action of CtBP1/2. Of interest is that dCtBP-interacting proteins are often, though not always, those involved in short-range repression (20). The long-range repressor Hairy has an intrinsic repression domain and binding sites of dCtBP and another corepressor, Groucho (21). How these coexistent multiple repression mechanisms allot the function of transcriptional repression associated with a single DNA binding protein represents an important problem in understanding the overall regulatory interactions between the activators and the repressors.
Correlation of the expression of δEF1 and CtBP1/2.
Two CtBP proteins, CtBP1 and CtBP2, with very similar amino acid sequences were identified as δEF1 binding proteins in the mouse. A search of the cDNA database confirms the existence of CtBP2 in addition to the original CtBP (human CtBP1) in humans as well. Individual CtBP proteins are highly conserved in amino acid sequence between the animal species, while the divergence between CtBP1 and CtBP2 is larger. Nevertheless, there are no appreciable differences in binding to δEF1 (Fig. 2) or in transrepression activity (Fig. 3 and 4). The only difference observed between CtBP1 and CtBP2 is the spatial and temporal regulation of their expression. CtBP1 is widely expressed throughout the developmental stages, but CtBP2 is primarily expressed during embryogenesis. It has been reported that in humans, CtBP2 is expressed at a level comparable to that of CtBP1 in multiple tissues (31), which may reflect a species-dependent variation.
Histological examination of CtBP1/2 gene expression in embryos revealed differences between CtBP1 and CtBP2 and a significant correlation with expression of δEF1. In 10.5-day embryos, CtBP2 is prominently expressed in the cranial ganglia, dorsal root ganglia, and posterior-distal portions of the limb bud mesenchyme, the major sites of δEF1 expression around this stage (Fig. 6 and reference 34). Expression of CtBP1 is weaker except in the spinal cord. Along the forming digits of 12.5-day embryos, δEF1 is expressed in the perichondria (34) concomitant with CtBP1/2 (Fig. 6E and F): CtBP1 is expressed through the whole length of digits, while CtBP2 expression is confined to the distal part.
In various functional assays done in this work, CtBP1 and CtBP2 were indistinguishable. Nevertheless, distinct expression specificities found between CtBP1 and CtBP2 could reflect unrecognized differences in their activities. In any event, the high correlation of expression sites of δEF1 and CtBP1/2 strongly argues that interaction with the corepressors is crucial for the regulatory functions of δEF1 in these tissues.
ACKNOWLEDGMENTS
We thank R. Sekido, Y. Kamachi, H. Sasaki, J. Remacle, D. Huylebroeck, and colleagues in this laboratory for stimulating discussions.
This work was supported by research grants from the Ministry of Education, Science and Culture of Japan. T.F. is a recipient of a fellowship for Junior Scientists from the Japan Society for Promotion of Sciences.
REFERENCES
- 1.Aronson B D, Fisher A J, Blechman K, Caudy M, Gergen J P. Groucho-dependent and -independent repression activities of Runt domain proteins. Mol Cell Biol. 1997;17:5581–5587. doi: 10.1128/mcb.17.9.5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cowell I G. Repression versus activation in the control of gene transcription. Trends Biochem Sci. 1994;19:38–42. doi: 10.1016/0968-0004(94)90172-4. [DOI] [PubMed] [Google Scholar]
- 4.Fortini M E, Lai Z, Rubin G M. The Drosophila zfh-1 and zfh-2 genes encode novel proteins containing both zinc-finger and homeodomain motifs. Mech Dev. 1991;34:113–122. doi: 10.1016/0925-4773(91)90048-b. [DOI] [PubMed] [Google Scholar]
- 5.Franklin A J, Jelton T L, Shelton K D, Magnuson M A. BZP, a novel serum-responsive zinc finger protein that inhibits gene transcription. Mol Cell Biol. 1994;14:6773–6788. doi: 10.1128/mcb.14.10.6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujisawa-Sehara A, Nabeshima Y, Komiya T, Uetsuki T, Asakura A, Nabeshima Y. Differential trans-activation of muscle-specific regulatory elements including the myosin light chain box chicken MyoD, myogenin and MRF4. J Biol Chem. 1992;267:10031–10038. [PubMed] [Google Scholar]
- 7.Funahashi J-I, Kamachi Y, Goto K, Kondoh H. Identification of nuclear factor δEF1 and its binding site essential for lens-specific activity of the δ1-crystallin enhancer. Nucleic Acids Res. 1991;19:3543–3547. doi: 10.1093/nar/19.13.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Funahashi J-I, Sekido R, Murai K, Kamachi Y, Kondoh H. δ-Crystallin enhancer binding protein δEF1 is a zinc finger homeodomain protein implicated in postgastrulation embryogenesis. Development. 1993;119:433–446. doi: 10.1242/dev.119.2.433. [DOI] [PubMed] [Google Scholar]
- 9.Genetta T, Ruezinsky D, Kadesch T. Displacement of an E-box-binding repressor by basic helix-loop-helix proteins: implication for B-cell specificity of the immunoglobulin heavy-chain enhancer. Mol Cell Biol. 1994;14:6153–6163. doi: 10.1128/mcb.14.9.6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gray S, Levine M. Transcriptional repression in development. Curr Opin Cell Biol. 1996;8:358–364. doi: 10.1016/s0955-0674(96)80010-x. [DOI] [PubMed] [Google Scholar]
- 11.Higashi Y, Moribe H, Takagi T, Sekido R, Kawakami K, Kikutani H, Kondoh H. Impairment of T cell development in δEF1 mutant mice. J Exp Med. 1997;185:1467–1480. doi: 10.1084/jem.185.8.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ikeda K, Halle J P, Stelzer G, Meisterernst M, Kawakami K. Involvement of negative cofactor NC2 in active repression by zinc finger-homeodomain transcription factor AREB6. Mol Cell Biol. 1998;18:10–18. doi: 10.1128/mcb.18.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ikeda K, Kawakami K. DNA binding through distinct domains of zinc-finger-homeodomain protein AREB6 has different effects on gene transcription. Eur J Biochem. 1995;233:73–82. doi: 10.1111/j.1432-1033.1995.073_1.x. [DOI] [PubMed] [Google Scholar]
- 14.Kamachi Y, Cheah K S E, Kondoh H. Mechanism of regulatory target selection by the SOX high-mobility-group domain proteins as revealed by comparison of SOX1/2/3 and SOX9. Mol Cell Biol. 1999;19:107–120. doi: 10.1128/mcb.19.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kamachi Y, Kondoh H. Overlapping positive and negative regulatory elements determine lens-specific activity of the δ1-crystallin enhancer. Mol Cell Biol. 1993;13:5206–5215. doi: 10.1128/mcb.13.9.5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kamachi Y, Sockanathan S, Liu Q, Breitman M, Lovell-Badge R, Kondoh H. Involvement of SOX proteins in lens-specific activation of crystallin genes. EMBO J. 1995;14:3510–3519. doi: 10.1002/j.1460-2075.1995.tb07357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamachi Y, Uchikawa M, Collignon J, Lovell-Badge R, Kondoh H. Involvement of Sox1, 2 and 3 in the early and subsequent molecular events of lens induction. Development. 1998;125:2521–2532. doi: 10.1242/dev.125.13.2521. [DOI] [PubMed] [Google Scholar]
- 18.Lai Z-C, Rushton E, Bate M, Rubin G M. Loss of function of the Drosophila zfh-1 gene results in abnormal development of mesodermally derived tissues. Proc Natl Acad Sci USA. 1993;90:4122–4126. doi: 10.1073/pnas.90.9.4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nibu Y, Zhang H, Bajor E, Barolo S, Small S, Levine M. dCtBP mediates transcriptional repression by knirps, Krüppel and snail in the Drosophila embryo. EMBO J. 1998;17:7009–7020. doi: 10.1093/emboj/17.23.7009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nibu Y, Zhang H, Levine M. Interaction of short-range repressors with Drosophila CtBP in the embryo. Science. 1998;280:101–103. doi: 10.1126/science.280.5360.101. [DOI] [PubMed] [Google Scholar]
- 21.Poortinga G, Watanabe M, Parkhurst S M. Drosophila CtBP: a Hairy-interacting protein required for embryonic segmentation and Hairy-mediated transcriptional repression. EMBO J. 1998;17:2067–2078. doi: 10.1093/emboj/17.7.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Postigo A A, Dean D C. ZEB represses transcription through interaction with the corepressor CtBP. Proc Natl Acad Sci USA. 1999;96:6683–6688. doi: 10.1073/pnas.96.12.6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Postigo A A, Dean D C. ZEB, a vertebrate homologue of Drosophila Zfh-1, is a negative regulator of muscle differentiation. EMBO J. 1997;16:3935–3943. doi: 10.1093/emboj/16.13.3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Proush Z, Finley R L, Jr, Kidd T, Wainwright S M, Ingham P, Brent R, Ish-Horowicz D. Groucho is required for Drosophila neurogenesis, segmentation, and Sex determination and interacts directly with Hairy-related bHLH protein. Cell. 1994;79:805–815. doi: 10.1016/0092-8674(94)90070-1. [DOI] [PubMed] [Google Scholar]
- 25.Reznikoff C A, Bertram J S, Brankow D W, Heidelberger C. Quantitative and qualitative studies of chemical transformation of cloned C3H mouse embryo cells sensitive to postconfluence inhibition of cell division. Cancer Res. 1973;33:3239–3249. [PubMed] [Google Scholar]
- 26.Rose M D, Winston F, Hieter P. Methods in yeast genetics: a laboratory course manual. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 27.Schaeper U, Boyd J M, Verma S, Uhlmann E, Subramanian T, Chinnadurai G. Molecular cloning and characterization of a cellular phosphoprotein that interacts with a conserved C-terminal domain of adenovirus E1A involved in negative modulation of oncogenid transformation. Proc Natl Acad Sci USA. 1995;92:10467–10471. doi: 10.1073/pnas.92.23.10467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sekido R, Murai K, Funahashi J, Kamachi Y, Fujisawa-Sehara A, Nabeshima Y, Kondoh H. The δ-crystallin enhancer-binding protein δEF1 is a repressor of E2-box-mediated gene activation. Mol Cell Biol. 1994;14:5692–5700. doi: 10.1128/mcb.14.9.5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sekido R, Murai K, Kamachi Y, Kondoh H. Two mechanisms in the action of repressor δEF1: binding site competition with an activator and active repression. Genes Cells. 1997;2:771–783. doi: 10.1046/j.1365-2443.1997.1570355.x. [DOI] [PubMed] [Google Scholar]
- 30.Sekido R, Takagi T, Okanami M, Moribe H, Yamamura M, Higashi Y, Kondoh H. Organization of the gene encoding transcriptional repressor δEF1 and cross-species conservation of its domains. Gene. 1996;173:227–232. doi: 10.1016/0378-1119(96)00185-0. [DOI] [PubMed] [Google Scholar]
- 31.Sewalt R A B, Gunster M J, van der Vlag J, Satijn D P E, Otte A P. C-terminal binding protein is a transcriptional repressor that interacts with a specific class of vertebrate Polycomb proteins. Mol Cell Biol. 1999;19:777–787. doi: 10.1128/mcb.19.1.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sollerbrant K, Chinnadurai G, Svensson C. The CtBP binding domain in the adenovirus E1A protein controls CR1-dependent transactivation. Nucleic Acids Res. 1996;24:2578–2584. doi: 10.1093/nar/24.13.2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sundqvist A, Sollerbrant K, Svensson C. The carboxy-terminal region of adenovirus E1A activates transcription through targeting of a C-terminal binding protein-histone deactylase complex. FEBS Lett. 1998;429:183–188. doi: 10.1016/s0014-5793(98)00588-2. [DOI] [PubMed] [Google Scholar]
- 34.Takagi T, Moribe H, Kondoh H, Higashi Y. δEF1, a zinc finger and homeodomain transcription factor, is required for skeleton patterning in multiple lineages. Development. 1998;125:21–31. doi: 10.1242/dev.125.1.21. [DOI] [PubMed] [Google Scholar]
- 35.Turner J, Crossley M. Cloning and characterization of mCtBP2, a co-repressor that associates with basic Krüppel-like factor and other mammalian transcriptional regulators. EMBO J. 1998;17:5129–5140. doi: 10.1093/emboj/17.17.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Verschueren K, Remacle J E, Collart C, Kraft H, Baker B S, Tylzanowski P, Nelles L, Wuytens G, Su M T, Bodmer R, Smith J C, Huylebroeck D. SIP1, a novel zinc finger/homeodomain repressor, interacts with smad proteins and binds to 5′-CACCT sequences in candidate target genes. J Biol Chem. 1999;274:20489–20498. doi: 10.1074/jbc.274.29.20489. [DOI] [PubMed] [Google Scholar]
- 37.Watanabe Y, Kawakami K, Hirayama Y, Nagano K. Transcription factors positively and negatively regulating the Na, K-ATPase α1 subunit gene. J Biochem. 1993;114:849–855. doi: 10.1093/oxfordjournals.jbchem.a124267. [DOI] [PubMed] [Google Scholar]
- 38.Wilkinson D G. Whole mount in-situ hybridization of vertebrate embryos. In: Wilkinson D G, editor. In situ hybridization: a practical approach. Oxford, England: IRL Press; 1992. pp. 75–84. [Google Scholar]