ABSTRACT
Fungal infections (mycoses) affect over a billion people per year. Approximately, two million of these infections are life-threatening, especially for patients with a compromised immune system. Fungi of the genera Aspergillus, Candida, Histoplasma and Cryptococcus are opportunistic pathogens that contribute to a substantial number of mycoses. To optimize the diagnosis and treatment of mycoses, we need to understand the complex fungal–host interplay during pathogenesis, the fungal attributes causing virulence and how the host resists infection via immunological defenses. In vitro models can be used to mimic fungal infections of various tissues and organs and the corresponding immune responses at near-physiological conditions. Furthermore, models can include fungal interactions with the host–microbiota to mimic the in vivo situation on skin and mucosal surfaces. This article reviews currently used in vitro models of fungal infections ranging from cell monolayers to microfluidic 3D organ-on-chip (OOC) platforms. We also discuss how OOC models can expand the toolbox for investigating interactions of fungi and their human hosts in the future.
Keywords: in vitro model, fungal–host interaction, Aspergillus, Candida, Histoplasma, Cryptococcus
From basic to complex: in vitro models to study interactions between human fungal pathogens and their host.
INTRODUCTION
Human fungal infections lead to approximately 1.5 million deaths worldwide each year, but receive little attention compared with malaria or tuberculosis, which kill a similar number of people on an annual basis (Brown et al. 2012; Bongomin et al. 2017). Over 70% of deaths resulting from fungal infections can be attributed to fungi of the genera Aspergillus, Candida, Cryptococcus and Histoplasma (Brown et al. 2012). These opportunistic fungal pathogens are either normal commensals of the human microbiota or reside in the environment, resulting in constant exposure to pathogenic fungi for humans. Even in immunocompetent human hosts, superficial fungal infections are widespread. Among them, fungal skin diseases are the most common health complications (Vos et al. 2012), and vulvovaginal candidiasis (VVC) affects approximately 70% of women (Gonçalves et al. 2016; Rosati et al. 2020). Such infections are often connected to an imbalance of the bacterial microbiota, for example, after the use of antibiotics that favor fungal overgrowth (Weiss and Hennet 2017)). In addition to superficial infections, opportunistic fungal pathogens can also cause severe life-threatening systemic infections under certain predispositions, like surgery, stem cell transplantation, chemotherapy or HIV/AIDS (Perlroth, Choi and Spellberg 2007; Polvi et al. 2015; Vallabhaneni and Chiller 2016). Considering their clinical significance, suitable models to study opportunistic fungal infections are essential for obtaining insights into disease pathogenesis. Ideally, these models allow the dissection of the molecular details of host–pathogen interactions under physiologically relevant conditions. They should provide sufficient complexity to mimic the different types and stages of infections and predispositions of the host. These models should also be suitable to test experimental therapeutic interventions and allow the evaluation of clinically relevant biomarkers. Here, we review currently used in vitro models to study molecular mechanisms of fungal infections caused by common fungal pathogens, including Aspergillus fumigatus, Candida spp., Cryptococcus neoformans and Histoplasma capsulatum, and provide an outlook about models that will likely expand our toolbox to study fungal–host interactions in the near future.
DISEASE MODELING
To study fungal pathogens and their related diseases, a wide range of models can be used. Commonly, host–pathogen interactions are investigated in animal model organisms such as mice, rats, fish, insects or worms. In vivo models offer the advantage to study host–pathogen interactions in a whole organism, providing the most complex interactions that can be achieved experimentally. However, in addition to critical ethical issues associated with the use of animal models (Robinson et al. 2019), the translation of results from animal experiments to human disease can be hampered by differences in physiology. Another approach is the use of tissue samples or organs from living organisms and their culture in an ex vivo environment that resembles in vivo conditions. These ex vivo models offer the advantage that conditions can be easily manipulated and are often easier to handle than living organisms. A broad overview of ex vivo models to study fungal infection is given by Maciel Quatrin et al. (2019). In vitro experiments are also performed outside of the natural biological environment. Primary cells isolated from tissues and biopsies can be cultured for a limited time or can be immortalized and cultured as cell lines. In vitro models may lack the complexity of in vivo models, but allow ample control over external growth conditions of cells concerning O2 and CO2 saturation, temperature, pH and nutrients. Moreover, it is relatively easy to manipulate as well as to quantitatively and qualitatively assess the metabolism, transcription and protein function of cells, making it possible to work in and test conditions that cannot be studied in in vivo models. It is also possible to introduce or omit different cell types to study the individual impact of different kinds of cells within the system. In vitro models (Fig. 1) range from monolayers in well plates, to transwell systems, 3D tissue structures and complex organ-on-chip (OOC) models (Mosig 2017), which are used to mimic several organs such as the liver (Groger et al. 2016; Jang et al. 2019), lung (Benam et al. 2016; Deinhardt-Emmer et al. 2020) and gut (Shin and Kim 2018; Maurer et al. 2019). OOC models represent the smallest functional entity of an organ as well as a versatile and promising resource to study host–pathogen interactions (Ahadian et al. 2018). However, each model has its specific advantages and disadvantages. The most suitable model is the one that meets the actual needs with high predictability and robustness, depending on the pathogen, the host and the questions to be answered.
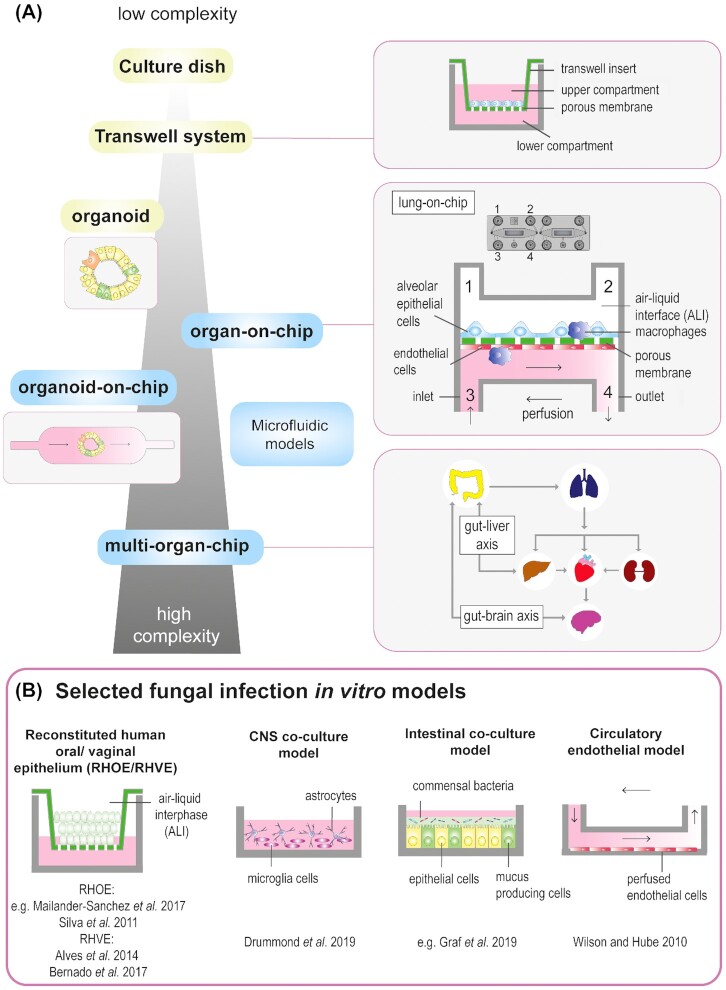
Figure 1.
(A)Evolution of in vitro models from low to high complexity. Culture dish: one cell type cultured in media. Transwell system: transwell inserts separate the culture area into an upper and lower compartment; cells are cultured under static conditions on a porous membrane allowing apical-basal polarization. Organoid: 3D miniature organ generated out of intestinal stem cells. Organ-on-chip (example): 3D lung on-chip model on a microfluidic biochip holding a porous membrane and two individually accessible channels with one inlet and outlet each; pulmonary epithelial cells are cultured in the upper compartment in an air–liquid interface; and endothelial cells in the lower compartment are perfused with cell culture medium enabling the removal of metabolites. Organoid-on-chip: maturation of organoids within a dynamic culture environment. Principle of a multi-organ-on-chip: interconnected organ-on-chip models of gut and liver, or gut and brain or other combinations of lung, intestine, liver, brain and/or kidneys. Such combinations can, for example, mimic certain steps of fungal dissemination throughout the body. The intestine and lung serve as primary infection sites. (B)Selected in vitro models to study host–fungal interactions. 3D reconstituted human oral (RHOE) or vaginal (RHVE) epithelium grown at an air–liquid interface. Central nervous system (CNS) co-culture model including microglia cells and astrocytes. Intestinal co-culture model including epithelial cells, goblet cells and bacteria. Circulatory model with perfused endothelial cells.
We discuss the fungal–host interactions in different biological niches (Fig. 2). We review in vitro models used to mimic infection routes and highlight relevant findings that contributed to expand our knowledge on fungal infections. Because the immune system plays a major role during fungal infections, the interplay of fungi and immune cells is discussed in the first part, followed by sections covering the respiratory tract, the gastrointestinal tract, the vaginal mucosa, the bloodstream and the blood–brain barrier (BBB).
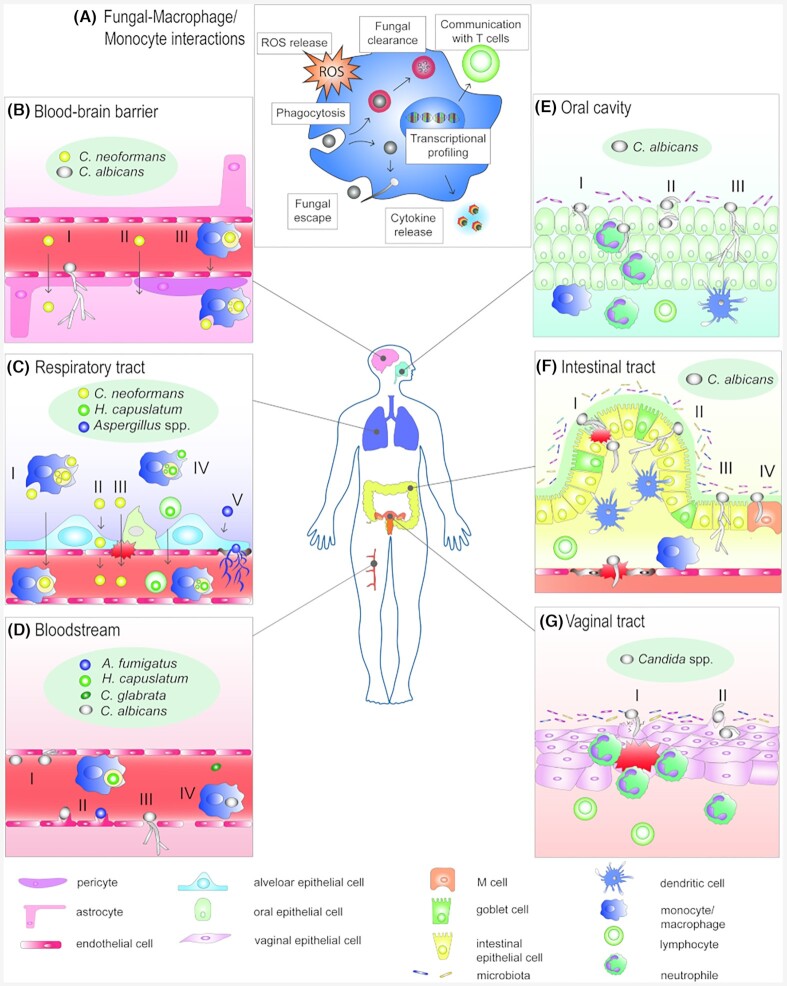
Figure 2.
Fungal–host interactions during fungal diseases that are mimicked by in vitro infection models discussed in this review. (A) Fungal–monocyte/macrophage interactions resulting in several effector mechanisms that contribute to immunity against fungal infections (ROS: reactive oxygen species). (B)C. neoformans and C. albicans can cross the BBB via transcytosis (I);C. neoformans can overcome the barrier paracellularly (II) or use macrophages as shuttles (macrophages as ‘Trojan horse’) (III). (C) In the lung, C. neoformans and H. capsulatum induce their own phagocytosis by innate immune cells; they can replicate intracellularly and use host cells as shuttles to reach the blood stream and subsequently escape (I and IV); evasion of C. neoformans via transcytosis (II) or crossing of C. neoformans through a compromised epithelium (III). Aspergillus spp. form hyphae, can invade endothelial cells and enter the bloodstream (V). (D)Candida spp. can escape the blood circulation after adhesion to endothelial cells (I). Candida spp. and A. fumigatus can be endocytosed (II); Candida spp. can also use fenestrated endothelium as an escape route (III) or use leukocytes as shuttles (IV). (E) In the oral cavity, C. albicans hyphae can actively penetrate the epithelium (I) and/or invade via induced endocytosis (II) or translocate paracellularly (III). (F) In the intestine, C. albicans can actively penetrate the epithelium by hyphal growth (I), translocate paracellularly (II), invade without damaging the host cell (III) or translocate via M cells by inducing endocytosis (IV). (G) In the vaginal tract, C. albicans hyphae can actively penetrate the epithelium (I) or invade via induced endocytosis (II), thereby attracting neutrophils.
STUDYING FUNGAL INTERACTIONS WITH THE IMMUNE SYSTEM
A properly functioning immune system is crucial for resistance against infections with fungal pathogens. Individuals with a compromised immune system are more susceptible to invasive fungal diseases, whereas detrimental, improper or hypersensitive immune reactions can also contribute to disease (Romani 2004; Wheeler, Limon and Underhill 2017). Thus, a protective host response against opportunistic fungal pathogens has to be specific, tightly regulated and effective. However, pathogenic fungi have evolved a series of mechanisms to deal with and evade the immune system. Knowledge of both aspects is crucial for the design of therapeutic strategies aiming to strengthen appropriate responses and suppress detrimental ones (Armstrong-James et al. 2017). We will discuss (i) the different immune cells involved in antifungal host defense, (ii) the different roles these cells play in antifungal immunity and (iii) different models and readouts that can be used to study the efficiency of the host response to pathogenic fungi.
Immune cells involved in antifungal host defense
A healthy and efficient immune system is fundamental to cope with the environmental fungi we encounter on a daily basis and to deal with the fungi we harbor as commensals. This antifungal immunity relies on the innate immune system represented by cells such as macrophages, monocytes, neutrophils, natural killer (NK) cells and dendritic cells (DCs) as well as the adaptive immune system, in particular on T helper cell responses. The importance of these different types of immune cells becomes apparent when they are dysfunctional or absent. For example, a compromised innate immune system due to immunosuppressive therapy predisposes not only to invasive candidiasis (Lionakis 2014) but also aspergillosis (Herbrecht et al. 2012). While the innate immune system plays a role in host defense against cryptococcosis (Voelz and May 2010), patients with a compromised adaptive immune response due to HIV infections are particularly susceptible (Warkentien and Crum-Cianflone 2010). In contrast to Candida,Aspergillus and Cryptococcus species, Histoplasma species more commonly cause infections in healthy individuals (Köhler et al. 2017). Nevertheless, a compromised innate as well as adaptive immune response increases the susceptibility to histoplasmosis (Akram and Koirala 2020).
Tissue-resident macrophages and monocyte-derived macrophages especially play an essential role against invasive candidiasis (Austermeier et al. 2020), whereas alveolar macrophages (AMs) are essential for clearance of fungi like Aspergillus, Cryptococcus or Histoplasma species that enter our body via the airways (Newman 2005; Xu and Shinohara 2017). Neutropenia is a common risk factor for aspergillosis and invasive candidiasis, showing the crucial role of neutrophils in antifungal host defense (Herbrecht et al. 2000). Dendritic cells (DCs) are crucial for activation of the adaptive immune system. Dysfunctions of the adaptive immune system like the reduced CD4+ T cell function in AIDS patients increase the susceptibility for infections with C. albicans, A. fumigatus, C. neoformans or H. capsulatum (van de Veerdonk and Netea 2010). Interestingly, this predisposition manifests as mucosal C. albicans infections, in particular oropharyngeal candidiasis (OPC), but systemic C. albicans infections are also observed under such conditions (Fidel 2011). This is believed to be closely connected to the crucial roles of T helper responses in orchestrating oral mucosal resistance to infection (Gaffen and Moutsopoulos 2020; Scheffold, Bacher and LeibundGut-Landmann 2020). Antifungal immunity in the brain is connected to microglia that are the resident macrophage-like cells of the central nervous system (CNS), which show strong responses to fungal species like C. albicans (Blasi et al. 1991) and C. neoformans (Barluzzi et al. 1998). The C-type lectin receptor signaling adaptor CARD9 is expressed by microglia cells and its deficiency is associated with fungal brain infections (Drummond and Lionakis 2019). NK cells also exhibit antifungal effects (Schmidt, Tramsen and Lehrnbecher 2017) and a delayed NK cell reconstitution (e.g. after allogeneic stem cell transplantation) is associated with a higher risk of invasive aspergillosis (Weiss et al. 2020).
Antifungal effector functions during host defense against fungal pathogens
After the recognition of pathogen-associated molecular patterns (PAMPs) via pathogen recognition receptors (PRRs), supported by opsonization, innate immune cells mount responses to counteract the invading fungi. At early stages of infection, macrophages detect and engulf fungal pathogens (Gilbert, Wheeler and May 2014) (Fig. 2A). In addition, through the release of cytokines and chemokines they recruit and activate other immune cells. When neutrophils migrate to the site of infection, they act against fungal pathogens through phagocytosis, oxidative bursts and NETosis (Gazendam et al. 2016; Urban and Nett 2019). The production of reactive oxygen species (ROS) by phagocytes can kill fungal pathogens, like C. albicans, directly (Grondman et al. 2019) or impact LC3-mediated phagocytosis during defense against A. fumigatus (Sprenkeler, Gresnigt and van de Veerdonk 2016). This is highlighted, for example, by the fact that chronic granulomatous disease (CGD) patients, incapable of producing ROS, are highly susceptible to aspergillosis (Segal et al. 2000). DCs represent the bridge to activate the adaptive immune system via antigen processing and presentation to T-cells (LeibundGut-Landmann et al. 2007). T-cell differentiation can influence infection in different ways. Th1 cells augment the innate immune function through the release of IFNγ (Lionakis and Levitz 2018), which increases the microbicidal capacity of macrophages (Netea et al. 2015). Th17 cells release proinflammatory cytokines such as IL-17 and IL-22, which mediate recruitment of neutrophils and induce production of antimicrobial peptides (Khader, Gaffen and Kolls 2009; Conti et al. 2016) (Fig. 2A). The importance of these T-cell types for antifungal defense is evident in corresponding knock-out mice that have an increased susceptibility to disseminated C. albicans infections (Balish et al. 1998; Huang et al. 2004), but also show a striking susceptibility to mucosal infections. Th2 responses can result in a detrimental immune response, manifesting in a higher susceptibly to disseminated C. albicans infections (Haraguchi et al. 2010) or an aberrant immune response to A. fumigatus spores connected to allergic bronchopulmonary aspergillosis (ABPA) (Knutsen and Slavin 2011). T regulatory cells can suppress inflammatory responses and are highly beneficial to prevent immunopathology in the case of ABPA (Montagnoli et al. 2006), but also allow C. albicans persistence in the gastrointestinal tract (De Luca et al. 2007).
In vitro models to study interactions between fungi and immune cells
The interactions between fungi and the different effector functions of the immune system can be easily studied in vitro using cell lines (Table S1A, Supporting Information) and primary immune cells (Table S1B, Supporting Information). Cell lines have the advantage of easy handling and provide highly reproducible results. The availability of many reporter cell lines and the possibilities to generate transgenic/knockout cell lines represent valuable resources that allow the study of highly conserved mechanisms in the immunology against fungal infections. Nevertheless, central cellular processes such as pyroptosis, apoptosis and autophagy are considerably different or modified in cancer cell lines. Over the past few years, the essential role of these processes in shaping antifungal immunity has become increasingly clear (Kanayama and Shinohara 2016; Sprenkeler, Gresnigt and van de Veerdonk 2016; Dominguez-Andres et al. 2017; Evans, Sundaramurthy and Frickel 2018; O'Meara and Cowen 2018; Gonçalves et al. 2020; Thak et al. 2020; Weerasinghe and Traven 2020). Therefore, primary cells offer the highest similarity to the physiological situation. Primary immune cells are commonly isolated from peripheral human blood. By density gradient centrifugation, peripheral blood mononuclear cells (PBMCs) can be separated from erythrocytes and granulocytes (Munoz and Leff 2006). An important aspect to consider when using primary cells is that strong donor variation and even seasonal differences can influence experimental outcomes (Ter Horst et al. 2016). However, genetic differences between donors can also be exploited to analyze the impact of specific genotypes on the antifungal immune response (Lionakis et al. 2013; Smeekens et al. 2013; Matzaraki et al. 2017; Gresnigt et al. 2018b; Jaeger et al. 2019a,b). In these functional genomic approaches, immune cells of large cohorts of volunteers are screened for variation in specific immunological effectors such as cytokine release, ROS release or fungal killing. After genotyping the donors, the results of immunological phenotypes can be stratified based on the corresponding genotype. This knowledge on the influence of common genetic variations on the antifungal host response can provide valuable information about the role of certain genes in antifungal host defense. Combined with genetic association studies, functional genomics can be used to validate the impact of identified variants on immune pathways and susceptibilities to infections. In this way, crucial roles have been identified for CX3CR1 and its role in host defense against of systemic candidiasis (Lionakis et al. 2013), as well as for the SIGLEC15 receptor in the susceptibility to vulvovaginal candidiasis (Jaeger et al. 2019b). Conversely, knowledge about genetic variations that influence critical antifungal host defense pathways can lead to the discovery of genetic susceptibilities. In this way NOD2 variants were found to increase resistance to invasive aspergillosis (Gresnigt et al. 2018b).
Macrophages
Interactions between macrophages/macrophage-like cells and fungal pathogens have been studied using cell lines like J774A.1, RAW, Ana-1, U937, BV-2 and THP-1 (Table S1A, Supporting Information). Such cell lines allow the generation of reporter constructs that can be used to monitor the activation of specific immune pathways. In this way, the importance of RAB-GTPases for maturation of C. albicans-containing phagosomes has been demonstrated (Bain et al. 2014; Okai et al. 2015). Another option is the use of macrophages derived from murine bone marrow cells and differentiated in vitro (BMDMs) (Table S1B, Supporting Information). A major advantage of this approach is the possibility to isolate BMDMs from mice with different genetic backgrounds (e.g. gene knockout or transgenic mice), thus providing a toolbox to obtain in-depth knowledge about key players of the host immune response during fungal infections. Such cells from knockout mice have been widely used to investigate, for example, inflammasome activation in the response to C. albicans (Kasper et al. 2018),C. neoformans (Guo et al. 2014) and A. fumigatus (Karki et al. 2015). In addition to BMDMs, human monocyte-derived macrophages (MDMs) can be used for in vitro studies. In such experiments, monocytes are isolated from PBMCs or whole blood and can be differentiated in vitro into a wide range of functionally different MDMs (Xue et al. 2014). MDMs have been used in numerous studies to dissect not only cytokine release, inflammasome activation, oxidative burst, phagocytosis and phagosome maturation after confrontation with fungi but also escape and survival mechanisms of fungi during these interactions (Smith, Dixon and May 2015; Gresnigt et al. 2018b; Kasper et al. 2018; O'Meara et al. 2018; Friedrich et al. 2019) (Table S1B, Supporting Information).
However, undifferentiated monocytes are also used to investigate how these cells are differentially activated (Halder et al. 2016; Dominguez-Andres et al. 2017; Klassert et al. 2017; Camilli et al. 2018; Leonhardt et al. 2018). The stimulation of monocytes using PAMPs such as β-glucan can induce epigenetic reprogramming, which alters the response to secondary C. albicans stimulation (Quintin et al. 2012), a concept known as innate immune memory or ‘trained immunity’. In contrast, the response to C. albicans can also be hampered by the induction of innate immune tolerance by PAMPs such as lipopolysaccharide (Grondman et al. 2019). Over the past years it has become increasingly evident that cell metabolism is linked with immune cell functionality. Global as well as targeted profiling of metabolic pathways in primary immune cells, especially monocytes and macrophages, have been used to uncover immunometabolism in response to fungi (Dominguez-Andres et al. 2017; Gonçalves et al. 2020; Weerasinghe and Traven 2020).
Since pathogenic fungi often colonize and infect specific organs, the corresponding tissue macrophages offer the highest physiological relevance. For example, specific cells lines such as the murine alveolar macrophage cell lines MH-S (Mattern et al. 2015) and AMJ2-C11 (Pitangui Nde et al. 2015) are used to study fungal pathogens that cause pulmonary infections (Table S1A, Supporting Information). Alternatively, primary alveolar macrophages can be used to study the immune response of pulmonary fungal infections ex vivo. Though, the limited availability of these cells makes it challenging to obtain sufficient numbers for experiments. Nevertheless, protocols are available to obtain large numbers of AMs from bronchoalveolar lavage (BAL) (Busch et al. 2019) or resected lung tissue (Nayak et al. 2018). Similarly, peritoneal macrophages have been used to study the interactions with Candida spp. (Ifrim et al. 2016; Shimamura et al. 2019). Because peritoneal macrophages are easier to obtain in larger quantities than AMs, they have also been used for interaction studies with H. capsulatum (primarily infecting the lung) (Youseff et al. 2012; Huang et al. 2018; Shen et al. 2018) (Table S1B, Supporting Information). To dissect fungal interactions with immune cells in the brain, BV-2 microglia cells (Blasi et al. 1990) (Table S1A, Supporting Information) were co-cultured with astrocytes to demonstrate that candidalysin induces IL-1β release, which in turn mediates neutrophil recruitment (Drummond et al. 2019) (Fig. 1B).
Interaction studies with macrophages revealed mechanisms enabling fungal cells to evade macrophage phagocytosis or to escape from phagosomes. Masking of cell wall epitopes can prevent the detection of A. fumigatus, C. albicans and H. capsulatum by macrophages (Rappleye, Eissenberg and Goldman 2007; Aimanianda et al. 2009; Ballou et al. 2016). Morphological changes such as titan cell formation by C. neoformans (Okagaki and Nielsen 2012) or filamentation by A. fumigatus and C. albicans influence phagocytosis efficiency (Lewis et al. 2012; Erwig and Gow 2016; Maxson et al. 2018). Additionally, these fungi can inhibit phagosome acidification or phagosome maturation to prevent intracellular killing. These processes are reviewed in detail by Gilbert, Wheeler and May (2014) and Seider et al. (2010).
Irrespectively of the immune cell type used, numerous readouts are available to study interactions between fungi and cells of the immune system. Transcriptional profiling has provided indispensable insights into the interplay between immune cells and fungal pathogens. Specifically, dual-species transcriptional profiling has helped to elucidate key features of the adaptations of fungal cells in response to immune cells and vice versa (Niemiec et al. 2017; Munoz et al. 2019). Given the crucial role of phagocytes in fungal clearance, protocols established to investigate phagocytosis and phagosome maturation are common (Fig. 2A). Using live-cell microscopy, phagocytosis and viability dynamics can be studied on a kinetic scale involving multiple phagocytes (Smith, Dixon and May 2015; Gresnigt et al. 2018a; Kasper et al. 2018; Lim et al. 2018; Guimaraes et al. 2019; Seoane et al. 2020). For example, a struggle for glucose availability between macrophages and C. albicans was demonstrated to be crucial in dictating inflammasome activation (Tucey et al. 2020).Candida albicans cells however, can filament thereby complicating clearance through phagocytosis (Erwig and Gow 2016). Phagocytosis and phagosome maturation can also be examined in detail on a single-cell level (Bain et al. 2014; Okai et al. 2015; Westman et al. 2018). Such studies have contributed to the understanding of the role of phagosome–lysosome fusion in maintaining phagosome integrity while fungal cells filament inside the phagosome (Westman et al. 2020). Apart from live cell imaging, phagocytes can also be fixed at specific time-points to investigate the co-localization of proteins to the phagosome using immunofluorescence staining. In this way, LC3-associated phagocytosis has been investigated as a crucial pathway to improve phagocytosis efficiency of H. capsulatum and A. fumigatus (Huang et al. 2018; Kyrmizi et al. 2018). Using a similar approach, a key role has been shown for flotillin-dependent microdomains or lipid rafts in phagosome formation for efficient host defense against A. fumigatus (Schmidt et al. 2020).
Natural killer (NK) cells
Primary NK cells can be obtained from PBMCs by different isolation kits (Wang et al. 2017). NK cells have been studied alone or in co-culture with other immune cells and have been observed to have direct antifungal capacity against C. neoformans through the release of perforins (Wiseman et al. 2007). The recognition of β1,3-glucan through the NKp30 receptor was identified to trigger and enhance the killing of C. albicans and C. neoformans by NK cells (Li et al. 2018). Other in vitro studies revealed an exhausted phenotype of NK cells, when they degranulate in contact with A. fumigatus (Santiago et al. 2018). NK cell activation in response to Candida species has been observed to occur indirectly by cross talk with monocytes (Marolda et al. 2020). Similarly, for A. fumigatus, crosstalk between NK cells and DCs was found to mediate DC activation (Weiss et al. 2018). Further, direct antifungal effects of NK-cells against A. fumigatus have been associated with release of IFNγ (Bouzani et al. 2011) (Table S1B, Supporting Information).
Neutrophils
Using hypotonic lysis of erythrocytes or other gradient solutions like PolymorphPrep© (Progen, Heidelberg, Germany) (Degel and Shokrani 2010), primary neutrophils can be isolated from PBMCs to investigate their interaction with fungi. Neutrophils can act as phagocytes, but can also form neutrophil extracellular traps (NETs) and release cytokines in the presence of fungal cells. These features were studied intensively in vitro (Urban et al. 2006; Bruns et al. 2010; Rocha et al. 2015; Sun and Shi 2016; Dasari et al. 2018; Thompson-Souza et al. 2020). By studying phagocytosis, killing, NETosis and cytokine release, spleen tyrosine kinase (Syk) was identified as a crucial mediator for inducing antifungal effector mechanisms against various Candida species (Negoro et al. 2020). Another aspect is to monitor how these phagocytes migrate to the site of infection. Chemotaxis assays using specialized in vitro systems (Richards et al. 2004; Chen 2005; Thunström Salzer et al. 2018) can be used to elucidate this process in the context of fungal infections (Coenjaerts et al. 2001; Drummond et al. 2015; Rieber et al. 2016) (Table S1B, Supporting Information). ROS release or oxidative bursts in response to fungal pathogens can be assessed not only in neutrophils (Boyle et al. 2011; Liu et al. 2018) but also in monocytes (Wellington, Dolan and Krysan 2009; Brunel et al. 2018) and macrophages (Wolf et al. 1987; Youseff et al. 2012; Sun et al. 2014; Arce Miranda et al. 2019) (Fig. 2A; Table S1B, Supporting Information). Using a modified model, in which C. albicans cells are grown in clusters on poly-l-lysine coated glass slides, neutrophils were observed to form ‘swarms’ to efficiently use oxidative stress mechanisms to attack C. albicans (Hopke et al. 2020).
Dendritic cells, T-cells and whole blood models
Virtually all immune cell types are being employed to study transcriptional responses to fungal pathogens (Smeekens et al. 2013; Hellwig et al. 2016; Van Prooyen et al. 2016; Niemiec et al. 2017) as well as cytokine and chemokine responses (Coady and Sil 2015; Becker et al. 2016; Marischen et al. 2018) to fungal pathogens (Fig. 2A). Often such studies involve crosstalk between different immune cell types such as antigen-presenting cells and cells of the adaptive immune system. PBMCs are frequently used due to their composition of innate and adaptive immune cells and allow the study of innate host responses (Becker et al. 2016; Alvarez-Rueda et al. 2020), but also T-cell mediated responses such as Th1, Th17, Th2 and Tregs (Zielinski et al. 2012; Gresnigt et al. 2013; Becker et al. 2015; Raijmakers et al. 2017; Page et al. 2018; Vogel et al. 2018) (Fig. 2A). For example, using PBMCs, the type I interferon pathway was identified to play a crucial role in C. albicans defense (Smeekens et al. 2013). Interactions between DCs and T-cells were used to investigate how the adaptive immune response is polarized through antigen presentation, co-stimulation and the cytokine environment (van der Does et al. 2012; Stephen-Victor et al. 2017). DC maturation can be examined in transwell systems (Lother et al. 2014) or by profiling maturation features via flow cytometry (Pietrella et al. 2005; Hefter et al. 2017; Vivas et al. 2019). For interaction studies including a wide range of immune cell types, whole blood models were used to gain information about fungal killing (Hunniger et al. 2014), transcriptional responses (Dix et al. 2015; Kämmer et al. 2020), cytokine release (Oesterreicher, Eberl and Zeitlinger 2019) and platelet interactions (Fréalle et al. 2018; Eberl et al. 2019) (Table S1B, Supporting Information).
STUDYING RESPIRATORY TRACT INFECTIONS WITH ASPERGILLUS, HISTOPLASMA AND CRYPTOCOCCUS SPP.
In the respiratory tract fungal pathogens such as A. fumigatus, H. capsulatum and C. neoformans can cause infections in predisposed hosts. Since the major biological niche of these fungi is the environment, fungal elements (mostly conidia or yeast) are frequently inhaled by the human host. The healthy immune system can clear these inhaled fungal elements, whereas immunocompromised individuals or patients with pre-existing pulmonary conditions may fail to clear fungi and have a higher risk to develop aspergillosis, histoplasmosis or cryptococcosis. The clinical manifestations of these fungal diseases, however, are very diverse. Infections with pathogenic Aspergillus species can develop differently, depending on the immune reaction and underlying lung pathology (Soubani and Chandrasekar 2002; van de Veerdonk et al. 2017). While a compromised immune response can result in invasive pulmonary aspergillosis, pre-existing lung injury can lead to the development of an aspergilloma and a chronic or hyper inflammatory response. Such responses can also provoke allergic bronchopulmonary aspergillosis (Kosmidis and Denning 2015). In immunocompromised patients, specifically patients suffering from AIDS, C. neoformans can cause either pulmonary cryptococcosis or can disseminate into other organs after an (asymptomatic) pulmonary infection (Setianingrum, Rautemaa-Richardson and Denning 2019). Cryptococcusneoformans cells can be engulfed by AMs and DCs and can survive within the phagolysosome, proliferate and eventually escape via non-lytic exocytosis (vomocytosis) (Fig. 2C I). Vomocytosis was also observed for C. albicans (Bain et al. 2012), C. krusei (García-Rodas et al. 2011), A. nidulans and A. fumigatus (Gresnigt et al. 2018a). Intracellular survival is one key strategy of C. neoformans to disseminate from the respiratory tract (Coelho, Bocca and Casadevall 2014). Other translocation routes involve fungal cells crossing the epithelial border via transcytosis (Fig. 2C II) or a direct migration through areas where the epithelial lining has been compromised (Fig. 2C III) (Denham and Brown 2018). Histoplasma capsulatum can cause pulmonary histoplasmosis, and similar to C. neoformans, it can evade the immune system by hiding inside AMs (Ray and Rappleye 2019). Following growth and replication, it can induce apoptosis facilitating further dissemination within the bloodstream and lymphatic organs (Fig. 2C IV) (Long et al. 2003; Mihu and Nosanchuk 2012; Pitangui Nde et al. 2015). In contrast to H. capsulatum and C. neoformans, which grow as yeast during infection, A. fumigatus proliferates as hyphae in the lung, allowing deep tissue invasion (Fig. 2C V).
Simple in vitro models mimicking lung infections
To mimic the alveolar environment, the pulmonary epithelial cell line A549, originating from a human alveolar cell carcinoma (Lieber et al. 1976), is frequently used to study pathogenicity attributes including adhesion (Gravelat et al. 2010; Pitangui et al. 2012; Teixeira et al. 2014), endocytosis (Liu et al. 2016), epithelial detachment (Kogan et al. 2004; Bertuzzi et al. 2014) and epithelial damage (Ejzykowicz et al. 2010; Bertuzzi et al. 2014). These studies revealed crucial roles for the A. fumigatus transcription factors PacC (Bertuzzi et al. 2014) and DvrA (Ejzykowicz et al. 2010) to mediate tissue invasion and damage. In addition, A549 cells were used to dissect pulmonary epithelial IL-8 responses to C. neoformans and H. capsulatum (Barbosa et al. 2007; Alcantara et al. 2020), and shed light on how different A. fumigatus isolates differentially regulate gene expression of epithelial cells (Watkins et al. 2018) (Table S2, Supporting Information). To examine the fungal translocation through the pulmonary epithelium, transwell models with different modifications have been employed (Fig. 1A).
Complex in vitro models mimicking lung infections
Models that combine A549 cells with DCs (Morton et al. 2018) or a bilayer of human pulmonary artery endothelial cells (HPAECs) with (Morton et al. 2014) or without DCs (Hope et al. 2007; Belic et al. 2018) were utilized to model the cellular complexity in the alveolus and the cellular cytokine response to fungal infections. The translational capacity of such a model was reflected in a study that validated the measurement of galactomannan as a biomarker of fungal infection and antifungal efficacy in vitro (Hope et al. 2007). These models have also been employed for microscopy-based analyses, gene expression analysis and analysis of immune activation to gain insights into the host–Aspergillus interactions at the alveolar epithelial interface (Table S2, Supporting Information).
To more closely resemble the physiological situation, primary human bronchial or small airway epithelial (HBE, SAE) cells were used to study proinflammatory epithelial cytokine responses to C. neoformans infections (Guillot et al. 2008). These cells differentiate when cultured at an air-liquid interphase (ALI) into lung epithelium and were also used to assess the host response to A. fumigatus conidia. Transcriptome and proteome analyses revealed the upregulation of apoptosis, autophagy, translation and cell cycle pathways as well as the downregulation of complement and coagulation pathways (Toor et al. 2018). The combination of differentiated pulmonary epithelial cells with DCs and macrophages provides an even more complex model, which allows the study of the interplay between fungal cells, the epithelium and the immune system (Chandorkar et al. 2017). As an alternative strategy to investigate Aspergillus spp. infections, bronchial mucosal tissue resected from cancer patients was used. Using this ex vivo model, adhesion, invasion, damage and structural changes of the epithelium were investigated (Amitani and Kawanami 2009). Although the latter model represents human physiology, its applicability is limited by the difficulty of obtaining patient material. Besides confounding factors, such as therapies and medication, inter-individual differences may impact the validity of this model and the ability to obtain reproducible results.
Lung-on-chip models
Most lung models used so far are cultured statically and thus are not subjected to shear stress. Further, these models rarely consider the impact of additional members of the microbial community, such as the lung microbiota in the infection process. A number of lung-on-chip models have been established that reflect additional physiological key features of the lung. A ‘breathing’ alveolus-on-chip is mimicked by stretching and contraction of a membrane using a vacuum, which leads to an increased uptake of nanoparticles of the epithelium and transport to the vasculature (Huh et al. 2010; Stucki et al. 2018). Mechanostimulation represents an important biophysical cue since the stretching of the lungs influences repair mechanisms in damaged epithelial cells and might also play a significant role during fungal invasion (Desai, Chapman and Waters 2008). Deinhardt-Emmer and colleagues established an alveolus-on-chip model that harbored immune cells and consisted of two compartments. In the upper compartment, lung epithelial cells differentiated into the two types of alveolar epithelial cells and were separated by a porous membrane from an endothelial lining, subjected to flow in the lower compartment (Deinhardt-Emmer et al. 2020) (Fig. 1A). Although this model was not used to dissect fungal–host interactions so far, it revealed new insights about the interplay of S. aureus and influenza virus at the alveolar–capillary interface. During co-infection, increased inflammatory responses were observed including cytokine expression and loss of barrier function similar to severe clinical outcomes of patients with bacterial-viral superinfections (Deinhardt-Emmer et al. 2020). Other platforms have used human alveolar epithelial cells (hAEpCs), and also integrated neutrophils (Huh et al. 2010; Benam et al. 2016; Jain et al. 2018; Zhang et al. 2018). Future models can be colonized with (additional) members of the pulmonary microbiome to investigate the interplay with fungi, which can contribute to progression of pulmonary fungal infections (Kolwijck and van de Veerdonk 2014). Taken together, current lung-on-chip models can produce a microenvironment resembling the in vivo physiology by imitating an ALI, mechanical strain and immune responses. This can facilitate the establishment of sophisticated pulmonary-infection models.
STUDYING COLONIZATION AND INFECTION OF THE ORAL CAVITY, THE INTESTINAL TRACT AND VAGINAL TRACT BY CANDIDA SPP.
In the oral cavity, the intestinal-, and vaginal tract, Candida spp. normally live as harmless commensal yeasts. However, some opportunistic Candida spp. can cause infections. These range from mucocutaneous infections such as OPC (Millsop and Fazel 2016) and VVC (Rosati et al. 2020) to invasive candidiasis (Pappas et al. 2018). Diverse predispositions, like immunosuppression, an impaired barrier function and an imbalanced microbiota, are prerequisites to enable infection of Candida species. However, both predisposition and protection by an adjusted immune response differ between the specific types of infections. In the following sections we discuss in vitro models used to study C. albicans and C. glabrata interactions with the host in three different niches of the human body.
Studying Candida spp. infections of the oral cavity
OPC occurs mostly in combination with the use of broad-spectrum antibiotic therapy and immune suppression, e.g. through HIV/AIDS, chemotherapy or radiation therapy. Further, neonates, diabetic and elderly individuals are more susceptible (Patil et al. 2015). Candida albicans is the most prevalent species, but also other Candida species like C. glabrata,C. dubliniensis,C. krusei,C. kefyr,C. parapsilosis,C. stellatoidea and C. tropicalis can be found in oral lesions (Millsop and Fazel 2016). Candida albicans mainly interacts with the oral epithelium by invading cells via active penetration (Fig. 2E I) and/or induced endocytosis (Fig. 2E II) (Phan et al. 2007; Dalle et al. 2010; Wachtler et al. 2011a; Sheppard and Filler 2014; Naglik et al. 2017), or invasion of the tissue by degradation of E-cadherin, thereby disrupting the epithelial barrier (Fig. 2E III) (Villar et al. 2007). In vivo, the uppermost layer of the oral epithelium consists of stratified squamous epithelium, followed by a basal membrane and fibroblasts in the lamina propria.
Simple in vitro models mimicking oral infections
To study Candida–host interactions of the oral cavity, oral epithelial cells are commonly used. TR146 cells are derived from a squamous cell carcinoma of the buccal mucosa (Rupniak et al. 1985) and used to investigate invasion (Puri et al. 2019), damage (Wilson et al. 2014; Meir et al. 2018) and gene expression (Schaller et al. 1998; McCall, Kumar and Edgerton 2018; Meir et al. 2018). The TR146 model has contributed significantly to the understanding of C. albicans pathogenicity by showing that the peptide toxin candidalysin is responsible for the capacity of C. albicans hyphae to cause damage (Moyes et al. 2016). The same model was used to demonstrate that candidalysin also activates epithelial proinflammatory responses through the epithelial growth factor receptor (Ho et al. 2019) and its synergistic signaling with IL-17 (Verma et al. 2017). Immortalized oral mucosal cells (OKF6/TERT-2) (Dickson et al. 2000) have also been used to study epithelial transcriptional responses (Liu et al. 2015), to visualize C. albicans invasion (Wollert et al. 2012) and to demonstrate that invasion is, in part, mediated through endocytosis (Solis et al. 2017; Swidergall et al. 2018). The same cell line was used to show that damage is mediated through white cells in contrast to opaque cells (Solis et al. 2018). Furthermore, Epha2 was identified as an epithelial cell pattern recognition receptor for fungal β-glucans, activating a signal cascade that results in a proinflammatory and antifungal response (Swidergall et al. 2018).
Tongue cells derived from a squamous cell carcinoma (SCC15) represent a third cell type used to dissect interactions of C. albicans with the oral epithelium (Lindberg and Rheinwald 1990). Similar to the studies discussed above, SCC15 cells were used to investigate epithelial damage (Kumar et al. 2015), invasion (Villar et al. 2007) and cytokine release (Dongari-Bagtzoglou and Kashleva 2003) (Table S3A, Supporting Information).
Complex in vitro models mimicking oral infections
In addition to monolayer models (Fig. 1A), organotypic 3D models known as reconstituted human oral epithelium (RHOE) are commonly used to study oral Candida spp. infections due to their histological similarity to physiological oral epithelium. In these RHOE models, TR146 cells are cultured on a polycarbonate filter at an ALI with culture medium on the basal side, resulting in a multilayer model with differentiated cells (Fig. 1B). This model has been used to study epithelial damage (Silva et al. 2011; Mailander-Sanchez et al. 2017) and fungal (Spiering et al. 2010) or host cell gene expression (Wagener, Mailander-Sanchez and Schaller 2012) (Table S3A, Supporting Information). In addition, the model was used to show enhanced invasion and tissue damage during co-infection of C. albicans and C. glabrata (Silva et al. 2011). Because fungal biofilm formation is crucial for the development of caries and OPC, the RHOE model has also been used to analyze the expression of C. albicans virulence genes associated with biofilm formation (Nailis et al. 2010). Similar RHOE models exist, containing collagen embedded fibroblasts from mice and oral mucosal cells OKF6/TERT-2 cells, differentiated at an ALI (Dongari-Bagtzoglou and Kashleva 2006a,b). Since the interplay with the oral microbiota plays an essential role for the maintenance of a commensal state of C. albicans or for development of OPC (Montelongo-Jauregui and Lopez-Ribot 2018), the organotypic 3D models were also used to study interactions between C. albicans and bacteria. For example, antagonistic interactions between Lactobacillus rhamnosus and C. albicans were dissected (Mailander-Sanchez et al. 2017). Furthermore, fungal-induced dysbiosis after chemotherapy (Bertolini et al. 2019) and synergistically increased tissue damage during interactions with S. mutans (Diaz et al. 2012) were observed. Additionally, biofilm formation of C. albicans and C. glabrata after chemotherapeutic treatment was examined in the latter organotypic 3D model (Sobue et al. 2018). The model was further ‘humanized’ by using human fibroblasts and spontaneously immortalized keratinocytes to analyze interactions between C. albicans and S. aureus (de Carvalho Dias et al. 2018) (Table S3A, Supporting Information).
In vitro modeling of C. albicans stomatitis
C. albicans mediated stomatitis, an inflammatory reaction of the oral mucosa, is a major complication for users of removable dental prostheses, but also common in smokers or patients suffering from diabetes mellitus (Salerno et al. 2011; Javed et al. 2017; Alzayer et al. 2018). To model this oral infection, primary human palate epithelial cells (HPECs) were used to study the host response to C. albicans in terms of apoptosis, nitric oxide production (Casaroto et al. 2019) and mucosal gene expression (Offenbacher et al. 2019). Similarly, a combination of TR146 cells and primary fibroblasts was used for adhesion and gene expression studies (Morse et al. 2018) (Table S3A, Supporting Information).
Mucosa-on-chip models
Monolayer or multilayered mucosal models commonly feature a perpendicular configuration. This vertical culture arrangement hampers the individual monitoring of different cell layers by microscopy, and resolution decreases in deeper layers. A horizontal organization of cell layers was applied in a mucosa-on-chip model (Rahimi et al. 2018) consisting of microchambers, which were aligned in parallel and interconnected by pores. A central subepithelial chamber harbored a collagen hydrogel with gingival fibroblasts, while keratinocytes were seeded into the pores connecting the luminal and subepithelial compartment. The luminal chamber can be microfluidically perfused to imitate saliva and saliva flow, which is an important contributor to epithelial barrier integrity. A further refinement for both static and microfluidic models can include an endothelial lining and immune cells such as dendritic Langerhans cells, which are almost exclusively found in stratified squamous epithelium and have been shown to react to Candida species (Upadhyay et al. 2013).
Studying Candida spp. colonization of the intestinal tract and intestinal translocation
Both C. albicans and C. glabrata colonize the human intestinal tract (Hallen-Adams and Suhr 2017). The gut represents the main reservoir of fungi, especially C. albicans, that can cause disseminated and systemic infections (Gouba and Drancourt 2015). In these life-threatening infections, the fungus overcomes the intestinal epithelium, which forms a barrier between the intestinal lumen and the sterile tissues of the human body. During this process, termed translocation, the fungus employs several mechanisms including active penetration (Fig. 2F I), paracellular translocation (Fig. 2F II) or migration through the intestinal epithelial layer without damaging the host cells (Fig. 2F III) (Allert et al. 2018; Basmaciyan et al. 2019). Certain predispositions favor fungal overgrowth and translocation: antibiotics induce an imbalance of the microbiota and cytostatic therapy or abdominal surgery, which compromise the barrier function (Pfaller and Diekema 2007). To better understand the conditions that keep C. albicans commensal or drive the commensal-to-pathogen shift, the interactions between C. albicans and the intestinal barrier are studied extensively to find ways to prevent or reverse this shift (Kumamoto, Gresnigt and Hube 2020).
Simple in vitro models mimicking intestinal infections
Monolayers of cell lines originating from colorectal adenocarcinomas are widely used (Fig. 1A). The most common cell lines are Caco-2 and HT-29. Caco-2 cells differentiate spontaneously into a polarized monolayer with characteristic villi and tight junctions after 12 days of culture (Fogh, Wright and Loveless 1977). These cells were used to demonstrate that damage to the intestinal epithelium induced by C. albicans relies on a combination of adhesion-mediated contact sensing, tissue invasion through hyphal extension and damage by the expression of pathogenicity factors (Wachtler et al. 2011a). Interactions with non-pathogenic yeast cells that can antagonize C. albicans pathogenicity were examined (Lohith and Anu-Appaiah 2018; Kunyeit et al. 2019). Furthermore, receptor signaling pathways (Mao et al. 2019), induction of defensins (Gacser et al. 2014), impact on tight junctions (Goyer et al. 2016) and the potential of epithelial cells to discriminate between yeast and hyphal morphologies (Schirbel et al. 2018) are processes that can be analyzed in this model. A subclone of the Caco-2 cell line, C2BBe1, was often used in in vitro systems due to its more homogeneous brush boarder expression (Peterson and Mooseker 1992). A model of C2BBe1 cells cultured in transwell systems (Fig. 1A) was instrumental to elucidate important virulence requirements of translocation through the epithelial barrier and revealed a key role for candidalysin by mediating necrotic cell damage that allowed transcellular translocation (Allert et al. 2018). Additionally, using this model, a MAPK/NFκB mediated epithelial response to C. albicans infection was shown to increase epithelial resistance (Bohringer et al. 2016) (Table S3B, Supporting Information).
Essential features of C. albicans pathogenicity like adhesion, invasion and damage were also studied using the HT-29 cell line (Deng et al. 2015; Garcia et al. 2018). A methotrexate treatment of HT-29 cells, transformed these cells into mucus-secreting goblet cells (HT-29-MTX) (Lesuffleur et al. 1990). These mucus-secreting cells were instrumental in demonstrating the role of mucus in suppressing virulence-associated attributes of C. albicans, such as hypha formation (Kavanaugh et al. 2014).
Complex in vitro models mimicking intestinal infections
As the intestinal epithelium consists of a myriad of cell types, combinations of different cell lines have been employed to more accurately mimic the in vivo situation. For example, a combination of Caco-2 cells and Raji B cells (human Burkitt's lymphoma) was used to study the interaction of C. albicans with an epithelial barrier including M-cells, which demonstrated M-cells as a preferred cell type for translocation via induced endocytosis (Fig. 2F IV) (Albac et al. 2016). In general, most in vitro models investigate C. albicans in its pathogenic state. To limit the pathogenicity of C. albicans and mimic commensalism, a mixture of C2BBe1 cells and the mucus-producing HT-29-MTX cells were colonized with L. rhamnosus to establish a basic ‘commensal’ model (Fig. 1B). Using this model, a damage reduction was observed in the presence of mucus and bacteria, both antagonizing C. albicans pathogenicity by reducing filamentation, proliferation and inducing shedding that physically separates hyphae from host cells (Graf et al. 2019) (Table S3B, Supporting Information).
Intestine-on-chip models
Although 2D intestinal models mimic the fundamental physiological structures of the intestinal tissue such as mucus production, M-cells and brush border epithelium, they do not reflect the unique 3D architecture of the intestinal epithelial tissue consisting of villi and crypts. Cells in these models are cultured statically and are not subjected to the peristaltic movement characteristic for the intestine. In addition, in vitro models often lack immune cells, which convey tolerance towards commensals and trigger inflammatory responses when pathogens inflict damage to the intestinal lining. A number of intestine-on-chip models have been developed that recapitulate some of these key physiological features (Bein et al. 2018). In these models, Caco-2 cells grow out and form villi-like structures when grown on a membrane and exposed to shear stress (Kim and Ingber 2013). Microfluidic intestine models often include endothelial cells adjacent to epithelial cells in an individually perfused compartment. The luminal and the vascular compartment are separated by a porous membrane to facilitate transmigration of cells and cell communication. Innate immune cells such as monocytes can be implemented in the endothelial layer and differentiated into macrophages and DC-like cells, which tolerate inflammatory triggers in the intestinal lumen, but elicit a strong inflammatory response when a systemic infection is mimicked (Maurer et al. 2019). In this model, C. albicans invasion of the epithelial layer and subsequent invasion of the bloodstream compartment in the presence and absence of the commensal bacterium L. rhamnosus were investigated. Patient-derived colon epithelial cells are difficult to access, but can sufficiently be maintained in microfluidic platforms and produce a mucus layer resembling the in vivo thickness (Sontheimer-Phelps et al. 2020). 3D intestine-on-chip models will be valuable tools to uncover the role of commensals and their products, as well as host immune responses in the yeast-to-hypha transition of C. albicans in the future (Table S3B, Supporting Information).
Intestinal organoids
Apart from intestine-on-chip models, human intestinal organoids have emerged as a valuable disease-modeling tool. Human intestinal organoids can be grown from adult stem cells extracted from intestine biopsies or induced pluripotent stem cells (iPSCs) (Rahmani et al. 2019) to form 3D organotypic structures by self-organization and resemblance of key embryonic signaling in vitro (Clevers 2016) (Fig. 1A). Intestinal organoids show a villus and crypt-like architecture with epithelial cells facing inwards, creating a lumen as an enclosed space (Sato et al. 2009; Spence et al. 2011). Organoid models face similar challenges like OOC platforms, such as additional cell types, immune cells, endothelial cells and extracellular matrix components that need to be incorporated to create a physiological microenvironment for cell differentiation and tissue development. However, mesenchymal cells and neural crest cells have already been successfully implemented in these models (Workman et al. 2017). Unlike microfluidic OOC models, stem cell-derived organoids currently lack perfusion and therefore deprive epithelial cells of shear stress and removal of metabolites. An idea has emerged that aims at combining self-assembling organoids with microfluidic OOC techniques, termed ‘Organoids-on-a-Chip’ (Park, Georgescu and Huh 2019) (Fig. 1A). The technique encompasses the maturation of organoids within a dynamic culture environment allowing the control of nutrient supply, establishment of biochemical gradients vital for self-organization of the organoids and the introduction of additional cell types.
Studying Candida spp. infections of the vaginal mucosa
The vaginal mucosa represents another commensal niche of Candida spp. in the human body. VVC affects 70–75% of women in their reproductive age (Sobel 2007). Antibiotic treatment is a strong predisposing factor for VVC (Shukla and Sobel 2019), most likely due to the induced dysbiosis of the vaginal microbiome. C.albicans is the most prominent species isolated from VVC, followed by C. glabrata (Makanjuola, Bongomin and Fayemiwo 2018). The interactions between Candida spp. and the vaginal epithelium, as well as the vaginal microbiota, are complex (Pekmezovic et al. 2019; Kalia, Singh and Kaur 2020), and invasion of the epithelium occurs through active penetration (Fig. 2G I) and induced endocytosis (Fig. 2G II), while neutrophils are attracted simultaneously.
Simple in vitro models mimicking vaginal infections
The VK2/E6E7 cell line originates from healthy human vaginal mucosal tissue and was immortalized by retroviral transduction (Fichorova, Rheinwald and Anderson 1997). This cell line was used to demonstrate synergistic interactions between C. albicans and streptococci (Pidwill et al. 2018) and a role for autophagy machinery in the survival of epithelial cells during C. albicans infection (Shroff and Reddy 2018). In addition, Type-I IFN signaling was elucidated to increase resistance of the epithelium to C. albicans infection (Li et al. 2017). By introducing high glucose conditions, this model has been used to demonstrate that the association of VVC in diabetes patients might be related to increased adhesion of C. albicans through a potential interaction with ICAM-1 (Mikamo et al. 2018). Another cell line, A431, originates from a vaginal epidermoid carcinoma. This cell line was used to investigate inflammatory cytokine responses and damage of A431 cells induced by candidalysin (Richardson et al. 2018). Additionally, the cell line was utilized to evaluate the impact of azole antifungal treatment on damage induced by C. albicans spp. (Wachtler, Wilson and Hube 2011b) (Table S3C, Supporting Information).
Complex in vitro models mimicking vaginal infections
A reconstituted vaginal epithelium (RHVE) is available as an alternative model. RHVE is based on A431 cells, cultivated at an ALI, similar to the previously described RHOE (Fig. 1B). RHVE was used to demonstrate that C. albicans facilitates interactions of C. glabrata with the vaginal epithelium by increasing fungal colonization, invasion and damage of epithelial cells during co-infection (Alves et al. 2014). Furthermore, the adaptation of C. glabrata to an acidic vaginal environment was investigated using RHVE (Bernardo et al. 2017) (Table S3C, Supporting Information).
Organ-on-chip models mimicking vaginal infections
Several OOC models for the female reproductive tract are available, predominantly to mimic the physiology of the endometrium, the uterus or the placenta (Mancini and Pensabene 2019). Possible OOC models of the vaginal mucosa should comprise stratified squamous epithelium and perfused endothelial cells, separated by a porous membrane. Immune cells can easily be integrated to recapitulate relevant inflammatory responses during hyphal invasion of the epithelium such as neutrophil recruitment.
In vivo, the vaginal tract harbors a microbiota that consist to a large extent of Lactobacillus species. Although predicted, it is not entirely clear whether the microbiota actually has a protective effect against Candida spp. infection and if so, whether diversity among microbial communities leads to a higher degree of protection (Cassone 2015).
STUDYING FUNGAL BLOODSTREAM INFECTION AND CROSSING OF THE BBB
Vascular infection models
Fungal dissemination into the bloodstream is a major driver for the development of multi-organ infections or sepsis. Aspergillus fumigatus, H. capsulatum and C. neoformans can enter the bloodstream after crossing the pulmonary alveolar epithelium (Fig. 2C), whereas C. albicans reaches the bloodstream mostly via the intestinal tract (Fig. 2F). Central venous catheters, surgery and parenteral nutrition represent additional entry routes, especially for Candida species (Hashemi Fesharaki et al. 2018). To exit the blood circulation and invade other organs, fungi interact with the endothelial lining of the blood vessels (Fig. 2D), which can be simulated by human umbilical vein endothelial cells (HUVECs) (Jaffe et al. 1973). Although access to umbilical cords is limited, high amounts of cells can be isolated from a single umbilical cord and stored frozen for several experiments (Crampton, Davis and Hughes 2007). HUVECs were used to dissect C. albicans adhesion to the endothelial lining (Fig. 2D I), for example, it was shown that a certain hyphal length is crucial for adhesion in a circulatory in vitro model that simulated physiological capillary blood pressure (Wilson and Hube 2010) (Fig. 1B). Following adhesion, three mechanisms to pass the endothelial barrier were discovered. Attached Candida cells can be endocytosed by endothelial cells (Phan et al. 2005; Liu et al. 2016) (Fig. 2D II), a process that depends on a complex formation including endothelial cell septin 7 (SEP7) and N-cadherin (Phan et al. 2013). Endocytosis was also described for A. fumigatus, independent of its morphology (Kamai et al. 2006) (Fig. 2D II). In addition, Candida spp. can cross the endothelial barrier via paracellular translocation (Fig. 2D III) or via leucocytes following engulfment (Fig. 2D IV) (Filler and Sheppard 2006; Grubb et al. 2008). It is likely that similar Trojan horse transport mechanisms following engulfment by mononuclear cells are exploited by intracellularly persistent H. capsulatum (Gilbert, Wheeler and May 2014) (Fig. 2D IV) as it already has been shown for C. neoformans (Coelho et al. 2019).
The ability of different C. albicans mutants to damage HUVECs was leveraged to identify virulence factors that are important for fungal dissemination (Sanchez et al. 2004). Similarly, the transcription factor DvrA was identified as crucial for endothelial damage induced by A. fumigatus (Ejzykowicz et al. 2010). Besides, the proteome profile of HUVECs was investigated during infection with A. fumigatus (Neves et al. 2017) and C. neoformans (Wang et al. 2011), indicating alterations that contribute to fungal invasion. Transcriptional profiling of HUVECs revealed the upregulation of genes involved in chemotaxis, stress response, angiogenesis and inhibition of apoptosis in response to C. albicans (Barker et al. 2008). A proinflammatory immune response associated with the release of TNF in HUVECs was reported after infections with C. albicans (Orozco, Zhou and Filler 2000) and A. fumigatus (Kamai et al. 2009; Neves et al. 2017). In addition, it was shown that neutrophils protect endothelial cells against C. albicans-induced damage in a co-culture model with HUVECs and neutrophils (Edwards et al. 1987) (Table S4, Supporting Information).
Blood–brain barrier
Whereas cerebral infections with Candida spp. (Drummond et al. 2015), Aspergillus spp. (Rieber et al. 2016) or Histoplasma spp. (Schestatsky et al. 2006) are rare, meningitis is the most prominent complication during cryptococcosis (Srikanta, Santiago-Tirado and Doering 2014). Cerebral infections are induced when fungi cross the BBB, a part of the neurovascular unit (NVU). Other than the endothelial lining, the NVU consists of pericytes, forming a scaffold for endothelial cells together with the basal lamina. Endfeet of astrocytes provide a connection to neurons and microglia (van der Helm et al. 2016). A physical barrier between the blood circulation and the brain tissue is maintained by an intact NVU via zona occludens proteins and claudins.
Simple in vitro models mimicking the BBB
Immortalized human brain vascular endothelial cells (HBMEC and HCMEC/D3) are commonly used for BBB models, whereas primary cells are not frequently used due to insufficient availability and loss of phenotype during culturing (Oddo et al. 2019). The HBMEC and HCMEC/D3 cell lines are especially suitable to model the BBB because of their expression of tight junction proteins, receptors and transporters (Weksler, Romero and Couraud 2013; Oddo et al. 2019). They can be cultured as monolayers on transwell inserts or cell culture plates and infected with C. albicans (Jong et al. 2001), A. fumigatus (Patel et al. 2018) or C. neoformans (Aaron et al. 2018) and used for transcytosis (Aaron et al. 2018), gene expression (Lahiri et al. 2019) and barrier integrity studies (Patel et al. 2018). For example, it was demonstrated that C. neoformans and C. albicans can pass the BBB via transcytosis (Fig. 2B I). True hyphae of C. albicans are associated with endocytosis by endothelial cells (Liu et al. 2011) (Fig. 2B I). Cryptococcus neoformans, however, was shown to also translocate paracellularly (Fig. 2B II) and use macrophages as a shuttle to cross the BBB using the Trojan horse mechanism mentioned above (Charlier et al. 2009; Santiago-Tirado et al. 2017) (Fig. 2B III). This mechanism was visualized and analyzed in detail using a co-culture model of HCMEC/D3 cells and THP-1 cells or primary monocytes (He et al. 2016; Santiago-Tirado et al. 2017) (Table S4, Supporting Information).
BBB-on-chip models
2D transwell models of the BBB can be valuable tools to gain insights into how fungi invade the CNS. However, current models lack some key properties of the NVU. For example, endothelial cells need to experience shear stress to trigger the establishment of a barrier that limits Na+ and Cl− ions efflux and influx (Oddo et al. 2019). Furthermore, to mimic the physiological situation more closely, the model should contain multiple cell types of the NVU such as astrocytes, pericytes and neurons since their communication influences each other's growth, differentiation and permeability (Abbott, Ronnback and Hansson 2006). A range of microfluidic BBB-on-Chip models has recently been developed, recapitulating the blood flow by perfusion of the endothelium in realistic dimensions and geometry and integration of various NVU cell types (Griep et al. 2013; Raasch et al. 2016; Maoz et al. 2018). In models using one cell type, HUVECs in astrocyte-conditioned medium or HCMEC/D3 cells have been cultured in a single perfused channel (Yeon et al. 2012; Griep et al. 2013; Englert et al. 2016). Using a CNS angiogenesis model comprising endothelial cells, pericytes, astrocytes and lung fibroblasts, it was demonstrated that a low vascular permeability can be achieved by co-culturing the different NVU cell types (Lee et al. 2020). These microfluidic BBB models can contribute to investigating the role of additional cell types of the NVU and shear stress in the transmigration of fungi across the BBB. Moreover, the implementation of innate immune cells would enable the simulation of inflammatory responses in the brain tissue following fungal invasion (Table S4, Supporting Information).
FUTURE DIRECTIONS
Interconnecting organ-on-chip systems to study fungal dissemination
Although the multiple infection models reviewed here have been and will be very useful tools to study fungal infections, we can expect a new generation of complex in vitro system based on OOC platforms. In fact, individual OOC systems can be combined to recapitulate multi-organ cross communication in an enclosed microfluidic network (Luni, Serena and Elvassore 2014). These platforms have the potential to investigate fungal infections not only at a single-organ level, but also at the multi-organ level, including systemic immune responses (Fig. 1A). The complexity of systemic immune reactions was only addressed in animal models until recently. Multi-organ-on-chip (MOC) models expand the toolbox with systems having a purely human genetic background to circumvent the problem of interspecies transferability. A range of MOC platforms have been developed that connect two or more organs such as the liver and intestine (Zhang et al. 2009; Chen, Miller and Shuler 2018; Ramme et al. 2019). MOC models provide the opportunity to study the dissemination of fungi throughout the body. It will allow (to mimic) tracking dissemination of Candida spp. from the intestine to the liver and kidney, the key target organs of disseminated candidiasis (Lionakis et al. 2011), or dissemination of A. fumigatus, C. neoformans and H. capsulatum from the lung to the brain, which has not been possible in vitro so far. An additional aspect to be elucidated using MOC models is the relationship between dysbiosis in the intestine resulting in overgrowth of C. albicans and concomitant biochemical changes in the brain or the liver (gut–brain axis and gut–liver axis, respectively) (Burrus 2012; Yang et al. 2017). However, MOC systems are still in their infancy and there are many obstacles to overcome. A current challenge is to scale the organs to their relative physiological size (Lee and Sung 2017; Rogal, Probst and Loskill 2017). Current MOC systems are mostly used for toxicity screening of drugs and chemicals and are constructed in a way to be suitable for this particular application (Rogal, Probst and Loskill 2017). MOC models dedicated for fungal studies may take into account other criteria, e.g. the distance between distinct tissues, the number of integrated immune cells, and possibilities to prevent adherence of fungi to tubing and subsequent clogging, to be applicable as tools.
Human induced pluripotent stem cells as another cell source for fungal in vitro systems
The in vitro models discussed in this review rely on primary cells and cell lines. Human induced pluripotent stem cells (hiPSC) are an alternative source of cells and are highly relevant for biomedical research (Raasch et al. 2019). hiPSC can be generated by reprogramming adult tissue cells, such as fibroblasts, to an embryonic-like pluripotent state (Takahashi and Yamanaka 2006). Once reprogrammed, they can be differentiated into virtually all cell types except extra-embryonic cell types. Therefore, they offer the opportunity to establish OOC systems containing various cell types originating from a single donor. However, current models often combine hiPSC with primary cells and cell lines. Taking the BBB as an example, Brown and colleagues cultured HBMEC, glutamatergic neurons differentiated from iPSC, primary pericytes and astrocytes in a two-chamber model. The resulting system consisted of a brain compartment, which is separated from perfused vasculature by a porous membrane (Brown et al. 2015).
hiPSC are also utilized for the establishment of ‘patient-on-chip’ models to mimic genetic predispositions. Aspergillosis is a common complication of patients suffering from asthma and cystic fibrosis (CF) (Knutsen and Slavin 2011) or CGD (Leiding and Holland 1993); CARD9 and STAT1 mutations predispose for C. albicans CNS (Drummond et al. 2019) and mucocutaneous infections (van de Veerdonk et al. 2011), respectively, and diabetes mellitus is a common predisposition for histoplasmosis (Lockhart and Guarner 2019). Furthermore, intestinal fungi have been tightly connected to inflammatory bowel diseases (Leonardi, Li and Iliev 2018). Future OOC models might be able to reflect these predispositions by implementing hiPSC generated from patients bearing these diseases. Alternatively, specific mutations associated with the disease can be reproduced in hiPSC. For example, they have been successfully differentiated into macrophages and lung epithelial cells that carry mutations associated with CF (Pollard and Pollard 2018) and CGD (Brault et al. 2017). Although there has been substantial progress in OOC systems incorporating hiPSC, caution should be exercised: Protocols for differentiation require optimization and standardization, especially the understanding of factors promoting differentiation needs improvement. Differentiation might differ under static and dynamic conditions (Luni, Serena and Elvassore 2014; Rogal, Probst and Loskill 2017). Standardization of these aspects is crucial to guarantee reproducibility of findings from different labs.
CONCLUDING REMARKS
To study human fungal infections on a higher level of complexity, expertise of fungal infection biology and the OOC platforms needs to be combined. This will ensure studies in the most suitable in vitro model, providing conditions akin to the in vivo situation. For example, 3D intestine-on-chip models will be valuable tools to uncover the role of microbial commensals and their products, as well as the host immune responses to a local yeast-to-hypha transition of C. albicans. In the future, it would be favorable to make use of experience gained with MOC systems to mimic and follow fungal dissemination throughout the body and evaluate novel therapeutic strategies addressing fungal infections.
ACKNOWLEDGEMENT
We thank Jakob Sprague for critical reading of the manuscript.
Supplementary Material
Contributor Information
Antonia Last, Department of Microbial Pathogenicity Mechanisms, Leibniz Institute for Natural Product Research and Infection Biology—Hans Knoell Institute, Beutenbergstrasse 11a, 07745, Jena, Germany.
Michelle Maurer, Center for Sepsis Control and Care (CSCC), Jena University Hospital, Am Klinikum 1, 07747, Jena, Germany; Institute of Biochemistry II, Jena University Hospital, Nonnenplan 2,07743, Jena, Germany.
Alexander S. Mosig, Center for Sepsis Control and Care (CSCC), Jena University Hospital, Am Klinikum 1, 07747, Jena, Germany; Institute of Biochemistry II, Jena University Hospital, Nonnenplan 2,07743, Jena, Germany.
Mark S. Gresnigt, Junior Research Group Adaptive Pathogenicity Strategies, Leibniz Institute for Natural Product Research and Infection Biology—Hans Knoell Institute, Beutenbergstrasse 11a, 07745, Jena, Germany.
Bernhard Hube, Department of Microbial Pathogenicity Mechanisms, Leibniz Institute for Natural Product Research and Infection Biology—Hans Knoell Institute, Beutenbergstrasse 11a, 07745, Jena, Germany; Institute of Microbiology, Friedrich Schiller University, Neugasse 24, 07743, Jena, Germany.
SUPPLEMENTARY DATA
Supplementary data are available at FEMSRE online.
FUNDING
MSG was supported by a Humboldt Research Fellowship for Postdoctoral Researchers by the Alexander von Humboldt Foundation, a Research Grant 2019 from the European Society of Clinical Microbiology and Infectious Diseases (ESCMID), and the German Research Foundation (Deutsche Forschungsgemeinschaft—DFG) Emmy Noether Programm (Project no. 434385622/GR5617/1-1). BH was supported by the European Union Horizon 2020 research and innovation program under the Marie Sklodowska-Curie grant agreement no. 812969 (FunHoMic), the DFG project Hu 532/20-1, project C1 within the Collaborative Research Centre (CRC)/Transregio (TRR) 124 FungiNet, the Leibniz Association Campus InfectoOptics SAS-2015-HKI-LWC, the Leibniz Research Alliance Infections'21 and the Wellcome Trust (grant 215599/Z/19/Z). BH, ASM, MM and AL were supported by the Center for Sepsis Control and Care (CSCC)/Bundesministerium für Bildung und Forschung (BMBF, grant no. 01EO1002). ASM received funding by the European Commission through Actions Marie Skłodowska-Curie (MSCA) Innovative Training Network EUROoC (grant no. 812954). BH and ASM were supported by funding through the Cluster of Excellence ‘Balance of the Microverse’, funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany's Excellence Strategy—EXC 2051, Project ID 390713860.
Conflict of Interest
None declared.
REFERENCES
- Aaron PA, Jamklang M, Uhrig JPet al. The blood–brain barrier internalises Cryptococcus neoformans via the EphA2-tyrosine kinase receptor. Cell Microbiol. 2018;20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbott NJ, Ronnback L, Hansson E. Astrocyte-endothelial interactions at the blood–brain barrier. Nat Rev Neurosci. 2006;7:41–53. [DOI] [PubMed] [Google Scholar]
- Ahadian S, Civitarese R, Bannerman Det al. Organ-on-a-chip platforms: a convergence of advanced materials, cells, and microscale technologies. Adv Healthc Mater. 2018;7. [DOI] [PubMed] [Google Scholar]
- Aimanianda V, Bayry J, Bozza Set al. Surface hydrophobin prevents immune recognition of airborne fungal spores. Nature. 2009;460:1117–21. [DOI] [PubMed] [Google Scholar]
- Akram SM, Koirala J. Histoplasmosis. Treasure Island, FL: StatPearls Publishing, 2020. [PubMed] [Google Scholar]
- Albac S, Schmitz A, Lopez-Alayon Cet al. Candida albicans is able to use M cells as a portal of entry across the intestinal barrier in vitro. Cell Microbiol. 2016;18:195–210. [DOI] [PubMed] [Google Scholar]
- Alcantara C, Almeida BR, Barros Bet al. Histoplasma capsulatum chemotypes I and II induce IL-8 secretion in lung epithelial cells in distinct manners. Med Mycol. 2020;58:1169–77. [DOI] [PubMed] [Google Scholar]
- Allert S, Forster TM, Svensson CMet al. Candida albicans-induced epithelial damage mediates translocation through intestinal barriers. mBio. 2018;9:e00915–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Rueda N, Rouges C, Touahri Aet al. In vitro immune responses of human PBMCs against Candida albicans reveals fungal and leucocyte phenotypes associated with fungal persistence. Sci Rep. 2020;10:6211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves CT, Wei XQ, Silva Set al. Candida albicans promotes invasion and colonisation of Candida glabrata in a reconstituted human vaginal epithelium. J Infect. 2014;69:396–407. [DOI] [PubMed] [Google Scholar]
- Alzayer YM, Gomez GF, Eckert GJet al. The impact of nicotine and cigarette smoke condensate on metabolic activity and biofilm formation of Candida albicans on acrylic denture material. J Prosthodont. 2018;29:173–8. [DOI] [PubMed] [Google Scholar]
- Amitani R, Kawanami R. Interaction of Aspergillus with human respiratory mucosa: a study with organ culture model. Med Mycol. 2009;47 Suppl 1:S127–31. [DOI] [PubMed] [Google Scholar]
- Arce Miranda JE, Baronetti JL, Sotomayor CEet al. Oxidative and nitrosative stress responses during macrophage–Candida albicans biofilm interaction. Med Mycol. 2019;57:101–13. [DOI] [PubMed] [Google Scholar]
- Armstrong-James D, Brown GD, Netea MGet al. Immunotherapeutic approaches to treatment of fungal diseases. Lancet Infect Dis. 2017;17:e393–402. [DOI] [PubMed] [Google Scholar]
- Austermeier S, Kasper L, Westman Jet al. I want to break free: macrophage strategies to recognize and kill Candida albicans, and fungal counter-strategies to escape. Curr Opin Microbiol. 2020;58:15–23. [DOI] [PubMed] [Google Scholar]
- Bain JM, Lewis LE, Okai Bet al. Non-lytic expulsion/exocytosis of Candida albicans from macrophages. Fungal Genet Biol. 2012;49:677–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain JM, Louw J, Lewis LEet al. Candida albicans hypha formation and mannan masking of β-glucan inhibit macrophage phagosome maturation. mBio. 2014;5:e01874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balish E, Wagner RD, Vázquez-Torres Aet al. Candidiasis in interferon-gamma knockout (IFN-gamma-/-) mice. J Infect Dis. 1998;178:478–87. [DOI] [PubMed] [Google Scholar]
- Ballou ER, Avelar GM, Childers DSet al. Lactate signalling regulates fungal β-glucan masking and immune evasion. Nat Microbiol. 2016;2:16238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa FM, Fonseca FL, Figueiredo RTet al. Binding of glucuronoxylomannan to the CD14 receptor in human A549 alveolar cells induces interleukin-8 production. Clin Vaccine Immunol. 2007;14:94–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker KS, Park H, Phan QTet al. Transcriptome profile of the vascular endothelial cell response to Candida albicans. J Infect Dis. 2008;198:193–202. [DOI] [PubMed] [Google Scholar]
- Barluzzi R, Brozzetti A, Delfino Det al. Role of the capsule in microglial cell–Cryptococcus neoformans interaction: impairment of antifungal activity but not of secretory functions. Med Mycol. 1998;36:189–97. [PubMed] [Google Scholar]
- Basmaciyan L, Bon F, Paradis Tet al. “Candida albicans interactions with the host: crossing the intestinal epithelial barrier”. Tissue Barriers. 2019;7:1612661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker KL, Aimanianda V, Wang Xet al. Aspergillus cell wall chitin induces anti- and proinflammatory cytokines in human PBMCs via the Fc-γ receptor/Syk/PI3K pathway. mBio. 2016;7:e01823–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker KL, Gresnigt MS, Smeekens SPet al. Pattern recognition pathways leading to a Th2 cytokine bias in allergic bronchopulmonary aspergillosis patients. Clin Exp Allergy. 2015;45:423–37. [DOI] [PubMed] [Google Scholar]
- Bein A, Shin W, Jalili-Firoozinezhad Set al. Microfluidic organ-on-a-chip models of human intestine. Cell Mol Gastroenterol Hepatol. 2018;5:659–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belic S, Page L, Lazariotou Met al. Comparative analysis of inflammatory cytokine release and alveolar epithelial barrier invasion in a transwell((R)) bilayer model of mucormycosis. Front Microbiol. 2018;9:3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benam KH, Villenave R, Lucchesi Cet al. Small airway-on-a-chip enables analysis of human lung inflammation and drug responses in vitro. Nat Methods. 2016;13:151–7. [DOI] [PubMed] [Google Scholar]
- Bernardo RT, Cunha DV, Wang Cet al. The CgHaa1-regulon mediates response and tolerance to acetic acid stress in the human pathogen Candida glabrata. G3 (Bethesda). 2017;7:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolini M, Ranjan A, Thompson Aet al. Candida albicans induces mucosal bacterial dysbiosis that promotes invasive infection. PLoS Pathog. 2019;15:e1007717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertuzzi M, Schrettl M, Alcazar-Fuoli Let al. The pH-responsive PacC transcription factor of Aspergillus fumigatus governs epithelial entry and tissue invasion during pulmonary aspergillosis. PLoS Pathog. 2014;10:e1004413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasi E, Barluzzi R, Bocchini Vet al. Immortalization of murine microglial cells by a v-raf/v-myc carrying retrovirus. J Neuroimmunol. 1990;27:229–37. [DOI] [PubMed] [Google Scholar]
- Blasi E, Mazzolla R, Barluzzi Ret al. Microglial cell-mediated anti-Candida activity: temperature, ions, protein kinase C as crucial elements. J Neuroimmunol. 1991;34:53–60. [DOI] [PubMed] [Google Scholar]
- Bohringer M, Pohlers S, Schulze Set al. Candida albicans infection leads to barrier breakdown and a MAPK/NF-κB mediated stress response in the intestinal epithelial cell line C2BBe1. Cell Microbiol. 2016;18:889–904. [DOI] [PubMed] [Google Scholar]
- Bongomin F, Gago S, Oladele ROet al. Global and multi-national prevalence of fungal diseases: estimate precision. J Fungi (Basel). 2017;3:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouzani M, Ok M, McCormick Aet al. Human NK cells display important antifungal activity against Aspergillus fumigatus, which is directly mediated by IFN-γ release. J Immunol. 2011;187:1369–76. [DOI] [PubMed] [Google Scholar]
- Boyle KB, Gyori D, Sindrilaru Aet al. Class IA phosphoinositide 3-kinase β and δ regulate neutrophil oxidase activation in response to Aspergillus fumigatus hyphae. J Immunol. 2011;186:2978–89. [DOI] [PubMed] [Google Scholar]
- Brault J, Vaganay G, Le Roy Aet al. Therapeutic effects of proteoliposomes on X-linked chronic granulomatous disease: proof of concept using macrophages differentiated from patient-specific induced pluripotent stem cells. Int J Nanomedicine. 2017;12:2161–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown GD, Denning DW, Gow NAet al. Hidden killers: human fungal infections. Sci Transl Med. 2012;4:165rv13. [DOI] [PubMed] [Google Scholar]
- Brown JA, Pensabene V, Markov DAet al. Recreating blood–brain barrier physiology and structure on chip: a novel neurovascular microfluidic bioreactor. Biomicrofluidics. 2015;9:054124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunel SF, Willment JA, Brown GDet al. Aspergillus-induced superoxide production by cystic fibrosis phagocytes is associated with disease severity. ERJ Open Res. 2018;4:00068–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruns S, Kniemeyer O, Hasenberg Met al. Production of extracellular traps against Aspergillus fumigatus in vitro and in infected lung tissue is dependent on invading neutrophils and influenced by hydrophobin RodA. PLoS Pathog. 2010;6:e1000873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrus CJ. A biochemical rationale for the interaction between gastrointestinal yeast and autism. Med Hypotheses. 2012;79:784–5. [DOI] [PubMed] [Google Scholar]
- Busch CJ, Favret J, Geirsdottir Let al. Isolation and long-term cultivation of mouse alveolar macrophages. Bio Protoc. 2019;9:e3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camilli G, Eren E, Williams DLet al. Impaired phagocytosis directs human monocyte activation in response to fungal derived β-glucan particles. Eur J Immunol. 2018;48:757–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casaroto AR, da Silva RA, Salmeron Set al. Candida albicans–cell interactions activate innate immune defense in human palate epithelial primary cells via nitric oxide (NO) and β-defensin 2 (hBD-2). Cells. 2019;8:707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassone A. Vulvovaginal Candida albicans infections: pathogenesis, immunity and vaccine prospects. BJOG. 2015;122:785–94. [DOI] [PubMed] [Google Scholar]
- Chandorkar P, Posch W, Zaderer Vet al. Fast-track development of anin vitro 3D lung/immune cell model to study Aspergillus infections. Sci Rep. 2017;7:11644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlier C, Nielsen K, Daou Set al. Evidence of a role for monocytes in dissemination and brain invasion by Cryptococcus neoformans. Infect Immun. 2009;77:120–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HC. Boyden chamber assay. Methods Mol Biol. 2005;294:15–22. [DOI] [PubMed] [Google Scholar]
- Chen HJ, Miller P, Shuler ML. A pumpless body-on-a-chip model using a primary culture of human intestinal cells and a 3D culture of liver cells. Lab Chip. 2018;18:2036–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H. Modeling development and disease with organoids. Cell. 2016;165:1586–97. [DOI] [PubMed] [Google Scholar]
- Coady A, Sil A. MyD88-dependent signaling drives host survival and early cytokine production during Histoplasma capsulatum infection. Infect Immun. 2015;83:1265–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho C, Bocca AL, Casadevall A. The tools for virulence of Cryptococcus neoformans. Adv Appl Microbiol. 2014;87:1–41. [DOI] [PubMed] [Google Scholar]
- Coelho C, Camacho E, Salas Aet al. Intranasal inoculation of Cryptococcus neoformans in mice produces nasal infection with rapid brain dissemination. mSphere. 2019;4:e00483–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coenjaerts FE, Walenkamp AM, Mwinzi PNet al. Potent inhibition of neutrophil migration by cryptococcal mannoprotein-4-induced desensitization. J Immunol. 2001;167:3988–95. [DOI] [PubMed] [Google Scholar]
- Conti HR, Bruno VM, Childs EEet al. IL-17 receptor signaling in oral epithelial cells is critical for protection against oropharyngeal candidiasis. Cell Host Microbe. 2016;20:606–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crampton SP, Davis J, Hughes CC. Isolation of human umbilical vein endothelial cells (HUVEC). J Vis Exp. 2007;3:183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalle F, Wachtler B, L'Ollivier Cet al. Cellular interactions of Candida albicans with human oral epithelial cells and enterocytes. Cell Microbiol. 2010;12:248–71. [DOI] [PubMed] [Google Scholar]
- Dasari P, Shopova IA, Stroe Met al. Aspf2 From Aspergillus fumigatus recruits human immune regulators for immune evasion and cell damage. Front Immunol. 2018;9:1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Carvalho Dias K, de Sousa DL, Barbugli PAet al. Development and characterization of a 3D oral mucosa model as a tool for host–pathogen interactions. J Microbiol Methods. 2018;152:52–60. [DOI] [PubMed] [Google Scholar]
- Degel J, Shokrani M. Validation of the efficacy of a practical method for neutrophils isolation from peripheral blood. Clin Lab Sci. 2010;23:94–8. [PubMed] [Google Scholar]
- Deinhardt-Emmer S, Rennert K, Schicke Eet al. Co-infection with Staphylococcus aureus after primary influenza virus infection leads to damage of the endothelium in a human alveolus-on-a-chip model. Biofabrication. 2020;12:025012. [DOI] [PubMed] [Google Scholar]
- De Luca A, Montagnoli C, Zelante Tet al. Functional yet balanced reactivity to Candida albicans requires TRIF, MyD88, and IDO-dependent inhibition of Rorc. J Immunol. 2007;179:5999–6008. [DOI] [PubMed] [Google Scholar]
- Deng K, Chen T, Wu Qet al. In vitro and in vivo examination of anticolonization of pathogens by Lactobacillus paracasei FJ861111.1. J Dairy Sci. 2015;98:6759–66. [DOI] [PubMed] [Google Scholar]
- Denham ST, Brown JCS. Mechanisms of pulmonary escape and dissemination by Cryptococcus neoformans. J Fungi (Basel). 2018;4:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai LP, Chapman KE, Waters CM. Mechanical stretch decreases migration of alveolar epithelial cells through mechanisms involving Rac1 and Tiam1. Am J Physiol Lung Cell Mol Physiol. 2008;295:L958–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz PI, Xie Z, Sobue Tet al. Synergistic interaction between Candida albicans and commensal oral streptococci in a novel in vitro mucosal model. Infect Immun. 2012;80:620–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson MA, Hahn WC, Ino Yet al. Human keratinocytes that express hTERT and also bypass a p16(INK4a)-enforced mechanism that limits life span become immortal yet retain normal growth and differentiation characteristics. Mol Cell Biol. 2000;20:1436–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dix A, Hunniger K, Weber Met al. Biomarker-based classification of bacterial and fungal whole-blood infections in a genome-wide expression study. Front Microbiol. 2015;6:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Andres J, Arts RJW, Ter Horst Ret al. Rewiring monocyte glucose metabolism via C-type lectin signaling protects against disseminated candidiasis. PLoS Pathog. 2017;13:e1006632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dongari-Bagtzoglou A, Kashleva H. Candida albicans triggers interleukin-8 secretion by oral epithelial cells. Microb Pathog. 2003;34:169–77. [DOI] [PubMed] [Google Scholar]
- Dongari-Bagtzoglou A, Kashleva H. Development of a highly reproducible three-dimensional organotypic model of the oral mucosa. Nat Protoc. 2006a;1:2012–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dongari-Bagtzoglou A, Kashleva H. Development of a novel three-dimensional in vitro model of oral Candida infection. Microb Pathog. 2006b;40:271–8. [DOI] [PubMed] [Google Scholar]
- Drummond RA, Collar AL, Swamydas Met al. CARD9-dependent neutrophil recruitment protects against fungal invasion of the central nervous system. PLoS Pathog. 2015;11:e1005293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond RA, Lionakis MS. Organ-specific mechanisms linking innate and adaptive antifungal immunity. Semin Cell Dev Biol. 2019;89:78–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond RA, Swamydas M, Oikonomou Vet al. CARD9(+) microglia promote antifungal immunity via IL-1β- and CXCL1-mediated neutrophil recruitment. Nat Immunol. 2019;20:559–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberl C, Speth C, Jacobsen IDet al. Candida: platelet interaction and platelet activityin vitro. J Innate Immun. 2019;11:52–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards JE Jr, Rotrosen D, Fontaine JWet al. Neutrophil-mediated protection of cultured human vascular endothelial cells from damage by growing Candida albicans hyphae. Blood. 1987;69:1450–7. [PubMed] [Google Scholar]
- Ejzykowicz DE, Solis NV, Gravelat FNet al. Role of Aspergillus fumigatus DvrA in host cell interactions and virulence. Eukaryot Cell. 2010;9:1432–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englert C, Trutzschler AK, Raasch Met al. Crossing the blood–brain barrier: glutathione-conjugated poly(ethylene imine) for gene delivery. J Control Release. 2016;241:1–14. [DOI] [PubMed] [Google Scholar]
- Erwig LP, Gow NA. Interactions of fungal pathogens with phagocytes. Nat Rev Microbiol. 2016;14:163–76. [DOI] [PubMed] [Google Scholar]
- Evans RJ, Sundaramurthy V, Frickel EM. The interplay of host autophagy and eukaryotic pathogens. Front Cell Dev Biol. 2018;6:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fichorova RN, Rheinwald JG, Anderson DJ. Generation of papillomavirus-immortalized cell lines from normal human ectocervical, endocervical, and vaginal epithelium that maintain expression of tissue-specific differentiation proteins. Biol Reprod. 1997;57:847–55. [DOI] [PubMed] [Google Scholar]
- Fidel PL Jr. Candida-host interactions in HIV disease: implications for oropharyngeal candidiasis. Adv Dent Res. 2011;23:45–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filler SG, Sheppard DC. Fungal invasion of normally non-phagocytic host cells. PLoS Pathog. 2006;2:e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogh J, Wright WC, Loveless JD. Absence of HeLa cell contamination in 169 cell lines derived from human tumors. J Natl Cancer Inst. 1977;58:209–14. [DOI] [PubMed] [Google Scholar]
- Friedrich D, Zapf D, Lohse Bet al. The HIF-1α/LC3-II axis impacts fungal immunity in human macrophages. Infect Immun. 2019;87:e00125–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fréalle E, Gosset P, Leroy Set al. In vitro coagulation triggers anti-Aspergillus fumigatus neutrophil response. Future Microbiol. 2018;13:659–69. [DOI] [PubMed] [Google Scholar]
- Gacser A, Tiszlavicz Z, Nemeth Tet al. Induction of human defensins by intestinal Caco-2 cells after interactions with opportunistic Candida species. Microbes Infect. 2014;16:80–5. [DOI] [PubMed] [Google Scholar]
- Gaffen SL, Moutsopoulos NM. Regulation of host–microbe interactions at oral mucosal barriers by type 17 immunity. Sci Immunol. 2020;5:eaau4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia C, Burgain A, Chaillot Jet al. A phenotypic small-molecule screen identifies halogenated salicylanilides as inhibitors of fungal morphogenesis, biofilm formation and host cell invasion. Sci Rep. 2018;8:11559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Rodas R, González-Camacho F, Rodríguez-Tudela JLet al. The interaction between Candida krusei and murine macrophages results in multiple outcomes, including intracellular survival and escape from killing. Infect Immun. 2011;79:2136–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazendam RP, van de Geer A, Roos Det al. How neutrophils kill fungi. Immunol Rev. 2016;273:299–311. [DOI] [PubMed] [Google Scholar]
- Gilbert AS, Wheeler RT, May RC. Fungal pathogens: survival and replication within macrophages. Cold Spring Harb Perspect Med. 2014;5:a019661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves B, Ferreira C, Alves CTet al. Vulvovaginal candidiasis: epidemiology, microbiology and risk factors. Crit Rev Microbiol. 2016;42:905–27. [DOI] [PubMed] [Google Scholar]
- Gonçalves SM, Duarte-Oliveira C, Campos CFet al. Phagosomal removal of fungal melanin reprograms macrophage metabolism to promote antifungal immunity. Nat Commun. 2020;11:2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouba N, Drancourt M. Digestive tract mycobiota: a source of infection. Med Mal Infect. 2015;45:9–16. [DOI] [PubMed] [Google Scholar]
- Goyer M, Loiselet A, Bon Fet al. Intestinal cell tight junctions limit invasion of Candida albicans through active penetration and endocytosis in the early stages of the interaction of the fungus with the intestinal barrier. PLoS One. 2016;11:e0149159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf K, Last A, Gratz Ret al. Keeping Candida commensal: how lactobacilli antagonize pathogenicity of Candida albicans in an in vitro gut model. Dis Model Mech. 2019;12:dmm039719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravelat FN, Ejzykowicz DE, Chiang LYet al. Aspergillus fumigatus MedA governs adherence, host cell interactions and virulence. Cell Microbiol. 2010;12:473–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gresnigt MS, Becker KL, Leenders Fet al. Differential kinetics of Aspergillus nidulans and Aspergillus fumigatus phagocytosis. J Innate Immun. 2018a;10:145–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gresnigt MS, Becker KL, Smeekens SPet al. Aspergillus fumigatus-induced IL-22 is not restricted to a specific Th cell subset and is dependent on complement receptor 3. J Immunol. 2013;190:5629–39. [DOI] [PubMed] [Google Scholar]
- Gresnigt MS, Cunha C, Jaeger Met al. Genetic deficiency of NOD2 confers resistance to invasive aspergillosis. Nat Commun. 2018b;9:2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griep LM, Wolbers F, de Wagenaar Bet al. BBB on chip: microfluidic platform to mechanically and biochemically modulate blood–brain barrier function. Biomed Microdevices. 2013;15:145–50. [DOI] [PubMed] [Google Scholar]
- Groger M, Rennert K, Giszas Bet al. Monocyte-induced recovery of inflammation-associated hepatocellular dysfunction in a biochip-based human liver model. Sci Rep. 2016;6:21868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grondman I, Arts RJW, Koch RMet al. Frontline science: endotoxin-induced immunotolerance is associated with loss of monocyte metabolic plasticity and reduction of oxidative burst. J Leukoc Biol. 2019;106:11–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubb SE, Murdoch C, Sudbery PEet al. Candida albicans–endothelial cell interactions: a key step in the pathogenesis of systemic candidiasis. Infect Immun. 2008;76:4370–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillot L, Carroll SF, Badawy Met al. Cryptococcus neoformans induces IL-8 secretion and CXCL1 expression by human bronchial epithelial cells. Respir Res. 2008;9:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimaraes AJ, de Cerqueira MD, Zamith-Miranda Det al. Host membrane glycosphingolipids and lipid microdomains facilitate Histoplasma capsulatum internalisation by macrophages. Cell Microbiol. 2019;21:e12976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, Chen M, Fa Zet al. Acapsular Cryptococcus neoformans activates the NLRP3 inflammasome. Microbes Infect. 2014;16:845–54. [DOI] [PubMed] [Google Scholar]
- Halder LD, Abdelfatah MA, Jo EAet al. Factor H binds to extracellular DNA traps released from human blood monocytes in response to Candida albicans. Front Immunol. 2016;7:671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallen-Adams HE, Suhr MJ. Fungi in the healthy human gastrointestinal tract. Virulence. 2017;8:352–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraguchi N, Ishii Y, Morishima Yet al. Impairment of host defense against disseminated candidiasis in mice overexpressing GATA-3. Infect Immun. 2010;78:2302–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashemi Fesharaki S, Aghili SR, Shokohi Tet al. Catheter-related candidemia and identification of causative Candida species in patients with cardiovascular disorder. Curr Med Mycol. 2018;4:7–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hefter M, Lother J, Weiss Eet al. Human primary myeloid dendritic cells interact with the opportunistic fungal pathogen Aspergillus fumigatus via the C-type lectin receptor Dectin-1. Med Mycol. 2017;55:573–8. [DOI] [PubMed] [Google Scholar]
- Hellwig D, Voigt J, Bouzani Met al. Candida albicans induces metabolic reprogramming in human NK cells and responds to perforin with a zinc depletion response. Front Microbiol. 2016;7:750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbrecht R, Bories P, Moulin JCet al. Risk stratification for invasive aspergillosis in immunocompromised patients. Ann N Y Acad Sci. 2012;1272:23–30. [DOI] [PubMed] [Google Scholar]
- Herbrecht R, Neuville S, Letscher-Bru Vet al. Fungal infections in patients with neutropenia: challenges in prophylaxis and treatment. Drugs Aging. 2000;17:339–51. [DOI] [PubMed] [Google Scholar]
- He X, Shi X, Puthiyakunnon Set al. CD44-mediated monocyte transmigration across Cryptococcus neoformans-infected brain microvascular endothelial cells is enhanced by HIV-1 gp41-I90 ectodomain. J Biomed Sci. 2016;23:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho J, Yang X, Nikou SAet al. Candidalysin activates innate epithelial immune responses via epidermal growth factor receptor. Nat Commun. 2019;10:2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope WW, Kruhlak MJ, Lyman CAet al. Pathogenesis of Aspergillus fumigatus and the kinetics of galactomannan in an in vitro model of early invasive pulmonary aspergillosis: implications for antifungal therapy. J Infect Dis. 2007;195:455–66. [DOI] [PubMed] [Google Scholar]
- Hopke A, Scherer A, Kreuzburg Set al. Neutrophil swarming delays the growth of clusters of pathogenic fungi. Nat Commun. 2020;11:2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JH, Liu CY, Wu SYet al. NLRX1 facilitates Histoplasma capsulatum-induced LC3-associated phagocytosis for cytokine production in macrophages. Front Immunol. 2018;9:2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Na L, Fidel PLet al. Requirement of interleukin-17A for systemic anti-Candida albicans host defense in mice. J Infect Dis. 2004;190:624–31. [DOI] [PubMed] [Google Scholar]
- Huh D, Matthews BD, Mammoto Aet al. Reconstituting organ-level lung functions on a chip. Science. 2010;328:1662–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunniger K, Lehnert T, Bieber Ket al. A virtual infection model quantifies innate effector mechanisms and Candida albicans immune escape in human blood. PLoS Comput Biol. 2014;10:e1003479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ifrim DC, Quintin J, Courjol Fet al. The role of Dectin-2 for host defense against disseminated candidiasis. J Interferon Cytokine Res. 2016;36:267–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger M, Matzaraki V, Aguirre-Gamboa Ret al. A genome-wide functional genomics approach identifies susceptibility pathways to fungal bloodstream infection in humans. J Infect Dis. 2019a;220:862–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger M, Pinelli M, Borghi Met al. A systems genomics approach identifies SIGLEC15 as a susceptibility factor in recurrent vulvovaginal candidiasis. Sci Transl Med. 2019b;11:eaar3558. [DOI] [PubMed] [Google Scholar]
- Jaffe EA, Nachman RL, Becker CGet al. Culture of human endothelial cells derived from umbilical veins. Identification by morphologic and immunologic criteria. J Clin Invest. 1973;52:2745–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A, Barrile R, van der Meer ADet al. Primary human lung alveolus-on-a-chip model of intravascular thrombosis for assessment of therapeutics. Clin Pharmacol Ther. 2018;103:332–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang KJ, Otieno MA, Ronxhi Jet al. Reproducing human and cross-species drug toxicities using a Liver-Chip. Sci Transl Med. 2019;11:eaax5516. [DOI] [PubMed] [Google Scholar]
- Javed F, Al-Kheraif AA, Kellesarian SVet al. Oral Candida carriage and species prevalence in denture stomatitis patients with and without diabetes. J Biol Regul Homeost Agents. 2017;31:343–6. [PubMed] [Google Scholar]
- Jong AY, Stins MF, Huang SHet al. Traversal of Candida albicans across human blood–brain barrier in vitro. Infect Immun. 2001;69:4536–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalia N, Singh J, Kaur M. Microbiota in vaginal health and pathogenesis of recurrent vulvovaginal infections: a critical review. Ann Clin Microbiol Antimicrob. 2020;19:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamai Y, Chiang LY, Lopes Bezerra LMet al. Interactions of Aspergillus fumigatus with vascular endothelial cells. Med Mycol. 2006;44 Suppl 1:S115–7. [DOI] [PubMed] [Google Scholar]
- Kamai Y, Lossinsky AS, Liu Het al. Polarized response of endothelial cells to invasion by Aspergillus fumigatus. Cell Microbiol. 2009;11:170–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanayama M, Shinohara ML. Roles of autophagy and autophagy-related proteins in antifungal immunity. Front Immunol. 2016;7:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karki R, Man SM, Malireddi RKet al. Concerted activation of the AIM2 and NLRP3 inflammasomes orchestrates host protection against Aspergillus infection. Cell Host Microbe. 2015;17:357–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasper L, Konig A, Koenig PAet al. The fungal peptide toxin Candidalysin activates the NLRP3 inflammasome and causes cytolysis in mononuclear phagocytes. Nat Commun. 2018;9:4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavanaugh NL, Zhang AQ, Nobile CJet al. Mucins suppress virulence traits of Candida albicans. mBio. 2014;5:e01911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khader SA, Gaffen SL, Kolls JK. Th17 cells at the crossroads of innate and adaptive immunity against infectious diseases at the mucosa. Mucosal Immunol. 2009;2:403–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Ingber DE. Gut-on-a-Chip microenvironment induces human intestinal cells to undergo villus differentiation. Integr Biol (Camb). 2013;5:1130–40. [DOI] [PubMed] [Google Scholar]
- Klassert TE, Brauer J, Holzer Met al. Differential effects of vitamins A and D on the transcriptional landscape of human monocytes during infection. Sci Rep. 2017;7:40599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutsen AP, Slavin RG. Allergic bronchopulmonary aspergillosis in asthma and cystic fibrosis. Clin Dev Immunol. 2011;2011:843763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogan TV, Jadoun J, Mittelman Let al. Involvement of secreted Aspergillus fumigatus proteases in disruption of the actin fiber cytoskeleton and loss of focal adhesion sites in infected A549 lung pneumocytes. J Infect Dis. 2004;189:1965–73. [DOI] [PubMed] [Google Scholar]
- Kolwijck E, van de Veerdonk FL. The potential impact of the pulmonary microbiome on immunopathogenesis of Aspergillus-related lung disease. Eur J Immunol. 2014;44:3156–65. [DOI] [PubMed] [Google Scholar]
- Kosmidis C, Denning DW. The clinical spectrum of pulmonary aspergillosis. Thorax. 2015;70:270–7. [DOI] [PubMed] [Google Scholar]
- Kumamoto CA, Gresnigt MS, Hube B. The gut, the bad and the harmless: Candida albicans as a commensal and opportunistic pathogen in the intestine. Curr Opin Microbiol. 2020;56:7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Saraswat D, Tati Set al. Novel aggregation properties of Candida albicans secreted aspartyl proteinase Sap6 mediate virulence in oral candidiasis. Infect Immun. 2015;83:2614–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunyeit L, Kurrey NK, Anu-Appaiah KAet al. Probiotic yeasts inhibit virulence of non-albicans Candida species. mBio. 2019;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrmizi I, Ferreira H, Carvalho Aet al. Calcium sequestration by fungal melanin inhibits calcium-calmodulin signalling to prevent LC3-associated phagocytosis. Nat Microbiol. 2018;3:791–803. [DOI] [PubMed] [Google Scholar]
- Kämmer P, McNamara S, Wolf Tet al. Survival strategies of pathogenic Candida species in human blood show independent and specific adaptations. mBio. 2020;11:e02435–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler JR, Hube B, Puccia Ret al. Fungi that infect humans. Microbiology Spectr. 2017;5:0014–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahiri S, Banerjee A, Bhutda Set al. In vitro expression of vital virulent genes of clinical and environmental isolates of Cryptococcus neoformans/gattii in endothelial cells of human blood–brain barrier. J Mycol Med. 2019;29:239–44. [DOI] [PubMed] [Google Scholar]
- Lee S, Chung M, Lee SRet al. 3D brain angiogenesis model to reconstitute functional human blood–brain barrier in vitro. Biotechnol Bioeng. 2020;117:748–62. [DOI] [PubMed] [Google Scholar]
- Lee SH, Sung JH. Microtechnology-based multi-organ models. Bioengineering (Basel). 2017;4:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeibundGut-Landmann S, Gross O, Robinson MJet al. Syk- and CARD9-dependent coupling of innate immunity to the induction of T helper cells that produce interleukin 17. Nat Immunol. 2007;8:630–8. [DOI] [PubMed] [Google Scholar]
- Leiding JW, Holland SM. Chronic granulomatous disease. In: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Mefford HC, Stephens K, Amemiya A, Ledbetter N (eds). GeneReviews(R). Seattle (WA), 1993. [PubMed] [Google Scholar]
- Leonardi I, Li X, Iliev ID. Macrophage interactions with fungi and bacteria in inflammatory bowel disease. Curr Opin Gastroenterol. 2018;34:392–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhardt J, Grosse S, Marx Cet al. Candida albicans β-glucan differentiates human monocytes into a specific subset of macrophages. Front Immunol. 2018;9:2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesuffleur T, Barbat A, Dussaulx Eet al. Growth adaptation to methotrexate of HT-29 human colon carcinoma cells is associated with their ability to differentiate into columnar absorptive and mucus-secreting cells. Cancer Res. 1990;50:6334–43. [PubMed] [Google Scholar]
- Lewis LE, Bain JM, Lowes Cet al. Stage specific assessment of Candida albicans phagocytosis by macrophages identifies cell wall composition and morphogenesis as key determinants. PLoS Pathog. 2012;8:e1002578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber M, Smith B, Szakal Aet al. A continuous tumor-cell line from a human lung carcinoma with properties of type II alveolar epithelial cells. Int J Cancer. 1976;17:62–70. [DOI] [PubMed] [Google Scholar]
- Lim J, Coates CJ, Seoane PIet al. Characterizing the mechanisms of nonopsonic uptake of cryptococci by macrophages. J Immunol. 2018;200:3539–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg K, Rheinwald JG. Three distinct keratinocyte subtypes identified in human oral epithelium by their patterns of keratin expression in culture and in xenografts. Differentiation. 1990;45:230–41. [DOI] [PubMed] [Google Scholar]
- Lionakis MS, Levitz SM. Host control of fungal infections: lessons from basic studies and human cohorts. Annu Rev Immunol. 2018;36:157–91. [DOI] [PubMed] [Google Scholar]
- Lionakis MS, Lim JK, Lee CCet al. Organ-specific innate immune responses in a mouse model of invasive candidiasis. J Innate Immun. 2011;3:180–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionakis MS, Swamydas M, Fischer BGet al. CX3CR1-dependent renal macrophage survival promotes Candida control and host survival. J Clin Invest. 2013;123:5035–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionakis MS. New insights into innate immune control of systemic candidiasis. Med Mycol. 2014;52:555–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SS, Ogbomo H, Mansour MKet al. Identification of the fungal ligand triggering cytotoxic PRR-mediated NK cell killing of Cryptococcus and Candida. Nat Commun. 2018;9:751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Niu X, Zhang Xet al. Recombinant human IFNα-2b response promotes vaginal epithelial cells defense against Candida albicans. Front Microbiol. 2017;8:697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Lee MJ, Solis NVet al. Aspergillus fumigatus CalA binds to integrin α5β1 and mediates host cell invasion. Nat Microbiol. 2016;2:16211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu NN, Uppuluri P, Broggi Aet al. Intersection of phosphate transport, oxidative stress and TOR signalling in Candida albicans virulence. PLoS Pathog. 2018;14:e1007076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Mittal R, Solis NVet al. Mechanisms of Candida albicans trafficking to the brain. PLoS Pathog. 2011;7:e1002305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Shetty AC, Schwartz JAet al. New signaling pathways govern the host response to C. albicans infection in various niches. Genome Res. 2015;25:679–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart SR, Guarner J. Emerging and reemerging fungal infections. Semin Diagn Pathol. 2019;36:177–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohith K, Anu-Appaiah KA. Antagonistic effect of Saccharomyces cerevisiae KTP and Issatchenkia occidentalis ApC on hyphal development and adhesion of Candida albicans. Med Mycol. 2018;56:1023–32. [DOI] [PubMed] [Google Scholar]
- Long KH, Gomez FJ, Morris REet al. Identification of heat shock protein 60 as the ligand on Histoplasma capsulatum that mediates binding to CD18 receptors on human macrophages. J Immunol. 2003;170:487–94. [DOI] [PubMed] [Google Scholar]
- Lother J, Breitschopf T, Krappmann Set al. Human dendritic cell subsets display distinct interactions with the pathogenic mould Aspergillus fumigatus. Int J Med Microbiol. 2014;304:1160–8. [DOI] [PubMed] [Google Scholar]
- Luni C, Serena E, Elvassore N. Human-on-chip for therapy development and fundamental science. Curr Opin Biotechnol. 2014;25:45–50. [DOI] [PubMed] [Google Scholar]
- Maciel Quatrin P, Flores Dalla Lana D, Andrzejewski Kaminski TFet al. Fungal infection models: current progress of ex vivo methods. Mycoses. 2019;62:860–73. [DOI] [PubMed] [Google Scholar]
- Mailander-Sanchez D, Braunsdorf C, Grumaz Cet al. Antifungal defense of probiotic Lactobacillus rhamnosus GG is mediated by blocking adhesion and nutrient depletion. PLoS One. 2017;12:e0184438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makanjuola O, Bongomin F, Fayemiwo SA. An update on the roles of non-albicans Candida species in vulvovaginitis. J Fungi (Basel). 2018;4:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini V, Pensabene V. Organs-on-chip models of the female reproductive system. Bioengineering (Basel). 2019;6:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X, Qiu X, Jiao Cet al. Candida albicans SC5314 inhibits NLRP3/NLRP6 inflammasome expression and dampens human intestinal barrier activity in Caco-2 cell monolayer model. Cytokine. 2019;126:154882. [DOI] [PubMed] [Google Scholar]
- Maoz BM, Herland A, FitzGerald EAet al. A linked organ-on-chip model of the human neurovascular unit reveals the metabolic coupling of endothelial and neuronal cells. Nat Biotechnol. 2018;36:865–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marischen L, Englert A, Schmitt ALet al. Human NK cells adapt their immune response towards increasing multiplicities of infection of Aspergillus fumigatus. BMC Immunol. 2018;19:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marolda A, Hünniger K, Böttcher Set al. Candida species-dependent release of IL-12 by dendritic cells induces different levels of NK cell stimulation. J Infect Dis. 2020, DOI: 10.1093/infdis/jiaa035. [DOI] [PubMed] [Google Scholar]
- Mattern DJ, Schoeler H, Weber Jet al. Identification of the antiphagocytic trypacidin gene cluster in the human-pathogenic fungus Aspergillus fumigatus. Appl Microbiol Biotechnol. 2015;99:10151–61. [DOI] [PubMed] [Google Scholar]
- Matzaraki V, Gresnigt MS, Jaeger Met al. An integrative genomics approach identifies novel pathways that influence candidaemia susceptibility. PLoS One. 2017;12:e0180824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer M, Gresnigt MS, Last Aet al. A three-dimensional immunocompetent intestine-on-chip model as in vitro platform for functional and microbial interaction studies. Biomaterials. 2019;220:119396. [DOI] [PubMed] [Google Scholar]
- Maxson ME, Naj X, O'Meara TRet al. Integrin-based diffusion barrier separates membrane domains enabling the formation of microbiostatic frustrated phagosomes. eLife. 2018;7:e34798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCall AD, Kumar R, Edgerton M. Candida albicans Sfl1/Sfl2 regulatory network drives the formation of pathogenic microcolonies. PLoS Pathog. 2018;14:e1007316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meir J, Hartmann E, Eckstein MTet al. Identification of Candida albicans regulatory genes governing mucosal infection. Cell Microbiol. 2018;20:e12841. [DOI] [PubMed] [Google Scholar]
- Mihu MR, Nosanchuk JD. Histoplasma virulence and host responses. Int J Microbiol. 2012;2012:268123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikamo H, Yamagishi Y, Sugiyama Het al. High glucose-mediated overexpression of ICAM-1 in human vaginal epithelial cells increases adhesion of Candida albicans. J Obstet Gynaecol. 2018;; 38:226–30. [DOI] [PubMed] [Google Scholar]
- Millsop JW, Fazel N. Oral candidiasis. Clin Dermatol. 2016;34:487–94. [DOI] [PubMed] [Google Scholar]
- Montagnoli C, Fallarino F, Gaziano Ret al. Immunity and tolerance to Aspergillus involve functionally distinct regulatory T cells and tryptophan catabolism. J Immunol. 2006;176:1712–23. [DOI] [PubMed] [Google Scholar]
- Montelongo-Jauregui D, Lopez-Ribot JL. Candida interactions with the oral bacterial microbiota. J Fungi (Basel). 2018;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse DJ, Wilson MJ, Wei Xet al. Denture-associated biofilm infection in three-dimensional oral mucosal tissue models. J Med Microbiol. 2018;67:364–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton CO, Fliesser M, Dittrich Met al. Gene expression profiles of human dendritic cells interacting with Aspergillus fumigatus in a bilayer model of the alveolar epithelium/endothelium interface. PLoS One. 2014;9:e98279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton CO, Wurster S, Fliesser Met al. Validation of a simplified in vitro Transwell((R)) model of the alveolar surface to assess host immunity induced by different morphotypes of Aspergillus fumigatus. Int J Med Microbiol. 2018;308:1009–17. [DOI] [PubMed] [Google Scholar]
- Mosig AS. Organ-on-chip models: new opportunities for biomedical research. Future Sci OA. 2017;3:Fso130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyes DL, Wilson D, Richardson JPet al. Candidalysin is a fungal peptide toxin critical for mucosal infection. Nature. 2016;532:64–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz JF, Delorey T, Ford CBet al. Coordinated host–pathogen transcriptional dynamics revealed using sorted subpopulations and single macrophages infected with Candida albicans. Nat Commun. 2019;10:1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz NM, Leff AR. Highly purified selective isolation of eosinophils from human peripheral blood by negative immunomagnetic selection. Nat Protoc. 2006;1:2613–20. [DOI] [PubMed] [Google Scholar]
- Naglik JR, Konig A, Hube Bet al. Candida albicans-epithelial interactions and induction of mucosal innate immunity. Curr Opin Microbiol. 2017;40:104–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nailis H, Kucharikova S, Ricicova Met al. Real-time PCR expression profiling of genes encoding potential virulence factors in Candida albicans biofilms: identification of model-dependent and -independent gene expression. BMC Microbiol. 2010;10:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak DK, Mendez O, Bowen Set al. Isolation and in vitro culture of murine and human alveolar macrophages. J Vis Exp. 2018;134:e57287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negoro PE, Xu S, Dagher Zet al. Spleen tyrosine kinase is a critical regulator of neutrophil responses to Candida species. mBio. 2020;11:e02043–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea MG, Joosten LA, van der Meer JWet al. Immune defence against Candida fungal infections. Nat Rev Immunol. 2015;15:630–42. [DOI] [PubMed] [Google Scholar]
- Neves GW, Curty NA, Kubitschek-Barreira PHet al. Modifications to the composition of the hyphal outer layer of Aspergillus fumigatus modulates HUVEC proteins related to inflammatory and stress responses. J Proteomics. 2017;151:83–96. [DOI] [PubMed] [Google Scholar]
- Newman SL. Interaction of Histoplasma capsulatum with human macrophages, dendritic cells, and neutrophils. Methods Mol Med. 2005;118:181–91. [DOI] [PubMed] [Google Scholar]
- Niemiec MJ, Grumaz C, Ermert Det al. Dual transcriptome of the immediate neutrophil and Candida albicans interplay. BMC Genomics. 2017;18:696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Meara TR, Cowen LE. Insights into the host–pathogen interaction: C. albicans manipulation of macrophage pyroptosis. Microb Cell. 2018;5:566–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Meara TR, Duah K, Guo CXet al. High-throughput screening identifies genes required for Candida albicans induction of macrophage pyroptosis. mBio. 2018;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oddo A, Peng B, Tong Zet al. Advances in microfluidic blood–brain barrier (BBB) models. Trends Biotechnol. 2019;37:1295–314. [DOI] [PubMed] [Google Scholar]
- Oesterreicher Z, Eberl S, Zeitlinger M. Impact of different antimycotics on cytokine levels in an in vitro aspergillosis model in human whole blood. Infection. 2019;48:65–73. [DOI] [PubMed] [Google Scholar]
- Offenbacher S, Barros SP, Bencharit Set al. Differential mucosal gene expression patterns in Candida-associated, chronic oral denture stomatitis. J Prosthodont. 2019;28:202–8. [DOI] [PubMed] [Google Scholar]
- Okagaki LH, Nielsen K. Titan cells confer protection from phagocytosis in Cryptococcus neoformans infections. Eukaryot Cell. 2012;11:820–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okai B, Lyall N, Gow NAet al. Rab14 regulates maturation of macrophage phagosomes containing the fungal pathogen Candida albicans and outcome of the host–pathogen interaction. Infect Immun. 2015;83:1523–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco AS, Zhou X, Filler SG. Mechanisms of the proinflammatory response of endothelial cells to Candida albicans infection. Infect Immun. 2000;68:1134–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page L, Weis P, Muller Tet al. Evaluation of Aspergillus and Mucorales specific T-cells and peripheral blood mononuclear cell cytokine signatures as biomarkers of environmental mold exposure. Int J Med Microbiol. 2018;308:1018–26. [DOI] [PubMed] [Google Scholar]
- Pappas PG, Lionakis MS, Arendrup MCet al. Invasive candidiasis. Nat Rev Dis Primers. 2018;4:18026. [DOI] [PubMed] [Google Scholar]
- Park SE, Georgescu A, Huh D. Organoids-on-a-chip. Science. 2019;364:960–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel R, Hossain MA, German Net al. Gliotoxin penetrates and impairs the integrity of the human blood–brain barrier in vitro. Mycotoxin Res. 2018;34:257–68. [DOI] [PubMed] [Google Scholar]
- Patil S, Rao RS, Majumdar Bet al. Clinical appearance of oral Candida infection and therapeutic strategies. Front Microbiol. 2015;6:1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekmezovic M, Mogavero S, Naglik JRet al. Host–pathogen interactions during female genital tract infections. Trends Microbiol. 2019;27:982–96. [DOI] [PubMed] [Google Scholar]
- Perlroth J, Choi B, Spellberg B. Nosocomial fungal infections: epidemiology, diagnosis, and treatment. Med Mycol. 2007;45:321–46. [DOI] [PubMed] [Google Scholar]
- Peterson MD, Mooseker MS. Characterization of the enterocyte-like brush border cytoskeleton of the C2BBe clones of the human intestinal cell line, Caco-2. J Cell Sci. 1992;102 (Pt 3):581–600. [DOI] [PubMed] [Google Scholar]
- Pfaller MA, Diekema DJ. Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev. 2007;20:133–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan QT, Eng DK, Mostowy Set al. Role of endothelial cell septin 7 in the endocytosis of Candida albicans. mBio. 2013;4:e00542–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan QT, Fratti RA, Prasadarao NVet al. N-cadherin mediates endocytosis of Candida albicans by endothelial cells. J Biol Chem. 2005;280:10455–61. [DOI] [PubMed] [Google Scholar]
- Phan QT, Myers CL, Fu Yet al. Als3 is a Candida albicans invasin that binds to cadherins and induces endocytosis by host cells. PLoS Biol. 2007;5:e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidwill GR, Rego S, Jenkinson HFet al. Coassociation between group B Streptococcus and Candida albicans promotes interactions with vaginal epithelium. Infect Immun. 2018;86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietrella D, Corbucci C, Perito Set al. Mannoproteins from Cryptococcus neoformans promote dendritic cell maturation and activation. Infect Immun. 2005;73:820–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitangui Nde S, Sardi Jde C, Voltan ARet al. An intracellular arrangement of Histoplasma capsulatum yeast-aggregates generates nuclear damage to the cultured murine alveolar macrophages. Front Microbiol. 2015;6:1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitangui NS, Sardi JC, Silva JFet al. Adhesion of Histoplasma capsulatum to pneumocytes and biofilm formation on an abiotic surface. Biofouling. 2012;28:711–8. [DOI] [PubMed] [Google Scholar]
- Pollard BS, Pollard HB. Induced pluripotent stem cells for treating cystic fibrosis: state of the science. Pediatr Pulmonol. 2018;53:S12–29. [DOI] [PubMed] [Google Scholar]
- Polvi EJ, Li X, O'Meara TRet al. Opportunistic yeast pathogens: reservoirs, virulence mechanisms, and therapeutic strategies. Cellular Mol Life Sci. 2015;72:2261–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri S, Kumar R, Rojas IGet al. Iron chelator deferasirox reduces Candida albicans invasion of oral epithelial cells and infection levels in murine oropharyngeal candidiasis. Antimicrob Agents Chemother. 2019;63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintin J, Saeed S, Martens JHAet al. Candida albicans infection affords protection against reinfection via functional reprogramming of monocytes. Cell Host Microbe. 2012;12:223–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raasch M, Fritsche E, Kurtz Aet al. Microphysiological systems meet hiPSC technology: new tools for disease modeling of liver infections in basic research and drug development. Adv Drug Deliv Rev. 2019;140:51–67. [DOI] [PubMed] [Google Scholar]
- Raasch M, Rennert K, Jahn Tet al. An integrative microfluidically supported in vitro model of an endothelial barrier combined with cortical spheroids simulates effects of neuroinflammation in neocortex development. Biomicrofluidics. 2016;10:044102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahimi C, Rahimi B, Padova Det al. Oral mucosa-on-a-chip to assess layer-specific responses to bacteria and dental materials. Biomicrofluidics. 2018;12:054106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmani S, Breyner NM, Su HMet al. Intestinal organoids: a new paradigm for engineering intestinal epithelium in vitro. Biomaterials. 2019;194:195–214. [DOI] [PubMed] [Google Scholar]
- Raijmakers RPH, Sprenkeler EGG, Aleva FEet al. Toll-like receptor 2 induced cytotoxic T-lymphocyte-associated protein 4 regulates Aspergillus-induced regulatory T-cells with pro-inflammatory characteristics. Sci Rep. 2017;7:11500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramme AP, Koenig L, Hasenberg Tet al. Autologous induced pluripotent stem cell-derived four-organ-chip. Future Sci OA. 2019;5:Fso413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappleye CA, Eissenberg LG, Goldman WE. Histoplasma capsulatum α-(1,3)-glucan blocks innate immune recognition by the β-glucan receptor. Proc Natl Acad Sci USA. 2007;104:1366–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray SC, Rappleye CA. Flying under the radar: Histoplasma capsulatum avoidance of innate immune recognition. Semin Cell Dev Biol. 2019;89:91–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards GR, Millard RM, Leveridge Met al. Quantitative assays of chemotaxis and chemokinesis for human neural cells. Assay Drug Dev Technol. 2004;2:465–72. [DOI] [PubMed] [Google Scholar]
- Richardson JP, Willems HME, Moyes DLet al. Candidalysin drives epithelial signaling, neutrophil recruitment, and immunopathology at the vaginal mucosa. Infect Immun. 2018;86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieber N, Gazendam RP, Freeman AFet al. Extrapulmonary Aspergillus infection in patients with CARD9 deficiency. JCI Insight. 2016;1:e89890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson NB, Krieger K, Khan FMet al. The current state of animal models in research: a review. Int J Surg. 2019;72:9–13. [DOI] [PubMed] [Google Scholar]
- Rocha JD, Nascimento MT, Decote-Ricardo Det al. Capsular polysaccharides from Cryptococcus neoformans modulate production of neutrophil extracellular traps (NETs) by human neutrophils. Sci Rep. 2015;5:8008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogal J, Probst C, Loskill P. Integration concepts for multi-organ chips: how to maintain flexibility?! Future Sci OA. 2017;3:Fso180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romani L. Immunity to fungal infections. Nat Rev Immunol. 2004;4:1–23. [DOI] [PubMed] [Google Scholar]
- Rosati D, Bruno M, Jaeger Met al. Recurrent vulvovaginal candidiasis: an immunological perspective. Microorganisms. 2020;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupniak HT, Rowlatt C, Lane EBet al. Characteristics of four new human cell lines derived from squamous cell carcinomas of the head and neck. J Natl Cancer Inst. 1985;75:621–35. [PubMed] [Google Scholar]
- Salerno C, Pascale M, Contaldo Met al. Candida-associated denture stomatitis. Med Oral Patol Oral Cir Bucal. 2011;16:e139–43. [DOI] [PubMed] [Google Scholar]
- Sanchez AA, Johnston DA, Myers Cet al. Relationship between Candida albicans virulence during experimental hematogenously disseminated infection and endothelial cell damage in vitro. Infect Immun. 2004;72:598–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago-Tirado FH, Onken MD, Cooper JAet al. Trojan horse transit contributes to blood–brain barrier crossing of a eukaryotic pathogen. mBio. 2017;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago V, Rezvani K, Sekine Tet al. Human NK cells develop an exhaustion phenotype during polar degranulation at the Aspergillus fumigatus hyphal synapse. Front Immunol. 2018;9:2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Vries RG, Snippert HJet al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–5. [DOI] [PubMed] [Google Scholar]
- Schaller M, Schafer W, Korting HCet al. Differential expression of secreted aspartyl proteinases in a model of human oral candidosis and in patient samples from the oral cavity. Mol Microbiol. 1998;29:605–15. [DOI] [PubMed] [Google Scholar]
- Scheffold A, Bacher P, LeibundGut-Landmann S. T cell immunity to commensal fungi. Curr Opin Microbiol. 2020;58:116–23. [DOI] [PubMed] [Google Scholar]
- Schestatsky P, Chedid MF, Amaral OBet al. Isolated central nervous system histoplasmosis in immunocompetent hosts: a series of 11 cases. Scand J Infect Dis. 2006;38:43–8. [DOI] [PubMed] [Google Scholar]
- Schirbel A, Shouval DS, Hebecker Bet al. Intestinal epithelial cells and T cells differentially recognize and respond to Candida albicans yeast and hypha. Eur J Immunol. 2018;48:1826–37. [DOI] [PubMed] [Google Scholar]
- Schmidt F, Thywißen A, Goldmann Met al. Flotillin-dependent membrane microdomains are required for functional phagolysosomes against fungal infections. Cell Rep. 2020;32:108017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt S, Tramsen L, Lehrnbecher T. Natural killer cells in antifungal immunity. Front Immunol. 2017;8:1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal BH, Leto TL, Gallin JIet al. Genetic, biochemical, and clinical features of chronic granulomatous disease. Medicine (Baltimore). 2000;79:170–200. [DOI] [PubMed] [Google Scholar]
- Seider K, Heyken A, Lüttich Aet al. Interaction of pathogenic yeasts with phagocytes: survival, persistence and escape. Curr Opin Microbiol. 2010;13:392–400. [DOI] [PubMed] [Google Scholar]
- Seoane PI, Taylor-Smith LM, Stirling Det al. Viral infection triggers interferon-induced expulsion of live Cryptococcus neoformans by macrophages. PLoS Pathog. 2020;16:e1008240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setianingrum F, Rautemaa-Richardson R, Denning DW. Pulmonary cryptococcosis: a review of pathobiology and clinical aspects. Med Mycol. 2019;57:133–50. [DOI] [PubMed] [Google Scholar]
- Shen Q, Beucler MJ, Ray SCet al. Macrophage activation by IFN-gamma triggers restriction of phagosomal copper from intracellular pathogens. PLoS Pathog. 2018;14:e1007444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard DC, Filler SG. Host cell invasion by medically important fungi. Cold Spring Harb Perspect Med. 2014;5:a019687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimamura S, Miyazaki T, Tashiro Met al. Autophagy-inducing factor Atg1 is required for virulence in the pathogenic fungus Candida glabrata. Front Microbiol. 2019;10:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin W, Kim HJ. Pathomimetic modeling of human intestinal diseases and underlying host–gut microbiome interactions in a gut-on-a-chip. Methods Cell Biol. 2018;146:135–48. [DOI] [PubMed] [Google Scholar]
- Shroff A, Reddy KVR. Autophagy gene ATG5 knockdown upregulates apoptotic cell death during Candida albicans infection in human vaginal epithelial cells. American J Reprod Immunol. 2018;80:e13056. [DOI] [PubMed] [Google Scholar]
- Shukla A, Sobel JD. Vulvovaginitis caused by Candida species following antibiotic exposure. Curr Infect Dis Rep. 2019;21:44. [DOI] [PubMed] [Google Scholar]
- Silva S, Henriques M, Hayes Aet al. Candida glabrata and Candida albicans co-infection of an in vitro oral epithelium. J Oral Pathol Med. 2011;40:421–7. [DOI] [PubMed] [Google Scholar]
- Smeekens SP, Ng A, Kumar Vet al. Functional genomics identifies type I interferon pathway as central for host defense against Candida albicans. Nat Commun. 2013;4:1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LM, Dixon EF, May RC. The fungal pathogen Cryptococcus neoformans manipulates macrophage phagosome maturation. Cell Microbiol. 2015;17:702–13. [DOI] [PubMed] [Google Scholar]
- Sobel JD. Vulvovaginal candidosis. Lancet (London, England). 2007;369:1961–71. [DOI] [PubMed] [Google Scholar]
- Sobue T, Bertolini M, Thompson Aet al. Chemotherapy-induced oral mucositis and associated infections in a novel organotypic model. Mol Oral Microbiol. 2018;33:212–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solis NV, Park YN, Swidergall Met al. Candida albicans white-opaque switching influences virulence but not mating during oropharyngeal candidiasis. Infect Immun. 2018;86:e00774–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solis NV, Swidergall M, Bruno VMet al. The aryl hydrocarbon receptor governs epithelial cell invasion during oropharyngeal candidiasis. mBio. 2017;8:e00025–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontheimer-Phelps A, Chou DB, Tovaglieri Aet al. Human colon-on-a-chip enables continuous in vitro analysis of colon mucus layer accumulation and physiology. Cell Mol Gastroenterol Hepatol. 2020;9:507–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soubani AO, Chandrasekar PH. The clinical spectrum of pulmonary aspergillosis. Chest. 2002;121:1988–99. [DOI] [PubMed] [Google Scholar]
- Spence JR, Mayhew CN, Rankin SAet al. Directed differentiation of human pluripotent stem cells into intestinal tissuein vitro. Nature. 2011;470:105–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiering MJ, Moran GP, Chauvel Met al. Comparative transcript profiling of Candida albicans and Candida dubliniensis identifies SFL2, a C. albicans gene required for virulence in a reconstituted epithelial infection model. Eukaryot Cell. 2010;9:251–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprenkeler EG, Gresnigt MS, van de Veerdonk FL. LC3-associated phagocytosis: a crucial mechanism for antifungal host defence against Aspergillus fumigatus. Cell Microbiol. 2016;18:1208–16. [DOI] [PubMed] [Google Scholar]
- Srikanta D, Santiago-Tirado FH, Doering TL. Cryptococcus neoformans: historical curiosity to modern pathogen. Yeast. 2014;31:47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephen-Victor E, Karnam A, Fontaine Tet al. Aspergillus fumigatus cell wall α-(1,3)-glucan stimulates regulatory T-cell polarization by inducing PD-L1 expression on human dendritic cells. J Infect Dis. 2017;216:1281–94. [DOI] [PubMed] [Google Scholar]
- Stucki JD, Hobi N, Galimov Aet al. Medium throughput breathing human primary cell alveolus-on-chip model. Sci Rep. 2018;8:14359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D, Shi M. Neutrophil swarming toward Cryptococcus neoformans is mediated by complement and leukotriene B4. Biochem Biophys Res Commun. 2016;477:945–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Xu XY, Tian XLet al. Activation of NF-κB and respiratory burst following Aspergillus fumigatus stimulation of macrophages. Immunobiology. 2014;219:25–36. [DOI] [PubMed] [Google Scholar]
- Swidergall M, Solis NV, Lionakis MSet al. EphA2 is an epithelial cell pattern recognition receptor for fungal β-glucans. Nat Microbiol. 2018;3:53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76. [DOI] [PubMed] [Google Scholar]
- Teixeira PA, Penha LL, Mendonca-Previato Let al. Mannoprotein MP84 mediates the adhesion of Cryptococcus neoformans to epithelial lung cells. Front Cell Infect Microbiol. 2014;4:106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ter Horst R, Jaeger M, Smeekens SPet al. Host and environmental factors influencing individual human cytokine responses. Cell. 2016;167:1111–24.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thak EJ, Lee SB, Xu-Vanpala Set al. Core N-glycan structures are critical for the pathogenicity of Cryptococcus neoformans by modulating host cell death. mBio. 2020;11:e00711–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson-Souza GA,Santos GMP, Silva JCet al. Histoplasma capsulatum-induced extracellular DNA trap release in human neutrophils. Cell Microbiol. 2020;22:e13195. [DOI] [PubMed] [Google Scholar]
- Thunström Salzer A, Niemiec MJ, Hosseinzadeh Aet al. Assessment of neutrophil chemotaxis upon G-CSF treatment of healthy stem cell donors and in allogeneic transplant recipients. Front Immunol. 2018;9:1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toor A, Culibrk L, Singhera GKet al. Transcriptomic and proteomic host response to Aspergillus fumigatus conidia in an air–liquid interface model of human bronchial epithelium. PLoS One. 2018;13:e0209652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucey TM, Verma J, Olivier FABet al. Metabolic competition between host and pathogen dictates inflammasome responses to fungal infection. PLoS Pathog. 2020;16:e1008695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay J, Upadhyay RB, Agrawal Pet al. Langerhans cells and their role in oral mucosal diseases. N Am J Med Sci. 2013;5: 505–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban CF, Nett JE. Neutrophil extracellular traps in fungal infection. Semin Cell Dev Biol. 2019;89:47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban CF, Reichard U, Brinkmann Vet al. Neutrophil extracellular traps capture and kill Candida albicans yeast and hyphal forms. Cell Microbiol. 2006;8:668–76. [DOI] [PubMed] [Google Scholar]
- Vallabhaneni S, Chiller TM. Fungal infections and new biologic therapies. Curr Rheumatol Rep. 2016;18:29. [DOI] [PubMed] [Google Scholar]
- van der Does AM, Joosten SA, Vroomans Eet al. The antimicrobial peptide hLF1-11 drives monocyte-dendritic cell differentiation toward dendritic cells that promote antifungal responses and enhance Th17 polarization. J Innate Immun. 2012;4:284–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Helm MW, van der Meer AD, Eijkel JCet al. Microfluidic organ-on-chip technology for blood–brain barrier research. Tissue Barriers. 2016;4:e1142493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Veerdonk FL, Gresnigt MS, Romani Let al. Aspergillus fumigatus morphology and dynamic host interactions. Nat Rev Microbiol. 2017;15:661–74. [DOI] [PubMed] [Google Scholar]
- van de Veerdonk FL, Netea MG. T-cell subsets and antifungal host defenses. Curr Fungal Infect Rep. 2010;4:238–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Veerdonk FL, Plantinga TS, Hoischen Aet al. STAT1 mutations in autosomal dominant chronic mucocutaneous candidiasis. N Engl J Med. 2011;365:54–61. [DOI] [PubMed] [Google Scholar]
- Van Prooyen N, Henderson CA, Hocking Murray Det al. CD103+ conventional dendritic cells are critical for TLR7/9-dependent host defense against Histoplasma capsulatum, an endemic fungal pathogen of humans. PLoS Pathog. 2016;12:e1005749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma AH, Richardson JP, Zhou Cet al. Oral epithelial cells orchestrate innate type 17 responses to Candida albicans through the virulence factor candidalysin. Sci Immunol. 2017;2:eaam8834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar CC, Kashleva H, Nobile CJet al. Mucosal tissue invasion by Candida albicans is associated with E-cadherin degradation, mediated by transcription factor Rim101p and protease Sap5p. Infect Immun. 2007;75:2126–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivas W, Leonhardt I, Hünniger Ket al. Multiple signaling pathways involved in human dendritic cell maturation are affected by the fungal quorum-sensing molecule farnesol. J Immunol. 2019;203:2959–69. [DOI] [PubMed] [Google Scholar]
- Voelz K, May RC. Cryptococcal interactions with the host immune system. Eukaryot Cell. 2010;9:835–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel K, Pierau M, Arra Aet al. Developmental induction of human T-cell responses against Candida albicans and Aspergillus fumigatus. Sci Rep. 2018;8:16904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos T, Flaxman AD, Naghavi Met al. Years lived with disability (YLDs) for 1160 sequelae of 289 diseases and injuries 1990–2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet (London, England). 2012;380:2163–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachtler B, Wilson D, Haedicke Ket al. From attachment to damage: defined genes of Candida albicans mediate adhesion, invasion and damage during interaction with oral epithelial cells. PLoS One. 2011a;6:e17046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachtler B, Wilson D, Hube B. Candida albicans adhesion to and invasion and damage of vaginal epithelial cells: stage-specific inhibition by clotrimazole and bifonazole. Antimicrob Agents Chemother. 2011b;55:4436–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagener J, Mailander-Sanchez D, Schaller M. Immune responses to Candida albicans in models of in vitro reconstituted human oral epithelium. Methods Mol Biol. 2012;845:333–44. [DOI] [PubMed] [Google Scholar]
- Wang G, Yu G, Wang Det al. Comparison of the purity and vitality of natural killer cells with different isolation kits. Exp Ther Med. 2017;13:1875–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XJ, Zhu YJ, Cui JGet al. Proteomic analysis of human umbilical vein endothelial cells incubated with Cryptococcus neoformans var. neoformans. Mycoses. 2011;54:e336–43. [DOI] [PubMed] [Google Scholar]
- Warkentien T, Crum-Cianflone NF. An update on Cryptococcus among HIV-infected patients. Int J STD AIDS. 2010;21:679–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins TN, Liu H, Chung Met al. Comparative transcriptomics of Aspergillus fumigatus strains upon exposure to human airway epithelial cells. Microb Genom. 2018;4:e000154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weerasinghe H, Traven A. Immunometabolism in fungal infections: the need to eat to compete. Curr Opin Microbiol. 2020;58:32–40. [DOI] [PubMed] [Google Scholar]
- Weiss E, Schlegel J, Terpitz Uet al. Reconstituting NK cells after allogeneic stem cell transplantation show impaired response to the fungal pathogen Aspergillus fumigatus. Front Immunol. 2020;11:2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss E, Ziegler S, Fliesser Met al. First insights in NK-DC cross-talk and the importance of soluble factors during infection with Aspergillus fumigatus. Front Cell Infect Microbiol. 2018;8:288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss GA, Hennet T. Mechanisms and consequences of intestinal dysbiosis. Cell Mol Life Sci. 2017;74:2959–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weksler B, Romero IA, Couraud PO. The hCMEC/D3 cell line as a model of the human blood brain barrier. Fluids Barriers CNS. 2013;10:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellington M, Dolan K, Krysan DJ. Live Candida albicans suppresses production of reactive oxygen species in phagocytes. Infect Immun. 2009;77:405–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westman J, Moran G, Mogavero Set al. Candida albicans hyphal expansion causes phagosomal membrane damage and luminal alkalinization. mBio. 2018;9:e01226–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westman J, Walpole GFW, Kasper Let al. Lysosome fusion maintains phagosome integrity during fungal infection. Cell Host Microbe. 2020;28:798–812.e6. [DOI] [PubMed] [Google Scholar]
- Wheeler ML, Limon JJ, Underhill DM. Immunity to commensal fungi: detente and disease. Annu Rev Pathol. 2017;12:359–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D, Hube B. Hgc1 mediates dynamic Candida albicans–endothelium adhesion events during circulation. Eukaryot Cell. 2010;9:278–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D, Mayer FL, Miramon Pet al. Distinct roles of Candida albicans-specific genes in host–pathogen interactions. Eukaryot Cell. 2014;13:977–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiseman JC, Ma LL, Marr KJet al. Perforin-dependent cryptococcal microbicidal activity in NK cells requires PI3K-dependent ERK1/2 signaling. J Immunol. 2007;178:6456–64. [DOI] [PubMed] [Google Scholar]
- Wolf JE, Kerchberger V, Kobayashi GSet al. Modulation of the macrophage oxidative burst by Histoplasma capsulatum. J Immunol. 1987;138:582–6. [PubMed] [Google Scholar]
- Wollert T, Rollenhagen C, Langford GMet al. Human oral keratinocytes: a model system to analyze host–pathogen interactions. Methods Mol Biol. 2012;845:289–302. [DOI] [PubMed] [Google Scholar]
- Workman MJ, Mahe MM, Trisno Set al. Engineered human pluripotent-stem-cell-derived intestinal tissues with a functional enteric nervous system. Nat Med. 2017;23:49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue J, Schmidt SV, Sander Jet al. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity. 2014;40:274–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Shinohara ML. Tissue-resident macrophages in fungal infections. Front Immunol. 2017;8:1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang AM, Inamine T, Hochrath Ket al. Intestinal fungi contribute to development of alcoholic liver disease. J Clin Invest. 2017;127:2829–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeon JH, Na D, Choi Ket al. Reliable permeability assay system in a microfluidic device mimicking cerebral vasculatures. Biomed Microdevices. 2012;14:1141–8. [DOI] [PubMed] [Google Scholar]
- Youseff BH, Holbrook ED, Smolnycki KAet al. Extracellular superoxide dismutase protects Histoplasma yeast cells from host-derived oxidative stress. PLoS Pathog. 2012;8:e1002713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Zhao Z, Abdul Rahim NAet al. Towards a human-on-chip: culturing multiple cell types on a chip with compartmentalized microenvironments. Lab Chip. 2009;9:3185–92. [DOI] [PubMed] [Google Scholar]
- Zhang M, Xu C, Jiang Let al. A 3D human lung-on-a-chip model for nanotoxicity testing. Toxicol Res (Camb). 2018;7:1048–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinski CE, Mele F, Aschenbrenner Det al. Pathogen-induced human TH17 cells produce IFN-γ or IL-10 and are regulated by IL-1β. Nature. 2012;484:514–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.