Abstract
Dissemination of extended-spectrum cephalosporin (ESC)-resistant Salmonella is a public health concern in the egg production industry. ESC-resistant Salmonella often acquires the bla gene via insertion sequences (ISs). Therefore, this study aimed to assess antimicrobial resistance in Salmonella from Japanese layer breeding chains and egg processing chains, and determine the genetic profiles of IS-like elements in ESC-resistant Salmonella. Antimicrobial susceptibility testing was performed on 224 isolates from 49 facilities involving layer breeder farms, hatcheries, pullet-rearing farms, and layer farms in breeding chains along with egg processing chains. ESC-resistant Salmonella strains were whole-genome sequenced. Among them, 40 (17.9%) were resistant to at least streptomycin, tetracycline, ampicillin, chloramphenicol, cefpodoxime, nalidixic acid, ciprofloxacin, and/or kanamycin despite lacking resistance to azithromycin and meropenem. Moreover, 15 were ESC-resistant Salmonella harboring blaCMY-2 (Salmonella enterica serovar Ohio, n=12; S. Braenderup, n=1; untypeable with O7:b:-, n=1) and blaCTX-M-14 (S. Cerro, n=1). IncA/C2 plasmids containing ISEcp1, IS26, and multiple antimicrobial resistance genes (including blaCMY-2) were identified in S. Ohio isolates from pullet-rearing and layer farms belonging to the same company. Chromosomal integration of partial or whole IncA/C2 plasmids was seen with two S. Ohio isolates via ISEcp1 or IS26, respectively. Antimicrobial resistance genes such as blaCMY-2 might be transmitted among the upper and the lower levels of layer breeding chains via the replicon type IncA/C2 plasmids containing ISEcp1 and IS26.
Keywords: blaCMY-2, chromosomally-integrated IncA/C2 plasmid, IS26, layer breeding chain, Salmonella
Investigations into antimicrobial-resistant Salmonella in the egg production industry are essential because dissemination of this bacterium in this industry is a public health concern. Extended-spectrum cephalosporin (ESC) treatment is required for severe infections with antimicrobial-resistant Salmonella in humans [10]. No data, however, are currently available about the incidence of chicken egg-borne salmonellosis in Japan despite the consumption of eggs and egg products being one of the major causes of human salmonellosis worldwide [38]. Therefore, better knowledge and understanding about the prevalence of antimicrobial-resistant Salmonella in contaminated eggs and egg products are important. Vertical transmission of Salmonella occurs in layer chickens in breeding chains [14, 15, 30]. The chains have upper levels (breeder farms, hatcheries, and pullet-rearing farms) and a lower level (layer farms) [7, 35]. To the best of our knowledge, only two studies have been conducted on antimicrobial-resistant Salmonella in layer breeding chains and shell and liquid egg processing chains (LB-EP chains) in Japan. These studies reported that the environments within the layer farms and the shell egg grading and packing centers (EGPs) contained Salmonella with resistance to antimicrobials such as ampicillin (ABPC), streptomycin (SM), and cephalothin [11, 32]. Moreover, the EGPs are key factors relating to Salmonella transmission in egg production. Therefore, investigating the prevalence of antimicrobial-resistant Salmonella, especially ESC-resistant Salmonella, in the LB-EP chains, including the upper levels of the layer breeding chains, is important.
ESC-resistant Salmonella is generated when bla genes encoding AmpC β-lactamase and extended-spectrum β-lactamase (ESBL) are acquired by Salmonella [8, 27,28,29]. bla genes translocate between plasmids and chromosomes containing insertion sequences (ISs) among Enterobacteriaceae, including Salmonella [8, 23, 27, 29]. In fact, blaCTX-M-14 was translocated by ISEcp1 from a plasmid into another plasmid in Salmonella enterica subsp. enterica serovar Senftenberg (S. Senftenberg), whereas blaCTX-M-15 was translocated by ISEcp1 from a plasmid into S. Haardt chromosomes [29]. Obtaining further knowledge about IS26 is also important because IS26 may generate multiple antimicrobial-resistant Salmonella via gene integration [9, 23, 27]. A field study indicated that nine antimicrobial resistance genes, including tet(A), sul2, floR, and blaCMY-2, in an IncA/C plasmid were simultaneously integrated into the S. Typhimurium chromosome by IS26 [27]. A more recent laboratory study showed that IS26 can translocate blaTEM-1, sul2, and strAB from the S. Typhimurium chromosome into its plasmid [23]. These studies suggest that IS26 may be associated with the generation of multiple antimicrobial-resistant Salmonella. Therefore, understanding the emergence and dissemination of ESC-resistant Salmonella requires the investigation of ISs such as ISEcp1 and IS26 on bla gene-carrying plasmids and chromosomes. The aims of this study were as follows: (i) assessing Salmonella-specific antimicrobial resistance patterns in LB-EP chains, especially ESC-resistant Salmonella; (ii) determining plasmid and chromosome encoded features in Salmonella including ISs and IS-related bla genes using whole-genome sequencing (WGS); and (iii) confirming whether IS26 generates a multiple antimicrobial resistance phenotype in Salmonella.
MATERIALS AND METHODS
Samples from eggshells and egg production environments
A total of 224 samples were collected from one breeder farm (n=4), two hatcheries (n=2), five pullet-rearing farms (n=20), 39 layer farms (n=194), and two EGPs (n=4) in Japanese LB-EP chains between 2009 and 2016, excluding 2014 (Table 1). From them, 51, 43, 41, 43, 40, 3, and 3 were gathered in 2009, 2010, 2011, 2012, 2013, 2015, and 2016, respectively. These 49 facilities are located in seven prefectures (A, B, C, D, E, F, and G), with six of the seven located in western Japan (prefectures A–F), and the remaining one in eastern Japan (prefecture G).
Table 1. Sample information for the Salmonella isolates obtained from upper a and lower b levels in layer breeding chains and egg shell grading and packing centers (EGPs).
| Facility type | No. of facilities |
No. of isolates |
No. of samples |
||||
|---|---|---|---|---|---|---|---|
| Pooled eggshell | Swab | Dust | Feces | Unidentified | |||
| Layer breeder farm | 1 | 4 | 4 | ||||
| Hatchery | 2 | 2 | 1 | 1 | |||
| Pullet-rearing farm | 5 | 20 | 14 | 5 | 1 | ||
| Layer farm | 39 | 194 | 89 | 58 | 45 | 2 | |
| EGP | 2 | 4 | 4 | ||||
| Total | 49 | 224 | 89 | 77 | 50 | 5 | 3 |
a Upper levels consist of layer breeder farms, hatcheries, and pullet-rearing farms. b Lower levels consist of layer farms.
The 224 samples were pooled eggshell samples (n=89) and environmental egg production samples (n=135; swab, n=77; dust, n=50; feces, n=5; unidentified, n=3) from facilities in the LB-EP chains. The types of facility where these samples were collected are shown in Table 1. Each pooled eggshell sample consisted of 100–150 eggshell remnants from liquid egg production on each farm. Environmental sampling of the egg production facilities was conducted as follows. Dust clouds were collected from the ventilators attached to the pullet-rearing farms and layer farms, and feces were collected from a layer breeder farm or a hatchery. Swabs were also obtained from the swabbing floors of all the facilities other than the EGPs, and from the swabbing floors and egg containers in the EGPs. The swabs were gathered using swabbing sheets (Kanto Chemical Co., Inc., Tokyo, Japan) and cotton gauze swabs (Eiken Chemical Co., Tokyo, Japan).
Salmonella were isolated from the samples in accordance with a previous description with minor revision [17, 28]. Each sample, except for the pooled eggshell samples, was mixed with 225 ml of buffered peptone water (BPW, Oxoid Ltd., Hampshire, UK), and then tapped by hand for 1 min. The mixed BPW was cultured (37°C, 20 ± 2 hr). The pooled eggshell samples were cultured in 1,500–2,000 ml of BPW (37°C, 20 ± 2 hr). An aliquot (0.1 ml) of the BPW culture was added to Rappaport-Vassiliadis enrichment broth (Oxoid Ltd.) and cultured (42°C, 20 ± 2 hr). The cultured broth was streaked onto mannitol, lysine, crystal violet and brilliant green agar (Oxoid Ltd.) and Rimler-Shotts-Maeda agar (Kanto Chemical Co.) at 37°C for 18–48 hr for bacterial isolation. Four putative Salmonella colonies were picked and then identified as Salmonella by biochemical tests following the previous reports [16, 28, 29]. Serological tests were conducted in accordance with our previous description [16, 28, 29].
Antimicrobial susceptibility and phenotype testing
Antimicrobial susceptibility tests were performed using the disk diffusion method in accordance with the Clinical and Laboratory Standards Institute (CLSI) guidelines [4]. The susceptibility test results were evaluated in accordance with the CLSI criteria for resistance breakpoints [5]. The following 10 antimicrobials were used: ABPC (10 µg), cefpodoxime (CPDX, 10 µg), chloramphenicol (CP, 30 µg), ciprofloxacin (CPFX, 5 µg), nalidixic acid (NA, 30 µg), kanamycin (KM, 30 µg), SM (10 µg), azithromycin (AZM, 15 µg), tetracycline (TC, 30 µg), and meropenem (MEPM, 10 µg). The antimicrobial disks came from Becton, Dickinson, and Co. (Franklin Lakes, NJ, USA). Regarding the isolates showing resistance to ABPC, ESBL-producing confirmatory tests were performed using ceftazidime (CAZ) (30 µg)-clavulanic acid (CVA) (Becton, Dickinson, and Co.), CTX (30 µg)-CVA (Becton, Dickinson, and Co.), and CPDX (30 µg)-CVA (Eiken Chemical Co.) disks, as per our previous reports [28, 29]. We also performed boronic acid tests using CPDX and cefoxitin disks as per our previous reports [28, 29]. Escherichia coli strain ATCC 25922 was used as the quality control strain in all the assays in accordance with CLSI guidelines [5]. Klebsiella pneumoniae ATCC 700603 and E. coli ATCC 35218 were also used as quality control strains.
The minimum inhibitory concentrations (MICs) of ABPC, ampicillin-sulbactam, CAZ, CPDX, CTX and ceftriaxone antimicrobials were evaluated against the four strains (three S. Ohio strains: SEOhiM1593, 1960, and 2008, and one S. Cerro strain: SECerM2017) using the Etest (bioMérieux, Marcy-l’Étoile, France), which was performed in accordance with the manufacturer’s instructions.
Detection and sequencing analysis of β-lactamase genes
Polymerase chain reactions (PCRs) were conducted as previously described to detect β-lactamase genes (blaCMY, blaCTX-M, blaSHV, and blaTEM) in the isolates that were resistant to ABPC [22, 28, 29]. The presence of the bla gene was determined by DNA sequencing as described in a previous study with minor revisions using the primers shown in Supplementary Table 1 [22]. PCR products were purified using the illustra ExoProStar enzymatic PCR and Sequencing Clean-Up kit (Global Life Sciences Solutions USA LLC., Marlborough, MA, USA). DNA sequencing was performed using the BigDye Terminator Cycle Sequencing Reaction v. 3.1 kit (Thermo Fisher Scientific Inc., Waltham, MA, USA), and the products were analyzed on the 3500 Genetic Analyzer (Thermo Fisher Scientific Inc.). The DNA sequences were analyzed by comparison with the reference sequences registered in the National Center for Biotechnology Information (NCBI) database [19].
Epidemiological information on bla gene-carrying Salmonella strains
To elucidate the relationship(s) involving the dissemination of ESC-resistant, bla gene-carrying Salmonella between the lower-level facilities and upper-level facilities in the layer breeding chains, we collected detailed information about the strains, such as the facility type and the isolation date (Supplementary Table 2).
WGS on Salmonella harboring blaCMY-2 or blaCTX-M-14 and Salmonella genome analysis
WGS was performed to obtain the complete chromosomal and plasmid DNA sequences of three S. Ohio strains harboring blaCMY-2 (SEOhiM1593, 1960, and 2008) and one S. Cerro strain harboring blaCTX-M-14 (SECerM2017) in accordance with the methods used in our previous study with minor revisions [18]. To confirm the genetic features of Salmonella harboring blaCMY-2 or blaCTX-M-14, we selected four strains for WGS analysis. Salmonella strains harboring blaCMY-2 were selected from the blaCMY-2-carrying strains derived from both layer farms and pullet-rearing farms belonging to the same layer breeding chains. Of the Salmonella strains harboring blaCMY-2, two S. Ohio strains were selected at random from the 11 blaCMY-2-carrying strains derived from pullet-rearing farms and one S. Ohio was selected from a layer farm. We also selected the S. Cerro strain (SECerM2017) for analysis because it was the only blaCTX-M-14-carrying strain. Total DNA was extracted from each strain using the MagAttract HMW DNA Kit (Qiagen, Venlo, Netherlands). The extracted DNA was barcoded (Native Barcoding Expansion 1–12; Oxford Nanopore, Oxford, UK), and made into a nanopore-sequencing library with Ligation Sequencing 1D Kit (Oxford Nanopore). The library was sequenced on R9.4.1 flowcells using the MinION portable DNA sequencer (Oxford Nanopore). We also performed Illumina sequencing on the extracted genomic DNA preparations remaining from the nanopore-sequencing. Libraries were prepared using the Nextra DNA Flex Library Prep Kit (Illumina, Inc., San Diego, CA, USA). Cycles of 300 or 500 dual-index paired-end sequencing were performed using Illumina MiSeq (Illumina, Inc.). Complete chromosomes and plasmids from the three S. Ohio strains and the single S. Cerro strain were generated from the Nanopore and Illumina Sequencing data using Unicycler v.0.4.4 or Flye v.2.4.2 for de novo assembling and Pilon v.1.23 for polishing [13, 36, 37]. The complete chromosomal sequences and plasmid sequences were analyzed using nucleotide BLAST (BLASTN) homology searches in PlasmidFinder and ResFinder to infer the plasmid replicon types and the antimicrobial resistance genes as per our previous study [1, 12, 28]. The DDBJ Fast Annotation and Submission Tool (DFAST) annotation pipeline in the DNA Data Bank of Japan (DDBJ) was used for genome annotation [20].
Comparative genome sequence analysis
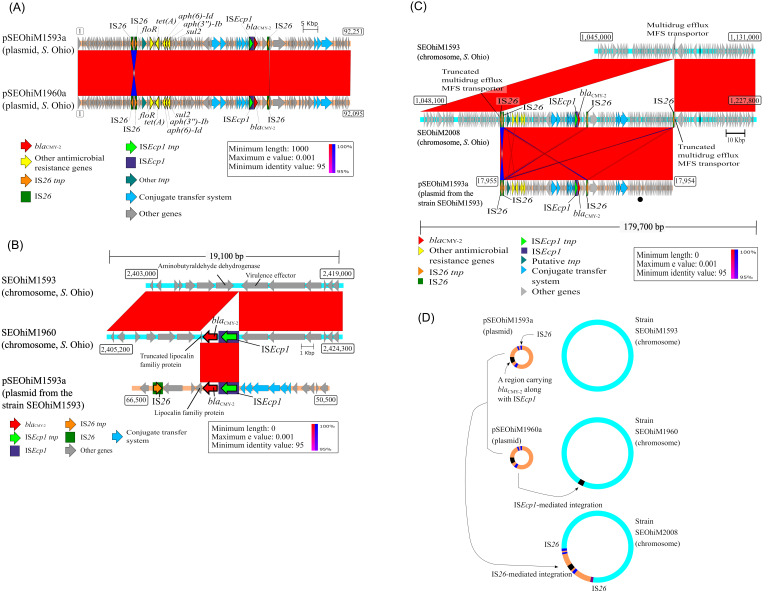
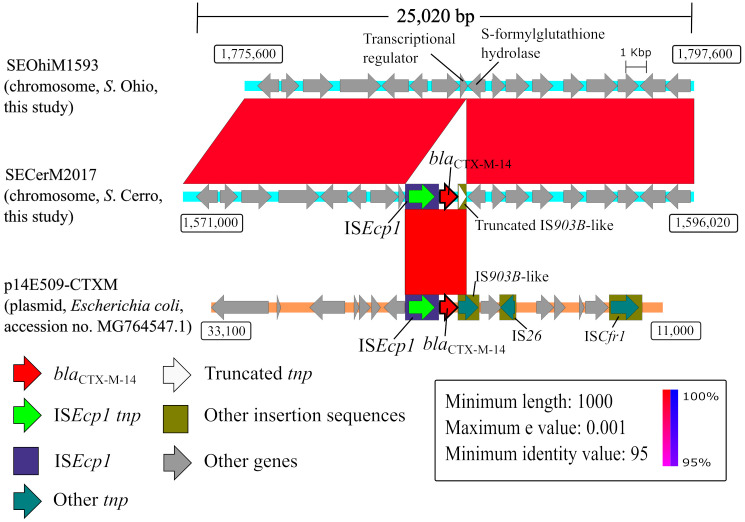
The nucleotide sequences from two blaCMY-2-carrying plasmids (pSEOhiM1593a and 1960a) and those from the chromosomes of the three S. Ohio strains (SEOhiM1593, 1960, and 2008) were used for structural genome comparisons (Fig. 1). The pSEOhiM1593a plasmid (recovered from strain SEOhiM1593) was compared with the pSEOhiM1960a plasmid (recovered from strain SEOhiM1960) (Fig. 1A). The blaCMY-2-containing SEOhiM1960 chromosome and the blaCMY-2-lacking SEOhiM1593 chromosome were used for comparison; they were also used for another comparison with the pSEOhiM1593a plasmid (Fig. 1B). The SEOhiM2008 chromosome was compared in the same manner as that of the SEOhiM1960 chromosome (Fig. 1C). Additionally, a genome structure comparison of the blaCTX-M-14-harboring S. Cerro (SECerM2017) chromosome was performed. The chromosomal nucleotide sequence from SECerM2017 was compared with that from SEOhiM1593 (Fig. 2). We also compared the SECerM2017 chromosome with the complete sequence of the p14E509-CTXM plasmid (GenBank accession no. MG764547.1) [39]. We did this because the BLASTN analysis revealed that this plasmid shares the highest nucleotide sequence similarity score (coverage 100%) with a blaCTX-M-14-containing region in SECerM2017. In common with the other genome comparisons, the sequence of the p14E509-CTXM reference plasmid was annotated using DFAST. Comparative genome sequence analysis was performed using BLASTN and the Artemis Comparison Tool [2], and the results were visualized using Easyfig [33]. ISs were confirmed in the analyzed plasmids using ISfinder [31].
Fig. 1.
(A) Structural comparison of pSEOhiM1593a and pSEOhiM1960a plasmids. pSEOhiM1960a is derived from a layer farm and shares high similarity with pSEOhiM1593a derived from a pullet-rearing farm in the same breeding chain of the same company in different years. These plasmids harbor six antimicrobial resistance genes, one ISEcp1, and three IS26 (Table 3). Numbers in rectangular boxes are nucleotide positions. (B) Structural comparison of the blaCMY-2-carrying-chromosome in strain SEOhiM1960. A chromosomal region carrying blaCMY-2 along with ISEcp1 is completely identical to a part of pSEOhiM1593a. Numbers in rectangular boxes are nucleotide positions. (C) Structural comparison of the Salmonella enterica subsp. enterica serovar Ohio chromosome from strain SEOhiM2008, which carries multiple antimicrobial-resistant genes (Table 3). The pSEOhiM1593a plasmid is drawn to show the rearrangement at the start position (from the rep gene-encoding plasmid’s replication protein). The black bullet point indicates the position of the rep gene. Numbers in rectangular boxes are nucleotide positions. A region similar to intact pSEOhiM1593a was detected on the SEOhiM2008 strain’s chromosome, although the region sandwiched between the two IS26 is inverted. This region is flanked by two IS26 elements, and a couple of target duplication sites of 8 bp in length were also detected, suggesting that IS26 integrated this region into the chromosome. (D) Horizontal transmission of positively related plasmids (pSEOhiM1593a and its highly related plasmids) between S. Ohio strains, and integration of these plasmids into the chromosomes of these strains via insertion sequences ISEcp1 or IS26. The plasmids were transmitted horizontally from the upper levels (pullet-rearing farm) to the lower level (layer farm) in a layer breeding egg production chain within the same company. A part of the whole plasmid or its entirety was integrated into the S. Ohio chromosomes by ISEcp1 or IS26, respectively.
Fig. 2.
Structural comparison of the blaCTX-M-14-carrying chromosome from Salmonella enterica subsp. enterica serovar Cerro (SECerM2017) versus the analogous chromosomal region from S. Ohio (SEOhiM1593). The region carrying blaCTX-M-14 along with ISEcp1 in the chromosome is identical to a blaCTX-M-15-carrying part of plasmid p14E509-CTXM from a clinical Escherichia coli isolate from China (GenBank accession no. MG764547.1). Numbers in rectangular boxes are nucleotide positions.
In silico comparative analysis of the pSEOhiM1593a plasmid and Enterobacteriaceae-derived plasmids
The genetic features of pSEOhiM1593a (e.g., antimicrobial resistance genes, plasmid replicon types, and ISs) were compared with those of plasmids derived from Enterobacteriaceae and Salmonella (Supplementary Table 3). The plasmids used for this comparison were identified by BLASTN searches based on their similarities (coverage >99%) with pSEOhiM1593a. The nucleotide sequence data for these plasmids were obtained from GenBank (accession nos. CP009564, MH760469, HQ023864, KP056256, KR559888, MF344573, and KJ909290). All sequence data from the plasmids were reanalyzed using PlasmidFinder and ResFinder [1, 12], and ISs in the plasmids were confirmed using ISfinder [31].
Ethics statement
This study was authorized by the Ethics Regulations Related to Epidemiological Research at the Fukuoka Institute of Health and Environmental Sciences (permission number R2-4) and its guidelines were observed.
Data availability
The complete, annotated genomic sequences from the S. Ohio and S. Cerro strains are deposited in the DDBJ under accession nos. AP024347–AP024353 and AP024345–AP024346, respectively. The obtained short- and long-read sequence data are deposited in the DDBJ as follows: S. Ohio (SEOhiM1593; BioProject PRJDB10937, BioSample SAMD00206786, DRA accession nos. DRR261749 and DRR261750); S. Ohio (SEOhiM1960; BioProject PRJDB10937, BioSample SAMD00206787, DRA accession nos. DRR261751 and DRR261752); S. Ohio (SEOhiM2008; BioProject PRJDB10937, BioSample SAMD00206788, DRA accession nos. DRR261753 and DRR261754); and S. Cerro (SECerM2017; BioProject PRJDB10937, BioSample SAMD00264093, DRA accession nos. DRR261747 and DRR261748).
RESULTS
Salmonella isolates and serotypes
Altogether, 224 Salmonella isolates were obtained from 224 samples. The 224 isolates were recovered from the 224 samples originating from one breeder farm (n=4), two hatcheries (n=2), five pullet-rearing farms (n=20), 39 layer farms (n=194), and two EGPs (n=4) (Table 1). The 224 Salmonella isolates derived from pooled eggshell samples and environmental egg production samples from the LB-EP chains were classified as the 36 serotypes listed in Supplementary Table 4.
Antimicrobial susceptibility tests and bla gene determination
The antimicrobial susceptibility tests showed that 40/224 Salmonella isolates were resistant to at least one antimicrobial (Table 2). In fact, 184 were susceptible to all 10 of the tested antimicrobials. None of the isolates (n=4) from a breeder farm displayed antimicrobial resistance, but one of the two isolates from the hatcheries did. The 40 antimicrobial-resistant Salmonella isolates generated 11 antimicrobial resistance patterns, with ABPC-CPDX-CP-SM-TC being the most frequently observed pattern (n=13). Among these 40 isolates, 30, 26, 20, 16, 15, 6, 4, and 1 were resistant to SM, TC, ABPC, CP, CPDX, NA, CPFX, and KM, respectively (Supplementary Table 5). No isolates were resistant to AZM or MEPM. Of the 20 isolates resistant to ABPC, 14 were AmpC β-lactamase-producers (S. Ohio; n=12, S. Braenderup; n=1, O7:b:- ; n=1) and carried blaCMY-2; one of the 20 isolates was an ESBL-producer (S. Cerro; n=1) and carried blaCTX-M-14 (Table 2). Five (S. Kentucky; n=4, S. Infantis; n=1) of the 20 isolates did not carry a bla gene other than the blaTEM-1 non-ESBL gene. The MICs of the ESCs from the four Salmonella strains (SEOhiM1593, 1960 and 2008, and SECerM2017) are shown in Supplementary Table 6. No significant differences were observed in the MICs of the ESCs between strain SEOhiM1960, which harbors multiple blaCMY-2 genes, and the other strains with a single blaCMY-2.
Table 2. Antimicrobial resistance patterns of Salmonella isolates and bla gene detection.
| Serotypes | Antimicrobial resistance patternsa | bla genes | Phenotypic tests | No. of isolates by sample source |
Total No. of isolates | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Eggshell | Environment |
|||||||||
| EGPb | Layer farm | Pullet-rearing farm | Hatchery | Breeder farm | ||||||
| Braenderup (n=36) | ABPC-CPDX | blaCMY-2 | AmpCc | 1 | 1 | |||||
| SM | 1 | 1 | ||||||||
| No resistance | 23 | 3 | 8 | 34 | ||||||
| Ohio (n=22) | ABPC-CPDX-CP-SM-TC | blaCMY-2 | AmpC | 1 | 11 | 12 | ||||
| SM-CP-TC | 1 | 1 | ||||||||
| No resistance | 9 | 9 | ||||||||
| O7:b:− (n=1) | ABPC-CPDX-CP-SM-TC | blaCMY-2 | AmpC | 1 | 1 | |||||
| Cerro (n=20) | ABPC-CPDX | blaCTX-M-14 | ESBLd | 1 | 1 | |||||
| SM | 3 | 3 | ||||||||
| No resistance | 6 | 7 | 1 | 2 | 16 | |||||
| Infantis (n=22) | ABPC-KM-NA-SM-TC | blaTEM-1 | NPe | 1 | 1 | |||||
| SM-CP | 1 | 1 | ||||||||
| SM | 1 | 1 | ||||||||
| No resistance | 14 | 5 | 19 | |||||||
| Kentucky (n=4) | ABPC-CPFX-NA-SM-TC | blaTEM-1 | NP | 2 | 2 | |||||
| ABPC-CPFX-NA | blaTEM-1 | NP | 2 | 2 | ||||||
| Agona (n=9) | TC | 6 | 6 | |||||||
| No resistance | 3 | 3 | ||||||||
| Alachua (n=21) | SM | 1 | 1 | 2 | ||||||
| No resistance | 5 | 14 | 19 | |||||||
| Livingstone (n=4) | SM-CP | 1 | 1 | |||||||
| No resistance | 1 | 2 | 3 | |||||||
| Manhattan (n=2) | SM-TC | 1 | 1 | 2 | ||||||
| Schwarzengrund (n=1) | SM-NA-TC | 1 | 1 | |||||||
| Tenessee (n=2) | SM | 1 | 1 | |||||||
| No resistance | 1 | 1 | ||||||||
| Othersf (n=80) | No resistance | 30 | 1 | 41 | 6 | 2 | 80 | |||
a ABPC: ampicillin, CP: chloramphenicol, CPFX: ciprofloxacin, CPDX: cefpodoxime, KM: kanamycin, NA: nalidixic acid, SM: streptomycin, TC: tetracycline. b EGP: egg shell grading and packing centers. c Isolates confirmed as AmpC β-lactamase producers. d An isolate confirmed as an extended-spectrum β-lactamase (ESBL) producer. e NP: these isolates produced neither ESBL nor AmpC β-lactamase. f Including the 24 serotypes listed in Supplementary Table 4.
Epidemiological information for the ESC-resistant strains carrying blaCMY-2 or blaCTX-M-14
ESC-resistant Salmonella strains harboring blaCMY-2 (n=14 in total: S. Ohio, n=12; S. Braenderup, n=1; untypeable with O7:b:-, n=1) and blaCTX-M-14 (S. Cerro, n=1) were isolated from two or three pullet-rearing farms and a layer farm belonging to egg-production company A in prefecture A, a layer farm belonging to company B in prefecture B, and a layer farm belonging to company Q in prefecture C (Supplementary Table 2). All three prefectures are contiguously located. One S. Cerro strain carrying the blaCTX-M-14 (SECerM2017) obtained from a layer farm belonging to company Q and three S. Ohio strains carrying blaCMY-2 (SEOhiM1593, 1960, and 2008) obtained from a pullet-rearing farm and a layer farm belonging to company A, were further analyzed as described below.
WGS identification of antimicrobial resistance genes, plasmid replicon types, and ISs
Table 3 shows the results of the analysis on antimicrobial resistance genes, plasmid replicon types, and ISs from the three S. Ohio strains (SEOhiM1593, 1960, and 2008) and the S. Cerro strain (SECerM2017). Based on this analysis, we classified pSEOhiM1593a and 1960a as IncA/C2 plasmids. The two IncA/C2 plasmids exhibit high-level nucleotide sequence identity with each other (Fig. 1A). Moreover, nucleotide sequences on the SEOhiM2008 chromosome were assigned as belonging to an IncA/C2 plasmid (Table 3). The chromosome region of SEOhiM2008 and the two IncA/C2 plasmids exhibit high nucleotide sequence identity with each other (Fig. 1A and 1C). Therefore, the evidence supports IncA/C2 plasmid integration into the SEOhiM2008 strain’s chromosome (Fig. 1D). The chromosomal region encompassing nucleotide position 1,090,811–1,183,061 surrounded by IS26 shares high sequence identity with the corresponding region of the plasmid belonging to pSEOhiM1593a (nucleotide position 1–92,251), except for an inverted region sandwiched (2,064 bp) between two IS26 elements. This chromosomal region is flanked by a target site duplication of 8 bp (5′-AAGAATAT-3′) suggesting that IS26 has integrated this region. pSEOhiM1593a, pSEOhiM1960a, and the chromosomal region of strain SEOhiM2008 assigned as belonging to an IncA/C2 plasmid harbored genes conferring resistance against aminoglycoside (aph(3”)-Ib and aph(6)-Ib), florfenicol/chloramphenicol (floR), sulfonamide (sul2), and tetracycline (tet(A)), as well as ESC (blaCMY-2). Strains SEOhiM1593, SEOhiM1960, and SEOhiM2008 were isolated from different production levels (pullet-rearing farm and layer farm strains) in the same company (Supplementary Table 2). blaCMY-2 was detected on the SEOhiM1960 strain’s chromosome (Table 3 and Fig. 1B). Furthermore, the chromosomal region of the nucleotide position between 2,412,761 and 2,415,912 is identical to the partial region carrying blaCMY-2 along with ISEcp1 in pSEOhiM1593a (nucleotide position 57,825–60,976).
Table 3. Chromosome and plasmid analysis on Salmonella enterica harboring blaCMY-2 or blaCTX-M-14 from whole genome sequencing.
| Strain name | Serotype | DNA type | Plasmid name | bla gene | Other antimicrobial resistant genes | No. of IS26 elements | No. of ISEcp1 elements within the region flanking the bla genea | Replicon type | Size (bp) | Strain origin |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Facility type | Sample type | Facility name | Isolation date | ||||||||||
| SEOhiM1593 | S. Ohiob | Chromosome | - | NDc | aac(6’)-Iaa | 0 | 0 | - | 4,867,732 | Pullet-rearing farm | Environment | a1 | January 2010 |
| Plasmid | pSEOhiM1593a | blaCMY-2 | aph(3”)-Ib, aph(6)-Id, floR, sul2, tet(A) | 3 | 1 | IncA/C2 | 92,251 | ||||||
| SEOhiM2008 | S. Ohio | Chromosome | - | blaCMY-2 | aph(3”)-Ib, aph(6)-Id, floR, sul2, tet(A) | 4 | 1 | IncA/C2d | 4,902,159 | Pullet-rearing farm | Environment | a1 | June 2012 |
| Plasmid | pSEOhiM2008a | ND | ND | 0 | 0 | Col440I | 2,264 | ||||||
| SEOhiM1960 | S. Ohio | Chromosome | - | blaCMY-2 | aac(6’)-Iaa | 0 | 1 | - | 4,872,199 | Layer farm | Eggshell | a5 | January 2012 |
| Plasmid | pSEOhiM1960a | blaCMY-2 | aph(3”)-Ib, aph(6)-Id, floR, sul2, tet(A) | 3 | 1 | IncA/C2 | 92,095 | ||||||
| Plasmid | pSEOhiM1960b | ND | ND | 0 | 0 | Col440I | 2,264 | ||||||
| SECerM2017 | S. Cerro | Chromosome | - | blaCTX-M-14 | ND | 0 | 1 | - | 4,479,959 | Layer farm | Eggshell | q1 | August 2012 |
| Plasmid | pSECerM2017a | ND | ND | 0 | 0 | Untypeable | 2,105 | ||||||
a Number of ISEcp1 elements within 5,000 bp of the bla gene. bSalmonella enterica subsp. enterica serovar Ohio. c ND: not-detected. d The integration region in this strain is structured almost identically to the IncA/C2 plasmids in the SEOhiM1593 and SEOhiM1960 strains.
Of note, blaCTX-M-14 is present on the SECerM2017 chromosome. We found that the region carrying blaCTX-M-14 along with ISEcp1 on this chromosome (3,006 bp, nucleotide position 1,581,874–1,584,879) is identical to the corresponding region in the p14E509-CTXM plasmid (nucleotide position 20,608–23,613) from a Chinese clinical E. coli strain in 2014 (GenBank accession no. MG764547.1) (Fig. 2).
In silico comparative analysis on the pSEOhiM1593a plasmid and Enterobacteriaceae-derived plasmids
We compared the genetic features of pSEOhiM1593a with those of Enterobacteriaceae family plasmids, including Salmonella from other studies (Supplementary Table 3). The plasmids used for the comparison were found in Enterobacteriaceae derived from humans and animals. All the plasmids were found to carry multiple antimicrobial resistance genes and more than one IS26. We identified antimicrobial resistance genes (blaCMY-2, aph(3”)-Ib, aph(6)-Id, floR, sul2, and tet(A)) sharing among all the plasmids, but the pKP-Gr642 plasmid carries blaCMY-4 instead of blaCMY-2.
DISCUSSION
Our study has three main findings. First, Salmonella harboring blaCMY-2 (S. Ohio; n=12, S. Braenderup; n=1, untypeable with O7:b:-; n=1) and harboring blaCTX-M-14 (S. Cerro; n=1) were found in LB-EP chains in Japan. Second, two S. Ohio strains, SEOhiM1593 and 1960, harbor IncA/C2 plasmids with multiple antimicrobial resistance genes including blaCMY-2, ISEcp1, and IS26. Third, strain SEOhiM1960 possesses a partial region of an IncA/C2 plasmid on its chromosome and strain SEOhiM2008 possesses the entire region of an IncA/C2 plasmid on its chromosome, and these chromosomal integrations involved ISEcp1 or IS26, respectively. Despite numerous reports of ESC-resistant Salmonella in broiler breeding chains [3, 6, 28], few studies have investigated it in any great depth in LB-EP chains. S. Virchow harboring blaCTX-M-9, S. Infantis harboring blaCTX-M-65, and S. Gallinarum harboring blaTEM were reported in LB-EP chains in Spain, Ecuador, and India, respectively [24,25,26]. Herein, we found that ESC-resistant Salmonella is present in Japanese LB-EP chains (Table 2 and Supplementary Table 2). Furthermore, because pSEOhiM1593a, pSEOhiM1960a, and the chromosomally integrated pSEOhiM1593a-related plasmid in strain SEOhiM2008, whose replicon type is classified as IncA/C2, were found in the upper- and lower-level strains (SEOhiM1593, 1960, and 2008), this indicates that antimicrobial-resistance gene transmission through IncA/C2 plasmids occurred among Salmonella in the layer chicken breeding chains. Better hygiene management in breeding chains, including the upper levels of layer chicken breeding chains, is needed to control egg contamination with antimicrobial-resistant Salmonella.
Obtaining pSEOhiM1593a-related plasmids with IncA/C2 from Enterobacteriaceae might enhance each recipient’s survival in various hosts via the acquisition of antimicrobial resistance genes. IncA/C2 plasmids carrying blaCMY are disseminated among Enterobacteriaceae from various sources and countries (Supplementary Table 3), and their dissemination might involve a specific antimicrobial resistance gene set. By acquiring IncA/C2 plasmids with the blaCMY-2, blaCMY-4, aph(3”)-Ib, aph(6)-Id, floR, sul2, and tet(A) antimicrobial-resistance gene set, Enterobacteriaceae will be resistant to β-lactams, aminoglycosides, florfenicol/chloramphenicol, sulfas, and tetracyclines. Harboring IncA/C2 plasmids containing this gene set would advantage Enterobacteriaceae survival in the presence of the above-named antimicrobials. According to the US Food and Drug Administration’s research in 2018, aminoglycosides, sulfas, and tetracyclines were used for infections in swine whereas cephalosporins, penicillins, aminoglycosides, sulfas, and tetracyclines were used for infections in cattle [34]. It is possible that Enterobacteriaceae including Salmonella recovered from swine and cattle in the US might have survived by obtaining the IncA/C2 pCVM21550 and pAR060302 plasmids listed in Supplementary Table 3. Japanese governmental research in 2018 indicated that penicillins, aminoglycosides, sulfonamides, and tetracyclines were used on layer chickens in Japan [21]. The present study might also show that acquisition of pSEOhiM1593a-related plasmids with IncA/C2 may benefit the persistence of S. Ohio in LB-EP chains. However, no data were available to this study about the use of antimicrobials on these Japanese farms; hence, we were unable to determine whether antimicrobial use was a relevant factor on these farms. Further investigation is required to confirm that IncA/C2 plasmids harboring the aforementioned antimicrobial resistance gene set might be more likely to spread among Enterobacteriaceae in various hosts and countries that have used antimicrobials.
ISEcp1 may spread the bla gene among Salmonella via the integration of a partial plasmid region, whereas IS26 may disseminate multiple antimicrobial resistance genes among Salmonella through incorporation of the whole plasmid or a broad range of plasmids. Herein, we confirmed that the plasmid-derived partial region containing blaCMY-2 or blaCTX-M-14 was integrated by ISEcp1 into the S. Ohio chromosome or S. Cerro chromosome, respectively. Furthermore, IS26 may disseminate multiple antimicrobial resistance genes (including the bla gene) within the plasmids present among Salmonella. IS26 reportedly occurred only after a partial regional integration of the IncY and IncA/C2 fusion plasmid, which carried blaCTX-M-15 into the S. Concord chromosome [8]. However, IS26 possibly integrated a large portion (~120 kbp) of an E. coli IncA/C plasmid containing multiple antimicrobial resistance genes (including blaCMY-2) into the S. Typhimurium chromosome [27]. Our own findings indicate that S. Ohio obtained multiple antimicrobial resistance genes (including blaCMY-2) via IS26-mediated integration of an intact pSEOhiM1593a-related plasmid. In fact, ISEcp1 integrated a partial bla gene-carrying region existing in pSEOhiM1593a-related plasmid into S. Ohio strain SEOhiM1960 (Fig. 1B), while IS26 integrated the whole region of the pSEOhiM1593a-related plasmid into S. Ohio strain SEOhiM2008 (Fig. 1C), which resulted in the dissemination of multiple antimicrobial resistance genes via IS26. Overall, it seems likely that antimicrobial resistance genes, including the bla gene, have dispersed in Salmonella within the layer breeding chains via ISs-mediated-integration of plasmids.
In conclusion, Salmonella obtains numerous antimicrobial resistance genes, especially bla genes on the IncA/C2 plasmid, through partial regional plasmid integration by ISEcp1 and the larger or whole regional integration of the plasmid by IS26. Antimicrobial resistance genes such as blaCMY-2 in Salmonella might be transmitted among the upper and the lower levels of layer breeding chains via the IncA/C2 plasmids containing ISEcp1 and IS26. Further studies are required to fully understand the IS-mediated-integration of plasmids in Salmonella in veterinary medicine.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supplementary
Acknowledgments
This study was supported in part by a Grant-in-aid (grant no. JP19K19428) from the Japan Society for the Promotion of Sciences (KAKENHI), and grants from the Japan Agency for Medical Research and Development (AMED) (grant nos. JP21fk0108120 and JP21fk0108103). The sponsors played no role in the study design or in the collection, analysis, or data interpretation, writing the report, or in the decision to submit the manuscript for publication. We would like to express our gratitude to Dr. Katsuki, Mr. Hamasaki, and Ms. Hirano (Fukuoka Institute of Health and Environmental Sciences) for their invaluable advice. We are grateful to Ms. Doi and Ms. Yamada (National Institute of Infectious Diseases, Japan) for their assistance.
REFERENCES
- 1.Carattoli A., Zankari E., García-Fernández A., Voldby Larsen M., Lund O., Villa L., Møller Aarestrup F., Hasman H.2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58: 3895–3903. doi: 10.1128/AAC.02412-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carver T. J., Rutherford K. M., Berriman M., Rajandream M. A., Barrell B. G., Parkhill J.2005. ACT: the Artemis comparison tool. Bioinformatics 21: 3422–3423. doi: 10.1093/bioinformatics/bti553 [DOI] [PubMed] [Google Scholar]
- 3.Chuma T., Miyasako D., Dahshan H., Takayama T., Nakamoto Y., Shahada F., Akiba M., Okamoto K.2013. Chronological change of resistance to β-lactams in Salmonella enterica serovar Infantis isolated from broilers in Japan. Front. Microbiol. 4: 113. doi: 10.3389/fmicb.2013.00113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clinical and Laboratory Standards Institute. 2012. Performance standards for antimicrobial disk susceptibility tests; approved standard, document M02-A11, Clinical and Laboratory Standards Institute, Wayne. [Google Scholar]
- 5.Clinical and Laboratory Standards Institute. 2020. Performance standards for antimicrobial susceptibility testing; document M100, 30th ed., Clinical and Laboratory Standards Institute, Wayne. [Google Scholar]
- 6.Dutil L., Irwin R., Finley R., Ng L. K., Avery B., Boerlin P., Bourgault A. M., Cole L., Daignault D., Desruisseau A., Demczuk W., Hoang L., Horsman G. B., Ismail J., Jamieson F., Maki A., Pacagnella A., Pillai D. R.2010. Ceftiofur resistance in Salmonella enterica serovar Heidelberg from chicken meat and humans, Canada. Emerg. Infect. Dis. 16: 48–54. doi: 10.3201/eid1601.090729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.European Parliament. 2019. The EU poultry meat and egg sector: Main features, challenges and prospects. https://www.europarl.europa.eu/RegData/etudes/IDAN/2019/644195/EPRS_IDA(2019)644195_EN.pdf [accessed on October 5, 2020].
- 8.Fabre L., Delauné A., Espié E., Nygard K., Pardos de la Gandara M., Polomack L., Guesnier F., Galimand M., Lassen J., Weill F. X.2009. Chromosomal integration of the extended-spectrum β-lactamase gene blaCTX-M-15 in Salmonella enterica serotype Concord isolates from internationally adopted children. Antimicrob. Agents Chemother. 53: 1808–1816. doi: 10.1128/AAC.00451-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harmer C. J., Hall R. M.2016. IS26-mediated formation of transposons carrying antibiotic resistance genes. MSphere 1: e00038–e00016. doi: 10.1128/mSphere.00038-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hohmann E. L.2001. Nontyphoidal salmonellosis. Clin. Infect. Dis. 32: 263–269. doi: 10.1086/318457 [DOI] [PubMed] [Google Scholar]
- 11.Iwabuchi E., Maruyama N., Hara A., Nishimura M., Muramatsu M., Ochiai T., Hirai K.2010. Nationwide survey of salmonella prevalence in environmental dust from layer farms in Japan. J. Food Prot. 73: 1993–2000. doi: 10.4315/0362-028X-73.11.1993 [DOI] [PubMed] [Google Scholar]
- 12.Kleinheinz K. A., Joensen K. G., Larsen M. V.2014. Applying the ResFinder and VirulenceFinder web-services for easy identification of acquired antibiotic resistance and E. coli virulence genes in bacteriophage and prophage nucleotide sequences. Bacteriophage 4: e27943. doi: 10.4161/bact.27943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kolmogorov M., Yuan J., Lin Y., Pevzner P. A.2019. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 37: 540–546. doi: 10.1038/s41587-019-0072-8 [DOI] [PubMed] [Google Scholar]
- 14.Lei C. W., Zhang Y., Kang Z. Z., Kong L. H., Tang Y. Z., Zhang A. Y., Yang X., Wang H. N.2020. Vertical transmission of Salmonella Enteritidis with heterogeneous antimicrobial resistance from breeding chickens to commercial chickens in China. Vet. Microbiol. 240: 108538. doi: 10.1016/j.vetmic.2019.108538 [DOI] [PubMed] [Google Scholar]
- 15.Liljebjelke K. A., Hofacre C. L., Liu T., White D. G., Ayers S., Young S., Maurer J. J.2005. Vertical and horizontal transmission of salmonella within integrated broiler production system. Foodborne Pathog. Dis. 2: 90–102. doi: 10.1089/fpd.2005.2.90 [DOI] [PubMed] [Google Scholar]
- 16.Murakami K., Horikawa K., Ito T., Otsuki K.2001. Environmental survey of salmonella and comparison of genotypic character with human isolates in Western Japan. Epidemiol. Infect. 126: 159–171. doi: 10.1017/S0950268801005283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murakami K., Ishihara T., Horikawa K., Oda T.2007. Features of Salmonella serovars among food handlers in Kyushu, Japan. New Microbiol. 30: 155–159. [PubMed] [Google Scholar]
- 18.Nagaoka H., Hirai S., Morinushi H., Mizumoto S., Suzuki K., Shigemura H., Takahashi N., Suzuki F., Mochizuki M., Asanuma M., Maehata T., Ogawa A., Ohkoshi K., Sekizuka T., Ishioka T., Suzuki S., Kimura H., Kuroda M., Suzuki M., Murakami K., Kanda T.2020. Coinfection with human Norovirus and Escherichia coli O25:H4 harboring two chromosomal blaCTX-M-14 genes in a foodborne Norovirus outbreak in Shizuoka prefecture, Japan. J. Food Prot. 83: 1584–1591. doi: 10.4315/JFP-20-042 [DOI] [PubMed] [Google Scholar]
- 19.National Center for Biotechnology Information. https://www.ncbi.nlm.nih.gov/ [accessed on February 10, 2021].
- 20.National Institute of Genetics. DNA Data Bank of Japan. https://www.ddbj.nig.ac.jp/index-e.html [accessed on January 22, 2021].
- 21.National Veterinary Assay Laboratory Ministry of Agriculture Forestry and Fisheries. 2018. Sales amounts and sales volumes (active substance) of antibiotics, synthetic antibacterials, anthelmintics and antiprotozoals. https://www.maff.go.jp/nval/iyakutou/hanbaidaka/pdf/h30_hanbaidaka.pdf [accessed on December 6, 2020].
- 22.Noda T., Murakami K., Etoh Y., Okamoto F., Yatsuyanagi J., Sera N., Furuta M., Onozuka D., Oda T., Asai T., Fujimoto S.2015. Increase in resistance to extended-spectrum cephalosporins in Salmonella isolated from retail chicken products in Japan. PLoS One 10: e0116927. doi: 10.1371/journal.pone.0116927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oliva M., Monno R., Addabbo P., Pesole G., Scrascia M., Calia C., Dionisi A. M., Chiara M., Horner D. S., Manzari C., Pazzani C.2018. IS26 mediated antimicrobial resistance gene shuffling from the chromosome to a mosaic conjugative FII plasmid. Plasmid 100: 22–30. doi: 10.1016/j.plasmid.2018.10.001 [DOI] [PubMed] [Google Scholar]
- 24.Riaño I., Moreno M. A., Teshager T., Sáenz Y., Domínguez L., Torres C.2006. Detection and characterization of extended-spectrum β-lactamases in Salmonella enterica strains of healthy food animals in Spain. J. Antimicrob. Chemother. 58: 844–847. doi: 10.1093/jac/dkl337 [DOI] [PubMed] [Google Scholar]
- 25.Sánchez-Salazar E., Gudiño M. E., Sevillano G., Zurita J., Guerrero-López R., Jaramillo K., Calero-Cáceres W.2020. Antibiotic resistance of Salmonella strains from layer poultry farms in central Ecuador. J. Appl. Microbiol. 128: 1347–1354. doi: 10.1111/jam.14562 [DOI] [PubMed] [Google Scholar]
- 26.Sannat C., Patyal A., Rawat N., Ghosh R. C., Jolhe D. K., Shende R. K., Hirpurkar S. D., Shakya S.2017. Characterization of Salmonella Gallinarum from an outbreak in Raigarh, Chhattisgarh. Vet. World 10: 144–148. doi: 10.14202/vetworld.2017.144-148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shahada F., Sekizuka T., Kuroda M., Kusumoto M., Ohishi D., Matsumoto A., Okazaki H., Tanaka K., Uchida I., Izumiya H., Watanabe H., Tamamura Y., Iwata T., Akiba M.2011. Characterization of Salmonella enterica serovar Typhimurium isolates harboring a chromosomally encoded CMY-2 β-lactamase gene located on a multidrug resistance genomic island. Antimicrob. Agents Chemother. 55: 4114–4121. doi: 10.1128/AAC.00560-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shigemura H., Matsui M., Sekizuka T., Onozuka D., Noda T., Yamashita A., Kuroda M., Suzuki S., Kimura H., Fujimoto S., Oishi K., Sera N., Inoshima Y., Murakami K.2018. Decrease in the prevalence of extended-spectrum cephalosporin-resistant Salmonella following cessation of ceftiofur use by the Japanese poultry industry. Int. J. Food Microbiol. 274: 45–51. doi: 10.1016/j.ijfoodmicro.2018.03.011 [DOI] [PubMed] [Google Scholar]
- 29.Shigemura H., Sakatsume E., Sekizuka T., Yokoyama H., Hamada K., Etoh Y., Carle Y., Mizumoto S., Hirai S., Matsui M., Kimura H., Suzuki M., Onozuka D., Kuroda M., Inoshima Y., Murakami K.2020. Food workers as a reservoir of extended-spectrum-cephalosporin-resistant Salmonella strains in Japan. Appl. Environ. Microbiol. 86: e00072–e20. doi: 10.1128/AEM.00072-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shirota K., Katoh H., Murase T., Ito T., Otsuki K.2001. Monitoring of layer feed and eggs for Salmonella in eastern Japan between 1993 and 1998. J. Food Prot. 64: 734–737. doi: 10.4315/0362-028X-64.5.734 [DOI] [PubMed] [Google Scholar]
- 31.Siguier P., Perochon J., Lestrade L., Mahillon J., Chandler M.2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34: D32–D36https://www-is.biotoul.fr/index.php [accessed on February 1, 2020]. doi: 10.1093/nar/gkj014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Someya A., Otsuki K., Murase T.2005. Antimicrobial susceptibilities of Salmonella isolates obtained from layer chicken houses on a commercial egg-producing farm in Japan, 1997 to 2002. J. Food Prot. 68: 2030–2034. doi: 10.4315/0362-028X-68.10.2030 [DOI] [PubMed] [Google Scholar]
- 33.Sullivan M. J., Petty N. K., Beatson S. A.2011. Easyfig: a genome comparison visualizer. Bioinformatics 27: 1009–1010. doi: 10.1093/bioinformatics/btr039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.U.S. Food and Drug Administration. 2019. 2018 Summary report on antimicrobials sold or distributed for use in food-producing animals. https://www.fda.gov/media/133411/download [accessed on December 6, 2020].
- 35.United States Department of Agriculture −Animal and Plant Health Inspection Service. 2013. Foreign Animal Disease Preparedness and Response Plan (FAD PReP)/National Animal Health Emergency Management System (NAHEMS) Guidelines, Poultry Industry Manual. https://www.aphis.usda.gov/animal_health/emergency_management/downloads/documents_manuals/poultry_ind_manual.pdf [accessed on October 5, 2020].
- 36.Walker B. J., Abeel T., Shea T., Priest M., Abouelliel A., Sakthikumar S., Cuomo C. A., Zeng Q., Wortman J., Young S. K., Earl A. M.2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9: e112963. doi: 10.1371/journal.pone.0112963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wick R. R., Judd L. M., Gorrie C. L., Holt K. E.2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLOS Comput. Biol. 13: e1005595. doi: 10.1371/journal.pcbi.1005595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.World Health Organization. 2018. Salmonella (non-typhoidal). https://www.who.int/news-room/fact-sheets/detail/salmonella-(non-typhoidal) [accessed on October 5, 2020].
- 39.Zhang D., Zhao Y., Feng J., Hu L., Jiang X., Zhan Z., Yang H., Yang W., Gao B., Wang J., Li J., Yin Z., Zhou D.2019. Replicon-based typing of IncI-complex plasmids, and comparative genomics analysis of IncIγ/K1 Plasmids. Front. Microbiol. 10: 48. doi: 10.3389/fmicb.2019.00048 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The complete, annotated genomic sequences from the S. Ohio and S. Cerro strains are deposited in the DDBJ under accession nos. AP024347–AP024353 and AP024345–AP024346, respectively. The obtained short- and long-read sequence data are deposited in the DDBJ as follows: S. Ohio (SEOhiM1593; BioProject PRJDB10937, BioSample SAMD00206786, DRA accession nos. DRR261749 and DRR261750); S. Ohio (SEOhiM1960; BioProject PRJDB10937, BioSample SAMD00206787, DRA accession nos. DRR261751 and DRR261752); S. Ohio (SEOhiM2008; BioProject PRJDB10937, BioSample SAMD00206788, DRA accession nos. DRR261753 and DRR261754); and S. Cerro (SECerM2017; BioProject PRJDB10937, BioSample SAMD00264093, DRA accession nos. DRR261747 and DRR261748).