Abstract
While the mechanotransduction-induced fate of adult neural stem/progenitor cells (NPCs) is relatively known, how substrate stiffness regulates the temporal evolution of the biomechanics and phenotype of developmentally relevant human fetal NPCs (hNPCs) and their mechanosensing pathways remain unknown. Here, we primed hNPCs on tissue-culture plastic (TCPS) for 3 days in non-differentiating medium before transferring to TCPS or Geltrex™ gels (< 1 kPa) for 9-day cultures post-priming, and regularly assessed stemness, differentiation, and cell mechanics (Young’s modulus, tether forces, apparent membrane tension, tether radius). hNPCs maintained stemness on TCPS while those on gels co-expressed stemness and neural/glial markers, 3-days post-priming. Biomechanical characteristics remained unchanged in cells on TCPS but were significantly altered in those on gels, 3-days post-priming. However, 9-days post-priming, hNPCs on gels differentiated, with significantly more neurons on softer gels and glia on stiffer gels, while those on TCPS maintained their native stemness. Withdrawal of bFGF and EGF in 9-day cultures induced hNPC differentiation and influenced cell mechanics. Cells on stiffer gels had higher biomechanical properties than those on softer gels throughout the culture period, with NPC-like > neural > glia subtypes. Higher stress fiber density in cells on stiffer gels explains their significantly different biomechanical properties on these gels. Blebbistatin treatment caused cell polarization, lowered elastic modulus, and enhanced tether forces, implicating the role of non-muscle myosin-II in hNPC mechanosensing, adaptability, and thereby mechanics. Such substrate-mediated temporal evolution of hNPCs guide design of smart scaffolds to investigate morphogenesis, disease modeling, stem cell biology, and biomaterials for tissue engineering.
Keywords: mechanotransduction, non-muscle myosin, atomic force microscopy, fetal neural stem cells, substrate stiffness
Graphical Abstract
We here report on the substrate stiffness dependent spatiotemporal evolution of mechanical properties of neural stem cells and their progenies.
Introduction
Cells continuously monitor their microenvironment which keeps them informed of their surroundings to maintain homeostasis1, 2. Mechanosensing and mechanotransduction allow cells to detect, decipher, and retort the physical properties of their niche by converting them into biochemical signals3. Such interplay between biochemical and biomechanical signals is more pronounced in stem and progenitor cells during various phases of development, starting with embryogenesis. For instance, neural progenitor cells (NPCs) migrate from their niche in various regions of the central nervous system (CNS) to proliferate and differentiate into neuronal and glial subtypes4, while simultaneously experiencing and responding to the mechanically heterogenous tissue microenvironment5. Outside their native niche, NPCs respond to substrate stiffness by biasing towards neuronal differentiation on soft substrates and glial lineages on relatively stiff substrates6, 7. Exploring the effects of mechanotransduction on stem cells and differentiated progenies could provide important insights in developmental biology and in formulating strategies (e.g., delivery vehicles, transplantation) to replace lost cells, promote in situ regeneration of degraded or injured tissues, or proffer immunotherapy, by enabling biological and mechanical mimicry of native tissues.
CNS formation and maturation is a mechanically-coordinated process8, 9. Brain tissue stiffness increases from neonatal to adult to old age, coinciding with the loss in regenerative ability of adult stem and progenitor CNS cells10, suggesting a strong link between cell function and matrix composition and rigidity. Brain stiffness decreases in neurodegenerative conditions due to compromised adult neurogenesis and changes in the ECM1. While ECM remodeling during injury or disorders could lead to abnormal mechanotransduction, the extent of contribution from such changes in mechanical characteristics on disease pathology and progression remains less known11. Translocation of transcriptional factors such as Yes-associated protein (YAP)12, 13 and transcriptional coactivator with PDZ-binding motif (TAZ)14, 15, expression of PIEZO110, 16 and other ion channels17, and RhoA activation18 during development and disorders have been shown to play important roles in converting ECM niche signals into functional outcomes. Elucidation of mechanisms underlying mechanobiology can provide greater insights into the progression of changes at the cellular level during normal CNS development versus pathologically aberrant conditions, opening new avenues for mechanotherapy.
Cell mechanics critically contributes to fate decisions by orchestrating biophysical changes-mediated development and remodeling of tissues19, 20. Biophysical changes due to dynamic evolution of cytoskeleton was shown as a hallmark of stem cell survival and differentiation21. Elucidating changes in cell deformability during and after differentiation is of great interest in the field of mechanotransduction22. Prior studies reported on the effects of substrate stiffness and composition on adult NPC fate at defined end points23–26. We have recently shown that alterations in biophysical and biomechanical properties could serve as markers for microenvironment induced changes in human fetal NSCs27. We here investigated the mechanosensing ability of developmentally relevant human fetal NPCs (hNPCs) on biological scaffolds constituted at different ECM protein concentrations and therefore scaffold stiffness. We report on the temporal evolution of biomechanical characteristics (Young’s modulus, tether force, membrane tension, tether radius) and fate of hNPCs over 12 days of culture, modulated by the underlying substrate characteristics. We propose mechanoadaptation as a possible mechanism and indicator of hNPC differentiation. Finally, using pre-processed RNA-sequencing data and immunolabeling, we identified the expression levels and roles of various mechanotransduction proteins (YAP, F-actin, non-muscle myosin II chains), and their contributions to the hNPC intrinsic mechanical properties and lineage commitment. Our results add to the growing body of literature on the role of substrate stiffness-mediated mechanotransduction in influencing the fate of various cell types, specifically that of soft biomimetic hydrogels, on the temporal evolution of fetal hNPC biomechanics and maturation.
Materials and methods
Substrate preparation
Geltrex™ LDEV-free reduced growth factor basement membrane matrix (12–18 mg/mL concentration; Thermo Fisher Scientific, Waltham, MA, USA) containing primarily laminin, collagen IV, entactin, and heparin sulfate proteoglycans was used. It is similar in composition to Matrigel™, but with different proportions of constituents. As-received matrix (hitherto referred as G-100) was diluted with serum-free ReNcell complete medium to result in 75%, 50%, and 25% Geltrex™ solutions (hitherto referred as G-75, G-50, G-25). Complete medium is defined as maintenance medium (SCM005; EMD Millipore, Burlington, MA, USA) added with 20 ng/mL each of bFGF and EGF (Thermo Fisher Scientific). The calculated volume of solutions was pipetted onto a 24-well plate and AFM specific petri dishes, maintained on ice. The cover glass coated with Sigmacote® (Sigma-Aldrich, St. Louis, MO, USA)28 was used to uniformly spread the small volumes in the well-plate or petri dish to obtain the desired gel height. Briefly, coverslips treated with Sigmacote® were placed on the solutions which were allowed to gel (30 min, 37 °C), and the coverslips were removed to obtain a ~ 70 μm thick gel. Tissue culture polystyrene (TCPS; Thermo Fisher Scientific) dishes were functionalized with laminin (Sigma-Aldrich) for two hours prior to use.
Cell culture
Human NPCs (ReNcell® VM; SCC008; EMD Millipore) cultured on laminin-coated flasks were maintained in an undifferentiated state using complete medium. These cells are an immortalized human NPC line derived from the ventral mesencephalon region of human fetal brain. In select cultures, hNPC differentiation on TCPS was induced by culturing in maintenance medium. Blebbistatin (Catalog # 72402, STEMCELL Technologies Inc., Cambridge, MA, USA) was added to the complete medium (5 μM) in select cases for non-muscle myosin II inhibition, as per concentrations and conditions optimized elsewhere for stem cells29.
Experimental design
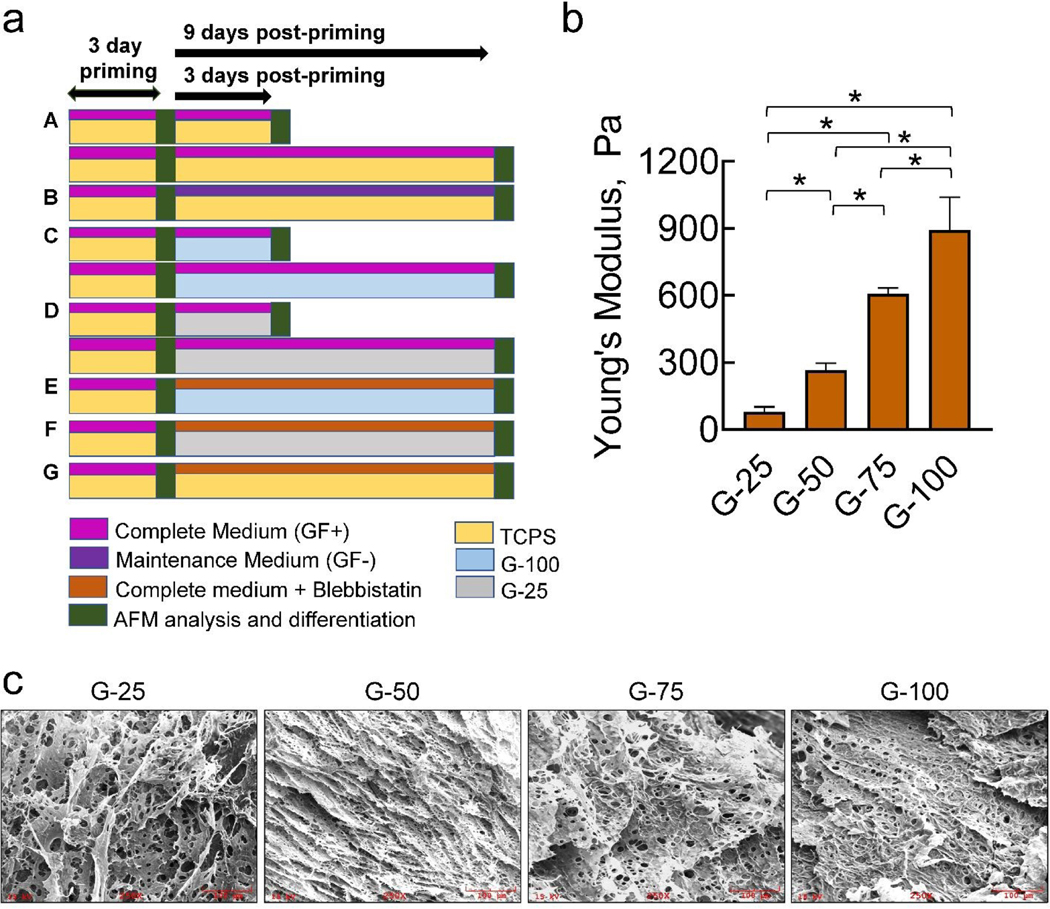
To elucidate the mechanosensing ability of hNPCs, they were first primed on TCPS for 3 days (Fig. 1a) in complete medium to keep them in undifferentiated state. These hNPCs were detached and reseeded on different substrates and cultured for further analysis as follows: hNPCs were cultured on TCPS for 3 days or 9 days with complete medium (Fig. 1a, A). At the end of 3 and 9 days, mechanical properties and expression of stemness marker were characterized. hNPCs were cultured on TCPS with maintenance medium. At the end of 9 days, mechanical properties and cell differentiation were evaluated (Fig. 1a, B). This experiment serves as a control to compare the changes in other culture conditions. hNPCs were cultured in complete medium on G-100 substrates for 3 or 9 days and evaluated for cell differentiation and mechanical properties at respective time points (Fig. 1a, C). hNPCs were cultured in complete medium on G-25 substrates for 3 or 9 days and evaluated for changes in mechanical properties and differentiation at respective time points (Fig. 1a, D). Cells were cultured in complete medium on G-100 (Fig. 1a, E), G-25 (Fig. 1a, F) and TCPS (Fig. 1a, G) substrates for 9 days, in the presence of blebbistatin, to elucidate the role of myosin II. At the end of 9 days, mechanical properties and differentiation were quantified.
Fig 1.
(a) Graphical summary of the experimental design to characterize substrate and culture effects on hNPC fate and mechanics. Post-priming on TCPS for 3 days, hNPCs were cultured on various substrates (TCPS, G-100, G-25) for 3 or 9 days, in the presence of complete medium (GF+) or maintenance medium (GF-). Changes in cell phenotype (differentiation) and biomechanical characteristics were assessed at regular intervals. (b) Young’s moduli values obtained from the force-indentation curves generated from at least 30 random locations on each gel specimen. Data was represented as average ± SD. * denotes p< 0.01. (c) Representative SEM images of the G-25, G-50, G-75, and G-100 gels, showing fenestrated 3D scaffold nature of these gels with significant porosity and pore-size. All images were at 250× magnification. Scale bar: 100 μm.
Scanning electron microscopy
G-25, G-50, G-75, and G-100 gels on coverslips were immersed in 2% paraformaldehyde solution, dehydrated in gradient-alcohol baths, and dried in desiccator overnight. The samples were mounted on a carbon tape, sputter-coated (SPI sputter model 13131) with gold for 30 sec (300 Å, 35 mA), and imaged at multiple magnifications (150×, 250×, 500×, 900×, 2700×) using a field emission scanning electron microscopy (SEM; FEI Inspect F50; 15 KeV source). The pore-size of the scaffolds was quantified from the SEM images (n = 5 per gel) in respective gels.
Atomic force microscopy
A high-performance MFP-3D-BIO atomic force microscope (Oxford Instruments, Santa Barbara, CA, USA) integrated with a Nikon Eclipse Ti inverted epi-fluorescence microscope (Melville, NY, USA) was used. The system has a temperature-controlled closed fluid cell (Bioheater®) for manipulating specimen temperatures with a precision of 0.1 °C and an accuracy to 0.2 °C, and a Himamatzu sCMOS Ocra FLASH 2.8 camera kit for excellent high-resolution video capture and display. Young’s moduli of the gels were obtained using tip-less AFM cantilevers (Arrow™ TL1, Nanoworld, Watsonville, CA, USA; nominal spring constant ~ 0.03 N/m), modified by attaching a 35-μm polystyrene bead using two-part epoxy (5 min setting time). Briefly, a small droplet of epoxy was placed on one end of the glass slide using a p10 pipette tip and an extremely diluted suspension of 35 μm polystyrene beads (Polybead® Microspheres, Polysciences, Inc., Warrington, PA) was placed on the other end of the slide. Using a cantilever-moving technique30, the cantilever mounted onto AFM head was first moved to pick tiny amount of glue and after 4 min, it was moved to pick the polystyrene bead. The beading process was observed using a 40× objective on an inverted optical microscope. The spring constants of beaded and regular cantilevers were obtained using thermal calibration method31. Geltrex™ solutions (G-100, G-75, G-50, G-25) for indentation assays were cold-pipetted on to custom-made PDMS wells (n=5 wells/concentration) and allowed to gel at 37 °C in an incubator. Force-indentation curves were obtained at random locations on the gels (rate ~ 5 μm/s, trigger force ~ 4 nN) and analyzed by fitting a Hertz model for a spherical indenter using Poisson’s ratio ≈ 0.4932, typically used for similar scaffolds.
hNPCs at a seeding density of 0.5 × 105 cells/well were cultured on three different substrates in AFM-specific petri dishes: 2D laminin-coated TCPS, G-100, and G-25 gel surfaces, for 3 and 9 days, in the presence or absence of growth factors (Fig. 1a). The cells were maintained at 37 °C throughout the live-cell indentation assay. For each experiment, 30 to 50 cells were randomly selected, and at least 50 to 60 force curves obtained between nuclei and cell margin at approach/ retraction velocity of 5 μm/s using non-beaded TR 400 PSA cantilever (Olympus Corp., Center Valley, PA; silicon nitride pyramidal tips; nominal spring constant ~0.05 N/m). Using Hertz’s contact model, the Young’s modulus was determined from the force–indentation curves at an indentation depth around 400 nm. The tether forces (FT) were directly calculated from the series of force steps in the retraction curve. The apparent membrane tension (TM) was calculated from tether forces using , where kB is the bending stiffness which lies in the range of 0.1 – 0.3 pN.μm33. Similarly, RT (tether radius) was calculated from the tether forces as .
Immunofluorescence labeling
Primed cells, and cultures on various substrates at days 3 and 9 post-priming, were processed to identify and assess the distinct neural and glial lineages. Cells were washed once in sterile 1×PBS, fixed with 4% PFA for 30 min, washed twice with 1×PBS (5 min each), and incubated with blocking buffer (0.5% Triton-X, 5% serum, 1× PBS) for 2 h at room temperature. Serum selection was based on primary and secondary antibody host species. After removing the blocking buffer, cells were incubated with these respective primary antibodies (4 °C, 24 h): mouse polyclonal anti-GFAP (Abcam, Cambridge, MA, USA) for astrocytes, mouse monoclonal anti-SOX2 (Thermo Fisher Scientific) for stemness, mouse monoclonal anti-β3 tubulin (Thermo Fisher Scientific) for neurons, mouse monoclonal YAP (Santa Cruz Biotechnology, Inc., Dallas, TX, USA) and PIEZO1 (Thermo Fisher Scientific) antibodies for mechanosensors, and F-actin staining Alexa Flour™ 488 Phalloidin for stress fibers (Thermo Fisher Scientific) visualization. Cells were washed four times in PBS for 10 min each, incubated with appropriate secondary antibodies (Santa Cruz Biotechnology) at room temperature for 4 h, washed again three times with PBS for 10 min each, and then counterstained with 4,6-diamidino-2-phenylindole (DAPI, Sigma-Aldrich). Cells were imaged using the Zeiss AxioVert A1 fluorescence microscope under both phase contrast and fluorescence channels using a digital camera (Axiocam C1, Carl Zeiss) and Axiovision data acquisition software. Cell density was quantified by manually counting the cells in each well emitting DAPI signal and comparing that to the total number of cells in that same well positively stained for the antibody markers (> 200 cells counted per condition).
The F-actin stained images (.tiff files) were quantified using a protocol developed by Zonderland et al.34 to quantify the intensity of F-actin distribution within the cytoplasm and over the nucleus. Multiple individual cells in each case (G-25 vs. G-100 substrates) were selected and processed as per the protocol, the mean ± SD was plotted, and the statistical differences between cases were calculated using GraphPad Prism 5.
RNA sequencing
Pre-processed RNA-sequencing (RNA-seq) data was downloaded from the gene expression omnibus (GSE 89623). Briefly, sequencing was performed using Illumina HiSeq 2000 and employed a paired-end, 50 base read length sequencing, with the RNA extraction, computational pipeline, and quality control steps reviewed elsewhere35. The data was explored using GREIN (GEO RNA-seq Experiments Interactive Navigator) which is an interactive web-based platform for re-analyzing subsets of GEO RNA-seq data36. Data for two cell lines, ReNcell CX and ReNcell VM (n = 2 for each group) were imported and volcano plot was created with log10 of the adjusted p-value (FDR < 0.01) on the y-axis and the log2 fold change on the x-axis using ggplot in R. The α-level for differential expression was set at 0.05. A separate plot of selected genes which had previously been reported in the literature or in the publicly available data base “genecards” (Crown Human Genome Center at the Weizmann Institute of Science) to affect nuclear mechanotransduction were then averaged and plotted independently in R using ggplot237–39.
Statistical analysis
Data were represented as mean ± standard error from at least n=4 wells/condition, with at least three independent repeats of each assay, and statistical analysis was performed using GraphPad Prism 5. Data analysis was performed using one-way ANOVA, followed by Tukey multiple comparison to find statistically significant differences between the groups (p < 0.05).
Results
Mechanical properties of Geltrex™ scaffolds
Since ECM mimicking scaffolds are preferable in mechanobiology studies exploring CNS development, we quantified the mechanical characteristics of Geltrex™ gels. AFM measurements (Fig. 1b) indicate that the average modulus of as-received Geltrex™ (G-100) was 892 ± 148 Pa (stiffness ≈ 7±1.4 nN/μm). Dilution to 75%, 50% and 25% concentrations drastically decreased average Young’s moduli to 605 ± 27 Pa, 265 ± 32 Pa, 78 ± 23 Pa, respectively (stiffness ≈ 0.7±0.1 nN/μm for G-25; p< 0.001 vs. G-100 for both modulus and stiffness). In comparison, the modulus of rigid TCPS dishes is typically in gigapascals range40, orders of magnitude higher than the gels (p< 0.001 vs. Geltrex™ gels). SEM images (Fig. 1c) reveal a fenestrated scaffold-like architecture of the gels with significant porosity and fibrillar structures, and an average pore-size ranging from 5 μm to 20 μm for these gels. The porosity monotonically decreased with increasing protein concentration in the gels, i.e., G-100 gels are denser with less porosity and collapsed 3D structure compared to G-25 gels. Therefore, G-100 and G-25 substrates were chosen for further studies based on their significant differences in Young’s moduli and topography, and as they represent lower and higher ends of the brain elasticity spectrum (0.1–1 kPa)41.
Substrate-dependent temporal evolution of differentiated cell phenotype
hNPCs were initially cultured and primed on TCPS for three days (Fig. 1a, A) with complete medium (i.e., non-differentiation medium). After priming, hNPCs maintained their phenotype (Fig. 2A), expressed stemness marker SOX2 (Fig. 3A), and did not express differentiation markers such as β-III tubulin and GFAP (Supplementary Figure 1).
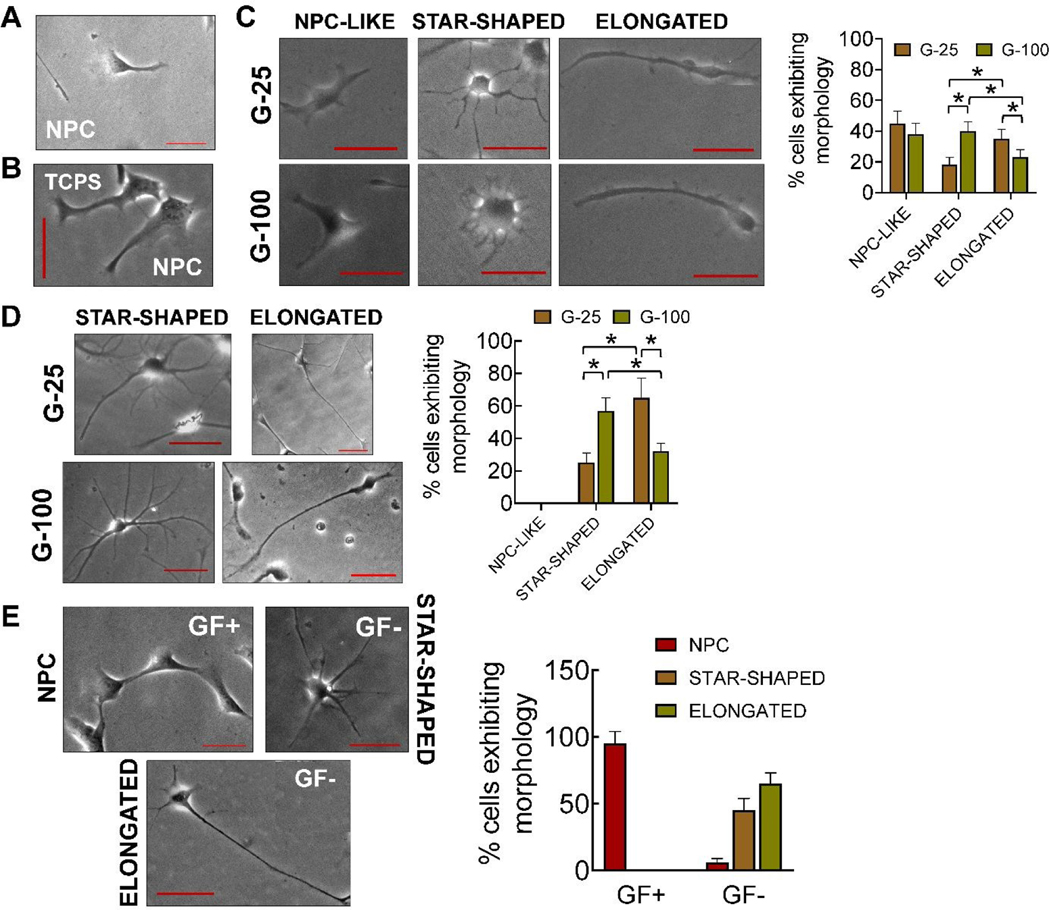
Fig 2.
Representative phase contrast images of (A) cells in complete medium (GF+), immediately after priming (three days in culture), (B) cells cultured on TCPS, three days post-priming, in the presence of complete medium, (C) NPC-like, star-shaped, and elongated cells on G-25 and G-100 after three days in culture post-priming, (D) star-shaped and elongated cells on G-25 and G-100 gels, nine days post-priming, in complete medium, and (E) cells after nine days of culture on laminin coated TCPS in the presence of complete medium or maintenance medium. Distinct morphologies of the cells in maintenance medium was evident unlike in the presence of complete medium. The number of cells exhibiting each morphology were counted manually from the images and normalized to the total cells counted in each case; n > 150 cells/case. * indicates p < 0.05 between respective cases. Scale bar: 50 μm.
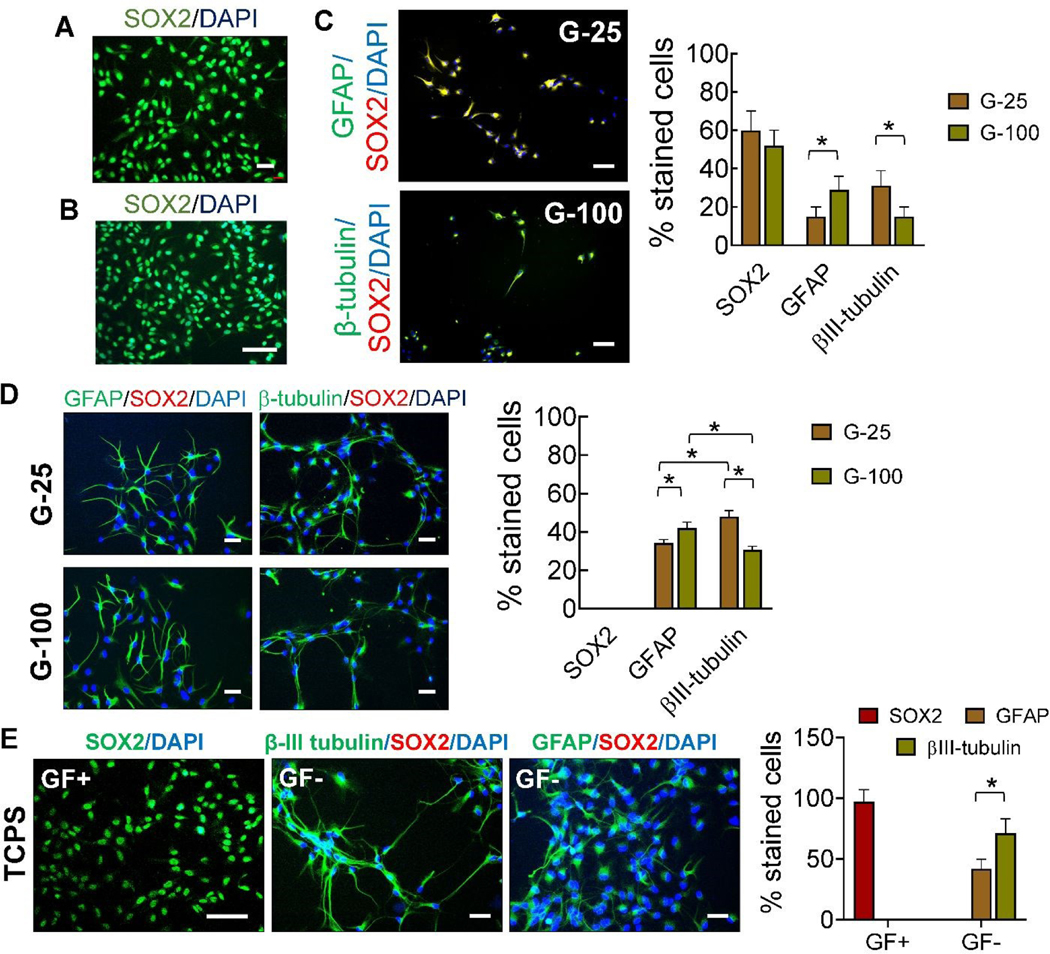
Fig. 3.
Representative SOX-2 labeled immunofluorescence images of cells cultured (A) in the presence of complete medium, after priming (three days in culture), and (B) on TCPS, three days post-priming, in the presence of complete medium. (C) Representative immunofluorescence images of NPC-like, star-shaped, and elongated cells on G-25 and G-100 after three days in culture post-priming, and stained for GFAP (astrocyte), βIII-tubulin (neuronal), and SOX2 (stemness) markers. (D) Representative immunofluorescence images of star-shaped and elongated cells on G-25 and G-100 gels, nine days post-priming, in complete medium. Primary antibodies for SOX2, β-III tubulin, and GFAP were used. Staining for SOX2 was not evident. (E) Representative immunofluorescence images of cells after nine days of culture on laminin coated TCPS in the presence of complete medium or maintenance medium. Primary antibodies for SOX2, β-III tubulin, and GFAP were used with appropriate secondary antibodies. Cultures were counterstained with DAPI for cell nuclei identification in all the cases. The number of stained cells for each marker were counted manually from the images and normalized to the total cells counted in each case; n > 200 cells/case. * indicates p < 0.05 between respective cases. Scale bar: 50 μm.
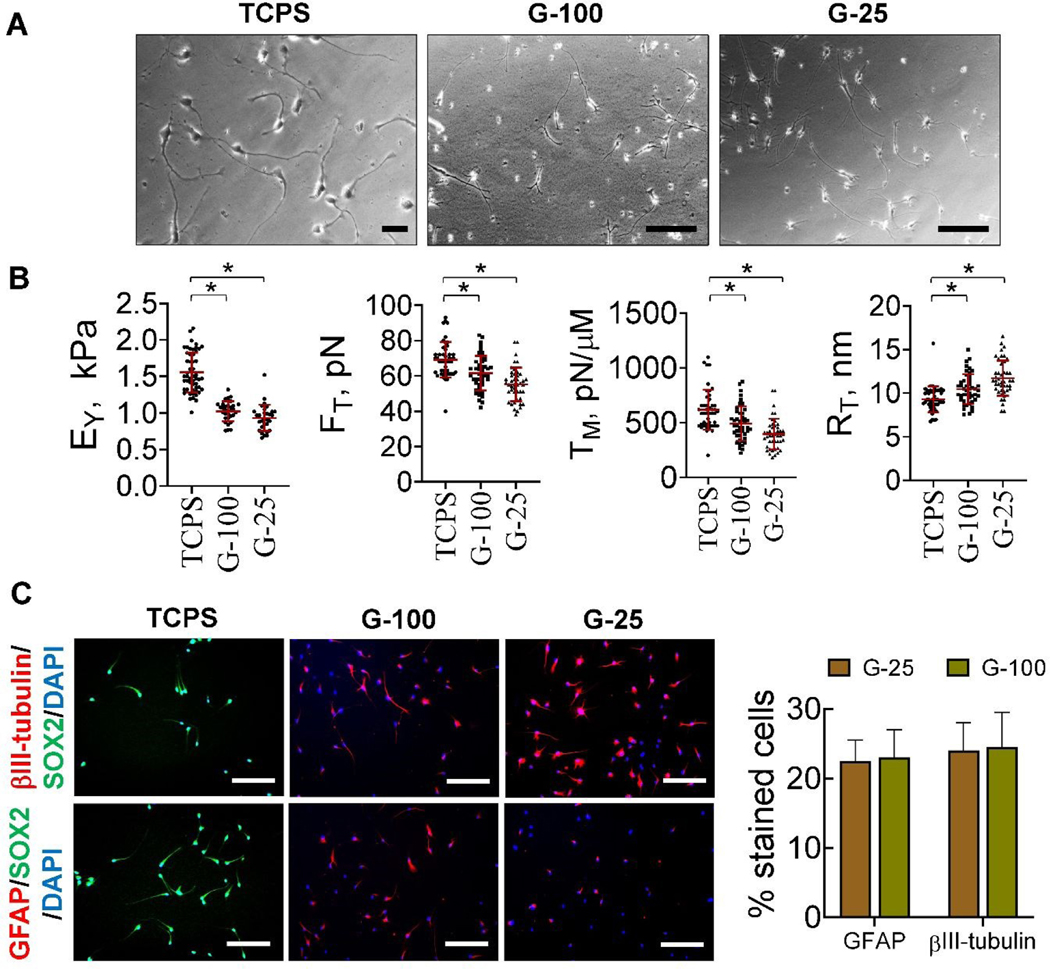
To establish the standalone effects of substrates on hNPCs, cells were cultured on gels or TCPS for 3 days post-priming, in the presence of complete medium (GF+). hNPCs cultured on TCPS showed homogenous stemness morphology (Fig. 2B) and SOX2 expression (Fig. 3B), similar to that observed during their priming. Interestingly, hNPCs on G-100 and G-25 gels started transforming to a heterogenous mixture of cells, which we classified into three distinct morphologies – NPC-like, star-shaped (representing glial cells), and elongated cells (representing neurons) (Fig. 2C). Image analysis suggested more star-shaped cells and fewer elongated cells on G-100 versus G-25 substrate. These cells expressed neural and astrocytic markers in addition to the stemness marker (Fig. 3C). GFAP expressing cells were significantly higher on G-100 gels while βIII-tubulin expressing cells were higher on G-25 gels, with no substrate-dependent differences noted in SOX2 expressing cells. Results from immunofluorescence images closely match trends from phase contrast images suggesting that star-shaped cells are more likely to be astrocyte phenotype while elongated cells could be neuronal in lineage. The individual channels of the fluorescence markers in each case was shown in Supplementary Figure 2.
To evaluate the evolution of maturation of differentiation markers, hNPCs were cultured in complete medium on G-100 and G-25 gels for nine days post-priming. In contrast to the three distinct morphologies noted after three days of culture on G-25 and G-100 substrates, only two morphologies (star-shaped cells, elongated cells) were observed after nine days of culture (Fig. 2D). Image analysis suggested significantly more star-shaped cells on G-100 gels but more elongated cells on G-25 gels. Higher number of cells expressed βIII-tubulin compared to GFAP on G-25, while the converse was true on G-100 gels (Fig. 3D), indicating the preferential differentiation of NPCs to neuronal lineage on softer matrices. SOX2 expression was absent in both these cultures indicating the absence of stemness in these cells and their complete maturation into progenies. These results attest to the biophysical role of substrates on hNPC phenotype even in the presence of complete medium (GF+).
hNPCs were cultured on laminin coated TCPS in the presence of complete medium (GF+) or maintenance medium (GF-) for nine days post-priming. hNPCs maintained their phenotype (Fig. 2E) and stemness (SOX2 expression, Fig. 3E) in the presence of complete medium (GF+), similar to their status after initial priming (Fig. 2A) and subsequent 3-day culture (Fig. 2B). Withdrawal of growth factors (i.e., maintenance medium; GF-) led to the formation of distinct elongated or star-shaped cells (Fig. 2E), positively staining for βIII-tubulin and GFAP and negatively staining for SOX2 (Fig. 3E), indicating their differentiation and lineage commitment.
Substrate-dependent temporal evolution in cell mechanics
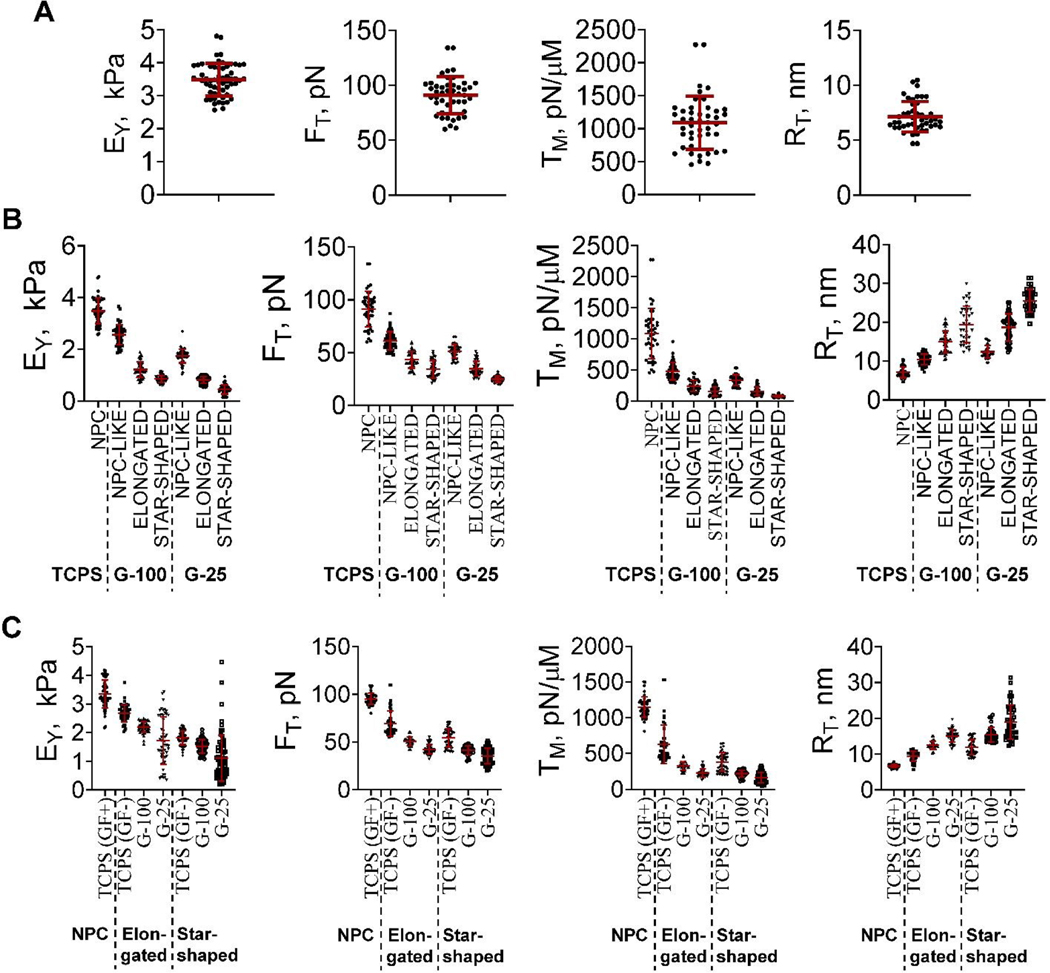
The biomechanical characteristics such as EY, FT, TM, and RT of hNPCs primed on TCPS for 3 days were 3.48 ± 0.06 kPa, 91 ± 1.12 pN, 1110 ± 28 pN/μm, and 6.1 ± 0.38 nm, respectively (Fig. 4A). These values establish the baseline mechanical properties of hNPCs cultured on TCPS for three days in the presence of growth factors (i.e., priming phase).
Fig. 4.
(A) Bee-swarm plots of mechanical characteristics (EY, FT, TM, RT) of cells (A) primed for three days in the presence of complete medium, (B) cultured on TCPS, G-100, and G-25, after three days in culture post-priming, in the presence of complete medium, and (C) cultured for nine days on TCPS, G-100 and G-25. Cells were cultured on TCPS in the presence or absence of growth factors, while those on G-100 and G-25 were cultured in the presence of growth factors. The red lines represent the mean and standard deviation of the data in each case while the symbols represent the data points. Notation: Young’s modulus (EY), tether force (FT), apparent membrane tension (TM), and tether radius (RT).
The mechanical properties of cells were quantified after three days in cultures post-priming, in the presence of complete medium (Fig. 4B). The average values of EY, FT, TM, and RT of cells on TCPS were similar to those of hNPCs primed for three days on TCPS (Fig. 4A), but significantly (p< 0.001) higher than the heterogenous phenotype of cells on G-100 and G-25. The average EYof NPC-like, elongated, and star-shaped cells on G-100 (stiffer matrix) was 1.4 to 1.9 -fold higher (p< 0.01 in all cases) compared to their counterparts on G-25 (softer matrix). Similarly, the average FT of NPC-like, elongated, and star-shaped cells on G-100 were 1.2 to 1.4 -fold higher (p< 0.05 in all cases) than their counterparts on G-25. Likewise, significant differences (p< 0.01) were noted in the average RT and TM values of cells cultured on G-100 vs. G-25. Finally, significant differences (p< 0.001) in EY, FT, and TM were noted among the various cell types (i.e., NPC-like, elongated and star-shaped) within each substrate conditions (i.e., G-100 or G-25), with the general trend being NPC-like > elongated > star-shaped.
The mechanical characteristics of resulting cells were quantified after nine days of culture post-priming (Fig. 4C) in complete medium (TCPS, G-100, G-25) or maintenance medium (TCPS). The biomechanical characteristics of cells on TCPS in complete medium (GF+) after nine days post-priming were (i) similar to those of hNPCs after three days under similar conditions (i.e., TCPS/GF+; Fig. 4B), and (ii) significantly higher (p< 0.01) than those of cells (elongated, star-shaped) on all three substrates (TCPS, G-100, G-25) in the presence of complete or maintenance medium. The mechanical properties of cells on TCPS in the absence of growth factors (i.e., GF-; Fig. 4C) were significantly different than those on G-25 and G-100 in the presence of growth factors. For elongated and star-shaped cells, the EY, FT and TM values are of the order TCPS(GF+) > TCPS(GF-) > G-100 > G-25. Finally, the EY, FT and TM values of elongated cells are higher than those of star-shaped cells on respective substrates (Table 1).
Table 1.
Average (± standard error) values of mechanical characteristics of cells as measured by AFM. Abbreviations: TCPS (tissue culture polystyrene), GF+ (with growth factors), GF- (without growth factors), D3 (day 3), D9 (day 9), B+ (blebbistatin-treated), EY (Young’s modulus), FT (tether force), TM (apparent membrane tension), RT (radius of tether).
| Culture conditions | Phenotype | EY, kPa | FT, pN | TM, pN/μm | RT, nm |
|---|---|---|---|---|---|
| D3 TCPS (GF+) | NPCs | 3.5 ± 0.06 | 91 ± 1.12 | 1087 ± 28 | 7.1 ± 0.38 |
| D9 TCPS (GF+) | NPCs | 3.3 ± 0.08 | 95 ± 1.86 | 1149 ± 12 | 6.6 ± 0.43 |
| D9 TCPS (GF-) | Elongated | 2.7 ± 0.04 | 69.3 ± 1.23 | 630 ± 35.2 | 9.4 ± 0.55 |
| D9 TCPS (GF-) | Star-shaped | 1.8 ± 0.06 | 54.2 ± 1.4 | 383 ± 30.3 | 12 ± 0.4 |
| D3 G-100 | NPC-like | 2.6 ± 0.05 | 61.2 ± 1.3 | 482.7 ± 24.7 | 10.5 ± 0.5 |
| D3 G-100 | Elongated | 1.2 ± 0.06 | 43.1 ± 1.2 | 245 ± 15 | 15.1 ± 0.55 |
| D3 G-100 | Star-shaped | 0.85 ± 0.04 | 34.1 ± 1.5 | 156.5 ± 31.9 | 19.5 ± 0.45 |
| D3 G-25 | NPC-like | 1.75 ± 0.06 | 51.3 ± 1.3 | 339 ± 30.1 | 12.4 ± 0.55 |
| D3 G-25 | Elongated | 0.82 ± 0.07 | 34.8 ± 1.2 | 159.8 ± 11.8 | 18.7 ± 1.2 |
| D3 G-25 | Star-shaped | 0.45 ± 0.05 | 24.9 ± 0.08 | 79.4 ± 18.9 | 25.5 ± 0.55 |
| D9 G-100 | Elongated | 2.2 ± 0.06 | 50.7 ± 1.13 | 327 ± 15.1 | 12.5 ± 0.55 |
| D9 G-100 | Star-shaped | 1.5 ± 0.04 | 41.3 ± 1.5 | 218.6 ± 31.2 | 15.4 ± 0.45 |
| D9 G-25 | Elongated | 1.7 ± 0.07 | 42.1 ± 1.25 | 228.7 ± 12.2 | 15.1 ± 1.2 |
| D9 G-25 | Star-shaped | 1.1 ± 0.05 | 35.1 ± 1.81 | 164.6 ± 19.8 | 19 ± 0.55 |
| D9 TCPS (B+) | Polarized | 1.56 ± 0.02 | 69.9 ± 1.20 | 617.5 ± 11.8 | 9.3 ± 0.62 |
| D9 G-100 (B+) | Polarized | 1.02 ± 0.22 | 61.3 ± 1.85 | 491.7 ± 22.5 | 10.5 ± 0.56 |
| D9 G-25 (B+) | Polarized | 0.93 ± 0.03 | 55.2 ± 1.31 | 397 ± 20.7 | 11.7 ± 0.86 |
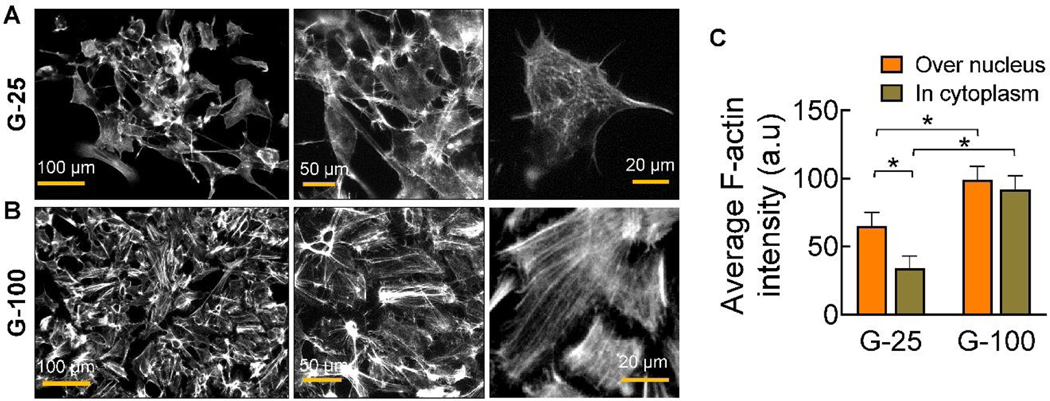
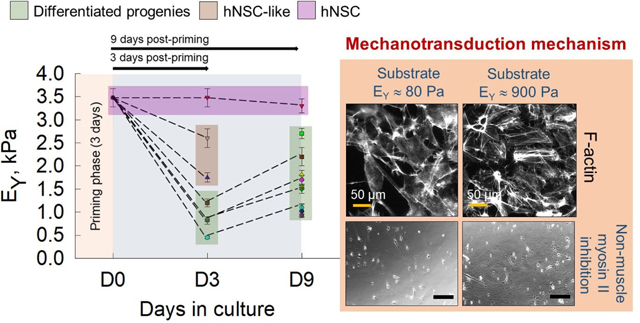
Role of stress fibers in cell mechanics
We observed prominent stress-fibers in cells cultured on G-100 (Fig. 5B) than on G-25 (Fig. 5A) on day 9 post-priming, which might explain the differences in their mechanical properties. The stress fibers seem to be highly organized and oriented in bundles within cells cultured on G-100 compared to the disorganized nature of their distribution in cells on G-25. The F-actin intensity was quantified in two regions within these cells – over the nucleus and in cytoplasm, and significant differences in each case were noted between G-100 and G-25 substrates (Fig. 5C). F-actin intensity also significantly varied within cells on G-25, with higher density near the nucleus. To explore other potential sensors dictating mechanosensing, we stained cells on both stiff (TCPS) and compliant (G-25, G-100) substrates for YAP and PIEZO1. However, their expression was not noted in the cells cultured on these substrates.
Fig. 5.
Representative immunofluorescence images of cells showing morphological changes, i.e., reorganization of F-actin (stress fibers) mesh network, on (A) G-25 and (B) G-100 substrates at 9-day time point, post-priming. Cultures were shown at various magnifications in each case to illustrate the organization of F-actin among the cell population and within each cell. Grey scale intensity of F-actin over the nucleus and in cytoplasm was quantified from these images (n = 6/case). * indicates statistically significant differences (p< 0.01) between the respective groups.
Role of non-muscle myosin II on hNPC characteristics
Blebbistatin is a potent inhibitor of non-muscle myosin-II and acts by blocking actomyosin crosslinking42. hNPCs were treated with blebbistatin throughout the 9-day cultures on TCPS and Geltrex™ substrates and assessed for changes in cell mechanics and differentiation. Compared to dense, network pattern formed by untreated cells on G-100 and G-25 substrates (Fig. 5, A–B), blebbistatin treatment led to the formation of polarized, independent cells (Fig. 6A). The Young’s modulus and tether forces of blebbistatin-treated cells on TCPS were significantly higher (p< 0.01) than cells on G-100 and G-25. There was no statistically significant difference in the EY and FT values of blebbistatin-treated cells on G-100 vs G-25 substrates (Table 1; Fig. 6B). The EY values of blebbistatin-treated cells were significantly lower than their non-treated counterparts on respective substrates, while the FT values were higher. This indicated that myosin-II plays an important role in sensing and adapting to mechanical cues thereby influencing cell mechanics. hNPCs partially maintained their stemness even after blebbistatin treatment for 9 days (SOX2 staining; Fig. 6C). No statistically significant differences were noted between the βIII-tubulinpositive and GFAP positive cells on G-100 vs. G-25, after blebbistatin treatment. Blebbistatin treatment led to reduced differentiation potential of hNPCs on Geltrex™ gels compared to their untreated counterparts (Fig. 3D), suggesting actomyosin-mediated mechanosensing as an important regulator of cell fate.
Fig. 6.
(A) Representative phase contrast images of blebbistatin-treated cells, cultured on TCPS, G-100, and G-25 substrates for nine days post-priming. Scale bar: 50 μm. (B) Biomechanical characteristics (EY, FT, TM, RT) of blebbistatin-treated cells on these substrates after nine days in culture. The red lines represent the mean ± standard deviation of the data while the symbols represent the data points in each case. * denotes p< 0.05. (C) Representative immunofluorescence images of blebbistatin-treated cells on TCPS, G-25, and G-100 substrates after nine days in culture. Primary antibodies for SOX2, β-III tubulin, and GFAP were used, with appropriate secondary antibodies, and cells counterstained with DAPI. Percentage of positively stained cells were manually counted in respective cultures and normalized to total cell density (n > 200 cells/condition). Scale bar: 50 μm.
Differential expression of mechanotransduction genes
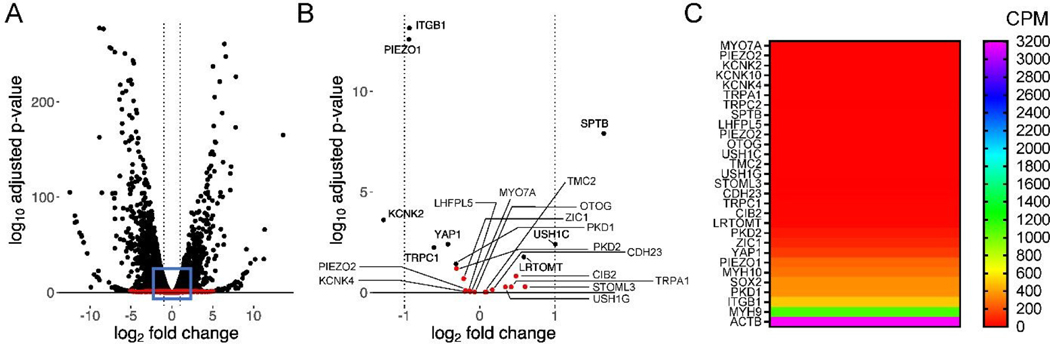
The RNA sequencing data shows robust difference in differential gene expression between ReNcell VM versus ReNcell CX (Fig. 7A). For the genes linked to mechanotransduction, we noted that most of these genes are not significantly differentially expressed (Fig. 7B; red dots). However, a few including KCNK2, PIEZ01, ITGB1, SPTB, YAP1, and USH1C were differentially expressed (black dots). Nevertheless, the data suggests a unique gene expression profile amongst these two cell populations. RNA sequencing data shows that most of the genes linked to mechanotransduction were not significantly expressed (< 10 CPM; Fig. 7C) in these fetal ReNcells, suggesting lowered protein expression in these cell types. Some genes (ZIC1, YAP1, PIEZO1, MYH10) were moderately expressed (100 – 300 CPM), while those for non-muscle myosin II (MYH9), integrin subunit β1 (ITGB1), and SOX2 were relatively higher (> 300 CPM). In comparison, ACTB gene encoding for cytoskeletal actin (stress fibers) was expressed in very high levels in these cells, which explains the strong staining noted for F-actin and non-muscle myosin II in cells cultured on Geltrex™ gels. On the other hand, PIEZO1 and YAP1 were expressed at 4-fold and 10-fold lower than MYH9, respectively, which possibly explains the absence of staining for these two markers in our immunofluorescence images. With low gene expression of some widely known sensors such as YAP and PIEZO1, it is worth exploring other transducers driving niche-dependent changes in phenotype and functional fate of these specific hNPCs (e.g., actin & non-muscle myosin), although it is difficult to decouple the effects of substrate mechanical properties and composition of biological scaffolds.
Fig. 7.
Gene expression comparison between ReNcell VM and ReNcell CX. (A) Volcano plot depicting differentially expressed genes between ReNcell VM and ReNcell CX from GSE89623 (n = 2 for both groups; culture time is 2 weeks), discriminated based on p-value and log2(foldchange) at an level of 0.05 and FDR adjusted p-value (≤ 0.1). Black dots correspond to individual genes whose expression differences were significant based on both p and logFC value, while red dots denote not significantly different genes. (B) Volcano plot of selected genes specifically associated with nuclear mechanotransduction show a majority number of such genes are differentially expressed between the two cell types. (C) Absolute levels of various genes linked to mechanotransduction in ReNcell VM. Data was denoted as counts per million (CPM), and from two separate cell lines of ReNcell VM which were averaged and plotted. Note: data shown here may not be comprehensive in depicting all known genes contributing to mechanotransduction.
Discussion
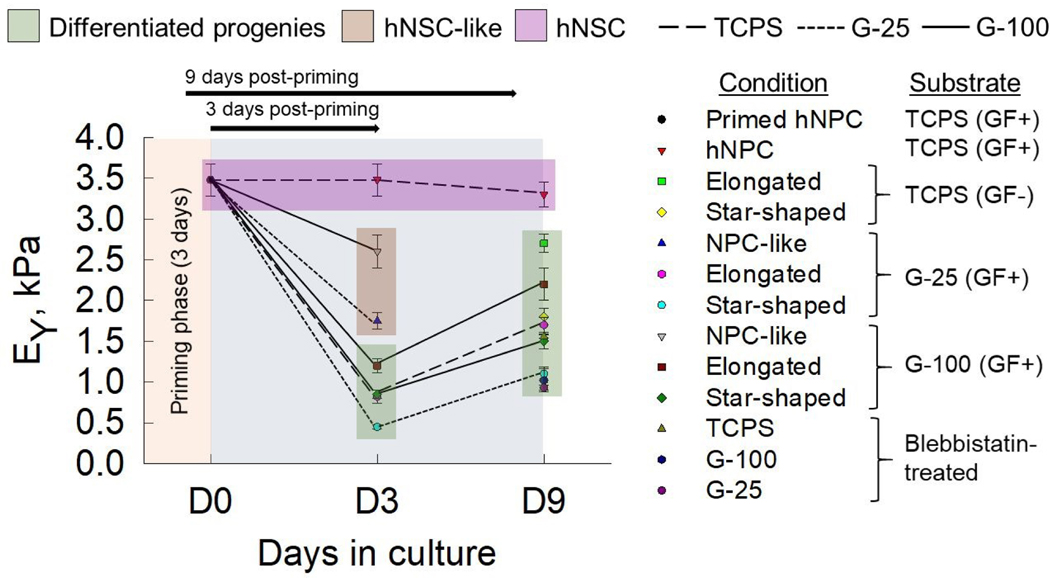
Mechanotransduction is regarded as one of the most important factors driving and guiding CNS development, and recent focus is geared towards elucidating the changes in biophysical and biomechanical properties during fate commitment of NPCs. During development, primitive cells such as NPCs are spatiotemporally regulated by mechanical cues27, 43. Any perturbation by intrinsic or extrinsic agents cause abnormal mechanotransduction in migrating cells, growing neurites, and neuronal growth cones, leading to neurological disorders. Towards understanding one of the many facets of mechanotransduction in CNS development, we here characterized and quantified the temporal evolution of mechanical properties of human fetal NPCs and differentiated progenies, facilitated primarily by mechanosensing. We explored the utility of AFM to elucidate the changes in biomechanical characteristics associated with substrate-mediated differentiation of hNPCs. Our key observations (Fig. 8) are: (i) neural progenitors (hNPCs & NPC-like) are significantly stiffer than their differentiated progenies (glial cells & neurons), (ii) differentiated neuronal-like cells (i.e., elongated phenotype) are stiffer than glial-like (i.e., star-shaped phenotype) cells on a given substrate at both time points, (iii) the stiffer the substrate (TCPS > G-100 > G-25), the higher the EY of the cells, (iv) growth factors removal not only induces differentiation but also drastically reduces EY of resulting progenies (~ 50%), and finally (v) substrate- and time-dependent trends in EY broadly reflect those in FT and therefore in TM. In this regard, this is the first study to evaluate the evolution of mechanics during hNPC differentiation.
Fig. 8.
Summary of evolution of Young’s modulus (EY) of cells over a 9-day culture period. Average EY of hNPCs (primed, D3, D9) on TCPS in complete medium (GF+), NPC-like cells post-priming (at D3) in complete medium on Geltrex™ substrates, elongated and star-shaped cells post-priming (D3, D9) on TCPS (maintenance medium) and Geltrex™ (complete medium) were plotted for comparison. The EY of blebbistatin-treated polarized cells on various substrates were also shown for comparison. Data was presented as average ± standard deviation for respective cases. Green box highlights the average EY values of progenies at D3 and D9. Solid lines represent G-100, short-dashed represent G-25, and medium-dashed represent TCPS substrates. Significant differences in EY values between NPCs and transforming progenies at D3, and between transforming vs. mature progenies at D3 vs. D9 are evident.
The topography of the substrates also affect mechanotransductive processes of cells possibly by regulating the spatial distribution of focal adhesion sites and cytoskeletal reorganization, thereby influencing cell rigidity44. The significant differences in the substrate topography (pore-size, 3D architecture, protein density) of G-25 versus G-100 gels, and thus their inherent substrate rigidities, could have significantly contributed to NPC differentiation, progeny morphology and mechanics (EY, FT, TM), and stress fiber organization and expression. We found that the differentiation process of NPCs has an intermediate state of softer cells at day 3, which transform into much stiffer and mature neural and glial lineages at day 9 (Fig. 8). NPCs and their differentiated progenies on softer substrate (G-25) were more compliant than cells on stiffer substrates (G-100, TCPS). We observed that NPCs on softer matrices (G-25) preferentially differentiate towards neural lineages compared to those on stiffer matrices (G-100). In line with our observations, murine NSCs have been reported to differentiate into neurons on softer substrates but towards glial lineages on stiffer substrates6, 18, while mesenchymal stem cells cultured on soft substrates mimicking brain stiffness committed to neurogenic phenotypes 45.
Translocation of transcriptional factors such as YAP and TAZ act as mechanosensors13, 46–48 by converting mechanical signals into functional fate. NSCs were shown to have maximum sensitivity towards substrate stiffness in the first 24 h, which drives their lineage commitment18, 22. Overexpression and silencing of YAP in the first 24 h led to inhibition and enhancement of neurogenesis, respectively22. Decline in regenerative capacity of stem cells and progenitor cells with aging could be linked to altered mechanical signaling from the ECM. PIEZO1 – a mechanoresponsive ion channel – was shown to mediate conversion of these mechanical signals from the ECM to NPC fate10, 49.
Our results indicate that the intrinsic mechanical properties play an important role during niche-mediated differentiation of hNPCs. We explored the role of non-muscle myosin in this process by its inhibition using blebbistatin, and found that myosin plays an critical role in regulating mechano-adaptability which ultimately influences cell fate50. Comparing differentiation and cell mechanics outcomes before and after blebbistatin treatment established substrate mechanosensing as one of the main factors driving cell fate. Intrinsic mechanical properties of cells have shown great potential as a label-free biomarker in many processes. The mechanical properties of stem cells have been explored to indicate their stemness. For instance, lower EY was correlated to greater differentiation potential, and pluripotent cells were found to be softer compared to multipotent and unipotent stem cells51. In another study, limbal stem cells were sorted from a heterogenous population of corneal cells using AFM-based mechanical characterization52. Extending these prior studies which utilized EY to mechanically sort cells, our study explored tether force, membrane tension, and tether radius to distinguish NPCs, glial cells, and neurons in a heterogenous cell population on substrates with varying stiffness. Trends in FT and TM of hNPCs and their progenies mirror their EY values suggesting that along with cytoskeleton, cell membranes also might be experiencing significant changes (e.g., blebs, ruffles, lamellae) in response to substrate mechanics and temporal evolution of their phenotype.
In support of our observation that differentiated progenies are more compliant than progenitor cells, others have reported on the formation of softer differentiated NPCs from stiffer iPSCs, and a positive correlation between EY and pluripotency20. Glial cells, being more compliant, serve as mobile immune cells in the CNS and provide protection during trauma. During development, stem cells experience heterogenous mechanical microenvironments which directly influence their biophysical characteristics, accompanied by genetic and epigenetic changes. We observed that cells on stiffer matrices possess higher EY compared to cells on softer matrices, elucidating potential mechano-adaptability of development relevant cells18, 45, 53.
Tension-based morphogenesis theory suggests that tension along glial cell extension, axons, and neurites is sufficient to generate folding patterns of the cortex, compactness of neural circuitry, and many other structural features of the CNS54. In support of this theory, application of tensile forces using glass microneedles on neurites caused active elongation when tension is maintained above a threshold, followed by active retraction upon tension withdrawal55. However, the intrinsic membrane tension of progenitor cells and their differentiated progenies forming mechanically-heterogenous CNS was not explored. We quantified for the first time the changes in membrane tension of hNPCs en route to differentiated progenies. We observed that membrane tension of hNPCs decreased after initial 3 days of differentiation, followed by an increase upon maturation at day 9.
In conclusion, we here investigated the effect of mechanosensing on hNPCs using substrates of varying stiffness. We highlighted the role of cell biomechanics as a label-free marker of NPC differentiation. We identified the role of mechano-adaptation as a possible mechanism of differentiation, specifically for these cells. We explored potential mechanosensors and elucidated the role of myosin motors in sensing and responding to the stiffness of their environment. Taken together, our findings establish the changes in the mechanical properties of hNPCs en route to differentiated progenies, trigged by substrate stiffness exploring niche-activated mechanosensors.
Supplementary Material
Acknowledgements:
This work was partially supported by funds from the US National Science Foundation (CBET, Award # 1337859) to C.K, and Graduate Student Research Award to G.M from Cleveland State University. K.F acknowledges partial support from the Ruth L. Kirschstein National Research Service Award Individual Postdoctoral Fellowship (F32 AG056098) from the National Institutes of Health (NIH).
Footnotes
Conflicts of interest/Competing interests: None
Publisher's Disclaimer: This is an Accepted Manuscript, which has been through the Royal Society of Chemistry peer review process and has been accepted for publication.
Publisher's Disclaimer: Accepted Manuscripts are published online shortly after acceptance, before technical editing, formatting and proof reading. Using this free service, authors can make their results available to the community, in citable form, before we publish the edited article. We will replace this Accepted Manuscript with the edited and formatted Advance Article as soon as it is available.
References
- 1.Barnes JM, Przybyla L. and Weaver VM, J Cell Sci, 2017, 130, 71–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kolahi KS and Mofrad MRK, WIRES Syst Biol Med, 2010, 2, 625–639. [DOI] [PubMed] [Google Scholar]
- 3.Chalfie M, Nat Rev Mol Cell Biol, 2009, 10, 44–52. [DOI] [PubMed] [Google Scholar]
- 4.Martino G, Pluchino S, Bonfanti L. and Schwartz M, Physiol Rev, 2011, 91, 1281–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Franze K, Development (Cambridge), 2013, 140, 3069–3077. [DOI] [PubMed] [Google Scholar]
- 6.Ulrich TA and Kumar S, in Mechanobiology Handbook, 2011, pp. 391–411. [Google Scholar]
- 7.Lourenço T. and Grãos M, Front Cell Neurosci, 2016, 10, 277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu W, Lakshman N. and Morshead CM, Neurogenesis, 2017, 4, e1300037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xue X, Sun Y, Resto-Irizarry AM, Yuan Y, Yong KMA, Zheng Y, Weng S, Shao Y, Chai y., Studer L. and Fu J, Nat Mater, 2018, 17, 633–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Segel M, Neumann B, Hill MFE, Weber IP, Viscomi C, Zhao C, Young A, Agley CC, Thompson AJ, Gonzalez GA, Sharma A, Holmqvist S, Rowitch DH, Franze K, Franklin RJM and Chalut KJ, Nature, 2019, 573, 130–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moeendarbary E, Weber IP, Sheridan GK, Koser DE, Soleman S, Haenzi B, Bradbury EJ, Fawcett J. and Franze K, Nat Commun, 2017, 8, 14787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nardone G, Oliver-De La Cruz J, Vrbsky J, Martini C, Pribyl J, Skládal P, Pešl M, Caluori G, Pagliari S, Martino F, Maceckova Z, Hajduch M, Sanz-Garcia A, Pugno NM, Stokin GB and Forte G, Nat Commun, 2017, 8, 15321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Panciera T, Azzolin L, Cordenonsi M. and Piccolo S, Nat Rev Mol Cell Biol, 2017, 18, 758–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, Elvassore N. and Piccolo S, Nature, 2011, 474, 179–184. [DOI] [PubMed] [Google Scholar]
- 15.Sorrentino G, Ruggeri N, Specchia V, Cordenonsi M, Mano M, Dupont S, Manfrin A, Ingallina E, Sommaggio R, Piazza S, Rosato A, Piccolo S. and Del Sal G, Nat Cell Biol, 2014, 16, 357–366. [DOI] [PubMed] [Google Scholar]
- 16.Koser DE, Thompson AJ, Foster SK, Dwivedy A, Pillai EK, Sheridan GK, Svoboda H, Viana M, Costa LF, Guck J, Holt CE and Franze K, Nature Neurosci, 2016, 19, 1592–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franze K, Gerdelmann J, Weick M, Betz T, Pawlizak S, Lakadamyali M, Bayer J, Rillich K, Gögler M, Lu YB, Reichenbach A, Janmey P. and Käs J, Biophys J, 2009, 97, 1883–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keung AJ, De Juan-Pardo EM, Schaffer DV and Kumar S, Stem Cells, 2011, 29, 1886–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ingber DE, Wang N. and Stamenović D,Rep Prog Phys, 2014, 77, 1–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Urbanska M, Winzi M, Neumann K, Abuhattum S, Rosendahl P, Müller P, Taubenberger A, Anastassiadis K. and Guck J, Development (Cambridge), 2017, 144, 4313–4321. [DOI] [PubMed] [Google Scholar]
- 21.Maloney JM, Nikova D, Lautenschläger F, Clarke E, Langer R, Guck J. and Van Vliet KJ, Biophys J, 2010, 99, 2479–2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rammensee S, Kang MS, Georgiou K, Kumar S. and Schaffer DV, Stem Cells, 2016, 35, 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arulmoli J, Pathak MM, McDonnell LP, Nourse JL, Tombola F, Earthman JC and Flanagan LA, Sci Rep, 2015, 5, 8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mori H, Takahashi A, Horimoto A. and Hara M, Neurosci Lett, 2013, 555, 1–6. [DOI] [PubMed] [Google Scholar]
- 25.Leipzig ND and Shoichet MS, Biomaterials, 2009, 30, 6867–6878. [DOI] [PubMed] [Google Scholar]
- 26.Saha K, Keung AJ, Irwin EF, Li Y, Little L, Schaffer DV and Healy KE, Biophys J, 2008, 95, 4426–4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mahajan G, Lee MY and Kothapalli CR, Arch Toxicol, 2019, 93, 2979–2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mullen CA, Vaughan TJ, Billiar KL and McNamara LM, Biophys J, 2015, 108, 1604–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walker A, Su H, Conti MA, Harb N, Adelstein RS and Sato N, Nat Commun, 2010, 1, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gan Y, Rev Sci Instrum, 2007, 78, 3–10. [DOI] [PubMed] [Google Scholar]
- 31.MacKay JL and Kumar S, in Cell Imaging Techniques: Methods and Protocols eds. Taatjes J. and Roth J, Humana Press, 2013, pp. 313–329 [Google Scholar]
- 32.Han W, Chen S, Yuan W, Fan Q, Tian J, Wang X, Chen L, Zhang X, Wei W, Liu R, Qu J, Jiao Y, Austin RH and Liu L, Proc Natl Acad Sci U S A, 2016, 113, 11208–11213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Diz-Muñoz A, Fletcher DA and Weiner OD, Trends Cell Biol, 2013, 23, 47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zonderland J, Wieringa P. and Moroni L, MethodsX, 2019, 6, 2562–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pollak J, Rai KG, Funk CC, Arora S, Lee E, Zhu J, Price ND, Paddison PJ, Ramirez JM and Rostomily RC, PLoS One, 2017, 12, e0172884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mahi NA, Najafabadi MF, Pilarczyk M, Kouril M. and Medvedovic M, Sci Rep, 2019, 9, 7580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chighizola M, Dini T, Lenardi C, Milani P, Podestà A. and Schulte C, Biophys Rev, 2019, 11, 701–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uhler C. and Shivashankar GV, Nat Rev Mol Cell Biol, 2017, 18, 717–727. [DOI] [PubMed] [Google Scholar]
- 39.Jin P, Jan LY and Jan YN, Annu Rev Neurosci, 2020, 43, 207–229. [DOI] [PubMed] [Google Scholar]
- 40.Gilbert PM, Havenstrite KL, Magnusson KE, Sacco A, Leonardi NA, Kraft P, Nguyen NK, Thrun S, Lutolf MP and Blau HM, Science 2010, 329, 1078–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Georges PC, Miller WJ, Meaney DF, Sawyer ES and Janmey PA, Biophys J, 2006, 90, 3012–3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kovács M, Tóth J, Hetényi C, Málnási-Csizmadia A. and Sellers JR, J Biol Chem, 2004, 279, 35557–35563. [DOI] [PubMed] [Google Scholar]
- 43.Vining KH and Mooney DJ, Nat Rev Mol Cell Biol, 2017, 18, 728–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schulte C, Rodighiero S, Cappelluti MA, Puricelli L, Maffioli E, Borghi F, Negri A, Sogne E, Galluzzi M, Piazzoni C, Tamplenizza M, Podestà A, Tedeschi G, Lenardi C. and Milani P, J Nanobiotechnology, 2016, 14, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Engler AJ, Sen S, Sweeney HL and Discher DE, Cell, 2006, 126, 677–689. [DOI] [PubMed] [Google Scholar]
- 46.Han D, Byun SH, Park S, Kim J, Kim I, Ha S, Kwon M. and Yoon K, Biochem Biophys Res Comm, 2015, 458, 110–116. [DOI] [PubMed] [Google Scholar]
- 47.Lee S, Stanton AE, Tong X. and Yang F, Biomaterials, 2019, 202, 26–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kang PH, Schaffer DV and Kumar S, Mol Biol Cell, 2020, 31, 386–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pathak MM, Nourse JL, Tran T, Hwe J, Arulmoli J, Le DT, Bernardis E, Flanagan LA and Tombola F, Proc Natl Acad Sci U S A, 2014, 111, 16148–16153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee S. and Kumar S, F1000Research, 2016, 5, 2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hammerick KE, Huang Z, Sun N, Lam MT, Prinz FB, Wu JC, Commons GW and Longaker MT, Tissue Eng Part A, 2011, 17, 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bongiorno T, Chojnowski JL, Lauderdale JD and Sulchek T, Biophys J, 2016, 111, 1761–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Overby DR, Zhou EH, Vargas-Pinto R, Pedrigi RM, Fuchshofer R, Braakman ST, Gupta R, Perkumas KM, Sherwood JM, Vahabikashi A, Dang Q, Kim JH, Ethier CR, Stamer WD, Fredberg JJ and Johnson M, Proc Natl Acad Sci U S A, 2014, 111, 13876–13881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van Essen DC, Nature, 1997, 385, 313–318. [DOI] [PubMed] [Google Scholar]
- 55.Zheng J, Lamoureux P, Santiago V, Dennerll T, Buxbaum RE and Heidemann SR, J Neurosci, 1991, 11, 1117–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.