SUMMARY
Polyamines, small organic polycations, are essential for cell viability and their physiological levels are homeostatically maintained by post-transcriptional regulation of key biosynthetic enzymes. In addition to de novo synthesis, cells can also take up polyamines; however, identifying cellular polyamine transporters has been challenging. Here, we identify that the S. cerevisiae HOL1 mRNA is under translational control by polyamines, and we reveal that the encoded membrane transporter Hol1 is a high-affinity polyamine transporter and is required for yeast growth under limiting polyamine conditions. Moreover, we show that polyamine inhibition of translation factor eIF5A impairs translation termination at a Pro-Ser-Stop motif in a conserved upstream open reading frame on the HOL1 mRNA to repress Hol1 synthesis under conditions of elevated polyamines. Taken together, our findings reveal that polyamine transport, like polyamine biosynthesis, is under translational autoregulation by polyamines in yeast, highlighting the extensive control cells impose on polyamine levels.
Keywords: polyamine, spermidine, HOL1, transporter, uORF, eIF5A, translational control, translation termination
Graphical Abstract
In Brief
Vindu et al. identify Hol1 as the major high-affinity polyamine transporter in S. cerevisiae. Polyamines autoregulate HOL1 expression through feedback inhibition of HOL1 mRNA translation mediated by polyamine inhibition of eIF5A-dependent translation termination on a conserved upstream open reading frame in the HOL1 mRNA leader.
INTRODUCTION
Polyamines, ubiquitous small organic cations, have been implicated in a variety of cell functions including transcription, translation, cell-cell communication, and ion channel gating (Igarashi and Kashiwagi, 2010b; Pegg, 2016). Owing to their cationic properties, polyamines interact with negatively charged nucleic acids and lipids. Though critical for cell viability, the precise function of polyamines has remained unclear. Manipulation of cellular polyamine levels via inhibition of biosynthesis and uptake are being tested as possible therapies for cancers (Casero et al., 2018), underscoring the critical nature of polyamines for cell growth and survival.
Polyamine synthesis in eukaryotes starts with the decarboxylation of ornithine by the enzyme ornithine decarboxylase (ODC) creating putrescine (1,4-diaminobutane). The key higher-order polyamines spermidine (SPD) and spermine are then synthesized in three steps (Pegg, 2009). First, the enzyme AdoMetDC (or AMD1) catalyzes decarboxylation of S-adenosylmethionine (SAM) to generate decarboxylated SAM (dcSAM). The enzyme SPD synthase then transfers an N-propylamine moiety from dcSAM to putrescine to form SPD. Finally, the tetraamine spermine is formed by transfer of an additional N-propylamine moiety from dcSAM to SPD by the enzyme spermine synthase.
Polyamine biosynthesis is under feedback control with the synthesis of multiple enzymes and regulators inhibited by polyamines at the translational level (reviewed in Dever and Ivanov, 2018; Ivanov et al., 2010a; Pegg, 2009). The protein antizyme (OAZ) binds to ODC and targets it for ubiquitin-independent degradation (Heller and Canellakis, 1981; Murakami et al., 1992). In yeast and higher eukaryotes, synthesis of OAZ is dependent on polyamine-regulated ribosomal frameshifting on the OAZ mRNA (Ivanov and Atkins, 2007; Kahana, 2009; Matsufuji et al., 1995; Palanimurugan et al., 2004). High polyamine levels are thought to impair termination and enable a fraction of the ribosomes translating the OAZ1 mRNA to shift on a slippery sequence into the +1 reading frame to synthesize the functional C-terminal portion of OAZ (reviewed in Dever and Ivanov, 2018). Thus, high polyamine levels induce synthesis of OAZ which targets ODC for degradation, blocking excess synthesis of polyamines when the levels are high.
In contrast to OAZ, synthesis of AdoMetDC is feedback inhibited by polyamines. In vertebrates, a short upstream open reading frame (uORF) in the AdoMetDC mRNA sensitizes translation to the level of polyamines (Ruan et al., 1996). Under low polyamine conditions, the uORF is not inhibitory and AdoMetDC synthesis is high. In contrast, under high polyamine conditions, ribosomes stall during termination on the uORF encoding the peptide sequence MAGDIS (Law et al., 2001; Raney et al., 2002), preventing ribosome access to the main ORF (mORF) and repressing AdoMetDC synthesis.
The ability of OAZ to target ODC is regulated by a protein called antizyme inhibitor (AZIN). AZIN is a catalytically defective form of ODC that still binds to OAZ (Murakami et al., 1996). Titration of OAZ by AZIN prevents OAZ from targeting ODC for degradation, thereby stabilizing ODC and enhancing polyamine synthesis. The synthesis of AZIN in vertebrates, as well as ODC in some invertebrates and fungi, is inhibited by polyamines. The leaders of these mRNAs contain an upstream conserved coding region (uCC) that confers polyamine control on AZIN mRNA translation (Ivanov et al., 2008). The uCC is a non-canonical uORF that initiates either at a near-cognate start codon that differs from AUG by a single nucleotide change or at an AUG codon with poor flanking nucleotide context sequence (Ivanov et al., 2008; Ivanov et al., 2018). The poor start codon of the uCC, which will allow a significant fraction of scanning ribosomes to leaky scan past without stopping (reviewed in Hinnebusch, 2011), is a critical feature for translational control of AZIN synthesis.
In accordance with the scanning mechanism of translation (reviewed in Hinnebusch, 2014), under conditions of low polyamines some ribosomes will leaky scan past the uCC without translating and then synthesize AZIN. The occasional ribosome that translates the uCC is likely to disengage from the mRNA upon termination at the uCC stop codon. Under conditions of high polyamines, ribosomes that initiate at the weak start site of the uCC pause when translating the highly conserved C-terminal PPWxxPS* motif (* = stop codon) because high polyamines inhibit the activity of the translation factor eIF5A that is required for PPW synthesis (Ivanov et al., 2018). The stalled ribosome is an impediment to scanning, and subsequent scanning ribosomes that leaky scan past the uCC start codon without initiating are proposed to form a queue behind the stalled elongating ribosome. Eventually the queue will extend back to the uCC start codon, poising a scanning ribosome over the uCC start codon for a longer time and enhancing initiation on the uCC. As ribosomes that translate the uCC do not reinitiate downstream, the increased translation of the uCC under conditions of high polyamines represses synthesis of AZIN (Ivanov et al., 2018).
In addition to synthesizing polyamines, cells also import polyamines. While several membrane transporters have been ascribed polyamine transport activity, the identity of the high-affinity polyamine transporter in yeast is unclear. The SAM transporter Sam3, as well as the urea transporter (Dur3), have both been proposed to serve as polyamine transporters in yeast (Uemura et al., 2007). In addition, the general amino acid transporter (Gap1) and Agp2, which belongs to the amino acid permease family, have also been proposed to play roles in polyamine uptake (Aouida et al., 2013; Uemura et al., 2005). However, none of these proteins appears to be responsible for high affinity uptake. Likewise, in mammalian cells, the identity of a dedicated high-affinity polyamine transporter has remained elusive (reviewed inCasero et al., 2018). Here, we identify Hol1 as the high-affinity polyamine transporter in yeast, and we show that expression of HOL1 is regulated at the translational level by polyamine inhibition of translation termination on a conserved uORF in the HOL1 mRNA leader.
RESULTS
HOL1 is Required for Polyamine Uptake and is Translationally Repressed by Polyamines
Whereas polyamines directly regulate the translation of three mRNAs in mammalian cells (encoding OAZ, AZIN and AdoMetDC), the only clear translational target of polyamines in the yeast S. cerevisiae is OAZ1, the gene encoding OAZ. To identify additional mRNAs whose translation is sensitive to changes in polyamine levels, we performed ribosome profiling in yeast grown in low or high levels of polyamines. To control polyamine levels, the yeast spe1 (ODC) spe2 (AdoMetDC) double mutant strain Y362 that is unable to synthesize polyamines and thus must acquire the essential polyamines exogenously was used for the study. In addition, to precisely control polyamine levels polyamine-deficient yeast nitrogen base was used when preparing media (see Methods). To define growth conditions for ribosome profiling, a polyamine-regulated yeast OAZ1 frameshift reporter was introduced into the strain. In the absence of added polyamines in the medium, where cell growth is maintained by the internal stores of polyamines accumulated during prior growth in medium containing polyamines, OAZ1 frameshifting was repressed, confirming a low concentration of intracellular polyamines (Figure S1A). As the SPD concentration in the medium was elevated, OAZ1 frameshifting increased reaching a maximal plateau at 5μM SPD (8.5-fold increase in frameshifting). Based on these findings, cells were first grown in medium containing 0 and 10 μM SPD for low and high polyamine conditions, respectively. Both RNAseq and RiboSeq analyses were performed on two independent cultures and the results were highly reproducible (Figure S1B; Pearson’s r ~ 0.99).
To identify translationally controlled mRNAs, the translation efficiency (TE) for each mRNA was calculated by normalizing the RiboSeq read density to the RNAseq read density. The change in TE for each mRNA upon shifting from 0 to 10 μM SPD was calculated and plotted relative to the adjusted p value (Figure 1A). Focusing on TE changes of at least 1.5-fold (either up or down) with a false discovery rate (FDR) < 0.1 revealed 400 mRNAs potentially under translational control by polyamines. Of note, the OAZ1 mRNA, as expected, was identified among the 212 translationally upregulated mRNAs in high polyamines (OAZ1 mRNA ΔTE ~2.3-fold [calculated for the coding sequence (CDS) after the frameshift]). While gene ontology (GO) analysis of the translationally upregulated mRNAs revealed several significantly enriched categories including catalytic activity and structural constituent of the ribosome (Figure S1C), transmembrane transporter and transport activity categories were enriched among mRNAs showing reduced TE in high polyamines (Figure S1D). Of the 188 translationally downregulated mRNAs, 23 encode transporters or proteins with membrane transport related functions (Figure S1E).
Figure 1. Translational Profiling and Uptake Assays Link Hol1 to Polyamine Transport.
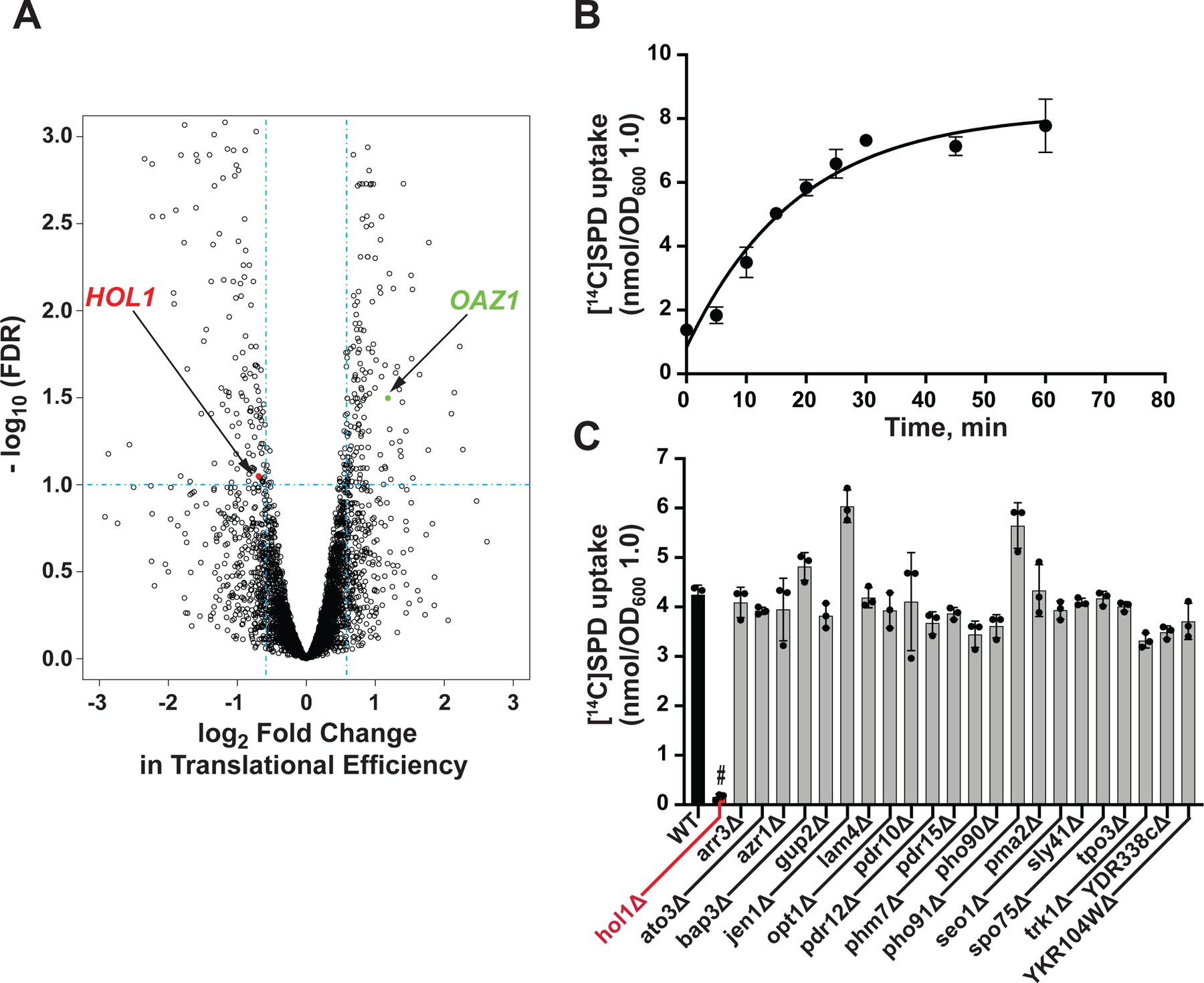
(A) Volcano plot showing translational efficiency (TE) changes versus FDR in spe1 spe2 mutant yeast strain Y362 grown in low (0 μM) and high (10 μM) SPD. Vertical dashed lines denote 1.5-fold changes in TE; horizontal dashed line denotes FDR = 0.1; translationally upregulated antizyme (OAZ1; green) and downregulated HOL1 (red) mRNAs are highlighted.
(B) Time-course of [14C]SPD uptake by log-phase WT BY4741 cells was fit to a single exponential curve. Error bars represent SD (n = 3).
(C) [14C]SPD uptake at 20 min in derivatives of BY4741 lacking the indicated gene encoding a TE-downregulated transmembrane transport-related protein. Error bars denote SD; #p < 10−5 relative to WT (Student’s two-tailed t test; n = 3).
As a dedicated transporter responsible for polyamine uptake in Saccharomyces cerevisiae has not been identified, we wondered if any of the 23 translationally controlled transporters had a role in polyamine uptake. A polyamine uptake assay was established by incubating [14C]-labelled SPD with log-phase WT BY4741 cells. As shown in Figure 1B, following an initial lag SPD uptake increased linearly until reaching a plateau at around 30 min. To test the possible role in polyamine uptake of the 23 translationally controlled transporters, single gene deletion mutants of BY4741 lacking one of the transporters were obtained from the yeast deletion collection and assayed for SPD uptake. Only the strain lacking HOL1 exhibited a profound defect in SPD uptake with a nearly complete loss of uptake activity (Figure 1C). The HOL1 gene was originally identified by gain-of-function mutations that enhanced uptake of the amino acid alcohol histidinol (Gaber et al., 1990). These HOL1 mutations were also found to enhance Ca2+ uptake in a manner that was sensitive to inhibition by other divalent cations (Wright et al., 1996), suggesting that Hol1 might transport other cations. However, HOL1 had not previously been linked to polyamine transport.
Hol1 is Essential for Growth in Low Polyamine Conditions
Deletion of HOL1 in a spe1 spe2 yeast strain lacking ODC and AdoMetDC, respectively, nearly abolished SPD uptake (Figure 2A). Importantly, re-introduction of HOL1 complemented the SPD transport defect in the spe1 spe2 hol1 triple mutant strain, confirming the critical HOL1 requirement for polyamine transport (Figure 2A). As the lack of ODC and AdoMetDC in the spe1 spe2 mutant blocks production of putrescine and the higher order polyamines SPD and spermine, the strain must acquire the essential polyamines from the medium. In contrast to the spe1 spe2 strain, which grows well on medium containing 10 nM SPD, deletion of HOL1 blocks cell growth under these conditions (Figure 2B, rows 1 and 2). Growth of the spe1 spe2 hol1 triple mutant was restored by either introducing a WT HOL1 plasmid or by increasing the SPD concentration to 1 mM (Figure 2B, rows 3 and 5). This synthetic growth defect observed upon deleting HOL1 in polyamine biosynthesis mutants is supported by synthetic genetic array studies showing negative genetic interactions between hol1 and spe1 or spe2 mutants (Costanzo et al., 2016; Usaj et al., 2017). Taken together, these results indicate that HOL1 is required for high-affinity polyamine uptake and that lower affinity transporters can compensate for the lack of HOL1 when a sufficiently high concentration of polyamines is present in the medium.
Figure 2. Hol1 is the Major High-affinity Polyamine Transporter in S. cerevisiae.
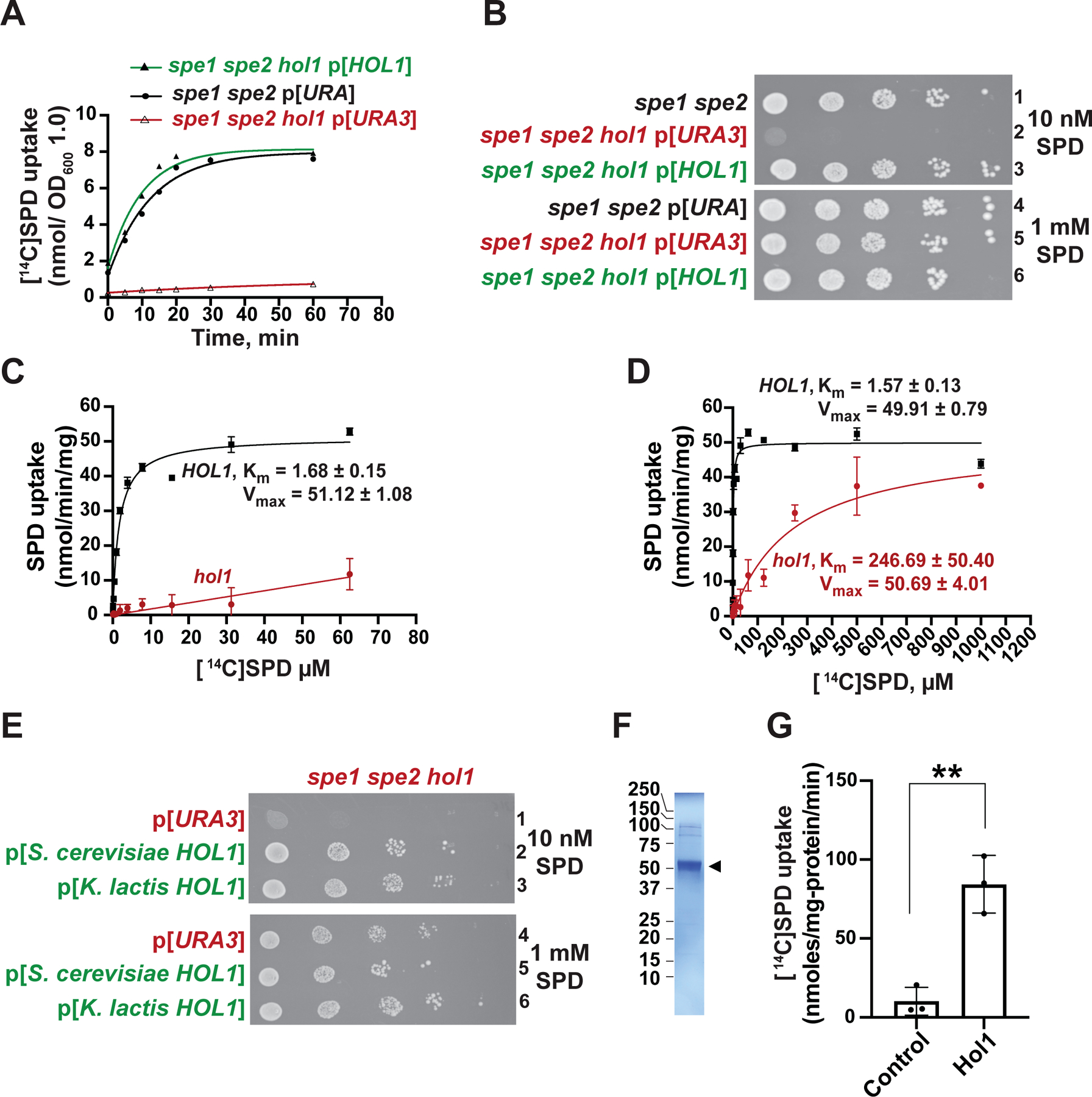
(A–B) The spe1 spe2 yeast strain Y362 carrying an empty vector (black) and the isogenic spe1 spe2 hol1 strain J1570 carrying empty vector YCplac33 (red) or HOL1 plasmid pC6464 (green) were tested (A) for [14C]SPD uptake and (B) for growth using spotting assays on SD plates containing low (10 nM, upper panel) or high (1 mM, lower panel) SPD. In (A), results were fit to a single exponential curve; error bars denote SD; n = 3.
(C–D) Rate of [14C]SPD uptake (nmol/min/mg protein) at the indicated SPD concentrations in spe1 spe2 hol1 strain J1570 carrying empty vector YCplac33 or HOL1 plasmid pC6464, as indicated, were fit to the Michaelis-Menten equation to calculate the Km (μM) and Vmax (nmol/min/mg) for uptake in the presence and absence of HOL1 (n = 3). Note the different SPD concentration ranges in the panels.
(E) Derivatives of the spe1 spe2 hol1 strain J1570 carrying empty vector pC6316 or a plasmid expressing S. cerevisiae (pC6455) or K. lactis (pC6639) HOL1 under the control of a weak constitutive promoter derived from the yeast HSF1 gene were tested for growth using spotting assays on SD plates containing low (10 nM, upper panel) or high (1 mM, lower panel) SPD.
(F) SDS-PAGE gel of detergent purified K. lactis Hol1 (arrowhead).
(G) [14C]SPD uptake by control or K. lactis Hol1 proteoliposomes; error bars denote SD, **p < 0.01 (Student’s two-tailed t test; n = 3).
Amino acid sequence analysis indicates that Hol1 belongs to the major facilitator superfamily (MFS1) of membrane transporters; more specifically, Hol1 is a member of the 12-spanner drug:proton (H+) antiporter (DHA1) family (Gbelska et al., 2006). Members of the DHA1 family include drug efflux pumps and detoxifying transporters, and these proteins have been localized to the vacuole, endoplasmic reticulum, plasma membrane, and other cellular membranes (Gbelska et al., 2006). To localize Hol1 in yeast cells, the chromosomal HOL1 gene was tagged with GFP. As shown in Figure S2A, the Hol1-GFP fusion protein localized to the plasma membrane, consistent with the notion that Hol1 functions as a plasma membrane polyamine transporter.
Hol1 is the Major High-affinity Polyamine Transporter in Yeast
While our studies clearly show the importance of Hol1 for polyamine uptake and growth of yeast on media containing low concentrations of polyamines, previous studies implicated Sam3, the S-adenosyl methionine (SAM) transporter, Dur3, the urea transporter, and Agp2, a polyamine transponder, in polyamine uptake (Aouida et al., 2013; Uemura et al., 2007). To assess the roles of these proteins in polyamine uptake, SAM3, DUR3 and AGP2 were individually deleted in the same spe1 spe2 strain used to assess HOL1 function. As expected, deletion of DUR3 impaired yeast cell growth on media containing 2 mM urea as the sole nitrogen source (Figure S3C), and deletion of SAM3 impaired SAM uptake (Figure S3D). As shown in Figure S3A, under the low polyamine growth conditions used for the assay, deletion of HOL1, but not SAM3, DUR3 or AGP2 impaired SPD uptake. Likewise, deletion of SAM3, DUR3, AGP2, the general amino acid permease GAP1, or the TPO1 – TPO5 genes, which were previously linked to polyamine export and intracellular transport (reviewed in Igarashi and Kashiwagi, 2010a), did not perturb polyamine uptake in the BY4741 background (Figure S3E). Consistent with the robust SPD uptake in the spe1 spe2 strains lacking SAM3, DUR3 or AGP2, these strains grew well on medium containing a low concentration (10 nM) of SPD (Figure S3B). In contrast, the hol1 mutant again exhibited a severe growth defect that was rescued by increasing the concentration of SPD in the medium (Figure S3B). Thus, under low polyamine conditions, where, as described below, HOL1 is strongly expressed, HOL1 is required, but SAM3, DUR3 and AGP2 are dispensable, for polyamine uptake and for growth.
To investigate the kinetics of polyamine uptake, the spe1 spe2 hol1 mutant was transformed with empty vector (hol1 mutant) or with a plasmid expressing WT HOL1 (HOL1 WT). The rate of SPD uptake was determined over a range of concentrations from 1 μM to 1 mM. Fitting the results (Figures 2C–D) to the Michaelis-Menten equation revealed a Km for SPD of ~1.6 μM and a Vmax of ~50 nmol SPD/min/mg protein in the HOL1 WT strain. In contrast, in the hol1 mutant cells the Km was ~250 μM and the Vmax was ~50 nmol/min/mg protein. The greater than 100-fold lower Km for SPD uptake in WT cells versus cells lacking HOL1 supports the conclusion that Hol1 is the major high-affinity polyamine transporter in S. cerevisiae.
To directly examine polyamine transport activity of Hol1 by in vitro transport assays, we tested several fungal homologs of Hol1 by fluorescence size-exclusion chromatography (FSEC) (Kawate and Gouaux, 2006) and found the Hol1 protein from Kluveromyces lactis to be amenable to preparative purification. When expressed in S. cerevisiae, K. lactis HOL1 functionally complemented the polyamine-dependent growth defect associated with the hol1 mutation (Figure 2E), confirming that K. lactis Hol1 is a homolog of the S. cerevisiae protein (see Figure S2B). The K. lactis Hol1 was expressed in Pichia pastoris with a C-terminal polyhistidine tag. Following purification by His-tag and size exclusion chromatography, the highly purified Hol1 (Figure 2F) was assembled into reconstituted proteoliposomes. Control liposomes lacking Hol1 exhibited very low SPD uptake activity (Figure 2G); whereas SPD uptake increased by at least 8-fold in the proteoliposomes containing Hol1, thus directly demonstrating polyamine transport activity by purified Hol1.
A Conserved uORF is Required for Polyamine Repression of HOL1 mRNA Translation
Having originally identified HOL1 based on its reduced TE under conditions of high polyamines (Figure 1A), we sought to determine how polyamines control HOL1 mRNA translation. Comparative sequence analysis of HOL1 across the Saccharomyces genus revealed a conserved uORF encoding the peptide MLLLPS* (Figure 3A). Interestingly, this uORF, which is located in the middle of the HOL1 mRNA 5’ leader (Figure 3A), shares features with the polyamine-controlled uCC elements in AZIN1 and some ODC mRNAs (Ivanov et al., 2008; Ivanov et al., 2018). First, the HOL1 uORF has a weak start site with a non-preferred U at position −3 rather than A as in the preferred yeast start codon context sequence (Dever et al., 2016; Dvir et al., 2013). This weak context is predicted to lead to increased leaky scanning by ribosomes over the uORF start codon without initiating. Second, the PS* motif in the HOL1 uORF is highly conserved in AZIN1 uCCs from vertebrates (Ivanov et al., 2018), and mutation of this motif was previously shown to partially impair polyamine regulation of uCC translation (Ivanov et al., 2018).
Figure 3. Conserved uORF Confers Polyamine Control of HOL1 mRNA Translation.
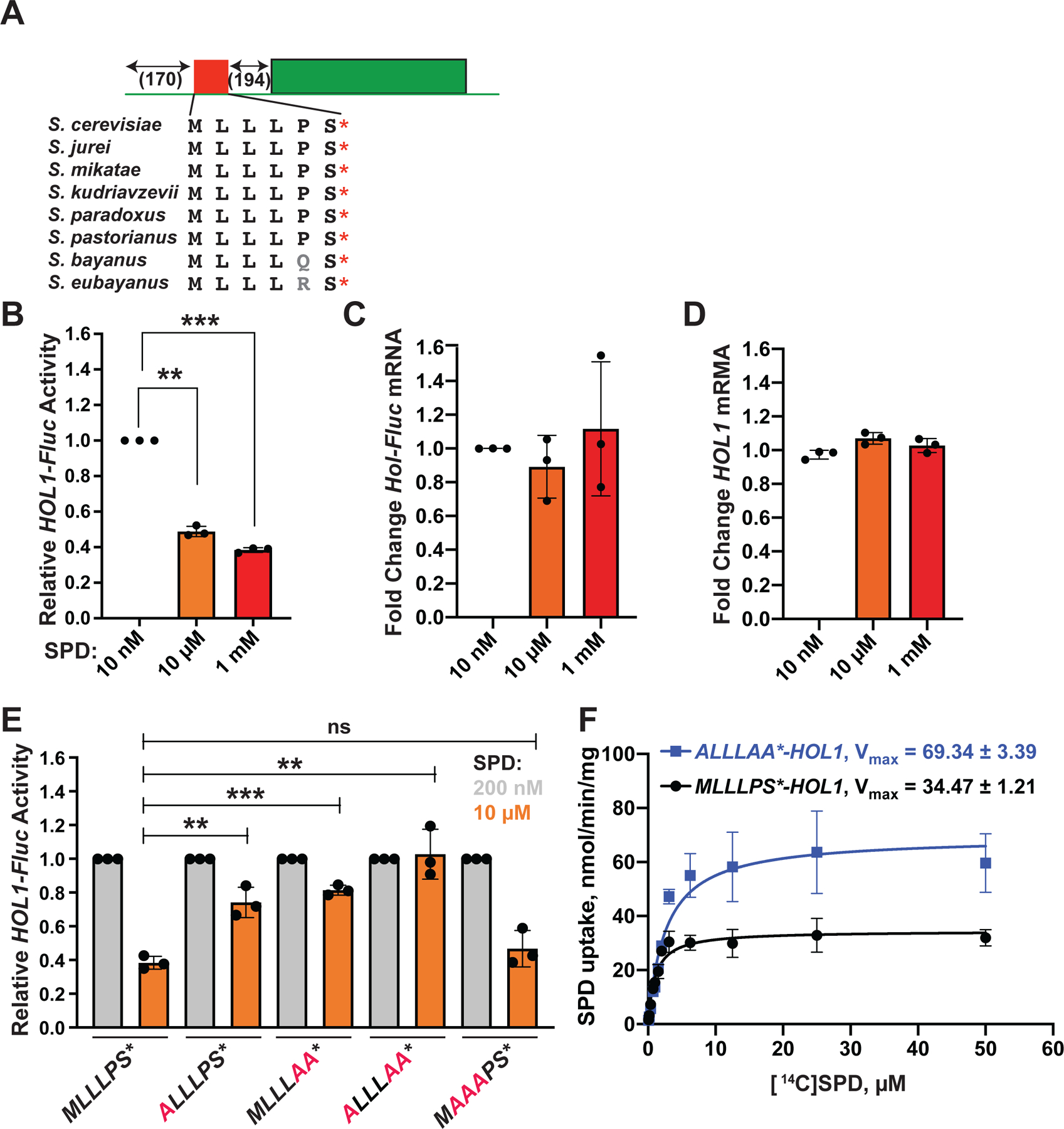
(A) Schematic representation of HOL1-Fluc reporter showing location of conserved uORF encoding MLLLPS peptide.
(B–E) Relative HOL1-Fluc expression (B,E), HOL1-Fluc mRNA levels (C), or endogenous HOL1 mRNA levels (D) in spe1 spe2 strain Y362 grown in polyamine-depleted SD medium in the presence of 10 nM (white), 200 nM (gray), 10 μM (orange), or 1 mM (red) additional SPD. In (E), reporters contained the indicated uORF mutations (red). mRNA levels were determined by RT-PCR and first normalized to actin mRNA; then, in each panel, measurements were normalized to samples containing 10 or 200 nM SPD (white or gray bars). Error bars denote SD; **p < 0.01, ***p < 0.001 (Student’s two-tailed t test; n = 3, assayed in duplicate).
(F) Rate of [14C]SPD uptake at the indicated SPD concentrations in spe1 spe2 hol1 strain J1570 expressing HOL1 with an intact (black, pC6464) or mutated (ALLLAA, blue, pC6465) uORF, as indicated, were fit to the Michaelis-Menten equation to calculate the Vmax (nmol/min/mg protein) for SPD uptake (n = 3).
To study polyamine control of HOL1 mRNA translation, the mORF of a plasmid borne HOL1 gene was replaced with the firefly luciferase coding sequence. In this HOL1-Fluc reporter (Figure 3A), the HOL1 promoter, 5’ leader and 3’ untranslated region are intact. Cells were first grown in low polyamine medium (10 nM SPD) to deplete free intracellular polyamines. The essential hypusine modification on translation factor eIF5A utilizes a 4-aminobutyl moiety from SPD (Park and Wolff, 2018) and is thus sensitive to changes in polyamine levels. At the low (10 nM) SPD concentrations used in these experiments, a normal cell growth rate is maintained even though the hypusine levels on eIF5A are substantially reduced (Figure S4A). These results are consistent with previous reports showing that low levels of hypusine are sufficient for yeast cell growth (Figure S4A, Chattopadhyay et al., 2008). Following polyamine depletion, cells were shifted to medium containing 10 nM, 10 μM, or 1 mM SPD and harvested during log-phase growth. As shown in Figure 3B, increasing the polyamine concentration inhibited HOL1-Fluc expression by up to 60%. Notably, these growth conditions had minimal impacts on the relative levels of the HOL1-Fluc and endogenous HOL1 mRNAs (Figures 3C–D), indicating that the HOL1-Fluc reporter recapitulates the polyamine control of HOL1 mRNA translation revealed in our ribosome profiling experiments (Figure 1A). Using a reversed protocol, nearly 5-fold derepression of HOL1-Fluc expression in low polyamines was observed by first growing cells in high (1 mM) SPD to repress HOL1 expression and then shifting to high (1 mM), medium (10 μM) or low (10 nM) levels of SPD (Figure S4B).
To assess whether the uORF in the HOL1 mRNA contributes to polyamine control of translation, mutations were introduced into the HOL1-Fluc reporter to change the AUG start codon to GCG (ALLLPS*), to change the PS* motif to AA* (MLLLAA*), and to combine both mutations (ALLLAA*). Reporters were introduced into the spe1 spe2 mutant strain and then grown in low (200 nM, where hypusine levels are normal, Figure S4A) or high (10 μM) SPD. As shown in Figure 3E, expression of the WT HOL1-Fluc reporter (MLLLPS*) was repressed by 2.5-fold in high polyamine medium. Mutating either the uORF start codon or the PS* motif partially derepressed HOL1-Fluc expression in high polyamines, while combining both mutations in the ALLLAA* mutant abolished repression of HOL1-Fluc expression by high polyamines. In contrast, mutation of the three Leu residues in the uORF to Ala resulted in regulation comparable to the WT uORF (Figure 3E). Thus, the start codon and two penultimate stop codon residues in the MLLLPS* uORF are necessary for polyamine control of HOL1 mRNA translation.
To assess the impact of the HOL1 uORF on polyamine uptake, the spe1 spe2 hol1 triple mutant strain was transformed with plasmids expressing HOL1 with an intact MLLLPS* uORF or the ALLLAA* mutant that fully derepresses HOL1 expression. Removal of the uORF increased the Vmax for polyamine uptake by ~2-fold (Figure 3F), consistent with the notion that removal of the uORF increases HOL1 expression resulting in increased rates of polyamine uptake. These results are consistent with the work of Gaber and colleagues (Gaber et al., 1990; Wright et al., 1996) showing that mutation of the uORF start codon increases Hol1 protein levels and enhances the histidinol and cation uptake activities associated with the HOL1-1 mutation.
MLLLPS* uORF is Sufficient to Confer Polyamine Regulation of Translation
To assess if the MLLLPS* uORF is sufficient for polyamine control of HOL1 translation, we took advantage of the S. cerevisiae CPA1 gene whose translation is controlled by an arginine-sensing uORF encoding the arginine attenuator peptide (AAP) (reviewed in Dever et al., 2020). Ribosomes translating the CPA1 uORF, or the analogous uORF on the N. crassa arg-2 mRNA, pause in the presence of excess arginine in a manner dependent on the amino acid sequence of the AAP (Fang et al., 2004; Wang and Sachs, 1997). Starting with a CPA1-Fluc reporter plasmid containing the CPA1 promoter and full 5’ leader (Figure 4A), the HOL1 MLLLPS* uORF was inserted in place of the CPA1 uORF leaving the rest of the CPA1 leader intact (HOL1/CPA1-Fluc, Figure 4A). In this chimeric construct, the uORF retains a poor start codon context with the upstream U−3AU nucleotides of the CPA1 uORF replacing the U−3UC context nucleotides of the native HOL1 uORF (Figure 5A). The WT HOL1-Fluc and CPA1-Fluc reporter constructs as well as the chimeric HOL1/CPA1-Fluc reporter were introduced into the spe1 spe2 mutant strain and grown in low polyamine medium or medium supplemented with 1 mM SPD or 1 mM Arg. As shown in Figure 4B, HOL1-Fluc expression was repressed by SPD but not Arg, and conversely expression of the CPA1-Fluc reporter was repressed by excess Arg but not SPD. Thus, the HOL1 and CPA1 reporters specifically respond to SPD and Arg, respectively.
Figure 4. The HOL1 uORF Confers Polyamine-specific Translational Control.
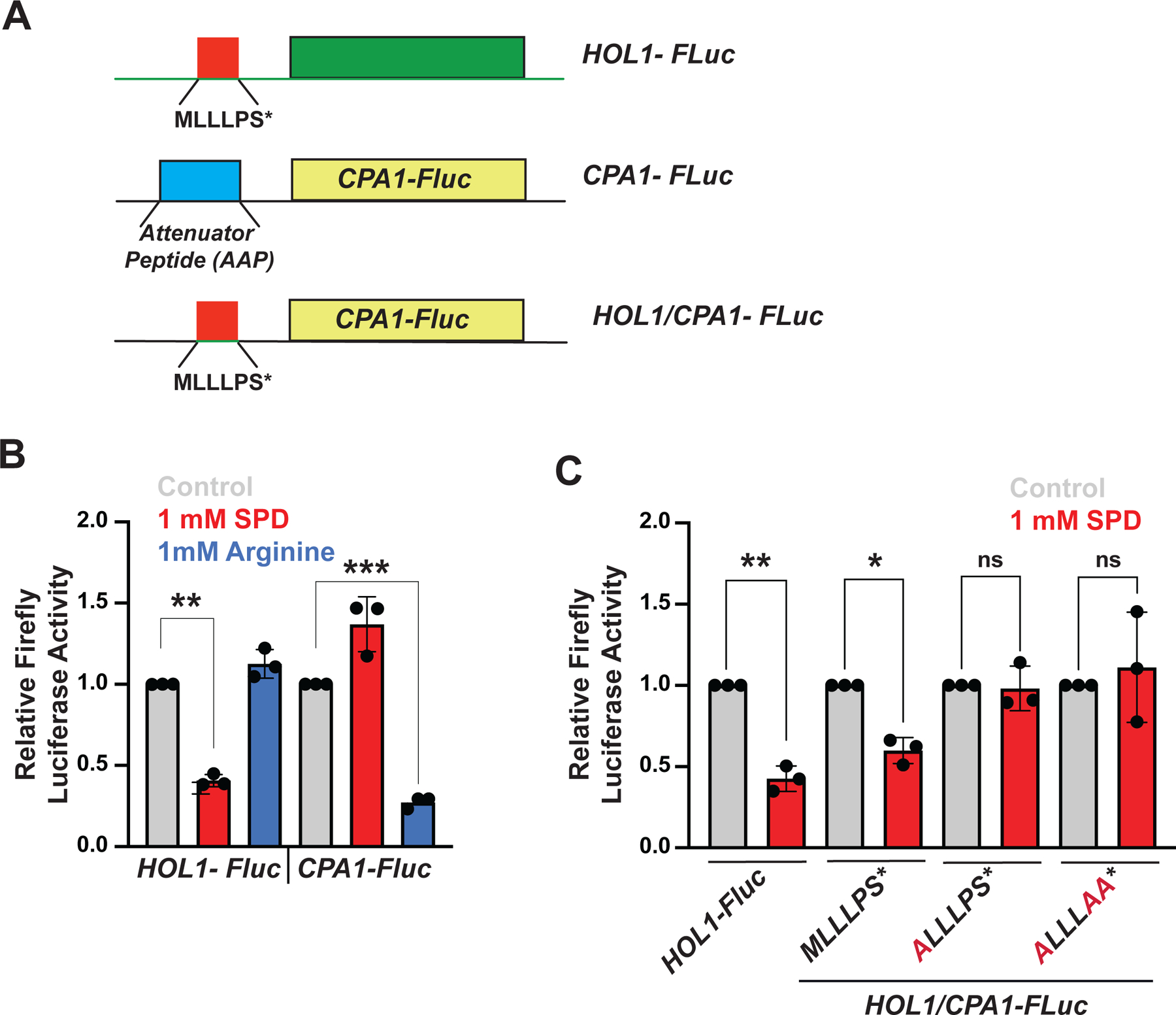
(A) Schematics of HOL1-Fluc, CPA1-Fluc and chimeric HOL1/CPA1-Fluc reporters. The CPA1 Arginine Attenuator Peptide (AAP) uORF is replaced by the HOL1 MLLLPS* uORF in the chimeric construct.
(B) Relative HOL1-Fluc and CPA1-Fluc expression in spe1 spe2 strain Y362 grown in polyamine-depleted SD medium in the presence of no additions (gray), 1 mM SPD (red) or 1 mM Arg (blue). Data were normalized to the no addition controls. Error bars denote SD; **p < 0.01, ***p<0.001 (Student’s two-tailed t test; n = 3, assayed in duplicate).
(C) Relative expression from HOL1-Fluc or HOL1/CPA1-Fluc reporters containing an intact or the indicated mutant uORF in spe1 spe2 strain Y362 grown in polyamine-depleted SD medium in the presence of no additions (gray) or 1 mM SPD (red). For each reporter, data were normalized to the no addition control. Error bars denote SD; *p < 0.05, **p<0.01 (Student’s two-tailed t test; n = 3, assayed in duplicate).
Figure 5. uORF Start Codon Context Sequence and Upstream Near Cognate Start Codons Contribute to Polyamine Control of HOL1 mRNA Translation.
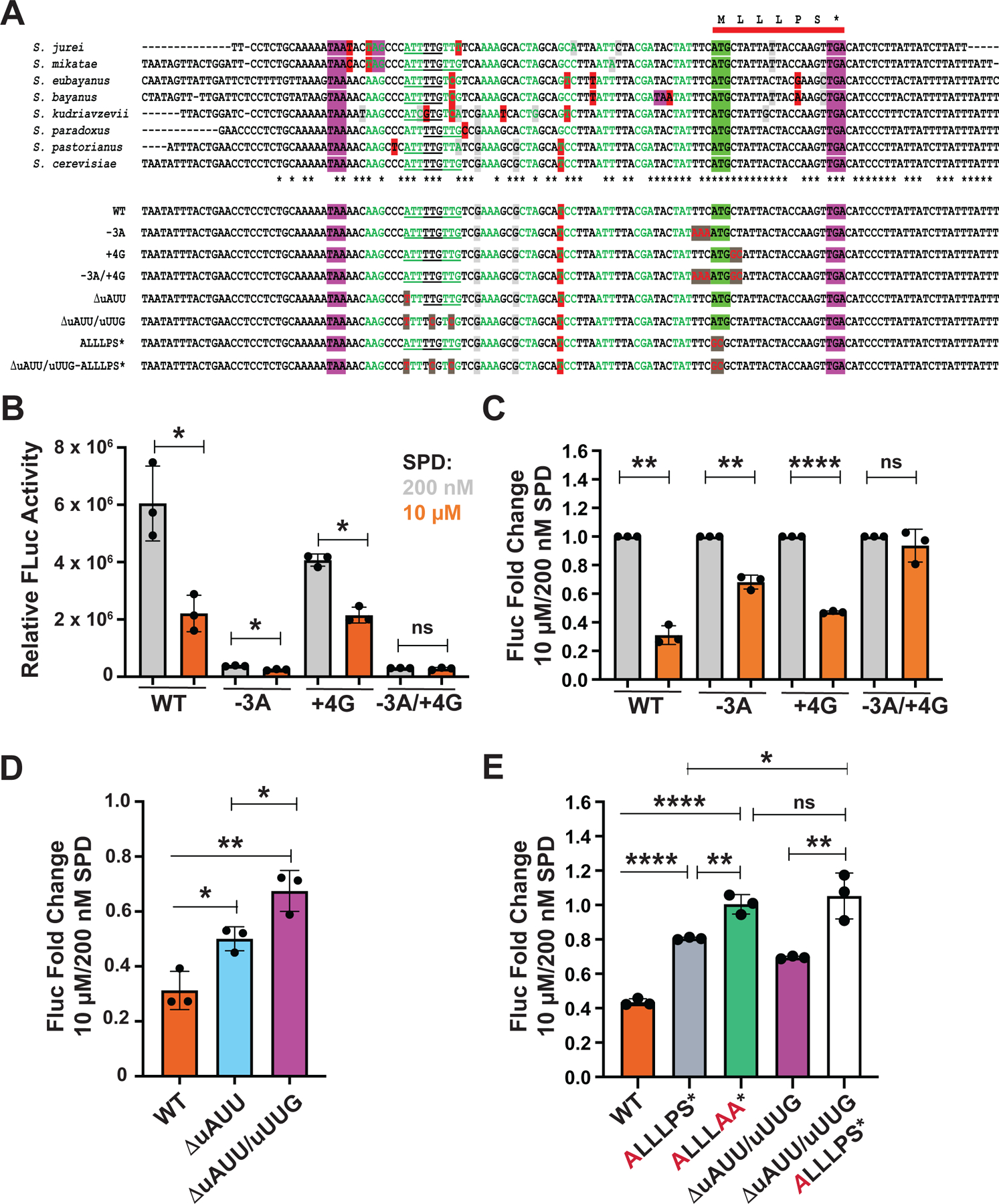
(A) (Upper panel) Multiple sequence alignment of a segment of HOL1 encoding the mRNA leader flanking the conserved uORF (red bar) from eight Saccharomyces species. The uORF start codon is highlighted in green and stop codons are highlighted in purple. The sequence upstream of the uORF is colored to highlight codons inframe with the uORF; nucleotide changes that alter the encoded amino acid are highlighted in red, while silent nucleotide changes that create synonymous codon substitutions are highlighted in light gray. Upstream inframe near cognate start codons are underlined; asterisks beneath alignment denote the positions that are conserved in all aligned sequences.
(Lower panel) Sequences of WT and mutant versions of S. cerevisiae HOL1 mRNA leader. Mutated residues are colored red and highlighted in dark gray.
(B) Relative HOL1-Fluc expression from the indicated WT or mutant reporters in spe1 spe2 strain Y362 grown in polyamine-depleted SD medium supplemented with 200 nM (gray) or 10 μM (orange) SPD. Data were normalized to total protein. Error bars denote SD; *p < 0.05 (Student’s two-tailed t test; n = 3, assayed in duplicate).
(C) Data from (B) normalized to the 200 nM SPD controls. Error bars denote SD; **p < 0.01; ****p < 0.0001; ns, not significant (Student’s two-tailed t test; n = 3).
(D–E) Fold change in relative HOL1-Fluc expression from the indicated WT or mutant reporters in spe1 spe2 strain Y362 grown in polyamine-depleted SD medium supplemented with 200 nM or 10 μM SPD. Data were normalized to total protein and then to each 200 nM SPD control. Error bars denote SD; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; ns, not significant (Student’s two-tailed t test; n = 3, assayed in duplicate).
Like the HOL1-Fluc reporter, expression from the HOL1/CPA1-Fluc reporter containing the WT MLLLPS* uORF was inhibited in the presence of excess SPD (Figure 4C, first four bars). Mutation of the uORF start codon (ALLLPS*) or the PS* motif (MLLLAA*) abolished polyamine regulation of HOL1/CPA1-Fluc expression (Figure 4C, last four bars). Thus, the HOL1 MLLLPS* uORF is sufficient to confer polyamine regulation on translation when present in a heterologous mRNA and can be added to the list of polyamine-responsive translational control elements.
Leaky Scanning and Upstream Near-cognate Initiation Contribute to Polyamine Regulation of HOL1 mRNA Translation
A key feature for translational control of the AZIN1 mRNA is the poor start codon of the uCC (Ivanov et al., 2008; Ivanov et al., 2018). Leaky scanning over the near-cognate start codon of the uCC is thought to enable translation of the mORF under low polyamine conditions. Whereas most AZIN1 and ODC uCCs initiate at a near cognate start codon, some uCCs start at an AUG codon in poor context that is also predicted to allow higher rates of leaky scanning. Multiple sequence alignment of the HOL1 5’ leaders from eight different Saccharomyces species (Figure 5A) revealed perfect conservation of the six nucleotides immediately upstream of the uORF start codon including the poor context U at position −3. To test whether the poor start codon context of the HOL1 uORF contributes to the regulation of HOL1 expression by polyamines, the non-preferred 5’ flanking sequence of the uORF (U−3UC) was changed to the preferred yeast sequence A−3AA (referred to as −3A, Figure 5A). Second, the +4 nucleotide, immediately downstream of the AUG start codon, was changed from C to G (+4G mutation, Figure 5A). Note that the +5 nucleotide was changed from U to C in this +4 nucleotide mutant (Figure 5A); this double mutation changes the second amino acid of the uORF from Leu to Ala, which as shown in Figure 3E does not affect polyamine regulation in an otherwise WT allele. In a third context mutant, the uORF start codon was placed in optimal context by introducing the preferred 5’ and 3’ flanking sequences (−3A/+4G).
Improving the 5’ context nucleotides dramatically reduced HOL1-Fluc expression under both low and high polyamine conditions (Figure 5B–C). This result is consistent with the idea that translation of the uORF prevents ribosomes from translating the mORF on the HOL1 mRNA. Thus, ribosomes typically access the mORF by leaky scanning past the uORF. In addition to reducing HOL1 expression in low SPD, the 5’ context mutations reduced regulation of HOL1 expression by high SPD. The 75% repression observed with the WT leader in high SPD was reduced to only 25% repression for the −3A mutant (Figure 5C). Mutating the +4 nucleotide modestly lowered HOL1-Fluc expression in low SPD but did not significantly impact expression in high SPD compared to the WT reporter (Figure 5B), thus polyamine regulation of HOL1-Fluc expression was maintained for the +4G mutant construct (Figure 5C). Finally, when the uORF start codon was placed in optimal context (−3A/+4G), HOL1-Fluc expression was strongly repressed under both low and high polyamine conditions (Figure 5B), resulting in loss of polyamine regulation (Figure 5C). Taken together, these data are consistent with the model that leaky scanning of the uORF under low polyamine conditions enables expression of HOL1, while ribosomes that translate the uORF fail to produce Hol1.
According to the uCC model for polyamine control of AZIN1 mRNA translation, pausing of elongating ribosomes translating the uCC triggers queuing of subsequent scanning ribosomes to promote initiation at an upstream weak translation start site (Ivanov et al., 2018). The observation that mutation of the start codon of the HOL1 uORF (ALLLPS*, Figure 3E) reduces, but does not eliminate, polyamine regulation, while mutation of the start codon and the final two residues of the uORF (ALLLAA*, Figure 3E) eliminates regulation, suggested that some ribosomes might translate the uORF by initiating at an upstream site. The multiple sequence alignment of the HOL1 5’ leaders (Figure 5A) revealed that the nucleotide sequence upstream of the uORF is generally conserved; however, the pattern of nucleotide changes indicate that the amino acid sequence of the encoded peptide (inframe with the uORF) is not conserved. Interestingly, with the exception of S. bayanus, the first upstream inframe stop codon (highlighted in purple in Figure 5A) is at least 51 nucleotides upstream of the uORF start codon, indicating that ribosomes could initiate at an upstream inframe near cognate start codon and translate the uORF. Of note, multiple inframe near cognate AUU/AUC and UUG start codons are present 16 codons upstream of the AUG codon of the uORF in the aligned Saccharomyces HOL1 sequences (Figure 5A, underlined nucleotides). To test whether ribosomes initiating from these upstream sites contribute to polyamine regulation of HOL1 expression and account for the residual polyamine regulation observed with the ALLLPS* HOL1-luc reporter (Figures 3E and 5E), the AUU codon was mutated to UUU in both the WT and ALLLPS* HOL1-luc reporters. As shown in Figure 5D, mutation of the AUU codon partially relaxed the repression of HOL1-luc expression in high polyamine conditions, and mutation of the AUU codon along with the adjacent UUG codons conferred an even greater, though still incomplete, derepression of HOL1-luc expression. These results are consistent with the idea that a subset of ribosomes initiate from these upstream near-cognate start sites, translate the extended uORF, and then fail to translate the HOL1 mORF. Introduction of these same mutations to eliminate all three upstream near-cognate start sites in the ALLLPS* reporter mimicked the ALLLAA* mutation and blocked repression of HOL1-luc expression in high polyamines (Figure 5E, 3rd versus 5th bar). This latter result is consistent with the notion that uORF translation initiating from these upstream near-cognate start codons accounts for the residual polyamine regulation when the uORF start codon is mutated. As mutation of these upstream near cognate start codons leads to partial derepression when the uORF is intact (Figure 5D), we propose that stalling of ribosomes that translate the uORF under high polyamine conditions triggers queuing of subsequent scanning ribosomes to promote initiation at the upstream near-cognate start codons. Accordingly, polyamine regulation of HOL1 mRNA translation follows, at least in part, the uCC mechanism that controls mammalian AZIN1 mRNA translation.
Polyamines Target eIF5A to Inhibit Termination on the MLLLPS* uORF and Control HOL1 mRNA Translation
As noted previously, the PS* motif in the HOL1 uORF is also found in the polyamine-responsive uCC elements in many vertebrate AZIN1 and some metazoan ODC mRNAs (Ivanov et al., 2008; Ivanov et al., 2018). The uCC elements also contain conserved PPW motifs, and we previously showed that translation of the PPW motif is highly dependent on translation factor eIF5A and that excess polyamines interfere with eIF5A function causing ribosomes to stall when translating the PPW motif (Ivanov et al., 2018). In addition to its critical role in synthesizing proline-rich motifs, eIF5A plays a global role in translation termination (Schuller et al., 2017). Based on these properties of eIF5A, we hypothesized that translation termination on the PS* motif is highly dependent on eIF5A and that polyamine inhibition of eIF5A impairs termination on this motif to control HOL1 mRNA translation. Using a reconstituted yeast in vitro translation assay system, peptide release triggered by termination factors eRF1 and eRF3 was monitored by electrophoretic thin-layer chromatography (TLC). As shown in Figure 6A, at 0.1 μM eIF5A, near the concentration (K1/2) required for half-maximal peptide synthesis (Ivanov et al., 2018), increasing the concentration of SPD from 1 mM to 4 mM inhibited both the rate and the maximal level of peptide release. At 4 mM SPD, the rate (kobs) of MPS peptide release was reduced to 15% of the rate observed at 1 mM SPD and the maximal level of peptide release was reduced by 50% (Figure 6B). While peptide release is generally dependent on eIF5A (Schuller et al., 2017), it is noteworthy that increasing the polyamine level was less inhibitory to release of an MFF peptide (Figures 6A–B). Importantly, increasing polyamine levels did not inhibit the yield of MPS or MFF peptide synthesized in assays that relied on base hydrolysis (KOH) rather than termination factors for peptide release (Figure S5A). Thus, the PS* motif is conferring a heightened sensitivity to polyamine inhibition of release factor- and eIF5A-dependent peptide release. Increasing the eIF5A concentration from 0.1 μM to 10 μM blocked the polyamine inhibition of peptide release (Figures 6A–B), indicating that polyamines competitively inhibit eIF5A.
Figure 6. Polyamine Inhibition of eIF5A Controls Translation Termination and HOL1-Fluc Expression.
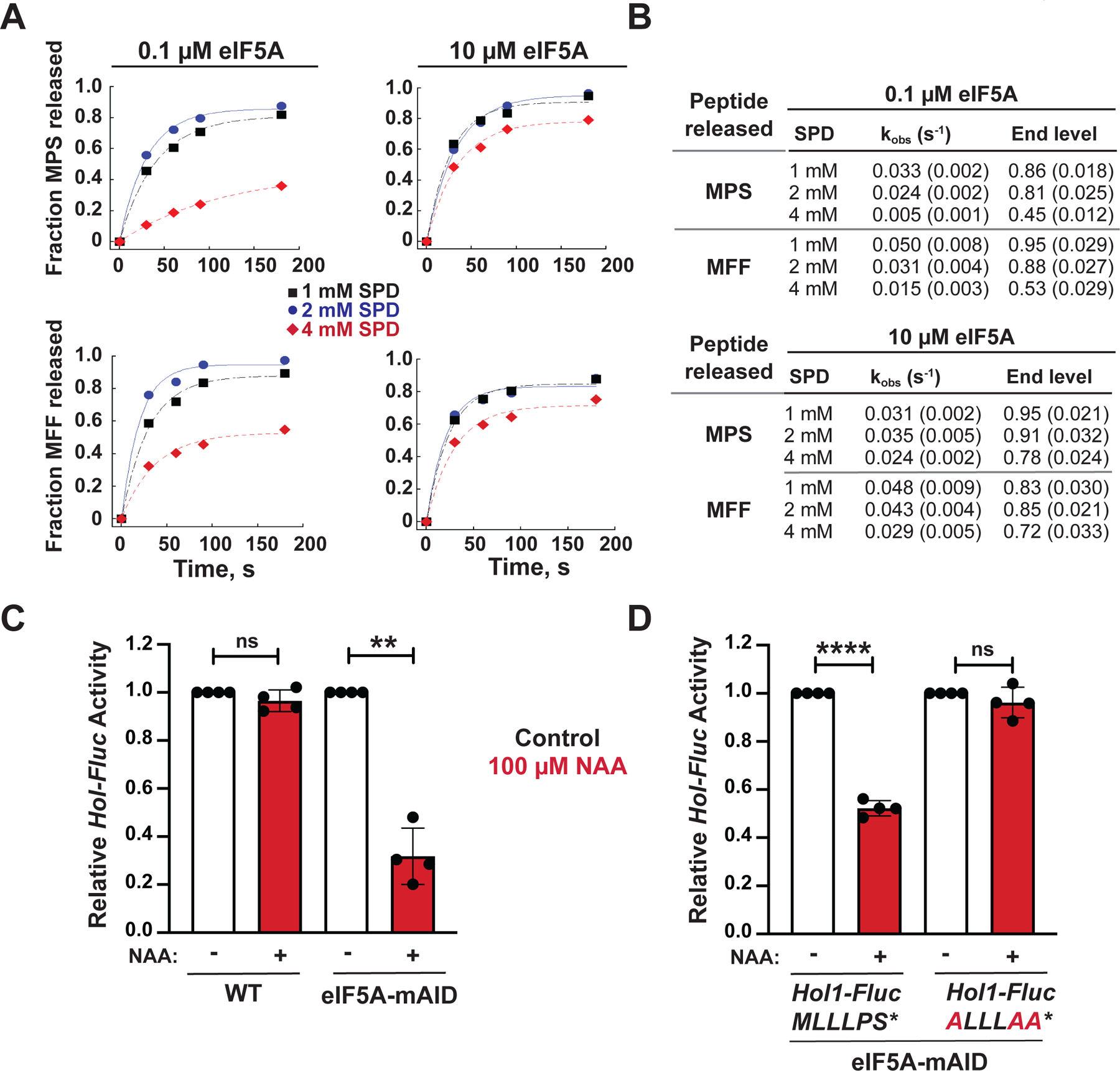
(A) Fractions of MPS* (upper panels) and MFF* (lower panels) peptide release out of total peptide formed in reconstituted translation termination assays performed in the presence of 1 (black squares), 2 (blue circles) or 4 (red diamonds) mM SPD and either 0.1 μM (left panels) or 10 μM (right panels) eIF5A were plotted and fit to a single exponential equation.
(B) Summary of observed rate constants (kobs) and maximum fractions (end levels) of peptide release calculated from the data in (A). Results are representative of three independent experiments; errors in rate constants and end levels are fitting errors from the curves in (A).
(C–D) Relative expression from HOL1-Fluc reporters containing an intact or mutant uORF, as indicated, in a WT strain expressing chromosomal eIF5A (HYP2 ANB1) or an isogenic anb1 hyp2 mutant strain expressing mini auxin-inducible degron-tagged eIF5A-mAID under control of the GAL1 promoter. Luciferase activities were measured from cells grown under control conditions in galactose medium (black) or following the switch to glucose medium and addition of 100 μM NAA to induce depletion of eIF5A-mAID. For each experiment data were normalized to the results obtained under control conditions. Error bars denote SD; **p < 0.01; ****p < 0.0001; ns, not significant (Student’s two-tailed t test; n = 3, assayed in duplicate).
Finally, to test whether eIF5A is involved in the control of HOL1 expression in vivo, the HOL1-Fluc reporter was introduced into isogenic strains expressing either WT eIF5A or eIF5A fused to a mini auxin-inducible degron (mAID-eIF5A) under the control of a galactose-inducible promoter (Schuller et al., 2017). Whereas mAID-eIF5A is expressed at levels comparable to WT eIF5A in medium containing galactose, mAID-eIF5A expression is repressed on glucose medium and the levels of mAID-eIF5A are reduced even further upon activation of the degron by addition of the synthetic auxin 1-naphthaleneacetic acid (NAA) (Figure S5B). Expression of the HOL1-Fluc reporter was high in WT and mAID-eIF5A cells grown in low polyamines (Figure 6C). Addition of NAA repressed HOL1-Fluc expression by 2.5-fold in the mAID-eIF5A strain but not in the strain expressing WT eIF5A (Figure 6C). Thus, depletion of eIF5A mimicked high polyamines and repressed HOL1 expression. Importantly, as was observed for polyamine repression, mutation of the MLLLPS* uORF sequence to ALLLAA* abolished NAA-triggered HOL1-Fluc repression in the mAID-eIF5A degron strain (Figure 6D). Taken together, these results support a model in which high polyamines inhibit eIF5A-dependent translation termination on the MLLLPS* uORF to repress synthesis of the polyamine transporter Hol1.
DISCUSSION
In this report we have identified Hol1 as a polyamine transporter and revealed that its expression is under translational feedback control by polyamines. We propose the following model (Figure 7) for the translational control of HOL1 expression. Under conditions of low polyamines (panel A), 43S ribosomal complexes consisting of the 40S subunit plus the eIF2–GTP–Met-tRNAiMet ternary complex and other translation factors binds near the cap of the mRNA and scans in search of a start codon. Some 43S complexes stop scanning at the start codon of the MLLLPS uORF. These ribosomes translate the uORF and then either disengage from the mRNA or resume scanning in order to reinitiate at the downstream start codon for the HOL1 mORF. Because the start codon of the uORF is in an imperfect context, some scanning ribosomes leaky scan past the uORF start codon without initiating translation and then translate the HOL1 mORF. Thus, under low polyamine conditions, Hol1 synthesis is high.
Figure 7. Schematic Model of Polyamine Autoregulation of HOL1 mRNA Translation.
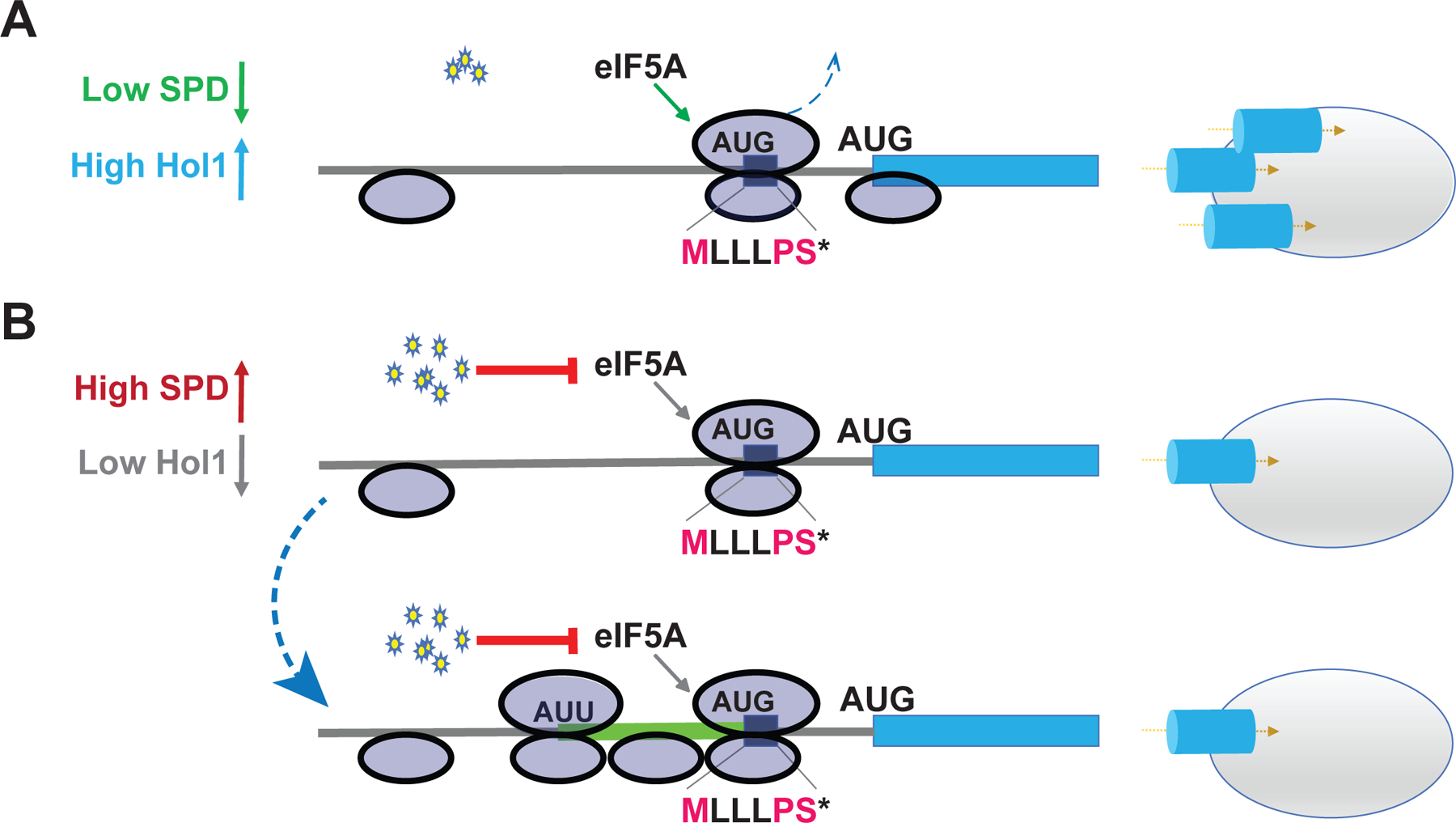
(A) Under low polyamine conditions, many scanning ribosomes skip over the uORF without translating (leaky scan). Some ribosomes translate the uORF, efficiently terminate with the aid of eIF5A, and then resume scanning. Both sets of ribosomes then efficiently initiate translation at the HOL1 start codon to increase expression of the transporter. (B) Under conditions of high polyamines, any ribosome that translates the uORF pauses at the stop codon because the high polyamines interfere with eIF5A activity and impair release of the uORF peptide ending in the conserved PS* motif. The termination pause decreases the likelihood that the ribosome will resume scanning following termination. Moreover (lower panel), the paused terminating ribosome serves as a roadblock, preventing other ribosomes from leaky scanning past the uORF. The subsequent queuing of scanning ribosomes behind the paused ribosome results in scanning ribosomes spending a longer time over the upstream near cognate start codons in the HOL1 leader, enabling increased translation of an extended uORF (green). The increased uORF translation, decreased leaky scanning over the uORF, and decreased reinitiation following uORF translation in high polyamine conditions decreases the number of ribosomes that translate the HOL1 mORF, resulting in reduced levels of the polyamine transporter.
Under conditions of elevated polyamines (Figure 7B), ribosomes translating the uORF pause at termination. As termination on the PS* motif is highly dependent on eIF5A, and polyamines competitively block eIF5A function, termination on the uORF is impaired. We propose that the extended pause at termination will have multiple effects leading to reduced Hol1 synthesis. First (Figure 7B, upper panel), the paused ribosome will be an impediment to subsequent scanning ribosomes and will thus prevent ribosomes from leaky scanning to the HOL1 mORF start codon. Second (Figure 7B, upper panel), the extended duration of the pause will decrease the likelihood of reinitiation following termination at the uORF stop codon. The probability of reinitiation following translation of a uORF is inversely correlated with the length of the uORF (Kozak, 2001; Luukkonen et al., 1995; Rajkowitsch et al., 2004) and this is thought to be related to the time-dependent release of some initiation factors following translation initiation. The slowed termination due to polyamine inhibition of eIF5A will cause the short uORF to mimic a longer uORF, allowing greater time for dissociation of factors like eIF3 (Bohlen et al., 2020; Mohammad et al., 2017; Wagner et al., 2020) and increasing the likelihood that the ribosome dissociates from the mRNA following termination at the uORF stop codon. Third (Figure 7B, lower panel), queuing of translating and scanning ribosomes upstream of the paused terminating ribosome will cause a scanning ribosome to spend a longer time over upstream near-cognate start codons providing greater opportunities for initiation at these sub-optimal sites. Consistent with this model, analysis of published disome (collided 80S ribosomes) and 40S ribosome profiling studies (Archer et al., 2016; Meydan and Guydosh, 2020) revealed the presence of queued ribosomes on the HOL1 mRNA leader (Figure S6).
Our model of HOL1 regulation resembles the polyamine-controlled translation of the uCC in the mammalian AZIN1 mRNA (Ivanov et al., 2018). In AZIN1, an elongation stall and queuing of subsequent scanning ribosomes enhances initiation at the upstream near-cognate start codon of the uCC leading to increased uCC translation and repression of AZIN1 mORF translation. We propose that translation of the HOL1 uORF inhibits mORF translation and acts as a seed to promote initiation at the upstream near-cognate start sites via ribosome queuing. Translation initiating from these upstream sites contributes to the repressive effects of polyamines on HOL1 expression. Consistent with this idea, mutation of the upstream near-cognate start sites partially derepressed expression of HOL1-luc containing an intact uORF and lead to full derepression when the uORF start codon was mutated (Figure 5D–E).
Our study adds another mRNA to the translationally controlled polyamine regulon. Cellular polyamine homeostasis is under tight control with key regulation occurring at the level of translation (reviewed in Dever and Ivanov, 2018). The polyamine-induced +1 ribosomal frameshift required for OAZ synthesis is conserved in eukaryotes from yeast to humans. Translation of the mammalian AZIN1 mRNA and some fungal and invertebrate ODC mRNAs is repressed under high polyamine conditions via polyamine-triggered ribosome queuing which, as described above, enhances translation of the repressive uCC element in the AZIN1 mRNA 5’ leader. This inverse regulation of OAZ1 and AZIN1 mRNA translation by polyamines is consistent with the opposing roles of their encoded proteins on control of ODC and polyamine synthesis. Polyamines also repress the translation of the mRNA encoding mammalian AdoMetDC. Under high polyamine conditions, stalling of a ribosome translating the short (MAGDIS) uORF located near the 5’ end of the AdoMetDC mRNA blocks other ribosomes from binding the mRNA and thereby inhibits AdoMetDC synthesis. We discovered that translation of the yeast HOL1 mRNA is also repressed by polyamines. The translational control of HOL1 shares the feature of ribosome stalling during translation elongation or termination with the AZIN1 and AdoMetDC mRNAs, and like the AZIN1 mRNA ribosome queuing promotes initiation at an upstream weak start site to potentiate the repression. The regulation of HOL1 synthesis by polyamines adds an additional mRNA to the polyamine translational regulon and extends the regulon from polyamine synthesis to polyamine transport. Moreover, the uORF-based mechanism controlling HOL1 mRNA translation further establishes the notion that uORFs have been selected during evolution for regulation of polyamine-related genes (Ivanov et al., 2010a).
The central role of eIF5A in translational control of the AZIN1 and HOL1 mRNAs reveals parallels to recent reports on regulation of translation start site selection by eIF5A in yeast and mammalian cells. We previously showed that polyamines inhibit the function of eIF5A in elongation on a polyproline (PPW) motif in the AZIN1 uCC (Ivanov et al., 2018) and here we show that polyamines block the eIF5A function in termination on the PS* motif of the HOL1 uORF. In both cases the paused ribosome triggers queuing of subsequent scanning ribosomes to promote initiation at upstream near-cognate start codons. Manjunath et al (Manjunath et al., 2019), working in mammalian cells, and Eisenberg et al (Eisenberg et al., 2020), working in yeast, found that knockdown of eIF5A or reduced levels of the factor during yeast meiosis, respectively, increased the levels of N-terminally extended protein products initiating from sub-optimal start codons. We propose that translation of these N-terminally extended products, like the extended uORF translation on the HOL1 mRNA and the uCC translation on the AZIN1 mRNA, is enabled by queuing of scanning ribosomes upstream of a ribosome paused at an eIF5A-dependent motif like polyproline. Based on the polyamine inhibition of eIF5A function, it will of interest to test whether elevated polyamines mimic downregulation of eIF5A levels and enhance translation of N-terminally extended proteins initiating at weak start sites.
HOL1 was originally identified based on gain-of-function mutations that enhanced uptake of histidinol as well as several cations including calcium and sodium (Gaber et al., 1990). Notably, these transport properties of HOL1 were dependent on the mutations in the gene and the transport function of the WT protein was not clear. Here, we demonstrate that Hol1 is the high affinity polyamine transporter in yeast. A question arises as to why Hol1 was not identified previously, and we propose that the polyamine regulation of HOL1 expression may have confounded its discovery. In typical yeast medium that contains some polyamines, HOL1 expression is at least partially repressed. Under these conditions, polyamine uptake is likely mediated by a mixture of limiting amounts of Hol1 plus various low affinity transporters. Upon switching to media depleted of polyamines, where HOL1 expression is induced, the role of HOL1 in polyamine uptake is readily observed.
The Hol1 protein belongs to the multifacilitator superfamily (MFS) of transport proteins and more precisely to the DHA1 family of transporters. The protein is predicted to contain twelve membrane-spanning domains and substrate transport is predicted to be driven by the proton-motive force. Consistent with this idea, we found that SPD transport by Hol1 in the proteoliposome assays (Figure 2G) was sensitive to pH. Phylogenetic analyses have revealed that orthologs of Hol1 are restricted to fungi (Figure S2B; see also the Transporter Classification Database [http://www.tcdb.org/]) and, interestingly, the MLLLPS* uORF is conserved in only a subset of the fungal HOL1 mRNAs (Figure S2B). Of note, all currently sequenced members of the Saccharomyces genus contain the HOL1 uORF, suggesting that it was already present in their last common ancestor (Figure S2B). The lack of a Hol1 homolog in mammals is consistent with previous reports linking the solute carriers SLC3A2 (Gamble et al., 2019; Uemura et al., 2010) and SLC18B1 (Hiasa et al., 2014), as well as the ATPases ATP13A2 (van Veen et al., 2020) and ATP13A3 (Hamouda et al., 2020), with polyamine transport in mammals. While SLC3A2 localizes to the plasma membrane like Hol1, the other transporters are proposed to act via endocytic pathways. As yet, polyamine control of mammalian polyamine transporter expression has not been described, and it will be interesting to learn whether the polyamine translational regulon in mammals, like that in yeast, extends beyond regulation of polyamine synthesis to regulation of transport.
Limitations of the Study
Although we showed that Hol1 is the high-affinity polyamine transporter in yeast and that Sam3, Dur3 and Agp2 do not contribute to high affinity polyamine uptake, these latter transporters as well as other transporters like Gap1 could contribute to the background low-affinity polyamine uptake in the cells lacking Hol1. In addition, while previous studies reported gain-of-function mutations in HOL1 that conferred enhanced uptake of histidinol, Na+, or Ca2+, we do not know if Hol1 plays a critical role in transporting substrates other than polyamines.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Please direct any requests for further information or reagents to the Lead Contact, Thomas E. Dever (thomas.dever@nih.gov).
Materials availability
The plasmids and yeast strains generated from this study will be available upon request. All reagents including antibodies and chemicals can be found in the key resources table.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Polyclonal Rabbit anti-GCD6 | (Bushman et al., 1993) | AB#34 |
| Polyclonal Rabbit anti-yeast eIF5A | (Saini et al., 2009) | HL5823 |
| Polyclonal Rabbit anti-hypusine | (Nishiki et al., 2013) | IU88 |
| Chemicals, peptides, and recombinant proteins | ||
| Spermidine | Sigma-Aldrich | S2626 |
| Spermidine [1,4-14C] trihydrochloride | American Radiolabeled Chemicals | ARC-3138 |
| Adenosyl-L-methionine, S-[methyl-14C] | American Radiolabeled Chemicals | ARC-0344 |
| L-Arginine monohydrochloride | Sigma-Aldrich | A5131 |
| Bradford protein assay dye reagent | Bio-Rad | 500-0006 |
| Cycloheximide | Sigma-Aldrich | C-7698 |
| D-Pantothenic acid hemicalcium salt | Sigma-Aldrich | 21210 |
| 1-Naphthaleneacetic acid (NAA) | Sigma-Aldrich | N0640 |
| Yeast nitrogen base w/o amino acids, w/o ammonium sulfate, w/o calcium pantothenate | Formedium | CYN3402 |
| Yeast nitrogen base w/o amino acids, w/o ammonium sulfate | BD Difco | 233520 |
| D-(+)-Dextrose, anhydrous | MP Biomedicals | 901521 |
| Ammonium sulphate | Macron Fine Chem. | 3512-12 |
| Whatman glass microfiber filters, Grade GF/C | Millipore Sigma | 1822-024 |
| Membrane Filter, 0.22 μm pore size | Millipore Sigma | GSWP02500 |
| Agar, yeast culture grade | Sunrise Science | 1910 |
| Critical commercial assays | ||
| Firefly Luciferase Assay System | Promega | E1501 |
| Dual Luciferase Reporter Assay System | Promega | E1980 |
| QuikChange Lightning Site-Directed Mutagenesis Kit | Agilent | 210518 |
| Ribo-Zero Gold rRNA (Yeast) | Illumina | MRZY1306 |
| SuperScript III First-Strand Synthesis SuperMix for qRT-PCR | Thermo Fisher | 11752050 |
| Brilliant II SYBR® Green QPCR Master Mix | Agilent | 600828 |
| Deposited data | ||
| Raw and Analyzed data | This paper | GEO: GSE171392 |
| R64-1-1 S288C Sac cer3 Genome Assembly | Saccharomyces Genome Database Project | http://sgd-archive.yeastgenome.org/sequence/S288C_reference/genome_releases/ |
| Yeast ncRNA Gene Database | Saccharomyces Genome Database Project | http://sgd-archive.yeastgenome.org/sequence/S288C_reference/rna/archive/rna_coding_R64-1-1_20110203.fasta.gz |
| Mendeley Database | This paper | https://doi.org/10.17632/3wg5js2n8j.1 |
| Experimental models: organisms/strains | ||
| S. cerevisiae: BY4741 MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Horizon Discovery | YSC1048 |
| S. cerevisiae: MATa ura3-52 leu2 spelΔ::LEU2 spe2Δ::LEU2 | (Balasundaram et al., 1994) | Y362 |
| S. cerevisiae: MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 AHD1p-OsTIR1-URA3 | (Schuller et al., 2017) | yCW30 - WT |
| S. cerevisiae: MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 Δanb1 Δhyp2 mAID-HYP2-pAG413GAL AHD1p-OsTIR1-URA3 | (Schuller et al., 2017) | yCW33 - eIF5Ad |
| S. cerevisiae: MATa ura3-52 leu2 spe1Δ::LEU2 spe2Δ::LEU2 hol1::KanMX4 | This study | J1570 |
| S. cerevisiae: MATa ura3-52 leu2 spe1Δ::LEU2 spe2Δ::LEU2 dur3Δ::kanMX4 | This study | J1525 |
| S. cerevisiae: MATa ura3-52 leu2 spe1Δ::LEU2 spe2Δ::LEU2 sam3Δ::kanMX4 | This study | J1527 |
| S. cerevisiae: MATa ura3-52 leu2 spe1Δ::LEU2 spe2Δ::LEU2 agp2Δ::kanMX4 | This study | J1529 |
| S. cerevisiae: MATa ura3-52 leu2 spe1Δ::LEU2 spe2Δ::LEU2 HOL1-GFP(S65T)-pFA6a-KanMX6 | This study | J1609 |
| Oligonucleotides | ||
| Please refer to Table S1 for oligonucleotides used in this study | This paper | N/A |
| Software and algorithms | ||
| Bowtie-1.2.2 | (Langmead et al., 2009) | http://bowtie-bio.sourceforge.net/index.shtml |
| Hisat2-2.1.0 | (Kim et al., 2015) | https://cloud.biohpc.swmed.edu/index.php/s/hisat2-210-OSX_x86_64/download |
| Cutadapt | (Martin, 2011) | https://cutadapt.readthedocs.io/en/stable/ |
| edgeR Bioconductor Package | (McCarthy et al., 2012; Robinson et al., 2010) | https://www.bioconductor.org |
| Gene Ontology Term Finder | Saccharomyces Genome Database Project | https://www.yeastgenome.org/goTermFinder |
| ImageJ | (Schneider et al., 2012) | https://imagej.nih.gov/ij/index.html |
| GraphPad Prism | GraphPad Software, Inc. | 8.4.3 (471) |
Data and code availability
Sequencing data were deposited in the GEO database under the accession number GSE171392. Gel images were deposited in the Mendeley Database link is: https://doi.org/10.17632/3wg5js2n8j.1
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Yeast strains used in this study and their genotypes are listed in the Key Resource Table.
To delete HOL1 in the strain Y362, the primers HOL1-F (5’-GACGTATTCGCATGATAGTATATACCAAAGAAGAACG-3’) and HOL1-R (5’-CCAGAATCTCAGAAGGGAGAAACCCAATCGC), complementary to sequences 1-kb upstream and 500-bp downstream, respectively, of the HOL1 CDS, were used to PCR amplify the hol1Δ::KANMX allele using genomic DNA obtained from yeast deletion collection hol1Δ strain. The PCR product was then used to transform strain Y362 and transformants were selected on YPD plates supplemented with 1mM SPD. Replacement of HOL1 by hol1Δ::KANMX was confirmed by PCR. The dur3Δ::KANMX, sam3Δ::KANMX, agp2Δ::KANMX deletion strains were generated using a similar strategy. The primers DUR3-F (5’-GAGATTAGCCAAGACCAAAGGTTCTCTTAAAGATG-3’) and DUR3-R (5’-CATACGTCGGTATGGACACTGTTGACATGATAG), SAM3-F (5’-CAAGCTATATTCGCATCACACAGAATGATGTGGCGTCTATG-3’) and SAM3-R (5’ GTTGCCGCACCGCCTGTTGCTTAACAATGCGAGCGCCAGGC-3’), AGP2-F (5’-AACTCTTCAATGGTAAATGGATCTCGACATGTTGCCTGCT) and AGP2-R (5’-TTATGGGACTTACAGGTTGACGTCTTACAGTTCTTGACTAGT), designed to anneal ~1-kb upstream and ~0.5-kb downstream of the respective genes, were used to PCR amplify the KANMX-tagged allele from the yeast deletion collection mutants.
In addition to PCR analyses, the deletions of DUR3 and SAM3 were confirmed by phenotypic analyses. Loss of DUR3 blocked cell growth on medium containing 2 mM urea as the sole nitrogen source (Figure S3C), and loss of SAM3 blocked uptake of [14C]SAM (Figure S3D).
For the construction of the C-terminal GFP-tagged HOL1 in strain Y362, the GFP KANMX cassette in the plasmid pFA6a-GFP(S65T)-kanMX6 (Longtine et al., 1998) was amplified by PCR using the HOL1-specific (underlined residues) primers GFP-F (5’-GAGATGGTATTTGCAATCTGTCAATTTGAGAGACGGTGTGCGGATCCCCGGGTTAAT TAA-3’) and GFP-R (5’-ATTCCTATCATCTACTAGATACAAGAAAGACGTCTCTGAAGAATTCGAGCTCGTTTA AAC-3’). The PCR product was used to transform strain Y362 and generate the HOL1-GFP allele.
For all experiments, unless specified, synthetic dextrose (SD) medium was prepared using yeast nitrogen base lacking pantothenic acid (Formedium, Norfolk, UK). Based on growth assays of polyamine auxotrophic strains, this YNB appears to contain lower levels of polyamines than other sources of YNB. As polyamines are required for biosynthesis of pantothenic acid, the medium was supplemented with 100 nM pantothenic acid. For experiments examining arginine repression of CPA1-luc expression or the urea-dependent growth of the ure2 mutant strain, YNB (Gibco), which apparently contains low levels of polyamines, was used to prepare SD medium.
METHOD DETAILS
SPD uptake assay
BY4741, isogenic derivatives obtained from the genome deletion collection (Giaever et al., 2002), and derivatives of strain Y362 were used for SPD uptake assays. PCR assays were used to confirm loss of the WT locus and insertion of the KANMX cassette in the strains from the yeast deletion collection and in the hol1 mutants derived from strain Y362. To measure SPD uptake, strains were grown in SD medium (supplemented with essential nutrients) at 30° C for 16 h, washed thrice in the same medium, and then used to inoculate a culture at OD600 = 0.20. The culture was then incubated for ~6 h until OD600 = 0.80–1.0. Cells were washed thrice in Uptake Buffer (50 mM sodium citrate pH 5.5 with 2% dextrose), and then OD600 = 1.0 units of cells were suspended in Uptake Buffer and incubated with 60 nM–1 mM [14C]SPD at 30° C for various times. The uptake reactions were stopped by adding 1 ml of ice-cold Uptake Buffer and transferring the reaction tubes quickly to an ice bath. Reaction mixtures were subjected to vacuum filtration on glass wool filter discs followed by washing cells with 1 ml ice-cold Uptake Buffer supplemented with 1 mM unlabeled SPD. Filter discs were transferred to scintillation vials, mixed with 5 mL Ecoscint A scintillation counting cocktail (National Diagnostics), and then counted in a liquid scintillation counter. Proteoliposome uptake assays were carried out using the same method except that the Uptake Buffer contained 50 mM sodium citrate pH 3.0 and 0.22 μm filters (Millipore) were used.
Hol1 expression and purification
Kluveromyces lactis Hol1 was heterologously expressed in Pichia pastoris as a carboxy-terminal eGFP and decahistidine fusion protein, separated by a 3C protease site for proteolytic tag removal. Protein purification was carried out similar to published protocols (Rana et al., 2018) except lauryl maltose neopentyl glycol (LMNG) and cholesteryl hemisuccinate (CHS) were used for extraction followed by subsequent purification in glyco-diosgenin (GDN), CHS and 1:1 1-palmitoyl-2-oleoylphosphatidylcholine (POPC) : yeast polar lipid extract ([w:w]; Avanti Polar Lipids) during TALON affinity and size-exclusion chromatography steps.
Proteoliposome preparation
Liposome reconstitutions in the presence or absence of detergent solubilized Hol1 were done using hydrophobic chromatography as previously described (Christenson et al., 2018). Protein-free liposomes were prepared using identical methods, except replacing the same volume of protein with the buffer in the final step of protein purification. 2:1 yeast polar lipid extract : POPC (by mass) dissolved in chloroform (25 mg/mL) was dried to a thin film. Lipid films were resuspended in 20 mM MOPS-KOH pH 7.4 to a final concentration of 20 mg/mL and subsequently sonicated to form large unilamellar vesicles (LUVs). KCl was added to a final concentration of 140 mM. LUVs were destabilized by ten consecutive additions of 10% Triton X-100 (dissolved in inside buffer, 20 mM MOPS-KOH, pH 7.4, 140 mM KCl) for a final lipid : detergent ratio of 10:6 by mass. To the destabilized LUVs was added 100 μg freshly purified Hol1 per 10 mg lipid (or the same volume of buffer for protein-free liposomes), and the reconstitution was brought to a total volume of 700 μl with inside buffer. After rocking the suspension at 4°C for 30 min, it was passed 15 times over a bed of 550 mg activated Amberlite XAD-2 (pre-equilibrated with ten bed volumes inside buffer) to adsorb the detergent. Reconstituted liposomes were extruded using a 400 nm filter prior to use in uptake assays.
Luciferase Reporter Assays
Individual yeast transformants were grown in SD medium containing 10 nM SPD and 100 nM pantothenic acid at 30° C for 24 hours, washed thrice in SD medium, and then used to inoculate cultures at OD600 = 0.025 in SD containing low (100 nM) or high (10 μM or 1mM) levels of SPD. Cells were grown to OD600 = 0.8–1.0, harvested, and the cell pellets were resuspended in 400 μl ice cold Breaking Buffer (1X PBS + 1mM PMSF + Complete Protease Inhibitor cocktail [Roche]), and vigorously mixed with 1 vol glass beads on a vortex for 8 min at 4° C. Lysates were cleared by centrifugation at 9,400 × g for 10 min at 4° C, and aliquots of the supernatant were assayed for firefly luciferase activity using a microplate luminometer (Berthold) and the Firefly Luciferase Reporter E1501 Assay System (Promega). Firefly luciferase readings were normalized to total protein, as determined by Bradford assays (Biorad).
For assays in the eIF5A degron strain, HOL1-Fluc transformants were first selected on synthetic complete galactose (SCGal) medium containing all amino acids plus 2% galactose and 2% raffinose. This medium was prepared using Gibco YNB. Transformants were grown in liquid SCGal-Leu medium at 30° C for 16 h. Cells were pelleted, washed thrice with SC-Leu (2% Glucose) medium, and then inoculated in the same medium at OD600 = 0.40 with and without 100 μM 1-Naphthaleneacetic acid (NAA). Cultures were grown to OD600 = 0.8–1.0 and then processed for measurement of firefly luciferase activity as described above.
Yeast growth spotting assay
Yeast strains were grown to saturation in SD medium with essential supplements, washed thrice in the same medium, and then inoculated at OD600 = 0.2. Following growth at 30° C for 6 h to OD600 = 0.8–1.0, 5-μl volumes of serial dilutions (OD600 = 1.0, 0.1, 0.01, 0.001, and 0.0001) were spotted on SD plates supplemented with various concentrations of polyamines and then incubated for 2 days at 30° C.
Imaging HOL1-GFP localization
The HOL1-GFP strain J1609 and the isogenic untagged WT HOL1 strain Y362 were grown in SD medium containing 10 nM SPD for 16 hr at 30° C, washed thrice in same medium, and then inoculated at OD600 = 0.20. Following further growth for 6 h to OD600 = 0.8 −1.0, cells in a 1-ml aliquot were pelleted and then resuspended in 100 μl filtered 1X PBS. 10 μl cell suspension was mounted on a 2% melted agarose patch on a clear glass slide and covered by a coverslip. Cells were visualized with the kind assistance of Latika Mathai and Orna Cohen-Fix (NIDDK, Bethesda, MD) using their Nikon Eclipse TE2000U spinning disc confocal microscope with IPLab4.0.8 (BioVision Technologies) software. The microscope was equipped with 100× 1.4 NA Apo objective, an LMM5 laser merge module with four diode lasers (excitation at 405, 491, 561 and 565 nM) from Applied research, a Yokogawa CSU10 spinning disc, and a Hamamatsu C9100-13 EM-CCD camera. Cells were visualized under GFP and bright field and images were processed using ImageJ software (National Institutes of Health).
Ribosome Profiling
The spe1 spe2 deletion strain Y362 transformed with the empty URA3 vector YCplac33 was grown in SD medium (10 nM SPD + 100 nM pantothenic acid) for 24 hr at 30° C and then washed thrice in the same medium. Cultures were inoculated at OD600 = 0.05 in SD medium containing 100 nM pantothenic acid and supplemented with zero or 10 μM SPD for low or high SPD conditions, respectively. Cells were harvested at OD600 = 0.60 after 16 h for low SPD and after 18 h for high SPD. Ribosome profiling methods were followed as described by McGlincy (McGlincy and Ingolia, 2017) with the following modifications. Cells were rapidly collected by filtration, flash frozen in liquid nitrogen, resuspended in Lysis Buffer (20 mM Tris pH 7.40, 150 mM NaCl, 5 mM MgCl2, 1mM DTT, 100 ug/ml cycloheximide, 1% v/v Triton X-100 and 25 U/ml Turbo DNase I), and then lysed using a freezer mill. The lysate was cleared by centrifugation at 3000 × g for 5 min at 4° C, then the supernatant was collected and subjected to centrifugation at 20,000 × g for 10 min at 4° C. The cleared lysate was collected, flash frozen in liquid nitrogen and stored at −80° C.
For Ribosome-protected mRNA fragment (footprint) library preparation, 30 μg total RNA was incubated with 1.5 μl RNase I (10 U/μl) for 45 min at 25 °C with gentle agitation, and then the reaction was stopped by adding RNase inhibitor (Superase-In, Thermo Fisher). Reaction mixtures were loaded on 0.9 ml of 1M sucrose cushions and subjected to ultracentrifugation at 287,000 × g for 1 h at 4 °C. The ribosome pellet was resuspended in 300 μl TRIzol, and the RNA was extracted using a Direct-zol kit (Zymo Research). The RNA was precipitated with a mixture containing 0.015 vol of GlycoBlue, 0.1 vol 3 M NaOAc pH 5.5, and 1.5 vol of isopropanol. The RNA was then separated on a 15% polyacrylamide TBE-Urea gel, and 15–34 nucleotide (RiboSeq) fragments were isolated and processed for dephosphorylation. For each library sample, barcoded 5’-pre-adenylated linkers were added to the 3’ ends of footprints and excess unligated linker was removed using 5’ deadenylase/RecJ exonuclease (Epicentre), followed by pooling and purification of ligated footprints using a Zymo-Spin column. rRNA was depleted using Yeast Ribo-Zero Gold rRNA Removal kit (Illumina) followed by reverse transcription, circularization of cDNA and PCR amplification for final cDNA library construction. DNA Sequencing was performed on Illumina HiSeq 2500 and single end reads were obtained at the NHLBI DNA Sequencing and Genomics Core at NIH (Bethesda, MD).
For RNAseq library preparation, 10 μg total RNA was incubated in Ambion Fragmentation Reagent at 70 °C for 15 min, and the reaction was stopped by adding Stop Solution (ThermoFisher). Fragments were separated by electrophoresis on a 15% polyacrylamide TBE-Urea gel; after column purification and precipitation, RNA fragments were processed for cDNA library preparation as described above for RiboSeq.
Analysis of sequence reads
After removal of the constant linker sequence (AGATCGGAAGAGCAC) using cutadapt 1.17, mixed sample sequences were separated by the sample barcodes followed by removal of PCR duplicates using a custom Python (3.7) script. The sequences aligned to yeast non-coding RNAs were removed using bowtie (1.1.2) with the option -v 3.
The remaining sequences were aligned against the reference genome (R64-1-1 S288C Sac cer3 Genome Assembly) using bowtie (1.1.2) with the option -v 2 -m 1 -y -a --best --strata. Uniquely mapped reads from the final genomic alignments were analyzed for TE using the Riborex package (Li et al., 2017) with edgeR option to identify genes with significant (FDR ≤ 0.1) change in TE in low and high SPD conditions, with a cutoff set at 1.5-fold.
Gene Ontology Analysis
The GO Term Finder (Version 0.86) was accessed via the Saccharomyces Genome Database (SGD) (SGD Project. https://www.yeastgenome.org/goTermFinder/ February 10, 2021). The ontology was based on gene function, and the default p-value was set to p ≤ 0.01.
Sequence Compilation and Phylogenetic Analysis
Sequences were obtained from GenBank using BLAST algorithm (Altschul et al., 1990) applying a strategy described previously (Ivanov et al., 2010b) by querying the Nucleotide collection (nr/nt), the NCBI Nucleotide collection (nr/nt), Transcriptome Shotgun Assembly (TSA) and Whole-genome shotgun contigs (wgs) databases. Sequences were aligned using ClustalX (Larkin et al., 2007). The phylogenetic figure was drawn using the browser-based tool iTOL (https://itol.embl.de/) (Letunic and Bork, 2019).
Cloning and reporter Constructs
The WT HOL1 plasmid was constructed by first amplifying the HOL1 locus by PCR using genomic DNA from strain BY4741 as a template and primers that anneal 1 kb upstream and 500 bp downstream of the HOL1 CDS. The PCR product was cloned between the SacI and PstI sites of the vector YCplac33 (Gietz and Sugino, 1988) generating the plasmid pC6464. The HOL1-Fluc reporter plasmid was generated in two steps. First, a BamHI restriction site was introduced in pC6464 at the codons encoding Hol1 residues Y4 and T5 and then an XbaI site was introduced 10 bp downstream of the HOL1 stop codon, generating the plasmid pC6775. Next, the firefly luciferase CDS (excluding the initiating Met codon) from the vector p2Luc (Grentzmann et al., 1998) was cloned between the BamHI and XbaI sites of pC6775 generating pC6548. Mutations in HOL1-Fluc construct pC6548 including the uORF, uORF start codon context nucleotides and upstream near cognate start codons were made using a QuikChange Lightning (Agilent) site-directed mutagenesis kit. All mutations were confirmed by sequencing and mutated fragments were subcloned into naive parental vectors to avoid possible second-site mutations in the plasmid.
Nucleotide sequences of the WT and mutant uORFs were as follows:
| M | L | L | L | P | S | ATG | CTA | TTA | CTA | CCA | AGT |
| A | L | L | L | P | S | GCG | CTA | TTA | CTA | CCA | AGT |
| M | L | L | L | A | A | ATG | CTA | TTA | CTA | GCA | GCT |
| A | L | L | L | A | A | GCG | CTA | TTA | CTA | GCA | GCT |
| M | A | A | A | P | S | ATG | GCA | GCA | GCA | CCA | AGT |
The CPA1 uORF in the CPA1-luc reporter plasmid pAG121 (a kind gift of Matt Sachs Texas A&M Gaba et al., 2005) was replaced by WT and mutant versions of the HOL1 uORF on NcoI-BglII DNA fragments. Next, the chimeric reporters were subcloned on HindIII-BamHI DNA fragments to the single-copy URA3 vector YCplac33 (Gietz and Sugino, 1988) generating the plasmids pC5791 (WT CPA1 uORF), pC6161 (WT HOL1 uORF MLLLPS*), and derivatives pC6162 (ALLLPS*) and pC6163 (ALLLAA*).
The low copy-number URA3 vector pYX111 (R&D Systems) containing the weak constitutive 786 promoter derived from the HSF1 promoter was modified to insert a triple hemagglutinin epitope (HA3) tag at the 3’ end of the polylinker creating vector pC6316. Synthesized DNA fragments encoding S. cerevisiae and K. lactis HOL1 were cloned into pC6316 generating plasmids pC6455 and pC6439, respectively.
MPS* Peptide release assay
Methionine, phenylalanine, proline, lysine and yeast tRNAPhe were purchased from Sigma. Yeast initiator tRNAMet and tRNALys were purchased from tRNA probes. The 3’-biotin-labeled RNA oligonucleotide used for affinity purification of yeast tRNAPro was purchased from Integrated DNA Technologies. His6-tagged hypusinated eIF5A was purified from BL21(DE3) CodonPlus-RIL transformed with the eIF5A, Dys1 (deoxyhypusine synthase), Lia1 (deoxyhypusine hydroxylase) co-expression plasmid pC4183 as described previously (Gutierrez et al., 2013).
Preparation of translation initiation factors eIF1, eIF1A, eIF2, eIF5 and eIF5B, and translation elongation factor eEF3 was performed using published protocols (Gutierrez et al., 2013). Native elongation factor eEF1A was purified from yeast strain YRP840 and polyhistidine tagged version of elongation factor eEF2 was purified from yeast strain TKY675 as described previously (Shin et al., 2017).
Eukaryotic release factor 1 (eRF1) was purified from E. coli BL21(DE3) CodonPlus-RIL transformed with the plasmid pPROEX-HTb-eRF1 (obtained from Rachel Green, Johns Hopkins University). Briefly, cultures were grown in 500 ml LB with 100 ug/ml of ampicillin to A600 = 0.5, and expression was induced by adding 0.2 mM IPTG and incubating at 20 °C for 14 h. Cells were harvested by centrifugation and resuspended in Lysis Buffer B (20 mM HEPES-KOH 7.4, 0.5 M KCl, 10 mM imidazole, 2 mM β-mercaptoethanol). After cells were lysed by sonication (30 sec sonication at 30% output with microtip followed by 30 sec cooling on ice for total time of 10 min), the lysate was clarified by centrifugation, add then mixed with 1 ml Ni-NTA resin on a nutator at 4 °C for 1 h. Resins were washed with 10 ml Lysis Buffer B and bound proteins were eluted with Lysis Buffer B containing 250 mM imidazole. The eluate was diluted to 100 mM KCl, applied to HitrapQ column, and bound proteins were eluted with a linear gradient to 1 M KCl. Fractions containing eRF1 were identified by SDS-PAGE, dialyzed against Storage Buffer (20 mM HEPES-KOH pH 7.4, 100 mM potassium acetate, 2 mM DTT and 10% glycerol) and concentrated.
To purify eukaryotic release factor 3 (eRF3), a DNA fragment encoding N-terminally truncated eRF3 (amino acids 166–685) was cloned in pGEX-6P-2 and transformed to E. coli BL21. Cells were grown in 500 ml LB with 100 ug/ml of ampicillin to A600 = 0.5, and expression was induced by adding 0.2 mM IPTG and incubating at 20° C for 14 h. The cell pellet was suspended in 25 ml of ice-cold 1X PBS, cells were disrupted by sonication as described for purifying eRF1. To solubilize the expressed protein, Triton X-100 (1% final concentration) was added to the cell lysate and then gently mixed for 30 min at room temperature. The lysate was clarified by centrifugation at 12,000 × g for 20 min at 4° C and then mixed with 1 ml of Glutathione Sepharose 4B resin (Amersham) and incubated at 4° C for 1 h. The resin was then transferred to a 5 ml disposable column (Qiagen), washed sequentially with 10 column volumes of cold 1X PBS buffer and then 10 column volumes of Cleavage Buffer (50 mM Tris-HCl pH 7.5, 100 mM NaCl, 1 mM EDTA, 1 mM DTT). Load Protease Mix (mix 160 units of PreScisson Protease with 920 ul of Cleavage Buffer) on the column, seal, and incubate at 4° C for 14 h. Protein was eluted by gravity and fractions containing eRF3 were identified by SDS-PAGE, dialyzed against Storage Buffer and concentrated.
For preparation and aminoacylation of yeast initiator tRNAiMet, tRNAPhe, tRNAPro, and tRNALys were done as described previously (Shin et al., 2017). For preparation of Seryl-tRNA synthetase, yeast SES1 CDS encoding cytosolic Seryl-tRNA synthetase was cloned in pQE-80L (Qiagen). Expression and purification of Ses1 protein were as described for eRF1 purification. Purified proteins were dialyzed against 50 mM KPO4 pH 7.5, 5 mM magnesium acetate, 2 mM DTT and 10% glycerol.
For reconstituted peptide release assays, 80S initiation complexes were assembled using uncapped model mRNA as previously described (Acker et al., 2007). The coding sequence of the mRNA for Met-Pro-Ser-Stop is AUGCCAUCUUAA. Final concentrations of each factor in the assay are: 4 nM initiation complex, 2 μM eEF1A, 1 μM eEF2, 1 μM eEF3, 0.1 or 10 μM eIF5A, 1 μM eRF1, 1 μM eRF3, 1 μM aminoacyl tRNA, 1 mM GTP-Mg2+, 1 mM ATP-Mg2+ in 1X Recon Buffer (30 mM HEPES pH 7.5, 100 mM potassium acetate, 1 mM magnesium acetate, 1 mM SPD, and 2 mM DTT). Reactions were started by adding aminoacyl tRNAs along with all elongation factors to the pre-assembled initiation complexes. After the peptide synthesis reactions reached plateau (usually after 4 min), peptide release reactions were started by adding eRF1 and eRF3, and reactions were quenched at different times by 30% formic acid. The progress of peptide release was monitored by electrophoretic thin-layer chromatography (TLC) as described previously (Gutierrez et al., 2013). The fractional yield of peptide release out of total peptide products in each reaction at different times was quantified and fit using Kaleidagraph (Synergy Software) to the first-order exponential equation A[1−exp(−kt)] where A is the amplitude and k is the observed rate constant.
QUANTIFICATION AND STATISTICAL ANALYSIS
Quantitative data represent the mean and standard deviation derived from at least three biological replicates. For SPD uptake measurements, Vmax and Km determinations (non-linear curve fitting using Michaelis-Menten equation), and luciferase reporter assays, statistical significance comparisons were made using Student’s two-tailed t tests with a p value threshold < 0.05, using the formula embedded in Graph Pad Prism 8.0 software. For peptide release assays, errors in rate constants and end levels are fitting errors from the curves as determined using Kaleidagraph software. The number of biological replicates (n) is indicated in the legend of each figure. Unless noted, all experiments not requiring quantitative analysis were performed three or more times to confirm reproducibility. Sample size estimates were not used. Studies were not conducted blind. For immunoblots, the reported images are representative of at least three independent experiments.
Supplementary Material
Highlights.
Hol1 is the major high-affinity polyamine transporter in S. cerevisiae
Polyamine levels autoregulate HOL1 mRNA translation through a conserved uORF
Polyamines inhibit eIF5A to impair translation termination on the uORF
Ribosome stalling on the uORF prevents HOL1 translation
ACKNOWLEDGMENTS
We thank members of our labs as well as Alan Hinnebusch, Jon Lorsch, Nick Guydosh, and members of their labs for helpful discussions. We thank Orna Cohen-Fix and Latika Mathai for instructions on microscopy and the use of their microscopes, Matt Sachs for the gift of plasmids, and Herb Tabor for the gift of yeast strains. This research was supported by the Intramural Research Program of the NIH, the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD, HD001010 to T.E.D. and HD008928 to A.B.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
T.E.D. is a member of the scientific advisory board at the journal Molecular Cell.
REFERENCES
- Acker MG, Kolitz SE, Mitchell SF, Nanda JS, and Lorsch JR (2007). Reconstitution of yeast translation initiation. Methods Enzymol 430, 111–145. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, and Lipman DJ (1990). Basic local alignment search tool. J Mol Biol 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Aouida M, Rubio-Texeira M, Thevelein JM, Poulin R, and Ramotar D (2013). Agp2, a member of the yeast amino acid permease family, positively regulates polyamine transport at the transcriptional level. PLoS One 8, e65717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer SK, Shirokikh NE, Beilharz TH, and Preiss T (2016). Dynamics of ribosome scanning and recycling revealed by translation complex profiling. Nature 535, 570–574. [DOI] [PubMed] [Google Scholar]
- Balasundaram D, Xie QW, Tabor CW, and Tabor H (1994). The presence of an active S-adenosylmethionine decarboxylase gene increases the growth defect observed in Saccharomyces cerevisiae mutants unable to synthesize putrescine, spermidine, and spermine. J Bacteriol 176, 6407–6409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohlen J, Fenzl K, Kramer G, Bukau B, and Teleman AA (2020). Selective 40S footprinting reveals cap-tethered ribosome scanning in human cells. Mol Cell 79, 561–574. [DOI] [PubMed] [Google Scholar]
- Bushman JL, Foiani M, Cigan AM, Paddon CJ, and Hinnebusch AG (1993). Guanine nucleotide exchange factor for eukaryotic translation initiation factor 2 in Saccharomyces cerevisiae: interactions between the essential subunits GCD2, GCD6, and GCD7 and the regulatory subunit GCN3. Mol Cell Biol 13, 4618–4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casero RA Jr., Murray Stewart T, and Pegg AE (2018). Polyamine metabolism and cancer: treatments, challenges and opportunities. Nat Rev Cancer 18, 681–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay MK, Park MH, and Tabor H (2008). Hypusine modification for growth is the major function of spermidine in Saccharomyces cerevisiae polyamine auxotrophs grown in limiting spermidine. Proc Natl Acad Sci USA 105, 6554–6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christenson ET, Gallegos AS, and Banerjee A (2018). In vitro reconstitution, functional dissection, and mutational analysis of metal ion transport by mitoferrin-1. J Biol Chem 293, 3819–3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M, VanderSluis B, Koch EN, Baryshnikova A, Pons C, Tan G, Wang W, Usaj M, Hanchard J, Lee SD, et al. (2016). A global genetic interaction network maps a wiring diagram of cellular function. Science 353, aaf1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever TE, and Ivanov IP (2018). Roles of polyamines in translation. J Biol Chem 293, 18719–18729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever TE, Ivanov IP, and Sachs MS (2020). Conserved upstream open reading frame nascent peptides that control translation. Annu Rev Genet 54, 237–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever TE, Kinzy TG, and Pavitt GD (2016). Mechanism and regulation of protein synthesis in Saccharomyces cerevisiae. Genetics 203, 65–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvir S, Velten L, Sharon E, Zeevi D, Carey LB, Weinberger A, and Segal E (2013). Deciphering the rules by which 5’-UTR sequences affect protein expression in yeast. Proc Natl Acad Sci U S A 110, E2792–2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg AR, Higdon AL, Hollerer I, Fields AP, Jungreis I, Diamond PD, Kellis M, Jovanovic M, and Brar GA (2020). Translation initiation site profiling reveals widespread synthesis of non-AUG-initiated protein isoforms in yeast. Cell Syst 11, 145–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang P, Spevak CC, Wu C, and Sachs MS (2004). A nascent polypeptide domain that can regulate translation elongation. Proc Natl Acad Sci U S A 101, 4059–4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaba A, Jacobson A, and Sachs MS (2005). Ribosome occupancy of the yeast CPA1 upstream open reading frame termination codon modulates nonsense-mediated mRNA decay. Mol Cell 20, 449–460. [DOI] [PubMed] [Google Scholar]
- Gaber RF, Kielland-Brandt MC, and Fink GR (1990). HOL1 mutations confer novel ion transport in Saccharomyces cerevisiae. Mol Cell Biol 10, 643–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble LD, Purgato S, Murray J, Xiao L, Yu DMT, Hanssen KM, Giorgi FM, Carter DR, Gifford AJ, Valli E, et al. (2019). Inhibition of polyamine synthesis and uptake reduces tumor progression and prolongs survival in mouse models of neuroblastoma. Sci Transl Med 11, eaau1099. [DOI] [PubMed] [Google Scholar]
- Gbelska Y, Krijger JJ, and Breunig KD (2006). Evolution of gene families: the multidrug resistance transporter genes in five related yeast species. FEMS Yeast Res 6, 345–355. [DOI] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, et al. (2002). Functional profiling of the Saccharomyces cerevisiae genome. Nature 418, 387–391. [DOI] [PubMed] [Google Scholar]
- Gietz RD, and Sugino A (1988). New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74, 527–534. [DOI] [PubMed] [Google Scholar]
- Grentzmann G, Ingram JA, Kelly PJ, Gesteland RF, and Atkins JF (1998). A dual-luciferase reporter system for studying recoding signals. RNA 4, 479–486. [PMC free article] [PubMed] [Google Scholar]
- Gutierrez E, Shin BS, Woolstenhulme CJ, Kim JR, Saini P, Buskirk AR, and Dever TE (2013). eIF5A promotes translation of polyproline motifs. Mol Cell 51, 35–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamouda NN, Van den Haute C, Vanhoutte R, Sannerud R, Azfar M, Mayer R, Cortes Calabuig A, Swinnen JV, Agostinis P, Baekelandt V, et al. (2020). ATP13A3 is a major component of the enigmatic mammalian polyamine transport system. J Biol Chem, doi: 10.1074/jbc.RA1120.013908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller JS, and Canellakis ES (1981). Cellular control of ornithine decarboxylase activity by its antizyme. J Cell Physiol 107, 209–217. [DOI] [PubMed] [Google Scholar]
- Hiasa M, Miyaji T, Haruna Y, Takeuchi T, Harada Y, Moriyama S, Yamamoto A, Omote H, and Moriyama Y (2014). Identification of a mammalian vesicular polyamine transporter. Sci Rep 4, 6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG (2011). Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiol Mol Biol Rev 75, 434–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG (2014). The scanning mechanism of eukaryotic translation initiation. Annu Rev Biochem 83, 779–812. [DOI] [PubMed] [Google Scholar]
- Igarashi K, and Kashiwagi K (2010a). Characteristics of cellular polyamine transport in prokaryotes and eukaryotes. Plant Physiol Biochem 48, 506–512. [DOI] [PubMed] [Google Scholar]
- Igarashi K, and Kashiwagi K (2010b). Modulation of cellular function by polyamines. Int J Biochem Cell Biol 42, 39–51. [DOI] [PubMed] [Google Scholar]
- Ivanov IP, and Atkins JF (2007). Ribosomal frameshifting in decoding antizyme mRNAs from yeast and protists to humans: close to 300 cases reveal remarkable diversity despite underlying conservation. Nucleic Acids Res 35, 1842–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov IP, Atkins JF, and Michael AJ (2010a). A profusion of upstream open reading frame mechanisms in polyamine-responsive translational regulation. Nucleic Acids Res 38, 353–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov IP, Firth AE, and Atkins JF (2010b). Recurrent emergence of catalytically inactive ornithine decarboxylase homologous forms that likely have regulatory function. J Mol Evol 70, 289–302. [DOI] [PubMed] [Google Scholar]
- Ivanov IP, Loughran G, and Atkins JF (2008). uORFs with unusual translational start codons autoregulate expression of eukaryotic ornithine decarboxylase homologs. Proc Natl Acad Sci U S A 105, 10079–10084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov IP, Shin BS, Loughran G, Tzani I, Young-Baird SK, Cao C, Atkins JF, and Dever TE (2018). Polyamine control of translation elongation regulates start site selection on antizyme inhibitor mRNA via ribosome queuing. Mol Cell 70, 254–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahana C (2009). Regulation of cellular polyamine levels and cellular proliferation by antizyme and antizyme inhibitor. Essays Biochem 46, 47–61. [DOI] [PubMed] [Google Scholar]
- Kawate T, and Gouaux E (2006). Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure 14, 673–681. [DOI] [PubMed] [Google Scholar]
- Kim D, Langmead B, and Salzberg SL (2015). HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12, 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M (2001). Constraints on reinitiation of translation in mammals. Nucleic Acids Res 29, 5226–5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, and Salzberg SL (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. [DOI] [PubMed] [Google Scholar]
- Law GL, Raney A, Heusner C, and Morris DR (2001). Polyamine regulation of ribosome pausing at the upstream open reading frame of S-adenosylmethionine decarboxylase. J Biol Chem 276, 38036–38043. [DOI] [PubMed] [Google Scholar]
- Letunic I, and Bork P (2019). Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47, W256–W259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Wang W, Uren PJ, Penalva LOF, and Smith AD (2017). Riborex: fast and flexible identification of differential translation from Ribo-seq data. Bioinformatics 33, 1735–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie III A, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, and Pringle JR (1998). Additonal modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961. [DOI] [PubMed] [Google Scholar]
- Luukkonen BG, Tan W, and Schwartz S (1995). Efficiency of reinitiation of translation on human immunodeficiency virus type 1 mRNAs is determined by the length of the upstream open reading frame and by intercistronic distance. J Virol 69, 4086–4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manjunath H, Zhang H, Rehfeld F, Han J, Chang TC, and Mendell JT (2019). Suppression of ribosomal pausing by eIF5A is necessary to maintain the fidelity of start codon selection. Cell Rep 29, 3134–3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10–12. [Google Scholar]
- Matsufuji S, Matsufuji T, Miyazaki Y, Murakami Y, Atkins JF, Gesteland RF, and Hayashi S (1995). Autoregulatory frameshifting in decoding mammalian ornithine decarboxylase antizyme. Cell 80, 51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy DJ, Chen Y, and Smyth GK (2012). Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res 40, 4288–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlincy NJ, and Ingolia NT (2017). Transcriptome-wide measurement of translation by ribosome profiling. Methods 126, 112–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meydan S, and Guydosh NR (2020). Disome and trisome profiling reveal genome-wide targets of ribosome quality control. Mol Cell 79, 588–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad MP, Munzarova Pondelickova V, Zeman J, Gunisova S, and Valasek LS (2017). In vivo evidence that eIF3 stays bound to ribosomes elongating and terminating on short upstream ORFs to promote reinitiation. Nucleic Acids Res 45, 2658–2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami Y, Ichiba T, Matsufuji S, and Hayashi S (1996). Cloning of antizyme inhibitor, a highly homologous protein to ornithine decarboxylase. J Biol Chem 271, 3340–3342. [DOI] [PubMed] [Google Scholar]
- Murakami Y, Matsufuji S, Kameji T, Hayashi S, Igarashi K, Tamura T, Tanaka K, and Ichihara A (1992). Ornithine decarboxylase is degraded by the 26S proteasome without ubiquitination. Nature 360, 597–599. [DOI] [PubMed] [Google Scholar]
- Nishiki Y, Farb TB, Friedrich J, Bokvist K, Mirmira RG, and Maier B (2013). Characterization of a novel polyclonal anti-hypusine antibody. Springerplus 2, 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palanimurugan R, Scheel H, Hofmann K, and Dohmen RJ (2004). Polyamines regulate their synthesis by inducing expression and blocking degradation of ODC antizyme. EMBO J 23, 4857–4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MH, and Wolff EC (2018). Hypusine, a polyamine-derived amino acid critical for eukaryotic translation. J Biol Chem 293, 18710–18718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegg AE (2009). Mammalian polyamine metabolism and function. IUBMB Life 61, 880–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegg AE (2016). Functions of Polyamines in Mammals. J Biol Chem 291, 14904–14912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajkowitsch L, Vilela C, Berthelot K, Ramirez CV, and McCarthy JE (2004). Reinitiation and recycling are distinct processes occurring downstream of translation termination in yeast. J Mol Biol 335, 71–85. [DOI] [PubMed] [Google Scholar]
- Raney A, Law GL, Mize GJ, and Morris DR (2002). Regulated translation termination at the upstream open reading frame in S-adenosylmethionine decarboxylase mRNA. J Biol Chem 277, 5988–5994. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan H, Shantz LM, Pegg AE, and Morris DR (1996). The upstream open reading frame of the mRNA encoding S-adenosylmethionine decarboxylase is a polyamine-responsive translational control element. J Biol Chem 271, 29576–29582. [DOI] [PubMed] [Google Scholar]
- Saini P, Eyler DE, Green R, and Dever TE (2009). Hypusine-containing protein eIF5A promotes translation elongation. Nature 459, 118–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller AP, Wu CC, Dever TE, Buskirk AR, and Green R (2017). eIF5A functions globally in translation elongation and termination. Mol Cell 66, 194–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin BS, Katoh T, Gutierrez E, Kim JR, Suga H, and Dever TE (2017). Amino acid substrates impose polyamine, eIF5A, or hypusine requirement for peptide synthesis. Nucleic Acids Res 45, 8392–8402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T, Kashiwagi K, and Igarashi K (2005). Uptake of putrescine and spermidine by Gap1p on the plasma membrane in Saccharomyces cerevisiae. Biochem Biophys Res Commun 328, 1028–1033. [DOI] [PubMed] [Google Scholar]
- Uemura T, Kashiwagi K, and Igarashi K (2007). Polyamine uptake by DUR3 and SAM3 in Saccharomyces cerevisiae. J Biol Chem 282, 7733–7741. [DOI] [PubMed] [Google Scholar]
- Uemura T, Stringer DE, Blohm-Mangone KA, and Gerner EW (2010). Polyamine transport is mediated by both endocytic and solute carrier transport mechanisms in the gastrointestinal tract. Am J Physiol Gastrointest Liver Physiol 299, G517–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usaj M, Tan Y, Wang W, VanderSluis B, Zou A, Myers CL, Costanzo M, Andrews B, and Boone C (2017). TheCellMap.org: A Web-Accessible Database for Visualizing and Mining the Global Yeast Genetic Interaction Network. G3 (Bethesda) 7, 1539–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Veen S, Martin S, Van den Haute C, Benoy V, Lyons J, Vanhoutte R, Kahler JP, Decuypere JP, Gelders G, Lambie E, et al. (2020). ATP13A2 deficiency disrupts lysosomal polyamine export. Nature 578, 419–424. [DOI] [PubMed] [Google Scholar]
- Wagner S, Herrmannova A, Hronova V, Gunisova S, Sen ND, Hannan RD, Hinnebusch AG, Shirokikh NE, Preiss T, and Valasek LS (2020). Selective translation complex profiling reveals staged initiation and co-translational assembly of initiation factor complexes. Mol Cell 79, 546–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, and Sachs MS (1997). Ribosome stalling is responsible for arginine-specific translational attenuation in Neurospora crassa. Mol Cell Biol 17, 4904–4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright MB, Howell EA, and Gaber RF (1996). Amino acid substitutions in membrane-spanning domains of Hol1, a member of the major facilitator superfamily of transporters, confer nonselective cation uptake in Saccharomyces cerevisiae. J Bacteriol 178, 7197–7205. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data were deposited in the GEO database under the accession number GSE171392. Gel images were deposited in the Mendeley Database link is: https://doi.org/10.17632/3wg5js2n8j.1
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


