Abstract
A high-quality dataset of 3289 complete SARS-CoV-2 genomes collected in Europe and European Economic Area (EAA) in the early phase of the first wave of the pandemic was analyzed. Among all single nucleotide mutations, 41 had a frequency ≥ 1%, and the phylogenetic analysis showed at least 6 clusters with a specific mutational profile. These clusters were differentially distributed in the EU/EEA, showing a statistically significant association with the geographic origin. The analysis highlighted that the mutations C14408T and C14805T played an important role in clusters selection and further virus spread. Moreover, the molecular analysis suggests that the SARS-CoV-2 strain responsible for the first Italian confirmed COVID-19 case was already circulating outside the country.
Keywords: COVID-19, Cluster analysis, SARS-CoV-2, SNVs analysis
1. Introduction
SARS-CoV-2 is an enveloped single stranded RNA virus with a genome of about 29.8 kilobases (kb). The current map of SARS-CoV-2 coding capacity is based on computational predictions and relies on homology with other coronaviruses. The reference sequence (an. NC_045512) has been annotated with 14 ORFs encoding 16 nonstructural proteins, 4 structural proteins and at least 7 accessory proteins (Wu et al., 2020). However, further studies have revealed additional ORFs in the SARS-CoV-2 genome, highlighting that the coding capacity has not yet been fully established (Michel et al., 2020; Finkel et al., 2021). Previous studies of SARS-CoV-2 genomes estimated the time of its emergence in China at the end of November 2020 (25th and 28th) (Benvenuto et al., 2020; Liu et al., 2020), approximately one month before the first confirmed cases. Since then, a huge number of genomes have been analyzed worldwide (Mercatelli and Giorgi, 2020). As previously reported (Mercatelli and Giorgi, 2020), amino acid changes through Single Nucleotide Variants (SNVs) are the predominant mutational events. Moreover, discontinuous transcription events have been described as a route to influence the translation process (Kim et al., 2020).
The primary goal of this study was to analyze SARS-CoV-2 genomes collected in Europe and in the European Economic Area (EAA) in the early phase of the pandemic in order to identify and characterize the main viral clusters. The data were collected until the second half of March 2020, a couple of weeks after the introduction of the first containment measures in Italy. To this end, a high-quality dataset of 3289 complete SARS-CoV-2 genomes available on GISAID, including 46 newly sequenced Italian SARS-CoV-2 genomes, was analyzed.
2. Methods
2.1. Whole genome analysis
In this study, 46 naso/oropharyngeal swabs provided by Scientific Department of Army Medical Center (Rome, Italy) and by the Istituto Superiore di Sanità (ISS, Rome, Italy), collected from February 5th, 2020 until March 22nd, 2020, were completely sequenced. All samples were confirmed positives as SARS-CoV-2 by the National Reference Laboratory (NRL) of the ISS in Rome. Samples were selected in accordance with the resulting of Real-Time PCR cycle threshold (Ct) value (Ct ranging from 16 to 22 cycles).
The viral RNA was extracted using the QIAMP VIRAL RNA Mini Kit or RNeasy Mini Kit. (Qiagen, Hilden, Germany). Genomic RNAs were retro-transcribed using the SuperScript III Reverse Transcriptase kit (Invitrogen, Carlsbad, CA, USA) and double-stranded DNAs were obtained by Klenow enzyme (Roche, Basel, Switzerland) according to the manufacturer's instructions. The Nextera XT kit was used for library preparations and whole genome sequencing was performed using the Illumina Miseq Reagent V2 (2 × 150 cycles) or the Illumina NextSeq 500 High Output Kit V 2.5 (2 × 150) (Illumina, San Diego, CA, USA) on the Illumina MiSeq or NextSeq 500 instruments, respectively. The reads were trimmed for quality (q score ≥ 20) and minimum length (= 100) using BBDuk trimmer (sourceforge.net/projects/bbmap /). High quality reads were assembled by mapping to the reference genome from Wuhan, China (GenBank an. NC_045512.2) with Bowtie2 mapping (Langmead and Salzberg, 2012). BBDuk and Bowtie2 algorithms were integrated in Geneious Prime software (www.geneious.com). Viral sequences were deposited in the Global Initiative on Sharing All Influenza Data (GISAID; https://www.gisaid.org). All European high-coverage SARS-CoV-2 complete genomes available on GISAID until March 22nd, 2020 were retrieved. The starting raw data-set consisted of 4428 complete genomes. The raw dataset was aligned to the reference genome (an. NC_045512) by using MAFFT v7.450 with default settings (Katoh et al., 2019). The flanking regions of the aligned sequences were trimmed to the consensus range 54 bps to 29,783 bps according to the reference genome. Sequences containing >0.1% of ambiguous nucleotides (N) were detected with an ad hoc script and removed from the dataset.
Single Nucleotide Variants (SNVs) and deletions were first identified by means of Geneious Prime software. The genomic locations and frequency of SNVs were then extracted using a custom script. To compare the SNVs frequency and distribution in each European country, the dataset was split to obtain a single subset for each nation, then the analysis was performed again for each dataset.
2.2. Phylogenetic trees and estimation of mutation rate
All the phylogenetic trees were obtained by using the maximum likelihood (ML) method using PhyML v3.3 (Guindon et al., 2010) with HKY + Γ 4 substitution model. Phylogenetic trees were constructed for the complete dataset as well as for each analyzed country. Due to the low number of genomes available from Bosnia, Hungary, Poland, Croatia, Slovakia, Latvia, Czech Republic, Slovenia and Lithuania, these genomes were merged into one dataset named “East Europe” to obtain a single phylogenetic tree.
Branch support values were estimated with the aRLT SH-like implemented in PhyML and/or with bootstrap analysis (bootstrap 200).
Each tree was manually inspected and relevant SNVs were assigned to each cluster. Mutation rate of the virus was first estimated on the entire dataset. Additionally, mutation rate was estimated in the subset of genomes which acquired the C14408T mutation in SS4 cluster and in the subset of genomes which acquired the C14805T mutation in the SS1 and SS2B clusters.
The graphical editing of the phylogenetic trees was performed using Megax (www.megasoftware.net) and Inkscape (https://inkscape.org).
2.3. Clusters analysis
To investigate the genetic diversity of SARS-CoV-2 among the EU/EEA countries, the mutation profile of each dataset was studied by means of the clusters scheme proposed by Yang et al. (2020).
2.4. Statistical analysis
Contingency tables among different categories (e.g., clusters and countries) were evaluated for unexpected frequencies with the Chi-square test. The collection date of the first cluster-representative genome, which had to fall within the month of February, was used as the selection criterion.
3. Results
3.1. SNVs analysis
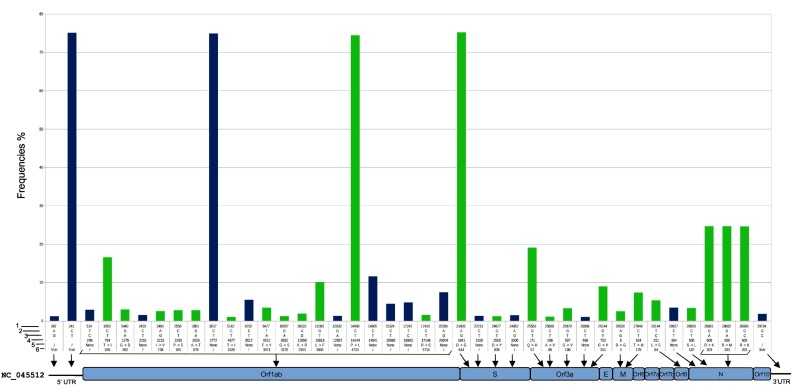
The final high-quality dataset consisted of 3289 genomic sequences of which 525 were from the UK, 352 from Iceland, 343 from the Netherlands, 265 from Spain, 260 from Denmark, 238 from Sweden, 225 from France, 173 from Belgium, 166 from Portugal, 165 from Austria, 110 from Italy, 99 from Germany, 78 from Switzerland, 70 from Greece, 68 from Luxembourg, 38 from Russia, 21 from Norway, 20 from Finland, 10 from Ireland, 15 from Turkey, and 48 from East Europe. Genome codes of the entire dataset together with 46 newly identified Italian genomes are listed in Supplementary Table S1. A total of 1897 SNVs, including 1199 (63%) rare mutations, were detected. Fig. 1 shows 41 mutations with frequency ≥ 1%, consisting of 16 silent and 25 missense mutations. These SNVs were distributed over the entire length of the genome, except for Env, Orf6, Orf7a, Orf7b, and Orf10 genes, where only sporadic mutations were detected.
Fig. 1.
Single nucleotide variants analysis (SNVs). A total of 41 SNVs detected on the high-quality dataset with a frequency > 1% are shown. The frequency of each mutation is represented by the bar height. At the bottom, the genomic organization of SARS-CoV-2 and the relative mutation localization. Blue bars = silent mutations; green bars = missense mutations; 1) = nucleotide position; 2) = ancestral nucleotide; 3) = variant nucleotide; 4) = CDS position; 5) = AA change; 6) codon number. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
A total of 10 deletions were also identified, 8 of which were previously described (Islam et al., 2020; Benedetti et al., 2020; Koyama et al., 2020). Two new deletions were detected, the first located in the 5’UTR region from position 185 to 218 (EPI_ISL_441377), and the second in the ORF1ab gene from position 19298 to 19551 (EPI_ISL_415435). The latter deletion involved 324 bps resulting in a stop codon in the nsp14 protein. Both sequences were from the UK.
3.2. Phylogenetic trees
The ML trees resulting from the entire dataset (Fig. 2, full resolution image Supplementary Fig. S1) and from each country (Supplementary Fig. S2, Supplementary Fig. S3, Supplementary Fig. S4, Supplementary Fig. S5, Supplementary Fig. S6, Supplementary Fig. S7, Supplementary Fig. S8, Supplementary Fig. S9, Supplementary Fig. S10, Supplementary Fig. S11, Supplementary Fig. S12, Supplementary Fig. S13, Supplementary Fig. S14, Supplementary Fig. S15, Supplementary Fig. S16, Supplementary Fig. S17, Supplementary Fig. S18, Supplementary Fig. S19, Supplementary Fig. S20, Supplementary Fig. S21, Supplementary Fig. S22), showed distinct clusters characterized by specific mutational profiles. The tree topology resulting from the entire dataset was fully supported by the aLRT SH-like analysis as well as in all the trees resulting from each specific geographic location, with the exception of Russia, Luxembourg, Portugal, Denmark and East Europe. The trees resulting from these dataset were not supported by the aLRT-SH-like most likely due to the absence of informative genomes, these datasets were mainly represented by genomes belonging to the SS4 cluster only. The topology of these trees was further evaluated by the bootstrap analysis (bootstrap = 200), which showed a value greater than 80% in the root branch of these clusters while minor values were obtained in the subclusters.
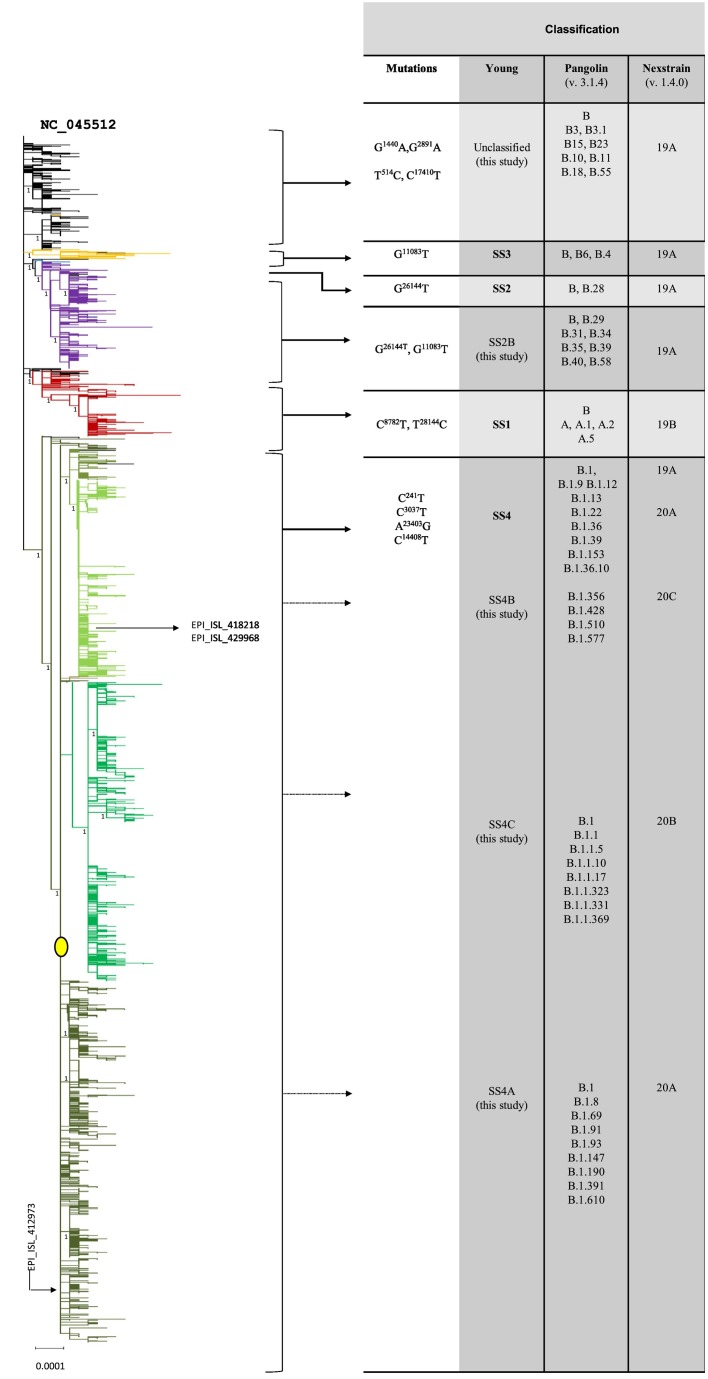
Fig. 2.
Phylogenetic tree of the entire dataset in collapsed form (rooted). The picture shows the phylogram of the entire dataset resulting from the ML analysis in collapsed form (full resolution image in Supplementary Fig. S1). The six main clusters are shown in different colors: black = unclassified; yellow = SS3; blue = SS2; violet = SS2B; red = SS1; green = SS4 (olive green SS4B, emerald green SS4C, dark green SS4A). The branch representing the central node at the root of the SS4 cluster is labeled by a filled yellow circle (see discussion and Fig. 4). The numbers next to the main nodes represent the output of the aRLT-SH-like analysis. On the right, the main mutations hosted by each cluster and the corresponding classifications according to Yang et al. (2020), Pangolin and Nexstrain nomenclatures are shown. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.3. Clusters analysis
As shown in Fig. 2, the two most used nomenclatures for SARS-CoV-2 classification, the Pangolin (Rambaut et al., 2020) and the Nexstrain (Hadfield et al., 2018), do not unequivocally describe the clusters resulting from the ML analysis. This is not surprising, because such nomenclatures were mainly conceived for fast detection and classification of new mutations. The classification of Yang et al. (2020) better fit to the ML analysis, therefore it has been adopted in this study. This classification identifies four main clusters defined as super-spreader 1 (SS1), 2 (SS2), 3 (SS3), and 4 (SS4), each one carrying a specific signature mutation. The ML analysis detected additional clusters. One cluster, here named as “SS2B,” originated by the merging of two clusters (SS2 and SS3) while others, here named as “unclassified,” included a subset of small clusters lacking the SS signature motif. The genomes which showed a mixed presence of SS signature mutations were defined as “ambiguous.”
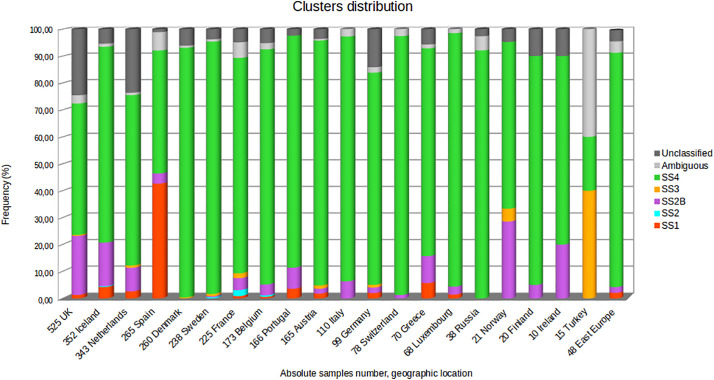
Fig. 3 shows the clusters distribution in EU/EEA for the study period.
Fig. 3.
Clusters distribution in EU/EEA area. The distributions and the relative frequencies of the six main clusters in the EU/EAA along with the ambiguous sequences are shown in different colors. The absolute genomes number for each country is reported.
3.3.1. SS1
SS1 carried the signature mutations C8782T and T28144C, the last resulting in the amino acid (AA) change L > S in the ORF8 protein. A total of 164 genomes from 12 EU countries belonged to SS1 cluster. This cluster was rarely identified in 10 out of the 12 countries. A high frequency was found in Spain where 113 genomes out of 265 (42,6%) belonged to SS1 cluster. The Spanish SS1 cluster has evolved in two sub-clusters (Supplementary Fig. S18). The first included 16 genomes and carried the C26088T mutation and the second comprised 88 genomes which carried five mutations (T9477A, C14805T, C28657T, C28863T and G25979T). In this sub-cluster two mutations resulted in AA changes, the C28863T transition resulting in S > L substitution in the nucleocapsid protein, and the G25979T transversion resulting in G > V substitution in the ORF3a protein.
SS1 genomes from Iceland showed a different profile carrying mainly the C17747T, A17858G, T17531C, C18060T and A24694T mutations (Supplementary Fig. S10). The first three transitions resulted in the AA substitutions P > L, Y > C and I > T, respectively, all affecting the nsp13 protein.
In Table 1 , the main mutations belonging to the SS1 clusters are reported.
Table 1.
SS1 clusters and additional mutations. On the bottom left column, the absolute harbored SS1 genomes number by countries are shown. On the top, additional mutations, their genomic positions, the effect on protein translation and the affected codons are reported.
| SS1 signature C8782T, T28144C | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Additional mutations bps position | 9477 | 14805 | 28657 | 28863 | 25979 | 26088 | 17747 | 17858 | 24694 | 18060 | 9445 | 17531 | 18756 | 25553 | 27801 | |
| ancestral/mutate | T/A | C/T | C/T | C/T | G/T | C/T | C/T | A/G | A/T | C/T | T/C | T/C | G/T | C/T | T/C | |
| AA change | F > Y | / | / | S > L | G > V | / | P > L | Y > C | / | / | / | I > T | / | A > V | F > L | |
| Gene | ORF1ab | ORF1ab | N | N | ORF3a | ORF3a | ORF1ab | ORF1ab | Spike | ORF1ab | ORF1ab | ORF1ab | ORF1ab | ORF3a | ORF7b | |
| product | nsp4 | / | / | Nucleocap. | orf3a | / | nsp13 | nsp13 | / | / | / | nsp13 | / | orf3a | orf7b | |
| Codon number | 3071 | / | / | 197 | 196 | / | 5828 | 5865 | / | / | / | 5756 | / | 54 | 16 | |
| Country | SS1 | |||||||||||||||
| AT | 3 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 |
| BE | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| FR | 2 | 2 | 2 | 2 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 |
| DE | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| GR | 4 | 2 | 2 | 2 | 2 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| IS | 15 | 0 | 0 | 0 | 0 | 0 | 0 | 15 | 15 | 15 | 14 | 13 | 10 | 10 | 0 | 0 |
| LU | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| NL | 9 | 4 | 4 | 4 | 4 | 4 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| PL | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| PT | 6 | 4 | 4 | 4 | 4 | 4 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ES | 113 | 94 | 94 | 93 | 93 | 90 | 16 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| UK | 7 | 2 | 2 | 2 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Tot. | 164 | 110 | 110 | 109 | 109 | 106 | 25 | 15 | 15 | 15 | 14 | 13 | 10 | 10 | 2 | 3 |
3.3.2. SS2
The SS2 cluster was characterized by the signature mutation G26144T which resulted in the AA change G > V in the ORF3a protein. This cluster was found in 8 genomes collected in 4 European countries (Sweden, Belgium, Iceland and France). Genomes from Belgium (EPI_ISL_417025) and from Iceland (EPI_ISL_417672) carried the C14805T mutation associated with T17247C mutation. The phylogenetic analysis suggested that the SS2 cluster evolved in the SS2B cluster through the acquisition of G11083T mutation (Supplementary Fig. S1).
In Table 2 , the main mutations belonging to the SS2 cluster are reported.
Table 2.
SS2 clusters and additional mutations. On the bottom left column, the absolute harbored SS2 genomes number by countries are shown. On the top, additional mutations, their genomic positions, the effect on protein translation and the affected codons are reported.
| SS2 signature G26144T | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Additional mutations bps position | 22661 | 14805 | 17247 | 1515 | 2668 | 2717 | 9223 | 9274 | 9438 | 13225 | 13226 | 17376 | 23952 | |
| ancestal/mutate | G /T | C /T | T/C | A/G | C/T | G/A | C/T | A/G | C/T | C/G | T/C | A/G | T/G | |
| AA change | V > F | / | / | H > R | / | G > S | / | / | T > I | / | F > L | / | F > C | |
| Gene | S | ORF1ab | ORF1ab | ORF1ab | ORF1ab | ORF1ab | ORF1ab | ORF1ab | ORF1ab | ORF1ab | ORF1ab | ORF1ab | S | |
| Product | Spike | / | / | nsp2 | / | nsp2 | / | / | nsp4 | / | nsp10 | / | Spike | |
| Codon number | 367 | / | / | 417 | / | 818 | / | / | 3058 | / | 4321 | / | 797 | |
| Country | SS2 | |||||||||||||
| BE | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| FR | 5 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| IS | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| SE | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 |
| Tot | 8 | 5 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
3.3.3. SS3
The SS3 cluster carried the signature mutation G11083T resulting in the AA substitution L > F in the nsp4 protein. It was identified in 22 genomes in 9 countries. With the exception of some genomes, the SS3 cluster in EU/EAA has been found always associated with the mutation G1397A, which resulted in the AA change V > I in the nsp2 protein, and with the silent mutations T28688C and G29742T. The same profile was detected in Turkey where the SS3 cluster was the predominant in our dataset. Additional mutations were found mainly in Turkey and to a lesser extent in France. In Turkey, this cluster spread efficiently with a mutation rate of 9.7 × 10−3 sub/site/year.
In Table 3 , the main mutations belonging to the SS3 cluster are reported.
Table 3.
SS3 clusters and additional mutations. On the bottom left column, the absolute harbored SS3 genomes number by countries are shown. On the top, additional mutations, their genomic positions, the effect on protein translation and the affected codons are reported.
| SS3 signature G11083T | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Additional mutations bps position | 1397 | 29742 | 28688 | 9479 | 9514 | 28835 | 26549 | 1190 | 9438 | 26720 | 228 | 1440 | 7011 | |
| ancestal/mutate | G /A | G/T | T/C | G/T | A/G | T/C | C/T | C/T | C/T | G/C | C/T | G/A | C/T | |
| AA change | V > I | / | / | G > C | / | S > P | / | P > S | T > I | / | / | G > D | A > V | |
| Gene | ORF1ab | 3’UTR | N | ORF1ab | ORF1ab | N | M | ORF1ab | ORF1ab | M | 5’UTR | ORF1ab | ORF1ab | |
| Product | nsp2 | / | / | nsp4 | / | nucleocaps. | / | nsp2 | nsp4 | / | / | nsp2 | nsp3 | |
| Codon number | 378 | / | / | 3072 | / | 188 | / | 309 | 3058 | / | / | 392 | 2249 | |
| Country | SS3 | |||||||||||||
| AT | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| DK | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| FR | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 3 | 0 | 0 | 0 | 0 |
| DE | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| NL | 3 | 3 | 3 | 3 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| NO | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| SE | 2 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| TR | 6 | 6 | 6 | 6 | 3 | 3 | 3 | 4 | 0 | 0 | 3 | 3 | 0 | 0 |
| UK | 2 | 2 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Tot | 22 | 15 | 15 | 14 | 4 | 4 | 4 | 4 | 3 | 3 | 3 | 3 | 2 | 2 |
3.3.4. SS2B
The G26144T (SS2) and G11083T (SS3) mutations were found associated in 276 genomes throughout 18 European countries. The cluster that harbored this mutational profile was named SS2B. It was not detected in Turkey, Russia, Denmark and East Europe, with the exception of Bosnia & Herzegovina and Slovenia.
The C14805T mutation, a silent transition detected also in SS1 and SS2 clusters, was found in 253 genomes out of 276. The mutation T17247C was found in 151 out of 276 genomes, always linked to the C14805T. The C2558T and A2480G mutations were found in 85 and 77 genomes respectively, mainly from UK and central Europe. The two mutations, which resulted in the AA change P > S and I > V in the nsp2 protein respectively, were associated with the C14805T. In France, the G24095T resulting in the AA substitution A > S in the spike protein, was found associated with the C14805T.
The C2558T/A2480G and the T17247C mutations were mutually exclusive in all SS2B clusters.
The mutation C5142T resulting in the AA change T > I in the nsp3 protein was detected exclusively in Iceland in more than 50% of the SS2B genomes.
The main mutations belonging to the SS2B cluster are reported in Table 4 .
Table 4.
SS2B clusters and additional mutations. On the bottom left column, the absolute harbored SS2B genomes number by countries are shown. On the top, additional mutations, their genomic positions, the effect on protein translation and the affected codons are reported.
| SS2B signature G26144T, G11083T | ||||||||
|---|---|---|---|---|---|---|---|---|
| Additional mutations bps position | 14805 | 17247 | 2558 | 2480 | 5142 | 1321 | 24095 | |
| ancestal/mutate | C/T | T/C | C/T | A/G | C/T | A/C | G/T | |
| AA change | / | / | P > S | I > V | T > I | E > D | A > S | |
| Gene | ORF1ab | ORF1ab | ORF1ab | ORF1ab | ORF1ab | ORF1ab | S | |
| Product | / | / | nsp2 | nsp2 | nsp3 | nsp2 | spike | |
| Codon number | / | / | 765 | 739 | 1626 | 352 | 845 | |
| Country | SS2B | |||||||
| AT | 3 | 3 | 2 | 0 | 0 | 0 | 0 | 0 |
| BE | 7 | 7 | 7 | 0 | 0 | 0 | 0 | 0 |
| BA | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| FI | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| FR | 10 | 8 | 1 | 7 | 7 | 0 | 0 | 7 |
| DE | 2 | 2 | 0 | 2 | 2 | 0 | 0 | 0 |
| GR | 7 | 7 | 3 | 3 | 3 | 0 | 0 | 0 |
| IS | 57 | 57 | 54 | 3 | 3 | 33 | 12 | 0 |
| IE | 2 | 2 | 1 | 1 | 1 | 0 | 0 | 0 |
| IT | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| LU | 2 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| NL | 30 | 30 | 26 | 2 | 2 | 0 | 0 | 0 |
| NO | 6 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| PT | 13 | 12 | 2 | 9 | 9 | 0 | 0 | 3 |
| SI | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ES | 10 | 2 | 1 | 0 | 0 | 0 | 0 | 0 |
| SE | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 0 |
| CH | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| UK | 115 | 115 | 50 | 57 | 49 | 0 | 0 | 0 |
| Tot | 276 | 253 | 151 | 85 | 77 | 33 | 12 | 1 |
3.3.5. SS4
The SS4 cluster carried the signature mutation C241T, C3037T and A23403G. The first two mutations were silent mutations harbored in the 5’UTR region and the ORF1ab gene respectively, while the last resulted in the AA substitution D > G in the spike protein (Becerra-Flores and Cardozo, 2020).
This cluster was found in all EU/EAA (Fig. 3) with different frequencies, and included 2425 genomes out of 3289 (75%).
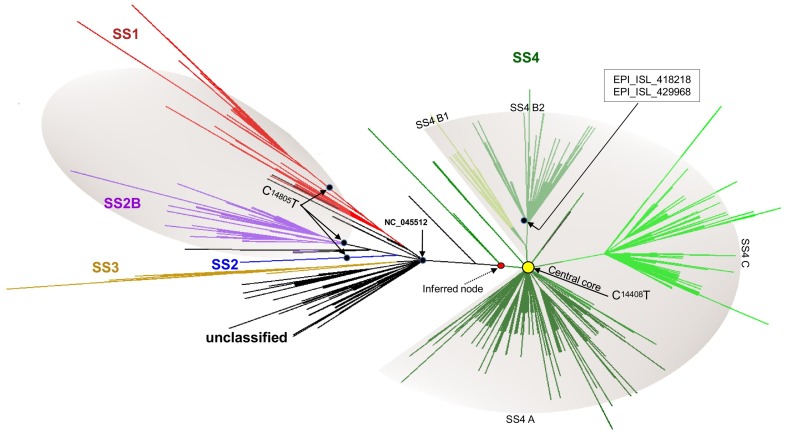
The first detection of the SS4 profile in Europe dates back to January 28th, 2020 in Bavaria, Germany (EPI_ISL_406862) (Rothe et al., 2020). This genome was found in a small cluster (Wölfel et al., 2020) from which arose a further sub-cluster that harbored the missense mutation G6446A (AA change V > I, nsp3 protein) (Supplementary Fig. S8, Supplementary Fig. S1). The ML analysis revealed that these German clusters arose from an inferred node from which the virus evolved mainly in a different direction through the acquisition of the missense mutation C14408T (AA change P > L, nsp12 protein). This epidemiological scenario is depicted in Fig. 4 which represents the European phylogenetic tree in radial form (unrooted).
Fig. 4.
Phylogenetic tree of the entire dataset in radial/collapsed form (unrooted). The picture shows the unrooted tree resulting from the ML analysis built from the entire dataset visualized in radial form. The main clusters (SS) are shown in different colors. The relative entry point of the C14805T and C14408T mutations in the tree, their area of influence (shaded area), the inferred node from which evolved the SS4 cluster and the central node at the root of the SS4 cluster (filled yellow circle) are shown. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The SS4 cluster with the acquired C14408T mutation only was found in 81 genomes gathered in a central node of the phylogenetic tree. These genomes were mainly from UK (n = 16), Italy (n = 14), Belgium (n = 10) and Sweden (n = 9). From the central node the virus evolved in three main groups, here named as SS4A, SS4B and SS4C (Fig. 4). Overall, the mutation C14408T was found in 2405 genomes out of 2425 of the SS4 profile.
SS4A included 905 genomes and comprised at least five groups, each one characterized by the following mutations: the silent C15324T, mainly harbored in western Europe; the silent A20268G, relevant in Spain and Iceland, (it is noteworthy that this mutation, with the exception of two genomes, was partially associated with the A10323G which resulted in the AA change K > R in the nsp5 protein, and exclusively detected in Iceland); the A26530G (AA change D > G, membrane protein) detected mainly in Belgium, Iceland, Sweden and Italy; the A24077G (AA change D > Y, spike protein) mostly identified in Portugal; the A187G (5’UTR) mostly identified in Luxembourg. The two associated mutations A12790G and C13568T, the latter resulting in the AA change A > V (nsp12 protein), were detected only in Sweden.
The SS4B group consisted of two subgroups widely distributed in Europe with the exception of western Europe and Ireland. The first sub-group included 83 genomes (SS4B1) of which 77 harbored the missense mutation G25563T (AA change Q > H, ORF3a protein). The silent C2416T mutation was found associated with this subgroup. The second sub-group (SS4B2) included 537 genomes and, with the exception of 6, evolved from the first by the acquisition of C1059T mutation, which resulted in the AA change T > I in the nsp2 protein. This second sub-group in Denmark represented the 72% of the dataset.
SS4C consisted of 814 genomes, of which 801 harbored the three variations A28881G, A28882G and G28883C. These three associated mutations resulted in two AA substitutions (RG > KR in the nucleocapsid protein). The missense mutation C27046 T (AA change T > M, membrane protein), with the exception of one genome, was fully associated with the triplet. Another associated mutation was the G12832A, detected exclusively in genomes from Austria.
Table 5 reports all the relevant mutations belonging to the SS4 cluster.
Table 5.
SS4 clusters and additional mutations. On the bottom left column, the absolute harbored SS4 genomes number by countries are shown. On the top, additional mutations, their genomic positions, the effect on protein translation and the affected codons are reported.
| SS4 signature C241T, C3037T, A23403G | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Additional mutations bp position | 14408 | 28881 | 28882 | 28883 | 25563 | 1059 | 27046 | 20268 | 15324 | 26530 | 10323 | 29734 | 24862 | 2416 | 12832 | 23731 | 187 | 24077 | 10097 | 25688 | 6446 | 12790 | 13568 | |
| ancestral/mutate | C/T | G/A | G/A | G/C | G/T | C/T | C/T | A/G | C/T | A/G | A/G | G/C | A/G | C/T | G/A | C/T | A/G | G/T | G/A | C/T | G/A | A/G | C/T | |
| AA change | P > L | RG > KR | Q > H | T > I | T > M | / | / | D > G | K > R | / | / | / | / | / | / | D > Y | G > S | A > V | V > I | / | A > V | |||
| gene | ORF1ab | N | ORF3a | ORF1ab | M | ORF1ab | ORF1ab | M | ORF1ab | 3’utr | S | ORF1ab | ORF1ab | S | 5’utr | S | ORF1ab | ORF3a | ORF1ab | ORF1ab | ORF1ab | |||
| product | nsp12 | nucleocaps. | orf3a | nsp2 | memb | / | / | memb | nsp5a | / | / | / | / | / | / | spike | nsp5 | orf3a | nsp3 | nsp9 | nsp12 | |||
| Codon number | 4715 | 203–204 | 57 | 265 | 175 | / | / | 3 | 3353 | / | / | / | / | / | / | 839 | 3278 | 99 | 2061 | / | 4435 | |||
| Country | SS4 | |||||||||||||||||||||||
| AT | 150 | 150 | 70 | 70 | 70 | 50 | 49 | 19 | 14 | 1 | 0 | 0 | 5 | 3 | 0 | 41 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| BE | 151 | 151 | 62 | 62 | 62 | 19 | 12 | 17 | 1 | 28 | 16 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 |
| BA | 10 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| HR | 3 | 3 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| CZ | 2 | 2 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| DK | 241 | 240 | 21 | 21 | 21 | 190 | 188 | 8 | 7 | 0 | 0 | 0 | 3 | 4 | 0 | 0 | 3 | 1 | 0 | 3 | 0 | 0 | 0 | 0 |
| FI | 17 | 17 | 3 | 3 | 3 | 8 | 2 | 3 | 2 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| FR | 180 | 180 | 11 | 11 | 11 | 101 | 65 | 2 | 1 | 61 | 0 | 1 | 0 | 0 | 28 | 0 | 0 | 2 | 0 | 0 | 7 | 0 | 0 | 0 |
| DE | 78 | 63 | 29 | 29 | 29 | 25 | 25 | 11 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 10 | 0 | 0 |
| GR | 54 | 53 | 44 | 44 | 44 | 6 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| HU | 12 | 12 | 6 | 6 | 6 | 2 | 2 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| IS | 256 | 256 | 76 | 76 | 76 | 53 | 52 | 42 | 68 | 5 | 15 | 58 | 3 | 8 | 1 | 0 | 6 | 5 | 4 | 6 | 3 | 0 | 0 | 0 |
| IE | 7 | 7 | 5 | 5 | 5 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| IT | 100 | 100 | 40 | 40 | 40 | 1 | 0 | 3 | 5 | 0 | 12 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 1 | 0 | 0 | 0 | 0 | 0 |
| LV | 3 | 3 | 2 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| LT | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| LU | 64 | 64 | 2 | 2 | 2 | 25 | 25 | 0 | 2 | 17 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 9 | 0 | 0 | 12 | 0 | 0 | 0 |
| NL | 217 | 217 | 79 | 79 | 79 | 35 | 33 | 62 | 15 | 1 | 5 | 0 | 13 | 28 | 0 | 0 | 2 | 5 | 5 | 2 | 8 | 0 | 0 | 0 |
| NO | 13 | 13 | 2 | 2 | 2 | 5 | 5 | 0 | 1 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| PL | 6 | 6 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| PT | 143 | 143 | 77 | 77 | 77 | 7 | 3 | 3 | 11 | 1 | 3 | 1 | 3 | 0 | 2 | 0 | 4 | 0 | 25 | 4 | 1 | 0 | 0 | 0 |
| RU | 35 | 35 | 17 | 17 | 17 | 7 | 6 | 6 | 1 | 1 | 4 | 0 | 0 | 0 | 1 | 0 | 3 | 1 | 0 | 3 | 0 | 0 | 0 | 0 |
| SK | 3 | 3 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| SI | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ES | 121 | 118 | 15 | 15 | 15 | 5 | 5 | 0 | 84 | 0 | 0 | 0 | 25 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| SE | 223 | 223 | 106 | 106 | 106 | 37 | 33 | 42 | 1 | 0 | 10 | 0 | 0 | 0 | 4 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 18 | 18 |
| CH | 75 | 75 | 31 | 31 | 31 | 5 | 5 | 0 | 5 | 25 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| TR | 3 | 3 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| UK | 256 | 256 | 100 | 100 | 100 | 21 | 19 | 20 | 15 | 2 | 4 | 0 | 4 | 0 | 5 | 0 | 18 | 5 | 4 | 16 | 0 | 0 | 0 | 0 |
| Tot | 2425 | 2405 | 801 | 801 | 801 | 608 | 531 | 241 | 239 | 146 | 81 | 60 | 58 | 47 | 45 | 41 | 40 | 39 | 39 | 38 | 35 | 10 | 18 | 18 |
3.3.6. Unclassified and ambiguous genomes
Those genomes that did not fall within the adopted classification were allocated into two groups named “unclassified” and “ambiguous”. In the phylogenetic trees, genomes with mixed signature motifs (ambiguous n = 72) were located in blind evolutive tips.
The genomes lacking the signature motif (unclassified n = 312) lie in the phylogenetic trees in several small clusters close to the reference sequence (an. NC_045512). These clusters were mainly represented by four mutations. The silent T514C mutation was mostly represented in genomes from the Netherlands (20%) and found partially associated with the missense mutation C17410T (AA change R > C, nsp13 protein).
The missense mutations G1440A (AA change G > D, nsp2 protein) and G2891A (AA change A > T, nsp3 protein) were found fully associated with the exception of five genomes. In Denmark and in Sweden the two latter mutations were found fully associated with the missense mutation C7011T (A > V, nsp3 protein).
In Table 6 , the main mutations belonging to the unclassified cluster are reported.
Table 6.
Unclassified clusters and additional mutations. On the bottom left column, the absolute harbored unclassified genomes number by countries are shown. On the top, additional mutations, their genomic positions, the effect on protein translation and the affected codons are reported.
| SS Unclassified | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Additional mutations bps position | 1440 | 514 | 2891 | 17410 | 7011 | 28851 | 20270 | 6696 | 11001 | 29683 | |
| ancestal/mutate | G/A | T/C | G/A | C/T | C/T | G/T | C/T | C/T | C/T | A/T | |
| AA change | G > D | / | A > T | R > C | A > V | S > I | A > V | P > L | T > I | / | |
| Gene | ORF1ab | ORF1ab | ORF1ab | ORF1ab | ORF1ab | N | ORF1ab | ORF1ab | ORF1ab | 3’utr | |
| Product | nsp2 | nsp1 | nsp3 | nsp13 | nsp3 | Nucleocap | nsp15 | nsp3 | nsp6 | / | |
| Codon number | 392 | / | 876 | 5716 | 2249 | 193 | 6669 | 2144 | 3579 | / | |
| Country | Unclassified | ||||||||||
| AT | 6 | 3 | 2 | 1 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| BE | 9 | 9 | 0 | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| DK | 16 | 13 | 2 | 13 | 1 | 13 | 0 | 0 | 0 | 0 | 0 |
| FI | 2 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| FR | 11 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| DE | 14 | 10 | 0 | 10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| GR | 4 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| IS | 19 | 7 | 1 | 7 | 0 | 1 | 0 | 4 | 0 | 0 | 2 |
| IE | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| NL | 81 | 6 | 69 | 6 | 38 | 1 | 0 | 0 | 0 | 0 | 0 |
| NO | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| PL | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| PT | 4 | 2 | 1 | 2 | 0 | 0 | 0 | 1 | 0 | 0 | 1 |
| RU | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| SK | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ES | 3 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| SE | 9 | 7 | 0 | 7 | 0 | 7 | 0 | 0 | 0 | 0 | 0 |
| UK | 129 | 32 | 13 | 30 | 2 | 1 | 24 | 15 | 17 | 17 | 8 |
| Tot | 312 | 93 | 92 | 89 | 48 | 26 | 24 | 20 | 17 | 17 | 11 |
3.4. Estimates of average mutation rates in SS clusters
The average mutation rate in the entire dataset has been estimated including a period of 126 days, from the time of the inferred appearance of SARS-CoV-2 (November 25th–28th, 2019) (Liu et al., 2020; Benvenuto et al., 2020) until March 22nd, 2020.
The average mutation rate was determined to be 6.3 × 10−4 (sub/site/year), (SE 5.7 × 10−5) in agreement to that already reported (6-8 × 10−4 sub/site/year) (Liu et al., 2020; van Dorp et al., 2020).
The mutation rate was also estimated in the subset of genomes which acquired the C14408T mutation in the SS4 cluster and in the subset of genomes which acquired the C14805T mutation in SS1 and SS2B clusters. Relating to the appearance of C14408T mutation in SS4 cluster (February 20th, 2020), the average mutation rate was 1.2 × 10−3 (SE 2.7 × 10−4) sub/site/year. Relating to the appearance of C14805T in SS1 (February 25th, 2020) and SS2B clusters (February 27th, 2020) the average mutation rates were 1.5 × 10−3 (SE 6 × 10−4) and 1.2 × 10−3 (SE 3.4 × 10−4) sub/site/year, respectively.
3.5. Statistical analysis
Table 7A, 7B and 7C show the results of the Chi-square test performed to evaluate the frequency distribution of clusters in the different EU/EEA countries Table 7. The SS1 cluster was found positively associated to Spain (p < 0.00001) and negatively correlated to the UK (p < 0.00001) and France (p < 0.00001). The SS2B cluster was found positively associated to the UK (p < 0.00001) and Norway (p < 0.05) and negatively associated to France (p < 0.002) and Switzerland (p < 0.05). The SS4 cluster was found positively associated to Denmark (p < 0.02) and negatively correlated to the UK (p < 0.0005) and Spain (p < 0.01).
Table 7.
| A. Contingency table for SS1 cluster by countries | |||||
|---|---|---|---|---|---|
| Countries | Cluster |
Total | S.R |
||
| SS1 | Others | SS1 | |||
| UK | Observed | 7 | 518 | 525 | (−) p < 0.00001 |
| Spain | Observed | 113 | 152 | 265 | (+) p < 0.00001 |
| France | Observed | 2 | 233 | 265 | (−) p < 0.00001 |
| Total | 122 | 893 | 1015 | ||
| χ2test | |||||
| Value | df | p | Cramer's V | ||
| χ2 | 318 | 2 | <0.00001 | 0.560 | |
| B. Contingency table for SS2B cluster by countries | |||||
|---|---|---|---|---|---|
| Countries | Cluster |
Total | S.R |
||
| SS2B | Others | SS2B | |||
| UK | Observed | 115 | 410 | 525 | (+) p < 0.00001 |
| Spain | Observed | 10 | 255 | 265 | (−) p < 0.0002 |
| France | Observed | 10 | 215 | 225 | (−) p < 0.002 |
| Netherland | Observed | 30 | 313 | 343 | n.s |
| Italy | Observed | 7 | 103 | 110 | n.s |
| Norway | Observed | 6 | 15 | 21 | (+) p < 0.05 |
| Switzerland | Observed | 1 | 77 | 78 | (−) p < 0.05 |
| Total | 179 | 1388 | 1567 | ||
| χ2test | |||||
| Value | df | p | Cramer's V | ||
| χ2 | 102 | 6 | <0.00001 | 0.256 | |
| C. Contingency table for SS4 cluster by countries | |||||
|---|---|---|---|---|---|
| Countries | Cluster |
Total | S.R |
||
| SS4 | Others | SS4 | |||
| UK | Observed | 256 | 269 | 525 | (−) p < 0.0005 |
| Netherland | Observed | 217 | 126 | 343 | n.s |
| Denmark | Observed | 241 | 19 | 260 | (+) p < 0.02 |
| Spain | Observed | 121 | 144 | 265 | (−) p < 0.01 |
| France | Observed | 180 | 45 | 225 | n.s |
| Belgium | Observed | 151 | 22 | 173 | n.s |
| Austria | Observed | 150 | 15 | 165 | n.s |
| Italy | Observed | 100 | 10 | 110 | n.s |
| Germany | Observed | 78 | 21 | 99 | n.s |
| Switzerland | Observed | 75 | 3 | 78 | n.s |
| Luxembourg | Observed | 64 | 4 | 68 | n.s |
| Greece | Observed | 54 | 16 | 70 | n.s |
| Total | 1687 | 694 | 2381 | ||
| χ2test | |||||
| Value | df | p | Cramer's V | ||
| χ2 | 470 | 11 | <0.00001 | 0.413 | |
Contingency Tables 7A, B, C for SS clusters by countries. At the bottom of each table, the Chi-square analysis related to the specific cluster. On the right column, the single country contribution to the Chi square value. S.R., = standardized residual; (+) = positive association; (−) = negative association; n.s = not significant.
4. Discussion
The aim of the study was to describe the mutational spectrum of SARS-CoV-2 virus during the first phase of the pandemic. Sequence alignment detected 41 mutations with a frequency ≥ 1%. The mutations were distributed over the entire length of the viral genome, with the exception of the coding genes for the Envelope, the ORF6, ORF7a, ORF7b, and the ORF10. The genetic conservation of these loci, with the exception of ORF10, is likely due to the important role of the encoded products in the life cycle of SARS-CoV-2 and, in particular, in the early phase of infection, when they act as immunosuppressors, contributing to escape the immune response (Yuen et al., 2020; Addetia et al., 2020; Gordon et al., 2020). Taking into account the short dimensions of these ORFs, however, the low number of mutations due to chance cannot be ruled out. Two new deletions were detected in two genomes from the UK, one of which involved 324 bps, resulting in a stop codon in the nsp14 protein.
Maximum likelihood analysis revealed six main clusters, of which four have been already described (SS1, SS2, SS3, SS4) (Yang et al., 2020). The phylogenetic trees suggest that one additional cluster, here named SS2B, arose from the fusion of two clusters, SS2 and SS3. An additional set of small clusters that did not fall in the adopted classification (unclassified) were also identified. Genomes carrying mixed mutation profiles were defined as “ambiguous.”
The inspection of the phylogenetic trees highlights that in some countries, such as the UK, France and Iceland, several genomes were identical to the Chinese reference genome (an. NC_045512). Furthermore, the unclassified clusters originated from these strains. The mutation rate in some nodes of these clusters (3 × 10−5 sub/site/year, Supplementary Fig. S1) suggests that they originated from an in-situ evolution. Noteworthy, important mutations contributing to the evolutionary success to some strains (i.e., with A23403G) were not present in these unclassified clusters.
The SS1 cluster was detected in several European countries. The dataset reveals its occurrence on January 29th 2020 in the UK (EPI_ISL_407071, EPI_ISL_407073). The small number of descendants indicates an evolutionary failure of this cluster in Europe (Fig. 3), with the exception of Spain and Iceland. In Spain, a high frequency of SS1 genomes was found (Table 1, Supplementary Fig. S18). This cluster accounted for about 43% of the entire Spanish dataset and evolved in two distinct subclusters. In Iceland, a SS1 cluster which harbored a different mutational profile from the Spanish cluster was identified (Table 1, Supplementary Fig. S10).
As already reported (Yang et al., 2020), the SS2 cluster did not spread successfully in the EU/EEA; however, the phylogenetic analysis suggests that this cluster evolved in the SS2B through the acquisition of the G11083T mutation (Supplementary Fig. S1).
The SS2B cluster spread efficiently through the EU/EEA, with the exception of Turkey, Russia, Denmark, and East Europe. It was first detected in Italy on January 29th, 2020 (EPI_ISL_412974, EPI_ISL_410546). Interestingly, the two samples were collected from two Chinese tourists at the end of January (Albarello et al., 2020), suggesting that the SS2B cluster evolved in China. Moreover, these two samples were already analyzed in other phylogenetic studies and did not show descendants (Giovanetti et al., 2020; Stefanelli et al., 2020; Lai et al., 2020). The phylogenetic analysis showed that these two genomes are at the root of the SS2B cluster that represented the origin of all the European genomes belonging to this cluster (Supplementary Fig. S1).
With only one exception, SS3 was not recovered in the EU/EEA. The exception was Turkey, where the SS3 cluster was predominant, representing 40% of the dataset.
Following its first detection in Europe (EPI_ISL_406862) (Rothe et al., 2020; Wölfel et al., 2020), the SS4 cluster evolved early, acquiring the mutation C14408T (Lai et al., 2020; Stefanelli et al., 2020). The SS4 genomes carrying this mutation only were found gathered in a central node of the phylogenetic tree and the first genome was detected on February 20th, in Italy (EPI_ISL_412973). From this node, the virus spread throughout the EU/EEA, evolving in three main groups, here named SS4A, SS4B and SS4C (Fig. 4).
On February 21st, 2020, two genomes collected in France that harbored the C14408T, already carried two additional mutations, the C1059T and the G25563T (EPI_ISL_418218, EPI_ISL_429968). These two genomes lie in the SS4B group and represent evolved strains arising from the central node (Fig. 4). On February 26th, 2020, additional genomes from France, carrying the same profile, were detected (EPI_ISL_414625, EPI_ISL_416502).
Taking into account the mutational rate of the SS4 cluster (1.,2 × 10–3, sub/site/year), the first appearance of C14408T mutation may be estimated at molecular level, to February 1st, 2020 (IC95 ± 9).
This hypothesis is further supported by the analysis of the SS4C cluster. This cluster harbored the triple A28881G, A28882G and G28883C mutations and, as well as the SS4B, it represents a further step in the evolution of the SS4 cluster. The first SS4C genome was detected in Austria on February 24th, 2020 (EPI_ISL_437932), then in Germany on February 25th (EPI_ISL_412912) and again in Austria and Denmark on February 26th, 2020.
It is therefore possible to hypothesize that, relative to the first detection in Europe, the SS4 cluster was already evolving in a different direction (inferred node, Fig. 4) through the acquisition of the C14408T mutation. It is not possible to assess where this happened. At the beginning of February, the virus, with this mutation, was present in several European countries; this is supported by the high number of SS4 genomes detected in several European countries that carried the additional C14408T mutation only (Fig. 4, central node). Then the virus evolved into two directions: the first one, through the acquisition of the C1059T and later of G25563T (SS4B), and another one with the triplet A28881G, A28882G and G28883C (SS4C). Other mutations occurred later, making up the SS4A group.
Most likely, in the initial stage, the virus was not detected due to mild or non-specific COVID-19 clinical signs.
The C14408T and the C14805T mutations could have played an important role in the evolution of SARS-CoV-2. The success of the SS4 cluster is linked to the acquisition of the C14408T (Mercatelli and Giorgi, 2020; Yuan et al., 2020); moreover, about 56% of SNVs with frequency ≥ 1% were related to this cluster, suggesting an acceleration of the mutation rate.
Similarly, the successful spread of the SS1 cluster in Spain appears to be linked to the acquisition of the C14805T mutation. The role of this mutation is also confirmed by the acquisition and positive selection encountered in different clusters. Indeed, C14805T mutation was found in SS2 cluster as well as in almost all the genomes belonging to the SS2B cluster.
The mutation rate of SARS-CoV-2 was estimated to be 6.2 × 10−4 sub/site/year; after the acquisition of C14408T and C14805T mutations, the mutation rates in SS4, SS1, and SS2B clusters were estimated to be double, with rates of 1.24 × 10−3, 1.48 × 10−3 and 1.25 × 10−3 sub/site/year values, respectively.
The C14408T and C14805T mutations fall in a region of the ORF1ab gene whose product is the nsp12 protein, the RNA-dependent RNA polymerase (RdRp). The exact role of these two mutations is difficult to explain. The C14805T is a silent mutation that could act to enhance the role of a cis-domain or to act specifically as a cis-site. Conversely, the C14408T resulted in the amino acid substitution P > L in the RdRp. This substitution falls in the interface domain that links the NiRAN domain to a subdomain of the RdRp. Such substitution might change the interactions between the two domains of RdRp and/or determine different interactions with the nsp8 or nsp14 (Peng et al., 2020; Romano et al., 2020).
The estimated mutation rate in each SS cluster was the resulting average from a highly heterogeneous group of samples. In each cluster, several samples carrying the specific mutations showed a very high mutation rate, highlighting a differential virus-host interaction.
The SS1 cluster included genomes more commonly detected in Spain, while a reverse correlation was found with the UK and France. Similarly, the SS2B cluster was found associated to UK and Norway while it was negatively correlated to France and Switzerland. The SS4 cluster evolved in a mutational profile that spread widely in all Europe. However, data analysis found a positive association to Denmark while negative correlations to the UK and Spain. Furthermore, several mutations in this cluster were restricted to specific countries (e.g., A12790G/C13568T found in genomes from Sweden, G12832A from Austria). The SS3 cluster did not meet the criteria for inclusion in the statistical analysis, however, a visual inspection of data immediately highlights a failure of the SS3 cluster in Europe while it was the predominant cluster in Turkey. Similarly, the SS1 cluster has spread with some success in Iceland, but there the cluster hosted a mutational profile completely different from that seen in Spain (Table 1).
A different frequency of missense mutations as well as the tendency to host different mutations among SARS-CoV-2 strains circulating in different continents has been already described (Islam et al., 2020). Recently, a significant association between SARS-CoV-2 mutations and their geographic origin has been reported (Goyal et al., 2021). In agreement with these studies, our data supports the hypothesis that an important contribution to clusters distribution is related to the geographic location of the outbreak. Anthropological studies suggest different susceptibility among the European population (Sanchez-Mazas et al., 2013). Geographical clines were mainly observed for HLA-A, B and C haplotypes along the European South-Southeast to North-Northwest axes, with the greatest difference found at geographic boundaries. Furthermore, a recent study showed a haplotypic HLA diversity in the Spanish population compared to the Northern European population (Montero-Martín et al., 2019). Overall, the data suggest that the mechanism underlying clusters distribution could be secondary to viral infection and related to the virus's ability to select a specific mutational profile compared to different HLA haplotypes. Therefore, as already observed in SARS-infected subjects in 2003 (Lin et al., 2003), it is possible to hypothesize that the immune escape strategy of SARS-CoV-2 might be to impair the antigen presentation by HLA class I molecules. This phenomenon was recently suggested to explain the different disease outcome following SARS-CoV-2 infection (Tomita et al., 2020).
Possible limits of this study could be due to the incompleteness of the dataset obtained from the genome available in GISAID. For this reason, a new high-quality dataset, obtained by retrieving the genomes available on GenBank that met the criteria used for the GISAID dataset, has been built. Cluster distribution and the relative frequencies were similar in both of the datasets (data not shown). Nevertheless, the limited number of genomes from some countries may not be adequately representative; furthermore, the sequence genomes uploaded into the databases represent a selection of those strains.
The authors of this article do believe that this molecular and epidemiological picture provides additional information that may be useful to help better characterize the first phase of the ongoing pandemic.
The following are the supplementary data related to this article.
Phylogenetic tree of the entire dataset. The full resolution image of the EU/EAA phylogram resulting from the ML analysis is shown. The signatures mutation (at the top-right) as well as the six main clusters are shown in different colors. The asterisk next to the nodes indicates supported branches. The mutation rate in each node is also shown.
The figure shows the phylogenetic tree resulting from the related dataset. The country name is provided at the top of the figure. The signatures mutation (at the top-right) and the six main clusters are shown in different colors. Nucleotide substitutions characterizing each cluster are shown. ABS = absence of substitution.
The figure shows the phylogenetic tree resulting from the related dataset. The country name is provided at the top of the figure. The signatures mutation (at the top-right) and the six main clusters are shown in different colors. Nucleotide substitutions characterizing each cluster are shown. ABS = absence of substitution.
The figure shows the phylogenetic tree resulting from the related dataset. The country name is provided at the top of the figure. The signatures mutation (at the top-right) and the six main clusters are shown in different colors. Nucleotide substitutions characterizing each cluster are shown. ABS = absence of substitution.
The figure shows the phylogenetic tree resulting from the related dataset. The country name is provided at the top of the figure. The signatures mutation (at the top-right) and the six main clusters are shown in different colors. Nucleotide substitutions characterizing each cluster are shown. ABS = absence of substitution.
The figure shows the phylogenetic tree resulting from the related dataset. The country name is provided at the top of the figure. The signatures mutation (at the top-right) and the six main clusters are shown in different colors. Nucleotide substitutions characterizing each cluster are shown. ABS = absence of substitution.
The figure shows the phylogenetic tree resulting from the related dataset. The country name is provided at the top of the figure. The signatures mutation (at the top-right) and the six main clusters are shown in different colors. Nucleotide substitutions characterizing each cluster are shown. ABS = absence of substitution.
The figure shows the phylogenetic tree resulting from the related dataset. The country name is provided at the top of the figure. The signatures mutation (at the top-right) and the six main clusters are shown in different colors. Nucleotide substitutions characterizing each cluster are shown. ABS = absence of substitution.
The figure shows the phylogenetic tree resulting from the related dataset. The country name is provided at the top of the figure. The signatures mutation (at the top-right) and the six main clusters are shown in different colors. Nucleotide substitutions characterizing each cluster are shown. ABS = absence of substitution.
The figure shows the phylogenetic tree resulting from the related dataset. The country name is provided at the top of the figure. The signatures mutation (at the top-right) and the six main clusters are shown in different colors. Nucleotide substitutions characterizing each cluster are shown. ABS = absence of substitution.
The figure shows the phylogenetic tree resulting from the related dataset. The country name is provided at the top of the figure. The signatures mutation (at the top-right) and the six main clusters are shown in different colors. Nucleotide substitutions characterizing each cluster are shown. ABS = absence of substitution.
The figure shows the phylogenetic tree resulting from the related dataset. The country name is provided at the top of the figure. The signatures mutation (at the top-right) and the six main clusters are shown in different colors. Nucleotide substitutions characterizing each cluster are shown. ABS = absence of substitution.
The figure shows the phylogenetic tree resulting from the related dataset. The country name is provided at the top of the figure. The signatures mutation (at the top-right) and the six main clusters are shown in different colors. Nucleotide substitutions characterizing each cluster are shown. ABS = absence of substitution.
The figure shows the phylogenetic tree resulting from the related dataset. The country name is provided at the top of the figure. The signatures mutation (at the top-right) and the six main clusters are shown in different colors. Nucleotide substitutions characterizing each cluster are shown. ABS = absence of substitution.
The figure shows the phylogenetic tree resulting from the related dataset. The country name is provided at the top of the figure. The signatures mutation (at the top-right) and the six main clusters are shown in different colors. Nucleotide substitutions characterizing each cluster are shown. ABS = absence of substitution.
The figure shows the phylogenetic tree resulting from the related dataset. The country name is provided at the top of the figure. The signatures mutation (at the top-right) and the six main clusters are shown in different colors. Nucleotide substitutions characterizing each cluster are shown. ABS = absence of substitution.
The figure shows the phylogenetic tree resulting from the related dataset. The country name is provided at the top of the figure. The signatures mutation (at the top-right) and the six main clusters are shown in different colors. Nucleotide substitutions characterizing each cluster are shown. ABS = absence of substitution.
The figure shows the phylogenetic tree resulting from the related dataset. The country name is provided at the top of the figure. The signatures mutation (at the top-right) and the six main clusters are shown in different colors. Nucleotide substitutions characterizing each cluster are shown. ABS = absence of substitution.
The figure shows the phylogenetic tree resulting from the related dataset. The country name is provided at the top of the figure. The signatures mutation (at the top-right) and the six main clusters are shown in different colors. Nucleotide substitutions characterizing each cluster are shown. ABS = absence of substitution.
The figure shows the phylogenetic tree resulting from the related dataset. The country name is provided at the top of the figure. The signatures mutation (at the top-right) and the six main clusters are shown in different colors. Nucleotide substitutions characterizing each cluster are shown. ABS = absence of substitution.
The figure shows the phylogenetic tree resulting from the related dataset. The country name is provided at the top of the figure. The signatures mutation (at the top-right) and the six main clusters are shown in different colors. Nucleotide substitutions characterizing each cluster are shown. ABS = absence of substitution.
The figure shows the phylogenetic tree resulting from the related dataset. The country name is provided at the top of the figure. The signatures mutation (at the top-right) and the six main clusters are shown in different colors. Nucleotide substitutions characterizing each cluster are shown. ABS = absence of substitution.
In the table the codes of the genome sequences retrieved from GISAID are listed.
Ethical approval
The viral RNA genome sequencing at ISS was approved by the ISS Ethical Committee (Prot. PRE BIO CE n.26259-29/07/2020-ISS).
Funding
This study was funded by the Italian Ministry of Defense.
Declaration of Competing Interest
None.
Acknowledgments
We gratefully acknowledge the Authors, the Originating and Submitting Laboratories for their sequences and metadata shared through GISAID (www.gisaid.org). The authors thank: Giulietta Venturi, Claudia Fortuna, Antonello Amendola, Giulia Marsili, Eleonora Benedetti, Laura Calzoletti, Giuseppina Di Mario, Dept. InfectiousDiseases, Istituto Superiore di Sanità, Rome, Italy; Gabriele Vaccari, Luca De Sabato, Ilaria Di Bartolo, Giovanni Ianiro, Giovanni Ianiro, Dept. of Food Safety, Nutrition and Veterinary Public Health, Istituto Superiore di Sanità, Rome, Italy; COVID-19 Italian Army Medical Center Study Group: Gaetano Alfano, Alessandra Amoroso, Anna Anselmo, Manfredo Bortone, Giandomenico Cerreto, Andrea Ciammaruconi, Angelo De Domenico, Nino D’Amore, De Santis Riccardo, Giovanni Faggioni, Silvia Fillo, Antonella Fortunato, Valeria Franchini, Francesco Giordani, Florigio Lista, Filippo Molinari, Anella Monte, Lucia Nicosia, Giancarlo Petralito, Maria Anna Spinelli, Stefano Palomba, Elisa Regarbuto, Vanessa Vera Fain.
References
- Addetia A., Xie H., Roychoudhury P., Shrestha L., Loprieno M., Huang M.L., Jerome K.R., Greninger A.L. Identification of multiple large deletions in ORF7a resulting in in-frame gene fusions in clinical SARS-CoV-2 isolates. J. Clin. Virol. 2020 Aug;129:104523. doi: 10.1016/j.jcv.2020.104523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albarello F., Pianura E., Di Stefano F., Cristofaro M., Petrone A., Marchioni L., Palazzolo C., Schininà V., Nicastri E., Petrosillo N., Campioni P., Eskild P., Zumla A. Ippolito G; COVID 19 INMI study group. 2019-novel coronavirus severe adult respiratory distress syndrome in two cases in Italy: an uncommon radiological presentation. Int. J. Infect. Dis. 2020 Apr;93:192–197. doi: 10.1016/j.ijid.2020.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becerra-Flores M., Cardozo T. SARS-CoV-2 viral spike G614 mutation exhibits higher case fatality rate. Int. J. Clin. Pract. 2020 Aug;74(8) doi: 10.1111/ijcp.13525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedetti F., Snyder G.A., Giovanetti M., Angeletti S., Gallo R.C., Ciccozzi M., Zella D. Emerging of a SARS-CoV-2 viral strain with a deletion in nsp1. J. Transl. Med. 2020 Aug 31;18(1):329. doi: 10.1186/s12967-020-02507-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benvenuto D., Giovanetti M., Salemi M., Prosperi M., De Flora C., Junior Alcantara L.C., Angeletti S., Ciccozzi M. The global spread of 2019-nCoV: a molecular evolutionary analysis. Pathog. Glob. Health. 2020 Mar;114(2):64–67. doi: 10.1080/20477724.2020.1725339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel Y., Mizrahi O., Nachshon A., Weingarten-Gabbay S., Morgenstern D., Yahalom-Ronen Y., Tamir H., Achdout H., Stein D., Israeli O., Beth-Din A., Melamed S., Weiss S., Israely T., Paran N., Schwartz M., Stern-Ginossar N. The coding capacity of SARS-CoV-2. Nature. 2021 Jan;589(7840):125–130. doi: 10.1038/s41586-020-2739-1. [DOI] [PubMed] [Google Scholar]
- Giovanetti M., Benvenuto D., Angeletti S., Ciccozzi M. The first two cases of 2019-nCoV in Italy: where they come from? J. Med. Virol. 2020 May;92(5):518–521. doi: 10.1002/jmv.25699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O’Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., Tummino T.A., Hüttenhain R., Kaake R.M., Richards A.L., Tutuncuoglu B., Foussard H., Batra J., Haas K., Modak M., Kim M., Haas P., Polacco B.J., Braberg H., Fabius J.M., Eckhardt M., Soucheray M., Bennett M.J., Cakir M., MJ McGregor, Li Q., Meyer B., Roesch F., Vallet T., Mac Kain A., Miorin L., Moreno E., ZZC Naing, Zhou Y., Peng S., Shi Y., Zhang Z., Shen W., Kirby I.T., Melnyk J.E., Chorba J.S., Lou K., Dai S.A., Barrio-Hernandez I., Memon D., Hernandez-Armenta C., Lyu J., CJP Mathy, Perica T., Pilla K.B., Ganesan S.J., Saltzberg D.J., Rakesh R., Liu X., Rosenthal S.B., Calviello L., Venkataramanan S., Liboy-Lugo J., Lin Y., Huang X.P., Liu Y., Wankowicz S.A., Bohn M., Safari M., Ugur F.S., Koh C., Savar N.S., Tran Q.D., Shengjuler D., Fletcher S.J., O’Neal M.C., Cai Y., JCJ Chang, Broadhurst D.J., Klippsten S., Sharp P.P., Wenzell N.A., Kuzuoglu-Ozturk D., Wang H.Y., Trenker R., Young J.M., Cavero D.A., Hiatt J., Roth T.L., Rathore U., Subramanian A., Noack J., Hubert M., Stroud R.M., Frankel A.D., Rosenberg O.S., Verba K.A., Agard D.A., Ott M., Emerman M., Jura N., von Zastrow M., Verdin E., Ashworth A., Schwartz O., d’Enfert C., Mukherjee S., Jacobson M., Malik H.S., Fujimori D.G., Ideker T., Craik C.S., Floor S.N., Fraser J.S., Gross J.D., Sali A., Roth B.L., Ruggero D., Taunton J., Kortemme T., Beltrao P., Vignuzzi M., García-Sastre A., Shokat K.M., Shoichet B.K., Krogan N.J. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020 Jul;583(7816):459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal M., De Bruyne K., van Belkum A., West B. Different SARS-CoV-2 haplotypes associate with geographic origin and case fatality rates of COVID-19 patients. Infect. Genet. Evol. 2021 Jun;90:104730. doi: 10.1016/j.meegid.2021.104730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S., Dufayard J.F., Lefort V., Anisimova M., Hordijk W., Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 2010 May;59(3):307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Hadfield J., Megill C., Bell S.M., Huddleston J., Potter B., Callender C., Sagulenko P., Bedford T., Neher R.A. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018 Dec 1;34(23):4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam M.R., Hoque M.N., Rahman M.S., Alam A.S.M.R.U., Akther M., Puspo J.A., Akter S., Sultana M., Crandall K.A., Hossain M.A. Genome-wide analysis of SARS-CoV-2 virus strains circulating worldwide implicates heterogeneity. Sci. Rep. 2020 Aug 19;10(1):14004. doi: 10.1038/s41598-020-70812-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Rozewicki J., Yamada K.D. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019 Jul 19;20(4):1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Lee J.Y., Yang J.S., Kim J.W., Kim V.N., Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. 2020 May 14;181(4):914–921. doi: 10.1016/j.cell.2020.04.011. e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T., Platt D., Parida L. Variant analysis of SARS-CoV-2 genomes. Bull. World Health Organ. 2020 Jul 1;98(7):495–504. doi: 10.2471/BLT.20.253591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai A., Bergna A., Caucci S., Clementi N., Vicenti I., Dragoni F., Cattelan A.M., Menzo S., Pan A., Callegaro A., Tagliabracci A., Caruso A., Caccuri F., Ronchiadin S., Balotta C., Zazzi M., Vaccher E., Clementi M., Galli M., Zehender G. Molecular tracing of SARS-CoV-2 in Italy in the first three months of the epidemic. Viruses. 2020 Jul 24;12(8):798. doi: 10.3390/v12080798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with bowtie 2. Nat. Methods. 2012 Mar 4;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M., Tseng H.K., Trejaut J.A., Lee H.L., Loo J.H., Chu C.C., Chen P.J., Su Y.W., Lim K.H., Tsai Z.U., Lin R.Y., Lin R.S., Huang C.H. Association of HLA class I with severe acute respiratory syndrome coronavirus infection. BMC Med. Genet. 2003 Sep 12;4:9. doi: 10.1186/1471-2350-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Zhao S., Shi C.M., Song S., Zhu S., Su Y., Zhao W., Li M., Bao Y., Xue Y., Chen H. Population genetics of SARS-CoV-2: disentangling effects of sampling bias and infection clusters. Genom. Proteom. Bioinform. 2020 Jul 12 doi: 10.1016/j.gpb.2020.06.001. S1672–0229(20)30062–0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercatelli D., Giorgi F.M. Geographic and genomic distribution of SARS-CoV-2 mutations. Front. Microbiol. 2020 Jul 22;11:1800. doi: 10.3389/fmicb.2020.01800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel C.J., Mayer C., Poch O., Thompson J.D. Characterization of accessory genes in coronavirus genomes. Virol. J. 2020 Aug 27;17(1):131. doi: 10.1186/s12985-020-01402-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero-Martín G., Mallempati K.C., Gangavarapu S., Sánchez-Gordo F., Herrero-Mata M.J., Balas A., Vicario J.L., Sánchez-García F., González-Escribano M.F., Muro M., Moya-Quiles M.R., González-Fernández R., Ocejo-Vinyals J.G., Marín L., Creary L.E., Osoegawa K., Vayntrub T., Caro-Oleas J.L., Vilches C., Planelles D. Fernández-Viña MA. High-resolution characterization of allelic and haplotypic HLA frequency distribution in a Spanish population using high-throughput next-generation sequencing. Hum. Immunol. 2019 Jul;80(7):429–436. doi: 10.1016/j.humimm.2019.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Q., Peng R., Yuan B., Zhao J., Wang M., Wang X., Wang Q., Sun Y., Fan Z., Qi J., Gao G.F., Shi Y. Structural and biochemical characterization of the nsp12-nsp7-nsp8 Core polymerase complex from SARS-CoV-2. Cell Rep. 2020 Jun 16;31(11):107774. doi: 10.1016/j.celrep.2020.107774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A., Holmes E.C., O’Toole Á., Hill V., McCrone J.T., Ruis C., du Plessis L., Pybus O.G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020 Nov;5(11):1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano M., Ruggiero A., Squeglia F., Maga G., Berisio R. A structural view of SARS-CoV-2 RNA replication machinery: RNA synthesis, proofreading and final capping. Cells. 2020 May 20;9(5):1267. doi: 10.3390/cells9051267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothe C., Schunk M., Sothmann P., Bretzel G., Froeschl G., Wallrauch C., Zimmer T., Thiel V., Janke C., Guggemos W., Seilmaier M., Drosten C., Vollmar P., Zwirglmaier K., Zange S., Wölfel R., Hoelscher M. Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N. Engl. J. Med. 2020 Mar 5;382(10):970–971. doi: 10.1056/NEJMc2001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Mazas A., Buhler S., Nunes J.M. A new HLA map of Europe: regional genetic variation and its implication for peopling history, disease-association studies and tissue transplantation. Hum. Hered. 2013;76(3–4):162–177. doi: 10.1159/000360855. [DOI] [PubMed] [Google Scholar]
- Stefanelli P., Faggioni G., Lo Presti A., Fiore S., Marchi A., Benedetti E., Fabiani C., Anselmo A., Ciammaruconi A., Fortunato A., De Santis R., Fillo S., Capobianchi M.R., Gismondo M.R., Ciervo A., Rezza G., Castrucci M.R., Lista F., On Behalf of ISS Covid-Study Group Whole genome and phylogenetic analysis of two SARS-CoV-2 strains isolated in Italy in January and February 2020: additional clues on multiple introductions and further circulation in Europe. Euro Surveill. 2020 Apr;25(13):2000305. doi: 10.2807/1560-7917.ES.2020.25.13.2000305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita Y., Ikeda T., Sato R., Sakagami T. Association between HLA gene polymorphisms and mortality of COVID-19: an in silico analysis. Immun. Inflamm. Dis. 2020 Dec;8(4):684–694. doi: 10.1002/iid3.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dorp L., Acman M., Richard D., Shaw L.P., Ford C.E., Ormond L., Owen C.J., Pang J., Tan C.C.S., Boshier F.A.T., Ortiz A.T., Balloux F. Emergence of genomic diversity and recurrent mutations in SARS-CoV-2. Infect. Genet. Evol. 2020 Sep;83:104351. doi: 10.1016/j.meegid.2020.104351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brünink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020 May;581(7809):465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020 Mar;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Dong N., Chan E.W., Chen S. Genetic cluster analysis of SARS-CoV-2 and the identification of those responsible for the major outbreaks in various countries. Emerg. Microb. Infect. 2020 Dec;9(1):1287–1299. doi: 10.1080/22221751.2020.1773745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan F., Wang L., Fang Y., Wang L. Global SNP analysis of 11,183 SARS-CoV-2 strains reveals high genetic diversity. Transbound. Emerg. Dis. 2020 Nov 18 doi: 10.1111/tbed.13931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen C.K., Lam J.Y., Wong W.M., Mak L.F., Wang X., Chu H., Cai J.P., Jin D.Y., To KK, Chan J.F., Yuen K.Y., Kok K.H. SARS-CoV-2 nsp13, nsp14, nsp15 and orf6 function as potent interferon antagonists. Emerg. Microb. Infect. 2020 Dec;9(1):1418–1428. doi: 10.1080/22221751.2020.1780953. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogenetic tree of the entire dataset. The full resolution image of the EU/EAA phylogram resulting from the ML analysis is shown. The signatures mutation (at the top-right) as well as the six main clusters are shown in different colors. The asterisk next to the nodes indicates supported branches. The mutation rate in each node is also shown.
The figure shows the phylogenetic tree resulting from the related dataset. The country name is provided at the top of the figure. The signatures mutation (at the top-right) and the six main clusters are shown in different colors. Nucleotide substitutions characterizing each cluster are shown. ABS = absence of substitution.
The figure shows the phylogenetic tree resulting from the related dataset. The country name is provided at the top of the figure. The signatures mutation (at the top-right) and the six main clusters are shown in different colors. Nucleotide substitutions characterizing each cluster are shown. ABS = absence of substitution.
The figure shows the phylogenetic tree resulting from the related dataset. The country name is provided at the top of the figure. The signatures mutation (at the top-right) and the six main clusters are shown in different colors. Nucleotide substitutions characterizing each cluster are shown. ABS = absence of substitution.
The figure shows the phylogenetic tree resulting from the related dataset. The country name is provided at the top of the figure. The signatures mutation (at the top-right) and the six main clusters are shown in different colors. Nucleotide substitutions characterizing each cluster are shown. ABS = absence of substitution.
The figure shows the phylogenetic tree resulting from the related dataset. The country name is provided at the top of the figure. The signatures mutation (at the top-right) and the six main clusters are shown in different colors. Nucleotide substitutions characterizing each cluster are shown. ABS = absence of substitution.
The figure shows the phylogenetic tree resulting from the related dataset. The country name is provided at the top of the figure. The signatures mutation (at the top-right) and the six main clusters are shown in different colors. Nucleotide substitutions characterizing each cluster are shown. ABS = absence of substitution.
The figure shows the phylogenetic tree resulting from the related dataset. The country name is provided at the top of the figure. The signatures mutation (at the top-right) and the six main clusters are shown in different colors. Nucleotide substitutions characterizing each cluster are shown. ABS = absence of substitution.
The figure shows the phylogenetic tree resulting from the related dataset. The country name is provided at the top of the figure. The signatures mutation (at the top-right) and the six main clusters are shown in different colors. Nucleotide substitutions characterizing each cluster are shown. ABS = absence of substitution.
The figure shows the phylogenetic tree resulting from the related dataset. The country name is provided at the top of the figure. The signatures mutation (at the top-right) and the six main clusters are shown in different colors. Nucleotide substitutions characterizing each cluster are shown. ABS = absence of substitution.
The figure shows the phylogenetic tree resulting from the related dataset. The country name is provided at the top of the figure. The signatures mutation (at the top-right) and the six main clusters are shown in different colors. Nucleotide substitutions characterizing each cluster are shown. ABS = absence of substitution.
The figure shows the phylogenetic tree resulting from the related dataset. The country name is provided at the top of the figure. The signatures mutation (at the top-right) and the six main clusters are shown in different colors. Nucleotide substitutions characterizing each cluster are shown. ABS = absence of substitution.
The figure shows the phylogenetic tree resulting from the related dataset. The country name is provided at the top of the figure. The signatures mutation (at the top-right) and the six main clusters are shown in different colors. Nucleotide substitutions characterizing each cluster are shown. ABS = absence of substitution.
The figure shows the phylogenetic tree resulting from the related dataset. The country name is provided at the top of the figure. The signatures mutation (at the top-right) and the six main clusters are shown in different colors. Nucleotide substitutions characterizing each cluster are shown. ABS = absence of substitution.
The figure shows the phylogenetic tree resulting from the related dataset. The country name is provided at the top of the figure. The signatures mutation (at the top-right) and the six main clusters are shown in different colors. Nucleotide substitutions characterizing each cluster are shown. ABS = absence of substitution.
The figure shows the phylogenetic tree resulting from the related dataset. The country name is provided at the top of the figure. The signatures mutation (at the top-right) and the six main clusters are shown in different colors. Nucleotide substitutions characterizing each cluster are shown. ABS = absence of substitution.
The figure shows the phylogenetic tree resulting from the related dataset. The country name is provided at the top of the figure. The signatures mutation (at the top-right) and the six main clusters are shown in different colors. Nucleotide substitutions characterizing each cluster are shown. ABS = absence of substitution.
The figure shows the phylogenetic tree resulting from the related dataset. The country name is provided at the top of the figure. The signatures mutation (at the top-right) and the six main clusters are shown in different colors. Nucleotide substitutions characterizing each cluster are shown. ABS = absence of substitution.
The figure shows the phylogenetic tree resulting from the related dataset. The country name is provided at the top of the figure. The signatures mutation (at the top-right) and the six main clusters are shown in different colors. Nucleotide substitutions characterizing each cluster are shown. ABS = absence of substitution.
The figure shows the phylogenetic tree resulting from the related dataset. The country name is provided at the top of the figure. The signatures mutation (at the top-right) and the six main clusters are shown in different colors. Nucleotide substitutions characterizing each cluster are shown. ABS = absence of substitution.
The figure shows the phylogenetic tree resulting from the related dataset. The country name is provided at the top of the figure. The signatures mutation (at the top-right) and the six main clusters are shown in different colors. Nucleotide substitutions characterizing each cluster are shown. ABS = absence of substitution.
The figure shows the phylogenetic tree resulting from the related dataset. The country name is provided at the top of the figure. The signatures mutation (at the top-right) and the six main clusters are shown in different colors. Nucleotide substitutions characterizing each cluster are shown. ABS = absence of substitution.
In the table the codes of the genome sequences retrieved from GISAID are listed.