Abstract
Staphylococcus aureus is an opportunistic human and animal pathogen with an arsenal of virulence factors that are tightly regulated during bacterial infection. The latter is achieved through a sophisticated network of regulatory proteins and regulatory RNAs. Here, we describe the involvement of a novel prophage-carried small regulatory S. aureus RNA, SprY, in the control of virulence genes. An MS2-affinity purification assay reveals that SprY forms a complex in vivo with RNAIII, a major regulator of S. aureus virulence genes. SprY binds to the 13th stem-loop of RNAIII, a key functional region involved in the repression of multiple mRNA targets. mRNAs encoding the repressor of toxins Rot and the extracellular complement binding protein Ecb are among the targets whose expression is increased by SprY binding to RNAIII. Moreover, SprY decreases S. aureus hemolytic activity and virulence. Our results indicate that SprY titrates RNAIII activity by targeting a specific stem loop. Thus, we demonstrate that a prophage-encoded sRNA reduces the pathogenicity of S. aureus through RNA sponge activity.
INTRODUCTION
Staphylococcus aureus is an opportunistic human and animal pathogen that can cause a wide range of illnesses, from food poisoning and superficial abscesses to more life-threatening diseases such as pneumonia, osteomyelitis, bacteremia, endocarditis and toxic shock syndrome (1,2). The infection process requires the controlled expression of virulence factors allowing bacteria to escape the host defense system, adapt to changing environmental conditions, and attack and destroy host cells. This process involves a wide range of wall-associated proteins and extracellular factors that are expressed during the different stages of infection. Global regulatory elements including transcription factors, sigma factors, two-component systems and regulatory RNAs assure the coordinated expression of these virulence factors.
Among the regulatory RNAs, small RNAs (sRNAs) are mostly non-coding, relatively short (50–550 nucleotides), and located within core genomes or mobile genetic elements. They are usually conditionally expressed, i.e. depending upon specific stress and growth phase. sRNAs control expression of their target genes by pairing with RNAs or forming complexes with proteins; in this way they usually modulate the stability and translation of mRNAs, and modify the activity of proteins (3). Over the past decade, hundreds of putative sRNAs discovered in S. aureus were compiled in the S. aureus RNA Database SRD (4). However, a recent analysis proposes that among them, only 50 are ‘bona fide’ sRNAs (5). Staphylococcus aureus sRNAs were demonstrated to contribute to regulation of dozens of functions. sRNAs contribute to regulation of bacterial metabolism, such as RsaE (6–8), or to antibiotic resistance, such as SprX (9). Several sRNAs were shown to be involved in the virulence of S. aureus such as SprD, which regulates expression of the immune evasion protein Sbi (10).
RNAIII is a paradigm for sRNA regulation of virulence (11,12). Besides encoding the PSM δ-hemolysin (known as Hld), RNAIII also positively regulates expression of hla, encoding the α-hemolysin (13), which leads to cell lysis (14–16), and intervenes in the expression switch between surface proteins and secreted toxins. Through direct binding to mRNA targets, RNAIII prevents the translation of major surface proteins, such as protein A (17), Sbi (10) and Ecb (18), which play key roles in adhesion and immune evasion. In addition, RNAIII inhibits translation of rot mRNA, encoding the repressor of toxins Rot (19–21), which blocks the transcription of exoproteins and toxins (22). By inhibiting Rot, RNAIII indirectly activates transcription of exotoxins and indirectly inhibits synthesis of protein A at the transcriptional level.
In this study, we report that SprY, an sRNA expressed from prophage φ12, is involved in the regulation of S. aureus virulence. By antisense pairing with RNAIII, SprY prevents RNAIII from regulating its targets and consequently decreases S. aureus hemolytic activity and virulence in a murine sepsis infection model. Together, our data reveal an sRNA acting as a sponge for RNAIII and further elucidate the regulation network controlling S. aureus virulence.
MATERIALS AND METHODS
Bacterial strains and growth condition
All bacterial strains and plasmids used in this work are listed in Supplementary Table S1. The DH5-α Escherichia coli strain was grown at 37°C in Luria-Bertani (LB) broth or LB agar plate supplemented with 50 μg/ml ampicillin if necessary. The S. aureus RN4220 strains was used to prepare phage-containing vectors expressing sRNA or target-gfp fusions. In this study, S. aureus HG003 strain was used to co-transform the target-gfp fusions with the sRNA expressing plasmid. Cultures of these co-transformed S. aureus strains were grown at 37°C either in brain heart infusion broth (BHI, Oxoid) or on BHI agar plates. When necessary, media were supplemented with 10 μg/ml of chloramphenicol and/or erythromycin. HG003 ΔrnaIII::tag004 (SaPhB618) and HG003 ΔsprY::tag112 (SaPhB980) were constructed using pMADΔrnaIII::tag004 and pIMsprY2::tag112, respectively, as described (23).
Plasmids constructions
Supplementary Table S2 lists all the primers used. To construct the sRNA-expressing vectors, we used pICS3 (24). To construct the pICS3-sprY, sprY with its endogenous promoter was amplified by PCR with primers 13–14 and cloned in pICS3 vector digested by PstI and NarI. To introduce mutations in sprY, primers 15–16 were used.
To construct the pCN33-PtufA-rot-gfp vector, which expresses rot under control of the PtufA promoter, we amplified 373 nucleotides rot with primers set 18–19. HG003 strains carrying each of the target-gfp fusions and the sRNA plasmids were grown on BHI agar plates supplemented with 10 μg/ml chloramphenicol and erythromycin. The fluorescence measurements of the co-transduced HG003 strains were performed as previously described (24).
PCR products corresponding to the sequence of ms2 tag, or ms2 fused with sprY were cloned into pRMC2 digested by KpnI / SacI (see Supplementary Table S2 for primers containing MS2 tag sequence). All cloning experiments were performed with Gibson Assembly Master Mix (New England Biolabs). The reactions were then transformed into E. coli DH5-α by heat shock at 42°C. The plasmids were purified from overnight cultures in LB broth supplemented with Ampicillin 100 μg/ml, extracted (Miniprep Extraction Kit, Qiagen) and Sanger sequenced by using BigDye Terminator v3.1 Cycle sequencing Kit, using a 3130 × 1 capillary electrophoresis genetic analyzer (Applied Biosystems). The purified plasmids were used for the transformation in S. aureus RN4220 strain by electroporation shock. The φ80 phages prepared from RN4220 were then used to transduce plasmids in HG003 strains. pMADΔrnaIII::tag004 is a pMAD derivative containing the PCR-amplified rnaIII upstream, tag004 and rnaIII downstream sequences cloned by Gibson assembly (using primers RNAIII UpF, RNAIII UpR, RNAIII DwF, RNAIII DwR for rnaIII adjacent sequences) as described . pIMsprY2::tag112 is a pIMAY derivative containing the PCR-amplified sprY upstream, tag004 and sprY downstream sequences cloned by Gibson assembly (using primers pIMAY-Up-SprY2-F, pIMAY-Up-SprY2-R, pIMAY-Down-SprY2-F and pIMAY-Down-SprY2-R for sprY adjacent sequences) as described (23).
Proteins extraction and Western blots
Staphylococcus aureus strains were grown until exponential phase (OD600 = 0.8) or stationary phase (OD600 = 10.1) in BHI at 37°C, with agitation at 160 rpm, and the cells were then pelleted for 10 min at 4°C (8000 × g). The total proteins extractions were prepared according to (25). Rot expression was visualized by anti-Rot antibodies (Benson, 2012, JB) and anti-rabbit IgG secondary antibodies (Jackson). Western blots were revealed using the Amersham ECL Plus detection Kit. Signals were visualized using LAS 4000 (GE Healthcare).
In vitro transcription, RNA labeling and Gel-shift assays
All RNAs were transcribed from PCR-generated DNA using MEGAscript T7 kit (Ambion). The template for transcription was amplified using HG003 genomic DNA and forward primers containing T7 promoter sequences (Supplementary Table S1). RNAs were labeled at 5′-end using [γ-32P] ATP (Amersham Biosciences) and T4 polynucleotide kinase (Invitrogen). Labeled and unlabeled RNAs were purified on a 5% acrylamide urea gel, eluted in Elution buffer (20 mM Tris-HCl pH 7.5, 250 mM NaCl, 1 mM EDTA, 1% SDS) at 37°C, eluted, ethanol precipitated, quantified by Qubit (Thermo Fisher Scientific) and stored at -80°C.
Gel-shift assays were performed as described in (26). RNAs were denaturated in 50 mM Tris/HEPES pH 7–7.5, 50 mM NaCl for 2 min at 80°C, followed by refolding for 10 min at 25°C after adding MgCl2 at final concentration of 5 mM. The binding reactions were performed in 50 mM Tris-HCl (pH 7.5), 50 mM NaCl, 5 mM MgCl2 for 20 min at 25°C. About 0.025 pmoles of labeled SprY or SprY mutants were incubated with various concentrations of RNAIII. The samples were supplemented with 10% glycerol and were loaded on a native 4% polyacrylamide gel containing 5% glycerol. The gels were dried and visualized by Typhoon FLA 9500 scanner (GE Healthcare).
RNA extractions, northern blots, RNA half-life determination and qPCR assay
The cells were collected at exponential and at stationary phases of growth, pelleted for 10 min at 4°C (4500 × g) and resuspended in RNA lysis buffer (0.5% SDS, 20 mM acetate of sodium, 1 mM EDTA, pH 5.5). Total RNA was extracted as previously described (7). The Northern blot assays were carried out as previously described (9). The membranes were hybridized with specific 32P-labeled probes (Supplementary Table S2) in ExpressHyb solution (Clontech) and were washed according manufacture recommendations. The membranes were then exposed and scanned with Typhoon FLA 9500 scanner (GE Healthcare). The images quantifications were realized with ImageQuant Tool 7.0.
For sRNA half-life determination, rifampicin was used as the most common treatment to stop the transcription (27). Staphylococcus aureus HG003 strain and its derivatives were cultured overnight, diluted to 1/100, grown for 5 h at 37°C, and incubated with 20 mg/ml rifampicin. About 8 ml of each strain was collected before and at 2, 5, 10, 20, 30, 40, 60 and 90 min after adding rifampicin. These samples were centrifuged, the pellets were frozen in liquid nitrogen then stored at -80°C. Total RNA was extracted. For the quantitative real-time PCR (qRT-PCR), total RNA extraction samples were treated with DNaseI Amplification grade Kit (Invitrogen). cDNAs preparations and qRT-PCR experiments were performed as previously described (24). The gyrB gene was used for normalization. As for absolute quantification by qPCR, instead of using gyrB, we prepared a calibration range with respective PCR products at concentrations of 1010/8/6/4/2/0/-2 copies/μl. Identification of TSS of sprY was performed as performed as described (28). For this reverse transcription (RT) was done on 5 μg of total RNA of HG003 strain with labeled primer 2.
Preparation of the MS2-affinity column
To prepare the 6His-MBP-MS2 protein, we used the pHMN plasmid (29) containing the 6His tag at the N-terminal and the MS2 tag at the C-terminal. The induction of protein production and bacterial lysis was performed as described by (30). The 6His tag allows a first purification on nickel resin, using an AKTÄ (GE healthcare). After being washed with water, the pumps and the column were equilibrated with lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 0.5% Tween 20, 10 mM Imidazole 10% glycerol). The bacterial lysate was added into the column and various fractions were recovered using the elution buffer (lysis buffer + 250 mM of imidazole). The fractions of interest were passed through a desalting column to remove traces of the elution buffer. A second purification was carried out using an amylose column washed and then equilibrated with buffer 2 (20 mM Tris-HCl, 0.2 mM NaCl, 0.5 mM EDTA). The fractions of interest were passed through the column and then eluted using a second elution buffer (buffer 2 + 10 mM maltose buffer). The fractions were also passed through a desalting column to remove all traces of maltose. For the preparation of the MS2-affinity column, 100 μl of amylose resin (NEB # E8021S) was added in a Bio-spin disposable chromatography column (Biorad # 732–6008). The column was then washed three times with 1 ml of buffer A (20 mM Tris-HCl pH 8, 150 mM KCl, 1 mM MgCl2, 1 mM DTT, 1 mM PSMF). A solution of 100 pmoles of 6His-MBP-MS2 protein was added to the column and was incubated for 5 min, and washed twice with 1 ml of buffer A.
Purification of bacterial lysates, RNA sequencing and Bioinformatic analysis
Staphylococcus aureus strains were grown until exponential phase (OD600 = 0.8) or stationary phase (OD600 = 10.1) in 50 ml of BHI (Oxoid). Then expressions of ms2-sprY and ms2 were induced with 1 μM of anhydrotetracycline for 10 min. The bacteria were placed in ice for 10 min and centrifuged for 5 min at 4000 × g. The pellets were then washed with 1 ml of buffer A, centrifuged for 1 min at 16 000 × g and stored at -80°C. Frozen pellets were thawed on ice and resuspended in 2 ml of buffer A. Mechanical lysis with Fastprep (Fastprep, MP Biomedicals) in the presence of 250 μl of glass beads was carried out for 3 × 30 s at 6500 × g. The samples were centrifuged for 10 min at 4°C at 16 000 × g. The bacterial lysates obtained were passed through an affinity column. Columns were washed 5 times with 1 ml of buffer A and then eluted with 1 ml of buffer A + 15 mM maltose. In order to carry out checks, we extracted the RNAs at different points in the experiment. We obtained four RNA extracts corresponding to the total RNAs taken before passage through the amylose column (input), RNA passed through a column (Flow-Through), RNA recovered after the last washing of the column (W) (Supplementary data). The RNAs from the fractions eluted from the columns are extracted with chloroform phenol as previously described (10). For MS2 and MS2-SprY, the northern blots were carried out using 5 μg of RNA according to the protocol as previously described (9). The primers used are listed in Supplementary Table S2. A bio-analyzer (2100-Agilent bio-analyzer) was used to quantify and verify the purity of the samples before being sequenced (following the manufacturer’s instructions [Agilent]). RNA sequencing was carried out as previously described (31). The cDNA libraries were prepared with the NEBNext® Ultra™ II Directional RNA Library Prep Kit for Illumina®, and then sequenced as paired-end reads (2 × 75 bp) using an Illumina MiSeq platform and the MiSeq reagent kit version 3. The reads were mapped against the genomic sequence of S. aureus HG003 strain (Genbank accession no. CP000253) and then counted using the CLC Genomics Workbench software v8.1 (Qiagen). Statistical analysis was performed using the DESeq2 R package (32). Raw and processed data generated in this study have been submitted to the Gene Expression Omnibus (GEO) repository at the National Center for Biotechnology Information (NCBI) and are available under accession no. GSE166499.
Hemolysis assays
Staphylococcus aureus strains grown overnight were diluted to OD600 = 0.1 in BHI. The supernatants were taken at 2 h of bacterial growth, filtered with 0.45-μm filter and stored at -20°C. For blood sample preparation, 1 ml of human or mouse blood was centrifuged for 10 min at 4000 × g at 25°C, followed by multiple washes with PBS 1X (qsp 1 ml) to eliminate the plasma and lysed red blood cells and re-suspended in 10 ml of PBS 1×. The prepared blood sample and the supernatants (diluted in 1/10) were added at a ratio 50:50 in 96 pointy wells plates to a final volume of 150 μl per well. After an incubation of 1 h at 37°C, the plate was centrifuged for 10 min at 4000 × g at 25°C and 100 μl of the supernatants were collected into a new 96 flat well plates and read at OD 540 nm. The mix of blood sample with PBS 1× or with 0.1% Triton were used respectively as negative and positive controls of hemolytic activity.
Animal infection model
HG003, HG003ΔsprY and complemented strains were used to study the virulence level in a murine intravenous sepsis model. All experimental protocols were approved by the Adaptive Therapeutics Animal Care and Use Committee (APAFIS#2123–2015100214568502v4). For the sepsis model, we used female Swiss mice (Janvier Labs), 6–8 weeks old and weighing ∼30 g. Groups of five female mice were inoculated i.v. with 200 μl of bacterial suspensions containing 2 × 108S. aureus cells in 0.9% NaCl. The survival of the mice was monitored for 12 days, and the statistical significance of differences between groups was evaluated by comparing Kaplan–Meier survival curves with the Mantel-Cox test. A P-value of 0.05 was considered significant.
RESULTS
SprY, a novel sRNA expressed from S. aureus srna cluster
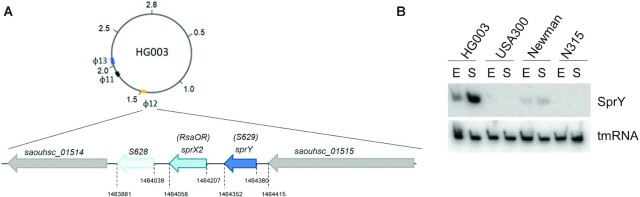
A small staphylococcal RNA expressed from prophage φ12 (33) in NCTC8325 derivatives was initially described as a putative 5′UTR of a small open reading frame SAOUHSC_A01455 of 60 amino acids, and called S629 (34). However, based on in-depth analysis of the HG003 strain genome, we designated S629 as a bona fide sRNA (5). Because the gene S629 is adjacent to the sprX2 sRNA gene (7,9), we renamed this sRNA SprY (Figure 1A and Supplementary Figure S1_A). Spr is an acronym coined for small pathogenicity island sRNA (35). The SprY 5′-end determined by reverse transcription (RT) (Supplementary Figure S1_B) corresponds to nucleotide position 1464380 of the NCTC8325 genome sequence (Supplementary Figure S1_A). Detection of SprY by northern blot (Figure 1B) and in silico identification of a Rho-independent transcriptional terminator upstream of SAOUHSC_A01455 (using ARNold; (36)) indicate that SprY is a bona fide sRNA of about 125 nucleotides. It is found in several S. aureus isolates with 95% sequence identity but is not present in other Staphylococcaceae (5). SprY is present only in strains containing the φ12 prophage; accordingly, it was detected by northern blot in Newman and HG003 strains but not in USA300 and N315 (Figure 1B).
Figure 1.
sprY and sprX2 genomic localization and expression in Staphylococcus aureus HG003 strain. (A) The genomic localization of sprY and sprX2 genes in S. aureus HG003 strain. sprY gene is located around 50 bp upstream of sprX2 gene (7,9) in the staphylococcal phage φ12. (B) Expression of SprY in different strains of S. aureus. RNA extraction was performed in HG003, USA300, Newman and N315 strains at exponential phase (E) and stationary phase (S). As loading controls, the blots were also probing for tmRNA.
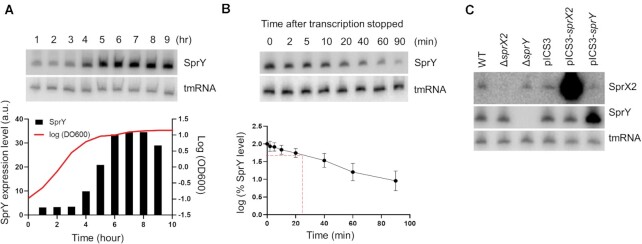
SprY amounts are highest in pre-stationary phase and slowly decrease during stationary phase (Figure 2A). SprY half-life is 23.3 ± 1.45 min in pre-stationary phase (Figure 2B), implying that SprY is stable RNA, compared to most mRNAs with half-life of 2–4 min (37,38). Since sprY and sprX2 are adjacent sRNA genes, we considered that their expression could be inter-dependent. For this purpose, we analyzed the amounts of both, SprY and SprX2 sRNAs in HG003 (39) and its derivatives deleted for either sprY (ΔsprY) or sprX2 (ΔsprX2). Despite their proximity, deletion of one sRNA gene did not affect the expression of the other (Figure 2C). In addition, overexpression of sprX2 (pICS3-sprX2) or sprY (pICS3-sprY) did not affect the expression level of the other sRNA (Figure 2C). We concluded that sprY and sprX2 are expressed independently from each other. Despite the close proximity of these genes, their expression patterns are different, since SprX2 accumulates during exponential phase while SprY accumulates in pre-stationary phase (Supplementary Figure S2, (9)).
Figure 2.
SprY expression in HG003 strain. Expression of sprY was determined by northern blot analysis using labeled DNA probes for SprY. As loading controls, the blots were also probed for tmRNA. (A) SprY expression profile during a 24 h growth of Staphylococcus aureus HG003 strain (WT). The growth curve of WT strain is presented by line, with the quantification of SprY expression level relative to the amount of tmRNA from the same RNA extraction in black chart (a.u., arbitrary units). (B) Stability of SprY. HG003 (WT) was grown until 6 h (t = 0) and the RNA extraction was performed at 2, 5, 10, 20, 40, 60 and 90 min after adding Rifampicin. The quantification of SprY stability, in semi-log plot, was performed by ImageQuant Tools 7.0. The data represent the mean of three different experiments ± standard error. The maximum value of rnaIII expression at time 0 is normalized to 100% and the time at which the sRNA reaches 50% of its original level is indicated with red dotted line. (C) The expression of sprX2 and sprY at 6 h of bacterial growth of HG003 wild-type strain (WT), strains deleted for sprX2 or sprY (ΔsprX2; ΔsprY) and HG003 overexpressing sprX2 or sprY (pICS3-sprX2, pICS3-sprY) by northern blots, using labeled DNA probes for SprY and SprX2. As loading control, the blots were also probing for tmRNA.
The SprY structure predicted by RNAfold (40,41) and LocaRNA softwares (42–44) revealed three stem loops, the last one being a Rho-independent transcriptional terminator (Supplementary Figure S3). The activity of several staphylococcal sRNAs involves single-stranded C-rich regions (8,45). However, this feature is not present in SprY, suggesting that it may not target G-rich regions associated with Shine-Dalgarno sequences but other sequences.
SprY interacts directly with RNAIII, a major regulator of virulence factors of S. aureus
Most sRNAs act by base-pairing to RNA targets. To identify potential direct targets of SprY, we performed in vivoMS2-RNA affinity purification coupled with RNA sequencing (MAPS) (46). For this purpose, an MS2 tagged version of SprY was expressed under the control of an inducible promoter in HG003 (Supplementary Figure S4_A and B). After 2 and 6 h of growth, each followed by 5 min of induction, RNAs in complex with MS2-SprY were isolated by MS2 affinity chromatography, eluted, and identified by RNAseq. RNAIII, rpmG1 mRNA and SAOUHSC_1342a mRNA were enriched 7.24-, 11.11- and 7.59-fold, respectively, with MS2-SprY compared to MS2 alone (Supplementary Figure S4_C and Supplementary Table S3). This significant enrichment suggests that these three RNAs are SprY targets. However, no base-pairing with a significant energy between SprY and rpmG1 mRNA was predicted by in silico analysis suggesting that rpmG1 mRNA enrichment with MS2 tagged SprY could be due to an indirect interaction. In contrast, IntaRNA software (47,48) indicated potential base-pairings of SprY with the 5′ UTR of SAOUHSC_1342a (Supplementary Figure S4_D) and with RNAIII (Figure 3A), supporting the MAPS results. As RNAIII is the major virulence regulator and effector of quorum sensing in S. aureus, we focus our study on the interaction between SprY and RNAIII.
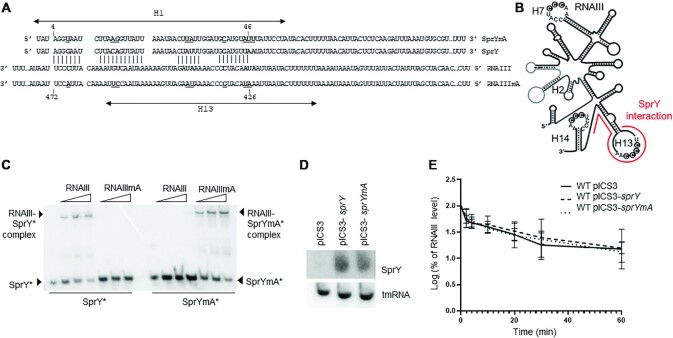
Figure 3.
SprY interacts with RNAIII. (A) Interaction between SprY and RNAIII was predicted by IntaRNA software (47). Mutant of SprY and compensatory mutant RNAIII were constructed (SprYmA and RNAIIImA). The nucleotides underlined and bolded correspond to the mutations in the sprY and rnaIII sequences. (B) Schematic presentation of RNAIII secondary structure and the region of RNAIII that interacts with SprY (shown by line). (C) Complex formations between SprY, SprYmA with RNAIII and RNAIIImA were analysed by native gel retardation assays. Shift assays of purified labeled SprY and SprYmA (SprY* and SprYmA*) were performed with increasing concentrations of RNAIII and RNAIIImA (0.25, 0.5 and 1.25 μM). (D) Verification of sprY expression in HG003 strain containing empty plasmid (pICS3) or over-expressing sprY (pICS3-sprY) or muted sprY (pCIS3-sprYmA) by northern blot using labeled DNA probes for SprY and tmRNA as control. (E) RNAIII stability was studied by northern blot using RNA total extraction in HG003 pICS3, HG003 pCIS3-sprY and HG003 pCIS3-sprYmA, at exponential growth phase (OD = 0.8). Time (min) corresponds to the time after adding Rifampicin. The RNAIII expression levels of in each sample were is normalized with the control gene (tmRNA). The data represent the mean of three different experiments ± standard error for RNAIII stability. The maximum value of rnaIII expression at time 0 is normalized to 100% and the time at which the sRNA reaches 50% of its original level is indicated with dotted line. All statistical analyses were performed using two-way ANOVA test and Bonferroni comparison test.
IntaRNA analysis indicates potential base-pairing between RNAIII and SprY involving the 1st hairpin of SprY (4th to 46th nucleotide) and the 13rd hairpin of RNAIII (Figure 3A and B). The predicted interaction between SprY and RNAIII was tested in vitro by EMSA using synthetic RNAs. A complex between SprY and RNAIII was observed (Figure 3C). Its specificity was challenged by SprY allele bearing point mutations in the 1st hairpin (SprYmA), which corresponds to the predicted RNAIII binding sequence (Figure 3A). Expectedly, SprYmA lost the ability to bind RNAIII (Figure 3C), implying that the 1st SprY hairpin is required for pairing. To bring further evidence to support the predicted pairing, an RNAIII derivative with compensatory mutations (RNAIIImA) restoring the pairing with SprYmA (Supplementary Figure S5_C) was synthesized and tested by EMSA. A gel retardation was observed between SprYmA and RNAIIImA but not between SprYmA and RNAIII (Figure 3C). In addition, RNAIII mutated in the 13th loop, which corresponds to a part of the predicted SprY binding region, noticeably lost the capacity to bind SprY (Supplementary Figure S5_A and B). Altogether, our in vivo (MAPS), in silico and in vitro (EMSA) experiments revealed an interaction between the 1st SprY hairpin and the 13th RNAIII loop.
In many cases, base-pairing with sRNAs affects RNA target stability. We therefore tested if binding of SprY would affect the stability of RNAIII. The stability of RNAIII was compared in HG003 harboring the plasmids pICS3-sprY, pICS3-sprYmA and the control vector pICS3 (Figure 3D). No significant difference was observed in RNAIII stability, whether SprY or SprYmA were overexpressed (Figure 3E). This result prompted us to further analyze the role of SprY toward RNAIII.
SprY affects expression of RNAIII targets
RNAIII is the effector of quorum sensing and a major regulator of a plethora of virulence factors. Interestingly, the 13th RNAIII stem-loop is involved in the repression of several among its numerous mRNA targets. By binding to the 5′ leader region of rot mRNA (for repressor of toxins), RNAIII, via the 13th stem-loop, negatively regulates the expression of this target at the translational level (49) and in consequence obstructs the expression of toxins (20,21). Another well-studied target of the 13th RNAIII stem-loop is ecb (for extracellular complement binding protein) (19,24), which is involved in blocking bacterial recognition by the host immune system (18).
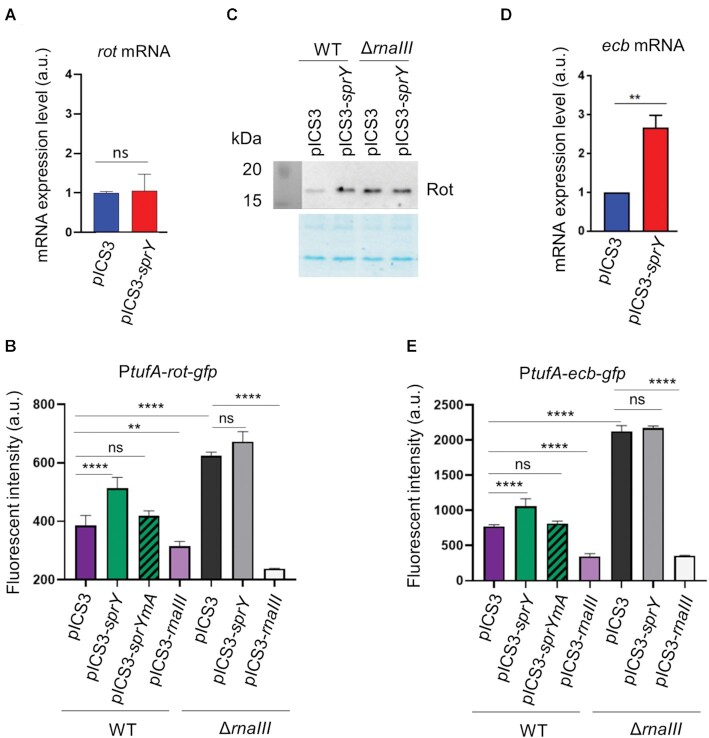
We therefore questioned if the SprY binding to the 13th RNAIII stem-loop could affect the activity of RNAIII against specific targets. Surprisingly, sprY overexpression showed no significant impact on rot mRNA levels (Figure 4A). We hypothesized that SprY could affect rot translational initiation rather than rot mRNA quantity. We constructed a rot-gfp translational gene fusion under the control of the constitutive PtufA promoter in pCN33 resulting in pCN33-PtufA-rot-gfp. HG003 and HG003ΔrnaIII containing empty pICS3 or pICS3-sprY were transformed with pCN33-PtufA-rot-gfp. In addition, a strain overexpressing rnaIII under constitutive promotor amiA (pICS3-PamiA-rnaIII) was used as a control. As expected, SprY overproduction significantly increased rot translation only in the presence of RNAIII (Figure 4B). Furthermore, the overexpression of SprYmA had no effect over the fluorescence of Rot-GFP fusion (Figure 4B), which implies that the regulation of rot expression by SprY involved the interaction between SprY 1st hairpin and RNAIII. In addition, deletion of sprY resulted in a small but significant decrease in fluorescence intensity of the Rot-GFP fusion, while strains overexpressing sprY showed increased fluorescence (Supplementary Figure S6). We also confirmed the effect of SprY on endogenous rot expression in an rnaIII dependent manner by Western blot (Figure 4C). Overexpression of SprY increases Rot level in the HG003 wild-type strain but not in the absence of RNAIII.
Figure 4.
SprY affects the expression of RNAIII targets. Analysis of rot (A) and ecb (D) transcript levels in HG003 strain containing empty plasmid (pICS3) or overexpressing sprY (pICS3-sprY) by qPCR, using RNA extraction of those strains at early exponential growth phase. The mRNA expression level of rot and ecb in qPCR is normalized with the control gene (gyrB) and is calculated with 2−ΔΔCt relative quantification. (C) Analysis of Rot protein level in HG003 strains (WT and ΔrnaIII) containing empty plasmid (pICS3) or over-expressing sprY (pICS3-sprY) by western blot using polyclonal antibodies against Rot. The translational initiation level of rot (B) and ecb (E) under SprY regulation were studied by using gfp gene reporter. Staphylococcus aureus strains (WT and ΔrnaIII) containing the pCN33-PtufA-rot-gfp or pCN33-PtufA-ecb-gfp fusion plasmids co–transformed with pICS3, pICS3-sprY, pICS3-sprYmA or pICS3-PamiA-rnaIII. The fluorescent intensity was measured every 10 min over 20 h in a Biotek microplate reader. Bacteria growth in different HG003 strains and GFP fluorescence intensity are shown respectively in the left panel at OD 600 and in right panel quantified by Biotek. All statistical analyses were performed using Student’s t test. The error bars correspond to the average values from three independent experiments. Statistical significance is indicated by bars and asterisks as follows: **, P < 0.01; ***, P < 0.005; ****, P < 0.001.
The effect of SprY was also tested on the expression of ecb, another target regulated by the 13th RNAIII stem-loop. Overexpression of SprY induced an increase in ecb mRNA levels (Figure 4D). As in the case for rot, SprY overproduction significantly increased ecb translation only in the presence of RNAIII (Figure 4E). Moreover, like for rot regulation, the overexpression of sprYmA had no effect on the fluorescence of Ecb-GFP (Figure 4E), which implies that the regulation of ecb expression by SprY requires the interaction between SprY and RNAIII. In addition, as in the case for rot, the absence of sprY resulted in decreased fluorescence of the Ecb-GFP fusion (Supplementary Figure S6). Bacterial growth of all strains used for this experiment was essentially equivalent (Supplementary Figure S7). As with Rot, we observed a significant decrease in fluorescence intensity of Ecb-GFP in the absence of SprY, and an increase of fluorescence of strains overexpressing sprY (Figure 4). Taken together, our results indicate that SprY affects the expression of RNAIII targets likely by titrating the regulation mediated by its 13th loop.
Abundance of RNAIII and SprY depends on growth conditions
As the regulatory activity of RNAIII controlled by SprY is based on RNAIII/SprY direct binding, the endogenous ratio of each partner likely controls the biological functions of this interaction. In rich medium, both SprY and RNAIII accumulate during growth and diminish in late stationary phase (Figure 5A). The expression profiles in different media were similar for both sRNAs (Figure 5B), even when bacterial growth rates were different (Supplementary Figure S8_A). Quantities of SprY and RNAIII were determined in different media in exponential (2 h) and stationary (6 h) phases by RT-qPCR (Supplementary Figure S8_B). Compared to RNAIII, the SprY copy number was 2-fold higher in BHI and RPMI, and 4-fold higher in TSB at 2 h of growth; however, it was significantly lower than RNAIII in NZM at 2 h (0.1-fold) (Figure 5C). The RNAIII copy number is far greater than that of SprY in all media at 6 h (RNAIII: SprY ratios are 5, 3, 1000, and 5 in BHI, TSB, NZM and RPMI, respectively). These results suggest that RNAIII regulation by SprY will likely be controlled by growth conditions.
Figure 5.
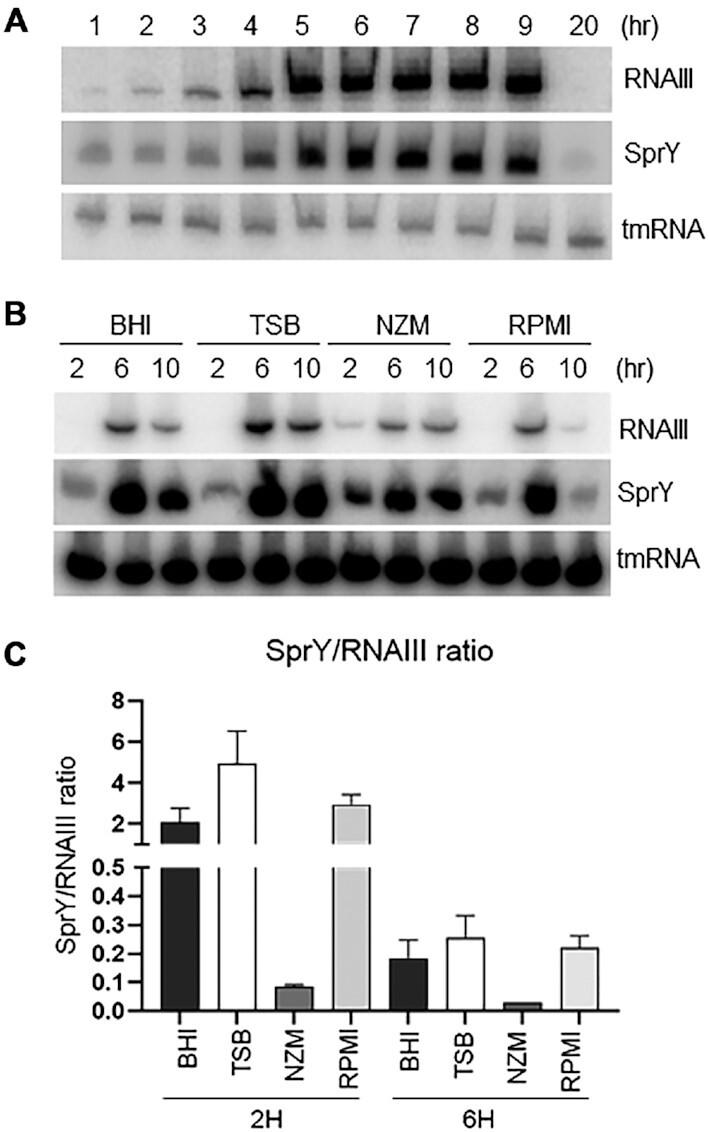
SprY and RNAIII ratio in different media. (A) HG003 wild-type strain was grown in BHI at 37°C. RNA was extracted at different times of growth. sprY and rnaIII expression profiles were analyzed by northern blot. (B) Analysis of sprY and rnaIII expressions in different media. HG003 wild-type strain was grown in different media (BHI, TSB, NZM and RPMI). Total RNA was extracted at 2, 6 and 10 h of growth and studied by northern blot using labeled DNA probes for SprY and RNAIII, and tmRNA as loading control. (C) The quantification of SprY and RNAIII by qPCR absolute. The RNA samples were prepared from the pellet of HG003 wild-type strain in BHI, TSB, NZM and RPMI at 2 and 6 h of growth. The chart represents the ratio SprY/RNAIII in these media. The error bars correspond to the average values from three independent experiments.
SprY limits hemolytic activity and decreases virulence of S. aureus in a mouse infection model
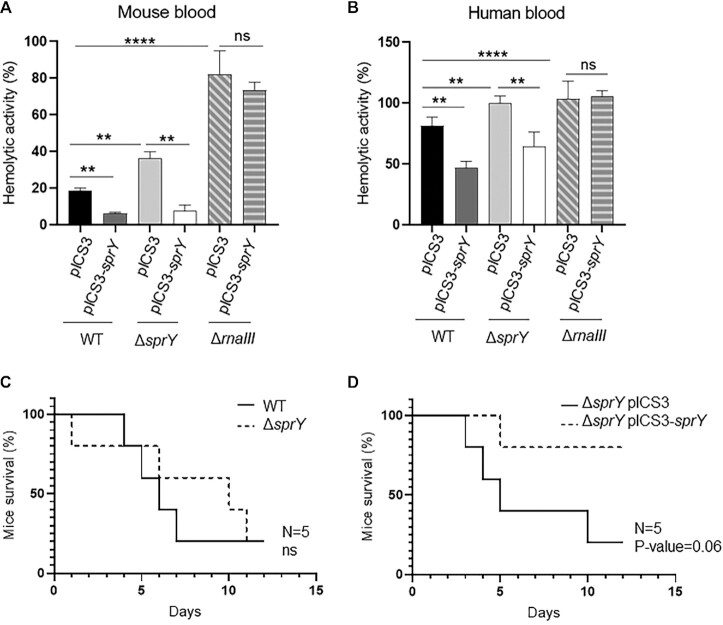
We showed above that SprY regulates the virulence factor Ecb as well as Rot through its effect on RNAIII. Moreover, Rot is known to repress toxins production (20). Since staphylococcal toxins are involved in hemolysis, the effect of SprY on S. aureus hemolytic activity was tested on mouse and human blood samples with HG003, HG003 ΔsprY, HG003 ΔrnaIII strains harboring either pICS3 or pICS3-sprY overexpressing sprY (Figure 6A and B). The accumulation of SprY led to a significant decrease in the S. aureus hemolytic activity in all strains except for HG003 ΔrnaIII, indicating that RNAIII is required for SprY-mediated hemolysis.
Figure 6.
Impact of SprY to hemolytic activity in Staphylococcus aureus in different models of infection. (A and B) The supernatants of different strains of HG003 (WT, ΔsprY, ΔrnaIII) containing empty plasmid (pICS3) or overexpressing sprY (pICS3-sprY) were collected after 6 h of growth in BHI at 37°C. Hemolysis was performed by incubating cell culture supernatants with mouse and human blood samples at a ratio 50:50 at 37°C for 1 h. The hemolytic activity was observed in a flat-bottom 96-well microtiter plate at OD 540 nm. The blood samples prepared in PBS1X represent as negative control and in Triton 0.1% as 100% hemolysis. (C and D) Survival of mice infected with S. aureus wild-type strain (WT), deleted strain for sprY (ΔsprY) and complemented strains for sprY (ΔsprY pICS3-sprY). Groups of 5 eight-week-old Swiss female mice were inoculated i.v. with 2 × 108 bacteria and monitored daily for 3 weeks. All statistical analyses were performed using Student’s t test. The error bars correspond to the average values from three independent experiments. Statistical significance is indicated by bars and asterisks as follows: **, P < 0.01; ****, P < 0.001.
Since SprY restricted hemolysis in mouse blood, we considered mice as a suitable model to test the role of sprY during staphylococcal infection. A murine intra-venous sepsis model was used with an inoculum of 2 × 108 bacteria per mouse. The survival rate of mice infected with either HG003 or HG003 ΔsprY was not statistically different (Figure 6C). We also tested the HG003 ΔsprY containing either the pICS3 (control plasmid) or overexpressing sprY (pICS3-sprY). The virulence of HG003 ΔsprY overexpressing sprY was drastically reduced compared to control the strain (pICS3) (Figure 6D). Taken together, our results show that SprY reduces S. aureus virulence likely by decreasing expression of virulence factors including RNAIII-activated hemolysin.
DISCUSSION
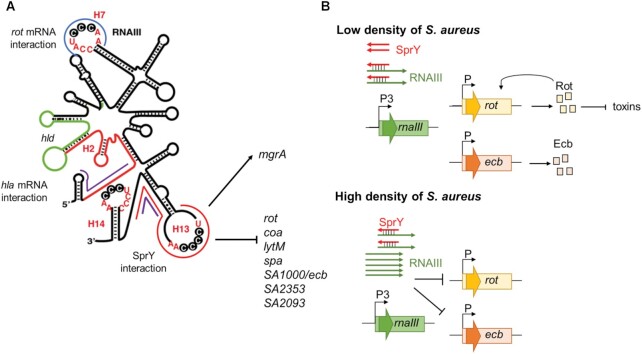
We report the function of SprY alias S629 (34), a recently identified bona fide sRNA (5), expressed from HG003 prophage φ12 (also known as bacteriophage Sa2 or φSa2) (33). Here, we identified SprY as an sRNA tuning the sophisticated RNAIII regulation network. Co-purification of in vivo RNAs associated to SprY revealed that SprY binds RNAIII. RNAIII is the key effector of the accessory gene regulator (agr) system and the major ribonucleic staphylococcal regulator of virulence, which controls translation and stability of several mRNA targets by antisense base-pairing involving its various stem-loop structures (17,19) (Figure 7A and Supplementary Figure S9). We demonstrated that the 5′ region of SprY binds to the 13th stem-loop of RNAIII. RNAIII sequence pairing with SprY is also the binding site for targets such as rot, spa, coa, lytM, ecb, SA2093 and SA2353 mRNAs (Supplementary Figure S9). SprY binding does not affect RNAIII stability but alters its function. We propose that SprY sterically prevents the formation of complexes between RNAIII and its targets, therefore affecting their expression. Indeed, SprY overproduction increases rot mRNA translation only when the interaction between SprY and RNAIII takes place. Moreover, SprY stimulates expression of the staphylococcal extracellular complement binding protein, Ecb (50) in an RNAIII-dependent manner. In accordance with positive regulation of rot, whose regulon includes genes encoding proteins with hemolytic activity such as hla, psmα and hlgACB (22,51), SprY reduces the hemolytic activity of S. aureus on human and mouse blood dependently from RNAIII presence.
Figure 7.
Schematic view of RNAIII and SprY actions. (A) Schematic view of the secondary structure of RNAIII (12) and base-pairings interaction regions with SprY and other mRNA targets. hla mRNA interacts with RNAIII at 2nd and 3rd hairpins (rnaIII sequence is colored in red). RNAIII encodes the delta-hemolysin (hld, in green). The interaction zone of RNAIII and SprY is indicated in red line and is overlapped with other RNAIII mRNA target listed here. rot mRNA interacts with RNAIII 7th hairpin (in blue line) and RNAIII 13rd stem-loop. mgrA mRNA interacts at two different sites of RNAIII (in purple line). (B) Model for SprY sponge function in RNAIII/ mRNA targets regulation. At low cell density (upper panel), SprY presents in a higher number of copies than RNAIII and bind to all RNAIII molecules produced. This interaction prevents RNAIII from binding to rot and ecb mRNA and favors the translation of Rot and Ecb proteins. At high cell density (bottom panel), RNAIII is produced in higher level than SprY and suppresses the sponge effect of SprY, therefore inhibits the translation of rot and ecb, among other targets.
The interactions we uncovered between SprY and RNAIII prompt us to hypothesize that SprY acts as an RNA sponge for RNAIII to prevent RNAIII-dependent regulations. The competition between mRNA targets and the mimicry of targeted RNA by other RNAs are essential mechanisms to adjust the action of regulatory sRNAs (reviewed in (52)). RNAs with sponge-like activity were recently described in different bacterial species such as E. coli, Bacillus subtilis and Pseudomonas aeruginosa (53–55) and reviewed in (52,56). These sponge-like RNAs use different mechanisms to control sRNA regulators. Their binding can induce the cleavage and/or destruction of sRNA, as reported for chb-ChiX (57). In addition, since degradation is not an obligate outcome of duplex formation, a sponge RNA can act by titrating a regulatory sRNA and by competing with their true targets, as occurs between the tRNA spacer sequence with RybB or RyhB sRNAs (31). Until now, only few cases of base pairings between two sRNAs were identified in S. aureus; however, no ‘sponge-like’ function was shown for these interactions (58,59). Thereby, the findings presented here identify a sRNA with an original sponge activity in pathogenic bacteria.
To detectably adjust RNAIII function, SprY has to accumulate in amounts likely comparable to those of RNAIII. SprY and RNAIII expression varies according to growth conditions. The differences in sRNA stoichiometry could push the balance to different outcomes according to the growth environments. In rich media, SprY abundance is comparable to that of RNAIII during growth at low cell density. However, in late exponential growth phase, the ratio between SprY and RNAIII reverses and amounts of RNAIII vastly dominate those of SprY. This result suggests that SprY actively interferes with RNAIII function at low cell density resulting in ablating RNAIII function and consequently allowing the production of surface proteins and inhibiting the production of extracellular toxins (Figure 7B). Early growth phase presumably represents the early stages of infection (reviewed in 60); therefore, the action of SprY is likely to be important during the beginning of infections, allowing S. aureus to colonize various niches without causing severe consequences for the host.
Recently, another sRNA, SprX, was also shown to modulate the pathogenicity of S. aureus through binding RNAIII. In contrast to SprY, SprX binds the RNAIII 5′-end near the hemolysin hld coding sequence (58). Although the significance of this pairing is unclear, the authors proposed that it might release the intramolecular base pairing between the RNAIII 5′ and 3′ ends, changing the secondary structure of RNAIII and facilitating the ribosomal binding for hld translation, resulting in an increase of Hld production and consequently a rise of hemolysis. Interestingly, SprY is expressed in immediate proximity to sprX2 (alias rsaOR), one of the two copies of sprX in HG003 (7), but the effect of SprY on hemolysis is opposite to SprX2. SprY and SprX2 also have opposite effects on the expression of ecb (24). Interestingly, the adjacent sprY and sprX2 genes show inverse expression profiles during growth, which might explain the contributions of sRNAs in controlling RNAIII expression and function in different points of S. aureus growth. Organization of genes in clusters is characteristic for genes expressing macromolecules with shared functions. In addition to protein gene clusters (61,62), sRNAs are also comprised in such clusters. For example, in S. aureus, an unusual condensed cluster was recently shown to contain several sRNA genes (63). Despite the discoveries of sRNAs in gene clusters, whether they possess similar or unrelated biological roles remains unclear (64). Although additional studies are needed to understand the collaboration between SprY and SprX, the current data suggest a functional relationship between these two sRNAs.
Prophages are known to have important roles in the pathogenicity of S. aureus either by encoding toxins and other accessory virulence factors or by interrupting chromosomal virulence genes such as for β-hemolysin (hlb) (reviewed in (65)). However, in some cases, phages not only carry virulence genes but also affect their expression. Induction of prophage was shown to also stimulate expression of some virulence determinants (66). Moreover, the presence of lysogenic phage deregulates expression of multiple genes (67). In this work, we report an example of phage encoded sRNA, acting through a novel mechanism to block an RNAIII regulatory domain. Although φ12 is present in different S. aureus strains such as HG003, MW2, MSSA476 and MRSA252, USA300, and RF122; however, the sprY gene is found in φSa2 phage of MSSA476, MRSA252, HG003 and Newman strains but not found neither in USA300 nor in MW2 (68–71). It is possible, that the acquisition of φ12 phage carrying sprY would fine tune RNAIII function and give the selective advantage to certain bacterial strains in some infection conditions. This sRNA-based regulation is one of the numerous pathways acquired by S. aureus to modulate RNAIII activity. In the absence of φ12-encoded sprY, this bacterium possesses alternative mechanisms to adjust expression and function of agr and its effector molecule, RNAIII, such as protein-based controls like SarA or CcpA (72–75), and RNA regulators like PSM-mec and SprX (58,76,77). Our work deciphers another layer in the multifaceted regulation of virulence factors during S. aureus infection and also raises a number of important evolutionary questions regarding regulatory control provided by the prophage.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Dr Victor J. Torres for antibodies against Rot. We acknowledge Mr. Stéphane Dréano from IGDR UMR 6290 CNRS-UR1 for DNA sequencing assistance. We are also grateful to Dr Alexandra Gruss for the critical reading of the manuscript. We also acknowledge the ‘plateform Génomique Santé’ Biogenouest Génomique Biosit core facility for their technical assistance. We thank the animal core facility ARCHE (ARCHE/SFR BIOSIT, Université Rennes 1, France).
Contributor Information
Kim Boi Le Huyen, Inserm, BRM [Bacterial Regulatory RNAs and Medicine] - UMR_S 1230, 35033 Rennes, France.
Cintia Daniela Gonzalez, Inserm, BRM [Bacterial Regulatory RNAs and Medicine] - UMR_S 1230, 35033 Rennes, France.
Gaëtan Pascreau, Inserm, BRM [Bacterial Regulatory RNAs and Medicine] - UMR_S 1230, 35033 Rennes, France.
Valérie Bordeau, Inserm, BRM [Bacterial Regulatory RNAs and Medicine] - UMR_S 1230, 35033 Rennes, France.
Vincent Cattoir, Inserm, BRM [Bacterial Regulatory RNAs and Medicine] - UMR_S 1230, 35033 Rennes, France.
Wenfeng Liu, Université Paris-Saclay, CEA, CNRS, Institute for Integrative Biology of the Cell (I2BC), 91198 Gif-sur-Yvette, France.
Philippe Bouloc, Université Paris-Saclay, CEA, CNRS, Institute for Integrative Biology of the Cell (I2BC), 91198 Gif-sur-Yvette, France.
Brice Felden, Inserm, BRM [Bacterial Regulatory RNAs and Medicine] - UMR_S 1230, 35033 Rennes, France.
Svetlana Chabelskaya, Inserm, BRM [Bacterial Regulatory RNAs and Medicine] - UMR_S 1230, 35033 Rennes, France.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Agence nationale de la recherche (ANR) [ANR-15-CE12-0003-01]; Fondation pour la Recherche Médicale (FRM) [DBF20160635724]; (INSERM) Institut National de la Santé et de la Recherche Médicale, the Région Bretagne (PhD thesis) (to K.B.L.H.); School of Pharmacy and Medical Sciences of the University of Rennes 1. Funding for open access charge: FRM [DBF20160635724].
Conflict of interest statement. None declared.
REFERENCES
- 1. FRANKLIN L, D.O.M.D. Staphylococcus aureus Infections. N. Engl. J. Med. 1998; 339:520–532. [DOI] [PubMed] [Google Scholar]
- 2. Archer GL., Climo MW.. Staphylococcus aureus bacteremia - consider the source. N. Engl. JMed. 2001; 344:55–56. [DOI] [PubMed] [Google Scholar]
- 3. Wagner E.G.H., Romby P.. Small RNAs in Bacteria and Archaea: Who They Are, What They Do, and How They Do It. Adv. Genet. 2015; 90:133–208. [DOI] [PubMed] [Google Scholar]
- 4. Sassi M., Augagneur Y., Mauro T., Ivain L., Chabelskaya S., Hallier M., Sallou O., Felden B.. SRD: A Staphylococcus regulatory RNA database. RNA. 2015; 21:1005–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liu W., Rochat T., Toffano-Nioche C., Le Lam T.N., Bouloc P., Morvan C.. Assessment of bona fide sRNAs in Staphylococcus aureus. Front. Microbiol. 2018; 9:228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Geissmann T., Chevalier C., Cros M.J., Boisset S., Fechter P., Noirot C., Schrenzel J., François P., Vandenesch F., Gaspin C.et al.. A search for small noncoding RNAs in Staphylococcus aureus reveals a conserved sequence motif for regulation. Nucleic Acids Res. 2009; 37:7239–7257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bohn C., Rigoulay C., Chabelskaya S., Sharma C.M., Marchais A., Skorski P., Borezée-Durant E., Barbet R., Jacquet E., Jacq A.et al.. Experimental discovery of small RNAs in Staphylococcus aureus reveals a riboregulator of central metabolism. Nucleic Acids Res. 2010; 38:6620–6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rochat T., Bohn C., Morvan C., Le Lam T.N., Razvi F., Pain A., Toffano-Nioche C., Ponien P., Jacq A., Jacquet E.et al.. The conserved regulatory RNA RsaE down-regulates the arginine degradation pathway in Staphylococcus aureus. Nucleic Acids Res. 2018; 46:8803–8816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Eyraud A., Tattevin P., Chabelskaya S., Felden B.. A small RNA controls a protein regulator involved in antibiotic resistance in Staphylococcus aureus. Nucleic Acids Res. 2014; 42:4892–4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chabelskaya S., Gaillot O., Felden B.. A Staphylococcus aureus small RNA is required for bacterial virulence and regulates the expression of an immune-evasion molecule. PLoS Pathog. 2010; 6:e1000927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bronesky D., Wu Z., Marzi S., Walter P., Geissmann T., Moreau K., Vandenesch F., Caldelari I., Romby P.. Staphylococcus aureus RNAIII and its regulon link quorum sensing, stress responses, metabolic adaptation, and regulation of virulence gene expression. Annu. Rev. Microbiol. 2016; 70:299–316. [DOI] [PubMed] [Google Scholar]
- 12. Benito Y., Kolb F.A., Romby P., Lina G., Etienne J., Vandenesch F.. Probing the structure of RNAIII, the Staphylococcus aureus agr regulatory RNA, and identification of the RNA domain involved in repression of protein A expression. RNA. 2000; 6:668–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Morfeldt E., Taylor D., Von Gabain A., Arvidson S.. Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. EMBO J. 1995; 14:4569–4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gray G.S., Kehoe M.. Primary sequence of the α-toxin gene from Staphylococcus aureus Wood 46. Infect. Immun. 1984; 46:615–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bramley A.J., Patel A.H., O’Reilly M., Foster R., Foster T.J.. Roles of alpha-toxin and beta-toxin in virulence of Staphylococcus aureus for the mouse mammary gland. Infect. Immun. 1989; 57:2489–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wilke G.A., Wardenburg J.B.. Role of a disintegrin and metalloprotease 10 in Staphylococcus aureus α-hemolysin - Mediated cellular injury. Proc. Natl. Acad. Sci. USA. 2010; 107:13473–13478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huntzinger E., Boisset S., Saveanu C., Benito Y., Geissmann T., Namane A., Lina G., Etienne J., Ehresmann B., Ehresmann C.et al.. Staphylococcus aureus RNAIII and the endoribonuclease III coordinately regulate spa gene expression. EMBO J. 2005; 24:824–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Amdahl H., Haapasalo K., Tan L., Meri T., Kuusela P.I., Van Strijp J.A., Rooijakkers S., Jokiranta T.S.. Staphylococcal protein Ecb impairs complement receptor-1 mediated recognition of opsonized bacteria. PLoS One. 2017; 12:e0172675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Boisset S., Geissmann T., Huntzinger E., Fechter P., Bendridi N., Possedko M., Chevalier C., Helfer A.C., Benito Y., Jacquier A.et al.. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev. 2007; 21:1353–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mcnamara P.J., Milligan-Monroe K.C., Khalili S., Proctor R.A.. Identification, cloning, and initial characterization of rot, a locus encoding a regulator of virulence factor expression in Staphylococcus aureus. J. Bacteriol. 2000; 182:3197–3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Oscarsson J., Tegmark-Wisell K., Arvidson S.. Coordinated and differential control of aureolysin (aur) and serine protease (sspA) transcription in Staphylococcus aureus by sarA, rot and agr (RNAIII). Int. J. Med. Microbiol. 2006; 296:365–380. [DOI] [PubMed] [Google Scholar]
- 22. Saïd-Salim B., Dunman P.M., McAleese F.M., Macapagal D., Murphy E., McNamara P.J., Arvidson S., Foster T.J., Projan S.J., Kreiswirth B.N.. Global regulation of Staphylococcus aureus genes by Rot. J. Bacteriol. 2003; 185:610–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Le Lam T.N., Morvan C., Liu W., Bohn C., Jaszczyszyn Y., Bouloc P.. Finding sRNA-associated phenotypes by competition assays: an example with Staphylococcus aureus. Methods. 2017; 117:21–27. [DOI] [PubMed] [Google Scholar]
- 24. Ivain L., Bordeau V., Eyraud A., Hallier M., Dreano S., Tattevin P., Felden B., Chabelskaya S.. An in vivo reporter assay for sRNA-directed gene control in Gram-positive bacteria: Identifying a novel sRNA target in Staphylococcus aureus. Nucleic Acids Res. 2017; 45:4994–5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chabelskaya S., Gaillot O., Felden B.. A Staphylococcus aureus small RNA is required for bacterial virulence and regulates the expression of an immune-evasion molecule. PLoS Pathog. 2010; 6:e1000927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Antal M., Bordeau V., Douchin V., Felden B.. A small bacterial RNA regulates a putative ABC transporter. J. Biol. Chem. 2005; 280:7901–7908. [DOI] [PubMed] [Google Scholar]
- 27. Steiner P.A., De Corte D., Geijo J., Mena C., Yokokawa T., Rattei T., Herndl G.J., Sintes E.. Highly variable mRNA half-life time within marine bacterial taxa and functional genes. Environ. Microbiol. 2019; 21:3873–3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Britton R.A., Wen T., Schaefer L., Pellegrini O., Uicker W.C., Mathy N., Tobin C., Daou R., Szyk J., Condon C.. Maturation of the 5′ end of Bacillus subtilis 16S rRNA by the essential ribonuclease YkqC/RNase J1. Mol. Microbiol. 2007; 63:127–138. [DOI] [PubMed] [Google Scholar]
- 29. Batey R.T., Kieft J.S.. Improved native affinity purification of RNA. RNA. 2007; 13:1384–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lalaouna D., Desgranges E., Caldelari I., Marzi S.. MS2-Affinity Purification Coupled With RNA Sequencing Approach in the Human Pathogen Staphylococcus aureus. Methods Enzymol. 2018; 612:393–411. [DOI] [PubMed] [Google Scholar]
- 31. Lalaouna D., Carrier M.C., Semsey S., Brouard J.S., Wang J., Wade J.T., Massé E.. A 3′ external transcribed spacer in a tRNA transcript acts as a sponge for small RNAs to prevent transcriptional noise. Mol. Cell. 2015; 58:393–405. [DOI] [PubMed] [Google Scholar]
- 32. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Iandolo J.J., Worrell V., Groicher K.H., Qian Y., Tian R., Kenton S., Dorman A., Ji H., Lin S., Loh P.et al.. Comparative analysis of the genomes of the temperate bacteriophages φ11, φ12 and φ13 of Staphylococcus aureus 8325. Gene. 2002; 289:109–118. [DOI] [PubMed] [Google Scholar]
- 34. Mäder U., Nicolas P., Depke M., Pané-Farré J., Debarbouille M., van der Kooi-Pol M.M., Guérin C., Dérozier S., Hiron A., Jarmer H.et al.. Staphylococcus aureus transcriptome architecture: from laboratory to infection-mimicking conditions. PLos Genet. 2016; 12:e1005962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pichon C., Felden B.. Small RNA genes expressed from Staphylococcus aureus genomic and pathogenicity islands with specific expression among pathogenic strains. Proc. Natl. Acad. Sci. USA. 2005; 102:14249–14254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Naville M., Ghuillot-Gaudeffroy A., Marchais A., Gautheret D.. ARNold: A web tool for the prediction of rho-independent transcription terminators. RNA Biol. 2011; 8:11–13. [DOI] [PubMed] [Google Scholar]
- 37. Massé E., Escorcia F.E., Gottesman S.. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 2003; 17:2374–2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Viegas S.C., Pfeiffer V., Sittka A., Silva I.J., Vogel J., Arraiano C.M.. Characterization of the role of ribonucleases in Salmonella small RNA decay. Nucleic Acids Res. 2007; 35:7651–7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Herbert S., Ziebandt A.K., Ohlsen K., Schäfer T., Hecker M., Albrecht D., Novick R., Götz F.. Repair of global regulators in Staphylococcus aureus 8325 and comparative analysis with other clinical isolates. Infect. Immun. 2010; 78:2877–2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rouillard J.M., Zuker M., Gulari E.. OligoArray 2.0: Design of oligonucleotide probes for DNA microarrays using a thermodynamic approach. Nucleic Acids Res. 2003; 31:3057–3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003; 31:3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Raden M., Ali S.M., Alkhnbashi O.S., Busch A., Costa F., Davis J.A., Eggenhofer F., Gelhausen R., Georg J., Heyne S.et al.. Freiburg RNA tools: A central online resource for RNA-focused research and teaching. Nucleic Acids Res. 2018; 46:W25–W29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Will S., Reiche K., Hofacker I.L., Stadler P.F., Backofen R.. Inferring noncoding RNA families and classes by means of genome-scale structure-based clustering. PLoS Comput. Biol. 2007; 3:680–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Will S., Joshi T., Hofacker I.L., Stadler P.F., Backofen R.. LocARNA-P: Accurate boundary prediction and improved detection of structural RNAs. RNA. 2012; 18:900–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chevalier C., Boisset S., Romilly C., Masquida B., Fechter P., Geissmann T., Vandenesch F., Romby P.. Staphylococcus aureus RNAIII binds to two distant regions of coa mRNA to arrest translation and promote mRNA degradation. PLoS Pathog. 2010; 6:e1000809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lalaouna D., Massé E.. Identification of sRNA interacting with a transcript of interest using MS2-affinity purification coupled with RNA sequencing (MAPS) technology. Genomics Data. 2015; 5:136–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Busch A., Richter A.S., Backofen R.. IntaRNA: Efficient prediction of bacterial sRNA targets incorporating target site accessibility and seed regions. Bioinformatics. 2008; 24:2849–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mann M., Wright P.R., Backofen R.. IntaRNA 2.0: Enhanced and customizable prediction of RNA-RNA interactions. Nucleic Acids Res. 2017; 45:W435–W439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Geisinger E., Adhikari R.P., Jin R., Ross H.F., Novick R.P.. Inhibition of rot translation by RNAIII, a key feature of agr function. Mol. Microbiol. 2006; 61:1038–1048. [DOI] [PubMed] [Google Scholar]
- 50. Amdahl H., Jongerius I., Meri T., Pasanen T., Hyvärinen S., Haapasalo K., van Strijp J.A., Rooijakkers S.H., Jokiranta T.S.. Staphylococcal Ecb protein and host complement regulator factor H enhance functions of each other in bacterial immune evasion. J. Immunol. 2013; 191:1775–1784. [DOI] [PubMed] [Google Scholar]
- 51. Tuffs S.W., Herfst C.A., Baroja M.L., Podskalniy V.A., DeJong E.N., Coleman C.E.M., McCormick J.K.. Regulation of toxic shock syndrome toxin-1 by the accessory gene regulator in Staphylococcus aureus is mediated by the repressor of toxins. Mol. Microbiol. 2019; 112:1163–1177. [DOI] [PubMed] [Google Scholar]
- 52. Figueroa-bossi N., Bossi L.. Sponges and predators in the small RNA World. Microbiol. Spectr. 2018; 6: 10.1128/microbiolspec.rwr-0021-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Figueroa-Bossi N., Valentini M., Malleret L., Bossi L.. Caught at its own game: Regulatory small RNA inactivated by an inducible transcript mimicking its target. Genes Dev. 2009; 23:2004–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Overgaard M., Johansen J., Møller-Jensen J., Valentin-Hansen P.. Switching off small RNA regulation with trap-mRNA. Mol. Microbiol. 2009; 73:790–800. [DOI] [PubMed] [Google Scholar]
- 55. Miyakoshi M., Chao Y., Vogel J.. Cross talk between ABC transporter m RNA s via a target m RNA -derived sponge of the G cv B small RNA. EMBO J. 2015; 34:1478–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Denham E.L. The Sponge RNAs of bacteria – How to find them and their role in regulating the post-transcriptional network. Biochim. Biophys. Acta - Gene Regul. Mech. 2020; 1863:194565. [DOI] [PubMed] [Google Scholar]
- 57. Bossi L., Figueroa-bossi N.. Competing endogenous RNAs: a target-centric view of small RNA regulation in bacteria. Nat. Rev. Microbiol. 2016; 14:775–784. [DOI] [PubMed] [Google Scholar]
- 58. Kathirvel M., Buchad H., Nair M.. Enhancement of the pathogenicity of Staphylococcus aureus strain Newman by a small noncoding RNA SprX1. Med. Microbiol. Immunol. 2016; 205:563–574. [DOI] [PubMed] [Google Scholar]
- 59. Bronesky D., Desgranges E., Corvaglia A., François P., Caballero C.J., Prado L., Toledo-Arana A., Lasa I., Moreau K., Vandenesch F.et al.. A multifaceted small RNA modulates gene expression upon glucose limitation in Staphylococcus aureus. EMBO J. 2019; 38:e99363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Repoila F., Darfeuille F.. Small regulatory non-coding RNAs in bacteria: physiology and mechanistic aspects. Biol. Cell. 2009; 101:117–131. [DOI] [PubMed] [Google Scholar]
- 61. Jongerius I., Köhl J., Pandey M.K., Ruyken M., Van Kessel K.P.M., Van Strijp J.A.G., Rooijakkers S.H.M.. Staphylococcal complement evasion by various convertase-blocking molecules. J. Exp. Med. 2007; 204:2461–2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. McCarthy A.J., Lindsay J.A.. Staphylococcus aureus innate immune evasion is lineage-specific: a bioinfomatics study. Infect. Genet. Evol. 2013; 19:7–14. [DOI] [PubMed] [Google Scholar]
- 63. Bronsard J., Pascreau G., Sassi M., Mauro T., Augagneur Y., Felden B.. SRNA and cis-Antisense sRNA identification in Staphylococcus aureus highlights an unusual sRNA gene cluster with one encoding a secreted peptide. Sci. Rep. 2017; 7:4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Felden B., Paillard L.. When eukaryotes and prokaryotes look alike: the case of regulatory RNAs. FEMS Microbiol. Rev. 2017; 41:624–639. [DOI] [PubMed] [Google Scholar]
- 65. Penadés J.R., Chen J., Quiles-Puchalt N., Carpena N., Novick R.P.. Bacteriophage-mediated spread of bacterial virulence genes. Curr. Opin. Microbiol. 2015; 23:171–178. [DOI] [PubMed] [Google Scholar]
- 66. Sumby P., Waldor M.K.. Transcription of the toxin genes present within the staphylococcal phage φsa3ms is intimately linked with the phage's life cycle. J. Bacteriol. 2003; 185:6841–6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Fernández L., González S., Quiles-Puchalt N., Gutiérrez D., Penadés J.R., García P., Rodríguez A.. Lysogenization of Staphylococcus aureus RN450 by phages ϕ11 and ϕ80α leads to the activation of the SigB regulon. Sci. Rep. 2018; 8:12662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Baba T., Takeuchi F., Kuroda M., Yuzawa H., Aoki K.I., Oguchi A., Nagai Y., Iwama N., Asano K., Naimi T.et al.. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet. 2002; 359:1819–1827. [DOI] [PubMed] [Google Scholar]
- 69. Holden M.T.G., Feil E.J., Lindsay J.A., Peacock S.J., Day N.P.J., Enright M.C., Foster T.J., Moore C.E., Hurst L., Atkin R.et al.. Complete genomes of two clinical Staphylococcus aureus strains: Evidence for the evolution of virulence and drug resistance. Proc. Natl. Acad. Sci. USA. 2004; 101:9786–9791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Diep B.A., Gill S.R., Chang R.F., Phan T.H.V, Chen J.H., Davidson M.G., Lin F., Lin J., Carleton H.A., Mongodin E.F.et al.. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet. 2006; 367:731–739. [DOI] [PubMed] [Google Scholar]
- 71. Herron-Olson L., Fitzgerald J.R., Musser J.M., Kapur V.. Molecular correlates of host specialization in Staphylococcus aureus. PLoS One. 2007; 2:e1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Reyes D., Andrey D.O., Monod A., Kelley W.L., Zhang G., Cheung A.L.. Coordinated regulation by AgrA, SarA, and SarR to control agr expression in Staphylococcus aureus. J. Bacteriol. 2011; 193:6020–6031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Jenul C., Horswill A.R.. Regulation of staphylococcus aureus virulence. Microbiol. Spectr. 2019; 7: 10.1128/microbiolspec.GPP3-0031-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Seidl K., Stucki M., Ruegg M., Goerke C., Wolz C., Harris L., Berger-Bächi B., Bischoff M.. Staphylococcus aureus CcpA affects virulence determinant production and antibiotic resistance. Antimicrob. Agents Chemother. 2006; 50:1183–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ueda T., Kaito C., Omae Y., Sekimizu K.. Sugar-responsive gene expression and the agr system are required for colony spreading in Staphylococcus aureus. Microb. Pathog. 2011; 51:178–185. [DOI] [PubMed] [Google Scholar]
- 76. Kaito C., Saito Y., Ikuo M., Omae Y., Mao H., Nagano G., Fujiyuki T., Numata S., Han X., Obata K.et al.. Mobile Genetic Element SCCmec-encoded psm-mec RNA Suppresses Translation of agrA and Attenuates MRSA Virulence. PLoS Pathog. 2013; 9:e1003269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Qin L., McCausland J.W., Cheung G.Y.C., Otto M.. PSM-Mec-A virulence determinant that connects transcriptional regulation, virulence, and antibiotic resistance in staphylococci. Front. Microbiol. 2016; 7:1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.