Abstract
Pervasive transcription of eukaryotic genomes results in expression of long non-coding RNAs (lncRNAs) most of which are poorly conserved in evolution and appear to be non-functional. However, some lncRNAs have been shown to perform specific functions, in particular, transcription regulation. Thousands of small open reading frames (smORFs, <100 codons) located on lncRNAs potentially might be translated into peptides or microproteins. We report a comprehensive analysis of the conservation and evolutionary trajectories of lncRNAs-smORFs from the moss Physcomitrium patens across transcriptomes of 479 plant species. Although thousands of smORFs are subject to substantial purifying selection, the majority of the smORFs appear to be evolutionary young and could represent a major pool for functional innovation. Using nanopore RNA sequencing, we show that, on average, the transcriptional level of conserved smORFs is higher than that of non-conserved smORFs. Proteomic analysis confirmed translation of 82 novel species-specific smORFs. Numerous conserved smORFs containing low complexity regions (LCRs) or transmembrane domains were identified, the biological functions of a selected LCR-smORF were demonstrated experimentally. Thus, microproteins encoded by smORFs are a major, functionally diverse component of the plant proteome.
INTRODUCTION
The progress of next generation RNA sequencing technologies has led to the striking discovery of pervasive transcription of eukaryotic genomes (1–3). Effectively, each base in animal and plant genomes is transcribed in some cell types, at some developmental stage(s), at some level. The great majority of these transcripts (98–99%) appear to be non-coding RNAs (4,5). A major, heterogeneous class of diverse long non-coding RNAs (lncRNAs) are traditionally defined as transcripts longer than 200 nucleotides (nt), without discernible coding potential (6,7). Only a minority of the lncRNAs have been shown to perform specific functions, primarily, in chromatin remodeling and regulation of gene expression (7).
An important question is, are all lncRNAs actually non-coding? Many recent studies have demonstrated that lncRNAs are frequently bound to ribosomes (8–10). The ribosome-associated lncRNAs could be translated to produce peptides or microproteins (11–13), and alternatively or additionally, might be involved in translation regulation (14); else, the interactions with ribosomes might play a role in the degradation of the lncRNAs (15). Potentially translated regions in lncRNAs are small ORFs (smORFs, from 10 to 100 codons) (16). Hundreds of peptides or microproteins have been identified by proteomics in mammals (17–19), fungi (20), plants (8,21,22) and bacteria (23,24). Some of the peptides or microproteins encoded by smORFs have been shown to perform diverse biological functions (22,25).
Transcription of non-coding portions of genomes can result in de novo emergence of new protein-coding genes (26,27). Because translation of (initially) spurious peptides encoded by lncRNAs can potentially be harmful to the cell, the primary selection has been suggested to be for the avoidance of aggregation (the ‘do no harm’ hypothesis). Alternatively, hydrophobic random peptides that can often emerge from T-rich sequences, given that T-rich codons largely encode hydrophobic amino acids, could readily become small transmembrane (TM) proteins, in a form of preadaptation, as suggested by the ‘TM-first’ model of gene birth (27–29). A specific case of evolution of a functional yeast membrane protein (YBR196C-A) originated from a thymine-rich intergenic sequence has been recently documented in detail (27). Other studies have shown that de novo genes may emerge from GC-rich sequences that have a low frequency of stop-codons (30,31).
Thus, there seem to be multiple evolutionary routes leading to the de novo emergence of small proteins. Several dedicated web resources and databases have been developed to catalog and annotate smORFs (32–35). However, the global role of lncRNAs as a source of functional peptides remains largely unknown. It seems likely that the majority of the lncRNAs that are inferred to be translatable from the ribosome profiling data actually are unannotated mRNAs. Furthermore, evolutionarily conserved regions in lncRNAs are significantly enriched in potentially translated smORFs and in protein–RNA interaction signatures (36). In addition, microproteins translated from lncRNAs with well-characterized non-coding functions have been identified as well (17), suggesting the existence of transcripts with dual functionality. Overall, the functions and evolutionary history of the ‘dark proteome’ hidden in genome regions that are currently considered non-coding still awaits a comprehensive analysis.
To our knowledge, the plant smORFome has not been systematically studied previously. To gain insight into the functions and evolution of smORFs present in lncRNAs, we performed a comprehensive analysis of the smORFs conservation across plant taxa, using as a reference set the lncRNAs of the moss Physcomitrella (Physcomitrium patens), a well-characterized plant model. Thousands of evolutionarily conserved smORFs were identified. The translation of numerous smORFs into peptides or proteins was validated by peptidomics, and the functions of selected small proteins in the moss were characterized experimentally.
MATERIALS AND METHODS
Analysis of publicly available long non-coding RNA datasets
The 1498 predicted lncRNAs from CANTATdb 2.0 database (37), 9416 high-confidence lncRNAs from GreeNC (38), 3018 lncRNAs from P. patens NCBI annotation (https://www.ncbi.nlm.nih.gov/genome/annotation_euk/Physcomitrella_patens/100), 1512 lncRNAs from Lung et al.'s paper (39) and 4648 from Simopoulos et al.'s paper (40) were brought together and redundant transcripts were excluded. Using bedtools (41) lncRNAs were intersect and combined into loci. Using TBLASTN search, we further filtered out lncRNAs having sequence similarity to annotated viridiplantae proteins from Uniprot database (E-value < 10–5; overlap > 30%). In addition, we also discarded lcnRNAs with sequence similarity to known noncoding RNAs from the Rfam database (E-value < 10–5). The control set of 16 178 mRNA transcripts was obtained from Phytozome v12.1 (https://phytozome.jgi.doe.gov/pz/portal.html). Only transcripts coding for annotated functional proteins and confirmed by nanopore analysis (see below) were included. The percent GC was calculated with EMBOSS infoseq (42).
smORFs prediction and classification
LncRNA transcripts and whole loci were scanned for possible AUG-started smORFs by the MiPepid tool (43). The NCBI finder (44) allows users to search for ORFs without limiting the length of the query sequence and using various genetic codes, including different start codons. The smORFs predicted by NCBI finder (https://www.ncbi.nlm.nih.gov/orffinder/) which intersected or nested in MiPepid-smORFs were then filtered out.
The conservation analysis
The transcriptomes of green algae, liverworts, mosses, hornworts, lycophytes, leptosporangiate ferns, conifers and basal eudicots species (Supplementary Table S1) from OneKP plant transcriptomes project (45) were downloaded from https://datacommons.cyverse.org/browse/iplant/home/shared/commons_repo/curated/oneKP_capstone_2019. We searched for significant sequence similarity hits between moss lncRNA loci and transcripts from different plant lineages using BLASTN (46) with the E-value 10−5 cut-off. Results were very similar for different E-value cut-offs: the percent of conserved lncRNAs was the same when making this parameter more stringent (E-value < 10−6) and only slightly increased by 0.1% when relaxing the E-value (E-value < 10−4).
To identify smORF orthologs in selected plant taxa, TBLASTN search with default parameters was performed using smORFs as queries and the transcriptome sequences as subjects. The alignments were filtered by E-value < 10−3 cut-off. The number of conserved smORFs only increased by 1.9% with a relaxed E-value cut-off (E-value < 10−2) and decreased by 1.64% in more stringent cut-off (E-value < 10−4).
Evolutionary analysis
The nucleotide sequence alignments of the orthologous transcripts and coding sequences were constructed using Owen (47) and Muscle (48) software and alignments of CDSs and ORFs sequences were guided by the amino acid sequence alignment (47–49). The selected coding sequence alignments should not contain internal stop codons and match a start codon. We also considered that the alignment length of orthologous coding sequences should be >80% of smORF or protein length.
The nucleotide sequence alignments of orthologous coding sequences were obtained by backtracking the amino acid sequence alignments using PAL2NAL (50). For the nucleotide sequences, the rates of divergence were calculated using Kimura's two parameter model (51). Evolutionary rates for synonymous and nonsynonymous positions (dS and dN, respectively) in coding regions were estimated using the PAML software and the Maximum Likelihood method for pairs of species (52). The dN and dS values of ambiguous alignments, leading to an unreliable estimation of evolutionary rates, were excluded from the analysis.
In addition to PAML, we also used HyPhy's BUSTED algorithm (53) to identify smORFs and proteins with evidence of positive selection. BUSTED (Branch-Site Unrestricted Statistical Test for Episodic Diversification) provides a test for positive selection in at least one site on at least one branch. The input trees for the tests of positive selection were constructed for each alignment with IQ-TREE 1.6.12 (54). We parsed all PAML and HyPhy results by custom Python scripts. We considered a P-value less than 0.05 as evidence for positive selection. We used the Benjamini-Hochberg (55) approach to correct for multiple testing. The phylogenetic trees were generated by the Toytree package (56).
Domains and motifs analysis
smORF sequences were scanned for domains using InterProScan 4.36 (57) with default settings. The web plant SSP-Prediction tool available at http://mtsspdb.noble.org/prediction/ was used (58). The potential SSPs were classified as Known SSP, Likely Known SSP, Putative SSP and Non-SSP (58). MEME (http://meme-suite.org/) tool was used to identify conserved motifs (59). Low-complexity regions (LCRs) were identified by SEG tool with default settings (60).
Cellular localization
SignalP-5.0 (61) was run with default parameters on all smORFs. TMHMM 2.0 (62) was run on the same set of smORFs with default parameters.
Plant material and mutant lines generation
Physcomitrella (Physcomitrium patens ‘Gransden 2004’, Frieburg) protonemata and gametophores were grown as described previously (22). The PSEP3 overexpression lines were generated based on the β-estradiol induction system (63). The plasmid pPGX8 (AB537482) was kindly provided by Prof. Mitsuyasu Hasebe. The resulting plasmid pPGX8_PSEP3 was used for transformation as described previously (22). Overexpression lines were screened using PCR and quantitative RT–PCR analyses. The primer sequences, plasmids and constructs used for the production of the overexpression lines can be found in Supplementary Figure S1.
The PSEP3 overexpression was induced by 1 μM β-estradiol (Sigma, USA) dissolved in ethanol. To confirm induction of PSEP3 transcription, RT-qPCR was used. Total RNA was isolated using TRIzol™ Reagent (Ambion, US) according to the manufacturer protocol and reverse transcribed by MMLV-RT kit (Evrogen, Russia). PCR qPCRmix-HS mix (Evrogen, Russia) was used for quantitative RT–qPCR analyses on a LightCycler® 96 (Roche, Mannheim, Germany). The representation of cDNA was normalized using stably transcribed reference gene actin 5 (Pp1s381_21V6.1). The 2-ddCT values were obtained using the LightCycler® 96 software. Control samples were used as a calibrator.
Peptide extraction
Endogenous peptides were extracted from 5-day old protonemata and 30-day old gametophores as described previously (22).
Protein extraction and trypsin digestion
Protein extraction and trypsin digestion we conducted as described previously (64,65). iTRAQ labelling (Applied Biosystems, Foster City, CA, USA) was conducted according to the manufacturer's manual. The experiments were conducted independently and samples were labelled and combined as follows: the wild type samples were labelled by 113, 114 and 116 isobaric tags and combined with mutant РSЕР3 ОЕ samples labelled by 117, 118 and 121 isobaric tags; the wild type samples labelled by 113, 114 and 115 isobaric tags were combined with PSEP3 КО mutant samples (116, 119 and 121 isobaric tags); wild type samples were labeled by 113, 114 and 116 isobaric tags and combined with samples from PSEP18 КО mutant plants labelled by 116,119 and 121 isobaric tags; iTRAQ reagents 113, 114 and 115 were used to label wild type samples that were combined with PSEP18 ОE samples labelled by 116, 119 and 121 isobaric tags. To increase the number of identified proteins, labelled peptides were fractionated by cation exchange chromatography. Peptides were eluted successively by 50, 75, 125, 200 and 300 mM ammonium acetate in 0.5% formic acid and 20% acetonitrile; 5% NH4OH in 80% acetonitrile; 10% NH4OH in 60% acetonitrile.
LC–MS/MS analysis and smORF identification
The LC–MS/MS analysis was performed as described earlier (22). For analysis of smORFs translation, five peptidomic data sets - PXD009532 (57), PXD007922 (17), PXD007923 (17), PXD025373 and PXD025267 were used. The PXD025373 and PXD025267 datasets were generated in this study. Tandem mass spectra from peptidomic samples were searched individually with PEAKS Studio version 8.0 software (Bioinfor Inc., CA, USA) and MaxQuant v1.6.14 (66) against a custom database containing 32 926 proteins from annotated genes in the latest version of the moss genome (V3.3) (39), 85 moss chloroplast proteins, 42 moss mitochondrial proteins, and predicted smORF peptides. MaxQuant's protein FDR filter was disabled, while 1% FDR was used to select high-confidence peptide-spectrum matches (PSMs), and ambiguous peptides were filtered out. The parameter ‘Digestion Mode’ was set to ‘unspecific’ and modifications were not permitted. All other parameters were left as default values. All other parameters were left as default values. After MaxQuant peptide searches, a more stringent FDR filtering strategy was used (67). A class specific FDR was calculated as the number of decoy smORF hits divided by the number of target smORF hits. 1% class specific FDR was applied to the smORF PSMs.
The search parameters of PEAKS 8.0 were a fragmentation mass tolerance of 0.05 Da; parent ion tolerance of 10 ppm; without modifications. The results were filtered by a 1% FDR, but with a significance threshold not less than 20 (equivalent is P-value < 0.01).
Protein quantification
The raw files were analyzed by PEAKS Studio version 8.0 software (Bioinfor Inc., CA, USA). The custom database was built from the Phytozome proteomic database combined with chloroplast and mitochondrial proteins. The database search was performed with the following parameters: a fragmentation mass tolerance of 0.05 Da; parent ion tolerance of 10 ppm; fixed modification—carbamidomethylation; variable modifications—oxidation (M), deamidation (NQ) and acetylation (Protein N-term). The results were filtered by a 1% false discovery rate (FDR). PEAKS Q was used for iTRAQ quantification. Normalization was performed by averaging the abundance of all peptides. Median values were used for averaging. Differentially expressed proteins were filtered if their fold change was greater than 1.2 and significance threshold 20 with a statistical P-value (ANOVA test with Benjamini and Hochberg FDR correction) below 0.05, variance homogeneity test (P-value > 0.05), and normal distribution test (P-value > 0.05).
Long-reading native RNA analysis
Total RNA from gametophores and protonemata was isolated by TRIzol™ Reagent (Ambion, USA). Four biological repeats of gametophores and three biological repeats of protonemata were used for analysis. RNA quality and quantity were evaluated via electrophoresis in an agarose gel with ethidium bromide staining. The precise concentration of total RNA in each sample was measured using a Quant-iT™ RNA Assay Kit, 5–100 ng on a Qubit 3.0 (Invitrogen, US) fluorometer. 100 μg aliquots of total RNA were diluted in 100 μl of nuclease-free water and poly(A) fraction was selected by Poly(A)Purist™-MAG Purification Kit (Thermo Fisher Scientific, USA). The Nanopore direct RNA sequencing kit (SQK-RNA002, Oxford Nanopore) was used to prepare libraries from the poly(A) RNA. About 200 ng of each prepared libraries were loaded onto FLO-MIN106 (ONT R9.4) flowcells and sequencing was performed in MinION sequencers. Each library was run for 48 h.
The obtained reads were basecalled by Guppy 4.0.15 (Oxford Nanopore Technologies). The resulting reads were mapped against the genome Physcomitrella patens V3.3 (68) by minimap 2.17 (69) with the following parameters: -ax splice -uf -k14 f -G2k. ONT reads with a primary alignment to the genome were retained. The obtained ‘SAM’ files were sorted and indexed with ‘SAMtools’(70).
To confirm lncRNAs and mRNAs transcription, we combined the results from StringTie (71) tool with -L parameter and Flair (72), both in the pipe with GffCompare (73). To estimate transcript abundance the default Flair (72) pipeline and featureCounts 2.0.0 (74) tools were used. Mono-exon features were quantified with featureCounts 2.0.0 (74) (parameters -s 1 -L -O –fracOverlapFeature 0.75 –fracOverlap 0.75), because Flair is suitable only for features with splice junctions. Both Flair and featureCounts produce quantification as simple read counts, which is appropriate for long reads. The full pipeline can be found on https://github.com/Liverworks/Ppatens_lncRNAs.
Fluorescent microscopy
To evaluate ROS induction and cell viability in the PSEP3 overexpressed lines, 6-day old moss protonemata were treated with 1 μM β-estradiol (Sigma, USA).
The detection of ROS was performed by fluorescent dye DCFH-DA (2′,7′-Dichlorofluorescin Diacetate, Sigma-Aldrich, USA) in 24-h after induction. The protonemal tissues were stained with 10 μM DCFH-DA during 10 min.
A cell viability assay was performed in 48-h after estradiol induction. The protonemal cells were stained by FDA (fluorescein diacetate, Sigma-Aldrich, USA) during 5 min and the ratio of live and dead cells was used to calculate cell viability. Fluorescence signal was detected by Axio Imager M2 microscope (Zeiss) with an AxioCam 506 mono digital camera (Zeiss) and Zen 2.6 pro software (Zeiss). Filter unit №44 (λex BP 475 nm/40 nm; λem BP 530 nm/50 nm) was used for DCFH-DA and FDA fluorescence detection.
Statistical analysis
Statistical analyses and visualization were made in Python v. 3.7.5 (75) using modules scipy 1.5.2 (76), seaborn 0.11.1 (77), numpy 1.20.1, pandas 1.2.3 (78) and upsetplot 0.5.0 (79).
RESULTS
Comprehensive bioinformatics characterization of the lncRNA-smORFome
Delineation of the set of smORFs in Physcomitrium patens
LncRNAs are a poorly characterized class of transcripts and their prediction is a challenging task (80). Therefore, we first defined a set of lncRNAs from the model plant - moss Physcomitrella (Physcomitrium patens) by combining several available datasets (37,38,40,68). Using the annotation from available databases (see Material and Methods), all lncRNAs were mapped to Physcomitrella genome v3.3 (39) and combined into 9168 loci (Supplementary Table S2) where alternative transcripts could occupy the same loci and overlap.
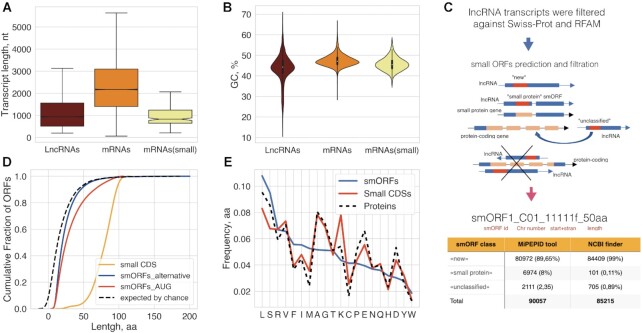
As a control set, we also selected 16 178 P. patens primary mRNA transcripts (Supplementary Table S3) coding for annotated functional proteins from Phytozome v12.1 and confirmed by nanopore RNA sequencing (see below). This set includes 252 mRNAs coding for proteins smaller than 100 aa (small coding sequences, small CDSs) that contain identifiable functional domains (median size = 81aa; small-mRNAs). In agreement with previous analyses (36,81), the length and GC content of lncRNAs significantly differed from those of the mRNAs and the small-mRNAs (Figure 1A and B, respectively; Kruskal–Wallis rank sum test, P < 10–15). In addition, lncRNAs contained significantly fewer exons (median = 1) than mRNAs (median = 6; Mann–Whitney U test, P< 0.000001).
Figure 1.
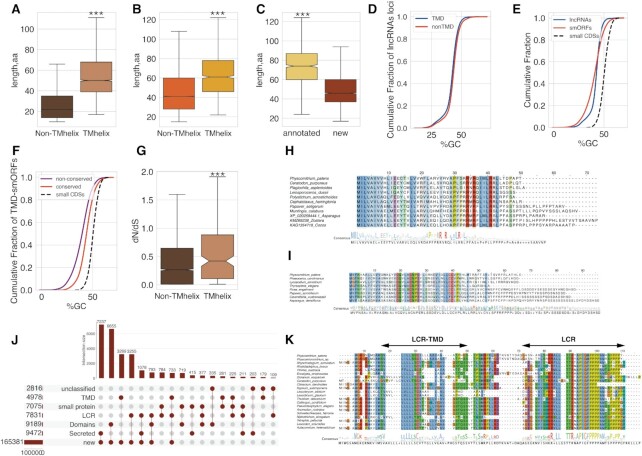
Prediction and classification of small ORFs. (A) Boxplot showing the length comparison of lncRNAs, mRNAs and small RNAs (code for functional proteins below 100aa). The median, quartiles, and 5th and 95th percentiles are shown. (B) Comparison of GC contents of lncRNAs, mRNAs and small RNAs. (C) Pipeline of smORFs prediction and classification. (D) Cumulative distribution of different CDS lengths; ‘random smORFs’ refer to smORFs expected to occur by chance; the median lengths are 25aa and 20aa for MiPEPID-smORF and NCBI ORF finder, respectively; ∼26% MiPEPID-smORFs had the length more than 40 aa. (E) Comparison of amino acid compositions of functionally characterized proteins (including small CDSs, <100aa) and AUG-started smORF-encoded peptides. smORFs were enriched in leucine (chi-square P-value < 10–15), isoleucine (chi-square P-value < 10–15), phenylalanine (chi-square P-value < 10–15) in comparison to functional proteins.
Having delineated a comprehensive set of moss lncRNAs, we then predicted smORFs (from 10 to 100 aa) starting with an AUG codon, using the MiPEPID tool (43). Because functional smORFs can start with alternative codons (18), we additionally used NCBI ORF finder (44) to predict smORFs with non-canonical starts, UUG and CUG. The ORF finder-predicted smORFs included ∼65% UUG-started and ∼35% CUG-started ones. The most common alternative translation initiation site (TIS) that has been identified in both plant and mammalian mRNAs by ribosome profiling is CUG (82,83). The set of MiPEPID-smORFs was then merged with the set of non-AUG starting ORFs identified with NCBI ORF finder to generate an unbiased set of smORFs.
The set of predicted smORFs was thoroughly filtered and classified (Figure 1C; Supplementary Table S4). First, smORFs containing sequences significantly similar to annotated proteins above 200 aa from Phytozome v12.1 (BLASTP, E-value < 0.00001; percent identity ≥ 80%) or overlapping exons of protein-coding genes on both strands by 50% of a smORF length or more were filtered out. The smORFs that did not meet the above criteria, but showing significant similarity to annotated P. patens proteins (E-value < 0.00001) were designated ‘unclassified’. The P. patens proteome includes 7028 predicted proteins smaller than 100 aa, of which many have no associated functional annotation and some could be incorrectly predicted (22). The smORFs that overlapped with such small proteins were designated ‘small protein’ smORFs. The smORFs that did not fit into the ‘small protein’ and ‘unclassified’ categories were designated ‘new’, and all three classes of smORFs were analyzed further (Figure 1C). Overall, ∼49% ‘new’, ∼99% ‘small protein’ and ∼76% ‘unclassified’ smORFs started with AUG. The percentage of smORFs with alternative start codons was ∼33% UUG and ∼18% CUG in ‘new’, ∼16% UUG and ∼8% CUG in ‘unclassified’, and ∼0.45% CUG and ∼0.55% UUG in ‘small protein’ smORFs.
The median sizes of the smORFs significantly differed from the median ORF size of 13 codons expected by chance for the P. patens genome (GC = 45.9%; Mood's median test P < 10–15; Figure 1C, D) (68). About 85% (76289/90057) of the MiPEPID-smORFs were predicted as coding using a logistic regression model (43). The BLASTP search of predicted smORFs against known smORF databases (E-value < 0.001, ≥ 50% of identity) revealed 16 smORFs shared by P. patens and Arabidopsis (84,85) and four homologs in smProt database (35).
Intrinsic features of transcripts, such as sequence conservation or nucleotide composition, are often used for the calculation of their coding potential (86). We found that, on average, the smORFs were significantly less GC-rich than protein-coding ORFs (Kolmogorov–Smirnov test, P < 10−20). It has been shown previously that different types of animal smORFs (lncRNAs-smORFs, upstreamORFs, downstreamORFs etc.) significantly differed from each other in the amino acid composition (25). The amino acid composition of the putative microproteins/peptides encoded by the AUG-started smORFs in our data set differed from the composition of functionally characterized proteins, especially in the content of some hydrophobic amino acids (Figure 1E). Increased frequencies of methionine and cysteine as well as decreased frequencies of alanine, glutamate and aspartate closely resembled the reported features of smORFs in mammalian lncRNAs (25). Overall, the observed composition of smORFs and small CDSs were concordant with the respective values calculated for mammals (25).
Numerous lncRNA-smORFs are conserved across different plant lineages
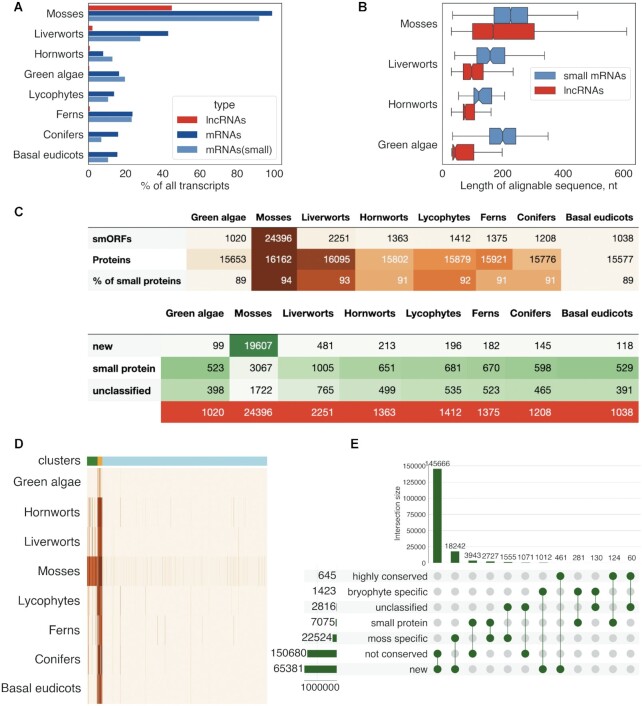
It has been previously shown that, for >70% of the lncRNAs, no homologs could be identified between animal species that diverged more than 50 million years ago (87). To explore the evolutionary conservation of the lncRNAs in plants, we performed BLASTN sequence similarity search (E-value < 0.00001) of the P. patens lncRNA set against transcriptomes from the 1000 (OneKP) plants project (45). The number of lncRNA matches precipitously dropped in more distant plant lineages (Figure 2A). In contrast to lncRNAs, the mRNA transcripts were far more strongly conserved across different plant lineages (Figure 2A). As expected, the conserved regions in mRNA transcripts were longer than those in the lncRNAs, with a median length of 753 and 168 nucleotides, respectively. These observations match the results obtained for mammalian mRNAs and lncRNAs (36,88). In small-mRNAs, the median length of the conserved regions was only slightly larger than in lncRNAs (226 versus 168, respectively; Mann–Whitney U-test, P < 10–15), but small-mRNAs showed a much stronger evolutionary conservation (Figure 2A, B). Thus, our results confirmed that the evolutionary conservation of lncRNAs at the nucleotide level is substantially lower than that of mRNAs.
Figure 2.
Evolutionary conservation of lncRNAs and smORFs. (A) Comparison of the sequence conservation between lncRNAs and mRNAs in different plant lineages; (B) Distributions of lengths of conserved regions of lncRNAs and small mRNAs (code for proteins below 100aa) in bryophytes and green algae. The median, quartiles, and 5th and 95th percentiles are shown; (C) the number of smORFs with detectable orthologs in different plant lineages; ‘% of small proteins'’ shows percent of small CDSs having orthologs in this plant lineage. (D) Heatmap showing the evolutionary conservation pattern of smORFs in different plant lineages. The scaled numbers of species with detectable smORF homologs are shown. Color bars indicate three conservation patterns: light blue—smORFs that are mostly conserved in a small number of moss species (lineage-specific, cluster 3), orange—widely conserved smORFs (highly conserved, cluster 1), green—smORFs that are preferably conserved in mosses and other bryophytes (bryophyte specific, cluster 2). (E) UpSet plot showing intersection between three conservation patterns and smORFs types.
We next analyzed smORFs conservation at the amino acid sequence level. Because increasing stringency of TBLASTN discriminates against short sequences (89), we used an E-value < 0.001 cut-off. In this search, 15167 MiPEPID-smORFs and 9425 ORFfinder-smORFs showed at least one match to 41 moss transcriptomes (see in Supplementary Table S5; Figure 2C), in particular, that of the closest moss species, Physcomitrium sp. (YEPO). In other moss species, the number of putative orthologs ranged from 1130 to 2887 (Supplementary Figure S2). Thus, nearly half of the lncRNA loci (4078 of the 9168 lncRNA loci) from our set contained at least one conserved smORFs.
The proportion of smORFs predicted with low ‘coding’ potential (based on MiPEPID tool classification) was significantly higher among the non-conserved smORFs compared to conserved ones (18% versus 6%, respectively, chi-square P < 10–15). The conserved moss smORFs were also found to be significantly longer than the non-conserved ones (Kruskal–Wallis, P < 0.00001; Supplementary Figure S3). The fraction of pairwise alignments between P. patens and YEPO containing internal stop codons was significantly higher in the MiPEPID-smORFs than in the small CDSs (11.6% versus 1.6%, respectively, chi-square P < 10–10), which is compatible with the lower evolutionary conservation of the smORFs.
The conserved smORFs were clustered according to their level of conservation in different plant lineages. Overall, three prominent patterns of smORF conservation were detected: (i) a high level of conservation among diverse plants, including bryophytes, lycophytes and ferns (n = 645); (ii) broad conservation in moss species and partial conservation in liverworts and hornworts (n = 1423); (iii) conservation in a small number of moss species (n = 22 524; 83% had orthologs in only one species; Figure 2D and E). The ‘new’ smORFs were significantly enriched in the third group and strongly depleted in the first and second groups compared to the other types of smORFs (chi-square P-value < 10–15; Figure 2E).
Thus, the conservation of smORFs rapidly dropped at the transition from mosses to other plant lineages (Figure 2C) and in particular, rapid depletion of ‘new’ smORFs was observed. As expected, annotated proteins (including small CDSs) were more widely conserved than smORFs, with the number of conserved proteins dropping only slightly in distant plant groups (Figure 2C). These findings are in line with previous results describing the fast turnover of smORFs in animal genomes (25,90).
Evolutionary rates and selection in lncRNAs and smORFs
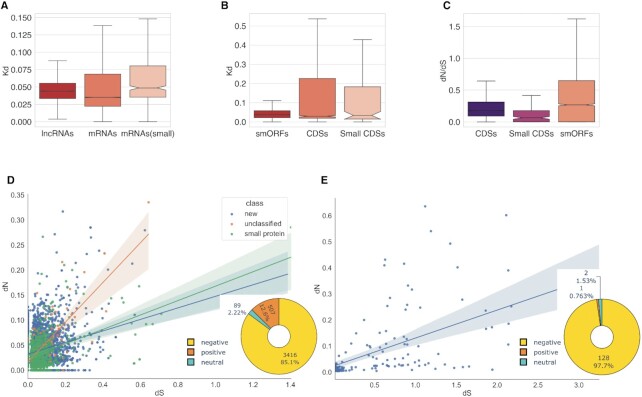
Comparative analysis of nucleotide pairwise alignments between P. patens and Physcomitrium sp. showed that the distributions of evolutionary rates, Kd (Kimura two-parametric model, K2P), differed substantially between lncRNAs and mRNAs (Kruskal–Wallis rank sum test, P < 10–15), with the mRNAs evolving significantly slower (Supplementary Table S6; Figure 3A). However, the median of Kd distributions of lncRNA hits and small-mRNAs were closely similar (median 0.046 versus 0.048, respectively) and differed significantly from another subset of mRNAs with longer CDSs (median = 0.035; Mann–Whitney U-test, P < 10–15 for both comparison). Thus, although plant lncRNAs are generally far less conserved at the nucleotide sequence level than mRNAs, some of lncRNAs contain conserved regions with the Kd values comparable to the short protein coding transcripts. This finding is in agreement with the results of comparative analyses of coding and non-coding RNAs in other eukaryotes (88,91,92).
Figure 3.
Evolutionary rates and selection regimes in lncRNAs, mRNAs, smORFs and protein-coding genes. (A) The evolutionary rates distribution in pairwise alignments of P. patens and Physcomitrium sp. lncRNA (n = 4078) and mRNA transcripts (n = 15 926 and n = 252 for mRNAs encoded proteins above and below 100aa, respectively); P < 10–15 by Kruskal–Wallis rank sum test. (B) The evolutionary rates distribution in pairwise alignments of P. patens and Physcomitrium sp. smORFs and protein CDSs; P = 0.2 by Kruskal–Wallis rank sum test. (C) The distribution of dN/dS ratios in smORFs (n = 4022) and functional proteins (n = 8203). Small CDSs are protein smaller than 100aa; P < 0.0001 by Kruskal–Wallis rank sum test. (D) The distribution of dN versus dS values from smORF tblastn alignments; pie chart shows the number of smORFs classified based on dN/dS values. (E) The distribution of dN versus dS values from small CDSs (proteins below 100aa) tblastn alignments; pie chart shows distribution of dN/dS values.
We then estimated the evolutionary rates and performed statistical tests for identification of purifying selection in the predicted smORFs (92,93). Based on the analysis of TBLASTN pairwise alignments, we concluded that the Kd values of smORFs were statistically indistinguishable from those for the CDSs, including small ones (Kruskal–Wallis rank sum test, P = 0.20; Figure 3B).
Because conserved regions in animal intergenic lncRNAs were enriched in translated smORFs (36), we next analyzed the evolutionary rates of 4022 smORFs overlapping conserved lncRNA regions between P. patens and Physcomitrium sp. Such lncRNA regions, overlapping >80% of smORF lengths, were found in ∼45% of the conserved lncRNA loci. We calculated the ratio of non-synonymous to synonymous substitution rates (dN/dS) to examine whether these lncRNAs contained regions with significant levels of protein selection pressure (Supplementary Table S7). As expected, protein-coding CDS (including small CDSs) had stronger purifying selection signatures compared to smORFs (Kruskal–Wallis rank sum test, P < 0.0001; Figure 3C). Overall, ∼76% of small CDSs displayed a robust signature of purifying selection (dN/dS < 0.20) compared to ∼45% of the smORFs (1771 smORFs). About 30% of both smORFs and small CDSs were ‘highly conserved’ (orthologs without mutations/substitutions or with single ones in YEPO). However, in contrast to the small CDSs, ∼83% of these ‘highly conserved’ smORFs were distributed in only a small number of moss species (cluster 3, see above). These observations suggest that about half of the evolutionary conserved smORF encodes lineage- and/or species-specific peptide/microproteins. On the contrary, smORFs with comparable levels of dN and dS values, could be under selection at the nucleotide level or have a signature of positive selection.
The number of putative orthologs of smORFs in more distant moss species drastically dropped, accompanied by a decline of the dN/dS ratio (Supplementary Figure S4). In more distant moss species the median dS values in smORFs were similar to those in functional proteins, whereas the dN values were twice as high in smORFs. Thus, numerous smORFs might have signatures of positive selection or could be subject to selection at the nucleotide level.
We next analyzed potential signatures of positive selection in the set of smORFs. About 12% (507, Figure 3E) of the analyzed alignments between P. patens and Physcomitrium sp. had dN/dS > 1. In contrast, no alignments with dN/dS > 1 were found in the set of small CDSs.
For further analysis, HyPhy's BUSTED algorithm has been used (53). We identified 125 smORFs as positively selected (LRT, P < 0.05; Supplementary Table S8), including ∼16% (20/125) of smORFs with dN/dS > 1. Thus, only ∼4% of the total set of 507 smORFs with dN/dS > 1 were supported by the BUSTED method as positively selected. We next ran HyPhy-BUSTED to test for positive selection in closely related moss species, using 398 smORFs and 146 small CDSs conserved from three to six moss species. About 12% of both smORFs and small CDSs contained evidence of episodic diversifying selection according to the BUSTED algorithm (LRT, P < 0.05) in both smORFs and small CDSs sets (Supplementary Table S8).
Thus, our findings suggest the existence of a group of smORFs, which may indeed encode small proteins that are broadly conserved, and smORF groups that are maintained by selection in groups of comparatively closely related organisms (species- and lineage-specific) as it has been shown in animals (94).
Structural features of predicted plant microproteins
Low complexity regions in lncORF-smORFs
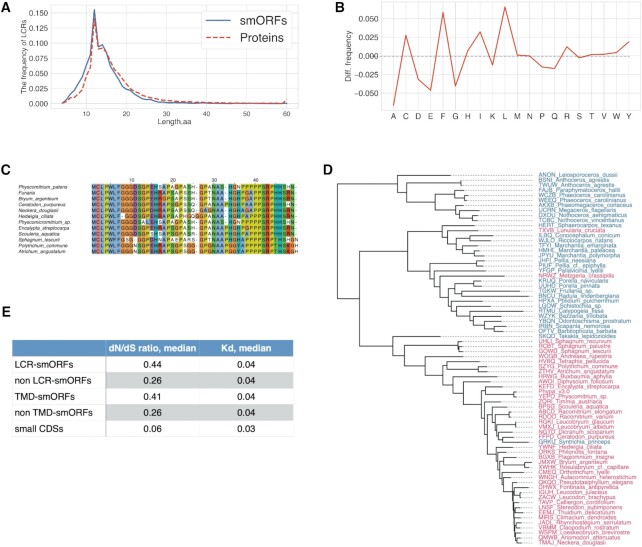
Numerous proteins, particularly in eukaryotes, contain Low sequence Complexity Regions (LCRs) of widely varying lengths. Despite suggestions that LCRs drive evolutionary changes in proteins, their functions remain obscure (95,96). In database searches for sequence similarity, the LCRs are either masked and excluded from further search or down weighed for the significance estimation (97). The exclusion of LCRs might result in underestimation of the number of potentially functional microproteins encoded by lncRNA-smORFs. To assess the prevalence of LCRs in smORFs, we used the SEG tool (98) and identified ∼10% AUG-started smORFs (7831 smORFs), containing predicted LCRs (average length = 14aa; Figure 4A, B; Supplementary Table S9). Overall, ∼4% of all amino acids constituted MiPEPID-smORFs were part of predicted LCRs.
Figure 4.
Low complexity regions in putative microproteins encoded by smORFs. (A) The length distribution of predicted LCRs in smORFs and functional proteins. (B) Amino acid propensities of LCRs in smORFs; difference from small CDS LCRs aa propenisties is shown. SmORF-LCRs were significantly enriched in leucine, phenylalanine and isoleucine (Fisher's exact test P < 0.00001). (C) The alignment of 47-aa smORF and selected orthologs. This smORFs is conserved among extant bryophytes. (D) the phylogenomic tree of extant bryophytes. The species with identified orthologs of smORF35290_C03_10088000r_47aa are shown in red. Tree was drawn based on OneKP data. (E) Evolutionary rates of LCR-smORFs and TM-containing smORFs compared to those of small CDSs.
We further reanalyzed the conservation of the smORFs without filtering out the LCRs (TBLASTN, E-value < 0.001, SEG = ‘no’). Overall, 2095 conserved smORFs were identified in this search compared to 1520 in the original search with LCR filtering. For example, for a proline-rich 47-aa ‘new’ translatable (see below) smORF that was previously considered ‘non-conserved’, apparent homologs were identified in transcriptomes of 43 species, including mosses, liverworts and ferns (Figure 4C, D). The transcriptomes of some moss species contained several transcripts encoding paralogs of this microprotein. Thus, a number of microproteins containing LCR can be overlooked in plant proteome annotations and our analysis of LCR patterns in smORF candidates showed that their numbers are likely underestimated. These findings suggest that the origin of new small proteins containing LCR is more widespread than previously thought.
Given that genome base composition could define the evolutionary trajectories of new ORFs (27), we next asked whether the evolutionary rates of smORFs originated from low complexity genome regions different from other smORFs. Although the distribution of the Kd evolutionary rates did not differ significantly, the dN/dS ratios were significantly different between the LCR-smORFs and smORFs without LCRs (Mann–Whitney U-test, P = 4 × 10–10; Figure 4E). In addition, the proportion of smORFs with dN/dS> 1 was significantly higher in LCR-smORFs than in non LCR smORFs (25% versus 12%, respectively; Fisher's exact test P < 0.00001). In contrast, the evolutionary rates (Kd and dN/dS ratios) did not differ significantly between small CDSs with and without LCRs. Thus, LCR-containing smORFs appear to evolve under weak purifying selection, and some might even be subject to positive selection, whereas small CDSs including those containing LCRs are subject to substantially stronger purifying selection (Figure 4E).
Many lncORF-smORFs contain transmembrane domains and signal peptides
Little is known about the gain of function by new genes that might originate from non-coding DNA. Novel ORFs, especially AT-rich ones, can have high propensity to form transmembrane domains (TMDs) (27), which function as protein sorting determinants (99). Therefore, we applied two algorithms, TMHHM 2.0 (62) and SignalP-5.0 (61) to predict transmembrane and signal peptides in the set of all smORFs. At first, 9472 smORFs were predicted to be secreted by TMHHM 2.0 (62) and/or SignalP-5.0 (61) tools (Supplementary Table S10). In addition, 4978 smORFs were predicted to be small transmembrane proteins. These TM-containing smORFs were designated as TMD-smORFs (Supplementary Table S10). The TMD-smORFs were significantly longer in comparison to other smORFs (Mann–Whitney U-test, P < 10–15, Figure 5A), including conserved smORFs (Mann–Whitney U-test, P < 10–10, Figure 5B).
Figure 5.
SmORFs with predicted transmembrane and/or signal peptides. (A) The length of all smORFs with/without predicted transmembrane domain. Plots indicate the median, quartiles, and 5th and 95th percentiles. (B) The length of conserved smORFs with/without predicted transmembrane domain. Plots indicate the median, quartiles, and 5th and 95th percentiles. (C) The length of ‘new’ and annotated (‘small protein’ and ‘unclassified’) TMD-containing conserved smORFs. Plots indicate the median, quartiles, and 5th and 95th percentiles. (D) Cumulative distribution of GC-content of lncRNAs loci with TMD-smORFs, TMD-smORFs and small CDSs. (E) Cumulative distribution of GC-content of lncRNAs loci with/without TMD-smORFs; (F) Cumulative distribution of GC-content of the conserved and non-conserved TMD-smORFs and small CDSs. (G) The distribution of dN/dS ratios in TMD-smORFs and smORFs without transmembrane domain. (H) Multiple sequence alignment of selected orthologs and ‘new’ TMD-smORF-smORF28298_C22_5645030r_50aa. (I) Multiple sequence alignment of potentially secreted smORF smORF32633_C27_3110343f_66aa and selected orthologs. (J) UpSet plot showing intersection between smORFs with predicted LCR, TM and functional domains, including ‘consensus disorder prediction’, and N-terminal secretion signal. (K) Multiple sequence alignment of 89-aa smORF smORF29906_C24_11564015f_89aa and orthologs from selected moss species. *** P < 10–10—Mann–Whitney U-test.
We found that 1182 TMD-smORFs had a putative ortholog in at least one examined species, and about 70% (821/1182) of them belonged to the ‘new’ class. The percent of TMD-smORFs in the conserved set was only slightly, albeit significantly, higher than in the entire smORFs set (∼5% TMD-smORFs versus ∼3% all smORFs; chi-square, P < 10–15). The ‘new’ TMD-smORFs were significantly shorter than the TMD-smORFs in other smORF types (Mann–Whitney U-test, P < 10–10, Figure 5C). One of the ‘new’ TMD-smORF identified in our previous work (22) as a 41-aa peptide (PSEP1) being overexpressed facilitated rapid growth of P. patens protonemata accompanied by earlier cell death; in contrast, the knock-out psep1 lines grew more slower compared with wild-type plants (22). Here, we found that PSEP1 is widely conserved in a range of land plant species. Thus, at least some of these ‘new’ TMD-smORFs could perform important functions in plants.
The TM-first model of gene birth suggests that emerging, adaptive new ORFs originate from AT-rich genome regions (27). In our set, the lncRNA loci containing TMD-smORFs had a slightly but significantly lower GC-content than other lncRNAs (Kolmogorov–Smirnov test, P < 10−20; Figure 5D). The GC-content of TMD-smORFs was even lower than that of the corresponding lncRNA loci, indicating that these smORFs were located in AU-rich regions of lncRNAs (Kolmogorov–Smirnov test, P < 10−20; Figure 5E). The TMD-smORFs without detected orthologs were found to be significantly less GC-rich than the conserved TMD-smORFs (Figure 5F). This observation might reflect selection against highly hydrophobic smORF, perhaps due to their aggregation potential, leading to increased GC-content. We next tested whether the evolutionary rates of TMD-smORFs differed from those of other smORFs. The dN/dS ratios were significantly higher in the set of TMD-smORFs than in other smORFs (Mann–Whitney U-test, P = 1.3 × 10–5; Figures 4E and 5G), suggesting that TMD-smORFs are subject to a weaker purifying selection and evolve faster than other smORFs.
Recently, ‘CDS elongation’ via a stop codon mutation was proposed as a model of de novo gene birth from ‘new’ smORFs (25). An example of a smORFs that is expanded in more distant species is a widely conserved 51-aa ‘new’ TMD-smORF (smORF28298_C22_5645030r_50aa), containing a predicted N-terminal signal sequence (4–26aa). This smORF contains a specific, conserved motif [P***R*R***LR] at the C-terminus that is shared with uncharacterized small proteins in the RefSeq database (Figure 5H). Another possible example of smORF with an expanded coding sequence at C-terminus is potentially secreted 66-aa microprotein smORF32633_C27_3110343f_66aa (Figure 5I). The C-terminal end of longer smORFs is similar to low complexity regions with stretches of identical amino acid (Figure 5I). These examples suggest possible evolution of this smORF by mutation in a stop codon that causes readthrough.
The role of low complexity regions in origin and evolution of secreted and transmembrane microproteins is poorly understood (96). Given that LCR-smORFs are enriched with nonpolar amino acids, identification of ∼32% LCR-smORFs that were predicted to comprise secreted or transmembrane peptides was not unexpected (Figure 5J). For example, a 89-aa ‘small protein’ smORF (smORF29906_C24_11564015f_89aa) contains one LCR that overlapped signal peptide (predicted by both TMHMM 2.0 and SignalP-5.0 tools) and another proline-rich LCR at the C-terminal end. A TBLASTN search without low-complexity filtration significantly expanded the number of identified orthologs in a range of moss species, suggesting wide conservation of this smORF (Figure 5K). Another example is a 96-aa secreted ‘small protein’ LCR-smORF (smORF10900_C12_8956284f_96aa) that had a match only to the Physcomitrium sp. transcriptome (TBLASTN, E-value < 0.001), but was found to be highly conserved in a range of moss and liverwort species in TBLASTN search against OneKP transcriptomes without filtering out the LCRs (Supplementary Figure S5). We detected the consecutive increase in the length of matched hits and performed a search for motifs in identified homologs by MEME software (59). This search revealed two proline-rich motifs, that are expanded in distant species (Supplementary Figure S6).
The lncORF-smORFs are enriched for intrinsically disordered regions
According to a previous study, novel short ORFs are substantially enriched in predicted intrinsically disordered regions than known small proteins (90). Therefore, we next queried smORFs against the InterProscan database to analyze possible domains and motifs in our set of smORFs (57). About 95% of smORFs were not assigned to any known functional domains or motifs. The most common type of motif identified was ‘consensus disorder prediction’, assigned to ∼93% (8595/9189) of all domain-assigned smORFs (Supplementary Table S11). As expected, the set of 3357 highly conserved smORFs (homologs found in at least 5 moss species) was significantly enriched for known domains and motifs compared to all conserved smORFs (Fisher's exact test, P < 10–5). About 80% of these highly conserved smORFs were also predicted to contain ‘consensus disorder prediction’ motifs. We next compared our set of smORFs with small functional proteins (small CDSs). Small CDSs were assigned a diverse range of known domains, with about ∼16% corresponding to different small ribosomal proteins (Supplementary Table S11). The prevalence of ribosomal proteins is in concordance with amino acid composition of small CDSs (Figure 1E).
Because precursors of bioactive peptides are often small proteins devoid of specific functions apart from the active peptide moiety, the corresponding genes can be mis-annotated as lncRNAs. Therefore, we next used the Small Secreted Peptide (SSP) prediction tool (58) that utilizes hidden Markov models (HMMs) of known SSP families to analyze the identified smORFs. Overall, we identified 45 smORFs with ‘known’ SSP domains in our set (Supplementary Table S12). The most highly conserved SSP motifs belonged to plant antimicrobial Cysteine-Rich Peptide (CRP) families, such as CRP5310 (Defensin-Like proteins) or CRP5660—glycine-rich proteins (GRP; Supplementary Figure S7).
Another identified conserved CRP family is TAXIMIN (TAX) which are involved in lateral organ separation in Arabidopsis (100). The detailed analysis of two smORFs containing a predicted CLE10 (CLAVATA3/ESR-related) domain revealed that these are precursors of PpCLE5 and PpCLE7 CLV3-like peptides, identified earlier in P. patens (101). We also identified three conserved smORFs with similarity to DEVIL/ROTUNDIFOLIA (DVL/ROT) peptides that are known to be encoded by short ORFs in plants (102). The orthologs of smORFs with DVL/ROT domain were identified in all plant lineages except green algae. The analysis of the set of small CDSs identified only four potential SSPs. These examples show that transcripts that encode small precursors of SSP can be predicted as lncRNAs and, therefore, the number of SSP families in plants could be underestimated.
Expression of lncRNAs and translation of smORFs
To analyze the expression of the diverse set of moss lncRNAs and further compare them with mRNAs, we performed Nanopore direct RNA sequencing of polyA(+) RNA fractions extracted from both P. patens protonemata (n = 3 biological repeats) and gametophores (n = 4 biological repeats). The nanopore sequencing allows full-length characterization of native RNA in transcriptomes (103,104), being an indispensable tool for the elucidation of lncRNA transcripts (105).
This analysis confirmed the transcription of ∼57% (5249/9168) of the loci encoding lncRNAs. The relatively low fraction of detected lncRNAs in these experiments can be explained by three, not mutually exclusive reasons: (i) lncRNAs expression is tissue- and/or condition-specific, so that many of these transcripts are not expressed at the two developmental stages and our experimental conditions; (ii) low-expressing lncRNAs are not detected; (iii) differences in read lengths and assembly protocols between RNA-seq and nanopore sequencing technologies. About 50% of the lncRNA nanopore-based transcripts exactly matched the intron chain of the set of lncRNAs and 20% were partially overlapped annotations. For further analysis the transcriptional level of 1678 lncRNA loci, which exactly matched the intron chain, was calculated (Supplementary Table S13).
It has been previously shown that the frequency of translation initiation at non-AUG codons is significantly lower when the corresponding ORF is located downstream of AUG codons, suggesting a bias in the distribution of conserved and potentially translated smORFs (106). To assess this trend, we explored the distribution of AUG- and non-AUG-started smORFs across the length of lncRNAs. The distribution of both types of non-conserved smORFs was found to be bimodal and significantly differed from conserved smORFs (Supplementary Figure S8; Kolmogorov–Smirnov test, P < 10−15). Specifically, the conserved ‘small protein’ AUG-smORFs were, typically, significantly closer to 5′-end of transcripts than ‘new’ and ‘unclassified’ ones (Kolmogorov–Smirnov test, P < 10−20). However, because the accurate prediction of smORFs with alternative start codons can be compromised by intersection with AUG-smORFs, these observations require further elucidation by ribosome profiling (106).
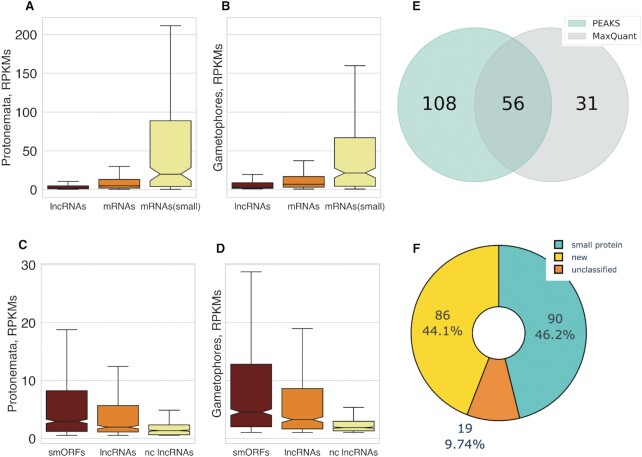
It has been shown that young and/or taxonomically restricted protein-coding genes as well as lncRNAs are on average shorter than conserved genes and are expressed at a lower level (7,107). In agreement with these observations, the transcriptional level of both mRNA subsets was significantly higher in protonemata (Kruskal–Wallis rank sum test, P < 10–15) and gametophores (Kruskal–Wallis rank sum test, P < 10–15) compared to the set of lncRNAs (Figure 6A, B). Due to the presence of the small ribosomal proteins in the small CDS set, the level of transcription in small-mRNAs subset was significantly higher than other mRNAs in protonemata (Mann–Whitney U-test, P < 10–15) and gametophores (Mann–Whitney U-test, P < 10–15).
Figure 6.
The analysis of smORFs transcription and translation. (A, B) the comparison of lncRNAs and mRNAs transcriptional level in gametophores and protonemata, respectively; mRNAs(small)—a subset of mRNAs, encoding proteins smaller than 100aa. (C, D) the transcriptional level of conserved smORFs (smORFs), conserved lncRNAs (lncRNAs) and non-conservative smORFs (nc smORFs) in gametophores and protonemata, respectively. (E) Venn diagram showing the comparison of smORFs identification by two search engines—PEAKS 8.0 and MaxQuant. 56 smORFs were identified by both search engines. (F) Pie chart depicting classification of identified translatable smORF types.
The transcriptional level of 1678 lncRNAs containing conserved smORFs (n = 629) was significantly higher than the expression level of both lncRNAs with conserved nucleotide regions other than smORFs (n = 451) and lncRNAs containing non-conserved smORFs (n = 598) in protonemata (Kruskal–Wallis rank sum test, P < 10–15) and gametophores (Kruskal–Wallis rank sum test, P < 10–15; Figure 6C, D). However, we did not find any significant differences between the transcriptional levels of lncRNAs containing broadly conserved (cluster 1 and 2) or lineage-specific (cluster 3) smORFs. Taken together, these findings show that, although the characteristic expression level of lncRNAs was expectedly lower than that of mRNAs, the transcriptional level of lncRNAs could be a determinant of smORF conservation as previously shown for proteins (108). Thus, the positive correlation between expression levels of transcripts and the evolutionary conservation of their coding regions is a universal trend. The lncRNAs that fit this trend could be considered as mRNAs with predicted low coding potential.
We then used mass-spectrometry analysis to identify translated smORFs. The peptidomic datasets from our previous studies (22,109) and additionally generated datasets were used. All datasets were searched against a custom database and thoroughly filtered using the target-decoy strategy. MS-based detection of smORFs is a challenging task due to the low expression and rapid turnover of lncRNA-encoded peptides (21,110). Overall, we obtained evidence of translation for 195 smORFs, including 56 identified by both search engines (Supplementary Table S14; Figure 6E). The number of peptides encoded by lncRNAs identified by MS analysis is consistent with results obtained on human cells (110,111).
Approximately 44% of the translated smORFs belonged to the ‘new’ smORFs class (Figure 6F). About 31% of the ‘new’ translated smORFs were not conserved, suggesting rapid turnover of microproteins. As expected, ‘new’ translatable smORFs were significantly overrepresented among non-conserved ones (chi-square test, P < 10–15).
Proteomic standards of identification (≥2 unique peptides for protein) can be expected to suffice to identify the microprotein products of smORF without false positives, but true smORF-encoded peptides (SEPs) could be lost (23). In our datasets, translation of 73 smORFs was confirmed by two and more unique peptides. Among these, there were 13 ‘new’ (8 non-conserved) smORFs, including the functional lncRNA-smORFs - PSEP1, PSEP3, PSEP18 identified in our previous study (22). This result can be considered as a validation of our identification strategy. It has been shown that mass-spectrometry can confirm the translation of microproteins from highly-expressed abundant transcripts (112). In both protonemata and gametophores, expression of the translated smORFs was significantly higher than the expression of ORFs without evidence of translation (Mann–Whitney U-test, P < 0.00001), implying that only smORFs from highly-expressed lncRNAs were detected in our proteomics analysis. Therefore, the number of translatable smORFs in our study is likely to be substantially underestimated. We next used these results to select smORFs for experimental validation.
Experimental validation of the functions of selected microproteins
The functions of smORF-encoded peptides (SEPs) are poorly studied in plants. We selected two SEPs: LCR-smORF PSEP3 and peptide with ‘consensus disorder prediction’ motif - PSEP18 both identified in our previous work (22) to examine their functional role in more detail. Knocking out PSEP18 resulted in a slight decrease in the moss plant diameter, whereas the PSEP3 KO lines displayed a severe decrease in growth rate and altered filament branching (22).
Here, we found that 40-aa ‘new’ peptide PSEP18, additionally confirmed by nanopore sequencing and MS analysis, is poorly conserved. Using iTRAQ (Isobaric tag for relative and absolute quantitation)-based quantitative proteomic analysis, we have not found significant changes in proteomes of both overexpression and knockout lines of PSEP18, suggesting that evolutionary conservation might be a marker of functional SEPs.
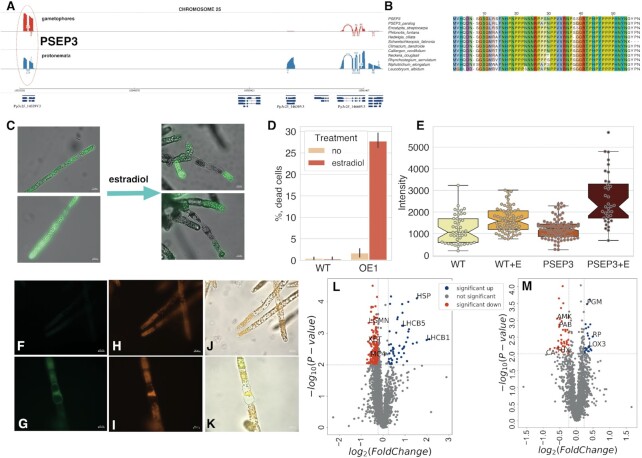
Hydroxyproline- and proline-rich peptides and proteins play pivotal roles in signal transduction cascades, plant development and stress tolerance (113,114). Our MS analysis confirmed the translation of 71 LCR-smORFs (36%), seven of which were enriched in prolines, including a previously identified 57-aa microprotein - PSEP3 (22) and its paralog (Figure 7A, B).
Figure 7.
Functional analysis of PSEP3 microprotein. (A) Sashimi plot showing nanopore-based transcription of PSEP3 and surrounding region on chromosome 25. (B) The multiple pairwise alignment of selected orthologs and translatable ‘new’ smORFs - PSEP3 and paralog XR_002972902.1_ORF5. (C) the influence of PSEP3 overexpression on cell viability measured by fluorescein diacetate (FDA) dye. The PSEP3 expression was induced by estradiol treatment in liquid culture; (D) The induction of cell death under estradiol treatment; WT - wild-type plants, OE1 - PSEP3 OE line. (E) Boxplot showing the difference in the intensity of DCFH-DA in PSEP3 OE mutant without/under estradiol treatment, respectively (P < 0.001 by two-way ANOVA followed by Tukey's multiple comparison test). (F, G) detection of ROS generation by DCFH-DA in PSEP3 OE mutant without/under estradiol treatment, respectively; (H, I) autofluorescence of chloroplasts without/under estradiol treatment, respectively. (J) the merged image of (F) and (H) pictures. (K) the merged image of (G) and (I) pictures. (L) Volcano plot of the entire set of proteins quantified during iTRAQ analysis in PSEP3 OE line. (M) Volcano plot of the entire set of proteins quantified during iTRAQ analysis in PSEP3 KO line. Proteins significantly changed in abundance are depicted in colour. Blue dots indicate up-regulated proteins and red dots indicate down-regulated proteins in PSEP3 mutants.
A TBLASTN search without low-complexity filtration identified PSEP3 orthologs in 27 moss and one liverwort species (Supplementary Figure S9). Due to severe growth inhibition and cell death in lines with PSEP3 overexpression (22), we additionally generated PSEP3 overexpressed lines (PSEP3 OE) using the β-estradiol induction system (63), to study the impact of PSEP3 translation on cell metabolism. The induction of PSEP3 expression resulted in cell death (Figure 7C, D), accompanied by an significant increase in reactive oxygen species (ROS) levels (ANOVA, P < 0.001) and changes in cell structure during 48-h (Figure 7F–K).
Using quantitative comparative proteomic analysis, we next identified 167 protein groups which were significantly changed in the proteome of induced PSEP3 OE line in comparison to wild-type plants (Supplementary Table S15; FC > 1.2, P < 0.01, Figure 7L). The most enriched up-regulated protein groups belonged to ‘photosynthesis’ (GO:0015979, P < 10–15) and ‘generation of precursor metabolites and energy’ (GO:0006091, P < 10–5) GO terms. We identified the Light-Harvesting Chlorophyll a/b Binding Proteins Lhcb1, Lhcb2 and Lhcb5 among the most up-regulated proteins in PSEP3 OE line. The down-regulated protein group included metacaspase-4-related (Pp3c2_20840V3) and xyloglucan endo-transglycosylase (Pp3c25_4050V3; Figure 7L). Thus, PSEP3 overexpression resulted in substantial changes in moss proteome and increased cell death.
The impact of PSEP3 knock-out on the protonema proteome was less pronounced. Overall, 56 differentially expressed protein groups (FC > 1.2, P < 0.01; Supplementary Table S15; Figure 7M). The down-regulated protein groups were mainly enriched in ‘oxidoreductase’ activity (GO:0016491, P < 0.0001). For example, ‘thioredoxin x’ and ‘thioredoxin m(mitochondrial)-type’ proteins were found. Among the most upregulated proteins were 60s ribosomal protein 10a-1 (Pp3c20_19190V3) and Phosphoglucomutase (Pp3c16_20760V3).
Thus, the PSEP3 overexpression was harmful for moss cells in contrast to PSEP3 knockout. It has been shown previously that overexpression of random peptides can generate visible phenotypes in plants (115). This could be a bona fide biological effect, but alternatively, might be an artefact of unphysiological peptide concentration. In the first case, tight regulation of the expression of such peptides in plant cells should be expected.
DISCUSSION
The properties and evolutionary fates of smORFs located on lncRNAs remain poorly understood. Although translation and functionality of thousands of smORFs have been validated in animals by different approaches (25), only a small fraction of these are widely conserved (90). In the present analysis of plant lncRNA-smORFs, we observed that the majority of the smORFs abruptly lost orthology even in close species, suggesting fast stochastic gain and loss of non-functional smORFs in plant genomes. Besides their possible roles in microprotein production, our result are also compatible with the possibility that some of the smORFs are located in the regions of lncRNA that are subject to RNA-level selection pressure and could be functionally important for RNA–RNA and/or RNA-protein interactions (36). Nevertheless, coding sequences in such regions could exist as well (116). Given that dS is usually lower in alternative and new eukaryotic exons compared to constitutive exons (117,118) and that the portions of proteins encoded by such exons also contain an increased fraction of intrinsically disordered regions, we hypothesize that many if not most of the lncRNA regions with low dS have dual functions, at the level of both transcripts and translated products (119). Well-known examples of transcripts combining coding and non-coding functions are plant pri-miRNAs that encode functional SEPs but also play roles in gene regulation (120,121). In addition, smORFs can contribute to the regulation of lncRNAs abundancy by engaging the corresponding transcripts in ribosomes (8) or triggering nonsense-mediated RNA decay (122). Arguably, such function would entail conservation of smORFs position in lncRNAs across species, which we indeed observed in many cases. Such position-specific smORFs can play regulatory roles, similar to uORFs (25) and dORFs (123) in mRNAs.
Additionally, we cannot rule out limitations in ortholog detection for smORFs. In particular, we found that the conservation of smORFs containing LCRs can be underestimated and requires new approaches for an adequate analysis (97). Moreover, these LCR-smORFs were enriched with AT-rich codons encoding hydrophobic amino acids. It has been recently shown that hydrophobic mutational ratchet entrenches protein molecular complexes (124), suggesting a new role for such AT-rich smORFs. Indeed, many functionally characterized SEPs have been shown to bind membrane proteins (25,125). We also identified a number of new conserved smORFs containing predicted export signal peptides and/or transmembrane domains, suggesting their role in cell-to-cell communication. Because the mutational process favours G/C to A/T transitions (126), lncRNAs can be prone to producing microproteins containing transmembrane domains (27). Novel TMD-containing peptides could escape in the membrane environment from degradation or deleterious interactions with cytoplasmic proteins.
The emergence of de novo genes from non-coding regions can be the first step in new gene birth although the routes through which emerging proteins become functional remain poorly understood. Such genes are taxonomically restricted and are often referred to as ‘orphans’ genes that constitute up to 30% of the genes in some eukaryotes (26,27,127–129). Our results support the hypothesis on the species-specific functions of the majority of smORFs. The most conserved smORFs were those that had similarity to annotated proteins (‘unclassified’ type) or intersect predicted small proteins (‘small protein’ type). These smORFs are enriched in known protein domains and come from already existing genome annotation, suggesting that these could be remnants of functional proteins. In contrast, new smORFs appear to be the main source of variability. Based on BLASTX search (E-value < 0.00001) against the Viridiplantae uniprot database, we roughly estimate the proportion of such remnants of ancestral protein-coding genes in our set of lncRNAs at ∼3%. This is an agreement with results obtained on mammals (130). These findings suggest that a relatively small fraction of the lncRNAs are potential pseudogenes or natural antisense transcripts of protein-coding genes. However, further analysis using whole genome alignments is needed to accurately estimate the contribution of pseudogenization of protein-coding genes to the evolution of lncRNAs in plants.
In conclusion, our analysis revealed several possible scenarios for the emergence and the subsequent evolution of the smORFs. First, some of the identified conserved smORFs are bona fide small functional proteins or precursors for secreted peptides. In this case, the corresponding transcripts were erroneously identified as long non-coding RNAs or represent transcripts with dual function. This finding is in agreement with our identification of many previously unnoticed, lineage-restricted or widely conserved secreted microproteins. The evolutionary conservation of such smORFs in a range of species could point at their functionality. Second, however, a major fraction of the smORFs either emerged by chance in a single species or have orthologs only in close species, suggesting rapid gain and loss. Some of these smORFs can be translated into SEPs, but both the expression level of the respective lncRNAs and the translation level are low. Such translatable but poorly conserved peptides might not have a well-defined function but could serve as a pool for the emergence of functional microproteins via positive selection. Third, there is a subset of smORFs that are conserved at the nucleotide level, but do not show protein selection signatures. The possible functions and evolution of such smORFs is of interest and require further elucidation.
In conclusion, we identified numerous, previously unknown, evolutionarily conserved smORFs, and validated the expression of a substantial subset of these by transcriptome and proteome analysis. By extension, many more smORFs in plants are likely to be functional and are candidates for future experimental study. Such studies on the plant smORFome should be expanded to more complex plant species, and will require large-scale pipelines to investigate the SEPs localization, toxicity, the potential for cellular uptake, and identification of protein-protein interactions.
DATA AVAILABILITY
Nanopore data were deposited in the BioProject with the accession number PRJNA681088. The mass spectrometry peptidomic data have been deposited to the ProteomeXchange Consortium via the PRIDE (131) partner repository with the dataset identifiers PXD025373 and PXD025267. All scripts are maintained in the GitHub code repository: https://github.com/IgorFesenko/smORF_analysis.
Supplementary Material
ACKNOWLEDGEMENTS
The research is supported by intramural funds of the US Department of Health and Human Services (to the National Library of Medicine, EVK and SAS).
This research was also supported in part by an appointment to the National Library of Medicine (NLM) National Center for Biotechnology Information (NCBI) Research Participation Program (IF). This program is administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy (DOE) and the National Library of Medicine (NLM). ORISE is managed by ORAU under DOE contract number DE-SC0014664. All opinions expressed in this paper are author's and do not necessarily reflect the policies and views of NLM, DOE or ORAU/ORISE. No conflict of interest declared.
We thank the Center for Precision Genome Editing and Genetic Technologies for Biomedicine, Federal Research and Clinical Center of Physical-Chemical Medicine of the Federal Medical Biological Agency for the expertise and guidance in genetic engineering.
Contributor Information
Igor Fesenko, Shemyakin and Ovchinnikov Institute of Bioorganic Chemistry of the Russian Academy of Sciences, Moscow 117997, Russian Federation.
Svetlana A Shabalina, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, MD 20894, USA.
Anna Mamaeva, Shemyakin and Ovchinnikov Institute of Bioorganic Chemistry of the Russian Academy of Sciences, Moscow 117997, Russian Federation.
Andrey Knyazev, Shemyakin and Ovchinnikov Institute of Bioorganic Chemistry of the Russian Academy of Sciences, Moscow 117997, Russian Federation.
Anna Glushkevich, Shemyakin and Ovchinnikov Institute of Bioorganic Chemistry of the Russian Academy of Sciences, Moscow 117997, Russian Federation.
Irina Lyapina, Shemyakin and Ovchinnikov Institute of Bioorganic Chemistry of the Russian Academy of Sciences, Moscow 117997, Russian Federation.
Rustam Ziganshin, Shemyakin and Ovchinnikov Institute of Bioorganic Chemistry of the Russian Academy of Sciences, Moscow 117997, Russian Federation.
Sergey Kovalchuk, Shemyakin and Ovchinnikov Institute of Bioorganic Chemistry of the Russian Academy of Sciences, Moscow 117997, Russian Federation.
Daria Kharlampieva, Department of Cell Biology, Federal Research and Clinical Center of Physical -Chemical Medicine of Federal Medical Biological Agency, Moscow 119435, Russian Federation.
Vassili Lazarev, Department of Cell Biology, Federal Research and Clinical Center of Physical -Chemical Medicine of Federal Medical Biological Agency, Moscow 119435, Russian Federation; Moscow Institute of Physics and Technology (National Research University), Dolgoprudny, Moscow region, 141701, Russian Federation.
Michael Taliansky, Shemyakin and Ovchinnikov Institute of Bioorganic Chemistry of the Russian Academy of Sciences, Moscow 117997, Russian Federation; The James Hutton Institute, Invergowrie, Dundee DD2 5DA, UK.
Eugene V Koonin, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, MD 20894, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Russian Science Foundation [17-14-01189]. Funding for open access charge: Intramural funds of the US Department of Health and Human Services (to the National Library of Medicine, the National Institutes of Health).
Conflict of interest statement. None declared.
REFERENCES
- 1. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007; 447:799–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kapranov P., Cheng J., Dike S., Nix D.A., Duttagupta R., Willingham A.T., Stadler P.F., Hertel J., Hackermüller J., Hofacker I.L.et al.. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007; 316:1484–1488. [DOI] [PubMed] [Google Scholar]
- 3. Wade J.T., Grainger D.C.. Pervasive transcription: illuminating the dark matter of bacterial transcriptomes. Nat. Rev. Microbiol. 2014; 12:647–653. [DOI] [PubMed] [Google Scholar]
- 4. Yu Y., Zhang Y., Chen X., Chen Y.. Plant Noncoding RNAs: hidden players in development and stress responses. Annu. Rev. Cell Dev. Biol. 2019; 35:407–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pauli A., Valen E., Lin M.F., Garber M., Vastenhouw N.L., Levin J.Z., Fan L., Sandelin A., Rinn J.L., Regev A.et al.. Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res. 2012; 22:577–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kopp F., Mendell J.T.. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018; 172:393–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wu H., Yang L., Chen L.-L.. The diversity of long noncoding RNAs and their generation. Trends Genet. 2017; 33:540–552. [DOI] [PubMed] [Google Scholar]
- 8. Bazin J., Baerenfaller K., Gosai S.J., Gregory B.D., Crespi M., Bailey-Serres J.. Global analysis of ribosome-associated noncoding RNAs unveils new modes of translational regulation. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:E10018–E10027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Carlevaro-Fita J., Rahim A., Guigó R., Vardy L.A., Johnson R.. Cytoplasmic long noncoding RNAs are frequently bound to and degraded at ribosomes in human cells. RNA. 2016; 22:867–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ingolia N.T., Brar G.A., Rouskin S., McGeachy A.M., Weissman J.S.. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat. Protoc. 2012; 7:1534–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dallagiovanna B., Pereira I.T., Origa-Alves A.C., Shigunov P., Naya H., Spangenberg L.. lncRNAs are associated with polysomes during adipose-derived stem cell differentiation. Gene. 2017; 610:103–111. [DOI] [PubMed] [Google Scholar]
- 12. Ji Z., Song R., Regev A., Struhl K.. Many lncRNAs, 5′UTRs, and pseudogenes are translated and some are likely to express functional proteins. Elife. 2015; 4:e08890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Martinez T.F., Chu Q., Donaldson C., Tan D., Shokhirev M.N., Saghatelian A.. Accurate annotation of human protein-coding small open reading frames. Nat. Chem. Biol. 2020; 16:458–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gong C., Maquat L.E.. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature. 2011; 470:284–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Minati L., Firrito C., Del Piano A., Peretti A., Sidoli S., Peroni D., Belli R., Gandolfi F., Romanel A., Bernabo P.et al.. One-shot analysis of translated mammalian lncRNAs with AHARIBO. Elife. 2021; 10:e59303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Brunet M.A., Leblanc S., Roucou X.. Reconsidering proteomic diversity with functional investigation of small ORFs and alternative ORFs. Exp. Cell Res. 2020; 393:112057. [DOI] [PubMed] [Google Scholar]
- 17. van Heesch S., Witte F., Schneider-Lunitz V., Schulz J.F., Adami E., Faber A.B., Kirchner M., Maatz H., Blachut S., Sandmann C.-L.et al.. The translational landscape of the human heart. Cell. 2019; 178:242–260. [DOI] [PubMed] [Google Scholar]
- 18. Slavoff S.A., Mitchell A.J., Schwaid A.G., Cabili M.N., Ma J., Levin J.Z., Karger A.D., Budnik B.A., Rinn J.L., Saghatelian A.. Peptidomic discovery of short open reading frame-encoded peptides in human cells. Nat. Chem. Biol. 2013; 9:59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ma J., Ward C.C., Jungreis I., Slavoff S.A., Schwaid A.G., Neveu J., Budnik B.A., Kellis M., Saghatelian A.. Discovery of human sORF-encoded polypeptides (SEPs) in cell lines and tissue. J. Proteome Res. 2014; 13:1757–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huraiova B., Kanovits J., Polakova S.B., Cipak L., Benko Z., Sevcovicova A., Anrather D., Ammerer G., Duncan C.D.S., Mata J.et al.. Proteomic analysis of meiosis and characterization of novel short open reading frames in the fission yeast Schizosaccharomyces pombe. Cell Cycle. 2020; 19:1777–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang S., Tian L., Liu H., Li X., Zhang J., Chen X., Jia X., Zheng X., Wu S., Chen Y.et al.. Large-scale discovery of non-conventional peptides in maize and Arabidopsis through an integrated peptidogenomic pipeline. Mol. Plant. 2020; 13:1078–1093. [DOI] [PubMed] [Google Scholar]
- 22. Fesenko I., Kirov I., Kniazev A., Khazigaleeva R., Lazarev V., Kharlampieva D., Grafskaia E., Zgoda V., Butenko I., Arapidi G.et al.. Distinct types of short open reading frames are translated in plant cells. Genome Res. 2019; 29:1464–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Miravet-Verde S., Ferrar T., Espadas-García G., Mazzolini R., Gharrab A., Sabido E., Serrano L., Lluch-Senar M.. Unraveling the hidden universe of small proteins in bacterial genomes. Mol. Syst. Biol. 2019; 15:e8290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sberro H., Fremin B.J., Zlitni S., Edfors F., Greenfield N., Snyder M.P., Pavlopoulos G.A., Kyrpides N.C., Bhatt A.S.. Large-scale analyses of human microbiomes reveal thousands of small, novel genes. Cell. 2019; 178:1245–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Couso J.-P., Patraquim P.. Classification and function of small open reading frames. Nat. Rev. Mol. Cell Biol. 2017; 18:575–589. [DOI] [PubMed] [Google Scholar]
- 26. Heames B., Schmitz J., Bornberg-Bauer E.. A continuum of evolving de novo genes drives protein-coding novelty in Drosophila. J. Mol. Evol. 2020; 88:382–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vakirlis N., Acar O., Hsu B., Castilho Coelho N., Van Oss S.B., Wacholder A., Medetgul-Ernar K., Bowman R.W. 2nd, Hines C.P., Iannotta J.et al.. De novo emergence of adaptive membrane proteins from thymine-rich genomic sequences. Nat. Commun. 2020; 11:781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wilson B.A., Foy S.G., Neme R., Masel J.. Young genes are highly disordered as predicted by the preadaptation hypothesis of de novo gene birth. Nat. Ecol. Evol. 2017; 1:0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Carvunis A.-R., Rolland T., Wapinski I., Calderwood M.A., Yildirim M.A., Simonis N., Charloteaux B., Hidalgo C.A., Barbette J., Santhanam B.et al.. Proto-genes and de novo gene birth. Nature. 2012; 487:370–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vakirlis N., Hebert A.S., Opulente D.A., Achaz G., Hittinger C.T., Fischer G., Coon J.J., Lafontaine I.. A molecular portrait of de novo genes in yeasts. Mol. Biol. Evol. 2018; 35:631–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Basile W., Sachenkova O., Light S., Elofsson A.. High GC content causes orphan proteins to be intrinsically disordered. PLoS Comput. Biol. 2017; 13:e1005375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dragomir M.P., Manyam G.C., Ott L.F., Berland L., Knutsen E., Ivan C., Lipovich L., Broom B.M., Calin G.A.. FuncPEP: a database of functional peptides encoded by non-coding RNAs. Noncoding RNA. 2020; 6:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ji X., Cui C., Cui Q.. smORFunction: a tool for predicting functions of small open reading frames and microproteins. BMC Bioinformatics. 2020; 21:455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Olexiouk V., Van Criekinge W., Menschaert G.. An update on sORFs.org: a repository of small ORFs identified by ribosome profiling. Nucleic Acids Res. 2018; 46:D497–D502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hao Y., Zhang L., Niu Y., Cai T., Luo J., He S., Zhang B., Zhang D., Qin Y., Yang F.et al.. SmProt: a database of small proteins encoded by annotated coding and non-coding RNA loci. Brief. Bioinform. 2018; 19:636–643. [DOI] [PubMed] [Google Scholar]
- 36. Ruiz-Orera J., Albà M.M.. Conserved regions in long non-coding RNAs contain abundant translation and protein–RNA interaction signatures. NAR Genom Bioinform. 2019; 1:e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Szcześniak M.W., Bryzghalov O., Ciomborowska-Basheer J., Makałowska I.. CANTATAdb 2.0: Expanding the Collection of Plant Long Noncoding RNAs. Methods Mol. Biol. 2019; 1933:415–429. [DOI] [PubMed] [Google Scholar]
- 38. Paytuvi-Gallart A., Sanseverino W., Aiese Cigliano R.. A walkthrough to the use of GreeNC: the plant lncRNA database. Methods Mol. Biol. 2019; 1933:397–414. [DOI] [PubMed] [Google Scholar]
- 39. Lang D., Ullrich K.K., Murat F., Fuchs J., Jenkins J., Haas F.B., Piednoel M., Gundlach H., Van Bel M., Meyberg R.et al.. The Physcomitrella patens chromosome-scale assembly reveals moss genome structure and evolution. Plant J. 2018; 93:515–533. [DOI] [PubMed] [Google Scholar]
- 40. Simopoulos C.M.A., Weretilnyk E.A., Golding G.B.. Molecular traits of long non-protein coding RNAs from diverse plant species show little evidence of phylogenetic relationships. G3. 2019; 9:2511–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Quinlan A.R. BEDTools: the Swiss-army tool for genome feature analysis. Curr. Protoc. Bioinformatics. 2014; 47:11–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rice P., Longden I., Bleasby A.. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000; 16:276–277. [DOI] [PubMed] [Google Scholar]
- 43. Zhu M., Gribskov M.. MiPepid: MicroPeptide identification tool using machine learning. BMC Bioinformatics. 2019; 20:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jenuth J.P. Misener S., Krawetz S.A.. The NCBI. Bioinformatics Methods and Protocols. 1999; Totowa, NJ: Humana Press; 301–312. [Google Scholar]
- 45. One Thousand Plant Transcriptomes Initiative One thousand plant transcriptomes and the phylogenomics of green plants. Nature. 2019; 574:679–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J.. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic. Acids. Res. 1997; 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ogurtsov A.Y., Roytberg M.A., Shabalina S.A., Kondrashov A.S.. OWEN: aligning long collinear regions of genomes. Bioinformatics. 2002; 18:1703–1704. [DOI] [PubMed] [Google Scholar]
- 48. Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004; 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kondrashov A.S., Shabalina S.A.. Classification of common conserved sequences in mammalian intergenic regions. Hum. Mol. Genet. 2002; 11:669–674. [DOI] [PubMed] [Google Scholar]
- 50. Suyama M., Torrents D., Bork P.. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006; 34:W609–W612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980; 16:111–120. [DOI] [PubMed] [Google Scholar]
- 52. Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007; 24:1586–1591. [DOI] [PubMed] [Google Scholar]
- 53. Murrell B., Weaver S., Smith M.D., Wertheim J.O., Murrell S., Aylward A., Eren K., Pollner T., Martin D.P., Smith D.M.et al.. Gene-wide identification of episodic selection. Mol. Biol. Evol. 2015; 32:1365–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nguyen L.-T., Schmidt H.A., Haeseler A., Minh B.Q.. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015; 32:268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Thissen D., Steinberg L., Kuang D. Quick and easy implementation of the Benjamini-Hochberg procedure for controlling the false positive rate in multiple comparisons. J. Educ. Behav. Stat. 2002; 27:77–83. [Google Scholar]
- 56. Eaton D.A.R. Toytree: a minimalist tree visualization and manipulation library for Python. Methods Ecol. Evol. 2020; 11:187–191. [Google Scholar]
- 57. Quevillon E., Silventoinen V., Pillai S., Harte N., Mulder N., Apweiler R., Lopez R.. InterProScan: protein domains identifier. Nucleic Acids Res. 2005; 33:W116–W120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Boschiero C., Lundquist P.K., Roy S., Dai X., Zhao P.X., Scheible W.. Identification and functional investigation of genome-encoded, small, secreted peptides in plants. Curr. Protoc. Plant Biol. 2019; 4:441. [DOI] [PubMed] [Google Scholar]
- 59. Bailey T.L., Elkan C.. Fitting a mixture model by expectation maximization to discover motifs in bipolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994; 2:28–36. [PubMed] [Google Scholar]
- 60. Wootton J.C., Federhen S.. [33]Analysis of compositionally biased regions in sequence databases. Methods in Enzymology. 1996; 266:Academic Press; 554–571. [DOI] [PubMed] [Google Scholar]
- 61. Armenteros J.J.A., Tsirigos K.D., Sønderby C.K., Petersen T.N., Winther O., Brunak S., von Heijne G., Nielsen H.. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019; 37:420–423. [DOI] [PubMed] [Google Scholar]
- 62. Krogh A., Larsson B., von Heijne G., Sonnhammer E.L.. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001; 305:567–580. [DOI] [PubMed] [Google Scholar]
- 63. Kubo M., Imai A., Nishiyama T., Ishikawa M., Sato Y., Kurata T., Hiwatashi Y., Reski R., Hasebe M.. System for stable β-estradiol-inducible gene expression in the moss Physcomitrella patens. PLoS One. 2013; 8:e77356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Fesenko I., Spechenkova N., Mamaeva A., Makhotenko A.V., Love A.J., Kalinina N.O., Taliansky M.. Role of the methionine cycle in the temperature-sensitive responses of potato plants to potato virus Y. Mol. Plant Pathol. 2021; 22:77–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Faurobert M., Pelpoir E., Chaïb J.. Phenol extraction of proteins for proteomic studies of recalcitrant plant tissues. Methods Mol. Biol. 2007; 355:9–14. [DOI] [PubMed] [Google Scholar]
- 66. Tyanova S., Temu T., Cox J.. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016; 11:2301–2319. [DOI] [PubMed] [Google Scholar]
- 67. Nesvizhskii A.I. Proteogenomics: concepts, applications and computational strategies. Nat. Methods. 2014; 11:1114–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lang D., Ullrich K.K., Murat F., Fuchs J., Jenkins J., Haas F.B., Piednoel M., Gundlach H., Van Bel M., Meyberg R.et al.. The Physcomitrella patens chromosome-scale assembly reveals moss genome structure and evolution. Plant J. 2018; 93:515–533. [DOI] [PubMed] [Google Scholar]
- 69. Li H. Minimap and miniasm: fast mapping and de novo assembly for noisy long sequences. Bioinformatics. 2016; 32:2103–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.1000 Genome Project Data Processing Subgroup . The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kovaka S., Zimin A.V., Pertea G.M., Razaghi R., Salzberg S.L., Pertea M.. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 2019; 20:278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Tang A.D., Soulette C.M., van Baren M.J., Hart K., Hrabeta-Robinson E., Wu C.J., Brooks A.N.. Full-length transcript characterization of SF3B1 mutation in chronic lymphocytic leukemia reveals downregulation of retained introns. Nat. Commun. 2020; 11:1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Pertea G., Pertea M.. GFF utilities: GffRead and GffCompare. F1000Res. 2020; 9:304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Liao Y., Smyth G.K., Shi W.. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014; 30:923–930. [DOI] [PubMed] [Google Scholar]
- 75. Van Rossum G., Drake F.L. Jr. 1995; Python tutorial Centrum voor Wiskunde en Informatica Amsterdam.
- 76. Virtanen P., Gommers R., Oliphant T.E., Haberland M., Reddy T., Cournapeau D., Burovski E., Peterson P., Weckesser W., Bright J.et al.. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods. 2020; 17:261–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Waskom M. seaborn: statistical data visualization. J. Open Source Softw. 2021; 6:3021. [Google Scholar]
- 78. McKinney W. 2012; Python for Data Analysis: Data Wrangling with Pandas, NumPy, and IPython ‘O’Reilly Media, Inc..
- 79. Lex A., Gehlenborg N., Strobelt H., Vuillemot R., Pfister H.. UpSet: visualization of intersecting sets. IEEE Trans. Vis. Comput. Graph. 2014; 20:1983–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Budak H., Kaya S.B., Cagirici H.B.. Long non-coding RNA in plants in the era of reference sequences. Front. Plant Sci. 2020; 11:276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Ulitsky I. Evolution to the rescue: using comparative genomics to understand long non-coding RNAs. Nat. Rev. Genet. 2016; 17:601–614. [DOI] [PubMed] [Google Scholar]
- 82. Li Y.-R., Liu M.-J.. Prevalence of alternative AUG and non-AUG translation initiators and their regulatory effects across plants. Genome Res. 2020; 30:1418–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Gao X., Wan J., Liu B., Ma M., Shen B., Qian S.-B.. Quantitative profiling of initiating ribosomes in vivo. Nat. Methods. 2015; 12:147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Hazarika R.R., De Coninck B., Yamamoto L.R., Martin L.R., Cammue B.P.A., van Noort V.. ARA-PEPs: a repository of putative sORF-encoded peptides in Arabidopsis thaliana. BMC Bioinformatics. 2017; 18:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Mergner J., Frejno M., List M., Papacek M., Chen X., Chaudhary A., Samaras P., Richter S., Shikata H., Messerer M.et al.. Mass-spectrometry-based draft of the Arabidopsis proteome. Nature. 2020; 579:409–414. [DOI] [PubMed] [Google Scholar]
- 86. Choi S.-W., Kim H.-W., Nam J.-W.. The small peptide world in long noncoding RNAs. Brief. Bioinform. 2019; 20:1853–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Hezroni H., Koppstein D., Schwartz M.G., Avrutin A., Bartel D.P., Ulitsky I.. Principles of long noncoding RNA evolution derived from direct comparison of transcriptomes in 17 species. Cell Rep. 2015; 11:1110–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Hartford C.C.R., Lal A.. When long noncoding becomes protein coding. Mol. Cell. Biol. 2020; 40:e00528-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Ladoukakis E., Pereira V., Magny E.G., Eyre-Walker A., Couso J.P.. Hundreds of putatively functional small open reading frames in Drosophila. Genome Biol. 2011; 12:R118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Mackowiak S.D., Zauber H., Bielow C., Thiel D., Kutz K., Calviello L., Mastrobuoni G., Rajewsky N., Kempa S., Selbach M.et al.. Extensive identification and analysis of conserved small ORFs in animals. Genome Biol. 2015; 16:179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Chaudhary R., Gryder B., Woods W.S., Subramanian M., Jones M.F., Li X.L., Jenkins L.M., Shabalina S.A., Mo M., Dasso M.et al.. Prosurvival long noncoding RNA PINCR regulates a subset of p53 targets in human colorectal cancer cells by binding to Matrin 3. Elife. 2017; 6:e23244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Managadze D., Lobkovsky A.E., Wolf Y.I., Shabalina S.A., Rogozin I.B., Koonin E.V.. The vast, conserved mammalian lincRNome. PLoS Comput. Biol. 2013; 9:e1002917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Resch A.M., Ogurtsov A.Y., Rogozin I.B., Shabalina S.A., Koonin E.V.. Evolution of alternative and constitutive regions of mammalian 5′UTRs. BMC Genomics. 2009; 10:162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Ruiz-Orera J., Verdaguer-Grau P., Villanueva-Cañas J.L., Messeguer X., Albà M.M.. Translation of neutrally evolving peptides provides a basis for de novo gene evolution. Nat. Ecol. Evol. 2018; 2:890–896. [DOI] [PubMed] [Google Scholar]
- 95. Toll-Riera M., Radó-Trilla N., Martys F., Albà M.M.. Role of low-complexity sequences in the formation of novel protein coding sequences. Mol. Biol. Evol. 2012; 29:883–886. [DOI] [PubMed] [Google Scholar]
- 96. Radó-Trilla N., Albà M.. Dissecting the role of low-complexity regions in the evolution of vertebrate proteins. BMC Evol. Biol. 2012; 12:155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Jarnot P., Ziemska-Legięcka J., Grynberg M., Gruca A.. LCR-BLAST—a new modification of BLAST to search for similar low complexity regions in protein sequences. Man-Machine Interactions 6. 2020; Springer International Publishing; 169–180. [Google Scholar]
- 98. Wootton J.C., Federhen S.. Statistics of local complexity in amino acid sequences and sequence databases. Comput. Chem. 1993; 17:149–163. [Google Scholar]
- 99. Singh S., Mittal A.. Transmembrane domain lengths serve as signatures of organismal complexity and viral transport mechanisms. Sci. Rep. 2016; 6:22352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Colling J., Tohge T., De Clercq R., Brunoud G., Vernoux T., Fernie A.R., Makunga N.P., Goossens A., Pauwels L.. Overexpression of the Arabidopsis thaliana signalling peptide TAXIMIN1 affects lateral organ development. J. Exp. Bot. 2015; 66:5337–5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Whitewoods C.D., Cammarata J., Nemec Venza Z., Sang S., Crook A.D., Aoyama T., Wang X.Y., Waller M., Kamisugi Y., Cuming A.C.et al.. CLAVATA was a genetic novelty for the morphological innovation of 3D growth in land plants. Curr. Biol. 2018; 28:2365–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Tavormina P., De Coninck B., Nikonorova N., De Smet I., Cammue B.P.A.. The plant peptidome: an expanding repertoire of structural features and biological functions. Plant Cell. 2015; 27:2095–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Parker M.T., Knop K., Sherwood A.V., Schurch N.J., Mackinnon K., Gould P.D., Hall A.J., Barton G.J., Simpson G.G.. Nanopore direct RNA sequencing maps the complexity of Arabidopsis mRNA processing and m6A modification. Elife. 2020; 9:e49658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Zhang S., Li R., Zhang L., Chen S., Xie M., Yang L., Xia Y., Foyer C.H., Zhao Z., Lam H.-M.. New insights into Arabidopsis transcriptome complexity revealed by direct sequencing of native RNAs. Nucleic Acids Res. 2020; 48:7700–7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Kirov I., Dudnikov M., Merkulov P., Shingaliev A., Omarov M., Kolganova E., Sigaeva A., Karlov G., Soloviev A.. Nanopore RNA sequencing revealed long non-coding and LTR retrotransposon-related RNAs expressed at early stages of triticale SEED development. Plants. 2020; 9:1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Michel A.M., Andreev D.E., Baranov P.V.. Computational approach for calculating the probability of eukaryotic translation initiation from ribo-seq data that takes into account leaky scanning. BMC Bioinformatics. 2014; 15:380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Palazzo A.F., Koonin E.V.. Functional long non-coding RNAs evolve from junk transcripts. Cell. 2020; 183:1151–1161. [DOI] [PubMed] [Google Scholar]
- 108. Zhang J., Yang J.-R.. Determinants of the rate of protein sequence evolution. Nat. Rev. Genet. 2015; 16:409–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Fesenko I., Azarkina R., Kirov I., Kniazev A., Filippova A., Grafskaia E., Lazarev V., Zgoda V., Butenko I., Bukato O.et al.. Phytohormone treatment induces generation of cryptic peptides with antimicrobial activity in the moss Physcomitrella patens. BMC Plant Biol. 2019; 19:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Chen J., Brunner A.-D., Cogan J.Z., Nuñez J.K., Fields A.P., Adamson B., Itzhak D.N., Li J.Y., Mann M., Leonetti M.D.et al.. Pervasive functional translation of noncanonical human open reading frames. Science. 2020; 367:1140–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Ruiz Cuevas M.V., Hardy M.-P., Hollý J., Bonneil É., Durette C., Courcelles M., Lanoix J., Côté C., Staudt L.M., Lemieux S.et al.. Most non-canonical proteins uniquely populate the proteome or immunopeptidome. Cell Rep. 2021; 34:108815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Aspden J.L., Eyre-Walker Y.C., Phillips R.J., Amin U., Mumtaz M.A.S., Brocard M., Couso J.-P.. Extensive translation of small Open Reading Frames revealed by Poly-Ribo-Seq. Elife. 2014; 3:e03528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Kavi Kishor P.B., Hima Kumari P., Sunita M.S.L., Sreenivasulu N.. Role of proline in cell wall synthesis and plant development and its implications in plant ontogeny. Front. Plant Sci. 2015; 6:544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Pearce G. Systemin, hydroxyproline-rich systemin and the induction of protease inhibitors. Curr. Protein Pept. Sci. 2011; 12:399–408. [DOI] [PubMed] [Google Scholar]
- 115. Bao Z., Clancy M.A., Carvalho R.F., Elliott K., Folta K.M.. Identification of novel growth regulators in plant populations expressing random peptides. Plant Physiol. 2017; 175:619–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Mortz M., Dégletagne C., Romestaing C., Duchamp C.. Comparative genomic analysis identifies small open reading frames (sORFs) with peptide-encoding features in avian 16S rDNA. Genomics. 2020; 112:1120–1127. [DOI] [PubMed] [Google Scholar]
- 117. Shabalina S.A., Ogurtsov A.Y., Spiridonov A.N., Novichkov P.S., Spiridonov N.A., Koonin E.V.. Distinct patterns of expression and evolution of intronless and intron-containing mammalian genes. Mol. Biol. Evol. 2010; 27:1745–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Shabalina S.A., Ogurtsov A.Y., Spiridonov N.A., Koonin E.V.. Evolution at protein ends: major contribution of alternative transcription initiation and termination to the transcriptome and proteome diversity in mammals. Nucleic. Acids. Res. 2014; 42:7132–7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Li X.L., Pongor L., Tang W., Das S., Muys B.R., Jones M.F., Lazar S.B., Dangelmaier E.A., Hartford C.C., Grammatikakis I.et al.. A small protein encoded by a putative lncRNA regulates apoptosis and tumorigenicity in human colorectal cancer cells. Elife. 2020; 9:e53734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Lauressergues D., Couzigou J.-M., Clemente H.S., Martinez Y., Dunand C., Bécard G., Combier J.-P.. Primary transcripts of microRNAs encode regulatory peptides. Nature. 2015; 520:90–93. [DOI] [PubMed] [Google Scholar]
- 121. Sharma A., Badola P.K., Bhatia C., Sharma D., Trivedi P.K.. Primary transcript of miR858 encodes regulatory peptide and controls flavonoid biosynthesis and development in Arabidopsis. Nat. Plants. 2020; 6:1262–1274. [DOI] [PubMed] [Google Scholar]
- 122. Zeng C., Fukunaga T., Hamada M.. Identification and analysis of ribosome-associated lncRNAs using ribosome profiling data. BMC Genomics. 2018; 19:414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Wu Q., Wright M., Gogol M.M., Bradford W.D., Zhang N., Bazzini A.A.. Translation of small downstream ORFs enhances translation of canonical main open reading frames. EMBO J. 2020; 39:e104763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Hochberg G.K.A., Liu Y., Marklund E.G., Metzger B.P.H., Laganowsky A., Thornton J.W.. A hydrophobic ratchet entrenches molecular complexes. Nature. 2020; 588:503–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Matsumoto A., Pasut A., Matsumoto M., Yamashita R., Fung J., Monteleone E., Saghatelian A., Nakayama K.I., Clohessy J.G., Pandolfi P.P.. mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide. Nature. 2017; 541:228–232. [DOI] [PubMed] [Google Scholar]
- 126. Hershberg R., Petrov D.A.. Evidence that mutation is universally biased towards AT in bacteria. PLoS Genet. 2010; 6:e1001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Van Oss S.B., Carvunis A.-R.. De novo gene birth. PLoS Genet. 2019; 15:e1008160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Wissler L., Godmann L.. Evolutionary dynamics of simple sequence repeats across long evolutionary time scale in genus Drosophila. Trends Evol. Biol. 2012; 4:e7. [Google Scholar]
- 129. Keeling D.M., Garza P., Nartey C.M., Carvunis A.-R.. Philosophy of Biology: The meanings of’function’in biology and the problematic case of de novo gene emergence. Elife. 2019; 8:e47014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Hezroni H., Ben-Tov Perry R., Meir Z., Housman G., Lubelsky Y., Ulitsky I.. A subset of conserved mammalian long non-coding RNAs are fossils of ancestral protein-coding genes. Genome Biol. 2017; 18:162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D.J., Inuganti A., Griss J., Mayer G., Eisenacher M.et al.. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019; 47:D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Nanopore data were deposited in the BioProject with the accession number PRJNA681088. The mass spectrometry peptidomic data have been deposited to the ProteomeXchange Consortium via the PRIDE (131) partner repository with the dataset identifiers PXD025373 and PXD025267. All scripts are maintained in the GitHub code repository: https://github.com/IgorFesenko/smORF_analysis.