ABSTRACT
Using scRNA-seq coupled with computational approaches, we studied transcriptional changes in cell states of sea urchin embryos during development to the larval stage. Eighteen closely spaced time points were taken during the first 24 h of development of Lytechinus variegatus (Lv). Developmental trajectories were constructed using Waddington-OT, a computational approach to ‘stitch’ together developmental time points. Skeletogenic and primordial germ cell trajectories diverged early in cleavage. Ectodermal progenitors were distinct from other lineages by the 6th cleavage, although a small percentage of ectoderm cells briefly co-expressed endoderm markers that indicated an early ecto-endoderm cell state, likely in cells originating from the equatorial region of the egg. Endomesoderm cells also originated at the 6th cleavage and this state persisted for more than two cleavages, then diverged into distinct endoderm and mesoderm fates asynchronously, with some cells retaining an intermediate specification status until gastrulation. Seventy-nine out of 80 genes (99%) examined, and included in published developmental gene regulatory networks (dGRNs), are present in the Lv-scRNA-seq dataset and are expressed in the correct lineages in which the dGRN circuits operate.
KEY WORDS: Sea urchin embryo, scRNA-seq, Gene regulatory networks, Cell lineage
Summary: A temporal study of sea urchin development captured using scRNA-seq. Gene expression during lineage trajectories matches known gene regulatory network changes, and demonstrates both synchronous and asynchronous lineage resolutions.
INTRODUCTION
Molecular specification of each cell lineage occurs during early development. The means by which these lineages diverge varies and it is of value to learn cell properties at the time cell fates separate.
Emerging technologies provide increased leverage to follow a developmental sequence and to explore the complex nature of cell diversification. High-throughput single-cell RNA sequencing (scRNA-seq) is one such approach. When combined with analytical approaches to infer developmental trajectories in mouse and zebrafish embryos (Farrell et al., 2018; Schiebinger et al., 2019), scRNA-seq was shown to reveal new details about embryonic cell types and their developmental trajectories. Because of its potential, use of this technology has rapidly expanded and is being applied to a growing number of embryos, tissues and disease states (Tintori et al., 2016; Cao et al., 2017; Karaiskos et al., 2017; Fincher et al., 2018; Han et al., 2018; Plass et al., 2018; Wagner et al., 2018; Foster et al., 2020). Owing to the destruction of the cell during scRNA-seq protocols, it is only possible to gain a snapshot of the cell's transcriptome. To measure temporal changes during development with scRNA-seq, a compromise solution is to capture cells at a series of time points that are relatively close together, then computationally stitch them together to infer a continuous sequence. The spacing of the time points must be sufficiently close for the temporal trajectory to be followed. If those conditions are satisfied, scRNA-seq enables the reconstruction of an atlas of transient cell states over time.
At a systems level, developmental gene regulatory networks (dGRNs) of transcription factors plus signaling inputs provide the necessary programming that directs cells toward their fates. Circuit motifs (feedback, feed forward, etc.) (Yeger-Lotem et al., 2004), confer a number of properties to dGRNs, including stability (Brandman and Meyer, 2008). Changes in these transcriptional and signaling circuits are hypothesized to be drivers of evolutionary diversification (Hinman et al., 2003; Hinman and Davidson, 2007; Erwin and Davidson, 2009; Erkenbrack and Davidson, 2015; Israel et al., 2016; Cary et al., 2020). The challenge is to find ways to explore the properties of dGRNs, including how they diverge to produce a chain of sublineages with new subcircuits and different functions.
To begin using scRNA-seq for inference into regulatory properties, a number of steps must be followed. The depth of sequencing must be sufficient to detect expression of the transcription factors and signals that constitute the dGRNs. To explore the properties of dGRNs we chose to analyze the sea urchin dGRN. Over the past two decades, a number of investigators have contributed to the assembly of a dGRN from fertilization to gastrulation in Strongycentrotus purpuratus (Sp) (Davidson et al., 2002; Cole et al., 2009; Ben-Tabou de-Leon et al., 2013; Andrikou et al., 2015; Peter and Davidson, 2015; McClay et al., 2020). Perturbation of the same transcription factors and signals have also been conducted on other species, especially on Lytechinus variegatus (Lv) and Paracentrotus lividus (Pl) (Logan et al., 1999; Sherwood and McClay, 1999; Angerer et al., 2000; Gross and McClay, 2001; Duboc et al., 2004, 2005; Oliveri et al., 2006; Röttinger et al., 2008; Saudemont et al., 2010; Croce and McClay, 2010; McIntyre et al., 2013; Saunders and McClay, 2014; Malik et al., 2017; Martik and McClay, 2017). Remarkably, the experimental evidence indicates that the dGRNs driving specification in those species are highly conserved despite a separation of up to 50 million years from a common ancestor. Recently, scRNA-seq was used to compile the first atlas of development for Sp (Foster et al., 2019), and that approach was further used to explore the development of pigment cells in Sp (Perillo et al., 2020). In those studies, selected dGRN genes were present and expressed in clusters associated with several lineages. That study also detected primordial germ cells (PGCs), which attested to the sensitivity of the approach. Given the extensive knowledge of dGRNs in the sea urchin embryo, our first goal was to determine the extent to which most known molecules in sea urchin dGRNs were ‘visible’ to scRNA-seq inquiries. We found that 99% of the molecules examined in the sea urchin dGRN models are present in our scRNA-seq database. Importantly, the signals and transcription factors that constitute those dGRNs are present both in the lineages expected and at the times the dGRN models indicate that their presence is required.
A recently developed computational tool, ‘Waddington-OT’ (Schiebinger et al., 2019), was adapted to explore cell trajectories. Novel methods were developed to use barycentric coordinates and illustrate developmental trajectories and dGRN changes over time. These provided an opportunity to examine the timing of divergence and novel properties of that diversification. The known existence of a seminal asymmetry event in separation of micromere/PMCs from other cell types is seen in the dataset. Other lineage separations, however, are gradual, implying a more-complex series of events necessary to resolve a cell type, and an ectoderm-endoderm state is observed in some cells, likely near the equator, before their lineage separation. A number of ‘signature’ genes representing known lineages were used for comparisons of synchronous versus asynchronous divergence of lineage trajectories. While PGCs, skeletogenic cells and some ectoderm become distinct early in cleavage, other cells retain an ecto-endoderm and endomesoderm status until daughter cells diverge into the distinct differentiated fates later and asynchronously.
RESULTS
Atlas of sea urchin development from fertilization to larval stage
Development of Lv from fertilization to larva occurs in 24 h at 23°C. To obtain an atlas of lineage trajectories over this time frame, we collected embryos at 18 time points initially at hourly intervals and then during later stages at 2- to 4-h intervals. Embryos were dissociated and processed using the 10x Genomics system as adapted to the sea urchin embryo (Perillo et al., 2020; Massri et al., 2021).
Reads were mapped to the Lv3.0 genome (Davidson et al., 2020), and low-quality cells filtered out (see Materials and Methods). In total, 50,935 cells remained and were used for the analysis. Fig. 1 shows the requisite overview UMAP visualizations of these 50,935 cells. Graph-based louvain clustering, implemented in Seurat, was used to obtain 63 distinct clusters of cells, and these were annotated using dGRN marker genes (Davidson et al., 2002; Peter and Davidson, 2015; McClay et al., 2020) to provide a preliminary identification of the clusters. Fig. S1A shows the major germ layers, Fig. S1B shows the 63 clusters and Fig. S1C-E shows dotplots of genes used to identify each of the endoderm, mesoderm and ectoderm clusters. The dynamics of the temporal progression of stages is shown in Movie 1.
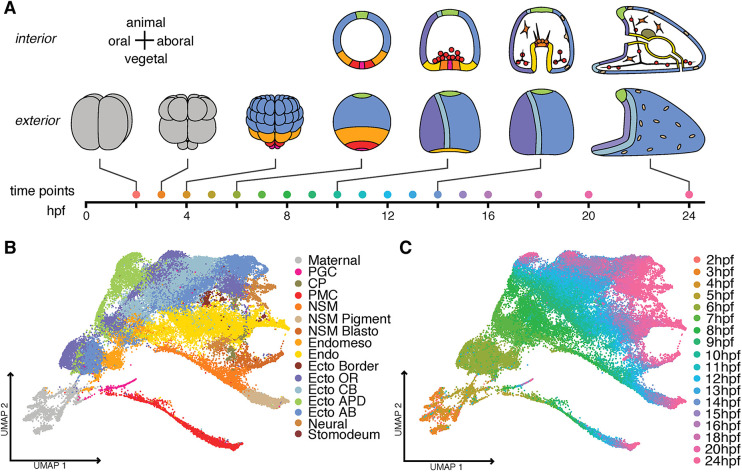
Fig. 1.
Temporal trajectory of scRNA-seq profiles during development of the sea urchin Lytechinus variagatus. (A) A timeline of development over 24 h, with representative stages of development illustrated. Colors represent lineages as in B. (B) UMAP plot mapped according to lineage domains. Sixty-three clusters were identified and grouped into 16 domains representing the lineages present at 24 hpf. (C) The UMAP plot colored according to time of development showing the position of the 18 time points collected during the first 24 h post-fertilization (hpf). PGC, primordial germ cells; CP, coelomic pouch; PMC, primary mesenchyme cell (also called skeletogenic cells); NSM, non-skeletogenic mesenchyme; Pigment, pigment cells; Blasto, blastocoelar cells; Endomeso, endomesoderm; Endo, endoderm; Ecto, ectoderm; Border, ectoderm at the border between ectoderm and endoderm; OR, oral; CB, ciliary band; APD, animal pole domain; AB, aboral.
The structure of the UMAP recapitulates the basic events of sea urchin development. It has long been established that the skeletogenic or primary mesenchyme cells (PMCs), and primordial germ cells (PGCs) become distinct from the other lineages as early as the 4th and 5th cleavages by two successive unequal cleavages (McClay, 2011). The UMAP displays that separation, and provides a continuous track of PMC and PGC trajectories as they are specified over time. The endomesoderm later splits into endoderm and mesoderm, and is reflected in the UMAP plot because, at 5 h post-fertilization (hpf; blastula stage), the endomesoderm is one cluster, indicating that the cells still have not yet diversified. By 7-8 hpf (hatched blastula) the endoderm and mesoderm cells have begun to separate, and later each further subdivides as diversification within the germ layers occurs. Meanwhile, specification of the ectoderm leads to regional anterior-posterior, dorsal-ventral and right-left differences in this lineage (Fig. 1B; Fig. S1). In each case, the diversification and timing fits with known results from earlier embryonic lineage analyses (Cameron et al., 1987; Logan and McClay, 1997, 1999; Martik and McClay, 2017; McClay et al., 2020).
The lineage analyses observed that a stereotypic cleavage sequence operates through the 8th cleavage (hatched blastula), and further observed later lineage doublings during gastrulation (10-16 hpf) (Table S1). These approximations of cell lineage numbers provided the necessary information to determine whether the pipeline from embryo to sequencing output introduced any bias in the relative abundance of cells of each lineage. Based on those analyses, the distribution of cells allocated to ectoderm, mesoderm, endoderm, primary mesenchyme cells (PMCs) and primordial germ cells (PGCs) were not biased by over- or under-selection once the lineages could be identified by genes expressed at 5-6 hpf (Fig. S2). Before 5 h, the prevalence of maternal transcripts distributed to all cells dominated and prevented lineages from being separately distinguished. At the 5 hpf time point, enough lineage markers had emerged to provide an imperfect approximation of lineages, and from 6 hpf until the larval stage, the number of cells for each major lineage closely matched the approximate cells expected based on the earlier lineage analyses.
Even relatively rare cell populations were detected. PMCs never constitute more than 4-5% of the cells, but a continuous track of that lineage is present. Four PGCs arise at 5th cleavage, later divide once and at gastrulation migrate to the coelomic pouches without dividing further (Voronina et al., 2008; Campanale et al., 2014; Martik and McClay, 2015). Despite contributing a maximum of eight cells per embryo, PGCs were present at each time point, providing a continuous record of that lineage (Fig. 1; Fig. S1). Only four or five serotonergic neurons are present in larvae at 24 hpf (Slota and McClay, 2018). Again, that rare cell type was detected (Fig. S1), giving us confidence that our developmental reconstruction reflected a detailed and accurate representation of the transcriptional history of each lineage over the first 24 h of development.
Detection of molecules contributing to developmental gene regulatory networks
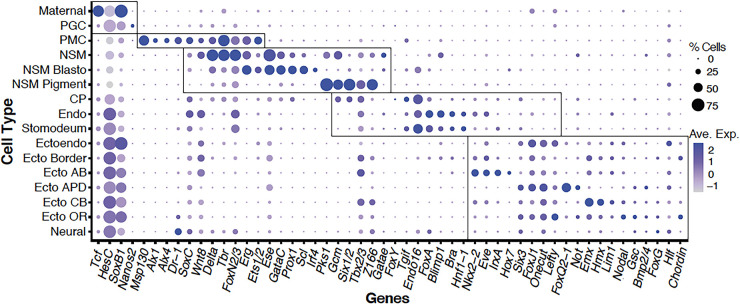
The scRNA-seq analysis faithfully reflected expression of known transcription factors in the networks of each lineage. We analyzed the expression of 80 genes that included the major transcription factors and signals plus 13 effector genes specific to several lineages from the published dGRN models. All but one gene in the dGRN models are detected in the relevant lineages, indicating that the scRNA-seq database provides information on 99% of the known dGRN genes [the missing gene is twist, a short gene (603 bp) expressed at very low levels (<21 copies per expressing cell) and not included in the Sp dGRN]. dGRN genes used as signatures for the germ layers are plotted in a quantitative dotplot shown in Fig. 2. Other dGRN genes and tissue-specific marker genes are plotted in Fig. S1C-E to reflect the genes used to identify the 63 selected clusters. dGRN genes are also individually displayed on the UMAP in Fig. S3A,B. As expected, transcription factors known to be expressed at low levels were detected in fewer cells of a cell type than genes expressed at higher levels in that same cell type. The scRNA-seq database has the sensitivity to examine the spatial and temporal patterns of expression of the vast majority of transcription factors and genes used in specification of embryonic cells, including, we suspect, transcription factors that contribute but are not yet included in the dGRNs.
Fig. 2.
Dotplot of genes expressed in the sea urchin (dGRN). Fifty-two of the more than 80 genes expressed in the dGRN models (Ben-Tabou de-Leon et al., 2013; Li et al., 2013; Peter and Davidson, 2015) were plotted according to expression in the clusters listed on the y axis. Each dot shows the relative level of expression in each cluster as well as the percentage of cells in each cluster that express that gene. Boxes surround the clusters assigned to PGC, PMC, NSM, endoderm and ectoderm. Genes expressed outside the boxes are expressed in more than one lineage, sometimes at the same time and in other cases at different times.
Inferring developmental trajectories with Waddington-OT
We next applied Waddington-OT methods to connect the scRNA-seq data from different time points and infer developmental trajectories (Schiebinger et al., 2019). Optimal transport connects cells sampled at one time point to their putative descendants at the next time point in a way that minimizes differences in expression over all genes. The algorithm requires as input an estimate of the proliferative ability of each cell, which it uses to allocate each cell a specific number of descendants. Based on the concordance between observed and expected change in abundance of each of the five primary lineages (Fig. S2), cell proliferation rates were assigned, and Waddington-OT run with the default parameter values (see the Materials and Methods for details). The resulting output is a time-course of transport matrices connecting each pair of time points. These matrices have a row for each early cell and a column for each late cell, and the i,j entry is interpreted as the number of descendants cell i would have of type j at the later time point.
The quality of inferred trajectories was tested by demonstrating that we could interpolate the distribution of cells at held-out time points, as described by Schiebinger et al. (2019). It established that our temporal resolution was sufficient to accurately stitch together cells from adjacent time points (Fig. S4A). In other tests, this result was robust to down-sampling cells or reads, to moderate changes in parameter values away from the default settings, or to moderate perturbations in division rates (Fig. S4B-F).
Visualizing lineage diversifications
The transport matrices allowed us to directly compute, for any cell from an early time point, the proportion of descendants that reach various fates at later time points. We developed a simple way to visualize these ‘fate probabilities’ for triples (or quadruples) of lineages in a triangular (or tetrahedral) plot (Fig. 3; Movie 1). The visualization employs barycentric coordinates to represent k-dimensional probability vectors in k-1 dimensional space. For example, suppose we wish to visualize the cells from 8 hpf (hatched blastula) according to their probability of giving rise to an ectodermal, endodermal or ‘Other’ descendants at 20 hpf (prism stage). We identify a vertex of the triangle for each of these possible fates, and position the cells according to their relative probabilities. Cells that have perfectly arrived at a single fate (as defined by all genes expressed by a cell type at 24 hpf), are positioned exactly at the corresponding vertex, while cells with as yet indeterminate fates are positioned in the interior of the triangle. Cells in the very center of the triangle are equally likely to choose any of the three fates, and cells along an edge have zero chance of reaching the opposite vertex. Fig. 3 illustrates examples of how selected pairs of lineages diverge in Lv over time. Movie 1 shows a tetrahedron visualization of the divergence of four fates simultaneously, in parallel with the progression of those same cells along the UMAP over time.
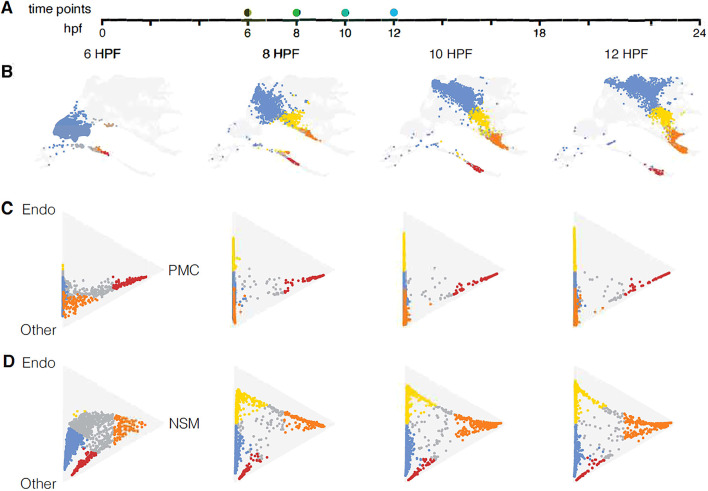
Fig. 3.
Diversification of cell lineages. (A) Timeline represented by the plots with dots to indicate the times shown. (B) UMAPs at those four time points showing presumptive ectoderm (blue), endoderm (yellow), NSM (orange) and PMCs (red). (C,D) The Waddington-OT optimal transport method-computed lineages graphically displayed using barycentric coordinates (Schiebinger et al., 2019). Triangle plots show progression of lineages at different time points, with (C) vertices designated endoderm, PMC and ‘other’ to represent all other lineages; and (D) vertices designated endoderm, NSM and ‘other’ to represent all other lineages. The colored cells in the triangle plots are those cells with at least a 50% probability of becoming one of the lineages represented by 20 hpf. Cells that have not reached a 50% probability towards any of those fates are colored gray. Cells located next to a side of the triangle have low to no chance of reaching the opposite vertex. Cells in the exact middle of the triangle have an equal chance of becoming a cell of any type. In C, pre-PMCs locate to the PMC side of the triangle from the earliest time point onwards. In D, many future NSM or endoderm cells remain intermediate with an apparent extended delay in reaching their final fates.
Embryonic lineages diversify in different ways, as seen using the transport matrices
We focused on analysis of five primary lineage diversifications [PGC, PMC, ectoderm, endoderm and non-skeletal mesoderm (NSM)] (Fig. 3), although a number of additional subdivisions can be detected both in the UMAP (Fig. 1) and in the 63 clusters (Fig. S1). A singular asymmetric event driving a lineage separation is the simplest kind of lineage divergence. In sea urchin, an example of this type occurs at fourth and fifth cleavage at the vegetal pole when skeletogenic cells diverge from the fates of other cells. The skeletogenic cells activate pmar1, a repressor that represses a repressor, hesC (Revilla-i-Domingo et al., 2007). All other lineages in the embryo fail to activate pmar1. Consequently, hesC is activated in all non-skeletogenic cells, resulting in repression of the skeletogenic fate in all other lineages. Fig. 3C shows the consequence of that diversification. By 6 hpf, cells destined to become PMCs (labeled red) are located along one edge of the triangle, the edge leading to the PMC or ‘other’ fate, indicating they have essentially a 0% probability of becoming endoderm. Those cells continue to remain near the PMC edge of the triangle until the PMC vertex is reached at 20 hpf, indicating that from very early on the future PMCs have a 100% probability of being directed toward the PMC fate. Notice also that almost all other cells locate along the opposite side of the triangle from the earliest time point, indicating that they have essentially a 0% chance of becoming PMCs. Fig. 3D, by contrast, shows the separation of endoderm and mesoderm from endomesoderm. That separation is far more delayed with a number of cells in the middle of the triangle between the endoderm or mesoderm vertices at 12 hpf (late mesenchyme blastula-early gastrula stage). Those cells move across the triangle over time, indicating that specification occurs, but they remain distant from either vertex. By 20 and 24 hpf, all endoderm and mesoderm cells are located at their respective vertices (Movie 1), but compared with the early movement of PMC precursors toward their final fate, the endomesoderm fate decision is non-uniform temporally, and often quite late, even though the cells are continually specified (the wave progression). Furthermore, the broad distribution of cells in the wave between the two final fate points suggests that the specification is variable in many cells for an extended time before they reach a committed state.
Some cells exhibit synchronous diversification towards their final fate
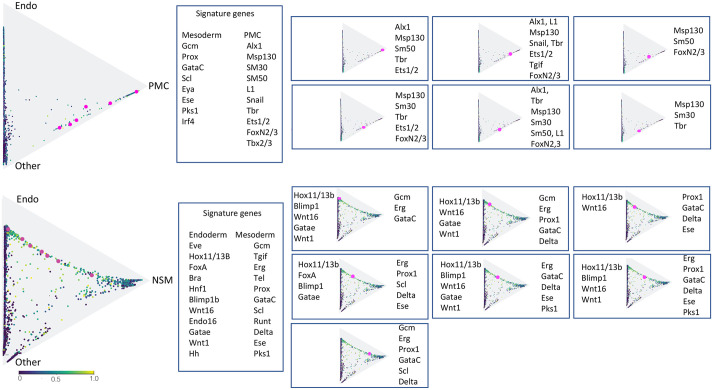
The transcriptomes of cells depicted as gray in Fig. 3C,D are cells that have not reached 50% probability of becoming one of the final cell types based on the genes expressed by that cell relative to genes expressed in a lineage at the 20 and 24 hpf time points. In Fig. 3C, gray cells are found along the side adjacent to cells colored red based on their transcriptomes, which indicate better than 50% probability of becoming PMCs. The gray cells along that side could become either PMCs or NSM (the two vertices on that side). The graphic indicates that, at 12 hpf, a few gray cells continue to be next to the red probable PMCs. This was puzzling because GRN models of PMCs indicate that all pre-PMCs are segregated at the 4th and 5th cleavage so we wondered why there was a difference in the optimal transit profiles. Collections of cell type/lineage-specific genes, or ‘signature’ genes, were used to examine the transcriptomes of the gray and red cells near the PMC side of the triangle. When randomly picked cells were tested for signature genes of both PMCs and NSM, the cells sampled expressed only PMC markers (Fig. 4, top), indicating that they too were pre-PMCs. We wondered why the transcriptomes of the lagging gray cells were not red (>50% probable PMCs based on genes expressed relative to cells at 20 hpf). We first asked whether perhaps those gray cells contained a smaller number of genes reported in their transcriptomes. That was ruled out as the cells sampled had, on average, as many genes sequenced as the red cells. We then turned to the literature, in which a large number of PMC genes were identified (Rafiq et al., 2014). That work showed that some PMC genes are expressed later than others in the process of skeletogenesis. We chose nine of those later-expressed genes based on their expression in only a subset of the PMCs, as seen with in situ hybridization (nk7, mmp16, mtmmpd, cbpdEL, clect, cp, Gabrb3 L, Fam20c and npnt) (Rafiq et al., 2014).
Fig. 4.
Signature plots of PMC versus endoderm-NSM specification trajectories. These triangles show the 12 hpf plots from Fig. 3C,D. The colors reflect known signature genes of the four major lineages. Cells that express a mix of lineage signature genes are colored blue-green, green and yellow, with yellow being cells with a 1:1 ratio of signature gene expression. The pink cells are individual cells with the signature genes shown in the boxes to the right. (Top) Cells at 12 hpf located next to the side between PMCs and ‘other’ (NSM and ectoderm) examined for expression of NSM versus PMC signature genes (they locate to this side because they do not express any endoderm signature genes). Each of the six cells express only PMC signature genes. (Bottom) Cells at 12 hpf examining endoderm versus NSM genes between the endoderm and NSM vertices. Six of the intermediate cells express signature genes of both fates, while the one closest to the NSM expresses only NSM signature genes.
When those genes were used as signature genes, the red PMCs expressed most of them while the gray cells along the PMC-edge expressed fewer of them. This suggested a possible explanation for the red versus gray designation. At 12 hpf, skeletogenesis is just beginning and only eight of the 64 PMCs are involved in producing the earliest triradiate spicules. As the skeleton grows, other PMCs become involved. Thus, we suspect that the gray cells express fewer mature PMC genes at these times because they have not yet begun participating in spicule growth.
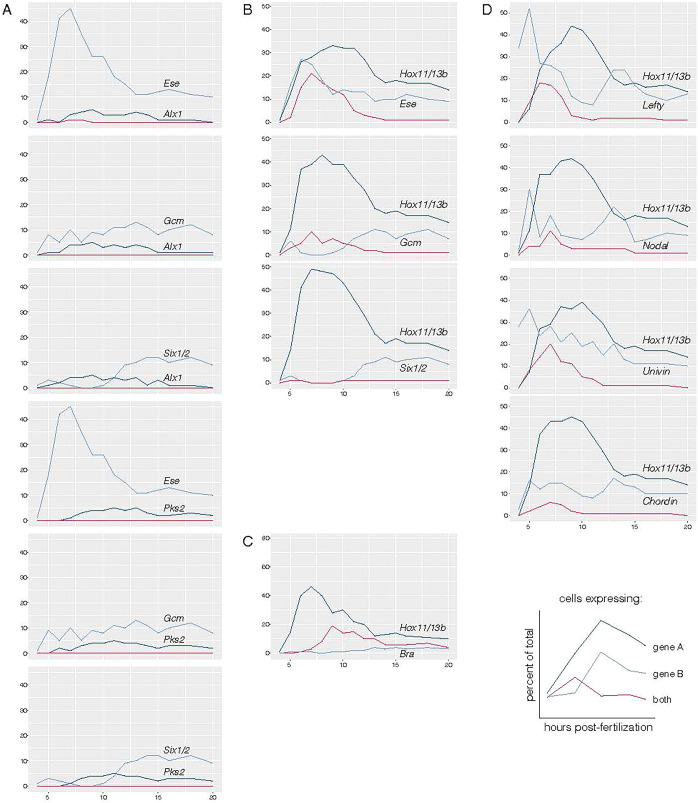
We then approached this question of committed red cells versus gray cells in a different way and asked how many signature gene pairs were co-expressed among all cells in the database. At fifth cleavage, only micromeres express alx1 as a signature gene (Ettensohn et al., 2003; Oliveri et al., 2008), and pks2 was identified in this analysis as an early expressed effector gene in only PMCs. When micromeres segregate from other lineages, cells of that lineage (pre-PMCs) should express PMC signature genes only, and no signature genes from other lineages from that point onward. Fig. 5A shows expression of Alx1 and pks2 in cells beginning at 5 hpf. No signature gene of any other lineage is co-expressed with alx1 or pks2 from the time of their first expression forward. Fig. 5A shows the comparison with signature genes from the NSM, the neighboring cells to the PMCs, while Fig. S5 shows additional comparisons between genes of each germ layer with the others. We expected to see a small amount of artifactual co-expression due to inclusion of multiplets (two or more cells artifactually sequenced as a single cell). The likelihood of obtaining multiplets was minimized by following the 10X protocol and by computationally filtering the database (see Materials and Methods); the data indicated that even potential multiplets were not detected in the alx1 or pks2 versus NSM signature genes. Thus, according to this criterion, micromeres definitively separate from other lineages at 4th and 5th cleavages. However, specification of the micromeres toward their differentiated PMC repertoire of molecules is uneven between progenitors (the gray versus red cells along the trajectory). Nevertheless, the signature gene test confirms that this lineage diversifies synchronously and completely from other lineages.
Fig. 5.
A comparison of pairs of signature genes expressed by cells over time with incidence of co-expression of two signature genes recorded. Blue lines show the percentage of all cells expressing a single selected signature gene as a lineage marker over time. Each graph reports expression of two such signature genes. The red line shows the percentage of all cells expressing both selected signature genes at those time points. If gene A is expressed in 30% of the cells on its own and co-expressed with the gene B in 20% of the cells, its total expression is detected in 50% of all cells at that time point. (A) alx1 expression (PMC lineage) is not co-expressed with signature genes of NSM (gcm, six1/2 and ese) at any time. (B) hox11/13b, a signature gene of endomesoderm and endoderm, is co-expressed with a population of NSM signature genes (gcm, six1/2 and ese) for an extended period of time in the endomesoderm state before the co-expression is lost as cells become definitive endoderm and mesoderm. (C) bra, an endoderm signature gene that is activated only in definitive endoderm, is co-expressed with hox11/13b in some cells, beginning at 8 hpf. By 9 hpf, most bra-expressing cells also express hox11/13b. (D) hox11/13b is co-expressed at early stages in some cells with ectodermal signatures nodal, lefty, univin and chordin between 5 and 9-10 hpf.
Endomesoderm segregation is asynchronous
The transcriptomes of cells in Fig. 3D report the trajectory toward endomesoderm segregation (Fig. 3D) and provide a different outcome. Signature genes of each cell type tested the hypothesis that the intermediate cells between the endoderm and mesoderm vertices (Fig. 3D) expressed dGRN genes of both the endoderm and NSM fates. If so, any of these cells might be capable of being directed toward either fate, as shown earlier in experiments on endomesoderm (Sherwood and McClay, 1999; Oliveri et al., 2006; Croce and McClay, 2010; Peter and Davidson, 2011; Sethi et al., 2012). The cells sampled from the mesoderm-endoderm side of the barycentric coordinate were, in fact, intermediate. In Fig. 4, bottom, most cells in the zone between endoderm and NSM are colored blue-green, green, yellow-green or yellow, indicating that both signatures were present (pure yellow indicated a 1:1 ratio of signature genes expressed in a cell). Thus, in separation of endomesoderm into mesoderm and endoderm, some cells proceed early toward a distinct fate (orange or yellow colored cells are those with >50% probability of becoming mesoderm or endoderm), but other cells continue to express transcription factors of both fates for an extended time. The number of intermediate cells declined considerably after 12 hpf, and by 20 hpf all cells had arrived at either the endoderm or mesoderm vertex.
To have confidence in the observation that the ‘intermediate’ class of cells existed, we ruled out several possible artifacts. First, the initial culture was carefully monitored so that embryos were phenotypically identical at each time point (i.e. there were no embryos developing with a significant delay). Second, multiplets were minimized as above (and see Materials and Methods). Third, the position of each cell in the triangle is based on algorithms that evaluate expression of all genes expressed by a cell according to the optimal transport method described above. The color of each cell in Fig. 4, bottom, however, is based only on the expression of signature genes from dGRN models. Fourth, as shown in the animated sequence of Movie 1, all cells end up at one of the points of the triangles or tetrahedrons by 20 and 24 h. Nevertheless, we randomly sampled individual cells at different positions along the ‘wave’ of intermediate cells, or along the sides of the triangle sides to determine which genes of the two signatures were present. Fig. 4, bottom, shows that six of the seven sampled cells in the intermediate zone between endoderm and NSM express signature genes of both dGRNs. Cells closer to one of the two vertices tended to express a larger number of signature genes of that nearby vertex. One cell, the cell closest to the mesoderm vertex, expressed only mesoderm signature genes. This observation illustrates that many cells of the mesoderm and endomesoderm spend extended time in an intermediate specification state before they finally diverge to one or the other fate. That decision likely occurs not as a consequence of a single event, as seen with skeletogenic cells, which were next to the PMC edge of the triangle by 5-6 hpf, but as a consequence of multiple inputs over time.
We next applied the pairwise signature gene comparison to endomesoderm to determine the percentage of cells that are intermediate (i.e. still endomesoderm) versus those cells that express exclusively one signature gene of either endoderm or mesoderm. This comparison reports trends rather than absolute numbers because the shallow depth of sequencing in scRNA-seq detects an incomplete cohort of genes expressed by each cell. Fig. 5B shows the outcome of that comparison. Several endoderm genes were tested and two were of particular interest. hox11/13b is known to be expressed early in the endomesoderm and later in definitive endoderm cells (Peter and Davidson, 2010). FoxA expression also begins in Veg2 endomesoderm cells, later than hox11/13b, and beginning at about 8th cleavage (8 hpf), is found increasingly in endoderm cells only. FoxA is also expressed in the stomodaeum, beginning at about 10 hpf (Oliveri et al., 2006). gcm, six1/2 and ese are expressed in endomesoderm and later in NSM, but not in endoderm (Ransick et al., 2002; Rizzo et al., 2006; Luo and Su, 2012; Materna and Davidson, 2012). Fig. 5B shows the percentage of cells of the endomesoderm, endoderm and mesoderm lineages over time using these signature genes. A number of cells remain as endomesoderm until gastrulation (12 hpf), whereas sister cells diverge to express exclusively endoderm or mesoderm markers over that same time period beginning at 8 hpf. After 12 hpf, the number of intermediate cells is small. These data indicate that endomesoderm divergence toward endoderm and mesoderm is asynchronous and extends from hatched blastula to the early gastrula stage.
While Fig. 5B provides evidence for the extended and asynchronous endomesoderm segregation, it does not provide a useful impression of when cells arrive at the definitive endoderm state. bra is expressed exclusively in the endoderm in the sea urchin (Gross and McClay, 2001; Croce et al., 2001). Thus, cells expressing bra in addition to hox11/13b are likely to be definitive endoderm cells. As shown in Fig. 5C at 7-8 hpf (8th cleavage-hatched blastula), most bra-expressing cells co-express hox11/13b, indicating that a number of the hox11/13b cells become definitive endoderm by that time (controls showed that bra is never co-expressed with NSM markers above background). Additional genes are compared in Fig. S5.
Fig. 5D examines ectoderm signature gene expression relative to other germ layers. nodal, lefty, univin, chordin and gsc are expressed early in ectoderm specification (Stenzel et al., 1994; Duboc et al., 2004, 2008, 2010; Bradham et al., 2009; Range and Lepage, 2011). Expression of these genes was compared with mesoderm, endoderm and PMC signature genes to determine whether co-expression ever occurs. As expected, ectoderm signature genes were never co-expressed with alx1. In addition, none of the ectoderm signature genes were co-expressed with gcm, indicating that mesoderm and ectoderm specification occur entirely independently. Surprisingly, when we examined co-expression of hox11/13b with the ectoderm signature genes, a number of cells co-express both the ectoderm and the endoderm gene between the fifth and ninth cleavage. Previous lineage analyses showed that Veg1 cells normally produce both endoderm and ectoderm, with a variable number of ectoderm cells arising from this vegetal tier depending on the position of the third cleavage (Henry et al., 1989; Logan and McClay, 1997, 1999). Based on the co-expression analysis, and the assumption that hox11/13b expression is restricted to blastomeres below the embryonic equator, the earliest expression of nodal, lefty, chordin, bmp2/4 and chordin, must also be in at least some Veg1 tier cells. This co-expression is lost by mesenchyme blastula (12 hpf), and earlier experiments examining the Wnt pathway show the progressive restriction of lineages along the A-P axis (Range et al., 2013). Presumably, that restriction confines expression of the ectoderm signature genes to definitive ectoderm, which includes some cells of the Veg1 lineage. Additional comparisons are shown in Fig. S5, which show that ectoderm signature genes are not co-expressed with NSM or PMCs at any time.
Matching cell diversification to developmental GRNs
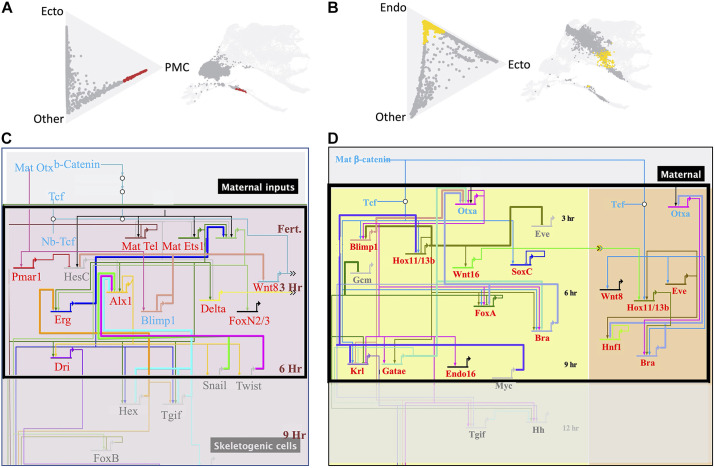
We next asked whether the Waddington-OT platform would identify populations of cells that correlate with the dGRN states established experimentally. If that were the case, cells in specification space should reflect the dGRN progression. This prediction was tested by examining collections of cells that had diverted toward their final fate by a defined probability. Fig. 6 shows two populations, predicted to become skeletogenic cells (red in Fig. 6A) or endoderm cells (yellow in Fig. 6B), both with 50% probability, or better, at 6 hpf or 9 hpf, respectively. The gray cells in each triangle of Fig. 6A,B are all ‘other’ cells that are not projected (at the >50% level) to become PMCs or endoderm, respectively, although some of those cells may have surpassed the 50% threshold for an alternative fate. The ratios of gene expressed in red (pre-PMC) versus gray cells, and in yellow (pre-endoderm) versus gray cells provided lists of the 200 genes with the highest differential expression for each lineage. This divided the transcriptome of each cell into two classes: a highly differentially expressed class and the remaining genes. Genes ubiquitously expressed, or expressed at a high level in cells of another lineage, were not expected to appear in the top 200 for a given lineage.
Fig. 6.
The predictive value of Waddington-OT plots relative to published dGRNs. (A) Future PMCs at 6 hpf. The cells in red have >50% probability of becoming PMCs, while those in gray have either not reached that level of probability or are specified towards other fates. Those cells also are shown on the UMAP plot. (B) Future endoderm seen at 9 hpf in yellow (>50% probability). All other cells are shown in gray. (C,D) dGRNs of PMCs (C) and endoderm (D) with the boxed areas showing genes expressed at 6 hpf and 9 hpf, respectively. Genes in red are highly differentially expressed in the top 200 list of genes when the genes in the red versus gray (or yellow versus gray) cells are compared. 73% of the genes in the PMC dGRN are in the highly differentially expressed list; 71% of the endoderm genes are in the top 200 gene list for endoderm. The remaining genes (27% and 29%) in blue are present in the PMC and endoderm gene lists, but not in the top 200 differentially expressed genes because either they are ubiquitously expressed or expressed in another cell type at that time. Genes in the dGRNs colored gray are not represented in the 6 hpf PMC or 9 hpf endoderm gene lists because those genes are either repressed or not yet expressed by the cells.
Fig. 6C,D show BioTapestry (Longabaugh et al., 2009) dGRN models of skeletogenic cells and endoderm cells at the same times as shown on the Triangle and UMAP plots. The highly differentially expressed genes in Fig. 6C and D are colored red, the other genes in the transcriptomes are colored blue, and genes not included in the transcriptomes at that time are colored gray. The data shows that 71% and 73% of the transcription factors (modeled at the 6 hpf and 9 hpf times, respectively) are in the highly differentially expressed class for each of the two cell types. The blue gene class includes genes known to be broadly expressed (β-catenin, TCF and Otxa), or also expressed at a high level in another tissue at that time (Blimp1, eve). The gray gene class includes genes known to be either repressed and therefore not expressed at the time points examined, or not yet activated at that time point. Thus, at the time points sampled, cells predicted by Waddington-OT to be cells destined toward a germ layer fate, matched known dGRN states at the given time points. As the differentially expressed lists include many genes with unidentified function, along with the validated dGRN genes, the dataset provides an excellent list of candidate genes for future study.
DISCUSSION
Here, we have learned that a high-quality scRNA-seq analysis detects almost all of the known transcription factors in a well-established dGRN. The 50,935 cells from 18 densely spaced time points provided an atlas of sea urchin development, and Waddington-OT provided a means of tracing lineage trajectories and divergences. It showed both rapid and protracted patterns of cell diversification. Methods were developed to follow cell fate probabilities based on the optimal transport methods. We found that 99% of the genes in the known dGRN are detected, readily quantifiable, spatiotemporally accurate and in agreement with the progression of dGRN models. The database was then explored to assess a number of properties of the lineages.
A number of examples exist in nature where a single molecular event directs a lineage divergence (Nüsslein-Volhard and Wieschaus, 1980; Driever and Nüsslein-Volhard, 1988; Kemphues et al., 1988; Guo and Kemphues, 1995; Nishida and Sawada, 2001). The skeletogenic cell divergence in the sea urchin embryo is one such example (Oliveri et al., 2002, 2003; Revilla-i-Domingo et al., 2007), and the analysis here supports that early synchronous diversification. We were curious about the other mode of divergence seen in this study, an asynchronous and an apparently delayed movement toward a final fate between the endoderm and mesoderm. Endomesoderm cells express dGRN signature genes of both endoderm and mesoderm fates for an extended time in specification space. This is in spite of the fact that the transcriptional state of these cells moves very far from the ancestral state over time, as seen by the progress across the triangles that represent specification space (Figs 3, 4), and yet the continuing expression of signature genes of two different fates suggests that some cells retain an intermediate state of specification for an extended period of time, whereas other cells express signature genes of only a single lineage, indicating that the endomesoderm divergence is asynchronous and extended. We also discovered that between fifth and ninth cleavage (early blastula to mesenchyme blastula), some cells co-express an endoderm signature gene along with cells expressing ectoderm signatures. This protracted movement of cells toward a final fate could be quite valuable for an embryo, e.g. by remaining conditional to assure correct proportional distribution of cells to tissues. Whether this is actually the case will require further study.
The published sea urchin dGRN was experimentally established in detail in Sp, with contributions from Lv and Pl, and most connections established in one of the three species were found to be identical in the others. Given that background, our analyses assessed the ability of scRNA-seq to reflect the spatiotemporal expression of the genes within the dGRN. At the time points examined in detail, 72% of the genes depicted in dGRN models were included in the top 200 highly differentially expressed genes of a lineage sampled at that time. The remaining 28% of the genes in the dGRN states sampled were also present in the single cell transcriptomes but not in the differentially expressed group. Those genes are known to be expressed ubiquitously, or expressed strongly in other lineages, thereby excluding them from the highly differentially expressed genes. We conclude that the optimal transport approach offers an efficient way to identify candidate transcription factors and effectors for a given cell lineage prior to carrying out perturbation studies. Among the many potential uses of scRNA-seq, the lineage trajectories and diversifications offer glimpses of candidate genes for participation in those cell lineage separations.
MATERIALS AND METHODS
Embryo spawning and culture
Six female urchins were spawned by injecting 1 ml 0.5 M KCl intracoelomically. Unfertilized eggs were allowed to settle and washed three times in artificial sea water (ASW). Eggs were then resuspended and fertilized by sperm from a single male in 0.03 g PABA/100 ml ASW. Following fertilization, eggs were washed three additional times in ASW to remove residual PABA. The fertilized embryos were then combined and co-cultured together at 22-23°C while gently being stirred using a motorized stir rod. Embryos were then sampled at various time points to be dissociated and fixed for scRNA-seq. At each stage, the embryos collected were very similar in stage to each other, an important consideration for temporally following development of the cells in this study.
Embryo dissociation and fixation
At each time point, approximately 106 embryos were dissociated to single cells and then fixed, using a published protocol we adapted (McClay, 1986; Juliano et al., 2014; Alles et al., 2017; Chen et al., 2018; Massri et al., 2021). The Chen protocol found both live and fixed cells to be similar in RNA integrity (RIN) so that fixation protocol was followed closely (Chen et al., 2018). For the dissociation, embryos were washed twice in calcium-free artificial seawater (CFASW) then resuspended in dissociation buffer [1.0 M glycine and 0.25 mM EDTA (pH 8.0)] at 4°C and gently rocked for 10 min. The embryos were then dissociated by gentle trituration. Following dissociation, cells were resuspended in CFASW, and fixed at a final concentration of 80% methanol in CFASW for 1 h, at 4°C. Following fixation, cells were stored at −20°C and all cell libraries were processed within 1 month of dissociation.
Rehydration of methanol-fixed single cells for library preparation and sequencing
Cells were centrifuged at 50 g, the supernatant was discarded, and 3 ml 3×SSC was added to the cell pellet, then the wash step was repeated. 20×SSC was purchased from Sigma (SKU SRE0068) and diluted with DNAse/RNAse free water to 3×. Single cell libraries for each time point were prepared using the 10x Genomics 3′ v3 gene expression kit and the 10x Chromium platform to encapsulate single cells within droplets. Library quality was verified using the Agilent 2100 Bioanalyzer. Libraries were pooled and Duke Genomics and Computational Biology Core facility sequenced samples across two NovaSeq6000 S1 flow cells with 28×8×91 bp sequencing performed. Samples were sequenced at >50,000 reads/cell to ensure read depth and coverage (Haque et al., 2017; Svensson et al., 2017; Ziegenhain et al., 2017).
Computational analysis
Data download, FastQ file generation, genome indexing, genome mapping and counting, and production of raw csv count files
Following sequencing, we used Cellranger 3.1.0 to convert Illumina-generated BCL files to fastq files using the Cellranger ‘mkfastq’ command. We then applied the ‘mkref’ command to index the most recent Lv3.0 Genome (Davidson et al., 2020). We then used the ‘count’ command to demultiplex and count reads mapping to the reference Lv genome. The ‘mat2csv’ command was used to obtain csv RNA count matrix files for each sample for further downstream analysis. In addition, we used the command ‘aggr’ on all samples to generate an automated 10x Cloupe browser that is easily accessible, and requires no coding experience to use.
Filtering and normalization
All csv RNA count matrix files were uploaded to R and a seurat object was generated for each sample. All seurat objects were then merged to undergo uniform quality control, normalization and data exploration with all 19 samples. The merged object was then filtered to remove low-quality cells with nFeature_RNA>200, nFeature_RNA<7500 and percent.mt<5. SCTransform was then applied to the merged filtered object to perform normalization and removal of technical variation, while preserving biological variation. The data were then scaled, and variable features among the cells were found.
Dimensionality reduction, visualization and clustering
We obtained a matrix of raw gene expression counts for each sample, which was normalized using SCTransform, a regularized negative binomial regression method that stabilizes variance across samples (Hafemeister and Satija, 2019), then visualized using UMAP (Uniform Manifold Approximation and Projection, Becht et al., 2018). UMAP was applied to multi-dimensional scRNA-seq data to visualize the cells in a two-dimensional space. Finally, clustering was performed using graph-based Louvain Clustering with resolution, res=2.4, resulting in 63 clusters. The 63 clusters were annotated using dGRN genes and published in situ hybridization patterns as markers (Fig. S1C-E).
Inferring developmental trajectories with Waddington-OT
We next applied Waddington-OT to infer developmental trajectories. As input, we used the SCTransform normalized expression matrix together with expansion rates estimated from expected changes in proportion of lineages over time (Fig. S2). Cell division rates were estimated by knowledge of lineages based on the expected number of cells of each lineage at key developmental time points (Table S1). Stereotypic cell divisions were assumed to be uniform between estimates of expected number of cells. Table S1 reports the approximate number of cells of each lineage at each hour although divisions occur asynchronously after the sixth cleavage so the nearest hour was chosen for each number given. Two additional cell division rates were fitted based on two cell cycle scores using a sigmoid function to smoothly fit cell division rates between the minimum and maximum expected at that time point. Validation values were very similar between the two cell division rates fitted on the cell cycle scores and the cell division rate based on expectation alone. For simplicity, the lineage expectation-based cell division rate was used throughout the analysis. Transport maps were calculated using the cell division rates described above, the optimization parameters ε=0.05, λ1=1 and λ2=50, and a single iteration of growth rate learning.
Validating trajectories with geodesic interpolation
The transport results were tested and validated by demonstrating that we can interpolate the distribution of cells at held-out time points (Fig. S4A). For each triplet of consecutive time points (e.g. 5,6,7 or 6,7,8 etc.), we held out the data from the middle time point and attempted to reconstruct it by connecting the first to the third. We then quantified our performance by comparing to the held-out midpoint. The blue curve shows the results from optimal transport, which is lower than various null models (yellow, orange, green and purple). The null models are: ‘Random’ (orange) – we randomly connect cells to descendants; ‘Random with cell divisions’ (yellow) – we randomly connect cells to descendants, incorporating the same estimate of cell division rate as for OT; ‘First’ (green) – we use the first time point in the triplet to estimate the second element of the triplet; ‘Last’ (purple) – we use the third element of the triplet to estimate the second element.
In order to test the stability of our results, we varied parameters of optimal transport over an order of magnitude in each direction (see Fig. S4B-D). Additionally, we repeated the analysis on datasets with downsampled cells and reads as low as 10% of cells and 500 UMI per cell, respectively. Downsampling cells, we found, using Waddington-OT, outperformed null methods for all proportions and saw only a gradual increase in validation values (Fig. S4E). When downsampling reads, we found Waddington-OT outperformed null methods as low as 500 UMI (Fig. S4F).
Visualizing divergence of fates with barycentric coordinates
In order to visualize the divergence of fates, we developed a simple way to visualize fate probabilities for triples (or quadruples) of lineages in a triangular (or tetrahedral) plot. The visualization employs barycentric coordinates to represent k-dimensional probability vectors in k-1 dimensional space. We identify a corner of the triangle for each of these possible fates, and position the cells according to their relative probabilities as follows. Let a, b, c denote the vertices of the triangle in R2 and let p=(p1, p2, p3) denote the probability vector we wish to visualize. The components of p are used as coefficients in a convex combination of the vertices. In other words, the probability vector p is mapped to p1a+p2b+p3c∈R2. Note that p1+p2+p3=1, so each probability vector is mapped to a point inside the triangle.
Cells perfectly fated to obtain a single fate are positioned exactly at the corresponding vertex, while cells with indeterminate fates are positioned in the interior of the triangle. The very center of the triangle corresponds to cells that are equally likely to choose any of the three fates, and cells along an edge have zero chance of reaching the opposite vertex. Fig. 3 illustrates examples of how selected pairs of lineages diverge in Lv over time. Movie 1 shows a tetrahedron visualization of the divergence of four fates simultaneously.
Visualizing gene regulatory networks
We visualized GRNs on the triangle plots and the UMAP plots using GRN score ratios. The illustration aimed to show how intermediate cells tend to express networks from both possible fates in nearly equal proportions. The GRN score was defined as the fraction of genes from a GRN that are expressed in a cell. Each cell was given two of these scores, each corresponding to a GRN of interest. To compare the expression of one regulatory network with the other, we defined a GRN score ratio as min(GRN 1 score/GRN 2score, GRN 2 score/GRN 1 score). The ratio produces values between 0 and 1. A value of 0 means only one of the networks is expressed in the cell. Meanwhile, a value of 1 means both networks are expressed in equal proportion. Fig. 4 shows cells colored by this ratio.
Visualizing signature gene expression of pairs of genes over time
Each of the 50,935 transcriptomes was screened for expression of a signature gene to determine the percent of all cells that express that gene. Cells that co-express two signature genes were also quantified. A simple graphic tool was produced that allows any two genes to be screened in this way.
Supplementary Material
Acknowledgements
We thank members of the McClay, Wray and Schiebinger labs for their critical input and feedback. We also appreciate the help provided by Nicolas Devos (PhD), and the Duke GCB core facility. We also appreciate the assistance provided by Philip Benfey (PhD) lab members, especially postdoctoral fellows Rachel Shahan (PhD) and Isaiah Taylor (PhD) in the Biology Department. We also thank Isabelle Peter at Caltech and William Longabaugh at the Institute of Systems Biology for their input on the classic BioTapestry model of endomesoderm.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: A.J.M., G.S., D.R.M.; Methodology: A.J.M., L.G., A.A., A.B., G.S., D.R.M.; Software: L.G., A.A., A.B., G.A.W., G.S.; Validation: A.J.M., L.G., A.A., D.R.M.; Formal analysis: A.J.M., L.G., A.A., A.B., G.A.W., G.S., D.R.M.; Investigation: A.J.M., G.S., D.R.M.; Resources: D.R.M.; Data curation: A.J.M., L.G., A.A., A.B., G.A.W., G.S.; Writing - original draft: D.R.M.; Writing - review & editing: A.J.M., A.B., G.S., D.R.M.; Visualization: G.A.W., D.R.M.; Supervision: D.R.M.; Project administration: D.R.M.; Funding acquisition: D.R.M.
Funding
Support for this project was provided by National Institutes of Health (RO1 HD 14483 and PO1 HD037105 to D.R.M.; predoctoral fellowship T32 HD040372 to A.J.M.) and by the National Science Foundation (IOS-1457305 to G.A.W.; predoctoral fellowship DGE-1644868 to A.J.M.). G.S. is supported by a Career Award at the Scientific Interface from the Burroughs Wellcome Fund, by funds from the Klarman Cell Observatory, by a Natural Sciences and Engineering Research Council of Canada Discovery grant, by the New Frontiers in Research Fund and by an Exploration grant. L.G. was supported by a Natural Sciences and Engineering Research Council of Canada Universities Space Research Association grant and A.A. was supported by a Genome Science and Technology summer student scholarship. Deposited in PMC for release after 12 months.
Data availability
The DNA sequencing data have been deposited in GEO under accession number GSE184538, and the hourly transcriptomes have been deposited under accession numbers GSM5592182 to GSM5592200.
References
- Alles, J., Karaiskos, N., Praktiknjo, S. D., Grosswendt, S., Wahle, P., Ruffault, P.-L., Ayoub, S., Schreyer, L., Boltengagen, A., Birchmeier, C.et al. (2017). Cell fixation and preservation for droplet-based single-cell transcriptomics. BMC Biol. 15, 44. 10.1186/s12915-017-0383-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrikou, C., Pai, C.-Y., Su, Y.-H. and Arnone, M. I. (2015). Logics and properties of a genetic regulatory program that drives embryonic muscle development in an echinoderm. eLife 4, e07343. 10.7554/eLife.07343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angerer, L. M., Oleksyn, D. W., Logan, C. Y., McClay, D. R., Dale, L. and Angerer, R. C. (2000). A BMP pathway regulates cell fate allocation along the sea urchin animal-vegetal embryonic axis. Development 127, 1105-1114. 10.1242/dev.127.5.1105 [DOI] [PubMed] [Google Scholar]
- Becht, E., McInnes, L., Healy, J., Dutertre, C.-A., Kwok, I. W. H., Ng, L. G., Ginhoux, F. and Newell, E. W. (2018). Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 37, 38-44. 10.1038/nbt.4314 [DOI] [PubMed] [Google Scholar]
- Ben-Tabou de-Leon, S., Su, Y.-H., Lin, K.-T., Li, E. and Davidson, E. H. (2013). Gene regulatory control in the sea urchin aboral ectoderm: spatial initiation, signaling inputs, and cell fate lockdown. Dev. Biol. 374, 245-254. 10.1016/j.ydbio.2012.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradham, C. A., Oikonomou, C., Kühn, A., Core, A. B., Modell, J. W., McClay, D. R. and Poustka, A. J. (2009). Chordin is required for neural but not axial development in sea urchin embryos. Dev. Biol. 328, 221-233. 10.1016/j.ydbio.2009.01.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandman, O. and Meyer, T. (2008). Feedback loops shape cellular signals in space and time. Science 322, 390-395. 10.1126/science.1160617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron, R. A., Hough-Evans, B. R., Britten, R. J. and Davidson, E. H. (1987). Lineage and fate of each blastomere of the eight-cell sea urchin embryo. Genes Dev. 1, 75-85. 10.1101/gad.1.1.75 [DOI] [PubMed] [Google Scholar]
- Campanale, J. P., Gökirmak, T., Espinoza, J. A., Oulhen, N., Wessel, G. M. and Hamdoun, A. (2014). Migration of sea urchin primordial germ cells. Dev. Dyn. 243, 917-927. 10.1002/dvdy.24133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, J., Packer, J. S., Ramani, V., Cusanovich, D. A., Huynh, C., Daza, R., Qiu, X., Lee, C., Furlan, S. N., Steemers, F. J.et al. (2017). Comprehensive single-cell transcriptional profiling of a multicellular organism. Science 357, 661-667. 10.1126/science.aam8940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cary, G. A., McCauley, B. S., Zueva, O., Pattinato, J., Longabaugh, W. and Hinman, V. F. (2020). Systematic comparison of sea urchin and sea star developmental gene regulatory networks explains how novelty is incorporated in early development. Nat. Commun. 11, 6235. 10.1038/s41467-020-20023-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J., Cheung, F., Shi, R., Zhou, H., Lu, W. and Consortium, C. H. I. (2018). PBMC fixation and processing for Chromium single-cell RNA sequencing. J. Transl. Med. 16, 198. 10.1186/s12967-018-1578-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole, A. G., Rizzo, F., Martinez, P., Fernandez-Serra, M. and Arnone, M. I. (2009). Two ParaHox genes, SpLox and SpCdx, interact to partition the posterior endoderm in the formation of a functional gut. Development 136, 541-549. 10.1242/dev.029959 [DOI] [PubMed] [Google Scholar]
- Croce, J. C. and McClay, D. R. (2010). Dynamics of Delta/Notch signaling on endomesoderm segregation in the sea urchin embryo. Development 137, 83-91. 10.1242/dev.044149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce, J., Lhomond, G. and Gache, C. (2001). Expression pattern of Brachyury in the embryo of the sea urchin Paracentrotus lividus. Dev. Genes Evol. 211, 617-619. 10.1007/s00427-001-0200-5 [DOI] [PubMed] [Google Scholar]
- Davidson, E. H., Rast, J. P., Oliveri, P., Ransick, A., Calestani, C., Yuh, C. H., Minokawa, T., Amore, G., Hinman, V., Arenas-Mena, C.et al. (2002). A genomic regulatory network for development. Science 295, 1669-1678. 10.1126/science.1069883 [DOI] [PubMed] [Google Scholar]
- Davidson, P. L., Guo, H., Wang, L., Berrio, A., Zhang, H., Chang, Y., Soborowski, A. L., McClay, D. R., Fan, G. and Wray, G. A. (2020). Chromosomal-level genome assembly of the Sea Urchin Lytechinus variegatus substantially improves functional genomic analyses. Genome Biol. Evol. 12, 1080-1086. 10.1093/gbe/evaa101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driever, W. and Nüsslein-Volhard, C. (1988). A gradient of bicoid protein in Drosophila embryos. Cell 54, 83-93. 10.1016/0092-8674(88)90182-1 [DOI] [PubMed] [Google Scholar]
- Duboc, V., Röttinger, E., Besnardeau, L. and Lepage, T. (2004). Nodal and BMP2/4 signaling organizes the oral-aboral axis of the sea urchin embryo. Dev. Cell 6, 397-410. 10.1016/S1534-5807(04)00056-5 [DOI] [PubMed] [Google Scholar]
- Duboc, V., Röttinger, E., Lapraz, F., Besnardeau, L. and Lepage, T. (2005). Left-right asymmetry in the sea urchin embryo is regulated by nodal signaling on the right side. Dev. Cell 9, 147-158. 10.1016/j.devcel.2005.05.008 [DOI] [PubMed] [Google Scholar]
- Duboc, V., Lapraz, F., Besnardeau, L. and Lepage, T. (2008). Lefty acts as an essential modulator of Nodal activity during sea urchin oral-aboral axis formation. Dev. Biol. 320, 49-59. 10.1016/j.ydbio.2008.04.012 [DOI] [PubMed] [Google Scholar]
- Duboc, V., Lapraz, F., Saudemont, A., Bessodes, N., Mekpoh, F., Haillot, E., Quirin, M. and Lepage, T. (2010). Nodal and BMP2/4 pattern the mesoderm and endoderm during development of the sea urchin embryo. Development 137, 223-235. 10.1242/dev.042531 [DOI] [PubMed] [Google Scholar]
- Erkenbrack, E. M. and Davidson, E. H. (2015). Evolutionary rewiring of gene regulatory network linkages at divergence of the echinoid subclasses. Proc. Natl. Acad. Sci. USA 112, E4075-E4084. 10.1073/pnas.1509845112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwin, D. H. and Davidson, E. H. (2009). The evolution of hierarchical gene regulatory networks. Nat. Rev. Genet. 10, 141-148. 10.1038/nrg2499 [DOI] [PubMed] [Google Scholar]
- Ettensohn, C. A., Illies, M. R., Oliveri, P. and De Jong, D. L. (2003). Alx1, a member of the Cart1/Alx3/Alx4 subfamily of Paired-class homeodomain proteins, is an essential component of the gene network controlling skeletogenic fate specification in the sea urchin embryo. Development 130, 2917-2928. 10.1242/dev.00511 [DOI] [PubMed] [Google Scholar]
- Farrell, J. A., Wang, Y., Riesenfeld, S. J., Shekhar, K., Regev, A. and Schier, A. F. (2018). Single-cell reconstruction of developmental trajectories during zebrafish embryogenesis. Science 360, eaar3131. 10.1126/science.aar3131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fincher, C. T., Wurtzel, O., de Hoog, T., Kravarik, K. M. and Reddien, P. W. (2018). Cell type transcriptome atlas for the planarian Schmidtea mediterranea. Science 360, eaaq1736. 10.1126/science.aaq1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster, S., Teo, Y. V., Neretti, N., Oulhen, N. and Wessel, G. M. (2019). Single cell RNA-seq in the sea urchin embryo show marked cell-type specificity in the Delta/Notch pathway. Mol. Reprod. Dev. 86, 931-934. 10.1002/mrd.23181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster, S., Oulhen, N. and Wessel, G. (2020). A single cell RNA sequencing resource for early sea urchin development. Development 147, dev191528. 10.1242/dev.191528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross, J. M. and McClay, D. R. (2001). The role of Brachyury (T) during gastrulation movements in the sea urchin Lytechinus variegatus. Dev. Biol. 239, 132-147. 10.1006/dbio.2001.0426 [DOI] [PubMed] [Google Scholar]
- Guo, S. and Kemphues, K. J. (1995). par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell 81, 611-620. 10.1016/0092-8674(95)90082-9 [DOI] [PubMed] [Google Scholar]
- Hafemeister, C. and Satija, R. (2019). Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 20, 296. 10.1186/s13059-019-1874-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, X., Wang, R., Zhou, Y., Fei, L., Sun, H., Lai, S., Saadatpour, A., Zhou, Z., Chen, H., Ye, F.et al. (2018). Mapping the mouse cell atlas by microwell-seq. Cell 172, 1091-1107.e1017. 10.1016/j.cell.2018.02.001 [DOI] [PubMed] [Google Scholar]
- Haque, A., Engel, J., Teichmann, S. A. and Lonnberg, T. (2017). A practical guide to single-cell RNA-sequencing for biomedical research and clinical applications. Genome Med. 9, 75. 10.1186/s13073-017-0467-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry, J. J., Amemiya, S., Wray, G. A. and Raff, R. A. (1989). Early inductive interactions are involved in restricting cell fates of mesomeres in sea urchin embryos. Dev. Biol. 136, 140-153. 10.1016/0012-1606(89)90137-1 [DOI] [PubMed] [Google Scholar]
- Hinman, V. F. and Davidson, E. H. (2007). Evolutionary plasticity of developmental gene regulatory network architecture. Proc. Natl. Acad. Sci. USA 104, 19404-19409. 10.1073/pnas.0709994104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinman, V. F., Nguyen, A. T., Cameron, R. A. and Davidson, E. H. (2003). Developmental gene regulatory network architecture across 500 million years of echinoderm evolution. Proc. Natl. Acad. Sci. USA 100, 13356-13361. 10.1073/pnas.2235868100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israel, J. W., Martik, M. L., Byrne, M., Raff, E. C., Raff, R. A., McClay, D. R. and Wray, G. A. (2016). Comparative developmental transcriptomics reveals rewiring of a highly conserved gene regulatory network during a major life history switch in the sea urchin genus heliocidaris. PLoS Biol. 14, e1002391. 10.1371/journal.pbio.1002391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juliano, C., Swartz, S. Z. and Wessel, G. (2014). Isolating specific embryonic cells of the sea urchin by FACS. Methods Mol. Biol. 1128, 187-196. 10.1007/978-1-62703-974-1_12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaiskos, N., Wahle, P., Alles, J., Boltengagen, A., Ayoub, S., Kipar, C., Kocks, C., Rajewsky, N. and Zinzen, R. P. (2017). The Drosophila embryo at single-cell transcriptome resolution. Science 358, 194-199. 10.1126/science.aan3235 [DOI] [PubMed] [Google Scholar]
- Kemphues, K. J., Priess, J. R., Morton, D. G. and Cheng, N. S. (1988). Identification of genes required for cytoplasmic localization in early C. elegans embryos. Cell 52, 311-320. 10.1016/S0092-8674(88)80024-2 [DOI] [PubMed] [Google Scholar]
- Li, E., Materna, S. C. and Davidson, E. H. (2013). New regulatory circuit controlling spatial and temporal gene expression in the sea urchin embryo oral ectoderm GRN. Dev. Biol. 382, 268-279. 10.1016/j.ydbio.2013.07.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan, C. Y. and McClay, D. R. (1997). The allocation of early blastomeres to the ectoderm and endoderm is variable in the sea urchin embryo. Development 124, 2213-2223. 10.1242/dev.124.11.2213 [DOI] [PubMed] [Google Scholar]
- Logan, C. Y. and McClay, D. R. (1999). Lineages that give rise to endoderm and Mesoderm in the sea urchin embryo. In: Cell Lineage and Fate Determination (ed. Moody S.), pp. 41-57. San Diego: Academic Press. [Google Scholar]
- Logan, C. Y., Miller, J. R., Ferkowicz, M. J. and McClay, D. R. (1999). Nuclear beta-catenin is required to specify vegetal cell fates in the sea urchin embryo. Development 126, 345-357. 10.1242/dev.126.2.345 [DOI] [PubMed] [Google Scholar]
- Longabaugh, W. J. R., Davidson, E. H. and Bolouri, H. (2009). Visualization, documentation, analysis, and communication of large-scale gene regulatory networks. Biochim. Biophys. Acta 1789, 363-374. 10.1016/j.bbagrm.2008.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, Y.-J. and Su, Y.-H. (2012). Opposing nodal and BMP signals regulate left-right asymmetry in the sea urchin larva. PLoS Biol. 10, e1001402. 10.1371/journal.pbio.1001402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik, A., Gildor, T., Sher, N., Layous, M. and Ben-Tabou de-Leon, S. (2017). Parallel embryonic transcriptional programs evolve under distinct constraints and may enable morphological conservation amidst adaptation. Dev. Biol. 430, 202-213. 10.1016/j.ydbio.2017.07.019 [DOI] [PubMed] [Google Scholar]
- Martik, M. L. and McClay, D. R. (2015). Deployment of a retinal determination gene network drives directed cell migration in the sea urchin embryo. eLife 4, e08827. 10.7554/eLife.08827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martik, M. L. and McClay, D. R. (2017). New insights from a high-resolution look at gastrulation in the sea urchin, Lytechinus variegatus. Mech. Dev. 148, 3-10. 10.1016/j.mod.2017.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massri, A. J., Schiebinger, G. R., Berrio, A., Wang, L., Wray, G. A. and McClay, D. R. (2021). Methodologies for following EMT in vivo at single cell resolution. Methods Mol. Biol. 2179, 303-314. 10.1007/978-1-0716-0779-4_23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Materna, S. C. and Davidson, E. H. (2012). A comprehensive analysis of Delta signaling in pre-gastrular sea urchin embryos. Dev. Biol. 364, 77-87. 10.1016/j.ydbio.2012.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClay, D. R. (1986). Embryo dissociation, cell isolation, and cell reassociation. Meth. Cell Biol. 27, 309-323. 10.1016/S0091-679X(08)60355-6 [DOI] [PubMed] [Google Scholar]
- McClay, D. R. (2011). Evolutionary crossroads in developmental biology: sea urchins. Development 138, 2639-2648. 10.1242/dev.048967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClay, D. R., Warner, J., Martik, M., Miranda, E. and Slota, L. (2020). Gastrulation in the sea urchin. Curr. Top. Dev. Biol. 136, 195-218. 10.1016/bs.ctdb.2019.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre, D. C., Seay, N. W., Croce, J. C. and McClay, D. R. (2013). Short-range Wnt5 signaling initiates specification of sea urchin posterior ectoderm. Development 140, 4881-4889. 10.1242/dev.095844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida, H. and Sawada, K. (2001). macho-1 encodes a localized mRNA in ascidian eggs that specifies muscle fate during embryogenesis. Nature 409, 724-729. 10.1038/35055568 [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard, C. and Wieschaus, E. (1980). Mutations affecting segment number and polarity in Drosophila. Nature 287, 795-801. 10.1038/287795a0 [DOI] [PubMed] [Google Scholar]
- Oliveri, P., Carrick, D. M. and Davidson, E. H. (2002). A regulatory gene network that directs micromere specification in the sea urchin embryo. Dev. Biol. 246, 209-228. 10.1006/dbio.2002.0627 [DOI] [PubMed] [Google Scholar]
- Oliveri, P., Davidson, E. H. and McClay, D. R. (2003). Activation of pmar1 controls specification of micromeres in the sea urchin embryo. Dev. Biol. 258, 32-43. 10.1016/S0012-1606(03)00108-8 [DOI] [PubMed] [Google Scholar]
- Oliveri, P., Walton, K. D., Davidson, E. H. and McClay, D. R. (2006). Repression of mesodermal fate by foxa, a key endoderm regulator of the sea urchin embryo. Development 133, 4173-4181. 10.1242/dev.02577 [DOI] [PubMed] [Google Scholar]
- Oliveri, P., Tu, Q. and Davidson, E. H. (2008). Global regulatory logic for specification of an embryonic cell lineage. Proc. Natl. Acad. Sci. USA 105, 5955-5962. 10.1073/pnas.0711220105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perillo, M., Oulhen, N., Foster, S., Spurrell, M., Calestani, C. and Wessel, G. (2020). Regulation of dynamic pigment cell states at single-cell resolution. eLife 9, e60388. 10.7554/eLife.60388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter, I. S. and Davidson, E. H. (2010). The endoderm gene regulatory network in sea urchin embryos up to mid-blastula stage. Dev. Biol. 340, 188-199. 10.1016/j.ydbio.2009.10.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter, I. S. and Davidson, E. H. (2011). A gene regulatory network controlling the embryonic specification of endoderm. Nature 474, 635-639. 10.1038/nature10100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter, I. S. and Davidson, E. H. (2015). Genomic Control Process. Development and Evolution. San Diego: Academic Press. [Google Scholar]
- Plass, M., Solana, J., Wolf, F. A., Ayoub, S., Misios, A., Glazar, P., Obermayer, B., Theis, F. J., Kocks, C. and Rajewsky, N. (2018). Cell type atlas and lineage tree of a whole complex animal by single-cell transcriptomics. Science 360, eaaq1723. 10.1126/science.aaq1723 [DOI] [PubMed] [Google Scholar]
- Rafiq, K., Shashikant, T., McManus, C. J. and Ettensohn, C. A. (2014). Genome-wide analysis of the skeletogenic gene regulatory network of sea urchins. Development 141, 950-961. 10.1242/dev.105585 [DOI] [PubMed] [Google Scholar]
- Range, R. and Lepage, T. (2011). Maternal Oct1/2 is required for Nodal and Vg1/Univin expression during dorsal-ventral axis specification in the sea urchin embryo. Dev. Biol. 357, 440-449. 10.1016/j.ydbio.2011.07.005 [DOI] [PubMed] [Google Scholar]
- Range, R. C., Angerer, R. C. and Angerer, L. M. (2013). Integration of canonical and noncanonical Wnt signaling pathways patterns the neuroectoderm along the anterior-posterior axis of sea urchin embryos. PLoS Biol. 11, e1001467. 10.1371/journal.pbio.1001467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransick, A., Rast, J. P., Minokawa, T., Calestani, C. and Davidson, E. H. (2002). New early zygotic regulators expressed in endomesoderm of sea urchin embryos discovered by differential array hybridization. Dev. Biol. 246, 132-147. 10.1006/dbio.2002.0607 [DOI] [PubMed] [Google Scholar]
- Revilla-i-Domingo, R., Oliveri, P. and Davidson, E. H. (2007). A missing link in the sea urchin embryo gene regulatory network: hesC and the double-negative specification of micromeres. Proc. Natl. Acad. Sci. USA 104, 12383-12388. 10.1073/pnas.0705324104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo, F., Fernandez-Serra, M., Squarzoni, P., Archimandritis, A. and Arnone, M. I. (2006). Identification and developmental expression of the ets gene family in the sea urchin (Strongylocentrotus purpuratus). Dev. Biol. 300, 35-48. 10.1016/j.ydbio.2006.08.012 [DOI] [PubMed] [Google Scholar]
- Röttinger, E., Saudemont, A., Duboc, V., Besnardeau, L., McClay, D. and Lepage, T. (2008). FGF signals guide migration of mesenchymal cells, control skeletal morphogenesis [corrected] and regulate gastrulation during sea urchin development. Development 135, 353-365. 10.1242/dev.014282 [DOI] [PubMed] [Google Scholar]
- Saudemont, A., Haillot, E., Mekpoh, F., Bessodes, N., Quirin, M., Lapraz, F., Duboc, V., Röttinger, E., Range, R., Oisel, A.et al. (2010). Ancestral regulatory circuits governing ectoderm patterning downstream of Nodal and BMP2/4 revealed by gene regulatory network analysis in an echinoderm. PLoS Genet. 6, e1001259. 10.1371/journal.pgen.1001259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders, L. R. and McClay, D. R. (2014). Sub-circuits of a gene regulatory network control a developmental epithelial-mesenchymal transition. Development 141, 1503-1513. 10.1242/dev.101436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiebinger, G., Shu, J., Tabaka, M., Cleary, B., Subramanian, V., Solomon, A., Gould, J., Liu, S., Lin, S., Berube, P.et al. (2019). Optimal-transport analysis of single-cell gene expression identifies developmental trajectories in reprogramming. Cell 176, 1517. 10.1016/j.cell.2019.02.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi, A. J., Wikramanayake, R. M., Angerer, R. C., Range, R. C. and Angerer, L. M. (2012). Sequential signaling crosstalk regulates endomesoderm segregation in sea urchin embryos. Science 335, 590-593. 10.1126/science.1212867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood, D. R. and McClay, D. R. (1999). LvNotch signaling mediates secondary mesenchyme specification in the sea urchin embryo. Development 126, 1703-1713. 10.1242/dev.126.8.1703 [DOI] [PubMed] [Google Scholar]
- Slota, L. A. and McClay, D. R. (2018). Identification of neural transcription factors required for the differentiation of three neuronal subtypes in the sea urchin embryo. Dev. Biol. 435, 138-149. 10.1016/j.ydbio.2017.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenzel, P., Angerer, L. M., Smith, B. J., Angerer, R. C. and Vale, W. W. (1994). The univin gene encodes a member of the transforming growth factor-beta superfamily with restricted expression in the sea urchin embryo. Dev. Biol. 166, 149-158. 10.1006/dbio.1994.1303 [DOI] [PubMed] [Google Scholar]
- Svensson, V., Natarajan, K. N., Ly, L.-H., Miragaia, R. J., Labalette, C., Macaulay, I. C., Cvejic, A. and Teichmann, S. A. (2017). Power analysis of single-cell RNA-sequencing experiments. Nat. Methods 14, 381-387. 10.1038/nmeth.4220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tintori, S. C., Osborne Nishimura, E., Golden, P., Lieb, J. D. and Goldstein, B. (2016). A transcriptional lineage of the early C. elegans embryo. Dev. Cell 38, 430-444. 10.1016/j.devcel.2016.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voronina, E., Lopez, M., Juliano, C. E., Gustafson, E., Song, J. L., Extavour, C., George, S., Oliveri, P., McClay, D. and Wessel, G. (2008). Vasa protein expression is restricted to the small micromeres of the sea urchin, but is inducible in other lineages early in development. Dev. Biol. 314, 276-286. 10.1016/j.ydbio.2007.11.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, D. E., Weinreb, C., Collins, Z. M., Briggs, J. A., Megason, S. G. and Klein, A. M. (2018). Single-cell mapping of gene expression landscapes and lineage in the zebrafish embryo. Science 360, 981-987. 10.1126/science.aar4362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeger-Lotem, E., Sattath, S., Kashtan, N., Itzkovitz, S., Milo, R., Pinter, R. Y., Alon, U. and Margalit, H. (2004). Network motifs in integrated cellular networks of transcription-regulation and protein-protein interaction. Proc. Natl. Acad. Sci. USA 101, 5934-5939. 10.1073/pnas.0306752101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegenhain, C., Vieth, B., Parekh, S., Reinius, B., Guillaumet-Adkins, A., Smets, M., Leonhardt, H., Heyn, H., Hellmann, I. and Enard, W. (2017). Comparative analysis of single-cell RNA sequencing methods. Mol. Cell 65, 631-643.e634. 10.1016/j.molcel.2017.01.023 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.