Abstract
Efficient penetration of the mucus layer is needed for respiratory viruses to avoid mucociliary clearance prior to infection. Many respiratory viruses bind to glycans on the heavily glycosylated mucins that give mucus its gel-like characteristics. Influenza viruses, some paramyxoviruses, and coronaviruses avoid becoming trapped in the mucus by releasing themselves by means of their envelope-embedded enzymes that destroy glycan receptors. For efficient infection, receptor binding and destruction need to be in balance with the host receptor repertoire. Establishment in a novel host species requires resetting of the balance to adapt to the different glycan repertoire encountered. Growing understanding of species-specific mucosal glycosylation patterns and the dynamic interaction with respiratory viruses identifies the mucus layer as a major host-range determinant and barrier for zoonotic transfer.
Respiratory mucus and viruses
All body surfaces exposed to the outside world are protected against environmental hazards by a layer of closely connected epithelial cells. The multiple layers of dead cells of the skin can be passed by viruses only via animal bites, wounds or needles. However, 95% of epithelial surfaces are mucosal surfaces consisting of a single layer of epithelial cells mostly covered by a gel-like layer of mucus (see Glossary). They form semipermeable barriers enabling nutrient absorption and waste secretion and provide a major route of entry and release for pathogens, including viruses. The mucus layer contains a range of mucin glycoproteins as the main components; they are secreted by specialized secretory epithelial cells. Mucus provides protection against dehydration, abrasion, toxins, and pathogens, and is a reservoir for antimicrobial molecules [1]. In the respiratory tract, heavily glycosylated and sialylated mucins form a barrier against glycan-receptor-binding viruses, which are at risk of being expelled by mucociliary clearance (MCC) after immobilization in the mucus layer [2]. This is counteracted by respiratory viruses – such as influenza viruses, some coronaviruses, and paramyxoviruses – which release themselves from decoy receptors by the action of their associated glycan-receptor-destroying enzymes (RDEs) [3].
Host species differ in the genetic makeup and expression of their glycan-modifying enzymes, resulting in a species-specific sialoglycan repertoire on functional cell surface receptors as well as soluble decoy receptors in the mucus [4., 5., 6.]. Glycan-binding viruses usually have a restricted host range invoked by optimal adaptation to the glycan repertoire of a specific host species amongst other factors. Host-specific evolution – leading to well-balanced binding to, and cleavage from, epithelial cell surface receptors – has been extensively documented for influenza viruses [7., 8., 9.] and others [10,11]. However, such a balance is also required to prevent immobilization on mucus decoy receptors leading to virus expulsion before reaching the epithelial cells. Thus, animal viruses that have successfully crossed the host species barrier to become human viruses, will not only have adjusted the specificity and/or affinity of their glycan-receptor-binding proteins (RBPs) and RDEs to match the sialoglycan repertoire of the epithelial cells but also to the rather different glycan repertoire of the mucus layer. The mucus layer can therefore be regarded as a major host-range determinant. In this opinion paper, we discuss our current understanding of the zoonotic barrier function of the mucus layer for glycan-binding respiratory viruses and indicate the major gaps in our knowledge. Table 1 summarizes the properties of respiratory viruses that bind to glycans, including all viruses that are discussed in more detail below.
Table 1.
Sialoglycan-binding respiratory viruses
| Family | Genus | Sub-genus | Virusa | Diameter (nm) | Receptorb | RBPc | RDEc | Host species | Zoonotic (Z) Enzootic (E) |
Refs | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Naked | Parvo | Dependoparvo | AAV1 | 25 | 2,3/2,6 SIA | Capsid | No | Human | [64] | ||
| AAV6 | 2,3/2,6 SIA | Capsid | No | Human | [64] | ||||||
| AAV5 | 2,3 SIA N-linked | Capsid | No | Human | [65] | ||||||
| AAV4 | 2,6 SIA O-linked | Capsid | No | Human | [65] | ||||||
| Picorna | Entero | EV-D68 | 30 | 2,3SIA/2,6 SIA | Capsid | No | Human | [66] | |||
| Adeno | Mastadeno | BAd3 | 90–100 | 2,3/2,6 SIA | Capsid | No | Bovine | [67] | |||
| Calici | Vesi | FCV | 40 | 2,6 SIA N-linked | Capsid | No | Feline | [68] | |||
| Enveloped | Paramyxo | Orthorubula | HPIV2 HPIV4 | 150–250 | 2,3 SIA | HN | HN | Human | [67] | ||
| MuV BatMuV | 100–600 | 2,3 SIA | HN | HN | Human Noctilonine |
[67] | |||||
| Orthoavula | NDV | 150–400 | 2,3 > 2,6 SIA | HN | HN | Avian | [69] | ||||
| Respiro | HPIV1 | 150–250 | 2,3 SIA | HN | HN | Human | [67] | ||||
| HPIV3 | 2,3 > 2,6 SIA | HN | HN | Human | [70] | ||||||
| BPIV-3 | 2,3 SIA | HN | HN | Bovine | E | [67] | |||||
| PPIV-1 | N.D. | HN | HN | Porcine | |||||||
| CPIV-3 | N.D. | HN | HN | Caprine | |||||||
| Sendai | 2,3/2,8 SIA | HN | HN | Murine | [67] | ||||||
| Corona | Beta | Embeco | HCoV-HKU1 | 80–120 | 9-OAc SIA | Spike HE | HE | Human | Z | [71] | |
| HCoV-OC43 | 9-OAc SIA | Spike HE | HE | Human | Z | [71] | |||||
| BCoV | 9-OAc/7,9-di-OAc SIA | Spike HE | HE | Bovine | [71] | ||||||
| PHEV | 9-OAc SIA | Spike HE | HE | Porcine | [71] | ||||||
| Sarbeco | SARS-CoV-2 | 2,3/2,6 SIA | Spike | No | Human | Z | [72] | ||||
| Merbeco | MERS-CoV | 2,3 SIA > 2,6 SIA | Spike | No | Human | Z | [73] | ||||
| Gamma | IBrV | 2,3/2,6 SIA | Spike | No | Avian | [74] | |||||
| Orthomyxo | Alphainfluenza | IAV | 90–110 | 2,3/2,6 SIA | HA | NA | Human Avian Porcine Canine Equine Otarine |
Z/E | [67] | ||
| Betainfluenza | IBV | 2,6 SIA > 2,3 SIA | HA | NA | Human | [67] | |||||
| Gammainfluenza | ICV | 4-OAc/9-OAc SIA | HEF | HEF | Human Porcine Canine |
Z/E | [67] | ||||
| Deltainfluenza | IDV | 4-OAc/9-OAc SIA | HEF | HEF | Bovine Ovine Caprine |
E | [67] |
Abbreviations: AAV, adeno-associated virus; EV, enterovirus; BAd3, bovine adenovirus 3; FCV, feline calicivirus; HPIV, human parainfluenzavirus; MuV, mumps virus; NDV, Newcastle disease virus; B/P/CPIV3, bovine/porcine/caprine parainfluenzavirus 3; H/BCoV, human/bovine coronavirus; PHEV, porcine hemagglutinating encephalomyelitis virus; IBrV, infectious bronchitis virus; IAV/IBV/ICV/IDV, influenza A/B/C/D virus. This table is not a complete list of respiratory viruses binding to sialoglycans. For more glycan-binding viruses, see https://sugarbind.expasy.org.
Abbreviations: SIA, sialic acid; 2,3 SIA, α2,3-linked SIA; 2,6 SIA, α2,6-linked SIA; 2,8 SIA, α2,8-linked SIA; N-linked and O-linked refers to SIAs attached to N-linked or O-linked glycan chains; N.D., not determined; 9-OAc SIA, 9-O-acetylated SIA; 7,9-di-OAc SIA, 7,9-di-O-acetylated SIA; 4-OAc SIA, 4-O-acetylated SIA.
Abbreviations: RBP, receptor-binding protein; RDE, receptor-destroying enzyme; HN, hemagglutinin-neuraminidase; HE, hemagglutinin-esterase; HA, hemagglutinin; HEF, hemagglutinin-esterase fusion protein; NA, neuraminidase.
The respiratory mucus layer and its protein constituents
Respiratory mucus, or airway surface liquid (ASL) is comprised of two distinct layers – the viscous gel-like mucus layer that is situated on top of the periciliary layer (PCL) – described by a 'gel-on-brush' model [12] (Figure 1 ). Mucins form the main constituents of both layers. They are large macromolecules (200–1500 kDa in their monomeric state) that assemble into networks of up to 200 MDa in size [2] that are conserved throughout the vertebrate kingdom. The protein backbone of mucins contains unique tandem repeats rich in proline, threonine, and serine – referred to as PTS domains – which are extensively modified with a heterogeneous array of O-glycans contributing to 50–80% of their molecular weight. Twenty-two mucins have been identified in humans, 16 of which have been found in the respiratory tract.
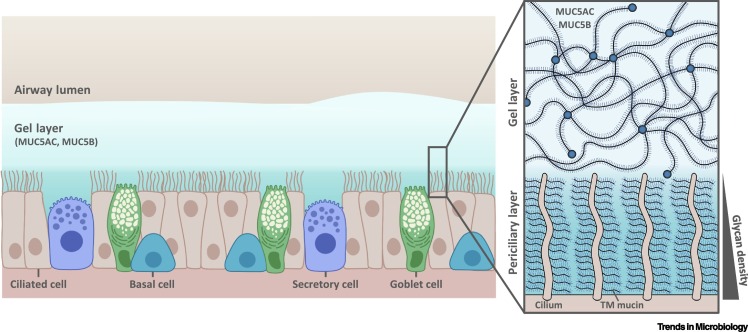
Figure 1.
Schematic diagram of the respiratory mucus layer.
The airway surface liquid (ASL) overlays the respiratory epithelia and consists of two layers: the gel layer and the periciliary layer (PCL). The gel layer contains soluble mucins MUC5AC and MUC5B, secreted primarily from goblet cells and mucous cells within submucosal glands (not shown), respectively. The soluble mucins are major contributors to the viscosity and gel-like properties of this layer which enables the impediment of airway pollutants to be cleared by mucociliary clearance. Compared to the gel layer, the PCL – the height of which is approximately that of outstretched cilia – is free of soluble mucins and is therefore less viscous, which provides favorable conditions for ciliary beating. Cilia present on the epithelial surface are rich in transmembrane (TM) mucins (MUC1, MUC4, MUC16, and MUC20) which create a glycan meshwork that increases in density closer to the cell surface, aiding the exclusion of molecules and invading pathogens. This figure represents a general schematic representation of the ciliated respiratory epithelium, thus the term 'secretory cell' may refer to different cell types including club or dense-core granulated cells of the airway epithelium [76].
The superficial gel layer (~2–10 μm) consists mainly of the large (~5800 amino acids) gel-forming mucins MUC5AC and MUC5B that are secreted from goblet cells and mucous cells of the submucosal glands, respectively (Figure 1). They multimerize into extended polymers (0.5–10 μm) by disulfide bonds between the N- and C-terminal cysteine-rich von Willebrand factor (vWF) domains (Figure 1, Figure 2 ), and along with mucin-interacting globular proteins form a dynamic, porous molecular network with pores ranging from 100 to 500 nm that impede the penetration of invading pathogens through the layer to the airway epithelia, thus facilitating removal by mucociliary clearance [13]. The underlaying PCL is as deep as cilia are in height (~7 μm) and is reduced in viscosity, providing a favorable environment for ciliary beating and cell surface lubrication. Membrane-spanning mucins (MUC1, MUC4, MUC16, and MUC20) and mucopolysaccharides present on cilia and airway epithelium form a meshwork that increases in density closer to the epithelial surface, enabling the exclusion of molecules and nanoparticles through this ASL layer [12]. Mucus covers the length of the respiratory tract, excluding the alveoli, with a thicker layer present in the upper respiratory tract that decreases moving down to the bronchi and bronchioles of the lower respiratory tract [14]. The mucus layer can vary in composition and properties due to changes in physiology and health, including virus and bacterial infections, which may affect its barrier function.
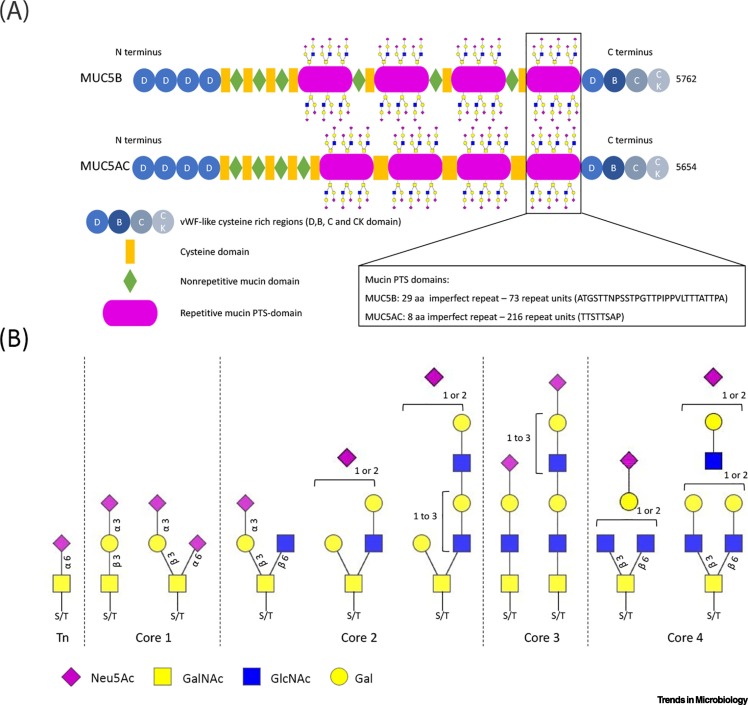
Figure 2.
Mucins and their glycosylation.
(A) The domain structure of the soluble mucins MUC5B (5762 amino acids) and MUC5AC (5654 amino acids). The N- and C-terminal von Willebrand factor (vWF)-like regions and cysteine-rich domains are highly conserved between MUC5B and MUC5AC as well as between species. The four central proline/threonine/serine-rich (PTS) regions consist of imperfect repeats (aa; amino acids). PTS repeats are densely decorated with O-linked glycans and their low sequence conservation between species will result in spatial differences in glycan presentation that could potentially affect the binding of a specific virus. (B) Diversity of sialylated O-linked glycan structures present on high-molecular-weight human mucins. Structures were interpreted from the glycan compositions reported in the most extensive analysis of glycans on mucus to date [39]. Note that di-sialylated bi-antennary structures with multiple LacNAc repeats are present. Such structures on mucins are likely to have differential effects on the binding of viruses as has been reported for N-linked glycans [77]. Sulfation of the sialoglycans shown here was also abundant [39] but is not indicated. Abbreviations: Gal, galactose; GalNAc, N-acetylgalactosamine; GlcNAc, N-acetylglucosamine; Neu5Ac, N-acetyl-neuraminic acid.
Although mucins serve as the major functional and structural component of mucus, respiratory epithelial cells secrete a myriad of nonmucin proteins, including enzymes, antimicrobial peptides, protease inhibitors and oxidants which accumulate and play important functional roles in the mucus layer [15]. Many of these mucus constituents have been identified from human bronchoalveolar lavage fluid, fluid from primary human airway epithelial cultures, fluid from the serous glands of the Calu-3 cell line, and nasal lavage fluid using proteomics mass spectrometry, chromatography, 2D PAGE and ELISA approaches [16,17]. Several of these proteins have been found to have antiviral effects – such as human defensin-β [18], lactoferrin [19], palate lung and nasal epithelium clone (PLUNC) [20], cathelicidins (specifically LL-37) [21], pulmonary surfactant proteins A and D [22], deleted in malignant brain tumors 1 (DMBT1) [23], and galectins [24] – although their ability to inhibit viruses when present at physiological levels within the mucus layer in vivo is unclear. In addition, mucus appears rich in exosomes, which may also interfere with virus infection of the epithelial cells [25,26]. Studies on the protein constituents of mucus of other animal species are limited but available data suggest a similar composition as found in human mucus [16,17,27., 28., 29.]. It is not clear whether putative differences between species in protein composition and/or antiviral activities of proteins are important for the mucus layer functioning as a host tropism barrier.
Penetration of mucus by nanoparticles
Permeability for nanoparticles and viruses is restricted by various means, including size exclusion, hydrogen bonding, electrostatic and hydrophobic interactions, and more specific binding interactions with (sialylated) glycans [30]. For virus particles that do not utilize glycans as receptors, the ability to penetrate mucus is probably governed by physicochemical properties of mucus such as pore size, viscoelasticity, pH, ionic strength, and charge. The mesh size of respiratory mucus is highly heterogeneous (~100–500 nm); thus, for virus particles larger than the pore size, mucus acts as a biological sieve, impeding the movement of particles regardless of surface chemistry [13]. In the PCL, glycans present on cilia create an even denser network than the overlaying gel layer. Particles greater than 40 nm cannot enter the PCL [12]. Smaller particles penetrate deeper with decreasing particle size, indicating an increase in glycan density closer to the cell surface. Particles with a net neutral charge have much greater mobility through the mucus layer compared to charged nanoparticles. A decrease in surface charge by PEGylation resulted in an 11-fold increase in mobility rate of the nonglycan-binding porcine respiratory virus [31]. While it seems clear that the mucus layer forms a formidable barrier against penetration of virus particles [13], the physiochemical properties of mucus per se may not form a host species-specific barrier.
Species-specific differences in respiratory mucus glycosylation
Species-specific differences in the sialoglycan repertoire probably contribute to mucus being a host-range determinant. The glycan repertoire of the respiratory tract mucus gel layer is present on secretory gel-forming mucins MUC5AC and MUC5B, secretory non-gel-forming mucins (MUC7, MUC8, MUC19), numerous globular proteins within the MUC5AC/5B meshwork, and secreted vesicles.
The serine and threonine residues within the repeated PTS domains of mucins, including the gel-forming MUC5AC and MUC5B, are extensively modified by a highly heterogeneous collection of O-glycans that contribute over 50% to the molecular weight (Figure 2). Also, N-linked glycosylation sites are predicted in the N- and C-terminal domains of MUC5AC (17 sites) and MUC5B (32 sites) but, only for MUC5B, at least 10 N-glycans were shown to be present [32]. Mucin glycosylation patterns are determined by cell type, tissue and species-dependent expression patterns of glycosyl transferases in the goblet cells and submucosal glands from which MUC5AC and MUC5B are secreted, respectively. The large array of globular proteins and vesicles present in mucus are derived from the many cell types in the respiratory epithelial layer, cells of the immune system present in the respiratory lumen or from cell leakages in the underlying tissue and vasculature. Their glycosylation pattern, as for mucins, depends on cell origin and is expected to be highly complex and variable between species [33., 34., 35., 36., 37.] or disease state [2,38,39] and to a large extent representing the glycome as determined for respiratory epithelium of several species. In contrast to mucins, a large amount of N-linked glycans is expected to be present.
Variations in glycan distribution in the respiratory tract between species and different locations/tissues/cells within a host have been revealed by lectin staining [40] but this technique is, at most, semiquantitative and misses a lot of detail on glycan structure. Glycan structures have been quantitatively determined by mass spectrometry glycomics analysis for several species, mostly for N-glycans. Species-specific modifications (Table 2 ) likely contribute to the potential of respiratory mucus as a host tropism barrier. Most of these modifications were present on glycoproteins from epithelial tissues, and their presence on glycoproteins within the mucus layer is inferred, but has not yet been convincingly demonstrated. Comprehensive studies describing the glycan composition of mucus are limited and mostly confined to humans. The potential complexity of O-linked glycans is extensive [41], and over 250 different O-linked glycans have been detected on mucins purified from human respiratory mucus [39]. About half of the total O-linked glycans were sialylated and/or sulfated. The masses of sialylated O-glycans corresponded to (extended) core 1 to core 4 structures (Figure 2B) [39]. Sialic acid (SIA) linkage type, number of LacNAc repeats, sulfation, fucosylation and other modifications of terminal and subterminal residues are known to affect binding and/or cleavage by respiratory viruses. These terminal glycotopes vary between species and tissues [35] and thereby affect the relative abundance and local density of each other (Table 2). Table 2 displays typical terminal structures identified on O- and N-linked glycans from mucus and respiratory tissues. SIA linkage type (α2,3 or α2,6) is the hallmark structural element governing binding affinity of influenza A viruses (α2,3SIA binding by avian viruses; α2,6SIA binding by human and swine viruses). Whereas mucus is assumed to be abundant in α2,3SIAs because of its high content of α2,3SIA-rich O-linked glycans [35,38], a detailed quantification of the α2,3/α2,6SIA ratio including the contribution of N-linked glycans is lacking. Sulfation of O-linked glycans (especially GlcNAc-6-Sul, Gal-6-Sul, and Gal-3-Sul) on human mucins accounts for more than 50% of charged glycans, being even more abundant than sialylation [39]. Sulfation has been shown to reduce or enhance the binding of specific viruses [42] and affects SIA density as addition of 3- and 6-O-linked sulfate is in competition with α2,3 and α2,6 sialylation at terminal Gal residues. Fucosylation is another abundant modification affecting specific virus binding affinity [43] and terminal SLeX and/or SLeA terminal glycotopes (containing fucose) are frequently found on O-linked glycans of human mucus and N-glycans of respiratory epithelium. Glycotopes, like the Sda antigen [blocking influenza A virus (IAV) binding to α2,3SIA [35]] and α2,6SIALacdiNAc, are abundant in ferrets whereas the terminal α-Gal epitope (Galα1,3Galβ1,4GlcNAc) that cannot be sialylated is abundant in swine, ferrets, and avian species. Other glycotopes concern modifications of the SIA itself such as Neu5Gc and O-acetylation at the C-4, -7, -8, and/or -9 positions. O-acetylation of SIA is known to vary considerably between species [6,44] and to affect virus binding to sialoglycans [4,10,11]. Viruses may also display preference for Neu5Ac or Neu5Gc [45]. Loss of function of CMAH, responsible for the production of Neu5Gc, occurred at least eight times in mammals – in humans and mustelids among others (but not in swine) – while also an entire avian lineage lacks Neu5Gc [46., 47., 48.]. The absence or presence of glycotopes mentioned above, including SIA modifications, are likely to affect the zoonotic potential of SIA-binding viruses.
Table 2.
Terminal glycotopes in the respiratory tract
| Glycotopea | Structurea | Mucus |
Respiratory tract |
Absent/N.D./traceb | |
|---|---|---|---|---|---|
| O-linked | O-linked | N-linked | |||
| sLeX | 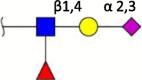 |
Human [39] | Human [38] Avian [37] |
Swine (N.D.) Ferret (N.D.) Horse (N.D.) Cow (N.D.) |
|
| Sda-epitope | 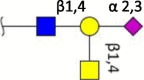 |
Ferret [35] | Ferret [35] | Human (trace) Swine (N.D.) Ferret (N.D.) Avian (N.D.) |
|
| NeuGc |  |
Cow [4] | Swine [34,36] | Human Avian Ferret (N.D.) |
|
| αGal-epitope |  |
Swine [34] | Swine [33,34,36] Ferret [35] Avian [75] |
Human | |
| Neu5Ac-O-Ac (4/7/8/9-O) |
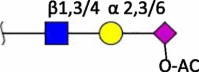 |
Human (9-O) [4] Horse (4-O/7-O/8-O/9-O/7,9-O) Cow (7-O/8-O/9-O/7,9-O) |
|||
| Neu5Gc-O-Ac (7/8-O) |
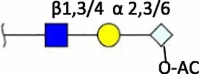 |
Swine (8-O) [4] Cow (7-O/8-O) |
|||
| 2,6SIA-LacdiNAc |  |
Ferret [35] | Human (N.D.) Swine (N.D.) |
||
| Sulfation (3S, 6S) | 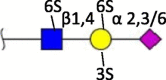 |
Human [39] | |||
Terminal structures at the non-reducing end of O- and N-linked glycan chains detected by mass spectrometry on epithelial cells or respiratory mucus in the indicated species. Differential expression at different locations along the respiratory tract has been analyzed in some cases but is not shown here. Neu5Ac (purple diamond), Neu5Gc (light blue diamond), N-acetylglucosamine (blue square), N-acetylgalactosamine (yellow square), galactose (yellow sphere), fucose (red triangle). S indicates sulfation.
Species in which a structure was not detectable (N.D.) or hardly detectable (trace) in extensive studies are indicated.
Respiratory mucus penetration by glycan-binding viruses
The mobility of viruses interacting with glycans is likely to be affected by heavily glycosylated mucus in a host species-specific manner. For viruses lacking RDE activity (Table 1) it is not well studied how they prevent being immobilized in the mucus layer. These viruses may bind at very low affinity and/or with very high specificity, thereby preventing high-avidity binding to glycans abundantly present in mucus. Viruses displaying RDE activity (Table 1), such as IAV, have been shown to require this activity to avoid immobilization in mucus [49., 50., 51.]. These also include members of Orthomyxo-, Paramyxo-, and Coronaviridae. Influenza A and B viruses and embeco-coronaviruses contain receptor-binding [hemagglutinin (HA) and spike (S)] and -cleaving [neuraminidase (NA) and hemagglutinin-esterase (HE)] functions in separate proteins. These activities are combined in a single protein for influenza C and D viruses and several paramyxoviruses [hemagglutinin-esterase fusion (HEF) and hemagglutinin-neuraminidase (HN) proteins]. Influenza A and B viruses and some paramyxoviruses prefer binding to either α2,3- or α2,6-linked SIAs, while embecoviruses, influenza C and D viruses bind to specifically O-acetylated SIAs (Table 1).
In view of the differences observed and anticipated between the sialoglycan repertoires of different host species, glycan-binding and -cleaving viruses likely need to adapt their sialoglycan interactions to successfully breach the host species barrier. Upon zoonotic transfer, animal IAVs not only adapt HA receptor binding (e.g., for avian viruses by switching receptor preference from α2,3- to α2,6-linked SIAs, the latter of which are abundantly present at the surface of epithelial cells in the upper respiratory tract of humans and swine) but also their NA-cleaving properties (reviewed in [7,52]). These latter adaptations may include changes that affect NA catalytic activity directly or indirectly, the latter for example by loss of a functional second SIA binding site, which is observed in all human viruses [53]. NA activity can also be adjusted by modification of the stalk length as frequently observed when viruses of wild waterfowl adapt to poultry [54]. Likewise, bovine coronaviruses that jumped into humans to become the novel human coronaviruses OC43 and HKU1 also reset their binding–cleaving balance to adapt to the human repertoire and/or densities of O-acetylated sialoglycans [11]. Analogous to the loss of a functional second SIA binding site in IAV NA, CoV-OC43 and -HKU1 lost the lectin function in HE as an adaptation to humans. Essentially nothing is known about the importance of balanced receptor-binding and -cleaving for the host range of paramyxoviruses.
For IAVs, the importance of well-balanced receptor-binding and -cleaving activity in enabling virus motility within respiratory mucus is demonstrated by several studies [49., 50., 51.]. Inhibition of NA activity decreased IAV infection of mucus-secreting human tracheobronchial epithelial cultures [55] and increased inhibition by mucus in other IAV infection assays [51]. NA activity was furthermore shown to drive mobility of IAV particles in/on mucus [49,50]. The extent to which N-glycan-rich exosomes [26] contribute to the inhibitory effect of mucus deserves greater attention. IAV binds, permanently but dynamically, via HA to receptor-coated surfaces, resulting in directional rolling-type motility that depends on NA activity [50,56,57]. Such motility has also been proposed for influenza C viruses [58] and coronaviruses [11,59,60], and the model can probably be extended to other viruses containing receptor-binding and -cleaving activities. We hypothesize that extended MUC5AC and MUC5B polymers provide a glycan track towards the epithelial cell surface for such motility [61]. The inhibition or support of virus transport by a mucus layer is fully dependent on the balance of receptor-binding and -destroying activity of a particular virus and on the mucus layer of a specific host. As such, the host- and organ-specific mucus glycome may facilitate infection by viruses that have coevolved with, and adapted to, their host for many years. Clearly, mucus can also function as an important barrier for zoonotic transfer that can only be overcome by much more rapid adaptations restoring the balance.
A comparison of inhibition of IAVs by mucus from different host species is mostly lacking. Human mucus was shown to inhibit swine and human viruses much more than swine mucus [62], which may be related to increased content of Neu5Gc and αGal epitope-containing glycans, at the expense of Neu5Ac-containing functional epitopes. The inhibitory potential of bovine submaxillary mucin was affected by the removal of O-acetyl groups from SIAs in an IAV subtype-specific manner [4], which demonstrates the importance of host-specific glycosylation of mucus for its inhibitory potential. Viruses that breach the host species barrier presumably evolve towards variants with decreased affinity for abundant mucus decoy sialoglycans as was similarly observed previously for acquiring serum resistance of IAVs. For example, swine-serum-resistant variants displayed a markedly decreased affinity for total swine serum sialylglycoproteins, while equine-serum-resistant viruses lost the ability to bind the NA-resistant 4-O-acetylated SIA moieties of equine α2-macroglobulin [63]. Similar studies on this topic using mucus from different species are absent thus far.
Concluding remarks
While there is ample evidence for the protective function of mucus against glycan-binding viruses, as well as for the counteracting function of viral RDEs, relatively little is still known on the species-specific properties of mucus against the establishment of zoonotic and enzootic infections. Numerous observations discussed in this paper are, however, indicative for such a function. With the advent of more sensitive and quantitative glycomic methods, as well as increasing knowledge and expanding techniques for the analysis of virus–receptor interaction dynamics and specificity, rapid progress on this issue can in principle be made. See Outstanding questions for the most urgent open questions. The principal importance of such studies lays in the acquisition of a better assessment of the zoonotic threats posed by animal viruses.
Outstanding questions.
To what extent does the mucus glycome differ between species? Detailed glycomic analysis of the mucins of species other than human is urgently required. Information on the SIA-linkage type (especially on LacNAc repeats present on O- and N-linked glycans) is highly relevant. The proteome and glycome of the globular proteins is largely unknown for any species. Virus-binding studies to the identified glycans, as well as to mucins and soluble proteins, should complement these studies.
How does virus motility in a mucus layer depend on the balance between mucus glycan receptors and viral RBP and RDE activity? To quantify this balance, the binding affinity/avidity and RDE activity/specificity of viruses with mucus of different species needs to be determined and correlated to the corresponding glycome as well as to motility and the dynamics of the interaction between viruses and mucin networks.
Is there a host-specific RBP/RDE/receptor balance to which viruses have to adapt to become established in a novel host species? In other words, can we prove that the mucus layer is a major zoonotic barrier? Adaptation of a virus to the functional cell-surface receptors in a novel host has been described in detail for influenza and coronaviruses. However, the extent to which adaptation to decoy receptors in the mucus layer, if at all, has taken place, and has actually been a requirement for establishment in another species, has hardly been described.
How do glycan-binding viruses that lack a RDE prevent immobilization in the mucus? Few data are available on the motility of such viruses in a mucus layer. It is expected that motility is mainly dependent on a receptor/RBP balance and therefore on the kinetic binding parameters (KD, koff of monomeric interactions) and binding polyvalency of such viruses. Whether there is any directionality in their movement is unknown.
What is the role of mucus in the formation and egress of infectious particles? Little is known about the role of mucus in respiratory virus transmission within and between species. Mucus may facilitate virus transmission by forming a protective layer that protects against inactivation, but might also constitute an additional barrier that a virus needs to overcome.
Alt-text: Outstanding questions
Declaration of interests
There are no interests to declare.
Glossary
- Glycan-receptor-binding protein (RBP)
a viral attachment protein capable of binding to glycotopes present on target cells. Enveloped viruses may contain hemagglutinin (HA; influenza A and B viruses), hemagglutinin-esterase (HE; some coronaviruses), hemagglutinin-esterase fusion (HEF; influenza C and D viruses), or hemagglutinin-neuraminidase (some paramyxoviruses) proteins. Many nonenveloped ('naked') viruses bind glycans via their capsid proteins.
- Glycan-receptor-destroying enzyme (RDE)
a viral protein which cleaves or modifies specific glycotopes that are recognized by the corresponding glycan-receptor-binding protein (RBP). Examples are neuraminidase (NA; influenza A and B viruses), hemagglutinin-esterase (HE; some coronaviruses), hemagglutinin-esterase fusion (HEF; influenza C and D viruses), and hemagglutinin-neuraminidase (some paramyxoviruses) proteins.
- Glycomics
the study of the entire complement of glycans (glycome).
- Glycotope
a glycan epitope that is recognized by glycan-binding proteins, including viral glycan-receptor-binding proteins (RBPs).
- Mucins
large network-forming, heavily sialylated macromolecules that are the main functional and structural component of mucus.
- Mucociliary clearance (MCC)
a process in the respiratory tract (bronchioles, bronchi, trachea, nasal cavity) in which the gel-like mucus layer on top of the periciliary layer (PCL) is transported by ciliary beating to the pharynx where it is then swallowed or coughed up.
- Mucus
a gel-like liquid overlaying epithelial surfaces. Air surface liquid (ASL) that overlays the respiratory epithelia consists of two layers: the gel layer and periciliary layer (PCL). The gel layer contains soluble mucins MUC5AC and MUC5B. The PCL contains cilia present on the epithelial surface that are rich in transmembrane mucins.
- Sialic acid (SIA)
a nine-carbon backbone saccharide that generally occupies a terminal position of an oligosaccharide (referred to as sialoglycan). SIAs are generally attached to a penultimate galactose via an α2,3- or an α2,6-linkage (referred to as α2,3SIA and α2,6SIA). SIAs may be differentially modified, for example by the addition of O-acetyl, N-glycolyl (Neu5Gc), or N-acetyl (Neu5Ac) groups.
- Sialoglycan
sialylated oligosaccharide.
References
- 1.Lillehoj E.P., et al. Cellular and molecular biology of airway mucins. Int. Rev. Cell Mol. Biol. 2013;303:139–202. doi: 10.1016/B978-0-12-407697-6.00004-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zanin M., et al. The interaction between respiratory pathogens and mucus. Cell Host Microbe. 2016;19:159–168. doi: 10.1016/j.chom.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matrosovich M., et al. In: SialoGlyco Chemistry and Biology II: Tools and Techniques to Identify and Capture Sialoglycans. Gerardy-Schahn R., et al., editors. Springer International Publishing; 2015. Sialic acid receptors of viruses; pp. 1–28. [Google Scholar]
- 4.Barnard K.N., et al. Modified sialic acids on mucus and erythrocytes inhibit influenza A virus hemagglutinin and neuraminidase functions. J. Virol. 2020;94 doi: 10.1128/JVI.01567-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Varki A., et al. In: Essentials of Glycobiology. Varki A., et al., editors. Cold Spring Harbor Laboratory Press; 2017. Sialic acids and other nonulosonic acids. [Google Scholar]
- 6.Langereis M.A., et al. Complexity and diversity of the mammalian sialome revealed by nidovirus virolectins. Cell Rep. 2015;11:1966–1978. doi: 10.1016/j.celrep.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Graaf M., Fouchier R.A. Role of receptor binding specificity in influenza A virus transmission and pathogenesis. EMBO J. 2014;33:823–841. doi: 10.1002/embj.201387442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Byrd-Leotis L., et al. The interplay between the host receptor and influenza virus hemagglutinin and neuraminidase. Int. J. Mol. Sci. 2017;18:1541. doi: 10.3390/ijms18071541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wagner R., et al. Functional balance between haemagglutinin and neuraminidase in influenza virus infections. Rev. Med. Virol. 2002;12:159–166. doi: 10.1002/rmv.352. [DOI] [PubMed] [Google Scholar]
- 10.Wasik B.R., et al. Effects of sialic acid modifications on virus binding and infection. Trends Microbiol. 2016;24:991–1001. doi: 10.1016/j.tim.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lang Y., et al. Coronavirus hemagglutinin-esterase and spike proteins coevolve for functional balance and optimal virion avidity. Proc. Natl. Acad. Sci. U. S. A. 2020;117:25759–25770. doi: 10.1073/pnas.2006299117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Button B., et al. A periciliary brush promotes the lung health by separating the mucus layer from airway epithelia. Science. 2012;337:937–941. doi: 10.1126/science.1223012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Witten J., et al. Selective permeability of mucus barriers. Curr. Opin. Biotechnol. 2018;52:124–133. doi: 10.1016/j.copbio.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fahy J.V., Dickey B.F. Airway mucus function and dysfunction. N. Engl. J. Med. 2010;363:2233–2247. doi: 10.1056/NEJMra0910061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tomazic P.V., et al. Nasal mucus proteome and its involvement in allergic rhinitis. Exp. Rev. Proteom. 2020;17:191–199. doi: 10.1080/14789450.2020.1748502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Atanasova K.R., Reznikov L.R. Strategies for measuring airway mucus and mucins. Respir. Res. 2019;20:261. doi: 10.1186/s12931-019-1239-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joo N.S., et al. Proteomic analysis of pure human airway gland mucus reveals a large component of protective proteins. PLoS One. 2015;10 doi: 10.1371/journal.pone.0116756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park M.S., et al. Towards the application of human defensins as antivirals. Biomol. Ther. (Seoul) 2018;26:242–254. doi: 10.4062/biomolther.2017.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wakabayashi H., et al. Lactoferrin for prevention of common viral infections. J. Infect. Chemother. 2014;20:666–671. doi: 10.1016/j.jiac.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 20.Akram K.M., et al. An innate defense peptide BPIFA1/SPLUNC1 restricts influenza A virus infection. Mucosal Immunol. 2018;11:71–81. doi: 10.1038/mi.2017.45. [DOI] [PubMed] [Google Scholar]
- 21.Tripathi S., et al. The human cathelicidin LL-37 inhibits influenza A viruses through a mechanism distinct from that of surfactant protein D or defensins. J. Gen. Virol. 2013;94:40–49. doi: 10.1099/vir.0.045013-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.LeVine A.M., et al. Surfactant protein D enhances clearance of influenza A virus from the lung in vivo. J. Immunol. 2001;167:5868–5873. doi: 10.4049/jimmunol.167.10.5868. [DOI] [PubMed] [Google Scholar]
- 23.Ligtenberg A.J., et al. Deleted in malignant brain tumors-1 protein (DMBT1): a pattern recognition receptor with multiple binding sites. Int. J. Mol. Sci. 2010;11:5212–5233. doi: 10.3390/ijms1112521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang M.L., et al. Galectin-1 binds to influenza virus and ameliorates influenza virus pathogenesis. J. Virol. 2011;85:10010–10020. doi: 10.1128/JVI.00301-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bedford J.G., et al. Airway exosomes released during influenza virus infection serve as a key component of the antiviral innate immune response. Front. Immunol. 2020;11:887. doi: 10.3389/fimmu.2020.00887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kesimer M., et al. Characterization of exosome-like vesicles released from human tracheobronchial ciliated epithelium: a possible role in innate defense. FASEB J. 2009;23:1858–1868. doi: 10.1096/fj.08-119131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bartlett J.A., et al. Protein composition of bronchoalveolar lavage fluid and airway surface liquid from newborn pigs. Am. J. Physiol. Lung Cell Mol. Physiol. 2013;305:L256–L266. doi: 10.1152/ajplung.00056.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo Y., et al. 1-DE MS and 2-D LC-MS analysis of the mouse bronchoalveolar lavage proteome. Proteomics. 2005;5:4608–4624. doi: 10.1002/pmic.200500052. [DOI] [PubMed] [Google Scholar]
- 29.Hägglund S., et al. Proteome analysis of bronchoalveolar lavage from calves infected with bovine respiratory syncytial virus. Insights in pathogenesis and perspectives for new treatments. PLoS One. 2017;12 doi: 10.1371/journal.pone.0186594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leal J., et al. Physicochemical properties of mucus and their impact on transmucosal drug delivery. Int. J. Pharm. 2017;532:555–572. doi: 10.1016/j.ijpharm.2017.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang X., et al. Immobilization of pseudorabies virus in porcine tracheal respiratory mucus revealed by single particle tracking. PLoS One. 2012;7 doi: 10.1371/journal.pone.0051054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramachandran P., et al. Identification of N-linked glycoproteins in human saliva by glycoprotein capture and mass spectrometry. J. Proteome Res. 2006;5:1493–1503. doi: 10.1021/pr050492k. [DOI] [PubMed] [Google Scholar]
- 33.Bateman A.C., et al. Glycan analysis and influenza A virus infection of primary swine respiratory epithelial cells: the importance of NeuAc{alpha}2-6 glycans. J. Biol. Chem. 2010;285:34016–34026. doi: 10.1074/jbc.M110.115998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chan R.W., et al. Infection of swine ex vivo tissues with avian viruses including H7N9 and correlation with glycomic analysis. Influenza Other Respir. Viruses. 2013;7:1269–1282. doi: 10.1111/irv.12144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jia N., et al. Glycomic characterization of respiratory tract tissues of ferrets: implications for its use in influenza virus infection studies. J. Biol. Chem. 2014;289:28489–28504. doi: 10.1074/jbc.M114.588541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sriwilaijaroen N., et al. N-glycans from porcine trachea and lung: predominant NeuAcα2-6Gal could be a selective pressure for influenza variants in favor of human-type receptor. PLoS One. 2011;6 doi: 10.1371/journal.pone.0016302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hiono T., et al. A chicken influenza virus recognizes fucosylated α2,3 sialoglycan receptors on the epithelial cells lining upper respiratory tracts of chickens. Virology. 2014;456–457:131–138. doi: 10.1016/j.virol.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 38.Morelle W., et al. FAB-MS characterization of sialyl Lewisx determinants on polylactosamine chains of human airway mucins secreted by patients suffering from cystic fibrosis or chronic bronchitis. Glycoconj. J. 2001;18:699–708. doi: 10.1023/a:1020871322769. [DOI] [PubMed] [Google Scholar]
- 39.Xia B., et al. Altered O-glycosylation and sulfation of airway mucins associated with cystic fibrosis. Glycobiology. 2005;15:747–775. doi: 10.1093/glycob/cwi061. [DOI] [PubMed] [Google Scholar]
- 40.Hirabayashi J., et al. Lectin-based structural glycomics: a practical approach to complex glycans. Electrophoresis. 2011;32:1118–1128. doi: 10.1002/elps.201000650. [DOI] [PubMed] [Google Scholar]
- 41.Hanisch F.G. O-glycosylation of the mucin type. Biol. Chem. 2001;382:143–149. doi: 10.1515/BC.2001.022. [DOI] [PubMed] [Google Scholar]
- 42.Gambaryan A.S., et al. Receptor-binding properties of influenza viruses isolated from gulls. Virology. 2018;522:37–45. doi: 10.1016/j.virol.2018.07.004. [DOI] [PubMed] [Google Scholar]
- 43.Guo H., et al. Highly pathogenic influenza A(H5Nx) viruses with altered H5 receptor-binding specificity. Emerg. Infect. Dis. 2017;23:220–231. doi: 10.3201/eid2302.161072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wasik B.R., et al. Distribution of O-acetylated sialic acids among target host tissues for influenza virus. mSphere. 2017;2 doi: 10.1128/mSphere.00379-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Broszeit F., et al. N-glycolylneuraminic acid as a receptor for influenza A viruses. Cell Rep. 2019;27:3284–3294. doi: 10.1016/j.celrep.2019.05.048. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Springer S.A., et al. Parallel evolution of a self-signal: humans and new world monkeys independently lost the cell surface sugar Neu5Gc. Immunogenetics. 2014;66:671–674. doi: 10.1007/s00251-014-0795-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ng P.S., et al. Ferrets exclusively synthesize Neu5Ac and express naturally humanized influenza A virus receptors. Nat. Commun. 2014;5:5750. doi: 10.1038/ncomms6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peri S., et al. Phylogenetic distribution of CMP-Neu5Ac hydroxylase (CMAH), the enzyme synthetizing the proinflammatory human xenoantigen Neu5Gc. Genome Biol. Evol. 2018;10:207–219. doi: 10.1093/gbe/evx251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang X., et al. A beneficiary role for neuraminidase in influenza virus penetration through the respiratory mucus. PLoS One. 2014;9 doi: 10.1371/journal.pone.0110026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vahey M.D., Fletcher D.A. Influenza A virus surface proteins are organized to help penetrate host mucus. eLife. 2019;8 doi: 10.7554/eLife.43764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cohen M., et al. Influenza A penetrates host mucus by cleaving sialic acids with neuraminidase. Virol. J. 2013;10:321. doi: 10.1186/1743-422X-10-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gambaryan A.S., Matrosovich M.N. What adaptive changes in hemagglutinin and neuraminidase are necessary for emergence of pandemic influenza virus from its avian precursor? Biochemistry (Mosc) 2015;80:872–880. doi: 10.1134/S000629791507007X. [DOI] [PubMed] [Google Scholar]
- 53.Du W., et al. Second sialic acid-binding site of influenza A virus neuraminidase: binding receptors for efficient release. FEBS J. 2020 doi: 10.1111/febs.15668. Published online December 14, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Long J.S., et al. One-way trip: influenza virus' adaptation to gallinaceous poultry may limit its pandemic potential. Bioessays. 2015;37:204–212. doi: 10.1002/bies.201400133. [DOI] [PubMed] [Google Scholar]
- 55.Matrosovich M.N., et al. Neuraminidase is important for the initiation of influenza virus infection in human airway epithelium. J. Virol. 2004;78:12665–12667. doi: 10.1128/JVI.78.22.12665-12667.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sakai T., et al. Influenza A virus hemagglutinin and neuraminidase act as novel motile machinery. Sci. Rep. 2017;7 doi: 10.1038/srep45043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guo H., et al. Kinetic analysis of the influenza A virus HA/NA balance reveals contribution of NA to virus-receptor binding and NA-dependent rolling on receptor-containing surfaces. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1007233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sakai T., et al. Unique directional motility of influenza C virus controlled by its filamentous morphology and short-range motions. J. Virol. 2018;92 doi: 10.1128/JVI.01522-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tortorici M.A., et al. Structural basis for human coronavirus attachment to sialic acid receptors. Nat. Struct. Mol. Biol. 2019;26:481–489. doi: 10.1038/s41594-019-0233-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schwegmann-Wessels C., Herrler G. Sialic acids as receptor determinants for coronaviruses. Glycoconj. J. 2006;23:51–58. doi: 10.1007/s10719-006-5437-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.de Vries E., et al. Influenza A virus hemagglutinin–neuraminidase–receptor balance: preserving virus motility. Trends Microbiol. 2020;28:57–67. doi: 10.1016/j.tim.2019.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zanin M., et al. Pandemic swine H1N1 influenza viruses with almost undetectable neuraminidase activity are not transmitted via aerosols in ferrets and are inhibited by human mucus but not swine mucus. J. Virol. 2015;89:5935–5948. doi: 10.1128/JVI.02537-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Matrosovich M., et al. Molecular mechanisms of serum resistance of human influenza H3N2 virus and their involvement in virus adaptation in a new host. J. Virol. 1998;72:6373–6380. doi: 10.1128/jvi.72.8.6373-6380.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang L.Y., et al. Characterization of the adeno-associated virus 1 and 6 sialic acid binding site. J. Virol. 2016;90:5219–5230. doi: 10.1128/JVI.00161-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kaludov N., et al. Adeno-associated virus serotype 4 (AAV4) and AAV5 both require sialic acid binding for hemagglutination and efficient transduction but differ in sialic acid linkage specificity. J. Virol. 2001;75:6884–6893. doi: 10.1128/JVI.75.15.6884-6893.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Baggen J., et al. Enterovirus D68 receptor requirements unveiled by haploid genetics. Proc. Natl. Acad. Sci. U. S. A. 2016;113:1399–1404. doi: 10.1073/pnas.1524498113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thompson A.J., et al. Virus recognition of glycan receptors. Curr. Opin. Virol. 2019;34:117–129. doi: 10.1016/j.coviro.2019.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stuart A.D., Brown T.D.K. Alpha2,6-linked sialic acid acts as a receptor for feline calicivirus. J. Gen. Virol. 2007;88:177–186. doi: 10.1099/vir.0.82158-0. [DOI] [PubMed] [Google Scholar]
- 69.Sánchez-Felipe L., et al. α2-3- and α2-6- N-linked sialic acids allow efficient interaction of Newcastle disease virus with target cells. Glycoconj. J. 2012;29:539–549. doi: 10.1007/s10719-012-9431-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fukushima K., et al. Terminal sialic acid linkages determine different cell infectivities of human parainfluenza virus type 1 and type 3. Virology. 2014;464–465:424–431. doi: 10.1016/j.virol.2014.07.033. [DOI] [PubMed] [Google Scholar]
- 71.Hulswit R.J.G., et al. Human coronaviruses OC43 and HKU1 bind to 9-O-acetylated sialic acids via a conserved receptor-binding site in spike protein domain A. Proc. Natl. Acad. Sci. U. S. A. 2019;116:2681–2690. doi: 10.1073/pnas.1809667116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Baker A.N., et al. The SARS-COV-2 spike protein binds sialic acids and enables rapid detection in a lateral flow point of care diagnostic device. ACS Cent. Sci. 2020;6:2046–2052. doi: 10.1021/acscentsci.0c00855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li W., et al. Identification of sialic acid-binding function for the Middle East respiratory syndrome coronavirus spike glycoprotein. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E8508–E8517. doi: 10.1073/pnas.1712592114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schultze B., et al. Neuraminidase treatment of avian infectious bronchitis coronavirus reveals a hemagglutinating activity that is dependent on sialic acid-containing receptors on erythrocytes. Virology. 1992;189:792–794. doi: 10.1016/0042-6822(92)90608-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hirose K., et al. Insight into glycan diversity and evolutionary lineage based on comparative Avio-N-glycomics and sialic acid analysis of 88 egg whites of Galloanserae. Biochemistry. 2011;50:4757–4774. doi: 10.1021/bi101940x. [DOI] [PubMed] [Google Scholar]
- 76.Jeffery P., Li D. Airway mucosa: secretory cells, mucus and mucin genes. Eur. Resp. J. 1997;10:1655–1662. doi: 10.1183/09031936.97.10071655. [DOI] [PubMed] [Google Scholar]
- 77.Peng W., et al. Recent H3N2 viruses have evolved specificity for extended, branched human-type receptors, conferring potential for increased avidity. Cell Host Microbe. 2017;21:23–34. doi: 10.1016/j.chom.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]