Abstract
Background and aims
Computed tomography (CT) scan is frequently used to detect hepatocellular carcinoma (HCC) in routine clinical practice. The aim of this study is to develop a deep-learning AI system to improve the diagnostic accuracy of HCC by analysing liver CT imaging data.
Methods
We developed a deep-learning AI system by training on CT images from 7512 patients at Henan Provincial Peoples’ Hospital. Its performance was validated on one internal test set (Henan Provincial Peoples’ Hospital, n = 385) and one external test set (Henan Provincial Cancer Hospital, n = 556). The area under the receiver-operating characteristic curve (AUROC) was used as the primary classification metric. Accuracy, sensitivity, specificity, precision, negative predictive value and F1 metric were used to measure the performance of AI systems and radiologists.
Results
AI system achieved high performance in identifying HCC patients, with AUROC of 0.887 (95% CI 0.855–0.919) on the internal test set and 0.883 (95% CI 0.855–0.911) on the external test set. For internal test set, accuracy was 81.0% (76.8–84.8%), sensitivity was 78.4% (72.4–83.7%), specificity was 84.4% (78.0–89.6%) and F1 (harmonic average of precision and recall rate) was 0.824. For external test set, accuracy was 81.3% (77.8–84.5%), sensitivity was 89.4% (85.0–92.8%), specificity was 74.0% (68.5–78.9%) and F1 was 0.819. Compared with radiologists, AI system achieved comparable accuracy and F1 metric on internal test set (0.853 versus 0.818, P = 0.107; 0.863 vs. 0.824, P = 0.082) and external test set (0.805 vs. 0.793, P = 0.663; 0.810 vs. 0.814, P = 0.866). The predicted HCC risk scores by AI system in HCC patients with multiple tumours and high fibrosis stage were higher than those with solitary tumour and low fibrosis stage (tumour number: 0.197 vs. 0.138, P = 0.006; fibrosis stage: 0.183 vs. 0.127, P < 0.001). Radiologists’ review showed that the accuracy of saliency heatmaps predicted by algorithms was 92.1% (95% CI: 89.2–95.0%).
Conclusions
AI system achieved high performance in the detection of HCC compared with a group of specialised radiologists. Further investigation by prospective clinical trials was necessitated to verify this model.
Subject terms: Cancer imaging, Cancer screening
Introduction
Hepatocellular carcinoma (HCC) is the sixth most common cancer type and the fourth leading cause of cancer-related death worldwide. The annual incidence of HCC is estimated to be 841,000 new cases in the world, 80% of which are in sub-Saharan Africa and eastern Asia [1, 2]. Computed tomography (CT) is a primary screening method for HCC surveillance. Contrast-enhanced multiphasic CT examination can detect dysplastic lesions and HCC nodules at early stages [3–8]. At present, all guidelines recommend multiphasic CT with extracellular agents as one of the first-line noninvasive modalities during the diagnosis and staging of HCC [9–11]. Interpretation of CT imaging data is conducted by radiologists according to liver imaging reporting and data system (LI-RADS) drafted by the American College of Radiology [12]. The LI-RADS guideline stratifies the detected lesions into five categories ranging from definitively benign (LR 1) to definitively HCC (LR 5). Subsequently, clinicians determine whether to proceed with invasive therapy or observational management of the lesions.
Clinically, HCC can be diagnosed by puncture biopsy or typical imaging examination according to the American Association for the Study of Liver Diseases (AASLD) guideline [9, 13, 14]. However, in routine clinical practice, precise biopsy for malignant lesions is not always possible. The discrepancy of puncture technique and variabilities of inter-pathologists can also affect the accuracy of biopsy [15]. Meanwhile, the performance of CT for HCC diagnosis is closely related to tumour size. In a meta-analysis including 33 comprehensive studies, the sensitivity was 0.70–0.86 in tumours larger than 2 cm. For tumours smaller than 1 cm, the sensitivity decreased to 0.34–0.62 [16]. Inaccurate diagnosis leads to inappropriate treatment, increased psychological burden and higher medical costs. Therefore, improving the diagnostic accuracy of suspicious lesions is imperative as lesions with high suspicion of HCC will be treated more appropriately.
Recently, a deep convolutional neural network (DCNN) has been increasingly investigated as an auxiliary technique in the field of medical imaging diagnosis [17–19]. Deep-learning algorithms enable-feature representation learning from a large volume of imaging data in an end-to-end manner to avoid the extensive labour of hand-crafted feature engineering. Previous studies reported on-par performance of deep-learning models as compared with specialists in diabetic retinopathy grading, skin lesion classification and thyroid cancer diagnosis [20–22]. Deep-learning models have also been applied to detect head CT scan abnormalities requiring urgent neurosurgical intervention and predict the risk of lung cancer using a patient’s current and prior CT volumes [23, 24]. From this point of view, an automated deep-learning model that can interpret liver CT imaging data to detect HCC is valuable for patients at high risk of HCC, especially in community hospitals.
In this study, we aimed to develop an end-to-end HCC diagnostic artificial intelligence (AI) system to differentiate HCC from other liver lesions. This clinically applicable AI system consisted of two deep-learning models: HCCNet and NoduleNet. Both HCCNet and NoduleNet models were deep residual convolutional networks trained by a large amount of CT imaging data. We used pathological examination as the golden standard to diagnose HCC and evaluate AI performance. We examined the performance of our AI system on one internal and one external test set.
Methods
Study design and data sources
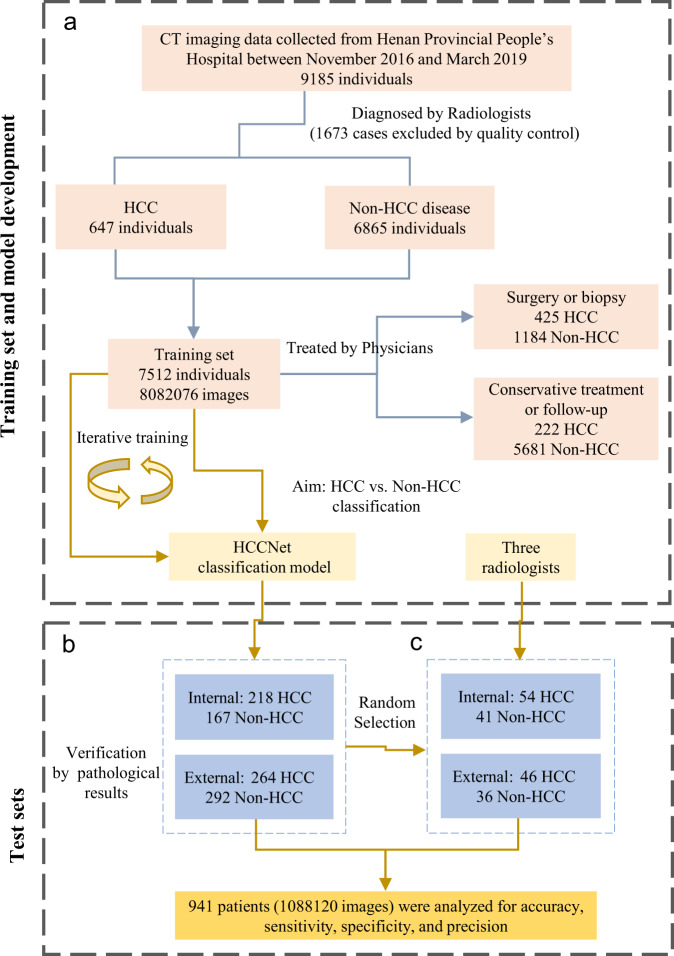
The liver CT imaging data of patients from in-hospital and outpatient radiology centres were retrospectively collected from two tertiary hospitals in China. We retrieved plain and contrast-enhanced CT imaging data from the picture archiving and communication system (PACS) at Henan Provincial Peoples’ Hospital between November 2016 and March 2019 as the training set. We used liver CT imaging data at Henan Provincial Peoples’ Hospital (internal test set) between December 2016 and September 2019 and at Henan Provincial Cancer Hospital (external test set) between February 2018 and April 2019 as test sets. All images and electronic clinical reports were anonymously processed before they were transferred to investigators. The HCC group consisted of patients not only treated by surgical resection but also treated by intervention, radiofrequency ablation, cryoablation, microwave therapy or any other invasive treatment therapy. Both solitary and multiple HCC tumour nodules were enrolled. Patients diagnosed with malignant lesions other than HCC such as hemangioendothelioma, sarcoma, intrahepatic cholangiocarcinoma and metastatic tumour were included in the control group. Patients diagnosed with benign lesions such as leiomyolipoma, hemangioma, cyst, abscess, adenoma and focal nodular hyperplasia were also included in the control group. All HCC patients in the validation sets had pathological examination after surgical resection or needle biopsy as the golden standard to evaluate AI performance. Surgically resected HCC tumours were staged according to the 7th edition of the TNM staging system drafted by the American Joint Committee on Cancer (AJCC). A flowchart illustrating this study is shown in Fig. 1.
Fig. 1. A flowchart depicting the procedures to develop and evaluate HCCNet.
a Model development procedure consisted of data acquisition and HCCNet training. b Evaluation of HCCNet on one internal and one external test set. c Comparison between HCCNet and three radiologists on subsets of randomly selected cases. 425 HCC patients, 1184 non-HCC patients of the training set and all patients of two test sets underwent surgery or biopsy for pathological examination.
This study was approved by the ethics committee of Henan Provincial Peoples’ Hospital (No. 2019068) and performed in accordance with principles of Good Clinical Practice and Declaration of Helsinki guidelines (1975, revised in 1983). All patients provided written informed consent before undergoing CT examinations.
Liver CT scanning
All selected patients underwent standard three-phase dynamic contrast-enhanced CT scan at initial diagnosis. Multiphase CT scans were performed using a 64-row scanner (General Electric Company Discovery CT750 HD, Milwaukee, Wisconsin, USA). The contrast agent was Ultravist 370 (Bayer AG, Berlin, Germany) and the flow rate was 3 mL/s. When the Hounsfield unit was set at +50, the arterial, portal venous and delayed phases were scanned 5 s after abdominal aorta enhancement, 30 s after the end of the arterial phase, and 90 s after the end of the portal venous phase, respectively. Scanning parameters were as follows: collimation, 64 rows × 0.625 mm; gantry rotation speed, 0.6 s; section thickness, 5 mm; image reconstruction increment, 1 mm; and tube voltage, 120 kV. Effective tube current was automatically set based on the weight of patients. Intense contrast uptake during the arterial phase and washout during the venous phase was defined as typical imaging features of HCC in cirrhotic patients.
Image classification procedures
All images obtained were transferred from DICOM format to jpeg format. The transaxial section from the apex to the bottom of the liver was used. Coronal or sagittal sections were excluded. Five senior radiologists (FFF, BJZ, YB, QXW and XYM) were asked to manually review all the images of HCC and non-HCC patients in the training set based on clinical radiology reports. If no consensual agreement was reached, the case was exempted. Images of each HCC patient in the training set featured by HCC tumour nodules were selected as positive samples, whereas those were not featured by HCC tumour nodules were discarded. All images from non-HCC patients were used as negative samples. Both plain CT scan images and enhanced CT images were included. Low-quality images such as artifacts, blank or blurred images were also excluded by these five radiologists. The performance of HCCNet model was evaluated by one internal test set and one external test set. At the patient level, to compare the HCCNet model with the expert group, a random subset of 80–100 cases were selected from these two test sets. We performed random selection via random sample function in R software. Three 8–10 year experienced radiologists (FFF, YB and QXW) reviewed all CT images from selected patients and interpreted these images according to LI-RADS system guidelines. Each radiologist read both test sets. These three senior radiologists were then asked to sort each patient as HCC or not HCC based on their clinical experience. Pathological reports were used to assess the prediction accuracy of both radiologists and HCCNet. Then the performance of radiologists was compared with HCCNet. Pathological results were obtained from surgically resected liver tissue specimen or ultrasound-guided liver biopsies.
Preliminary data processing
Four radiologists (FFF, QXW, XYM and JQW) manually reviewed 115,876 images of 331 patients to differentiate images with nodule from those without nodule. The consensual interpretation outputs of these four radiologists were used to develop an AI model NoduleNet to identify nodule images, which functioned as a subsidiary to HCCNet. Subsequently, we applied NoduleNet to identify images with HCC nodules in the training set. Together with the clinical report, images with HCC nodules predicted by NoduleNet were grouped into HCC, whereas, images without HCC nodules predicted by NoduleNet and images from non-HCC patients were placed in the non-HCC group.
AI model development
Two deep-learning AI models NoduleNet and HCCNet were developed in this study. NoduleNet acted as an assistant to HCCNet and the results of its analysis were directly integrated into HCCNet. Both NoduleNet and HCCNet are two deep residual convolutional network of 34 layers. The prominent feature of the residual network is the use of shortcut connection, which can speed up convergence at the early training stage [25] (Supplementary Fig. 1). We initialised the weights of NoduleNet and HCCNet from the same network that has been trained on the ImageNet data set except the last fully connected layer [26] (Supplementary Fig. 2). The output unit of the last fully connected layer was set to two to match the number of classes in this study and its weight was randomly initialised. We trained NoduleNet and HCCNet in an end-to-end fashion with stochastic gradient descent for 120 epochs by using cosine learning rate decay scheduling and a learning rate warmup scheme. We set an initial learning rate of 0.2, a momentum of 0.9 and a minibatch of 256. Data augmentation included random resize and crop, perspective, horizontal flip, rotation, colour jittering and mixup [27]. In addition, label-smoothing was enabled during training. We used a random subset of images as test set, which was not included during training, to calculate the loss of the model at the end of each epoch. Finally, we evaluated its performance on test sets. This procedure was developed with Python (version 3.7.1), MxNet (version 1.5.1), GluonCV (version 0.8.0), PyTorch (version 1.3.0) and torchvision (version 0.5.0).
Visual explanation
We used Saliency Map Order Equivalence (SMOE) algorithm to assess the importance of the spatial locations in convolutional layers [28]. Quantified pixel was also calculated by SMOE algorithm to describe contribution to the final prediction results. We sketched saliency heatmaps to highlight features most influenced NoduleNet prediction in malignant hepatocellular carcinoma images (Supplementary Fig. 3).
Calculation of HCC risk score
For each patient, we used the weighted mean of predicted probabilities of all images from that patient to calculate a HCC risk score. Specifically, we denoted the number of all images from that patient as n and the probability of each image predicted to be HCC as p = [p1, p2, …, pn]. The predicted HCC risk score for that individual patient was calculated as = −[w1 × log10(1 − p1) + w2 × log10(1 − p2) + ··· + wn × log10 (1 − pn)]/n, where wi is calculated as wi = pi/(p1 + p2 + … + pn). The correlation between pathological parameters (tumour size, AJCC tumour stage, tumour number, METAVIR fibrosis stage [29], major vascular invasion and histologic grade) and HCC risk scores were evaluated.
Statistical analysis
We used the area under the receiver-operating characteristic curve (AUROC) as the primary metric to describe the classification performance of HCCNet. The operating characteristic curve (ROC) was generated by plotting sensitivity against specificity for different thresholds. The other metrics used to measure the performance of HCCNet included accuracy, sensitivity, specificity, positive predictive rate, negative predictive rate, kappa coefficient and F1 metric. The kappa coefficient measures the inter-rater agreement among radiologists, an agreement between prediction results and pathological examination. The F1 metric is calculated as F1 = 2 × precision × recall/(precision + recall). The Clopper–Pearson method was used to calculate sensitivity, specificity, positive predictive rate, and negative predictive rate. We used R package pROC (version 1.3.1) to plot the ROC curve and calculate AUROC. Inter-radiologists agreement rate and Fleiss’ kappa were calculated by R package irr (version 0.84). Statistical analysis was conducted with R software (version 3.4.3).
Results
Baseline characteristics of training and test datasets
Between November 2016 and March 2019, we obtained CT images of 9185 patients from Henan Provincial Peoples’ Hospital as the training set. After quality control, the ultimate training set consisted of 8,082,076 images from 7512 individuals: 647 patients with HCC and 6865 controls. The internal test set consisted of 385 individuals (413,251 images) from Henan Provincial Peoples’ Hospital. The external test set consisted of 556 individuals (674,869 images) from Henan Provincial Cancer Hospital. In the training set, 425 (65.7%) HCC cases and 1184 (17.2%) non-HCC cases underwent pathological examination. All patients in two test sets had pathological examination results. The non-HCC patients in the training set (n = 6865) consisted of malignant tumours such as sarcoma (0.1%, n = 9), intrahepatic cholangiocarcinoma (0.8%, n = 59), metastatic tumour (3.6%, n = 271) and neuroendocrine neoplasm (0.3%, n = 24); benign tumours such as angioleiomyolipoma (0.2%, n = 16), hemangioma (25.2%, n = 1896), cyst (14.6%, n = 1097), abscess (0.5%, n = 36), adenoma (0.2%, n = 13) and focal nodular hyperplasia (0.3%, n = 23). The rest of the patients (3421, 45.5%) are normal liver cases. Detailed characteristics of patients concerning gender, age and disease subtype are shown in Table 1.
Table 1.
Baseline characteristics of the training set and two test sets.
| HPPH training set | HPPH test set | HPCH test set | ||||
|---|---|---|---|---|---|---|
| HCC | Non-HCC | HCC | Non-HCC | HCC | Non-HCC | |
| Patients | 647 | 6865 | 218 | 167 | 264 | 292 |
| Male | 509 | 3710 | 168 | 45 | 214 | 101 |
| Female | 138 | 3155 | 50 | 122 | 50 | 191 |
| Age (years) | 57.36 (8–86) | 54.19 (2–93) | 56.38 (17–87) | 52.42 (20–84) | 55.43 (28–82) | 53.74 (17–90) |
| Age ≤60 years male | 320 (49.46%) | 2314 (33.71%) | 115 (52.75%) | 38 (22.75%) | 149 (56.44%) | 56 (19.18%) |
| Age >60 years male | 189 (29.21%) | 1396 (20.34%) | 53 (24.31%) | 7 (4.19%) | 65 (24.62%) | 45 (15.41%) |
| Age ≤60 years female | 58 (8.96%) | 2045 (29.79%) | 27 (12.39%) | 92 (55.09%) | 24 (9.09%) | 142 (48.63%) |
| Age >60 years female | 80 (12.36%) | 1110 (16.17%) | 23 (10.55%) | 30 (17.96%) | 26 (9.85%) | 49 (16.78%) |
| Non-HCC disease variety | ||||||
| Intrahepatic cholangiocarcinoma | -- | 59 | -- | 0 | -- | 0 |
| Liver metastasis | -- | 271 | -- | 5 | -- | 205 |
| Sarcoma | -- | 9 | -- | 0 | -- | 0 |
| Hepatic hemangioma | -- | 1896 | -- | 119 | -- | 49 |
| Focal nodular hyperplasia | -- | 23 | -- | 0 | -- | 1 |
| Angioleiomyolipoma | -- | 16 | -- | 0 | -- | 0 |
| Hepatic adenoma | -- | 13 | -- | 0 | -- | 0 |
| Liver abscess | -- | 36 | -- | 7 | -- | 3 |
| Hepatic cyst | -- | 1097 | -- | 36 | -- | 16 |
| Neuroendocrine neoplasm | -- | 24 | -- | 0 | -- | 18 |
| Normal | -- | 3421 | -- | 0 | -- | 0 |
| Surgery or biopsy pathology | 425 (65.7%) | 1184 (17.2%) | 218 (100.00%) | 167 (100.00%) | 264 (100.00%) | 292 (100.00%) |
Performance of AI model HCCNet on two test sets
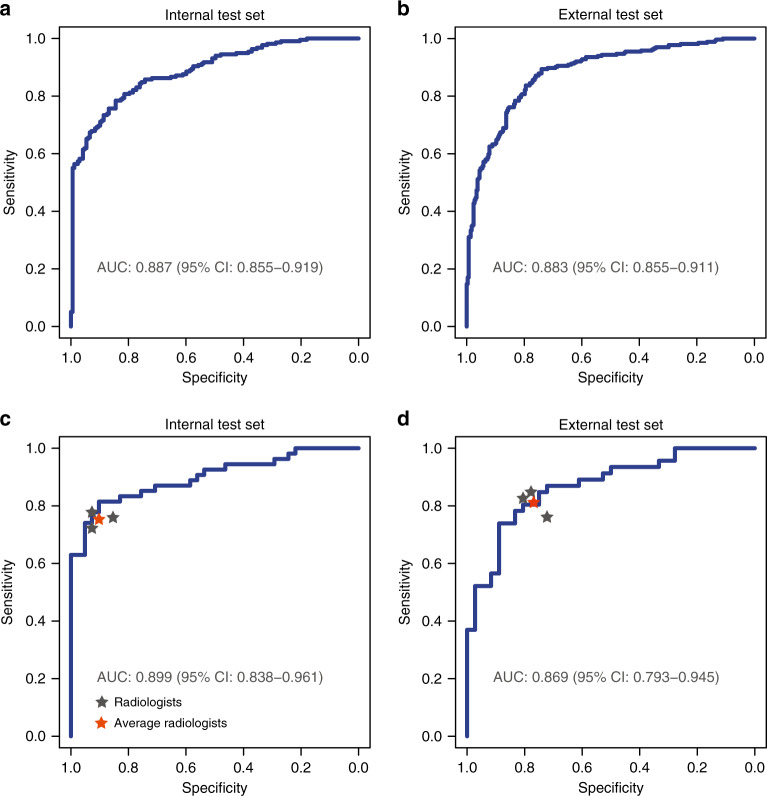
To verify the general applicability of our AI diagnostic model, HCCNet performance was tested in two different hospitals using different datasets. In the two retrospectively collected cohorts, the AI model HCCNet achieved high performance in identifying hepatocellular carcinoma patients on the internal and external test sets. The classification metrics of HCCNet are provided in Table 2 and Fig. 2. For the internal test set, AUROC was 0.887 (95% CI: 0.855–0.919), accuracy was 81.0% (76.8–84.8%), sensitivity was 78.4% (72.4–83.7%), specificity was 84.4% (78.0–89.6%) and F1 was 0.824. For external test set, AUROC was 0.883 (0.855–0.911), accuracy was 81.3% (77.8–84.5%), sensitivity was 89.4% (85.0–92.8%), specificity was 74.0% (68.5–78.9%), and F1 was 0.819. (Table 2). The ROC curves are shown in Fig. 2.
Table 2.
Classification performance of HCCNet on test sets.
| Performance metrics | The performance of deep-learning model on two test sets | |
|---|---|---|
| Internal test set (n = 385; HCC = 218, non-HCC = 167) | External test set (n = 556; HCC = 264, non-HCC = 292) | |
| Accuracy (95% CI) | 0.810 (0.768–0.848) | 0.813 (0.778–0.845) |
| Sensitivity (95% CI) | 0.784 (0.724–0.837) | 0.894 (0.850–0.928) |
| Specificity (95% CI) | 0.844 (0.780–0.896) | 0.740 (0.685–0.789) |
| Precision (95% CI) | 0.868 (0.813–0.912) | 0.756 (0.705–0.803) |
| Negative predictive value (95% CI) | 0.750 (0.682–0.810) | 0.885 (0.838–0.922) |
| Kappaa | 0.620 | 0.628 |
| F1b | 0.824 | 0.819 |
aMeasures the agreement between predicted classification and pathological report.
bHarmonic average of the precision and recall rate.
Fig. 2. Performance of the AI model HCCNet and radiologists on two test sets.
The receiver-operating curve of HCCNet on test sets were displayed in blue lines. Classification performance of each radiologist (grey star) and their average values (red star) were provided. Area under the curve and corresponding confidence interval are shown. a Internal test set, n = 385, b external test set, n = 556, c subset of the internal test set, n = 95, d subset of the external test set, n = 82.
The performance of NoduleNet was evaluated by comparing its classification results with the consensus interpretation of radiologists. In total, 14,778 images of 31 patients were randomly selected. NoduleNet achieved an AUROC of 0.901 (95% CI 0.893–0.910), sensitivity of 91.5% (89.5–93.2%) and specificity of 76.0% (74.7–77.2%) (Supplementary Fig. 4).
HCC risk scores predicted by AI model
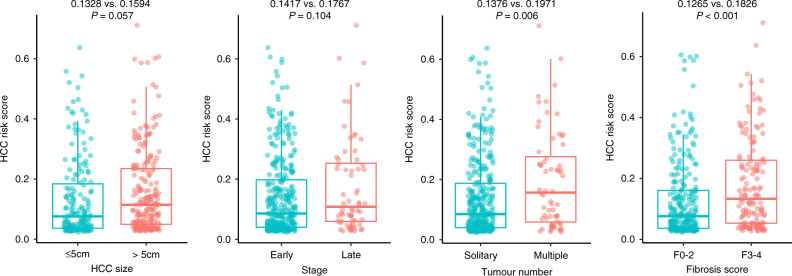
Among both internal and external test sets, except biopsy cases, 204 HCC patients of the internal test set and 184 HCC patients of external test set underwent surgical resection. Pathological parameters were further evaluated and the relationship with HCC risk score was assessed (Table 3). For HCC patients with tumour size >5 cm, the predicted HCC risk scores by AI model were higher than those ≤5 cm, but not statistically different (0.1594 ± 0.1404 vs. 0.1328 ± 0.1332, P = 0.057). Meanwhile, in terms of AJCC tumour stage, the predicted HCC risk scores of Stage III, IV patients were also slightly higher than Stage I, II patients (0.1767 ± 0.1596 vs. 0.1417 ± 0.1324, P = 0.104) (Fig. 3).
Table 3.
HCC risk scores predicted by AI model HCCNet on test sets.
| Pathological variables | The predicted HCC risk scores on two test sets | |||||
|---|---|---|---|---|---|---|
| Internal test set (HCC = 204) | P | External test set (HCC = 184) | P | Total | P | |
| Tumour size | 0.123 | 0.111 | 0.057 | |||
| ≤5 cm | 0.1398 ± 0.1429 | 0.1201 ± 0.1136 | 0.1328 ± 0.1332 | |||
| >5 cm | 0.1712 ± 0.1446 | 0.1507 ± 0.1370 | 0.1594 ± 0.1404 | |||
| TNM stage | 0.041 | 0.838 | 0.104 | |||
| Stage I + II | 0.1435 ± 0.1324 | 0.1394 ± 0.1329 | 0.1417 ± 0.1324 | |||
| Stage III + IV | 0.2393 ± 0.2031 | 0.1438 ± 0.1216 | 0.1767 ± 0.1596 | |||
| Tumour number | 0.007 | 0.341 | 0.006 | |||
| Solitary tumour | 0.1390 ± 0.1319 | 0.1361 ± 0.1298 | 0.1376 ± 0.1307 | |||
| Multiple tumours | 0.2334 ± 0.1800 | 0.1607 ± 0.1316 | 0.1971 ± 0.1606 | |||
| Major vascular invasion | 0.979 | 0.017 | 0.124 | |||
| Non-portal or hepatic vein invasion | 0.1546 ± 0.1506 | 0.1208 ± 0.1169 | 0.1390 ± 0.1359 | |||
| Portal or hepatic vein invasion | 0.1534 ± 0.1368 | 0.1689 ± 0.1433 | 0.1613 ± 0.1399 | |||
| Fibrosis stage | <0.001 | 0.826 | 0.001 | |||
| F0: no fibrosis | 0.0892 ± 0.0817 | 0.1454 ± 0.1400 | 0.1228 ± 0.1225 | |||
| F1: portal fibrosis without septa | 0.1434 ± 0.1381 | 0.1540 ± 0.1385 | 0.1498 ± 0.1378 | |||
| F2: portal fibrosis with few septa | 0.0512 ± 0.0368 | 0.1031 ± 0.1081 | 0.0858 ± 0.0936 | |||
| F3: numerous septa without cirrhosis | 0.1997 ± 0.2003 | 0.0989 ± 0.1160 | 0.1637 ± 0.1794 | |||
| F4: cirrhosis | 0.1929 ± 0.1478 | 0.1659 ± 0.1159 | 0.1871 ± 0.1416 | |||
| Histologic Grade | 0.507 | 0.395 | 0.285 | |||
| G1 | 0.1298 ± 0.1496 | 0.1656 ± 0.1381 | 0.1439 ± 0.1447 | |||
| G2 | 0.1577 ± 0.1353 | 0.1291 ± 0.1236 | 0.1427 ± 0.1298 | |||
| G3 | 0.1502 ± 0.1647 | 0.1769 ± 0.1495 | 0.1580 ± 0.1581 | |||
| G4 | 0.2269 ± 0.1742 | NA | 0.2511 ± 0.1603 | |||
Fig. 3. The predicted HCC risk score by AI model HCCNet on test sets stratified by HCC tumour size, AJCC TNM stage, tumour number and METAVIR fibrosis stage.
Boxplot representation of HCC risk score stratified by tumour size, TNM stage, tumour number and METAVIR fibrosis stage. Early stage = Stage I and II; late stage = Stage III and IV.
However, for HCC patients with multiple tumours, the predicted HCC risk scores by AI model were remarkably higher than those with solitary tumour (0.1971 ± 0.1606 vs. 0.1376 ± 0.1307, P = 0.006). When separated by METAVIR fibrosis stage, patients with severe fibrosis or cirrhosis had significant higher HCC risk scores than patients with none to moderate fibrosis (F3-4: 0.1826 ± 0.1492 vs. F0-2: 0.1265 ± 0.1260, P < 0.001). For major vascular invasion and histologic grade, no significant difference was observed (P = 0.124 and 0.285).
AI model HCCNet versus radiologists
We randomly selected 95 individuals from the internal test set and 82 individuals from the external test set for manual interpretation by radiologists. The entire image set of each selected patient was presented to three radiologists. Every radiologist read all the 177 patients. The total number of images read and interpreted by each radiologist were 192,772 (Table 4).
Table 4.
Classification performance of HCCNet and radiologists on subsets of two test sets.
| Performance metrics | Internal test set (n = 95; HCC = 54, non-HCC = 41) | External test set (n = 82; HCC = 46, non-HCC = 36) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Radiologist 1 | Radiologist 2 | Radiologist 3 | AI | Radiologist 1 | Radiologist 2 | Radiologist 3 | AI | ||
| Accuracy (95% CI) | 0.811 (0.717–0.884) | 0.842 (0.753–0.909) | 0.800 (0.705–0.875) | 0.853 (0.765–0.917) | 0.744 (0.636–0.834) | 0.817 (0.716–0.894) | 0.817 (0.716–0.894) | 0.805 (0.703–0.884) | |
| Sensitivity (95% CI) | 0.722 (0.584–0.835) | 0.778 (0.644–0.880) | 0.759 (0.624–0.865) | 0.815 (0.686–0.907) | 0.761 (0.612–0.874) | 0.848 (0.711–0.937) | 0.826 (0.686–0.922) | 0.739 (0.589–0.857) | |
| Specificity (95% CI) | 0.927 (0.801–0.985) | 0.927 (0.801–0.985) | 0.854 (0.708–0.944) | 0.902 (0.769–0.973) | 0.722 (0.548–0.858) | 0.778 (0.608–0.899) | 0.806 (0.640–0.918) | 0.889 (0.739–0.969) | |
| Precision (95% CI) | 0.929 (0.805–0.985) | 0.933 (0.817–0.986) | 0.872 (0.743–0.952) | 0.917 (0.800–0.977) | 0.778 (0.629–0.888) | 0.830 (0.692–0.924) | 0.844 (0.705–0.935) | 0.895 (0.752–0.971) | |
| Negative predictive value (95% CI) | 0.717 (0.577–0.832) | 0.760 (0.618–0.869) | 0.729 (0.582–0.847) | 0.787 (0.643–0.893) | 0.703 (0.530–0.841) | 0.800 (0.631–0.916) | 0.784 (0.618–0.902) | 0.727 (0.572–0.850) | |
| Kappa | 0.627 | 0.686 | 0.601 | 0.705 | 0.482 | 0.627 | 0.630 | 0.613 | |
| F1 | 0.813 | 0.848 | 0.812 | 0.863 | 0.769 | 0.839 | 0.835 | 0.810 | |
Among these radiologists, for internal test set, accuracy ranged from 80.0% (95% CI 70.5–87.5) to 84.2% (75.3–90.9), sensitivity from 72.2% (58.4–83.5) to 77.8% (64.4–88.0), and specificity from 85.4% (70.8–94.4) to 92.7% (80.1–98.5). For the external test set, accuracy ranged from 74.4% (95% CI 63.6–83.4) to 81.7% (71.6–89.4), sensitivity from 76.1% (61.2–87.4) to 84.8% (71.1–93.7), and specificity from 72.2% (54.8–85.8) to 80.6% (64.0–91.8). HCCNet achieved an AUROC of 0.899 (95% CI 0.838–0.961) for internal test set, 0.869 (95% CI 0.793–0.945) for external test set (Fig. 2).
As compared with radiologists, the accuracy, sensitivity, specificity, precision, negative predictive value and F1 score of HCCNet were similar on internal test set (0.853 versus 0.818, P = 0.107; 0.815 vs. 0.753, P = 0.064; 0.902 vs. 0.903, P = 0.981; 0.917 vs. 0.911, P = 0.801; 0.787 vs. 0.735, P = 0.056; 0.863 vs. 0.824, P = 0.082). On external test set, besides a slightly higher specificity (0.889 vs. 0.769, P = 0.039), HCCNet also achieved comparable accuracy, sensitivity, precision, negative predictive value, and F1 score with radiologists (0.805 vs. 0.793, P = 0.663; 0.739 vs. 0.812, P = 0.109; 0.895 vs. 0.817, P = 0.061; 0.727 vs. 0.762, P = 0.360; 0.810 vs. 0.814, P = 0.866). Detailed classification metrics for each radiologist were provided in Table 4, Supplementary Tables 1 and 2. Inter-rater agreement rate for this group of three experienced radiologists was 82.1% (78/95, Fleiss’ Kappa 0.761; two-sided z test, P < 0.001) in internal test set, and 62.2% (51/82, Fleiss’ Kappa 0.489; two-sided z test, P < 0.001) in external test set.
Radiologists versus radiologists with AI assistance
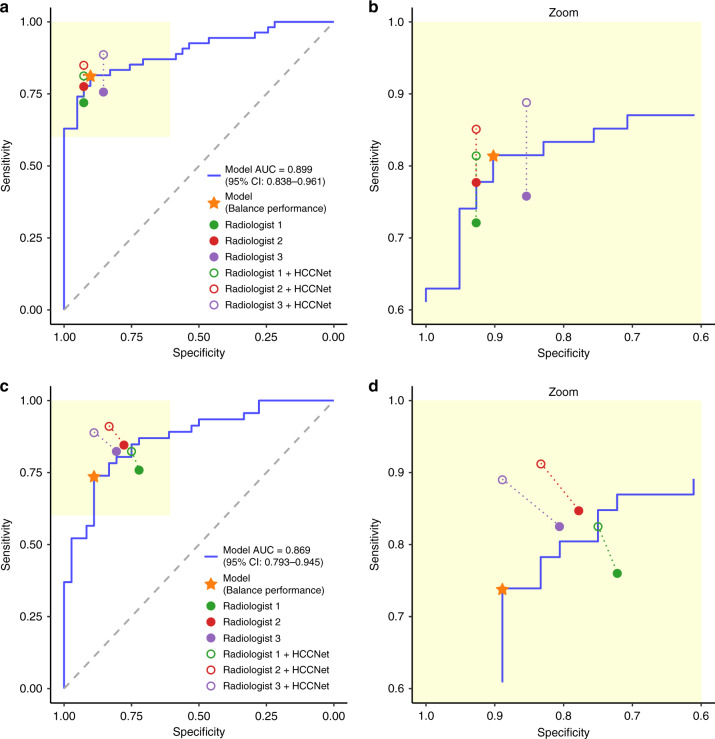
To investigate whether our AI model HCCNet could help radiologists to improve their diagnostic performance, every radiologist was given diagnostic probability result on each case by HCCNet model. These three radiologists were asked to make a diagnosis with the assistance of HCCNet-generated results. The subsequent HCCNet-assisted diagnostic test was performed 12 months after the primary test. Compared with previous results, the follow-up performance by radiologists was significantly improved. Among these radiologists, for the internal test set, the mean accuracy of radiologists was 0.873, which was significantly higher than the previous one (0.873 vs. 0.818 P = 0.026). For the external test set, the mean accuracy of radiologists was also significantly better than the previous one (0.854 vs. 0.793 P = 0.017) (Fig. 4). The classification performance of radiologists with HCCNet assistance is shown in Table 5.
Fig. 4. Comparisons of diagnostic performance by radiologists with and without AI assistance.
The performance of AI model HCCNet and three experienced radiologists on the internal test set (a, b) and external test set (c, d). Three radiologists’ original performances are denoted by filled dots, and the performances with HCCNet assistance are denoted by hollow dots. Dashed lines connected paired performance points of each radiologist. The asterisk denoted the performance of our model in the ‘balanced performance’ setting.
Table 5.
Classification performance of radiologists on subsets of two test sets with AI assistance.
| Performance metrics | Internal test set (n = 95; HCC = 54, non-HCC = 41) | External test set (n = 82; HCC = 46, non-HCC = 36) | ||||
|---|---|---|---|---|---|---|
| Radiologist 1 | Radiologist 2 | Radiologist 3 | Radiologist 1 | Radiologist 2 | Radiologist 3 | |
| Accuracy (95% CI) | 0.863 (0.777–0.925) | 0.884 (0.802–0.941) | 0.874 (0.790–0.933) | 0.793 (0.689–0.874) | 0.878 (0.787–0.940) | 0.890 (0.802–0.949) |
| Sensitivity (95% CI) | 0.927 (0.686–0.907) | 0.927 (0.729–0.934) | 0.854 (0.774–0.958) | 0.750 (0.686–0.922) | 0.833 (0.792–0.976) | 0.889 (0.764–0.964) |
| Specificity (95% CI) | 0.815 (0.801–0.985) | 0.852 (0.801–0.985) | 0.889 (0.708–0.944) | 0.826 (0.578–0.879) | 0.913 (0.672–0.936) | 0.891 (0.739–0.969) |
| Precision (95% CI) | 0.792 (0.825–0.987) | 0.826 (0.831–0.987) | 0.854 (0.774–0.958) | 0.771 (0.667–0.909) | 0.882 (0.748–0.953) | 0.865 (0.788–0.975) |
| Negative predictive value (95% CI) | 0.936 (0.650–0.895) | 0.939 (0.686–0.922) | 0.889 (0.708–0.944) | 0.809 (0.599–0.896) | 0.875 (0.725–0.967) | 0.911 (0.712–0.955) |
| Kappa | 0.727 | 0.767 | 0.743 | 0.578 | 0.751 | 0.778 |
| F1 | 0.854 | 0.874 | 0.854 | 0.761 | 0.857 | 0.877 |
Furthermore, to avoid a potential memorisation bias, we selected dozes of new cases, which did not overlap with the previous two subsets data. These new cases consisted of 42 patients (26 HCC, 16 non-HCC) from internal test set, and 50 patients (23 HCC, 27 non-HCC) from external test set. The performance of radiologists was improved compared to the previous one, but not significantly. Among these radiologists, for internal test set, the mean accuracy of radiologists on new cases was 0.826, which is slightly higher than previous cases, but not significantly different (0.826 vs. 0.818 P = 0.712). For external test set, the mean accuracy of radiologists on new cases was 0.820, also slightly higher, but not remarkably different from previous cases (0.820 vs. 0.793 P = 0.673). Detailed classification metrics for each radiologist are provided in Supplementary Table 3 and Supplementary Fig. 5. Classification performance of radiologists on new cases with HCCNet assistance is shown in Supplementary Tables 4 and 5.
SMOE heatmaps
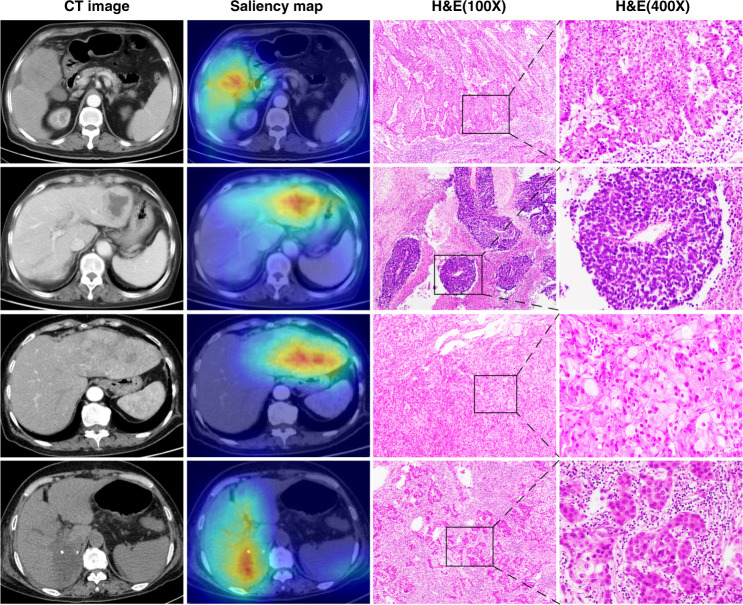
Saliency Map Order Equivalence algorithm was applied to identify image areas that contributed mostly to the prediction results of NoduleNet. In total, 13,868 images of 63 HCC patients were randomly selected to sketch saliency heatmaps. Examples of HCC with accompanying heatmaps and haematoxylin–eosin staining images are shown in Fig. 5 and Supplementary Fig. 6. Non-HCC images such as focal nodular hyperplasia, hemangioma, intrahepatic cholangiocarcinoma, angioleiomyolipoma are displayed in Supplementary Fig. 7. Meanwhile, all the 13,868 randomly selected HCC images and accompanying saliency heatmaps were inspected by five radiologists respectively. The accuracy of heatmaps capturing the main area of HCC tumour nodules was assessed, and the comprehensive percentage was 92.1% (95% CI: 89.2–95.0%). Results of each radiologist are shown in Supplementary Table 6.
Fig. 5. Exemplified images of HCC tumours.
CT images, saliency heatmaps, and haematoxylin–eosin (HE) staining images are displayed correspondingly.
Discussion
To our knowledge, this study consisted of by far the largest number of liver CT images to train a deep-learning model for HCC detection. The developed deep-learning AI system can be a valuable tool for HCC diagnosis and clinical decision-making in high-risk patients. It achieved robust performance across two test sets and comparable accuracy versus a group of three radiologists. Specifically, as the diagnosis of HCC is usually carried out with LI-RADS standard, the standardisation of this model has great potential in reducing the interobserver variability of HCC evaluation.
Deep learning is currently widely used in medical imaging, including disease diagnosis, risk management and clinical decision-making. Radiologists often assess medical images and report their findings based on education level and clinical experience, which sometimes may be subjective. In contrast to physicians’ judgement, the deep-learning model can automatically assess imaging data in a quantitative mode. Efforts have been delivered to the exploration of using deep-learning models for emergency diagnosis, cancer screening and evaluation of tumour treatment effect [23, 24, 30–35]. Chilamkurthy et al. developed a deep-learning approach to automatically identify head CT scan abnormalities in patients with head trauma [23]. Ardila et al. reported high AUC in predicting the risk of lung cancer by using patients’ current and previous computed tomography volumes [24]. More recently, Sun and colleagues used the contrast-enhanced CT images and RNA sequencing data to develop a radiomic signature of CD8 cells to predict immunotherapy response in patients treated with anti-programmed cell death protein (PD)-1 [35].
This study indicated that artificial intelligence might not only provide a potential for standardisation of HCC risk stratification but also can supplement the LI-RADS system. The AASLD guidelines proposed hypervascular arterial profile demonstrated by two dynamic imaging techniques as HCC in patients with cirrhosis [9, 13, 14]. This diagnostic criterion is only restricted to tumour nodules larger than 2 cm in a cirrhotic liver. For nodules smaller than 2 cm, a fine needle aspiration biopsy is recommended. Nevertheless, biopsy usually is not an optimal strategy due to a series of limitations such as pain, bleeding, tumour needle track seeding and repeated biopsies due to negative results [15, 36]. Meanwhile, in patients with cirrhosis, only 14–23% of 1 to 2 cm indeterminate nodules monitored by ultrasound are confirmed as malignant [37]. Although the AASLD suggests several other options other than biopsies, such as follow-up imaging, a different imaging modality and an alternative contrast agent, but could not recommend the best option. Therefore, it is important to classify liver nodules correctly and evaluate their risk accurately, as different nodules warranting different treatment therapies. At present, the LI-RADS categories classify nodules into the different likelihood of HCC in patients with cirrhosis and can be used as a reference in some clinical circumstances [38]. Nonetheless, LI-RADS system is subjectively assessed by the radiologists and the variability of results is inevitable. Therefore, HCCNet model may provide an alternative way to quantify the risk of HCC.
Our study is unique as it integrated the contrast-enhanced CT based HCC diagnostic algorithm into a fully automated processing pipeline which allows radiologists to overcome the bottleneck effect due to un-quantitative analysis. This study consisted of the largest number of liver CT images used for deep-learning algorithm development so far. All patients in the two test sets underwent surgical resection or biopsy and had pathological examination results, which enabled objective evaluation of HCCNet. Our HCCNet produced fairly good and comparable performance on both test sets (AUROC = 0.887 and 0.883). Compared with the expert group, the accuracy of HCCNet was comparable on the internal test set (0.853 vs. 0.818) and external test set (0.805 vs. 0.793). On the image level, NoduleNet also showed relatively high degrees of fidelity, stability and consistency in distinguishing images of tumour nodules from normal images. In addition, the trained NoduleNet model correctly pinpointed malignant HCC tumour nodules through saliency analysis, suggesting that it can serve as an auxiliary tool to help radiologists speed up the interpretation process.
Although the prognosis of HCC patients is closely related to tumour stage, liver function and treatment effect, tumour size is still an inevitable factor associated with clinical outcome. This model demonstrated an unbiased performance in assessing HCC patients with different tumour diameters and AJCC stages. For both the internal and external test sets, the HCC risk scores of HCC patients with tumour size ≤5 cm were comparable with those tumour sizes>5 cm (P = 0.273 and 0.111). This point suggested that HCCNet performed equally well in detecting small tumour nodules as those large tumour nodules. To some extent, it could reduce the probability of unnecessary biopsy for smaller neoplasm masses. Further clinical trials are required to validate the aforementioned HCCNet diagnostic advantage.
Integration of our AI system into the PACS system can assist radiologists in speeding up the interpretation process. However, HCCNet cannot replace radiologists in diagnosing HCC. It only uses CT images for analysis but does not take into account other auxiliary diagnostic parameters, such as viral hepatitis, liver cirrhosis, long-term alcohol intake and aflatoxin. Future deep-learning model taking into account radiological image data, laboratory reports, medical history and pathological graphics will potentially increase the performance of artificial intelligence diagnosis.
Our study has several limitations. Firstly, this AI system was trained based on plain and contrast-enhanced CT images. Theoretically, compared with CT, MRI demonstrates a higher superiority in the diagnosis of liver tumour nodules. In daily clinical practice, CT scan has features of more efficiency, lower cost and higher popularity in rural Chinese hospitals, which may increase the generalisability of HCCNet. With the increasing installation of MRI appliances among Chinese community hospitals, algorithm models based on MRI images will display better performance in future clinical trials. Secondly, our current DCNN model can only distinguish between HCC and non-HCC cases. Although other malignant and benign tumours were included in the training set, this model can only classify two categories, not multiple categories. In the future, we will expand this model to further discern liver metastases, hemangioma, focal nodular hyperplasia, hepatic cyst and other rare neoplasms. Thirdly, patients in the two cohorts are mainly from central China. Variation of patients’ lifestyle and ethics may affect model accuracy and generalisability. Multiregional investigations could potentially mitigate these shortcomings. Fourthly, the present HCCNet did not incorporate other staging parameters, such as hepatic or portal vein invasion, solitary or multiple tumours, regional lymph node status, and distant metastasis. Integrating these variables will provide a promising way to stage HCC tumour accurately and grosso modo, predict the prognosis of patients.
Our results showed that HCCNet can detect hepatocellular carcinoma on liver CT scans with improved accuracy, sensitivity, and specificity at levels similar to a group of experienced radiologists. Given the unbalanced medical resources between community hospitals and large general hospitals, such a model could be a helpful adjunct in underdeveloped areas. We established a website to provide free access to HCCNet. Prospective randomised clinical trials are necessary to further verify the efficacy of HCCNet.
Supplementary information
Author contributions
MW, XL and FT take full responsibility for the integrity of the data and the accuracy of the analysis. Concept and design were contributed by FT, XL and DK. Drafting of the manuscript is attributed to FT, XL and MW. Clinical record data were obtained and reviewed by FF, BZ, YB and XM. Statistical analysis is attributed to YY and HS. FF, BZ, YB, QW, JW and XM read and interpreted CT images. LS, QL and ML extracted and reviewed pathological data. Technical and material support is attributed to PY, XL and YY. We authors thank Genevieve Nemeth of Harvard Medical School for editing the language of this manuscript.
Funding information
This work was supported by the natural science foundation of Tianjin Education Committee (Project No. 2018KJ072), Tianjin Science and Technology Committee (Project No. 18JCQNJC80800), Henan Provincial Science and Technology Research Projects (Project No. 212102310689) and National Natural Science Foundation of China (Project No. 31801117).
Data availability
The original CT data that consist of the training sets and test sets are available on request from the corresponding author. They are not publicly available due to the privacy of research participants.
Code availability
The code used in this study to train AI models is freely accessed and uploaded in supplementary files.
Ethics approval and consent to participate
Not applicable.
Consent to publish
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Meiyun Wang, Fangfang Fu, Bingjie Zheng, Yan Bai.
These authors jointly supervised this work: Meiyun Wang, Fangfang Fu, Bingjie Zheng, Yan Bai.
Contributor Information
Xiangchun Li, Email: lixiangchun@tmu.edu.cn.
Fei Tian, Email: tianfei@tmu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41416-021-01511-w.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492.. [DOI] [PubMed] [Google Scholar]
- 2.El-Serag HB. Hepatocellular carcinoma. N Engl J Med. 2011;365:1118–27. doi: 10.1056/NEJMra1001683.. [DOI] [PubMed] [Google Scholar]
- 3.Lim JH, Kim MJ, Park CK, Kang SS, Lee WJ, Lim HK. Dysplastic nodules in liver cirrhosis: detection with triple phase helical dynamic CT. Br. J Radiol. 2004;77:911–6. doi: 10.1259/bjr/56713551.. [DOI] [PubMed] [Google Scholar]
- 4.Choi JY, Lee JM, Sirlin CB. CT and MR imaging diagnosis and staging of hepatocellular carcinoma: part I. Development, growth, and spread: key pathologic and imaging aspects. Radiology. 2014;272:635–54. doi: 10.1148/radiol.14132361.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laghi A, Iannaccone R, Rossi P, Carbone L, Ferrari R, Mangiapane F, et al. Hepatocellular carcinoma: detection with triple-phase multi-detector row helical CT in patients with chronic hepatitis. Radiology. 2003;226:543–9. doi: 10.1148/radiol.2262012043.. [DOI] [PubMed] [Google Scholar]
- 6.Ippolito D, Sironi S, Pozzi M, Antolini L, Invernizzi F, Ratti L, et al. Perfusion CT in cirrhotic patients with early stage hepatocellular carcinoma: assessment of tumor-related vascularization. Eur J Radiol. 2010;73:148–52. doi: 10.1016/j.ejrad.2008.10.014.. [DOI] [PubMed] [Google Scholar]
- 7.Sahani DV, Holalkere NS, Mueller PR, Zhu AX. Advanced hepatocellular carcinoma: CT perfusion of liver and tumor tissue-initial experience. Radiology. 2007;243:736–43. doi: 10.1148/radiol.2433052020.. [DOI] [PubMed] [Google Scholar]
- 8.Khalili K, Kim TK, Jang HJ, Haider MA, Khan L, Guindi M, et al. Optimization of imaging diagnosis of 1-2 cm hepatocellular carcinoma: an analysis of diagnostic performance and resource utilization. J Hepatol. 2011;54:723–8. doi: 10.1016/j.jhep.2010.07.025.. [DOI] [PubMed] [Google Scholar]
- 9.Bruix J, Sherman M. Management of hepatocellular carcinoma: an update. American Association for the Study of Liver Diseases. Hepatology. 2011;53:1020–2. doi: 10.1002/hep.24199.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.European Association For The Study Of The Liver; European Organisation For Research And Treatment Of Cancer. EASL-EORTC clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol. 2012;56:908–43. doi: 10.1016/j.jhep.2011.12.001.. [DOI] [PubMed] [Google Scholar]
- 11.Omata M, Lesmana LA, Tateishi R, Chen PJ, Lin SM, Yoshida H, et al. Asian Pacific Association for the Study of the Liver consensus recommendations on hepatocellular carcinoma. Hepatol Int. 2010;4:439–74. doi: 10.1007/s12072-010-9165-7.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang A, Bashir MR, Corwin MT, Cruite I, Dietrich CF, Do RKG, et al. Evidence supporting LI-RADS major features for CT- and MR imaging-based diagnosis of hepatocellular carcinoma: a systematic review. Radiology. 2018;286:29–48. doi: 10.1148/radiol.2017170554.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bruix J, Sherman M. Management of hepatocellular carcinoma. Practice Guidelines Committee, American Association for the Study of Liver Diseases. Hepatology. 2005;42:1208–36. doi: 10.1002/hep.20933.. [DOI] [PubMed] [Google Scholar]
- 14.Heimbach JK, Kulik LM, Finn RS, Sirlin CB, Abecassis MM, Roberts LR, et al. AASLD guidelines for the treatment of hepatocellular carcinoma. Hepatology. 2018;67:358–80. doi: 10.1002/hep.29086.. [DOI] [PubMed] [Google Scholar]
- 15.Russo FP, Imondi A, Lynch EN, Farinati F. When and how should we perform a biopsy for HCC in patients with liver cirrhosis in 2018? A review. Dig Liver Dis. 2018;50:640–6. doi: 10.1016/j.dld.2018.03.014.. [DOI] [PubMed] [Google Scholar]
- 16.Roberts LR, Sirlin CB, Zaiem F, Almasri J, Prokop LJ, Heimbach JK, et al. Imaging for the diagnosis of hepatocellular carcinoma: a systematic review and meta-analysis. Hepatology. 2018;67:401–21. doi: 10.1002/hep.29487.. [DOI] [PubMed] [Google Scholar]
- 17.Hosny A, Parmar C, Quackenbush J, Schwartz LH, Aerts HJWL. Artificial intelligence in radiology. Nat Rev Cancer. 2018;18:500–10. doi: 10.1038/s41568-018-0016-5.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bi WL, Hosny A, Schabath MB, Giger ML, Birkbak NJ, Mehrtash A, et al. Artificial intelligence in cancer imaging: Clinical challenges and applications. CA Cancer J Clin. 2019;69:127–57. doi: 10.3322/caac.21552.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rajkomar A, Dean J, Kohane I. Machine learning in medicine. N Engl J Med. 2019;380:1347–58. doi: 10.1056/NEJMra1814259.. [DOI] [PubMed] [Google Scholar]
- 20.Gulshan V, Peng L, Coram M, Stumpe MC, Wu D, Narayanaswamy A, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. J Am Med Assoc. 2016;316:2402–10. doi: 10.1001/jama.2016.17216.. [DOI] [PubMed] [Google Scholar]
- 21.Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542:115–8. doi: 10.1038/nature21056.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li XC, Zhang S, Zhang Q, Wei X, Pan Y, Zhao J, et al. Diagnosis of thyroid cancer using deep convolutional neural network models applied to sonographic images: a retrospective, multicohort, diagnostic study. Lancet Oncol. 2019;20:193–201. doi: 10.1016/S1470-2045(18)30762-9.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chilamkurthy S, Ghosh R, Tanamala S, Biviji M, Campeau NG, Venugopal VK, et al. Deep learning algorithms for detection of critical findings in head CT scans: a retrospective study. Lancet. 2018;392:2388–96. doi: 10.1016/S0140-6736(18)31645-3.. [DOI] [PubMed] [Google Scholar]
- 24.Ardila D, Kiraly AP, Bharadwaj S, Choi B, Reicher JJ, Peng L, et al. End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat Med. 2019;25:954–61. doi: 10.1038/s41591-019-0447-x.. [DOI] [PubMed] [Google Scholar]
- 25.He KM, Zhang XY, Ren SQ, Sun J. Deep residual learning for image recognition. IEEE conference on computer vision and pattern recognition, 2016. Las Vegas, NV, USA; 2016. p. 770–8. 10.1109/CVPR.2016.90
- 26.Russakovsky O, Deng J, Su H, Krause J, Satheesh S, Ma S, et al. ImageNet large scale visual recognition challenge. Int J Comput Vis. 2015;115:211–52. doi: 10.1007/s11263-015-0816-y. [DOI] [Google Scholar]
- 27.Zhang HY, Cisse M, Dauphin YN, Lopez-Paz D. Mixup: beyond empirical risk minimization. International Conference on Learning Representations, 2018. arXiv:1710.09412v2. 2018. Available from: https://arxiv.org/abs/1710.09412
- 28.Mundhenk TN, Chen B, Friedland G. Efficient saliency maps for explainable AI. International Conference on Learning Representations, 2020. arXiv:1911.11293v2. 2019. Available from: https://arxiv.org/abs/1911.11293
- 29.Bedossa P, Poynard T. An algorithm for the grading of activity in chronic hepatitis C. The METAVIR Cooperative Study Group. Hepatology. 1996;24:289–93. doi: 10.1002/hep.510240201.. [DOI] [PubMed] [Google Scholar]
- 30.Hannun AY, Rajpurkar P, Haghpanahi M, Tison GH, Bourn C, Turakhia MP, et al. Cardiologist-level arrhythmia detection and classification in ambulatory electrocardiograms using a deep neural network. Nat Med. 2019;25:65–69. doi: 10.1038/s41591-018-0268-3.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou DJ, Tian F, Tian XD, Sun L, Huang XH, Zhao F, et al. Diagnostic evaluation of a deep learning model for optical diagnosis of colorectal cancer. Nat Commun. 2020;11:2961. doi: 10.1038/s41467-020-16777-6.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ahmad OF, Soares AS, Mazomenos E, Brandao P, Vega R, Seward E, et al. Artificial intelligence and computer-aided diagnosis in colonoscopy: current evidence and future directions. Lancet Gastroenterol Hepatol. 2019;4:71–80. doi: 10.1016/S2468-1253(18)30282-6.. [DOI] [PubMed] [Google Scholar]
- 33.Ehteshami Bejnordi B, Veta M, Johannes van Diest P, Ginneken BV, Karssemeijer N, Litjens G, et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. J Am Med Assoc. 2017;318:2199–210. doi: 10.1001/jama.2017.14585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Byrne MF, Chapados N, Soudan F, Oertel C, Pérez ML, Kelly R, et al. Real-time differentiation of adenomatous and hyperplastic diminutive colorectal polyps during analysis of unaltered videos of standard colonoscopy using a deep learning model. Gut. 2019;68:94–100. doi: 10.1136/gutjnl-2017-314547.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun R, Limkin EJ, Vakalopoulou M, Dercle L, Champiat S, Han SR, et al. A radiomics approach to assess tumor-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: an imaging biomarker, retrospective multicohort study. Lancet Oncol. 2018;19:1180–91. doi: 10.1016/S1470-2045(18)30413-3.. [DOI] [PubMed] [Google Scholar]
- 36.Silva MA, Hegab B, Hyde C, Guo B, Buckels JAC, Mirza DF. Needle track seeding following biopsy of liver lesions in the diagnosis of hepatocellular cancer: a systematic review and meta-analysis. Gut. 2008;57:1592–6. doi: 10.1136/gut.2008.149062.. [DOI] [PubMed] [Google Scholar]
- 37.Khalili K, Kim TK, Jang HJ, Yazdi LK, Guindi M, Sherman M. Indeterminate 1-2-cm nodules found on hepatocellular carcinoma surveillance: biopsy for all, some, or none? Hepatology. 2011;54:2048–54. doi: 10.1002/hep.24638.. [DOI] [PubMed] [Google Scholar]
- 38.Chernyak V, Fowler KJ, Kamaya A, Kielar AZ, Elsayes KM, Bashir MR, et al. Liver imaging reporting and data system (LI-RADS) version 2018: imaging of hepatocellular carcinoma in at-risk patients. Radiology. 2018;289:816–30. doi: 10.1148/radiol.2018181494.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original CT data that consist of the training sets and test sets are available on request from the corresponding author. They are not publicly available due to the privacy of research participants.
The code used in this study to train AI models is freely accessed and uploaded in supplementary files.