Abstract
The emergence of drug-resistant strains of Mycobacterium tuberculosis (Mtb) impedes the End TB Strategy by the World Health Organization aiming for zero deaths, disease, and suffering at the hands of tuberculosis (TB). Mutations within anti-TB drug targets play a major role in conferring drug resistance within Mtb; hence, computational methods and tools are being used to understand the mechanisms by which they facilitate drug resistance. In this article, computational techniques such as molecular docking and molecular dynamics are applied to explore point mutations and their roles in affecting binding affinities for anti-TB drugs, often times lowering the protein’s affinity for the drug. Advances and adoption of computational techniques, chemoinformatics, and bioinformatics in molecular biosciences and resources supporting machine learning techniques are in abundance, and this has seen a spike in its use to predict mutations in Mtb. This article highlights the importance of molecular modeling in deducing how point mutations in proteins confer resistance through destabilizing binding sites of drugs and effectively inhibiting the drug action.
Keywords: mutations, drug resistance, computational tools, Mycobacterium tuberculosis, molecular modeling
Introduction
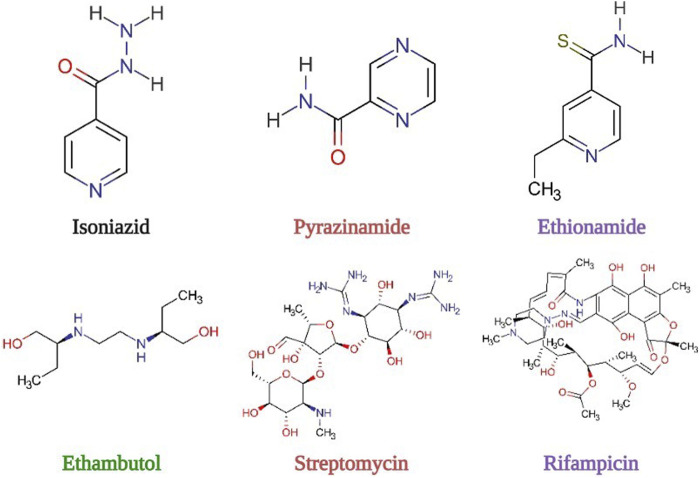
Drug resistance in tuberculosis chemotherapy is fast becoming a health crisis on a global scale. The emergence of multidrug-resistant (MDR), extensively drug-resistant (XDR), and totally drug-resistant (TDR) strains of Mycobacterium tuberculosis (Mtb) has been observed as a result of ineffective directly observed treatment short-course (DOTS) (Bihari et al., 2008; Whalen, 2006) among a myriad of other factors. MDR is due to resistance to at least one first-line drug (Figure 1) including isoniazid (INH) which inhibits mycolic acid synthesis (Bollela et al., 2016) and rifampicin (RIF) that inhibits RNA synthesis (Zhang et al., 2019). Other TB drugs facing resistance include ethambutol (EMB) that targets the arabinogalactan synthesis (Zhang and Yew, 2009), streptomycin (STR) that inhibits protein synthesis (Ruiz et al., 2002), and pyrazinamide (PZA) that inhibits pantothenate and CoA synthesis, disrupting plasma membrane and energy metabolism (Zhang et al., 2014).
FIGURE 1.
Structures of first-line drugs and ethionamide, a second-line drug.
Resistance to first-line drugs leads to the implementation of treatment regiments belonging to the second-line drugs which are fluoroquinolones, kanamycin/amikacin and capreomycin/viomycin, and ethionamide whose mechanisms of action involve introducing negative supercoils in DNA molecules, inhibiting protein synthesis, and disrupting cell wall biosynthesis by inhibiting mycolic acid synthesis, respectively (Table 1). XDR and TDR are, therefore, due to resistance to several second-line drugs including fluoroquinolones in conjunction with MDR. For better management of drug resistance and rapid detection of resistance, knowledge of the mechanism of resistance at the molecular level is extremely important for an effective treatment regimen to be prescribed.
TABLE 1.
Drug targets and the mode of action (Louw et al., 2009; Zhang and Yew, 2009).
| Drug | Target | Gene | Drug mode of action |
|---|---|---|---|
| Ethambutol | Arabinosyl transferase | embCAB | Inhibits arabinogalactan synthesis |
| Streptomycin | Ribosomal protein S12 | rpsL | Inhibits protein synthesis |
| 16S rRNA | rrs | ||
| 7-Methylguanosine methyltransferase | gidB | ||
| Pyrazinamide | Pyrazinamidase | pncA | Disrupts plasma membrane and energy metabolism (inhibits pantothenate and CoA synthesis) |
| Rifampicin | β subunit of RNA polymerase | rpoB | Inhibits RNA synthesis |
| Isoniazid | Fatty acid enoyl acyl carrier protein reductase A | InhA | Inhibits mycolic acid synthesis |
| Catalase peroxidase | katG | ||
| β-Ketoacyl-ACP synthase | kasA | ||
| NADH dehydrogenase | ndh | ||
| Alkyl hydroperoxidase reductase | ahpC | ||
| Ethionamide | Flavin monooxygenase | ethA | Disrupts cell wall biosynthesis by inhibition of mycolic acid synthesis |
| Fatty acid enoyl acyl carrier protein reductase A | InhA | ||
| Transcriptional repressor | ethR | ||
| Kanamycin/Amikacin | 16S rRNA | rrs | Inhibits protein synthesis |
| Capreomycin/Viomycin | rRNA methyltransferase | tlyA | |
| 16S rRNA | rrs | ||
| Fluoroquinolones | DNA gyrase | gyrA | Introduces negative supercoils in DNA molecules |
| gyrB |
More often, drug resistance in Mtb is associated with mutations within the drug targets; however, not all mutations within the organism are associated with resistance. Drug resistance mechanisms are driven mainly by single-nucleotide polymorphisms or other polymorphisms resulting in the modification of drug targets (Palomino and Martin, 2014). Therefore, understanding the mechanism of action and resistance of the drugs is of paramount importance. Of the first-line drugs, ethambutol, which is active against fast-multiplying bacteria, disrupts the synthesis of arabinogalactan in the cell wall by targeting the mycobacterial arabinosyl transferase enzyme encoded by the gene embB, encapsulated in the embCAB operon, and mutations in the embB306 gene confers ethambutol resistance (Zhang and Yew, 2009). On the other hand, streptomycin, a drug active against slow-growing bacteria, irreversibly binds to the 30S ribosome subunit, blocking translation thereby inhibiting protein synthesis. Chromosomally acquired streptomycin resistance is associated with mutations in the rpsL, rrs, and gidB encoding for ribosomal protein S12, 16S rRNA, and 7-methylguanosine methyl transferase, respectively (Zhang and Yew, 2009). Similarly, resistance to rifampicin, a key component in the first-line treatment of TB that binds to the β subunit of RNA polymerase, has been linked to mutations in a region of the 81 bp region of the rpoB gene. Whilst the gene encodes for the β subunit of RNA polymerase, rifampicin resistance is mostly due to mutations at positions 516, 526, and 531 (Goldstein, 2014; Uddin et al., 2020). This is achieved by inhibition of elongation of the messenger RNA, which interferes with transcription (Uddin et al., 2020).
Pyrazinamide is also a key antituberculosis (TB) drug that substantially enhances the activity of novel agents bedaquiline (BDQ) and pretomanid (PA50) in murine models of TB. A vital attribute of this prodrug is its ability to inhibit semidormant bacteria in acidic environments. In its activity, the prodrug is converted by pyrazinamidase/nicotinamidase to its active form, pyrazinoic acid which inhibits membrane transport by disrupting the bacterial membrane energetics. Resistance to pyrazinamide is mainly characterized by mutations clustered at positions 3–17, 61–85, and 132–142 in the pncA gene that codes for mycobacterial enzyme pyrazinamidase (PZase) (Zhang et al., 2014). The association of multiple mutations throughout the pncA gene with PZA resistance makes it difficult to develop a test for detecting PZA resistance (Piersimoni et al., 2013). In most instances, molecular methods are applied to investigate PZA resistance by screening mutations in pncA genes in distinct epidemiological regions offering a much more rapid alternative method compared to that of conventional bacteriology (Khan et al., 2019). Miotto identified 280 mutations in 1950 clinical strains (Miotto et al., 2014), which were categorized into four groups: very high–confidence resistance mutations, high-confidence resistance mutations, mutations with an unclear role, and mutations not associated with phenotypic resistance based on the confidence level.
Isoniazid and ethionamide are effective drugs for the treatment of TB; however, several clinical MDR-TB strains have shown high levels of resistance (Machado et al., 2012). Structurally, INH and ETH are highly similar, both containing the pyridine ring; however, ETH is a second-line drug primarily used to treat MDR-TB, and just like INH, it is a prodrug that requires metabolic activation (DeBarber et al., 2000). Although the active metabolites of both drugs inhibit an NADH-enoyl acyl protein reductase, InhA, the drugs have independent activation pathways. The validated drug target InhA is an enzyme involved in fatty acid biosynthesis II, which is important in the bio-production of mycolic acids. These long-chain fatty acids are responsible for the unique impermeable nature of the Mycobacterium tuberculosis cell wall (Dover et al., 2004; Timmins and Voja, 2006).
INH is activated by the catalase-peroxidase KatG to INH-NAD and INH-NADP adducts that effectively inhibit InhA (Timmins and Voja, 2006). Resistance to INH has been attributed to mutations or deletion in the active site of the katG gene, which encodes the enzyme, KatG (Hameed et al., 2018), at position S315 and position 15 in the InhA promoter region. Also, mutations in ahpC, kasA, and ndh encoding for alkyl hydroperoxidase reductase, β-ketoacyl ACP synthase, and NADH dehydrogenase, respectively, are associated with INH resistance (Nayak et al., 2017). Cross-resistance occurs between INH and its structural analog, and ETH has been attributed to mutations in the InhA promoter.
On the contrary, ETH is activated by the enzyme EthA encoded by the gene Rv3854c to the toxic S-oxide then to 2-ethyl-4-aminopyrimidine (DeBarber et al., 2000; Baulard et al., 2000). The transcription of the FAD-containing monooxygenase, EthA, is controlled by another gene ethR that encodes the protein, EthR. Earlier studies of the resistance mechanism of ethionamide revealed that an increase in the amount of EthR, a member of the TetR repressors, reduces the amount of EthA and results in ethionamide resistance by mycobacterium tuberculosis (DeBarber et al., 2000; Baulard, 2000). Mutation studies on MDR-TB isolates revealed the presence of EthR F110L mutants implicated in resistance to ETH. The residue F110 occupies a central position in the long cylindrical and hydrophobic ligand-binding site of EthR.
Similar to INH, ethionamide (ETH) is a second-line prodrug activated by the monooxygenase encoded by the ethA gene. Once activated, it forms an adduct with NAD, which inhibits the enzyme enoyl-ACP reductase, thus disrupting mycolic acid synthesis. Transcription of the monooxygenase, ethA is negatively regulated by ethR; hence, allosteric inhibition of ethR would enhance activation of ETH and computer some of the mutation processes.
The advances in computational techniques and expansions in bioinformatics and chemoinformatics have brought a sigh of relief in the study of mutations and provided a rapid drug susceptibility testing important in the detection and control of MDR/XDR TB (Shinnick et al., 2005). Therefore, in this article, we analyze the effective application of computational techniques and tools in the study and understanding of molecular target mutations in conferring drug resistance to first-line drugs and also analyze how we are applying these methods to identify inhibitors that would circumvent resistance in ethR, a gene implicated in the resistance of ethionamide as well as highlight prospects in fast and cost-effective advances to understand drug resistance of antituberculosis drugs.
Method
To give a detailed account of how the computational techniques have been applied in the study of the contributions of mutations to the emergence of drug-resistant Mycobacterium tuberculosis, an extensive literature search was performed. A description of the mechanisms of action of the first-line drugs rifampicin and isoniazid as well as ethionamide, a second-line drug is given. An analysis of the common computational methods used to study the mutations in relevant genes for each drug was performed. Lastly, a detailed account of the importance of F110 in ethR, a transcription regulator implicated in the resistance of ethionamide, is presented. Modeling of the proteome for mycobacteria, and identification of the hotspots and druggability of the proteins are given.
Computational Approaches
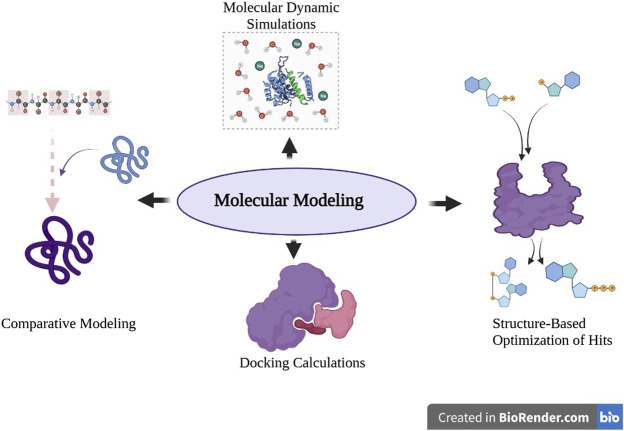
A variety of computational techniques that include comparative (homology) modeling, molecular dynamics, protein–ligand docking, and structure-based optimization of ligands (Figure 2) have been successfully used to study the impact of mutations at atomic levels on protein–ligand binding and interactions and how they negatively affect ligand affinity by the mutant proteins (Phelan et al., 2016; Zhang et al., 2019; Jamal et al., 2020). Advanced approaches that include machine learning alongside artificial intelligence, bioinformatics, and cheminformatics databases have also been successfully used to build models and tools that can predict mutation and determine their capabilities in conferring resistance (Jamal et al., 2020; Ghosh et al., 2020; Sandgren et al., 2009).
FIGURE 2.
Common computational approaches applied to study the effect of mutations on protein–ligand interactions.
Effect of Mutation in rpoB on Protein–RIF Interactions
Pang and co-workers approached RIF mutations with a computational approach. They used homology modeling to generate a three-dimensional structure of the wild type rpoB based on the crystal structure of Thermus aquaticus (Taq) core RNAP complexed with RIF. Discovery Studio 3.1 was used for this structural analysis exercise. The protein was modeled using a Build Homology module within the Protein Data Bank; a loop refinement module from Modeller was used to perform structural refinements, and energy minimizations were performed with the Smart Minimizer algorithm. The Build Mutants module was used for building mutants Ser531Leu, His526Asp, His526Gly, His526Leu, His526Arg, and Leu533Pro, and the Align and Superimpose Proteins module was used to compare the wild-type and mutant structures. Their study sought to evaluate the effects of mutating specific amino acid residues involved in the binding of RIF on protein–ligand interactions. The mutated protein–ligand interactions are evaluated subsequently using the Analyze Ligand Interactions and Structure Monitor module. They discovered that the mutated target protein had some level of resistance for RIF as it showed a decrease in its binding affinity. Mutations in His526Asp and Ser531Leu significantly reduced the affinity of rpoB for RIF by introducing charge repulsion and conformational changes in rpoB, respectively. The other strains with mutations His526Gly, His526Leu, His526Arg, and Leu533Pro exhibited low-level resistance (Pang et al., 2013). On the other hand, Zhang approached this challenge in exploring resistance mechanisms by combining the molecular dynamics simulation, molecular mechanics generalized-Born surface area calculation, dynamic network analysis, and residue interaction network analysis. Molecular dynamics simulations were all performed with the Amber14 package, and it was observed that the binding free energies of RIF with the three mutants H451D/Y/R decreased with molecular mechanics generalized-Born surface area calculations. Dynamic network analysis and residue interaction network analysis indicated increased flexibility within the binding pocket due to mutation of residue 451 which in turn weakened Q438, F439, M440, D441, and S447 residue interactions within the binding pocket. Such flexibility allowed for residues meant that a hydrogen bond to RIF was lost, thus accounting for decreased RIF binding in the mutant RNA polymerase. Changes within the binding pocket in the H451R mutant are extensive, giving too much freedom for RIF to move within the pocket (Zhang et al., 2019). Therefore, H451D/Y/R mutations increased the flexibility of the active pocket which in turn weakened the binding ability of Mtb RNA polymerase with RIF. Thus, the H451D/Y/R mutations weaken the interaction of the mutated residue with its adjacent residues. In similar work involving homology modeling of rpoB and docking calculations of RIF, Kumar and Jena have shown that two mutants S450L and H445Y exhibit low binding affinity toward the wild type rpoB, which has high affinity for the RIF molecule (Kumar and Jena, 2014).
Singh and co-workers investigated mutations of H451. The Mtb rpoB sequence was obtained from UniProt, the structure was built through comparative modeling with Modeller, and it was mutated computationally at position 451 using PyMol. GROMACS version 5.0 molecular dynamics simulation was performed on all the structures to obtain stable structures at 40ns. On the stable structures, RIF was docked onto them with AutoDock 4.2, and ligand–RIF complexes were subjected to molecular dynamics and molecular mechanics for estimation of free binding energies in wild-type and mutant systems. Resistance in the mutants arises due to changes within the binding pocket when polar and hydrophobic amino acids were replaced, which affected packing and folding in the vicinity, and relocation of the binding site itself rendering the RNA exit channel inaccessible to the drug (Singh et al., 2017).
The aforementioned studies on RIF resistance all have a consensus on the conference of resistance by the mutations in the target protein. They showed that mutations in rpoB cause structural changes within the binding pocket and its vicinity. They also indicated that interactions between the binding pocket residues are changed as a result of a mutation within the binding pocket and its vicinity greatly affecting the location and structure of the binding pocket. Most of these studies concluded that mutations that cause extensive structural changes will affect the way RIF sits in the binding pocket and increase freedom for the ligand in the pocket, which greatly decreases its affinity. Mutations that specifically occur within the binding pocket starve the RIF ligands of residues that contribute to a better binding affinity.
Effect of Mutation in InhA, and katG on INH Binding
Computational studies of INH resistance in Mtb have been extensively studied (Jena et al., 2014). INH is activated by katG and converted to an active intermediate displaying antimycobacterial properties; in the presence of NADH, an INH-NAD adduct is formed. It is the adduct that inhibits InhA (2-trans-enoyl-acyl carrier protein reductase), blocking the synthesis of mycolic acid (Dookie et al., 2018). In one study, homology modeling was employed to predict the 3D structure of Mtb UDP-galactopyranose mutase (Glf) and NADH Dehydrogenase (Ndh) with Modeller9v14, and the sequence in the FASTA format was obtained from the NCBI database. The NAD binder server was used in the identification of the binding site; docking studies and visualization were performed with AutoDock Vina Tool 1.5.4 and Pymol, respectively. FADH2 and NADH were both found to have a high affinity for Glf; thus, overexpression of Glf utilizes more NADH reducing its concentration. This results in decreased INH-NAD adduct formation thereby causing INH resistance (Nayak et al., 2017)
In another study, on the influence of mutation in INH, katG mutations S315T/S315N were modeled with Modeller9v10 and compared with the wild-type katG. It was observed that INH was forming a hydrogen bond with the mutant katG which hindered radical formation. AutoDock Tool 1.5.4 docking calculation indicated INH-NAD is more effective at inhibiting InhA compared to INH (Jena et al., 2014). The katG mutation S315T was computationally observed to decrease the flexibility of binding site residues, and katG mutants at His276Met, Gln295His, and Ser315Thr decreased the stability and flexibility of the mutant protein associated with INH resistance. Mutation of the arylamine N-acetyltransferase (NAT) enzyme increases the stability and catalytic activity of the enzyme making the NAT-INH interaction ineffective. Mutations in the ahpC result in overexpression of the protein, which is a compensatory mechanism for loss of activity due to the katG mutation; thus, the ability to defend against oxidative stress is maintained within the system (Jena and Wankhade, 2016; Waghmare, and Harinath, 2016). Just as in the RIF studies, conformational changes and pocket flexibility changes greatly affect the atomic-level interactions between the target protein and the drug compound, and the trend shows a decreased affinity for the drug by mutant protein targets.
Other Studies on the Effect of Mutation Mtb Drug Resistance
Deedler applied machine learning approaches to Mtb isolates that had undergone whole-genome sequencing. Nonparametric classification tree and gradient-boosted tree models were used to predict drug resistance alongside uncovering any associated new mutations. Resistance markers to drugs other than the drug of interest was used in fitting separate drug models for each drug based on the presence and absence of the co-occurrent resistance markers. Predictive performance testing was performed alongside laboratory drug-susceptibility testing. The performance was highest for resistance to first-line drugs, amikacin, kanamycin, ciprofloxacin, moxifloxacin, and multidrug-resistant tuberculosis. The inclusion of resistance markers led to improved results (Deelder et al., 2019).
In a bid to understand the molecular consequences of polymorphisms within loci associated with antituberculosis drugs, Portelli and co-workers employed computational methods to quantify point mutations in conferring resistance. Homology models of target proteins were built with UCSF Chimera 1.1, and protein–ligand docking and protein–ligand interactions were carried out with GLIDE and Arpeggio, respectively. Portelli et al., 2018 concluded that mutational effects are mostly imparted via steric or electrostatic changes within the protein, leading to functional changes and affecting target–drug interactions. They also noted that most phenotypically resistant mutations act allosterically, and the introduction of variants affects the drug–protein complex stability, leading to resistance. Frequently occurring mutations do not confer extreme changes in parameters; the protein retains its functionality, but the drug–protein complex is weakened. Mildly stabilizing mutations may confer local fitness advantages. Drug-resistant mutations within the protein are enhanced while maintaining stability within the protein function. It was also concluded that concurrent mutations in close topological proximity enable localized effects of the mutation, and their combination with external mutations ensures different mechanisms that lead to drug resistance.
Nakatani and Helen, 2017 predicted that the alr M319T mutation observed in an XDR strain of Mtb would likely confer resistance to D-cycloserine (DSC) as it had been noted that the acquisition of this mutation occurred with treatment of DSC suggesting that the mutation is sufficient and necessary to confer resistance. Molecular modeling of the C-8T, M319T, Y364D, and R373L mutations provided insights into how resistance is conferred upon treatment with DSC. DSC covalently binds to an alr cofactor pyridoxal 5′-phosphate (PLP); this act irreversibly inhibits alr through disruption of the alr-PLP covalent bond (Fenn et al., 2003). A generated model of Mtb, alr, and DSC highlighted the residues 319 and 364 located directly in the active site. A mutation to aspartic acid at residue 364 introduced a shorter negatively charged side chain. Such a change affects the positioning of the phosphate moiety in PLP, potentially affecting PLP orientation in the active site. The location of the residue 319 mutation could alter the interactions with 364, likely affecting DSC inhibition. Alr functions as a homodimer, and the R373L mutation is not located directly in the active site; however, it is close to M319 and D320 and the dimer interface. Such a mutation is most likely going to disrupt molecular interactions at the dimer interface and greatly destabilizing the DSC binding site. This study was strategic to the pharmaceutical sector in 2015 amidst a Global Drug Facility declaration of a price reduction of the DSC drug. Understanding the resistance mechanisms was important for facilitating phenotypic and genotypic drug susceptibility testing (Stop, 2015).
Malik et al. (2012) provided insights into fluoroquinolone resistance through functional genetic analyses and structural modeling techniques. Crystal structures of the N-terminal and C-terminal domains for gyrA and gyrB were superimposed on the crystal structure of the complex of Streptococcus pneumoniae gyrase with a DNA substrate and levofloxacin, all obtained from the Protein Data Bank using the tool Coot (Emsley and Kevin, 2004). This study highlights that gyrB mutations M330I, V340L, R485C, D500A, D533A, A543T, A543V, and T546M are not sufficient to confer drug resistance. N538D, E540V, and R485C + T539N mutations did confer resistance to all fluoroquinolones whilst N538K and E540D conferred resistance to moxifloxacin only, and D500H and D500N mutations conferred resistance only to levofloxacin and ofloxacin. The importance of this study was in explaining minimum inhibitory concentrations as observed in experimental work; molecular modeling explained how resistance came about to be through a 3D spatial orientation of substitute residues in the mutant proteins.
Effect of Mutation on ethR-Ligand Binding Affinity
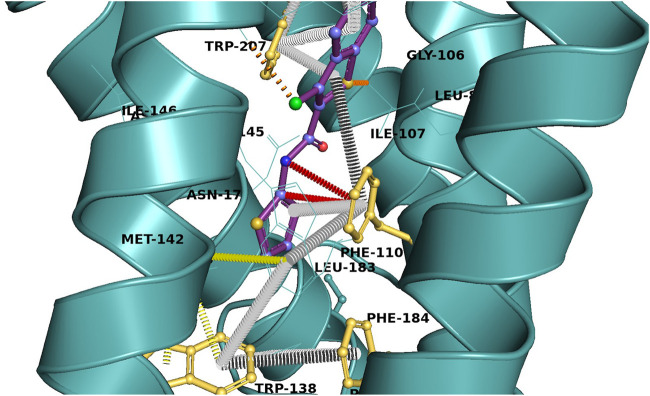
Resistance to ETH has been linked to mutations in the ethR, ethA, and inhA genes (Hameed et al., 2018) that collectively play crucial roles in the activity of the drug. As a regulator, the N-terminus helix-turn-helix (HTH) domains of the dimeric EthR bind DNA sequences responsible for the transcription of EthA and suppress its expression (Wolff and Nguyen, 2012). This process is controlled by small–molecular weight ligands that bind to the allosteric binding pocket of EthR located in the C-terminal end (Mugumbate et al., 2015). Binding of the ligands induces molecular conformational changes that increase the distances between DNA binding domains of the enzyme, inhibit DNA binding, and hence increase the transcription of EthA. For this reason, EthR has been validated as a suitable drug target for a new collection of antituberculosis compounds that would boost the activity of ETH. Targeting the resistance pathway of antituberculosis drugs has long been proposed (Wolff and Nguyen, 2012); therefore, independent research groups have deposited the apo and bound structures of EthR into the Protein Data Bank (PDB, https://www.rcsb.org/). These structures reveal that the protein is characterized by a long hydrophobic and promiscuous pocket that binds to structurally diverse small molecules like dioxane and long molecular chains with more than 30 atoms. The residue F110 is centrally positioned in the binding pocket with its aromatic side chain strategically positioned to participate in protein–ligand interactions (Figure 3).
FIGURE 3.
The residue F110 facilitates the pi–pi cascade (grey rings) between aromatic residues in the binding pocket of EthR (yellow) and the ligand (purple), which later translates into a structural modification of the HTH motif and inhibition of DNA binding. Analysis of the interactions was performed using Aperggio (http://bleoberis.bioc.cam.ac.uk/arpeggioweb/) and viewed using PyMol.
In a previous study (Bishi, et al.), we carried out docking calculations of a Maybridge dataset containing more than 200 drug-like compounds to investigate binding modes and protein–ligand interactions. The results indicated that F110 played a crucial role in ligand binding, supporting the observation that F110L drastically reduces ligand affinity (Brossier et al., 2011). Most ligands were stabilized by a cascade of pi–pi interactions, where F110 played a central role by linking pi–pi interactions from the ligand to Phe114 (Figure 3) in a way that will stabilize the bound ligand and increase ligand affinity. This implies that the F110L mutation disrupts the pi–pi cascade and reduces the ligand–binding affinity.
Application of Machine Learning and Artificial Intelligence Approaches
Bioinformatics was employed for studies concerned with mutations focusing on Mtb. Ghosh and co-workers developed a Drug Resistance–Associated Genes database (DRAGdb) which is a repository of mutational data of drug resistance–associated genes (DRAGs) across ESKAPE (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp.). Homoplasy is observed in six genes namely gidB, gyrA, gyrB, rpoB, rpsL, and rrs with mutations related to drug resistance being observed at the codon level. A single-nucleotide mutation that was associated with resistance to amikacin, gentamicin, rifampicin, and vancomycin in Staphylococcus aureus was an indication of pleiotropy. The database compiles Mtb drug-resistance genes across bacterial species allowing for homoplasy and pleiotropy identification in genes (Ghosh et al., 2020).
In their recent efforts, Jamal and co-workers developed machine learning algorithms alongside artificial intelligence to study and predict resistance in the genes rpoB, inhA, katG, pncA, gyrA, and gyrB for the drugs rifampicin, isoniazid, pyrazinamide, and fluoroquinolones. Machine learning algorithms naïve Bayes, k nearest neighbor, support vector machine, and artificial neural network were used to build the prediction models. Further molecular docking and molecular dynamics simulations were carried out on predicted resistance causing mutant proteins and their wild-type counterparts. This study evaluated protein conformation and its impact to confirm the observed phenotype (Jamal et al., 2020).
Discussion
The application of machine learning and artificial intelligence in mutation studies is a fast-growing trend in computational research. At the center of it all, bioinformatics and cheminformatics databases are contributing a lot of information that is needed by machine learning algorithms to predict drug resistance–conferring mutations. Information was gathered across species in the experimental work, where the previously mentioned mutations and their influence on drug resistance were observed in detail, and the lack of hereafter is used to train machine learning algorithms in identifying possible novel mutations that might occur and probe their potential in conferring resistance. Usage of multiple layers or algorithms (deep learning) and artificial intelligence has greatly improved the accuracy of drug resistance and mutation prediction tools that have been made available to researchers (Deelder et al., 2019; Jamal et al., 2020).
Structure-guided drug discovery has lately become paramount to combat the emergence of Mtb drug-resistant strains which pose a concern to global public health. The rapid expansion of genome sequencing and pathway annotations has shown a positive impact on the progress of drug discovery. Computational tools have been developed to address the effect of mutations on the structure and function of proteins. The mutation cutoff scanning matrix (mCSM) is a machine learning approach which predicts the structural and functional effects of mutations on the target proteins. Its variants are capable of predicting the effects of mutations on protein stability, protein–protein interaction, and protein–ligand interactions (Pandurangan and Blundell 2020). EnCOM and FoldX are tools that are capable of predicting the effects of mutations on flexible protein conformations (Schymkowitz et al., 2005; Frappier et al., 2015). Rapid assessment of many mutations that are difficult to access with experimental methods has been made possible through predictive learning with machine learning algorithms (Waman et al., 2019).
Machine learning techniques have also been developed to address the need to improve TB resistance prediction in less-studied drugs. Rapid detection of antimicrobial resistance is vital in the prevention of existing drug resistance amplification, given that resistance markers are known; machine learning techniques are capable of timely prediction of resistance for a given Mtb drug. Machine learning methods are capable of ranking mutations regarded as important and mutations from other genes associated with resistance to other drugs; this reflects on multidrug resistance from taking second-line drugs after taking first-line drugs, which is a huge advantage over experimental methods (Kouchaki et al., 2019).
DeepAMR has been developed with the task of identifying co-occurrent resistance within anti-TB drugs. This machine learning technique had a high performance with mean AUROC (Area Under the Receiver Operating Characteristics) from 94.4 to 98.7% for predicting resistance to four first-line drugs, RIF, EMB, INH, and PZA multi-drug resistant TB (MDR-TB) and pan-susceptible TB (PANS-TB: MTB that is susceptible to all four first-line anti-TB drugs). DeepAMR achieved its best mean sensitivity of 94.3, 91.5, 87.3, and 96.3% for INH, EMB, PZA, and MDR-TB, respectively. High-performance machine learning models have made the predictions of co-occurrent drug resistance to be performed timely and prevented amplification of existing resistance (Yang et al., 2019).
The use of machine learning and artificial intelligence makes them possible to identify novel resistance markers which are very difficult and costly to investigate with experimental methods. The timely and rapid prediction of drug resistance has made it possible for drugs to be returned to the discovery pipeline for optimization in a structure-guided drug design approach. To this end, the application of these techniques makes it possible for scientists to comprehensively study the protein–drug interactions at very little cost and shorter time frames.
Proposed Computational Protocol
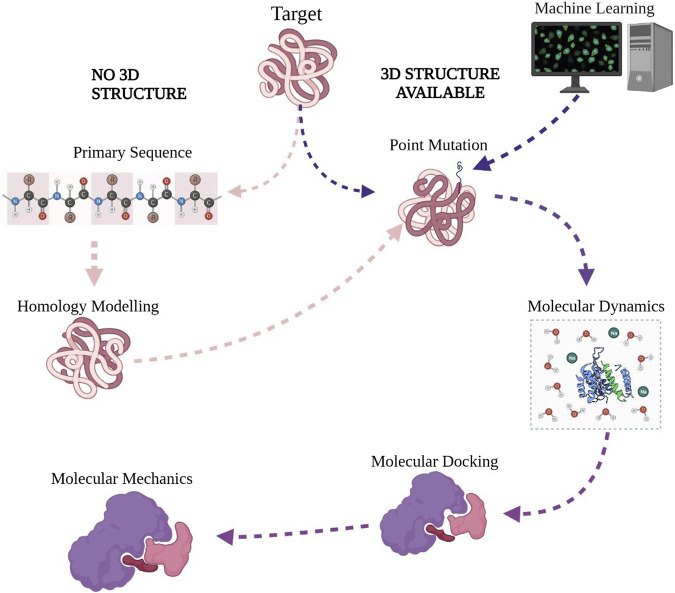
The Application of computational tools (Table 2) in understanding mutations that confer drug resistance in Mycobacterium tuberculosis still require a canonization of the process for a standard result output. Initially, 3D structures of drug targets are obtained from the Protein Data Bank followed by point mutations which may be performed by changing an amino acid in a protein sequence with Pymol (Figure 4). In the absence of a 3D structure, the primary sequence of the protein is obtained from UniProt (a freely accessible database of protein sequences and functional information). 3D structures are modeled through a process known as homology/comparative modeling of proteins with a standalone program such as Modeller or, alternatively, an online server such as SWISS-MODEL (expasy.org). Molecular dynamics simulations are performed for energy minimizations of the wild-type and mutant drug targets obtaining the most stable protein structures; standalone programs such as GROMACS and Amber are used for performing the task (Singh et al., 2017; Zhang et al., 2019). In computational chemistry, energy minimizations which may also be referred to as geometry optimization entail the exploration of the conformational space for a collection of atoms to find a proper molecular arrangement in space which is energy favorable and stable; it is also referred to as the global energy minimum (Jabeen et al., 2019). The resultant structures are then subjected to molecular docking, where the position of the ligand when bound to a protein receptor is predicted for the drug’s wild type and mutated targets. AutoDockTools and Glide among other standalone software packages may be used for this task (Kumar and Jena 2014; Jamal et al., 2020). Protein–ligand complex structures may also undergo energy minimization with molecular dynamics (Kumar and Jena, 2014; Portelli et al., 2018). In the presence of a 3D structure complexed with the preferred drug, molecular mechanics is employed to probe free binding energies and compare protein–ligand complexes for wild-type and mutated drug targets (Zhang et al., 2019). Recent trends that are being explored in the field of computational work include the usage of machine learning algorithms to build prediction tools (Lee et al., 2020). Studies that make use of mathematical models alongside bioinformatics for drug resistance mutations have also been reported (Fonseca et al., 2015). There has also been an exploration of artificial intelligence alongside machine learning algorithms for drug resistance mutation predictive tests (Deelder et al., 2019).
TABLE 2.
Computational tools used in the study of mutations.
| Computational tool | Use | References |
|---|---|---|
| AutoDock | Molecular docking and visualization | Morris et al. (2009) |
| Glide | Molecular docking and visualization | Friesner and Mainz (2006) |
| Pymol | Molecular visualization | DeLano (2002) |
| Gromacs | Molecular dynamics simulations | Van Der Spoel et al. (2005) |
| Amber | Molecular dynamics simulations | Case et al. (2005) |
| Modeller | Homology or comparative modeling of protein 3D structures | Webb and Sali (2016) |
| Discovery Studio | Molecular visualization | Studio (2008) |
| Arpeggio | A web server for calculating and visualizing interatomic interactions in protein structures | Jubb et al. (2017) |
| UCSF Chimera | Interactive visualization and analysis of molecular structures and related data | Pettersen et al. (2004) |
| DeepAMR | Predicting co-occurrent resistance of Mycobacterium tuberculosis | Yang et al. (2019) |
| EnCom | Predicting the effects of mutations on flexible protein conformations | Frappier et al. (2015) |
| FoldX | Schymkowitz et al. (2005) |
FIGURE 4.
Proposed protocol for studying the effect of mutations on protein–ligand interaction.
Concluding Remarks
With the increase in the number of drug-resistant and multidrug-resistant strains of Mtb, a need has arisen for techniques that are rapid for extensive studies of the previously mentioned mutations. Computational methods (Figure 4) present us with the opportunity to rapidly carry out these studies in silico with outputs comparable with experimental work at high confidence at even lower costs. Such methods have been extensively employed in exploring drug resistance in rifampicin, isoniazid, and ethionamide with the findings correlating to what is observed in experimental work; structural changes within the mutant protein drastically reduce protein–ligand binding affinity.
Machine learning and artificial intelligence have brought about massive changes and advancements in studying mutations and drug resistance in Mtb and other diseases. These techniques have made it possible to identify resistance markers within the whole genome Muzondiwa et al., 2020, to predict drug resistance for a given molecule, and to predict co-occurrent drug resistance between two or more drugs. The techniques are driven by big data (Table 3), and to that effect, smaller specific repositories/databases (Drug Resistance–Associated Genes database) have been created for the sole purpose of helping researchers who are studying mutations. Computational tools have also been created for the identification of resistance markers and prediction of drug resistance (DeepAMR). Predictive learning makes it possible for scientists to identify potentially unwanted drug characteristics that may not be picked up with experimental methods, greatly reducing the risk for drug failure and saving time and money in the process.
TABLE 3.
Databases used alongside computational packages.
| Database | Information contained | References |
|---|---|---|
| UniProt | Protein sequence and functional information | Consortium (2015) |
| Protein Databank | Protein 3D Structures | Berman et al. (2000) |
| DRAGdb | Mutational data of drug resistance–associated genes | Ghosh et al. (2020) |
| NCBI | Biological data and small-molecule database | Wheeler et al. (2006) |
| ChEMBL | Binding, functional, and ADMET information for a large number of drug-like bioactive compounds | Gaulton and LouisaBellis (2012) |
Author Contributions
BN and AM wrote the manuscript, and GrM and GaM were involved in concept development and editing the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Baulard A. R., Joanna C. B., Jean E.-N., Selwyn Q., Ruth A. M., Patrick J. B., et al. (2000). Activation of the pro-drug ethionamide is regulated in mycobacteria. J. Biol. Chem. 275 (36), 28326–28331. 10.1074/jbc.M003744200 [DOI] [PubMed] [Google Scholar]
- Berman H. M., Westbrook J., Feng Z., Gilliland G., Bhat T. N., Weissig H., et al. (2000). The Protein Data Bank. Nucleic Acids Res. 28 (1), 235–242. 10.1093/nar/28.1.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bihari S., Arun S., Rawat T., Katiyar S. (2008). An Analysis of Failure of Category II DOTS Therapy. Indian J. Community Med. 33 (2), 129. 10.4103/0970-0218.40886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brossier F., Veziris N., Truffot-Pernot C., Jarlier V., Sougakoff W. (2011). Molecular investigation of resistance to the antituberculous drug ethionamide in multidrug-resistant clinical isolates of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 55 (1), 355. 10.1128/AAC.01030-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollela V. R., Namburete E. I., Feliciano C. S., Macheque D., Harrison L. H., Caminero J. A. (2016). Detection of KatG and InhA Mutations to Guide Isoniazid and Ethionamide Use for Drug-Resistant Tuberculosis. Int. J Tuberc. Lung Dis. 20 (8), 1099–1104. 10.5588/ijtld.15.0864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case D. A., Cheatham T. E., III, Darden T., Gohlke H., Luo R., Merz K. M., et al. (2005). The Amber Biomolecular Simulation Programs. J. Comput. Chem. 26 (16), 1668–1688. 10.1002/jcc.20290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium U. P. (2015). UniProt: a Hub for Protein Information. Nucleic Acids Res. 43, D204–D212. 10.1093/nar/gku989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBarber A. E., Mdluli K., Bosman M., Bekker L.-G., Barry C. E. (2000). Ethionamide Activation and Sensitivity in Multidrug-Resistant Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. 97 (17), 9677–9682. 10.1073/pnas.97.17.9677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deelder W., Christakoudi S., Phelan J., Benavente E. D., Campino S., McNerney R., et al. (2019). Machine Learning Predicts Accurately Mycobacterium Tuberculosis Drug Resistance from Whole Genome Sequencing Data. Front. Genet. 10 (SEP), 1–9. 10.3389/fgene.2019.00922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano W. L. (2002). Pymol: An Open-Source Molecular Graphics Tool. CCP4 Newsl. Protein Crystallogr. 40 (1), 82–92. [Google Scholar]
- Dookie N., Rambaran S., Padayatchi N., Mahomed S., Naidoo K. (2018). Evolution of Drug Resistance in Mycobacterium Tuberculosis: A Review on the Molecular Determinants of Resistance and Implications for Personalized Care. J. Antimicrob. Chemother. 73 (5), 1138–1151. 10.1093/jac/dkx506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dover L. G., Cerdeño-Tárraga A. M., Pallen M. J., Parkhill J., Besra G. S. (2004). Comparative Cell wall Core Biosynthesis in the Mycolated Pathogens,Mycobacterium tuberculosisandCorynebacterium Diphtheriae. FEMS Microbiol. Rev. 28 (2), 225–250. 10.1016/j.femsre.2003.10.001 [DOI] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. (2004). Coot: Model-Building Tools for Molecular Graphics. Acta Crystallogr. D Biol. Cryst. 60 (12), 2126–2132. 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- Fenn T. D., Stamper G. F., Morollo A. A., Ringe. D. (2003). A Side Reaction of Alanine Racemase: Transamination of Cycloserine. Biochemistry 42 (19), 5775–5783. 10.1021/bi027022d [DOI] [PubMed] [Google Scholar]
- Fonseca J. D., Knight G. M., McHugh T. D. (2015). The Complex Evolution of Antibiotic Resistance in Mycobacterium Tuberculosis. Int. J. Infect. Dis. 32, 94–100. 10.1016/j.ijid.2015.01.014 [DOI] [PubMed] [Google Scholar]
- Frappier V., Chartier M., Najmanovich R. J. (2015). ENCoM Server: Exploring Protein Conformational Space and the Effect of Mutations on Protein Function and Stability. Nucleic Acids Res. 43, W395–W400. 10.1093/nar/gkv343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesner R. A., Murphy R. B., Repasky M. P., Frye L. L., Greenwood J. R., Halgren T. A., et al. (2006). Extra Precision Glide: Docking and Scoring Incorporating a Model of Hydrophobic Enclosure for Protein−Ligand Complexes. J. Med. Chem. 49 (21), 6177–6196. 10.1021/jm051256o [DOI] [PubMed] [Google Scholar]
- Gaulton A., BellisBellis L. J. A., Bento A. P., Chambers J., Davies M., Hersey A., et al. (2012). ChEMBL: a Large-Scale Bioactivity Database for Drug Discovery. Nucleic Acids Res. 40, D1100–D1107. 10.1093/nar/gkr777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A., N. S., Saha. S. (2020). Survey of Drug Resistance Associated Gene Mutations in Mycobacterium Tuberculosis, ESKAPE and Other Bacterial Species. Sci. Rep. 10 (1), 1–11. 10.1038/s41598-020-65766-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein B. P. (2014). Resistance to Rifampicin: A Review. J. Antibiot. 67, 625–630. Nature Publishing Group. 10.1038/ja.2014.107 [DOI] [PubMed] [Google Scholar]
- Hameed H. M. A., Islam M. M., Chhotaray C., Wang C., Liu Y., Tan Y., et al. (2018). Molecular Targets Related Drug Resistance Mechanisms in MDR-, XDR-, and TDR-Mycobacterium Tuberculosis Strains. Front. Cel. Infect. Microbiol. 8, 114. 10.3389/fcimb.2018.00114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabeen A., Mohamedali A., Ranganathan S. (2019). “Protocol for Protein Structure Modelling,” in Encyclopedia of Bioinformatics and Computational Biology. Oxford: Academic, 252–272. 10.1016/B978-0-12-809633-8.20477-9 [DOI] [Google Scholar]
- Jamal S., Khubaib M., Gangwar R., Grover S., Grover A., Hasnain S. E., et al. (2020). Artificial Intelligence and Machine Learning Based Prediction of Resistant and Susceptible Mutations in Mycobacterium Tuberculosis. Sci. Rep. 10 (1), 1–16. 10.1038/s41598-020-62368-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jena L., Waghmare P., Kashikar S., Kumar S., Harinath B. C. (2014). Computational Approach to Understanding the Mechanism of Action of Isoniazid, an Anti-TB Drug. Int. J. Mycobacteriology 3 (4), 276–282. 10.1016/j.ijmyco.2014.08.003 [DOI] [PubMed] [Google Scholar]
- Jubb H. C., Higueruelo A. P., Ochoa-Montaño B., Pitt W. R., Ascher D. B., Blundell T. L. (2017). Arpeggio: a Web Server for Calculating and Visualising Interatomic Interactions in Protein Structures. J. Mol. Biol. 429 (3), 365–371. 10.1016/j.jmb.2016.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M. T., Malik S. I., Ali S., Masood N., Nadeem T., Khan A. S., et al. (2019). Pyrazinamide Resistance and Mutations in PncA Among Isolates of Mycobacterium Tuberculosis from Khyber Pakhtunkhwa, Pakistan. BMC Infect. Dis. 19 (1), 1–7. 10.1186/s12879-019-3764-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouchaki S., Yang Y., Walker T. M., Sarah Walker A., Wilson D. J., Peto T. E. A., et al. (2019). Application of Machine Learning Techniques to Tuberculosis Drug Resistance Analysis. Bioinformatics 35 (13), 2276–2282. 10.1093/bioinformatics/bty949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Jena L. (2014). Understanding Rifampicin Resistance in Tuberculosis through a Computational Approach. Genomics Inform. 12 (4), 276. 10.5808/gi.2014.12.4.276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- L J., G W. (2016). Computational Approach in Understanding Mechanism of Action of Isoniazid and Drug Resistance. Mycobact Dis. 06 (01), 1–3. 10.4172/2161-1068.1000202 [DOI] [Google Scholar]
- Lee B. M., Harold L. K., Almeida D. V., Aung H. L., Forde B. M., Hards K., et al. (2020). Predicting Nitroimidazole Antibiotic Resistance Mutations in Mycobacterium Tuberculosis with Protein Engineering. Plos Pathog. 16 (2), e1008287. 10.1371/journal.ppat.1008287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louw G. E., Warren R. M., Gey Van Pittius N. C., McEvoy C. R. E., Van Helden P. D., Victor T. C. (2009). A Balancing Act: Efflux/Influx in Mycobacterial Drug Resistance. Antimicrob. Agents Chemother. 53 (8), 3181–3189. 10.1128/AAC.01577-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado D., Isabel C. B., João P., Liliana P., Pedro B., Isabel J. B., Bruno V., et al. (2012). Contribution of efflux to the emergence of isoniazid and multidrug resistance in Mycobacterium tuberculosis. PLoS One 7 (4), e34538. 10.1371/journal.pone.0034538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S., Willby M., Sikes D., Tsodikov O. V., Posey J. E. (2012). New Insights into Fluoroquinolone Resistance in Mycobacterium tuberculosis: Functional Genetic Analysis of gyrA and gyrB Mutations. PloS one 7 (6), e39754. 10.1371/journal.pone.0039754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miotto P., Cabibbe A. M., Feuerriegel S., Casali N., Drobniewski F., Rodionova Y., et al. (2014). Mycobacterium tuberculosis Pyrazinamide Resistance Determinants: a Multicenter Study. MBio 5 (5), e01819. 10.1128/mBio.01819-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris G. M., Huey R., Lindstrom W., Sanner M. F., Belew R. K., Goodsell D. S., et al. (2009). AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 30 (16), 2785–2791. 10.1002/jcc.21256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugumbate G., Abrahams K. A., Cox J. A. G., Papadatos G., van Westen G., Lelièvre J., et al. (2015). Mycobacterial Dihydrofolate Reductase Inhibitors Identified Using Chemogenomic Methods and In Vitro Validation. PloS one 10, 3e0121492. 10.1371/journal.pone.0121492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzondiwa D., Mutshembele A., Pierneef R. E., Reva O. N. (2020). Resistance Sniffer: an Online Tool for Prediction of Drug Resistance Patterns of Mycobacterium tuberculosis Isolates Using Next Generation Sequencing Data. Int. J. Med. Microbiol. 310 (2), 151399. 10.1016/j.ijmm.2020.151399 [DOI] [PubMed] [Google Scholar]
- Nakatani Y., Opel-Reading H. K., Merker M., Machado D., Andres S., Kumar S. S., et al. (2017). Role of Alanine Racemase Mutations in Mycobacterium tuberculosis D -Cycloserine Resistance. Antimicrob. Agents Chemother. 61, e01575–17. 10.1128/AAC.01575-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak T., Jena L., Bc H. (2017). “Austin Tuberculosis : Research & Treatment Isoniazid Drug Resistance : Computational Study to Understand the Mechanism of over Expressed UDP- Galactopyranose Mutase Enzyme in Causing Drug Resistance in Tuberculosis. Austin Tuberculosis: Res. Treat. 2 (1), 2–7. Available at: https://austinpublishinggroup.com/tuberculosis/fulltext/atrt-v2-id1006.php . 10.4103/ijmy.ijmy_174_17 [DOI] [Google Scholar]
- Palomino J. C., Martin A. (2014). Drug Resistance Mechanisms in Mycobacterium Tuberculosis. Antibiotics 3 (3), 317–340. 10.3390/antibiotics3030317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandurangan A. P., Blundell T. L. (2020). Prediction of Impacts of Mutations on Protein Structure and Interactions: SDM, a Statistical Approach, and mCSM, Using Machine Learning. Protein Sci. 29 (1), 247–257. 10.1002/pro.3774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Y., Lu J., Wang Y., Song Y., Wang S., Zhao Y. (2013). Study of the Rifampin Monoresistance Mechanism in Mycobacterium tuberculosis . Antimicrob. Agents Chemother. 57 (2), 893–900. 10.1128/AAC.01024-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., et al. (2004). UCSF Chimera? A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 25 (13), 1605–1612. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Phelan J., Coll F., McNerney R., Ascher D. B., Pires D. E. V., Furnham N., et al. (2016). Mycobacterium tuberculosis Whole Genome Sequencing and Protein Structure Modelling Provides Insights into Anti-tuberculosis Drug Resistance. BMC Med. 14 (1), 1–13. 10.1186/s12916-016-0575-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piersimoni C., Mustazzolu A., Giannoni F., Bornigia S., Gherardi G., Fattorini L. (2013). Prevention of False Resistance Results Obtained in Testing the Susceptibility of Mycobacterium Tuberculosis to Pyrazinamide with the Bactec MGIT 960 System Using a Reduced Inoculum. J. Clin. Microbiol. 51 (1), 291–294. 10.1128/JCM.01838-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portelli S., Phelan J. E., Ascher D. B., Clark T. G., Furnham N. (2018). Understanding Molecular Consequences of Putative Drug Resistant Mutations in Mycobacterium Tuberculosis. Sci. Rep. 8 (1), 1–12. 10.1038/s41598-018-33370-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz P., Rodríguez-Cano F., Zerolo F. J., Casal M. (2002). Investigation of the In Vitro Activity of Streptomycin Against Mycobacterium Tuberculosis. Microb. Drug Resist. 8 (2), 147–149. 10.1089/107662902760190707 [DOI] [PubMed] [Google Scholar]
- Sandgren A., Michael S., Preetika M., Brian K. W., George M. C., Megan B. M., et al. (2009). Tuberculosis drug resistance mutation database. PLoS Med. 6 (2), e1000002. 10.1371/journal.pmed.1000002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schymkowitz J., Borg J., Stricher F., Nys R., Rousseau F., Serrano L. (2005). The FoldX Web Server: an Online Force Field. Nucleic Acids Res. 33, W382–W388. 10.1093/nar/gki387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinnick T. M., Iademarco M. F., Ridderhof J. C. (2005). National Plan for Reliable Tuberculosis Laboratory Services Using a Systems Approach. Recommendations from CDC and the Association of Public Health Laboratories Task Force on Tuberculosis Laboratory Services. MMWR. Recomm. Rep: Morbidity Mortality Weekly Rep. 54 (RR-6), 1–12. Recommendations and Reports/Centers for Disease Control. Available at: http://europepmc.org/abstract/MED/15829862 . [PubMed] [Google Scholar]
- Singh A., Grover S., Sinha S., Das M., Somvanshi P., Grover A. (2017). Mechanistic Principles behind Molecular Mechanism of Rifampicin Resistance in Mutant RNA Polymerase Beta Subunit of Mycobacterium Tuberculosis. J. Cel. Biochem. 118 (12), 4594–4606. 10.1002/jcb.26124 [DOI] [PubMed] [Google Scholar]
- Stop T. B. (2015). Stop TB Partnership’s Global Drug Facility (GDF) Achieves Historic price Reduction for MDR-TB Drug Cycloserine. Geneva, Switzerland: Stop TB Partnership. [Google Scholar]
- Studio, Discovery (2008). Dassault systemes BIOVIA, Discovery studio modelling environment, Release 4.5. Accelrys Softw Inc, 98–104. 10.4016/8372.01 [DOI] [Google Scholar]
- Timmins G. S., Deretic V. (2006). Mechanisms of Action of Isoniazid. Mol. Microbiol. 62 (5), 1220–1227. 10.1111/j.1365-2958.2006.05467.x [DOI] [PubMed] [Google Scholar]
- Uddin M. K. M., Rahman A., Ather M. F., Ahmed T., Rahman S. M. M., Ahmed S., et al. (2020). Distribution and Frequency of rpoB Mutations Detected by Xpert MTB/RIF Assay Among Beijing and Non-Beijing Rifampicin Resistant Mycobacterium tuberculosis Isolates in Bangladesh. Idr 13, 789–797. 10.2147/IDR.S240408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A. E., Berendsen H. J. C. (2005). GROMACS: Fast, Flexible, and Free. J. Comput. Chem. 26 (16), 1701–1718. 10.1002/jcc.20291 [DOI] [PubMed] [Google Scholar]
- Waman V. P., Vedithi S. C., Thomas S. E., Bannerman B. P., Munir A., Skwark M. J., et al. (2019). Mycobacterial Genomics and Structural Bioinformatics: Opportunities and Challenges in Drug Discovery. Emerg. Microbes.Infect. 8, 109–118. 10.1080/22221751.2018.1561158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb B., Sali A. (2016). Comparative Protein Structure Modeling Using MODELLER. Curr. Protoc. Bioinformatics 54 (1), 5–6. 10.1002/cpbi.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalen C. C. (2006). Failure of Directly Observed Treatment for Tuberculosis in Africa: A Call for New Approaches. Clin. Infect. Dis. 42 (7), 1048–1050. 10.1086/501022 [DOI] [PubMed] [Google Scholar]
- Wheeler D. L., Barrett T., Benson D. A., Bryant S. H., Canese K., Chetvernin V., et al. (2006). Database Resources of the National center for Biotechnology Information. Nucleic Acids Res. 34, D173–D180. 10.1093/nar/gkj158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff K. A., Nguyen. L. (2012). Strategies for Potentiation of Ethionamide and Folate Antagonists against Mycobacterium Tuberculosis. Expert Rev. anti-infective Ther. 10, 971–981. 10.1586/eri.12.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Walker T. M., Walker A. S., Wilson D. J., Peto T. E. A., Crook D. W., et al. (2019). DeepAMR for Predicting Co-occurrent Resistance of Mycobacterium tuberculosis . Bioinformatics 35, 3240–32493249. 10.1093/bioinformatics/btz067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., An X., Liu H., Wang S., Xiao T., Liu H. (2019). Uncovering the Resistance Mechanism of Mycobacterium Tuberculosis to Rifampicin Due to RNA Polymerase H451D/Y/R Mutations from Computational Perspective. Front. Chem. 7 (December), 1–13. 10.3389/fchem.2019.00819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Shi W., Zhang W., Mitchison D. (2014). Mechanisms of Pyrazinamide Action and Resistance. Microbiol. Spectr. 2 (4), 1–12. 10.1128/microbiolspec.mgm2-0023-2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Yew W.-W. (2015). Mechanisms of Drug Resistance in Mycobacterium tuberculosis: Update 2015. Int. J. Tuberculosis Lung Dis. 19, 1276–1289. 10.5588/ijtld.15.0389 [DOI] [PubMed] [Google Scholar]