Abstract
The aquatic Lemnaceae family, commonly called duckweed, comprises some of the smallest and fastest growing angiosperms known on Earth. Their tiny size, rapid growth by clonal propagation, and facile uptake of labeled compounds from the media were attractive features that made them a well-known model for plant biology from 1950 to 1990. Interest in duckweed has steadily regained momentum over the past decade, driven in part by the growing need to identify alternative plants from traditional agricultural crops that can help tackle urgent societal challenges, such as climate change and rapid population expansion. Propelled by rapid advances in genomic technologies, recent studies with duckweed again highlight the potential of these small plants to enable discoveries in diverse fields from ecology to chronobiology. Building on established community resources, duckweed is reemerging as a platform to study plant processes at the systems level and to translate knowledge gained for field deployment to address some of society’s pressing needs. This review details the anatomy, development, physiology, and molecular characteristics of the Lemnaceae to introduce them to the broader plant research community. We highlight recent research enabled by Lemnaceae to demonstrate how these plants can be used for quantitative studies of complex processes and for revealing potentially novel strategies in plant defense and genome maintenance.
The characteristics of the Lemnaceae are introduced to the plant biology community, and recent studies are described showing how duckweed represents an ideal model for systems-level investigations.
Introduction
The duckweed family Lemnaceae belongs to the monocot order Alismatales (Figure 1) and consists of 36 recognized species representing five genera: Spirodela (Sp.), Landoltia (La.), Lemna (Le.), Wolffiella (We.), and Wolffia (Wo.) (Les et al., 2002; Bog et al., 2020b). The common name duckweed derives from their global distribution along with waterfowl such as ducks (Silva et al., 2018) and their prodigious growth rates. Some duckweeds are commonly referred to as water lentil (Lemna spp.) or water meal (Wolffia spp.). The individual plant can range in size from 1.5 cm (Sp. polyrhiza) to <1 mm (Wo. angusta) and is composed of a leaf–stem structure called a frond, with some genera having roots, such as Spirodela, Landoltia, and Lemna (Figure 2).
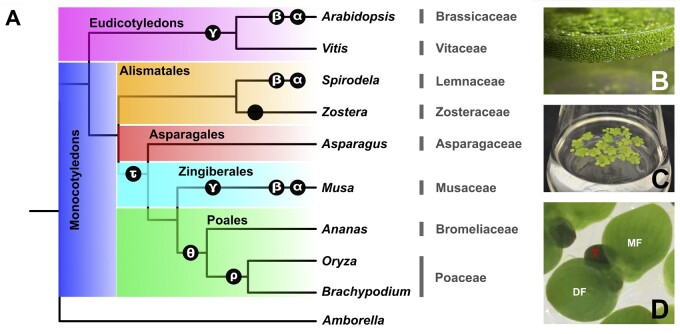
Figure 1.
The Lemnaceae family is a sister lineage to other extant monocot families that has readapted to an aquatic lifestyle. (A) Phylogenetic relationship between the greater duckweed (Spirodela, Lemnaceae, Alismatales) and other branches of the angiosperms. The genus names are shown immediately to the right of the tree diagram, followed by the family names. WGD events are shown in black circles at the approximate time during their evolutionary history. “A field in a flask”: a culture of Wo. globosa (B) or Sp. polyrhiza (C) plants in a flask of growth medium. (D) Turions are a dormant form of many Lemnaceae species that enable these simple plants to overwinter at the bottom of their resident water bodies. Pictured is a clone of Sp. polyrhiza under low phosphate conditions in the growth medium. A typical growing frond cluster is shown in the middle, surrounded by turions. DF, daughter frond; MF, mother frond; T, turion.
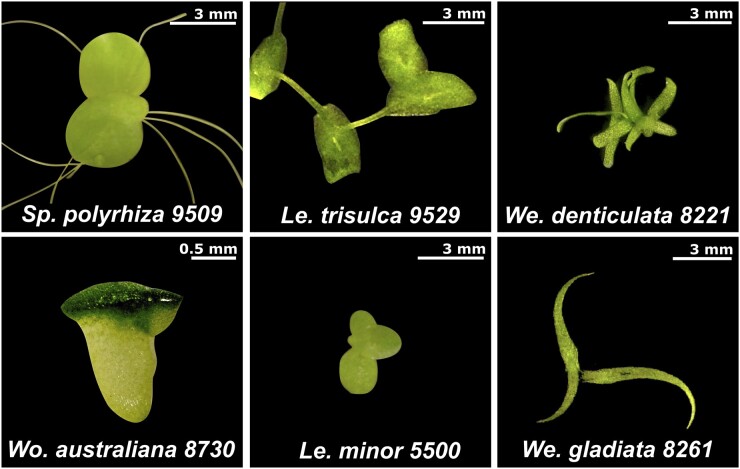
Figure 2.
Morphological variations among diverse genera and species of duckweed. Six different species from four genera of duckweeds are shown to illustrate the various sizes and shapes of these aquatic plants. The genome sequences of three of these clones (Sp. polyrhiza 9509, Le. minor 5500, and Wo. australiana 8730) are currently available.
Before Arabidopsis thaliana was adopted as a model plant in the genomics era, duckweed was an important experimental system for plant physiology and biochemistry. While a core group of researchers has continued to study duckweeds since the 1950s, the era of genomics has opened new opportunities to build tools for the broader community. Like Arabidopsis, Sp. polyrhiza has a small genome of ∼158 Mb, yet it only has ∼19,000 annotated genes, which represent a conserved set of angiosperm genes without large paralog expansions (Wang et al., 2014; Michael et al., 2017). Moreover, the further loss of genetic redundancy in the 354 Mb Wo. australiana genome, which has even fewer genes (∼15,000) with remarkable attrition in pathways such as disease resistance and organogenesis, provides a unique opportunity to define gene functions in a minimalist plant (Michael et al., 2021). As aquatic plants, duckweeds also present an important opportunity to define the molecular mechanisms underlying the biochemistry, metabolism, and interactions with microbial symbionts (Acosta et al., 2020). The development of tools such as robust transformation methods and multiomics database resources will make the duckweed platform more accessible and a versatile component of the plant molecular biology toolkit once again.
As we show in this model system review, the types of experiments which unique features of duckweeds will enable and facilitate, as well as their growing commercial applications, make it an exciting model plant in the postgenomic era. For instance, the reduced anatomical features of duckweed coupled to its clonal reproduction should make it possible to track all cells with current single-cell capture methods and high-throughput omics technologies. There is also growing interest in the commercial sector to farm duckweed as a staple food and as a source of protein for plant-based ingredients that are sustainable and resilient to climate change. In this review, we provide an overview of the species in the Lemnaceae family, how they can be used for commercial and research applications, current and future tool development, and available resources that again make these aquatic monocots an attractive model for plant research.
Phylogenetic history and traits
Taxonomic position
Closely connected with the simple morphology of duckweeds is the question of whether these aquatic plants represent the end of an evolutionary reduction process (Hegelmaier, 1868). The emergence of molecular taxonomy revolutionized the systematics of duckweeds and their position within angiosperms (Les et al., 2002), and demonstrated that duckweeds are closely related to the Araceae (Nauheimer et al., 2012). Extending these studies, several reports deployed plastidic barcodes on a larger number of clones for each species to determine their phylogenetic relationships within the Lemnaceae (Wang et al., 2010; Borisjuk et al., 2015; Tippery et al., 2015; Bog et al., 2019). More recently, amplified fragment length polymorphisms (AFLPs) and the application of genotyping-by-sequencing helped to resolve problematic species that were difficult to distinguish based on plastidic sequences alone (Bog et al., 2020a, 2020b, 2020c). In contrast to the Angiosperm Phylogeny Group III definition (APG, 1998), which characterizes duckweeds as a subfamily (Lemnoideae) of the Araceae, many researchers in the field now consider duckweeds to be a separate family (Lemnaceae), which is in agreement with general taxonomic rules (Bog et al., 2020a; Tippery and Les, 2020).
Phylogenetic analysis with both nuclear and plastidic markers showed that Spirodela and Landoltia represent older phylogenetic branches from the common ancestor than the more recently derived genera of Lemna, Wolffiella, and Wolffia. When comparing species belonging to each of the five genera of Spirodela (2 species), Landoltia (1), Lemna (12), Wolffiella (10), and Wolffia (11), a reduction of organismic complexity from Spirodela toward Wolffia is observed, which is quantified by the so-called “degree of primitivity” (Landolt, 1986), and is generally accompanied by a higher nuclear DNA content (Figure 3). These observations suggest an evolutionary trajectory from a more to less differentiated plant body during adaptation to the aquatic lifestyle through a series of morphological reductions (Landolt, 1986). While there is general concordance at the genus level between taxonomic trees created with morphological characteristics (Landolt, 1986) and those generated from molecular data (Borisjuk et al., 2015; Bog et al., 2019), the latter has a much higher resolving power at the species level.
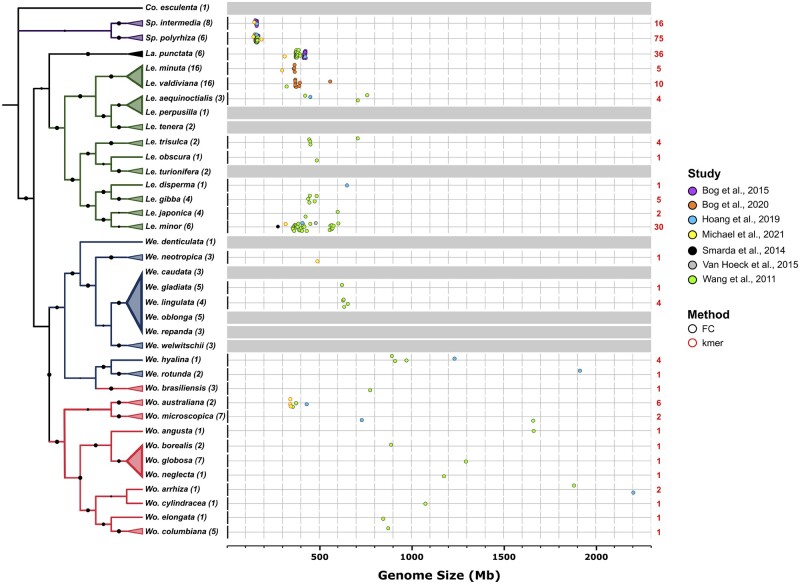
Figure 3.
Phylogeny and variations in genome size of different Lemnaceae species. Left: Evolutionary relationships between Lemnaceae species based on maximum likelihood analysis of concatenated alignment of 139 atpF-atpH and psbK-psbI intergenic spacer sequences from all 36 Lemnaceae species with taro (Colocasia esculenta) as an outgroup. Numbers in parentheses represent the number of clones analyzed. Species that could not be confidently resolved into a single clade were collapsed into a multispecies clade. One interesting observation is that the plastidic barcode sequences of Wo. brasiliensis consistently showed higher similarity to those of We. hyalina and We. rotunda, while morphologically it is distinctly a Wolffia species. This apparent discrepancy could be due to potential hybridization events in the past that resulted in the transfer of plastid genome sequences from a Wolffiella ancestor to a Wolffia lineage. Future genome sequencing of relevant species that may be involved will help clarify this issue. For a detailed methods description, see https://github.com/kenscripts/tpc_dw_review/. Right: The genome sizes for 28 selected species from six groups representing all five genera were estimated using several methods, and in some cases the genome sizes for a significant number of clones from the same species were measured (Sp. polyrhiza, Sp. Intermedia, La. punctata, and Le. minor). Genome size estimates were carried out by flow cytometry (FC, black outline), which requires the inclusion of accurate controls, or by K-mer frequency analysis (kmer, red outline), which relies on high quality short-read sequencing data. Numbers in red depict the number of clones used for each species in genome size estimations. The genome size of We. neotropica was estimated based on K-mer frequency.
Anatomy, morphology, and growth characteristics
The adaptation of duckweeds to a floating aquatic lifestyle apparently led to morphological and biochemical properties distinct from those normally found in land plants (Figure 2). Examples of such features are their meristem structure and low lignin content in the cell wall. Endemic species such as We. denticulata and We. gladiata have evolved distinctive morphologies compared to the more cosmopolitan pioneer species, such as Sp. polyrhiza and Le. minor (Figure 2). The simplified vegetative body of a duckweed is called a frond or thallus (Hillman, 1961). Depending on the genus, duckweed fronds can be roundish to obovate (Spirodela, Landoltia, and Lemna), hemispherical or boat-shaped (Wolffia) or sickle-shaped, tongue-shaped or ovate (Wolffiella). The fronds of subsequent generations are held together, in some cases even after maturity, thus resulting in colonies of connected fronds (see Le. trisulca in Figure 2). The smallest colonies consist of two fronds (Wolffia and some Wolffiella) and the largest range up to 50, such as in We. gladiata and Le. trisulca (Landolt, 1986; Bog et al., 2020a). The fronds are physically held together with the help of an elongated stipe. Abscission zones (two in Sp. polyrhiza and one in Wo. microscopica) are present on the stipe, facilitating the separation of daughter fronds from the mother frond (Landolt, 1986; Kim, 2016). The stipe originates from the base at the ventral side of the mother frond from where the cells further divide and grow (Landolt, 1986; Sree et al., 2015a). This could be considered intercalary growth at the base of the frond. The stipe has been speculated to function in the transport of nutrients and substances from the mother frond to the daughter frond (Kim, 2016).
A transparent waxy cuticle encloses the entire duckweed body and fortifies its epidermis against mechanical injury and solar radiation (Borisjuk et al., 2018). It may also serve as a barrier for gas and solute exchange controlled by the epidermis. The cell walls of epidermal cells in duckweed have distinct morphologies depending on the genus and are bent (Spirodela), undulated (Landoltia, Lemna), or straight (Wolffiella, Wolffia). While stomata are found on the epidermis of duckweeds, these aquatic plants do not form trichomes or root hairs (Landolt, 1986). Differentiation of stomata is restricted to the dorsal epidermis and may depend on growth conditions, such as light and temperature (Klich et al., 1986). In contrast to land plants, stomata remain open in Lemna (McLaren and Smith, 1976) even upon prolonged exposure to the phytohormone abscisic acid (ABA). The ventral epidermis is involved in nutrient uptake (Cedergreen and Madsen, 2002) and may provide an active surface for interactions with aquatic bacteria (Duong and Tiedje, 1985).
The bulk of the frond consists of parenchyma cells with a central vacuole, contributing to the high (up to 95%) water content of the tissues. Dorsal cell layers contain a higher density of chloroplasts (Figure 2) and perform the function of the chlorenchyma (White and Wise, 1998; Kwak and Kim, 2008). The photosynthetic properties of chloroplasts and their rearrangements in response to light can be associated with distinct light-utilization strategies in different species (Paolacci et al., 2018a). A loose tissue structure, increased cell size, and formation of gas/air spaces (Jones et al., 2021) are indicative of the aerenchyma. The aerenchyma in duckweed supports the exchange of gases between the dorsal and ventral portion of the body. Importantly, the presence of the aerenchyma may also allow fronds to control their degree of flotation on or under the water surface by regulating the air space volume within fronds (Landolt and Kandeler, 1987).
The duckweed root is an adventitious organ found in Spirodela, Landoltia, and Lemna species (Bellini et al., 2014) that develops on the lower side of the frond, next to the budding pouches, and is subtended by both the epidermal sheath at the junction and by the root-cap at the root tip. Apical root growth is followed by the differentiation of epidermis, cortex, tracheary elements, and phloem cells (Melaragno and Walsh, 1976; Echlin et al., 1982; Landolt, 1986; Kim, 2007). Because of the considerable length and the thread-like structure with high cytoplasmic density at the root tip, the root may act like a pendulum to attenuate dynamic loads from water and wind motion.
The frond meristem is composed of densely packed, proliferating mitotic cells that are significantly smaller than their neighboring parenchyma cells. Frond meristem cells contain small vacuoles and proplastids with only a few thylakoids (McCormac and Greenberg, 1992; Kim, 2011). Meristems usually localize to the ventral side of the frond body inside a small cavity (vegetative pouch) where clonal daughters bud and detach (Landolt, 1986; Lemon and Posluszny, 2000). Wolffia and Wolffiella contain only one basal pouch, whereas Spirodela, Landoltia, and Lemna possess two lateral pouches. At the abscission stage, the young daughter frond usually contains at least two successive generations of vegetative buds (Rimon and Galun, 1968; Sree et al., 2015a). While differentiation of the meristem is less understood in duckweeds and rather unlike the differentiation of the canonical shoot apical meristem of land plants, the frond itself can be botanically described as a juvenile tissue (Landolt, 1986). Interactions between meristem cells and functionally differentiated tissues of the frond mainly occur via symplastic connections. The arrangement of plasmodesmata and their dynamic features appear to be sufficient for communication and metabolite transport. In general, a vascular system is completely absent (Wolffia and Wolffiella) or fairly simplified (Spirodela, Landoltia, and Lemna) in duckweeds (Landolt, 1986).
A prominent morphological feature that facilitates the survival of many duckweed species under unfavorable environments is the formation of turions (Landolt, 1986). Turions are thought to represent overwintering, dormant duckweed derived from the meristematic pocket in place of normal proliferating daughter fronds (Figure 1D). As it matures, the color of a dormant turion can change from green to purple due to hyperaccumulation of anthocyanin; the mature turion eventually detaches from the mother frond colony and sinks to the bottom of the water body. When proper light, temperature, and nutrient conditions return, such as in spring, turions can germinate and resume metabolic activities (Landolt and Kandeler, 1987; Appenroth and Augsten, 1990). During the early phase of germination, the low-molecular weight carbohydrate reserve is consumed, but starch degradation is not observed. Storage starch, however, is used to support the rapid growth of newly germinated fronds, allowing them to cover the water surface quickly in spring (Appenroth et al., 2013).
Compared to fronds, turions have smaller cells, lack aerenchyma and plasmodesmata, and have thicker cell walls (Jacobs, 1947; Kim, 2013). Turion cells are densely packed with starch grains (Smart and Trewavas, 1983b), with starch content in turions exceeding 70% dry weight (Dolger et al., 1997). In addition to the Lemnaceae, turion-producing species have been reported in 11 genera of aquatic vascular plants (Adamec 2018), indicating that this may be a common strategy for adaptation to aquatic habitats. In the Lemnaceae, turions are observed in almost all Wolffia species (not reported for Wo. microscopica), Le. turionifera, Le. perpusilla, Le. aequinoctialis (Landolt, 1986), and Sp. polyrhiza (Figure 1D). Abiotic turion-inducing factors include limiting nutrient levels (phosphate, nitrate, sulfate) and low temperatures (Appenroth et al., 1989; Appenroth, 2002; Appenroth and Nickel, 2010). Turion formation rates for different clones of Sp. polyrhiza were shown to be linked to local climatic conditions (Kuehdorf et al., 2014). The ‘specific turion yield’ is a quantitative trait that can be predicted for an adapted population of Sp. polyrhiza by using five different local climatic parameters. This remarkable observation indicates that the production of turions in Sp. polyrhiza is a key trait for the species’ survival in a particular climate. In addition to nutrient limitation in the growth medium, ABA is a powerful experimental tool to induce the formation of turions (Smart and Trewavas, 1983a) and could function as an intracellular mediator of other turion-inducing factors (Smart et al., 1995).
Vegetative reproduction, such as the budding of clonal daughter fronds from a mother frond (Figure 1), is the most common mode of duckweed propagation. Fronds display a limited lifespan of a few weeks and produce a finite number of daughter fronds whose area progressively diminishes (Ashby et al., 1949). Therefore, growth in duckweeds includes an increase in cell size, the number of individual plants, and the number of daughter fronds produced by each plant. Growth can thus be measured in terms of either biomass (such as fresh weight and dry weight) or the number of fronds. A standard measure of relative yield for duckweed was recently introduced (Ziegler et al., 2015). In nature, growth responses of different duckweed species to abiotic factors such as light and temperature are closely linked to the range of their geographical distribution. Consistent with geobotanical data, some S. polyrhiza clones can withstand temperatures of up to 38°C, unlike Le. minor clones, which typically display growth arrest at 32°C. Similarly, Le. minor clones can often withstand temperatures down to 5°C, while Sp. polyrhiza clones typically only tolerate temperatures as low as 12°C before growth arrest (Docauer, 1983; Landolt, 1986). Like turion formation, growth rate also displays a high level of clonal dependence (Sree et al., 2015b; Ziegler et al., 2015).
Although vegetative clonal division is most common in duckweeds, they can also propagate generatively through sexual reproduction. When flowering occurs, the floral organs are located in a cavity on the dorsal surface of the frond in Wolffiella and Wolffia spp. or in a membranous, sac-like spathe within a lateral budding pouch in Spirodela, Landoltia, and Lemna spp. (Landolt, 1986; Lemon and Posluszny, 2000). Flower size and morphology are minimized to a male (androecium) and female (gynoecium) floral organ, while a corolla and calyx are absent. Flowers are bisexual, usually protogynous, and smallest in Wolffia and Wolffiella. Modulation of various abiotic factors and the addition of different chemical molecules to duckweeds have been used as inducers of flowering under in vitro conditions. Exposure to low temperature (22°C) was found to induce flowering in Wo. microscopica (Rimon and Galun, 1968), while the effects of phytohormones, chelators, heavy metal ions, and photosynthetic products on flowering initiation have been investigated in other species (Landolt and Kandeler, 1987). Among these, ethylenediamine-di-o-hydroxyphenylacetic acid (EDDHA) and salicylic acid (SA) were reported to be floral inducers in duckweeds (Maheshwari and Seth, 1966; Cleland and Ajami, 1974), even under noninductive conditions in terms of day length (Landolt and Kandeler, 1987). For example, EDDHA could induce flowering in short-day plants (Maheshwari and Seth, 1966), long-day plants (Pieterse and Müller, 1977), and day-neutral plants (Khurana and Maheshwari, 1986). EDDHA was initially thought to act by chelating metal ions that might be required for floral induction in duckweeds. It was later suggested that the breakdown of EDDHA may release SA-like active molecules (Tanaka et al., 1979; Pieterse, 2013). In line with the current understanding of the role of SA in plant defense, it was suggested that flowering could be a stress response and that endogenous SA produced in stressed plants leads to flower induction (Pieterse, 2013).
Chromosomes and genomics of duckweeds
Variations in chromosome number and genome size
While the most common diploid chromosome number in duckweeds is 2n = 40 (Hoang et al., 2019), highly variable chromosome numbers have been reported within some duckweed species. For example, 20–84 chromosomes have been reported for Le. aequinoctialis (Urbanska, 1980; Geber, 1989; Wang et al., 2011; Hoang et al., 2019). Even considering the predominantly asexual propagation of duckweed species, such high variability in chromosome number appears unusual. However, in cases where previously variable numbers were reported and the corresponding clones are still available, deviating chromosome numbers could not always be confirmed (Hoang et al., 2019). Studies with some of the cultured clones indicated apparent autotetraploidy (e.g. Le. aequinoctialis, 2n = 42 and 84), which might have occurred spontaneously or could be chemically induced, such as in La. punctata 5562 with 2n = 46 and 92 (Vunsh et al., 2015; Hoang et al., 2019).
Genome size across 28 duckweed species with available data (Figure 3) ranges from ∼158 Mb in the phylogenetically oldest genus Spirodela up to 2,203 Mb in Wo. arrhiza, a species in the phylogenetically most recent genus (Wang et al., 2011; Šmarda et al., 2014; Van Hoeck et al., 2015; Bog et al., 2015, 2020c; Hoang et al., 2019; Michael et al., 2021). The largest intrageneric variation was found in the most derived genus Wolffia, spanning from 354 to 2,203 Mb. Among eleven species sampled across the five genera of duckweed, genome size was positively correlated with guard cell and nucleus volume, in parallel with progressive organ size reduction. In contrast, no correlation between genome size and the number of chromosomes was observed (Hoang et al., 2019). More surprising is the considerable variation in estimated genome size for clones within some species. For 27 Le. minor clones, genome size varied from 356 to 604 Mb (Wang et al., 2011), while two verified diploid genomes from this species displayed size estimates of 409 and 481 Mbp by K-mer analysis for clones 5500 and 8623, respectively (Van Hoeck et al., 2015; Hoang et al., 2019). With Le. aequinoctialis and La. punctata, a possible explanation is spontaneous whole-genome duplication (WGD) in clones of these species (Hoang et al., 2019). Further, cytogenomic investigations and whole-genome resequencing would be helpful for resolving whether clone-specific WGD occurred, followed by an extremely rapid genome size reduction, or whether a subset of clones comprise interspecific hybrids or cryptic species.
Genome sequences and features suggest novel modes of regulation
The first duckweed genome sequenced was that of Sp. polyrhiza due to its small genome size of ∼158 Mb and basal position in the Lemnaceae. The initial Sp. polyrhiza reference genome draft (×20 coverage) was carried out with clone 7498 (Sp7498) from Durham, NC, USA. The genome sequence revealed two lineage-specific ancient WGD events (Figure 1A) and the smallest gene repertoire of any plant sequenced at the time of its publication, with 19,623 protein-coding genes (Wang et al., 2014), which is ∼30% fewer than reported for A. thaliana. While the Sp. polyrhiza genome shares most of the core gene families found in plants, its reduced gene content reflects the lowest gene expansion and copy number, which is consistent with the aggressive removal of duplicated genes after the two WGD events. Therefore, the Sp. polyrhiza genome is ideal for gene discovery and functional analysis of highly conserved core pathways in plants, since it likely contains fewer redundancies that often confound reverse genetic approaches.
To improve the Sp. polyrhiza genome assembly and to help uncover the genetic basis for variations in turion formation, clone 9509 (Sp9509) from Lotschen, Germany was chosen for assembly of a high-quality genome due to its lower specific turion yield compared to Sp7498 (Kuehdorf et al., 2014). Sp9509 was sequenced using a combination of high coverage (×100) Illumina short read libraries containing small (∼500 bp) to large (∼20 kbp) genome fragment inserts and single-molecule BioNano Genomics optical maps for additional chromosome scaffolding (Michael et al., 2017). The higher resolution Sp9509 reference genome revealed the highest solo-to-intact ratio of any plant genome tested to date at the time, indicating that this species is actively purging active long terminal repeat retrotransposons as well as other sequences to maintain or further decrease its genome size. This finding is consistent with the reduced protein-coding gene content after two WGD events and is further supported by the observation that Sp. polyrhiza has retained only 20% of the ribosomal DNA repeats found in similarly sized plant genomes. In addition, the Sp9509 genome has the lowest DNA methylation level of any known flowering plant, specifically in the syntenic regions with retained paralogs involved in growth control and photosynthesis having little to no DNA methylation (Michael et al., 2017). Comparison of the two Sp. polyrhiza reference genomes also revealed a surprisingly low level of nucleotide diversity and highly conserved chromosomal structure. Resequencing of additional Sp. polyrhiza populations from across the globe further extended this observation and revealed a very low level of nucleotide variation in this species yet a large population size, which possibly reflects its rapid clonal propagation and local adaptation (Ho et al., 2019; Xu et al., 2019). More recently, additional orthogonal technologies have been applied to further improve the end-to-end assemblies and gene annotation for the 20 chromosomes in these two Sp. polyrhiza reference genomes by several groups (Hoang et al., 2018; An et al., 2019; Harkess et al., 2021). These are excellent resources for duckweed research specifically and more broadly for comparative genomics in plant research.
Lemna minor (Lm5500) was the first duckweed species to be sequenced after Sp. polyrhiza, revealing a slightly larger gene repertoire of 22,382 protein-coding genes and a much higher repeat content of 61% (Van Hoeck et al., 2015). Lm5500 is diploid, in contrast to another sequenced Le. minor clone (Lm8627), which is polyploid. The assembled sequences for both Lm8627 and Le. gibba clone 7742 (Lg7742) are currently available to the community (lemna.org). The intraspecific differences in genome size and chromosome number that have been observed in Le. minor clones (Figure 3; Wang et al., 2011) contrast with the relatively stable and highly conservative genome of Sp. polyrhiza. Since Wo. australiana has the smallest genome of the Wolffia genus at 354 Mb, reference genomes were recently completed for two clones (Wa7733 and Wa8730). These high-quality genome assemblies revealed that Wa7733 and Wa8730 have a further reduction in the number of predicted protein-coding genes compared to Sp. polyrhiza, with 15,312 and 14,324, respectively. This finding is consistent with the minimal tissue types and lack of roots in Wo. australiana (Michael et al., 2021). The Wo. australiana genome also contains ∼50% repetitive sequences, which is less than that of Le. minor (61%) but approximately twice that of Sp. polyrhiza (25%). Consistent with having the smallest genome in the Wolffia genus, transposable elements have been actively purged from Wo. australiana, resulting in an even higher solo-to-intact ratio than in Sp. polyrhiza. The reduction in circadian, light signaling, developmental, root-related, and disease resistance genes in the Wo. australiana genome is apparently linked to the proliferation and purging of transposable elements, which provides a unique opportunity to explore the genesis and genomic architecture of a highly derived plant.
These initial genomics studies in the Lemnaceae represent an important resource for the plant biology community. With their basal position in the monocot lineage, the Lemnaceae provide valuable information about the genome structures of the early common ancestors of grasses and other grain crops. In addition, as an angiosperm family that has adapted to an aquatic habitat, the distinct developmental attributes in the Lemnaceae offer unique opportunities to associate gene presence or absence to the gain and loss of traits. Thus, high-quality genome assemblies are being completed for additional duckweed species in diverse genera. For instance, the genomes of two Sp. intermedia clones (Si8410 and Si7747) were recently sequenced, shedding light on chromosomal dynamics between Sp. intermedia with 18 chromosomes (2n = 36) and Sp. polyrhiza with 20 chromosomes (2n = 40) (Hoang et al., 2020). Also, draft genomes for Le. minuta, Le. turionifera, Le. japonica, La. punctata, and We. neotropica have been generated: requests can be made for the data from the corresponding authors. These resources and additional genome assemblies covering the remaining duckweed species should provide an essential foundation for understanding the relationships between genera and species in Lemnaceae, as well as the evolutionary path that enables these tiny plants to adapt to aquatic niches in a wide geographic range.
Ecology and biogeography
Duckweed biota
Duckweeds typically inhabit relatively small and shallow water bodies in areas ranging from tropical to boreal regions (Supplemental Table S1). The freshwater ecosystems in which Lemnaceae can be found include small rivers, lakes, ditches, and wetlands. Lemnaceae represent cornerstone species in aquatic food webs, as they comprise an essential food resource for many organisms (Landolt, 1986). On the other hand, the growth of Lemnaceae can also reduce the abundance of other macrophytes and microphytes due to their competition for nutrients and light in the water column (Scheffer et al., 2003). Thus, changes in duckweed growth and distribution can drastically influence the diversity and stability of freshwater ecosystems. Because of anthropogenic activities, nitrogen and phosphorus levels in the water column as well as average water temperature have increased globally. The combination of these factors changes the growth dynamics and equilibrium between duckweeds and their biota. Using mesocosms (controlled outdoor experimental systems), Feuchtmayr et al. (2009) showed that both warming water and increased nutrient levels in the water column favor duckweeds over phytoplankton, one of the major competitors of duckweeds (Scheffer et al., 2003). Studies on the interactions between Le. minor and moth larvae (Cataclysta lemnata), a natural herbivore of duckweeds, showed that increased temperature reduced the grazing pressure of Le. minor by the insect (Van Der Heide et al., 2006). Consistently, long-term monitoring of Dutch ditches showed that higher temperatures and increased water nutrient levels increased the risk of duckweed dominance, which can result in a reduction in biodiversity (Peeters et al., 2013). Therefore, models that can forecast different stable states between duckweeds and their biota can be useful for developing sustainable strategies that prevent the loss of biodiversity in freshwater ecosystems in the future (Scheffer et al., 2003). To this end, systematic and quantitative measures of the ecological consequences of duckweed dominance are needed. Also, the current long-term and global freshwater ecosystems monitoring system (https://www.sdg661.app/home) would benefit from the inclusion of Lemnaceae.
Duckweed dispersal and distribution
Lemnaceae, which rarely undergo sexual reproduction, are model species to study the dispersal of vegetative propagules. The local dispersal of Lemnaceae can be facilitated by streaming water and occasionally by strong winds, while dispersal over longer distances (>10 km), as well as dispersal between separate, isolated waterbodies, is often facilitated by a dispersing organism. Birds, in particular, have long been flagged as epizoochorous dispersers of many aquatic plants, including the Lemnaceae (Darwin, 1859; Coughlan et al., 2017). The mechanism underlying the attachment to a disperser species is rarely studied, but nonspecific entanglement is often assumed. It was recently suggested that the stickiness of roots may play a key role in attaching colonies of rooted Lemnaceae species to disperser surfaces (Cross, 2017). Once attached to a disperser, dehydration and the associated loss of viability of the propagule are major constraints for the survival of the dispersed plant (Landolt and Kandeler, 1987). However, mallard ducks can effectively transfer Le. minuta across significant distances (up to 250 km), likely due to the humid microclimate that exists between the feathers of dispersing birds (Coughlan et al., 2015; Coughlan et al., 2017). Perhaps unexpectedly, endozoochorous dispersal has also been reported. Viable Wo. columbiana propagules were identified in the feces of ducks and swans, indicating that plants can survive passage through the guts of some waterfowls (Silva et al., 2018). Endozoochory may also be relevant for the dispersal of relatively well-protected turions (Landolt and Kandeler, 1987). Thus, epi- and endozoochorous transport can realistically facilitate the dispersal of Lemnaceae over distances of several hundred kilometers. The dispersal of Lemnaceae over longer distances most likely occurs through a “step-by-step” process involving a series of intermediate waterbodies with birds acting as dispersing agents (Coughlan et al., 2017), which might explain the disjointed population distribution for some species of Lemnaceae (Les et al., 2003). While the likelihood that a propagule survives a long flight may be low, staggering numbers of birds covering large distances as part of their annual migration (sometimes in excess of 10,000 km) could provide a potential transport highway, with a significant number of successful dispersals of viable individuals.
The extent of natural dispersal of Lemnaceae is debated, and contradictory information can be found in the literature. It might be surmised that the extent of dispersal could potentially be revealed by the genetic structure of a population. Studies of the intraspecific genetic variation within a local population of Le. minor showed that such populations are made up of just a small number of genetically distinct individuals, while substantial intraspecific diversity has been observed between closely located populations of Le. minor (Cole and Voskuil, 1996; Martirosyan et al., 2008; El-Kholy et al., 2015). These data suggest low levels of gene flow between populations. In some cases, such limitations in gene flow were attributed to geographic barriers. Resequencing and comparative study of 68 genomes of Sp. polyrhiza suggested that the Himalayas represent one of the barriers preventing the dispersal of this species in Southeast Asia (Xu et al., 2019), while a smaller study of 23 clones of Sp. polyrhiza suggested that the Hungarian population is isolated from other European genotypes, possibly due to the mountainous borders surrounding this region (Chu et al., 2018).
Alien, invasive duckweeds
Plants, animals, and other organisms that disperse to new locations and negatively affect the ecosystems and ecosystem services of their new environment are defined as alien invasive species. Many species of Lemnaceae have dispersed widely beyond their natural distribution range and are considered to be more invasive than other species based on traits such as rapid vegetative propagation (Moodley et al., 2016). A prime example of an alien invasive species is Le. minuta. This species is native throughout the temperate zones of the Americas, but it dispersed widely throughout Eurasia in the 1950–1960s through natural means, such as bird-mediated dispersal (Ceschin et al., 2018; Lucey, 2003; Mifsud, 2010). Le. minuta is not the only alien Lemnaceae in Europe, since evidence is emerging that both Le. turionifera and Le. valdiviana are invasive in parts of Eurasia (Iberite et al., 2011), as is Wo. columbiana (Ardenghi et al., 2017). La. punctata is another alien species in both Europe and North America. Florida in the United States, with its extensive aquatic habitats, is home to at least six nonnative species of Lemnaceae (Ward and Hall, 2010). However, La. punctata is the only species to exert a strong enough impact on ecosystems to be considered an alien, invasive species. Comparative analysis of congeneric plant species with similar morphological structures and lifecycles is a powerful tool to identify plant traits related to invasiveness. This makes the Lemnaceae excellent model organisms for the study of dispersal and invasiveness, two processes that are particularly relevant in a world experiencing climate change.
What are duckweeds good for?
The potential commercial applications of duckweed have attracted investigators from both the basic and applied sectors for more than 50 years. Aside from their prodigious growth rates, other unique qualities of duckweed that compare favorably to traditional crop plants are their natural aquatic habitat (which obviates the need for arable land), their small size, and a floating lifestyle that enables easy harvesting. Duckweeds can produce relative yields (i.e. the amount of biomass after 7 days of cultivation starting with 1 g of initial biomass) of up to 50 g in the case of some clones of Wo. microscopica under optimized conditions (Sree et al., 2015b). With these attractive qualities and an urgent need for additional crops that can be produced sustainably as well as economically, there are great opportunities to develop the Lemnaceae into a novel agriculture platform that can augment traditional farming systems. While there are engineering challenges for growing duckweeds reliably at scale, in addition to potential hurdles involved in creating a market for duckweed-related products, there are also encouraging advances in delineating numerous applications that duckweeds are well suited for. Here, we summarize these applications to illustrate how basic research in duckweed could have societal impacts in the near term.
Human nutrition and animal feed
Duckweeds, especially Wo. globosa, have traditionally been used as human food source in some Asian countries such as Thailand, Laos, and Cambodia (Bhanthumnavin and McGarry, 1971). Rusoff et al. (1980) reported four duckweed species with the remarkably high protein content of 35%–40% of dry weight and an essential amino acid spectrum for the human diet that compares well with soybean (Glycine max). Representatives from all five known genera (Appenroth et al., 2017), and especially from Wolffia (Edelman and Colt, 2016), including all 11 species of the genus (Appenroth et al., 2018), were analyzed for their starch, protein, fat, mineral, vitamin, and phytosterol content, as well as amino acid and fatty acid spectra. All data showed that their contents in duckweeds were in good agreement with the recommended levels for human nutrition by the World Health Organization. Moreover, no toxic effects on three different human cell lines were detected in extracts of different duckweeds spanning all five genera (Sree et al., 2019). While some duckweed species such as those in the Lemna genus have been known to contain significant levels of calcium oxalate in the form of raphides (Landolt, 1986), which may be linked to health issues such as kidney stones, raphides are not found in the rootless duckweeds such as Wolffiella spp. (Appenroth et al., 2017). Over the past decade, several companies (e.g. Parabel, Hinoman, GreenOnyx, and Plantible) have emerged that aim to popularize duckweed as a food and protein source, while products derived from Lemna and Wolffia species have been granted Generally Recognized as Safe status by the US Food and Drug Administration. These activities are thus paving the way for the large-scale use of duckweed-derived products for human consumption.
The use of duckweed as animal feed also has a long tradition. Animals such as cows, chicken, pigs, ram, sheep, horses, and especially a broad spectrum of fishes were reported to feed on duckweeds (Landolt and Kandeler, 1987; Sonta et al., 2019). Comparisons between duckweed and corn (Zea mays) revealed that duckweed can be a better feedstock for animals than corn kernels due to its high protein content of up to 30% (Lee et al., 2016). However, the commercial application of duckweeds for livestock feeding might be a challenge since the current price of animal feed is generally low. Nonetheless, duckweed could attract a higher price when used as a nutritious pet food, making this a more realistic possibility. As a source of either food or animal feed, a high protein content in duckweed biomass is desirable, since adequate protein intake is a major requirement for the proper development of young animals as well as proper health in adults.
Feedstocks for biofuel and biogas production
Duckweeds can accumulate up to 50% starch on a dry-weight basis, with increased starch accumulation during wastewater cleaning (Cheng and Stomp, 2009), making them a potential feedstock for bioenergy production (Ma et al., 2018). Several treatments can induce starch accumulation in duckweed, such as abiotic stressors, nutrient limitation (Cui and Cheng, 2015; Guo et al., 2020), the addition of the phytohormone ABA (Liu et al., 2018), and treatment with heavy metals or salt (Sree et al., 2015c; Shao et al., 2020). Duckweed starch can then be degraded to sugars and fermented to bioethanol or higher alcohols, such as butanol (Cui and Cheng, 2015). In addition to starch and soluble sugars, the cell wall material from duckweed, comprising more than 30% of its biomass, is more easily converted to fermentable sugars compared to the cell wall material of the energy plant sugarcane due to its low lignin content (Sowinski et al., 2019; Pagliuso et al., 2020). Duckweed biomass can also be used to produce biogas via anaerobic digestion (Ren et al., 2018) and can be combined with saccharification and fermentation to bioalcohols to significantly enhance the total energy output (Calicioglu and Brennan, 2018; Kaur et al., 2019). In addition, wastewater purification could be readily combined, at least in principle, with the production of bioalcohols or biogas (Xu et al., 2012; Cui and Cheng, 2015). The high rate of biomass production by duckweed, assimilation of CO2, and its benefit for carbon credit should be economically relevant in view of its potential to mitigate climate change via carbon sequestration.
Phytoremediation
Phytoremediation refers to the cleaning of the environment via the uptake or degradation of pollutants using plants. Duckweeds are especially useful for this purpose in aquatic locales because most of the surfaces of these fast-growing plants are in direct contact with water. Water purification by duckweeds can be facilitated by the uptake of heavy metals, uptake and metabolism of xenobiotics and pharmaceutical drugs, or uptake of macroelements such as nitrates and phosphates from eutrophic water (Ziegler et al., 2016). Experiments on nutrient removal by duckweed cultivation have been carried out in several pilot scale studies with promising results (Xu et al., 2012; Zhao et al., 2015). More recently, the applications of duckweed for remediation of crude oil contaminants and polyester manufacturing wastewater have been reported (Ekperusi et al., 2020; Osama et al., 2020). While these studies have documented the effectiveness of duckweed to aid in water remediation, how duckweed biomass is utilized after harvest will be determined by the particular contaminant’s lifecycle in duckweed tissues. Effective solutions to manage and utilize the duckweed harvested from contaminated sources would open this platform for large-scale applications in many communities.
Phytotoxicity testing
The toxic effects on duckweeds upon exposure to different concentrations of heavy metals can be measured based on common physiological parameters, such as relative growth rate, chlorophyll, or carotenoid content under standardized conditions. Using dose–response curves, toxicity parameters for effective doses at an effect level of 50%, 20%, or 10% can be calculated (Naumann et al., 2007). This procedure is called biomonitoring, which can be used to quantify the toxicity of the substances present or to evaluate the health of water bodies in environmental monitoring (Ziegler et al., 2016). The well-known phenotyping company LemnaTec developed an automated method for monitoring the phytotoxic effects of substances by visually tracking duckweed growth (Perera et al., 2019). While Le. minor growth has been deployed as a standardized assay by the Environmental Protection Agency of the United States and the International Organization for Standardization to monitor the presence of toxic substances (USEPA, 1996; ISO, 2005), the nature of a causal agent is difficult to ascertain without any predetermined target(s). One proposed approach is to identify highly specific plant responses for each toxic substance of interest, making it possible to recognize the type of contaminants based on a number of chosen markers to monitor (Ziegler et al., 2018). This may be possible using metabolomics to generate chemically induced metabolite fingerprints in duckweed that could be used to evaluate the causal agent for the stress responses (Kostopoulou et al., 2020). However, resources would have to be invested to create a comprehensive reference data library in order to test this possibility.
Production of biopolymers, proteins, and vaccines
Biopolymers such as polylactic acid and polyhydroxybutyrate can be produced from different plant components. Duckweed powder made from complete fronds of Lemna species has been used to produce bioplastics by mixing it with glycerol and polyethylene (Zeller et al., 2013). Genetically modified duckweed could be used as an expression system for valuable products such as high-value monoclonal antibodies or antibodies with humanized glycosylation patterns, as well as virus-like particles for vaccine platforms (reviewed in Cross, 2015). While products from transgenic duckweed have yet to make it into the commercial space, genetic modification of duckweed for commercial applications remains an interesting prospect. It has been shown that the E1 gene from the bacterium A. cellulolyticus, encoding a hydrolytic enzyme used in fuel production (Sun et al., 2007), as well as therapeutic monoclonal antibodies against CD20, CD30, and a 2b interferon (Cox et al., 2006) can be successfully expressed in Le. minor.
One benefit of duckweed’s aquatic growth habitat is the secretion of target proteins directly into the growth medium for easier purification. Aprotinin, a medically important protease inhibitor, has been expressed in La. punctata, secreted into the medium, and successfully purified using an immunoaffinity column (Rival et al., 2008). Adequate vaccine production is a growing challenge in a globalized world where pathogens can spread with greater ease than ever before. Duckweeds may provide an efficient and safe system for vaccine production to help tackle the rising demand for vaccines, especially in the developing world. Protective antigens have been developed against porcine epidemic diarrhea virus (Ko et al., 2011) and tuberculosis (Peterson et al., 2015). Several studies have shown the potential of using duckweeds to express antigens from the avian influenza virus H5N1 (Thu et al., 2015; Bertran et al., 2015), while Firsov et al. (2018) also successfully expressed a part of the M2e surface protein of H5N1 with the mucosal adjuvant ricin lectin subunit B (RTB) in duckweed. Mice orally immunized with the RTB-M130 protein produced specific antibodies against M2e peptide. Transgenic duckweed plants thus appear to be a promising route for producing quality antigens as edible vaccines that may provide affordable control of H5N1 in animals.
History of duckweed in plant biology research
The first recorded scientific studies of the Lemnaceae concentrated on describing their morphology and histology. A monograph focused on the Lemnaceae was published in 1839 by the botanist Matthias J. Schleiden (Schleiden, 1839), one of the originators of the cell theory. Later, Christoph F. Hegelmaier published “The Lemnaceae: a monographic study” (Hegelmaier, 1868) with detailed drawings that are still relevant today. By the turn of the 20th century, it had been established that the family Lemnaceae comprised “four well-defined genera and about twenty-eight species, distributed throughout the torrid and temperate zones” (Thompson, 1898). The family has since been expanded to 5 genera with 36 species (Bog et al., 2020a, 2020b; Figure 3).
Modern studies of the Lemnaceae can be dated to the mid-1950s, with the movement of William Hillman and others away from the descriptive toward more quantitative biochemical mechanisms (Hillman, 1957). As pointed out by Hillman (1961) and expanded on by Landolt and Kandeler (1987), the use of duckweed as a model system for physiological and photosynthetic studies is based on its ease of manipulation and maintenance. Since duckweeds are small, morphologically reduced, fast-growing, easily cultivated under aseptic conditions, and particularly suited to biochemical studies involving isotope labeling, they were considered to be an ideal system for plant research (Hillman, 1976). For example, much of what we know about photoperiodic flowering responses came from fundamental studies conducted with Lemna by Hillman at the Brookhaven National Laboratory. This included diversification of the time measurement systems among different photoperiodic species both within the Lemnaceae and among different geographical isolates of a single species. In the era of plant molecular biology (since the 1980s), this valuable physiological information from early studies involving duckweed served to inspire researchers utilizing model plants with better genetic tools or stronger commercial interests, such as A. thaliana and rice.
The Lemnaceae are especially well suited for in vivo biochemical research compared to other plants. Their facile uptake of solutes from defined growth media and easy handling under controlled conditions have made them attractive for whole-plant biochemical research (Figure 1, B and C). Anthony J. Trewavas, one of the first to recognize this, used radiolabeled compounds to study nucleic acid and protein turnover in Le. minor plants (Trewavas, 1970, 1972). In a series of papers, Trewavas et al. also explored the effects of ABA on turion development, including changes in protein and mRNA synthesis (Smart and Trewavas, 1984) and the downregulation of cell wall polysaccharide synthesis by UDP-apiose/UDP-xylose synthase during turion development (Longland et al., 1989). Similarly, Datko et al. (1978a) used Le. perpusilla 6746 to carry out in vivo labeling studies with radiolabeled sulfate and other sulfur-containing compounds to quantify sulfur assimilation by plants into various products under defined conditions. A phytostat system (analogous to the powerful chemostat platform for the study of microbial metabolism networks) was established for the first time with plants to enable quantitative measurement of the effects from changing concentrations of a single component in the system (Datko et al., 1978b).
Another exceptional achievement leveraging the in vivo labeling capacity of the duckweed system was the early characterization of auxin biosynthesis pathways in plants. Using intact duckweed plants to carry out isotopic compound loading to avoid confounding issues of other studies that rely on excised plant parts, the first clear biochemical evidence for the existence of a tryptophan (Trp)-independent indole-3-acetic acid (IAA) biosynthesis pathway in Le. gibba G3 was demonstrated (Baldi et al., 1991). This conclusion was soon supported by the isolation and study of Trp-auxotrophs from A. thaliana (Normanly et al., 1993). The relative importance of the two IAA biosynthesis pathways was also shown to be influenced by temperature in Le. gibba G3, with the Trp-independent pathway predominating at higher growth temperatures, such as 30°C (Rapparini et al., 2002). While the Trp-dependent pathway has been well characterized over the past two decades (Mano and Nemoto, 2012), the first breakthrough for the Trp-independent pathway finally came with the report of indole synthase (INS) as a key branchpoint toward IAA from indole-3 glycerol phosphate (Wang et al., 2015). However, the pathway from indole produced by INS in the cytosol to IAA remains to be elucidated.
Finally, perhaps the most impactful contribution of duckweed to plant biology is the discovery of the function and roles of the D1 protein in photosystem II (PSII) of the thylakoids, also known as the 32 kDa herbicide-binding protein. Mattoo et al. in the Edelman laboratory produced seminal results describing the lifecycle (Mattoo and Edelman, 1987) of this highly unstable protein that acts as a critical primary electron acceptor in the PSII complex (Mattoo et al., 1984). Pulse labeling of La. punctata plants with 35S-methionine enabled the discovery of the translocation of precursor D1 protein from its synthesis on the stromal lamellae to its site of action in the granal lamellae of the thylakoids, among other findings (Mattoo and Edelman, 1987).
Coinciding with the rise of Arabidopsis genetics and molecular biology beginning in the mid-1980s, basic research using duckweeds subsided over the period of 1990–2010. While some of the first cloned plant genes were from Lemna, and the dissection of promoter functions had been carried out in duckweed using transient assays (Stiekema et al., 1983; Kehoe et al., 1994), the duckweed research community remained small during this boom time of plant molecular genetics. Over the past decade, however, the rapid accumulation of genomics tools coupled with advances in analytical and computational technologies have set the stage for a renaissance of duckweed research (Lam et al., 2014). The many advantages of duckweed as an experimental platform, such as its aquatic habitat and rapid clonal propagation, coupled with its smaller gene repertoire, should enable it once again to help illuminate novel pathways and paradigms in plant biology.
Hot topics in duckweed research
Propelled in large part by recent advances in genomics tools, several areas of duckweed research are beginning to contribute new knowledge to key areas of plant biology. In the following sections, we describe research topics that exemplify how the physical characteristics of duckweeds help to simplify experimental systems. We also illustrate how Lemnaceae could provide a novel window into the diversity of evolved strategies that have helped shape genome structures as well as their associated metabolic, physiological, and evolutionary processes.
Plant–microbiota interactions
A rapidly growing area of plant biology is plant microbiome research, which ultimately aims to understand and manipulate plant-associated microbial communities toward beneficial agricultural outcomes (Busby et al., 2017). This research is hindered in terrestrial plant model systems, where both biological factors such as complex plant development (Beilsmith et al., 2021) and technical factors such as the need to manipulate soil conditions (Kremer et al., 2021) complicate reductionist approaches used to gain a mechanistic understanding of plant microbiome processes (Vorholt et al., 2017). In contrast, duckweed presents a facile model plant microbiome system for dissecting mechanisms underlying plant–microbiota interactions. Compared to soil, the liquid growth medium used for duckweed allows for an experimental system with minimal heterogeneity and ready access to exudates and microbe(s) of interest. An exudate trapping system (Lu et al., 2014) and whole microbial community capture method (Ishizawa et al., 2017) have been developed to analyze these exudates and microbial communities. When testing interactions between bacteria isolates and gnotobiotic plants, the aquatic lifestyle and small size of duckweed greatly facilitate the functional analysis of plant–microbe interactions under defined conditions. To begin to dissect the mode of interaction between duckweed associated bacteria (DABs) and duckweed, a PCR-based attachment assay and methods for reintroduction of microbes with gnotobiotic plants have been reported (Huang et al., 2020; Acosta et al., 2020; Ishizawa et al., 2020). These studies should help pave the way for the application of reductionist approaches to the duckweed system, as have been shown in Arabidopsis (Durán et al., 2018). Together, these tools could be leveraged to build a tractable high-throughput model plant microbiome system with duckweed.
One question that arises is whether the microbiome profiles found in terrestrial plants are related to those found in duckweed. To this end, several high-throughput amplicon sequencing studies have generated community profiles and characterized the assembly of the DAB community from field sites and reconstitution experiments using gnotobiotic plants. The DAB community is primarily composed of Proteobacteria, followed by Bacteroidetes (Xie et al., 2015; Acosta et al., 2020), and most closely resembles the profile from terrestrial leaf microbiomes (Acosta et al., 2020). In addition, duckweed serves as a distinct habitat for bacteria compared to the surrounding environmental community, demonstrating the selection of particular bacteria, as observed in terrestrial plants (Xie et al., 2015; Acosta et al., 2020; Fitzpatrick et al., 2020; Huang et al., 2020). Furthermore, many of the bacterial taxa associated with duckweed appear to be enriched in the surrounding water when compared to the original inoculum (Acosta et al., 2020), as has been observed for the recruitment of root-associated communities (Edwards et al., 2015). Therefore, similar structuring principles may be found between the duckweed microbiome and terrestrial plant microbiome.
While community profiling studies are essential for providing a comprehensive understanding of the ecological processes and structural aspects of the plant microbiome, reductionist-based approaches involving gnotobiotic plants and synthetic microbial consortia are used to establish causality and gain a mechanistic understanding of the roles various microbial community members may play in this ecosystem (Vorholt et al., 2017; Durán et al., 2018). Prerequisites for this approach include being able to isolate representative microbial community members as well as having reference genomes for these community members and the plant host. Fortunately, high-quality genomes are available for different duckweed species and a significant portion of the DAB community can be cultivated (Matsuzawa et al., 2010; Tanaka et al., 2018), with an increasing number of DAB genomes being sequenced. Functional studies with DAB isolates have shown that they affect duckweed growth under defined conditions (Toyama et al., 2017; Ishizawa et al., 2019b), facilitate the removal of nutrients (Zhao et al., 2015), enhance bioremediation of pollutants (Toyama et al., 2006; Yamaga et al., 2010; Ogata et al., 2013; Xie et al., 2014), produce phytohormones (Gilbert et al., 2018), and affect plant development (Huang et al., 2020). The ability to readily analyze duckweed exudates and collect microbial community members from the growth medium allows researchers to examine which classes of exudates play key roles in community assembly and to characterize how microbiota respond to these compounds. For example, certain duckweed exudates, such as fatty acid esters and amides, were found to stimulate the nitrogen-removal efficiency of bacteria by activating nitrate and nitrite reductases (Sun et al., 2016), the methane oxidation activity of methanotrophs (Iguchi et al., 2019), or the pollutant degradation activity of bacteria (Xu et al., 2015). Analysis of the community dynamics revealed by inoculating different plant-growth-promoting bacteria onto duckweed demonstrated that the colonization of bacteria can change in the presence of other microbes even though these bacteria occupy different plant niches (Yamakawa et al., 2018) and irrespective of inoculation density or inoculation order (Ishizawa et al., 2019a). Furthermore, exogenous DABs with plant-growth-promoting ability can be displaced from the host by native DAB communities (Ishizawa et al., 2020), highlighting the resiliency of the indigenous community against invaders.
The emergence of duckweed genomics will advance the exploration of the mechanistic underpinnings from the plant host perspective using transcriptomic approaches. These advances, together with the ability to precisely define media conditions and construct synthetic bacterial communities, should promote our understanding of DAB community establishment at the systems level. Future directions for duckweed microbiome research should include characterizing communities of other microorganisms associated with duckweed, including fungi and algae (Watanabe et al., 2016). While this floating macrophyte is evolutionarily distant from terrestrial models, investigation of the duckweed microbiome may assist in the discovery of conserved principles underlying plant microbiota interactions that can be translated to economically important crops.
Disease resistance genes in duckweed
As duckweeds are aquatic plants that have adapted to thrive in organics-rich environments with high microbial loads, these plants might be expected to have heightened disease resistance functions. It is thus surprising that the number of nucleotide-binding leucine-rich repeat domain genes (NLRs), which encode many of the proteins involved in disease resistance, was found to be significantly lower in the Sp. polyrhiza 9509 (Sp9509) genome than in other model plants (Michael et al., 2017). Interestingly, similar attrition of these NLRs was observed in the eelgrass Zostera marina and in carnivorous plants of the Utricularia and Genlisea genera (Baggs et al., 2020). These observations raise this intriguing question: What is the basis for the broad-spectrum resistance in the Lemnaceae? In a recent study of the Sp7498 genome, it was suggested that there may be an amplification of antimicrobial protein (AMP)-encoding genes due in part to the presence of many of these loci with duplicated AMPs (An et al., 2019). The heightened expression of some of these AMPs in Sp7498 was hypothesized to provide enhanced immunity in this duckweed.
Using the latest available annotations from high-quality reference genome assemblies for two clones each of Sp. polyrhiza (Hoang et al., 2018; Harkess et al., 2021) and Wo. australiana (Michael et al., 2021), NLR genes were curated using the NB-ARC (nucleotide-binding adaptor shared by APAF-1, certain R gene products, and CED-4) conserved motif and their gene numbers compared to those from well-curated genomes of Brachypodium distachyon BD21, rice (Oryza sativa), A. thaliana Col-0, and alfalfa (Medicago truncatula). Genes encoding pattern-recognition receptors (PRRs), which include genes involved in microbial-associated molecular pattern-triggered immunity (PTI), were also identified. Those that contain a kinase domain (LRR-RK, LysM-RK) were separately curated from those that do not (LRR-RP, LysM-RP). Lastly, genes encoding members from eight families of known plant AMPs (Hammami et al., 2009) in these same genomes were identified as well. Using a relatively stringent informatics pipeline, 169 NLR genes were found in A. thaliana with a genome size of ∼150 Mb (Table 1). The number of NLR-related genes increased in the other model terrestrial plant genomes with larger genome sizes. In contrast, both Sp. polyrhiza genome assemblies produced sets of 20–35 NLR genes. Since Sp. polyrhiza has approximately the same genome size as A. thaliana, this lower NLR gene count is unlikely to be due to a smaller genome even when factoring in the smaller gene set of the Sp. polyrhiza genome (Michael et al., 2017). A more dramatic decrease to only 3–4 NLR-related genes is observed in the two clones of Wo. australiana (Michael et al., 2021). Since the Wo. australiana genomes are ∼375 Mb in size, similar to the rice genome, this low number of NLR genes indicates that a significant repertoire of varied NLRs is not needed by Wo. australiana to provide immune functions for the survival of this species. Interestingly, examining the number of PRR genes encoding proteins with a kinase domain, which likely include genes in the PTI pathway, both Wo. australiana genomes exhibit an increase in the number of genes in this category compared to the two Spirodela genomes. In contrast, the number of PRR-related genes without a kinase domain showed the opposite trend with the two Wolffia genomes displaying two- to three-fold lower numbers, which reflects a similar trend for most of the other gene families in this species compared to other sequenced Lemnaceae species (Michael et al., 2021). Strikingly, while both Spirodela genomes are missing LysM-type PRRs, they can be found in the Wo. australiana genomes. This observation indicates that these LysM-type PRRs must have been present in the common ancestor of Sp. polyrhiza and Wo. australiana. Altogether, these results suggest that in Wo. australiana, the basal immunity functions mediated by PRR genes could play a more dominant role in the pathogen response in contrast to Sp. polyrhiza, where the NLR genes could play a more significant role.
Table 1.
Comparison of defense-related genes between sequenced duckweeds and those of other model plant species. Genes encoding AMPs, PRRs containing LRR (LRR-RK, LRR-RP) or LysM (LysM-RK, LysM-RP) domains, and NLR proteins were curated for Sp9509 (Hoang et al., 2018), Sp7498 (Harkess et al., 2021), Wo. australiana 8730, and Wo. australiana 7733 (Michael et al., 2021). For comparison, B. distachyon, O. sativa, A. thaliana, and M. truncatula were analyzed with the same pipeline using proteomes from the Monocot and Dicot PLAZA 4.5 database (Van Bel et al., 2018). For a detailed methods description, see https://github.com/kenscripts/tpc_dw_review/.
| Defense-Related Gene Category | Gene Type | Sp. polyrhiza 9509 | Sp. polyrhiza 7498 | Wo. australiana 8730 | Wo. australiana 7733 | B. distachyon | O. sativa | A. thaliana | M. truncatula |
|---|---|---|---|---|---|---|---|---|---|
| NLRs | NB-ARC | 20 | 35 | 3 | 4 | 346 | 483 | 169 | 797 |
|
| |||||||||
| PRRs | LRR-RK | 56 | 36 | 95 | 78 | 401 | 375 | 356 | 434 |
| LRR-RP | 78 | 80 | 20 | 32 | 111 | 143 | 162 | 332 | |
| LysM-RK | 0 | 0 | 2 | 2 | 3 | 3 | 5 | 19 | |
| LysM-RP | 0 | 0 | 5 | 5 | 3 | 7 | 4 | 9 | |
|
| |||||||||
| AMPs | Cyclotide | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 |
| Defensin | 5 | 5 | 9 | 9 | 44 | 23 | 60 | 85 | |
| Hevein | 0 | 0 | 6 | 5 | 16 | 10 | 10 | 8 | |
| Knottins | 0 | 0 | 0 | 0 | 2 | 2 | 0 | 0 | |
| LTP | 25 | 26 | 28 | 28 | 67 | 79 | 61 | 91 | |
| Snakin | 9 | 8 | 12 | 13 | 15 | 15 | 19 | 28 | |
| Thionin | 2 | 1 | 0 | 0 | 12 | 12 | 4 | 1 | |
| Vicilin | 3 | 2 | 0 | 0 | 2 | 0 | 2 | 1 | |
Finally, of the eight classes of AMPs that were examined, the class of AMPs with the highest number of genes in duckweeds is in the lipid transfer protein (LTP) category, followed by the Snakin and Defensin classes (Table 1). While this analysis did not reveal a significant increase in the number of AMP-encoding genes in these duckweed genomes, the persistent numbers of LTP, Snakin, and Defensin genes in Wo. australiana suggest that they are indispensable for this minimalist species where the total gene set has decreased to ∼15,000 genes. These observations raise the possibility that these AMPs could be downstream effectors from the LRR-RK-type PRRs in Wolffia to provide major immune functions in comparison to Spirodela. It would be interesting to test predictions of this hypothesis, such as the induction of AMP-encoding genes upon the challenge of Wolffia with microbial pathogens. Elucidating the mechanisms of induced immunity to potential invaders in duckweeds is likely to uncover novel disease resistance strategies and could be a key advance for a successful cropping system to be designed using these versatile plants.
Cell autonomy and transcriptome studies of circadian rhythms in duckweeds
The small size, relatively flat surface of the fronds, and simple architecture of intact duckweed are ideal for long-term, automated microscopic observations, which can be used to track and quantify plant physiological processes at high spatial resolution. One such process is the circadian clock, an internal biological timing device for adjusting various functions to the cyclic natural environment. Once entrained, physiological outputs of the clock display circadian rhythms even under constant conditions. These rhythms are usually synchronized to the day–night cycles in the natural environment. Core circadian clock genes encoding single MYB domain transcription factors form conserved transcription–translation feedback loops that generate circadian oscillations in plants. While circadian regulation of plant genes has been well studied at the organismal and tissue levels, whether the clock can function cell autonomously was unknown. To observe circadian rhythms of individual cells in live duckweed plants, bioluminescence imaging of Le. gibba fronds that were transfected with the AtCCA1:LUC reporter by particle bombardment was used to monitor rhythmic gene expression of multiple cells simultaneously (Muranaka et al., 2013). Analyses of the circadian clock’s behavior at a single-cell level in these intact plants revealed quantitative parameters for the heterogeneity and stability of individual cellular clocks, spontaneous local coupling of cellular clocks, and the stochastic synchronization patterns of cellular clocks under non-24-h light/dark cycles (reviewed in Muranaka and Oyama, 2018). Using the experience and tools generated from these studies, bioluminescence monitoring at single-cell resolution was successfully applied to detached Arabidopsis leaves (Kanesaka et al., 2019). Although the gradual growth with 3D distortion of Arabidopsis leaves was a disadvantage for long-term studies of more than several days, the bioluminescence of individual cells was traceable to detect cellular circadian rhythms. Thus, the bioluminescence monitoring system originally established in duckweed could be applied to other plants of interest to examine cell autonomous behaviors in different species.
Plant growth is controlled by the coordination of time-of-day (TOD) pathways to specific times in a day by the circadian clock (Michael et al., 2008a). In fact, global TOD gene regulation is a common feature in plants, with 30%–50% and 10%–20% of genes displaying peak expression or phased expression at specific times over the day under diurnal and circadian free-run conditions, respectively (Michael et al., 2008b; Filichkin et al., 2011). All core circadian clock orthologs cycle in Spirodela with the expected phases as in other plants, and most fundamental biological processes such as photosynthesis, the cell cycle, protein synthesis, and metabolism are phased to a conserved TOD. Strikingly, only 13% of genes in Wo. australiana were found to display TOD expression under the same diurnal LDHH conditions (Michael et al., 2021). While all the core circadian clock orthologs have conserved TOD expression, many pathways are not phased to a specific time of day in Wolffia. However, the core photosynthetic and chloroplast-associated pathways still cycle in a TOD fashion, which is consistent with the notion that anticipation of the light–dark transitions to optimize these light-dependent, energy-related functions is still being maintained. The loss of TOD regulation for most pathways in Wolffia apparently results from the loss of key developmental and light-regulated pathways, which parallels its highly reduced body plan and loss of roots. The contrasts between the control of global gene expression patterns among duckweeds should provide an exciting model to further understand growth-related pathways in plants as well as mechanisms for energy use optimization via strategic minimization of gene control.
Duckweed metabolomics: plant secondary metabolites
Duckweeds have been recognized as an excellent model system to study biosynthetic pathways and metabolic mechanisms in large part due to their simplified structure and the rapid uptake of labeled precursor compounds under defined growth conditions. With the rapid advance in analytical instrumentation and growth of databases for compound identification over the past decades, the time is ripe for using the duckweed platform to delineate the intricate metabolite network of a complete plant at the systems level. Characterizing these metabolites will also help to explore and optimize the use of duckweed beyond its role as a model organism in the laboratory, since these plants could have commercial potential.
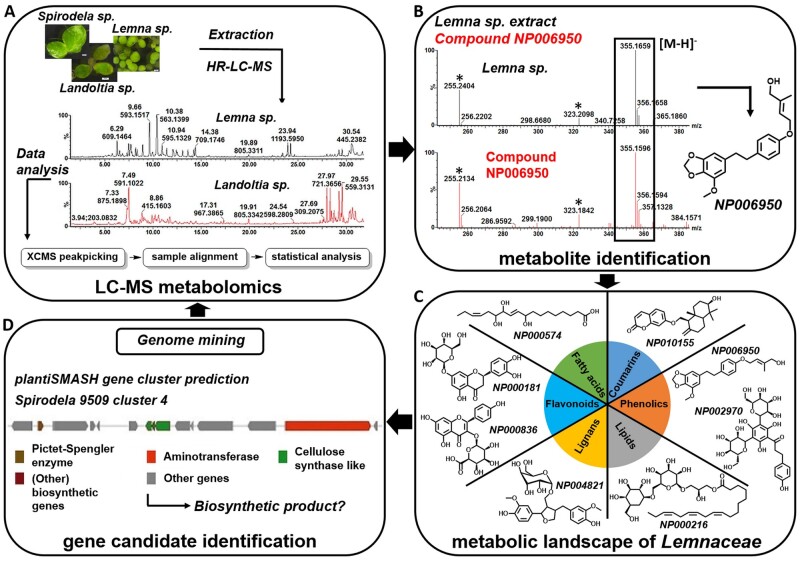
Secondary metabolites and small molecules have been less studied in the Lemnaceae than in other model plants, but this is now rapidly changing. Using gas-chromatography coupled with mass spectrometry (GC–MS ), the profile of epicuticular waxes was analyzed in Sp. polyrhiza (Borisjuk et al., 2018). While in land plants, sterols are not present on the surface of leaves or stems but in the cuticle, high levels of sterols were found on the surface of Spirodela fronds, which is thought to function as a sunscreen for these plants. Furthermore, GC–MS analysis of exudates of Sp. polyrhiza also led to the identification of several fatty acids and fatty acid amides that might be involved in plant/bacteria interactions (Sun et al., 2016). Semipolar metabolites from duckweeds have been analyzed by liquid chromatography coupled with mass spectrometry (LC–MS) techniques, and distinct patterns of metabolite profiles have been found between different genera and species (Figure 4A). Flavonoids, compounds used for nutraceutical, medicinal, and cosmetic purposes, have also been characterized in some duckweeds (Qiao et al., 2011; Ren et al., 2016; Böttner et al., 2020). Currently, the major bottleneck in mass spectrometry-based metabolomics is metabolite identification. Shahaf et al. (2016) developed a unique analysis platform that enables matching of mass spectra from plant extracts against a natural product library containing several thousand entries of secondary metabolite standards (Figure 4B). This technique was used for analysis of LC–MS chromatograms from Le. gibba, La. punctata, Sp. polyrhiza, and Wo. globosa, which led to the identification of 29 metabolites from 8 different natural product classes with high confidence, providing the first glimpse of the “natural product repertoire” for the Lemnaceae (Shahaf et al., 2016; Figure 4C). While several of the identified molecule classes are typically found in monocots, such as flavonoids related to the apigenin and luteolin type, the Lemnaceae metabolite profile also showed several molecules/metabolite classes previously not described in these plants, such as gingerglycolipid “A” and its derivatives. These compounds were only previously described in ginger (Zingiber officinale) and were believed to be specialized metabolites unique to this plant.
Figure 4.
Integration of metabolomics and genomics analysis in the Lemnaceae. (A) Comprehensive mass spectrometry-based metabolomics as a tool for pathway elucidation and characterization of biosynthetic enzymes and genes. Plant extracts are analyzed using high-resolution LC–MS (HR-LC–MS). Raw chromatograms typically consist of several thousand mass signals, and data can be deconvoluted using computation tools. (B) One method for compound identification from complex mixtures involves matching the mass spectra from an extract corresponding to a peak onto an entry in spectral libraries. An example for metabolite NP006950 (Weizmass library) is shown, illustrating that chromatographic retention, accurate mass, and mass fragmentation of a peak fraction in a Lemna gibba extract (upper black peak) matched a library entry (lower red peak) that can be assigned with high confidence (Shahaf et al., 2016). (C) These techniques, in combination with chemical classification, allow the metabolic landscape of Lemnaceae to be defined. (D) This knowledge can help identify metabolic genes/enzymes using either structure-based reaction prediction or genome mining approaches (such as metabolic gene cluster prediction that hints at possible biosynthetic pathways) as well as possible secondary metabolites (such as by correlation with the metabolomics dataset).
Metabolite identification not only provides leads for the search and characterization of derivatives for these compounds, but it also facilitates the prediction of metabolic pathways as well as biosynthetic genes/enzymes, and hence the metabolic network of duckweed. One approach to defining metabolic pathways involving a biosynthetic precursor of interest is called DLEMMA (Feldberg et al., 2009). In this method, the duckweed is fed different stable isotope-labeled variants of a precursor compound to facilitate the subsequent use of the labeling pattern of detected compounds for intermediate identification. For example, feeding of Sp. polyrhiza with differentially labeled tyrosine led to the identification of 59 tyrosine-derived metabolites, all of them new for Lemnaceae (Feldberg et al., 2018). Furthermore, the identification of these derivatives enabled the construction of entirely new metabolic pathway models based only on metabolite data without using genetic information. Knowledge about the metabolic landscape of duckweed, especially metabolites identified with high confidence, allows one to predict enzymatic reactions necessary to form a specific structure. These hypothetical reactions can in turn hint at enzymes and genes that are involved in biosynthesis and for which candidate genes with the predicted enzyme activity may be identified from transcriptome and/or genome data.
The production of secondary metabolites is often under environmental control, with a particular stress or condition modulating the pathway for the metabolite. The use of metabolite extraction techniques under a single environmental condition will likely not be sufficient to characterize all the potential compounds a duckweed clone can produce. Mining the genome for potential metabolic pathways (i.e. the gene space) can help establish the repertoire of potential enzymes in one duckweed clone needed to generate a specific secondary metabolite. Genes encoding enzymes involved in successive steps of biosynthetic pathways that form secondary metabolites in plants are sometimes found in clusters (Osbourn, 2010), termed biosynthetic gene clusters (BGCs). Identifying duckweed BGCs by analyzing well-annotated reference genomes in combination with metabolite extraction and identification creates a powerful tool that can be used to help discover biosynthetic pathways in duckweed. As described earlier, reference genomes are rapidly being completed for the Lemnaceae (see “Chromosomes and genomics of duckweeds”). These resources enable the use of strategies for linking metabolites to their biosynthetic genes/enzymes. Figure 4D illustrates how whole-genome data can be used by the PlantiSMASH analysis pipeline to identify plant BGCs (Kautsar et al., 2017). Candidate BGC contents were compared using reference genomes of Sp. polyrhiza and Wo. australiana. Sp. polyrhiza had a lower number of candidate clusters (five), while the Wo. australiana genome showed eleven candidate BGCs. Interestingly, this analysis revealed more unique gene clusters between the two duckweed genera than common ones. Although the data are preliminary, the occurrence of clusters and the resulting metabolites that can be produced from them might help explain important phenotypic differences between the genera. In summary, the Lemnaceae display a diverse and unique metabolite profile between genera. Their metabolites and pathways show potential for varied biological activity and might help explain the lifestyles of the different members of the Lemnaceae family in their natural aquatic environment. The combination of metabolic profiling and gene discovery could be a powerful tool to characterize the chemical space in these aquatic plants and to investigate how metabolites might mediate the interactions of duckweeds with their aquatic environment.
Experimental evolution and eco-evolutionary dynamics
Their short generation times and small size also make duckweed an ideal model system to tackle problems in evolutionary ecology (Laird and Barks, 2018). Here, we highlight two emerging fields that can greatly benefit from using duckweed as a model system. Experimental evolution, in which phenotypic and genotypic changes are measured under controlled selection regimes, is a powerful approach to study the processes and mechanisms of adaptive evolution in real-time (Teotonio et al., 2009; Schlotterer et al., 2015). Plant scientists have used long-term experiments to investigate the phenotypic responses of plants to herbivory under natural conditions (either herbivore exclusion or addition to the plant population) for decades (Detling and Painter, 1983). They have convincingly demonstrated that herbivores can drive the contemporary evolution of plant traits, such as the levels of defense metabolites (Züst et al., 2012), plant phenology (Agrawal et al., 2012), and growth rates (Turley et al., 2013). However, due to the long lifecycles of many plants, most of these studies only investigated the evolutionary process during a few generations, which limits the power of detecting traits under selection (Kofler and Schlotterer, 2014). In addition, such studies are difficult to replicate in many other laboratories, as they require a large planting area and long durations. The short generation times (within a few days) and small size (1–15 mm) of duckweeds are excellent attributes for experimental evolution studies to address many questions that are not as tractable with other systems. For example, under indoor conditions, one can use the “select and resequence” approach to study >100 generations of plants that are evolving under different stress conditions within 1 year (Burghardt et al., 2018). A critical component for such studies is to have diverse genotypes at the beginning of the study, as evolution in such experiments will likely be affected by initial genetic variations in the population. To this end, the currently available duckweed collections and their ongoing sequencing efforts described here should provide the resources and reference material critical for such studies.
It is now apparent that evolution can take place within ecological timescales and alter population dynamics (Lavergne et al., 2010), which in turn can modify evolutionary trajectories and create eco-evolutionary feedbacks (Kinnison and Hairston, 2007; Pelletier et al., 2009). Understanding how these feedbacks alter the evolution of species interactions is one of the major challenges in evolutionary ecology. Addressing this challenge requires the interacting organisms to be specifically manipulated at the ecological and evolutionary levels (Bailey et al., 2009; Hendry, 2019), which is challenging for many study systems. However, recent work has demonstrated the potential of using duckweeds to study eco-evolutionary feedbacks. Using a combination of theoretical modeling and experimental manipulation of duckweed evolution, rapid evolution was shown to affect the dynamics of competing species and eco-evolutionary interactions to shape the trajectory of species coexistence (Hart et al., 2019). In another recent study, eco-evolutionary dynamics were characterized for duckweed–microbiome interactions where microbiome presence apparently led to rapid evolutionary change in bacterial community members, which in turn affected the bacterial species composition and host fitness (Tan et al., 2021). The advent of duckweed genomics will put these tiny plants in a position to further advance research on eco-evolutionary dynamics by providing quantitative genomic data to help unravel the genomic underpinnings and the underlying molecular mechanisms of these complex processes at the systems level (Rudman et al., 2018).
Duckweed research tools, technologies, and resources
Critical for successful model plant systems are tools, technologies, and resources that allow researchers to fully explore their questions. Key technologies include a genotyping platform, transformation system, and genetic methods. Also at the heart of a vibrant community is an organizing committee and international meetings that provide researchers opportunities to share research ideas and findings. Finally, resources such as genomics databases (and, in the case of duckweed, clone collections) will allow interested laboratories to quickly adopt a new model and explore its potential.
Genotyping
Several universal plastidic barcode sequences, among those recommended by the Consortium for the Barcode of Life plant-working group (Hollingsworth et al., 2009), have been used to distinguish 31 of the 37 duckweed species recognized at the time (Borisjuk et al., 2015), with more challenging species found within the Lemna, Wolffiella, and Wolffia genera. AFLP technology, although technically challenging and more expensive, was used to further resolve most of the species within these genera (Bog et al., 2010, 2013). More recently, genotyping-by-sequencing (Bog et al., 2020c) and a tubulin-based polymorphism method (Braglia et al., 2021) demonstrated the feasibility of resolving interspecific differences amongst closely related duckweed species. As more genome sequences become available, these technologies can be refined and serve as high-resolution methods for rapid species identification in duckweed.
Besides distinguishing duckweed species, biotechnological applications will require identification at the clonal level, since significant physiological and biochemical differences can exist between clones of the same species (Sree et al., 2015b). Facile genotyping methods with the capability of resolving intraspecific variations would be invaluable. A novel bioinformatic pipeline using so-called polymorphic NB-ARC-related genes was successfully tested using a training set of genome sequences from nine clones of Sp. polyrhiza to identify primer sets with the ability to distinguish these nine clones from each other as well as 11 additional Sp. polyrhiza clones, illustrating this strategy’s efficacy (Chu et al., 2018). Sp. polyrhiza was selected for this proof-of-concept study due to its low intraspecific variations based on whole-genome sequencing studies (Michael et al., 2017; Ho et al., 2019; Xu et al., 2019). A limitation of this technique is that it requires the generation of multiple whole-genome assemblies for the investigated species as a training set for the algorithm. Fortunately, more and more high-quality genome assemblies for numerous duckweed species and clones are being generated and made available through large-scale sequencing studies.
Transformation and its application in duckweed
Transformation has been demonstrated for almost all duckweed genera, with the fastest methods taking 2–3 months, like in other model crops. The first demonstration of transgenic gene expression in duckweed was reported in 1991 by transfecting Le. gibba fronds with a transgene using the biolistic particle gun system to demonstrate phytochrome regulation of a sequence upstream of the SSU5B gene by transient assays (Rolfe and Tobin, 1991). Conditions for transient expression using Agrobacterium-mediated gene transfer were subsequently described for Wo. columbiana (Boehm et al., 2001). Stable transformation was demonstrated in the late 1990s (Stomp and Rajbhandari 1999; Edelman et al., 1999). Some of the key challenges for development of stable transformation systems were the high inter- and intraspecific variability observed in attempts to standardize tissue culture conditions for plant regeneration from callus tissue (Chang and Chiu, 1978; Chang and Hsing, 1978; Liu et al., 2019). The effects of medium composition and light intensity on callus induction, growth, and frond regeneration were optimized for Le. gibba (Moon and Stomp,1997; Li et al., 2004), while carbohydrate and phytohormone requirements for callus induction, callus growth, and frond regeneration were established for clones of La. punctata 5562 (Li et al., 2004; formerly named Sp. oligorrhiza) and more recently, for Wo. arrhiza (Khvatkov et al., 2015).
Duckweed can be transformed through calli or directly using fronds or roots. Duckweed callus transformation (DCT) protocols generally involve the selection/treatment of an explant, callus induction, callus maintenance and growth, callus transformation, transformant selection, and frond regeneration (Li et al., 2004), with many of these steps requiring a different set of conditions for different clones (Huang et al., 2016; Yang et al., 2017). Different DCT protocols require specific conditions because duckweed clones, even within the same species, can respond differently to transformation conditions (Chang and Hsing, 1978; Liu et al., 2019). Furthermore, only certain clones produce callus under a particular condition, which also limits the generality of each DCT protocol (Yang et al., 2018a; Liu et al., 2019). As an alternative, direct in planta transformation (DIPT) of fronds or roots has been used to produce stable transformants of duckweed (Ko et al., 2011; Balaji et al., 2016; Yang et al., 2018b). Duckweed DIPT involves the selection/treatment of explants, Agrobacterium inoculation, cocultivation, selection, and frond regeneration. DIPT protocols with Lemna have used wounded fronds and separated mother and daughter fronds as explants (Ko et al., 2011; Yang et al., 2018b), while unwounded Spirodela fronds may be required for transformation (Balaji et al., 2016). Like DCT protocols, Agrobacterium inoculation and cocultivation in DIPT protocols involve the addition of acetosyringone (Ko et al., 2011; Balaji et al., 2016; Yang et al., 2018b), while selection and frond regeneration can also be combined in DIPT protocols to speed up transformation (Ko et al., 2011; Yang et al., 2018b). Monitoring transgene expression for a period of 10 months showed stable transformation with a DIPT protocol (Balaji et al., 2016). Transformation protocols also exist for Wolffia species, albeit with lower transformation efficiency compared to protocols for other species (Khvatkov et al., 2015; Heenatigala et al., 2018).
In summary, the short transformation time required in DIPT protocols makes them an attractive alternative to DCT protocols for duckweed research. Currently, researchers can adopt two transformation strategies depending on their research objective: (1) transformation conditions can be optimized for a particular clone from a species of interest that has already been identified as amenable to transformation or (2) several clones from a species of interest can be screened using a particular transformation protocol to identify ones with a higher frequency of success. With the introduction of miRNA silencing (Cantó‐Pastor et al., 2015) and precise genome-editing technologies using the CRISPR/Cas9 method (Liu et al., 2019), a routine transformation protocol should allow for hypothesis-driven studies in the duckweed model system.
Genetics and crossing systems
Flower-induction and cross-pollination protocols will help to facilitate genetic studies in the Lemnaceae. While duckweed propagation is mainly vegetative, many studies have investigated flowering induction in duckweed to reveal triggers of sexual reproduction in these plants (see “Anatomy, morphology, and growth characteristics”). Among these, SA and the chelating agent EDDHA can promote flowering in many duckweed species when added to the culture medium (Pieterse, 2013). A recent systematic comparison of the effects of these chemicals, daylength, and other culture conditions on the frequency of flower induction and seed production was reported for clones of Sp. polyrhiza, Le. gibba, and Wo. microscopica (Fourounjian et al., 2021). Although floral induction is possible in duckweed, flowers are often aborted and do not form seeds, as observed in some clones of Le. gibba. Fu et al. (2017) found that the application of SA can prevent seed abortion in Le. gibba clone 7741 but not in clone 5504. The anthers did not dehisce in clone 5504, and artificially released pollen grains did not germinate, suggesting male sterility. This hypothesis of male sterility was supported by cross-pollination between the two clones, since pollination of clone 5504 with pollen grains from clone 7741 yielded seeds. Analysis of the resulting progeny using intersimple sequence repeat markers suggested that cross-pollination occurred, resulting in the production of intraspecific hybrids (Fu et al., 2017). While it would be important to verify the hybrid nature of these putative F1 plants by examining the F2 lines for segregation of the two sets of chromosomes, the data so far indicate that outcrossing between duckweed clones might be possible using this strategy. If validated, this approach could open the gateway for facile methods of transformation and genetic studies that are commonly performed in other model plants.
Missing duckweed technologies
To further enhance the field of duckweed research as a model plant system, the development and advances in several key technologies are highly desirable. One such missing technology is a reliable genetic system where routine flowering induction and hybridization between strains or backcrosses can be performed. As discussed above, there are several species that have clones known to flower routinely, and good progress is being made to establish the necessary knowledge base for environmental and hormonal cues that induce flowering in a few key duckweed species. The current lack of such a reliable and facile genetic protocol has hindered the development of basic research in duckweed. While mutant induction and identification in duckweed were shown to be feasible, predominantly via the deployment of X-ray bombardment or treatment with nitrosomethyl urea as a mutagen to generate so-called “aberrants” that have auxotrophic or growth phenotypes (Smith and Castle, 1960; Posner, 1962; Tam et al., 1995), the genetic basis of the phenotypes observed in these lines has never been resolved. For example, Strain 1073 derived from Lemna perpusilla 6746 was found to exhibit differences in its flowering response to light and is blocked in its photosynthetic activities (Eames and Posner, 1974). This was subsequently traced to a loss of expression of the nuclear gene encoding the Rieske Fe–S protein (Lam and Malkin, 1985), which led to rapid turnover of the other proteins that make up the cytochrome b6/f complex in the thylakoid membrane (Bruce and Malkin, 1991). The question of whether the flowering phenotype in the Le. perpusilla 1073 mutant is related to its photosynthesis defect could potentially be dissected using classical genetic approaches such as backcrossing to wild-type parents. Unfortunately, this clone has not been maintained.
Another important technology that is missing is systematic mutant library generation to facilitate functional analysis of all genes encoded in the genomes of a representative species. With the completion of multiple high-quality assemblies for a growing number of duckweed species, the availability of indexed insertion libraries in these species would provide a powerful resource to query gene function rapidly, as in Arabidopsis (Matus et al., 2014) and the green alga Chlamydomonas reinhardtii (Li et al., 2016). To enable the deployment of this approach, several accessory technologies are needed: (1) a facile transformation technology that is easily scalable; (2) robust and reliable transformant identification and curation; (3) high-throughput insert sequencing and indexing; and (4) protocols for selfing of transformants to produce homozygotes for phenotyping. To generate a large number of random insertions, an efficient transformation system would need to be deployed, and a more rapid method such as DIPT would be preferable (Yang et al., 2018b). The recent advent of in planta transformation using nanomaterials as DNA carriers could also be an attractive method to develop for duckweeds to further improve the throughput of transgenic plant line production (Wang et al., 2019). In addition to generating stable transformants in large numbers, this approach could also be applied for rapid, high-throughput transient expression assays to test gene functions.
Creating and maintaining a large library of transgenic duckweed lines presents another challenge. While the incorporation of a transgene can be readily detected using PCR-based screening strategies after selecting duckweed plants on the appropriate antibiotics, continuously propagating thousands of transgenic duckweed lines as living plant clones in culture would be laborious and costly. A better approach for maintaining the selected transgenic lines may be to produce seeds from the identified transgenic lines and store them until they are needed. Thus, the selection of a proper duckweed clone that is amenable to viable seed production is an important consideration. Systematic attempts at cryopreservation of duckweeds have only been marginally successful (Sauter 1993; https://patents.google.com/patent/WO2011005502A3), so it is not a viable option at this time .
Insertion site identification is facilitated by rapid advances in high-throughput sequencing technologies that reduce costs while increasing the accuracy of DNA sequencing. Techniques such as TAILED-PCR (Liu and Chen, 2007) can be used to amplify DNA from regions adjacent to the inserts embedded in the genome of the transformed duckweed. A next generation approach would be to employ CRISPR coupled to long read sequencing such as Oxford Nanopore Technologies to define the sites of transgene insertions. Sequencing should reveal the genome locations of the insertion events and whether they are likely to disrupt the function of a duckweed gene.
Lastly, in order to use insertion lines to study gene function, it is important to keep in mind that most insertions are initially hemizygous. Unless the insertion resulted in a dominant phenotype, which will probably be a rare event, the transgenic lines will not show a mutant phenotype in the T0 primary transformants. Thus, readily induced flowering and setting of viable seeds will be highly desirable traits for users of this resource as well.
Community organization, newsletter, and international meetings
Steering committees have been instrumental in guiding, coordinating, and communicating research for successful plant model systems such as Arabidopsis, maize, and Brachypodium (Parry et al., 2020). The International Steering Committee on Duckweed Research and Applications (ISCDRA) was founded in 2013 to provide similar functions for the duckweed community. The ISCDRA releases a quarterly newsletter (Duckweed Forum) that represents a comprehensive resource for new and current researchers as well as application specialists, providing announcements, research updates, guidelines, protocols, and discussion topics such as nomenclature standards that are relevant for the community. Past and present issues of the Duckweed Forum can be freely accessed via the website of the Rutgers Duckweed Stock Cooperative (RDSC; ruduckweed.org). The ISCDRA organizes a biennial international meeting that brings together researchers from all over the world, with the next meeting scheduled for 2022 in Germany (previous meetings were held in China, the United States, Japan, India, and Israel). In addition, each year at the Plant and Animal Genome meeting in San Diego, CA, there is a workshop titled “Duckweed Research and Applications,” which has been running since 2018.
Clone collections
The first major duckweed clone collection in the community was amassed and maintained from 1953 to 2012 by Elias Landolt at the Geobotanical Institute of SFIT Zurich. At its peak, this collection comprised over 1,000 clones and served as a source of duckweed germplasm for duckweed researchers around the world as well as starting material for later collections (Lämmler and Bogner, 2014). Currently, several duckweed clone collections exist around the world, including the United States, Germany, Switzerland, and China (see Lam et al., 2020, for locations and contacts for these facilities). Researchers can request duckweed clones from these collections by directly contacting the respective director of the collection. To identify and track duckweed clones, a 4-digit numbering system was first adopted by Elias Landolt. Clones from the original Landolt Collection maintain this identification system, with numbers for more recent clone isolates assigned by the RDSC. However, with the increasing number of investigators interested in population studies involving duckweed over the past decade, newer clones in collections can also adopt an individualized identification system consisting of an investigator’s initials followed by digits (Lam et al., 2020). An accepted clone identification system is critical for replicating experimental results between laboratories and extending prior work. It thus serves as a foundation for the community in which results from various reported studies can be integrated and leveraged together based on knowledge of the biological material being tested. As noted earlier in this review, duckweeds have historically been used to study various aspects of plant biology. In some cases, keynote clones have been extensively used in a particular field of study. This includes Le. minor 8627 and Le. aequinoctialis 6002 for transformation studies (Cantó‐Pastor et al., 2015; Liu et al., 2019); Le. perpusilla 6746 (now renamed as Le. aequinoctialis 6746) and Le. gibba G3 to study flowering (Hillman, 1959; Fu et al., 2017); Le. minor 5512 as well as Le. minor 5576 to study plant–microbe interactions (Ishizawa et al., 2020; Acosta et al., 2020); Le. minor 5500 for ecological and toxicological studies (Paolacci et al., 2018b); La. punctata 5562 (previously named Sp. oligorrhiza) to study the D1 protein of PSII (Mattoo and Edelman, 1987); and Sp. polyrhiza clones 7498 and 9509 for genomic studies (Wang et al., 2014; Michael et al., 2017; Hoang et al., 2018).
Genomic resources
The duckweed community is growing and generating valuable resources for genome discovery at an increasing rate. A growing number of datasets are available at the National Library of Medicine (NCBI) and the Short Read Archive (SRA). Currently, the Sp. polyrhiza reference genomes (Sp7498v2; Sp9509v3) can be accessed from multiple locations including phytozome (https://phytozome-next.jgi.doe.gov/), PLAZA (https://bioinformatics.psb.ugent.be/plaza/), SpirodelaGenome (http://spirodelagenome.org/), and NCBI. The reference genomes for two Sp. intermedia clones, Si7747 and Si8410, can be obtained from the European Nucleotide Archive and raw data from NCBI SRA (Hoang et al., 2020). Additional Lemna and Wolffia genomes can also be found at lemna.org and CoGe (https://genomevolution.org/coge/). One of the first duckweed datasets at NCBI contains full-length cDNA clones for Wo. arrhiza 8872a, Wo. australiana 7733, and La. punctata 9264 sequenced by the Waksman Student Scholars Program at Rutgers University in New Jersey since 2009 (https://wssp.rutgers.edu/research/previous). In addition to genome sequencing data, multiple transcriptome, proteome, and bacterial DNA amplicon datasets from studies involving duckweeds have been deposited in publicly accessible databases. These duckweed-related resources are summarized in Table 2, along with their access information and references.
Table 2.
Lemnaceae germplasm and data resources
| Germplasm | Affiliation | Location | Director | Number of Clones | Contact | ||
|---|---|---|---|---|---|---|---|
| University of Jena | Jena, Germany | Klaus J. Appenroth | ∼550 | Klaus.Appenroth@uni-jena.de | |||
| Landolt Stock Collection | Zurich, Switzerland | Walter Laemmler | ∼400 | wlaemmler@duckweed.ch | |||
| RDSC | New Brunswick, NJ, USA | Eric Lam | ∼740 | ruduckweed.org | |||
| University of Münster | Münster, Germany | Shuqing Xu | ∼260 | shuqing.xu@uni-muenster.de | |||
| Chengdu Institute of Biology | Chengdu, China | Hai Zhao | ∼800 | zhaohai@cib.ac.cn | |||
|
| |||||||
| Genomes | Type | Clone | Technology | Assembly | Protein Coding Genes | Identifier | Reference |
|
| |||||||
| Nuclear | Sp. polyrhiza 7498 | PacBio | 139 Mb | 18,708 | CoGe: 55812 | Harkess et al. (2021); An et al. (2019) | |
| Sp. polyrhiza 9509 | ONT, mcFISH, BioNano | 138 Mb | 20,661 | CoGe: 51364 | Hoang et al. (2018); Michael et al. (2017) | ||
| Sp. intermedia 7747 | PacBio | 148 Mb | 22,245 | PRJEB35514 | Hoang et al. (2020) | ||
| Sp. intermedia 8410 | ONT, Illumina | 137 Mb | 21,594 | PRJEB35634 | Hoang et al. (2020) | ||
| Le. minor 5500 | Illumina | 472 Mb | 22,382 | CoGe: 27408 | Van Hoeck et al. (2015) | ||
| Wo. australiana 8730 | PacBio, Illumina | 354 Mb | 14,324 | CoGe: 56605 | Michael et al. (2021) | ||
| Wo. australiana 7733 | PacBio, Illumina | 393 Mb | 15,312 | CoGe: 56606 | Michael et al. (2021) | ||
| Mitochondrial | Sp. polyrhiza 7498 | SOLiD | 228 Kb | 57 | NC_017840.1 | Wang et al. (2012) | |
| Chloroplast | Sp. polyrhiza 7498 | PacBio | 168 Kb | 78 | MN419335.1 | Zhang et al. (2020) | |
| Le. minor | ABI | 166 Kb | 78 | NC_010109.1 | Mardanov et al. (2008) | ||
| We. lingulata 7289 | SOLiD | 169 Kb | – | JN160604.1 | Wang and Messing (2011) | ||
| Wo. australiana 7733 | SOLiD | 168 Kb | – | JN160605.1 | Wang and Messing (2011) | ||
| La. punctata (5 clones) | Illumina | 166–171 Kb | 80 | (See reference) | Ding et al. (2017) | ||
|
| |||||||
| Datasets | Type | Clone | Technology | Description | Data size | Identifier | Reference |
|
| |||||||
| Transcriptome | Sp. polyrhiza 7498 | PacBio | Multiple conditions | 492,435 isoforms | SRX5321175 | An et al. (2019) | |
| Sp. polyrhiza 7498 | Illumina | ABA treatment | ∼196 million reads | PRJNA557001 | An et al. (2019) | ||
| Le. aequinoctialis 6000 | Illumina | Nitrogen starvation | ∼202 million reads | PRJNA368628 | Yu et al. (2017) | ||
| Le. gibba 7741 | Illumina | SA-induced flowering | ∼953 million reads | PRJNA631155 | Fu et al. (2020b) | ||
| La. punctata 6001 | Illumina | Cadmium treatment | ∼419 million reads | PRJNA361433 | Xu et al. (2018) | ||
| sRNA and degradome | Sp. polyrhiza (2 clones) | Illumina | miRNA identification | ∼57 million reads | PRJNA473779 | Fourounjian et al. (2019) | |
| Sp. polyrhiza 7498 | Illumina | Degradome sequencing | ∼911 million reads | PRJNA473779 | Fourounjian et al. (2019) | ||
| Population | 69 Sp. polryhiza ecotypes | Illumina | Genetic diversity | ∼3,491 million reads | PRJNA476302 | Xu et al. (2019) | |
| 39 Sp. polyrhiza ecotypes | Illumina | Genetic diversity | ∼532 million reads | PRJNA552613 | Ho et al. (2019) | ||
| ssRNA sequencing | Sp. polyrhiza 7498 | Illumina | Salt stress | ∼459 million reads | PRJNA563960 | Fu et al. (2020a) | |
| Proteomics | La. punctata 0202 | iTRAQ-LC–MS/MS | Nutrient starvation | 6955 unique spectra | – | Huang et al. (2015) | |
| Bacterial community | Wo. australiana | Illumina | Reconstituted | 34 samples | PRJNA219151 | Xie et al. (2015) | |
| Spirodela, Lemna, Wolffia | Illumina | Natural and reconstituted | 64 samples | PRJNA561628 | Acosta et al. (2020) | ||
| Le. aequinoctialis, Rice | Illumina | Natural | 57 samples | PRJNA545325 | Huang et al. (2020) | ||
Outlook
There is a need for new models to tackle complex molecular and ecological processes in plant biology using multidisciplinary approaches. Duckweeds are well-suited to play an important role in these endeavors, as their clonal mode of reproduction, transformation systems, minimal core gene content, rapid growth, small size, easy uptake of labeled compounds, fewer cell types, and unique lifestyle provide new opportunities for the discovery of novel traits and pathways. Two advanced technologies that we believe will be especially exciting to apply to the duckweed model system are single-cell genomics and X-ray computed microtomography (microCT). Both of these methodologies have recently been successfully applied to excised plant organs or tissues to demonstrate their ability to reveal new information on root cell biology in the case of single-cell transcriptomics (Rhee et al., 2019) and leaf air-space quantification in intact leaf samples by microCT (Mathers et al., 2018). In fact, as this review was being prepared, the first report describing the application of microCT to duckweed was published (Jones et al., 2021). Because of its diminutive size, the duckweed model system should allow these approaches to be applied at the whole plant level and provide quantitative information on discrete lineages of cell populations or 3D parameters for plant architecture as a function of growth and the environment. These types of discoveries will be required for a future in which improved crops will have to withstand large temperature swings, droughts, and floods and there will be an increasing need for higher yields and nutritional qualities. As aquatic plants that have adapted to habitats that are distinct from those of land plants, the Lemnaceae appears to deploy different strategies for defense and growth regulation. Elucidating these alternative pathways may provide insights on novel strategies for designing future crops by the next generation of farmers, scientists, and climate warriors.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Table S1 . Examples of habitats and growth habits of duckweeds.
Supplementary Material
Acknowledgments
We would like to acknowledge the amazing work of Elias Landolt who amassed and organized the first large-scale duckweed collection in the community, as well as his monumental monographs that summarized all-things-duckweed up to the 1980s, which served as his legacy to our community. These are truly the cornerstones on which progress has been built upon up to the current time. We apologize to researchers whose relevant work we have not cited due to space limitations.
Funding
Duckweed research at the Lam laboratory is supported in part by a grant from the Department of Energy (DE-SC0018244), a Hatch project (#12116), and a Multi-State Capacity project (#NJ12710) from the New Jersey Agricultural Experiment Station at Rutgers University. Duckweed research at the Xu laboratory is funded by the German Science Foundation (DFG, #427577435 and #438887884) and the University of Münster.
Conflict of interest statement. None declared.
Contributor Information
Kenneth Acosta, Department of Plant Biology, Rutgers the State University of New Jersey, New Brunswick, NJ 08901, USA.
Klaus J Appenroth, Plant Physiology, Matthias Schleiden Institute, University of Jena, Jena 07737, Germany.
Ljudmilla Borisjuk, The Leibniz Institute of Plant Genetics and Crop Plant Research, Gatersleben D-06466, Germany.
Marvin Edelman, Department of Plant and Environmental Sciences, Weizmann Institute of Science, Rehovot 7610001, Israel.
Uwe Heinig, Department of Plant and Environmental Sciences, Weizmann Institute of Science, Rehovot 7610001, Israel.
Marcel A K Jansen, School of Biological, Earth and Environmental Sciences, Environmental Research Institute, University College Cork, Cork T23 TK30, Ireland.
Tokitaka Oyama, Department of Botany, Graduate School of Science, Kyoto University, Kyoto 606-8502, Japan.
Buntora Pasaribu, Department of Plant Biology, Rutgers the State University of New Jersey, New Brunswick, NJ 08901, USA.
Ingo Schubert, The Leibniz Institute of Plant Genetics and Crop Plant Research, Gatersleben D-06466, Germany.
Shawn Sorrels, Department of Plant Biology, Rutgers the State University of New Jersey, New Brunswick, NJ 08901, USA.
K Sowjanya Sree, Department of Environmental Science, Central University of Kerala, Periye 671320, India.
Shuqing Xu, Institute for Evolution and Biodiversity, University of Münster, Münster 48149, Germany.
Todd P Michael, Plant Molecular and Cellular Biology Laboratory, The Salk Institute of Biological Studies, La Jolla, California 92037, USA.
Eric Lam, Department of Plant Biology, Rutgers the State University of New Jersey, New Brunswick, NJ 08901, USA.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) are: Eric Lam (eric.lam@rutgers.edu) and Todd P. Michael (toddpmichael@gmail.com)
References
- Acosta K, Xu J, Gilbert S, Denison E, Brinkman T, Lebeis S, Lam E (2020) Duckweed hosts a taxonomically similar bacterial assemblage as the terrestrial leaf microbiome. PloS One 15: p.e0228560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamec L (2018) Ecophysiological characteristics of turions of aquatic plants: a review. Aquat Bot 148: 64–77 [Google Scholar]
- Agrawal AA, Hastings AP, Johnson MT, Maron JL, Salminen JP (2012) Insect herbivores drive real-time ecological and evolutionary change in plant populations. Science 338: 113–116 [DOI] [PubMed] [Google Scholar]
- An D, Zhou Y, Li C, Xiao Q, Wang T, Zhang Y, Wu Y, Li Y, Chao D-Y, Messing J et al. (2019) Plant evolution and environmental adaptation unveiled by long-read whole-genome sequencing of Spirodela. Proc Natl Acad Sci U S A 116: 18893–18899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- APG (1998) An ordinal classification for the families of flowering plants. Ann Missouri Bot Gard 85: 531–553 [Google Scholar]
- Appenroth KJ, Hertel W, Jugnickel F, Augsten H (1989) Influence of nutrient deficiency and light on turion formation in Spirodela polyrhiza (L.) Schleiden. Biochem Physiol Pflanzen 184: 395–403 [Google Scholar]
- Appenroth KJ, Augsten H (1990) Photophysiology of turion germination in Spirodela polyrhiza (L.) Schleiden-V. Demonstration of a calcium-requiring phase during phytochrome-mediated germination. Photochem Photobiol 52: 61–65 [Google Scholar]
- Appenroth KJ (2002) Co‐action of temperature and phosphate in inducing turion formation in Spirodela polyrhiza (greater duckweed). Plant Cell Environ 25: 1079–1085 [Google Scholar]
- Appenroth KJ, Nickel G (2010) Induction of turion formation in Spirodela polyrhiza under close-to-nature conditions: the environmental signals that induce the developmental process in nature. Physiol Plant 138: 312–320 [DOI] [PubMed] [Google Scholar]
- Appenroth KJ, Palharini L, Ziegler P (2013) Low-molecular weight carbohydrates modulate dormancy and are required for post-germination growth in turions of Spirodela polyrhiza. Plant Biol 15: 284–291 [DOI] [PubMed] [Google Scholar]
- Appenroth KJ, Sree KS, Böhm V, Hammann S, Vetter W, Leiterer M, Jahreis G (2017) Nutritional value of duckweeds (Lemnaceae) as human food. Food Chem 217: 266–273 [DOI] [PubMed] [Google Scholar]
- Appenroth KJ, Sree KS, Bog M, Ecker J, Seeliger C, Bohm V, Lorkowski S, Sommer K, Veeter W, Tolzin-Banasch K, et al. (2018) Nutritional value of the duckweed species of the genus Wolffia (Lemnaceae) as human food. Front Chem 6: 483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardenghi NM, Armstrong WP, Paganelli D (2017) Wolffia columbiana (Araceae, Lemnoideae): first record of the smallest alien flowering plant in southern Europe and Italy. Bot Lett 164: 121–127 [Google Scholar]
- Ashby E, Wangermann E, Winter EJ (1949) Studies in the morphogenesis of leaves. III. Preliminary observations on vegetative growth in Lemna minor. New Phytol 48: 374–381 [Google Scholar]
- Baggs EL, Monroe GJ, Thanki AS, O’Grady R, Schudoma C, Haerty W, Krasileva KV (2020) Convergent loss of an EDS1/PAD4 signaling pathway in several plant lineages reveals coevolved components of plant immunity and drought response. Plant Cell 32: 2158–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JK, Hendry AP, Kinnison MT, Post DM, Palkovacs EP, Pelletier F, Harmon LJ, Schweitzer JA (2009) From genes to ecosystems: an emerging synthesis of eco-evolutionary dynamics. New Phytol 184: 746–749 [DOI] [PubMed] [Google Scholar]
- Balaji P, Satheeshkumar PK, Venkataraman K, Vijayalakshmi MA (2016) Expression of anti‐tumor necrosis factor alpha (TNFα) single‐chain variable fragment (scFv) in Spirodela punctata plants transformed with Agrobacterium tumefaciens. Biotechnol Appl Biochem 63: 354–361 [DOI] [PubMed] [Google Scholar]
- Baldi BG, Maher BR, Slovin JP, Cohen JD (1991) Stable isotope labeling, in vivo, of D- and L-tryptophan pools in Lemna gibba and the low incorporation of label into IAA. Plant Physiol 95: 1203–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beilsmith K, Perisin M, Bergelson J (2021) Natural bacterial assemblages in Arabidopsis thaliana tissues become more distinguishable and diverse during host development. Mbio 12: e02723–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellini C, Pacurar DI, Perrone I (2014) Adventitious roots and lateral roots: similarities and differences. Annu Rev Plant Biol 65: 639–666 [DOI] [PubMed] [Google Scholar]
- Bertran K, Thomas G, Guo X, Bublot M, Pritchard N, Regan JT, Cox KM, Gasdaska JR, Dickey LF, Kapczynski DR, et al. (2015) Expression of H5 hemagglutinin vaccine antigen in common duckweed (Lemna minor) protects against H5N1 high pathogenicity avian influenza virus challenge in immunized chickens. Vaccine 33: 3456–3462 [DOI] [PubMed] [Google Scholar]
- Bhanthumnavin K, McGarry MG (1971) Wolffia arrhiza as a possible source of inexpensive protein. Nature 232: 495. [DOI] [PubMed] [Google Scholar]
- Boehm R, Kruse C, Voeste D, Barth S, Schnabl H (2001) A transient transformation system for duckweed (Wolffia columbiana) using Agrobacterium-mediated gene transfer. J Appl Bot 75: 107–111 [Google Scholar]
- Bog M, Baumbach H, Schween U, Hellwig F, Landolt E, Appenroth KJ (2010) Genetic structure of the genus Lemna L. (Lemnaceae) as revealed by amplified fragment length polymorphism. Planta 232: 609–619 [DOI] [PubMed] [Google Scholar]
- Bog M, Schneider P, Hellwig F, Sachse S, Kochieva EZ, Martyrosian E, Landolt E, Appenroth KJ (2013) Genetic characterization and barcoding of taxa in the genus Wolffia Horkel ex Schleid. (Lemnaceae) as revealed by two plastidic markers and amplified fragment length polymorphism (AFLP). Planta 237: 1–13 [DOI] [PubMed] [Google Scholar]
- Bog M, Lautenschläger U, Landrock M F., Landolt E, Fuchs J, Sree K S., Oberprieler C, Appenroth KJ (2015) Genetic characterization and barcoding of taxa in the genera Landoltia and Spirodela (Lemnaceae) by three plastidic markers and amplified fragment length polymorphism (AFLP). Hydrobiologia 749: 169–182 [Google Scholar]
- Bog M, Appenroth KJ, Sree KS (2019) Duckweed (Lemnaceae): its molecular taxonomy. Front Sustain Food Syst 3: 117 [Google Scholar]
- Bog M, Appenroth KJ, KS Sree (2020a) Key to the determination of taxa of Lemnaceae: an update. Nord J Bot 38: e02658 [Google Scholar]
- Bog M, Sree KS, Fuchs J, Hoang PTN., Schubert I, Kuever J, Rabenstein A, Paolacci S, Jansen MAK, Appenroth KJ (2020b) A taxonomic revision of Lemna sect. Uninerves (Lemnaceae). Taxon 69: 56–66 [Google Scholar]
- Bog M, Xu S, Himmelbach A, Brandt R, Wagner F, Appenroth KJ, Sree KS (2020c) Genotyping-by-sequencing for species delimitation in Lemna section Uninerves Hegelm. (Lemnaceae). In Cao XH, Fourounjian P, Wang W, eds, The Duckweed Genomes. Springer, Berlin, Heidelberg, pp. 115–123 [Google Scholar]
- Borisjuk N, Chu P, Gutierrez R, Zhang H, Acosta K, Friesen N, Sree KS, Garcia C, Appenroth KJ, Lam E (2015) Assessment, validation and deployment strategy of a two‐barcode protocol for facile genotyping of duckweed species. Plant Biol 17: 42–49 [DOI] [PubMed] [Google Scholar]
- Borisjuk N, Peterson AA, Lv J, Qu G, Luo G, Shi L, Chen G, Kishchenko O, Zhou Y, Shi J (2018) Structural and biochemical properties of duckweed surface cuticle. Front Chem 6: 317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttner L, Grabe V, Gablenz S, Böhme N, Appenroth KJ, Gershenzon J, Huber M (2020) Differential localization of flavonoid glucosides in an aquatic plant implicates different functions under abiotic stress. Plant Cell Environ 44: 900–914 [DOI] [PubMed] [Google Scholar]
- Braglia L, Lauria M, Appenroth KJ, Bog M, Breviario D, Grasso A, Gavazzi F, Morello L (2021) Duckweed species genotyping and interspecific hybrid discovery by tubulin-based polymorphism fingerprinting. Front Plant Sci 12: 625670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce BD, Malkin R (1991) Biosynthesis of the chloroplast cytochrome b6-f complex: studies in a photosynthetic mutant of Lemna. Plant Cell 3: 203–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burghardt LT, Epstein B, Guhlin J, Nelson MS, Taylor MR, Young ND, Sadowsky MJ, Tiffin P (2018) Select and resequence reveals relative fitness of bacteria in symbiotic and free-living environments. Proc Natl Acad Sci U S A 115: 2425–2430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busby PE, Soman C, Wagner MR, Friesen ML, Kremer J, Bennett A, Morsy M, Eisen JA, Leach JE, Dangl JL (2017) Research priorities for harnessing plant microbiomes in sustainable agriculture. PLoS Biol 15: p.e2001793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calicioglu O, Brennan RA (2018) Sequential ethanol fermentation and anaerobic digestion increases bioenergy yields from duckweed. Bioresour Technol 257: 344–348 [DOI] [PubMed] [Google Scholar]
- Cantó‐Pastor A, Mollá‐Morales A, Ernst E, Dahl W, Zhai J, Yan Y, Meyers BC, Shanklin J, Martienssen R (2015) Efficient transformation and artificial miRNA gene silencing in Lemna minor. Plant Biol 17: 59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedergreen N, Madsen TV (2002) Nitrogen uptake by the floating macrophyte Lemna minor. New Phytol 155: 285–292 [DOI] [PubMed] [Google Scholar]
- Ceschin S, Abati S, Ellwood NTW, Zuccarello V (2018) Riding invasion waves: spatial and temporal patterns of the invasive Lemna minuta from its arrival to its spread across Europe. Aquat Bot 150: 1–8 [Google Scholar]
- Chang WC, Chiu PL (1978) Regeneration of Lemna gibba G3 through callus culture. Z Pflanzenphysiol 89: 91–94 [Google Scholar]
- Chang WC, Hsing YI (1978) Callus formation and regeneration of frond-like structures in Lemna perpusilla 6746 on a defined medium. Plant Sci Lett 13: 133–136 [Google Scholar]
- Cheng JJ, Stomp AM (2009) Growing duckweed to recover nutrients from wastewaters and for production of fuel ethanol and animal feed. Clean Soil Air Water 37: 17–26 [Google Scholar]
- Chu P, Wilson GM, Michael TP, Vaiciunas J, Honig J, Lam E (2018) Sequence-guided approach to genotyping plant clones and species using polymorphic NB-ARC-related genes. Plant Mol Biol 98: 219–231 [DOI] [PubMed] [Google Scholar]
- Cleland CF, Ajami A (1974) Isolation and identification of the flower-inducing factor from aphid honeydew as being salicylic acid. Plant Physiol 54: 904–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole CT, Voskuil MI (1996) Population genetic structure in duckweed (Lemna minor, Lemnaceae). Can J Bot 74: 222–230 [Google Scholar]
- Coughlan NE, Kelly TC, Jansen MAK (2015) Mallard duck (Anas platyrhynchos)‐mediated dispersal of Lemnaceae: a contributing factor in the spread of invasive Lemna minuta? Plant Biol 17: 108–114 [DOI] [PubMed] [Google Scholar]
- Coughlan NE, Kelly TC, Jansen MAK (2017) “Step by step”: high frequency short-distance epizoochorous dispersal of aquatic macrophytes. Biol Invasions 19: 625–634 [Google Scholar]
- Cox KM, Sterling JD, Regan JT, Gasdaska JR, Frantz KK, Peele CG, Black A, Passmore D, Moldovan-Loomis C, Srinivasan M. et al. (2006) Glycan optimization of a human monoclonal antibody in the aquatic plant Lemna minor. Nat Biotechnol 24: 1591–1597 [DOI] [PubMed] [Google Scholar]
- Cross JW (2015) The rise and fall of a duckweed biotechnology firm: what can it tell us? ISCDRA Duckweed Forum 3: 83–88 [Google Scholar]
- Cross JW (2017) Duckweed roots: their role in vegetative dispersal. ISCDRA Duckweed Forum 5: 58–59 [Google Scholar]
- Cui W, Cheng JJ (2015) Growing duckweed for biofuel production: a review. Plant Biol 17: 16–23 [DOI] [PubMed] [Google Scholar]
- Darwin C (1859) On the Origin of Species by Means of Natural Selection, Vol 167. John Murray, London [Google Scholar]
- Datko AH, Mudd HS, Giovanelli J, MacNicol PK (1978a) Sulfur-containing compounds in Lemna perpusilla 6746 grown at a range of sulfate concentrations. Plant Physiol 62: 629–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datko AH, Mudd HS, MacNicol PK, Giovanelli J (1978b) Phytostat for the growth of Lemna in semicontinuous culture with low sulfate. Plant Physiol 62: 622–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detling JK, Painter EL (1983) Defoliation responses of western wheatgrass populations with diverse histories of prairie dog grazing. Oecologia 57: 65–71 [DOI] [PubMed] [Google Scholar]
- Ding Y, Fang Y, Guo L, Li Z, He K, Zhao Y, Zhao H (2017) Phylogenetic study of Lemnoideae (duckweeds) through complete chloroplast genomes for eight accessions. PeerJ 5: e4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Docauer DM (1983) A nutrient basis for the distribution of the Lemnaceae. PhD thesis. University of Michigan, USA
- Dolger C, Tirlapur UK, Appenroth KJ (1997) Phytochrome-regulated starch degradation in germinating turions of Spirodela polyrhiza. Photochem Photobiol 66: 124–127 [DOI] [PubMed] [Google Scholar]
- Duong TP, Tiedje JM (1985) Nitrogen fixation by naturally occurring duckweed—cyanobacterial associations. Can J Microbiol 31: 327–330 [Google Scholar]
- Durán P, Thiergart T, Garrido-Oter R, Agler M, Kemen E, Schulze-Lefert P, Hacquard S (2018) Microbial interkingdom interactions in roots promote Arabidopsis survival. Cell 175: 973–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eames C, Posner HB (1974) Abnormal flowering responses and a lack of photosynthesis in a mutant of Lemna perpusilla. Plant Physiol 55: S11 [Google Scholar]
- Echlin P, Lai CE, Hayes TL (1982) Low-temperature X-ray microanalysis of the differentiating vascular tissue in root tips of Lemna minor L. J Microsc 126: 285–306 [Google Scholar]
- Edelman M, Perl A, Flaishman M, Blumenthal A (1999) Transgenic Lemnaceae. WO 99/19497
- Edelman M, Colt M (2016) Nutrient value of leaf vs. seed. Front Chem 4: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards J, Johnson C, Santos-Medellín C, Lurie E, Podishetty NK, Bhatnagar S, Eisen JA, Sundaresan V (2015) Structure, variation, and assembly of the root-associated microbiomes of rice. Proc Natl Acad Sci U S A 112: E911–E920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekperusi AO, Nwachukwu EO, Sikoki FD (2020) Assessing and modeling the efficacy of Lemna paucicostata for the phytoremediation of petroleum hydrocarbons in crude oil-contaminated wetlands. Sci Rep 10: 8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Kholy AS, Youssef MS, Eid EM (2015) Genetic diversity of Lemna gibba L. and L. minor L. populations in Nile delta based on biochemical and ISSR markers. Egypt J Exp Biol Bot 11: 11–19 [Google Scholar]
- Feldberg L, Venger I, Malitsky S, Rogachev I, Aharoni A (2009) Dual labeling of metabolites for metabolome analysis (DLEMMA): a new approach for the identification and relative quantification of metabolites by means of dual isotope labeling and liquid chromatography-mass spectrometry. Anal Chem 81: 9257–9266 [DOI] [PubMed] [Google Scholar]
- Feldberg L, Dong Y, Heinig U, Rogachev I, Aharoni A (2018) DLEMMA-MS-imaging for identification of spatially localized metabolites and metabolic network map reconstruction. Anal Chem 90: 10231–10238 [DOI] [PubMed] [Google Scholar]
- Feuchtmayr H, Moran R, Hatton K, Connor L, Heyes T, Moss B, Harvey I, Atkinson D (2009) Global warming and eutrophication: effects on water chemistry and autotrophic communities in experimental hypertrophic shallow lake mesocosms. J Appl Ecol 46: 713–723 [Google Scholar]
- Filichkin SA, Breton G, Priest HD, Dharmawardhana P, Jaiswal P, Fox SE, Michael TP, Chory J, Kay SA, Mockler TC (2011) Global profiling of rice and poplar transcriptomes highlights key conserved circadian-controlled pathways and cis-regulatory modules. PLoS One 6: e16907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firsov A, Tarasenko I, Mitiouchkina T, Shaloiko L, Kozlov O, Vinokurov L, Rasskazova E, Murashev A, Vainstein A, Dolgov S (2018) Expression and immunogenicity of M2e peptide of avian influenza virus H5N1 fused to ricin toxin B chain produced in duckweed plants. Front Chem 6: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick CR, Salas-González I, Conway JM, Finkel OM, Gilbert S, Russ D, Teixeira PJPL, Dangl JL (2020). The plant microbiome: from ecology to reductionism and beyond. Ann Rev Microbiol 74: 81–100 [DOI] [PubMed] [Google Scholar]
- Fourounjian P, Tang J, Tanyolac B, Feng Y, Gelfand B, Kakrana A, Tu M, Wakim C, Meyers BC, Ma J, Messing J (2019) Post‐transcriptional adaptation of the aquatic plant Spirodela polyrhiza under stress and hormonal stimuli. Plant J 98: 1120–1133 [DOI] [PubMed] [Google Scholar]
- Fourounjian P, Slovin J, Messing J (2021) Flowering and seed production across the Lemnaceae. Int J Mol Sci 22: 2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L, Huang M, Han B, Sun X, Sree KS, Appenroth KJ, Zhang J (2017). Flower induction, microscope-aided cross-pollination, and seed production in the duckweed Lemna gibba with discovery of a male-sterile clone. Sci Rep 7: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L, Ding Z, Tan D, Han B, Sun X, Zhang J (2020a) Genome-wide discovery and functional prediction of salt-responsive lncRNAs in duckweed. BMC Genomics 21: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L, Tan D, Sun X, Ding Z, Zhang J (2020b) Transcriptional analysis reveals potential genes and regulatory networks involved in salicylic acid-induced flowering in duckweed (Lemna gibba). Plant Physiol Biochem 155: 512–522 [DOI] [PubMed] [Google Scholar]
- Geber G (1989) Zur Karyosystematik der Lemnaceae. PhD thesis. University of Vienna, Vienna, Austria
- Gilbert S, Xu J, Acosta K, Poulev A, Lebeis S, Lam E (2018) Bacterial production of indole related compounds reveals their role in association between duckweeds and endophytes. Front Chem 6: 265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Jin Y, Xiao Y, Tan L, Tian X, Ding Y, He K, Du A, Li J, Yi Z, et al. (2020) Energy-efficient and environmentally friendly production of starch-rich duckweed biomass using nitrogen-limited cultivation. J Clean Prod 251: 119726 [Google Scholar]
- Hammami R, Ben Hamida J, Vergoten G, Fliss I (2009) PhytAMP: a database dedicated to antimicrobial plant peptides. Nucleic Acids Res 37: D963–D968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harkess A, McLaughlin F, Bilkey N, Elliot K, Emenecker R, Mattoon E, Miller K, Czymmek K, Vierstra R, Meyers BCet al. (2021) Improved Spirodela polyrhiza genome and proteomic analyses reveal a conserved chromosomal structure with high abundance of chloroplastic proteins favoring energy production. J Exp Bot 72: 2491–2500 [DOI] [PubMed] [Google Scholar]
- Hart SP, Turcotte MM, Levine JM (2019) Effects of rapid evolution on species coexistence. Proc Natl Acad Sci 116: 2112–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heenatigala PPM., Yang J, Bishopp A, Sun Z, Li G, Kumar S, Hu S, Wu Z, Lin W, Yao L, Duan P (2018) Development of efficient protocols for stable and transient gene transformation for Wolffia globosa using Agrobacterium. Front Chem 6: 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegelmaier F (1868) Die Lemnaceen. Eine monographische Untersuchung. Engelmann, Leipzig, Germany [Google Scholar]
- Hendry AP (2019) A critique for eco-evolutionary dynamics. Funct Ecol 33: 84–94 [Google Scholar]
- Hillman WS (1957) Nonphotosynthetic light requirement in Lemna minor and its partial satisfaction by kinetin. Science 126: 165–166 [DOI] [PubMed] [Google Scholar]
- Hillman WS (1959) Experimental control of flowering in Lemna I. General methods. Photoperiodism in Lemna perpusilla 6746. Am J Bot 46: 466–473 [Google Scholar]
- Hillman WS (1961) The Lemnaceae, or duckweeds. A review of the descriptive and experimental literature. Bot Rev 27: 221–287 [Google Scholar]
- Hillman WS (1976) Calibrating duckweeds—light, clocks, metabolism, flowering. Science 193: 453–458 [DOI] [PubMed] [Google Scholar]
- Ho EKH., Bartkowska M, Wright SI, Agrawal AF (2019) Population genomics of the facultatively asexual duckweed Spirodela polyrhiza. New Phytol 224: 1361–1371 [DOI] [PubMed] [Google Scholar]
- Hoang PTN., Michael TP, Gilbert S, Chu P, Motley ST, Appenroth KJ, Schubert I, Lam E (2018) Generating a high-confidence reference genome map of the Greater Duckweed by integration of cytogenomic, optical mapping, and Oxford Nanopore technologies. Plant J 96: 670–684 [DOI] [PubMed] [Google Scholar]
- Hoang PTN, Schubert V, Meister A, Fuchs J, Schubert I (2019) Variation in genome size, cell and nucleus volume, chromosome number and rDNA loci among duckweeds. Sci Rep 9: 3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang PTN, Fiebig A, Novak P, Macas J, Cao HX, Stepaneko A, Chen G, Borisjuk N, Scholz U, Schubert I (2020) Chromosome-scale genome assembly for the duckweed Spirodela intermedia, integrating cytogenetic maps, PacBio and Oxford Nanopore libraries. Sci Rep 10: 19230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth PM, Forrest LL, Spouge JL, Hajibabaei M, Ratnasingham S, van der Bank M, Chase MW, Cowan RS, Erickson DL, Fazekas AJ, et al. (2009) A DNA barcode for land plants. Proc Natl Acad Sci U S A 106: 12794–12797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Fang Y, Liu Y, Jin Y, Sun J, Tao X, Ma X, He K, Zhao H (2015) Using proteomic analysis to investigate uniconazole-induced phytohormone variation and starch accumulation in duckweed (Landoltia punctata). BMC Biotechnol 15: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Fu L, Sun X, Di R, Zhang J (2016) Rapid and highly efficient callus induction and plant regeneration in the starch-rich duckweed strains of Landoltia punctata. Acta Physiol Plant 38: 122 [Google Scholar]
- Huang W, Gilbert S, Poulev A, Acosta K, Lebeis S, Long C, Lam E (2020) Host-specific and tissue-dependent orchestration of microbiome community structure in traditional rice paddy ecosystems. Plant Soil 452: 379–395 [Google Scholar]
- Iberite M, Iamonico D, Abati S, Abbate G (2011) Lemna valdiviana Phil. (Araceae) as a potential invasive species in Italy and Europe: taxonomic study and first observations on its ecology and distribution. Plant Biosyst 145: 751–757 [Google Scholar]
- Iguchi H, Umeda R, Taga H, Oyama T, Yurimoto H, Sakai Y (2019) Community composition and methane oxidation activity of methanotrophs associated with duckweeds in a freshwater lake. J Biosci Bioeng 128: 450–455 [DOI] [PubMed] [Google Scholar]
- Ishizawa H, Kuroda M, Morikawa M, Ike M (2017) Evaluation of environmental bacterial communities as a factor affecting the growth of duckweed Lemna minor. Biotechnol Biofuels 10: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizawa H, Kuroda M, Inoue K, Inoue D, Morikawa M, Ike M (2019a) Colonization and competition dynamics of plant growth-promoting/inhibiting bacteria in the phytosphere of the duckweed Lemna minor. Microbiol Ecol 77: 440–450 [DOI] [PubMed] [Google Scholar]
- Ishizawa H, Tada M, Kuroda M, Inoue D, Ike M (2019b) Performance of plant growth-promoting bacterium of duckweed under different kinds of abiotic stress factors. Biocatal Agric Biotechnol 19: 101146 [Google Scholar]
- Ishizawa H, Kuroda M, Inoue D, Morikawa M, Ike M (2020) Community dynamics of duckweed-associated bacteria upon inoculation of plant growth-promoting bacteria. FEMS Microbiol Ecol 96: fiaa101. [DOI] [PubMed] [Google Scholar]
- ISO (2005) Water quality – determination of the toxic effect of water constituents and waste to duckweed (Lemna minor) – Duckweed growth inhibition test. International Organization for Standardization, Geneva, Switzerland
- Jacobs DL (1947) An ecological life-history of Spirodela polyrrhiza (greater duckweed) with emphasis on the turion phase. Ecol Monogr 17: 437–469 [Google Scholar]
- Jones DH, Atkinson BS, Ware A, Sturrock CJ, Bishopp A, Wells D.M. (2021) Preparation, scanning and analysis of duckweed using X-Ray computed microtomography Front. Plant Sci 11: 2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanesaka Y, Okada M, Ito S, Oyama T (2019) Monitoring single-cell bioluminescence of Arabidopsis leaves to quantitatively evaluate the efficiency of a transiently introduced CRISPR/Cas9 system targeting the circadian clock gene ELF3. Plant Biotechnol 36: 187–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur M, Kumar M, Singh D, Sachdeva S, Puri SK (2019) A sustainable biorefinery approach for efficient conversion of aquatic weeds into bioethanol and biomethane. Energy Convers Manag 187: 133–147 [Google Scholar]
- Kautsar SA, Duran HGS, Blin K, Osbourn A, Medema MH (2017) plantiSMASH: automated identification, annotation and expression analysis of plant biosynthetic gene clusters. Nucleic Acids Res 45: W55–W63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehoe DM, Degenhardt J, Winicov I, Tobin EM (1994) Two 10-bp regions are critical for phytochrome regulation of a Lemna gibba Lhcb gene promoter. Plant Cell 6: 1123–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana JP, Maheshwari SC (1986) A comparison of the effects of chelates, salicylic acid and benzoic acid on growth and flowering of Spirodela polyrrhiza. Plant Cell Physiol 27: 919–924 [Google Scholar]
- Khvatkov P, Chernobrovkina M, Okuneva A, Pushin A, Dolgov S (2015) Transformation of Wolffia arrhiza (L.) Horkel ex Wimm. Plant Cell Tissue Organ Cult 123: 299–307 [Google Scholar]
- Kim I (2007) Development of the root system in Spirodela polyrhiza (L.) schleiden (Lemnaceae). J Plant Biol 50: 540–547 [Google Scholar]
- Kim I (2011) Features of plastids within Spirodela polyrhiza. Korean J Microsc 41: 55–60 [Google Scholar]
- Kim I (2013) Cellular features of the fronds and turions in Spirodela polyrhiza. Korean Soc Microsc 43:140–145 [Google Scholar]
- Kim I (2016) Structural differentiation of the connective stalk in Spirodela polyrhiza (L.) Schleiden. Korean Soc Microsc Appl Microsc 46: 83–88 [Google Scholar]
- Kinnison MT, Hairston NG (2007) Eco-evolutionary conservation biology: contemporary evolution and the dynamics of persistence. Funct Ecol 21: 444–454 [Google Scholar]
- Klich G, Mújica MB, Fernández OA (1986) Stomatal morphology and ontogeny in Spirodela intermedia W. Koch. Aquat Bot 26: 155–164 [Google Scholar]
- Ko SM, Sun HJ, Oh MJ, Song IJ, Kim MJ, Sin HS, Goh CH, Kim YW, Lim PO, Lee HY, Kim SW (2011) Expression of the protective antigen for PEDV in transgenic duckweed, Lemna minor. Horticult Environ Biotechnol 52: 511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler R, Schlotterer C (2014) A guide for the design of evolve and resequencing studies. Mol Biol Evol 31: 474–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostopoulou S, Ntatsi G, Arapis G, Aliferis KA (2020) Assessment of the effects of metribuzin, glyphosate, and their mixtures on the metabolism of the model plant Lemna minor L. applying metabolomics. Chemosphere 239: 124582. [DOI] [PubMed] [Google Scholar]
- Kremer JM, Sohrabi R, Paasch BC, Rhodes D, Thireault C, Schulze-Lefert P, Tiedje JM, He SY (2021) Peat-based gnotobiotic plant growth systems for Arabidopsis microbiome research. Nat Protoc 16: 2450–2470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehdorf K, Jetschke G, Ballani L, Appenroth KJ (2014) The clonal dependence of turion formation in the duckweed Spirodela polyrhiza - an ecogeographical approach. Physiol Plant 150: 46–54 [DOI] [PubMed] [Google Scholar]
- Kwak M, Kim I (2008) Turion as dormant structure in Spirodela polyrhiza. Korean J Microsc 38: 307–314 [Google Scholar]
- Laird RA, Barks PM (2018) Skimming the surface: duckweed as a model system in ecology and evolution. Am J Bot 105: 1962–1966 [DOI] [PubMed] [Google Scholar]
- Lam E, Malkin R (1985) Characterization of a photosynthetic mutant of Lemna lacking the cytochrome b6-f complex. Biochim Biophys Acta 810: 106–109 [DOI] [PubMed] [Google Scholar]
- Lam E, Appenroth KJ, Michael T, Mori K, Fakhoorian T (2014) Duckweed in bloom: the 2nd International Conference on Duckweed Research and Applications heralds the return of a plant model for plant biology. Plant Mol Biol 84: 737–742 [DOI] [PubMed] [Google Scholar]
- Lam E, Appenroth KJ, Ma Y, Shoham T, Sree KS (2020) Registration of duckweed clones—future approach. Duckweed Forum 8: 35–37 [Google Scholar]
- Lä, mmler W, Bogner J (2014) Elias Landolt and the duckweeds. Aroideana 37: 81–88 [Google Scholar]
- Landolt E (1986) Biosystematic investigations in the family of duckweeds (Lemnaceae) (Vol 2). The family of Lemnaceae—a monographic study. Veroeffentlichungen des Geobotanischen Instituts der ETH, Vol 1. Stiftung Ruebel, Switzerland [Google Scholar]
- Landolt E, Kandeler R (1987) The family of Lemnaceae—a monographic study, 2. Biosystematic Investigations in the Family of Duckweeds (Lemnaceae). Veröffentlichungen des Geobotanischen Instutes der ETH. Stiftung Rübel, Zurich.
- Lavergne S, Mouquet N, Thuiller W, Ronce O (2010) Biodiversity and climate change: integrating evolutionary and ecological responses of species and communities. Annu Rev Ecol Evol Syst 41: 321–350 [Google Scholar]
- Lee CJ, Yangcheng HY, Cheng JJ, Jane JL (2016) Starch characterization and ethanol production of duckweed and corn kernel. Starch-Staerke 68: 348–354 [Google Scholar]
- Lemon C D, Posluszny U (2000) Comparative shoot development and evolution in the Lemnaceae. Int J Plant Sci 161: 733–748 [Google Scholar]
- Les DH, Crawford DJ, Landolt E, Gabel JD, Kimball RT (2002) Phylogeny and systematics of Lemnaceae, the duckweed family. Syst Bot 27: 221–240 [Google Scholar]
- Les DH, Crawford DJ, Kimball RT, Moody ML, Landolt E (2003) Biogeography of discontinuously distributed hydrophytes: a molecular appraisal of intercontinental disjunctions. Int J of Plant Sci 164: 917–932 [Google Scholar]
- Li J, Jain M, Vunsh R, Vishnevetsky J, Hanania U, Flaishman M, Perl A, Edelman M (2004) Callus induction and regeneration in Spirodela and Lemna. Plant Cell Rep 22: 457–464 [DOI] [PubMed] [Google Scholar]
- Li X, Zhang R, Patena W, Gang SS, Blum SR, Ivanova N, Yue R, Robertson JM, Lefebvre PA, Fitz-Gibbon ST, Grossman AR, Jonikas MC (2016) An indexed, mapped mutant library enables reverse genetics studies of biological processes in Chlamydomonas reinhardtii. Plant Cell 28: 367–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YG, Chen Y (2007) High-efficiency thermal asymmetric interlaced PCR for amplification of unknown flanking sequences. Biotechniques 43: 649–650 [DOI] [PubMed] [Google Scholar]
- Liu Y, Chen X, Wang X, Fang Y, Huang M, Guo L, Zhang Y, Zhao H (2018) Improving biomass and starch accumulation of bioenergy crop duckweed (Landoltia punctata) by abscisic acid application. Sci Rep 8: 9544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Wang Y, Xu S, Tang X, Zhao J, Yu C, He G, Xu H, Wang S, Tang Y, et al. (2019) Efficient genetic transformation and CRISPR/Cas9‐mediated genome editing in Lemna aequinoctialis. Plant Biotechnol J 17: 2143–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longland JM, Fry SC, Trewavas AJ (1989) Developmental control of apiogalacturonan biosynthesis and UDP-apiose production in a duckweed. Plant Physiol 90: 972–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Zhou Y, Nakai S, Hosomi M, Zhang H, Kronzucker HJ, Shi W (2014) Stimulation of nitrogen removal in the rhizosphere of aquatic duckweed by root exudate components. Planta 239: 591–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucey J (2003) Lemna minuta Kunth (Least Duckweed) in E. Cork (VC H5). Irish Bot News 13: 5–8 [Google Scholar]
- Ma YB, Zhu M, Yu CJ, Wang Y, Liu Y, Li ML, Sun YD, Zhao JS, Zhou GK (2018) Large-scale screening and characterization of Lemna aequinoctialis and Spirodela polyrhiza strains for starch production. Plant Biol 20: 357–364 [DOI] [PubMed] [Google Scholar]
- Mano Y, Nemoto K (2012) The pathway of auxin biosynthesis in plants. J Exp Bot 63: 2853–2872 [DOI] [PubMed] [Google Scholar]
- Maheshwari SC, Seth PN (1966) Induction of flowering in Wolffia microsporica by iron salt by ethylenediamine-di-O-hydroxyphenilacetic acid (FE-EDDHA). Z Pl Physiol 55: 89–91 [Google Scholar]
- Mardanov AV, Ravin NV, Kuznetsov BB, Samigullin TH, Antonov AS, Kolganova TV, Skyabin KG (2008) Complete sequence of the duckweed (Lemna minor) chloroplast genome: structural organization and phylogenetic relationships to other angiosperms. J Mol Evol 66: 555–564 [DOI] [PubMed] [Google Scholar]
- Martirosyan EV, Ryzhova NN, Skryabin KG, Kochieva EZ (2008) RAPD analysis of genome polymorphism in the family Lemnaceae. Russ J Genet 44: 360–364 [PubMed] [Google Scholar]
- Mathers AW, Hepworth C, Baillie AL, Sloan J, Jones H, Lundgren M, Fleming AJ, Mooney S, Sturrock CJ (2018) Investigating the microstructure of plant leaves in 3D with lab-based X-ray computed tomography. Plant Methods 14: 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzawa H, Tanaka Y, Tamaki H, Kamagata Y, Mori K (2010) Culture-dependent and independent analyses of the microbial communities inhabiting the giant duckweed (Spirodela polyrrhiza) rhizoplane and isolation of a variety of rarely cultivated organisms within the phylum Verrucomicrobia. Microbes Environ 25: 302–308 [DOI] [PubMed] [Google Scholar]
- Mattoo AK, Hoffman-Falk H, Marder JB, Edelman M (1984) Regulation of protein metabolism: Coupling of photosynthetic electron transport to in vivo degradation of the rapidly-metabolized 32-kilodalton protein of the chloroplast membranes. Proc Natl Acad Sci U S A 81: 1380–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattoo KA, Edelman M (1987) Intramembrane translocation and posttranslational palmitoylation of the chloroplast 32kDa herbicide binding protein. Proc Natl Acad Sci U S A 84: 1497–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matus JT, Ferrier T, Riechmann JL (2014) Identification of Arabidopsis knockout lines for genes of interest. Methods Mol Biol 1110: 347–362 [DOI] [PubMed] [Google Scholar]
- McLaren JS, Smith H (1976) The effect of abscisic acid on growth, photosynthetic rate and carbohydrate metabolism in Lemna minor L. New Phytol 76: 11–20 [Google Scholar]
- McCormac DJ, Greenberg BM (1992) Differential synthesis of photosystem cores and light-harvesting antenna during proplastid to chloroplast development in Spirodela oligorrhiza. Plant Physiol 98: 1011–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melaragno JE, Walsh MA (1976) Ultrastructural features of developing sieve elements in Lemna minor L.—the protoplast. Am J Bot 63: 1145–1157 [Google Scholar]
- Michael TP, Breton G, Hazen SP, Priest H, Mockler TC, Kay SA, Chory J (2008a) A morning-specific phytohormone gene expression program underlying rhythmic plant growth. PLoS Biol 6: e225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael TP, Mockler TC, Breton G, McEntee C, Byer A, Trout JD, Hazen SP, Shen R, Priest HD, Sullivan CM, et al. (2008b) Network discovery pipeline elucidates conserved time-of-day–specific cis-regulatory modules. PLoS Genet 4: e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael TP, Bryant D, Gutierrez R, Borisjuk N, Chu P, Zhang H, Xia J, Zhou J, Peng H, El Baidouri M, et al. (2017) Comprehensive definition of genome features in Spirodela polyrhiza by high-depth physical mapping and short-read DNA sequencing strategies. Plant J 89: 617–635 [DOI] [PubMed] [Google Scholar]
- Michael TP, Ernst E, Hartwick N, Chu P, Bryant D, Gilbert S, Ortleb S, Baggs EL, Sree KS, Appenroth KJ, et al. (2021) Genome and time-of-day transcriptome of Wolffia australiana link morphological minimization with gene loss and less growth control. Genome Res 31: 225–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mifsud S (2010) First occurences of Lemna minuta Kunth (fam. Lemnaceae) in the Maltese islands. Cent Medit Nat 5: 1–4 [Google Scholar]
- Moodley D, Procheş Ş, Wilson JR (2016) A global assessment of a large monocot family highlights the need for group-specific analyses of invasiveness. AoB Plants 8: plw009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon HK, Stomp AM (1997) Effects of medium components and light on callus induction, growth, and frond regeneration in Lemna gibba (Duckweed). In Vitro Cell Dev Biol Plant 33: 20–25 [Google Scholar]
- Muranaka T, Kubota S, Oyama T (2013) A single-cell bioluminescence imaging system for monitoring cellular gene expression in a plant body. Plant Cell Physiol 54: 2085–2093 [DOI] [PubMed] [Google Scholar]
- Muranaka T, Oyama T (2018) Monitoring circadian rhythms of individual cells in plants. J Plant Res 131: 15–21 [DOI] [PubMed] [Google Scholar]
- Nauheimer L, Metzler D, Renner SS (2012) Global history of the ancient monocot family Araceae inferred with models accounting for past continental positions and previous ranges based on fossils. New Phytol 195: 938–950 [DOI] [PubMed] [Google Scholar]
- Naumann B, Eberius M, Appenroth KJ (2007) Growth rate based dose-response relationships and EC-values of ten heavy metals using the duckweed growth inhibition test (ISO 20079) with Lemna minor L. clone St. J Plant Physiol 164: 1656–1664 [DOI] [PubMed] [Google Scholar]
- Normanly J, Cohen JD, Fink GR (1993) Arabidopsis thaliana auxotrophs reveal a tryptophan-independent biosynthetic pathway for indole-3-acetic acid. Proc Natl Acad Sci U S A 90: 10355–10359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata Y, Toyama T, Yu N, Wang X, Sei K, Ike M (2013) Occurrence of 4-tert-butylphenol (4-t-BP) biodegradation in an aquatic sample caused by the presence of Spirodela polyrhiza and isolation of a 4-t-BP-utilizing bacterium. Biodegradation 24: 191–202 [DOI] [PubMed] [Google Scholar]
- Osama R, Awadc HM, Ibrahima MG, Tawfikd A (2020) Mechanistic and economic assessment of polyester wastewater treatment via baffled duckweed pond. J Water Process Eng 35: 101179 [Google Scholar]
- Osbourn A (2010) Secondary metabolic gene clusters: evolutionary toolkits for chemical innovation. Trends Genet 26: 449–457 [DOI] [PubMed] [Google Scholar]
- Pagliuso D, Grandis A, Lam E, Buckeridge MS (2020) High saccharification, low lignin, and high sustainability potential make duckweeds adequate as bioenergy feedstocks. BioEnergy Res DOI:10.1007/s12155-020-10211-x [Online publication date: November 2, 2020] [Google Scholar]
- Paolacci S, Harrison S, Jansen MA (2018a) The invasive duckweed Lemna minuta Kunth displays a different light utilization strategy than native Lemna minor Linnaeus. Aquat Bot 146: 8–14 [Google Scholar]
- Paolacci S, Jansen MAK., Harrison S (2018b) Competition between Lemna minuta, Lemna minor, and Azolla filiculoides. Growing fast or being steadfast? Front Chem 6: 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry G, Provart NJ, Brady SM, Uzilday BMultinational Arabidopsis Steering CommitteeAdams K, Araújo W, Aubourg S, Baginsky S, Bakker Eet al. (2020) Current status of the multinational Arabidopsis community. Plant Direct 4: p.e00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters ETH.M., van Zuidam JP, van Zuidam BG, Van Nes EH, Kosten S, Heuts PGM, Roijackers RMM, Netten JJC, Scheffer M (2013) Changing weather conditions and floating plants in temperate drainage ditches. J Appl Ecol 50: 585–593 [Google Scholar]
- Pelletier F, Garant D, Hendry AP (2009) Eco-evolutionary dynamics. Philos T R Soc B 364: 1483–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera WH, Meepagala KM, Fronczek FR, Cook DD, Wedge DE, Duke SO (2019) Bioassay-guided isolation and structure elucidation of fungicidal and herbicidal compounds from Ambrosia salsola (Asteraceae). Molecules 24: 835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson AA, Vasylenko MY, Matvieieva NA, Kuchuk MV (2015) Accumulation of recombinant fusion protein-secretory analog of Ag85B and ESAT6 Mycobacterium tuberculosis proteins-in transgenic Lemna minor L. plants. Biotechnol Acta 8: 39–48 [Google Scholar]
- Pieterse AH, Müller LJ (1977) Induction of flowering in Lemna gibba G3 under short-day conditions. Plant Cell Physiol 18: 45–53 [Google Scholar]
- Pieterse AH (2013) Is flowering in Lemnaceae stress-induced? A review. Aquat Bot 104: 1–4 [Google Scholar]
- Posner HB (1962) Characteristics of X-ray-induced aberrants of Lemna perpusilla 6746. Plant Cell Physiol 3: 275–284 [Google Scholar]
- Qiao X, He W, Xiang C, Han J, Wu L, Guo D, Ye M (2011) Qualitative and quantitative analyses of flavonoids in Spirodela polyrrhiza by high-performance liquid chromatography coupled with mass spectrometry. Phytochem Anal 22: 475–483 [DOI] [PubMed] [Google Scholar]
- Rapparini F, Tam YY, Cohen JD, Slovin JP (2002) Indole-3-acetic acid metabolism in Lemna gibba undergoes dynamic changes in response to growth temperature. Plant Physiol 128: 1410–1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D, Han B, Xin Z, Liu W, Ma S, Liang Y, Yi L (2016) Computation-aided separation of seven components from Spirodela polyrrhiza (L.) via counter-current chromatography. Sep Purif Technol 165: 160–165 [Google Scholar]
- Ren H, Jiang N, Wang T, Omar MM, Ruan W, Ghafoor A (2018) Enhanced biogas production in the duckweed anaerobic digestion process. J Energy Res Technol Trans ASME 140: 041805 [Google Scholar]
- Rhee SY, Birnbaum KD, Ehrhardt DW (2019) Toward building a plant cell atlas. Trends Plant Sci 24: 303–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimon D, Galun E (1968) Morphogenesis of Wolffia microscopica: frond and flower development. Phytomorphology 18: 364–372 [Google Scholar]
- Rival S, Wisniewski JP, Langlais A, Kaplan H, Freyssinet G, Vancanneyt G, Vunsh R, Perl A, Edelman M (2008) Spirodela (duckweed) as an alternative production system for pharmaceuticals: a case study, aprotinin. Transgenic Res 17: 503–513 [DOI] [PubMed] [Google Scholar]
- Rolfe SA, Tobin EM (1991) Deletion analysis of a phytochrome-regulated monocot RBCS promotor in a transient assay system. Proc Natl Acad Sci 88: 2683–2686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudman SM, Barbour MA, Csillery K, Gienapp P, Guillaume F, Hairston Jr NG, Hendry AP, Lasky JR, Rafajlovic M, Rasanen K, et al. (2018) What genomic data can reveal about eco-evolutionary dynamics. Nat Ecol Evol 2: 9–15 [DOI] [PubMed] [Google Scholar]
- Rusoff LL, Blakeney Jr.EW, CulleyJr. DD (1980) Duckweeds (Lemnaceae family): a potential source of protein and amino acids. J Agric Food Chem 28: 848–850 [DOI] [PubMed] [Google Scholar]
- Sauter PR (1993) Cryopreservation of Lemnaceae (in German). Veroeffentlichungen des Geobotanischen Instituts der ETH. Stiftung Ruebel, Zurich, 112 Heft
- Scheffer M, Szabo S, Gragnani A, Van Nes EH, Rinaldi S, Kautsky N, Norberg J, Roijackers RM, Franken RJ (2003) Floating plant dominance as a stable state. Proc Natl Acad Sci U S A 100: 4040–4045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleiden MJ (1839) Prodromus Monographiae Lemnacearum oder Conspectus generum atque specierum. Linnaea 13: 385–392 [Google Scholar]
- Schlotterer C, Kofler R, Versace E, Tobler R, Franssen SU (2015) Combining experimental evolution with next-generation sequencing: a powerful tool to study adaptation from standing genetic variation. Heredity (Edinb) 114: 431–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahaf N, Rogachev I, Heinig U, Meir S, Malitsky S, Battat M, Wyner H, Zheng S, Wehrens R, Aharoni A (2016) The WEIZMASS spectral library for high-confidence metabolite identification. Nat Commun 7:12423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao J, Liu Z, Ding YQ, Wang JM, Li XF, Yang Y (2020) Biosynthesis of the starch is improved by the supplement of nickel (Ni2+) in duckweed (Landoltia punctata). J Plant Res 133: 587–596 [DOI] [PubMed] [Google Scholar]
- Silva GG, Green AJ, Weber V, Hoffmann P, Lovas-Kiss Á, Stenert C, Maltchik L (2018) Whole angiosperms Wolffia columbiana disperse by gut passage through wildfowl in South America. Biol Lett 14: p.20180703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šmarda P, Bureš P, Horová L, Leitch IJ, Mucina L, Pacini E, Tichý L, Grulich V, Rotreklová O (2014) Ecological and evolutionary significance of genomic GC content diversity in monocots. Proc Natl Acad Sci 111: E4096–E4102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart CC, Trewavas AJ (1983a) Abscisic acid‐induced turion formation in Spirodela polyrrhiza. I. Production and development of the turion. Plant Cell Environ 6: 507–514 [Google Scholar]
- Smart CC, Trewavas AJ (1983b) Abscisic acid-induced turion formation in Spirodela polyrrhiza L. II. Ultrastructure of the turion; a stereological analysis. Plant Cell Environ 6: 515–522 [Google Scholar]
- Smart CC, Trewavas AJ (1984) Abscisic acid-induced turion formation in Spirodela polyrrhiza L. III. Specific changes in protein synthesis and translatable RNA during turion development. Plant Cell Environ 7: 121–132 [Google Scholar]
- Smart CC, Fleming AJ, Chaloupkova K, Hanke DE (1995) The physiological role of abscisic acid in eliciting turion morphogenesis. Plant Physiol 108: 623–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DH, Castle JE (1960) Production of auxotrophs in a duckweed, Spirodela polyrhiza. Plant Physiol 35: 809–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonta M, Rekiel A, Batorska M (2019) Use of duckweed (Lemna L.) in sustainable livestock production and aquaculture – a review. Ann Anim Sci 19: 257–271 [Google Scholar]
- Sowinski EE, Gilbert S, Lam E, Carpita NC (2019) Linkage structure of cell-wall polysaccharides from three duckweed species. Carbohydr Polym 223: 115119. [DOI] [PubMed] [Google Scholar]
- Sree KS, Maheshwari SC, Boka K, Khurana JP, Keresztes A, Appenroth KJ (2015a) The duckweed Wolffia microscopica: a unique aquatic monocot. Flora 210: 31–39 [Google Scholar]
- Sree KS, Sudakaran S, Appenroth KJ (2015b) How fast can angiosperm grow? Species and clonal diversity of growth rates in the genus Wolffia (Lemnaceae). Acta Physiol Plant 37: 204 [Google Scholar]
- Sree KS, Adelmann K, Garcia C, Lam E, Appenroth KJ (2015c) Natural variance in salt tolerance and induction of starch accumulation in duckweeds. Planta 241: 1395–1404 [DOI] [PubMed] [Google Scholar]
- Sree KS, Dahse HM, Chandran JN, Schneider B, Jahreis G, Appenroth KJ (2019) Duckweed for human nutrition: no cytotoxic and no anti-proliferative effects on human cell lines. Plant Foods Hum Nutr 74: 223–224 [DOI] [PubMed] [Google Scholar]
- Stiekema WJ, Wimpee CF, Tobin EM (1983) Nucleotide sequence encoding the precursor of the small subunit of ribulose 1,5-bisphosphate carboxylase from Lemna gibba L. G3. Nucleic Acid Res 11: 8051–8061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stomp AM, Rajbhandari N (1999) Genetically engineered duckweed. WO 1999007210
- Sun Y, Cheng JJ, Himmel ME, Skory CD, Adney WS, Thomas SR, Tisserat B, Nishimura Y, Yamamoto YT (2007) Expression and characterization of Acidothermus cellulolyticus E1 endoglucanase in transgenic duckweed Lemna minor 8627. Bioresour Technol 98: 2866–2872 [DOI] [PubMed] [Google Scholar]
- Sun L, Lu Y, Kronzucker HJ, Shi W (2016) Quantification and enzyme targets of fatty acid amides from duckweed root exudates involved in the stimulation of denitrification. J Plant Physiol 198: 81–88 [DOI] [PubMed] [Google Scholar]
- Tam YY, Slovin JP, Cohen JD (1995) Selection and characterization of a-methyltryptophan-resistant lines of Lemna gibba showing a rapid rate of indole-3-acetic acid turnover. Plant Physiol 107: 77–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan JQ, Kerstetter JE, Turcotte MM (2021) Eco-evolutionary interaction between microbiome presence and rapid biofilm evolution determines plant host fitness. Nat Ecol Evol 5: 670–676 [DOI] [PubMed] [Google Scholar]
- Tanaka O, Cleland CF, Hillman WS (1979) Inhibition of flowering in the long-day plant Lemna gibba G3 by Hutner’s medium and its reversal by salicylic acid. Plant Cell Physiol 20: 839–846 [Google Scholar]
- Tanaka Y, Tamaki H, Tanaka K, Tozawa E, Matsuzawa H, Toyama T, Kamagata Y, Mori K (2018) “Duckweed-microbe co-cultivation method” for isolating a wide variety of microbes including taxonomically novel microbes. Microbes Environ 33: 402–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teotonio H, Chelo IM, Bradic M, Rose MR, Long AD (2009) Experimental evolution reveals natural selection on standing genetic variation. Nat Genet 41: 251–257 [DOI] [PubMed] [Google Scholar]
- Thompson CH (1898) A revision of the American Lemnaceae occurring North of Mexico. Ann Rep Miss Bot Gard 9: 21–42 [Google Scholar]
- Thu PTL., Huong PT, Tien VV, Ham L, Khanh T (2015) Regeneration and transformation of gene encoding the hemagglutinin antigen of the H5N1 virus in frond of duckweed (Spirodela polyrhiza L.). J Agric Stud 3: 48–59. [Google Scholar]
- Tippery NP, Les DH, Crawford DJ (2015) Evaluation of phylogenetic relationships in Lemnaceae using nuclear ribosomal data. Plant Biol 17: 50–58 [DOI] [PubMed] [Google Scholar]
- Tippery NP, Les DH (2020) Tiny plants with enormous potential: phylogeny and evolution of duckweeds. In Cao XH, Fourounjian P, Wang W, eds, The Duckweed Genomes. Springer, Berlin, Heidelberg, pp. 19–38 [Google Scholar]
- Toyama T, Yu N, Kumada H, Sei K, Ike M, Fujita M (2006) Accelerated aromatic compounds degradation in aquatic environment by use of interaction between Spirodela polyrhiza and bacteria in its rhizosphere. J Biosci Bioeng 101: 346–353. [DOI] [PubMed] [Google Scholar]
- Toyama T, Kuroda M, Ogata Y, Hachiya Y, Quach A, Tokura K, Tanaka Y, Mori K, Morikawa M, Ike M (2017) Enhanced biomass production of duckweeds by inoculating a plant growth-promoting bacterium, Acinetobacter calcoaceticus P23, in sterile medium and non-sterile environmental waters. Water Sci Technol 76: 1418–1428 [DOI] [PubMed] [Google Scholar]
- Trewavas AJ (1970) The turnover of nucleic acids in Lemna minor. Plant Physiol 45: 742–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trewavas AJ (1972) Determination of the rates of protein synthesis and degradation in Lemna minor. Plant Physiol 49: 40–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turley NE, Odell WC, Schaefer H, Everwand G, Crawley MJ, Johnson MT (2013) Contemporary evolution of plant growth rate following experimental removal of herbivores. Am Nat 181: S21–S34 [DOI] [PubMed] [Google Scholar]
- Urbanska WK (1980) Cytological variation within the family of "Lemnaceae". Veröffentlichungen des Geobotanischen Institutes der Eidg. Tech. Hochschule, Stiftung Rübel, Zürich doi:10.5169/seals-308615
- USEPA (1996) Aquatic plants toxicity test using Lemna spp. United States Environmental Protection Agency, EPA 712-C-96-156
- Van Bel M, Diels T, Vancaester E, Kreft LBotzki A, Van de Peer Y, Coppens F, Vandepoele K (2018) P LAZA 4.0: an integrative resource for functional, evolutionary and comparative plant genomics. Nucleic acids research. 46: D1190–D1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Heide TJ, Roijackers RM, Peeters ET, Van Nes EH (2006) Experiments with duckweed–moth systems suggest that global warming may reduce rather than promote herbivory. Freshw Biol 51: 110–116 [Google Scholar]
- Van Hoeck A, Horemans N, Monsieurs P, Cao HX, Vandenhove H, Blust R (2015) The first draft genome of the aquatic model plant Lemna minor opens the route for future stress physiology research and biotechnological applications. Biotechnol Biofuels 8: 188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorholt JA, Vogel C, Carlström CI, Müller DB (2017). Establishing causality: opportunities of synthetic communities for plant microbiome research. Cell Host Microbe 22: 142–155 [DOI] [PubMed] [Google Scholar]
- Vunsh R, Heinig U, Malitsky S, Aharoni A, Avidov A, Lerner A, Edelman M (2015) Manipulating duckweed through genome duplication. Plant Biol 17: 115–119 [DOI] [PubMed] [Google Scholar]
- Wang WQ, Wu YR, Yan YH, Ermakova M, Kerstetter R, Messing J (2010) DNA barcoding of the Lemnaceae, a family of aquatic monocots. BMC Plant Biol 10: 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Kerstetter RA, Michael TP (2011) Evolution of genome size in Duckweeds (Lemnaceae). J Bot 2011: 1–9 [Google Scholar]
- Wang W, Messing J (2011) High-throughput sequencing of three Lemnoideae (duckweeds) chloroplast genomes from total DNA. PLoS One 6: e24670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Wu Y, Messing J (2012) The mitochondrial genome of an aquatic plant, Spirodela polyrhiza. PLoS One 7: p.e46747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Haberer G, Gundlach H, Glaber C, Nussbaumer TC, Luo MC, Lomsadze A, Borodovsky M, Kerstetter RA, Shanklin J, et al. (2014) The Spirodela polyrhiza genome reveals insights into its neotenous reduction fast growth and aquatic lifestyle. Nat Commun 5: 3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Chu J, Yu T, Xu Q, Sun X, Yuan J, Xiong G, Wang G, Wang Y, Li J (2015) Tryptophan-independent auxin biosynthesis contributes to early embryogenesis in Arabidopsis. Proc Natl Acad Sci U S A 112: 4821–4826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Zhao FJ, Kopittke PM (2019) Engineering crops without genome integration using nanotechnology. Trends Plant Sci 24: 574–577 [DOI] [PubMed] [Google Scholar]
- Ward DB, Hall DW (2010) Keys to the flora of Florida–25, Lemnaceae. Phytologia 92: 241–248 [Google Scholar]
- Watanabe S, Fučíková K, Lewis LA, Lewis PO (2016) Hiding in plain sight: Koshicola spirodelophila gen. et sp. nov. (Chaetopeltidales, Chlorophyceae), a novel green alga associated with the aquatic angiosperm Spirodela polyrhiza. Am J Bot 103: 865–875 [DOI] [PubMed] [Google Scholar]
- White SL, Wise RR (1998) Anatomy and ultrastructure of Wolffia columbiana and Wolffia borealis, two nonvascular aquatic angiosperms. Int J Plant Sci 159: 297–304 [Google Scholar]
- Xie WY, Su JQ, Zhu YG (2014) Arsenite oxidation by the phyllosphere bacterial community associated with Wolffia australiana. Environ Sci Technol 48: 9668–9674 [DOI] [PubMed] [Google Scholar]
- Xie WY, Su JQ, Zhu YG (2015) Phyllosphere bacterial community of floating macrophytes in paddy soil environments as revealed by Illumina high-throughput sequencing. Appl Environ Microbiol 81: 522–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu JL, Cheng JJ, Stomp AM (2012) Growing Spirodela polyrrhiza in swine wastewater for the production of animal feed and fuel ethanol: a pilot study. Clean Soil Air Water 40: 760–765 [Google Scholar]
- Xu XJ, Sun JQ, Nie Y, Wu XL (2015) Spirodela polyrhiza stimulates the growth of its endophytes but differentially increases their fenpropathrin-degradation capabilities. Chemosphere 125: 33–40 [DOI] [PubMed] [Google Scholar]
- Xu H, Yu C, Xia X, Li M, Li H, Wang Y, Wang S, Wang C, Ma Y, Zhou G (2018) Comparative transcriptome analysis of duckweed (Landoltia punctata) in response to cadmium provides insights into molecular mechanisms underlying hyperaccumulation. Chemosphere 190: 154–165 [DOI] [PubMed] [Google Scholar]
- Xu S, Stapley J, Gablenz S, Boyer J, Appenroth KJ, Sree KS, Gershenzon J, Widmer A, Huber M (2019) Low genetic variation is associated with low mutation rate in the giant duckweed. Nat Commun 10: 1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaga F, Washio K, Morikawa M (2010) Sustainable biodegradation of phenol by Acinetobacter calcoaceticus P23 isolated from the rhizosphere of duckweed Lemna aoukikusa. Environ Sci Technol 44: 6470–6474 [DOI] [PubMed] [Google Scholar]
- Yamakawa Y, Jog R, Morikawa M (2018) Effects of co-inoculation of two different plant growth-promoting bacteria on duckweed. Plant Growth Regul 86: 287–296 [Google Scholar]
- Yang L, Han Y, Wu D, Yong W, Liu M, Wang S, Liu W, Lu M, Wei Y, Sun J (2017) Salt and cadmium stress tolerance caused by overexpression of the Glycine max Na+/H+ Antiporter (GmNHX1) gene in duckweed (Lemna turionifera 5511). Aquat Toxicol 192: 127–135 [DOI] [PubMed] [Google Scholar]
- Yang J, Li G, Hu S, Bishopp A, Heenatigala PPM., Kumar S, Duan P, Yao L, Hou H (2018a) A protocol for efficient callus induction and stable transformation of Spirodela polyrhiza (L.) Schleiden using Agrobacterium tumefaciens. Aquat Bot 151: 80–86 [Google Scholar]
- Yang GL, Fang Y, Xu YL, Tan L, Li Q, Liu Y, Lai F, Jin YL, Du AP, He KZ, Ma XR, Zhao H (2018b) Frond transformation system mediated by Agrobacterium tumefaciens for Lemna minor. Plant Mol Biol 98: 319–331 [DOI] [PubMed] [Google Scholar]
- Yu C, Zhao X, Qi G, Bai Z, Wang Y, Wang S, Ma Y, Liu Q, Hu R, Zhou G (2017) Integrated analysis of transcriptome and metabolites reveals an essential role of metabolic flux in starch accumulation under nitrogen starvation in duckweed. Biotechnol Biofuels 10: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller MA, Hunt R, Sharma S (2013) Sustainable bioderived polymeric materials and thermoplastic blends made from floating aquatic macrophytes such as ‘‘duckweed’’. J Appl Polym Sci 127: 375–386 [Google Scholar]
- Zhang Y, An D, Li C, Zhao Z, Wang W (2020) The complete chloroplast genome of greater duckweed (Spirodela polyrhiza 7498) using PacBio long reads: insights into the chloroplast evolution and transcription regulation. BMC Genomics 21: 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Fang Y, Jin Y, Huang J, Ma X, He K, He Z, Wang F, Zhao H (2015) Microbial community and removal of nitrogen via the addition of a carrier in a pilot-scale duckweed-based wastewater treatment system. Bioresour Technol 179: 549–558 [DOI] [PubMed] [Google Scholar]
- Ziegler P, Adelmann K, Zimmer S, Schmidt C, Appenroth KJ (2015) Relative in vitro growth rates of duckweeds (Lemnaceae)—the most rapidly growing higher plants. Plant Biol 17: 33–41 [DOI] [PubMed] [Google Scholar]
- Ziegler P, Sree KS, Appenroth KJ (2016) Duckweed for water remediation and toxicity testing. Toxicol Environ Chem 98: 1127–1154 [Google Scholar]
- Ziegler P, Sree KS, Appenroth KJ (2018) Duckweed biomarkers for identifying toxic water contaminants? Environ Sci Pollut Res 26: 14797–14822 [DOI] [PubMed] [Google Scholar]
- Züst T, Heichinger C, Grossniklaus U, Harrington R, Kliebenstein DJ, Turnbull LA (2012) Natural enemies drive geographic variation in plant defenses. Science 338: 116–119 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.