Abstract
Background
KCNC1 encodes Kv3.1, a subunit of the Kv3 voltage-gated potassium channels. It is predominantly expressed in inhibitory GABAergic interneurons and cerebellar neurons. Kv3.1 channelopathy has been linked to a variety of human diseases including epilepsy, developmental delay, and ataxia. Characterization of structural and functional disturbances of this channel, and its relationship to a heterogenous group of clinical phenotypes, is a current topic of research. We herein characterize the clinical phenotype as well as the functional and structural consequences of the novel KCNC1 p.R317S variant. We further set out to explore the mechanistic basis for the spectrum of KCNC1 related channelopathies.
Methods
Variant was identified via whole-exome sequencing and its functional impact was determined using two-electrode voltage clamp recordings in Xenopus laevis oocytes. Homolog modeling and in silico structural analysis were performed on the p.R317S variant and other KCNC1 related variants.
Results
We identified a novel loss-of-function KCNC1 variant c.949C>A (p.R317S) presenting with symptoms similar to myoclonic epilepsy and ataxia due to potassium channel (MEAK), but with distinct radiological features. Functional analysis in the Xenopus laevis oocyte’s expression system revealed that the current amplitudes were significantly decreased in the p.R317S variant compared to the wild type, indicating a dominant-negative effect. Atomic structural analysis of the KCNC1 related variants provided a possible mechanistic explanation for the heterogeneity in the clinical spectrum.
Conclusions
We have identified the p.R317S loss-of-function variant in the KCNC1 gene, expanded the spectrum of potassium channelopathy and provided mechanistic insights into KCNC1 related disorders.
Keywords: Potassium ion-channel, epilepsy, channelopathy, exome, ataxia
Introduction
Voltage-gated potassium channels (Kv channel) are a specific subgroup of potassium channels that open and close in response to the voltage change on the cell membrane. Hence, they are crucial for action potential repolarization and electrical signal conduction (1). They are further divided into twelve subfamilies: Kv1–Kv12, each playing a distinctive physiological role (2-4). Among these, Kv3 channels are specifically known for their positively shifted voltage-dependent activation as well as fast activation and deactivation. Dysfunction of Kv3 channels disrupts the firing of the fast-spiking neurons and results in a dysregulated release of neurotransmitters, which may lead to cell death (5-8).
In the human genome, Kv3 channels have four isoforms, Kv3.1, Kv3.2, Kv3.3 and Kv3.4, encoded by KCNC1, KCNC2, KCNC3 and KCNC4 genes, respectively. They can assemble into homo- and heterotetrameric voltage-gated potassium channels. Within each subunit, there are 6 transmembrane segments (S1–S6). While S5–S6 segments form the ion-conducting pore domain (PD), S1–S4 segments are known as the voltage-sensing domain (VSD). S4, the central voltage sensing unit, has six positively charged arginine/lysine residues located at every third position along the amino acid sequence. They are highly conserved among different subfamilies and even across species (9). The positively charged residues can sense changes in the membrane voltage potential and allow for conformational change that leads to opening and closing of the channel (5,10,11).
In the past few years, various types of rare variants within KCNC genes have been linked to human diseases. Interestingly, several variants alter the voltage sensing Arginine residues. KCNC3 p.R420H and p.R423H variants were found to be the cause of spinocerebellar ataxia type 13 (12-15). KCNC1 p.R320H variant was found in patients with myoclonic epilepsy and ataxia due to potassium channel mutation (MEAK), which is a subtype of progressive myoclonic epilepsy (PME) with milder presentation and better prognosis (9,16-19). p.A421V was identified in patients with severe developmental and epileptic encephalopathy (DEE) (19-21). Subsequently, a wider spectrum of phenotypes designated as DE (developmental encephalopathy without seizures) has been reported to be associated with KCNC1. Two nonsense variants p.R339*, p.Q492* and two missense variants p.R317H, p.T399M were described in individuals with intellectual disability and development delay without epilepsy (20-22). Missense variant p.C208Ywas reported in isolated non-progressive myoclonus without intellectual disability (21).
Here we report a novel p.R317S variant that resembles the clinical phenotype of MEAK, but with unique radiological features. We further set out to understand the mechanistic basis of the spectrum of KCNC1 related potassium channelopathy phenotypes.
We present the following article in accordance with the MDAR reporting checklist (available at https://dx.doi.org/10.21037/atm-21-1885).
Methods
Standard protocol approvals, registrations, and patient consents
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Medical Ethics Committee of Huashan Hospital, Shanghai Medical College, Fudan University (No. 2008-71). A written informed consent was obtained from the patient’s legal guardian.
Exome sequencing
After blood was collected from patient’s trio family, genomic DNA was extracted using the Solpure Blood DNA kit (Magen) and fragmented to 300–500 bp size by Q800R Sonicator (Qsonica). After library preparation according to protocol (Illumina, San Diego, CA), target regions were enriched using custom-designed NGS capture library with in-solution hybridization probes (Roche NimbleGen, Madison, WI). The enriched sample was sequenced on a NextSeq500 sequencer (Illumina, San Diego, CA) with 100–150 cycles of pair-end reads.
Mitochondrial genome sequencing
DNA collected from the patient and his mother were extracted using the same method as described above. Long-range PCR was performed to acquire the whole mitochondrial genome according to methods described in a previous publication (23). After library preparation, sample was sequenced using the same method as described above.
Variants interpretation
Sequencing data were cleaned with Illumina Pipeline to improve the data quality by removing low-quality reads. The genome alignment and variant call were carried out by NextGENe software with HG19 as reference genome (SoftGenetics, State College, PA). Some nucleotide changes were reviewed manually by NextGENe viewer. Coverage-based algorithm developed in-house, eCNVscan, was used to examine the CNVs. Variant annotation was carried out by using population and mutation related databases including 1000 Genomes, dbSNP, GnomAD, Clinvar, HGMD, and OMIM. Computational algorithms SIFT, polyphen-2, and so on were used to analyze missense variant effects. Variant classification was done according to the American College of Medical Genetics (ACMG) guidelines.
Mutagenesis and RNA preparation
Functional analysis of c.949C>A (p.R317S) was performed as previously described (20). Briefly, the variant was introduced into cDNA encoding KCNC1 transcript variant 1 (NM_001112741; Kv3.1). The cDNA cloned into a pCMV‐Entry vector was obtained from OriGene Technologies (Rockville, MD). Quik Change Lightning Site-directed mutagenesis kit (Agilent, Santa Clara, CA) was used according to the manufacturer’s instructions (20). Sanger sequencing was performed to verify the insertion of the variant and exclude additional changes. In vitro transcription was performed using T7 mMessage mMachine kit (Ambion, Austin, TX).
Oocyte injection and data collection
The use of animals and all experimental procedures were approved by the Animal Ethics Committee at the Florey Institute of Neuroscience and Mental Health. Ovary pieces were extracted from Xenopus laevis, prepared as previously described (24), and kept in ND96 solution on 17 °C. On the following day, oocytes were injected with 50 nl cRNA (0.2 µg/µL) of wild-type (WT) or mutant cRNA using Robooinject (Multi Channel Systems, Reutlingen, Germany). In the coexpression experiments, total amount of WT and R317S cRNA mixed in 1:1 ratio was the same as above and compared to the WT mixed with H2O. Thus, the amount of WT cRNA was kept the same enabling us to determine if R317S exerts a dominant-negative effect. Recordings were performed at days 2–3 after injection. To determine current amplitudes, we analyzed the end of current traces elicited with the voltage step to +60 mV. All amplitudes were normalized to the wild type mean current amplitude recorded on the same day. This enabled us to pull the data from different experiments together. We did not predetermine the sample sizes.
Automated oocyte two-microelectrode voltage clamp
Potassium ion currents in oocytes were recorded on Roboocyte2 (Multi Channel Systems, Reutlingen, Germany) using intracellular glass microelectrodes with a resistance of 0.3–1 MΩ when filled with a solution of 1 M KCl and 1.5 M potassium acetate. Oocytes were perfused with bath solution containing (in mmol/L) 96 NaCl, 2 KCl, 1.8 CaCl2, 1 MgCl2 and 5 HEPES, pH 7.5. Currents were sampled at 5 kHz. For the analysis of channel activation, we used 0.5‐s long depolarizing steps from the holding potential of −90 mV applied at 10 mV increments, from −60 to +60 mV, followed by a step to −90 mV for 0.5 sec to analyze tail currents (9).
Statistical analysis
Data were analyzed using Roboocyte2+ (Multi Channel Systems, Reutlingen, Germany) and AxoGraph (AxoGraph Scientific, Sydney, Australia). Plotting of graphs and statistical analysis were performed using GraphPad Prism 7 (GraphPad Software, La Jolla, CA). A Boltzmann function was fit to the current‐voltage relationships, I(V) = I max/(1 + exp[(V − V0.5)/k]) + C, where I max is the maximum tail current amplitude at test potential V, V0.5 the half‐maximal activation potential, k a slope factor reflecting characteristics of voltage‐dependent channel gating and C a constant. All data are shown as mean values ± s.e.m. The normality of distribution was assessed using Anderson-Darling test and the statistically significant differences were determined using non-parametric Mann-Whitney test, t-test or One-way ANOVA, with Dunnett’s multiple comparisons test.
Atomic structural modeling and analysis
The protein structure and amino acid changes in the 3-dimensional structure of the Kv3.1 potassium channel was based on the X-ray diffraction structure of Shaker family voltage dependent potassium channel (Kv1.2–Kv2.1 paddle chimera channel) at resolution 2.4 Å. The overall sequence similarity of these full-length proteins of between Kv3.1 and Kv1.2-Kv2.1 is about 40%. The selective filter regions around p.R317 position had about 80% similarity, with the complete conservation of these key Arginine (R2-R6) residues. The homology model was built using the SWISS-MODEL Homology Modeling program. In silico structural analysis and visualization of Kv3.1 potassium channel were performed by using PyMOL (DeLano Scientific, Palo Alto, CA).
Results
Clinical characteristics
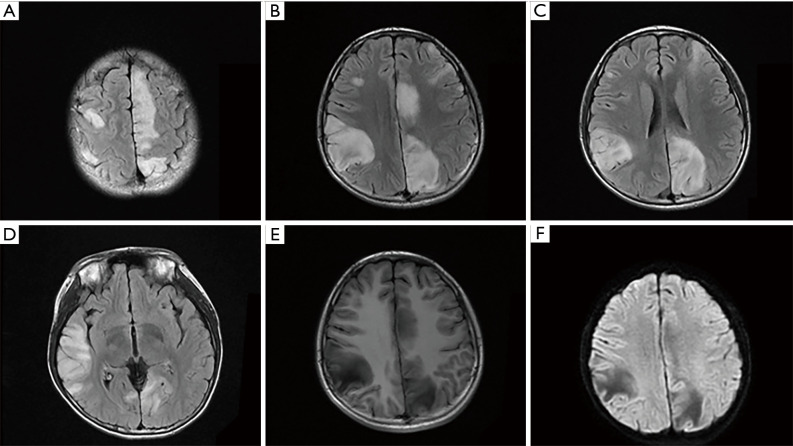
A 14-year-old male with a history of mild developmental delay presented with new-onset seizures one month prior to admission. His clinical events were characterized by focal onset right-hand myoclonic jerking, gaze deviation, followed by secondary generalization. Initial MRI of the brain showed multiple T2/fluid attenuated inversion recovery (FLAIR) cortical signal abnormalities in bilateral frontal, temporal, parietal and occipital lobes, without gadolinium contrast enhancement or restricted diffusion. (Figure 1, Figure S1). Laboratory tests including complete blood count, basic metabolic panel, antinuclear antibodies, extractable nuclear antigen antibodies, antineutrophil cytoplasmic antibodies, lactate, autoimmune encephalopathy panel were unremarkable. Lumbar puncture revealed nucleated cells 7×106/L (0–8×106/L), protein 214 mg/L (150–450 mg/L) and glucose 3.34 mmol/L (2.20–3.90 mmol/L), which were within normal limits. Electromyography and nerve conduction studies were normal. Routine electroencephalogram revealed bilateral scattered sharp waves and spike-and-wave discharges. Although his generalized tonic-clonic seizures were controlled with multiple antiepileptic agents including valproic acid, levetiracetam, clonazepam and oxcarbazepine, he continued to have episodes of myoclonic jerking of his right lower extremity. Intravenous immunoglobulins and methylprednisolone had also been trialed given the suspicion for autoimmune encephalitis and CNS vasculitis, however no clinical or radiological improvement was observed. Serial MRI scans were obtained in 2-week intervals, which demonstrated increasing size and burden of the cortical lesions (Figure S1A-S1C). MR angiogram and MR venogram were negative for vascular abnormalities. Cerebral angiogram and brain biopsy were declined.
Figure 1.
The MRI features of the novel R317S variant. (A-D) The axial FLAIR showed multiple signal abnormalities in bilateral frontal, temporal, parietal occipital lobes. Hypointense lesions were seen on T1-weighted imaging (E) and on DWI (F).
The patient was born to non-consanguineous parents with uneventful pregnancy and delivery. He had no overt congenital anomaly or stigmata at birth but was delayed in reaching developmental milestones. His height and weight were below average compared to peers. Family history was notable for short stature and mild gait ataxia in his mother since her childhood, although there was no history of seizure. Unfortunately, patient’s mother was not able to return for brain MRI. His maternal grandparents were not genetically tested. However, there was no clinical signs or symptoms of seizure, gait instability, intellectual disability, or short stature in his maternal grandparents. He had no siblings.
On neurological examination, he was able to answer orientation questions and follow commands but was slow to respond. He had significant trouble with serial subtraction of 7 s. His cranial nerve exam was normal. Motor exam was notable for decreased strength in the proximal right lower extremity (MRC scale 4-) and distal bilateral lower extremities (MRC scale 1). Sensory exam was intact. Deep tendon reflexes were brisk in the bilateral lower extremities. Plantar reflexes were flexor. He did have ataxic gait on coordination exam.
Mitochondrial and exome sequencing
This proband presented with myoclonic seizures and his brain MRI showed multiple signal abnormalities. He was diagnosed with epileptic encephalopathy. Mitochondrial and exome sequencing were performed to determine the molecular genetic etiology. No pathogenic single nucleotide variants or copy number variants were identified from mitochondrial sequencing data. Mitochondrial encephalopathy such as MELAS (Mitochondrial encephalomyopathy, lactic acidosis, and stroke-like episodes) was excluded. Using trio exome sequencing, 12,862 SNVs were detected in the proband. Trio sequencing quality scores were shown in Table S1. The genetic analysis process was shown as follows. First, rare variants were selected by population frequency. 298 variants with population frequency in GnomAD database <0.005 and population frequency <0.1 in Amcare internal database (East Asian population) were identified. Second, candidate variants were labeled using two co-segregation models. The first model (dominant inheritance) assumed that the proband and his mother had the same disease. We only labeled maternal variants with dominant inheritance and 21 variants were marked out of 298 variants. The second model (proband only) assumed that the proband did not have the same disease as his mother. 6 variants, which were consistent with co-segregation (AD-de novo heterozygous, AR-compound heterozygous/homozygous, XL/YL-hemizygous), were identified from 298 variants. In the second step, we identified a total of 27 variants. Third, the variants were narrowed down according to the proband’s phenotype, including epilepsy and related disorders, by matching Human Phenotype Ontology. Six variants were selected, including 4 maternal inherited AD variants: ARID1A c.3760G>A (p.G1254S), MACF1 c.6341A>T (p.D2114V), KCNC1 c.949C>A (p.R317S), RELN c.3190G>A (p.A1064T) and 2 AR compound heterozygous: paternal CNTNAP2 c.2190C>T (p.C730C) and maternal CNTNAP2 c.2240C>T (p.A747V). Fourth, the disease-causing variant was determined by combining information from prevalence/population frequency, variant prediction results and literature review (phenotype, age of onset, etc.). ARID1A c.3760G>A (p.G1254S), MACF1 c.6341A>T (p.D2114V) and RELN c.3190G>A (p.A1064T) variants were excluded because the population frequency of these 3 variants were higher than disease prevalence and most software prediction results were benign. Since CNTNAP2 synonymous variant c.2190C>T (p.C730C) was predicted to affect neither coding sequence nor mRNA splicing, CNTNAP2 compound heterozygous variants were also excluded. KCNC1 c.949C>A (p.R317S) variant was absent in GnomAD and Amcare internal population databases and predicted as deleterious, and KCNC1 phenotype was consistent with the patient’s clinical diagnosis. Therefore, KCNC1 c.949C>A (p.R317S) was most likely to be the causative mutation of the proband and his mother.
KCNC1 c.949C>A (p.R317S) variant of this family was shown in Figure S2. The proband’s KCNC1 c.949C>A (p.R317S) variant quality scores were shown as follows. Coverage: 489×, mutant allele frequency: 50.1 (wild type/mutant =242/245), zygosity: heterozygous, read balance: 0.88. His mother’s KCNC1 c.949C>A (p.R317S) variant quality scores: coverage: 529×, mutant allele frequency: 50.1 (wild type/mutant =262/265), zygosity: heterozygous, read balance: 0.87.
Functional analysis of p.R317S variant
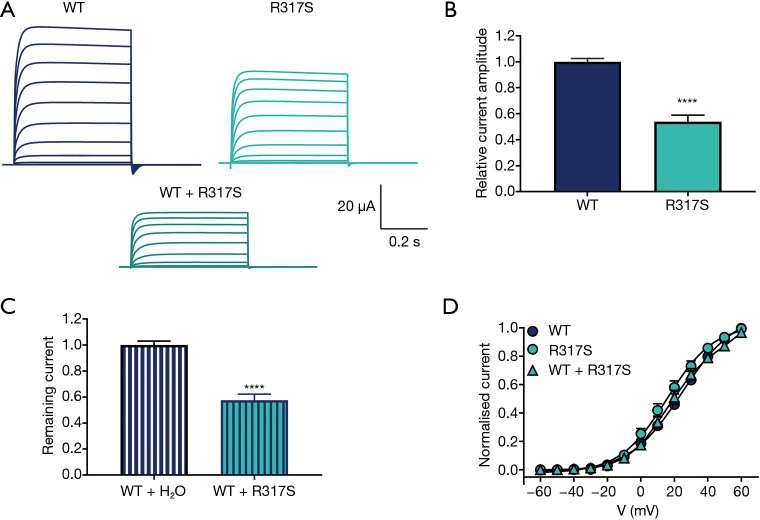
To investigate the functional impact of p.R317S variant on channel function, we used the Xenopus laevis oocyte expression system. Potassium currents were recorded from oocytes injected with wild-type or mutant KCNC1 cRNA (Figure 2A). Current amplitudes, analyzed at the end of the voltage step to +60 mV and normalized to the mean current amplitude of the WT (n=136) recorded on the same day, revealed significantly smaller potassium currents for the mutant p.R317S (n=45) indicating this variant causes a loss of function (Figure 2B).
Figure 2.
Functional characterization of R317S variant. (A) Representative whole-cell current traces recorded from Xenopus laevis oocytes injected with the same amount of cRNA encoding Kv3.1 wild-type (WT) or mutant, R317S, or their 1:1 mixture mimicking the heterozygous expression. Currents were recorded during 0.5 s voltage steps ranging from −60 to +60 mV. (B) Current amplitudes assessed at the end of the voltage step to +60 mV and normalized to the mean current amplitude of WT recorded on the same experimental day. WT (n=136), R317S (n=45), ****, P<0.0001, using Mann-Whitney test. (C) Current amplitudes of WT + H2O (n=111) and WT + R317S (n=26) analyzed at the end of the voltage step to +60 mV and normalized to the mean current amplitude of WT + H2O recorded on the same day. ****, P<0.0001, using unpaired t-test. (D) Conductance-voltage relationships for the WT transcripts and R317S variant expressed alone or together with the WT. V0.5 values in mV: WT 22.6±1.9 (n=36), R317S 16.1±2.6 (n=17) and R317S + WT 21.7±2.9 (n=17) were not significantly different (one-way ANOVA with Dunnett’s multiple comparisons test).
Since Kv3 channels assemble into tetramers, we further investigated the interaction of mutant and wild-type subunits, by co-expressing the corresponding cRNAs at a 1:1 ratio. The current amplitude of WT + R317S (n=26) was significantly decreased compared with WT + H2O (n=111), P<0.0001, indicating a dominant-negative effect (Figure 2C). Conductance-voltage relationship curves obtained from the tail currents showed no significant differences in values for V0.5 or the slope factor for WT, R317S and WT + R317S (Figure 2D).
Structural analysis of MEAK, DEE, DE and myoclonus related variants
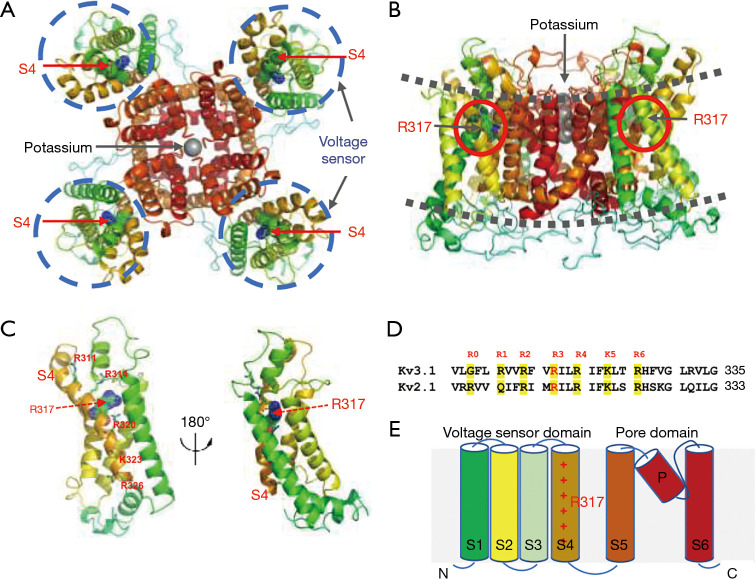
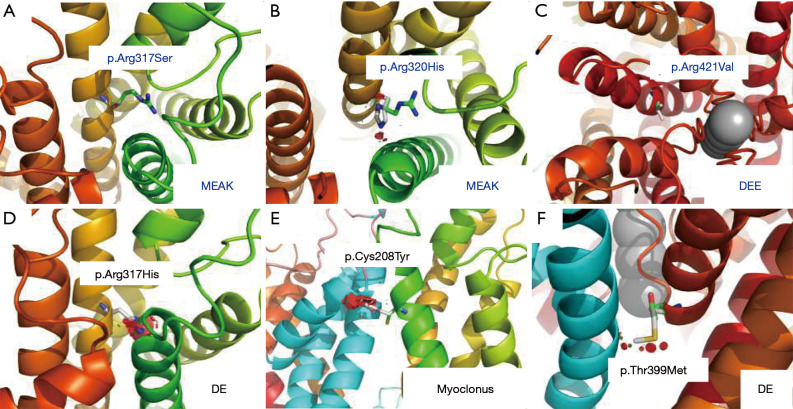
Variant p.R317S is located within the S4 helix of voltage sensing domain (VSD). The voltage sensing of VSD is critically dependent on six positively charged Arginine/Lysine residues (R311, R314, R317, R320, K323, R326), which are highly conserved among different subfamilies and even across species (Figure 3). On close examination of the atomic model of Kv3.1 channel, the replacement of Arginine at position 317 with Serine caused minimal disturbance (Figure 4A). We further examined the two recurrent variants, p.R320H associated with MEAK, and p.A421V linked to DEE. The MEAK variant p.R320H showed good tolerance at the site of S4 helix, with only slight disturbance (Figure 4B). The second mutant, p.A421V, is located in the middle of S6 helix, and is adjacent to the pore domain. The side of p.A421 is about 9.5 Å distance away from the closest potassium ion and has no direct contact with them. The replacement of Ala at amino acid position 421 with Val can result in the addition of two hydrophobic carbon atoms in the core region of the Kv3.1 channel. The structural analysis did not reveal obvious steric hindrance effect, but it was difficult to predict the electronic effect on the gating and selectivity filter (Figure 4C). In addition, p.R317H was found to be a variant associated with DE (20). The replacement with Histidine at this position can lead to severe steric hindrance, which may likely destabilize the protein structure (Figure 4D). Structure analysis of p.C208Y and p.T399M, the other two known variants associated with myoclonus or DE phenotypes, also revealed severe steric hindrance. The clashes were not predicted to affect the monomeric subunit, but rather its interaction with adjacent subunit of the symmetric pair (Figure 4E,4F).
Figure 3.
In silico analysis and structural analysis of Kv3.1 potassium channel involved p.R317S mutation. (A) Top view of ribbon representation of the Kv3.1 potassium channel model and the voltage sensor domains were highlighted in dash-line circle colored in blue. (B) Side view of ribbon representation of the Kv3.1 potassium channel model and lipid bilayer were highlighted in dash-line colored in grey. (C) Rotational view of ribbon representation of the selective filter region of Kv3.1 potassium channel model. (D) The amino acid sequence alignment of selective filter regions showed the high degree conservation of these key Arginine (R0–R6) residues. (E) The schematic representation of Kv3.1 potassium channel model by cartoon diagram.
Figure 4.
Structural analysis of Kv3.1 potassium channel involved known variants for MEAK, DEE, DE and myoclonus phenotypes. (A-C) The close view of different variants at positions of p.Arg317Ser, p.Arg320His and p.Arg421Val showed minimal disturbance for the adjacent configuration. (D-F) The close view of different variants at positions of p.Arg317His, p.Cys208Tyr and p.Thr399Met showed large steric hinderance, which likely disturb the protein structure or stability.
Discussion
As a member of the Kv3 subfamily, Kv3.1 is predominantly expressed in inhibitory GABAergic interneurons and cerebellar neurons (25). Variants that alter the voltage-sensing domain of Shaker K+ channel may affect the channel opening upon depolarization (10,26). Therefore, it is plausible that the functional impairment or structural perturbation of Kv3.1 protein would result in neurological abnormalities, such as epilepsy, myoclonus, and ataxia.
A pathogenic variant of KCNC1 gene was first reported in PME patients by Muona et al. (9). This subtype was designated as myoclonic epilepsy and ataxia due to potassium channel mutation (MEAK). The age onset of MEAK patients was between 3–15 years. Clinical manifestations include myoclonus, ataxia, cognitive decline, and generalized tonic-clonic seizures. Global symmetrical cerebellar atrophy can be seen on brain MRI. Patients become wheelchair dependent by their mid to late teens. A recurrent variant p.R320H was identified in all MEAK patients (9,16-19). KCNC1 variants have been recently reported in another cohort of patients with DE or isolated non-progressive myoclonus. The identified variants include p.R339*, p.Q492*, p.C208Y, p.R317H and p.T399M (20-22). The case which carried p.R317H variant presented with mild intellectual disability and autism without seizure onset. A 23-hour EEG further revealed rare irregular generalized spike-wave discharges and brain MRI revealed cerebellar and posterior pontine atrophy.
In this report, we identified a 14-year-old male carrying KCNC1 p.R317S variant who presented with myoclonus, generalized tonic-clonic seizures and ataxia, which closely resembled the phenotype of MEAK. The similarity of the clinical presentation may be explained by the variant occurring at the evolutionary conserved voltage sensing sites within the voltage-gated potassium channel (R311, R314, R317, R320, K323, R326). However, our patient had unique radiological findings manifested by multiple cortical signal abnormalities on the brain MRI, whereas the previously reported p.R320H and p.R317H cases mainly manifested with cerebellar atrophy or even normal MRI (16,20). The proband’s mother who carried the same p.R317S variant, presented with short stature and ataxia. But she had normal intellectual development. There were no seizures reported in this female. Therefore, the expressivity of p.R317S variant seemed to be variable in this mother-son pair.
Functional assessments of KCNC1 variants in heterologous expression systems showed loss-of-function combined with the dominant-negative effect. However, there was no clear correlation between the functional properties of different variants and the severity of clinical symptoms. For example, although both p.R320H and p.R317S were detected in MEAK patients, the current amplitudes of p.R320H were much lower than p.R317S in Xenopus laevis oocytes. Moreover, the currents of p.R317H and p.R320H were all significantly decreased, but the phenotype of p.R317H patients was milder than the one caused by p.R320H (9,20). Another recurrent variant p.A421V was also identified in ten unrelated patients, and these patients had a variety of clinical features. The onset of the disease in these patients was in the first 10 months of life. They presented with myoclonic and other types of refractory seizures, combined with ataxia, hypotonia, developmental delay, and moderate to severe intellectual disability. Three patients had similar dysmorphic features including large mouth, smooth philtrum, up-slanting palpebral fissures, and dental enlargement (19-21).
Furthermore, the substitution of Arginine with other residues may change the conformation of the channel, generating leaky current through the gating pore (27). The substitution of the first Arginine residue with Histidine was reported to result in a proton pore within the voltage-sensing domain of the Shaker K+ channel (28,29). Changing the Arginine to a small non-charged amino acid such as Serine in the Shaker K+ channel would allow permeation of cations through the voltage-sensing domain, known as ω-current (30). These leaky currents may potentially contribute to the pathogenesis of the analogous Arginine variant within the Kv3 channel. In this report, we found that atomic structural modeling has the potential to predict the severity of clinical symptoms. Based on atomic structural modeling results, p.R317S and p.R320H related to MEAK as well as p.A421V in DEE patients, all revealed only slight steric hindrance. Conversely, variants identified in DE or myoclonus patients, including p.R317H, p.T399M and p.C208Y, showed severe disturbance.
Thus, we hypothesize that Kv3.1 subunits with slight steric hindrance can form tetramers with other normal subunits, but the mutant subunit may impair the function of the whole potassium channel complex and lead to a severe phenotype with epilepsy and ataxia. On the other hand, severe steric hindrance may affect the propensity of mutant subunits to compose potassium channel tetramers with the wild-type subunits. This may result in a decreased amount of tetramer complexes, with the majority of tetramers composed of normal subunits. Decreased quantity but preserved functionality of the potassium channels could lead to a milder phenotype without seizures.
On the other hand, nonsense mediated decay of the p.R339* variant supported our hypothesis of haploinsufficiency being related to milder symptoms (22). However, the available data from the functional studies in Xenopus oocytes indicates a clear dominant-negative effect of the mutant subunits on the wild type in three out of the five reported DE variants (20,21). Therefore, further functional studies in more sophisticated models may be required to test our hypothesis. Further confirmation in future cases would also be helpful. If confirmed, our structure model may have the potential to predict disease progression and prognosis according to genotype and may be conducive to clinical diagnosis and treatment.
Conclusions
In summary, the discovery of the novel KCNC1 p.R317S variant, along with its unique radiological features and its loss-of-function effect determined by functional assessment, further expands our knowledge about the spectrum of potassium channel variant associated diseases. The epileptic encephalopathy caused by this KCNC1 p.R317S variant should be considered in the differential when evaluating diseases with similar clinical and radiological presentations, such as mitochondrial diseases.
We have identified the p.R317S loss-of-function variant in the KCNC1 gene, expanded the clinical spectrum of potassium channelopathy, and provided mechanistic insights into the KCNC1 related disorders.
Supplementary
The article’s supplementary files as
Acknowledgments
We thank Ye Htet Aung, Florey Institute of Neuroscience and Mental health, Australia, for excellent technical assistance. We thank Carolyn Tsai and Michael Miao, University of North Carolina at Chapel Hill, for manuscript review and language editing.
Funding: This work was supported by Shanghai Municipal Science and Technology Major Project (2018SHZDZX01) to JL; ZJLab, Major Medical Collaboration and Innovation Program of Guangzhou Science Technology and Innovation Commission (201604020020, 201604020009) to VWZ; Science and Technology Planning Project of Guanzhou (2018-1202-SF-0019) to VWZ; National Health and Medical Research Council of Australia (10915693) to SP.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Medical Ethics Committee of Huashan Hospital, Shanghai Medical College, Fudan University (No. 2008-71). Informed consent was obtained.
Footnotes
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://dx.doi.org/10.21037/atm-21-1885
Data Sharing Statement: Available at https://dx.doi.org/10.21037/atm-21-1885
Peer Review File: Available at https://dx.doi.org/10.21037/atm-21-1885
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-1885). Dr. SP reports grants from NHMRC, during the conduct of the study; personal fees and other from Praxis Precision Medicine, grants from RogCon, outside the submitted work. The other authors have no conflicts of interest to declare.
References
- 1.Coetzee WA, Amarillo Y, Chiu J, et al. Molecular diversity of K+ channels. Ann N Y Acad Sci 1999;868:233-85. 10.1111/j.1749-6632.1999.tb11293.x [DOI] [PubMed] [Google Scholar]
- 2.Ranjan R, Logette E, Marani M, et al. A Kinetic Map of the Homomeric Voltage-Gated Potassium Channel (Kv) Family. Front Cell Neurosci 2019;13:358. 10.3389/fncel.2019.00358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gutman GA, Chandy KG, Grissmer S, et al. International Union of Pharmacology. LIII. Nomenclature and molecular relationships of voltage-gated potassium channels. Pharmacol Rev 2005;57:473-508. 10.1124/pr.57.4.10 [DOI] [PubMed] [Google Scholar]
- 4.Allen NM, Weckhuysen S, Gorman K, et al. Genetic potassium channel-associated epilepsies: Clinical review of the Kv family. Eur J Paediatr Neurol 2020;24:105-16. 10.1016/j.ejpn.2019.12.002 [DOI] [PubMed] [Google Scholar]
- 5.Rudy B, McBain CJ. Kv3 channels: voltage-gated K+ channels designed for high-frequency repetitive firing. Trends Neurosci 2001;24:517-26. 10.1016/S0166-2236(00)01892-0 [DOI] [PubMed] [Google Scholar]
- 6.Erisir A, Lau D, Rudy B, et al. Function of specific K(+) channels in sustained high-frequency firing of fast-spiking neocortical interneurons. J Neurophysiol 1999;82:2476-89. 10.1152/jn.1999.82.5.2476 [DOI] [PubMed] [Google Scholar]
- 7.Sabatini BL, Regehr WG. Control of neurotransmitter release by presynaptic waveform at the granule cell to Purkinje cell synapse. J Neurosci 1997;17:3425-35. 10.1523/JNEUROSCI.17-10-03425.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Irie T, Matsuzaki Y, Sekino Y, et al. Kv3.3 channels harbouring a mutation of spinocerebellar ataxia type 13 alter excitability and induce cell death in cultured cerebellar Purkinje cells. J Physiol 2014;592:229-47. 10.1113/jphysiol.2013.264309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muona M, Berkovic SF, Dibbens LM, et al. A recurrent de novo mutation in KCNC1 causes progressive myoclonus epilepsy. Nat Genet 2015;47:39-46. 10.1038/ng.3144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aggarwal SK, MacKinnon R. Contribution of the S4 segment to gating charge in the Shaker K+ channel. Neuron 1996;16:1169-77. 10.1016/S0896-6273(00)80143-9 [DOI] [PubMed] [Google Scholar]
- 11.Chanda B, Bezanilla F. A common pathway for charge transport through voltage-sensing domains. Neuron 2008;57:345-51. 10.1016/j.neuron.2008.01.015 [DOI] [PubMed] [Google Scholar]
- 12.Herman-Bert A, Stevanin G, Netter JC, et al. Mapping of spinocerebellar ataxia 13 to chromosome 19q13.3-q13.4 in a family with autosomal dominant cerebellar ataxia and mental retardation. Am J Hum Genet 2000;67:229-35. 10.1086/302958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Waters MF, Minassian NA, Stevanin G, et al. Mutations in voltage-gated potassium channel KCNC3 cause degenerative and developmental central nervous system phenotypes. Nat Genet 2006;38:447-51. 10.1038/ng1758 [DOI] [PubMed] [Google Scholar]
- 14.Figueroa KP, Minassian NA, Stevanin G, et al. KCNC3: phenotype, mutations, channel biophysics-a study of 260 familial ataxia patients. Hum Mutat 2010;31:191-6. 10.1002/humu.21165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Figueroa KP, Waters MF, Garibyan V, et al. Frequency of KCNC3 DNA variants as causes of spinocerebellar ataxia 13 (SCA13). PLoS One 2011;6:e17811. 10.1371/journal.pone.0017811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nascimento FA, Andrade DM. Myoclonus epilepsy and ataxia due to potassium channel mutation (MEAK) is caused by heterozygous KCNC1 mutations. Epileptic Disord 2016;18:135-8. 10.1684/epd.2016.0859 [DOI] [PubMed] [Google Scholar]
- 17.Oliver KL, Franceschetti S, Milligan CJ, et al. Myoclonus epilepsy and ataxia due to KCNC1 mutation: Analysis of 20 cases and K+ channel properties. Ann Neurol 2017;81:677-89. 10.1002/ana.24929 [DOI] [PubMed] [Google Scholar]
- 18.Kim H, Lee S, Choi M, et al. Familial cases of progressive myoclonic epilepsy caused by maternal somatic mosaicism of a recurrent KCNC1 p.Arg320His mutation. Brain Dev 2018;40:429-32. 10.1016/j.braindev.2018.01.006 [DOI] [PubMed] [Google Scholar]
- 19.Zhang J, Zhang YH, Chen JY, et al. Pathogenic gene variants and clinical phenotype features of 26 children with progressive myoclonic epilepsy. Zhonghua Er Ke Za Zhi 2019;57:458-64. [DOI] [PubMed] [Google Scholar]
- 20.Cameron JM, Maljevic S, Nair U, et al. Encephalopathies with KCNC1 variants: genotype-phenotype-functional correlations. Ann Clin Transl Neurol 2019;6:1263-72. 10.1002/acn3.50822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park J, Koko M, Hedrich UBS, et al. KCNC1-related disorders: new de novo variants expand the phenotypic spectrum. Ann Clin Transl Neurol 2019;6:1319-26. 10.1002/acn3.50799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poirier K, Viot G, Lombardi L, et al. Loss of Function of KCNC1 is associated with intellectual disability without seizures. Eur J Hum Genet 2017;25:560-4. 10.1038/ejhg.2017.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang W, Cui H, Wong LJ. Comprehensive one-step molecular analyses of mitochondrial genome by massively parallel sequencing. Clin Chem 2012;58:1322-31. 10.1373/clinchem.2011.181438 [DOI] [PubMed] [Google Scholar]
- 24.Petrou S, Ugur M, Drummond RM, et al. P2X7 purinoceptor expression in Xenopus oocytes is not sufficient to produce a pore-forming P2Z-like phenotype. FEBS Lett 1997;411:339-45. 10.1016/S0014-5793(97)00700-X [DOI] [PubMed] [Google Scholar]
- 25.Gan L, Kaczmarek LK. When, where, and how much? Expression of the Kv3.1 potassium channel in high-frequency firing neurons. J Neurobiol 1998;37:69-79. [DOI] [PubMed] [Google Scholar]
- 26.Seoh SA, Sigg D, Papazian DM, et al. Voltage-sensing residues in the S2 and S4 segments of the Shaker K+ channel. Neuron 1996;16:1159-67. 10.1016/S0896-6273(00)80142-7 [DOI] [PubMed] [Google Scholar]
- 27.Moreau A, Gosselin-Badaroudine P, Chahine M. Biophysics, pathophysiology, and pharmacology of ion channel gating pores. Front Pharmacol 2014;5:53. 10.3389/fphar.2014.00053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Starace DM, Bezanilla F. Histidine scanning mutagenesis of basic residues of the S4 segment of the shaker k+ channel. J Gen Physiol 2001;117:469-90. 10.1085/jgp.117.5.469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Starace DM, Bezanilla F. A proton pore in a potassium channel voltage sensor reveals a focused electric field. Nature 2004;427:548-53. 10.1038/nature02270 [DOI] [PubMed] [Google Scholar]
- 30.Khalili-Araghi F, Tajkhorshid E, Roux B, et al. Molecular dynamics investigation of the ω-current in the Kv1.2 voltage sensor domains. Biophys J 2012;102:258-67. 10.1016/j.bpj.2011.10.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as